SUMMARY
Negative-sense RNA virus populations are composed of diverse viral components that interact to form a community and shape the outcome of virus infections. At the genomic level, RNA virus populations consist not only of a homogeneous population of standard viral genomes but also of an extremely large number of genome variants, termed viral quasispecies, and nonstandard viral genomes, which include copy-back viral genomes, deletion viral genomes, mini viral RNAs, and hypermutated RNAs. At the particle level, RNA virus populations are composed of pleomorphic particles, particles missing or having additional genomes, and single particles or particle aggregates. As we continue discovering more about the components of negative-sense RNA virus populations and their crucial functions during virus infection, it will become more important to study RNA virus populations as a whole rather than their individual parts. In this review, we will discuss what is known about the components of negative-sense RNA virus communities, speculate how the components of the virus community interact, and summarize what vaccines and antiviral therapies are being currently developed to target or harness these components.
KEYWORDS: negative-sense RNA virus, virus community, viral quasispecies, nonstandard viral genomes, copy-back viral genomes, deletion viral genomes, filamentous particles, semi-infectious particles, multiple genome packaging, virion aggregation, antiviral immunity, copy-backs, virus
INTRODUCTION
Decades of research have enhanced our understanding of RNA virus biology and provided a solid ground for the development of therapies to fight important human pathogens and reduce disease burden. However, the diversity within a virus population and how this diversity shapes the outcome of RNA virus infections is often overlooked. As molecular virology methods have improved over the years, we are beginning to appreciate how incredibly diverse RNA virus populations are in vitro and in the clinic. RNA virus populations consist not only of identical viral genomes packaged in uniform particles, but also of large numbers of genome variants and nonstandard viral genomes packaged in pleomorphic particles that may contain several genomes. Although many studies have focused on understanding the role these viral components play during infection, they have done so by looking at these roles individually and not as a whole virus community. We propose that by integrating the role these various viral components have during infection, we can enhance our understanding of how RNA viruses establish infections, promote transmission to other hosts, and are maintained within the host population. In this review, we will summarize the functions of each component of the population, speculate how all components could be acting together as a community, and discuss how we could manipulate this community to our benefit. Although diversity in virus populations has been described to exist in many virus families, we will restrict this review to discussing negative-sense RNA viruses.
DIVERSITY OF VIRAL GENOMES WITHIN NEGATIVE-SENSE RNA VIRUS POPULATIONS
Negative-sense RNA viruses generate a diverse population of viral genomes (Fig. 1). This population can consist of replication-competent standard viral genome variants that differ from the consensus sequence only by a few nucleotides and can include nonstandard viral genomes which are viral RNAs unable to complete full viral life cycles in the absence of a standard viral genome. Nonstandard viral genomes include copy-back viral genomes, deletion viral genomes, mini viral RNAs (mvRNAs), and hypermutated RNAs. In this section, we will review how diversity at the genome level within negative-sense RNA virus populations can affect replication, host interactions, antiviral responses, and infection outcomes.
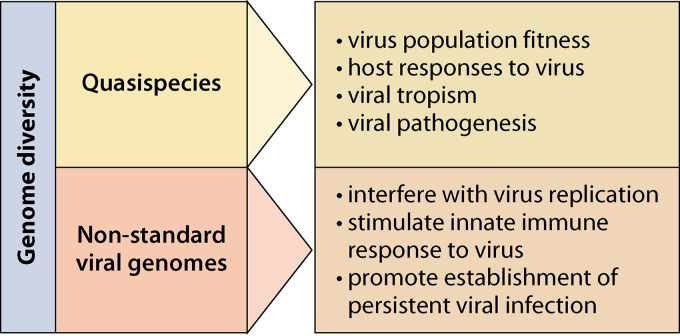
FIG 1.
Functions of diverse viral genomes found in negative-sense RNA virus populations. Overview of the known effects of quasispecies and nonstandard viral genomes, such as copy-back viral genomes, deletion viral genomes, mini viral RNAs, and hypermutated RNAs, on the standard virus, host cells, and pathogenesis.
Viral Quasispecies
A common component of RNA virus populations is viral quasispecies, which consist of closely related but nonidentical standard viral genome variants, also referred to as mutant spectra, mutant swarms, or mutant clouds (1, 2). Viral quasispecies result from the high error rate and lack of proofreading capacity of the RNA-dependent RNA polymerase encoded by RNA viruses (3, 4). Studies over several decades have established that the fitness of viral populations is often not determined by the predominant genome variant but by viral quasispecies, underpinning the influence of nonpredominant genome variants on viral populations. Viral quasispecies can shape infection outcomes and affect viral fitness through two types of interactions between variants: (i) negative interactions, such as interference/suppression and (ii) positive interactions, such as complementation or cooperation (1, 2).
Early studies using vesicular stomatitis virus (VSV) show that variants that replicate worse at an individual level can suppress variants that replicate better at a population level, demonstrating that variants can affect each other’s fitness within a virus population (5). Additionally, negative interactions can also impact host responses, as it is in the case of VSV, where viral populations that do not induce interferons (IFNs) can contain IFN-inducing VSV variants. These data suggest that non-IFN-inducing VSV variants suppress IFN-inducing VSV variants (6). Negative interactions between variants in a population can also drive disease outcome, as it is in the case of lymphocytic choriomeningitis virus (LCMV) (7). LCMV infections can lead to growth hormone deficiency syndrome or persistent infections (8, 9). Although LCMV populations will usually contain variants that are more pathogenic, they will only cause severe disease when they are the predominant variant within the population (9, 10). These data show that negative-sense RNA viruses can maintain pathogenic variants within virus populations without causing disease. Collectively, these studies suggest that viral quasispecies keep pathogenic variants in check, although one could imagine that transmission bottlenecks could lead to stochastic selection for pathogenic variants. Why a virus population would maintain a pathogenic variant is not known, but it suggests that pathogenic variants could provide an advantage to the virus population.
Positive interactions between variants within viral populations were also described for negative-sense RNA viruses. A measles virus (MeV) study showed that two different MeV variants that cannot mediate membrane fusion individually can do so when present together (11). Interestingly, these genomes were both packaged in the same viral particle, suggesting that, in addition to a cooperative mechanism at the genome level, there is cooperation at the particle level (discussed later) (11). Cooperation between two H3N2 influenza variants was also shown to increase viral population fitness (12). This increase in fitness was attributed to one variant improving cell entry, whereas the other variant improved budding from the cell. The presence of both variants in the viral population led to more efficient viral growth in cell culture. These data demonstrate that cooperation in the presence of variants conferring fitness at different stages of the virus life cycle is important for successful infections.
Viral quasispecies also allow for quick adaptation of the virus to new environments. This has been shown, for example, during MeV infections. MeV is known for initiating replication in lymphatic tissues and later transitioning to epithelial cells. This transition in tropism is driven by a switch from variants deficient in the V protein dominating the infection to V-competent variants (13). Similar results were observed with rabies virus, where certain variants accumulated when the virus was passaged in kidney cells, whereas others accumulated when the virus was passaged in brain cells (14). This switch of variant accumulation in different host environments could be a mechanism that allows negative-sense RNA viruses to adapt and overcome barriers that affect spread and overall survival, both within the host and between hosts. Viral quasispecies could therefore serve as a “toolbox” consisting of a large group of variants necessary for adaptation to the different environments a virus might encounter.
Copy-Back and Deletion Viral Genomes
Negative-stranded RNA virus populations also consist of nonstandard viral genomes, viral RNAs unable to complete full viral life cycles in the absence of a standard viral genome. The most studied nonstandard viral genomes are copy-back viral genomes (cbVGs) and deletion viral genomes (delVGs) (15, 16). cbVGs are generated when the polymerase detaches from the template and resumes elongation downstream by copying the 5′ end of the nascent strand (17). Some cbVGs consist of complementary regions representing almost the entire sequence, sometimes referred to as snap-back viral genomes (15, 16). delVGs have internal truncations while retaining the viral 3′ and 5′ untranslated regions (17). Interestingly, mosaic nonstandard viral genomes consisting of combinations of cbVGs and delVGs have been reported (18). Viral particles containing cbVGs and delVGs were first termed defective interfering particles (DIPs) due to their observed interference with the replication of the standard virus. cbVGs and delVGs have also received other names through the years, including defective interfering RNA or defective viral genomes (DVGs) (19). Although cbVGs have only been identified in negative-sense RNA viruses, delVGs can be generated by both negative- and positive-stranded RNA viruses, double-stranded RNA viruses, and retroviruses (17). Within the negative-sense RNA virus phylum, cbVGs and delVGs have been identified in every order (recently reviewed in reference 17) and are continually being identified in additional negative-sense RNA viruses (20–24). The presence and maintenance of the nonstandard viral genome population within so many RNA viruses, along with the functions that have been attributed to nonstandard viral genomes (discussed below), suggests that they play a pivotal role during RNA virus infections.
Early studies identified cbVGs and delVGs by passaging viruses multiple times at a high multiplicity of infection (MOI) in vitro. cbVGs and delVGs were later detected in vaccines and natural infections, demonstrating that, contrary to what was previously thought, they are not cell culture artifacts (23, 25–33). Furthermore, the presence of identical delVGs in individuals in close contact suggests that delVGs are transmitted (31). In turkeys, transmission efficiency of influenza A virus (IAV) is affected by delVGs (34). Although it is well established that high replication levels are associated with accumulation of cbVGs and delVGs, the specific mechanism that leads to this accumulation is unknown. Interestingly, delVGs have been identified in negative-sense segmented viruses such as arenaviruses and orthomyxoviruses, while cbVGs are observed in negative-sense nonsegmented viruses of the order Mononegavirales (17). These differences in the class of nonstandard viral genomes detected in different RNA viruses may reflect a natural preference by different virus families to produce one or another type of nonstandard viral genomes. However, better analysis of the whole population of nonstandard viral genomes using unbiased tools is needed to support this claim, given that most of the work reported focuses on one type of nonstandard viral genome or another. Regardless, the reason for and the consequences of generating either predominantly cbVGs or delVGs, or a combination of both, are unknown. Moreover, segmented negative-sense RNA viruses such as IAV preferentially generate delVGs in specific segments, particularly PB2, PB1, and PA, and the reason and consequence of this selectivity remain unclear (35–37). There can also be differences in the deleted segments of IAV when comparing intracellular versus supernatant samples, suggesting that differences in the packaging of delVGs can affect the detection of nonstandard viral genomes (38).
Nonstandard viral genomes have three well-described functions: interfering with the standard virus replication, stimulating the innate immune response, and promoting the establishment of persistent infections (17). Interference with the standard virus replication was demonstrated in studies that compare virus titers of stocks with or without nonstandard viral genomes. These studies observed a reduction in the titer of the standard virus primarily during infections in the presence of cbVGs and delVGs. This reduction in virus titer is thought to occur due to competition for the replication machinery among standard and nonstandard viral genomes (39–43). Nonstandard viral genomes are usually shorter than the standard viral genome, and in the case of cbVGs, their flanking trailer promoters have a higher affinity for the viral polymerase than the leader promoter present at the 3′ end of the standard genome, thereby providing a replication advantage to cbVGs (44, 45). In segmented negative-sense RNA viruses, such as IAV, it has been shown that specific noncoding sequences on each segment lead to a segment-specific competition for packaging between delVGs and standard segments (e.g., PA delVGs interfere with PA segments) (46). These results imply that packaging of IAV viral RNA is not random and shows that delVGs have an impact intracellularly and influence transmission as they affect the content of viral particles (discussed later). It is possible that generation and packaging of certain IAV delVGs attenuates the virus, which increases its ability to persist in the host, or it could affect reassortment with other IAV strains. These mechanisms remain to be studied.
Nonstandard viral genomes also potently induce the antiviral response in cells, contributing to the replication interference observed during infections with high contents of nonstandard viral genomes (47). Multiple studies have consistently demonstrated that the IFN response is induced when cells are infected with virus stocks that contain high levels of nonstandard genomes. This has been especially true for negative-sense RNA viruses that generate cbVGs, such as human metapneumovirus (HMPV) (48), Sendai virus (SeV) (49, 50), respiratory syncytial virus (RSV) (47), parainfluenza virus 5 (51, 52), MeV (53), and VSV (54, 55). The dependence on cbVGs to trigger the antiviral response was also observed in mice and in humans (47, 56, 57). The immunostimulatory ability of cbVGs also leads to the activation of dendritic cells, a crucial player in linking innate and adaptive immune responses, further establishing the role of cbVGs in driving not only the host innate immune response but also the adaptive immune response to the virus (53, 57, 58). For example, SeV cbVGs enhance activation of T cells by increasing the antigen presentation capacity of dendritic cells (59). A role for IAV delVGs in inducing a potent adaptive immune response has also been suggested (60, 61).
In both cells and mice infected with stocks containing high levels of nonstandard genomes, nonstandard genomes are protective by inducing a robust immune response, which leads to faster control of the virus (47, 56, 62). This effect was also observed in patients infected with IAV, where strains that generate fewer delVGs were found to be more pathogenic (32). However, our own study of RSV revealed that disease outcome in patients depends on when and how long cbVGs are present (33). If cbVGs were present early after infection (such as with in vitro and mouse experiments when virus stocks with high levels of cbVGs are used), we observed reduced viral titers and better clinical outcomes (33). However, if cbVGs were present late after infection or over several days, we observed higher viral loads, overexpression of many inflammatory genes, and worse outcomes in patients (33). Because the latter patients were symptomatic and sicker for longer, one could speculate that late and sustained generation of cbVGs might be advantageous for the virus, as it has more time to establish the infection within the host, transmit to other people, and potentially persist within the host.
