Abstract
Methyltransferases (MTases) of procaryotes affect general cellular processes such as mismatch repair, regulation of transcription, replication, and transposition, and in some cases may be essential for viability. As components of restriction-modification systems, they contribute to bacterial genetic diversity. The genome of Helicobacter pylori strain 26695 contains 25 open reading frames encoding putative DNA MTases. To assess which MTase genes are active, strain 26695 genomic DNA was tested for cleavage by 147 restriction endonucleases; 24 were found that did not cleave this DNA. The specificities of 11 expressed MTases and the genes encoding them were identified from this restriction data, combined with the known sensitivities of restriction endonucleases to specific DNA modification, homology searches, gene cloning and genomic mapping of the methylated bases m4C, m5C, and m6A.
The bacterium Helicobacter pylori chronically infects the upper gastrointestinal tract of the majority of people worldwide and is major cause of chronic gastritis and peptic ulcer disease and an early risk factor for gastric cancer (39). H. pylori strains are diverse genetically (1, 2, 4, 16, 21, 44), and certain strain differences may contribute to the diversity of clinical outcome of infection (13, 15, 47).
Analyses of genome sequences of H. pylori strains 26695 (46) and J99 (3) revealed that each strain contained more than two dozen genes likely to encode DNA methyltransferases (MTases), far more than have been detected in other bacterial genomes to date (20). These MTase genes comprise a large fraction of all genes that were specific to one or the other strain. Bacterial MTases exist either as components of restriction-modification (R-M) systems or as separate enzymes. The former are strain specific, and their main function is to protect host DNA from damage by a cognate restriction enzyme (RE). Separate MTases, which tend to be present in all strains of a species (species-specific MTases), are involved in general cellular processes such as regulation of transcription of specific genes (5, 48), DNA replication (32), mismatch repair (33), or DNA transposition (40). Certain of them can be essential for viability (43) or contribute to specificity in bacterium-host interactions or virulence in pathogens (8, 17).
Most MTase genes found in the genomes of H. pylori strains 26695 and J99 probably belong to R-M systems. That certain R-M systems might also affect bacterium-host interactions was suggested by induction of transcription of iceA1, a homolog of RE NlaIII, by H. pylori-host cell contact (36). Although iceA1 does not encode a functional protein in most H. pylori strains (14), it is adjacent to a generally active MTase gene (hpyIM) specific for sites recognized by NlaIII. Thus, the hpyI-iceA1 pair may represent a vestige of an R-M system that has evolved some special function relevant to host-pathogen interactions (14). By extrapolation, the same might be true for the MTases of other putative R-M systems in H. pylori. The identification and study of both species-specific and strain-specific MTases of H. pylori may improve our understanding of pathogenic mechanisms of this organism. Here we have addressed two issues: (i) which putative MTases of H. pylori 26695 are expressed by bacteria cultivated in vitro and (ii) the specificity of the encoded MTases.
MATERIALS AND METHODS
General microbial and molecular methods.
The H. pylori strain used here was 26695 (46). Tissue culture flasks containing 200 ml of brucella broth (BBL) with 5% (vol/vol) fetal calf serum and Dent selective supplement (Oxoid SR147E) (50) were inoculated and then incubated horizontally for 72 to 96 h at 37°C in a microaerobic chamber containing 6% O2, 10% CO2, and 84% N2 (Flow Laboratories CO2 Incubator 220). Total genomic DNA was prepared as described elsewhere (29), with minor modifications. Briefly, 1 g of cells was suspended in 10 ml of TE buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA) and incubated for 2 h at 37°C with pronase E (0.5 mg/ml) and sodium dodecyl sulfate (0.25%). The cell lysate was deproteinized by extraction once with Tris-buffered phenol-chloroform (1:1) and twice with chloroform. DNA was precipitated by adding 0.1 volume of 2.5 M NaCl and 1 volume of isopropanol, washed in 75% ethanol, dissolved in TE buffer, treated with RNase A (300 μg/ml) for 1 h at 37°C, reextracted, precipitated with isopropanol, and dissolved in 3 ml of TE buffer. The DNA concentration was determined spectrophotometrically and adjusted to 400 μg/ml with TE buffer. DNA samples were stored at 4°C.
Escherichia coli K-12 strain AL2268 [genotype F′ 128zzf-13::mTn10 (Tcr) lacZΔm12/e14− (mcrA) endA1 thi-1 Δ(mcrC-mrr)114::IS10 Δ(argF-lac)U169 recA1], derived from ER2267 (New England Biolabs; A. Lubys, personal communication), was used as host for an H. pylori 26695 DNA library and to select plasmids that were resistant to PstI. E. coli strain RR1 (27) was used to select plasmids that were resistant to Hin6I and TaqI. The positive selection vector pBR-R (25), containing an intact cfr9IR gene (24), was used to generate a library of strain 26695 DNAs by ligation into its unique Eco47III site. Transformants were obtained by electroporation and selection on Luria-Bertani agar with ampicillin and chloramphenicol. E. coli cells carrying pBR-R or related plasmids were grown in Luria-Bertani medium containing ampicillin (60 μg/ml) and chloramphenicol (30 μg/ml).
Plasmid DNAs were prepared by alkaline lysis (6) and further purified by binding to glass powder (28). Restriction, agarose gel electrophoresis, isolation of DNA fragments from agarose gels, and other DNA manipulations were carried out by standard procedures (42).
RE cleavage tests.
All REs and buffers were from MBI Fermentas, Vilnius, Lithuania. All REs in the MBI Fermentas catalog (30) except for ApaI, AloI, BseXI, PpiI, and SalI were tested. The FauI and BcefI isoschizomers used here were isolated from unidentified bacteria (unpublished data). DdeI, CviRI, and Tsp4CI isoschizomers and Hpy8I were isolated from H. pylori strains.
