Abstract
Objective
Human immunodeficiency virus (HIV) persistence in blood and tissue reservoirs, including the brain, is a major barrier to HIV cure and possible cause of comorbid disease. However, the size and replication competent nature of the central nervous system (CNS) reservoir is unclear. Here, we used the intact proviral DNA assay (IPDA) to provide the first quantitative assessment of the intact and defective HIV reservoir in the brain of people with HIV (PWH).
Methods
Total, intact, and defective HIV proviruses were measured in autopsy frontal lobe tissue from viremic (n = 18) or virologically suppressed (n = 12) PWH. Total or intact/defective proviruses were measured by detection of HIV pol or the IPDA, respectively, through use of droplet digital polymerase chain reaction (ddPCR). HIV‐seronegative individuals were included as controls (n = 6).
Results
Total HIV DNA was present at similar levels in brain tissues from untreated viremic and antiretroviral (ART)‐suppressed individuals (median = 22.3 vs 26.2 HIV pol copies/106 cells), reflecting a stable CNS reservoir of HIV that persists despite therapy. Furthermore, 8 of 10 viremic and 6 of 9 virally suppressed PWH also harbored intact proviruses in the CNS (4.63 vs 12.7 intact copies/106 cells). Viral reservoirs in CNS and matched lymphoid tissue were similar in the composition of intact and/or defective proviruses, albeit at lower levels in the brain. Importantly, CNS resident CD68+ myeloid cells in virally suppressed individuals harbored HIV DNA, directly showing the presence of a CNS resident HIV reservoir.
Interpretation
Our results demonstrate the first evidence for an intact, potentially replication competent HIV reservoir in the CNS of virally suppressed PWH. ANN NEUROL 2022;92:532–544
The major barrier to human immunodeficiency virus (HIV) cure is the persistence of latent virus in reservoirs, which is unable to be eradicated by antiretroviral therapies (ARTs). Tissue reservoirs of HIV are difficult to access (both therapeutically and physically) and potentially exhibit different viral profiles and dynamics compared to blood CD4+ T cell reservoirs. 1 Therefore, characterizing HIV persistence in tissue sites, including the brain, is critical to HIV treatment and cure approaches.
HIV infects the central nervous system (CNS) during acute infection, likely via infiltration of infected monocytes and/or T cells across the blood brain barrier, 2 which can spread to CNS resident myeloid cells (perivascular macrophages and microglia), 3 , 4 , 5 astrocytes, 6 , 7 and pericytes, 8 thus, establishing a viral reservoir. Whereas early infection events are relatively well‐defined, the location, frequency, and proviral landscape of the CNS reservoir in ART‐suppressed people with HIV (PWH) is unclear. This is problematic as HIV persistence in the CNS may act as a source and site of ongoing replication. Furthermore, CNS reservoirs of HIV may contribute to the pathogenesis of HIV‐associated neurocognitive disorders, which affect approximately 30% of ART‐suppressed PWH. 9 , 10 , 11 Previous quantitative polymerase chain reaction (qPCR)‐based studies of humans and animal models have detected viral DNA or RNA in the CNS of ART‐suppressed individuals/animals, 1 , 12 , 13 supporting a stable viral reservoir in the brain. However, these studies do not define the proportion/magnitude of proviruses that are “intact” and potentially replication competent, which is <2 to 11.7% of all proviruses in CD4+ T cells (approximately 88.3–98% of total proviruses). 14 , 15 , 16 Cerebrospinal fluid (CSF) escape studies, 17 and in vivo non‐human primate (NHP) latency studies 18 suggest that the brain, particularly resident myeloid cells, may harbor replication competent genomes. However, the size of the intact proviral reservoir in the brain of ART‐suppressed PWH is unclear.
Typical qPCR‐based assays for HIV DNA cannot distinguish intact from defective proviruses, thus overestimating the size of the intact, replication competent reservoir. The intact proviral DNA assay (IPDA) is a novel multiplex droplet digital PCR (ddPCR) approach to distinguish and separately quantify intact versus defective HIV proviruses. The assay defines proviruses by a lack of fatal defects, such as large deletions in the 5′ region of the packaging signal (ψ) of the provirus as well as APOBEC3G‐mediated hypermutations or deletions in the Rev response element (RRE) of HIV envelope (env). 16 The IPDA has been validated as an accurate measure of intact, replication competent HIV genomes in the blood 16 , 19 and lymph nodes. 20 Furthermore, intact proviruses detected by IPDA correlate with total integrated HIV 21 and with quantitative viral outgrowth assays (qVOAs), 16 , 19 supporting the use of this technology as a reliable surrogate measure of intact, potentially replication competent HIV proviruses in tissues where it is difficult to perform qVOA. Here, we utilized the IPDA to quantify intact and defective proviruses in brain tissue from PWH to characterize the proviral landscape in the CNS.
Methods
Tissue Cohort
Fresh frozen and formalin‐fixed paraffin embedded (FFPE) human autopsy brain tissue (frontal lobe deep white matter) and matched lymphoid tissue (spleen, lymph node, or gut‐associated lymphoid tissue) from PWH or HIV‐seronegative individuals were provided by the National NeuroAIDS Tissue Consortium (NNTC, USA; https://nntc.org 22 ). All tissues were acquired and processed with ethics approval (RMIT University, Australia; HREC#20843).
Genomic DNA Extractions
The gDNA/RNA from fresh frozen brain and matched lymphoid tissues (approximately 10 mg pieces) were extracted using the AllPrep DNA/RNA/miRNA universal kit (QIAGEN, Hilden, Germany), where isolated gDNA was used for HIV pol DNA quantification and RNA for T cell receptor (TCR) quantification, or the DNA extraction kit (Agilent, Santa Clara, CA) for the IPDA, as per manufacturer's instructions. DNA shearing was minimized during DNA extraction for the IPDA (Agilent) by excluding mechanical homogenization (wide‐bore pipetting was used instead).
Droplet Digital PCR
The ddPCR (QX200; BioRad, Hercules, CA) was used for HIV pol DNA quantification and IPDA analyses, as previously described. 16 , 23 Positive droplets per sample were defined by examining negative control wells (no template controls and gDNA from HIV‐seronegative biological controls) for false‐positive HIV droplets above the set threshold to determine a false‐positive rate.
HIV pol DNA Quantification
HIV pol and ribonuclease P/MRP subunit P30 (RPP30) for total input gDNA quantification was quantified, as previously described 24 (Table S1). A median (interquartile range [IQR]) of 482,026 (452,543–571,480) cells were screened per sample.
