Abstract
Acute myeloid leukemia (AML) is the most heterogeneous hematological disorder and blast cells need to fight against immune system. Natural killer (NK) cells can elicit fast anti-tumor responses in response to surface receptors of tumor cells. NK-cell activity is often impaired in the disease, and there is a risk of insufficient tumor suppression and progression. The aim of this study is to assess the dysfunction of NK cells in AML patients via focusing on two important pathways. We obtained single-cell RNA-sequencing data from NK cells obtained from healthy donors and AML patients. The data were used to perform a wide variety of approaches, including DESeq2 (version 3.9), limma (version 3.26.8) power differential expression analyses, hierarchical clustering, gene set enrichment, and pathway analysis. ATP6AP2, LNPEP, PREP, IGF2R, CTSA, and THOP1 genes were found to be related to the renin–angiotensin system (RAS) family, while DPP3, GLRA3, CRCP, CHRNA5, CHRNE, and CHRNB1 genes were associated with the neurotransmitter pathways. The determined genes are expressed within different patterns in the AML and healthy groups. The relevant molecular pathways and clusters of genes were identified, as well. The cross-talks of NK-cell dysfunction in relation to the RAS and neurotransmitters seem to be important in the genesis of AML.
Keywords: leukemia, natural killer cells, dysfunction
1. Introduction
Acute myeloid leukemia (AML) is a phenotypically and genetically heterogeneous malignancy with an extremely poor prognosis due to disease recurrence being the major reason for therapy failure [1,2]. Comprehensive immunological profiling of newly diagnosed patients with AML reveals that those abnormalities in T-cell and natural killer (NK)-cell activity are the primary contributors to immune dysfunction, although B-cell function remains unaltered. Immunosuppression that leads to anti-leukemia immunity evasion is caused by T-cell maturity and depletion, as well as increased NK maturation and reduced T-cell activity. Effective therapeutic response to chemotherapy is associated with the restoration of T and NK function, and specific immunological markers are associated with overall survival [3,4].
Blood pressure, vascular resistance, and fluid/electrolyte balance are controlled by the renin–angiotensin system (RAS), which is a complex set of proteins that regulate these parameters. Also, RAS pathway genes have been identified in tissues, such as the bone marrow, where they have been demonstrated to be involved in leukemic hematopoiesis, or the generation of new blood cells. Studies have found that exposure to RAS inhibitors (RASi) can suppress the development of cancers via multimodal mechanisms and has attracted increased attention in the recent past. Due to their ability to limit tumor growth, proliferation, and metastasis, RASi are thought to be promising options for enhancing the effectiveness of chemoradiotherapy and targeted therapy [5].
Because of the variability and intricacy of the tumor microenvironment, AML remains untreatable. To develop successful treatment methods, it is critical to clarify the etiology of AML and to discover biomarkers related to leukemia. NK cells are a kind of cytotoxic immune cell that is capable of promptly recognizing and eliminating cancer cells. Analyzing tumor cells’ expression and kinds of NK-associated molecules might help predict how well patients will fare after being diagnosed with acute lymphoblastic leukemia (ND-AML).
A growing amount of research indicates that NK cells may be very effective against CML, AML, and MDS. But autoimmune condition processes generally prevent endogenous NK cells from performing their functions properly, resulting in poor tumor management and an increased risk of disease progression. While allogeneic NK cells have been shown to prevent leukemia recurrence in some circumstances, including stem cell transplantation, this therapy is not suitable for all patients. In addition, adoptively infused NK-cell-induced remissions are temporary and need repeated treatment to sustain persistent responses. As a result, novel techniques for inducing complete and lasting anti-leukemia responses in individuals with myeloid malignancies are required [6].
On the other hand, in recent years, scientists have focused increasing emphasis on traditional neurotransmitter receptors to get a deeper knowledge of the biology of leukemia and the cellular response to it [7]. The pharmacological suppression of dopamine and serotonin receptors, which both affect AML viability in relevant preclinical models, is used as a prognostic, diagnostic, and therapeutic target in AML [8]. Inhibition of both neurotransmitter receptors caused AML cells to undergo terminal differentiation, which had a significant impact on the leukemic cells at the very beginning of their differentiation process [9]. Differentiation-based treatments, which are more susceptible to chemo and lose their ability to self-renew, are appealing since they are frequent in all AML subtypes [10]. NK dysfunction has been seen in many hematological malignancies, including AML. However, in many of the AML cases, it is observed that NK cells cannot perform their anticancer functions appropriately. This might be related either to the number or dysfunction of NK cells.
The aim of the work was to compare the whole-genome data of NK cells isolated from AML patients and healthy individuals, to determine the genes, gene sets, and pathways responsible for the dysfunction of NK cells. In this study, we identified significant alterations in the expression of RAS and neurotransmitter genes in NK cells from AML patients and demonstrated how these changes could influence the anti-tumor capabilities of NK cells.
2. Methods
2.1. Obtaining and normalizing whole-genome expression of NK cells from healthy and AML patients
The Gene expression omnibus (GSE159624) was used to obtain data on human NK cells. The single-cell RNA profiling data of human bone marrow NK cells from healthy donors and AML patients were generated using the Illumina NextSeq 500 platform. The data were normalized in R (version 3.6.3) with the DESeq2 (version 3.9) package. The DESeq2 (version 3.9) package was designed for the purpose of normalizing, visualizing, and performing differential analysis on high-dimensional count data. To estimate log fold change and dispersion priors, and to construct posterior estimates for these values, it uses empirical Bayes methods [11]. Each sample has over 4,000 single cells with data on whole-genome expression. The average expression levels for each gene in the samples were determined.
2.2. Identification of RAS and neurotransmitter families related genes with differential expression in NK cells in healthy and AML patients
Using the limma (version 3.26.8) powers differential expression analysis, the whole normalized gene expression data of NK cells from healthy and AML patients were compared to identify the significance and differentially expressed RAS family and neurotransmitter pathway-related genes. Limma (version 3.26.8) is an R/Bioconductor (version 3.6.3) software tool for evaluating data from gene expression studies. It has a multitude of options for managing complicated experimental designs and borrowing information to overcome the issue of small sample numbers [12].
2.3. Hierarchical cluster analysis
A hierarchical clustering analysis was carried out to assess if the discovered genes could differentiate between the healthy and AML groups. Differentially expressed RAS genes and neurotransmitter pathways were grouped hierarchically using the similarity metric parameter and the Euclidean distance Gene Cluster v3.0 software as a full link.
2.4. Gene set enrichment analysis (GSEA)
GSEA was performed in accordance with the GSEA guideline method (http://software.broadinstitute.org/gsea/docGSEAUserGuideFrame.html). The study was conducted using GSE159624 data.
2.5. Pathway analysis
The “Database for Annotation, Visualization, and Integrated Discovery” software was used to deduce the biological relationships between these differentially expressed genes. We identified the functional pathways underlying the 100 most significantly differentially expressed genes between the healthy and AML groups.
