Abstract
Cross-reactive immunity occurs when infection with or vaccination against one virus protects against another related family member. A search for homologues of the HIV-1 envelope glycoprotein revealed that it is composed of thousands of intercalating and overlapping viral matches of pentapeptide or longer gapped consensi, belonging to over 70% of the currently sequenced virome, infecting all kingdoms from bacteria to man. It was also highly homologous to proteins from the Visna/Maedi and other ovine viruses, while other proteins (nef/tat/gag/pol) were homologous to proteins from the equine infectious anaemia virus and HTLV-2/HTLV-3 viruses. This phenomenon suggests that horizontal gene transfer from coinfecting RNA and DNA viruses to retroviruses is extensive, providing a route for the subsequent insertion of non-retroviral genes into human and other genomes via retroviral integration. This homology includes all viruses for which vaccines already exist. Cross-reactive immunity may be operative in AIDS, as Vaccinia vaccination decreases viral replication in HIV-1 infected patients’ cells, for the CCR5 tropic form. Measles, Dengue virus, or GB virus C infections also decrease the HIV-1 viral load. A resumption of Vaccinia/smallpox vaccination might be expected to have a significant effect on the AIDS pandemic, and a careful study of the potential uses of other existing viral and bacterial vaccines merits close attention. This phenomenon may also be relevant to other recalcitrant viruses, bacteria, and parasites for which no vaccine exists and the armory of existing vaccines may have a role to play in diseases other than those for which they were designed.
Keywords: AIDS, HIV-1, smallpox, vaccine, vaccinia
Introduction
The birth of immunology, over 200 years ago, noted that smallpox could be prevented by inoculation with cowpox,(1) a principle of immunity leading to the development of vaccines that have eliminated smallpox(2) and which combat many other viral and bacterial diseases. Many viruses are however, recalcitrant to vaccination, particularly the AIDS virus, HIV-1.(3) However it has recently been shown that Vaccinia virus vaccination reduces CCR5 tropic HIV-1 replication of the cells of infected patients.(4) In HIV-1 infected patients the viral load has also been reported to be reduced in patients infected with measles or Dengue fever.(5,6) The suppression of HIV-1 replication by measles infection is concurrent with intense immune activation.(7) It has also been shown that GB virus type C infection prolongs the survival of HIV-1 infected patients and that this effect is related to antibodies raised to the GB virus envelope protein, that cross-react with HIV-1 particles.(8) This latter effect suggests cross-reactive immunity. These apparent protective effects of other viral infections could also be related to a general activation of defense networks such as the protein kinase R or retinoic acid inducible gene (RIG-1) pathways leading to interferon production and the activation of antiviral signaling programs, although some viruses, including herpes simplex and influenza are able to subvert these and other pathways.(9,10)
If cross-reactive immunity is also involved in such effects, one would expect a degree of homology between HIV-1 and other viral proteins, within antigenic regions. In an attempt to find homologous viruses that might serve as the cowpox equivalent to HIV-1, the HIV-1 envelope glycoprotein (env) was compared to all other viral proteomes. Short contiguous amino acid stretches (pentapeptides or longer gapped sequences) belonging to proteins from almost the entire current virome are encased within the env protein and include those for which vaccines are available. These could perhaps play a role in the development of cross-reactive immunity to HIV-1.
Methods
B-cell epitopes for the HIV-1 env glycoprotein (P04578: Human immunodeficiency virus type 1 group M subtype B (isolate HXB2)) were retrieved from the BepiPred server(11) (http://www.cbs.dtu.dk/services/BepiPred/) and examples of immunogenic regions compared to all viral proteomes using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) server (BLASTp). This env sequence was derived from the reference env gene in the NCBI gene database (NC_001802.1). To detect small intraprotein consensi, the E value was set to 100,000. HIV-1, HIV-2, and other immunodeficiency viruses (Bovine, Feline, and Simian, the HIV-like cancer virus or the Aids-associated retrovirus, and the Murine AIDS virus-related provirus) were eliminated from the search due to evident homology. A list of available viral vaccines was obtained from the Center for disease control website at http://www.cdc.gov/vaccines/vpd-vac/vaccines-list.htm. BLASTS against these specific viruses (Table 1) were also undertaken. The env protein epitopes registered in the Immune epitope database (http://www.immuneepitope.org)(12) were also compared to these viruses. In some cases, the amino acid sequences of these env protein epitopes differed from that of the chosen example, due to viral strain differences. Viral matches (vatches) of five contiguous amino acids or more or longer gapped sequences were identified by eye and copied to a table in the appropriate position relative to the HIV-1 env amino acid sequence (Supplementary Table 1). The entire protein was not processed, but the many results illustrated the principles involved. To the author’s knowledge or ability, there is currently no way of automating this process (every single pentapeptide of the env glycoprotein tested shares similarity with several other viruses) and it is hoped that this illustration will stimulate work in this direction, which is also applicable to millions of vatches within the human proteome. Finally, the env protein from various HIV-1 strains was screened by BLAST analysis (BlastP) versus various Vaccinia viruses. The common peptides identified were analyzed for potential B cell immunogenicity using the Bepipred server. The server-set index of 0.35 was used as the immunogenicity index threshold.
Table 1. .
