SUMMARY
Dietary protein restriction (PR) has rapid effects on metabolism including improved glucose and lipid homeostasis, via multiple mechanisms. Here, we investigate responses of fecal microbiome, hepatic transcriptome, and hepatic metabolome to six diets with protein from 18% to 0% of energy in mice. PR alters fecal microbial composition, but metabolic effects are not transferable via fecal transplantation. Hepatic transcriptome and metabolome are significantly altered in diets with lower than 10% energy from protein. Changes upon PR correlate with calorie restriction but with a larger magnitude and specific changes in amino acid (AA) metabolism. PR increases steady-state aspartate, serine, and glutamate and decreases glucose and gluconeogenic intermediates. 13C6 glucose and glycerol tracing reveal increased fractional enrichment in aspartate, serine, and glutamate. Changes remain intact in hepatic ATF4 knockout mice. Together, this demonstrates an ATF4-independent shift in gluconeogenic substrate utilization toward specific AAs, with compensation from glycerol to promote a protein-sparing response.
Graphical abstract
In brief
MacArthur et al. use multiple unbiased methods to investigate metabolic and molecular responses across six levels of dietary protein restriction in mice. The authors show that many key hepatic responses occur below 10% protein energy and promote ATF4-independent rewiring of glucose metabolism to support serine, aspartate, and glutamate synthesis.
INTRODUCTION
Protein restriction (PR), also known as protein dilution, is defined as reduced protein/essential amino acid (EAA) intake without malnutrition or enforced energy restriction. PR is associated with a range of health benefits in animal models including improved glucose and lipid homeostasis (Laeger et al., 2014; Fontana et al., 2016; Maida et al., 2016; Treviño-Villarreal et al., 2018; MacArthur et al., 2021), increased resilience against acute stressors (Harputlugil et al., 2014; Robertson et al., 2015), reduced cancer incidence (Dunaif and Campbell 1987; Fontana et al., 2013; Levine et al., 2014), and increased longevity (Leto et al., 1976; Goodrick 1978; Yu et al., 1985; Solon-Biet et al., 2014). In humans, low protein intake is associated with reduced all-cause mortality and improved cardiovascular risk factor profiles, including improved glucose and lipid homeostasis (Fontana et al., 2016; Maida et al., 2016; Treviño-Villarreal et al., 2018). In most experimental systems, PR is achieved by isocalorically replacing protein with carbohydrate, hence increasing the dietary carbohydrate:protein ratio.
Benefits of PR overlap those of individual EAA restriction, including sulfur AA restriction (Orentreich et al., 1993; Orgeron et al., 2014), branched-chain AA restriction (Fontana et al., 2016), leucine restriction (Xiao et al., 2011), and tryptophan restriction (Ooka et al., 1988; Peng et al., 2012; Zapata et al., 2018), suggesting that restriction of any EAA may be sufficient to trigger health benefits without a requirement for reduced overall nitrogen intake. Benefits of PR also overlap those of calorie restriction (CR) but without enforcement of energy restriction and are thus distinct from energy restriction per se (Solon-Biet et al., 2015).
Because dietary protein is essential, beneficial effects of PR are dependent on several factors including magnitude and duration of restriction, developmental stage, and baseline metabolic state. For example, in sedentary adult mice, 1 week of a protein-free diet can impart metabolic benefits and stress resistance without irreversible side effects (Peng et al., 2012). On the other hand, early post-natal PR can limit growth, and even mild PR (8%–−9% of energy) during gestation in rodents can have detrimental consequences on offspring (Langley et al., 1994; Fernandez-Twinn et al., 2005).
Due to the potential risks of PR, an understanding of the molecular mechanisms underlying benefits could potentiate safe clinical translation and shed light on pathways targetable by pharmacological agents. While a number of studies have identified non-mutually exclusive mechanisms that can contribute to health benefits of PR, including changes in energy expenditure, energy intake, and adiposity, each of these mechanisms displays distinctly non-linear responses and interactions across a range of protein intakes, complicating causal inference regarding health benefits. For example, one study comparing PR at 8% versus 22% in rats attributed metabolic benefits to reduced feeding efficiency without a change in energy intake (Rothwell et al., 1983). However, a similar study across a range of protein concentrations (0%–15%) found increased energy expenditure and reduced energy intake at 0% and 5% versus the 15% control group; while at 10%, energy intake, but not expenditure, increased (Pezeshki et al., 2016). Similar non-linear effects are found with adiposity. Protein or AA restriction up to a certain threshold tends to increase total energy intake (Stirling and Stock 1968; Tulp et al., 1979; Rothwell et al., 1983), but excessive restriction or total deprivation can result in food aversion (Kamata et al., 2014; Wu et al., 2021). In most cases of non-aversive restriction, this correlates with increased adiposity but not necessarily with poor metabolic outcome. For example, diets with carbohydrate:protein ratio increased to 5% of energy from protein increase lifespan in mice despite increased adiposity (Solon-Biet et al., 2014). Although food intake is increased in low-protein diets in an FGF21-dependent manner (Hill et al., 2019), severe restriction of protein below 5% or of individual AAs can result in food aversion despite increased circulating FGF21 (Kamata et al., 2014; Wu et al., 2021), potentially due to inhibition of hypothalamic mTOR signaling (Wu et al., 2021). Similarly, in humans, protein leverage acts to increase intake of low-protein food to meet a protein target while simultaneously improving metabolic health parameters but appears to function only above 10% protein (Gosby et al., 2011; Gosby et al., 2016; Maida et al., 2016). On the other hand, high-protein diets (>20%) can suppress food intake with concomitant weight loss but without the expected improvements in insulin sensitivity (Smith et al., 2016; Te Morenga et al., 2017).
Studies using targeted molecular approaches to understand the mechanistic underpinnings of PR-mediated health effects have uncovered important roles for the AA-deprivation-sensing pathway. This has been proposed to start with the activation of integrated stress response kinases GCN2 and PERK and signal through ATF4-responsive hormones such as FGF21 to produce metabolic effects (Guo and Cavener 2007; Laeger et al., 2014; Wanders et al., 2016; Hill et al., 2019; Patel et al., 2019). These studies have shown that FGF21 signaling is a key mediator of changes in food intake, body composition, and energy expenditure. However, the mechanisms underlying other metabolic benefits of PR, such as improved glucose and lipid homeostasis, remain unclear with uncertain results on both the roles and mechanisms of regulation of these important downstream mediators (Maida et al., 2016; Wanders et al., 2016; Cooke et al., 2020).
The limited number of studies that use unbiased approaches to understand the mechanisms of PR-mediated health effects through molecular changes in the transcriptome and metabolome point to adaptive protein-sparing pathways including reduced urea synthesis, increased biosynthesis of non-EAAs (NEAAs), and perturbations in fat metabolism. For example, rats fed 6% versus 24% casein for 7 to 10 days (Kalhan et al., 2011) display increased levels of serine and glycine, reduced sulfur-containing AAs in multiple tissues, and gene-expression changes consistent with reduced fatty acid synthesis and increased fatty acid oxidation. However, it is still unknown how these marker profiles respond across a range of protein intakes and if they are causally related to PR-mediated health effects.
The host microbiome also rapidly responds to changes in diet (Carmody et al., 2015), including caloric restriction (Tanca et al., 2018), protein nitrogen (Holmes et al., 2017), and individual EAA restriction (Zapata et al., 2018) and can causally contribute to metabolic improvements resulting from dietary interventions including CR (Fabbiano et al., 2018). In the context of PR, although antibiotic ablation of the host microbiome has little effect on the metabolic adaptation to a 7% versus 22% protein diet (Pak et al., 2019), whether functional benefits of 7 days of PR can be transferred by fecal microbiota transplantation to diet-naive mice remains untested.
The physiologic response to long-term (>2 months) low-protein-diet feeding in mice has been thoroughly characterized (Solon-Biet et al., 2014; Solon-Biet et al., 2015; Maida et al., 2016; Pak et al., 2019). These results show that diets with increased carbohydrate:protein ratio to ~5%–6% of energy from protein cause improved glucose and lipid homeostasis and can increase longevity. Here, we show that most of these metabolic changes are attained to the same degree as long-term PR within 7 days. As such, we used this short-term model to investigate the mechanisms underlying metabolic improvements.
We report that short-term PR with increased carbohydrate:protein ratio results in dose-dependent host transcriptomic and metabolomic signatures that are associated with improvements in glucose and lipid homeostasis, with a comparatively limited role for the microbiome. These PR signatures correlated with signatures of CR but were of greater magnitude and included changes in AAs and redox homeostasis not seen in CR, including increased steady-state levels of specific AAs including aspartate, serine, and glutamate and the glutathione analog tripeptide ophthalmic acid. Tracing experiments with 13C-labeled glucose and glycerol revealed increased glucose incorporation into these specific AAs and increased glycerol usage as a gluconeogenic substrate. Changes in metabolic parameters and hepatic gene expression were independent of hepatic ATF4. Together, these data suggest that 7 days of low-protein diet cause altered hepatic glucose utilization to spare protein/AAs and protect against oxidative stress and that these mechanisms may contribute to health benefits of PR.
RESULTS
Titration of dietary protein results in linear and non-linear phenotypic responses
Sixteen-week-old B6D2F1 male mice were fed ad libitum for 7 days with semi-purified diets with protein content varying from 18% (control) down to 0% of energy (Figure 1). Dietary protein was provided using purified AAs with a profile mimicking casein. All diets were isocaloric with sucrose replacing protein (Table S1).
Figure 1. Phenotypic effects of dietary amino acid titration.
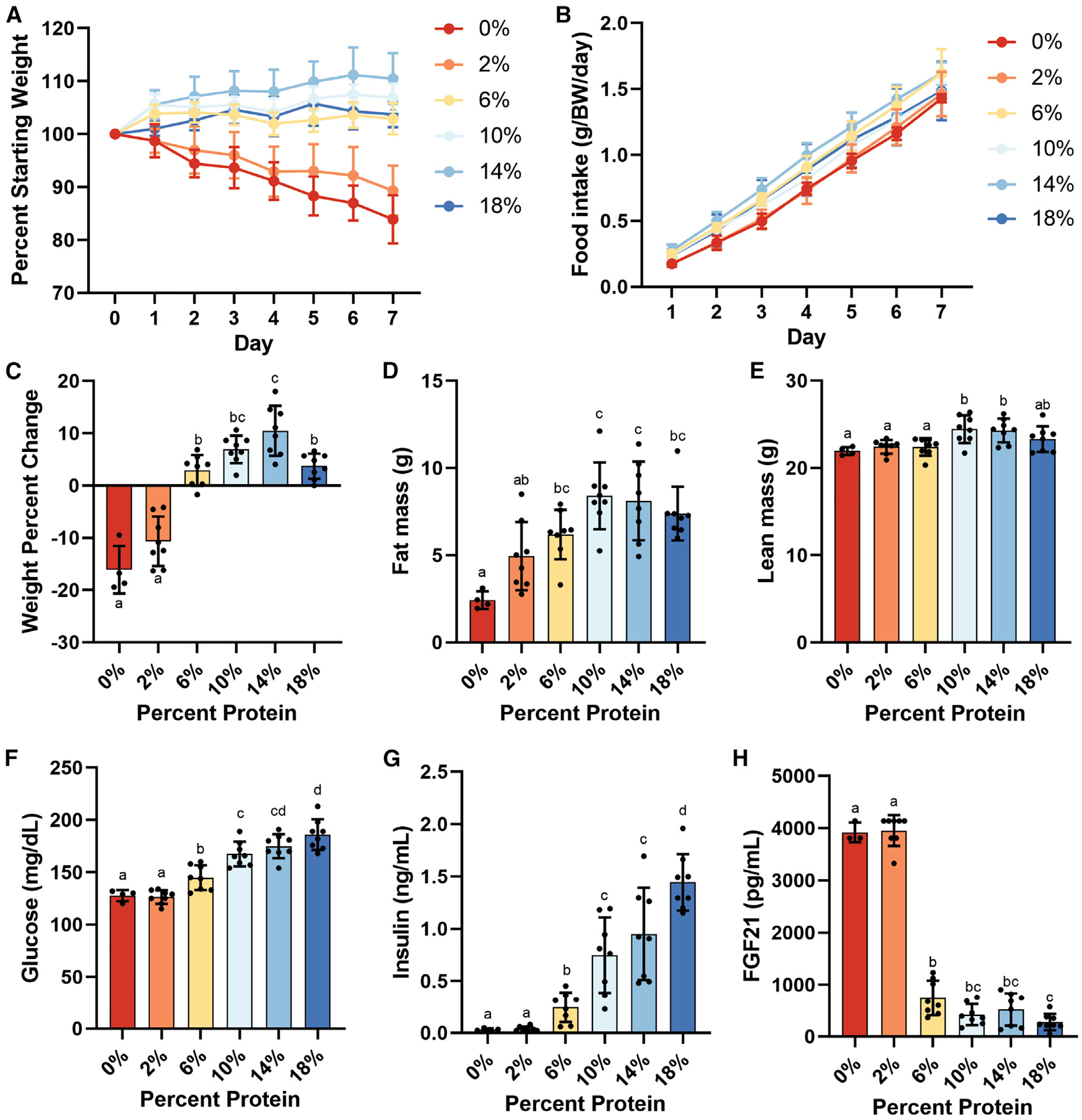
Adult male B6D2F1 mice (4/group) were fed ad libitum for 1 week on an isocaloric semi-purified diet of the indicated protein calorie percentage. (A) Body weight change expressed as percentage of starting weight.