3. Results
3.1. RAS and neurotransmitter gene families differentially expressed in NK cells from healthy and AML patients
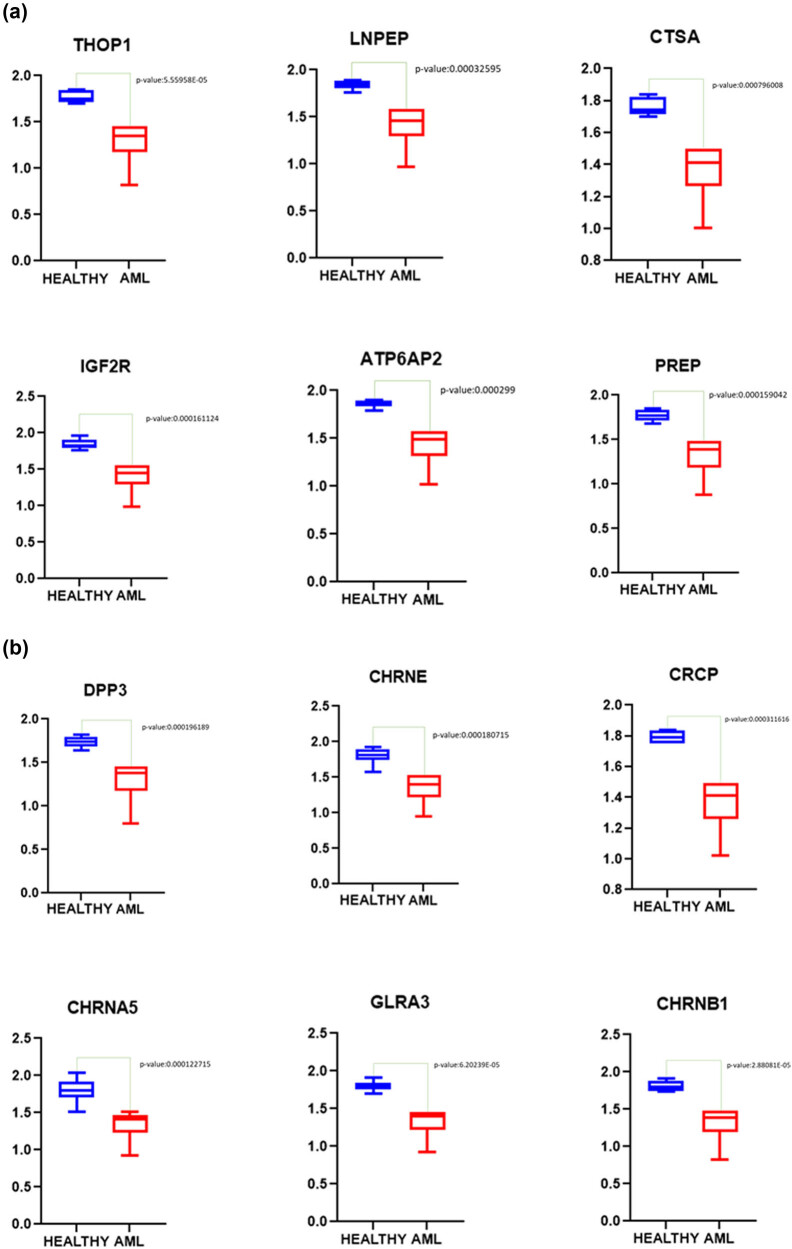
Following normalization of the data, 6 out of 16 samples were discarded due to 0 values for several genes in the samples. To ascertain gene expression alterations in AML patients’ NK cells, whole-genome expression data of NK cells from healthy and AML patient groups were compared. According to the findings, considering the adjusted p-values of the genes, 1,834 genes were found to be expressed statistically different between the two groups (adj p-value <0.05). All of these genes had log fold change values greater than 0.2 (Figure 1). We determined that six of these genes are connected with the RAS gene family (ATP6AP2, LNPEP, PREP, IGF2R, CTSA, and THOP1) and six are related to neurotransmitter pathways (DPP3, GLRA3, CRCP, CHRNA5, CHRNE, and CHRNB1). Each of these 12 genes was discovered to be downregulated in the NK cells of AML patients [p-values were calculated using the limma (version 3.26.8) package in the R (version 3.6.3)] (Figure 2a and b). As shown in Figure 3, these genes can distinguish well between healthy and AML patients (Figure 3).
Figure 1.
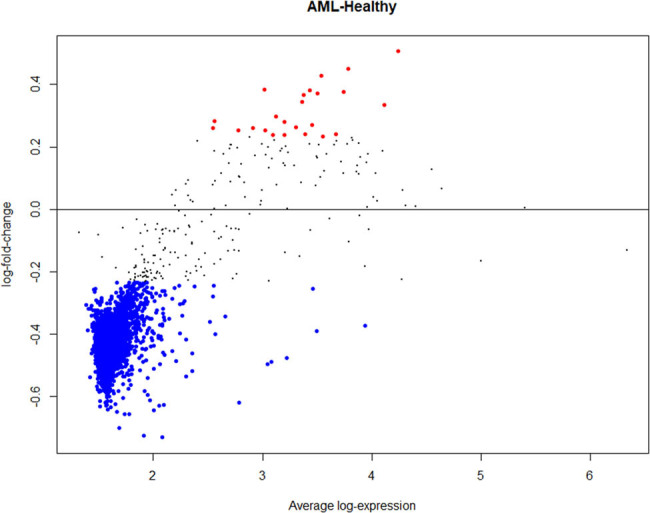
Differentially expressed genes in NK cells isolated from AML patients. In total, 1,834 genes were expressed statistically different between the two groups (adj p-value <0.05). Red dots indicate significantly upregulated genes. Blue dots indicate significantly downregulated genes.
Figure 2.
Significantly downregulated genes. Twelve genes were downregulated in the NK cells of AML patients. Six belong to RAS family genes that are significantly downregulated (AML patients) (a). The other six genes belong to Neurotransmitter gene family that are significantly downregulated (AML patients) (b).
Figure 3.
