Summary
Culturing eukaryotic cells has widespread applications in research and industry, including the emerging field of cell-cultured meat production colloquially referred to as “cellular agriculture”. These applications are often restricted by the high cost of growth medium necessary for cell growth. Mitogenic protein growth factors (GFs) are essential components of growth medium and account for upwards of 90% of the total costs. Here, we present a set of expression constructs and a simplified protocol for recombinant production of functionally active GFs, including FGF2, IGF1, PDGF-BB, and TGF-β1 in Escherichia coli. Using this E. coli expression system, we produced soluble GF orthologs from species including bovine, chicken, and salmon. Bioactivity analysis revealed orthologs with improved performance compared to commercially available alternatives. We estimated that the production cost of GFs using our methodology will significantly reduce the cost of cell culture medium, facilitating low-cost protocols tailored for cultured meat production and tissue engineering.
Subject areas: Biological sciences, Biochemistry, Biomolecules
Graphical abstract
Highlights
-
•
Developed methodology for low-cost production of soluble, bioactive GFs
-
•
Purified GFs were active on NIH-3T3 and bovine satellite cells
-
•
Some GF orthologs outperformed commercially sourced GFs
-
•
Production of GFs using these methods can foster significant cost savings
Biological sciences; Biochemistry; Biomolecules.
Introduction
Cellular agriculture is a rapidly emerging field that seeks to leverage biotechnology to produce agricultural products from cell cultures (Post et al., 2020). Study in cellular agriculture has intensified in recent years because of the proposed environmental and ethical benefits compared to traditional agriculture (Mattick et al., 2015). The field has spurred the development of a range of products such as milk and egg alternatives produced using recombinantly produced protein (Geistlinger et al., 2017; Dance, 2017); however, the primary focus of research in cellular agriculture relates to the development of cell-cultured meat (CCM). Also referred to as cell-based meat or cultivated meat, these products marry cell culture, tissue engineering, and bioprocessing to develop three-dimensional “slab” meat products independent of animals (save for the initial biopsy used for cell line development or primary cell isolation) (Melzener et al., 2021). Direct comparison between CCM and traditionally produced meat products predict that an animal-free approach to meat production can reduce greenhouse gas emissions by up to 95%, conserve land and water usage, attenuate the overuse of antibiotics in agricultural settings, and reduce the incidence of zoonotic infections (Godfray et al., 2018). Furthermore, the development of CCM as a bona fide food source has the potential to decentralize supply chains and address issues related to food insecurity and sustainability. The potential benefits of CCM have been well studied; however, for these advantages to be actualized there are several technological challenges that continue to remain as significant hurdles.
The core technology at the foundation of CCM production is eukaryotic cell culture. Cell lines or primary cells of bovine, avian, and teleost origin are used to produce cell-cultured beef, chicken, and fish, respectively. Within the field of cellular agriculture, and eukaryotic cell culture more broadly, cells can be supported with the addition of 5–20% fetal bovine serum (FBS). FBS is obtained from blood and used as a growth supplement to sustain in vitro cell culture by providing hormones, lipids, micronutrients, and, most critically, mitogenic growth factors (GFs) that promote cell division (van der Valk et al., 2018). The use of FBS is near-ubiquitous in research settings; however, its continued use within cellular agriculture is seen as problematic for a number of reasons. FBS is costly; it can suffer from batch-to-batch variation that can result in issues of reproducibility or reduced yield, and the fact that it is sourced from fetal calves undermines the ethical advantage afforded by CCM compared to meat produced using traditional agriculture.
Serum-free alternatives to FBS have been commercially available for decades and in the case of chemically defined serum-free media (SFM), they offer improved consistency more compatible with the controlled manufacturing processes adopted in CCM production. Commercially available serum-free options include Essential 8 (Chen et al., 2011), TeSR, and FBM and recent efforts have endeavored to use these as templates for creating low-cost serum-free formulations tailored for iPSC cells (Kuo et al., 2020) and bovine satellite cells (BSC) (Stout et al., 2022). Although SFM removes the ethical concerns related with the use of FBS and offers improved consistency in cell culture performance, it can be even more cost prohibitive than FBS-containing media. At current prices, the use of SFM for the production of CCM creates a significant barrier for competitive entry into consumer markets.
Mitogenic GFs, such as basic fibroblast growth factor (FGF2) and transforming growth factor β1 (TGF-β1), are the main cost drivers of SFM, estimated to contribute more than 95% of the total cost (Specht, 2020). These signaling proteins bind to cell surface receptors activating several downstream pathways to result in cell proliferation, cell migration, differentiation, and apoptosis, and are added as a supplement to SFM to mimic the GF composition provided by traditional serum additives. Although a wide array of GFs is used to support eukaryotic cell culture, many of which are tissue/cell-type specific, there are a subset which are considered essential for supporting cell proliferation in general (Yao and Asayama, 2017). These GFs are key components of SFM used to support cell growth for academic and cellular agriculture applications.
Recombinant production of these GFs is challenging and is a major contributor to their high cost. They are of eukaryotic origin and typically require some level of post-translational modification, such as oxidation or glycosylation, and proper disulfide bond formation is essential for protein folding and maturation into their biologically active conformation. Consequently, production of these proteins in bacterial expression systems has been challenging. Strategies to address this issue include refolding GFs expressed in bacteria from inclusion bodies or using mammalian cell culture systems (e.g., CHO, HEK293) for their production (Alexander et al., 1992; Zou and Sun, 2004). Both strategies result in high costs and render SFM cost-prohibitive for use in the production of CCM (Tripathi and Shrivastava, 2019).
Escherichia coli remains a staple of recombinant protein production and when compatible with a target protein, offers advantages in terms of speed, tractability, and cost (Rosano and Ceccarelli, 2014). Several advancements in strain engineering have made E. coli an improved host for production of disulfide-containing proteins. Mutation of trxB and gor genes (Bessette et al., 1999), as well as cytoplasmic overexpression of disulfide bond isomerases (e.g., DsbC) (Lobstein et al., 2012) are strategies that have facilitated the expression of disulfide-containing proteins in E. coli, culminating in the development of commercial strains such as Rosetta-gami B (Novagen) and SHuffle T7 (NEB), respectively. In addition, the use of redox fusion partners such as thioredoxin (Trx) (LaVallie et al., 1993), glutathione S-transferase (GST) (Schafer et al., 2015), and disulfide bond isomerases (DsbA, DsbC) (Nozach et al., 2013) have greatly improved soluble expression of proteins that contain disulfide bonds in E. coli. Despite these advancements, the use of E. coli to produce soluble mitogenic GFs remains under documented as commercial production still relies on refolding in instances where E. coli is the chosen expression host.
To facilitate cost reductions in SFM and to advance its adoption in the field of cellular agriculture, we screened a myriad of GF genes, fusion partner combinations and E. coli strains to develop a collection of expression systems that allow for production of biologically active mitogenic GFs in E. coli. As a result, we were able to identify expression conditions to successfully produce several GFs from a variety of species including those useful in CCM production (bovine, avian, and fish). Expressed GFs can be purified using a one-step immobilized metal affinity chromatography (IMAC) purification procedure with an optional additional “clean-up” step. This methodology is feasible for academic labs as well as CCM start-up ventures and results in biologically active GFs able to promote cellular proliferation. Using this methodology, we identified GF orthologs that provide higher potency than commercially available counterparts. Finally, we provide a techno-economic assessment to demonstrate that our methodology provides a strategy for significant cost reductions associated with the recombinant production of mitogenic GFs. These results have the potential to significantly reduce the cost of SFM formulations in the field of cellular agriculture and eukaryotic cell culture applications more broadly.
Results
Selection of growth factors and expression system components
GFs to target for cost effective production were selected based on their prevalence in FBS or in current SFM-based formulations, and their likelihood of being widely adopted in CCM production. Based on these criteria, we focused on four major GF families represented by fibroblast growth factor (FGF1 and FGF2), platelet-derived growth factor (PDGF-BB), insulin-like growth factor (IGF1 and IGF2) and transforming growth factor beta 1 (TGF-β1). For each of these families we selected multiple representatives from diverse species within available genome sequences to sample sequence diversity and to expand the repertoire of GFs able to support the growth of cells relevant for cultured meat production. Furthermore, we tested our most successful expression strategies on additional GFs including leukemia inhibitory factor (LIF), epidermal growth factor (EGF) and myostatin/growth differentiation factor 8 (MSTN/GDF8).
We chose to express the GFs in E. coli, which remains the most widely used, versatile, and inexpensive recombinant expression system. Our selection of specific E. coli strains and expression vectors was driven by the general goal of maximizing the production of GFs in a soluble and biologically active form that would also allow for simplified purification using a standard IMAC protocol. Considering the prevalence of intramolecular disulfide bonds in many GF proteins, we selected commercially available E. coli strains altered to accommodate the expression of such proteins including Rosetta-gami 2(DE3) (Novagen), Origami B(DE3) (Novagen) and SHuffle T7 express (NEB) in addition to BL21-Gold (DE3) (Agilent) as the hosts to test for recombinant GF production.
For our first attempt, each of the selected GFs was expressed with an N-terminal polyhistidine (6xHis) tag using the pMCSG53 vector or the redox fusion partner, thioredoxin A (TrxA) via pET-TrxA. To assess the success of these protein production strategies we performed small-scale expression tests of proteins for each family of GF in the selected E. coli host strains and screened for the presence of soluble intracellular protein expression using SDS-PAGE (Figures 1A and S1). We achieved limited success with these two fusion partners for some GF families, so we expanded our screen to include additional redox and non-redox fusion partner sequences. For non-redox fusion partners, we used the 97 amino acid SUMO ubiquitin-like protein from Saccharomyces cerevisiae (Addgene plasmid #29711) and the B1 domain of the Streptococcal protein G (GB1) (Cheng and Patel, 2004), the latter of which has been successfully used to facilitate soluble expression of human EGF in E. coli (Zheng et al., 2016).
