Abstract
Acknowledging the importance of studies toward the development of measures against terrorism and bioterrorism, this study aims to contribute to the design of new prototypes of potential drugs against smallpox. Based on a former study, nine synthetic feasible prototypes of selective inhibitors for thymidylate kinase from Variola virus (VarTMPK) were designed and submitted to molecular docking, molecular dynamics simulations and binding energy calculations. The compounds are simplifications of two more complex scaffolds, with a guanine connected to an amide or alcohol through a spacer containing ether and/or amide groups, formerly suggested as promising for the design of selective inhibitors of VarTMPK. Our study showed that, despite the structural simplifications, the compounds presented effective energy values in interactions with VarTMPK and HssTMPK and that the guanine could be replaced by a simpler imidazole ring linked to a –NH2 group, without compromising the affinity for VarTMPK. It was also observed that a positive charge in the imidazole ring is important for the selectivity toward VarTMPK and that an amide group in the spacer does not contribute to selectivity. Finally, prototype 3 was pointed as the most promising to be synthesized and experimentally evaluated.
Communicated by Ramaswamy H. Sarma
Keywords: Drug design, Variola virus, thymidylate kinase, smallpox, docking, molecular dynamics simulations
Introduction
Chemical and biological weapons can be defined as warfare agents belonging to the class of unconventional weapons, which comprises substances or organisms of complex identification and control, which can be produced in more economically viable production systems than those of conventional weapons (Taylor & Junior, 1992). Generally speaking, these warfare agents are feared throughout the world due to their high lethality, causing panic and emotional instability in superior levels when compared to the psychological impact of conventional weapons (Bismuth, Borron, Baud, & Barriot, 2004; Byrnes, King, & Junior, 2003). It is important to point out that few countries have ideal resources to resist to chemical or biological attacks today. It is under this context that studies on the field of chemical and biological defense become paramount, needing continuous development and updating, as they are the best strategy for professional training and the developing of countermeasures against such weapons (Lindler, Lebeda, & Korch, 2004).
With regard to biological warfare agents, a prominent threat is the use of viruses with high fatality rates – such as smallpox – in terrorist attacks (Chapman, Nichols, Martinez, & Raymond, 2010; Davenport, Satchell, & Shaw-Taylor, 2018; Hammarlund et al, 2010; Kennedy, Ovsyannikova, Jacobson, & Poland, 2009). Smallpox is an infectious disease transmitted among human beings, mainly through contact with droplets containing the virus in suspension, expelled by infected individuals. It is caused by the Variola virus, a virus rich in DNA-type genetic material, which genus and family are, respectively, Orthopoxvirus and poxviridae (Hendrickson, Wang, Hatcher, & Lefkowitz, 2010; Liszewski et al., 2006; Sakhatskyy et al., 2008). This virus replicates itself in the cytoplasm of host cells and codifies essential enzymes for the replication and transcription of the genome, such as thymidylate and thymidine kinases (TMPK and TK). Conversely, it can also be employed in the rational development of agents used in the treatment of illnesses such as cancer, functioning as a base for chemotherapies using the TMPK gene as a reference (Caillat et al., 2008).
Despite the eradication of Variola virus declared by the World Health Organization (WHO) in the early 1980s, the risk of a bioterrorist attack deploying smallpox in the future is recognized as real and imminent, since there is evidence that some laboratories illicitly possess strains of this biological agent (Lindler et al., 2004). Regarding immunization, it should be mentioned that vaccination against smallpox was interrupted worldwide soon after its eradication in the 1980s. Besides, there are a few restrictions to its use, such as in the case of people who are HIV-positive, or have neoplasms and are under treatment with immunosuppressants. Furthermore, the side effects of the vaccine are numerous and cannot be neglected (Greenberg et al., 2013). This has motivated efforts for the development of new and more effective vaccines against smallpox (Kennedy et al., 2016; Lee, Kumar, Jhan, & Bishop, 2018; Pittman et al., 2015). However, no new and more effective vaccine is available yet.
Recently, the first drug against smallpox, tecoviramat, was approved by the FDA (U.S. Food and Drug Administration) representing a great advance, as until recently, no efficient treatment for this disease was known (Grosenbach et al., 2018; Manus, 2018). Despite encouraging, this is not enough, and the search for more drugs against this disease should continue in order to increase the therapeutic arsenal available. If we consider that almost the totality of the world population aged less than 40 is not immunized, a return of smallpox dissemination, either as a natural pandemic or in a terrorist attack, would cause catastrophic consequences. In face of this scenario, studies for developing as much as possible chemicals against molecular targets of Variola virus (Chaudhuri, Symons, & Deval, 2018; Chittick, Morrison, Brundage, & Nichols, 2017; Crump, Korom, Buller, & Parker, 2017; Grossi et al., 2017; Trost et al., 2015) are imperative (Damon, Damaso, & Mcfadden, 2014; Prichard, & Kern, 2012).
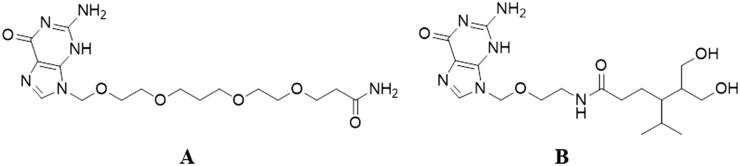
Recently, our research group started an innovative molecular modeling study proposing the enzyme TMPK from Variola virus (VarTMPK) as the molecular target for antivirals like cidofovir, acyclovir and its derivatives, with the aim of developing potential new inhibitors of this enzyme as drugs against smallpox (Guimarães, Ramalho, & França, 2014; Guimarães et al., 2015). Based on our first results, 10 structures were proposed and nine were highlighted as potential lead compounds for selective inhibitors of VarTMPK. The most promising leads pointed on these studies (Figure 1), however, are quite complex molecules, needing some structural simplifications to become more feasible from the synthetic point of view. Therefore, in order to move forward on this project, we proposed as the main objective of the present study, to minimize the structural complexity of these compounds in order to make its synthesis more viable and less costly, with a minimal impact on their potential affinity and selectivity for VarTMPK. For this, nine new prototypes derived from leads A and B (Figure 1) were designed and submitted to docking and molecular dynamics (MD) simulations in order to analyze their modes of interaction and, consequently, assess their selectivity within the active sites of VarTMPK and human (Homo sapiens sapiens) TMPK (HssTMPK). Later, for the most promising results, free energy calculations were performed using the Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) technique (Almeida et al., 2015; Bastos et al., 2016; Jayaram, Sprous, Young, & Beveridge, 1998; Kar, Lipowsky, & Knecht, 2013; Shao et al., 2006; Souza et al., 2017; Vorobjev, Almagro, & Hermans, 1998), a method of free energy simulation known for its high efficiency and reliability in the study of protein–ligand interactions (Wang, Greene, Xiao, Qi, & Luo, 2018).