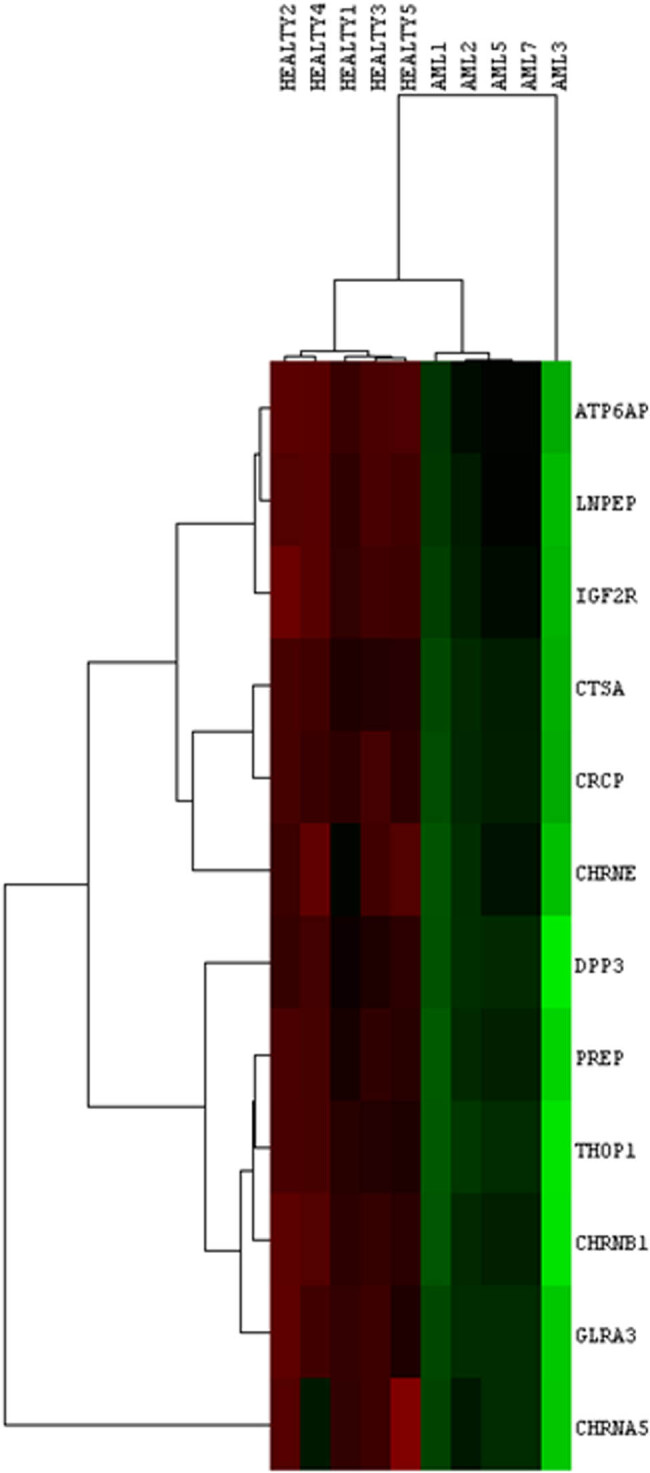
Hierarchical cluster analysis. Clusters of 12 differentially expressed genes with the 5 AML and healthy NK cells. All these genes are downregulated in NK cells isolated from AML patients.
3.2. Discovery of enriched gene sets in the NK cells of healthy and AML patients
We used GSEA to discover which gene sets were enriched in the NK cells of healthy and AML patients. According to our findings, gene sets involved in tumor necrosis factor (TNF) superfamily cytokine production, hematopoietic stem cell (HSC) differentiation, scaffold protein binding, protein kinase holoenzyme complex construction, and B-cell proliferation are enriched in NK cells from AML patients (p-value 0.05, sizes 71, 61, 29, 27, and 24, respectively). The gene sets enriched in the AML patient group are listed in Table 1. On the other hand, GSEA findings indicate that gene sets associated with cellular response to TGF-β stimuli, nucleotide metabolic processes, and ATP metabolic processes are enriched in the healthy group (p-value <0.05, size 18). The most significant gene sets enriched in the healthy group are presented in Table 2.
Table 1.
Gene sets enriched in NK cells from AML patients
| Gene sets | Size | ES | NES | Nom p-val | FDR q-val | FWER p-val | Rank at max |
|---|---|---|---|---|---|---|---|
| GOBP_TUMOR_NECROSIS_FACTOR_SUPERFAMILY_CYTOKINE_PRODUCTION | 71 | 0.35 | 2.13 | 0.006 | 1 | 0.229 | 4,265 |
| GOCC_RESPIRATORY_CHAIN_COMPLEX | 75 | 0.45 | 2.07 | 0 | 1 | 0.254 | 2,812 |
| GOCC_PROTEASOME_CORE_COMPLEX | 18 | 0.78 | 2.01 | 0 | 1 | 0.281 | 2,147 |
| GOBP_PROTEASOMAL_UBIQUITIN_INDEPENDENT_PROTEIN_CATABOLIC_PROCESS | 20 | 0.69 | 2.01 | 0 | 1 | 0.281 | 2,948 |
| GOBP_REGULATION_OF_TRANSCRIPTION_FROM_RNA_POLYMERASE_II_PROMOTER | 67 | 0.38 | 2 | 0 | 1 | 0.281 | 2,402 |
| GOBP_ATP_SYNTHESIS_COUPLED_ELECTRON_TRANSPORT | 89 | 0.43 | 2 | 0 | 1 | 0.281 | 2,812 |
| GOCC_NADH_DEHYDROGENASE_COMPLEX | 48 | 0.52 | 1.97 | 0 | 1 | 0.308 | 2,811 |
| GOCC_RESPIRASOME | 88 | 0.4 | 1.94 | 0 | 1 | 0.323 | 3,222 |
| GOBP_ANAPHASE_PROMOTING_COMPLEX_DEPENDENT_CATABOLIC_PROCESS | 71 | 0.35 | 1.94 | 0.02 | 1 | 0.323 | 2,290 |
| GOBP_NEGATIVE_REGULATION_OF_TUMOR_NECROSIS_FACTOR_SUPERFAMILY_CY | 23 | 0.35 | 1.92 | 0.033 | 1 | 0.359 | 4,265 |
| GOBP_MITOCHONDRIAL_ELECTRON_TRANSPORT_NADH_TO_UBIQUINONE | 52 | 0.49 | 1.89 | 0 | 1 | 0.418 | 2,811 |
| GOBP_OXIDATIVE_PHOSPHORYLATION | 122 | 0.36 | 1.87 | 0 | 1 | 0.435 | 2,812 |
| GOBP_REGULATION_OF_HEMATOPOIETIC_STEM_CELL_DIFFERENTIATION | 61 | 0.37 | 1.86 | 0 | 1 | 0.448 | 4,010 |
| GOBP_REGULATION_OF_CELLULAR_AMINO_ACID_METABOLIC_PROCESS | 51 | 0.4 | 1.85 | 0.022 | 1 | 0.448 | 2,290 |
| GOBP_REGULATION_OF_CELLULAR_AMINE_METABOLIC_PROCESS | 58 | 0.36 | 1.85 | 0.026 | 1 | 0.448 | 2,290 |
| GOMF_SCAFFOLD_PROTEIN_BINDING | 29 | 0.36 | 1.85 | 0 | 1 | 0.448 | 5,778 |
| GOCC_CYCLIN_DEPENDENT_PROTEIN_KINASE_HOLOENZYME_COMPLEX | 27 | 0.39 | 1.85 | 0.018 | 1 | 0.451 | 3,005 |
| GOBP_POSITIVE_REGULATION_OF_B_CELL_PROLIFERATION | 24 | 0.47 | 1.84 | 0 | 1 | 0.451 | 4,122 |
| GOCC_ENDOPEPTIDASE_COMPLEX | 64 | 0.35 | 1.84 | 0 | 1 | 0.457 | 2,363 |
| HP_LIMB_GIRDLE_MUSCLE_ATROPHY | 18 | 0.46 | 1.83 | 0.01 | 1 | 0.49 | 2,462 |
Table 2.
