Abstract
Older adults experience a higher prevalence of multiple chronic conditions (MCCs). Establishing the presence and pattern of MCCs in individuals or populations is important for healthcare delivery, research, and policy. This report describes four emerging approaches and discusses their potential applications for enhancing assessment, treatment, and policy for the aging population. The National Institutes of Health convened a 2‐day panel workshop of experts in 2018. Four emerging models were identified by the panel, including classification and regression tree (CART), qualifying comorbidity sets (QCS), the multimorbidity index (MMI), and the application of omics to network medicine. Future research into models of multiple chronic condition assessment may improve understanding of the epidemiology, diagnosis, and treatment of older persons.
Keywords: measurement, multimorbidity, multiple chronic conditions, older people
Key points
New approaches are needed to better understand multiple chronic conditions (MCCs) in older adults.
Machine learning, multivariate matching, decision algorithms, and omics (genomics, transcriptonomics, proteomics, and metabolomics) and their application to the study of MCCs are described.
These approaches can improve identification of population subgroups at risk for MCCs, benchmark the care of MCC patients for quality improvement, and describe the intersections and multi‐layered genomic, biologic, and behavioral pathways resulting in multimorbidity.
Why does this paper matter?
The prevalence of multiple chronic conditions (MCCs) and the challenges they pose for older persons have motivated the development of four emerging approaches for MCC assessment to improve the precision and comprehensiveness of epidemiology, diagnosis, and treatment.
INTRODUCTION
Older adults experience a higher prevalence of multiple chronic conditions than having a single condition in isolation. 1 Establishing the presence and pattern of MCCs in individuals or populations has importance for clinicians, researchers and health policy makers. For example, researchers studying the determinants or effects of MCCs need valid, comprehensive measurement instruments. Health policy makers require valid indices to evaluate programs and quality of care for these complex and vulnerable patients. Finding the most appropriate MCC measure or instrument can be challenging because, although many are available, each has limitations. 2 For example, often only a relatively narrow range of chronic conditions is captured in many existing MCC measures.
To assess MCCs, researchers and clinicians have relied on five major data sources: medical records and clinical assessments, administrative claims data, public health surveys, patient reports, and electronic health records (EHRs) (the last with the potential to merge the other sources). 2 Selection of appropriate data sources and instruments for MCC measurement should be guided by the purpose for which MCCs is being assessed. For example, MCCs may be measured to describe the overall health of a population or subpopulation, or to assess risk of mortality, hospitalization, disability, or other important outcomes in populations or individuals. We previously summarized several of the instruments or tools that have been developed to measure MCCs for these purposes and using one or more of these data sources. 2 Limitations of some instruments include failure to capture undiagnosed or rare conditions, which are not part of diagnostic lists; not assessing illness severity or stage; and having weak clinical specificity. Patient reports also are subject to potential recall and social desirability biases, and low‐literacy populations may be less able to accurately report their health conditions. Especially for clinicians, assessing MCCs can be important for identifying patients in need of extra care or support; one tool developed for this purpose is the Care Assessment Need (CAN) score that is used in the Veterans Administration healthcare system. 3 CAN and similar scores also may be used by payors to adjust payments for more complex patients requiring more involved care, such as those with MCCs. 4
METHODS
A special 2‐day expert workshop on the measurement of MCCs was sponsored by the National Institutes of Health in the Fall of 2018. A planning committee consisting of scientific staff from the Office of Disease Prevention (ODP), National Institute on Aging (NIA), National Cancer Institute (NCI), National Institute for Minority Health and Health Disparities (NIMHHD), and Office of Behavioral and Social Sciences Research (OBSSR), invited 40 experts from academic medical centers, healthcare institutes and departments, who held MDs, PhDs or MPHs and were acknowledged for their research and experience about MCCs, multimorbidity, comorbidity and their measurement. The experts were drawn from several fields and specialties, including internal medicine, family medicine, geriatrics and gerontology, pediatrics, epidemiology, public health, health scientist administration, nutrition science, health statistics, informatics and systems biology, behavioral medicine and clinical health psychology. Scientific staff from NIH, Agency for Health Research Quality, the Centers for Disease Control, and Center for Medicare and Medicaid Services also participated. The workshop involved individual presentations, instrument review, Q&A, and extensive discussion. Conclusions and guidance were determined by verbal agreement among those present.
One report arising from the workshop reviews available instruments. 2 A second report describes research needs and gaps related to MCC/multimorbidity assessment. 5 A segment of the workshop devoted to emerging approaches to improve the validity, reliability, generalizability, and breadth of assessment of multimorbidity was not addressed in either of the prior reports. The present article describes four emerging measurement approaches that were highlighted at the workshop, and discusses their potential applications for improving prevention, treatment and policy for the aging population.