Intact Proviral DNA Assay
Intact (Ψ+ and env +) and defective (Ψ+ or env +) HIV genomes were measured by the IPDA using high molecular weight gDNA from homogenized fresh frozen brain and matched lymphoid tissues, as previously described. 16 NL4‐3 control plasmids were generated by restriction enzyme excision to remove Ψ and/or env regions, where 5′ overhangs were filled‐in with DNA Polymerase I, Large (Klenow) Fragment (NEB) and blunt ends ligated with Blunt/TA ligase master mix (NEB). Additionally, a commercially synthesized gBlock fragment of a hypermutated env sequence was used for env probe validation experiments (Integrated DNA Technologies, Coralville, IA). Intact HIV copies in PWH were corrected for DNA shearing by calculating the fraction of gDNA that has been sheared between the RPP30 amplicons. Prior to droplet formation, high molecular weight gDNA was digested with the restriction enzyme Xhol (NEB), due to the absence of recognition sites within and between genomic regions of interest. HIV and human RPP30 reactions were conducted independently and in parallel, where copies were normalized to total RPP30 detected, as a measure of input cell number. In each ddPCR reaction, a median of 320 ng (300 – 350 ng, HIV) or a median of 40 ng (37.5–43.75 ng, RPP30) of Xhol digested gDNA was combined with ddPCR mastermix. Ten technical replicates were performed for HIV detection and 2 technical replicates for RPP30 quantification for each patient sample, where a median (IQR) of 549,576 (518,430 – 636,372) cells were assayed in total. For samples that exhibited low levels of env via IPDA, likely due to HIV polymorphisms within the RRE IPDA target region, a secondary env primer/probe set was used as a single‐plex reaction. 23 Primers/probes are listed in Table S1.
Amplification of HIV V3 env from CD68+ Cells Isolated by Laser Capture Microdissection
The nuclei of CD68+ myeloid cells (>50 μm from observed vessels) were isolated from FFPE frontal lobe tissue, as previously described. 3 Greater than 500 cells/sample were isolated per sample and DNA purified (FFPE DNA extraction kit; QIAGEN, Germany) according to the manufacturer's protocol. HIV V3 env sequences were amplified from DNA prepared from the nuclei of CD68+ cell populations, as previously described. 6 Five to 10 cell equivalents/μl were used in multiples of 8 independent PCR reactions (V3 primers; Table S1). V3 env was purified from PCR positive reactions (<2+ per 8 PCR reactions performed) and Sanger sequenced. CLC Main Workbench version 20 was used to align sequences.
DNAscope Detection of HIV vDNA+CD68+ Myeloid Cells
HIV DNA+ myeloid cells in FFPE fixed human frontal cortex tissue sections (10 μm) were identified using DNAscope (V‐HIV1‐CladeB‐sense) and CD68 antibody (KP‐1, DAKO, Glostrup, Denmark; 1:400), as previously described, 25 with the following modifications. Nuclei was labeled with Hoechst (Invitrogen, Waltham, MA; 1:750 in TBS; 15 minutes), and autofluorescence quenched with TrueBlack Lipofuscin Quencher (Biotium, Fremont, CA; 1:50 in 70% EtOH; 30 seconds). Slides were cover slipped and imaged using the PALM microbeam (Zeiss, Oberkochen, Germany).
Quantification of CD3+ T Cells in Brain Tissue
CD3+ T cells were identified in FFPE frontal cortex tissue section (6 μm) by chromogenic immunohistochemistry analysis using anti‐CD3 (rabbit polyclonal, 1:100; DAKO), as previously described. 3 , 6 Whole tissue samples were imaged (VS120 slide scanner; Olympus, Shinjuku, Japan) and CD3+ T cells (relative to total nuclei) were quantified using HALO imaging software (Indica Labs version 3.3).
Quantification of TCR in Brain Tissue
Relative expression of TCRbC mRNA in brain tissue was quantified by qPCR. The cDNA was synthesized (SuperScript III First‐Strand Synthesis System; Thermo Fisher) and quantified by SYBR‐based qPCR (Quantinova; QIAGEN) using a QuantStudio 5 real‐time PCR machine (Thermo Fisher). Cycle settings: 95°C for 2 minutes, 95°C for 5 seconds, 60°C for 18 seconds. Relative gene expression was standardized using the ΔΔCt method to 60S ribosomal protein L13 (RPL13). Primers are listed in Table S1.
Statistics
Comparisons made using nonparametric Mann–Whitney U (unpaired) and Wilcoxon tests (paired). Pearson analyses used for correlative analyses. All analysis was performed using GraphPad Prism (version 9.2.0; GraphPad Software, La Jolla, CA).
Results
HIV DNA Persists in the CNS of PWH despite Viral Suppression with ART
To first quantify and characterize the presence of HIV in the CNS, ddPCR for HIV pol and the RPP30 gene to determine total cell input were performed using DNA extracted from frontal lobe brain tissue. Fresh frozen brain tissue was obtained from HIV‐seronegative individuals (n = 6), viremic PWH (median plasma viral load: 56,129 HIV RNA copies/ml; median CSF viral load: 595 HIV RNA copies/ml; n = 18) and virally suppressed PWH (VS PWH; all undetectable plasma and CSF HIV viral load; CD4 T cell count [median]: 355 cells/mm3; n = 12) provided by the NNTC (Table 1). Viral suppression was defined as a minimum of a 2‐year period of ART‐controlled HIV viremia with undetectable plasma and CSF viral loads (a single blip of 250 HIV RNA copies/ml >6 months from death was allowed). The median time of viral suppression of VS PWH was 6.2 years (IQR = 2.6–9.1).
TABLE 1.
Clinical cohort characteristics
| HIV+ | |||
|---|---|---|---|
| HIV− | Viremic | Virally suppressed | |
| N | 6 | 18 | 12 |
| Age | 55 (39–74) | 44 (35–62) | 59 (44–67) |
| Male sex (%) | 2 (33.3) | 11 (61.1) | 12 (100) |
| HIV parameters | |||
| Viral load (plasma) a | ‐ | 56,129 (15,090–207,526) | UD |
| Viral load (CSF) a | ‐ | 594.5 (340.3–5,973) | UD b |
| CD4 T cells (mm3) | 72.0 (5.50–185) c | 355 (239–785) d | |
| Nadir CD4 T cells (mm3) | 8.00 (2.00–72.0) c | 78.0 (54.0–161) d | |
| Current ART (%) | 0 (0%) | 12 (100%) | |
| Viral suppression (yr) e | N/A | 6.2 (2.6–9.1) | |
ART = antiretroviral therapy; CSF = cerebrospinal fluid; N/A = not applicable; UD = undetectable.
HIV RNA copies/ml.
Data missing for 4 donors.
Data missing for 13 donors.
Data missing for 1 donor.
Viral suppression defined by undetectable plasma viral load for >2 years (1 blip <250 HIV RNA copies/ml >6 months from death permitted).
