Abstract
The ςS (or ς38) subunit of RNA polymerase, encoded by the rpoS gene, is a crucial regulator in the transcriptional control of a set of genes under stressful conditions, such as nutrient starvation. The expression of rpoS is regulated in a complex manner at the levels of transcription, translation, and stability of the product. Although a number of factors involved in the regulation of rpoS expression have been identified, the underlying molecular mechanisms are not fully understood. In this study, we identified the Crr (or EIIAGlc) protein as a novel factor that plays an important role not only in the transcriptional control but also in the translational control of rpoS expression. Crr is an important component in glucose uptake through the well-characterized phosphoenolpyruvate:carbohydrate phosphotransferase system. The results of a series of genetic analyses revealed that Crr negatively controls rpoS translation and transcription. The observed transcriptional control by Crr appears to be mediated by cyclic AMP. However, it was found that Crr negatively controls rpoS translation rather directly. These results suggest a possible linkage between the control of rpoS expression and carbon metabolism.
When Escherichia coli cells are exposed to several stressful conditions preventing rapid growth, such as nutrient starvation, the expression of a set of genes is induced to allow the cells to survive under the harsh conditions (8). The ςS (or ς38) subunit of RNA polymerase, encoded by the rpoS gene, has been shown to function as a central regulator in the transcriptional control of such genes (11). Since the gene expression is, at least, mainly dependent on the cellular ςS content, it is important to determine how ςS content is regulated during cell growth.
rpoS expression is severely repressed at the logarithmic growth phase, while it is induced during entry into the stationary phase (5, 23). The regulation is conducted in a complex manner at the levels of transcription, translation, and stability of ςS (10). Recent extensive studies have revealed that several proteinaceous factors as well as small molecules are involved in the regulation. rpoS transcription was proposed to be negatively regulated by a cyclic AMP (cAMP) receptor protein-cAMP complex, based on the fact that ςS is significantly accumulated in the cells of cya and crp mutants (9, 10). Posttranscriptional mechanisms also, and more importantly under certain conditions, determine the cellular ςS content. It has been revealed that an RNA-binding protein, HF-I, encoded by the hfq gene, and a nucleoid protein, H-NS, are involved in rpoS translation positively and negatively, respectively (1, 15, 27). In rapidly growing cells, ςS is markedly unstable, being degraded by an ATP-dependent protease complex, ClpPX (18). This rapid turnover requires the functions of both H-NS and RssB (or SprE) (1, 14, 17, 27), the latter protein belonging to the response regulator family in a two-component signal transduction system. Although the genes involved in the regulation of rpoS expression have been identified to some extent, the underlying molecular mechanisms remain to be elucidated. The questions of how cells sense the external and internal growth conditions and how such signals are transduced in the cells and then regulate rpoS expression through the factors described above have not been answered, in spite of their biological significance.
In this study, we identified the Crr protein as a novel factor that is crucial for the translational as well as transcriptional control of rpoS expression. The Crr protein (or EIIAGlc) is an important component in glucose uptake in the phosphoenolpyruvate:carbohydrate phosphotransferase system (PEP:PTS) (16). In this system, the phosphate group of PEP is transferred through successive phosphorelay reactions involving enzyme I (EI), histidine protein (HPr), and glucose-specific enzyme II (EIIGlc), and then extracellular glucose is phosphorylated by EIIGlc concomitantly with its uptake. Crr is a cytoplasmic component of EIIGlc and is known to be crucial not only for glucose uptake but also for the regulation of several cellular functions in carbon metabolism, such as inducer exclusion and modulation of adenylate cyclase activity. We first isolated an E. coli mutant in which the expression of rpoS is derepressed even at the logarithmic growth phase. The following genetic analyses revealed that the function of the crr gene is impaired by a transposon insertion in the mutant, suggesting that Crr is deeply involved in the negative control of rpoS expression. In this case, the phosphorylation of Crr is also crucial in glucose uptake. Crr appears to be involved in both the translational and the transcriptional control of rpoS expression. We will discuss the underlying molecular mechanisms and the physiological role of Crr in the control of rpoS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. They are all derivatives of MC4100 (2). Several mutant strains were constructed by P1 transduction (13). Cells were grown at 37°C mainly in Luria broth (13). An overnight culture was inoculated into fresh medium, and cells were grown until logarithmic phase, whereupon the culture was appropriately diluted with fresh medium and further grown (see Fig. 2) and then used for all experiments at the mid-logarithmic phase. Antibiotics were added as necessary.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype |
|---|---|
| MC4100 | F− Δ(arg-lac)U169 araD139 rpsL150 ptsF25 flbB5301 rbsR deoC relA1 |
| CU263 | MC4100(λrpoS-lacZ PF977) |
| CU264 | MC4100(λrpoS-lacZ PF212) |
| CU329 | CU263 crr2-3::mini-Tn10cam |
| CU330 | CU263 crr::kan |
| CU334 | CU263 clpX::kan |
| CU344 | CU263 nupC510::Tn10 |
| CU345 | CU263 nupC510::Tn10 ΔptsHI |
| CU348 | CU263 ptsG::Tn5 |
| CU351 | CU263 crr::kan rssB::cam |
| CH11 | MC4100(λrpoS-lacZ OFa) |
| CH12 | MC4100(λkatE-lacZ) |
| NM5 | CH12 crr::kan |
| NM7 | CH11 crr::kan |
| NM9 | CU264 crr::kan |
| NM11 | CU264 hfq1::Ω |
| NM12 | CU264 rssB::kan |
| NM13 | CU264 clpX::kan |
| NM20 | CU264 crr2-3::mini-Tn10cam |
| NM21 | CU264 crr2-3::mini-Tn10cam hfq1::Ω |
| NM23 | CU263 cya::kan |
| NM24 | CU264 cya::kan |
| NM25 | CH11 cya::kan |
OF, operon fusion.
