SUMMARY
Despite the recent decrease in overall prevalence of Helicobacter pylori infection, morbidity and mortality rates associated with gastric cancer remain high. The antimicrobial resistance developments and treatment failure are fueling the global burden of H. pylori-associated gastric complications. Accurate diagnosis remains the opening move for treatment and eradication of infections caused by microorganisms. Although several reports have been published on diagnostic approaches for H. pylori infection, most lack the data regarding diagnosis from a clinical perspective. Therefore, we provide an intensive, comprehensive, and updated description of the currently available diagnostic methods that can help clinicians, infection diagnosis professionals, and H. pylori researchers working on infection epidemiology to broaden their understanding and to select appropriate diagnostic methods. We also emphasize appropriate diagnostic approaches based on clinical settings (either clinical diagnosis or mass screening), patient factors (either age or other predisposing factors), and clinical factors (either upper gastrointestinal bleeding or partial gastrectomy) and appropriate methods to be considered for evaluating eradication efficacy. Furthermore, to cope with the increasing trend of antimicrobial resistance, a better understanding of its emergence and current diagnostic approaches for resistance detection remain inevitable.
KEYWORDS: Helicobacter pylori, laboratory diagnosis, urea breath test, stool antigen test, rapid urease test, RT-PCR, antimicrobial resistance
INTRODUCTION
Helicobacter pylori is a bacterial pathogen that was classified as a type 1 carcinogen by the International Agency for Research on Cancer in 1994 (1). Subsequently, its carcinogenic behavior and association with cancer development were reinforced in 2001, when a study showed the association of H. pylori infection with gastric cancer. In this study, none of the individuals who were not infected with H. pylori were found to develop gastric cancer after a median follow-up of 8 to 10 years (2). The persistent infection established by this pathogen has been associated with the development of severe gastric complications (3, 4). This pathogen has been found as the major etiologic factor responsible for the development of gastric adenocarcinoma and is considered responsible for more cancer cases worldwide than hepatitis B and C viruses combined (5). Although a recent decline in gastric cancer incidence has been observed, it remains one of the leading causes of cancer-related deaths worldwide (6). According to the GLOBOCAN 2020 report, gastric cancer ranked as the fourth most common cause of cancer-related mortality, leading to estimated deaths of 769,000 individuals in 2020 (7).
Being a bacterial complication, the eradication therapy for H. pylori requires appropriate antibiotic regimens, which are recommended for all patients who are positive for infection by this pathogen. Successful eradication of this pathogen decreases the risk of developing severe gastric complications (8–15). However, to treat the infection, accurate diagnosis is of utmost importance.
MICROBIOLOGICAL ASPECTS (VIRULENCE FACTORS) OF H. PYLORI
H. pylori is a Gram-negative bacterium with a helical shape that chronically infects the human gastric epithelium (16). For successful infection of H. pylori, multiple factors, such as host factors, environmental conditions of the stomach, and bacterial virulence factors, play important roles. Among bacterial virulence factors, some factors, such as bacterial shape, polar-sheathed flagella, motility, chemotaxis, and adherence (reviewed by Ansari and Yamaoka [17]), render bacterial colonization in the gastric epithelium successful, whereas pathogenic factors, such as cagPAI and cytotoxin-associated gene A (CagA), vacuolating cytotoxin A (VacA), blood group antigen-binding adhesin (BabA), outer inflammatory protein A (OipA), duodenal ulcer promoting gene A product (DupA), and sialic acid-binding adhesin (SabA), are associated with increased virulence and pathogenicity of H. pylori (reviewed by Ansari and Yamaoka [18]). The virulence-associated proteins CagA and VacA are the two most studied factors that are closely involved in epithelial cell apoptosis and development of severe gastric complications such as peptic ulcer disease (PUD), gastric cancer, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (19, 20).
H. pylori, after its transmission and passage to the gastric lumen, localizes to a specific location and binds with specific host cell receptors. Although the gastric lumen consists of a harsh acidic condition for its survival, the H. pylori possesses well-established urease-dependent and -independent mechanisms to survive in the gastric lumen (reviewed by Ansari and Yamaoka [17]). In addition to the acid neutralization function, urease induces angiogenesis, which involves the formation of new blood vessels from preexisting vasculature. Angiogenesis is important for tumor growth and metastatic dissemination, which plays an important role in the progression of gastric cancer (21, 22). The bacterial attachment mediated by the binding of BabA with the host epithelial cells protects the bacteria from gastric washing, thereby mediating the development of persistent infection (reviewed by Ansari and Yamaoka [23]). In addition to protection from bacterial washing, the BabA-mediated bacterial attachment with Leb to the gastric epithelial cells aids in the induction of double-stranded DNA breaks in the host cells (24), triggering the production of proinflammatory cytokines involved in cancer development and enhancement of type IV secretion system (T4SS)-mediated direct translocation of CagA and gastric inflammation (25). Moreover, the strains possessing functional triple-positive status (CagA, VacA, and BabA) are associated with a higher colonizing bacterial density, an increased level of gastric inflammation, and an increased incidence of intestinal metaplasia in patients compared with the strains possessing only CagA and VacA (26, 27).
More than 70% of strains isolated globally demonstrate cagPAI, a region with a size of approximately 40 kb in the chromosomal DNA, with a regional variation in 95% of strains isolated from East Asian countries and 60% of strains isolated from Western Hemisphere (28, 29). The cagPAI open reading frames (ORFs) encode the effector protein CagA and other proteins that are involved as components of the bacterial T4SS, which forms a syringe-like structure to deliver the CagA protein directly to the gastric epithelial cells (30, 31). Among the cagPAI ORFs, at least 17 CagPAI proteins are required for synthesizing functional and intact T4SS (30, 32). H. pylori strains harboring intact cagPAI have been associated with increased risks for developing gastric cancer and peptic ulcer (28, 33, 34). CagA is an oncogenic effector protein demonstrating one or multiple specialized regions termed EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs (reviewed by Ansari and Yamaoka [35]). Depending on the geographic variation, the EPIYA motifs can be of four different types, i.e., EPIYA-A, -B, -C, and -D. The CagA of H. pylori strains isolated from East Asian regions possesses EPIYA-A, -B, and -D types, whereas the CagA of strains isolated from western regions typically demonstrates EPIYA-A, -B, and -C types, and one to five EPIYA-C motifs (ABCCCCC) can be observed (36). The third type of EPIYA motif, i.e., EPIYA-C or EPIYA-D, possesses geographic, genotypic, and pathogenic properties determining the virulence characteristics of the protein associated with the increased risk for developing gastric cancer (36). CagA, after its synthesis, is directly translocated to the gastric epithelial cells, and the tyrosine (Y) residue present in the EPIYA motifs undergoes phosphorylation by several types of cellular kinases, including Csk, Src family kinases, and c-Abl, leading to the dysregulation of cell signaling, which induces alterations in cellular physiology (reviewed by Ansari and Yamaoka [35]). The CagA protein possessing the D type of its third EPIYA motif (EPIYA-D) mediates a higher level of dysregulation of cellular functions than CagA harboring the C type (EPIYA-C). Thus, the strains that possess the EPIYA motifs, namely, EPIYA-A, EPIYA-B, and EPIYA-D types, are considered more virulent, as they are frequently associated with gastric cancer than strains possessing EPIYA-A, EPIYA-B, and EPIYA-C motifs, which are considered less virulent (37). However, a large number of EPIYA-C sequences increase the strength of virulence, inducing a significant increase in the risk for developing PUD in populations of Asian countries and an increased risk for developing gastric cancer in the United States and European populations (36).
After its synthesis as a 140-kDa protoxin, VacA undergoes enzymatic degradation to produce mature VacA consisting of an 88-kDa monomer (possesses two proteolytic fragments of the N-terminal p33 domain and the C-terminal p55 domain). The tryptophan-rich region of the p33 domain is involved in host cell membrane binding, whereas the entire p33 domain together with the 111 N-terminal amino acid residues in the p55 domain is involved in the efficient formation of vacuoles (reviewed by Ansari and Yamaoka [38]). After its intracellular transportation, VacA accumulates inside different cellular compartments, induces the formation of vacuoles inside host epithelial cells (39), distorts the function of mitochondria (40, 41), inhibits the activity of T cells (42, 43), activates mitogen-mediated protein kinase pathways (44), and mediates apoptosis and cell death (45–47).
Other virulence proteins, such as OipA, DupA, SabA, and a protein that is activated on contact with the epithelium (IceA), are involved in the stimulation of gastric epithelial cell apoptosis and development of severe gastric complications, including peptic ulcers and gastric cancer (48–52). The H. pylori strains that harbor these proteins are considered more pathogenic than strains that lack these proteins.
EPIDEMIOLOGY OF H. PYLORI INFECTION AND ASSOCIATED DISEASES
Typically, H. pylori infection is contracted during childhood after an infant is weaned (53). Although the exact mode of bacterial transmission remains unclear, epidemiological studies suggest its transmission via an oral-oral or fecal-oral route from person to person, particularly among family members, such as from mother to child (54–56). After transmission, H. pylori neutralizes the gastric acidic conditions and survives in harsh environments, leading to the colonization and persistent infection of the gastric epithelium (17) (Fig. 1). It is estimated that at least half of the world’s population is infected by H. pylori (57, 58). Factors such as geographic variation, socioeconomic status, urbanization level, and poor sanitation during childhood play a key role in determining the prevalence of H. pylori infection in countries. According to a meta-analysis conducted examining its global burden in 2018, the highest prevalence of 89.7% was found in Nigeria and the lowest prevalence of 8.9% was found in Yemen (57); however, a more recent study conducted in 2021 found an even lower prevalence of 2.5% in Sri Lanka (59). The persistent infection leads to the development of severe gastroduodenal complications, including chronic gastritis, peptic ulcer, gastric ulcer, gastric cancer, and gastric MALT lymphoma (3, 4). However, the frequency of developing these complications among infected patients is very low, and it has been estimated that 100 to 1,000 patients, 10 to 300 patients, and less than one patient develop PUD, gastric cancer, and gastric MALT lymphoma, respectively, among every 10,000 patients infected with H. pylori (60). H. pylori infection is thought to be associated with approximately 70% of all gastric ulcers and up to 80% of all duodenal ulcers, and the risk increases with a history of H. pylori infection even after its successful elimination compared with that observed in noninfected individuals (60). According to a cohort study, 1 to 2% of H. pylori-infected individuals develop gastric cancer (2).
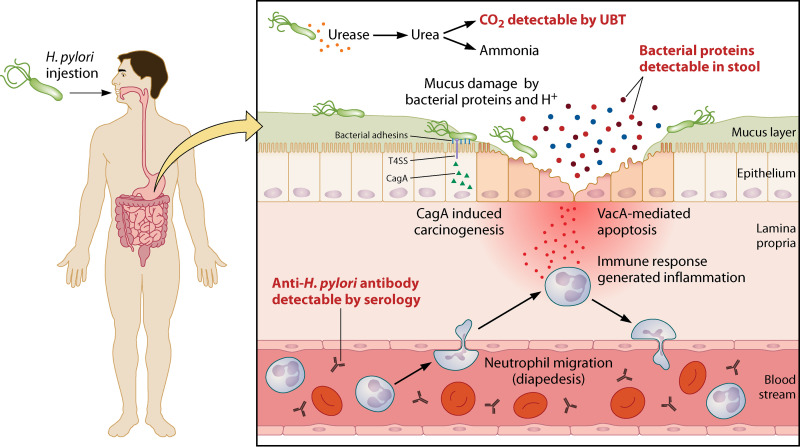
FIG 1.
H. pylori-associated pathogenicity. After ingestion, H. pylori enters the stomach, and the urease produced by the bacteria hydrolyzes the urea, thereby generating CO2, which can be detected by the urea breath test (UBT), and ammonia. Ammonia neutralizes the acidic pH, creating an almost neutral microenvironment around bacterial cells that enables the bacteria to survive under adverse gastric conditions. Later, the bacteria find their way into the mucus layer owing to multifactorial mechanisms such as their helical shape, the presence of flagella, and chemotaxis. Several proteins (such as BabA, SabA, and OipA) produced by bacteria help in the colonization and persistence of infection. Moreover, these proteins are detected in stool specimens of infected patients by stool antigen tests (SATs). The immune response targeting numerous immunogenic proteins is evaluated by identifying antibodies using serological tests. The protein CagA is directly translocated into the gastric epithelium, and CagA-mediated carcinogenesis is triggered, whereas the VacA protein contributes to apoptosis and epithelial cell death.
“TEST-AND-TREAT” STRATEGY
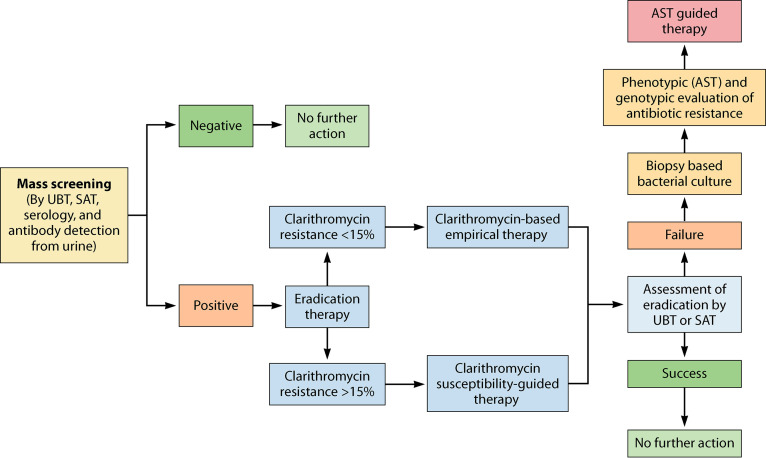
Eradication therapy for H. pylori has been significantly associated with reduced risks for gastric cancer development if administered to healthy and asymptomatic patients before the development of preneoplastic lesions (8, 12, 61, 62). However, some studies have reported significant improvement and reversal of atrophy and even intestinal metaplasia, to a lesser degree, after successful eradication therapy (63–65). Moreover, a recent clinical trial conducted in South Korea found that endoscopic removal of early gastric cancer lesions can significantly prevent the development of gastric cancer (10). A systematic review and meta-analysis reported that eradication therapy administered to infected healthy adults was associated with a 46% reduction in the incidence of gastric cancer and a 39% reduction in the mortality associated with gastric cancer (13). A significant reduction has been observed in the incidence of gastric cancer among participants accurately diagnosed with successful eradication of the bacterium (8, 10, 12, 66, 67). Furthermore, the successful eradication of H. pylori also reduces the risk of transmission of infection, and thus, the financial burden associated with this infection may be avoided. Therefore, screening should be performed with the intention to recommend eradication therapy if the test results are positive for H. pylori infection, also known as the “test-and-treat” strategy (68).
Simple, noninvasive, and relatively low-cost tests are recommended for the test-and-treat strategy. A locally validated serology test to detect anti-H. pylori antibody in serum could be an optimal approach if sufficient accuracy could be achieved (69). However, since IgG detection cannot differentiate between past and current infection status, serology as a single test may not be appropriate, especially in countries with high prevalence (70, 71). In countries with high prevalence, individuals who are positive for the presence of IgG should also be tested by other suitable tests capable of differentiating between past and present infections. The urea breath test (UBT) is a simple test and one of the most examined tests capable of detecting active H. pylori infection. Hence, it is the most widely recommended noninvasive test for the test-and-treat strategy (69, 72). This test is also recommended for assessing the success of eradication after antibiotic therapy (72). In the test-and-treat strategy, the monoclonal antibody-based stool antigen test (SAT) can also be used after its local validation; it is also a recommended test (69). However, if patients are subjected to proton-pump inhibitor (PPI) treatment, the sensitivity of these tests decreases to an unacceptable level. Hence, the PPI treatment should be discontinued before 14 days of performing the tests (69, 73). Irrespective of the diagnostic methods used, the eradication therapy based on the antibiotic resistance rate of that geographic region should be offered to all patients positive for H. pylori infection.
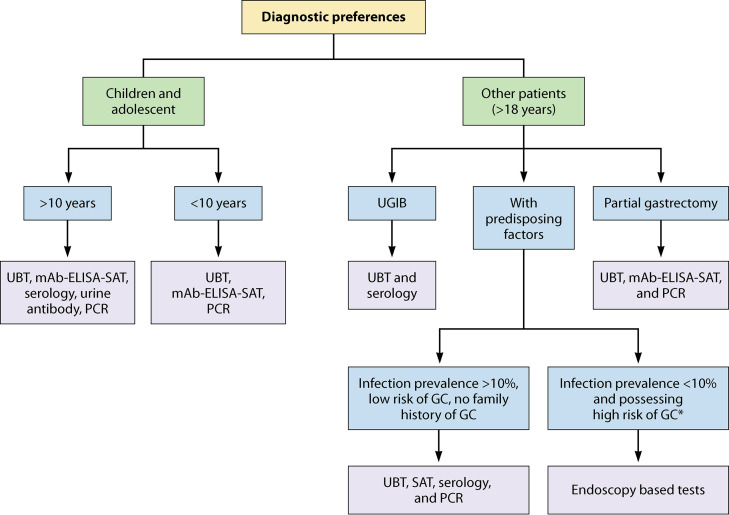
The Maastricht IV/Florence consensus report in 2012 recommended the test-and-treat strategy for H. pylori infection, particularly in populations comprising communities with a high burden of gastric cancer (69). The Kyoto global consensus report published in 2015 recommended the test-and-treat strategy and suggested screening for H. pylori infection after 12 years of age and administration of eradication therapy for all positive cases even in the absence of any related symptoms or conditions (74). Regarding the Kyoto global consensus recommendation, the Maastricht V/Florence consensus report in 2016 recommended the test-and-treat strategy for patients showing dyspeptic symptoms and even for patients with hematological disorders such as iron deficiency anemia, immune-thrombocytopenic purpura, and vitamin B12 deficiency because of the considerable evidence of the association of H. pylori infection with these hematological disorders (72). Similarly, the Houston consensus report on testing for H. pylori infection in the United States. in 2018, also recommended an antibiotic therapy to eradicate H. pylori infection in all individuals with proven infection (75). However, because of the low incidence of H. pylori-related gastric cancer in the United States, the American College of Gastroenterology (ACG) suggests testing for H. pylori infection in patients with predisposing factors, such as a current case or a history of PUD, low-grade gastric MALT lymphoma, and history of endoscopic resection of early gastric cancer lesions (68). The American Gastroenterological Association (AGA) strongly recommends the test-and-treat strategy for individuals with confirmed gastrointestinal metaplasia (76). The Bangkok consensus report for the Association of Southeast Asian Nation countries (Indonesia, Thailand, the Philippines, Malaysia, Singapore, Vietnam, Myanmar, Cambodia, Laos, and Brunei) published in 2018 recommended testing for H. pylori infection in patients with chronic dyspeptic symptoms and not in asymptomatic patients, owing to the infection being more commonly observed among patients with dyspepsia (77). In 2020, the Taipei global consensus report recommended screening and eradication therapy for H. pylori infection in populations with a high incidence or high risk for gastric cancer (78). The test-and-treat strategy should be routinely implemented for individuals belonging to high-risk populations. Although the eradication stops the progression of infection, the genetic instability is not completely reversed. Therefore, the early screening and treatment of H. pylori infection are performed before developing irreversible genetic instability histologically reflected in atrophic gastritis and intestinal metaplasia (63–65).
LABORATORY DIAGNOSIS
Given the association and the causative role of H. pylori in PUD and gastric cancer, finding the best diagnostic method is of utmost importance for clinicians and microbiologists. Since the presence or absence of current infection provides information for determining the type of treatment to be administered, testing for H. pylori infection is crucial for the monitoring of the effectiveness of treatment and disease management (79, 80).
Currently, H. pylori infection can be diagnosed by several methods, such as noninvasive tests, which do not require endoscopy or biopsy specimens (antibody detection from serum and urine, UBT, SAT, and PCR from stool), and invasive tests, which require biopsy specimens collected via endoscopy (histopathology, rapid urease test [RUT], culture, and PCR from biopsy specimen) (81, 82). All these methods have their own merits and limitations. Some of these methods demonstrate superiority over other methods depending on the clinical setting. Therefore, promising diagnostic tests for H. pylori infection with high sensitivity, high specificity, cost-effectiveness, rapid performance, and noninvasiveness are recommended depending on clinical circumstances (81, 83). Although noninvasive tests provide added advantages of cost-effectiveness, convenience of sample collection, and rapid results, the antibody detection methods have relatively lower reliability owing to their low specificity compared with invasive tests (83). It is universally accepted that no single test is considered the gold standard for infection diagnosis, and the reliability and accuracy of the diagnosis strengthen when multiple diagnostic tests are performed (79, 80, 82, 84). Furthermore, all the diagnostic tests should be validated to achieve high diagnostic accuracy in a specific region. The performance of different diagnostic methods according to the latest published reports is summarized in Tables 1 to 5.
TABLE 1.
Performance of stool antigen-based diagnostic kits (compared with the gold standard reference method) currently in clinical use, according to recent reportsa
| Test | Gold standard method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Reference |
|---|---|---|---|---|---|---|---|
| Testmate Pylori antigen EIA (Wakamoto Pharmaceutical Co., Ltd., Tokyo, Japan) | Culture alone or histology and RUT combined | 99.6 | 100 | 127 | |||
| Amplified IDEIA HpStAR (Thermo Fisher Scientific, Waltham, MA, USA) | Culture alone or histology and RUT combined | 93.6 | 100 | 100 | 87.3 | 96 | 128 |
| Diagnostec H. pylori antigen EIA kit (Reininghun Diagnostics Biomedical, Inc, Taiwan) | UBT | 92.9 | 98.3 | 95.8 | 97.1 | 96.7 | 129 |
| Diagnostec H. pylori antigen rapid test kit (Reininghun Diagnostics Biomedical, Inc, Taiwan) | UBT | 92.9 | 95.8 | 90.1 | 97.0 | 94.9 | 129 |
| H. pylori Quik Chek test (TechLab Inc., Blacksburg, VA, USA) | At least two of histology, culture, and RUT positive | 91 | 100 | 98 | 97 | 130 | |
| H. pylori Chek test (TechLab Inc., Blacksburg, VA, USA | At least two of histology, culture, and RUT positive | 92 | 91 | 76 | 97 | 130 | |
| Wondfo one-step H. pylori feces test | RUT | 65.1 | 70.2 | 62.2 | 72.7 | 68 | 131 |
| Uni-Gold H. pylori antigen test (Trinity Biotech, Ireland) | At least two of RUT, histology, and UBT positive | 83.2 | 89.3 | 87.6 | 85.4 | 132 | |
| RAPID Hp StAR (Oxoid Ltd., United Kingdom) | At least two of RUT, histology, and UBT positive | 95.0 | 84.7 | 85.0 | 94.9 | 132 | |
| ImmunoCard STAT! HpSA (Meridian Diagnostics, USA) | At least two of RUT, histology, and UBT positive | 81.5 | 91.6 | 88.7 | 89.8 | 132 | |
| Genx H. pylori card test (Genx Bioresearch GOSB Teknopark A.S., Gebze, Kocaeli, Turkey) | Histology and RUT | 51.2 | 95.0 | 91.5 | 65.5 | 72.8 | 134 |
| CerTest H. pylori blister test (CerTest Biotec S.L., Zaragoza, Spain) | UBT | 68.7 | 97.6 | 88.5 | 92.0 | 91.5 | 135 |
| Quick Chaser H. pylori, QCP (Misuho Medy, Tosu, Japan) | RUT and culture | 92.3 | 100 | 136 | |||
| Liaison H. pylori SA assay (DiaSorin, Stillwater, MN, USA) | At least two of histology, culture, and RUT positive | 90.5 | 97.6 | 92.8 | 98.6 | 139 |
PPV, positive predictive value; NPV, negative predictive value.
TABLE 2.
Performance of antibody detection kits (compared with the gold standard reference method) currently in clinical use, according to recent reportsa
| Test | Gold standard method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Reference(s) |
|---|---|---|---|---|---|---|---|
| H. pylori IgG EIA test kit (Rapid Labs, Ltd., Little Bentley, Essex, United Kingdom) | RUT | 90.5 | 39.6 | 52.1 | 85.2 | 61 | 131 |
| E-plate Eiken H. pylori Antibody II (Eiken Chemical Co., Ltd., Tokyo, Japan) | UBT | 97.4 | 76.3 | 91.5 | 91.8 | 91.6 | 145 |
| H. pylori IgG Seiken (Denka Seiken Co., Ltd., Tokyo, Japan) | UBT | 99.4 | 74.6 | 91.1 | 97.8 | 92.5 | 145 |
| GastroPanel (Biohit Oyj, Helsinki, Finland) | Histology | 74.7 | 95.6 | 91 | 86 | 160, 161 | |
| Unified GastroPanel (Biohit Oyj, Helsinki, Finland) | Histology | 93.8 | 88.9 | 93.8 | 88.9 | 165 | |
| URINELISA (Otsuka Pharmaceuticals Co., Ltd., Tokyo, Japan) | UBT or SAT | 86.5 | 85.8 | 167 | |||
| Rapirun (Otsuka Pharmaceutical Co., Ltd., Tokyo, Japan) | Culture alone or histopathology and RUT combined | 86.2 | 90.8 | 80.6 | 93.7 | 89.4 | 171 |
| gabControl H. pylori (gabmed GmbH, Cologne, Germany) | UBT | 91.4 | 76.7 | 65.3 | 94.9 | 174 | |
| Eiken H. pylori antibody (Eiken Chemical Co., Ltd., Tokyo, Japan) | UBT | 98.1 | 78.0 | 92.1 | 93.9 | 92.5 | 145 |
| H. pylori Latex Seiken (Denka Seiken Co., Ltd., Tokyo, Japan) | UBT | 98.1 | 71.2 | 89.9 | 93.3 | 90.6 | 145 |
| Pyloriset Dry (Orion Diagnostica, Espoo, Finland) | 95 | 82 | 181 | ||||
| RecomLine H. pylori IgG (Mikrogen Diagnostik, Germany) with 6 antigens | Histology | 98.3 | 95.5 | 146 | |||
| RecomLine H. pylori IgG (Mikrogen Diagnostik, Germany) with 4 antigens | Histology | 96.1 | 20.9 | 69.7 | 73.7 | 191 |
PPV, positive predictive value; NPV, negative predictive value.
TABLE 3.
Performance of rapid urease tests (compared with the gold standard reference method) currently in clinical use, according to recent reportsa
| Test | Gold standard method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Reference(s) |
|---|---|---|---|---|---|---|---|
| PyloriTek (Serim Research Corp., Elkhart, IN, USA) | Histology | >96 | >97 | 203 | |||
| Pronto Dry (Medical Instruments Corporation, Solothurn, Switzerland) | Histology | 100 | 96.1 | 96.7 | 100 | 204 | |
| CLO (Ballard Medical Products, Draper, UT, USA) | Culture | 94 | 88 | 89 | 93 | 205 | |
| Hp-Fast and Hp-One (GI Supply, Mechanicsburg, PA, USA) | Histology | 92.3 | 100 | 100 | 91.2 | 206 | |
| Endosc-Hp (Cambridge Life Sciences, Ltd., United Kingdom) | CLO test | 94.4 | 98.4 | 97.1 | 96.9 | 97.0 | 207 |
| UFT 300 (Biohit Oyj, Helsinki, Finland) | Histology and UBT | 94.5 | 100 | 208, 209 |
PPV, positive predictive value; NPV, negative predictive value.
TABLE 4.
Performance of histopathological diagnostic methods (compared with the respective gold standard reference method) currently in clinical use, according to recent reportsa
| Test | Gold standard method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Reference |
|---|---|---|---|---|---|---|
| IHC | Histology | 98.8 | 99.2 | 98.0 | 99.6 | 250 |
| MTB | IHC | 99 | 96 | 95 | 99 | 258 |
| Imprint cytology stained with: | ||||||
| Toluidine blue | Histology | 57.1 | 97.9 | 80.0 | 94.0 | 259 |
| Giemsa stain | Histology | 42.9 | 97.9 | 75.0 | 92.2 | 259 |
PPV, positive predictive value; NPV, negative predictive value. Accuracy was not available for these tests.
TABLE 5.
Performance of molecular methods (compared with the gold standard reference method) currently in clinical use for H. pylori detection, according to recent reportsa
| Test | Gold standard method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Reference |
|---|---|---|---|---|---|---|---|
| Nested PCR | UBT | 93.8 | 95.9 | 299 | |||
| Multiplex PCR | CIM and CLO | 100 | 59.6 | 73.8 | 100 | 81.1 | 300 |
| BACTfish H. pylori Combi test (Izinta Kft., Budapest, Hungary) | Histology | 98 | 99.6 | 98.9 | 99.2 | 250 | |
| RIDA GENE H. pylori (R-Biopharm AG, Darmstadt, Germany) | Culture alone or histopathology and RUT combined | 100 | 99 | 94 | 100 | 318 | |
| Amplidiag H. pylori+ClariR (Mobidiag, Espoo, Finland) | Culture or TaqMan RT-PCR | 96.3 | 98.7 | 92.2 | 99.3 | 319 | |
| MutaREAL H pylori (Inmundiagnostik, Bensheim, Germany) | Culture | 93.3 | 86.9 | 90.3 | 90.9 | 320 | |
| ViaSure H. pylori real-time PCR detection kit (CerTest Biotec S.L., Zaragoza, Spain) | SAT | 85.7 | 100 | 100 | 84.6 | 92 | 321 |
PPV, positive predictive value; NPV, negative predictive value.
Urea Breath Test
The UBT is a noninvasive and highly reliable diagnostic test widely used for diagnosing H. pylori infection. The test is based on the hydrolysis of 13C or 14C isotope-labeled urea (85). The orally administered urea is hydrolyzed by bacterial urease into ammonia and isotope-labeled carbon dioxide in the stomach. The isotope-labeled carbon dioxide is diffused into the blood circulation and expelled during exhalation. The exhaled carbon dioxide is measured using an isotope ratio mass spectrometer (82, 86–88). Since its introduction in clinical settings, the 13C-labeled UBT has garnered significant interest, and thus, it is recommended by several national and international guidelines and several expert consensus reports over 14C-labeled urea because 13C is a stable, nonradioactive carbon isotope with a natural abundance of approximately 1% among all carbon isotopes (72, 89–91). The measurement of 13C-labeled carbon dioxide by conventional methods is more expensive, and it requires skilled personnel. Therefore, other alternatives with less expensive methods, such as infrared spectroscope and laser-assisted ratio analyzer, have been developed to measure the 13C-labeled carbon dioxide activity. The increase in the 13C-labeled carbon dioxide activity before and 30 min after consuming the urea-based meal indicates the urease activity exploited by live H. pylori in the stomach (92). The differences in the 13C isotope/12C (in normal breath) ratio between the value observed at 30 min and the baseline value are determined, which are then expressed as delta over baseline (DOB, per mille).
The cutoff DOB value of 5.0‰ originally determined and recommended by standard European protocol is the most widely used value to discriminate between positive and negative H. pylori infection (93). However, several factors affect the results, and using this DOB cutoff of 5.0‰, the test showed a lower accuracy in different clinical settings to determine positive infections. Therefore, several attempts were made to validate a new cutoff value according to clinical settings. In a validation study, the diagnostic accuracy was improved using DOB cutoffs of 7.0‰ for children less than 6 years old and 4.0‰ for children over 6 years old (94). In other studies, lower cutoff values, between 3.0 and 4.0‰, enhanced the diagnostic accuracy without compromising the performance (95, 96). Therefore, no consensus has been established with respect to the precise cutoff value for DOB; hence, it is difficult to determine the optimum cutoff value. Precise cutoff values should be validated according to the populations targeted.
UBT and test duration.
Besides patient characteristics, various other test characteristics, such as the test duration and test meal, have been determined to influence the cutoff value. For instance, several studies have validated the precise cutoff by shortening the test duration. Malaty (97) et al. decreased the test duration to 20 min in their study, omitted the test meal, and used 125 mg of [13C]urea. A cutoff DOB value of 2.4‰ showed 96% sensitivity and 100% specificity (97). Ohara (98) et al. used a test duration of 20 min, omitted the test meal, and used 100 mg of 13C-labeled urea. Sensitivity and specificity of 98.1% and 97.9%, respectively, were observed when a cutoff DOB value of 2.5‰ was used, which is widely accepted in Japan (98). Similarly, by shortening the test duration to 15 min, using fresh milk as the test meal, and using 100 mg of [13C]urea, a DOB cutoff of 2.8‰ demonstrated 99% sensitivity and 93% specificity (99).
UBT and test meal.
Other studies have also attempted to validate the cutoff values by altering the test meals. Leodolter (100) et al. used a test meal of 200 mL of 0.1 mol/L citric acids with a test duration of 30 min as originally recommended. The optimum DOB cutoff was determined to be 4.0‰ (100). With this result, it is evident that the manufacturer’s recommended cutoff values are not applicable in every clinical setting. The UBT values are significantly higher for females than for males. The values increase significantly with increasing age, decrease with increasing BMI, increase in patients with low socioeconomic status, and decrease with smoking habits (101, 102). Minor adjustments in the [13C]UBT cutoff value to achieve better diagnostic accuracy can have major public health benefits. The upward adjustment has the potential to decrease the unnecessary antimicrobial exposure, leading to a decrease in microbial resistance and antibiotic-associated mortality, whereas the downward adjustment has the potential to increase the diagnosis of positive cases, leading to decreased gastric cancer burden in high-prevalence and high-risk regions (103).
Although the UBT is highly sensitive and specific in detecting infection, it is not recommended for patients consuming PPI because of high rates of false-negative results (104–107). Therefore, it is recommended by current guidelines to discontinue PPI medications for 14 days before the UBT is performed (69, 74, 108–110); however, according to one study, a withdrawal of 7 days was found to be sufficient (111). PPI medications inhibit acid secretion and urease activity, which can consequently reduce the number of bacteria in the stomach, especially in the antrum, thereby raising the possibility of false-negative UBT results (104, 112). In addition to PPI medications, other factors, such as antimicrobial medication, bleeding ulcers, and corpus predominant gastritis, may give false-negative UBT results, whereas the presence of other pathogens that also synthesize urease in the stomach, such as Helicobacter heilmannii may give false-positive UBT results (113–117). Therefore, antimicrobial consumption should be stopped 4 weeks before conducting UBTs, and bleeding should be resolved before the UBT is performed. According to previous studies, false-negative results may be partially reversed if stomach acidification is complete (118, 119). Based on these findings, a new acidification test meal was developed to overcome the effect of PPI medications. This novel acidified test meal (Refex) contains a mixture of three organic acids, i.e., citric acid, malic acid, and tartaric acid (120). These acids bind to many trace elements, such as nickel, and increase urease activity by lowering the pH and mediating the activation and opening of urea channels by H. pylori. In a study examining the effects of PPI with ingestion of the new test meal, three cutoff values of 3.0, 2.5, and 2.0 DOB were set for testing results, and a PPI medication intake was discontinued for 1 day. Interestingly, with these modifications, high sensitivity of 92.5% and specificity of 97.96% were achieved using cutoff values of 2.5 and 2.0 DOB (120). This result indicates that the performance of the UBT with the new test meal is safe for patients consuming PPI medication, and a comparable accuracy can be achieved using a more precise cutoff value.
Because of its high performance with over 95% sensitivity and specificity, safety, and minimal or no radiation exposure, the [13C]UBT is the most preferred test for diagnosing H. pylori infection in children and pregnant women (88, 121). Moreover, the collected breath samples can be sent by post to commercial laboratories that have mass spectrometers for analysis (122). However, the requirement for expensive equipment, lack of infrastructure, need for skilled personnel, requirement of multiple office visits to complete testing, and high cost of testing limit the widespread use of the [13C]UBT in clinical practice (82). Moreover, to obtain the highest diagnostic accuracy, [13C]UBT has to be validated and adjusted in terms of cutoff value, pretest meal, and urea dose.
Stool Antigen Test
The detection of H. pylori-specific antigen from stool samples is a reliable noninvasive method (123–125). SATs detecting bacterial antigens can diagnose active infections and thus are prone to give fewer false-positive results than serological tests. SATs are easy to perform and are recommended for diagnosing H. pylori infection in pediatric patients (123–126). In addition, this test is recommended in cases for which the UBT cannot be performed, such as for patients with asthma or achlorhydria and after gastrectomy. The currently available SATs are based on the enzyme-linked immunosorbent assay (ELISA), immunochromatographic assay, and chemiluminescence immunoassay (CLIA).
Enzyme immunoassay.
The ELISA-based SAT utilizes mono- or polyclonal antibodies against H. pylori antigens to detect H. pylori-specific antigens in stool samples (82). ELISAs utilizing monoclonal antibodies captured on the surfaces of microplate wells are widely used in epidemiological studies and assessment of eradication therapies. The Testmate Pylori antigen enzyme immunoassay (EIA) (Wakamoto Pharmaceutical Co., Ltd., Tokyo, Japan) (127) and amplified IDEIA HpStAR (Thermo Fisher Scientific, Waltham, MA, USA) (128) are currently available monoclonal antibody-based ELISAs capable of providing a sensitivity over 93% and a specificity up to 100% (Table 1). The Diagnostec H. pylori antigen EIA kit (Reininghun Diagnostics Biomedical, Inc., Taiwan) utilizes polyclonal antibodies, is a highly sensitive test, and demonstrates better diagnostic performance. A study evaluated the diagnostic performance of this test compared with the immunochromatography-based Diagnostec H. pylori antigen rapid test kit (Reininghun Diagnostics Biomedical, Inc., Taiwan) in the Chinese population to assess the updated age-standardized prevalence of H. pylori infection in symptomatic and dyspeptic patients. The study found that the performance of the ELISA-based test in the diagnosis of H. pylori infection designed for updated age-standardized prevalence was superior to the chromatography-based test. Here, the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of these tests were 92.9% versus 92.9%, 98.3% versus 95.8%, 95.8% versus 90.1%, 97.1% versus 97.0%, and 96.7% versus 94.9%, respectively (129). Moreover, the newly introduced H. pylori Quik Chek test (TechLab Inc., Blacksburg, VA, USA), a rapid membrane EIA (rapid EIA), and the H. pylori Chek test, a microwell EIA (TechLab Inc., Blacksburg, VA, USA) have demonstrated excellent performance for initial diagnosis recently examined in patients recruited from diverse geographic regions, including USA, Germany, and Bangladesh (130). The sensitivity, specificity, PPV, and NPV were 91%, 100%, 98%, and 97%, respectively, for H. pylori Quik Chek and 92%, 91%, 76%, and 97%, respectively, for H. pylori Chek when positive results obtained with at least two of three tests (histology, culture, and RUT) were used as references (130).