To test for RE sensitivity, generally 0.8 μg of DNA was digested in 30 μl of the recommended buffer for 2 to 4 h with 20 to 30 U of an RE under conditions recommended by MBI Fermentas. When H. pylori DNA was not digested, a mixed digest was set up with control DNA of phage λ along with DNA of strain 26695; the λ DNA was cleaved in every case, as expected. Digested DNAs were electrophoresed at 80 to 100 V for 2 to 3 h in 0.7 or 1.0% agarose gels in 0.1 M sodium borate (pH 8.2) buffer containing 2 mM EDTA and 0.5 μg of ethidium bromide per ml. The gels were then UV illuminated and photographed with the ULTRA LUM gel documentation system.
PFGE analysis.
REs to be assessed by pulsed-field gel electrophoresis (PFGE) were selected, and the sizes and numbers of fragments that they would generate (if no sites were modified) were inferred from the strain 26695 genome sequence (46). For PFGE, cells of strain 26695 were first suspended in TE buffer at an A600 of 10 and embedded in low-melting-point agarose (1.4%) blocks in a 1:1 ratio (final volume, 30 μl). The blocks were placed in lysis solution containing 0.25 M EDTA (pH 9.0), 0.5% lauroyl sarcosyl, and 0.5 mg of proteinase K per ml (10). Thin slices were cut from the DNA agarose blocks, washed at least three times with phenylmethylsulfonyl fluoride solution (0.175 mg/ml; 15 min each time), and then washed three more times with TE buffer. The slices were preincubated with 100 μl of the appropriate buffer and then incubated overnight in fresh buffer with 30 to 40 U of the corresponding RE (AatII, Bsp120I, Bsp1407I, Bst1107I, Cfr42I, Eco72I, Eco105I, KpnI, NotI, Pfl23II, ScaI, SdaI, TatI, or XhoI) at the appropriate temperature. DNA fragments were separated in 1% agarose gels in TBE buffer (89 mM Tris-borate [pH 8.3], 0.25 mM EDTA) for 44 h at 14°C and 190 to 150 V, using a GeneLine system apparatus (Beckman). The pulse times were varied from 2 min to 45 s to detect DNA fragments of various sizes.
Deduction of sequence specificities of MTases.
The DNA sequence specificities of MTases were deduced from the sensitivity or resistance of the DNAs to various REs, based on known effects of site-specific methylation on their activities (30, 31), essentially as described elsewhere (9, 34, 54). H. pylori MTases were named as suggested in REBASE, version 007: the acronym of the MTase includes the open reading frame (ORF) (The Institute for Genomic Research database [TDB] annotation 46) that encodes it (e.g., M.HpyAORF51 encoded by HP51). Suffix “P” (putative) denotes a protein with a conserved MTase motif for which MTase activity has not been shown directly (e.g., M.HpyAORF263P).
Detection of strand-specific type IIS MTases.
The bisulfite method (38, 49) was used to map m5C and m4C in the recognition sequences of M.HpyAORF51 (CCTC) and M.HpyAORF1368 (TCTTC), respectively. Strain 26695 DNA was treated with sodium bisulfite (49) and purified by adsorption on glass beads using a DNA extraction kit (MBI Fermentas). A 400-bp DNA segment containing TCTTC (genome position 585647) and CCTC (genome position 585660) was then PCR amplified using direct and reverse primers (5′-tgtgttggattttaaaatattaagg and 5′-ctccacttttaatttaccacaaaca, respectively), purified in an agarose gel, and subcloned into the pTZ57R/T vector using the InsT/Aclone PCR product cloning kit (MBI Fermentas). Cloned inserts were sequenced using the same direct primer and a Cycle Reader DNA sequencing kit (MBI Fermentas).
A test for methylated adenines in sequences modified by M.HpyAORF1367 and M.HpyAORF50 (5′-GAAGm6A and 5′-Gm6AGG, the complements of TCTTC and CCTC, respectively) was carried out with REs HindIII and XbaI (recognition sequences 5′-AAGCTT and 5′-TCTAGA; inhibited by methylation of underlined bases). First, a 250-bp DNA segment containing overlapping M.HpyAORF1367 and HindIII recognition sequences 5′-GAAGAAGCTT (position 1008166) and the separate HindIII site 5′-AAGCTT (position 1008050) was PCR amplified using 5′-ggtgttcggtgcaaagcca and 5′-gggatcgataaaaaccttattttaa (direct and reverse primers, respectively), and a 280-bp DNA segment containing overlapping M.HpyAORF50 and XbaI recognition sequences 5′-TCTAGAGGA (position 141547) and a separate XbaI site (position 141557) was amplified using 5′-ccgaagcgatcaaactcactc and 5′-tccctcaaaaatttcaagctc (direct and reverse, respectively). Aliquots of total genomic DNA and appropriate PCR fragments were digested separately with HindIII or XbaI, and digested DNAs were used as templates in primer extension reactions with Taq polymerase, using 5′-radiolabeled direct primers. Aliquots of PCR fragments were sequenced using the same 5′-radiolabeled primers and a Cycle Reader DNA sequencing kit (MBI Fermentas). The 5′-radiolabeled extension reaction products and dideoxy sequencing reaction products were loaded on a standard sequencing gel. Methylated bases within recognition sequences of respective MTases were identified by comparing of 5′-radiolabeled products of extension reactions of digests of genomic DNAs (methylated) and of PCR fragments (nonmethylated) with dideoxy sequence ladders of corresponding PCR fragments.
Library construction and selection of clones conferring resistance to PstI, Hin6I, or TaqI.
To find DNA segments encoding MTases, H. pylori 26695 genomic DNA was fragmented by sonication, and a 2- to 9-kb size fraction was selected by agarose gel electrophoresis. DNA ends were polished with T4 DNA polymerase, and 5 μg of DNA was ligated with 1 μg of Eco47III-cleaved, calf intestinal alkaline phosphatase-dephosphorylated pBR-R vector DNA in 40 μl, using T4 DNA ligase at 16°C for 24 h. The ligation mixture was extracted with an equal volume of chloroform, ethanol precipitated, and dissolved in 40 μl of H2O, and 12 μl was then used to transform competent E. coli AL2268 cells by electroporation. Plasmid DNA was isolated from pools of 75,000 ampicillin- and chloramphenicol-resistant transformants; 6 μg was digested with excess PstI, Hin6I, or TaqI (30 U, each in a separate tube) for 6 h and then transformed back into AL2268 cells. Plasmid DNAs of 40 randomly picked transformants from each selection were then screened for resistance to PstI, Hin6I, and TaqI digestion. Resistant recombinant plasmids containing cloned DNA fragments of different lengths were sequenced using vector-specific primers.