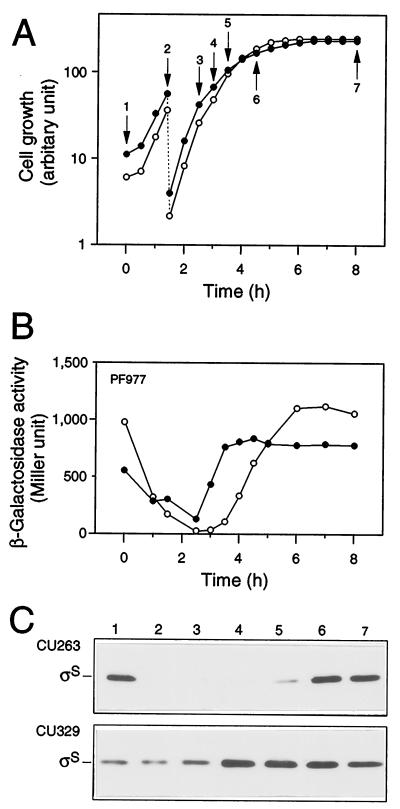
FIG. 2.
Expression of rpoS during cell growth. Strains CU263 (open circles) and CU329 (closed circles), each carrying rpoS-lacZ PF977, were grown at 37°C in Luria broth. Both cell growth (A) and β-galactosidase activity expressed by PF977 (B) were measured. Protein samples were prepared at the indicated time points in panel A, and then each 20 μg of total proteins was subjected to immunoblotting with anti-ςS antiserum (C).
Construction of rpoS-lacZ fusion genes.
rpoS-lacZ fusion genes were constructed essentially by the method of Hirano et al. (6). The rpoS-lacZ operon fusion was constructed as follows. A 1.6-kb ClaI-DraI fragment encompassing rpoS promoters was purified from pKTF101 (22). After treatment with T4 DNA polymerase, the resultant fragment was inserted into the previously blunt-ended HindIII site of pMS434HS. An E. coli strain harboring λpF13 was transformed with the resultant plasmid carrying the rpoS-lacZ operon fusion, and a lambda phage lysate was prepared from the transformant by UV irradiation. MC4100 was infected with the phage lysate, and then lysogens were selected from blue plaques on plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as candidates carrying the fusion gene on the chromosome. The lysogens thus purified were further scored for the Lac+ phenotype, and then one such lysogen, named CH11, was used in this study.
rpoS-lacZ PF977 was constructed as follows. A 2.4-kb ClaI-Eco47III fragment encompassing the rpoS gene lacking the C-terminal portion, as well as the promoter region, was first purified from pKTF101 (22). After treatment with T4 DNA polymerase, the fragment was inserted into the HincII site of pUC119 (25) to construct the rpoS-lacZα protein fusion. From the resultant plasmid, a 2.6-kb HindIII-BglI fragment encompassing the protein fusion was purified by partial digestion with BglI (the BglI site is located within lacZα), and then the fragment was inserted between the HindIII and BglI sites of pMS434HS (the BglI site corresponds to the identical site of the rpoS-lacZα protein fusion) to yield pCU71. The resultant rpoS-lacZ protein fusion, including the N-terminal 325 codons of the rpoS open reading frame, was named PF977 and subsequently transferred to the chromosome of MC4100, as described above. rpoS-lacZ PF212 was constructed by means of the same procedures except that a 1.6-kb ClaI-HincII fragment encompassing the N-terminal 70 codons of the rpoS open reading frame as well as the promoter region was used for the construction.
The katE-lacZ operon fusion was constructed as for the rpoS-lacZ operon fusion, except that a 1.25-kb HindIII-PstI fragment encompassing the katE promoter was used.
Transposon insertion mutagenesis.
Transposon insertion using mini-Tn10cam was carried out essentially according to the method of Kleckner et al. (7). Cells of MC4100 were grown in Luria broth at 37°C to the mid-logarithmic phase. A portion of the culture was infected with a lambda phage lysate of λNK1324 and then plated onto a Luria agar plate containing 25 μg of chloramphenicol per ml. Chloramphenicol-resistant (Cmr) transductants were pooled and infected with the P1vir phage to prepare a P1 phage lysate. The resultant phage lysate was stored and used for P1 transduction.
Assaying of β-galactosidase activity.
Assaying of β-galactosidase activity was carried out by the method of Miller (13).
Immunoblotting analysis.
