SUMMARY
Antimicrobial resistance (AMR) is a global health crisis that poses a great threat to modern medicine. Effective prevention strategies are urgently required to slow the emergence and further dissemination of AMR. Given the availability of data sets encompassing hundreds or thousands of pathogen genomes, machine learning (ML) is increasingly being used to predict resistance to different antibiotics in pathogens based on gene content and genome composition. A key objective of this work is to advocate for the incorporation of ML into front-line settings but also highlight the further refinements that are necessary to safely and confidently incorporate these methods. The question of what to predict is not trivial given the existence of different quantitative and qualitative laboratory measures of AMR. ML models typically treat genes as independent predictors, with no consideration of structural and functional linkages; they also may not be accurate when new mutational variants of known AMR genes emerge. Finally, to have the technology trusted by end users in public health settings, ML models need to be transparent and explainable to ensure that the basis for prediction is clear. We strongly advocate that the next set of AMR-ML studies should focus on the refinement of these limitations to be able to bridge the gap to diagnostic implementation.
KEYWORDS: antimicrobial resistance, machine learning
INTRODUCTION
The antimicrobial resistance (AMR) crisis is responsible for more than 1.27 million deaths per year worldwide (1). It is accompanied by high economic costs due to associated morbidity and mortality, the need for additional diagnostics and treatments for drug-resistant infections (DRI), and prolonged hospital admissions (2). Such impacts have driven governing authorities to identify several key objectives to rectify the urgent issue, including improved surveillance and the development of rapid diagnostics (3–5). Surveillance will inform when and where AMR is occurring to improve policies on antimicrobial use (AMU) in human and animal health, and improved diagnostics will aid in the effective use and stewardship of the already limited armory of antimicrobials. Depending on the purpose of AMR detection, different requirements are necessary in terms of speed, accuracy, and mechanistic understanding. For example, clinical diagnostic AMR detection requires high speed and accuracy to guide urgent treatment protocols, whereas research and surveillance are generally less time critical.
Current laboratory-based diagnostic and characterization methods for priority pathogens do not provide all information needed for effective surveillance. Moreover, routine procedures for antibiotic susceptibility testing (AST) do not always yield consistent results between different phenotypic methods or across different laboratories (6, 7). The data extracted from high sequencing, in conjunction with lab-based methods, can help overcome such obstacles (8). Specifically, whole-genome sequence (WGS) data can be used to monitor which genetic variants associated with AMR are widespread. One can also infer an organism’s functional potential from sequence information, making rapid diagnostics possible.
Different strategies have been used to link genomic information with AMR phenotype. A common set of genomic methods to study functional potential are genome-wide association studies (GWAS), which test the statistical associations between genetic variants in whole genomes and the corresponding AMR phenotypes to potentially discover new resistance determinants (9–11). Classical GWAS often takes a single-locus approach, providing interpretable results with P values for each identified variant (12). However, given that phenotype is often the product of epistatic interactions of variants rather than the product of individual loci (13, 14), this approach can lead to reduced prediction accuracy. Microbial genomes provide extra difficulties for the single-locus approach in the form of increased heterogeneity due to factors such as the potential for high rates of recombination and horizontal gene transfer (HGT) events. This is especially relevant for AMR genes, as they are often horizontally transferred and are part of a dynamic accessory genome (15, 16). To deal with such limitations, microbial GWAS have increasingly adopted multilocus approaches and methods derived from machine learning (ML) (17, 18).
ML methods employ algorithms to learn and predict AMR phenotypes directly from sequenced bacterial genomes, which can be rapid and highly accurate. However, much as with GWAS adopting ML influenced approaches, ML methods can also incorporate elements traditionally associated with GWAS, such as association tests (19) and correction for population structure (20). This overlap between ML and GWAS approaches and the influence they exert on one another can make it increasingly difficult and contentious to differentiate between them (much as with ML and statistics [21]). Arguably the main difference in these approaches is a matter of prioritization: GWAS focuses on detection of strong (putatively causal) associations between genomic features and phenotype, whereas ML attempts to maximize the predictive value of genomic features.
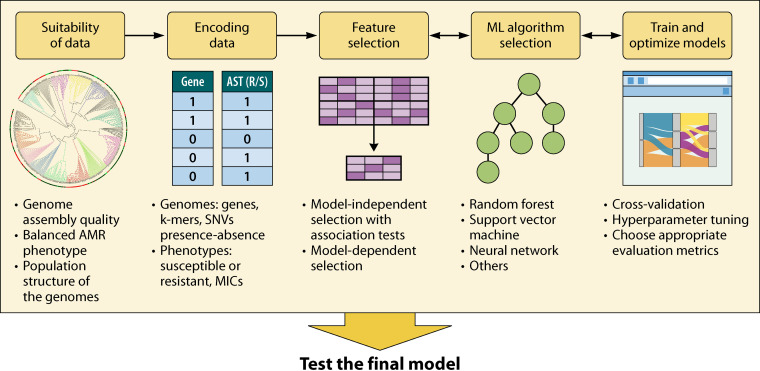
ML implementation has gained popularity for AMR phenotype prediction studies, which can support surveillance and precise diagnosis as well as offering the chance to explore the mechanisms driving AMR (22–24). In spite of the promise of ML, relatively few methods have been adopted in clinical or broader public-health settings (25). A recent review by Anahtar and colleagues (26) summarizes the recent findings in the field with a focus on implementing the ML technology in electronic health records to aid antimicrobial stewardship and combat AMR. In this review, we present key developments in AMR prediction using ML and suggest appropriate practices (Fig. 1), examine current limitations, and propose future directions. Specifically, we advocate for collaborative and interdisciplinary efforts with basic science research, which will push ML applications to fully integrate into public health and clinical usage.
FIG 1.
Workflow for ML prediction of AMR.
MACHINE LEARNING FOR AMR PREDICTION
ML methods for AMR prediction typically use supervised learning, where a set of data with known labels (e.g., genome assemblies and their corresponding antimicrobial susceptibility profiles) are used to train and test a predictive model (27). The goal is to learn a set of rules or functions that can transform input genetic data or “features” (e.g., genes or single nucleotide variants [SNVs] in genomic context) into output predictions (i.e., labels) that are interpreted as phenotypes. ML has been applied to AMR prediction in pathogens such as nontyphoidal Salmonella and Mycobacterium tuberculosis with well-characterized resistance mechanisms. For example, the abundance of nontyphoidal Salmonella genomes and the corresponding antimicrobial susceptibility profiles obtained through programs like the National Antimicrobial Resistance Monitoring System (NARMS) led to the implementation of ML methods to predict the MICs within ±1 2-fold dilution range, which resulted in a model with an overall average prediction accuracy of 95% (28). The aim of the investigation was to develop ML models that can predict MIC values that potentially can guide responses to outbreaks and inform antibiotic stewardship decisions. As NARMS tracks select enteric bacteria found in ill people, retail meats, and food animals in the United States, the MIC prediction models based on such data may have wider applicability for various sectors, like medicine and agriculture.
For multidrug-resistant M. tuberculosis, well-defined SNVs associated with AMR were used to yield multidrug prediction models with the highest sensitivity of 96.3% (29). The investigation’s aim was to apply a recently developed ML technique, the deep denoising auto-encoder, to learn the M. tuberculosis isolates collected from 16 countries and predict their multidrug resistance. Multiple-drug classification is not yet as common as single antibiotic resistance prediction models, and the model was able to outperform other more commonly used classifiers. As most pathogens are now resistant to multiple antibiotics, multidrug-resistant models better emulate the current state of AMR.
In addition to showing high accuracy in resistance phenotype prediction, ML models can be further dissected to investigate the drivers of AMR. Maguire et al. applied an ML model to Salmonella WGSs isolated from broiler chickens (30). The authors were able to construct models with average precision ranging from 0.91 to 0.98, achieved through learning the known AMR genes in WGSs. The authors also identified the main genetic drivers for resistance in the studied group of Salmonella, which corresponded with previously identified AMR mechanisms for respective antibiotics (30). In Kavvas et al. the authors used ML models to find known AMR-implicated genes of M. tuberculosis, uncover new putative AMR determinants, and resolve potential epistatic interactions driving AMR evolution (31). The goal of the study was to extract key insights from AMR genomic data instead of predicting resistance with high accuracy. Tsang et al. combined explainable ML models with Escherichia coli targeted gene expression experiments of the selected features from the model. This uncovered previously unknown substrate activity for known β-lactamases (32).
These examples demonstrate that trained ML models can potentially answer a wide array of research questions beyond phenotypic resistance prediction. However, an essential predicate to the application of ML techniques is their ability to generalize (i.e., to make accurate predictions on data sets beyond those that were used to train and test the model).
Suitability of Genomic Data Sets
A critical first question surrounds the amount of data required for ML analysis. An appropriate but unsatisfying answer is that the genomic data set size required to implement ML depends on the prediction problem in question, although “sufficiently large” is often implicitly defined based on the availability of data rather than any theoretical lower bound of required numbers of genomes. In general, larger data sets offer more information to support the identification of key predictors, especially when certain predictors are relatively rare, but complex resistance mechanisms and nonrandom sampling can limit the effectiveness of additional genomes within a data set.
If the bacterial species of interest exhibits low DNA sequence and gene content diversity between isolates (e.g., highly clonal M. tuberculosis) with well-known high-penetrance AMR genes, fewer genomes will suffice to identify key genes or sequence features that predict the phenotype of interest. The reality is that there are many factors interfering with the linear process of resistance genotype to phenotype translation. That is, resistance phenotypes are the product of multiple genes of various penetrance. Only accounting for the high-penetrance AMR genes with a limited number of genomes is not a suitable approach for a number of species. Organisms with higher genomic diversity (e.g., highly recombining Helicobacter pylori) (33) will have many differences that are not associated with the AMR phenotype. In such instances, a larger data set is required to identify the key AMR features against a large backdrop of noise that may or may not be relevant to AMR.
Recent AMR prediction studies vary widely in the number of genomes used, from 97 nontyphoidal Salmonella genomes isolated from broiler chickens (30) to a multispecies bacterial study containing more than 7,000 genomes (34). Each study resolved ML models with high precision (>0.90) (30) and readily differentiated resistant from susceptible phenotypes (34). Regardless of the number of sampled genomes, nonrandom sampling of pathogen diversity induces correlation structure within the set of sampled pathogens, which can reduce the effective size of the data set. Samples from similar locations, times, and habitat types can produce large sets of very similar genomes, and the resulting population structure can be a significant confounder in the ML model. This structure can also provide less information to a prediction model than a more evenly sampled data set, resulting in a prediction model that is constrained to the characteristics of the data set and not broadly applicable. Models are more likely to associate features that are common in the background of closely related strains to the AMR phenotype, and controlling for this effect in studies with unevenly sampled data is vital (35, 36).