DNA sequence determination.
Sequencing was performed by the dideoxynucleotide chain termination method (11) using an MBI Fermentas DNA sequencing kit, [α-33P]dATP and double-stranded, supercoiled plasmid DNA as the template, and electrophoresis on wedge-shaped gels. The primers used, 5′-gctctgcacgtaaaccgtatgttgcg and 5′-cactgcttgaaacagttcgtgatatgg, are specific to DNA sequences flanking the Eco47III cloning site of the pBR-R vector. At least 150 bp from each end of each cloned DNA fragment of interest was determined, and their positions and ORF content were inferred by comparison with the strain 26695 genome sequence (46).
Amino acid sequence analysis.
The amino acid sequences of putative MTases of strains 26695 and J99 were taken from the EMBL database (accession no. AE000511 and AE01439, respectively); A. Lubys provided the sequences of MnlIA and MnlIC MTases (personal communication); those for M.Hpy84-183I and M.Hpy8I were determined by K. Stankevicius (unpublished) and J. Minkute (personal communication), respectively. Inferred homologies of putative MTases of strain 26695 were initially taken from the TDB annotation (http://www.tigr.org/). MTase conserved motifs, identified using C-SEARCH software and methods (45), were used to predict the base modifications (m4C, m5C, and m6A) generated by a given MTase. Full-length sequence alignments were produced using MULTALIN software (12). The percent similarity for full-length putative MTase proteins was calculated from complete amino acid sequences including gaps introduced into sequence alignments. The identical amino acid pairs within sequence alignments were calculated. For prediction of the sequence specificity of MTase, variable amino acid sequence regions (putative target recognition domain [TRD]) were identified as recommended elsewhere (18, 19, 26, 35, 37, 45). These amino acid sequences were searched for similarities to all protein sequences available in sequence databases (EMBL and SWISS PROT) using the FASTA procedure (22). Alignments characterized by FASTA E values below 0.01 were taken as indicating statistically significant similarity and extracted for further analysis. Sequence specificities of putative 26995 MTases were inferred only if significant similarity between a putative TRD and that of a known MTase was observed and if their identities exceeded 40%. We also regarded cases of lower (but statistically significant) similarities as indicating the same specificity only in cases of similarities of putative TRDs exceeding 20% which at the same time were higher than those of full-length alignments of the corresponding MTases.
RESULTS
Four approaches were used to identify the specificities of the DNA MTases of H. pylori strain 26695 and the genes encoding them: (i) the pattern of resistance or sensitivity of chromosomal DNA to digestion with a large battery of REs; (ii) a detailed search for homology with known MTases; (iii) activities of certain MTase genes after cloning in E. coli; and (iv) genomic mapping of m4C, m5C, and m6A modifications generated by certain MTases.
RE resistance/sensitivity patterns of strain 26695 DNA.
Genomic DNA of strain 26695 was resistant to cleavage by 24 of the 147 different REs tested (Table 1), indicating sequence-specific modification by numerous MTases during growth in culture. In some cases, these data either confirmed specificities of MTases predicted from homology searches (Table 2) or allowed MTase specificities to be deduced (Table 1; Materials and Methods). In certain other cases (especially predicted type IIS MTases), MTase specificity was determined by additional genome mapping of m4C, m5C, and m6A (see below). Eleven different sequence-specific MTases were identified in this way (Table 1).
TABLE 1.
Empirically determined sequence specificities of H. pylori 26695 MTases
| No. | RE | Recognition sequence of RE and sensitivity to modificationa | Cleavageb | Specificity of MTasec | H. pylori 26695 MTase |
|---|---|---|---|---|---|
| 1 | MnlI | G A G G | − | G A G G (m6A) | M.HpyAORF50 |
| + | C C T C (m5C) | M.HpyAORF51 | |||
| C T C C | |||||
| + | |||||
| 2 | MboI | G A T C | − | G A T C (m6A) | M.HpyAORF92 |
| + − | |||||
| Bsp143I | G A T C | + | |||
| − + | |||||
| PvuI | C G A T C G | + | |||
| − + | |||||
| BamHI | G G A T C C | + | |||
| − + | |||||
| BglII | A G A T C T | + | |||
| − + | |||||
| BclI | T G A T C A | − | |||
| + − | |||||
| 3 | TaqI | T C G A | − | T C G A(m6A) | M.HpyAORF260 |
| − + | |||||
| XhoI | C T C G A G | − | |||
| + − + | |||||
| Bsu15I | A T C G A T | − | |||
| + + | |||||
| Bsp119I | T T C G A A | − | |||
| + | |||||
| 4 | Tru1I | T T A A | + | ||
| BspTI (AflII) | C T T A A G | + | |||
| + + | |||||
| KspAI (HpaI) | G T T A A C | + | |||
| +− | |||||
| VspI | A T T A A T | − | A T T A A T (m6A) | M.HpyAORF478 | |
| DraI | T T T A A A | + | |||
| − | |||||
| 5 | CviRI | T G C A | + | ||
| ++ | |||||
| PstI | C T G C A G | − | C T G C A G (m6A) | M.HpyAORF593 | |
| + ++ | |||||
| Alw44I | G T G C A C | + | |||
| +− | |||||
| Mph1103I | T T G C A A | + | |||
| SdaI | C C T G C A G G | − | |||
| 6 | Hin6I | G C G C | − | G C G C (m5C) | M.HpyAORF1121 |
| + | |||||
| EheI | G G C G C C | − | |||
| + +− | |||||
| Eco47III | A G C G C T | − | |||
| − + | |||||
| NsbI | T G C G C A | − | |||
| + − | |||||
| Bsp143II | R G C G C Y | − | |||
| + − | |||||
| 7 | Hin1II (NlaIII) | C A T G | − | C A T G (m6A) | M.HpyAORF1208 |
| ++ | |||||
| NcoI | C C A T G G | + | |||
| + − | |||||
| PaeI | G C A T G C | − | |||
| + − | |||||
| PagI (RspXI) | T C A T G A | − | |||
| + + | |||||
| XceI (NspI) | R C A T G Y | − | |||
| ++ | |||||
| 8 | HinfI | G A N T C | − | G A N T C (m6A) | M.HpyAORF1352 |
| + − | |||||
| PleI | G A G T C | − | |||
| 9 | MboII | G A A G A | − | G A A G A(m6A) | M.HpyAORF1367 |
| −+ + | |||||
| T C T T C (m4C) | M.HpyAORF1368 | ||||
| C T T C T | |||||
| BpiI | G A A G A C | − | |||
| Eam1104I | G A A G A G | − | |||
| − |
+ or − indicates that methylation (m6A or m5C) at this position either prevents or has no effect on RE cleavage, respectively. When such data are not available for an RE used in experiments, it is given for the isoschizomer, indicated in parentheses in the RE column. For type IIS enzymes, both strands of the recognition sequence are provided when necessary.
