Abstract
LC–MS/MS is a major analytical platform for metabolomics, which has become a recent hotspot in the research fields of life and environmental sciences. By contrast, structure elucidation of small molecules based on LC–MS/MS data remains a major challenge in the chemical and biological interpretation of untargeted metabolomics datasets. In recent years, several strategies for structure elucidation using LC–MS/MS data from complex biological samples have been proposed, these strategies can be simply categorized into two types, one based on structure annotation of mass spectra and for the other on retention time prediction. These strategies have helped many scientists conduct research in metabolite-related fields and are indispensable for the development of future tools. Here, we summarized the characteristics of the current tools and strategies for structure elucidation of small molecules based on LC–MS/MS data, and further discussed the directions and perspectives to improve the power of the tools or strategies for structure elucidation.
Keywords: LC–MS/MS, Structure elucidation, Complex biological samples
1. Introduction
The metabolome, first introduced in 1998, refers to the complement of small molecules in biological samples [1]. Small molecules have been studied extensively, as many of them have special biological significance for cell biology, physiology and medicine [2]. For plants, small molecules can act as defense compounds (glucosinolate in Brassicaceae, gossypol in cotton, etc.), and plant developmental and growth regulators [3]. In addition, small molecules can also function as signaling molecules, immune modulators, endogenous toxins, and environmental sensors [4]. As the chemicals and physical properties of small molecules are very diverse, the detection and identification of small molecules accordingly becomes a major bottleneck for metabolomics [5], [6], [7].
Liquid chromatography–tandem mass spectrometry (LC–MS/MS) is one of the major analytical platforms used in the small molecule identification process [8]. Liquid chromatography separates mixtures with multiple components can be mainly divided into three parts: reversed-phase liquid chromatography (RPLC), hydrophilic interaction liquid chromatography (HILIC) and ion chromatography (IC) [9]. Tandem mass spectrometry provides mass spectral information that can be used for identifying separated components [10]. The mass spectrometry can be classified into in-time (ion traps, FTICR) and in-space (quadrupoles, TOFs) mass analyzers based on the principle of different platforms [11]. The mass spectrometry for LC–MS/MS commonly produces weak or absent in-source fragment ions (MS1) with electrospray ionization, compared to the mass spectrometry for GC–MS/MS using electron ionization [12]. The precursor ions are selected to generate MS/MS spectra in collision-induced dissociation (CID) or higher energy collisional dissociation (HCD) modes which can produce complementary fragments for further detection and structure annotation [10], [13]. Compared to nuclear magnetic resonance (NMR) and gas chromatography–tandem mass spectrometry (GC–MS/MS), LC–MS/MS can produce more data and requires relatively simple extraction steps, which makes it more popular for exploring the metabolites in complex biological samples, especially for metabolomics [3], [14]. For example, many features, defined as unique ions with MS1 and retention time information [15], can be routinely detected in biological samples, and follow-up studies were conducted to uncover the regulatory mechanism of some identified metabolites [16], [17], [18], [19], [20], [21], [22], [23], [24]. Regrettably, most features reported in these studies remains unknown due to the vast diversity of metabolites in biological samples, the lack of corresponding spectra in MS/MS spectral libraries, the disunity of collision energy with different platform, the existence of noise signal, the complexity of LC conditions optimization and the lack of large and diverse RT training sets [8], [14]. For the past twenty years, many tools and methods have been developed for annotating the features from LC–MS/MS analysis [10], [25]. These tools all involved in the metabolite identification are based on the information of mass spectra and retention time from LC–MS/MS [10], [14], [25]. In this review, we will discuss the characteristics of the various tools and strategies for metabolite identification based on LC–MS/MS data, and analyzes the ways to further improve the power of the tools or strategies for structure elucidation.
2. Acquisition and curation of LC–MS/MS data from complex biological samples
The features, with information of chromatographic peak (retention time) and mass spectral peak (m/z), can be detected by multiple software packages or frameworks such as MetAlign [26], OpenMS [27], MZmine [28], XCMS [29] and CAMERA [30]. In a biological sample, over ten thousand features can be detected, while there are a large number of artifactual peaks, chemical contaminants, and signal redundancies, which hamper the identification of “true” metabolites [31], [32], [33], [34], [35]. Those peaks are mainly arising from background, isotopes, adducts, homodimers, heterodimers and in source fragments [31], [32], [33], [36], which can be identified based on retention time grouping, correlations between features, features clustering by retention time and calculating pairwise correlations, chromatographic peak-shape similarity, relative adduct frequency, isotope detection and specifying common adducts and neutral loss events [30], [32], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48]. The filtered features can be then identified via MS/MS spectra [36], [38].
LC–MS/MS data from complex biological samples is produced by liquid chromatography coupled with tandem mass spectrometry in targeted/untargeted MS based-metabolomics [10]. Targeted metabolomics involves multiplexed analysis of a set of defined metabolites using multiple reaction monitoring (MRM) and is limited with metabolites coverage compared to untargeted metabolomics. In untargeted metabolomics, data-independent acquisition (DIA) and data-dependent acquisition (DDA) are two approaches to acquire MS/MS spectra [49]. In DDA acquisition workflows, the precursor ions exceed a predefined threshold of intensity or other predefined criteria are selected from a full scan analysis for further fragments obtaining [50]. DDA generally produces relatively good quality tandem mass spectra, which are friendly to the subsequent structure elucidation [50]. In DIA acquisition workflows, precursor ion and fragments information are obtained from alternating scans acquired at either low or high collision energy in the collision chamber [51]. For conventional DIA, including All-Ion Fragmentation (AIF), MSALL and MSE approaches, it is much difficulty to deduce the physical relationship between the precursor ions and their fragments for the wide scan range and the diversity of metabolites in biological samples [49], [51]. To enhance selectivity, SWATH (sequential window acquisition of all theoretical fragment-ion spectra) narrows the precursor ion selection range to 20–50 Da consecutive isolation windows and gives a higher quality spectra [52]. SWATH can obtain a similar quantitative result with MRM, while deconvolution algorithms or tools are also indispensably to deduce the assignment of precursors to the corresponding fragment ions and improve the MS2 spectral quality, such as MS-Dial, Progenesis QI (Waters Co., Manchester, UK) and MasterView (AB Sciex, USA) [53], [54].
3. Strategies for structure annotation of mass spectra
Strategies for structure annotation of mass spectra can mainly classified into three categories which are separately based on authentic standard compounds, public/commercial reference spectral libraries and an in-silico approach. The strategy based on authentic standard compounds is the earliest developed road to illustrate the molecular structures of mass spectra, and is sufficient for ‘Level 1′ annotations (confident 2D structure annotations) in metabolomics. However, a certain amount of pure chemical standards is essential for this strategy and most metabolites are not commercially available, which make it often difficult and time-consuming in many instances [55], [56]. Structure annotation of mass spectra relies on the searches of public/commercial reference spectral libraries can result in ‘Level 2′ annotations (probable structures) in metabolomics. This strategy could provide more information for mass spectra, while the number and reliability are extremely reliant on the reference spectral libraries, which are still limited compared with the number of potential metabolites in complex biological samples [25], [56]. The third strategy utilizes quantum chemistry, heuristic-based methods, chemical reaction-based methods, machine learning to predict the in-silico mass spectra of a molecular library, or annotate the substructures of query mass spectra and only requires a molecular structure library, rather than reference spectral libraries. This strategy can provide a large number of annotations, however, the accuracy of the identification is relatively low to ‘Level 3′ annotations (tentative structure candidates or putatively characterized compound classes), or even equal to ‘Level 4′ annotations (formula determined) [8], [25], [56].
3.1. Structure elucidation based on authentic standard compounds
Authentic standard compounds can be used for targeted metabolomics, which focuses on several metabolites or a specific category of metabolites. As a comparison of retention times with references limits the range of candidates, a low-resolution LC–MS/MS analyzer is sufficient for targeted metabolomics. In addition, the metabolites in untargeted metabolomics can also be identified with unique 2D structures based on authentic standard compounds [57], [58]. The key step of this strategy is to compare the query mass spectra and retention time to that of purified authentic standards, which can be obtained through purchases from chemical companies or isolation from complex biological samples or via enzyme-based synthesis from other purified metabolites by enzymes [25].
Purchasing authentic standard compounds is the most convenient way to conduct targeted metabolomics based on LC–MS/MS, while this strategy limits targeted metabolomics to common metabolites. For example, some amino acids, catecholamines, lipids and steroids were detected in urine samples in a targeted manner [59]. In addition to common metabolites, some species-characteristic metabolites were also identified and detected by this strategy. As an example, glucosinolates are distinctively present in nearly all members of the plant order Capparales, and are well studied as a model for research on secondary metabolism. The qualitative and quantitative analysis of glucosinolates is relatively easier than that of some other secondary metabolites [60]. In recent years following the development of synthesis and separation technology, some vendors can provide thousands of standards for researchers, such as IROA Technologies LLC (https://www.iroatech.com/), Sigma–Aldrich (https://www.sigmaaldrich.cn/), and Agilent (https://www.agilent.com.cn/).
3.2. Structure elucidation based on public/commercial reference spectral libraries
The query mass spectra can be additionally identified by searching the public/commercial reference spectral libraries, which are built with authentic reference standards by institutions and companies around the world. Thanks to the great progress that mass spectrometry technology and chemical synthesis/isolation techniques have made in the past two decades, many mass spectral databases have been established and developed to extend millions of reference mass spectral data for diverse instruments and different collision energies, such as MassBank of North America (MoNA) (https://mona.fiehnlab.ucdavis.edu/), the Golm Metabolome database (https://gmd.mpimp-golm.mpg.de/), MassBank (https://massbank.eu/MassBank/), METLIN (https://metlin.scripps.edu/), mzCloud (https://www.mzcloud.org/), GNPS (https://gnps.ucsd.edu/), NIST 20 (https://www.nist.gov/) and Wiley Science Solutions (https://sciencesolutions.wiley.com/) [61]. In addition, some special spectral libraries were established for particular researches, such as ReSpect (https://spectra.psc.riken.jp/) for phytochemicals-related researches, HMDB (https://hmdb.ca/) for the human metabolomics.
MassBank is the first public repository of mass spectra of small chemical compounds for life sciences, and the mass spectra it houses are from different instruments and multiple contributors [62]. With the efforts that global chemists and computer scientists have made, many more sharing platforms have been established for research from various fields and have greatly promoted the development of metabolomics. MoNA is a centralized repository with 695,425 spectra, including 145,316 MS/MS spectra, with the spectra contained is mainly being contributed by MassBank, ReSpect, HMDB, GNPS, LipidBlast, Vaniya/Fiehn Natural Products Library and RIKEN PlaSMA. In addition to the free public spectral libraries, several commercial spectral libraries were established with well curated spectra and enriched contents. HMDB is a comprehensive database on small molecules from Homo sapiens and contains 64,923 experimental MS/MS spectra of 4,064 metabolites and 1,440,324 in-silico MS/MS spectra of 217,920 metabolites [63]. GNPS is a web-based mass spectrometry ecosystem, which also collect the MS/MS spectra from public spectral libraries, including Massbank, ReSpect, HMDB, CASMI [64]. METLIN is the largest library of mass spectra among all of the public and commercial spectral libraries, and hosts over 850,000 molecular standards with over 4,000,000 curated high-resolution tandem mass spectra [65]. NIST 20 is another commercial spectral library contributed by many researchers currently containing 1,320,389 spectra of 185,608 precursor ions from 30,999 chemical compounds. Among all of the spectra in the library, both High-Resolution, Accurate-Mass (HRAM) MS/MS (1,026,712 spectra for 27,840 chemical compounds) and Low-Resolution MS/MS (215,649 spectra for 28,559 chemical compounds) are represented. In addition to the small molecule, 90,244 spectra of 6,803 precursor ions from 1,904 peptides were also included in this library. mzCloud is specialized with high quality spectral trees of MSn spectra, which are generated with various collision energies. As each tree represents a molecule in this library, 19,515 molecules are contained with 2,310,148 positive mass spectra, and 7,875 molecules are contained with 850,580 negative mass spectra.
