Abstract
Objective
To study the role of m6A and lncRNAs in the prognosis and immunotherapy of hepatocellular carcinoma, construct the risk score of overall survival of hepatocellular carcinoma, and search for new therapeutic targets and drugs.
Methods
The data used in this study are obtained from The Cancer Genome Atlas (TCGA) database. A total of 424 HCC samples were included. The co-expression of lncRNAs and M6A-related genes in HCC was analyzed, COX regression analysis was conducted to construct the risk score for HCC prognosis, and the model's validity was further verified in different clinical trials subtypes and principal component analysis. GO enrichment analysis and immune function analysis were performed for the differential genes in the high-risk group and the low-risk group divided by risk score and analyzed the prognostic effect of TMB on the two groups. Based on the results, potential therapeutic agents for HCC were screened.
Results
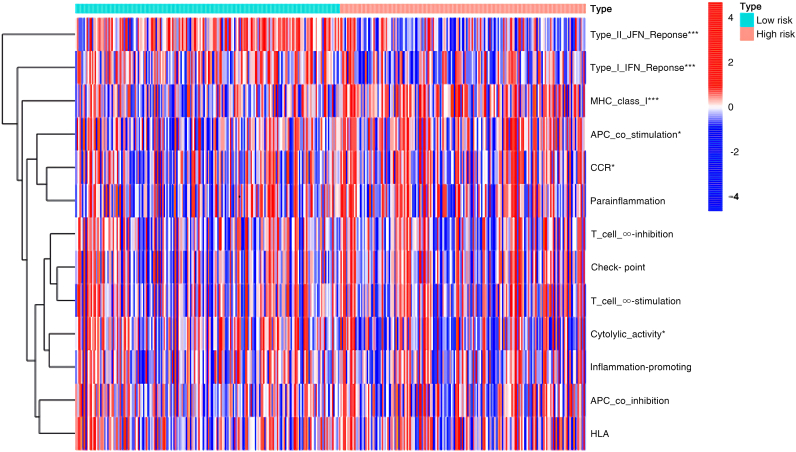
The risk score can better predict the prognosis of HCC, the area under the ROC curve is 0.727. Differential genes were mainly located in the extracellular matrix and chromosomal regions and may play regulatory roles in binding sites and catalytic enzymes, thereby affecting the chromosome division and cell proliferation of cells. Type Ⅱ IFN response, type Ⅰ IFN response and MHC class Ⅰ were the three most different functions in terms of immune function between the high-risk group and the low-risk group. Type II IFN response, type I IFN response was significantly down-regulated in the high-risk group, while MHC class I was up-regulated. 14 potential therapeutic drugs were screened out.
Conclusions
The risk score constructed with NRAV and AL031985.3 had a good predictive effect on the prognosis of HCC. Differences in genes and immune function between high-risk and low-risk groups promoted the occurrence and progression of HCC.
Keywords: NRAV, AL031985.3, Hepatocellular carcinoma, Risk score, m6A, Immune function
NRAV; AL031985.3; Hepatocellular carcinoma; Risk score; m6A; Immune function.
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world and the second leading cause of cancer-related deaths [1]. There are approximately 841,000 new cases and 782,000 deaths of HCC each year [2]. There are many methods to treat HCC, including surgery, radiofrequency ablation, transarterial chemoembolization (TACE), hepatic arterial infusion chemotherapy (HAIC), and drug therapy [3, 4, 5]. But the current treatment results of these methods are not satisfactory. The 3-year survival rate of HCC was only 12.7% and the median survival was 9 months [6].
Sorafenib is the first targeted drug used in the treatment of advanced HCC. It is a blocker of vascular endothelial growth factor receptors (VEGFRs). Although Sorafenib was established as a first-line drug for advanced HCC in 2017, it only prolonged the median overall survival of patients by 2.8 months [7]. After sorafenib, there are many clinical trials aimed at the treatment of advanced liver cancer, including Sunitinib [8], Brivanib [9], Linifanib [10], Lenvatinib [11]. It is worth noting that even lenvatinib (which has been approved for advanced HCC) did not show an advantage in the efficacy comparison with sorafenib [12]. Therefore, we need to develop better therapeutic targets and drugs for HCC.
N6-methyladenosine (m6A) is the most extensive and abundant transcription modification among eukaryotic messenger RNAs (mRNAs) [13], microRNAs (miRNAs) [14], and long non-coding RNAs (lncRNAs) [15]. M6A is mainly divided into three parts: “writers”, “erasers”, and “readers”. The main role of “writers” is to catalyze the production of m6A, including Methyltransferase-like 3 (METTL3) [16], KIAA1429 [17], METTL14 [18], RBM15/15B [19] and Wilms tumor 1-associated protein (WTAP) [20]. The core function of “erasers” is to remove the methyl code from mRNA, including alkB homologue 5 (ALKBH5) [21] and fat mass and obesity-associated protein (FTO) [22]. “Reader” is responsible for decoding m6A methylation, including nuclear ribonucleoprotein (HNRNP) protein [23], IGF2 mRNA binding proteins (IGF2BP) [24], eukaryotic initiation factor (eIF) 3[25] and YT521-B homology (YTH) domain-containing protein [26]. Studies have found that m6A modification plays an important role in tumor proliferation, differentiation, invasion, and metastasis, and can also be used as a tumor oncogene or anti-oncogene [27, 28, 29]. M6A regulates many aspects of the development of HCC. METTL3 is related to the poor prognosis of HCC patients and promotes the proliferation, migration, and colony formation of HCC cells [30]. In addition, METTL14 is an anti-metastatic factor, and its down-regulation will also promote the progression and metastasis of HCC [31]. Therefore, it is of great significance to study the regulation of m6A in liver cancer, to identify new therapeutic targets and potential therapeutic drugs, and to improve the prognosis of patients with liver cancer.
LncRNAs contain more than 200 nucleotides, interacts with DNA, RNA, or protein, regulate gene expression at the transcription or post-transcription level, and are closely related to the progression of gastric cancer, HCC, and colon cancer [32, 33, 34, 35]. Studies have shown that many lncRNAs, including MIAT [36], CDKN2BAS [37], and PCNAP1 [38], promote the proliferation and invasion of HCC cells, and lncRNA-D16366 can be used as a biological marker for the diagnosis and prognosis of HCC [39]. Therefore, studying the interaction between lncRNA and m6A in the occurrence and development of HCC can further discover its influence on the development of HCC, find potential therapeutic targets and drugs, and improve the prognosis of HCC patients.
2. Methods
2.1. Data of the research
The data used in this study are obtained from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/). A total of 424 HCC samples were included, including 50 normal samples and 374 tumor samples. Clinical data mainly included age, survival status, TMN stage, tumor stage, grade, and gender.
2.2. Co-expression analysis of m6A and lncRNA
mRNAs and lncRNAs were screened from TCGA transcriptome data. Subsequently, m6A gene expression was extracted from transcriptome data. A total of 23 m6A genes were used in this paper. (Writers: METTL3, METTL14, METTL16, WTAP, VIRMA, ZC3H13, RBM15, RBM15B. Readers: YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, HNRNPC, FMR1, LRPPRC, HNRNPA2B1, IGFBP1, IGFBP2, IGFBP3, RBMX. Erasers: FTO, ALKBH5.) In the analysis of co-expression of the m6A genes and lncRNAs, the correlation test was adopted. The filter criterion for the correlation coefficient was set as corFilter = 0.4, and the filter criterion for the p-value of the correlation test was set as pvalueFilter = 0.001. Drawing the Sankey diagram based on the co-expression relationship of m6A and lncRNAs.
2.3. Construct a prognostic model
Clinical data of the samples were combined with m6A-related lncRNA data, then divided into a training group and test group by 1:1. The training group is used to build the model, and the text group is used to validate the model. Construct predictive models using the “survival”, “caret”, “glmnet”, “survminer”, and “timeROC” packages. When constructing the prediction model, univariate COX regression was adopted, coxPfilter was set to 0.05, and then the forest map was drawn. Univariate COX regression was performed for the training group, followed by Lasso regression and cross-validation. Then the COX model is constructed. The text group works the same way as the training group. The samples of the two groups were divided into the high-risk group and the low-risk group with the median risk score as the boundary, and the ROC curves of the high-risk group and the low-risk group were plotted.
Used the “pheatmap” package to draw the risk curve, sorted the samples according to the patient risk score, and then drew the risk curve, survival status map, and risk heat map of the overall sample, the training group, and the text group, respectively. Univariate COX analysis and multivariate COX analysis were performed for the risk score, followed by independent prognostic analysis to obtain the forest map of the risk score. The predictive function of the model was calculated by the area under the ROC curve. The validity of the model was evaluated by the c-index curve. Used the “survival”, “regplot”, and “rms” packages to draw a nomogram of the risk score.
2.4. Model validation for clinical grouping
Used the “survival” and “survminer” packages to plot the survival curves of the high-risk group and the low-risk group in subgroups of age, sex, and tumor stage, respectively, to verify the validity of the risk score.
2.5. Principal component analysis
Principal component analysis was performed using the “limma” and “scatterplot3d” packages, and PCA plots of all gene expression levels, m6A-related gene expression levels, m6A-related lncRNA expression levels, and model-related lncRNA expression levels were plotted.
2.6. Risk differential analysis and GO enrichment analysis
The wilcoxon test was used to analyze the genes that differ between the high-risk and low-risk groups, and the filter conditions were set as logFCfilter = 1 and fdrFilter = 0.05. GO enrichment analysis of differential genes in high-risk group and low-risk group was performed using “clusterProfiler”, “org.Hs.eg.db”, “enrichplot”, “ggplot2”, “ggpubr”, and “dplyr” packages. (pvalueFilter = 0.05, qvalueFilter = 0.05).
2.7. Immune function analysis
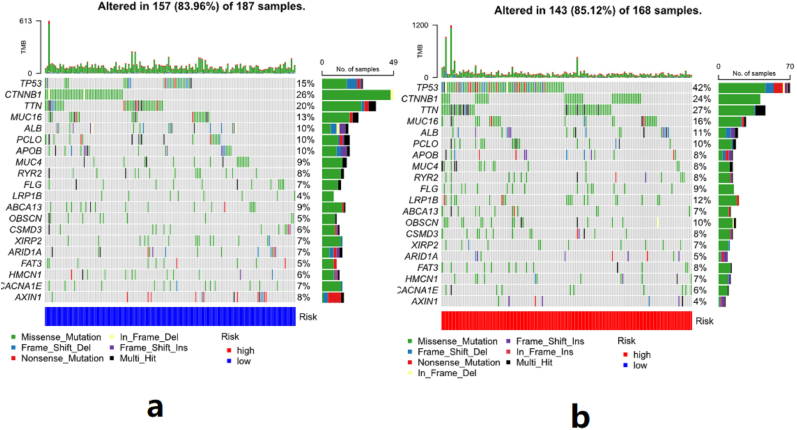
Used “limma”, “GSVA”, “GSEABase”, “pheatmap”, and “reshape2” to perform differential analysis of immune function between the high-risk group and low-risk group, and drew a heat map. The mutation dataset of HCC was divided into the high-risk group and low-risk group by risk score, and then drew the waterfall chart of gene mutations in the high-risk group and low-risk group, respectively.
2.8. Tumor mutational burden (TMB)
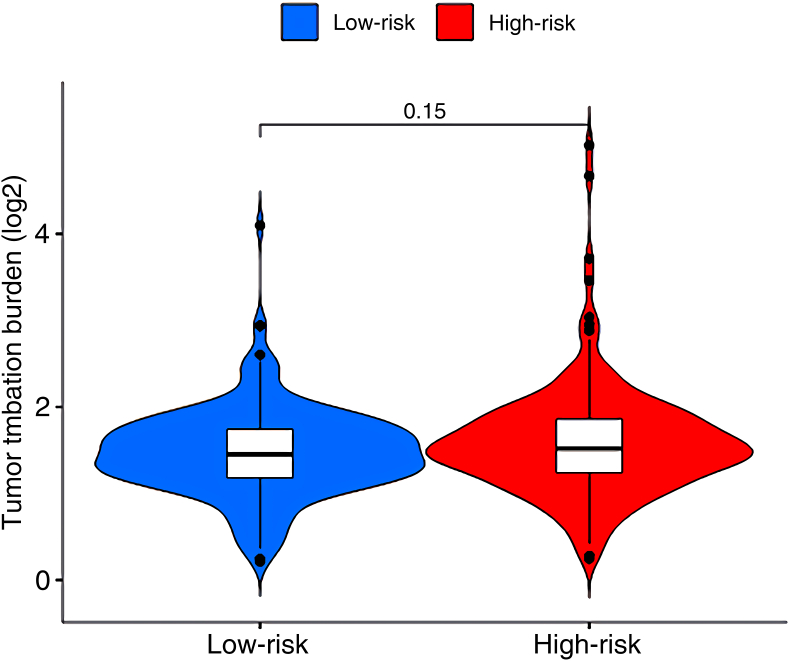
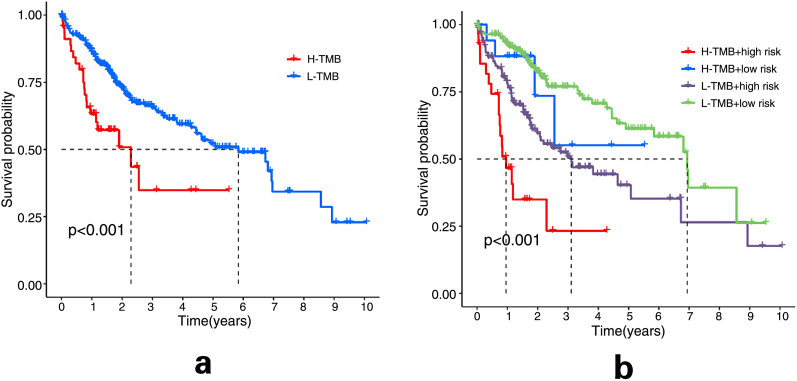
Get TMB data from the TCGA database and selected the results of varscan for analysis. TMB data for each sample was extracted using Strawberry Perl software. Using the “limma” and “ggpubr” packages in R software to merge TMB data with high-risk group data and low-risk group data, and draw a violin plot. Then use the “survival” and “survminer” packages to draw the survival curves of TMB and TMB combined with the risk score.
2.9. Screening for potential drugs
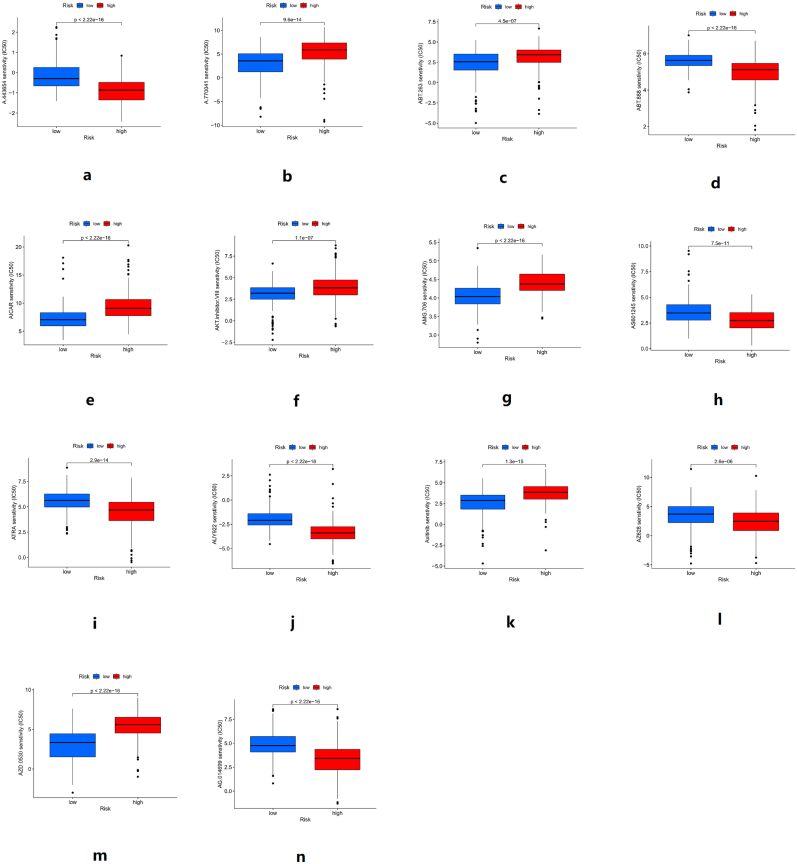
The “pRRophetic” package was used to predict potential therapeutic drugs for HCC. Combined the data of drug sensitivity with the data of the high-risk group and low-risk group, calculated the IC50 value of the drug in the high-risk group and low-risk group, and then drew the box plot.
3. Statistics
Analysis was performed using R 4.1.2 and Strawberry Perl.
4. Results
4.1. Co-expression analysis of m6A and lncRNA
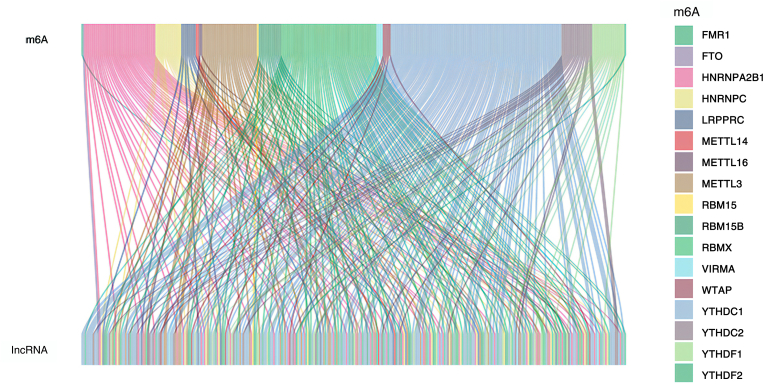
A total of 424 HCC samples were included, including 50 normal samples and 374 tumor samples. The co-expression of the m6A genes and lncRNAs can be seen in Table 1. LncRNAs were positively regulated by the m6A genes in HCC samples. YTHDC1 is involved in the most extensive regulation of lncRNAs in HCC. The correlation between the m6A genes and lncRNAs can be seen in Figure 1.
Table 1.
Co-expression of m6A and lncRNA.
| m6A | lncRNA | Cor | pvalue | Regulation |
|---|---|---|---|---|
| YTHDF2 | AL031985.3 | 0.51148 | 2.54E-26 | positive |
| YTHDF1 | NORAD | 0.550622 | 4.93E-31 | positive |
| YTHDF1 | AC091057.1 | 0.547303 | 1.31E-30 | positive |
| YTHDF1 | PTOV1-AS1 | 0.531677 | 1.12E-28 | positive |
| YTHDF1 | AC125257.1 | 0.526077 | 5.24E-28 | positive |
| YTHDF1 | AL035461.2 | 0.525615 | 5.94E-28 | positive |
| YTHDF1 | RNF216P1 | 0.523227 | 1.13E-27 | positive |
| YTHDF1 | AC099850.3 | 0.522158 | 1.51E-27 | positive |
| YTHDF1 | LINC00205 | 0.521027 | 2.05E-27 | positive |
| YTHDF1 | HCG18 | 0.519164 | 3.37E-27 | positive |
| YTHDF1 | AC093227.1 | 0.517472 | 5.28E-27 | positive |
| YTHDF1 | AL078644.1 | 0.5164 | 7.02E-27 | positive |
| YTHDF1 | BACE1-AS | 0.514815 | 1.06E-26 | positive |
| YTHDF1 | SREBF2-AS1 | 0.514512 | 1.15E-26 | positive |
| YTHDF1 | WAC-AS1 | 0.508586 | 5.37E-26 | positive |
| YTHDF1 | AC005288.1 | 0.505018 | 1.34E-25 | positive |
| YTHDF1 | AC145207.5 | 0.504318 | 1.60E-25 | positive |
| YTHDF1 | MCM3AP-AS1 | 0.50412 | 1.68E-25 | positive |
| YTHDF1 | NRAV | 0.500804 | 3.87E-25 | positive |
| YTHDF1 | SNHG1 | 0.500649 | 4.03E-25 | positive |
| YTHDF1 | AC102953.2 | 0.500525 | 4.15E-25 | positive |
| YTHDC2 | AC005288.1 | 0.594952 | 3.52E-37 | positive |
| YTHDC2 | PAXIP1-AS2 | 0.590209 | 1.78E-36 | positive |
| YTHDC2 | Z68871.1 | 0.590141 | 1.82E-36 | positive |
| YTHDC2 | EBLN3P | 0.589931 | 1.96E-36 | positive |
| YTHDC2 | AC008906.1 | 0.560293 | 2.69E-32 | positive |
| YTHDC2 | AC005034.5 | 0.558154 | 5.16E-32 | positive |
| YTHDC2 | FGD5-AS1 | 0.539001 | 1.43E-29 | positive |
| YTHDC2 | AC108010.1 | 0.537957 | 1.93E-29 | positive |
| YTHDC2 | AC024075.1 | 0.535953 | 3.40E-29 | positive |
| YTHDC2 | AC104532.2 | 0.527398 | 3.65E-28 | positive |
| YTHDC2 | AC025917.1 | 0.525789 | 5.67E-28 | positive |
| YTHDC2 | AC073046.1 | 0.523695 | 1.00E-27 | positive |
| YTHDC2 | OIP5-AS1 | 0.523133 | 1.16E-27 | positive |
| YTHDC2 | AC010834.3 | 0.514932 | 1.03E-26 | positive |
| YTHDC2 | AL109614.1 | 0.51457 | 1.14E-26 | positive |
| YTHDC2 | LINC00630 | 0.510046 | 3.69E-26 | positive |
| YTHDC2 | AC021078.1 | 0.509839 | 3.89E-26 | positive |
| YTHDC2 | Z83843.1 | 0.508917 | 4.93E-26 | positive |
| YTHDC2 | AL031846.2 | 0.501408 | 3.33E-25 | positive |
| YTHDC1 | AC005288.1 | 0.742167 | 1.25E-66 | positive |
| YTHDC1 | AC007406.4 | 0.722877 | 1.09E-61 | positive |
| YTHDC1 | AC010834.3 | 0.6921 | 1.28E-54 | positive |
| YTHDC1 | LINC00630 | 0.683689 | 7.69E-53 | positive |
| YTHDC1 | ZNF32-AS2 | 0.664701 | 4.91E-49 | positive |
| YTHDC1 | EBLN3P | 0.656172 | 2.05E-47 | positive |
| YTHDC1 | FAM111A-DT | 0.643089 | 4.96E-45 | positive |
| YTHDC1 | ANKRD10-IT1 | 0.642009 | 7.71E-45 | positive |
| YTHDC1 | LINC02035 | 0.636429 | 7.33E-44 | positive |
| YTHDC1 | CR936218.1 | 0.631009 | 6.26E-43 | positive |
| YTHDC1 | THAP9-AS1 | 0.625603 | 5.09E-42 | positive |
| YTHDC1 | MCM3AP-AS1 | 0.625419 | 5.46E-42 | positive |
| YTHDC1 | LINC01560 | 0.621005 | 2.93E-41 | positive |
| YTHDC1 | AL031714.1 | 0.62024 | 3.91E-41 | positive |
| YTHDC1 | FGD5-AS1 | 0.618292 | 8.13E-41 | positive |
| YTHDC1 | AL157392.3 | 0.617146 | 1.25E-40 | positive |
| YTHDC1 | EIF3J-DT | 0.615715 | 2.12E-40 | positive |
| YTHDC1 | ABALON | 0.615207 | 2.56E-40 | positive |
| YTHDC1 | AC004771.2 | 0.611763 | 9.08E-40 | positive |
| YTHDC1 | AC073046.1 | 0.605194 | 9.73E-39 | positive |
| YTHDC1 | HCG18 | 0.601879 | 3.15E-38 | positive |
| YTHDC1 | AC093227.1 | 0.600514 | 5.10E-38 | positive |
| YTHDC1 | AC091057.1 | 0.599273 | 7.87E-38 | positive |
| YTHDC1 | AC037459.2 | 0.598336 | 1.09E-37 | positive |
| YTHDC1 | WARS2-AS1 | 0.595721 | 2.70E-37 | positive |
| YTHDC1 | AC009120.3 | 0.595252 | 3.18E-37 | positive |
| YTHDC1 | LINC00909 | 0.594674 | 3.88E-37 | positive |
| YTHDC1 | NORAD | 0.594225 | 4.52E-37 | positive |
| YTHDC1 | AC129510.1 | 0.592677 | 7.69E-37 | positive |
| YTHDC1 | AC109460.2 | 0.591656 | 1.09E-36 | positive |
| YTHDC1 | AC091185.1 | 0.588817 | 2.85E-36 | positive |
| YTHDC1 | AC008124.1 | 0.588412 | 3.26E-36 | positive |
| YTHDC1 | LINC00265 | 0.586132 | 7.01E-36 | positive |
| YTHDC1 | Z68871.1 | 0.580921 | 3.93E-35 | positive |
| YTHDC1 | AL132989.1 | 0.578257 | 9.37E-35 | positive |
| YTHDC1 | PSMA3-AS1 | 0.575882 | 2.02E-34 | positive |
| YTHDC1 | AL133355.1 | 0.570903 | 9.92E-34 | positive |
| YTHDC1 | AC092614.1 | 0.569092 | 1.76E-33 | positive |
| YTHDC1 | AC092910.3 | 0.568975 | 1.82E-33 | positive |
| YTHDC1 | AP005899.1 | 0.566982 | 3.41E-33 | positive |
| YTHDC1 | C2orf49-DT | 0.566697 | 3.72E-33 | positive |
| YTHDC1 | TAPT1-AS1 | 0.560589 | 2.46E-32 | positive |
| YTHDC1 | LENG8-AS1 | 0.560318 | 2.67E-32 | positive |
| YTHDC1 | AC138028.4 | 0.559864 | 3.07E-32 | positive |
| YTHDC1 | MIR4453HG | 0.557289 | 6.71E-32 | positive |
| YTHDC1 | AC112220.2 | 0.557286 | 6.72E-32 | positive |
| YTHDC1 | ZEB1-AS1 | 0.556724 | 7.96E-32 | positive |
| YTHDC1 | CTBP1-DT | 0.5535 | 2.10E-31 | positive |
| YTHDC1 | NFYC-AS1 | 0.552178 | 3.11E-31 | positive |
| YTHDC1 | AC004067.1 | 0.551588 | 3.70E-31 | positive |
| YTHDC1 | UBA6-AS1 | 0.550578 | 4.99E-31 | positive |
| YTHDC1 | INE1 | 0.548871 | 8.26E-31 | positive |
| YTHDC1 | NRAV | 0.54811 | 1.03E-30 | positive |
| YTHDC1 | UBE2Q1-AS1 | 0.547881 | 1.10E-30 | positive |
| YTHDC1 | AC106037.2 | 0.54751 | 1.23E-30 | positive |
| YTHDC1 | AF131215.5 | 0.546567 | 1.62E-30 | positive |
| YTHDC1 | LINC00641 | 0.545955 | 1.94E-30 | positive |
| YTHDC1 | MAP3K14-AS1 | 0.543844 | 3.58E-30 | positive |
| YTHDC1 | AC008969.1 | 0.543399 | 4.07E-30 | positive |
| YTHDC1 | AC107027.3 | 0.542611 | 5.10E-30 | positive |
| YTHDC1 | AC008760.1 | 0.539962 | 1.09E-29 | positive |
| YTHDC1 | AC005104.1 | 0.539632 | 1.20E-29 | positive |
| YTHDC1 | AC096586.2 | 0.53862 | 1.60E-29 | positive |
| YTHDC1 | HMGN3-AS1 | 0.537345 | 2.29E-29 | positive |
