Abstract
A homologue of the grmA spore germination gene of Bacillus megaterium and of a NaH-antiporter gene (napA) of Enterococcus hirae has been identified in Bacillus cereus 569 (ATCC 10876). The putative protein product has 58 and 43% amino acid identity with GrmA and NapA, respectively. Insertional inactivation of this B. cereus gene, named gerN, did not affect vegetative growth or sporulation. The null mutant spores were 30-fold slower to germinate in inosine (5 mM) but germinated almost normally in response to l-alanine (10 mM). The null mutant spores germinated after several hours with inosine as the sole germinant, but germination was asynchronous and the normal order of germination events was perturbed. At a suboptimal germinant concentration (50 μM), inosine germination was completely blocked in the mutant, while the rate of germination in 50 μM l-alanine was reduced to one-third of that of the wild type. The requirement for GerN function in the response to a particular germinant suggests that a germination receptor may have a specifically associated antiporter, which is required at the initiation of germination and which, in the case of the inosine receptor, is GerN. Since germination in suboptimal concentrations of l-alanine shows a delay, additional germination transporters may be required for optimal response at low germinant concentrations.
Under certain nutrient stresses Bacillus species undergo a complex differentiation process resulting in the formation of highly resistant endospores, which subsequently, when favorable growth conditions return, germinate back to vegetative cells and grow and divide. The molecular genetics of sporulation (7, 25) and germination (14, 15) of Bacillus species have been reviewed.
Analysis of Bacillus subtilis mutants which are defective in response to one or both of their germinants—l-alanine or asparagine with glucose, fructose, and K+ (AGFK)—suggests that the germinants interact with separate germinant-specific receptor complexes within the spore (14). Mutations within the gerA operon of B. subtilis specifically block germination initiated by l-alanine (34). The predicted amino acid sequences of the three GerA proteins encoded by the gerA operon suggest that they may be membrane associated, and they are the most likely candidates for the germinant receptor for l-alanine. Mutations in the gerB operon (18), responsible for AGFK germination in B. subtilis, allowed recognition of the novel germinant d-alanine; this strongly reinforces the argument that such gerA homologues encode the germinant receptor complexes.
B. cereus endospores have been shown to germinate in response to inosine and l-alanine; a combination of these germinants elicits the most rapid response (32). Operons encoding putative receptor complexes for the germinants have been named gerI (6) and gerL (3), respectively, and these operons are members of the gerA family identified in B. subtilis. How the germinant receptors are activated and how this leads to the global changes of spore germination have, however, not been determined.
Cation transport may play an important role in spore germination; a rise in the internal pH of germinating spores and release of Na+ and K+ have been shown to be early events, preceding dipicolinic acid (DPA) release. At least 80% of the spore's Na+ and K+ is released early in spore germination, the K+ being subsequently reabsorbed by an energy-dependent process (27).
Tani et al. (28) showed that a NaH antiporter homologue, named grmA, was important for the germination of spores of B. megaterium ATCC 12872 in any of its germinants (glucose, l-proline, l-leucine, or KNO3). The mutant spores appeared to be blocked at an early stage of germination, as they did not lose heat resistance or release DPA, both relatively early events in spore germination. GrmA has a 47% amino acid identity to NapA of Enterococcus hirae (33), which has been shown to mediate NaH antiport activity (26). It was proposed that GrmA plays a critical role in the early stages of spore germination because of an effect on cation transport.
In this study a homologue of grmA was identified in B. cereus. The gene was designated gerN due to the effects of its disruption on spore germination. The gerN gene was shown to be important for the germination of B. cereus 569 UM20.1 spores in response to inosine, but not to l-alanine at optimal concentrations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Escherichia coli and B. cereus were routinely cultured in or on L broth and L agar containing the appropriate antibiotics (for E. coli, ampicillin at 50 μg ml−1; for B. cereus, erythromycin and lincomycin at 1 and 25 μg ml−1, respectively). CCY medium (24) was used for spore preparation. Nutrient agar no. 1 (NA; Oxoid) was used for spore enumeration. Some germination and outgrowth experiments were carried out in Oxoid nutrient broth (NB).
Spore preparation.
A culture of B. cereus in CCY broth (700 ml inoculated with 7 ml of a mid-log-phase PAB [Penassay broth] culture) was incubated with shaking, at 37°C for 2 days, until>90% free spores were present. Spores were harvested and washed 10 times by repeated centrifugation and resuspension in distilled water, discarding the upper layer of cellular debris in the pellet from early washing steps. The spores were stored in distilled water at −20°C. Synchronous sporulation was carried out by the procedure of Sterlini and Mandelstam (22).