Total cellular proteins were prepared by precipitation with trichloroacetic acid (final concentration, 5%) and then collected by centrifugation. After a wash with ice-cold acetone, the precipitate was dissolved in 1% (wt/vol) sodium dodecyl sulfate (SDS)–50 mM Tris-HCl (pH 8)–1 mM EDTA buffer. The protein concentration was accurately determined for each sample using a Micro BCA protein assay reagent kit (Pierce Chemical Co., Rockford, Ill.). Appropriate amounts of total cellular proteins were separated by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting with anti-ςS and anti-CbpA polyclonal antisera.
RESULTS
Isolation of transposon-insertional mutations that result in derepression of rpoS
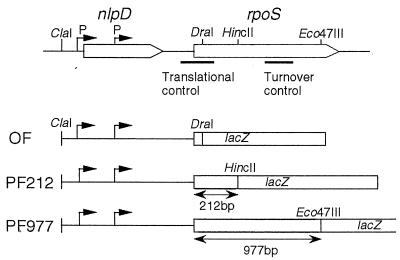
To monitor rpoS expression appropriately in different genetic backgrounds, in this study we constructed three types of rpoS-lacZ fusion genes on the chromosome (Fig. 1). An rpoS-lacZ operon (transcriptional) fusion contains only the rpoS promoter region, fused to the lacZ gene, so it can be used to monitor the transcriptional control of rpoS. Two protein (translational) fusion genes, named PF212 and PF977, were also constructed. Note that these fusions include the N-terminal 70 and 325 codons of the rpoS open reading frame, respectively. The former contains a cis-acting element required for translational control in addition to the rpoS promoter region, whereas the latter contains all regulatory elements required for rpoS control at the levels of transcription, translation, and stability of the gene product (15). These two protein fusions, PF212 and PF977, were used appropriately to evaluate the translational efficiency and overall output of rpoS expression, respectively.
FIG. 1.
Schematic representation of a set of rpoS-lacZ fusion genes used in this study. The rpoS operon is diagrammed at the top. The rpoS open reading frame is indicated. P, promoter of rpoS. cis-acting elements required for translational control and turnover control and restriction sites used for the construction of the rpoS-lacZ fusion genes are indicated. The structures of three types of rpoS-lacZ fusion genes are diagrammed below.
Using PF977 as a monitoring probe, an attempt was first made to isolate mutants exhibiting derepressed expression for rpoS in order to identify a novel factor(s) involved in the regulatory mechanism for rpoS expression. PF977 should be advantageous for comprehensively isolating mutants affecting rpoS expression at any regulatory step(s). Indeed, on tetrazolium-lactose plates (19), the wild-type strain carrying PF977 on the chromosome gives red colonies (indicating low β-galactosidase activity), whereas the hns::neo derivative, in which rpoS expression is known to be derepressed, gives white ones (indicating high β-galactosidase activity). Thus, mutants exhibiting derepressed expression of rpoS should give white or pink colonies on tetrazolium-lactose plates due to the enhanced β-galactosidase activity.
Cells of CU263, carrying PF977 on the chromosome, were mutagenized by random insertion of mini-Tn10cam carrying a Cmr marker (7). From among ∼7 × 104 Cmr transductants, we selected 20 white or pink colonies on tetrazolium-lactose plates containing chloramphenicol. The following immunoblotting analysis revealed that 2 of these 20 mutants significantly accumulated ςS in their cells even at the logarithmic phase, whereas in the remaining 18 mutants the ςS content showed only a three- to fourfold induction compared to that in the parental wild-type strain at the logarithmic phase. These two mutants were genetically purified by repeated P1 transduction into the fresh CU263 background and designated CU328 and CU329. In this study, we focus our attention on the latter mutant, CU329, to investigate the underlying regulatory mechanism for rpoS expression (the former mutant will be dealt with elsewhere).
ςS is accumulated in CU329 even at the logarithmic phase.
To clarify the phenotype more precisely, we monitored the expression of PF977 in CU329 by measuring β-galactosidase activity during cell growth. Cells were grown at 37°C in Luria broth, and β-galactosidase activity was measured at appropriate intervals. The expression of PF977 in the wild-type background decreased once after inoculation of an overnight culture and then increased concomitantly with cell growth, reaching the maximum level at the onset of the stationary phase (Fig. 2A and B). PF977 expression in CU329 was found to be ∼20-fold higher than that in wild-type cells only around the logarithmic phase (Fig. 2A and B). This kinetic profile was similar to that observed for a clpX::kan mutant (data not shown), in which ςS is known to be accumulated (18). Immunoblotting analysis also confirmed this particular phenotype; that is, ςS content increased approximately 10- to 20-fold in CU329 around the logarithmic phase (Fig. 2C). These results indicated that rpoS expression in CU329 is indeed derepressed even at the logarithmic phase. It should be noted that the accumulation of CU329 at the logarithmic phase was less significant, i.e., three- to fourfold accumulation, when cells were grown in M9 synthetic medium (data not shown). The following analyses were thus carried out using Luria broth exclusively as the culture medium.
The crr gene is involved in the control of rpoS expression.
