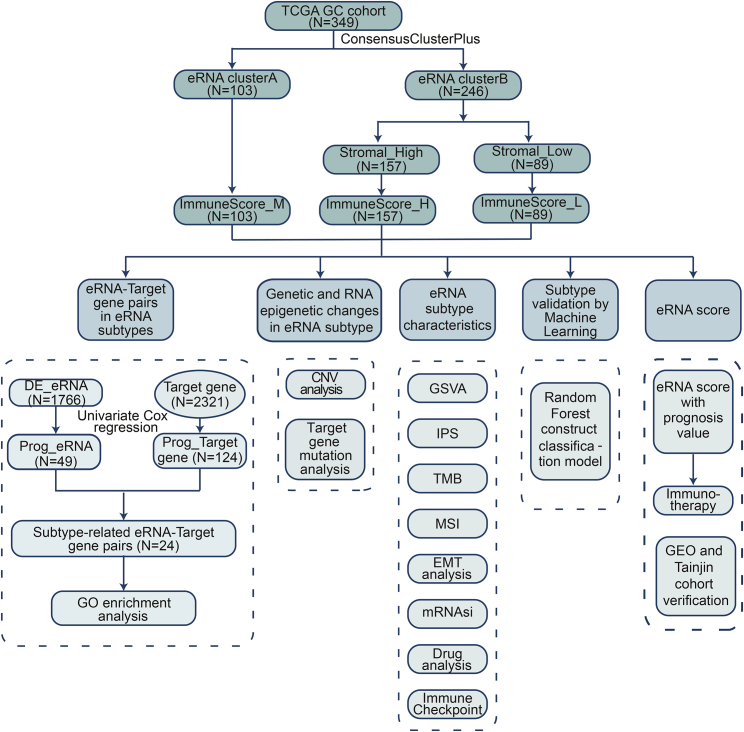
Summary
The comprehensive regulation effect of eRNA on tumor immune cell infiltration and the outcome remains obscure. We comprehensively identify the eRNA-mediated immune infiltration patterns of gastric cancer (GC) samples. We creatively proposed a random forest machine-learning (ML) algorithm to map eRNA to mRNA expression patterns. The eRNA score was constructed using principal component analysis algorithms and validated in an independent cohort. Three subtypes with distinct eRNA expression patterns were determined in GC. There were significant differences between the three subtypes in the overall survival rate, immune cell infiltration characteristics, and immunotherapy response indicators. The patients in the high eRNA score group have a higher overall survival rate and might benefit from immunotherapy. This work revealed that eRNA regulation might be a new prognostic index and might offer a potential biomarker in the response of immunotherapy. Evaluating the eRNA regulation manner of GC will contribute to guiding more effective immunotherapy strategies.
Subject areas: Biological sciences, Bioinformatics, Systems biology, Cancer
Graphical abstract
Highlights
-
•
eRNA is enigmatic “oasis” composed of lncRNA transcribed from enhancer in gene desert
-
•
We discovered eRNA-mediated immune subtypes with distinct biological properties of GC
-
•
We proposed a machine-learning (ML) algorithm to map eRNA to mRNA expression patterns
-
•
The eRNA score is a promising biomarker to predict GC immunotherapy response
Biological sciences; Bioinformatics; Systems biology; Cancer
Introduction
Gastric cancer (GC) is the fifth most frequently diagnosed malignancy worldwide and the third leading cause of cancer-associated death. According to epidemiologic data, in regions of East Asia GC has a high incidence rate (Bray et al., 2018). The emerging therapeutic strategies, including immunotherapy and targeted therapy, have brought hope to patients with cancer, such as gastrointestinal tumors (Kaufman et al., 2019). However, the response to existing immune-based treatments varies between individuals. Therefore, exploring more effective targets for improving patient outcomes is essential.
There are noncoding RNAs transcribed from enhancers, named enhancer RNAs (eRNAs), which can mediate other gene expressions by affecting chromatin accessibility, chromatin cyclization, and transcription of transcription cofactors to promoters (Sartorelli and Lauberth, 2020). In recent years, several eRNA-related public databases have been established, laying the foundation for the eRNA function in cancer development (Jin et al., 2022; Yao et al., 2022; Zhang et al., 2019, 2021). Many studies have shown that the activation of oncogenes or oncogenic signaling pathways is often associated with enhancer activation and the production of eRNAs. For example, KLK3e, an androgen-induced eRNA regulating gene KLK3, can scaffold the androgen receptor (AR)-associated protein complex to control AR-dependent gene expression in prostate cancer (Hsieh et al., 2014). A recent study shows that AR can be the transcriptional activator for oncogenic miR-125b in GC (Liu et al., 2021a). eRNA may also affect drug response by within-pathway or cross-pathway models. For example, Zhang et al. characterized the oncogenic potential and therapeutic liability of one eRNA, NET1e, supporting the clinical feasibility of eRNA-targeted therapy (Zhang et al., 2019).
GC is a heterogeneous disease reflected by various molecular and histological subtypes. For example, The Cancer Genome Atlas (TCGA) gastric subtype and the Asian Cancer Research Group (ACRG) subtype present various kinds of GC molecular patterns (Cancer Genome Atlas Research, 2014; Cristescu et al., 2015). A comprehensive molecular subtype can lead to a better understanding of cancer biology and more effective cancer treatment. Although enhancers are critical to all cell types, eRNA enrichment varies dramatically across tissues. The previous study (Zhang et al., 2019) showed that the number of eRNAs in GC is abundant. Therefore, it warrants the need to explore eRNA-mediated regulation patterns in GC.
Of additional concern is that the eRNAs participate in a variety of immune responses. For example, the knockdown of IL1β-eRNA attenuates the release of bacterial lipopolysaccharide (LPS)-induced inflammatory mediators such as IL1β and CXCL8 (Iiott et al., 2014). The interferon-induced apoptosis can be repressed through targeted reduction of eRNAs (Kim et al., 2018b). A previous study indicated interactions between eRNAs and immune checkpoints (Zhang et al., 2019). This result suggested that eRNAs have emerged as a potential target in cancer immunotherapy. However, no studies comprehensively analyze eRNAs and immune infiltration in GC.
So far, the tumor microenvironment has proved of great significance for immunotherapy. EMT (epithelial-mesenchymal transition) and mRNAsi (mRNA-expression-based stemness index) have been associated with various aspects of the tumor immune microenvironment (Becht et al., 2016; Malta et al., 2018; Miranda et al., 2019; Suarez-Carmona et al., 2017). In addition, tumor mutation burden (TMB), neoantigen load, immune checkpoint molecule levels (PD-L1), impaired DNA mismatch repair (MMR), and microsatellite instability (MSI) have been found to affect the response of cancer cells to immunotherapy (Chen et al., 2019b; Choi et al., 2014; Dong et al., 2018). Based on these complicated and mixed immunotherapy factors, Charoentong et al. identified an immunophenoscore (IPS) to the responsivity of immunotherapy (Charoentong et al., 2017). Although the aforementioned factors are widely recognized as biomarkers for immune checkpoint blockade (ICB), predictive effects are still not satisfactory for tumor-induced immunosuppression in the tumor microenvironment (TME), resulting in intrinsic immunotherapy resistance. Therefore, the hitherto unrecognized biomarkers, such as eRNAs, with greater efficacy in discriminating potential responders to immune therapies remain an urgent priority (Kim et al., 2018a; Mariathasan et al., 2018). In addition, the development of potent immunotherapy targets requires a comprehensive understanding of the regulatory relationship between eRNA and TME.
The present study identified three distinct eRNA subtypes of GC based on GC-related 2267 eRNAs (Zhang et al., 2019) and calculated the subtype-related eRNAs between these three eRNA subtypes. Meanwhile, we also explored the indexes related to the effect of immunotherapy between these three eRNA subtypes. Finally, we used the random forest algorithm to identify eRNA subtypes in the GEO cohort. Multiple analyses strongly suggest that patients with a high eRNA score group benefit more from immunotherapy.