Immunochromatographic assay.
In immunochromatographic assay methods for SATs, H. pylori antibodies are immobilized in a test line on a nitrocellulose membrane, and the antigens present in the stool samples migrate upward from the sample well and form an antigen-antibody complex at the test line. The antigen-antibody complex bound at the test line is detected based on the dye-antibody conjugate that produces the colored band at the test line, thereby indicating positive results. In case of the absence of antigens in stool samples, no colored band would be observed, indicating negative results (131). Currently available commercial immunochromatography-based tests for stool antigens include the Wondfo one-step H. pylori feces test (Guangzhou Wondfo Biotech Co., Ltd., China) (131), the Uni-Gold H. pylori antigen test (Trinity Biotech, Ireland), RAPID Hp StAR (Oxoid Ltd., United Kingdom), ImmunoCard STAT! HpSA (Meridian Diagnostics, USA) (132), the Testmate Rapid Pylori antigen test (Wakamoto Pharmaceutical Co., Ltd., Tokyo, Japan) (127), Immunocard ST HpSA (FujiRebio Co., Ltd., Tokyo, Japan) (133), the Genx H. pylori card test (Genx Bioresearch, GOSB Teknopark A.S., Gebze, Kocaeli, Turkey) (134), and the Diagnostec H. pylori antigen rapid test kit (Reininghun Diagnostics Biomedical, Inc., Taiwan) (129). These tests demonstrate high accuracy for detecting H. pylori antigens in the stool (Table 1).
The CerTest H. pylori blister test (CerTest Biotec S.L. Zaragoza, Spain) is a one-step immunochromatographic assay that has been evaluated for diagnostic accuracy in patients over 65 years old. The test demonstrates overall sensitivity, specificity, PPV, NPV, and accuracy of 68.7%, 97.6%, 88.5%, 92.0%, and 91.5%, respectively (135). When the sensitivity of the test was compared between patients with constipation and those without constipation and patients with colorectal polyps and those without colorectal polyps, the test showed significantly lower sensitivity in patients with constipation and those without colorectal polyps (135). Recently, Kakiuchi et al. (136) evaluated the diagnostic performance of a novel stool antigen rapid kit, Quick Chaser H. pylori (QCP; Misuho Medy, Tosu, Japan), that targets the detection of H. pylori-specific flagellar protein (i.e., flagellin). The newly developed QCP test was found to be a very promising test, with a sensitivity of 92.3% when the RUT and bacterial culture were considered the gold standard reference test (136). The QCP test showed a positive concordance rate, negative concordance rate, and overall concordance rate of 100%, 92.9%, and 98.6%, respectively (136).
Chemiluminescence immunoassay.
CLIA is another novel one-step sandwich assay that utilizes monoclonal antibodies intended for the qualitative detection of H. pylori antigen in stool samples. This assay consists of paramagnetic particles captured with antibodies against H. pylori antigens. The enzymatic cleavage of the mouse monoclonal antibody conjugated with luminol, which is a very common chemiluminescent substrate, results in the production of flashes of visible light signals that are measured by a photomultiplier as relative light units (RLUs). The number of RLUs is a measure of the proportion of the concentration of H. pylori antigen in the stool sample (137). The Liaison H. pylori SA assay (DiaSorin, Stillwater, MN, USA) is a recently introduced method for detecting H. pylori antigen based on CLIA (137, 138). For adult patients, the company has reported a sensitivity of 95.5%, specificity of 98.6%, positive agreement of 100%, negative agreement of 98.2%, and an overall agreement of 98.8% compared with the results of the histopathological evaluation, bacterial culture, and urease detection test used as the composite reference (137). However, the company has not evaluated the performance of this test in pediatric patients. The diagnostic performance of this test has been also evaluated by several authors, and the sensitivity, specificity, PPV, and NPV were 95.5%, 97.6%, 92.8%, and 98.6%, respectively, when evaluated in adult patients in the United States (139) and 90.1%, 92.4%, 91.6%, and 90.1%, respectively, when evaluated in adults with dyspeptic symptoms in Spain (140).
SATs are considered reliable tests that can achieve sensitivity and specificity up to 99% (127). However, the diagnostic accuracy of SATs is influenced by some gastrointestinal factors, such as bleeding ulcers and treatments with PPIs, antibiotics, bismuth-containing compounds, and N-acetylcysteine (NAC) (116). The intake of PPIs, antibiotics, and bismuth leads to a decrease in the bacterial load in the stomach that may result in false-negative results obtained with SATs, similar to those obtained with the UBT (141). In addition to these factors, storage and handling of collected stool samples may influence test results. If the testing for stool antigen is not possible within a short period of time, the stool samples should be kept frozen to keep the antigens intact. Therefore, the test results may be severely affected in a setting where there is a lack of resources (92). Furthermore, the tests need to be validated locally to select a proper cutoff value to achieve higher sensitivity and specificity, which may vary according to different populations (92).
Antibody Detection Tests
H. pylori infection induces immune responses, and antibodies (IgM, IgA, and IgG) are produced against immunogenic proteins (142). Similar to other infections, IgM can be detected in the acute phase of infection, whereas IgA and IgG are detected in the chronic phase of infection. Several antibody detection methods, such as enzyme immunoassay, immunochromatographic assay, latex agglutination immunoassay, immunoblotting assay, and multiplex immunoassay, are available for detecting these antibodies from serum (serological methods), whole blood, saliva, and urine samples (143, 144). Although IgA, IgG, and IgM are produced and can be detected by several tests, only the detection of IgG is performed owing to its reliability in initial screening of infections, which can achieve sensitivity and specificity up to 99% and 96%, respectively (145, 146). High rates of false-positive results are attributed to IgA and IgM; therefore, the detection of these antibodies is not reliable to confirm the infection (147). Moreover, IgG can be continuously found in the serum for a long duration even after successful eradication of bacteria; hence, serological testing of IgG detection is not reliable to assess the success of eradication therapy (75, 148). These tests are considered initial screening tests for excluding the necessity of diagnosis of H. pylori infection in populations with low disease prevalence, and positive results should be confirmed by other tests with higher specificity (68, 149–151).
Despite these contraindications, these tests do have several advantages. For instance, if patients are subjected to treatment with colloidal bismuth, antibiotics, and PPIs, the detection of IgG by these methods could be beneficial, as it does not require discontinuing these medications because of the persistence of this antibody for a long duration (69, 152, 153). Furthermore, in special clinical cases, such as in patients with gastrointestinal bleeding, gastric carcinoma, gastric MALT lymphoma, and atrophic gastritis, antibody detection tests could be the method of choice for diagnosis (154). The bacterial density is significantly decreased under these conditions, and therefore, false-negative results can be obtained with other tests. These tests are also widely used because they require less expensive materials and they demonstrate high reproducibility and sensitivity for initial diagnosis. Currently, several antibody detection kits are commercially available (Table 2), and the tests are easy to perform; however, the performance of these kits depends on several factors, such as age, sex, and ethnicity, and local validation of their good performance is therefore required (149, 155). Compared with other noninvasive tests, such as UBT and SAT, antibody detection tests for detecting IgG have lower diagnostic accuracy owing to continuous persistence of IgG for several months even after successful eradication, and hence, the tests are unable to differentiate past infection from present infection (75, 148). Furthermore, the performance of antibody detection tests is severely affected if strains that were used for isolation of antigens bound on the serology kits are different from strains circulating in a specific locality. Therefore, only locally validated antibody detection tests with a reliable cutoff value for that region should be considered in clinical settings (149, 156).
Enzyme immunoassay.
The antibody detection test based on ELISA is an indirect solid-phase enzyme immunoassay (EIA) that is based on the qualitative and quantitative detection of IgG antibodies against H. pylori. In this method, the antigens are used to coat and are immobilized on the inner surface of the microwell plate, and after the addition of the samples to the well, the IgG antibody present in the sample binds with these coating antigens, forming antigen-antibody complexes. These complexes are subsequently treated with enzyme-conjugated anti-human IgG antibodies, and the detection of the presence or absence of the antibody in the sample is performed by the addition of a suitable substrate in the reaction well followed by the reading of the results with an EIA plate reader at 450 nm (131). Currently, several ELISA-based tests, such as the H. pylori IgG EIA test kit (Rapid Labs, Ltd., Little Bentley, Essex, United Kingdom) (131), E-plate Eiken H. pylori Antibody II (Eiken Chemical Co., Ltd., Tokyo, Japan) (EP), and H. pylori IgG Seiken (Denka Seiken Co., Ltd., Tokyo, Japan) (EIA) are commercially available (145, 157). These tests are widely used for detecting H. pylori infection owing to their sufficient diagnostic performance in diverse clinical settings.
A noninvasive serological biomarker assay, the GastroPanel test (Biohit Oyj, Helsinki, Finland), which is based on ELISA and uses monoclonal antibodies, is capable of detecting anti-H. pylori antibodies and three additional biomarkers, including pepsinogen I, pepsinogen II, and gastrin 17, that are measured in the same serum or plasma samples (158). This test is considered a reliable method for the serological detection of H. pylori infection and the examination of the physiology of gastric mucosa owing to its high sensitivity and specificity (159). The pooled sensitivity and specificity of this method have been reported to be over 70% and 95%, respectively (160, 161). Because of its high reliability for detecting H. pylori-associated atrophic gastritis, the GastroPanel has been extensively used in large-scale gastroscopy-referral studies (symptomatic patients), screening of infection (asymptomatic subjects), and longitudinal (prospective) studies (159, 160, 162, 163).
The new-generation Unified GastroPanel capable of detecting the same four biomarkers was introduced in 2018 and has been approved with clinical validation studies (164, 165), and outstanding diagnostic performance has been observed, with increased sensitivity and specificity. The best advantage of this new generation test is that the biomarker results can be classified into five categories morphologically equivalent in the updated Sydney system classification of gastritis (normal mucosa, nonatrophic H. pylori gastritis, atrophic gastritis in the corpus, atrophic gastritis in the antrum, and atrophic gastritis in both the antrum and corpus) (166). The sensitivity, specificity, PPV, and NPV of the new-generation (unified) GastroPanel test are 93.8%, 88.9%, 93.8%, and 88.9%, respectively, when H. pylori is present in the antrum, whereas the values are 95.3%, 60.7%, 65.1%, and 94.4%, respectively, when H. pylori is present in the corpus (165).
Similarly, URINELISA (Otsuka Pharmaceuticals Co. Ltd., Tokyo, Japan) is an ELISA method that can detect anti-H. pylori IgG from urine samples (167). The sensitivity and specificity of URINELISA have been found to be 86.5% and 85.8%, respectively (167). ELISA methods for detecting anti-H. pylori IgG are considered the third best noninvasive methods next to UBT and SAT for the screening of H. pylori infection (92). Moreover, a better diagnostic accuracy could be achieved with ELISA methods using antigens from local strains than with commercial tests that incorporate antigens from nonlocal strains. To enhance the diagnostic accuracy of ELISA-based methods, various antigen preparations have been utilized for coating ELISA wells, including crude antigens such as whole-cell extracts and sonicated cell extracts, glycine extracts, heat-stable antigens, and recombinant antigens (168, 169). The utilization of antigen pools prepared from multiple strains can enhance the diagnostic performance of serology-based tests.
Immunochromatographic assay.
The immunochromatographic method of anti-H. pylori IgG antibody detection is widely used for urine samples (170). The test assay comprises a test stick where H. pylori antigen and dried anti-human IgG antibody are immobilized on the nitrocellulose membrane. To detect IgG in the urine, the test stick is dipped in the mixture of urine and a diluent. The mixture diffuses through the membrane, and the anti-H. pylori IgG, if present in the urine, forms a complex with anti-human IgG, thereby demonstrating a red band (171). Rapirun (Otsuka Pharmaceutical Co. Ltd., Tokyo, Japan) is a rapid urine test that uses an immunochromatography-based assay for the qualitative detection of H. pylori IgG in urine samples. The Rapirun kit is a point-of-care testing method that can provide rapid results. Moreover, it has the added advantage over serum ELISA of being able to be performed at a physician’s office, and a rapid result with high performance can be obtained (171). The test demonstrated comparable results with a sensitivity, specificity, and accuracy of 83.3%, 94.7%, and 93.2%, respectively, in Indonesia and 84.7%, 89.9%, and 87.0%, respectively, in Vietnam (172, 173). Recently, the immunochromatographic assay-based rapid whole-blood immunoassay gabControl H. pylori (gabmed GmbH, Cologne, Germany) was introduced for the qualitative detection of H. pylori IgG antibodies in whole blood, serum, or plasma (174). The performance evaluation of gabControl H. pylori demonstrated a sensitivity, specificity, PPV, and NPV of 91.4%, 76.7%, 65.3%, and 94.9%, respectively, when the UBT was considered the noninvasive gold standard reference method (174).
The current infection marker (CIM) is an H. pylori-specific novel recombinant protein that is identified from the cDNA library. The CIM test involves an indirect immunochromatographic assay that detects antibodies against the CIM present in blood samples (serum, plasma, or whole blood). Currently, the Assure H. pylori rapid test kit (MP Biomedicals Asia Pacific, Co. Ltd., Singapore) is used to diagnose H. pylori infection (175). The test is noninvasive and easy to perform. Moreover, this test differentiates past infection from present infection (176). However, it needs a duration of more than 6 months after the eradication therapy to accurately differentiate past and present infections (177).
Latex agglutination immunoassay.
The method of antibody detection using the latex agglutination immunoassay requires the mixing of specimens with a small amount of reagent. Currently, two latex agglutination assay-based kits, i.e., the Eiken H. pylori antibody (Eiken Chemical Co., Ltd., Tokyo, Japan) (LZ) test and H. pylori latex Seiken (Denka Seiken Co., Ltd., Tokyo, Japan) (LIA) test are being widely used for diagnosing H. pylori infection (145, 178). These latex agglutination-based tests are capable of detecting IgA and IgM in addition to IgG (145). In these tests, the antigens derived from the Japanese strains are bound to the surface of latex particles that react with antibodies present in the test samples, thereby inducing agglutination of the latex particles (145, 178). The positive agglutination reaction is perceived with a change in turbidity that is measured at a given wavelength (178). Pyloriset Dry (Orion Diagnostica, Espoo, Finland) is another latex agglutination-based kit that has been examined in adults in a number of studies, with a sensitivity in the range of 87 to 93.3% and specificity in the range of 65 to 95.6% (179–181). Although Pyloriset Dry has demonstrated excellent sensitivity in detecting H. pylori antibodies in adults, its detection sensitivity is poor among children, with up to 36% of sensitivity observed (182). As latex immunoassay-based testing methods can provide comparable and rapid results (usually taking only 10 min) and are easier to perform with a general automatic analyzer, whereas ELISA methods require a spectrometer and longer times to obtain results, latex immunoassay methods are being widely used (183).
Immuno-dot blot assay.
The method based on the immuno-dot blot assay can be used for detecting antibodies in serum, urine, stool, and saliva samples that demonstrate antibodies against some specific proteins such as CagA and VacA (184, 185). This test, which utilizes monoclonal antibodies specific for H. pylori antigens, leads to the rapid and highly specific identification of H. pylori infection. Furthermore, the dot blot assay is regarded as a more specific test capable of identifying strains on biotype level, thereby eliminating the need for biochemical tests that are used for the typing of bacterial isolates. The diagnostic performance (sensitivity and specificity) of dot blot assay methods is comparable to that of serum ELISA (184, 186, 187).
Multiplex immunoassay.
The application of various immunodominant antigens of H. pylori can enhance the diagnostic yield of antibody detection tests such as multiplex immunoblotting and ELISA (188–190). Several immunodominant proteins, such as UreA, UreB, catalase, GroEL, NapA, CagA, CagM, CagD, HP0231, VacA, HpaA, Cad, HyuA, Omp, HcpC, HP0305, gGT, Tip-α, HP0175, and FliD, are currently being used in combination (189–192). In 2009, a multiplex ELISA incorporating 15 recombinant glutathione S-transferase (GST) H. pylori fusion proteins, namely, UreA, catalase, GroEL, NapA, CagA, CagM, CagD, HP0231, VacA, HpaA, Cad, HyuA, Omp, HcpC, and HP0305, was introduced for diagnosing H. pylori infection (190). A novel multiplex immunoblotting method, RecomLine H. pylori IgG (Mikrogen Diagnostik, Germany), utilizing six highly immunogenic virulence factors that include CagA, VacA, GroEL, gGT, HcpC, and UreA was introduced as a serological method for diagnosing H. pylori infection (193). The diagnostic performance of this test was determined clinically in a German cohort study, which showed a sensitivity and specificity of 97.6% and 96.2%, respectively, when the histological analysis was considered the gold standard reference test (146). Moreover, this method could distinguish between past and current infections. This test can also be used to identify specific H. pylori virulence factors (189, 194) and gastrointestinal diseases, such as atrophic gastritis and gastric cancer (146, 195). A recent multiplex immunoblotting method that incorporates four immunodominant proteins, i.e., UreB, CagA, Tip-α, and HP0175, is a simple and cost-effective method to detect current H. pylori infection (191). The performance of this method was determined, and the reported sensitivity, specificity, PPV, and NPV were 96.1%, 20.9%, 69.7%, and 73.7%, respectively, to discriminate current infection from past infection. The method detected atrophic gastritis with a sensitivity, specificity, PPV, and NPV of 96.4%, 21.6%, 36.8%, and 92.6%, respectively (191).
Rapid Urease Test
The RUT is a simple and reliable method for detecting H. pylori infection, which is based on the detection of the activity of bacterial enzyme urease present in biopsy specimens (196). The biopsy tissues containing the urease produced by H. pylori are placed in a medium containing urea, which is hydrolyzed to carbon dioxide and ammonia. The produced ammonia increases the overall pH to an alkaline condition, which is detected by a pH indicator, manifesting a color change in the medium (197). The RUT is the most commonly used invasive method for detecting H. pylori infection and requires minutes to hours to provide results, enabling immediate treatment. Furthermore, the biopsy specimens used for the RUT can be reused for bacterial evaluation by molecular testing (198). The diagnostic performance of the test depends on the bacterial numbers (load) in the biopsy samples, and it may give false-negative results if the biopsy specimens contain a low concentration of the bacteria; therefore, the biopsy specimens should be collected from a site where the bacteria are present in sufficient numbers (199, 200). Though the diagnostic performance of the RUT is high and can be enhanced if biopsy specimens are collected from multiple sites (201, 202), it increases the risk of mucosal damage and bleeding. In clinical practice, the RUT is recommended as the first-line diagnostic test when endoscopy is performed for biopsy specimen collection. For enhanced accuracy of RUTs, two biopsy specimens (one each from the antrum and the corpus) are collected (72).
Several commercial RUT kits, including paper-based tests, gel-based tests, and liquid-based tests, are available (Table 3). Paper-based tests such as PyloriTek (Serim Research Corp., Elkhart, IN, USA) give results within 15 min with a sensitivity and specificity of over 96% and 97%, respectively (203), and Pronto Dry (Medical Instruments Corporation, Solothurn, Switzerland) can provide accurate results within 20 min (204). Gel-based tests such as the Campylobacter-like organism (CLO) test (Ballard Medical Products, Draper, UT, USA) can give positive results after 30 min with a sensitivity, specificity, PPV, and NPV of 94%, 88%, 89%, and 93%, respectively (205), and the Hp-Fast and Hp-One tests (GI Supply, Mechanicsburg, PA, USA) can provide results within 24 h and 1 h, respectively, with sensitivity and specificity over 90% (206). Liquid-based tests, such as the Endosc-Hp test (Cambridge Life Sciences Ltd., United Kingdom), can provide quick results within 30 min with a sensitivity of 94.4% and specificity of 98.4% (207). A new RUT kit with improved utility and speed, i.e., the ultrafast UFT300 test (Biohit Oyj, Helsinki, Finland), can read results in 5 min with a sensitivity of 94.5% and specificity of 100% (208, 209). The fast and accurate results of the UFT300 can simplify clinical management further, allowing treatment to be prescribed in the endoscopy unit before patients leave the unit (209). In a study comparing UFT300 with the RUT, the results of the RUT and UFT300 with respect to PPI intake were accurate in 93% and 97% of patients, respectively (210).
Recently, a new method for sample collection known as the sweeping method was shown to provide enhanced results in the RUT (CLO test) (211). In the sweeping method, an absorbent swab held with forceps is used to collect specimens by swabbing the mucosa of the great curvature of the antrum and the corpus using a sweeping motion, and the specimens collected with the swabs are used to perform the CLO test. In the study, the sensitivity of the sweeping method was higher than that of the conventional method (0.941 versus 0.685); however, the specificity was lower than that of the conventional method (0.826 versus 0.859). The overall accuracy of the sweeping method for detecting H. pylori infection was 0.903 (95% confidence interval [CI], 0.862 to 0.935), versus 0.742 (95% CI, 0.686 to 0.792) for the conventional method (211). The biopsy specimens collected using forceps typically contain both mucosal and submucosal tissues and a small amount of the mucus that lies on the mucosa. As H. pylori can survive and reside in the mucus layer owing to the characteristics of the mucus layer and the helical shape of bacteria (17), the sweeping method can acquire more H. pylori organisms, as it involves the collection of the gastric mucus from a larger gastric surface area than the conventional method of obtaining biopsy specimens. The sweeping method of specimen collection is safe, without a risk of mucosal damage and bleeding; however, it does not provide histological information, which can be derived from biopsy specimens.
Since H. pylori urease is absent in a healthy stomach, the RUT rarely gives false-positive results (83). False-positive results are unusual; however, such results may be obtained with infections caused by other urease-producing gastric non-H. pylori helicobacters, such as H. heilmannii (212), and other urease-producing bacterial pathogens, such as Proteus mirabilis, Citrobacter freundii, Klebsiella pneumoniae, Enterobacter cloacae, Staphylococcus aureus, Staphylococcus capitis subsp. urealyticus (117, 213). Some of these pathogens, including C. freundii, K. pneumoniae, E. cloacae, and S. aureus, are commensal oropharyngeal flora; however, they are not sufficient to affect test results, as the quantity of urease produced in the oral cavity is not sufficient. Therefore, biopsy specimen-based tests are most accurate for assessing the quantity of bacterial load in the stomach (214). False-negative results are obtained in cases involving the use of antibiotics, PPIs, bismuth-containing compounds, gastric atrophy, intestinal metaplasia, and peptic ulcer bleeding where the bacterial number is reduced (82, 197, 199, 201, 215–218). In cases where using the RUT is unavoidable, antibiotics or bismuth-containing compounds should be discontinued for 4 weeks and PPI therapy for 2 weeks before collecting biopsy specimens (104, 219, 220).
Bacterial Culture
The indication for bacterial culture from gastric biopsy specimens is usually restricted to phenotypic drug susceptibility testing to detect strains that are resistant to antibiotics in the case of failure of first-line antibiotic treatment (221–223). However, recent reports indicate that antimicrobial susceptibility testing (AST)-guided treatment improves the bacterial eradication rates (224, 225). Therefore, in the current scenario, it is wise to perform bacterial culture and AST prior to starting the antimicrobial therapy to improve its efficacy. Furthermore, bacterial culture allows the isolation of H. pylori in pure form for its identification and further microbiological studies, such as determination of biological and virulence properties (226). Bacterial culture is not included in routine testing for H. pylori infection because the method requires an invasive process (i.e., endoscopy) of biopsy specimen collection, and several molecular methods are now available that can be performed with specimens collected noninvasively (86, 227). Moreover, the culture and isolation of bacterial strains are time-consuming, requiring skilled personnel and significant resources that render the method expensive. However, bacterial culture derived from a gastric biopsy specimen is one of the most reliable methods, providing specificity up to 100%, although a lower sensitivity (around 90%) compared to that observed with histology and the RUT is observed when performed under optimal conditions (82, 228–230).
H. pylori is a fastidious pathogen that is difficult to grow under in vitro conditions. It requires rapid transport in a special transport medium (to keep the bacterium alive during transportation) and a growth medium to support the multiplication of the bacteria present in the collected samples (231). Transport media such as Portagerm pylori, Stuart’s transport medium, urea-containing saline, and normal saline keep the bacteria (present in samples) alive for up to 24 h at 4ºC. The H. pylori isolates can be kept frozen at −80°C in brucella broth containing 10% dimethyl sulfoxide and 10% horse serum to keep the pure isolates alive (231–234). Semisolid GESA transport medium can be used to keep the bacterium alive for up to 10 days at 4°C (235). The growth media that support the growth of H. pylori include Pylori agar, Skirrow agar, Wang media, Wilkins-Chalgren agar, Columbia blood agar, brucella agar, brain heart infusion agar, and Trypticase soy agar supplemented with sheep blood (86, 232, 233). Antibiotics can be included in the culture medium to make it selective for H. pylori, thereby preventing growth of or contamination with other bacteria (236). However, it should be noted that antibiotics in the culture media can delay the growth of some strains or can completely inhibit the growth in such media. Therefore, the use of a nonselective and selective culture medium is likely to increase the sensitivity of the culture (237). Several factors have been found to influence the growth of bacteria, such as the skills of microbiologists, bacterial load in biopsy samples, quality of specimens, presence of microbial normal flora in specimens, bacterial load in the gastric biopsy specimen, degree of gastritis (reduction of the bacterial load with advancing gastritis), alcohol consumption, bleeding ulcers, use of antibiotics, H2 receptor antagonists, PPIs, quality and composition of transport media, transport duration, air exposure, and transport temperature (229, 232, 238–240).
In addition to gastric biopsy samples collected invasively, several attempts have been made to successfully recover H. pylori from other samples obtained by noninvasive methods, including testing of gastric juice, saliva, and stool and the string test. However, because of the very low sensitivity of the recovery of H. pylori from these specimens, the culture method utilizing these specimens is not recommended either in routine diagnostics or in phenotypic drug susceptibility testing (116, 197, 238, 241). The tissues left over after RUT have been also used for the culture and recovery of bacteria (242–244). The tissue left over after RUT interpretation is discarded. The reuse of this tissue reduces the number of biopsy specimens collected during endoscopy, which can be cost-saving in practice (245, 246). Bacterial culture has demonstrated a good recovery of H. pylori from biopsy specimens left over after RUT if culture processing is performed within 4 h of interpretation of RUT results (242–244).
Histopathological Examinations
Histological examination is usually considered a gold standard method for diagnosing H. pylori infection that enables the direct detection of causative agents. Histology also allows the evaluation of the degree of pathological lesions such as gastritis, gastric atrophy, intestinal metaplasia, and cancer. A histological examination can be conducted by applying several staining techniques such as hematoxylin and eosin (HE), Giemsa, Warthin-Starry, H. pylori silver, toluidine blue, acridine orange, McMullen, Genta, Dieterle, Romanowski, and immunohistochemical (IHC) staining to evaluate pathological lesions and detect the presence of H. pylori (92, 143, 152, 228, 247–249). Although IHC staining demonstrates the best sensitivity and specificity, it is not recommended as the first choice for routine clinical practice owing to its high cost and time-consuming nature (228). HE and Giemsa staining methods are the first choice in routine clinical practice and are the recommended methods for assessing the level of inflammation and the detection of H. pylori, respectively (92, 143, 228). In cases of unclear results, other methods, such as toluidine blue, acridine orange, Genta, Romanowski, or McMullen staining, can be performed to verify results (152, 249). The Giemsa staining method has several drawbacks, including higher cost, time-consuming nature, and interobserver variability, and its performance is also strongly affected by the presence of inflammatory activity (228, 250). The recently developed modified Giemsa staining method has demonstrated improvements with respect to time consumption, and it requires fewer organic chemicals such as methanol and acetic acid to be used in the washing step while providing the same accuracy as that observed with traditional Giemsa staining (251). Moreover, the diagnostic performance of modified Giemsa is better than that of the RUT.
The IHC staining method that utilizes anti-H. pylori antibodies is now considered a gold standard method for detecting H. pylori in tissues and provides a sensitivity and specificity of approximately 100% (152, 250, 252). When the standard HE staining method is used in biopsy samples, H. pylori can be often detected in most cases (249, 253–255). Therefore, further staining by Giemsa or IHC may be omitted unless chronic active gastritis is noted without H. pylori identification by standard staining methods owing to low bacterial load or atypical localization of the pathogen, which necessitates histological evaluation by a pathologist (256, 257). Modified toluidine blue (MTB) can be used in histological preparations to detect H. pylori, which demonstrates better performance than the HE staining method (258, 259). MTB staining is capable of visualizing the bacteria even in specimens with a low density of bacteria when collected posteradication, in small biopsy specimens with few glands, and in cases of abundant mucus debris on the surface of pits. MTB staining is also simple, needs less time (provides results within 20 min), and highlights the neutrophil infiltration highly associated with the presence of H. pylori in the tissue. The sensitivity, specificity, PPV, and NPV of MTB staining were 99%, 96%, 95%, and 99% when IHC was considered the gold standard (258).
Imprint cytology, although rarely used in H. pylori diagnosis, is a method for obtaining imprints from biopsy specimen by placing the specimens on clean glass slides, which can then be stained by toluidine blue or Giemsa staining methods (259). Imprint cytology enables microbiologists to detect H. pylori is tissue using simple staining methods, thereby facilitating an early diagnosis. Toluidine blue and Giemsa staining can be performed on imprints, which demonstrate the unique morphology of curved or spiral rods of H. pylori that are strongly stained in the gastric mucosa (259). The imprint cytology method with toluidine blue demonstrates 100% agreement if the tissue contains a high density of H. pylori (259); however, the agreement is poor when the tissue contains a low density of H. pylori (260). A study reported a sensitivity and specificity of 83% and 100%, respectively, for diagnosing H. pylori (261), and the diagnostic accuracy can be improved to 100% if imprint cytology is combined with histology (260, 262, 263). Other studies have also demonstrated high sensitivity and specificity of imprint cytology (263, 264). Considering histology the gold standard method for diagnosing H. pylori, the sensitivity, specificity, PPV, and NPV of imprint cytology combined with toluidine blue were 57.1%, 97.9%, 80.0%, and 94.0%, respectively, whereas the values were 42.9%, 97.9%, 75.0%, and 92.2%, respectively, when it was combined with Giemsa staining (259), as shown in Table 4. This method offers a rapid, cost-effective, and simple method of H. pylori diagnosis in clinical routine practice; however, this method may show false-negative results if the specimen contains low bacterial density, which results in poor transfer of bacteria from specimens during imprint smear preparation.
Although histology is regarded as a gold standard method with several merits, the results of histology-based methods are affected by several factors, such as the site of specimen collection, size and number of biopsy specimens, staining methods, PPI and antibiotic treatment, experience of pathologists, and peptic ulcer bleeding (69, 72, 86, 92). Furthermore, the correct orientation of the biopsy specimens used for histological evaluation improves the accurate assessment of gastric atrophy (265). The treatment of gastritis and consumption of PPI have negative effects, and they affect the diagnostic accuracy of HE and Giemsa staining methods in clinical practice to a greater degree than the IHC method, which demonstrates high sensitivity and high diagnostic reliability (152, 228, 248). Therefore, it is recommended to discontinue PPI consumption for 2 weeks and antibiotic consumption for 4 weeks before performing histological investigations (69, 75, 82). The treatment of atrophic gastritis with acid-suppressing agents such as PPIs can induce migration of the bacteria from the gastric antrum to the proximal stomach. Therefore, the Sydney system protocol of multiple specimen (a minimum of 5) collection, including one each from the lesser and greater curvature of the antrum, the lesser curvature of the corpus, the middle of the greater curvature, and the incisura angularis, is recommended by the AGA to increase the maximum possibility of detecting H. pylori (162, 256). Other reports have recommended the collection of a reduced number of biopsy specimens, including four biopsy specimens (two from the antrum, including the greater and lesser curvature, and two from the corpus, including the greater and lesser curvature) (211, 266), three biopsy specimens (the greater curvature of the antrum and the corpus and the incisura) (256), and two biopsy specimens (from the antrum and the corpus) (82, 267) for maximum diagnostic yield; however, the results have not proven to be equivalent to the system utilizing five biopsy specimens.
Molecular Methods
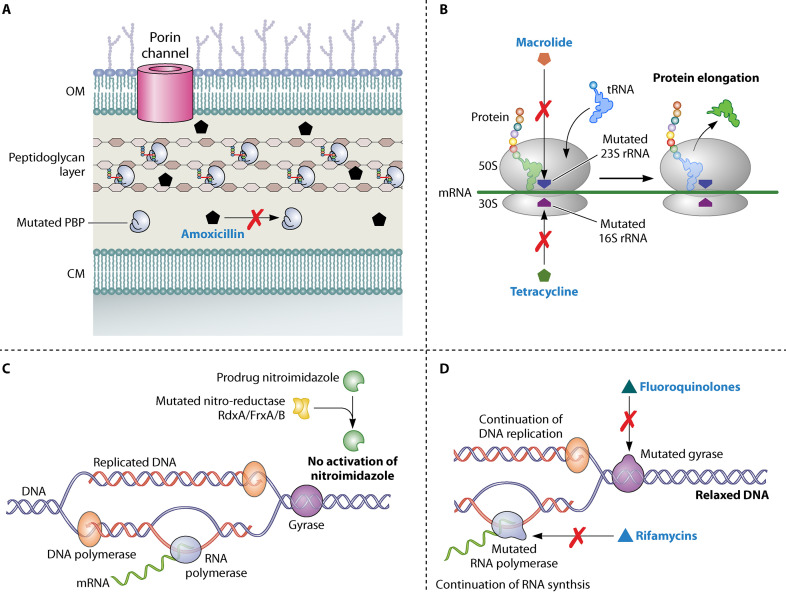
In the last few years, the use of molecular methods in the detection of infection has dramatically changed the landscape of clinical management of several infectious diseases (268). Molecular methods are useful for broad-spectrum infection detection, evaluation of emerging infections, epidemiological studies, genotyping, and assessment of antibiotic resistance trends (269, 270). However, a notable disadvantage of such methods is that false-positive results may be observed as a consequence of residual genetic elements following antimicrobial therapy or the presence of other flora that produce urease or mimic genetic information (271). PCR is one of the best molecular methods with a wide range of clinical applications (269, 270). PCR-based detection of H. pylori infection could be classified as invasive when it involves specimens collected invasively, such as gastric juice and gastric biopsy specimen, or as noninvasive when it involves specimens collected noninvasively, such as saliva and stool specimens (269, 270, 272, 273). Being highly sensitive, PCR-based methods can help detect H. pylori infection in patients with peptic ulcer bleeding, gastric cancer, or gastric MALT lymphoma for whom the diagnosis of H. pylori is important but difficult to obtain by other nonmolecular methods; these methods have a sensitivity and specificity close to 100% (274, 275). Moreover, previous studies have found that PCR-based methods can detect low-density infection in a considerable number of patients with dyspepsia compared with nonmolecular conventional methods (276, 277). PCR has been found to detect active infection in a proportion of healthy individuals who were diagnosed as negative according to conventional nonmolecular methods (278, 279). Furthermore, a recent study showed that 49% of patients with chronic mucosal inflammation who tested H. pylori negative according to histological methods were found to be positive for the infection by PCR-based methods (280). Currently, there are several molecular-method-based commercial kits that are easy to perform and provide results with high accuracy (Table 5). Molecular methods for H. pylori detection include conventional PCR, nested PCR, multiplex PCR, fluorescence in situ hybridization (FISH), real-time PCR (RT-PCR), and digital PCR.
Conventional PCR.
In conventional PCR, several gene candidates such as vacA, cagA, ureA, glmM, hsp60, 16S rRNA, 23S rRNA, ureC, and flaA are amplified using specifically designed primers (143, 273, 281–283). However, the amplification of the 23S rRNA gene demonstrates the highest performance with respect to the detection of H. pylori infection (283). Conventional PCR is usually performed with bacterial isolates recovered from biopsy specimens. However, other specimens, such as gastric biopsy specimen or gastric juice, can be also used for amplifying specific genes. In a study performed with gastric juice specimens, PCR involving amplification of ureA and cagA demonstrated sensitivity of 92.7%, in contrast to culture, which showed a sensitivity of 70.2%, and the areas under the curve (AUC) of the gastric juice PCR and bacterial culture were 96.7% and 91.3%, respectively (284). The results indicate that gastric juice PCR is a better diagnostic method than the culture method, with results having high reliability and stability even in patients with more rounds of treatment (284, 285). However, caution should be exercised when ureA-based PCR is used, in case false-positive results arise from the presence of other urease-producing bacteria (286). The detection of H. pylori from saliva by PCR amplification of 16S rRNA genes has demonstrated a reliable method with sensitivity and specificity of 80.0% and 77.7%, respectively (287). However, the positivity rate of H. pylori from saliva specimens (18.0%) remains lower than that from stool specimens (50.4%) (288).
Nested PCR.
Several modifications have been added to conventional PCR to achieve increased sensitivity and specificity for the detection of H. pylori infection (285, 289–291). However, these modified molecular methods are very expensive, require a high level of technical skill, and demonstrate an increased risk of false-negative results owing to technical and human errors (289, 292, 293). One of these modifications is nested PCR, which involves two rounds of PCR. In the first round of PCR, a larger DNA region is targeted, whereas in the second round of PCR, a smaller subregion of the product from the first round that serves as the template is targeted (136, 294, 295). Nested PCR has been successfully applied to identify resistant mutations in 23S rRNA in stool specimens without any false-positive results (296). Compared to conventional PCR, nested PCR is more sensitive because it can amplify target DNA with severalfold-lower concentrations than those required for conventional PCR (295, 297, 298). Nested PCR can identify H. pylori from controversial specimens, such as those derived from patients with bleeding or those consuming PPIs, and can be performed in laboratories with basic equipment. In a previous study, the sensitivity and specificity of nested PCR in the amplification of target genes from gastric biopsy specimens were found to be 93.8% and 95.9%, respectively, compared with results obtained using [13C]UBT, which was used as the gold standard method (299). Nested PCR was found to be at least 10-fold more effective than conventional PCR. However, false-positive results due to the contamination are also possible with this method (295, 299).
