Abstract
Type 1 fimbriae are proteinaceous surface appendages that carry adhesins specific for mannosylated glycoproteins. These fimbriae are found on most members of the family Enterobacteriaceae and are known to facilitate binding to a variety of eukaryotic cells, including those found on the mucosal surfaces of the alimentary tract. We have shown that the regulation of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium is controlled, in part, by the products of four genes found within the fim gene cluster: fimZ, fimY, fimW, and fimU. To better understand the specific role of FimW in fimbrial expression, a mutation was constructed in this gene by the insertion of a kanamycin resistance DNA cassette into the chromosome. The resulting fimW mutation was characterized by mannose-sensitive hemagglutination and agglutination with fimbria-specific antiserum. Assays suggested that this mutant was more strongly fimbriate than the parental strain, exhibiting a four- to eightfold increase in fimbrial production. The fimW mutation was introduced into a second strain of Salmonella enterica serovar Typhimurium, and this mutant was also found to be strongly fimbriate compared to the parental strain. Consistent with the role of this protein as a negative regulator, fimA-lacZ expression in serovar Typhimurium, as well as in Escherichia coli, was increased twofold in the absence of functional FimW. Primer extension analysis determined that fimW transcription is initiated from its own promoter 31 bp upstream of the translation start site. Analysis using a fimW-lacZ reporter indicated that fimW expression in serovar Typhimurium was increased under conditions that select for poorly fimbriate bacteria and low fimA expression. FimW also appears to act as an autoregulator, since expression from the fimW-lacZ reporter was increased in a fimW mutant. FimW was partially purified by fusion with the E. coli maltose-binding protein. Use of this FimW protein extract, as well as others, in DNA-binding assays was unable to identify a specific binding site for FimW in the fimA, fimZ, fimY, or fimW promoter regions. To analyze protein-protein interactions, FimW was expressed in a LexA-based two-hybrid system in E. coli. A significant interaction between FimW and the DNA-binding activator protein, FimZ, was detected using this system. These results indicate that FimW is a negative regulator of serovar Typhimurium type 1 fimbrial expression and may function by interfering with FimZ-mediated activation of fimA expression.
Salmonellae are important pathogens belonging to the family of gram-negative bacilli, the Enterobacteriaceae. The virulence of these bacteria, in humans, has been attributed to their ability to invade and survive within host macrophages or enterocytes. Fimbriae are believed to play a critical role in this process by facilitating the initial attachment to specific host cells and tissues. Type 1 fimbriae are associated with most Salmonella enterica serovars, as well as other members of the family Enterobacteriaceae, and are characterized by their ability to mediate the mannose-sensitive agglutination of red blood cells in vitro (7, 18, 38). Despite numerous studies indicating that Salmonella type 1 fimbriae facilitate the adhesion to, and invasion of, human epithelial cell lines, the specific role of these surface appendages in pathogenesis remains controversial (3, 21, 31, 35, 61). A report analyzing the mouse model of infection recently suggested that the presence of fimbriae inhibits proliferation in the bloodstream during systemic infection (42). However, studies examining the colonization of host animals, such as chickens and pigs, indicate that type 1 fimbriae may be important in establishing persistent infections within these animals (2, 32, 33, 47).
The expression of type 1 fimbriae is known to phase vary, or alternate between a fimbriate and a nonfimbriate phenotype. This variation is affected by environmental conditions, and in vitro, growth in static liquid media promotes the expression of fimbriae, whereas growth on solid media inhibits expression (18, 50). Regulation of variation at the genetic level has been closely examined in Escherichia coli. The E. coli fim gene cluster is composed of seven structural genes transcribed from the promoter upstream of the gene encoding the major fimbrial subunit, fimA. The fimA promoter is flanked by two inverted repeats whose site-specific recombination results in the inversion of a 314-bp DNA fragment (1, 36). Inversion of this sequence to an orientation allowing transcription, or the opposite orientation blocking transcription, is mediated by two site-specific recombinases, FimB and FimE (24, 37). Inversion of the promoter to the “off” orientation is largely dependent upon FimE, while FimB mediates recombination in either orientation at a lower frequency (23, 43). In addition, inversion-independent mechanisms of regulation, as well as global regulators involved in DNA topology, such as the leucine responsive regulatory protein (LRP) and integration host factor (IHF), affect type 1 fimbrial expression in E. coli (4, 17, 20, 25, 44).
Despite significant homology between the structural genes, expression of Salmonella enterica serovar Typhimurium type 1 fimbriae is regulated in a manner distinct from that for E. coli. The Salmonella fim gene cluster is located on a different region of the chromosome and does not possess homologs of the E. coli recombinases FimB and FimE (14, 57). In addition, the serovar Typhimurium fimA promoter region was found to be in the orientation promoting transcription regardless of fimbrial phenotype, indicating that phase variation in this bacterium is not absolutely dependent upon promoter inversion (10). Studies analyzing expression from the serovar Typhimurium fimA promoter in E. coli have shown that regulators encoded by E. coli alone are unable to activate transcription from this promoter (66). Instead, the serovar Typhimurium FimZ and FimY proteins are necessary for serovar Typhimurium fimA transcription (62, 66). These proteins are located downstream of the fim structural genes and are transcribed independently in the opposite orientation (57, 62, 66). FimZ reveals significant amino acid homology to a two component response regulator from Bordetella pertussis, and previous studies have demonstrated the ability of FimZ to bind independently to a region upstream of fimA to promote transcription from this promoter (66; unpublished data). In contrast, FimY contains only limited sequence homology to prokaryotic DNA binding proteins; however, we have shown that it is essential for type 1 fimbrial production and functions as a coactivator with FimZ (62).