Results
Three distinct immune subtypes mediated by eRNAs
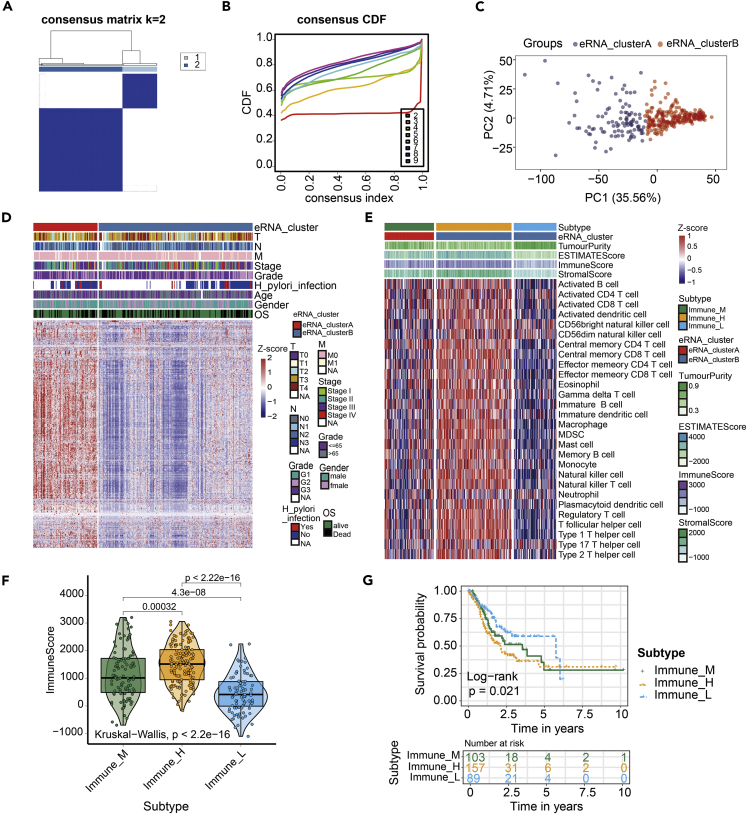
We extracted TCGA GC-related 2267 eRNA expression data from eRic database (Zhang et al., 2019). A total of 349 TCGA GC samples accompanied with complete survival information were retained for consensus cluster analysis (Figure 1). Based on eRNA expression data, consensus cluster analysis indicated that the optimal number of clusters was two, which was defined by cumulative distribution function (CDF) curves (Figures 2A–2C). According to the clustering result, GC samples were clustered into the eRNA cluster groups A and B (N = 103 and 246, respectively). Heatmap showed the whole eRNA expression between eRNA clusters (Figure 2D). Meanwhile, the immune cells infiltration results of ssGSEA showed that the patients in the eRNA cluster B seem to have two immune cell infiltration state: "Hot tumor" and "Cold tumor" (Figure S2). As stromal cells play an important role in modeling tumor immune evasion (Chen and Mellman, 2017), we further dissected the gene expression profiles of the patients with the eRNA cluster B, in which 64% (157/246) was characterized with an activated stromal response. Therefore, according to the stromal score, GC patients in the eRNA cluster B were clustered into the high- and low-stromal score group (N = 157 and 89, respectively) (Figure 2E). So far, we have got three eRNA groups. They are eRNA cluster A, high-stromal score group (patients originate from eRNA cluster B group, 64% [157/246]), and low-stromal score group (patients originate from eRNA cluster B group, 36% [89/246]), respectively. For convenience, according to the immune score, we describe eRNA cluster A to be "medium-immune score subtype" (Immune_M), high-stromal score group to be "high-immune score subtype" (Immune_H), and low-stromal score group to be "low-immune score subtype" (Immune_L), respectively (Figure 2F). The survival analysis demonstrated that the overall survival in the Immune_L group was markedly higher in contrast with the Immune_H group and Immune_M group, indicating that eRNA-mediated immune infiltration subtypes had prognostic values in GC patients (p = 0.021) (Figure 2G).
Figure 1.
Flowchart illustrating the workflow of this study
Figure 2.
Construction of the eRNA subtype with distinct immune infiltration and prognosis of GC
Cluster analysis identified eRNA subtypes with the relative expression of 2267 eRNAs.
(A and B) The optimal number of clusters (K = 2) was determined from cumulative distribution function (CDF) curves, and the classification effect is the best.
(C) Principal component analysis (PCA) plot of GC samples.
(D) The patients were divided into two groups: eRNA_clusterA and eRNA_clusterB. The eRNA expression was normalized into Z score.
(E) Single-sample gene set enrichment analysis (ssGSEA) identified the relative infiltration of 28 immune cell types subpopulations with different eRNA subtypes. The relative infiltration of each cell type was normalized into Z score.
(F) The boxplot showed a statistical difference in immune score between the three eRNA subtypes (p < 2.2 × 10−16).
(G) Survival analysis of three eRNA subtypes in the TCGA GC cohort was created using Kaplan-Meier curves (p=0.021).
Identification of subtype-related eRNA-target gene pairs
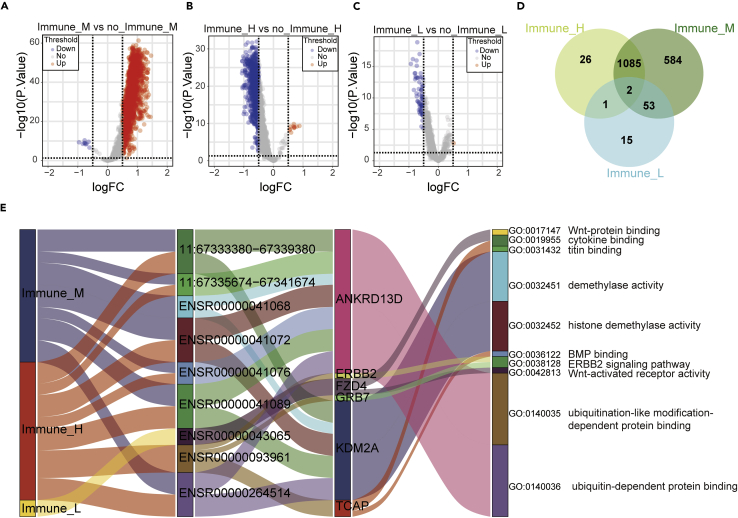
We fetched 2267 eRNA and 2335 eRNA-target gene information from the eRic database. A total of 2,321 eRNA-target genes can be found in the TCGA database. Results of differential expression analysis showed 1,724 different expression eRNA between Immune_M and no_Immune_M subtypes (Figure 3A, Table S3), 1,114 different expression eRNA between Immune_H and no_Immune_H subtypes (Figure 3B and Table S4), and 71 different expression eRNA between Immune_L and no_Immune_L subtypes (Figure 3C and Table S5). Then, we obtained 1,766 unique differentially expressed eRNAs among three subtypes (Figure 3D and Table S6). Univariate cox regression analyses showed 49 eRNA and 124 eRNA target genes with prognostic value. A total of 6,144 pairs of effective eRNA-target gene relationships were derived from the eRic database. We acquired 24 subtype-related eRNA pairs with prognostic values (Table S7). The GO enrichment analyses further suggested that nine eRNA pairs played critical roles in ten biological processes (Figure 3E).
Figure 3.
Analysis of differentially expressed eRNAs and identification of subtype-related eRNA-target gene pairs in GC
(A) The volcano plot showed 1,724 differentially expressed eRNAs between Immune_M and no_Immune_M subtypes. Each red dot showed an upregulated eRNA, and each blue dot showed a downregulated eRNA (p<0.05).
(B) The volcano plot showed 1,114 differentially expressed eRNAs between Immune_H and no_Immune_H subtypes. Each red dot showed an upregulated eRNA, and each blue dot showed a downregulated eRNA (p<0.05).
(C) The volcano plot showed 71 differentially expressed eRNAs between Immune_L and no_Immune_L subtypes. Each red dot showed an upregulated eRNA, and each blue dot showed a downregulated eRNA (p<0.05).
(D) The Venn plot showed 1,766 unique differentially expressed eRNAs between the three eRNA subtypes.
(E) Sankey plot showed subtype-related eRNA-target gene pairs in three eRNA subtypes. The GO terms were regulated by the target gene, also shown in this plot.
