Abstract
Numerous genes have been associated with multiple morphological abnormalities of the sperm flagella (MMAF), which cause severe asthenozoospermia and lead to male infertility, while the causes of approximately 50% of MMAF cases remain unclear. To reveal the genetic causes of MMAF in an infertile patient, whole-exome sequencing was performed to screen for pathogenic genes, and electron microscope was used to reveal the sperm flagellar ultrastructure. A novel heterozygous missense mutation in the outer dense fiber protein 2 (ODF2) gene was detected, which was inherited from the patient's mother and predicted to be potentially damaging. Transmission electron microscopy revealed that the outer dense fibers were defective in the patient's sperm tail, which was similar to that of the reported heterozygous Odf2 mutation mouse. Immunostaining of ODF2 showed severe ODF2 expression defects in the patient's sperm. Therefore, it was concluded that the heterozygous mutation in ODF2 caused MMAF in this case. To evaluate the possibility of assisted reproductive technology (ART) treatment for this patient, intracytoplasmic sperm injection (ICSI) was performed, with the help of a hypo-osmotic swelling test and laser-assisted immotile sperm selection (LAISS) for available sperm screening, and artificial oocyte activation with ionomycin was applied to improve the fertilization rate. Four ICSI cycles were performed, and live birth was achieved in the LAISS-applied cycle, suggesting that LAISS would be valuable in ART treatment for MMAF.
Keywords: intracytoplasmic sperm injection, laser-assisted immotile sperm selection, multiple morphological abnormalities of the sperm flagella, outer dense fiber protein 2 (ODF2)
INTRODUCTION
It has been reported that infertility occurs in approximately 8%–12% of couples. Approximately 40%–50% of infertility cases are caused by male factors.1 Oligozoospermia, asthenozoospermia, teratozoospermia, and azoospermia are common causes of male infertility. Semen from asthenozoospermic patients is characterized by reduced sperm motility. Ultrastructural defects in sperm flagella are one of the main causes of severe asthenozoospermia.2
Sperm flagellum and motile cilia share a conserved core structure named axoneme, which consists of nine microtubular doublets and a central pair of microtubules, in a classical arrangement of 9 + 2 microtubules. Moreover, the sperm tail contains accessory structures that do not appear in other cilia, such as the mitochondrial sheath, fibrous sheath, and outer dense fibers.
Primary ciliary dyskinesia (PCD) is a genetic disease caused by motile ciliary malfunction, with symptoms including chronic nasal discharge, ear, nose, and chest infections, and pulmonary disease, which lead to malformation of sperm flagella.3 Furthermore, a sperm-specific phenotype has been distinguished, characterized by five morphological defects of sperm flagella, namely short, absent, bent, coiled, or irregular-caliber sperm flagella.4 Generally, unassembled sperm fibrous sheaths and a lack of central microtubules or dynein arms can be observed when transmission electron microscopy (TEM) is employed. Moreover, commonly observed PCD symptoms, such as chronic nasal discharge, ear, nose, and chest infections, and pulmonary disease, are absent in some of the patients with these sperm-specific phenotypes, which distinguish them from those of PCD. This phenotype is termed multiple morphological abnormalities of the sperm flagella (MMAF).5 Serious structural defects in sperm flagella can cause low sperm motility and even lead to completely immotile sperm. Thus, individuals with MMAF can hardly achieve spontaneous pregnancy, and no success of conventional in vitro fertilization has been reported. Intracytoplasmic sperm injection (ICSI) is the only option for these patients to produce a child.
Previous studies have shown that immotile spermatozoa from the ejaculate or testis can fertilize oocytes and successfully produce viable pregnancies.6 However, the quality of embryos created by immotile sperm is commonly poor, resulting in lower pregnancy rates.7 It was reported that 70% of immotile spermatozoa have higher degrees of DNA fragmentation,8 which may contribute to lower embryo quality. Therefore, selection of available sperm via hypo-osmotic swelling (HOS) test, activating substances, or laser-assisted immotile sperm selection (LAISS) before ICSI is necessary.6
Using whole-exome sequencing (WES), an increasing number of genes related to clinical MMAF have been identified. The products of these genes are components of the inner and outer dynein arm complex (such as dynein axonemal heavy chain [DNAH] family genes), components of the dynein arm-associated complex (such as cilia- and flagella-associated protein 43 [CFAP43], cilia- and flagella-associated protein 44 [CFAP44], and cilia- and flagella-associated protein 70 [CFAP70]), components of the peri-axonemal structure (such as fibrous sheath-interacting protein 2 [FSIP2]), components of the centrosome (such as centrosomal protein 135 [CEP135]), components of the intraflagellar transport complex (such as tetratricopeptide repeat domain 21A [TTC21A], tetratricopeptide repeat domain 29 [TTC29], and sperm flagellar 2 [SPEF2]), and components of the protein degradation complex (such as glutamine rich 2 [QRICH2]).9 Additionally, genes with undefined localization or associated processes, such as adenylate kinase 7 (AK7) and androgen-regulated protein 2 (ARM2), were reported to be involved in the occurrence of MMAF. However, due to the genetic heterogeneity of MMAF, the causes in approximately 50% of MMAF cases remain unclear. Other genes involved in clinical MMAF remain to be identified.
ODF2 encodes outer dense fiber protein 2 (ODF2), an important component of the mammalian sperm tail, and is involved in the assembly of outer dense fibers. Therefore, defects in this protein are closely related to abnormal sperm flagellar structure. It has been reported that conditional knockout of Odf2 in mice could lead to sperm tail deformity and infertility; however, no human ODF2 mutation has been reported.10,11
Herein, a clinical infertile patient with sperm tail deformity caused by ODF2 mutation was reported, along with the successful treatment of this patient with assisted reproductive technology (ART).
PATIENT AND METHODS
Case presentation
This study was approved by the Institutional Review Board of Shanghai General Hospital (Shanghai, China; license number of ethics statement: 2016KY196). Informed consent was obtained from the patient. The patient provided his semen and blood samples, along with his parents' blood samples, for research purposes. The patient was 39 years old and presented with infertility. The patient and his wife failed to conceive following regular, contraception-free intercourse for 2 years, and attended Shanghai General Hospital for treatment in April 2017. General physical examination of the patient was normal, with normal scrotum appearance and normal pubic hair distribution. The bilateral testicular sizes were 12 ml with normal texture, and the bilateral epididymis and vas deferens could be touched and were of normal size. No abnormalities were observed in the hormone tests. No other complaints commonly observed in PCD were presented.
Semen analysis
Semen analysis was performed according to the World Health Organization (WHO) Laboratory Manual for the Examination and Processing of Human Semen (5th edition).12
The viability of sperm was detected by the HOS test using a sperm viability testing kit (Solarbio Technology Co., Ltd., Beijing, China). Briefly, liquefied semen was mixed with the hypo-osmic agent provided in the kit at a 1:10 ratio and incubated for 30 min. Sperm were then counted under a microscope. The tail of viable sperm was swollen in the hypo-osmic agent.
Sperm staining was performed using a Diff-Quik staining kit (Baso Diagnostics Inc., Zhuhai, China). Briefly, a sperm smear was prepared and fixed in Diff-Quik Fix for 30 s, and then the sperm smear was stained with Diff-Quik I and Diff-Quik II for 30 s. After the stained smear was washed with fresh water and air-dried, sperm morphology analysis was performed according to the criteria recommended by the WHO. Semen specimens from healthy donors who had signed informed consents served as controls.
Electron microscope observation
TEM (H-7650, Hitachi, Tokyo, Japan) was used to observe the structure of the sperm flagella. Scanning electron microscopy (SEM; Quanta 200, FEI, Hillsboro, OR, USA) was employed for general morphology observation. Semen from the patient was centrifuged at 600g (Centrifuge 5415 R, Eppendorf AG, Hamburg, Germany) for 30 min. After the precipitate was washed with phosphate-buffered saline (PBS) three times, the precipitate was suspended in 2.5% glutaraldehyde for fixing. A portion of the fixed sample was loaded onto polylysine-coated coverslips for SEM observation, and the remainder was sent for TEM observation.
For SEM, the fixed sample on the coverslip was washed twice with PBS. Postfixation was performed using 1% osmium tetroxide for 1 h, followed by washing twice with PBS. The sample was then dehydrated using a gradient alcohol and dried in a critical point dryer. The dried sample was placed in a vacuum evaporator and sputter coated with gold–palladium.
For TEM, the fixed samples were washed twice with PBS. Postfixation, dehydration, and drying were performed as described in the SEM procedure. Resin was then used to embed the dried precipitate, and a section was prepared. Semen specimens from health donors served as controls.
WES
The blood samples were provided by the patient and his parents. Genomic DNA was extracted from the blood using the TIANamp Blood DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China). DNA was fragmented using Covaris-focused ultrasonication. Known exons and exon–intron boundary sequences were captured using xGen Exome Research Panel (Integrated DNA Technology, Inc., Coralville, IA, USA), and DNA libraries were prepared according to the manufacturer's instructions. Sequencing was performed using the Illumina HiSeq X10 platform (Illumina Inc., San Diego, CA, USA). The number of sequencing reads was 6.05 M. Sequencing reads were aligned to the human genome (GRCh37/hg19) using Burrows–Wheeler Aligner. Both single-nucleotide variants and indels within the captured coding exonic intervals were called using GATK, Platypus, VarScan, LoFreq, FreeBayes, SNVer, SAMtools, and VarDict. Moreover, the variants were filtered and annotated following the analysis pipeline, as reported previously.4 Sequencing and analysis were conducted with the help of Nuprobe Company (Shanghai, China).
Sanger sequencing
Sanger sequencing was performed to verify mutations identified by WES. First, primers flanking the mutation site were designed. The target sequence was amplified by PCR using the genomic DNAs as templates. The PCR products were sent for sequencing. Sanger sequencing service was provided by Sangon Biotech Co., Ltd. (Shanghai, China).
Detecting the expression of transcripts that could be affected by the mutation in sperm
Twenty-three transcript variations of ODF2 genes were recorded in GenBank, therefore, it was essential to determine whether the mutations affecting transcripts were expressed in sperm. The exon distributions of all recorded ODF2 transcript variations were annotated (Supplementary Table 1). Primers that could distinguish the transcript variations with or without the sequence that could be affected by the mutation were designed with the intron spanned (Figure 1a and 1b), and the corresponding product sizes were checked using the Primer-BLAST tool provided by the National Center for Biotechnology Information (taking primer pair for detecting glyceraldehyde-3-phosphate dehydrogenase [GAPDH] as an example; Supplementary Figure 1 (798.8KB, tif) ).