RESULTS
Four emerging approaches for measuring MCCs were identified in individual presentations, group discussion, and verbal agreement. Each approach is defined and discussed below (see Table 1).
TABLE 1.
Description of four measurement approaches
Classification and regression tree analysis (CART):
|
Qualifying comorbidity sets (QCSs):
|
Multimorbidity index:
|
Omics and network medicine:
|
LR indicates how much the odds of mortality change when the patient has the disease. LR's are available as a look‐up table; free of cost at http://openonlinecourses.com/464/default.asp.
Classification and regression tree analysis
Most studies of MCCs have analyzed outcomes in relation to one condition at a time. However, chronic conditions often co‐occur, and their impact relative to the outcome(s) of interest may vary depending on the individual's sociodemographic characteristics, the specific condition combinations, and/or the presence of other health‐related factors, such as functional limitations, sensory impairment, or other geriatric syndromes (e.g., cognitive impairment and urinary incontinence). 6 In addition, multivariable models are usually developed based on a priori hypotheses, and are limited in their ability to incorporate several independent variables, which may be highly correlated.
Classification and regression tree (CART) analysis may provide a superior alternative to traditional multivariable models in MCC research. Although CART has been used since the early 1980s, 7 , 8 only recently has it been applied to MCC research 9 ; and differs from traditional measures 2 and other recently developed instruments, such as Wei et al., 10 that list and provide weighting to conditions constituting MCCs. CART allows us to identify combinations of MCC conditions that are associated with a certain outcome, regardless of the instrument from which these conditions originate.
CART is an artifical intelligence model, 11 using recursive partitioning, which considers all possible splits from all possible variables (e.g., marital status, <85 years vs >85 years, lung disease [yes/no]); it selects the variable that creates the most homogeneous clusters when split relative to a study outcome (e.g., self‐reported health, mortality). The variable that is most strongly associated with the outcome is selected for the next split, and so on (hence, the term “recursive”). Thus, rather than testing a priori hypotheses, CART provides a way for researchers to learn “what the data say.” For example, a CART model developed to predict fair or poor self‐rated health status identified visual impairment as a condition strongly associated with the outcome and ahead of many major chronic conditions. 9 Yet, one may not have hypothesized a priori that visual impairment would be one of the top predictors. CART is also a nonparametric model, meaning that the distribution of the variables under investigation does not matter, and it can accommodate continuous, binary, multinomial, and even time‐to‐event outcomes. Use of other measurement approaches are typically constrained by these features. 2
CART begins with the “parent” node, which includes the entire sample, then branches into binary child nodes by examining all independent variables and selecting the ones that yield the most unique groups in terms of the outcome variable. Each child node becomes a parent node itself, and the partitioning continues until a terminal node is reached. Figure 1 presents a hypothetical CART to predict 2‐year worse self‐rated health. The top splitting variable was difficulty walking several blocks, reflecting its importance in predicting the outcome. The combination of variables associated with the highest percentage of 2‐year self‐rated worse health includes difficulty walking several blocks, visual impairment, and age 68.5 years or older (see the first bar at the right, with a percentage of nearly 60%). Conversely, the lowest percentage of 2‐year self‐rated worse health is observed among individuals with no difficulty walking several blocks, no self‐rated fair/poor health, and no urinary incontinence.
FIGURE 1.
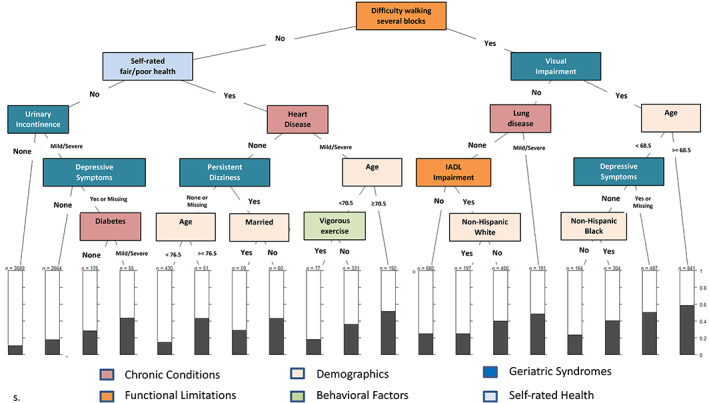
Hypothetical classification and regression tree analysis to predict 2‐year self‐rated worse health. Chronic conditions may include such conditions as heart or lung disease, diabetes, and cancer; functional limitations may include limitations in upper/lower body strength and limitations, and/or limitations in activities of daily living or instrumental activities of daily living (IADL); demographic variables may include age, race/ethnicity, sex, and marital status; behavioral factors may include smoking, alcohol consumption, and physical activity; geriatric syndromes may include such conditions as urinary incontinence, sensory impairment, and depressive sympoms.