DNA cleavage by RE (+) and resistance to RE (−). Data for H. pylori DNA sensitivity to REs is given only for the members of those families of related enzymes among which at least one RE did not cleave the DNA.
Bases shown or predicted to be modified by corresponding MTases (see text) are underlined.
TABLE 2.
Homology search for putative H. pylori 26695 MTasesa
| No. | HP ORF | Annotation
|
Sequence similarity (%)b
|
RE testedc | r/sd | MTase | J99 homolog JHP ORFe | ||
|---|---|---|---|---|---|---|---|---|---|
| TDB | SSD | TDB | SSD | ||||||
| 1 | 50 | M.MnlIA, m6A | 52 (57) | MnlI | − | M.HpyAORF50 | 43 (−) | ||
| M.DpnA, m6A | 37 | 32 (29) | |||||||
| 2 | 51 | M.MnlIC, m5C | 56 (55) | MnlI | − | M.HpyAORF51 | |||
| M.DdeI, m5C | 39 | 32 (23) | DdeI | + | |||||
| 3 | 92 | M.MboIC | M.MboIC, m6A | 55 | 52 (61) | MboI | − | M.HpyAORF92 | 85 (−) |
| 4 | 260 | M.EcoP15I, m6A | M.EcoP15I, m6A | 34 | 15 (24) | TaqI* | − | M.HpyAORF260 | 244 (−) |
| 5 | 263 | M.Hpy84-183I, m4C | 90 (96) | MspI | + | M.HpyAORF263P | 248 (−) | ||
| M.HpaI, m6A | 34 | 14 (15) | KspAI | + | |||||
| 6 | 478 | M.VspI, m6A | M.VspI, m6A | 42 | 40 (41) | VspI | − | M.HpyAORF478 | 430 (−) |
| 7 | 483 | M.HphIC, m5C | 37 | 29 (22) | HphI | + | |||
| M.Hpy99XI, m5C | 94 (100) | TaiI | + | M.HpyAORF483P | 435 (−) | ||||
| 8 | 593 | M.EcoP15I, m6A | ?, m6A | 38 | 33 (20) | PstI* | − | M.HpyAORF593 | |
| 9 | 910 | M.Hpy8I, m6A | 97 (97) | Hpy8I | + | M.HpyAORF910P | 846 (+) | ||
| M.HindII, m6A | 33 | 19 (16) | HincII | + | |||||
| 10 | 1121 | ?, m5C | Hin6I* | − | M.HpyAORF1121 | 1050 (−) | |||
| M.Bsp6I, m5C | 36 | 35 (13) | Bsp6I | + | |||||
| 11 | 1208 | M.HpyI, m6A | M.HpyI, m6A | 93 | 94 (94) | NlaIII | − | M.HpyAORF1208 | 1131 (−) |
| 12 | 1352 | M.HinfI, m6A | M.HinfI, m6A | 63 | 60 (65) | HinfI | − | M.HpyAORF1352 | 1271 (−) |
| 13 | 1367 | M.MboII | M.MboII, m6A | 59 | 56 (56) | MboII | − | M.HpyAORF1367 | |
| 14 | 1368 | M.MthZI | ?, m4C | 33 | 24 (16) | M.HpyAORF1368 | |||
SSD, our similarity search data; m4C, m6A, or m5C, nature of modified base that could be generated by MTase (45). The active MTases and expressed ORFs are in boldface.
Numbers indicate percent identity of aligned sequences of MTases. The first number in the SSD column represents full-length MTases. In the TDB column, in most cases numbers indicate percent identity only between the most similar parts of amino acid sequences of aligned MTases. Numbers in parentheses indicate percent identity of putative TRDs.
RE sequences isospecific to the homologs of putative MTases given in columns TDB and/or SSD. Asterisks mark RE sequences isospecific to putative MTases, specificity of which was deduced by testing resistance or sensitivity of the DNA to REs (Table 1).
r/s, DNA resistance (−)/sensitivity (+) to REs described in footnote c.
ORF of putative H. pylori J99 MTase homologous to the H. pylori 26695 counterpart. (−) and (+) denote DNA resistance and DNA sensitivity, respectively, to REs described in footnote c (unpublished data).
Homology search.
Twenty-five ORFs encoding putative MTases were identified in the 26695 genome (46). Table 2 summarizes results of homology searches for 14 of these ORFs listed in the TDB and as found here, plus our experimental findings. First, our analyses agreed with TDB assignments in six cases: HP92 (M.MboIC), HP260 (M.EcoP151), HP478 (M.VspI), HP1208 (M.HpyI), HP1352 (M.HinfI), and HP1367 (M.MboII). However, the TDB annotations for remaining eight putative MTases [HP50 (M.DpnA), HP51 (M.DdeI), HP263 (M.HpaI), HP483 (M.HphI), HP593 (M.EcoP15I), HP910 (M.HindII) HP1121 (M.Bsp6I), and HP1368 (M.MthZI)] were not supported by comparison of MTase variable regions, which correspond to likely DNA TRDs. The TDB conclusions were also not supported in these eight cases by tests of resistance/sensitivity of 26695 genomic DNA to RE cleavage (Table 2).