Gene sets enriched in NK cells from healthy donors
| Gene sets | Size | ES | NES | Nom p-Val | FDR q-val | FWER p-Val | Rank at max |
|---|---|---|---|---|---|---|---|
| HP_MOTOR_AXONAL_NEUROPATHY | 20 | −0.45 | −1.73 | 0.01 | 1 | 0.695 | 3,963 |
| HP_3_METHYLGLUTACONIC_ACIDURIA | 16 | −0.43 | −1.7 | 0.004 | 1 | 0.8 | 4,503 |
| HP_ABNORMAL_CIRCULATING_FATTY_ACID_ANION_CONCENTRATION | 15 | −0.54 | −1.68 | 0 | 1 | 0.832 | 2,057 |
| GOMF_LIGASE_ACTIVITY_FORMING_CARBON_OXYGEN_BONDS | 15 | −0.53 | −1.62 | 0.016 | 1 | 0.938 | 3,341 |
| GOBP_POSITIVE_REGULATION_OF_CELLULAR_RESPONSE_TO_TRANSFORMING_GROWTH_FACTOR_BETA_STIMULUS | 18 | −0.39 | −1.58 | 0.048 | 1 | 0.955 | 4,843 |
| GOBP_TETRAPYRROLE_BIOSYNTHETIC_PROCESS | 22 | −0.38 | −1.57 | 0.045 | 1 | 0.959 | 2,901 |
| HP_ABNORMAL_URINE_CARBOXYLIC_ACID_LEVEL | 30 | −0.38 | −1.52 | 0.041 | 1 | 0.977 | 3,589 |
| GOMF_SODIUM_ION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 28 | −0.33 | −1.51 | 0.087 | 1 | 0.989 | 1,979 |
| GOBP_PIGMENT_BIOSYNTHETIC_PROCESS | 35 | −0.29 | −1.49 | 0.109 | 1 | 0.989 | 2,901 |
| GOBP_REGULATION_OF_NUCLEOTIDE_BIOSYNTHETIC_PROCESS | 16 | −0.37 | −1.49 | 0.094 | 1 | 0.989 | 1,704 |
| GOMF_LIGAND_GATED_CATION_CHANNEL_ACTIVITY | 17 | −0.44 | −1.48 | 0.123 | 1 | 0.991 | 3,292 |
| GOMF_CATALYTIC_ACTIVITY_ACTING_ON_A_TRNA | 75 | −0.34 | −1.47 | 0.135 | 1 | 0.991 | 4,263 |
| GOMF_LIGAND_GATED_ION_CHANNEL_ACTIVITY | 19 | −0.42 | −1.47 | 0.134 | 1 | 0.993 | 3,292 |
| HP_TYPE_1_MUSCLE_FIBER_PREDOMINANCE | 19 | −0.36 | −1.46 | 0.138 | 1 | 0.993 | 2,667 |
| GOBP_POSITIVE_REGULATION_OF_NUCLEOTIDE_METABOLIC_PROCESS | 18 | −0.35 | −1.46 | 0.102 | 1 | 0.993 | 1,357 |
| GOBP_POSITIVE_REGULATION_OF_ATP_METABOLIC_PROCESS | 18 | −0.35 | −1.46 | 0.102 | 1 | 0.993 | 1,357 |
| HP_ABNORMAL_SPERM_MOTILITY | 17 | −0.39 | −1.45 | 0.061 | 1 | 0.993 | 2,822 |
| GOBP_GLUTAMATE_METABOLIC_PROCESS | 16 | −0.41 | −1.45 | 0.09 | 1 | 0.993 | 4,643 |
| HP_BILATERAL_TONIC_CLONIC_SEIZURE_WITH_GENERALIZED_ONSET | 23 | −0.34 | −1.45 | 0.067 | 1 | 0.994 | 3,068 |
| GOMF_TRNA_METHYLTRANSFERASE_ACTIVITY | 25 | −0.35 | −1.43 | 0.172 | 1 | 0.994 | 4,166 |
To determine molecular mechanisms underlying differential response of NK cells of AML and healthy donors, we analyzed GSEA results. Several gene sets were significantly enriched among NK cells of AML and healthy donors (FDR q-value <0.25). But especially, gene sets enriched in AML NK cells with an FDR q-value of lower than 0.25 included TNF superfamily cytokine production; while gene sets, such as stimuli of TGF-β, were enriched in healthy NK cells (Figure 4a and b).
Figure 4.
GESA of AML and healthy NK cells. Gene sets enriched in AML NK cells with an FDR q-value of lower than 0.25 included TNF superfamily cytokine production (a). Gene sets enriched in healthy NK cells with an FDR q-value of lower than 0.25 included TGF-β stimuli (b).
3.3. Seven distinct groups from the most differentially expressed genes
Clustering analysis of functional annotations for the most differentially expressed 100 genes revealed seven distinct groupings. According to their enrichment scores, the first cluster (ES: 2.05) contains the majority of genes involved in neurotransmitter-related pathways, the second cluster (ES: 1.93) contains genes associated with RAS and RAS-related pathways, and the third cluster (ES: 0.83) contains genes associated with zinc- and metal-binding pathways (Table 3). All seven clusters regarding functional annotation analysis are presented in Table S1.
Table 3.
