SUMMARY
Class C β-lactamases or cephalosporinases can be classified into two functional groups (1, 1e) with considerable molecular variability (≤20% sequence identity). These enzymes are mostly encoded by chromosomal and inducible genes and are widespread among bacteria, including Proteobacteria in particular. Molecular identification is based principally on three catalytic motifs (64SXSK, 150YXN, 315KTG), but more than 70 conserved amino-acid residues (≥90%) have been identified, many close to these catalytic motifs. Nevertheless, the identification of a tiny, phylogenetically distant cluster (including enzymes from the genera Legionella, Bradyrhizobium, and Parachlamydia) has raised questions about the possible existence of a C2 subclass of β-lactamases, previously identified as serine hydrolases. In a context of the clinical emergence of extended-spectrum AmpC β-lactamases (ESACs), the genetic modifications observed in vivo and in vitro (point mutations, insertions, or deletions) during the evolution of these enzymes have mostly involved the Ω- and H-10/R2-loops, which vary considerably between genera, and, in some cases, the conserved triplet 150YXN. Furthermore, the conserved deletion of several amino-acid residues in opportunistic pathogenic species of Acinetobacter, such as A. baumannii, A. calcoaceticus, A. pittii and A. nosocomialis (deletion of residues 304–306), and in Hafnia alvei and H. paralvei (deletion of residues 289–290), provides support for the notion of natural ESACs. The emergence of higher levels of resistance to β-lactams, including carbapenems, and to inhibitors such as avibactam is a reality, as the enzymes responsible are subject to complex regulation encompassing several other genes (ampR, ampD, ampG, etc.). Combinations of resistance mechanisms may therefore be at work, including overproduction or change in permeability, with the loss of porins and/or activation of efflux systems.
KEYWORDS: AmpC β-lactamases, cephalosporinases, ESAC, extended-spectrum, phylogeny, primary structure
INTRODUCTION
β-lactamases remain an important natural or acquired mechanism of resistance to β-lactam antibiotics. New enzymes of this type, belonging to the molecular classes defined by Ambler and then completed by several authors (1–3), have been regularly discovered since the 1980s. The class C β-lactamases (BLCs), also known as AmpC or cephalosporinases, have a long history marked by the gradual loss of efficacy for the treatment of many bacterial infections, due initially to their large inactivation spectrum, including penicillins, the first cephalosporins (e.g., cephalothin), and cephamycins (e.g., cefoxitin), together with the general absence of an inhibitory effect of clavulanic acid, sulbactam, and tazobactam (4–6). The next problem encountered was the emergence of constitutive or overproduced mutants, eventually overcome by the development of oxyiminocephalosporins, such as cefotaxime and ceftazidime (7). However, plasmid-borne cephalosporinases were subsequently discovered, particularly in species without chromosomal ampC genes (e.g., Klebsiella pneumoniae, Salmonella enterica, and Proteus mirabilis) and in Escherichia coli, which possesses an intrinsic ampC gene usually not expressed. These enzymes are derived from chromosomally encoded enzymes specific to other species, such as Enterobacter cloacae and Citrobacter freundii, and their discovery raised new fears, allayed by the discovery of cefepime and cefpirome (4, 8). However, a new step in resistance development was then detected, with the discovery of extended-spectrum β-lactamases AmpC (ESAC), mutants or variants with an extended inactivation spectrum for oxyiminocephalosporins (ceftazidime, cefotaxime, cefepime, and cefpirome) due to the mutation of certain sites, in the R2-loop (9, 10), for example, by substitution, deletion, or insertion. Carbapenem resistance mediated by a combination of mechanisms, including a constitutive species-specific cephalosporinase and porin loss, has emerged more recently (11, 12). Finally, the recent development of novel enzyme inhibitors, such as avibactam, has elicited considerable medical interest due to its ability to inhibit serine β-lactamases (classes A, C, and D). However, this has already led to the selection of clinical mutants resistant to ceftazidime–avibactam combinations, most of them bearing deletions of various sizes in the Ω-loop region of AmpC (13, 14), similarly to β-lactamases from other classes (e.g., KPC-3).
Various methods for detecting resistant clinical isolates have been proposed, particularly for resistance to oxyiminocephalosporins and carbapenems. These methods include phenotypic tests, enzymatic methods based on hydrolysis, immunochromatographic assays, and molecular tests designed to test for the presence of particular genes, encoding class C β-lactamases, for example (15). Whole-genome sequencing (WGS) is a very useful approach for the precise identification of mechanisms of resistance to β-lactams, and has provided a large body of sequence data. Nevertheless, genotype-to-phenotype extrapolations are not straightforward, due to the variability of gene expression and polymorphisms linked to silent mutations at diverse sites, depending on the β-lactamase considered (16). Improvements in our knowledge should make it possible to improve analyses of the resistance mechanisms detected, particularly against β-lactams, in the future. These mechanisms are numerous and differ considerably between species. The current classification of β-lactamases into four molecular classes is based on motifs involved in binding and hydrolysis, such as 70SXXK, 130SDN, and 234KTG for class A (17), and on diverse residues involved in determining affinity, either increasing the inactivation spectrum (e.g., ESBLs) or decreasing it (e.g., IRT) (18–20). A more detailed comparative analysis of the primary structure of a large number of proteins would facilitate the classification of enzymes into groups of clusters displaying common structural features, as for the enzymes of class A (21). BLCs seem to have a lower level of structural diversity, but the abundance of data now available in databases (e.g., Beta-Lactamase DataBase [BLDB, http://bldb.eu/] [22], Bacterial Antimicrobial Resistance Reference Gene Database [https://www.ncbi.nlm.nih.gov/bioproject/313047] [23], CARD [https://card.mcmaster.ca/] [24]) suggests that a more detailed analytical approach, based on several thousand sequences, would now be justified.
When we began this analysis in 2019, several numbering schemes had been proposed for class C β-lactamases, which were unified in a standardized structure-based numbering scheme published in 2020 (25). Accurate comparisons of protein sequences have improved structural classification, by providing a clearer identification of polymorphisms by species, a more precise identification of the bacterium updated in line with the continual changes in taxonomy, a better understanding of the residues or zones involved in the possible extension of the inactivation spectrum by species or bacterial group, and with respect to both substrates and enzyme inhibitors, such as avibactam. The ACC-type β-lactamases appeared to be plasmid-encoded enzymes originating from Hafnia alvei with an unusual susceptibility pattern characterized by resistance to expanded-spectrum cephalosporins such as ceftazidime, cefotaxime, and, sometimes, cefpirome, and by susceptibility to cefepime and inhibition by cefoxitin (see the section on Natural ESACs for more details).
PHYLOGENETIC COMPARISON
The serine β-lactamases have been divided into three classes (A, C, and D) based on sequence similarity (1, 2, 26). A protein structure-based phylogeny clearly distinguished between these classes (27–29). The amino acid-based phylogenetic tree based on 3,943 class C sequences reveals large differences between the major groups (Acinetobacter, Aeromonadales, Burkholderiales, Enterobacterales, Pseudomonadales, Rhizobiales), each of which displays at least 24% amino-acid sequence identity (Fig. 1).
FIG 1.
(A) Phylogram for representative and putative class C β-lactamases, compared with β-lactamases from classes A, B, and D. (B) Focused view on the phylogram of class C β-lactamases. The protein sequences of representative enzymes are listed in references 33, 47, 54, and 64. The sequences were filtered using CD-HIT (https://github.com/weizhongli/cdhit) at 90% sequence identity, then aligned with Clustal Omega (319). The tree was constructed using RAxML (320), and the phylogram generated using FigTree (version 1.4.3). The tree was unrooted.
Highly conserved and major clusters, including many variants displaying >75% sequence identity with each other, were found for the following genera: Escherichia/Shigella, Citrobacter, Enterobacter, Hafnia, Klebsiella, Morganella, Serratia. Other clusters (Acinetobacter, Aeromonas, Burkholderia, Erwinia, Pseudomonas, Rhizobium, Yersinia, etc.) are less conserved, with a percent amino-acid identity between sequences of less than 50%. These findings highlight the need for more accurate taxonomic approaches in the future. Sequences from the Proteobacteria were particularly prevalent, but BLCs were widely distributed between bacterial groups, with the exception of Gram-positive bacteria, in which they were rare (Table 1) (28, 29). Finally, a separate cluster including several genera (e.g., Bradyrhizobium, Legionella, Parachlamydia) was identified and found to contain generic serine hydrolases and some carboxylesterases VIII with weak β-lactamase activity (Fig. 1).
TABLE 1.
Overview of bacteria producing class C β-lactamases
| Class | Order | Genus |
|---|---|---|
| Alphaproteobacteria | Rhizobiales | Agrobacterium, Bosea, Bradyrhizobium, Inorhizobium, Mesorhizobium, Methylobacterium, Microvirga, Ochrobactrum, Phyllobacterium, Pseudorhodoplanes, Rhizobium |
| Rhodobacterales | Rhodobacter, Ruegeria, Silicibacter, Sulfitobacter | |
| Rhodospirillales | Dongia | |
| Betaproteobacteria | Burkholderiales | Achromobacter, Bordetella, Burkholderia, Caballeronia, Collimonas, Cupriavidus, Herbaspirillum, Janthinobacterium, Massilia, Noviherbaspirillum, Pandoraea, Paraburkholderia |
| Neisseriales | Chromobacterium, Laribacter, Snodgrassella | |
| Rhodocyclales | Thauera | |
| Gammaproteobacteria | Aeromonadales | Aeromonas |
| Alteromonadales | Shewanella | |
| Cellvibrionales | Microbulbifer | |
| Enterobacterales | Budvicia, Buttiauxella, Cedecea, Citrobacter, Cronobacter, Edwardsiella, Enterobacter, Erwinia, Escherichia:Shigella, Ewingella, Hafnia, Klebsiella, Lelliottia, Morganella, Pantoea, Photorhabdus, Pluralibacter, Pragia, Providencia, Regiella, Rouxiella, Serratia, Siccibacter, Xenorhabdus, Yersinia | |
| Legionellales | Legionella | |
| Oceanospirillales | Aidingimonas, Chromohalobacter, Halomonas, Salinicola | |
| Pseudomonadales | Acinetobacter, Pseudomonas, Psychrobacter | |
| Vibrionales | Vibrio | |
| Xanthomonadales | Dyella, Lysobacter, Xanthomonas | |
| Deltaproteobacteria | Myxococcales | Myxococcus |
| Terrabacteria | Actinobacteria | Mycobacterium |
| Negativicutes | Pelosinus | |
| FCB group | Chitinophagales | Sediminibacterium |
| Cytophagales | Dyadobacter, Emticicia, Siphonobacter | |
| Flavobacteriales | Chryseobacterium | |
| Sphingobacteriales | Sphingobacterium | |
| PVC group | Chlamydiales | Chlamydia |
| Parachlamydiales | Parachlamydia | |
| Unclassified | Dependentiae |
PRIMARY STRUCTURE/SEQUENCE ANALYSIS
Highly Conserved Motifs and Residues
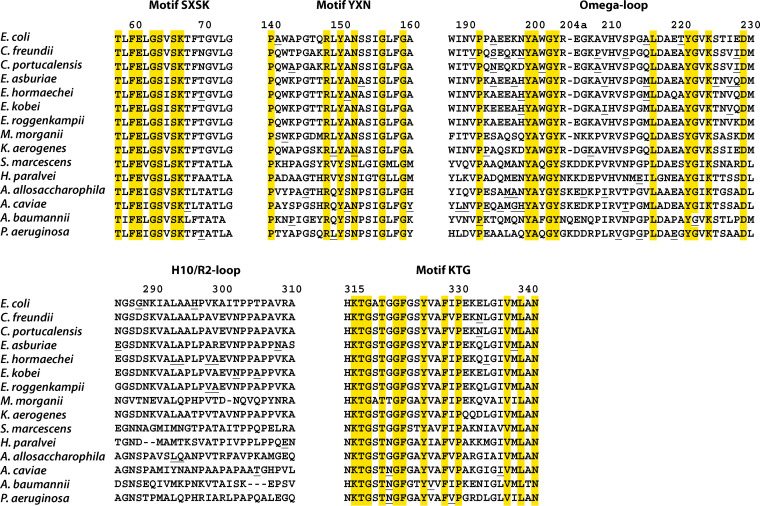
A standardized amino-acid numbering scheme was recently developed, through collaboration, for class C β-lactamases (25). This numbering scheme has greatly facilitated molecular comparisons between class C enzymes. Indeed, “SANC” (structural alignment-based numbering of class C β-lactamases) preserves the usual numbering of the major catalytic residues (64S, 67K, 150Y, and 315K). Three highly conserved motifs are currently used to characterize this molecular class (64SXSK, 150YXN, and 315KTG) (17, 28), but other conserved residues have been characterized and can be used for the more accurate identification of AmpC enzymes (Tables 2 and 3). They include 37 residues strictly conserved (100%) and 33 residues highly conserved (90–97%) in a data set of 32 representative class C β-lactamases examined in a previous study (25). Proline (P) and aromatic (F, W, Y) residues are overrepresented at these positions compared to class A β-lactamases. Indeed, prolines are present at positions 18, 26, 94, 118, 122, 140, 192, 213, 277, 330, and 345 for class C β-lactamases, but only at positions 107, 183, and 226 for class A β-lactamases. Similarly, strictly conserved aromatic residues are present at positions 60, 138, 150, 199, 203, 221, 271, 322, 325, and 328 for class C β-lactamases, but only at positions 66, 210, and 229 for class A β-lactamases. If aromatic amino acids (F, Y and W) are considered together, as a single category, additional conserved positions emerge for BLCs: 43, 69, 134, 135, 170, 188, 233, 260, 276, and 344 (Table 3).
TABLE 2.
