ABSTRACT.
This review describes the current understanding of Clostridium (Clostridioides) difficile infection (CDI) in southeast Asia regarding the prevalence of CDI, C. difficile detection methods, antimicrobial susceptibility profiles, and the potential significance of a One Health approach to prevention and control. Our initial focus had been the Indochina region, however, due to limited studies/surveillance of CDI in Indochina, other studies in southeast Asian countries and neighboring Chinese provinces are presented here for comparison. Clostridium (Clostridioides) difficile infection is one of the most common causes of hospital-acquired gastroenteritis worldwide. Since its discovery as a cause of pseudomembranous colitis in 1978, C. difficile-related disease has been more prevalent in high-income rather than low-income countries. This may be because of a lack of knowledge and awareness about the significance of C. difficile and CDI, resulting in underreporting of true rates. Moreover, the abuse of antimicrobials and paucity of education regarding appropriate usage remain important driving factors in the evolution of CDI worldwide. The combination of underreporting of true CDI rates, along with continued misuse of antimicrobial agents, poses an alarming threat for regions like Indochina. C. difficile ribotype (RT) 027 has caused outbreaks in North America and European countries, however, C. difficile RT 017 commonly occurs in Asia. Toxin A-negative/toxin B-positive (A−B+) strains of RT 017 have circulated widely and caused outbreaks throughout the world and, in southeast Asia, this strain is endemic.
INTRODUCTION
Clostridium (Clostridioides) difficile commonly causes toxin-mediated intestinal disease in humans after hospitalization and/or antimicrobial treatment. The outcomes of infection range from asymptomatic colonization to severe conditions such as pseudomembranous colitis (PMC), bowel perforation, toxic megacolon, sepsis, septic shock, and death.1 Hardy spores are usually the infectious form of C. difficile and are commonly found in a variety of environments both in hospital and community settings. These include surfaces in hospitals and long-term care facilities,2 soil and water, and the intestinal tracts of humans and food production animals such as pigs and cattle, often with higher numbers in younger animals.3,4 Spores can persist in the environment for many months and cannot be eradicated with regular disinfectants.2
Toxins A and B produced by C. difficile play a major role in its pathogenicity. The main risk factor for Clostridium (Clostridioides) difficile infection (CDI) is antimicrobial exposure,5 and antimicrobial use plays an important role in the development of CDI-related diseases. To date, CDI has occurred most commonly in high-income countries, where the management of antimicrobial use can be achieved. The regulation of antimicrobials in both medical and agricultural settings is extremely challenging in parts of Asia;6 specifically the Indochina region has poor regulation of antimicrobial usage in both humans and animals.7–10 Furthermore, the level of awareness of CDI among Asian physicians remains limited.11 Hence, the prevalence of CDI in both humans and animals in Asia is assumed to be relatively high.
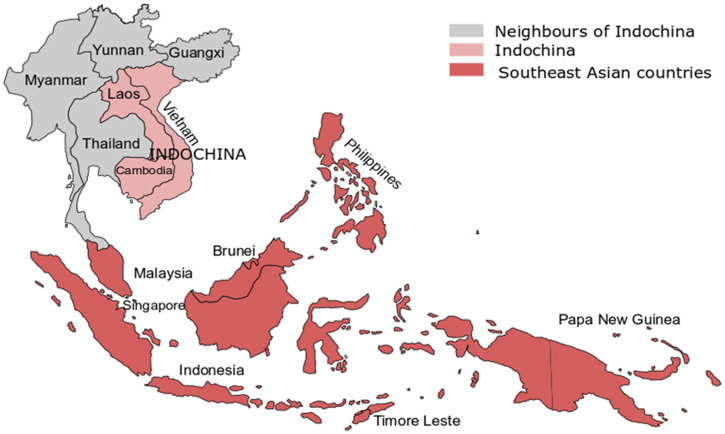
This review describes the epidemiology of CDI in southeast Asia, with a focus on Indochina, a region within that lies between India and China, and includes Cambodia, Lao PDR, and Vietnam. Reports of cases of CDI or CDI outbreaks in this region have been limited. Geographically, neighboring Indochina are Thailand, Myanmar, and two southern Chinese provinces (Yunnan and Guangxi), as illustrated in Figure 1. Other than from Thailand, there have been few published studies from these countries on the incidence or prevalence of C. difficile in hospital or community environments.12–15 Thus, additional information from these southeast Asian countries surrounding Indochina has been included in the review for comparative purposes.
Figure 1.
Indochina includes Cambodia, Laos, and Vietnam. The closest neighbors of Indochina are Myanmar, Thailand, and two Chinese provinces (Yunnan and Guangxi). Southeast Asian countries include all countries on this map, except two Chinese provinces. This figure appears in color at www.ajtmh.org.
HISTORICAL BACKGROUND OF CDI
The first case of the gastrointestinal disease most likely caused by C. difficile was reported at the Johns Hopkins Hospital in Baltimore in 1893 by Finney who described PMC in a 22-year-old patient with hemorrhagic diarrhea after surgery and a diphtheritic membrane in the large bowel at autopsy.16 Clostridium (Clostridioides) difficile was first identified from the meconium and feces of healthy neonates, and named Bacillus difficilis, by Hall and O’Toole in 1935.17 Shortly after it was renamed C. difficile by Prevot18 and, initially, was of little interest as a pathogen. Indeed, in 1940, Snyder described C. difficile as normal intestinal flora, based on his finding that C. difficile comprised 15.4% of the flora of infants from 2 weeks to 1 year of age.19 Clostridium (Clostridioides) difficile was ignored for some 30 years when the toxins were shown to cause a number of biological effects in the early 1970s,20 as well as a cytopathic effect (CPE) in cultured cells (although the investigators thought the CPE was viral rather than a bacterial toxin).21 Finally, C. difficile was shown to be the cause of PMC in a series of papers published in 1978.22–25
For the next 20 years, C. difficile was not often thought of as a major infectious diseases issue; for example, the prevalence of toxigenic C. difficile in diarrheal stool samples in Sweden from 1980 to 1982 was only 1.8%.26 However, between 1980 and 1984, C. difficile accounted for approximately 45% of nosocomial diarrhea in the United States, with the incidence rate of nosocomial gastroenteritis a relatively low 1.3 per 10,000 discharges.27 During the 1980s and 1990s, a significant correlation between an increase of the use of antimicrobials, such as third-generation cephalosporins, and an increase of CDI cases was demonstrated.28,29 In Australia, the prevalence of C. difficile in stool samples rose when third-generation cephalosporins were increasingly used from 1983 to 1992.28 The incidence of community-acquired C. difficile infection (CA-CDI) in the United States was 7.7 cases per 100,000 person-years from 1988 to 199030 and, in 1989, the prevalence of C. difficile associated hospital-acquired infection in the United States was 21%.31 However, from 1996 to 2003, the prevalence of discharges with CDI in the United States almost doubled from 31/100,000 population to 61/100,000 population.32 Similar increases were seen elsewhere in the world. Between 1994 and 2004, the incidence rate of CDI in the United Kingdom increased from below one case per 100,000 persons to 22 cases per 100,000 persons.33 Polymerase chain reaction (PCR) for C. difficile toxin genes became popular for C. difficile detection in clinical samples from the early 1990s,34,35 and this generated more interest in CDI, however, it also came with the problem of overdiagnosis that remains today.