Examples of viral vatches within the HIV-1 envelope protein, for viruses where vaccines are available.
| Viruses and Alignments with the HIV-1 env protein All are within a predicted B cell epitope region (or within an experimentally described IEDB epitope) | ||||
|---|---|---|---|---|
| Chicken pox (Human Herpesvirus 3) 73: A +PTDP+ 108: IIS W+ 121: KLTP LC TL 142: SSSGR 214: PIHY APA 252: RP V +LL 276: NFT NA 305: KR R IG 307: KRIH and KR RI 313: P RAF+ PGRA 413: TITLP 500: KAKRRV 502: KRRVV 573: GI QLQ 574: IK QLQA 690: GGLVG |
Hepatitis A 38: VYYGV 477: +NWRS+ +KI 492: EPLGV 575: QL VLA 608: VP NAS |
Hepatitis B 39: Y VPV WK 74: CVPTD 75: VPTDP P 108: IS W SL and IISL 141: NSSSG 142: SSSGR 214: PIH CA 218: CAPA F 237: GPCT+ 252: RP V QL 253: PIVST 255: ST+LL 293: B +INCT 307: KR H PGRA 314: GR AFYT 359: QSS GD 362: KQSSG 441: GQ RCS S I 494: LGVAP 495: GVAPT 575: QL ARV 583: VE YLKD ++L LG GC KL+C 584: E YLKD 607: AV WNA and AV WN S 678: W LW +I IF 685: FI++V 690: GGL GL 708: VRQ YS LS 712: YSPLS 802: YW QEL 803: W QELK |
Influenza A virus ( many different strains) 32: DT+VHN 42: VPVW 108: I SLWDQ and IISLW 110: SLWDQ 121: K T PLCV L 124: PLCV L 125: LCVTL 131: CTDLK 139: NTNSS 142: SSSGR 167: GKVQK 205: CPKVS 207: KI FEP IP and KI SFE IP 214: P+HYC 234: NGTGP 252: R IVSTQ and RPIV Q 254: IVSTQ 263: GSLAE+ 294: INCTR 298: RPNNN 302: NNTRK and NYNK KRI I and NYNKR 303: YNK KR 304: TRKRI 305: KRIRI and KR R+ I PG and KRKR 312: GPGR F+ and GPG F+ 349: LREQF 350: REQFG 356: NKTII 357: KTIIF 358: TIIFK 362: KQSSG 364: SSGGD 369: PEIVT 370: EIVTH 371: IVTHS 372: VTHSF 373: THSFN 405: SNNTE 407: NTEGS 412: DTITL 445: CSSNI 490: KIEPL 493: PLGVA 571: VWGI AR 573: QL ARV 576: L+ARVL 605: TTAVP 608: +PW NASW 679: LWYI K F and LW IKI 685: FI+ IV GLV 686: IMIV 687: MIV G V L and MI+GG 710: QG YS LSFQ 711: GYS LSF 725: RGPDR 730: PEG+EE 744: RDRSI 805: +ELKN 806: ELKNS and ELK+ AV 807: LKNS V 825: G DRVI 828: RV E LQR and RV E+LQR 830: IEVLQ |
Japanese encephalitis virus 59: K YDTE 110: SLWD 240: T++STV 241: NVSTV 306: RKRI+ 497: APTKA 526: AGST+G A S TL R 576: L ARV Y LK 688: IVGGL L I 690: GGLV 742: RDRSI 829: VIEVL R and VIE VLQR 830: IEVLQR |
| Measles virus (repeat motifs in Bold) 55: ASDAKA 109: ISL WD SL 110: SLWD 252: RPI S QL 253: PI S QL 263: GSLA EE 304: NKRK 308: RIH IGPG 312: GPGRA 314: GRAF T 401: STEGS 493: PLGVA 573: IKQL QA V 581: LAVE LK 647: E SQ+QQ+ 685: F+M LVGL 688: IVGG V 706: NRVRQ 707: RVRQG 715: L F+ LPTPR 716: NRVRQ 719: THLPT 721: LPTPR 732: GI EE+G + R DRDR 801: +Y SQEL 828: R EVVQ |
Mumps virus 112: W DQSL 135: N NT SSS 140: NT SSS 254: I STQL 305: KRI IG 443: QI CS NI 493: PLGVA 573: IK QLQA 577: QAR LA 688: IV GLV 823:AEG RVI |
Papillomavirus(several strains) 35: WV V YGV 36: VTV PV 58: AYDT+ 60: AY+T HN+ 70: ATHAC 74: CVPTD P P 77: TDPNP 82: QE+VLV 142: SSSGR 208: ISF+P 218: CAPA F 234: NGTGP 238: PCTNV 239: CT N STVQC 240: TNVST 253: PI STQ 264: SLAEE 300: NNNTR 301: NNTRK 302: NTRKR 305: KR RI I and KRKR+ 349: LREQF 363: QSSGG 364: SSGGD 401: STEGS 406: NNTEG 407: NTEGS 410: GSDTI 413: TITLP 440: SGQIR 493: PLGVA 497: APTKA 500: KAKRR 502: KRRVV 570: VWGIK L+ 574: KQLQ 576: LQ VLA 605: TTAVP 633: REIN Y S 635: I+NYTS 644: SLIEES 658: QELLE 688: IVGG 690: GG+VG 721: LPTP GP 730: PEG ++EGG 731: EG+EEE 734: EEEG E +R D S R 799: LL W QEL 806: ELKNS V 807: LKNSA 820: IAVAE D IE 828: R IEVL |
Poliovirus (1,2 and 3) 36: V VYYG 252: RP + TQ 255: VSTQ 314: GRA YT 336: AKW++ and AK NN 529: TMGAA 531: GAAS+ 836: AC I IP IRQG |