Multiplex PCR.
Multiplex PCR is the modification of conventional PCR where multiple genes can be amplified simultaneously in the same PCR. Multiplex PCR is used to simultaneously detect positive infection and genotypes, such as cagA and vacA genotypes (s1/s2 and m1/m2 fragments) (300, 301). The presence of H. pylori is considered positive by multiplex PCR if at least one of the specific gene products, including cagA, vacA s1, vacA s2, vacA m1, or vacA m2, is detected in the specimens in the agarose gel (301). Multiplex PCR studies targeting three genes for amplification, such as a conserved region flanked by genus-specific primer-binding sites in Helicobacter 16S rRNA and species-specific sequences (ureA and hpaA), result in a clear distinction of H. pylori from Campylobacter and other bacterial genera (302–304). Multiplex PCR is more accurate and demonstrates a higher H. pylori detection rate (305). The sensitivity, specificity, PPV, NPV, and accuracy of multiplex PCR were 100%, 59.57%, 73.79%, 100%, and 81.09%, respectively, when CIM and CLO tests were considered the gold standard reference methods (300). Multiplex PCR demonstrates excellent performance for the detection of H. pylori from stool specimens and can identify a small amount of H. pylori nucleic acid (306).
Fluorescence in situ hybridization.
FISH is a highly sensitive and specific molecular cytogenetic technique, in which fluorescent-labeled oligonucleotide probes bind to a specific complementary target sequence of DNA or RNA, enabling its detection and quantification when exposed to light of specific wavelengths (250, 307). FISH can detect infection with coccoid forms of H. pylori (308). Along with the detection of H. pylori infection, FISH has the advantage of being able to determine susceptibility or resistance to clarithromycin in bacteria (116, 308, 309). Currently, several H. pylori FISH commercial kits, such as the BACTfish H. pylori Combi test (Izinta Kft., Budapest, Hungary) and Probe4Pylori (Biomode SA, Caldas das Taipas, Portugal), are available. BACTfish H. pylori Combi is highly sensitive and maintains specificity as shown in Table 5 (250). Being highly accurate, the BACTfish H. pylori Combi test has the potential to be the gold standard test in the evaluation of H. pylori infection.
Real-time PCR.
RT-PCR is gaining popularity and is more widely used in clinical laboratories for diagnosing H. pylori infections because of its short working time, high sensitivity and accuracy, and low risk of cross-contamination. RT-PCR can be conducted with various specimens, such as fresh or frozen biopsy specimens, paraffin-embedded biopsy specimens, and stool specimens. The most commonly targeted genes in RT-PCR involve small segments of 16S rRNA genes, ureA, and 23S rRNA genes that are well preserved in embedded tissues (310–312). RT-PCR is a highly sensitive and specific method to detect H. pylori from fresh or frozen biopsy specimens derived from patients with nonbleeding peptic ulcers (275, 313). It also shows better results than histology with fresh or frozen gastric samples collected via biopsies of patients with peptic ulcer bleeding (314). Freshly collected or frozen biopsy specimens are most commonly used to extract nucleic acids. Since viable bacteria are not required in the RT-PCR method, which is based on DNA detection, it is a useful method when the patient has already started antimicrobial therapy (315). RT-PCR can detect infection in a significant percentage of histologically negative biopsy specimens, and it demonstrates a 5% increase in the positivity rate compared with bacterial culture methods (316, 317).
There are several commercially available RT-PCR kits, such as RIDA GENE H. pylori (R-Biopharm AG, Darmstadt, Germany), with sensitivity, specificity, PPV, and NPV of 100%, 99.0%, 94.0%, and 100%, respectively, when culture alone or histology together with the RUT is considered the gold standard reference method (318). Amplidiag H. pylori+ClariR (Mobidiag, Espoo, Finland) can simultaneously detect H. pylori infections and mutations conferring clarithromycin susceptibility. It also demonstrates high performance, with sensitivity, specificity, PPV, and NPV of 96.3%, 98.7%, 92.2%, and 99.3%, respectively (319). MutaREAL H. pylori (Inmundiagnostik, Bensheim, Germany), when performed with biopsy specimens collected from children, demonstrates sensitivity, specificity, PPV, and NPV of 93.3%, 86.9%, 90.3%, and 90.9%, respectively (320). The ViaSure H. pylori real-time PCR detection kit (CerTest Biotec S.L. Zaragoza, Spain) has exhibited better performance when performed in patients above 18 years of age, with sensitivity, specificity, PPV, NPV, and accuracy of 85.7%, 100%, 100%, 84.6%, and 92.0%, respectively (321). Recently, Leonardi et al. developed and evaluated the highly sensitive and specific (sensitivity, 94.12%, and specificity, 93.75%) RT-PCR based assay for the detection of H. pylori specific ureC and cagA, respectively, to assess their presence in stool specimens coinfected with protozoal or helminthic parasites (322).
Detection of H. pylori by 23S rRNA RT-PCR provides 100% concordance results compared with reference methods of bacterial culture, histopathology, or RUT (323, 324). In clinical or research settings, formalin-fixed and paraffin-embedded tissues are the most widely available specimens that provide highly efficient and specific results for detecting H. pylori (325, 326). RT-PCR is capable of detecting H. pylori infection when paraffin-embedded tissues collected from patients with peptic ulcer bleeding are assessed and therefore can be used to identify occult infection in patients who test negative according to histopathological analyses (323). A sufficient amount of genomic DNA can be obtained from 7 to 8 sections of tissues with 10-μm thickness (243). The DNA extracted after deparaffinization and rehydration of paraffin-embedded sections provides a well-preserved template for RT-PCR. In a study of paraffin-embedded tissues, the sensitivity of RT-PCR in the detection of H. pylori infection was 95.6%, compared with histological results, which showed a sensitivity of only 69.9% (327).
Stool specimens provide a noninvasive method ideal for patients who do not meet the criteria for invasive specimen collection. However, PCR-based tests with stool specimens can be challenging because of the small amount of target DNA, presence of inhibitory substances, and presence of other Helicobacter species with homologous sequences (238, 328). The results of RT-PCR performed with stool specimens depend on several factors, such as the quality and amount of genomic DNA recovered, target sequences, differences in sensitivity and specificity of primers used, and nature of amplification protocols (329). Therefore, a promising PCR method uses a customized extraction kit employing a method to remove inhibitors present and maximize the genomic DNA extraction from stool specimens. Furthermore, an ideal PCR method for stool specimens should possess advantages of H. pylori detection and genotypic clarithromycin resistance screening, stability of stored specimen, detection of heteroresistance when more than one H. pylori strain is present in the stool specimen, a high degree of standardization, and good reproducibility (330). The number of bacteria present in clinical specimens can influence the results of PCR-based tests. The minimum number of H. pylori CFU/mL should be 1.5 × 10 in pure bacterial suspension, 1.5 × 103 in stool and mixed specimens, and 100 in gastric biopsy specimen to obtain clear positive results using PCR-based methods (286, 331).
Digital PCR.
Digital PCR (dPCR) is the latest commercially available refinement of PCR technology with increased sensitivity compared with conventional PCR and a sensitivity comparable to that of RT-PCR, while the specificity of the method is maintained (332–334). dPCR is useful in detecting infectious agents in various specimen types, and it is also useful in detecting H. pylori infection and its genotyping of resistance genes (335–337). Droplet digital PCR (ddPCR) is a method for performing dPCR that is based on the generation of water-oil emulsion droplets. In this method, the PCR solution of a sample is fractioned into thousands of droplets (10,000 to 100,000) and then subjected to complete PCR. The amplification of the template strand is observed in each individual droplet, and the droplets showing positive amplification are counted using a fluorescence detector. The fractioning of a small amount of PCR solution into thousands of parts within droplets results in absolute quantification of target sequences, thereby enhancing the sensitivity of this method compared with other PCR. ddPCR can detect occult H. pylori infection, especially in patients with very low density of bacteria, as is found in conditions such as peptic ulcer bleeding, gastric MALT lymphoma, or atrophic gastritis, when the standard test results are suboptimal. In a previous study, Raderer et al. reported six patients having H. pylori infection with gastric MALT lymphoma who were negative according to several H. pylori conventional tests (338). Similarly, Güell et al. reported that 79% of patients with peptic ulcer bleeding reported to be H. pylori negative had an active infection and were found to be positive only when retested a few weeks after the bleeding episode (339).
PREFERENCES FOR DIAGNOSTIC METHODS ACCORDING TO CLINICAL CONDITIONS AND AGE
Preferences for appropriate diagnostic tests for H. pylori infection depend on many factors, such as the prevalence of infection, age-related gastric cancer incidence, patient’s choice, accuracy of the test, availability, and cost-effectiveness (Fig. 2). Noninvasive tests are preferred mostly in areas where the incidence of gastric cancer is low, whereas endoscopy-based diagnostic tests are recommended for patients who are highly likely to develop gastric cancer, such as those belonging to a geographic region with a high incidence of gastric cancer, patients over 50 years of age, and patients having a family history of gastric cancer (78). According to the Maastricht V/Florence consensus report, noninvasive tests, such as locally validated serological tests, should be recommended over endoscopy-based tests for the diagnosis of H. pylori infection in patients with dyspepsia (72). The guidelines of the Japanese Society for Helicobacter Research suggest the use of at least one of several invasive and noninvasive tests available for the diagnosis of H. pylori infection; however, multiple diagnostic tests can be used to obtain increased diagnostic accuracy (108). Furthermore, the ACG and the Canadian Association of Gastroenterology (CAG), taking into account the probability of adverse effects attributed to endoscopy, suggest the use of upper gastrointestinal endoscopy-based diagnosis for patients with dyspepsia and for those who have active or a history of PUD, low-grade gastric MALT lymphoma, a history of endoscopic resection of early gastric cancer, an age over 60 years, or a high risk of developing gastric cancer, such as a patient belonging to a family with a history of gastric cancer or to a geographic region with a high incidence of gastric cancer (68, 109).
FIG 2.
Preferences regarding diagnostic methods. Age and clinical conditions (if any) should be considered when diagnostic methods are being selected. For children more than 10 years old, noninvasive tests, such as the urea breath test (UBT), monoclonal antibody (MAb) ELISA-based stool antigen test (SAT), serological tests, and antibody detection performed with urine specimens, are considered. For children less than 10 years old, due to immature immune response, antibody detection methods (serology and antibody detection in urine) are not considered; in such cases, the UBT or the monoclonal antibody ELISA-based SAT is the most appropriate test. The UBT and serology are considered for patients with upper gastrointestinal bleeding (UGIB), whereas for patients with partial gastrectomy, monoclonal antibody ELISA-based SAT is recommended. In patients with high risk of developing gastric cancers (*, those having active or history of PUD, low-grade gastric MALT lymphoma, history of endoscopic resection of early gastric cancer, or age over 60 years and those belonging to a family with a history of gastric cancer or a population at high risk for gastric cancer), endoscopy-based detection methods are recommended. Molecular methods, such as RT-PCR, can be conducted on specimens such as biopsy specimens, gastric juice, and stool.
In patients with upper gastrointestinal bleeding (UGIB), although invasive tests such as the RUT, histology, and bacterial culture demonstrate high specificity, the sensitivity is low. Therefore, in patients with UGIB, the performance of endoscopy-based tests can be delayed until the bleeding stops. Among noninvasive tests, the UBT is considered a reliable test, and SAT provides less accurate results in these patients. Although the serological test is not influenced by UGIB, it is not recommended as the diagnostic test for H. pylori infection owing to its inability to discriminate between present and past infection (340). For patients with UGIB for whom endoscopy is unavoidable, the histology test, which is less likely to be influenced by the presence of blood, may be preferred over other invasive tests (341). In patients with partial gastrectomy, the best results are obtained with histology, followed by intermediate results obtained with the RUT and poor results obtained with the UBT (342). Among noninvasive tests, the SAT may be considered a reliable test to detect H. pylori infection in patients with distal gastrectomy owing to its high performance in these patients (343).
Although histology-based tests provide accurate diagnostic results for H. pylori infection and comprehensive assessment of the gastric mucosa, the test is not generally performed for children or is less recommended because of its invasiveness (344). According to the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) and North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition (NASPGHAN) guidelines, the diagnosis of H. pylori infection in children using endoscopy-based tests (such as RUT, culture, and histology) is performed when the patient has first-degree relatives with gastric cancer and refractory iron deficiency anemia without a well-known reason (110). The patient is considered positive if the results of both RUT and histology are positive or only the culture is positive. In case of inconsistent results, a noninvasive test, such as the SAT or the UBT, is considered. Noninvasive tests such as the UBT that show high sensitivity and specificity for the diagnosis of H. pylori infection could be methods of choice for children with dyspepsia (88, 345, 346). However, studies have also found a reduced accuracy of this method in children less than 6 years old (347, 348). Since the age of patients is a major factor influencing the optimal threshold value of the [13C]UBT, this test could be a valuable diagnostic test even for children less than 6 years old if used after its local validation for the optimal threshold value (88, 94). Although the [14C]UBT has been proposed because of its cost-effectiveness, it is not recommended for children and pregnant women due to exposure of patients to radiation (349).
The SAT is another noninvasive method preferred for the detection of H. pylori infection in children (350); however, its performance is compromised with low bacterial load in the stool and in the case of peptic ulcer bleeding (351). Other studies have observed good accuracy with the ELISA-based SAT using monoclonal antibodies, which may be considered an efficient noninvasive test for diagnosing H. pylori infection in children (330, 352–354). Moreover, the monoclonal antibody ELISA-based SAT is convenient and can be used in children of different age groups and in patients with prior use of PPIs (126, 329, 355, 356). Studies have also shown that the monoclonal antibody ELISA-based SAT can be used for children aged below 3 years with a reliable performance (354, 357). H. pylori antibody-based tests are not useful for the detection of H. pylori infection in children younger than 10 years because of their immature immune response (358, 359). A poor sensitivity (54.5%) of the ELISA in the detection of H. pylori antibody has been observed in children younger than 10 years, whereas higher sensitivity (up to 100%) and specificity (93.8%) have been observed in those 10 years old or above (360). Thus, among noninvasive tests, the H. pylori antibody-based serology or urine-based ELISA is unacceptable for young children (361, 362). Since molecular tests (PCR) can be performed with various specimens such as gastric biopsy specimen, gastric juice, and stool (325, 363), such tests are acceptable and demonstrate appropriate sensitivity in the diagnosis of H. pylori infection in children. Furthermore, PCR can detect a very small amount of DNA in case of infection with low bacterial load and demonstrates acceptable sensitivity in the case of patients receiving PPI medications (325, 364–366).
MASS SCREENING FOR H. PYLORI INFECTION
In 2014, the International Agency for Research on Cancer (IARC), a working expert group of WHO recommended the implementation of population-based screening of H. pylori infection and treatment in order to control gastric cancer (367). However, mass screening is not a usual activity in many countries due to the lack of resources; it is implemented mostly in developed countries, where the prevalence of H. pylori infection and the incidence of gastric cancer are relatively low (58). Biopsy specimens collected for endoscopy-based diagnostic tests are highly accurate for diagnosing H. pylori infection; however, these methods are not recommended for screening because of their invasiveness, expensiveness, and unavailability (368). In this context, noninvasive diagnostic methods are preferred and recommended for mass screening for H. pylori infection (Fig. 3).
FIG 3.
Mass screening for H. pylori infection. Noninvasive tests, including the urea breath test (UBT), the stool antigen test (SAT), serology, or antibody detection from urine samples, are preferred methods for mass screening for Helicobacter pylori infection among communities. The positive serology results should be further confirmed by other specific methods (such as RT-PCR) on gastric biopsy or stool specimens. Patients who test positive are subjected to clarithromycin susceptibility-guided eradication therapy (when clarithromycin resistance is over 15% in that population) or clarithromycin-based empirical therapy (when clarithromycin resistance is below 15% in that population). After the completion of eradication therapy, patients are subjected to the assessment of successful eradication with either the UBT or SAT. In case of the failure of eradication therapy, a biopsy specimen-based bacterial culture is suggested to evaluate the antibiotic resistance by phenotypic (antimicrobial susceptibility testing [AST]) or genotypic (evaluation of the mutations conferring the resistance) methods, which allow selection of AST-guided therapy.
Among noninvasive methods, locally validated [13C]UBT, H. pylori SAT, and serological testing are currently available and are the most sensitive and cost-effective mass screening methods for the detection of H. pylori infection in the community (78). In the Cochrane review of hospital-based studies, the indirect comparison demonstrated that the [13C]UBT was the most accurate among these three tests (87). Because of its high accuracy, the UBT is currently recommended as the best approach for screening H. pylori infection (72). However, this test also has several drawbacks, including its high cost, need for mass-spectrometric analysis, which may not be available at remote or resource-limited centers, and false-positive and false-negative results, as described above (82, 115, 117). The SAT is also a preferred method for epidemiological study and screening tests and is currently adopted because it is noninvasive, cost-effective, easy to perform, and more suitable (129, 369). In Japan, the screening for H. pylori infection by SAT among children has increased in recent years to develop a preventive strategy for gastric cancer (370–372). However, a previous study found a low accuracy of the SAT compared with that of a serological test in patients with severe atrophic gastritis, and so this approach needs to be evaluated for screening H. pylori-associated diseases such as gastric cancer (373). However, other studies found no significant differences in the results when using polyclonal antibody ELISA-based SAT conducted in patients with atrophic gastritis and/or intestinal metaplasia (374).
H. pylori antibody-based serological tests are also frequently used for the screening of H. pylori infection for epidemiological purposes because of their rapid results and cost-effectiveness as well as wide acceptance by patients. The latex agglutination-based LZ and ELISA-based E-plate serological methods were used to screen for infections in junior high school students in Japan (178). The serological test is more useful in screening for H. pylori infection in children. The serology-positive cases should be confirmed by other more specific tests before commencing eradication therapy or confirmed by RT-PCR methods, if available, on gastric biopsy or stool specimens, as this method provides the simultaneous detection of clarithromycin susceptibility results and thus treatment guidance (375). Recently, a urine-based immunochromatographic method that detects antibodies in urine was used for the primary screening of H. pylori infection in Japanese school children (376, 377).
The cost-effectiveness of the test is one of several factors that determine which test is suitable for screening purposes. Among the three most commonly preferred screening methods, i.e., [13C]UBT, SAT, and serological testing, the UBT is a highly accurate but expensive test, followed by the SAT, which is less expensive than the UBT. The serological test is the least expensive. Several factors should be considered to make mass screening tests cost-effective, including the prevalence of H. pylori infection, patient adherence, purchase and running costs of the test, incidence rate of gastric cancer, cost of gastric cancer treatment, additional benefits of testing, and estimated cancer reduction (378, 379). Despite the cost-effectiveness and acceptability of the SAT for mass screening, the delayed delivery of stool specimens might lead to a deterioration of antigens in the samples, thereby leading to false-negative results.
ASSESSMENT OF SUCCESSFUL BACTERIAL ERADICATION
In the last few years, the effectiveness of antimicrobial treatments has been declining owing to growing antimicrobial resistance. Therefore, posttreatment assessment of the antimicrobial activity for H. pylori infection is of utmost importance (Fig. 3). The assessment for antimicrobial treatment activity is especially important because treatment failure can be attributed to antimicrobial resistance, and refractory cases may pose an increased risk for severe complications, such as PUD and gastric cancer (74, 75). In this context, currently, all consensus reports, such as the Kyoto global consensus report (74), Maastricht V/Florence consensus report (72), Japanese Society for Helicobacter Research report (108), ACG/CAG report (109), ESPGHAN/NASPGHAN report (110), and Taipei global consensus report (78), recommend assessment for successful eradication by either the UBT or SAT (75, 82). The UBT shows excellent performance in the assessment of H. pylori infection before and after antimicrobial treatment. Monoclonal antibody ELISA-based SAT is also a reliable test that is widely recommended by various guidelines to assess the efficacy of the eradication treatment; however, testing for successful eradication should be conducted at least 4 weeks after completing therapy (69, 108). Study reports have found sensitivity in the range of 91.6% to 100% and specificity in the range of 93.6% to 98.4% for monoclonal antibody ELISA-based SAT, which is used to confirm successful eradication after antimicrobial treatment (380–382).
No recommendations have been made indicating preferences or selection of a particular test; however, some considerations are required in order to select the best method depending on the prevalence of H. pylori infection. For instance, in geographic regions where the prevalence of H. pylori infection is above 30%, SAT may prove to be the best method for diagnosis (383), whereas for a region with lower prevalence, the confirmation of successful eradication would be appropriate with UBT (383, 384). As antimicrobials, bismuth-containing compounds, and PPIs affect the performance of both noninvasive tests, it is wise to perform testing for successful eradication at least 4 to 6 weeks after completing therapy. The sensitivity of polyclonal antibody-based SAT is poorer than that of monoclonal antibody-based SAT in the diagnosis of H. pylori infection and assessment of successful eradication (149, 385, 386). Once the assessment indicates failure of the eradication therapy, an endoscopic biopsy is performed for bacterial culture and phenotypic susceptibility testing to guide further antimicrobial therapy. However, the bacterial culture method is slow, does not always succeed, and is not widely available (68, 72).
Serological tests are not considered for the assessment of successful eradication because of the inability to distinguish between present and past infections. Anti-H. pylori IgG is found in the blood for a long duration even after successful bacterial eradication (69, 387). Therefore, serological tests are not useful for confirming effective eradication treatment; however, they are useful for epidemiological surveys (388). Since molecular methods are highly sensitive for the detection of H. pylori infections, these tests are not considered appropriate for assessing successful bacterial eradication, since they are capable of detecting genetic material remaining from killed bacteria (389).
MAGIC BULLET CONCEPT AND ANTIMICROBIAL RESISTANCE
Paul Ehrlich’s “magic-bullet” concept of killing specific microbes, which uses the analogy of a bullet fired from a gun, by hitting a specific target without harming the body itself was a brilliant idea that led him to discover the antimicrobial properties of several compounds (390). In 1929, Alexander Fleming discovered the antimicrobial compound penicillin, a β-lactam antibiotic (391). This discovery proved to be a milestone in the field of antimicrobial therapy, and soon after this, many other antimicrobial compounds were used to treat human infections. After the introduction of antimicrobials for treatment, it was believed that the evolution of resistance was unlikely to occur. Previously, it was assumed that the frequency of mutations resulting in bacterial resistance is negligible (392). However, owing to the emerging circumstances, magic bullets (antimicrobials) are losing their magic (antibacterial activity), and more contradicting and disappointing outcomes have been seen over time. The emergence of antimicrobial resistance in H. pylori owing to the widespread use of antimicrobials is an important concern in the community following mass screening and eradication in asymptomatic individuals. In bacteria, magic bullets can lose their magic through several mechanisms, such as modification of antibiotic targets, enzymatic degradation of antimicrobial agents, multidrug efflux systems, changes in the bacterial cell wall permeability, acquisition of alternative metabolic pathways, overproduction of target proteins, and biofilm formation. However, several mechanisms have not been reported or studied in detail in H. pylori. Therefore, we discuss the commonly observed mechanisms that are becoming a bottleneck for eradication therapy.
Modification of Antibiotic Targets
Modifications in molecules commonly targeted by antibiotics are the most common mechanism underlying antibiotic resistance in H. pylori. Spontaneous mutations in antibiotic target genes that neutralize the activity of antibiotics pose a major problem for the eradication of H. pylori infections.
Amoxicillin resistance.
Amoxicillin, a derivative of ampicillin and a β-lactam antibiotic, was discovered in 1958 and first used in medicine in 1972 (393, 394). The World Health Organization has placed this antibiotic on the list of the most effective and safe medicines for the treatment of human infections (395). β-Lactam antibiotics are among the most effective bactericidal antibiotics used to treat infections caused by Gram-positive and Gram-negative bacteria. It is a drug of choice because it is better absorbed following oral administration than other β-lactam antibiotics (396).
Penicillin-binding proteins (PBPs) are bacterial peptidoglycan-synthesizing enzymes with transpeptidase activity associated with the C-terminal region (397). In H. pylori, nine different types of PBPs, three with high molecular weight and six with low molecular weight, have been identified (398). However, only three PBPs (PBP1, PBP2, and PBP3) are involved in amoxicillin resistance (399, 400). During the synthesis of the cell wall (peptidoglycan layer), PBPs catalyze the synthesis of cross-linking bridges between the linear peptidoglycan polymer chains and ultimately the synthesis of the peptidoglycan layer (Fig. 4A). Beta-lactam antibiotics (for example, amoxicillin) inhibit the synthesis of cell walls by binding PBPs and halting their transpeptidase activity (Fig. 4A) (401).
FIG 4.
Mechanism of antimicrobial activity in H. pylori. (A) During bacterial multiplication, the bacterial cell wall component (i.e., the multisheet peptidoglycan layer) is synthesized by penicillin-binding proteins (PBPs), which act as transpeptidases, causing the cross-linking of the peptidoglycan polymer chains. Beta-lactams (e.g., amoxicillin) binding with PBPs via penicillin binding motifs inhibit their action, preventing the synthesis of peptidoglycan layer and leading to bacterial cell lysis and cell death. (B) The bacterial ribosomes translate the mRNA to proteins. However, the macrolides (e.g., clarithromycin) and tetracyclines bind with 50S and 30S ribosomal subunits, respectively, inhibiting protein synthesis and causing bacterial cell death. (C) In the case of nitroimidazole (e.g., metronidazole), a prodrug is activated to its active form by reductases such as RdxA, FrxA, and FrxB. The activated metronidazole damages helicoidal DNA, causing bacterial cell death. (D) DNA replication and transcription of gene initiates with the formation of a replication fork, which creates supercoiled DNA with high tension in DNA strands. DNA gyrases (A and B) cause the unwinding of supercoiled DNA which is important for the normal DNA replication and transcription by RNA polymerases. Fluoroquinolones (e.g., levofloxacin) bind with DNA gyrases (A and B), whereas the rifamycins (e.g., rifabutin) bind with RNA polymerases and inhibit their respective functions, leading to bacterial cell death.
To exert bactericidal effects, amoxicillin needs to bind penicillin-binding motifs, highly conserved amino acid sequences which comprise STGK338–341, SAIK368–371, SKN402–404, SLN433–435, KTG555–557, and SNN559–561 (402). The inability of antibiotics to bind penicillin-binding motifs due to amino acid variations caused by mutational changes in or around these regions renders the bacteria resistant to penicillin (Fig. 5A). The amino acid variations in and around the penicillin-binding motif sequences of PBP1 due to point mutations prevent the binding of amoxicillin to PBP1, which is a major mechanism underlying amoxicillin resistance in H. pylori (399, 400). The substitutions I259T in PBP1, T498S in PBP2, and D2N, A50S, F490Y, and A541T in PBP3 were found in strains showing remarkably high MICs (256 μg/mL). The substitution N564Y in PBP1 moderately increased the MIC (2.0 μg/mL). Substitutions such as N107R, A201V, V250I, and S543T in PBP1 and V374I in PBP3 moderately increase the MIC (0.25 μg/mL), inducing amoxicillin resistance (399, 402). The substitutions V469M, F473L, S543R, N562Y, T556S, A369T, V374, L423F, T593T, V45I, S414R, V414R, D465K, V471M, and N564Y in PBP1, A296V, S494H, A541M, and E572G in PBP2, and A499V and E536K in PBP3 affect the susceptibility of H. pylori to amoxicillin (400, 402–404). Furthermore, mutations in the multidrug efflux protein-encoding gene hefC, the porin protein-encoding gene hopC, and the putative outer membrane protein-encoding gene hofH that probably alter the porin channels, preventing the entry of amoxicillin into the bacterial cells, have also been suggested to increase the MICs (405).
FIG 5.
Antimicrobial resistance in H. pylori. (A) Mutations leading to amino acid alterations in or around the penicillin binding motifs cause the inability of β-lactams (e.g., amoxicillin) to bind with altered PBPs. Therefore, in the presence of beta-lactams, cell wall synthesis continues, leading to bacterial resistance to beta-lactams. (B) Point mutations in the specific regions of domain V of 23S rRNA lead to the inhibition of binding of macrolides (e.g., clarithromycin) to the 50S ribosomal subunit. Therefore, even in the presence of macrolides, the synthesis of protein continues, constituting bacterial resistance to macrolides. Similarly, mutations in 16S rRNA prevent the binding of the tetracyclines to the 30S ribosomal subunit. In this way, bacterial protein synthesis continues in the presence of tetracyclines, indicating bacterial resistance to tetracyclines. (C) Due to the mutational change in the nitroreductases (RdxA, FrxA, and FrxB) the nitroreduction activity is inhibited and the nitroimidazole (e.g., metronidazole) prodrug is not activated to its functional form. Therefore, the bacterial cells become unsusceptible to the nitroimidazole antibiotics. (D) Mutations in DNA gyrases (specifically in the QRDR) prevent the binding of fluoroquinolones (e.g., levofloxacin) to the target sites. Therefore, bacterial DNA replication continues in the presence of fluoroquinolones, leading to bacterial resistance to fluoroquinolones. Similarly, mutations in rpoB leading to alterations in RNA polymerases prevent the action of rifamycins (e.g., rifabutin), resulting in bacterial resistance to rifamycins.
Clarithromycin resistance.
Clarithromycin is a bacteriostatic antibiotic belonging to the group of macrolides that inhibits the growth of bacteria by restricting protein synthesis. Macrolides bind to the 50S ribosomal subunit (via 23S rRNA) and prevent protein synthesis (Fig. 4B). The peptidyl-transferase region in the domain V of 23S rRNA is responsible for the binding of antibiotics; therefore, point mutations in this region result in the inhibition of binding of macrolide antibiotics and ribosomal subunits, leading to bacterial resistance to macrolides (Fig. 5B) (406, 407). In H. pylori, two 23S rRNA genes have been found, and mutations in any of these genes confer clarithromycin resistance (408). Major mutations implicated in clarithromycin resistance are A2143G, A2142G, and A2142C, as shown in several studies (409–411). These common mutations which are also referred to as A2147G, A2146G, and A2146C, respectively, in some studies confer resistance in 80–90%, 16–17%, and 2–4% of clinical isolates, respectively. However, clarithromycin resistance is primarily mediated by A2143G, followed by A2142G point mutations. Molecular pathways associated with these mutations are the most frequently studied mechanisms (399, 407, 409, 412, 413, 414). Several other mutations, such as G1939A, A2115G, G2141A, A2144G, A2144T, C2147G, T2182C, G2224A, T2215C, T1958G, A1957G, G1964T, and A1968T, have been reported in diverse geographical regions and are associated with the clarithromycin resistance phenotype (408, 415–417).
Metronidazole resistance.
The antimicrobial activity of azomycin, a nitroimidazole derived from an extract of Streptomyces spp., against Trichomonas vaginalis (a causative agent of vaginal itching) was examined at the Rhone-Poulenc laboratory in France. Metronidazole, a synthetic derivative of nitroimidazole, was used to treat chronic infections caused by Trichomonas spp. in 1959 (418). The antibacterial activity of metronidazole was discovered accidentally when a patient with trichomonas vaginitis and bacterial gingivitis was cured with metronidazole in 1962 (419). Metronidazole, a prodrug that contains a nitro-group in its imidazole ring, is activated by the reduction of its nitro-group, which leads to the production of helicoidal DNA-damaging compounds such as nitroso- and hydroxylamine (Fig. 4C). Oxygen-insensitive NADPH nitro-reductase (RdxA), NADPH-flavin-oxidoreductase (FrxA), and ferredoxin-like enzymes (FrxB) perform the reducing activity of the nitro-group of nitroimidazole, converting it to the active form, in H. pylori (420). Metronidazole resistance is attributed to mutations in RdxA and FrxA, which reduce the potency of their reductase activity, leading to insufficient activation of metronidazole, and mutations in rdxA are involved in the primary underlying mechanism (Fig. 5C) (399, 403, 421, 422). Mutations such as frameshifts, missense mutations, and premature termination in rdxA and frxA have been reported in metronidazole-resistant H. pylori (423, 424). Substitutions in rdxA, such as R16H, H99R, V57A, N14T, W209Q, L210N, V175I, S91P, R16C, R16P, H25R, H53R, H53A, D59N, L62V, A68T, A68V, A68S, A68N, G98S, G163V, G163D, V204I, and A206T have been detected, which confer metronidazole resistance to H. pylori isolates derived from diverse geographical regions (399, 403, 421). Although the high prevalence of metronidazole resistance is conferred by mutational sequence variation in rdxA and frxA, premature termination inducing inactivation of these enzymes has also been recently reported in clinical isolates (399, 403, 421). Premature inactivation attributed to the stop codon in the frxA sequence, together with the intact rdxA gene in some metronidazole-susceptible strains, suggests that inactivation of rdxA is more important for conferring metronidazole resistance (421). Furthermore, while searching for novel genetic mutations associated with metronidazole resistance using a next-generation sequencing approach, we detected a single nucleotide polymorphism, in addition to the insertion-deletion in rdxA in metronidazole-resistant strains, suggesting that mutations in frxA confer metronidazole resistance only in the presence of mutations in rdxA (425).
A novel mechanism attributed to a mutation in the ferric-uptake regulator (Fur) has also been linked with metronidazole resistance. Superoxide dismutase, which is important for protection against superoxide stress, is regulated by Fur, and a fur mutant expressing elevated levels of superoxide dismutase demonstrates metronidazole resistance (426, 427). The rpsU gene encoding 30S ribosomal protein S1, which is involved in protein synthesis, contributes to metronidazole resistance (425). However, the mechanism by which rpsU mediates metronidazole resistance is unknown. Recently, Hanafi et al. (428), who induced resistance in metronidazole-sensitive strains by exposing the strains to metronidazole and compared the protein expression in metronidazole-sensitive and -resistant strains, reported the enhanced expression of aminoacyl-tRNA synthetases, such as ProS, IleS, and CysS, which are involved in the synthesis of aminoacyl-tRNAs, ribosomal proteins such as RpsS, RplF, and RpsI, and elongation factor P (EFP) in metronidazole-resistant strains. Bacterial cells with enhanced protein expression are better at countering the action of metronidazole to maintain the energy balance and fitness that may represent alternate mechanisms for metronidazole resistance in the absence of mutations in rdxA and frxA (428).
Levofloxacin resistance.
Levofloxacin is a fluoroquinolone antibiotic with potent and broad-spectrum activity against Gram-positive and Gram-negative bacterial pathogens. It targets the bacterial DNA gyrase and topoisomerase IV, resulting in impairments in DNA replication (Fig. 4D) (429, 430). The targeting and impairment of either DNA gyrase or topoisomerase IV depend on the type of bacterial pathogen. Typically, DNA gyrase is targeted in Gram-negative organisms, whereas topoisomerase IV is targeted in Gram-positive organisms (431). In bacteria, double-stranded DNA exists in the supercoiling state, which enables large DNA to be packed in bacterial cells (432). During DNA replication, the formation of replication forks, which results in overtwisting, introduces stress in double-stranded DNA. Such DNA strands are unwound by topoisomerases II (DNA gyrases), which can introduce negative supercoils to overtwisted DNA, inducing a relaxed state, which is of utmost importance for DNA replication (433). DNA gyrase is a tetramer consisting of two (A) and two (B) subunits, encoded by gyrA and gyrB (434). The topoisomerase IV-encoding gene has not been reported in H. pylori, which is a major antibiotic target in Gram-positive organisms. Hence, the most common mechanism of high-level levofloxacin resistance in H. pylori is point mutations in DNA gyrase (gyrA and gyrB) (403).
The region in the short sequence of DNA gyrase where point mutations originate, leading to levofloxacin resistance, is known as the quinolone-resistance-determining region (QRDR), which prevents the binding of levofloxacin to DNA gyrase and ultimately confers quinolone (or fluoroquinolone) resistance to bacteria (Fig. 5D) (435, 436). Point mutations in QRDR sequence of gyrA at amino acid 87 and 91, such as N87I, N87K, N87T, N87Y, D91Y, D91N, D91G, and D91H, are the most common mutations reported that lead to resistance to fluoroquinolones in H. pylori (399, 421, 436–438). Other less common mutations reported are D34N, D34Y, A129T, R140K, D161N, D192N, S63P, A88P, D99V, R130K, V172I, and P188S (437, 438). The S63P and R130K substitutions have been associated with high MICs (438). The gyrB mutation is not considered a common mechanism; however, a novel mutation at position 463 leading to fluoroquinolone resistance was suggested by Rimbara et al. (439). Other mutations in gyrB, such as S479T, R484K, and E483K, have also recently been reported in levofloxacin-resistant isolates (437, 438). Mutations at 479 and 484 in gyrB are associated with the most common mutations at 87 and 91 in gyrA; therefore, it is difficult to determine whether the mutations at 479 and 483 in gyrB are responsible for levofloxacin resistance (437). Novel mutations in gyrA (D91A) and gyrB (E381G) have also been reported in levofloxacin-resistant strains; however, their role in mediating resistance needs to be evaluated (440).
Tetracycline resistance.
Tetracycline is a broad-spectrum tetracycline antibiotic (441) that exhibits bacteriostatic activity against a wide range of Gram-positive and Gram-negative bacteria. Tetracyclines (such as tetracycline and minocycline) exhibit antibacterial activity by binding with high-affinity specific pockets involving 16S rRNA on the 30S ribosomal subunit of bacteria and inhibiting protein synthesis by preventing the attachment of aminoacyl-tRNA to the ribosomal acceptor site (Fig. 4B) (442). Tetracycline resistance is not frequently observed in H. pylori, as has been found in other bacteria. A recent meta-analysis reported tetracycline resistance in 10 to 14% of H. pylori isolates worldwide (443). Therefore, the molecular mechanisms leading to tetracycline resistance in H. pylori have not been fully explored. However, mutations in the 16S rRNA at positions 965 to 967 (AGA codon) are responsible for tetracycline resistance (Fig. 5B) (444, 445). One study demonstrated that tetracycline-resistant H. pylori strains carried single nucleotide substitutions, such as A965C, A965G, A965T, A967C, or A967T, with slightly increased MICs (446). Nonaka et al. suggested that guanine (G) at position 966 plays an important role as the primary site of tetracycline binding, and its substitution with other nucleotides results in a high MIC (447). Single and double mutations are associated with low and intermediate MICs, whereas simultaneous triple mutations are associated with high MICs (447). In recent studies, mutations in the 16S rRNA gene at position 926 to 928, such as A926G, TCT926-928AAG, T926A/C927A, T926A/T928G, T926C/C927A, T926C/T928G, and C927A/T928G, which are putative drug-binding sites, were found in tetracycline-resistant strains (411, 421).