A significant change in CDI epidemiology occurred at the beginning of the current millennium.36 Clostridium (Clostridioides) difficile infection associated with the C. difficile ribotype (RT) 027 strain was first reported as a cause of major outbreaks in South-Eastern Canada in the early 2000s.37 This strain later (or perhaps concurrently) caused outbreaks in the North East of the United States38 spreading throughout North America and crossing the Atlantic to the United Kingdom and then to other European countries where numerous outbreaks were recorded39–42 The prevalence of C. difficile RT 027 infection in many countries was high, particularly in the United Kingdom; RT 027 accounted for 41.3% and 27.5% of all toxigenic C. difficile strains in England and France, respectively.40,42 Most countries invested in improved laboratory diagnostics and infection prevention and control43–46 and a hospital-based survey across 34 European countries finished in 2008 (but published in 2011) recorded a decline in the prevalence of C. difficile RT 027 to as low as 5%, while an increased prevalence was seen for RTs 014/020, 001, and 078.41 Clostridium (Clostridioides) difficile RT 078 had been reported as an emerging strain in The Netherlands around the same time.47 Interestingly, small clusters of C. difficile RT 027 infections occurred in several countries outside North America and Europe such as Australia and Singapore between 2008 and 2009, but the strain did not appear to establish in these countries.48,49 A report of a RT 027 outbreak in Japan was incorrect as it was neither an outbreak nor a “hypervirulent” strain, rather a historical RT 027 strain.50 Over the past two decades, CDI in North America, Europe, and Australia has been better understood, while CDI data from Asia is still limited.
CLOSTRIDIUM DIFFICILE INFECTION IN SOUTHEAST ASIA
In the Asian region in general, the number of publications about C. difficile and CDI has increased substantially over the last 10–15 years; however, given this is where nearly 60% of the world’s population live, a lack of data remains an important problem. A recent publication pertaining to the Asia-Pacific region confirmed that A−B+ strains of C. difficile, predominantly RT 017 (16.4%), and A+B+ strains of RT 014/020 (10.9%) were the most common strains of C. difficile causing CDI from 2014 to 2015. Binary toxin-positive (CDT+) strains of C. difficile were rare (5.3%).51
The prevalence of CDI in southeast Asia appears variable, most likely due to a lack of awareness or a lack of testing (Table 1).52 In the Philippines, for example, an ELISA method detected C. difficile in 43.6% of diarrheal stool samples from patients with colitis. At the time, this prevalence was the highest reported in southeast Asian countries,53 and suggested that CDI was common and might be overlooked in settings where colitis due to Entamoeba histolytica and other intestinal parasites was common.53 In addition, metronidazole therapy for parasitic infections would likely have had some impact on CDI. The following sections contain information from several other southeast Asian countries for comparative purposes.
Table 1.
Summary of the prevalence of Clostridium difficile infection in southeast Asia
| Country | Prevalence % (n) or incidence | Year | Toxigenic C. difficile | Testing method | Reference |
|---|---|---|---|---|---|
| Indonesia | 20.6% (70/340) | 2014–2015 | 10.9% (37/340) | EIA, PCR ribotyping, and toxin profiling | 57 |
| Malaysia | 22.9% (100/437) | 2015–2016 | 10.3% (45/437) | EIA, PCR ribotyping, and toxin profiling | 62 |
| Singapore | 10.7/10,000 patient-days | 2011–2012 | 92.4% (61/66) | EIA, PCR ribotyping | 69 |
| Thailand | 23% (100/422) | 2015 | 9.2% (39/422) | BD MAX CDiff assay, PCR ribotyping, and toxin gene profiling | 75 |
| China | 14.1% (138/978) | 2013–2016 | 14.1% (138/978) | MLST, PCR ribotyping, and toxin gene profiling | 84 |
| Vietnam | 24.9% (95/382) | 2013–2015 | 24.9% (95/382) | Nested PCR, and slpA sequence typing | 12 |
| Cambodia | 3.75% (3/80) | 2005 | 3.75% (3/80) | Toxin A-EIA | 14 |
| Lao PDR | 8.6% (6/70) | 2013 | 60% (3/5) | EIA, PCR ribotyping, and toxin gene profiling | 15 |
EIA = enzyme immunoassay; PCR = polymerase chain reaction.
Indonesia.
Publications on CDI in Indonesia are not common. In a study on the etiology of pediatric diarrheal diseases in Jakarta published in 2002, 1.3% (2/154) of samples were positive for toxin A using the Premier C. difficile Toxin A Enzyme Immunoassay (EIA) (Meridian Diagnostic Inc., USA).54 As this EIA detected toxin A only, the true prevalence of C. difficile was likely to have been higher. Later in 2014, a rapid membrane EIA (C. Diff Quik Chek Complete, Techlab) was used in a small cross-sectional study of elderly patients with hepatocellular carcinoma in Banten. The prevalence of nosocomial CDI in those patients was 25% (4/16 patients) and stool samples were positive for both glutamate dehydrogenase (GDH) and toxins A/B.55 A more recent study combined multiple methods, including C. difficile selective agar or chromogenic agar culture, MALDI-ToF mass spectrometry, EIA (Quik Chek Complete) or Serazym® C. difficile toxin A+B immunoassay, multiplex PCR, and PCR ribotyping.56 The prevalence of symptomatic CDI was 14.7% (25/170).56 A total of 14 different RTs was identified among C. difficile isolates with SLO160 and RT 017 dominating (45.5%). Furthermore, toxin profiling showed tcdA+/tcdB+ accounted for 63.6% of C. difficile strains; the prevalence of RT 017 (tcdA−/tcdB+) was 18.2%.56
Between July 2014 and February 2015 in four hospitals in Central Java, diarrheal stool samples were tested with an EIA for GDH and toxins A/B.57 The prevalence of samples positive for GDH alone and for both GDH and toxins A/B was 20.6% (70/340) and 5.6% (19/340), respectively.57 After culture, 74 C. difficile strains were isolated and the prevalence of toxigenic C. difficile strains was 10.9% (37/340). The most common toxigenic C. difficile strains were RT 017 (A−B+) (24.3%), QX134 (A−B+) (4.1%), RT 053 (A+B+) (4.1%), and QX215 (A+B+) (4.1%), however, there were no CDT+ strains.57 In a recent small study, 1/31 diarrheal patients (3%) who received at least 2 days of antimicrobial therapy was positive for both GDH and toxins A/B by a rapid lateral flow immunoassay (IMMUNOQUICK Tox A/B).58 There was no further investigation in this study.
Malaysia.
Like Indonesia, few reports of CDI in Malaysia have been published. The earliest of these, in 1998, was a series of seven C. difficile-associated diarrhea (CDAD) cases with comorbidities.59 Clostridium (Clostridioides) difficile isolates from these cases were phenotypically identified by colonial morphology on C. difficile agar, Gram staining, chartreuse-green fluorescence under long-wavelength ultraviolet light and fatty acid profiles. Additionally, C. difficile toxins were detected by tissue culture assay.59 In a 2008 study from northeastern Malaysia, an immunochromatographic kit (Remel Xpect, Oxoid, United Kingdom) and cell cytotoxicity assays were used to test 175 stool samples from antimicrobial-associated diarrhea (AAD) inpatients. The prevalence of C. difficile toxins was 13.7% and CDI was most common in ethnic Malays;60 molecular typing was not performed. In 2015, a molecular component was added to a study conducted on inpatients in a hospital in Kota Bharu and showed the prevalence of CDAD in inpatients was 13%. The predominant toxigenic strains of C. difficile were RT 043 and RT 017 (both 14%, 3/22 strains).61 In a more recent report from 2018, across four hospitals in Kuala Lumpur and Kota Bharu, the prevalence of toxigenic strains from diarrheal stool samples was 10.3%.62 Of the toxigenic strains, RTs 017 (A−B+CDT−) and 043 (A+B+CDT-) accounted for 20% (20 isolates) and 10% (10), respectively. Furthermore, the remaining four toxigenic C. difficile strains producing both toxins A and B were RT 053, QX 026, and RT 014/020.62 No binary toxin-producing strains were reported.62
Singapore.
