Abstract
Smallpox was one of the most devastating diseases in the human history and still represents a serious menace today due to its potential use by bioterrorists. Considering this threat and the non-existence of effective chemotherapy, we propose the enzyme thymidylate kinase from Variola virus (VarTMPK) as a potential target to the drug design against smallpox. We first built a homology model for VarTMPK and performed molecular docking studies on it in order to investigate the interactions with inhibitors of Vaccinia virus TMPK (VacTMPK). Subsequently, molecular dynamics (MD) simulations of these compounds inside VarTMPK and human TMPK (HssTMPK) were carried out in order to select the most promising and selective compounds as leads for the design of potential VarTMPK inhibitors. Results of the docking and MD simulations corroborated to each other, suggesting selectivity towards VarTMPK and, also, a good correlation with the experimental data.
Keywords: smallpox, Variola virus, thymidylate kinase, docking, molecular dynamics
1. Introduction
Smallpox is an acute disease, highly contagious, and exclusive of human beings. It is caused by two variants of viruses from the genera Orthopoxvirus known as Variola major and Variola minor and presents lethality around 30% (Behbehani, 1983; Gileva et al., 2006; Hendrickson, Wang, Hatcher, & Lefkowitz, 2010; Liszewski et al., 2006; Ryan & Ray, 2004; Sakhatskyy et al., 2008). As one of the most devastating diseases affecting human kind, smallpox had been present in all continents for centuries, decimating populations in several pandemics, and changing the course of history (Chapman, Nichols, Martinez, & Raymond, 2010; Hammarlund et al., 2010; Kennedy, Ovsyannikova, Jacobson, & Poland, 2009; Kennedy, Ovsyannikova, & Poland, 2009). Despite declared eradicated by the World Health Organization (WHO) in 1980, there are no guaranties that this disease would not come back in future because orthopoxvirus are capable of surviving for decades crystallized in nature at low temperature and humidity (Harper, 1961; Huq, 1976). Besides, due to the interruption of the vaccination programs, the world population is now more susceptible to infections with orthopoxvirus. This is a serious menace to the public health either by the risk of natural outbreaks, like the monkeypox and cowpox episodes occurred in Germany and France in 2002 and 2009 respectively, or by the use of smallpox as a biological weapon by the terrorist groups (Alzhanova & Früh, 2010; Campe et al., 2009; Handley, Buller, Frey, Bellone, & Parker, 2009; Johnston et al., 2012; Kennedy, Ovsyannikova, Jacobson, et al., 2009; Mätz-Rensing et al., 2006; Ninove et al., 2009; Schmiedeknecht et al., 2010).
Soon after the eradication of smallpox, all countries members of WHO agreed in destroying eventual strains or sending it to the two unique laboratories in the world accredited to keep strains of these viruses: the Center of Disease Control in Atlanta, USA, and the Institute of Viruses Preparations in Moscow, Russia. The remaining strains in these labs should have been destroyed by January of 2002 but the fear of smallpox being used as a biological agent has postponed this decision (Baker, Bray, & Huggins, 2003; Mätz-Rensing et al., 2010). Besides, WHO also admits the existence of these viruses in clandestine labs around the world that could be accidentally or intentionally spread out (Hammarlund et al., 2003; Lindler, Lebeda, & Korch, 2005).
Considering the high infectiveness and lethality of smallpox, allied to the potential risk represented by its eventual use by bioterrorists, it is imperative to search for new drugs to combat this disease. This situation is aggravated by the fact that there is no chemotherapy against smallpox approved by the Food and Drugs Administration (FDA). The proposition of molecular targets for the drug design against smallpox is, therefore, a worldwide emergency. Thus, we proposed in the present work the enzyme thymidylate kinase from Variola virus (VarTMPK) as a potential target. A homology model was built and further submitted to docking and molecular dynamics (MD) studies in order to investigate the interactions of known inhibitors of Vaccinia virus TMPK (VacTMPK) inside VarTMPK active site. Also, the selectivity of each compound was investigated by performing the same studies inside human TMPK (HssTMPK). These studies led to the selection of the most promising and selective compounds as lead for the design of potential VarTMPK inhibitors. Results of the docking and MD studies corroborated to each other and, also, suggested a good correlation with the experimental data. We also observed the main interactions responsible for molecular recognition and suggested insights to the design of efficient and selective VarTMPK inhibitors.
2. Materials and methods
2.1. Homology modeling
The primary sequence of VarTMPK was downloaded from Expasy (http://au.expasy.org/) using the UniProtKB database (http://www.uniprot.org/). The search for templates with 3D structures available in the Protein Data Bank (PDB) (Berman et al., 2000) was performed with the basic local alignment search tool program (BLAST)(Altschul, Gish, Miller, Myers, & Lipman, 1990; Altschul et al., 1997). The dimeric crystallographic structure of VacTMPK at 2.40 Å resolution with factor R = .216 (PDB code: 2V54) (Caillat et al., 2008) was the one presenting the biggest sequential identity with VarTMPK being, for this reason, chosen as template to build the 3D model for VarTMPK. The alignment of the monomeric sequences was done with the software Swiss-Pdb Viewer (Guex & Peitsch, 1997) and, after some manual adjustments, submitted to the SWISS-MODEL server (Guex & Peitsch, 1997) in order to generate the final monomer model.
In order to check for the quality of our model, it was submitted to the validation server of the PDB (Berman et al., 2000) and to the Verify3D structure evaluation server (Bowie, Luthy, & Eisenberg, 1991; Luthy, Bowie, & Eisenberg, 1992), to check for the chemical environment of each residue.
After validation, the model was superposed to the 3D structure of VacTMPK with the program Swiss-Pdb Viewer (Guex & Peitsch, 1997) in order to copy the coordinates of thymidine-5′-diphosphate (TDP), the cofactor Mg2+, and the co-crystallized water molecules from the VacTMPK active site to build the complex used in our docking studies.