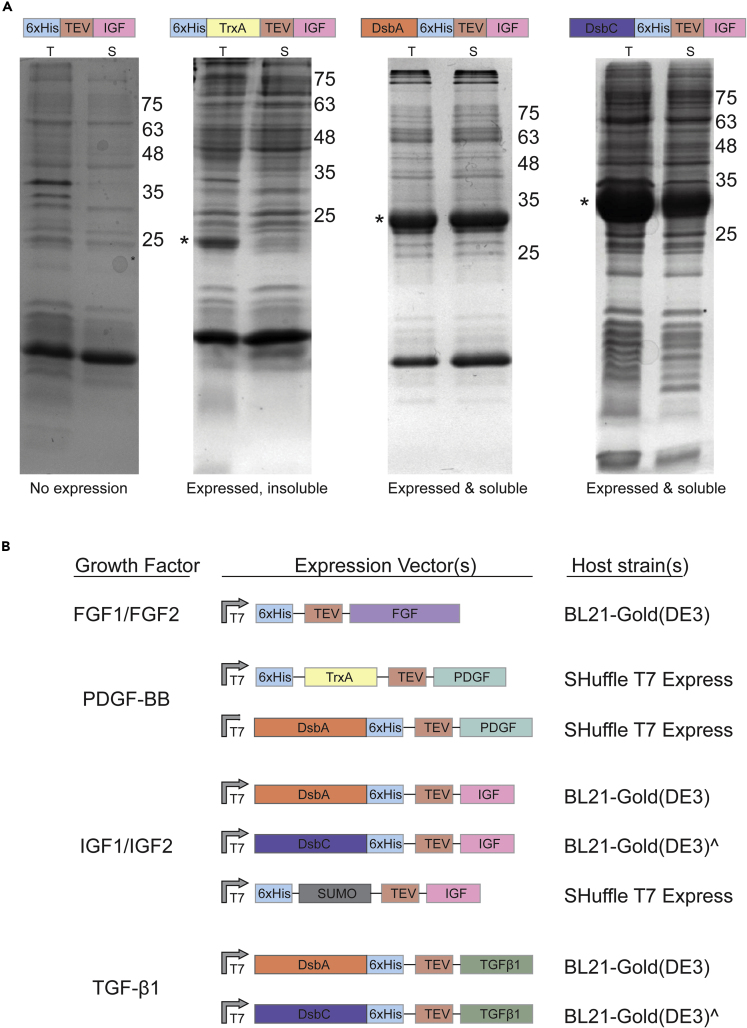
Figure 1.
Expression systems for recombinant GF production
(A) Small-scale protein expression screening used to identify the expression vector and host strain combination capable of facilitating cytoplasmic soluble protein expression. The band corresponding to the protein of interest is marked with (∗). T - total cell lysate; S - soluble fraction. Molecular weight markers are labeled to the right of each gel image.
(B) Expression vector and host strain combinations for successful expression and purification of soluble, bioactive growth factors. (ˆ) denotes instances where the use of SHuffle T7 Express was required for soluble expression of some orthologs.
For additional redox fusion partners, we incorporated disulfide bond isomerases (E. coli DsbA or DsbC) that promote formation of disulfide bonds and have been previously described as potent facilitators of soluble expression of disulfide-bond rich proteins in E. coli. For this, we modified a previously constructed T7-based expression vector, pMCSG53 (Biancucci et al., 2017; Eschenfeldt et al., 2013), to include either the leaderless DsbA or DsbC protein as an N-terminal fusion partner for the GF protein. These new vectors, pMCSG53-DsbA and pMCSG53-DsbC, are LIC and Gibson Assembly compatible, allowing for high-throughput entry of GF sequences to promote investigation of amino acid sequence diversity within these protein families. Two variants of the DsbA and DsbC vectors were tested; Dsb[A/C]-6His-TEV and 6His-Dsb[A/C]-TEV; however, we observed no functional differences between variants. All expression constructs included a Tobacco Etch Virus (TEV) protease cleavage site (ENLYFQG) between the tag and GF sequence to allow for removal of the fusion partner after purification.
The expression level of GFs was tested on a small scale (see STAR Methods for details) to identify the most promising strain and construct combinations for soluble recombinant GF expression (Figure 1A). An overview of the most successful expression combination(s) for each GF is summarized in Figure 1B. Below we discuss the outcome of these strategies for each of the selected growth factor families in detail.
Expression and purification strategy for fibroblast growth factor (FGF1 and FGF2)
FGF1 (acidic FGF) and FGF2 (basic FGF) are 155 amino acid proteins that exert mitogenic and angiogenic activities on target cells through interaction with fibroblast growth factor receptor (FGFR) and subsequent activation of the downstream RAS-MAPK, P13K-AKT, STAT, and PLCγ signaling pathways (Brewer et al., 2016). FGF2 also plays a role in tissue development and repair by binding to heparin sulfate cofactor (Koledova et al., 2019). FGFs are a critical component of cell culture medium, particularly for maintaining pluripotency of stem cells and their proliferative capacity (Mossahebi-Mohammadi et al., 2020). It is also a major mitogen in FBS, required for proliferative activity of cells and is typically present in the range of 10–100 ng/mL in cell culture medium (Yang and Xiong, 2012).
Both FGF1 and FGF2 have been shown to be prone to proteolytic degradation and denaturation in cell culture, contributing to their relatively short half-life and present a major cost driver for cell culture medium (Caldwell et al., 2004; Chen et al., 2012). These undesirable properties can be mitigated through addition of heparin to the growth medium; however, this strategy adds to the overall medium costs and may not meet food safety requirements for CCM applications. FGF variants offering improved thermostability have been identified through protein engineering efforts (Zakrzewska et al., 2004; Benington et al., 2020), but their approval for application in CCM products may be complicated because of their “genetically modified” categorization. With these shortcomings of currently available FGF in mind, we developed a workflow to allow for low-cost bacterial expression and purification of an array of FGF orthologs.
FGF1 and FGF2 have been previously produced recombinantly in E. coli with a variety of fusion partners (e.g., TrxA, 6HFh8, 6His (Kim et al., 2021; Gasparian et al., 2009; Kuo et al., 2020); however, some of these approaches require multiple purification steps and the use of specialized chromatography equipment. We tested the production of these GFs by a single step IMAC protocol followed by a TEV protease tag cleavage and removal using the same affinity resin used for initial purification as shown in Figure 2A (see details in STAR Methods). Additional aspects of our methodology include the use of the synthetic codon-optimized genes and sampling of 21 sequence orthologs (5 FGF1, 16 FGF2) to capture a representative level of amino acid sequence diversity within this protein family. Our strategy allowed for successful expression and purification of 5 FGF1 and 16 FGF2 proteins from terrestrial (bovine, chicken, sheep, and human) and aquatic (Atlantic salmon, Nile tilapia, Chinook salmon) species (Tables S1 and S2). The protein yields of purified FGF2 ranged from 7 mg/L for Bos taurus (bovine) to 36 mg/L for Oryzias latipes (rice fish), highlighting significant variation in FGF ortholog yield while expressed under the same general conditions (Figure S2).
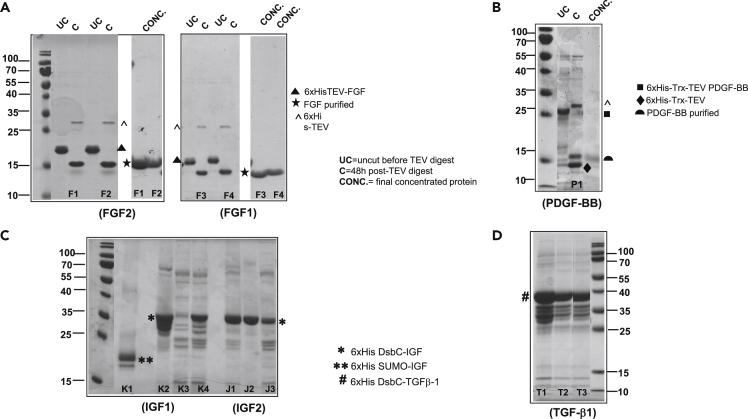
Figure 2.
Recombinant GF production
Scale up of protein expressions for (A) FGF2 AND FGF1 cloned in pMCSG53 vector with N-terminal His6x tag and expressed in BL21(DE3) Gold cells. Targets include F1 (FGF2_Atlantic salmon); F2 (FGF2_Pufferfish); F3 (FGF1_Sheep); F4 (FGF1_Bovine) (B) PDGF-BB expressed in SHuffle T7 express cells. Target shown is P1 (PDGF-BB_Cormorant) (C) IGF1/IGF2 cloned in pMCSG53-His6x-DsbC/pMCSG53-His6x-SUMO and expressed in SHuffle T7 express cells. Targets include (K1) IGF1_Bovine (SUMO-His6x tag); (K2) IGF1_Bovine (DsbC-His6x tag); (K3) IGF1_Goose; (K4) IGF1_Frog; (J1) IGF2_Human; (J2) IGF2_Bovine; (J3) IGF2_Nile tilapia (D) TGFβ-1 cloned in pMCSG53-His6x-DsbC and expressed in SHuffle T7 express cells. Targets shown are TGFβ1_human (T1); TGFβ−1_bovine (T2); TGFβ−1_chicken (T3); TGFβ−1_little egret (T4). UC = uncut before TEV digest;C = 48h post-TEV digest; TEV protease runs at 25 kDa (marked with ˆ). After the TEV digest and a second Ni-NTA affinity chromatography step, the concentrated, purified FGF2/FGF1 runs at 15 kDa on an SDS-PAGE (marked with ★) shown in (A). PDGF-BB runs at 15 kDa corresponding to the monomer (marked with  ) shown in (B). DsbC fusion IGF1/IGF2 runs at 35 kDa (marked with ∗). IGF1-SUMO runs at 20 kDa (marked with ∗∗), as seen in (C). DsbC-TGFβ-1 runs at 40 kDa (marked with #).
) shown in (B). DsbC fusion IGF1/IGF2 runs at 35 kDa (marked with ∗). IGF1-SUMO runs at 20 kDa (marked with ∗∗), as seen in (C). DsbC-TGFβ-1 runs at 40 kDa (marked with #).
Expression and purification strategy for platelet-derived growth factor (PDGF-BB)
Platelet-derived growth factors (PDGF) are potent mitogens for a variety of cells including fibroblasts, smooth muscle cells, connective tissue cells, and bone and cartilage cells. PDGF has been shown to increase proliferation of mouse myoblasts and is a key component for the production of CCM (Albrecht and Tidball, 1997). Isoforms (A, B, C, and D) of PDGF form homo- or heterodimers (e.g., PDGF-BB and PDGF-AB) stabilized by intermolecular disulfide bonding. All four PDGF isoforms feature a highly conserved 100 amino acid GF domain (Breitkopf et al., 2005). Dimerized PDGF binds to two cognate tyrosine kinase receptors, platelet-derived growth factor receptor α (PDGFRα) and PDGFRβ, initiating intracellular signal transduction pathways. Examples of pathways involved in response to PDGF-BB activation include MEK/ERK, Src, and PI3K/AKT (Fredriksson et al., 2004).
Commercially available human PDGF-BB is produced recombinantly in E. coli; however, previously published protocols are plagued by low yields, or rely on purification from inclusion bodies under denaturing conditions with the requirement of subsequent refolding to obtain bioactive PDGF-BB (Alexander et al., 1992). To improve on existing PDGF expression procedures we tested expression of 25 PDGF-BB orthologs (avian, mammalian, reptilian, and aquatic) in fusion with N-terminal 6xHis tag and 6xHis-TrxA or 6xHis-DsbA double tags. The 6xHis-TEV-PDGF-BB fusions were individually expressed in BL21(DE3)-Gold, Rosetta-gami B, and SHuffle T7 E. coli host strains, but only 1 out of 25 GFs showed soluble expression under these conditions. In contrast, the use of an N-terminal 6xHis-TrxA double fusion resulted in 21 out of 25 PDGF-BB produced in a cytoplasmic soluble form (Table S2). We scaled up protein expression and purified 12 of these PDGF-BB orthologs ranging from bovine, bony-tongue fish, cormorant, eagle, cobra, green anole, and sea turtle and achieving protein yields ranging from 2–5 mg/L (Figure S2). The 6xHis-TrxA tag was efficiently cleaved from PDGF-BB using TEV protease (Figure 2B); however, in a subset of the orthologs that were purified we observed co-elution of TrxA and PDGF-BB after the second Ni-NTA purification step. Adding a size-exclusion chromatography step was successful in separation of PDGF-BB from TrxA in cases where this tag remained associated with PDGF-BB after TEV protease digestion (Figure S3).