Representative class C β-lactamases
| Bla | Origin of namec | Accession no. | Genomic localizationa | Organism | No. of residues | References |
|---|---|---|---|---|---|---|
| ACC-1 | Ambler Class C-1 | AJ133121 | P | Klebsiella pneumoniae | 386 | 321 |
| ACT-1 | AmpC Type | U58495 | P | Klebsiella pneumoniae | 381 | 322 |
| ABA-1 (ADC-2) | Acinetobacter baumannii | AY177427 | IS | Oligella urethralis | 383 | 323 |
| ABAC-1 (ADC-3) | Acinetobacter baumannii Class C | AY178995 | Chr | Acinetobacter baumannii | 383 | 323 |
| ADC-1 | Acinetobacter-derived cephalosporinase | AJ009979 | Chr | Acinetobacter baumannii | 383 | 247 |
| AQU-1 | Aeromonas aquariorum | AB765393 | Chr | Aeromonas dhakensis | 380 | 249 |
| AsbA1 | Aeromonas so bria | U10250 | In | Aeromonas jandaei | 381 | 324 |
| BIL-1 (CMY-2-like) | Name of patient (Bilal) | X74512 | P | Escherichia coli | 383 | 325, 326 |
| BlaE | Gene name | AY442183 | Chr | Mycobacterium smegmatis | 380 | 327, 328 |
| BUT-1 | Buttiauxella sp. | AJ415568 | Chr | Buttiauxella sp. | 383 | 329 |
| CAV-1 | Aeromonas caviae | AF462690 | Chr | Aeromonas caviae | 382 | 44 |
| CDA-1 | Cedecea davisae | KJ650399 | Chr | Cedecea davisae | 382 | 330 |
| CepH | Cephalosporinase hydrophila | AJ276030 | Chr | Aeromonas hydrophila | 382 | 41 |
| CepS | Cephalosporinase sobria | X80277 | Chr | Aeromonas sobria | 382 | 331 |
| CFE-1 | Citrobacter fr eundii | AB107899 | P | Escherichia coli | 381 | 208 |
| CHR-1 | Chromohalobacter sp. | AB070219 | Chr | Chromohalobacter sp. | 396 | 169, 332 |
| CMA-1 | Cronobacter malonaticus | KF640251 | Chr | Cronobacter malonaticus | 375 | 333 |
| CMH-1 | Chi Mei Hospital | JQ673557 | P | Enterobacter cloacae | 381 | 334 |
| CMY-1 | Active on cephamycins | X92508 | P | Klebsiella pneumoniae | 382 | 233 |
| CMY-2 | Active on cephamycins | X91840 | P | Klebsiella pneumoniae | 381 | 335 |
| CSA-1 | Cronobacter sakazakii | KF623543 | Chr | Cronobacter sakazakii | 375 | 333 |
| DHA-1 | Dhahran (Saudi Arabia) | Y16410 | P | Salmonella enteritidis | 379 | 336, 337 |
| Ear-1 | Enterobacter ae rogenes | AJ544162 | Chr | Enterobacter aerogenes | 381 | 338 |
| EDC-1 | Edwardsiella AmpC | EF467366 | Chr | Edwardsiella tarda | 386 | – |
| ENT-1 | Buttiauxella agrestis CF01Ent1 | AJ489827 | Chr | Buttiauxella agrestis | 390 | 339 |
| ERH-1 | Erwinia rhapontici | AY288518 | Chr | Erwinia rhapontici | 379 | 340 |
| FOX-1 | Active on cefoxitin | X77455 | P | Klebsiella pneumoniae | 382 | 341 |
| IDC-1 | Integron derived cephalosporinase | MN985649 | In | sediment metagenome | 395 | 31 |
| K12 (EC-1) | Escherichia coli K12 | J01611 | Chr | Escherichia coli | 377 | 342, 343 |
| LAT-1 | Active on latamoxef | X78117 | P | Klebsiella pneumoniae | 381 | 344, 345 |
| LHK-1 | Laribacter hongkongensis | AY632070 | Chr | Laribacter hongkongensis | 388 | 346 |
| LRA10-1 | β-lactam resistance from Alaska | EU408357 | – | uncultured bacteria (soil) | 375 | 30 |
| LRA13-1b | β-lactam resistance from Alaska | EU408352 | – | uncultured bacteria (soil) | 609b | 30 |
| LRA18-1 | β-lactam resistance from Alaska | EU408355 | – | uncultured bacteria (soil) | 386 | 30 |
| LYL-1 | Lysobacter lactamgenus | X56660 | Chr | Lysobacter lactamgenus | 385 | 347 |
| MIR-1 | Miriam hospital | M37839 | P | Klebsiella pneumoniae | 381 | 348, 349 |
| MOX-1 | Active on moxalactam | D13304 | P | Klebsiella pneumoniae | 382 | 232, 350 |
| OCH-1 | Ochrobactrum anthropi | AJ401618 | Chr | Ochrobactrum anthropi | 390 | 351 |
| P99 (ACT-89) | Enterobacter hormaechei P99 | X07274 | Chr | Enterobacter hormaechei | 397 | 352 |
| PAO-1 (PDC-1) | Pseudomonas aeruginosa (Pseudomonas-derived cephalosporinase) | AY083595 | Chr | Pseudomonas aeruginosa | 397 | 353, 354 |
| PAC-1 | Pseudomonas aeruginosa Class C | KY285014 | Tn | Pseudomonas aeruginosa | 381 | 75 |
| SLC-1 | Serratia liquefaciens Class C | DQ022079 | – | uncultured bacteria (soil) | 379 | 76 |
| PSI-1 | Psychrobacter immobilis | X83586 | Chr | Psychrobacter immobilis | 401 | 355, 356 |
| RHO-1 | Rhodobacter sphaeroides | CP000144 | Chr | Rhodobacter sphaeroides | 380 | 357 |
| SR50 (SRT-1-like) | Serratia marcescens SR50, Serratia resistant | X52964 | Chr | Serratia marcescens | 376 | 358 |
| SST-1 | Susceptible strain | AB008455 | Chr | Serratia marcescens | 378 | 359 |
| TRU-1 | Formerly Aeromonas tructi | EU046614 | Chr | Aeromonas enteropelogenes | 382 | 360 |
| YEC-1 | Yersinia enterocolitica cephalosporinase | X63149 | Chr | Yersinia enterocolitica | 388 | 361 |
| YRC-1 | Yersinia ruckeri cephalosporinase | DQ185144 | Chr | Yersinia ruckeri | 383 | 362 |
Chr, chromosome; In, integron; P, plasmid; Tn, transposon; IS, insertion sequence; –, unknown.
Fusion between two β-lactamases (class C and class D).
The underlined letters form the name of the β-lactamase.
TABLE 3.
Conserved residues in class C β-lactamases
| Positiona | Residue (>90 % conserved) | Secondary structureb | % conservedb |
|---|---|---|---|
| 18 | P | H1 | 94 |
| 26 | P | S | 97 |
| 27 | G | E | 100 |
| 29 | A | E | 97 |
| 36 | G | T | 97 |
| 43 | F/Y/W | E | 100 |
| 44 | G | E | 100 |
| 54 | V | 91 | |
| 58 | T | 100 | |
| 60 | F | E | 100 |
| 61 | E | E | 100 |
| 63 | G | G | 100 |
| 64 | S | G | 100 |
| 66 | S | H2 | 100 |
| 67 | K | H2 | 100 |
| 71 | G/A | H2 | 100 |
| 73 | L | H2 | 94 |
| 77 | A | H2 | 91 |
| 94 | P | G | 94 |
| 96 | L | G | 100 |
| 109 | L | H3 | 97 |
| 110 | A/G | H3 | 100 |
| 111 | T | T | 100 |
| 113 | T/S | 100 | |
| 115 | G | S | 100 |
| 116 | G | 96 | |
| 118 | P | 94 | |
| 119 | L | S | 97 |
| 122 | P | 100 | |
| 123 | D/E | T | 100 |
| 134 | F/Y/W | H4 | 100 |
| 135 | Y/F | H4 | 97 |
| 138 | W | 97 | |
| 140 | P | 100 | |
| 145 | G | T | 97 |
| 148 | R | E | 100 |
| 150 | Y | 100 | |
| 152 | N | H5 | 100 |
| 155 | I | H5 | 91 |
| 156 | G | H5 | 97 |
| 159 | G | H5 | 100 |
| 170 | F/Y | H6 | 100 |
| 187 | T/S | Ω | 100 |
| 188 | Y/W/F | Ω | 100 |
| 191 | V | Ω | 97 |
| 192 | P | Ω | 97 |
| 199 | Y | Ω | 97 |
| 200 | A | Ω | 100 |
| 202 | G | Ω | 100 |
| 203 | Y | Ω | 100 |
| 210 | R/H | Ω | 100 |
| 211 | V | Ω | 91 |
| 213 | P | Ω | 94 |
| 214 | G | Ω | 94 |
| 221 | Y | Ω H7 | 100 |
| 222 | G | Ω | 100 |
| 224 | K | Ω | 94 |
| 229 | D | H8 | 100 |
| 260 | Y/W/F | E | 97 |
| 267 | Q | E | 100 |
| 269 | L | S | 97 |
| 271 | W | 100 | |
| 272 | E | E | 100 |
| 277 | P | S | 97 |
| 286 | G | H10 R2 | 97 |
| 315 | K | E | 100 |
| 316 | T | E | 100 |
| 317 | G | E | 100 |
| 319 | T | 97 | |
| 321 | G | S | 97 |
| 322 | F | 100 | |
| 325 | Y | E | 100 |
| 328 | F | E | 97 |
| 330 | P | E | 100 |
| 335 | G/A | E | 100 |
| 337 | V | E | 91 |
| 339 | L | E | 100 |
| 340 | A | E | 97 |
| 341 | N | S | 100 |
| 345 | P | 97 | |
| 349 | R | H11 | 100 |
| 353 | A | H11 | 100 |
According to the “SANC” class C β-lactamases numbering scheme described in reference 25. Residues in boldface type are involved in the catalytic mechanism and/or in substrate binding.
From the alignment of 32 representative class C β-lactamases examined in reference 25. H, α-Helix; S/T, bend or turn; E/B, β-strand or β-bridge; G, 310-Helix; Ω, Ω-loop; R2, R2-loop.
In a representative set of 50 typical AmpC (see Table 2), the total number of amino acids in these enzymes ranged from 358 (P. fluorescens TAE4) to 397 (PDC-1) with a mean value of 383 ± 5.2, and the mean of molecular mass was estimated at 41.64 ± 0.58 kDa. The number of amino acids may be much higher for enzymes with fused domains (29, 30). AmpC enzymes typically have alkaline isoelectric points (pI), with a mean pI of 8.48 ± 1.22 for this representative set. Nevertheless, low predicted pI values were obtained for some enzymes (4.37 for CHR-1, 5.27 for BlaE, 4.59 for RHO-1, and 5.59 for IDC-1) (4, 31).
Enterobacterales
The Enterobacterales (EB) are one of the major groups emerging from the phylogram in Fig. 1. This group was clearly separated into two major clusters based on the analysis of 413 protein sequences: EB1, including Buttiauxella, Cedecea, Citrobacter, Edwardsiella, Erwinia, Escherichia, Enterobacter, Klebsiella aerogenes, Lelliottia, Morganella, Pluralibacter, Serratia fonticola, Yersinia, and Xenorhabdus; and EB2, including Budvicia, Cronobacter, Erwinia, Hafnia, Pantoea, Photorhabdus, Pragia, Providencia, Regiella, Rouxiella, Serratia, and Siccibacter (32). Most species generally cluster together in the same genus or cluster, but, surprisingly, several species (e.g., Erwinia teleogrylli, S. fonticola and Y. ruckeri) were located at some distance from their main clusters. The following conserved residues distinguished between the EB1 and EB2 groups: D47K, V65L, G71A, V72T, G74A, W101F, Y112H, Q120F, S154G, F158L, W201Q, A255T, R258G, G270M, and H314N. A deletion in position 116 and an insertion in position 204a were also identified in EB2 (25, 33). These groups displayed a high degree of diversity (e.g., 55–100% sequence identity for the whole EB1 group). Consensus sequences (CS) were therefore determined for the main species.
For Escherichia species (E. coli/Shigella, E. albertii, and E. fergusonii), sequence identity ranged from 88% to 100% and 11 consensus sequences were obtained from 367 protein sequences. The E. coli sensu stricto population is now considered to have a strong phylogenetic structure, with the identification of at least 12 phylogroups (A, B1, B2, C, D, E, F, G, H, clade 1, clade III, and clade IV) (34).
Several conserved residues were found to be specific for phylogroups A, A1, and B1 (235R, 238M, 239N, 241R, 245N), whereas others were specific for phylogroups B2 and D (235Q, 238L, 239K, 241L, 245N/T) (16). All four clusters (A–D) carry RMNRE residues in the 235–245 region (33), and this key feature was observed in clinical isolates able to generate extended-spectrum AmpC β-lactamases (ESACs), as described above. The 287SGN triplet is present in the sequence of such enzymes (35). Phylogroup B2 contains the specific conserved residues 175K, 193S, 282I, 288D, 296R, and 300P, and is separated from phylogroup D by the residues 185T and 244T, in particular.
Finally, 78 protein sequences from uncultured bacteria obtained by metagenomic analyses on human fecal and environmental samples, originally found in the NCBI database as class C CMY-LAT-MOX-ACT-MIR-FOX β-lactamases, belong to this major cluster. They have been assigned to E. coli, Shigella, E. fergusonii, or E. albertii, and are characterized by a typical triplet (6HSE) (36–38).
One particular feature of interest is that all the protein sequences obtained from the genus Hafnia (H. alvei, H. paralvei) carry a two-residue deletion (residues 289–290) in H-10 and the R2-loop. The plasmid-encoded enzymes ACC-1 and ACC-4 are more than 99.4% identical to the AmpC of H. paralvei, a newly identified bacterial species (39, 40).
Aeromonas
The second major group identified included 318 protein sequences from Aeromonas, a genus for which many new species have been discovered over the last 2 decades. This is also the group in which the first inducible cephalosporinases (AsbA1, CAV-1, CepH, CepS) were detected, in A. hydophila (41), A. jandei (42), A. sobria (43), and A. caviae (44). The group is highly diverse, with a sequence identity ranging from 50% to 100%. Thirty-six species from this group have been identified, including about 20 species pathogenic to humans, such as A. caviae, A. dhakensis, A. veronii, and A. hydrophila (45). Phylogenetic analysis identified two CS for A. caviae, A. dhakensis, A. hydrophila, and A. salmonicida, with sequence identities of 80% to 100%, suggesting that new species may be discovered in the future (46, 47). Some of these species displayed a particular primary structure modification: the deletion of two or three residues (positions 301 to 303) in the R2-loop, as in A. caviae and in A. dhakensis (25).
Acinetobacter
Genus Acinetobacter has a long history of taxonomic changes, but is dominated by A. baumannii, the principal genomic species, which plays a major role in nosocomial infections, particularly in intensive care units (48, 49). This major group is heterogeneous, with 500 protein sequences and 26 species, and with sequence identities ranging from 42% to 100% (50). The denomination ADC for “Acinetobacter-derived cephalosporinase,” followed by a number to distinguish between individual enzymes, was proposed because of the large numbers of A. baumannii variants or members of the Abc complex (51) (http://bldb.eu/BLDB.php?prot=C#ADC). In a previous analysis of 103 genomes, the blaADC-like gene was found to be present in 13 validly named species, eight genomic species, and six taxa (52). Three major clusters were identified: one cluster including the A. baumannii–A. calcoaceticus or Abc complex and a number of other species: A. lactucae (formerly known as A. dijkshoorniae), A. nosocomialis, A. oleivorans, A. pittii, and A. seifertii; a second cluster containing nine species: A. apis, A. albensis, A. bereziniae, A. bohemicus, A. celticus, A. guillouiae, A. johnsonii, A. kyonggiensis, and A. rudis; and a third cluster including a number of other species: A. baylyi, A. beijerinckii, A. gyllenbergii, A. haemolyticus, A. junii, A. proteolyticus, A. parvus, A. soli, A. ursingii, A. tjernbergiae, and A. venetianus (50). A large number of protein sequences were initially misidentified, for two reasons. The first was the substantial taxonomic modifications that had taken place, and the second was the preferential use of standard biochemical methods and automated systems or devices widely used in clinical bacteriology laboratories, such as API 20NE, Vitek 2, Phoenix, Biolog, and MicroScan WalkAway, potentially leading to incorrect identification. MALDI-TOF can facilitate correct identification of the members of the Abc complex to species level, provided that an accurate database is used (15, 49). Accurate identification of the clinically important members of the Abc complex (A. baumannii, A. pittii, A. nosocomialis, A. seifertti and A. dijkshoorniae) and A. calcoaceticus, which is considered pathogenic, is possible only by molecular methods such as DNA-DNA hybridization (gold standard method) and DNA sequence-based analysis on various types of DNA sequences (16S rRNA, rpoB, 16S-23S intergenic spacer) (15). The 16S rRNA sequencing method is highly reliable at genus level, but discriminates poorly between species. By contrast, rpoB genes are highly variable and considered appropriate for Acinetobacter species identification. The multiplex PCR based on species-specific genes, such as gyrB, is simple, rapid (results obtained within 2 h), and reproducible, but limited to major species: A. baumannii, A. nosocomialis, A. pittii and A. calcoaceticus.