To verify the similarity between the residues of the active sites of VarTMPK and HssTMPK and to determine the degree of identity between them, we aligned the VarTMPK and HssTMPK (PDB code: 1E2G) structures using Swiss-Pdb Viewer (Guex & Peitsch, 1997).
2.2. Docking energy calculations
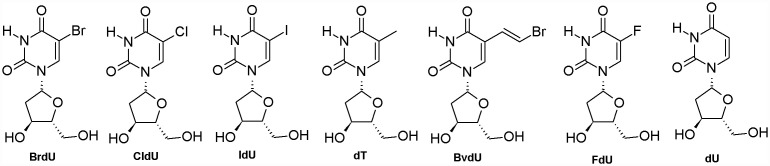
The 3D structure of HssTMPK with resolution of 1.7 Å, complexed with TDP and Mg2+ (Ostermann et al., 2000) was obtained from the PDB (Berman et al., 2000) under the code 1E2G, and the 3D structures of the compounds used in this study (Figure 1) were built using the program PC Spartan® (Hehre, Deppmeier, & Klunzinger, 1999) and had their partial atomic charges calculated by the RM1 semiempirical molecular orbital method (Rocha, Freire, & Simas, 2006). These compounds are potential inhibitors of VacTMPK and had its activities investigated by Caillat et al. (2008). Calculations of their docking energies inside the actives sites of VarTMPK and HssTMPK were performed using the software Molegro Virtual Docker® (Thomsen & Christensen, 2006) (MVD). The binding sites of VarTMPK and HssTMPK were restricted into spheres with radius of 6 Å around TDP and all the residues inside these spheres were set to be flexible. The influence of the water molecules was also considered during the docking studies because the solvent has an important role for protein–ligand interactions as it can modify the characteristics of the active site, mediate the formation of H-bonds, and also increase the entropy of the system due to the hydrophobic effect. To validate the docking protocol used, we first performed the re-docking of TDP over the active sites of VarTMPK and HssTMPK. Due to the stochastic nature of the docking algorithm, about 20 runs were performed for each compound with 30 poses (conformation and orientation of the ligand) returned to the analysis of the overlap with TDP inside VarTMPK and HssTMPK and, also, the ligand–protein interactions. The best pose of each compound was selected for the further steps of MD simulations.
Figure 1.
Structures of the compounds studied.
2.3. MD simulations
In order to check the docking results, we chose the best conformations obtained from the docking studies to run further steps of MD simulations. Before performing the simulations it was necessary to parameterize the ligands, so they could be recognized by the forcefield GROMOS 96 53a6 (Oostenbrink, Villa, Mark, & Van Gunsteren, 2004) from the program GROMACS 4.5.4 (Hess, Kutzner, Van Der Spoel, & Lindahl, 2008). The parameterization was carried out in the Dundee PRODRG Server (Van Aalten et al., 1996) and the charges distributions were calculated by the CHELPG (Brenaman & Wiberg, 1990) method using the Gaussian® (Frisch et al., 2001) program at DFT method (Hohenberg & Kohn, 1964) with functional B3LYP and basis function 6-31G (d,p). The dynamic behaviors of the complexes VarTMPK–ligand and HssTMPK–ligand were simulated using the GROMACS 4.5.4 package (Hess et al., 2008), in cubic boxes of approximately 640 and 940 nm3, containing around 7800 and 8000 water molecules of the SPC type, respectively, using periodic boundary conditions (Berendsen, Postma, Van Gunsteren, & Hermans, 1981). In this study, for both pressure and temperature, we used the Berendsen’s weak-coupling algorithm scheme which maintained temperature and pressure at 300 K and 1 bar. These systems were submitted to four steps of energy minimization as follows: steepest descent with position restrained of the ligands and convergence criterion of 50.00 kcal mol−1 nm−1, followed by steepest descent without restriction, conjugate gradients and, finally, quasi-Newton–Raphson until an energy of 10.00 kcal mol−1 nm−1.
The minimized complexes were then submitted to MD simulations in two steps. Initially, we performed 500 ps of MD at 300 K with PR for the entire system, except the water molecules, in order to ensure a balance of the solvent molecules around protein and ligand. Subsequently, there were performed 20 ns of MD without any restriction, using 2 fs of integration time and a cutoff of 10 Å for long-distance interactions. The electrostatic interactions were calculated using the Particle Mesh Ewald method and a total of 1000 conformations were obtained during each simulation. In this step, the lists of pairs (pairlists) were updated every 500 steps, all Arg and Lys residues were assigned with positive charges and the residues Glu and Asp were assigned with negative charges.
To analyze the structures generated after the optimization and MD steps, we used the visual molecular dynamics (VMD) (Humphrey, Dalke, & Schulten, 1996) and Swiss-Pdb Viewer (Guex & Peitsch, 1997) programs. Plots of variation of total energy, distance, random mean-square deviation (RMSD), and H-bonds formed during the MD simulation were generated with the Origin® program (Edwards, 2002). Qualitative spatial RMSD pictures were generated in MolMol program (Koradi, Billeter, & Wüthrich, 1996) and the figures of the frames of MD simulations were generated in the PyMOL program (DeLano & Bromberg, 2004).
3. Results and discussion
3.1. Homology modeling
The alignment between the sequences of VacTMPK and VarTMPK (Figure 2) shows that these enzymes share 98.5% of sequential identity with each other as reported before (Caillat et al., 2008). This is an excellent result considering literature reports that high quality models can be achieved when the similarity to the template is superior to 50%. Such models are comparable to structures determined by NMR and X-ray techniques and, therefore, suitable to the drug design. It was also observed that only four residues (Thr30, Thr103, Ala148, and Glu165) are different and none of them belong to the active site. This result corroborates the premise that potential inhibitors of VacTMPK could, eventually, become inhibitors of VarTMPK.
Figure 2.