(B) Cumulative food intake normalized to body weight.
(C–E) Body weight percentage change (C), fat mass (D), and lean mass (E) after 1 week on the indicated diet.
(F–H) Four-hour-fasted values for (F) blood glucose, (G) serum insulin, and (H) FGF21.
Error bars are standard deviations, different letters indicate p < 0.05 in Tukey post-hoc test following significant one-way ANOVA F test. See also Figure S1 and Table S1.
Titration of protein produced a non-linear response in body mass with 0% and 2% groups losing body mass, 10% and 14% groups tending to gain body mass, and the 6% group remaining unchanged versus the 18% control group (Figures 1A and 1C). The 0% and 2% groups also consumed significantly less food, and the 14% group consumed significantly more food, than the 18% control group on a gram-per-mouse basis (Figures 1B and S1C). Reduced food intake in the 0% and 2% groups was largely explained by a transient decrease during the first 3 days (Figures S1A and S1B), and intake between groups was not significantly different when normalized to body weight (Figure S1D). Similar non-linear changes in fat mass and lean mass were observed with the 0% and 2% groups having significantly less lean mass, fat mass, and lower body fat percent versus the 18% group (Figures 1D, 1E, and S1E).
PR resulted in a linear decrease in fasting blood glucose and insulin (Figures 1F and 1G), non-linear increases in circulating FGF21 and adiponectin (Figures 1H and S1F), and non-linear decreases in triglycerides and cholesterol (Figure S1G and S1H).
PR alters the fecal microbiota, but these changes contribute minimally to metabolic phenotype
Dietary interventions including protein or EAA restriction can rapidly alter the fecal microbiome (Zapata et al., 2018; Pak et al., 2019), but whether this contributes to health outcomes remains unclear. To determine the potential contribution of the fecal microbiome to PR metabolic phenotypes, we performed fecal sampling on male 16-week-old B6D2F1 mice fed 18% (control), 6%, or 0% protein diets for 8 days. These protein levels were selected because of our observation of significant changes in glucose and lipid homeostasis that were coupled to either minimal (6%) or significant effects (0%) on body composition (Figure S2). Day 8 fecal communities were then transplanted into germ-free recipient mice.
Over 8 days, gut microbial communities of donors fed 0% and 6% protein grew distinct from controls in both within- (alpha) and between-sample (beta) diversity. PR led to increased Shannon alpha diversity at day 4 (p = 0.048) and trended at day 8 (p = 0.08; Figure S2A). PR also caused significant shifts in beta diversity (Figure 2A), with significant effects on non-metric multidimensional scaling dimension 1 (NMDS1) at days 4 and 8 (p = 0.048 and 0.007, respectively; Figure S2C). PERMANOVA analysis showed significant group differences at 8, but not 4, days (F = 3.48, p = 0.004). Similar trends were apparent in the recipient mice. Low-protein recipients had significantly higher Shannon alpha diversity at day 14 and trended higher at day 21 (p = 0.008 and 0.08, respectively; Figure S2B). Significant group discrimination along NMDS1 was observed at day 14 with a trend at day 7 (p = 0.035 and 0.087, respectively; Figures 2L and S2D). PERMANOVA analysis showed significant group differences in beta diversity at 14 and 19 days after inoculation (p < 0.001, F = 3.38 and p < 0.001, F = 3.20, respectively). A trend toward increased relative abundance of Firmicutes in low-protein donors on day 4 (Figures 2B and S2E) was also observed in low-protein recipients at days 7 and 14, with significance reached at day 19 (Figures 2M and S2F). Other changes that were not consistent between the donors and recipients included a significant decrease in Proteobacteria in low-protein donors (Figures 2B and S2E) and significant decreases in Verrucomicrobia in the lowest protein recipients at days 14 and 19 (Figures 2M and S2F). PERMANOVA analysis showed significant group differences in beta diversity at 14 and 19 days after inoculation (p < 0.001, F = 3.38 and p < 0.001, F = 3.20, respectively). While cohoused recipients (2/cage) exhibited cage effects due to inoculation from a common donor, coprophagy, and environmental influences, ADONIS analysis validated that these effects were dependent on the original donor diet through to 19 days post-inoculation (p = 0.001, F = 7.08).
Figure 2. Effects of dietary protein titration on fecal microbiome composition.
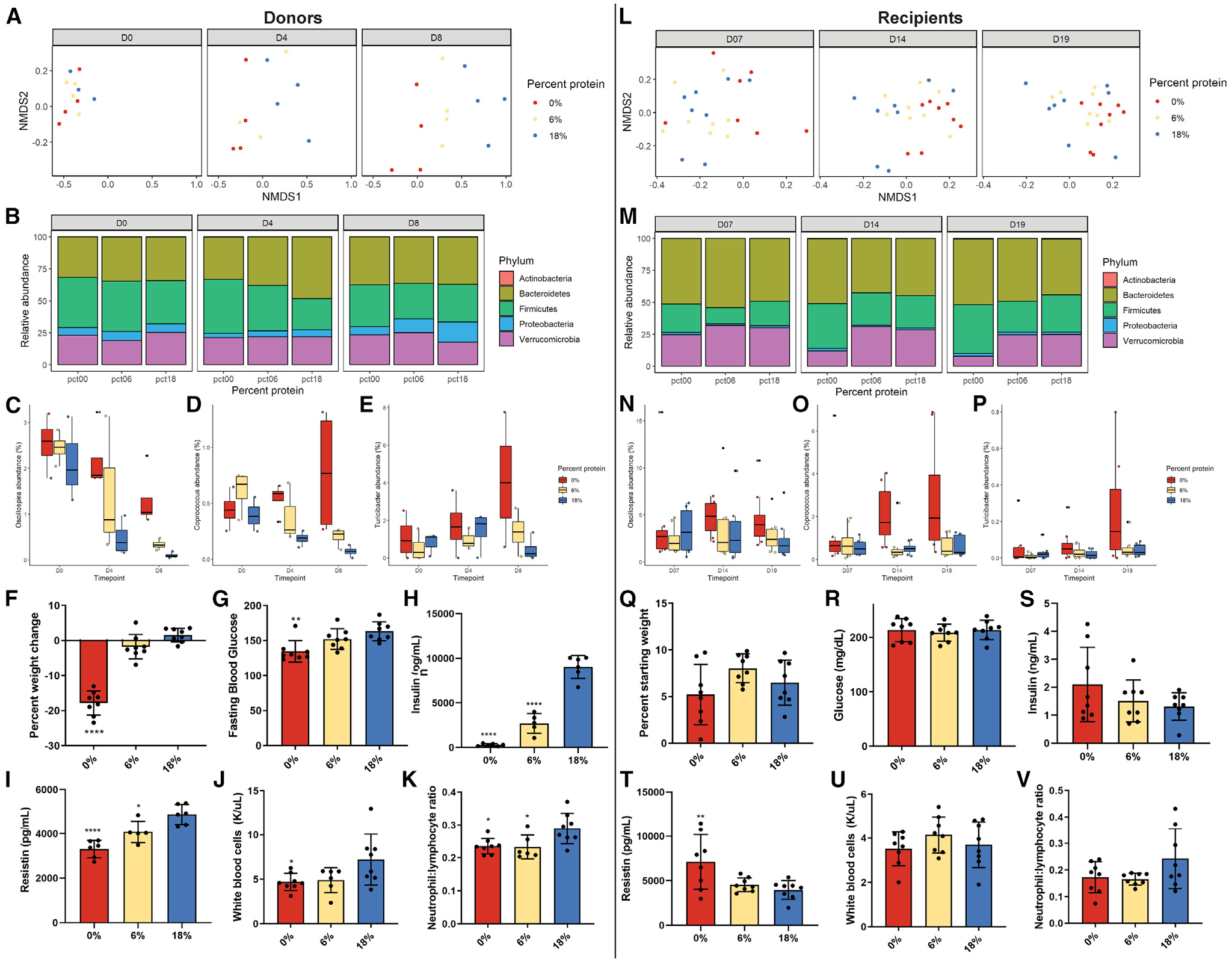
(A and L) NMDS plots after (A) 0, 4, and 8 days of diet in donor mice and (L) 7, 14, and 19 days after inoculation of gnotobiotic recipient mice.
(B and M) Relative abundance of the 5 most abundant phyla in fecal samples from (B) donor mice and (M) recipient mice across the time courses.
(C–E and N–P) Relative abundances of the genera Oscillospira, Coprococcus, and Turicibacter across the time course in (C–E) donor and (N–P) recipient mice. (F–K) Percentage of weight change, fasting blood glucose, insulin, resistin, white blood cell count, and neutrophil:lymphocyte ratio in donor mice after 8 days of diet.
(Q–V) the same parameters as (F)–(K) in recipient mice 19 days after inoculation.
*p < 0.05, **p < 0.01, ***p < 0.001 Dunnett’s post-hoc test versus 18% control group following significant one-way ANOVA F test. Error bars are standard deviations.
We performed differential abundance analysis to identify bacterial taxa associated with the 0% and 6% versus 18% groups for donors and recipients. In donors, amplicon sequence variants (ASVs) mapping to genera Turicibacter and Oscillospira were associated with the 0% group (Figures 2C, 2E, and S2G), while in recipients, ASVs mapping to genera Coprococcus and Oscillospira were associated with the 0% group. In donors, single ASVs from each Allobaculum and Sutterella were significantly associated with the 18% group (Figure S2G), while in the recipients, only Akkermansia was significantly associated with the 18% group (Figure S2H). In donors, no ASVs were significantly associated with the 6% group (Figure S2I and S2J).
One week of PR in donor mice caused decreased body mass, fasting blood glucose, fasting insulin, and resistin (Figures 2F–2I). The low-protein diets also tended to reduce white blood cell count and decrease circulating neutrophil:lymphocyte ratio (Figures 2J and 2K). In contrast, 19 days after inoculation, the recipient mice did not have any significant differences in body weight, fasting glucose, or insulin (Figures 2Q–2S). Unlike donors, the recipients that received fecal material from the 0% donors had significantly higher resistin levels versus control (Figures 2T). No differences were observed in circulating white blood cell count, but the neutrophil:lymphocyte ratio trended to be lower in the low-protein recipients (Figures 2U and 2 V).
Dietary protein titration produces linear and non-linear responses in hepatic transcriptome
To explore host-mediated mechanisms, we looked at the response of the hepatic transcriptome to 1 week of dietary protein titration.
Multidimensional scaling distances revealed relatively few differences in hepatic transcriptomes among 18%, 14%, and 10% groups (Figure 3A), with no more than 105 differentially expressed (DE) genes in either the 14% or 10% group compared with the 18% group (Figure S3A). There were increasing numbers of DE genes in the 6%, 2%, and 0% groups compared with the 18% group (Figure 3B), with each having over 1,000 DE genes (Figure S3A).
Figure 3. Effects of dietary protein titration on hepatic transcriptome.