For example, Hicks et al. demonstrated skewed resistance model performance based on sampling bias (37). Gonococcal models from one clinical source versus aggregated data from various clinical sources showed substantial variation in predictive performance, which was attributed to the population structure (37). Consideration of population effects is especially relevant for studies developing prediction models from a multisectoral One-Health perspective with genomic data sets of various origins. Although there is overlap in the genomic content of samples from different environments, clinical and agricultural settings can possess distinct sets of AMR determinants and genomic backgrounds (38–40). This habitat sorting of determinants is enhanced by horizontal transfer of genes present on mobile genetic elements. Given such evidence, it is important to consider that an ML model trained on samples from one source may be ineffective in predicting the resistance phenotype for isolates from different environments, and universal ML models for AMR may not be feasible. Rather than focusing on constructing universal ML models, adapting ML models based on the genomic epidemiology of the concerned species could create more generalizable ML models that reduce the confounding effect of source location.
Another common concern is class imbalance, where the majority of the data are comprised of one phenotype. In many AMR phenotype prediction studies, representation of resistant (R) and susceptible (S) phenotypes is often uneven, in part due to focus of sequencing efforts on isolates associated with poor clinical outcomes or just the rarity of certain types of resistance in a population. If a classifier naïvely learns from an imbalanced data set containing predominantly resistant instances, it may predict all genomes as resistant and be inaccurate for identifying susceptible phenotypes. Resampling techniques such as the Synthetic Minority Oversampling Technique (SMOTE) (41, 42) aim to increase the size of the underrepresented set by interpolating between existing samples or otherwise generating new points from an inferred distribution. An alternative is to reweight cases to increase the importance of the minority class relative to the majority phenotype or to undersample the majority class. However, the use of such methods in AMR studies has been limited, and the utility of resampling does not always guarantee improved model accuracy, especially when data sets are very small (43), such as in the case of recently emerged resistant strains. Expanding genomic surveillance efforts would benefit ML studies by sequencing a broad diversity of antimicrobial-susceptible and -resistant isolates.
It is also imperative to explore and evaluate genome assembly quality. Poor assemblies can confound the learning algorithm and lead to inaccurate prediction if key features are missing or false features are introduced. For example, an assembled genome with a high contamination rate may associate the contaminated sequences as the key contributor to the phenotype of interest. To establish quality control metrics such as number of reads, average read length, and depth of coverage of assemblies (44), tools like QUAST (45) are used. The quantity and quality of assembled genomes increase as sequencing and genome assembly technologies advance. For example, hybrid short- and long-read assembly approaches have been used to reconstruct complete genomes and identify determinants in pathogens with highly diverse and mosaic resistance content in clinically relevant Neisseria gonorrhoeae (46) and agriculturally important Mannheimia haemolytica (47). Similarly, long-read sequencing and hybrid assembly have shown high efficacy in resolving the genetic context and plasmid associations of mobile AMR genes that are difficult to resolve with short-read WGS (48, 49). Nonetheless, sharing of raw sequencing results (e.g., FASTQ for short-read data deposited in an NCBI BioProject) along with assemblies and phenotypic data are critical for assessment of assembly quality as well as development of assembly-free ML approaches as described below.
Representing Genomes and Phenotype Labels
Once the suitability of genomic data sets has been confirmed, representations of the sampled genomes and associated AMR phenotypes must be created using an appropriate encoding scheme. In terms of phenotype encoding, the key consideration is whether we are interested in a regression approach to predict specific quantitative measures of resistance (e.g., MIC) from genomes or a classification approach to predict discrete categorical labels derived from MICs (e.g., susceptible/intermediate/resistant, or SIR, interpretive categories). SIR categories are based on a series of cutoffs determined by standard-setting organizations such as the Clinical and Laboratory Standards (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST; https://www.eucast.org/clinical_breakpoints/), with the exact cutoffs dependent on context, e.g., clinical versus veterinary. It is important to note that the cutoff guidelines do not exist for all antimicrobials, hence sometimes only MICs can be determined for epidemiological purposes.
The SIR interpretive categories are used to help guide antibiotic choice and dosing (50); however, there are differences between guidelines that have implications for AMR prediction applicability (51). The majority of ML AMR studies have focused on the classification of isolates into binary S/R phenotypes and bypass information that can be drawn from the intermediate, or I, category, as discussed in detail in “Limitations of ML analysis in AMR research,” below. On the other hand, while creating predictive models for the underlying MICs is possible (28, 52–54) and potentially provides models less prone to biases from categorical cutoffs derived from different guidelines, this approach is more difficult, as MICs cannot be interpreted based on single absolute values. To predict MICs, larger antimicrobial susceptibility test (AST) data sets are required, and we need to consider the complexities of MIC measurements (e.g., 2-fold serial dilutions, left and right censorship, etc.) and the suitability of ML methods for MIC interval regression (55).
In preparation for ML, genomes can be represented in several different ways, including gene presence and absence, mutations in specific genes, and compositional properties. Gene-based approaches can focus on known AMR genes by finding closest-matching known resistance genes using curated AMR databases such as the Comprehensive Antibiotic Resistance Database (CARD) and its Resistance Gene Identifier (RGI) software (56) or the PathoSystems Resource Integration Center (PATRIC)’s AMR-focused tools (57), among others (see, e.g., reference 58). Using different databases can lead to fluctuating results in identifying AMR genes and predicting AMR (59–61); hence, a harmonized approach incorporating more than one database may be necessary. Alternatively, more comprehensive functional assignments can be made using general-purpose genome annotation tools such as the Prokaryotic Genome Annotation Pipeline (62, 63) or Prokka (64), which draw on multiple reference databases (65–67).
Even simple gene-focused genomic encoding approaches contain a lot of nuance. For example, Nguyen et al. (68) implemented a feature-encoding approach that used only core, non-AMR-associated genes from partial genome sequence data of Klebsiella pneumoniae, Mycobacterium tuberculosis, Salmonella enterica, and Staphylococcus aureus to construct ML models with incomplete WGS. Random nonoverlapping subsets of core genes were chosen to construct different models to eliminate the confounding effects of population structure. Although the accuracy of this approach was lower than that of models built from assembled genomes (52), the outcomes demonstrated that incomplete genome sequences, like those obtained without culturing from shotgun metagenomics or PCR-based amplification, can still contain discriminating information for S/R phenotype prediction. The exclusion of well-known AMR genes, many of which are often or always plasmid associated, also hints that other highly scored core gene features are associated with resistance phenotypes. The associated core genes may contribute to the clinically resistant phenotype, or they may correlate with the phenotype without themselves being causal; experimental work must be done to differentiate these two hypotheses. Such studies illustrate that alternative approaches to encoding genetic information can be used to generate new hypotheses for investigating AMR mechanisms.
Rather than considering the presence or absence of entire genes (known AMR determinants or otherwise), a genome can also be represented by the presence of all mutations, including SNVs, insertions, and deletions, relative to a known reference genome. Such encodings are especially suitable for bacterial species that show relatively low sequence diversity and limited evidence of HGT, such as clinically relevant M. tuberculosis (69) and agriculturally relevant Mycoplasma bovis (70). Many studies on drug-resistant M. tuberculosis have genomes at the level of SNVs (31, 71, 72). For example, the presence and absence of SNVs can be seen by comparing every nucleotide site different from the reference genome that contributes to amino acid substitution (71). Kavvas et al. (31) used a similar approach but avoided the need for a single reference genome by representing strain-to-strain variation within each protein-coding cluster. However, focusing only on genes, their nonsynonymous mutations, and their functional annotations can overlook the role of noncoding sequences and potentially unknown or poorly annotated resistance determinants. Instead, combining information from annotated genes and noncoding regulatory sequences may contribute significant insights on biological mechanisms underlying the predicted resistance phenotype.
Composition-based approaches offer a promising alternative to gene-centric feature-encoding methods by examining variation across the entire genome. In this approach, a genome is decomposed into fixed-length nucleotide sequences, referred to as k-mers, where k represents the nucleotide length. The presence and absence patterns of k-mers among resistant and susceptible isolates can then be used as features for ML. Analyzing the features used by trained models can help identify k-mers derived from resistance-associated genes or noncoding elements (73, 74). For example, 31-mer-based prediction analysis with 12 different pathogenic species yielded models that in 95% of cases had an accuracy of >80% and in nearly 50% of cases had accuracy of >95% (24), demonstrating the broad efficacy of this approach in resistance prediction. Notably, predictions using 31-mers could identify mutations previously known to confer resistance, increasing confidence in the method. A k value of 31 has been shown to be suitable for bacterial genome assembly (75) and reference-free bacterial genome comparisons (35, 73). However, investigators can examine the predictive ability of alternative lengths or even a range of k-mer sizes. k-mer methods additionally provide some benefits over other feature-encoding methods, as k-mer sets can be generated without alignments or reference genomes and can even be used on raw sequencing reads without genome assembly.
One can also combine different encoding approaches. Davies et al. chose several feature-encoding strategies to improve the challenging prediction of amoxicillin-clavulanate resistance in E. coli, including the presence/absence of well-known β-lactamase genes (76). Models based only on the presence of β-lactamase genes did not perform well, but the addition of features including promoter mutations and copy number estimates associated with β-lactamase hyperproduction improved performance. Although the authors used random-effect models and not ML, Davies and authors suggest that a broader set of genetic features improves the prediction outcome.
Feature Selection for Interpretable Models
The encoding methods described above can produce huge numbers of genetic features. For example, Drouin et al. generated up to 123 million k-mers from pathogen genomes (73). Large numbers of genetic features can lead to extremely long learning times and a failure to identify meaningful associations among candidate predictors. In general, only a very small subset of genes, k-mers, or mutations in a genome will correlate with or contribute to the resistance phenotype of bacteria. To simplify the learning process, a first pass of feature evaluation and selection can be conducted. Feature selection techniques often apply filter methods (77) that statistically evaluate the association of features with the phenotype of interest (similar to classical GWAS approaches), either one by one or in small subsets, prior to training an ML model. A benefit of this type of filtering is that it is computationally feasible even for very high-dimensional data. Pairwise association tests, such as the chi-square test, that eliminate features prior to constructing a model can help select those features with the highest predictive power.
Other types of feature selection methods exist, such as the model-dependent wrapper methods like the recursive feature elimination (RFE), which selects features based on weights (e.g., the coefficients of a linear model) and recursively eliminates the least important features, or the sequential feature selection (SFS), which can start with either zero or all features included in the model, with features added or removed until an optimal set is obtained (78, 79). If features are being added, the process stops when additional features yield no improvement to the model, whereas if features are being removed, the process stops when removal of additional features starts to reduce model accuracy. Lastly, some model types have built-in feature selection known as embedded methods. Feature selection can drastically reduce the complexity of the model and the learning time, making it more comprehensible to users when the model is dissected.
One disadvantage of conducting feature selection is the potential of losing prediction information based on rare resistant genetic variants. Therefore, it is recommended that investigators benchmark prediction performance with and without feature selection methods and thoroughly examine the features ranked highly by the selection method rather than following a completely automated process without human intervention.