Alignment of VarTMPK and VacTMPK sequences. Non-matching residues are shown in red.
After building the 3D structure of VarTMPK, we superimposed model and template using Swiss-Pdb Viewer (Guex & Peitsch, 1997) and copied the structures of TDP, Mg2+, and the water molecules from the template to the active site of our model in order to build the system for the docking and MD studies. The RMSD of .18 Å obtained on the superposition evidences the high sequential identity between model and template.
The Ramachandram plot (Ramachandran & Sasisekharan, 1968) of our model (data not shown) presented 99.5% of the residues in the most favored regions; a result considered very good for a homology model (Laskowski, Macarthur, Moss, & Thornton, 1993). Regarding the main chain properties, there were not found any bad contacts neither distortions of Cα nor energy problems with H-bonds. Also, there were not found distortions on the torsion angles of the side chains. The 3D–1D averaged score for each residue was calculated by the Verify3D program (Bowie et al., 1991) and presented a variation between .03 and .58. The Z-scores calculated using PROVE (Pontius, Richelle, & Wodak, 1996) showed that our model is comparable to a typical native structure. Besides, G factor (one measure of the normality degree of the protein properties) obtained of −.24 was below the maximum value allowed for homology models (Laskowski et al., 1993).
The 3D structure of HssTMPK was superimposed to our model, using SPDBViewer, in order to verify the degree of similarity among the active site residues and the possibility of designing selective VarTMPK inhibitors. The superposition presented a RMSD of 1.19 Å and the sequential identity observed between these two enzymes was of 43.1%. The active site residues of VarTMPK were determined by analogy with the VacTMPK active site residues former identified by Caillat et al. (2008). Analyzing the alignment with HssTMPK (data not shown), we verified that among 14 residues of the active site 7 are different. This means that there is a similarity of 50% between the active sites and suggests that there is room for the drug design of selective inhibitors.
3.2. Docking studies
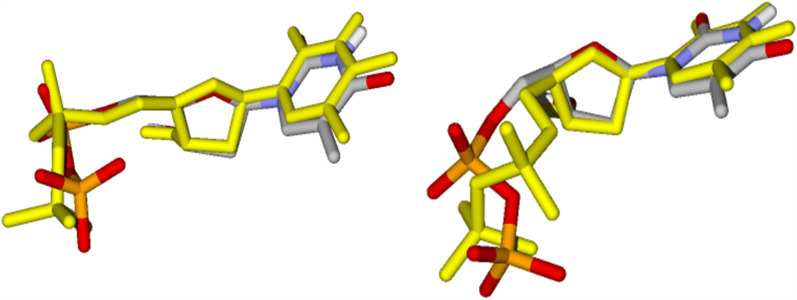
Figure 3 shows the re-docking results of TDP inside the active sites of VarTMPK and HssTMPK. The RMSD values observed were of 1.58 and .86 Å, respectively. These values validate the docking methodology considering that, according to literature, a RMSD lower than 2.00 Å is considered acceptable (Kontoyanni, McClellan, & Sokol, 2004; Leach, Shoichet, & Peishoff, 2006; Warren et al., 2006).
Figure 3.
Re-docking of TDP in the active sites of VarTMPK (right) and HssTMPK (left), TDP from crystal is colored by element, while the TDP conformation obtained by docking is colored in yellow.
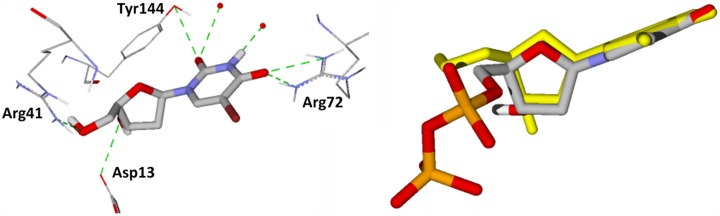
The cavities of the active sites of VarTMPK and HssTMPK were predicted by MVD software (Thomsen & Christensen, 2006) as having 78,336 and 90,112 Å3, respectively. The inhibitors (Figure 1) were docked into these cavities, according to the procedure described in the methodology, in order to evaluate their interactions to provide insights to the design of selective inhibitors. The best poses for each compound were selected on the basis of the interaction energies with the enzymes (intermolecular/electrostatic and H-bond) and best superposition to TDP, as illustrated in Figure 4 for the system BrdU/VarTMPK).
Figure 4.
Left: Interactions of BrdU with aminoacids and water molecules inside VarTMPK. Right: superposition of the best conformation of BrdU (in CPK) with TDP (in yellow) after docking.
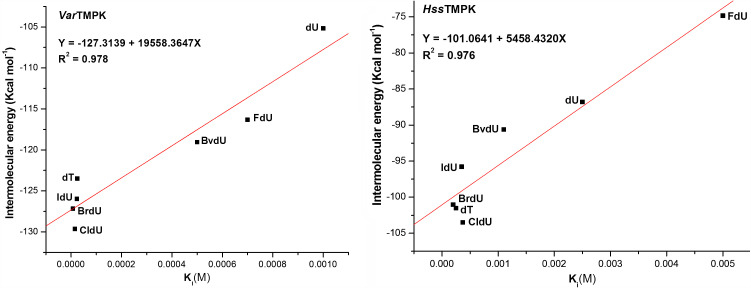
Tables12 present the active site residues of both enzymes participating in H-bonds with each compound studied, the energy values obtained, and the respective experimental values of Ki for these compounds inside VacTMPK, reported by Caillat et al. (2008). Our theoretical results present a good correlation to the experimental values with R2 of .978 and .976 for VarTMPK and HssTMPK, respectively (see Tables12 and Figure 5).