One structural feature distinguishing these clusters was a conserved deletion of three residues in the R2-loop (residues 304–306) for the first two clusters (53, 54). The third cluster, including A. baylyi producing ADC-8 with a low susceptibility pattern, particularly for cephalosporins, did not display this deletion (54, 55). ADC-1 and ADC-68, naturally produced by A. baumannii, carry this deletion of three residues in the R2-loop (residues 304–306) and were considered to be extended-spectrum AmpC (ESACs). Such enzymes typically hydrolyze penicillins and narrow- and expanded-spectrum cephalosporins and aztreonam, but not cefepime. Acquired resistance to cefepime or cefpirome has been reported to be associated with amino-acid substitutions—R148Q (close to YXN), V211A (within the Ω-loop), and N287S (in helix H-10)—also conferring higher levels of resistance (4 to 64 times higher) to ceftazidime and cefotaxime (56, 57). Ceftazidime resistance resulted from overproduction of the ADC β-lactamase and the provision of strong promoter sequences by the insertion sequence ISAba1 (56, 58).
In conclusion, the complexity of this bacterial group suggests that other species are likely to be identified, particularly given the very small size of some of the clusters identified.
Pseudomonas
A phylogenetic analysis of 582 class C protein sequences from Pseudomonas yielded a broad distribution, with at least 36 clusters. Sequence identity varied considerably, from 42% to 100%, between the 33 species producing class C β-lactamases from this group. Heterogeneity levels were particularly high for some species, such as P. putida, for which at least 11 clusters were observed, and for P. fluorescens, suggesting misidentification, as reported by subsequent comparative studies of genomes (59–64). Indeed, the Pseudomonas group is among the most diverse in terms of the species it contains, but, with the exception of P. aeruginosa, for which 430 sequences have been analyzed, its taxonomy is still under revision.
P. aeruginosa is one of the principal pathogens isolated from immunosuppressed patients and cases of hospital-acquired infections associated with multiresistance (65). It also causes chronic lung infections in patients with cystic fibrosis (66). The effective treatment of P. aeruginosa infections is challenging, because several mechanisms of resistance to antibiotics, including β-lactamases and efflux pump overexpression, have evolved in this species (67). An inducible chromosomal AmpC-type enzyme was identified, with a wild-type inactivation spectrum including various β-lactams, such as aminopenicillins, the oldest cephalosporins, and cephamycins. Following its overproduction due to mutations altering the peptidoglycan recycling process, this cephalosporinase is a major source of resistance to ticarcillin, piperacillin, ceftazidime, and aztreonam (68, 69). However, additional missense mutations have extended its inactivation spectrum to cefepime and cefpirome. Many variants, also observed as natural polymorphisms, have been named according to a nomenclature specifically developed for P. aeruginosa, as PDCs (Pseudomonas-derived cephalosporinases) (68, 69). A phylogenetic analysis of 430 sequences displaying 87% to 100% identity yielded two clusters (C1, C2). The main cluster (C1), containing 416 protein sequences, had a consensus sequence with several variants (70). The second cluster (C2) was very small, comprising only 14 sequences. The polymorphism observed in cluster C1 had already been described in 44 antibiotic-susceptible strains with amino-acid substitutions at positions 53, 71, 79, 149, 178, and 365 (69). However, other variants were observed, in particular in positions 121, 153, 174, 178, 213, 216, 219, and 293, which served as mutation hot spots, thus extending the substrate specificity of PDC β-lactamases (see the section on ESACs).
Plasmid-encoded Enzymes
Chromosomal class C β-lactamases predominate, but plasmid-mediated enzymes are nevertheless important, and the protein sequences of 256 such enzymes were examined. Since their emergence in 1989, several types of plasmid-encoded AmpCs and their variants have been identified: ACC, ACT, CFE, CMY-2/BIL/LAT, DHA, FOX, MIR, and MOX/CMY-1 (4, 8, 22). CMY-2 is the most common plasmid-encoded enzyme family, detected worldwide in isolates that do not naturally produce a cephalosporinase (P. mirabilis, K. pneumoniae, and S. enterica, in particular), and also in E. coli, which has a weak expression of the natural cephalosporinase. This family displays little variability, with only about 8% of amino acids varying in a comparison of 92 sequences, including BIL-1 and LAT-1. Other families varied in terms of the number of protein sequences and percent identity: ACC (2 and 99%), ACT (10 and 86%), CFE (2 and 92%), DHA (15 and 97%), FOX (13 and 94%), MIR (5 and 99%), and MOX/CMY-1 (10 and 75%). Since the 1990s, many modifications have been made to bacterial taxonomy, with the definition of new bacterial species, particularly in the genera Citrobacter, Enterobacter, and Aeromonas. Finally, phylogenetic analysis confirmed several changes in the identification of the progenitors for chromosomal and plasmid-encoded cephalosporinases (25, 71–74) (Table 4). The rare class C β-lactamases SLC-1 and PAC-1, encoded by a gene in a chromosome-inserted Tn1721-like transposon, probably originate from an enterobacterium, but the origins of other enzymes (integron-encoded IDC-types, Alaska soil metagenomes LRA10-1, LRA13-1, LRA18-1) remain unknown (30, 31, 75, 76).
TABLE 4.
Class C β-lactamases and species-specific progenitors
| Enzyme/straina | First identification | Updated identification | %b | References |
|---|---|---|---|---|
| ACC-1*c | Hafnia alvei | Hafnia paralvei | 99.7 | 321, this review |
| ACT-1*c | Enterobacter cloacae | Enterobacter asburiae | 98.4 | 322, this review |
| Aer-1 | Enterobacter aerogenes | Klebsiella aerogenes | 98.9 | 338, 363 |
| AQU-1 | Aeromonas aquariorum | Aeromonas dhakensis | 99.2 | 249, 364 |
| AsbA1 | Aeromonas sobria | Aeromonas jandaei | 94.5 | 324, this review |
| BIL-1* | Citrobacter freundii | Citrobacter portucalensis | 96.8 | 326, this review |
| BUT-1 | Buttiauxella sp. | Scandinavium goeteborgense | 99.0 | 329, 365 |
| BUT-2 | Buttiauxella agrestis | Scandinavium goeteborgense | 99.5 | 339, 365 |
| CAV-1 | Aeromonas caviae | Aeromonas allosaccharophila | 97.1 | 44, 71 |
| CFE-1* | Citrobacter freundii | Citrobacter europaeus | 99.2 | 208, this review |
| CFE-2* | Citrobacter freundii | Citrobacter werkmanii | 95.0 | 366, this review |
| CMY-1*c | Aeromonas hydrophila | Aeromonas sanarellii | 95.3 | 233, this review |
| CMY-2* | Citrobacter freundii | Citrobacter portucalensis | 98.4 | 335, this review |
| EDC-1 | Edwardsiella tarda | Edwardsiella piscicida | 99.1 | This review |
| FOX-1* | Aeromonas caviae | Aeromonas allosaccharophila | 94-98 | 71, 341 |
| LAT-1* | Citrobacter freundii | Citrobacter portucalensis | 97.4 | 345, this review |
| MIR-1*c | Enterobacter cloacae | Enterobacter roggenkampii | 99.7 | 25, 352 |
| MOX-1* | Aeromonas hydrophila | Aeromonas sanarellii | 94.5 | 72, 232 |
| MOX-2* | Aeromonas sp. | Aeromonas caviae | 98.9 | 72, 234 |
| MOX-9* | Aeromonas caviae | Aeromonas media | 98.0 | 72, 367 |
| P99 (ACT-89) | Enterobacter cloacae | Enterobacter hormaechei | 97.9 | 25, 352, this review |
| TRU-1 | Aeromonas tructi | Aeromonas enteropelogenes | 97.1 | 360, this review |
Plasmid-encoded enzymes are labeled by an asterisk.
Percentage of identity with consensus sequence.
Other plasmid-encoded types: ACC-4 (99.5% for H. paralvei); ACT-3, ACT-6, ACT-8, and ACT-10 (≥97.6% for E. asburiae); CMY-17, CMY-55, CMY-132, and CMY-161 (≥ 98.1% for A. sanarellii); MIR-4 (99.4% for E. roggenkampii). Their respective phylogenies can be found in references 32, 46, 50, and 63; and 73 and 74.
Serine Hydrolase, a Class C β-lactamase or a Carboxylesterase VIII?
BLCs are usually genus-specific and chromosomally encoded, found mostly in Gram-negative bacteria, and more specifically in Alpha-, Beta- and Gammaproteobacteria. However, several clusters (e.g., Legionella, Bradyrhizobium, Parachlamydia) are clearly divergent, with low levels of sequence identity (between 10 and 20%) (28) (Fig. 1). This early divergence is related to a limited number of highly conserved motifs or residues, most reported in Table 3: 43Y/G, 54V/T, 58T, 60F/E, 63A/G, 64SXXK, 94P, 96L, 109L, 111T, 138W, 140P, 145G, 148R/Y, 150YXN/H, 187T, 202G/Y, 229D, 271W, 315KXG, 330P, 335G, 337V, 339L, 341N, 357L, and 360L. Several bacteria from Bacteroidetes and Firmicutes (e.g., Pelosinus fermentans, Sediminibacterium salmoneum) also carry enzymes with some of these highly conserved residues and motifs: 27G, 44G, 58T, 60F, 61E/F, 63G, 64SXXK, 67K, 94P, 115G, 144G, 150YXN, 200A, 202G, 220A/S, 222G/S, 229D, 315K/HT/SG, 319T, 321GF, 337V, and 341N. These enzymes are generally identified as serine hydrolases in databases, but with no additional bacteriological and enzymatic details that could be used to determine whether or not they should be considered as class C β-lactamases.
One family of enzymes (microbial carboxylesterases VIII, EC 3.1.1.1) mostly identified in metagenomic libraries from various environmental samples, including EstC, EstM-N1/N2, Est22, EstU1, and EstSRT1, appears to be phylogenetically and structurally related to BLCs (Fig. 1). These enzymes generally hydrolyze nitrocefin (EstC), cephalothin (Est22), cephaloridine, cefazolin (EstU1), and oxyiminocephalosporins (EstSRT1) (77–83). The EstB and Est22 enzymes selectively deacetylate cephalosporin-based substrates leaving the amide bond of the β-lactam ring intact (82, 83), while for EstU1 it was not clear from the HPLC data if the observed pattern against cephalosporin substrates was due to deacetylation or amide bond hydrolysis of the β-lactam ring (77). The catalytic efficiencies of EstSRT1 for cephalothin, cefotaxime, and cefepime are similar to that of EstU1 for cefazolin (77). These enzymes bear the active site residues 100S, 103K, and 218Y (64S, 67K, and 150Y according to SANC numbering), as well as other residues highly conserved in BLCs (Table 3). They have an overall structure consisting of a mixed α/β domain and a small helical domain, similar to that of class C β-lactamases (80, 84–86).
The classification of these carboxylesterases as BLCs does not appear to be appropriate, given the total absence of β-lactamase activity for some of these enzymes, their low levels of in vitro antibacterial activity against β-lactams, including cephalosporins, their primary structure, and their low levels of sequence identity to genuine BLCs. Nevertheless, these findings suggest that the promiscuous β-lactamase activity of some of these family VIII esterases may have evolved from BLCs or vice versa, and that some may have closer evolutionary relationships to BLCs (79).
SECONDARY AND TERTIARY STRUCTURES
The first crystallographic structure of an AmpC was reported in 1994, for the covalent complex between a phosphonate transition-state analog and the E. hormaechei P99 (formerly known as E. cloacae P99) cephalosporinase, which belongs to the ACT family (87). Many structures from different families of class C β-lactamases have since been published, including ACC (88, 89), ACT (87, 90–102), ADC (53, 103–108), CMH (109), CMY (100, 110–114), EC (111, 115–152), FOX (153–155), MOX (156, 157), PDC (158–167) and TRU (168) enzymes, together with the halophilic β-lactamase (HaBLA) from Chromohalobacter sp. 560 (169) and the class C β-lactamase from a psychrophilic organism, Pseudomonas fluorescens (170). More than 200 structures of class C β-lactamases have been described, and an updated list of these structures can be found in the Beta-Lactamase DataBase (BLDB; http://bldb.eu/S-BLDB.php) (22).
The typical three-dimensional structure of class C β-lactamase contains two mixed α/β domains: one composed of nine antiparallel β-sheets and three α-helices (H1, H10 and H11) on one side, and the other, composed of three small antiparallel β-sheets and eight α-helices (H2 to H9) on the other side (Fig. 2). Two additional structural elements important for the interaction with substrates, Ω-loop and R2-loop, are located between H6 and H8 (residues 189–225), and close to H10 (residues 288–309), respectively (see reference 25 for a detailed analysis of residues involved in these regions). Structurally, class C β-lactamases belong to the Beta-lactamase PFAM family (PF00144) and share the same overall fold with class A and class D β-lactamases, penicillin-binding proteins (PBPs), and family VIII carboxylesterases.
FIG 2.
Representative three-dimensional structure for class C β-lactamases. The structure of E. hormaechei P99 (formerly known as E. cloacae P99; PDB code 1BLS) (87) is colored in orange (α-helixes) and purple (β-strands). The Ω- and R2-loops are colored in green and blue, respectively. The most conserved residues (see Table 3) are represented as sticks and colored in cyan. The numbering of residues follows the SANC nomenclature (25).
REGULATION AND EXPRESSION OF CLASS C β-LACTAMASES
In some Enterobacterales (other than E. coli) and P. aeruginosa, ampC gene expression is weak and inducible. This induction involves several proteins (AmpR, AmpD, and AmpG) and two muropeptides (4, 171–174). Nevertheless, the molecular mechanisms mediating AmpC overproduction in P. aeruginosa appear to be more complex than initially thought, as other proteins, such as AmpDh2, AmpDh3, and DacB (PBP4), have since been identified as involved in these mechanisms (175–177).
Genetic Context and Regulation
The ampR and ampC genes are linked. AmpR is encoded by a gene located immediately upstream from ampC and acts as a transcriptional activator, binding to the intercistronic region upstream from the ampC gene promoter. The ampD gene encodes an N-acetyl-anhydromuramyl-L-alanine amidase involved in recycling the products of peptidoglycan catabolism, including, in particular, the 1,6-anhydro-N-acetylmuramyl-tripeptide (anhNAM-tripeptide). AmpD releases the tripeptide, which is then recycled for synthesis of the UDP-NAM pentapeptide (uridine 5′-pyrophosphoryl-N-acetylmuramic acid-pentapeptide), for integration into the neo-peptidoglycan. There is therefore a permanent balance between these two components in the cytoplasm.
Under normal conditions, AmpR associates with the UDM-NAM pentapeptide, and the resulting complex binds to the intercistronic region, thereby preventing transcription from both the ampR and ampC promoters. However, the presence of excess anhNAM-tripeptide alters this binding and promotes an increase in ampC transcription. Two clinical situations can result in an excessive increase in anhNAM-tripeptide levels: substantial degradation of the peptidoglycan due to the presence of β-lactams in the periplasmic space, and changes in the AmpD amidase preventing the recycling of this component.
In E. cloacae complex and C. freundii, the main cause of ampC overexpression is represented by amino-acid substitutions in AmpD, resulting in the constitutive production of large amounts of AmpC and increasing resistance to cefotaxime and ceftazidime. Several resistant clinical isolates or in vitro mutants displaying AmpC overproduction have been identified in the Enterobacterales, including C. freundii and E. cloacae with various amino-acid substitutions (e.g., W7G, H34A, S37R, Y63F, R80H, G82C, A94P/V, Y102, E116A, L117R, R108 D121G, D127G, N150I, H154N, A158D, K162H/Q, D164E/A, W171, A172L) capable of triggering constitutive β-lactamase production (178–182). In some cases, the ampD gene is truncated by a premature stop codon or an IS1 insertion, or an IS4321 element may be inserted into the promoter (178, 183, 184).