Genetic and RNA epigenetic changes affecting eRNA expression
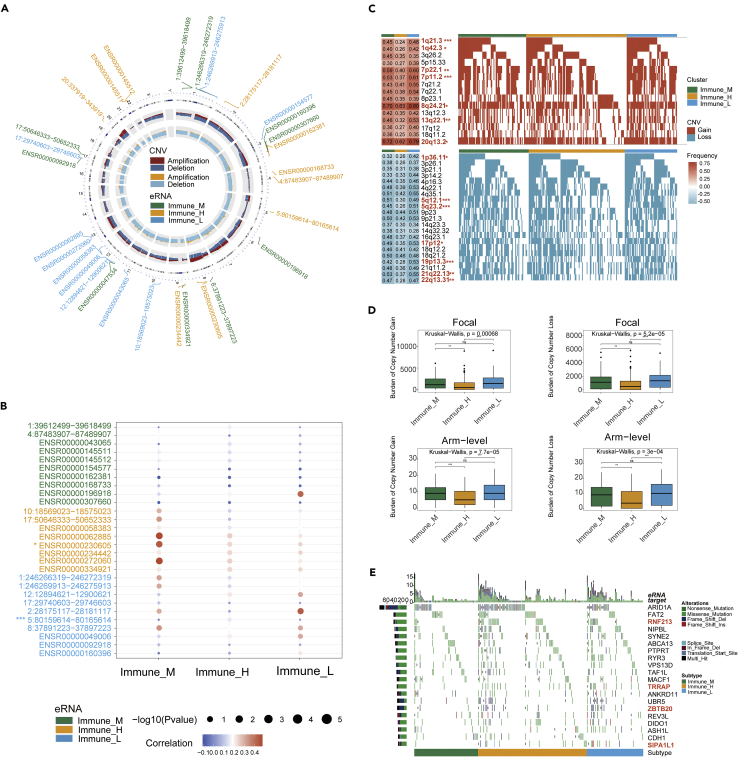
For copy number variation (CNV) may affect eRNA expression, we evaluated CNV situation in regions of eRNA positions among three subtypes. The overall frequency and gistic score showed the CNV pattern in a circle diagram (Figure 4A). The top-ranked ten differentially expressed eRNAs in each subtype were marked in their chromosomal locations in the circos plot. The gistic analysis of CNV frequency showed that most differential eRNAs had CNV alteration (Figure 4A). ENSR00000168733 in chromosome 4 observed the deletion in copy number, whereas ENSR00000230605 in chromosome 8 had a high-level frequency of CNV amplification (Figure 4A). Spearman correlation analysis was assessed between eRNA expression and CNV (Figure 4B). The results showed that in the Immune_M subtype, the expression of most eRNAs was distinctly correlated with CNV, indicating that the abnormality of copy number in this subtype could affect the expression of eRNAs. Then we also compared the CNV frequency alteration of differential eRNAs and nondifferential eRNAs between three eRNA subtypes. The results indicated that two eRNAs had statistically significant differences in CNV frequency among 28 differential eRNAs (Figure 4B). In nondifferential eRNAs, 197 eRNAs had statistically significant differences in CNV frequency among 2,163 nondifferential eRNAs. The result implied that the CNV might not be the predominant reason for its differential expression between subtypes. We further investigated the differential CNV alteration among three eRNA subtypes. The results showed that three eRNA clusters had significant different CNV alteration, including copy number amplifications (1q21.3 [Fisher test, p < 0.001], 1q42.3 [Fisher test, p < 0.05] and 7p22.1 [Fisher test, p < 0.01]) and deletions (1p36.11 [Fisher test, p < 0.05], 5q12.1 [Fisher test, p < 0.001], and 5q23.2 [Fisher test, p < 0.001]) (Figure 4C). When comparing the focal and arm level CNV through gistic 2.0 approach, the Immune_L subtype showed the overall highest burden of copy number gain and loss. In contrast, the Immune_H subtype was observed in the lowest CNV burden within all three eRNA groups (Figure 4D). It suggested that focal copy number alterations contribute to the difference in eRNA expression in GC. The waterfall plot showed the target gene mutation ratio in GC samples among eRNA subtypes, and we determined several significantly differentially mutation target genes including RNF213 (Fisher test, p = 0.01427), TRRAP (Fisher test, p = 0.02085), ZBTB20 (Fisher test, p = 0.02214), and SIPA1L1 (Fisher test, p = 0.003773) (Figure 4E).
Figure 4.
Genomic mutation changes affect eRNA expression
(A) The circle plot showed a distribution of 30 eRNAs (top ten differentially expressed in three eRNA subtypes, respectively). The circles from outside to inside stand for chromosome location, copy-number frequency (dark red: amplification, dark blue: deletion), and copy number gistic score (yellow: amplification, blue: deletion), respectively. Each green font showed an Immune_M differentially expressed eRNA, each yellow font showed an Immune_H differentially expressed eRNA, and each blue font showed an Immune_L differentially expressed eRNA.
(B) The bubble plot displayed the Spearman’s correlation between the copy number alterations and the expression of differential eRNAs. Asterisks (∗) in the upper left corner of eRNAs indicate significant differences in CNV distributions between three eRNA subtypes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Detailed plots with copy number gain (up) and copy number loss (down) between three eRNA subtypes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Distribution of focal and broad copy number alterations between three eRNA subtypes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = not statistically significant.
(E) Mutational landscape of target gene between three eRNA subtypes.
Significant differences of TME-related characteristics and biological features in eRNA subtypes
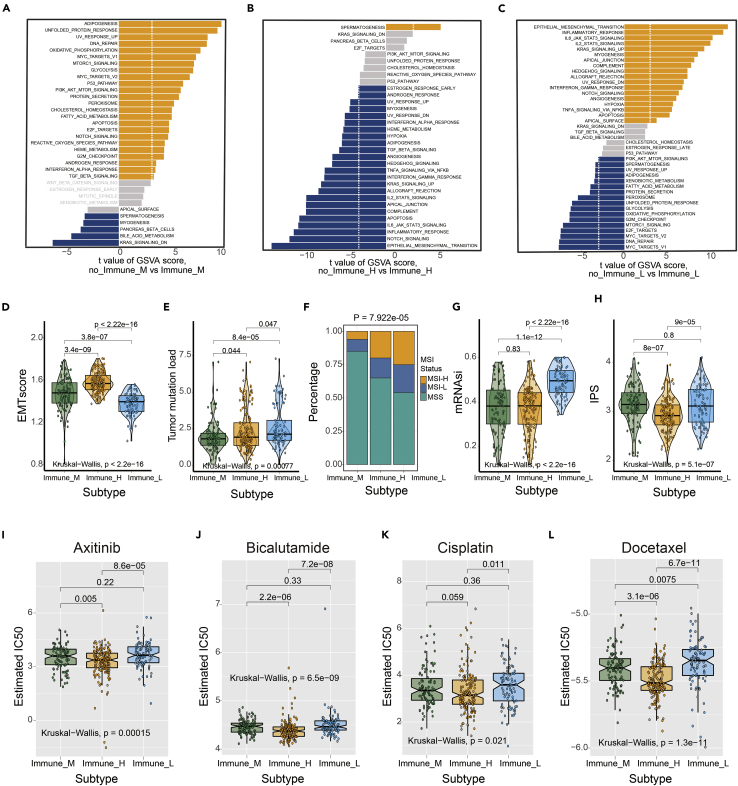
Gene set variation analysis (GSVA) showed that spermatogenesis and myogenesis were activated in the Immune_M group. In the Immune_L group, KRAS signaling was downregulated, which was contrary to the results of the Immune_M group (Figures 5A–5C).
Figure 5.
Significant differences in TME-related characteristics and biological features in eRNA subtypes
(A–C) The GSVA plot showed a differentially activated pathway between the three eRNA subtypes. Each yellow bar showed an activated pathway, and each blue bar showed an inactivated pathway. (|t value| > 3 and p < 0.05).
(D) The boxplot showed a statistical difference in EMT score between three eRNA subtypes (p < 2.2 × 10−16).
(E) The boxplot showed a statistical difference in TMB between three eRNA subtypes (p = 0.00077).
(F) The proportion of MSI status between three eRNA subtypes (Fisher test, p = 7.922 × 10−5).
(G) The boxplot showed a statistical difference in mRNAsi between three eRNA subtypes (p < 2.2 × 10−16).
(H) The boxplot showed a statistical difference in IPS between three eRNA subtypes (p = 5.1 × 10−7).
(I–L) The IC50s of chemotherapeutic agents with eRNA subtypes. (I) Axitinib; (J) bicalutamide; (K) cisplatin; (L) doxorubicin (p<0.05).
To evaluate the differences in the abundance of tumor-infiltrating immune cells among the three eRNA subtypes, we calculated the EMT, TMB, and mRNAsi scores to reflect the status of the immune microenvironment. The result showed that the Immune_H group had the highest EMT score, but Immune_L had the lowest EMT score (p < 0.0001) (Figure 5D). The analysis of the TMB pattern demonstrated that the Immune_L group had the highest TMB, the Immune_H group had intermediate TMB, and Immune_M had the lowest TMB (p < 0.001) (Figure 5E). Chi-square test analyses showed that the Immune_L group and Immune_H group had more MSI-H status than the Immune_M group, which had more MSS status (p < 0.001) (Figure 5F). Furthermore, mRNAsi score in the Immune_L group was higher than that in Immune_M and Immune_H groups (p < 0.0001) (Figure 5G). The aforementioned results show significant differences in TME-related characteristics and biological features among eRNA subtypes.
The situation of immune cell infiltration in the TME is closely related to the response to tumor checkpoint blockade therapy. For immunophenoscore (IPS) to determine the tumor immunogenicity and predict response to ICI therapy in multitypes of tumors, we utilized IPS to investigate the immune response among the eRNA clusters. The result showed that the Immune_L and Immune_M group had the higher IPS, but the Immune_H group had the lower IPS (p < 0.0001) (Figure 5H), which suggested that the patients in Immune_L and Immune_M group might benefit from ICI therapy.
We further performed a prediction analysis of response to chemotherapy by applying the pRRophetic method. In contrast with patients in the Immune_M and Immune_H group, patients in the Immune_L group had a lower estimated IC50 in the following chemotherapy drugs: axitinib, bicalutamide, cisplatin, and dasatinib (p < 0.05) (Figures 5I–5L).