The role of a third polypeptide, FimW, in the production of type 1 fimbriae has, until this time, remained undefined. fimW is located between the regulatory gene, fimY, and an arginine tRNA molecule, encoded by fimU, that has also been found to regulate fimbrial expression (13, 60). Similar to FimY, FimW reveals little amino acid homology to prokaryotic proteins; however, it appears to be related to transcriptional activators. Thus, based upon chromosomal location and amino acid sequence similarity, it was hypothesized that FimW is also involved in fimbrial regulation (12). We describe here the construction of a fimW mutant in serovar Typhimurium and the characterization of this protein as a negative regulator of type 1 fimbrial expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
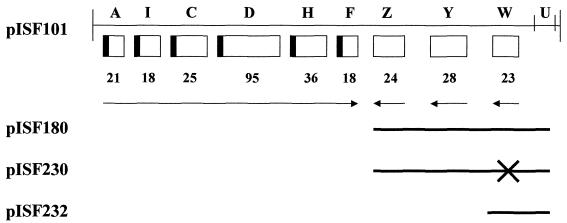
The strains and recombinant molecules used in this study are shown in Table 1. The fimbriate strain serovar Typhimurium LB5010 (8) was used to construct the fimW mutant, LBW100. The mutation was subsequently introduced into the strongly fimbriate and invasive serovar Typhimurium strain, SL1344 (30), by P22 transduction using lysates of serovar Typhimurium LBW100, and this strain is referred to as SL1344JTW. Serovar Typhimurium IS145 is a λfimA-lacZ lysogen used as a single-copy reporter of fimA expression, and its construction has been described previously (58). All strains were cultured on Luria-Bertani (LB) media and incubated at 37°C, or 30°C for lysogens, for 24 or 48 h. Plasmids were prepared by standard techniques, and manipulation of recombinant DNA was performed by using conventional procedures (52). Plasmids pISF180, pISF230, and pISF232 used in this study are derivatives of pISF101 carrying the serovar Typhimurium fim gene cluster cloned into pACYC184 (New England Biolabs, Beverly, Mass.), as shown in Fig. 1. The plasmid pISF180 carries fimZ, fimY, and fimW and was constructed by isolation of a SphI-BamHI fragment containing these genes from pISF101 followed by ligation into pACYC184 to generate a 10.7-kb plasmid. pISF230 is a derivative of pISF180 with a universal translation terminator (Pharmacia Biotech, Inc., Piscataway, N.J.) inserted into a unique BspHI site 64 bp from the translation start site of fimW to effectively disrupt this gene. The plasmid pISF232 is 4.4 kb and possesses only the fimW gene of the fim gene cluster. pISF232 was constructed following digestion of pISF180 with PpuMI and AvaII and religation to remove all fim genes except fimW. The construction of a multicopy fimA-lacZ reporter (pISF145) in the promoterless lacZYA vector, pMC1403 (9), has been described previously (58). A multicopy fimW-lacZ reporter (pISF252) was constructed by PCR amplification of a 407-bp region upstream of fimW encompassing the determined transcription initiation site and ligation of this fragment into the EcoRI and BamHI sites of pMC1403. The primers JT47 (5′-GATACCGGGAATTCCCATATGGAAAATAAGGAGG-3′) and JT38 (5′-CGAAATCTGGATCCCCTTAATAGCGATACGC-3′) were used for this PCR. A single-copy fimW-lacZ (pISF253) reporter was constructed by subcloning the isolated EcoRI/BamHI promoter-containing fragment from the multicopy reporter and ligation into the plasmid pGS375 (kindly supplied by George Stauffer, University of Iowa). pGS375 is an ampicillin-resistant derivative of the single-copy pDF41 plasmid ligated to the promoterless lacZ, lacY, and lacA genes from pMC1403 (28). Construction of the fimY-lacZ and fimZ-lacZ reporters was accomplished by a similar mechanism and has been described previously (62). All plasmids were analyzed by DNA sequencing to confirm the identity of the constructs (University of Iowa DNA Sequencing Facility, Iowa City).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant features | Reference or source |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| LB5010 | Wild type; fimbriate with complete fim cluster | 8 |
| LBW100 | LB5010 fimW::kan Kanr | This study |
| ISF145 | LB5010 λfimA::lacZ lysogen | 58 |
| ISF145W | LBW100 λfimA::lacZ lysogen | This study |
| SL1344 | Wild type; fimbriate with complete fim cluster | 30 |
| SL1344JTW | SL1344 fimW::kan Kanr | This study |
| E. coli | ||
| SY327 | Host for suicide vector pGP704 | 46 |
| JM109λ145 | JM109 λfimA::lacZ lysogen | 66 |
| ER2508 | lon mutant for maltose-binding protein fusion expression | 39 |
| SU202 | JL1434λsulAlacZ lysogen | 15 |
| Plasmids | ||
| pISF101 | fimAICDHFZYWU, pACYC184 | 59 |
| pISF145 | fimA::lacZ reporter fusion, pMC1403 | 58 |
| pISF180 | fimZYW, pACYC184 | This study |
| pISF230 | fimZYW, pACYC184 | This study |
| pISF232 | fimW, pACYC184 | This study |
| pISF242 | malE::fimW fusion for FimW purification, pMalc2 | This study |
| pISF245 | lexA::fimZ fusion for two-hybrid system, pDP804 | This study |
| pISF248 | lexA::fimW fusion for two-hybrid system, pMS604 | This study |
| pISF252 | fimW::lacZ reporter fusion, pMC1403 | This study |
| pISF253 | fimW::lacZ reporter fusion single copy, pGS375 | This study |
FIG. 1.
Genetic organization of the Salmonella fim gene cluster and plasmids used in this study. The sizes of the polypeptides encoded by the genes are shown below the boxes. fimA is the gene encoding the major fimbrial subunit, whereas fimZ, fimY, and fimW are as described in the text. The arrows indicate direction of transcription, and the black boxes represent the presence of an N-terminal signal sequence. The derivatives of pISF101 utilized in this study are indicated below the map, with solid lines representing the DNA retained by each derivative. For pISF230, the cross indicates the location of the inserted translation terminator.
Detection of type 1 fimbriae.
Bacteria were serially subcultured in 10 ml of LB broth and incubated without shaking for 48 h to select for highly fimbriate cultures. Alternatively, cultures were grown on solid LB agar for 24 h to select for poorly fimbriate bacteria. Cells were collected by centrifugation and gently resuspended in the residual fluid as described previously (18, 49). Subsequently, 50 μl of bacterial suspension was mixed with 50 μl of a 3% (vol/vol) suspension of guinea pig erythrocytes in phosphate-buffered saline (PBS). Mannose-sensitive hemagglutination was determined by incubation of the bacterial suspension with cells resuspended in PBS containing 3% (wt/vol) α-methyl-d-mannoside. The mannose-sensitive adhesin was considered to be present if the red blood cells agglutinated only in the absence of mannose within 1 min. Fimbrial antigens were detected using monospecific serovar Typhimurium antifimbrial serum as described previously (29). Titers of the hemagglutination and antibody agglutination reactions were determined as the reciprocal of the highest bacterial or serum dilution resulting in hemagglutination or bacterial agglutination, respectively, and is described in detail elsewhere (11). For transmission electron microscopy, aliquots of 48-h bacterial suspensions were placed on carbon-coated grids and stained for 1 min with phosphotungstic acid before visualization at a ×50,000 magnification with a Hitachi H-600 electron microscope.