Rifabutin resistance.
Rifabutin is structurally similar to other rifamycins, such as rifampicin (448). Rifamycins bind to the bacterial DNA-dependent RNA-polymerase enzyme and inhibit its function. Rifabutin binds to the catalytic center of the enzyme, which is a β-subunit encoded by rpoB. Therefore, rifabutin inhibits prokaryotic RNA synthesis (Fig. 4D) (449). A mutation in the rpoB gene encoding the target site for rifabutin, the β-subunit of DNA-dependent RNA-polymerase, confers resistance against rifabutin (Fig. 5D) (448). Although there is a very low rate of resistance, mutations in codon 149, codons 525 to 545, and codon 586 in the rpoB gene have been reported in rifabutin-resistant strains (411, 450). The point mutation with the substitution D530N is a common mechanism conferring rifabutin resistance (451), whereas other mutations, such as D530N, D530G, D530Y, D530V, D530E, H540N, and S545L, have been reported to have increased MICs (452). A link between the past consumption of rifampicin for the treatment of pulmonary tuberculosis and increased rifabutin MICs has been found, suggesting a cross-resistance between the two antibiotics (448, 453); thus, rifabutin should be recommended based on the history of rifampicin use.
Furazolidone resistance.
Furazolidone is a nitrofuran antibiotic that is structurally similar to metronidazole (454). Furazolidone acts by inhibiting the enzyme monoamine oxidase and lowering bacterial oxidation. Its secondary derivative damages the RNA. Although the mechanism of furazolidone resistance is not clear, it is supposed that mutations in porD and oorD genes, which encode the δ-subunits of reductases such as pyruvate-flavodoxin oxidoreductase and 2-oxoglutarate reductase, respectively, are responsible for furazolidone resistance in H. pylori (455, 456). The mutations G353A, A356G, and C357T were reported in porD and other mutations, such as A041G, A122G, and C349A/G, were reported in oorD in furazolidone resistance isolates, suggesting that these mutations in six different positions of the two genes may be associated with furazolidone resistance in H. pylori (456).
Enzymatic Degradation of Antimicrobial Agents
In addition to target site alteration, the inactivation or destruction of antibiotics by antibiotic-destroying enzymes is another common mechanism of antibiotic resistance. Although this mechanism is uncommon in H. pylori, the involvement of antibiotic-degrading enzymes in mediating resistance has been reported. Beta-lactamases are enzymes that can destroy beta-lactam antibiotics and are the most common mechanism of beta-lactam antibiotic resistance in Gram-negative bacteria; however, such resistance is typically uncommon in H. pylori (457). The use of a beta-lactamase inhibitor together with amoxicillin was found to enhance the eradication rate, suggesting enzymatic degradation of amoxicillin (458). Tseng et al. first reported the role of beta-lactamase in conferring high levels of amoxicillin resistance (MIC ≥ 256 mg/L) in H. pylori (459). Sequence analysis of the β-lactamase PCR product was identical to that of blaTEM-1. Furthermore, dot blot hybridization confirmed the presence of the blaTEM-1 β-lactamase in H. pylori.
Upregulation of the Multidrug Efflux Pump System
Bacterial efflux proteins are plasma (cytoplasmic) membrane proteins that recognize noxious agents, including antibiotics, entering bacterial cells and pump them outside the bacterial cell before they reach their targets (460). Of the five families of multidrug efflux pumps, the resistance nodulation-cell-division (RND) family consists of three membrane fusion proteins (AcrA), an inner membrane protein (AcrB), and an outer membrane protein (TolC) and is one of the efflux pumps originally discovered in Gram-negative bacteria (461, 462). Four RND families of efflux pumps have been recognized in H. pylori: HP0605 to HP0607 (HefABC), HP0969 to HP0971 (HefDEF), HP1327 to HP1329 (HefGHI), and HP1487 to HP1489, which are involved in multidrug resistance in H. pylori (463–465). Mehrabadi et al. (466) reported the involvement of TolC homologs in metronidazole resistance and found that HP0605 and HP0971 could be expressed in the absence of metronidazole. However, overexpression was found in the presence of high-level metronidazole, whereas HP1327 and HP1489 were not expressed under common conditions (466). Hirata et al. also reported the role of efflux pump proteins in the development of clarithromycin resistance in H. pylori (467). A recent study demonstrating the use of the efflux protein inhibitor Phe-Arg-naphthylamide (PAβN) showed reversal of antibacterial potency from 8- to 128-fold against multidrug-resistant strains (468). Recently, the roles of other transporter proteins, such as HP0939, HP0497, HP0471, HP1174 (GluP), HP1017, HP0497, HP0471, HP1165, and HefA, have been depicted in single-drug or multidrug efflux-mediated resistance (469–471). During stress and under nutrient-limiting conditions, the enzyme SpoT, which is a bifunctional enzyme with synthetase and hydrolase activities in H. pylori, regulates bacterial adaptation (472). In recent studies, SpoT was found to induce high levels of expression of transporter proteins, such as HP0939, HP0471, HP0497, HP1017, and HP1174, which are involved in clarithromycin and multidrug efflux-mediated resistance (470, 473).
Biofilm Formation
Biofilms, or microbial communities, are characterized by bacterial cells embedded in a matrix of extracellular polymeric materials produced by microbial cells (474). The biofilm matrix prevents antibiotics from reaching the bacteria and protects them from antibiotic action, aiding the development of antibiotic resistance (475). In vivo production of biofilms by H. pylori in a mouse model was demonstrated by Attaran et al., who concluded that the same potential may be reflected in humans (476). In another recent study, Attaran et al. found a 2- to 4-fold increase in the MIC level of amoxicillin, tetracycline, and metronidazole by biofilm-forming strains than their planktonic counterparts (471). Although most studies have evaluated the in vitro formation of biofilms, several recent studies have shown the capacity for biofilm formation in vivo on human gastric mucosa (477–479). The expression of efflux proteins, such as HP0939, HP0497, HP0471, HP1174, HP1165, and HefA, by biofilm-forming strains in comparison to their planktonic counterparts suggests the enhancement of efflux-mediated resistance by biofilm formation (469–471).
HETERORESISTANCE
In one study, two different H. pylori strains were isolated from two antral biopsy specimens collected from a single patient that significantly differed in their antibiotic susceptibility, showing MICs of amoxicillin between 2 and 0.06 μg/mL, respectively (480). This finding provides new insights into the importance of heteroresistance in H. pylori, which is defined as the coexistence of susceptible and resistant strains in the same patient at the same anatomical site for the same antimicrobial agent (481). Heteroresistance can occur as a result of multiple infections with genetically different strains or the presence of susceptible and resistant variants of genetically related strains. Genetically different heteroresistant strains, if present at the same time at the same anatomical site (i.e., the gastric mucosa), are described as intradistrict, whereas if susceptible and resistant strains are present in different areas of the stomach, they are described as interdistrict (480, 482, 483).
Heteroresistance can be either polyclonal or monoclonal, based on the behavior exhibited by bacterial populations in the presence of antibiotics. Polyclonal heteroresistance describes the presence of stable resistant or susceptible phenotypes exhibited by genetically distinct clones. For example, two or more isolates causing coinfections show different MICs for the same antibiotic or the emergence of stable resistant mutant clones during antibiotic treatment. Monoclonal heteroresistance describes the presence of subpopulations of genetically identical isolates displaying transiently increased resistance, and during this interval, any resistant or susceptible cell may give rise to a new heteroresistant population (484, 485). The importance of heteroresistance implies that no single biopsy site should be considered representative of antimicrobial susceptibility testing (480). Therefore, to obtain a full view of the inflammation status and to identify the presence of heteroresistance to antibiotics, biopsy specimens from all gastric regions should be collected from patients with pangastritis (486). Furthermore, the importance of the presence of heteroresistant H. pylori strains suggests the clinical consequence of the possibility of further propagation of resistant clones despite antibiotic therapy (487). Heteroresistance can be detected by E-tests or the disc diffusion method of antimicrobial susceptibility testing against different antibiotics. In the presence of heteroresistance, colonies of resistant strains appear within the clearing zone around E-test strips or antibiotic-impregnated discs (488). Heteroresistance can also be detected more accurately by comparing the growth rates of many single cells at various antibiotic concentrations (489). Furthermore, the recently developed droplet digital PCR has also been successfully employed for the detection of clarithromycin resistance in the context of heteroresistance (336).
DETECTION OF ANTIMICROBIAL RESISTANCE
The detection of antimicrobial resistance is important because the proportion of bacterial constituents exhibiting resistance to antibiotics continues to change over time, which is critical for designing and optimizing the most effective therapy (490). Injudicious consumption of antibiotics empirically causes the development of antimicrobial resistance, generates a financial burden, or may cause adverse events (491). Therefore, the application of antimicrobial susceptibility testing prior to the initiation of antimicrobial therapy needs to be evaluated. The detection of antimicrobial resistance can be evaluated either by phenotypic methods that utilize the bacterial isolate obtained by culture growth or by genotypic methods that detect resistance in pure culture isolates or directly in specimens such as biopsy, gastric juice, or stool specimens.
Phenotypic Detection of Antimicrobial Resistance
Phenotypic detection of antimicrobial resistance can be carried out using the dilution method (agar dilution and broth dilution methods) and the diffusion method (E-test and disc diffusion methods). Agar dilution, broth dilution, and E-test methods provide the MICs of antibiotics, whereas the disc diffusion method does not provide MICs. However, it provides a resistance cutoff based on the zone diameter of inhibition. Unfortunately, as described in “Bacterial Culture,” culture-based susceptibility results may not be obtained in all cases because the sensitivity of bacterial culture is not 100% and isolation of bacterial strains depends on several technical factors. According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST), the MIC breakpoints that define the cutoff values for resistance are >0.125 mg/L for amoxicillin, >1 mg/L for levofloxacin, >0.5 mg/L for clarithromycin, >1 mg/L for tetracycline, >8 mg/L for metronidazole, and >1 mg/L for rifampicin (492). The Clinical and Laboratory Standard Institute (CLSI) recommendation describes resistance breakpoints as ≥1.0 mg/L for clarithromycin (sensitive cutoff of ≤0.25 and intermediate cutoff of 0.5) (493). The cutoff MIC for furazolidone has not been described by EUCAST; however, several authors have used a resistance cutoff MIC of ≥2 mg/L (494, 495). Different cutoff MICs have been used in different studies, for example, >0.125 and 2 mg/L for amoxicillin, >1 mg/L and 2 mg/L for levofloxacin, >0.5 and 1 mg/L for clarithromycin, >1 mg/L and 4 mg/L for tetracycline, and >8 mg/L and 8 mg/L for metronidazole (421, 492–494, 496, 497).
Dilution methods (agar and broth dilutions).
The agar dilution method of susceptibility testing is the gold standard and is recommended by the CLSI (493). This method has been widely used in epidemiological studies on a large number of stored clinical strains. However, its use in clinical practice is unrealistic and not suited for individual testing of a single strain in everyday practice, because it requires laborious preparation, takes a long time, is technically demanding, and may not be cost-effective (498). In this method,1 to 3 μL of 2.0 McFarland-adjusted bacterial suspension (containing 1 × 107 to 1 × 108 CFU/mL) is spot inoculated on Mueller-Hinton agar supplemented with 5 to 10% sheep or horse blood (aged >2 weeks) and containing 2-fold dilutions of the antibiotics. After incubation for 72 h at 35°C ± 2°C under microaerophilic conditions, the plates are read for any bacterial growth (421, 493, 499). The maximum dilution of antibiotic resulting in no growth on the agar plate is considered the MIC of that antibiotic. The broth microdilution method of susceptibility testing is not recommended and is seldom used because of the difficulty in growing H. pylori strains in broth without supplementation with serum or defibrinated blood. However, a few studies using supplemented broth have reported acceptable MIC results (500–503). Recent studies have reported discrepancies in the results between agar dilution and E-test for antibiotics, such as metronidazole (494, 504). Considering these discrepancies, some authors have recently used the broth dilution method to monitor drug resistance rates and obtained reliable results (494, 505). Agar and broth dilution are quantitative methods that can provide more accurate results (506).
Diffusion method (E-test and disc diffusion).
The E-test, an alternative method of agar dilution, is relatively simple and easy to perform. In this method, the bacterial suspension adjusted to 3.0 McFarland standard is spread on Mueller-Hinton agar supplemented with 5 to 10% sheep blood. An E-test strip impregnated with dried antibiotics at increasing concentrations from one end to the other is placed on the inoculated plate and incubated for 72 h at 37°C under microaerophilic conditions. After incubation, the elliptical zone of inhibition around the strip is observed in the inoculated plate, indicating the MIC at the intersection point between the zone of inhibition and the strip edge (507, 508). This method has been found to correlate closely with the results of the agar dilution method (504, 509–511). However, according to recent studies, the E-test has a tendency to overestimate the resistance to metronidazole compared with the gold standard agar dilution method (494, 504).
The disc diffusion method is the simplest method for antimicrobial susceptibility testing, with the additional benefit of being the least expensive method. However, because of the inadequate studies on its performance and because it is semiquantitative and thus unreliable, this method is not recommended for slow-growing bacteria, including H. pylori (506). During the late 1990s and the early 2000s, several studies performed susceptibility testing using this method and validated the results for macrolides such as erythromycin and metronidazole (512, 513). Recently, a few studies, such as those performed by Ducournau et al. in 2016 (317) and Zhong et al. in 2021 (514) conducted susceptibility testing using the disc diffusion method. Ducournau et al. used resistance cutoff diameters of <17 mm, <14 mm, and <17 mm for tetracycline, rifamycin, and levofloxacin discs, respectively, and found good concordance for rifamycin-resistant strains, which was subsequently confirmed by sequencing the rpoB gene that possessed the mutations (317). Zhong et al. used resistance cutoffs of ≤13 mm, <13 mm, <14 mm, ≤14 mm, ≤14 mm, and <16 mm for clarithromycin, levofloxacin, amoxicillin, furazolidone, tetracycline, and metronidazole, respectively, and found good concordance between phenotypic and genotypic detection of resistance to clarithromycin and levofloxacin (514).
For susceptibility testing by diffusion methods, Mueller-Hinton agar supplemented with 5 to 10% sheep or horse blood is most frequently used, and recommended medium with Columbia agar supplemented with 5 to 10% sheep or horse blood is used as a second choice (515). However, its performance has been found to vary according to the commercial sources and brands of Mueller-Hinton agar used for susceptibility testing (516, 517). E-test and disc diffusion methods provide the additional benefit of allowing the visualization of resistant subpopulations in the case of heteroresistance, as they form colonies within the clear zone of inhibition (488).
Genotypic Detection of Antimicrobial Resistance
Although culture followed by MIC detection-based antimicrobial susceptibility testing is the gold standard method, it is not widely used because the method requires highly trained laboratory personnel and usually takes up to 14 days to obtain results due to the fastidious nature of H. pylori. Therefore, rapid and highly accurate molecular methods are becoming popular for evaluating antimicrobial resistance. A recent meta-analysis by Gong et al. emphasizes that the PCR-based detection of clarithromycin resistance conducted on stool specimens is a reliable and accurate method with high sensitivity (91%) and specificity (97%) (518). Molecular methods such as conventional PCR followed by Sanger sequencing, RT-PCR, and whole-genome sequencing (WGS) are commonly used for the detection of mutations conferring resistance.
Conventional PCR-based detection.
Advances in molecular biology techniques, such as conventional PCR and Sanger sequencing, have enabled the identification of mutation-based molecular mechanisms that cause observed phenotypic antimicrobial resistance. In conventional PCR-based methods, a small gene region exploiting specific mutations is targeted; therefore, this method is insufficient for the discovery of novel or rare resistance mechanisms (519). In the conventional genotypic method, targeted genes, such as pbp1, pbp2, and pbp3 for amoxicillin, 23S rRNA gene for clarithromycin, rdxA and frxA for metronidazole, gyrA and gyrB for levofloxacin, 16S RNA gene for tetracycline, rpoB for rifabutin, and oorD and porD for furazolidone, are amplified using specific primers targeting the regions that frequently show the mutations. The amplified PCR products are subjected to Sanger sequencing and compared with consensus sequences for the presence of any mutations in the genes (421, 500, 514). Conventional PCR-based detection of mutations shows good agreement with phenotypic drug resistance (421, 500, 514). High agreement with the phenotypic method and easy operational management suggest that the conventional PCR-based method can be extended to the detection of mutation-based drug resistance (500, 514).
RT-PCR-based detection.
Clarithromycin resistance attributed to well-known single mutations, such as A2142C, A2142G, and A2143G in the 23S rRNA gene, is a major factor that contributes to the treatment failure of standard clarithromycin-based triple therapy (520, 521). These mutations have been associated with a high level of clarithromycin resistance in Europe, Asia, South America, North America, and Africa, whereas other mutations, including T2182C, C2611A, T2717C, and T2142C, have been reported to confer low levels of clarithromycin resistance (317, 522, 523). With increasing clarithromycin resistance rates in H. pylori, the use of molecular methods that offer rapid and accurate detection of H. pylori infection and clarithromycin resistance simultaneously in real time is being emphasized. In line with this, modern molecular methods based on RT-PCR techniques can provide results directly from biopsy specimens or stool specimens within a few hours.
Currently, several molecular methods for simultaneous detection of H. pylori infection and clarithromycin resistance are commercially available, including H. pylori ClariRes (Ingenetix, Vienna, Austria) (524), Allplex H. pylori and ClariR (Seegene, South Korea) (525), Lightmix H. pylori (TIBMolbiol, Germany) (526), H. pylori TaqMan real-time PCR assay (Meridian Bioscience, United States) (328), Amplidiag H. pylori + ClariR (Mobidiag, Espoo, Finland) (319, 527), and RIDA GENE H. pylori (r-Biopharm, Darmstadt, Germany) (318). These methods have shown excellent performance for simultaneous detection of H. pylori infection and clarithromycin susceptibility with a sensitivity and specificity up to 94% and 100%, respectively, from biopsy or stool specimens. Furthermore, several other RT-PCR assays have been developed for the detection of resistance to clarithromycin and other antimicrobials (528, 529). A one-tube multiplex PCR that applies the amplification refractory mutation system (ARMS) combined with RT-PCR assay was developed and validated for the simultaneous detection of H. pylori, clarithromycin resistance, and levofloxacin resistance (528). RT-PCR was performed with relevant ARMS-PCR primers specific for the 16S rRNA, 23S rRNA, and gyrA genes in a single tube. The ARMS-PCR-specific products were detected using fluorescent TaqMan gene probes specific for these genes. The diagnostic performance of this assay was very high, with sensitivity and specificity for H. pylori, clarithromycin resistance, levofloxacin resistance, and double resistance of 100% and 95%, 100% and 100%, 98% and 95%, 100%, and 97%, respectively (528). A novel assay for wild-type and QRDR-specific mutations leading to quinolone resistance detection using Forster resonance energy transfer (FRET)-labeled probes was evaluated for quick detection of quinolone-resistant H. pylori strains (529).
The RT-PCR-based assays are simple, highly accurate, fast, and cost-effective and provide results for infection as well as for antimicrobial resistance in real time that can be easily applied even in resource-limited, small and medium-sized hospital laboratories and will foster AST-guided therapy.
Whole-genome sequencing-based detection.
Recently, whole-genome sequencing technology has advanced considerably in terms of its affordability with regard to the technology as well as the sample running, which has provided a good opportunity for laboratories to conduct mutation-based drug resistance evaluation (526, 530). Next-generation sequencing (NGS) technology-based bacterial WGS is currently emerging as a high-throughput, cost-effective, and faster technology that offers a more comprehensive and accurate tool for the detection of infection as well as antimicrobial resistance (531, 532). NGS-based WGS has been found to be highly successful and employed for the detection of mutations resulting in phenotypic drug resistance, enabling its potential for designing and implementing local treatment policies (436, 499, 533, 534). The performance of NGS over conventional Sanger sequencing is evident from the fact that alleles with frequencies of 2 to 10% can be detected by NGS, whereas those with frequencies of 15 to 25% can be detected by Sanger sequencing (535). However, the requirement of culture isolates remains the major limitation of WGS, as the application of metagenomics approaches directly on clinical specimens, such as gastric biopsy specimens, is hampered due to the low bacterial genome and high human genome background. A good concordance of WGS has been reported in detecting mutation-based phenotypic antimicrobial resistance to amoxicillin, clarithromycin, and levofloxacin compared with Sanger sequencing (534, 536–540). Furthermore, the results of NGS-based testing of formalin-fixed paraffin-embedded tissues for resistance mutations have been correlated with those of agar dilution method for clarithromycin, levofloxacin, rifabutin, and tetracycline resistance with fair concordance for metronidazole and amoxicillin (411, 541). Nevertheless, NGS-based sequencing performed with formalin-fixed paraffin-embedded tissues has also enabled the detection of mutations that correlate with treatment failure (521, 541).
Owing to their high concordance in mutations and phenotypic drug resistance, WGS-based methods are an attractive alternative to phenotypic methods. Recently, PacBio technology was used to evaluate mutations causing resistance to antibiotics, including amoxicillin, clarithromycin, levofloxacin, metronidazole, tetracycline, and rifampicin (421, 507). Although good agreement between PacBio technology-based mutations and the mutations evidenced by targeted PCR and Sanger sequencing has been found (421), moderate agreement for levofloxacin and an unusual number of SNPs showing no agreement for metronidazole have also been demonstrated (507). However, discrepancies in the agreement between the NGS-based genotype mutation and phenotypic drug resistance for clarithromycin (542) and metronidazole (534) have also been reported. The metronidazole resistance-conferring genes, rdxA and frxA, turned out to be highly variable genes with an unusually high number of SNPs without any clear association with phenotype resistance, resulting in discrepant agreements (507). Furthermore, it is worth considering other mechanisms of metronidazole resistance, including mutations in dapF and efflux system proteins, to understand the discrepant concordance with metronidazole (437). Moreover, NGS could resolve and detect novel, rare, and complex drug resistance mechanisms, such as deletions, large insertions ending with stop codons, or no-stop mutations, making it challenging to detect by conventional PCR followed by Sanger sequencing (430).
Other genotypic detection methods of antimicrobial resistance.
Other novel methods that have been successfully employed for the detection of antimicrobial resistance in H. pylori include droplet digital PCR (336, 337), DNA microarray technology (543), and multiplex quantitative PCR (544). The droplet digital PCR-based detection of antimicrobial resistance, especially to clarithromycin, has shown a high concordance with phenotypic detection by E-test or genetic detection by Sanger sequencing (336, 337). Moreover, droplet digital PCR is efficient in detecting clarithromycin resistance alleles in DNA extracted from formalin-fixed paraffin-embedded tissues (337). Furthermore, studies have found a high prevalence of mixed drug-resistant isolates, and their accurate detection of culture-based antimicrobial resistance testing of a single clone provides a misleading result (480, 481) that could lead to the failure of eradication therapy. However, with the advent of droplet digital PCR for detecting heteroresistance, eradication therapy can be performed more effectively. DNA microarray technology is a reliable, cost-effective, high-throughput, and rapid method that can provide clarithromycin and levofloxacin results within 6 h with a sensitivity to detect 103 CFU/mL and specificity of 97.5% (543). Quantitative PCR has provided perfect concordance with the E-test for detecting clarithromycin and levofloxacin resistance (544). The sensitivity, specificity, PPV, NPV, and accuracy for detecting clarithromycin resistance were 98.7%, 100%, 75.0%, 100%, and 98.8%, respectively, whereas they were 99.8%, 100%, 93.8%, 100%, and 99.8%, respectively, for detecting levofloxacin resistance (544). The line probe assay GenoType HelicoDR (Hain Life Sciences, Germany), a commercially available assay based on multiplex PCR with strip hybridization requiring operator interpretation, is capable of simultaneously detecting the most common point mutations in 23S rRNA (A2143G, A2142G, and A2142C) for clarithromycin resistance and in the gyrA gene (N87K, D91G, D91N, and D91Y) for levofloxacin resistance from biopsy specimens (545).
SUSCEPTIBILITY-GUIDED TREATMENT FOR EFFECTIVE ERADICATION
Typically, the rates of resistance to clarithromycin and metronidazole in a particular region determine the constituents and predict the success rate of eradication therapy. However, resistance to these antibiotics is frequently observed. A meta-analysis conducted on the resistance pattern of primary antibiotics in the Asia-Pacific region reported resistance rates of 17% for clarithromycin, 44% for metronidazole, and 18% for levofloxacin; the rate was below 5% for amoxicillin and tetracycline (546). In most countries, the prevalence of resistance to clarithromycin, metronidazole, and levofloxacin has increased to a level that renders their empirical use in triple therapies unsuccessful (443). Based on the threat that may be imposed, clarithromycin-resistant H. pylori was listed in the WHO’s priority list of antibiotic-resistant bacteria, and it was ranked as the most common cause of community-acquired infection (547). Given the importance of AST in eradication therapy, recently published reports indicate that AST-guided therapy improves the efficacy of clarithromycin-based first-line triple therapy for H. pylori eradication (224, 225, 541, 548). Current consensus guidelines, including the Maastricht V/Florence and Taipei consensus report, also recommend AST for clarithromycin before the initiation of therapy when it is included as a constituent of the first-line standard therapy, unless a low resistance rate of clarithromycin is documented in populations or regions for empirical therapy (72, 78). However, currently, AST for H. pylori is largely unavailable, and only a few major reference laboratories offer culture-based or molecular-based antimicrobial susceptibility testing because the cultivation of H. pylori to determine susceptibility can be very difficult, as it can demonstrate poor growth and be time-consuming and expensive (493, 549).
In the absence of AST results, inappropriate prescription and use of resistant antimicrobial constituents are the most common factors contributing to eradication failure and fueling the development of clarithromycin resistance. To increase the efficacy of standard eradication therapy and decrease the spread of antimicrobial resistance, the application of properly articulated and carefully implemented hospital-wide H. pylori therapy using antimicrobial stewardship principles should be considered, which is based on the use of optimized and direct or indirect AST-guided therapy (550). Furthermore, the coronavirus disease 2019 (COVID-19) pandemic has resulted in universal accessibility of PCR-based diagnostic approaches in hospitals and led health care policy makers to speed up the collection and dissemination of clinical evidence, including diagnostics, treatments, vaccine developments, and health-related technology advancements, which could be repurposed to implement the readily available molecular approaches to prevent the emergence of antimicrobial resistance and its dissemination. For example, due to the COVID-19 pandemic, almost all of the local hospitals and testing laboratories in Europe adopted RT-PCR-based testing for simultaneous detection of H. pylori infection and clarithromycin resistance from biopsy and stool samples (551).
CONCLUSIONS
Increasing clarithromycin resistance and lack of AST-guided therapy are key factors contributing to the failure of eradication therapy and worsening of H. pylori-associated mortality. Accurate diagnosis of an infection is a critical step in the successful eradication and curbing of the development of antimicrobial resistance. The performance of the diagnostic methods depends on several factors, such as clinical setting, skills of laboratory personnel, patient factors, and H. pylori strains used for preparing diagnostic kits. Therefore, methods achieving excellent performance after local validation usually provide the most accurate infection diagnosis. Moreover, the real-time assessment of clarithromycin susceptibility by RT-PCR-based methods is valuable for the judicious use of appropriate and effective therapy to achieve the highest eradication rate. Furthermore, AST-guided therapy using the antimicrobial stewardship principle might play a role in minimizing eradication failure and resistance development.
ACKNOWLEDGMENTS
This study was supported in part by grants from the National Institutes of Health (DK62813) and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (26640114, 15H02657, 16H05191, 16H06279, 18KK0266, 19H03473). This work was also supported by Japan Agency for Medical Research and Development (AMED) [e-ASIA JRP, Science and Technology Research Partnership for Sustainable Development (SATREPS), Global Alliance for Chronic Diseases (GACD)] and Japan International Cooperation Agency (JICA) [SATREPS]. This study was also supported by Thailand Science Research and Innovation Fundamental Fund, Bualuang ASEAN Chair Professorship at Thammasat University, and Center of Excellence in Digestive Diseases, Thammasat University, Thailand. We also express our sincere thanks to Patrick Lane of ScEYEnce Studios for enhancing the graphics.
We declare that there are no conflicts of interest.
Biographies

Shamshul Ansari, M.Sc., Ph.D., completed his M.Sc. in clinical microbiology in 2012 at the Institute of Medicine, Tribhuvan University, Kathmandu, Nepal, and his Ph.D. in medical sciences in 2019 at the Oita University Faculty of Medicine, Oita, Japan. He has worked in both academia and research for over 8 years. Currently, he is an assistant professor in the Department of Environmental and Preventive Medicine, Oita University Faculty of Medicine, Oita, Japan. His research interests focus on molecular epidemiology, antimicrobial resistance, and diagnostic approaches for H. pylori infection.

Yoshio Yamaoka, M.D., Ph.D., is full professor and head of the Department of Environmental and Preventive Medicine, Oita University Faculty of Medicine, Japan, and professor of medicine (gastroenterology) at Baylor College of Medicine, Houston, TX, USA. Professor Yamaoka is also a vice-president and trustee of Oita University. Professor Yamaoka has research interest in Helicobacter pylori-related gastroduodenal diseases, molecular epidemiology, virulence factors of Helicobacter pylori, antimicrobial resistance, immunology of gastroduodenal diseases, gastric cancer, and diagnostic approaches for gastroduodenal diseases.