Spore germination assays.
Spores were activated by heating in distilled water at 70°C for 30 min prior to germination.
OD fall.
Spores were resuspended in germination buffer (10 mM Tris-HCl [pH 8.4]–10 mM salts) at an optical density at 580 nm (OD580) 0.5. For l-alanine germination, 5 μg of O-carbamyl-d-serine ml−1 was added to inhibit l-alanine racemase activity. Germination was carried out at 37°C in inosine and at 30°C in l-alanine. After a 15-min preincubation of spores with buffer, germination was initiated by the addition of inosine (to 5 mM or 50 μM) or l-alanine (to 10 mM or 50 μM). The OD580 of the spore suspension was monitored, and the percentage of the initial OD lost was calculated. Maximum rates of OD loss per minute quoted in the text were derived from the steepest part of the germination curve. Germination in calcium DPA was carried out by the method of Keynan and Halvorson (10).
DPA release.
Heat-shocked spores were germinated at 1 mg (dry weight) ml−1. At intervals, 3-ml samples were removed and filtered (with a Millipore 0.45-μm-pore-size filter), and the filtrate was stored on ice. The DPA content of the filtrates was determined by a modification of the method of Janssen et al. (9).
Heat resistance loss.
Spores were germinated in l-alanine or inosine. Samples (100 μl) were removed at intervals, serially diluted 100-fold to 10−4 in sterile distilled water, preheated to 70 or 80°C, and incubated for up to 40 min at this temperature. Aliquots were plated in 3-ml overlays on NA plates. After overnight incubation at 37°C, colonies were counted and compared with a control prepared using spores that were not exposed to germinant but were subjected to heat treatment.
Measurement of ions released from germinating spores.
Spores were heat activated and then washed 3 times with distilled water to remove any released ions. They were then germinated, at 10 mg (dry weight) of spores ml−1. No salts were added externally; samples were taken at intervals and pelleted by centrifugation (21,000 × g for 2 min at 4°C), and the ion content of the supernatant was assayed by atomic absorption spectrophotometry. Ions were released from dormant spores by autoclaving spores (1 mg [dry weight] per ml) in a total volume of 5 ml.
Measurement of ATP produced by germinating spores.
Spores were germinated in 5 mM inosine. At intervals a 50-μl sample of the germinating spores was removed, added to 100 μl of lysis solution (Celsis International, Cambridge, United Kingdom), and incubated at room temperature for 3 min. Then 100 μl of luciferase-luciferin (10 mg ml−1; Sigma) was added to the sample, and the luminescence was measured for 10 s in a Celsis Opticomp luminometer.
Outgrowth of germinating spores.
Germination and outgrowth were carried out in NB or, alternatively, the spores were germinated in Tris-HCl buffer (pH 8.4) plus l-alanine (10 mM) or inosine (5 mM), and then 2 volumes of NB were added to stimulate outgrowth. Germination and outgrowth were monitored by OD580 measurement and phase-contrast microscopy.
Molecular biology methods.
Electroporation of B. cereus was performed by the procedure of Bone and Ellar (5). PCR and inverse PCR, were carried out by standard procedures (17), using High Fidelity Taq Extend (Boehringer Mannheim). DNA sequencing was performed with the BigDye Terminator cycle sequencing Ready Reaction kit (Applied Biosystems) and an Applied Biosystems DNA sequencer. The DNA sequence was analyzed and assembled by using the Staden suite of programs (21).
Nucleotide sequence accession number.
The 1,733-bp region including B. cereus gerN that was completely sequenced on both strands has been submitted to GenBank (accession number AF246294).
RESULTS AND DISCUSSION
Identification and sequencing of gerN, a homologue of the B. megaterium grmA gene.
Degenerate PCR primers were designed to internal regions of GrmA conserved in NapA, using a codon usage table for B. cereus. The regions used (4) were from residue 65 (MFLAGLE) for the forward primer and from residue 266 (YAVFVPVFFV) for the reverse primer. Primers were designed with EcoRI and BamHI restriction sites at each end. PCR on B. cereus 569 UM20.1 chromosomal DNA yielded a single fragment of the expected size. This fragment was cloned into the integrational vector pMUTIN4 (31), and the inserts in two clones were sequenced (the plasmid was named pMNAP). They both contained an identical open reading frame (ORF), which would encode a product with 42% identity to the corresponding region of the GrmA protein.