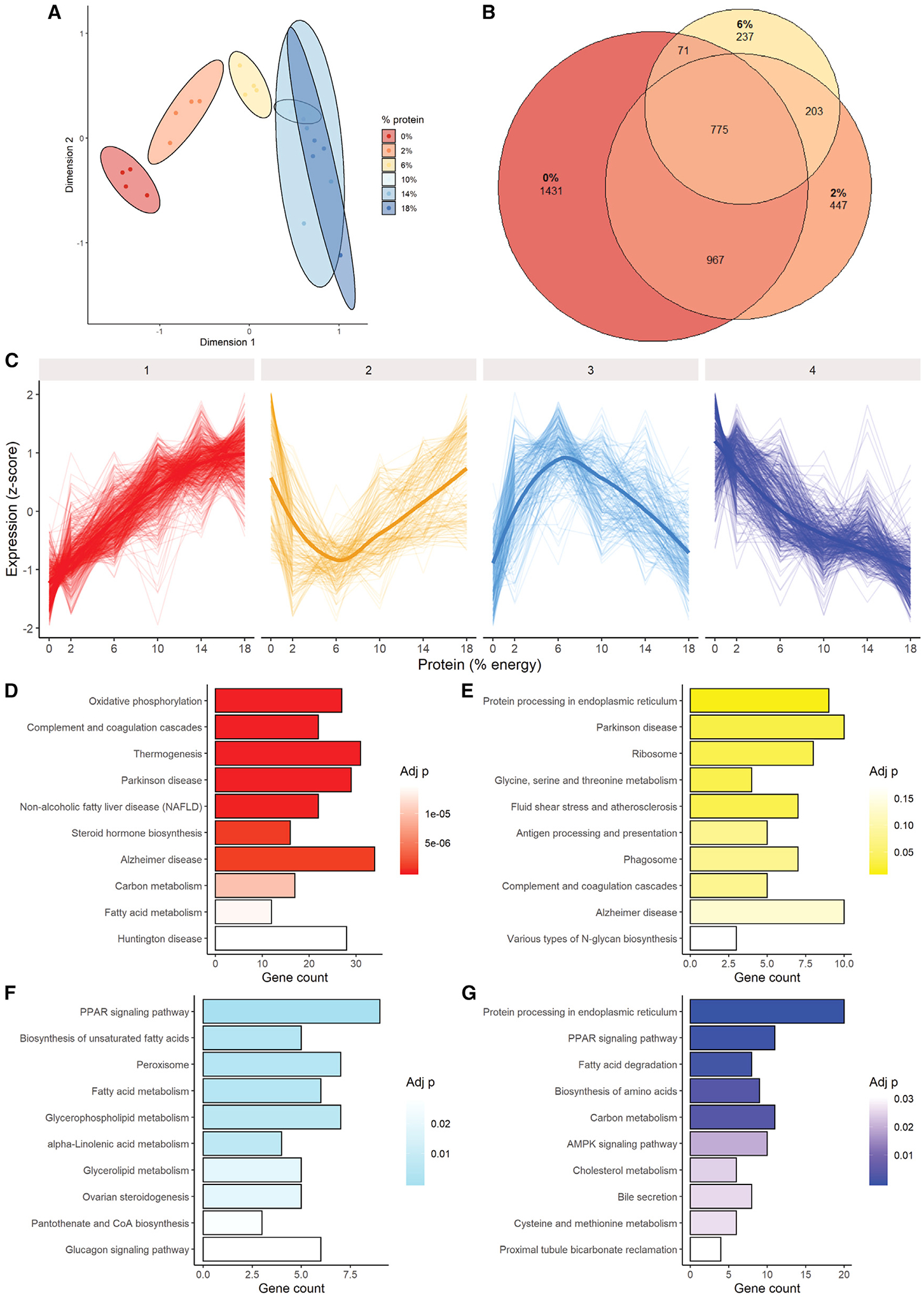
Adult male B6D2F1 mice (4/group) were fed ad libitum for 1 week on an isocaloric semi-purified diet of the indicated protein calorie percentage, and liver transcriptomes were analyzed.
(A) Multidimensional scaling distances.
(B) Venn diagram of significantly differentially expressed genes in 0%, 2%, and 6% protein groups versus 18% control.
(C) Clustering analysis separated into four major cluster patterns.
(D–G) Pathway enrichment in the 4 major clusters identified in (C).
See also Figure S3.
To understand the biological relevance of these DEs, a clustering analysis was performed. Four major clusters were identified as a function of decreasing dietary protein content: linear decreasing, non-linear decreasing, non-linear increasing, and linear increasing genes (Figure 3C, clusters 1–4, respectively). The major signature in the linear-decreasing cluster was related to energy metabolism, including oxidative phosphorylation and thermogenesis, in addition to complement and coagulation cascade (Figure 3D). In the non-linear-decreasing cluster, which tended to decrease until 6% and increase again at 2% and 0%, the top enriched pathways were related to protein and AA homeostasis, including protein processing in endoplasmic reticulum, ribosome and glycine, and serine and threonine metabolism (Figure 3E). In the non-linear-increasing cluster, the dominant signature related to lipid metabolism, including PPAR signaling pathway, biosynthesis of unsaturated fatty acids, peroxisome, and fatty acid metabolism (Figure 3F). In the linear-increasing cluster, both lipid and protein/AA metabolism were enriched, with protein processing in endoplasmic reticulum, PPAR signaling pathway, fatty acid degradation, and biosynthesis of AAs as the top pathways hits (Figure 3G). Similar trends were apparent when assessing pathway enrichment in up- and downregulated gene sets versus18% protein control (Figure S3B).
Dietary protein titration below 10% produces distinct effects on hepatic metabolome
An assessment of the polar metabolome across the 0%–18% range of protein intakes revealed similar trends, with the three highest protein diets grouping together and the three lowest groups diverging (Figure 4A). Although some changes were evident within the three highest (data not shown) and three lowest groups (Figure S4), the main changes were observed between the high and low groups, which formed the first branch in k-means clustering (Figure 4B). The major signature among the 6%, 2%, and 0% metabolomes was related to AA metabolism and included D-glutamine and D-glutamate metabolism, tyrosine metabolism, alanine, aspartate and glutamate metabolism, valine leucine and isoleucine biosynthesis, histidine metabolism, arginine and proline metabolism, and aminoacyl-tRNA biosynthesis.
Figure 4. Effects of dietary protein titration on hepatic metabolome.
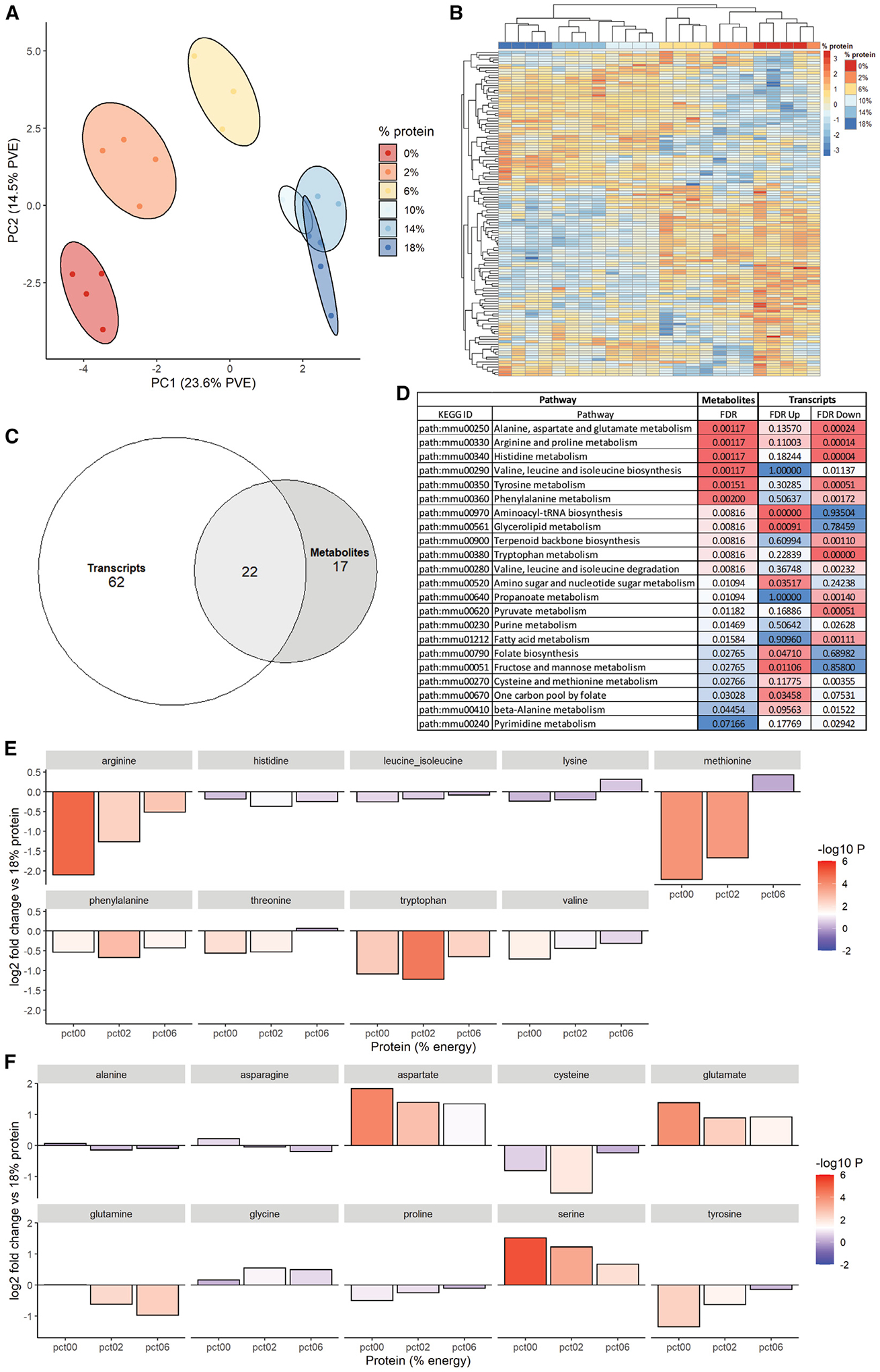
(A and B) Principal component analysis (A) and unbiased k-means clustering (B) of hepatic polar metabolites from the indicated diet groups.
(C and D) Pathway enrichment analysis (C) encompassing both hepatic metabolite and transcript datasets comparing high- (18%, 14%, and 10%) versus low- (6%, 2%, and 0%) protein groups represented as a Venn diagram and (D) showing the 22 commonly altered pathways in the overlap.
(E and F) Log2 fold changes in (E) essential and (F) non-essential amino acids in livers of the low protein groups (0%, 2%, and 6%) versus 18% control group.
See also Figure S4.
We next performed pathway enrichment analysis between the high (18%, 14%, and 10%) and low (6%, 2%, and 0%) groups for both metabolite and transcript datasets and compared the overlap. Between the transcripts and metabolites, 22 common pathways were differentially enriched between the high- and low-protein groups (Figure 4C). Among these, the top pathways were related to AA metabolism including alanine, aspartate, and glutamate metabolism, arginine and proline metabolism, and histidine metabolism (Figure 4D). These pathways tended to decrease on the transcript level in the low-protein group, with some notable exceptions such as aminoacyl-tRNA biosynthesis. To determine the directionality in the metabolite group, we specifically looked at changes in EAAs (Figure 4E) and NEAAs (Figure 4F) in the low-protein groups versus the 18% group. In the EAA group, all AAs tended to decrease, with arginine, methionine, phenylalanine, threonine, tryptophan, and valine reaching significance (Figure 4E). In the NEAA group, cysteine, glutamine, and tyrosine decreased, while aspartate, glutamate, and serine increased significantly at 6%, 2%, and 0% (Figure 4F).
PR induces similar effects on hepatic transcriptome and metabolome as CR but with greater magnitudes of change and specific AA-related signatures
Due to the non-linear effects of PR on food intake combined with the potential of self-imposed CR to contribute to metabolic benefits, we analyzed hepatic omics upon 7 days of ad libitum (AL) feeding versus 40% CR at 18% and 0% protein. Both PR and CR resulted in significant reductions in body weight, though PR groups lost significantly more (Figures 5A and S5A). As mentioned previously, the PR group self-imposed food restriction during the first 4 days of feeding, resulting in ~20% reduction in calorie intake over 7 days (Figures 5B and S5B). Both CR and PR significantly reduced body fat percentage and lean mass versus AL control and tended to reduce fat mass (Figures 5C, S5C, and S5D). CR and PR also both significantly reduced fasting blood glucose and insulin versus AL control (Figures 5D and 5E). However, only PR significantly reduced triglycerides and cholesterol (Figure S5E and S5F) and significantly increased FGF21 versus AL control (Figure 5F).
Figure 5. Phenotypic effects of dietary protein and calorie restriction.