Supplementary Table 1.
The distribution of the exons of recorded outer dense fiber protein 2 alternative splicing transcripts
| Transcript variation | Accession number | Bk1 | Bk2 | Bk3 | Bk4 | Bk5 | Bk6 | Bk7 | Bk8 | Bk9 | Bk10 | Bk11-Bk20 | Bk21 | Bk22 | Bk23 | Bk24-Bk26 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NM_153435.1 | a | # | b | a | # | # | b | d | a | # | |||||
| 2 | NM_153433.2 | # | # | # | b | # | # | b | d | a | # | |||||
| 3 | NM_153432.1 | a | # | b | # | # | b | b | ||||||||
| 4 | NM_153436.2 | # | # | b | # | # | b | a | ||||||||
| 5 | NM_153439.1 | a | # | a | # | # | a | c | ||||||||
| 6 | NM_153437.3 | # | # | # | a | # | # | b | a | |||||||
| 7 | NM_153440.2 | # | # | a | # | # | b | a | ||||||||
| 8 | NM_001242354.2 | b | # | a | a | # | b | a | ||||||||
| 9 | NM_002540.5 | b | # | # | # | b | a | # | # | b | d | a | # | |||
| 10 | NM_001242353.2 | b | # | # | b | # | # | b | d | a | # | |||||
| 11 | NM_001242352.2 | b | # | a | b | # | # | b | d | a | # | |||||
| 12 | NM_001351577.1 | a | # | b | b | # | # | b | d | b | # | |||||
| 13 | NM_001351578.2 | a | # | b | b | # | # | b | d | a | # | |||||
| 14 | NM_001351579.2 | b | # | a | b | # | # | b | d | b | # | |||||
| 15 | NM_001351580.2 | b | # | # | a | b | # | # | b | d | b | # | ||||
| 16 | NM_001351581.1 | a | # | b | # | # | b | d | a | # | ||||||
| 17 | NM_001351582.2 | b | # | # | b | b | # | # | b | d | a | # | ||||
| 18 | NM_001351583.2 | b | # | a | a | # | # | b | d | b | # | |||||
| 19 | NM_001351584.2 | # | # | a | b | # | # | b | d | a | # | |||||
| 20 | NM_001351585.2 | b | # | # | a | b | # | # | b | d | a | # | ||||
| 21 | NM_001351586.2 | b | # | a | a | # | # | b | d | a | # | |||||
| 22 | NM_001351587.2 | b | # | a | b | # | b | d | a | # | ||||||
| 23 | NM_001351588.2 | b | # | a | a | # | b | d | a | # |
Bk, abbreviation of “Block;” which was used for informal numbering of the exons in this work; “#” suggests that the block is identical in all transcript variations; “a,” “b,” “c,” and “d” suggest that the block is different among the transcript variations; The mutated nucleotide located in Bk 9b
Figure 1.
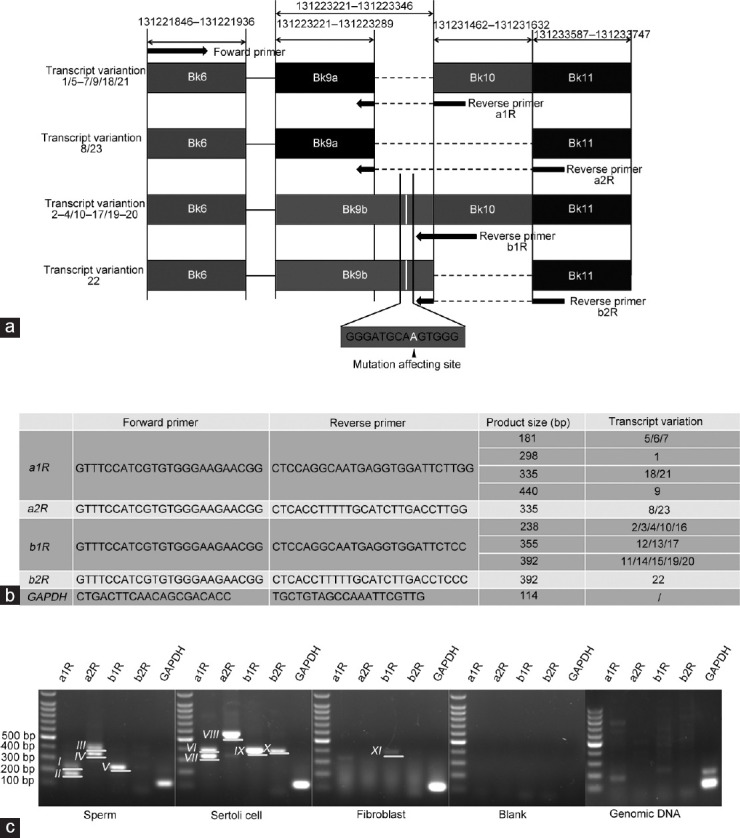
PCR detection of the transcripts that could be affected by the mutation in sperm. (a) The positions of the primers designed to distinguish the ODF2 transcripts with or without the sequence that could be affected by the mutation are indicated by the arrows. The mutated site is located in Bk9b (the location of the mutation affecting site is indicated in white, and sequence flanked the mutation affecting site is indicated below). The forward primer was shared. Reverse primer a1R and a2R were employed to detect transcripts containing Bk9a which does not contain the mutation affecting site. Reverse primer b1R and b2R were employed to detect transcripts containing Bk9b which contains the mutation affecting site. Only 4–5 nucleotides at the 3' terminal of these reverse primers match the sequence located in Bk9, indicating the annealing temperature is extremely low for these primers to bind to Bk9 directly unless incorporated with the rest of the primer sequences that match the sequence located in Bk10 or Bk11, avoiding the nonspecific amplification when common PCR conditions were used. (b) The primer sequences employed for detecting the transcripts. The product sizes of corresponding transcript variations are also listed. (c) PCR results for detecting the transcripts that could be affected by the mutation. cDNA derived from purified sperm was used as a template. cDNAs derived from Sertoli cells and fibroblast served as somatic controls. Water served as a negative control. Genomic DNA was also used as a PCR template for comparison, determining whether genomic DNA contamination existed in these cDNA specimens. The reverse primers used for each reaction are indicated above the lanes. GAPDH was detected as an internal control. It was confirmed that some of the transcripts that could be affected by the mutation were highly expressed in sperm (these transcripts generated product approximately 238 bp in size, numbered Band V), and the others were expressed in Sertoli cells and fibroblast (these transcripts generated products approximately 355 bp or 392 bp in size, numbered Band IX and Band X, respectively). Besides, unexpected products were generated (Band I, Band III, Band VI, and Band VIII. The sizes of these products do not match the expected ones), PCR amplification using genomic DNA as a template could not generate products with the same sizes as those generated from cDNA specimens, indicating there was no contamination of genomic DNA in these cDNA specimens. Bk: block, used for informal numbering of the exons in this work; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; cDNA: complementary DNA; ODF2: outer dense fiber protein 2.
Spermatozoa were purified from the semen of healthy donors via a modified swim-up method. Briefly, approximately 1–2 ml of liquefied semen was centrifuged at 600g for 10 min, the upper semen plasma was removed, and approximately 200 μl of liquid was retained. The precipitate was resuspended and the suspension was added at the bottom of a 1.5-ml tube, and then approximately 1-ml modified human tubal fluid (mHTF) supplied with 5% human serum albumin was carefully covered on the suspension. The tube was incubated in a 37°C incubator for at least 1 h, and then the upper mHTF (approximately 500–700 μl) was collected, avoiding the inclusion of somatic cells. Fibroblasts isolated from foreskin collected in circumcision served as somatic cell controls, along with Sertoli cells isolated from testis biopsy in our previous work.13
RNA was extracted from the screened sperm and somatic cells using the RNAiso Plus Kit (#9109, TaKaRa Bio, Shiga, Japan), and cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (K1631, Thermo Fisher Scientific, Boston, MA, USA), according to the manufacturer's instructions. PCR was performed using the mentioned primers and the cDNAs, water served as a blank control, genomic DNA served as the comparison template, determining whether there was contamination of genomic DNA in cDNA specimens, and GAPDH was detected as the internal reference (primer pair for detecting GAPDH transcripts was employed as positive control primers for genomic DNA, since it was predicted to generate specific products from genomic DNA; Supplementary Figure 1 (798.8KB, tif) ). The PCR products were separated by agarose gel electrophoresis. The results were recorded using a gel-imaging system (FluorChem E, ProteinSimple, San Jose, CA, USA).
To further determine the potential sources of the PCR products, PCR products were sent for sequencing. PCR products generating multiple bands were separated using agarose gel electrophoresis. The bands were recovered using a gel extraction kit (MA0017, Meilun Biotech Co., Ltd., Dalian, China), followed by TA cloning using a TOPO-TA cloning kit (10907ES20, Yeasen Biotech Co., Ltd., Shanghai, China) before being sent for sequencing. The sequencing results were aligned with the reference RNA sequence (Ref RNA) database to determine the product sources.
To further determine which transcript variations were expressed in sperm, primers were designed to detect each recorded transcript variation of ODF2, and the corresponding product sizes were checked using the Primer-BLAST tool (Supplementary Tables 2 and 3). PCR was performed using these primers. Sperm cDNA and genomic DNA were used as templates, and water served as a blank control. Specific products with expected product size and high abundance were sent for sequencing, further confirming the product sources.
Supplementary Table 2.