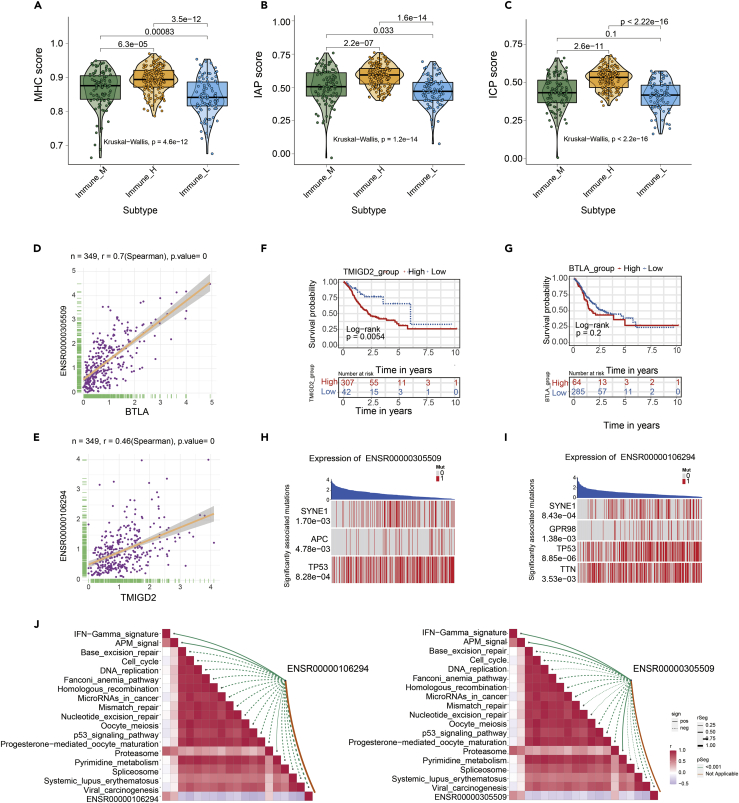
Then we employed single-sample gene set enrichment analysis (ssGSEA) to generate three types of immune checkpoints score and compare the score between different eRNA clusters (including MHC, IAP, and ICP). The immune checkpoints score of MHC, IAP, and ICP were different among the three eRNA subtypes (p < 0.05) (Figures 6A–6C). The results indicated that three eRNA clusters were characterized by distinct immune checkpoint patterns. Consistent with the previous conclusion, the Immune_H group had the highest immune checkpoints score, perhaps illustrating their role in shaping the tumor microenvironment. The observed differences in immune checkpoints score between three clusters may have implications for immune therapy development.
Figure 6.
The expression and survival analysis of eRNA-mediated immune checkpoint score
(A–C) The immune checkpoints score of three types of gene sets, including major histocompatibility complex (MHC), immune co-inhibitory checkpoints (IAP), and immune co-stimulator checkpoints (ICP), were all significant differences in the three eRNA subtypes (p < 0.05).
(D) The correlation of BTLA and ENSR00000305509 (p < 0.05).
(E) The correlation of TMIGD2 and ENSR00000106294 (p < 0.05).
(F) K-M analysis of the high versus low TMIGD2 expression group in the TCGA GC cohort. (p = 0.0054).
(G) K-M analysis of the high versus low BTLA expression group in the TCGA GC cohort. (p = 0.2).
(H) ENSR00000305509 corresponding mutation driver genes (SYNE1, APC, TP53) (p<0.05).
(I) ENSR00000106294 corresponding mutation driver genes (SYNE1, GPR98, TP53, TNN) (p<0.05).
(J) The correlation of ENSR00000106294 and immunotherapy-related pathway (left). The correlation of ENSR00000305509 and immunotherapy-related pathway (right).
Special attention was paid to two immune checkpoint molecules, BTLA and TMIGD2, and also eRNA targets. Their expression was positively correlated with eRNA by Spearman correlation analysis (BTLA & ENSR00000305509, r = 0.7, p = 0; TMIGD2 & ENSR00000106294, r = 0.46, p = 0) (Figures 6D and 6E). Kaplan-Meier survival analyses showed GC patients with TMIGD2 high expression had a worse prognosis than those with TMIGD2 low expression (p = 0.0054) (Figure 6F). However, the expression of BTLA has not been associated with the outcome of GC patients (p = 0.2) (Figure 6G). In addition, BTLA and TMIGD2 genes were more highly expressed in the Immune_H group. This result suggested that the patients in the Immune_H group may benefit more from the immunotherapy targeting BTLA and TMIGD2. Then, we obtained pharmacogenomic data of two immune checkpoints and their corresponding eRNA from the eRic database, and we only found 19 kinds of target drugs of ENSR00000305509 (Table S8). Furthermore, the permutation test analysis showed that the expression of ENSR00000305509 and ENSR00000106294 associated with APC (p = 4.78 × 10−3) and SYNE1 gene mutation (p = 8.43 × 10−4), respectively (Figures 6H and 6I). Finally, we found that ENSR00000305509 and ENSR00000106294 strongly correlated with immunotherapy-related pathways (Figure 6J) (Mariathasan et al., 2018).
Validation of prognosis and immune cell infiltration characteristics in the validation cohort by a developed machine learning method
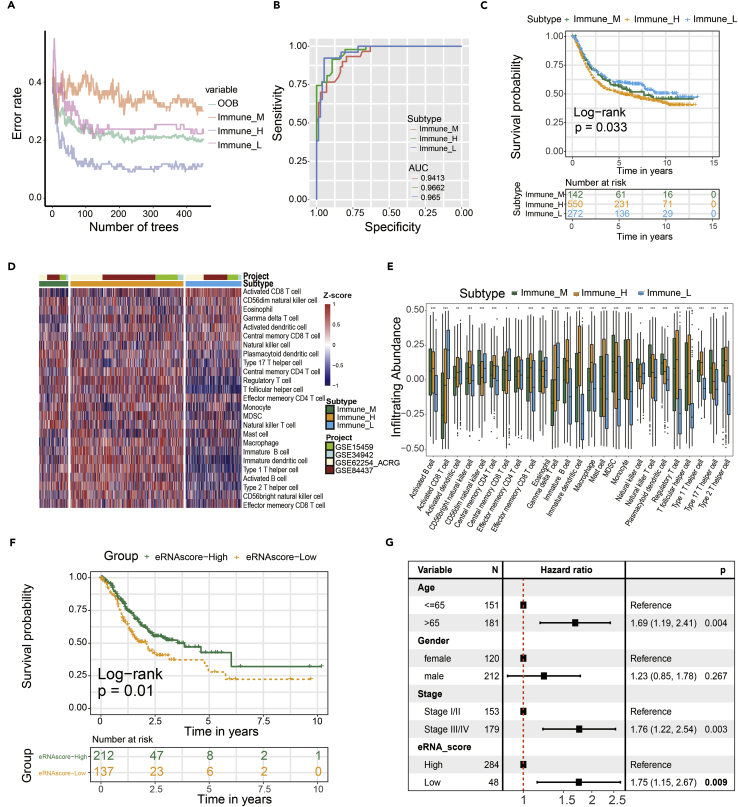
We identified 271 differentially expressed target genes, i.e., subtype-related target gene, between three eRNA subtypes. Based on the characteristics of 271 subtype-related target gene expressions (Table S9), we could predict the eRNA subtypes in the GEO cohort. We used 450 decision trees to train the classification model (Figure 7A). ROC analyses result showed that classification model can effectively recognize three eRNA subtypes (Immune_M, AUC = 0.9413; Immune_H, AUC = 0.9662; Immune_L, AUC = 0.965) (Figure 7B). Our data demonstrated that the overall survival in the Immune_L group was markedly higher in contrast with the Immune_H and Immune_M group in the validation cohort (p = 0.033) (Figure 7C). A similar situation of immune cell infiltration was observed in GEO cohorts (Figures 7D and 7E). These analyses evaluated the effect of the eRNA cluster in prognostic and immune cell infiltration.
Figure 7.
Validation of the eRNA subtypes on an independent GC cohort by a random forest prediction model
(A) Error rate of random forest model.
(B) ROC analyses result showed that classification model can effectively recognize three eRNA subtypes (Immune_M, AUC = 0.9413; Immune_H, AUC = 0.9662; Immune_L, AUC = 0.965).
(C) Survival analysis of three eRNA subtypes in the GEO GC cohort was created using Kaplan-Meier curves (p=0.033).
(D) Single sample gene set enrichment analysis (ssGSEA) identified the relative infiltration of immune cell subpopulations with different eRNA subtypes in the GEO GC cohort. The relative infiltration of each cell type was normalized into Z score.
(E) The relative infiltration level of the immune cell was all significant differences in the three eRNA subtypes (p < 0.05).
(F) K-M analysis of the high- versus low-eRNA-score group in the TCGA GC cohort (p = 0.01).
(G) Multivariate Cox regression analyses of the association between clinicopathological factors and overall survival of GC patients in TCGA GC cohort.