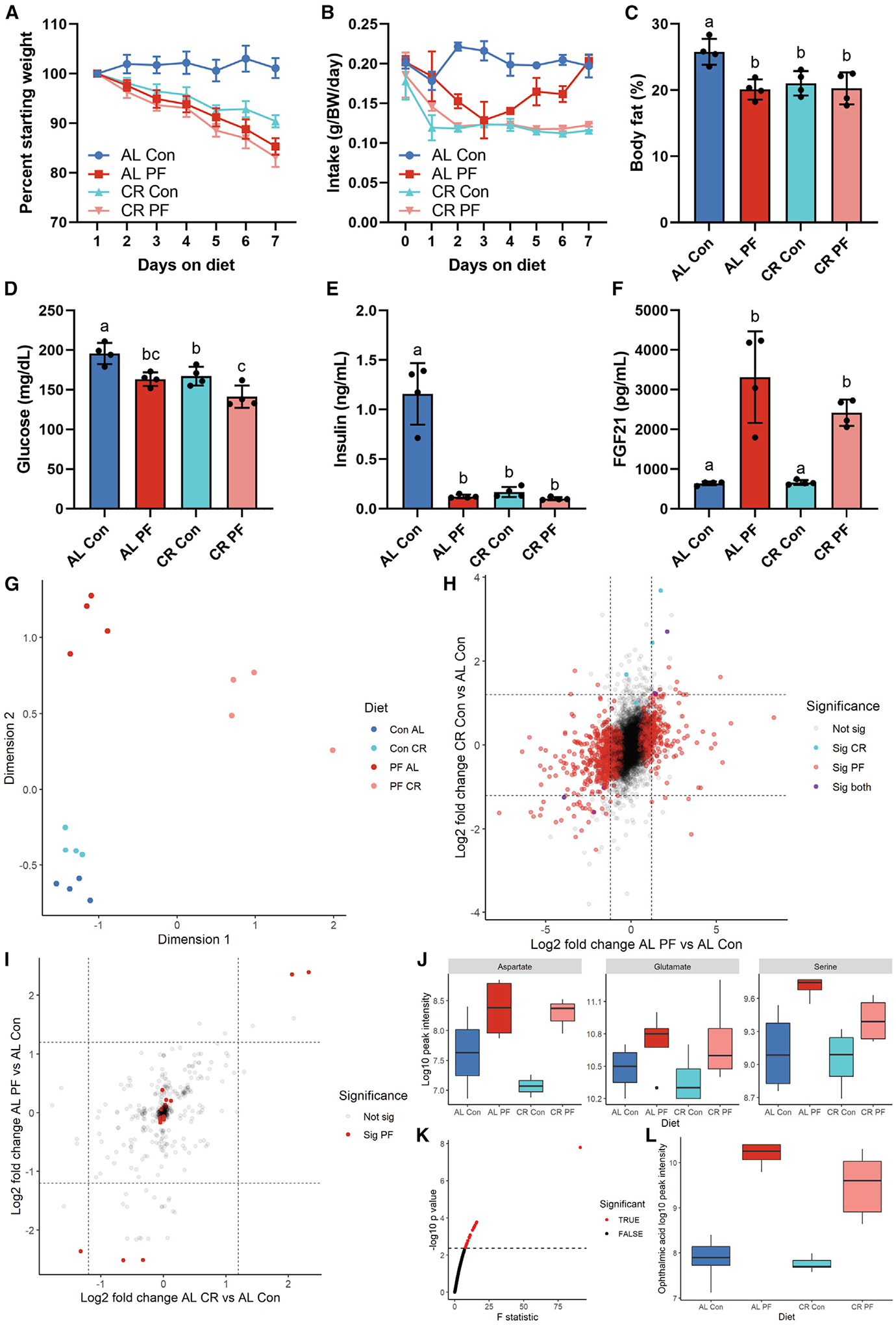
Adult male B6D2F1 mice (n = 4/group) were fed ad libitum (AL) or 40% calorie restricted (CR) on a control 18% protein (Con) or 0% protein (PF) diet for one week.
(A) Daily body weight expressed as percentage of starting weight.
(B) Daily food intake normalized to body weight.
(C–F) Body fat percentage (C), blood glucose (D), serum insulin (E), and FGF21 (F) in the fasted state after 1 week on diet.
(G) MDS plot of hepatic transcriptome data.
(H) Scatterplot of log2 fold changes of hepatic transcripts versus AL Con of AL PF (x axis) and CR Con (y axis).
(I) Scatterplot of log2 fold changes in hepatic metabolites versus AL Con of AL PF (x axis) and CR Con (y axis).
(J) Boxplots of non-essential amino acids aspartate, glutamate, and serine across diets.
(K) One way ANOVA F-statistic values for hepatic metabolites; red indicates FDR-adjusted p <0.1.
(L) Boxplots of ophthalmic acid, the metabolite with the largest ANOVA F statistic, across diets. Error bars are standard deviations; different letters indicate p < 0.05 in Tukey post-hoc test following significant one-way ANOVA F test.
See also Figure S5.
While PR produced robust global changes in the hepatic transcriptome, CR effects were comparatively limited (Figure 5G). Combining CR and PR produced some specific changes but largely mirrored the effects of PR alone (Figure 5G). Changes upon CR showed moderate positive correlation with the changes upon PR (r = 0.4, p < 0.001; Figure 5H). The changes upon protein deprivation with CR were strongly positively correlated with the changes upon PR alone (r = 0.76, p < 0.001; Figure S5G) but only moderately positively correlated with the changes upon CR alone (r = 0.36, p < 0.001; Figure S5H). Despite correlation between CR and PR, pathway overrepresentation tests of significantly up- and downregulated genes upon CR versus control diet were lacking the changes in gene sets related to AA metabolism (Figure S5K and S5L) observed in both PR groups (Figure S5I, S5J, S5M, and S5N).
Similar trends were observed when comparing hepatic polar metabolite profiles between PR versus CR. Similar global changes versus control were evident for both CR and PR (r = 0.5, p < 0.001; Figure 5I). However, changes in specific NEAAs upon PR were not evident in the CR condition. Specifically, while aspartate, glutamate, and serine tended to increase upon PR with or without CR, in CR alone, they remained unchanged (Figure 5J). When looking across all metabolites, there were 25 metabolites that had a significant false discovery rate (FDR)-corrected ANOVA p value (Figure 5K). Of these, the glutathione-like tripeptide ophthalmic acid (OPA) stood out, with an F statistic nearly triple the next closest metabolite. OPA increased upon PR but not CR (Figure 5L).
PR increases synthesis of aspartate, serine, and glutamate from glucose
Due to the robust decrease in blood glucose levels despite isocaloric replacement of dietary protein with sucrose (Figure 1F), we next tested the possibility of increased glucose utilization for biosynthesis of specific NEAA using stable isotope tracing experiments. To trace glucose-derived carbon into aspartate, glutamate, and serine in the liver, mice fed 18% or 0% protein diets for 7 days were injected intravenously with a bolus of 13C6 glucose (Figure 6A). After 30 min, mice were euthanized, and polar metabolites were extracted from the liver. Total metabolite pool sizes (labeled and unlabeled; Figures 6B and 6C) confirmed significant increases in aspartate, serine, and glutamate as well as reductions in glucose, glucose-6P, DHAP, and glyceraldehyde-3P in the PR group. In addition to the increased total pool size in PR, there was a significant increase in tracing into all observed species of aspartate (Figure 6D), into the m+1 species of serine (Figure 6E), and into the m+1, +2, and +3 species of glutamate (Figure 6F). No significant differences in the proportions of labeled glucose were observed between diet groups, with m+6 being the predominant species, consistent with similar label uptake independent of diet (Figure 6G).
Figure 6. Effects of dietary protein restriction on hepatic glucose and glycerol flux.
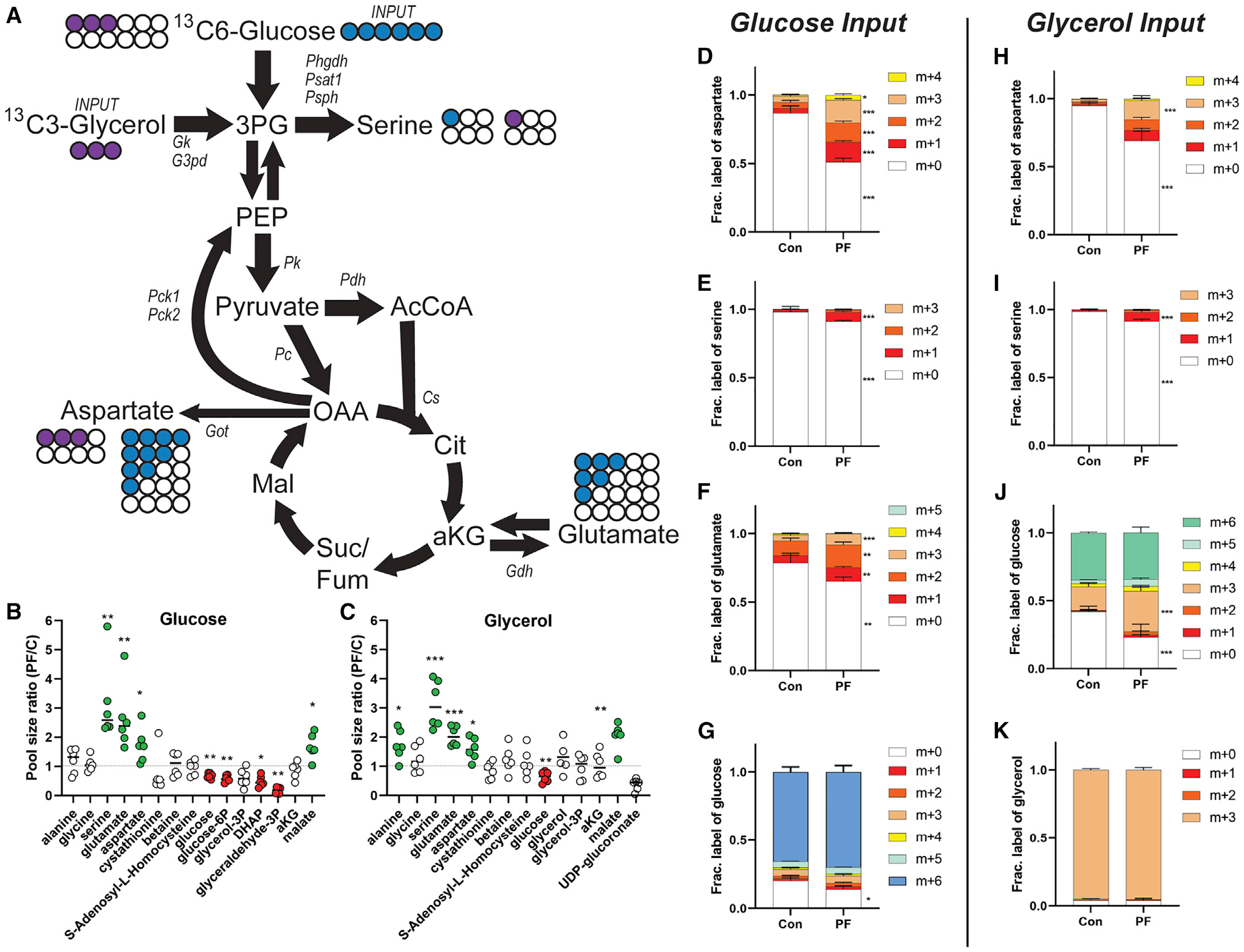
(A) Diagram showing unlabeled (open circle) and labeled carbon from glucose (blue) or glycerol (purple) in fractionally enriched glycolytic/gluconeogenic and citric acid cycle metabolites relevant to amino acid metabolism.
(B and C) Total pool sizes (labeled plus unlabeled) of the indicated metabolite from the glucose (B) or glycerol (C) labeling experiment.
(D–K) Mice subject edto 1 week on 18% protein (Con) or 0% protein (PF) diet and injected with a bolus of 13C6 glucose (D–G) or 13C3 glycerol (H–K) and polar metabolites extracted from the liver after 30 min. Fractional enrichment of heavy carbon derived from 13C6 glucose or 13C3 glycerol incorporated into aspartate (D and H), serine (E and I), glutamate (F), or glucose (J) are indicated. (G and K) Tracer enrichment of 13C6 glucose (G) or 13C3 glycerol (K) in liver.
*p < 0.05, **p < 0.01, ***p < 0.001 between diet groups using FDR-corrected Student’s t test. Error bars are standard deviations, n = 6 per group.
To test for cell autonomy of these relative increases in specific NEAA synthesis in response to reduced AA availability, we performed stable isotope tracing experiments using 13C6 glucose in cultured HEPA1–6 cells in either control or AA-deficient media for 6 h (Figure S6A). Similar to the in vivo result, tracing of glucose into aspartate and serine was significantly increased in the AA-deficient conditions (Figures S6B and S6C). However, the labeling patterns differed between the in vivo and in vitro conditions, with m+2 showing the greatest increase in aspartate upon AA deprivation and m+3 showing the greatest increase in serine. Tracing of glucose into glutamate was not increased in vitro (Figure S6D). Tracing of glucose into phosphoenolpyruvate was increased in the m+2 species (Figure S6E), into oxaloacetate in the m+3 and +4 species (Figure S6F), and into 3-phophoglycerate in the m+2 and +3 species (Figure S6G) upon AA deprivation.