Training and Testing Machine Learning Models
Processed input data with resistance phenotypes are typically divided into different subsets: training, validation, and a holdout test set. The training set is used to optimize the parameters of the predictive model with respect to some function measuring how well the desired output phenotypes are predicted. The validation set is used to evaluate the relative effectiveness of different types of models and model hyperparameters. Hyperparameters are important external variables related to the model (e.g., splitting criterion in a decision tree model) that must be set before a model can be trained and cannot be directly optimized during training. Careful tuning of hyperparameters can greatly increase the prediction performance of individual models. The test set is used only once to evaluate the performance of the final optimized predictive model, hence it should not be used by the classifier in any of the previous training and optimization steps. The performance on the test set expresses the ability of the model to generalize its predictions to new or larger data sets. More directly, the test set performance allows us to speculate how well the trained model is likely to work when used outside the specific study.
Typically, to maximize the amount of data available to train the model, a separate validation set is dispensed with in favor of a k-fold cross-validation approach over the training set. Cross-validation splits the training set into k equal-sized subgroups, or folds, with all but one of these folds used to fit the model, and the remaining fold then used as a validation set to evaluate the model’s performance. This process is repeated for all folds and used to select the models and hyperparameter values that achieve best overall performance. Ensuring that the test set is only used once, and being methodical in training and validation, is vital to reduce the risk of producing models that cannot be generalized and are low in utility.
Given the range of current ML algorithms, the speed at which new ones are developed, and the variety in strengths and weaknesses these algorithms encompass, it is imperative to robustly evaluate a range of models and carefully optimize each one. A simple, well-trained model with carefully optimized hyperparameters can outperform even highly complex models on some data sets (80). Investigators can pursue a comprehensive approach and implement a huge number of model types before selecting for the best cross-validation results. However, structured implementation of a smaller number of models with optimized hyperparameters may be a more effective use of resources and less prone to overfitting to the training data (see below for more details).
Choosing the appropriate classifier/algorithm.
A key decision in the choice of classification algorithm is the expected complexity of the classification task. Generally, relatively simple models (in terms of numbers of learned parameters) have a higher bias, while more complex models have higher variance (81). Simple models can be more interpretable (i.e., easier to determine which genetic features are important for prediction), and have shorter run times, but may be unable to learn more-complex classification rules leading to a model with higher bias (“underfitting”). Alternatively, higher-complexity models can capture more complex associations among input features but are more prone to creating higher-variance models that are excessively tailored to the training set (“overfitting”), leading to poor predictions on the test set and unseen/new data. However, the overfitting associated with more complex models can sometimes be mitigated by using techniques like regularization that penalize model complexity. Overall, it is important to assess the parameters in an algorithm to consider the trade-off between bias and variance and the types of feature relationships a model can represent when choosing an algorithm.
It is often necessary to examine more than one type of algorithm to carefully assess their interpretability (also referred to as explainability) and to train performance metrics in the context of a specific problem to make an appropriate choice. Especially in a public health workflow, the ML classifiers’ prediction process should be transparent to develop a highly interpretable model. Interpretability can be expressed in many ways (82, 83) and is a very active area of current ML research (84, 85).
Three aspects of interpretability are highly relevant to AMR prediction: (i) ability to evaluate individual input features, (ii) traceability, and (iii) ability to assess the interactions of features. First, certain methods like logistic regression (LG)- and decision tree (DT)-based classifiers explicitly evaluate individual input features: LG associates a weight to each feature, while DT can rank the importance of features by identifying features that reduce the variance. DT and its derivatives, such as random forest (RF), contain hierarchically structured sets of internal nodes that apply explicit decision criteria until a final prediction is made and a class label assigned (86). Hence, each node is traceable (82) for DT-based algorithms, i.e., one can backtrack from the decision class to understand the decision logic. DTs are flexible, as they can handle classification and regression, such as the maximum margin interval trees designed for the specific difficulties of MIC predictions (55). DT ensemble classifiers such as gradient-boosted decision methods are increasingly being applied to AMR prediction problems (e.g., XGBoost used to predict MICs by Nguyen et al. [28]). The ensemble of decision trees is created by sequentially adding trees and correcting errors at every iteration based on previously grown trees (87) and has been found to outperform other learning methods in AMR prediction studies (88). It is of high popularity due to its faster learning speed, ability to handle sparse data sets (i.e., missing values), and avoidance of overfitting via regularization (89). Another traceable set of algorithms is rule-based ML methods that stem from the traditional but rigid rule-based system that uses IF-THEN statements with conditions and a prediction, but these can only handle classification problems (unless combined with another model type) (90). Rule-based algorithms like the set-covering machine (91), which uses a set of Boolean rules (conjunction or disjunction of features), can construct models that generalize well to different data sets as the rules identify the smallest number of features that maximize prediction performance. Finally, some algorithms integrate the calculation of feature interaction, e.g., addition of an interaction term in the prediction model after considering the individual feature effects to examine dependency. However, choosing a classifier that can model increased interactions among features can reduce traceability. The studies highlighted in Table 1 illustrate the use of different ML methods and demonstrate successful design of ML models for classifying antibiotic resistance within genomes and identifying the relevant genetic attributes.
TABLE 1.
Commonly used ML algorithms in AMR investigationsa
| Algorithm | Learning method | Feature evaluation | Traceable | Interaction | AMR investigation |
|---|---|---|---|---|---|
| Logistic regression | Regression algorithm with logistic curve that associates weights to each input features (134) | Yes | No | Nob | Maguire et al. (30), investigated primary AMR drivers of nontyphoidal Salmonella with known AMR determinants as features |
| Support vector machines | Separates labeled training data via constructing an optimal hyperplane, grouping appropriate genes, k-mer, or SNV features together (135) | Noc | No | Yes | Niehaus et al. (136) used M. tuberculosis SNVs to develop resistance prediction models for the 4 common first-line drugs |
| Random forest | Set of decision trees with internal nodes containing a series of questions about relevant features; different answers are directed to separate child nodes until reaching the final class label (86) | Yes | Yes | Yes | Moradigaravand et al. (88) predicted AMR from E. coli pangenome with various feature representations including the presence-absence of accessory genomes and the population structure inferred from the core genome |
| Rule based | Set of IF-THEN statements with condition and a prediction | Yes | Yes | Yes | Drouin et al. (73) predicted resistance of Gram-negative and -positive pathogens using k-mer features and the optimized set covering machine |
| Neural networks | Models loosely inspired by the structure of human brains, including deep learning (DL) models, and capable of modeling complex nonlinear relationships but require large amounts of data | Yes | No | Yes | Yang et al. (29) used M. tuberculosis SNV with DL to predict resistance to the four first-line antituberculosis drugs |
Feature evaluation is when the algorithm can weigh or rank the features’ impact on the prediction. A traceable algorithm allows visualization of the logical flow that leads to a prediction. Interaction indicates whether the algorithm can represent feature interdependencies.
Without additional data processing.
Unless using a linear kernel.
Evaluating machine-learning models.
Several evaluation metrics are available to assess the different characteristics of a model, such as its ability to accurately discriminate classes and generalizability to unseen data (92). Two-class problems are often expressed in terms of positive and negative sets; in the context of AMR, the positive set usually encompasses the resistant isolates and the negative set the susceptible isolates for each antibiotic examined. One should choose an appropriate metric based on the implementation purpose of the prediction model. For example, misclassification is costly in clinical laboratories where a false-negative diagnosis (i.e., a resistant case incorrectly classified as a sensitive case; also known as a very major error) can lead to treatment failure. Therefore, metrics that consider the effectiveness of the classifier on each class separately are necessary. False-positive diagnosis, known often as major error, is also a high risk that leads to misuse of antibiotics increasing the selective pressure for AMR pathogens. Based on the intended use of the model, whether that be clinical or surveillance oriented, the error threshold can change and the choice of metric may differ.
Many accuracy measures are based on correct and incorrect assignments to each class, which are typically organized into a confusion matrix (Table 2). True positives are correct predictions of resistance, while true negatives are correct predictions of susceptibility to an antibiotic. Multiclass classification can also be handled without denoting the classes as positive or negative but instead using the class name. For example, with the inclusion of intermediately resistant class of pathogens, the classes could simply be resistant, intermediate, and susceptible, without designating a preference for any class as positive.
TABLE 2.
Confusion matrix
| True label | Predicted label |
|
|---|---|---|
| Positive | Negative | |
| Positive | True positive | False negative |
| Negative | False positive | True negative |
Many evaluation metrics base their calculations on values in the underlying confusion matrix, detailed in Table 3. A classifier’s error rate (E) can be calculated by dividing the incorrectly predicted samples by the total number of samples (93). The accuracy measure is the total number of correctly predicted samples divided by the total number of samples (i.e., 1 − E). Many studies report accuracy due to its simplicity and applicability in binary or multiclass classification problems. However, if the data set is imbalanced, the model training and accuracy will be more strongly influenced by the abundant class, which will give a skewed representation of the model’s performance. In such cases, metrics such as precision, recall/sensitivity, and specificity are better measures to assess model performance. Precision measures which proportion of a model’s positive predictions were correct, recall measures the proportion of actual positives correctly identified, and specificity measures the proportion of actual negatives correctly identified. The best choice of evaluation method can be application specific. For example, in the context of AMR diagnostics, a high-precision model can reduce the overuse of antibiotics stemming from false-positive results. Conversely, high-recall models can help reduce morbidity caused by treatment failure due to false-negative results. Following model optimization, a desired balance between these needs can be determined with metrics such as the F1 score and precision/recall (PR) plot. The F1 score is the harmonic mean of precision and recall, allowing simultaneous evaluation of both measures, while the PR plot illustrates precision versus recall to allow model-wide evaluation. The PR plot has a baseline that caters to the class distribution and is well suited for imbalanced data (94). Overall, these evaluation metrics are commonly used as performance measures in AMR studies, aiding model selection based on performance with test/held-out sets.
TABLE 3.
Evaluation metrics based on positive and negative predictions of a modela
| Evaluation metric | Equation |
|---|---|
| Error rate | |
| Accuracy | |
| Precision | |
| Recall/Sensitivity | |
| Specificity | |
| F1 |
TP, true positive; FP, false positive; TN, true negative; FN, false negative; P, precision; R, recall.