The immunostimulatory ability of cbVGs and delVGs is dependent on triggering the intracellular viral sensors Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), as inhibition of melanoma differentiation-associated protein 5 (MDA5) (57), RIG-I (63–65), or mitochondrial antiviral-signaling protein (MAVS) (47, 63) inhibits induction of IFN in infections containing nonstandard viral genomes. Direct interaction of cbVGs with RIG-I was shown for SeV, MeV, and VSV (63, 66–68). MeV cbVGs were also shown to interact with Laboratory of Genetics and Physiology 2 (LGP2), another RLR, and PACT, a double-stranded RNA-binding protein that functions as a cellular activator of RIG-I (65, 69). Initially, the double-stranded RNA (dsRNA) part (stem) of cbVGs was thought to be critical for the immunostimulatory ability of cbVGs. However, this has been disproved by the lack of dsRNA detection during SeV infections, along with unaffected immunostimulatory activity when SeV cbVGs lacking the complementary ends are transfected into cells, suggesting the dsRNA stem of cbVGs does not play a major role in stimulating innate immune responses (54, 63, 67, 70, 71). Of note, the 5′ triphosphate RNA of SeV cbVGs is important for RIG-I activation, as SeV cbVGs treated with phosphatase lose their immunostimulatory potential (56). This is consistent with data demonstrating that 5′ triphosphate RNA is a ligand for RIG-I and is crucial to induce type I IFN signaling (72, 73). Moreover, it was shown that in a strongly stimulatory SeV cbVG, a unique stem-loop secondary structure that is formed where the poylmerase detaches and re-attaches facilitates its binding to RIG-I and MDA5, inducing their polymerization and signaling. Activation of RIG-I and MDA5 occurs even in the absence of complementary ends but requires the 5′ triphosphate for potent immune stimulation (63).
cbVGs from many negative-sense RNA viruses, including VSV, HMPV, and MeV, were found to be highly edited (48, 74–76). How this editing affects cbVGs’ immunostimulatory ability and which host proteins edit the RNA is not fully understood. Changes from adenosine (A) to inosine (I), which are made by the host protein adenosine deaminases acting on RNA 1 (ADAR1), are often found in cbVGs, suggesting a role of ADAR1 in editing cbVGs (48, 74–76). A direct interaction of cbVGs with ADAR1 has not been shown yet, but a recent study showed that MeV cbVGs were more edited after replication in ADAR1 expressing than in ADAR1 KO cells (76). Although RNA editing by ADAR1 was shown to have both proviral and antiviral effects (77), how ADAR1 editing of cbVGs could affect cbVGs’ functions remains unknown. It is possible that ADAR1 editing of cbVGs affects their immunostimulatory ability or leads to more diverse cbVG populations with different immunostimulatory profiles. Further research is necessary to assess how ADAR1 affects cbVGs’ diversity and functions during infection and whether there are other RNA editing enzymes with similar roles.
Nonstandard viral genomes also play a role in establishing viral persistence. In vitro, this has been observed for many negative-sense RNA viruses, including IAV (78), SeV (79, 80), Ebola virus (EBOV) (81), LCMV (82, 83), human parainfluenza virus 3 (84, 85), MeV (86), mumps virus (87, 88), RSV (89), VSV (90, 91), and rabies virus (92, 93). In vivo, MeV cbVGs were found in brain tissues of patients with subacute sclerosing panencephalitis (94, 95), and EBOV cbVGs were found in testes of rhesus macaques, where EBOV seems to persist in humans (20, 96). The mechanisms by which nonstandard viral genomes drive persistence are poorly understood. In the context of SeV infections in vitro, cells that harbor predominantly cbVGs are protected from tumor necrosis factor (TNF)-mediated cell death by expressing TNF receptor-2 (TNFR2) and TNF receptor-associated factor 1 (TRAF1), allowing for the survival of standard viruses within these cells and the establishment of persistence (97). Cells that harbor predominantly standard virus instead express TNFR1, resulting in TNF-mediated cell death. Both the early antiviral response (IFN response) and late proviral response (TRNFR2/TRAF1) triggered by SeV cbVGs are dependent on RLR/MAVS signaling (47, 97). These observations suggest that the antiviral response induced by cbVGs, although counterintuitive, aids in maintaining an infected cell subpopulation alive. The surviving cells are specifically those that accumulate high levels of cbVGs, while those accumulating high levels of standard genomes and actively producing virus die. In vivo, cbVGs could ensure the survival of the host by limiting the amount of cell death, while also maintaining the virus within the host. Because many RNA viruses have been shown to persist in patients (98), understanding how cbVGs lead to persistent infections could allow us to combat persistent infections in the clinic.
Other Nonstandard Viral Genomes
Although cbVGs and delVGs have been extensively studied, other nonstandard viral genomes have been described, albeit with limited literature only focused on IAV. IAV generates short aberrant RNAs (mini viral RNAs, or mvRNAs) during replication (99). IAV mvRNAs differ from other IAV nonstandard viral genomes by their shorter size and ability to replicate in the absence of IAV nucleoprotein. IAV mvRNAs do not form canonical ribonucleoprotein complexes (99). Another novel type of nonstandard genome was found during IAV infection, called OP7. OP7 is a hypermutated RNA with 37 point mutations in the genomic viral RNA of segment 7, which affects promoter regions, encoded proteins, and genome packaging signals; hence, it cannot replicate without the presence of a standard viral genome (100). Further research on these types of nonstandard viral genomes in IAV and other negative-sense RNA viruses will be required to better understand what function they play during infection and how they affect RNA virus populations. As tools and methods become more accurate and sensitive, we will likely discover new nonstandard viral genomes and their contributions to negative-sense RNA virus communities.
DIVERSITY OF VIRAL PARTICLES WITHIN NEGATIVE-SENSE RNA VIRUS POPULATIONS
Heterogeneity at the viral genome level often translates into heterogeneity at the viral particle level, influencing host adaptation and infectivity due to the virus’ increased ability to enter cells and resist immune pressure. For negative-sense RNA viruses, differences in particle morphology, genomic packaging, and particle aggregation modulate the outcome of the infection and can ensure that the virus survives within the host (Fig. 2). Below, we compile evidence demonstrating mechanisms that negative-sense RNA viruses employ at the particle level to enhance overall fitness and survival.
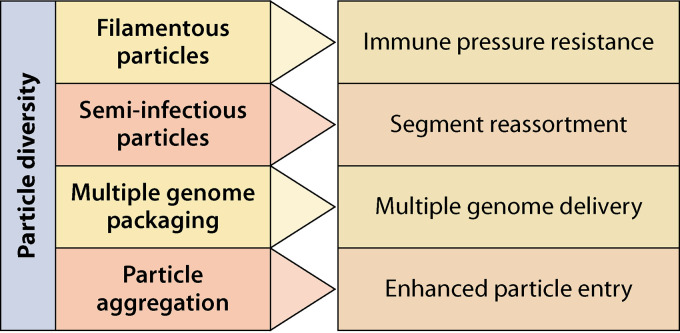
FIG 2.
Functions of diverse particles found in negative-sense RNA virus populations. Overview of the known effects of filamentous particles, semi-infectious particles, multiple-genome packaging, and particle aggregation on the virus population.
Filamentous Particles
Many negative-sense RNA viruses are pleomorphic and are described to form both spherical and filamentous particles. Within these viruses are some clinically relevant respiratory viruses such as RSV, influenza A, B, and C viruses, and human parainfluenza virus type 2 (HPIV2), as well as filoviruses such as EBOV and Marburg virus (101–106). Differences in virus particle morphology are often observed between strains isolated from different hosts, with filamentous particles commonly identified in clinical isolates and spherical particles in cell culture or mouse-adapted strains (107). Differences in particle morphology are also observed at a cellular level, where polarized epithelial cells favor the production of filamentous particles, while nonpolarized cells favor spherical particles (108). Because filamentous particles are predominant in clinical isolates, it is thought that filamentous particles confer an advantage over spherical particles in vivo. Indeed, studies have demonstrated that isolates dominated by IAV filamentous particles have better growth when passaged in guinea pigs compared to isolates dominated by spherical particles (107). However, this view has been challenged by the identification of an avian IAV H9N7 isolate from chicken outbreaks in China with predominantly spherical particles that showed increased infectivity and disease severity in chickens (109). Thus, the advantage conferred by filamentous particles could be driven primarily by the host environment. Nevertheless, the selection of either form of particle morphology in different host environments highlights the importance of maintaining diversity in particle morphology within the virus community.
Filamentous particles confer an advantage to the virus by allowing resistance to the host immune pressure and facilitating cell entry (110). An example of this advantage was recently demonstrated in the context of complement activation during RSV infection. In this study, the researchers found that opsonization of viral particles through complement activation, an important immune response activated during RSV infections, mainly affects spherical particles over complement-binding-resistant filamentous particles (111). This difference is suggested to be due to the conformation of the viral F protein, spherical particles having more of the postfusion F conformation than the filamentous particles, which contain more prefusion F. It was demonstrated that the postfusion F conformation induced higher complement activation (111). This suggests that changes in the morphology of the particles could serve different roles: spherical particles being beneficial during infections where the virus has adapted to the host, and filamentous particles being beneficial when the virus is less adapted to the host and immune responses are stronger (110, 112). A balance between spherical and filamentous particles then needs to be established as the infection proceeds, maintaining a filamentous population to combat immune pressure but also a spherical population to enhance replication, as it was demonstrated that IAV strains containing more filamentous particles have growth disadvantages compared to spherical particles (113). In this context, it was shown that the differences in growth were related to the ability of the IAV M protein to bind more tightly to the ribonucleoprotein in strains dominated by spherical particles than in strains dominated by filamentous particles (113). Additionally, the motility of influenza C virus filamentous particles is also reduced compared to spherical particles, which could have implications on the infectivity rates (114).
It has also been demonstrated that the mechanism of entry for IAV filamentous and spherical particles is different (115, 116). While spherical particles can enter the cell via endocytosis and macropinocytosis, filamentous particles seem to be restricted to enter only via macropinocytosis (115). How these differences confer advantages to the virus remains to be elucidated, but it could provide further understanding of viral entry during natural infections. Because filamentous particles are commonly found in clinical samples, the restriction of cell entry via macropinocytosis could be selected to enhance overall fitness. The selection for this type of cell entry by filamentous particles also provides a route for therapeutic intervention via macropinocytosis inhibition (115).
Most of the work to determine what controls the particle morphology has been done with influenza viruses, and several mutations in the M1 and M2 protein have been shown to be involved in controlling these different phenotypes (117–122). However, the accumulation of these mutations, together with the speed at which the switch from either spherical or filamentous morphology occurs during passaging in different hosts, demonstrates that control of particle morphology is a complex process that involves adaptation to the host (107). Interestingly, the M segment of the highly pathogenic and transmissible IAV H5N8 strain isolated from the 2016 European clade 2.3.4.4B correlated with increased formation of filamentous particles, suggesting that filamentous particle formation could be a determinant for virulence (123). Additionally, introducing the M segment of an IAV H1N1 isolate from the 2009 pandemic into a IAV PR8 strain was sufficient to increase filamentous morphology and enhance transmission in a guinea pig model (124). Because the M segment introduced in the PR8 strain also conferred increased neuraminidase activity, whether the increase in transmissibility was due to filamentous morphology, neuraminidase activity, or both is yet to be determined.
Host factors are also involved in the formation of filamentous particles, as cells infected with IAV and HPIV2 treated with actin polymerization inhibitors show low levels of filamentous particles being produced (105, 108, 125). The involvement of actin in producing filamentous particles is most likely controlled by the M protein, which has roles in recruiting actin within the virion (126). However, in the context of RSV infection, actin polymerization is dispensable for filament formation during infection (127). Instead, RhoA, a GTPase involved in many essential biological processes, is an important cellular factor for filamentous particle formation, as inhibition of RhoA reduces the ratio of filamentous to spherical particles produced during infection (128).
Variability in particle morphology is also shown to be dependent on the particle surface proteins, as the amounts of hemagglutinin (HA) to neuraminidase (NA) protein present in IAV particles influence the size of the virion. The differences in protein composition are accompanied by functional differences where viruses with higher NA contents are smaller and detach from the cell easily, while viruses containing higher HA contents are bigger and are deemed to be better at adhering to cells (129). Additionally, changes in pH correlate with less filamentous particle formation and cause changes in the matrix layer of the viral particle (130). The ability of IAV to confer flexibility in particle assembly and shape that is not driven by genetic changes could be beneficial, specifically at earlier times of infection in a new host, where genetic changes might be limited due to low replication levels (129). In the context of RSV infection, change in particle morphology was demonstrated to be controlled by the attachment of the matrix protein to the viral membrane (111).