Human Rotavirus A 63: TEVHN 121: KL LCV 133: DLKND 208: ISF P +Y 252: A R I VSTQ 303: YNKR 307: KRIH and KR RI 308: A RIHI 313: PG AF+ 337: KW +TL 369: PEIVT 413: TITLP 576: LQ V L VE YLK 583: VER L D QLL I G and VE YLK and VE Y+K 686: IM V GL V L 691: GLVG+ 822: VAEGT+ 829: VIEVL 831: EV LQRA |
| Rabies virus 122: L LC+TL 218: CAPA F 220: PAG AI 337: KWNN 354: GNNKT 584: ER+LK 687: MI GGL L 800: LQ WSQ 805: QELKN |
Rubella 70: AT ACV PTD 738: GGE DR 825: G DRV+ V Q |
Vaccinia virus or Vaccinia virus Tian Tan 34: K W+TV 35: LW YYGV 36: VT+ Y GVPV 37: T+YYG 55: V LNAT IA 57: DAKAY 82: Q VVLV 108: II LW+ |
Yellow fever virus 108: IIS DQ 131: KC L D SSS 231: KTF GTG CT 259: LLLN E+ SV T N T I + S E NC PN R 276: N D KTI V L T P 354: GNNKT 417: PCRI I+ 484: YK KVVK+ L AP KA V+ R+ R G 632: + EI NY S H |
|
| |
|
112: W DQSL 122: LTPL V 143: SSGRM 153: EIKNC 267: EEEVV 276: NFTD A 312: GPGR 313: A PG AF+ 355: NNKTI 359: IIFKQ 360: IFKQS 524: GAAGST 577: QARV AV 579: RVLA+ R and RV AVE and RVL AV RY 587: L +QQ LL 635: I NYTS 645: LI EE +QE E Q L+E 646: +EE N+++ K EQELL 712: YSPLS 714: P SFQT 719: TH P T + PE I 720: HL TP GP 741: DR+R IR 804: SQELK 807: LK+SA+ |
687: M VGG V L 691: GLVG 826: TDR IE V+G 842: H RIR GL+ |
|
| Viruses that modulate HIV-1 infection (for measles see above) | ||||
| Dengue virus (1, 2 or 3) 36: VT Y+GV H and VT Y GV V WK 108: IIS DQ 208: I EPIP 242: VSTVQ 252: RP V LL 254: IVST LL 256: STQLL 264: SL EE+ 264: SLAEE 265: LAE EV 302: NTRKR 302: NKRKR 313: PGR TT 335: RAK NT 348: ESQ +QE 363: QSSGG 402: TEGSN 523: LG GSTM 570: VWGI AR 580: VLAVE 606: TA PWN 685: F +VGG VG 686: IM V GLV L 691: VGGL 722: PTPRG |
|
GB virus C 70: ATHAC D P+++ 122: LTPL CV 140: T SSSG EK 209: SFE IP 213: IPI AG A 252: RP+VS 253: PIVS 306: RKR + PG 468: FR GGG D W 521: FLG T A LT L G V 523: LGAAG TM A M 531: GA S+T T QA 574: K L ARVL 581: LAVE LK 582: AVE LK 610: VPW AS 686: IMI GL 689: VG LVG and VG L GL 729: R G GER DR 738: GER IRLV 807: LK VSLLNA 828: RVIE 830: IE QRA 831: EVL RA |
|
|
| Position within env protein |
Epitope from IEDB |
Type |
||
| 33 | KLWVTVYYGV | MHC binding | ||
| 36 | VTVYYGVPVWK | T cell/MHC binding | ||
| 108 | IISLWDQSL | MHC binding | ||
| 121 | KLTPLCVTL | T cell/MHC binding | ||
| 206 | PKISFEPIPIHYCAPAGFA | MHC binding | ||
| 252 | RPIVSTQLL | MHC binding | ||
| 302 | NYNKRKRIHIGPGRAFYTTKNII | B cell | ||
| 311 | IGPGRAFHT | T cell | ||
| 312 | GPGRAFYTT | MHC binding | ||
| 335 | RAKWNNTLK | MHC binding | ||
| 570 | VWGIKQLQARVLAVERYLKD | MHC binding | ||
| 606 | TAVPWNASW | MHC binding | ||
| 678 | WLWYIKIFI | MHC binding | ||
| 685 | FIMIVGGLV | MHC binding | ||
| 686 | IMIVGGLVGL | MHC binding | ||
| 799 | LLQYWSQEL | MHC binding | ||
| 828 | RVIEVLQRA | MHC binding | ||
Their start position (within the env protein of 856 amino acids) is marked as is their position with respect to predicted B-cell epitopes within the env protein (these are all within regions with an antigenicity index of greater than the server-set threshold of 0.35: see supplementary Table 1). Spaces within the sequences indicate nonidentical amino acids and + signs an amino acid with similar physicochemical properties. The gray shaded sequences are within sequences that have been described as epitopes in experimental studies (B cell, T cell, or MHC binding from IEDB: The amino acid sequences of these experimentally verified epitopes are appended at the bottom of the table). Note that these sequences often overlap within consecutive regions of the env protein. In the majority of cases shown, contiguous sequences were of pentapeptides, although longer gapped sequences are also illustrated.