Singapore has the most advanced medical technology and research institutions in southeast Asia.63 The earliest finding of C. difficile in Singapore was in an etiological study of diarrhea in which 4,508 diarrheal stool samples were examined over a 50-month period from 1985 to 1989. Clostridium (Clostridioides) difficile was recovered from 9.6% (35/365) of stool samples that were requested for C. difficile investigation.64
According to Lim et al., between 2001 and 2006, CDAD incidence in a 1,200-bed general hospital in Singapore dramatically increased from 1.49 cases per 10,000 patient-days to 6.64 cases per 10,000 patient-days, with a simultaneous increase in toxin detection in stool samples.65 The incidence of CDI in a larger hospital was 5.38 cases per 10,000 patient-days from 2002 to 2003, and CDI was more common in ethnic Malays and patients aged above 50 years. Of 118 C. difficile isolates, 14 (11.8%) produced toxin B only and were presumably RT 017.66 Based on several publications and the Network for Antibiotic Resistance Surveillance in Singapore, the incidence of CDI in inpatients gradually dropped until 2010.49,67,68 The detection of C. difficile RT 027 in Singapore was reported in 2008.49 Since then, molecular and ribotyping techniques have been widely implemented and, consequently, more data on CDI in Singapore hospitals has been available.
From December 2011 to May 2012, the incidence of CDI significantly increased to 10.7 cases per 10,000 patient-days. Clostridium (Clostridioides) difficile RTs 053 (21.3%) and 012 (18%) were common, followed by RTs 014, 020, 017, 002, 087, and 064-like that ranged in prevalence from 1.6% to 9.8%; however, 31.1% of RTs remained unclassified.69 Between 2013 and 2014, nucleic acid tests for the C. difficile toxin B gene and immunochromatography kits were combined to identify the causes of community-acquired diarrhea in adult patients admitted to the hospital for acute gastroenteritis. Of 100 samples examined, only 2% contained toxigenic C. difficile strains.70
Thailand.
Thailand has a longer history of C. difficile research than other countries in southeast Asia. Siriraj Hospital, which is the oldest and largest hospital in Thailand, was responsible for conducting several of these studies. The first CDAD research in Thailand was carried out at Siriraj Hospital, Ramathibodi Hospital, and a children’s hospital in 1990 using culture on selective media (CCFA, Oxoid) and a cytotoxin assay. The prevalence of cytotoxin-positive samples in patients aged < 60 years was 52.5% (106 of 203 samples), but the proportion of C. difficile in diarrheal stool samples from all age groups was only 4.8% (13/269) by culture.71 Cytotoxin positivity increased to 61% in samples from patients who received antimicrobial treatment, compared with 51% in samples from patients without antimicrobial treatment.71 A later study in Siriraj Hospital reported that the prevalence of toxin A-positive C. difficile in patients with clindamycin (10.7%) or beta-lactams (10%) treatment was higher than the control group (1.4%).72 Between 2000 and 2001, 574 fecal samples from in-patients with suspected AAD in Siriraj hospital and other hospitals in Thailand were analyzed.73 The prevalence of C. difficile culture-positivity was 18.6% (107/574 stool samples).73 Of C. difficile isolates, the proportions of detection of tcdA and tcdB by PCR and toxin A/B by EIA were 44.9% and 46.7%, respectively.73 A combination of EIA for toxins A and B and direct stool PCR for tcdB more than doubled the rate of detection compared with when EIA alone was used.74
In Siriraj Hospital in 2015, the prevalence of toxigenic C. difficile in samples was 9.2% (39/422).75 Toxin profiles of toxigenic C. difficile strains, A+B+ and A−B+, were 69.2% (27/39) and 30.8% (12/39), respectively. By PCR ribotyping, RTs 014/020, 010, 017, 039, and 009 (53.3%) were predominant RTs among 38 RTs, noting that RTs 009, 010, and 039 are nontoxigenic.75 In a 2019 study which covered over 13 provinces in Thailand, 51% (74/145) of all C. difficile isolates were toxigenic. Of those toxigenic strains, the most common were RTs 017 (A−B+CDT−) and 014/020 (A+B+CDT−), which accounted for 19% (28/145) and 7% (10/145), respectively.76
The first publication on CDI among immunosuppressed patients was in 1998.77 Based on an ELISA method, toxin A was present in 36.7% (11/30) of stool samples from pediatric patients with febrile neutropenia at Siriraj Hospital. Chemotherapy and cephalosporins were the risk factors for C. difficile colonization in neutropenic malignancy pediatric patients.77 At King Chulalongkorn Memorial Hospital from 2002 to 2005, toxin A-positive stool samples from hospitalized patients with CDAD were analyzed.78 Approximately 90% of positive samples were from patients with comorbidities, mostly immunosuppression, such as those with hematologic malignancies, solid tumors, diabetes, HIV infection, cirrhosis, and chronic renal failure. All patients received antimicrobial therapy within 60 days of being diagnosed with CDAD, and oral metronidazole was commonly prescribed for treatment. Furthermore, the study also concluded chemotherapy and gastric anti-acid agents were risk factors for CDAD.78
CLOSTRIDIUM DIFFICILE INFECTION IN INDOCHINA AND ITS NEIGHBORS
Cambodia.
There have been no studies specifically looking at C. difficile in Cambodia. In 2005, there was an investigation of chronic diarrhea in Cambodian patients with HIV/AIDS that identified toxin A-producing C. difficile in 3.75% (3/80) of stool samples with an immunoassay.14 Chronic diarrhea was defined as > 3 loose stools per day for at least 30 days. Of C. difficile positive stool samples, 5% (2/40) were from chronic diarrhea patients, and 2.5% (1/40) were from nonchronic diarrhea patients.14 The study was limited to toxin A identification only and groups of HIV/AIDS patients with chronic and nonchronic diarrhea and suffered from a lack of clarity in defining CDI. There was no molecular typing and therefore no information on the strains of C. difficile circulating in Cambodia.
Lao PDR.
Like Cambodia, very little has been published about CDI in Lao PDR. In 2013, a small survey was undertaken on diarrheal stool samples being routinely examined. ChromID C. difficile agar (BioMerieux, France) was used for anaerobic culture, latex agglutination (C. difficile latex test kit, Oxoid, United Kingdom) for identification of isolates, and ribotyping and toxin profiling were performed at a reference laboratory in Australia.15 By culture and latex agglutination, five of 70 fecal samples were positive for C. difficile. Of the five isolates, three were positive for tcdB and two were positive for tcdA by PCR. The five isolates were identified as five different RTs: 014 (A+B+CDT−), 020 (A+B+CDT−), 017 (A−B+CDT−), QX107 (A−B−CDT−), and QX574 (A−B−CDT−). They were from patients aged 1 to 46 years most of whom had diarrheal symptoms.15 However, two cases whose stool samples were positive for toxin B-producing C. difficile were concurrently infected with Salmonella species, and/or Burkholderia pseudomallei, and had a history of antimicrobial exposure.15
Vietnam.