CART is based on the concept of reducing impurity, so that the child nodes are “purer” (i.e., more homogeneous) than the parent node. 12 A node that has no impurity is one in which there is no variability in the dependent variable. 13 The splitting criterion ensures that the split is based on the largest difference in impurity between the impurity of the parent node and the weighted average of the impurity of the child nodes. 13 Consequently, the subgroups of the population that are identified in terminal nodes are more homogeneous relative to the outcome of interest than the population average.
CART models have several advantages. They allow investigators to characterize phenotypes consisting of the combinations of MCCs that are most closely associated with the study outcome. CART modeling also facilitates identification of empirically emerging population subgroups, highlighting specific combinations of variables that are strongly associated with the outcome of interest. In addition, when other variables such as demographics are included, the tree produced by the CART model may show combinations that include demographic variables, if they are important enough for the tree to base its partition, which may provide a way of showing substantial racial or ethnic disparities and inequities. Lastly, visual presentation of CART results in a tree diagram that affords easier interpretation of the complex associations among variables.
CART modeling also has limitations. Trees may be susceptible to change structure even with small changes in the data, especially when the decision tree model fails to generalize to new data, small samples, predictors that are weakly related to the outcomes, or when predictors are too strongly correlated with each other. 14 In addition, while certain variables may be quite useful in predicting the outcome in a small partition of the sample, a single tree may fail to identify these variables if the preceding splits were not optimal. 14 These limitations can be overcome by using ensembles of trees (referred to as a random forest), which use subsets of the data to identify the most important predictors. 11 The random forest approach yields more robust models, but it may not allow us to visually identify combinations of variables, which is of paramount importance when studying outcomes in the context of multimorbidity. To remedy this problem, the data can be divided into subsets, and the tree is derived in all but one of the subsets. This tree is then applied to the remaining subsets to estimate the cost of misclassification. 13
Qualifying comorbidity sets
Qualifying comorbidity sets (QCS) is a tool developed to enable hospitals and healthcare systems to evaluate how their patients with multiple health conditions fare, in terms of mortality, morbidity, length of hospitalization, expense, and so forth, relative to other hospitals and health systems (a process known as “benchmarking”). Benchmarking hinges on having an appropriate definition of multimorbidity and requires the comparison of similar patients with similar risk. 15 , 16 , 17 Although conventional approaches rely on a set (often arbitrary) number of comorbid conditions 2 , 5 or define groups with increased risk of specific outcomes through multivariate models, 2 a model that just identifies which patients are high‐risk does not convey the same depth of information as establishing that patients are high‐risk because of specific co‐occurring health conditions. Furthermore, decision‐making about quality improvement actions for specific types of patients is challenging without the ability to match specific combinations of health conditions.
The approach developed by Silber et al. applies multivariate matching algorithms to Medicare claims data to create QCSs. 18 , 19 In one application comparing mortality of hospitalized general surgery patients, multivariate matching identified QCSs composed of at most three comorbid conditions. Each QCS was required to exceed a doubling of the odds of 30‐day mortality (95% confidence interval). 19 Sixty‐seven candidate comorbid conditions and 50,183 potential QCSs were evaluated. Rigorous out‐of‐sample validation was applied to mitigate spurious findings. The algorithm identified a total of 576 QCSs; after removing redundant sets, 113 QCSs remained; 25 comorbid conditions were represented in these QCSs.
The importance of the specific QCSs identified by the algorithm can be discerned when one compares patients without qualifying but with multiple co‐occurring conditions, to patients with a similar number of co‐occurring conditions who met the QCS definition. In a separate data set, using the identified 25 comorbidities, the mortality rates of patients who did not have any of the QCSs, but who had concurrent conditions, were identified. Using four comorbidities as an example, 30‐day mortality was examined in elderly surgical patients with 4 of the 25 comorbidities that comprise QCSs but without any QCSs defined by the algorithm. There were 13,673 non‐multimorbid patients who still had four MCCs, but just not any combinations that were qualifying by the Silber et al. algorithm. 18 Their mortality odds were one‐half that of multimorbid patients who had the same number of conditions but who had at least 1 QCS (OR = 0.49 (95% CI 0.41, 0.58). If the reference population consisted of all patients with multiple conditions, the odds ratio was even lower at 0.34 (0.30, 0.39). (See look‐up tables, 19, Appendix Table 7c.) Lower odds of mortality were similarly found in the non‐multimorbid group for as many as eight qualifying comorbidities. In short, it is not just the number of comorbidities, not even the number of qualifying comorbidities, that elevates risk. Instead, it is the specific combination of co‐occurring conditions that places elderly patients at elevated risk of mortality after surgery.