HIV DNA was detected in brain tissue from all PWH, as measured by ddPCR for HIV pol (Fig 1A). Importantly, the levels of HIV DNA in brain tissue from VS PWH were similar to those from viremic PWH (median = 26.2 vs 22.3 HIV pol DNA copies/106 cells, p > 0.05), indicating that viral suppression of HIV in the blood does not influence HIV DNA levels in the CNS. HIV DNA levels in the CNS of viremic PWH correlated with CSF viral load (ρ = 0.655, p = 0.03; Fig 1B), but not with plasma HIV RNA levels (ρ = 0.143, p = 0.58; Fig 1C), suggesting that the CNS is a unique tissue compartment of HIV. Due to the definition of “viral suppression” as an undetectable viral load (ie, below the level of detection of clinical assays), no associations between HIV viremia in plasma and HIV DNA in brain tissue of VS PWH were made. Together, these findings predict the presence of an HIV reservoir in the CNS which is not associated with plasma HIV viral load levels or substantially influenced by long‐term viral suppression in the blood by ART.
FIGURE 1.
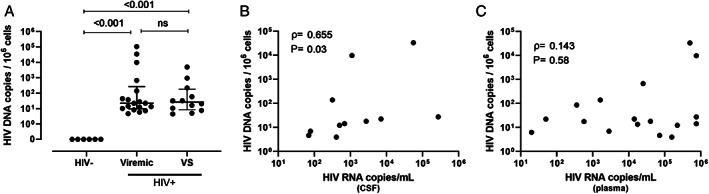
HIV DNA persists in the CNS of people with HIV PWH despite long term suppression with ART. (A) HIV pol DNA copy number in human frontal lobe brain tissue from viremic (n = 18) or virally suppressed (n = 12) PWH or HIV‐uninfected individuals (n = 6) as measured by droplet digital PCR. (B) Correlative analysis of HIV pol DNA copy number in human brain tissue relative to HIV RNA copy number in cerebrospinal fluid or (C) plasma in untreated viremic PWH only (n = 18). Median and interquartile ranges shown; comparisons made using Mann–Whitney U tests. Correlative analyses assessed by non‐parametric Pearson's correlations with Pearson's rho and p values shown. The p < 0.05 considered statistically significant. Note: CSF viral load data unavailable for n = 4. ART = antiretroviral; CNS = central nervous system; ns = not significant; PCR = polymerase chain reaction; PWH = people with HIV; VS = virally suppressed.
CNS Tissues from VS PWH Harbor Intact HIV Proviral Genomes
To date, identifying the presence of intact proviruses in the CNS of large cohorts of VS PWH has been difficult due to (i) the requirement of a large amount of high quality, high molecular weight DNA for advanced sequencing procedures, including full‐length individual proviral sequencing (FLIPS) and/or (ii) the inability to perform qVOA analyses using autopsy material. To overcome these obstacles we optimized the IPDA to quantify both intact and defective HIV genomes in the CNS via multiplex ddPCR. The IPDA is a duplex ddPCR assay which targets HIV proviral DNA that is integrated in genomic DNA of host cells during viral infection. HIV integration is an essential step for HIV replication. Using two primer probe sets that target either the 5′ or 3′ end of the integrated HIV provirus, the simultaneous detection of both probe sets (separated by 7,564 bp base pairs) in a single lipid droplet demonstrates the presence of an “intact” near full length HIV provirus integrated into host DNA which may be capable of viral replication. This assay differs from other single or dual target ddPCR assays that can only detect and quantify short regions of integrated HIV DNA (ie, HIV gag, pol, etc.) as it simultaneously detects 2 regions of HIV that are commonly deleted or hypermutated in defective proviruses. 12 , 26 The IPDA was validated using HIV NL4‐3 plasmid controls that mimic intact or defective proviruses; generated via restriction enzyme digestion, as previously described (Fig 2A, 16 ). As expected, only intact (ψ+env+), 3′ defective (ψ+ only) or 5′ defective proviruses (env+) were present in respective controls, validating primer specificity in our hands (representative 2D amplitude ddPCR plots shown; Fig 2B). To remove the possibility of hypermutated sequences in HIV env being amplified and mistakenly quantified by the IPDA, amplification of env involved a VIC‐labeled probe that bound to intact/nonhypermutated sequences, and an unlabeled competitor probe that favorably binds to hypermutated sequences in the RRE, as previously described 16 (Fig 2C). The ddPCR was performed using varying mixed ratios of both full‐length HIV NL4‐3 and a commercially synthesized gBlock fragment of a hypermutated env sequence. HIV env copies correlated with NL4‐3 input, but not levels of hypermutated gBlock fragment DNA (see Fig 2C), confirming the specificity and sensitivity of these primers to effectively discriminate and quantify nonhypermutated HIV env. To further confirm the sensitivity of the assay, the IPDA was performed on gDNA from peripheral blood mononuclear cells of uninfected donors (n = 3) spiked with a series of ACH‐2 cell equivalents, which contains a single provirus per cell (Fig 2D). As expected, the HIV DNA copy number of HIV ψ (ρ = 0.98, p = 0.006) or env (ρ = 0.99, p = 0.004) measured by the IPDA strongly correlated with input HIV (based on ACH‐2 cell input; see Fig 2D), directly validating the sensitivity and specificity of the IPDA in our hands.
FIGURE 2.