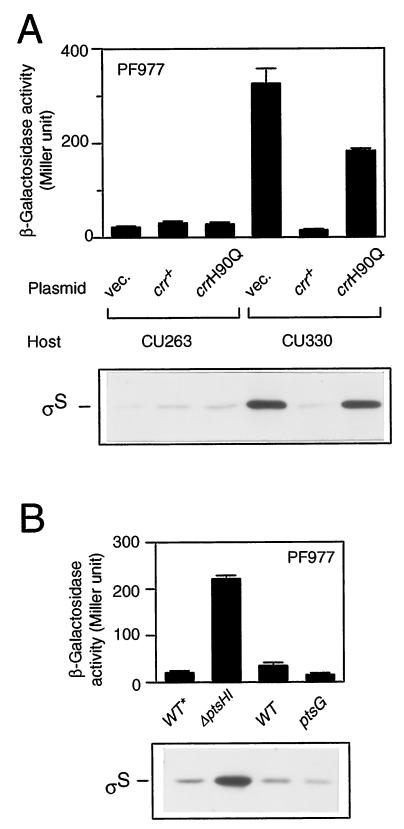
To clarify the gene disrupted by the mini-Tn10cam insertion in CU329, we first cloned a DNA segment encompassing the insertion from the chromosomal DNA using the cam (Cmr) gene of the transposon as a selective marker. The following DNA sequencing of the flanking region of the cam gene revealed that the transposon is inserted just within the crr gene encoding the A subunit of glucose-specific enzyme II (EIIAGlc). The insertion point is the 95th codon of the open reading frame consisting of 169 codons. We thus named the relevant insertional mutation crr2–3::mini-Tn10cam. To further confirm that the crr mutation indeed affects rpoS expression, we constructed a derivative of CU263, named CU330, carrying a well-characterized crr::kan mutation (21). The resultant strain also exhibited both enhanced expression of PF977 and accumulation of ςS at the mid-logarithmic phase (Fig. 3A; compare vector controls). In addition, the wild-type crr gene was able to complement the mutational phenotype in CU330 with regard to both the enhanced expression of PF977 and the accumulation of ςS (Fig. 3A). Thus, we concluded that the crr gene product somehow negatively controls rpoS expression at the logarithmic phase.
FIG. 3.
Phosphorylation of Crr is crucial for rpoS regulation. (A) Complementation assay of the crr::kan mutant. Strains CU263 (wild type) and CU330 (crr::kan) carrying plasmid pTSV28 (vector [vec.]), pSTCRR (crr+), or pST172 (crrH90Q) were grown at 37°C in Luria broth supplemented with 25 μg of chloramphenicol per ml. At the mid-logarithmic phase, β-galactosidase activity (upper panel) and ςS content (lower panel) were measured. (B) Effect of the ptsHI or ptsG mutation on rpoS expression. Strains CU344 (wild type [WT*]), CU345 (ΔptsHI), CU263 (wild type), and CU348 (ptsG::Tn5) were grown at 37°C in Luria broth. Note that CU344 and CU345 carry the nupC510::Tn10 allele, which was used as a selectable marker to construct the ΔptsHI mutant. At the mid-logarithmic phase, β-galactosidase activity expressed by PF977 (upper panel) and ςS content (lower panel) were measured. β-Galactosidase activity data are means with standard deviations for four independent assays. Each 20 μg of total proteins was used for immunoblotting.
Phosphorylation of the Crr protein is crucial for negative regulation of rpoS expression.
The Crr protein (EIIAGlc) plays an important role in glucose uptake through the PEP:PTS system (16). In this system, the phosphate group of PEP is transferred to glucose through successive phosphorelay reactions involving enzyme I (EI), histidine protein (HPr), and glucose-specific enzyme II (EIIGlc). Extracellular glucose is transported into cells through a coupled phosphorylation reaction catalyzed by EIIGlc. To determine whether the phosphorylation of Crr is crucial for the negative regulation of rpoS expression, we carried out the following two lines of experiments. First, we examined the complementation ability of a mutant crr gene (crrH90Q), in which the phosphorylated His residue is replaced by Gln so that the mutant Crr protein is no longer phosphorylated (21). The introduction of the crrH90Q allele into CU330 failed to fully complement the mutational phenotype of CU330, although both the expression of PF977 and the cellular content of ςS decreased slightly (Fig. 3A). Second, we examined whether a certain lesion of ptsHI affects rpoS expression. Since the phosphorylation of Crr is dependent on both EI and HPr, which are encoded by the ptsI and ptsH genes, respectively, the Crr protein cannot be phosphorylated in ptsHI mutant cells (16). Both the expression of PF977 and the ςS content appeared to increase in the ΔptsHI mutant cells (Fig. 3B, left pair). This result also supported the view that the phosphorylation of Crr is important for regulation of rpoS expression. These lines of evidence demonstrated that the phosphorylation of the Crr protein is crucial for the negative regulation of rpoS expression.
Since during glucose uptake the function of the Crr protein (EIIAGlc) is coupled to EIICBGlc, which is encoded by the ptsG gene (16), both subunits of EIIGlc could be required for the negative regulation of rpoS expression. To examine this possibility, rpoS expression in a ptsG mutant was investigated. Neither the expression of PF977 nor the ςS content were ever increased by the ptsG::kan mutation, and instead, both were reduced twofold (Fig. 3B; right pair), indicating that only the A subunit, i.e., not the CB subunit, of EIIGlc is involved in the negative regulation of rpoS expression.
ςS accumulated due to the crr mutation is transcriptionally active.