Results
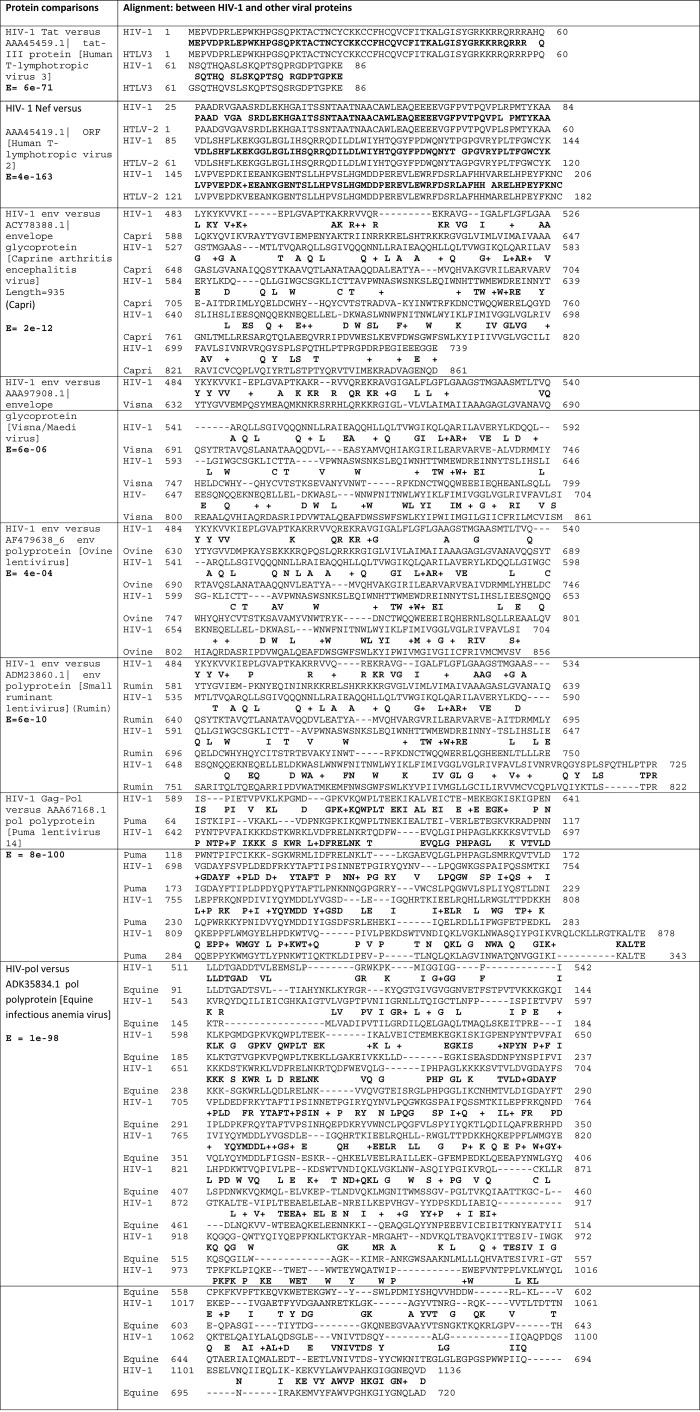
The env glycoprotein shows significant overall homology with proteins from four other viruses, the env proteins of the Caprine arthritis encephalitis virus (E = 3e-12), the small ruminant lentivirus (E = 6e-10), the visna/maedi virus (E = 6e-06) and the ovine lentivirus (E = 4e-04) (Figure 1). The HIV-1 nef protein showed significant overall homology with an ORF protein from HTLV-2 (E = 2e-45), while the HIV-1 tat protein showed significant overall homology with the HTLV-3 tat protein (E = 1e-35) (not shown). The HIV-1 gag protein is highly homologous to a protein from the puma lentivirus (E = 1E-98) and to a gag protein from the equine infectious anaemia virus (E = 3e-42) (Figure 1).
Figure 1. .
Significant overall homologies of HIV-1 viral proteins with proteins from other viruses. Consensi and e values are shown in bold.
The results in relation to other viruses are shown in supplementary Table 1, where viral vatches are aligned with the env sequence, which is also characterized in relation to the B cell epitope index. Even though only ∼70% of the env protein was processed, HIV-1 vatches were observed in 1827 RNA and DNA viruses and phages, known to infect all kingdoms from bacteria to man. These were majoritarily species rather than strains. At the time of writing, there are 3753 reference sequences for 2565 viral genomes in the NCBI Entrez Genomes database, and the viruses containing HIV-1 env sequences account for ∼72% of the known current virome. Examples of such alignments, for viruses where vaccines are available, are shown in Table 1. All of these viruses contain HIV-1 vatches in both B cell epitope and non-epitope regions and within epitopes that have been experimentally verified.
A BLAST analysis of the env protein from several HIV-1 viral strains compared with Vaccinia viruses revealed a further layer of complexity. While certain identical Vaccinia/HIV-1 sequences were maintained across several HIV-1 viral strains, for example, the hexapeptides GAAGST or VVKIEP, these were often in differing positions of the env protein (e.g. GAAGST at positions 386, 510, 512, 524,529, or 531). Otherwise, the profile of matching peptides derived from this sweep appears to be distinct for each strain of the HIV-1 virus. The viral matches shown in Tables 1 and 2 were predominantly pentapeptides, but longer contiguous or gapped sequences as well as frequent tetrapeptides were also observed (see supplementary Table 1).
Table 2. .