Because we saw robust changes in transcriptional pathways related to lipid homeostasis and observed robust changes in circulating lipids, we also tested the non-mutually exclusive hypothesis that carbon from glycerol could be the substrate supporting increased synthesis of specific NEAAs. To test this, we performed glycerol tracing in vivo as above. Similar to the glucose study, there were no significant differences in the proportions of labeled glycerol between diet groups, consistent with similar label uptake, with m+3 being the predominant species (Figure 6K). Pool sizes of serine, glutamate, and aspartate were significantly increased in the livers of the 0% protein group (Figure 6C). Glycerol carbons were fractionally increased in aspartate (Figure 6H) and serine (Figure 6I), which could be explained by the observation that glycerol carbons were also fractionally enriched in glucose (Figure 6J). Interestingly, this fractional enrichment of glycerol in glucose occurred despite a significant reduction in overall pool size of glucose (and glucose-6P) upon PR (Figures 6B and 6C), suggesting altered gluconeogenic substrate utilization upon PR.
Hepatic ATF4 is not required for metabolic or transcriptional changes upon PR
Because the transcription factor ATF4 regulates many of the genes related to the transport and biosynthesis of serine, glutamate, and aspartate, we tested whether hepatic ATF4 was required for the observed transcriptional or metabolic changes. Mice harboring an ATF4 floxed allele were crossed with mice expressing albumin Cre to generate liver-specific ATF4 knockouts or wild-type littermate controls (lacking albumin Cre expression) and fed either a zero protein (0%), low-protein (6%), or control diet (18%) for 7 days.
There were no significant differences between the genotypes in body weight changes (Figure 7B) or food intake (Figure 7C). There was no significant genotype by diet interaction effects on adiposity, serum FGF21 level, fasting blood glucose, or fasting insulin (Figures 7E–7H). The key ATF4 target genes Asns, Psat1, and Fgf21 were still significantly upregulated in the liver of ATF4 liver-specific knockout (LKO) mice, suggesting redundant regulators (Figure 7I).
Figure 7. Phenotypic effects of dietary protein restriction in hepatic ATF4 knockout mice.
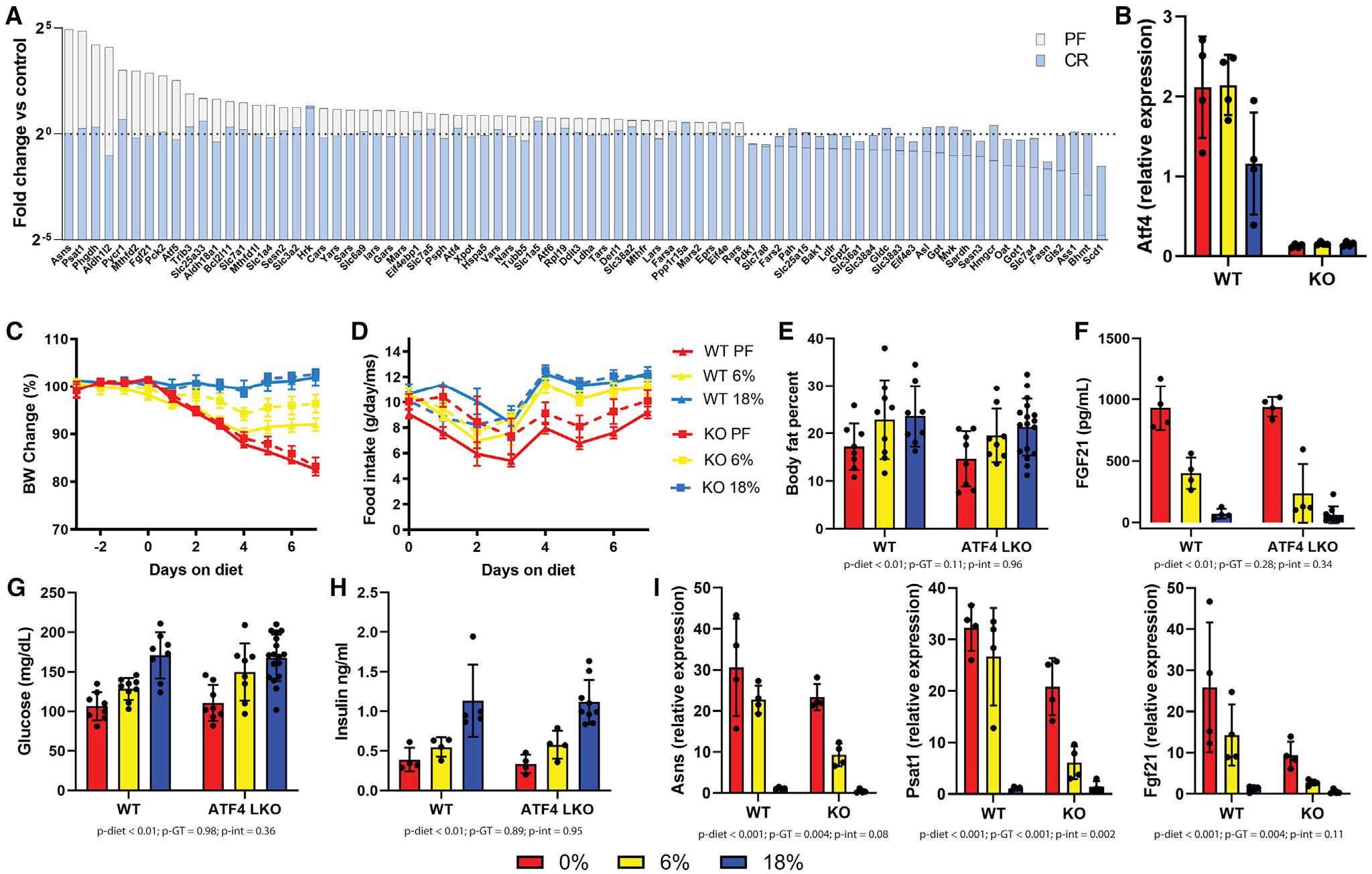
Adult male wild-type or hepatic ATF4 knockout (LKO) littermates were fed AL isocaloric diets for 1 week with 18%, 6%, or 0% protein with protein calories replaced with sucrose.
(A) ATF4 target gene expression from livers of wild-type mice fed AL 0% (white) or 40% CR 18% protein (blue) as log2 fold change versus AL 18% protein control.
(B) Atf4 transcript expression in the liver.
(C) Body weight changes expressed as percentage of starting weight for wild-type (solid lines) or ATF4 LKO (dashed lines) mice.
(D) Food intake in grams per mouse per day.
(E) Body fat percentage after 1 week of diets.
(F) Serum FGF21 concentration after 1 week of diets.
(G and H) Blood glucose (G) and insulin (H) levels after 1 week of diets.
(I) Hepatic gene expression of selected ATF4 target genes.
(E–I) were analyzed using two-way ANOVA, p-diet: p value for main effect of diet, p-GT, p value for main effect of genotype, p-int, p value for diet by genotype interaction effect.
DISCUSSION
Dietary AA titration produced linear and non-linear changes in metabolic phenotypes and transcriptional and metabolite profiles
Most studies of PR-mediated benefits compare only a single restriction group to a single control group. As many of the responses to PR are non-linear, this limits causal inference. Here, we used a PR dose response in conjunction with unbiased omics methods to investigate underlying mechanisms of PR-mediated metabolic benefits. We hypothesized that linear molecular responses would more likely explain linear metabolic changes, including improved glucose homeostasis. Two such gene-expression patterns were an increase in NEAA biosynthesis and PPARa signaling (Figure 3G) and an unexpected decrease in oxidative phosphorylation and thermogenesis in the liver (Figure 3D). Other non-linear gene-expression clusters tended to increase or decrease up to 6% protein and then change in opposite directions. While these non-linear expression patterns are harder to interpret, we note that many of the metabolic phenotypes of PR were also non-linear. These included changes in food intake, body weight and composition, circulating triglycerides, and hormones like FGF21, which also increased significantly below 6% protein. These expression patterns may represent longevity-associated changes, as the 6% diet is most closely associated with extended lifespan in the literature. Importantly, mouse sex and strain also robustly modify the response to PR, thus this 6% “inflection point” is likely to differ in females or different strains (Green et al., 2022).
AA titration altered the fecal microbiota, but PR metabolic phenotypes were not transferrable via fecal microbiota transplantation
Diet can have rapid and profound effects on the microbiome, which can, in certain contexts, be causally linked to the effects of dietary interventions using microbiota transplant experiments. In the context of PR, there is evidence of profound changes in the fecal microbiome but limited data on causality. In one study, antibiotic ablation of the microbiome with antibiotics did not alter the metabolic response to PR (Pak et al., 2019). However, another study showed that gnotobiotic mice had a blunted FGF21 response to a marginally low-protein diet (8%) (Martin et al., 2021). Thus, it is unclear whether a microbiome is necessary to mount a metabolic response to low protein diets.
The goal of our gnotobiotic study was to determine the extent to which gut microbial conditioning is sufficient to confer metabolic benefits of PR. By placing all gnotobiotic recipients on control (18% protein) diet, our study design aimed to isolate the potential microbial contributions to metabolic phenotype by divorcing them from the effects of host-driven responses to the PR diet. Conventionalization of germ-free mice with PR-conditioned donor inocula did not result in the transfer of metabolic phenotypes. Although gnotobiotic mice exhibit physiological differences compared with conventional laboratory mice that may alter metabolic response (including differences in body fat and immune function), our results lead us to conclude that changes in gut microbial structure are likely not a causative factor in metabolic changes upon PR.
A caveat of this interpretation is that the lack of transferability of such effects does not rule out their importance but makes causality difficult to determine. There may be interactions between the transplanted microbiome and the recipient diet that were undetected because all recipient mice received the control 18% protein diet. A final caveat is the short time frame of the study. Although we see rapid effects on metabolic parameters such as glucose homeostasis, some phenotypes of PR, including increased lifespan, may take longer to manifest and be affected differentially by changes in microbial composition. Future testing of a wider protein titration in germ-free mice would help to confirm the role of the gut microbiota in the metabolic effects of PR.
Low AA diets selectively increased hepatic-free AA levels of aspartate (Asp), serine (Ser), and glutamate (Glu) while significantly lowering others
While an increase in several NEAAs in the liver upon PR is consistent with previous reports, it remains at odds with the conventional notion that PR drives a global increase in ATF4-activated NEAA biosynthetic genes. Instead, several genes were either not activated or were significantly repressed, including the terminal transsulfuration gene Cth (also known as Cgl or Cse) involved in cysteine biosynthesis from cystathionine, Shmt1 involved in glycine synthesis from Ser, and the cytoplasmic transaminase Got1 involved in Glu biosynthesis from Asp or cysteine. Furthermore, despite the increase in Asns gene expression, Asn levels were not increased, which could be due to increase flux of this AA into other metabolites. Interestingly, in cancer cells auxotrophic for arginine, ATF4-dependent Asns activation upon Arg deprivation depletes Asp, resulting in mitochondrial dysfunction and cell death (Cheng et al., 2018), suggesting that Asns gene expression and Asn production are not necessarily linked to directly to Asn depletion.
A limitation of our study is that it is unclear why steady-state levels of these specific AAs were increased or the metabolic consequences of increased glucose incorporation into Asp, Glu, and Ser on glucose homeostasis. Nonetheless, the increase in OPA, which follows a similar pattern with respect to PR, is intriguing due to its sizable increase and potential direct or indirect association with increased Asp, Glu, and Ser production. OPA is a glutathione (GSH)-like tripeptide of glycine, Glu, and 2-aminobutyrate (2AB) synthesized in response to GSH depletion upon acetaminophen challenge (Soga et al., 2006), doxorubicin treatment (Irino et al., 2016), fasting (Kobayashi et al., 2017), low-protein diet (Yap et al., 2020), or cysteine deprivation in hepatocytes (Lee et al., 2017) by the GSH synthesis pathway but using 2AB instead of cysteine. 2AB is produced by transamination of 2-oxobutyric acid (2OB) by Asp transaminase (Ast), which has been shown to use Glu as an amine donor in vitro (Irino et al., 2016), but could also potentially use Asp. 2OB is produced in equimolar amounts with cysteine (Cys) by CTH in the final step of the transsulfuration pathway or via threonine catabolism. Whether or not Ser contributes to Cys biosynthesis under PR or CR conditions remains unclear. Here, we observed a decrease in CTH expression and Cys levels upon PR, while in CR and methionine restriction (MR), we have previously observed an increase in CTH expression (Hine et al., 2015; Longchamp et al., 2018). Future flux experiments will be required to answer this question under these different dietary conditions as well as to determine the potential contribution of Ser to OPA via cystathionine or glycine. Interestingly, GSH depletion is another common feature of CR and MR, where redox imbalance drives a prominent hepatic NRF2 activation signature (Hine et al., 2015; Ghosh et al., 2017), while here we observed a distinctly non-linear response of NRF2 target genes, peaking at 6% protein and returning to control levels at 0%. While the functional role of OPA remains unclear, 2AB administration in mice elevates circulating and hepatic GSH and exerts cardioprotective effects against doxorubicin-induced cardiomyopathy (Irino et al., 2016) and may have a beneficial role in the case of GSH insufficiency.