Table 1. Docking results for the compounds docked in VarTMPK and their Ki values, obtained by Caillat et al. (2008), with VacTMPK.
| Compound | Residue | Dist. (Å) | Energy (kcal mol−1) | Intermolecular energy (kcal mol−1) | H-bond energy (kcal mol−1) | a K i |
|---|---|---|---|---|---|---|
| BrdU | Asp13 | 3.15 | −2.25 | −127.16 | −10.49 | 7 μM |
| Arg41 | 3.10 | −1.71 | ||||
| Arg72 | 2.87 | −2.50 | ||||
| 3.09 | −2.09 | |||||
| Tyr144 | 3.21 | −1.94 | ||||
| H2O | 3.09 | −2.50 | ||||
| 3.26 | −1.70 | |||||
| CldU | Asp13 | 3.04 | −2.50 | −129.63 | −8.11 | 15 μM |
| Arg41 | 2.74 | −1.55 | ||||
| Arg72 | 2.60 | −2.34 | ||||
| 2.95 | −1.69 | |||||
| Gly98 | 3.35 | −.02 | ||||
| IdU | Asp13 | 3.16 | −2.18 | −125.97 | −10.43 | 23 μM |
| Arg41 | 3.10 | −1.75 | ||||
| Arg72 | 2.79 | −2.50 | ||||
| 2.99 | −2.05 | |||||
| Tyr144 | 3.21 | −1.94 | ||||
| H2O | 3.10 | −2.50 | ||||
| 3.26 | −1.68 | |||||
| dT | Asp13 | 3.32 | −1.41 | −123.49 | −9.18 | 25 μM |
| Arg41 | 3.10 | −1.59 | ||||
| Arg72 | 2.82 | −1.97 | ||||
| 2.63 | −2.44 | |||||
| Tyr101 | 3.47 | −.62 | ||||
| Tyr144 | 3.37 | −1.14 | ||||
| H2O | 3.07 | −2.50 | ||||
| 3.39 | −1.04 | |||||
| 3.19 | −2.04 | |||||
| BvdU | Arg72 | 2.60 | −1.99 | −119.06 | −4.11 | .5 mM |
| 2.60 | −2.01 | |||||
| Tyr94 | 3.57 | −.11 | ||||
| H2O | 2.61 | −2.50 | ||||
| 3.10 | −2.50 | |||||
| FdU | Asp13 | 3.11 | −2.45 | −116.32 | −9.49 | .7 mM |
| Arg41 | 2.74 | −1.57 | ||||
| Arg72 | 3.10 | −2.09 | ||||
| 2.94 | −2.47 | |||||
| Tyr101 | 3.42 | −.90 | ||||
| H2O | 3.54 | −.31 | ||||
| dU | Asp13 | 3.10 | −2.49 | −105.17 | −11.20 | 1.0 mM |
| Arg41 | 3.10 | −1.15 | ||||
| Arg72 | 3.09 | −2.32 | ||||
| 3.00 | −2.50 | |||||
| Tyr101 | 3.45 | −.77 | ||||
| Tyr144 | 3.20 | −1.98 | ||||
| H2O | 3.31 | −1.43 | ||||
| 3.41 | −.95 | |||||
| 2.86 | −2.50 |
aExperimental data.27
Table 2. Docking results for the compounds docked in HssTMPK and their Ki values obtained by Caillat et al. (2008).
| Compound | Residue | Dist. (Å) | Energy (kcal mol−1) | Intermolecular energy (kcal mol−1) | H-bond energy (kcal mol−1) | a K i |
|---|---|---|---|---|---|---|
| BrdU | Asp15 | 3.09 | −2.50 | −101.04 | −7.66 | .2 mM |
| 2.73 | −2.50 | |||||
| Arg76 | 3.27 | −.99 | ||||
| 2.48 | −1.50 | |||||
| Arg97 | 3.38 | −.17 | ||||
| H2O | 3.42 | −.90 | ||||
| 2.96 | −2.50 | |||||
| 2.80 | −2.50 | |||||
| 2.61 | −2.50 | |||||
| 2.77 | −2.50 | |||||
| 3.10 | −2.50 | |||||
| dT | Asp15 | 3.29 | −1.56 | −101.53 | −6.00 | .25 mM |
| Arg76 | 3.54 | −.20 | ||||
| 2.83 | −2.50 | |||||
| Arg97 | 2.94 | −.73 | ||||
| 2.62 | −1.02 | |||||
| H2O | 3.10 | −2.50 | ||||
| 2.93 | −2.50 | |||||
| 3.10 | −2.50 | |||||
| 2.68 | −2.50 | |||||
| IdU | Asp15 | 3.17 | −2.11 | −95.79 | −8.47 | .35 mM |
| 3.10 | −2.49 | |||||
| Arg76 | 2.90 | −2.50 | ||||
| Arg97 | 3.26 | −.49 | ||||
| 2.74 | −.89 | |||||
| H2O | 3.07 | −2.50 | ||||
| 2.10 | −2.50 | |||||
| 3.24 | −1.77 | |||||
| 2.30 | −.03 | |||||
| CldU | Asp15 | 3.17 | −2.50 | −103.52 | −7.77 | .37 mM |
| 2.87 | −2.50 | |||||
| Arg76 | 3.54 | −.16 | ||||
| 2.57 | −2.29 | |||||
| Arg97 | 3.34 | −.33 | ||||
| H2O | 3.10 | −2.50 | ||||
| 2.80 | −2.50 | |||||
| 2.62 | −2.50 | |||||
| 2.60 | −2.50 | |||||
| 2.82 | −2.50 | |||||
| 3.10 | −2.50 | |||||
| BvdU | Pro43 | 3.56 | −.10 | −90.61 | −7.43 | 1.1 mM |
| Arg45 | 2.29 | .07 | ||||
| Arg76 | 2.88 | −1.76 | ||||
| 2.60 | −2.31 | |||||
| Tyr98 | 3.51 | −.43 | ||||
| Ser101 | 3.45 | −.74 | ||||
| 2.54 | −2.04 | |||||
| Phe105 | 3.54 | −.011 | ||||
| H2O | 3.05 | −2.50 | ||||
| dU | Asp15 | 3.01 | −2.50 | −86.80 | −8.71 | 2.5 mM |
| Arg76 | 3.08 | −2.13 | ||||
| 2.92 | −2.50 | |||||
| Arg97 | 2.64 | −1.07 | ||||
| 2.84 | −.51 | |||||
| H2O | 3.06 | −2.50 | ||||
| 3.10 | −2.50 | |||||
| 2.60 | −2.50 | |||||
| FdU | Asp15 | 3.10 | −2.50 | −74.83 | −8.88 | >5 mM |
| Arg76 | 2.75 | −2.04 | ||||
| 2.85 | −1.88 | |||||
| Arg97 | 2.67 | −1.27 | ||||
| H2O | 3.10 | −2.49 | ||||
| 3.26 | −1.67 | |||||
| 3.40 | −1.01 | |||||
| 3.10 | −2.50 | |||||
| 3.07 | −2.50 | |||||
| 3.22 | −1.89 |
aExperimental data.27
Figure 5.