The TDB annotation had suggested that HP50 and HP51 were similar to M.DpnA and M.DdeI, respectively, whereas our analyses revealed much higher homology to M.MnlIA and M.MnlIC, respectively. In addition, the putative MTases encoded by HP263 (TDB; homolog of M.HpaI), HP483 (TDB; homolog of M.HphI), and HP910 (TDB; homolog of M.HindII) shared more than 90% amino acid sequence identity with M.Hpy84-183I (recognition sequence CCGG), M.Hpy99XI (ACGT), and M.Hpy8I (GTNNAC), the genes for which were cloned from H. pylori strains other than 26695 (Stankevicius, unpublished; Minkute, personal communication; 20).
Specificities of H. pylori 26695 DNA MTases.
The specificity of a DNA MTase is characterized by a 2- to 8-bp recognition sequence (sequence specificity), position of the methylated base in it (site specificity), and nature of modified base (m5C, m6A, or m4C).
Recognition sequence.
Homology searches identified the following 11 H. pylori 26695 putative MTases as having amino acid sequence similarity to MTases of known specificity sufficient to predict that they would recognize the same site and methylate the same base (isomethylomers): M.HpyAORF50 (M.MnlIA), M.HpyAORF51 (M.MnlIC), M.HpyAORF92 (M.MboI), M.HpyAORF260 (M.EcoP15I), M.HpyAORF263 (M.Hpy84-183; renders DNA resistant to MspI RE), M.HpyAORF478 (M.VspI), M.HpyAORF483 (M.Hpy99XI; renders DNA resistant to TaiI), M.HpyAORF910 (M.Hpy8I), M.Hpy AORF1208 (M.HpyI), M.HpyAORF1352 (M.HinfI), and M.HpyAORF1367 (M.MboII) (Table 2). In the case of M.HpyAORF260, the type III EcoP15I RE was the only one not available for testing of the predicted specificities of these MTases, but later we found that HP260 encodes an M.TaqI-isospecific MTase, not one specific for EcoP15I sites (see below). All remaining 10 sequence-isospecific REs tested, except for MspI, TaiI, and Hpy8I, did not cleave 26695 genomic DNA (Tables 1 and 2). This resistance to REs provided evidence that all type II MTases of the predicted different sequence specificities that were inferred from the genome sequence, except for M.HpyAORF263P, M.HpyAORF483P, and M.HpyAORF910P, are produced in H. pylori 26695 grown in culture.
Two REs that did not cleave strain 26695 DNA, MnlI and MboII, are type IIS and recognize asymmetric DNA sequences (CCTC and GAAGA, respectively). Modification of an asymmetric target duplex requires two separate MTases, each specific for a different DNA strand, and type IIS MTases either consist of joint bifunctional enzymes or are represented by two separate MTases (23, 25). The homologs of both strand-specific MnlI MTases were found in the strain 26695 genome (Table 2). Also found was a homolog of the m6A MboII MTase (M.HpyAORF1367), which modifies the last adenine in the GAAGA sequence but cannot act on the complementary strand (TCTTC) because it is adenine specific. Adjacent to hpyAORF1367 is HP1368, which encodes a putative m4C MTase of unknown specificity. Perhaps this MTase (M.HpyAORF1368P) modifies the TCTTC strand. However, because modification of just one strand protects DNA against the cognate RE (31), resistance to MnlI and MboII did not prove that each of the MnlI- and MboII-specific MTases was produced.
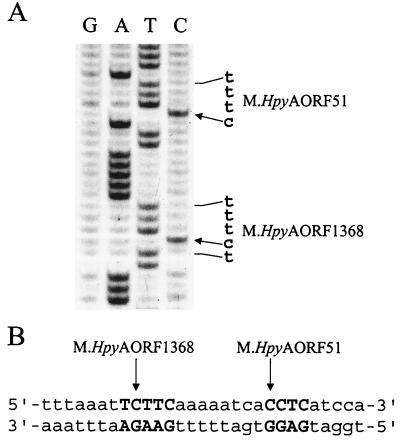
Production of M.HpyAORF51 (homolog of MnlIC, recognition sequence CCTC) and M.HpyAORF1368P (TCTTC) MTases was shown using the bisulfite sequencing protocol for mapping of m5C and m4C in genomic DNA (38, 49). As illustrated in Fig. 1, strain 26695 DNA contains methylated cytosines in m5CCTC and Tm4CTTC sequences. Because bisulfite sequencing does not differentiate m4C and m5C, the nature of methylated cytosines was inferred based on amino acid sequences of corresponding MTases.
FIG. 1.
Detection of methylated cytosines (m4C and m5C) within 5′-TCTTC and 5′-CCTC sequences of H. pylori 26695 genomic DNA, recognized by M.HpyAORF1368 and M.HpyAORF51 methylases, respectively. (A) Sequencing data after bisulfite treatment of the H. pylori 26695 genomic DNA. The methylated cytosines are indicated by arrows. Sequences on the right correspond to the bisulfite-converted methylated M.HpyAORF1368 and M.HpyAORF51 sites. (B) The recognition sequences of M.HpyAORF1368 and M.HpyAORF51 are shown in the bold; locations of modified cytosines are indicated by arrows.
The m4C MTase, which modifies TCTTC sequences (M.HpyAORF1368), represents an MTase whose specificity was not previously identified. It is noteworthy that in the cloned MboII R-M system, only an m6A MboII MTase (7) homolog of M.HpyAORF1367 was found. An MboII type IIS R-M system whose MTase modifies only one strand of a recognition sequence would be exceptional among type IIS R-M systems. This R-M system has not been thoroughly studied and most likely also contains two MTases, one of which (TCTTC specific) remains to be identified.