LIMITATIONS OF MACHINE LEARNING ANALYSIS IN AMR RESEARCH
ML has great potential to replace a significant portion of conventional bench work to determine AMR, streamlining and accelerating surveillance, diagnosis, and treatment. However, further refinements are necessary to safely and confidently incorporate these methods for applications beyond research. Global recognition of the AMR crisis has increased the research and clinical attention being paid to AMR, with a corresponding aggressive search for new antibiotics, yet our knowledge of the underlying mechanisms of (and corresponding determinants of) AMR and modes of evolution still needs to be expanded. The underlying mechanisms of many observed drug-resistant infections are still poorly understood. This is especially acute for new, rapidly emerging, resistant infections. A mechanistic understanding of AMR becomes even more challenging when resistance arises from changes at the cellular or microbial community level. For example, subpopulations of bacteria can become persister cells in response to antibiotics or other cellular stressors (95), and genomic capacity for resistance can be rapidly augmented via HGT during biofilm formation (96). These complex processes are challenging to predict even when the entire genome sequence is available. Consequently, ML models can struggle to learn and generalize with incomplete information and highly complex cellular and evolutionary mechanisms of resistance. However, ML can be used to generate hypotheses to aid in the elucidation of AMR genes or mechanisms. For example, if sequences previously unrelated to resistance are highly scored during a feature selection step, one can postulate that there is some connection to resistance that can be further examined via experimentation.
Another problem is that many current ML investigations treat each gene or sequence independently. While such models tend to be more interpretable, phenotypes are often the product of several genes working in concert in a nonlinear manner. For example, AMR is often associated with tolerance to heavy metals stemming from coselection of metal and antibiotic resistance genes (97). The association has been suggested to enhance the maintenance and spread of AMR in the environment (98). Metal resistance genes do not directly confer resistance to antimicrobials, except in cases of a shared efflux mechanism, but they improve the fitness of the bacteria to tolerate a higher level of antibiotics, prolonging bacterial survival and persistence of AMR genes. Current ML models using univariate features cannot capture such variations of gene interplay. Techniques to better resolve the interplay of gene features in ML studies have been investigated, although they are not yet commonly adopted. A study optimizing a classical LG algorithm to measure the interaction effects of multivariate features is an example (99). However, the abundance of features in genomic studies makes it very challenging and complex to consider the large potential number of interactions among features. Some aspects of classical multivariate statistics simply do not translate well to rich genomic data sets and ML algorithms.
Kavvas et al. (31) used M. tuberculosis SNVs and incorporated an improved support vector machine (SVM) classifier (Table 1) to postulate genetic interactions of multiple alleles and identify potentially new resistance genes not directly related to drug targets but involved in the regulation of the resistance mechanisms. The candidate alleles were mapped to homologous protein structures to validate their role in AMR. A positive control using a known allele selected by the modified SVM confirmed the identified mutation mapped to a known location of AMR-conferring mutations, indicating that the new method could associate newly identified alleles with the AMR phenotype. Another recent study by Benkwitz-Bedford et al. presented a reverse genetics approach and paired ML models to predict bacterial growth and doubling time under subinhibitory concentrations of various antimicrobials from E. coli genomic data. Although the prediction performance was not at the level of applicability, the features from the model provided insight into the resistant-specific and housekeeping-related genes that bacterial cells incorporate to evolve AMR (100). Such approaches better resemble the biological reality of resistance mechanisms that utilize many genes that participate in epistatic interactions. However, these computational correlation studies cannot replace experimental validation to confirm the causality of resistance by the deduced features. Pairing of ML with high-throughput chemical-genomic screens, Tn-Seq knockout libraries, and antibiotic selection thus could hold great promise.
Finally, to date, most AMR prediction methods have been classifying susceptible and resistant binary categories based on clinical guidelines. These models correspond to a snapshot in time that is useful for diagnostic purposes (50) but will not recognize low-level resistance that may become fully resistant with selection pressure and misuse of antibiotics. The use of an intermediate category (between susceptible and resistant) could address this limitation. Isolates with an intermediate phenotype have often been included in the resistant category in AMR ML studies, but considering them a separate class could provide further insights into emerging resistance. However, there are several challenges that arise from the inclusion of an intermediate class. First, the unclear definition of the term requires standardized data collection and clear guidelines on the boundary between resistant, intermediate, and sensitive. To complicate things further, the EUCAST definition of I was recently revised, and what was traditionally grouped with resistant is now referred to as susceptible, increased exposure, denoting a high likelihood of therapeutic success from increasing the exposure to the antimicrobial agent (https://www.eucast.org/newsiandr/). Combining CLSI and EUCAST definitions, a pathogen is said to exhibit intermediate phenotype when a drug exerts a certain level of antimicrobial activity without a definitive therapeutic effect and when an infection can be treated with a concentrated or high dosage of drug (51). The complications and consequences of inconsistent phenotypic testing protocols and guidelines have been reported numerous times in the literature and summarized in Cusack et al. (101). This is an important issue that will need to be tackled as ML begins to get implemented into real-life applications.
Another challenge is that intermediate isolates are relatively rare in genomic data sets, partially attributable to the lack of a standardized definition, which leads to imbalanced data sets and limits the effectiveness of training and generalization. Finally, there is an added complexity that stems from developing multiclass classification, relative to the binary approach, that leads to less interpretable models and imbalanced data sets.
A few investigations have used regression-based approaches to predict MICs rather than a category-based approach, allowing the results to be interpreted under CLSI or EUCAST definitions. Nguyen et al. generated nontyphoidal Salmonella MIC predictions with genomes encoded as 10-mers, using a modified tree-based algorithm (28). The predictions in the study had an overall average accuracy of 95%. The study defined accuracy as the model’s ability to predict the correct MIC within ±1 2-fold dilution step of the laboratory-derived MIC. MICs are qualitative outputs that can be indicated as intervals, which makes ML implementation more complicated compared to binary interpretive categories, but ML prediction of MICs is increasingly being investigated (55).
Overcoming the Limitations and Bridging the Knowledge Gap
An important hurdle for AMR-ML prediction is that there are knowledge gaps in the molecular understanding of evolving AMR mechanisms. While ML methods can deduce and narrow down associations of sequences with resistance phenotypes, identifying causation is not possible without experimental validation. All areas of AMR investigations, from AMR evolutionary research to clinical diagnostics, will benefit from having a better mechanistic understanding of resistance mechanisms. Therefore, we advocate for the inclusion of follow-on validation steps such as transcriptomic analysis and experimental expression of the putative AMR determinants used as key features by the ML model. The validation step will require periodic updates as AMR mechanisms emerge and evolve.
Transcriptomic analysis can be used to investigate how gene expression varies with environmental changes; this can include the induction of resistance genes in response to antibiotics, host immune cells, or other environmental selective pressures (102–104). This method provides quantifiable gene expression results that offer insights into how relevant genes may be responding. For less well-studied pathogens that do not yet have an extensive library of known AMR mechanisms, expression studies will increase the chance of identifying new resistance determinants. However, the global transcriptome assessed in laboratory settings cannot be an exact reflection of how bacteria respond to an antibiotic because, in reality, an environment contains a mixture of factors that influence the expression profile. Given the central roles of promoters and transcription factors in the regulation of gene expression, an appealing option would be to use ML to predict the expression profiles of genes based solely on genomic DNA sequence. The highly challenging problem of gene expression prediction from genetic sequences is in its very early days and shows exciting potential (105, 106).
Transcriptome profiling alone is insufficient to validate ML predictions and must be accompanied by direct validation of putative AMR genes and their activity against antimicrobials. A recent example of AMR ML work that incorporates experimental validation is that of Tsang et al. (32). By evaluating LG models, the authors used targeted gene expression experiments via the Antibiotic Resistance Platform (ARP) (107) to validate novel genotype-phenotype relationships between known β-lactamases and resistance phenotypes. This approach generated the first experimental validation that the β-lactamase CTX-M-15 inactivates cefazolin.
The few comprehensive ML investigations paired with experimental validation have demonstrated their effectiveness in confirming the accuracy of ML predictions as well as their ability to postulate previously unknown AMR determinants or substrate activities. Genetic sequences that are validated by gene expression and experimental studies can be further used to optimize the initial ML model, improving prediction performance and increasing interpretability. This level of understanding will be necessary to support the development of ML models that are grounded in known mechanisms and less susceptible to genomic variation that correlates with the resistance phenotype but does not contribute to it.
TRANSLATING ML-AMR PREDICTION FROM RESEARCH TO PRACTICE
Most AMR-ML models constructed to date are not yet ready to be implemented in real-life settings. Some models were created with the intention of deducing hypotheses about potential new AMR genes or mutational variants to further expand our understanding of AMR mechanisms (108), while other models were designed to achieve the highest prediction performance for clinical diagnostics (28). To integrate these models beyond research, we need to precisely define the intended use and design the ML methods accordingly.
ML for Public Health AMR Surveillance
AMR surveillance programs focus on select top-priority organisms, such as the extended-spectrum β-lactamase (ESBL)- and carbapenemase-producing organisms Staphylococcus aureus, Salmonella spp., and Enterococcus (109). Integration of genomics to improve AMR surveillance is an active topic of discussion, with systems and analysis pipelines around resistomes and metagenomics data being rapidly developed for implementation in the near future (110). Monitoring known causal resistance genes can show emerging AMR trends, identify new variants, and reveal transmission patterns that can help with the identification and control of outbreaks of multidrug-resistant pathogens. With the expanding library of WGS data from pathogens and their corresponding AST profiles, ML can increase the sensitivity and efficiency of the current surveillance process (111–113).
The focal point of ML implementation in surveillance is the features of high importance that the models base their predictions on (i.e., causal genes that contribute to the phenotype). Knowing the important features, ML models can be used to conduct the initial monitoring and highlighting of the potentially significant AMR genes. One possible way to implement and refine ML models to streamline the surveillance process is to construct an initial model based on current genomic epidemiological information or species, systematically test model performance as new data are acquired, and update the model as necessary. When the prediction performance deviates beyond a set error threshold, the training sets should then be reevaluated and division of training data can be assessed. The ML performance could be used as a gauge to determine the relevancy of the data.
Timely integration of data from hot spots of AMR transmission and growing inventories of specific priority organisms will allow longitudinal monitoring of AMR evolution in a manner that is highly useful for public health. The monitoring of AMR genes has already been initiated with metagenomic analysis, like the investigation of untreated sewage from 60 countries that showed AMR gene abundance correlating with socioeconomic, health, and environmental factors rather than antimicrobial use (114). Such an approach is being advocated as a feasible strategy for continuous global surveillance of AMR genes, and we believe the integration of ML with the metagenomes can further enhance surveillance efforts. While AMR research in basic science would use ML to discover and explain a wide array of AMR determinants, public health genomic surveillance with ML would focus on the select few AMR genes known to be of high risk. Key k-mers identified through a composition-based approach can also provide the potential to monitor mutational variants that arise within well-defined AMR genes of interest or elsewhere.
Timely identification of emerging AMR genes with ML can also accelerate the bridging of the surveillance data to individual care via diagnostic stewardship programs. Based on AMR surveillance data, the treatment guidelines and AMR control strategies are regularly updated (115). Consolidating such observations into diagnostic stewardship programs allows improved therapeutic decisions for better patient outcomes. ML has the potential to contribute to and accelerate this process.
In some parts of the world, priority organisms already have an established library of WGS and ASTs along with a defined set of AMR genes of concern (116, 117), hence ML models could be constructed readily. However, policies and guidelines on updating and monitoring the models with appropriate isolates should be detailed first. When the ML model implementation reaches the stage of producing reliable surveillance data, there is a potential to significantly accelerate the routine update procedure of diagnostic stewardship programs and influence the diagnostic process in clinical microbiology and laboratory management.