Construction of the serovar Typhimurium fimW mutant.
A plasmid possessing an intact fimW was constructed by PCR using primers LC94 (5′-CATCTGGTGGATCCCTTCGTGTAGACGAAACG-3′) and LC93 (5′-CGTACTGAGGATCCGCCTGTAGGTATCGTTAC-3′). This product was then cloned into pACYC184 and linearized at a unique EcoRV site within fimW. A HincII digest of a DNA cassette containing a kanamycin resistance determinant, isolated from the plasmid pUC4K (Pharmacia Biotech), was prepared and subsequently ligated into the fimW gene at the EcoRV site. After isolation of kanamycin-resistant E. coli HB101 (6) transformants, the plasmid carrying the insertionally inactive fimW gene was isolated by standard techniques (52). The disrupted fimW determinant was then cloned into the BamHI site of the suicide vector pGP704 (kindly supplied by John Mekalanos, Harvard Medical School) and maintained in the permissive E. coli host, SY327 (46). Recombinant DNA was prepared from kanamycin- and ampicillin-resistant transformants and analyzed by restriction digest. The appropriate construct was then introduced into serovar Typhimurium LB5010, and kanamycin-resistant but ampicillin-sensitive transformants were selected. Further analysis of putative fimW mutants was completed by Southern hybridization using random-primed dUTP-labeled DNA probes (Genius Kit; Boehringer Mannheim, Indianapolis, Ind.) specific for the fimW gene or the pUC4K kanamycin resistance determinant. Chromosomal DNA from serovar Typhimurium LB5010 and LBW100 was isolated by standard techniques, digested to completion with BglII, and transferred to nitrocellulose. All hybridizations were performed under high-stringency conditions as described elsewhere (27).
Primer extension.
Total RNA was isolated from E. coli HB101 carrying the fim gene cluster on pISF101 by phenol extraction and digestion with DNase I. A primer that anneals 129 bp downstream of the fimW translation initiation site (LC82, 5′-GGCATTATCTATCTCTTCTGGCGG-3′) was end labeled by incubation with [γ-32P]ATP and T4 polynucleotide kinase. RNA (5 ng) was subjected to reverse transcriptase PCR using the above labeled probe and according to the manufacturer's instructions of the Primer Extension System (Promega, Madison, Wis.). φX174 HinfI molecular weight markers were also labeled with [γ-32P]ATP and analyzed by electrophoresis alongside the resulting reverse transcriptase PCR product and DNA sequence through a 6% (wt/vol) acrylamide–42% (wt/vol) urea DNA sequencing gel at 200 V. Double-stranded DNA sequencing of the region encompassing the fimW promoter was performed by the dideoxy chain termination method (53), utilizing the above-labeled primer.
β-Galactosidase assays.
Assays for β-galactosidase were performed in triplicate by the method of Miller (45), using the chloroform-sodium dodecyl sulfate (SDS) lysis procedure, and λfimA-lacZ lysogens or fimA-lacZ and fimW-lacZ plasmid transformants. Strains were grown on LB agar for 24 h or static liquid LB broth for 48 h before analysis. The data represent the means of cultures assayed in triplicate. All assays were performed with independent cultures at least three times with <20% variability.
Construction of the maltose-binding protein–FimW and FimW-His6 fusions and gel mobility shift assays.
The plasmid pISF242 (Table 1) was used to purify a maltose binding protein-FimW fusion. pISF242 was constructed from the vector pMal-c2 (New England Biolabs), which contains the β-galactosidase coding region fused to the maltose-binding protein of E. coli. The β-galactosidase gene was disrupted by digestion with BamHI and PstI, and the remaining vector was ligated to a PCR product of the fimW coding region digested with BamHI and PstI. Primers JT28 (5′-GATACCGGGGATCCCTGCGTATCGCTATTAAG-3′) and JT8 (5′-AATACCGTCTGCAGGCATCATTGTGGCAGCGTTA-3′) were used to isolate fimW. The resulting construct was confirmed by sequencing through the junction. pISF242 was introduced into the lon protease mutant, E. coli ER2508 (39), and grown at room temperature to an optical density at 600 nm of ∼0.5 before induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The culture was allowed to grow for an additional 12 h at room temperature before the cells were collected and harvested following sonication and then resuspended in Column Buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA). The maltose-binding protein–FimW fusion was separated from the crude extract by binding to an amylose-agarose bead resin and eluted from the resin by washing with Column Buffer plus 10 mM maltose according to the manufacturer's instructions. A FimW-His6 fusion was constructed by ligation of the fimW PCR product into pQE31 (Qiagen, Valencia, Calif.), transformation of this plasmid into E. coli JM109, and subsequent separation of proteins using a Ni-nitrilotriacetic acid matrix according to the manufacturer's instructions (Qiagen).
Gel mobility shift assays were performed using dilutions of the above FimW fusion preparations or extracts of E. coli JM109 (64) transformed with pISF232 or pACYC184 (described above). The preparation of the 452-bp fimA and 564-bp fimY promoter-containing regions used as target DNA has been described elsewhere (59, 62). A 409-bp PCR fragment encompassing the fimW promoter region and upstream fimU coding region, as well as a 755-bp PCR fragment encompassing the fimZ promoter region, was also amplified and used as target DNAs for gel mobility shift assays. Primers JT47 and JT38 were used to make the fimW promoter fragment, and primers LC95 (5′-CATGTGGTGAATTCCAGGATAAGTGCGCAGAT-3′) and LC96 (5′-GATCTGTGGAATTCCGAAGTAACGTTTTGGTG-3′) were used to make the fimZ promoter fragment. End labeling was performed by removing the 5′ phosphate from the promoter containing DNA fragments with calf intestine alkaline phosphatase followed by incubation with T4 polynucleotide kinase and [γ-32P]ATP. Assays were performed using standard techniques (26), except that 0.25 μg of unlabeled single-stranded sperm carrier DNA was added to each incubation mixture and no bovine serum albumin was added. The DNA was subsequently mixed with appropriate twofold dilutions (up to 5 μg) of FimW-containing extracts, and all volumes were adjusted with sterile distilled water. The samples were loaded onto a 5% (wt/vol) nondenaturing polyacrylamide gel and, following electrophoresis at 200 V, the mobility of the DNA fragments was analyzed by autoradiography as previously described (66). In all experiments, the concentration of protein was determined by the use of a commercially available Bradford protein assay kit (Pierce, Rockford, Ill.).