Examples of Vaccinia virus homologues (from BLASTS of the relevant HIV-1 env proteins versus Ankara, GLV-1h68, Tian Tian, and L-IPV Vaccinia strains) compared with the env glycoprotein from a selection of HIV-1 viral strains (various subtypes from groups M, N, and O).
| Group M subtype A (isolate Z321) (Accession # P05881) | Group M subtype B (isolate BRU/LAI) (Accession # P03377) | Group M subtype B (isolate BH10)(Accession # P03375) | Group M subtype C (isolate ETH2220) (Accession # Q75008) |
|---|---|---|---|
| 340: DTLSKV 388: TSGLF 524: GAAGST 488: VVKIEP 617: KSQSD 644: NLIEE 702:LL LSIIN 703: SIINR 841: LNIPR |
187: CSFNIS S 529: GAAGST 303: KLDII ID 493: VVKIEP 705: A LSIVN |
524: GAAGST |
154: CSFNI 391: LELFN 429: GIIMC 512: GAAGST 574: HLRDQ 629: IIYNL 690: LSIVN 685: IIFAV |
| Group M subtype D (isolate Z84) (Accession # P05882) |
Group M subtype F1 (isolate VI850) (Accession # Q9QSQ7) |
Group M subtype G (isolate 92NG083) (Accession # O41803) |
Group M subtype H (isolate 90CF056) (Accession #O70902) |
| 63:EAHNI 197: NTNYT Y 291: NNVKTII 364: LNQTT 495: VV IEP 531: GAAGST |
377: TSGLF 386: SNNGT 678: LSIVN 696: LIPSP 753: IAARI 768: ALKYL 771: YLGNL 817: LNIPR |
329: NVSRI 352: NKNIT 383: TSGLF 392: SNINN 473: KTVK+K 510: GAAGST 688: LSIVN |
661: WFDIS 690: LSIVN 752: LSLFS |
| Group M subtype J (isolate SE9173) (Accession # Q9WC69) |
Group M subtype K (isolate 96CM-MP535) (Accession # Q9QBY2) |
Group N (isolate YBF106) (Accession # Q9IDV2) |
Group O (isolate ANT70)(Accession #Q77377) |
| 146: SPEIM N 180: INSDN 194: TSVIK 482: VVELEP 615: DIWEN 691: IIFAV |
187: NNSST 448: NTHNE 510: GAAGST 732: VRLVS 797: AISLL |
74: LLTNV 79: TEYFN 134: +RTEDL 156: RDRKK 250: QLILN 476: VSREK 513: RTLLS 787: LKDSAI |
254: QLILN 544: HTLLK 696: RVIMI 698: IMIVL 704: IVKNIR+G |
Identical peptides (HIV-1 = Vaccinia) were analyzed for B cell antigenicity using the BepiPred server and those predicted as epitopes are highlighted in bold.
Discussion
The close homology of the env, nef, tat, and gag/pol proteins with caprine, ovine, visna/Maedi, equine, and small ruminant viruses and particularly with HTLV-2 and HTLV-3 is of evolutionary interest as it suggests a source of the AIDS virus and its relatives, prior to simian integration and passage to man. However this is not the subject of this article.
In terms of cross-reactive immunity, no vaccines for HTLV-2 or HTLV-3 yet exist,(13) although interestingly, HTLV-2 infection appears to have a protective influence on HIV-1 infection.(14) Should HTLV vaccines be developed, they may also have a role to play in relation to HIV-1.
As regards the shorter contiguous sequences and matches, the extensive homology of a single HIV-1 protein (env) with numerous phage and viral proteins (∼72% of the currently sequenced virome) suggests that horizontal partial gene transfer from coinfecting DNA and RNA viruses to retrovirus, and/or vice versa, has proceeded on a massive scale during the evolutionary history of the AIDS virus and its ancestors. These include sequences from viruses infecting all kingdoms (e.g. bacteria, amoeba, fungi, plants, molluscs, insects, invertebrates, fish, birds, reptiles, and mammals) suggesting that these have at some time hosted the HIV-1 virus or its ancestors, along with other viruses, whose partial gene sequences have somehow been incorporated into the HIV-1 viral genome. There is no reason to suppose that this is not a feature of other retroviruses. As such sequences can subsequently be transferred to other genomes via retroviral insertion, this may partly explain the presence of phage and viral partial gene sequences within the genomes of plants, arthropods, fungi, nematodes, protozoa,(15) mammals and man.(16–18) The human proteome also contains multiple peptide consensi from bacterial, plant, and animal viruses.(19)
Horizontal gene transfer from virus to retrovirus does not appear to have been specifically studied in the laboratory. However, gene exchange is common between viruses,(20,21) and also between retroviruses(22) where, for example, recombination can lead to the development of novel HIV-1 viral strains.(23) However, horizontal gene transfer has been reported from phages to bacteria,(24) between bacteria,(25) or from man to bacteria(26) and indeed appears to be a common feature of all living matter.(27) The acquisition of genomic DNA or RNA from infected higher species, by viruses, has also been proposed as a driving force in the evolution of viruses in general.(28) Plant, arthropod, fungal, nematode, and protozoan(15) as well as animal and human genomes also contain multiple retroviral and non-retroviral sequences.(16,18) Clearly, this provides many potential routes for an interviral melange of genomic material. The direction or route of transfer cannot be imputed from a simple bioinformatics alignment, and the reasons for this homology require further laboratory testing. Again this evolutionary aspect is not the central theme of this analysis, and does not alter the implications ensuing from this homology.
