Abstract
l‐Lactate oxidase (LOx) is a flavin mononucleotide (FMN)‐dependent triose phosphate isomerase (TIM) barrel fold enzyme that catalyzes the oxidation of l‐lactate using oxygen as a primary electron acceptor. Although reductive half‐reaction mechanism of LOx has been studied by structure‐based kinetic studies, oxidative half‐reaction and substrate/product‐inhibition mechanisms were yet to be elucidated. In this study, the structure and enzymatic properties of wild‐type and mutant LOxs from Enterococcus hirae (EhLOx) were investigated. EhLOx structure showed the common TIM‐barrel fold with flexible loop region. Noteworthy observations were that the EhLOx crystal structures prepared by co‐crystallization with product, pyruvate, revealed the complex structures with “d‐lactate form ligand,” which was covalently bonded with a Tyr211 side chain. This observation provided direct evidence to suggest the product‐inhibition mode of EhLOx. Moreover, this structure also revealed a flip motion of Met207 side chain, which is located on the flexible loop region as well as Tyr211. Through a saturation mutagenesis study of Met207, one of the mutants Met207Leu showed the drastically decreased oxidase activity but maintained dye‐mediated dehydrogenase activity. The structure analysis of EhLOx Met207Leu revealed the absence of flipping in the vicinity of FMN, unlike the wild‐type Met207 side chain. Together with the simulation of the oxygen‐accessible channel prediction, Met207 may play as an oxygen gatekeeper residue, which contributes oxygen uptake from external enzyme to FMN. Three clades of LOxs are proposed based on the difference of the Met207 position and they have different oxygen migration pathway from external enzyme to active center FMN.
Keywords: crystal structure, dehydrogenase, flavin, lactate oxidase, lactate sensor, l‐lactate, oxidase, oxygen, product inhibition, substrate inhibition
Short abstract
Abbreviations
- 4‐AA
4‐aminoantipyrine
- AvLOx
LOx from Aerococcus viridans
- DCIP
2,6‐dichloroindophenol
- EhLOx
LOx from Enterococcus hirae
- Fcb2
flavocytochrome b 2
- FMN
flavin mononucleotide
- FNR
FMN reduced form
- GOx
glycolate oxidase
- LCHO
long chain hydroxy acid oxidase
- LMO
lactate monooxygenase
- LOx
lactate oxidase
- MDH
mandelate oxidase
- PMS
phenazine methosulfate
- PPB
potassium phosphate buffer
- TIM
triose‐phosphate isomerase.
- TOOS
N‐Ethyl‐N‐(2‐hydroxy‐3‐sulfopropyl)‐3‐methylaniline sodium salt
1. INTRODUCTION
l‐Lactate oxidase (LOx; EC 1.1.3.15) is a member of flavin mononucleotide (FMN)‐dependent α‐hydroxyacid oxidizing flavoproteins family. Many LOx structures and mutagenesis studies have been reported and elucidated its substrate recognition scheme and reductive half‐reaction mechanisms, but its substrate/product inhibition and oxidative half‐reaction mechanisms are yet to be elucidated. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 The well‐characterized proteins of this family members, LOx, 9 glycolate oxidase (GOx), 10 , 11 long‐chain hydroxy acid oxidase (LCHO), 12 , 13 , 14 , 15 mandelate dehydrogenase (MDH), 16 , 17 flavocytochrome b 2 (Fcb2), 18 , 19 , 20 , 21 , 22 and lactate monooxygenase (LMO) 23 , 24 have a (β/α)8 triose phosphate isomerase (TIM) barrel fold with the FMN bound at the C‐terminal end of the β‐sheets. 1 , 7 , 25 , 26 , 27 , 28 , 29 , 30 LOx catalyzes a reductive half‐reaction in which l‐lactate is oxidized to pyruvate by FMN reduction (Equation (1)) as well as an oxidative half‐reaction in which the reduced FMN is reoxidized by molecular oxygen coupled with H2O2 production (Equation (2)). 3
| (1) |
| (2) |
Due to its stability and high specificity, LOx has been utilized as l‐lactate biosensor element. 31 , 32 , 33 In the bioelectrochemical application using LOx, an artificial electron acceptor or redox dye, such as potassium ferricyanide, is used as an electron mediator between enzyme and electrode instead of oxygen (Equations (3) and (4)).
| (3) |
| (4) |
Substrate and product inhibitions were reported as a potential issue when LOx is utilized as the molecular recognition element in the continuous lactate sensor, 34 , 35 and also as the biocatalyst for the biochemical conversion processes. 36 , 37 Substrate and product inhibitions are considered to be caused by reoccupation of the active site with substrate or product after the reductive half‐reaction. 38 Although it has previously been demonstrated that the mutational approach can minimize substrate inhibition, 34 the mechanisms of the product inhibition have not been elucidated. One of the reasons is the lack of structure–function relationship study about LOx, especially focused on an active site flexible loop region. This flexible loop region, between strand β4 and helix α4 of TIM barrel, is called “loop 4.” 26 This loop 4 is a non‐conserved region in the FMN‐dependent α‐hydroxyacid oxidizing flavoproteins family, and its function is still unknown, since this loop 4 is often disordered in X‐ray crystal structure analysis due to its high flexibility (Figure S1). In an attempt to determine its function, Stoisser et al. investigated the LOx loop 4 function through mutagenesis studies. 4 , 5 They reported that two tyrosine residues in loop 4 strongly related to product release and substrate entry. Further studies have revealed a wild‐type LMO and a membrane‐associated MDH whole structures including loop 4 regions. 26 , 29 Those loop 4 regions are longer than that of other FMN‐dependent α‐hydroxyacid oxidizing flavoproteins. Kean and Karplus proposed this loop 4 performs an important role for the slow product release and contributes to the monooxygenation reaction of LMO. 26 On the other hand, Sukumar et al. suggested one of the three helices of MDH loop 4 takes part in electron acceptor binding or gating, and the other hydrophobic helices associate with the membrane. 29 Our group also showed the LOx loop 4 region plays a significant role in the dye‐mediated dehydrogenase activity. 39 Therefore, this loop 4 is likely to be related to both reductive and oxidative half‐reactions.
In this study, LOx derived from Enteroccus hirae (EhLOx) was examined to elucidate product inhibition and the role of loop 4. This was done by preparing protein crystals and determining the crystal structures of the wild‐type and a mutant enzyme harboring amino acid substitution in loop 4, which are co‐crystallized with pyruvate. EhLOx structure was determined to be a TIM‐barrel fold which is commonly observed in other FMN‐dependent α‐hydroxyacid oxidizing flavoproteins structure. Although X‐ray structural study was performed by co‐crystallizing samples with high concentration of pyruvate, the resulting structures were determined to be a complex structure with “d‐lactate form ligand,” which was covalently bonded with Tyr211. This finding is the direct evidence to support a new hypothesis of the product‐inhibition mechanism of LOx. It was also remarkable that the EhLOx structure captured Met207 side chain flip motion in the vicinity of FMN. To investigate the Met207 function, a saturated mutagenesis study was performed on Met207. One of the mutants, Met207Leu showed a drastically decreased oxidative half‐reaction using oxygen as an electron acceptor, while maintaining dye‐mediated dehydrogenase activity using artificial electron acceptors. Remarkably, both Met207 and Tyr211 were located on the loop 4 region. These results suggested an involvement of flexible loop region of LOx in product inhibition and the oxidative half‐reaction. Those suggestions should help us to understand the LOx reaction mechanisms and to solve the problems faced in future industrial applications.