ML for Clinical Diagnostics
Clinical diagnostics of AMR must produce a rapid and highly accurate result without necessarily needing to know the causality of the prediction. The categorical agreement rate of commonly used phenotypic AST methods to inocula prepared from the same subculture can range from 89.6% with Phoenix (118) to 98.9% with Vitek 2 (119). ML could in the future complement the current diagnostic tests to potentially improve accuracy and speed. The focal point of ML implementation for diagnostics would not be to dissect the internal processing of the ML models but to achieve the most reliable predictions of resistance and susceptibility.
A tool leading to personalized medicine based on rapid detection of a bacterial pathogen and its resistance profile directly from clinical samples would be revolutionary for antimicrobial stewardship. One key application would be in sepsis, where delayed effective antibiotic therapy adversely impacts mortality by up to 20% (120). There is increasing interest for molecular rapid diagnostic tests in bloodstream infections that, combined with antimicrobial stewardship programs, have the potential to improve patient outcomes (121). Metagenomics is in its infancy for the identification of pathogens from blood, but a protocol (122) was recently combined with ML methods to produce a commercial assay (123). This test does not currently provide susceptibility results but illustrates how the commercial sector could produce a viable product. Any new tool will need to demonstrate added value and cost-effectiveness compared to existing rapid tools for identification (e.g., matrix-assisted laser desorption ionization time-of-flight mass spectrometry) and susceptibility testing from positive blood cultures (124) and ought to concentrate on the limited number of the most common causative bacteria and drug-pathogen combinations. New tools must also be supported by robust validated databases for each drug-pathogen combination that are curated and used to retrain the models periodically.
In some public health laboratories, the use of whole-genome sequencing is already established for specific target organisms (125, 126), which is likely to become increasingly used in routine diagnostic laboratories. Sequencing can be cost-effective as it replaces multiple phenotypic and genotypic tests at scale to provide a range of information (e.g., lineage, isolate relatedness, resistance mechanism surveillance, and detection of toxin genes), although prediction of AST is not a primary output. The addition of ML methods would only be accepted with further improvement in accuracy at a reduced financial cost (127). Mycobacterium tuberculosis is one example where WGS has resulted in a paradigm shift in phenotypic susceptibility testing, with tools such as Mykrobe (128) being used to infer susceptibility to first-line and some second-line agents and in defined cases obviating the need for phenotypic susceptibility testing altogether (129, 130). For very slow-growing organisms such as M. tuberculosis, which can take up to 8 weeks to culture (131, 132), the WGS-based method that significantly cuts the detection time is highly favored. In the case of other bacterial pathogens, phenotypic or PCR tests targeting specific resistance markers remain gold standards as they provide a cheap, reliable result to clinicians in an actionable turnaround time.
Making the leap from a research tool to routine clinical practice remains challenging. Existing wet- and dry-lab workflows will require extensive updates and optimization and will need to undergo a robust clinical evaluation for accuracy. These protocols will need to be underpinned by defined quality control requirements for preanalytical, analytical, and postanalytical components. A report should be easily interpretable by clinicians with no specialist knowledge of genomics or ML, which could include predicted susceptibility/resistance together with a measure for degree of uncertainty. External quality assessment schemes and accreditation standards will need to be developed. A risk of very major errors will remain an issue due to the emergence of novel resistance mechanisms, and phenotypic testing for surveillance or diagnostic purposes will continue to be necessary, especially for novel agents or those where models perform poorly. As technologies advance to overcome current limitations, shotgun metagenomic sequencing directly from clinical samples provides the most feasible opportunity for culture-free identification and antimicrobial susceptibility prediction (124). However, AMR genes identified in a metagenomic sample may not be assignable to a specific organism, especially in cases where mobile genetic elements are frequently shared between different pathogen species. The clinical relevance of identified AMR genes may be unclear, especially when pathogenic organisms are present at very low frequencies. The ML methodologies will be translated and accepted in the clinical settings for the appropriate end users upon resolving the outlined barriers.
CONCLUDING REMARKS
The global resistome of AMR constantly evolves in human populations, animal populations, and the environment (133), and as such the suite of AMR threats is ever changing via both novel mutation and transmission of resistance via HGT. The selection pressure from unchecked antibiotic use and misuse has led to the current AMR crisis. ML has become a popular choice to predict the resistance potential of high-risk pathogens from genome sequences that are now readily available, but many prediction models are based primarily on well-defined resistance genes for molecular diagnostics purposes. The focus on well-defined genes does not recognize that genes may not be expressed or may play a different role in a given isolate and ignores new and potentially high-risk resistance genes and mutations.
Integrating AMR phenotype prediction into surveillance and diagnostic pipelines will require several activities. Data availability and quality is a significant concern, and newly sequenced pathogens should include associated metadata with MIC/AST results and experimental details. The range of antibiotics tested in the published literature varies widely, and the community should consider standards when describing new AMR genes, e.g., a standardized panel of β-lactams, when describing a new β-lactamase. This standardization will allow greater harmonization of data sets, models, and predictions.
We also advocate for associated experimental workflows with hypotheses obtained from ML models experimentally tested to increase the breadth, depth, and scope of ML activities and to provide confidence in ML methods for real-life public health and diagnostics implementation. Enhanced efforts to obtain genome sequences and antimicrobial susceptibility information from various niches beyond clinical settings to make successful predictions across the One-Health continuum will significantly contribute to the risk assessment of hot spot reservoirs that accelerate AMR evolution.
Biographies

Jee In Kim is a Ph.D. candidate in the Interdisciplinary Ph.D. program at Dalhousie University, Canada. Jee is investigating antimicrobial resistance genomics of Enterococcus species via combining molecular science and computer science techniques. Jee previously worked as a consultant at the Pan American Health Organization Antimicrobial Resistance Special Program and holds an M.Sc. in molecular science (Ryerson University) and B.Sc. in biology (Western University).

Finlay Maguire is an Assistant Professor in Genomic Epidemiology based in the Faculty of Medicine's Department of Community Health & Epidemiology and the Faculty of Computer Science at Dalhousie University. Additionally, they are the Pathogenomics Bioinformatics lead for Toronto’s Shared Hospital Laboratory and a visiting scientist at Sunnybrook Research Institute. Dr. Maguire studied for a B.A. in Life Sciences at Oxford University before undertaking a Ph.D. in bioinformatics on the evolution of endosymbiotic systems at University College London/Natural History Museum London. Through postdoctoral positions at the University of Exeter and Dalhousie University, they worked on computational methods for the use of genomic and metagenomic data in the surveillance and diagnosis of antimicrobial resistance and SARS-CoV-2. Their current research program focuses on the challenges of effective genomically informed clinical and public health epidemiology of infectious diseases.

Kara K. Tsang is a computational microbiologist with expertise in antimicrobial resistance prediction from genotype to phenotype. Kara is currently a Research Fellow at the London School of Hygiene and Tropical Medicine. In this role, she aims to integrate and extend existing tools to develop a comprehensive framework for Klebsiella pneumoniae genomic surveillance, the KlebNET Genomic Surveillance Platform. Kara earned a B.HSc. in Biomedical Discovery and Commercialization in 2016 and a Ph.D. in Biochemistry and Biomedical Sciences in 2021 (McMaster University).

Theodore Gouliouris is a Consultant in Microbiology and Infectious Diseases at Cambridge University Hospitals, UK. He obtained his Ph.D. as part of a Wellcome Research Training Fellowship at the University of Cambridge where he studied the hospital and community spread of vancomycin-resistant Enterococcus faecium using bacterial genomics. His research interests include the clinical and public health applications of genomics in relation to antimicrobial-resistant and healthcare-associated infections.

Sharon J. Peacock is currently Professor of Public Health and Microbiology at the University of Cambridge; Executive Director of the COVID-19 Genomics UK (COG-UK) consortium; and a Non-Executive Director on the board of Cambridge University Hospitals NHS Foundation Trust. She has raised around £60 million in science funding, published more than 500 peer-reviewed papers, and has trained a generation of scientists in the UK and elsewhere. Sharon is a Fellow of the Academy of Medical Sciences, a Fellow of the American Academy of Microbiology, and an elected Member of the European Molecular Biology Organization (EMBO). In 2015, she received a CBE for services to medical microbiology, and in 2018 she won the Unilever Colworth Prize for her outstanding contribution to translational microbiology. Sharon was recently awarded the MRC Millennium Medal 2021.

Tim A. McAllister was raised on a mixed cow-calf operation in Innisfail, Alberta. He obtained his M.Sc. in Animal Biochemistry at the University of Alberta and his Ph.D. in microbiology and nutrition from the University of Guelph in 1991. He is presently a principal research scientist with Agriculture and Agri-Food in Lethbridge, Alberta, Canada. Tim leads a diverse research team that has been studying antimicrobial resistance in beef cattle production systems since 1997. The team’s recent work has focused on studying AMR from a “One Health” perspective using enterococci as AMR indicators in beef production, human sewage, and clinical settings. He is also investigating the role of integrative conjugative elements in the transfer of antimicrobial resistance genes within the bacterial bovine respiratory disease complex. Tim has authored over 850 scientific papers, is the recipient of several national and international awards, and holds adjunct professorship appointments at several universities in Canada and abroad.

Andrew G. McArthur is an Associate Professor and the inaugural David Braley Chair in Computational Biology in the Michael G. DeGroote Institute for Infectious Disease Research and Department of Biochemistry & Biomedical Sciences at McMaster University. Dr. McArthur has had a 20+ year research career in the United States and Canada, including postdoctoral experience at the National Museum of Natural History and NIH-funded faculty positions at the Marine Biological Laboratory (Woods Hole, MA) and Brown University, where he led the computational biology of sequencing the genome of the diarrheal pathogen Giardia intestinalis, plus 10 years experience in the private sector. As part of the McMaster Global Nexus for Pandemics and Biological Threats, his research team focuses on building tools, databases, and algorithms for genomic surveillance of infectious pathogens and he leads the globally recognized Comprehensive Antibiotic Resistance Database (card.mcmaster.ca).

Robert G. Beiko is a full Professor and Associate Dean Research in the Faculty of Computer Science at Dalhousie University. Prior to taking up his faculty position, he obtained an honors degree in biology at Dalhousie and a Ph.D. in biology (bioinformatics) at the University of Ottawa. He also held a postdoctoral fellowship at the University of Queensland, Brisbane, Australia. Dr. Beiko’s research focuses on microbial evolution and ecology, with a particular focus on the role played by lateral gene transfer in the evolution of human commensals and pathogens. His research group has developed software for the analysis, comparison, and prediction of microbial communities, and inference of gene transfer from large datasets. Dr. Beiko’s recent work is centered on the emergence, transmission, and ecology of antimicrobial resistance in pathogenic organisms, including Enterococcus and Salmonella.