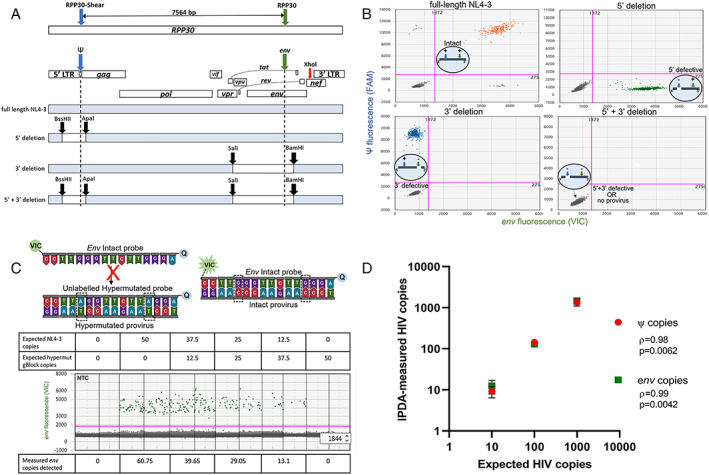
Validation of IPDA specificity. (A) Primer binding map for HIV Ψ (blue), HIV env (green) and RPP30 (blue and green arrows; same spacing as the Ψ and env amplicons (7,564 bp) to correct for DNA shearing in a separate multiplex ddPCR). XhoI (red arrow) binding site for gDNA digestion for the IPDA is shown. Maps of NL4‐3 plasmid controls including restriction enzyme cut sites (black arrow): (i) full‐length NL4‐3, (ii) 5′ deletion, (iii) 3′ deletion, or (iv) 5′ + 3′ deletion. (B) Representative IPDA 2D‐amplitude plots using 1,000 DNA copies of NL4‐3 plasmid controls shown in A assayed on a background of PBMC gDNA from HIV seronegative donor. Intact (HIV Ψ + and env+; orange), 5′ deleted (HIV env + only; green), 3′ deleted (HIV Ψ + only, blue), and 5′ + 3′ deleted NL4‐3 plasmid controls (HIV ψ‐ and env‐; grey). Bottom left quadrants also represents droplets with no provirus. (C) Map of env probes specific for the RRE region consisting of 2 adjacent G‐>A APOBEC3G sites (dashed boxes). An unlabeled hypermutated competitor probe preferentially binds to hypermutated proviruses, preventing the binding of the VIC‐labelled intact probe, which only hybridizes to intact proviruses to release fluorescent signal away from 3′ minor‐groove binder quencher (Q). Specificity of the env probes was validated using varying copies of HIV NL4‐3 spiked with a synthetic gBlock fragment of a hypermutated env sequence (average env copies from n = 3 experiments shown). (D) Correlative analysis of expected and IPDA‐measured frequencies of ψ and env copies of HIV. The gDNA from PBMCs of HIV seronegative donors was spiked with cell equivalent concentrations of ACH‐2 cells and subjected to the IPDA. Nonparametric Pearson's rho and significant p values (p < 0.05) shown. There were 3 experiments performed. ddPCR = droplet digital polymerase chain reaction; IPDA = intact proviral DNA assay; NNTC = National NeuroAIDS Tissue Consortium; PBMC = peripheral blood mononuclear cell; RRE = Rev response element.
The IPDA was performed on brain tissue from PWH where adequate CNS tissue (>4.5 × 105 cells) was available (n = 10 VS PWH; n = 11 viremic PWH) and 6 HIV‐seronegative individuals as biological controls. IPDA results were corrected for DNA shearing based on the frequency of RPP30 double positive droplets using a separate multiplex ddPCR for 2 regions of the RPP30 gene with similar genomic spacing to the Ψ and env amplicons (approximately 7.5 kb; see Fig 2, 16 ). Minimal gDNA shearing was observed in all samples and within expected levels 16 (median droplet shearing index [IQR]: brain gDNA 0.17 [0.13–0.23] and lymphoid tissue gDNA: 0.24 [0.16–0.38]; Fig 3A). Intact proviral genomes were present in brain tissue from the majority of viremic PWH (8/10 individuals, median [IQR] copies/106 cells: 4.63 [1.93–350.7]; Fig 3B, C), accounting for approximately 11.1% of all proviruses (Fig 3D). The major proportion of proviral DNA in brain tissue from viremic PWH was either 3′ defective (45.0 [21.2–448] copies/106 cells, approximately 43.1% of total proviruses) or 5′ defective (36.9 [12.3–807] copies/106 cells, approximately 45.8% of proviruses; see Fig 3C, D). Importantly, intact proviral genomes were also present in CNS tissue from the majority of VS PWH (6/9 individuals, 12.7 [0–67.8] copies/106 cells; approximately 15.5% of total proviruses; see Fig 3C). Similar to findings in viremic PWH, the majority of proviruses detected in VS PWH were either 3′ (24.7 [7.74–107] copies/106 cells; see Fig 3C, D; approximately 37.2% of proviruses) or 5′ defective (59.4 [11.2–162] copies/106 cells; see Fig 3C, D; approximately 47.1% of proviruses).
FIGURE 3.

Intact proviral genomes persist in the CNS despite long term viral suppression as measured by the IPDA. (A) The DSI was determined in brain (n = 27) or lymphoid (n = 7) tissue by duplex ddPCR for two regions of the RPP30 gene and represents the fraction of gDNA that has been sheared between the amplicons. Median and interquartile range shown. (B) Representative 2D‐amplitude ddPCR plot of proviruses in brain tissue from a virally suppressed PWH. (C) Copy number (standardized to 106 cells) or (D) frequency of proviral genomes defined as intact (HIV ψ + and env +; orange), 3′ defective (ψ +; blue) or 5′ defective (env +, green) from CNS frontal lobe tissue from viremic (n = 10), or virally suppressed (VS; n = 9) PWH or HIV‐uninfected individuals (n = 6) as measured by the IPDA. Median and interquartile ranges shown. (E) Correlative analysis of HIV pol DNA and intact, (F) 3′ defective or (G) 5′ defective proviruses in frontal lobe brain tissue from PWH. Nonparametric Pearson's rho and p values shown. p values <0.05 considered statistically significant. CNS = central nervous system; ddPCR = droplet digital polymerase chain reaction; DSI = droplet shearing index; IPDA = intact proviral DNA assay; PWH = people with HIV.
Notably, the IPDA was also performed for an additional viremic PWH and an additional VS PWH. As low HIV env signal was detected for these individuals, a phenomenon known to occur in a small number of cases due to polymorphisms within the env primer/probe region, 23 an additional round of ddPCR was performed using previously published alternate env primers (data not shown). This confirmed failure of env amplification and these samples were removed from analysis. HIV‐seronegative controls (n = 6) showed no indicators of intact, 3′ defective or 5′ defective proviruses, ruling out that the levels detected were due to experimental contamination.
The levels of intact, 3′, and 5′ defective proviruses in the CNS all positively correlated with levels of HIV pol DNA (p < 0.05 for all; Fig 3E–G), indicating that the level of intact and defective proviruses in the CNS is proportionally associated with reservoir size. Together, these findings provide the first evidence for an intact proviral HIV reservoir in the CNS of a subset of VS PWH.
CNS and Lymphoid Tissue Reservoirs Exhibit a Similar Frequency of Intact and Defective Proviruses
To compare the relative size and nature of the intact and defective HIV reservoir in the CNS versus lymphoid tissue reservoirs, the IPDA was performed on lymphoid tissues of a subset of individuals where lymphoid tissue was available (n = 7; 2 individuals were excluded from analysis due to inefficient env amplification, as noted above). Intact proviral genomes were detected in lymphoid tissues of all individuals tested (5/5), with a trend to higher intact HIV copy number present in lymphoid tissue versus CNS tissue (median HIV intact copy number CNS vs lymphoid tissue: 3.78 vs 110 copies/106 cells; Fig 4). Similar to findings in the CNS (see Fig 3C), the majority of proviruses in lymphoid tissues were either 3′ (233 [120–6,757] copies/106 cells) or 5′ defective (302 [154–1,483] copies/106 cells; see Fig 4A). A tread to higher absolute levels of all forms of HIV proviruses in lymphoid tissues relative to matched CNS tissues was observed, however, this did not reach statistical significance. No difference was observed in the frequency of intact proviruses between the CNS and matched lymphoid tissues (Fig 4B), indicating that the composition of the proviral landscape in the CNS reflects that in the peripheral lymphoid tissues.