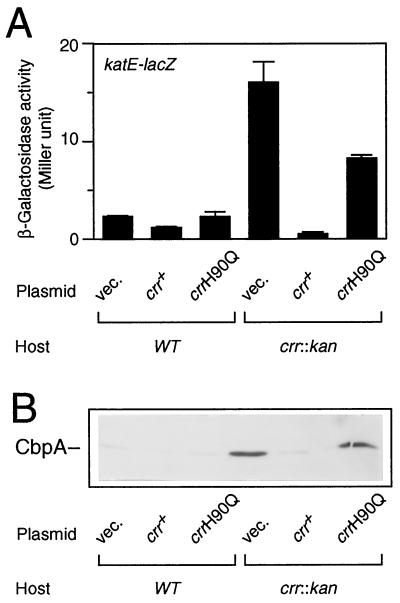
Since ςS positively controls the expression of a set of genes whose functions are induced under certain harsh conditions, it is important to determine whether the ςS accumulated in the crr mutant is transcriptionally active or not. In the crr::kan background, we thus examined the expression of the katE and cbpA genes, whose transcription is known to be dependent on the ςS function (12, 26). A set of strains (Fig. 3A) carrying a katE-lacZ operon fusion on the chromosome was constructed, and katE-lacZ expression was measured at the logarithmic phase. The results show that katE promoter activity was increased fivefold by the crr::kan mutation (Fig. 4A). The wild-type crr gene was able to complement this particular phenotype, but the crrH90Q allele was not. cbpA expression was also induced by the crr::kan mutation (Fig. 4B). Thus, these results of immunoblotting analysis of the CbpA protein essentially led to the same conclusion; that is, the ςS accumulated in the crr mutant cells indeed has the ability to allow the transcription of its target genes.
FIG. 4.
Expression of katE and cbpA in the crr::kan mutant. (A) Strains CH12 (wild type [WT], katE-lacZ) and NM5 (crr::kan katE-lacZ), each harboring plasmids as described for Fig. 3A, were grown at 37°C in Luria broth supplemented with chloramphenicol (25 μg/ml). At the mid-logarithmic phase, β-galactosidase activity expressed by the katE-lacZ operon fusion was measured. The data are means with standard deviations for four independent assays. (B) A set of transformants as described for Fig. 3A was grown at 37°C in Luria broth supplemented with 25 μg of chloramphenicol per ml. Immunoblotting with anti-CbpA antiserum was carried out as described for Fig. 3A.
The crr mutation affects rpoS expression at the level of transcription.
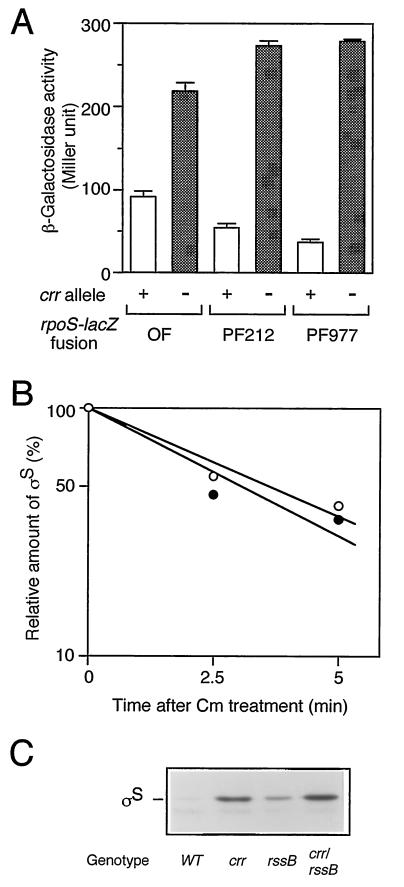
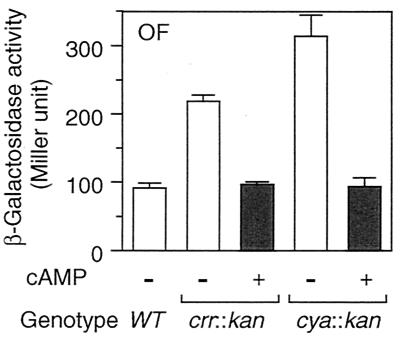
rpoS expression is regulated at the levels of transcription, translation, and stability of the protein (10). Which step(s) does Crr negatively regulate? To answer this question, we examined the phenotypic effects of the crr mutation on three types of rpoS-lacZ fusion genes: operon fusion, PF212, and PF977. As mentioned above (Fig. 1), PF212 allows the monitoring of the effects of certain mutations on translational efficiency, other than the stabilization of ςS. Indeed, the expression of PF212 was affected by the hfq1::Ω mutation (24) but not by the clpX::kan or rssB::kan mutation (data not shown). While the crr::kan mutation affected the rpoS promoter activity slightly (twofold or less), it significantly induced expression of PF212 (fivefold) as well as that of PF977 (eightfold) (Fig. 5A). The half-life of ςS in the crr mutant, as determined by immunoblotting analysis of chloramphenicol-treated cells, was approximately 2.5 min, which is quite similar to that observed for wild-type cells (Fig. 5B). Moreover, the crr::kan rssB::cam double mutant showed higher ςS content than that in either single mutant (Fig. 5C), suggesting that the crr::kan mutation does not affect the stability of ςS. Thus, Crr seems to be partly involved in the negative regulation of rpoS expression at the level of its transcription. However, it should be noted that phosphorylated Crr is able to activate the enzymatic activity of adenylate cyclase and that the cellular concentration of cAMP is positively controlled by Crr (16). Therefore, the effect of the crr mutation on rpoS transcription described above is not surprising, since the cAMP receptor protein-cAMP complex has been shown to negatively regulate rpoS transcription in vivo (9, 10). To confirm such an indirect effect of the crr mutation on rpoS transcription through adenylate cyclase activity, we examined the effect of the addition of cAMP on the derepressed expression of rpoS in the crr mutant. Cells harboring the rpoS-lacZ operon fusion were grown in Luria broth in either the absence or the presence of 5 mM cAMP, and then β-galactosidase activity was measured at the mid-logarithmic phase. As shown in Fig. 6, the expression of the operon fusion was completely restored to the wild-type level in both the crr and the cya backgrounds by the addition of cAMP. This indicated that the effect of the crr mutation on rpoS transcription is rather indirect due to modulation of adenylate cyclase activity.