The regions of the gerN gene flanking this internal fragment were obtained by inverse PCR from the wild-type chromosomal DNA, using a HindIII digest to obtain the downstream region and an HpaII digest to obtain the upstream region. The sequences from the extreme ends were then used to design new primers for direct PCR. Each region was sequenced from at least two independent PCR products.
The gerN gene is predicted to start at base 475 of GenBank sequence AF246294 and has an appropriately located ribosome binding site. The gerN ORF is preceded by an intergenic region of 333 bp, separating it from a divergently transcribed ORF that encodes a homologue of B. subtilis YkqC (T.W. Southworth, unpublished data). The gerN stop codon is followed by a potential rho-independent terminator (coordinates 1651 to 1691), with a predicted ΔG of −26.4 kcal mol−1 (30). The gerN gene therefore appears to be monocistronic. Southern blotting suggested that it is chromosomally located.
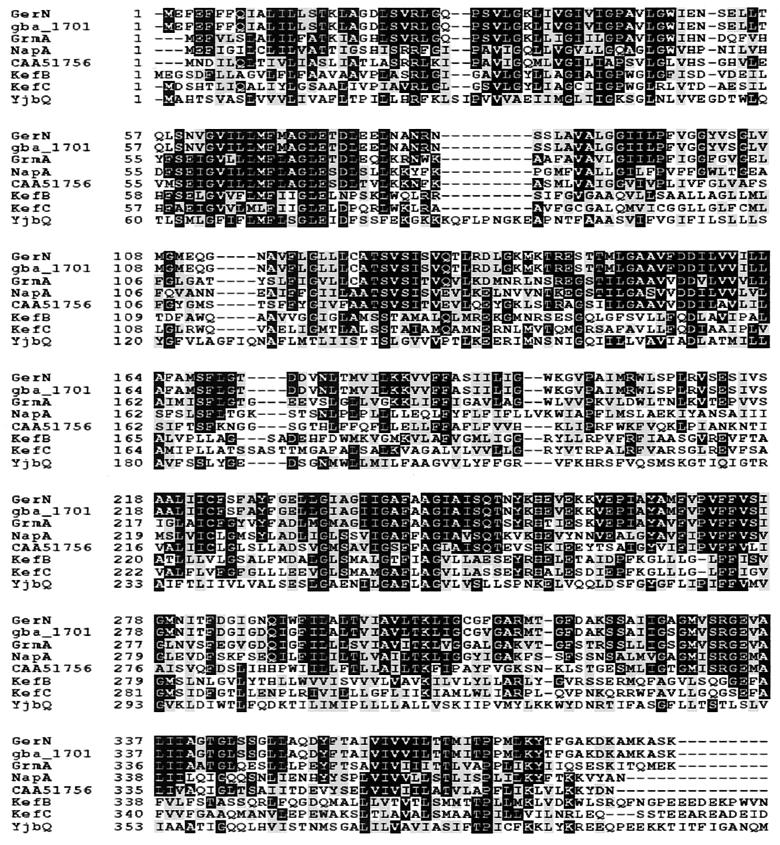
The putative GerN protein is highly hydrophobic and would be an integral membrane protein, with 14 predicted membrane-spanning regions on the basis of a hydropathy profile (12). It has a close homologue (98% amino acid identity) in the B. anthracis genome, estimated by tBLASTn searches of the sequence information from the TIGR Unfinished Genomes database of the Institute for Genomic Research (TIGR) (http://www.tigr.org). A BLAST search (1) of protein databases revealed a 58% amino acid identity to GrmA of B. megaterium (28) and a 43% amino acid identity to NapA of E. hirae (33). Locus CAA51756 of Lactococcus lactis (13) also encodes a NaH antiporter homologue. The kefB and kefC genes of E. coli (2, 16) encode two domain proteins, whose N-terminal domains are thought to carry out a KH antiport function, regulated by the C-terminal glutathione-binding domain. The B. subtilis genome encodes two more distant homologues, YhaU and YjbQ (11). A multiple sequence alignment of these with GerN was complied using CLUSTAL-W (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html) (Fig. 1). The monovalent cation: proton antiporter-2 (CPA-2) family (20), which includes GerN, GrmA, NapA, KefB, and KefC, is moderately large, with more than 20 sequenced members from bacteria, archaebacteria, and eukaryotes. As gerN is a member of this family, and is most closely related to a demonstrated NaH antiporter in this family, it is probable that the GerN protein carries out some function in cation (probably sodium or potassium) transfer during spore germination.
FIG. 1.