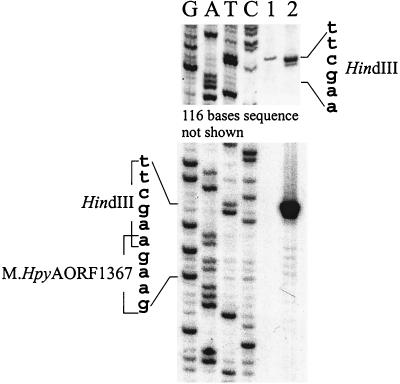
The high similarity of M.HpyAORF50 and M.HpyAORF1367 to MnlIA and MboII m6A MTases, respectively, suggested that they were specific for GmAGG and GAAGmA, respectively. The activity of M.HpyAORF1367 in vivo was tested using HindIII, which is sensitive to methylation of the 5′ outer A in its AAGCTT recognition sequence. M.MboII-specific DNA methylation should render DNA resistant to HindIII, where recognition sequences of MboII and HindIII overlap (GAAGm6AAGCTT). The 26695 genome contains such a nucleotide sequence at position 1008166, and close to it is a separate HindIII site (position 1008050). The product of a primer extension reaction through the segment containing these sequences after pretreatment of template DNA with HindIII was compared with the sequence of the corresponding segment. Just one fragment of the expected size, corresponding to cleavage at the individual HindIII site (Fig. 2, lane 1) was detected. The absence of the other fragment indicates that M.HpyAORF1367 is produced and protects the DNA against HindIII at the MboII/HindIII overlap site. In control experiments with a PCR fragment (which is not methylated), both fragments of the expected size were generated, showing that each of these sequences is indeed potentially available to HindIII (Fig. 2, lane 2).
FIG. 2.
Detection of the modified adenine in the M.HpyAORF1367 recognition sequence 5′-GAAGA. Lanes G, A, T, and C, sequence ladders through the overlapping M.HpyAORF1367/HindIII sites, 5′-GAAGAAGCTT (position 1008166), and the individual HindIII site 5′-AAGCTT (position 1008050). Lanes: 1, primer extension reaction of H. pylori 26695 DNA after digestion with HindIII; 2, primer extension reaction of PCR fragment digested with HindIII.
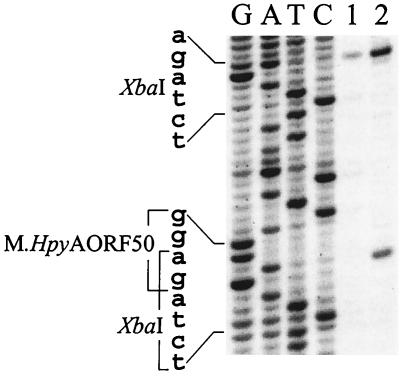
Production of M.HpyAORF50 was shown by an approach analogous to that described above for M.HpyAORF1367 and was based on sensitivity of XbaI to modification of 3′A in recognition sequence TCTAGA. Specifically, DNA was protected against XbaI at the site of overlap between its recognition sequence and that of M.HpyAORF50 (GAGG) (TCTAGm6AGG; genomic position 141547) (Fig. 3).
FIG. 3.
Detection of the modified adenine in the M.HpyAORF50 recognition sequence 5′-GAGG. Lanes G, A, T, and C, sequence ladders through the overlapping XbaI/M.HpyAORF50 site, 5′-CTCAGAGG (position 141547), and the individual XbaI site, 5′-TCTAGA (position 141557). Lanes: 1, primer extension reaction of H. pylori 26695 DNA after digestion with XbaI; 2, primer extension reaction of PCR fragment digested with XbaI.
Our results (Table 1) indicated that strain 26695 also produces MTases isospecific with M.TaqI (TCGA), M.PstI (CTGCAG), and M.Hin6I (GCGC). The deduced sequence specificities of these MTases could not be supported by amino acid sequence comparisons and thus should be regarded as preliminary. This conclusion is especially true for the CTGCAG-specific MTase. In this case (in contrast to TCGA and GCGC-specific MTases), analysis of the data on the resistance or sensitivity to 147 REs used in our experiments does not exclude the possibility of resistance to PstI being due to a hypothetical MTase of a lower specificity (CTGCA, TGCAG, etc.).
Cloning of MTases specific for TCGA, CTGCAG, and GCGC.
Strain 26695 genomic DNA cloning and RE selection were used to identify ORFs encoding TCGA-, CTGCAG-, and GCGC-specific MTases. Recombinant plasmids resistant to TaqI, PstI, and Hin6I were isolated, DNA sequences at both ends of the cloned fragments (150 to 200 nucleotides long) were determined, and the gene contents of the cloned DNA segments were determined using the strain 26695 genome sequence (46). Such analysis was continued until cloned inserts that contained only one intact ORF were found. Recombinant plasmid pHpyTaqIM2.5, resistant to TaqI, contained HP260; pHpyPstIM2.5 contained HP593; and pHpyHin6IM3.1 contained HP1121. Each of these ORFs had also been identified as encoding DNA MTases by amino acid sequence analysis (Table 2). The recombinant plasmids containing these MTase genes were also resistant to the expected set of related enzymes that had been identified in RE tests of strain 26695 genomic DNA.
Site specificity of MTases and nature of modified bases.
The amino acid sequence of the catalytic motif is characterized by well-defined differences between m4C, m6A, and m5C MTases (18, 45), which can be used to predict the modified bases they generate. Corresponding data for H. pylori 26695 MTases are given in Tables 1 and 2. In the cases of M.HpyAORF50 (M.MnlIA), M.HpyAORF51 (M.MnlIC), and M.HpyAORF1367 (M.MboII), predictions on the nature of the modified base and its position within a defined recognition sequence (based on high similarities of the MTases to their isomethylomers, shown in parentheses) were confirmed by direct experiments (see above). The position of m4C within the recognition sequence of the unique M.HpyAORF1368 MTase was also determined in the same experiments.
The positions and base specificities of modification were inferred easily for M.HpyAORF92, M.HpyAORF1208, and M.HpyAORF1352 as follows: (i) each is predicted to be an m6A MTase; (ii) each enzyme's recognition sequence contains only one adenine base; and (iii) these MTases share high similarity with their isomethylomers (Table 2). The first two arguments also apply for M.HpyAORF260 and M.HpyAORF593, which most likely are also m6A MTases.