| YTHDC1 | ASH1L-AS1 | 0.537236 | 2.37E-29 | positive |
| YTHDC1 | SEC24B-AS1 | 0.536885 | 2.61E-29 | positive |
| YTHDC1 | AC004076.2 | 0.535602 | 3.75E-29 | positive |
| YTHDC1 | AL031670.1 | 0.534471 | 5.15E-29 | positive |
| YTHDC1 | OIP5-AS1 | 0.533589 | 6.59E-29 | positive |
| YTHDC1 | AL078644.1 | 0.533381 | 6.99E-29 | positive |
| YTHDC1 | AC005253.1 | 0.530689 | 1.48E-28 | positive |
| YTHDC1 | AC022150.2 | 0.530433 | 1.58E-28 | positive |
| YTHDC1 | AP003392.1 | 0.529425 | 2.09E-28 | positive |
| YTHDC1 | AC087289.5 | 0.528915 | 2.41E-28 | positive |
| YTHDC1 | FBXL19-AS1 | 0.528801 | 2.49E-28 | positive |
| YTHDC1 | TRAPPC12-AS1 | 0.527675 | 3.39E-28 | positive |
| YTHDC1 | XPC-AS1 | 0.525204 | 6.64E-28 | positive |
| YTHDC1 | AC026412.3 | 0.52382 | 9.67E-28 | positive |
| YTHDC1 | PKD1P6-NPIPP1 | 0.52277 | 1.28E-27 | positive |
| YTHDC1 | AL049840.4 | 0.5219 | 1.62E-27 | positive |
| YTHDC1 | LINC01521 | 0.520677 | 2.25E-27 | positive |
| YTHDC1 | AL049840.3 | 0.520507 | 2.36E-27 | positive |
| YTHDC1 | LINC00342 | 0.520418 | 2.41E-27 | positive |
| YTHDC1 | AP001469.2 | 0.518913 | 3.60E-27 | positive |
| YTHDC1 | AC232271.1 | 0.51881 | 3.70E-27 | positive |
| YTHDC1 | WAC-AS1 | 0.518746 | 3.77E-27 | positive |
| YTHDC1 | AC004596.1 | 0.518099 | 4.48E-27 | positive |
| YTHDC1 | N4BP2L2-IT2 | 0.516555 | 6.73E-27 | positive |
| YTHDC1 | AC108010.1 | 0.51638 | 7.05E-27 | positive |
| YTHDC1 | ADNP-AS1 | 0.516281 | 7.24E-27 | positive |
| YTHDC1 | PTOV1-AS1 | 0.51493 | 1.03E-26 | positive |
| YTHDC1 | AL031282.2 | 0.514642 | 1.11E-26 | positive |
| YTHDC1 | AL355102.4 | 0.514274 | 1.23E-26 | positive |
| YTHDC1 | AC004908.1 | 0.514027 | 1.31E-26 | positive |
| YTHDC1 | AC015849.3 | 0.510056 | 3.68E-26 | positive |
| YTHDC1 | KANSL1L-AS1 | 0.509829 | 3.90E-26 | positive |
| YTHDC1 | AC245060.2 | 0.507945 | 6.33E-26 | positive |
| YTHDC1 | NIFK-AS1 | 0.507335 | 7.40E-26 | positive |
| YTHDC1 | CTBP1-AS | 0.506378 | 9.46E-26 | positive |
| YTHDC1 | THBS3-AS1 | 0.504239 | 1.63E-25 | positive |
| YTHDC1 | LAMC1-AS1 | 0.503713 | 1.86E-25 | positive |
| YTHDC1 | AC067852.3 | 0.503548 | 1.94E-25 | positive |
| YTHDC1 | AC099850.3 | 0.502613 | 2.46E-25 | positive |
| YTHDC1 | AC127024.4 | 0.502497 | 2.53E-25 | positive |
| YTHDC1 | AC114956.2 | 0.501877 | 2.96E-25 | positive |
| YTHDC1 | AC025287.3 | 0.501436 | 3.30E-25 | positive |
| WTAP | AC026356.1 | 0.582071 | 2.69E-35 | positive |
| WTAP | TRAF3IP2-AS1 | 0.569147 | 1.73E-33 | positive |
| WTAP | AC099850.3 | 0.554761 | 1.44E-31 | positive |
| WTAP | AC091057.1 | 0.527277 | 3.78E-28 | positive |
| WTAP | AL031714.1 | 0.521811 | 1.66E-27 | positive |
| VIRMA | OTUD6B-AS1 | 0.645582 | 1.78E-45 | positive |
| VIRMA | UBR5-AS1 | 0.586982 | 5.27E-36 | positive |
| VIRMA | AF117829.1 | 0.53939 | 1.28E-29 | positive |
| VIRMA | AC064807.1 | 0.511552 | 2.50E-26 | positive |
| RBMX | HCG18 | 0.698662 | 4.73E-56 | positive |
| RBMX | AC091057.1 | 0.668986 | 7.20E-50 | positive |
| RBMX | AC099850.3 | 0.66859 | 8.61E-50 | positive |
| RBMX | PTOV1-AS1 | 0.611412 | 1.03E-39 | positive |
| RBMX | AC007406.4 | 0.602737 | 2.33E-38 | positive |
| RBMX | LINC00630 | 0.595027 | 3.43E-37 | positive |
| RBMX | MCM3AP-AS1 | 0.593656 | 5.50E-37 | positive |
| RBMX | SNHG1 | 0.589773 | 2.06E-36 | positive |
| RBMX | WAC-AS1 | 0.586376 | 6.46E-36 | positive |
| RBMX | AC005288.1 | 0.584444 | 1.23E-35 | positive |
| RBMX | LINC00909 | 0.582578 | 2.28E-35 | positive |
| RBMX | TRAF3IP2-AS1 | 0.579985 | 5.34E-35 | positive |
| RBMX | NIFK-AS1 | 0.57686 | 1.47E-34 | positive |
| RBMX | ASH1L-AS1 | 0.572963 | 5.15E-34 | positive |
| RBMX | ZNF32-AS2 | 0.572703 | 5.60E-34 | positive |
| RBMX | AC093227.1 | 0.570616 | 1.09E-33 | positive |
| RBMX | WARS2-AS1 | 0.568393 | 2.19E-33 | positive |
| RBMX | EIF3J-DT | 0.566846 | 3.55E-33 | positive |
| RBMX | AL078644.1 | 0.562215 | 1.49E-32 | positive |
| RBMX | NRAV | 0.560055 | 2.90E-32 | positive |
| RBMX | DNAJC9-AS1 | 0.559791 | 3.14E-32 | positive |
| RBMX | LINC00205 | 0.558976 | 4.02E-32 | positive |
| RBMX | MIR600HG | 0.54688 | 1.48E-30 | positive |
| RBMX | NUTM2A-AS1 | 0.546598 | 1.61E-30 | positive |
| RBMX | LINC02035 | 0.545136 | 2.46E-30 | positive |
| RBMX | LENG8-AS1 | 0.545093 | 2.49E-30 | positive |
| RBMX | LINC00641 | 0.539012 | 1.43E-29 | positive |
| RBMX | AC025176.1 | 0.536152 | 3.21E-29 | positive |
| RBMX | SBF2-AS1 | 0.534498 | 5.11E-29 | positive |
| RBMX | LINC00665 | 0.532905 | 7.98E-29 | positive |
| RBMX | AC107027.3 | 0.531993 | 1.03E-28 | positive |
| RBMX | FAM111A-DT | 0.530129 | 1.72E-28 | positive |
| RBMX | AL050341.2 | 0.529013 | 2.34E-28 | positive |
| RBMX | AL157392.3 | 0.526427 | 4.76E-28 | positive |
| RBMX | AC004076.2 | 0.525023 | 6.98E-28 | positive |
| RBMX | AL008729.1 | 0.524459 | 8.13E-28 | positive |
| RBMX | SNHG32 | 0.523448 | 1.07E-27 | positive |
| RBMX | AL592148.3 | 0.518266 | 4.28E-27 | positive |
| RBMX | DDX11-AS1 | 0.51823 | 4.32E-27 | positive |
| RBMX | TMEM147-AS1 | 0.517423 | 5.35E-27 | positive |
| RBMX | AC008969.1 | 0.515879 | 8.05E-27 | positive |
| RBMX | AC007066.2 | 0.514552 | 1.14E-26 | positive |
| RBMX | AC232271.1 | 0.514447 | 1.17E-26 | positive |
| RBMX | AC099850.1 | 0.512819 | 1.79E-26 | positive |
| RBMX | PSMA3-AS1 | 0.509336 | 4.43E-26 | positive |
| RBMX | AL356481.1 | 0.509116 | 4.69E-26 | positive |
| RBMX | AC127024.5 | 0.508843 | 5.03E-26 | positive |
| RBMX | SNHG4 | 0.508719 | 5.19E-26 | positive |
| RBMX | TMCO1-AS1 | 0.507927 | 6.36E-26 | positive |
| RBMX | LINC01560 | 0.507793 | 6.58E-26 | positive |
| RBMX | ZNF529-AS1 | 0.507117 | 7.83E-26 | positive |
| RBMX | SEC24B-AS1 | 0.505458 | 1.20E-25 | positive |
| RBMX | AC125257.1 | 0.505072 | 1.32E-25 | positive |
| RBMX | CAPN10-DT | 0.50388 | 1.78E-25 | positive |
| RBMX | AL121772.3 | 0.50385 | 1.80E-25 | positive |
| RBMX | AP000873.1 | 0.503369 | 2.03E-25 | positive |
| RBMX | SNHG20 | 0.502945 | 2.26E-25 | positive |
| RBMX | LINC00653 | 0.502135 | 2.77E-25 | positive |
| RBMX | AC018690.1 | 0.500309 | 4.38E-25 | positive |
| RBM15B | AC091057.1 | 0.560355 | 2.64E-32 | positive |
| RBM15B | FGD5-AS1 | 0.553376 | 2.17E-31 | positive |
| RBM15B | HCG18 | 0.552725 | 2.64E-31 | positive |
| RBM15B | BACE1-AS | 0.546771 | 1.53E-30 | positive |
| RBM15B | LINC00205 | 0.540933 | 8.26E-30 | positive |
| RBM15B | AC099850.3 | 0.530714 | 1.47E-28 | positive |
| RBM15B | CTBP1-DT | 0.530306 | 1.64E-28 | positive |
| RBM15B | AC125257.1 | 0.52832 | 2.84E-28 | positive |
| RBM15B | AC005288.1 | 0.527695 | 3.37E-28 | positive |
| RBM15B | SREBF2-AS1 | 0.522054 | 1.56E-27 | positive |
| RBM15B | MCM3AP-AS1 | 0.520187 | 2.57E-27 | positive |
| RBM15B | PTOV1-AS1 | 0.519661 | 2.95E-27 | positive |
| RBM15B | AL078644.1 | 0.510507 | 3.27E-26 | positive |
| RBM15B | NRAV | 0.50048 | 4.20E-25 | positive |
| RBM15 | AC125257.1 | 0.505343 | 1.23E-25 | positive |
| METTL3 | AC007406.4 | 0.574483 | 3.17E-34 | positive |
| METTL3 | TMEM147-AS1 | 0.573369 | 4.53E-34 | positive |
| METTL3 | LINC00641 | 0.559358 | 3.58E-32 | positive |
| METTL3 | AC007066.2 | 0.556181 | 9.38E-32 | positive |
| METTL3 | PTOV1-AS2 | 0.555605 | 1.12E-31 | positive |
| METTL3 | MCM3AP-AS1 | 0.552766 | 2.61E-31 | positive |
| METTL3 | AC093227.1 | 0.542113 | 5.89E-30 | positive |
| METTL3 | PKD1P6-NPIPP1 | 0.541276 | 7.49E-30 | positive |
| METTL3 | AL512770.1 | 0.53459 | 4.98E-29 | positive |
| METTL3 | AC091057.1 | 0.531741 | 1.10E-28 | positive |
| METTL3 | TRAF3IP2-AS1 | 0.531243 | 1.27E-28 | positive |
| METTL3 | AC107068.1 | 0.530405 | 1.60E-28 | positive |
| METTL3 | AC127024.4 | 0.526424 | 4.77E-28 | positive |
| METTL3 | AC027601.1 | 0.52485 | 7.31E-28 | positive |
| METTL3 | DNAJC9-AS1 | 0.522 | 1.58E-27 | positive |
| METTL3 | AC084018.1 | 0.521196 | 1.96E-27 | positive |
| METTL3 | AL355075.2 | 0.521169 | 1.97E-27 | positive |
| METTL3 | AL049840.4 | 0.521 | 2.07E-27 | positive |
| METTL3 | LENG8-AS1 | 0.520857 | 2.15E-27 | positive |
| METTL3 | AC008735.2 | 0.51703 | 5.94E-27 | positive |
| METTL3 | AC124045.1 | 0.511815 | 2.33E-26 | positive |
| METTL3 | AC018690.1 | 0.511586 | 2.47E-26 | positive |
| METTL3 | HCG18 | 0.509198 | 4.59E-26 | positive |
| METTL3 | SEMA3F-AS1 | 0.508958 | 4.88E-26 | positive |
| METTL3 | AL161452.1 | 0.507631 | 6.86E-26 | positive |
| METTL3 | AL008729.1 | 0.506437 | 9.31E-26 | positive |
| METTL3 | KDM4A-AS1 | 0.504539 | 1.51E-25 | positive |
| METTL3 | AL157392.3 | 0.503974 | 1.74E-25 | positive |
| METTL3 | AL031670.1 | 0.5036 | 1.92E-25 | positive |
| METTL3 | AC074117.1 | 0.501311 | 3.41E-25 | positive |
| METTL3 | AC037459.2 | 0.501077 | 3.62E-25 | positive |
| METTL3 | AC073575.2 | 0.500937 | 3.75E-25 | positive |
| METTL3 | C2orf49-DT | 0.500632 | 4.04E-25 | positive |
| METTL3 | MED8-AS1 | 0.500274 | 4.42E-25 | positive |
| METTL16 | STARD7-AS1 | 0.565025 | 6.27E-33 | positive |
| METTL16 | AC020915.1 | 0.500863 | 3.82E-25 | positive |
| METTL14 | FGD5-AS1 | 0.534167 | 5.61E-29 | positive |
| METTL14 | EBLN3P | 0.521662 | 1.73E-27 | positive |
| LRPPRC | AC005288.1 | 0.566461 | 4.01E-33 | positive |
| LRPPRC | AC092614.1 | 0.544749 | 2.75E-30 | positive |
| LRPPRC | MCM3AP-AS1 | 0.537938 | 1.94E-29 | positive |
| LRPPRC | HCG18 | 0.532317 | 9.40E-29 | positive |
| LRPPRC | AL133243.2 | 0.51537 | 9.20E-27 | positive |
| LRPPRC | AC016747.1 | 0.50876 | 5.14E-26 | positive |
| LRPPRC | FGD5-AS1 | 0.5064 | 9.40E-26 | positive |
| LRPPRC | SNHG16 | 0.503641 | 1.90E-25 | positive |
| LRPPRC | AC099850.3 | 0.50047 | 4.21E-25 | positive |
| HNRNPC | HCG18 | 0.627855 | 2.14E-42 | positive |
| HNRNPC | AC091057.1 | 0.585151 | 9.72E-36 | positive |
| HNRNPC | SNHG1 | 0.564401 | 7.61E-33 | positive |
| HNRNPC | AC099850.3 | 0.559551 | 3.38E-32 | positive |
| HNRNPC | LINC00909 | 0.554289 | 1.66E-31 | positive |
| HNRNPC | AC007406.4 | 0.539936 | 1.10E-29 | positive |
| HNRNPC | AC005288.1 | 0.529579 | 2.01E-28 | positive |
| HNRNPC | LINC00205 | 0.526993 | 4.08E-28 | positive |
| HNRNPC | MCM3AP-AS1 | 0.526508 | 4.66E-28 | positive |
| HNRNPC | NRAV | 0.525897 | 5.50E-28 | positive |
| HNRNPC | TRAF3IP2-AS1 | 0.512055 | 2.19E-26 | positive |
| HNRNPC | LINC00641 | 0.507041 | 7.98E-26 | positive |
| HNRNPC | AL078644.1 | 0.502616 | 2.46E-25 | positive |
| HNRNPC | PTOV1-AS1 | 0.502459 | 2.55E-25 | positive |
| HNRNPC | AC093227.1 | 0.500813 | 3.86E-25 | positive |
| HNRNPC | WARS2-AS1 | 0.500654 | 4.02E-25 | positive |
| HNRNPA2B1 | AC091057.1 | 0.697174 | 1.01E-55 | positive |
| HNRNPA2B1 | AC099850.3 | 0.641693 | 8.77E-45 | positive |
| HNRNPA2B1 | MCM3AP-AS1 | 0.610744 | 1.32E-39 | positive |
| HNRNPA2B1 | TRAF3IP2-AS1 | 0.5818 | 2.94E-35 | positive |
| HNRNPA2B1 | LINC00205 | 0.576077 | 1.90E-34 | positive |
| HNRNPA2B1 | AL355488.1 | 0.571513 | 8.18E-34 | positive |
| HNRNPA2B1 | PTOV1-AS1 | 0.567864 | 2.59E-33 | positive |
| HNRNPA2B1 | LINC00909 | 0.565523 | 5.37E-33 | positive |
| HNRNPA2B1 | ZNF32-AS2 | 0.561768 | 1.71E-32 | positive |
| HNRNPA2B1 | C2orf49-DT | 0.559212 | 3.74E-32 | positive |
| HNRNPA2B1 | AC005288.1 | 0.558661 | 4.43E-32 | positive |
| HNRNPA2B1 | AC007406.4 | 0.55855 | 4.58E-32 | positive |
| HNRNPA2B1 | HCG18 | 0.558452 | 4.72E-32 | positive |
| HNRNPA2B1 | NCK1-DT | 0.554035 | 1.79E-31 | positive |
| HNRNPA2B1 | AC018690.1 | 0.547897 | 1.10E-30 | positive |
| HNRNPA2B1 | WARS2-AS1 | 0.539057 | 1.41E-29 | positive |
| HNRNPA2B1 | LENG8-AS1 | 0.535455 | 3.91E-29 | positive |
| HNRNPA2B1 | AC025176.1 | 0.53232 | 9.39E-29 | positive |
| HNRNPA2B1 | LINC00630 | 0.530991 | 1.36E-28 | positive |
| HNRNPA2B1 | TMCO1-AS1 | 0.529289 | 2.17E-28 | positive |
| HNRNPA2B1 | SNHG21 | 0.527344 | 3.71E-28 | positive |
| HNRNPA2B1 | WAC-AS1 | 0.526861 | 4.23E-28 | positive |
| HNRNPA2B1 | AL050341.2 | 0.526771 | 4.33E-28 | positive |
| HNRNPA2B1 | SNHG1 | 0.526728 | 4.39E-28 | positive |
| HNRNPA2B1 | FAM111A-DT | 0.52572 | 5.77E-28 | positive |
| HNRNPA2B1 | RNF216P1 | 0.521357 | 1.88E-27 | positive |
| HNRNPA2B1 | AC125257.1 | 0.520696 | 2.24E-27 | positive |
| HNRNPA2B1 | AC102953.2 | 0.520271 | 2.51E-27 | positive |
| HNRNPA2B1 | SNHG4 | 0.517178 | 5.71E-27 | positive |
| HNRNPA2B1 | CAPN10-DT | 0.516675 | 6.52E-27 | positive |
| HNRNPA2B1 | AL031714.1 | 0.516285 | 7.23E-27 | positive |
| HNRNPA2B1 | DDX11-AS1 | 0.51562 | 8.62E-27 | positive |
| HNRNPA2B1 | AC093227.1 | 0.514662 | 1.11E-26 | positive |
| HNRNPA2B1 | AC012467.2 | 0.513989 | 1.32E-26 | positive |
| HNRNPA2B1 | THAP9-AS1 | 0.512563 | 1.92E-26 | positive |
| HNRNPA2B1 | ADNP-AS1 | 0.51172 | 2.39E-26 | positive |
| HNRNPA2B1 | LINC00294 | 0.5112 | 2.73E-26 | positive |
| HNRNPA2B1 | PTOV1-AS2 | 0.510187 | 3.55E-26 | positive |
| HNRNPA2B1 | UBE2Q1-AS1 | 0.508405 | 5.63E-26 | positive |
| HNRNPA2B1 | DLEU2 | 0.50722 | 7.63E-26 | positive |
| HNRNPA2B1 | AC073842.2 | 0.506719 | 8.67E-26 | positive |
| HNRNPA2B1 | AP005136.3 | 0.506053 | 1.03E-25 | positive |
| HNRNPA2B1 | AL031985.3 | 0.504822 | 1.41E-25 | positive |
| HNRNPA2B1 | AC092910.3 | 0.500986 | 3.70E-25 | positive |
| FTO | AC005288.1 | 0.536076 | 3.28E-29 | positive |
| FMR1 | FMR1-IT1 | 0.553239 | 2.27E-31 | positive |
Figure 1.
Sankey diagram of m6A and lncRNA. All m6A genes are shown above, and all m6a-related lncRNAs in HCC are shown below. The line indicates that there is the correlation between the two.
4.2. Construct a prognostic model
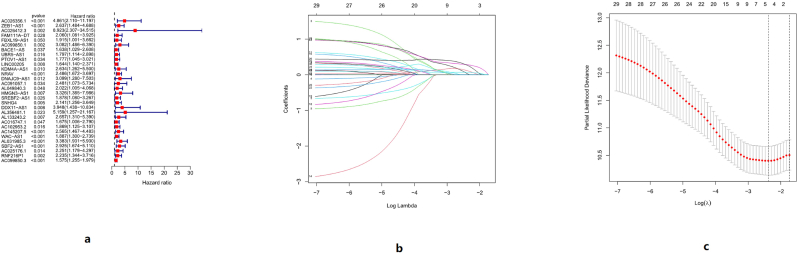
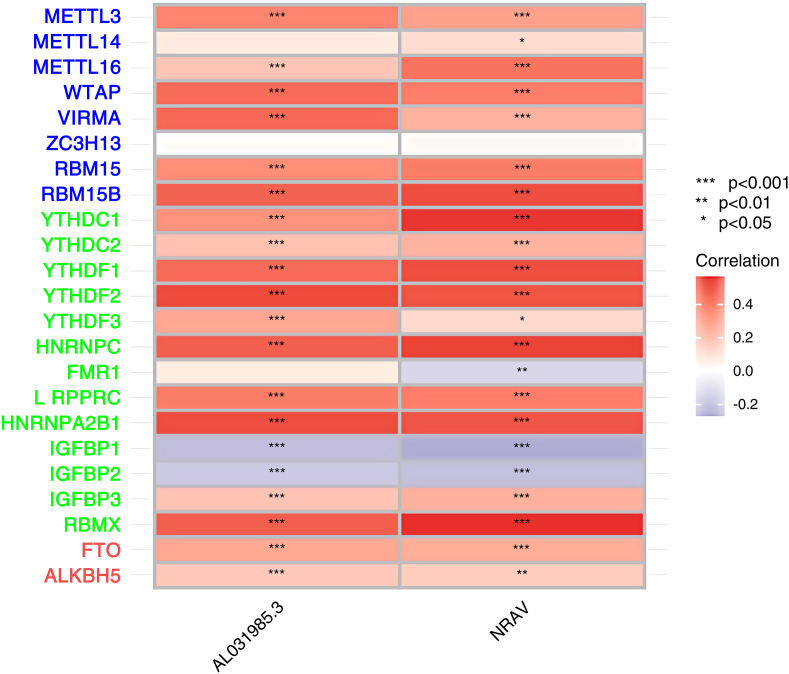
Cox regression analysis was performed on the m6A-related lncRNA on HCC, and the results were shown in Figure 2. Univariate Cox regression analysis of m6A-related lncRNAs showed that the HR of AL031985.3 was 3.383 [1.931, 5.930], and the HR of NRAV was 2.468, [1.672, 3.697]. After univariate Cox regression-related lncRNAs were incorporated into the prognostic model and optimized, the included lncRNAs were NRAV and AL031985.3. The correlation between m6A-related genes and two lncRNAs was shown in Figure 3. ZC3H13 had no correlation with the two lncRNAs, FMR1 had no correlation with AL031985.3, and was negatively correlated with NRAV. IGFBP1 and IGFBP2 were negatively correlated with the two lncRNAs, while the remaining genes showed positive correlations. The risk score of HCC prognosis constructed by m6A-related lncRNA was: Risk score = 0.706559118591986∗ NRAV+ 0.766983203593444∗ AL031985.3.
Figure 2.
a: COX regression analysis was performed on the influence of m6A-related lncRNAs on HCC. b: Lasso regression of m6A-related lncRNAs. c: Cross-validation results of prediction models.
Figure 3.
Heatmap of correlations between lncRNAs (NRAV, AL031985.3) and m6A-related genes in HCC. Among the m6A-related genes, blue represents “writers”, green represents “Reader”, and red represents “erasers”. In the correlation heatmap, red represents a positive correlation between lncRNAs and m6A-related genes, and blue represents a negative correlation. The darker the color, the stronger the correlation. “∗” means Pvalue<0.05, “∗∗” means Pvalue<0.01, “∗∗∗” means Pvalue<0.001.
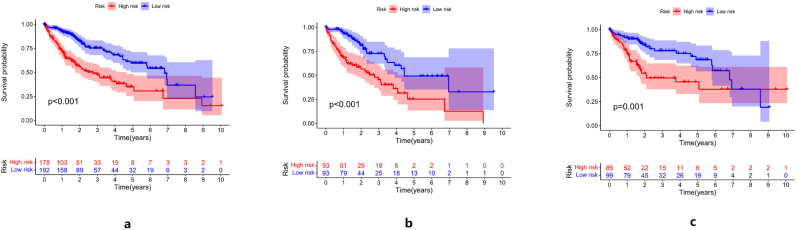
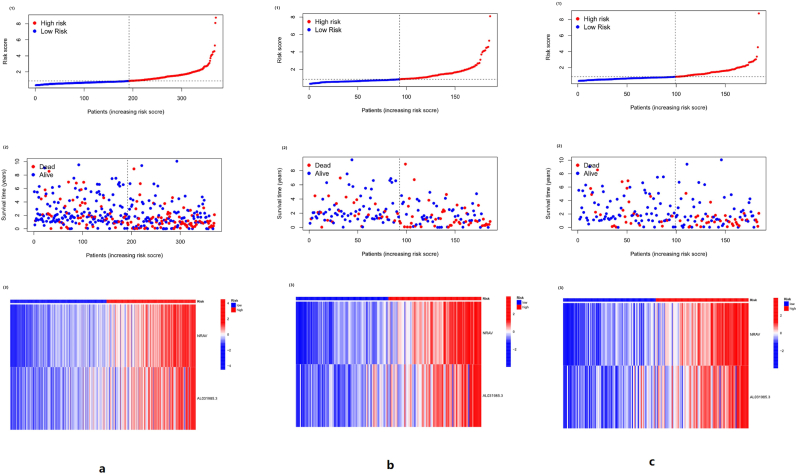
In the survival curves of all sample group, training group and text group, the overall survival time of the low-risk group was longer than that of the high-risk group, as shown in Figure 4. The risk curve of the prediction model can be seen in Figure 5. No matter in the whole sample group, training group, or text group, with the increase in risk score, the death of patients increased, and the expression of NRAV and AL031985.3 increased.
Figure 4.
a: the survival curve of the whole sample group, b: the survival curve of the training group, c: the survival curve of the text group. The high-risk group and the low-risk group are divided by the median value of the risk score, with red representing the survival curve of the high-risk group and blue representing the survival curve of the low-risk group. Among the three groups, the high-risk group had a worse prognosis than the low-risk group.
Figure 5.
The risk curve of the three groups. In Figures 5a, (1) and (2) are the same in abscissa. They are the samples arranged according to the risk score from small to large, and the ordinate is the risk score of the samples. The patient risk score increases from left to right, with the median risk score dividing patients into high-risk (red) and low-risk (blue) groups. (2) The far point in red represents the death of the patient, the blue represents the survival of the patient, and the ordinate represents the survival time of the patient. As the patient's risk score increases, the number of patients who died increases. (3) Red represents the high expression of lncRNA, and blue represents low expression. In the high-risk group, both NRAV and AL031985.3 were highly expressed. The same trend as Figure 5a can be seen in Figure 5b and Figure 5c. As the risk score increased, the number of deaths increased, and NRAV and AL031985.3 were highly expressed in the high-risk group.
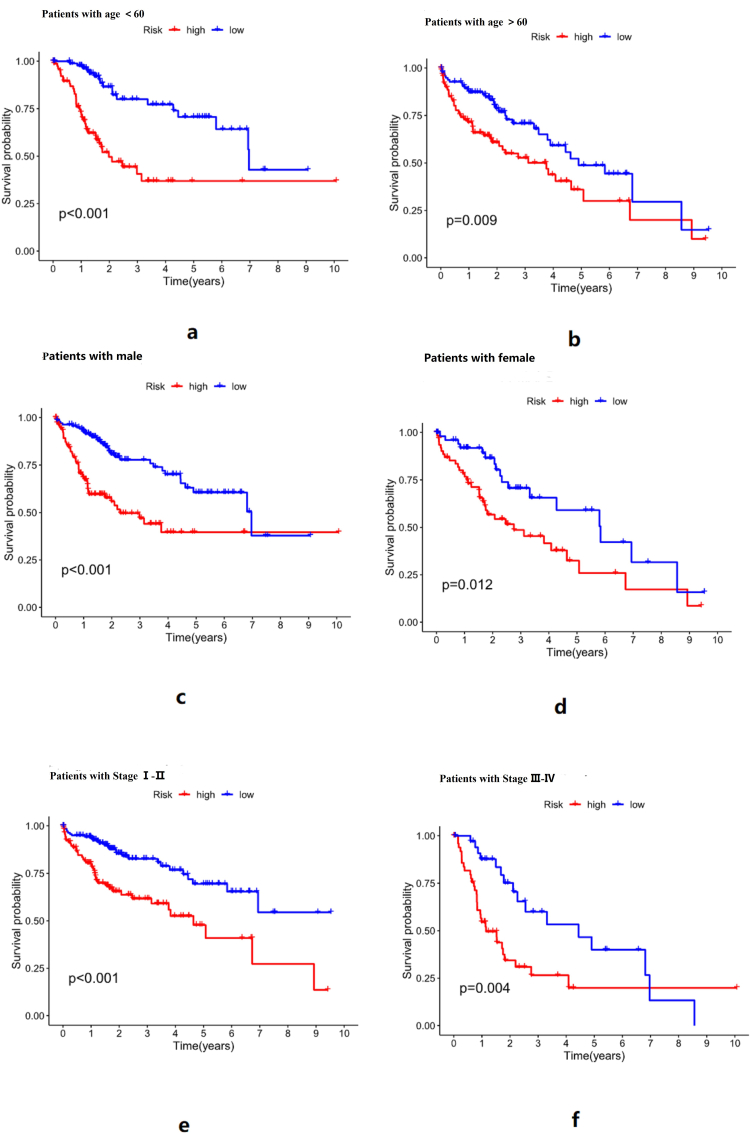
In subgroups of age, gender, and tumor stage, overall survival was longer in the low-risk group than in the high-risk group, as shown in Figure 6. The chi-square test of clinical data between groups can be seen in Table 2, Pvalue >0.05 in each group, so there is no difference in age, gender, grade, and stage among all groups.