Figure 1.
Selected structures proposed by Guimarães and coworkers (2015) as inhibitors for VarTMPK.
Methodology
Investigated structures
The structures of the prototypes proposed in this study are shown in Figure 2. They are derivatives of leads A (prototypes 1–5) and B (prototypes 6–9), proposed by Guimarães et al. (2015) and shown in Figure 1. As mentioned before, these prototypes were proposed to be more feasible from the synthetic point of view, without losing the important selective interactions inside VarTMPK mentioned by Guimarães et al. (2015). In all cases, the guanine group was replaced by the simpler imidazole group, and the spacers were simplified to two types. The first type (in prototypes 1–5) contained only ethers, and the second type (in prototypes 6 and 7) contained one ether and one amide group. The isopropyl substituent in lead B was eliminated, since it had no relevant interaction, as reported by Guimarães et al. (2015). The guanine group on the other hand was reduced to the imidazole ring because its six-membered ring was not found by Guimarães et al. (2015) to be relevant for the selectivity related to HssTMPK. With the purpose of determining the predominant species of these ligands under physiologic pH (7.4), the ionization states of each one were calculated with the aid of the Chemicalize database (Swain, 2012). This software was also used to check if the leads meet the druggability criteria established by Lipinski’s rule of five (Lipinski, 2004). The three-dimensional (3D) structures of the leads were constructed using the Spartan 08® Suite software (Shao et al., 2006), and the optimization and calculation of its atomic charges were performed employing the RM1 semi-empirical method (Gonçalves, Franca, Villar, & Pascutti, 2010; Rocha, Freire, Simas, & Stewart, 2006).
Figure 2.
Structures of the proposed prototypes.
Docking studies
The structure of VarTMPK used in this study was the homology model compiled and validated in a previous study (Guimarães et al., 2014), while the structure of HssTMPK was the one complexed with thymidine-5-diphosphate (TDP) and Mg2+, available in the Protein Data Bank (PDB) under the code 1E2G (Berman et al., 2000). Both 3D structures of VarTMPK and HssTMPK, were inspected and optimized before the docking studies using the software Swiss-Pdb Viewer (Guex & Peitsch, 1997) and Molegro Virtual Docker (MVD®) (Thomsen & Christensen, 2006).
The docking energy calculations, with the MolDock algorithm, were performed in the MVD® software (Thomsen & Christensen, 2006). The water molecules inside VarTMPK and HssTMPK were maintained during the energy calculations, so that the interaction between ligands and solvents, when present, could be assessed. The validation of the docking protocol through re-docking studies was previously done by Guimarães et al. (2014). The dockings of the ligands were performed in the area where the substrate TDP and the cofactor Mg2+ are found. Only the Mg2+ was kept during docking calculations, since its interaction energies with the proposed prototypes were taken into consideration. As before (Guimarães et al., 2014), the binding sites inside VarTMPK and HssTMPK were restricted into spheres, with radii of 6 and 10 Å for VarTMPK and HssTMPK, respectively, around TDP, and all the residues inside these spheres were set to be flexible. The coordinates were centered on x = 8.95, y = 22.41 and z = 0.69 for VarTMPK, and x = 13.92, y = 75.19 and z = 25.05 for HssTMPK.
Due to the stochastic nature of the anchoring algorithm, about 10 repetitions of the docking protocol were performed for each compound. In turn, 30 poses for the conformation and orientation of each ligand were generated for each analysis assessing the active sites of HssTMPK and VarTMPK. The best poses for each ligand in the viral and human enzymes were selected for further MD simulations. They were chosen according to the stability of each complex formed between enzyme and ligand, the interaction energy between enzyme and cofactor, the number of hydrogen bonds observed in each TMPK/ligand complex and the overlap between each ligand and TDP.
MD studies
For the MD simulations, each ligand had to be parameterized (Berendsen, Van Der Spoel, & Van Drunen, 1995; Pronk et al., 2013) in order to be recognized by the OPLS-AA force field (Kaminski, Friesner, Tirado-Rives, & Jorgensen, 2001) used in this study. Thus, the parameters of topologies and coordinates were obtained using the AnteChamber PYthon Parser InterfacE (AcPype) software (Sousa da Silva, & Vranken, 2012).
The HssTMPK/ligand and VarTMPK/ligand complexes were constructed within cubic boxes of approximately 450 nm3, containing around 13,000 Tip4P type water molecules and with periodic boundary conditions (PBCs), using the GROMACS 5.1.4 software (Abraham et al., 2015). These complexes were submitted to four steps of energy minimization, with the algorithms: (1) steepest descent with position restraint (PR) of the ligand; (2) steepest descent without PR; (3) conjugate gradients; and, lastly, (4) quasi Newton Broyden–Fletcher–Goldfarb–Shanno (L-BFGS) algorithm (Byrd, Lu, Nocedal, & Zhu, 1995), with a minimal energy of 1 kcal.mol−1.
After the energy minimization process, the systems underwent two temperature and pressure balancing phases in order to attain equilibrium. Temperature equilibrium was reached by using the NVT ensemble (constant volume and temperature) for 100 ps, while pressure equilibrium was reached with the NPT ensemble (constant pressure and temperature), also for 100 ps. Both phases kept their particle numbers fixed.
After attaining system equilibrium, the MD simulations were performed in two stages. Firstly, the complexes were submitted to 500 ps of MD at 310 K with PR for the entire system, except for water molecules, in order to ensure the equilibrium of solvent molecules around the enzyme residues. Lastly, the systems were submitted to 100,000 ps of MD simulation at 310 K without PR, with 2 fs of integration time, and a cutting radius of 10 Å for long-distance interactions. This study protocol was previously tested so as to ascertain that the proposed conditions would suffice for the conduction of the systems toward equilibrium. With the aim of reproducing the protonation of some residues under physiological conditions, the Glu and Asp residues were assigned with negative charges, and the Lys and Arg residues were assigned with positive charges.
The Visual Molecular Dynamics (VMD) software (Humphrey, Dalke, & Schulten, 1996) was used to analyze the MD results of the systems, and plots of variation of total energy, root-mean-square-deviation (RMSD) and average number of hydrogen bonds were drawn with the Origin® software (Edwards, 2002).