2. RESULTS
2.1. The overall structure of EhLOx
In this study, a structure of EhLOx wild‐type in complex with ligand was elucidated by preparing crystal sample by co‐crystallization with pyruvate. The overall structure of EhLOx wild‐type was homotetramer (Figure 1a). Each monomer of EhLOx has (β/α)8 TIM‐barrel fold with FMN, same as other α‐hydroxyacid oxidizing flavoproteins. The FMN in this structure was observed to have a slightly bent structure of isoalloxazine ring. The oxidized form of isoalloxazine shows a planar conformation which are in an sp 2 configuration while the reduced form of isoalloxazine shows bent structure at the N5 and N10 of FMN which are in an sp 3 configuration. 40 Therefore, the FMN in this structure was refined as the reduced form (FNR). Triethylene glycol, 2‐amino‐2‐hydroxymethyl‐propane‐1,3‐diol, and 1,2‐ethanediol were observed in the structure, and these molecules were derived from the reservoir solution at protein crystallization procedure. A pyruvate molecule was also seen around Arg362. In addition, d‐lactate‐like ligand was identified in the vicinity of the isoalloxazine ring of FMN. Comparing with LOx from Aerococcus viridans (AvLOx) structure (PDB code: 2E77), the EhLOx is structurally similar with AvLOx (52% identity, 0.8 Å r.m.s.d., 60.6 Z‐score) as calculated by Dali search 41 and almost all of the amino acid residues around isoalloxazine ring of FMN are conserved except for Phe189 and Met207 which corresponded to AvLOx Tyr191 and Leu211 (Figure 1b, c). In the complex structure of EhLOx wild‐type, the whole flexible loop region (187–216, so‐called loop 4) was ordered. The simulated‐annealing omit maps (sa‐omit maps) revealed the presence of the strong and continuous electron density maps from Cα of the ligand to the hydroxyl group of Tyr211 (Figure 2). These observations revealed that Cα of the ligand was mainly bonded to Tyr211 in this complex structure, and the bound ligand does not fit an sp 2 configuration of pyruvate (Figure 2h), but instead adapts sp 3 configuration of a Cα of d‐lactate (Figure 2g). Therefore, the bound ligand was refined as d‐lactate form. The distance between the hydroxyl group of Tyr211 and Cα of the d‐lactate form ligand was determined 1.4 Å. Flexibility of Met207 is noteworthy as this residue located on the α‐helix of the loop 4 region. Loop 4 is known as “active‐site lid” between strand β4 and helix α4 of TIM‐barrel fold which can be open or closed state with or without the substrate. 1 , 5 This Met207 was flipped in closed state and this motion has not been reported in previous studies about LOxs.
FIGURE 1.
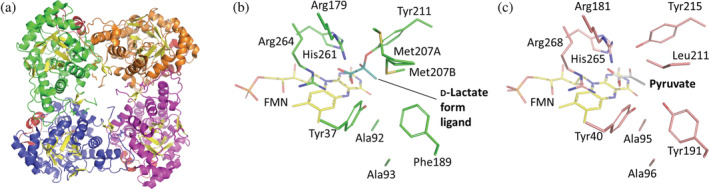
X‐ray structure of EhLOx wild‐type and comparison with AvLOx structure. (a) Overall structure of EhLOx wild‐type. (b) Active site of the EhLOx structure bound with d‐lactate form ligand. Met207 showed alternative conformation, A and B, indicating its flexibility. Each monomer was shown in green, orange, magenta, and blue. The flexible loop regions (187–218) are colored in red and all β‐strands are colored in yellow. (c) Active site of the AvLOx structure bound with pyruvate from PDB ID 2E77 chain D
FIGURE 2.
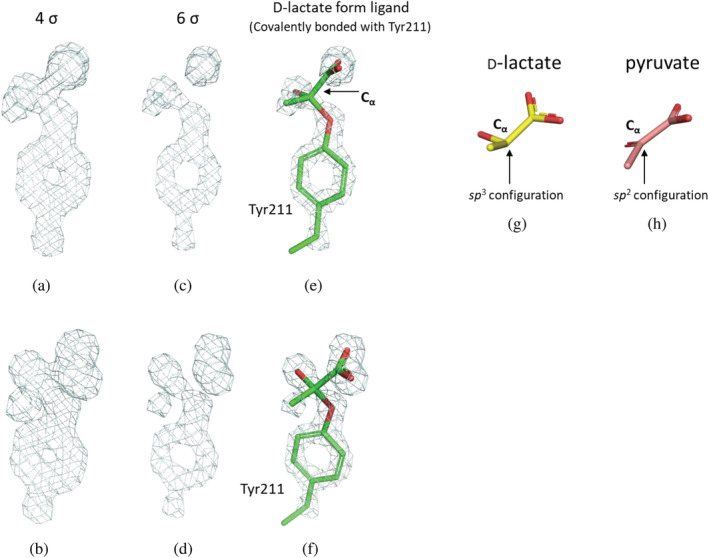
Sa‐omit maps of the side chain of EhLOx wild‐type Tyr211 and ligand. These sa‐omit maps were contoured with two different orientations at 4 σ ((a) and (b)) and 6 σ ((c) and (d)), and structures of d‐lactate (g) and pyruvate (h) for the references. Stick models of Tyr211 and d‐lactate form ligand with green are shown in the sa‐omit maps contoured at 6 σ ((e) and (f)). These images were prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC
2.2. Mutagenesis study
Focusing on the structurally characteristic observation of Met207, the saturation mutagenesis at EhLOx Met207 was carried out as shown in Table 1. As a result, EhLOx wild‐type showed the highest oxidase activity and dehydrogenase activity, which dehydrogenase activity was 170% of the oxidase activity (Dh/Ox ratio). All mutants at Met207 showed a drastic decrease of oxidase activity. Among them, Met207Leu mutant showed a slight decrease of dehydrogenase activity but showed only 1.5% of the wild‐type oxidase activity which results in a Dh/Ox ratio at approximately 10,000% which is 59‐fold higher value than wild‐type. Met207Ile mutant also increased Dh/Ox ratio and maintained dehydrogenase activity comparatively. With introducing small and nonpolar amino acid residues (Ala, Val, and Pro), both the oxidase and dehydrogenase activities were decreased. Moreover, large nonpolar amino acid residue substitution (Phe and Trp) lost activity altogether. Polar uncharged amino acid (Cys, Ser, Thr, Tyr, Gln, and Asn) and Gly substitution were shown to have much lower oxidase activity and dehydrogenase activity. Polar charged amino acid substitution (Asp, Glu, Lys, Arg, and His) did not show the oxidase activity clearly and only His mutation showed little dehydrogenase activity. From these results, Leu and Ile residues, which have similar size and polarity to Methionine residue, showed high dehydrogenase activity and quite low oxidase activity. The other uncharged amino acid substitution showed a decrease in both oxidase and dehydrogenase activities and almost all charged amino acid substitution showed a loss of activity. Relative expression level was calculated from SDS‐PAGE (Figure S2) band intensity as a wild‐type is 100%, revealing there were no significant impact of mutation on the expression level, except Met207Phe, which was lower than 50% of wild‐type. Therefore, the above observations in decreasing activity are due to the impact on mutagenesis on the enzyme structure and kinetics, rather than on the expression level.
TABLE 1.
The oxidase and dehydrogenase activities of EhLOx mutants
| Met207 substitutions | Oxidase activity (Ox) | Dehydrogenase activity (Dh) | Dh/Ox (%) | Relative expression Level (%)* | ||
|---|---|---|---|---|---|---|
| U/mg | % (vs. wild‐type) | U/mg | % (vs. wild‐type) | |||
| Gly | 1.2 ± 0.0 | 8.9% | 4.6 ± 0.0 | 20% | 400% | 83 |
| Ala | 3.0 ± 0.0 | 26% | 7.1 ± 0.1 | 36% | 240% | 73 |
| Val | 1.3 ± 0.0 | 9.1% | 6.2 ± 0.2 | 26% | 490% | 88 |
| Leu | 0.24 ± 0.00 | 1.6% | 25 ± 1 | 96% | 10,000% | 95 |
| Ile | 0.68 ± 0.02 | 4.3% | 20 ± 1 | 83% | 3,300% | 86 |
| Met (wild‐type) | 16 ± 0 | 100% | 27 ± 2 | 100% | 170% | 100 |
| Pro | 1.2 ± 0.0 | 9.2% | 2.0 ± 0.4 | 8.9% | 170% | 83 |
| Phe | 0.0024 ± 0.0001 | 0.037% | 0.038 ± 0.003 | 0.34% | 1,600% | 41 |
| Trp | 0.032 ± 0.001 | 0.23% | 0.093 ± 0.004 | 0.37% | 290% | 91 |
| Ser | 2.8 ± 0.1 | 19% | 6.0 ± 0.2 | 23% | 210% | 94 |
| Thr | 0.99 ± 0.01 | 7.3% | 5.2 ± 0.0 | 22% | 520% | 87 |
| Cys | 1.7 ± 0.1 | 11% | 8.1 ± 0.20 | 31% | 480% | 95 |
| Asn | 0.30 ± 0.01 | 1.7% | 2.2 ± 0.1 | 7.1% | 740% | 120 |
| Gln | 1.5 ± 0.0 | 11% | 5.2 ± 0.1 | 20% | 340% | 93 |
| Tyr | 0.93 ± 0.02 | 6.5% | 5.2 ± 0.1 | 21% | 560% | 91 |
| Asp | 0.048 ± 0.002 | 0.32% | 0.15 ± 0.00 | 0.58% | 320% | 96 |
| Glu | 0.13 ± 0.00 | 0.94% | 0.34 ± 0.02 | 1.4% | 260% | 86 |
| Lys | 0.018 ± 0.000 | 0.11% | 0.22 ± 0.00 | 0.82% | 1,200% | 98 |
| Arg | 0.021 ± 0.000 | 0.13% | 0.33 ± 0.00 | 1.2% | 1,600% | 100 |
| His | 0.12 ± 0.00 | 0.83% | 5.8 ± 0.2 | 23% | 4,800% | 93 |
Note: These expression levels were calculated from SDS‐PAGE band intensity (*Figure S2).