Multiple sequence alignment of the GerN protein against its homologues Gba_1701 of B. anthracis, GrmA of B. megaterium, NapA of E. hirae, locus CAA51756 of L. lactis, the N-terminal domains of KefB and KefC of E. coli, and the N-terminal domain of YjbQ of B. subtilis. Solid boxes indicate identical amino acids, and shaded boxes indicate conserved amino acids, for 50% of the sequences.
Disruption of the gerN gene.
The PCR fragment internal to the gerN gene (corresponding to codons 65 to 275) cloned into pMUTIN4 was introduced into B. cereus by electroporation, and the transformants were selected on erythromycin and lincomycin. Two transformants were characterized; AM1419 and AM1420. Intergration of the plasmid by homologous recombination into the chromosome within the gerN gene was confirmed by Southern blotting and the gene disruption mutant was named gerN1. This allele would encode a truncated product lacking the final 112 amino acids of the 387-residue protein.
Characterization of the gerN1 mutant. (i) Growth and sporulation.
There is no difference between the growth rates of the wild-type and mutant strains in either NB or NB plus 0.5 M NaCl (data not shown). The gerN1 mutant showed normal sporulation in CCY medium and synchronous sporulation experiments (data not shown).
The cortex of dormant spores of the mutant was normal, as estimated by high-performance liquid chromatography (HPLC) of muropeptide fragments (A. Atrih, personal communication), and gerN mutant spores had the same level of heat resistance (at 70, 80, or 90°C) as the wild type (29).
(ii) Measurement of spore germination in inosine by OD fall.
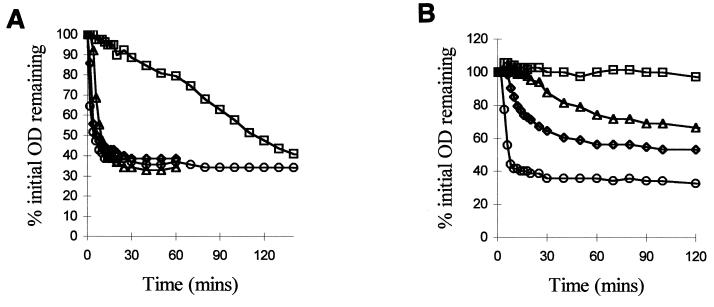
Wild-type B. cereus 569 UM20.1 spores germinate rapidly in 5 mM inosine plus 10 mM NaCl in Tris-HCl buffer (maximum rate, 18% OD loss/min) (Fig. 2); OD fall, DPA release, loss of heat resistance, and phase darkening of the spores were 90% complete 10 min after the addition of germinant. In contrast, the rate of germination of the gerN mutant was severely inhibited under these conditions (maximum rate, 0.55% OD loss/min) (Fig. 2). The OD of the mutant spore suspensions did eventually fall as low as that of the wild type, but only after 3 h. The mutation blocked spore germination at an early stage; after 90 min, only 50 to 55% of the potential OD fall had occurred, 55 to 60% of the total DPA had been released, and 55 to 60% of the mutant spores still retained their heat resistance (data not shown). Surprisingly, at this time ca. 90% of the spores had become phase dark. Heat resistance loss and DPA release therefore occur at approximately the same rate as OD fall, but it appears that phase darkening of these spores can occur without the completion of the other events.
FIG. 2.
Effect of a gerN mutation on OD loss during germination. (A) Germination of B. cereus 569 and gerN1 mutant spore suspensions in response to 5 mM inosine plus 10 mM NaCl (circles and squares, wild-type and mutant spores, respectively) or 10 mM l-alanine plus 10 mM NaCl (diamonds and triangles, wild-type and mutant spores, respectively). (B) Germination of B. cereus and gerN1 mutant spore suspensions in response to 50 μM inosine plus 10 mM NaCl (circles and squares, wild-type and mutant spores, respectively) or 50 μM l-alanine plus 10 mM NaCl (diamonds and triangles, wild-type and mutant spores, respectively).
Substitution of K+, Ca2+, or NH4+ for Na+ (29) had the same general effect on the germination of the gerN mutant spores as the effect on wild-type spores described by Clements and Moir (6); the rate of germination for the mutant was generally 30-fold lower than that for the wild type under equivalent conditions. Use of K+ as the cation adjunct dramatically slows wild-type spore germination in inosine and completely blocked the germination of the gerN mutant spores. Inclusion of a subgerminal concentration of l-alanine (10 μM) increased the germination rate of the gerN mutant in inosine threefold (maximum rate 1.75% OD fall/min), but the stimulated rate was still very low in comparison to that of the wild type (which increased 1.5-fold upon addition of subgerminal l-alanine, giving a maximum rate of 26.5% OD fall/min).