In P. aeruginosa, acquired resistance to amino- and ureido-penicillins, cephamycins, and, to a lesser extent, to oxyiminocephalosporins (ceftazidime, cefotaxime, ceftriaxone) and aztreonam, is very often related to overproduction of the PDC β-lactamase, which is also linked to virulence (185). Several variants with ampD mutations displaying resistance in vivo and in vitro have been reported (186–189). As in the Enterobacterales, the principal genetic mechanisms involved missense mutations creating amino-acid substitutions (A84G, D61Y, G148A, A136V, G148A, S175L) or insertion sequences (IS1669).
The LysR-type transcriptional regulator AmpR was initially identified as involved in regulating the inducible class C β-lactamases produced by various Gram-negative rods, such as C. freundii, E. cloacae, S. marcescens, and P. aeruginosa. However, recent results indicate that in P. aeruginosa AmpR regulates the expression of many genes involved in other pathways, such as quinolone resistance, quorum sensing, and associated virulence phenotypes (190). Phylogenetic analyses revealed the presence of AmpR homologs in many Alpha-, Beta-, and Gammaproteobacteria, and several highly conserved residues were found in C. freundii, E. cloacae, and P. aeruginosa (190, 191). AmpR loss leads to susceptibility to β-lactams, whereas constitutively high levels of ampC expression were reported for clinical isolates and in vitro strains with mutations resulting in AmpR amino-acid substitutions: C. freundii (S35F, G102D/E, D135A, Y264N) (4, 192, 193), E. cloacae (T84I, R86C, D135N/V, E274K) (178, 194), and P. aeruginosa (A12R, D135N, G154R, G237A) (186, 195–197). Transposon mutagenesis of P. aeruginosa strain PAO-1 and complementation experiments have generated two mutants with stronger β-lactam resistance due to transposition into two new genes (mpl, nuoN) (198).
AmpG, an intrinsic membrane protein, displays permease activity mediating the transport of muropeptides from the periplasm to the cytoplasm, such transport being essential for the induction of class C β-lactamases (199–201). Deletion of the gene encoding this protein in P. aeruginosa results in a lower level of bacterial resistance to ampicillin. AmpG proteins are widespread and highly variable in Gram-negative bacteria (e.g., Enterobacterales, A. baumannii, P. aeruginosa). Genetic experiments showed that ampG genes from E. coli and A. baumannii can complement AmpG function in P. aeruginosa (200). In this species, the site-directed mutagenesis of some highly conserved AmpG residues (G29A, G29V, A129V, A197S, and A197D mutants) results in a loss of resistance to ampicillin; ampG mRNA levels were found to be normal for two other mutants (A129T and A129D), but the proteins encoded had much lower levels of activity (200).
AmpC Overproduction in E. coli
In E. coli, in the absence of the ampR gene, the expression of ampC is usually weak, under the control of a naturally nonefficient promoter (4). In this species, acquired resistance to old or narrow-spectrum cephalosporins, cephamycins (e.g., cefoxitin), or even oxyiminocephalosporins, is related to ampC overexpression due to the selection of a stronger promoter by mutation, or, less often, by insertion (202, 203). The most important factors responsible for strengthening the ampC promoter are mutations creating a consensus -35 box (TTGACA) by T/A transversion at position -32 or C/T transition at position -42, and base-pair insertions increasing the distance between the -35 and -10 boxes to 17 or 18 bp (4, 203, 204). Mutations have also been identified within the attenuator region and the -10 box, but such mutations have little effect on ampC expression. Similar promoter mutations have been reported in strains from livestock, such as pigs or calves (205, 206).
AmpC Overproduction in A. baumannii
Overexpression of the chromosomal ampC gene in A. baumannii was observed following the acquisition of an IS element, mostly ISAba1, resulting in a strong promoter and resistance to expanded-spectrum cephalosporins (cefotaxime, ceftazidime) and aztreonam (207).
Transferable β-lactamases
Most plasmid-encoded class C β-lactamases mobilized by transposons and insertion sequence elements, with the exception of the ACT-1, CFE-1, and DHA-types, are in a “derepressed” state (31, 208–210). The genes encoding the ACC-1 and CMY-2 types are transcribed under the control of a strong promoter in the mobile element (ISEcp1 or IS26) (211, 212). The high level of blaMIR-1 gene expression is due to the presence of a more efficient hybrid promoter upstream from the natural promoter (213). All these genes are often associated with integrons inserted in close proximity and forming composite structures with other transposable elements, including a gene cassette in a class 1 integron characterized by rapid spread (31). Gene cassettes are under the control of a constitutive promoter specific for the gene cassette array. Their expression level is determined not only by promoter strength, but also by distance from the promoter (214). The removal of ampC gene cassettes from the integron may constitute a less costly control mechanism than the continuous overproduction of many plasmid-borne class C enzymes (215). However, three plasmid-encoded enzyme types with an ampR gene (ACT-1, CFE-1, DHA-1) and a genetic organization identical to those of chromosomal enzymes rarely cause resistance to oxyiminocephalosporins despite constitutively high levels of AmpC activity due to an amino-acid substitution (D135A) (190, 193).
In conclusion, given the complexity of the regulation of ampC expression, the variability of these genes in databases and the small number of clinical examples of acquired resistance with the identification of hot spots for at least ampR, ampD, and ampG, it is not currently appropriate to analyze such mutants by genomic procedures, particularly in the absence of the initial susceptible clinical isolate. Classical methodologies are preferable for the detection of AmpC β-lactamase overproduction. These methods include phenotypic approaches assessing synergy between a cephamycin or an oxyiminocephalosporin and an inhibitor, such as cloxacillin or boronic acid (4). Nevertheless, the determination of β-lactamase activity with or without imipenem or cefoxitin induction, together with determinations of the mRNA levels for these genes by real-time RT-PCR, is probably much more accurate (69, 200, 216–218).
ACQUIRED EXTENDED-SPECTRUM CEPHALOSPORINASES (ESACs)
In Gram-negative bacteria, such as the Enterobacterales, various genetic events may increase the level of resistance, particularly to oxyiminocephalosporins, such as ceftazidime, cefotaxime, and, to a lesser extent, cefepime and cefpirome. The evolution of such resistance led to a new denomination: ESAC, for extended spectrum AmpC β-lactamases (219). The first ESAC was identified in an E. cloacae isolate (GC1) in Japan in 1992, based on the duplication of three amino acids at positions 208–210 (Ω-loop) and constitutive production (10).
In E. coli, AmpC is constitutively produced in small amounts in the absence of the ampR gene, under the control of a weak promoter (4). Within this species, acquired resistance to old or narrow-spectrum cephalosporins (e.g., cefoxitin) is related to the overproduction of AmpC due to the selection of a strong promoter generated by mutation, or, less often, by insertion (202, 203). A second step in the development of resistance involved the acquisition of plasmid-encoded AmpCs with a higher level of resistance to ceftazidime and aztreonam, for example (8). Finally, an even higher level of resistance (e.g., to cefepime or cefpirome) was developed following the emergence of an ESAC at low prevalence (≤1%) in human clinical isolates and even isolates from cattle (16, 220–222).
The genetic determinism of ESACs, leading to an increase in the catalytic efficiency of these enzymes against extended-spectrum cephalosporins, is based principally on missense mutations resulting in amino-acid substitutions, with structural modifications to the R1 (Ω-loop between residues 189 and 225) or R2 (H10 helix between residues 280 and 292 and/or R2-loop between residues 286 and 310) binding sites. Insertions or duplications in H10 or the R2 loop were also observed (Table 5).
TABLE 5.
Localization of ESAC-associated mutations in chromosomal or plasmid-encoded class C β-lactamases, the exact position of each mutation and the specific regions associated with each cluster
| Bla or straina | Location | First numbering | Updated numberingb | References |
|---|---|---|---|---|
| E. coli | ||||
| MEV | H10-helix | S282 duplication | idem | 368 |
| ECB33 | H10-helix | I283 duplication | I284 | 223 |
| HKY28 | H10-helix | 286GSD deletion | idem | 225 |
| EC16 | H10-helix | S287C | idem | 16 |
| EC13 | H10-helix | S287N | idem | 16, 35 |
| 8009162 | H10-helix, R2-loop | A292V | idem | 221 |
| EC80 | H10-helix, R2-loop | L293P | idem | 369 |
| BER | R2-loop | 293AA insertion | idem | 370 |
| 7014517 | R2-loop | 295ALA insertion | idem | 221 |
| EC15 | R2-loop | H296P | idem | 16, 240 |
| EC14 | R2-loop | V298L | idem | 16, 240 |
| KL | H11-helix | V350F | idem | 224 |
| Citrobacter | ||||
| CHA | close to YSN | R148H | idem + Q196H | 35, 371 |
| CMY-107 | Ω-loop | Y199C | idem | 372 |
| CMY-27 | Ω-loop | W221C | W201C | 373 |
| CMY-30 | Ω-loop | V231G | V211G | 374 |
| CMY-42 | Ω-loop | V231S | V211S | 260, 375 |
| CMY-95 | Ω-loop | V211A | idem | 369 |
| CMY-32 | Ω-loop | G214E | idem | 376 |
| CMY-54 | Ω-loop | 217EL insertion | 216aEL insertion | 377 |
| GN346 | Ω-loop | E219K | idem | 378 |
| CMY-136 | Ω-loop | Y221H | idem | 113 |
| CMY-172 | R2-loop | 290KVA deletion+ N346I | idem | 295 |
| CMY-2 | R2-loop | A292P or L293P | idem | 379 |
| CMY-33 | R2-loop | 293LA deletion | idem | 235, 380 |
| CMY-44 | R2-loop | 293LAAL deletion | idem | 235 |
| CMY-69 | R2-loop | A295P | A294P | 381 |
| CMY-99 | R2-loop | P306T | idem | 382 |
| CMY-37 | R2-loop | L316I | L296I | 383 |
| Enterobacter | ||||
| LN04004SS1 | Ω-loop | 213K-226G deletion | idem | 384 |
| GC1 | Ω-loop | 208AVR duplication | idem | 10, 97 |
| CHE | R2-loop | 289SKVALA294 deletion | idem | 9 |
| Ent630 | R2-loop | 292-293 AL deletion | idem | 294 |
| P99 (ACT-89) | R2-loop | L293P | idem | 385 |
| MHN | R2-loop | V298E | idem | 386 |
| LN04004SS1 | H11-helix | N366H | N346H | 384 |
| K. aerogenes | ||||
| EA6/13/17/20 | H3-helix | Q90H, W101C, L107Y | idem | 387 |
| Ea595 | H10-helix, R2-loop | V291G | idem | 388 |
| Ear2 | R2-loop | L293P | idem | 338 |
| S. marcescens | ||||
| 520R | H2-helix | T64I | T70I | 389 |
| SRT-1 | Ω-loop | E213K | E219K | 359 |
| ES46, ES71 | Ω-loop | E235K | E219K | 390 |
| SMSA | Ω-loop | S220Y | idem | 391 |
| HD | R2-loop | 287MNGT deletion | 293MNGT deletion | 392 |
| Hafnia | ||||
| ACC-4g | Ω-loop | V211G + 289-290 deletion | idem | 393, this review |
| ACC-1g | R2-loop | 289-290 deletion | idem | 25, 250, this review |
| ACC-2 | R2-loop | 289-290 deletionc | idem | 250, this review |
| Aeromonas | ||||
| CMY-9 | R2-loop | E85D + 299-301 deletion | E61D + 301-303 deletion | 394 – 396 |
| CMY-19 | R2-loop | I292S + 299-301 deletion | I292S + 301-303 deletion | 236, 395, 396 |
| CMY-1 | R2-loop | 299-301 deletion | 301-303 deletiond | 110, 236, 397 |
| MOX-1 | R2-loop | 303-305 deletion | 301-303 deletiond | 156 |
| MOX-2 | R2-loop | 303-305 deletion | 301-303 deletione | 234, this review |
| MOX-13 | R2-loop | 303-305 deletion + N346I | 301-303 deletione | 398, this review |
| CMY-10 | R2-loop, H11-helix | E85D + 299-301 deletion | E61D + 301-303 deletion | 110, 237, 395, 397 |
| A. baumannii | ||||
| ADC-56 | H5-helix | R148Q | idem | 57 |
| ADC-53 | Ω-loop | V208A | V211A | 56 |
| ADC-33 | Ω-loop | P210R + 215A duplication | P213R + 218aA duplication | 248 |
| ADC-51 | R2-loop | N283S | N287S | 56 |
| ADC-1 | R2-loop | 304-306 deletion | idem deletionf | 53 |
| ADC-68 | Ω-loop | G220D + R320G | G217D + R321G | 108 |
| P. aeruginosa | ||||
| PDC-222 | H2-helix | T96I | T70I | 230 |
| PDC-78 | H2-H3-loop, Ω-loop | R100H + G216R | R100H + R215G | 69 |
| PDC-82 | H3-H4-loop | F121L + M175L | F121L + M174L | 69, 229 |
| PDC-73 | H5-helix | P154L | P153L | 69 |
| PDC-82 | H6-helix | M175L | M174L | 69 |
| PDC-50 | Ω-loop | V213A | V211A | 69 |
| PDC-74 | Ω-loop | G216R | G215R | 69 |
| PDC-86 | Ω-loop, H7-helix | E221K | E219K | 69 |
| PDC-80 | Ω-loop, H7-helix | E221G | E219G | 69 |
| PDC-85 | Ω-loop, H7-helix | Y223H | Y221H | 69 |
| PDC-223 | Ω-loop | 229G-247E deletion | 202G-219E deletion | 230 |
| PDC-221 | Ω-loop | E247K | E219K | 69 |
| PDC-88 | H10-helix, R2-loop | 290TP deletion | 289TP deletion | 69 |
| PDC-89 | H10-helix, R2-loop | 290TPM deletion | 289TPM deletion | 69 |
| PDC-91 | H10-helix, R2-loop | 290TPMA deletion | 289TPMA deletion | 69 |
| PDC-44 | R2-loop | L294P | L293P | 69 |
| PDC-92 | R2-loop | 294LQ deletion | 293LQ deletion | 69 |
| PDC-76 | H11-helix | N347I | N346I | 69 |
Strain names are underlined.
According to the Standard Numbering Scheme (25).
All AmpC sequences of H. paralvei and H. alvei examined have this deletion.
This cluster includes CMY-1, MOX-1, CMY-8, CMY-10, CMY-11, CMY-19, and MOX-14.
These enzymes were located in different clusters (72).
This deletion was observed for all A. baumannii sequences, and particularly for ADC-7 (51).
Plasmid-encoded.
Most ESACs belong to phylogenetic group A, but others belong to phylogenetic group B1 and are characterized by the presence of the following conserved residues: 235R, 238M, 239N, 241R, 245D, 281S, 288G, and 296H (16, 35, 221, 223). Nevertheless, two isolates with different residues in these positions have been reported (224). A single E. coli strain (HKY28) isolated from urine in Japan was found to produce an ESAC with a tripeptide deletion (286GS289D) altering binding to extended-spectrum cephalosporins, but also to sulbactam and tazobactam (225) (Table 5). Reversion of the deletion through a nine-base insertion restored the typical inhibitor-resistant phenotype of class C enzymes and decreased the level of resistance to cefepime and cefpirome. Finally, other positions affecting the ESAC phenotype were identified by the selection of mutants in vitro or by site-directed mutagenesis, resulting in a greater structural flexibility or affinity for extended-spectrum cephalosporins (141, 226).