A potential role for PR in metabolic benefits of CR
CR appears to increase longevity in rodents to a greater extent than PR (Speakman et al., 2016), which is consistent with reports in humans that low-protein diets may be unhealthy beyond a certain age (Levine et al., 2014). Nevertheless, the independent role of PR in the metabolic benefits of CR remain unclear. Here, we found evidence of a significant overlap in gene signatures between CR and PR but generally larger magnitudes of change in the PR group. Interestingly, changes in NEAA synthetic genes (commonly associated with ATF4 activity) were not among these common changes. However, ATF4 was not required for upregulation of these genes upon PR. This stands in contrast to previous findings, which showed ATF4 to be required for the regulation of these genes in response to other stimuli, including insulin (Torrence et al., 2021). Thus, although ATF4 has been hypothesized to be a key mediator of metabolic effects of PR, here, we show it is dispensable for many of the hallmark effects. This finding is consistent with recent work showing that ATF4 is not required for many of the metabolic effects of MR (Jonsson et al., 2021). In concurrent work, we have performed transcription factor binding site analysis on this group of genes in an independent dataset and identified a number of candidate regulators, but these observations will require experimental validation (Kalafut et al., 2022).
The role of altered hepatic glucose utilization and gluconeogenic substrate preference in metabolic benefits of PR
Here, we found increased incorporation of glucose and glycerol into Ser, Asp, and Glu. These changes in flux could influence glucose homeostasis as a potential sink for glucose and may also be a link to lipid metabolism via glycerol. Relative to the 18% group, the percentage of labeled carbon from glucose incorporated into Asp, Ser, and Glu was significantly elevated in the 0% protein group. The significant increases in relative abundance of m+3 Glu and m+3 Asp are consistent with direct entry of fully labeled pyruvate into m+3 oxalacetate (OAA) via anaplerosis, while significant increases in m+2 and m+4 Asp and Glu are consistent with multiple rounds through the citric acid cycle (Figures 6D, 6H, 6F, and 6J).
Ser 3C backbones can either be derived directly from glucose or glycerol via the glycolytic intermediate 3PG and the de novo Ser synthesis pathway, which is not normally active in liver, or indirectly via PEP following decarboxylation of OAA during gluconeogenesis. Surprisingly, m+1 Ser, rather than m+3, was significantly elevated with both glucose and glycerol in vivo (Figures 6E and 6I). This m+1 Ser can be derived from OAA (Figure 6A) during gluconeogenesis.
Finally, the finding that glycerol, which is released from triglycerides during PR and is known to spare protein independently of fatty acid, was increased in fractional abundance in glucose is evidence toward altered gluconeogenic substrate utilization upon PR, which could potentially spare AAs by reducing the need for use of gluconeogenic AAs such as alanine (Brennan et al., 1975). Whether or not an overall decrease in gluconeogenesis occurs upon PR remains unclear, as increases in fractional abundance of glycerol into glucose, and increased m+1 tracing from glucose into Ser in this study points to maintenance, if not increased gluconeogenic potential. Thus, our study raises the interesting hypothesis that altered flux of glucose and glycerol upon PR may represent a mechanism of euglycemia. This mechanism may be one of many pleiotropic effects of PR working in concert with improved insulin sensitivity (Maida et al., 2016) and improved fatty acid clearance (Maida et al., 2018). Future studies using genetic models will be required to determine if either reduced gluconeogenesis and/or altered glucogenic substrate utilization contribute significantly to the benefits of PR on glucose homeostasis.
Limitations of the study
While we have shown interesting shifts in hepatic metabolism in response to PR, more mechanistic work is needed to determine whether these changes causally impact PR-associated improvements in glucose or lipid homeostasis. Additionally, although transcriptomic and metabolomic profiles are highly consistent with profiles reported by Green and colleagues after 3 months (Green et al., 2022), we did not specifically show whether changes in flux were durable beyond 1 week, and longer-term follow ups may be necessary, including females and additional strains. Regarding the microbiome, while we showed that transplantation of the PR fecal microbiome does not transfer metabolic benefits, more work remains to understand the role of the microbiome in the host response to the diet. Follow-up studies profiling responses to a protein titration in gnotobiotic mice would help resolve open questions in this area. Finally, we have focused primarily on the liver as a major metabolic organ, but more work profiling flux in other organ systems in response to PR may yield additional interesting insights.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Requests for further information, resources or reagents should be directed to Lead Contact Michael R. MacArthur (mmacarthur@ethz.ch).
Resource availability
This study did not generate any new reagents.
Data and code availability
RNA sequencing data are available under the SRA accession PRJNA851959. An interactive app with sequencing data is available at https://macarthur.shinyapps.io/multiomicspr/.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
For all experiments except for the gnotobiotic transfer and ATF4 LKO, 16 wk old male B6D2F1 hybrids were purchased from The Jackson Laboratory (strain no. 100006, Bar Harbor ME). Mice were acclimated in the facility for at least one week prior to experiments and allowed ad libitum access to food (Purina 5053 chow) and water unless otherwise noted. Mice were housed in conventional microisolator static cages (Lab Products LLC, Seaford DE) and were changed biweekly under a laminar flow hood. All food, bedding and enrichment was autoclaved prior to introduction to mice. The facility housing the mice had SPF status and SPF out-bred sentinel mice on every rack were checked on a rotating quarterly schedule. Liver specific ATF4 knockout mice were generated by crossing heterozygous ATF4 floxed mice (Atf4tm1.1Cmad) (Ebert et al., 2012) with heterozygous ATF4 floxed mice expressing cre recombinase under the albumin promoter (Speer6-ps1Tg(Alb-cre)21Mgn). Mice homozygous for the ATF4 wild type allele and expressing cre were used as controls and mice homozygous for the ATF4 floxed allele and expressing cre were used as experimental KOs. Mice were maintained at 22°C with 12 h light-dark cycles and 30–50% relative humidity. For all experiments mice were house in cages of 2. All experiments were approved by the respective IACUC of the Harvard Medical School and the Brigham and Women’s Hospital.
Gnotobiotic experiment
The gnotobiotic study was performed at the Massachusetts Host-Microbiome Center at Brigham & Women’s Hospital. A total of 24 8-week old male C57BL/6 germ-free mice were sourced for the gnotobiotic colonization study. For 7 days prior to colonization, all germ-free mice were acclimated to an irradiated semi-purified 18% protein diet (Research Diets D12450B) while in germ-free isolators. This same diet was fed throughout the study. After the 7 days of diet acclimation, baseline 4-h fasting glucose measurements, duplicate samples of feces, and measurements of body mass and food in hopper were collected from all germ-free animals. Following baseline sample and data collection, mice were colonized with fecal inocula. Fecal samples from 4 different conventional B6D2F1/J adult mice reared on each of the 3 diet treatments (0%, 6%, and 18% protein) were collected, flash-frozen, and reconstituted in reduced PBS for use as separate inocula. This resulted in 4 unique inocula per dietary condition. Each of the unique inocula were used to colonize 2 germ-free animals (cagemates), resulting in n = 8 per dietary condition. Colonized gnotobiotic animals were then cohoused in OptiMice cages. Every 7 days post-colonization, samples of feces, as well as data on body mass and food intake were collected. On Day 19 post-colonization, mice were sacrificed and harvested for tissue and blood samples. The facility housing the mice had SPF status and SPF out-bred sentinel mice were checked on a regular schedule.
Diets
For all experiments except the gnotobiotic transfer, semi-purified, isocaloric diets were made with crystalline AA with casein profile and Research Diets D12450BSpx base in final 1% agar to form a homogeneous solid diet as described previously (MacArthur et al., 2021). The Research Diets D12450BSpx formulation is similar to the D12450B formulation but contains no casein/cystine and is soluble allowing custom formulation of diets with differing protein contents. For protein titration studies, the crystalline AAs representing the protein component for control animals was isocalorically replaced with sucrose (see Table S1 for ingredients). For caloric restriction studies ad libitum food intake normalized to body weight was monitored daily for 5 days prior to starting the study in all mice. 40% calorie restriction was calculated based on these values. Mice were fed the restricted amount daily between ZT22 and ZT23. For the gnotobiotic transfer experiment, mice were kept on Research Diets D12450B with approximately 18% of calories from protein (hydrolyzed casein), 10% from fat and 72% from carbohydrate. Food intake was measured daily at approximately ZT22. For all studies, mice were fasted for four hours prior to sacrifice and tissue collection which took place at approximately ZT20.
METHOD DETAILS
Body composition
Body mass was determined by daily measurement at approximately ZT22. Lean and fat mass were measured in awake mice using an EchoMRI analyzer system (EchoMRI LLC, Houston TX).
Healthspan markers
Glucose was measured using a Clarity BG1000 handheld glucometer (VWR, Radnor PA). (Insulin was measured using by ELISA following manufacturer protocol (Crystal Chem #90080). FGF21 was measured by ELISA following manufacturer protocol (R&D Systems #MF2100). Adiponectin was measured by ELISA following manufacturer protocol (ALPCO #47-ADPMS-E01). Serum triglycerides were measured using a colorimetric kit according to manufacturer protocol (MilliporeSigma TRO100–1KT). Serum cholesterol was measured using a colorimetric assay according to manufacturer protocol (BioVision #K603–100). Resistin was measured using a Luminex multiplex assay according to manufacturer protocol (EMD Millipore MMHMAG-44K). Complete blood cell counts were measured using a Hemavet blood analyzer according to manufacturer’s protocol.
Microbiome analysis
Fresh fecal pellets were collected daily at approximately ZT22, flash frozen in liquid nitrogen and stored at −80°C until analysis. Bacterial DNA was isolated from fecal samples using the DNeasy PowerSoil kit (Qiagen) and PCR-amplified using barcoded universal bacterial primers targeting the V4 region of the 16S rRNA gene (515F/806R). The following thermocycler protocol was used for PCR amplification: 94°C for 3 min, followed by 35 cycles of 94°C for 45 s, 50°C for 30 s and 72°C for 90 s, and a final extension at 72°C for 10 min. Triplicate amplicons for each sample were pooled and checked for amplification using 1.5% gel electrophoresis. Amplicons were then cleaned using AMPure XP beads (Beckman Coulter) and quantified using the Quant-iT Picogreen dsDNA Assay Kit (Invitrogen). Amplicons were pooled so as to contribute equal amounts of DNA and sequenced using the Illumina HiSeq platform, generating 1 × 150 bp sequences. 16S sequences were analyzed on the Harvard Odyssey computational cluster using the QIIME 2 (Quantitative Insights into Microbial Ecology) software package version 2020.2 (Bolyen et al., 2019). Sequences were demultiplexed using the Demux plugin, and denoised for quality control using DADA2. This produced feature tables comprised of amplicon sequence variants (ASVs) that were mapped to characterized bacterial taxa. Taxonomy was assigned using the Naive Bayes classifier trained on Greengenes 13_8 99% OTUs from the 515F/806R region.
Downstream analyses were performed in R (4.0.2) using the phyloseq package (1.32.0) (McMurdie and Holmes 2013). Phyloseq objects were created by loading feature tables, taxonomy files, rooted trees and metadata through the qza_to_phyloseq function from the qiime2R package (0.99.34). Species from uncharacterized phyla or which had no counts in any samples were removed. Filtering was performed by pruning taxa which did not appear in at least 5 samples. Sample counts were then transformed to relative abundances for subsequent analyses. Ordination plots were generated using Bray distances and NMDS reduction. PERMANOVA was performed using the adonis function from the vegan package (2.5.6) and Bray distances. Shannon diversity analyses were performed using the phyloseq package plot_richness function. Differential abundance analysis was performed using the DESeq2 package (1.28.1) and models were fit using the voom and lmFit functions from the limma package (3.44.3). Significantly differentially expressed species were considered those with Benjamini-Hochberg FDR adjusted p value <0.05.