Two-hybrid system for the analysis of FimW-FimZ protein interactions.
E. coli SU202 and plasmids pMS604 and pDP804 for the LexA two hybrid system were generously donated by M. Granger-Schnarr and M. Schnarr (Institut de Biologie Moleculaire et Cellulaire, Strasbourg, France) (15). To construct the lexA-fimZ fusion pISF245, pDP804 was digested with BglII and XhoI and religated to a BglII-XhoI PCR fragment encompassing the fimZ coding region. Primers JT34 (5′-GGTAAGCTCTCGAGAAACCTGCATCTGTTATC-3′) and JT35 (5′-GCGTTGCTAGATCTGGGAGTACATTTACAATAA-3′) were used to amplify fimZ. To construct the lexA-fimW fusion pISF248, pMS604 was digested with XhoI and PstI and religated to an XhoI-PstI PCR fragment encompassing the fimW coding region. Primers JT39B (5′-AATAAGCTCTGCAGCTGCGTATCGCTATTAAG-3′) and JT40 (5′-GGTTGTGCCTCGAGGCATCATTGTGGCAGCGTTA-3′) were used to amplify fimW. All fusions were confirmed to be correct and in frame by DNA sequencing at the University of Iowa DNA Sequencing Facility. E. coli SU202 lysogens, containing the fusion constructs, were grown at 30°C in static liquid LB medium plus 1 mM IPTG for 48 h before analysis of the β-galactosidase as described above.
RESULTS
Construction of the fimW mutant of serovar Typhimurium LB5010.
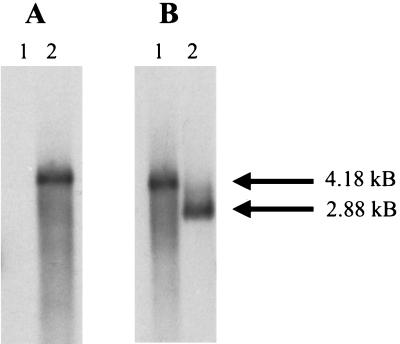
The fimW mutant, serovar Typhimurium LBW100, was constructed following transformation of serovar Typhimurium LB5010 with the suicide vector, pGP704, carrying an inactive fimW::Km gene. Kanamycin-resistant and ampicillin-sensitive bacteria that had retained the inactivated gene but lost the plasmid vector were isolated and further analyzed. Genomic DNA was prepared from both the parental and the mutant strain and used in Southern hybridization analysis to confirm the location of the mutated allele (Fig. 2). DNA preparations were restricted with BglII and hybridized to a 1,300-bp DNA probe possessing the gene encoding kanamycin resistance. In addition, the restricted DNA was probed with a 160-bp DNA fragment comprising nucleotides of the fimW gene itself. The probe possessing the kanamycin resistance determinant hybridized to a 4,180-bp BglII genomic DNA fragment only found in serovar Typhimurium LBW100, and no sequences homologous to the probe were detected in the parental strain. The fimW DNA probe hybridized to a 4,180-bp BglII DNA fragment from serovar Typhimurium LBW100 and a 2,880-bp fragment from serovar Typhimurium LB5010. The sizes of these fragments are consistent with insertion of the 1.3-kb kanamycin resistance cassette, which lacks a BglII restriction site, into the chromosome of serovar Typhimurium LBW100 and replacement of the intact fimW gene by allelic exchange. Confirmation of the location of the mutant allele and orientation of the Km cassette in fimW was performed by additional restriction analysis using several endonucleases (data not shown).
FIG. 2.
Southern hybridization profiles of genomic DNA isolated from serovar Typhimurium strains LB5010 (wild type) and LBW100 (fimW). (A) LB5010 and LBW100 genomic DNA (lanes 1 and 2, respectively) digested with BglII and probed with sequences from the Kanr cassette. (B) LBW100 and LB5010 genomic DNA (lanes 1 and 2, respectively) digested with BglII and probed with sequences from fimW. The sizes of the DNA fragments are as shown.
Characterization of the serovar Typhimurium fimW mutant.
The fimW mutant was analyzed for the phenotypic expression of type 1 fimbriae on its surface. Serovar Typhimurium strains LB5010 and LBW100 were grown under conditions optimal for the expression of fimbriae and were examined for their ability mediate mannose-sensitive hemagglutination of guinea pig erythrocytes and agglutination by type 1 fimbria-specific antisera. In addition, these strains were visually examined for fimbrial expression by transmission electron microscopy. Initially, serovar Typhimurium LBW100 was found to be phenotypically fimbriate and indistinguishable from the parental strain. However, as shown in Table 2, quantitative analysis determined that the fimW mutant exhibited a higher titer of both agglutination reactions, indicating that this strain produces more surface-associated fimbriae than the wild-type strain. Serum agglutination titers for serovar Typhimurium LBW100 were eightfold higher and hemagglutination titers were fourfold higher than those for serovar Typhimurium LB5010. A low-copy-number plasmid carrying only fimW (pISF232) was used to transform the fimW mutant and was found to decrease the phenotypic expression of fimbriae (Table 2). Transformation with the vector alone (pACYC184) did not result in a significant decrease in fimbrial expression (data not shown). The chromosomal fimW mutation of strain LBW100 was introduced into a second strain of serovar Typhimurium, SL1344, by P22 phage transduction. This strain, SL1344JTW, was also found to produce greater amounts of fimbriae compared to SL1344. Transformation of SL1344JTW with pISF232 similarly resulted in a decrease in fimbrial production (Table 2).
TABLE 2.
Phenotypic expression of type 1 fimbriae by the serovar Typhimurium LB5010 and SL1344 fimW mutants
| Strain | Plasmid (relevant genotype) | Serum titera | Slide agglutinationb | Hemagglutination titerc | Presence of fimbriae on bacteriad |
|---|---|---|---|---|---|
| LB5010 | None | 160 | + | 16 | + |
| LBW100 | None | 1,280 | + | 64 | + |
| pISF232 (fimW+) | 160 | + | 8 | ND | |
| SL1344 | None | 160 | + | 16 | + |
| SL1344JTW | None | 1,280 | + | 128 | + |
| pISF232 (fimW+) | 40 | + | 8 | ND |
Reciprocal of the highest serum dilution causing bacterial agglutination.
Agglutination by fimbria-specific polyclonal antisera observed after 60 s.
Reciprocal of the highest bacterial dilution causing hemagglutination.