Functional annotation analysis of first 100 differentially expressed genes
| Annotation cluster 1 | Pathways enrichment score: 2.05 | p-Value | Benjamini |
|---|---|---|---|
| INTERPRO | Neurotransmitter-gated ion-channel. conserved site | 8.80 × 10−4 | 6.00 × 10−2 |
| INTERPRO | Neurotransmitter-gated ion-channel transmembrane domain | 1.00 × 10−3 | 6.00 × 10−2 |
| INTERPRO | Neurotransmitter-gated ion-channel | 1.00 × 10−3 | 6.00 × 10−2 |
| INTERPRO | Neurotransmitter-gated ion-channel ligand binding | 1.00 × 10−3 | 6.00 × 10−2 |
| INTERPRO | Nicotinic acetylcholine receptor | 2.40 × 10−3 | 1.20 × 10−1 |
| UP_KEYWORDS | Synapse | 2.80 × 10−3 | 2.20 × 10−1 |
| UP_KEYWORDS | Ion channel | 2.90 × 10−3 | 2.20 × 10−1 |
| UP_KEYWORDS | Ligand-gated ion channel | 3.90 × 10−3 | 2.20 × 10−1 |
| GOTERM_MF_DIRECT | Acetylcholine receptor activity | 4.20 × 10−3 | 3.50 × 10−1 |
| GOTERM_MF_DIRECT | Acetylcholine-activated cation-selective channel activity | 4.20 × 10−3 | 3.50 × 10−1 |
| INTERPRO | Nicotinic acetylcholine-gated receptor, transmembrane domain | 4.30 × 10−3 | 1.70 × 10−1 |
| GOTERM_CC_DIRECT | Acetylcholine-gated channel complex | 4.30 × 10−3 | 6.10 × 10−10 |
| GOTERM_MF_DIRECT | Acetylcholine binding | 5.30 × 10−3 | 3.50 × 10−1 |
| UP KEYWORDS | Postsynaptic cell membrane | 5.50 × 10−3 | 2.30 × 10−1 |
| GOTERM_BP_DIRECT | Neuromuscular synaptic transmission | 5.50 × 10−3 | 1.00 × 1000 |
| GOTERM MF_DIRECT | Ligand-gated ion channel activity | 1.00 × 10−2 | 4.90 × 10−1 |
| GOTERM_BP_DIRECT | Synaptic transmission, cholinergic | 1.10 × 10−2 | 1.00 × 1000 |
| Annotation cluster 2 | Pathways enrichment score: 1.93 | p-value | Benjamini |
|---|---|---|---|
| KEGG_PATHWAY | RAS | 4.60 × 10−6 | 3.60 × 10−4 |
| GOTERM_BP_DIRECT | Proteolysis | 2.00 × 10−2 | 1.00 × 1000 |
| GOTERM_MF_DIRECT RT | Metallopeptidase activity | 4.60 × 10−2 | 1.00 × 1000 |
| UP_KEYWORDS | Protease | 6.30 × 10−2 | 8.00 × 10−1 |
| Annotation cluster 3 | Pathways enrichment score: 0.83 | p-value | Benjamini |
|---|---|---|---|
| UP_KEYWORDS | Zinc | 4.50 × 10−1 | 1.00 × 1000 |
| GOTERM_MF DIRECT | Zinc ion binding | 2.20 × 10−1 | 1.00 × 1000 |
| UP_KEYWORDS | Metal binding | 3.60 × 10−1 | 1.00 × 1000 |
| INTERPRO | Zinc finger C2H2-type/integrase DNA binding domain | 7.90 × 10−1 | 1.00 × 1000 |
| SMART | ZnF C2H2 | 7.90 × 10−1 | 1.00 × 1000 |
| INTERPRO | Zinc finger, C2H2-like | 8.20 × 10−1 | 1.00 × 1000 |
| UP_KEYWORDS | Zinc finger | 8.40 × 10−1 | 1.00 × 1000 |
| UP_SEQ_FEATURE | Metal ion-binding site: zinc; catalytic | 0.092 | 1 |
| INTERPRO | Zinc finger, C2H2 | 8.40 × 10−1 | 1.00 × 1000 |
4. Discussion
The bioinformatic analyses undertaken in this work focused on two critical pathways and associated genes to identify NK-cell dysfunction in AML patients. In this study, we showed that the downregulation of some critical genes related to the RAS gene family and neurotransmitter-associated pathways might be responsible for the inappropriate and dysfunctional NK cells in AML patients. Novel pathways and genes related to RAS and neurotransmitters, which play a significant role in NK biology and proper function in patients with AML, were also focused and directed in this work.
In addition to immunological responses and inflammation, the RAS is made up of a variety of substrates and enzymes that may work together or independently [13]. The identification of local RAS in the heart, brain, pancreas, lymphatic, and adipose tissue was a real advance in the area. Local RAS may work alone or in conjunction with circulating RAS. A functioning intracellular RAS has also been found, showing angiotensin II’s (AngII) pro-inflammatory, proliferative, and fibrotic properties [14,15]. It regulates the generation of free radicals and the cellular synthesis of cytokines, chemokines, and transcription factors and modulates cell proliferation and differentiation via these mechanisms. RAS and its primary effector molecule, AngII, are implicated in inflammation and immunity [16]. AngII enhances their chemotaxis, perhaps amplifying inflammation [17]. It is yet unknown if AngII has a direct influence on the function and cellular location of certain immune cells in humans. T and NK cells have been shown to be capable of generating and transporting AngII to inflammatory areas. AngII therapies enhanced the proliferation of T and NK cells activated by mitogens and anti-CD3. This research implies that NK and T cells are capable of producing functional renin and angiotensin-converting enzyme (ACE) activity.
According to the findings of the current study, ATP6AP2, LNPEP, PREP, IGF2R, CTSA, and THOP1 genes belonging to the RAS gene family were discovered to be downregulated in the NK cells of AML patients.
As supported, ATP6AP2 is involved in the regulation of organellar, cellular, systemic homeostasis, and activating this receptor is essential for the conversion of angiotensinogen to angiotensin [18]. ATP6AP2 is upregulated in NK cells compared to CD8+ T cells [19]. LNPEP is capable of degrading peptide hormones, such as oxytocin, vasopressin, and angiotensin III (AngIII) [20]. AngIII, like AngII, increases bvasopressin release and promotes the production of pro-inflammatory regulators [21].
The IGF2R activates proton rechanneling into the mitochondrial intermembrane area, allowing for persistent OXPHOS [22,23]. L-IGF2 increases DNA methylation by activating GSK3. This sequestered v-ATPase assembly prevents protons from reaching lysosomes and redirects them to mitochondria [24].
CTSA is required for proper lysosomal routing, stability, and activation of β-galactosidase and alpha-neuraminidase. It also inactivates bioactive peptides. Cytotoxic chemicals are stored in secretory lysosomes, an exocytic organelle present in NK cells [25]. Thus, changes in the expression of genes that control or affect lysosomal activity may affect the cytotoxicity of NK cells, as well.
THOP1 assists with present MHC-I antigens [26]. It is involved in short-term neuropeptide metabolism. Cytoplasmic peptides degrade amyloid-β precursor protein fragments. Peptide transport disruption appears to impact NK cells [26]. NK cells interact with dendritic cells to destroy target cells and modulate innate and adaptive immunity [27]. Any MHC1 presentation issue may impact NK-cell function. Based on our findings, DPP3, GLRA3, CRCP, CHRNA5, CHRNE, and CHRNB1 genes that are involved in neurotransmitter pathways were found to be downregulated in the NK cells of AML patients as well.
Dipeptidyl peptidase (DPP4), a member of the dipeptidyl peptidase family, is present in trace concentrations on newly isolated human NK cells and is increased in a small subpopulation in response to interleukin (IL)-2 stimulation [28]. In addition, it was shown that inhibiting DPP4 inhibits DNA synthesis and cell cycle progression in NK cells. Interestingly, in a model of lung metastasis, NK cytolytic ability against tumor cells was decreased in DPP4-deficient mice [29]. Glycine (Gly) is a neurotransmitter that acts mostly in the central nervous system’s caudal region. Flow cytometry has shown the presence of GlyR in human NK cells. This receptor may influence NK-cell activity in AML patients [30]. It was shown that calcitonin gene-related peptide, or CGRP, has a dose-dependent effect on the NK activity of spleen cells in mice [31]. So, CGRP may be involved in regulating NK function. It has also been established that CGRP may influence the activity of macrophages in sensory neuron nerve terminals during the onset and maintenance of inflammation. CHRNA5, CHRNB1, and CHRNE from the cholinergic receptor nicotinic gene family were found to be downregulated in the NK cells of AML patients in our study. Both adaptive and innate immune cells are controlled by the cholinergic system, which mediates their mobilization, differentiation, and antigen presentation. NK activation with cytokines increases mRNA and protein levels of the nicotinic receptor 7 have been demonstrated [32]. It has been suggested that the expression level of nicotinic receptors is critical to the effective activity of NK cells. NK cells perform their cytotoxic function by secreting cytokines, which cause malignant or infected cells to rupture and death.