In addition to the size and quality of spectral libraries, the algorithms of the search systems employed affect the outcomes of the structure annotation of querying mass spectra against public/commercial reference spectral libraries. In contrast to EI-based spectra (produced by GC–MS), ESI-based spectra (produced by LC–MS/MS) tend to be less reproducible, especially for the cross-instrument comparisons, which raise higher requirements for the search system [66], [67], [68]. In this regard, the traditional mass spectral library search algorithms were firstly utilized for EI-based spectra, such as dot-product, Euclidean distance and probability-based matching (PBM) system. For dot-product and Euclidean distance, each mass spectrum can be considered as a point in a multidimensional hyperspace, with the axis presented by the mass (m/z) and the position of the point presented by the intensities of those masses [69]. Dot-product, as the name implies, calculates the dot product of two vectors, of which one is the vector from the coordinate origin to the point of a query mass spectrum and the other one is a library of mass spectrum. By analogy, Euclidean distance is the algorithm that calculates the Euclidean distance between the two points of the query mass spectrum and a library mass spectrum. Comparing the two algorithms mentioned above, the PBM system is relatively complex and cannot be described as an analytic function. However, the PBM system can to some extent avoid mismatch results when the metabolite with the query spectrum is not in the reference library [70]. Among those algorithms, dot-product derived algorithm has gained widespread use for ESI-based spectra [71], [72]. To overcome the great differences of ESI-based spectra from a range of instruments, some sophisticated matching algorithms different from the routine algorithms were created [73], [74], [75]. In an independent approach, X-Rank is based on statistical relations between mass over charge values, ordered by intensities, rather than taking into account absolute or relative intensities, which makes it more effective in supporting cross platform identification [75]. Recently, spectral entropy similarity has been developed in addition to the previous approaches, and has been proven to outperform all classic algorithms by adapting concepts from information theory [76].
3.3. Structure elucidation based on an in-silico approach
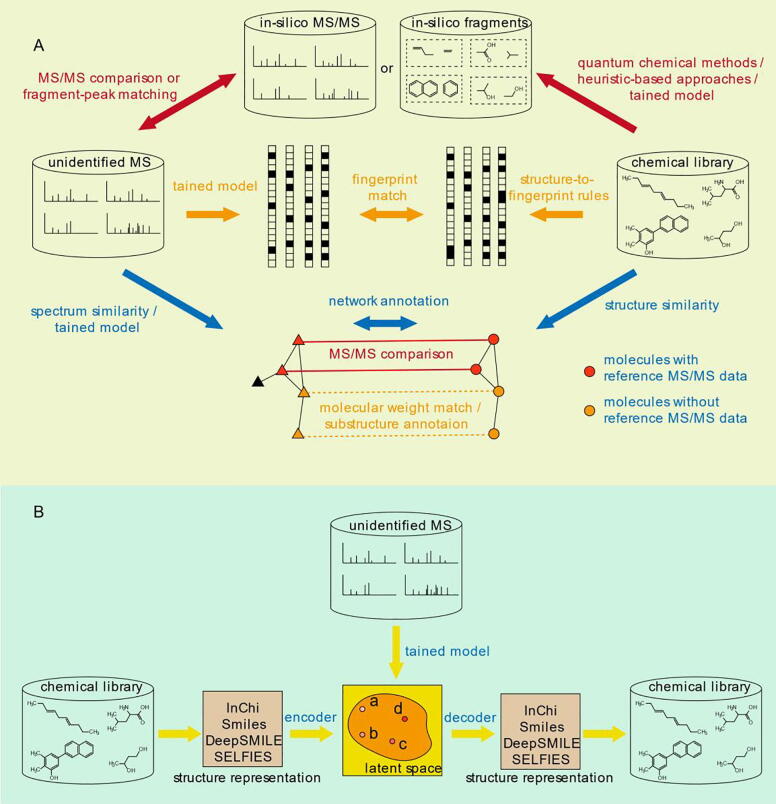
The range of metabolites in complex biological samples is daunting, with hundreds of thousands of metabolites representing the lower end of estimates, and this number is continually growing [2], [77], [78], [79]. PubChem (https://pubchem.ncbi.nlm.nih.gov/) [80] and ChemSpider (https://www.chemspider.com/) [81] are the world’s two largest collections of freely accessible chemical information. While the number of compounds contained in PubChem and ChemSpider has reached 111 million and 114 million, respectively, the number of metabolites with MS data collected in the reference library is considerably more limited, with the METLIN database, which is the largest experimental spectral library, covers a mere 850,000 metabolites, which represents less than 1 % of the compounds found in PubChem or ChemSpider [65]. To overcome this difficulty, in-silico qualitative tools have been developed in the last decade [82], [83], [84], [85], [86], [87]. Early tools were mainly developed by commercial companies, such as Mass Frontier by Thermo Fisher Scientific. Afterwards, many open-source mass spectrometry identification software programs appeared, and most of those tools could be divided into four categories (Fig. 1).
Fig. 1.
The strategies for metabolite identification based on in-silico approach. (A) From top to bottom workflow represent, in order, the first strategy indicated by the red arrows (generation of in-silico spectral libraries), the second strategy indicated by the orange arrows (substructure annotation for ESI-based spectra), the third strategy indicated by the blue arrows (network-based strategies in metabolite identification for spectrum), (B) The workflow of the fourth strategy indicated by the yellow arrows (metabolite identification for mass spectra with generative methods). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.1. Generation of in-silico spectral or fragmental libraries
The earliest strategy for structure elucidation using MS/MS data based on an in-silico approach is to predict mass spectra or fragments from structures of chemical species subjected to a fragmentation process. Generally, a SMILES string or other molecular string is utilized for representing the structure of an individual chemical [88], [89], [90], [91]. The fragmentation process can be distinguished as quantum chemistry, heuristic-based methods, and machine learning [8], [82].
Fragmentation methods based on quantum chemistry (QC) were first established for structure annotation of EI-based spectra [92], [93], [94]. Then, a quantum mechanical and molecular mechanical combined approach was built for the structure annotation of ESI-based spectra of large polypeptides [95]. Similarly, QCMS2 is another approach that combines QC with a heuristic approach, and can also simulate and understand the mass spectra of peptides [96]. QC-FPT takes the dominant fragment peaks and 3D candidate structures from a particular experiment and attempts to model the collision-induced dissociation [97]. In contrast to the QC-FPT, ChemFrag supports fragment ion annotations of an entire spectrum, rather than predicting the dominant mass spectrum [98]. Although fragmentation methods based on quantum chemistry have been developed for nearly twenty years, a large amount of computational power is required to implement this strategy, especially for large molecules, compared with tools based on other strategies, such as machine learning approaches and heuristic-based approaches [82].
Heuristic-based methods generate in-silico fragments relying on a collection of general heuristic rules of fragmentation. For this strategy, tools were developed for structure elucidation based on the match scores between in-silico fragments and experimental peaks. Among those tools, Mass Frontier (HighChem Ltd., Bratislava, Slovakia) was one of the earliest developed tools and involves cleaving the bond in the structure based on reactions described in the literature. In addition, MetFrag first generates all possible fragments of the candidate structures using simple bond-breaking rules and combinatorial fragmentation, and then the fragmentation trees were traversed by breadth-first search (BFS) [99]. Compared to MetFrag, MIDAS traversed the fragmentation trees by depth-first search (DFS), which was more memory-efficient than BFS [100]. MAGMA performed the fragmentation of a structure or a substructure by removing each heavy atom sequentially (i.e. non-hydrogen atoms), and can be used for structure annotation of MSn data [101], [102]. As hydrogens always rearrange during bond cleavage in low-energy CID, MS-FINDER applied nine hydrogen rearrangement (HR) rules to generate in-silico fragments [103]. DEREPLICATOR + produced in-silico fragments by disconnecting bridges and 2-cuts at N—C, O—C and C—C bonds, and could identify many variants within spectral networks [104]. It was recently demonstrated that DEREPLICATOR + improves the structure elucidation of general metabolites and natural products above that of the previous developed DEREPLICATOR, which was a Heuristic-based algorithm that generated fragments for peptidic natural products [104], [105]. Heuristic-based methods are also utilized for structure elucidation for a group of substances. For example, the two heuristic-based tools, LipidBlast and LipidMatch, are popular tools for lipid identification [106], [107]. The in-silico spectra of LipidBlast contains 212,516 spectra covering 119,200 compounds from 26 lipid compound classes, were calculated with this strategy [108]. In addition, heuristic-based methods can be worth “adjusting” the relative abundances of fragment ions within in-silico generated tandem mass spectra to facilitate metabolite identification [109]. Thus, heuristic-based methods are considered to be very helpful strategies for the structure annotation of fragment ions in the field of biochemistry.
The advent of machine learning has expedited the structure annotation of experimental mass spectra. ISIS is one of the earliest tools based on machine learning to find accurate bond cleavage rates for collision-induced dissociation in an ESI-based spectrum, and targets the identification of lipids [110]. CFM-ID, arguably the most popular machine learning approach based on the Markov module, was published to identify multiple collision energy spectra [111]. The results showed that CFM-ID obtained substantially better rankings for the correct candidate than existing methods (MetFrag and FingerID) on tripeptide and other metabolite data, when querying PubChem [80] or KEGG [112] for candidate structures of similar mass [111]. A new version of CFM-ID (version 4.0) has been published recently, and it has been proven to be significantly more accurate than previous versions [113]. With the advance of artificial neural networks (ANNs), deep learning has been utilized for the structure annotation of a considerable amount of experimental mass spectra, such as NEIMS for EI-based spectra [114]. By contrast, there has not been any deep learning method for directly predicting ESI-based spectra of a given molecular. However, given the rapid development of public ESI-based spectral libraries, it should be possible to develop such tools in the future.