The choosing of the sequences for primer designing
| Transcript variation | Bk1 | Bk2 (U) | Bk3 | Bk4 | Bk5 | Bk6 | Bk7 | Bk8 (U) | Bk9 (D) | Bk10 | Bk11 | Bk21 | Bk22 (U/D) | Bk23 (D) | Bk24 | Forward primer | Reverse primer | Expected product size (bp) | Size (s) of the predicted additional product (s) generated by the primers (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | af | # | b | ar | #r | # | b | d | a | # | 1F | 1R | 438 | T5-321 | |||||
| 2 | #f | # | # | br | # | # | b | d | a | # | 2F | 2R | 505 | / | |||||
| 3 | af | # | b | # | # | b | br | 1F | 3R | 2193 | T5-2049 | ||||||||
| 4 | #f | # | b | # | # | b | ar | 3F | 4R | 2060 | / | ||||||||
| 5 | af | # | a | # | # | ar | cr | 1F | 5R | 1782 | / | ||||||||
| 6 | #f | # | # | a | # | # | b | ar | 4F | 4R | 2267 | / | |||||||
| 7 | #f | # | ar | #r | # | b | a | 2F | 1R | 227 | / | ||||||||
| 8 | b | # | af | a | # | b | ar | 5F | 4R | 1856 | / | ||||||||
| 9 | b | # | # | #f | b | ar | #r | # | b | d | a | # | 6F | 1R | 259 | / | |||
| 10 | bf | #f | #r | br | # | # | b | d | a | # | 7F | 6R | 359 | / | |||||
| 11 | bf | #f | a | b | # | # | b | d | ar | #r | 8F | 7R | 2160 | T13-2123/T23-1932/T21-2103/T22-1989/T16-2206/T1-2066 | |||||
| 12 | af | # | b | b | # | # | b | d | br | #r | 1F | 8R | 2408 | / | |||||
| 13 | af | # | b | b | # | # | b | d | ar | #r | 1F | 7R | 2251 | T16-2134/T1-2194 | |||||
| 14 | bf | #f | a | b | # | # | b | d | br | #r | 8F | 8R | 2317 | T18-2260/T12-2280 | |||||
| 15 | bf | #f | # | a | b | # | # | b | d | br | #r | 7F | 8R | 2554 | / | ||||
| 16 | af | # | b | # | # | b | d | ar | #r | 1F | 7R | 2134 | T13-2251/T1-2194 | ||||||
| 17 | bf | #f | #r | br | b | # | # | b | d | a | # | 7F | 9R | 357 | / | ||||
| 18 | b | # | a | af | #f | # | b | d | br | #r | 9F | 8R | 1949 | / | |||||
| 19 | #f | # | ar | b | # | # | b | d | a | # | 3F | 10R | 154 | / | |||||
| 20 | bf | #f | # | a | b | # | # | b | d | ar | #r | 7F | 7R | 2397 | T17-2360/T9-2445/T10-2243 | ||||
| 21 | bf | #f | a | a | # | # | b | d | ar | #r | 8F | 7R | 2103 | T11-2160/T13-2123/T23-1932/T22-1989/T16-2206/T1-2066 | |||||
| 22 | b | # | a | bf | #f | b | d | ar | #r | 10F | 7R | 1619 | / | ||||||
| 23 | b | # | a | af | #f | b | d | ar | #r | 11F | 7R | 1620 | / |
All the blocks that were valuable for specific primer designing were listed in the table; The positions of the variant sequences in some blocks which are different among the transcript variations were indicated by “U” (upstream, 5’terminal) or “D” (downstream, 3’ terminal) in the brackets following the block numbers; The blocks with superscript “f” indicated the positions of the sequences used for forward primers designing, and the blocks with superscript “r” indicated the positions of the sequences used for reverse primers designing; In some transcripts, the superscript “f” or “r” may appeared in two neighbored blocks, indicated the primers spanned the introns between the two exons to enhance the specificity; Primer names for each transcript variation and the corresponding product sizes were listed in the table. Transcript variation 11 and 21 shared the primer pair, and transcript 13 and 16 shared the primer pair; Due to the similarity of these transcript variations, many sequences are shared among transcripts. Thus, some of the primer pairs were predicted to produce multiple products. The sizes of the additional products predicted were listed in the table in the format of “Tx-xxx.” “Tx” suggested the source of the product and “xxx” suggested the size of the product. For example, “T5-321” means the product was derived from transcript variation 5 and was 321 bp in size
Supplementary Table 3.
The sequences of the primers designed for detecting the transcript variations
| Primer | Sequence |
|---|---|
| 1F | GTCGGAAAAGGAGGCGGAAG |
| 1R | CCAGGCAATGAGGTGGATTCTTG |
| 2F | CTTCACCTTTCTCTCTATGGGCAG |
| 2R | CTCCCACTTGCATCCCACAG |
| 3R | GGCATTTGGCACACCAGAC |
| 3F | GAGAAGATGGCGGACCAGCAAG |
| 4R | CAGCACTGATGATAGCTCACTCC |
| 5R | TGATCTTCAGGATCTTGTTC |
| 4F | TGGGGCCGAGCGGTTCTCAC |
| 5F | TCAACCATTTTTGTGTTGTCCTCC |
| 6F | GAGATCCAGCCCTAAGAACCCT |
| 7F | TGTCGCTCCTGGACAGTTGC |
| 6R | CCTCGCTTGTGAGATTTCGTC |
| 8F | CCGTGTCGCTCCTGGTTTC |
| 7R | GCCTCCAGTTTCCTCTCCAG |
| 8R | CGCCTCCAGTTTCCACTGAAATG |
| 9R | CTTCATGGTTGGCTTCGTCAC |
| 9F | TCCGGGTGAAAACCAAGAATC |
| 10R | GGAGGACAACACAAAAATGGTTGA |
| 10F | GGATGCAAGTGGGAGGTCAAG |
| 11F | CCGGGTGAAAACCAAGGTCAAG |
Protein variation effect analysis
To evaluate whether the mutation would affect target gene function, protein variation effect analysis was performed based on Polyphen2 and Sorting Intolerant from Tolerant (SIFT) algorithms.14,15 The sequence information of the target gene was uploaded to the Polyphen2 (http://genetics.bwh.harvard.edu/pph2) and Protein Variation Effect Analyzer (PROVEAN; http://provean.jcvi.org/index.php) platforms to obtain the score and corresponding annotation of the variation.
Immunofluorescence staining determining the expression and localization of ODF2 in sperm
Immunofluorescence staining was performed to identify the expression and localization of ODF2 in sperm derived from the patient and healthy donors. Both anti-ODF2 antibodies provided by Abcam (Cambridge, UK) and Proteintech Group (Rosemont, IL, USA) were employed, since ODF2 isoforms with different antigenic epitopes that are recognized by antibodies from different sources may be located at distinct positions in sperm.
The semen specimens were washed twice with PBS. The sperm precipitates were resuspended and fixed in 4% paraformaldehyde solution, and smears were prepared using the suspension. Antigen retrieval was performed using citrate antigen retrieval buffer (Yeasen Biotech Co., Ltd.) by microwaving the slides for 10 min. Permeabilization was performed using 0.3% Triton X-100. The slides were then incubated with 5% bovine serum albumin blocking buffer for 1 h, followed by incubation with rabbit anti-ODF2 antibody (anti-Cenexin1/ODF2, ab43840, Abcam), or ODF2 rabbit polyclonal antibody (12058-1-AP, Proteintech Group), which was diluted in the blocking buffer (1:400 and 1:100, respectively), at 4°C overnight. Normal rabbit IgG (Sangon Biotech Co., Ltd.) was used as an isotype control for control slide incubation. The following day, the slides were washed three times with PBS. Alexa Fluor 488 donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA, USA) diluted in blocking buffer at a 1:400 ratio was added and incubated at 25°C for 1 h. After incubation, the slides were washed twice in PBS and stained with 4',6-diamidino-2-phenylindole (DAPI) staining solution (Sangon Biotech Co., Ltd.) for 5 min. The slides were then washed twice and mounted with coverslips using a mounting medium (Vector Laboratories Inc., Burlingame, CA, USA). The stained spermatozoa were observed under a fluorescence microscope.
Available sperm selection, ICSI, and artificial oocyte activation (AOA) procedure
Four oocyte retrieval procedures were performed with different sperm origins, or available sperm selection methods (Table 1). Since no motile spermatozoa could be retrieved in ejaculated semen, microdissection testicular sperm aspiration was performed to determine whether testicular spermatozoa were available for the first ICSI cycle. Surgery was performed on the day of oocyte retrieval. However, no motile sperm were retrieved from the testicular tissue following incubation with pentoxifylline. The HOS test16 was used to determine available sperm with tail swelling. Considering this, direct centrifugation combined with HOS test, isodensity centrifugation combined with HOS test, and isodensity centrifugation combined with LAISS17 were employed in the following three ICSI cycles to select available sperm from semen. AOA was performed 1 h after ICSI with 10 μmol l−1 ionomycin (Merck KGaA, Darmstadt, Germany) treatment for 10 min. The fertilization rate was calculated 16–18 h after ICSI. The blastocyst rate (the number of blastocysts accounting for the number of meiosis stage II [MII] oocytes) was evaluated 5–6 days after fertilization.
Table 1.
Characteristics and outcomes in the present couple that underwent different sperm selection protocols in in vitro fertilization therapy
| Cycle | Spermatozoan origin (semen or testis) | Semen procedure | Available sperm selection | AOA (ionomycin, μmol l−1) | Available spermatozoa/selected spermatozoa, n/total (%) | MII oocyte/oocyte (n) | 2PN fertilization, n (%) | Embryo cleaved, n (%) | Available embryo, n (grade) | Outcome (pregnancy date) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Testis | General centrifugation | HOS | 10 | 4/51 (7.8) | 4/5 | 2 (50.0) | 2 (100.0) | 1 (6II) | No pregnancy |
| 2 | Semen | General centrifugation | HOS | 10 | 2/122 (1.6)* | 2/2 | 1 (50.0) | 1 (100.0) | 1 (6II) | No pregnancy |
| 3 | Semen | Isodensity centrifugation | HOS | 10 | 3/32 (9.4) | 3/3 | 0 (0) | 0 (0) | 0 | No embryo transfer |
| 4 | Semen | Isodensity centrifugation | LAISS | 10 | 6/78 (7.7) | 6/6 | 6 (100.0) | 5 (83.3) | 4 (7III, 10III, 10IIa, and 8IIIa) | Single live birth |
*Significant differences existed in this group compared with others (P<0.05). aLive birth derived from the transfer cycle using these embryos. HOS: hypo-osmotic swelling test; LAISS: laser-assisted immotile sperm selection; AOA: artificial oocyte activation; IVF: in vitro fertilization; 2PN: two pronuclei; MII: meiosis stage II
Ovarian stimulation and pregnancy outcome
The woman received stimulation treatment with recombinant follicle-stimulating hormone (Merck KGaA), human menopausal gonadotropins (Livzon Pharmaceutical Group Inc.), and gonadotropin-releasing hormone antagonist (Merck KGaA). Transvaginal sonography-guided follicular puncture was performed 36 h after human chorionic gonadotropin (hCG) injection (Livzon Pharmaceutical Group Inc.) injection. The harvested oocytes were denudated enzymatically with recombinant human hyaluronidase and mechanically by pipetting with glass pipettes. The retrieved available spermatozoa were injected into MII oocytes according to a previous description.18 Fertilized embryos were cultured in G1 plus medium (Vitrolife AB, Gothenburg, Sweden) at 37°C in 6% CO2, 5% O2, and 89% N2. Embryo transfer was performed on day 3.