When inosine was used at a suboptimal concentration of 50 μM, the wild-type rate of spore germination was twofold lower than that in 5 mM inosine, but the mutants showed no OD loss at all in this concentration of inosine (Fig. 2), and no other germination-associated changes occurred. Therefore, a higher threshold concentration of inosine may be required to stimulate the residual changes that occur in the mutant spores.
(iii) Other changes during inosine germination.
The gerN mutant and wild-type dormant spores were found to contain similar levels of Na+, K+, Ca2+, Mg2+, Fe2+, and Mn2+ (29). Calcium and magnesium release from wild-type spores germinating in inosine was complete in 10 min, whereas the equivalent release in the gerN mutant was delayed (Table 1). Potassium ions are released and readsorbed (27), so the concentration in the supernatant rises (to less than the real maximum) and then decreases; the kinetics of these fluxes were also delayed in the mutant.
TABLE 1.
Release of cations during the germination of B. cereus 569 and gerN null spores in 5 mM inosine
| Time (min) | Cation release (mg/g of spores)a during germination of:
|
|||||
|---|---|---|---|---|---|---|
|
B. cereus 569
|
gerN1 mutant
|
|||||
| Ca2+ | Mg2+ | K+ | Ca2+ | Mg2+ | K+ | |
| 1 | 0.1 | 0.013 | 0.02 | 0.1 | 0.018 | 0.0 |
| 2 | 0.63 | 0.09 | 0.052 | ND | ND | ND |
| 10 | 2.74 | 0.379 | 0.0 | 0.12 | 0.01 | 0.029 |
| 30 | 3.26 | 0.689 | 0.0 | 0.23 | 0.031 | 0.033 |
| 90 | ND | ND | ND | 3.02 | 0.327 | 0.0 |
ND, not determined.
ATP levels in germinating wild-type spores increased between 2 and 10 min after addition of germinant, to 170 pmol/mg of spores, whereas in the gerN mutant spores ATP synthesis began between 10 and 30 min after initiation of germination and reached the wild-type 10-min level only after 60 min of germination. This is consistent with the delay in other germination events measured.
(iv) Germination in l-alanine.
When germinated in 10 mM l-alanine and 10 mM NaCl, the gerN mutant spores show no significant defect (Fig. 2). Changing the cation adjunct had very little effect on the germination rate of either the wild-type or mutant spores (reference 6; also data not shown). However, in suboptimal concentrations of l-alanine (50 μM), the germination rate of the mutant, measured as OD fall, was reduced nearly threefold compared to that of the wild type (Fig. 2). This is different from the slow germination seen in 5 mM inosine, because, although the rate of response to germinant was reduced, the spores showed the normal pattern of germination behavior (29).
GerN is dispensable at optimal concentrations of l-alanine; an alternative protein must therefore be able to substitute for its function. However, a higher level of l-alanine is required to stimulate germination to maximum rates when gerN has been disrupted.
(v) Germination in calcium DPA.
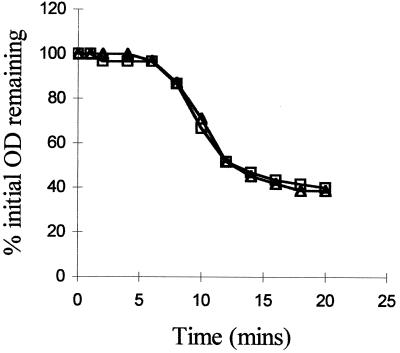
The rate of germination of the gerN mutant in the nonnutrient germinant, calcium DPA, was the same as that of the wild type (Fig. 3). Hence, germination which does not use the germinant receptors is unaffected and does not appear to require the function of the GerN protein.
FIG. 3.
Germination of spore suspensions of B. cereus 569 (squares) and the gerN mutant (triangles) in calcium DPA.
(vi) Cortex degradation.
The peptidoglycan composition of spores was analyzed during germination. After germination for 90 min in 10mM inosine and 10 mM NaCl, only primordial cell wall remains in germinated wild-type spores, whereas the gerN null mutant had 60 to 70% of its cortex remaining (A. Atrih, personal communication). The peptidoglycan fragments released into the medium from the gerN mutant spores had the same structure as those from the wild-type spores, though they were released in lower quantities.