Correlation between the intermolecular energies and the Ki, obtained by Caillat et al. (2008), and values of each compound for VarTMPK (left) and HssTMPK (right).
The lower values of energies observed in Table 1 related to the interactions of the ligands with VarTMPK suggest that these compounds could have more affinity for this enzyme. It was also observed that, except for fluxuridine (FdU), the insertion of halogen groups in the position 5′ of the pyrimidine ring reduces the energy values. Brivudin (BvdU) presented lower energy inside VarTMPK than in HssTMPK (−119.06 kcal mol−1 versus – 90.61 kcal mol−1). This could be related to the narrowest cavity of HssTMPK that disable this enzyme to accommodate bulkier molecules inside its active site, allowing a major specificity to substrates, like pirimidines with larger groups in the position 5′ or purine nucleotides like dGMP, as reported by Caillat et al. (2008).
3.3. MD studies
The best poses obtained in the docking studies were submitted to 20 ns of MD simulations inside VarTMPK and HssTMPK in order to corroborate the docking results and assess the dynamical behavior of the compounds.
The energy plots for the 14 systems simulated showed that the total energy tends to stability since the first ps of simulation.
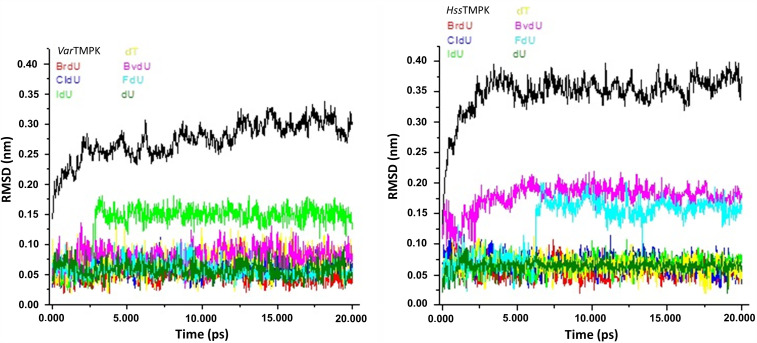
In the plots of temporal RMSD presented in Figure 6, it is also possible to observe the equilibration of the systems since the first ps. The RMSD values never overpassed .10 nm during the simulations except for IdU inside VarTMPK that stabilized at .15 nm, and for BvdU and FdU inside HssTMPK that stabilized around .20 nm.
Figure 6.
Temporal RMSD values for VarTMPK (left) and HssTMPK (right) and the compounds studied. Black lines refer to the enzymes without ligands.
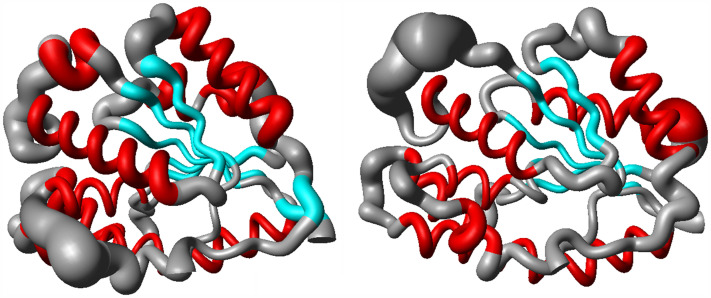
The spatial RMSD for the systems VarTMPK/BrdU and HssTMPK/BrdU are shown in Figure 7. As can be seen, the regions with major fluctuations during the simulation (larger tubes in the picture) correspond to the extremities and loops. On the other hand, the residues in the active site region and in the α-helix and β-sheets presented lower fluctuations (lower thickness in the tubes). This behavior was commom to all the 14 systems studied.
Figure 7.
Spatial RMSD of the systems VarTMPK/BrdU (right) and HssTMPK/BrdU (left). BrdU was omitted in the figure for better clarity.