3.3.2. Molecular fingerprints prediction for ESI-based spectra
Predicting molecular fingerprints for ESI-based spectra is another strategy for metabolite identification based on in-silico methods. Commonly, the molecular fingerprint can be represented by a vector, in which each number represents the possibility for the presence of the molecular property. The predicted fingerprint of the unknown compound can be compared against the fingerprint of each candidate molecular structure to produce a similarity score, and then candidate structures are sorted according to the similarity scores. In the case of a molecular fingerprint, the prediction of the substructures or fingerprints for each query spectrum is the key step. For example, FingerID is the first molecular identification tool that predicts the molecular fingerprints for each query spectrum with a support vector machine (SVM) model trained by a large set of tandem mass spectra in MassBank [115]. In addition, CSI:FingerID performs molecular fingerprint prediction using multiple kernel learning with fragmentation trees inputted [116], [117], [118]. Instead of fragmentation trees, SIMPLE formulates a sparse interaction model for metabolite peaks to predict fingerprints and is lighter and more readily interpretable than CSI:FingerID [119]. The IOKRreverse model maps molecular structures into the MS/MS feature space and then solves a pre-image problem to find the molecule with the most similar fingerprint [120], [121]. By contrast, ADAPTIVE is an IOKR-derivative tool that learns a model to generate fingerprints for metabolites [122]. As a result, all of these mentioned fingerprints are specific to both data and the task of metabolite identification and are therefore nonredundant [122]. In addition to the above mentioned SVM-based methods, SF-Matching achieved similar performance to CSI:FingerID with a random forest model used for fingerprint prediction [123]. Compared to common machine learning, deep learning is another strategy for predicting the fingerprints of query mass spectra. MetFID applied an artificial neural network with two hidden layers to predict a composite vector comprising of 528 binary entries [124]. McSearch applied core structure-based search (CSS) algorithm based on hypothetical neutral loss values to predict the core substructure of the query MS [125].
3.3.3. Network-based strategies in structure elucidation for spectrum
Network-based strategies could be not only used for predicting the functional activity or category of metabolites directly from spectral features [126], [127], but also for metabolite identification per se [128], [129]. iMet was the first metabolite identification tool based on spectral similarity and structure similarity for MS data [130]. Regarding this method, neighbor metabolites (with similar MS/MS spectra) share structural similarities, so the unknown metabolites could be identified according to the identification of the neighbor metabolites. This strategy can be easily integrated with other methods to improve the accuracy of metabolite identification. As an example, NAP integrated MetFrag into network-based strategy, which improved the ranking of the correct spectra from a mean ranking position of 14.7 to a mean ranking position of 4.7 [131]. This network-based strategy embraces the construction of two kinds of networks: one is a network usually based on spectral similarity, the other is a network usually based on structural similarity. Compared with NAP, MetDNA uses the reaction in KEGG instead of the fingerprint similarity to represent the structure similarity and cumulatively annotated approximately 2000 metabolites from one experiment [132]. DeepMASS and MS2DeepScore both train deep learning models to predict structural similarity scores for spectral pairs instead of other spectral similarity scores, such as dot-product, and then construct a network for metabolite identification [133], [134]. MS similarity scores can also be presented by the fingerprint similarities predicted from mass spectra. For example, Spec2Vec is a novel spectral similarity score inspired by Word2Vec, which is a natural language processing algorithm. Word2Vec learned fragmental relationships within a large set of spectral data to derive abstract spectral embeddings that can be used to assess spectral similarities [135]. The feature information (e.g., isotope patterns, adduct formation, chromatographic retention times, and fragmentation patterns) can also be used for aiding the construction of metabolite networks [44], [136], [137].Compared to the above-mentioned methods, NetID connected MS peaks based on mass differences reflecting adduction, fragmentation, isotopes, or feasible biochemical transformations, and performed the global network optimization to produce an optimal and consistent network annotation by linear programming [138].
3.3.4. Structure elucidation for mass spectra with generative methods
The above-mentioned strategies for metabolite identification based on in-silico approach are seriously dependent on the structure libraries, and the identification results must be included in those libraries. For the diversity of chemical modification in complex biological individuals, especially for plants, there are still many metabolites not included in public libraries. Meanwhile, it is immensely time-consuming to predict the in-silico spectra or calculate all of the fingerprints for all metabolites. The direct approach is to translate the query high-resolution mass spectral peak to a representation (for instance, in SMILES) of the annotated metabolite rather than to predict the in-silico spectrum from the structure or intermediate fingerprints from the query spectrum, which requires another comparison process. In a similar approach MassGenie utilizes a transformer-based deep neural network coupled with VAE-Sim, a variational autoencoder (VAE)-based model, to directly predict the structure of a molecule from the query spectrum [139]. VAE-Sim is a variational autoencoder that is the backbone of MassGenie, which is used to generate ‘true’ molecules [140]. This strategy for direct prediction of a molecule from the query MS has been studied recently, proving it is convincing to provide valuable clues to expedite structure annotation of experimental mass spectra. However, in some ways, the accuracy of identification could be further improved. For instance, SMILES, a regular molecular representation, could be replaced by SELFIES [141] and DeepSMILES [142], which are more convenient methods for representing a valid molecule.
3.3.5. Other in-silico methods assisting the structure elucidation
Besides the above four classes of methods, some other machine learning approaches were developed to assist the metabolite identification. MS2LDA adopted Latent Dirichlet Allocation, an algorithm originally used for text mining, to extract the co-occurring molecular fragments and neutral losses [143]. With structure annotation for the extracted motifs by additional methods, the compounds can be identified with candidate structures or classified. MESSAR (MEtabolite SubStructure Auto-Recommender) extracted spectral features of spectra and generated substructures of metabolites in the spectral library, then generated the association rules linking spectral features (with exact masses) with specific substructures based on the concept of association rule mining (ARM) [144]. With the association rules applied, the structure elucidation or classification of metabolites can be conducted automatically. It is a very complex and time-consuming process to identify metabolites with a unique structure, while it can be a solution to perform automatic classification of compounds into compound classes based on machine learning methods [145], [146]. In particular, CANOPUS used a deep neural network to classify the metabolites based on mass spectrometry [147].
4. Retention time prediction methods assisting structure elucidation based on LC–MS/MS data
In addition to the MS information, chromatographic behavior, reflecting the physicochemical properties (molecular weight, hydrophobicity, polarity, molecular shape etc.) of metabolites, should also provide structural information. As an example, many isomeric compounds are indistinguishable in both MS^1 and tandem MS analyses, and must be resolved chromatographically if separate quantitation and identification is desired. Retention time prediction commonly are not utilized for structure elucidation separately, while the results of retention time prediction can be combined to screening the candidates and/or improve candidates rankings based on mass spectra information [148], [149]. Empirically, the octanol/water partition coefficients (log P values) of each metabolite were believed to determine the chromatographic retention time, and the linear solvation energy relationships (LSERs) equation was utilized for predicting retention data in early researches [150], [151]. Such technique, relating the variations in one response variables (chromatographic retention time) to the variations of several descriptors, is called Quantitative structure–retention relationships (QSRRs) [151], [152]. The chromatographic retention time always varies considerably depending on the experimental conditions such as column packing, flow rate, elution gradient, and PH of the mobile phase. Afterwards, much more complex models and larger training sets were used for the prediction of LC retention time (Table 1). Among those methods, Molecular descriptors (MDs) are the most common variables for LC retention time prediction, as MDs encode structural information and chemical information, such as the type of atoms and bonds, number of rings, charge, and stereochemical configuration, through mathematical and statistical approaches [153]. MDs have been widely utilized for training common machine learning models, such as multiple linear regression (MLR), random forest (RF) regression, support vector machine (SVM), and partial least squares (PLS) regression. With the advance of artificial neural networks (ANNs), it has been proven that deep neural networks (DNNs), convolutional neural networks (CNNs), recurrent neural networks (RNNs) and graph neural networks (GNNs) models (neural network types of ANNs) show robust performance for RT prediction in both RP and HILIC chromatography, compared to XGBOOST, BRNN, RF and LIGHTGBM [154], [155], [156], [157], [158], [159]. For the METLIN small molecule retention time (SMRT) dataset, the artificial neural networks (ANNs) had lower mean absolute error (MAE, MAE = ∼0.5 min) than the traditional machine learning model (MAE = ∼1 min) [156], [160]. While the training datasets of thousand metabolites were still not sufficient for the ANNs models, and with more MDs and more complex models considered, a larger database was needed to overcome the overfitting problem [154]. As the chromatographic retention time always varies considerably depending on the experimental conditions, it is a priority to make community sharing of RT information possible across laboratories and chromatographic systems [161], [162]. To obtain more databases, an approach directly mapping RTs between different systems was developed, and a sufficient number of common compounds were needed in both systems for this approach [161]. In addition to directly mapping RTs, a retention order prediction model can also be trained using retention time measurements from different LC systems and configurations, and this can be an effective way to learn the retention behavior of molecules from heterogeneous retention time data [148]. In addition to the large databases, transfer learning in combination with self-supervised pre-training is another option to overcome the limitation of the training data required for training ANNs models [159], [163], [164].
Table 1.
Publications relevant to RT prediction.