All of the viruses for which vaccines are available, or which are known to favorably modulate HIV-1 viral load (Vaccinia, Dengue viruses, GB virus C, and measles) contain sequences matching those of the env protein. It is not possible to predict whether any particular sequence would potentially create cross-reactive anti-HIV-1 antibodies, but the Vaccinia virus as well as Dengue viruses, measles, and GB virus C contain several vatches in B cell epitope regions of the env protein. Field work is necessary to define whether any of these epitopes are able to modify HIV-1 infection. In addition, theoretical T cell epitopes were not examined and are likely to reveal a yet more complex picture that may also depend upon the HLA genetic composition of the host. However, many of the matching sequences are within epitopes known to be able to label the AIDS virus in experimental studies, as cataloged by the immune epitope database. In addition, it is unlikely that all possible epitopes have been reported or characterized. While many of the viral matches were of pentapeptides or greater, multiple tetrapeptide matches were also observed. Antibodies are quite capable of recognizing such short sequences(29) A further point to be considered is that this homology may enable different viruses to share the same binding partners in relation to the host proteome. Viruses also demonstrate this type of homology with human proteins,(18,30,31) an ability that no doubt enables them to compete with their human counterparts as binding partners in the numerous host/pathogen interactomes that they use during their life cycles.(17,19) HIV-1 and pox viruses both use the CCR5 chemokine receptor and such sharing may also influence the outcome of co-infection.(4)
The differing matching peptide profiles for different HIV-1 viral strains also highlights the underlying complexity and shows that matching sequences will depend upon the HIV-1 strain, and presumably also the strain of the homologous virus.
In relation to AIDS, this homology may have clinical application as infection or vaccination in relation to these viral homologues might be expected, in some cases, to confer cross-reactive immunity to HIV-1. There is indeed some evidence that this may be operative. For example, Vaccinia vaccination in HIV-1 infected subjects has been shown to inhibit HIV-1 viral replication in subsequent in vitro tests, but only in the CCR5 tropic HIV-1 Major M strain.(4)These authors noted that the increase in the incidence of AIDS correlated with the successful eradication of smallpox and cessation of the use of Vaccinia vaccination. However, even within the M group of HIV-1 viruses, which displays tropism for the CCR5 chemokine receptor,(32) considerable variation exists between the Vaccinia/HIV-1 peptide matches.
In a small study (four patients), hyperimmunization with the killed poliomyelitis (Salk) vaccine was also shown to increase the T cell count and to improve symptoms in HIV-1 infected patients.(33) Influenza vaccination in non-HIV-1 patients also results in the suppression of HIV-1 replication in vitro. However, this was not observed in HIV-1 infected patients, and influenza vaccination has also been reported to increase HIV-1 replication in some patients,(34) perhaps due to the ability of the influenza virus to inhibit viral defense pathways.(10) Both measles or GB virus C infection are also known to decrease the HIV-1 viral load in infected patients(7,8) although other coinfections may perhaps worsen the effects of each other.
Vaccination can be a double-edged sword. For example, a lower titre of hepatitis B antibodies has been observed in several autoimmune disorders, including multiple sclerosis, suggesting a protective effect of infection.(35) However, hepatitis B vaccination can have the opposite effect and provoke demyelinating lesions in certain cases.(36) It has been shown that the HIV-1 proteome displays a similar type of homology with the human proteome and the problems of autoimmunity in relation to certain of these vaccines need to be addressed.(31) Nevertheless, Vaccinia virus vaccination does reduce the HIV-1 viral load for the common CCR5 tropic strain, in vitro, and a resumption of smallpox vaccination might be expected to be of benefit in certain cases, as already suggested(4) It would be premature to suggest the immediate use of other available vaccines as preventive agents without further research into the question. A more in-depth analysis of the viral homology of the env glycoprotein and of other HIV-1 proteins and strains is also necessary, and, given the scale of the phenomenon, which also applies to millions of viral/human and bacterial/human short consensi.(17,37) It is clear that the development of powerful algorithms is necessary for this purpose. However, the results with the Vaccinia virus are promising and suggest that this homology may be harnessed to good effect. Whether other available vaccines could confer cross-reactive immunity remains to be assessed. These are often used in HIV-1 positive patients, once HIV-1 is present,(38) but their use as potential preventive agents, given prior to HIV-1 infection, merits further study.
A further point to consider is the microbiome in AIDS patients. If so many viruses, and probably also bacteria and other pathogens, resemble HIV-1 viral proteins, it is possible that certain species could exert beneficial (or deleterious) effects. Sequencing of the various microbiota in AIDS resistant and nonresistant patients may, thus, be of value as such analyses may well be able to identify protective viral or bacterial strains. Microbiomes can exert a powerful influence on disease. For example, the intestinal microbiome is able to influence obesity, cardiovascular disease, and inflammatory bowel disease,(39) and its manipulation in relation to HIV-1 is already attracting attention.(40) Indeed, probiotic yoghurt containing Lactobacillus rhamnosus Fiti is able to increase the CD4+ cell count in HIV-1 infected patients.(41)
HIV-1 vaccine development using attenuated Ankara Vaccinia strains, containing HIV-1 proteins, is already under development.(42) The most immediately relevant conclusion of this study is that the beneficial effects of unmodified Vaccinia vaccination in HIV-1 infected patients, in vitro, may well be related to cross-reactive immunity due to Vaccinia/HIV-1 homology, and that, as previously suggested(4) a resumption of Vaccinia/smallpox vaccination might have a significant effect on the AIDS pandemic, even if only effective against certain strains.