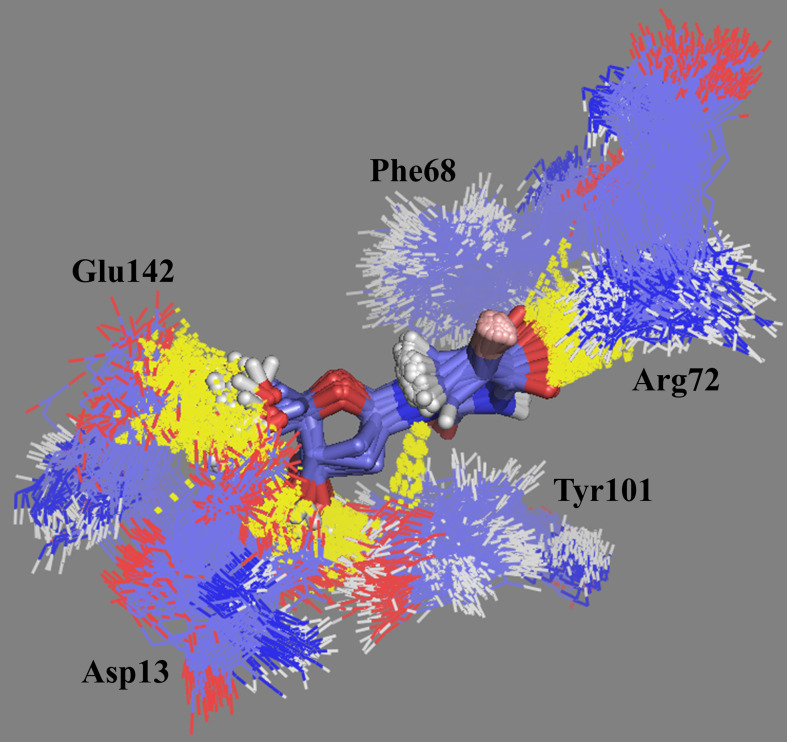
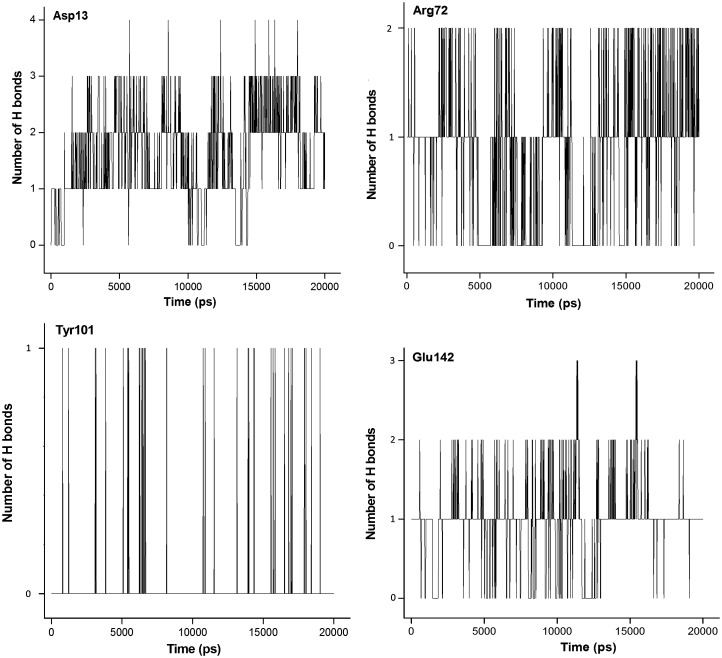
The docking studies showed that BrdU was able to establish H-bonds with residues Asp13, Ar41, Arg72, Tyr144, and two water molecules (Figure 3). The MD simulations showed that this compound was able to establish until 6 H-bond during the simulation and keep between 2 and 4. The frames of the MD simulation for this compound inside VarTMPK (Figure 8) show the interactions with residues Asp13, Phe68, Arg72, Tyr101, Glu142, and water molecules as suggested in the docking studies, except for the H-bond with Tyr144.
Figure 8.
Frames of BrdU in the binding site of VarTMPK during the 20.0 ns of MD simulation. The H-bonds formed along the simulation are shown in yellow.
Regarding CldU, MD showed the formation of 4 H-bonds with VarTMPK during the simulation, including H-bonds with residues Glu142 and Tyr144 not observed in the dockings. The frames extracted for this compound also showed the possibility of hydrophobic interactions with Phe68 (Figure 9). This interaction was observed to all compounds inside VarTMPK but not inside HssTMPK and, therefore, can contribute for an additional stabilization and permanence of them inside VarTMPK.
Figure 9.
Hydrophobic interactions observed along the MD simulation between CldU (in tubes) and the side chain of Phe68.
In the same way as for CldU, IdU formed about 4 H-bonds with VarTMPK during the simulation and an additional interaction with Tyr101 not plotted in the docking study.
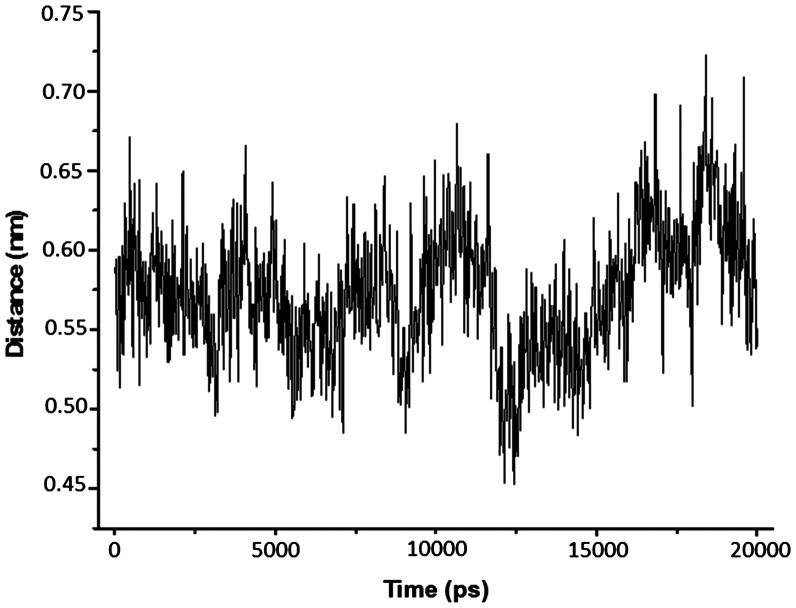
The analysis described above were performed for all compounds studied inside both enzymes and, in general, good concordances with the docking results were observed. The results obtained point the interactions with residues Asp13, Phe68, Arg72, Tyr101, Glu142, and Tyr144 as essential for the drug design. As BrdU was the ligand with the most promissing results, we show in plots of Figures1011 the H-bonds formed among this compound and residues Asp13, Arg72, Tyr101, and Glu142 during the MD simulations and, also, the variation of distance between the aromatic rings of BrdU and Phe68. As can be seen, BrdU was able to form and keep until 3 H-bonds with Asp13, while its aromatic ring was kept at about 6.0 Å from the aromatic ring of Phe68, suggesting the formation of the hydrophobic interaction.