Vietnam has conducted more studies on the epidemiology of CDI than the other two countries in Indochina, although some have been reported in Vietnamese. Diarrhea was usually defined as ≥ 3 loose stools per day based on the definition of the WHO.79 In one investigation of diarrhea, 9% (45/479) of stool samples from inpatients were positive for toxigenic C. difficile, using the Luminex xTAG Gastrointestinal Pathogen Panel assay.13 Of the toxigenic strains, the prevalence of toxin A-positive C. difficile strains was twice as high as toxin B-positive C. difficile strains. The prevalence of nontoxigenic or binary toxin-positive C. difficile strains was not reported, and there was no further analysis, such as molecular typing.
A study of CDI among patients with AAD in northern Vietnam used culture on cycloserine–cefoxitin–mannitol agar, nested PCR, PCR ribotyping, and surface-layer protein A (slpA) typing. Antimicrobial-associated diarrhea was defined as loose stools and a history of antimicrobial exposure within 4 weeks. The prevalence of CDI among patients with AAD was 24.9% (95/382), and stool samples from seven fatal cases were positive for toxigenic C. difficile.12 Of the toxigenic isolates, toxin A- and B-producing C. difficile strains were found in 8.1% (31/382) of all diarrheal stool samples and 10.2% (39/382) were only toxin B-producing strains. Clostridium (Clostridioides) difficile RTs 017 A−B+ and trf A−B+ (53.5%) were common among the eight RTs identified by PCR ribotyping. By slpA typing, the most common strain was fr-01, followed by kr-03.1 and og39-01.12
From 2013 to 2015, C. difficile RT 017 was the most common (9/30) of six RTs, including trf, og39, cc835, 001 and cr (using Japanese nomenclature).80 Research in Bach Mai Hospital from 2013 to 2017 identified 107 toxigenic C. difficile strains from 101 adult patients with diarrhea.81–83 The definition of CDI included detection of toxigenic C. difficile and diarrhea ≥ 3 times per day. Various acceptable methods were used including GDH EIA (Nissui, Tokyo, Japan), multiplex PCR for tcdA, tcdB, and triosephosphate isomerase (tpi) genes, and PCR ribotyping. The PCR RTs were aligned with other RTs in the database of the Department of Bacteriology II, National Institute of Infectious Diseases, Japan.82 The prevalence of CDI patients positive for toxins A+B+ and A−B+ was 49.5% and 44.6%, respectively. Six cases (5.9%) had two types of toxigenic C. difficile strains (A+B+ and A−B+).82,83 Among eight PCR RTs, RTs trf, 017, and cc835 were 24.5%, 23.5%, and 22.6% prevalent, respectively,82,83 RTs 027 and 078 were not detected.
Thus, although studies of CDI in Vietnam are still limited, there has been some information published on the prevalence of CDI in diarrheal patients together with ribotyping and toxin gene data.
China.
China is immediately north of the Indochina region and two provinces, Yunnan and Guangxi, share borders with Indochina. There have no published reports on CDI in Guangxi, however, some work has been done in Yunnan. Yunnan province shares its border with Laos and Vietnam. From 2013 to 2016, a retrospective study of community-acquired CDI in Yunnan used a variety of techniques, including PCR (Tiangen, Beijing), PCR ribotyping, toxin gene profiling, and MLST. By PCR, the prevalence of tcdA and/or tcdB in 978 fecal samples was 14.11% (138 samples).84 However, from those 138 fecal samples, only 55 C. difficile strains were cultured, 87.3% of which (48/55) were toxigenic. Nine RTs were identified; RTs 001, 009, 010, 012, 046, 085, 140, 207, and 220, and the remaining isolates were unclassified.84 Strains ST3, ST35, and ST54 were the most common among 15 ST types, by MLST.84 Although there is only one available study, it provides a broad picture of CDI in Yunnan province.
SUMMARY OF C. DIFFICILE RIBOTYPING DATA FROM THE INDOCHINA REGION
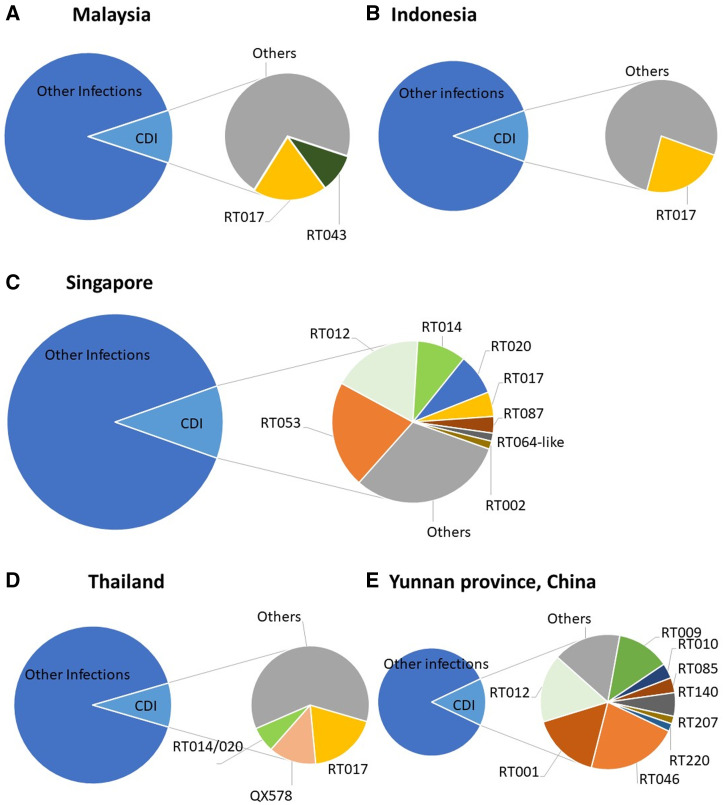
The most common C. difficile RTs seen in southeast Asia were RTs 017, 014/020, 043, and 053. Other RTs, 009, 010, and 012 (009 and 010 being nontoxigenic) were reported in Indonesia, Malaysia, Singapore, Thailand, and Yunnan province, as shown in Figure 2. Although C. difficile RT 027 strains have caused many outbreaks in high-income countries,85 and were introduced in Singapore in 2008,49 they have not established nor caused epidemics in southeast Asia.
Figure 2.
(A) Malaysia: N = 100 isolates, from July 2015 to July 2016; over 32 ribotypes.62 (B) Indonesia: N = 74 isolates, from July 2014 to February 2015; 26 ribotypes.57 (C) Singapore: N = 61 isolates, from December 2011 to May 2012; 8 ribotypes.69 (D) Thailand: For the prevalence of CDI (N = 105 isolates, April–June 2015; 38 ribotypes),75 for ribotyping analysis (N = 145 isolates, from 2006 to 2018; 40 ribotypes).76 (E) Yunnan province, China: N = 55 isolates, from 2013–2016; 9 ribotypes.84 This figure appears in color at www.ajtmh.org.