2.3. Enzymatic property of EhLOx wild‐type and Met207Leu mutant
The enzymatic properties of purified EhLOx wild‐type and Met207Leu mutant were investigated (Table 2). These purified levels were confirmed by SDS‐PAGE analysis after gel filtration chromatography (Figure S3). The V max value of wild‐type EhLOx oxidase activity was more than twice as high as that of AvLOx wild‐type, though the K m value was similar to the AvLOx wild‐type. The oxidase activity of Met207Leu mutant was almost lost, and then, substrate inhibition was observed (Figure 3a,b). This result was also observed in AvLOx Ala96Leu mutants. 42 This phenomenon seems to be caused by undersupplying molecular oxygen to the active site, indicating blocking of the oxygen migration by mutation. The V max value for dehydrogenase activity of Met207Leu mutant was 72% of that of wild‐type while also displaying a slightly lower K m value. However, the V max value of the dehydrogenase activity of Met207Leu was still higher than AvLOx wild‐type or Ala96Leu mutant. Substrate specificity was investigated about l‐lactate, d‐lactate, glycolate, and dl‐2‐hydroxybutyrate but Met207Leu mutation did not affect the substrate specificity (Figure 3c). Thermal stability investigation revealed that there was no remarkable difference between wild‐type and Met207Leu mutant EhLOx (Figure 3d). From these results, Met207 has an important role in facilitating the access of oxygen from outside enzyme to the catalytic site. Therefore, Met207 is strongly related to the oxidative half‐reaction of EhLOx.
TABLE 2.
The kinetic parameters of wild‐type EhLOx and Met207Leu mutant for l‐lactate
| Oxidase activity | Dehydrogenase activity | |||||
|---|---|---|---|---|---|---|
| K m (mM) | V max (U/mg) | V max/K m (U/mg·mM) | K m (mM) | V max (U/mg) | V max/K m (U/mg·mM) | |
| Wild‐type | 0.55 ± 0.02 | 200 ± 2 | 370 | 0.48 ± 0.03 | 290 ± 3 | 600 |
| Met207Leu | 0.013 ± 0.002 | 2.0 ± 0.0 | 150 | 0.45 ± 0.04 | 200 ± 3 | 450 |
FIGURE 3.
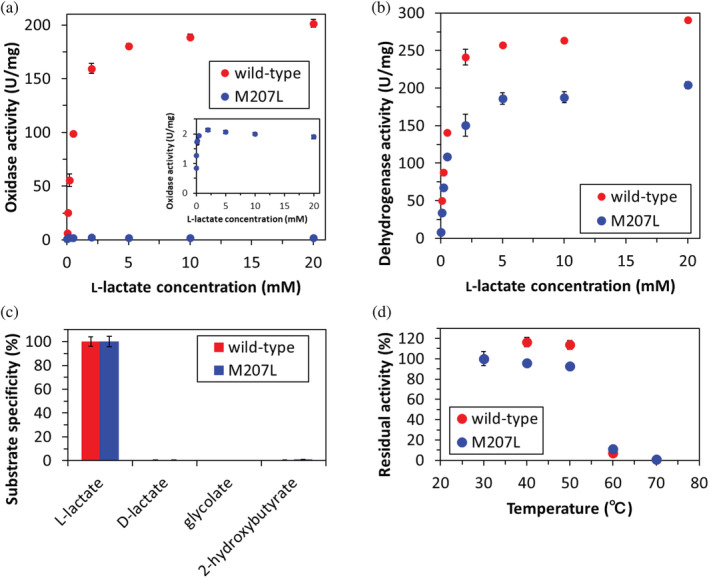
Enzymatic property of EhLOx wild‐type and Met207Leu mutant. (a) Oxidase activity of wild‐type (red) and Met207Leu mutant (blue). (b) Dye‐mediated dehydrogenase activity of wild‐type (red) and Met207Leu mutant (blue). (c) Substrate specificity of wild‐type and Met207Leu mutant evaluated by dehydrogenase activity using 20 mM each substrate. (d) Thermal stability of wild‐type and Met207Leu mutant evaluated by dehydrogenase activity assay after 10 min incubation at each temperature. All assays were conducted using 20 mM PPB pH 7.0 at 25°C
2.4. Comparison of EhLOx wild‐type and Met207Leu mutant structure
Following the structural analysis of EhLOx wild‐type, the structure of EhLOx Met207Leu mutant was also elucidated. Crystallographic data collection and refinement statistics are summarized in Table 3. The overall structure was almost the same as the structure of the wild‐type, including the loop 4 region with a covalently bonded d‐lactate form ligand as a homotetramic structure (Figures 4 and S4). In the mutant structure, an alternative conformation of the 207th amino acid residue side chain was not observed (Figures 4c and S5), suggesting the orientation of the Leu207 side chain fluctuation was fixed. Considering that Met207Leu showed drastic decrease in oxidase activity, and also that the observed Leu substitution mitigated the side chain fluctuation, the flip of Met207 may relate to the reactivity for oxygen. The other amino acid side chains oriented to FMN were not fluctuated in these structures which also indicates that Met207 movement is a critical function for oxidative half‐reaction with oxygen.
TABLE 3.
Data collection and refinement statistics
| Wild‐type LOx | Met207Leu | |
|---|---|---|
| Data collection | ||
| Beamline | PF BL‐5A | PF BL‐5A |
| Temperature (K) | 100 | 100 |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Resolution range (Å) | 19.53–1.70 (1.74–1.70) | 19.54–1.52 (1.56–1.52) |
| No. of measured refs. | 889,509 (67,513) | 1,230,530 (89,913) |
| No. of unique refs. | 65,713 (4,953) | 93,906 (6,875) |
| Redundancy | 13.2 (13.7) | 13.1 (13.1) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.9) |
| Mean I o/(I o) | 22.2 (4.3) | 27.0 (4.4) |
| R merge (%) | 8.5 (76.2) | 6.1 (70.3) |
| CC 1/2 | 0.999 (0.930) | 0.999 (0.942) |
| Space group | I422 | I422 |
| Unit cell parameters | a = 137.96 | a = 138.13 |
| a, b, c (Å) | b = 137.96 | b = 138.13 |
| β (°) | c = 127.80 | c = 127.36 |
| Refinement | ||
| Resolution range (Å) | 19.53–1.70 (1.74–1.70) | 19.54–1.52 (1.56–1.52) |
| No. of refs. | 64,118 (4,710) | 89,192 (6,527) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.9) |
| R factor (%) | 13.2 (18.5) | 12.8 (20.1) |
| R free (%) | 15.4 (22.3) | 14.4 (23.8) |
| RMSD bond lengths (Å) | 0.005 | 0.006 |
| RMSD bond angles (°) | 1.3 | 1.4 |
| Ramachandran plot | ||
| Preferred region (%) | 95.5 | 95.5 |
| Allowed region (%) | 4.2 | 4.2 |
| B‐factor (Å2) | ||
| Protein | 18.0 | 17.1 |
| FNR | 14.3 | 13.8 |
| Lactate (active site) | 17.1 | 14.8 |
| Pyruvate (surface) | 36.5 | 33.5 |
| Water | 27.9 | 28.2 |
| PDB code | 6M73 | 6M74 |
Note: Values in parentheses are of the high‐resolution bin. R merge = ∑ h ∑ i [|I i (h) − <I(h)>|/∑ h ∑ i I i (h)], in which I i is the ith measurement and <I(h)> is the weighted mean of all measurements of I(h).
FIGURE 4.