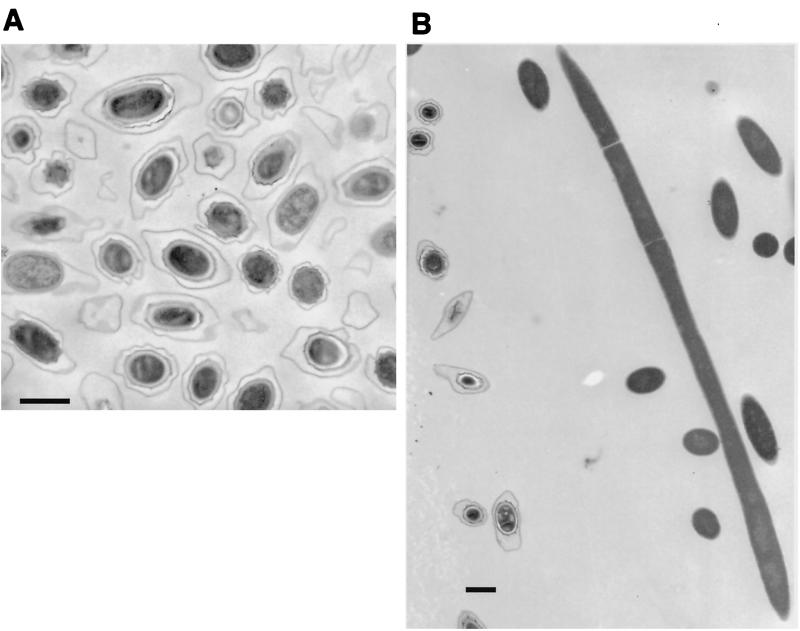
(vii) Electron microscopy of germinated and outgrowing spores.
After 90 min of germination in 5 mM inosine (Fig. 4), cortex degradation and swelling of the spore core were apparent in only 50% of the gerN mutant spores, which appeared as an asynchronous mixture of spores at different stages of germination.
FIG. 4.
(A) Electron micrograph of B. cereus gerN1 mutant spores after germination in 5 mM inosine and 10 mM NaCl for 90 min. (B) Electron micrograph of B. cereus gerN1 mutant spores after germination and outgrowth in NB for 210 min. Bar, 1 μm.
Some germination-associated events had occurred in most of the spores, as 90% had turned phase dark and the majority of ion movements were complete, although OD loss, DPA release, and loss of heat resistance were not complete, as discussed earlier for inosine germination. Some spore changes, not visible by electron microscopy, had occurred in the spores that were phase dark; this would suggest that phase darkening of some spores may have preceded the loss of heat resistance, which is a very early event in normal spore germination.
When germination and outgrowth experiments were carried out in NB, germination of the mutant spores was very slow and asynchronous. Outgrowing cells did not appear until 3 h after the initiation of germination and were very elongated with irregular septa (Fig. 4). By 5 h the elongated cells had septated to give chains of 15 to 20 cells of normal cell length, and after 6 h 80% of the culture was made up of vegetative cells in chains of 4 to 15 cells.
This mutant outgrowth phenotype could be reversed by fully germinating the spores in Tris-HCl plus germinant (l-alanine [15 min] or inosine [3 h]) before addition of NB to stimulate outgrowth. As the septation defects are not apparent during vegetative growth or if the spores are fully germinated before outgrowth, it is apparent that these abnormalities are a consequence of the abnormal germination of the gerN mutant spores. Wild-type spores outgrown at low OD did not show the elongated or aseptate phenotype, and hence lack of competition in the asynchronously germinating mutant spore culture could not account for the mutant phenotype. It has been shown using germinated and outgrown spores of B. subtilis that when normal initiation of DNA replication is blocked, acentral septa will eventually form (8). As the normal germination sequence in the spore is perturbed, the onset of DNA replication may have been affected by a delay in a late germination event, such as the removal of small acid-soluble proteins (SASPs) from the DNA.
The gerN spore germination defect.
Disruption of the gerN gene of B. cereus 569 causes a major defect in inosine germination of the mutated spores. The normal germination process is blocked at an early stage, before loss of heat resistance; instead, there is a slow, residual germination response, which is asynchronous and grossly abnormal. As the gerN gene product is not required for germination in high concentrations of the alternative germinant l-alanine, an alternative protein must be able to carry out this or some analogous role. At limiting concentrations of l-alanine, the germination rate of mutant spores is slowed, suggesting GerN's involvement, at least under these conditions.