These findings were supported by a large surveillance study that found C. difficile RTs 017, 018/QX239, 014/020, and 002 were the most common RTs among 79 RTs in the Asia-Pacific region. Of the toxigenic strains, toxin A-negative and toxin B-positive C. difficile strains mostly belonged to RT 017.51 Clostridium (Clostridioides) difficile RT 017 A−B+ strains have circulated in southeast Asia for many years and caused epidemics in other countries in the world.86,87 There is still controversy about the origin of RT 017 strains in southeast Asia. It was thought that these strains had arisen in Asia due to their high prevalence in the region, belonged to clade 4 and had spread to other parts of the world causing outbreaks.88 However, based on whole genome sequencing (WGS) and single-nucleotide polymorphism analysis,87 Cairns et al. suggested that C. difficile RT 017 originated in North America and then spread to Europe, Asia, and Australia. A more recent analysis has again supported the theory of an origin in Asia.89
C. DIFFICILE IN ANIMALS IN SOUTHEAST ASIA
Studies on C. difficile in animals in southeast Asia are minimal. In 2015/2016, a molecular epidemiology study of the prevalence of C. difficile in piglets from Thailand and Malaysia was performed using culture on ChromID agar and selective enrichment culture, followed by toxin profiling, PCR ribotyping, and cgSNV analysis following WGS. The prevalence of C. difficile in the gastrointestinal tracts of piglets from Malaysia was 91.6%, while the prevalence of C. difficile in Thai piglets was 35.1%.90
On average, approximately 90% of environmental samples (soils and water) from pig farms were positive for C. difficile. Clostridium (Clostridioides) difficile RT 038 was the most prevalent RT among six RTs recovered. All C. difficile strains from both piglets and the environment were nontoxigenic.90 Clostridium (Clostridioides) difficile RT 038 strains from piglets in Thailand and Malaysia belonged to clade 1 ST48 by in silico MLST. In addition, RT 038 strains from humans selected from Thailand and Indonesia for comparison differed by a relatively small 30 cgSNVs compared with animal strains.90
In a recent animal study from Yunnan province, two toxigenic C. difficile strains (RT 126, A+B+CDT+ and ICDC 094, A+B+CDT−) were recovered from 200 stool samples from adult sheep.91 In another animal study in Japan, 57.5% of fecal samples from neonatal piglets were positive for C. difficile. Among C. difficile isolates, 61% were toxin A and B− producing strains and 42.6% were binary toxin-producing.92 Clostridium (Clostridioides) difficile RT 078 was the third-most prevalent strain among 14 RTs and was genetically related to European RT 078 strains, which caused outbreaks in humans and pigs in Europe. The ancestors of these pigs most likely originated in Europe and North America and were brought to Japan and other Asian countries for breeding.92
ANTIMICROBIAL SUSCEPTIBILITY PROFILE OF C. DIFFICILE
While there is little data on the antimicrobial susceptibility of C. difficile in Indochina specifically, there are some good data from the southeast Asian region in general, and particularly from Thailand. In a 2015 Thai study, 53 C. difficile human strains, isolated from 2006 to 2008, were tested using the gradient strip method against metronidazole (100% susceptible), vancomycin (98.2%), moxifloxacin (54.8%), tigecycline (100%), and daptomycin (100%).93 In a bigger/longer study of 100 strains isolated from 2006 to 2015 used an agar dilution method, the antimicrobial susceptibility of C. difficile remained stable for antimicrobials such as metronidazole and vancomycin used for CDI treatment. However, susceptibility decreased for clindamycin (4% susceptible), moxifloxacin (61%), tetracycline (82%), and chloramphenicol (88%). For the 22 C. difficile RT 017 strains, the most common RT, the proportions susceptible were clindamycin (14%), moxifloxacin (23%), tetracycline (55%), and chloramphenicol (100%).76
In Singapore, all C. difficile isolates were susceptible to metronidazole,64,66 however, a vancomycin-resistant C. difficile strain was reported in 2007.66 After C. difficile RT 027 emerged in Singapore, multidrug resistance in RT 027 was observed; resistant to clindamycin and levofloxacin, but susceptible to erythromycin and moxifloxacin.49 Last, reports from Laos and Indonesia were examined; five C. difficile isolates from Laos were susceptible to moxifloxacin, metronidazole, and vancomycin, but not clindamycin15 whereas, in Indonesia, 24.2% of C. difficile isolates, mainly RT 017, were resistant to moxifloxacin.56 The gradient strip method was commonly used in these studies.
In a recent report from 12 Asia-Pacific countries, all C. difficile strains, which were tested using agar dilution, were susceptible to metronidazole, vancomycin, fidaxomicin, and amoxicillin-clavulanate, whereas most were resistant to clindamycin, erythromycin, and moxifloxacin. The highest rates of resistance to at least three antimicrobials were found in RT 017 (66.1%), RT 018 (92.7%), RT 369 (100%), and RT QX239 (100%).94
ONE HEALTH
“One Health” focuses on the relationships between the health of humans, animals, and the environment, and encompasses food safety, the control of zoonotic diseases and combatting antimicrobial resistance.95 Physicians in many parts of the world think of CDI as a hospital-acquired infection only and neglect community-acquired infections, however, a link between some toxigenic C. difficile RTs seen in humans and those in animals and the environment has been observed.96–100 There is emerging evidence suggesting sources of CDI from the community and environment play an important role in transmission. In a United Kingdom study based on WGS and core-genome single nucleotide variant differences, there were only 35% (333/957) of all isolates that were genetically associated with previous cases, while 45% (428/957) of isolates were genetically different from previous cases.98
Clostridium (Clostridioides) difficile RT 078 and RT 014 are considered to be zoonotic strains as they and RT 027 have been detected in food, animals, vegetables, households, and seafood.99 Based on the comparison of WGS and MLST of C. difficile strains from human and animal samples, 42% of C. difficile RT 014 strains from human samples in Australia have a clonal relationship with at least one porcine strain.100 Australian piglets predominantly carry C. difficile RT 014, which also commonly causes CDI in humans in many parts of the world.100 Furthermore, based on outbreaks of C. difficile RT 078 infection in humans in Europe and the presence of this strain in pigs and cattle, C. difficile RT 078 has been suggested as a potentially zoonotic pathogen.96 Furthermore, some southeast Asian countries where the prevalence of both toxigenic C. difficile in hospitalized patients is high are popular for medical tourism. Therefore, there is a risk of C. difficile transmission to many parts of the world.86
Apart from C. difficile RT 078, other binary toxin-positive C. difficile strains were also thought to be zoonotic. Rupnik reported the prevalence of binary toxin-positive C. difficile strains in animals, including horses, piglets, and cattle, ranged from 23% to 100%. Those binary toxin-positive strains were probably spread from animals to humans as the prevalence of HA-CDI and CA-CDI caused by binary toxin-positive strains in humans has increased since the 1990s.101 In addition to this, some studies in Scotland and the United States reported the presence of C. difficile RTs 017, 027, and 078 in food such as salads, uncooked meat, and several types of sausage.102,103
In terms of a One Health perspective, the most important requirement is to closely monitor production animals and antimicrobial use in these animals, particularly for infection prophylaxis.86
CONCLUSION
Very little is known about the epidemiology of C. difficile in humans, animals, and the environment in southeast Asia. A lack of diagnostics and awareness of physicians are probably the main factors that restrict reports on CDI from this region. In addition, inappropriate use of antimicrobials in animals and humans is a significant concern in southeast Asia. Consequently, this may increase the prevalence of CDI. Although RTs vary from one country to another in southeast Asia, toxin A-negative/toxin B-positive strains of C. difficile RT 017 are endemic throughout the region and have caused outbreaks in other parts of the world. Virulent strains of C. difficile such as RT 078 have been detected from animals that were brought from Europe to northern Asia, however, these strains do not appear to have caused CDI in humans or to be circulating in animals in southeast Asia. One exception is Taiwan where C. difficile RT 078 strains have been found in piglets and caused CDI in humans.104 Improvement in laboratory capacity and surveillance in the region is essential to reduce morbidity and mortality from CDI. More important, regulation of antimicrobial use should be improved to decrease both multidrug resistance and CDI.