Overall, the ability of negative-sense RNA viruses to shape particle morphology to increase fitness under different host pressures demonstrates how maintaining a diverse virus community confers evolutionary advantages that allow the virus to prevail despite the environment in which it exists. These changes in particle morphology were attributed to bet-hedging-like strategies, where the virus utilizes multiple strategies that, although they can compromise fitness in permissive conditions, enhance fitness during more stressful conditions (112). More work is needed to characterize the mechanisms that control particle morphology and the effect this has during infection with other negative-sense RNA viruses. The fact that pleomorphic particles are observed for so many of these important human pathogens points to the relevance particle morphology has in ensuring the success of the virus.
Semi-Infectious Particles
Semi-infectious (also called incomplete) particles arise due to inefficient packaging of all the genomes of segmented viruses. The prevalence of semi-infectious particles in IAV stocks has been debated and is mostly due to differences in experimental approaches and interpretations. Some groups have consistently shown that semi-infectious particles are predominant during IAV infection by looking at viral protein expression or the presence of intracellular RNA segments. By doing this, they observe that most cells do not express all viral proteins, nor do they contain all eight segments (129, 131–133). This is contrary to the observation that only a minority of viral particles are missing segments, and most IAV virions contain all eight segments as seen through electron microscopy and fluorescent in situ hybridization (FISH) (134, 135). The absence of certain segments and protein expression intracellularly could be due to the loss of the genomes as the ribonucleoprotein travels to the nucleus (132). Nevertheless, whether the segments were absent in the particle or were lost after cell entry, complementation by more than one particle during IAV infection is widely reported and is suggested to be an integral part of the IAV life cycle.
Semi-infectious particles may provide an evolutionary advantage that allows host adaptation of segmented viruses by enhancing genome reassortment (132, 136). Indeed, IAV is characterized by the ability of different strains to combine and exchange the HA or NA genomes that give rise to a new strain, known as antigenic shift (137). This recombination of segments has been proposed to be controlled and promoted by the complementation of semi-infectious particles (136, 138).
Incomplete genome packaging has also been described for other segmented viruses of the Bunyavirales order, such as Rift Valley fever virus (RVFV) and Schmallenberg virus (139). For both viruses, complete packaging of all segments was observed in less than 10% of the virus particles produced during infections in mammalian cells, with the remaining percent being empty and incomplete particles (139). Studies with the tri-segmented virus RVFV have demonstrated great flexibility in the packaging processes, showing successful rescue of virus stocks containing particles with more or less than 3 segments (140, 141). Interestingly, packaging of RVFV was more efficient in insect cells than in mammalian cells, with particles containing all three segments occurring three times more often in insect cells (139). The higher packaging efficiency could be selected in insect cells to enhance viral replication and facilitate transmission from insects to vertebrates. Although it is unknown what drives this difference in genome packaging, it suggests that genome packaging efficiency depends on the virus adaptation to the host environment, which could potentially have implications on the virus life cycle.
Multiple-Genome Packaging
Many nonsegmented negative-sense RNA viruses can carry multiple genomes in their virions to ensure productive infection. This phenomenon has been observed for various viruses, including Newcastle disease virus (NDV), MeV, Junin virus, EBOV, and SeV (142–148). Multiple-genome packaging is often thought of as a mechanism to ensure establishment of infections, enhance efficient packaging, and promote maintenance of nonstandard genomes within the virus population (149). The ability of a virus to introduce multiple genomes into a single cell can ensure that a cell is infected, either by competing with the negative effects of the antiviral responses induced in the cell or by allowing at least one genome to overcome the chances of degradation inside the cell (149, 150). This mechanism also overcomes the detrimental effects replication defective genomes have during infection if copackaging with a replication competent genome occurs (11, 151). This co-packaging mechanism enhances the number of genomes entering a cell leading to a more productive infection.
Multiple-genome packaging can also ensure that nonstandard viral genomes are maintained in the virus population by ensuring that at least one standard viral genome is present in the cell to support nonstandard viral genome replication. Multiple-virion packaging should then be regulated to prevent the detrimental effects nonstandard viral genomes can have during infection (149). Regulation of multiple genome packaging would also be important because larger particles are less stable and require more viral proteins and genomic material to form (150). Indeed, studies in NDV have shown that although multiple-genome packaging is observed, it is not the majority of the particle population, revealing that a balance between virions carrying single and multiple genomes exists (150).
Virion Aggregation
It is thought that, during viral infections, each viral particle acts as an independent entity able to enter the cell and carry out replication. However, this view has been challenged by the observation that viral particles can come together to form aggregates to increase the number of viral particles that enter the cell and promote cotransmission of virus variants (152–154). Studies that combine wild-type viruses with VSV tagged viruses lacking surface proteins required for cell entry showed that the deficient viruses can successfully enter the cell, demonstrating that the deficient virus gained entry through aggregation with the wild-type virus (152). Aggregation of VSV is driven by the presence of saliva (152, 155), which is also a route of transmission for the virus. It is thus tempting to hypothesize that virion aggregation occurs in vivo and that this facilitates the establishment of infection and transmission.
Virion aggregation is a mechanism used by viruses to ensure initial infection and to enhance the chance of complementation. Virion aggregation may be important for virus genome mixing, introduction of multiple virions into the same cell, and maintenance of nonstandard viral genomes in the virus population. Indeed, aggregation during VSV infection causes higher infection rates and leads to the accumulation of nonstandard particles (156). Whether virion aggregation is a process that must be controlled to prevent overaccumulation of nonstandard particles and how this process is controlled is still an understudied question. Additional studies must be done to determine if the aggregation of other negative-sense RNA viruses has similar roles, if the aggregation state of the virions changes depending on the environment the virus is physically present in, and how this mechanism could be regulated as the virus shifts from tissues within the host.
HOW DO NEGATIVE-SENSE RNA VIRUS POPULATIONS INTERACT AS A COMMUNITY?
Much work has been done to demonstrate the heterogeneity of negative-sense RNA virus populations and how each component of the population may influence the outcome of the infection. However, how all the components of the virus community work together to shape the establishment of infection, adaptation, transmission, and overall fitness is less studied. Below, we will piece together the knowledge gathered from the previous sections to hypothesize how the different components of a negative-sense RNA virus population could be interacting as a community.
The presence of multiple types of viral genomes during infection could be a mechanism for the virus to self-regulate and control the accumulation kinetics of each of its genomic components, facilitating each step of the viral life cycle. For example, during infections with VSV stocks that contain high levels of nonstandard viral genomes, variants resistant to the replication inhibitory effects of nonstandard viral genomes are selected for (157, 158). As the infection continues, emergence of new nonstandard viral genomes with stronger replication inhibitory effects can then suppress the variants (157, 158). This cycling ensures a balance between the proviral and antiviral effects nonstandard viral genomes have during infection (i.e., induction of the immune response, replication interference, and establishment of persistence). What drives the resistance of emerging variants to the inhibitory effects of nonstandard viral genomes has not been explored. This resistance could be mediated by changes in the new variants that have higher affinity for the viral polymerase or that exhibit increased packaging efficiency. These resistant variants could also harbor mutations that improve the virus-encoded antagonists of the immune response and therefore reduce inhibition of viral replication. The inverse could also be true, where newly generated nonstandard viral genomes could be more efficient at replicating due to changes in size or higher affinity for the polymerase. It would also be interesting to determine if host factors such as ADAR1, which has been proposed to edit nonstandard viral genomes, have a role in introducing changes that affect the efficiency of nonstandard viral genome replication, activate the antiviral response, and/or are packaged (75, 76). The ability of the virus to regulate the content of standard and nonstandard viral genomes during infection makes the virus adaptable and more likely to survive within the host. These observations suggest that when we evaluate how variants interact with each other, we must also consider how each variant affects the generation of nonstandard viral genomes.
In the case of influenza virus, the reported total viral particle to PFU ratio can range from 10 to 100 (159–162). The fact that the noninfectious particles are consistently maintained in virus populations during infections suggests that this component of the population provides an evolutionary advantage to the virus. For semi-infectious particles, their roles have been associated with enhancing reassortment events (136), which can result in fitter virus populations. These reassortment events, however, can be reduced in the presence of nonstandard viral genomes (136). The accumulation of nonstandard genomes could be a mechanism to control the number of semi-infectious particles being produced during infection. Although semi-infectious particles enhance the reassortment events and allow within-host spread, they can negatively affect transmission to other hosts (132). In the context of transmission to other hosts, fully infectious particles would be required for transmission to occur, as they would increase the chance that a fully replicating genome will establish infection. One could then imagine that, as replication levels and coinfection events increase within the host, nonstandard viral genomes also begin to accumulate, reducing the chance for reassortment and accumulation of semi-infectious particles. This would then lead to infected cells that will begin generating fully infectious particles, allowing for transmission and establishment of infection in other hosts.
Coinfection of semi-infectious particles may be facilitated by virion aggregation (163), as it has been reported that aggregation increases coinfection and complementation in the context of VSV and IAV infection (156, 164). Enhancing virion aggregation also enhances the accumulation of nonstandard viral particles, most likely by facilitating coinfection with a standard viral particle and ensuring that the nonstandard viral genomes are in the presence of the necessary complementing viral proteins (156). Aggregation then could be a mechanism employed by the virus to maintain nonstandard viral genomes in the virus population, as well as to facilitate reassortment by increasing coinfection of semi-infectious particles. This could also be applied in the context of maintaining virus variants with different functions in the virus population. Although virion aggregation has been well studied during VSV infection in the presence of saliva, it is tempting to hypothesize that respiratory viruses take advantage of the mucosal environment in the lung to facilitate aggregation, as has been proposed previously (163). Determining how the environment affects the kinetics of nonstandard particle accumulation by enhancing virion aggregation could provide new insights into how the host environment promotes changes in the virus population and possibly enhances its fitness.
Multiple-genome packaging is also a mechanism thought to enhance delivery of more than one genome in a cell. This mechanism, in addition to increasing the MOI, could also be employed to deliver both standard and nonstandard genomes into a cell. Although no studies have investigated this possibility, it is tempting to envision this mechanism being employed to maintain nonstandard viral genomes in the virus population. Combined packaging of a standard and nonstandard genome might be more efficient if the nonstandard genome is smaller in size, as this would require fewer resources to form the particle. This mechanism would also increase the chance for coinfection of a nonstandard particle with a standard particle for efficient replication, as the particle containing the nonstandard genome would also contain the standard genome, which provides the necessary genes to undergo full cycles of replication.
Filamentous morphology could also be involved in allowing multiple-genome packaging. This has been speculated in studies demonstrating that filamentous particles are five times more resistant to UV inactivation than spherical particles during IAV infection, suggesting that filamentous particle preparations have more genomic content than spherical particle preparations (165). It has also been demonstrated that IAV filamentous particles have higher contents of the nucleoprotein than spherical particles, further suggesting that filamentous particles have higher genome contents (121). This could also have implications during infections where filamentous particles are enriched and may give insights into why these are selected for in vivo. As mentioned earlier, if filamentous particles are prone to packaging more than one genome, then selection for this phenotype earlier during infection would be advantageous for the virus to enhance the MOI. This might be especially important at the initiation of infection in a new host, where a higher MOI will ensure successful establishment of infection. Although a study analyzing fluorescent and electron microscopy images could not determine if filamentous particles from IAV (A/Udorn/72) packaged extra copies of segments compared to the spherical particles, it did observe differences in the structure and organization of the ribonucleoproteins (166). Similar studies looking at the genome content of filamentous particles have also been unable to identify multiple-genome packaging (166). More follow-up studies will be required to determine if particle morphology can provide differences in genome packaging and how this mechanism of genome delivery could be impacting the outcome of the infection.
More interesting questions emerge when we think about how selection of these different mechanisms could be impacting coinfection with other pathogens in the same host. For instance, if a virus actively infecting a host generates nonstandard viral genomes that induce the host immune responses, it could establish an antiviral environment that will prevent incoming viruses from establishing an infection. Generation of immunostimulatory nonstandard viral genomes could be a mechanism that evolved to allow the virus to prevent competition with other viruses and dominate the infection. Later during the infection, the accumulation of nonstandard viral genomes could be controlled by emergence of variants that are resistant to the replication inhibitory effects of nonstandard viral genomes, which may prevent overstimulation and the detrimental effects the immune response could have on the virus population. This is interesting to consider, for example, when thinking about how the emergence of COVID-19 prevented the occurrence of the typical annual influenza season. Although this is commonly attributed to changes in social behavior such as isolation and mask wearing, it would be interesting to determine if components of the SARS-CoV-2 viral population could have played a role in decreasing influenza infections at the beginning of the COVID-19 pandemic.
Although our current knowledge of the diversity of negative-sense RNA virus populations allows us to grasp the importance the components of this population have in shaping the outcome of the infection, we need more understanding of the kinetics of their appearance together with how they interact with each other. This will give us a broader understanding of how negative-sense RNA viruses successfully infect their host and will provide insights to how we can target these components to reduce disease burden.
HOW COULD WE MANIPULATE VIRAL POPULATIONS TO OUR BENEFIT?
The more we learn about the different components of virus populations, what they do, and how they interact with each other, the more we can manipulate these components (e.g., inhibit, enhance, or manipulate ratios) for biomedical purposes. In this section, we will review current vaccine approaches or therapies under development that use the knowledge of the different components of negative-sense RNA virus populations to fight viral infections, along with how they work and their limitations.