The four PDGF-BB orthologs that were not produced as soluble proteins using the pET-TrxA fusion were all sourced from Teleost (ray-finned fish) species. Fusion of these orthologs to an N-terminal DsbA tag allowed for their soluble expression. Using this approach, the PDGF-BB from climbing perch (Anabas testudineus) was purified at a yield of 7 mg/L. Analysis of all recombinantly purified PDGF-BB proteins via non-reducing SDS-PAGE confirmed the expected pattern of protein dimerization (Figure S4).
Expression and purification strategy for insulin-like growth factor (IGF1 and IGF2)
IGF1 and IGF2 are 70 amino acid disulfide-rich proteins with a similar structure to the peptide hormone insulin (Vajdos et al., 2001; Brown et al., 2008). IGF signaling is central to pathways that control cell growth, maturation, and proliferation. IGF1 is a major component of FBS and is widely used in cell culture media including many different formulations used to promote myogenesis and the development of skeletal muscle (McCubrey et al., 1991; Hakuno and Takahashi, 2018; Yu et al., 2015). IGF1 and IGF2 function on cells by binding to receptor-tyrosine kinases (RTKs), the IGF1 receptor (IGF1R) and IGF2 receptor (IGF2R), respectively. IGF1 and IGF2 can also bind the insulin receptor, though they do so with lower affinity compared to their interactions with their cognate receptors. IGF1 and IGF2 initiate signal transduction through the PI3K/AKT and ERK1/2 signaling pathways resulting in stimulation of cell growth and proliferation (Peruzzi et al., 1999). Commercially available IGF1 is typically produced in E. coli as inclusion bodies, purified under denaturing conditions and then refolded into the bioactive form. Recent studies have reported that DsbA (Emamipour et al., 2019) or 6HFh8 (a six histidine tag followed by the Fh8 peptide from Fasciola hepatica) (Kim et al., 2021) fusion partners promote the soluble expression of mammalian IGF1 in E. coli.
Based on our success using the TrxA fusion tag to facilitate PDGF-BB expression, we tested the same approach for expression of 15 IGF1 and IGF2 orthologs derived from mammalian, avian, and fish species. This strategy did not result in soluble expression of IGF based on small-scale expression testing (Figure 1A). Next, we assessed the ability of DsbC, DsbA, and SUMO fusion tags to promote soluble expression of IGF in E. coli. Expression of IGF as a DsbC-IGF fusion resulted in significant increases in the production of soluble GF protein (Figures 1A and 2C). With this expression system, all selected IGF orthologs (human, bovine, and fish) were produced primarily as soluble fusion protein (6 IGF1 and 3 IGF2), (Figure S2). Yields of DsbC-IGF1 fusion protein ranged from 6 to 10 mg/L. The DsbA-IGF2 fusion had similar results in terms of protein solubility, but the overall expression level and yield was generally less than compared to expression of DsbC-IGF1 fusions. The expression of SUMO-IGF1 fusions also resulted in production of soluble IGF protein but only in the SHuffle T7 E. coli host strain. TEV protease cleavage of the DsbA/DsbC fusion tags was efficient; however, similar to what we observed for some TrxA-PDGF-BB fusion proteins, the cleaved fusion partner co-eluted with IGF1 after the second Ni-NTA chromatography step. Anion exchange chromatography (MonoQ) resulted in some degree of separation, but the addition of this step may be problematic at production scales necessary for cellular agriculture applications. To determine if separation of the fusion DsbA/DsbC tag from IGF1/IGF2 was a necessary step for bioactivity, we assayed the ability of these fusion GFs to support cell growth and to ascertain if the presence of an N-terminal DsbA/DsbC/SUMO tag had an impact on their functional bioactivity (discussed in sections below).
Expression and purification strategy for transforming growth factor beta 1 (TGF-β1)
TGF-β1 belongs to the transforming growth factor superfamily which consists of isoforms TGF-β1-3 and of other signaling proteins such as myostatin (MSTN), growth differentiation factor (GDF8), and bone morphogenetic factor (BMP) (Poniatowski et al., 2015). TGF-β GF are synthesized as a larger precursor protein and then proteolytically cleaved into a 112 amino acid active GF (Derynck et al., 1985). They are rich in cysteine residues and contain multiple intramolecular disulfide bonds participating in formation of a cysteine knot (Hinck et al., 1996). In addition, disulfide bonding results in the formation of a covalently linked homodimer that is critical for biological activity. TGF-β1 is a key component of the growth medium used for skeletal muscle growth and differentiation and is critical for maintaining the pluripotency of stem cells (Mullen and Wrana, 2017; Scharenberg et al., 2014). For these reasons, it is a major component of growth medium optimized for production of CCM (Boudreault et al., 2001).
Recombinant production of TGF-β1 has traditionally relied on expression using eukaryotic expression systems such as Chinese hamster ovary (CHO) or human embryonic kidney (HEK293) cells, contributing to its high cost and role as a major cost driver in cell culture media (Zou and Sun, 2004). Commercial sources of TGF-β1 derived from E. coli are available (e.g., QKine) but remain costly compared with in-house production. A recent report detailed successful soluble expression of TGF-β3 in E. coli in fusion with a TrxA tag but reported low yields of soluble TGF-β1 using the same approach (Kuo et al., 2020). Based on our success in using the TrxA fusion partner to facilitate soluble expression of PDGF-BB, and reports detailing soluble expression of TGF-β3 using this fusion partner, we attempted to express 10 TGF-β1 orthologs (human, bovine, fish, and avian) with an N-terminal TrxA fusion. Although we did observe expression of a protein species of appropriate size, none of the TrxA-TGF-β1 orthologs were expressed as a soluble protein. Next, we tested GB1 tag as the TGF-β1 fusion partner, but the results were similar with the expressed protein found only in the insoluble fraction. In contrast, expression of TGF-β1 as a DsbA or DsbC fusion protein resulted in several TGF-β1 orthologs (human, bovine, fish, and avian) expressed in cytoplasmic soluble fraction. Yields of purified DsbC-TGF-β1 fusion protein ranged from 4.5 to 6 mg/L (Figure 2D). As observed with the IGF1/IGF2 GFs, separation of DsbC/DsbA from TGF-β1 after TEV-digest required an anion exchange chromatography step.
Expression and purification of additional growth factor families
Our success with recombinant expression of GF that are the key components of SFM prompted us to try similar strategies for expression of additional GFs that have utility in CCM production. Epidermal growth factor (EGF) is used in differentiation of satellite cells into myotubes and supports the proliferative capacity of mammalian adipose-derived stem cells (Ai et al., 2017). A previous study showed that a GB1 fusion supported soluble expression of human EGF in E. coli (Zheng et al., 2016). We tested this system on an EGF ortholog from Betta splendens and confirmed its ability to promote soluble expression of EGF from a Teleost fish species (Figure S5). We were able to produce soluble GB1-EGF from B. splendens with a yield of 13.5 mg/L.
Leukemia inhibitory factor (LIF) is used in supporting cell culture of BSC (Spangenburg andBooth, 2002). We had success in achieving soluble expression of LIF orthologs from human, bovine, fish, and avian species by using an N-terminal 6xHis-GB1 double fusion partner (Figure S5). Myostatin (GDF8) is a member of the TGF-β superfamily and is important in regulating muscle development and skeletal muscle mass. Similar to our results with TGF-β1, an N-terminal DsbC fusion facilitated soluble cytoplasmic expression of myostatin (GDF8) orthologs from human and bovine species (Figure S5). These results further validate our developed methodology for GFs and its application to several GF families relevant to cell culture media formulation.
Validating the functional bioactivity of the recombinantly purified growth factors
We tested the purified recombinant GFs for their ability to stimulate the proliferation of NIH-3T3 mouse fibroblast cells in a dose-dependent manner using a colorimetric MTT assay. This assay follows the cellular metabolic activity as a measure of viable proliferating cells. To assess the dose-dependent behavior, serum-starved (0.11% FBS) fibroblast NIH-3T3 cells were individually treated with varying concentrations of each purified GF. All of the recombinantly purified GFs tested demonstrated the ability to support similar and, in some cases higher, cell proliferation than their commercially available mammalian counterparts. We report the cell proliferation capacity of cultures containing each recombinantly produced GF as a fold-change when compared with that of the non-supplemented, serum-starved cells (negative control) under similar experimental conditions (Figure 3). Cells grown in culture medium supplemented with 10% commercial FBS served as the positive control.
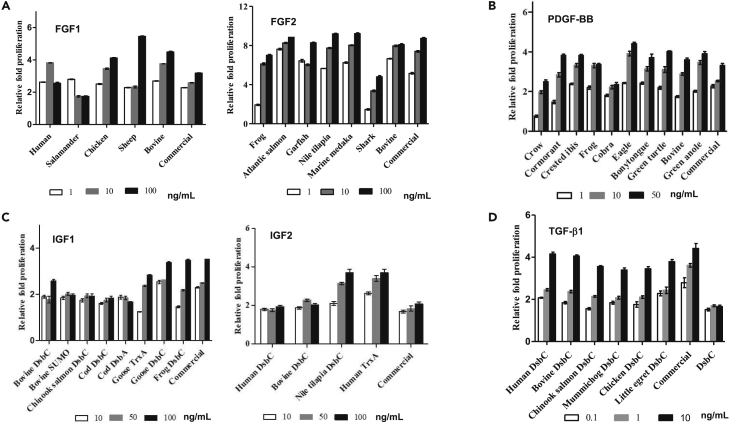
Figure 3.
Functional bioactivity validation by colorimetric MTT assay
For each growth factor target: (A) FGF; (B) PDGF-BB; (C) IGF; (D) TGF-β1, cell proliferation is plotted as relative fold-change when compared with that of the untreated serum-starved cells under similar experimental conditions. Absorbance readings at 540 nm were recorded in six replicates and averaged. Data plotted as mean ± standard deviation (SD) using Graphpad Prism v5.0. Error bars represent the SD calculated for the replicates. Growth factor concentrations are in ng/mL.