RNA seq
Livers were collected from euthanized mice and immediately flash frozen in liquid nitrogen and stored at −80°C until analysis. Livers were homogenized using a handheld homogenizer and RNA was extracted using a Qiagen RNeasy Plus Mini Kit (Qiagen #74134). The concentration and purity of RNA was determined using a Nanodrop Spectrophotometer and confirmed using an Agilent 2100 Bio-analyzer. Libraries were prepared using the Illumina TruSeq Stranded Total RNA Sample Preparation protocol. RNA was sequenced on an Illumina NovaSeq 6000 with 20 million paired end reads per sample.
Reads were aligned to the mouse GRCm38.p6 assembly using the align function and annotated using the featureCounts function from the Rsubread package (version 2.3.7). Differential expression analysis was performed using the edgeR (3.30.3) and limma (3.44.3) packages. Gene symbols were mapped to Entrez IDs using the mapIds function from the AnnotationDbi package (1.51.1). Cluster analysis was performed using the degPatterns function from the DEGreport (1.24.1) package. In total, reads were mapped to 27,179 genes. Read counts were normalized to counts per million reads and genes that did not have at least one read in at least two samples were filtered, leaving 13,667 genes. Normalization was performed using the trimmed mean of M-values method as implemented in the calcNormFactors from edgeR. Data were modeled and differential expression was determined using the limma voom pipeline to generate linear models with empirical Bayes moderation. Differential expression was determined using a Benjamini-Hochberg adjusted p value less than 0.05. Once differentially expressed genes or gene clusters were determined, gene set enrichment analysis was determined using the enrichKEGG function from the clusterProfiler package (3.16.0).
Hepatic metabolite profiling
After 7 days on diet with either 18% or 0% energy from protein, mice were anesthetized under isoflurane/oxygen gas and 2 mg/g body weight of 13C6 glucose was given in a total volume of 200uL of phosphate buffered saline via retroorbital injection. After approximately 30 min mice were sacrificed and livers were collected and immediately flash frozen in liquid nitrogen and stored at −80C until analysis. Approximately 30 mg of frozen tissue was homogenized in 1 mL of 80% methanol using a handheld homogenizer on dry ice to keep samples frozen throughout the process. For tracing samples, an internal standard was added after homogenization. Samples were vortex and incubated for 4 hours at −80C. After incubation samples were centrifuged at 15,000xg for 10 min at 4C. Supernatant was collected and dried in a Speedvac.
For stable isotope labeling LC/MS analyses were conducted on a QExactive bench top orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific, San Jose, CA). External mass calibration was performed using the standard calibration mixture every 7 days. Typically, dried samples were reconstituted in 100 uL water and 2 uL were injected onto a SeQuant ZIC-pHILIC 150 × 2.1 mm analytical column equipped with a 2.1 × 20 mm guard column (both 5 mm particle size; MilliporeSigma). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; Buffer B was acetonitrile. The column oven and autosampler tray were held at 25°C and 4°C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–20 min: linear gradient from 80–20% B; 20–20.5 min: linear gradient form 20–80% B; 20.5–28 min: hold at 80% B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275°C, and the HESI probe held at 350°C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1000, with the resolution set at 70,000, the AGC target at 1×106, and the maximum injection time at 20 msec. Relative quantitation of 13C label in polar metabolites was performed using El Maven v07.1 (Elucidata) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards. Data were corrected for natural isotopic abundance using the R package IsoCorrectoRGUI (Heinrich et al., 2018).
For steady state metabolite measurements samples were analyzed by LC-MS as previously as previously described (Ben-Sahra et al., 2013; Longchamp et al., 2018). Briefly, dried pellets were resuspended using 20 μL HPLC grade water for mass spectrometry. 10 μL were injected and analyzed using a 5500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (SRM). Some metabolites were targeted in both positive and negative ion mode for a total of 287 SRM transitions using pos/neg polarity switching. ESI voltage was +4900 V in positive ion mode and −4500 V in negative ion mode. The dwell time was 3 ms per SRM transition and the total cycle time was 1.55 seconds. Approximately 10–14 data points were acquired per detected metabolite. Samples were delivered to the MS via normal phase chromatography using a 4.6 mm i.d. × 10 cm Amide Xbridge HILIC column (Waters Corp.) at 350 μL/min. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 42% B from 0–5 min; 42% B to 0% B from 5–16 min; 0% B was held from 16–24 min; 0% B to 85% B from 24–25 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A was comprised of 20 mM ammonium hydroxide/20 mM ammonium acetate (pH = 9.0) in 95:5 water:acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/SCIEX). For stable isotope labeling experiments, custom SRMs were created for expected 13C incorporation in various forms for targeted LC/MS/MS.
RT-qPCR
RNA was isolated from flash-frozen liver by homogenization in TRIzol Reagent (Thermo Fisher) followed by chloroform extraction and isopropanol precipitation. The concentration of RNA was determined using a Nanodrop Spectrophotometer. RNA (1 μg) was reverse transcribed using the Advanced cDNA Synthesis Kit (Bio-Rad). qPCR was performed using Power Up SYBR green (Applied Biosystems) with duplicate technical replicates using the QuantStudio 5 Real-Time PCR system. ΔΔCt values were normalized to B2m and 18s and relative expression was plotted. Primer sequences were: B2m forward: CGGCCTGTATGCTATCCAGA, B2m re verse: GGGTGAATTCAGTGTGAGCC, 18s forward: CATGCAGAACCCACGACAGTA, 18s reverse: CCTCACGCAGCTTGTTGTCTA, Asns forward: GTCAAGAACTCCTGGTTCAAG, Asns reverse: GATCTGACGGTAGAAGTAGC, Atf4 forward: TCGATGCTCTGTTTCGAATG, Atf4 reverse: AGAATGTAAAGGGGGCAACC, Fgf21 forward: CTGCTGGGGGTCTACCAAG, Fgf21 reverse: CTGCGCCTACCACTGTTCC, Psat1 forward: CAGTGGAGCGCCAGAATAGAA, Psat1 reverse: CCTGTGCCCCTTCAAGGAG.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed in Prism (version 8, GraphPad Software). All plots are means +/− standard deviations unless otherwise specified. One-way ANOVAs were followed by either Tukey’s or Dunnett’s post-hoc test as indicated. RT-qPCR data were analyzed using the delta-delta CT method. Transcript and metabolite data were analyzed using R (version 4.0.2) and multiple comparisons in omics data were corrected using the Benjamini-Hochberg false detection rate correction. All graph error bars represent 95% confidence intervals.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Power SYBR Green Master Mix | Applied Biosystems | Cat# 43-679-59 |
| Critical commercial assays | ||
| FGF21 ELISA | R&D Systems | Cat# MF2100 |
| Adiponectin ELISA | ALPCO | Cat# 47-ADPMS-E01 |
| Triglyceride Quantification Kit | Millipore Sigma | Cat# TRO100-1KT |
| Cholesterol Quantification Kit | BioVision | Cat# K603-100 |
| Milliplex metabolic hormone multiple assay | Millipore | Cat# MMHMAG-44K |
| Deposited data | ||
| Hepatic RNA seq data | NIH SRA Database | PRJNA851959 |
| Experimental models: Organisms/strains | ||
| Mouse strain: B6D2/F1 | The Jackson Laboratory | Cat# 100006 |
| Mouse strain: ATF4 flox | Chris Adams | Atf4tm1.1Cmad |
| Mouse strain: C57BL6/J | The Jackson Laboratory | Cat# 000664 |
| Mouse strain: Albumin-Cre | The Jackson Laboratory | Cat# 003574 |
| Oligonucleotides – see also Table S4 | ||
| 18s: F: CATGCAGAACCCACGACAGTA | This paper | N/A |
| 18s: R: CCTCACGCAGCTTGTTGTCTA | This paper | N/A |
| Asns: F: GTCAAGAACTCCTGGTTCAAG | This paper | N/A |
| Asns: R: GATCTGACGGTAGAAGTAGC | This paper | N/A |
| Atf4: F: TCGATGCTCTGTTTCGAATG | This paper | N/A |
| Atf4: R: AGAATGTAAAGGGGGCAACC | This paper | N/A |
| Fgf21: F: CTGCTGGGGGTCTACCAAG | This paper | N/A |
| Fgf21: R: CTGCGCCTACCACTGTTCC | This paper | N/A |
| Psat1: F: CAGTGGAGCGCCAGAATAGAA | This paper | N/A |
| Psat1: R: CCTGTGCCCCTTCAAGGAG | This paper | N/A |
| Software and algorithms | ||
| R (4.0.2) | N/A | N/A |
| QIIME 2 (2020.2) | (Bolyen et al., 2019) | N/A |
| phyloseq (1.32.0) | (McMurdie and Holmes 2013) | N/A |
| qiime2R (0.99.34) | N/A | https://github.com/jbisanz/qiime2R |
| DEseq2 (1.28.1) | (Love et al., 2014) | N/A |
| Rsubread (2.3.7) | (Liao et al., 2019) | N/A |
| edgeR (3.30.3) | (Robinson et al., 2010) | N/A |
| clusterProfiler (3.16.0) | (Yu et al., 2012) | N/A |
| IsoCorrectoRGUI | (Heinrich et al., 2018) | N/A |
| Other | ||
| Normal maintenance mouse chow | Purina | Cat# 5053 |
| Protein-free base diet | Research Diets | Cat# D12450BSpx |
Highlights.
Physiologic and molecular responses to protein restriction (PR) are dose responsive
PR below 10% of energy produces the strongest responses
Glucose is used for serine, aspartate, and glutamate synthesis upon PR
Metabolic and molecular changes upon PR are independent of hepatic ATF4
ACKNOWLEDGMENTS
We dedicate this manuscript to the memory of Jay Mitchell. We thank John Asara and Brendan Manning for assistance with metabolomics and thank members of the Mitchell Lab as well as Dudley Lamming and Cara Green for helpful discussions and feedback. We also thank David MacArthur for help with data management logistics. This project was supported by grants from the NIH (RO1DK090629 and R01AG036712) and Charoen Pokphand Group to J.R.M. and M.R.M.
DECLARATION OF INTERESTS
T.-Z.A.C. and A.K. are employees of the Charoen Pokphand Group. J.R.M. received research funding from the Charoen Pokphand Group.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111187.