REFERENCES
- 1.Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrestha P, Cooper BS, Coast J, Oppong R, Do Thi Thuy N, Phodha T, Celhay O, Guerin PJ, Wertheim H, Lubell Y. 2018. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob Resist Infect Control 7:98. 10.1186/s13756-018-0384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Federal Government of Canada. 2014. Antimicrobial resistance and use in Canada: a federal framework for action. Federal Government of Canada, Ottawa, Canada. [Google Scholar]
- 4.World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.UK Review on AMR. 2016. Tackling drug-resistant infections globally: final report and recommendations. https://amr-review.org/home.html.
- 6.Soares A, Pestel-Caron M, de Rohello FL, Bourgoin G, Boyer S, Caron F. 2020. Area of technical uncertainty for susceptibility testing of amoxicillin/clavulanate against Escherichia coli: analysis of automated system, Etest and disk diffusion methods compared to the broth microdilution reference. Clin Microbiol Infect 26:1685–1691. 10.1016/j.cmi.2020.02.038. [DOI] [PubMed] [Google Scholar]
- 7.Hendriksen RS, Seyfarth AM, Jensen AB, Whichard J, Karlsmose S, Joyce K, Mikoleit M, Delong SM, Weill FX, Aidara-Kane A, Lo Fo Wong DMA, Angulo FJ, Wegener HC, Aarestrup FM. 2009. Results of use of WHO Global Salm-Surv external quality assurance system for antimicrobial susceptibility testing of Salmonella Isolates from 2000 to 2007. J Clin Microbiol 47:79–85. 10.1128/JCM.00894-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su M, Satola SW, Read TD. 2019. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57:e01405-18. 10.1128/JCM.01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma KC, Mortimer TD, Duckett MA, Hicks AL, Wheeler NE, Sánchez-Busó L, Grad YH. 2020. Increased power from conditional bacterial genome-wide association identifies macrolide resistance mutations in Neisseria gonorrhoeae. Nat Commun 11:5374. 10.1038/s41467-020-19250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volkman SK, Herman J, Lukens AK, Hartl DL. 2017. Genome-wide association studies of drug-resistance determinants. Trends Parasitol 33:214–230. 10.1016/j.pt.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Alam MT, Petit RA, Crispell EK, Thornton TA, Conneely KN, Jiang Y, Satola SW, Read TD. 2014. Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol Evol 6:1174–1185. 10.1093/gbe/evu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.San JE, Baichoo S, Kanzi A, Moosa Y, Lessells R, Fonseca V, Mogaka J, Power R, de Oliveira T. 2020. Current affairs of microbial genome-wide association studies: approaches, bottlenecks and analytical pitfalls. Front Microbiol 10:3119. 10.3389/fmicb.2019.03119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu M, Arnold R, Bundalovic-Torma C, Gagarinova A, Wong KS, Kumar A, Stewart G, Samanfar B, Aoki H, Wagih O, Vlasblom J, Phanse S, Lad K, Yeou Hsiung Yu A, Graham C, Jin K, Brown E, Golshani A, Kim P, Moreno-Hagelsieb G, Greenblatt J, Houry WA, Parkinson J, Emili A. 2014. Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli. PLoS Genet 10:e1004120-15. 10.1371/journal.pgen.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong A. 2017. Epistasis and the evolution of antimicrobial resistance. Front Microbiol 8:246. 10.3389/fmicb.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevenson C, Hall JP, Harrison E, Wood A, Brockhurst MA. 2017. Gene mobility promotes the spread of resistance in bacterial populations. ISME J 11:1930–1932. 10.1038/ismej.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gahlaut V, Jaiswal V, Singh S, Balyan HS, Gupta PK. 2019. Multi-locus genome wide association mapping for yield and its contributing traits in hexaploid wheat under different water regimes. Sci Rep 9:19486. 10.1038/s41598-019-55520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saber MM, Shapiro BJ. 2020. Benchmarking bacterial genome-wide association study methods using simulated genomes and phenotypes. Microb Genom 10.1099/mgen.0.000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazer CL, Al-Mamun MA, Kaniyamattam K, Love WJ, Booth JG, Lanzas C, Gröhn YT. 2019. Shared multidrug resistance patterns in chicken-associated Escherichia coli identified by association rule mining. Front Microbiol 10:687. 10.3389/fmicb.2019.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyun JC, Kavvas ES, Monk JM, Palsson BO. 2020. Machine learning with random subspace ensembles identifies antimicrobial resistance determinants from pan-genomes of three pathogens. PLoS Comput Biol 16:e1007608. 10.1371/journal.pcbi.1007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bzdok D, Altman N, Krzywinski M. 2018. Statistics versus machine learning. Nat Methods 15:233–234. 10.1038/nmeth.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaillard M, Palmieri M, van Belkum A, Mahé P. 2020. Interpreting k-mer–based signatures for antibiotic resistance prediction. GigaScience 9:giaa110. 10.1093/gigascience/giaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macesic N, Bear Don’t Walk OJ, Pe’er I, Tatonetti NP, Peleg AY, Uhlemann AC. 2020. Predicting phenotypic polymyxin resistance in Klebsiella pneumoniae through machine learning analysis of genomic data. mSystems 5:e00656-19. 10.1128/mSystems.00656-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drouin A, Letarte G, Raymond F, Marchand M, Corbeil J, Laviolette F. 2019. Interpretable genotype-to-phenotype classifiers with performance guarantees. Sci Rep 9:4071. 10.1038/s41598-019-40561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewin-Epstein O, Baruch S, Hadany L, Stein GY, Obolski U. 2021. Predicting antibiotic resistance in hospitalized patients by applying machine learning to electronic medical records. Clin Infect Dis 72:e848–e855. 10.1093/cid/ciaa1576. [DOI] [PubMed] [Google Scholar]
- 26.Anahtar MN, Yang JH, Kanjilal S. 2021. Applications of machine learning to the problem of antimicrobial resistance: an emerging model for translational research. J Clin Microbiol 59:e01260-20. 10.1128/JCM.01260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bzdok D, Krzywinski M, Altman N. 2018. Machine learning: supervised methods. Nat Methods 15:5–6. 10.1038/nmeth.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, Davis JJ. 2019. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J Clin Microbiol 57:e01260-18. 10.1128/JCM.01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Walker TM, Walker AS, Wilson DJ, Peto TEA, Crook DW, Shamout F, Arandjelovic I, Comas I, Farhat MR, Gao Q, Sintchenko V, van Soolingen D, Hoosdally S, Gibertoni CA, Carter J, Grazian C, Earle SG, Kouchaki S, Yang Y, Walker TM, Fowler PW, Clifton DA, Iqbal Z, Hunt M, Smith EG, Rathod P, Jarrett L, Matias D, Cirillo DM, Borroni E, Battaglia S, Ghodousi A, Spitaleri A, Cabibbe A, Tahseen S, Nilgiriwala K, Shah S, Rodrigues C, Kambli P, Surve U, Khot R, Niemann S, Kohl T, Merker M, Hoffmann H, Molodtsov N, Plesnik S, Ismail N, Thwaites G, CRyPTIC Consortium, et al. 2019. DeepAMR for predicting co-occurrent resistance of Mycobacterium tuberculosis. Bioinformatics 35:3240–3249. 10.1093/bioinformatics/btz067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maguire F, Rehman MA, Carrillo C, Diarra MS, Beiko RG. 2019. Identification of primary antimicrobial resistance drivers in agricultural nontyphoidal Salmonella enterica serovars by using machine learning. mSystems 4:e00211-19. 10.1128/mSystems.00211-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, Heckmann D, Anand A, Yang L, Nizet V, Monk JM, Palsson BO. 2018. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun 9:4306. 10.1038/s41467-018-06634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang KK, Maguire F, Zubyk HL, Chou S, Edalatmand A, Wright GD, Beiko RG, McArthur AG. 2021. Identifying novel β-lactamase substrate activity through in silico prediction of antimicrobial resistance. Microb Genom 10.1099/mgen.0.000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suerbaum S, Josenhans C. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol 5:441–452. 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 34.Aytan-Aktug D, Clausen PTLC, Bortolaia V, Aarestrup FM, Lund O. 2020. Prediction of acquired antimicrobial resistance for multiple bacterial species using neural networks. mSystems 5:e00774-19. 10.1128/mSystems.00774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earle SG, Wu CH, Charlesworth J, Stoesser N, Gordon NC, Walker TM, Spencer CCA, Iqbal Z, Clifton DA, Hopkins KL, Woodford N, Smith EG, Ismail N, Llewelyn MJ, Peto TE, Crook DW, McVean G, Walker AS, Wilson DJ. 2016. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol 1:16041. 10.1038/nmicrobiol.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. 2018. pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 34:4310–4312. 10.1093/bioinformatics/bty539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hicks AL, Wheeler N, Sánchez-Busó L, Rakeman JL, Harris SR, Grad YH. 2019. Evaluation of parameters affecting performance and reliability of machine learning-based antibiotic susceptibility testing from whole genome sequencing data. PLoS Comput Biol 15:e1007349. 10.1371/journal.pcbi.1007349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludden C, Raven KE, Jamrozy D, Gouliouris T, Blane B, Coll F, de Goffau M, Naydenova P, Horner C, Hernandez-Garcia J, Wood P, Hadjirin N, Radakovic M, Brown NM, Holmes M, Parkhill J, Peacock SJ, Sansonetti PJ. 2019. One Health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 10:e02693-18. 10.1128/mBio.02693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouliouris T, Raven KE, Ludden C, Blane B, Corander J, Horner CS, Hernandez-Garcia J, Wood P, Hadjirin NF, Radakovic M, Holmes MA, de Goffau M, Brown NM, Parkhill J, Peacock SJ, Schaik WV, Hughes JM. 2018. Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. mBio 9:e01780-18. 10.1128/mBio.01780-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaheer R, Cook SR, Barbieri R, Goji N, Cameron A, Petkau A, Polo RO, Tymensen L, Stamm C, Song J, Hannon S, Jones T, Church D, Booker CW, Amoako K, Van Domselaar G, Read RR, McAllister TA. 2020. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci Rep 10:3937. 10.1038/s41598-020-61002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. 2002. SMOTE: synthetic minority over-sampling technique. J Artificial Intell Res 16:321–357. 10.1613/jair.953. [DOI] [Google Scholar]
- 42.Lemaitre G, Nogueira F, Aridas CK. 2016. Imbalanced-learn: a Python toolbox to tackle the curse of imbalanced datasets in machine learning. arXiv 1609.06570. https://arxiv.org/abs/1609.06570.