Aside from identifying specific genes whose expression differs between healthy and AML NK cells, the second important component of our work was the identification of critical pathways and gene sets associated with NK dysfunction. As revealed by our findings, TNF superfamily cytokine production, HSC differentiation, scaffold protein binding, kinase holoenzyme complex construction, and B-cell proliferation are enriched in NK cells from patients with AML. Gene sets related with cellular response to TGF-β stimuli, nucleotide metabolic activities, and ATP metabolic processes, on the other hand, were shown to be enriched in the healthy group, according to GSEA results.
Tumor development and metastasis are slowed or stopped entirely when interferon and TNF-α are released or membrane bound by NK cells. NK cells are the primary immune effectors that play a role in this process. It was discovered that, in addition to TNF alpha, NK cells relied on additional members of the TNF family. It has been revealed that the TNF superfamily member TRAIL plays a significant role in tumor defense by NK cells or CD8+ T lymphocytes [33]. It has been shown that TNF receptors are involved in the formation of lymphokine-activated killer cell activity and the multiplication of NK cells [34].
On the other hand, TNF production is elevated in the presence of the angiotensin subtype-1 receptor, which has been demonstrated to affect inflammatory processes by increasing the signal transduction and activator of transcription proteins 3 (STAT3) signaling [35]. In addition, studies suggest that increased AT2R expression may play a role in the observed decrease in the inflammatory pathway activation through decreased TNF-α production and STAT3 signaling. AT2R expression and/or activation are the possible therapeutic target for the control of inflammation [36]. Meanwhile, TNF superfamily members have also been demonstrated to have an impact on the production and function of a variety of neurotransmitters [37,38,39,40].
Cytokine stimuli activate HSCs. Self-renewing multipotent HSCs commit to common lymphoid progenitor status in response to certain stimuli. Progenitors of NK cells are generated by non-hematopoietic cells, such as mesenchymal stem cells, that release IL-7 or IL-15, which play a variety of functions in programming CLPs (NK-cell progenitors). It is possible to functionally develop immature NK cells into well-defined subsets of adult NK cells using chimeric antigen receptor cells. The sinusoidal blood channels carry mature NK cells to the secondary lymphoid organs. The formation, development, and function of NK cells will be affected if this process is disrupted. In addition, ACE and its major enzymatic product, Ang II, are recognized as critical determinants of angiogenesis, inflammation, tumor progression, and hematopoiesis [41]. Studies also identified ACE as a marker of both adult and embryonic human hematopoietic stem/progenitor cells [42]. On the other hand, HSC niche is regulated in part by the neurological system and neurotransmitters [43].
Scaffolding proteins play role in stabilizing the structure of other enriched gene sets inside the NK of AML patients. Multiple protein–protein interaction modules are common among the many scaffolding proteins that exist in nature [44]. A number of critical signaling pathways rely on scaffold proteins, which are proteins that interact with numerous elements of a route and bind them together form complexes [45]. It is possible that the alterations in the gene sets that regulate the binding of the scaffold proteins may contribute to the degradation of NK-cell activities by disrupting numerous essential signal pathways, such as RAS and neurotransmitter.
Furthermore, gene sets involved in B-cell proliferation were found to be enriched in the AML group. Circulating RAS and local paracrine–autocrine–intracrine tissue-based RAS participates in numerous pathobiological events [46]. Multiple components of the RAS signaling cascade influence inflammatory cell phenotype and function with unpredictable and context-specific effects on innate and adaptive immunities [13].
However, in NK cells from healthy donors, TGF-β stimulates an increased response. Anti-inflammatory cytokines like TGF-β may be produced in response to the pro-inflammatory cellular and molecular environment. Activated lymphocytes, macrophages, and dendritic cells create these anti-inflammatory cytokines, which may reduce the immune cell activity. When it comes to preventing the spread of infectious diseases, the immune system is the most important weapon. It is founded that tumor cell death produced by NK cells was reduced in vitro by metastasis-associated lung macrophages, which were shown to be membrane bound and reliant on the growth factor TGF-β [47]. This process is important for the prompt arrest of NK cells. TGF-β synthesis is boosted by RAS components. RAS and TGF-β have crucial clinical consequences for each other’s relationship [13]. Reduction in RAS inhibitory effects is directly linked to the reduction of TGF-β production in both experimental and clinical renal disorders [48].
NK cells of normal individuals are also rich in two additional gene sets, nucleotide metabolic activities and ATP metabolic processes, both of which seem to be linked to a wide range of critical cell processes.
In addition, we determined that the nucleotide and metabolic processes in healthy donors’ NK cells are favorably regulated. The importance of immunometabolism in the control of NK-cell activity is becoming more widely recognized. An imbalanced metabolic profile and defective mitochondria in NK cells resulted in an inability to destroy them. As a result, NK-cell metabolic pathways are altered, resulting in tumor-promoting settings with NK-cell malfunction. Furthermore, we identified seven clusters of genes that differed significantly between healthy and AML NK cells. According to our findings, the best clusters were linked to neurotransmitter pathways, RAS, and zinc-related pathways. There is a connection with the neurological and immune systems because each of the systems expresses receptors for neurotransmitters. Because autocrine and paracrine systems allow immune cells to produce and release neurotransmitters themselves, neurotransmitter-mediated pathways are used. The neurological system and several neurotransmitter-related pathways have a significant impact on NK-cell function. The immune system may be modulated by zinc, a vital trace element. Zinc supplementation increases perforin expression, which promotes NK-cell function. The activity of NK cells was also raised by zinc supplementation, which also elevated IL-2. Zinc deficiency, on the other hand, decreased NK-cell activity and dampened the IL-2 medication’s boosting impact on NK cells [49,50].
The RAS regulates immune and cancer cells in fundamental ways [48]. All of the many RAS molecules play an important role in controlling inflammation and other immune responses [51]. Most innate and adaptive immune cells exhibit components of the RAS pathway. RAS signaling seems to influence the activity of numerous immune cells in both in vitro and in vivo.
The in silico design may be considered the limitation of our study. However, based on the results of this present study, the expression of RAS and neurotransmitter genes in NK cells from AML patients could influence the (patho)biological course of anti-tumor NK responses.
The immune and neurological systems are dependent on one another for their fine tuning and functioning, and they work together to preserve physiological homeostasis and prevent infection as well as the development of cancer, respectively. According to our research, neurotransmitters have been proven to have a significant influence on the biology and functioning of NK cells. On the other hand, it has been shown that the RAS gene family plays a crucial role in the development of NK cells. Important genes belonging to these two gene families were highlighted in this work for the first time, and the interaction between RAS and neurotransmitters was investigated. In the context of these data, the next step in research planning should be the experiments in the laboratory and in the clinic for in vitro and clinical validation.