FIGURE 4.

CNS reservoir size relative to lymphoid reservoirs in PWH. (A) Copy number (standardized to 106 cells) or (B) frequency of proviral genomes measured by the IPDA from CNS frontal lobe tissue and matched lymphoid tissue (the spleen, lymph node, or gut) from viremic (n = 3, closed symbols) or virally suppressed PWH (n = 2, open symbols), where proviruses are defined as intact (ψ+ and env +; orange), 3′ defective (ψ +; blue), or 5′ defective (env +, green). Median and interquartile ranges shown; comparisons made using paired Wilcoxon tests, p < 0.05 statistically significant. CNS = central nervous system; IPDA = intact proviral DNA assay; PWH = people with HIV.
CNS Resident Myeloid Cells Harbour HIV DNA in VS PWH
To confirm cellular reservoirs of HIV in the brain, CNS cells containing HIV viral DNA (vDNA) were assessed in FFPE tissue from viremic and VS PWH by in situ hybridization DNAscope. HIV vDNA+ myeloid cells (CD68+) were present in frontal lobe tissue from both viremic and VS PWH (representative images shown in Fig 5). HIV vDNA+ CD68+ cells were >50 μm from blood vessels indicating infection of parenchymal cells, and not perivascular macrophages, directly demonstrating the presence of HIV‐infected resident myeloid cells in the brain. No HIV DNA signal was detected in HIV‐seronegative biological controls. As HIV‐infected T cells passaging into the brain may also contribute, in part, to CNS reservoir measures by qPCR/ddPCR, 27 CD3+ T cells were quantified in matched FFPE brain tissues by immunohistochemistry. CD3+ T cells were observed in CNS tissue from both viremic and VS PWH at similar levels, contributing to approximately 0.5% of total cells in frontal lobe (median = 6,251 CD3+ T cells/106 CNS cells in VS PWH). Importantly, the frequency and number of T cells in the CNS (Fig 6A; n = 20), or gene expression levels of TCR RNA levels in matched frozen frontal lobe tissue of PWH (n = 11) did not correlate with HIV pol measures in the CNS (Fig 6B; p > 0.05 for all), suggesting that the level of T cells in the brain did not predict HIV DNA levels in the brain.
FIGURE 5.
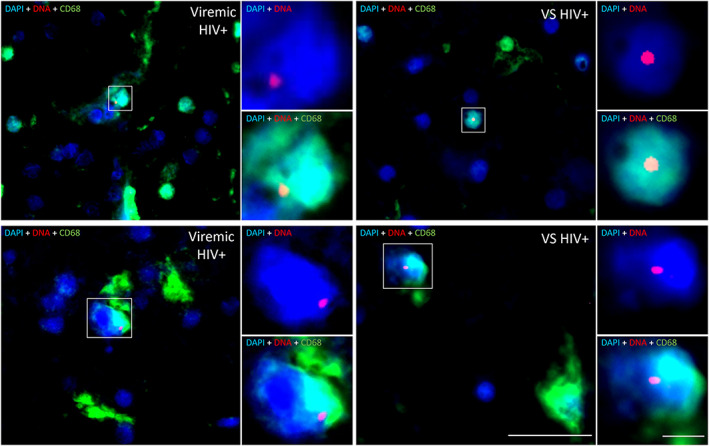
CNS resident myeloid cells in virally suppressed PWH harbour HIV DNA. Representative images of HIV DNA+ CD68+ myeloid cells in frontal brain tissue from virally suppressed (VS HIV+; n = 2) or viremic PWH (n = 2) as determined by in situ hybridization for HIV DNA. HIV DNA (red), CD68 (green), or nuclei (blue) shown. Images acquired on a PALM Robo 4.2 at × 63 magnification. HIV DNA myeloid cells identified by white boxes. HIV DNA+DAPI+ and HIV DNA+CD68+DAPI+ insets of colocalization shown. Scale bars = 20 μm (×63 image) and 5 μm (×3.5 digital zoom ‐ insets). CNS = central nervous system; PWH = people with HIV; VS = virally suppressed.
FIGURE 6.
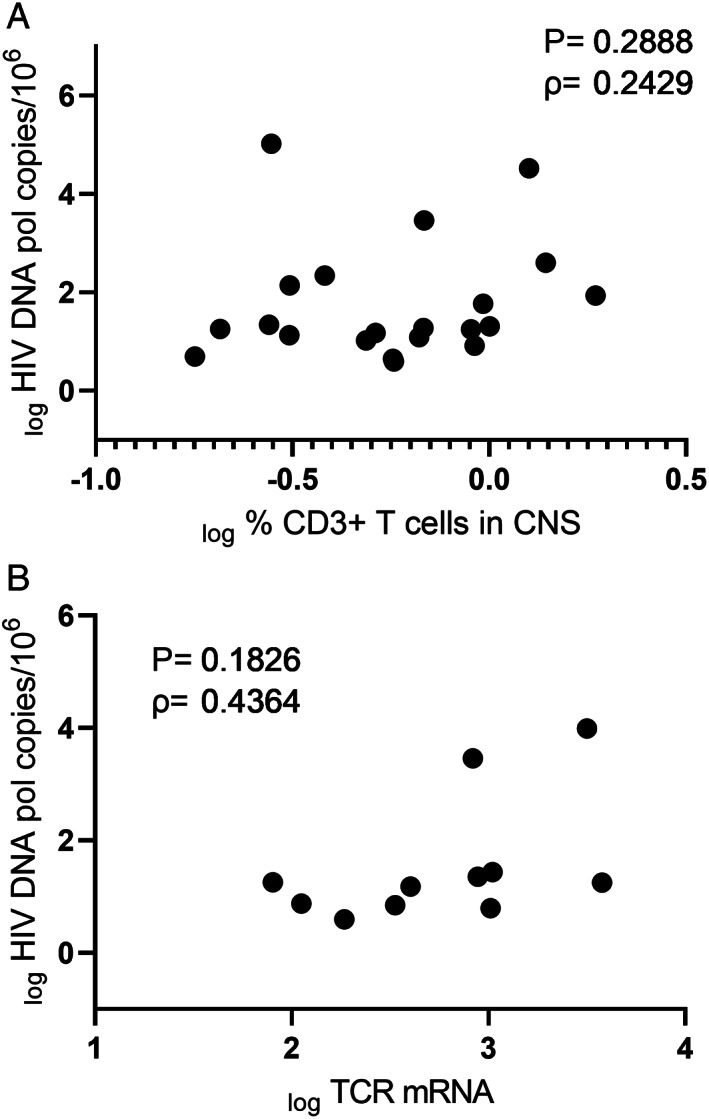
T cells infiltrating the CNS are not predictive of HIV DNA levels in the brain. (A) Correlative analysis of frequency of infiltrating CD3+ T cells (as measured by immunohistochemistry) and HIV pol DNA levels (as determined by droplet digital PCR) in frontal brain tissue from virally suppressed (n = 10) or viremic PWH (n = 10; groups combined are shown; n = 20). (B) Correlative analysis of relative expression of TCR mRNA and HIV pol DNA levels from fresh frozen human brain of PWH (n = 11). Parameters log transformed and spearman rho and p value shown. p < 0.05 considered statistically significant. CNS = central nervous system; PCR = polymerase chain reaction; PWH = people with HIV; TCR = T cell receptor.