In addition, we constructed the eRNA score by applying 271 target genes that were differentially expressed between eRNA clusters. K-M analysis showed that the patients with high eRNA scores have a higher overall survival rate (Figure 7F). In the GEO and Tianjin cohorts, we attained similar results (Figure S3). Multivariate Cox regression analysis showed that eRNA score was an independent predictor of prognosis (HR = 1.75, 95% CI: 1.15–2.67, p = 0.009) (Figure 7G).
eRNA score can predict the response of immunotherapy
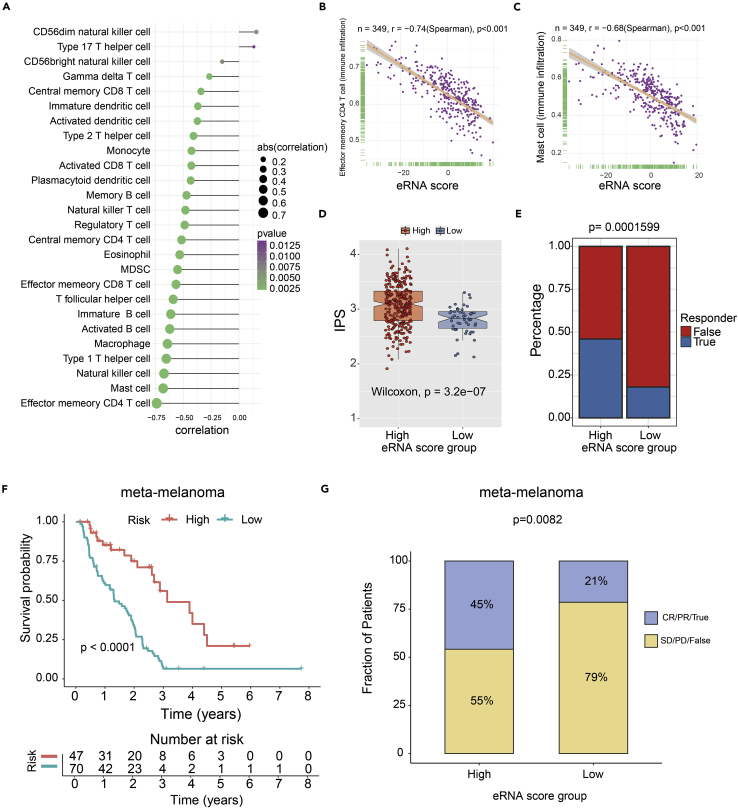
The spearman analysis results showed a negative correlation between eRNA score and infiltration abundances of the majority of immune cells (Figure 8A). And among these immune cells, the top two negative correlation occurred between eRNA score with the infiltration of effector memory CD4 T cell (r = −0.74, p < 0.001) and mast cell (r = −0.68, p < 0.001) (Figures 8B and 8C). The IPS score in the high-eRNA-score group was significantly higher compared with that in the low-eRNA-score group (p < 0.001) (Figure 8D). These results strongly suggest that patients in the high-eRNA-score group may benefit from immunotherapy. To further validate this result, we applied the Tumor Immune Dysfunction and Exclusion (TIDE) tool (Jiang et al., 2018) to predict the response of patients to immune checkpoint blockade (CTLA4 and PD1 therapy). TIDE analysis result further confirmed our results (p < 0.001) (Figure 8E). Finally, we tested the predictor efficiency of the eRNA score in non–small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma cohort with immunotherapy (GSE93157). K-M analysis showed that the patients with high eRNA scores have better progression-free survival (PFS) (log rank test, p = 0.0072). The results of the GSE93157 cohort showed that the high-eRNA-score group patients had a higher immunotherapy response rate than the patients in the low-eRNA-score group (p < 0.05) (Figure S4). Due to small numbers in each independent dataset, we added a meta-melanoma cohort treated with immunotherapy for eRNA score validation (including GSE78220, GSE91061, GSE100797, a melanoma cohort from Nathanson et al.) (Hugo et al., 2016; Lauss et al., 2017; Nathanson et al., 2017; Riaz et al., 2017). The conclusion agreed with the previous result that the high-eRNA-score subgroup showed a favorable prognosis (log rank test, p < 0.0001) Figure 8F) and improved response to immunotherapy compared with the low eRNA score subgroup (Fisher test, p = 0.0082) Figure 8G).
Figure 8.
Predicted eRNA score associated with immunotherapy response and outcomes in GC
(A) The correlation of 28 immune cell types and eRNA score (p < 0.05).
(B) The correlation of effector memory CD4 T cell and eRNA score (p < 0.001).
(C) The correlation of mast cell and eRNA score (p < 0.001).
(D) The boxplot showed a statistical difference in IPS between high- and low-eRNA-score groups (p = 3.2 × 10−7).
(E) The proportion of immunotherapy response rate between three eRNA subtypes by TIDE (Fisher test, p < 0.001).
(F) Kaplan-Meier analysis of the high- versus low-eRNA-score subgroup in the meta-melanoma cohort (p<0.0001).
(G) The proportion of immune response to immunotherapy in high- versus low-eRNA-score subgroups. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; False, no response; True, response (p=0.082).
Discussion
Although immunotherapy is remarkably successful in many tumor cases, it remains unclear why a subset of GC patients fails to respond to it (Hanahan and Weinberg, 2011; Marconcini et al., 2018; Santoni et al., 2018). Intratumoral genetic heterogeneity might answer the above question. There have recently been numerous reports on tumor immunomodulation of eRNA, but none comprehensively analyses the association between eRNAs and immune infiltration in GC (Wu and Shen, 2019). Here, based on 2267 GC-related eRNAs, we revealed three eRNA-mediated distinct immune subtypes with prognostic and therapeutic relevance.
In the GO enrichment of target genes in nine eRNA pairs, we identified several biological processes related to the regulation of GC, such as ERBB2 signaling pathway and demethylase activity. There is previous evidence that the ERBB2/ERBB3 signaling pathway is involved in regulating EMT-related migration and invasion of GC. Specifically, miR-200b suppressed ERBB2/ERBB3 signaling pathway via targeting NRG1(Xu et al., 2021). It has been previously reported that the histone demethylase JMJD1A may have therapeutic intervention potential for patients in GC (Ning et al., 2020). The three eRNA subtypes vary significantly in specific molecular characteristics, immune infiltration features, and immunotherapy indicators. In the present study, GSVA analyses suggested that the active situation of the KRAS signaling pathway was different in ImmuneH patients and Immune_L patients. The result implied that the KRAS signaling pathway might be considered in the personalized antitumor immunotherapy for Immune_H patients and Immune_L patients. Our results observed statistically significant differences in prognosis between eRNA subtypes. The immune_L group had the best prognosis and lowest immune score. The activation of the EMT signaling pathway may increase the incidence of tumor invasion and metastasis, resulting in drug resistance and poor prognosis (Iwadate, 2016). The results suggested that the highest EMT score may explain the poorer prognosis of patients in the Immune_H group. In addition, genetic alterations have been implicated in immune therapy response, including high TMB and high MSI status (which also results in high TMB). A high TMB enables tumors to develop somatic-mutation-derived neoantigens and trigger a T-cell-dependent immune response (McGranahan et al., 2016; Sha et al., 2020). Kim et al. showed that TMB might be a prognostic marker of survival in patients with advanced GC who received immunotherapy. TMB-H GC patients had longer progression-free survival (Kim et al., 2020). Furthermore, MSI-H tumor could induce an immune response characterized by a large number of tumor-infiltrating lymphocytes (Chang et al., 2018), and MSI was a prognostic marker for immunotherapy (Dudley et al., 2016). Our results would account for the patients in the Immune_L group (with a better prognosis) having higher TMB scores and the highest rate of MSI-H subtype. The conclusions from these prior studies are consistent with the results presented here. mRNAsi score is a refreshing index for estimating the statement of dedifferentiation. A higher mRNAsi score is negatively correlated with immune infiltration, accounting for the lowest immune infiltration level in the Immune_L group (Malta et al., 2018). We calculated the IPS to predict the response to checkpoint blockers between three eRNA subtypes. The results showed that the Immune_L group might ideally suit for immunotherapy.
We observed an apparent difference in the above indicator between the three eRNA subtypes. As widely known, the immune checkpoint was also an important indicator of response to immunotherapy. We found that BTLA and TMIGD2, two immune checkpoints, were regulated by eRNA. Increased levels of BTLA led to adverse outcomes in GC, which was confirmed in the study of specimens from patients with GC (Lan et al., 2017). In light of the research of Chen et al., inhibition of BTLA combined with chemotherapy could improve immune activation and produce potent antitumor effects (Chen et al., 2019a). Overexpression of HHLA2 was a prognostic marker of GC (Wei et al., 2020), and TMIGD2 as a receptor protein of HHLA2 could become a target for cancer therapy (Janakiram et al., 2015). In addition, we also found that the corresponding eRNA has an association with the immunotherapy pathway.
We evaluated correlations between the chemotherapy drug and eRNA subtypes. The current research found that Immune_H patients benefit more from four chemotherapy drugs (axitinib, bicalutamide, cisplatin, and docetaxel). A study from China confirmed that immunotherapy combined with cisplatin in the treatment of GC patients had good progression-free survival and overall one-year survival (Qiao et al., 2019).
It is noteworthy that subtype-related eRNA pairs with prognostic value. Remarkably, we found ERBB2 was explicitly regulated by eRNA in Immune_H patients. ERBB2, also known as HER2, was associated with poor outcomes in GC (Boku, 2014). Four innovative anti-ErbB2-antibody-based therapies, including trastuzumab, are currently available. The result implied Immune_H patients might be considered in the personalized antitumor immunotherapy for ERBB2 expression.