| Publication | Year | LC type | Model type | Size of training data | Molecular type | Variables |
|---|---|---|---|---|---|---|
| Hagiwara et al. [175] | 2010 | RP-LC | SVR and MLR | 150 authentic compounds | 9 MDs | |
| Creek et al. [176] | 2011 | HILIC | MLR | 120 authentic compounds | 6 MDs | |
| D'Archivio, Maggi and Ruggieri [177] | 2014 | RP-LC | MLR and PLS regression | 47 authentic compounds | butyl esters of 47 acylcarnitines | 73 MDs |
| Kouskoura, Hadjipavlou-Litina and Markopoulou [178] | 2014 | RP-LC | PLS regression | 100 authentic compounds | 66 MDs | |
| D'Archivio et al. [179] | 2014 | RP-LC | DNNs | 24 authentic compounds | s-triazines | 5 MDs |
| Cao et al. [180] | 2015 | HILIC | MLR and RF | 93 authentic compounds | 346 MDs | |
| Aicheler et al. [181] | 2015 | RP-LC | SVR | 201 authentic compounds | lipid | 11 MDs |
| Munro et al. [182] | 2015 | RP-LC | DNNs | 166 authentic compounds | drugs | 17 MDs |
| Falchi et al. [183] | 2016 | RP-LC | Four combined (fingerprints + ordinary) KPLS models | 1383 authentic compounds | molecular and fingerprints descriptors | |
| Ovcacikova et al. [184] | 2016 | RP-LC | The second degree polynomial regression | 400 authentic compounds | lipid | The carbon number (CN) and the double bonds (DB) number |
| Aalizadeh et al. [185] | 2016 | RP-LC | MLR, DNNs, and SVM | 528 and 298 compounds for positive and negative electrospray ionization mode respectively | 6 MDs | |
| Wolfer et al. [186] | 2016 | RP-LC | Combination of RF and SVR models | 442 authentic compounds | 97 MDs | |
| Kubik and Wiczling [187] | 2016 | RP-LC | Lasso, Stepwise and PLS regressions | 115 authentic compounds | drugs | 50 MDs |
| Barron and McEneff [188] | 2016 | RP-LC | DNNs | 1,117 authentic compounds | 16 MDs | |
| Randazzo et al. [189] | 2016 | RP-LC | PLS regression | 91 authentic compounds | steroids | 97 MDs |
| Taraji et al. [190] | 2017 | HILIC | PLS regression | 16 authentic compounds | β-adrenergic agonists and related compounds | 321 MDs |
| Taraji et al. [191] | 2017 | HILIC | PLS regression | 98 authentic compounds | pharmaceutical compounds | 321 MDs |
| Zhang et al. [192] | 2017 | RP-LC | MLR | 24 authentic compounds | 16-membered ring macrolides | 8 MDs |
| Park et al. [193] | 2017 | RP-LC | MLR | 41 authentic compounds | drugs | 10 MDs |
| Wen et al. [194] | 2018 | RP-LC | PLS regression | 148 authentic compounds | 126 MDs | |
| Wen et al. [195] | 2018 | RP-LC | PLS regression | 191 authentic compounds | 128 MDs | |
| McEachran et al. [196] | 2018 | RP-LC | PLS regression | 97 authentic compounds | 7 MDs | |
| Hall et al. [197] | 2018 | RP-LC | DNNs | 1,955 authentic compounds | 47 MDs | |
| Bouwmeester, Martens and Degroeve [198] | 2019 | RPLC (33) & HILIC (3) | Bayesian Ridge Regression (BRR), Least Absolute Shrinkage and Selection Operator (LASSO), DNNs, Adaptive Boosting (AB), Gradient Boosting (GB), RF and SVR | 6,759 authentic compounds | 151 MDs | |
| Bonini et al. [154] | 2020 | HILIC & RP-LC | XGBoost, Bayesian-regularized Neural Network (BRNN), RF, Light Gradient-Boosting Machine (LightGBM), DNNs | 1,023 (HILIC) & 494 (RP-LC) authentic compounds | 286 MDs | |
| Ju et al. [163] | 2021 | HILIC & RP-LC | DNNs + TL | 77,898 authentic compounds (DNNs), and 17 data sets (Transfer Learning) | 1,470 MDs | |
| Osipenko et al. [159] | 2021 | HILIC & RP-LC | RNNs + TL | 1 million molecules (pre-training) and 269–457 authentic compounds (transfer Learning) | SMILES | |
| Kensert et al. [156] | 2021 | HILIC & RP-LC | Graph Convolutional Networks (GCNs) | 77,980 (SMRT), 852(RIKEN) and 1,400 (Fiehn HILIC) authentic molecules | Graph and 25 atom and bond features | |
| Yang et al. [157] | 2021 | HILIC | GNNs + TL | in silico HILIC RT dataset with about 306 K molecules for GNNs, 100∼200 molecules for TL | Graph, 16 kinds of atoms and 4 kinds of bonds | |
| Yang et al. [158] | 2021 | RP-LC | GNNs + TL | 80,038 authentic molecules (SMRT) for Graph Neural Network, and the MoNA and PredRet datasets for Transfer Learning | Graph | |
| Souihi et al. [199] | 2022 | HILIC & RP-LC | RF regression | 78 authentic compounds | 153 MDs | |
| Liapikos et al. [200] | 2022 | RP-LC | Bayesian Ridge Regression (BRidgeR), Extreme Gradient Boosting Regression (XGBR) and SVR | 26–350 authentic compounds | 70–92 MDs | |
| Fedorova et al. [155] | 2022 | RP-LC | 1D CNN + TL | 77,983 authentic molecules (SMRT) for 1D CNN, 5 data sets for Transfer Learning | SMILES |
5. Fusion tools for metabolite identification based on LC–MS/MS data
Metabolite identification based on LC–MS/MS data is a sophisticated subject that involves analysis of authentic standard compound LC–MS/MS data, comparison between query spectra and reference/in-silico spectra, sub-annotating of the features, and prediction of retention time of metabolites. In this regard, the development of fusion tools, in the form of client software or web servers, has greatly boosted the utilization of LC–MS/MS in the metabolism of complex biological samples (Table 2). ChemDistiller is a fusion software that combines a fingerprint prediction algorithm (FingerScorer) inspired by CSI:FingerID with an in-silico fragmentation algorithm (FragScorer) inspired by CFM-ID and MetFrag, and can retrieve and rank candidates from multiple target databases [165]. SIRIUS is a java-based software framework integrating a collection of tools, including ‘SIRIUS’ (the core function of SIRIUS), CSI:FingerID (with COSMIC), ZODIAC and CANOPUS [166], [167], [168]. Besides CSI:FingerID (a fingerprint predictor) and CANOPUS (a metabolites classifier) mentioned above, COSMIC [167] provides a confidence score for every structure annotated by CSI:FingerID and ZODIAC [166] performs de novo molecular formula annotation. Additionally, MS-DIAL combines MS-FINDER, LipidBlast with reference library search engine to solve the comprehensive identification of metabolites further in complex biological extracts [146], [169]. Strategies based on public/commercial spectral libraries and in-silico approaches can be combined to improve the accuracy and rate of metabolite identification. As a concept of network-based identification, NAP and MetDNA also build their own repositories of mass spectra for the first seed identification, and in-silico structure annotation is then propagated through the network [131], [132]. GNPS is a data repository and collection of different web services of computational methods, including NAP and DEREPLICATOR+, and aims to build an open-access mass spectrometry ecosystem for sharing of raw, processed, or annotated fragmentation mass spectrometry data (MS/MS) [64]. Combined with mass spectrometry data, the RT can also be utilized as additional and orthogonal information for the putative identification of small molecules [170]. MetFrag combines compound database searching, retention times prediction, and in-silico fragments prediction for small molecule identification from tandem mass spectrometry data [99], [171], [172]. Although, the fusion tools involving RT prediction are still limited compared to those tools based on both in-silico and experimental spectral libraries, retention time prediction and utilizing artificial neural networks, was proven to reduce structure isomer hit lists when used prior to in-silico spectral prediction software [149]. Retention order prediction and spectrum-based scores can also be combined for more accurate metabolite identifications in a LC–MS/MS experiment [148]. All these studies have thus triggered new initiatives for developing fusion tools based on RT to promote the use of LC–MS/MS.
Table 2.
Fusion tools for metabolite identification based on LC–MS/MS.
| Name | Function | Availability |
|---|---|---|
| ChemDistiller | FingerScorer + FragScorer | https://bitbucket.org/iAnalytica/chemdistillerpython/src/master/ |
| SIRIUS | “Sirius”, CSI:FingerID (with COSMIC), ZODIAC and CANOPUS | https://bio.informatik.uni-jena.de/software/sirius/ |
| msms_rt_score_integration | Mass spectrum and retention time prediction | https://github.com/aalto-ics-kepaco/msms_rt_score_integration |
| MetFrag | MetFrag (algorithm) + reference library search + retention times prediction | https://msbi.ipb-halle.de/MetFragBeta/ |
| MetDNA | Structure elucidation from knowns to unknowns | https://metdna.zhulab.cn/metdna/analysis |
| MS-DIAL | MS-FINDER + LipidBlast + reference library search | https://prime.psc.riken.jp/compms/msdial/main.html |
| GNPS | mass spectrometry ecosystem for sharing of MS data and metabolites identification | https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp |
| NAP | spectral networks to propagate information from spectral library matching | https://proteomics2.ucsd.edu/ProteoSAFe/?params=%7B%22workflow%22:%22NAP_CCMS2%22%7D |
6. Conclusions and perspectives
Commonly, the strategy based on authentic standard compounds can provide relative credible structure elucidation, whereas the number of the structure annotations is almost too small compared to the number of metabolites in complex biological samples. Recently, the strategy based on public/commercial reference spectral libraries is developing rapidly, as the reference spectral libraries can be collected from the institutions and companies around the world in a simple way. The in-silico approaches can produce numerous complement structure annotations for the results of the above strategies, whereas the accuracy of the annotation based on in-silico approaches is quite lower than that of strategies based on authentic standard compounds or public/commercial reference spectral libraries. Although many tools for structure elucidation based on LC–MS/MS data have been developed in recent years, most of them have been shown to lack a degree of reliability that needs to be evaluated by a third-party organization. In particular, the Critical Assessment of Small Molecule Identification (CASMI) is an open contest on the identification of small molecules from mass spectrometry data and has been held five times since 2012 (https://casmi-contest.org/) [83], [173]. With top or top n candidates used for evaluating the approaches, the individual in-silico approaches were proved to produce structure elucidation with low accuracy (17–25 %), while the combination of the strategy based on public/commercial reference spectral libraries and in-silico approaches can correctly identified up-to 87–93 % [83]. It is believed that the prediction models with different strategies combined are more suitable for real-world applications, at least, more training MS data and fragment rules need to be achieved to optimize the prediction models of in-silico methods, especially for deep learning models. In addition, no third-party organization appeared heretofore to host an open contest on the RT prediction. Though the critical assessment for RT prediction models from different researches is usually difficult, as the chromatography conditions are much diverse across different platforms, the prediction models can be optimized by larger and more diverse training datasets. With the continuing and developing collection of RT datasets, we believe that complex deep learning models, like transfer learning, could accurately predict RT across different platforms.
In addition to the algorithms for structure elucidation of metabolites based on LC–MS/MS data, the construction and preservation of public data are also crucial to metabolomics. Although a large number of reference spectral databases have been built, most of the databases are still insufficient to train a complex model, especially for deep learning models. With the rapid development of public MS libraries, like GNPS and MassBank, we are convinced that a much larger, comprehensive and user-friendly library will be established for the researchers in the world. Such a resource, alongside faithful adherence to recently published standards for metabolomics reporting [174], allow us to be confident that the large coverage gap between annotated and non-annotated metabolites will ultimately be closed.
CRediT authorship contribution statement
Zhitao Tian: Writing – original draft. Fangzhou Liu: Data curation. Dongqin Li: Data curation. Alisdair R. Fernie: Writing – review & editing. Wei Chen: Funding acquisition, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Natural Science Foundation for Distinguished Young Scientists of Hubei Province (No. 2021CFA058), the Young Top-notch Talent Cultivation Program of Hubei Province, the Fundamental Research Funds for the Central Universities (No. 2662019PY008).