Clearly further work is needed, both in vitro and in vivo to analyze these effects. Rather than suggest specific proposals for vaccine development or the use of already available vaccines, the main purpose of this article is to draw attention to this extensive protein homology, which may have far-reaching implications in this and other diseases. A similar bioinformatics approach may be relevant to other recalcitrant viruses, bacteria, and pathogens and the current treasury of available vaccines may well find uses in diseases other than those for which they were designed. Other HIV-1 proteins and numerous strains of both the HIV-1 and other viruses also require analysis perhaps enabling the construction of more effective epitopes. The wheel has turned full circle since Edward Jenner’s observation over 200 years ago that cowpox prevented smallpox, as, if this is effective, the same phenomenon and the same viruses may have a role to play in relation to today’s viral scourge.
Declaration of interest
The author declares no conflict of interest.
References
- 1.Jenner E. (1798). An inquiry into the causes and effects of the Variolæ Vaccinæ, or cow-pox. London. [Google Scholar]
- 2.Arita, I. Farewell to smallpox vaccination. Dev. Biol. Stand. 1979, 43, 283–296. [PubMed] [Google Scholar]
- 3.Bulatao RA. Population and mortality after AIDS In: Disease and Mortality in Sub-Saharan Africa. 2nd edition. Washington (DC): World Bank. Jamison DT, Feachem RG, Makgoba MW, Bos ER, Baingana FK, Hofman KJ, Rogo KO, eds. [PubMed] [Google Scholar]
- 4.Weinstein, R.S., Weinstein, M.M., Alibek, K., Bukrinsky, M.I., Brichacek, B.. Significantly reduced CCR5-tropic HIV-1 replication in vitro in cells from subjects previously immunized with Vaccinia Virus. BMC Immunol. 2010, 11, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moss, W.J., Scott, S., Ndhlovu, Z., Monze, M., Cutts, F.T., Quinn, T.C., Griffin, D.E.. Suppression of human immunodeficiency virus type 1 viral load during acute measles. Pediatr. Infect. Dis. J. 2009, 28, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watt, G., Kantipong, P., Jongsakul, K.. Decrease in human immunodeficiency virus type 1 load during acute dengue fever. Clin. Infect. Dis. 2003, 36, 1067–1069. [DOI] [PubMed] [Google Scholar]
- 7.Moss, W.J., Ryon, J.J., Monze, M., Cutts, F., Quinn, T.C., Griffin, D.E.. Suppression of human immunodeficiency virus replication during acute measles. J. Infect. Dis. 2002, 185, 1035–1042. [DOI] [PubMed] [Google Scholar]
- 8.Mohr, E.L., Xiang, J., McLinden, J.H., Kaufman, T.M., Chang, Q., Montefiori, D.C., Klinzman, D., Stapleton, J.T.. GB virus type C envelope protein E2 elicits antibodies that react with a cellular antigen on HIV-1 particles and neutralize diverse HIV-1 isolates. J. Immunol. 2010, 185, 4496–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters, G.A., Khoo, D., Mohr, I., Sen, G.C.. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 2002, 76, 11054–11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff, T., Ludwig, S.. Influenza viruses control the vertebrate type I interferon system: factors, mechanisms, and consequences. J. Interferon Cytokine Res. 2009, 29, 549–557. [DOI] [PubMed] [Google Scholar]
- 11.Larsen, J.E., Lund, O., Nielsen, M.. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathiamurthy, M., Peters, B., Bui, H.H., Sidney, J., Mokili, J., Wilson, S.S., Fleri, W., McGuinness, D.L., Bourne, P.E., Sette, A.. An ontology for immune epitopes: application to the design of a broad scope database of immune reactivities. Immunome Res. 2005, 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carneiro-Proietti, A.B., Catalan-Soares, B.C., Castro-Costa, C.M., Murphy, E.L., Sabino, E.C., Hisada, M., Galvão-Castro, B., Alcantara, L.C., Remondegui, C., Verdonck, K., Proietti, F.A.. HTLV in the Americas: challenges and perspectives. Rev. Panam. Salud Publica 2006, 19, 44–53. [DOI] [PubMed] [Google Scholar]
- 14.Brites, C., Sampalo, J., Oliveira, A.. HIV/human T-cell lymphotropic virus coinfection revisited: impact on AIDS progression. AIDS Rev. 2009, 11, 8–16. [PubMed] [Google Scholar]
- 15.Liu, H., Fu, Y., Jiang, D., Li, G., Xie, J., Cheng, J., Peng, Y., Ghabrial, S.A., Yi, X.. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol. 2010, 84, 11876–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzourakis, A., Gifford, R.J.. Endogenous viral elements in animal genomes. Plos Genet 2010, 6(11), e1001191. doi:10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter, C.J. Alzheimer’s disease: a pathogenetic autoimmune disorder caused by herpes simplex in a gene-dependent manner. Int. J. Alzheimers. Dis. 2010, 2010, 140539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanduc, D., Stufano, A., Lucchese, G., Kusalik, A.. Massive peptide sharing between viral and human proteomes. Peptides 2008, 29, 1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter, C.J. Epstein-Barr and other viral mimicry of autoantigens, myelin and vitamin D related proteins, and of EIF2B, the cause of vanishing white matter disease: Massive mimicry of multiple sclerosis relevant proteins by the Synechococcus phage. Immunopharmacol Immunotoxicol (in press) 2011 Apr 12. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 20.Alejska, M., Kurzyniska-Kokorniak, A., Broda, M., Kierzek, R., Figlerowicz, M.. How RNA viruses exchange their genetic material. Acta Biochim. Pol. 2001, 48, 391–407. [PubMed] [Google Scholar]