Figure 10.
H-bonds formed among BrdU and residues Asp13, Arg72, Tyr101, and Glu142 during 20,000 ps of MD simulation.
Figure 11.
Variation of distance between the aromatic rings of BrdU and Phe68 during 20,000 ps of MD simulation.
Our studies suggest that, despite of its larger size, the cavity of HssTMPK is narrower and have no empty space to accommodate the modifications in the pyrimidine rings of the compounds. This could be observed analyzing the results for BvdU. So, the derivatives of the compounds studied should present modifications in the position 5′ of their pyrimidine rings in order to be more promising as selective inhibitors.
4. Conclusion
The docking and MD results obtained in the present work corroborated to each other and suggested a good selectivity of the compounds face to HssTMPK. Also, the good correlations with experimental data suggest our docking protocol as a suitable tool for an initial prediction of affinities of potential new inhibitors.
Our results pointed out CldU and BrdU as the most promising inhibitors of VarTMPK. These compounds should, therefore, be submitted to experimental studies. Additionally, we observed that the interactions with residues Asp13, Phe68, Arg72, Tyr101, Glu142, and Tyr144 and the differences in the shapes of the active sites of VarTMPK and HssTMPK should be taken in account in the drug design of selective inhibitors for VarTMPK, through modifications on the positions 3′ and 5′ of the pyrimidine rings of CldU and BrdU.
Acknowledgments
The authors wish to thank the Military Institute of Engineering for the infrastructure, the Brazilian financial agencies CNPq, FAPERJ, and CAPES for the financial support. TCCF thanks CNPq and FAPERJ for the fellowships.
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology , 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Mille, W., & Lipman, D. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research , 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzhanova, D., & Früh, K. (2010). Modulation of the host immune response by cowpox virus. Microbes and Infection , 12, 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R. O., Bray, M., & Huggins, J. W. (2003). Potential antiviral therapeutics for smallpox, monkeypox and other orthopoxvirus infections. Antiviral Research , 57, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbehani, A. M. (1983). The smallpox story: Life and death of an old disease. Microbiological Reviews , 47, 455–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., Van Gunsteren, W. F., & Hermans, J. (1981). Interaction models for water in relation to protein hydration. Dordrecht: Reidel. [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., … Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research , 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie, J. U., Luthy, R., & Eisenberg, D. (1991). A method to identify protein sequences that fold into a known three-dimensional structure. Science , 253, 164–170. [DOI] [PubMed] [Google Scholar]
- Brenaman, C. M., & Wiberg, K. B. (1990). Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. Journal of Computational Chemistry , 11, 361–373. [Google Scholar]
- Caillat, C., Topalis, D., Agrofoglio, L. A., Pochet, S., Balzarini, J., Deville-Bonne, D., & Meyer, P. (2008). An unexpected dimerization has implications for antiviral therapy. Proceedings of National Academy of Science USA , 105, 16900–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campe, H., Zimmermann, P., Glos, K., Bayer, M., Bergemann, H., Dreweck, C., … Sing, A. (2009). Cowpox virus transmission from pet rats to humans. Germany, Emerging Infectious Diseases , 15, 777–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, J. L., Nichols, D. K., Martinez, M. J., & Raymond, J. W. (2010). Animal models of orthopoxvirus infection. Veterinary Pathology , 47, 852–870. [DOI] [PubMed] [Google Scholar]
- DeLano, W. L., & Bromberg, S. (2004). PyMOL user’s guide. San Francisco, CA: DeLano Scientific LLC. [Google Scholar]
- Edwards, P. M. (2002). Origin 7.0: Scientific graphing and data analysis software. Journal of Chemical Information and Computer Sciences , 42, 1270–1271. [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, E. S., & Pople, J. A. (2001). Gaussian 98 (Revision A.11). Pittsburgh, PA: Gaussian. [Google Scholar]
- Gileva, I. P., Nepomnyashchikh, T. S., Antonets, D. V., Lebadev, L. R., Kochneva, G. V., Grazhdanrseva, A. V., & Shchelkunov, S. N. (2006). Properties of the recombinant TNF-binding proteins from variola, monkeypox, and cowpox viruses are different. Biochimica et Biophysica Acta , 1764, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis , 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hammarlund, E., Lewis, M. W., Hanifin, J. M., Mori, M., Koudelka, C. W., & Slifka, M. K. (2010). Antiviral immunity following smallpox virus infection: A case-control study. Journal of Virology , 84, 12754–12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund, E., Lewis, M. W., Hansen, S. G., Strelow, L. I., Nelson, J. A., Sexton, G. J., … Slifka, M. K. (2003). Duration of antiviral immunity after smallpox vaccination. Nature Medicine , 9, 1131–1137. [DOI] [PubMed] [Google Scholar]
- Handley, L., Buller, R. M., Frey, S. E., Bellone, C., & Parker, S. (2009). The new ACAM2000™ vaccine and other therapies to control orthopoxvirus outbreaks and bioterror attacks. Expert Review of Vaccines , 8, 841–850. [DOI] [PubMed] [Google Scholar]
- Harper, G. J. (1961). Airborne micro-organisms: Survival tests with four viruses. Journal of Hygiene (London) , 59, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehre, W. J., Deppmeier, B. J., & Klunzinger, P. E. (1999). PC SPARTAN Pro. Irvine, CA: Wavefunction. [Google Scholar]