The recognition sequence of m5C-specific M.HpyAORF1121 MTase (GCGC) contains two modifiable cytosines. From the data provided in Table 1, we conclude that this MTase modifies the inner cytosine (Gm5CGC), thereby conferring resistance to all tested REs of the GCGC family. Modification of the outer cytosine could not interfere with DNA cleavage by Bsp143II, contrary to what is observed. The site specificity of M.HpyAORF478 (m6A) cannot be predicted from available data. Its recognition sequence, ATTAAT, contains more then one modifiable base, and the site specificity of its homolog M.VspI has not yet been determined.
DISCUSSION
This study reports the specificities of 11 MTases produced by H. pylori strain 26695 cultivated in vitro and the genes encoding them. Unpublished data on specificity of some cloned H. pylori 26695 MTases (L. F. Lin and H. Kong, unpublished data cited in REBASE, version 007) (41) confirm our conclusions on the specificity of MTases encoded by HP92, HP1352, and HP1367. Evidence was also obtained that a cloned MTase encoded by HP54 recognizes GACNNNNNGTC (Lin and Kong, unpublished data cited in REBASE, version 007). We could not confirm production of a DNA MTase of such sequence specificity by H. pylori 26695, however, because genomic DNA was susceptible to digestion by the RE Eam1105I.
Recent data on digestion of DNAs from 16 H. pylori strains including 26695 with 15 REs (52) support our conclusions on expression of type II MTases M.HpyAORF1121, M.HpyAORF1208, and M.HpyAORF1352. Protection of MnlI and MboII sites was also detected. Resistance to Tsp45I cleavage was also found in that study but not in ours. No homolog of H. pylori J99 ORF 45, which encodes an isomethylomer of Tsp45I MTase (20), exists in the 26695 genomic sequence. Therefore, either strain 26695 cannot produce an isomethylomer of Tsp45I MTase or the enzyme may be encoded by a nonhomologous gene whose expression we did not observe.
The putative MTases encoded by HP263, HP483, and HP910 share more than 90% identity with M.Hpy84-183I (Stankevicius, unpublished), M.Hpy99XI (20), and M.Hpy8I (Minkute, personal communication), respectively. However, MspI, TaiI, and Hpy8I REs did cleave H. pylori 26695 DNA, indicating that these putative MTases (M.HpyAORF263P, M.HpyAORF483P, and M.HpyAORF910P) are not produced by strain 26695 or that the genes for them are mutant.
The DNA MTases of procaryotes may exist as separate enzymes or as components of R-M systems. The latter are strain specific, whereas some of the former MTases are species specific and involved in general cellular processes (5, 32, 33, 40, 48), including the regulation of gene expression. The HpyI-specific MTase has been found in all 60 H. pylori strains from various parts of the world that were tested, including strain 26695 (51, 52; J. Vitkute, unpublished data). Therefore, M.HpyI may be considered a candidate for a species-specific MTase (52).
Strain-specific DNA modification genes might conceivably affect strain-specific phenotypic traits, host specificity, adaptability to changing gastric conditions, or virulence. The amino acid sequence comparison of putative MTases encoded by H. pylori 26695 and H. pylori J99 ORFs has revealed both similarities and differences (3). We found that 13 MTases are expressed by strain J99, as assessed by resistance of its DNA to REs (unpublished data). Seven of them correspond to MTases that are expressed in 26695 as well (Table 2). We also found that strain J99 expresses MTases that render DNA resistant to MspI, TaiI, Tsp45I, XbaI, BseDI, and RsaI. These modification activities were not detected in strain 26695. MTases protecting DNA against MspI, TaiI, Tsp45I, and BseDI are encoded by JHP248, JHP435, JHP45, and JHP629, respectively (20). Of them, only JHP248 and JHP435 have homologs in strain 26695 (HP263 and HP483, respectively), where they are not expressed or inactivated. The genes rendering J99 DNA resistant to XbaI and RsaI remain to be determined. The data presented here illustrate the diversity of the two H. pylori strains with respect to MTase content and expression profile. Screening of DNA resistance against REs for numerous H. pylori strains provides a simple means for evaluating DNA modification potential in this species. Such a study would provide preliminary data allowing species-specific and strain-specific MTases to be distinguished; this would be useful in epidiomological studies and would help elucidate the mechanism of genome evolution.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants AI38166, DK53727, and P30DK2574.
We are grateful to MBI Fermentas for generous gifts of restriction endonucleases and other enzymes. We thank M. Nelson for critically reading the manuscript and for valuable comments and suggestions, A. Lubys for providing plasmid pBR-R and strain AL2268, A. Lubys and J. Minkute for unpublished sequences of M.MnlI and M.Hpy8I MTases, and J. Lubiene for DNA preparation.