Negative-sense RNA virus quasispecies arise due to the error-prone RNA-dependent RNA polymerase, making them adaptable to new environments. However, mutation rates that go beyond the error threshold (167) lead to lethal mutagenesis, where the number of mutations accumulated can be detrimental to the virus population (168). This has led to the development of therapies using drugs that increase mutation rates, which have been tested on many negative-sense RNA viruses, including IAV (169, 170), LCMV (171), and VSV (172). A major class of these drugs is the nucleoside analogs such as ribavirin, 5-azacytidine, and 5-fluorouracil (173, 174). Other drugs, such as the substituted pyrazine compound T-705, have also been tested (173). A cause of concern with nucleoside analog therapy is the development of drug-resistant viruses. Different reports show conflicting data in this regard. While some studies demonstrate no selection of drug-resistant mutants during treatment (175), others suggest the development of antiviral drug resistance to be possible (176, 177). These discrepancies could be due to differences in the viruses and MOIs used to test the effects of these drugs. For example, LCMV and IAV have shown higher mutation rates at a low MOI, whereas positive-sense RNA viruses showed higher mutation rates at a high MOI when treated with nucleoside analogs (178). Complete understanding of the mechanisms that drive antiviral drug resistance together with the standardization of methods to test this resistance will be critical.
Although the nucleoside analog ribavirin is being currently used in the clinic against some viruses, such as RSV, the American Academy of Pediatrics does not recommend the routine use of ribavirin in children with RSV due to its complex delivery system, high cost, and controversial efficacy (179). It will be important to continue looking for novel nucleoside analogs, or other drugs, that can provide better safety features to be used in the clinic (180). Some studies have also speculated that treatment with nucleoside analogs, in addition to forcing viral populations toward extinction by enriching for deleterious mutations, might also increase the generation of nonstandard viral genomes such as delVGs and cbVGs, which can further lead the viral population to extinction (169).
Other promising therapies to combat negative-sense RNA virus disease burden are based on using nonstandard particles (commonly known as defective interfering particles [DIPs]) containing cbVGs or delVGs, which have been extensively evaluated as potential antivirals (181). Although early studies focused on naturally occurring nonstandard particles, recently, most studies have developed synthetically engineered nonstandard particles with more interfering potential, sometimes referred to as therapeutic interfering particles (182, 183). This therapy relies on the ability of nonstandard particles to compete for the replication machinery of the virus and/or stimulate strong immune responses (181–183). Because nonstandard particles depend on the standard virus to replicate, they would be cleared out of the system once patients are cleared of the virus. It has also been suggested that nonstandard particles could be a transmissible antiviral therapy that could selectively target patients infected with a specific virus (184). Nonstandard particles from many negative-sense RNA viruses are currently being tested such as for IAV (185–191), EBOV (192), and NiV (21).
Preparation of nonstandard particles stocks can be a challenge because they often contain remainders of standard virus particles even after purification methods are used, which can be problematic at a biosafety level. To overcome this challenge, methods using cell culture have been developed where only nonstandard particles are generated (182, 185, 193, 194). For example, coculturing 293T and MDCK cells stably expressing codon-optimized IAV PB2 allowed production of nonstandard particles from plasmids without the need of a standard virus (193). Additionally, in vitro-transcribed cbVGs and delVGs can be packaged in lipid nanoparticles instead of the natural virus particle (182). The complex interaction of standard viral genomes with cbVGs and delVGs brings other challenges, such as determining the dose of nonstandard viral particles necessary for an effective therapy. Determining the correct doses of nonstandard viral particles will require mathematical models and predictions, such as the ones developed to determine the ideal ratio of IAV nonstandard particles to standard virus (195, 196). One of these models, for example, predicted that to restrict standard virus production, a ratio of about 104 nonstandard particles to 1 standard virus is needed (195). Although most of the studies only use one type of nonstandard particle, one research group also tested if using two types would improve the antiviral effect and showed no differences (197). Whether it is best to find the most interfering nonstandard viral particle or to generate cocktails of different nonstandard particle species remains to be investigated.
Nonstandard particle therapy assumes that, in patients, cbVGs and delVGs will preferentially be replicated and packaged. However, some studies show that delVGs are not efficiently packaged relative to the standard viral genomes (38, 192). This observation suggests that there might be viral mechanisms to counteract interference of delVGs. Determining the timing of nonstandard particle treatment is also a factor to be considered, as RSV patients that generate cbVGs early during infection were protected from severe disease, whereas patients that generate cbVGs later and/or for several days were sicker (33). How to use this information to enhance patient treatment and whether patients can make it to the hospital in time for this therapy to work remains to be determined. All these challenges are important to keep in mind when developing these new therapies. It would also be important to address whether nonstandard particle therapy would lead to establishment of persistent infections, selection of resistant virus mutants, or generation of new infectious viruses by complementation.
Other proposed applications of cbVG or delVG containing nonstandard particles are their use as adjuvants for vaccines due to their potent immunostimulatory ability (198, 199). For example, SeV cbVGs are potent activators of dendritic cells and lead to a strong induction of CD4+ and CD8+ T cell responses. When SeV cbVGs are used as adjuvants with experimental vaccines against influenza virus, an improvement in antibody production and increased protection from virus challenge was observed (59, 200). Furthermore, our lab has developed SeV DVG-derived oligonucleotides (DDOs) to improve vaccine efficacy (201). DDOs are shorter and consist of the major immunostimulatory motif found in a cbVG isolated from SeV (201). Use of DDOs as adjuvants biases the humoral and cellular responses toward type I immune responses against both influenza inactivated and HA-subunit vaccines, causing an increase in antibodies of the IgG2a/c isotypes, Th1 CD4+ T cells, and cytotoxic CD8+ T cells in mice (201). Influenza nonstandard particles have also been shown to protect against the genetically unrelated respiratory virus, pneumonia virus of mice (202). These studies suggest that using nonstandard particles or DDOs as adjuvants for vaccines could enhance protection against a wide array of viruses. Interestingly, nonstandard viral particles have been found in some live attenuated vaccines (25–27). Although their role in these vaccines remains unknown, it is possible that they might improve vaccine efficacy. If that is the case, we could develop methods to regulate nonstandard particle content within vaccine preparations (203, 204). SeV nonstandard particles have also been proposed to be used as antitumor therapies by inducing antitumor immunity and selectively killing prostate cancer cells through apoptosis (205, 206). It has also been suggested that nonstandard particles activate dendritic cells (DCs) and alert T cells of the presence of tumor cells within the host (205, 206).
The use of nucleoside analogs and nonstandard viral particles in medicine so far focuses mainly on changing intracellular viral RNA content to a point that is detrimental to the virus population or to shape host responses. Less attention has been given to the particles themselves and how they could be harnessed to develop therapies or improve vaccines. One study, for example, addressed whether virus particle aggregation improves vaccine efficacy and, indeed, when an IAV vaccine was used in combination with an aggregating peptide, an enhancement in cell-mediated responses was observed (207). It has also been suggested that as we learn more about the different entry mechanisms of filamentous and spherical particles, we can develop new therapies to target specific host proteins, depending on the particle characteristics of a pathogen (110). For example, because evidence suggests filamentous particles use macropinocytosis as a cell entry pathway during IAV infection, this could serve as a target to block particle entry and perhaps treat infections with strains that generate predominantly filamentous particles in natural infections (115). It is also important to keep in mind that different vaccine and nonstandard viral particle stock preparations could affect the ratio of filamentous to spherical particles, ratios of multiple genome packaging and semi-infectious particles. This will need to be addressed in the future, as we might need to standardize or at least characterize these stocks to improve their efficacy.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
Natural selection has allowed negative-sense RNA viruses to develop multiple strategies to combat host pressure, and perhaps these strategies have led to the diversification of virus populations. It is now more evident than ever that negative-sense RNA virus populations do not consist of homogeneous viral particles packaging identical viral genomes but rather they consist of diverse viral particles and viral RNAs that interact as a community to enhance virus survival within the host (Fig. 3). We need to continue focusing on understanding how all the different components of negative-sense RNA virus populations influence the outcome of the infection and even rethink previous studies where this diversity was not appreciated. The development of new techniques will also be vital, as it will help us address more questions about the complex interactions that occur within a virus population.
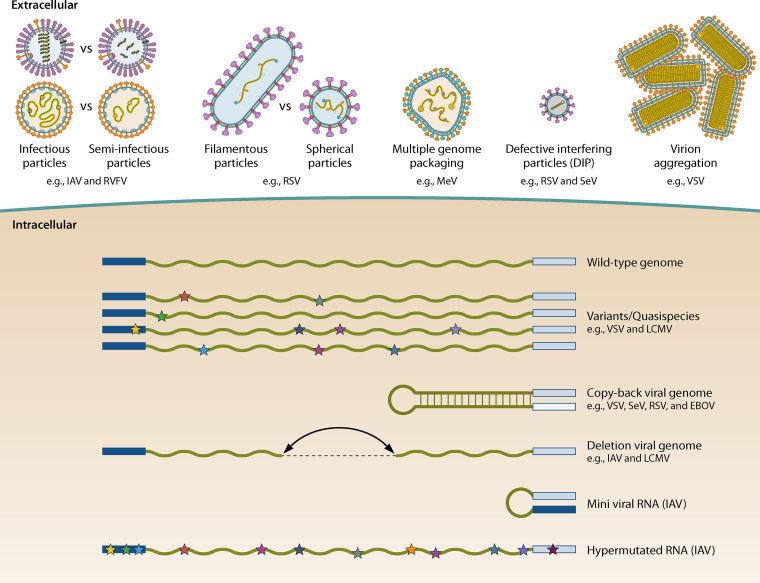
FIG 3.
Viral particles and viral genomes found in negative-sense RNA virus populations. On top, outside the cell, all types of viral particles discussed in the review are represented. At the bottom, within the cells, all types of viral RNAs discussed in the review are represented. The stars represent point mutations. The beginning and end structures of viral RNAs represent 3′ and 5′ sequences, respectively. Mini viral RNAs have very long deleted sequences and hence are similar to deletion viral genomes but smaller. As mini viral RNAs have only been found in IAV infections, we represented it as a hairpin loop, as IAV 3′ and 5′ ends are complementary in IAV.
Recently, and with the help of new methods, it was possible to assess the diversity of viral genomes at a population level and at a single-cell level. Techniques such as RNA fluorescent in situ hybridization and single-cell sequencing have shown that cbVGs and delVGs are spread heterogeneously across a cell population (208–211). We have previously demonstrated that SeV has distinct intracellular distributions of standard genomes and cbVGs, which drives functional heterogeneity during infection. High content of cbVGs within a cell can affect intracellular cargo trafficking and prevent the virus from generating particles (208, 209). Cells with predominant levels of standard viral genomes, on the other hand, can utilize the cellular trafficking machinery, making them the main producers of virions during infection (208, 209). Heterogeneity of non-standard virus segment expression across the cell population has also been observed for IAV (210, 211).
Our ability to enhance our resolution and look at negative-sense RNA virus infections at a single-cell level has uncovered a level of understanding that is often lost when we perform population analyses. Analysis at a single-cell level will become pivotal to understanding how the heterogenous virus population impacts the overall outcome of the infection. Indeed, we have demonstrated that many genes and pathways are differentially induced in cells with high cbVG content compared to cells with high contents of standard viral genomes (97). Similar and deeper analyses can be performed to look at how other genome components (i.e., variants, semi-infectious particles, etc.) affect gene expression in cells and how this expression influences the infection. The use and development of bioinformatic pipelines that allow us to detect nonstandard viral genomes will also be important. Our own lab has developed Viral Opensource DVG Key Algorithm (VODKA), which can detect cbVGs from RNAseq data (33, 212). VODKA allowed us to detect RSV cbVGs both in in vitro samples and in nasal washes of RSV-infected patients (33, 212). Similar tools such as VireMa and DI-detector, which can detect cbVGs, delVGs, and other recombination events, have been developed (213, 214). By continuing developing tools that allow us to identify the different components of the virus population, we will be better equipped to deepen our understanding of the interactions that occur within a virus population and how this leads to differences in disease burden.
Although the focus of this review is on negative-sense RNA viruses, diversity at the genome and particle levels is also observed in positive-sense RNA virus populations. For example, poliovirus forms quasispecies, and different polymerase mutants associate with more or less quasispecies diversity. Interestingly, both high-fidelity and low-fidelity mutations lead to attenuation of the virus and less pathogenesis (215, 216). These studies suggest that an optimal quasispecies diversity for a positive-sense RNA virus population is required for the virus to thrive. It would be interesting to determine how these mutations that affect quasispecies diversity also affect generation of nonstandard viral genomes and, if so, whether these are leading to an effect on the fitness of the virus. Nonstandard viral genomes of the deletion type have been identified in positive-sense RNA virus populations (e.g., Dengue virus, West Nile virus, and poliovirus), and these are associated with standard viral genome interference and establishment of persistent infections (17). At a particle level, mechanisms to improve viral fitness have also been observed. Enterovirus particles use phosphatidylserine lipid-enriched vesicles to aggregate and enhance infectivity (217). Poliovirus particles have also been shown to aggregate in specific environments (218, 219). In the case of flaviviruses, differences in particle morphology have been observed, and these differences are associated with adaptability to different host environments (220–222). Taken together, positive-sense RNA virus populations also act as a community by driving different functions that shape the outcome of infection.