We are further devoted to investigating the genomic changes affecting eRNA expression. Large-scale CNVs change the proportion of gene RNA expression in the acquired or lost genome region, which changes gene expression by changing the copy number and dose of genes in the genome region (Hou et al., 2016). Taken together, CNVs might affect eRNA expression. This study appropriately evaluated the above aspects.
We developed a random forest algorithm to evaluate the eRNA cluster in the GEO cohort. Our results suggested that the random forest algorithm has high discrimination performance, so it may be the first choice for researchers to establish noncoding RNA classification models. Many studies have shown that the random forest model shows superior predictive ability when using multicenter clinical data (Deist et al., 2018; Li et al., 2014; Toth et al., 2019). Lastly, and most importantly, we establish eRNA score to evaluate the eRNA activation degree of the individual patient. In this study, we believe that the predictive value of eRNA score in response to immunotherapy should be tested in GC cohorts treated with immunotherapy firstly. However, there were no available immunotherapy datasets of GC in the existing database. As a complementary measure, a meta-melanoma cohort that received immunotherapy was used to test the immunotherapy benefit of the eRNA score. Interestingly, the predictive value of the eRNA score was validated in the meta-melanoma cohort. Unfortunately, there were no confirmed responses in the remaining two cohorts (non–small cell lung carcinoma and head and neck squamous cell carcinoma; data not shown). In addition to the present small cohort size, we speculate that the main reason is eRNA-tissue-specific expression. Previous studies suggested that there was a cancer-/lineage-specificity of eRNAs, which may be largely driven by tissue-specific TFs. Different human tissues were characterized by the distinct landscapes and regulatory networks of eRNAs (Kopp and Mendell, 2018; Wu et al., 2014; Zhang et al., 2019, 2021). The result indicated that the eRNA score might act as a robust classifier with good predictive performance in immunotherapy response in several kinds of tumor types for tissue context specificity of eRNAs.
Conclusion
In conclusion, our findings highlight the potential association between the tumor immune infiltration landscape and eRNA. Here, we hypothesized that identifying the eRNA-related subtype of GCs might reveal valuable molecular subgroups and may be beneficial in predicting the prognosis and response to immunotherapy. We discovered many aspects that those in three eRNA subtypes differ phenotypically or genetically, which provided exciting insights into the biology of GC. Furthermore, we shed light on establishing an eRNA score to predict the clinical outcome and immunotherapy responses.
Limitations of the study
Several limitations of this research are addressed as follows. The main limitation of our study is the modest sample size of GC. We still need more GC samples to verify the reliability of our conclusions. Another pitfall is that the immune-based therapy data of GC patients are not available now. More validation datasets of received immunotherapy are needed to verify the stability of eRNA subtypes and eRNA scores. Finally, more functional experiments are desired to elucidate the underlying mechanism of these eRNAs in tumor progression and immune escape. A combination of GRO-seq/PRO-seq and RNA-seq may likely apply to identifying and quantifying the eRNAs robustly, thereby contributing to post-transcriptional regulation in GC (Kim et al., 2010; Kristjansdottir et al., 2020; Wang et al., 2011).
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and materials should be directed to and will be fulfilled by the lead contact, Ben Liu (benliu100@tmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Method details
Data collection and pre-processing
The workflow of our study is shown in Figure 1. This study used public gene expression data from TCGA and Gene Expression Omnibus (GEO) database. Corresponding clinical data of TCGA gastric cancer were extracted from the UCSC Xena web data resource (https://xenabrowser.net/datapages/). Patients without survival information were removed from further evaluation. As to datasets in TCGA, the FPKM (fragments per kilobase of per million) value of GC RNA-seq was extracted from the TCGA program (https://portal.gdc.cancer.gov/). Then FPKM values were transformed into transcripts per kilobase million (TPM) values. For GEO microarray data from Affymetrix®, we downloaded the raw "CEL" files and adopted a robust multiarray averaging method with the affy package to perform background adjustment and quantile normalization. For GEO microarray data from other platforms, the normalized matrix files were directly downloaded. Based on the mRNA information in the GENECODE data resource V22 (https://www.gencodegenes.org/), we extracted the mRNA expression profiles from the RNA-seq cohort. In total, five eligible GC cohorts, GSE15459 (N=192), GSE34942 (N=56), GSE62254 (N=283), GSE84437 (N=433), and TCGA-STAD (The Cancer Genome Atlas-Stomach Adenocarcinoma) (N=349), were integrated into our study for further analysis. The mRNA expression data of the TCGA and GEO cohorts were normalized, respectively. The baseline information of all eligible GC datasets was summarized in Tables S1 and S2. The somatic mutation data were acquired from the UCSC Xena database. Genome-wide somatic copy number profiles of gastric cancer were extracted from Broad GDAC Firehouse (https://gdac.broadinstitute.org/). eRNA expression data and eRNA target genes information were extracted from enhancer RNA in cancers (eRic) (https://hanlab.uth.edu/eRic/).
Consensus cluster of GC-related eRNAs
We performed the consensus cluster analysis to identify distinct eRNA transcriptional regulation patterns based on the expression of GC-related 2267 eRNAs. The consensus clustering algorithm determined the number of clusters and their stability. We used the ConsensusClusterPlus package to perform the above steps, and 100 times repetitions were conducted to guarantee the stability of classification.
Estimation of TME cell infiltration between eRNA subtypes
We used the single sample gene set enrichment analysis (ssGSEA) algorithm to quantify the relative abundance of each cell infiltration in the GC TME. A set of biomarker genes for 28 types of immune cells was acquired from a past study. A total of 28 immune cell subtypes included MDSC, activated dendritic cell, macrophage, natural killer T cell, and regulatory T cell (Barbie et al., 2009; Charoentong et al., 2017). The enrichment scores calculated by ssGSEA analysis were utilized to represent the relative abundance of each TME infiltrating cell in each sample. ESTIMATE R package was used to calculate Stromal Score, Immune Score, ESTIMATE Score, and Tumor Purity based on TPM values of RNA-seq data. Stromal Score and Immune Score were used to further stratify the eRNA clusters. The Kaplan-Meier method was used to assess overall survival (OS) between the different eRNA subtypes. The p-value of the log rank test less than 0.05 was considered significant.
Analyses of subtype-related eRNA-target gene pairs between eRNA distinct subtypes
We identified the subtype-related eRNA-target gene pairs following two steps: First, the Bioconductor "limma" package was employed to calculate differential expression eRNA based on RNA-seq data of three eRNA subtypes. Differentially expressed eRNAs (DEeRNAs) were determined using the cutoff thresholds of p < 0.05 and |log2 fold change| > 0.5. Second, we adopted a univariate Cox regression to calculate the prognostic value of DEeRNAs and target genes in gastric cancer. The p-value of univariate Cox regression less than 0.05 was considered significant. After the above filtering steps, the remaining eRNA-target gene pairs were termed subtype-related eRNA-target gene pairs. In addition, we performed GO (gene ontology) enrichment analyses of target genes that remained.
Gene set variation analysis (GSVA)
To investigate the difference in the biological pathway between eRNA subtypes, we performed GSVA enrichment analysis using "GSVA" R packages. GSVA, in a non-parametric and unsupervised method, is commonly employed for estimating the variation in the pathway and biological process activity in the samples of an expression dataset(Hänzelmann et al., 2013). The gene sets of "Hallmark gene sets" were downloaded from the MSigDB database for running GSVA analysis. A p-value less than 0.05 was considered statistically significant. Furthermore, considering that mutations may affect RNA-RNA binding, we recognized the significantly mutated eRNA-target genes by applying the R package "maftools".
Immunophenoscore (IPS)
Immunophenoscore (IPS), the superior immune response molecular marker, was used to characterize the intratumoral immune landscapes and the cancer antigenomes. The scoring scheme was created from a panel of immune-related genes belonging to the four clusters: major histocompatibility complex (MHC)-related molecules, checkpoints or immunomodulators, effector cells, and suppressor cells. Higher IPS represents higher levels of immunotherapy response rate (Charoentong et al., 2017).
Copy number variation (CNV)
We evaluated copy number variation (CNV) in regions of eRNA positions. "OmicCircos" R package was used to draw a Circle plot showing the genome-wide somatic copy number profiles of GC patients. We downloaded the CNV data of STAD from The Cancer Genome Atlas (TCGA, http://gdac.broadinstitute.org/) and the CNVs-associated peak regions of eRNAs and the CNVs-associated peak regions of eRNAs were assigned with BEDtools (Quinlan and Hall, 2010). We also evaluated significant variation CNV segments (alternation frequency >0.3) in GC. "ComplexHeatmap" R package was used to draw a waterfall plot showing significant CNV segments between three eRNA subtypes. The heatmap was used to show the frequency of CNV between three eRNA subtypes.
Epithelial-mesenchymal transition (EMT) and mRNA expression-based stemness index (mRNAsi)
EMT-related genes were obtained from published literature (Tao et al., 2020). We used ssGSEA to evaluate EMT scores by the EMT-related genes. We got the mRNA expression-based stemness index (mRNAsi) from published literature (Malta et al., 2018).