Biochemical pregnancy was confirmed by a positive β-hCG level in the blood or urine 2 weeks after embryo transfer. Ultrasonography was performed at 6-week gestation to confirm the fetal viability. Clinical pregnancy was defined as the presence of a gestational sac in the uterine cavity detected by ultrasound at 4–6 weeks after transfer.
Determining the gene type of the offspring
To determine whether the offspring of the patient were carriers of the mutation, approximately 20 μl of saliva was collected from the newborn baby with parental consent. Genomic DNA was extracted using an Ezup Saliva/Urine DNA Extraction Kit (Sangon Biotech Co., Ltd.). Sanger sequencing was performed to determine the genetic type of the baby.
RESULTS
Semen analysis revealed serious tail deformity in the patient
The semen of the patient revealed a normal semen volume, with a normal pH value (7.2). Sperm density was above the reference value set by the WHO, while no motile sperm could be observed. The HOS test revealed that only 2.0% of the sperm were alive.
Two-hundred spermatozoa were analyzed after Diff-Quik staining, of which 2.0% were normal, 4.0% were head deformed, 3.0% were neck and midpiece deformed, 20.0% were principle piece deformed, and 71.0% were mixed malformed (more than one type of deformity appears in a single spermatozoon). In particular, tail deformity appeared in almost all spermatozoa with mixed malformation (Figure 2a). Compared with that obtained from the healthy donors (Figure 2b), the majority of the tails were short, and some were even absent (Figure 2c–2e).
Figure 2.
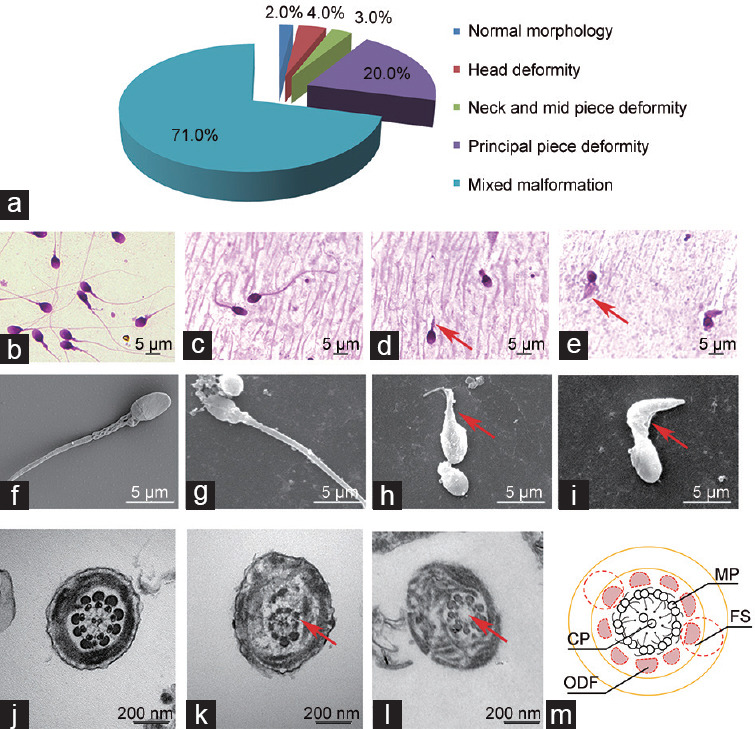
Typical morphology of spermatozoa observed in the current case. (a) Morphological analysis results of the patient's spermatozoa. Tail deformity was the dominant deformity type. (b) Typical morphology of the spermatozoa derived from healthy donors stained with Diff-Quik kit under a microscope. (c) Sperm with normal morphology retrieved in the patient's semen stained with Diff-Quik. (d) Sperm with shortened tail (indicated by the arrow) in the patient's semen stained with Diff-Quik. (e) Sperm with shortened and bulked tail (indicated by the arrows) in the patient's semen stained with Diff-Quik. Besides, sperm with tail absence could also be observed in this field. (f) Typical morphology of the spermatozoa derived from healthy donors observed using an SEM. (g) Sperm with normal morphology retrieved in the patient's semen observed using an SEM. (h) Sperm with shortened tail (indicated by the arrows) in the patient's semen observed using an SEM. Besides, sperm with tail absence could also be observed in this field. (i) Sperm with shortened and bulked tail (indicated by the arrows) in the patient's semen observed using an SEM. The spermatozoa with normal morphology retrieved in the patient's semen like those shown in c and g are rare. Sperm with severe malformed tails like those shown in d, e, f, and i represent the morphology of the most spermatozoa observed in the patient's semen. (j) Typical sperm tail structure in a cross section of the sperm derived from healthy donors, with complete structure of axoneme and accessory. (k) Missing outer dense fibers are observed in the cross section of patient-derived malformed sperm tail derived. An arrow indicates the missing outer fibers. (l) Although no outer dense fiber is missing, in some sperm tails of the patient's, central microtubules are observed to be absent (indicated by the arrow). (m) The schematic diagram shows the normal sperm tail structure. Nine outer dense fibers (two of which are replaced by fibrous sheaths in the principle piece) paralleled with the outer doublets can be observed. ODF: outer dense fiber; FS: fibrous sheath; MP: microtubule doublet; CP: center microtubule pair; SEM: scanning electron microscopy.
A high proportion of outer dense fiber deficiencies were observed in patient's sperm tails
An electron microscope was utilized to check the ultrastructure of the malformed sperm, particularly focusing on the tail. Although few tails with normal structure could be observed, loss of outer dense fibers in varying degrees was detected in the vast majority of cases, and in certain cases, the central microtubules were absent (Figure 2f–2l).
A heterozygous mutation was found in ODF2 gene
WES was performed to screen for potential disease-causing mutations, and a heterozygous mutation in the ODF2 gene was identified, which was inherited from the patient's mother, who was heterozygous at the same site. No related mutations were identified in the patient's father, and no other infertility-related mutations were detected. The existence of the ODF2 mutation was further confirmed by Sanger sequencing (Figure 3a and 3b).
Figure 3.
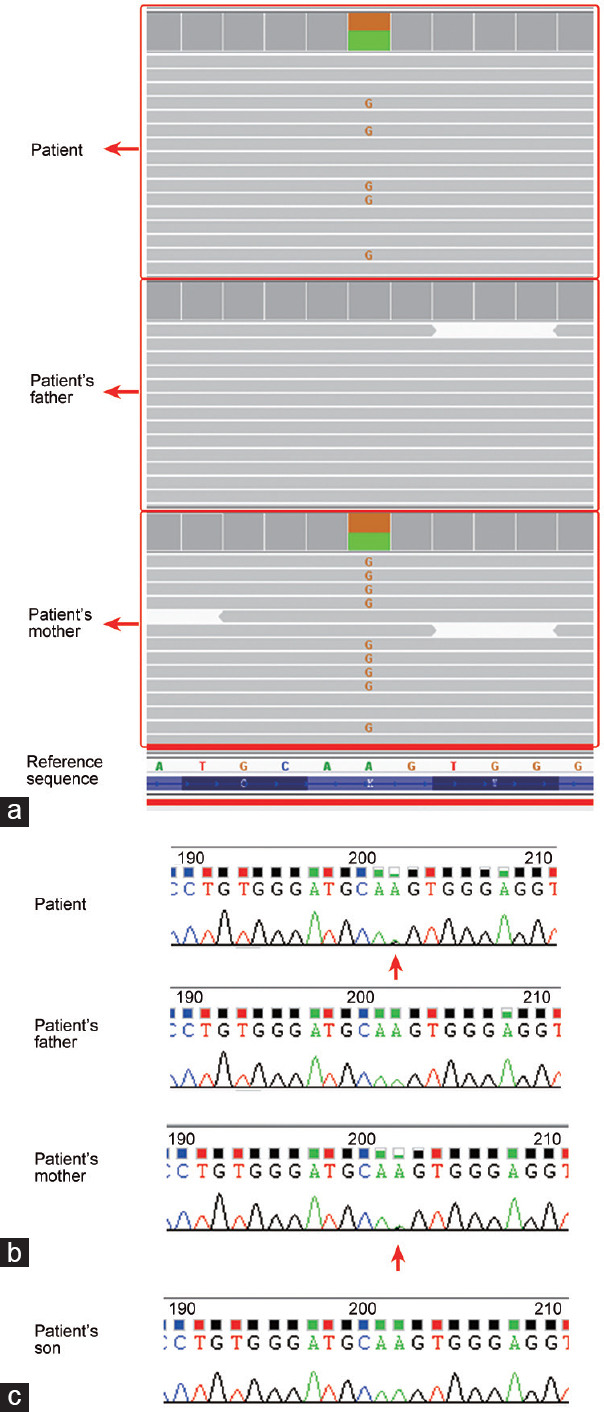
Sequencing revealed a heterozygous mutation in ODF2 gene in the patient. (a) Whole-exome sequencing results show that a heterozygous mutation is detected in ODF2 gene in the patient, which was inherited from the patient's mother (the red G appearing in some of the fragments indicates heterozygous mutation of A>G); (b) Sanger sequencing confirmed the existence of the heterozygous mutation in ODF2 genes in the patient and his mother (mutated site indicated by red arrows). (c) Sanger sequencing determined that the son of the patient does not carry the mutation in ODF2. ODF2: outer dense fiber protein 2.
The mutation was located at Chr9: 131223339 (GRCh37/hg19). The adenine at this site was replaced with guanine. The recorded mutant allele frequency was 0. No existing records on this mutation could be retrieved in the single-nucleotide polymorphism (SNP) database.
Since there were 23 transcript variations of the ODF2 gene recorded in GenBank, it was necessary to identify the influence of the mutation on each transcript variation. Exon distributions of all recorded ODF2 transcript variations were annotated, and it was confirmed that the nucleotide alteration caused missense mutations in 14/23 transcript variations (Supplementary Table 1).
Protein variation effect analysis was performed, and the mutation was predicted to be damaging in the products of 13 involved transcript variations (Supplementary Table 4); only in transcript variation 17, the mutation was predicted to be benigh.