- 43.Van Hulse J, Khoshgoftaar TM, Napolitano A. 2007. Experimental perspectives on learning from imbalanced data, p 935–942. In Proc 24th Int Conf Machine Learning ICML ’07. Association for Computing Machinery, New York, NY. 10.1145/1273496.1273614. [DOI] [Google Scholar]
- 44.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain J-M, Samuelsen Ø, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golparian D, Donà V, Sánchez-Busó L, Foerster S, Harris S, Endimiani A, Low N, Unemo M. 2018. Antimicrobial resistance prediction and phylogenetic analysis of Neisseria gonorrhoeae isolates using the Oxford Nanopore MinION sequencer. Sci Rep 8:17596. 10.1038/s41598-018-35750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim A, Naidenov B, Bates H, Willyerd K, Snider T, Couger MB, Chen C, Ramachandran A. 2019. Nanopore ultra-long read sequencing technology for antimicrobial resistance detection in Mannheimia haemolytica. J Microbiol Methods 159:138–147. 10.1016/j.mimet.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Berbers B, Saltykova A, Garcia-Graells C, Philipp P, Arella F, Marchal K, Winand R, Vanneste K, Roosens NHC, De Keersmaecker SCJ. 2020. Combining short and long read sequencing to characterize antimicrobial resistance genes on plasmids applied to an unauthorized genetically modified Bacillus,. Sci Rep 10: 4310. 10.1038/s41598-020-61158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arredondo-Alonso S, Willems RJ, van Schaik W, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom. 10.1099/mgen.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jorgensen JH, Turnidge JD. 2015. Susceptibility test methods: dilution and disk diffusion methods, p 1253–1273. In Manual of clinical microbiology. ASM Press, Washington, DC. 10.1128/9781555817381.ch71. [DOI] [Google Scholar]
- 51.Kahlmeter G, Giske CG, Kirn TJ, Sharp SE. 2019. Point-counterpoint: differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J Clin Microbiol 57:e01129-19. 10.1128/JCM.01129-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen M, Brettin T, Long SW, Musser JM, Olsen RJ, Olson R, Shukla M, Stevens RL, Xia F, Yoo H, Davis JJ. 2018. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci Rep 8:421. 10.1038/s41598-017-18972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eyre DW, De Silva D, Cole K, Peters J, Cole MJ, Grad YH, Demczuk W, Martin I, Mulvey MR, Crook DW, Walker AS, Peto TEA, Paul J. 2017. WGS to predict antibiotic MICs for Neisseria gonorrhoeae. J Antimicrob Chemother 72:1937–1947. 10.1093/jac/dkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pataki B, Matamoros S, van der Putten BCL, Remondini D, Giampieri E, Aytan-Aktug D, Hendriksen RS, Lund O, Csabai I, Schultsz C. 2020. Understanding and predicting ciprofloxacin minimum inhibitory concentration in Escherichia coli with machine learning. Sci Rep 10:15026. 10.1038/s41598-020-71693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drouin A, Hocking TD, Laviolette F. 2017. Maximum margin interval trees. In 31st Conference on Neural Information Processing Systems. Association for Computing Machinery, New York, NY. [Google Scholar]
- 56.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen ALV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2019. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antonopoulos DA, Assaf R, Aziz RK, Brettin T, Bun C, Conrad N, Davis JJ, Dietrich EM, Disz T, Gerdes S, Kenyon RW, Machi D, Mao C, Murphy- Olson DE, Nordberg EK, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Santerre J, Shukla M, Stevens RL, VanOeffelen M, Vonstein V, Warren AS, Wattam AR, Xia F, Yoo H. 2019. PATRIC as a unique resource for studying antimicrobial resistance. Brief Bioinform 20:1094–1102. 10.1093/bib/bbx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McArthur AG, Tsang KK. 2017. Antimicrobial resistance surveillance in the genomic age. Ann N Y Acad Sci 1388:78–91. 10.1111/nyas.13289. [DOI] [PubMed] [Google Scholar]
- 59.Doyle RM, O’Sullivan DM, Aller SD, Bruchmann S, Clark T, Coello PA, Cormican M, Diez BE, Ellington MJ, McGrath E, Motro Y, Phuong Thuy Nguyen T, Phelan J, Shaw LP, Stabler RA, van Belkum A, van Dorp L, Woodford N, Moran-Gilad J, Huggett JF, Harris KA. 2020. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb Genom. 10.1099/mgen.0.000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xavier BB, Das AJ, Cochrane G, De Ganck S, Kumar-Singh S, Aarestrup FM, Goossens H, Malhotra-Kumar S. 2016. Consolidating and exploring antibiotic resistance gene data resources. J Clin Microbiol 54:851–859. 10.1128/JCM.02717-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahfouz N, Ferreira I, Beisken S, von Haeseler A, Posch AE. 2020. Large-scale assessment of antimicrobial resistance marker databases for genetic phenotype prediction: a systematic review. J Antimicrob Chemother 75:3099–3108. 10.1093/jac/dkaa257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O'Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatusova T, Dicuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 65.Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. 2019. GenBank. Nucleic Acids Res 47:D94–D99. 10.1093/nar/gky989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, Bye-A-Jee H, Coetzee R, Cukura A, Da Silva A, Denny P, Dogan T, Ebenezer T, Fan J, Castro LG, Garmiri P, Georghiou G, Gonzales L, Hatton-Ellis E, Hussein A, Ignatchenko A, Insana G, Ishtiaq R, Jokinen P, Joshi V, Jyothi D, Lock A, Lopez R, Luciani A, Luo J, Lussi Y, MacDougall A, Madeira F, Mahmoudy M, Menchi M, Mishra A, Moulang K, Nightingale A, Oliveira CS, Pundir S, Qi G, Raj S, Rice D, Lopez MR, Saidi R, Sampson J, The UniProt Consortium, et al. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen M, Olson R, Shukla M, VanOeffelen M, Davis JJ. 2020. Predicting antimicrobial resistance using conserved genes. PLoS Comput Biol 16:e1008319. 10.1371/journal.pcbi.1008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cohen KA, Manson AL, Desjardins CA, Abeel T, Earl AM. 2019. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med 11:45. 10.1186/s13073-019-0660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ledger L, Eidt J, Cai HY. 2020. Identification of antimicrobial resistance-associated genes through whole genome sequencing of Mycoplasma bovis isolates with different antimicrobial resistances. Pathogens 9:588. 10.3390/pathogens9070588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y, Niehaus KE, Walker TM, Iqbal Z, Walker AS, Wilson DJ, Peto TEA, Crook DW, Smith EG, Zhu T, Clifton DA. 2018. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics 34:1666–1671. 10.1093/bioinformatics/btx801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen ML, Doddi A, Royer J, Freschi L, Schito M, Ezewudo M, Kohane IS, Beam A, Farhat M. 2019. Beyond multidrug resistance: leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction. EBioMedicine 43:356–369. 10.1016/j.ebiom.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drouin A, Giguère S, Déraspe M, Marchand M, Tyers M, Loo VG, Bourgault AM, Laviolette F, Corbeil J. 2016. Predictive computational phenotyping and biomarker discovery using reference-free genome comparisons. BMC Genomics 17:754. 10.1186/s12864-016-2889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lees JA, Mai TT, Galardini M, Wheeler NE, Horsfield ST, Parkhill J, Corander J. 2020. Improved prediction of bacterial genotype-phenotype associations using interpretable pangenome-spanning regressions. mBio 11:e01344-20. 10.1128/mBio.01344-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boisvert S, Raymond F, Godzaridis É, Laviolette F, Corbeil J. 2012. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 13:R122. 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies TJ, Stoesser N, Sheppard AE, Abuoun M, Fowler P, Swann J, Quan TP, Griffiths D, Vaughan A, Morgan M, Phan HTT, Jeffery KJ, Andersson M, Ellington MJ, Ekelund O, Woodford N, Mathers AJ, Bonomo RA, Crook DW, Peto TEA, Anjum MF, Walker AS. 2020. Reconciling the potentially irreconcilable? Genotypic and phenotypic amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother 64:e02026-19. 10.1128/AAC.02026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bommert A, Sun X, Bischl B, Rahnenführer J, Lang M. 2020. Benchmark for filter methods for feature selection in high-dimensional classification data. Comput Stat Data Anal 143:106839. 10.1016/j.csda.2019.106839. [DOI] [Google Scholar]
- 78.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 79.Ferri FJ, Pudil P, Hatef M, Kittler J. 1994. Comparative study of techniques for large-scale feature selection, p 403–413. In Machine intelligence and pattern recognition, vol 16. Elsevier, New York, NY. [Google Scholar]
- 80.Koutsoukas A, Monaghan KJ, Li X, Huan J. 2017. Deep-learning: investigating deep neural networks hyper-parameters and comparison of performance to shallow methods for modeling bioactivity data. J Cheminform 9:1–13. 10.1186/s13321-017-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lever J, Krzywinski M, Altman N. 2016. Model selection and overfitting. Nat Methods 13:703–704. 10.1038/nmeth.3968. [DOI] [Google Scholar]
- 82.Molnar C. 2019. Interpretable machine learning. A guide for making black box models explainable. https://christophm.github.io/interpretable-ml-book/.
- 83.Doshi-Velez F, Kim B. 2017. Towards a rigorous science of interpretable machine learning. arXiv 1702.08608. https://arxiv.org/abs/1702.08608.