Derivation of QCSs and matching should be specific to the application. In this way, hospitals wishing to examine how they treat patients cannot only closely compare their multimorbid patient outcomes to similar patients treated at other hospitals but can also closely examine those QCSs that define the hospital's population of patients with multiple health conditions. For example, a hospital may find that they are especially deficient with respect to patients who have respiratory, in addition to other conditions, and with this knowledge may gain insights into changing their care for such patients. Further, recognizing the growing prevalence of Alzheimer's Disease and Related Dementias (ADRD) in the United States and the care challenges that it presents for hospitals and healthcare systems, Jain et al. recently developed a claims‐based validated tool for defining ADRD, 20 which should further advance the explication of MCC/multimorbidity definitions with QCSs.
The QCS approach using multivariate matching of electronic records can provide considerable specificity for benchmarking and identify areas of improvement in care of patients with MCCs. It is somewhat dependent on the dataset available and utilized. Additional clinical data may become available and care patterns may change in the future requiring algorithms to be re‐run. Also, any approach that uses claims data may result in some misclassification of medical conditions and their severity. The decision of Silber et al. to use the more severe state for some MCCs may have overestimated the severity of these conditions for some patients. Like any system relying on administrative claims, records are vulnerable to compiling, transcribing and coding errors.
Multimorbidity index
The multimorbidity index (MMI) was developed by a group of data scientists taking advantage of the massive data available within EHRs from the Veterans Affairs data warehouse and the Healthcare Cost and Utilization Project of the Agency for Health Care Research and Quality. 21 , 22 , 23 , 24 The MMI assumes that every illness worsens a patient's prognosis and identifies these illnesses from the ICD‐9 and ICD‐10 codes in the EHR. Recent versions of the index organize patients' diagnoses into 13 body systems, such as “circulatory” or “endocrine/nutrition/metabolic.” The index is scored in three steps. First, a likelihood ratio (LR) is estimated for each diagnosis. 25 A LR greater than 1 worsens, and a value below 1 improves, the patient's prognosis. These ratios are made avaiable by Farrokh Alemi at http://openonlinecourses.com/464/default.asp. Second, within each body system the most serious diagnosis is identified. This corresponds to the diagnosis that has the highest LR. Third, the patient's prognosis is calculated as product of the LRs.
For example, consider a patient with iatrogenic hypotension (LR = 0), cardiac arrest (LR = 2.26), tumor lysis syndrome (LR = 1.58), and methemoglobinemia (LR = 0.31). In step 1, the LRs are obtained from the URL. In step 2, the most serious condition within each body system is identified. This step leads us to ignore hypotension because cardiac arrest is the most serious disease within the circulatory system. In step 3, under assumption of independence of body systems, the overall odds of mortality are calculated as the product of LRs associated with the most serious conditions within all body systems: Posterior Odds = Prior Odds × ∑s(Likelihood Ratios)]. In this example, we assume a prior odds of 1 to 1. The product of 0.31 × 2.26 × 1.58 estimates a posterior mortality odds of 1.11. The probability of mortality can be calculated from the odds of mortality as: Probability = Odds/(1 + Odds) = 1.11/(1 + 1.11) = 0.52.
Unlike comorbidity measures such as the Charlson index, 26 the MMI is not based on a pre‐determined subset of conditions that are the most significant predictors of mortality. 24 Instead, it considers all diagnoses of the patient, including repeated, similar, and related diagnoses; within these diagnoses it empirically finds the most serious diagnoses. For some patients, if the less severe diagnosis is the only diagnosis, then it is scored and the LR may reduce the probability of mortality. For other patients, less severe diagnoses are ignored when a more serious diagnosis in the same body system exists. The more serious diagnosis has a higher LR and it increases the probability of mortality.