Determining free energy
The MM-PBSA method is employed to predict the free energy of binding processes, where the concepts of molecular mechanics and continuous solvent models are combined (Evertts, Zee, & Garcia, 2010; Genheden & Ryde, 2015; Kumari, Kumar, & Lynn, 2014; Luscombe, Austin, Berman, & Thornton, 2000; Norambuena & Melo, 2010).
The study of the free energy of a system is the measure of the amount of energy usable by this system, consisting of a process by which one can indicate, for instance, whether the formation of bonds in a complex at constant temperature and pressure is spontaneous (freeing energy) or nonspontaneous (requiring energy). Because of that, this phase is extremely important for the purposes of this study, since through the average values of free binding energy, we will be able to infer which prototype is more interesting and promising in relation to the process of selectively inhibiting VarTMPK.
Lastly, the determination of the free energy of formation of the complexes, in association to the MD simulations, takes into consideration three energetic terms in its calculation of binding energy: (1) changes in the potential energy of the system in vacuum; (2) polar and apolar solvation of the different species; and (3) the entropy related to the formation of the complexes during the gaseous phase (Almeida et al., 2018; Genheden & Ryde, 2015; Kumari et al., 2014).
For all the enzyme/ligand complexes, MM-PBSA calculations were performed using the g_mmpbsa tool (Kumari et al., 2014) from the GROMACS package. In order to consider non-correlated frames, the structures for the free energy calculations were obtained at 500 ps each.
Results and discussion
Docking studies
Prediction of the ionization states using Chemicalize (Swain, 2012) showed that only prototypes 2 and 3 are ionized under physiologic pH, while the others are neutral. The predicted species shown in Table 1 were, then, used in the docking studies inside VarTMPK and HssTMPK, complexed with TDP and Mg2+, which results are shown in Table 2. Chemicalize (Swain, 2012) results also showed that all prototypes meet the druggability criteria established by the Lipinski rule of five (Lipinski, 2004).
Table 1.
Molecular structures of ligands predicted through protonation analysis under physiologic pH (7.4).
| Prototype | Structurea | % of micro speciesa | Meet the druggability criteria of the Lipinski rule?b |
|---|---|---|---|
| 1 |
|
87.09 | Yes |
| 2 |
|
78.37 | Yes |
| 3 |
|
78.25 | Yes |
| 4 |
|
89.04 | Yes |
| 5 |
|
89.04 | Yes |
| 6 |
|
87.08 | Yes |
| 7 |
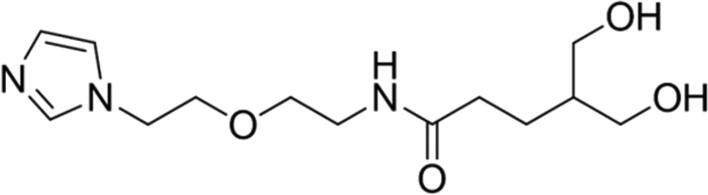
|
89.04 | Yes |
| 8 |
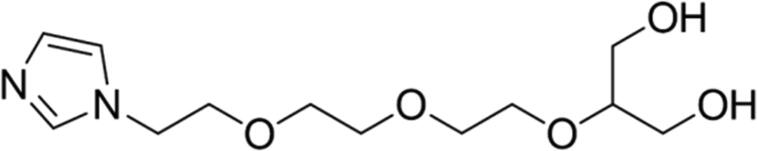
|
89.04 | Yes |
| 9 |
|
89.04 | Yes |
Calculated through Chemicalize web-based resource (https://chemicalize.com/welcome) (Swain, 2012) as the highest percentage of this micro species.
(Lipinski, 2004).
Table 2.
Docking results for the prototypes inside the active sites of VarTMPK and HssTMPK.
| Prototype |
Einteraction (kcal.mol−1) |
Ecofactor (kcal.mol−1) |
H-bond energy (kcal.mol−1) |
H-bond interactions |
ΔEinteractiona (kcal.mol−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VarTMPK | HssTMPK | VarTMPK | HssTMPK | VarTMPK | HssTMPK | VarTMPK | HssTMPK | ||||
| 1 | −151.60 | −124.33 | −1.03 | −2.07 | −13.57 | −10.44 | Arg41 Arg72 Tyr144 | Lys14 Glu142 H2O | Arg45 Arg97 | Arg76 H2O | 27.27 |
| 2 | −152.53 | −116.43 | −2.24 | −2.86 | −12.76 | −11.34 | Arg41 Tyr144 Arg72 | Glu145 Asp13 | Arg76 Ser101 Arg97 | Asp15 H2O | 36.10 |
| 3 | −123.68 | −62.92 | −5.60 | −0.08 | −8.90 | −5.51 | Asp13 Lys14 Arg72 | Tyr94 H2O | Pro43 Arg16 H2O | 60.76 | |
| 4 | −133.18 | −101.31 | −4.63 | −3.42 | −8.90 | −5.51 | Asp13 Lys14 Arg41 | Arg72 Ser97 H2O | Arg76 Ser101 | Arg97 H2O | 31.87 |
| 5 | −132.57 | −110.51 | −2.67 | −4.97 | −9.74 | −11.15 | Asp13 Lys14 Arg72 | Tyr144 H2O | Asp15 Arg97 | Arg76 H2O | 22.06 |
| 6 | −106.83 | −126.01 | −3.39 | −1.87 | −10.15 | −16.26 | Arg72 Asp92 Asn37 | Phe38 H2O | Arg76 Glu152 Asp15 | Arg97 H2O | −19.18 |
| 7 | −120.45 | −116.91 | −0.71 | −0.25 | −16.18 | −10.96 | Lys14 Arg72 Glu142 | Arg97 H2O | Arg76 Arg45 H2O | 3.54 | |
| 8 | −118.20 | −102.38 | −5.26 | −2.28 | −12.25 | −11.16 | Arg72 Phe38 Arg41 | Lys17 Asp13 H2O | Arg76 Asp15 | Arg97 H2O | 15.82 |
| 9 | −123.47 | −92.84 | −2.86 | −2.07 | −15.51 | −15.24 | Arg72 Phe38 Asp92 | Lys17 Asp13 | Arg76 Arg16 Asp15 | Arg97 H2O | 30.63 |
| TDPb | −228.18 | −195.60 | −50.38 | −18.13 | −3.82 | −13.97 | Asp13 Lys17 Asn37 | Arg41 Arg72 Arg93 | Asp15 Arg45 Arg76 | Arg97 H2O | – |
| Prototype Ab | −202.46 | −187.77 | −34.65 | −40.72 | −12.54 | −12.94 | Lys14 Lys17 Phe38 | Asp92 Arg93 H2O | Arg16 Lys19 Phe42 Arg76 | Arg97 Ser101 H2O | 14.69 |
| Prototype Bb | −222.55 | −161.09 | −38.43 | −27.72 | −18.71 | −13.16 | Asp13 Lys17 Thr18 Phe38 | Arg72 Asp92 Tyr94H2O | Asp15 Arg16 Lys19 Pro43 | Arg45 Arg76H2O | 61.46 |
Einteraction (VarTMPK) – Einteraction (HssTMPK).