FIG. 5.
(A) Effect of the crr::kan mutation on a set of rpoS-lacZ fusion genes. Strains carrying the three types of rpoS-lacZ genes described for Fig. 1 in either crr+ (+) or crr::kan (−) background were grown at 37°C in Luria broth. At the mid-logarithmic phase, β-galactosidase activity was measured. The data are means with standard deviations for four independent assays. OF, operon fusion. (B) Effect of the crr::kan mutation on the half-life of ςS. Strains CU263 (wild type; open circles) and CU330 (crr::kan; closed circles) were grown at 37°C in Luria broth. At the mid-logarithmic phase, cells were treated with 25 μg of chloramphenicol per ml. At the indicated intervals, cells were harvested and subjected to immunoblotting using anti-ςS antiserum. The amount of ςS was expressed relative to that determined for the sample at time zero. (C) Cellular ςS content in the crr::kan rssB::cam double mutant. Strains carrying the indicated mutations were grown at 37°C in Luria broth. At the mid-logarithmic phase, cells were harvested and subjected to immunoblotting using anti-ςS antiserum. WT, wild type.
FIG. 6.
Effect of cAMP on rpoS transcription. Strains CH11 (wild type [WT]), NM7 (crr::kan), and NM25 (cya::kan), each carrying the rpoS-lacZ operon fusion (OF), were grown at 37°C in Luria broth either in the absence (−) or the presence (+) of 5 mM cAMP. At the mid-logarithmic phase, β-galactosidase activity expressed by the rpoS-lacZ operon fusion was measured. The data are means with standard deviations for four independent assays.
Phosphorylated Crr is involved in translational control of rpoS
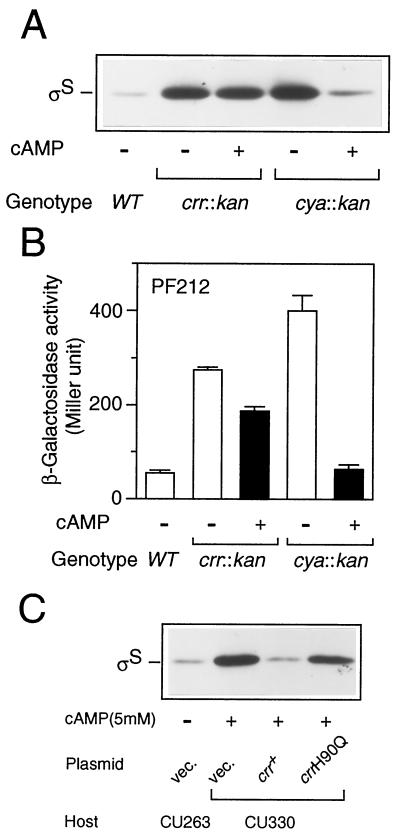
We next examined whether Crr is involved in rpoS translation indirectly through modulation of the cellular concentration of cAMP. CU330 cells were grown in Luria broth at 37°C in either the presence or the absence of 5 mM cAMP, and then ςS content was determined by immunoblotting analysis. As shown in Fig. 7A, ςS content decreased only slightly (∼10% or less) in the crr mutant upon the addition of cAMP. In contrast, the ςS content of the cya::kan mutant decreased to the normal level upon the addition of cAMP. Moreover, expression of PF212 in the crr::kan background only slightly decreased upon the addition of cAMP, whereas that in the cya::kan background clearly decreased to the normal level (Fig. 7B). These results strongly suggested that Crr is involved in rpoS translation rather directly or, at least, not via the modulation of adenylate cyclase activity.
FIG. 7.