Fimbriae observed by transmission electron micrography. ND, not determined.
Expression of fimA from a fimA-lacZ reporter in the presence or absence of a functional fimW.
The serovar Typhimurium LB5010 λfimA-lacZ lysogen (IS145), which has been described previously (58), was used as a source of recombinant phage to generate a λfimA-lacZ lysogen of the serovar Typhimurium LBW100 mutant (IS145W). Table 3 shows the results of β-galactosidase expression by the serovar Typhimurium LBW100 lysogen grown as static liquid broth cultures favoring optimal fimbrial expression. Expression of fimA was two- to threefold higher in the fimW mutant strain. In addition, fimA expression was analyzed in an E. coli JM109λfimA-lacZ background after 48 h of growth in static liquid broth. As previously reported, there was no detectable fimA expression in E. coli unless the serovar Typhimurium fim regulatory genes, fimZ and fimY, are present (66). As shown in Table 3, use of a plasmid carrying fimZ, fimY, and fimW resulted in the production of a large amount of β-galactosidase by transformants. However, this fimA expression was further increased two- to threefold after transformation with the same plasmid carrying fimZ, fimY, and a disrupted fimW containing a translation terminator within the N terminus.
TABLE 3.
Expression of β-galactosidase by λfimA-lacZ reporters in serovar Typhimurium and E. coli
| Strain | Plasmid (relevant genotype) | β-Galactosidase expression (Miller units)a |
|---|---|---|
| S. enterica serovar Typhimurium | ||
| IS145 | None | 30 |
| IS145W | None | 74 |
| E. coli | ||
| JM109λfimA-lacZ | None | 0 |
| JM109λfimA-lacZ | pISF180 (fimZ+Y+W+) | 570 |
| JM109λfimA-lacZ | pISF230 (fimZ+Y+W) | 1,400 |
β-Galactosidase activity is reported in Miller units (45), and all assays were performed at least three times with ≤20% variability.
Primer extension analysis to determine the fimW transcriptional start site.
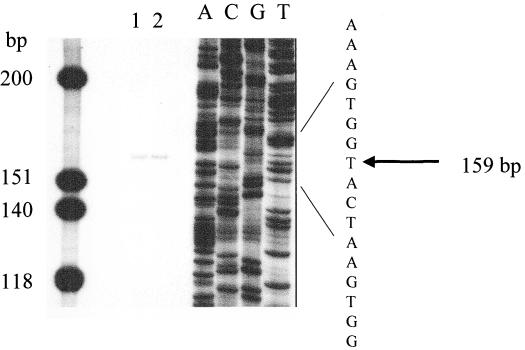
It has been previously demonstrated that fimY and fimZ can be independently expressed using promoters immediately upstream of these genes (62, 65). To determine if fimW is also transcribed independently and to identify the transcriptional start site, primer extension analysis was performed using RNA isolated from E. coli expressing the serovar Typhimurium fim gene cluster from the multicopy plasmid pISF101. Amplification of the RNA using reverse transcriptase and a primer that annealed to a region of 129 bp within the fimW coding region isolated a fragment of approximately 160 bp, as shown in Fig. 3. Sequence analysis of the putative fimW promoter region indicated that the site of transcription initiation is 31 bp upstream of the translation start site. The determined fimW transcription initiation site is illustrated in Fig. 3.
FIG. 3.
Identification of the fimW transcription initiation start site. Primer extension was performed with 5 μg (lane 1) or 10 μg (lane 2) of E. coli HB101 (pISF101) total cellular RNA and a primer that anneals 129 bp downstream of the fimW translation initiation. Lanes A, C, G, and T correspond to a dideoxy sequencing reaction performed with plasmid pISF101 and the same primer used to map transcription. The molecular size standards are as shown.
Analysis of fimW expression from a fimW-lacZ reporter fusion.
A fimW-lacZ fusion (pISF253) was constructed by ligation of a PCR product, encompassing the fimW transcription initiation site, into a single-copy promoterless lacZ vector, pGS375 (28). Analysis of expression from this reporter in serovar Typhimurium LB5010 revealed that fimW expression was consistently twofold greater when strains were grown on solid agar plates compared to bacteria grown in static liquid broth (Table 4). In contrast, growth on solid agar promoted low fimA expression in serovar Typhimurium LB5010 compared to fimA expression under static liquid conditions, as shown in Table 4. This is consistent with the phenotypic expression of fimbriae that occurs under these environmental conditions. In addition, as shown in Table 4, expression from the fimW-lacZ reporter was increased approximately threefold in serovar Typhimurium LBW100, compared to the parental strain, when cells are grown in static liquid broth. These studies indicate that fimW expression is increased under conditions that select for poorly fimbriate bacteria.
TABLE 4.
Analysis of fimW expression in serovar Typhimurium strains LB5010 and LBW100 under different growth conditions
| Conditions | Straina | β-Galactosidase expression (Miller units)b |
|---|---|---|
| 24-h plate | LB5010fimW-lacZ | 39 |
| LB5010fimA-lacZ | 24 | |
| LBW100fimW-lacZ | 33 | |
| LBW100fimA-lacZ | 42 | |
| 48-h static broth | LB5010fimW-lacZ | 20 |
| LBW5010fimA-lacZ | 81 | |
| LBW100fimW-lacZ | 59 | |
| LBW100fimA-lacZ | 77 |
The fimW-lacZ reporter is encoded on a single-copy plasmid (pISF253); the fimA-lacZ reporter is located on the chromosome as a lambda phage lysogen (ISF145).
β-Galactosidase expression is in Miller units (45), and at least three independent cultures were assayed with ≤20% variation.
Partial purification of FimW for use in in vitro DNA-binding assays.
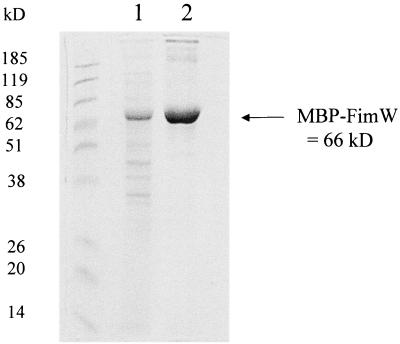
FimW was partially purified by construction of a fusion with the E. coli maltose-binding protein, and separation of this fusion protein from bacterial extracts on an amylose-agarose bead resin. Figure 4 shows the SDS-polyacrylamide gel electrophoresis (PAGE) analysis of the resulting protein preparation after elution from the resin. The major protein eluting from the column was approximately 66 kDa, a result consistent with fusion of the 23-kDa FimW protein and the 43-kDa maltose-binding protein. Up to 5 μg of this extract was combined with radiolabeled DNA fragments of fimA, fimY, fimZ, or fimW containing their respective promoter regions in gel mobility shift assays, and no altered mobility was observed (not shown). Additional DNA-binding assays were performed using a partially purified FimW-His6 fusion protein, as well as bacterial extracts prepared from E. coli transformed with pISF232 (fimW+) or pACYC184 alone. These assays also indicated no interaction between any of the fim-specific promoter regions and cell extracts possessing FimW.