REFERENCES
- 1. Rupnik M Wilcox MH Gerding DN , 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7: 526–536. [DOI] [PubMed] [Google Scholar]
- 2. Vonberg RP et al. 2008. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect 14: 2–20. [DOI] [PubMed] [Google Scholar]
- 3. Knight DR Thean S Putsathit P Fenwick S Riley TV , 2013. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol 79: 2630–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knight DR Squire MM Riley TV , 2015. Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes. Appl Environ Microbiol 81: 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jawa RS Mercer DW , 2012. Clostridium difficile-associated infection: a disease of varying severity. Am J Surg 204: 836–842. [DOI] [PubMed] [Google Scholar]
- 6. Zellweger RM Carrique-Mas J Limmathurotsakul D Day NPJ Thwaites GE Baker S , 2017. A current perspective on antimicrobial resistance in Southeast Asia. J Antimicrob Chemother 72: 2963–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Om C Daily F Vlieghe E McLaughlin JC McLaws ML , 2017. Pervasive antibiotic misuse in the Cambodian community: antibiotic-seeking behaviour with unrestricted access. Antimicrob Resist Infect Control 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quet F et al. 2015. Antibiotic prescription behaviours in Lao People’s Democratic Republic: a knowledge, attitude and practice survey. Bull World Health Organ 93: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Om C McLaws ML , 2016. Antibiotics: practice and opinions of Cambodian commercial farmers, animal feed retailers and veterinarians. Antimicrob Resist Infect Control 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thu TA Rahman M Coffin S Harun-Or-Rashid M Sakamoto J Hung NV , 2012. Antibiotic use in Vietnamese hospitals: a multicenter point-prevalence study. Am J Infect Control 40: 840–844. [DOI] [PubMed] [Google Scholar]
- 11. Mavros MN Alexiou VG Vardakas KZ Tsokali K Sardi TA Falagas ME , 2012. Underestimation of Clostridium difficile infection among clinicians: an international survey. Eur J Clin Microbiol Infect Dis 31: 2439–2444. [DOI] [PubMed] [Google Scholar]
- 12. Duong DTT , 2017. Determination of Infection Rates and Some Molecular Epidemiological Characteristics of Toxin Gene Carried Clostridium difficile Strains Isolated from Patients with Diarrhea after Antibiotic Treatment in Four Hospitals in Hanoi from 2013 to 2015. Hanoi, Vietnam: Department of Microbiology, Vietnam University of Science.
- 13. Duong VT et al. 2016. Evaluation of Luminex xTAG gastrointestinal pathogen panel assay for detection of multiple diarrheal pathogens in fecal samples in Vietnam. J Clin Microbiol 54: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chhin S Harwell JI Bell JD Rozycki G Ellman T Barnett JM Ward H Reinert SE Pugatch D , 2006. Etiology of chronic diarrhea in antiretroviral-naive patients with HIV infection admitted to Norodom Sihanouk Hospital, Phnom Penh, Cambodia. Clin Infect Dis 43: 925–932. [DOI] [PubMed] [Google Scholar]
- 15. Cheong E Roberts T Rattanavong S Riley TV Newton PN Dance DAB , 2017. Clostridium difficile infection in the Lao People’s Democratic Republic: first isolation and review of the literature. BMC Infect Dis 17: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finney JMT , 1893. Gastroenterostomy for cicatrizing ulcer of the pylorus. Bull Johns Hopkins Hosp 4: 53–55. [Google Scholar]
- 17. Hall IC O’Toole E , 1935. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. AMA Am J Dis Child 49: 390–402. [Google Scholar]
- 18. Prévot AR , 1938. Études de systématique bactérienne. IV. Critique de la conception actuelle du genre Clostridium. Ann Inst Pasteur (Paris) 61: 72–91. [Google Scholar]
- 19. Snyder ML , 1940. The normal fecal flora of infants between two weeks and one year of age. J Infect Dis 66: 1–16. [Google Scholar]
- 20. Hafiz S , 1974. Clostridium difficile and Its Toxins. Leeds, United Kingdom: Department of Microbiology, Biological Sciences, University of Leeds.
- 21. Green RH , 1974. The association of viral activation with penicillin toxicity in guinea pigs and hamsters. Yale J Biol Med 47: 166–181. [PMC free article] [PubMed] [Google Scholar]
- 22. Larson HE Price AB , 1977. Pseudomembranous colitis: presence of clostridial toxin. Lancet 310: 1312–1314. [DOI] [PubMed] [Google Scholar]
- 23. Bartlett JG Onderdonk AB Cisneros RL Kasper DL , 1977. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis 136: 701–705. [DOI] [PubMed] [Google Scholar]
- 24. Bartlett JG Chang TW Gurwith M Gorbach SL Onderdonk AB , 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med 298: 531–534. [DOI] [PubMed] [Google Scholar]
- 25. George WL Goldstein EJC Sutter VL Ludwig SL Finegold SM , 1978. Etiology of antimicrobial agent associated colitis. Lancet 311: 802–803. [DOI] [PubMed] [Google Scholar]
- 26. Aronsson B Mollby R Nord CE , 1985. Antimicrobial agents and Clostridium difficile in acute enteric disease: epidemiological data from Sweden, 1980–1982. J Infect Dis 151: 476–481. [DOI] [PubMed] [Google Scholar]
- 27. Hughes JM Jarvis WR , 1987. Nosocomial gastrointestinal infections. In: Wenzel RP. (ed.), Prevention and Control of Nosocomial Infections. Baltimore, MD: Williams & Wilkins, 405–439. [Google Scholar]
- 28. Riley TV O’Neill GL Bowman RA Golledge CL , 1994. Clostridium difficile-associated diarrhoea: epidemiological data from Western Australia. Epidemiol Infect 113: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomas C Stevenson M Williamson DJ Riley TV , 2002. Clostridium difficile-associated diarrhea: epidemiological data from Western Australia associated with a modified antibiotic policy. Clin Infect Dis 35: 1457–1462. [DOI] [PubMed] [Google Scholar]
- 30. Hirschhorn LR Trnka Y Onderdonk A Lee MLT Platt R , 1994. Epidemiology of community-acquired Clostridium difficile-associated diarrhea. J Infect Dis 169: 127–133. [DOI] [PubMed] [Google Scholar]
- 31. McFarland LV Mulligan ME Kwok RY Stamm WE , 1989. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med 320: 204–210. [DOI] [PubMed] [Google Scholar]