Figure 6.
a: Survival curves of the high-risk and low-risk groups in the age group <60 years, b: Survival curves of the high-risk and low-risk groups in the age group>60 years, c: Survival curves of the high-risk and low-risk groups in the male group, d: Survival curves of the high-risk and low-risk groups in the female group, e: Survival curves of the high-risk and low-risk groups in the stage Ⅰ-Ⅱ group, f: Survival curves of the high-risk and low-risk groups in the stage Ⅲ-Ⅳ group. It can be seen from the above survival curve results of different groups that the risk score has a good predictive effect on the survival of HCC patients of different genders, ages, tumor stages, and grades.
Table 2.
The comparison of clinical data between train group and text group.
| Covariates | Type | Total | Test | Train | Pvalue |
|---|---|---|---|---|---|
| Age | ≤65 | 232(62.7%) | 116(63.04%) | 116(62.37%) | 0.9782 |
| Age | >65 | 138(37.3%) | 68(36.96%) | 70(37.63%) | |
| Gender | FEMALE | 121(32.7%) | 69(37.5%) | 52(27.96%) | 0.065 |
| Gender | MALE | 249(67.3%) | 115(62.5%) | 134(72.04%) | |
| Grade | G1 | 55(14.86%) | 21(11.41%) | 34(18.28%) | 0.3224 |
| Grade | G2 | 177(47.84%) | 91(49.46%) | 86(46.24%) | |
| Grade | G3 | 121(32.7%) | 61(33.15%) | 60(32.26%) | |
| Grade | G4 | 12(3.24%) | 7(3.8%) | 5(2.69%) | |
| Grade | unknown | 5(1.35%) | 4(2.17%) | 1(0.54%) | |
| Stage | Stage I | 171(46.22%) | 86(46.74%) | 85(45.7%) | 0.9747 |
| Stage | Stage II | 85(22.97%) | 42(22.83%) | 43(23.12%) | |
| Stage | Stage III | 85(22.97%) | 43(23.37%) | 42(22.58%) | |
| Stage | Stage IV | 5(1.35%) | 3(1.63%) | 2(1.08%) | |
| Stage | unknown | 24(6.49%) | 10(5.43%) | 14(7.53%) | |
| T | T1 | 181(48.92%) | 91(49.46%) | 90(48.39%) | 0.7669 |
| T | T2 | 93(25.14%) | 44(23.91%) | 49(26.34%) | |
| T | T3 | 80(21.62%) | 42(22.83%) | 38(20.43%) | |
| T | T4 | 13(3.51%) | 5(2.72%) | 8(4.3%) | |
| T | unknown | 3(0.81%) | 2(1.09%) | 1(0.54%) | |
| M | M0 | 266(71.89%) | 136(73.91%) | 130(69.89%) | 1 |
| M | M1 | 4(1.08%) | 2(1.09%) | 2(1.08%) | |
| M | unknown | 100(27.03%) | 46(25%) | 54(29.03%) | |
| N | N0 | 252(68.11%) | 124(67.39%) | 128(68.82%) | 0.6033 |
| N | N1 | 4(1.08%) | 3(1.63%) | 1(0.54%) | |
| N | unknown | 114(30.81%) | 57(30.98%) | 57(30.65%) |
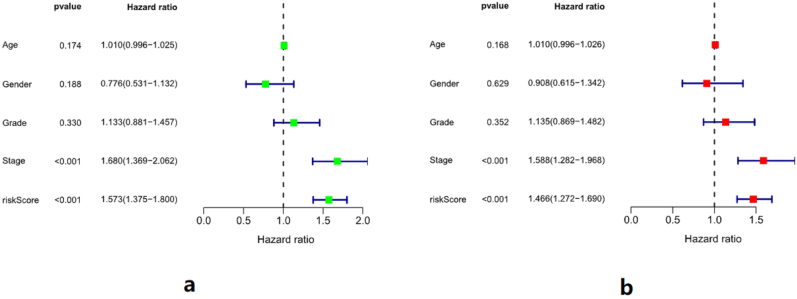
In univariate analysis, Hazard ratio (HR) of the risk score was 1.573, 95%IC [1.375,1.8], P < 0.001, and in multivariate analysis, HR of the risk score was 1.466, 95%IC: [1.272,1.169], P < 0.001. Univariate and multivariate analyses of the risk score showed that the risk score was an independent predictor of HCC prognosis (Figure 7).
Figure 7.
Independent prognostic analysis of risk scores. a: In univariate analysis, Hazard ratio (HR) of the prediction model was 1.573, 95%IC [1.375,1.8], P < 0.001. b: In multivariate analysis, HR of the prediction model was 1.466, 95%IC: [1.272,1.169], P < 0.001. Therefore, the risk score is an independent predictor of prognosis in HCC patients.
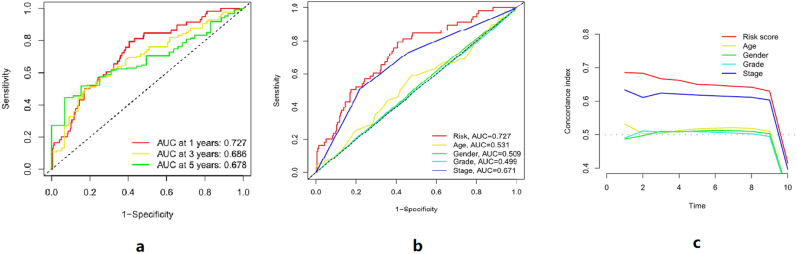
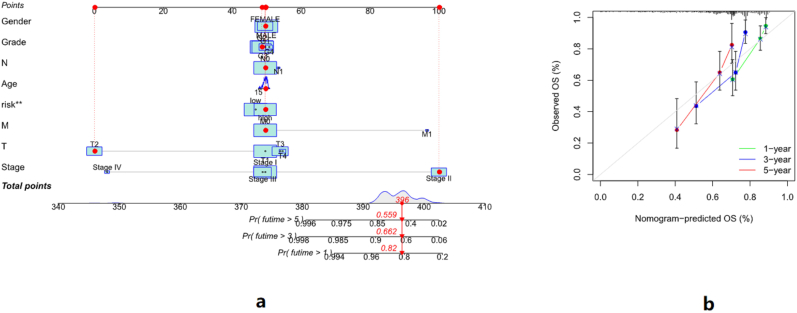
The areas under the ROC curve for predicting 1-year, 3-year, and 5-year survival of patients with HCC by risk score were 0.727, 0.686, and 0.678, respectively. This shows that the risk score we constructed has a better prediction effect on the survival of HCC patients. To further verify the prediction effect of the risk score, we compared the prediction effect of the risk score with age, gender, tumor stage, and grade. The results showed that the area under the ROC curve of the risk score was 0.727, which was significantly better than age, sex, tumor stage, and grade. Besides that, the C-index of risk score was also significantly better than age, sex, tumor stage, and grade (Figure 8). The above results indicate that risk score can be used as an effective method for prognosis prediction of HCC patients.
Figure 8.
a: The area under the ROC curve of 1 year, 3 years, and 5 years. The area under the ROC curve of the risk score for predicting 1-year survival of HCC patients was 0.727, that for predicting 3-year survival was 0.686, and that for predicting 5-year survival was 0.678. b: The area under the ROC curve of the risk score, age, gender, grade, and stage, c: The C-index curve of risk score, age, gender, grade, and stage. As can be seen from Figure 8b and Figure 8c, the risk score can better predict the prognosis of HCC patients.
Based on the risk score constructed by lncRNAs, we drew the nomogram (Figure 9a). We took the 15th sample as an example. The patient had a total score of 396 for all items, and the probability of survival over 1 year was 82%, the probability of survival over 3 years was 66.2%, and the probability of survival over 5 years was 55.9%. The results of the correction curve of the nomogram also showed that the nomogram had a better prediction effect on the 1-, 3-, and 5-year survival rates of HCC patients (Figure 9b).
Figure 9.
a: The nomogram of the risk score. We take the 15th sample as an example, with a total score of 396 points for each item. Therefore, the probability of his survival over 1 year is 82%, the probability of his survival over 3 years is 66.2%, and the probability of his survival over 5 years is 55.9%. b: Calibration curves for risk scores predicting patients at 1, 3, and 5 years.
4.3. Principal component analysis
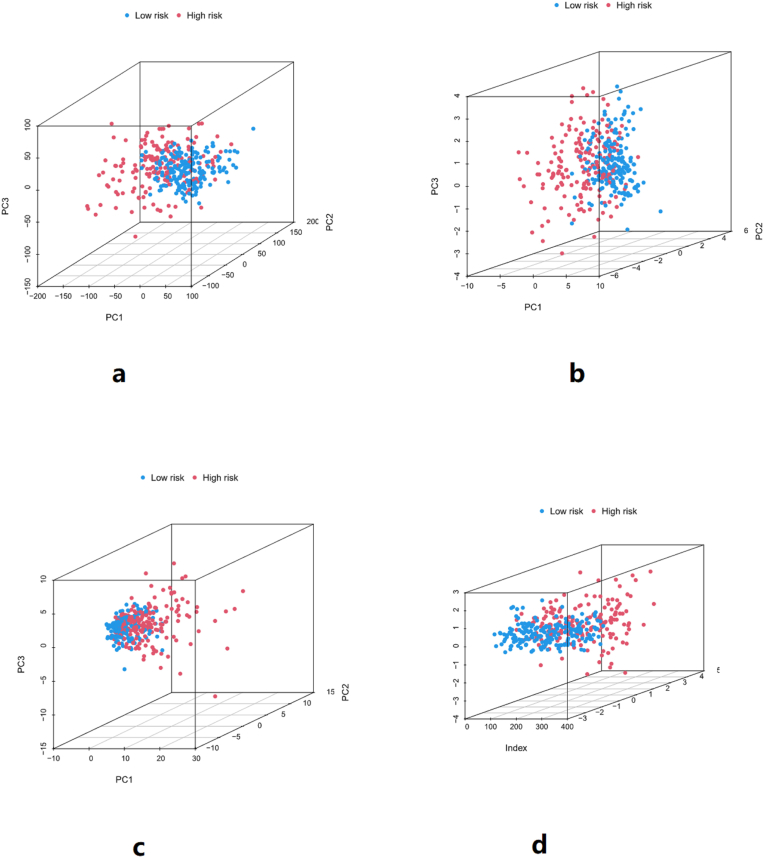
Principal component analysis (PCA) was performed on the whole gene, m6A genes, m6A-related lncRNAs, and lncRNAs used for risk scores construction, respectively, as shown in Figure 10. The results showed that compared with m6A genes, m6A-related lncRNAs, and other genes, the lncRNAs used for risk score construction had the best discrimination between high-risk group and low-risk group samples. This means that the lncRNAs used to construct the risk score can well distinguish samples from the high-risk group and the low-risk group.
Figure 10.
a: PCA of the whole gene, b: PCA of m6A genes, c: PCA of m6A-related lncRNAs, d: PCA of lncRNAs used for risk scores construction. The results showed that compared with m6A genes, m6A-related lncRNAs, and other genes, the lncRNAs used for risk score construction had the best discrimination between high-risk group and low-risk group samples. This means that the lncRNAs used to construct the risk score can well distinguish samples from the high-risk group and the low-risk group.
4.4. Risk differential analysis and GO enrichment analysis
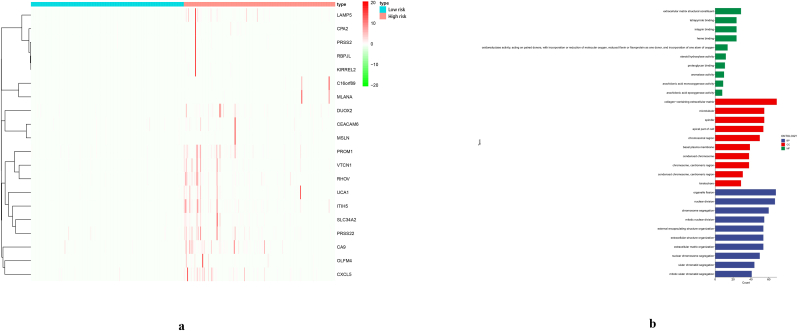
Genes that differ between high-risk and low-risk groups were shown in Table 3. Differential genes were highly expressed in the high-risk group (Figure 11a). The GO enrichment analysis of the differential genes showed that the differential genes were mainly located in the extracellular matrix and chromosomal regions, and may play regulatory roles in binding sites and catalytic enzymes, thereby affecting the chromosome division and cell proliferation of cells (Figure 11b).
Table 3.
Genes that differ between high-risk and low-risk groups.
| gene | lowMean | highMean | logFC | pValue | fdr |
|---|---|---|---|---|---|
| SOGA1 | 0.762441385 | 1.824778388 | 1.259022929 | 4.72E-23 | 1.93E-21 |
| CYS1 | 0.278046091 | 2.053921639 | 2.88498518 | 0.033887537 | 0.0408347 |
| CTNND2 | 0.577150807 | 3.06400425 | 2.408398055 | 1.86E-07 | 4.03E-07 |
| C4orf48 | 2.310128195 | 5.155930001 | 1.158259764 | 0.00010396 | 0.0001662 |
| SOX12 | 4.084522532 | 8.348446134 | 1.031340257 | 1.47E-15 | 1.01E-14 |
| AL662795.2 | 0.748358071 | 1.565414701 | 1.064744264 | 6.32E-17 | 5.75E-16 |
| FTCD | 115.976255 | 54.82683524 | -1.08087535 | 1.10E-19 | 1.81E-18 |
| RHOV | 0.146416731 | 2.746931604 | 4.229668667 | 1.41E-11 | 5.17E-11 |
| CDCA8 | 1.959628819 | 6.02557972 | 1.620519636 | 3.72E-25 | 3.14E-23 |
| CYP4F2 | 39.10312697 | 18.70670726 | -1.06372834 | 1.73E-15 | 1.18E-14 |
| NPTX2 | 2.192038303 | 8.332143897 | 1.926414747 | 6.22E-05 | 0.0001019 |
| OLFML3 | 2.444922906 | 6.730261952 | 1.460873683 | 1.82E-06 | 3.55E-06 |
| AC137723.1 | 1.625261897 | 0.633187447 | -1.35996766 | 1.47E-10 | 4.68E-10 |
| RNF157 | 1.597150798 | 3.241197133 | 1.021026235 | 1.96E-07 | 4.24E-07 |
| RMI2 | 1.863554698 | 3.761635296 | 1.013302816 | 1.82E-16 | 1.49E-15 |
| RHNO1 | 3.221034348 | 6.507791967 | 1.014644089 | 2.18E-27 | 4.63E-25 |
| IFT57 | 1.434889536 | 3.045737501 | 1.085851931 | 6.87E-13 | 3.02E-12 |
| DYRK2 | 1.242583961 | 2.579993887 | 1.05402431 | 3.39E-23 | 1.45E-21 |
| IL2RG | 4.706857184 | 10.81368313 | 1.200022002 | 2.51E-06 | 4.82E-06 |
| AP000769.1 | 0.909100227 | 2.172440052 | 1.256805103 | 6.69E-10 | 1.95E-09 |
| SCRN1 | 0.910306529 | 3.704860497 | 2.024994888 | 1.23E-10 | 3.95E-10 |
| CD24 | 25.24229194 | 71.66716996 | 1.505469476 | 9.37E-14 | 4.73E-13 |
| CSGALNACT1 | 0.841990382 | 2.007520658 | 1.253539175 | 0.002754124 | 0.0037626 |
| FZD7 | 0.475086959 | 1.868792166 | 1.975842621 | 2.95E-14 | 1.62E-13 |
| CSPP1 | 1.310948933 | 2.81857042 | 1.104352125 | 1.15E-14 | 6.76E-14 |
| PPP1R14D | 0.713302608 | 1.799713889 | 1.335181418 | 4.72E-07 | 9.79E-07 |
| FAM133A | 0.608962332 | 1.742805546 | 1.516986712 | 1.77E-06 | 3.45E-06 |
| SOX9 | 5.212905911 | 15.20305852 | 1.544201866 | 1.09E-12 | 4.68E-12 |
| MRC2 | 1.545081071 | 4.085646627 | 1.40288189 | 8.57E-05 | 0.0001382 |
| CD44 | 3.564727589 | 8.493284304 | 1.252530711 | 1.77E-05 | 3.07E-05 |
| TUBA1A | 5.82656021 | 11.92201192 | 1.032911396 | 1.03E-05 | 1.84E-05 |
| SPATS2 | 1.658761467 | 3.811684859 | 1.200322406 | 3.07E-34 | 9.14E-31 |
| CXCL5 | 0.367517406 | 6.772840097 | 4.20387645 | 1.71E-09 | 4.74E-09 |
| P3H4 | 2.466147173 | 5.80461829 | 1.234942301 | 1.13E-15 | 7.96E-15 |
| BACE2 | 1.650550936 | 7.986519449 | 2.274619248 | 3.95E-08 | 9.24E-08 |
| ANKRD10-IT1 | 1.49081584 | 3.09610523 | 1.054352454 | 3.62E-11 | 1.26E-10 |
| ABCC1 | 1.061173667 | 3.843910247 | 1.85691387 | 5.95E-18 | 6.73E-17 |
| CYP4A11 | 99.73993045 | 43.74599058 | -1.1890204 | 7.55E-18 | 8.26E-17 |
| IDO1 | 0.807892825 | 1.62669382 | 1.009706906 | 0.012429087 | 0.0158238 |
| RHPN1 | 1.13536993 | 2.778093943 | 1.290932949 | 1.94E-11 | 7.01E-11 |
| PDCD1 | 0.654982552 | 1.966524444 | 1.586119738 | 1.11E-05 | 1.97E-05 |
| CFTR | 0.231759152 | 3.99978452 | 4.109224061 | 6.93E-05 | 0.000113 |
| NDC80 | 1.421201834 | 3.521709558 | 1.309164477 | 1.97E-17 | 1.98E-16 |
| CRYAB | 4.817038411 | 11.36279722 | 1.238099699 | 0.017037426 | 0.0213031 |
| IER5L | 2.234043739 | 5.44235659 | 1.284574055 | 1.57E-12 | 6.59E-12 |
| CYP2C8 | 141.833323 | 57.30456804 | -1.30747447 | 7.27E-12 | 2.77E-11 |
| SPRED1 | 0.931632471 | 2.361691086 | 1.34198744 | 1.33E-18 | 1.74E-17 |
| CBX2 | 0.509733628 | 1.663612838 | 1.706504287 | 1.58E-16 | 1.31E-15 |
| EPO | 0.988229782 | 4.975480087 | 2.331917299 | 8.62E-11 | 2.83E-10 |
| PITX1 | 0.654483775 | 2.906078947 | 2.150644563 | 6.62E-12 | 2.53E-11 |
| ALOX5 | 1.094513511 | 3.41969985 | 1.64357994 | 1.02E-09 | 2.91E-09 |
| MCUB | 0.732285309 | 1.841648823 | 1.330520227 | 1.52E-10 | 4.82E-10 |
| ANG | 369.2250878 | 177.8023201 | -1.05422643 | 4.14E-17 | 3.87E-16 |
| MUC13 | 27.40074782 | 56.08700444 | 1.033451264 | 0.001610715 | 0.0022677 |
| FXYD2 | 1.912891571 | 7.283101564 | 1.928797865 | 9.30E-06 | 1.67E-05 |
| AC009005.1 | 0.92748876 | 1.906386183 | 1.039438697 | 2.24E-11 | 8.00E-11 |
| COL1A2 | 14.90655905 | 38.58812017 | 1.372209492 | 8.31E-06 | 1.50E-05 |
| AC016885.3 | 0.741704892 | 1.667776384 | 1.169008675 | 0.004262725 | 0.0057043 |
| PLXNA3 | 0.876452565 | 2.027489136 | 1.209946268 | 1.43E-14 | 8.30E-14 |
| MMP7 | 2.727885941 | 13.34629389 | 2.290583949 | 2.27E-06 | 4.37E-06 |
| CXCL8 | 4.109531976 | 11.95328047 | 1.540360604 | 4.06E-08 | 9.48E-08 |
| TTC22 | 0.908201636 | 1.863023412 | 1.036561264 | 2.19E-10 | 6.81E-10 |
| TINAG | 1.619376624 | 3.379565451 | 1.061399197 | 0.001255914 | 0.0017894 |
| NAA40 | 1.5047136 | 3.323470003 | 1.143201415 | 1.23E-30 | 1.04E-27 |
| ZDHHC1 | 0.945212521 | 2.14730752 | 1.183818171 | 0.000907024 | 0.0013155 |
| ANTXR1 | 1.504406927 | 3.163700235 | 1.072418054 | 0.001112216 | 0.0015963 |
| NBL1 | 1.214368075 | 3.352736791 | 1.465133459 | 0.000275858 | 0.0004226 |
| NCEH1 | 1.048580674 | 4.761677062 | 2.183031918 | 2.95E-16 | 2.30E-15 |
| CRYBB1 | 0.292442019 | 4.467851366 | 3.933358669 | 7.81E-05 | 0.0001265 |
| BCL9L | 1.49257711 | 3.124001458 | 1.065589661 | 3.02E-12 | 1.22E-11 |
| ITGAM | 0.98673375 | 2.654476031 | 1.427694354 | 4.33E-11 | 1.49E-10 |
| KCNE4 | 0.711908812 | 1.936080485 | 1.443374564 | 2.13E-05 | 3.67E-05 |
| CDKN2A | 2.503256906 | 5.829550124 | 1.219578192 | 3.16E-09 | 8.50E-09 |
| S100A11 | 52.02634995 | 186.4531412 | 1.841498703 | 5.05E-13 | 2.27E-12 |
| BHMT | 123.112429 | 54.03195865 | -1.18809153 | 4.22E-13 | 1.92E-12 |
| MGAT5 | 1.844473089 | 4.07332581 | 1.142998475 | 2.42E-15 | 1.60E-14 |
| MARCKSL1 | 28.67798548 | 59.56908508 | 1.054620117 | 5.20E-13 | 2.33E-12 |
| LAMP5 | 0.118305757 | 2.825986048 | 4.578162157 | 7.16E-10 | 2.08E-09 |
| CSF1 | 3.897025635 | 9.406648984 | 1.271307448 | 2.89E-14 | 1.58E-13 |
| PAGE2 | 5.166092308 | 11.79718005 | 1.191296718 | 5.74E-05 | 9.44E-05 |
| PNMA1 | 3.142605462 | 7.946675346 | 1.338390245 | 1.26E-22 | 4.40E-21 |
| MUC1 | 0.364885758 | 3.933870129 | 3.430432585 | 2.49E-08 | 5.98E-08 |
| ARL4C | 3.434925189 | 9.020185591 | 1.392878439 | 1.38E-12 | 5.86E-12 |
| LAMA5 | 2.996030302 | 7.651097661 | 1.352614523 | 4.52E-15 | 2.84E-14 |
| ADGRD1 | 0.680772244 | 1.551612724 | 1.188524389 | 0.000222974 | 0.0003445 |
| CEACAM6 | 0.130061269 | 3.876690804 | 4.897562362 | 0.017010877 | 0.0212766 |
| NFE2L3 | 1.010001383 | 3.307987133 | 1.711596354 | 1.47E-19 | 2.33E-18 |
| TREM2 | 3.421131511 | 7.52300552 | 1.136835585 | 7.00E-09 | 1.80E-08 |
| PTAFR | 0.837020261 | 2.100965963 | 1.32771834 | 6.78E-08 | 1.55E-07 |
| RBL1 | 0.622736543 | 1.509901489 | 1.277760579 | 1.21E-25 | 1.28E-23 |
| CASC9 | 1.504257938 | 3.267509472 | 1.119139448 | 9.73E-05 | 0.0001561 |
| SPINT1-AS1 | 0.63408306 | 1.656854973 | 1.385703588 | 9.38E-07 | 1.88E-06 |
| SLC2A5 | 0.92351958 | 2.20700133 | 1.256873045 | 9.24E-09 | 2.34E-08 |
| AC245100.3 | 7.308543701 | 23.46461986 | 1.68283122 | 0.002155181 | 0.0029865 |
| NCAPH | 1.031887546 | 3.132277879 | 1.60192645 | 2.58E-23 | 1.16E-21 |
| MAEL | 0.564197602 | 1.752532919 | 1.635169104 | 1.59E-05 | 2.79E-05 |
| MEX3A | 0.58633949 | 1.927560044 | 1.71696767 | 8.36E-16 | 6.01E-15 |
| PCSK1 | 1.106614147 | 3.651671457 | 1.722404699 | 0.000384745 | 0.0005807 |
| SUSD1 | 0.849965003 | 1.765125976 | 1.054295805 | 1.31E-17 | 1.35E-16 |
| TMIE | 0.76224025 | 1.703827402 | 1.1604615 | 0.000149059 | 0.0002345 |
| SFRP2 | 0.37720339 | 1.833153558 | 2.280913092 | 0.01709177 | 0.0213688 |
| TNC | 1.319199686 | 4.249673981 | 1.687689207 | 0.000101086 | 0.0001619 |
| UPP2 | 5.137791942 | 1.85707062 | -1.46811979 | 2.80E-10 | 8.57E-10 |
| LMNB1 | 4.297094176 | 10.30835286 | 1.262380526 | 1.08E-21 | 2.90E-20 |
| TPPP3 | 1.600259314 | 3.705888837 | 1.211513901 | 0.000532583 | 0.0007916 |
| MARCKS | 14.25561745 | 30.13037156 | 1.079687936 | 3.34E-17 | 3.18E-16 |
| PRAME | 1.078533735 | 5.188271469 | 2.266182665 | 5.72E-09 | 1.49E-08 |
| PLBD1 | 1.535586868 | 5.514195996 | 1.84436042 | 1.27E-13 | 6.26E-13 |
| HAUS6 | 0.956745937 | 2.05700371 | 1.104336621 | 1.10E-25 | 1.21E-23 |
| STK39 | 1.675403225 | 4.629913925 | 1.466477017 | 1.33E-14 | 7.77E-14 |
| ETNPPL | 30.70566778 | 11.90737614 | -1.36664944 | 3.93E-12 | 1.55E-11 |
| DCAF16 | 1.776379287 | 4.022862323 | 1.17928271 | 3.87E-29 | 1.92E-26 |
| PLAUR | 0.893045845 | 2.909205969 | 1.703819297 | 1.77E-14 | 1.01E-13 |
| CYBB | 2.457813617 | 6.034106715 | 1.295764695 | 7.56E-07 | 1.53E-06 |
| CIDEC | 1.535843994 | 3.402754763 | 1.1476715 | 0.000369288 | 0.0005584 |
| COL6A3 | 2.651730971 | 6.612948247 | 1.318359196 | 8.01E-06 | 1.45E-05 |
| TMC4 | 3.739501577 | 10.23691171 | 1.452862649 | 0.034143993 | 0.0411229 |
| TUBA3C | 0.724125494 | 4.895085467 | 2.757022399 | 0.000280582 | 0.0004295 |
| CHST3 | 0.524242128 | 1.60205389 | 1.611617479 | 0.000114407 | 0.0001821 |
| MIR3189 | 0.994954027 | 2.066691879 | 1.054621544 | 0.000623817 | 0.0009199 |
| INAVA | 0.825246571 | 2.279663829 | 1.465923949 | 9.64E-14 | 4.85E-13 |
| LINC02768 | 1.996761296 | 0.867513644 | -1.20270352 | 1.30E-10 | 4.16E-10 |
| LINC00665 | 0.694312631 | 2.17954562 | 1.650370079 | 2.12E-11 | 7.60E-11 |
| BCL2A1 | 1.017610078 | 2.493149564 | 1.29278457 | 5.87E-06 | 1.08E-05 |
| SPRING1 | 1.580663752 | 4.16176875 | 1.396666302 | 8.52E-28 | 2.20E-25 |
| AC079466.1 | 2.161833365 | 7.295315891 | 1.754715125 | 0.000148131 | 0.0002331 |
| CCNF | 0.91008703 | 2.105970381 | 1.210408727 | 4.70E-21 | 1.07E-19 |
| RASA3 | 1.355274983 | 2.96174721 | 1.127862908 | 8.30E-10 | 2.39E-09 |
| RAI1 | 1.063265537 | 2.377915615 | 1.161195582 | 1.36E-20 | 2.78E-19 |
| MKI67 | 1.452944653 | 4.246575351 | 1.547320101 | 1.15E-22 | 4.04E-21 |
| NUMBL | 0.818716157 | 1.699158373 | 1.053385056 | 1.85E-14 | 1.05E-13 |
| FUT4 | 0.540782043 | 1.868107848 | 1.788458595 | 3.53E-11 | 1.23E-10 |
| PPP1R13L | 2.168525724 | 4.342274085 | 1.001736235 | 9.24E-09 | 2.34E-08 |
| ZIC2 | 1.055950719 | 2.802653293 | 1.408250778 | 9.73E-12 | 3.65E-11 |
| DUOXA2 | 0.476067745 | 5.096430155 | 3.420248261 | 0.000178726 | 0.0002787 |
| CNTRL | 0.68093086 | 1.466969208 | 1.107258366 | 2.84E-20 | 5.27E-19 |
| UGT1A3 | 2.816408087 | 1.363620684 | -1.046414 | 6.52E-06 | 1.19E-05 |
| OIP5 | 1.163328888 | 2.771809111 | 1.252568882 | 3.39E-18 | 4.05E-17 |
| CHRNA4 | 1.883688122 | 0.853278197 | -1.14247203 | 0.000495952 | 0.0007397 |
| SEC14L2 | 34.52708866 | 14.4776505 | -1.2539012 | 4.56E-14 | 2.42E-13 |
| WFDC2 | 2.651178178 | 9.450155931 | 1.833704502 | 0.003609464 | 0.004869 |
| ARHGAP11A | 0.792833721 | 2.136166767 | 1.429934051 | 4.58E-23 | 1.88E-21 |
| ADH4 | 310.2887243 | 141.4941644 | -1.13286872 | 4.65E-10 | 1.38E-09 |
| TM6SF2 | 8.636530557 | 3.986073897 | -1.11548342 | 1.95E-14 | 1.10E-13 |
| ARL6IP6 | 1.271372429 | 2.647399558 | 1.058189242 | 2.24E-20 | 4.25E-19 |
| HMOX1 | 17.90904514 | 38.61737129 | 1.108561543 | 5.95E-08 | 1.36E-07 |
| NCS1 | 1.222775519 | 2.729887382 | 1.158681863 | 3.71E-10 | 1.12E-09 |
| DBN1 | 2.909053687 | 9.760042116 | 1.746337451 | 5.23E-11 | 1.78E-10 |
| CRMP1 | 0.683845294 | 1.519211912 | 1.151581237 | 7.41E-07 | 1.50E-06 |