References
- 1.Oliver S.G., Winson M.K., Kell D.B., Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16(9):373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 2.Patti G.J., Yanes O., Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang C., Fernie A.R., Luo J. Exploring the diversity of plant metabolism. Trends Plant Sci. 2019;24(1):83–98. doi: 10.1016/j.tplants.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Wishart D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev. 2019;99(4):1819–1875. doi: 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- 5.da Silva R.R., Dorrestein P.C., Quinn R.A. Illuminating the dark matter in metabolomics. PNAS. 2015;112(41):12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson S.R., Lange B.M. Open-access metabolomics databases for natural product research: present capabilities and future potential. Front Bioeng Biotechnol. 2015;3 doi: 10.3389/fbioe.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bocker S. Searching molecular structure databases using tandem MS data: are we there yet? Curr Opin Biotechnol. 2017;36:1–6. doi: 10.1016/j.cbpa.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Blazenovic I., Kind T., Ji J., Fiehn O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites. 2018;8(2) doi: 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddad P.R., Taraji M., Szucs R. Prediction of analyte retention time in liquid chromatography. Anal Chem. 2021;93(1):228–256. doi: 10.1021/acs.analchem.0c04190. [DOI] [PubMed] [Google Scholar]
- 10.Kind T., Tsugawa H., Cajka T., Ma Y., Lai Z., et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom Rev. 2018;37(4):513–532. doi: 10.1002/mas.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glish G.L., Burinsky D.J. Hybrid mass spectrometers for tandem mass spectrometry. J Am Soc Mass Spectrom. 2008;19(2):161–172. doi: 10.1016/j.jasms.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Werner E., Heilier J.F., Ducruix C., Ezan E., Junot C., et al. Mass spectrometry for the identification of the discriminating signals from metabolomics: Current status and future trends. J Chromatogr B-Anal Technol Biomed Life Sci. 2008;871(2):143–163. doi: 10.1016/j.jchromb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Ichou F., Schwarzenberg A., Lesage D., Alves S., Junot C., et al. Comparison of the activation time effects and the internal energy distributions for the CID, PQD and HCD excitation modes. J Mass Spectrom. 2014;49(6):498–508. doi: 10.1002/jms.3365. [DOI] [PubMed] [Google Scholar]
- 14.Chaleckis R., Meister I., Zhang P., Wheelock C.E. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr Opin Biotechnol. 2019;55:44–50. doi: 10.1016/j.copbio.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Tautenhahn R., Bottcher C., Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinf. 2008;9 doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu G., Wang S., Huang Z., Zhang S., Liao Q., et al. Rewiring of the fruit metabolome in tomato breeding. Cell. 2018;172(1–2):249–261 e12. doi: 10.1016/j.cell.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Wen W., Li D., Li X., Gao Y., Li W., et al. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W., Gao Y., Xie W., Gong L., Lu K., et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46(7):714–721. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- 19.Chen J., Hu X., Shi T., Yin H., Sun D., et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol J. 2020;18(8):1722–1735. doi: 10.1111/pbi.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi T., Zhu A., Jia J., Hu X., Chen J., et al. Metabolomics analysis and metabolite-agronomic trait associations using kernels of wheat (Triticum aestivum) recombinant inbred lines. Plant J. 2020;103(1):279–292. doi: 10.1111/tpj.14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marie B., Gallet A. Fish metabolome from sub-urban lakes of the Paris area (France) and potential influence of noxious metabolites produced by cyanobacteria. Chemosphere. 2022;296 doi: 10.1016/j.chemosphere.2022.134035. [DOI] [PubMed] [Google Scholar]
- 22.Zhalnina K., Louie K.B., Hao Z., Mansoori N., da Rocha U.N., et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3(4):470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 23.Li J.X., Wang Z.Z., Zhai G.T., Chen C.L., Zhu K.Z., et al. Untargeted metabolomic profiling identifies disease-specific and outcome-related signatures in chronic rhinosinusitis. J Allergy Clin Immunol. 2022 doi: 10.1016/j.jaci.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Moreau R., Claria J., Aguilar F., Fenaille F., Lozano J.J., et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol. 2020;72(4):688–701. doi: 10.1016/j.jhep.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Tsugawa H., Rai A., Saito K., Nakabayashi R. Metabolomics and complementary techniques to investigate the plant phytochemical cosmos. Nat Prod Rep. 2021;38(10):1729–1759. doi: 10.1039/d1np00014d. [DOI] [PubMed] [Google Scholar]
- 26.Tikunov Y., Lommen A., de Vos C.H.R., Verhoeven H.A., Bino R.J., et al. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005;139(3):1125–1137. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sturm M., Bertsch A., Gropl C., Hildebrandt A., Hussong R., et al. OpenMS-An open-source software framework for mass spectrometry. BMC Bioinf. 2008;9 doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11 doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C.A., Want E.J., O'Maille G., Abagyan R., Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 30.Kuhl C., Tautenhahn R., Bottcher C., Larson T.R., Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem. 2012;84(1):283–289. doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahieu N.G., Patti G.J. Systems-level annotation of a metabolomics data set reduces 25 000 features to fewer than 1000 unique metabolites. Anal Chem. 2017;89(19):10397–10406. doi: 10.1021/acs.analchem.7b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu W.Y., Xing X., Wang L., Chen L., Zhang S.S., et al. Improved annotation of untargeted metabolomics data through buffer modifications that shift adduct mass and intensity. Anal Chem. 2020;92(17):11573–11581. doi: 10.1021/acs.analchem.0c00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domingo-Almenara X., Montenegro-Burke J.R., Benton H.P., Siuzdak G. Annotation: A computational solution for streamlining metabolomics analysis. Anal Chem. 2018;90(1):480–489. doi: 10.1021/acs.analchem.7b03929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sindelar M., Patti G.J. Chemical discovery in the era of metabolomics. J Am Chem Soc. 2020;142(20):9097–9105. doi: 10.1021/jacs.9b13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P.W., Ang I.L., Lam M.M.T., Wei R., Lei K.M.K., et al. Susceptibility to false discovery in biomarker research using liquid chromatography-high resolution mass spectrometry based untargeted metabolomics profiling. Clin Transl Med. 2021;11(6) doi: 10.1002/ctm2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Xing X., Chen L., Yang L.F., Su X.Y., et al. Peak annotation and verification engine for untargeted LC-MS metabolomics. Anal Chem. 2019;91(3):1838–1846. doi: 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso A., Julia A., Beltran A., Vinaixa M., Diaz M., et al. AStream: an R package for annotating LC/MS metabolomic data. Bioinformatics. 2011;27(9):1339–1340. doi: 10.1093/bioinformatics/btr138. [DOI] [PubMed] [Google Scholar]
- 38.Broeckling C.D., Afsar F.A., Neumann S., Ben-Hur A., Prenni J.E. RAMClust: a novel feature clustering method enables spectral-matching-based annotation for metabolomics data. Anal Chem. 2014;86(14):6812–6817. doi: 10.1021/ac501530d. [DOI] [PubMed] [Google Scholar]
- 39.Bueschl C., Kluger B., Lemmens M., Adam G., Wiesenberger G., et al. A novel stable isotope labelling assisted workflow for improved untargeted LC-HRMS based metabolomics research. Metabolomics. 2014;10(4):754–769. doi: 10.1007/s11306-013-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daly R., Rogers S., Wandy J., Jankevics A., Burgess K.E.V., et al. MetAssign: probabilistic annotation of metabolites from LC-MS data using a Bayesian clustering approach. Bioinformatics. 2014;30(19):2764–2771. doi: 10.1093/bioinformatics/btu370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeFelice B.C., Mehta S.S., Samra S., Cajka T., Wancewicz B., et al. Mass Spectral Feature List Optimizer (MS-FLO): A tool to minimize false positive peak reports in untargeted liquid chromatography-mass spectroscopy (LC-MS) data processing. Anal Chem. 2017;89(6):3250–3255. doi: 10.1021/acs.analchem.6b04372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva R.R., Jourdan F., Salvanha D.M., Letisse F., Jamin E.L., et al. ProbMetab: an R package for Bayesian probabilistic annotation of LC-MS-based metabolomics. Bioinformatics. 2014;30(9):1336–1337. doi: 10.1093/bioinformatics/btu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tikunov Y.M., Laptenok S., Hall R.D., Bovy A., de Vos R.C.H. MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics. 2012;8(4):714–718. doi: 10.1007/s11306-011-0368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uppal K., Walker D.I., Jones D.P. xMSannotator: An R package for network-based annotation of high-resolution metabolomics data. Anal Chem. 2017;89(2):1063–1067. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senan O., Aguilar-Mogas A., Navarro M., Capellades J., Noon L., et al. CliqueMS: a computational tool for annotating in-source metabolite ions from LC-MS untargeted metabolomics data based on a coelution similarity network. Bioinformatics. 2019;35(20):4089–4097. doi: 10.1093/bioinformatics/btz207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kachman M., Habra H., Duren W., Wigginton J., Sajjakulnukit P., et al. Deep annotation of untargeted LC-MS metabolomics data with Binner. Bioinformatics. 2020;36(6):1801–1806. doi: 10.1093/bioinformatics/btz798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonner R., Hopfgartner G. Annotation of complex mass spectra by multi-layered analysis. Anal Chim Acta. 2022;1193 doi: 10.1016/j.aca.2021.339317. [DOI] [PubMed] [Google Scholar]
- 48.Kofeler H.C., Eichmann T.O., Ahrends R., Bowden J.A., Danne-Rasche N., et al. Quality control requirements for the correct annotation of lipidomics data. Nature Communications. 2021;12(1) doi: 10.1038/s41467-021-24984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenaille F., Saint-Hilaire P.B., Rousseau K., Junot C. Data acquisition workflows in liquid chromatography coupled to high resolution mass spectrometry-based metabolomics: Where do we stand? J Chromatogr A. 2017;1526:1–12. doi: 10.1016/j.chroma.2017.10.043. [DOI] [PubMed] [Google Scholar]
- 50.Defossez E., Bourquin J., Reuss S., Rasmann S., Glauser G. Eight key rules for successful data-dependent acquisition in mass spectrometry-based metabolomics. Mass Spectrom Rev. 2021 doi: 10.1002/mas.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bilbao A., Varesio E., Luban J., Strambio-De-Castillia C., Hopfgartner G., et al. Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics. 2015;15(5–6):964–980. doi: 10.1002/pmic.201400323. [DOI] [PubMed] [Google Scholar]
- 52.Gillet L.C., Navarro P., Tate S., Rost H., Selevsek N., et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11(6) doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raetz M., Bonner R., Hopfgartner G. SWATH-MS for metabolomics and lipidomics: critical aspects of qualitative and quantitative analysis. Metabolomics. 2020;16(6) doi: 10.1007/s11306-020-01692-0. [DOI] [PubMed] [Google Scholar]
- 54.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernie A.R., Aharoni A., Willmitzer L., Stitt M., Tohge T., et al. Recommendations for reporting metabolite data. Plant Cell. 2011;23(7):2477–2482. doi: 10.1105/tpc.111.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blazenovic I., Kind T., Sa M.R., Ji J., Vaniya A., et al. Structure annotation of all mass spectra in untargeted metabolomics. Anal Chem. 2019;91(3):2155–2162. doi: 10.1021/acs.analchem.8b04698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Folberth J., Begemann K., Johren O., Schwaninger M., Othman A. MS2 and LC libraries for untargeted metabolomics: Enhancing method development and identification confidence. J Chromatogr B-Anal Technol Biomed Life Sci. 2020;1145 doi: 10.1016/j.jchromb.2020.122105. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Morato J., Pozo O.J., Marcos J. Targeting human urinary metabolome by LC-MS/MS: a review. Bioanalysis. 2018;10(7):489–516. doi: 10.4155/bio-2017-0285. [DOI] [PubMed] [Google Scholar]
- 60.Bennett R.N., Mellon F.A., Kroon P.A. Screening crucifer seeds as sources of specific intact glucosinolates using ion-pair high-performance liquid chromatography negative ion electrospray mass spectrometry. J Agric Food Chem. 2004;52(3):428–438. doi: 10.1021/jf030530p. [DOI] [PubMed] [Google Scholar]
- 61.Vinaixa M., Schymanski E.L., Neumann S., Navarro M., Salek R.M., et al. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC-Trends Anal Chem. 2016;78:23–35. doi: 10.1016/j.trac.2015.09.005. [DOI] [Google Scholar]
- 62.Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 63.Wishart D.S., Guo A.C., Oler E., Wang F., Anjum A., et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50(D1):D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue J., Guijas C., Benton H.P., Warth B., Siuzdak G. METLIN MS(2) molecular standards database: a broad chemical and biological resource. Nat Methods. 2020;17(10):953–954. doi: 10.1038/s41592-020-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogusz M.J., Maier R.D., Kruger K.D., Webb K.S., Romeril J., et al. Poor reproducibility of in-source collisional atmospheric pressure ionization mass spectra of toxicologically relevant drugs. J Chromatogr A. 1999;844(1–2):409–418. doi: 10.1016/s0021-9673(99)00312-x. [DOI] [PubMed] [Google Scholar]
- 67.Bristow A.W., Webb K.S., Lubben A.T., Halket J. Reproducible product-ion tandem mass spectra on various liquid chromatography/mass spectrometry instruments for the development of spectral libraries. Rapid Commun Mass Spectrom. 2004;18(13):1447–1454. doi: 10.1002/rcm.1492. [DOI] [PubMed] [Google Scholar]
- 68.Oberacher H., Pavlic M., Libiseller K., Schubert B., Sulyok M., et al. On the inter-instrument and inter-laboratory transferability of a tandem mass spectral reference library: 1. Results of an Austrian multicenter study. J Mass Spectrom. 2009;44(4):485–493. doi: 10.1002/jms.1545. [DOI] [PubMed] [Google Scholar]
- 69.Stein S.E., Scott D.R. Optimization and testing of mass spectral library search algorithms for compound identification. J Am Soc Mass Spectrom. 1994;5(9):859–866. doi: 10.1016/1044-0305(94)87009-8. [DOI] [PubMed] [Google Scholar]
- 70.Atwater BL, Stauffer DB, Mclafferty FW, Peterson DW. Reliability ranking and scaling improvements to the probability based matching system for unknown mass-spectra. Anal Chem 1985;57(4): 899-903. doi:10.1021/ac00281a028.