Results from Guimarães et al., 2015.
As can be seen in Table 2, all compounds but prototype 6 presented lower energies of interaction with VarTMPK, in relation to HssTMPK. Besides, from the values of ΔEinteraction (Table 2), it is noticeable that prototypes 2, 3, 4 and 9 are the compounds with the higher energy differences between VarTMPK and HssTMPK. This suggests that they are the most selective compounds regarding VarTMPK. Therefore, these prototypes were selected for further MD studies.
It is important to mention that prototype 3 was the one with the greatest ΔEinteraction value (60.76 kcal.mol−1) and that it also presented the lowest affinity for HssTMPK, establishing hydrogen bonds only with residues Arg16 and Pro43, which are not part of the active site. This can also be related to the greater distance between the cofactor Mg2+ of HssTMPK and the carbonyl group of prototype 3 (4.87 Å), leading to an interaction energy with the cofactor (−0.08 kcal.mol−1) inferior to the one found for VarTMPK (−5.60 kcal.mol−1). These results point to prototype 3 as the most promising selective inhibitor, based on the docking studies. The best poses obtained for prototype 3 inside VarTMPK and HssTMPK are shown in Figure 3.
Figure 3.
Best molecular docking poses for prototype 3 inside (a) VarTMPK and (b) HssTMPK. Interacting residues are shown in different colors, and residues belonging to the active site are labeled in red..
MD studies
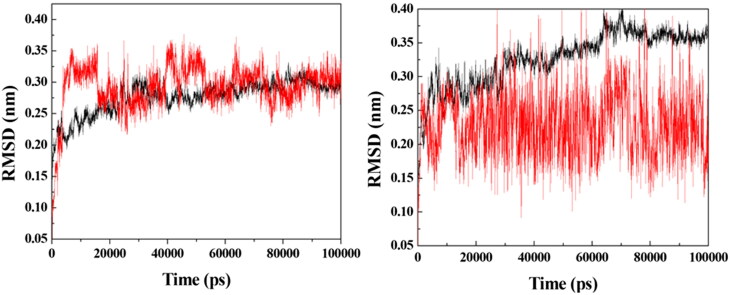
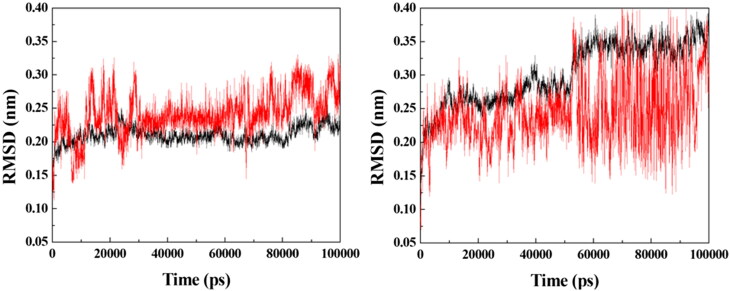
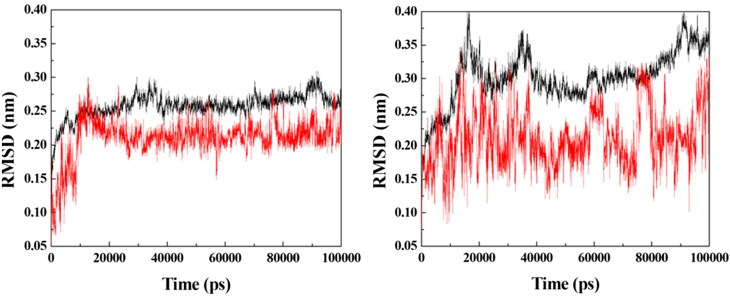
Based on the docking results, prototypes 2, 3, 4 and 9 were selected for additional MD simulation studies with the purpose of investigating their dynamic behaviors inside VarTMPK and HssTMPK. Energy plots of the simulations (data not shown) showed stabilization before 50 ns of simulation for all the systems studied. Next, RMSD analyses were performed for each system in order to verify and compare the variations of positions of the prototypes within the active sites of VarTMPK and HssTMPK and thus to determine the most promising prototypes as selective inhibitors on the basis of their dynamical behavior. The RMSD plots obtained are shown in Figures 4–7.
Figure 4.
RMSD for the systems formed between the enzymes (in black) and prototype 2 (in red). Left: VarTMPK/prototype 2; right: HssTMPK/prototype 2.
Figure 5.
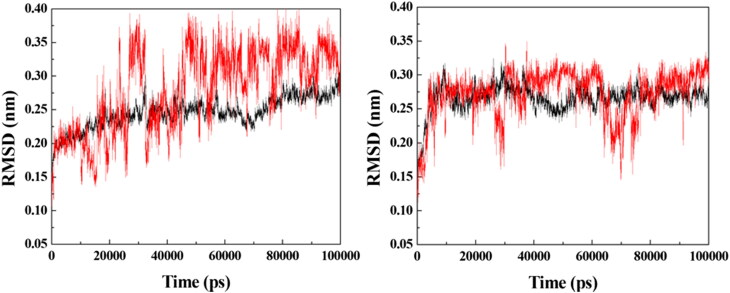
RMSD for the systems formed between the enzymes (in black) and prototype 3 (in red). Left: VarTMPK/prototype 3; right: HssTMPK/prototype 3.
Figure 6.
RMSD for the systems formed between the enzymes (in black) and prototype 4 (in red). Left: VarTMPK/prototype 4; right: HssTMPK/prototype 4.
Figure 7.
RMSD for the systems formed between the enzymes (in black) and prototype 9 (in red). Left: VarTMPK/prototype 9; right: HssTMPK/prototype 9.