REFERENCES
- 1.ARC Working Group. 1994. World Health Organization, International Agency for Research on Cancer. Schistosomes, liver flukes and Helicobacter pylori. WHO, Lyon, France. [Google Scholar]
- 2.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. 2001. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345:784–789. 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 3.Jiang J, Chen Y, Shi J, Song C, Zhang J, Wang K. 2017. Population attributable burden of Helicobacter pylori related gastric cancer, coronary heart disease, and ischemic stroke in China. Eur J Clin Microbiol Infect Dis 36:199–212. 10.1007/s10096-016-2810-x. [DOI] [PubMed] [Google Scholar]
- 4.Backert S, Neddermann M, Maubach G, Naumann M. 2016. Pathogenesis of Helicobacter pylori infection. Helicobacter 21:19–25. 10.1111/hel.12335. [DOI] [PubMed] [Google Scholar]
- 5.de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. 2020. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 8:e180–90–e190. 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 6.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 7.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 8.Chiang TH, Chang WJ, Chen SL, Yen AM, Fann JC, Chiu SY, Chen YR, Chuang SL, Shieh CF, Liu CY, Chiu HM, Chiang H, Shun CT, Lin MW, Wu MS, Lin JT, Chan CC, Graham DY, Chen HH, Lee YC. 2020. Mass eradication of Helicobacter pylori to reduce gastric cancer incidence and mortality: a long-term cohort study on Matsu Islands. Gut 70:243–250. 10.1136/gutjnl-2020-322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen TH, Cheng HT, Yeh CT. 2021. Epidemiology changes in peptic ulcer diseases 18 years apart explored from the genetic aspects of Helicobacter pylori. Transl Res 232:115–120. 10.1016/j.trsl.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Choi IJ, Kook MC, Kim YI, Cho SJ, Lee JY, Kim CG, Park B, Nam BH. 2018. Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N Engl J Med 378:1085–1095. 10.1056/NEJMoa1708423. [DOI] [PubMed] [Google Scholar]
- 11.Choi IJ, Kim CG, Lee JY, Kim YI, Kook MC, Park B, Joo J. 2020. Family history of gastric cancer and Helicobacter pylori treatment. N Engl J Med 382:427–436. 10.1056/NEJMoa1909666. [DOI] [PubMed] [Google Scholar]
- 12.Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. 2016. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology 150:1113–1124.E5. 10.1053/j.gastro.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Ford AC, Yuan Y, Moayyedi P. 2020. Helicobacter pylori eradication therapy to prevent gastric cancer: systematic review and meta-analysis. Gut 69:2113–2121. 10.1136/gutjnl-2020-320839. [DOI] [PubMed] [Google Scholar]
- 14.Crowe SE. 2019. Helicobacter pylori infection. N Engl J Med 380:1158–1165. 10.1056/NEJMcp1710945. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura S, Sugiyama T, Matsumoto T, Iijima K, Ono S, Tajika M, Tari A, Kitadai Y, Matsumoto H, Nagaya T, Kamoshida T, Watanabe N, Chiba T, Origasa H, Asaka M, JAPAN GAST Study Group. 2012. Long-term clinical outcome of gastric MALT lymphoma after eradication of Helicobacter pylori: a multicentre cohort follow-up study of 420 patients in Japan. Gut 61:507–513. 10.1136/gutjnl-2011-300495. [DOI] [PubMed] [Google Scholar]
- 16.Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19:449–490. 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansari S, Yamaoka Y. 2017. Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 22:e12386. 10.1111/hel.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansari S, Yamaoka Y. 2019. Helicobacter pylori virulence factors exploiting gastric colonization and its pathogenicity. Toxins 11:677. 10.3390/toxins11110677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wandler AM, Guillemin K. 2012. Transgenic expression of the Helicobacter pylori virulence factor CagA promotes apoptosis or tumorigenesis through JNK activation in Drosophila. PLoS Pathog 8:e1002939. 10.1371/journal.ppat.1002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akazawa Y, Isomoto H, Matsushima K, Kanda T, Minami H, Yamaghchi N, Taura N, Shiozawa K, Ohnita K, Takeshima F, Nakano M, Moss J, Hirayama T, Nakao K. 2013. Endoplasmic reticulum stress contributes to Helicobacter pylori VacA-induced apoptosis. PLoS One 8:e82322. 10.1371/journal.pone.0082322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Palma M, Biziato D, Petrova TV. 2017. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer 17:457–474. 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 22.Macedo F, Ladeira K, Longatto-Filho A, Martins SF. 2017. Gastric cancer and angiogenesis: is VEGF a useful biomarker to assess progression and remission? J Gastric Cancer 17:1–10. 10.5230/jgc.2017.17.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ansari S, Yamaoka Y. 2017. Helicobacter pylori BabA in adaptation for gastric colonization. World J Gastroenterol 23:4158–4169. 10.3748/wjg.v23.i23.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, Kalali B, Gerhard M, Sartori AA, Lopes M, Müller A. 2011. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci USA 108:14944–14949. 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Borén T, Haas R, Sasakawa C, Mimuro H. 2011. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem 286:25256–25264. 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rad R, Gerhard M, Lang R, Schöniger M, Rösch T, Schepp W, Becker I, Wagner H, Prinz C. 2002. The Helicobacter pylori blood group antigen-binding adhesin facilitates bacterial colonization and augments a nonspecific immune response. J Immunol 168:3033–3041. 10.4049/jimmunol.168.6.3033. [DOI] [PubMed] [Google Scholar]
- 27.Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA 96:12778–12783. 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaoka Y. 2010. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol 7:629–641. 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. 2010. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet 6:e1001069. 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Backert S, Tegtmeyer N, Fischer W. 2015. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol 10:955–965. 10.2217/fmb.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung JM, Sheedlo MJ, Campbell AM, Sawhney N, Frick-Cheng AE, Lacy DB, Cover TL, Ohi MD. 2019. Structure of the Helicobacter pylori Cag type IV secretion system. Elife 8:e47644. 10.7554/eLife.47644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Backert S, Haas R, Gerhard M, Naumann M. 2017. The Helicobacter pylori type IV secretion system encoded by the cag pathogenicity island: architecture, function, and signaling. Curr Top Microbiol Immunol 413:187–220. 10.1007/978-3-319-75241-9_8. [DOI] [PubMed] [Google Scholar]
- 33.Cover TL. 2016. Helicobacter pylori diversity and gastric cancer risk. mBio 7:e01869-15. 10.1128/mBio.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miernyk KM, Bruden D, Rudolph KM, Hurlburt DA, Sacco F, McMahon BJ, Bruce MG. 2020. Presence of cagPAI genes and characterization of vacA s, i and m regions in Helicobacter pylori isolated from Alaskans and their association with clinical pathologies. J Med Microbiol 69:218–227. 10.1099/jmm.0.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansari S, Yamaoka Y. 2020. Helicobacter pylori virulence factor cytotoxin associated gene A (CagA)-mediated gastric pathogenicity. Int J Mol Sci 21:7430. 10.3390/ijms21197430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Liu J, Gong Y, Yuan Y. 2017. Association of CagA EPIYA-D or EPIYA-C phosphorylation sites with peptic ulcer and gastric cancer risks: a meta-analysis. Medicine 96:e6620. 10.1097/MD.0000000000006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA 99:14428–14433. 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ansari S, Yamaoka Y. 2020. Role of vacuolating cytotoxin A in Helicobacter pylori infection and its impact on gastric pathogenesis. Expert Rev Anti Infect Ther 18:987–996. 10.1080/14787210.2020.1782739. [DOI] [PubMed] [Google Scholar]
- 39.Foegeding NJ, Raghunathan K, Campbell AM, Kim SW, Lau KS, Kenworthy AK, Cover TL, Ohi MD. 2019. Intracellular degradation of Helicobacter pylori VacA toxin as a determinant of gastric epithelial cell viability. Infect Immun 87:e00783-18. 10.1128/IAI.00783-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teng Y, Liu X, Han B, Ma Q, Liu Y, Kong H, Lv Y, Mao F, Cheng P, Hao C, Yang S, Zhang J, Peng L, Zou Q, Zhuang Y. 2019. Helicobacter pylori-downregulated tumor necrosis factor receptor-associated protein 1 mediates apoptosis of human gastric epithelial cells. J Cell Physiol 234:15698–15707. 10.1002/jcp.28223. [DOI] [PubMed] [Google Scholar]
- 41.Zhu P, Xue J, Zhang ZJ, Jia YP, Tong YN, Han D, Li Q, Xiang Y, Mao XH, Tang B. 2017. Helicobacter pylori VacA induces autophagic cell death in gastric epithelial cells via the endoplasmic reticulum stress pathway. Cell Death Dis 8:3207. 10.1038/s41419-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres VJ, VanCompernolle SE, Sundrud MS, Unutmaz D, Cover TL. 2007. Helicobacter pylori vacuolating cytotoxin inhibits activation-induced proliferation of human T and B lymphocyte subsets. J Immunol 179:5433–5440. 10.4049/jimmunol.179.8.5433. [DOI] [PubMed] [Google Scholar]
- 43.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. 2004. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc Natl Acad Sci USA 101:7727–7732. 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim IJ, Blanke SR. 2012. Remodeling the host environment: modulation of the gastric epithelium by the Helicobacter pylori vacuolating toxin (VacA). Front Cell Infect Microbiol 2:37. 10.3389/fcimb.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain P, Luo ZQ, Blanke SR. 2011. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc Natl Acad Sci USA 108:16032–16037. 10.1073/pnas.1105175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calore F, Genisset C, Casellato A, Rossato M, Codolo G, Esposti MD, Scorrano L, de Bernard M. 2010. Endosome-mitochondria juxtaposition during apoptosis induced by H. pylori VacA. Cell Death Differ 17:1707–1716. 10.1038/cdd.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radin JN, González-Rivera C, Ivie SE, McClain MS, Cover TL. 2011. Helicobacter pylori VacA induces programmed necrosis in gastric epithelial cells. Infect Immun 79:2535–2543. 10.1128/IAI.01370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teymournejad O, Mobarez AM, Hassan ZM, Talebi Bezmin Abadi A. 2017. Binding of the Helicobacter pylori OipA causes apoptosis of host cells via modulation of Bax/Bcl-2 levels. Sci Rep 7:8036. 10.1038/s41598-017-08176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi A, Shiota S, Matsunari O, Watada M, Suzuki R, Nakachi S, Kinjo N, Kinjo F, Yamaoka Y. 2013. Intact long-type dupA as a marker for gastroduodenal diseases in Okinawan subpopulation, Japan. Helicobacter 18:66–72. 10.1111/j.1523-5378.2012.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J, Leung WK, Go MY, Chan MC, To KF, Ng EK, Chan FK, Ling TK, Chung SC, Sung JJ. 2002. Relationship between Helicobacter pylori babA2 status with gastric epithelial cell turnover and premalignant gastric lesions. Gut 51:480–484. 10.1136/gut.51.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaoka Y. 2008. Increasing evidence of the role of Helicobacter pylori SabA in the pathogenesis of gastroduodenal disease. J Infect Dev Ctries 2:174–181. 10.3855/jidc.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma YJ, Duan GC, Zhang RG, Fan QT, Zhang WD. 2010. Mutation of iceA in Helicobacter pylori compromised IL-8 induction from human gastric epithelial cells. J Basic Microbiol 50(Suppl 1):S83–S88. 10.1002/jobm.200900410. [DOI] [PubMed] [Google Scholar]
- 53.Yokota S, Konno M, Fujiwara S, Toita N, Takahashi M, Yamamoto S, Ogasawara N, Shiraishi T. 2015. Intrafamilial, preferentially mother-to-child and intraspousal, Helicobacter pylori infection in Japan determined by mutilocus [sic] sequence typing and random amplified polymorphic DNA fingerprinting. Helicobacter 20:334–342. 10.1111/hel.12217. [DOI] [PubMed] [Google Scholar]
- 54.Bui D, Brown H, Harris R, Oren E. 2016. Serologic evidence for fecal-oral transmission of Helicobacter pylori. Am J Trop Med Hyg 94:82–88. 10.4269/ajtmh.15-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mamishi S, Eshaghi H, Mahmoudi S, Bahador A, Hosseinpour Sadeghi R, Najafi M, Farahmand F, Khodadad A, Pourakbari B. 2016. Intrafamilial transmission of Helicobacter pylori: genotyping of faecal samples. Br J Biomed Sci 73:38–43. 10.1080/09674845.2016.1150666. [DOI] [PubMed] [Google Scholar]
- 56.Osaki T, Konno M, Yonezawa H, Hojo F, Zaman C, Takahashi M, Fujiwara S, Kamiya S. 2015. Analysis of intra-familial transmission of Helicobacter pylori in Japanese families. J Med Microbiol 64:67–73. 10.1099/jmm.0.080507-0. [DOI] [PubMed] [Google Scholar]
- 57.Zamani M, Ebrahimtabar F, Zamani V, Miller WH, Alizadeh-Navaei R, Shokri-Shirvani J, Derakhshan MH. 2018. Systematic review with meta-analysis: the worldwide prevalence of Helicobacter pylori infection. Aliment Pharmacol Ther 47:868–876. 10.1111/apt.14561. [DOI] [PubMed] [Google Scholar]
- 58.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global Prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterol 153:420–429. 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 59.Doohan D, Fauzia KA, Rathnayake J, Lamawansa MD, Waskito LA, Tuan VP, Dashdorj A, Kabamba ET, Phuc BH, Ansari S, Akada J, Matsumoto T, Uchida T, Matsuhisa T, Yamaoka Y. 2021. Pepsinogen and serum IgG detection is a valuable diagnostic method for Helicobacter pylori infection in a low-prevalence country: a report from Sri Lanka. Diagnostics (Basel) 11:1364. 10.3390/diagnostics11081364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuipers EJ, Thijs JC, Festen HP. 1995. The prevalence of Helicobacter pylori in peptic ulcer disease. Aliment Pharmacol Ther 9(Suppl 2):59–69. [PubMed] [Google Scholar]
- 61.Rokkas T, Rokka A, Portincasa P. 2017. A systematic review and meta-analysis of the role of Helicobacter pylori eradication in preventing gastric cancer. Ann Gastroenterol 30:414–423. 10.20524/aog.2017.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ford AC, Forman D, Hunt RH, Yuan Y, Moayyedi P. 2014. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. BMJ 348:g3174. 10.1136/bmj.g3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kodama M, Murakami K, Okimoto T, Sato R, Uchida M, Abe T, Shiota S, Nakagawa Y, Mizukami K, Fujioka T. 2012. Ten-year prospective follow-up of histological changes at five points on the gastric mucosa as recommended by the updated Sydney system after Helicobacter pylori eradication. J Gastroenterol 47:394–403. 10.1007/s00535-011-0504-9. [DOI] [PubMed] [Google Scholar]
- 64.Mera RM, Bravo LE, Camargo MC, Bravo JC, Delgado AG, Romero-Gallo J, Yepez MC, Realpe JL, Schneider BG, Morgan DR, Peek RM, Jr, Correa P, Wilson KT, Piazuelo MB. 2018. Dynamics of Helicobacter pylori infection as a determinant of progression of gastric precancerous lesions: 16-year follow-up of an eradication trial. Gut 67:1239–1246. 10.1136/gutjnl-2016-311685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwang YJ, Kim N, Lee HS, Lee JB, Choi YJ, Yoon H, Shin CM, Park YS, Lee DH. 2018. Reversibility of atrophic gastritis and intestinal metaplasia after Helicobacter pylori eradication—a prospective study for up to 10 years. Aliment Pharmacol Ther 47:380–390. 10.1111/apt.14424. [DOI] [PubMed] [Google Scholar]
- 66.Kumar S, Metz DC, Ellenberg S, Kaplan DE, Goldberg DS. 2020. Risk factors and incidence of gastric cancer after detection of Helicobacter pylori infection: a large cohort study. Gastroenterol 158:527–536.e7. 10.1053/j.gastro.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi JM, Kim SG, Choi J, Park JY, Oh S, Yang HJ, Lim JH, Im JP, Kim JS, Jung HC. 2018. Effects of Helicobacter pylori eradication for metachronous gastric cancer prevention: a randomized controlled trial. Gastrointest Endosc 88:475–485.E2. 10.1016/j.gie.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Chey WD, Leontiadis GI, Howden CW, Moss SF. 2017. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol 112:212–239. 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 69.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ, European Helicobacter Study Group. 2012. Management of Helicobacter pylori infection—the Maastricht IV/Florence consensus report. Gut 61:646–664. 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 70.Leja M, Cine E, Rudzite D, Vilkoite I, Huttunen T, Daugule I, Rumba-Rozenfelde I, Pimanov S, Liepniece-Karele I, Pahomova J, Purmalis K, Eglitis J, Pirags V, Dzerve V, Erglis A. 2012. Prevalence of Helicobacter pylori infection and atrophic gastritis in Latvia. Eur J Gastroenterol Hepatol 24:1410–1417. 10.1097/MEG.0b013e3283583ca5. [DOI] [PubMed] [Google Scholar]
- 71.Skrebinska S, Daugule I, Santare D, Isajevs S, Liepniece-Karele I, Rudzite D, Kikuste I, Vanags A, Tolmanis I, Atstupens J, Park JY, Herrero R, Leja M. 2018. Accuracy of two plasma antibody tests and faecal antigen test for non-invasive detection of H. pylori in middle-aged Caucasian general population sample. Scand J Gastroenterol 53:777–783. 10.1080/00365521.2018.1476909. [DOI] [PubMed] [Google Scholar]
- 72.Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM, European Helicobacter and Microbiota Study Group and Consensus panel. 2017. Management of Helicobacter pylori infection-the Maastricht V/Florence consensus report. Gut 66:6–30. 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 73.Shiotani A, Graham DY. 2015. Helicobacter pylori: new thoughts and practices. Gastroenterol Clin North Am 44:15–16. 10.1016/S0889-8553(15)00082-5. [DOI] [PubMed] [Google Scholar]
- 74.Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P, faculty members of Kyoto Global Consensus Conference. 2015. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64:1353–1367. 10.1136/gutjnl-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.El-Serag HB, Kao JY, Kanwal F, Gilger M, LoVecchio F, Moss SF, Crowe SE, Elfant A, Haas T, Hapke RJ, Graham DY. 2018. Houston consensus conference on testing for Helicobacter pylori infection in the United States. Clin Gastroenterol Hepatol 16:992–1002.E6. 10.1016/j.cgh.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta S, Li D, El Serag HB, Davitkov P, Altayar O, Sultan S, Falck-Ytter Y, Mustafa RA. 2020. AGA clinical practice guidelines on management of gastric intestinal metaplasia. Gastroenterology 158:693–702. 10.1053/j.gastro.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mahachai V, Vilaichone R-K, Pittayanon R, Rojborwonwitaya J, Leelakusolvong S, Maneerattanaporn M, Chotivitayatarakorn P, Treeprasertsuk S, Kositchaiwat C, Pisespongsa P, Mairiang P, Rani A, Leow A, Mya SM, Lee Y-C, Vannarath S, Rasachak B, Chakravuth O, Aung MM, Ang T-L, Sollano JD, Trong Quach D, Sansak I, Wiwattanachang O, Harnsomburana P, Syam AF, Yamaoka Y, Fock K-M, Goh K-L, Sugano K, Graham D. 2018. Helicobacter pylori management in ASEAN: the Bangkok consensus report. J Gastroenterol Hepatol 33:37–56. 10.1111/jgh.13911. [DOI] [PubMed] [Google Scholar]
- 78.Liou JM, Malfertheiner P, Lee YC, Sheu BS, Sugano K, Cheng HC, Yeoh KG, Hsu PI, Goh KL, Mahachai V, Gotoda T, Chang WL, Chen MJ, Chiang TH, Chen CC, Wu CY, Leow AH, Wu JY, Wu DC, Hong TC, Lu H, Yamaoka Y, Megraud F, Chan FKL, Sung JJ, Lin JT, Graham DY, Wu MS, El-Omar EM, Asian Pacific Alliance on Helicobacter and Microbiota (APAHAM). 2020. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut 69:2093–2112. 10.1136/gutjnl-2020-322368. [DOI] [PubMed] [Google Scholar]
- 79.Oluwasola AO, Okolo CA, Otegbayo JA, Adeniyi B, Kehinde AO, Akere A, Ola SO, Lawal TO, Idowu PA, Odaibo G, Lawan AI. 2011. Comparative study of methods of diagnosis of Helicobacter pylori infection in Ibadan, Nigeria Niger J Gastroenterol Hepatol 3:31–38. [Google Scholar]
- 80.Kabir S. 2001. Detection of Helicobacter pylori in faeces by culture, PCR and enzyme immunoassay. J Med Microbiol 50:1021–1029. 10.1099/0022-1317-50-12-1021. [DOI] [PubMed] [Google Scholar]
- 81.Hunt RH, Xiao SD, Megraud F, Leon-Barua R, Bazzoli F, van der Merwe S, Vaz Coelho LG, Fock M, Fedail S, Cohen H, Malfertheiner P, Vakil N, Hamid S, Goh KL, Wong BC, Krabshuis J, Le Mair A, World Gastroenterology Organization. 2011. Helicobacter pylori in developing countries. World Gastroenterology Organisation global guideline. J Gastrointestin Liver Dis 20:299–304. [PubMed] [Google Scholar]
- 82.Chey WD, Wong BC, Practice Parameters Committee of the American College of Gastroenterology. 2007. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol 102:1808–1825. 10.1111/j.1572-0241.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 83.Iwańczak F, Pytrus T, Iwańczak B, Gościniak G, Witkowska-Vogtt E, Dzierzanowska D, Laszewicz W. 2005. Assessment of selected tests in diagnostics of Helicobacter pylori infections. Pol Merkur Lekarski 19:52–56. (In Polish.) [PubMed] [Google Scholar]
- 84.Ansari S, Yamaoka Y. 2018. Current understanding and management of Helicobacter pylori infection: an updated appraisal. F1000Res 7:721. 10.12688/f1000research.14149.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Savarino V, Mela GS, Zentilin P, Bisso G, Pivari M, Mansi C, Mele MR, Bilardi C, Vigneri S, Celle G. 1999. Comparison of isotope ratio mass spectrometry and nondispersive isotope-selective infrared spectroscopy for 13C-urea breath test. Am J Gastroenterol 94:1203–1208. 10.1111/j.1572-0241.1999.01067.x. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y-K, Kuo F-C, Liu C-J, Wu M-C, Shih H-Y, Wang SSW, Wu J-Y, Kuo C-H, Huang Y-K, Wu D-C. 2015. Diagnosis of Helicobacter pylori infection: current options and developments. World J Gastroenterol 21:11221–11235. 10.3748/wjg.v21.i40.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Best LM, Takwoingi Y, Siddique S, Selladurai A, Gandhi A, Low B, Yaghoobi M, Gurusamy KS. 2018. Non-invasive diagnostic tests for Helicobacter pylori infection. Cochrane Database Syst Rev 3:CD012080. 10.1002/14651858.CD012080.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leal YA, Flores LL, Fuentes-Pananá EM, Cedillo-Rivera R, Torres J. 2011. 13C-urea breath test for the diagnosis of Helicobacter pylori infection in children: a systematic review and meta-analysis. Helicobacter 16:327–337. 10.1111/j.1523-5378.2011.00863.x. [DOI] [PubMed] [Google Scholar]
- 89.Jones NL, Koletzko S, Goodman K, Bontems P, Cadranel S, Casswall T, Czinn S, Gold BD, Guarner J, Elitsur Y, Homan M, Kalach N, Kori M, Madrazo A, Megraud F, Papadopoulou A, Rowland M, ESPGHAN, NASPGHAN. 2017. Joint ESPGHAN/NASPGHAN guidelines for the management of Helicobacter pylori in children and adolescents (update). J Pediatr Gastroenterol Nutr 64:991–1003. 10.1097/MPG.0000000000001594. [DOI] [PubMed] [Google Scholar]
- 90.Keller J, Bassotti G, Clarke J, Dinning P, Fox M, Grover M, Hellström PM, Ke M, Layer P, Malagelada C, Parkman HP, Scott SM, Tack J, Simren M, Törnblom H, Camilleri M, International Working Group for Disorders of Gastrointestinal Motility and Function. 2018. Expert consensus document: advances in the diagnosis and classification of gastric and intestinal motility disorders. Nat Rev Gastroenterol Hepatol 15:291–308. 10.1038/nrgastro.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Löhr JM, Dominguez-Munoz E, Rosendahl J, Besselink M, Mayerle J, Lerch MM, Haas S, Akisik F, Kartalis N, Iglesias-Garcia J, Keller J, Boermeester M, Werner J, Dumonceau JM, Fockens P, Drewes A, Ceyhan G, Lindkvist B, Drenth J, Ewald N, Hardt P, de Madaria E, Witt H, Schneider A, Manfredi R, Brøndum FJ, Rudolf S, Bollen T, Bruno M, HaPanEU/UEG Working Group. 2017. United European Gastroenterology evidence-based guidelines for the diagnosis and therapy of chronic pancreatitis (HaPanEU). United European Gastroenterol J 5:153–199. 10.1177/2050640616684695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Braden B. 2012. Diagnosis of Helicobacter pylori infection. BMJ 344:e828. 10.1136/bmj.e828. [DOI] [PubMed] [Google Scholar]
- 93.Eggers RH, Kulp A, Tegeler R. 1990. A methodological analysis of the 13C urea breath test for detection of Helicobacter pylori infections: high sensitivity and specificity within 30 min using 75 mg of 13C-urea. Eur J Gastro Hepatol 2:437–444. [Google Scholar]
- 94.Yang HR, Seo JK. 2005. Diagnostic accuracy of the C-urea breath test in children: adjustment of the cut-off value according to age. J Gastroenterol Hepatol 20:264–269. 10.1111/j.1440-1746.2004.03541.x. [DOI] [PubMed] [Google Scholar]
- 95.Li ZX, Huang LL, Liu C, Formichella L, Zhang Y, Wang YM, Zhang L, Ma JL, Liu WD, Ulm K, Wang JX, Zhang L, Bajbouj M, Li M, Vieth M, Quante M, Zhou T, Wang LH, Suchanek S, Soutschek E, Schmid R, Classen M, You WC, Gerhard M, Pan KF. 2017. Cut-off optimization for 13C-urea breath test in a community-based trial by mathematic, histology and serology approach. Sci Rep 7:2072. 10.1038/s41598-017-02180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gisbert JP, Pajares JM. 2004. 13C-urea breath test in the diagnosis of Helicobacter pylori infection—a critical review. Aliment Pharmacol Ther 20:1001–1017. 10.1111/j.1365-2036.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- 97.Malaty HM, el-Zimaity HM, Genta RM, Klein PD, Graham DY. 1996. Twenty-minute fasting version of the US 13C-urea breath test for the diagnosis of H. pylori infection. Helicobacter 1:165–167. 10.1111/j.1523-5378.1996.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 98.Ohara S, Kato M, Asaka M, Toyota T. 1998. Studies of 13C-urea breath test for diagnosis of Helicobacter pylori infection in Japan. J Gastroenterol 33:6–13. 10.1007/pl00009968. [DOI] [PubMed] [Google Scholar]
- 99.Wang WM, Lee SC, Ding HJ, Jan CM, Chen LT, Wu DC, Liu CS, Peng CF, Chen YW, Huang YF, Chen CY. 1998. Quantification of Helicobacter pylori infection: simple and rapid 13C-urea breath test in Taiwan. J Gastroenterol 33:330–335. 10.1007/s005350050092. [DOI] [PubMed] [Google Scholar]
- 100.Leodolter A, Domínguez-Muñoz JE, von Arnim U, Kahl S, Peitz U, Malfertheiner P. 1999. Validity of a modified 13C-urea breath test for pre- and posttreatment diagnosis of Helicobacter pylori infection in the routine clinical setting. Am J Gastroenterol 94:2100–2104. 10.1111/j.1572-0241.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 101.Perets TT, Gingold-Belfer R, Leibovitzh H, Itskoviz D, Schmilovitz-Weiss H, Snir Y, Dickman R, Dotan I, Levi Z, Boltin D. 2019. Optimization of 13 C-urea breath test threshold levels for the detection of Helicobacter pylori infection in a national referral laboratory. J Clin Lab Anal 33:e22674. 10.1002/jcla.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eisdorfer I, Shalev V, Goren S, Chodick G, Muhsen K. 2018. Sex differences in urea breath test results for the diagnosis of Helicobacter pylori infection: a large cross-sectional study. Biol Sex Differ 9:1. 10.1186/s13293-017-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, Lai KC, Hu WH, Yuen ST, Leung SY, Fong DY, Ho J, Ching CK, Chen JS, China Gastric Cancer Study Group. 2004. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA 291:187–194. 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 104.Gatta L, Vakil N, Ricci C, Osborn JF, Tampieri A, Perna F, Miglioli M, Vaira D. 2004. Effect of proton pump inhibitors and antacid therapy on 13C urea breath tests and stool test for Helicobacter pylori infection. Am J Gastroenterol 99:823–829. 10.1111/j.1572-0241.2004.30162.x. [DOI] [PubMed] [Google Scholar]
- 105.Levine A, Shevah O, Shabat-Sehayek V, Aeed H, Boaz M, Moss SF, Niv Y, Avni Y, Shirin H. 2004. Masking of 13C urea breath test by proton pump inhibitors is dependent on type of medication: comparison between omeprazole, pantoprazole, lansoprazole and esomeprazole. Aliment Pharmacol Ther 20:117–122. 10.1111/j.1365-2036.2004.02021.x. [DOI] [PubMed] [Google Scholar]
- 106.Oztürk E, Yeşilova Z, Ilgan S, Ozgüven M, Dağalp K. 2009. Performance of acidified 14C-urea capsule breath test during pantoprazole and ranitidine treatment. J Gastroenterol Hepatol 24:1248–1251. 10.1111/j.1440-1746.2009.05845.x. [DOI] [PubMed] [Google Scholar]
- 107.Shirin H, Frenkel D, Shevah O, Levine A, Bruck R, Moss SF, Niv Y, Avni Y. 2003. Effect of proton pump inhibitors on the continuous real time (13)C-urea breath test. Am J Gastroenterol 98:46–50. 10.1111/j.1572-0241.2003.07187.x. [DOI] [PubMed] [Google Scholar]
- 108.Asaka M, Kato M, Takahashi S, Fukuda Y, Sugiyama T, Ota H, Uemura N, Murakami K, Satoh K, Sugano K, Japanese Society for Helicobacter Research. 2010. Guidelines for the management of Helicobacter pylori infection in Japan: 2009 revised edition. Helicobacter 15:1–20. 10.1111/j.1523-5378.2009.00738.x. [DOI] [PubMed] [Google Scholar]
- 109.Moayyedi P, Lacy BE, Andrews CN, Enns RA, Howden CW, Vakil N. 2017. ACG and CAG clinical guideline: management of dyspepsia. Am J Gastroenterol 112:988–1013. 10.1038/ajg.2017.154. [DOI] [PubMed] [Google Scholar]
- 110.Koletzko S, Jones NL, Goodman KJ, Gold B, Rowland M, Cadranel S, Chong S, Colletti RB, Casswall T, Elitsur Y, Guarner J, Kalach N, Madrazo A, Megraud F, Oderda G, H. pylori Working Groups of ESPGHAN and NASPGHAN. 2011. Evidence-based guidelines from ESPGHAN and NASPGHAN for Helicobacter pylori infection in children. J Pediatr Gastroenterol Nutr 53:230–243. 10.1097/MPG.0b013e3182227e90. [DOI] [PubMed] [Google Scholar]
- 111.Malfertheiner P. 2015. Diagnostic methods for H. pylori infection: choices, opportunities and pitfalls. United European Gastroenterol J 3:429–431. 10.1177/2050640615600968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Graham DY, Opekun AR, Hammoud F, Yamaoka Y, Reddy R, Osato MS, El-Zimaity HM. 2003. Studies regarding the mechanism of false negative urea breath tests with proton pump inhibitors. Am J Gastroenterol 98:1005–1009. 10.1111/j.1572-0241.2003.07426.x. [DOI] [PubMed] [Google Scholar]
- 113.McColl KE. 2010. Helicobacter pylori infection. N Engl J Med 362:1597–1604. 10.1056/NEJMcp1001110. [DOI] [PubMed] [Google Scholar]
- 114.Ontsira Ngoyi EN, Atipo Ibara BI, Moyen R, Ahoui Apendi PC, Ibara JR, Obengui O, Ossibi Ibara RB, Nguimbi E, Niama RF, Ouamba JM, Yala F, Abena AA, Vadivelu J, Goh KL, Menard A, Benejat L, Sifre E, Lehours P, Megraud F. 2015. Molecular detection of Helicobacter pylori and its antimicrobial resistance in Brazzaville, Congo. Helicobacter 20:316–320. 10.1111/hel.12204. [DOI] [PubMed] [Google Scholar]
- 115.Capurso G, Carnuccio A, Lahner E, Panzuto F, Baccini F, Delle Fave G, Annibale B. 2006. Corpus-predominant gastritis as a risk factor for false-negative 13C-urea breath test results. Aliment Pharmacol Ther 24:1453–1460. 10.1111/j.1365-2036.2006.03143.x. [DOI] [PubMed] [Google Scholar]
- 116.Garza-González E, Perez-Perez GI, Maldonado-Garza HJ, Bosques-Padilla FJ. 2014. A review of Helicobacter pylori diagnosis, treatment, and methods to detect eradication. World J Gastroenterol 20:1438–1449. 10.3748/wjg.v20.i6.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Osaki T, Mabe K, Hanawa T, Kamiya S. 2008. Urease-positive bacteria in the stomach induce a false-positive reaction in a urea breath test for diagnosis of Helicobacter pylori infection. J Med Microbiol 57:814–819. 10.1099/jmm.0.47768-0. [DOI] [PubMed] [Google Scholar]
- 118.Chey WD, Chathadi KV, Montague J, Ahmed F, Murthy U. 2001. Intragastric acidification reduces the occurrence of false-negative urea breath test results in patients taking a proton pump inhibitor. Am J Gastroenterol 96:1028–1032. 10.1111/j.1572-0241.2001.03687.x. [DOI] [PubMed] [Google Scholar]
- 119.Agha A, Opekun AR, Abudayyeh S, Graham DY. 2005. Effect of different organic acids (citric, malic and ascorbic) on intragastric urease activity. Aliment Pharmacol Ther 21:1145–1148. 10.1111/j.1365-2036.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- 120.Tepeš B, Malfertheiner P, Labenz J, Aygen S. 2017. Modified Helicobacter test using a new test meal and a 13C-urea breath test in Helicobacter pylori positive and negative dyspepsia patients on proton pump inhibitors. World J Gastroenterol 23:5954–5961. 10.3748/wjg.v23.i32.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gal E, Abuksis G, Fraser G, Koren R, Shmueli C, Yahav Y, Niv Y. 2003. 13C-urea breath test to validate eradication of Helicobacter pylori in an Israeli population. Isr Med Assoc J 5:98–100. [PubMed] [Google Scholar]
- 122.Logan RPH. 2018. Breath test to detect Helicobacter pylori. In Goodwin CS, Worsley BW (ed), Helicobacter pylori biology and clinical practice. CRC Press, Boca Raton, FL. [Google Scholar]
- 123.de Carvalho Costa Cardinali L, Rocha GA, Rocha AM, de Moura SB, de Figueiredo Soares T, Esteves AM, Nogueira AM, Cabral MM, de Carvalho AS, Bitencourt P, Ferreira A, Queiroz DM. 2003. Evaluation of [13C]urea breath test and Helicobacter pylori stool antigen test for diagnosis of H. pylori infection in children from a developing country. J Clin Microbiol 41:3334–3335. 10.1128/JCM.41.7.3334-3335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kato S, Ozawa K, Okuda M, Fujisawa T, Kagimoto S, Konno M, Maisawa S, Iinuma K. 2003. Accuracy of the stool antigen test for the diagnosis of childhood Helicobacter pylori infection: a multicenter Japanese study. Am J Gastroenterol 98:296–300. 10.1111/j.1572-0241.2003.07263.x. [DOI] [PubMed] [Google Scholar]
- 125.Oderda G, Rapa A, Ronchi B, Lerro P, Pastore M, Staiano A, de'Angelis GL, Strisciuglio P. 2000. Detection of Helicobacter pylori in stool specimens by non-invasive antigen enzyme immunoassay in children: multicentre Italian study. BMJ 320:347–348. 10.1136/bmj.320.7231.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang HR, Seo JK. 2008. Helicobacter pylori stool antigen (HpSA) tests in children before and after eradication therapy: comparison of rapid immunochromatographic assay and HpSA ELISA. Dig Dis Sci 53:2053–2058. 10.1007/s10620-007-0131-8. [DOI] [PubMed] [Google Scholar]
- 127.Sato M, Shimoyama T, Takahashi R, Kajiyama H, Sano Y, Sakaedani N, Kato A, Hirata H, Fukuda Y. 2012. Characterization and usefulness of stool antigen tests using a monoclonal antibody to Helicobacter pylori catalase. J Gastroenterol Hepatol 27(Suppl 3):23–28. 10.1111/j.1440-1746.2012.07066.x. [DOI] [PubMed] [Google Scholar]
- 128.Moubri M, Burucoa C, Kalach N, Larras RR, Nouar N, Mouffok F, Arrada Z. 2019. Performances of the IDEIA HpStAR stool antigen test in detection of Helicobacter pylori infection before and after eradication treatment in Algerian children. J Trop Pediatr 65:210–216. 10.1093/tropej/fmy035. [DOI] [PubMed] [Google Scholar]
- 129.Chen MJ, Fang YJ, Wu MS, Chen CC, Chen YN, Yu CC, Kuo CC, Chiu MC, Hu WH, Tsai MH, Hsieh CL, Chen HH, Bair MJ, Liou JM, Taiwan Gastrointestinal Disease and Helicobacter Consortium. 2020. Application of Helicobacter pylori stool antigen test to survey the updated prevalence of Helicobacter pylori infection in Taiwan. J Gastroenterol Hepatol 35:233–240. 10.1111/jgh.14828. [DOI] [PubMed] [Google Scholar]
- 130.Halland M, Haque R, Langhorst J, Boone JH, Petri WA. 2021. Clinical performance of the H. PYLORI QUIK CHEK and H. PYLORI CHEK assays, novel stool antigen tests for diagnosis of Helicobacter pylori. Eur J Clin Microbiol Infect Dis 40:1023–1028. 10.1007/s10096-020-04137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kouitcheu Mabeku LB, Bello Epesse M, Fotsing S, Kamgang R, Tchidjo M. 2021. Stool antigen testing, a reliable noninvasive method of assessment of Helicobacter pylori infection among patients with gastro-duodenal disorders in Cameroon. Dig Dis Sci 66:511–520. 10.1007/s10620-020-06219-0. [DOI] [PubMed] [Google Scholar]
- 132.Lario S, Ramírez-Lázaro MJ, Montserrat A, Quílez ME, Junquera F, Martínez-Bauer E, Sanfeliu I, Brullet E, Campo R, Segura F, Calvet X. 2016. Diagnostic accuracy of three monoclonal stool tests in a large series of untreated Helicobacter pylori infected patients. Clin Biochem 49:682–687. 10.1016/j.clinbiochem.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 133.Mori N, Nishiura Y, Suga D, Moritani I, Yamanaka Y, Ooya Y, Inoue H, Takase K, Hioki M, Shiraki K. 2018. Second-line triple therapy in failures with vonoprazan-based triple therapy for eradication of Helicobacter pylori. Biomed Rep 9:169–174. 10.3892/br.2018.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Korkmaz H, Findik D, Ugurluoglu C, Terzi Y. 2015. Reliability of stool antigen tests: investigation of the diagnostic value of a new immunochromatographic Helicobacter pylori approach in dyspeptic patients. Asian Pac J Cancer Prev 16:657–660. 10.7314/apjcp.2015.16.2.657. [DOI] [PubMed] [Google Scholar]
- 135.Han Y, Dai W, Meng F, Gan X, Liu M, Deng X, Li Y, Wang G. 2020. Diagnosis of Helicobacter pylori infection in the elderly using an immunochromatographic assay-based stool antigen test. Microbiologyopen 9:e1102. 10.1002/mbo3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kakiuchi T, Okuda M, Hashiguchi K, Imamura I, Nakayama A, Matsuo M. 2019. Evaluation of a novel stool antigen rapid test kit for detection of Helicobacter pylori infection. J Clin Microbiol 57:e01825-18. 10.1128/JCM.01825-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.DiaSorin. LIAISON Meridian H. pylori SA (REF: 318200). https://www.meridianbioscience.com/uploads/318200_pi.pdf.
- 138.DiaSorin. H. pylori DLH. LIAISON H. pylori SA. https://www.diasorin.com/sites/default/files/allegati_prodotti/ese_flyer_helicobacter_pilori_inf_dis_low.pdf. Accessed 28 July 2015.
- 139.Opekun AR, Zierold C, Rode A, Blocki FA, Fiorini G, Saracino IM, Vaira D, Sutton FM. 2020. Clinical performance of the automated LIAISON Meridian H. pylori SA stool antigen test. Biomed Res Int 2020:7189519. 10.1155/2020/7189519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramírez-Lázaro MJ, Lite J, Lario S, Pérez-Jové P, Montserrat A, Quílez ME, Martínez-Bauer E, Calvet X. 2016. Good diagnostic accuracy of a chemiluminescent immunoassay in stool samples for diagnosis of Helicobacter pylori infection in patients with dyspepsia. J Invest Med 64:388–391. 10.1136/jim-2015-000004. [DOI] [PubMed] [Google Scholar]
- 141.Manes G, Balzano A, Iaquinto G, Ricci C, Piccirillo MM, Giardullo N, Todisco A, Lioniello M, Vaira D. 2001. Accuracy of the stool antigen test in the diagnosis of Helicobacter pylori infection before treatment and in patients on omeprazole therapy. Aliment Pharmacol Ther 15:73–79. 10.1046/j.1365-2036.2001.00907.x. [DOI] [PubMed] [Google Scholar]
- 142.Czinn SJ. 2005. Helicobacter pylori infection: detection, investigation, and management. J Pediatr 146(3 Suppl):S21–S26. 10.1016/j.jpeds.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 143.Thaker Y, Moon A, Afzali A. 2016. Helicobacter pylori: a review of epidemiology, treatment, and management. J Clin Gastroenterol Treat 2:019. 10.23937/2469-584X/1510019. [DOI] [Google Scholar]
- 144.Vaira D, Holton J, Menegatti M, Ricci C, Landi F, Ali' A, Gatta L, Acciardi C, Farinelli S, Crosatti M, Berardi S, Miglioli M. 1999. New immunological assays for the diagnosis of Helicobacter pylori infection. Gut 45(Suppl 1):I23–127. 10.1136/gut.45.2008.i23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kawai S, Arai K, Lin Y, Nishiyama T, Sasakabe T, Wang C, Miwa H, Kikuchi S. 2019. Comparison of the detection of Helicobacter pylori infection by commercially available serological testing kits and the 13C-urea breath test. J Infect Chemother 25:769–773. 10.1016/j.jiac.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 146.Formichella L, Romberg L, Meyer H, Bolz C, Vieth M, Geppert M, Göttner G, Nölting C, Schepp W, Schneider A, Ulm K, Wolf P, Holster IL, Kuipers EJ, Birkner B, Soutschek E, Gerhard M. 2017. Validation of a novel immunoline assay for patient stratification according to virulence of the infecting Helicobacter pylori strain and eradication status. J Immunol Res 2017:8394593. 10.1155/2017/8394593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Day AS, Sherman PM. 2002. Accuracy of office-based immunoassays for the diagnosis of Helicobacter pylori infection in children. Helicobacter 7:205–209. 10.1046/j.1523-5378.2002.00082.x. [DOI] [PubMed] [Google Scholar]
- 148.Guarner J, Kalach N, Elitsur Y, Koletzko S. 2010. Helicobacter pylori diagnostic tests in children: review of the literature from 1999 to 2009. Eur J Pediatr 169:15–25. 10.1007/s00431-009-1033-x. [DOI] [PubMed] [Google Scholar]
- 149.Burucoa C, Delchier JC, Courillon-Mallet A, de Korwin JD, Mégraud F, Zerbib F, Raymond J, Fauchère JL. 2013. Comparative evaluation of 29 commercial Helicobacter pylori serological kits. Helicobacter 18:169–179. 10.1111/hel.12030. [DOI] [PubMed] [Google Scholar]
- 150.McNicholl AG, Forné M, Barrio J, De la Coba C, González B, Rivera R, Esteve M, Fernandez-Bañares F, Madrigal B, Gras-Miralles B, Perez-Aisa A, Viver-Pi-Sunyer JM, Bory F, Rosinach M, Loras C, Esteban C, Santolaria S, Gomollon F, Valle J, Gisbert JP, Helicobacter pylori Study Group of Asociación Española de Gastroenterología (AEG). 2014. Accuracy of GastroPanel for the diagnosis of atrophic gastritis. Eur J Gastroenterol Hepatol 26:941–948. 10.1097/MEG.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Redeen S, Petersson F, Tornkrantz E, Levander H, Mardh E, Borch K. 2011. Reliability of diagnostic tests for Helicobacter pylori infection. Gastroenterol Res Pract 2011:940650. 10.1155/2011/940650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Batts KP, Ketover S, Kakar S, Krasinskas AM, Mitchell KA, Wilcox R, Westerhoff M, Rank J, Gibson J, Mattia AR, Cummings OW, Davison JM, Naini BV, Dry SM, Yantiss RK, Rodger C Haggitt Gastrointestinal Pathology Society. 2013. Appropriate use of special stains for identifying Helicobacter pylori: recommendations from the Rodger C. Haggitt Gastrointestinal Pathology Society. Am J Surg Pathol 37:e12–e22. 10.1097/PAS.0000000000000097. [DOI] [PubMed] [Google Scholar]
- 153.She RC, Wilson AR, Litwin CM. 2009. Evaluation of Helicobacter pylori immunoglobulin G (IgG), IgA, and IgM serologic testing compared to stool antigen testing. Clin Vaccine Immunol 16:1253–1255. 10.1128/CVI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Makristathis A, Hirschl AM, Mégraud F, Bessède E. 2019. Review: Diagnosis of Helicobacter pylori infection. Helicobacter 24(Suppl 1):e12641. 10.1111/hel.12641. [DOI] [PubMed] [Google Scholar]
- 155.Biranjia-Hurdoyal SD, Seetulsingh-Goorah SP. 2016. Performances of four Helicobacter pylori serological detection kits using stool antigen test as gold standard. PLoS One 11:e0163834. 10.1371/journal.pone.0163834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hoang TT, Wheeldon TU, Bengtsson C, Phung DC, Sörberg M, Granström M. 2004. Enzyme-linked immunosorbent assay for Helicobacter pylori needs adjustment for the population investigated. J Clin Microbiol 42:627–630. 10.1128/JCM.42.2.627-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ueda J, Okuda M, Nishiyama T, Lin Y, Fukuda Y, Kikuchi S. 2014. Diagnostic accuracy of the E-plate serum antibody test kit in detecting Helicobacter pylori infection among Japanese children. J Epidemiol 24:47–51. 10.2188/jea.je20130078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.GastroPanel ELISA. Test panel for stomach health check. https://www.biohithealthcare.com/en/products/diagnostic-tests/gastropanel/.