- 71.Gan F, Yang JH, Liang YZ. Library search of mass spectra with a new matching algorithm based on substructure similarity. Anal Sci 2001;17(5):635-638. doi:10.2116/analsci.17.635. [DOI] [PubMed]
- 72.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 2007;7(5): 655-667. doi:10.1002/pmic.200600625. [DOI] [PubMed]
- 73.Pavlic M., Libiseller K., Oberacher H. Combined use of ESI-QqTOF-MS and ESI-QqTOF-MS/MS with mass-spectral library search for qualitative analysis of drugs. Anal Bioanal Chem. 2006;386(1):69–82. doi: 10.1007/s00216-006-0634-8. [DOI] [PubMed] [Google Scholar]
- 74.Oberacher H., Pavlic M., Libiseller K., Schubert B., Sulyok M., et al. On the inter-instrument and the inter-laboratory transferability of a tandem mass spectral reference library: 2. Optimization and characterization of the search algorithm. J Mass Spectrom. 2009;44(4):494–502. doi: 10.1002/jms.1525. [DOI] [PubMed] [Google Scholar]
- 75.Mylonas R., Mauron Y., Masselot A., Binz P.A., Budin N., et al. X-Rank: a robust algorithm for small molecule identification using tandem mass spectrometry. Anal Chem. 2009;81(18):7604–7610. doi: 10.1021/ac900954d. [DOI] [PubMed] [Google Scholar]
- 76.Li Y., Kind T., Folz J., Vaniya A., Mehta S.S., et al. Spectral entropy outperforms MS/MS dot product similarity for small-molecule compound identification. Nat Methods. 2021;18(12):1524–1531. doi: 10.1038/s41592-021-01331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newgard C.B. Metabolomics and metabolic diseases: Where do we stand? Cell Metab. 2017;25(1):43–56. doi: 10.1016/j.cmet.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang C., Luo J. Metabolic GWAS-based dissection of genetic bases underlying the diversity of plant metabolism. Plant J. 2019;97(1):91–100. doi: 10.1111/tpj.14097. [DOI] [PubMed] [Google Scholar]
- 79.Wurtzel E.T., Kutchan T.M. Plant metabolism, the diverse chemistry set of the future. Science. 2016;353(6305):1232–1236. doi: 10.1126/science.aad2062. [DOI] [PubMed] [Google Scholar]
- 80.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pence H.E., Williams A. ChemSpider: an online chemical information resource. J Chem Educ. 2010;87(11):1123–1124. doi: 10.1021/ed100697w. [DOI] [Google Scholar]
- 82.Krettler C.A., Thallinger G.G. A map of mass spectrometry-based in silico fragmentation prediction and compound identification in metabolomics. Brief Bioinf. 2021;22(5) doi: 10.1093/bib/bbab073. [DOI] [PubMed] [Google Scholar]
- 83.Blazenovic I., Kind T., Torbasinovic H., Obrenovic S., Mehta S.S., et al. Comprehensive comparison of in silico MS/MS fragmentation tools of the CASMI contest: database boosting is needed to achieve 93% accuracy. J Cheminf. 2017;9(1):32. doi: 10.1186/s13321-017-0219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liebal U.W., Phan A.N.T., Sudhakar M., Raman K., Blank L.M. Machine learning applications for mass spectrometry-based metabolomics. Metabolites. 2020;10(6) doi: 10.3390/metabo10060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scheubert K., Hufsky F., Bocker S. Computational mass spectrometry for small molecules. J Cheminf. 2013;5 doi: 10.1186/1758-2946-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hufsky F., Bocker S. Mining molecular structure databases: identification of small molecules based on fragmentation mass spectrometry data. Mass Spectrom Rev. 2017;36(5):624–633. doi: 10.1002/mas.21489. [DOI] [PubMed] [Google Scholar]
- 87.O'Shea K, Misra BB. Software tools, databases and resources in metabolomics: updates from 2018 to 2019. Metabolomics 2020;16(3). doi:10.1007/s11306-020-01657-3. [DOI] [PubMed]
- 88.Carhart RE, Smith DH, Venkataraghavan R. Atom pairs as molecular-features in structure activity studies - definition and applications. J Chem Inf Comput Sci 1985;25(2):64-73. doi:DOI 10.1021/ci00046a002.
- 89.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28(1):31–36. doi: 10.1021/ci00057a005. [DOI] [Google Scholar]
- 90.Capecchi A., Probst D., Reymond J.L. One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J Cheminf. 2020;12(1):43. doi: 10.1186/s13321-020-00445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morgan H.L. The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J Chem Doc. 2002;5(2):107–113. doi: 10.1021/c160017a018. [DOI] [Google Scholar]
- 92.Grimme S. Towards first principles calculation of electron impact mass spectra of molecules. Angew Chem Int Ed. 2013;52(24):6306–6312. doi: 10.1002/anie.201300158. [DOI] [PubMed] [Google Scholar]
- 93.Bauer C.A., Grimme S. Elucidation of electron ionization induced fragmentations of adenine by semiempirical and density functional molecular dynamics. J Phys Chem A. 2014;118(49):11479–11484. doi: 10.1021/jp5096618. [DOI] [PubMed] [Google Scholar]
- 94.Wang S., Kind T., Tantillo D.J., Fiehn O. Predicting in silico electron ionization mass spectra using quantum chemistry. J Cheminf. 2020;12(1):63. doi: 10.1186/s13321-020-00470-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spezia R., Lee S.B., Cho A., Song K. Collision-induced dissociation mechanisms of protonated penta- and octa-glycine as revealed by chemical dynamics simulations. Int J Mass Spectrom. 2015;392:125–138. doi: 10.1016/j.ijms.2015.10.001. [DOI] [Google Scholar]
- 96.Cautereels J., Blockhuys F. Quantum chemical mass spectrometry: Verification and extension of the mobile proton model for histidine. J Am Soc Mass Spectrom. 2017;28(6):1227–1235. doi: 10.1007/s13361-017-1636-9. [DOI] [PubMed] [Google Scholar]
- 97.Janesko B.G., Li L., Mensing R. Quantum chemical fragment precursor tests: Accelerating de novo annotation of tandem mass spectra. Anal Chim Acta. 2017;995:52–64. doi: 10.1016/j.aca.2017.09.034. [DOI] [PubMed] [Google Scholar]
- 98.Schuler J.A., Neumann S., Muller-Hannemann M., Brandt W. ChemFrag: Chemically meaningful annotation of fragment ion mass spectra. J Mass Spectrom. 2018;53(11):1104–1115. doi: 10.1002/jms.4278. [DOI] [PubMed] [Google Scholar]
- 99.Wolf S., Schmidt S., Muller-Hannemann M., Neumann S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinf. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y., Kora G., Bowen B.P., Pan C. MIDAS: a database-searching algorithm for metabolite identification in metabolomics. Anal Chem. 2014;86(19):9496–9503. doi: 10.1021/ac5014783. [DOI] [PubMed] [Google Scholar]
- 101.Ridder L., van der Hooft J.J., Verhoeven S. Automatic compound annotation from mass spectrometry data using MAGMa. Mass Spectrom. 2014;3(Spec Iss 2):S0033. doi: 10.5702/massspectrometry.S0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ridder L., van der Hooft J.J., Verhoeven S., de Vos R.C., van Schaik R., et al. Substructure-based annotation of high-resolution multistage MS(n) spectral trees. Rapid Commun Mass Spectrom. 2012;26(20):2461–2471. doi: 10.1002/rcm.6364. [DOI] [PubMed] [Google Scholar]
- 103.Tsugawa H., Kind T., Nakabayashi R., Yukihira D., Tanaka W., et al. Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal Chem. 2016;88(16):7946–7958. doi: 10.1021/acs.analchem.6b00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mohimani H., Gurevich A., Shlemov A., Mikheenko A., Korobeynikov A., et al. Dereplication of microbial metabolites through database search of mass spectra. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mohimani H., Gurevich A., Mikheenko A., Garg N., Nothias L.F., et al. Dereplication of peptidic natural products through database search of mass spectra. Nat Chem Biol. 2017;13(1):30–37. doi: 10.1038/Nchembio.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kind T., Liu K.H., Lee D.Y., DeFelice B., Meissen J.K., et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10(8):755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koelmel J.P., Kroeger N.M., Ulmer C.Z., Bowden J.A., Patterson R.E., et al. LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinf. 2017;18(1):331. doi: 10.1186/s12859-017-1744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Theodoridis G.A., Gika H.G., Want E.J., Wilson I.D. Liquid chromatography-mass spectrometry based global metabolite profiling: a review. Anal Chim Acta. 2012;711:7–16. doi: 10.1016/j.aca.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 109.Keshet U., Kind T., Lu X.C., Devi S., Fiehn O. Acyl-CoA identification in mouse liver samples using the in silico CoA-Blast tandem mass spectral library. Anal Chem. 2022;94(6):2732–2739. doi: 10.1021/acs.analchem.1c03272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kangas L.J., Metz T.O., Isaac G., Schrom B.T., Ginovska-Pangovska B., et al. In silico identification software (ISIS): a machine learning approach to tandem mass spectral identification of lipids. Bioinformatics. 2012;28(13):1705–1713. doi: 10.1093/bioinformatics/bts194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Allen F., Greiner R., Wishart D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics. 2014;11(1):98–110. doi: 10.1007/s11306-014-0676-4. [DOI] [Google Scholar]
- 112.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;28(1): 27-30. doi:DOI 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed]
- 113.Wang F., Liigand J., Tian S., Arndt D., Greiner R., et al. CFM-ID 4.0: More accurate ESI-MS/MS spectral prediction and compound identification. Anal Chem. 2021;93(34):11692–11700. doi: 10.1021/acs.analchem.1c01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wei J.N., Belanger D., Adams R.P., Sculley D. Rapid prediction of electron-ionization mass spectrometry using neural networks. ACS Cent Sci. 2019;5(4):700–708. doi: 10.1021/acscentsci.9b00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Heinonen M., Shen H., Zamboni N., Rousu J. Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics. 2012;28(18):2333–2341. doi: 10.1093/bioinformatics/bts437. [DOI] [PubMed] [Google Scholar]