As can be observed in Figure 4, the VarTMPK/prototype 2 and HssTMPK/prototype 2 complexes did not attain equilibrium during the entire simulation time. A large RMSD variation of more than 0.20 nm was observed for prototype 2, inside VarTMPK, since the beginning of the simulation, suggesting that this compound does not stabilize itself inside the enzyme. Regarding HssTMPK, the behavior of this prototype showed some stabilization during most of the simulation, but it also showed large variations around 30 ns and between 60 and 80 ns.
The RMSD plots for the VarTMPK/prototype 3 and HssTMPK/prototype 3 complexes (Figure 5) show that, inside VarTMPK, this prototype presented no variation larger than 0.10 nm, tending toward stabilization after 50 ns of simulation. Inside HssTMPK, on the other hand, one can see that this prototype could not achieve stability, with variations of around 0.30 nm, since the beginning of the simulation. This is indicative of the absence of stabilizing interactions inside HssTMPK, suggesting some selectivity toward VarTMPK.
Figure 6 shows the RMSD plots for the VarTMPK/prototype 4 and HssTMPK/prototype 4 complexes. Inside VarTPMK, this prototype shows some stabilization after 30 ns of simulation, but destabilizes again after 60 ns, until the end of the simulation, with variations close to 0.15 nm. Inside HssTMPK, this behavior was even more aggravated, with variations after 50 ns of around 0.25 nm, until the end of simulation. This result suggests that, despite having more affinity for VarTMPK, this prototype is unable to become stable inside neither of the enzymes during the simulated time.
The RMSD plots for the VarTMPK/prototype 9 and HssTMPK/prototype 9 complexes (Figure 7) show that this prototype stabilizes itself inside VarTMPK, starting around 15 ns of simulation, with variations close to 0.05 nm until the end of the simulation, while inside HssTMPK, stabilization never happens, with variations of around 0.20 nm since the beginning of the simulation.
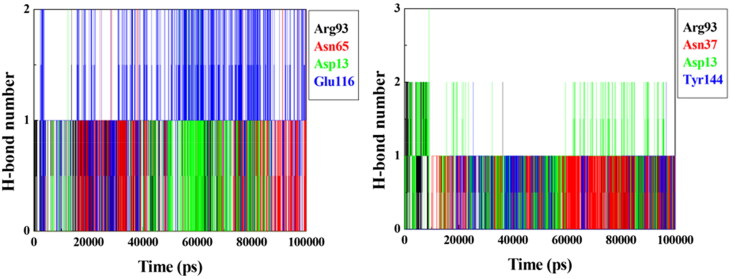
Results of the RMSD analysis suggest that, among the compounds chosen for the MD simulations, only prototypes 3 and 9 are possible inhibitors for VarTMPK once they were the only ones capable of stabilizing well inside the active site of the viral enzyme. These compounds also did not stabilize well inside HssTMPK, corroborating with the docking results pointing them as selective inhibitors. The plots of H-bonds observed for these prototypes during the 100 ns of MD simulations, shown in Figure 8, confirm the H-bond with Asp13 observed for both prototypes during the docking process. They also show additional H-bonds formed with residues Asn65, Arg93 and Glu116 for prototype 3, and with Asn37, Arg93 and Tyr144 for prototype 9. It is important to mention that Asp13, Asn65 and Arg93 are residues belonging to the active site of VarTMPK.
Figure 8.
Plots of mean H-bonds formed by prototypes 3 (left) and 9 (right) inside VarTMPK during the 100 ns of MD simulation.
Because the MD process is the assessment of the dynamic behavior of the ligands inside a complex, i.e. an analysis involving the movement of ligand and protein structures, its results may often not corroborate the docking results. According to Chen (2015), during an MD simulation, a ligand can leave the active site of the protein, which cannot be observed in a docking study due to the movement restriction imposed by this procedure. Therefore, our docking and MD simulation results are complimentary and helps to refine the investigation on the capacity of the prototypes to selectively inhibit VarTMPK.
Free energy determination
Table 3 shows the results of the MM-PBSA calculations for prototypes 3 and 9 inside VarTMPK and HssTMPK. As one can see, prototype 3 presented a binding energy of −142.22 kJ.mol−1 inside VarTMPK versus −68.77 kJ.mol−1 inside HssTMPK, while prototype 9 presented −29.16 kJ.mol−1 versus −53.79 kJ.mol−1.
Table 3.
MM-PBSA results for prototypes 3 and 9.
| Structures | Binding energies kJ.mol−1 |
|
|---|---|---|
| VarTMPK | HssTMPK | |
| 3 | −142.22 | −68.77 |
| 9 | −29.16 | −53.79 |
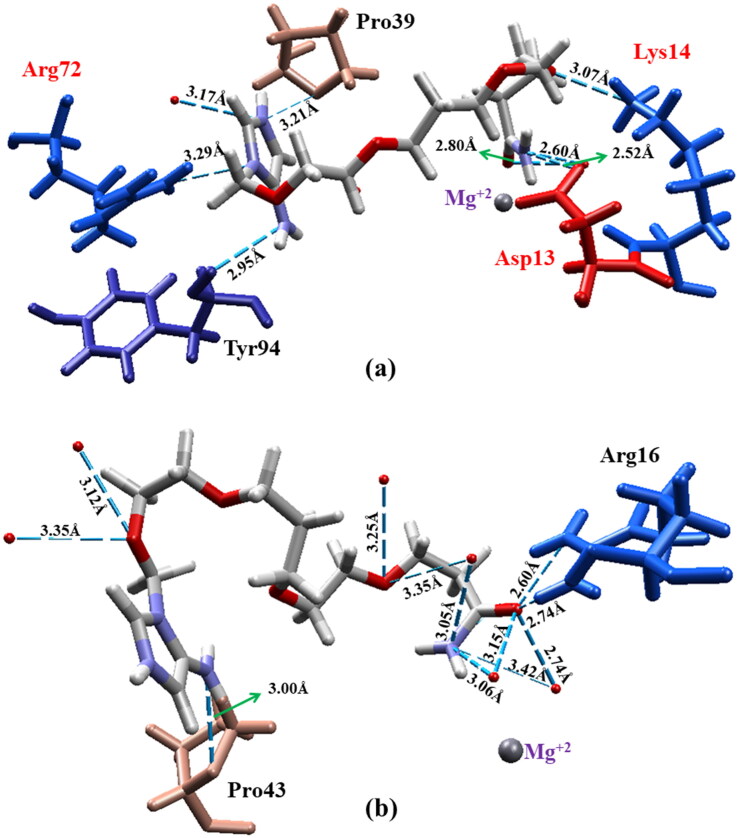
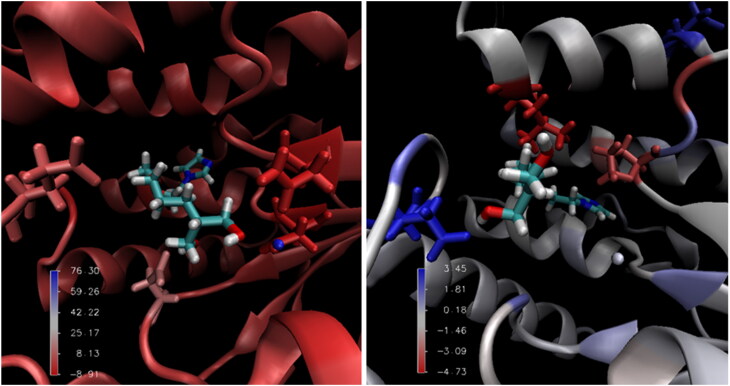
Figures 9 and 10 present an illustration of the favorable and unfavorable energetic contributions of some residues to the enzyme/ligand complexes for prototypes 3 and 9, respectively. Figure 9 shows that the residues that contributed to the binding energy of the HssTMPK/prototype 3 complex are located outside the active site of HssTMPK, indicating that prototype 3 did not become stable within it, remaining outside the active site.