Supplementary Table 4.
The damaging prediction of the mutant in involved outer dense fiber protein 2 transcripts
| Transcript variation | Nucleic acid change | Amino acid change | Polyphen2 score | SIFT score | Prediction |
|---|---|---|---|---|---|
| 2 | c.242A>G | p.K81R | 0.999 | 0.007 | Damaging |
| 3 | c.374A>G | p.K125R | 0.998 | 0.011 | Damaging |
| 4 | c.242A>G | p.K81R | 1.000 | 0.012 | Damaging |
| 10 | c.242A>G | p.K81R | 0.999 | 0.007 | Damaging |
| 11 | c.227A>G | p.K76R | 0.999 | 0.007 | Damaging |
| 12 | c.491A>G | p.K164R | 0.862 | N/A* | Damaging |
| 13 | c.491A>G | p.K164R | 0.563 | N/A* | Damaging |
| 14 | c.227A>G | p.K76R | 0.733 | N/A* | Damaging |
| 15 | c.227A>G | p.K76R | 0.733 | N/A* | Damaging |
| 16 | c.374A>G | p.K125R | 0.998 | N/A* | Damaging |
| 17 | c.359A>G | p.K120R | 0.165 | N/A* | Benign |
| 19 | c.227A>G | p.K76R | 0.999 | 0.007 | Damaging |
| 20 | c.227A>G | p.K76R | 0.999 | 0.007 | Damaging |
| 22 | c.227A>G | p.K76R | 0.994 | N/A* | Damaging |
*SIFT algorithm provided by PROVEAN platform could not find the matching protein accession ID and returned no results. N/A: not available; PROVEAN: protein variation effect analyzer; SIFT: sorting intolerant from tolerant
The transcripts that could be affected by the mutation were expressed in sperm at a high level
Primers that could distinguish the transcripts that could be affected by mutations were employed. PCR results showed that certain transcripts were expressed at high levels in the sperm (Figure 1c). Judging from the product size and sequencing results, these transcripts may be transcript variations 2, 3, 4, 10, or 16 (approximately 238 bp in product size), while transcript variations 11, 12, 13, 14, 15, 17, 19, 20, and 22 (approximately 355 bp or 392 bp in product size) were undetected (Supplementary Table 5 and Supplementary Figure 2 (3.6MB, tif) ). Contrastingly, transcript variations 2, 3, 4, 10, and 16 were undetected in Sertoli cells and fibroblasts. Transcript variations 11, 12, 13, 14, 15, 17, 19, and 20 may be expressed in fibroblasts and Sertoli cells at a low and high level, respectively, and transcript variation 22 was expressed in Sertoli cells at a high level. Furthermore, certain unexpected products were generated in sperm and Sertoli cells when reverse primers a1R and a2R were employed. Sanger sequencing revealed that they were generated by mismatching of the reverse primers, while all of these products were derived from known ODF2 transcripts (Supplementary Table 5 and Supplementary Figure 2 (3.6MB, tif) ).
Supplementary Table 5.
Sequencing revealing the sources of the PCR products
| Products | Actual size | Probably sources (transcript variant) | Reverse primer | Matching of the reverse primer | Dismatching details |
|---|---|---|---|---|---|
| Band I | 238bp | 10/2/16/4/3 | a1R | Mismatching | CTCCAGGCAATGAGGTGGATTCTTGGa |
| Band II | 181bp | 7/6/5 | a1R | Matching | / |
| Band III | 409bp | 10/2/16/4/3 | a2R | Mismatching | CTCACCTTTTTGCATCTTGACCTTGGb |
| Band IV* | 352bp | 7/6/5 | a2R | Mismatching | CTCACCTTTTTGCATCTTGACCTTGGb |
| Band IV* | 335bp | 8/23 | a2R | Matching | / |
| Band V | 238bp | 10/2/16/4/3 | b1R | Matching | / |
| Band VI | 392bp | 19/15/14/20/11 | a1R | Mismatching | CTCCAGGCAATGAGGTGGATTCTTGGa |
| Band VII | 335bp | 21/18 | a1R | Matching | / |
| Band VIII | 563bp | 19/15/14/20/11 | a2R | Mismatching | CTCACCTTTTTGCATCTTGACCTTGGb |
| Band IX | 392bp | 19/15/14/20/11 | b1R | Matching | / |
| Band X | 392bp | 22 | b2R | Matching | / |
| Band XI | 392bp | 19/15/14/20/11 | b1R | Matching | / |
*Multiple fragments were detected in the clones generated from band IV due to the similarity in product sizes; aThe underlined “T” was mismatched with “C,” and “GG” following the underlined “T” was missing from the primer according to the sequencing results; bThe underlined “TG” was mismatched with “CA.” PCR: polymerase chain reaction; A: Adenine; T: Thymine; C: Cytosine; G: Guanine
Additionally, almost all the products detected from cDNAs were unobserved in PCR products amplified from genomic DNA, which indicated that there was no contamination of genomic DNA in these cDNA specimens.
PCR was performed to detect the expression of each transcript variation of ODF2 in sperm. However, because of the limited sequences that could be used for primer design and the poor specificity of some primers, only transcript variation 10 and GAPDH were specifically detected to be highly expressed in sperm, with the product of transcript variation 10 being further confirmed by Sanger sequencing (Supplementary Figure 3 (660.4KB, tif) ).
ODF2 expression was altered in the patient's sperm
Two antibodies for ODF2 were used in this study. Specific positive signals were observed in the sperm tail of healthy donors. The location of the positive signal likely overlapped with the structure of the sperm annulus when the Abcam antibody was used (Figure 4a), while almost the whole sperm tail was positive when the Proteintech Group antibody was used (Figure 4b). No positive signal could be observed when normal IgG was employed which confirmed the specific staining of these antibodies (Figure 4c). Regarding the patient's sperm, positive signals were rare when the Abcam antibody was used (Figure 4d). These signals were clearly weakened compared to that of the sperm from healthy donors, and the location was adjoined to the sperm head. Meanwhile, when the Proteintech Group antibody was used for sperm staining, >50% of the sperm was negative (Figure 4e), and approximately 40% of the sperm was positively stained, most of which showed a short or severe coiled tail with disordered structures. The tails of ≤2% positively stained sperm were relatively normal in length. Like that in the health donor's sperm, no positive signal could be observed when normal IgG was employed (Figure 4f).
Figure 4.
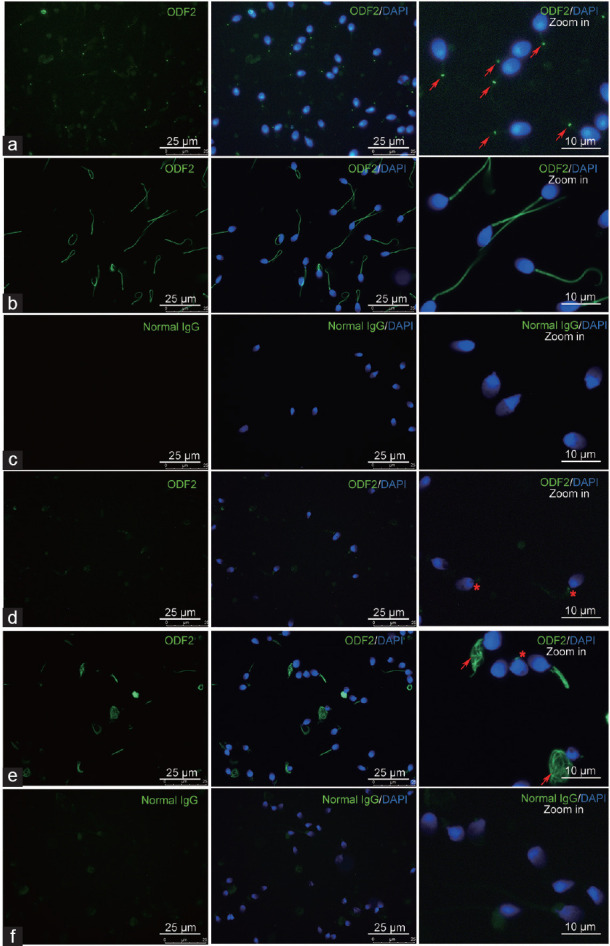
Immunostaining of sperm using anti-ODF2 antibodies. (a) Sperm derived from healthy donors was stained using an Abcam antibody. Arrows indicate the positive signals in the sperm, which are located at the position likely overlapped with sperm annulus. (b) Sperm derived from healthy donors was stained using a Proteintech Group antibody. Almost the whole sperm tail was positive. (c) Sperm derived from healthy donors was stained using normal IgG which served as a control. (d) Sperm derived from the patient was stained using an Abcam antibody. Asterisks indicate the positive signals in the sperm. Rare spermatozoa are positive, and the signals are weakened. (e) Sperm derived from the patient was stained using a Proteintech Group antibody. Arrows indicate the staining of severe coiled sperm tails and an asterisk indicates the staining of sperm tail residue. (f) Sperm derived from the patient was stained using normal IgG which served as a control. DAPI: 4',6-diamidino-2-phenylindole; ODF2: outer dense fiber protein 2.
Clinical result of ART treatment
Four oocyte retrieval procedures were performed in this couple (Table 1) with different sperm selection methods. The available sperm determined by general centrifugation combined with HOS in semen were significantly less compared with those of general centrifugation combined with HOS in testis specimen, isodensity centrifugation combined with HOS in semen, and isodensity centrifugation combined with LAISS in semen (1.6% vs 7.8%, 9.4%, and 7.7%, respectively). The available sperm selected by isodensity centrifugation combined with LAISS obtained higher normal fertilization rates, and may have a considerably higher embryo developmental potential, resulting in higher grade embryos and a single live birth (a healthy boy).
The baby does not carry the mutation
Since the heterozygous mutation in ODF2 was related to the MMAF phenotype in the patient, whether the male offspring carries the mutation was of great concern. Fortunately, Sanger sequencing revealed that the baby does not carry the mutation (Figure 3c).
DISCUSSION
MMAF causes severe asthenozoospermia and leads to male infertility. Although numerous genes have been related to clinical MMAF, the causes of approximately 50% of MMAF cases remain to be elucidated.