- 116.Duhrkop K., Shen H., Meusel M., Rousu J., Bocker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. PNAS. 2015;112(41):12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bocker S., Duhrkop K. Fragmentation trees reloaded. J Cheminf. 2016;8:5. doi: 10.1186/s13321-016-0116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vaniya A., Fiehn O. Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics. TrAC, Trends Anal Chem. 2015;69:52–61. doi: 10.1016/j.trac.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nguyen D.H., Nguyen C.H., Mamitsuka H. SIMPLE: Sparse interaction model over peaks of molecules for fast, interpretable metabolite identification from tandem mass spectra. Bioinformatics. 2018;34(13):i323–i332. doi: 10.1093/bioinformatics/bty252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brouard C., Shen H., Duhrkop K., d'Alche-Buc F., Bocker S., et al. Fast metabolite identification with Input Output Kernel Regression. Bioinformatics. 2016;32(12):i28–i36. doi: 10.1093/bioinformatics/btw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brouard C., Basse A., d'Alche-Buc F., Rousu J. Improved small molecule identification through learning combinations of kernel regression models. Metabolites. 2019;9(8) doi: 10.3390/metabo9080160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nguyen D.H., Nguyen C.H., Mamitsuka H. ADAPTIVE: Learning data-dependent, concise molecular vectors for fast, accurate metabolite identification from tandem mass spectra. Bioinformatics. 2019;35(14):i164–i172. doi: 10.1093/bioinformatics/btz319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Y., Kuhn M., Gavin A.C., Bork P. Identification of metabolites from tandem mass spectra with a machine learning approach utilizing structural features. Bioinformatics. 2020;36(4):1213–1218. doi: 10.1093/bioinformatics/btz736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fan Z., Alley A., Ghaffari K., Ressom H.W. MetFID: artificial neural network-based compound fingerprint prediction for metabolite annotation. Metabolomics. 2020;16(10):104. doi: 10.1007/s11306-020-01726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xing S., Hu Y., Yin Z., Liu M., Tang X., et al. Retrieving and utilizing hypothetical neutral losses from tandem mass spectra for spectral similarity analysis and unknown metabolite annotation. Anal Chem. 2020;92(21):14476–14483. doi: 10.1021/acs.analchem.0c02521. [DOI] [PubMed] [Google Scholar]
- 126.Li S., Park Y., Duraisingham S., Strobel F.H., Khan N., et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pirhaji L., Milani P., Leidl M., Curran T., Avila-Pacheco J., et al. Revealing disease-associated pathways by network integration of untargeted metabolomics. Nat Methods. 2016;13(9):770–776. doi: 10.1038/nmeth.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Watrous J., Roach P., Alexandrov T., Heath B.S., Yang J.Y., et al. Mass spectral molecular networking of living microbial colonies. PNAS. 2012;109(26):E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Morreel K., Saeys Y., Dima O., Lu F.C., Van de Peer Y., et al. Systematic structural characterization of metabolites in Arabidopsis via candidate substrate-product pair networks. Plant Cell. 2014;26(3):929–945. doi: 10.1105/tpc.113.122242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aguilar-Mogas A., Sales-Pardo M., Navarro M., Guimera R., Yanes O. iMet: A network-based computational tool to assist in the annotation of metabolites from tandem mass spectra. Anal Chem. 2017;89(6):3474–3482. doi: 10.1021/acs.analchem.6b04512. [DOI] [PubMed] [Google Scholar]
- 131.da Silva R.R., Wang M., Nothias L.F., van der Hooft J.J.J., Caraballo-Rodriguez A.M., et al. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput Biol. 2018;14(4):e1006089. doi: 10.1371/journal.pcbi.1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shen X., Wang R., Xiong X., Yin Y., Cai Y., et al. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat Commun. 2019;10(1):1516. doi: 10.1038/s41467-019-09550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ji H., Xu Y., Lu H., Zhang Z. Deep MS/MS-aided structural-similarity scoring for unknown metabolite identification. Anal Chem. 2019;91(9):5629–5637. doi: 10.1021/acs.analchem.8b05405. [DOI] [PubMed] [Google Scholar]
- 134.Huber F., van der Burg S., van der Hooft J.J.J., Ridder L. MS2DeepScore: a novel deep learning similarity measure to compare tandem mass spectra. J Cheminf. 2021;13(1):84. doi: 10.1186/s13321-021-00558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huber F., Ridder L., Verhoeven S., Spaaks J.H., Diblen F., et al. Spec2Vec: Improved mass spectral similarity scoring through learning of structural relationships. PLoS Comput Biol. 2021;17(2) doi: 10.1371/journal.pcbi.1008724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Del Carratore F., Schmidt K., Vinaixa M., Hollywood K.A., Greenland-Bews C., et al. Integrated probabilistic annotation: A Bayesian-based annotation method for metabolomic profiles integrating biochemical connections, isotope patterns, and adduct relationships. Anal Chem. 2019;91(20):12799–12807. doi: 10.1021/acs.analchem.9b02354. [DOI] [PubMed] [Google Scholar]
- 137.Yu M., Petrick L. Untargeted high-resolution paired mass distance data mining for retrieving general chemical relationships. Commun Chem. 2020;3(1) doi: 10.1038/s42004-020-00403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen L., Lu W.Y., Wang L., Xing X., Chen Z.Y., et al. Metabolite discovery through global annotation of untargeted metabolomics data. Nat Methods. 2021;18(11):1377. doi: 10.1038/s41592-021-01303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shrivastava A.D., Swainston N., Samanta S., Roberts I., Wright Muelas M., et al. MassGenie: A transformer-based deep learning method for identifying small molecules from their mass spectra. Biomolecules. 2021;11(12) doi: 10.3390/biom11121793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Samanta S., O'Hagan S., Swainston N., Roberts T.J., Kell D.B. VAE-Sim: A novel molecular similarity measure based on a variational autoencoder. Molecules. 2020;25(15) doi: 10.3390/molecules25153446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krenn M., Häse F., Nigam A., Friederich P., Aspuru-Guzik A. Self-referencing embedded strings (SELFIES): A 100% robust molecular string representation. Mach Learn: Sci Technol. 2020;1(4):045024. doi: 10.1088/2632-2153/aba947. [DOI] [Google Scholar]
- 142.Berenger F., Tsuda K. Molecular generation by Fast Assembly of (Deep)SMILES fragments. J Cheminf. 2021;13(1):88. doi: 10.1186/s13321-021-00566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.van der Hooft J.J., Wandy J., Barrett M.P., Burgess K.E., Rogers S. Topic modeling for untargeted substructure exploration in metabolomics. PNAS. 2016;113(48):13738–13743. doi: 10.1073/pnas.1608041113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu Y., Mrzic A., Meysman P., De Vijlder T., Romijn E.P., et al. MESSAR: Automated recommendation of metabolite substructures from tandem mass spectra. PLoS ONE. 2020;15(1):e0226770. doi: 10.1371/journal.pone.0226770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Peters K., Treutler H., Doll S., Kindt A.S.D., Hankemeier T., et al. Chemical diversity and classification of secondary metabolites in nine Bryophyte species. Metabolites. 2019;9(10) doi: 10.3390/metabo9100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tsugawa H., Nakabayashi R., Mori T., Yamada Y., Takahashi M., et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat Methods. 2019;16(4):295. doi: 10.1038/s41592-019-0358-2. [DOI] [PubMed] [Google Scholar]
- 147.Duhrkop K., Nothias L.F., Fleischauer M., Reher R., Ludwig M., et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat Biotechnol. 2021;39(4):462. doi: 10.1038/s41587-020-0740-8. [DOI] [PubMed] [Google Scholar]
- 148.Bach E., Szedmak S., Brouard C., Bocker S., Rousu J. Liquid-chromatography retention order prediction for metabolite identification. Bioinformatics. 2018;34(17):i875–i883. doi: 10.1093/bioinformatics/bty590. [DOI] [PubMed] [Google Scholar]
- 149.Samaraweera M.A., Hall L.M., Hill D.W., Grant D.F. Evaluation of an artificial neural network retention index model for chemical structure identification in nontargeted metabolomics. Anal Chem. 2018;90(21):12752–12760. doi: 10.1021/acs.analchem.8b03118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Abraham M.H., Ibrahim A., Zissimos A.M. Determination of sets of solute descriptors from chromatographic measurements. J Chromatogr A. 2004;1037(1–2):29–47. doi: 10.1016/j.chroma.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 151.Heberger K. Quantitative structure-(chromatographic) retention relationships. J Chromatogr A. 2007;1158(1–2):273–305. doi: 10.1016/j.chroma.2007.03.108. [DOI] [PubMed] [Google Scholar]
- 152.Witting M., Bocker S. Current status of retention time prediction in metabolite identification. J Sep Sci. 2020;43(9–10):1746–1754. doi: 10.1002/jssc.202000060. [DOI] [PubMed] [Google Scholar]
- 153.Xue L., Bajorath J. Molecular descriptors in chemoinformatics, computational combinatorial chemistry, and virtual screening. Comb Chem High Throughput Screening. 2000;3(5):363–372. doi: 10.2174/1386207003331454. [DOI] [PubMed] [Google Scholar]
- 154.Bonini P., Kind T., Tsugawa H., Barupal D.K., Fiehn O. Retip: Retention time prediction for compound annotation in untargeted metabolomics. Anal Chem. 2020;92(11):7515–7522. doi: 10.1021/acs.analchem.9b05765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fedorova E.S., Matyushin D.D., Plyushchenko I.V., Stavrianidi A.N., Buryak A.K. Deep learning for retention time prediction in reversed-phase liquid chromatography. J Chromatogr A. 2022;1664 doi: 10.1016/j.chroma.2021.462792. [DOI] [PubMed] [Google Scholar]
- 156.Kensert A., Bouwmeester R., Efthymiadis K., Van Broeck P., Desmet G., et al. Graph convolutional networks for improved prediction and interpretability of chromatographic retention data. Anal Chem. 2021;93(47):15633–15641. doi: 10.1021/acs.analchem.1c02988. [DOI] [PubMed] [Google Scholar]
- 157.Yang Q., Ji H., Fan X., Zhang Z., Lu H. Retention time prediction in hydrophilic interaction liquid chromatography with graph neural network and transfer learning. J Chromatogr A. 2021;1656 doi: 10.1016/j.chroma.2021.462536. [DOI] [PubMed] [Google Scholar]
- 158.Yang Q., Ji H., Lu H., Zhang Z. Prediction of liquid chromatographic retention time with graph neural networks to assist in small molecule identification. Anal Chem. 2021;93(4):2200–2206. doi: 10.1021/acs.analchem.0c04071. [DOI] [PubMed] [Google Scholar]
- 159.Osipenko S., Botashev K., Nikolaev E., Kostyukevich Y. Transfer learning for small molecule retention predictions. J Chromatogr A. 2021;1644 doi: 10.1016/j.chroma.2021.462119. [DOI] [PubMed] [Google Scholar]
- 160.Domingo-Almenara X., Guijas C., Billings E., Montenegro-Burke J.R., Uritboonthai W., et al. The METLIN small molecule dataset for machine learning-based retention time prediction. Nat Commun. 2019;10 doi: 10.1038/s41467-019-13680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stanstrup J., Neumann S., Vrhovsek U. PredRet: prediction of retention time by direct mapping between multiple chromatographic systems. Anal Chem. 2015;87(18):9421–9428. doi: 10.1021/acs.analchem.5b02287. [DOI] [PubMed] [Google Scholar]
- 162.Bouwmeester R., Martens L., Degroeve S. Generalized calibration across liquid chromatography setups for generic prediction of small-molecule retention times. Anal Chem. 2020;92(9):6571–6578. doi: 10.1021/acs.analchem.0c00233. [DOI] [PubMed] [Google Scholar]