The culture medium treated with FGF2 orthologs derived from bovine (B. taurus), Atlantic salmon (Salmo salar), marine medaka (Oryzias melastigma), and Nile tilapia (Oreochromis niloticus), at 10 ng/mL, all showed similar fold proliferation (8.0–8.3-fold for the aforementioned FGF2 orthologs versus 7.4-fold proliferation with commercial human FGF2). In the case of FGF2 orthologs derived from bovine and Atlantic salmon, cells treated with these FGF2 variants showed 1.5× higher proliferation (6.8-fold and 7.6-fold, respectively), at a much lower (1 ng/mL) FGF2 concentration compared to cells grown in culture medium supplemented with commercial human FGF2 (5-fold change compared to serum starvation). If this fold-change could be equivalently achieved in bovine myoblast cells or fish cells, it could have a significant impact on the cost incurred for culture media formulation in CCM production. According to our results, the FGF2 orthologs we tested can accommodate up to a 10X reduction in the amount of FGF2 added while still promoting similar levels of cell proliferation.
We observed a similar phenomenon when assaying recombinant FGF1. FGF1 orthologs derived from human (Homo sapiens) and bovine (B. taurus) showed 1.6× higher proliferative capacity when compared to the commercial human FGF1 at 10 ng/mL. The FGF1 ortholog derived from chicken (Gallus gallus) showed 3.5-fold proliferation at a concentration of 10 ng/mL in comparison to 2.5-fold with commercial human FGF1. At 100 ng/mL, FGF1 from sheep achieved 1.8× higher proliferative capacity as compared to commercial human FGF1.
PDGF-BB orthologs derived from bovine (B. taurus), crested ibis (Nipponia nippon), frog (Xenopus tropicalis), eagle (Haliaeetus albicilla), and green anole (Anolis carolinensis) showed comparatively higher fold-proliferation (3.5- to 4.0-fold) at a much lower concentration of 10 ng/mL in comparison to commercial PDGF-BB (3.5-fold) proliferation at 50 ng/mL, which is the typical culture medium concentration for PDGF-BB. The ability to produce these PDGF-BB orthologs at a low cost and their potential to sufficiently stimulate proliferation at lower concentrations makes them key candidates for advancing cost reductions in CCM production.
All orthologs of IGFs derived from expression with the different fusion partners (SUMO, DsbC, DsbA) showed similar fold-proliferation as the commercial human IGF1/IGF2. Recombinantly purified bovine IGF1 (DsbC fusion) showed 2.6-fold versus 3.5-fold proliferation for commercial human IGF1 at 100 ng/mL concentration when compared to untreated serum-starved cells. Purified IGF2 orthologs derived from human and bovine both showed similar 2-fold proliferation as the commercial human IGF2 at a concentration of 100 ng/mL. Interestingly, IGF2 ortholog from Nile tilapia (O. niloticus) showed 1.8× higher fold proliferation than the commercial human IGF2 (3.6-fold and 2-fold respectively at a concentration of 100 ng/mL).
TGF-β1 orthologs from human and bovine both showed 4-fold proliferation as compared to 4.4-fold from commercial human TGF-β1 at a concentration of 10 ng/mL. TGF-β1 from chicken (G. gallus), northern pike (Egretta garzetta), Chinook salmon (Oncorhynchus tshawytscha) and mummichog (Fundulus heteroclitus) showed 3.5-fold, 3.8-fold, 3.6-fold, and 3.4-fold respectively, proliferation at a concentration of 10 ng/mL when compared with that of untreated cells. Taken together, our results with IGFs and TGF-β1 demonstrate that although it is possible to remove the 6xHis-DsbC, DsbA or SUMO fusion partners, the presence of the fusion tags does not interfere with bioactivity of IGF1/IGF2 or TGF-β1 in cell culture medium.
In addition to measuring the ability of our recombinantly produced GFs to stimulate cell proliferation, we conducted western blot analysis to observe activation of the downstream cell signaling pathways that regulate cellular proliferation. Activation of ERK1/2 through phosphorylation is the key determinant of the cellular response to FGF, PDGF-BB, and IGF growth factor signaling (Zhu et al., 2010). NIH-3T3 cells treated with the recombinantly purified FGF2, PDGF-BB, and DsbC-IGF1/IGF2 fusion orthologs were analyzed for the detection of phosphorylated ERK1/2 (p-ERK1/2). ERK1/2 requires a dual phosphorylation of conserved threonine and tyrosine residues to be activated. Our western blot results showed bands at 42 and 44 kDa, indicative of phosphorylated ERK1/2, thus complementing the results of our cell proliferation assays and validating that our recombinant GFs activate the expected intracellular signaling pathways (Figures 4 and S6).
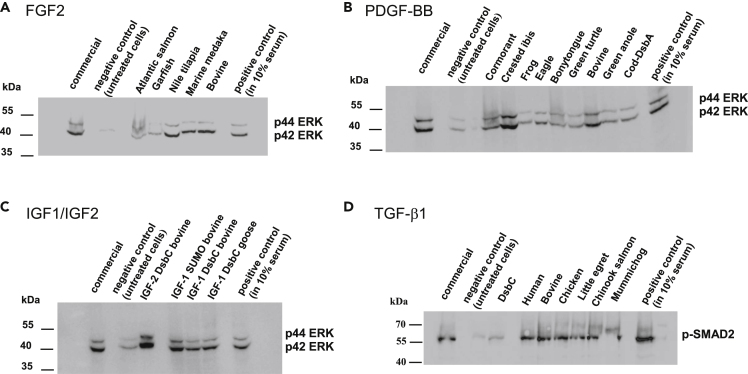
Figure 4.
Western blot analysis of cell extracts from NIH-3T3 cells treated GFs
Cells treated with (A) FGF2 targets at 10 ng/mL for 24 h at 37°C (B) PDGF-BB targets at 50 ng/mL for 24 h at 37°C (C) IGF1/IGF2 targets at 100 ng/mL for 24 h at 37°C. All the lysed cell extracts were analyzed for phosphorylated p44/p42 ERK1/2 (p-ERK1/2, 42, 44 kDa). Negative control (untreated cells) shows no detection of phosphorylated ERK1/2, whereas all recombinantly purified FGF2, PDGF-BB, IGF1, IGF2 targets and their commercial counterparts show the presence of bands approximately at 42 kDa and 44 kDa (D) TGF-β1 targets at 10 ng/mL for 45 min at 37°C and analyzed for phosphorylated SMAD2 (p-smad2; 60 kDa). Negative control (untreated cells) shows no detection of p-smad2 band whereas all recombinantly purified TGF-β1 targets and commercial TGF-β1 show the presence of a band at 60 kDa.
Phosphorylation of SMAD proteins is an event that occurs after TGF-β1 binds to the TGFβ-receptor at the cell surface, thereby promoting cell proliferation (Schmierer and Hill, 2007). To confirm that our DsbC-TGF-β1 fusion proteins were functionally active, we performed western blotting against phosphorylated SMAD2 (p-SMAD2). Our results showed that the recombinantly purified DsbC-TGF-β1 GF were capable of inducing phosphorylation of SMAD2, validating this GF retains the proper signaling functionality even with the DsbC fusion tag. (Figures 4 and S6).
Exploring cultured meat applicability by testing bioactivity on BSC
To assess the suitability of the recombinantly-produced GF for use in cultured meat applications we assayed for bioactivity on BSC. The BSC used were isolated from three different donor animals and have been previously shown to express myogenic markers and to form myosin heavy chain-positive myotubes (Skrivergaard et al., 2021). Using these cells, we screened multiple orthologs from each GF family in seven different concentrations ranging from 1.56 to 100 ng/mL. Similar to our results with NIH-3T3 cells, we observed clear evidence that the recombinantly produced GF promote cell proliferation in BSC. The immediate effect after 48 h was less pronounced than what we observed in NIH-3T3 cells; however, this is likely because of the slower growth and/or more complex biology of primary cells. These intrinsic differences associated with primary cells are evident even in the 10% FBS “optimal” condition tested, which produced a relatively modest 1.43-fold increase in proliferation (Figure 5).
Figure 5.
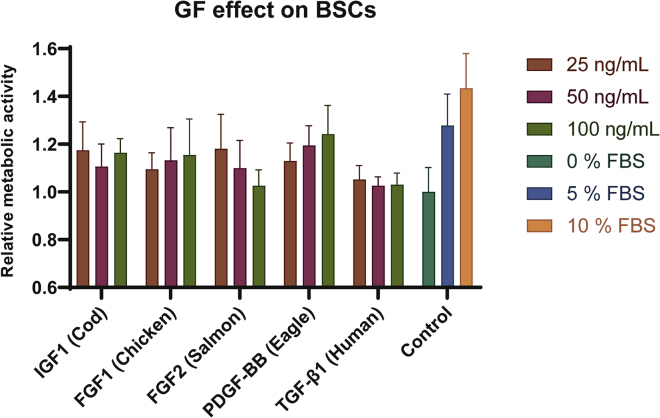
Growth factor effect on Bovine Satellite Cells (BSCs)
Proliferation data from the best performing GF orthologs summarized with the three highest concentrations compared to FBS controls. All values are relative to 0% FBS (serum-free) control. WST-1 absorbance readings at 450 nm were recorded in duplicate wells of the three biological replicates (n = 3) for the GF samples, whereas the FBS controls were measured in quadruplicate wells and averaged from five independent experiments (each GF group) of the three biological replicates (n = 15). Data plotted as mean ± standard deviation (SD), with error bars depicting SD.
Variation in their ability to promote BSC proliferation was observed between GF orthologs and the best performing ortholog for each GF family is shown in Figure 5. Atlantic salmon FGF2 was the most potent, promoting the highest level of proliferation among the FGF2 orthologs tested. The effect was highest at 25 ng/mL and notably decreased at higher concentrations (Figure S7). Atlantic salmon FGF2 was also the most potent when tested on NIH-3T3 cells, highlighting the potential for use of this protein in broader cell culture applications. The most pronounced effect was observed when BSC were treated with PDGF-BB from Eagle at 100 ng/mL. At this concentration, the GF promoted BSC proliferation at a level (1.24-fold increase) comparable to that observed during treatment with 5% FBS (1.28-fold increase). Overall, all GF families tested showed a positive effect on the BSC, validating their bioactivity in a cell system highly relevant to cultured meat production (Figure S7). The GFs produced using the E. coli systems outlined demonstrate clear potential to aid in development of SFM for cultured beef applications.
Techno-economic analysis of “in-house” growth factor production
We sought to develop an economic assessment for production of GFs using the methodology in this study. An estimate based on laboratory consumables costs and labour cost showed that we could achieve recombinant production of GFs for CAD$ 10.22 per milligram of purified protein (Table 1). A one-time cloning cost incurred for each GF target protein (inclusive of gene synthesis, sequencing primers and PCR) was calculated as approximately CAD$ 60.00. A more rigorous breakdown for the total cost estimation analysis is presented in the supplemental information (Table S3).
Table 1.