The MMI relies on thousands of diseases within all body systems, and therefore is based on a comprehensive set of diagnoses. The scoring includes rare diseases, which are often ignored by other measures. Rare diseases, however, can radically change the patient's prognosis. In hospitals, patients with one rare disease are 1.80 times and those with two or more rare diseases are 2.78 times more likely to die compared with patients who do not have a rare disease. 27 Rare diseases are almost never part of statistical prognostic indices. One does not need to look at extremely rare diseases. For example, coma is a relatively uncommon condition, that can be fatal. Yet, because coma is uncommon, it is missing in almost all prognostic indices. “Although rare diseases are individually rare by definition, they are collectively common.” 28 When thousands of infrequent, but serious, conditions are missing from a prognostic index, the effect may be substantial. MMI can improve accuracy of predictions by including rare but serious diseases in its calculations. 24
A recent review 24 reported that the MMI was 15% more accurate in predicting mortality than the Quan variant of the Charlson index 29 ; 27% more accurate than the Deyo variant of the Charlson index 30 ; and 22% more accurate than the von Walraven variant of the Elixhauser Index. 31 These margins of improvement in cross‐validated accuracy are not small and show the value of the MMI, compared with conventional assessments, may be large enough to change reported treatment effects in many studies.
A limitation or impediment to uptake of MMI is it scores every diagnosis so it consequently takes more effort and therefore may be difficult for many clinicians to understand and implement. The MMI score can, however, be computed in the background of the EHR and provided to clinicians. The index and the scoring methods are described 22 , 24 and the look‐up LRs available at http://openonlinecourses.com/464/default.asp. Like methods based on administrative claims data, the MMI's reliance on the EHR does not eliminate the possibility of coding errors and missing entries.
Omics and network medicine
Omics refers to biological entities, such as the genome, its epigenomic modifications and transcription products (transcriptome), protein products (proteome), and metabolic products (metabolome)—biological molecules involved in the structure, function and dynamics of a cell, tissue or organism. Network medicine refers to construction of a complex system of interconnected elements to visualize and understand the functions and interactions of these biomolecules, which underly health and disease. 32 Application of omics and network medicine to MCC assessment has the potential to radically improve diagnosis and better understand pathogenesis by going beyond conventional clinical, epidemiological, and medical records data. Network science can provide easily explorable maps of disease co‐occurrence networks, where diseases or disease phenotypes are displayed as nodes and edges, showing the relationships between two nodes/diseases. Depicting these pairwise relationships within a map or network gives a more comprehensive picture of the problem of MCCs (Figure 2). For example, a database summarizing correlations obtained from the medical records for the disease history of more than 30 million patients resulted in a large Phenotypic Disease Network (PDN) of disease phenotypes with unique ICD9 codes. 33 Overlaying the PDN structure with longitudinal information on patient disease trajectories showed that patients, over time, develop diseases more highly connected in the PDN. Moreover, patients diagnosed with a disease that is highly connected in the network died sooner than those affected with less connected diseases, demonstrating the value of such data‐driven disease maps for understanding progression to MCCs.
FIGURE 2.
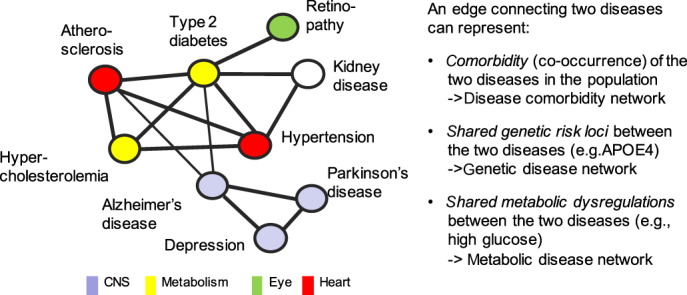
Disease co‐occurrence networks. Disease networks depict and combine pairwise relationships between diseases and allow visualization and analysis of more complex interrelationships between multiple diseases. While each node represents a disease or disease phenotype, an edge between two diseases can represent different types of relationships in different types of disease networks; depicted relationships can range from mere comorbidity (co‐occurrence) of the two diseases as observed in the population (comorbidity network) to molecular links (e.g., genetic, metabolic) between the diseases, derived from omics studies (genetic disease network; metabolic disease network). For example, the APOE‐ε4 genotype, a genetic risk factor for both Alzheimer's diseases and for atherosclerosis, suggests shared aspects in pathomechanisms.
Big omics data sets also facilitate creation of disease maps based on shared genetic associations or shared metabolic processes, to show why some diseases co‐occur. For example, a network of Mendelian gene‐disease associations was created by connecting diseases that have been associated with the same genes. 34 (See Figure 2) Another network was created in which two diseases were linked if their associated genes encode enzymes that catalyze adjacent metabolic reactions. 32 In addition to gene‐ and metabolism‐based links, diseases can be related via shared protein signals because disease‐associated proteins act on the same pathways 32 or are strongly correlated in proteomic analyses. 35 As these gene‐, metabolism‐, and protein‐based disease networks typically resemble the structure of disease co‐occurrence networks, the revealed molecular connections can help elucidate the interrelated origins of many diseases. 32
Conceptually, network medicine/systems biology treats disease phenotypes as the result of various pathobiological processes represented in a complex layered network of the organism's “omics.” Disturbances in these complex interactions within‐ and between layers of disease‐associated genes, proteins, and metabolites can result in physiological failures that eventually lead to functional, molecular and causal relationships among apparently distinct (disease) phenotypes. 30 Until recently, disease networks built on these types of associations could not differentiate among direct, potentially causal, and indirect, probably mediated, links between molecular entities and diseases.