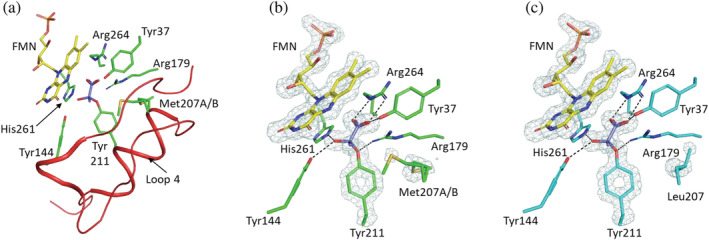
Structures of EhLOx active site in complex with d‐lactate form ligand. The structures of EhLOx wild‐type (a, b) and Met207Leu mutant (c) in complex with d‐lactate form ligand are shown. Met207 and Tyr211 are located in a common flexible region (Phe187‐Leu218, loop4) colored with red which approaches and covers the active site like a lid (a). The simulated‐annealing omit maps of FMN, bound ligand, Tyr211, Met207, and Leu207 in EhLOxs contoured at 4 σ are shown in light blue. Met207 showed alternative conformation (b), revealing the movement of side chain in complex structure, while such an alternative conformation was not observed in Met207Leu mutant (c). The bound substrate is also recognized by Tyr37, Tyr144, Arg179, His261, and Arg264, and forms hydrogen bond with them. The selected hydrogen bonds within a distance of 3.0 Å are indicated by the dotted lines
3. DISCUSSION
This is the first report of the complex structure of LOx which was covalently bonded with loop 4 amino acid side chains and d‐lactate form ligand in the active site. The loop 4 is known as a flexible region because this loop can form an open or closed state depending on the existence of ligands in the FMN‐dependent α‐hydroxyacid oxidizing enzymes. 43 , 44 In the absence of ligands, the loop 4 was often disordered in structural analysis. 1 EhLOx structure without ligand was also evaluated, however, we were not able to obtain clear electron density maps around loop 4 including Tyr211, which is consistent with the cases in the other FMN‐dependent oxidizing enzymes. This observation suggested that the loop4 of EhLOx is also highly flexible, as was reported from the structure of AvLOx 1 (data not shown). Therefore, it was considered that pyruvate enters the active site at the open state and Tyr211 approaches the pyruvate and transition the EhLOx to the closed state. Once in the closed state, the pyruvate binds to the active site and upon the activation, the Tyr211 side chain attacks the pyruvate. This reaction was also aided by the distance between Tyr211 side chain and the ligand Cα. Other amino acids should not interact with this reaction due to the size of their side chain and nucleophilicity, resulting in only the Tyr211 being able to form this d‐lactate form ligand complexed structure. The role of EhLOx Tyr211 in the reductive half‐reaction can be expected to be similar with AvLOx Tyr215 due to the structural similarity between EhLOx and AvLOx. AvLOx Tyr215 was investigated by mutagenesis and kinetic studies and Tyr215 substitution showed decreasing activities because both the l‐lactate dissociation constant and pyruvate release constant were reduced by mutation. 4 From the report, this residue takes part in substrate entry and product release. As well as AvLOx Tyr215, mutations into EhLOx Tyr211 are anticipated to decrease the activity. The hydroxyl group of Tyr211 bound to Cα of the ligand with the length of 1.4 Å. FNR (reduced FMN) and d‐lactate form ligand are shown in yellow and purple stick models of Figure 4, respectively. Because the high concentration of pyruvate (0.4 M) was used to prepare enzyme crystal complexed with ligand, the observed product‐binding mode may show a product‐inhibition mode or an intermediate to a reduced mimic form in the collected data in Synchrotron. d‐Lactate is known as an inhibitor of LOx, 9 therefore, the elucidated structure might be stabilized by d‐lactate form ligand binding as a potential inhibitor of EhLOx (Figure 5), which explains the mechanism of product inhibition. The hydroxyl group of Tyr211 might be activated by the catalytic base His261 (Figure 5a), which is similar to a catalytic acid/base couple of Asp172 and His261. The deprotonated Tyr211 could attack pyruvate Cα as nucleophile and form a hemiacetal (hemiketal) structure due to its proximity between Tyr211 and pyruvate (Figure 5b), as well as a common serine protease reaction scheme. 45 EhLOx Asp172, His261, and Tyr211 aligned like a catalytic triad (Asp‐His‐Ser/Cys) which stabilized each amino acid side chains via hydrogen bonds. The covalently bound d‐lactate form ligand might be stabilized by Tyr144 and His261 side chain which behaves similar to an oxyanion hole of serine protease (Figure 5c). However, as there were no water molecules around the ligand, this only mimicked the oxyanion hole and not the entirety of the serine protease. Furthermore, the His261 and Tyr144 form a hydrogen bond which aides in stabilizing the bound ligand. In addition, the bound ligand is recognized and stacked by active site residues, such as Tyr37, Tyr144, Arg179, His261, and Arg264. The hydrophobic residues of Phe189 and Met207 may also aid in stabilizing the methyl group of d‐lactate form ligand. The tyrosine residue is known as a nucleophile in active sites and formed a hemiacetal structure in sialidase 46 so that the proposed mechanism should be possible. In this binding mode, the nucleophilic sp 3 N5 of reduced FMN cannot attack on α‐proton of d‐lactate form ligand because it orients to methyl group. Therefore, the observed d‐lactate form ligand could bind in the active site as an inhibitor in EhLOx (Figure 5d). According to the report about serine protease inhibitors, there are two types of covalent inhibitors. One is reversible inhibitors which form reversible tetrahedral transition‐state‐like complexes with active site residue of serine proteases. The second is irreversible inhibitors which have a strong terminal electrophilic group, such as sulfonyl fluorides, chloromethyl ketones, and alkyl fluorophosphate. 47 Given that pyruvate is not strong electrophile as found in irreversible protease inhibitors, it may be a covalent reversible inhibitor of LOx.
FIGURE 5.
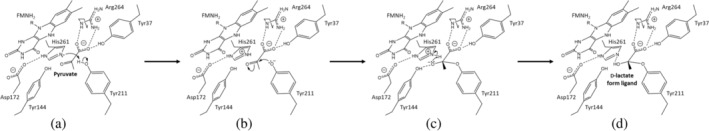
Proposed mechanism for the inhibition mode by d‐lactate form ligand in the presence of high concentration of pyruvate in EhLOx at pH 8.0. (a) Pyruvate entering to the active site and Tyr211 hydroxyl group activation by the catalytic base His261. (b) The deprotonated Tyr211 attacking pyruvate Cα as nucleophile. (c) Oxyanion hole generation and proton transfer from the catalytic base His261. (d) Hemiacetal (hemiketal) structure, d‐lactate form
The reaction mechanism of oxygen in flavin‐dependent oxidases were investigated in many kinds of enzymes but is still not fully elucidated. However, the key to the alteration of the reactivity for oxygen can be divided into three methods by accumulated literature: (a) mutation to the putative residues that compose the cavity where oxygen is located, (b) mutation into the vicinities where the reaction with oxygen takes place, and (c) mutation around possible oxygen‐access routes to the isoalloxazine ring. 48 EhLOx Met207 is far from FMN isoalloxazine C4a, where is the reaction site with molecular oxygen (distance between Cβ and FMN C4a is 10.8 Å in state A and 10.3 Å in state B), or the corresponding oxygen binding site of AvLOx. 49 Many flavin‐dependent oxidases and dehydrogenases had their reactivity for oxygen altered by the second method, mutation into the vicinities of flavin, which is in 3.7–7.3 Å from isoalloxazine C4a. Common examples include: glucose oxidase from Penicillium amagasakiense Val564; 7.1 Å (PDB code: 1GPE), 50 glucose oxidase from Aspergillus niger Val560; 7.3 Å (PDB code: 1CF3), 51 pyranose oxidase Leu547; 7.1 Å and Asn593; 5.2 Å (PDB code: 1TT0), 52 choline oxidase Val464; 5.9 Å (PDB code: 2JBV), 53 aryl‐alcohol oxidase Phe501; 6.0 Å (PDB code: 5OC1), 54 berberine bridge enzyme G164; 3.7 Å and V169; 6.1 Å (PDB code: 4PZF), 55 and pollen allergen Phl p4 I153; 5.8 Å (PDB code: 4PVE), 55 but do exclude proton relay system‐based enzymes (D‐amino acid oxidase family). In these enzymes, the environments around flavin such as hydrophobicity or cavity size were changed by the mutation. On the other hand, EhLOx Met207Leu mutation did not change the hydrophobicity or cavity size around flavin. Considering the above, the Met207Leu mutation might not change the environment around FMN directly but impacted the predicted oxygen‐accessible pathway.