Production cost of “in house” growth factors: All pricing in CAD and per milligram purified protein
| Growth Factor | Protein production costs (“in house”) (CAD) | Cost “in house”/mg protein (CAD) | Cost PeproTech/mg protein (CAD) | Cost R&D Systems/mg protein (CAD) | Expression system used by commercial suppliers |
|---|---|---|---|---|---|
| FGF1/FGF2 | $1225.90/120 mg protein | $10.22 | $1043.00 | $2,600.00 | E. coli |
| PDGF-BB | $1225.90/60 mg protein | $20.43 | $5,087.00 | $5,500.00 | E. coli (from inclusion bodies) |
| IGF1/IGF2 | $1225.90/120 mg protein | $10.22 | $326.00 | $766.00 | E. coli (from inclusion bodies) |
| TGF-β1 | $1225.90/120 mg protein | $10.22 | $6,782.00 | $7,150.00 | CHO cells, HEK293 cells |
Total production cost of 1225.90 CAD includes all lab consumables and labor costs (see Table S3) and estimated for 12 L production scale of E. coli culture growth. Calculations based on total yield of 120 mg purified protein for all except PDGFBB for which the total yield is 60 mg from a 12L culture.
Earlier studies had estimated that FGF2 and TGF-β1 can account for as much as 95% of the costs of SFM when sourced from traditional commercial suppliers (Specht, 2020). Using our “in-house” methodology, the overall cost contribution of GFs (FGF2 and TGF-β1) to the total cost of SFM can be substantially reduced (Table 2). Table 3 summarizes a price comparison in the contribution of GF to total costs, specifically shown for Essential 8 medium demonstrating a 7-fold reduction in culture medium costs and a substantial reduction in the GF cost contribution to the total media cost from 76–95% (depending on commercial supplier) to 2%.
Table 2.
Pricing comparison of FGF2 and TGF-β1 produced “in-house” versus commercially sourced (R&D Systems commercial source as reference)
| Growth Factor | Final conc. (mg/L) in media | “In house” Cost/L (CAD) |
R&D Systems Cost/L (CAD) |
Pricing Comparison (fold-change) |
|---|---|---|---|---|
| FGF2 (100 ng/mL) | 0.1 | $1.02 | $260.00 | 255× reduction |
| TGF-β1 (10 ng/mL) | 0.01 | $0.10 | $72.00 | 720× reduction |
GF concentrations calculated based on TGF-β1 and FGF2 usage levels in Essential 8 media.
Table 3.
Price comparison of the total cost of cell culture medium
| Medium Component | “In house” cost/L | PeproTech cost/L | R&D Systems cost/L |
|---|---|---|---|
| FGF2 | $1.02 | $104.30 | $260.00 |
| TGF-β1 | $0.10 | $67.82 | $72.00 |
| DMEM basal medium Thermo Fisher (cat. 1195065) |
$31.06 | $31.06 | $31.06 |
| Ascorbic Acid 2-phosphate Sigma (cat. A8960) |
$1.13 | $1.13 | $1.13 |
| NaHCO3 Sigma (cat. S5761) |
$0.04 | $0.04 | $0.04 |
| Insulin Sigma (cat. 91077C) |
$10.74 | $10.74 | $10.74 |
| Transferrin Sigma (cat. T8158) |
$11.47 | $11.47 | $11.47 |
| Sodium selenite Sigma (cat. S5261) |
<$0.01 | <$0.01 | <$0.01 |
| Total Cost | $55.57 | $226.57 | $386.45 |
| Percentage of Cost Attributed to FGF2 and TGF-β1 | 2.0% | 76% | 86% |
Media pricing was estimated per liter of E8.
Beyond the cost of producing the individual GFs, our discovery of specific non-mammalian orthologs that provide comparable or improved bioactivity at lower concentrations also allows for a further reduction in the overall cost of growth medium. Specifically, the ability to achieve comparable cell proliferation with reduced amounts (ng/mL) of FGF2 (10-fold reduction as in FGF2 Atlantic salmon ortholog) can result in even further reductions in media costs.
Discussion
The presence of mitogenic GFs is a primary requirement for cell culture medium essential for various applications both in academia and bioindustry including the emerging fields of tissue engineering and cellular agriculture. To address the need for cost effective serum-free eukaryotic cell culture medium, we pursued the development of protocols for producing scalable, low-cost recombinantly expressed GFs commonly used in medium compositions. This new data will help to facilitate cost reductions in basic tissue engineering research applications as well as the large-scale applications anticipated for future CCM production.
Environmental and ethical concerns surrounding traditional animal agriculture have brought increased focus on the development of alternative sources for dietary protein. CCM represents a promising approach for augmenting existing food production in a land, water, and carbon efficient manner while eliminating the need to harvest from whole animals. Despite its promise, CCM production is currently challenged by the high cost associated with in vitro cell culture, in particular the costs associated with growth medium. Although SFM is effective in supporting in vitro cell culture, its cost is often prohibitive for large scale applications, and this is driven primarily by the cost of mitogenic GF components. Addressing this challenge, we have developed an array of expression protocols that facilitate high levels of soluble bioactive GF protein production in E. coli, a tractable host that can be cultured with minimal specialized equipment and in BSL1 laboratory facilities.
Our methodology outlined is standardized and based on classic IMAC affinity chromatography techniques, allowing for the same procedure to be used in preparation of bioactive FGF1 and FGF2, IGF1 and IGF2, PDGF-BB and TGF-β1. The modest demands for laboratory equipment and technical expertise allow for this methodology to be suitable for both academic and industrial settings with minimal equipment and experience in recombinant protein production technology that should facilitate significant reductions in the costs associated with procuring bioactive GFs (Figure 6). Furthermore, we have demonstrated that many of the GFs retain bioactivity as fusion proteins, allowing for purification to proceed via a single chromatography step and without the need for fusion partner cleavage and separation. Despite this observation, we have also outlined and validated strategies for efficient removal of fusion partner tags should it be deemed necessary by end users of these expression systems (Figure S3). The output of these systems is also compatible with further purification steps, such as for the removal of endotoxins (Reichelt et al., 2006), should they be required in the final application of the GFs. The expression systems we have developed represent significant improvements in the expression of soluble and bioactive PDGF-BB and TGF-β1 in E. coli.
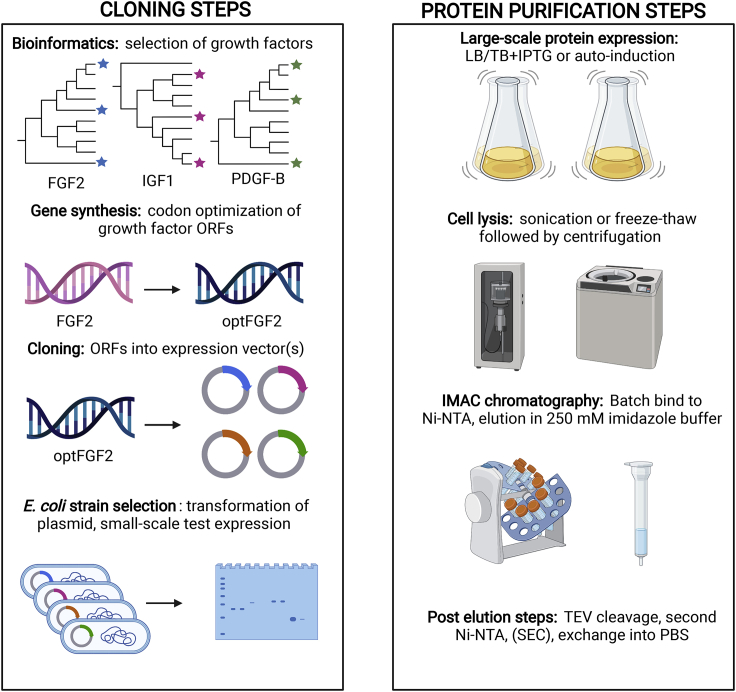
Figure 6.
Generalized GF cloning and protein expression strategy
Cloning strategy allows for high-throughput screening of desired growth factor sequences. The protein purification flowchart highlights the ease of purification using IMAC affinity chromatography, requiring the use of Ni-NTA resin and a gravity column. This generalized strategy was used for purification of all GF described in this study. Image was made using BioRender.
The presented approach allowed for expression of multiple orthologs from each of the GF families, including sequences from mammalian, avian, and Teleost fish origin in accordance with the CCM industry focus on growing skeletal muscle cells belonging to these groups. Though we did not investigate or rationalize why sequence variations among GF orthologs dramatically affects their purification and bioactivity characteristics, the ultimate outcome is projected to be a significant cost reduction as decreased amounts of FGF2, PDGF-BB or TGF-β1 are suitable for promoting similar fold cell proliferation as the commercial GFs. Further exploration of existing amino acid sequence diversity within these GF families is warranted and may uncover variants that provide improved performance for meeting the unique needs of CCM production.
In screening GF orthologs for bioactivity, we found several FGF2 orthologs that induced higher levels of cell proliferation on NIH-3T3 cells compared to commercial mammalian FGF2. This effect was observed not only at the standard working concentration of FGF2 (10 ng/mL) but also at reduced concentrations of 0.1–1 ng/mL, raising the possibility that FGF2 orthologs may provide improved proliferation while using less material. Critically, this result was consistent in experiments conducted to test the GF on BSC, a highly relevant cell line for CCM applications. Our functional bioactivity results thus prove a significant cost reduction strategy by using decreased concentrations of FGF2, PDGF-BB or TGF-β1 to promote similar fold cell proliferation as the commercially available GFs.
Analysis of the cost of production showed that GFs generated using the protocols we have outlined provide substantial cost savings compared to commercially available alternatives, significantly reducing the cost contribution of GFs to approximately 2% of the culture medium. Of note, our pricing is based on yields obtained using a bench-scale shake flask approach. It can reasonably be assumed that optimizing growth conditions and transitioning to large-scale bioreactor fermentation could further improve GF yields and facilitate greater cost reductions.
The methodology outlined for soluble expression of key GF components is designed to allow for academic laboratories as well as small-scale cellular agriculture start-ups to bring GF production “in-house”, thereby allowing for significant cost savings. Most of the expression protocols presented in our study are based on a single IMAC purification and require no in vitro protein refolding steps. Although we have documented additional steps that can improve GF purity (e.g., TEV cleavage, size exclusion chromatography, anion-exchange chromatography), in many cases these are optional and may not be required for GFs aimed to be used in cell culture medium. Because the initial purification steps for all these GFs are identical, a standardized workflow that requires minimal specialized equipment or technical expertise can be adopted to facilitate production of the major cost drivers of SFM (Figure 6).
The expression systems we have developed proved to be robust for a variety of mitogenic GFs and represent a key advancement to meeting the increasing demands for these critical components in cell culture media. The set of expression vectors generated in this study also provide a toolset for bacterial expression of other GF families not tested here, and in general, for disulfide-rich proteins more broadly that may have been recalcitrant to bacterial expression system.