Predict chemotherapeutic response
The R package "pRRophetic" (Geeleher et al., 2014a) was employed to predict chemotherapeutic response in GC patients, of which the half-maximal inhibitory concentration (IC50) of the samples was demonstrated using ridge regression, and the prediction accuracy was assessed using 10-fold cross-validation based on the GDSC training set (Geeleher et al., 2014b).
Immune checkpoint molecules
The gene sets of immune checkpoint molecules were obtained from previous research, which include members of the major histocompatibility complex (MHC), immune co-stimulator checkpoint (ICP), and immune co-inhibitory checkpoint (IAP) (Huang and Fu, 2019). To assess the immune checkpoints score in GC, we performed ssGSEA algorithm to evaluate three types of immune checkpoints score by the above gene sets. The pharmacogenomic data were obtained from the Genomics of Drug Sensitivity in eRic (Kong et al., 2020).
Machine learning algorithms recognized eRNA subtypes in the validation cohort
Independent validation for eRNA subtype was performed in a meta-cohort combining four GEO cohorts (N=964). Batch effects from non-biological technical biases were corrected using the "ComBat" algorithm of sva package. Due to the absence of eRNA expression data in the GEO cohort, we used the random forest (RF) model, a machine learning algorithm, to evaluate the eRNA subtype based on the characteristics of subtype-related target gene expression in the TCGA cohort. We identified the differentially expressed target gene, i.e., subtype-related target gene, between three eRNA subtypes based on Bioconductor "limma" package with the cutoff thresholds of p < 0.05 and |log2 fold change| > 0.5. The R package "randomForest" was used to construct the random forest model. Random forests are widely used for data prediction and interpretation purposes (Han et al., 2020; Jeong et al., 2020). The workflow of the random forest model was shown in Figure S1.
Generation of eRNA signature in gastric patients
To quantify the eRNA expression patterns of an individual tumor, we constructed a scoring system to evaluate the eRNA regulatory degree of the individual patient.
We named this eRNA signature as an eRNA score. The procedures for the establishment of eRNA signature were as follows:
We performed subtype-related target genes to construct an eRNA signature. Principal component analysis (PCA) was used to construct an eRNA-relevant gene signature. Both principal components 1 and 2 were selected to act as signature scores. We defined the eRNA score using a method similar to GGI (Sotiriou et al., 2006; Zeng et al., 2019):
| eRNA score = Σ (PC1i + PC2i) |
where i is the expression of a subtype-related target gene.
Independent validation for eRNA score was performed in three GEO (GSE13861 (N = 64), GSE26901 (N = 109), GSE84437 (N = 433)) and Tianjin (N = 90) cohorts.
Quantification and statistical analysis
Statistical analysis was done on R version 4.0.0 (https://www.r-project.org). Kruskal-Wallis test was used to conduct difference comparisons of three or more groups. p < 0.05 (two-tailed) represented statistical significance. Clustering of eRNA was performed using the consensus clustering method to robustly identify structure over many clustering realizations(Liu et al., 2021b, 2021c, 2021d). The ssGSEA method is an extension of the GSEA method (Mootha et al., 2003; Subramanian et al., 2005), working at a single sample level rather than a sample population as in the original GSEA application. The score derived from ssGSEA reflects how the input gene signature is coordinately up- or down-regulated within a sample. The survival curves for the prognostic analysis were generated via the Kaplan-Meier method. The Log rank test p < 0.05 was considered statistically significant. The patients were assigned to a high- and a low-gene expression group, respectively, based on the best cutoff values calculated using the survminer package in R.
Additional resources
This study did not generate additional data.
Acknowledgments
This work was supported by the following grants: National Key R&D Program of China (2021YFC2500400) to KC. National Natural Science Foundation of China (82073028, 81572445) to BL; National Natural Science Foundation of China (82172894) to KC; National Key R&D Program of China (2017YFC0908300) to BL; The 14th five-year Special Project of Cancer Prevention and Treatment for the Youth Talents of Tianjin Cancer Institute and Hospital (YQ-04) to BL; Beijing-Tianjin-Hebei Basic Research Cooperation Special Project (20JCZXJC00090) to KC; Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-009A) to KC. We would like to thank Dr. Leng Han from McGovern Medical School at The University of Texas Health Science Center who established eRic database that inspired our research. We thank Dr. Mulin Jun Li for the helpful discussions. I thank my friend Huan Wang for her support in my daily life. Graphical abstract partially created by BioRender (https://biorender.com/).
Author contributions
Ben Liu and Kexin Chen designed and supervised the study; Xin Hu, Liuxing Wu, and Yanxin Yao performed data collection, analysis and wrote the manuscript; Junfu Ma, Hongru Shen, Wei Wang, Xinlei Chu, Hongji Dai, and Luyang Liu collected data; Hong Zheng, Meng Yang, and Fengju Song interpreted the data; Xin Hu, Liuxing Wu, Yanxin Yao, Ben Liu, and Xiangchun Li revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105075.
Contributor Information
Kexin Chen, Email: chenkexin@tmu.edu.cn.
Ben Liu, Email: benliu100@tmu.edu.cn.
Supplemental information
Data and code availability
-
•
The datasets analyzed during the current study are available from open access database. These accession numbers for the datasets are listed in the key resources table
-
•
No custom codes were used in the study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., Giraldo N.A., Germain C., de Reyniès A., Laurent-Puig P., Zucman-Rossi J., Dieu-Nosjean M.-C., Sautès-Fridman C., Fridman W.H. Immune contexture, immunoscore, and malignant cell molecular subgroups for prognostic and theranostic classifications of cancers. Adv. Immunol. 2016;130:95–190. doi: 10.1016/bs.ai.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Boku N. HER2-positive gastric cancer. Gastric Cancer. 2014;17:1–12. doi: 10.1007/s10120-013-0252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Chang M., Chang H.M., Chang F. Microsatellite instability: a predictive biomarker for cancer immunotherapy. Appl. Immunohistochem. Mol. Morphol. 2018;26:e15–e21. doi: 10.1097/PAI.0000000000000575. [DOI] [PubMed] [Google Scholar]
- Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Chen Y.-L., Lin H.-W., Chien C.-L., Lai Y.-L., Sun W.-Z., Chen C.-A., Cheng W.-F. BTLA blockade enhances Cancer therapy by inhibiting IL-6/IL-10-induced CD19 B lymphocytes. J. Immunother. Cancer. 2019;7:313. doi: 10.1186/s40425-019-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Peng C., Chen J., Chen D., Yang B., He B., Hu W., Zhang Y., Liu H., Dai L., et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.Y., Bae J.M., An J.Y., Kwon I.G., Cho I., Shin H.B., Eiji T., Aburahmah M., Kim H.-I., Cheong J.-H., et al. Is microsatellite instability a prognostic marker in gastric cancer? A systematic review with meta-analysis. J. Surg. Oncol. 2014;110:129–135. doi: 10.1002/jso.23618. [DOI] [PubMed] [Google Scholar]
- Cristescu R., Lee J., Nebozhyn M., Kim K.M., Ting J.C., Wong S.S., Liu J., Yue Y.G., Wang J., Yu K., et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- Deist T.M., Dankers F.J.W.M., Valdes G., Wijsman R., Hsu I.C., Oberije C., Lustberg T., van Soest J., Hoebers F., Jochems A., et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: an empirical comparison of classifiers. Med. Phys. 2018;45:3449–3459. doi: 10.1002/mp.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z.-Y., Zhang C., Li Y.-F., Su J., Xie Z., Liu S.-Y., Yan L.-X., Chen Z.-H., Yang X.-N., Lin J.-T., et al. Genetic and immune profiles of solid predominant Lung adenocarcinoma reveal potential immunotherapeutic strategies. J. Thorac. Oncol. 2018;13:85–96. doi: 10.1016/j.jtho.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Dudley J.C., Lin M.-T., Le D.T., Eshleman J.R. Microsatellite instability as a biomarker for PD-1 blockade. Clin. Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P., Cox N.J., Huang R.S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15:R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Chen H., Yao Y., Liu X., Nie C., Min J., Zeng Y., Lutz M.W. Genetic and non-genetic factors associated with the phenotype of exceptional longevity & normal cognition. Sci. Rep. 2020;10:19140. doi: 10.1038/s41598-020-75446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Guo H., Cao C., Li X., Hu B., Zhu P., Wu X., Wen L., Tang F., Huang Y., Peng J. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Fei T., Chen Y., Li T., Gao Y., Wang X., Sun T., Sweeney C.J., Lee G.-S.M., Chen S., et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. USA. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.-X., Fu L. The immune landscape of esophageal cancer. Cancer Commun. 2019;39:79. doi: 10.1186/s40880-019-0427-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiott N.E., Heward J.A., Roux B., Tsitsiou E., Fenwick P.S., Lenzi L., Goodhead I., Hertz-Fowler C., Heger A., Hall N., et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwadate Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol. Lett. 2016;11:1615–1620. doi: 10.3892/ol.2016.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiram M., Chinai J.M., Zhao A., Sparano J.A., Zang X. HHLA2 and TMIGD2: new immunotherapeutic targets of the B7 and CD28 families. OncoImmunology. 