Many BLCs (e.g., those of Pseudomonas, Citrobacter, Enterobacter, Morganella, and Serratia) are inducible, opening up possibilities for higher levels of production, resulting in stronger resistance to aminopenicillins, old cephalosporins, and ceftazidime or cefotaxime, and even to cefepime if the mutated enzyme is overproduced (227). Additionally, a significant proportion of strains has constitutive overproduction of class C enzymes, often after selection with β-lactam antibiotics. However, the risk of transitions between susceptibility and intermediate resistance (S/I) and between susceptibility and resistance (S/R) to cefepime is species-dependent. It is particularly high for the E. cloacae complex (66.3%), non-negligible for H. alvei (36.4%), moderate for Citrobacter and Proteus (18.1–21.9%) and S. marcescens (12.3%), low for K. aerogenes (1.1%), and inexistent for M. morganii (0%) (228). As summarized in Table 5, expansion of the inactivation spectrum results from missense mutations at several mutation hot spots (around triplet 150YAN, Ω-loop, H10-helix, and R2-loop), from duplications or deletions of 2–4 amino acids in the H10 helix (position 293), or insertions of amino acids into the Ω-loop. In some cases, two such events may be combined (Table 5). Various examples of increased catalytic efficiency (kcat/Km) of these enzymes, particularly against extended-spectrum cephalosporins (ceftazidime, cefotaxime, cefepime), are illustrated in Table 6.
TABLE 6.
ESAC and effects on kinetic constants for groups of enzymes with different phenotypesa
| Bla or strainb | Mutation | Ceftazidime (CAZ) |
Cefotaxime (CTX) |
Cefepime (FEP) |
References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM · s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM · s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM · s−1) | |||
| CMY-2-like | |||||||||||
| CMY-2 | 0.004 | 0.5 | 0.008c | 0.007 | 0.001 | 7 | 371 | ||||
| CMY-2 | R148H | 0.67 | 0.6 | 1.12c | NM | 0.003 | NM | 371 | |||
| CMY-2 | 0.01 | 0.02 | 0.5 | <0.01 | 0.005 | <2 | 372, 399 | ||||
| CMY-107 | 0.14 | 0.15 | 0.9 | 0.8 | 0.075 | 10.7 | 372 | ||||
| CMY-30 | 0.4 | 0.14 | 2.9 | 1.7 | 0.3 | 5.7 | 399 | ||||
| CMY-42 | 0.5 | 0.3 | 1.7 | 0.2 | 0.08 | 2.5c | 375 | ||||
| CMY-2 | 0.005 | 0.15 | 0.033 | 0.007 | 0.001 | 7 | 0.37 | 412 | 9 · 10−4 | 113, 296, 371 | |
| CMY-136 | 6.26 | 2360 | 0.003 | 4.71 | 20 | 0.24 | 1.79 | 3588 | 5 · 10−4 | 113 | |
| CMY-2 | NM | 1.8 | NM | NM | 108.1 | NM | 376 | ||||
| CMY-32 | 0.9 | 4.05 | 0.22 | NM | 988.9 | NM | 376 | ||||
| ACC | |||||||||||
| ACC-2 | 0.03 | 5.2 | 0.006c | 0.02 | 19 | 0.001 | 3.6 | 147 | 0.024 | 250 | |
| ACC-4 | 1.5 | 15 | 0.1 | 2.7 | 9.4 | 0.29 | 0.14 | 73 | 0.002 | 393 | |
| SRT | |||||||||||
| SerS | <0.5 | 50d | <0.01 | 2 | 4 | 0.5 | <0.5 | 100d | <0.005c | 391 | |
| SerR | S220Y | 520 | 570 | 0.9 | 800 | 980 | 0.8 | 330 | 1000 | 0.33 | 391 |
| FOX | |||||||||||
| FOX-3 | 0.273 | 1.18 | 0.231 | 0.081 | 0.076 | 1.06 | ND | ND | 3.32 · 10−3 | 241 | |
| FOX-8 | 2.6 · 10−3 | 0.382 | 6.8 · 10−3 | 18.2 · 10−3 | 10.95 · 10−3 | 1.66c | ND | ND | 1.36 · 10−3 | 241 | |
| FOX-4 | 1.33 | 13 | 10 | 1.33 | 0.23 | 5.69 | 11.01 | 1071 | 0.01 | 245 | |
| FOX-4 | 306GNSΔ | 0.84 | 6.44 | 0.13 | 0.24 | 0.087 | 2.75 | 2.5 | 103 | 0.02 | 245 |
| MOX-1/CMY-1-like | |||||||||||
| CMY-9 | 1.8 | 560 | 3.2 · 10−3 | 0.27 | 0.28 | 0.96 | NM | 950 | ND | 236 | |
| CMY-19 | 0.085 | 3.7 | 0.023 | 0.33 | 31 | 0.011 | 1.8 | 630 | 2.9 · 10−3 | 236 | |
| CMY-8 | 0.091 | 48 | 1.9 · 10−3 | 0.36 | 2.3 | 0.16 | 394 | ||||
| CMY-9 | 0.53 | 120 | 4.4 · 10−3 | 0.48 | 3.4 | 0.14 | 394 | ||||
| MOX-1 | ND | 311 | ND | ND | 211 | ND | 232 | ||||
| CMY-1 | 0.01 | 0.015 | 0.67 | 400 | |||||||
| CMY-10 | 5 | 33.9 | 0.15 | 110 | |||||||
| ACT | |||||||||||
| ACT-89 (P99) | 6.1 · 10−3 | 18.4 | 3.2 · 10−4 | 110 | |||||||
| ACT-89 (P99) | <1 | 20d | ND | 0.5 | 0.5d | 1 | 1 | 15d | 0.067 | 9 | |
| CHE | 289-294Δ | <1 | 1d | ND | 0.5 | 0.05d | 10 | 2 | 3d | 0.67 | 9 |
| ACT-89 (P99) | 0.065 | 28 | 2.3 · 10−3 | 401 | |||||||
| ACT-89 (P99) | L293C | 0.041 | 7 | 5.9 · 10−3 | 401 | ||||||
| ACT-89 (P99) | 0.013 | 15 | 8.7 · 10−4 | 0.5 | 100 | 4.7 · 10−3 | 385 | ||||
| ACT-89 (P99) | L293P | 0.10 | 10 | 0.01 | 3.1 | 24 | 0.13 | 385 | |||
| Ear | |||||||||||
| Ear | ND | 16d | ND | 0.15 | >500 | ND | 0.4 | 126 | 0.003 | 338 | |
| Ear2 | ND | 9.8d | ND | 0.15 | 10d | 0.015 | 0.4 | 9.1 | 0.044 | 338 | |
| ADC | |||||||||||
| ADC-1 | 0.7 | 16.0 | 0.044 | 0.16 | 0.5 | 0.32 | 53, 108 | ||||
| ADC-68 | 1.66 | 147.7 | 0.01 | 18.5 | 117.5 | 0.16 | 108 | ||||
| ADC-11 | 0.01 | 10 | 0.001 | 0.2 | 2.5d | 0.1 | 1 | 1800 | 5.5 · 10−4 | 248 | |
| ADC-33 | 4 | 30 | 0.13 | 1 | 0.5d | 2 | 10 | 1300 | 7.7 · 10−3 | 248 | |
| ADC-1 | 1.255 | 265 | 4.7 · 10−3 | 207 | |||||||
| ADC-5 | 0.011 | 232 | 4.7 · 10−5 | 207 | |||||||
| ADC-5 | P167S | 2.5 · 10−3 | 120 | 2.1 · 10−5 | 207 | ||||||
| ADC-5 | P167S/D242G/ Q163K/G342R | 1.235 | 90 | 0.014 | 207 | ||||||
| ADC-30 | 0.05 | 1.39 | 0.04 | 0.18 | 0.51 | 0.32 | 57 | ||||
| ADC-56 | 0.1 | 1.42 | 0.07 | 0.27 | 1 | 0.27 | 0.2 | 17.17 | 0.011 | 57 | |
| PDC | |||||||||||
| PDC-1 | 0.004 | 20 | 2 · 10−4 | 0.02 | 6 | 3.3 · 10−3 | 0.08 | 800 | 1 · 10−4 | 68 | |
| PDC-2 | 0.01 | 20 | 5 · 10−4 | 0.15 | 5 | 0.03 | 2 | 850 | 2.4 · 10−3 | 68 | |
| PDC-3 | 0.02 | 35 | 5.7 · 10−4 | 0.15 | 8 | 0.019 | 2 | 1300 | 1.5 · 10−3 | 68 | |
| PDC-5 | 0.015 | 30 | 5 · 10−4 | 0.1 | 5 | 0.02 | 2.5 | 1700 | 1.5 · 10−3 | 68 | |
| PDC-5 | 0.01 | 7.3 | 1.3 · 10−3 | 0.07 | 0.14 | 0.5 | >0.15 | >250 | 6 · 10−4 | 402 | |
| PDC-5 | N346Y | 0.06 | 19 | 3.2 · 10−3 | 0.2 | 1.2 | 0.17 | >0.17 | >300 | 5.5 · 10−4 | 402 |
NM, kcat not measurable; ND, not determined.
Strain names are underlined.
Value computed from kcat and Km, which is different from the value reported in the original paper.
Ki values (μM) were determined instead of Km values, using cephalothin as a reporter substrate.
In P. aeruginosa, acquired resistance to amino- and ureidopenicillins, cephamycins, and, at low levels, to oxyiminocephalosporins (ceftazidime, cefotaxime, ceftriaxone) and monobactams (aztreonam), is frequently related to overproduction of the AmpC β-lactamase (14, 68, 185, 187, 229). ESACs emerged in P. aeruginosa several years after Enterobacterales, and such enzymes were identified mainly from clinical sources, but also from in vitro studies. They differ from the wild-type AmpC of P. aeruginosa by various amino-acid substitutions, deletions, or insertions in four regions in the vicinity of the active site: the Ω-loop, the H10-helix, the H2-helix, and the C-terminal end of the protein (69, 229, 230) (Table 5).
Genus Aeromonas may have been the origin of several chromosomal (e.g., MOX-3, MOX-10) or plasmid-encoded β-lactamases (e.g., MOX-1, MOX-2, CMY-1, CMY-8, CMY-19) identified on several occasions from clinical isolates, mostly in Enterobacterales (e.g., K. pneumoniae and E. coli). The first plasmid-encoded enzyme, MOX-1, was identified in a K. pneumoniae isolate with a high level of resistance to various broad-spectrum β-lactams, including moxalactam, flomoxef, ceftizoxime, cefotaxime, and ceftazidime (231). According to its kinetic parameters, cephalothin was its ideal substrate, and it had good activity against benzylpenicillin, but poor activity against cloxacillin and piperacillin. Moxalactam and cefoxitin were also hydrolyzed, but ceftazidime and cefepime were poor substrates, with very high Km values (Table 6). Finally, aztreonam was found to inhibit MOX-1 (232). The X-ray crystallographic structure of MOX-1 suggested that residues 303–306 show a significant structural flexibility, possibly underlying the unique substrate profile of this enzyme, which can hydrolyze penicillins, cephalothin, expanded-spectrum cephalosporins, cefepime, and moxalactam (156). The position of the H10-helix in both MOX-1 and CMY-10 is shifted further away from the catalytic serine residue compared with the AmpCs from E. coli, E. hormaechei P99 (formerly known as E. cloacae P99), and E. cloacae GC1, whereas that of the AmpC from P. aeruginosa occupies an intermediate position. Such structural features may underlie the extended substrate profile of MOX-1 and CMY-10, which are also considered to be ESBLs, but have never been called ESACs (110, 156). Finally, the susceptibility profile of these enzymes is characterized by a high level of hydrolysis for cephamycins, oxacephems (e.g., cefoxitin), and moxalactam, increased resistance to cefotaxime compared with ceftazidime and aztreonam, and resistance to cefpirome and/or cefepime (233–239). The deletion of three residues in the R2-loop (positions 301–303) appears to be responsible for the expanded spectrum activity of CMY-10, and further mutation around this deletion in the P99 enzyme extended its substrate spectrum by widening the R2 region (Table 5) (110). Globally, all chromosomal (e.g., MOX-3, MOX-10, MOX-11) or plasmid-encoded (e.g., MOX-1, MOX-2, MOX-14, CMY-1, CMY-8, CMY-8b, CMY-9, CMY-10, CMY-11, CMY-19) β-lactamases from this family feature the sequence 289AKVILEANPTAAΔΔΔPRESG309S in the conserved R2 region (25). An increase in resistance to cefepime was also achieved by an additional amino-acid substitution (I292S) in the H10-helix region, as observed in CMY-11 and CMY-19 relative to CMY-9 (236) (Table 5).
The other enzymes from Aeromonas include a cluster of plasmid-encoded FOX-type enzymes without the amino-acid deletion at positions 301–303. These enzymes have a susceptibility profile characterized by a higher degree of resistance to cefoxitin (the origin of the family name, FOX) and to ceftazidime compared with cefotaxime (240–244). No ESACs have been identified among FOX variants (22). A tripeptide deletion (286GN288S) in the R2-loop of E. coli HKY28 led to an extended hydrolysis spectrum, but the same deletion by site-directed mutagenesis in FOX-4 did not increase catalytic efficiency for ceftazidime, cefotaxime, or cefepime, despite large differences in Km and kcat values (Table 6). A decrease in the MIC for cefoxitin was obtained in both E. coli HKY28 and FOX-4 Δ286-288, together with a slight increase in susceptibility to clavulanate, sulbactam, and tazobactam (245).
Structural studies have confirmed the importance of the Ω- and R2-loops for modulating the catalytic activity of class C β-lactamases. In CMY-10, a three-amino acid deletion in the R2-loop appears to underlie the extended spectrum activity (110), whereas a two-amino-acid deletion in the R2-loop of CMH-family AmpC enzymes from E. cloacae Ent385 leads to reduced susceptibility to ceftazidime-avibactam and cefiderocol (109). Similarly, the Y221H mutation in CMY-136 induces an important change in the confirmation of the Ω-loop, widening the active site cavity and conferring resistance to many β-lactams and combinations, including ceftolozane-tazobactam (113).
NATURAL ESACs
The term “naturally occurring ESAC” has been proposed for the enzymes in E. coli isolates with increased hydrolysis of oxyiminocephalosporins, including cefepime and cefpirome (MICs greater than or equal to 16 μg/mL and 0.5 μg/mL for ceftazidime and cefepime, respectively, without a positive synergy test with clavulanic acid), and resistance to amoxicillin and to amoxicillin-clavulanic acid (16). As for class A ESBLs, which are sometimes encoded by chromosomal genes (e.g., OXY-types for K. oxytoca) (246), both chromosomal and natural ESACs have been identified in A. baumannii (53), in which these β-lactamases are the principal source of resistance (49). The genes encoding ADC-type β-lactamases are non-inducible and generally expressed at low levels, resulting in resistance to penicillins and narrow-spectrum cephalosporins (51, 247). However, overexpression may be observed following the acquisition of an IS element, principally ISAba1, providing a strong promoter, and this may lead to resistance to expanded-spectrum cephalosporins (cefotaxime, ceftazidime) and aztreonam (207). All ADC-type β-lactamases (about 450 protein sequences examined) are characterized by a deletion of three residues (positions 304–306) in the R2-loop that enhances their catalytic efficiency against these clinically important drugs compared with enzymes without this deletion (53, 54, 108). Clinical variants with higher levels of resistance (4- to 64-fold increase) have been detected, with V211A (Ω-loop) or N287S (H10-helix) substitutions (56) (Table 5). Moreover, acquired resistance to cefepime or cefpirome and increased hydrolysis efficiency (ESAC) were induced by the P210R substitution and duplication of A215 in the Ω-loop (ADC-33) or by the substitution R148Q in the P2-loop (ADC-56) (57, 248). ADC-68 has seven amino-acid substitutions compared with ADC-1, one of which (321G) is located in the C-loop and another two (192A and 217D) in the Ω-loop. The overall structures of ADC-68 and ADC-1 are conserved, but there are marked structural differences in the Ω-loop and the C-loop. In particular, residues 217D and 321G in ADC-68 make a major contribution to the structural differences between these two ADC-type β-lactamases (108).