The sperm tail, also known as the flagellum, provides the motile force for the spermatozoa. The sperm tail can be divided into four major segments, namely the connecting, mid, principle, and end pieces, which share a common innermost structure but differ in their external substructure. The structure of the sperm tail is based on a 9 + 2 arrangement of microtubules. The 9 + 2 arrangement refers to nine peripheral, asymmetrically arranged microtubule doublets connected by doublet to doublet by dynein arms and to the sheath of the central pair of microtubules by radial spokes. The outer doublets were parallel to nine outer dense fibers.19 Moreover, two fibrous sheaths composed of three longitudinal columns attached to microtubule doublets 3 and 8 run along the principal piece, replacing the corresponding outer dense fibers (Figure 2m). The end of the outer dense fibers and the fibrous sheaths terminates in the end piece. The integrity of this structure ensures the proper sperm tail function, along with its normal appearance. Sperm tail component defects would lead to MMAF.
In the current case, a typical MMAF phenotype was observed in the patient's spermatozoa, and a potentially damaging mutation was detected in the ODF2 gene, which is a major component of outer dense fibers. Outer dense fibers are sperm tail-specific cytoskeletal structures composed of 10 major proteins and at least 15 small proteins.20 They provide flexibility along with firm support during flagellar movement.21
Previous clinical studies have reported a deficiency in outer dense fibers with idiopathic male infertility and asthenozoospermia.22,23,24,25 However, no ODF2 gene mutation has been previously reported. Could this MMAF case be caused by the mutation in ODF2? Considering the heterozygous gene type in this case, was heterozygous mutation in ODF2 sufficient to cause pathological changes in spermatozoa?
Outer dense fiber proteins primarily include ODF1, ODF2, ODF3, and ODF4, which belong to a class of macromolecular functional proteins widely present in animal cell centrosomes and sperm tails. It has been reported that ODF2 is expressed in spermatocytes in the testis and is the most expressed postmeiosis.26,27 Single-cell sequencing revealed that human ODF2 expression in the testis is initiated at the meiosis stage with a very low expression level. It increased rapidly after meiosis and reached its highest level in late spermatid.28
In addition to the formation of sperm flagella, ODF2 is involved in sperm capacitation. Mariappa et al.29 treated hamster sperm with a tyrosine phosphorylation inhibitor and found a decrease in tyrosine phosphorylation in ODF2 and tektin-2, with impaired sperm flagellar function, and in turn affected sperm capacitation. The ODF2 phosphorylation site is located in the C-terminal region, which acts on the testis-specific serine/threonine-protein kinase-4 (Tssk-4). Tssk-4 is involved in sperm structure organization and motility regulation. It can alter ODF2 phosphorylation, while ODF2 enhances the autophosphorylation of Tssk-4. These proteins interact with each other during spermatogenesis.30,31
Twenty-three ODF2 transcript variations were recorded in GenBank, therefore, before discussing whether the mutation could affect sperm phenotype, it was necessary to determine whether the transcripts affected by the mutation were expressed in sperm. Primers that could distinguish transcripts with or without the sequence that could be affected by the mutation were designed, and PCR was performed to detect the expression of these transcripts in sperm. It was determined that certain transcripts containing the sequence that could be affected by the mutation were highly expressed in sperm, while some were unexpressed in sperm. The unexpressed transcripts included transcript variation 17, the only transcript variation that achieved a benign score in protein variation effect analysis of the mutation. This means that the mutation is damaging to the products of all the affected ODF2 transcript variations expressed in sperm. In particular, ODF2 transcript variation 10 was determined to be highly expressed in sperm, and was likely the dominant ODF2 transcript variation expressed in sperm, although it requires further confirmation via different methods. Although the mentioned transcript variations expressed in sperm were undetected in Sertoli cells, certain transcript variations were highly expressed, and some were expressed in fibroblasts, indicating differential expression of ODF2 transcript variations among cells, which may be related to the differential roles of ODF2 isoforms. Although more samples from different tissues would be required to determine whether these transcripts were selectively expressed in specific cell types, the high expression level of the transcripts whose products would be damaged by the mutation in sperm indicated that the mutation could impair sperm functions.
To determine whether the heterozygous mutation in ODF2 resulted in the MMAF phenotype in the current case, immunostaining and electron microscopy were employed to detect ODF2 expression in the patient's sperm and the alteration of the sperm flagellum structure. Immunostaining revealed that ODF2 expression and localization were severely disturbed in the patient's sperm. ODF2 expression was absent in a high proportion of the patient's sperm, and the sperm tail structures were commonly disordered. Notably, the two ODF2 antibodies used in this study stained differently. These differences may be because these antibodies were generated using peptides with different antigenic epitopes, while the varied splicing of ODF2 transcripts generates protein isoforms with different antigenic epitopes that are differentially recognized by the antibody. These staining results suggested that the products of different ODF2 transcript variations may be localized at different positions in sperm, and thus play different roles, which were also indicated by Rivkin et al.32 Notably, the single amino acid alteration was not involved in the portion recognized by these antibodies. The high proportion of negative staining in the patient's spermatozoa indicated the loss of ODF2 products posttranscriptionally or posttranslationally, which may disturb the formation of a normal sperm flagellum.
To evaluate how the alteration in ODF2 contributes to deformed sperm tail formation, the sperm flagellum structure was analyzed using an electron microscope. The flagellum ultrastructure exhibited obvious defects in the outer dense fibers in most of the spermatozoa. Varying degrees of outer dense fiber defects could be observed in spermatozoa, coinciding with the varying appearances of sperm tails in the current case. However, certain spermatozoa with normal morphology could also be retrieved, along with a normal tail structure without missing outer dense fibers.
Dose-dependent effects caused by heterozygous mutations in pathogenic genes could explain the heterogeneity of spermatozoan appearance. It is well known that allele separation occurs at meiosis stage I during spermatogenesis, whereas germ cells are not completely divided into individuals. They connect with each other via intercellular bridges, which help cells to communicate with each other and keep developing synchronously.33 The gene products expressed after meiosis are distributed to neighboring cells through intercellular bridges. It is likely that products derived from different alleles of heterozygotes expressed after allele separation would be distributed at a concentration gradient in these cells, leading to varying phenotypes (Figure 5).
Figure 5.
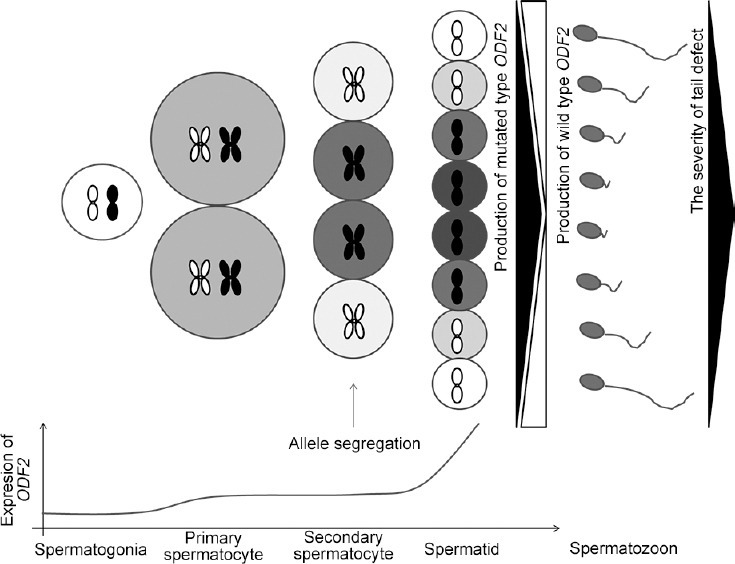
Schematic diagram shows the possible mechanism of how heterozygous mutation in ODF2 could result in malformed sperm tails at different degrees in the same semen specimen. ODF2 expression initiated at the spermatocyte stage during spermatogenesis at a very low level. Allele segregation occurred at meiosis stage I. Then different products of ODF2 gene accumulated in germ cells possessing different gene alleles. However, due to the existence of intercellular bridges, these products would be redistributed in these germ cells at a gradient, leading to a variety of morphology appearance in the spermatozoa finally. The chromosome in white indicates that the cell possesses a chromosome with a wild-type ODF2 allele, while the chromosome in black indicates that the cell possesses a chromosome with a mutated type ODF2 allele. ODF2: outer dense fiber protein 2.
In order to provide direct evidence that the heterozygous mutation contributes to the MMAF phenotype, an animal model that causes functional disturbance on ODF2 is required, while such animal models have been previously reported in ODF2 function research.
Tarnasky et al.10 demonstrated the role of ODF2 in sperm motility through gene knockout. The authors used XL169 embryonic stem cells to show that ODF2+/− chimeric mice were severely infertile due to poor sperm motility. Lee11 observed a severe defect in mouse sperm tail development in a Cenexin1 S796A mutant-expressing mouse in the ODF2+/− background.
Surprisingly, the deformed sperm tail ultrastructure reported in their work was highly similar to that in our study, which showed defects in outer dense fibers in the sperm tails. These findings support that the heterozygous mutation in ODF2 may lead to MMAF in the current case.
Furthermore, it was reported that the introduction of human Cenexin1 to heterozygous mutant mice could rescue the deformed spermatozoan phenotype, while ectopic expression of the Cexinxin1 S796A may disrupt the correct localization or function of the endogenous ODF2 protein during sperm tail differentiation, suggesting that Cenexin1 plays a role in sperm tail formation.11
Cenexins are believed to be the cleavage variants or isoforms derived from transcript variants of ODF2, which are abundantly expressed in somatic cells as centriole components, and are involved in the assembly of primary cilia and mitosis.34,35,36 The human Cenexin1 variant, for example, the product of ODF2 transcript variant 11, was affected by the mutation in the current case.
As a centriole protein, Cenexin1 plays a role in primary ciliogenesis. It was demonstrated that Cenexin1 helps recruit Chibby (CBY), a distal centriole protein required for ciliogenesis.36 CBY was shown to be located at the tip of the elongating centrioles from early spermatocytes to spermatids in Drosophila, suggesting that it plays a role in spermatogenesis.37
Moreover, Cenexins play a role in primary ciliogenesis and motile cilia. Homozygous knockout exons 6 and 7 of Cenexin/Odf2 in mice did not disturb ciliary formation. However, it resulted in basal feet lacking in the basal body, disturbing the coordination of ciliary beating and leading to a phenotype of coughing/sneezing in the mice.38
Recently, a mouse line with heterozygous knockout exon 6 and exon 7 of Cenexin/Odf2 was reported to be male infertile with decapitated and decaudated spermatozoa.39 Haploinsufficiency of the disrupted gene did not cause outer dense fiber defects in the mouse line, which was different from that observed in our study. We believe that the knockout of exon 6 and exon 7 of Odf2 mainly disrupted the function of Cenexin variants, which are located at the appendage of the basal body and recruit distal centriole proteins required for ciliogenesis.36 The decapitated phenotype could be explained by a defect in the connecting piece where the basal body is located. As for the ODF variants, the haploinsufficiency of Cenexin/Odf2 would not disrupt the assembly of outer dense fibers, as all the produced ODF2 products were normal and functional. In the current case, the mutation damages ODF2 product function. The defect is introduced into the outer dense fibers when these abnormal ODF2 products are incorporated into them.