- 32. McDonald LC Owings M Jernigan DB , 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg Infect Dis 12: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dial S Delaney JA Barkun AN Suissa S , 2005. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA 294: 2989–2995. [DOI] [PubMed] [Google Scholar]
- 34. Wren B Clayton C Tabaqchali S , 1990. Rapid identification of toxigenic Clostridium difficile by polymerase chain reaction. Lancet 335: 1. [DOI] [PubMed] [Google Scholar]
- 35. Kato N Ou CY Kato H Bartley SL Brown VK Dowell VR Ueno K , 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction. J Clin Microbiol 29: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clements ACA Magalhaes RJS Tatem AJ Paterson DL Riley TV , 2010. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect Dis 10: 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pepin J Valiquette L Alary ME Villemure P Pelletier A Forget K Pepin K Chouinard D , 2004. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171: 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muto CA et al. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect Control Hosp Epidemiol 26: 273–280. [DOI] [PubMed] [Google Scholar]
- 39. He M et al. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brazier JS Raybould R Patel B Duckworth G Pearson A Charlett A Duerden BI Network HPARM , 2008. Distribution and antimicrobial susceptibility patterns of Clostridium difficile PCR ribotypes in English hospitals, 2007–08. Euro Surveill 13: 13. [DOI] [PubMed] [Google Scholar]
- 41. Bauer MP Notermans DW van Benthem BHB Brazier JS Wilcox MH Rupnik M Monnet DL van Dissel JT Kuijper EJ , 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377: 63–73. [DOI] [PubMed] [Google Scholar]
- 42. Kuijper EJ et al. 2008. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill 13: 1–7. [PubMed] [Google Scholar]
- 43. Adler A Schwartzberg Y Samra Z Schwartz O Carmeli Y Schwaber MJ , 2014. Trends and changes in Clostridium difficile diagnostic policies and their impact on the proportion of positive samples: a national survey. Clin Microbiol Infect 20: O904–O910. [DOI] [PubMed] [Google Scholar]
- 44. Riley TV Huovinen P , 2008. Infection control measures to limit the spread of Clostridium difficile. Introduction. Clin Microbiol Infect 14: 1. [DOI] [PubMed] [Google Scholar]
- 45. Katz KC et al. 2018. The evolving epidemiology of Clostridium difficile infection in Canadian hospitals during a postepidemic period (2009–2015). CMAJ 190: E758–E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gessel HV , 2008. Measuring the incidence of Clostridium difficile-associated diarrhoea in a group of Western Australian hospitals. Healthc Infect 13: 56–62. [Google Scholar]
- 47. Goorhuis A Bakker D Corver J Debast SB Harmanus C Notermans DW Bergwerff AA Dekker FW Kuijper EJ , 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 47: 1162–1170. [DOI] [PubMed] [Google Scholar]
- 48. Riley TV Thean S Hool G Golledge CL , 2009. First Australian isolation of epidemic Clostridium difficile PCR ribotype 027. Med J Aust 190: 706–708. [DOI] [PubMed] [Google Scholar]
- 49. Lim PL et al. 2011. Isolation of the first three cases of Clostridium difficile polymerase chain reaction ribotype 027 in Singapore. Singapore Med J 52: 361–364. [PubMed] [Google Scholar]
- 50. Collins DA Riley TV , 2018. Clostridium difficile guidelines. Clin Infect Dis 67: 1639. [DOI] [PubMed] [Google Scholar]
- 51. Collins DA Sohn KM Wu Y Ouchi K Ishii Y Elliott B Riley TV Tateda K , 2020. Clostridioides difficile infection in the Asia-Pacific region. Emerg Microbes Infect 9: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collins DA Hawkey PM Riley TV , 2013. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Warren CA Labio E Destura R Sevilleja JE Jamias JD Daez ML , 2012. Clostridium difficile and Entamoeba histolytica infections in patients with colitis in the Philippines. Trans R Soc Trop Med Hyg 106: 424–428. [DOI] [PubMed] [Google Scholar]
- 54. Oyofo BA et al. 2002. Enteropathogens associated with acute diarrhea in community and hospital patients in Jakarta, Indonesia. FEMS Immunol Med Microbiol 34: 139–146. [DOI] [PubMed] [Google Scholar]
- 55. Kurniawan A Lugito NPH Yanto TA Tjiang MM Setiadinata R Wijaya I Soemantri S , 2014. Clostridium difficile infection in elderly hepatocellular carcinoma patients in general hospital, Karawaci, Tangerang, Banten, Indonesia. J Geriatr Oncol 5: S17. [Google Scholar]
- 56. Seugendo M et al. 2018. Prevalence and strain characterization of Clostridioides (Clostridium) difficile in representative regions of Germany, Ghana, Tanzania and Indonesia - a comparative multi-center cross-sectional study. Front Microbiol 9: 1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Collins DA Gasem MH Habibie TH Arinton IG Hendriyanto P Hartana AP Riley TV , 2017. Prevalence and molecular epidemiology of Clostridium difficile infection in Indonesia. New Microbes New Infect 18: 34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chrisnanda RH Wardhani P , 2019. The incidence of Clostridium difficile infection in diarrhea patients after receiving antibiotics at Dr. Soetomo Hospital Surabaya. Bali Med J 8: 342–346. [Google Scholar]
- 59. Parasakthi N Puthucheary SD Goh KL Sivanesaratnam V , 1988. Clostridium difficile associated diarrhoea: a report of seven cases. Singapore Med J 29: 504–507. [PubMed] [Google Scholar]
- 60. Hassan SA Othman N Idris FM Abdul Rahman Z Maning N Abdul Rahman R Tiong CG , 2012. Prevalence of Clostridium difficile toxin in diarhoeal stool samples of patients from a tertiary hospital in North Eastern Penisular Malaysia. Med J Malaysia 67: 402–405. [PubMed] [Google Scholar]
- 61. Zainul NH Ma ZF Besari A Siti Asma H Rahman RA Collins DA Hamid N Riley TV Lee YY , 2017. Prevalence of Clostridium difficile infection and colonization in a tertiary hospital and elderly community of north-eastern Peninsular Malaysia. Epidemiol Infect 145: 3012–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Riley TV et al. 2018. High prevalence of toxigenic and nontoxigenic Clostridium difficile strains in Malaysia. J Clin Microbiol 56: e00170–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Theseira L , 2021. Singapore-Country Commercial Guide. Available at: https://www.trade.gov/country-commercial-guides/singapore-healthcare. Accessed February 3, 2022.