To further demonstrate the presence of CD68 + vDNA+ cells in the brain, LCM of the nuclei of parenchymal myeloid cells from viremic (n = 2) and VS PWH (n = 5) was performed. FFPE tissue was stained by immunohistochemistry for CD68, and the nuclei of >500 CD68+ cells that were >50 μm from blood vessels were collected. Nuclei from cells were isolated, DNA purified, and triple nested PCR analysis was performed using primers specific for HIV V3 prior to sequence analysis. HIV DNA was detected in the nuclei of CD68+ cells isolated from both viremic (2/2 individuals) and VS PWH (4/5 individuals; sequences shown; Fig 7). HIV V3 sequences showed diversity between isolates, each of which were distinct from the laboratory strain NL4.3, indicating that sequences were true clinical isolates and not laboratory introduced contamination. Collectively, these findings directly demonstrate the presence of a resident CNS reservoir in the brain of VS PWH.
FIGURE 7.

HIV V3 protein sequences derived from CD68+ myeloid cells isolated from brain tissue of VS or viremic PWH. The nuclei of CD68+ myeloid cells (labelled by immunohistochemistry) were isolated from FFPE brain tissue from viremic (n = 2) or VS PWH (vs; n = 5) by laser capture microdissection prior to lysis, PCR, and sequence analysis. Two sequences per patient shown. Changes in amino acids relative to HIV laboratory strain NL4.3 shown. FFPE = formalin‐fixed paraffin embedded; PCR = polymerase chain reaction; PWH = people with HIV; VS = virally suppressed
Discussion
The size and proviral landscape of the HIV reservoir in the CNS is presently unclear. Here, we utilized state‐of‐the‐art molecular approaches coupled with a well‐characterized cross‐sectional tissue bank to provide the most comprehensive evidence for an intact proviral HIV reservoir in the CNS of PWH. HIV DNA was present at similar levels in brain tissue from both viremic and VS PWH, reflecting findings in NHP studies that the HIV reservoir in the brain is not dramatically affected by long‐term viral suppression with ART. 1 , 28 Importantly, intact proviral genomes were detected in brain tissue from both viremic and VS PWH, supporting the presence of an intact HIV reservoir in the brain. Finally, HIV DNA was identified in CD68+ myeloid cells in brain tissue, directly demonstrating the presence of a CNS resident reservoir in the CNS of VS PWH.
Our observation of intact proviral genomes in brain tissue, identified by the IPDA, strongly suggests the presence of a replication competent reservoir in the CNS. These data support the results of previous studies that have shown HIV RNA 29 , 30 and anti‐HIV antibodies in the CSF despite up to 10 years of suppressive ART, 31 which has previously been the most convincing evidence for ongoing HIV replication in the CNS. Furthermore, cell‐associated HIV DNA and/or RNA in the CSF has been associated with poorer neurocognitive outcomes, implying a role for ongoing viral replication/persistence in the CNS with the development of neurocognitive disorders. 32 However, analysis of CSF has raised questions as to whether CSF viremia is directly related to ongoing CNS replication or possible infiltration from the periphery due to incomplete viral suppression due to drug resistance. 33 , 34 Therefore, our findings within brain tissue, and more specifically, myeloid cells provide further evidence for a localized CNS reservoir in the brain. Intact proviral genomes were detected in the CNS of 6 of 9 VS PWH, whereas intact genomes were detected in peripheral lymphoid tissues of all individuals. One explanation is that individuals without detectable intact proviruses in the CNS may have harbored a smaller reservoir prior to ART initiation, which is supported by our findings that the number of intact proviruses directly correlated with HIV pol DNA levels (see Fig 3). Alternatively, as others have shown the preferential decay of cells containing intact proviruses with time, 35 , 36 it is possible that these individuals harbored a more effective immune response within the CNS. Together, our findings support the presence of an intact, potentially replication competent HIV reservoir in the CNS of a proportion of VS PWH.
Similar to studies in peripheral sites, such as blood cells and lymph node, 14 , 16 , 19 , 20 the majority of proviruses identified in the CNS were either 5′ or 3′ defective (approximately 47 vs 37% of total proviruses, respectively) which may have important implications on CNS health. Whereas defective HIV proviruses are incapable of generating replication competent virions, some can still generate viral proteins including Nef and Tat 37 , 38 which are neurotoxic and well known to induce CNS cellular activation, neuronal damage, and are associated with adverse cognitive outcomes. 39 , 40 , 41 , 42 Many of these proteins have been identified in the CNS of virally suppressed human and NHPs, 43 supporting a role for defective proviruses in contributing to the generation of neurotoxic proteins. Additionally, studies in blood have shown that cells containing defective proviruses may be more prone to immune targeting by cytotoxic T cells, 38 , 44 suggesting that similar immune activation phenomena may occur in the CNS. In our study, no inverse association between 5′ or 3′ defective proviruses and years of viral suppression was observed (data not shown), albeit limited by small patient numbers, suggesting that the presence of defective proviruses may not necessarily result in increased immune clearance of infected cells in the brain. Therefore, the persistence of high levels of defective HIV proviruses in the CNS, capable of generating neurotoxic proteins despite viral suppression, may contribute to ongoing immune activation in the CNS.
Our findings of HIV vDNA+ cells in the CNS by both in situ hybridization methods and PCR of HIV in isolated myeloid cells provides direct evidence for a CNS HIV reservoir in VS PWH. These findings confirm and extend previous studies by us and others who have identified HIV‐infected myeloid cells, 4 , 5 astrocytes, 6 , 7 and pericytes 8 in the CNS of both viremic and VS PWH. Of note, in vivo NHP studies have shown that HIV‐infected T cells can also passage into the CNS which may also contribute to HIV DNA levels in the brain, particularly in the meninges during acute, untreated infection. 45 Whereas T cells undoubtably traffic into the brain and may, in part, contribute to HIV viral persistence in the CNS, we did not find any association between T cell infiltration and HIV pol DNA or IPDA measures. Therefore, our findings directly demonstrate the presence of a CNS resident HIV reservoir, and whereas infected T cells may contribute to HIV DNA levels in the CNS, T cells are not the sole reservoir in frontal white matter of VS PWH.