For further investigation of the function of Met207, an oxygen‐accessible channel was analyzed with structural data. Many flavin‐dependent oxidase studies have already revealed the availability of the prediction of an oxygen‐accessible channel. 48 The software CAVER was used for analysis, which is a powerful tool for tunnel analysis in proteins based on reciprocal distance function grid. 56 CAVER analysis was performed using the CAVER 3.0.3 plugin of PyMOL and only the minimum probe radius was changed to 0.95 Å from default value referred to in the previous report from Collazo and Klinman. 57 The initial starting point was set to FMN C4a. For the fluctuation of Met207 of wild‐type EhLOx, the structure was divided into two states depending on the Met207 position. The Met207 side chain of State A oriented to enzyme surface, but State B Met207 side chain oriented to FMN. Analyzing the State A of wild‐type EhLOx, two tunnels were observed at CAVER analysis. One of the tunnels passes by Tyr37, Asp182, Phe187, Thr206, and Met207, called “channel A,” and the other tunnel passes by Ala93, Leu96, Ser123, Phe189, Pro190, Leu191, and Met193, called “channel B” (Figure 6a). The channel B is observed in not only the State B of the wild‐type but also in Met207Leu mutant (Figure 6b and c). However, the channel A is not observed in both wild‐type State B and Met207Leu mutant. Consequently, methionine fluctuation is considered to be responsible for the channel A opening and leucine mutation lost this fluctuation, which can shut channel A stably. Loop 4 is known as a flexible loop region because it can form an open or closed state depending on the substrate or product existence in FMN‐dependent α‐hydroxyacid oxidizing enzymes. The EhLOx structure was obtained as a closed state because of the d‐lactate form ligand may support and stabilize the closed state via covalent bonding. Though other FMN‐dependent α‐hydroxyacid oxidizing enzyme structures including AvLOx showed ambiguous electron density maps of loop 4 region, EhLOx structures showed clear electron density maps. Nevertheless, the flexibility of Met207 side chain was still observed without a steric clash with the ligand. This flexibility can be due to the unbranched methionine side chain which is more flexible than that of other branched hydrophobic amino acid side chain, such as leucine or isoleucine. 58 An amino acid side chain flexibility at the Met207 position has not been reported in other α‐hydroxyacid oxidizing enzymes including AvLOx, thus, it cannot be determined if this phenomenon is unique. However, this methionine side chain flexibility may be required for oxidative half‐reaction with oxygen in EhLOx, considering the difference between the wild‐type and Met207Leu mutant. AvLOx showed only channel B and AvLOx Ala96 is close to this channel (the closest part is 2.6 Å from alanine Cβ), which the side chain orients to the channel (Figure S6). Leucine mutation should occupy the channel due to the larger size of side chain than that of alanine, which assumed that the AvLOx Ala96Leu mutation blocks the bottom channel B due to the steric hindrance of leucine side chain and showed drastically low oxidase activity. EhLOx Ala93Leu mutant, which is the corresponding mutation to the oxygen insensitive mutant of AvLOx Ala96Leu, 42 showed 56% oxidase activity, 45% dehydrogenase activity, and 150% of Dh/Ox ratio compared with wild‐type EhLOx (Table S1). Although AvLOx Leu211Met mutant was also evaluated by activity assay, this mutant also showed decreasing of both oxidase and dehydrogenase activities (20% and 26% of wild‐type AvLOx activities; Table S2). From these results, it is hypothesized that EhLOx and AvLOx employ different oxygen‐accessible channel.
FIGURE 6.
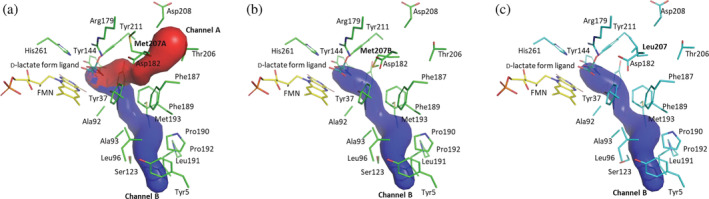
Predicted oxygen‐accessible channels of EhLOx. (a) EhLOx wild‐type state A. (b) EhLOx wild‐type state B. (c) EhLOx Met207Leu mutant. These images were prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. Oxygen‐accessible channel was predicted using PyMOL plugin CAVER 3.0.3. The minimum probe radius, shell depth, shell radius, and clustering threshold were 0.95 Å, 4 Å, 3 Å, and 3.5 respectively. The initial starting point was the FMN C4a. The maximum distance and desired radius were 3 and 5 Å, respectively. d‐lactate form ligand and water molecule were removed in this operation
To support the hypothesis of the difference between EhLOx and AvLOx, a phylogenetic study was conducted. Outputted phylogenetic tree with representative 42 putative LOx sequences including EhLOx and AvLOx can be divided into three major clades based on their amino acid sequences (Figure 7). The first clade is containing LOx from Enterococcus hirae and LOx from Aerococcus viridans belongs to the third clade. The second clade includes putative LOx sequences from Pediococcus but this putative LOx from Pediococcus acidilactici has already studied and did not show lactate oxidase activity. 59 Therefore, the second clade proteins have not been reported as lactate oxidase yet. When compared with the phylogenetic tree and amino acid sequence alignment of representative sequences, Met207 position of EhLOx was completely different depending on each clade. The first clade preserves methionine residue, the second clade shows isoleucine residue, and leucine residue is preserved in third clade. Therefore, the difference between EhLOx and AvLOx can be explained by oxygen‐accessible channel and loop 4 sequence, especially, the position of Met207 in EhLOx.
FIGURE 7.
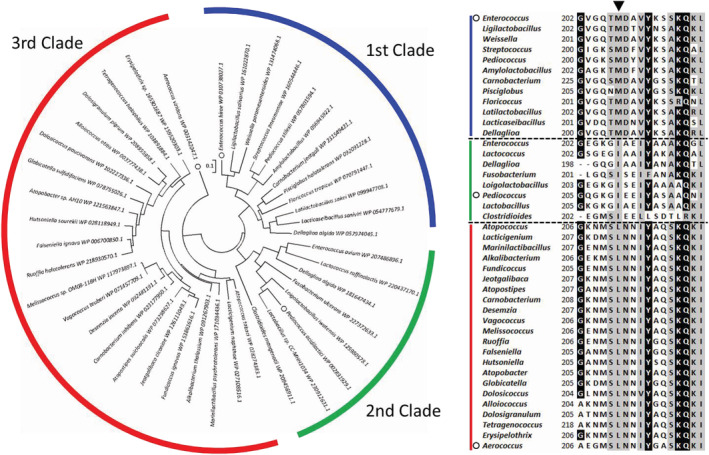
Phylogenetic study of putative LOxs including AvLOx and EhLOx. Phylogenetic tree was constructed using maximum likelihood method using 42 representative lactate oxidase sequences. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There was a total of 352 positions in the final dataset. This analysis was conducted in MEGA7 software. Sequence alignment was prepared by ClustalW
4. CONCLUSION
In this study, the wild‐type and Met207Leu mutant LOxs from Enterococcus hirae were investigated to assess their structures and enzymatic properties. EhLOx structure showed the common TIM‐barrel fold with flexible loop region as observed in other FMN‐dependent α‐hydroxyacid oxidizing flavoproteins. As a result of the structural studies of wild‐type EhLOx, the hydroxyl group of Tyr211 bound to Cα of the d‐lactate‐like ligand has a length of 1.4 Å. Considering the additional covalent bond, the bound ligand was refined as “d‐lactate” and hence called the ligand the “d‐lactate form ligand.” This hemiacetal structure was indicated to be formed and stabilized by the covalent bond between Tyr211 and pyruvate which was tightly fixed by active site amino acids. The reduced FMN cannot transfer N5 protons to the ligand and completely stacked in the active site. EhLOx Asp172, His261, and Tyr211 aligned like a catalytic triad (Asp‐His‐Ser/Cys) which stabilized each amino acid side chains via hydrogen bonds. The covalently bound d‐lactate form ligand is believed to be stabilized by Tyr144 and His261 side chain which behaves similar to an oxyanion hole of serine protease. Therefore, this observation provided direct evidence to suggest the product‐inhibition mode of LOxs. Moreover, this structure also revealed a flip motion of Met207 side chain, which is located on the flexible loop region, as well as Tyr211. Through a saturation mutagenesis study of Met207, one of the mutant Met207Leu showed drastically decreased oxidase activity but maintained dye‐mediated dehydrogenase activity. The other amino acids substitution decreased or lost both oxidase and dehydrogenase activities. The oxidase activity of Met207Leu mutant was almost lost but the V max value for dehydrogenase activity of Met207Leu mutant was maintained 72% of that of wild‐type while also displaying a slightly lower K m value. EhLOx Met207Leu mutant structure revealed the absence of flipping of Leu207 side chain, unlike the wild‐type Met207 side chain. The oxygen‐accessible channel prediction showed two channels in the wild‐type structure and one of the channels can be opened or closed by Met207 flipping, but this channel was not identified in Met207Leu mutant. Therefore, Met207 may play a role as an oxygen gatekeeper residue, which contributes to the oxygen uptake from external enzyme to FMN. Finally, we propose that LOxs can be divided into three clades based on the difference of the Met207 position and the subsequent different oxygen migration pathway from external enzyme to active center FMN. This division is also supported by the result of the EhLOx Ala93Leu and AvLOx Leu211Met mutant activities, which the substitution decreased both oxidase activity and dehydrogenase activities, unlike the corresponding mutation of AvLOx (Ala96Leu mutant) and EhLOx (Met207Leu mutant). Through a phylogenetic study and oxygen‐accessible pathway prediction, AvLOx and EhLOx were different in the key amino acid residue for the reaction with oxygen. These structural features were not captured in previously reported AvLOx structural studies and have unveiled new aspects of the function of the LOx active site loop. These findings also can help us to engineer LOxs for industrial applications.