| APOBEC3C | 2.104884297 | 5.059488479 | 1.265250602 | 4.97E-06 | 9.20E-06 |
| CD109 | 1.041496609 | 2.100820995 | 1.012295097 | 4.92E-06 | 9.12E-06 |
| PROM1 | 0.120128762 | 2.130148215 | 4.1483003 | 1.05E-05 | 1.86E-05 |
| ADGRE5 | 4.623528453 | 9.496789304 | 1.038445579 | 3.35E-09 | 9.00E-09 |
| BIRC5 | 4.133098801 | 10.09889651 | 1.288901904 | 4.19E-18 | 4.90E-17 |
| UNC13D | 0.649607581 | 2.235198362 | 1.782762495 | 1.08E-07 | 2.40E-07 |
| VSIG4 | 2.76585398 | 5.558859787 | 1.007064 | 0.010291586 | 0.0132128 |
| SLC39A10 | 1.058144507 | 2.22103423 | 1.069694963 | 1.37E-19 | 2.19E-18 |
| SLC13A5 | 36.30341246 | 18.11451919 | -1.00295865 | 2.37E-13 | 1.12E-12 |
| CENPA | 0.921740335 | 2.795630206 | 1.600741251 | 2.97E-22 | 9.00E-21 |
| RPP25 | 1.779760066 | 3.746931907 | 1.074026999 | 7.52E-10 | 2.18E-09 |
| DCXR-DT | 7.275206925 | 2.981725281 | -1.28684094 | 7.55E-09 | 1.94E-08 |
| SERPINH1 | 14.20940324 | 31.42917225 | 1.14525831 | 7.96E-14 | 4.06E-13 |
| ZFPM2-AS1 | 0.812098673 | 2.215064465 | 1.44762175 | 1.75E-06 | 3.42E-06 |
| CCND2 | 0.932096126 | 2.460944945 | 1.400661731 | 4.40E-05 | 7.34E-05 |
| GOLM1 | 14.02535343 | 33.26803878 | 1.246099693 | 2.01E-11 | 7.24E-11 |
| SLC1A5 | 3.42422531 | 14.68339004 | 2.100335552 | 3.93E-20 | 7.06E-19 |
| ABR | 0.843665592 | 2.314316583 | 1.455843059 | 3.80E-08 | 8.90E-08 |
| TLCD3A | 1.185360493 | 2.649798278 | 1.160556656 | 1.19E-15 | 8.39E-15 |
| FMO1 | 0.939561762 | 2.382556155 | 1.342450313 | 4.47E-06 | 8.31E-06 |
| SERPINE2 | 1.577624798 | 5.286019533 | 1.744427622 | 2.88E-08 | 6.85E-08 |
| TUBB4A | 1.985751636 | 5.669002405 | 1.51340969 | 4.82E-07 | 9.98E-07 |
| CDK1 | 2.366377086 | 6.398031135 | 1.434948025 | 7.39E-21 | 1.62E-19 |
| KCNK5 | 2.741613018 | 5.834185055 | 1.0895062 | 0.001668011 | 0.0023409 |
| DDR1 | 2.138462895 | 8.647424744 | 2.015696379 | 2.03E-16 | 1.66E-15 |
| C16orf89 | 0.123468345 | 4.583089729 | 5.214107418 | 4.67E-06 | 8.67E-06 |
| TMEM132A | 0.703676427 | 4.720306382 | 2.745896414 | 1.67E-13 | 8.11E-13 |
| CLDN10 | 0.55626587 | 3.604859095 | 2.696096374 | 0.01675755 | 0.0209862 |
| MT2A | 516.2996407 | 182.6353283 | -1.49924273 | 0.00096892 | 0.0014013 |
| EZH2 | 1.794428018 | 4.16935424 | 1.216299901 | 4.12E-24 | 2.54E-22 |
| DTX3 | 1.161957788 | 2.609732906 | 1.167344502 | 1.34E-05 | 2.36E-05 |
| HLA-DQB2 | 2.056099011 | 5.136764138 | 1.320950093 | 8.73E-07 | 1.76E-06 |
| SLC22A1 | 120.2348974 | 40.55850728 | -1.56777923 | 1.91E-14 | 1.08E-13 |
| TTC36 | 19.99192077 | 4.091237301 | -2.28880796 | 3.50E-15 | 2.25E-14 |
| MEP1A | 1.748810821 | 5.45922804 | 1.642322729 | 5.88E-05 | 9.65E-05 |
| ARHGAP33 | 0.822655609 | 1.659503558 | 1.01239122 | 5.85E-18 | 6.62E-17 |
| F13A1 | 1.586746125 | 3.747369646 | 1.239806975 | 4.84E-05 | 8.04E-05 |
| UCA1 | 0.123690551 | 4.733081744 | 5.257972637 | 0.001110218 | 0.0015942 |
| XXYLT1 | 0.792611213 | 1.718810911 | 1.116725558 | 8.22E-20 | 1.39E-18 |
| TPX2 | 4.990859927 | 13.74896077 | 1.461962256 | 1.24E-21 | 3.29E-20 |
| SFRP4 | 1.044703012 | 2.192162494 | 1.069261871 | 0.001205296 | 0.0017214 |
| TMEM165 | 2.67775801 | 5.379646751 | 1.006485853 | 4.93E-24 | 2.91E-22 |
| ADORA2BP1 | 2.808905036 | 1.136344822 | -1.30560717 | 1.48E-10 | 4.71E-10 |
| GPX8 | 0.711548147 | 2.086803637 | 1.552261568 | 2.19E-08 | 5.30E-08 |
| ARL14 | 0.399767559 | 3.439272032 | 3.104869924 | 3.23E-06 | 6.10E-06 |
| MAD2L1 | 0.899762746 | 2.295468521 | 1.351172108 | 4.70E-25 | 3.83E-23 |
| LINC00844 | 32.59893743 | 9.371669622 | -1.79844694 | 9.62E-11 | 3.14E-10 |
| HAVCR2 | 1.188691923 | 2.846952538 | 1.260043585 | 4.58E-08 | 1.06E-07 |
| PAFAH1B3 | 8.039837001 | 20.44194487 | 1.346294305 | 1.42E-16 | 1.20E-15 |
| PLEK | 2.419877075 | 5.401234999 | 1.158355556 | 5.21E-07 | 1.07E-06 |
| ITGB1-DT | 0.724568839 | 2.476247098 | 1.772960618 | 1.05E-07 | 2.35E-07 |
| CHD3 | 2.249726351 | 4.678443698 | 1.056279164 | 1.29E-08 | 3.22E-08 |
| APOC3 | 5547.85486 | 2292.035526 | -1.27530064 | 2.99E-26 | 4.14E-24 |
| AC016735.1 | 1.04813775 | 3.512311889 | 1.744592626 | 1.38E-05 | 2.42E-05 |
| COL5A1 | 3.409245622 | 7.861512209 | 1.205354307 | 3.22E-05 | 5.45E-05 |
| NCAPD2 | 2.895043205 | 7.395360895 | 1.353035674 | 7.07E-26 | 8.95E-24 |
| LTO1 | 1.83379779 | 3.955177724 | 1.108907959 | 2.95E-13 | 1.37E-12 |
| CHIT1 | 0.345042837 | 1.808129652 | 2.389650743 | 0.010714597 | 0.0137263 |
| CDK19 | 1.389641153 | 2.794193882 | 1.007719745 | 5.74E-15 | 3.55E-14 |
| ITGAV | 5.403178116 | 13.06780039 | 1.274136177 | 6.33E-14 | 3.27E-13 |
| CREB3L1 | 0.784379864 | 3.775772772 | 2.26714754 | 0.000176272 | 0.0002751 |
| SLC6A8 | 2.690748065 | 10.15256803 | 1.915765471 | 5.00E-11 | 1.70E-10 |
| LRFN4 | 0.525042518 | 1.6547338 | 1.656092986 | 6.32E-06 | 1.15E-05 |
| HAPLN3 | 0.668856601 | 1.465169161 | 1.131298396 | 9.79E-07 | 1.96E-06 |
| LIF | 0.657630788 | 3.1636277 | 2.266230084 | 6.43E-12 | 2.47E-11 |
| MCM6 | 5.159295547 | 11.57631843 | 1.165930514 | 9.02E-23 | 3.24E-21 |
| RUNX1 | 0.882099282 | 1.802243529 | 1.030781021 | 1.21E-14 | 7.10E-14 |
| FCGBP | 0.362939615 | 2.05203354 | 2.49925287 | 3.62E-12 | 1.44E-11 |
| SLC9A1 | 1.079368145 | 2.523309628 | 1.225130231 | 5.45E-14 | 2.85E-13 |
| AGR2 | 3.323233751 | 21.98725652 | 2.726007924 | 1.94E-05 | 3.37E-05 |
| B3GNT7 | 0.510409195 | 2.072584878 | 2.021704959 | 2.20E-07 | 4.73E-07 |
| VGLL4 | 1.245002262 | 2.64819166 | 1.088859176 | 7.99E-19 | 1.09E-17 |
| SKA1 | 1.069656551 | 3.039736972 | 1.506798847 | 4.40E-19 | 6.34E-18 |
| EMB | 0.703305378 | 1.571828733 | 1.160220876 | 6.36E-05 | 0.0001042 |
| MTF2 | 1.252107445 | 2.566799125 | 1.035612029 | 1.10E-27 | 2.67E-25 |
| PLEKHB1 | 0.421965603 | 3.052130447 | 2.854619319 | 2.36E-10 | 7.31E-10 |
| MEIS2 | 0.973616928 | 2.209390183 | 1.182222067 | 2.15E-10 | 6.69E-10 |
| AC111000.4 | 0.747461623 | 1.531562105 | 1.034932457 | 9.30E-06 | 1.67E-05 |
| AC012065.2 | 15.7593154 | 5.453554886 | -1.53093601 | 1.09E-10 | 3.52E-10 |
| GTSE1 | 0.66432363 | 2.251478484 | 1.760914554 | 1.58E-24 | 1.10E-22 |
| HOXB7 | 0.60623601 | 1.80197447 | 1.571627116 | 0.012395001 | 0.0157821 |
| PRKX | 0.77749003 | 1.775889247 | 1.19164553 | 2.54E-05 | 4.35E-05 |
| DBF4 | 0.886852256 | 1.828913225 | 1.044220941 | 7.76E-26 | 9.42E-24 |
| POF1B | 0.475416688 | 1.867725013 | 1.97401761 | 3.84E-08 | 9.00E-08 |
| PI3 | 3.283544345 | 10.34227382 | 1.655227563 | 0.000123462 | 0.000196 |
| RBPJL | 0.009656708 | 2.504139394 | 8.018567731 | 0.028012913 | 0.0340769 |
| WFDC21P | 1.176705498 | 3.051879881 | 1.374944887 | 0.001878844 | 0.0026247 |
| COL3A1 | 30.50422594 | 80.12475329 | 1.393238887 | 0.000138341 | 0.0002184 |
| ZWINT | 4.728890567 | 11.31832343 | 1.259086608 | 5.92E-21 | 1.33E-19 |
| MNDA | 0.932901964 | 2.035911039 | 1.125877138 | 8.15E-07 | 1.65E-06 |
| BIRC3 | 5.986421013 | 12.3721305 | 1.047328307 | 8.73E-06 | 1.57E-05 |
| KRT19 | 2.549848708 | 27.2336082 | 3.416904585 | 0.000147873 | 0.0002327 |
| OSMR | 2.290136699 | 6.325041649 | 1.465641264 | 2.55E-13 | 1.20E-12 |
| GNMT | 104.497557 | 31.77421135 | -1.71754099 | 3.06E-15 | 2.00E-14 |
| NPAS2 | 1.10908399 | 2.23704005 | 1.012222461 | 5.51E-18 | 6.29E-17 |
| AL161668.4 | 3.406264282 | 1.575286954 | -1.11257572 | 4.45E-10 | 1.33E-09 |
| INCENP | 1.454755728 | 3.060517683 | 1.072998777 | 2.41E-20 | 4.55E-19 |
| THSD4 | 0.726220464 | 1.721366867 | 1.245075115 | 0.0368668 | 0.0442275 |
| MACROH2A2 | 5.94819219 | 12.54651326 | 1.07676332 | 1.29E-08 | 3.20E-08 |
| ISLR | 3.242726836 | 8.936778116 | 1.462547307 | 0.003171214 | 0.0043047 |
| CYP2C9 | 169.0238079 | 61.62565952 | -1.45562339 | 2.11E-17 | 2.10E-16 |
| PLK1 | 1.351478327 | 4.276531245 | 1.661902704 | 2.45E-24 | 1.65E-22 |
| CPA2 | 0.070217762 | 7.424496107 | 6.724313295 | 1.54E-05 | 2.69E-05 |
| ETV4 | 4.298901441 | 8.963634751 | 1.060115829 | 5.70E-10 | 1.68E-09 |
| TPM4 | 12.02328621 | 27.20711003 | 1.178152451 | 9.64E-16 | 6.87E-15 |
| UHRF1 | 0.671316103 | 2.179420344 | 1.698880321 | 5.72E-24 | 3.27E-22 |
| MFAP2 | 0.396864444 | 1.983989389 | 2.321686092 | 4.10E-10 | 1.23E-09 |
| FAM83A-AS1 | 9.962472875 | 3.514503289 | -1.50318309 | 1.82E-06 | 3.55E-06 |
| LINC01702 | 19.91832353 | 8.678032009 | -1.19865641 | 2.76E-09 | 7.49E-09 |
| HPX | 778.5962644 | 333.4381475 | -1.22345626 | 1.64E-24 | 1.14E-22 |
| TMEM54 | 6.767982511 | 14.28154917 | 1.077354737 | 3.21E-09 | 8.64E-09 |
| CD7 | 2.233613534 | 6.610223724 | 1.565319513 | 0.000327866 | 0.0004982 |
| PTGES | 1.674796239 | 4.911651864 | 1.552222723 | 6.65E-08 | 1.52E-07 |
| CDCA5 | 2.15583613 | 4.998936444 | 1.213373665 | 2.91E-20 | 5.39E-19 |
| MCM4 | 4.788695019 | 10.68098397 | 1.157340097 | 3.66E-21 | 8.67E-20 |
| CDKN3 | 3.519248945 | 7.104568951 | 1.01347955 | 2.73E-11 | 9.66E-11 |
| LINC02041 | 0.464983671 | 1.793959412 | 1.947895292 | 2.09E-09 | 5.71E-09 |
| PTGFRN | 4.511081522 | 9.843033793 | 1.12562969 | 4.85E-19 | 6.90E-18 |
| CCL25 | 39.60654341 | 9.669980021 | -2.03415398 | 0.000379945 | 0.0005739 |
| AC092868.1 | 0.61648605 | 1.647775301 | 1.418379368 | 1.04E-07 | 2.33E-07 |
| SLC6A9 | 0.650626829 | 1.655263859 | 1.347158991 | 3.18E-11 | 1.11E-10 |
| PKP3 | 0.227095297 | 2.365755751 | 3.3809314 | 1.30E-09 | 3.66E-09 |
| ZNF667-AS1 | 0.814833609 | 1.650936259 | 1.018707028 | 0.000794076 | 0.0011587 |
| CYP2B6 | 40.07238038 | 16.38337363 | -1.29037575 | 7.16E-10 | 2.08E-09 |
| TUBA1C | 5.847423549 | 12.45277656 | 1.090594452 | 9.52E-22 | 2.60E-20 |
| SOX4 | 3.115068136 | 11.65990706 | 1.904220665 | 2.14E-16 | 1.73E-15 |
| MIR210HG | 0.716541838 | 2.191159641 | 1.612571748 | 5.14E-14 | 2.70E-13 |
| PTTG1 | 5.895142943 | 12.51997581 | 1.086633074 | 2.76E-14 | 1.52E-13 |
| MXRA8 | 2.734894077 | 7.063376206 | 1.36887298 | 0.000599797 | 0.0008862 |
| CTSC | 4.691214305 | 10.49160455 | 1.161202023 | 4.53E-14 | 2.41E-13 |
| SEPTIN5 | 0.985285361 | 2.031347338 | 1.043823418 | 5.82E-09 | 1.51E-08 |
| SPP1 | 153.2336432 | 540.8368222 | 1.819460297 | 5.13E-13 | 2.30E-12 |
| ZNF468 | 1.022169514 | 2.38293582 | 1.221105627 | 1.32E-14 | 7.72E-14 |
| FDCSP | 3.459184914 | 20.2190758 | 2.547213013 | 2.73E-06 | 5.22E-06 |
| MTHFD2 | 0.788328273 | 1.754005683 | 1.153785 | 1.18E-08 | 2.95E-08 |
| MIR4292 | 1.233677587 | 2.892175406 | 1.229189646 | 2.75E-12 | 1.11E-11 |
| FZD1 | 0.624029408 | 2.314887724 | 1.891256298 | 1.28E-15 | 8.93E-15 |
| LOXL1 | 1.113627139 | 3.64806161 | 1.711863819 | 0.009350889 | 0.0120624 |
| PFKP | 1.71667649 | 8.33828591 | 2.280132654 | 1.93E-09 | 5.30E-09 |
| PYCR1 | 4.428659467 | 12.20195983 | 1.462170913 | 8.35E-11 | 2.75E-10 |
| TMEM237 | 0.632782778 | 1.437310358 | 1.183589375 | 3.20E-27 | 6.44E-25 |
| ADH1B | 297.5167939 | 137.0766887 | -1.11798786 | 2.27E-14 | 1.27E-13 |
| TROAP | 1.445574533 | 3.829598929 | 1.405550313 | 3.45E-18 | 4.11E-17 |
| SPINT2 | 4.184393182 | 13.52231457 | 1.69225179 | 8.03E-05 | 0.0001299 |
| UNC5B | 1.511955959 | 3.208337924 | 1.085409987 | 1.50E-11 | 5.48E-11 |
| FCER1G | 13.91700924 | 32.3845239 | 1.218455324 | 5.40E-07 | 1.11E-06 |
| ZNF731P | 2.459721675 | 5.387506402 | 1.131122599 | 8.03E-13 | 3.50E-12 |
| CCNB2 | 2.228830605 | 6.025262183 | 1.434737047 | 2.39E-20 | 4.52E-19 |
| AQP8 | 4.407179136 | 2.053486558 | -1.10178003 | 0.004686372 | 0.0062396 |
| PLEKHA2 | 1.241028914 | 2.676129269 | 1.108611078 | 2.35E-12 | 9.67E-12 |
| SLC27A5 | 71.47095298 | 25.40754009 | -1.49210032 | 1.60E-23 | 7.77E-22 |
| SCD5 | 0.408239527 | 3.937443364 | 3.269771393 | 0.000628381 | 0.0009265 |
| RASSF8 | 0.637071654 | 1.892830877 | 1.57101796 | 2.52E-15 | 1.66E-14 |
| AC239800.2 | 1.95778209 | 0.873779435 | -1.16387914 | 1.24E-08 | 3.09E-08 |
| AREG | 0.465910787 | 1.879882018 | 2.012516482 | 3.13E-08 | 7.42E-08 |
| HKDC1 | 5.914846344 | 15.38335481 | 1.378957565 | 1.48E-13 | 7.24E-13 |
| HSPA7 | 1.184613434 | 3.233501269 | 1.448680824 | 1.03E-05 | 1.83E-05 |
| DMPK | 1.626946658 | 3.329891578 | 1.033308253 | 1.99E-13 | 9.52E-13 |
| TMEM45A | 4.17081155 | 8.446351048 | 1.018000081 | 0.000825351 | 0.0012017 |
| ENDOD1 | 1.760182053 | 3.928093454 | 1.158104601 | 0.008271441 | 0.0107223 |
| KRT17 | 0.667611179 | 3.502840055 | 2.391445096 | 3.65E-07 | 7.65E-07 |
| HROB | 0.715209502 | 1.584664979 | 1.147740058 | 1.19E-16 | 1.01E-15 |
| REN | 2.234571773 | 0.813463765 | -1.4578484 | 4.46E-07 | 9.28E-07 |
| FBXO32 | 1.239771322 | 2.565231763 | 1.049015139 | 9.48E-05 | 0.0001522 |
| MAFA-AS1 | 1.109634022 | 2.502862876 | 1.173495325 | 0.000416811 | 0.0006266 |
| LINC00205 | 0.905113996 | 1.950869348 | 1.107945751 | 4.93E-17 | 4.56E-16 |
| CTSK | 3.828560333 | 26.39328678 | 2.785297124 | 1.85E-07 | 4.01E-07 |
| RNF145 | 3.368278279 | 7.377983449 | 1.131215218 | 1.20E-20 | 2.51E-19 |
| FNBP1L | 3.591897529 | 7.279042871 | 1.019002569 | 1.15E-21 | 3.06E-20 |
| ETV5 | 1.797391551 | 3.917106949 | 1.123883793 | 7.19E-16 | 5.21E-15 |
| TMEM65 | 1.360242305 | 3.014335169 | 1.147976175 | 1.90E-18 | 2.38E-17 |
| ENAH | 3.672603915 | 7.464534635 | 1.023249009 | 1.61E-20 | 3.23E-19 |
| FAM241B | 2.252294427 | 4.662047976 | 1.049568418 | 1.22E-11 | 4.50E-11 |
| RAVER2 | 0.697204228 | 1.725333272 | 1.307221843 | 7.80E-10 | 2.25E-09 |
| LPCAT4 | 0.865316614 | 2.535079796 | 1.550731152 | 6.65E-19 | 9.26E-18 |
| GCA | 1.886367828 | 3.819348327 | 1.017715482 | 2.18E-14 | 1.22E-13 |
| SLC1A7 | 1.023863046 | 3.269039488 | 1.674844053 | 0.015564277 | 0.0195764 |
| AL355102.4 | 1.118981906 | 4.06460321 | 1.860927815 | 1.28E-08 | 3.18E-08 |
| TREH | 1.783453595 | 0.764504452 | -1.22207687 | 0.000175586 | 0.0002741 |
| OGFRL1 | 0.794115329 | 1.679971825 | 1.081016588 | 6.01E-15 | 3.70E-14 |
| APOF | 46.53064897 | 21.40930421 | -1.1199434 | 2.78E-11 | 9.83E-11 |
| PFKFB3 | 3.064625998 | 11.21052113 | 1.871070419 | 1.42E-10 | 4.55E-10 |
| NRAV | 1.233467892 | 3.036729337 | 1.299798165 | 8.40E-51 | 9.99E-47 |
| KNTC1 | 0.711345288 | 1.821579742 | 1.356568232 | 3.56E-21 | 8.46E-20 |
| ASF1B | 2.412406435 | 6.83208781 | 1.501853529 | 2.72E-22 | 8.45E-21 |
| CERCAM | 0.802464496 | 2.835374465 | 1.821029815 | 1.64E-08 | 4.04E-08 |
| DAB2 | 3.771358863 | 9.221588854 | 1.289930908 | 1.66E-11 | 6.02E-11 |
| SCAMP5 | 2.675680505 | 5.500549563 | 1.039669908 | 2.48E-08 | 5.95E-08 |
| LINC01287 | 1.928090352 | 4.042614187 | 1.068115865 | 0.001065955 | 0.0015336 |
| CAPN6 | 0.18805603 | 3.042621037 | 4.016080186 | 2.21E-06 | 4.26E-06 |
| E2F5 | 0.623995665 | 1.416791808 | 1.183019865 | 7.82E-17 | 6.97E-16 |
| CENPO | 0.772177198 | 1.841970922 | 1.25424643 | 4.35E-26 | 5.75E-24 |
| BHLHE41 | 0.441232288 | 1.944560294 | 2.139833696 | 5.78E-09 | 1.50E-08 |
| HAGLR | 0.69150872 | 2.365820164 | 1.774521065 | 2.14E-05 | 3.69E-05 |
| HK2 | 0.861592875 | 3.200526229 | 1.893230907 | 1.16E-15 | 8.15E-15 |
| RHOQ | 2.994419086 | 6.038657933 | 1.011951803 | 1.10E-19 | 1.81E-18 |
| HIF1A | 8.477794192 | 19.55008276 | 1.205413866 | 3.31E-14 | 1.80E-13 |
| IGLV7-46 | 1.081626254 | 6.857903535 | 2.664565535 | 0.020645467 | 0.0255246 |
| CDC6 | 1.549584904 | 4.410060847 | 1.508916757 | 4.59E-22 | 1.34E-20 |
| IGHV3-43 | 1.017299232 | 3.943423848 | 1.954704682 | 0.033009359 | 0.0398168 |
| ABHD17C | 1.875160062 | 3.870855413 | 1.045638672 | 5.49E-14 | 2.87E-13 |
| PDX1 | 1.322156688 | 2.701541097 | 1.030889469 | 0.002337012 | 0.0032231 |
| JAG1 | 2.656742436 | 6.910552942 | 1.379142782 | 3.06E-07 | 6.47E-07 |
| CLIC3 | 0.544309931 | 2.109142272 | 1.954156151 | 3.42E-05 | 5.77E-05 |
| KCTD17 | 2.675743383 | 7.491394807 | 1.485294595 | 1.84E-14 | 1.04E-13 |
| MSC | 2.94283375 | 15.84409574 | 2.428667376 | 8.72E-09 | 2.22E-08 |
| TRAIP | 0.791321916 | 1.811085785 | 1.194518264 | 1.44E-18 | 1.85E-17 |
| FBXO5 | 0.668359075 | 1.560230311 | 1.223063703 | 1.31E-21 | 3.47E-20 |
| LPCAT1 | 3.778173822 | 10.43563396 | 1.465757263 | 6.83E-24 | 3.81E-22 |
| TOP2A | 3.801539376 | 12.12654655 | 1.673513113 | 2.63E-23 | 1.18E-21 |
| F2RL1 | 3.863199674 | 7.853850723 | 1.02360393 | 6.52E-06 | 1.19E-05 |
| NR1I3 | 23.37646096 | 11.41336266 | -1.03433262 | 1.85E-13 | 8.90E-13 |
| SGPP2 | 0.221575467 | 2.275127088 | 3.360077076 | 2.76E-17 | 2.65E-16 |
| IGFBP6 | 1.958618775 | 4.670783839 | 1.25382806 | 0.006633178 | 0.0086876 |
| EIF5A2 | 0.765778534 | 1.950869134 | 1.349117879 | 2.59E-13 | 1.21E-12 |
| PPT1 | 12.18349134 | 25.06045081 | 1.040484752 | 9.57E-27 | 1.65E-24 |
| WASF1 | 2.028930168 | 4.23232839 | 1.060732361 | 1.25E-15 | 8.73E-15 |
| MAP3K21 | 0.708193477 | 1.774574936 | 1.325258036 | 1.85E-09 | 5.09E-09 |
| SLC6A6 | 0.759756697 | 2.541676118 | 1.74217081 | 7.10E-11 | 2.36E-10 |
| TSPAN15 | 3.776135856 | 11.74749267 | 1.637370292 | 4.48E-10 | 1.34E-09 |
| ADAMTS9 | 0.741903753 | 1.807609999 | 1.284779499 | 6.33E-14 | 3.27E-13 |
| PODXL2 | 3.384594712 | 6.854369822 | 1.018040945 | 0.001139223 | 0.0016327 |
| CYP2A7 | 40.46244486 | 9.076428402 | -2.15638688 | 4.31E-15 | 2.73E-14 |
| P3H3 | 0.786514061 | 1.817629329 | 1.208513556 | 0.003830688 | 0.0051528 |
| CHST11 | 0.997230043 | 2.936177505 | 1.557940936 | 1.43E-12 | 6.05E-12 |
| DDX11 | 0.976846962 | 2.147816669 | 1.13666639 | 2.60E-17 | 2.52E-16 |
| TNFRSF21 | 5.063633586 | 15.24614356 | 1.590199448 | 1.18E-15 | 8.27E-15 |
| FXYD3 | 1.561314522 | 4.380903818 | 1.488467348 | 2.01E-05 | 3.47E-05 |
| RELN | 3.968348171 | 8.456546862 | 1.091530066 | 0.013975611 | 0.0176867 |
| SPDEF | 0.51150466 | 2.483619381 | 2.279624807 | 7.74E-07 | 1.57E-06 |
| SLC7A7 | 1.196793773 | 3.020806203 | 1.335759059 | 2.93E-11 | 1.03E-10 |
| ACTL8 | 0.637215731 | 1.909552709 | 1.583380954 | 0.000536226 | 0.0007968 |
| HSD11B1 | 264.5586111 | 116.8643563 | -1.1787524 | 4.29E-09 | 1.14E-08 |
| INHA | 0.585054368 | 2.433967081 | 2.056667053 | 0.001237089 | 0.0017642 |
| TMEM51 | 1.740132714 | 6.118222507 | 1.813915236 | 1.91E-13 | 9.15E-13 |
| DSG2 | 3.340466088 | 11.01479472 | 1.72132129 | 8.49E-17 | 7.48E-16 |
| CDKN1C | 1.977896239 | 4.953883486 | 1.324593194 | 0.03539813 | 0.0425472 |
| PIGS | 2.386049772 | 5.050667467 | 1.081849922 | 7.88E-23 | 2.93E-21 |
| STK17B | 1.011570412 | 2.183166292 | 1.10982528 | 2.42E-12 | 9.91E-12 |
| NIBAN1 | 0.775779827 | 1.914655748 | 1.303365855 | 1.46E-05 | 2.57E-05 |
| PLAU | 2.025639526 | 6.142584033 | 1.600468228 | 3.86E-10 | 1.16E-09 |
| CDH24 | 0.632456701 | 1.410050431 | 1.156708143 | 3.55E-09 | 9.53E-09 |
| SMC4 | 1.452840056 | 3.436403043 | 1.24202337 | 1.33E-16 | 1.12E-15 |
| GPLD1 | 9.629323296 | 3.773653209 | -1.35147257 | 4.73E-12 | 1.85E-11 |