Supplementary Material
Footnotes
Funding information: None.
Author contributions: Seyhan Turk: conception or design of the work; data acquisition; data analysis and interpretation; drafting the article; critically revising the article; final approval of the version to be published; and accountability for all aspects of the work. Ayriana Safari Baesmat: data acquisition and data analysis and interpretation. Aysegul Yılmaz: conception or design of the work and drafting the article. Can Turk: conception or design of the work; drafting the article; data acquisition; and data analysis and interpretation. Umit Yavuz Malkan: conception or design of the work; drafting the article; critically revising the article; final approval of the version to be published; and accountability for all aspects of the work. Gulberk Ucar: conception or design of the work; drafting the article; critically revising the article; final approval of the version to be published; and accountability for all aspects of the work. Ibrahim Celalettin Haznedaroglu: conception or design of the work; drafting the article; critically revising the article; final approval of the version to be published; and accountability for all aspects of the work.
Conflict of interest: The authors declare that there is no conflict of interest regarding the publication of this article.
Data availability statement: The data supporting this study are from previously reported studies and datasets, which have been cited. The processed data are available from the corresponding author upon request.
References
- [1].Sánchez-Corrales YE, Pohle RVC, Castellano S, Giustacchini A. Taming cell-to-cell heterogeneity in acute myeloid leukaemia with machine learning. Front Oncol. 2021;11:666829. [DOI] [PMC free article] [PubMed]
- [2].Xu J, Niu T. Natural killer cell-based immunotherapy for acute myeloid leukemia. J Hematol Oncol. 2020;13(1):167. [DOI] [PMC free article] [PubMed]
- [3].Tang L, Wu J, Li CG, Jiang HW, Xu M, Du M, et al. Characterization of immune dysfunction and identification of prognostic immune-related risk factors in acute myeloid leukemia. Clin Cancer Res. 2020;26(7):1763–72. [DOI] [PubMed]
- [4].Jamal E, Azmy E, Ayed M, Aref S, Eisa N. Clinical impact of percentage of natural killer cells and natural killer-like T cell population in acute myeloid leukemia. J Hematol. 2020;9(3):62–70. [DOI] [PMC free article] [PubMed]
- [5].Fujita M, Hayashi I, Yamashina S, Itoman M, Majima M. Blockade of angiotensin AT1a receptor signaling reduces tumor growth, angiogenesis, and metastasis. Biochem Biophys Res Commun. 2002;294(2):441–7. [DOI] [PubMed]
- [6].Carlsten M, Järås M. Natural killer cells in myeloid malignancies: Immune surveillance, NK cell dysfunction, and pharmacological opportunities to bolster the endogenous NK cells. Front Immunology. 2019;10:2357. [DOI] [PMC free article] [PubMed]
- [7].Ganor Y, Levite M. The neurotransmitter glutamate and human T cells: glutamate receptors and glutamate-induced direct and potent effects on normal human T cells, cancerous human leukemia and lymphoma T cells, and autoimmune human T cells. J Neural Transm. 2014;121(8):983–1006. [DOI] [PubMed]
- [8].Aslostovar L, Boyd AL, Benoit YD, Di Lu J, Garcia Rodriguez JL, Nakanishi M, et al. Abnormal dopamine receptor signaling allows selective therapeutic targeting of neoplastic progenitors in AML patients. Cell Rep Med. 2021;2(2):100202. [DOI] [PMC free article] [PubMed]
- [9].Banús-Mulet A, Etxabe A, Cornet-Masana JM, Torrente M, Lara-Castillo MC, Palomo L, et al. Serotonin receptor type 1B constitutes a therapeutic target for MDS and CMML. Sci Rep. 2018;8(1):13883. [DOI] [PMC free article] [PubMed]
- [10].Risueño RM, Cuesta-Casanovas L, Carbó JM, Cornet-Masana JM. New therapeutic approaches for acute myeloid leukemia. EMJ. 2021;6:82–9.
- [11].Dündar F, Skrabanek L, Zumbo P. Introduction to differential gene expression analysis using RNA-seq. Switzerland: Zenodo; 2020.
- [12].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed]
- [13].Crowley SD, Rudemiller NP. Immunologic effects of the renin-angiotensin system. J Am Soc Nephrol. 2017;28(5):1350–61. [DOI] [PMC free article] [PubMed]
- [14].Remuzzi G, Perico N, Macia M, Ruggenenti P. The role of renin-angiotensin-aldosterone system in the progression of chronic kidney disease. Kidney Int. 2005;68:S57–65. [DOI] [PubMed]
- [15].Singh KD, Karnik SS. Angiotensin receptors: Structure, function, signaling and clinical applications. J Cell Signal. 2016;1(2):111. [DOI] [PMC free article] [PubMed]
- [16].Capettini LS, Montecucco F, Mach F, Stergiopulos N, Santos RA, da Silva RF. Role of renin-angiotensin system in inflammation, immunity and aging. Curr Pharm Des. 2012;18(7):963–70. [DOI] [PubMed]
- [17].Jurewicz M, McDermott DH, Sechler JM, Tinckam K, Takakura A, Carpenter CB, et al. Human T and natural killer cells possess a functional renin-angiotensin system: Further mechanisms of angiotensin II–induced inflammation. J Am Soc Nephrol. 2007;18(4):1093–102. [DOI] [PubMed]
- [18].Wendling O, Champy M-F, Jaubert S, Pavlovic G, Dubos A, Lindner L, et al. Atp6ap2 ablation in adult mice impairs viability through multiple organ deficiencies. Sci Rep. 2017;7(1):9618. [DOI] [PMC free article] [PubMed]
- [19].Dybkaer K, Iqbal J, Zhou G, Geng H, Xiao L, Schmitz A, et al. Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: Gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics. 2007;8(1):230. [DOI] [PMC free article] [PubMed]
- [20].Paladini F, Fiorillo MT, Tedeschi V, Mattorre B, Sorrentino R. The multifaceted nature of aminopeptidases ERAP1, ERAP2, and LNPEP: From evolution to disease. Front Immunology. 2020;11:1576. [DOI] [PMC free article] [PubMed]
- [21].Ruiz-Ortega M, Lorenzo O, Egido J. Angiotensin III increases MCP-1 and activates NF-kappaB and AP-1 in cultured mesangial and mononuclear cells. Kidney Int. 2000;57(6):2285–98. [DOI] [PubMed]
- [22].Oshima A, Nolan CM, Kyle JW, Grubb JH, Sly WS. The human cation-independent mannose 6-phosphate receptor. Cloning and sequence of the full-length cDNA and expression of functional receptor in COS cells. J Biol Chem. 1988;263(5):2553–62. [PubMed]