Nevertheless, the alteration in Cenexin1 protein may have contributed to sperm tail defects in the current case, particularly because central microtubules missing were also observed in a portion of the patient's spermatozoa.
Taken together, based on the following facts: 1) the mutation in the ODF2 gene was predicted to be damaging in all the affected transcripts expressed in sperm; 2) the outer dense fibers were defective in the patient's sperm; and 3) the phenotype of Odf2 heterozygous mutated mice was highly similar to that of this patient, it could be preliminarily concluded that the heterozygous mutation in ODF2 causes the MMAF phenotype in the patient. To our knowledge, this is the first report of an infertile male suffering from MMAF with an ODF2 mutation. A novel mutation contributing to the clinical MMAF was revealed. However, there could be mutations in novel genes or mutations that lie outside of exons, which could not be detected by WES. Besides, though prior mouse studies provide good validation for the function of ODF2 in spermatogenesis, it was not the specific mutation as being causative for the phenotype. Therefore, further studies are needed to validate the functional change of ODF2 caused by specific mutations in the current case.
Since almost all the spermatozoa in the ejaculate were immotile, available sperm selection was essential for the patient to produce a child. Viable but immotile spermatozoa could be identified by the appearance of curling or coiling at the tip of the flagellum following treatment with a single laser shot.40 The primary advantage of laser usage is that it carries no risk of genetic mutation or embryotic toxicity and only requires a laser instrument used for embryo-assisted hatching or biopsy. The fertilization rate injected with immotile but vital spermatozoa by ICSI significantly improved when applying LAISS compared with the control protocol, and the take-home infant rate increased significantly.17 Remarkably similar results were obtained in our own trial using chemical substance activation combined with laser testing. We found that these methods are quick, easy, and feasible for sperm viability testing. More importantly, the immotile spermatozoa selected via LAISS may have a better development potential compared with other methods from our study results. As in the current case, the available sperm selected by isodensity centrifugation combined with LAISS obtained higher fertilization rates and higher grade embryos, and achieved a single live birth, suggesting the great value of LAISS in clinical application for immotile sperm selection.
It was also noted that homozygous mutation in Odf2 would be lethal to mouse fetuses, showing the critical role of Odf2 products in embryonic development.41 It remains unclear whether homozygous mutations in ODF2 would disturb embryonic development in humans. Although the patient successfully fathered a healthy child with the help of ART in this case, it is highly recommended that genetic counseling and preimplantation genetic diagnosis be conducted to avoid the risks of fetal arrest and abortion in future practice.
CONCLUSION
In conclusion, a novel heterozygous mutation in the ODF2 gene was associated with MMAF in an infertile male, and successful pregnancy was derived from the patient with the assistance of LAISS, showing the great value of LAISS in the ART treatment of MMAF.
AUTHOR CONTRIBUTIONS
ZJZ and ZL designed the work; ZJZ, YZW, and WC drafted the manuscript; ZJZ, YZW, and XBW collected the clinical data of the patient; ZJZ, CCY, and LYZ performed the molecular biological experiments and bioinformatic analysis; YW, ZBZ, and WC performed the ART treatment. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
Predicted product sizes of primer pair for detecting GAPDH. Primer-BLAST tool provided by NCBI was employed to predict the product size of primer pair for detecting GAPDH. (a) Predicted product sizes of product generated from cDNA (primer pair specificity checked using Ref RNA as a searching database). The primer pair used for detecting GAPDH transcripts in our work was predicted to generate a specific product about 114 bp in size from cDNA; (b) Predicted product sizes of product generated from genomic DNA (primer pair specificity checked using genomes for selected organisms (primary reference assembly only) as a searching database). The primer pair used for detecting GAPDH transcripts in our work was predicted to generate a specific product about 218 bp in size from genomic DNA. Besides, it was also predicted to have the ability to generate additional products about 114 bp and 120 bp in size via mismatching. The specificity of all the other primers used in this work was checked in the same way before PCR was performed.
Aligning of sequencing results with Ref RNA database. Transcripts with 100% Query Cover and over 99% in percent identity (Per. Ident) shown in aligning results are listed in the table. Alignment illustrations show detailed alignment results of the sequencing results with typical transcripts. Primer sequences matched with reference transcripts are highlighted in green. Mismatched nucleotides in primers are highlighted in red. Altered nucleotides in primers are highlighted in blue. (a) Aligning of sequencing result of product Band I. Primer mismatching and altering were detected; (b) Aligning of sequencing result of product Band II; (c) Aligning of sequencing result of product Band III. Primer mismatching was detected; (d) Aligning of sequencing result of product Band IV. Two different fragments were cloned from product Band IV, one of which was the result of primer mismatching; (e) Aligning of sequencing result of product Band V; (f) Aligning of sequencing result of product Band VI. Primer mismatching and altering were detected; (g) Aligning of sequencing result of product Band VII; (h) Aligning of sequencing result of product Band VIII. Primer mismatching was detected; (i) Aligning of sequencing result of product Band IX; (j) Aligning of sequencing result of product Band X; (k) Aligning of sequencing result of product Band XI.
The results of the PCR amplification detecting each transcript variation in sperm. (a) Gel electrophoresis of PCR products. Water served as blank control. Genomic DNA was used as the comparison template to determine whether there was contamination of genomic DNA in the sperm cDNA specimen. ODF2 transcript variation 11 and 21 shared the primer pair for detecting, and ODF2 transcript variation 13 and 16 shared the primer pair for detecting. Only ODF2 transcript variation 10 and GAPDH were specifically detected to be expressed at a high level in sperm (indicated by the rectangle frames). Primer pair for detecting ODF2 transcript variation 4 generated a nonspecific product with strong band which was different from the expected product in size (the expected product should be 2060 bp in size). Some of the other primer pairs, such as those for detecting transcript variant 3 and transcript variant 6, generated products that matched expected products in size (indicated by the underlines), while the bands were pretty weak, indicating the low abundances of these transcripts expressed in sperm. Based on the result provided in Figure 3, it could be determined that transcript variations 9, 11, 12, 13, 14, 15, 17, 18, 19, 20, 21, and 22 were not expressed in sperm, and no expected products could be observed when primer pairs for detecting them were employed. However, due to the limited sequences that could be used for primer designing, the specificity of some primers was not satisfying. Many unexpected products could be observed. Besides, the low abundances of some transcripts made the bands unclear in the gel. It was unable to determine whether the other transcript variations were expressed in sperm from these results. Almost no product in PCR products generated from sperm cDNA could be detected in that generated from genomic DNA, determining that there was no contamination of genomic DNA in sperm cDNA specimen; (b) Aligning of sequencing result of the PCR product generated by primer pair for detecting ODF2 transcript variation 10. Matched primer sequences were highlighted in green. Only ODF2 transcript variation 10 matches the sequencing result perfectly.