REFERENCES
- 1.Akopyants N S, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopyants N S, Fradkov A, Diatchenko L, Hill J E, Siebert P D, Lukyanov S A, Sverdlov E D, Berg D E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Atherton J C, Cao P, Peek R M, Tummuru K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 5.Barras F, Marinus M G. The great GATC:DNA methylation in E. coli. Trends Genet. 1989;5:138–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bocklage H, Heeger K, Muller-Hill B. Cloning and characterization of the MboII restriction-modification system. Nucleic Acids Res. 1991;19:1007–1013. doi: 10.1093/nar/19.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten B A, Nou X, Kaltenbach L S, Low D A. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 9.Brooks J E, Roberts R J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982;10:913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang N, Taylor D E. Use of pulsed-field agarose gel electrophoresis to size genomes of Campylobacter species and to construct a SalI map of Campylobacter jejuni UA580. J Bacteriol. 1990;172:5200–5217. doi: 10.1128/jb.172.9.5211-5217.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen E Y, Seeburg P H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 12.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128 kDa immunodominant antigen of Helicobacter pylori association with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donahue, J. P., R. M. Peek, L. J. van Doorn, S. A. Thompson, Q. Xu, M. J. Blaser, and G. G. Miller. Analysis of iceAI transcription Helicobacter pylori. Helicobacter 5:1–12. [DOI] [PMC free article] [PubMed]
- 15.Dubois A, Berg D E, Incecik E T, Fiala N, Heman-Ackah L M, Perez-Perez G I, Blaser M J. Transient and persistent experimental infection of nonhuman primates with Helicobacter pylori: implications for human disease. Infect Immun. 1996;64:2885–2891. doi: 10.1128/iai.64.8.2885-2891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 17.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 18.Klimasauskas S, Timinskas A, Menkevicius S, Butkiene D, Butkus V, Janulaitis A. Sequence motifs characteristic of DNA [cytosine-N4] methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989;17:9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimasauskas S, Nelson J L, Roberts R J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991;19:6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong H, Lin L F, Porter N, Stickel S, Byrd D, Posfai J, Roberts R J. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 2000;28:3216–3223. doi: 10.1093/nar/28.17.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labigne A, de Reuse H. Determination of Helicobacter pylori pathogenicity. Infect Agents Dis. 1996;5:191–202. [PubMed] [Google Scholar]
- 22.Lipman W R, Pearson D J. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Looney M C, Moran L S, Jack W E, Feehery G R, Benner J S, Slatko B E, Wilson G G. Nucleotide sequence of the FokI restriction-modification system: separate strand specificity domains in the methyltransferase. Gene. 1989;80:193–208. doi: 10.1016/0378-1119(89)90284-9. [DOI] [PubMed] [Google Scholar]
- 24.Lubys A, Menkevicius S, Timinskas A, Butkus V, Janulaitis A. Cloning and analysis of translational control for genes encoding the Cfr9I restriction-modification system. Gene. 1994;141:85–89. doi: 10.1016/0378-1119(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 25.Lubys A, Lubiene J, Kulakauskas S, Stankevicius K, Timinskas A, Janulaitis A. Cloning and analysis of the genes encoding the type IIs restriction-modification system HphI from Haemophilus parahaemolyticus. Nucleic Acids Res. 1996;24:2760–2766. doi: 10.1093/nar/24.14.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 28.Marko M A, Chipperfield R, Birnboim H C. A procedure for the large scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982;121:382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- 29.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 30.MBI Fermentas. MBI Fermentas catalog, 2000/2001. 2000. MBI Fermentas, Vilnius, Lithuania. [Google Scholar]
- 31.McClelland M, Nelson M, Raschke E. Effect of site-specific modification on restriction nucleases and DNA modification methyltransferases. Nucleic Acids Res. 1994;22:3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messer W, Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 33.Modrich P. Methyl-directed DNA mismatch repair. J Biol Chem. 1989;264:6579–6600. [PubMed] [Google Scholar]
- 34.Nelson M, Burbank D, Van Etten J L. Chlorella viruses encode multiple DNA methyltransferases. Biol Chem. 1998;379:423–428. doi: 10.1515/bchm.1998.379.4-5.423. [DOI] [PubMed] [Google Scholar]
- 35.Noyer-Weidner M, Trautner T A. Methylation of DNA in prokaryotes. In: Jost J P, Saluz H P, editors. DNA methylation: molecular biology and biological significance. Basel, Switzerland: Birkhauser Verlag; 1993. pp. 39–108. [Google Scholar]
- 36.Peek R M, Thompson S A, Donahue J P, Tham K T, Atherton J C, Blaser M J, Miller G G. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physic. 1998;110:531–544. [PubMed] [Google Scholar]
- 37.Posfai J, Bhagwat A S, Posfai G, Roberts R J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17:2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raizis A M, Schmitt F, Jost J P. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal Bioch. 1995;226:161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- 39.Riegg S J, Dunn B E, Blaser M J. Microbiology and pathogenesis of Helicobacter pylori. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. Philadelphia, Pa: Raven Press; 1995. pp. 535–50. [Google Scholar]
- 40.Roberts D, Hoopes B C, McClure W R, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 41.Roberts R J, Macelis D. REBASE—restriction enzymes and methylases. Nucleic Acids Res. 1999;27:312–313. doi: 10.1093/nar/27.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor D E, Eaton M, Chang N, Salama S M. Construction of a Helicobacter pylori genome map and demonstration of diversity at the genome level. J Bacteriol. 1992;174:6800–6806. doi: 10.1128/jb.174.21.6800-6806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timinskas A, Butkus V, Janulaitis A. Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene. 1995;157:3–11. doi: 10.1016/0378-1119(94)00783-o. [DOI] [PubMed] [Google Scholar]
- 46.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleishmann R D, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 47.Tummuru M K R, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Woude M, Hale W B, Low D A. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J Bacteriol. 1998;180:5913–5920. doi: 10.1128/jb.180.22.5913-5920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vilkaitis G, Klimasauskas S. Bisulfite sequencing protocol displays both 5-methylcytosine and N4 methylcytosine. Anal Biochem. 1999;271:116–119. doi: 10.1006/abio.1999.4116. [DOI] [PubMed] [Google Scholar]
- 50.Walsh E J, Moran A P. Influence of medium composition on the growth and antigen expression of Helicobacter pylori. J Appl Microbiol. 1997;83:67–75. doi: 10.1046/j.1365-2672.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 51.Xu Q, Peek R M, Miller G G, Blaser M J. The Helicobacter pylori genome is modified at GATC by the product of hpyIM. J Bacteriol. 1997;179:6807–6815. doi: 10.1128/jb.179.21.6807-6815.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Q, Morgan R D, Roberts R J, Blaser M J. Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc Natl Acad Sci USA. 2000;97:9671–9676. doi: 10.1073/pnas.97.17.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Q, Stickel S, Roberts R J, Blaser M J, Morgan R D. Purification of the novel endonuclease, Hpy1881, and cloning of its restriction-modification genes reveal evidence of its horizontal transfer to the Helicobacter pylori genome. J Biol Chem. 2000;275:17086–17093. doi: 10.1074/jbc.M910303199. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Nelson M, Nietfeldt J, Xia Y, Burbank D, Ropp S, Van Etten J L. Chlorella virus NY-2A encodes at least 12 DNA endonuclease/methyltransferase genes. Virology. 1998;240:366–375. doi: 10.1006/viro.1997.8936. [DOI] [PubMed] [Google Scholar]