- 159.Paloheimo L, Tiusanen T, Suovaniemi O, Syrjänen K. 2021. Serological biomarker test (GastroPanel) in the diagnosis of functional gastric disorders, Helicobacter pylori and atrophic gastritis in patients examined for dyspeptic symptoms. Anticancer Res 41:811–819. 10.21873/anticanres.14833. [DOI] [PubMed] [Google Scholar]
- 160.Syrjänen K. 2016. A panel of serum biomarkers (GastroPanel) in non-invasive diagnosis of atrophic gastritis. Systematic review and meta-analysis. Anticancer Res 36:5133–5144. 10.21873/anticanres.11083. [DOI] [PubMed] [Google Scholar]
- 161.Zagari RM, Rabitti S, Greenwood DC, Eusebi LH, Vestito A, Bazzoli F. 2017. Systematic review with meta-analysis: diagnostic performance of the combination of pepsinogen, gastrin-17 and anti-Helicobacter pylori antibodies serum assays for the diagnosis of atrophic gastritis. Aliment Pharmacol Ther 46:657–611. 10.1111/apt.14248. [DOI] [PubMed] [Google Scholar]
- 162.Syrjänen KJ, Sipponen P, Härkönen M, Peetsalu A, Korpela S. 2015. Accuracy of GastroPanel testing in detection of atrophic gastritis. Eur J Gastroenterol Hepatol 27:102–104. 10.1097/MEG.0000000000000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Telaranta-Keerie A, Kara R, Paloheimo L, Härkönen M, Sipponen P. 2010. Prevalence of undiagnosed advanced atrophic corpus gastritis in Finland: an observational study among 4,256 volunteers without specific complaints. Scand J Gastroenterol 45:1036–1041. 10.3109/00365521.2010.487918. [DOI] [PubMed] [Google Scholar]
- 164.Koivurova O-P, Ukkola O, Koivikko M, Ebeling T, Yliaska I, Koskela R, Blomster T, Ala-Rämi A, Kettunen O, Karttunen TJ, Mäkinen M, Ronkainen J, Syrjänen K. 2020. Screening of the patients with autoimmune thyroid disease (AITD) and type 1 diabetes mellitus (DM1) for atrophic gastritis (AG) by serological biomarker testing (GastroPanel). EC Gastroenterol Digest Syst 7:181–195. https://www.ecronicon.com/ecgds/ECGDS-07-00621.php. [Google Scholar]
- 165.Mäki M, Söderström D, Paloheimo L, Hendolin P, Suovaniemi O, Syrjänen K. 2020. Helicobacter pylori (Hp) IgG ELISA of the new-generation GastroPanel is highly accurate in diagnosis of Hp infection in gastroscopy referral patients. Anticancer Res 40:6387–6398. 10.21873/anticanres.14660. [DOI] [PubMed] [Google Scholar]
- 166.Dixon MF, Genta RM, Yardley JH and Correa P. 1996. Classification and grading of gastritis. the updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston, 1994. Am J Surg Pathol 20:1161–1181. 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 167.Shimoyama T, Sawada Y, Sawada N, Chinda D, Fukuda S. 2017. Accuracy of a stick-type kit and enzyme-linked immunosorbent assay in detecting Helicobacter pylori antibodies in urine of people living in the Japan Sea region of northern Japan. Jpn J Infect Dis 70:207–209. 10.7883/yoken.JJID.2015.642. [DOI] [PubMed] [Google Scholar]
- 168.Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin Microbiol Rev 10:720–741. 10.1128/CMR.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Basset C, Holton J, Ricci C, Gatta L, Tampieri A, Perna F, Miglioli M, Vaira D. 2003. Diagnosis and treatment of Helicobacter: a 2002 updated review. Aliment Pharmacol Ther 17:89–97. 10.1046/j.1365-2036.17.s2.6.x. [DOI] [PubMed] [Google Scholar]
- 170.Graham DY, Reddy S. 2001. Rapid detection of anti-Helicobacter pylori IgG in urine using immunochromatography. Aliment Pharmacol Ther 15:699–702. 10.1046/j.1365-2036.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- 171.Aumpan N, Vilaichone RK, Chotivitayatarakorn P, Pornthisarn B, Cholprasertsuk S, Bhanthumkomol P, Kanokwanvimol A, Siramolpiwat S, Mahachai V. 2019. High efficacy of rapid urine test for diagnosis of Helicobacter pylori infection in Thai people. Asian Pac J Cancer Prev 20:1525–1529. 10.31557/APJCP.2019.20.5.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Syam AF, Miftahussurur M, Uwan WB, Simanjuntak D, Uchida T, Yamaoka Y. 2015. Validation of urine test for detection of Helicobacter pylori infection in Indonesian population. Biomed Res Int 2015:152823. 10.1155/2015/152823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Quach DT, Hiyama T, Shimamoto F, Le QD, Ho LX, Vu NH, Yoshihara M, Uemura N. 2014. Value of a new stick-type rapid urine test for the diagnosis of Helicobacter pylori infection in the Vietnamese population. World J Gastroenterol 20:5087–5091. 10.3748/wjg.v20.i17.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Enko D, Halwachs-Baumann G, Stolba R, Rössler O, Kriegshäuser G. 2016. Performance evaluation of a rapid whole-blood immunoassay for the detection of IgG antibodies against Helicobacter pylori in daily clinical practice. Ann Clin Microbiol Antimicrob 15:47. 10.1186/s12941-016-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.MP Biomedicals. Assure H. pylori rapid test (CE). https://www.mpbio.com/jp/0743490020-assure-hp-rt-20t.
- 176.Hung CT, Leung WK, Chan FK, Sung JJ. 2002. Comparison of two new rapid serology tests for diagnosis of Helicobacter pylori infection in Chinese patients. Dig Liver Dis 34:111–115. 10.1016/s1590-8658(02)80239-0. [DOI] [PubMed] [Google Scholar]
- 177.Wang XY, Yang Y, Shi RH, Ho B, Wang HD, Zhang GX. 2008. An evaluation of a serologic test with a current infection marker of Helicobacter pylori before and after eradication therapy in Chinese. Helicobacter 13:49–55. 10.1111/j.1523-5378.2008.00578.x. [DOI] [PubMed] [Google Scholar]
- 178.Tsutsumi K, Kusano C, Suzuki S, Gotoda T, Murakami K. 2018. Diagnostic accuracy of aatex agglutination turbidimetric immunoassay in screening adolescents for Helicobacter pylori infection in Japan. Digestion 98:75–80. 10.1159/000487184. [DOI] [PubMed] [Google Scholar]
- 179.Midolo PD, Lambert JR, Russell EG, Lin SK. 1995. A practical single sample dry latex agglutination test for Helicobacter pylori antibody detection. J Clin Pathol 48:969–971. 10.1136/jcp.48.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lozniewski A, De Korwin JD, Conroy MC, Plenat F, Weber M. 1996. Evaluation of pyloriset dry, a new rapid agglutination test for Helicobacter pylori antibody detection. J Clin Microbiol 34:1773–1775. 10.1128/JCM.34.7.1773-1775.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen L-W, Chien C-Y, Yang K-J, Kuo S-F, Chen C-H, Chien R-N. 2015. Helicobacter pylori infection increases insulin resistance and metabolic syndrome in residents younger than 50 years old: a community-based study. PLoS One 10:e0128671. 10.1371/journal.pone.0128671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Westblom TU, Madan E, Gudipati S, Midkiff BR, Czinn SJ. 1992. Diagnosis of Helicobacter pylori infection in adult and pediatric patients by using Pyloriset, a rapid latex agglutination test. J Clin Microbiol 30:96–98. 10.1128/jcm.30.1.96-98.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ohara N. 2017. Is it possible to treat high negative titers in the new latex turbidimetric immunoassay as in ELISA? J Gastrointest Cancer Screen 55:1045–1052. [Google Scholar]
- 184.Amini Najafabadi H, Paknejad M, Farshad S, Mohammadian TS, Ebrahimi SS, Amini Najafabadi A. 2012. Immunodot blot assay to detect Helicobacter pylori using monoclonal antibodies against the 26 kDa protein. Hybridoma (Larchmt) 31:403–410. 10.1089/hyb.2012.0066. [DOI] [PubMed] [Google Scholar]
- 185.Gatta L, Ricci C, Tampieri A, Vaira D. 2003. Non-invasive techniques for the diagnosis of Helicobacter pylori infection. Clin Microbiol Infect 9:489–496. 10.1046/j.1469-0691.2003.00707.x. [DOI] [PubMed] [Google Scholar]
- 186.Kabir S. 2003. Clinic-based testing for Helicobacter pylori infection by enzyme immunoassay of faeces, urine and saliva. Aliment Pharmacol Ther 17:1345–1354. 10.1046/j.1365-2036.2003.01577.x. [DOI] [PubMed] [Google Scholar]
- 187.Leung WK, Chow TP, Ng EK, Chan FK, Chung SC, Sung JJ. 2001. Validation of a new immunoblot assay for the diagnosis of Helicobacter pylori in the Asian population. Aliment Pharmacol Ther 15:423–428. 10.1046/j.1365-2036.2001.00899.x. [DOI] [PubMed] [Google Scholar]
- 188.Yamaoka Y, Kodama T, Graham DY, Kashima K. 1998. Search for putative virulence factors of Helicobacter pylori: the low-molecular-weight (33–35 K) antigen. Dig Dis Sci 43:1482–1487. 10.1023/A:1018850412148. [DOI] [PubMed] [Google Scholar]
- 189.Formichella L, Romberg L, Bolz C, Vieth M, Geppert M, Göttner G, Nölting C, Walter D, Schepp W, Schneider A, Ulm K, Wolf P, Busch DH, Soutschek E, Gerhard M. 2013. A novel line immunoassay based on recombinant virulence factors enables highly specific and sensitive serologic diagnosis of Helicobacter pylori infection. Clin Vaccine Immunol 20:1703–1710. 10.1128/CVI.00433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Michel A, Waterboer T, Kist M, Pawlita M. 2009. Helicobacter pylori multiplex serology. Helicobacter 14:525–535. 10.1111/j.1523-5378.2009.00723.x. [DOI] [PubMed] [Google Scholar]
- 191.Shafaie E, Saberi S, Esmaeili M, Karimi Z, Najafi S, Tashakoripoor M, Abdirad A, Hosseini ME, Mohagheghi MA, Khalaj V, Mohammadi M. 2018. Multiplex serology of Helicobacter pylori antigens in detection of current infection and atrophic gastritis—a simple and cost-efficient method. Microb Pathog 119:137–144. 10.1016/j.micpath.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 192.Khalifeh Gholi M, Kalali B, Formichella L, Göttner G, Shamsipour F, Zarnani AH, Hosseini M, Busch DH, Shirazi MH, Gerhard M. 2013. Helicobacter pylori FliD protein is a highly sensitive and specific marker for serologic diagnosis of H. pylori infection. Int J Med Microbiol 303:618–623. 10.1016/j.ijmm.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 193.Mikrogen Diagnostik. RecomLine H. pylri IgG. https://www.mikrogen.de/english/products/product-overview/testsystem/helicobacter-igg-20.html.
- 194.Pan KF, Formichella L, Zhang L, Zhang Y, Ma JL, Li ZX, Liu C, Wang YM, Goettner G, Ulm K, Classen M, You WC, Gerhard M. 2014. Helicobacter pylori antibody responses and evolution of precancerous gastric lesions in a Chinese population. Int J Cancer 134:2118–2125. 10.1002/ijc.28560. [DOI] [PubMed] [Google Scholar]
- 195.Liu C, Wang YM, Li ZX, Zhang L, Ma JL, Zhou T, You WC, Pan KF. 2013. Serological assessment of Helicobacter pylori-specific antibodies and their association with gastric lesions in a high-risk population. Zhonghua Zhong Liu Za Zhi 35:547–551. (In Chinese.) [PubMed] [Google Scholar]
- 196.Graham DY, Miftahussurur M. 2018. Helicobacter pylori urease for diagnosis of Helicobacter pylori infection: a mini review. J Adv Res 13:51–57. 10.1016/j.jare.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ferguson DA, Li C, Patel NR, Mayberry WR, Chi DS, Thomas E. 1993. Isolation of Helicobacter pylori from saliva. J Clin Microbiol 31:2802–2804. 10.1128/jcm.31.10.2802-2804.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dore MP, Pes GM. 2021. What is new in Helicobacter pylori diagnosis. An overview. J Clin Med 10:2091. 10.3390/jcm10102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mégraud F, Bessède E, Lehours P. 2014. Helicobacter pylori: a worldwide perspective, p 234–258. In Miklós BG (ed), Current methods used for the diagnosis of Helicobacter pylori infection. Bentham Science Publisher, Sharjah, United Arab Emirates. [Google Scholar]
- 200.Uotani T, Graham DY. 2015. Diagnosis of Helicobacter pylori using the rapid urease test. Ann Transl Med 3:9. 10.3978/j.issn.2305-5839.2014.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lee TH, Lin CC, Chung CS, Lin CK, Liang CC, Tsai KC. 2015. Increasing biopsy number and sampling from gastric body improve the sensitivity of rapid urease test in patients with peptic ulcer bleeding. Dig Dis Sci 60:454–457. 10.1007/s10620-014-3351-8. [DOI] [PubMed] [Google Scholar]
- 202.Moon SW, Kim TH, Kim HS, Ju JH, Ahn YJ, Jang HJ, Shim SG, Kim HJ, Jung WT, Lee OJ. 2012. United rapid urease test is superior than separate test in detecting Helicobacter pylori at the gastric antrum and body specimens. Clin Endosc 45:392–396. 10.5946/ce.2012.45.4.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Serim Research. PyloriTek for H. pylori. https://www.serim.com/h-pylori.
- 204.Njoroge S, Mwangi C, Nyerere K, Revathi G, Devani S, Rajula A, Kamenwa R, Onkoba N, Odhiambo F. 2019. Comparing Diagnostic Performance of Pronto Dry Rapid Urease and Culture to Histopathology among Endoscopy Patients at the Aga Khan University Hospital, Nairobi-Kenya. JAMMR 29:1–7. 10.9734/jammr/2019/v29i1230147. [DOI] [Google Scholar]
- 205.Northern Devon Healthcare NHS Trust. CLO (campylobacter-like organism) test. https://www.northdevonhealth.nhs.uk/wp-content/uploads/2012/04/Biohit-CLO-Test.pdf.
- 206.GI Supply. HpFast and HpOne H. pylori Tests. https://www.instrumedsurgical.com/pdf/H.Pylori-Sell-Sheet.01.pdf.
- 207.Cambridge Life Sciences, Ltd. Endosc-Hp test: Helicobacter pylori urease test. https://www.clsdiagnostics.com/files/endosc-hp-test-data-sheet-v02.pdf.
- 208.BIOHIT Health Care. Helicobacter pylori UFT300 Quick Test. https://www.biohithealthcare.com/en/products/diagnostic-tests/helicobacter-pylori-uft300-quick-test/.
- 209.Vaira D, Vakil N, Gatta L, Ricci C, Perna F, Saracino I, Fiorini G, Holton J. 2010. Accuracy of a new ultrafast rapid urease test to diagnose Helicobacter pylori infection in 1000 consecutive dyspeptic patients. Aliment Pharmacol Ther 31:331–338. 10.1111/j.1365-2036.2009.04196.x. [DOI] [PubMed] [Google Scholar]
- 210.McNicholl AG, Ducons J, Barrio J, Bujanda L, Forné-Bardera M, Aparcero R, Ponce J, Rivera R, Dedeu-Cuso JM, Garcia-Iglesias P, Montoro M, Bejerano A, Ber-Nieto Y, Madrigal B, Zapata E, Loras-Alastruey C, Castro M, Nevarez A, Mendez I, Bory-Ros F, Miquel-Planas M, Vera I, Nyssen OP, Gisbert JP, Helicobacter pylori Study Group of the Asociación Española de Gastroenterología (AEG). 2017. Accuracy of the ultra-rapid urease test for diagnosis of Helicobacter pylori infection. Gastroenterol Hepatol 40:651–657. 10.1016/j.gastrohep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 211.Noh CK, Lee GH, Park JW, Roh J, Han JH, Lee E, Park B, Lim SG, Shin SJ, Cheong JY, Kim JH, Lee KM. 2020. Diagnostic accuracy of “sweeping” method compared to conventional sampling in rapid urease test for Helicobacter pylori detection in atrophic mucosa. Sci Rep 10:18483. 10.1038/s41598-020-75528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fox JG. 2002. The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50:273–283. 10.1136/gut.50.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Brandi G, Biavati B, Calabrese C, Granata M, Nannetti A, Mattarelli P, Di Febo G, Saccoccio G, Biasco G. 2006. Urease-positive bacteria other than Helicobacter pylori in human gastric juice and mucosa. Am J Gastroenterol 101:1756–1761. 10.1111/j.1572-0241.2006.00698.x. [DOI] [PubMed] [Google Scholar]
- 214.Debongnie JC, Pauwels S, Raat A, de Meeus Y, Haot J, Mainguet P. 1991. Quantification of Helicobacter pylori infection in gastritis and ulcer disease using a simple and rapid carbon-14-urea breath test. J Nucl Med 32:1192–1198. PMID: 2045933. [PubMed] [Google Scholar]
- 215.Siavoshi F, Saniee P, Khalili-Samani S, Hosseini F, Malakutikhah F, Mamivand M, Shahreza S, Sharifi AH. 2015. Evaluation of methods for H. pylori detection in PPI consumption using culture, rapid urease test and smear examination. Ann Transl Med 3:11. 10.3978/j.issn.2305-5839.2014.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schilling D, Demel A, Adamek HE, Nüsse T, Weidmann E, Riemann JF. 2003. A negative rapid urease test is unreliable for exclusion of Helicobacter pylori infection during acute phase of ulcer bleeding. A prospective case control study. Dig Liver Dis 35:217–221. 10.1016/S1590-8658(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 217.Lee JM, Breslin NP, Fallon C, O'Morain CA. 2000. Rapid urease tests lack sensitivity in Helicobacter pylori diagnosis when peptic ulcer disease presents with bleeding. Am J Gastroenterol 95:1166–1670. 10.1111/j.1572-0241.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- 218.Yoo JY, Kim N, Park YS, Hwang JH, Kim JW, Jeong SH, Lee HS, Choe C, Lee DH, Jung HC, Song IS. 2007. Detection rate of Helicobacter pylori against a background of atrophic gastritis and/or intestinal metaplasia. J Clin Gastroenterol 41:751–755. 10.1097/MCG.0b013e31802c347d. [DOI] [PubMed] [Google Scholar]
- 219.Lan HC, Chen TS, Li AF, Chang FY, Lin HC. 2012. Additional corpus biopsy enhances the detection of Helicobacter pylori infection in a background of gastritis with atrophy. BMC Gastroenterol 12:182. 10.1186/1471-230X-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Weston AP, Campbell DR, Hassanein RS, Cherian R, Dixon A, McGregor DH. 1997. Prospective, multivariate evaluation of CLO test performance. Am J Gastroenterol 92:1310–1315. [PubMed] [Google Scholar]
- 221.Yu L, Luo L, Long X, Liang X, Ji Y, Chen Q, Song Y, Li X, Graham DY, Lu H. 2019. Susceptibility-guided therapy for Helicobacter pylori infection treatment failures. Therap Adv Gastroenterol 12:1756284819874922. 10.1177/1756284819874922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Holmes KP, Fang JC, Jackson BR. 2010. Cost-effectiveness of six strategies for Helicobacter pylori diagnosis and management in uninvestigated dyspepsia assuming a high resource intensity practice pattern. BMC Health Serv Res 10:344. 10.1186/1472-6963-10-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.McNulty CAM, Lehours P, Mégraud F. 2011. Diagnosis of Helicobacter pylori infection. Helicobacter 16(Suppl 1):10–18. 10.1111/j.1523-5378.2011.00875.x. [DOI] [PubMed] [Google Scholar]
- 224.Bonoso Criado R, Pérez Citores L, Pérez Millán AG, Montero Moretón Á, González de Castro E, Cabezudo Molleda L, García Castro MA, Moreira Da Silva BA, Maestro Antolín S, Santos Santamarta F, Barcenilla Laguna J. 2021. Prospective comparative study of the treatment of Helicobacter pylori with antibiotic susceptibility testing-guided triple therapy compared to quadruple therapy with bismuth-metronidazole-tetracycline subcitrate. Rev Esp Enferm Dig 113:597–601. 10.17235/reed.2020.7395/2020. [DOI] [PubMed] [Google Scholar]
- 225.Kang S, Kim Y, Ahn JY, Jung HY, Kim N, Na HK, Lee JH, Jung KW, Kim DH, Choi KD, Song HJ, Lee GH. 2021. Role of antimicrobial susceptibility testing before first-line treatment containing clarithromycin for Helicobacter pylori eradication in the clinical setting. Antibiotics (Basel) 10:214. 10.3390/antibiotics10020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tserentogtokh T, Gantuya B, Subsomwong P, Oyuntsetseg K, Bolor D, Erdene-Ochir Y, Azzaya D, Davaadorj D, Uchida T, Matsuhisa T, Yamaoka Y. 2019. Western-type Helicobacter pylori CagA are the most frequent type in Mongolian patients. Cancers (Basel) 11:725. 10.3390/cancers11050725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Perez-Perez GI. 2000. Accurate diagnosis of Helicobacter pylori. Culture, including transport. Gastroenterol Clin North Am 29:879–884. 10.1016/s0889-8553(05)70155-2. [DOI] [PubMed] [Google Scholar]
- 228.Lee JY, Kim N. 2015. Diagnosis of Helicobacter pylori by invasive test: histology. Ann Transl Med 3:10. 10.3978/j.issn.2305-5839.2014.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hirschl AM, Makristathis A. 2007. Methods to detect Helicobacter pylori: from culture to molecular biology. Helicobacter 12(Suppl 2):6–11. 10.1111/j.1523-5378.2007.00560.x. [DOI] [PubMed] [Google Scholar]
- 230.Ramis IB, de Moraes EP, Fernandes MS, Mendoza-Sassi R, Rodrigues O, Juliano CR, Scaini CJ, da Silva PE. 2012. Evaluation of diagnostic methods for the detection of Helicobacter pylori in gastric biopsy specimens of dyspeptic patients. Braz J Microbiol 43:903–908. 10.1590/S1517-83822012000300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gong YN, Li YM, Yang NM, Li HZ, Guo F, Lin L, Wang QY, Zhang JK, Ji ZZ, Mao JB, Mao JL, Shi ZC, Tang WH, Zhu XJ, Shao W, Zhang XF, Wang XH, Tong YF, Jiang MZ, Chen GL, Wang ZY, Tu HM, Jiang GF, Wu JS, Chen XP, Ding QL, Ouyang H, Jin FZ, Xu YL, Zhang JZ. 2015. Centralized isolation of Helicobacter pylori from multiple centers and transport condition influences. World J Gastroenterol 21:944–952. 10.3748/wjg.v21.i3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ndip RN, MacKay WG, Farthing MJ, Weaver LT. 2003. Culturing Helicobacter pylori from clinical specimens: review of microbiologic methods. J Pediatr Gastroenterol Nutr 36:616–622. 10.1097/00005176-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 233.Park SA, Ko A, Lee NG. 2011. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol 11:96. 10.1186/1471-2180-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Grove DI, McLeay RA, Byron KE, Koutsouridis G. 2001. Isolation of Helicobacter pylori after transport from a regional laboratory of gastric biopsy specimens in saline, Portagerm pylori or cultured on chocolate agar. Pathology 33:362–364. 10.1080/00313020126308. [DOI] [PubMed] [Google Scholar]
- 235.Cellini L, Di Campli E, Di Bartolomeo S, Bessa LJ, Baffoni M, Di Giulio M. 2014. New transport medium for cultural recovery of Helicobacter pylori. J Clin Microbiol 52:4325–4329. 10.1128/JCM.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kalali B, Formichella L, Gerhard M. 2015. Diagnosis of Helicobacter pylori: changes towards the future. Diseases 3:122–135. 10.3390/diseases3030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Peretz A, On A, Koifman A, Brodsky D, Isakovich N, Glyatman T, Paritsky M. 2015. An efficiency comparison between three invasive methods for the diagnosis of Helicobacter pylori infections: culture from stomach biopsy, rapid urease test (CUTest), and histologic examination of gastric biopsy. Ann Clin Lab Sci 45:148–151. [PubMed] [Google Scholar]
- 238.Mégraud F, Lehours P. 2007. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev 20:280–322. 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hutton ML, Kaparakis-Liaskos M, Ferrero RL. 2012. The use of AlbuMAX II as a blood or serum alternative for the culture of Helicobacter pylori. Helicobacter 17:68–76. 10.1111/j.1523-5378.2011.00914.x. [DOI] [PubMed] [Google Scholar]
- 240.Leszczyńska K, Namiot A, Namiot Z, Leszczyńska JK, Jakoniuk P, Chilewicz M, Namiot DB, Kemona A, Milewski R, Bucki R. 2010. Patient factors affecting culture of Helicobacter pylori isolated from gastric mucosal specimens. Adv Med Sci 55:161–166. 10.2478/v10039-010-0028-1. [DOI] [PubMed] [Google Scholar]
- 241.Thomas JE, Gibson GR, Darboe MK, Dale A, Weaver LT. 1992. Isolation of Helicobacter pylori from human faeces. Lancet 340:1194–1195. 10.1016/0140-6736(92)92894-l. [DOI] [PubMed] [Google Scholar]
- 242.Gong EJ, Ahn JY, Jung DK, Lee SM, Pih GY, Kim GH, Na HK, Lee JH, Jung H-Y, Kim JM. 2020. Isolation of Helicobacter pylori using leftover tissue in the rapid urease test kit. Helicobacter 25:e12733. 10.1111/hel.12733. [DOI] [PubMed] [Google Scholar]
- 243.Rautelin H, Seppälä K, Nuutinen H, Kärkkäinen P, Sipponen P, Kosunen TU. 1997. Culture of Helicobacter pylori from gastric biopsies transported in biopsy urease test tubes. Eur J Clin Microbiol Infect Dis 16:380–383. 10.1007/BF01726367. [DOI] [PubMed] [Google Scholar]
- 244.Windsor HM, Ho GY, Marshall BJ. 1999. Successful recovery of H. pylori from rapid urease tests (CLO tests). Am J Gastroenterol 94:3181–3183. 10.1111/j.1572-0241.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 245.Li Y, Rimbara E, Thirumurthi S, Trespalacios A, Reddy R, Sabounchi S, Attumi TA, Graham DY. 2012. Detection of clarithromycin resistance in Helicobacter pylori following noncryogenic storage of rapid urease tests for 30 days. J Dig Dis 13:54–59. 10.1111/j.1751-2980.2011.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Furuta T, Sagehashi Y, Shirai N, Sugimoto M, Nakamura A, Kodaira M, Kenmotsu K, Nagano M, Egashira T, Ueda K, Yoneyama M, Ohashi K, Ishizaki T, Hishida A. 2005. Influence of CYP2C19 polymorphism and Helicobacter pylori genotype determined from gastric tissue samples on response to triple therapy for H. pylori infection. Clin Gastroenterol Hepatol 3:564–573. 10.1016/S1542-3565(04)00779-7. [DOI] [PubMed] [Google Scholar]
- 247.Janulaityte-Günther D, Günther T, Pavilonis A, Kupčinskas L. 2003. What Bizzozero never could image—Helicobacter pylori today and tomorrow. Medicina (Kaunas) 39:542–549. [PubMed] [Google Scholar]
- 248.Ashton-Key M, Diss TC, Isaacson PG. 1996. Detection of Helicobacter pylori in gastric biopsy and resection specimens. J Clin Pathol 49:107–111. 10.1136/jcp.49.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hartman DJ, Owens SR. 2012. Are routine ancillary stains required to diagnose Helicobacter infection in gastric biopsy specimens? An institutional quality assurance review. Am J Clin Pathol 137:255–260. 10.1309/AJCPD8FFBJ5LSLTE. [DOI] [PubMed] [Google Scholar]
- 250.Kocsmár É, Szirtes I, Kramer Z, Szijártó A, Bene L, Buzás GM, Kenessey I, Bronsert P, Csanadi A, Lutz L, Werner M, Wellner UF, Kiss A, Schaff Z, Lotz G. 2017. Sensitivity of Helicobacter pylori detection by Giemsa staining is poor in comparison with immunohistochemistry and fluorescent in situ hybridization and strongly depends on inflammatory activity. Helicobacter 22:e12387. 10.1111/hel.12387. [DOI] [PubMed] [Google Scholar]
- 251.Fan CC, Chen CH, Chou C, Kao TY, Cheng AN, Lee AY, Kuo CL. 2020. A time-saving-modified Giemsa stain is a better diagnostic method of Helicobacter pylori infection compared with the rapid urease test. J Clin Lab Anal 34:e23110. 10.1002/jcla.23110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Rotimi O, Cairns A, Gray S, Moayyedi P, Dixon MF. 2000. Histological identification of Helicobacter pylori: comparison of staining methods. J Clin Pathol 53:756–759. 10.1136/jcp.53.10.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yantiss RK, Lamps LW. 2012. To stain or not to stain. That remains the question. Am J Clin Pathol 137:343–345. 10.1309/AJCPXC4K9IOVUEKT. [DOI] [PubMed] [Google Scholar]
- 254.Smith SB, Snow AN, Perry RL, Qasem SA. 2012. Helicobacter pylori: to stain or not to stain? Am J Clin Pathol 137:733–738. 10.1309/AJCP8DGTAVG7MBMT. [DOI] [PubMed] [Google Scholar]
- 255.Panarelli NC, Ross DS, Bernheim OE, Landzberg ZB, Schuetz AN, Jenkins SG, Landzberg BR, Jessurun J, Yantiss RK. 2015. Utility of ancillary stains for Helicobacter pylori in near-normal gastric biopsies. Hum Pathol 46:397–403. 10.1016/j.humpath.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 256.Yang YX, Brill J, Krishnan P, Leontiadis G, American Gastroenterological Association Clinical Practice Guidelines Committee. 2015. American Gastroenterological Association Institute guideline on the role of upper gastrointestinal biopsy to evaluate dyspepsia in the adult patient in the absence of visible mucosal lesions. Gastroenterology 149:1082–1087. 10.1053/j.gastro.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 257.Benoit A, Hoyeau N, Fléjou JF. 2018. Diagnosis of Helicobacter pylori infection on gastric biopsies: standard stain, special stain or immunohistochemistry? Ann Pathol 38:363–369. 10.1016/j.annpat.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 258.Sakonlaya D, Apisarnthanarak A, Yamada N, Tomtitchong P. 2014. Modified toluidine blue: an alternative stain for Helicobacter pylori detection in routine diagnostic use and post-eradication confirmation for gastric cancer prevention. Asian Pac J Cancer Prev 15:6983–6987. 10.7314/apjcp.2014.15.16.6983. [DOI] [PubMed] [Google Scholar]
- 259.Arachchi PS, Weerasekera MM, Seneviratne B, Weerasekera D, Fernando N, Gunasekara CP. 2018. Imprint cytology: a useful screening test for diagnosis of Helicobacter pylori in resource poor settings. BMC Res Notes 11:481. 10.1186/s13104-018-3592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rahbar M, Mardanpur K, Tavafzadeh R. 2012. Imprint cutology: a simple cost efectiveness analysis for diagnosing Helicobacter pylori, in west of Iran. Med J Islam Repub Iran 26:12–16. [PMC free article] [PubMed] [Google Scholar]
- 261.Kaur G, Madhavan M, Basri AH, Sain AH, Hussain MS, Yatiban MK, Naing NN. 2004. Rapid diagnosis of Helicobacter pylori infection in gastric imprint smears. Southeast Asian J Trop Med Public Health 35:676–680. [PubMed] [Google Scholar]
- 262.Jafari M, Khalilian A, Rostampour F. 2013. Comparison the sensitivity of stomach mucus touching cytology and urease rapid test in Helicobacter pylori diagnosis in endoscopies patients with gastritis or peptic ulcer. Jundishapur J Microbiol 6:e7446. 10.5812/jjm.7446. [DOI] [Google Scholar]
- 263.Misra SP, Dwivedi M, Misra V, Gupta SC. 1993. Imprint cytology—a cheap, rapid and effective method for diagnosing Helicobacter pylori. Postgrad Med J 69:291–295. 10.1136/pgmj.69.810.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Vijayanarasimha D, Mahadevappa A, Manjunath GV, Sunila R. 2014. Imprint cytology: a diagnostic aid in interpretation of upper gastrointestinal endoscopic biopsies. J Dig Endosc 5:144–148. [Google Scholar]
- 265.Cotruta B, Gheorghe C, Iacob R, Dumbrava M, Radu C, Bancila I, Becheanu G. 2017. The orientation of gastric biopsy samples improves the inter-observer agreement of the OLGA staging system. J Gastrointestin Liver Dis 26:351–356. 10.15403/jgld.2014.1121.264.olg. [DOI] [PubMed] [Google Scholar]
- 266.Clayton CL, Kleanthous H, Coates PJ, Morgan DD, Tabaqchali S. 1992. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol 30:192–200. 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lash JG, Genta RM. 2013. Adherence to the Sydney System guidelines increases the detection of Helicobacter gastritis and intestinal metaplasia in 400738 sets of gastric biopsies. Aliment Pharmacol Ther 38:424–431. 10.1111/apt.12383. [DOI] [PubMed] [Google Scholar]
- 268.Yang S, Rothman RE. 2004. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis 4:337–348. 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Duś I, Dobosz T, Manzin A, Loi G, Serra C, Radwan-Oczko M. 2013. Role of PCR in Helicobacter pylori diagnostics and research—new approaches for study of coccoid and spiral forms of the bacteria. Postepy Hig Med Dosw (Online) 67:261–268. 10.5604/17322693.1044005. [DOI] [PubMed] [Google Scholar]
- 270.Rimbara E, Sasatsu M, Graham DY. 2013. PCR detection of Helicobacter pylori in clinical samples. Methods Mol Biol 943:279–287. 10.1007/978-1-60327-353-4_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Gingras BA, Maggiore JA. 2020. Performance of a new molecular assay for the detection of gastrointestinal pathogens. Access Microbiol 2:acmi000160. 10.1099/acmi.0.000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Saez J, Belda S, Santibáñez M, Rodríguez JC, Sola-Vera J, Galiana A, Ruiz-García M, Brotons A, López-Girona E, Girona E, Sillero C, Royo G. 2012. Real-time PCR for diagnosing Helicobacter pylori infection in patients with upper gastrointestinal bleeding: comparison with other classical diagnostic methods. J Clin Microbiol 50:3233–3237. 10.1128/JCM.01205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lehours P, Mégraud F. 2011. Helicobacter pylori molecular diagnosis. Expert Rev Mol Diagn 11:351–355. 10.1586/erm.11.17. [DOI] [PubMed] [Google Scholar]
- 274.Lin HJ, Lo WC, Perng CL, Tseng GY, Li AF, Ou YH. 2005. Mucosal polymerase chain reaction for diagnosing Helicobacter pylori infection in patients with bleeding peptic ulcers. World J Gastroenterol 11:382–385. 10.3748/wjg.v11.i3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.He Q, Wang JP, Osato M, Lachman LB. 2002. Real-time quantitative PCR for detection of Helicobacter pylori. J Clin Microbiol 40:3720–3728. 10.1128/JCM.40.10.3720-3728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Weiss J, Tsang TK, Meng X, Zhang H, Kilner E, Wang E, Watkin W. 2008. Detection of Helicobacter pylori gastritis by PCR: correlation with inflammation scores and immunohistochemical and CLOtest findings. Am J Clin Pathol 129:89–96. 10.1309/APMPEP54G7PN958G. [DOI] [PubMed] [Google Scholar]
- 277.Chen T, Meng X, Zhang H, Tsang RW, Tsang TK. 2012. Comparing multiplex PCR and rapid urease test in the detection of H. pylori in patients on proton pump inhibitors. Gastroenterol Res Pract 2012:898276. 10.1155/2012/898276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA 103:732–737. 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Thorell K, Bengtsson-Palme J, Liu OH, Palacios Gonzales RV, Nookaew I, Rabeneck L, Paszat L, Graham DY, Nielsen J, Lundin SB, Sjöling Å. 2017. In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infect Immun 85:e00031-17. 10.1128/IAI.00031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kiss S, Zsikla V, Frank A, Willi N, Cathomas G. 2016. Helicobacter-negative gastritis: polymerase chain reaction for Helicobacter DNA is a valuable tool to elucidate the diagnosis. Aliment Pharmacol Ther 43:924–932. 10.1111/apt.13564. [DOI] [PubMed] [Google Scholar]
- 281.Calvet X, Lehours P, Lario S, Mégraud F. 2010. Diagnosis of Helicobacter pylori infection. Helicobacter 15(Suppl 1):7–13. 10.1111/j.1523-5378.2010.00784.x. [DOI] [PubMed] [Google Scholar]
- 282.Ricci C, Holton J, Vaira D. 2007. Diagnosis of Helicobacter pylori: invasive and non-invasive tests. Best Pract Res Clin Gastroenterol 21:299–313. 10.1016/j.bpg.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 283.Khadangi F, Yassi M, Kerachian MA. 2017. Diagnostic accuracy of PCR-based detection tests for Helicobacter pylori in stool samples. Helicobacter 22:e12444. 10.1111/hel.12444. [DOI] [PubMed] [Google Scholar]
- 284.Hsieh MS, Liu CJ, Hsu WH, Li CJ, Tsai PY, Hu HM, Shih HY, Lu CY, Yu FJ, Kuo FC, Wu DC, Kuo CH. 2019. Gastric juice-based PCR assay: an alternative testing method to aid in the management of previously treated Helicobacter pylori infection. Helicobacter 24:e12568. 10.1111/hel.12568. [DOI] [PubMed] [Google Scholar]
- 285.Skrebinska S, Mégraud F, Bessède E. 2018. Diagnosis of Helicobacter pylori infection. Helicobacter 23(Suppl 1):e12515. 10.1111/hel.12515. [DOI] [PubMed] [Google Scholar]
- 286.Sugimoto M, Wu JY, Abudayyeh S, Hoffman J, Brahem H, Al-Khatib K, Yamaoka Y, Graham DY. 2009. Unreliability of results of PCR detection of Helicobacter pylori in clinical or environmental samples. J Clin Microbiol 47:738–742. 10.1128/JCM.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Sekhar Goud EVS, Kannan R, Rao UK, Joshua E, Tavaraja R, Jain Y. 2019. Identification of Helicobacter pylori in saliva of patients with and without gastritis by polymerase chain reaction. J Pharm Bioallied Sci 11:S523–S529. 10.4103/jpbs.JPBS_260_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Martínez-Santos VI, Hernández Catalán M, Ojeda Salazar LO, Orozco Gómez OA, Lorenzo SI, Santos Gómez R, Romero-Castro NS, Reyes Ríos R, Martinez Carrillo DN, Fernández-Tilapa G. 2021. Helicobacter pylori prevalence in healthy Mexican children: comparison between two non-invasive methods. PeerJ 9:e11546. 10.7717/peerj.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Klein D. 2002. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med 8:257–260. 10.1016/s1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 290.Mikula M, Dzwonek A, Jagusztyn-Krynicka K, Ostrowski J. 2003. Quantitative detection for low levels of Helicobacter pylori infection in experimentally infected mice by real-time PCR. J Microbiol Methods 55:351–359. 10.1016/s0167-7012(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 291.Shukla SK, Prasad KN, Tripathi A, Ghoshal UC, Krishnani N, Nuzhat H. 2011. Quantitation of Helicobacter pylori ureC gene and its comparison with different diagnostic techniques and gastric histopathology. J Microbiol Methods 86:231–237. 10.1016/j.mimet.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 292.Valasek MA, Repa JJ. 2005. The power of real-time PCR. Adv Physiol Educ 29:151–159. 10.1152/advan.00019.2005. [DOI] [PubMed] [Google Scholar]