Limitations of the study
The functional bioactivity of the recombinant GFs was assayed using a fibroblast cell line and bovine satellite cells. Further testing against additional cell types such as adipocytes will help to further explore their potential for use in cellular agriculture. It was beyond the scope of the work presented to investigate the molecular mechanisms behind the variation in functional activity observed between different growth factor orthologs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-smad2-rabbit mAb | Cell Signaling Technologies | cat. no. 3108; RRID: AB_490941 |
| Smad2 rabbit mAb | Cell Signaling Technologies | cat. no. 5339; RRID: AB_10626777 |
| Phospho-p44/42 MAPK ERK1/2 rabbit mAb | Cell Signaling Technologies | cat. no. 9101; RRID: AB_331646 |
| P44/42 MAPK ERK1/2 rabbit mAb | Cell Signaling Technologies | cat. no. 4695; RRID: AB_390779 |
| Bacterial and virus strains | ||
| E. coli DH5α | CGSC | CGSC# 14231 |
| E. coli BL21-Gold (DE3) Competent Cells | Agilent | cat. no. 230132 |
| E. coli SHuffle T7 Express Competent Cells | NEB | cat. no. C3029J |
| E. coli Rosetta-gami B (DE3) Competent Cells | Millapore Sigma | SKU 71136–3 |
| Chemicals, peptides, and recombinant proteins | ||
| Tryptone (Bacteriological) | BioShop | cat. no. TRP402 |
| Yeast extract | BioShop | cat. no. YEX401 |
| Sodium chloride | VWR | Cat. no. BDH9286 |
| Ampicillin, sodium salt | BioShop | Cat. no. AMP201 |
| HEPES, biotechnology grade | BioShop | Cat. no. HEP001 |
| Imidazole | Sigma | Cat. no. 15513 |
| HisPur Ni-NTA resin | Thermo Fisher | Cat. no. 88221 |
| 30% Acrylamid/Bis solution, 37.5:1 | BioRad | Cat. no. 1610159 |
| TEMED | BioRad | Cat. no. 1610800 |
| Ammonium persulfate, electrophoresis grade | BioShop | Cat. no. AMP001 |
| PBS | Wisent | Cat. no. 311-010-CL |
| PBS (BSCs) | Gibco | Cat. no. 14190144 |
| Trypsin-0.25% EDTA | Wisent | Cat. no. 325-042-CL |
| Trypsin 2.5% (BSCs) | Gibco | Cat. no. 15090046 |
| Human FGF1 | PeproTech | Cat. no. 100-17A |
| Human FGF2 | PeproTech | Cat. no. 100-18B |
| Human PDGF-BB | PeproTech | Cat. no. 100-14B |
| Human IGF1 | PeproTech | Cat. no. 100-11 |
| Human IGF2 | PeproTech | Cat. no. 100-12 |
| Human TGF-β1 | PeproTech | Cat. no. 100-21C |
| Tobacco Etch Virus (TEV) protease | This study | N/A |
| 1X RIPA Buffer | Cell Signaling Technologies | Cat. no. 9806 |
| DMEM | Gibco | Cat. no. 11995-065 |
| DMEM + Glutamax (BSCs) | Gibco | Cat. no. 61965-026 |
| Newborn calf serum | Gibco | Cat. no. 26010-074 |
| Fetal bovine serum (BSCs) | Gibco | Cat. no. 10100-147 |
| Horse Serum (BSCs) | Gibco | Cat. no. 16050122 |
| Sodium pyruvate (BSCs) | Gibco | Cat. no. S8636 |
| Matrigel Matrix (BSCs) | Corning | Cat. no. 356237 |
| Pen-Strep (BSCs) | Gibco | Cat. no. 15140122 |
| Amphotericin B (BSCs) | Gibco | Cat. no. 15290026 |
| Gentamicin (BSCs) | Sigma | Cat. no. G1397 |
| Critical commercial assays | ||
| MTT Assay | Invitrogen | cat. no. M6494 |
| Coomassie Plus (Bradford) Protein Assay | Thermo Fisher | Cat. no. 23236 |
| Clarity Western ECL | BioRad | Cat. no. 170-5060 |
| WST-1 (BSCs) | Roche | Cat. no. 11 644 807001 |
| Experimental models: Cell lines | ||
| NIH-3T3 Mouse fibroblasts | ATCC | CRL-1658 |
| Experimental models: Organisms/strains | ||
| Bovine Satellite Cells (from 3 donors) | Previous study (see reference) | N/A |
| Oligonucleotides | ||
| T7, promoter TAATACGACTCACTATAGGG |
IDT DNA | N/A |
| T7, terminator | IDT DNA | N/A |
| pMCSG-FWD TACTTCCAATCCAATGCC |
IDT DNA | N/A |
| pMCSG-REV TTATCCACTTCCAATGTTA |
IDT DNA | N/A |
| Recombinant DNA | ||
| pMCSG53 | N/A | |
| pET-TrxA | AddGene | Cat. no. 29712 |
| pET-SUMO | AddGene | Cat. no. 29711 |
| pMCSG53-6His-DsbC | This study | N/A |
| pMCSG53-DsbC-6His | This study | N/A |
| pMCSG53-6His-DsbA | This study | N/A |
| pMCSG53-DsbA-6His | This study | N/A |
| pMCSG53-GB1 | This study | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be filled by the lead contact, Alexei Savchenko (alexei.savchenko@ucalgary.ca).
Materials availability
Plasmids pMCSG53-DsbC (no. 186623), pMCSG53-DsbA (no. 186624) and pMCSG53-GB1 (no. 186786) have been deposited to AddGene. Additional materials used in this study are available upon request.
Experimental model and subject details
Bacterial strains used in this study are outlined in the Key Resources Table. E. coli was cultured at 37°C in LB or TB at 200 rpm. Induction for protein expression occurred at 20°C.
Satellite cells were isolated from three different bovine donors as described in (Skrivergaard et al., 2021). The donors (3–4 yearold Holstein dairy cows) were transported to a commercial plant (Danish Crown, Aalborg, Denmark), and slaughtered within 1 h after arrival by stunning with captive bolt followed by bleeding and electrical stunning at 100 V for 400 s. The Musculus semitendinosus was removed within 20 min postmortem and transported on ice to the cell laboratory at department of Food Science, Aarhus University.
Method details
Bacterial strains, vector construction
E. coli DH5α were used for cloning and vector construction. For protein expression, BL21-Gold (DE3) (Agilent), Shuffle T7 express (NEB), Origami B (DE3), Rosetta Gami B (DE3) (EMD-Millipore) competent cells were used. Commercially used vectors include pMCSG53-His6x-TEV-LIC (MCSG) with LIC cloning sites, pET-His6x-TrxA-TEV-LIC (Addgene 29712), pET- His6x-SUMO-TEV-LIC (Addgene 29711). The vectors pMCSG53-His6x-DsbC-TEV and pMCSG53-His6x-DsbA-TEV were designed “in-house” using the pMCSG53-His6x-TEV-LIC (Biancucci et al., 2017) backbone and introducing the DsbC/DsbA gene in frame downstream of His6x tag.
All the insert gene sequences were codon optimized for E. coli expression system and synthesized from Twist BioSciences or Codex DNA. The insert genes were cloned into the vector pMCSG53 or pET-TrxA or pET-SUMO or pMCSG53-DsbC, pMCSG53-DsbA, coding for a fusion protein with an N-terminal 6× histidine tag cleavable by TEV protease. DNA sequencing in the cloned plasmid was verified at the TCAG DNA sequencing facility, SickKids, Toronto.
Protein expression and purification
E. coli competent cells were transformed with the plasmids. 20 mL of overnight culture (16h approx.) in Luria Bertani (LB) broth was diluted into 1 L of LB or terrific broth (TB) containing selected antibiotics (100 ug/mL ampicillin) and grown at 37°C using the shaker flask approach. Expression was induced with 0.6 mM IPTG at 17°C when optical density, OD600 reached 0.8–0.9 absorbance units and allowed to grow overnight for 16–18h. The large overnight cell cultures were then collected by centrifugation at 7000g.
Cells were resuspended in binding buffer [pH 7.5, 100 mM HEPES, 500 mM NaCl, 5 mM imidazole, and 5% glycerol (v/v)] and lysed. The cell debris was removed by centrifugation at 20,000g. Ni-NTA affinity chromatography was used for protein purification and performed in a Econo-column chromatography column (Bio-Rad). The soluble cytoplasmic fraction containing the GF protein was purified by batch-binding to Ni-NTA resin (performed at 4°C for 1 h), washed with wash buffer [pH 7.5, 100 mM HEPES, 500 mM NaCl, 30 mM imidazole, and 5% glycerol (v/v)], and the protein was eluted with elution buffer [pH 7.5, 100 mM HEPES, 500 mM NaCl, 250 mM imidazole, and 5% glycerol (v/v)]. The His6-tagged protein was then subjected to overnight (16–24h approx.) TEV protease digest using 50 μg of “in house” TEV protease per mg of His6x-tagged protein and simultaneously dialyzed overnight against phosphate-buffered saline (PBS), pH 7.4 buffer containing no imidazole. The 6xHis-tag and TEV was removed by binding the protein onto a Ni-NTA column again, whereby the flow through contained the His-tag cleaved protein of interest. For plasmid constructs where the TEV protease digest step was skipped, the final proteins were dialyzed with a minimum of 2 x 2L dilution cycles in (PBS) pH 7.4.
For PDGF-BB, the His6x-Trx tag was separated from PDGF-BB using size-exclusion chromatography (Superdex 75 16/60). The flowthrough from the second Ni-NTA was applied to Superdex 75 16/60 column (AKTA systems) using PBS pH 7.4 at a flow rate of 1 mL/min and 0.5 mL fractions were collected over 1.5× column volume. Fractions were analyzed via SDS-PAGE, and those containing PDGF-BB were pooled and concentrated.
For TGF-β1 and IGF1/IGF2, the His6x-DsbC tag was separated from the GF using anion exchange chromatography (MonoQ 5/5). The flowthrough of the second Ni-NTA was applied to an anion exchange chromatography (IEX), MonoQ HR 5/5 (AKTA systems) using 50 mM CAPS pH 10.65, 10 mM NaCl (Buffer A) and 50 mM CAPS pH 10.65, 1M NaCl. A stepwise gradient of 30%B resulted in separation from the DsbC tag to a significant extent (data not shown).
The purity of all the recombinantly purified proteins were analyzed by SDS-polyacrylamide gel electrophoresis. Non-reducing gel electrophoresis was used to confirm the presence of dimeric state of the proteins. The purified proteins were also subjected to size exclusion chromatography (Superdex 200 16/60) analysis for determination of their oligomeric state. The proteins were concentrated using a Vivaspin concentrator (GE Healthcare) and passed through a 0.2 μm Ultrafree-MC centrifugal filter (EMD Millipore) before storing in aliquots at −80°C.
Cell culture – NIH 3T3
NIH 3T3 mouse fibroblast cell lines (ATCC CRL-1658) were cultured in 100 × 20 mm and 150 × 25 mm culture dishes (Falcon Corning cat. no. 353003 and 353025) using Dulbecco’s modified Eagle’s medium containing 4.5 g/L D-glucose, L-glutamine, 110 mg/L sodium pyruvate (DMEM, GIBCO 11995-065) with 10% heat inactivated newborn calf serum, NBCS (GIBCO 26010-074). Primary pancreatic stellate cells (PSC) were cultured until passage 8 in DMEM with 10% heat inactivated fetal bovine serum and penicillin-streptomycin. Cultures were allowed to reach 80% confluency before passaging them into new media. Briefly, cells were washed twice with phosphate-buffered saline (PBS) without calcium and magnesium, Wisent 311-010-CL, cells detached using Trypsin-0.25% EDTA (Wisent) and split at a seeding density of 5,000 cells/cm2. Commercial GFs used in this study included the following: human FGF1 (PeproTech, cat. no. 100–17A), human FGF2 (PeproTech, cat. no. 100–18B), human PDGF-BB (PeproTech, cat. no. 100–14B), human IGF1 (PeproTech, cat. no. 100-11), human IGF2 (PeproTech, cat. no. 100-12), human TGF-β1(PeproTech, cat. no. 100–21C).