The phosphorylated Crr is involved in rpoS translation. (A) Effect of cAMP on cellular ςS content. Strains CU263 (wild type [WT]), CU330 (crr::kan), and NM23 (cya::kan) were grown at 37°C in Luria broth in either the absence (−) or the presence (+) of 5 mM cAMP. At the mid-logarithmic phase, total protein samples were prepared and each 20 μg was subjected to immunoblotting with anti-ςS antiserum. (B) Effect of the addition of cAMP on rpoS-lacZ PF212 expression. Strains CU264 (wild type), NM9 (crr::kan), and NM24 (cya::kan), each carrying rpoS-lacZ PF212, were grown, and then β-galactosidase activity expressed by rpoS-lacZ PF212 was measured. The data are means with standard deviations for four independent assays. (C) Strains CU263 (wild type) and CU330 (crr::kan) harboring plasmid pTSV28 (vector [vec.]), pSTCRR (crr+), or pST172 (crrH90Q) were grown at 37°C in Luria broth supplemented with 25 μg of chloramphenicol per ml in either the absence (−) or the presence (+) of 5 mM cAMP. At the mid-logarithmic phase, ςS content was measured by immunoblotting.
We further addressed the issue of whether the translational control of rpoS requires the phosphorylation of Crr. The strains harboring crr plasmids described in the legend to Fig. 3 were grown in the presence of 5 mM cAMP, and then ςS contents were determined by immunoblotting. It should be noted that with the addition of cAMP one is able only to estimate precisely the effect of a mutation on translational control by eliminating the effect on transcriptional control. As shown in Fig. 7C, the crrH90Q allele was unable to complement the mutational phenotype of CU330. The slight reduction in ςS content, as shown in Fig. 3A, would be due to the overproduction of the mutated Crr by the multicopy plasmid. The results demonstrated that the phosphorylation of Crr is crucial for translational control of rpoS.
Epistasis analysis with the hfq mutation.
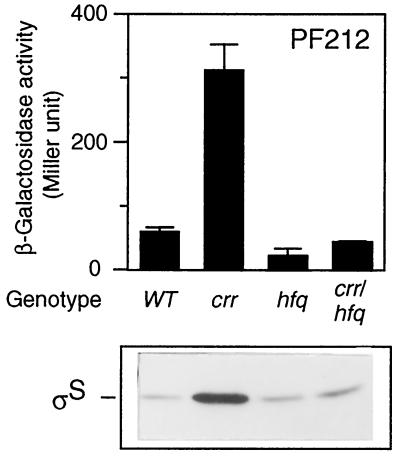
The results described above suggested that Crr is involved in translational regulation of ςS. It was reported that an RNA-binding protein, HF-I, the hfq gene product, is required for rpoS translation (15). We thus finally examined the genetic relationship between the hfq and crr genes with respect to rpoS translation. A set of strains carrying the hfq1::Ω and/or crr2–3::mini-Tn10cam mutations were constructed, and the expression of PF212 was measured. The expression of PF212 in the hfq1::Ω background was apparently lower than that in wild-type cells, a finding which is in good agreement with the previous report (15), whereas that in the crr2–3::mini-Tn10cam background was fivefold higher. In hfq crr double-mutant cells, the β-galactosidase activity was slightly lower than that in wild-type cells and similar to that in the hfq single mutant (Fig. 8). This particular phenotype was also confirmed by immunoblotting (Fig. 8). There are two possible explanations. First, the hfq mutation is epistatic to the crr mutation because the strong effect of the crr mutation was diminished. Second, the two gene products act independently because the phenotype exhibited by the double mutant was similar to that of the wild-type strain. It is difficult to determine which explanation is correct.
FIG. 8.
Effect of an hfq mutation on rpoS expression in crr background. Strains CU264 (wild type [WT]), NM20 (crr2–3::mini-Tn10cam), NM11 (hfq1::Ω), and NM21 (crr2–3::mini-Tn10cam hfq1::Ω), each carrying rpoS-lacZ PF212 on the chromosome, were grown at 37°C in Luria broth. At the mid-logarithmic phase, β-galactosidase activity expressed by rpoS-lacZ PF212 (upper panel) and ςS content (lower panel) were measured. β-Galactosidase activity data are means with standard deviations for four independent assays.
DISCUSSION
In this study, we demonstrated that Crr negatively controls rpoS expression. The crr mutation affects rpoS expression mainly at the level of translation rather than that of transcription, since the addition of cAMP did not drastically affect the ςS content of the crr mutant even while rpoS transcription was decreased to the wild-type level (Fig. 6 and 7). Although Crr has been shown to be able to modulate several cellular functions in carbon metabolism, such as inducer exclusion and adenylate cyclase activity, this is the first evidence that Crr participates in the translational process.
How does Crr negatively control rpoS translation? Two proteins, HF-I and H-NS, have previously been reported to be factors involved in the translational control of ςS (1, 15, 27). HF-I is required for efficient rpoS translation, probably through melting of the preformed complex secondary structure of rpoS mRNA inhibiting the translation (3, 15). H-NS is, instead, a negative regulator of rpoS translation (1, 27), and we have no information as to the underlying molecular mechanism. One possible explanation is that Crr covalently modifies HF-I to modulate its function, although no modification of HF-I has been reported. Alternatively, Crr may somehow prevent the formation of the translational initiation complex required for rpoS translation. Since Crr is known to interact with many other proteins, such as sugar transporters and adenylate cyclase, in carbon metabolism, it is likely that Crr is also able to interact with the translational machinery or HF-I. In this regard, it is notable that ribosomal protein S7 has a consensus sequence implicated in the interaction with Crr (20). Crr may repress rpoS translation by modulating the function of S7, since S7 is known to repress its own translation through direct binding to mRNA (4). Of course, more complicated explanations cannot be excluded at present. In any event, further extensive genetic and biochemical analyses are necessary to clarify how Crr negatively regulates rpoS translation.