There is a two-amino acid deletion (residues 301–302) in the R2-loop of AmpC from two species, A. dhakensis and A. caviae (25), but an equivalent deletion in AQU-1 does not seem to affect the usual β-lactam resistance phenotype. However, cefotaxime monotherapy should be used with caution for severe A. dhakensis infections, because such treatment could lead to the selection of variants with constitutively high levels of β-lactamase production. Nevertheless, cefepime susceptibility was conserved in all cases studied (249).
A specific structural alteration to the R2-loop (deletion of two amino acids in positions 289–290) was observed in all Hafnia protein sequences, and this feature is unique among Enterobacterales (25, 33, 88) (Table 5). Furthermore, ACC-1 confers an unusual and unique pattern of susceptibility to β-lactams, with higher MICs for ceftazidime and cefotaxime than for cefoxitin (4, 240). In addition to the two-amino-acid deletion in positions 289–290 mentioned above, ACC-1 features two other remarkable structural alterations, one in the Ω-loop (213ME instead of 213PG) and another along the active-site rim (120F instead of 120Q) (25, 88). Moreover, ACC-2, a cephalosporinase encoded by an inducible chromosomal gene, displays marked hydrolysis activity against cefpirome in particular (250), in opposition to the widely accepted view that cefpirome is resistant to hydrolysis by class C enzymes, even when they are overproduced (251). ACC-2 is also strongly inhibited by cefoxitin (250).
In conclusion, these natural ESACs constitute a threat because of their resistance to cefepime and/or cefpirome, or even to carbapenems if they are overproduced, combined with porin loss and/or efflux pump activation.
CARBAPENEM RESISTANCE
Carbapenem resistance has become a serious threat to public health (252). It has spread worldwide in Gram-negative bacteria, and is continuing to increase due to the production of plasmid-encoded carbapenemases by Enterobacterales and by nonfermenting organisms (mainly A. baumannii and P. aeruginosa) (253, 254). The main mechanism underlying carbapenem resistance is the production of highly transmissible carbapenemases, such as KPC-types (class A), IMP-, NDM-, or VIM-types (class B), and OXA-types (class D). Carbapenems, which diffuse efficiently across the native outer membrane of Gram-negative bacteria, are poor substrates for class C β-lactamases (4). However, they may act as inhibitors in some cases, due to the high affinity of AmpCs for these antibiotics, suggesting that resistance may be acquired by a mechanism known and sometimes described as “trapping” (255), in which the cephalosporinase has a good affinity for the substrate combined with a very slow hydrolysis (256–259).
Nevertheless, several clinical failures and in vitro studies have been published concerning chromosomally encoded and transmissible class C β-lactamases with weak carbapenem hydrolysis activity, suggesting poor carbapenemase activity (253, 260, 261). Such resistance was always obtained from the combination of a high production of β-lactamase (a hyperproducing mutant of a chromosomally-encoded enzyme or a plasmid-mediated enzyme) with another mutation-driven mechanism, such as porin loss (OmpC, OmpF, OmpG, OmpK35, OmpK36) or efflux overexpression, which was essential for carbapenem resistance, preferentially against ertapenem, then imipenem, and finally meropenem (240, 262) (Table 7). The prevalence of such nonenzymatic mechanisms is low and differs between bacteria, with relatively few occurrences among Enterobacterales (mostly in K. pneumoniae, but also in E. coli, P. mirabilis, and S. enterica).
TABLE 7.
Chromosomal or plasmid-encoded class C β-lactamases: mechanisms of acquired resistance to carbapenems
| Fold-increase MICsc |
Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bla or straina | Species | Locationb | Mechanisms | CAZ | FEP | IMP | ERT | MER | |
| MEV | E. coli | Chr | ESAC + OmpC decrease + OmpF loss | >128 | >16 | 8 | >16 | 8 | 368 |
| ACC-1 | E. coli | P | + OmpC/OmpF loss | >64 | 64 | 4 | >128 | 4 | 240 |
| ACT-1 | E. coli | P | + OmpC/OmpF loss | >256 | >16 | 128 | >512 | 32 | 240 |
| CMY-2 | E. coli | P | + OmpC/OmpF loss | >256 | >16 | 256 | >512 | 256 | 240 |
| DHA-1 | E. coli | P | + OmpC/OmpF loss | >512 | 64 | 16 | 256 | 8 | 240 |
| FOX-1 | E. coli | P | + OmpC/OmpF loss | >512 | >128 | 4 | >256 | 16 | 240 |
| CMY-2 | E. coli | P | Overproduction + OmpC/OmpF loss | >256 | >16 | >64 | >256 | >32 | 403 |
| CMY-2 | E. coli | P | Overproduction + OmpC insertion IS1 | >64 | >256 | 260 | |||
| CMY-2 | E. coli | P | Overproduction + OmpC/OmpF loss | >256 | >128 | >256 | >512 | 258 | |
| EC14 | E. coli | Chr | ESAC + OmpC/OmpF loss | >256 | >128 | 16 | >32 | 1 | 404 |
| CMY-4 | S. enterica | P | + OmpF loss | >512 | >64 | >16 | 405 | ||
| ACT-1 | K. pneumoniae | P | + Omp42-kDa loss | >128 | 8-32 | 8->16 | 322 | ||
| ACT-1 | K. pneumoniae | P | + OmpK35/36 insertion + PhoE decrease | 64-256 | 128 | 256 | 406 | ||
| DHA-1 | K. pneumoniae | P | + OmpK36 loss | >128 | >32 | >32 | 407 | ||
| DHA-1 | K. pneumoniae | P | + OmpK35/36 loss + AcrAB/OqxAB | 32 | >32 | 277 | |||
| ACC-1 | K. pneumoniae | P | + OmpK35/36 loss | >128 | 16 | 8 | 32 | 16 | 262 |
| FOX-1 | K. pneumoniae | P | + OmpK35/36 loss | >512 | 32 | 64 | 128 | 64 | 262 |
| MOX-1 | K. pneumoniae | P | + OmpK35/36 loss | 64 | 8 | 32 | 32 | 32 | 262 |
| EA-Z | K. aerogenes | Chr | + Omp40-kDa loss | 16 | 408 | ||||
| E15 | K. aerogenes | Chr | + OmpK35/36 loss | >256 | >16 | >16 | 409 | ||
| E11 | E. cloacae | Chr | Overproduction + OmpK35/36 loss | >256 | >16 | >16 | 409 | ||
| 144 | E. cloacae | Chr | Overproduction + porin loss | 64 | 16 | >128 | 410 | ||
| 213 | P. rettgeri | Chr | Overproduction + porin loss | 4 | 8 | >128 | 410 | ||
| ACT-28 | E. kobei | Chr | Overproduction + OmpC-like protein | >256 | >64 | >128 | >512 | >512 | 264 |
| A-1 | A. baumannii | Chr | Overproduction + Omp33/36 kDa decrease | 32 | >64 | 265 | |||
| A(4) or B(4) | A. baumannii | Chr | Overproduction + Omp37/44/47 kDa decrease | >32 | >32 | 8-16 | >16 | 411 | |
| Paeβ-04 | P. aeruginosa | Chr | Overproduction ESAC + mexB increase | 32 | 16 | 16 | 16 | 275 | |
| 3-D8 | P. aeruginosa | Chr | Overproduction + mexB increase + OprD loss + dacB | 128 | 64 | 32 | >256 | 273 | |
| AM339 | P. aeruginosa | Chr | Overproduction + mexA and mexC and mexX increase + OprD loss | >32 | >128 | >16 | >16 | >16 | 216 |
Strain names are underlined.
Chr, chromosome; P, plasmid.
CAZ, ceftazidime; FEP, cefepime; IMP, imipenem; ERT, ertapenem; MER, meropenem.
Within the genus Enterobacter, carbapenem resistance is primarily due to the hyperproduction of chromosomal ampC associated with defective penetration of the carbapenemases in the bacterium (263). However, E. kobei from the E. cloacae complex group naturally produces an ACT-28-type AmpC β-lactamase that hydrolyzes imipenem more efficiently (264) and several plasmid-encoded AmpCs show intrinsically low carbapenemase activity (CMY-10, and to a lesser extent, CMY-2 and ACT-1) (240, 261).
Although in A. baumannii carbapenem resistance is mainly related to class B (VIM-, IMP-, and NDM-type) and class D (OXA-type) carbapenemases, the combination of overproduced class C β-lactamase and porin loss was also proposed for this species, which produces a natural ESAC with a deletion in the R2-loop and a low binding affinity for imipenem (53, 108, 265–267). No carbapenemase activity was detected in crude extracts, for example, so it was assumed that porin loss (CarO, Omp22-33, Omp33-36, Omp37, Omp43, Omp44, and Omp47) was the most likely mechanism (253, 265, 268–270).
Carbapenems (e.g., imipenem and meropenem) are important drugs to treat P. aeruginosa infections. Carbapenem resistance is mediated by various mechanisms, including the production of a carbapenemase (mostly class B enzymes), the overproduction of efflux pumps (mostly MexA-MexB), the overproduction of class C enzymes (e.g., PDC-type β-lactamases), and decreases in porin expression (mostly OprD), with combinations of these mechanisms observed in many cases (69, 216, 271, 272). In P. aeruginosa, transmissible class B carbapenemases or MBLs are considered to be the most clinically relevant source of resistance. Class D (oxacillinases) enzymes are also frequently encountered, and sometimes even class C β-lactamases, which are not real carbapenemases and have a low carbapenem hydrolysis capacity. The combination of PDC overproduction with a decrease in outer membrane permeability (OprD) and/or the overexpression of efflux systems (MexA-MexB-OprM) has been shown to lead to carbapenem resistance (69, 216, 271, 273–275) (Table 7). Various studies on clinical isolates have highlighted the difficulty of clearly defining the contribution of each mechanism, as resistance is generally a multifactorial phenomenon. However, most imipenem- and meropenem-resistant isolates display high levels of PDC-type enzymes and increased efflux pump expression (216).
RESISTANCE TO INHIBITORS
Early Inhibitors
The first inhibitors to be developed (e.g., clavulanate, sulbactam, tazobactam) were inactive against a majority of class C β-lactamases, but some older combinations have been revamped (e.g., ceftolozane-tazobactam) because ceftolozane has a reduced affinity for PDC enzymes and is not, therefore, hydrolyzed (276). Ceftolozane also has a high potency against P. aeruginosa when used alone. Resistance may be acquired during treatment and is mediated principally by transmissible β-lactamases (e.g., ESBLs and serine carbapenemases, such as KPCs). Several missense mutations or deletions in PDC genes have also been reported in P. aeruginosa isolates displaying overexpression of these genes and in studies performed in vitro, but the underlying mechanisms of resistance (such as combination with porin deficiency and overexpression of various efflux pumps) remain poorly defined (69, 229, 230, 277–282).
Avibactam
Novel non-β-lactam (diazabicyclooctane or DBO) β-lactamase inhibitors, such as avibactam, with a high affinity for a wide range of Ambler class A (e.g., ESBLs and KPC-types), class C, and some D β-lactamases (e.g., OXA-48), have been developed and showed encouraging results in clinical trials (276, 283). Avibactam is currently available in combination with ceftazidime, and is being used to combat multidrug-resistant Gram-negative bacterial infections, which are on the rise worldwide, including carbapenem-resistant Enterobacterales in particular. These inhibitors are not active against class B metallo-β-lactamases (e.g., NDM, VIM), and resistance to the ceftazidime–avibactam combination or combinations of avibactam with other antibiotics have been reported in patients treated since 2015 and in screens in vitro (283–289). This type of resistance has yet to be fully elucidated, but several possible underlying mechanisms have been reported. The most frequently encountered resistance mechanism is enzymatic, due to mutations identified mostly in transmissible class A (e.g., KPC and ESBLs) and some class D (e.g., OXA-48) β-lactamases. Other combined mechanisms have been reported, including membrane impermeability and efflux (277, 290), and there have been several reports of PBP3 mutations leading to ceftazidime-avibactam resistance (291–293). Finally, the overexpression of chromosomally- or plasmid-encoded AmpCs, with or without mutations and even in combination with porin deficiency, appears to make only a small contribution to resistance (13, 14, 160). Avibactam inhibits the E. cloacae CHE ESAC less efficiently than other E. cloacae AmpC proteins, due to a subtle rearrangement of the binding site (13). In vitro studies with P. aeruginosa showed that high-level resistance to combinations of avibactam with ceftazidime or aztreonam occurs with low frequency. In all cases, the mutant displayed alterations to the chromosomal ampC gene, mostly deletions of various sizes (5 to 19 residues) in the Ω-loop region of AmpC (14). Eight highly conserved residues made key contributions to binding interactions with avibactam (64S, 67K, 120Q, 150Y, 152N, 315K, 316T, and 346N). The PAC-1 enzyme, recently identified in four P. aeruginosa isolates and encoded by a gene on a chromosome-inserted Tn1721-like transposon, mediates very high levels of resistance to β-lactams, such as ceftazidime, cefepime, and ceftolozane alone or in combination with avibactam or tazobactam (75). Resistance to the ceftazidime–avibactam combination has been attributed to a deletion of two amino acids in the R2 loop of the AmpC β-lactamase produced independently by Enterobacter, which simultaneously causes resistance to cefepime and carbapenems and reduced susceptibility to cefiderocol, a novel siderophore cephalosporin (294).
Six carbapenem-resistant clinical isolates of K. pneumoniae in China were recently shown to produce KPC-2, a class A carbapenemase, and CMY-172, a new CMY-2-like class C β-lactamase (295). CMY-172 mediates high levels of resistance to β-lactams, including ceftazidime, cefotaxime, and cefepime, and to the ceftazidime–avibactam combination. This resistance results from a major modification (290KVA deletion) to the R2-loop (ESAC) and an amino-acid substitution, N346I (Table 5). The variant retains all the highly conserved residues for binding to avibactam (64S, 67K, 120Q, 150Y, 152N, 315K, 316T) except for 346N (162). In addition, the residue in position 346, which is well conserved among AmpC-type enzymes, modulates the hydrolysis spectrum of cephalosporinases (296). More generally, in vitro resistance to ceftazidime-avibactam and aztreonam-avibactam was examined among Enterobacterales, showing that resistance associated with changes to β-lactamases was seen only for mutants of AmpC. The mutants R148H, G156R/D, N346Y, or small deletions at positions 289–294, were obtained with ceftazidime-avibactam, whereas with aztreonam-avibactam the Y150C or N346Y substitutions were observed (297).
Relebactam
The newer imipenem-relebactam combination recently approved by the FDA has antimicrobial activity against Enterobacterales and P. aeruginosa strains producing class A and C β-lactamases, through the strong inhibition of KPC and AmpC enzymes (276, 298). Isolates resistant to this combination, due to porin loss, have been reported in Enterobacteriaceae (299, 300). There are in vivo and in vitro examples of resistance to this combination in P. aeruginosa mediated by class C enzymes, but resistance was again multifactorial, dependent on OprD loss, cephalosporinase overproduction, and mutations of the genes encoding the MexAB-OprM efflux system pump and the peptidoglycan recycling machinery (276, 301, 302).