5. MATERIALS AND METHODS
5.1. Chemicals and materials
Sodium l‐lactate was purchased from Sigma‐Aldrich (S. Louis, MO). 4‐aminoantipyrine (4‐AA) and phenazine methosulfate (PMS) were obtained from Kanto Kagaku (Tokyo, Japan). 2,6‐dichloroindophenol (DCIP) (Merck, Darmstadt, Germany), Peroxidase (Amano Enzyme, Gifu, Japan), N‐Ethyl‐N‐(2‐hydroxy‐3‐sulfopropyl)‐3‐methylaniline sodium salt (TOOS, Dojindo Laboratories, Kumamoto, Japan), 140 mM NaCl, 10 mM phosphate buffer, and 3 mM KCl (PBS) tablets (Calbiochem, NJ) were purchased. All chemicals were reagent grade.
5.2. Bacterial strains and plasmids
The EhLOx wild‐type gene (NCBI Reference Sequence: WP_010738027.1) was initially cloned from Enterococcus hirae NBRC 3181, which was obtained from the National Institute of Technology and Evaluation of Japan. This gene was inserted into the multi‐cloning site of the expression vector pET30c(+) (Merck KGaA, Darmstadt, Germany). For mutagenesis study, site‐directed mutagenesis was performed using the Quikchange method with two primers (Table S3). The plasmid sequences were confirmed by ABI Prism 3,100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
5.3. Recombinant production and purification of EhLOx
The recombinant wild‐type or Met207Leu mutant EhLOx preparation procedure, saturation mutagenesis study, and enzyme activity assay followed previously reported procedures. 42 Briefly, E. coli BL21 (DE3) were transformed with the constructed expression vector and cultivated aerobically in 3 ml (for mutagenesis study) or 2 L (for structure analysis and characterization) of ZYP‐5052 medium (0.5% glycerol, 0.05% glucose, 0.2% lactose, 50 mM KH2PO4, 25 mM [NH4]2SO4, 50 mM Na2HPO4, and 1 mM MgSO4) containing 50 μg/ml kanamycin 60 for 24 hr at 30°C. Cells were harvested via centrifugation and disrupted in 20 mM potassium phosphate buffer (PPB [pH 7.0]). For structure analysis and characterization, crude samples were centrifuged and the resulting supernatants were dialyzed overnight in 20 mM PPB (pH 7.0). EhLOx was purified with an Äkta FPLC system. The crude enzymes were loaded onto a ResourceQ column (GE Healthcare UK Ltd, Buckinghamshire, England; 6 ml), equilibrated with 20 mM PPB (pH 7.0), and the purified fraction was eluted with a linearly increasing gradient of 0.0–0.5 M KCl in the same buffer. The partial purified fractions, characterized by an oxidized FMN absorbance peak at 455 nm, were concentrated four‐fold by ultrafiltration and loaded onto a HiLoad 26/600 Superdex 200 pg column (GE Healthcare; 320 ml), equilibrated with 20 mM PPB (pH 7.0) containing 0.2 M KCl. The purified samples were stored at 4°C until evaluation of enzymatic property and structure analysis. Protein purity was confirmed by SDS‐PAGE using a 10–17% gradient polyacrylamide gel (SuperSep™; Wako Pure Chemical Industries, Osaka, Japan).
5.4. Crystallization and X‐ray crystallography
The initial crystal screening was performed using the sitting‐drop vapor diffusion method with the Mosquito system (TTP LabTech, Hertfordshire, UK) at 20°C. The protein concentration of wild‐type LOx and Met207Leu was prepared to 11.7 mg/ml and 11.2 mg/ml in 20 mM PPB pH 7.0. Using Crystal Screen kits 1 & 2; PEGRx 1 & 2 (Hampton Research Corp., CA), yellow crystals were observed in several conditions. For further optimization, the conditions were manually set up by the sitting‐drop method using a 96‐well plate (Corning Inc., Corning, NY) at 20°C. Well‐diffracting crystals were obtained in a droplet containing 0.8 μl of protein solution (10–12 mg/ml in 20 mM PPB pH 7.0) and 0.8 μl of reservoir solution (0.1 M Tris–HCl, pH 8.0, 30% (w/v) Polyethylene glycol monomethyl ether 2,000) in a well containing 50 μl of the reservoir solution. To obtain the structure in complex with substrate, each protein solution containing 0.4 M sodium pyruvate was used in the same crystallization condition.
Each crystal of wild‐type LOx and Met207Leu in complex with pyruvate was mounted in a cryoloop using 30% (v/v) ethylene glycol as cryo‐protectant, and was flash‐cooled in a stream of nitrogen gas at 100 K. X‐ray diffraction data were collected using the DECTRIS Pilatus3 S 6M detector with a wavelength of 1.0 Å on the Photon Factory BL‐5A at the High Energy Accelerator Research Organization (KEK; Tsukuba, Japan). The collected data were processed using the XDS program, 61 and the CCP4 program suite. 62 The initial phases of the wild‐type LOx were determined by molecular replacement using the program MOLREP 63 with the structure of l‐lactate oxidase from Aerococcus viridans (PDB code: 4YL2) 5 as a search model. Further model building was performed with the program Coot, 64 and the structure was refined using the program Refmac5. 65 Data collection and refinement statistics are listed in Table 3. Figures 1, 2, 4, and 6 were prepared using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
5.5. Enzyme assays
LOx activities were measured in 20 mM PPB (pH 7.0) containing 1.5 mM 4‐AA, 1.5 mM TOOS, 2.0 U/ml peroxidase, and various concentrations of sodium l‐lactate by monitoring the formation of quinonimine dye at 555 nm with 39.2 mM−1 cm−1 as the molar absorption coefficient of TOOS (pH 7.0). One unit of oxidase activity was defined as the amount of enzyme catalyzing the production of 1 μmol H2O2 per minute at 25°C. Dye‐mediated lactate dehydrogenase activities were measured in 20 mM PPB (pH 7.0) containing 0.06 mM DCIP, 4 mM PMS and various concentrations of sodium l‐lactate by monitoring the reduced DCIP at 600 nm with 16.3 mM−1 cm−1 as the molar absorption coefficient of DCIP (pH 7.0). One unit of dehydrogenase activity was defined as the amount of enzyme catalyzing the reduction of 1 μmol DCIP per minute at 25°C. The kinetic parameters were calculated by SigmaPlot 14 (Systat Software, Inc.). Substrate specificity was evaluated by dehydrogenase activity using 20 mM l‐lactate, d‐lactate, glycolate, and dl‐2‐hydroxybutyrate. Thermal stability was tested by evaluating dehydrogenase activity assay after 10 min incubation at 30, 40, 50, 60, and 70°C. These assays were performed in triplicates. Protein concentrations were determined by the DC Protein Assay Kit (Bio‐Rad Laboratories, Hercules, CA), and expression levels were calculated by image analysis (Band %) with Image Lab 4.0 software (Bio‐Rad Laboratories).
5.6. Amino acid sequence comparison and phylogenetic studies
Sequence alignment of FMN‐dependent α‐hydroxyacid oxidizing flavoproteins family was prepared by ClustalW 66 using the software, BioEdit version 7.2.6. 67 Amino acid sequences were chosen from the structures of known enzymes registered in Protein Data Bank (PDB). Phylogenetic study was carried out using the 1,000 putative LOx sequences. These sequences were searched in BLAST search 68 in April 2022 with EhLOx amino acid sequence as query. Some sequences containing gaps or missing data were eliminated from the 1,000 sequences. The phylogenetic tree and evolutionary distances with 941 selected putative LOx sequences were calculated by Maximum Likelihood method 69 using the software, MEGA7. 70 As the first generated tree can be divided into two main clades, the representative sequences were selected from different genera in each clade. Then, the phylogenetic tree was generated using the selected 42 sequences.
5.7. Data deposition
The atomic coordinates and structure factors of EhLOx wild‐type and Met207Leu mutant in complexes with substrates have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB), 6M73 (EhLOx wild‐type in complex with d‐lactate; Data S1) and 6M74 (Met207Leu mutant in complex with d‐lactate; Data S2), respectively.
AUTHOR CONTRIBUTIONS
Kentaro Hiraka: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); validation (equal); visualization (equal); writing – original draft (lead); writing – review and editing (equal). Hiromi Yoshida: Data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); project administration (equal); resources (equal); writing – original draft (equal); writing – review and editing (equal). Wakako Tsugawa: Writing – original draft (equal); writing – review and editing (equal). Ryutaro Asano: Supervision (equal); writing – original draft (equal); writing – review and editing (equal). Jeffrey T. La Belle: Supervision (equal); writing – review and editing (equal). Kazunori Ikebukuro: Supervision (equal); writing – review and editing (equal). Koji Sode: Conceptualization (equal); funding acquisition (lead); investigation (equal); project administration (equal); supervision (lead); writing – original draft (equal); writing – review and editing (equal).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1: The oxidase and dehydrogenase activities of EhLOx wild‐type and Ala93Leu mutant.