Figure 9.
Main interactions of the complexes of VarTMPK/prototype 3 (left) and HssTMPK/prototype 3 (right) calculated by MM-PBSA.
Figure 10.
Main interactions of the complexes of VarTMPK/prototype 9 (left) and HssTMPK/prototype 9 (right) calculated by MM-PBSA.
The behavior demonstrated by each compound during the MD simulations allowed us to assess the affinity of the enzymes toward the structures; however, the calculation of the free binding energy permitted the assessment of the differences in affinity among the species integrating the complexes. The differences in interaction energy show that, very probably, the quaternary nitrogen in prototype 3 is of great importance to its affinity for the viral enzyme.
Despite the favorable MD results for the VarTMPK/prototype 9 complex, the binding energy calculations showed that prototype 9 did not demonstrate enough affinity toward the interaction site of the viral enzyme to remain stable, as presented in Table 3. Such instability is probably related to the fact that prototype 9 does not contain the imidazole ring on its protonated form neither the –NH2 substituent in the imidazole ring, in comparison with prototype 3. The –NH2 group showed in the docking studies to be able to establish H-bonds inside VarTMPK, while the positive charge in the imidazole ring helps the molecule to stabilize inside the more negative pocket of VarTMPK. The absence of these groups in the structure can hamper interactions with the amino acids in the protein, destabilizing prototype 9 inside the viral enzyme.
Conclusion
From the two selective inhibitors for VarTMPK proposed by Guimarães et al (2015), we were able to design structures of nine new prototypes that are much simpler from a synthetic point of view, but still able to keep effective energy values in interactions with VarTMPK and HssTMPK. Docking studies pointed to prototypes 2, 3, 4, and 9 as potential selective inhibitors of VarTMPK, with prototype 3 as the most promising. In the MD simulations, prototypes 3 and 9 demonstrated the highest stabilities inside VarTMPK, with the smallest RMSD variations. However, the MM-PBSA results did not corroborate prototype 9 as a selective inhibitor of VarTMPK. We believe that this is due to the fact that prototype 9 is neutral under physiological conditions and does not contain the –NH2 substituent in the imidazole ring. This could compromise its stabilization inside VarTMPK. Among the structural modifications proposed on prototypes A and B from Guimarães et al. (2015), we observed that the simplification of the guanine to an imidazole ring should not compromise the affinity of the ligand for VarTMPK, since a H-bond donor group, like an –NH2, is kept as substituent in the imidazole ring. It was also observed that a positive charge in the imidazole ring is important for the selectivity toward VarTMPK. However, the inclusion of an amide group in the middle or the extremity of the spacer was not observed to contribute for any selectivity. Based on our results, we believe that prototype 3 has a great potential as a selective inhibitor of VarTMPK and, therefore, a new drug against smallpox. In order to validate and scale up this work, we think that it is worth synthesizing and experimentally evaluating this molecule.
Funding Statement
The authors are grateful to the Brazilian agencies CNPq (grant 306156/2015-6 – T.C.C.F.) and FAPERJ (grant number 02/202.961/2017 – T.C.C.F.), for financial support and to the Military Institute of Engineering, Federal University of Lavras, Federal University of Viçosa and University of Hradec Kralové for the infrastructure. This work was also supported by the excelence FIM project.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abraham, M. J., Murtola, T., Schulz, R., Pall, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X, 1-2,19–25. doi: 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Almeida, J. S. D., Guizado, T. R. C., Guimarães, A. P., Ramallho, T. C., Goncalves, A. S., Koning, M. C., & Franca, T. C. C. (2015). Docking and molecular dynamics studies of peripheral site ligand-oximes as reactivators of sarin-inhibited human acetylcholinesterase. Journal of Biomolecular Structure and Dynamics, 34,2632–2642. doi: 10.1080/07391102.2015.1124807 [DOI] [PubMed] [Google Scholar]
- Almeida, J. S. F. D., Cavalcante, S. F. A., Dolezal, R., Kuca, K., Musilek, K., Jun, D., & Franca, T. C. C. (2018). Molecular modeling studies on the interactions of aflatoxin B1 and its metabolites with the peripheral anionic site (PAS) of human acetylcholinesterase. Journal of Biomolecular Structure and Dynamics, 1–8 [published online]. doi: 10.1080/07391102.2018.1475259 [DOI] [PubMed] [Google Scholar]
- Bastos, L. C., Souza, F. R., Guimarães, A. P., Sirouspour, M., Guizado, T. R. C., Forgione, P., … Franca, T. C. C. (2016). Virtual screening, docking and dynamics of potential new inhibitors of dihydrofolate reductase from Yersinia pestis. Journal of Biomolecular Structure and Dynamics, 34(10),2184–2198. 2016. doi: 10.1080/07391102.2015.1110832 [DOI] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Van Der Spoel, D., & Van Drunen, R. (1995). GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications, 91(1-3),43–56. doi: 10.1016/0010-4655(95)00042-E [DOI] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliand, G., Bhat, T. N., Weissig, H., … Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research, 28(1),235–242. doi: 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bismuth, C., Borron, S. W., Baud, F. J., & Barriot, P. (2004). Chemical weapons: Documented use and compounds on the horizon. Toxicology Letters, 149(1-3),11–18. doi: 10.1016/j.toxlet.2003.12.016 [DOI] [PubMed] [Google Scholar]
- Byrd, R. H., Lu, P., Nocedal, J., & Zhu, C. (1995). A limited memory algorithm for bound constrained optimization. SIAM Journal on Scientific Computing, 16(5),1190–1208. doi: 10.1137/0916069 [DOI] [Google Scholar]