Advanced computational modeling, however, allows integration into network models of different kinds of data, such as metabolites, gene transcripts, and clinical parameters (e.g., diagnostics, questionnaires) available for the same individuals. 36 By focusing on direct relationships, network models can help to elucidate why certain health conditions confer risk for others.
Use of longitudinal disease data can also disentangle direct and indirect relationships in co‐occurrence or molecular disease networks. Overlaying disease maps with data from individual disease trajectories and associations of non‐genetic molecular entities, such as transcript, proteins or metabolites, with future (incident) disease reveal which molecular signatures precede one or multiple diseases. For example, a study with 11,000 participants found that 65.5% of 640 significant associations between metabolites and incident disease within >20 years were shared between at least two of 27 incident noncommunicable diseases (NCD). 37 Integration of over 50 clinical risk factors demonstrated that shared metabolitic signals, such as low‐grade inflammation, decline in liver and kidney function and lipid and glucose metabolism and specific health‐related behaviors represented antecedents of common NCD MCCs.
Combining the vast molecular (omics), clinical and disease phenotypes data within disease networks provides the opportunity to cross barriers of current disease definition based on symptoms and organ systems and eventually move to a more mechanism‐based definition of disease. This task is massive, ongoing, and requires integration of genetic, proteomic, metabolic and phenotypic datasets from genetic testing, assays, clinical diagnosis, medical records, and so forth. Network development and computational modeling also require large data sets and the collection and testing of numerous potential confounding variables. These approaches also are expensive, and may miss confounders. Nonetheless, omics/network medicine approaches have potential to estimate the likelihood that a patient will develop a particular pattern of MCCs, identify pathogenesis, and suggest appropriate preventive measures—beyond what current measurement approaches provide.
POTENTIAL APPLICATIONS
Each approach to MCC assessment adds to the toolbox for improving the epidemiology, diagnosis and treatment of persons with co‐occuring medical conditions. Three of the approaches (CART, QCS, and MMI) already are available for implementation; the fourth (omics and network medicine) has contributed new understanding about why different health conditions co‐occur, and is likely to lead to clinical translation.
As a descriptive tool, CART offers the unique advantage of identifying subgroups of the patient population that are at highest (or lowest) risk for the outcome of interest—based not only on single factors or the interaction of two variables, but on combinations of multiple variables, besides medical conditions per se, that are deemed important by the model (e.g., functional limitations, age, SES). CART models also can identify older patients who would benefit from comprehensive geriatric assessment, close monitoring, and targeted interventions to improve quality of life, paving the way for precision geriatric medicine.
The ability of clinicians and hospital systems to assess their success (or failure) relative to other clinicians and hospitals in treating patients with MCCs has been limited by multiple definitions of multimorbidity and difficulties identifying and comparing similar patients with similar risk. QCS, derived from multivariate matching of EHR data, provides a valid, empirically‐based metric that overcomes these limitations. The approach enables hospitals and clinicians to benchmark the care of their MCC patients. It also facilitates identification of the MCCs representing specific disease components, that pose the highest mortality risk and costs across caregivers, hospitals, and hospital systems, thereby providing information to improve systems of care and referral patterns within healthcare organizations.
Compared to conventional measures, the MMI extracts more comprehensive diagnostic information (even about conditions that may be rare, but serious) from EHRs and applies risk algorithms to provide detailed prognostic information. The complex computations required by MMI can be performed in the background, so that user‐friendly information is provided for MCC patients and their clinicans for planning the most appropriate care decisions, evaluating comparative effectiveness of treatments, and anticipating patients' acuity and nursing needs by administrators. Successful translations of MMI have been reported by several projects with Veterans Administration ICU and nursing home patients. 21 , 23
Omics and network medicine permit the mapping of phenotypic, genetic and biological overlap among health conditions and identify common pathogenic pathways. As omics are integrated successfully with EHRs, clinicians and researchers may be able to select treatments that are complementary in their effects and improve prediction and prognosis for MCCs. The multi‐level nature of the omics offers the radical possibility that current diagnostic systems that tend to treat one condition at a time will be replaced by constellations of MCCs and interconnected pathogenic processes to guide preventive measures and medical procedures.