Table S2: The oxidase and dehydrogenase activities of AvLOx wild‐type, Ala96Leu, and Leu211Met mutants.
Table S3: Primers used for cloning and site‐directed mutagenesis study.
Figure S1: Amino acid sequence alignment of FMN dependent α‐hydroxyacid oxidizing flavoproteins family members.
Figure S2: SDS‐PAGE analysis at the site‐directed mutagenesis study for evaluation of soluble expression level.
Figure S3: SDS‐PAGE analysis at the protein purification.
Figure S4: Sa‐omit maps of the side chain of EhLOx Met207Leu mutant Tyr211 and ligand.
Figure S5: Sa‐omit maps of the side chains of Met207 and Met207Leu.
Figure S6: Predicted oxygen‐accessible pathway of AvLOx (PDB code: 2E77 chain D).
Figure S7: SDS‐PAGE analysis at the site‐directed mutagenesis study of AvLOx wild‐type, Ala96Leu mutant, and Leu211Met mutant.
Data S1 6M73
Data S2 6M74
ACKNOWLEDGMENTS
We greatly acknowledge supporting from the Japan Public–Private Partnership Student Study Abroad Program (TOBITATE! Young Ambassador Program) by the Japan Student Services Organization (JASSO). Kentaro Hiraka was financially supported by the Nakatani Foundation for Advancement of Measuring Technologies in Biomedical Engineering and Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowships. The authors also thank Mr. Blake Morrow for English, his proofreading efforts on this manuscript.
Hiraka K, Yoshida H, Tsugawa W, Asano R, La Belle JT, Ikebukuro K, et al. Structure of lactate oxidase from Enterococcus hirae revealed new aspects of active site loop function: Product‐inhibition mechanism and oxygen gatekeeper. Protein Science. 2022;31(10):e4434. 10.1002/pro.4434
Review Editor: John Kuriyan
Funding information Japan Society for the Promotion of Science; Japan Student Services Organization; Nakatani Foundation for Advancement of Measuring Technologies in Biomedical Engineering
REFERENCES
- 1. Leiros I, Wang E, Rasmussen T, et al. The 2.1 Å structure of Aerococcus viridans l‐lactate oxidase (LOX). Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li SJ, Umena Y, Yorita K, et al. Crystallographic study on the interaction of l‐lactate oxidase with pyruvate at 1.9 Å resolution. Biochem Biophys Res Commun. 2007;358:1002–1007. [DOI] [PubMed] [Google Scholar]
- 3. Maeda‐Yorita K, Aki K, Sagai H, Misaki H, Massey V. l‐lactate oxidase and l‐lactate monooxygenase: Mechanistic variations on a common structural theme. Biochimie. 1995;77:631–642. [DOI] [PubMed] [Google Scholar]
- 4. Stoisser T, Brunsteiner M, Wilson DK, Nidetzky B. Conformational flexibility related to enzyme activity: Evidence for a dynamic active‐site gatekeeper function of Tyr215 in Aerococcus viridans lactate oxidase. Sci Rep. 2016;6:27892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stoisser T, Klimacek M, Wilson DK, Nidetzky B. Speeding up the product release: A second‐sphere contribution from Tyr191 to the reactivity of l‐lactate oxidase revealed in crystallographic and kinetic studies of site‐directed variants. FEBS J. 2015;282:4130–4140. [DOI] [PubMed] [Google Scholar]
- 6. Stoisser T, Rainer D, Leitgeb S, Wilson DK, Nidetzky B. The Ala95‐to‐Gly substitution in Aerococcus viridans l‐lactate oxidase revisited—Structural consequences at the catalytic site and effect on reactivity with O2 and other electron acceptors. FEBS J. 2015;282:562–578. [DOI] [PubMed] [Google Scholar]
- 7. Umena Y, Yorita K, Matsuoka T, Kita A, Fukui K, Morimoto Y. The crystal structure of l‐lactate oxidase from Aerococcus viridans at 2.1 Å resolution reveals the mechanism of strict substrate recognition. Biochem Biophys Res Commun. 2006;350:249–256. [DOI] [PubMed] [Google Scholar]
- 8. Yorita K, Aki K, Ohkuma‐Soyejima T, Kokubo T, Misaki H, Massey V. Conversion of l‐lactate oxidase to a long chain α‐hydroxyacid oxidase by site‐directed mutagenesis of alanine 95 to glycine. J Biol Chem. 1996;271:28300–28305. [DOI] [PubMed] [Google Scholar]
- 9. Duncan JD, Wallis JO, Azari MR. Purification and properties of Aerococcus viridans lactate oxidase. Biochem Biophys Res Commun. 1989;164:919–926. [DOI] [PubMed] [Google Scholar]
- 10. Frigerio NA, Harbury HA. Preparation and some properties of crystalline glycolic acid oxidase of spinach. J Biol Chem. 1958;231:135–157. [PubMed] [Google Scholar]
- 11. Zelitch I, Ochoa S. Oxidation and reduction of glycolic and glyoxylic acids in plants. I. Glycolic and oxidase. J Biol Chem. 1953;201:707–718. [PubMed] [Google Scholar]
- 12. Allen JM, Beard ME. α‐Hydroxy acid oxidase: Localization in renal microbodies. Science. 1965;149:1507–1509. [DOI] [PubMed] [Google Scholar]
- 13. Duley JA, Holmes RS. L‐α‐hydroxyacid oxidase isozymes: Purification and molecular properties. Eur J Biochem. 1976;63:163–173. [DOI] [PubMed] [Google Scholar]
- 14. Lê KD, Lederer F. Amino acid sequence of long chain alpha‐hydroxy acid oxidase from rat kidney, a member of the family of FMN‐dependent alpha‐hydroxy acid‐oxidizing enzymes. J Biol Chem. 1991;266:20877–20881. [PubMed] [Google Scholar]
- 15. Nakano M, Ushijima Y, Saga M, Tsutsumi Y, Asami H. Aliphatic L‐α‐hydroxyacid oxidase from rat livers purification and properties. Biochim Biophys Acta Enzymol. 1968;167:9–22. [DOI] [PubMed] [Google Scholar]
- 16. Mitra B, Gerlt JA, Babbitt PC, et al. A novel structural basis for membrane association of a protein: Construction of a chimeric soluble mutant of (S)‐mandelate dehydrogenase from Pseudomonas putida . Biochemistry. 1993;32:12959–12967. [DOI] [PubMed] [Google Scholar]
- 17. Tsou AY, Ransom SC, Gerlt JA, Buechter DD, Babbitt PC, Kenyon GL. Mandelate pathway of Pseudomonas putida: Sequence relationships involving mandelate racemase, (S)‐mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli . Biochemistry. 1990;29:9856–9862. [DOI] [PubMed] [Google Scholar]
- 18. Appleby CA, Morton RK. Lactic dehydrogenase and cytochrome b 2 of baker's yeast. Enzymic and chemical properties of the crystalline enzyme. Biochem J. 1959;73:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bach SJ, Dixon M, Keilin D. A new soluble cytochrome component from yeast. Nature. 1942;149:21. [Google Scholar]
- 20. Bach SJ, Dixon M, Zerfas LG. Lactic dehydrogenase of yeast. Nature. 1942;149:48–49. [Google Scholar]
- 21. Bach SJ, Dixon M, Zerfas LG. Yeast lactic dehydrogenase and cytochrome b 2 . Biochem J. 1946;40:229–239. [PMC free article] [PubMed] [Google Scholar]
- 22. Boeri E, Cutolo E, Luzzati M, Tosi L. Preparation and properties of cytochrome b 2 from yeast. Arch Biochem Biophys. 1955;56:487–499. [DOI] [PubMed] [Google Scholar]
- 23. Edson N. The oxidation of lactic acid by Mycobacterium phlei . Biochem J. 1947;41:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sutton W. Mechanism of action and crystallization of lactic oxidative decarboxylase from Mycobacterium phlei . J Biol Chem. 1957;226:395–405. [PubMed] [Google Scholar]
- 25. Cunane LM, Barton JD, Chen Z‐w, et al. Crystal structure analysis of recombinant rat kidney long chain hydroxy acid oxidase. Biochemistry. 2005;44:1521–1531. [DOI] [PubMed] [Google Scholar]
- 26. Kean KM, Karplus PA. Structure and role for active site lid of lactate monooxygenase from Mycobacterium smegmatis . Protein Sci. 2019;28:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindqvist Y. Refined structure of spinach glycolate oxidase at 2 Å resolution. J Mol Biol. 1989;209:151–166. [DOI] [PubMed] [Google Scholar]
- 28. Sukumar N, Dewanti AR, Mitra B, Mathews FS. High resolution structures of an oxidized and reduced flavoprotein. The water switch in a soluble form of (S)‐mandelate dehydrogenase. J Biol Chem. 2004;279:3749–3757. [DOI] [PubMed] [Google Scholar]
- 29. Sukumar N, Liu S, Li W, Mathews FS, Mitra B, Kandavelu P. Structure of the monotopic membrane protein (S)‐mandelate dehydrogenase at 2.2 Å resolution. Biochimie. 2018;154:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia Z‐x, Mathews FS. Molecular structure of flavocytochrome b 2 at 2.4 Å resolution. J Mol Biol. 1990;212:837–863. [DOI] [PubMed] [Google Scholar]