| LAPTM4B | 26.27050428 | 55.88174315 | 1.08893313 | 5.39E-13 | 2.40E-12 |
| SH3BP1 | 0.882736976 | 1.96921352 | 1.157564014 | 7.18E-14 | 3.68E-13 |
| CMTM3 | 3.580551011 | 7.364443496 | 1.040394889 | 1.62E-09 | 4.50E-09 |
| LAMB1 | 6.199246528 | 15.89080979 | 1.358027863 | 4.57E-13 | 2.07E-12 |
| PRR11 | 0.863673141 | 2.569425865 | 1.572888697 | 2.16E-23 | 9.96E-22 |
| C1orf116 | 0.458835241 | 2.057567115 | 2.164891382 | 8.81E-08 | 1.98E-07 |
| S100A6 | 51.13331181 | 155.5831539 | 1.605350483 | 4.34E-07 | 9.03E-07 |
| BICC1 | 1.452105166 | 6.565477177 | 2.176753928 | 1.08E-08 | 2.72E-08 |
| GUCY1A1 | 0.760898776 | 1.663912874 | 1.128803446 | 5.55E-05 | 9.15E-05 |
| SHCBP1 | 0.480971595 | 1.630904685 | 1.761648868 | 7.62E-24 | 4.17E-22 |
| PRC1 | 2.170466316 | 5.482352889 | 1.336790162 | 6.95E-23 | 2.66E-21 |
| HILPDA | 1.477722129 | 5.335455173 | 1.852236344 | 3.79E-33 | 9.01E-30 |
| FABP5 | 2.112097388 | 5.085173787 | 1.267620721 | 1.23E-07 | 2.72E-07 |
| LYPD1 | 1.57965677 | 4.807612278 | 1.60570943 | 2.53E-13 | 1.19E-12 |
| UBE2T | 5.85236276 | 12.49292856 | 1.094020607 | 1.08E-17 | 1.14E-16 |
| GCNT3 | 0.342017791 | 2.639351582 | 2.948040262 | 1.10E-17 | 1.16E-16 |
| QSOX1 | 7.578203387 | 16.89161111 | 1.156379172 | 2.89E-05 | 4.92E-05 |
| SPINDOC | 1.949850692 | 4.93780187 | 1.340505294 | 2.75E-26 | 3.94E-24 |
| RBMS2 | 0.797283653 | 1.613397707 | 1.016937116 | 2.01E-07 | 4.34E-07 |
| GAL3ST1 | 2.547079074 | 11.12100207 | 2.126371136 | 9.68E-14 | 4.87E-13 |
| BASP1 | 2.538883647 | 8.526005185 | 1.747675653 | 0.00023612 | 0.000364 |
| FLNC | 1.806447809 | 3.872817031 | 1.10022777 | 5.55E-08 | 1.27E-07 |
| SLC2A6 | 2.062849964 | 4.269082437 | 1.049287127 | 2.28E-11 | 8.15E-11 |
| SLC29A4 | 2.999509087 | 6.814807271 | 1.183946455 | 1.62E-08 | 3.97E-08 |
| C6orf223 | 0.712189866 | 2.834419486 | 1.992719476 | 4.98E-16 | 3.72E-15 |
| IGFBP3 | 26.2782954 | 52.79217193 | 1.006452326 | 3.21E-12 | 1.29E-11 |
| EXO1 | 0.728520841 | 1.900988693 | 1.383707802 | 1.60E-21 | 4.12E-20 |
| NT5DC2 | 3.286921947 | 9.494195578 | 1.530308571 | 4.08E-15 | 2.60E-14 |
| CHST1 | 0.797754158 | 1.997115784 | 1.323901848 | 1.08E-07 | 2.40E-07 |
| GBP5 | 0.838743293 | 2.716871329 | 1.695645012 | 0.001154926 | 0.001654 |
| MSI2 | 0.962184261 | 1.948493284 | 1.017973854 | 2.86E-19 | 4.28E-18 |
| GLIS2 | 1.665136642 | 4.456675171 | 1.420327242 | 5.96E-13 | 2.64E-12 |
| ALOX5AP | 1.301505291 | 3.444493071 | 1.404110499 | 5.89E-08 | 1.35E-07 |
| SLC10A1 | 98.8214734 | 26.99211934 | -1.87228631 | 1.03E-22 | 3.68E-21 |
| LIMCH1 | 0.726809375 | 2.058976565 | 1.502278475 | 6.18E-05 | 0.0001013 |
| SYT13 | 0.238879736 | 3.691406439 | 3.94981421 | 1.99E-07 | 4.31E-07 |
| DNASE1L3 | 6.674977871 | 2.728724008 | -1.29053657 | 1.01E-16 | 8.71E-16 |
| CFHR4 | 19.201294 | 6.714445828 | -1.5158633 | 8.15E-17 | 7.23E-16 |
| STC2 | 1.027600177 | 2.999976985 | 1.54567239 | 3.81E-09 | 1.02E-08 |
| MT1XP1 | 2.108372892 | 0.801786848 | -1.39483939 | 0.016011949 | 0.0201181 |
| MT2P1 | 7.265677076 | 2.697948233 | -1.42923458 | 0.008794561 | 0.0113744 |
| CDT1 | 2.746364845 | 5.584856353 | 1.023996879 | 3.41E-19 | 5.01E-18 |
| TTK | 0.702914439 | 1.948516242 | 1.470954961 | 3.60E-18 | 4.27E-17 |
| CSAG1 | 2.366426605 | 7.215651279 | 1.60841944 | 0.005598311 | 0.0073876 |
| TMSB4XP8 | 4.098889837 | 9.007215102 | 1.135847899 | 5.37E-07 | 1.11E-06 |
| FOXQ1 | 2.811327012 | 5.807802464 | 1.046741108 | 1.41E-06 | 2.78E-06 |
| FOLR1 | 0.341645737 | 1.965899013 | 2.524616181 | 1.89E-07 | 4.10E-07 |
| CHAF1B | 0.767581926 | 2.142364005 | 1.480810981 | 1.61E-20 | 3.23E-19 |
| SKA3 | 0.937692705 | 2.213072061 | 1.238863315 | 1.06E-17 | 1.12E-16 |
| AC105118.1 | 0.542066191 | 2.562660512 | 2.241101436 | 0.000127205 | 0.0002017 |
| BICDL1 | 1.634291292 | 3.540835597 | 1.115424711 | 2.39E-11 | 8.51E-11 |
| NUF2 | 1.129605089 | 3.397835756 | 1.588797624 | 8.58E-22 | 2.38E-20 |
| TUBB6 | 2.624976479 | 5.789401856 | 1.141109805 | 0.000251896 | 0.0003871 |
| FKBP10 | 3.443701605 | 13.67520329 | 1.989530235 | 9.27E-07 | 1.86E-06 |
| IGKV1OR2-108 | 0.523116924 | 1.950644777 | 1.898745729 | 0.019323313 | 0.0240043 |
| B3GNT5 | 0.895881778 | 2.046474489 | 1.191760412 | 8.09E-09 | 2.07E-08 |
| LINC02608 | 7.88758801 | 3.924100893 | -1.00722206 | 1.29E-09 | 3.62E-09 |
| SSX1 | 5.089912624 | 11.16378263 | 1.133113144 | 0.004417436 | 0.0058967 |
| LTB | 5.308374896 | 12.12050577 | 1.191107734 | 5.67E-05 | 9.34E-05 |
| GNPDA1 | 4.780988847 | 10.08050943 | 1.076187603 | 8.79E-29 | 3.60E-26 |
| LINC01436 | 0.50443838 | 2.997287506 | 2.570907525 | 4.15E-07 | 8.65E-07 |
| DRAM1 | 2.327840645 | 5.4996557 | 1.240349003 | 1.09E-25 | 1.21E-23 |
| PROK1 | 2.26710684 | 0.241616859 | -3.23005935 | 0.003916045 | 0.0052617 |
| ANKRD1 | 1.022082984 | 3.414624893 | 1.740214768 | 0.002065759 | 0.0028736 |
| LPAR2 | 0.805586739 | 3.040008417 | 1.915963478 | 3.21E-12 | 1.29E-11 |
| SYK | 0.986857668 | 2.4754022 | 1.326749022 | 1.26E-08 | 3.13E-08 |
| CYP3A43 | 1.967438733 | 0.817137699 | -1.26766759 | 5.60E-10 | 1.65E-09 |
| HID1 | 1.584045216 | 3.873319434 | 1.289956968 | 3.27E-07 | 6.90E-07 |
| SLC25A47 | 79.97957502 | 30.58917863 | -1.38661025 | 5.49E-10 | 1.62E-09 |
| AC008549.1 | 14.71651529 | 7.208044496 | -1.02975627 | 1.89E-07 | 4.09E-07 |
| CYP1A2 | 34.46191091 | 14.87232956 | -1.21237206 | 5.39E-06 | 9.93E-06 |
| ST6GALNAC4 | 2.120431178 | 4.359233584 | 1.039716852 | 2.35E-17 | 2.31E-16 |
| RCC2 | 7.16660893 | 18.16305531 | 1.341644371 | 6.22E-35 | 2.47E-31 |
| VCAN | 1.371602901 | 4.433431806 | 1.692561024 | 5.08E-07 | 1.05E-06 |
| ZBTB12 | 1.060583481 | 2.910264461 | 1.456292075 | 2.04E-25 | 2.00E-23 |
| FABP3 | 3.337262972 | 13.21956285 | 1.985937191 | 7.62E-05 | 0.0001237 |
| LTBP1 | 2.222838595 | 4.583550257 | 1.044062297 | 8.73E-06 | 1.57E-05 |
| MACIR | 0.564333778 | 1.759011774 | 1.640144531 | 1.00E-13 | 5.02E-13 |
| ENTPD2 | 1.054641704 | 2.972674928 | 1.495008756 | 5.55E-09 | 1.44E-08 |
| ICAM1 | 10.77119825 | 23.22327694 | 1.108392807 | 3.78E-10 | 1.14E-09 |
| S100A9 | 14.96286044 | 83.28613269 | 2.476690304 | 2.94E-07 | 6.23E-07 |
| RNF144A | 0.637643734 | 1.608392152 | 1.334796713 | 3.08E-12 | 1.24E-11 |
| BAK1 | 4.75037241 | 10.38551153 | 1.128459752 | 3.76E-28 | 1.32E-25 |
| TTR | 1237.440583 | 569.8336548 | -1.11874652 | 3.66E-17 | 3.46E-16 |
| KCNQ1 | 1.201690312 | 4.120362121 | 1.777705988 | 3.88E-06 | 7.29E-06 |
| LPA | 4.817584386 | 2.202713641 | -1.12902798 | 5.59E-13 | 2.49E-12 |
| KIF2C | 1.404464139 | 5.068041331 | 1.851408503 | 1.00E-25 | 1.15E-23 |
| MT1A | 49.06528349 | 16.44213568 | -1.57730489 | 0.001249168 | 0.0017804 |
| EMILIN2 | 0.740410435 | 1.686324541 | 1.187485082 | 2.54E-11 | 9.00E-11 |
| CDK4 | 9.371139413 | 19.3549388 | 1.046405369 | 1.33E-26 | 2.16E-24 |
| TFF1 | 2.645030417 | 9.198796543 | 1.798160816 | 3.65E-05 | 6.14E-05 |
| ASNS | 1.393188136 | 3.885504715 | 1.47971192 | 1.42E-10 | 4.55E-10 |
| GMIP | 1.796678182 | 3.603422519 | 1.004035806 | 2.24E-17 | 2.21E-16 |
| CDCA3 | 0.878836856 | 2.134034631 | 1.279916309 | 2.34E-18 | 2.87E-17 |
| AKR1B1 | 4.849224231 | 11.43499423 | 1.237629767 | 3.39E-09 | 9.10E-09 |
| TTYH3 | 7.981481401 | 19.74955987 | 1.307092054 | 5.60E-24 | 3.22E-22 |
| PRTFDC1 | 0.877559085 | 1.884147014 | 1.10234337 | 8.57E-11 | 2.82E-10 |
| CACNG4 | 1.036746738 | 2.708186217 | 1.385263435 | 0.002743895 | 0.0037499 |
| MAGEB2 | 0.965472535 | 3.63318247 | 1.9119267 | 0.000629794 | 0.0009285 |
| ZNF28 | 0.726035997 | 1.655288186 | 1.188969429 | 3.21E-14 | 1.75E-13 |
| PKDCC | 3.60018815 | 7.667230256 | 1.090633201 | 4.09E-09 | 1.09E-08 |
| ANKRD52 | 2.596645585 | 5.345578793 | 1.04169705 | 5.73E-28 | 1.70E-25 |
| FANCE | 1.060125881 | 2.281496357 | 1.105744768 | 7.45E-20 | 1.27E-18 |
| MT1X | 144.6116349 | 37.05118762 | -1.96459194 | 1.19E-05 | 2.10E-05 |
| FCN3 | 5.409702287 | 2.676969182 | -1.01494867 | 0.005245222 | 0.0069432 |
| KLF5 | 1.568159992 | 4.399675356 | 1.488324315 | 1.26E-05 | 2.23E-05 |
| ITIH5 | 0.230960622 | 4.517544807 | 4.289820104 | 0.000273782 | 0.0004195 |
| ITGA3 | 0.799281245 | 4.898168867 | 2.615467371 | 8.69E-08 | 1.96E-07 |
| WWTR1 | 3.026242252 | 6.316116861 | 1.061510384 | 2.07E-08 | 5.03E-08 |
| SLAMF8 | 1.63654421 | 3.507894613 | 1.099952829 | 3.14E-06 | 5.95E-06 |
| MMP9 | 4.859248072 | 13.70751313 | 1.496161865 | 1.48E-09 | 4.12E-09 |
| CRABP2 | 0.664487146 | 2.749148816 | 2.048671808 | 4.22E-05 | 7.06E-05 |
| RRAD | 1.249388208 | 2.769866115 | 1.148594425 | 0.000839941 | 0.0012222 |
| ZNF385A | 2.963170555 | 6.322763669 | 1.093413628 | 1.17E-14 | 6.91E-14 |
| PIP4P2 | 1.271487559 | 3.080145728 | 1.276481263 | 2.57E-13 | 1.21E-12 |
| HDAC7 | 2.112893945 | 4.33071494 | 1.035384859 | 4.36E-14 | 2.32E-13 |
| LRRC8B | 0.624000282 | 1.713877874 | 1.457645725 | 7.39E-21 | 1.62E-19 |
| MFSD2A | 14.19102219 | 6.46081198 | -1.13519112 | 7.48E-06 | 1.36E-05 |
| TM4SF20 | 1.988884468 | 7.675899268 | 1.948376305 | 4.93E-05 | 8.18E-05 |
| WDR72 | 1.727965895 | 4.212267163 | 1.285522199 | 1.02E-06 | 2.03E-06 |
| SOD3 | 5.242306863 | 13.87916483 | 1.404647047 | 0.003206348 | 0.0043499 |
| PHF19 | 1.229207214 | 2.788758101 | 1.18189466 | 6.69E-23 | 2.57E-21 |
| KITLG | 0.859319545 | 1.937741894 | 1.173109804 | 1.79E-12 | 7.49E-12 |
| FMNL2 | 1.240110466 | 3.433995297 | 1.469419426 | 5.35E-13 | 2.39E-12 |
| SDS | 232.0839493 | 89.69024365 | -1.37162379 | 4.93E-07 | 1.02E-06 |
| B4GALNT4 | 0.457747493 | 1.78229265 | 1.961110355 | 0.001170825 | 0.0016752 |
| USH1C | 1.767534462 | 5.144361071 | 1.541253562 | 1.19E-10 | 3.85E-10 |
| ERP27 | 0.482055747 | 1.767366753 | 1.874329551 | 1.67E-06 | 3.26E-06 |
| NEO1 | 2.488961833 | 5.298371337 | 1.090004851 | 3.94E-16 | 3.00E-15 |
| MMD | 3.618325947 | 8.585179747 | 1.246525965 | 5.15E-25 | 4.14E-23 |
| DEPDC1B | 0.690687808 | 2.805287347 | 2.022042892 | 2.68E-24 | 1.78E-22 |
| CENPW | 5.408123256 | 11.82020085 | 1.128054613 | 4.10E-13 | 1.87E-12 |
| FJX1 | 1.059198484 | 2.55081584 | 1.267985783 | 0.000975658 | 0.0014105 |
| GPD2 | 1.102896379 | 2.500662238 | 1.181012956 | 3.72E-28 | 1.32E-25 |
| PLXNA1 | 1.221960775 | 3.168665164 | 1.374677242 | 5.05E-23 | 2.02E-21 |
| TTLL4 | 2.166614368 | 4.564746725 | 1.075092421 | 4.49E-18 | 5.20E-17 |
| S100A8 | 2.106548368 | 9.320217705 | 2.145482612 | 0.019527169 | 0.0242276 |
| SIPA1L3 | 1.895670397 | 3.871331896 | 1.030121855 | 2.76E-17 | 2.65E-16 |
| LRCOL1 | 3.937731949 | 1.951089028 | -1.0130853 | 4.30E-08 | 1.00E-07 |
| DTL | 1.542127235 | 3.609069135 | 1.226704979 | 7.12E-20 | 1.22E-18 |
| NDRG1 | 16.59639833 | 35.49800628 | 1.09686781 | 8.73E-11 | 2.87E-10 |
| ALDH3B1 | 1.519818733 | 3.620674353 | 1.25235916 | 1.52E-12 | 6.38E-12 |
| F9 | 92.97811661 | 39.03211419 | -1.25222959 | 3.34E-13 | 1.54E-12 |
| FZD6 | 1.905579786 | 4.059472293 | 1.091062184 | 7.07E-13 | 3.10E-12 |
| PBK | 1.783046926 | 4.470063561 | 1.325950674 | 2.32E-18 | 2.85E-17 |
| RFLNA | 0.256567352 | 2.10759909 | 3.038190955 | 0.000717646 | 0.0010524 |
| CNDP1 | 1.579449578 | 0.392994687 | -2.00684017 | 7.31E-06 | 1.33E-05 |
| LILRB4 | 0.697499772 | 1.582921534 | 1.182325093 | 4.88E-07 | 1.01E-06 |
| CKAP2L | 0.555466718 | 1.59194536 | 1.519018443 | 3.16E-23 | 1.37E-21 |
| OLFML2B | 1.990150622 | 5.228052876 | 1.393396109 | 2.06E-10 | 6.42E-10 |
| CCNE1 | 1.934778429 | 5.169430495 | 1.417836993 | 1.32E-20 | 2.71E-19 |
| SLC7A10 | 0.193137123 | 1.905836106 | 3.302726665 | 4.56E-07 | 9.47E-07 |
| COL16A1 | 0.725455629 | 1.923704397 | 1.406927844 | 3.45E-07 | 7.25E-07 |
| BUB1B | 0.740996211 | 2.573734703 | 1.79632528 | 9.57E-27 | 1.65E-24 |
| MFSD10 | 5.157719532 | 13.39259871 | 1.376630701 | 1.42E-18 | 1.83E-17 |
| MOGAT2 | 8.752677084 | 3.597652768 | -1.2826684 | 4.59E-10 | 1.37E-09 |
| VNN2 | 3.592147267 | 9.714258366 | 1.43525736 | 5.14E-08 | 1.18E-07 |
| C11orf49 | 0.975349037 | 2.053108487 | 1.073819365 | 2.61E-16 | 2.06E-15 |
| VLDLR | 0.48867081 | 1.605026606 | 1.715662379 | 3.70E-08 | 8.68E-08 |
| GUCA2A | 0.691065352 | 2.027061682 | 1.552495936 | 0.00439018 | 0.0058643 |
| ADAM8 | 0.889338739 | 1.922793208 | 1.112398678 | 2.77E-10 | 8.47E-10 |
| PLP2 | 20.33237089 | 47.71337652 | 1.230615332 | 1.67E-10 | 5.27E-10 |
| SERPINC1 | 2060.690685 | 870.9342536 | -1.24249225 | 1.16E-24 | 8.33E-23 |
| KIF18B | 0.73166824 | 2.490500043 | 1.767173894 | 5.56E-23 | 2.19E-21 |
| SMARCD3 | 0.997210312 | 2.683385754 | 1.428084759 | 9.58E-08 | 2.15E-07 |
| SCTR | 0.332201283 | 5.707567113 | 4.102746369 | 6.11E-07 | 1.25E-06 |
| ZSWIM5 | 0.835310658 | 1.817199178 | 1.121331807 | 4.33E-14 | 2.31E-13 |
| TACC3 | 2.812949903 | 6.858681152 | 1.28584733 | 9.16E-22 | 2.52E-20 |
| ARNTL2 | 0.967987577 | 2.274592358 | 1.232547578 | 8.62E-13 | 3.74E-12 |
| IQGAP3 | 1.785035749 | 3.958679343 | 1.149066245 | 4.58E-17 | 4.24E-16 |
| HMMR | 1.57945492 | 3.436153631 | 1.121367781 | 2.56E-14 | 1.42E-13 |
| FCN2 | 2.739530247 | 1.211759733 | -1.17682486 | 4.65E-05 | 7.74E-05 |
| CYP39A1 | 6.301191961 | 2.664690707 | -1.24165667 | 1.45E-06 | 2.86E-06 |
| MCM8 | 0.720166923 | 1.698596868 | 1.237940251 | 2.60E-23 | 1.17E-21 |
| CLEC11A | 3.146621141 | 6.801555639 | 1.112061269 | 0.002205604 | 0.0030518 |
| MCM2 | 4.158312021 | 11.37663222 | 1.452003625 | 3.04E-23 | 1.33E-21 |
| ITPR3 | 0.473521954 | 2.804073392 | 2.566020889 | 1.74E-12 | 7.28E-12 |
| CEP55 | 0.587135168 | 2.441350738 | 2.055914998 | 2.33E-26 | 3.42E-24 |
| PHLDA2 | 5.626041107 | 11.54832276 | 1.037491335 | 6.11E-11 | 2.05E-10 |
| ANXA13 | 4.260807834 | 13.0103642 | 1.610462457 | 3.93E-06 | 7.37E-06 |
| APOC1P1 | 91.19091263 | 28.18276143 | -1.69407709 | 3.77E-11 | 1.31E-10 |
| TAT | 223.4565137 | 75.14001954 | -1.5723407 | 5.33E-18 | 6.09E-17 |
| SFXN3 | 1.995151291 | 4.862679303 | 1.285253299 | 6.34E-12 | 2.44E-11 |
| PTK7 | 1.046929201 | 2.718679801 | 1.376742361 | 2.37E-05 | 4.07E-05 |
| MSLN | 0.098129695 | 2.067691031 | 4.397187036 | 0.002421659 | 0.0033332 |
| AMIGO2 | 0.888620182 | 2.040481341 | 1.199270705 | 3.05E-07 | 6.44E-07 |
| WDR76 | 1.122815442 | 2.4677984 | 1.136103731 | 8.94E-18 | 9.61E-17 |
| LECT2 | 47.76647012 | 21.56888241 | -1.14704684 | 2.55E-14 | 1.41E-13 |
| CHST2 | 0.640067169 | 1.550442578 | 1.27638488 | 2.91E-07 | 6.17E-07 |
| LINC01093 | 6.373255501 | 1.988212938 | -1.68055822 | 2.08E-06 | 4.03E-06 |
| TNFRSF19 | 1.620055202 | 3.343278462 | 1.04522055 | 0.009350889 | 0.0120624 |
| DES | 0.465574583 | 2.325749673 | 2.32061162 | 0.029546129 | 0.0358467 |
| GP2 | 1.294940026 | 4.281925162 | 1.725374298 | 0.026033356 | 0.0318055 |
| IGSF23 | 17.62609313 | 6.446796873 | -1.4510583 | 2.01E-15 | 1.35E-14 |
| FMNL1 | 1.101697754 | 2.251410013 | 1.031100334 | 1.43E-09 | 4.01E-09 |
| TYMS | 5.91652986 | 12.02118158 | 1.022755543 | 1.13E-14 | 6.68E-14 |
| MAGEA12 | 1.523400503 | 4.700922899 | 1.625648742 | 0.00070302 | 0.0010318 |
| ATP1B3 | 7.881347234 | 17.07328873 | 1.115226814 | 2.02E-18 | 2.51E-17 |
| FBXL19 | 1.413048145 | 2.952027617 | 1.062895597 | 5.61E-29 | 2.47E-26 |
| NCDN | 2.096088898 | 4.614817938 | 1.138573828 | 8.51E-30 | 5.17E-27 |
| CYP4A22 | 28.20823886 | 10.86706028 | -1.37615488 | 4.81E-18 | 5.54E-17 |
| NXPH4 | 1.509634883 | 5.175257432 | 1.777430964 | 2.23E-14 | 1.25E-13 |
| SLC25A24 | 0.492358443 | 1.694507411 | 1.783085044 | 1.19E-12 | 5.11E-12 |
| RRM2 | 4.036027681 | 8.807663435 | 1.125823271 | 7.88E-18 | 8.56E-17 |
| CTSV | 0.312796228 | 2.603413233 | 3.057109303 | 2.24E-16 | 1.80E-15 |
| FIBIN | 0.78250499 | 2.111155555 | 1.431861027 | 6.01E-05 | 9.85E-05 |
| MDFI | 0.791218271 | 2.468964993 | 1.641758734 | 0.000408793 | 0.0006149 |
| PAEP | 2.00022586 | 21.18295082 | 3.404668753 | 1.78E-08 | 4.35E-08 |
| SULF1 | 1.052563852 | 3.373377297 | 1.680285931 | 1.82E-08 | 4.45E-08 |
| CPNE7 | 0.611433642 | 1.770428451 | 1.533830701 | 0.000359848 | 0.0005447 |
| HJURP | 1.263483256 | 3.616622101 | 1.517236314 | 2.85E-22 | 8.77E-21 |
| SLC41A1 | 1.521060108 | 3.430553561 | 1.173364226 | 2.30E-16 | 1.83E-15 |
| SFRP5 | 0.926727006 | 14.67491147 | 3.985063576 | 0.001647289 | 0.0023151 |
| TMC6 | 1.556604492 | 4.145718321 | 1.413219671 | 5.56E-10 | 1.64E-09 |
| AC136601.2 | 2.044484106 | 0.956327002 | -1.09616093 | 3.21E-08 | 7.60E-08 |
| PCK1 | 132.9858515 | 59.95370077 | -1.14935205 | 4.78E-16 | 3.59E-15 |
| NDN | 2.604970436 | 7.091816795 | 1.444888268 | 0.00453586 | 0.0060466 |
| AC006329.1 | 3.824379603 | 1.701556959 | -1.16837028 | 3.47E-14 | 1.88E-13 |
| CLDN4 | 4.698047234 | 14.42551328 | 1.61848953 | 8.52E-09 | 2.17E-08 |
| KRT80 | 0.258642798 | 2.677350573 | 3.371773134 | 1.65E-08 | 4.04E-08 |
| CDCA7L | 0.890631231 | 2.707623911 | 1.604127254 | 1.02E-18 | 1.36E-17 |
| LINC01554 | 28.87353625 | 9.929266032 | -1.53998883 | 3.54E-08 | 8.32E-08 |
| PDE4A | 0.840183829 | 1.826735136 | 1.120490545 | 3.24E-08 | 7.66E-08 |
| PCLAF | 1.523515964 | 3.983511258 | 1.386636035 | 1.37E-21 | 3.60E-20 |
| CYP2A13 | 3.943499094 | 0.583964867 | -2.75552283 | 1.68E-13 | 8.14E-13 |
| ARHGEF25 | 0.707096235 | 1.465638482 | 1.051550805 | 0.020144953 | 0.0249499 |
| MT1E | 166.8333022 | 49.87607975 | -1.74198732 | 0.002981826 | 0.0040573 |
| RCAN3 | 0.7005272 | 1.49896803 | 1.09745664 | 7.11E-18 | 7.85E-17 |
| KIF20A | 1.530817994 | 4.533725499 | 1.56639428 | 5.15E-22 | 1.49E-20 |
| FOXJ1 | 0.356983704 | 2.571392036 | 2.848619458 | 5.00E-13 | 2.25E-12 |
| LINC00942 | 0.496991553 | 2.732225945 | 2.458783558 | 9.20E-10 | 2.63E-09 |
| MMP1 | 0.412451325 | 3.7161184 | 3.171500692 | 4.00E-10 | 1.20E-09 |
| LINC01018 | 15.34634161 | 7.613101589 | -1.01133854 | 1.00E-08 | 2.52E-08 |
| DCLRE1B | 0.9535369 | 1.924650921 | 1.01323613 | 1.52E-26 | 2.41E-24 |
| RACGAP1 | 2.209263936 | 5.352581979 | 1.276669202 | 3.04E-23 | 1.33E-21 |
| GPD1L | 1.171834579 | 2.700198223 | 1.204296393 | 2.66E-15 | 1.75E-14 |
| MAGEA3 | 3.555433908 | 8.36529844 | 1.234391373 | 3.78E-05 | 6.35E-05 |
| SLCO3A1 | 0.974019948 | 2.01418321 | 1.048171692 | 2.16E-05 | 3.72E-05 |
| PLPP2 | 1.677511591 | 6.26593044 | 1.901206019 | 2.15E-08 | 5.22E-08 |
| LAMC2 | 0.236178135 | 3.093514326 | 3.711299399 | 6.11E-05 | 0.0001001 |
| PLCD3 | 0.546386652 | 1.975646556 | 1.854330726 | 1.31E-13 | 6.48E-13 |
| TES | 3.750483569 | 7.941026307 | 1.082248853 | 1.26E-17 | 1.32E-16 |
| SH3RF1 | 1.553139394 | 3.376483781 | 1.12033431 | 2.50E-11 | 8.89E-11 |
| EPS8L3 | 3.392616651 | 8.23331938 | 1.279075769 | 3.32E-10 | 1.01E-09 |
| AC011591.1 | 1.890094803 | 0.874295948 | -1.11226498 | 1.35E-06 | 2.66E-06 |