- 293.Kralik P, Ricchi M. 2017. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol 8:108. 10.3389/fmicb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. 2014. Diagnosis of Helicobacter pylori: what should be the gold standard? World J Gastroenterol 20:12847–12859. 10.3748/wjg.v20.i36.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Yu G, Fadrosh D, Goedert JJ, Ravel J, Goldstein AM. 2015. Nested PCR biases in interpreting microbial community structure in 16S rRNA gene sequence datasets. PLoS One 10:e0132253. 10.1371/journal.pone.0132253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Noguchi N, Rimbara E, Kato A, Tanaka A, Tokunaga K, Kawai T, Takahashi S, Sasatsu M. 2007. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J Med Microbiol 56:1174–1180. 10.1099/jmm.0.47302-0. [DOI] [PubMed] [Google Scholar]
- 297.Singh V, Mishra S, Rao GR, Jain AK, Dixit VK, Gulati AK, Mahajan D, McClelland M, Nath G. 2008. Evaluation of nested PCR in detection of Helicobacter pylori targeting a highly conserved gene: HSP60. Helicobacter 13:30–34. 10.1111/j.1523-5378.2008.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Silva DG, Tinoco EM, Rocha GA, Rocha AM, Guerra JB, Saraiva IE, Queiroz DM. 2010. Helicobacter pylori transiently in the mouth may participate in the transmission of infection. Mem Inst Oswaldo Cruz 105:657–660. 10.1590/s0074-02762010000500009. [DOI] [PubMed] [Google Scholar]
- 299.Šeligová B, Lukáč Ľ, Bábelová M, Vávrová S, Sulo P. 2020. Diagnostic reliability of nested PCR depends on the primer design and threshold abundance of Helicobacter pylori in biopsy, stool, and saliva samples. Helicobacter 25:e12680. 10.1111/hel.12680. [DOI] [PubMed] [Google Scholar]
- 300.Trung TT, Minh TA, Anh NT. 2019. Value of CIM, CLO test and multiplex PCR for the diagnosis of Helicobacter pylori infection status in patients with gastritis and gastric ulcer. Asian Pac J Cancer Prev 20:3497–3503. 10.31557/APJCP.2019.20.11.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Oktem-Okullu S, Tiftikci A, Saruc M, Cicek B, Vardareli E, Tozun N, Kocagoz T, Sezerman U, Yavuz AS, Sayi-Yazgan A. 2015. Multiplex-PCR-based screening and computational modeling of virulence factors and T-Cell mediated immunity in Helicobacter pylori infections for accurate clinical diagnosis. PLoS ONE 10(8):e0136212. 10.1371/journal.pone.0136212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Clayton CL, Pallen MJ, Kleanthous H, Wren BW, Tabaqchali S. 1990. Nucleotide sequence of two genes from Helicobacter pylori encoding for urease subunits. Nucleic Acids Res 18:362. 10.1093/nar/18.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Goodwin CS, Armstrong A, Chilvers T, Peters M, Collins MD, Sly L, McConnell W, Harper WES. 1989. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustalae comb. nov., respectively. Int J Syst Bacteriol 39:397–405. 10.1099/00207713-39-4-397. [DOI] [Google Scholar]
- 304.Thompson LM, Smibert RM, Johnson JL, Krieg NR. 1988. Phylogenetic study of the genus campylobacter. Int J Syst Bacteriol 38:190–200. 10.1099/00207713-38-2-190. [DOI] [Google Scholar]
- 305.Fadilah N, Hanafiah A, Razlan H, Wong ZQ, Mohamed Rose I, Rahman MM. 2016. Multiplex PCR for detection of Helicobacter pylori infection in gastric biopsies with lower inflammatory score. Br J Biomed Sci 73:180–187. 10.1080/09674845.2016.1220705. [DOI] [PubMed] [Google Scholar]
- 306.Beer-Davidson G, Hindiyeh M, Muhsen K. 2018. Detection of Helicobacter pylori in stool samples of young children using real-time polymerase chain reaction. Helicobacter 23:e12450. 10.1111/hel.12450. [DOI] [PubMed] [Google Scholar]
- 307.Fontenete S, Leite M, Figueiredo C, Cos P, Azevedo NF. 2017. Detection of Helicobacter pylori in the gastric mucosa by fluorescence in vivo hybridization. Methods Mol Biol 1616:137–146. 10.1007/978-1-4939-7037-7_8. [DOI] [PubMed] [Google Scholar]
- 308.Tajbakhsh S, Samarbaf-Zadeh AR, Moosavian M. 2008. Comparison of fluorescent in situ hybridization and histological method for the diagnosis of Helicobacter pylori in gastric biopsy samples. Med Sci Monit 14:18758410. [PubMed] [Google Scholar]
- 309.Lopes AI, Vale FF, Oleastro M. 2014. Helicobacter pylori infection—recent developments in diagnosis. World J Gastroenterol 20:9299–93313. 10.3748/wjg.v20.i28.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kobayashi D, Eishi Y, Ohkusa T, Ishige I, Suzuki T, Minami J, Yamada T, Takizawa T, Koike M. 2002. Gastric mucosal density of Helicobacter pylori estimated by real-time PCR compared with results of urea breath test and histological grading. J Med Microbiol 51:305–311. 10.1099/0022-1317-51-4-305. [DOI] [PubMed] [Google Scholar]
- 311.Schabereiter-Gurtner C, Hirschl AM, Dragosics B, Hufnagl P, Puz S, Kovách Z, Rotter M, Makristathis A. 2004. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol 42:4512–4518. 10.1128/JCM.42.10.4512-4518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lascols C, Lamarque D, Costa JM, Copie-Bergman C, Le Glaunec JM, Deforges L, Soussy CJ, Petit JC, Delchier JC, Tankovic J. 2003. Fast and accurate quantitative detection of Helicobacter pylori and identification of clarithromycin resistance mutations in H. pylori isolates from gastric biopsy specimens by real-time PCR. J Clin Microbiol 41:4573–4577. 10.1128/JCM.41.10.4573-4577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Tankovic J, Chaumette-Planckaert MT, Deforges L, Launay N, Le Glaunec JM, Soussy CJ, Delchier JC. 2007. Routine use of real-time PCR for detection of Helicobacter pylori and of clarithromycin resistance mutations. Gastroenterol Clin Biol 31:792–795. 10.1016/S0399-8320(07)73967-2. [DOI] [PubMed] [Google Scholar]
- 314.Lo CC, Lai KH, Peng NJ, Lo GH, Tseng HH, Lin CK, Shie CB, Wu CM, Chen YS, Huang WK, Chen A, Hsu PI. 2005. Polymerase chain reaction: a sensitive method for detecting Helicobacter pylori infection in bleeding peptic ulcers. World J Gastroenterol 11:3909–3914. 10.3748/wjg.v11.i25.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Resti M, Micheli A, Moriondo M, Becciolini L, Cortimiglia M, Canessa C, Indolfi G, Bartolini E, de Martino M, Azzari C. 2009. Comparison of the effect of antibiotic treatment on the possibility of diagnosing invasive pneumococcal disease by culture or molecular methods: a prospective, observational study of children and adolescents with proven pneumococcal infection. Clin Ther 31:1266–1273. 10.1016/j.clinthera.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 316.Bénéjat L, Ducournau A, Lehours P, Mégraud F. 2018. Real-time PCR for Helicobacter pylori diagnosis. The best tools available. Helicobacter 23:e12512. 10.1111/hel.12512. [DOI] [PubMed] [Google Scholar]
- 317.Ducournau A, Bénéjat L, Sifré E, Bessède E, Lehours P, Mégraud F. 2016. Helicobacter pylori resistance to antibiotics in 2014 in France detected by phenotypic and genotypic methods. Clin Microbiol Infect 22:715–718. 10.1016/j.cmi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 318.Van den Poel B, Gils S, Micalessi I, Carton S, Christiaens P, Cuyle PJ, Moons V, Van Olmen G, Smismans A, Bourgain C, Bossuyt P, Frans J. 2021. Molecular detection of Helicobacter pylori and clarithromycin resistance in gastric biopsies: a prospective evaluation of RIDA GENE Helicobacter pylori assay. Acta Clin Belg 76:177–183. 10.1080/17843286.2019.1685741. [DOI] [PubMed] [Google Scholar]
- 319.Pichon M, Pichard B, Barrioz T, Plouzeau C, Croquet V, Fotsing G, Chéron A, Vuillemin É, Wangermez M, Haineaux PA, Vasseur P, Thiebault Q, Lefèvre C, de Singly A, Cremniter J, Broutin L, Michaud A, Silvain C, Burucoa C. 2020. Diagnostic accuracy of a noninvasive test for detection of Helicobacter pylori and resistance to clarithromycin in stool by the Amplidiag H. pylori+ClariR real-time PCR assay. J Clin Microbiol 58:e01787-19. 10.1128/JCM.01787-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Agudo S, Alarcón T, Urruzuno P, Martínez MJ, López-Brea M. 2010. Detection of Helicobacter pylori and clarithromycin resistance in gastric biopsies of pediatric patients by using a commercially available real-time polymerase chain reaction after NucliSens semiautomated DNA extraction. Diagn Microbiol Infect Dis 67:213–219. 10.1016/j.diagmicrobio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 321.Kovacheva-Slavova M, Valkov H, Angelov T, Tropcheva R, Vladimirov B. 2021. Screening for Helicobacter pylori infection and clarithromycin resistance using real-time polymerase chain reaction. Eur Rev Med Pharmacol Sci 25:5042–5046. 10.26355/eurrev_202108_26461. [DOI] [PubMed] [Google Scholar]
- 322.Leonardi M, La Marca G, Pajola B, Perandin F, Ligozzi M, Pomari E. 2020. Assessment of real-time PCR for Helicobacter pylori DNA detection in stool with co-infection of intestinal parasites: a comparative study of DNA extraction methods. BMC Microbiol 20:131. 10.1186/s12866-020-01824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Ramírez-Lázaro MJ, Lario S, Casalots A, Sanfeliu E, Boix L, García-Iglesias P, Sánchez-Delgado J, Montserrat A, Bella-Cueto MR, Gallach M, Sanfeliu I, Segura F, Calvet X. 2011. Real-time PCR improves Helicobacter pylori detection in patients with peptic ulcer bleeding. PLoS One 6:e20009. 10.1371/journal.pone.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Bilgilier C, Thannesberger J, Ojeda Cisneros M, Boehnke K, Wu J, Xi C, Bussalleu Rivera A, Steininger C. 2021. Antimicrobial resistance of Helicobacter pylori in gastric biopsy samples from Lima/Peru. Microb Drug Resist 27:951–955. 10.1089/mdr.2020.0241. [DOI] [PubMed] [Google Scholar]
- 325.Kalach N, Gosset P, Dehecq E, Decoster A, Spyckerelle C, Papadopolos S, Dupont C, Raymond J. 2015. Usefulness of gastric biopsy-based real-time polymerase chain reaction for the diagnosis of Helicobacter pylori infection in children. J Pediatr Gastroenterol Nutr 61:307–312. 10.1097/MPG.0000000000000787. [DOI] [PubMed] [Google Scholar]
- 326.Ciesielska U, Jagoda E, Marciniak Z. 2002. Value of PCR technique in detection of Helicobacter pylori in paraffin-embedded material. Folia Histochem Cytobiol 40:129–130. [PubMed] [Google Scholar]
- 327.Liu Q, Qi D, Kang J, Jin Y, Liu W, Gao W, Hou P, Lu J. 2015. Efficacy of real-time PCR-based detection of Helicobacter pylori infection and genotypic resistance-guided quadruple therapy as the first-line treatment for functional dyspepsia with Helicobacter pylori infection. Eur J Gastroenterol Hepatol 27:221–225. 10.1097/MEG.0000000000000186. [DOI] [PubMed] [Google Scholar]
- 328.Clines N, Beckman E. 2019. Development of a high throughput human stool specimen processing method for a molecular Helicobacter pylori clarithromycin resistance assay. PLoS One 14:e0224356. 10.1371/journal.pone.0224356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Leal YA, Cedillo-Rivera R, Simón JA, Velázquez JR, Flores LL, Torres J. 2011. Utility of stool sample-based tests for the diagnosis of Helicobacter pylori infection in children. J Pediatr Gastroenterol Nutr 52:718–728. 10.1097/MPG.0b013e3182077d33. [DOI] [PubMed] [Google Scholar]
- 330.Pohl D, Keller PM, Bordier V, Wagner K. 2019. Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World J Gastroenterol 25:4629–4660. 10.3748/wjg.v25.i32.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Giorgio F, Ierardi E, Sorrentino C, Principi M, Barone M, Losurdo G, Iannone A, Giangaspero A, Monno R, Di Leo A. 2016. Helicobacter pylori DNA isolation in the stool: an essential pre-requisite for bacterial noninvasive molecular analysis. Scand J Gastroenterol 51:1429–1432. 10.1080/00365521.2016.1216592. [DOI] [PubMed] [Google Scholar]
- 332.Quan PL, Sauzade M, Brouzes E. 2018. dPCR: a technology review. Sensors (Basel) 18:1271. 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10:1003–1005. 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Cao L, Cui X, Hu J, Li Z, Choi JR, Yang Q, Lin M, Ying HL, Xu F. 2017. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron 90:459–474. 10.1016/j.bios.2016.09.082. [DOI] [PubMed] [Google Scholar]
- 335.Talarico S, Safaeian M, Gonzalez P, Hildesheim A, Herrero R, Porras C, Cortes B, Larson A, Fang FC, Salama NR. 2016. Quantitative detection and genotyping of Helicobacter pylori from stool using droplet digital PCR reveals variation in bacterial loads that correlates with cagA virulence gene carriage. Helicobacter 21:325–333. 10.1111/hel.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Sun L, Talarico S, Yao L, He L, Self S, You Y, Zhang H, Zhang Y, Guo Y, Liu G, Salama NR, Zhang J. 2018. Droplet digital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J Clin Microbiol 56:e00019-18. 10.1128/JCM.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Talarico S, Korson AS, Leverich CK, Park S, Jalikis FG, Upton MP, Broussard E, Salama NR. 2018. High prevalence of Helicobacter pylori clarithromycin resistance mutations among Seattle patients measured by droplet digital PCR. Helicobacter 23:e12472. 10.1111/hel.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Raderer M, Streubel B, Wöhrer S, Häfner M, Chott A. 2006. Successful antibiotic treatment of Helicobacter pylori negative gastric mucosa associated lymphoid tissue lymphomas. Gut 55:616–618. 10.1136/gut.2005.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Güell M, Artigau E, Esteve V, Sánchez-Delgado J, Junquera F, Calvet X. 2006. Usefulness of a delayed test for the diagnosis of Helicobacter pylori infection in bleeding peptic ulcer. Aliment Pharmacol Ther 23:53–59. 10.1111/j.1365-2036.2006.02726.x. [DOI] [PubMed] [Google Scholar]
- 340.Gisbert JP, Abraira V. 2006. Accuracy of Helicobacter pylori diagnostic tests in patients with bleeding peptic ulcer: a systematic review and meta-analysis. Am J Gastroenterol 101:848–863. 10.1111/j.1572-0241.2006.00528.x. [DOI] [PubMed] [Google Scholar]
- 341.Choi YJ, Kim N, Lim J, Jo SY, Shin CM, Lee HS, Lee SH, Park YS, Hwang JH, Kim JW, Jeong SH, Lee DH, Jung HC. 2012. Accuracy of diagnostic tests for Helicobacter pylori in patients with peptic ulcer bleeding. Helicobacter 17:77–85. 10.1111/j.1523-5378.2011.00915.x. [DOI] [PubMed] [Google Scholar]
- 342.Tian XY, Zhu H, Zhao J, She Q, Zhang GX. 2012. Diagnostic performance of urea breath test, rapid urea test, and histology for Helicobacter pylori infection in patients with partial gastrectomy: a meta-analysis. J Clin Gastroenterol 46:285–292. 10.1097/MCG.0b013e318249c4cd. [DOI] [PubMed] [Google Scholar]
- 343.Yan J, Yamaguchi T, Odaka T, Suzuki T, Ohyama N, Hara T, Sudo K, Nakamura K, Denda T, Takiguchi N, Yokosuka O, Nomura F. 2010. Stool antigen test is a reliable method to detect Helicobacter pylori in the gastric remnant after distal gastrectomy for gastric cancer. J Clin Gastroenterol 44:73–74. 10.1097/MCG.0b013e3181aae65e. [DOI] [PubMed] [Google Scholar]
- 344.Walker WA, Goulet O, Kleinman RE, Sherman PM, Shneider BL, Sanderson IR. 2004. Helicobacter pylori and peptic ulcer disease, p 491–512. In: Rowland M, Bourke B, Drumm B (ed), Pediatric gastrointestinal disease. BC Decker, Ontario, Canada. [Google Scholar]
- 345.Kawakami E, Machado RS, Reber M, Patrício FR. 2002. 13C-urea breath test with infrared spectroscopy for diagnosing Helicobacter pylori infection in children and adolescents. J Pediatr Gastroenterol Nutr 35:39–43. 10.1097/00005176-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 346.Mégraud F, European Paediatric Task Force on Helicobacter pylori. 2005. Comparison of non-invasive tests to detect Helicobacter pylori infection in children and adolescents: results of a multicenter European study. J Pediatr 146:198–203. 10.1016/j.jpeds.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 347.Drumm B, Koletzko S, Oderda G. 2000. Helicobacter pylori infection in children: a consensus statement. European Paediatric Task Force on Helicobacter pylori. J Pediatr Gastroenterol Nutr 30:207–213. 10.1097/00005176-200002000-00020. [DOI] [PubMed] [Google Scholar]
- 348.Pathak CM, Bhasin DK, Khanduja KL. 2004. Urea breath test for Helicobacter pylori detection: present status. Trop Gastroenterol 25:156–161. PMID: 15912972. [PubMed] [Google Scholar]
- 349.Ferwana M, Abdulmajeed I, Alhajiahmed A, Madani W, Firwana B, Hasan R, Altayar O, Limburg PJ, Murad MH, Knawy B. 2015. Accuracy of urea breath test in Helicobacter pylori infection: meta-analysis. World J Gastroenterol 21:1305–1314. 10.3748/wjg.v21.i4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Prell C, Osterrieder S, Lottspeich C, Schwarzer A, Rüssmann H, Ossiander G, Koletzko S. 2009. Improved performance of a rapid office-based stool test for detection of Helicobacter pylori in children before and after therapy. J Clin Microbiol 47:3980–3984. 10.1128/JCM.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Lin HJ, Lo WC, Perng CL, Li AF, Tseng GY, Sun IC, Ou YH. 2004. Helicobacter pylori stool antigen test in patients with bleeding peptic ulcers. Helicobacter 9:663–668. 10.1111/j.1083-4389.2004.00276.x. [DOI] [PubMed] [Google Scholar]
- 352.Crone J, Gold BD. 2004. Helicobacter pylori infection in pediatrics. Helicobacter 9(Suppl 1):49–56. 10.1111/j.1083-4389.2004.00253.x. [DOI] [PubMed] [Google Scholar]
- 353.Tonkic A, Tonkic M, Lehours P, Mégraud F. 2012. Epidemiology and diagnosis of Helicobacter pylori infection. Helicobacter 17(Suppl 1):1–8. 10.1111/j.1523-5378.2012.00975.x. [DOI] [PubMed] [Google Scholar]
- 354.Zhou X, Su J, Xu G, Zhang G. 2014. Accuracy of stool antigen test for the diagnosis of Helicobacter pylori infection in children: a meta-analysis. Clin Res Hepatol Gastroenterol 38:629–638. 10.1016/j.clinre.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 355.Kuloğlu Z, Kansu A, Kirsaçlioğlu CT, Ustündağ G, Aysev D, Ensari A, Küçük NO, Girgin N. 2008. A rapid lateral flow stool antigen immunoassay and (14)C-urea breath test for the diagnosis and eradication of Helicobacter pylori infection in children. Diagn Microbiol Infect Dis 62:351–356. 10.1016/j.diagmicrobio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 356.Nares-Cisneros J, Jaramillo-Rodríguez Y, Martínez-Ordaz VA, Velasco-Rodríguez VM, Madero A, Mena-Arias G, Manriquez-Covarrubias L. 2007. Immunochromatographic monoclonal test for detection of Helicobacter pylori antigen in stool is useful in children from high-prevalence developing country. Helicobacter 12:354–358. 10.1111/j.1523-5378.2007.00514.x. [DOI] [PubMed] [Google Scholar]
- 357.Queiroz DM, Saito M, Rocha GA, Rocha AM, Melo FF, Checkley W, Braga LL, Silva IS, Gilman RH, Crabtree JE. 2013. Helicobacter pylori infection in infants and toddlers in South America: concordance between [13C]urea breath test and monoclonal H. pylori stool antigen test. J Clin Microbiol 51:3735–3740. 10.1128/JCM.01752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Leal YA, Flores LL, García-Cortés LB, Cedillo-Rivera R, Torres J. 2008. Antibody-based detection tests for the diagnosis of Helicobacter pylori infection in children: a meta-analysis. PLoS One 3:e3751. 10.1371/journal.pone.0003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Okuda M, Miyashiro E, Koike M, Tanaka T, Bouoka M, Okuda S, Yoshikawa N. 2002. Serodiagnosis of Helicobacter pylori infection is not accurate for children aged below 10. Pediatr Int 44:387–390. 10.1046/j.1442-200X.2002.01585.x. [DOI] [PubMed] [Google Scholar]
- 360.Oderda G, Rapa A, Bona G. 2004. Diagnostic tests for childhood Helicobacter pylori infection: invasive, noninvasive or both? J Pediatr Gastroenterol Nutr 39:482–484. 10.1097/00005176-200411000-00006. [DOI] [PubMed] [Google Scholar]
- 361.Kato S, Nakayama K, Minoura T, Konno M, Tajiri H, Matsuhisa T, Iinuma K. 2004. Comparison between the 13C-urea breath test and stool antigen test for the diagnosis of childhood Helicobacter pylori infection. J Gastroenterol 39:1045–1050. 10.1007/s00535-004-1442-6. [DOI] [PubMed] [Google Scholar]
- 362.Hino B, Eliakim R, Levine A, Sprecher H, Berkowitz D, Hartman C, Eshach-Adiv O, Shamir R. 2004. Comparison of invasive and non-invasive tests diagnosis and monitoring of Helicobacter pylori infection in children. J Pediatr Gastroenterol Nutr 39:519–523. 10.1097/00005176-200411000-00013. [DOI] [PubMed] [Google Scholar]
- 363.Sabbagh P, Mohammadnia-Afrouzi M, Javanian M, Babazadeh A, Koppolu V, Vasigala VR, Nouri HR, Ebrahimpour S. 2019. Diagnostic methods for Helicobacter pylori infection: ideals, options, and limitations. Eur J Clin Microbiol Infect Dis 38:55–66. 10.1007/s10096-018-3414-4. [DOI] [PubMed] [Google Scholar]
- 364.Chung WC, Jung SH, Oh JH, Kim TH, Cheung DY, Kim BW, Kim SS, Kim JI, Sin EY. 2014. Dual-priming oligonucleotide-based multiplex PCR using tissue samples in rapid urease test in the detection of Helicobacter pylori infection. World J Gastroenterol 20:6547–6553. 10.3748/wjg.v20.i21.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Bazin T, Nchare Mfondi A, Julie C, Émile JF, Raymond J, Lamarque D. 2018. Contribution of genetic amplification by PCR for the diagnosis of Helicobacter pylori infection in patients receiving proton pump inhibitors. United European Gastroenterol J 6:1267–1273. 10.1177/2050640618787055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Yakoob J, Jafri W, Abbas Z, Abid S, Islam M, Ahmed Z. 2008. The diagnostic yield of various tests for Helicobacter pylori infection in patients on acid-reducing drugs. Dig Dis Sci 53:95–100. 10.1007/s10620-007-9828-y. [DOI] [PubMed] [Google Scholar]
- 367.International Agency for Research on Cancer, World Health Organization. 2014. Helicobacter pylori rradication as a strategy for preventing gastric cancer. In Working group report, vol 8. International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- 368.Siddique I, Al-Mekhaizeem K, Alateeqi N, Memon A, Hasan F. 2008. Diagnosis of Helicobacter pylori: improving the sensitivity of CLO test by increasing the number of gastric antral biopsies. J Clin Gastroenterol 42:356–360. 10.1097/MCG.0b013e31802b650d. [DOI] [PubMed] [Google Scholar]
- 369.Lee YC, Tseng PH, Liou JM, Chen MJ, Chen CC, Tu CH, Chiang TH, Chiu HM, Lai CF, Ho JC, Wu MS. 2014. Performance of a onestep fecal sample-based test for diagnosis of Helicobacter pylori infection in primary care and mass screening settings. J Formos Med Assoc 113:899–907. 10.1016/j.jfma.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 370.Akamatsu T, Ichikawa S, Okudaira S, Yokosawa S, Iwaya Y, Suga T, Ota H, Tanaka E. 2011. Introduction of an examination and treatment for Helicobacter pylori infection in high school health screening. J Gastroenterol 46:1353–1360. 10.1007/s00535-011-0450-6. [DOI] [PubMed] [Google Scholar]
- 371.Nakayama Y, Lin Y, Hongo M, Hidaka H, Kikuchi S. 2017. Helicobacter pylori infection and its related factors in junior high school students in Nagano Prefecture, Japan. Helicobacter 22:e12363. 10.1111/hel.12363. [DOI] [PubMed] [Google Scholar]
- 372.Inoue M. 2017. Changing epidemiology of Helicobacter pylori in Japan. Gastric Cancer 20:3–7. 10.1007/s10120-016-0658-5. [DOI] [PubMed] [Google Scholar]
- 373.Shimoyama T, Oyama T, Matsuzaka M, Danjo K, Nakaji S, Fukuda S. 2009. Comparison of a stool antigen test and serology for the diagnosis of Helicobacter pylori infection in mass survey. Helicobacter 14:87–90. 10.1111/j.1523-5378.2009.00672.x. [DOI] [PubMed] [Google Scholar]
- 374.Choi J, Kim CH, Kim D, Chung SJ, Song JH, Kang JM, Yang JI, Park MJ, Kim YS, Yim JY, Lim SH, Kim JS, Jung HC, Song IS. 2011. Prospective evaluation of a new stool antigen test for the detection of Helicobacter pylori, in comparison with histology, rapid urease test, (13)C-urea breath test, and serology. J Gastroenterol Hepatol 26:1053–1059. 10.1111/j.1440-1746.2011.06705.x. [DOI] [PubMed] [Google Scholar]
- 375.Skrebinska S, Megraud F, Daugule I, Santare D, Isajevs S, Liepniece-Karele I, Bogdanova I, Rudzite D, Vangravs R, Kikuste I, Vanags A, Tolmanis I, Savcenko S, Alix C, Herrero R, Park JY, Leja M. 2022. Who could be blamed in the case of discrepant histology and serology results for Helicobacter pylori detection? Diagnostics (Basel) 12:133. 10.3390/diagnostics12010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Kusano C, Gotoda T, Ishikawa H, Moriyama M. 2017. The administrative project of Helicobacter pylori infection screening among junior high school students in an area of Japan with a high incidence of gastric cancer. Gastric Cancer 20:16–19. 10.1007/s10120-017-0688-7. [DOI] [PubMed] [Google Scholar]
- 377.Mabe K, Kikuchi S, Okuda M, Takamasa M, Kato M, Asaka M. 2016. Diagnostic accuracy of urine Helicobacter pylori antibody test in junior and senior high school students in Japan. Helicobacter 22:e12329. 10.1111/hel.12329. [DOI] [PubMed] [Google Scholar]
- 378.Areia M, Carvalho R, Cadime AT, Rocha Gonçalves F, Dinis-Ribeiro M. 2013. Screening for gastric cancer and surveillance of premalignant lesions: a systematic review of cost-effectiveness studies. Helicobacter 18:325–337. 10.1111/hel.12050. [DOI] [PubMed] [Google Scholar]
- 379.Boklage SH, Mangel AW, Ramamohan V, Mladsi D, Wang T. 2016. Impact of patient adherence on the cost-effectiveness of noninvasive tests for the initial diagnosis of Helicobacter pylori infection in the United States. Patient Prefer Adherence 10:45–55. 10.2147/PPA.S93320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Deguchi R, Matsushima M, Suzuki T, Mine T, Fukuda R, Nishina M, Ozawa H, Takagi A. 2009. Comparison of a monoclonal with a polyclonal antibody-based enzyme immunoassay stool test in diagnosing Helicobacter pylori infection after eradication therapy. J Gastroenterol 44:713–716. 10.1007/s00535-009-0069-z. [DOI] [PubMed] [Google Scholar]
- 381.Calvet X, Lario S, Ramírez-Lázaro MJ, Montserrat A, Quesada M, Reeves L, Masters H, Suárez-Lamas D, Gallach M, Miquel M, Martínez-Bauer E, Sanfeliu I, Segura F. 2010. Accuracy of monoclonal stool tests for determining cure of Helicobacter pylori infection after treatment. Helicobacter 15:201–205. 10.1111/j.1523-5378.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 382.Gisbert JP, de la Morena F, Abraira V. 2006. Accuracy of monoclonal stool antigen test for the diagnosis of H. pylori infection: a systematic review and meta-analysis. Am J Gastroenterol 101:1921–1930. 10.1111/j.1572-0241.2006.00668.x. [DOI] [PubMed] [Google Scholar]
- 383.Veijola L. 2005. Stool antigen tests in the diagnosis of Helicobacter pylori infection before and after eradication therapy. World J Gastroenterol 11:7340. 10.3748/wjg.v11.i46.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Vaira D, Vakil N, Menegatti M, van't Hoff B, Ricci C, Gatta L, Gasbarrini G, Quina M, Pajares Garcia JM, van Der Ende A, van Der Hulst R, Anti M, Duarte C, Gisbert JP, Miglioli M, Tytgat G. 2002. The stool antigen test for detection of Helicobacter pylori after eradication therapy. Ann Intern Med 136:280–287. 10.7326/0003-4819-136-4-200202190-00007. [DOI] [PubMed] [Google Scholar]
- 385.Manes G, Zanetti MV, Piccirillo MM, Lombardi G, Balzano A, Pieramico O. 2005. Accuracy of a new monoclonal stool antigen test in post-eradication assessment of Helicobacter pylori infection: comparison with the polyclonal stool antigen test and urea breath test. Dig Liver Dis 37:751–755. 10.1016/j.dld.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 386.Graham DY, Klein PD, Evans DJ, Jr, Evans DG, Alpert LC, Opekun AR, Boutton TW. 1987. Campylobacter pylori detected noninvasively by the 13C-urea breath test. Lancet i:1174–1177. 10.1016/s0140-6736(87)92145-3. [DOI] [PubMed] [Google Scholar]
- 387.Jemilohun AC, Otegbayo JA. 2016. Helicobacter pylori infection: past, present and future. Pan Afr Med J 23:216. 10.11604/pamj.2016.23.216.8852. [DOI] [Google Scholar]
- 388.Monteiro L, de Mascarel A, Sarrasqueta AM, Bergey B, Barberis C, Talby P, Roux D, Shouler L, Goldfain D, Lamouliatte H, Mégraud F. 2001. Diagnosis of Helicobacter pylori infection: noninvasive methods compared to invasive methods and evaluation of two new tests. Am J Gastroenterol 96:353–358. 10.1111/j.1572-0241.2001.03518.x. [DOI] [PubMed] [Google Scholar]
- 389.Calvet X. 2015. Diagnosis of Helicobacter pylori infection in the proton pump inhibitor era. Gastroenterol Clin North Am 44:507–518. 10.1016/j.gtc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 390.Tan SY, Grimes S. 2010. Paul Ehrlich (1854–1915): man with the magic bullet. Singapore Med J 51:842–843. [PubMed] [Google Scholar]
- 391.Flemming A. 1929. Classics in infectious diseases: on the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol 10:226–236. [PubMed] [Google Scholar]
- 392.Davies J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 64:375–382. [DOI] [PubMed] [Google Scholar]
- 393.Mégraud F. 1997. Resistance of Helicobacter pylori to antibiotics. Aliment Pharmacol Ther 11(Suppl 1):43–53. 10.1046/j.1365-2036.11.s1.11.x. [DOI] [PubMed] [Google Scholar]
- 394.Roy J. 2012. An introduction to pharmaceutical sciences production: chemistry, techniques and technology. Woodhead Publishing, Cambridge, United Kingdom. [Google Scholar]
- 395.World Health Organization. 2015. WHO model list of essential medicines (19th list). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 396.Akhavan BJ, Khanna NR, Vijhani P. 2022. Amoxicillin. In StatPearls. StatPearls Publishing, Treasure Island (FL). https://www.ncbi.nlm.nih.gov/books/NBK482250/. [PubMed] [Google Scholar]
- 397.Lovering AL, Safadi SS, Strynadka NCJ. 2012. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem 81:451–478. 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 398.Zanotti G, Cendron L. 2019. Structural aspects of Helicobacter pylori antibiotic resistance. Adv Exp Med Biol 1149:227–241. 10.1007/5584_2019_368. [DOI] [PubMed] [Google Scholar]
- 399.Saranathan R, Levi MH, Wattam AR, Malek A, Asare E, Behin DS, Pan DH, Jacobs WR, Jr, Szymczak WA. 2020. Helicobacter pylori infections in the Bronx, New York: surveying antibiotic susceptibility and strain lineage by whole-genome sequencing. J Clin Microbiol 58:e01591-19. 10.1128/JCM.01591-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2008. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J Antimicrob Chemother 61:995–998. 10.1093/jac/dkn051. [DOI] [PubMed] [Google Scholar]
- 401.Cho H, Uehara T, Bernhardt TG. 2014. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159:1300–1311. 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Qureshi NN, Morikis D, Schiller NL. 2011. Contribution of specific amino acid changes in penicillin binding protein 1 to amoxicillin resistance in clinical Helicobacter pylori isolates. Antimicrob Agents Chemother 55:101–109. 10.1128/AAC.00545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Subsomwong P, Doohan D, Fauzia KA, Akada J, Matsumoto T, Yee TT, Htet K, Waskito LA, Tuan VP, Uchida T, Matsuhisa T, Yamaoka Y. 2022. Next-generation sequencing-based study of Helicobacter pylori isolates from Myanmar and their susceptibility to antibiotics. Microorganisms 10:196. 10.3390/microorganisms10010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2007. Correlation between substitutions in penicillin-binding protein 1 and amoxicillin resistance in Helicobacter pylori. Microbiol Immunol 51:939–944. 10.1111/j.1348-0421.2007.tb03990.x. [DOI] [PubMed] [Google Scholar]
- 405.Qureshi NN, Gallaher B, Schiller NL. 2014. Evolution of amoxicillin resistance of Helicobacter pylori in vitro: characterization of resistance mechanisms. Microb Drug Resist 20:509–516. 10.1089/mdr.2014.0019. [DOI] [PubMed] [Google Scholar]
- 406.Anis S, Farooqi SR, Niaz SK. 2021. Characterization of domain V mutations in clinical isolates of Helicobacter pylori in Pakistan and their effect on clarithromycin MIC. Infect Drug Resist 14:3393–3403. 10.2147/IDR.S306878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Tran VH, Ha TMT, Le PTQ, Phan TN, Tran TNH. 2019. Characterisation of point mutations in domain V of the 23S rRNA gene of clinical Helicobacter pylori strains and clarithromycin-resistant phenotype in central Vietnam. J Glob Antimicrob Resist 16:87–91. 10.1016/j.jgar.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 408.Marques AT, Vítor JMB, Santos A, Oleastro M, Vale FF. 2020. Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microb Genom 6:e000344. 10.1099/mgen.0.000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Chen D, Cunningham SA, Cole NC, Kohner PC, Mandrekar JN, Patel R. 2017. Phenotypic and molecular antimicrobial susceptibility of Helicobacter pylori. Antimicrob Agents Chemother 61:e02530-16. 10.1128/AAC.02530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Aguilera-Correa JJ, Urruzuno P, Barrio J, Martinez MJ, Agudo S, Somodevilla A, Llorca L, Alarcón T. 2017. Detection of Helicobacter pylori and the genotypes of resistance to clarithromycin and the heterogeneous genotype to this antibiotic in biopsies obtained from symptomatic children. Diagn Microbiol Infect Dis 87:150–153. 10.1016/j.diagmicrobio.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 411.Hulten KG, Genta RM, Kalfus IN, Zhou Y, Zhang H, Graham DY. 2021. Comparison of culture with antibiogram to next-generation sequencing using bacterial isolates and formalin-fixed, paraffin-embedded gastric biopsies. Gastroenterol 161:1433–1442.E2. 10.1053/j.gastro.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Vazirzadeh J, Falahi J, Moghim S, Narimani T, Rafiei R, Karbasizadeh V. 2020. Molecular assessment of resistance to clarithromycin in Helicobacter pylori strains isolated from patients with dyspepsia by fluorescent in situ hybridization in the center of Iran. Biomed Res Int 2020:2304173. 10.1155/2020/2304173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Tamayo E, Montes M, Fernández-Reyes M, Lizasoain J, Ibarra B, Mendarte U, Zapata E, Mendiola J, Pérez-Trallero E. 2017. Clarithromycin resistance in Helicobacter pylori and its molecular determinants in Northern Spain, 2013–2015. J Glob Antimicrob Resist 9:43–46. 10.1016/j.jgar.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 414.Sanches BS, Martins GM, Lima K, Cota B, Moretzsohn LD, Ribeiro LT, Breyer HP, Maguilnik I, Maia AB, Rezende-Filho J, Meira AC, Pinto H, Alves E, Mascarenhas R, Passos R, de Souza JD, Trindade OR, Coelho LG. 2016. Detection of Helicobacter pylori resistance to clarithromycin and fluoroquinolones in Brazil: a national survey. World J Gastroenterol 22:7587–7594. 10.3748/wjg.v22.i33.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Ng HK, Goh KL, Chuah KH, Thalha AM, Kee BP, Por LY, Chua KH, Puah SM. 2020. Sequencing-based detection of 23S rRNA domain V mutations in treatment-naïve Helicobacter pylori patients from Malaysia. J Glob Antimicrob Resist 23:345–348. 10.1016/j.jgar.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 416.Kocazeybek B, Sakli MK, Yuksel P, Demirci M, Caliskan R, Sarp TZ, Saribas S, Demiryas S, Kalayci F, Cakan H, Uysal HK, Gareayaghi N, Ergin S, Erzin YZ, Bal K, Tascı İ, Tokman HB. 2019. Comparison of new and classical point mutations associated with clarithromycin resistance in Helicobacter pylori strains isolated from dyspeptic patients and their effects on phenotypic clarithromycin resistance. J Med Microbiol 68:566–573. 10.1099/jmm.0.000944. [DOI] [PubMed] [Google Scholar]