- 31. Fu L, Liu J, Hu Z, Zhou M. Recent advances in the construction of biofuel cells based self‐powered electrochemical biosensors: A review. Electroanalysis. 2018;30:2535–2550. [Google Scholar]
- 32. Nikolaus N, Strehlitz B. Amperometric lactate biosensors and their application in (sports) medicine, for life quality and wellbeing. Microchim Acta. 2008;160:15–55. [Google Scholar]
- 33. Rathee K, Dhull V, Dhull R, Singh S. Biosensors based on electrochemical lactate detection: A comprehensive review. Biochem Biophys Rep. 2016;5:35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hiraka K, Tsugawa W, Asano R, et al. Rational design of direct electron transfer type l‐lactate dehydrogenase for the development of multiplexed biosensor. Biosens Bioelectron. 2021;176:112933. [DOI] [PubMed] [Google Scholar]
- 35. Taurino I, Reiss R, Richter M, et al. Comparative study of three lactate oxidases from Aerococcus viridans for biosensing applications. Electrochim Acta. 2013;93:72–79. [Google Scholar]
- 36. Li G, Lian J, Xue H, et al. Enzymatic preparation of pyruvate by a whole‐cell biocatalyst coexpressing l‐lactate oxidase and catalase. Process Biochem. 2020;96:113–121. [Google Scholar]
- 37. Li G, Lian J, Xue H, et al. Biocascade synthesis of L‐tyrosine derivatives by coupling a thermophilic tyrosine phenol‐lyase and l‐lactate oxidase. Eur J Org Chem. 2020;2020:1050–1054. [Google Scholar]
- 38. Kühl PW. Excess‐substrate inhibition in enzymology and high‐dose inhibition in pharmacology: A reinterpretation. Biochem J. 1994;298:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hiraka K, Kojima K, Tsugawa W, Asano R, Ikebukuro K, Sode K. Rational engineering of Aerococcus viridans l‐lactate oxidase for the mediator modification to achieve quasi‐direct electron transfer type lactate sensor. Biosens Bioelectron. 2020;151:111974. [DOI] [PubMed] [Google Scholar]
- 40. Dixon DA, Lindner DL, Branchaud B, Lipscomb WN. Conformations and electronic structures of oxidized and reduced isoalloxazine. Biochemistry. 1979;18:5770–5775. [DOI] [PubMed] [Google Scholar]
- 41. Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hiraka K, Kojima K, Lin C‐E, et al. Minimizing the effects of oxygen interference on l‐lactate sensors by a single amino acid mutation in Aerococcus viridans l‐lactate oxidase. Biosens Bioelectron. 2018;103:163–170. [DOI] [PubMed] [Google Scholar]
- 43. Furubayashi N, Inaka K, Kamo M, Umena Y, Matsuoka T, Morimoto Y. Dynamic interactions in the l‐lactate oxidase active site facilitate substrate binding at pH4.5. Biochem Biophys Ress Commun. 2021;568:131–135. [DOI] [PubMed] [Google Scholar]
- 44. Murray MS, Holmes RP, Todd Lowther W. Activesite and loop 4 movements within human glycolate oxidase: Implications for substrate specificity and drug design. Biochemistry. 2008;47:2439–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu B, Schofield CJ, Wilmouth RC. Structural analyses on intermediates in serine protease catalysis. J Biol Chem. 2006;281:24024–24035. [DOI] [PubMed] [Google Scholar]
- 46. Watts AG, Damager I, Amaya ML, et al. Trypanosoma cruzi trans‐sialidase operates through a covalent sialyl‐enzyme intermediate: Tyrosine is the catalytic nucleophile. J Am Chem Soc. 2003;125:7532–7533. [DOI] [PubMed] [Google Scholar]
- 47. Leung D, Abbenante G, Fairlie DP. Protease inhibitors: Current status and future prospects. J Med Chem. 2000;43:305–341. [DOI] [PubMed] [Google Scholar]
- 48. Hiraka K, Tsugawa W, Sode K. Alteration of electron acceptor preferences in the oxidative half‐reaction of flavin‐dependent oxidases and dehydrogenases. Int J Mol Sci. 2020;21:3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Furuichi M, Suzuki N, Dhakshnamoorhty B, et al. X‐ray structures of Aerococcus viridans lactate oxidase and its complex with d‐lactate at pH 4.5 show an α‐hydroxyacid oxidation mechanism. J Mol Biol. 2008;378:436–446. [DOI] [PubMed] [Google Scholar]
- 50. Tremey E, Stines‐Chaumeil C, Gounel S, Mano N. Designing an O2‐insensitive glucose oxidase for improved electrochemical applications. ChemElectroChem. 2017;4:2520–2526. [Google Scholar]
- 51. Gutierrez EA, Mundhada H, Meier T, Duefel H, Bocola M, Schwaneberg U. Reengineered glucose oxidase for amperometric glucose determination in diabetes analytics. Biosens Bioelectron. 2013;50:84–90. [DOI] [PubMed] [Google Scholar]
- 52. Brugger D, Krondorfer I, Shelswell C, Huber‐Dittes B, Haltrich D, Peterbauer CK. Engineering pyranose 2‐oxidase for modified oxygen reactivity. PLoS One. 2014;9:e109242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Finnegan S, Agniswamy J, Weber IT, Gadda G. Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase. Biochemistry. 2010;49:2952–2961. [DOI] [PubMed] [Google Scholar]
- 54. Hernández‐Ortega A, Lucas F, Ferreira P, Medina M, Guallar V, Martínez AT. Modulating O2 reactivity in a fungal flavoenzyme involvement of aryl‐alcohol oxidase Phe‐501 contiguous to catalytic histidine. J Biol Chem. 2011;286:41105–41114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zafred D, Steiner B, Teufelberger AR, et al. Rationally engineered flavin‐dependent oxidase reveals steric control of dioxygen reduction. FEBS J. 2015;282:3060–3074. [DOI] [PubMed] [Google Scholar]
- 56. Chovancova E, Pavelka A, Benes P, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8(10):e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Collazo L, Klinman JP. Control of the position of oxygen delivery in soybean lipoxygenase‐1 by amino acid side chains within a gas migration channel. J Biol Chem. 2016;291:9052–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aledo JC. Methionine in proteins: The Cinderella of the proteinogenic amino acids. Protein Sci. 2019;28:1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ashok Y, Maksimainen MM, Kallio T, Kilpeläinen P, Lehtiö L. FMN‐dependent oligomerization of putative lactate oxidase from Pediococcus acidilactici . PLoS One. 2020;15:e0223870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Studier FW. Protein production by auto‐induction in high‐density shaking cultures. Protein Expr Purif. 2005;41:207–234. [DOI] [PubMed] [Google Scholar]
- 61. Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Struct Biol. 2010;66:22–25. [DOI] [PubMed] [Google Scholar]
- 64. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. [DOI] [PubMed] [Google Scholar]
- 66. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hall T. BioEdit: A user‐friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 68. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. [DOI] [PubMed] [Google Scholar]
- 69. Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8:275–282. [DOI] [PubMed] [Google Scholar]
- 70. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: The oxidase and dehydrogenase activities of EhLOx wild‐type and Ala93Leu mutant.
Table S2: The oxidase and dehydrogenase activities of AvLOx wild‐type, Ala96Leu, and Leu211Met mutants.
Table S3: Primers used for cloning and site‐directed mutagenesis study.
Figure S1: Amino acid sequence alignment of FMN dependent α‐hydroxyacid oxidizing flavoproteins family members.
Figure S2: SDS‐PAGE analysis at the site‐directed mutagenesis study for evaluation of soluble expression level.
Figure S3: SDS‐PAGE analysis at the protein purification.
Figure S4: Sa‐omit maps of the side chain of EhLOx Met207Leu mutant Tyr211 and ligand.
Figure S5: Sa‐omit maps of the side chains of Met207 and Met207Leu.
Figure S6: Predicted oxygen‐accessible pathway of AvLOx (PDB code: 2E77 chain D).
Figure S7: SDS‐PAGE analysis at the site‐directed mutagenesis study of AvLOx wild‐type, Ala96Leu mutant, and Leu211Met mutant.
Data S1 6M73
Data S2 6M74