- 84.Loyola-Gonzalez O. 2019. Black-box vs. white-box: understanding their advantages and weaknesses from a practical point of view. IEEE Access 7:154096–154113. 10.1109/ACCESS.2019.2949286. [DOI] [Google Scholar]
- 85.Azodi CB, Tang J, Shiu SH. 2020. Opening the black box: interpretable machine learning for geneticists. Trends Genet 36:442–455. 10.1016/j.tig.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 86.Kingsford C, Salzberg SL. 2008. What are decision trees? Nat Biotechnol 26:1011–1013. 10.1038/nbt0908-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friedman JH. 2001. Greedy function approximation: a gradient boosting machine. Ann Stat 29:1189–1232. https://www.jstor.org/stable/2699986. [Google Scholar]
- 88.Moradigaravand D, Palm M, Farewell A, Mustonen V, Warringer J, Parts L. 2018. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput Biol 14:e1006258. 10.1371/journal.pcbi.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen T, Guestrin C. 2016. XGBoost: a scalable tree boosting system, p 785–794. In KDD ’16 Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 10.1145/2939672.2939785. [DOI] [Google Scholar]
- 90.Fürnkranz J, Gamberger D, Lavrač N. 2012. Rule learning in a nutshell. Springer, Heidelberg. 10.1007/978-3-540-75197-7{\_}2. [DOI] [Google Scholar]
- 91.Marchand M, Shawe-Taylor J. 2002. The set covering machine. J Mach Learn Res. 10.1162/jmlr.2003.3.4-5.723. [DOI] [Google Scholar]
- 92.Hossin M, Sulaiman MN. 2015. A review on evaluation metrics for data classification evaluations. IJDKP 5:1–11. 10.5121/ijdkp.2015.5201. [DOI] [Google Scholar]
- 93.Kubat M. 2017. An introduction to machine learning. Springer International Publishing, Cham, Switzerland. 10.1007/978-3-319-63913-0. [DOI] [Google Scholar]
- 94.Saito T, Rehmsmeier M. 2015. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 10:e0118432. 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 96.Olsen I. 2015. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis 34:877–886. 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 97.Seiler C, Berendonk TU. 2012. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:399. 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 99.Xu EL, Qian X, Yu Q, Zhang H, Cui S. 2018. Feature selection with interactions in logistic regression models using multivariate synergies for a GWAS application. BMC Genomics 19:170. 10.1186/s12864-018-4552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Benkwitz-Bedford S, Palm M, Demirtas TY, Mustonen V, Farewell A, Warringer J, Parts L, Moradigaravand D. 2021. Machine learning prediction of resistance to subinhibitory antimicrobial concentrations from Escherichia coli genomes. mSystems 6:e00346-21. 10.1128/mSystems.00346-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cusack T, Ashley E, Ling C, Rattanavong S, Roberts T, Turner P, Wangrangsimakul T, Dance D. 2019. Impact of CLSI and EUCAST breakpoint discrepancies on reporting of antimicrobial susceptibility and AMR surveillance. Clin Microbiol Infect 25:910–911. 10.1016/j.cmi.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alkasir R, Ma Y, Liu F, Li J, Lv N, Xue Y, Hu Y, Zhu B. 2018. Characterization and transcriptome analysis of Acinetobacter baumannii persister cells. Microb Drug Resist 24:1466–1474. 10.1089/mdr.2017.0341. [DOI] [PubMed] [Google Scholar]
- 103.Nudel K, McClure R, Moreau M, Briars E, Abrams AJ, Tjaden B, Su XH, Trees D, Rice PA, Massari P, Genco CA. 2018. Transcriptome analysis of Neisseria gonorrhoeae during natural infection reveals differential expression of antibiotic resistance determinants between men and women. mSphere 3:312–330. 10.1128/mSphereDirect.00312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhattacharyya R, Liu J, Ma P, Bandyopadhyay N, Livny J, Hung D. 2017. Rapid phenotypic antibiotic susceptibility testing through RNA detection. Open Forum Infect Dis 4:S33. 10.1093/ofid/ofx162.082. [DOI] [Google Scholar]
- 105.Zhao L, Abedpour N, Blum C, Kolkhof P, Beller M, Kollmann M, Capriotti E. 2019. Predicting gene expression level in E. coli from mRNA sequence information. 2019 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB). IEEE, Piscataway, NJ. [Google Scholar]
- 106.Zrimec J, Börlin CS, Buric F, Muhammad AS, Chen R, Siewers V, Verendel V, Nielsen J, Töpel M, Zelezniak A. 2020. Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure. Nat Commun 11:6141. 10.1038/s41467-020-19921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cox G, Sieron A, King AM, De Pascale G, Pawlowski AC, Koteva K, Wright GD. 2017. A common platform for antibiotic dereplication and adjuvant discovery. Cell Chem Biol 24:98–109. 10.1016/j.chembiol.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 108.Kavvas ES, Yang L, Monk JM, Heckmann D, Palsson BO. 2020. A biochemically-interpretable machine learning classifier for microbial GWAS. Nat Commun 11:2580. 10.1038/s41467-020-16310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Public Health Agency of Canada. 2016. Canadian antimicrobial resistance surveillance system report–report 2016. Public Health Agency of Canada, Ottawa, Canada. [Google Scholar]
- 110.Genome Alberta. 2021. UK–Canada One Health workshop on antimicrobial resistance in agriculture and the environment–report 2021. Genome Alberta, Alberta, Canada. [Google Scholar]
- 111.Deng X, den Bakker HC, Hendriksen RS. 2016. Genomic epidemiology: whole-genome-sequencing–powered surveillance and outbreak investigation of foodborne bacterial pathogens. Annu Rev Food Sci Technol 7:353–374. 10.1146/annurev-food-041715-033259. [DOI] [PubMed] [Google Scholar]
- 112.Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, Tewolde R, Schaefer U, Jenkins C, Dallman TJ, de Pinna EM, Grant KA, Salmonella Whole Genome Sequencing Implementation Group. 2016. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4:e1752. 10.7717/peerj.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Argimón S, Masim MAL, Gayeta JM, Lagrada ML, Macaranas PKV, Cohen V, Limas MT, Espiritu HO, Palarca JC, Chilam J, Jamoralin MC, Villamin AS, Borlasa JB, Olorosa AM, Hernandez LFT, Boehme KD, Jeffrey B, Abudahab K, Hufano CM, Sia SB, Stelling J, Holden MTG, Aanensen DM, Carlos CC. 2020. Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nat Commun 11:2719. 10.1038/s41467-020-16322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hendriksen RS, Munk P, Njage P, van Bunnik B, McNally L, Lukjancenko O, Röder T, Nieuwenhuijse D, Pedersen SK, Kjeldgaard J, Kaas RS, Clausen PTLC, Vogt JK, Leekitcharoenphon P, van de Schans MGM, Zuidema T, de Roda Husman AM, Rasmussen S, Petersen B, Bego A, Rees C, Cassar S, Coventry K, Collignon P, Allerberger F, Rahube TO, Oliveira G, Ivanov I, Vuthy Y, Sopheak T, Yost CK, Ke C, Zheng H, Baisheng L, Jiao X, Donado-Godoy P, Coulibaly KJ, Jergović M, Hrenovic J, Karpíšková R, Villacis JE, Legesse M, Eguale T, Heikinheimo A, Malania L, Nitsche A, Brinkmann A, Saba CKS, Kocsis B, Solymosi N, Thorsteinsdottir TR, The Global Sewage Surveillance Project Consortium, et al. 2019. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun 10:1–12. 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.World Health Organization. 2016. Diagnostic stewardship: a guide to implementation in antimicrobial resistance surveillance sites. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 116.Karp BE, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, Hale KR, Wilson W, Friedman CR, Griffin PM, McDermott PF. 2017. National antimicrobial resistance monitoring system: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Dis 14:545–557. 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brolund A, Lagerqvist N, Byfors S, Struelens MJ, Monnet DL, Albiger B, Kohlenberg A, European Antimicrobial Resistance Genes Surveillance Network (EURGen-Net) Capacity Survey Group. 2019. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Eurosurveillance 24:1900123. 10.2807/1560-7917.ES.2019.24.9.1900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tenover FC, Williams PP, Stocker S, Thompson A, Clark LA, Limbago B, Carey RB, Poppe SM, Shinabarger D, McGowan JE. 2007. Accuracy of six antimicrobial susceptibility methods for testing linezolid against staphylococci and enterococci. J Clin Microbiol 45:2917–2922. 10.1128/JCM.00913-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bobenchik AM, Hindler JA, Giltner CL, Saeki S, Humphries RM. 2014. Performance of Vitek 2 for antimicrobial susceptibility testing of Staphylococcus spp. and Enterococcus spp. J Clin Microbiol 52:392–397. 10.1128/JCM.02432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zasowski EJ, Bassetti M, Blasi F, Goossens H, Rello J, Sotgiu G, Tavoschi L, Arber MR, McCool R, Patterson JV, Longshaw CM, Lopes S, Manissero D, Nguyen ST, Tone K, Aliberti S. 2020. A systematic review of the effect of delayed appropriate antibiotic treatment on the outcomes of patients with severe bacterial infections. Chest 158:929–938. 10.1016/j.chest.2020.03.087. [DOI] [PubMed] [Google Scholar]
- 121.Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. 2017. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 64:15–23. 10.1093/cid/ciw649. [DOI] [PubMed] [Google Scholar]
- 122.Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD, Cheung A, Rogers ZN, Meshulam-Simon G, Huijse L, Balakrishnan S, Quinn JV, Hollemon D, Hong DK, Vaughn ML, Kertesz M, Bercovici S, Wilber JC, Yang S. 2019. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 4:663–674. 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 123.Hogan CA, Yang S, Garner OB, Green DA, Gomez CA, Dien Bard J, Pinsky BA, Banaei N. 2021. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis 72:239–245. 10.1093/cid/ciaa035. [DOI] [PubMed] [Google Scholar]
- 124.Peker N, Couto N, Sinha B, Rossen JW. 2018. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin Microbiol Infect 24:944–955. 10.1016/j.cmi.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 125.Grant K, Jenkins C, Arnold C, Green J, Zambon M. 2018. Implementing pathogen genomics: a case study. Public Health England, London, United Kingdom. [Google Scholar]
- 126.Armstrong GL, MacCannell DR, Taylor J, Carleton HA, Neuhaus EB, Bradbury RS, Posey JE, Gwinn M. 2019. Pathogen genomics in public health. N Engl J Med 381:2569–2580. 10.1056/NEJMsr1813907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gröschel MI, Owens M, Freschi L, Vargas R, Marin MG, Phelan J, Iqbal Z, Dixit A, Farhat MR. 2021. GenTB: a user-friendly genome-based predictor for tuberculosis resistance powered by machine learning. Genome Med 13:1–14. 10.1186/s13073-021-00953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hunt M, Bradley P, Lapierre SG, Heys S, Thomsit M, Hall MB, Malone KM, Wintringer P, Walker TM, Cirillo DM, Comas I, Farhat MR, Fowler P, Gardy J, Ismail N, Kohl TA, Mathys V, Merker M, Niemann S, Omar SV, Sintchenko V, Smith G, Soolingen D, Supply P, Tahseen S, Wilcox M, Arandjelovic I, Peto TEA, Crook DW, Iqbal Z. 2019. Antibiotic resistance prediction for Mycobacterium tuberculosis from genome sequence data with Mykrobe. Wellcome Open Res 4:191. 10.12688/wellcomeopenres.15603.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.CRyPTIC Consortium and the 100000 Genomes Project. 2018. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. New Engl J Med 379:1403–1415. 10.1056/NEJMoa1800474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Public Health England. 2018. National mycobacterium reference service–South (NMRS-South): user handbook. Public Health England, London, United Kingdom. [Google Scholar]
- 131.Pfyffer GE, Wittwer F. 2012. Incubation time of mycobacterial cultures: how long is long enough to issue a final negative report to the clinician? J Clin Microbiol 50:4188–4189. 10.1128/JCM.02283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.World Health Organization. 2020. Global tuberculosis report 2020. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 133.Wright GD. 2019. Environmental and clinical antibiotic resistomes, same only different. Curr Opin Microbiol 51:57–63. 10.1016/j.mib.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 134.Lever J, Krzywinski M, Altman N. 2016. Logistic regression. Nat Methods 13:541–542. 10.1038/nmeth.3904. [DOI] [Google Scholar]
- 135.Noble WS. 2006. What is a support vector machine? Nat Biotechnol 24:1565–1567. 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 136.Niehaus KE, Walker TM, Crook DW, Peto TEA, Clifton DA. 2014. Machine learning for the prediction of antibacterial susceptibility in Mycobacterium tuberculosis, p 618–621. 2014 IEEE-EMBS Int Conf on Biomed Health Informatics. 10.1109/BHI.2014.6864440. [DOI]