Vaborbactam
Meropenem-vaborbactam is a fixed-dose combination of a carbapenem antibiotic and a novel boronic acid-based β-lactamase inhibitor. It has in vitro activity against Enterobacterales (e.g., K. pneumoniae) producing class A (e.g., ESBL, KPC), class C (mostly plasmid-encoded enzymes), and also some class D enzymes (103, 105, 121, 303). The impact of vaborbactam on class C enzymes appears to be limited, but resistance may nevertheless result from combinations of mechanisms, including the overexpression of ESAC mutants after cefepime treatment, associated with OmpK35 and OmpK36 porin deficiencies due to the insertion of an IS903-like element, as recently reported in a clinical isolate of E. hormaechei (294). This isolate was multidrug-resistant, with resistance even against ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam combinations. This resistance should serve as an important warning that cephalosporins and carbapenems should be used in a judicious manner, whether used alone or in combination with these novel inhibitors.
NEW CEPHALOSPORINS
Cefiderocol
Cefiderocol is an advanced injectable siderophore cephalosporin active against MDR and XDR Gram-negative rods, including producers of various β-lactamases, including members of Enterobacterales, P. aeruginosa, A. baumannii, Stenotrophomonas maltophila, B. cepacia, and B. pseudomallei (304–307). Through its catechol siderophore moiety, cefiderocol exploits iron-transport systems via a Trojan-horse strategy, navigating the bacterial periplasm and evading various β-lactamases, including ESBLs, KPC (class A), NDM, IMP, VIM (class B), and various class C and D enzymes (OXA-23, OXA-24, OXA-48, and OXA-51), as well as other mechanisms of resistance (308–310). Resistance to cefiderocol may be due to a two-amino-acid deletion in the R2 loop of the AmpC of E. cloacae complex (109, 294), but can also be acquired through the loss of energy-transducing proteins or catecholate receptors, such as PirA, PiuA, PiuD, and TonB in Enterobacterales, P. aeruginosa and A. baumannii (309, 311–313).
CONCLUSIONS
BLCs have a highly variable primary structure, with minimum sequence identity levels as low as 20% (28). However, the three principal catalytic motifs that are characteristic for this molecular class (64SXSK, 150YXN, and 315KTG) are highly conserved, together with at least 70 other residues displaying >90% conservation. Some of these other residues, including 60F, 61E, 63G, 145G, 148R, 318T, 321G, 322F, 325Y, and 328F, are located around the conserved catalytic motifs. Generally, with the exception of a few sequences corresponding to serine hydrolases often identified by BLAST analysis, structural classification should be based on the notion of “genus.” Genera contain highly variable numbers of species, and future taxonomic reorganizations are possible, as already observed for the genus Erwinia, for example, with the discovery of E. teleogrylli, or for the genus Pseudomonas, with definition of the species P. putida and P. fluorescens.
The considerable increase in the number of protein sequences present in databases for several species has made it possible to confirm the existence of a certain degree of polymorphism, the large number of clusters observed (more than 10 for E. coli/Shigella) making it possible to define consensus sequences for each cluster or phylogroup (16). By contrast, enzymes of the DHA- and FOX-types, encoded by plasmid-borne genes, and the species from which they originate, M. morganii and A. allosaccharophila, respectively, display little polymorphism.
With the development of high-throughput sequencing, this notion of ampC gene polymorphism complicates the detection of variants responsible for a significant increase in the MICs of β-lactams, particularly for oxyiminocephalosporins (e.g., cefotaxime, ceftazidime, cefepime, and cefpirome), monobactams (e.g., aztreonam), and carbapenems. However, most infections are identified in hospital inpatients, from whom several isolates of the same species can be obtained, therefore facilitating the detection of such variants. Nevertheless, in the absence of an isolate obtained at the start of treatment, certain variants, such as insertions, duplications, or deletions, appear to be easy to detect, whereas others, such as the substitution of a single residue in a hot spot, may be much harder to identify. The diverse examples of ESAC-producing strains observed in clinical practice demonstrate that genetic modifications similar to those described above essentially affect three regions (around the 150YXN motif, the Ω-loop, and the H10-helix/R2-loop). Surprisingly, these regions vary to different extents in different species, with a limited number of conserved residues. The notion of a “consensus sequence” should therefore favor the possible detection of ESACs in the framework of increasingly frequent genotype/phenotype analyses (Fig. 3). Such diversity at the heart of species, particularly for the H10-helix/R2-loop region, could account for the considerable variation of MICs between substrates, or the diversity of the β-lactam resistance and kinetic constants reported. Wild-type bacterial species have variable MICs for β-lactams (https://mic.eucast.org/), suggesting that combinations of other, non-enzymatic resistance mechanisms may be at work.
FIG 3.
Consensus partial protein sequences of species or their progenitors susceptible to expand their spectrum of inactivation. For E. coli, the consensus sequence was calculated from protein sequences of clusters A, B, C and D (32, 46, 50, 63). Residue boxed in yellow indicates 100% conserved. Underlined positions indicate at least two different residues (polymorphism).
It should not be forgotten that the chromosomal ampC gene is subject to regulation, and thus is dependent on other genes, particularly as it is inducible in most of the bacterial species producing the corresponding enzyme. Diverse genetic events can, therefore, occur in the intercistronic space, generating a strong promoter, or in other genes, such as ampR, ampD, and ampG, to generate mutants displaying derepression or overexpression. The analysis would not really be complete without an examination of several systems involved in the transfer of β-lactams across the cell wall, such as porins (e.g., OmpC, OmpF, OmpK35, OmpK36), and efflux systems (e.g., AcrAB-TolC, MexAB-OprM) (293, 314–318).
For medical biologists, the identification of a β-lactamase from one of the four known molecular classes is based on the presence of at least three motifs (17). The accumulation of a large number of protein sequences in current databases, with the development of WGS in particular, has made it possible to compare almost 4,000 protein sequences from class C β-lactamases, facilitating classification within clusters or taxa. This class is as diverse as class A, with about 20% identity between sequences, but its genetic footprint may be enriched in fewer than 80 highly conserved residues, justifying the exclusion of certain carboxylesterases with β-lactamase activity. In addition, the existence of major taxa in bacterial genera, such as Citrobacter, Enterobacter, Acinetobacter, and Aeromonas, and of recent and more long-standing taxonomic reorganizations, provides an indispensable aid to diagnosis at species level, particularly in light of the potential failings of the phenotypic methods currently used in laboratories. This type of analysis, based on primary structure, makes it possible to propose one or several consensus sequences for a species, potentially improving the characterization of possible polymorphisms of the β-lactamase. In the absence of the susceptible strain of the bacterium at the start of infection, prior knowledge of the diverse genetic modifications likely to lead to an extended spectrum of inactivation, which vary between bacterial species, should make it possible to detect these modifications (substitutions, duplications, insertions, deletions). The recent adaptation of a numbering system for this molecular class of enzymes has led to comparisons of protein sequences and emergence of the notion of a “natural ESAC.” Finally, medical biologists should not ignore possible interactions with other genes likely to lead to hyperproduction, which are particularly important for this molecular class.
ACKNOWLEDGMENTS
This work was supported, in part, by grants from the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT, ANR-10-LABX-33), the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR, ANR-14-JAMR-0002), and the PPR Antibioresistance (ANR‐20‐PAMR‐0010).
The authors declare no potential conflicts of interest.
Biographies

Alain Philippon is a veterinarian (Ecole Nationale Vétérinaire d’Alfort, 1963) graduated in biochemistry (University of Orsay, 1965) and bacteriology-immunology (Institut Pasteur, 1966) who prepared his Ph. D. thesis at the Commissariat à l’Energie Atomique (CEA). Initially trained on Brucella and experimental bovine brucellosis at Nouzilly (Institut National de la Recherche Agronomique/INRA) between 1966 and 1970, he started to analyze bacterial resistance to antibiotics, mostly to β-lactams (susceptibility patterns, ESBL, and plasmid-encoded AmpC) at CHU Cochin, Paris, France, and in collaboration with François Le Goffic and Roger Labia (ENS Ulm) in 1972. As professor emeritus of Microbiology at the University of Paris Descartes and previous head of the Bacteriology Laboratory at Hopital Cochin, he published more than 200 scientific papers and mentored 20 doctoral and postdoctoral researchers. He was also codirector of a course on medical bacteriology at the Institut Pasteur of Paris for a decade.

Guillaume Arlet is a doctor (René Descartes University, Paris, 1979), graduate in Medical Biology in 1984, in Biochemistry and Molecular Biology in 1986 (Denis Diderot University, Paris), and Bacteriology (Institut Pasteur, 1986). He did his PhD (University Paris-Sud, 1989–1992) under the supervision of Alain Philippon. Associate professor at Saint-Louis Hospital and Denis Diderot University (1991–1999), he became professor at Pierre & Marie Curie University (now Sorbonne University), and head of the medical bacteriology department, first at Tenon Hospital (1999–2012) and then within the Eastern Parisian Hospitals Group (2012–2018). He headed the teaching department of Clinical Bacteriology (2007–2018). He joined in 2014 the Center for Immunology and Infectious Diseases (INSERM U1135) at Sorbonne Université. He supervised ten PhD students and published more than 210 scientific papers mainly on β-lactamases (ESBL, AmpC β-lactamases, carbapenemases), their genetic supports, and in relation to virulence of their bacterial hosts. He is currently professor emeritus.

Roger Labia graduated from the world-renowned Ecole Polytechnique in Paris, France (1962 to 1964), with specialization in mathematics, physics, and chemistry. In relation to his high and early interest in chemical and biological problems, he joined scientific research, first at the Pasteur Institute (starting in 1965). In 1969, he received a PhD, studying the organic synthesis of natural compounds. Subsequently, he spent one postdoctoral year at Ottawa University, Canada (1969 to 1970), where he began studying biochemistry and bacteriology. Back in France, he started a research program on antibiotics, including their mode of action and mechanisms of resistance. This allowed him to develop multiple collaborations with chemists and bacteriologists from France and other countries. He has published more than 320 scientific papers, mostly in high-impact international journals. He has been involved in teaching and directed about 50 theses.

Bogdan I. Iorga received in 2001 a PhD in molecular chemistry from the Ecole Polytechnique, France. He is currently CNRS Research Director and heads the Molecular Modeling and Structural Crystallography team at the Institut de Chimie des Substances Naturelles (CNRS, Université Paris-Saclay, Gif-sur-Yvette, France). He is author of one book, six book chapters, four patents, and more than 100 publications. He is the developer of the Beta-Lactamase DataBase (BLDB, http://bldb.eu) and is involved in several French and European projects related to the antimicrobial resistance. His main research interests include the design of biologically active compounds, the study of structure–function relationships in different classes of β-lactamases, and the development of innovative protocols in molecular modeling. His recent work focuses on the development of tools for in silico prediction of antibiotic susceptibility from genomic data using machine learning and deep learning approaches.