- Hendrickson, R. C., Wang, C., Hatcher, E. L., & Lefkowitz, E. J. (2010). Orthopoxvirus genome evolution: The role of gene loss. Viruses , 2, 1933–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., Kutzner, C., Van Der Spoel, D., & Lindahl, E. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation , 4, 435–447. [DOI] [PubMed] [Google Scholar]
- Hohenberg, P., & Kohn, W. (1964). Inhomogeneous electron gas. Physical Reviews B , 136, B864–B871. [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics and Modelling , 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Huq, F. (1976). Effect of temperature and relative humidity on Variola virus in crusts. Bulletin of the World Health Organization , 54, 710–712. [PMC free article] [PubMed] [Google Scholar]
- Johnston, C. S., Lin, K. L., Connor, J. H., Ruthel, G., Goff, A., & Hensley, L. E. (2012). In vitro inhibition of monkeypox virus production and spread by Interferon-β. Virology Journal , 9(5), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, R. B., Ovsyannikova, I. G., Jacobson, R. M., & Poland, G. A. (2009). The immunology of smallpox vaccines. Current Opinion in Immunology , 21, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, R. B., Ovsyannikova, I., & Poland, G. A. (2009). Smallpox vaccines for biodefense. Vaccine , 27, D73–D79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyanni, M., McClellan, L. M., & Sokol, G. S. (2004). Evaluation of docking performance: Comparative data on docking algorithms. Journal of Medicinal Chemistry , 47, 558–565. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., & Wüthrich, K. (1996). MOLMOL: A program for display and analysis of macromolecular structures. Journal of Molecular Graphics and Modelling , 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., Macarthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography , 26, 283–291. [Google Scholar]
- Leach, A. R., Shoichet, B. K., & Peishoff, C. E. (2006). Prediction of protein − Ligand interactions. Docking and scoring: Successes and gaps. Journal of Medicinal Chemistry , 49, 5851–5855. [DOI] [PubMed] [Google Scholar]
- Lindler, L. E., Lebeda, F. J., & Korch, G. W. (2005). Biological weapons defense: Infectious diseases and counter bioterrorism (1st ed.). Totowa, NJ: Human. [Google Scholar]
- Liszewski, M. K., Leung, M. K., Hauhart, R., Buller, R. M., Bertram, P., Wang, X., … Atkinson, J. P. (2006). Structure and regulatory profile of the monkeypox inhibitor of complement: Comparison to homologs in Vaccinia and variola and evidence for dimer formation. Journal of Immunology , 176, 3725–3734. [DOI] [PubMed] [Google Scholar]
- Luthy, R., Bowie, J. U., & Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature , 356, 83–85. [DOI] [PubMed] [Google Scholar]
- Mätz-Rensing, K., Ellerbrok, H., Ehlers, B., Pauli, G., Floto, A., Alex, M., … McFadden, G. (2010). Killing a killer: What next for smallpox? PLoS Pathogens , 6, e1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mätz-Rensing, K., Ellerbrok, H., Ehlers, B., Pauli, G., Floto, A., Alex, M., … Kaup, F. J. (2006). Fatal poxvirus outbreak in a colony of new world monkeys. Veterinary Pathology , 43, 212–218. [DOI] [PubMed] [Google Scholar]
- Ninove, L., Domart, Y., Vervel, C., Voinot, C., Salez, N., Raoult, D., … Charrel, R. N. (2009). Cowpox virus transmission from pet rats to humans, France. Emerging Infectious Diseases , 15, 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink, C., Villa, A., Mark, A. E., & Van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. Journal of Computational Chemistry , 25, 1656–1676. [DOI] [PubMed] [Google Scholar]
- Ostermann, N., Schlichting, I., Brundiers, R., Konrad, M, Reinstein, J., Veit, T., … Lavie, A. (2000). Insights into the phosphoryltransfer mechanism of human thymidylate kinase gained from crystal structures of enzyme complexes along the reaction coordinate. Structure, 8, 629–642. [DOI] [PubMed] [Google Scholar]
- Pontius, J., Richelle, J., & Wodak, S. J. (1996). Deviations from standard atomic volumes as a quality measure for protein crystal structures. Journal of Molecular Biology , 264, 121–136. [DOI] [PubMed] [Google Scholar]
- Ramachandran, G. N., & Sasisekharan, V. (1968). Conformation of polypeptides and proteins. Advances in Protein Chemistry , 23, 283–438. [DOI] [PubMed] [Google Scholar]
- Rocha, G. B., Freire, R. O., & Simas, A. M. (2006). RM1: A reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. Journal of Computational Chemistry , 27, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Ryan, K. J., & Ray, C. G. (2004). Sherris medical microbiology (4th ed.). New York: McGraw Hill. [Google Scholar]
- Sakhatskyy, P., Wang, S., Zhang, C., Chou, T., Kishko, M., & Lu, S. (2008). Immunogenicity and protection efficacy of subunit-based smallpox vaccines using Variola major antigens. Virology , 371, 98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeknecht, G., Eickmann, M., Köhler, K., Herden, C. E., Kolesnikova, L., Forster, C., … Thiel, M. (2010). Fatal cowpox virus infection in captive banded mongooses (Mungos mungo). Veterinary Pathology , 47, 547–552. [DOI] [PubMed] [Google Scholar]
- Thomsen, R., & Christensen, M. H. (2006). MolDock: A new technique for high-accuracy molecular docking. Journal of Medicinal Chemistry , 49, 3315–3321. [DOI] [PubMed] [Google Scholar]
- Van Aalten, D. M. F., Bywater, R., Findlay, J. B. C., Hendlich, M., Hooft, R. W. W., & Vriend, G. (1996). PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. Journal of Computer-Aided Molecular Design , 10, 255–262. [DOI] [PubMed] [Google Scholar]
- Warren, G. L., Andrews, C. W., Capelli, A. M., Clarke, B., LaLonde, J., Lambert, M. H., … Head, M. S. (2006). A critical assessment of docking programs and scoring functions. Journal of Medicinal Chemistry , 49, 5912–5931. [DOI] [PubMed] [Google Scholar]