- Byrnes, M. E., King, D. A., & Junior, P. M. T. (2003). Nuclear, chemical, and biological terrorism: Emergency response and public protection. Boca Raton, FL: CRC Press. [Google Scholar]
- Caillat, C., Topalis, D., Agrofoglio, L. A., Pochet, S., Balzarini, J., Deville-Bonne, D., & Meyer, P. (2008). Crystal structure of poxvirus thymidylate kinase: An unexpected dimerization has implications for antiviral therapy. Proceedings of the National Academy of Sciences of the United States of America, 105(44),16900–16905. doi: 10.1073/pnas.0804525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, J. L., Nichols, D. K., Martinez, M. J., & Raymond, J. W. (2010). Animal models of orthopoxvirus infection. Veterinary Pathology, 47(5),852–870. doi: 10.1177/0300985810378649 [DOI] [PubMed] [Google Scholar]
- Chaudhuri, S., Symons, J. A., & Deval, J. (2018). Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antiviral Research, 155,76–88. doi: 10.1016/j.antiviral.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-C. (2015). Beware of docking!. Trends in Pharmacological Sciences, 36(2),78–95. doi: 10.1016/j.tips.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Chittick, G., Morrison, M., Brundage, T., & Nichols, W. G. (2017). Short-term clinical safety profile of brincidofovir: A favorable benefit risk proposition in the treatment of smallpox. Antiviral Research, 143,269–277. doi: 10.1016/j.antiviral.2017.01.009 [DOI] [PubMed] [Google Scholar]
- Crump, R., Korom, M., Buller, R. M., & Parker, S. (2017). Buccal viral DNA as a trigger for brincidofovir therapy in the mousepox model of smallpox. Antiviral Research, 139,112–116. doi: 10.1016/j.antiviral.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon, I. K., Damaso, C. R., & Mcfadden, G. (2014). Are we there yet? The smallpox research agenda using variola virus. PLoS Pathogens, 10,1–3. doi: 10.1371/journal.ppat.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, R. J., Satchell, M., & Shaw-Taylor, L. M. W. (2018). The geography of smallpox in England before vaccination: A conundrum resolve. Social Science & Medicine, 206,75–85. doi: 10.1016/j.socscimed.2018.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M. (2002). Origin 7.0: Scientific graphing and data analysis software. Journal of Chemical Information and Computer Sciences, 42,1270–1271. doi: 10.1021/ci0255432 [DOI] [Google Scholar]
- Evertts, A. G., Zee, B. M., & Garcia, B. A. (2010). Modern approaches for investigating epigenetic signaling pathways. Journal of Applied Physiology, 109(3),927–933. doi: 10.1152/japplphysiol.00007.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5),449–461. doi: 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves, A. S., Franca, T. C. C., Villar, J. D. F., & Pascutti, P. G. (2010). Conformational analysis of toxogonine, TMB-4 and HI-6 using PM6 and RM1 methods. Journal of the Brazilian Chemical Society, 21(1),179–184. doi: 10.1590/S0103-50532010000100025 [DOI] [Google Scholar]
- Greenberg, R. N., Overton, E. T., Haas, D. W., Frank, I., Goldman, M., von Krempelhuber, A., … Chaplin, P. (2013). Safety, immunogenicity, and surrogate markers of clinical efficacy for modified vaccinia Ankara as a smallpox vaccine in HIV-infected subjects. The Journal of Infectious Diseases, 207(5),749–758. doi: 10.1093/infdis/jis753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosenbach, D. W., Honeychurch, K., Rose, E. A., Chinsangaram, J., Frimm, A., Maiti, B., … Hruby, D. E. (2018). Oral tecovirimat for the treatment of smallpox. New England Journal of Medicine, 379(1),44–53. doi: 10.1056/NEJMoa1705688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi, I. M., Foster, S. A., Gainey, M. R., Krile, R. T., Dunn, J. A., Brundage, T., & Khouri, J. M. (2017). Efficacy of delayed brincidofovir treatment against a lethal rabbitpox virus challenge in New Zealand White rabbits. Antiviral Research, 143,278–286. doi: 10.1016/j.antiviral.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18(15),2714–2723. doi: 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Guimarães, A. P., de Souza, F. R., Oliveira, A. A., Gonçalves, A. S., de Alencastro, R. B., Ramalho, T. C., & França, T. C. C. (2015). Design of inhibitors of thymidylate kinase from Variola virus as new selective drugs against smallpox. European Journal of Medicinal Chemistry, 91,72–90. doi: 10.1016/j.ejmech.2014.09.099 [DOI] [PubMed] [Google Scholar]
- Guimarães, A. P., Ramalho, T. C., & França, T. C. C. (2014). Preventing the return of smallpox: Molecular modeling studies on thymidylate kinase from Variola virus. Journal of Biomolecular Structure and Dynamics, 32(10),1601–1612. doi: 10.1080/07391102.2013.830578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund, E., Lewis, M. W., Hanifin, J. M., Mori, M., Koudelka, C. W., & Slifka, M. K. (2010). Antiviral immunity following smallpox virus infection: A case–control study. Journal of Virology, 84(24),12754–12760. doi: 10.1128/JVI.01763-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, R. C., Wang, C., Hatcher, E. L., & Lefkowitz, E. J. (2010). Orthopoxvirus genome evolution: The role of gene loss. Viruses, 2(9),1933–1967. doi: 10.3390/v2091933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics and Modelling, 14(1),33–38. doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jayaram, B., Sprous, D., Young, M. A., & Beveridge, D. L. (1998). Free energy analysis of the conformational preferences of A and B forms of DNA in solution. Journal of the American Chemical Society, 120(41),10629–10633. doi: 10.1021/ja981307p [DOI] [Google Scholar]
- Kaminski, G. A., Friesner, R. A., Tirado-Rives, J., & Jorgensen, W. L. (2001). Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. The Journal of Physical Chemistry B, 105(28),6474–6487. doi: 10.1021/jp003919d [DOI] [Google Scholar]