Bioactivity MTT assay
NIH-3T3 cells were cultured in 96-well plates at a seeding density of 6000 cells/well and allowed to attach for 24h. Cells were subsequently serum starved for 24 h in 0.11% serum medium. GFs were added to individual wells at specific serial dilution concentrations and incubated at 37°C with 5% CO2. After 48 h, the metabolic activity of viable cells was tested by a colorimetric dimethyl-thiazole-diphenyltetrazolium bromide (MTT) assay (cat. no. M6494, Invitrogen). Briefly, the steps involved aspirating the culture media, washing the wells with PBS to remove any dead cells, and then adding 100 μL of DMEM media containing 10 μL MTT (5 mg/mL in sterile PBS) into each well. The 96 well plates were incubated at 37°C for 4 h. The media was then aspirated, wells washed with 100 μL PBS and the insoluble MTT crystals were solubilized with 50 μL of DMSO by incubating in an orbital plate shaker at 700 rpm, 37°C for 20–30 min. Absorbance was recorded immediately on a Tecan plate reader at a wavelength of 540 nm. Three independent experiments were conducted for each set of GFs with different passage number cells. Triplicate readings from each set were averaged and data was presented as mean ± SD (SD = standard deviation) using Graphpad Prism v5.0.
Western blotting
Cells were cultured in 6-well plates at a seeding density of 100,000 cells/well. After 24 h, culture medium in the wells was changed into 0.11% serum. Cells were serum starved for 24 h before adding the respective GFs. Cell lysis was done after 45 min for TGF-β1, 24h for FGF2, PDGFBB, IGF1. To extract the total protein, cells were lysed by washing the cells twice in ice-cold PBS, then lysed with 1X RIPA buffer (Cell Signalling Tech. cat. no. 9806), incubated at 4°C for 1 h and centrifuged at 14,000 rpm for 20 min at 4°C. The supernatant was mixed with 1X SDS-sample buffer containing β-mercaptoethanol, boiled at 95°C for 10 min and samples loaded onto a 15% SDS-PAGE protein gel. Proteins were transferred to a nitrocellulose membrane using Trans-Blot apparatus (Bio-Rad). After blocking the membrane with 5% w/v BSA and 0.1% v/v Tween-20 in TBS (Tris buffered-saline) for 1 h, the membrane was washed 5x with TBST (Tris buffered-saline containing 0.1% v/v Tween20). The membrane was incubated with the following primary antibodies overnight at 4°C on a rotating shaker: phospho-smad2 rabbit mAb (Cell Signalling Tech. cat. no. 3108; 1:1000), smad2 rabbit mAb (Cell Signalling Tech. cat. no. 5339; 1:1000), phospho-p44/42 MAPK ERK1/2 rabbit mAb (Cell Signalling Tech., cat. no. 9101; 1:1000), p44/42 MAPK ERK1/2 rabbit mAb (Cell Signalling Tech., cat. no. 4695; 1:1000). The membrane was then incubated with Anti-rabbit IgG, HRP-conjugated secondary antibody (Cell Signalling Tech., cat. no. 7074; 1:2000) with gentle agitation for 1h at room temperature. Immunoactivity was detected using Bio-Rad Clarity Western ECL peroxide-enhancer chemiluminescence kit (cat. no. 170–5060). Chemiluminescent bands were visualized with a Chemidoc XRS+ system (BioRad).
Cell culture – Bovine satellite cells
Satellite cells that were previously isolated from three different bovine donors (Skrivergaard et al., 2021) were expanded in Matrigel coated T75 flasks (Nunc cat. no. 156499) using growth media consisting of DMEM (Gibco, 61965-026) with 10% FBS (fetal bovine serum), 10% HS (horse serum), 1mM sodium pyruvate and 1 × antibiotics (penicillin 100 units/mL, streptomycin 0.1 mg/mL, amphotericin B 2.5 μg/mL and gentamicin 0.1 mg/mL) before being cryopreserved in growth media with 10% DMSO in liquid nitrogen. Cryopreserved cell samples (passage 3) were quickly thawed and diluted in growth media, before being pelleted at 500 × g for 10 min at 4°C and resuspended in 37°C growth media to remove DMSO-containing freeze media. Satellite cells were then initially cultured in Matrigel coated T25 flasks (Nunc cat. no. 156367) for 4 days using growth media at 37°C in a 5% CO2 humidified atmosphere before cell experiments.
WST-1 metabolic assay
Bovine satellite cells were washed twice with PBS without calcium and magnesium (Gibco, 14190144) and detached with 0.25% Trypsin (Gibco, 15090046) in PBS before being centrifuged at 500 × g for 10 min at 4°C and counted using a Burker-Turk hemacytometer. Satellite cells were cultured in Matrigel coated 96-well plates (Nunc cat. no. 167008) at a cell density of 1000 cells/well and were allowed to attach in DMEM with 10% FBS for 24 h. Cells were then washed with PBS and 100 μL serum-free DMEM with the different GFs at various concentrations was added. After 48 h of incubation the metabolic activity was tested by adding 30 μL/well WST-1 reagent (Roche) diluted in serum-free DMEM (1:10 final concentration) and incubating for 2 h before reading the absorbance at 450 nm (600 nm background subtraction). WST-1 has a similar metabolic reaction as MTT used in the NIH 3T3 cell line. Serum-free control (0%) and serum control samples (5 and 10%) were included in all plates and all GF concentrations were analyzed in duplicate wells for the three different biological replicates which were averaged and presented as mean ± SD in relative values compared to the serum-free control.
Quantification and statistical analysis
Standard deviation was calculated using Graphpad Prism v5.0 and the SD data is found in Figures 3 and 5.
Acknowledgments
We thank Dr. Alison McGuigan and members of the McGuigan lab for allowing us to use their cell culture facility and for helpful input on methodology. We also thank Dr. Tom Ben-Arye, Senior Scientist, The Good Food Institute (GFI) for scientific input and advice. This work has been funded by a research grant from GFI awarded to Peter J Stogios, a New Harvest Postdoctoral Fellowship Award awarded to Cameron Semper, a grant from the Animal Advocacy Research Fund awarded to Cameron Semper and grants from the Danish Agricultural Agency (33010-NIFA-19-716), Ministry of Food, Agriculture and Fisheries of Denmark, The Graduate School of Technical Sciences (GSTS), Aarhus University and the Center of Innovative Food Research (CiFOOD), Aarhus University, Denmark. The graphical abstract and Figure 6 were created with BioRender.com
Author contributions
Conceptualization, M.V., C.S., P.J.S., and A.S.; Methodology, M.V., C.S., and S.S.; Investigation, M.V., C.S., S.S, R.D., and N.M.; Writing – Original Draft, M.V., C.S., and S.S.; Writing – Review and Editing, M.V., C.S., P.J.S., and A.S.; Supervision – M.K.R, J.F.Y, M.T, P.J.S., and A.S.; Funding Acquisition – C.S., P.J.S., and A.S.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105054.
Contributor Information
Peter J. Stogios, Email: p.stogios@utoronto.ca.
Alexei Savchenko, Email: alexei.savchenko@ucalgary.ca.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ai G., Shao X., Meng M., Song L., Qiu J., Wu Y., Zhou J., Cheng J., Tong X. Epidermal growth factor promotes proliferation and maintains multipotency of continuous cultured adipose stem cells via activating STAT signal pathway in vitro. Medicine. 2017;96:e7607. doi: 10.1097/MD.0000000000007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D.E., Tidball J.G. Platelet-derived growth factor-stimulated secretion of basement membrane proteins by skeletal muscle occurs by tyrosine kinase-dependent and -independent pathways. J. Biol. Chem. 1997;272:2236–2244. doi: 10.1074/jbc.272.4.2236. [DOI] [PubMed] [Google Scholar]
- Alexander D.M., Hesson T., Mannarino A., Cable M., Dalie B.L. Isolation and purification of a biologically active human platelet-derived growth factor BB expressed in Escherichia coli. Protein Expr. Purif. 1992;3:204–211. doi: 10.1016/1046-5928(92)90016-p. [DOI] [PubMed] [Google Scholar]
- Benington L., Rajan G., Locher C., Lim L.Y. Fibroblast growth factor 2-A Review of stabilisation approaches for clinical applications. Pharmaceutics. 2020;12:E508. doi: 10.3390/pharmaceutics12060508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette P.H., Aslund F., Beckwith J., Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancucci M., Dolores J.S., Wong J., Grimshaw S., Anderson W.F., Satchell K.J.F., Kwon K. New ligation independent cloning vectors for expression of recombinant proteins with a self-cleaving CPD/6xHis-tag. BMC Biotechnol. 2017;17:1. doi: 10.1186/s12896-016-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault P., Tremblay J.P., Pépin M.F., Garnier A. Scale-up of a myoblast culture process. J. Biotechnol. 2001;91:63–74. doi: 10.1016/s0168-1656(01)00291-7. [DOI] [PubMed] [Google Scholar]
- Breitkopf K., Roeyen C.v., Sawitza I., Wickert L., Floege J., Gressner A.M. Expression patterns of PDGF-A, -B, -C and -D and the PDGF-receptors alpha and beta in activated rat hepatic stellate cells (HSC) Cytokine. 2005;31:349–357. doi: 10.1016/j.cyto.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Brewer J.R., Mazot P., Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30:751–771. doi: 10.1101/gad.277137.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Delaine C., Zaccheo O.J., Siebold C., Gilbert R.J., Van Boxel G., Denley A., Wallace J.C., Hassan A.B., Forbes B.E., Jones E.Y. Structure and functional analysis of the IGF-II/IGF2R interaction. EMBO J. 2008;27:265–276. doi: 10.1038/sj.emboj.7601938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell M.A., Garcion E., Terborg M.G., He X., Svendsen C.N. Heparin stabilizes FGF-2 and modulates striatal precursor cell behavior in response to EGF. Exp. Neurol. 2004;188:408–420. doi: 10.1016/j.expneurol.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Yu P., Hou Z., Thomson J.A. Thermal stability of fibroblast growth factor protein is a determinant factor in regulating self-renewal, differentiation, and reprogramming in human pluripotent stem cells. Stem Cell. 2012;30:623–630. doi: 10.1002/stem.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Patel D.J. An efficient system for small protein expression and refolding. Biochem. Biophys. Res. Commun. 2004;317:401–405. doi: 10.1016/j.bbrc.2004.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance A. Engineering the animal out of animal products. Nat. Biotechnol. 2017;35:704–707. doi: 10.1038/nbt.3933. [DOI] [PubMed] [Google Scholar]