- 163.Ju R., Liu X., Zheng F., Lu X., Xu G., et al. Deep neural network pretrained by weighted autoencoders and transfer learning for retention time prediction of small molecules. Anal Chem. 2021;93(47):15651–15658. doi: 10.1021/acs.analchem.1c03250. [DOI] [PubMed] [Google Scholar]
- 164.Osipenko S., Bashkirova I., Sosnin S., Kovaleva O., Fedorov M., et al. Machine learning to predict retention time of small molecules in nano-HPLC. Anal Bioanal Chem. 2020;412(28):7767–7776. doi: 10.1007/s00216-020-02905-0. [DOI] [PubMed] [Google Scholar]
- 165.Laponogov I., Sadawi N., Galea D., Mirnezami R., Veselkov K.A. ChemDistiller: an engine for metabolite annotation in mass spectrometry. Bioinformatics. 2018;34(12):2096–2102. doi: 10.1093/bioinformatics/bty080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ludwig M., Nothias L.F., Duhrkop K., Koester I., Fleischauer M., et al. Database-independent molecular formula annotation using Gibbs sampling through ZODIAC. Nat Mach Intell. 2020;2(10):629. doi: 10.1038/s42256-020-00234-6. [DOI] [Google Scholar]
- 167.Hoffmann M.A., Nothias L.F., Ludwig M., Fleischauer M., Gentry E.C., et al. High-confidence structural annotation of metabolites absent from spectral libraries. Nat Biotechnol. 2022;40(3):411. doi: 10.1038/s41587-021-01045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duhrkop K., Fleischauer M., Ludwig M., Aksenov A.A., Melnik A.V., et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods. 2019;16(4):299. doi: 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- 169.Tsugawa H., Ikeda K., Takahashi M., Satoh A., Mori Y., et al. A lipidome atlas in MS-DIAL 4. Nat Biotechnol. 2020;38(10):1159. doi: 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- 170.Bach E., Rogers S., Williamson J., Rousu J. Probabilistic framework for integration of mass spectrum and retention time information in small molecule identification. Bioinformatics. 2021;37(12):1724–1731. doi: 10.1093/bioinformatics/btaa998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gerlich M., Neumann S. MetFusion: integration of compound identification strategies. J Mass Spectrom. 2013;48(3):291–298. doi: 10.1002/jms.3123. [DOI] [PubMed] [Google Scholar]
- 172.Ruttkies C., Schymanski E.L., Wolf S., Hollender J., Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminf. 2016;8 doi: 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nikolic D. CASMI 2016: A manual approach for dereplication of natural products using tandem mass spectrometry. Phytochem Lett. 2017;21:292–296. doi: 10.1016/j.phytol.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Alseekh S., Aharoni A., Brotman Y., Contrepois K., D'Auria J., et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat Methods. 2021;18(7):747–756. doi: 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hagiwara T., Saito S., Ujiie Y., Imai K., Kakuta M., et al. HPLC Retention time prediction for metabolome analysis. Bioinformation. 2010;5(6):255–258. doi: 10.6026/97320630005255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Creek D.J., Jankevics A., Breitling R., Watson D.G., Barrett M.P., et al. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal Chem. 2011;83(22):8703–8710. doi: 10.1021/ac2021823. [DOI] [PubMed] [Google Scholar]
- 177.D'Archivio A.A., Maggi M.A., Ruggieri F. Modelling of UPLC behaviour of acylcarnitines by quantitative structure-retention relationships. J Pharm Biomed Anal. 2014;96:224–230. doi: 10.1016/j.jpba.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 178.Kouskoura M.G., Hadjipavlou-Litina D., Markopoulou C.K. Elucidation of the retention mechanism on a reverse-phase cyano column by modeling. J Sep Sci. 2014;37(15):1919–1929. doi: 10.1002/jssc.201400057. [DOI] [PubMed] [Google Scholar]
- 179.D'Archivio A.A., Maggi M.A., Ruggieri F. Prediction of the retention of s-triazines in reversed-phase high-performance liquid chromatography under linear gradient-elution conditions. J Sep Sci. 2014;37(15):1930–1936. doi: 10.1002/jssc.201400346. [DOI] [PubMed] [Google Scholar]
- 180.Cao M., Fraser K., Huege J., Featonby T., Rasmussen S., et al. Predicting retention time in hydrophilic interaction liquid chromatography mass spectrometry and its use for peak annotation in metabolomics. Metabolomics. 2015;11(3):696–706. doi: 10.1007/s11306-014-0727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Aicheler F., Li J., Hoene M., Lehmann R., Xu G., et al. Retention time prediction improves identification in nontargeted lipidomics approaches. Anal Chem. 2015;87(15):7698–7704. doi: 10.1021/acs.analchem.5b01139. [DOI] [PubMed] [Google Scholar]
- 182.Munro K., Miller T.H., Martins C.P., Edge A.M., Cowan D.A., et al. Artificial neural network modelling of pharmaceutical residue retention times in wastewater extracts using gradient liquid chromatography-high resolution mass spectrometry data. J Chromatogr A. 2015;1396:34–44. doi: 10.1016/j.chroma.2015.03.063. [DOI] [PubMed] [Google Scholar]
- 183.Falchi F., Bertozzi S.M., Ottonello G., Ruda G.F., Colombano G., et al. Kernel-based, partial least squares quantitative structure-retention relationship model for UPLC retention time prediction: A useful tool for metabolite identification. Anal Chem. 2016;88(19):9510–9517. doi: 10.1021/acs.analchem.6b02075. [DOI] [PubMed] [Google Scholar]
- 184.Ovcacikova M., Lisa M., Cifkova E., Holcapek M. Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography-electrospray ionization mass spectrometry. J Chromatogr A. 2016;1450:76–85. doi: 10.1016/j.chroma.2016.04.082. [DOI] [PubMed] [Google Scholar]
- 185.Aalizadeh R., Thomaidis N.S., Bletsou A.A., Gago-Ferrero P. Quantitative structure-retention relationship models to support nontarget high-resolution mass spectrometric screening of emerging contaminants in environmental samples. J Chem Inf Model. 2016;56(7):1384–1398. doi: 10.1021/acs.jcim.5b00752. [DOI] [PubMed] [Google Scholar]
- 186.Wolfer A.M., Lozano S., Umbdenstock T., Croixmarie V., Arrault A., et al. UPLC-MS retention time prediction: a machine learning approach to metabolite identification in untargeted profiling. Metabolomics. 2016;12(1) doi: 10.1007/s11306-015-0888-2. [DOI] [Google Scholar]
- 187.Kubik L., Wiczling P. Quantitative structure-(chromatographic) retention relationship models for dissociating compounds. J Pharm Biomed Anal. 2016;127:176–183. doi: 10.1016/j.jpba.2016.02.050. [DOI] [PubMed] [Google Scholar]
- 188.Barron L.P., McEneff G.L. Gradient liquid chromatographic retention time prediction for suspect screening applications: A critical assessment of a generalised artificial neural network-based approach across 10 multi-residue reversed-phase analytical methods. Talanta. 2016;147:261–270. doi: 10.1016/j.talanta.2015.09.065. [DOI] [PubMed] [Google Scholar]
- 189.Randazzo G.M., Tonoli D., Hambye S., Guillarme D., Jeanneret F., et al. Prediction of retention time in reversed-phase liquid chromatography as a tool for steroid identification. Anal Chim Acta. 2016;916:8–16. doi: 10.1016/j.aca.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 190.Taraji M., Haddad P.R., Amos R.I., Talebi M., Szucs R., et al. Prediction of retention in hydrophilic interaction liquid chromatography using solute molecular descriptors based on chemical structures. J Chromatogr A. 2017;1486:59–67. doi: 10.1016/j.chroma.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 191.Taraji M., Haddad P.R., Amos R.I.J., Talebi M., Szucs R., et al. Use of dual-filtering to create training sets leading to improved accuracy in quantitative structure-retention relationships modelling for hydrophilic interaction liquid chromatographic systems. J Chromatogr A. 2017;1507:53–62. doi: 10.1016/j.chroma.2017.05.044. [DOI] [PubMed] [Google Scholar]
- 192.Zhang X., Li J., Wang C., Song D., Hu C. Identification of impurities in macrolides by liquid chromatography-mass spectrometric detection and prediction of retention times of impurities by constructing quantitative structure-retention relationship (QSRR) J Pharm Biomed Anal. 2017;145:262–272. doi: 10.1016/j.jpba.2017.06.069. [DOI] [PubMed] [Google Scholar]
- 193.Park H., Lee J.M., Kim J.Y., Hong J., Oh H.B. Prediction of liquid chromatography retention times of erectile dysfunction drugs and analogues using chemometric approaches. J Liq Chromatogr Relat Technol. 2017;40(15):790–797. doi: 10.1080/10826076.2017.1364264. [DOI] [Google Scholar]
- 194.Wen Y., Talebi M., Amos R.I.J., Szucs R., Dolan J.W., et al. Retention prediction in reversed phase high performance liquid chromatography using quantitative structure-retention relationships applied to the Hydrophobic Subtraction Model. J Chromatogr A. 2018;1541:1–11. doi: 10.1016/j.chroma.2018.01.053. [DOI] [PubMed] [Google Scholar]
- 195.Wen Y., Amos R.I.J., Talebi M., Szucs R., Dolan J.W., et al. Retention index prediction using quantitative structure-retention relationships for improving structure identification in nontargeted metabolomics. Anal Chem. 2018;90(15):9434–9440. doi: 10.1021/acs.analchem.8b02084. [DOI] [PubMed] [Google Scholar]
- 196.McEachran A.D., Mansouri K., Newton S.R., Beverly B.E.J., Sobus J.R., et al. A comparison of three liquid chromatography (LC) retention time prediction models. Talanta. 2018;182:371–379. doi: 10.1016/j.talanta.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hall L.M., Hill D.W., Bugden K., Cawley S., Hall L.H., et al. Development of a reverse phase HPLC retention index model for nontargeted metabolomics using synthetic compounds. J Chem Inf Model. 2018;58(3):591–604. doi: 10.1021/acs.jcim.7b00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bouwmeester R., Martens L., Degroeve S. Comprehensive and empirical evaluation of machine learning algorithms for small molecule LC retention time prediction. Anal Chem. 2019;91(5):3694–3703. doi: 10.1021/acs.analchem.8b05820. [DOI] [PubMed] [Google Scholar]
- 199.Souihi A., Mohai M.P., Palm E., Malm L., Kruve A. MultiConditionRT: Predicting liquid chromatography retention time for emerging contaminants for a wide range of eluent compositions and stationary phases. J Chromatogr A. 2022;1666 doi: 10.1016/j.chroma.2022.462867. [DOI] [PubMed] [Google Scholar]
- 200.Liapikos T., Zisi C., Kodra D., Kademoglou K., Diamantidou D., et al. Quantitative structure retention relationship (QSRR) modelling for Analytes' retention prediction in LC-HRMS by applying different Machine Learning algorithms and evaluating their performance. J Chromatogr, B: Anal Technol Biomed Life Sci. 2022;1191 doi: 10.1016/j.jchromb.2022.123132. [DOI] [PubMed] [Google Scholar]