FIG. 4.
SDS-PAGE of the partially purified MBP-FimW fusion. Lanes: 1 and 2, amylose resin column after first and second wash with Column Buffer; 3 and 4, samples from first two column volumes after elution with Column Buffer plus 10 mM maltose. Approximately 5 μg of protein was loaded per well. The molecular size standards and predicted molecular sizes are as shown.
Analysis of FimW-FimZ protein interactions using the LexA-based two-hybrid system in E. coli.
The LexA based two-hybrid system (15) utilizes the ability of the E. coli LexA repressor protein to dimerize on specific operator sequences. The construction of fusion proteins with the DNA-binding N terminus of wild-type LexA, as well as a mutant LexA that recognizes a unique operator sequence, allows the detection of protein heterodimerization. Protein interactions are identified as a repression of β-galactosidase from a lacZ reporter on the E. coli chromosome. A PCR product of the entire fimW coding region was cloned into pMS604, and the entire fimZ coding region was cloned into pDP804 to make the in-frame LexA fusions, pISF248 and pISF245, respectively. These fusions were introduced into the E. coli SU202 lysogen, and β-galactosidase expression was analyzed after a 48-h incubation in static liquid broth plus IPTG. Table 5 shows β-galactosidase expression from the E. coli lysogens carrying the Jun and Fos control fusions, as well as the FimW and FimZ fusions. Strains carrying either the Jun or Fos fusion alone produce high levels of β-galactosidase, indicating that there is little homodimerization resulting in repression of lacZ expression. However, strains carrying both fusions produced 40- to 50-fold less β-galactosidase, thus confirming that the control fusions strongly interact in a specific manner. The E. coli lysogen transformed with the individual FimW or FimZ fusions similarly produced high levels of β-galactosidase. However, transformation of the lysogen with both FimW and FimZ fusion plasmids generated a strain that consistently expressed 10- to 15-fold less β-galactosidase than strains carrying either plasmid alone, indicating that FimW and FimZ are interacting proteins. The LexA-based system was also used to construct a fimY fusion, and no interaction of FimY with FimW was detected using this pair of molecules (Table 5).
TABLE 5.
Expression of β-galactosidase by E. coli SU202 using the LexA two-hybrid system
| Plasmid (relevant genotype) | β-Galactosidase expression (Miller units)a |
|---|---|
| pDP804 (jun-lexA408) | 6,093 |
| pMS604 (fos-lexA) | 6,092 |
| pDP804 + pMS604 (jun-lexA408 + fos-lexA) | 133 |
| pISF245 (fimZ-lexA408) | 5,829 |
| pISF248 (fimW-lexA) | 4,607 |
| pISF244 (fimY-lexA408) | 6,302 |
| pISF245 + pISF248 (fimZ-lexA408 + fimW-lexA) | 395 |
| pISF244 + pISF248 (fimY-lexA408 + fimW-lexA) | 6,393 |
β-Galactosidase activity is reported in Miller units (45), and all assays were performed at least three times with <20% variability.
DISCUSSION
Analysis of the regulation of type 1 fimbrial expression in S. enterica serovar Typhimurium has determined that it is dependent upon multiple regulators and is distinct from the mechanism described in E. coli. Two proteins, FimZ and FimY, have been described as transcriptional coactivators and were found to be necessary for serovar Typhimurium type 1 fimbrial expression (62, 66). In addition, an arginine tRNA molecule encoded by fimU was determined to be involved in translational regulation of the serovar Typhimurium fim operon (13, 60). The present report describes the functional analysis of FimW. This protein is encoded by a gene on the fim cluster located between the regulators fimY and fimU, and the studies presented here indicate that FimW is also involved in serovar Typhimurium type 1 fimbrial regulation.
Amino acid sequence analysis of FimW suggests that this protein may be related to prokaryotic transcriptional regulators. The most closely related proteins include the response regulator, BpdT from Rhodococcus (40) and an uncharacterized response regulator from Pseudomonas putida (41). Both response regulators possess approximately 30% identity to FimW over a 50-amino-acid region in the C terminus of the protein. This region of the BpdT response regulator contains strong homology to a helix-turn-helix DNA-binding domain; however, FimW exhibits only weak homology to this structural motif (16).
Disruption of fimW on the serovar Typhimurium chromosome resulted in a strain that, by electron microscopy, appeared to be fimbriate and not significantly altered from the parental strain. However, analysis using serum agglutination and hemagglutination assays to quantify the expression of surface-associated fimbriae suggested that the fimW mutant expresses four- to eightfold more type 1 fimbriae compared to the parental strain. In addition, fimA expression from a fimA-lacZ reporter was increased in the absence of a functional FimW, in both an E. coli and a serovar Typhimurium background. These results are consistent with the role of FimW as a negative regulator of fimbrial expression, acting either directly on the fimA promoter to repress transcription or indirectly through the activity of other regulatory molecules. In addition, growth on solid agar, which is known to select for poorly fimbriate bacteria and low fimA expression, was found to stimulate a twofold increase in fimW expression. FimW may also act as an autoregulator, since this protein was found to negatively regulate its own expression under static liquid conditions. These results, using bacteria grown under conditions influencing phenotypic expression of fimbriae in salmonellae (49), indicate that FimW functions as a negative regulator that is expressed and acts to reduce fimbrial expression when serovar Typhimurium is subcultured in aerobic static broth.
To better understand the role of FimW in fimbrial regulation, FimW was partially purified by the construction of maltose-binding protein and histidine-tag fusions. These protein extracts were used in DNA-binding assays in which no interactions were observed between FimW and the fimA promoter as well as the fimZ, fimY, and fimW promoters. The inability to demonstrate a specific DNA-protein interaction in these studies may be due to several factors. The conditions used in these in vitro assays may not reflect the proper in vivo conditions necessary to demonstrate binding. Alternatively, FimW may be unable to bind to DNA or affect fimA transcription without the presence of a second regulatory molecule, such as FimZ or FimY. The use of fusion proteins may also inhibit the DNA-binding activity of FimW. However, the substitution of extracts containing native FimW for the fusions did not indicate that FimW could bind on its own to DNA fragments. In addition, a plasmid encoding the MBP-FimW fusion was found to decrease fimA expression in vivo, suggesting that the fusion retains negative regulatory activity.