- Derynck R., Jarrett J.A., Chen E.Y., Eaton D.H., Bell J.R., Assoian R.K., Roberts A.B., Sporn M.B., Goeddel D.V. Human transforming growth factor-beta complementary DNA sequence and expression in normal and transformed cells. Nature. 1985;316:701–705. doi: 10.1038/316701a0. [DOI] [PubMed] [Google Scholar]
- Emamipour N., Vossoughi M., Mahboudi F., Golkar M., Fard-Esfahani P. Soluble expression of IGF1 fused to DsbA in SHuffle T7 strain: optimization of expression and purification by Box-Behnken design. Appl. Microbiol. Biotechnol. 2019;103:3393–3406. doi: 10.1007/s00253-019-09719-w. [DOI] [PubMed] [Google Scholar]
- Eschenfeldt W.H., Makowska-Grzyska M., Stols L., Donnelly M.I., Jedrzejczak R., Joachimiak A. New LIC vectors for production of proteins from genes containing rare codons. J. Struct. Funct. Genom. 2013;14:135–144. doi: 10.1007/s10969-013-9163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson L., Li H., Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Gasparian M.E., Elistratov P.A., Drize N.I., Nifontova I.N., Dolgikh D.A., Kirpichnikov M.P. Overexpression in Escherichia coli and purification of human fibroblast growth factor (FGF-2) Biochemistry. 2009;74:221–225. doi: 10.1134/s000629790902014x. [DOI] [PubMed] [Google Scholar]
- Geistlinger T., Jhala R., Krueger K., Ramesh B. 2017. Food Products Comprising Milk Proteins and Non-animal Proteins, and Methods of Producing the Same. United States patent application PCT/US20 17/048730. [Google Scholar]
- Godfray H.C.J., Aveyard P., Garnett T., Hall J.W., Key T.J., Lorimer J., Pierrehumbert R.T., Scarborough P., Springmann M., Jebb S.A. Meat consumption, health, and the environment. Science. 2018:eaam5324. doi: 10.1126/science.aam5324. [DOI] [PubMed] [Google Scholar]
- Hakuno F., Takahashi S.I. IGF1 receptor signaling pathways. J. Mol. Endocrinol. 2018;61:T69–T86. doi: 10.1530/JME-17-0311. [DOI] [PubMed] [Google Scholar]
- Hinck A.P., Archer S.J., Qian S.W., Roberts A.B., Sporn M.B., Weatherbee J.A., Tsang M.L., Lucas R., Zhang B.L., Wenker J., Torchia D.A. Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2. Biochemistry. 1996;35:8517–8534. doi: 10.1021/bi9604946. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Lee H.J., Han M.H., Yoon N.K., Kim Y.C., Ahn J. Effective production of human growth factors in Escherichia coli by fusing with small protein 6HFh8. Microb. Cell Factories. 2021;20:9. doi: 10.1186/s12934-020-01502-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koledova Z., Sumbal J., Rabata A., De La Bourdonnaye G., Chaloupkova R., Hrdlickova B., Damborsky J., Stepankova V. Fibroblast growth factor 2 protein stability provides decreased dependence on heparin for induction of FGFR signaling and alters ERK signaling dynamics. Front. Cell Dev. Biol. 2019;7:331. doi: 10.3389/fcell.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.H., Gao X., Dekeyser J.M., Fetterman K.A., Pinheiro E.A., Weddle C.J., Fonoudi H., Orman M.V., Romero-Tejeda M., Jouni M., et al. Negligible-cost and weekend-free chemically defined human iPSC culture. Stem Cell Rep. 2020;14:256–270. doi: 10.1016/j.stemcr.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie E.R., Diblasio E.A., Kovacic S., Grant K.L., Schendel P.F., Mccoy J.M. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- Lobstein J., Emrich C.A., Jeans C., Faulkner M., Riggs P., Berkmen M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Factories. 2012;11:56. doi: 10.1186/1475-2859-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick C.S., Landis A.E., Allenby B.R., Genovese N.J. Anticipatory life cycle analysis of in vitro biomass cultivation for cultured meat production in the United States. Environ. Sci. Technol. 2015;49:11941–11949. doi: 10.1021/acs.est.5b01614. [DOI] [PubMed] [Google Scholar]
- McCubrey J.A., Steelman L.S., Mayo M.W., Algate P.A., Dellow R.A., Kaleko M. Growth-promoting effects of insulin-like growth factor-1 (IGF-1) on hematopoietic cells: overexpression of introduced IGF-1 receptor abrogates interleukin-3 dependency of murine factor-dependent cells by a ligand-dependent mechanism. Blood. 1991;78:921–929. [PubMed] [Google Scholar]
- Melzener L., Verzijden K.E., Buijs A.J., Post M.J., Flack J.E. Cultured beef: from small biopsy to substantial quantity. J. Sci. Food Agric. 2021;101:7–14. doi: 10.1002/jsfa.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossahebi-Mohammadi M., Quan M., Zhang J.S., Li X. FGF signaling pathway: a key regulator of stem cell pluripotency. Front. Cell Dev. Biol. 2020;8:79. doi: 10.3389/fcell.2020.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Wrana J.L. TGF-Beta family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harbor Perspect. Biol. 2017;9:a022186. doi: 10.1101/cshperspect.a022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozach H., Fruchart-Gaillard C., Fenaille F., Beau F., Ramos O.H.P., Douzi B., Saez N.J., Moutiez M., Servent D., Gondry M., et al. High throughput screening identifies disulfide isomerase DsbC as a very efficient partner for recombinant expression of small disulfide-rich proteins in E. coli. Microb. Cell Factories. 2013;12:37. doi: 10.1186/1475-2859-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi F., Prisco M., Dews M., Salomoni P., Grassilli E., Romano G., Calabretta B., Baserga R. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol. Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poniatowski Ł.A., Wojdasiewicz P., Gasik R., Szukiewicz D. Transforming growth factor Beta family: insight into the role of growth factors in regulation of fracture healing biology and potential clinical applications. Mediat. Inflamm. 2015;2015:137823. doi: 10.1155/2015/137823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post M.J., Levenberg S., Kaplan D.L., Genovese N., Fu J., Bryant C.J., Negowetti N., Verzijden K., Moutsatsou P. Scientific, sustainability and regulatory challenges of cultured meat. Nat. Food. 2020;1:403–415. [Google Scholar]
- Reichelt P., Schwarz C., Donzeau M. Single step protocol to purify recombinant proteins with low endotoxin contents. Protein Expr. Purif. 2006;46:483–488. doi: 10.1016/j.pep.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Rosano G.L., Ceccarelli E.A. Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer F., Seip N., Maertens B., Block H., Kubicek J. Purification of GST-tagged proteins. Methods Enzymol. 2015;559:127–139. doi: 10.1016/bs.mie.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Scharenberg M.A., Pippenger B.E., Sack R., Zingg D., Ferralli J., Schenk S., Martin I., Chiquet-Ehrismann R. TGF-beta-induced differentiation into myofibroblasts involves specific regulation of two MKL1 isoforms. J. Cell Sci. 2014;127:1079–1091. doi: 10.1242/jcs.142075. [DOI] [PubMed] [Google Scholar]
- Schmierer B., Hill C.S. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- Skrivergaard S., Rasmussen M.K., Therkildsen M., Young J.F. Bovine satellite cells isolated after 2 and 5 Days of tissue storage maintain the proliferative and myogenic capacity needed for cultured meat production. Int. J. Mol. Sci. 2021;22:8376. doi: 10.3390/ijms22168376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenburg E.E., Booth F.W. Multiple signaling pathways mediate LIF-induced skeletal muscle satellite cell proliferation. Am. J. Physiol. Cell Physiol. 2002;283:C204–C211. doi: 10.1152/ajpcell.00574.2001. [DOI] [PubMed] [Google Scholar]
- Specht L. Good Food Institute; 2020. An Analysis of Culture Medium Costs and Production Volumes for Cultivated Meat.https://gfi.org/wp-content/uploads/2021/01/clean-meat-production-volume-and-medium-cost.pdf [Google Scholar]
- Stout A.J., Mirliani A.B., Rittenberg M.L., Shub M., White E.C., Yuen J.S.K., Kaplan D.L. Simple and effective serum-free medium for sustained expansion of bovine satellite cells for cell cultured meat. Commun. Biol. 2022;5:466. doi: 10.1038/s42003-022-03423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi N.K., Shrivastava A. Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front. Bioeng. Biotechnol. 2019;7:420. doi: 10.3389/fbioe.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajdos F.F., Ultsch M., Schaffer M.L., Deshayes K.D., Liu J., Skelton N.J., De Vos A.M. Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions. Biochemistry. 2001;40:11022–11029. doi: 10.1021/bi0109111. [DOI] [PubMed] [Google Scholar]
- van der Valk J., Bieback K., Buta C., Cochrane B., Dirks W.G., Fu J., Hickman J.J., Hohensee C., Kolar R., Liebsch M., et al. Fetal bovine serum (FBS): past - present - future. ALTEX. 2018;35:99–118. doi: 10.14573/altex.1705101. [DOI] [PubMed] [Google Scholar]
- Yang Z., Xiong H.-R. In: Biomedical Tissue Culture. Ceccherini-Nelli L., Matteoli B., editors. IntechOpen; 2012. Culture conditions and types of growth media for mammalian cells. [DOI] [Google Scholar]
- Yao T., Asayama Y. Animal-cell culture media: history, characteristics, and current issues. Reprod. Med. Biol. 2017;16:99–117. doi: 10.1002/rmb2.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wang H., Xu Y., Yu D., Li D., Liu X., Du W. Insulin-like growth factor-1 (IGF-1) promotes myoblast proliferation and skeletal muscle growth of embryonic chickens via the PI3K/Akt signalling pathway. Cell Biol. Int. 2015;39:910–922. doi: 10.1002/cbin.10466. [DOI] [PubMed] [Google Scholar]
- Zakrzewska M., Krowarsch D., Wiedlocha A., Otlewski J. Design of fully active FGF-1 variants with increased stability. Protein Eng. Des. Sel. 2004;17:603–611. doi: 10.1093/protein/gzh076. [DOI] [PubMed] [Google Scholar]
- Zheng X., Wu X., Fu X., Dai D., Wang F. Expression and purification of human epidermal growth factor (hEGF) fused with GB1. Biotechnol. Biotechnol. Equip. 2016;30:813–818. [Google Scholar]
- Zhu H., Duchesne L., Rudland P.S., Fernig D.G. The heparan sulfate co-receptor and the concentration of fibroblast growth factor-2 independently elicit different signalling patterns from the fibroblast growth factor receptor. Cell Commun. Signal. 2010;8:14. doi: 10.1186/1478-811X-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Sun P.D. Overexpression of human transforming growth factor-beta1 using a recombinant CHO cell expression system. Protein Expr. Purif. 2004;37:265–272. doi: 10.1016/j.pep.2003.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.