Levels of HIV pol DNA, intact, or defective proviruses did not correlate with HIV plasma load in this study, supporting the CNS as a unique tissue compartment. It is noteworthy that plasma viral loads were determined by clinically available assays prior to death, so no single copy HIV RNA analyses were performed on either plasma or CSF. This may be of interest in future studies, as HIV RNA levels below the traditional limit of detection of clinical assays (ie, 50 HIV RNA copies/ml plasma) could potentially contribute to reservoir size in blood in some cases. 46
This study has some limitations which need to be considered. Although the IPDA is a validated surrogate measure of intact HIV proviruses and correlates well with qVOA outgrowth assays, 16 , 19 we acknowledge that not all proviruses deemed “intact” in this study are likely to be replication competent, as the IPDA primers do not assess the full genome. Future studies with techniques, such as FLIPs, may offer additional insight of the intact nature of the CNS reservoir. However, due to the low throughput nature of FLIPS, our use of the IPDA in this study allowed for the assessment of the near‐intact reservoir in a large number of patients which would not have been feasible by other approaches. As this study utilized an autopsy tissue bank, it is possible that DNA fragmentation may have occurred postmortem leading to a higher proportion of proviruses deemed 5′ or 3′ defective than present in vivo. We acknowledge that it is possible that a gap in treatment may exist between last viral load test and death for some individuals. Although treatment history was not provided at the time of death, we found no association between the time between last viral load test (performed while subjects were alive) and autopsy and HIV pol DNA levels in the brain (p = 0.203, ρ = −0.418). This study characterized the HIV reservoir in the frontal cortex as cognitive impairment in PWH is associated with this site. Future studies assessing the size and composition of HIV reservoirs in different regions within the CNS would be of interest.
In summary, we utilized a well‐characterized cross‐sectional bank of brain tissue to provide the first evidence for an intact proviral reservoir in the brains of PWH which, importantly, appears to persist despite long‐term viral suppression. These findings highlight the importance of the CNS as a known and substantial reservoir of HIV that must be considered in both HIV treatment and cure strategies.
Author Contributions
M.J.C., M.R., J.D.E., C.R.C., T.A.A., P.R.G., B.J.B, and S.R.L. contributed to the conception and design of the study. C.R.C., A.C.G., S.J.B., E.W., G.S.T., J.V., J.Z., T.A.A., J.J.E., M.G., E.W., and L.S. contributed to the acquisition and analysis of data. T.A.A., C.R.C., M.R., and M.J.C. contributed to drafting the text or preparing the figures.
Potential Conflicts of Interest
S.R.L. has received investigator‐initiated grant funding from Gilead, Merck, and ViiV Healthcare. She has provided paid scientific advice to Abivax, Abbvie, and Gilead. P.R.G. previously received investigator‐initiated grant funding from ViiV Healthcare. <zbmrule>
Supporting information
Table S1 PCR primers and probes
Acknowledgments
This work was supported by the National Health and Medical Research Council, Australia 1183032 (to M.J.C., J.D.E., M.R., and P.R.G.), The Jack Brockhoff Foundation JBF Grant number 4501 ‐ 2018 (to T.A.A. and M.J.C.), and the Melbourne HIV Cure Consortium (to M.R. and C.R.C.). Research reported in this publication was supported by the National Institute On Drug Abuse of the National Institutes of Health under Award Number R21DA055489 (to M.J.C., M.R., and T.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. S.R.L. is supported by a National Health and Medical Research Council, Australia Practitioner Fellowship (S.R.L.). All authors gratefully acknowledge the support from the NNTC and this publication was made possible from NIH funding through the NIMH and NINDS Institutes by the following grants: Texas NeuroAIDS Research Center: U24MH100930, California NeuroAIDS Tissue Network: U24MH100928, National Neurological AIDS Bank: U24MH100929, Manhattan HIV Brain Bank: U24MH100931, and Data Coordinating Center: U24MH100925. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NNTC or NIH. The authors thank Dr Anna Hearps and Dr Michelle Wong (Burnet Institute, Australia) for providing reagents. Open access publishing facilitated by RMIT University, as part of the Wiley ‐ RMIT University agreement via the Council of Australian University Librarians.
References
- 1. Estes JD, Kityo C, Ssali F, et al. Defining total‐body AIDS‐virus burden with implications for curative strategies. Nat Med 2017;23:1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valcour V, Chalermchai T, Sailasuta N, et al. Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis 2012;206:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Churchill MJ, Gorry PR, Cowley D, et al. Use of laser capture microdissection to detect integrated HIV‐1 DNA in macrophages and astrocytes from autopsy brain tissues. J Neurovirol 2006;12:146–152. [DOI] [PubMed] [Google Scholar]
- 4. Ko A, Kang G, Hattler JB, et al. Macrophages but not Astrocytes Harbor HIV DNA in the brains of HIV‐1‐infected Aviremic individuals on suppressive antiretroviral therapy. J Neuroimmune Pharmacol 2019;14:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thompson KA, Cherry CL, Bell JE, McLean CA. Brain cell reservoirs of latent virus in presymptomatic HIV‐infected individuals. Am J Pathol 2011;179:1623–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Churchill MJ, Wesselingh SL, Cowley D, et al. Extensive astrocyte infection is prominent in human immunodeficiency virus‐associated dementia. Ann Neurol 2009;66:253–258. [DOI] [PubMed] [Google Scholar]
- 7. Lutgen V, Narasipura SD, Barbian HJ, et al. HIV infects astrocytes in vivo and egresses from the brain to the periphery. PLoS Pathog 2020;16:e1008381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho HJ, Bertrand L, Toborek M. Blood–brain barrier pericytes as a target for HIV‐1 infection. Brain 2019;142:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cysique LA, Brew BJ. Prevalence of non‐confounded HIV‐associated neurocognitive impairment in the context of plasma HIV RNA suppression. J Neurovirol 2011;17:176–183. [DOI] [PubMed] [Google Scholar]