- Kar, P., Lipowsky, R., & Knecht, V. (2013). Importance of polar solvation and configurational entropy for design of antiretroviral drugs targeting HIV-1 protease. The Journal of Physical Chemistry B, 117(19),5793–5805. doi: 10.1021/jp3085292 [DOI] [PubMed] [Google Scholar]
- Kennedy, R. B., Ovsyannikova, I. G., Jacobson, R. M., & Poland, G. A. (2009). The immunology of smallpox vaccines. Current Opinion in Immunology, 21(3),314–320. doi: 10.1016/j.coi.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, R. B., Poland, G. A., Ovsyannikova, I. G., Oberg, A. L., Asmann, Y. W., Grill, D. E., … Jacobson, R. M. (2016). Impaired innate, humoral, and cellular immunity despite a take in smallpox vaccine recipients. Vaccine, 34(28),3283–3290. doi: 10.1016/j.vaccine.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A. (2014). G_mmpbsa: A GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7),1951–1962. doi: 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lee, J., Kumar, S. A., Jhan, Y. Y., & Bishop, C. J. (2018). Engineering DNA vaccines against infectious diseases. Acta Biomaterialia, 80,31–47. doi: 10.1016/j.actbio.2018.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindler, L. E., Lebeda, F. J., & Korch, G. W. (2004). Biological weapons defense: Infectious diseases and counter bioterrorism. New York, NY: Humana Press. [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today: Technologies, 1(4),337–341. doi: 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Liszewski, M. K., Leung, M. K., Hauhart, R., Buller, R. M. L., Bertram, P., Wang, X., … Atkinson, J. P. (2006). Structure and regulatory profile of the monkeypox inhibitor of complement: Comparison to homologs in Vaccinia and variola and evidence for dimer formation. Journal of Immunology, 176(6),3725–3734. doi: 10.4049/jimmunol.176.6.3725 [DOI] [PubMed] [Google Scholar]
- Luscombe, N. M., Austin, S. E., Berman, H. M., & Thornton, J. M. (2000). An overview of the structures of protein–DNA complexes. Genome Biology, 1(1), reviews001. doi: 10.1186/gb-2000-1-1-reviews001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manus, J.-M. (2018). La FDA approuve le premier traitement de la variole arme de guerre. Revue Francophone Des Laboratoires, 2018,11. doi: 10.1016/S1773-035X(18)30230-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena, T., & Melo, F. (2010). The protein–DNA interface database. BMC Bioinformatics, 11,261. doi: 10.1186/1471-2105-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman, P. R., Garman, P. M., Kim, S.-H., Schmader, T. J., Nieding, W. J., Pike, J. G., … Meyers, M. S. (2015). Smallpox vaccine, ACAM2000: Sites and duration of viral shedding and effect of povidone iodine on scarification site shedding and immune response. Vaccine, 33(26),2990–2996. 2015. doi: 10.1016/j.vaccine.2015.04.062 [DOI] [PubMed] [Google Scholar]
- Prichard, M. N., & Kern, E. R. (2012). Orthopoxvirus targets for the development of new antiviral agents. Antiviral Research, 94(2),111–125. doi: 10.1016/j.antiviral.2012.02.012. doi: 10.1016/j.antiviral.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., … Lindahl, E. (2013). GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics, 29(7),845–854. doi: 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, G. B., Freire, R. O., Simas, A. M., & Stewart, J. J. P. (2006). RM1: A reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. Journal of Computational Chemistry, 27(10),1101–1111. doi: 10.1002/jcc.20425 [DOI] [PubMed] [Google Scholar]
- Sakhatskyy, P., Wang, S., Zhang, C., Chou, T.-H., Kishko, M., & Lu, S. (2008). Immunogenicity and protection efficacy of subunit-based smallpox vaccines using Variola major antigens. Virology, 371(1),98–107. doi: 10.1016/j.virol.2007.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y., Molnar, L. F., Jung, Y., Kussmann, J., Ochsenfeld, C., Brown, S. T., … Head-Gordon, M. (2006). Advances in methods and algorithms in a modern quantum chemistry program package. Physical Chemistry Chemical Physics, 8(27),3172–3319. doi: 10.1039/B517914A [DOI] [PubMed] [Google Scholar]
- Silva, A. W. S., & Vranken, W. F. (2012). ACPYPE – AnteChamber Python Parser interfacE. BMC Research Notes, 5,367. doi: 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, F. R., Guimarães, A. P., Cuya, T., Freitas, M. P., Gonçalves, A. S., Forgione, P., & França, T. C. C. (2017). Analysis of Coxiela burnetti dihydrofolate reductase via in silico docking with inhibitors and molecular dynamics simulation. Journal of Biomolecular Structure and Dynamics, 35(13),2975–2986. doi: 10.1080/07391102.2016.1239550 [DOI] [PubMed] [Google Scholar]
- Swain, M. (2012). Chemicalize.org. Journal of Chemical Information and Modeling, 52(2),613–615. doi: 10.1021/ci300046g [DOI] [Google Scholar]
- Taylor, C. L., & Junior, L. B. T. (1992). Chemical and biological warfare. New York, NY: Franklin Watts. [Google Scholar]
- Thomsen, R., & Christensen, M. H. (2006). MolDock: A new technique for high- accuracy molecular docking. Journal of Medicinal Chemistry, 49(11),3315–3321. doi: 10.1021/jm051197e [DOI] [PubMed] [Google Scholar]
- Trost, L. C., Rose, M. L., Khouri, J., Keilholz, L., Long, J., Godin, S. J., & Foster, S. A. (2015). The efficacy and pharmacokinetics of brincidofovir for the treatment of lethal rabbitpox virus infection: A model of smallpox disease. Antiviral Research, 117,115–121. doi: 10.1016/j.antiviral.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Vorobjev, Y. N., Almagro, J. C., & Hermans, J. (1998). Discrimination between native and intentionally misfolded conformations of proteins: ES/IS, a new method for calculating conformational free energy that uses both dynamics simulations with an explicit solvent and an implicit solvent continuum model. Proteins: Structure, Function, and Genetics, 32(4),399–413. doi: [DOI] [PubMed] [Google Scholar]
- Wang, C., Greene, D., Xiao, L., Qi, R., & Luo, R. (2018). Recent developments and applications of the MMPBSA method. Frontiers in Molecular Biosciences, 4,1–18. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]