- 21.Filée, J., Chandler, M.. Gene exchange and the origin of giant viruses. Intervirology 2010, 53, 354–361. [DOI] [PubMed] [Google Scholar]
- 22.McClure, M.A., Johnson, M.S., Doolittle, R.F.. Relocation of a protease-like gene segment between two retroviruses. Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 2693–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onafuwa-Nuga, A., Telesnitsky, A.. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009, 73, 451–80, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent, B.N., Salichos, L., Gibbons, J.G., Rokas, A., Newton, I.L., Clark, M.E., Bordenstein, S.R.. Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol. Evol. 2011, 3, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDaniel, L.D., Young, E., Delaney, J., Ruhnau, F., Ritchie, K.B., Paul, J.H.. High frequency of horizontal gene transfer in the oceans. Science 2010, 330, 50. [DOI] [PubMed] [Google Scholar]
- 26.Anderson, M.T., Seifert, H.S.. Opportunity and means: Horizontal gene transfer from the human host to a bacterial pathogen. MBio 2011, 2(1):e00005–11. doi: 10.1128/mBio.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinkovics, J.G. Horizontal Gene Transfers with or without Cell Fusions in All Categories of the Living Matter. Adv. Exp. Med. Biol. 2011, 714, 5–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker, Y. Evolution of viruses by acquisition of cellular RNA or DNA nucleotide sequences and genes: an introduction. Virus Genes 2000, 21, 7–12. [PubMed] [Google Scholar]
- 29.Agarwal, A., Sarkar, S., Nazabal, C., Balasundaram, G., Rao, K.V.. B cell responses to a peptide epitope. I. The cellular basis for restricted recognition. J. Immunol. 1996, 157, 2779–2788. [PubMed] [Google Scholar]
- 30.Kanduc, D. Describing the hexapeptide identity platform between the influenza A H5N1 and Homo sapiens proteomes. Biologics 2010, 4, 245–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucchese, G., Stufano, A., Calabro, M., Kanduc, D.. Charting the peptide crossreactome between HIV-1 and the human proteome. Front. Biosci. (Elite Ed). 2011, 3, 1385–1400. [DOI] [PubMed] [Google Scholar]
- 32.Coakley, E., Petropoulos, C.J., Whitcomb, J.M.. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 2005, 18, 9–15. [DOI] [PubMed] [Google Scholar]
- 33.Pitts, F.N. Jr, Allen, R.E., Haraszti, J.S., Allen, A.D.. Improvement of four patients with HIV-1 related symptoms after hyperimmunization with killed poliomyelitis (Salk) vaccine. Clin. Immunol. Immunopathol. 1988, 46, 167–168. [DOI] [PubMed] [Google Scholar]
- 34.Pinto, L.A., Blazevic, V., Anderson, S.A., Venzon, D.J., Trubey, C.M., Rowe, T., Katz, J.M., Liewehr, D., Dolan, M.J., Shearer, G.M.. Influenza virus-stimulated generation of anti-human immunodeficiency virus (HIV) activity after influenza vaccination in HIV-infected individuals and healthy control subjects. J. Infect. Dis. 2001, 183, 1000–1008. [DOI] [PubMed] [Google Scholar]
- 35.Ram, M., Anaya, J.M., Barzilai, O., Izhaky, D., Porat Katz, B.S., Blank, M., Shoenfeld, Y.. The putative protective role of hepatitis B virus (HBV) infection from autoimmune disorders. Autoimmun. Rev. 2008, 7, 621–625. [DOI] [PubMed] [Google Scholar]
- 36.Mikaeloff, Y., Caridade, G., Suissa, S., Tardieu, M.. Hepatitis B vaccine and the risk of CNS inflammatory demyelination in childhood. Neurology 2009, 72, 873–880. [DOI] [PubMed] [Google Scholar]
- 37.Trost, B., Lucchese, G., Stufano, A., Bickis, M., Kusalik, A., Kanduc, D.. No human protein is exempt from bacterial motifs. Not even one. Self Nonself 2011, 1, 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horster, S., Laubender, R.P., Lehmeyer, L., Ankerst, D.P., Eberle, J., Reinert, R., Imöhl, M., van der Linden, M., Schweiger, B., Bogner, J.R.. Influence of antiretroviral therapy on immunogenicity of simultaneous vaccinations against influenza, pneumococcal disease and hepatitis A and B in human immunodeficiency virus positive individuals. J. Infect. 2010, 61, 484–491. [DOI] [PubMed] [Google Scholar]
- 39.Kinross, J.M., Darzi, A.W., Nicholson, J.K.. Gut microbiome-host interactions in health and disease. Genome Med. 2011, 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hummelen, R., Vos, A.P., van’t Land, B., van Norren, K., Reid, G.. Altered host-microbe interaction in HIV: a target for intervention with pro- and prebiotics. Int. Rev. Immunol. 2010, 29, 485–513. [DOI] [PubMed] [Google Scholar]
- 41.Irvine, S.L., Hummelen, R., Hekmat, S., Looman, C.W., Habbema, J.D., Reid, G.. Probiotic yogurt consumption is associated with an increase of CD4 count among people living with HIV/AIDS. J. Clin. Gastroenterol. 2010, 44, e201–e205. [DOI] [PubMed] [Google Scholar]
- 42.Watkins, D.I. HIV vaccine development. Top. HIV Med. 2010, 18, 35–36. [PubMed] [Google Scholar]