The complexity of RNA virus populations makes it a challenge to advance the development of therapies to reduce disease burden caused by pathogenic viruses, but it demonstrates the importance of studying viruses as a community and not as homogeneous entities. As a research community, we need to find ways to standardize and characterize the virus stock preparations we use in our laboratories because variations in the virus population in the stocks lead to misinterpretations of the results and affect reproducibility. The more we expand our knowledge of the components of viral populations and their functions, the more we will learn about viral population dynamics, virus-host interactions, and evolutionary processes. With this knowledge, we will be able to develop more targeted therapies against viral pathogens but also use viruses as tools to understand basic biological processes and advance the medical field.
ACKNOWLEDGMENTS
We are grateful to López lab members David Holthausen, Matthew Hackbart, Munyaradzi Tambo, Italo Araujo Castro, Sydney Faber, Hanaa Saleh, Nicole Rivera-Espinal, and Sarah Pye for critical review of the manuscript.
Biographies

Lavinia J. González Aparicio received her bachelor’s degree in molecular biology from the University of Puerto Rico, Rio Piedras Campus. There, she worked in the laboratory of Reginald Morales and Orestes Quesada studying the enzyme phospholipase A2 (PLA2) found in cobra venom, with the goal of modifying it to target and kill cancer cells. She is currently obtaining her Ph.D. in microbiology and microbial pathogenesis at Washington University in St. Louis under the laboratory of Carolina López. At the López lab she studies the role stress granules play during negative-sense RNA virus infections containing high levels of copy-back viral genomes (cbVGs). Additional to her research, Lavinia spends her time mentoring and participating in outreach programs that foster interest in STEM careers to middle and high school students.

Carolina B. López obtained a Ph.D. from the Mount Sinai School of Medicine in New York. She joined the University of Pennsylvania in 2010 as an assistant professor and relocated to the Washington University in St Louis as a professor in 2020. Her laboratory studies how different components of a virus community affect the infected organism and how this interaction influences the infection outcome. She was elected to the American Academy of Microbiology in 2022. She is a member of the American Society for Microbiology, the American Association of Immunologists, and the American Society for Virology. She serves as an editor for several scientific journals, is a member of the NIH VIRB study section, a fellow of the Professional Mentoring Skills Enhancing Diversity Program funded by the NIH-National Research Mentoring Network, a WashU BJC investigator, and a 2018–2019 U.S. Fulbright scholar.

Sébastien A. Felt received his Ph.D. in 2017 from the University of North Carolina at Charlotte. His dissertation in Dr. Valery Grdzelishvili’s laboratory was focused on mole-cular mechanisms of resistance of pancreatic ductal adenocarcinoma cells to oncolytic vesicular stomatitis virus. He then spent 3 years as a postdoctoral researcher at the University of Pennsylvania in Dr. Carolina López’s laboratory to study the role of copy-back viral genomes (cbVGs) in respiratory syncytial virus infection (RSV) outcome. Felt then relocated with the López laboratory in 2020 to Washington University in St. Louis, where he has continued his postdoctoral and work with RSV. He recently established that detection of RSV cbVGs in nasal secretions is associated with distinct clinical outcomes. Felt is now investigating how different components of RSV populations, such as cbVGs, deletion viral genomes, and genome variants, interact with each other and shape the evolution of the viral population.
Contributor Information
Carolina B. López, Email: clopezzalaquett@wustl.edu.
Sébastien A. Felt, Email: felt@wustl.edu.
REFERENCES
- 1.Domingo E, Perales C. 2019. Viral quasispecies. PLoS Genet 15:e1008271. 10.1371/journal.pgen.1008271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo E, Garcia-Crespo C, Perales C. 2021. Historical perspective on the discovery of the quasispecies concept. Annu Rev Virol 8:51–72. 10.1146/annurev-virology-091919-105900. [DOI] [PubMed] [Google Scholar]
- 3.Steinhauer DA, Domingo E, Holland JJ. 1992. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 122:281–288. 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 4.Borderia AV, Rozen-Gagnon K, Vignuzzi M. 2016. Fidelity variants and RNA quasispecies. Curr Top Microbiol Immunol 392:303–322. 10.1007/82_2015_483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Torre JC, Holland JJ. 1990. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J Virol 64:6278–6281. 10.1128/JVI.64.12.6278-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcus PI, Rodriguez LL, Sekellick MJ. 1998. Interferon induction as a quasispecies marker of vesicular stomatitis virus populations. J Virol 72:542–549. 10.1128/JVI.72.1.542-549.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grande-Perez A, Martin V, Moreno H, de la Torre JC. 2016. Arenavirus quasispecies and their biological implications. Curr Top Microbiol Immunol 392:231–276. 10.1007/82_2015_468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buesa-Gomez J, Teng MN, Oldstone CE, Oldstone MB, de la Torre JC. 1996. Variants able to cause growth hormone deficiency syndrome are present within the disease-nil WE strain of lymphocytic choriomeningitis virus. J Virol 70:8988–8992. 10.1128/JVI.70.12.8988-8992.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med 160:521–540. 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng MN, Oldstone MB, de la Torre JC. 1996. Suppression of lymphocytic choriomeningitis virus: induced growth hormone deficiency syndrome by disease-negative virus variants. Virology 223:113–119. 10.1006/viro.1996.0460. [DOI] [PubMed] [Google Scholar]
- 11.Shirogane Y, Watanabe S, Yanagi Y. 2012. Cooperation between different RNA virus genomes produces a new phenotype. Nat Commun 3:1235. 10.1038/ncomms2252. [DOI] [PubMed] [Google Scholar]
- 12.Xue KS, Hooper KA, Ollodart AR, Dingens AS, Bloom JD. 2016. Cooperation between distinct viral variants promotes growth of H3N2 influenza in cell culture. Elife 5:e13974. 10.7554/eLife.13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donohue RC, Pfaller CK, Cattaneo R. 2019. Cyclical adaptation of measles virus quasispecies to epithelial and lymphocytic cells: to V, or not to V. PLoS Pathog 15:e1007605. 10.1371/journal.ppat.1007605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morimoto K, Hooper DC, Carbaugh H, Fu ZF, Koprowski H, Dietzschold B. 1998. Rabies virus quasispecies: implications for pathogenesis. Proc Natl Acad Sci USA 95:3152–3156. 10.1073/pnas.95.6.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazzarini RA, Keene JD, Schubert M. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26:145–154. 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 16.Ziegler CM, Botten JW. 2020. Defective interfering particles of negative-strand RNA viruses. Trends Microbiol 28:554–565. 10.1016/j.tim.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vignuzzi M, Lopez CB. 2019. Defective viral genomes are key drivers of the virus-host interaction. Nat Microbiol 4:1075–1087. 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosma TJ, Karagiannis K, Santana-Quintero L, Ilyushina N, Zagorodnyaya T, Petrovskaya S, Laassri M, Donnelly RP, Rubin S, Simonyan V, Sauder CJ. 2019. Identification and quantification of defective virus genomes in high throughput sequencing data using DVG-profiler, a novel post-sequence alignment processing algorithm. PLoS One 14:e0216944. 10.1371/journal.pone.0216944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang AS, Baltimore D. 1970. Defective viral particles and viral disease processes. Nature 226:325–327. 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RI, Boczkowska B, Alfson K, Weary T, Menzie H, Delgado J, Rodriguez G, Carrion R, Jr, Griffiths A. 2021. Identification and characterization of defective viral genomes in Ebola virus-infected rhesus macaques. J Virol 95:e0071421. 10.1128/JVI.00714-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch SR, Tilston NL, Lo MK, Whitmer SLM, Harmon JR, Scholte FEM, Spengler JR, Duprex WP, Nichol ST, Spiropoulou CF. 2020. Inhibition of Nipah virus by defective interfering particles. J Infect Dis 221:S460–S470. 10.1093/infdis/jiz564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilston-Lunel NL, Welch SR, Nambulli S, de Vries RD, Ho GW, Wentworth DE, Shabman R, Nichol ST, Spiropoulou CF, de Swart RL, Rennick LJ, Duprex WP. 2021. Sustained replication of synthetic canine distemper virus defective genomes in vitro and in vivo. mSphere 13:e00537-21. 10.1128/mSphere.00537-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson DM, Cubitt B, Pfeffer TL, de la Torre JC, Lukashevich IS. 2021. Lassa virus vaccine candidate ML29 generates truncated viral RNAs which contribute to interfering activity and attenuation. Viruses 13:214. 10.3390/v13020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng Z, Liu R, Yu J, Ran Z, Newkirk SJ, An W, Li F, Wang D. 2018. Identification and characterization of viral defective RNA genomes in influenza B virus. J Gen Virol 99:475–488. 10.1099/jgv.0.001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calain P, Roux L. 1988. Generation of measles virus defective interfering particles and their presence in a preparation of attenuated live-virus vaccine. J Virol 62:2859–2866. 10.1128/JVI.62.8.2859-2866.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellocq C, Mottet G, Roux L. 1990. Wide occurrence of measles virus subgenomic RNAs in attenuated live-virus vaccines. Biologicals 18:337–343. 10.1016/1045-1056(90)90039-3. [DOI] [PubMed] [Google Scholar]
- 27.Gould PS, Easton AJ, Dimmock NJ. 2017. Live attenuated influenza vaccine contains substantial and unexpected amounts of defective viral genomic RNA. Viruses 9:269. 10.3390/v9100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santak M, Markusic M, Balija ML, Kopac SK, Jug R, Orvell C, Tomac J, Forcic D. 2015. Accumulation of defective interfering viral particles in only a few passages in Vero cells attenuates mumps virus neurovirulence. Microbes Infect 17:228–236. 10.1016/j.micinf.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Bean WJ, Kawaoka Y, Wood JM, Pearson JE, Webster RG. 1985. Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J Virol 54:151–160. 10.1128/JVI.54.1.151-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chambers TM, Webster RG. 1987. Defective interfering virus associated with A/Chicken/Pennsylvania/83 influenza virus. J Virol 61:1517–1523. 10.1128/JVI.61.5.1517-1523.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saira K, Lin X, DePasse JV, Halpin R, Twaddle A, Stockwell T, Angus B, Cozzi-Lepri A, Delfino M, Dugan V, Dwyer DE, Freiberg M, Horban A, Losso M, Lynfield R, Wentworth DN, Holmes EC, Davey R, Wentworth DE, Ghedin E, INSIGHT FLU002 Study Group, INSIGHT FLU003 Study Group. 2013. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J Virol 87:8064–8074. 10.1128/JVI.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasilijevic J, Zamarreno N, Oliveros JC, Rodriguez-Frandsen A, Gomez G, Rodriguez G, Perez-Ruiz M, Rey S, Barba I, Pozo F, Casas I, Nieto A, Falcon A. 2017. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog 13:e1006650. 10.1371/journal.ppat.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felt SA, Sun Y, Jozwik A, Paras A, Habibi MS, Nickle D, Anderson L, Achouri E, Feemster KA, Cardenas AM, Turi KN, Chang M, Hartert TV, Sengupta S, Chiu C, Lopez CB. 2021. Detection of respiratory syncytial virus defective genomes in nasal secretions is associated with distinct clinical outcomes. Nat Microbiol 6:672–681. 10.1038/s41564-021-00882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Świętoń E, Tarasiuk K, Śmietanka K. 2020. Low pathogenic avian influenza virus isolates with different levels of defective genome segments vary in pathogenicity and transmission efficiency. Vet Res 51:108. 10.1186/s13567-020-00833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis AR, Hiti AL, Nayak DP. 1980. Influenza defective interfering viral RNA is formed by internal deletion of genomic RNA. Proc Natl Acad Sci USA 77:215–219. 10.1073/pnas.77.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alnaji FG, Holmes JR, Rendon G, Vera JC, Fields CJ, Martin BE, Brooke CB. 2019. Sequencing framework for the sensitive detection and precise mapping of defective interfering particle-associated deletions across influenza A and B viruses. J Virol 93:e00354-19. 10.1128/JVI.00354-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duhaut SD, Dimmock NJ. 1998. Heterologous protection of mice from a lethal human H1N1 influenza A virus infection by H3N8 equine defective interfering virus: comparison of defective RNA sequences isolated from the DI inoculum and mouse lung. Virology 248:241–253. 10.1006/viro.1998.9267. [DOI] [PubMed] [Google Scholar]
- 38.Alnaji FG, Reiser WK, Rivera-Cardona J, Te Velthuis AJW, Brooke CB. 2021. Influenza A virus defective viral genomes are inefficiently packaged into virions relative to wild-type genomic RNAs. mBio 12:e0295921. 10.1128/mBio.02959-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portner A, Kingsbury DW. 1971. Homologous interference by incomplete Sendai virus particles: changes in virus-specific ribonucleic acid synthesis. J Virol 8:388–394. 10.1128/JVI.8.4.388-394.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stampfer M, Baltimore D, Huang AS. 1969. Ribonucleic acid synthesis of vesicular stomatitis virus. I. Species of ribonucleic acid found in Chinese hamster ovary cells infected with plaque-forming and defective particles. J Virol 4:154–161. 10.1128/JVI.4.2.154-161.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang AS, Manders EK. 1972. Ribonucleic acid synthesis of vesicular stomatitis virus. IV. Transcription by standard virus in the presence of defective interfering particles. J Virol 9:909–916. 10.1128/JVI.9.6.909-916.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giachetti C, Holland JJ. 1989. Vesicular stomatitis virus and its defective interfering particles exhibit in vitro transcriptional and replicative competition for purified L-NS polymerase molecules. Virology 170:264–267. 10.1016/0042-6822(89)90375-9. [DOI] [PubMed] [Google Scholar]