The germination defects of the gerN mutant of B. cereus are slightly different from those reported for the B. megaterium grmA null mutant (28). The mutant B. megaterium spores germinated poorly in any germinant (glucose, l-proline, l-leucine, or KNO3), suggesting that either the putative NaH antiporter encoded by grmA is required by all the germinant receptors in B. megaterium or, at least formally, there is a pleiotropic defect in the mutant spores that affects germination. Like the gerN mutant, the grmA spores appeared to be blocked at an early stage of germination, as they did not lose heat resistance or release DPA. As no detailed characterization of the germination of the mutant spores was presented, and as the duration of the germination experiment was not stated, we do not know whether there was a residual, slow, and aberrant response, as described here for B. cereus. In the case of gerN spore germination, the defect is specific to inosine-dependent germination; germination in l-alanine can be completed with near-normal kinetics, and the order of germination events is not perturbed. As the defect is germinant specific, it is likely that GerN has a specific role in germination; a pleiotropic defect in the mutant spores might be expected to affect the germination response to all, rather than some, organic germinants. Assay of the time of gerN expression will reveal whether its product is spore specific and therefore likely to have been recruited for a role exclusively in germination.
As GerN is a member of the CPA-2 family of sodium and potassium transporters, and is a homologue of NapA, a proven sodium-proton antiporter, it is likely that it, too, is an ion transporter of related function. It is coupled, at least functionally, to the inosine germination receptor of B. cereus. It may be that all germinant receptors require the function of a separate ion transport protein. In this model the l-alanine receptor is associated with a different ion antiporter and may be driven by a different monovalent cation, for example, K+, as l-alanine-initiated germination is not inhibited by disruption of the gerN gene. As expected by this model, calcium DPA-induced germination, which does not involve a nutrient germinant receptor complex (19), did not require the gerN gene to be intact.
Several models for germination would invoke such a requirement for an ion transporter. For example, the ion transporter could be activated to induce the local transfer of a small number of ions, changing local properties in the membrane, that could be propagated across its surface—during germination, the membrane changes rapidly from a semicrystalline to fluid state (23). Alternatively, the activity of a coupled ion transporter might be required to restore ion or charge balance if the germinant association with the receptor itself involved linked movement of the germinant with or counter to an ion species. Such models would implicate local, small-scale ion movements in an early stage of the germination response, prior to the bulk movement of ions that we could measure experimentally; movement of all the bulk ions, not only Na+, was blocked.
In response to a stronger germinant stimulus, more ions could be released, causing a greater germination signal. At low germinant concentrations the stimulus would be weak. Barlass (3) has shown that the gerL operon of B. cereus encodes the major l-alanine receptor system but that a minor contribution to the germination rate in l-alanine, particularly at limiting conditions, is provided by the inosine (GerI) germinant receptor. Hence, germination in suboptimal concentrations of l-alanine may show the germination defect, as optimal response in all the pathways is required at low germinant concentrations.
Not all sporulating bacteria contain a close grmA/gerN homologue. Although a GrmA protein appears essential for germination in B. megaterium, in B. cereus some germination receptor activity is not dependent on GerN. The molecular events associated with the GerN protein during germination remain to be established. Although there are multiple operons of the germination receptor (gerA) family in B. cereus (P. J. Barlass, M. O. Clements, C. W. Houston, and A. Moir, unpublished data) and B. anthracis (http://www.tigr.org), there is no evidence for multiple gerN-like genes in B. cereus (from our PCR screening) or in B. anthracis, from the database of incomplete sequences. There are however, candidates for transporters of monovalent cations in the genomes of these species, and presumably some of these are involved in bulk ion movements in germinating and vegetative bacilli. Spore germination in B. subtilis, for example, has a different ion specificity; germination is stimulated by K+, whereas this ion inhibits inosine germination in B. cereus. Mutations in the distant homologues of gerN in B. subtilis, yhaU and yjbQ, do not affect germination under standard germination conditions in either l-alanine or the asparagine-glucose-fructose combination, with either Na+ or K+ as the stimulating ion (T. W. Southworth and A. Moir, unpublished data).