2015;4:e1026534. doi: 10.1080/2162402X.2015.1026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Kim J.Y., Kim N. GMStool: GWAS-based marker selection tool for genomic prediction from genomic data. Sci. Rep. 2020;10:19653. doi: 10.1038/s41598-020-76759-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Jiang G., Yang Y., Yang J., Yang W., Wang D., Niu X., Zhong R., Zhang Z., Gong J. Animal-eRNAdb: a comprehensive animal enhancer RNA database. Nucleic Acids Res. 2022;50:D46–D53. doi: 10.1093/nar/gkab832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman H.L., Atkins M.B., Subedi P., Wu J., Chambers J., Joseph Mattingly T., Campbell J.D., Allen J., Ferris A.E., Schilsky R.L., et al. The promise of Immuno-oncology: implications for defining the value of cancer treatment. J. Immunother. Cancer. 2019;7:129. doi: 10.1186/s40425-019-0594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim B., Kang S.Y., Heo Y.J., Park S.H., Kim S.T., Kang W.K., Lee J., Kim K.-M. Tumor mutational burden determined by panel sequencing predicts survival after immunotherapy in patients with advanced gastric cancer. Front. Oncol. 2020;10:314. doi: 10.3389/fonc.2020.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.T., Cristescu R., Bass A.J., Kim K.-M., Odegaard J.I., Kim K., Liu X.Q., Sher X., Jung H., Lee M., et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018;24:1449–1458. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Xie P., Cao L., Zhang M.Q., Kim T.H. Global transcriptional activity dynamics reveal functional enhancer RNAs. Genome Res. 2018;28:1799–1811. doi: 10.1101/gr.233486.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., Lee H., Kim D., Han S.K., Ha D., Shin K., Kim S. Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat. Commun. 2020;11:5485. doi: 10.1038/s41467-020-19313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjánsdóttir K., Dziubek A., Kang H.M., Kwak H. Population-scale study of eRNA transcription reveals bipartite functional enhancer architecture. Nat. Commun. 2020;11:5963. doi: 10.1038/s41467-020-19829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X., Li S., Gao H., Nanding A., Quan L., Yang C., Ding S., Xue Y. Increased BTLA and HVEM in gastric cancer are associated with progression and poor prognosis. OncoTargets Ther. 2017;10:919–926. doi: 10.2147/OTT.S128825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauss M., Donia M., Harbst K., Andersen R., Mitra S., Rosengren F., Salim M., Vallon-Christersson J., Törngren T., Kvist A., et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017;8:1738. doi: 10.1038/s41467-017-01460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen Z., Tian L., Zhou C., He M.Y., Gao Y., Wang S., Zhou F., Shi S., Feng X., et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhou M., Li X., Zhang X., Wang Q., Liu L., Yang M., Yang D., Guo Y., Zhang Q., et al. Interrogation of gender disparity uncovers androgen receptor as the transcriptional activator for oncogenic miR-125b in gastric cancer. Cell Death Dis. 2021;12:441. doi: 10.1038/s41419-021-03727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang L., Liu L., Lu T., Jiao D., Sun Y., Han X. The identification and validation of two heterogenous subtypes and a risk signature based on ferroptosis in hepatocellular carcinoma. Front. Oncol. 2021;11:619242. doi: 10.3389/fonc.2021.619242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang Y., Dang Q., Wu K., Jiao D., Li Z., Sun Z., Han X. Genomic alteration characterization in colorectal cancer identifies a prognostic and metastasis biomarker: fam83A|Ido1. Front. Oncol. 2021;11:632430. doi: 10.3389/fonc.2021.632430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang Y., Shi C., Zhou X., Xu K., Jiao D., Sun Z., Han X. A novel immune classification reveals distinct immune escape mechanism and genomic alterations: implications for immunotherapy in hepatocellular carcinoma. J. Transl. Med. 2021;19:5. doi: 10.1186/s12967-020-02697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N., Kamińska B., Huelsken J., Omberg L., Gevaert O., et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconcini R., Spagnolo F., Stucci L.S., Ribero S., Marra E., Rosa F.D., Picasso V., Di Guardo L., Cimminiello C., Cavalieri S., et al. Current status and perspectives in immunotherapy for metastatic melanoma. Oncotarget. 2018;9:12452–12470. doi: 10.18632/oncotarget.23746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel E.E., Koeppen H., Astarita J.L., Cubas R., et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N., Furness A.J.S., Rosenthal R., Ramskov S., Lyngaa R., Saini S.K., Jamal-Hanjani M., Wilson G.A., Birkbak N.J., Hiley C.T., et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A., Hamilton P.T., Zhang A.W., Pattnaik S., Becht E., Mezheyeuski A., Bruun J., Micke P., de Reynies A., Nelson B.H. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. USA. 2019;116:9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nathanson T., Ahuja A., Rubinsteyn A., Aksoy B.A., Hellmann M.D., Miao D., Van Allen E., Merghoub T., Wolchok J.D., Snyder A., Hammerbacher J. Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol. Res. 2017;5:84–91. doi: 10.1158/2326-6066.CIR-16-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning K., Shao Y., He Y., Wang F., Cui X., Liu F., Li D., Li F. Histone demethylase Jumonji domain-containing 1A inhibits proliferation and progression of gastric cancer by upregulating runt-related transcription factor 3. Cancer Sci. 2020;111:3679–3692. doi: 10.1111/cas.14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao G., Wang X., Zhou L., Zhou X., Song Y., Wang S., Zhao L., Morse M.A., Hobeika A., Song J., et al. Autologous dendritic cell-cytokine induced killer cell immunotherapy combined with S-1 plus cisplatin in patients with advanced gastric cancer: a prospective study. Clin. Cancer Res. 2019;25:1494–1504. doi: 10.1158/1078-0432.CCR-18-2360. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S., Hodi F.S., Martín-Algarra S., Mandal R., Sharfman W.H., et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni M., Massari F., Di Nunno V., Conti A., Cimadamore A., Scarpelli M., Montironi R., Cheng L., Battelli N., Lopez-Beltran A. Immunotherapy in renal cell carcinoma: latest evidence and clinical implications. Drugs Context. 2018;7:212528. doi: 10.7573/dic.212528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Lauberth S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020;27:521–528. doi: 10.1038/s41594-020-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha D., Jin Z., Budczies J., Kluck K., Stenzinger A., Sinicrope F.A. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 2020;10:1808–1825. doi: 10.1158/2159-8290.CD-20-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B., et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Suarez-Carmona M., Lesage J., Cataldo D., Gilles C. EMT and inflammation: inseparable actors of cancer progression. Mol. Oncol. 2017;11:805–823. doi: 10.1002/1878-0261.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao C., Huang K., Shi J., Hu Q., Li K., Zhu X. Genomics and prognosis analysis of epithelial-mesenchymal transition in glioma. Front. Oncol. 2020;10:183. doi: 10.3389/fonc.2020.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R., Schiffmann H., Hube-Magg C., Büscheck F., Höflmayer D., Weidemann S., Lebok P., Fraune C., Minner S., Schlomm T., et al. Random forest-based modelling to detect biomarkers for prostate cancer progression. Clin. Epigenetics. 2019;11:148. doi: 10.1186/s13148-019-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A., et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Tang L., Chang H., Huo S., Li Y. HHLA2 overexpression is a novel biomarker of malignant status and poor prognosis in gastric cancer. Hum. Cell. 2020;33:116–122. doi: 10.1007/s13577-019-00280-2. [DOI] [PubMed] [Google Scholar]
- Wu H., Nord A.S., Akiyama J.A., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Visel A. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Shen J. From Super-enhancer non-coding RNA to immune checkpoint: frameworks to functions. Front. Oncol. 2019;9:1307. doi: 10.3389/fonc.2019.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Xie F., Xu D., Xu W., Ge X., Lv S., Li S., Yuchi A. MiR-200b suppresses gastric cancer cell migration and invasion by inhibiting NRG1 through ERBB2/ERBB3 signaling. J. Oncol. 2021;2021:4470778–4470810. doi: 10.1155/2021/4470778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L., Liang J., Ozer A., Leung A.K.Y., Lis J.T., Yu H. A comparison of experimental assays and analytical methods for genome-wide identification of active enhancers. Nat. Biotechnol. 2022;40:1056–1065. doi: 10.1038/s41587-022-01211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Li M., Zhou R., Zhang J., Sun H., Shi M., Bin J., Liao Y., Rao J., Liao W. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 2019;7:737–750. doi: 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hong W., Ruan H., Jing Y., Li S., Liu Y., Wang J., Li W., Diao L., Han L. HeRA: an atlas of enhancer RNAs across human tissues. Nucleic Acids Res. 2021;49:D932–D938. doi: 10.1093/nar/gkaa940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lee J.H., Ruan H., Ye Y., Krakowiak J., Hu Q., Xiang Y., Gong J., Zhou B., Wang L., et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019;10:4562. doi: 10.1038/s41467-019-12543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets analyzed during the current study are available from open access database. These accession numbers for the datasets are listed in the key resources table
-
•
No custom codes were used in the study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.