- 43.Duhaut SD, McCauley JW. 1996. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology 216:326–337. 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 44.Li T, Pattnaik AK. 1997. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology 232:248–259. 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 45.Rao DD, Huang AS. 1982. Interference among defective interfering particles of vesicular stomatitis virus. J Virol 41:210–221. 10.1128/JVI.41.1.210-221.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odagiri T, Tashiro M. 1997. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J Virol 71:2138–2145. 10.1128/JVI.71.3.2138-2145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Jain D, Koziol-White CJ, Genoyer E, Gilbert M, Tapia K, Panettieri RA, Jr, Hodinka RL, Lopez CB. 2015. Immunostimulatory defective viral genomes from respiratory syncytial virus promote a strong innate antiviral response during infection in mice and humans. PLoS Pathog 11:e1005122. 10.1371/journal.ppat.1005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Hoogen BG, van Boheemen S, de Rijck J, van Nieuwkoop S, Smith DJ, Laksono B, Gultyaev A, Osterhaus A, Fouchier RAM. 2014. Excessive production and extreme editing of human metapneumovirus defective interfering RNA is associated with type I IFN induction. J Gen Virol 95:1625–1633. 10.1099/vir.0.066100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston MD. 1981. The characteristics required for a Sendai virus preparation to induce high levels of interferon in human lymphoblastoid cells. J Gen Virol 56:175–184. 10.1099/0022-1317-56-1-175. [DOI] [PubMed] [Google Scholar]
- 50.Strahle L, Garcin D, Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111. 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 51.Killip MJ, Young DF, Gatherer D, Ross CS, Short JA, Davison AJ, Goodbourn S, Randall RE. 2013. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J Virol 87:4798–4807. 10.1128/JVI.03383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wignall-Fleming EB, Vasou A, Young D, Short JAL, Hughes DJ, Goodbourn S, Randall RE. 2020. Innate intracellular antiviral responses restrict the amplification of defective virus genomes of parainfluenza virus 5. J Virol 94:e00246-20. 10.1128/JVI.00246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shivakoti R, Siwek M, Hauer D, Schultz KL, Griffin DE. 2013. Induction of dendritic cell production of type I and type III interferons by wild-type and vaccine strains of measles virus: role of defective interfering RNAs. J Virol 87:7816–7827. 10.1128/JVI.00261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcus PI, Sekellick MJ. 1977. Defective interfering particles with covalently linked [+/-]RNA induce interferon. Nature 266:815–819. 10.1038/266815a0. [DOI] [PubMed] [Google Scholar]
- 55.Marcus PI, Gaccione C. 1989. Interferon induction by viruses. XIX. Vesicular stomatitis virus—New Jersey: high multiplicity passages generate interferon-inducing, defective-interfering particles. Virology 171:630–633. 10.1016/0042-6822(89)90637-5. [DOI] [PubMed] [Google Scholar]
- 56.Tapia K, Kim WK, Sun Y, Mercado-Lopez X, Dunay E, Wise M, Adu M, Lopez CB. 2013. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog 9:e1003703. 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yount JS, Gitlin L, Moran TM, Lopez CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J Immunol 180:4910–4918. 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 58.Yount JS, Kraus TA, Horvath CM, Moran TM, Lopez CB. 2006. A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J Immunol 177:4503–4513. 10.4049/jimmunol.177.7.4503. [DOI] [PubMed] [Google Scholar]
- 59.Mercado-Lopez X, Cotter CR, Kim WK, Sun Y, Munoz L, Tapia K, Lopez CB. 2013. Highly immunostimulatory RNA derived from a Sendai virus defective viral genome. Vaccine 31:5713–5721. 10.1016/j.vaccine.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan DJ, Dimmock NJ. 1992. Defective interfering influenza virus inhibits immunopathological effects of infectious virus in the mouse. J Virol 66:1188–1192. 10.1128/JVI.66.2.1188-1192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott PD, Meng B, Marriott AC, Easton AJ, Dimmock NJ. 2011. Defective interfering influenza virus confers only short-lived protection against influenza virus disease: evidence for a role for adaptive immunity in DI virus-mediated protection in vivo. Vaccine 29:6584–6591. 10.1016/j.vaccine.2011.06.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabinowitz SG, Huprikar J. 1979. The influence of defective-interfering particles of the PR-8 strain of influenza A virus on the pathogenesis of pulmonary infection in mice. J Infect Dis 140:305–315. 10.1093/infdis/140.3.305. [DOI] [PubMed] [Google Scholar]
- 63.Xu J, Mercado-Lopez X, Grier JT, Kim WK, Chun LF, Irvine EB, Del Toro Duany Y, Kell A, Hur S, Gale M, Jr, Raj A, Lopez CB. 2015. Identification of a natural viral RNA motif that optimizes sensing of Viral RNA by RIG-I. mBio 6:e01265-15–e01215. 10.1128/mBio.01265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strahle L, Marq JB, Brini A, Hausmann S, Kolakofsky D, Garcin D. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J Virol 81:12227–12237. 10.1128/JVI.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho TH, Kew C, Lui PY, Chan CP, Satoh T, Akira S, Jin DY, Kok KH. 2016. PACT- and RIG-I-dependent activation of type I interferon production by a defective interfering RNA derived from measles virus vaccine. J Virol 90:1557–1568. 10.1128/JVI.02161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA 107:16303–16308. 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Runge S, Sparrer KM, Lassig C, Hembach K, Baum A, Garcia-Sastre A, Soding J, Conzelmann KK, Hopfner KP. 2014. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog 10:e1004081. 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Linder A, Bothe V, Linder N, Schwarzlmueller P, Dahlstrom F, Bartenhagen C, Dugas M, Pandey D, Thorn-Seshold J, Boehmer DFR, Koenig LM, Kobold S, Schnurr M, Raedler J, Spielmann G, Karimzadeh H, Schmidt A, Endres S, Rothenfusser S. 2021. Defective interfering genomes and the full-length viral genome trigger RIG-I after infection with vesicular stomatitis virus in a replication dependent manner. Front Immunol 12:595390. 10.3389/fimmu.2021.595390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mura M, Combredet C, Najburg V, Sanchez David RY, Tangy F, Komarova AV. 2017. Nonencapsidated 5' copy-back defective interfering genomes produced by recombinant measles viruses are recognized by RIG-I and LGP2 but not MDA5. J Virol 91:e00643-17. 10.1128/JVI.00643-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. 2013. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep 14:780–787. 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80:5059–5064. 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5'-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 73.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. 2009. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34. 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Hara PJ, Nichol ST, Horodyski FM, Holland JJ. 1984. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell 36:915–924. 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- 75.Pfaller CK, Mastorakos GM, Matchett WE, Ma X, Samuel CE, Cattaneo R. 2015. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J Virol 89:7735–7747. 10.1128/JVI.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pfaller CK, Donohue RC, Nersisyan S, Brodsky L, Cattaneo R. 2018. Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLoS Biol 16:e2006577. 10.1371/journal.pbio.2006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samuel CE. 2011. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411:180–193. 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De BK, Nayak DP. 1980. Defective interfering influenza viruses and host cells: establishment and maintenance of persistent influenza virus infection in MDBK and HeLa cells. J Virol 36:847–859. 10.1128/JVI.36.3.847-859.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roux L, Waldvogel FA. 1981. Establishment of Sendai virus persistent infection: biochemical analysis of the early phase of a standard plus defective interfering virus infection of BHK cells. Virology 112:400–410. 10.1016/0042-6822(81)90287-7. [DOI] [PubMed] [Google Scholar]
- 80.Roux L, Holland JJ. 1979. Role of defective interfering particles of Sendai virus in persistent infections. Virology 93:91–103. 10.1016/0042-6822(79)90278-2. [DOI] [PubMed] [Google Scholar]
- 81.Calain P, Monroe MC, Nichol ST. 1999. Ebola virus defective interfering particles and persistent infection. Virology 262:114–128. 10.1006/viro.1999.9915. [DOI] [PubMed] [Google Scholar]
- 82.Lehmann-Grube F, Slenczka W, Tees R. 1969. A persistent and inapparent infection of L cells with the virus of lymphocytic choriomeningitis. J Gen Virol 5:63–81. 10.1099/0022-1317-5-1-63. [DOI] [PubMed] [Google Scholar]
- 83.Meyer BJ, Southern PJ. 1997. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J Virol 71:6757–6764. 10.1128/JVI.71.9.6757-6764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moscona A. 1991. Defective interfering particles of human parainfluenza virus type 3 are associated with persistent infection in cell culture. Virology 183:821–824. 10.1016/0042-6822(91)91018-c. [DOI] [PubMed] [Google Scholar]
- 85.Moscona A, Peluso RW. 1993. Persistent infection with human parainfluenza virus 3 in CV-1 cells: analysis of the role of defective interfering particles. Virology 194:399–402. 10.1006/viro.1993.1275. [DOI] [PubMed] [Google Scholar]
- 86.Rima BK, Davidson WB, Martin SJ. 1977. The role of defective interfering particles in persistent infection of Vero cells by measles virus. J Gen Virol 35:89–97. 10.1099/0022-1317-35-1-89. [DOI] [PubMed] [Google Scholar]
- 87.Andzhaparidze OG, Boriskin YS, Bogomolova NN, Drynov ID. 1982. Mumps virus-persistently infected cell cultures release defective interfering virus particles. J Gen Virol 63:499–503. 10.1099/0022-1317-63-2-499. [DOI] [PubMed] [Google Scholar]
- 88.Andzhaparidze OG, Bogomolova NN, Boriskin YS, Drynov ID. 1983. Chronic non-cytopathic infection of human continuous cell lines with mumps virus. Acta Virol 27:318–328. [PubMed] [Google Scholar]
- 89.Sarmiento RE, Tirado R, Gomez B. 2002. Characteristics of a respiratory syncytial virus persistently infected macrophage-like culture. Virus Res 84:45–58. 10.1016/s0168-1702(01)00420-8. [DOI] [PubMed] [Google Scholar]
- 90.Sekellick MJ, Marcus PI. 1978. Persistent infection. I Interferon-inducing defective-interfering particles as mediators of cell sparing: possible role in persistent infection by vesicular stomatitis virus. Virology 85:175–186. 10.1016/0042-6822(78)90422-1. [DOI] [PubMed] [Google Scholar]
- 91.DePolo NJ, Holland JJ. 1986. Very rapid generation/amplification of defective interfering particles by vesicular stomatitis virus variants isolated from persistent infection. J Gen Virol 67:1195–1198. 10.1099/0022-1317-67-6-1195. [DOI] [PubMed] [Google Scholar]
- 92.Andzhaparidze OG, Bogomolova NN, Boriskin YS, Bektemirova MS, Drynov ID. 1981. Comparative study of rabies virus persistence in human and hamster cell lines. J Virol 37:1–6. 10.1128/JVI.37.1.1-6.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawai A, Matsumoto S, Tanabe K. 1975. Characterization of rabies viruses recovered from persistently infected BHK cells. Virology 67:520–533. 10.1016/0042-6822(75)90452-3. [DOI] [PubMed] [Google Scholar]
- 94.Baczko K, Liebert UG, Billeter M, Cattaneo R, Budka H, ter Meulen V. 1986. Expression of defective measles virus genes in brain tissues of patients with subacute sclerosing panencephalitis. J Virol 59:472–478. 10.1128/JVI.59.2.472-478.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sidhu MS, Crowley J, Lowenthal A, Karcher D, Menonna J, Cook S, Udem S, Dowling P. 1994. Defective measles virus in human subacute sclerosing panencephalitis brain. Virology 202:631–641. 10.1006/viro.1994.1384. [DOI] [PubMed] [Google Scholar]
- 96.Calain P, Roux L, Kolakofsky D. 2016. Defective interfering genomes and Ebola virus persistence. Lancet 388:659–660. 10.1016/S0140-6736(16)31272-7. [DOI] [PubMed] [Google Scholar]
- 97.Xu J, Sun Y, Li Y, Ruthel G, Weiss SR, Raj A, Beiting D, Lopez CB. 2017. Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence. Nat Commun 8:799. 10.1038/s41467-017-00909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McCarthy MK, Morrison TE. 2017. Persistent RNA virus infections: do PAMPS drive chronic disease? Curr Opin Virol 23:8–15. 10.1016/j.coviro.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Te Velthuis AJW, Long JC, Bauer DLV, Fan RLY, Yen HL, Sharps J, Siegers JY, Killip MJ, French H, Oliva-Martin MJ, Randall RE, de Wit E, van Riel D, Poon LLM, Fodor E. 2018. Mini viral RNAs act as innate immune agonists during influenza virus infection. Nat Microbiol 3:1234–1242. 10.1038/s41564-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kupke SY, Riedel D, Frensing T, Zmora P, Reichl U. 2019. A novel type of influenza A virus-derived defective interfering particle with nucleotide substitutions in its genome. J Virol 93:e01786-18. 10.1128/JVI.01786-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. 2013. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci USA 110:11133–11138. 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bharat TA, Noda T, Riches JD, Kraehling V, Kolesnikova L, Becker S, Kawaoka Y, Briggs JA. 2012. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci USA 109:4275–4280. 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bharat TA, Riches JD, Kolesnikova L, Welsch S, Krahling V, Davey N, Parsy ML, Becker S, Briggs JA. 2011. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol 9:e1001196. 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mosley VM, Wyckoff RWG. 1946. Nature 157:263. 10.1038/157263a0. [DOI] [PubMed] [Google Scholar]