| TMPRSS3 | 0.980130858 | 2.911992123 | 1.57096017 | 1.00E-07 | 2.25E-07 |
| GRAMD1B | 0.610173215 | 1.550932638 | 1.34584527 | 1.04E-08 | 2.62E-08 |
| PTPRS | 1.048286382 | 2.389381331 | 1.188604216 | 0.010883469 | 0.0139351 |
| UBE2C | 6.927108672 | 20.73463099 | 1.58171716 | 6.99E-18 | 7.74E-17 |
| CCL28 | 0.935982083 | 2.207016835 | 1.237544816 | 2.60E-08 | 6.24E-08 |
| MOB3B | 0.947041232 | 2.084490659 | 1.138195762 | 4.74E-05 | 7.88E-05 |
| CXCL6 | 1.674757628 | 7.578877783 | 2.178031919 | 2.70E-05 | 4.61E-05 |
| AC021146.9 | 14.29035041 | 5.917827812 | -1.27190167 | 4.68E-14 | 2.48E-13 |
| NKD2 | 0.650193205 | 1.445605359 | 1.152733376 | 0.012774443 | 0.0162202 |
| BUB1 | 0.764343944 | 2.32884243 | 1.607319148 | 2.10E-25 | 2.03E-23 |
| NUP210 | 5.049615177 | 10.26708442 | 1.0237812 | 1.65E-22 | 5.56E-21 |
| DPYSL3 | 1.559520668 | 3.490608887 | 1.162376044 | 0.000357186 | 0.0005411 |
| KIRREL2 | 0.025128506 | 2.93219843 | 6.866514004 | 2.00E-07 | 4.32E-07 |
| CLSTN1 | 6.220024741 | 16.46536509 | 1.404442278 | 2.25E-19 | 3.44E-18 |
| MISP | 2.12081692 | 4.585267056 | 1.112385676 | 1.16E-06 | 2.31E-06 |
| HHIPL2 | 0.988339826 | 2.294716056 | 1.215236567 | 0.001429814 | 0.0020255 |
| MMP2 | 5.274654201 | 13.64490245 | 1.37121366 | 0.005437551 | 0.0071842 |
| RAD51AP1 | 1.003933596 | 2.583212013 | 1.363502209 | 5.10E-22 | 1.48E-20 |
| F3 | 0.693787034 | 1.755877666 | 1.33962755 | 3.62E-09 | 9.68E-09 |
| SPDL1 | 0.994512957 | 2.09653461 | 1.075944574 | 2.14E-23 | 9.96E-22 |
| TMEM201 | 1.088505532 | 2.250448196 | 1.047863615 | 4.02E-22 | 1.19E-20 |
| RAVER1 | 0.943852648 | 2.169528648 | 1.200748085 | 1.21E-15 | 8.51E-15 |
| ITGB4 | 1.384860979 | 5.987328837 | 2.112171351 | 1.13E-09 | 3.21E-09 |
| GPC4 | 0.85659542 | 2.962596078 | 1.790176073 | 5.41E-07 | 1.11E-06 |
| AC079360.1 | 2.022514021 | 0.765451872 | -1.40176613 | 6.27E-09 | 1.62E-08 |
| CABYR | 1.411857791 | 2.931254374 | 1.05392339 | 8.69E-07 | 1.75E-06 |
| RBP2 | 0.646034907 | 2.268796706 | 1.812243318 | 0.000203877 | 0.0003164 |
| STK26 | 1.476139389 | 3.051054587 | 1.047479033 | 1.25E-12 | 5.35E-12 |
| TPM2 | 8.232690345 | 16.61335424 | 1.012907515 | 0.000544256 | 0.000808 |
| HSD17B13 | 71.23076279 | 33.13393212 | -1.10419102 | 1.03E-05 | 1.84E-05 |
| ARMCX1 | 0.984601774 | 2.47252248 | 1.328371394 | 7.08E-07 | 1.44E-06 |
| APOA5 | 153.2167519 | 65.87362752 | -1.21780114 | 1.19E-19 | 1.94E-18 |
| ROBO1 | 4.40580932 | 9.360246935 | 1.087139533 | 1.77E-09 | 4.89E-09 |
| FOXM1 | 2.083811788 | 6.028895847 | 1.532668828 | 2.75E-22 | 8.49E-21 |
| VANGL1 | 0.837054277 | 1.71864552 | 1.037878932 | 3.77E-23 | 1.59E-21 |
| LINC00221 | 0.663833377 | 1.600720846 | 1.269828661 | 0.000760651 | 0.0011127 |
| KIF4A | 1.380760899 | 4.345625012 | 1.654100173 | 1.74E-23 | 8.35E-22 |
| FCGR2A | 1.56101762 | 3.335676926 | 1.095492743 | 7.98E-11 | 2.64E-10 |
| CA9 | 1.282150169 | 23.45336591 | 4.193157836 | 1.81E-23 | 8.65E-22 |
| CENPM | 2.479174913 | 6.138758827 | 1.308086931 | 6.40E-20 | 1.11E-18 |
| TNFAIP2 | 8.003255848 | 16.92747011 | 1.080707436 | 1.63E-08 | 4.01E-08 |
| NMB | 3.154512179 | 6.961575132 | 1.141996847 | 9.81E-11 | 3.20E-10 |
| RTL8B | 1.020430338 | 2.433294122 | 1.253733019 | 0.000127846 | 0.0002026 |
| WNK2 | 0.400754493 | 2.442022762 | 2.607286048 | 9.21E-10 | 2.63E-09 |
| C2CD4A | 0.76155848 | 2.833271858 | 1.895442307 | 1.66E-07 | 3.63E-07 |
| RAB25 | 1.95969367 | 6.809745134 | 1.796972647 | 0.040143566 | 0.0479457 |
| FLNA | 11.72480016 | 33.15760393 | 1.499776424 | 5.30E-09 | 1.38E-08 |
| KRT23 | 8.645093136 | 19.6694981 | 1.186006734 | 0.0102728 | 0.013193 |
| FNDC5 | 14.71227059 | 4.727985832 | -1.6377223 | 5.88E-09 | 1.53E-08 |
| FAM117B | 0.643434303 | 1.808957038 | 1.491293387 | 3.11E-22 | 9.39E-21 |
| LAMC1 | 12.60036578 | 25.46800327 | 1.01522024 | 4.66E-15 | 2.92E-14 |
| CLIP2 | 1.728641085 | 4.44493539 | 1.362524093 | 1.30E-14 | 7.62E-14 |
| ANLN | 1.047692443 | 3.69084861 | 1.816737296 | 1.14E-25 | 1.24E-23 |
| IMPDH1 | 2.111284929 | 6.587299158 | 1.641565779 | 6.03E-16 | 4.42E-15 |
| MAGEC2 | 3.156360583 | 7.207833834 | 1.191303724 | 0.018879587 | 0.0234805 |
| SEL1L3 | 2.316729462 | 9.640088351 | 2.056956787 | 5.28E-12 | 2.05E-11 |
| MXRA5 | 0.502759764 | 1.57650962 | 1.648792873 | 0.008176929 | 0.010609 |
| TNFRSF11B | 1.710386195 | 4.827479091 | 1.496947897 | 9.02E-07 | 1.81E-06 |
| CXCL1 | 2.949650768 | 15.05914531 | 2.352023833 | 2.90E-08 | 6.89E-08 |
| TMC5 | 0.597803931 | 3.238503614 | 2.437583066 | 1.62E-09 | 4.51E-09 |
| PKM | 7.984789813 | 44.22628061 | 2.469577581 | 6.18E-25 | 4.83E-23 |
| AC010547.2 | 0.236780575 | 2.122033402 | 3.163824729 | 4.82E-05 | 8.00E-05 |
| ANXA2P2 | 1.528443945 | 3.309747372 | 1.11465746 | 1.85E-14 | 1.05E-13 |
| PAPLN | 0.931573918 | 3.565689469 | 1.936438913 | 5.63E-10 | 1.66E-09 |
| CYP8B1 | 97.75362979 | 37.21709778 | -1.39318472 | 4.63E-14 | 2.46E-13 |
| HPR | 251.3515068 | 109.4280816 | -1.19972333 | 1.02E-16 | 8.77E-16 |
| CYBA | 13.59713983 | 27.96502592 | 1.040320457 | 2.44E-07 | 5.23E-07 |
| OSBPL3 | 0.715950317 | 1.726675022 | 1.270065198 | 2.33E-17 | 2.29E-16 |
| CHGA | 0.287707916 | 4.154755352 | 3.852086705 | 0.018491621 | 0.0230269 |
| PRSS2 | 0.538876755 | 15.94776985 | 4.887255525 | 0.000363268 | 0.0005496 |
| BCL9 | 2.319450868 | 4.807110684 | 1.051386736 | 2.76E-19 | 4.15E-18 |
| LINGO1 | 1.178675923 | 2.422029641 | 1.039049417 | 1.30E-05 | 2.29E-05 |
| GSTP1 | 12.8884454 | 35.15601786 | 1.447693408 | 0.004864141 | 0.0064603 |
| LHFPL2 | 1.050698304 | 2.539885807 | 1.27341516 | 2.92E-15 | 1.91E-14 |
| RASEF | 0.900176454 | 2.296095758 | 1.350903076 | 1.99E-05 | 3.45E-05 |
| PLTP | 8.829201059 | 23.22371512 | 1.395243978 | 4.57E-06 | 8.50E-06 |
| WDR54 | 1.007901789 | 2.342129897 | 1.216466023 | 1.13E-15 | 7.96E-15 |
| EPCAM | 13.33681586 | 31.0687255 | 1.220048794 | 0.000443177 | 0.0006641 |
| BAMBI | 11.02923 | 23.36579671 | 1.083066157 | 3.82E-12 | 1.52E-11 |
| B4GALT5 | 6.859948313 | 13.8483172 | 1.013441064 | 7.79E-20 | 1.33E-18 |
| GLYATL1 | 19.54299934 | 7.936262886 | -1.30012018 | 2.94E-19 | 4.36E-18 |
| H2AC7 | 0.667513986 | 1.373532488 | 1.041021064 | 1.05E-06 | 2.09E-06 |
| CTH | 39.35070407 | 19.42141986 | -1.01874077 | 9.40E-08 | 2.11E-07 |
| IL2RB | 1.092382808 | 2.245721772 | 1.039700686 | 0.000432749 | 0.0006495 |
| IGHV4-39 | 8.351142607 | 38.18702814 | 2.193037142 | 0.004321298 | 0.0057774 |
| FMOD | 3.08178588 | 6.409816996 | 1.05651654 | 0.00121976 | 0.0017408 |
| ARHGEF2 | 2.154037406 | 4.863302612 | 1.174893061 | 7.94E-18 | 8.62E-17 |
| MICAL1 | 1.536731073 | 3.258635083 | 1.084403085 | 1.57E-11 | 5.73E-11 |
| MAGEA6 | 2.799655404 | 6.847895139 | 1.290411346 | 3.12E-06 | 5.91E-06 |
| SAA1 | 1663.310761 | 770.2018143 | -1.11074931 | 0.001199147 | 0.0017136 |
| POSTN | 2.178208668 | 6.68480026 | 1.617742285 | 1.58E-07 | 3.45E-07 |
| AC007423.1 | 4.245302445 | 1.704951015 | -1.31613705 | 0.009942972 | 0.0127943 |
| LAD1 | 15.7104891 | 33.34500834 | 1.085742714 | 6.97E-05 | 0.0001135 |
| COLCA2 | 0.814394139 | 2.419432075 | 1.570869353 | 2.28E-13 | 1.08E-12 |
| RDH16 | 59.94166922 | 25.58666655 | -1.22816706 | 1.38E-11 | 5.09E-11 |
| SH3YL1 | 0.810711701 | 1.730102987 | 1.093597048 | 0.020513148 | 0.0253742 |
| TC2N | 1.123958654 | 3.005953506 | 1.419233729 | 1.64E-06 | 3.20E-06 |
| FBLN1 | 3.63412805 | 12.14267568 | 1.740405199 | 0.000130913 | 0.0002071 |
| C4orf46 | 0.667025478 | 1.497736314 | 1.166969876 | 2.66E-30 | 1.86E-27 |
| FCGR3A | 7.273749111 | 15.06410316 | 1.050343715 | 4.83E-06 | 8.96E-06 |
| H2AW | 1.795635376 | 4.387402185 | 1.288872537 | 8.14E-09 | 2.08E-08 |
| ATP1A1 | 46.6862051 | 103.1068441 | 1.143071872 | 2.02E-17 | 2.02E-16 |
| ELOVL7 | 0.973682531 | 4.064293195 | 2.061481117 | 1.54E-10 | 4.90E-10 |
| ABCC4 | 1.187405293 | 2.499839498 | 1.074023021 | 1.35E-09 | 3.79E-09 |
| YBX3 | 2.604403257 | 5.429675863 | 1.059913228 | 2.30E-05 | 3.95E-05 |
| CCDC9B | 0.818273985 | 2.134911924 | 1.383520661 | 1.51E-06 | 2.98E-06 |
| PLEKHB2 | 3.75336848 | 8.093695736 | 1.108612684 | 2.12E-22 | 6.82E-21 |
| GINS1 | 1.26835561 | 3.763457057 | 1.569099218 | 7.62E-24 | 4.17E-22 |
| CTHRC1 | 2.414051697 | 7.689371655 | 1.67140914 | 1.18E-10 | 3.81E-10 |
| KEL | 0.251761452 | 2.067650333 | 3.037862918 | 0.001305337 | 0.0018569 |
| AC022784.1 | 0.622474181 | 1.450136015 | 1.220102321 | 0.001565547 | 0.0022082 |
| PLCB1 | 0.717044323 | 1.484963103 | 1.050292881 | 4.95E-13 | 2.23E-12 |
| PTPRC | 1.361791484 | 2.961287595 | 1.120718793 | 8.85E-05 | 0.0001425 |
| SMIM10 | 0.62978046 | 1.581025873 | 1.327940076 | 8.96E-05 | 0.0001442 |
| LAIR1 | 0.984054585 | 2.053134374 | 1.061017804 | 2.98E-08 | 7.07E-08 |
| GPRIN1 | 0.586928024 | 1.569933652 | 1.419448091 | 3.59E-23 | 1.52E-21 |
| CDC7 | 0.74194361 | 2.032728513 | 1.454036099 | 1.08E-23 | 5.52E-22 |
| CTAG2 | 2.799859541 | 9.502421868 | 1.762940804 | 0.000225277 | 0.0003479 |
| KIF11 | 1.045191944 | 2.934767775 | 1.489478439 | 1.19E-24 | 8.53E-23 |
| E2F3 | 1.743441133 | 3.987426433 | 1.193520249 | 3.40E-27 | 6.75E-25 |
| CPVL | 5.127042703 | 10.76807662 | 1.070561761 | 6.80E-06 | 1.24E-05 |
| WIPF1 | 1.49765821 | 3.041380319 | 1.022017819 | 2.64E-07 | 5.62E-07 |
| TACSTD2 | 0.953710912 | 4.925605281 | 2.368677091 | 0.039960032 | 0.0477315 |
| ZSWIM4 | 0.881326434 | 2.037882838 | 1.209322728 | 3.91E-16 | 2.98E-15 |
| NAALADL1 | 1.118325344 | 2.583365381 | 1.207911748 | 0.000526834 | 0.0007837 |
| HES4 | 2.219322947 | 5.055287297 | 1.187673466 | 4.31E-08 | 1.00E-07 |
| NCK2 | 4.366113169 | 11.94082857 | 1.451481518 | 9.68E-11 | 3.16E-10 |
| RAB31 | 1.740745591 | 3.681118936 | 1.080438994 | 7.21E-09 | 1.85E-08 |
| MUC20 | 1.413744959 | 3.634206123 | 1.362118368 | 0.012225805 | 0.0155783 |
| MUC6 | 1.907107271 | 6.704086244 | 1.813654714 | 0.007144751 | 0.0093289 |
| PNMA5 | 0.482597189 | 2.123632912 | 2.137642989 | 0.012550677 | 0.0159683 |
| MAMLD1 | 0.917466312 | 1.898412017 | 1.049066048 | 0.000172213 | 0.000269 |
| CCNB1 | 5.434333107 | 13.71317493 | 1.335387723 | 5.11E-21 | 1.16E-19 |
| MPZL1 | 6.789253276 | 13.8940544 | 1.03314284 | 6.42E-27 | 1.16E-24 |
| MAB21L2 | 0.426217229 | 1.837552757 | 2.108124855 | 0.000552425 | 0.0008195 |
| SINHCAF | 1.236250555 | 3.471653739 | 1.489651892 | 7.95E-17 | 7.07E-16 |
| FANCD2 | 0.567575161 | 1.495982576 | 1.398210014 | 1.14E-22 | 4.01E-21 |
| LINC02506 | 1.496656197 | 3.105803053 | 1.053223495 | 0.005650716 | 0.0074518 |
| B3GNT9 | 0.636420046 | 1.784470977 | 1.487445253 | 9.91E-15 | 5.93E-14 |
| SLC12A2 | 1.394471138 | 3.188703158 | 1.193251726 | 2.20E-08 | 5.33E-08 |
| LINC02362 | 3.358075462 | 1.561193037 | -1.10498572 | 3.27E-09 | 8.80E-09 |
| ADCY1 | 1.748204112 | 0.795043258 | -1.13676837 | 2.92E-05 | 4.96E-05 |
| HENMT1 | 0.752111543 | 1.587507419 | 1.07774479 | 5.58E-05 | 9.19E-05 |
| LCAT | 22.50708933 | 10.8917337 | -1.04714588 | 1.67E-11 | 6.06E-11 |
| YEATS2 | 1.096401926 | 2.369497615 | 1.11180444 | 2.45E-32 | 3.65E-29 |
| STX3 | 2.37924109 | 5.626275861 | 1.241678822 | 1.42E-19 | 2.26E-18 |
| PSRC1 | 1.194910053 | 2.85682236 | 1.257509308 | 2.15E-19 | 3.32E-18 |
| TMED3 | 1.292560231 | 2.759378282 | 1.09411174 | 5.01E-10 | 1.49E-09 |
| NEK2 | 1.674595172 | 4.318924313 | 1.366859663 | 3.59E-20 | 6.55E-19 |
| LRRC1 | 1.523137968 | 3.355726648 | 1.139578572 | 7.45E-17 | 6.68E-16 |
| MLLT11 | 1.104879783 | 2.352214517 | 1.090130232 | 0.000170216 | 0.000266 |
| NCF2 | 2.595653199 | 5.659345768 | 1.124537644 | 2.06E-10 | 6.42E-10 |
| ADAP1 | 0.616716472 | 1.444227626 | 1.22761886 | 6.65E-06 | 1.21E-05 |
| GPRC5B | 1.833202388 | 3.94750436 | 1.106574789 | 0.000388944 | 0.0005868 |
| GRB7 | 2.840067986 | 6.42864761 | 1.178589805 | 4.34E-08 | 1.01E-07 |
| NDST1-AS1 | 1.725319053 | 0.669094058 | -1.36658224 | 6.41E-06 | 1.17E-05 |
| SLC7A5 | 3.279134455 | 7.001072704 | 1.094260931 | 1.31E-08 | 3.25E-08 |
| NDUFA4L2 | 5.702258116 | 16.26585042 | 1.512241002 | 8.28E-08 | 1.87E-07 |
| MLANA | 0.099599843 | 2.1071894 | 4.403032715 | 0.001127575 | 0.0016172 |
| COMP | 0.533316393 | 1.877355246 | 1.815638093 | 0.000992777 | 0.0014337 |
| EGLN3 | 0.627488559 | 2.624802925 | 2.064548047 | 2.74E-12 | 1.11E-11 |
| AGRN | 10.6736508 | 24.00624289 | 1.169355914 | 3.55E-16 | 2.72E-15 |
| PRR15L | 3.394785974 | 8.342061606 | 1.297083345 | 1.77E-05 | 3.09E-05 |
| PKMYT1 | 0.81171693 | 1.916572362 | 1.23947986 | 1.07E-20 | 2.26E-19 |
| MFSD6 | 1.346386151 | 2.94100896 | 1.127218937 | 1.27E-09 | 3.59E-09 |
| G6PC | 206.8615047 | 91.6913541 | -1.17380759 | 6.21E-18 | 7.00E-17 |
| C15orf48 | 2.991941813 | 8.31304792 | 1.47429541 | 4.79E-10 | 1.43E-09 |
| CDR2L | 0.945959603 | 2.932599022 | 1.632329344 | 8.16E-05 | 0.0001319 |
| RAD51 | 0.712902404 | 1.730822812 | 1.279681548 | 1.55E-20 | 3.13E-19 |
| MARVELD1 | 1.79315935 | 3.693960055 | 1.042664566 | 4.47E-06 | 8.31E-06 |
| TUSC3 | 1.226148474 | 3.827437505 | 1.642245136 | 6.60E-07 | 1.35E-06 |
| UAP1L1 | 0.673683219 | 3.313172203 | 2.298070921 | 2.21E-15 | 1.47E-14 |
| CTSE | 0.634666395 | 3.1806548 | 2.325253443 | 7.12E-05 | 0.000116 |
| FGFR1 | 0.623770525 | 2.147981462 | 1.783894255 | 2.69E-05 | 4.59E-05 |
| SPC25 | 1.146117957 | 2.748988849 | 1.262145522 | 2.16E-20 | 4.15E-19 |
| CAD | 2.869295297 | 5.768452489 | 1.007487885 | 2.93E-23 | 1.29E-21 |
| CYP1B1 | 3.815962219 | 8.364953503 | 1.132310633 | 1.40E-05 | 2.47E-05 |
| ASRGL1 | 0.99002363 | 2.491745767 | 1.331622012 | 1.16E-14 | 6.86E-14 |
| TEAD2 | 4.180454503 | 11.2695979 | 1.430704334 | 2.53E-19 | 3.81E-18 |
| TMEM74B | 1.256465766 | 3.072764865 | 1.290166009 | 1.26E-07 | 2.79E-07 |
| ASAP1 | 2.077355391 | 4.688727234 | 1.174448301 | 5.83E-19 | 8.17E-18 |
| LOX | 0.921727427 | 2.839564836 | 1.623257767 | 7.79E-14 | 3.97E-13 |
| RHBDF2 | 2.381643871 | 4.820602706 | 1.017255832 | 2.86E-15 | 1.87E-14 |
| PAQR4 | 1.562690557 | 3.921793995 | 1.32748163 | 1.53E-20 | 3.09E-19 |
| COL1A1 | 29.15642712 | 74.17811896 | 1.347179744 | 4.86E-06 | 9.00E-06 |
| TRIM47 | 6.974430073 | 14.46114595 | 1.052034646 | 1.50E-13 | 7.33E-13 |
| FERMT1 | 0.62857588 | 1.448159102 | 1.204061294 | 5.98E-10 | 1.75E-09 |
| LGALS9 | 3.700400332 | 7.588280583 | 1.036091666 | 1.38E-07 | 3.05E-07 |
| RGS2 | 4.732105562 | 16.64024542 | 1.814122549 | 6.17E-12 | 2.37E-11 |
| ANO1 | 14.27514056 | 6.395939747 | -1.1582767 | 5.48E-07 | 1.13E-06 |
| LRIG3 | 1.703888369 | 3.675807637 | 1.109230449 | 1.53E-12 | 6.42E-12 |
| NCAPG | 1.190000376 | 3.372570629 | 1.502886627 | 1.45E-21 | 3.80E-20 |
| GPNMB | 5.659566511 | 17.62743106 | 1.639058777 | 1.96E-07 | 4.24E-07 |
| KIFC1 | 2.720050313 | 7.784091467 | 1.516895325 | 8.35E-23 | 3.06E-21 |
| RENBP | 3.654499646 | 10.44566102 | 1.515157993 | 5.37E-07 | 1.11E-06 |
| KIF23 | 0.541861042 | 1.903720198 | 1.812826622 | 1.91E-26 | 2.88E-24 |
| SERPINI1 | 1.626982143 | 5.059861798 | 1.636899564 | 7.84E-12 | 2.98E-11 |
| IFFO2 | 1.3613032 | 3.019617693 | 1.149377474 | 7.38E-20 | 1.26E-18 |
| CMTM4 | 1.153109585 | 2.346418862 | 1.024930948 | 2.68E-10 | 8.24E-10 |
| AGAP2-AS1 | 0.836322548 | 1.932401796 | 1.208263733 | 0.005806763 | 0.0076491 |
| SAA2 | 204.587123 | 88.23878713 | -1.21323048 | 0.007674173 | 0.0099851 |
| HSD17B6 | 222.6620503 | 109.7435786 | -1.02071916 | 1.99E-17 | 1.99E-16 |
| TRIP13 | 0.832927506 | 2.876511885 | 1.78805759 | 5.87E-25 | 4.63E-23 |
| B3GNT3 | 3.727614578 | 12.10245963 | 1.698975678 | 1.14E-06 | 2.28E-06 |
| MELK | 1.346911266 | 3.854865149 | 1.517025582 | 1.49E-23 | 7.32E-22 |
| C19orf33 | 1.985111867 | 5.322886916 | 1.422988607 | 0.001078361 | 0.0015496 |
| NCAPG2 | 1.236334073 | 2.707611675 | 1.130952212 | 3.49E-21 | 8.34E-20 |
| SAA2-SAA4 | 54.9473459 | 22.76048135 | -1.27151873 | 0.002650126 | 0.0036282 |
| VSIG1 | 0.466795694 | 1.797176315 | 1.944868797 | 0.000233207 | 0.0003596 |
| EVC | 0.901327207 | 2.110008562 | 1.227126009 | 4.17E-06 | 7.80E-06 |
| SMOX | 2.473520881 | 6.197421545 | 1.325102024 | 4.21E-16 | 3.19E-15 |
| IL4I1 | 0.786999433 | 3.097442826 | 1.976643151 | 9.42E-16 | 6.71E-15 |
| HS1BP3-IT1 | 7.247961125 | 3.619053986 | -1.00196259 | 1.30E-09 | 3.66E-09 |
| ADAM17 | 1.031387723 | 2.336882693 | 1.179998537 | 1.30E-19 | 2.09E-18 |
| KIF12 | 5.830518003 | 14.41629351 | 1.306004321 | 3.54E-08 | 8.32E-08 |
| ZNF320 | 0.540765657 | 1.599266027 | 1.564334505 | 3.21E-13 | 1.49E-12 |
| ISYNA1 | 5.448709107 | 13.60230977 | 1.319865276 | 0.004480861 | 0.0059773 |
| SRPX2 | 1.043119518 | 4.068401825 | 1.963557709 | 1.08E-05 | 1.92E-05 |
| BSPRY | 1.146394303 | 2.448972624 | 1.095073301 | 4.45E-05 | 7.42E-05 |
| KIRREL1 | 0.941752519 | 2.050654785 | 1.122664751 | 0.000105636 | 0.0001688 |
| SCPEP1 | 8.146094589 | 18.24118771 | 1.163019197 | 2.30E-13 | 1.09E-12 |
| NEURL3 | 1.145732917 | 4.634319647 | 2.01608678 | 5.40E-11 | 1.83E-10 |
| POLA1 | 1.03262919 | 2.091070445 | 1.017919379 | 5.84E-23 | 2.28E-21 |
| FANCI | 1.054717903 | 2.662157584 | 1.335738789 | 8.83E-24 | 4.71E-22 |
| TRIM16 | 2.111524299 | 4.641239387 | 1.136225261 | 1.69E-10 | 5.33E-10 |
| AL162582.1 | 2.759466113 | 0.863154397 | -1.67669862 | 2.01E-10 | 6.27E-10 |
| UROC1 | 16.99271454 | 6.651649996 | -1.35313218 | 1.66E-10 | 5.24E-10 |
| TGFB1 | 7.96789167 | 17.65262929 | 1.147613145 | 8.56E-07 | 1.73E-06 |
| CORO2A | 1.336777309 | 2.797645122 | 1.065453821 | 1.29E-09 | 3.64E-09 |
| CRYGS | 0.719704798 | 2.51240403 | 1.803591308 | 7.08E-07 | 1.44E-06 |
| STMN1 | 8.496105099 | 18.88927736 | 1.152693992 | 2.88E-19 | 4.30E-18 |
| FBLIM1 | 3.801091089 | 8.680870279 | 1.191426085 | 2.08E-11 | 7.46E-11 |
| HLA-DOA | 1.89294888 | 4.26185117 | 1.170844762 | 0.004480861 | 0.0059773 |
| ASPDH | 33.6850447 | 13.46154716 | -1.32326398 | 8.35E-19 | 1.14E-17 |
| CDC20 | 5.026303415 | 16.60192642 | 1.723780988 | 7.66E-22 | 2.16E-20 |
| PLA2G7 | 2.56551112 | 5.62623881 | 1.132924512 | 8.68E-05 | 0.0001398 |
| OXCT1 | 0.933996781 | 2.122374893 | 1.184190032 | 1.19E-06 | 2.37E-06 |
| PPP1R18 | 6.011572358 | 12.56742343 | 1.06387461 | 8.22E-17 | 7.28E-16 |
| IGHV1-3 | 0.285192023 | 1.809460048 | 2.665553721 | 0.012611728 | 0.0160392 |
| PLEKHG2 | 1.159788049 | 2.353208372 | 1.020767896 | 6.10E-18 | 6.89E-17 |
| GBA3 | 12.16814765 | 5.822792763 | -1.06332639 | 1.46E-09 | 4.08E-09 |
| PAQR5 | 1.135232714 | 3.419658267 | 1.590864092 | 2.44E-12 | 9.97E-12 |
| IGSF3 | 1.14125313 | 3.724290486 | 1.706346787 | 5.70E-14 | 2.97E-13 |
| CDC45 | 1.365998489 | 3.430590333 | 1.328500967 | 1.26E-20 | 2.60E-19 |