All four approaches to multimorbidity stem from advances in statistics and computing, omics and informatics and “Big Data” extracted from EHRs and administrative claims data. In addition, researchers using CART have also availed themselves of data on functional limitations, geriatric sydromes, and so forth, such as collected in the U.S. Health & Retirement Study. The reader may be concerned that these new tools' reliance on algorithms, artifical intelligence, interactions among different omics, and psychosocial and behavioral variables introduces a level of complexity that will discourage clinical uptake. Fortunately, the developers of these approaches have from the outset kept in mind accessibility, time and effort (often with computation “working in the background”), and usability, which will facilitate clinical translation and increase the ability to meet the ongoing challenges of multimorbidity in older adults.
It has been said that if you cannot measure it, you cannot change it. The four emerging measurement approaches described in this article have the potential to advance research and help caregivers treat patients and policy makers evaluating care for these complex and vulnerable patients.
AUTHOR CONTRIBUTIONS
All authors contributed to concept, design, interpretation, and preparation of manuscript.
CONFLICT OF INTEREST
None.
SPONSOR'S ROLE
None.
FINANCIAL DISCLOSURE
Funding support for the NIH 2018 workshop on which this article is based was provided by the National Institute on Aging, National Cancer Institute, National Institute on Minority Health and Health Disparities, NIH Office of Disease Prevention, and NIH Office of Behavioral and Social Sciences Research.
ACKNOWLEDGMENTS
Acknowledge Dr. Nicholas K. Schiltz of Frances Payne Bolton School of Nursing, Case Western Reserve University, and Mr. Wyatt Bensken of the Department of Population and Quantitative Health Sciences, School of Medicine, Case Western Reserve University, for their careful review of this manuscript, and useful comments.
Suls J, Salive ME, Koroukian SM, et al. Emerging approaches to multiple chronic condition assessment. J Am Geriatr Soc. 2022;70(9):2498‐2507. doi: 10.1111/jgs.17914
Portions of this article were presented at the workshop. This material should not be interpreted as representing the official viewpoint of the U.S. Department of Health and Human Services, the National Institutes of Health, or its represented agencies.
Funding information National Cancer Institute; National Institute on Aging; National Institute on Minority Health and Health Disparities; National Institutes of Health Office of Behavioral and Social Sciences Research; National Institutes of Health Office of Disease Prevention
Contributor Information
Jerry Suls, Email: jsuls@northwell.edu.
Carrie N. Klabunde, Email: klabundc@mail.nih.gov.
REFERENCES
- 1. King DE, Xiang J, Pilkerton CS. Multimorbidity trends in United States, 1988–2014. J Am Board Fam Med. 2018;31:503‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suls J, Bayliss E, Berry J, et al. Measuring multimorbidity: selecting the right instrument for the purpose and the data source. Med Care. 2021;59(8):743‐756. doi: 10.1097/MLR.0000000000001566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang L, Porter B, Maynard C, et al. Predicting risk of hospitalization or death among patients receiving primary care in the Veterans Health Administration. Med Care. 2013;51(4):368‐373. doi: 10.1097/MLR.0b013e31827da95a [DOI] [PubMed] [Google Scholar]
- 4. Zink A, Rose S. Identifying undercompensated groups defined by multiple attributes in risk adjustment. BMJ Health Care Inform. 2021;28(1):e100414. doi: 10.1136/bmjhci-2021-100414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salive M, Suls J, Farhat T, Klabunde C. Commentary: National Institutes of Health advancing multimorbidity research. Med Care. 2021;59(7):622‐624. doi: 10.1097/MLR.0000000000001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koroukian SM, Warner DF, Owusu C, Given CW. Multimorbidity redefined: prospective health outcomes and the cumulative effect of co‐occurring conditions. Prev Chronic Dis. 2015;12:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldman L, Weinberg M, Weisberg M, et al. A computer‐derived protocol to aid in the diagnosis of emergency room patients with acute chest pain. New Engl J Med. 1982;307:588‐596. [DOI] [PubMed] [Google Scholar]
- 8. Goldman L, Cook EF, Brand DA, et al. A computer protocol to predict myocardial infarction in emergency department patients with chest pain. New Engl J Med. 1988;318:797‐803. [DOI] [PubMed] [Google Scholar]