ACKNOWLEDGMENTS
This work was funded by a BBSRC grant to A.M., BBSRC studentships to P.D.T. and T.W.S., and a postgraduate studentship to J.B. from the Ministry of Health and Medical Education of Iran.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakker E P, Booth I R, Dinnbier U, Epstein W, Gajewska A. Evidence for multiple K+ export systems in Escherichia coli. J Bacteriol. 1987;169:3743–3749. doi: 10.1128/jb.169.8.3743-3749.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlass P J. Ph.D. thesis. Sheffield, United Kingdom: University of Sheffield; 1998. [Google Scholar]
- 4.Behravan J. Ph.D. thesis. Sheffield, United Kingdom: University of Sheffield; 1998. [Google Scholar]
- 5.Bone E J, Ellar D J. Transformation of Bacillus thuringiensis by electroporation. FEMS Lett. 1989;58:171–178. doi: 10.1016/0378-1097(89)90033-5. [DOI] [PubMed] [Google Scholar]
- 6.Clements M O, Moir A. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J Bacteriol. 1998;180:6729–6735. doi: 10.1128/jb.180.24.6729-6735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harry E J, Rodwell J, Wake R G. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 9.Janssen F W, Lund A J, Anderson L E. Colorimetric assay for dipicolinic acid in bacterial spores. Science. 1958;127:26–27. doi: 10.1126/science.127.3288.26. [DOI] [PubMed] [Google Scholar]
- 10.Keynan A, Halvorson H O. Calcium dipicolinic acid-induced germination of Bacillus cereus spores. J Bacteriol. 1962;83:100–105. doi: 10.1128/jb.83.1.100-105.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 12.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 13.Martinussen J, Hammer K. Cloning and characterisation of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J Bacteriol. 1994;176:6457–6463. doi: 10.1128/jb.176.21.6457-6463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 15.Moir A, Kemp E H, Robinson C, Corfe B M. The genetic analysis of bacterial spore germination. J Appl Bacteriol. 1994;76(Suppl.):S9–S16. [PubMed] [Google Scholar]
- 16.Munro A W, Ritchie G Y, Lamb A J, Douglas R M, Booth I R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium efflux system KefC of Escherichia coli. Mol Microbiol. 1991;5:607–616. doi: 10.1111/j.1365-2958.1991.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 17.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innes M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 222–227. [Google Scholar]
- 18.Paidhungat M, Setlow P. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J Bacteriol. 1999;181:3341–3350. doi: 10.1128/jb.181.11.3341-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paidhungat M, Setlow P. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol. 2000;182:2513–2519. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saier M H, Eng B H, Fard S, Garg J, Haggerty D A, Hutchinson W J, Jack D L, Lai E C, Liu H J, Nusinew D P, Omar A M, Pao S S, Paulsen I T, Quan J A, Sliwinski M, Tseng T-T, Wachi S, Young G B. Phylogenetic characterisation of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 21.Staden R. Finding protein coding regions in genomic sequences. Methods Enzymol. 1990;183:163–180. doi: 10.1016/0076-6879(90)83012-x. [DOI] [PubMed] [Google Scholar]
- 22.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart G S A B, Eaton M W, Johnstone K, Barrett M D, Ellar D J. An investigation of membrane fluidity changes during sporulation and germination of Bacillus megaterium KM measured by electron spin and nuclear magnetic resonance spectroscopy. Biochim Biophys Acta. 1980;600:270–290. doi: 10.1016/0005-2736(80)90432-0. [DOI] [PubMed] [Google Scholar]
- 24.Stewart G S A B, Johnstone K, Hagelberg E, Ellar D J. Commitment of bacterial spores to germinate—a measure of the trigger reaction. Biochem J. 1981;198:101–106. doi: 10.1042/bj1980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 26.Strausak D, Waser M, Solioz M. Functional expression of the Enterococcus hirae NaH-antiporter in Escherichia coli. J Biol Chem. 1993;268:26334–26337. [PubMed] [Google Scholar]
- 27.Swerdlow B M, Setlow B, Setlow P. Levels of H+ and other monovalent cations in dormant and germinating spores of Bacillus megaterium. J Bacteriol. 1981;148:20–29. doi: 10.1128/jb.148.1.20-29.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tani K, Watanabe T, Matsuda H, Nasu M, Kondo M. Cloning and sequencing of the spore germination gene of Bacillus megaterium ATCC 12872: similarities to the NaH-antiporter gene of Enterococcus hirae. Microbiol Immunol. 1996;40:99–105. doi: 10.1111/j.1348-0421.1996.tb03323.x. [DOI] [PubMed] [Google Scholar]
- 29.Thackray P D. Ph.D. thesis. Sheffield, United Kingdom: University of Sheffield; 1999. [Google Scholar]
- 30.Tinoco I, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nature. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 31.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 32.Warren S C, Gould G W. Bacillus cereus spore germination: absolute requirement for an amino acid. Biochim Biophys Acta. 1968;170:341–350. doi: 10.1016/0304-4165(68)90014-7. [DOI] [PubMed] [Google Scholar]
- 33.Waser M, Hess-Bienz D, Davies K, Solioz M. Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J Biol Chem. 1992;267:5396–5400. [PubMed] [Google Scholar]
- 34.Zuberi A R, Moir A, Feavers I M. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene. 1987;51:1–11. doi: 10.1016/0378-1119(87)90468-9. [DOI] [PubMed] [Google Scholar]