- 64. Kumarasinghe G Lim YS Chow C Bassett DC , 1992. Prevalence of bacterial agents of diarrhoeal disease in the National University Hospital, Singapore and their resistance to antimicrobial a gents. Trop Geogr Med 44: 229–232. [PubMed] [Google Scholar]
- 65. Lim PL Barkham TM Ling LM Dimatatac F Alfred T Ang B , 2008. Increasing incidence of Clostridium difficile-associated disease, Singapore. Emerg Infect Dis 14: 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koh TH Tan AL Tan ML Wang G Song KP , 2007. Epidemiology of Clostridium difficile infection in a large teaching hospital in Singapore. Pathology 39: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hsu LY Tan TY Koh TH Kwa AL Krishnan P Tee NW Jureen R , 2011. Decline in Clostridium difficile-associated disease rates in Singapore public hospitals, 2006 to 2008. BMC Res Notes 4: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chan M Lim PL Chow A Win MK Barkham TM , 2011. Surveillance for Clostridium difficile infection: ICD-9 coding has poor sensitivity compared to laboratory diagnosis in hospital patients, Singapore. PLOS One 6: e15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tan XQ Verrall AJ Jureen R Riley TV Collins DA Lin RT Balm MN Chan D Tambyah PA , 2014. The emergence of community-onset Clostridium difficile infection in a tertiary hospital in Singapore: a cause for concern. Int J Antimicrob Agents 43: 47–51. [DOI] [PubMed] [Google Scholar]
- 70. Chau ML Hartantyo SH Yap M Kang JS Aung KT Gutierrez RA Ng LC Tam CC Barkham T , 2016. Diarrheagenic pathogens in adults attending a hospital in Singapore. BMC Infect Dis 16: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wongwanich S Ramsiri S Vanasin B Khowsaphit P Tantipatayangkul P Phan-urai R , 1990. Clostridium difficile-associated disease in Thailand. Southeast Asian J Trop Med Public Health 21: 367–372. [PubMed] [Google Scholar]
- 72. Thamlikitkul V Danpakdi K Chokloikaew S , 1996. Incidence of diarrhea and Clostridium difficile toxin in stools from hospitalized patients receiving clindamycin, beta-lactams, or nonantibiotic medications. J Clin Gastroenterol 22: 161–163. [DOI] [PubMed] [Google Scholar]
- 73. Wongwanich S Rugdeekha S Pongpech P Dhiraputra C , 2003. Detection of Clostridium difficile toxin A and B genes from stool samples of Thai diarrheal patients by polymerase chain reaction technique. J Med Assoc Thai 86: 970–975. [PubMed] [Google Scholar]
- 74. Chotiprasitsakul D Janvilisri T Kiertiburanakul S Watcharananun S Chankhamhaengdecha S Hadpanus P Malathum K , 2012. A superior test for diagnosis of Clostridium difficile-associated diarrhea in resource-limited settings. Jpn J Infect Dis 65: 326–329. [DOI] [PubMed] [Google Scholar]
- 75. Putsathit P Maneerattanaporn M Piewngam P Kiratisin P Riley TV , 2017. Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand. New Microbes New Infect 15: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Imwattana K Wangroongsarb P Riley TV , 2019. High prevalence and diversity of tcdA-negative and tcdB-positive, and non-toxigenic, Clostridium difficile in Thailand. Anaerobe 57: 4–10. [DOI] [PubMed] [Google Scholar]
- 77. Aanpreung P Veerakul G Chaichanwatanakul K , 1998. Clostridium difficile infection in febrile neutropenic malignancy children. Siriraj Hosp Gaz 50: 588–593. [Google Scholar]
- 78. Pupaibool J Khantipong M Suankratay C , 2008. A study of Clostridium difficile-associated disease in King Chulalongkorn Memorial Hospital, Thailand. J Med Assoc Thai 91: 37–43. [PubMed] [Google Scholar]
- 79. World Health Organization , 2005. The Treatment of Diarrhoea: A Manual for Physicians and Other Senior Health Workers, 4th Revision. Geneva, Switzerland: WHO. [Google Scholar]
- 80. Huong VTT Hang PTT Nga TT Trang LT Duong DTT Canh TQ , 2018. Molecular characterization of Clostridium difficile isolates from patients with antibiotics-associated diarrhea in Hanoi, Vietnam. VJPM 28: 70. [Google Scholar]
- 81. Giang NTH Thuy PTH Huong VTT Duong TN , 2019. Epidemiology and clinical manifestations of Clostridium difficile-associated diarrhea in adults in Bach Mai Hospital from 2013 to 2017. VJPM 29: 9. [Google Scholar]
- 82. Giang NTH , 2020. Characteristics of Epidemiology, Clinical Manifestations, and Risk Factors of Clostridium difficile Infection Among Adult Patients with Diarrhoea in Bach Mai Hospital from 2013 to 2017. Hanoi, Vietnam: Department of Epidemiology, National Institute of Hygiene & Epidemiology. [Google Scholar]
- 83. Giang NTH Huong VTT Thuy PTTT Duong TN , 2019. Genotypic distribution characteristics of Clostridium difficile associated diarrhea in adult patients in Bach Mai Hospital from 2013 to 2017. VJPM 29: 18. [Google Scholar]
- 84. Liao F Li W Gu W Zhang W Liu X Fu X Xu W Wu Y Lu J , 2018. A retrospective study of community-acquired Clostridium difficile infection in southwest China. Sci Rep 8: 3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Warny M Pepin J Fang A Killgore G Thompson A Brazier J Frost E McDonald LC , 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366: 1079–1084. [DOI] [PubMed] [Google Scholar]
- 86. Collins DA Riley TV , 2018. Clostridium difficile in Asia: opportunities for One Health management. Trop Med Infect Dis 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cairns MD et al. 2017. Comparative genome analysis and global phylogeny of the toxin variant Clostridium difficile PCR ribotype 017 reveals the evolution of two independent sublineages. J Clin Microbiol 55: 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Imwattana K Knight DR Kullin B Collins DA Putsathit P Kiratisin P Riley TV , 2019. Clostridium difficile ribotype 017 - characterization, evolution and epidemiology of the dominant strain in Asia. Emerg Microbes Infect 8: 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Imwattana K Putsathit P Collins DA Leepattarakit T Kiratisin P Riley TV Knight DR , 2022. Global evolutionary dynamics and resistome analysis of Clostridioides difficile ribotype 017. Microb Genom 8. 10.1099/mgen.0.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Putsathit P Neela VK Joseph NMS Ooi PT Ngamwongsatit B Knight DR Riley TV , 2019. Molecular epidemiology of Clostridium difficile isolated from piglets. Vet Microbiol 237: 108408. [DOI] [PubMed] [Google Scholar]
- 91. Zhang WZ Li WG Liu YQ Gu WP Zhang Q Li H Liu ZJ Zhang X Wu Y Lu JX , 2020. The molecular characters and antibiotic resistance of Clostridioides difficile from economic animals in China. BMC Microbiol 20: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Usui M Nanbu Y Oka K Takahashi M Inamatsu T Asai T Kamiya S Tamura Y , 2014. Genetic relatedness between Japanese and European isolates of Clostridium difficile originating from piglets and their risk associated with human health. Front Microbiol 5: 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ngamskulrungroj P Sanmee S Putsathit P Piewngam P Elliott B Riley TV Kiratisin P , 2015. Molecular epidemiology of Clostridium difficile infection in a large teaching hospital in Thailand. PLOS One 10: e0127026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lew T Putsathit P Sohn KM Wu Y Ouchi K Ishii Y Tateda K Riley TV Collins DA , 2020. Antimicrobial susceptibilities of Clostridium difficile isolates from 12 Asia-Pacific countries in 2014 and 2015. Antimicrob Agents Chemother 64: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. World Health Organization , 2017. One Health. Available at: https://www.who.int/features/qa/one-health/en/. Accessed December 10, 2020.
- 96. Squire MM Riley TV , 2013. Clostridium difficile infection in humans and piglets: a ‘One Health’ opportunity. Curr Top Microbiol Immunol 365: 299–314. [DOI] [PubMed] [Google Scholar]
- 97. Niwa H et al. 2013. Postoperative Clostridium difficile infection with PCR ribotype 078 strain identified at necropsy in five thoroughbred racehorses. Vet Rec 173: 607. [DOI] [PubMed] [Google Scholar]
- 98. Eyre DW et al. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369: 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lim SC Knight DR Riley TV , 2020. Clostridium difficile and One Health. Clin Microbiol Infect 26: 857–863. [DOI] [PubMed] [Google Scholar]
- 100. Knight DR Squire MM Collins DA Riley TV , 2016. Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front Microbiol 7: 2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rupnik M , 2007. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin Microbiol Infect 13: 457–459. [DOI] [PubMed] [Google Scholar]
- 102. Songer JG Trinh HT Killgore GE Thompson AD McDonald LC Limbago BM , 2009. Clostridium difficile in retail meat products, USA, 2007. Emerg Infect Dis 15: 819–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bakri MM Brown DJ Butcher JP Sutherland AD , 2009. Clostridium difficile in ready-to-eat salads, Scotland. Emerg Infect Dis 15: 817–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tsai BY Ko WC Chen TH Wu YC Lan PH Chen YH Hung YP Tsai PJ , 2016. Zoonotic potential of the Clostridium difficile RT078 family in Taiwan. Anaerobe 41: 125–130. [DOI] [PubMed] [Google Scholar]