To investigate the possibility of protein-protein interactions between FimW and other fim regulatory molecules, we utilized a two-hybrid system in E. coli based upon the repressor protein, LexA. This system demonstrated significant repression in the presence of the FimW and FimZ fusion molecules. This repression was not observed in the presence of FimY and FimW. These results suggest that FimW and FimZ interact in vivo and that the regulatory effect of FimW is not due to the binding of FimW alone at the fimA promoter. Instead, FimW may function by influencing the ability of FimZ to activate transcription from this promoter, either by inhibiting the binding of FimZ to the fimA promoter or by inhibiting activation by FimZ once this protein has bound. Currently, we are purifying both FimZ and FimW in order to investigate the effect of combining these proteins in gel mobility shift assays. FimZ is related to the response regulator BvgA of B. pertussis (55, 63). The regions of homology between these proteins include conserved phosphorylated residues, indicating that the action of FimZ is dependent upon phosphorylation. FimW may be involved in this phosphorylation cascade, such that it is able to inactivate FimZ. Recently, small proteins involved in unique His-Asp-His-Asp phosphorelays, containing phosphotransfer modules or HPt domains, have been described (22, 34, 51). In addition, regulatory proteins, such as PhoU and SixA of E. coli, have been identified that function to dephosphorylate members of two-component systems (48, 56).
To date, we have described four regulatory genes, located on the fim gene cluster, that are involved in serovar Typhimurium type 1 fimbrial expression. In addition, a number of global regulators have been implicated in type 1 fimbrial expression in E. coli, including LRP, H-NS, and IHF (4, 5, 17, 20, 25, 54), and these proteins may be involved in serovar Typhimurium fimbrial regulation as well. Therefore, the expression of these appendages in Salmonella serovars is likely to be controlled by a complex regulatory cascade. In 1966, Duguid et al. (18) extensively characterized the fimbriae and adhesive properties of salmonellae. From their results, it was observed that Salmonella phase variation is distinct from that of E. coli and Shigella spp. such that most Salmonella strains remain detectably, although poorly, fimbriate after serial subculture on agar. Thus, the highly controlled and complex cascade of serovar Typhimurium type 1 fimbrial regulation may be essential for facilitating intermediate levels of fimbrial expression that do not occur with the relatively tight on-off switching mechanism found in E. coli. It is likely that these unique mechanisms of regulation are the result of divergent host adaptation and play an important role in the host environment. It remains to be determined what the specific environmental signals are that promote Salmonella fimbrial phase variation and how this may relate specifically to this organism's ability to cause disease.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Initiative of the USDA (97-35204-4616) and a predoctoral fellowship to J.K.T. from a National Institutes of Health Parasitism Training Grant (TE AI07511).
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA that controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba E, Tsukamoto Y, Fukata T, Sasai K, Arakawa A. Increase in mannose residues, as Salmonella typhimurium-adhering factor, on the cecal mucosa of germ-free chickens infected with Eimeria tenella. Am J Vet Res. 1993;54:1471–1475. [PubMed] [Google Scholar]
- 3.Baumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomfield I C, Calie P J, Eberhardt K J, McClain M S, Eisenstein B I. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomfield I C, Kulasekara D H, Eisenstein B I. Integration host factor stimulates both FimB and FimE mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol Microbiol. 1997;23:705–717. doi: 10.1046/j.1365-2958.1997.2241615.x. [DOI] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix S. A complementary analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan K, Falkow S, Hull R A, Hull S I. Frequency among Enterobacteriaceae of the DNA sequences encoding type 1 pili. J Bacteriol. 1985;162:799–803. doi: 10.1128/jb.162.2.799-803.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullas L R, Ryu J. Salmonella typhimurium LT2 strains are r-m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban M J, Chou J, Cohen S. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clegg S, Hancox L S, Yeh K-S. Salmonella typhimurium phase variation and FimA expression. J Bacteriol. 1996;178:542–545. doi: 10.1128/jb.178.2.542-545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clegg S, Purcell B K, Pruckler J. Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect Immun. 1987;55:281–287. doi: 10.1128/iai.55.2.281-287.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clegg S, Swenson D L. Salmonella fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 105–113. [Google Scholar]
- 13.Clouthier S C, Collinson S K, White A P, Banser P A, Kay W W. tRNA(Arg) (fimU) and expression of SEF 14 and SEF 21 in Salmonella enteritidis. J Bacteriol. 1998;180:840–845. doi: 10.1128/jb.180.4.840-845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collinson S K, Liu S, Clouthier S C, Banser P A, Doran J L, Sanderson K E, Kay W W. The location of four fimbrin-encoding genes, agfA, fimA, sefA, and sefD, on the Salmonella enteritidis and/or S. typhimurium XbaI-BinI genomic restriction maps. Gene. 1996;169:75–80. doi: 10.1016/0378-1119(95)00763-6. [DOI] [PubMed] [Google Scholar]
- 15.Dmitrova M, Younes-Cauet G, Oertel-Buchheit P, Porte D, Schnarr M, Granger-Schnarr M. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol Gen Genet. 1998;257:205–212. doi: 10.1007/s004380050640. [DOI] [PubMed] [Google Scholar]
- 16.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorman C J, Higgins C F. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J Bacteriol. 1987;169:3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duguid J P, Anderson E S, Campbell I. Fimbriae and adhesive properties in Salmonellae. J Pathol Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- 19.Eisenstein B I. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science. 1981;214:337–338. doi: 10.1126/science.6116279. [DOI] [PubMed] [Google Scholar]