- [23].McCormick PJ, Dumaresq-Doiron K, Pluviose AS, Pichette V, Tosato G, Lefrancois S. Palmitoylation controls recycling in lysosomal sorting and trafficking. Traffic. 2008;9(11):1984–97. [DOI] [PMC free article] [PubMed]
- [24].Wang X, Lin L, Lan B, Wang Y, Du L, Chen X, et al. IGF2R-initiated proton rechanneling dictates an anti-inflammatory property in macrophages. Sci Adv. 2020;6(48):eabb7389. [DOI] [PMC free article] [PubMed]
- [25].Topham NJ, Hewitt EW. Natural killer cell cytotoxicity: how do they pull the trigger? Immunology. 2009;128(1):7–15. [DOI] [PMC free article] [PubMed]
- [26].Ferro ES, Gewehr MCF, Navon A. Thimet oligopeptidase biochemical and biological significances: Past, present, and future directions. Biomolecules. 2020;10(9):1229. [DOI] [PMC free article] [PubMed]
- [27].Peterson EE, Barry KC. The natural killer–dendritic cell immune axis in anti-cancer immunity and immunotherapy. Front Immunol. 2021;11:621254. [DOI] [PMC free article] [PubMed]
- [28].Prajapati SC, Chauhan SS. Dipeptidyl peptidase III: A multifaceted oligopeptide N-end cutter. FEBS J. 2011;278(18):3256–76. [DOI] [PubMed]
- [29].Waumans Y, Baerts L, Kehoe K, Lambeir A-M, De Meester I. The dipeptidyl peptidase family, prolyl oligopeptidase and prolyl carboxypeptidase in the immune system and inflammatory disease, including atherosclerosis. Front Immunol. 2015;6:387. [DOI] [PMC free article] [PubMed]
- [30].Van Den Eynden J, SahebAli S, Horwood N, Carmans S, Brône B, Hellings N, et al. Glycine and glycine receptor signalling in non-neuronal cells. Front Mol Neurosci. 2009;2:9. [DOI] [PMC free article] [PubMed]
- [31].Umeda Y, Arisawa M. Inhibition of natural killer activity by calcitonin gene-related peptide. Immunopharmacol Immunotoxicol. 1989;11(2–3):309–20. [DOI] [PubMed]
- [32].Zanetti SR, Ziblat A, Torres NI, Zwirner NW, Bouzat C. Expression and functional role of α7 nicotinic receptor in human cytokine-stimulated natural killer (NK) cells. J Biol Chem. 2016;291(32):16541–52. [DOI] [PMC free article] [PubMed]
- [33].Mirandola P, Ponti C, Gobbi G, Sponzilli I, Vaccarezza M, Cocco L, et al. Activated human NK and CD8 + T cells express both TNF-related apoptosis-inducing ligand (TRAIL) and TRAIL receptors but are resistant to TRAIL-mediated cytotoxicity. Blood. 2004;104(8):2418–24. [DOI] [PubMed]
- [34].Naume B, Shalaby R, Lesslauer W, Espevik T. Involvement of the 55- and 75-kDa tumor necrosis factor receptors in the generation of lymphokine-activated killer cell activity and proliferation of natural killer cells. J Immunol. 1991;146(9):3045–8. [PubMed]
- [35].Suzuki Y, Ruiz-Ortega M, Lorenzo O, Ruperez M, Esteban V, Egido J. Inflammation and angiotensin II. Int J Biochem Cell Biol. 2003;35(6):881–900. [DOI] [PubMed]
- [36].Angiotensin II Type-2 receptors modulate inflammation through signal transducer and activator of transcription proteins 3 phosphorylation and TNFα production. J Interferon Cytokine Res. 2011;31(6):471–4. [DOI] [PMC free article] [PubMed]
- [37].Pribiag H, Stellwagen D. TNF- downregulates inhibitory neurotransmission through protein phosphatase 1-dependent trafficking of GABAA receptors. J Neurosci off J Soc Neurosci. 2013;33:15879–93. [DOI] [PMC free article] [PubMed]
- [38].Twohig JP, Cuff SM, Yong AA, Wang EC. The role of tumor necrosis factor receptor superfamily members in mammalian brain development, function and homeostasis. Rev Neurosci. 2011;22(5):509–33. [DOI] [PMC free article] [PubMed]
- [39].Ren S, Breuillaud L, Yao W, Yin T, Norris KA, Zehntner SP, et al. TNF-α–mediated reduction in inhibitory neurotransmission precedes sporadic Alzheimer’s disease pathology in young Trem2R47H rats. J Biol Chem. 2021;296:100089. [DOI] [PMC free article] [PubMed]
- [40].Fu X-Q, Peng J, Wang A-H, Luo Z-G. Tumor necrosis factor alpha mediates neuromuscular synapse elimination. Cell Discovery. 2020;6(1):9. [DOI] [PMC free article] [PubMed]
- [41].Park TS, Zambidis ET. A role for the renin-angiotensin system in hematopoiesis. Haematologica. 2009;94(6):745–7. [DOI] [PMC free article] [PubMed]
- [42].Julien E, Biasch K, El Omar R, Freund J-N, Gachet C, Lanza F, et al. Renin-angiotensin system is involved in embryonic emergence of hematopoietic stem/progenitor cells. Stem Cell. 2021;39(5):636–49. [DOI] [PubMed]
- [43].Agarwala S, Tamplin OJ. Neural crossroads in the hematopoietic stem cell niche. Trends Cell Biol. 2018;28(12):987–98. [DOI] [PMC free article] [PubMed]
- [44].Garbett D, Bretscher A. The surprising dynamics of scaffolding proteins. Mol Biol Cell. 2014;25(16):2315–9. [DOI] [PMC free article] [PubMed]
- [45].Bellazzo A, Collavin L. A mechanism for cell non-autonomous inactivation of the tumor suppressor DAB2IP. Oncoscience. 2018;5(5–6):177–8. [DOI] [PMC free article] [PubMed]
- [46].Ciftciler R, Haznedaroglu IC. Pathobiological interactions of local bone marrow renin-angiotensin system and central nervous system in systemic arterial hypertension. Front Endocrinol (Lausanne). 2020;11:425. [DOI] [PMC free article] [PubMed]
- [47].Brownlie D, Doughty-Shenton D, Soong DY, Nixon C, Carragher NO, Carlin LM, et al. Metastasis-associated macrophages constrain antitumor capability of natural killer cells in the metastatic site at least partially by membrane bound transforming growth factor β. J Immunother Cancer. 2021;9(1). [DOI] [PMC free article] [PubMed]
- [48].Wolf G. Renal injury due to renin–angiotensin–aldosterone system activation of the transforming growth factor-β pathway. Kidney Int. 2006;70(11):1914–9. [DOI] [PubMed]
- [49].Rolles B, Maywald M, Rink L. Influence of zinc deficiency and supplementation on NK cell cytotoxicity. J Funct Foods. 2018;48:322–8.
- [50].Allen JI, Perri RT, McClain CJ, Kay NE. Alterations in human natural killer cell activity and monocyte cytotoxicity induced by zinc deficiency. J Lab Clin Med. 1983;102(4):577–89. [PubMed]
- [51].Johnson DS, Chen YH. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol. 2012;12(4):458–63. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.