- 105.Yao QCR. 2000. Filamentous particle formation by human parainfluenza virus type 2. J Gen Virol 81:1305–1312. 10.1099/0022-1317-81-5-1305. [DOI] [PubMed] [Google Scholar]
- 106.Nishimura H, Hara M, Sugawara K, Kitame F, Takiguchi K, Umetsu Y, Tonosaki A, Nakamura K. 1990. Characterization of the cord-like structures emerging from the surface of Influenza C virus-infected cells. Virology 179:179–188. 10.1016/0042-6822(90)90287-2. [DOI] [PubMed] [Google Scholar]
- 107.Seladi-Schulman J, Steel J, Lowen AC. 2013. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J Virol 87:13343–13353. 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts PCC, Compans RW. 1998. Host cell dependence of viral morphology. [DOI] [PMC free article] [PubMed]
- 109.Pu J, Sun H, Qu Y, Wang C, Gao W, Zhu J, Sun Y, Bi Y, Huang Y, Chang KC, Cui J, Liu J. 2017. M gene reassortment in H9N2 influenza virus promotes early infection and replication: contribution to rising virus prevalence in chickens in China. J Virol 91:e02055-16. 10.1128/JVI.02055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li T, Li Z, Deans EE, Mittler E, Liu M, Chandran K, Ivanovic T. 2021. The shape of pleomorphic virions determines resistance to cell-entry pressure. Nat Microbiol 6:617–629. 10.1038/s41564-021-00877-0. [DOI] [PubMed] [Google Scholar]
- 111.Kuppan JP, Mitrovich MD, Vahey MD. 2021. A morphological transformation in respiratory syncytial virus leads to enhanced complement deposition. Elife 10:e70575. 10.7554/eLife.70575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lowen AC. 2021. Filamentous viruses prevail under pressure. Nat Microbiol 6:536–537. 10.1038/s41564-021-00903-1. [DOI] [PubMed] [Google Scholar]
- 113.Liu T, Muller J, Ye Z. 2002. Association of influenza virus matrix protein with ribonucleoproteins may control viral growth and morphology. Virology 304:89–96. 10.1006/viro.2002.1669. [DOI] [PubMed] [Google Scholar]
- 114.Sakai T, Takagi H, Muraki Y, Saito M. 2018. Unique directional motility of influenza C virus controlled by its filamentous morphology and short-range motions. J Virol 92:e01522-17. 10.1128/JVI.01522-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rossman JS, Leser GP, Lamb RA. 2012. Filamentous influenza virus enters cells via macropinocytosis. J Virol 86:10950–10960. 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sieczkarski SB, Whittaker GR. 2005. Characterization of the host cell entry of filamentous influenza virus. Arch Virol 150:1783–1796. 10.1007/s00705-005-0558-1. [DOI] [PubMed] [Google Scholar]
- 117.Elleman CJ, Barclay WS. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144–153. 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 118.Seladi-Schulman J, Campbell PJ, Suppiah S, Steel J, Lowen AC. 2014. Filament-producing mutants of influenza A/Puerto Rico/8/1934 (H1N1) virus have higher neuraminidase activities than the spherical wild-type. PLoS One 9:e112462. 10.1371/journal.pone.0112462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nishimura H, Hongo S, Sugawara K, Muraki Y, Kitame F, Washioka H, Tonosaki A, Nakamura K. 1994. The ability of influenza C virus to generate cord-like structures is influenced by the gene coding for M protein. Virology 200:140–147. 10.1006/viro.1994.1172. [DOI] [PubMed] [Google Scholar]
- 120.Campbell PJ, Kyriakis CS, Marshall N, Suppiah S, Seladi-Schulman J, Danzy S, Lowen AC, Steel J. 2014. Residue 41 of the Eurasian avian-like swine influenza a virus matrix protein modulates virion filament length and efficiency of contact transmission. J Virol 88:7569–7577. 10.1128/JVI.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roberts PCL, Lamb RA, Compans RW. 1998. The M1 and M2 proteins of influenza a virus are important determinants in filamentous particle formation. Virology 240:127–137. 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 122.Roberts KL, Leser GP, Ma C, Lamb RA. 2013. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J Virol 87:9973–9982. 10.1128/JVI.01363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leyson CM, Youk S, Ferreira HL, Suarez DL, Pantin-Jackwood M. 2021. Multiple gene segments are associated with enhanced virulence of clade 2.3.4.4 H5N8 highly pathogenic avian influenza virus in mallards. J Virol 95:e00955-21. 10.1128/JVI.00955-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, Steel J. 2014. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J Virol 88:3802–3814. 10.1128/JVI.03607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Simpson-Holley M, Ellis D, Fisher D, Elton D, McCauley J, Digard P. 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301:212–225. 10.1006/viro.2002.1595. [DOI] [PubMed] [Google Scholar]
- 126.Giuffre RM, Tovell DR, Kay CM, Tyrrell DL. 1982. Evidence for an interaction between the membrane protein of a paramyxovirus and actin. J Virol 42:963–968. 10.1128/jvi.42.3.963-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shaikh FY, Utley TJ, Craven RE, Rogers MC, Lapierre LA, Goldenring JR, Crowe JE, Jr.. 2012. Respiratory syncytial virus assembles into structured filamentous virion particles independently of host cytoskeleton and related proteins. PLoS One 7:e40826. 10.1371/journal.pone.0040826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gower TL, Pastey MK, Peeples ME, Collins PL, McCurdy LH, Hart TK, Guth A, Johnson TR, Graham BS. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J Virol 79:5326–5336. 10.1128/JVI.79.9.5326-5336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vahey MD, Fletcher DA. 2019. Low-fidelity assembly of influenza A virus promotes escape from host cells. Cell 176:281–294.e19. 10.1016/j.cell.2018.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci USA 107:10685–10690. 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW. 2013. Most influenza A virions fail to express at least one essential viral protein. J Virol 87:3155–3162. 10.1128/JVI.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jacobs NT, Onuoha NO, Antia A, Steel J, Antia R, Lowen AC. 2019. Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread. Nat Commun 10:3526. 10.1038/s41467-019-11428-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Russell AB, Trapnell C, Bloom JD. 2018. Extreme heterogeneity of influenza virus infection in single cells. Elife 7:e32303. 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakatsu S, Sagara H, Sakai-Tagawa Y, Sugaya N, Noda T, Kawaoka Y. 2016. Complete and incomplete genome packaging of influenza A and B viruses. mBio 7:e01248-16. 10.1128/mBio.01248-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chou Y-Y, Vafabakhsh R, Doğanay S, Gao Q, Ha T, Palese P. 2012. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc Natl Acad Sci USA 109:9101–9106. 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fonville JM, Marshall N, Tao H, Steel J, Lowen AC. 2015. Influenza virus reassortment is enhanced by semi-infectious particles but can be suppressed by defective interfering particles. PLoS Pathog 11:e1005204. 10.1371/journal.ppat.1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Webster RGL, Laver WG, Air GM, Schild GC. 1982. Molecular mechanisms of variation in influenza viruses. Nature 296:115–121. 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 138.Diefenbacher M, Sun J, Brooke CB. 2018. The parts are greater than the whole: the role of semi-infectious particles in influenza A virus biology. Curr Opin Virol 33:42–46. 10.1016/j.coviro.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bermudez-Mendez E, Katrukha EA, Spruit CM, Kortekaas J, Wichgers Schreur PJ. 2021. Visualizing the ribonucleoprotein content of single bunyavirus virions reveals more efficient genome packaging in the arthropod host. Commun Biol 4:345. 10.1038/s42003-021-01821-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brennan B, Welch SR, McLees A, Elliott RM. 2011. Creation of a recombinant Rift Valley fever virus with a two-segmented genome. J Virol 85:10310–10318. 10.1128/JVI.05252-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wichgers Schreur PJ, Oreshkova N, Moormann RJ, Kortekaas J. 2014. Creation of Rift Valley fever viruses with four-segmented genomes reveals flexibility in bunyavirus genome packaging. J Virol 88:10883–10893. 10.1128/JVI.00961-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hosaka Y, Kitano H, Ikeguchi S. 1966. Studies on the pleomorphism of HVJ virions. Virology 29:205–221. 10.1016/0042-6822(66)90027-4. [DOI] [PubMed] [Google Scholar]
- 143.Dhalberg JES, Simon EH. 1969. Physical and genetic studies of Newcastle disease virus: evidence for multiploid particles. Virology 38:666–678. 10.1016/0042-6822(69)90185-8. [DOI] [PubMed] [Google Scholar]
- 144.Cattaneo R, Donohue RC, Generous AR, Navaratnarajah CK, Pfaller CK. 2019. Stronger together: multi-genome transmission of measles virus. Virus Res 265:74–79. 10.1016/j.virusres.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gaudin R, Barteneva NS. 2015. Sorting of small infectious virus particles by flow virometry reveals distinct infectivity profiles. Nat Commun 6:6022. 10.1038/ncomms7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rager M, Vongpunsawad S, Duprex WP, Cattaneo R. 2002. Polyploid measles virus with hexameric genome length. EMBO J 21:2364–2372. 10.1093/emboj/21.10.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. 2012. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One 7:e29608. 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Loney C, Mottet-Osman G, Roux L, Bhella D. 2009. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of Sendai virus. J Virol 83:8191–8197. 10.1128/JVI.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leeks A, Sanjuan R, West SA. 2019. The evolution of collective infectious units in viruses. Virus Res 265:94–101. 10.1016/j.virusres.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goff PH, Gao Q, Palese P. 2012. A majority of infectious Newcastle disease virus particles contain a single genome, while a minority contain multiple genomes. J Virol 86:10852–10856. 10.1128/JVI.01298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sanjuan R. 2018. Collective properties of viral infectivity. Curr Opin Virol 33:1–6. 10.1016/j.coviro.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cuevas JM, Duran-Moreno M, Sanjuan R. 2017. Multi-virion infectious units arise from free viral particles in an enveloped virus. Nat Microbiol 2:17078. 10.1038/nmicrobiol.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wallis CM, Melnick JL. 1967. Virus aggregation as the cause of the nonneutralizable persistent fraction. J Virol 1:478–488. 10.1128/jvi.1.3.478-488.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Andreu-Moreno I, Sanjuan R. 2018. Collective infection of cells by viral aggregates promotes early viral proliferation and reveals a cellular-level Allee effect. Curr Biol 28:3212–3219 e4. 10.1016/j.cub.2018.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Anschau V, Sanjuan R. 2020. Fibrinogen gamma chain promotes aggregation of vesicular stomatitis virus in saliva. Viruses 12:282. 10.3390/v12030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Andreu-Moreno I, Sanjuan R. 2020. Collective viral spread mediated by virion aggregates promotes the evolution of defective interfering particles. mBio 11:e02156-19. 10.1128/mBio.02156-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.DePolo NJ, Giachetti C, Holland JJ. 1987. Continuing coevolution of virus and defective interfering particles and of viral genome sequences during undiluted passages: virus mutants exhibiting nearly complete resistance to formerly dominant defective interfering particles. J Virol 61:454–464. 10.1128/JVI.61.2.454-464.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Horodyski FM, Nichol ST, Spindler KR, Holland JJ. 1983. Properties of DI particle resistant mutants of vesicular stomatitis virus isolated from persistent infections and from undiluted passages. Cell 33:801–810. 10.1016/0092-8674(83)90022-3. [DOI] [PubMed] [Google Scholar]
- 159.Donald HB, Isaacs A. 1954. Counts of influenza virus particles. J Gen Microbiol 10:457–464. 10.1099/00221287-10-3-457. [DOI] [PubMed] [Google Scholar]
- 160.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J Gen Virol 91:313–328. 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 161.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J Virol 82:11869–11879. 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nakajima K, Sugiura A. 1977. Three-factor cross of influenza virus. Virology 81:486–489. 10.1016/0042-6822(77)90165-9. [DOI] [PubMed] [Google Scholar]
- 163.Brooke CB. 2014. Biological activities of ‘noninfectious’ influenza A virus particles. Future Virol 9:41–51. 10.2217/fvl.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hirst GK, Pons MW. 1973. Mechanism of influenza recombination. II. Virus aggregation and its effect on plaque formation by so-called noninfective virus. Virology 56:620–631. 10.1016/0042-6822(73)90063-9. [DOI] [PubMed] [Google Scholar]
- 165.Smirnov YA, Kuznetsova MA, Kaverin NV. 1991. The genetic aspects of influenza virus filamentous particle formation. Arch Virol 118:279–284. 10.1007/BF01314038. [DOI] [PubMed] [Google Scholar]