ACKNOWLEDGMENTS
The work was supported by grant from the National Key Research and Development Program of China (No. 2017YFC1002003). We appreciated Prof. Feng Zhang (Obstetrics and Gynecology Hospital, State Key Laboratory of Genetic Engineering at School of Life Sciences, Institute of reproduction and Development, Fudan University, Shanghai, China) for kindly providing the Proteintech Group anti-ODF2 antibody. We appreciated Dr. Yu-Xiang Zhang and Mr. Cun-Zhong Deng (Department of Andrology, Urological Medical Center, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China) for their helps on control sperm preparing.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Kumar N, Singh AK. Trends of male factor infertility, an important cause of infertility: a review of literature. J Hum Reprod Sci. 2015;8:191–6. doi: 10.4103/0974-1208.170370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SM, Li HB, Wang JX, Shi YC, Cheng HB, et al. Morphological characteristics and initial genetic study of multiple morphological anomalies of the flagella in China. Asian J Androl. 2015;17:513–5. doi: 10.4103/1008-682X.146100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sironen A, Shoemark A, Patel M, Loebinger MR, Mitchison HM. Sperm defects in primary ciliary dyskinesia and related causes of male infertility. Cell Mol Life Sci. 2020;77:2029–48. doi: 10.1007/s00018-019-03389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang S, Wang X, Li W, Yang X, Li Z, et al. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2017;100:854–64. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordhoff V. How to select immotile but viable spermatozoa on the day of intracytoplasmic sperm injection? An embryologist's view. Andrology. 2015;3:156–62. doi: 10.1111/andr.286. [DOI] [PubMed] [Google Scholar]
- 7.Stalf T, Mehnert C, Hajimohammad A, Manolopoulos K, Shen Y, et al. Influence of motility and vitality in intracytoplasmic sperm injection with ejaculated and testicular sperm. Andrologia. 2005;37:125–30. doi: 10.1111/j.1439-0272.2005.00665.x. [DOI] [PubMed] [Google Scholar]
- 8.Derijck AA, van der Heijden GW, Ramos L, Giele M, Kremer JA, et al. Motile human normozoospermic and oligozoospermic semen samples show a difference in double-strand DNA break incidence. Hum Reprod. 2007;22:2368–76. doi: 10.1093/humrep/dem166. [DOI] [PubMed] [Google Scholar]
- 9.Toure A, Martinez G, Kherraf ZE, Cazin C, Beurois J, et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 10.Tarnasky H, Cheng M, Ou Y, Thundathil JC, Oko R, et al. Gene trap mutation of murine outer dense fiber protein-2 gene can result in sperm tail abnormalities in mice with high percentage chimaerism. BMC Dev Biol. 2010;10:67. doi: 10.1186/1471-213X-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KH. Ectopic expression of Cenexin1 S796A mutant in ODF2+/- knockout background causes a sperm tail development defect. Dev Reprod. 2012;16:363–70. doi: 10.12717/DR.2012.16.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. Geneva: World Health Organization; 2010. [Google Scholar]
- 13.Tian RH, Yang S, Zhu ZJ, Wang JL, Liu Y, et al. NODAL secreted by male germ cells regulates the proliferation and function of human Sertoli cells from obstructive azoospermia and nonobstructive azoospermia patients. Asian J Androl. 2015;17:996–1005. doi: 10.4103/1008-682X.159722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteves SC, Varghese AC. Laboratory handling of epididymal and testicular spermatozoa: what can be done to improve sperm injections outcome. J Hum Reprod Sci. 2012;5:233–43. doi: 10.4103/0974-1208.106333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordhoff V, Schuring AN, Krallmann C, Zitzmann M, Schlatt S, et al. Optimizing TESE-ICSI by laser-assisted selection of immotile spermatozoa and polarization microscopy for selection of oocytes. Andrology. 2013;1:67–74. doi: 10.1111/j.2047-2927.2012.00020.x. [DOI] [PubMed] [Google Scholar]
- 18.Greenblatt EM, Meriano JS, Casper RF. Type of stimulation protocol affects oocyte maturity, fertilization rate, and cleavage rate after intracytoplasmic sperm injection. Fertil Steril. 1995;64:557–63. doi: 10.1016/s0015-0282(16)57792-9. [DOI] [PubMed] [Google Scholar]
- 19.Sutovsky P, Manandhar G. Mammalian spermatogenesis and sperm structure: anatomical and compartmental analysis. In: Jonge CD, Barratt C, editors. The Sperm Cell: Production, Maturation, Fertilization, Regeneration. New York: Cambridge University Press; 2006. pp. 1–30. [Google Scholar]
- 20.Linck RW, Chemes H, Albertini DF. The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J Assist Reprod Genet. 2016;33:141–56. doi: 10.1007/s10815-016-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao W, Li Z, Ping P, Wang G, Yuan X, et al. Outer dense fibers stabilize the axoneme to maintain sperm motility. J Cell Mol Med. 2018;22:1755–68. doi: 10.1111/jcmm.13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetherington L, Schneider EK, Scott C, DeKretser D, Muller CH, et al. Deficiency in outer dense fiber 1 is a marker and potential driver of idiopathic male infertility. Mol Cell Proteomics. 2016;15:3685–93. doi: 10.1074/mcp.M116.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haidl G, Becker A, Henkel R. Poor development of outer dense fibers as a major cause of tail abnormalities in the spermatozoa of asthenoteratozoospermic men. Hum Reprod. 1991;6:1431–8. doi: 10.1093/oxfordjournals.humrep.a137283. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Wang Y, Xu X, Yu Z, Gui YT, et al. Differential expression of ODF1 in human ejaculated spermatozoa and its clinical significance. Zhonghua Nan Ke Xue. 2009;15:891–4. Article in Chinese. [PubMed] [Google Scholar]
- 25.Lestari SW, Pujianto DA, Soeharso P, Loanda E. Evaluation of outer dense fiber-1 and -2 protein expression in asthenozoospermic infertile men. Med J Indones. 2015;24:79–83. [Google Scholar]
- 26.Horowitz E, Zhang ZB, Jones BH, Moss SB, Ho C, et al. Patterns of expression of sperm flagellar genes: early expression of genes encoding axonemal proteins during the spermatogenic cycle and shared features of promoters of genes encoding central apparatus proteins. Mol Hum Reprod. 2005;11:307–17. doi: 10.1093/molehr/gah163. [DOI] [PubMed] [Google Scholar]
- 27.Turner KJ, Sharpe RM, Gaughan J, Millar MR, Foster PM, et al. Expression cloning of a rat testicular transcript abundant in germ cells, which contains two leucine zipper motifs. Biol Reprod. 1997;57:1223–32. doi: 10.1095/biolreprod57.5.1223. [DOI] [PubMed] [Google Scholar]
- 28.Zhao L, Yao C, Xing X, Jing T, Li P, et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat Commun. 2020;11:5683. doi: 10.1038/s41467-020-19414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariappa D, Aladakatti RH, Dasari SK, Sreekumar A, Wolkowicz M, et al. Inhibition of tyrosine phosphorylation of sperm flagellar proteins, outer dense fiber protein-2 and tektin-2, is associated with impaired motility during capacitation of hamster spermatozoa. Mol Reprod Dev. 2010;77:182–93. doi: 10.1002/mrd.21131. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Wei Y, Fu G, Li H, Saiyin H, et al. Tssk4 is essential for maintaining the structural integrity of sperm flagellum. Mol Hum Reprod. 2015;21:136–45. doi: 10.1093/molehr/gau097. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Li H, Fu G, Wang Y, Du S, et al. Testis-specific serine/threonine protein kinase 4 (Tssk4) phosphorylates Odf2 at Ser-76. Sci Rep. 2016;6:22861. doi: 10.1038/srep22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivkin E, Tres LL, Kierszenbaum AL. Genomic origin, processing and developmental expression of testicular outer dense fiber 2 (ODF2) transcripts and a novel nucleolar localization of ODF2 protein. Mol Reprod Dev. 2008;75:1591–606. doi: 10.1002/mrd.20911. [DOI] [PubMed] [Google Scholar]
- 33.Greenbaum MP, Yan W, Wu MH, Lin YN, Agno JE, et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci U S A. 2006;103:4982–7. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soung NK, Kang YH, Kim K, Kamijo K, Yoon H, et al. Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes. Mol Cell Biol. 2006;26:8316–35. doi: 10.1128/MCB.00671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soung NK, Park JE, Yu LR, Lee KH, Lee JM, et al. Plk1-dependent and -independent roles of an ODF2 splice variant, hCenexin1, at the centrosome of somatic cells. Dev Cell. 2009;16:539–50. doi: 10.1016/j.devcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang J, Seo SG, Lee KH, Nagashima K, Bang JK, et al. Essential role of Cenexin1, but not Odf2, in ciliogenesis. Cell Cycle. 2013;12:655–62. doi: 10.4161/cc.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat JL, et al. Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol. 2012;197:313–25. doi: 10.1083/jcb.201109148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, et al. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 2012;148:189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
- 39.Ito C, Akutsu H, Yao R, Yoshida K, Yamatoya K, et al. Odf2 haploinsufficiency causes a new type of decapitated and decaudated spermatozoa, Odf2-DDS, in mice. Sci Rep. 2019;9:14249. doi: 10.1038/s41598-019-50516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aktan TM, Montag M, Duman S, Gorkemli H, Rink K, et al. Use of a laser to detect viable but immotile spermatozoa. Andrologia. 2004;36:366–9. doi: 10.1111/j.1439-0272.2004.00636.x. [DOI] [PubMed] [Google Scholar]
- 41.Salmon NA, Pera RA, Xu EY. A gene trap knockout of the abundant sperm tail protein, outer dense fiber 2, results in preimplantation lethality. Genesis. 2006;44:515–22. doi: 10.1002/dvg.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted product sizes of primer pair for detecting GAPDH. Primer-BLAST tool provided by NCBI was employed to predict the product size of primer pair for detecting GAPDH. (a) Predicted product sizes of product generated from cDNA (primer pair specificity checked using Ref RNA as a searching database). The primer pair used for detecting GAPDH transcripts in our work was predicted to generate a specific product about 114 bp in size from cDNA; (b) Predicted product sizes of product generated from genomic DNA (primer pair specificity checked using genomes for selected organisms (primary reference assembly only) as a searching database). The primer pair used for detecting GAPDH transcripts in our work was predicted to generate a specific product about 218 bp in size from genomic DNA. Besides, it was also predicted to have the ability to generate additional products about 114 bp and 120 bp in size via mismatching. The specificity of all the other primers used in this work was checked in the same way before PCR was performed.
Aligning of sequencing results with Ref RNA database. Transcripts with 100% Query Cover and over 99% in percent identity (Per. Ident) shown in aligning results are listed in the table. Alignment illustrations show detailed alignment results of the sequencing results with typical transcripts. Primer sequences matched with reference transcripts are highlighted in green. Mismatched nucleotides in primers are highlighted in red. Altered nucleotides in primers are highlighted in blue. (a) Aligning of sequencing result of product Band I. Primer mismatching and altering were detected; (b) Aligning of sequencing result of product Band II; (c) Aligning of sequencing result of product Band III. Primer mismatching was detected; (d) Aligning of sequencing result of product Band IV. Two different fragments were cloned from product Band IV, one of which was the result of primer mismatching; (e) Aligning of sequencing result of product Band V; (f) Aligning of sequencing result of product Band VI. Primer mismatching and altering were detected; (g) Aligning of sequencing result of product Band VII; (h) Aligning of sequencing result of product Band VIII. Primer mismatching was detected; (i) Aligning of sequencing result of product Band IX; (j) Aligning of sequencing result of product Band X; (k) Aligning of sequencing result of product Band XI.
The results of the PCR amplification detecting each transcript variation in sperm. (a) Gel electrophoresis of PCR products. Water served as blank control. Genomic DNA was used as the comparison template to determine whether there was contamination of genomic DNA in the sperm cDNA specimen. ODF2 transcript variation 11 and 21 shared the primer pair for detecting, and ODF2 transcript variation 13 and 16 shared the primer pair for detecting. Only ODF2 transcript variation 10 and GAPDH were specifically detected to be expressed at a high level in sperm (indicated by the rectangle frames). Primer pair for detecting ODF2 transcript variation 4 generated a nonspecific product with strong band which was different from the expected product in size (the expected product should be 2060 bp in size). Some of the other primer pairs, such as those for detecting transcript variant 3 and transcript variant 6, generated products that matched expected products in size (indicated by the underlines), while the bands were pretty weak, indicating the low abundances of these transcripts expressed in sperm. Based on the result provided in Figure 3, it could be determined that transcript variations 9, 11, 12, 13, 14, 15, 17, 18, 19, 20, 21, and 22 were not expressed in sperm, and no expected products could be observed when primer pairs for detecting them were employed. However, due to the limited sequences that could be used for primer designing, the specificity of some primers was not satisfying. Many unexpected products could be observed. Besides, the low abundances of some transcripts made the bands unclear in the gel. It was unable to determine whether the other transcript variations were expressed in sperm from these results. Almost no product in PCR products generated from sperm cDNA could be detected in that generated from genomic DNA, determining that there was no contamination of genomic DNA in sperm cDNA specimen; (b) Aligning of sequencing result of the PCR product generated by primer pair for detecting ODF2 transcript variation 10. Matched primer sequences were highlighted in green. Only ODF2 transcript variation 10 matches the sequencing result perfectly.