| AP001065.3 | 3.758197335 | 1.524818848 | -1.30140296 | 2.13E-20 | 4.12E-19 |
| AC026401.3 | 2.140118871 | 4.50124617 | 1.072633535 | 3.60E-18 | 4.27E-17 |
| CRHBP | 1.782535474 | 0.758628294 | -1.2324657 | 1.63E-06 | 3.19E-06 |
| ADRA2A | 0.489887565 | 1.566905283 | 1.677395396 | 0.016156368 | 0.0202829 |
| ACSS1 | 1.477464119 | 3.153910439 | 1.094018598 | 2.11E-08 | 5.11E-08 |
| FAAP24 | 0.755043718 | 1.576218117 | 1.061835103 | 1.44E-27 | 3.29E-25 |
| IGF2BP1 | 1.162457531 | 2.643876364 | 1.185476704 | 3.57E-05 | 6.01E-05 |
| RTL6 | 0.870500541 | 2.124599395 | 1.287273739 | 4.21E-10 | 1.26E-09 |
| TRNP1 | 7.150933275 | 16.21608864 | 1.181222433 | 1.64E-10 | 5.18E-10 |
| ALPI | 2.929743538 | 6.541219263 | 1.158785194 | 0.014656966 | 0.0184978 |
| PDGFRA | 0.891198114 | 1.874455561 | 1.072653538 | 0.02828746 | 0.0343933 |
| DUSP9 | 6.33900341 | 16.2421334 | 1.357413194 | 1.84E-05 | 3.19E-05 |
| NRM | 4.260981681 | 10.52148478 | 1.304080557 | 7.06E-21 | 1.56E-19 |
| PTGDS | 25.39593323 | 160.2730313 | 2.657862293 | 0.021267179 | 0.0262441 |
| FGD6 | 0.771487832 | 2.026793471 | 1.39348378 | 1.18E-17 | 1.24E-16 |
| CHST4 | 0.358924084 | 2.882127743 | 3.005383644 | 8.84E-06 | 1.59E-05 |
| CARMIL1 | 0.778902117 | 2.046160786 | 1.39340557 | 1.46E-18 | 1.87E-17 |
| FYB1 | 0.909512257 | 2.030553265 | 1.158707886 | 1.06E-05 | 1.90E-05 |
| TUBA1B | 17.6856021 | 38.75260907 | 1.131718109 | 8.94E-23 | 3.23E-21 |
| AC004832.5 | 1.519642013 | 0.745203306 | -1.02802552 | 1.40E-11 | 5.15E-11 |
| OLFM4 | 0.086887058 | 5.488698157 | 5.981178889 | 3.00E-06 | 5.70E-06 |
| SLC7A1 | 0.734210594 | 2.208629055 | 1.588885297 | 2.09E-14 | 1.17E-13 |
| SNAP25 | 0.570993076 | 3.221696416 | 2.496275396 | 4.63E-12 | 1.82E-11 |
| LMNB2 | 3.051458072 | 8.258303365 | 1.436346649 | 1.19E-27 | 2.82E-25 |
| IFT140 | 0.680126251 | 1.40148577 | 1.043082615 | 1.94E-14 | 1.09E-13 |
| CHEK1 | 0.820907074 | 1.867953665 | 1.186167845 | 3.66E-21 | 8.67E-20 |
| PTHLH | 0.305860786 | 2.239110849 | 2.871978894 | 3.30E-10 | 1.00E-09 |
| CLGN | 2.730404033 | 6.690187163 | 1.292932121 | 1.64E-10 | 5.18E-10 |
| ZG16 | 4.696420252 | 2.005288251 | -1.22775188 | 6.94E-10 | 2.02E-09 |
| ARMCX2 | 0.589872916 | 1.59124058 | 1.431675898 | 0.000112152 | 0.0001787 |
| SLC34A2 | 0.181030529 | 11.21109249 | 5.952550049 | 4.66E-06 | 8.65E-06 |
| PRSS22 | 0.093895989 | 2.033897269 | 4.437039466 | 3.49E-05 | 5.89E-05 |
| EMP3 | 5.242534605 | 11.55796444 | 1.140550952 | 3.63E-07 | 7.61E-07 |
| MMP14 | 10.96727525 | 36.33563819 | 1.728180103 | 1.95E-15 | 1.31E-14 |
| SLC16A3 | 1.424356898 | 4.874028442 | 1.774803985 | 7.89E-17 | 7.02E-16 |
| PAQR8 | 0.901493498 | 2.094754411 | 1.216392121 | 2.85E-10 | 8.72E-10 |
| NUSAP1 | 5.323551476 | 12.35950914 | 1.215160517 | 7.32E-20 | 1.25E-18 |
| MYBL2 | 4.239831113 | 14.78759023 | 1.802308268 | 7.96E-22 | 2.23E-20 |
| ATP13A2 | 2.47524825 | 5.164071988 | 1.060935888 | 4.81E-23 | 1.95E-21 |
| ACSM2A | 30.11097873 | 14.20418415 | -1.08397363 | 1.51E-19 | 2.38E-18 |
| TMEM156 | 0.970716132 | 3.526115883 | 1.860958515 | 1.42E-06 | 2.79E-06 |
| ADRA2C | 1.875430502 | 4.194074256 | 1.161130603 | 0.001314404 | 0.0018682 |
| PFN2 | 2.266865907 | 7.363058974 | 1.699606204 | 2.24E-08 | 5.41E-08 |
| SLC36A1 | 0.925549816 | 1.852285086 | 1.000923614 | 3.10E-21 | 7.48E-20 |
| KIF2A | 1.135761441 | 2.413373677 | 1.087391475 | 1.23E-26 | 2.03E-24 |
| SLC38A1 | 2.113447855 | 6.854965234 | 1.697550832 | 1.14E-16 | 9.71E-16 |
| GLS | 2.654380543 | 5.842100258 | 1.138111901 | 1.48E-15 | 1.02E-14 |
| TGFB3 | 1.567622014 | 3.370268008 | 1.104285583 | 2.35E-05 | 4.04E-05 |
| MAPK13 | 1.723350361 | 4.183423462 | 1.279468006 | 7.24E-11 | 2.41E-10 |
| CASP2 | 1.413617896 | 3.171634216 | 1.165834186 | 9.04E-32 | 1.19E-28 |
| ELF4 | 0.704116506 | 2.497068644 | 1.826349413 | 4.27E-13 | 1.94E-12 |
| TCF19 | 3.558432533 | 7.971545823 | 1.163617632 | 2.88E-19 | 4.30E-18 |
| SAMSN1 | 0.702023503 | 1.483688404 | 1.0795969 | 3.17E-07 | 6.70E-07 |
| MIR23AHG | 1.281877255 | 2.892952537 | 1.17428453 | 1.28E-06 | 2.53E-06 |
| NEMP1 | 1.618112207 | 3.424529942 | 1.081594321 | 2.62E-22 | 8.18E-21 |
| LINC02428 | 5.323676422 | 2.646121573 | -1.00854354 | 0.00046413 | 0.0006941 |
| COL4A5 | 0.600816094 | 1.558144136 | 1.374833332 | 8.08E-08 | 1.83E-07 |
| PCSK1N | 1.272454998 | 7.664120871 | 2.59050568 | 6.40E-05 | 0.0001047 |
| ASPM | 1.23064423 | 2.893469331 | 1.233386603 | 1.84E-15 | 1.25E-14 |
| MCM3 | 13.61353202 | 29.09929594 | 1.095942826 | 8.41E-24 | 4.54E-22 |
| MAPRE1 | 12.28492105 | 25.12339098 | 1.032142616 | 1.08E-25 | 1.21E-23 |
| AL137798.1 | 2.65089651 | 1.123724114 | -1.23819247 | 0.000102066 | 0.0001633 |
| TEDC2 | 1.211170778 | 2.590696321 | 1.096937611 | 3.81E-17 | 3.59E-16 |
| FOLH1B | 1.51210468 | 0.519534122 | -1.54126761 | 9.26E-06 | 1.66E-05 |
| DCXR | 420.4422837 | 176.9769336 | -1.24834643 | 5.52E-16 | 4.08E-15 |
| CENPH | 1.383119342 | 2.893086643 | 1.064683886 | 3.95E-22 | 1.17E-20 |
| NTS | 12.25950821 | 151.6044749 | 3.628339327 | 1.02E-05 | 1.82E-05 |
| TTC39A | 0.615580321 | 2.424157396 | 1.977464356 | 7.45E-17 | 6.68E-16 |
| MELTF | 0.794933105 | 2.322944744 | 1.547049472 | 2.99E-09 | 8.08E-09 |
| SNHG3 | 2.070233742 | 4.665190713 | 1.172142393 | 9.51E-14 | 4.79E-13 |
| RAB3IL1 | 1.808474365 | 5.008448824 | 1.469590705 | 2.43E-14 | 1.35E-13 |
| AACS | 0.641124971 | 1.471496019 | 1.198606133 | 8.17E-26 | 9.72E-24 |
| GSDME | 0.741367747 | 1.563470906 | 1.076491116 | 1.89E-10 | 5.94E-10 |
| DLGAP5 | 0.949460485 | 2.904900967 | 1.613309117 | 3.17E-22 | 9.52E-21 |
| SLAMF7 | 0.990225748 | 2.32681696 | 1.232528357 | 0.00305824 | 0.004157 |
| TYRO3 | 0.496302074 | 1.696488505 | 1.773261266 | 1.19E-16 | 1.01E-15 |
| OPN1SW | 0.945498194 | 1.956630942 | 1.049225054 | 9.37E-17 | 8.18E-16 |
| TNFSF13B | 0.931455232 | 1.911990728 | 1.03751719 | 1.22E-05 | 2.16E-05 |
| NBEAL2 | 1.389860866 | 2.97761603 | 1.09921726 | 5.07E-19 | 7.19E-18 |
| DNER | 0.25232066 | 2.165143213 | 3.101132212 | 1.97E-05 | 3.42E-05 |
| IQGAP1 | 2.23829944 | 5.397626494 | 1.269922095 | 1.78E-14 | 1.02E-13 |
| AL035461.2 | 1.377707857 | 2.794791502 | 1.020470662 | 9.37E-11 | 3.07E-10 |
| AC090204.1 | 0.849813445 | 1.952478921 | 1.200088899 | 8.62E-05 | 0.000139 |
| DLG3 | 0.526990969 | 1.639386858 | 1.637306193 | 4.39E-10 | 1.31E-09 |
| PLGLA | 2.6846561 | 1.128560126 | -1.25025401 | 1.18E-09 | 3.33E-09 |
| CENPU | 2.368273461 | 4.943545908 | 1.061710554 | 3.12E-19 | 4.63E-18 |
| MAFG-DT | 0.977759758 | 1.993344461 | 1.027639104 | 4.88E-13 | 2.20E-12 |
| AZGP1 | 358.0799149 | 178.0887726 | -1.00768503 | 3.88E-18 | 4.56E-17 |
| SERPINA11 | 123.7130299 | 60.9909264 | -1.02033092 | 5.55E-09 | 1.44E-08 |
| HLA-U | 1.128925814 | 2.280999276 | 1.014715304 | 0.000796251 | 0.0011617 |
| STMN2 | 0.757125544 | 1.99702923 | 1.399251001 | 0.008113915 | 0.010533 |
| KCNJ11 | 0.710805443 | 1.473555282 | 1.051774551 | 4.88E-10 | 1.45E-09 |
| CHML | 1.373440158 | 3.013111129 | 1.13345983 | 1.28E-16 | 1.08E-15 |
| PDE7A | 0.876612602 | 2.082644053 | 1.248404964 | 3.56E-23 | 1.51E-21 |
| ALDOA | 43.11331274 | 104.0724784 | 1.271383278 | 1.87E-20 | 3.69E-19 |
| CA12 | 2.025027305 | 5.662726808 | 1.483555569 | 2.59E-06 | 4.95E-06 |
| TRIM71 | 0.654816742 | 1.467037805 | 1.163742935 | 1.97E-06 | 3.82E-06 |
| TMEM158 | 0.312584947 | 1.762651584 | 2.495427122 | 3.37E-05 | 5.69E-05 |
| IGLV3-25 | 9.307933338 | 21.40672192 | 1.201531105 | 0.016811631 | 0.0210517 |
| NRSN2 | 2.742072734 | 7.874632026 | 1.52194567 | 3.05E-11 | 1.07E-10 |
| TLDC2 | 0.664686215 | 1.825084231 | 1.457217709 | 1.69E-10 | 5.33E-10 |
| CDCA4 | 1.77069613 | 4.040684343 | 1.190283001 | 3.57E-25 | 3.06E-23 |
| RARRES1 | 3.188856581 | 7.513007122 | 1.236351256 | 1.19E-05 | 2.11E-05 |
| DUOX2 | 0.488749303 | 8.900316419 | 4.186690076 | 2.22E-05 | 3.82E-05 |
| RGS1 | 2.742806529 | 7.831951537 | 1.513718976 | 2.13E-12 | 8.82E-12 |
| DCDC2 | 3.190442217 | 13.25176194 | 2.054355882 | 1.98E-05 | 3.42E-05 |
| PTGFR | 0.996117187 | 2.621149451 | 1.395812233 | 0.008680876 | 0.0112384 |
| MMP12 | 0.52384234 | 3.207957262 | 2.614450346 | 1.10E-11 | 4.09E-11 |
| PGAP4 | 0.932536446 | 2.613944554 | 1.486996524 | 1.33E-10 | 4.25E-10 |
| ARMCX6 | 0.855917116 | 1.999229272 | 1.223900927 | 2.84E-10 | 8.67E-10 |
| IL18 | 1.294635487 | 3.332752122 | 1.364168064 | 3.88E-05 | 6.51E-05 |
| ESPL1 | 0.868845948 | 1.897952231 | 1.127271376 | 2.72E-15 | 1.79E-14 |
| EPHB6 | 0.699308739 | 2.443604652 | 1.805009452 | 6.28E-07 | 1.28E-06 |
| CTTNBP2NL | 0.810962945 | 1.812022794 | 1.159893202 | 2.40E-18 | 2.94E-17 |
| CYRIB | 2.725228925 | 5.489073259 | 1.01018517 | 2.72E-22 | 8.45E-21 |
| NQO1 | 38.28129917 | 79.35985816 | 1.051769655 | 0.002567007 | 0.0035223 |
| LINC01980 | 1.145185364 | 2.449194029 | 1.096725933 | 0.001180295 | 0.0016875 |
| GLS2 | 2.477955915 | 0.991931943 | -1.32083748 | 9.87E-05 | 0.0001582 |
| ITGA2 | 0.677916555 | 2.031457956 | 1.583335899 | 4.95E-13 | 2.23E-12 |
| ORC1 | 0.720348643 | 2.168108049 | 1.589669422 | 2.16E-23 | 9.96E-22 |
| C12orf75 | 1.771335555 | 10.56879535 | 2.576901504 | 1.18E-20 | 2.47E-19 |
| GCGR | 14.58060975 | 5.691348639 | -1.35720859 | 2.24E-08 | 5.41E-08 |
| KIAA1522 | 9.070285966 | 18.32459229 | 1.014561158 | 9.03E-19 | 1.22E-17 |
| TMEM159 | 0.654399043 | 2.050664581 | 1.647848992 | 7.87E-07 | 1.59E-06 |
| SMIM22 | 0.168725358 | 2.560089008 | 3.923445252 | 1.02E-07 | 2.28E-07 |
| HPD | 718.9159057 | 332.6224488 | -1.11193748 | 1.40E-12 | 5.93E-12 |
| NDE1 | 0.741491839 | 1.532054301 | 1.046964712 | 1.14E-24 | 8.29E-23 |
| APLP1 | 0.454625676 | 2.467908269 | 2.440537699 | 3.16E-09 | 8.50E-09 |
| TUBB2B | 0.849740475 | 1.943484681 | 1.193551547 | 6.77E-06 | 1.23E-05 |
| CDC25A | 0.653444546 | 1.628765261 | 1.317641983 | 5.83E-19 | 8.17E-18 |
| CYP3A4 | 505.9259451 | 175.1266907 | -1.53052725 | 1.78E-09 | 4.92E-09 |
| ZNF213 | 0.782500225 | 1.66590578 | 1.090143735 | 2.22E-20 | 4.23E-19 |
| HNF1B | 2.227577062 | 6.283552799 | 1.496105166 | 1.28E-07 | 2.82E-07 |
| TOR4A | 1.102418897 | 3.054173494 | 1.470109494 | 3.06E-10 | 9.32E-10 |
| HRG | 550.9675112 | 241.5937742 | -1.18938397 | 1.28E-15 | 8.93E-15 |
| IER3 | 14.69091713 | 38.84321829 | 1.402738274 | 7.75E-15 | 4.71E-14 |
| GLYAT | 31.29027123 | 10.68630063 | -1.54995165 | 3.12E-16 | 2.42E-15 |
| TMEM44 | 0.883613473 | 1.863458068 | 1.076495034 | 1.54E-16 | 1.29E-15 |
| ZNF292 | 0.799530137 | 1.686044016 | 1.07641788 | 5.11E-19 | 7.24E-18 |
| LDHB | 7.711137835 | 18.41531368 | 1.255890311 | 0.002807275 | 0.0038321 |
| NSD2 | 1.236739893 | 2.584843376 | 1.063534756 | 1.22E-23 | 6.16E-22 |
| ABCA3 | 0.901105165 | 2.057605282 | 1.191198857 | 0.025219305 | 0.0308363 |
| TGFA | 0.555029552 | 2.286992275 | 2.042814999 | 6.84E-09 | 1.76E-08 |
| MMP11 | 2.413420683 | 5.841909563 | 1.275360611 | 1.86E-10 | 5.84E-10 |
| PKIB | 1.471593012 | 3.641774763 | 1.307262967 | 4.71E-08 | 1.09E-07 |
| CYP2A6 | 417.5544684 | 127.6948302 | -1.70926429 | 7.48E-18 | 8.21E-17 |
| CENPF | 1.230017397 | 3.576550297 | 1.53989001 | 3.76E-21 | 8.90E-20 |
| PDP1 | 0.846266066 | 1.950573298 | 1.204714989 | 1.24E-08 | 3.10E-08 |
| NIBAN2 | 8.035694646 | 19.65502482 | 1.290403537 | 2.06E-14 | 1.16E-13 |
| GYS2 | 17.26614447 | 7.441144493 | -1.21434953 | 3.99E-13 | 1.82E-12 |
| ASAH1 | 10.92761003 | 22.31292489 | 1.029901737 | 8.24E-11 | 2.72E-10 |
| DMKN | 0.836952572 | 2.578091463 | 1.623085671 | 0.000277882 | 0.0004256 |
| LINC02381 | 1.209575361 | 3.476578574 | 1.523167539 | 3.45E-07 | 7.25E-07 |
| KRT20 | 1.044289423 | 7.827202982 | 2.905975251 | 0.001525481 | 0.0021538 |
| AP001783.1 | 13.51548822 | 3.625467447 | -1.8983747 | 5.07E-11 | 1.73E-10 |
| AC099850.3 | 0.936043601 | 3.280274104 | 1.809168736 | 6.45E-23 | 2.51E-21 |
| SAMD5 | 0.729623137 | 1.545742343 | 1.083076475 | 0.000242962 | 0.0003741 |
| PLAGL2 | 1.305376405 | 2.727839375 | 1.063292828 | 7.07E-19 | 9.79E-18 |
| IKBKE | 1.222921098 | 2.843840074 | 1.217509011 | 9.57E-18 | 1.02E-16 |
| HMGB2 | 11.85112986 | 23.84253369 | 1.008512947 | 4.30E-18 | 5.02E-17 |
| HOMER3 | 2.176931685 | 6.067612403 | 1.478832795 | 1.28E-14 | 7.52E-14 |
| CDC25C | 1.096483815 | 2.263780931 | 1.045849836 | 3.50E-13 | 1.61E-12 |
| SH3PXD2B | 1.234530979 | 2.622388318 | 1.086918293 | 3.07E-16 | 2.39E-15 |
| CEP170 | 0.687202941 | 1.439594967 | 1.066854848 | 2.57E-21 | 6.34E-20 |
| MYLIP | 1.686398316 | 3.651657635 | 1.114606179 | 2.12E-14 | 1.19E-13 |
| ADAM9 | 3.094173578 | 8.156478074 | 1.398392207 | 4.62E-15 | 2.90E-14 |
| SLC6A13 | 2.334961034 | 1.118318622 | -1.06206719 | 5.20E-12 | 2.02E-11 |
| AFAP1-AS1 | 0.629904457 | 2.100292707 | 1.737385479 | 7.99E-05 | 0.0001293 |
| DNMT1 | 2.771726365 | 6.095945087 | 1.13706507 | 2.84E-23 | 1.26E-21 |
| SRC | 3.485091293 | 7.442819484 | 1.094652797 | 2.50E-14 | 1.39E-13 |
| UCHL1 | 2.15791425 | 13.23880687 | 2.617063665 | 5.32E-07 | 1.10E-06 |
| PPM1H | 0.935639643 | 2.646872467 | 1.500263788 | 2.84E-12 | 1.15E-11 |
| RNU6-850P | 0.710762104 | 1.579920475 | 1.152413275 | 1.78E-12 | 7.45E-12 |
| MYOF | 1.091664302 | 3.095820768 | 1.503792669 | 1.28E-07 | 2.82E-07 |
| AC112206.2 | 1.694244047 | 0.665020782 | -1.34917037 | 3.65E-08 | 8.58E-08 |
| TRIM16L | 3.983602125 | 8.613913401 | 1.112595259 | 0.000142203 | 0.0002241 |
| TIMP2 | 11.96057606 | 28.98386677 | 1.276963203 | 0.000372059 | 0.0005623 |
| ANXA4 | 13.00207078 | 28.86044419 | 1.150352092 | 5.00E-07 | 1.03E-06 |
| MUC5B | 0.9866622 | 6.852834003 | 2.796072597 | 1.87E-05 | 3.25E-05 |
| RNF24 | 0.694810026 | 1.651814277 | 1.249361007 | 1.49E-23 | 7.32E-22 |
| SPINT1 | 4.406293766 | 18.5597592 | 2.074540405 | 4.76E-08 | 1.10E-07 |
| G6PD | 5.127073376 | 22.64686307 | 2.143103779 | 4.57E-28 | 1.43E-25 |
| HGFAC | 36.68660018 | 15.47748709 | -1.24508196 | 1.54E-07 | 3.37E-07 |
| ECT2 | 1.347096838 | 4.239324846 | 1.653980955 | 3.55E-27 | 6.92E-25 |
| FGFR3 | 11.53870451 | 23.87537433 | 1.049042095 | 2.52E-07 | 5.40E-07 |
| SEMA6A | 0.691647627 | 2.964373314 | 2.099618022 | 2.72E-08 | 6.50E-08 |
| LINC01549 | 2.719826203 | 1.304912128 | -1.05956181 | 0.002935599 | 0.0039972 |
| AL035446.1 | 0.959154845 | 2.332977765 | 1.282336911 | 0.000549073 | 0.0008147 |
| ZNF83 | 1.207418888 | 2.63247865 | 1.124495555 | 4.44E-09 | 1.17E-08 |
| TM4SF1 | 21.84704512 | 49.80957376 | 1.188984901 | 5.70E-08 | 1.31E-07 |
| GTSF1 | 1.443152259 | 3.966616397 | 1.458685366 | 1.30E-05 | 2.29E-05 |
| TFF2 | 0.813748011 | 8.78795831 | 3.432874009 | 7.50E-06 | 1.36E-05 |
| TRIM45 | 0.534058777 | 1.6695602 | 1.64439768 | 5.78E-23 | 2.27E-21 |
| TESC | 8.373724253 | 24.13972186 | 1.527467737 | 7.36E-08 | 1.67E-07 |
| HTRA3 | 1.465823407 | 4.1126326 | 1.488350888 | 1.53E-06 | 3.00E-06 |
| CKS2 | 20.60586523 | 41.47885629 | 1.009321072 | 7.48E-16 | 5.40E-15 |
| WDR91 | 2.358778163 | 4.889640981 | 1.051688797 | 2.56E-17 | 2.49E-16 |
| NUAK2 | 2.411727212 | 5.421907664 | 1.168733809 | 1.80E-10 | 5.67E-10 |
| LIMK1 | 1.476797033 | 4.385565855 | 1.570291442 | 8.16E-28 | 2.16E-25 |
| AC083809.1 | 1.322670419 | 4.106382519 | 1.634414406 | 3.24E-07 | 6.84E-07 |
| SPHK1 | 2.495495983 | 8.284510697 | 1.731089909 | 4.52E-15 | 2.84E-14 |
| MROH2A | 1.913211068 | 0.511769408 | -1.90243023 | 0.006630058 | 0.0086855 |
| RAP1GAP | 5.470403451 | 14.52306823 | 1.408627134 | 3.02E-15 | 1.97E-14 |
| RHEX | 0.700020292 | 1.997838334 | 1.512971195 | 0.000881652 | 0.0012798 |
| TRAV30 | 0.769917323 | 1.596104527 | 1.051779698 | 0.000813167 | 0.001185 |
| SPP2 | 91.93488936 | 36.12592556 | -1.34757792 | 7.93E-15 | 4.81E-14 |
| MT1CP | 6.817395704 | 0.231762958 | -4.87849882 | 6.06E-05 | 9.94E-05 |
| SLC44A3 | 4.074064106 | 8.791460703 | 1.109634208 | 7.80E-10 | 2.25E-09 |
| COL9A2 | 0.377370674 | 2.238401595 | 2.568414677 | 7.57E-17 | 6.77E-16 |
| PMEPA1 | 1.652735971 | 7.939652003 | 2.264219507 | 0.000101899 | 0.0001631 |
| ANXA5 | 39.56572746 | 81.97613026 | 1.050952605 | 2.08E-19 | 3.22E-18 |
| SYT8 | 0.561914946 | 2.604639072 | 2.212659791 | 2.23E-08 | 5.40E-08 |
| TMX2P1 | 0.793098194 | 1.702620777 | 1.102185737 | 2.31E-16 | 1.84E-15 |
| PELI1 | 2.835953519 | 5.825879156 | 1.038641889 | 2.12E-15 | 1.42E-14 |
| CDCA7 | 0.453413617 | 2.243204441 | 2.306661487 | 2.73E-11 | 9.66E-11 |
| VTCN1 | 0.164212328 | 5.468735533 | 5.057572947 | 1.56E-06 | 3.06E-06 |
| SMIM6 | 1.842730189 | 3.971668922 | 1.107900517 | 0.00114111 | 0.0016352 |
| SLC7A8 | 0.674403788 | 1.830944678 | 1.440903657 | 9.42E-08 | 2.11E-07 |
| FSCN1 | 5.859355989 | 15.62549882 | 1.415088237 | 4.58E-08 | 1.06E-07 |
| EHF | 1.14026879 | 3.075169282 | 1.431291886 | 2.86E-07 | 6.06E-07 |
| AURKB | 2.459040195 | 6.074374662 | 1.304640578 | 2.02E-17 | 2.02E-16 |
| TEAD4 | 1.639197963 | 3.624541497 | 1.14480841 | 1.94E-06 | 3.76E-06 |
| ADGRG1 | 1.912657804 | 5.170296002 | 1.434668095 | 6.06E-05 | 9.93E-05 |
| ESRP1 | 0.971853733 | 2.742129269 | 1.49648548 | 0.013765555 | 0.0174338 |
| RBBP8 | 1.999888448 | 4.043542169 | 1.015700127 | 6.42E-19 | 8.95E-18 |
| GRAMD1A | 5.57342807 | 11.4416061 | 1.037652713 | 1.22E-15 | 8.56E-15 |
| ENO2 | 0.439070556 | 2.89879361 | 2.722927925 | 1.01E-18 | 1.35E-17 |
| SLC2A1 | 1.012764415 | 4.209320461 | 2.055288728 | 1.60E-10 | 5.06E-10 |
| UGT2B10 | 125.3522526 | 58.06017755 | -1.11036703 | 2.48E-13 | 1.17E-12 |
| CLIP3 | 0.907678849 | 2.054577202 | 1.178587698 | 4.90E-05 | 8.14E-05 |
| NPNT | 1.495150224 | 5.524254868 | 1.885489436 | 0.005893764 | 0.0077577 |
| HMGA1 | 23.92692421 | 55.31608865 | 1.209064197 | 1.23E-19 | 2.00E-18 |
| MTMR2 | 1.18672992 | 2.692358311 | 1.181878784 | 4.08E-24 | 2.54E-22 |
| MYRF | 3.792317853 | 8.95836084 | 1.24015489 | 5.46E-12 | 2.12E-11 |
| MYO10 | 0.635324523 | 1.926339281 | 1.600296213 | 0.031594939 | 0.0382154 |
| DKK1 | 4.594915934 | 14.09200111 | 1.616766122 | 4.40E-07 | 9.14E-07 |
Figure 11.
a: Heatmap of differential genes between high-risk and low-risk groups. Differential genes are highly expressed in high-risk groups b: GO enrichment analysis of differential genes in high-risk and low-risk groups. The GO enrichment analysis of the differential genes showed that the differential genes were mainly located in the extracellular matrix and chromosomal regions, and may play regulatory roles in binding sites and catalytic enzymes, thereby affecting the chromosome division and cell proliferation. CC: cellular Component; BP: biological Process; MF: molecular Function.