REFERENCES
- Ben-Sahra I, Howell JJ, Asara JM, and Manning BD (2013). Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MF, Fitzpatrick GF, Cohen KH, and Moore FD (1975). Glycerol: major contributor to the short term protein sparing effect of fat emulsions in normal man. Ann. Surg 182, 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM Jr., Gatti DM, Somes L, Svenson KL, and Turnbaugh PJ (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CT, Qi Y, Wang YC, Chi KK, Chung Y, Ouyang C, Chen YR, Oh ME, Sheng X, Tang Y, et al. (2018). Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Commun. Biol 1, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke D, Mattocks D, Nichenametla SN, Anunciado-Koza RP, Koza RA, and Ables GP (2020). Weight loss and concomitant adipose autophagy in methionine-restricted obese mice is not dependent on adiponectin or FGF21. Obesity 28, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaif GE, and Campbell TC (1987). Dietary protein level and aflatoxin B1-induced preneoplastic hepatic lesions in the rat. J. Nutr 117, 1298–1302. [DOI] [PubMed] [Google Scholar]
- Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, Dierdorff JM, Foster ED, and Adams CM (2012). Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J. Biol. Chem 287, 27290–27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbiano S, Suárez-Zamorano N, Chevalier C, Lazarević V, Kieser S, Rigo D, Leo S, Veyrat-Durebex C, Gaïa N, Maresca M, et al. (2018). Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab 28, 907–921.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Twinn DS, Wayman A, Ekizoglou S, Martin MS, Hales CN, and Ozanne SE (2005). Maternal protein restriction leads to hyperinsulinemia and reduced insulin-signaling protein expression in 21-mo-old female rat offspring. Am. J. Physiol. Regul. Integr. Comp. Physiol 288, R368–R373. [DOI] [PubMed] [Google Scholar]
- Fontana L, Adelaiye RM, Rastelli AL, Miles KM, Ciamporcero E, Longo VD, Nguyen H, Vessella R, and Pili R (2013). Dietary protein restriction inhibits tumor growth in human xenograft models. Oncotarget 4, 2451–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. (2016). Decreased consumption of branched-chain amino acids improves metabolic health. Cell Rep 16, 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Forney LA, Wanders D, Stone KP, and Gettys TW (2017). An integrative analysis of tissue-specific transcriptomic and metabolomic responses to short-term dietary methionine restriction in mice. PLoS One 12, e0177513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrick CL (1978). Body weight increment and length of life: the effect of genetic constitution and dietary protein. J. Gerontol 33, 184–190. [DOI] [PubMed] [Google Scholar]
- Gosby AK, Conigrave AD, Lau NS, Iglesias MA, Hall RM, Jebb SA, Brand-Miller J, Caterson ID, Raubenheimer D, and Simpson SJ (2011). Testing protein leverage in lean humans: a randomised controlled experimental study. PLoS One 6, e25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosby AK, Lau NS, Tam CS, Iglesias MA, Morrison CD, Caterson ID, Brand-Miller J, Conigrave AD, Raubenheimer D, and Simpson SJ (2016). Raised FGF-21 and triglycerides accompany increased energy intake driven by protein leverage in lean, Healthy individuals: a randomised trial. PLoS One 11, e0161003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CL, Pak HH, Richardson NE, Flores V, Yu D, Tomasiewicz JL, Dumas SN, Kredell K, Fan JW, Kirsh C, et al. (2022). Sex and genetic background define the metabolic, physiologic, and molecular response to protein restriction. Cell Metab 34, 209–226.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, and Cavener DR (2007). The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab 5, 103–114. [DOI] [PubMed] [Google Scholar]
- Harputlugil E, Hine C, Vargas D, Robertson L, Manning BD, and Mitchell JR (2014). The TSC complex is required for the benefits of dietary protein restriction on stress resistance in vivo. Cell Rep 8, 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich P, Kohler C, Ellmann L, Kuerner P, Spang R, Oefner PJ, and Dettmer K (2018). Correcting for natural isotope abundance and tracer impurity in MS-MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci. Rep 8, 17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Laeger T, Dehner M, Albarado DC, Clarke B, Wanders D, Burke SJ, Collier JJ, Qualls-Creekmore E, Solon-Biet SM, et al. (2019). FGF21 signals protein status to the brain and adaptively regulates food choice and metabolism. Cell Rep 27, 2934–2947.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine C, Harputlugil E, Zhang Y, Ruckenstuhl C, Lee BC, Brace L, Longchamp A, Treviño-Villarreal JH, Mejia P, Ozaki CK, et al. (2015). Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 160, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Chew YV, Colakoglu F, Cliff JB, Klaassens E, Read MN, Solon-Biet SM, McMahon AC, Cogger VC, Ruohonen K, et al. (2017). Diet-microbiome interactions in health are controlled by intestinal nitrogen source constraints. Cell Metab 25, 140–151. [DOI] [PubMed] [Google Scholar]
- Irino Y, Toh R, Nagao M, Mori T, Honjo T, Shinohara M, Tsuda S, Nakajima H, Satomi-Kobayashi S, Shinke T, et al. (2016). 2-Aminobutyric acid modulates glutathione homeostasis in the myocardium. Sci. Rep 6, 36749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson WO, Margolies NS, Mirek ET, Zhang Q, Linden MA, Hill CM, Link C, Bithi N, Zalma B, Levy JL, et al. (2021). Physiologic responses to dietary sulfur amino acid restriction in mice are influenced by Atf4 status and biological sex. J. Nutr 151, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafut KC, Mitchell SJ, MacArthur MR, and Mitchell JR (2022). Short-term ketogenic diet induces a molecular response that is distinct from dietary protein restriction. Front. Nutr 9, 839341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Uppal SO, Moorman JL, Bennett C, Gruca LL, Parimi PS, Dasarathy S, Serre D, and Hanson RW (2011). Metabolic and genomic response to dietary isocaloric protein restriction in the rat. J. Biol. Chem 286, 5266–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata S, Yamamoto J, Kamijo K, Ochiai T, Morita T, Yoshitomi Y, Hagiya Y, Kubota M, Ohkubo R, Kawaguchi M, et al. (2014). Dietary deprivation of each essential amino acid induces differential systemic adaptive responses in mice. Mol. Nutr. Food Res 58, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Lee J, Takao T, and Fujii J (2017). Increased ophthalmic acid production is supported by amino acid catabolism under fasting conditions in mice. Biochem. Biophys. Res. Commun 491, 649–655. [DOI] [PubMed] [Google Scholar]
- Laeger T, Henagan TM, Albarado DC, Redman LM, Bray GA, Noland RC, Münzberg H, Hutson SM, Gettys TW, Schwartz MW, and Morrison CD (2014). FGF21 is an endocrine signal of protein restriction. J. Clin. Invest 124, 3913–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley SC, Browne RF, and Jackson AA (1994). Altered glucose tolerance in rats exposed to maternal low protein diets in utero. Comp. Biochem. Physiol. Physiol 109, 223–229. [DOI] [PubMed] [Google Scholar]
- Lee J, Kang ES, Kobayashi S, Homma T, Sato H, Seo HG, and Fujii J (2017). The viability of primary hepatocytes is maintained under a low cysteine-glutathione redox state with a marked elevation in ophthalmic acid production. Exp. Cell Res 361, 178–191. [DOI] [PubMed] [Google Scholar]
- Leto S, Kokkonen GC, and Barrows CH (1976). Dietary protein life-span, and physiological variables in female mice. J. Gerontol 31, 149–154. [DOI] [PubMed] [Google Scholar]
- Levine ME, Suarez JA, Brandhorst S, Balasubramanian P, Cheng CW, Madia F, Fontana L, Mirisola MG, Guevara-Aguirre J, Wan J, et al. (2014). Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 19, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longchamp A, Mirabella T, Arduini A, MacArthur MR, Das A, Treviño-Villarreal JH, Hine C, Ben-Sahra I, Knudsen NH, Brace LE, et al. (2018). Amino acid restriction triggers angiogenesis via GCN2/ATF4 regulation of VEGF and H(2)S production. Cell 173, 117–129.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur MR, Mitchell SJ, Treviño-Villarreal JH, Grondin Y, Reynolds JS, Kip P, Jung J, Trocha KM, Ozaki CK, and Mitchell JR (2021). Total protein, not amino acid composition, differs in plant-based versus omnivorous dietary patterns and determines metabolic health effects in mice. Cell Metab 33, 1808–1819.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida A, Zota A, Sjøberg KA, Schumacher J, Sijmonsma TP, Pfenninger A, Christensen MM, Gantert T, Fuhrmeister J, Rothermel U, et al. (2016). A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J. Clin. Invest 126, 3263–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida A, Zota A, Vegiopoulos A, Appak-Baskoy S, Augustin HG, Heikenwalder M, Herzig S, and Rose AJ (2018). Dietary protein dilution limits dyslipidemia in obesity through FGF21-driven fatty acid clearance. J. Nutr. Biochem 57, 189–196. [DOI] [PubMed] [Google Scholar]
- Martin A, Ecklu-Mensah G, Ha CWY, Hendrick G, Layman DK, Gilbert J, and Devkota S (2021). Gut microbiota mediate the FGF21 adaptive stress response to chronic dietary protein-restriction in mice. Nat. Commun 12, 3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H, Segall PE, and Timiras PS (1988). Histology and survival in age-delayed low-tryptophan-fed rats. Mech. Ageing Dev 43, 79–98. [DOI] [PubMed] [Google Scholar]
- Orentreich N, Matias JR, DeFelice A, and Zimmerman JA (1993). Low methionine ingestion by rats extends life span. J. Nutr 123, 269–274. [DOI] [PubMed] [Google Scholar]
- Orgeron ML, Stone KP, Wanders D, Cortez CC, Van NT, and Gettys TW (2014). The impact of dietary methionine restriction on biomarkers of metabolic health. Prog. Mol. Biol. Transl. Sci 121, 351–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak HH, Cummings NE, Green CL, Brinkman JA, Yu D, Tomasiewicz JL, Yang SE, Boyle C, Konon EN, Ong IM, and Lamming DW (2019). The metabolic response to a low amino acid diet is independent of diet-induced shifts in the composition of the gut microbiome. Sci. Rep 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Alvarez-Guaita A, Melvin A, Rimmington D, Dattilo A, Miedzybrodzka EL, Cimino I, Maurin AC, Roberts GP, Meek CL, et al. (2019). GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metab 29, 707–718.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Robertson L, Gallinetti J, Mejia P, Vose S, Charlip A, Chu T, and Mitchell JR (2012). Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci. Transl. Med 4, 118ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezeshki A, Zapata RC, Singh A, Yee NJ, and Chelikani PK (2016). Low protein diets produce divergent effects on energy balance. Sci. Rep 6, 25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LT, Treviño-Villarreal JH, Mejia P, Grondin Y, Harputlugil E, Hine C, Vargas D, Zheng H, Ozaki CK, Kristal BS, et al. (2015). Protein and calorie restriction contribute additively to protection from renal ischemia reperfusion injury partly via leptin reduction in male mice. J. Nutr 145, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell NJ, Stock MJ, and Tyzbir RS (1983). Mechanisms of thermogenesis induced by low protein diets. Metabolism 32, 257–261. [DOI] [PubMed] [Google Scholar]
- Smith GI, Yoshino J, Kelly SC, Reeds DN, Okunade A, Patterson BW, Klein S, and Mittendorfer B (2016). High-protein intake during weight loss therapy eliminates the weight-loss-induced improvement in insulin action in obese postmenopausal women. Cell Rep 17, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, and Tomita M (2006). Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem 281, 16768–16776. [DOI] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JWO, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. (2014). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19, 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Mitchell SJ, Coogan SCP, Cogger VC, Gokarn R, McMahon AC, Raubenheimer D, de Cabo R, Simpson SJ, and Le Couteur DG (2015). Dietary protein to carbohydrate ratio and caloric restriction: comparing metabolic outcomes in mice. Cell Rep 11, 1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Mitchell SE, and Mazidi M (2016). Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp. Gerontol 86, 28–38. [DOI] [PubMed] [Google Scholar]
- Stirling JL, and Stock MJ (1968). Metabolic origins of thermogenesis induced by diet. Nature 220, 801–802. [DOI] [PubMed] [Google Scholar]
- Tanca A, Abbondio M, Palomba A, Fraumene C, Marongiu F, Serra M, Pagnozzi D, Laconi E, and Uzzau S (2018). Caloric restriction promotes functional changes involving short-chain fatty acid biosynthesis in the rat gut microbiota. Sci. Rep 8, 14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Morenga L, Docherty P, Williams S, and Mann J (2017). The effect of a diet moderately high in protein and fiber on insulin sensitivity measured using the dynamic insulin sensitivity and secretion test (DISST). Nutrients 9, E1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, and Manning BD (2021). The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife 10, e63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treviño-Villarreal JH, Reynolds JS, Bartelt A, Langston PK, MacArthur MR, Arduini A, Tosti V, Veronese N, Bertozzi B, Brace LE, et al. (2018). Dietary protein restriction reduces circulating VLDL triglyceride levels via CREBH-APOA5-dependent and -independent mechanisms. JCI Insight 3, 99470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulp OL, Krupp PP, Danforth E Jr., and Horton ES (1979). Characteristics of thyroid function in experimental protein malnutrition. J. Nutr 109, 1321–1332. [DOI] [PubMed] [Google Scholar]
- Wanders D, Stone KP, Forney LA, Cortez CC, Dille KN, Simon J, Xu M, Hotard EC, Nikonorova IA, Pettit AP, et al. (2016). Role of GCN2-independent signaling through a noncanonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes 65, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li B, Li L, Mitchell SE, Green CL, D’Agostino G, Wang G, Wang L, Li M, Li J, et al. (2021). Very-low-protein diets lead to reduced food intake and weight loss, linked to inhibition of hypothalamic mTOR signaling, in mice. Cell Metab 33, 1264–1266.e6. [DOI] [PubMed] [Google Scholar]
- Xiao F, Huang Z, Li H, Yu J, Wang C, Chen S, Meng Q, Cheng Y, Gao X, Li J, et al. (2011). Leucine deprivation increases hepatic insulin sensitivity via GCN2/mTOR/S6K1 and AMPK pathways. Diabetes 60, 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap YW, Rusu PM, Chan AY, Fam BC, Jungmann A, Solon-Biet SM, Barlow CK, Creek DJ, Huang C, Schittenhelm RB, et al. (2020). Restriction of essential amino acids dictates the systemic metabolic response to dietary protein dilution. Nat. Commun 11, 2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BP, Masoro EJ, and McMahan CA (1985). Nutritional influences on aging of Fischer 344 rats: I. Physical, metabolic, and longevity characteristics. J. Gerontol 40, 657–670. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata RC, Singh A, Ajdari NM, and Chelikani PK (2018). Dietary tryptophan restriction dose-dependently modulates energy balance, gut hormones, and microbiota in obesity-prone rats. Obesity 26, 730–739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available under the SRA accession PRJNA851959. An interactive app with sequencing data is available at https://macarthur.shinyapps.io/multiomicspr/.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