- 166.Vijayakrishnan S, Loney C, Jackson D, Suphamungmee W, Rixon FJ, Bhella D. 2013. Cryotomography of budding influenza A virus reveals filaments with diverse morphologies that mostly do not bear a genome at their distal end. PLoS Pathog 9:e1003413. 10.1371/journal.ppat.1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Biebricher CK, Eigen M. 2005. The error threshold. Virus Res 107:117–127. 10.1016/j.virusres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 168.Bull JJ, Sanjuan R, Wilke CO. 2007. Theory of lethal mutagenesis for viruses. J Virol 81:2930–2939. 10.1128/JVI.01624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pauly MD, Lauring AS. 2015. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J Virol 89:3584–3597. 10.1128/JVI.03483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Baranovich T, Wong SS, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA. 2013. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol 87:3741–3751. 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 308:37–47. 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 172.Lee CH, Gilbertson DL, Novella IS, Huerta R, Domingo E, Holland JJ. 1997. Negative effects of chemical mutagenesis on the adaptive behavior of vesicular stomatitis virus. J Virol 71:3636–3640. 10.1128/JVI.71.5.3636-3640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Domingo E. 2016. Virus as populations: composition, complexity, dynamics, and biological implications. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 174.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med 6:1375–1379. 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 175.Martin V, Grande-Perez A, Domingo E. 2008. No evidence of selection for mutational robustness during lethal mutagenesis of lymphocytic choriomeningitis virus. Virology 378:185–192. 10.1016/j.virol.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 176.Sanjuan R, Cuevas JM, Furio V, Holmes EC, Moya A. 2007. Selection for robustness in mutagenized RNA viruses. PLoS Genet 3:e93. 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Codoner FM, Daros JA, Sole RV, Elena SF. 2006. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog 2:e136. 10.1371/journal.ppat.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Moreno H, Tejero H, de la Torre JC, Domingo E, Martin V. 2012. Mutagenesis-mediated virus extinction: virus-dependent effect of viral load on sensitivity to lethal defection. PLoS One 7:e32550. 10.1371/journal.pone.0032550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Turner TL, Kopp BT, Paul G, Landgrave LC, Hayes D, Jr, Thompson R. 2014. Respiratory syncytial virus: current and emerging treatment options. Clinicoecon Outcomes Res 6:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Graci JD, Cameron CE. 2004. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antivir Chem Chemother 15:1–13. 10.1177/095632020401500101. [DOI] [PubMed] [Google Scholar]
- 181.Dimmock NJ, Easton AJ. 2014. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J Virol 88:5217–5227. 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xiao Y, Lidsky PV, Shirogane Y, Aviner R, Wu CT, Li W, Zheng W, Talbot D, Catching A, Doitsh G, Su W, Gekko CE, Nayak A, Ernst JD, Brodsky L, Brodsky E, Rousseau E, Capponi S, Bianco S, Nakamura R, Jackson PK, Frydman J, Andino R. 2021. A defective viral genome strategy elicits broad protective immunity against respiratory viruses. Cell 184:6037–6051.e14. 10.1016/j.cell.2021.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chaturvedi S, Vasen G, Pablo M, Chen X, Beutler N, Kumar A, Tanner E, Illouz S, Rahgoshay D, Burnett J, Holguin L, Chen PY, Ndjamen B, Ott M, Rodick R, Rogers T, Smith DM, Weinberger LS. 2021. Identification of a therapeutic interfering particle: a single-dose SARS-CoV-2 antiviral intervention with a high barrier to resistance. Cell 184:6022–6036 e18. 10.1016/j.cell.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Notton T, Sardanyes J, Weinberger AD, Weinberger LS. 2014. The case for transmissible antivirals to control population-wide infectious disease. Trends Biotechnol 32:400–405. 10.1016/j.tibtech.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 185.Wasik MA, Eichwald L, Genzel Y, Reichl U. 2018. Cell culture-based production of defective interfering particles for influenza antiviral therapy. Appl Microbiol Biotechnol 102:1167–1177. 10.1007/s00253-017-8660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pelz L, Rudiger D, Dogra T, Alnaji FG, Genzel Y, Brooke CB, Kupke SY, Reichl U. 2021. Semi-continuous propagation of influenza A virus and its defective interfering particles: analyzing the dynamic competition to select candidates for antiviral therapy. J Virol 95:e01174-21. 10.1128/JVI.01174-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Smith C, Scott P, O’Callaghan C, Easton A, Dimmock N. 2016. A defective interfering influenza RNA inhibits infectious influenza virus replication in human respiratory tract cells: a potential new human antiviral. Viruses 8:237. 10.3390/v8080237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rand U, Kupke SY, Shkarlet H, Hein MD, Hirsch T, Marichal-Gallardo P, Cicin-Sain L, Reichl U, Bruder D. 2021. Antiviral activity of influenza A virus defective interfering particles against SARS-CoV-2 replication in vitro through stimulation of innate immunity. Cells 10:1756. 10.3390/cells10071756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hein MD, Kollmus H, Marichal-Gallardo P, Puttker S, Benndorf D, Genzel Y, Schughart K, Kupke SY, Reichl U. 2021. OP7, a novel influenza A virus defective interfering particle: production, purification, and animal experiments demonstrating antiviral potential. Appl Microbiol Biotechnol 105:129–146. 10.1007/s00253-020-11029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dimmock NJ, Dove BK, Meng B, Scott PD, Taylor I, Cheung L, Hallis B, Marriott AC, Carroll MW, Easton AJ. 2012. Comparison of the protection of ferrets against pandemic 2009 influenza A virus (H1N1) by 244 DI influenza virus and oseltamivir. Antiviral Res 96:376–385. 10.1016/j.antiviral.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dimmock NJ, Dove BK, Scott PD, Meng B, Taylor I, Cheung L, Hallis B, Marriott AC, Carroll MW, Easton AJ. 2012. Cloned defective interfering influenza virus protects ferrets from pandemic 2009 influenza A virus and allows protective immunity to be established. PLoS One 7:e49394. 10.1371/journal.pone.0049394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Smither SJ, Garcia-Dorival I, Eastaugh L, Findlay JS, O’Brien LM, Carruthers J, Williamson ED, Molina-Paris C, Hiscox JA, Laws TR. 2020. An investigation of the effect of transfected defective, Ebola virus genomes on Ebola replication. Front Cell Infect Microbiol 10:159. 10.3389/fcimb.2020.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bdeir N, Arora P, Gartner S, Hoffmann M, Reichl U, Pohlmann S, Winkler M. 2019. A system for production of defective interfering particles in the absence of infectious influenza A virus. PLoS One 14:e0212757. 10.1371/journal.pone.0212757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yamagata Y, Muramoto Y, Miyamoto S, Shindo K, Nakano M, Noda T. 2019. Generation of a purely clonal defective interfering influenza virus. Microbiol Immunol 63:164–171. 10.1111/1348-0421.12681. [DOI] [PubMed] [Google Scholar]
- 195.Rudiger D, Pelz L, Hein MD, Kupke SY, Reichl U. 2021. Multiscale model of defective interfering particle replication for influenza A virus infection in animal cell culture. PLoS Comput Biol 17:e1009357. 10.1371/journal.pcbi.1009357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tapia F, Laske T, Wasik MA, Rammhold M, Genzel Y, Reichl U. 2019. Production of defective interfering particles of influenza A virus in parallel continuous cultures at two residence times-insights from qPCR measurements and viral dynamics modeling. Front Bioeng Biotechnol 7:275. 10.3389/fbioe.2019.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bdeir N, Arora P, Gartner S, Pohlmann S, Winkler M. 2021. Evidence that two instead of one defective interfering RNA in influenza A virus-derived defective interfering particles (DIPs) does not enhance antiviral activity. Sci Rep 11:20477. 10.1038/s41598-021-99691-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vasou A, Sultanoglu N, Goodbourn S, Randall RE, Kostrikis LG. 2017. Targeting pattern recognition receptors (PRR) for vaccine adjuvantation: from synthetic PRR agonists to the potential of defective interfering particles of viruses. Viruses 9 9:186. 10.3390/v9070186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mann A, Marriott AC, Balasingam S, Lambkin R, Oxford JS, Dimmock NJ. 2006. Interfering vaccine (defective interfering influenza A virus) protects ferrets from influenza, and allows them to develop solid immunity to reinfection. Vaccine 24:4290–4296. 10.1016/j.vaccine.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 200.Martinez-Gil L, Goff PH, Hai R, Garcia-Sastre A, Shaw ML, Palese P. 2013. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J Virol 87:1290–1300. 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fisher DG, Coppock GM, Lopez CB. 2018. Virus-derived immunostimulatory RNA induces type I IFN-dependent antibodies and T-cell responses during vaccination. Vaccine 36:4039–4045. 10.1016/j.vaccine.2018.05.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Easton AJ, Scott PD, Edworthy NL, Meng B, Marriott AC, Dimmock NJ. 2011. A novel broad-spectrum treatment for respiratory virus infections: influenza-based defective interfering virus provides protection against pneumovirus infection in vivo. Vaccine 29:2777–2784. 10.1016/j.vaccine.2011.01.102. [DOI] [PubMed] [Google Scholar]
- 203.Xue J, Chambers BS, Hensley SE, Lopez CB. 2016. Propagation and characterization of influenza virus stocks that lack high levels of defective viral genomes and hemagglutinin mutations. Front Microbiol 7:326. 10.3389/fmicb.2016.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sun Y, Lopez CB. 2016. Preparation of respiratory syncytial virus with high or low content of defective viral particles and their purification from viral stocks. Bio Protoc 6:e1820. 10.21769/BioProtoc.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yang Y, Lyu T, Zhou R, He X, Ye K, Xie Q, Zhu L, Chen T, Shen C, Wu Q, Zhang B, Zhao W. 2019. The antiviral and antitumor effects of defective interfering particles/genomes and their mechanisms. Front Microbiol 10:1852. 10.3389/fmicb.2019.01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu LW, Nishikawa T, Kaneda Y. 2016. An RNA molecule derived from Sendai virus DI particles induces antitumor immunity and cancer cell-selective apoptosis. Mol Ther 24:135–145. 10.1038/mt.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jones JC, Settles EW, Brandt CR, Schultz-Cherry S. 2011. Virus aggregating peptide enhances the cell-mediated response to influenza virus vaccine. Vaccine 29:7696–7703. 10.1016/j.vaccine.2011.07.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Genoyer E, Kulej K, Hung CT, Thibault PA, Azarm K, Takimoto T, Garcia BA, Lee B, Lakdawala S, Weitzman MD, Lopez CB. 2020. The viral polymerase complex mediates the interaction of viral ribonucleoprotein complexes with recycling endosomes during Sendai virus assembly. mBio 11:e02028-20. 10.1128/mBio.02028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Genoyer E, Lopez CB. 2019. Defective viral genomes alter how Sendai virus interacts with cellular trafficking machinery, leading to heterogeneity in the production of viral particles among infected cells. J Virol 93:e01579-18. 10.1128/JVI.01579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang C, Forst CV, Chou TW, Geber A, Wang M, Hamou W, Smith M, Sebra R, Zhang B, Zhou B, Ghedin E. 2020. Cell-to-cell variation in defective virus expression and effects on host responses during influenza virus infection. mBio 11:e02880-19. 10.1128/mBio.02880-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kupke SY, Ly LH, Borno ST, Ruff A, Timmermann B, Vingron M, Haas S, Reichl U. 2020. Single-cell analysis uncovers a vast diversity in intracellular viral defective interfering RNA Content affecting the large cell-to-cell heterogeneity in influenza A virus replication. Viruses 12:71. 10.3390/v12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sun Y, Kim EJ, Felt SA, Taylor LJ, Agarwal D, Grant GR, Lopez CB. 2019. A specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes. PLoS Pathog 15:e1007707. 10.1371/journal.ppat.1007707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Routh A, Johnson JE. 2014. Discovery of functional genomic motifs in viruses with ViReMa: a virus recombination mapper-for analysis of next-generation sequencing data. Nucleic Acids Res 42:e11. 10.1093/nar/gkt916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Beauclair G, Mura M, Combredet C, Tangy F, Jouvenet N, Komarova AV. 2018. DI-tector: defective interfering viral genomes’ detector for next-generation sequencing data. RNA 24:1285–1296. 10.1261/rna.066910.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Korboukh VK, Lee CA, Acevedo A, Vignuzzi M, Xiao Y, Arnold JJ, Hemperly S, Graci JD, August A, Andino R, Cameron CE. 2014. RNA virus population diversity, an optimum for maximal fitness and virulence. J Biol Chem 289:29531–29544. 10.1074/jbc.M114.592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Young DCS, Sharp DG. 1977. Poliovirus aggregates and their survival in water. Appl Environ Microbiol 33:168–177. 10.1128/aem.33.1.168-177.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Floyd RS, Sharp DG. 1977. Aggregation of poliovirus and reovirus by dilution in water. Appl Environ Microl 33:159–167. 10.1128/aem.33.1.159-167.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Morrone SR, Chew VSY, Lim XN, Ng TS, Kostyuchenko VA, Zhang S, Wirawan M, Chew PL, Lee J, Tan JL, Wang J, Tan TY, Shi J, Screaton G, Morais MC, Lok SM. 2020. High flavivirus structural plasticity demonstrated by a non-spherical morphological variant. Nat Commun 11:3112. 10.1038/s41467-020-16925-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci USA 110:6795–6799. 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. 2013. Structural changes in dengue virus when exposed to a temperature of 37° C. J Virol 87:7585–7592. 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]