The phosphorylation of Crr appears to be important for the negative control of rpoS translation (Fig. 3 and 7). Since phosphorylated Crr is produced during successive phosphorelay reactions in PEP:PTS, not only Crr but also the overall system is apparently important for the control. The fact that the system absolutely depends on PEP, an important intermediate in carbon metabolism, leads us to the attractive idea that PEP:PTS regulates rpoS expression by monitoring the amounts of available nutrients throughout cell growth. However, this possibility seems unlikely because the phosphorylation state of Crr did not drastically fluctuate with the growth phase but rather with the amount of extracellular glucose. Indeed, ςS content in wild-type cells was not affected significantly by the addition of glucose (data not shown), which is known to dramatically decrease the levels of phosphorelated species of Crr (21). Therefore, alternatively, the intactness of PEP:PTS may be crucial for maintaining a lower level of rpoS expression. To ensure rapid proliferation at the logarithmic growth phase, the cells require a large amount of energy and several metabolic intermediates. In this situation, efficient transport of extracellular carbohydrates into cells by means of PEP:PTS should be necessary. When PEP:PTS is impaired by certain mutations, the cells may be unable to satisfy the requirement for adaptation to rapid growth conditions and subsequently rpoS expression is induced. If this is the case, the effect of the crr mutation on rpoS expression should be more significant under conditions permitting a higher growth rate, a hypothesis which is consistent with our observation that the effect of the crr mutation in a rich medium was stronger than that in a synthetic medium.
rpoS expression is enhanced when cells enter conditions preventing efficient proliferation, such as nutrient starvation, high osmolarity, and low pH, while it is repressed at the fast-growth logarithmic phase. Many proteinaceous factors involved in regulation of rpoS expression have been identified, and all of them except HF-I were found to negatively control rpoS expression (see the introduction), suggesting that the control of rpoS expression is achieved mainly through negative regulation at the logarithmic phase. When the growth conditions are sufficient for rapid proliferation, these negative regulators are activated by individual signals and subsequently repress rpoS expression at the transcription, translation, or protein stabilization step. If the growth conditions are not appropriate for rapid proliferation, a part of the negative regulatory mechanism may be unable to work well, resulting in derepressed expression of rpoS to some extent. The intactness of PEP:PTS may be recognized by cells as one of such cues for repression of rpoS expression.
ACKNOWLEDGMENTS
We are grateful to H. Aiba (Nagoya University) for the kind gifts of mutant strains and plasmids of PEP:PTS and the helpful discussion. We also thank A. Ishihama (National Institute of Genetics) and M. Kitagawa (Nara Institute of Science and Technology) for providing the hfq1::Ω and clpX::kan mutants, respectively. We also thank C. Seto for her technical assistance (i.e., construction of the rpoS-lacZ operon fusion and katE-lacZ fusion).
This work was supported by a grant from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Barth M, Marschall C A, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of ςS and many ςS-dependent genes in Escherichia coli. J Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 3.Cunning C, Brown L, Elliott T. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J Bacteriol. 1998;180:4564–4570. doi: 10.1128/jb.180.17.4564-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean D, Yates J L, Nomura M. Identification of ribosomal protein S7 as a repressor of translation within the str operon of E. coli. Cell. 1981;24:413–419. doi: 10.1016/0092-8674(81)90331-7. [DOI] [PubMed] [Google Scholar]
- 5.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ςS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano M, Shigesada K, Imai M. Construction and characterization of plasmid and lambda phage vector systems for study of transcriptional control in Escherichia coli. Gene. 1987;57:89–99. doi: 10.1016/0378-1119(87)90180-6. [DOI] [PubMed] [Google Scholar]
- 7.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 8.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 9.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 10.Lange R, Hengge-Aronis R. The cellular concentration of the ςS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 11.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 12.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the systhesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 14.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςS subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 15.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 16.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1149–1174. [Google Scholar]
- 17.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweder T, Lee K-H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor (ςS) by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 20.Sondej M, Sun J, Seok Y-J, Kaback H R, Peterkofsky A. Deduction of consensus binding sequences on proteins that bind IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system by cysteine scanning mutagenesis of Escherichia coli lactose permease. Proc Natl Acad Sci USA. 1999;96:3525–3530. doi: 10.1073/pnas.96.7.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi H, Inada T, Postma P, Aiba H. CRP down-regulates adenylate cyclase activity by reducing the level of phosphorylated IIAGlc, the glucose-specific phosphotransferase protein, in Escherichia coli. Mol Gen Genet. 1998;259:317–326. doi: 10.1007/s004380050818. [DOI] [PubMed] [Google Scholar]
- 22.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal ς factor in Escherichia coli: the rpoS gene product, ς38, is a second principal ς factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci USA. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsui H-C T, Leung H-C E, Winkler M E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 25.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamashino T, Kakeda M, Ueguchi C, Mizuno T. An analogue of the DnaJ molecular chaperone whose expression is controlled by ςS during the stationary phase and phosphate starvation in Escherichia coli. Mol Microbiol. 1994;13:475–483. doi: 10.1111/j.1365-2958.1994.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 27.Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, ςS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]