4.5. Immune function analysis
Type Ⅱ IFN response, type Ⅰ IFN response, and MHC class Ⅰ were significantly different between the high-risk group and low-risk group. Type Ⅱ IFN response and type Ⅰ IFN Response were highly expressed in the low-risk group, while MHC class Ⅰ was highly expressed in the high-risk group. Besides that, APC co-stimulation, CCR, and Cytolytic activity were different, APC co-stimulation and CCR had high expression in the high-risk group, while Cytolytic activity had high expression in the low-risk group (Figure 12). The results suggest that changes in immune function in the high-risk group may contribute to the progression of HCC.
Figure 12.
Analysis of differences in immune function between the high-risk group and low-risk group. Blue represents low-risk, red represents high-risk, “∗” represents the difference in immune function between the two groups, and “∗∗∗” represents the significant difference. The results of immune function analysis showed that, compared with the high-risk group and the low-risk group, the two immune functions of type II IFN response and type I IFN response were suppressed, and the function of major histocompatibility complex I (MHC I) was enhanced.
4.6. TMB analysis
Mutations in the high-risk group and low-risk group were analyzed, with the higher frequency of mutations in the high-risk group, as shown in Figure 13. But the comparison of TMB between the high-risk group and the low-risk group found no difference (P = 0.15) (Figure 14). The overall survival of the low TMB group was better than that of the high TMB group (Figure 15 a). When the TMB was the same, the high-risk group had a worse prognosis, and when the risk score was the same, the high TMB had a worse prognosis. Furthermore, the results showed that, in HCC, patients with low TMB in the low-risk group had the best prognosis (Figure 15 b). However, the Low-risk group seems to converge around year 7. This may be due to the smaller sample size of the high-TMB group in the low-risk group, especially 7 years later, no samples were included in the low-risk-high TMB group.
Figure 13.
a: Frequency of mutations in the low-risk group, b: Frequency of mutations in the high-risk group. The high-risk group has a higher proportion of gene mutations, and the proportion of TP53 mutations in the high-risk group is significantly higher.
Figure 14.
The comparison of TMB between the high-risk group and the low-risk group. There was no difference in TMB between the high-risk and low-risk groups.
Figure 15.
a: survival curves of the high TMB group and the low TMB group. The high TMB group had a worse prognosis and shorter survival than the low TMB group; b: Comparison of survival curves of TBM combined with the risk score for HCC patients.
4.7. Screening for potential drugs
Based on previous studies, we found a total of 14 potential drugs, including A.443654, A.770041, ABT.263 (Navitoclax), ABT.888 (Veliparib), AICAR (Acadesine), Akt. inhibitor, AMG-706 (Motesanib), AS601245, ATRA, AUY922 (Luminespib), Axitinib, AZ628, AZD.0530, Rucaparib. A.443654, ABT.888 (Veliparib), AS601245, ATRA, AUY922 (Luminespib), AZ628, and Rucaparib were more effective for the high-risk group, while A.770041, ABT.263 (Navitoclax), AICAR (Acadesine), Akt. inhibitor, AMG-706 (Motesanib), Axitinib and AZD.0530 were more effective in the low-risk group (Figure 16).
Figure 16.
Drug sensitivity analysis of potential drugs for HCC. a: A.443654, b: A.770041, c: ABT.263 (Navitoclax), d: ABT.888 (Veliparib), e: AICAR (Acadesine), f: Akt. inhibitor, g: AMG-706 (Motesanib), h: AS601245, i: ATRA, j: AUY922 (Luminespib), k:Axitinib, l:AZ628, m: AZD.0530, n: Rucaparib.
5. Discussion
In our research, we found that NRAV and AL031985.3 are the most closely related lncRNAs in HCC. NRAV is a negative regulator of antiviral response. There are still few studies on its function. Existing research shows that it mainly stimulates gene transcription of immune response by influencing IFITM3 and MxA [40]. Besides that, NRAV may promote the development of HCC by regulating immune cell infiltration and immune checkpoint blockade (ICB) immunotherapy-related molecules [41]. Recent studies have shown that NRAV is up-regulated in HCC samples and is associated with a poor prognosis [41, 42, 43, 44, 45]. Possible mechanisms include the involvement of NRAV in regulating acetyltransferase activity, mRNA binding, and transport of GLUT4 to plasma membrane [42]. Similar to NRAV, many studies have shown that AL031985.3 is associated with the prognosis of HCC and can be used to construct a prediction model for the prognosis of HCC [46, 47, 48]. The results of our study were also consistent with those of previous studies, indicating that NRAV and AL031985.3 were correlated with the prognosis of HCC.
Studies have shown that NRAV and AL031985.3 were highly expressed in HCC tissues, modified by m6A, and were associated with prognosis [49, 50]. However, the mechanism by which m6A modifies these two lncRNAs remains unclear. METTL3, an important m6A methyltransferase, is significantly upregulated in HCC and is involved in the proliferation, migration, and colony formation of HCC cells [51]. The study by Zuo et al [52]. also demonstrated that METTL3 can up-regulate some lncRNAs, which is associated with the poor prognosis in HCC. Our results also showed that METTL3 up-regulated NRAV and AL031985.3 in tumor tissues, which was associated with poor prognosis of HCC. Yang et al [53] found that METTL14 is associated with the occurrence and metastasis of colorectal cancer. However, we found that in HCC, METTL14 was not significantly associated with NRAV and AL031985.3. High expression of IGFBP1 can inhibit the development of liver disease [54, 55], and our study also found that IGFBP1 can negatively regulate two lncRNAs. The above studies have shown that m6A is an important regulatory mode of lncRNAs, which regulates the occurrence and progression of HCC by affecting the expression of lncRNAs.
Our research found that type Ⅱ IFN response, type Ⅰ IFN response, and MHC class Ⅰ were the three most different functions in terms of immune function between the high-risk group and the low-risk group. Type II IFN response, type I IFN response was significantly down-regulated in the high-risk group, while MHC class I was up-regulated. Although it was believed that the main role of type I IFN was to play an antiviral immune role, existing studies have shown that type I IFN can directly act on tumors by blocking cell cycle progression and inducing apoptosis. They also have indirect antitumor activity, promoting clearance and preventing metastasis by priming immune cells [56, 57, 58]. It is worth noting that hepatitis virus infection is an important pathogenic factor for liver cancer. Compared with other tumors, the influencing factors of type I IFN response in liver cancer are more complicated. Our results suggested that the type I IFN response is functionally enhanced in the low-risk group, which may be related to the involvement of type I IFN in the naturally occurring protective immune response of primary tumors [59]. Our results are also consistent with previous studies showing that downregulation of type I IFN enhances tumor growth in a paracrine manner, reduces T lymphocyte cytotoxicity, and the ability of dendritic cells (DC) to process and present antigens [59, 60]. Type II IFN is mainly produced by immune cells (activated T cells, natural killer cells, and macrophages) and plays an important role in anti-tumor proliferation and cellular immunity [61, 62]. The low expression of type II IFN response and type I IFN response in the high-risk group may be due to an immunodeficiency of the tumor, which is present in a variety of tumors including breast cancer, melanoma, and gastrointestinal tumors [63]. Liver cancer cells all express MHC class I (MHC-I) antigens, but not in normal liver cells [64]. MHC-I plays an important role in controlling the phagocytosis of macrophages and can protect tumor cells from phagocytosis [65]. Differences in immune function create a favorable immune environment for cell proliferation and immune escape for HCC patients.
In our results, the GO enrichment analysis of the differential genes showed that the differential genes were mainly located in the extracellular matrix and chromosomal regions and may play regulatory roles in binding sites and catalytic enzymes, thereby affecting the chromosome division and cell proliferation of cells. In the high-risk group and the low-risk group, the most mutated genes were TP53, CTNNB1, TTN, and MUC16. About 11–41% of liver malignancies had CTNNB1 mutations, and 13–48% had TP53 mutations [66, 67]. CTNNB1 and TP53 mutations promoted glycolysis and cell proliferation through ALDOA phosphorylation [68]. It was worth noting that CTNNB1 and TP53 mutations were mutually exclusive, and the two mutations can cause different characteristics of HCC. HCC patients with CTNNB1 mutations had large tumors, well-differentiated, bile accumulation, and no inflammatory infiltration, while HCC patients with P53 mutations had poorly differentiated tumors, dense cells, and frequent vascular infiltration [69]. Our results also showed that in the high-risk group compared to the low-risk group, TP53 mutations increased and CTNNB1 decreased, which may mean that the patient’s tumor cells had a worse differentiated and a higher risk of vascular invasion, which meant a worse prognosis.
We screened 14 potential therapeutic agents for HCC in our study. There were already some studies on these drugs. Navitoclax made liver cancer cells sensitive to sorafenib and regorafenib through a mitochondrial caspase-dependent mechanism [70]. However, the current problem was that due to the low binding affinity of Navitoclax and Mcl-1, the anti-cancer activity of Navitoclax was often weakened by the increased expression of Mcl-1 in HCC and other cancers [71]. In addition, the phase III clinical trials of veliparib combined with temozolomide in advanced HCC showed that it was well tolerated in patients with advanced HCC, but the patients did not show a significant survival benefit [72]. AICAR regulated the redox status of liver cancer cells through nuclear factor erythroid 2-related factor signaling pathway and AMP-activated protein kinase independent mechanism [73]. Akt inhibitor, as an ATP-competitive AKT kinase inhibitor, blocked the phosphorylation of downstream molecules of the AKT signaling pathway in a dose and time-dependent manner, inhibited the proliferation, and induces apoptosis of hepatoma cells [74, 75]. A-443654 was also an Akt inhibitor and has antitumor effects in various tumors [76, 77]. A-770041 was a potent Src family kinase (Lck and Src) inhibitor with antitumor activity in osteosarcoma [78]. The phase III, randomized clinical trial of motesanib in non-small cell lung cancer showed that its objective response rate was 60.1% [79]. In thyroid papilloma, the objective response rate was 14%. 67% of patients had stable disease [80]. AS601245 was a selective c-Jun-terminal kinase inhibitor, the strongly inhibits cell adhesion and migration, with antitumor potential [81, 82, 83]. All-trans retinoic acid (ATRA) was an active metabolite of vitamin A and played an important role in tumor cell proliferation, differentiation, and apoptosis [84]. AUY922, an inhibitor of heat shock protein 90, reduced the proliferation and viability of HCC cells in a dose-dependent manner [85]. Axitinib combined with radiotherapy for advanced HCC was well tolerated, with ORR of 66.7%, 1-year OS of 66.7%, and median PFS of 7.4 months [86]. AZ628 was a hydrophobic Raf-kinase inhibitor that may play a therapeutic role in breast cancer [87], adrenocortical carcinoma [88]. Saracatinib (AZD.0530) treatment abolished the activation of Fyn, down-regulated the Fyn/FAK/N-WASP signaling pathway in hepatic stellate cells, and significantly reduced the degree of CCl4-induced liver fibrosis in mice [89]. It was expected to be a therapeutic drug for liver fibrosis and HCC. Rucaparib, a poly (ADP-ribose) polymerase inhibitor, had been approved for the treatment of recurrent ovarian and prostate cancer, and clinical trial results had shown that moderate liver injury did not require dose adjustment [90]. Although many potential drugs for the treatment of HCC had been discovered through studies, we should also be aware that the mechanism of HCC development is very complex, and the effect of a single targeted drug was [91] not ideal. In the future, we should study the combination of different drugs to block the progression of HCC, to improve the overall survival of HCC patients.
There were also some limits in this study. Firstly, the samples of this study are from the TCGA database, the sample size of some groups may be small. For example, in Figure 15b, the Low-risk group seems to converge around year 7. This may be due to the smaller sample size of the high-TMB group in the low-risk group, especially 7 years later, no samples were included in the low-risk-high TMB group. Secondly, the risk score constructed by the study requires further validation by external validation and prospective studies. However, our study found the role of m6A-related lncRNAs in HCC prognosis and immunotherapy, established an effective risk score, and identified 14 potential therapeutic drugs for HCC, which is significant for further improving the prognosis of HCC patients.
6. Conclusion
The risk score constructed with NRAV and AL031985.3 had a good predictive effect on the Prognosis of HCC. Differences in genes and immune function between high-risk groups and low-risk groups promoted the occurrence and progression of HCC.
Declarations
Author contribution statement
Yan Xu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Rong LIU: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at Figshare under the URL https://doi.org/10.6084/m9.figshare.20575707.v1.
Acknowledgements
None.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Peck-Radosavljevic M., Bota S., Hucke F. Time to stop using hepatic arterial infusion chemotherapy (HAIC) for advanced hepatocellular carcinoma?-the SCOOP-2 trial experience. Ann. Transl. Med. 2020;8(21):1340. doi: 10.21037/atm-2020-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsurusaki M., Murakami T. Surgical and locoregional therapy of HCC: TACE. Liver Cancer. 2015;4(3):165–175. doi: 10.1159/000367739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couri T., Pillai A. Goals and targets for personalized therapy for HCC. Hepatol. Int. 2019;13(2):125–137. doi: 10.1007/s12072-018-9919-1. [DOI] [PubMed] [Google Scholar]
- 6.Giannini E.G., Farinati F., Ciccarese F., et al. Prognosis of untreated hepatocellular carcinoma. Hepatology. 2015;61(1):184–190. doi: 10.1002/hep.27443. [DOI] [PubMed] [Google Scholar]
- 7.Llovet J.M., Ricci S., Mazzaferro V., et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Cheng A.L., Kang Y.K., Lin D.Y., et al. Sunitinib versus sorafenib in advanced hepatocellular cancer: results of a randomized phase III trial. J. Clin. Oncol. 2013;31(32):4067–4075. doi: 10.1200/JCO.2012.45.8372. [DOI] [PubMed] [Google Scholar]
- 9.Johnson P.J., Qin S., Park J.W., et al. Brivanib versus sorafenib as first-line therapy in patients with unresectable, advanced hepatocellular carcinoma: results from the randomized phase III BRISK-FL study. J. Clin. Oncol. 2013;31(28):3517–3524. doi: 10.1200/JCO.2012.48.4410. [DOI] [PubMed] [Google Scholar]
- 10.Cainap C., Qin S., Huang W.T., et al. Linifanib versus Sorafenib in patients with advanced hepatocellular carcinoma: results of a randomized phase III trial. J. Clin. Oncol. 2015;33(2):172–179. doi: 10.1200/JCO.2013.54.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda K., Kudo M., Kawazoe S., et al. Phase 2 study of lenvatinib in patients with advanced hepatocellular carcinoma. J. Gastroenterol. 2017;52(4):512–519. doi: 10.1007/s00535-016-1263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda M., Morizane C., Ueno M., et al. Chemotherapy for hepatocellular carcinoma: current status and future perspectives. Jpn. J. Clin. Oncol. 2018;48(2):103–114. doi: 10.1093/jjco/hyx180. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alarcon C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patil D.P., Chen C.K., Pickering B.F., et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumann U., Shafik A., Preiss T. METTL3 gains R/W access to the epitranscriptome. Mol. Cell. 2016;62(3):323–324. doi: 10.1016/j.molcel.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz S., Mumbach M.R., Jovanovic M., et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8(1):284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Yue Y., Han D., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer K.D., Jaffrey S.R. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ping X.L., Sun B.F., Wang L., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G., Dahl J.A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia G., Fu Y., Zhao X., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller S., Glass M., Singh A.K., et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic. Acids Res. 2019;47(1):375–390. doi: 10.1093/nar/gky1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer K.D., Jaffrey S.R. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haussmann I.U., Bodi Z., Sanchez-Moran E., et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 27.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J., Eckert M.A., Harada B.T., et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20(9):1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J.Z., Yang F., Zhou C.C., et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 30.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 31.Ma J.Z., Yang F., Zhou C.C., et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 32.Yang F., Bi J., Xue X., et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279(17):3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 33.Xu Z.Y., Yu Q.M., Du Y.A., et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 2013;9(6):587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao W.J., Wu H.L., He B.S., Zhang Y.S., Zhang Z.Y. Analysis of long non-coding RNA expression profiles in gastric cancer. World J. Gastroenterol. 2013;19(23):3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C., Yang M., Tian J., Wang X., Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int. J. Oncol. 2011;39(1):169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 36.Huang X., Gao Y., Qin J., Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;314(5):G559–G565. doi: 10.1152/ajpgi.00242.2017. [DOI] [PubMed] [Google Scholar]
- 37.Chen J., Huang X., Wang W., et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY) 2018;10(11):3371–3381. doi: 10.18632/aging.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng J., Yang G., Liu Y., et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9(18):5227–5245. doi: 10.7150/thno.34273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao Y., Zhou D. lncRNA-D16366 is a potential biomarker for diagnosis and prognosis of hepatocellular carcinoma. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2019;25:6581–6586. doi: 10.12659/MSM.915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouyang J., Zhu X., Chen Y., et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16(5):616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Q., Wang Y., Huang W. Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int. Immunopharm. 2021;92 doi: 10.1016/j.intimp.2020.107333. [DOI] [PubMed] [Google Scholar]
- 42.Feng Y., Hu X., Ma K., Zhang B., Sun C. Genome-wide screening identifies prognostic long noncoding RNAs in hepatocellular carcinoma. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/6640652. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Chen Z.A., Tian H., Yao D.M., et al. Identification of a ferroptosis-related signature model including mRNAs and lncRNAs for predicting prognosis and immune activity in hepatocellular carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.738477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Z.H., Li Z.W., Yang D.L., Liu J. Development and validation of a pyroptosis-related long non-coding RNA signature for hepatocellular carcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.713925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou P., Lu Y., Zhang Y., Wang L. Construction of an immune-related six-lncRNA signature to predict the outcomes, immune cell infiltration, and immunotherapy response in patients with hepatocellular carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.661758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia Y., Chen Y., Liu J. Prognosis-predictive signature and nomogram based on autophagy-related long non-coding RNAs for hepatocellular carcinoma. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.608668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng B., Yang M., Wang M., Liu Z. Development and validation of 9-long Non-coding RNA signature to predicting survival in hepatocellular carcinoma. Medicine (Baltim.) 2020;99(21) doi: 10.1097/MD.0000000000020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong W., Wang X., Zuo X., et al. Development and validation of an immune-related lncRNA signature for predicting the prognosis of hepatocellular carcinoma. Front. Genet. 2020;11:1037. doi: 10.3389/fgene.2020.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin L., Zhou L., Gao S., et al. Classification of hepatocellular carcinoma based on N6-methylandenosine-related lncRNAs profiling. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.807418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai T., Li J., Ye L., et al. Prognostic role and potential mechanisms of N6-methyladenosine-related long noncoding RNAs in hepatocellular carcinoma. J. Clin. Transl. Hepatol. 2022;10(2):308–320. doi: 10.14218/JCTH.2021.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 52.Zuo X., Chen Z., Gao W., et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13(1):5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X., Zhang S., He C., et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19(1):46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L.J., Tang Q., Wu J., et al. Inter-regulation of IGFBP1 and FOXO3a unveils novel mechanism in ursolic acid-inhibited growth of hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2016;35:59. doi: 10.1186/s13046-016-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adamek A., Kasprzak A. Insulin-like growth factor (IGF) system in liver diseases. Int. J. Mol. Sci. 2018;19(5) doi: 10.3390/ijms19051308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booy S., Hofland L., van Eijck C. Potentials of interferon therapy in the treatment of pancreatic cancer. J. Interferon Cytokine Res. 2015;35(5):327–339. doi: 10.1089/jir.2014.0157. [DOI] [PubMed] [Google Scholar]
- 57.Borden E.C. Interferons alpha and beta in cancer: therapeutic opportunities from new insights. Nat. Rev. Drug Discov. 2019;18(3):219–234. doi: 10.1038/s41573-018-0011-2. [DOI] [PubMed] [Google Scholar]
- 58.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50(4):907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6(11):836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 60.Miar A., Arnaiz E., Bridges E., et al. Hypoxia induces transcriptional and translational downregulation of the type I IFN pathway in multiple cancer cell types. Cancer Res. 2020;80(23):5245–5256. doi: 10.1158/0008-5472.CAN-19-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagiwara M., Fushimi A., Bhattacharya A., et al. MUC1-C integrates type II interferon and chromatin remodeling pathways in immunosuppression of prostate cancer. OncoImmunology. 2022;11(1) doi: 10.1080/2162402X.2022.2029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6(11):836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 63.Critchley-Thorne R.J., Simons D.L., Yan N., et al. Impaired interferon signaling is a common immune defect in human cancer. Proc. Natl. Acad. Sci. U. S. A. 2009;106(22):9010–9015. doi: 10.1073/pnas.0901329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sung C.H., Hu C.P., Hsu H.C., et al. Expression of class I and class II major histocompatibility antigens on human hepatocellular carcinoma. J. Clin. Invest. 1989;83(2):421–429. doi: 10.1172/JCI113900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barkal A.A., Weiskopf K., Kao K.S., et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 2018;19(1):76–84. doi: 10.1038/s41590-017-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guichard C., Amaddeo G., Imbeaud S., et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahn S.M., Jang S.J., Shim J.H., et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60(6):1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 68.Gao Q., Zhu H., Dong L., et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179(2):561–577. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 69.Calderaro J., Couchy G., Imbeaud S., et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 2017;67(4):727–738. doi: 10.1016/j.jhep.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Tutusaus A., Stefanovic M., Boix L., et al. Antiapoptotic BCL-2 proteins determine sorafenib/regorafenib resistance and BH3-mimetic efficacy in hepatocellular carcinoma. Oncotarget. 2018;9(24):16701–16717. doi: 10.18632/oncotarget.24673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G., Zhang S., Fang H., et al. Aspirin overcomes Navitoclax-resistance in hepatocellular carcinoma cells through suppression of Mcl-1. Biochem. Biophys. Res. Commun. 2013;434(4):809–814. doi: 10.1016/j.bbrc.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 72.Gabrielson A., Tesfaye A.A., Marshall J.L., et al. Phase II study of temozolomide and veliparib combination therapy for sorafenib-refractory advanced hepatocellular carcinoma. Cancer Chemother. Pharmacol. 2015;76(5):1073–1079. doi: 10.1007/s00280-015-2852-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sid B., Glorieux C., Valenzuela M., et al. AICAR induces Nrf2 activation by an AMPK-independent mechanism in hepatocarcinoma cells. Biochem. Pharmacol. 2014;91(2):168–180. doi: 10.1016/j.bcp.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 74.Yang F., Deng R., Qian X.J., et al. Feedback loops blockade potentiates apoptosis induction and antitumor activity of a novel AKT inhibitor DC120 in human liver cancer. Cell Death Dis. 2014;5:e1114. doi: 10.1038/cddis.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crouthamel M.C., Kahana J.A., Korenchuk S., et al. Mechanism and management of AKT inhibitor-induced hyperglycemia. Clin. Cancer Res. 2009;15(1):217–225. doi: 10.1158/1078-0432.CCR-08-1253. [DOI] [PubMed] [Google Scholar]
- 76.Zheng J., Hudder A., Zukowski K., Novak R.F. Rapamycin sensitizes Akt inhibition in malignant human breast epithelial cells. Cancer Lett. 2010;296(1):74–87. doi: 10.1016/j.canlet.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Frias M., Iglesias-Serret D., Cosialls A.M., et al. Akt inhibitors induce apoptosis in chronic lymphocytic leukemia cells. Haematologica. 2009;94(12):1698–1707. doi: 10.3324/haematol.2008.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duan Z., Zhang J., Ye S., et al. A-770041 reverses paclitaxel and doxorubicin resistance in osteosarcoma cells. BMC Cancer. 2014;14:681. doi: 10.1186/1471-2407-14-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kubota K., Yoshioka H., Oshita F., et al. Phase III, randomized, placebo-controlled, double-blind trial of motesanib (AMG-706) in combination with paclitaxel and carboplatin in east asian patients with advanced nonsquamous non-small-cell lung cancer. J. Clin. Oncol. 2017;35(32):3662–3670. doi: 10.1200/JCO.2017.72.7297. [DOI] [PubMed] [Google Scholar]
- 80.Sherman S.I., Wirth L.J., Droz J.P., et al. Motesanib diphosphate in progressive differentiated thyroid cancer. N. Engl. J. Med. 2008;359(1):31–42. doi: 10.1056/NEJMoa075853. [DOI] [PubMed] [Google Scholar]
- 81.Cerbone A., Toaldo C., Minelli R., et al. Rosiglitazone and AS601245 decrease cell adhesion and migration through modulation of specific gene expression in human colon cancer cells. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0040149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bubici C., Papa S. JNK signalling in cancer: in need of new, smarter therapeutic targets. Br. J. Pharmacol. 2014;171(1):24–37. doi: 10.1111/bph.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Q., Wu W., Jacevic V., et al. Selective inhibitors for JNK signalling: a potential targeted therapy in cancer. J. Enzym. Inhib. Med. Chem. 2020;35(1):574–583. doi: 10.1080/14756366.2020.1720013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ni X., Hu G., Cai X. The success and the challenge of all-trans retinoic acid in the treatment of cancer. Crit. Rev. Food Sci. Nutr. 2019;59(sup1):S71–S80. doi: 10.1080/10408398.2018.1509201. [DOI] [PubMed] [Google Scholar]
- 85.Augello G., Emma M.R., Cusimano A., et al. Targeting HSP90 with the small molecule inhibitor AUY922 (luminespib) as a treatment strategy against hepatocellular carcinoma. Int. J. Cancer. 2019;144(10):2613–2624. doi: 10.1002/ijc.31963. [DOI] [PubMed] [Google Scholar]
- 86.Yang K.L., Chi M.S., Ko H.L., et al. Axitinib in combination with radiotherapy for advanced hepatocellular carcinoma: a phase I clinical trial. Radiat. Oncol. 2021;16(1):18. doi: 10.1186/s13014-020-01742-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hossain S.M., Shetty J., Tha K.K., Chowdhury E.H. Alpha-ketoglutaric acid-modified carbonate apatite enhances cellular uptake and cytotoxicity of a Raf- kinase inhibitor in breast cancer cells through inhibition of MAPK and PI-3 kinase pathways. Biomedicines. 2019;7(1) doi: 10.3390/biomedicines7010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma C., Xiong J., Su H., Li H. The underlying molecular mechanism and drugs for treatment in adrenal cortical carcinoma. Int. J. Med. Sci. 2021;18(13):3026–3038. doi: 10.7150/ijms.60261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du G., Wang J., Zhang T., et al. Targeting Src family kinase member Fyn by Saracatinib attenuated liver fibrosis in vitro and in vivo. Cell Death Dis. 2020;11(2):118. doi: 10.1038/s41419-020-2229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grechko N., Skarbova V., Tomaszewska-Kiecana M., et al. Pharmacokinetics and safety of rucaparib in patients with advanced solid tumors and hepatic impairment. Cancer Chemother. Pharmacol. 2021;88(2):259–270. doi: 10.1007/s00280-021-04278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng W., Ainiwaer A., Xiao L., et al. Role of the novel HSP90 inhibitor AUY922 in hepatocellular carcinoma: potential for therapy. Mol. Med. Rep. 2015;12(2):2451–2456. doi: 10.3892/mmr.2015.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.