- 417.Miftahussurur M, Cruz M, Subsomwong P, Jiménez Abreu JA, Hosking C, Nagashima H, Akada J, Yamaoka Y. 2017. Clarithromycin-based triple therapy is still useful as an initial treatment for Helicobacter pylori infection in the Dominican Republic. Am J Trop Med Hyg 96:1050–1059. 10.4269/ajtmh.16-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Petrin D, Delgaty K, Bhatt R, Garber G. 1998. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev 11:300–317. 10.1128/CMR.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Shinn DLS. 1962. Metronidazole in acute ulcerative gingivitis. Lancet 279:1191. 10.1016/S0140-6736(62)92243-2. [DOI] [Google Scholar]
- 420.Francesco VD, Zullo A, Hassan C, Giorgio F, Rosania R, Ierardi E. 2011. Mechanisms of Helicobacter pylori antibiotic resistance: an updated appraisal. World J Gastrointest Pathophysiol 2:35–41. 10.4291/wjgp.v2.i3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Mannion A, Dzink-Fox JAnn, Shen Z, Piazuelo MB, Wilson KT, Correa P, Peek RM, Camargo MC, Fox JG. 2021. Helicobacter pylori antimicrobial resistance and gene variants in high- and low-gastric-cancer-risk populations. J Clin Microbiol 59:e03203-20. 10.1128/JCM.03203-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Tanih NF, Ndip LM, Ndip RN. 2011. Characterisation of the genes encoding resistance to metronidazole (rdxA and frxA) and clarithromycin (the 23S-rRNA genes) in South African isolates of Helicobacter pylori. Ann Trop Med Parasitol 105:251–259. 10.1179/136485911X12899838683485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. 2014. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One 9:e101481. 10.1371/journal.pone.0101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Butlop TR, Mungkote NT, Chaichanawongsaroj NT. 2016. Analysis of allelic variants of rdxA associated with metronidazole resistance in Helicobacter pylori: detection of common genotypes in rdxA by multiplex allele-specific polymerase chain reaction. Genet Mol Res 15:gmr.15038674. 10.4238/gmr.15038674. [DOI] [PubMed] [Google Scholar]
- 425.Binh TT, Suzuki R, Trang TTH, Kwon DH, Yamaoka Y. 2015. Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob Agents Chemother 59:2343–2348. 10.1128/AAC.04852-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Tsugawa H, Suzuki H, Satoh K, Hirata K, Matsuzaki J, Saito Y, Suematsu M, Hibi T. 2011. Two amino acids mutation of ferric uptake regulator determines Helicobacter pylori resistance to metronidazole. Antioxid Redox Signal 14:15–23. 10.1089/ars.2010.3146. [DOI] [PubMed] [Google Scholar]
- 427.Choi SS, Chivers PT, Berg DE. 2011. Point mutations in Helicobacter pylori's fur regulatory gene that alter resistance to metronidazole, a prodrug activated by chemical reduction. PLoS One 6:e18236. 10.1371/journal.pone.0018236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Hanafi A, Lee WC, Loke MF, Teh X, Shaari A, Dinarvand M, Lehours P, Mégraud F, Leow AH, Vadivelu J, Goh KL. 2016. Molecular and proteomic analysis of levofloxacin and metronidazole resistant Helicobacter pylori. Front Microbiol 7:2015. 10.3389/fmicb.2016.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. 2009. Quinolones: action and resistance updated. Curr Top Med Chem 9:981–998. 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Saracino IM, Pavoni M, Zullo A, Fiorini G, Lazzarotto T, Borghi C, Vaira D. 2021. Next generation sequencing for the prediction of the antibiotic resistance in Helicobacter pylori: a literature review. Antibiotics (Basel) 10:437. 10.3390/antibiotics10040437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob Agents Chemother 52:385–392. 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Webber MA, Ricci V, Whitehead R, Patel M, Fookes M, Ivens A, Piddock LJ. 2013. Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. mBio 4:e00273-13. 10.1128/mBio.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Baranello L, Levens D, Gupta A, Kouzine F. 2012. The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim Biophys Acta 1819:632–638. 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Matsuzaki J, Suzuki H, Tsugawa H, Nishizawa T, Hibi T. 2010. Homology model of the DNA gyrase enzyme of Helicobacter pylori, a target of quinolone-based eradication therapy. J Gastroenterol Hepatol 25(Suppl 1):S7–S10. 10.1111/j.1440-1746.2010.06245.x. [DOI] [PubMed] [Google Scholar]
- 435.Nishizawa T, Suzuki H, Hibi T. 2009. Quinolone-based third-line therapy for Helicobacter pylori eradication. J Clin Biochem Nutr 44:119–124. 10.3164/jcbn.08-220R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Tshibangu-Kabamba E, Ngoma-Kisoko PJ, Tuan VP, Matsumoto T, Akada J, Kido Y, Tshimpi-Wola A, Tshiamala-Kashala P, Ahuka-Mundeke S, Ngoy DM, Disashi-Tumba G, Yamaoka Y. 2020. Next-generation sequencing of the whole bacterial genome for tracking molecular insight into the broad-spectrum antimicrobial resistance of Helicobacter pylori clinical isolates from the Democratic Republic of Congo. Microorganisms 8:887. 10.3390/microorganisms8060887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Miftahussurur M, Syam AF, Nusi IA, Makmun D, Waskito LA, Zein LH, Akil F, Uwan WB, Simanjuntak D, Wibawa ID, Waleleng JB, Saudale AM, Yusuf F, Mustika S, Maimunah AP, Maulahela U, Rezkitha H, Subsomwong YA, Nasronudin P, Rahardjo D, Suzuki R, Akada J, Yamaoka Y. 2016. Surveillance of Helicobacter pylori antibiotic susceptibility in Indonesia: different resistance types among regions and with novel genetic mutations. PLoS One 11:e0166199. 10.1371/journal.pone.0166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Miftahussurur M, Shrestha PK, Subsomwong P, Sharma RP, Yamaoka Y. 2016. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol 16:256. 10.1186/s12866-016-0873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2012. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter 17:36–42. 10.1111/j.1523-5378.2011.00912.x. [DOI] [PubMed] [Google Scholar]
- 440.Zerbetto De Palma G, Mendiondo N, Wonaga A, Viola L, Ibarra D, Campitelli E, Salim N, Corti R, Goldman C, Catalano M. 2017. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires City. Microb Drug Resist 23:351–358. 10.1089/mdr.2015.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Fuoco D. 2012. Classification framework and chemical biology of tetracycline-structure-based drugs. Antibiotics (Basel) 1:1–13. 10.3390/antibiotics1010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Rusu A, Buta EL. 2021. The development of third-generation tetracycline antibiotics and new perspectives. Pharmaceutics 13:2085. 10.3390/pharmaceutics13122085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. 2018. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology 155:1372–1382.E17. 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Dailidiene D, Bertoli MT, Miciuleviciene J, Mukhopadhyay AK, Dailide G, Pascasio MA, Kupcinskas L, Berg DE. 2002. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob Agents Chemother 46:3940–3946. 10.1128/AAC.46.12.3940-3946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Trieber CA, Taylor DE. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol 184:2131–2140. 10.1128/JB.184.8.2131-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Wu JY, Kim JJ, Reddy R, Wang WM, Graham DY, Kwon DH. 2005. Tetracycline-resistant clinical Helicobacter pylori isolates with and without mutations in 16S rRNA-encoding genes. Antimicrob Agents Chemother 49:578–583. 10.1128/AAC.49.2.578-583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Nonaka L, Connell SR, Taylor DE. 2005. 16S rRNA mutations that confer tetracycline resistance in Helicobacter pylori decrease drug binding in Escherichia coli ribosomes. J Bacteriol 187:3708–3712. 10.1128/JB.187.11.3708-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Gisbert JP, Calvet X. 2012. Review article: rifabutin in the treatment of refractory Helicobacter pylori infection. Aliment Pharmacol Ther 35:209–221. 10.1111/j.1365-2036.2011.04937.x. [DOI] [PubMed] [Google Scholar]
- 449.Gisbert JP. 2020. Rifabutin for the treatment of Helicobacter pylori infection: a review. Pathogens 10:15. 10.3390/pathogens10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Heep M, Lehn N, Brandstätter B, Rieger U, Senzenberger S, Wehrl W. 2002. Detection of rifabutin resistance and association of rpoB mutations with resistance to four rifamycin derivatives in Helicobacter pylori. Eur J Clin Microbiol Infect Dis 21:143–145. 10.1007/s10096-001-0672-2. [DOI] [PubMed] [Google Scholar]
- 451.Nishizawa T, Suzuki H, Matsuzaki J, Muraoka H, Tsugawa H, Hirata K, Hibi T. 2011. Helicobacter pylori resistance to rifabutin in the last 7 years. Antimicrob Agents Chemother 55:5374–5375. 10.1128/AAC.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Glocker E, Bogdan C, Kist M. 2007. Characterization of rifampicin-resistant clinical Helicobacter pylori isolates from Germany. J Antimicrob Chemother 59:874–879. 10.1093/jac/dkm039. [DOI] [PubMed] [Google Scholar]
- 453.Suzuki S, Suzuki H, Nishizawa T, Kaneko F, Ootani S, Muraoka H, Saito Y, Kobayashi I, Hibi T. 2009. Past rifampicin dosing determines rifabutin resistance of Helicobacter pylori. Digestion 79:1–4. 10.1159/000191204. [DOI] [PubMed] [Google Scholar]
- 454.Vass M, Hruska K, Franek M. 2008. Nitrofuran antibiotics: a review on the application, prohibition and residual analysis. Vet Med 53:469–500. 10.17221/1979-VETMED. [DOI] [Google Scholar]
- 455.David Martínez J, Henao SC, Lizarazo JI. 2014. Antibiotic resistance of Helicobacter pylori in Latin America and the Caribbean. Rev Col Gastroenterol 29:217–226. [Google Scholar]
- 456.Su Z, Xu H, Zhang C, Shao S, Li L, Wang H, Wang H, Qiu G. 2006. Mutations in Helicobacter pylori porD and oorD genes may contribute to furazolidone resistance. Croat Med J 47:410–415. [PMC free article] [PubMed] [Google Scholar]
- 457.Zeng X, Lin J. 2013. Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front Microbiol 4:128. 10.3389/fmicb.2013.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Ojetti V, Migneco A, Zocco MA, Nista EC, Gasbarrini G, Gasbarrini A. 2004. Beta-lactamase inhibitor enhances Helicobacter pylori eradication rate. J Intern Med 255:125–129. 10.1046/j.0954-6820.2003.01239.x. [DOI] [PubMed] [Google Scholar]
- 459.Tseng YS, Wu DC, Chang CY, Kuo CH, Yang YC, Jan CM, Su YC, Kuo FC, Chang LL. 2009. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur J Clin Invest 39:807–812. 10.1111/j.1365-2362.2009.02166.x. [DOI] [PubMed] [Google Scholar]
- 460.Amaral L, Cerca P, Spengler G, Machado L, Martins A, Couto I, Viveiros M, Fanning S, Pagès JM. 2011. Ethidium bromide efflux by Salmonella: modulation by metabolic energy, pH, ions and phenothiazines. Int J Antimicrob Agents 38:140–145. 10.1016/j.ijantimicag.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 461.Mahamoud A, Chevalier J, Alibert-Franco S, Kern WV, Pagès JM. 2007. Antibiotic efflux pumps in Gram-negative bacteria: the inhibitor response strategy. J Antimicrob Chemother 59:1223–1229. 10.1093/jac/dkl493. [DOI] [PubMed] [Google Scholar]
- 462.Misra R, Bavro VN. 2009. Assembly and transport mechanism of tripartite drug efflux systems. Biochim Biophys Acta 1794:817–825. 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.van Amsterdam K, Bart A, van der Ende A. 2005. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother 49:1477–1482. 10.1128/AAC.49.4.1477-1482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Bina JE, Alm RA, Uria-Nickelsen M, Thomas SR, Trust TJ, Hancock RE. 2000. Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother 44:248–254. 10.1128/AAC.44.2.248-254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Liu ZQ, Zheng PY, Yang PC. 2008. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol 14:5217–5222. 10.3748/wjg.14.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Mehrabadi JF, Sirous M, Daryani NE, Eshraghi S, Akbari B, Shirazi MH. 2011. Assessing the role of the RND efflux pump in metronidazole resistance of Helicobacter pylori by RT-PCR assay. J Infect Dev Ctries 5:88–93. 10.3855/jidc.1187. [DOI] [PubMed] [Google Scholar]
- 467.Hirata K, Suzuki H, Nishizawa T, Tsugawa H, Muraoka H, Saito Y, Matsuzaki J, Hibi T. 2010. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol 25(Suppl 1):S75–S79. 10.1111/j.1440-1746.2009.06220.x. [DOI] [PubMed] [Google Scholar]
- 468.Kouitcheu Mabeku LB, Eyoum Bille B, Tepap Zemnou C, Tali Nguefack LD, Leundji H. 2019. Broad spectrum resistance in Helicobacter pylori isolated from gastric biopsies of patients with dyspepsia in Cameroon and efflux-mediated multi-resistance detection in MDR isolates. BMC Infect Dis 19:880. 10.1186/s12879-019-4536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Cai Y, Wang C, Chen Z, Xu Z, Li H, Li W, Sun Y. 2020. Transporters HP0939, HP0497, and HP0471 participate in intrinsic multidrug resistance and biofilm formation in Helicobacter pylori by enhancing drug efflux. Helicobacter 25:e12715. 10.1111/hel.12715. [DOI] [PubMed] [Google Scholar]
- 470.Ge X, Cai Y, Chen Z, Gao S, Geng X, Li Y, Li Y, Jia J, Sun Y. 2018. Bifunctional enzyme SpoT is involved in biofilm formation of Helicobacter pylori with multidrug resistance by upregulating efflux pump Hp1174 (gluP). Antimicrob Agents Chemother 62:e00957-18. 10.1128/AAC.00957-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Attaran B, Falsafi T, Ghorbanmehr N. 2017. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J Gastroenterol 23:1163–1170. 10.3748/wjg.v23.i7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Zhou YN, Coleman WJ, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. 2008. Regulation of cell growth during serum starvation and bacterial survival in macrophages by the bifunctional enzyme SpoT in Helicobacter pylori. J Bacteriol 190:8025–8032. 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Geng X, Li W, Chen Z, Gao S, Hong W, Ge X, Hou G, Hu Z, Zhou Y, Zeng B, Li W, Jia J, Sun Y. 2017. The bifunctional enzyme SpoT is involved in the clarithromycin tolerance of Helicobacter pylori by upregulating the transporters HP0939, HP1017, HP0497, and HP0471. Antimicrob Agents Chemother 61:e02011-16. 10.1128/AAC.02011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Yonezawa H, Osaki T, Kamiya S. 2015. Biofilm formation by Helicobacter pylori and its involvement for antibiotic resistance. Biomed Res Int 2015:914791. 10.1155/2015/914791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Krzyżek P, Grande R, Migdał P, Paluch E, Gościniak G. 2020. Biofilm formation as a complex result of virulence and adaptive responses of Helicobacter pylori. Pathogens 9:1062. 10.3390/pathogens9121062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Attaran B, Falsafi T, Moghaddam AN. 2016. Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J Gastroenterol 22:161–168. 10.4103/1319-3767.178529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Carron MA, Tran VR, Sugawa C, Coticchia JM. 2006. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg 10:712–717. 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 478.Cellini L, Grande R, Di Campli E, Traini T, Di Giulio M, Lannutti SN, Lattanzio R. 2008. Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand J Gastroenterol 43:178–185. 10.1080/00365520701675965. [DOI] [PubMed] [Google Scholar]
- 479.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. 2006. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg 10:883–889. 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 480.Matteo MJ, Granados G, Olmos M, Wonaga A, Catalano M. 2008. Helicobacter pylori amoxicillin heteroresistance due to point mutations in PBP-1A in isogenic isolates. J Antimicrob Chemother 61:474–477. 10.1093/jac/dkm504. [DOI] [PubMed] [Google Scholar]
- 481.Selgrad M, Tammer I, Langner C, Bornschein J, Meißle J, Kandulski A, Varbanova M, Wex T, Schlüter D, Malfertheiner P. 2014. Different antibiotic susceptibility between antrum and corpus of the stomach, a possible reason for treatment failure of Helicobacter pylori infection. World J Gastroenterol 20:16245–16251. 10.3748/wjg.v20.i43.16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Amitrano M, Spezzaferro M, Sacco F, Serio M, Cerasa B, Grossi L, Marzio L. 2008. H. pylori isolates from proximal and distal stomach of patients with H. pylori infection exhibit resistance and sensitivity to the same antibiotic. Gastroenterology 134:A-335. 10.1016/S0016-5085(08)61563-2. [DOI] [Google Scholar]
- 483.de la Obra P, Alarcon T, Domingo D, Garcia J, Lopez-Brea M. 2001. Heteroresistance to metronidazole and genetic relationship of Helicobacter pylori clinical isolates. Gut 49:A4. [Google Scholar]
- 484.Andersson DI, Nicoloff H, Hjort K. 2019. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol 17:479–496. 10.1038/s41579-019-0218-1. [DOI] [PubMed] [Google Scholar]
- 485.Dewachter L, Fauvart M, Michiels J. 2019. Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol Cell 76:255–267. 10.1016/j.molcel.2019.09.028. [DOI] [PubMed] [Google Scholar]
- 486.Mascellino MT, Oliva A, De Angelis M, Porowska B. 2015. Helicobacter pylori infection: susceptibility to antimicrobials and eradication rate in pluritreated pangastritis patients. Indian J Appl Res 5:30–32. 10.36106/ijar. [DOI] [Google Scholar]
- 487.Gravina A, Federico A, Dallio M, Sgambato D, Miranda A, Tuccillo C, Loguercio C, Romano M. 2016. Intrafamilial spread of Helicobacter pylori infection. J Gastroenterol Hepatol Endosc 1:1003. [Google Scholar]
- 488.Nicoloff H, Hjort K, Levin BR, Andersson DI. 2019. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4:504–514. 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 489.Balaji V, Jeremiah SS, Baliga PR. 2011. Polymyxins: antimicrobial susceptibility concerns and therapeutic options. Indian J Med Microbiol 29:230–242. 10.4103/0255-0857.83905. [DOI] [PubMed] [Google Scholar]
- 490.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 13:1057–1098. 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 491.Dang BN, Graham DY. 2017. Helicobacter pylori infection and antibiotic resistance: a WHO high priority? Nat Rev Gastroenterol Hepatol 14:383–384. 10.1038/nrgastro.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.The European Committee on Antimicrobial Susceptibility Testing. 2022. Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf.
- 493.Clinical and Laboratory Standard Institute. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed. CLSI guideline M45. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 494.Huang X, Liu Y, Lin Z, Wu B, Nong G, Chen Y, Lu Y, Ji X, Zhou X, Suo B, Chen Q, Wei J. 2021. Minimum inhibitory concentrations of commonly used antibiotics against Helicobacter pylori: a multicenter study in South China. PLoS One 16:e0256225. 10.1371/journal.pone.0256225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Bai P, Zhou LY, Xiao XM, Luo Y, Ding Y. 2015. Susceptibility of Helicobacter pylori to antibiotics in Chinese patients. J Dig Dis 16:464–470. 10.1111/1751-2980.12271. [DOI] [PubMed] [Google Scholar]
- 496.Su P, Li Y, Li H, Zhang J, Lin L, Wang Q, Guo F, Ji Z, Mao J, Tang W, Shi Z, Shao W, Mao J, Zhu X, Zhang X, Tong Y, Tu H, Jiang M, Wang Z, Jin F, Yang N, Zhang J. 2013. Antibiotic resistance of Helicobacter pylori isolated in the southeast coastal region of China. Helicobacter 18:274–279. 10.1111/hel.12046. [DOI] [PubMed] [Google Scholar]
- 497.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y, Study Group participants. 2013. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 62:34–42. 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 498.Valdivieso-García A, Imgrund R, Deckert A, Varughese BM, Harris K, Bunimov N, Reid-Smith R, McEwen S. 2009. Cost analysis and antimicrobial susceptibility testing comparing the E-test and the agar dilution method in Campylobacter jejuni and Campylobacter coli. Diagn Microbiol Infect Dis 65:168–174. 10.1016/j.diagmicrobio.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 499.Azzaya D, Gantuya B, Oyuntsetseg K, Davaadorj D, Matsumoto T, Akada J, Yamaoka Y. 2020. High antibiotic resistance of Helicobacter pylori and its associated novel gene mutations among the Mongolian population. Microorganisms 8:1062. 10.3390/microorganisms8071062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Lok CH, Zhu D, Wang J, Ren YT, Jiang X, Li SJ, Zhao XY. 2020. Phenotype and molecular detection of clarithromycin and levofloxacin resistance in Helicobacter pylori clinical isolates in Beijing. Infect Drug Resist 13:2145–2153. 10.2147/IDR.S249370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Coudron PE, Stratton CW. 1995. Factors affecting growth and susceptibility testing of Helicobacter pylori in liquid media. J Clin Microbiol 33:1028–1030. 10.1128/jcm.33.4.1028-1030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Hachem CY, Clarridge JE, Reddy R, Flamm R, Evans DG, Tanaka SK, Graham DY. 1996. Antimicrobial susceptibility testing of Helicobacter pylori. Comparison of E-test, broth microdilution, and disk diffusion for ampicillin, clarithromycin, and metronidazole. Diagn Microbiol Infect Dis 24:37–41. 10.1016/0732-8893(95)00252-9. [DOI] [PubMed] [Google Scholar]
- 503.Piccolomini R, Di Bonaventura G, Catamo G, Carbone F, Neri M. 1997. Comparative evaluation of the E-test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J Clin Microbiol 35:1842–1846. 10.1128/jcm.35.7.1842-1846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Miftahussurur M, Fauzia KA, Nusi IA, Setiawan PB, Syam AF, Waskito LA, Doohan D, Ratnasari N, Khomsan A, Adnyana IK, Akada J, Yamaoka Y. 2020. E-test versus agar dilution for antibiotic susceptibility testing of Helicobacter pylori: a comparison study. BMC Res Notes 13:22. 10.1186/s13104-019-4877-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Grande R, Sisto F, Puca V, Carradori S, Ronci M, Aceto A, Muraro R, Mincione G, Scotti L. 2020. Antimicrobial and antibiofilm activities of new synthesized silver ultra-nanoclusters (SUNCs) against Helicobacter pylori. Front Microbiol 11:1705. 10.3389/fmicb.2020.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 507.Camorlinga-Ponce M, Gó Mez-Delgado A, Aguilar-Zamora E, Torres RC, Giono-Cerezo S, Escobar-Ogaz A, Torres J. 2021. Phenotypic and genotypic antibiotic resistance patterns in Helicobacter pylori strains from ethnically diverse population in Mexico. Front Cell Infect Microbiol 10:539115. 10.3389/fcimb.2020.539115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Fauzia KA, Miftahussurur M, Syam AF, Waskito LA, Doohan D, Rezkitha YAA, Matsumoto T, Tuan VP, Akada J, Yonezawa H, Kamiya S, Yamaoka Y. 2020. Biofilm formation and antibiotic resistance phenotype of Helicobacter pylori clinical isolates. Toxins (Basel) 12:473. 10.3390/toxins12080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Glupczynski Y, Labbé M, Hansen W, Crokaert F, Yourassowsky E. 1991. Evaluation of the E test for quantitative antimicrobial susceptibility testing of Helicobacter pylori. J Clin Microbiol 29:2072–2075. 10.1128/jcm.29.9.2072-2075.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Hirschl AM, Hirschl MM, Rotter ML. 1993. Comparison of three methods for the determination of the sensitivity of Helicobacter pylori to metronidazole. J Antimicrob Chemother 32:45–49. 10.1093/jac/32.1.45. [DOI] [PubMed] [Google Scholar]
- 511.Glupczynski Y, Broutet N, Cantagrel A, Andersen LP, Alarcon T, López-Brea M, Mégraud F. 2002. Comparison of the E-test and agar dilution method for antimicrobial suceptibility testing of Helicobacter pylori. Eur J Clin Microbiol Infect Dis 21:549–552. 10.1007/s10096-002-0757-6. [DOI] [PubMed] [Google Scholar]
- 512.Perna F, Gatta L, Figura N, Ricci C, Tampieri A, Holton J, Miglioli M, Vaira D. 2003. Susceptibility of Helicobacter pylori to metronidazole. Am J Gastroenterol 98:2157–2161. 10.1111/j.1572-0241.2003.07681.x. [DOI] [PubMed] [Google Scholar]
- 513.Lang L, García F. 2004. Comparison of E-test and disk diffusion assay to evaluate resistance of Helicobacter pylori isolates to amoxicillin, clarithromycin, metronidazole and tetracycline in Costa Rica. Int J Antimicrob Agents 24:572–577. 10.1016/j.ijantimicag.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 514.Zhong Z, Zhang Z, Wang J, Hu Y, Mi Y, He B, Zhang Y, Zhang X, Xia X, Huang H, Lai Y, Lin M, Su C, Zhang Z, Wu Z, Lu L, Zhang B, Huang S, Zhong C, Zeng X, Peng Y, Chen G, Zhang H, Zhou G, Liu S, Yang C, Yan L, Chen A, Zhang G, Xu P, Wang S, Zheng P, Xu S, Gao H. 2021. A retrospective study of the antibiotic-resistant phenotypes and genotypes of Helicobacter pylori strains in China. Am J Cancer Res 11:5027–5037. [PMC free article] [PubMed] [Google Scholar]
- 515.McNulty C, Owen R, Tompkins D, Hawtin P, McColl K, Price A, Smith G, Teare L, PHLS Helicobacter Working Group. 2002. Helicobacter pylori susceptibility testing by disc diffusion. J Antimicrob Chemother 49:601–609. 10.1093/jac/49.4.601. [DOI] [PubMed] [Google Scholar]
- 516.Nakashima H, Takahashi H, Kameko M, Saito H. 2012. Daptomycin E-test MICs for methicillin-resistant Staphylococcus aureus vary among different media. J Infect Chemother 18:970–972. 10.1007/s10156-012-0424-5. [DOI] [PubMed] [Google Scholar]
- 517.Miller SA, Karichu J, Kohner P, Cole N, Hindler JA, Patel R, Richter S, Humphries RM. 2017. Multicenter evaluation of a modified cefoxitin disk diffusion method and PBP2a testing to predict mecA-mediated oxacillin resistance in atypical Staphylococcus aureus. J Clin Microbiol 55:485–494. 10.1128/JCM.02211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Gong RJ, Xu CX, Li H, Liu XM. 2021. Polymerase chain reaction-based tests for detecting Helicobacter pylori clarithromycin resistance in stool samples: a meta-analysis. World J Clin Cases 9:133–147. 10.12998/wjcc.v9.i1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Anjum MF, Zankari E, Hasman H. 2017. Molecular methods for detection of antimicrobial resistance. Microbiol Spectr 5:ARBA-0011-2017. 10.1128/microbiolspec.ARBA-0011-2017. [DOI] [PubMed] [Google Scholar]
- 520.Jenks PJ. 2002. Causes of failure of eradication of Helicobacter pylori. BMJ 325:3–4. 10.1136/bmj.325.7354.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Nezami BG, Jani M, Alouani D, Rhoads DD, Sadri N. 2019. Helicobacter pylori mutations detected by next-generation sequencing in formalin-fixed, paraffin-embedded gastric biopsy specimens are associated with treatment failure. J Clin Microbiol 57:e01834-18. 10.1128/JCM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Palmitessa V, Monno R, Panarese A, Cuppone R, Burattini O, Marangi S, Curlo M, Fumarola L, Petrosillo A, Parisi A, Capozzi L, Bianco A, Lippolis A. 2020. Evaluation of antibiotic resistance of Helicobacter pylori strains isolated in Bari, southern Italy, in 2017–2018 by phenotypic and genotyping methods. Microb Drug Resist 26:909–917. 10.1089/mdr.2019.0262. [DOI] [PubMed] [Google Scholar]
- 523.Xuan SH, Wu LP, Zhou YG, Xiao MB. 2016. Detection of clarithromycin-resistant Helicobacter pylori in clinical specimens by molecular methods: a review. J Glob Antimicrob Resist 4:35–41. 10.1016/j.jgar.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 524.Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. 2010. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol 8:309–312. 10.1016/j.cgh.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 525.Jehanne Q, Bénéjat L, Mégraud F, Bessède E, Lehours P. 2020. Evaluation of the Allplex H. pylori and ClariR PCR assay for Helicobacter pylori detection on gastric biopsies. Helicobacter 25:e12702. 10.1111/hel.12702. [DOI] [PubMed] [Google Scholar]
- 526.Redondo JJ, Keller PM, Zbinden R, Wagner K. 2018. A novel RT-PCR for the detection of Helicobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. Diagn Microbiol Infect Dis 90:1–6. 10.1016/j.diagmicrobio.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 527.Hays C, Delerue T, Lamarque D, Burucoa C, Collobert G, Billöet A, Kalach N, Raymond J. 2019. Molecular diagnosis of Helicobacter pylori infection in gastric biopsies: evaluation of the Amplidiag H. pylori + ClariR assay. Helicobacter 24:e12560. 10.1111/hel.12560. [DOI] [PubMed] [Google Scholar]
- 528.Li Y, Lv T, He C, Wang H, Cram DS, Zhou L, Zhang J, Jiang W. 2020. Evaluation of multiplex ARMS-PCR for detection of Helicobacter pylori mutations conferring resistance to clarithromycin and levofloxacin. Gut Pathog 12:35. 10.1186/s13099-020-00373-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Haumaier F, Schneider-Fuchs A, Backert S, Vieth M, Sterlacci W, Wöhrl BM. 2022. Rapid detection of quinolone resistance mutations in gyrA of Helicobacter pylori by real-time PCR. Pathogens 11:59. 10.3390/pathogens11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Lauener FN, Imkamp F, Lehours P, Buissonnière A, Benejat L, Zbinden R, Keller PM, Wagner K. 2019. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. J Clin Med 8:53. 10.3390/jcm8010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Jiang F, Wu Z, Zheng Y, Frana TS, Sahin O, Zhang Q, Li G. 2019. Genotypes and antimicrobial susceptibility profiles of hemolytic Escherichia coli from diarrheic piglets. Foodborne Pathog Dis 16:94–103. 10.1089/fpd.2018.2480. [DOI] [PubMed] [Google Scholar]
- 532.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain JM, Samuelsen Ø, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 533.Boyanova L, Nikolov R, Gergova G, Evstatiev I, Lazarova E, Kamburov V, Panteleeva E, Spassova Z, Mitov I. 2010. Two-decade trends in primary Helicobacter pylori resistance to antibiotics in Bulgaria. Diagn Microbiol Infect Dis 67:319–326. 10.1016/j.diagmicrobio.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 534.Tuan VP, Narith D, Tshibangu-Kabamba E, Dung HDQ, Viet PT, Sokomoth S, Binh TT, Sokhem S, Tri TD, Ngov S, Tung PH, Thuan NPM, Truc TC, Phuc BH, Matsumoto T, Fauzia KA, Akada J, Trang TTH, Yamaoka Y. 2019. A next-generation sequencing-based approach to identify genetic determinants of antibiotic resistance in Cambodian Helicobacter pylori clinical isolates. J Clin Med 8:858. 10.3390/jcm8060858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Serratì S, De Summa S, Pilato B, Petriella D, Lacalamita R, Tommasi S, Pinto R. 2016. Next-generation sequencing: advances and applications in cancer diagnosis. Oncotargets Ther 9:7355–7365. 10.2147/OTT.S99807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Egli K, Wagner K, Keller PM, Risch L, Risch M, Bodmer T. 2020. Comparison of the diagnostic performance of qPCR, Sanger sequencing, and whole-genome sequencing in determining clarithromycin and levofloxacin resistance in Helicobacter pylori. Front Cell Infect Microbiol 10:596371. 10.3389/fcimb.2020.596371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Hendriksen RS, Bortolaia V, Tate H, Tyson GH, Aarestrup FM, McDermott PF. 2019. Using genomics to track global antimicrobial resistance. Front Public Health 7:242. 10.3389/fpubh.2019.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Su M, Satola SW, Read TD. 2019. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57:e01405-18. 10.1128/JCM.01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. 2015. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio 6:e01888-15. 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.MacCannell D. 2016. Next generation sequencing in clinical and public health microbiology. Clin Microbiol Newsl 38:169–176. 10.1016/j.clinmicnews.2016.10.001. [DOI] [Google Scholar]
- 541.Argueta EA, Alsamman MA, Moss SF, D’Agata EMC. 2021. Impact of antimicrobial resistance rates on eradication of Helicobacter pylori in a US population. Gastroenterol 160:2181–2183.E1. 10.1053/j.gastro.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Mascellino MT, Oliva A, Miele MC, De Angelis M, Bruno G, Severi C. 2020. Secondary antibiotic resistance, correlation between genotypic and phenotypic methods and treatment in Helicobacter pylori infected patients: a retrospective study. Antibiotics (Basel) 9:549. 10.3390/antibiotics9090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Song Y, Dou F, Zhou Z, Yang N, Zhong J, Pan J, Liu Q, Zhang J, Wang S. 2018. Microarray-based detection and clinical evaluation for Helicobacter pylori resistance to clarithromycin or levofloxacin and the genotype of CYP2C19 in 1083 patients. Biomed Res Int 2018:2684836. 10.1155/2018/2684836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Binmaeil H, Hanafiah A, Mohamed Rose I, Raja Ali RA. 2021. Development and validation of multiplex quantitative PCR assay for detection of Helicobacter pylori and mutations conferring resistance to clarithromycin and levofloxacin in gastric biopsy. Infect Drug Resist 14:4129–4145. 10.2147/IDR.S325056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Pastukh N, Binyamin D, On A, Paritsky M, Peretz A. 2017. GenoType HelicoDR test in comparison with histology and culture for Helicobacter pylori detection and identification of resistance mutations to clarithromycin and fluoroquinolones. Helicobacter 22:e12447. 10.1111/hel.12447. [DOI] [PubMed] [Google Scholar]
- 546.Kuo YT, Liou JM, El-Omar EM, Wu JY, Leow AHR, Goh KL, Das R, Lu H, Lin JT, Tu YK, Yamaoka Y, Wu MS, Asian Pacific Alliance on Helicobacter and Microbiota. 2017. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2:707–715. 10.1016/S2468-1253(17)30219-4. [DOI] [PubMed] [Google Scholar]
- 547.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 548.Gingold-Belfer R, Niv Y, Schmilovitz-Weiss H, Levi Z, Boltin D. 2021. Susceptibility-guided versus empirical treatment for Helicobacter pylori infection: a systematic review and meta-analysis. J Gastroenterol Hepatol 36:2649–2658. 10.1111/jgh.15575. [DOI] [PubMed] [Google Scholar]
- 549.Lee JH, Ahn JY, Choi KD, Jung HY, Kim JM, Baik GH, Kim BW, Park JC, Jung HK, Cho SJ, Shin CM, Choi YJ, Lee SH, Kim JH, Lee WS, Sung JK, Chung JW, Cheung DY, Lee H, Min YW, Kim JJ, Kim SY, Korean College of Helicobacter, Upper Gastrointestinal Research. 2019. Nationwide antibiotic resistance mapping of Helicobacter pylori in Korea: a prospective multicenter study. Helicobacter 24:e12592. 10.1111/hel.12592. [DOI] [PubMed] [Google Scholar]
- 550.Graham DY, Liou JM. 2021. Primer for development of guidelines for Helicobacter pylori therapy using antimicrobial stewardship. Clin Gastroenterol Hepatol 3565:e00336. 10.1016/j.cgh.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Shiotani A, Roy P, Lu H, Graham DY. 2021. Helicobacter pylori diagnosis and therapy in the era of antimicrobial stewardship. Therap Adv Gastroenterol 14:17562848211064080. 10.1177/17562848211064080. [DOI] [PMC free article] [PubMed] [Google Scholar]