- 9. Koroukian SM, Schiltz N, Warner DF, et al. Combinations of chronic conditions, functional limitations, and geriatric syndromes that predict health outcomes. J Gen Intern Med. 2016;31(6):630‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei MY, Kawachi I, Okereke OI, Mukamal KJ. Diverse cumulative impact of chronic diseases on physical health‐related quality of life: implications for a measure of multimorbidity. Am J Epidemiol. 2016;184(5):357‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Breiman L. Random forests. Mach Learn. 2001;45(1):5‐32. [Google Scholar]
- 12. Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods. 2009;14(4):323‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemon SC, Roy J, Clark MA, Friedmann PD, Rakowski W. Classification and regression tree analysis in public health: methodological review and comparison with logistic regression. Ann Behav Med. 2003;26(3):172‐181. [DOI] [PubMed] [Google Scholar]
- 14. King MW, Resick PA. Data mining in psychological treatment research: a primer on classification and regression trees. J Consult Clin Psychol. 2014;82(5):895‐905. [DOI] [PubMed] [Google Scholar]
- 15. Silber JH, Rosenbaum PR, Ross RN, et al. Template matching for auditing hospital cost and quality. Health Serv Res. 2014;49:1446‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silber JH, Rosenbaum PR, Ross RN, et al. A hospital‐specific template for benchmarking its cost and quality. Health Serv Res. 2014;49:1475‐1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silber JH, Rosenbaum PR, Ross RN, et al. Indirect standardization matching: assessing specific advantage and risk synergy. Health Serv Res. 2016;51:2330‐2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosenbaum PR. Design of Observational Studies. Springer; 2010. [Google Scholar]
- 19. Silber JH, Reiter JG, Rosenbaum PR, et al. Defining multimorbidity in older surgical patients. Med Care. 2018;56:701‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain S, Rosenbaum PR, Reiter JG, et al. Using Medicare claims in identifying Alzheimer's disease and related dementias. Alzheimer Dementia. 2021;17:515‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Min H, Avramovic S, Wojtusiak J, et al. A comprehensive multimorbidity index for predicting mortality in intensive care unit patients. J Palliat Med. 2017;20(1):35‐41. [DOI] [PubMed] [Google Scholar]
- 22. Kheirbek RE, Alemi F, Fletcher R. Heart failure prognosis: comorbidities matter. J Palliat Med. 2015;18(5):447‐452. [DOI] [PubMed] [Google Scholar]
- 23. Levy C, Kheirbek R, Alemi F, et al. Predictors of six‐month mortality among nursing home residents: diagnoses may be more predictive than functional disability. J Palliat Med. 2015;18(2):100‐106. [DOI] [PubMed] [Google Scholar]
- 24. Alemi F, Levy CR, Kheirbek RE. The multimorbidity index: a tool for assessing the prognosis of patients from their history of illness. EGEMS (Wash DC). 2016;4(1):1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McGee S. Simplifying likelihood ratios. J Gen Intern Med. 2002;17(8):646‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373‐383. [DOI] [PubMed] [Google Scholar]
- 27. Blazsik RM, Beeler PE, Tarcak K, Cheetham M, von Wyl V, Dressel H. Impact of single and combined rare diseases on adult inpatient outcomes: a retrospective, cross‐sectional study of a large inpatient population. Orphanet J Rare Dis. 2021;16(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferreira CR. The burden of rare diseases. Am J Med Genet A. 2019;179(6):885‐892. doi: 10.1002/ajmg.a.61124 [DOI] [PubMed] [Google Scholar]
- 29. Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD‐9‐CM and ICD‐10 administrative data. Med Care. 2005;43:1130‐1139. [DOI] [PubMed] [Google Scholar]
- 30. Deyo R, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD‐9‐CM administrative databases. J Clin Epidemiol. 1992;45:613‐619. [DOI] [PubMed] [Google Scholar]
- 31. von Walraven C, Austin PC, Jennings A, et al. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Med Care. 2009;47:626‐633. [DOI] [PubMed] [Google Scholar]
- 32. Barabási A‐L, Gulbahce N, Loscalzo J. Network medicine: a network‐based approach to human disease. Nat Rev Genet. 2011;12(1):56‐68. doi: 10.1038/nrg2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hidalgo CA, Blumm N, Barabási A‐L, Christakis NA. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5(4):e1000353. doi: 10.1371/journal.pcbi.1000353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685‐8690. doi: 10.1073/pnas.0701361104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zierer J, Pallister T, Tsai PC, et al. Exploring the molecular basis of age‐related disease comorbidities using a multi‐omics graphical model. Sci Rep. 2016;6:37646. doi: 10.1038/srep37646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pietzner M, Stewart ID, Raffler J, et al. Plasma metabolites to profile pathways in noncommunicable disease multimorbidity. Nat Med. 2021;27(3):471‐479. doi: 10.1038/s41591-021-01266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]