- 20.Eisenstein B L, Sweet D S, Vaughn V, Friedman D I. Integration host factor is required for the inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of Hep-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman J A, Bassler B L. Sequence and function of LuxU: a two-component phosphorelay protein that regulated quorum sensing in Vibrio harveyi. J Bacteriol. 1999;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of media and temperature. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in E. coli K-12. Mol Microbiol. 1996;21:752–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 25.Gally D L, Rucker T J, Blomfield I C. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli. J Bacteriol. 1994;176:5665–5672. doi: 10.1128/jb.176.18.5665-5672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garner M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the E. coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerlach G-F, Clegg S, Ness N J, Swenson D L, Allen B L, Nichols W A. Expression of type 1 fimbriae and mannose-sensitive hemagglutinin by recombinant plasmids. Infect Immun. 1989;57:764–770. doi: 10.1128/iai.57.3.764-770.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghrist A C, Stauffer G V. Characterization of the Escherichia coli gvcR gene encoding a negative regulator of gcv expression. J Bacteriol. 1995;177:4980–4984. doi: 10.1128/jb.177.17.4980-4984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hancox L S, Yeh K-S, Clegg S. Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol Med Microbiol. 1998;19:289–296. doi: 10.1111/j.1574-695X.1997.tb01099.x. [DOI] [PubMed] [Google Scholar]
- 30.Hoiseth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 31.Horiuchi S, Inagaki Y, Okamura N, Nakaya R, Yamamoto N. Type 1 pili enhance the invasion of Salmonella braenderup and Salmonella typhimurium to HeLa cells. Microbiol Immunol. 1992;36:593–602. doi: 10.1111/j.1348-0421.1992.tb02059.x. [DOI] [PubMed] [Google Scholar]
- 32.Isaacson R E, Argyilan C, Kwan L, Patterson S, Yoshinaga K. Phase variable switching of in vivo and environmental phenotypes of Salmonella typhimurium. Adv Exp Med Biol. 1999;473:281–289. doi: 10.1007/978-1-4615-4143-1_30. [DOI] [PubMed] [Google Scholar]
- 33.Isaacson R E, Kinsel M. Adhesion of Salmonella typhimurium to porcine intestinal epithelial surfaces: identification and characterization of two phenotypes. Infect Immun. 1992;60:3193–3200. doi: 10.1128/iai.60.8.3193-3200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishige K, Nagasawa S, Tokishita S, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones G W, Richardson L A. The attachment to, and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose resistant haemagglutinating activities. J Gen Microbiol. 1981;127:361–370. doi: 10.1099/00221287-127-2-361. [DOI] [PubMed] [Google Scholar]
- 36.Klemm P. The fimA gene encoding the type 1 fimbrial subunit of Escherichia coli. Eur J Biochem. 1984;143:395–399. doi: 10.1111/j.1432-1033.1984.tb08386.x. [DOI] [PubMed] [Google Scholar]
- 37.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in E. coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klemm P, Krogfelt K A. Type 1 fimbriae of Escherichia coli. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 9–26. [Google Scholar]
- 39.Kowit J D, Goldberg A L. Intermediate steps in the degradation of a specific abnormal protein in E. coli. J Biol Chem. 1977;252:8350–8357. [PubMed] [Google Scholar]
- 40.Labbe D, Garon J, Lau P C. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl degrading bacterium, Rhodococcus strain M5. J Bacteriol. 1997;179:2772–2776. doi: 10.1128/jb.179.8.2772-2776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau P C, Wang Y, Patel A, Labbe D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lockman H A, Curtiss R., III Virulence of non-type 1 fimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect Immun. 1992;60:491–496. doi: 10.1128/iai.60.2.491-496.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClain M S, Blomfield I C, Eisenstein B I. Roles of fimB and fimE in site-specific inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClain M S, Blomfield I C, Eberhardt K J, Eisenstein B I. Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1993;175:4335–4344. doi: 10.1128/jb.175.14.4335-4344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 46.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolan L K, Wooley R E, Brown J B, Payeur J B. Comparison of phenotypic characteristics of Salmonella spp isolated from healthy and ill (infected) chickens. Am J Vet Res. 1991;52:1512–1517. [PubMed] [Google Scholar]
- 48.Ogino T, Matsubara M, Kato N, Nakamura Y, Mizuno T. An Escherichia coli protein that exhibits phosphohistine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol Microbiol. 1998;27:573–585. doi: 10.1046/j.1365-2958.1998.00703.x. [DOI] [PubMed] [Google Scholar]
- 49.Old D C. Inhibition of the interaction between fimbrial haemagglutinins and erythrocytes by d-mannose and other carbohydrates. J Gen Microbiol. 1972;71:149–157. doi: 10.1099/00221287-71-1-149. [DOI] [PubMed] [Google Scholar]
- 50.Old D C, Duguid J P. Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol. 1970;103:447–456. doi: 10.1128/jb.103.2.447-456.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perraud A L, Wiess V, Gross R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 1999;7:115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Sanger R, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schembri M A, Olsen P B, Klemm P. Orientation-dependent enhancement by H-NS of the activity of the type 1 fimbrial phase switch promoter in Escherichia coli. Mol Gen Genet. 1998;259:336–344. doi: 10.1007/s004380050820. [DOI] [PubMed] [Google Scholar]
- 55.Stibitz S. Mutations in the bvgA gene of Bordetella pertussis that differentially affect regulation of virulence determinants. J Bacteriol. 1994;176:5615–5621. doi: 10.1128/jb.176.18.5615-5621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swenson D L. Ph.D. thesis. Iowa City: The University of Iowa; 1993. [Google Scholar]
- 58.Swenson D L, Clegg S. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J Bacteriol. 1992;174:7697–7704. doi: 10.1128/jb.174.23.7697-7704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swenson D L, Clegg S, Old D C. The frequency of fim genes among Salmonella serovars. Microb Pathog. 1991;10:487–492. doi: 10.1016/0882-4010(91)90115-q. [DOI] [PubMed] [Google Scholar]
- 60.Swenson D L, Kim K-J, Six E W, Clegg S. The gene fimU affects expression of Salmonella type 1 fimbriae and is related to the E. coli tRNA gene argU. Mol Gen Genet. 1994;244:216–218. doi: 10.1007/BF00283525. [DOI] [PubMed] [Google Scholar]
- 61.Tavendale A, Jardine C K H, Old D C, Duguid J P. Haemagglutinins and adhesion of Salmonella typhimurium to Hep-2 and Hela cells. J Med Microbiol. 1983;16:371–380. doi: 10.1099/00222615-16-3-371. [DOI] [PubMed] [Google Scholar]
- 62.Tinker J K, Clegg S. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica Serovar Typhimurium. Infect Immun. 2000;68:3305–3313. doi: 10.1128/iai.68.6.3305-3313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 64.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 65.Yeh K-S. Ph.D. thesis. Iowa City: The University of Iowa; 1997. [Google Scholar]
- 66.Yeh K-S, Hancox L S, Clegg S. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J Bacteriol. 1995;177:6861–6865. doi: 10.1128/jb.177.23.6861-6865.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]