- 10. Antinori A, Arendt G, Becker JT, et al. Updated research nosology for HIV‐associated neurocognitive disorders. Neurology 2007;69:1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heaton RK, Clifford DB, Franklin DR, et al. HIV‐associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER study. Neurology 2010;75:2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamers SL, Rose R, Maidji E, et al. HIV DNA is frequently present within pathologic tissues evaluated at autopsy from cART‐treated patients with undetectable viral load. J Virol 2016;90:8968–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaillon A, Gianella S, Dellicour S, et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 2020;130:1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bruner KM, Murray AJ, Pollack RA, et al. Defective proviruses rapidly accumulate during acute HIV‐1 infection. Nat Med 2016;22:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ho Y‐C, Shan L, Hosmane NN, et al. Replication‐competent noninduced proviruses in the latent reservoir increase barrier to HIV‐1 cure. Cell 2013;155:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruner KM, Wang Z, Simonetti FR, et al. A quantitative approach for measuring the reservoir of latent HIV‐1 proviruses. Nature 2019;566:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joseph SB, Kincer LP, Bowman NM, et al. HIV‐1 RNA detected in the CNS after years of suppressive antiretroviral therapy can originate from a replicating CNS reservoir or clonally expanded cells. Clin Infect Dis 2018;69:1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avalos CR, Abreu CM, Queen SE, et al. Brain macrophages in simian immunodeficiency virus‐infected, antiretroviral‐suppressed macaques: a functional latent reservoir. MBio 2017 2017;8:e01186–e01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Falcinelli SD, Kilpatrick KW, Read J, et al. Longitudinal dynamics of intact HIV Proviral DNA and outgrowth virus frequencies in a cohort of individuals receiving antiretroviral therapy. J Infect Dis 2020;224:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin AR, Bender AM, Hackman J, et al. Similar frequency and inducibility of intact HIV‐1 proviruses in blood and lymph nodes. J Infect Dis 2020;224:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papasavvas E, Azzoni L, Ross BN, et al. Intact human immunodeficiency virus (HIV) reservoir estimated by the intact Proviral DNA assay correlates with levels of Total and integrated DNA in the blood during suppressive antiretroviral therapy. Clin Infect Dis 2020;72:495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morgello S, Gelman BB, Kozlowski PB, et al. The national NeuroAIDS tissue consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol 2001;27:326–335. [DOI] [PubMed] [Google Scholar]
- 23. Kinloch NN, Ren Y, Conce Alberto WD, et al. HIV‐1 diversity considerations in the application of the intact Proviral DNA assay (IPDA). Nat Commun 2021;12:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strain MC, Lada SM, Luong T, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 2013;8:e55943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deleage C, Wietgrefe SW, Del Prete G, et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathogens Immunity 2016;1:68–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chung HK, Hattler JB, Narola J, et al. Development of droplet digital PCR‐based assays to quantify HIV Proviral and integrated DNA in brain tissues from Viremic individuals with encephalitis and virally suppressed Aviremic individuals. Microbiol Spectr 2022;10:e0085321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee CA, Beasley E, Sundar K, et al. Simian immunodeficiency virus‐infected memory CD4(+) T cells infiltrate to the site of infected macrophages in the Neuroparenchyma of a chronic macaque model of neurological complications of AIDS. MBio 2020;11:e00602–e00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perez S, Johnson A‐M, Xiang S‐H, et al. Persistence of SIV in the brain of SIV‐infected Chinese rhesus macaques with or without antiretroviral therapy. J Neurovirol 2018;24:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dahl V, Peterson J, Fuchs D, et al. Low levels of HIV‐1 RNA detected in the cerebrospinal fluid after up to 10 years of suppressive therapy are associated with local immune activation. AIDS 2014;28:2251–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson AM, Muñoz‐Moreno JA, McClernon DR, et al. Prevalence and correlates of persistent HIV‐1 RNA in cerebrospinal fluid during antiretroviral therapy. J Infect Dis 2016;215:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burbelo PD, Price RW, Hagberg L, et al. Anti‐human immunodeficiency virus antibodies in the cerebrospinal fluid: evidence of early treatment impact on central nervous system reservoir? J Infect Dis 2018;217:1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spudich S, Robertson KR, Bosch RJ, et al. Persistent HIV‐infected cells in cerebrospinal fluid are associated with poorer neurocognitive performance. J Clin Invest 2019;129:3339–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edén A, Nilsson S, Hagberg L, et al. Asymptomatic cerebrospinal fluid HIV‐1 viral blips and viral escape during antiretroviral therapy: a longitudinal study. J Infect Dis 2016;214:1822–1825. [DOI] [PubMed] [Google Scholar]
- 34. Mukerji SS, Misra V, Lorenz DR, et al. Impact of antiretroviral regimens on cerebrospinal fluid viral escape in a prospective multicohort study of antiretroviral therapy‐experienced human immunodeficiency Virus‐1‐infected adults in the United States. Clin Infect Dis 2018;67:1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gandhi RT, Cyktor JC, Bosch RJ, et al. Selective decay of intact HIV‐1 Proviral DNA on antiretroviral therapy. J Infect Dis 2021;223:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peluso MJ, Bacchetti P, Ritter KD, et al. Differential decay of intact and defective proviral DNA in HIV‐1–infected individuals on suppressive antiretroviral therapy. JCI. Insight 2020;5:e132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imamichi H, Smith M, Adelsberger JW, et al. Defective HIV‐1 proviruses produce viral proteins. Proc Natl Acad Sci U S A 2020;117:3704–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pollack RA, Jones RB, Pertea M, et al. Defective HIV‐1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the Proviral landscape. Cell Host Microbe 2017;21:494–506.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sami Saribas A, Cicalese S, Ahooyi TM, et al. HIV‐1 Nef is released in extracellular vesicles derived from astrocytes: evidence for Nef‐mediated neurotoxicity. Cell Death Dis 2017;8:e2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haughey NJ, Holden CP, Nath A, Geiger JD. Involvement of inositol 1,4,5‐trisphosphate‐regulated Stores of Intracellular Calcium in calcium dysregulation and neuron cell death caused by HIV‐1 protein tat. J Neurochem 1999;73:1363–1374. [DOI] [PubMed] [Google Scholar]
- 41. Carvallo L, Lopez L, Fajardo JE, et al. HIV‐tat regulates macrophage gene expression in the context of neuroAIDS. PLoS One 2017;12:e0179882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hudson L, Liu J, Nath A, et al. Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol 2000;6:145–155. [DOI] [PubMed] [Google Scholar]
- 43. Yarandi SS, Robinson JA, Vakili S, et al. Characterization of Nef expression in different brain regions of SIV‐infected macaques. PLoS One 2020;15:e0241667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Casartelli N, Guivel‐Benhassine F, Bouziat R, et al. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV‐infected T cells. J Exp Med 2010;207:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsu DC, Sunyakumthorn P, Wegner M, et al. Central nervous system inflammation and infection during early, nonaccelerated simian‐human immunodeficiency virus infection in rhesus macaques. J Virol 2018;92:e00222–e00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chun TW, Murray D, Justement JS, et al. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis 2011;204:135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 PCR primers and probes