REFERENCES
- 1.Ambler RP. 1980. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci 289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 2.Jaurin B, Grundström T. 1981. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc Natl Acad Sci USA 78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huovinen P, Huovinen S, Jacoby GA. 1988. Sequence of PSE-2 β-lactamase. Antimicrob Agents Chemother 32:134–136. doi: 10.1128/AAC.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K, Bradford PA. 2016. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med 6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Bradford PA. 2019. Interplay between β-lactamases and new β-lactamase inhibitors. Nat Rev Microbiol 17:295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 7.Sanders CC. 1987. Chromosomal cephalosporinases responsible for multiple resistance to newer β-lactam antibiotics. Annu Rev Microbiol 41:573–593. doi: 10.1146/annurev.mi.41.100187.003041. [DOI] [PubMed] [Google Scholar]
- 8.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnaud G, Labia R, Raskine L, Sanson-Le Pors MJ, Philippon A, Arlet G. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol Lett 195:185–190. doi: 10.1111/j.1574-6968.2001.tb10519.x. [DOI] [PubMed] [Google Scholar]
- 10.Nukaga M, Haruta S, Tanimoto K, Kogure K, Taniguchi K, Tamaki M, Sawai T. 1995. Molecular evolution of a class C β-lactamase extending its substrate specificity. J Biol Chem 270:5729–5735. doi: 10.1074/jbc.270.11.5729. [DOI] [PubMed] [Google Scholar]
- 11.Nikaido H, Liu W, Rosenberg EY. 1990. Outer membrane permeability and β-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob Agents Chemother 34:337–342. doi: 10.1128/AAC.34.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HY, Livermore DM. 1993. Activity of cefepime and other β-lactam antibiotics against permeability mutants of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother 32:63–74. doi: 10.1093/jac/32.suppl_B.63. [DOI] [PubMed] [Google Scholar]
- 13.Lahiri SD, Giacobbe RA, Johnstone MR, Alm RA. 2014. Activity of avibactam against Enterobacter cloacae producing an extended-spectrum class C β-lactamase enzyme. J Antimicrob Chemother 69:2942–2946. doi: 10.1093/jac/dku237. [DOI] [PubMed] [Google Scholar]
- 14.Lahiri SD, Walkup GK, Whiteaker JD, Palmer T, McCormack K, Tanudra MA, Nash TJ, Thresher J, Johnstone MR, Hajec L, Livchak S, McLaughlin RE, Alm RA. 2015. Selection and molecular characterization of ceftazidime/avibactam-resistant mutants in Pseudomonas aeruginosa strains containing derepressed ampC. J Antimicrob Chemother 70:1650–1658. doi: 10.1093/jac/dkv004. [DOI] [PubMed] [Google Scholar]
- 15.Vijayakumar S, Biswas I, Veeraraghavan B. 2019. Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci OA 5:FSO395. doi: 10.2144/fsoa-2018-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammeri H, Poirel L, Fortineau N, Nordmann P. 2006. Naturally occurring extended-spectrum cephalosporinases in Escherichia coli. Antimicrob Agents Chemother 50:2573–2576. doi: 10.1128/AAC.01633-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush K. 2013. The ABCD's of β-lactamase nomenclature. J Infect Chemother 19:549–559. doi: 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- 18.Bradford PA. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaïbi EB, Sirot D, Paul G, Labia R. 1999. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic and biochemical characteristics. J Antimicrob Chemother 43:447–458. doi: 10.1093/jac/43.4.447. [DOI] [PubMed] [Google Scholar]
- 20.Philippon A, Labia R, Jacoby G. 1989. Extended-spectrum β-lactamases. Antimicrob Agents Chemother 33:1131–1136. doi: 10.1128/AAC.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philippon A, Slama P, Dény P, Labia R. 2016. A structure-based classification of class A β-lactamases, a broadly diverse family of enzymes. Clin Microbiol Rev 29:29–57. doi: 10.1128/CMR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-lactamase database (BLDB)—structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack AR, Barnes MD, Taracila MA, Hujer AM, Hujer KM, Cabot G, Feldgarden M, Haft DH, Klimke W, van den Akker F, Vila AJ, Smania A, Haider S, Papp-Wallace KM, Bradford PA, Rossolini GM, Docquier JD, Frère JM, Galleni M, Hanson ND, Oliver A, Plésiat P, Poirel L, Nordmann P, Palzkill TG, Jacoby GA, Bush K, Bonomo RA. 2020. A standard numbering scheme for class C β-lactamases. Antimicrob Agents Chemother 64:e01841-19. doi: 10.1128/AAC.01841-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouellette M, Bissonnette L, Roy PH. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc Natl Acad Sci USA 84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall BG, Barlow M. 2003. Structure-based phylogenies of the serine β-lactamases. J Mol Evol 57:255–260. doi: 10.1007/s00239-003-2473-y. [DOI] [PubMed] [Google Scholar]
- 28.Brandt C, Braun SD, Stein C, Slickers P, Ehricht R, Pletz MW, Makarewicz O. 2017. In silico serine β-lactamases analysis reveals a huge potential resistome in environmental and pathogenic species. Sci Rep 7:43232. doi: 10.1038/srep43232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silveira MC, Azevedo da Silva R, Faria da Mota F, Catanho M, Jardim R, R Guimarães AC, de Miranda AB. 2018. Systematic identification and classification of β-lactamases based on sequence similarity criteria: β-lactamase annotation. Evol Bioinform Online 14:1176934318797351. doi: 10.1177/1176934318797351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2009. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J 3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 31.Böhm ME, Razavi M, Flach CF, Larsson DGJ. 2020. A novel, integron-regulated, class C β-lactamase. Antibiotics (Basel) 9:123. doi: 10.3390/antibiotics9030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philippon A, Arlet G, Labia R, Iorga B. 2021. Unrooted phylogram of class C β-lactamases from Enterobacterales. Figshare. doi: 10.6084/m9.figshare.16869195. [DOI] [Google Scholar]
- 33.Philippon A, Arlet G, Labia R, Iorga B. 2021. Amino acid sequence alignments of class C β-lactamases of Enterobacterales. Figshare. doi: 10.6084/m9.figshare.16915675. [DOI] [Google Scholar]
- 34.Denamur E, Clermont O, Bonacorsi S, Gordon D. 2021. The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol 19:37–54. doi: 10.1038/s41579-020-0416-x. [DOI] [PubMed] [Google Scholar]
- 35.Mammeri H, Galleni M, Nordmann P. 2009. Role of the Ser-287-Asn replacement in the hydrolysis spectrum extension of AmpC β-lactamases in Escherichia coli. Antimicrob Agents Chemother 53:323–326. doi: 10.1128/AAC.00608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore AM, Ahmadi S, Patel S, Gibson MK, Wang B, Ndao MI, Deych E, Shannon W, Tarr PI, Warner BB, Dantas G. 2015. Gut resistome development in healthy twin pairs in the first year of life. Microbiome 3:27. doi: 10.1186/s40168-015-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pehrsson EC, Tsukayama P, Patel S, Mejía-Bautista M, Sosa-Soto G, Navarrete KM, Calderon M, Cabrera L, Hoyos-Arango W, Bertoli MT, Berg DE, Gilman RH, Dantas G. 2016. Interconnected microbiomes and resistomes in low-income human habitats. Nature 533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbott SL, Moler S, Green N, Tran RK, Wainwright K, Janda JM. 2011. Clinical and laboratory diagnostic characteristics and cytotoxigenic potential of Hafnia alvei and Hafnia paralvei strains. J Clin Microbiol 49:3122–3126. doi: 10.1128/JCM.00866-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huys G, Cnockaert M, Abbott SL, Janda JM, Vandamme P. 2010. Hafnia paralvei sp. nov., formerly known as Hafnia alvei hybridization group 2. Int J Syst Evol Microbiol 60:1725–1728. doi: 10.1099/ijs.0.018606-0. [DOI] [PubMed] [Google Scholar]
- 41.Avison MB, Niumsup P, Walsh TR, Bennett PM. 2000. Aeromonas hydrophila AmpH and CepH β-lactamases: derepressed expression in mutants of Escherichia coli lacking creB. J Antimicrob Chemother 46:695–702. doi: 10.1093/jac/46.5.695. [DOI] [PubMed] [Google Scholar]
- 42.Alksne LE, Rasmussen BA. 1997. Expression of the AsbA1, OXA-12, and AsbM1 β-lactamases in Aeromonas jandaei AER 14 is coordinated by a two-component regulon. J Bacteriol 179:2006–2013. doi: 10.1128/jb.179.6.2006-2013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh TR, Payne DJ, MacGowan AP, Bennett PM. 1995. A clinical isolate of Aeromonas sobria with three chromosomally mediated inducible β-lactamases: a cephalosporinase, a penicillinase and a third enzyme, displaying carbapenemase activity. J Antimicrob Chemother 35:271–279. doi: 10.1093/jac/35.2.271. [DOI] [PubMed] [Google Scholar]
- 44.Fosse T, Giraud-Morin C, Madinier I, Labia R. 2003. Sequence analysis and biochemical characterisation of chromosomal CAV-1 (Aeromonas caviae), the parental cephalosporinase of plasmid-mediated AmpC “FOX” cluster. FEMS Microbiol Lett 222:93–98. doi: 10.1016/S0378-1097(03)00253-2. [DOI] [PubMed] [Google Scholar]
- 45.Fernández-Bravo A, Figueras MJ. 2020. An update on the genus Aeromonas: taxonomy, epidemiology, and pathogenicity. Microorganisms 8:129. doi: 10.3390/microorganisms8010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philippon A, Arlet G, Labia R, Iorga B. 2021. Unrooted phylogram of class C β-lactamases from Aeromonas. Figshare. doi: 10.6084/m9.figshare.16869443. [DOI] [Google Scholar]
- 47.Philippon A, Arlet G, Labia R, Iorga B. 2021. Amino acid sequence alignments of class C β-lactamases of Aeromonas. Figshare. doi: 10.6084/m9.figshare.16915678. [DOI] [Google Scholar]
- 48.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philippon A, Arlet G, Labia R, Iorga B. 2021. Unrooted phylogram of class C β-lactamases from Acinetobacter. Figshare. doi: 10.6084/m9.figshare.16869455. [DOI] [Google Scholar]
- 51.Hujer KM, Hamza NS, Hujer AM, Perez F, Helfand MS, Bethel CR, Thomson JM, Anderson VE, Barlow M, Rice LB, Tenover FC, Bonomo RA. 2005. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 β-lactamase: defining a unique family of class C enzymes. Antimicrob Agents Chemother 49:2941–2948. doi: 10.1128/AAC.49.7.2941-2948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Périchon B, Goussard S, Walewski V, Krizova L, Cerqueira G, Murphy C, Feldgarden M, Wortman J, Clermont D, Nemec A, Courvalin P. 2014. Identification of 50 class D β-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob Agents Chemother 58:936–949. doi: 10.1128/AAC.01261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhattacharya M, Toth M, Antunes NT, Smith CA, Vakulenko SB. 2014. Structure of the extended-spectrum class C β-lactamase ADC-1 from Acinetobacter baumannii. Acta Crystallogr D Biol Crystallogr 70:760–771. doi: 10.1107/S1399004713033014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philippon A, Arlet G, Labia R, Iorga B. 2021. Amino acid sequence alignments of class C β-lactamases of Acinetobacter. Figshare. doi: 10.6084/m9.figshare.16915681. [DOI] [Google Scholar]
- 55.Beceiro A, Pérez-Llarena FJ, Pérez A, Tomás MDM, Fernández A, Mallo S, Villanueva R, Bou G. 2007. Molecular characterization of the gene encoding a new AmpC β-lactamase in Acinetobacter baylyi. J Antimicrob Chemother 59:996–1000. doi: 10.1093/jac/dkm070. [DOI] [PubMed] [Google Scholar]
- 56.Rodríguez-Martínez JM, Poirel L, Nordmann P. 2010. Genetic and functional variability of AmpC-type β-lactamases from Acinetobacter baumannii. Antimicrob Agents Chemother 54:4930–4933. doi: 10.1128/AAC.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian G-B, Adams-Haduch JM, Taracila M, Bonomo RA, Wang H-N, Doi Y. 2011. Extended-spectrum AmpC cephalosporinase in Acinetobacter baumannii: ADC-56 confers resistance to cefepime. Antimicrob Agents Chemother 55:4922–4925. doi: 10.1128/AAC.00704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Héritier C, Poirel L, Nordmann P. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect 12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 59.Hesse C, Schulz F, Bull CT, Shaffer BT, Yan Q, Shapiro N, Hassan KA, Varghese N, Elbourne LDH, Paulsen IT, Kyrpides N, Woyke T, Loper JE. 2018. Genome-based evolutionary history of Pseudomonas spp. Environ Microbiol 20:2142–2159. doi: 10.1111/1462-2920.14130. [DOI] [PubMed] [Google Scholar]
- 60.Kidd TJ, Ramsay KA, Hu H, Bye PT, Elkins MR, Grimwood K, Harbour C, Marks GB, Nissen MD, Robinson PJ, Rose BR, Sloots TP, Wainwright CE, Bell SC, ACPinCF Investigators. 2009. Low rates of Pseudomonas aeruginosa misidentification in isolates from cystic fibrosis patients. J Clin Microbiol 47:1503–1509. doi: 10.1128/JCM.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scales BS, Dickson RP, LiPuma JJ, Huffnagle GB. 2014. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin Microbiol Rev 27:927–948. doi: 10.1128/CMR.00044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tran PN, Savka MA, Gan HM. 2017. In-silico taxonomic classification of 373 genomes reveals species misidentification and new genospecies within the genus Pseudomonas. Front Microbiol 8:1296. doi: 10.3389/fmicb.2017.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Philippon A, Arlet G, Labia R, Iorga B. 2021. Unrooted phylogram of class C β-lactamases from Pseudomonas. Figshare. doi: 10.6084/m9.figshare.16869463. [DOI] [Google Scholar]
- 64.Philippon A, Arlet G, Labia R, Iorga B. 2021. Amino acid sequence alignments of class C β-lactamases of Pseudomonas. Figshare. doi: 10.6084/m9.figshare.16915684. [DOI] [Google Scholar]
- 65.Weinstein RA, Gaynes R, Edwards JR, National Nosocomial Infections Surveillance System. 2005. Overview of nosocomial infections caused by Gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 66.Malhotra S, Hayes D, Jr, Wozniak DJ. 2019. Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe interface. Clin Microbiol Rev 32:e00138-18. doi: 10.1128/CMR.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother 51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodríguez-Martínez JM, Poirel L, Nordmann P. 2009. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:1766–1771. doi: 10.1128/AAC.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Philippon A, Arlet G, Labia R, Iorga B. 2021. Consensus sequences of class C β-lactamases of Pseudomonas aeruginosa. Figshare. doi: 10.6084/m9.figshare.16915696. [DOI] [Google Scholar]
- 71.Ebmeyer S, Kristiansson E, Larsson DGJ. 2019. The mobile FOX AmpC β-lactamases originated in Aeromonas allosaccharophila. Int J Antimicrob Agents 54:798–802. doi: 10.1016/j.ijantimicag.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 72.Ebmeyer S, Kristiansson E, Larsson DGJ. 2019. CMY-1/MOX-family AmpC β-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three Aeromonas species. J Antimicrob Chemother 74:1202–1206. doi: 10.1093/jac/dkz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Philippon A, Arlet G, Labia R, Iorga B. 2021. Class C plasmid-encoded β-lactamases and progenitors: unrooted phylogram for selected class C β-lactamases of Enterobacterales. Figshare. doi: 10.6084/m9.figshare.16915654. [DOI] [Google Scholar]
- 74.Philippon A, Arlet G, Labia R, Iorga B. 2021. Class C plasmid-encoded β-lactamases and progenitors: unrooted phylogram for selected class C β-lactamases of Aeromonas. Figshare. doi: 10.6084/m9.figshare.16915666. [DOI] [Google Scholar]
- 75.Bour M, Fournier D, Jové T, Pouzol A, Miltgen G, Janvier F, Jeannot K, Plésiat P. 2019. Acquisition of class C β-lactamase PAC-1 by ST664 strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e01375-19. doi: 10.1128/AAC.01375-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, Steele HL, Reymond JL, Jaeger KE, Streit WR. 2006. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol 72:3637–3645. doi: 10.1128/AEM.72.5.3637-3645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeon JH, Kim SJ, Lee HS, Cha SS, Lee JH, Yoon SH, Koo BS, Lee CM, Choi SH, Lee SH, Kang SG, Lee JH. 2011. Novel metagenome-derived carboxylesterase that hydrolyzes β-lactam antibiotics. Appl Environ Microbiol 77:7830–7836. doi: 10.1128/AEM.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeon JH, Lee HS, Lee JH, Koo BS, Lee CM, Lee SH, Kang SG, Lee JH. 2016. A novel family VIII carboxylesterase hydrolysing third- and fourth-generation cephalosporins. Springerplus 5:525. doi: 10.1186/s40064-016-2172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu EY, Kwon MA, Lee M, Oh JY, Choi JE, Lee JY, Song BK, Hahm DH, Song JK. 2011. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl Microbiol Biotechnol 90:573–581. doi: 10.1007/s00253-011-3132-7. [DOI] [PubMed] [Google Scholar]
- 80.Wagner UG, Petersen EI, Schwab H, Kratky C. 2002. EstB from Burkholderia gladioli: a novel esterase with a β-lactamase fold reveals steric factors to discriminate between esterolytic and β-lactam cleaving activity. Protein Sci 11:467–478. doi: 10.1110/ps.33002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rashamuse K, Magomani V, Ronneburg T, Brady D. 2009. A novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous β-lactamase activity on nitrocefin. Appl Microbiol Biotechnol 83:491–500. doi: 10.1007/s00253-009-1895-x. [DOI] [PubMed] [Google Scholar]
- 82.Petersen EI, Valinger G, Sölkner B, Stubenrauch G, Schwab H. 2001. A novel esterase from Burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to β-lactamases and DD-peptidases. J Biotechnol 89:11–25. doi: 10.1016/s0168-1656(01)00284-x. [DOI] [PubMed] [Google Scholar]
- 83.Mokoena N, Mathiba K, Tsekoa T, Steenkamp P, Rashamuse K. 2013. Functional characterisation of a metagenome derived family VIII esterase with a deacetylation activity on β-lactam antibiotics. Biochem Biophys Res Commun 437:342–348. doi: 10.1016/j.bbrc.2013.06.076. [DOI] [PubMed] [Google Scholar]
- 84.Cha SS, An YJ, Jeong CS, Kim MK, Jeon JH, Lee CM, Lee HS, Kang SG, Lee JH. 2013. Structural basis for the β-lactamase activity of EstU1, a family VIII carboxylesterase. Proteins 81:2045–2051. doi: 10.1002/prot.24334. [DOI] [PubMed] [Google Scholar]
- 85.Cha SS, An YJ. 2016. Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins. Biochem Biophys Res Commun 478:818–824. doi: 10.1016/j.bbrc.2016.08.031. [DOI] [PubMed] [Google Scholar]
- 86.Knox JR. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother 39:2593–2601. doi: 10.1128/AAC.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lobkovsky E, Billings EM, Moews PC, Rahil J, Pratt RF, Knox JR. 1994. Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a β-lactamase transition-state analog. Biochemistry 33:6762–6772. doi: 10.1021/bi00188a004. [DOI] [PubMed] [Google Scholar]
- 88.Bae DW, Jung YE, An YJ, Na JH, Cha SS. 2019. Structural insights into catalytic relevances of substrate poses in ACC-1. Antimicrob Agents Chemother 63:e01411-19. doi: 10.1128/AAC.01411-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bae DW, Jung YE, Jeong BG, Cha SS. 2020. Novel inhibition mechanism of carbapenems on the ACC-1 class C β-lactamase. Arch Biochem Biophys 693:108570. doi: 10.1016/j.abb.2020.108570. [DOI] [PubMed] [Google Scholar]
- 90.Shimizu-Ibuka A, Bauvois C, Sakai H, Galleni M. 2008. Structure of the plasmid-mediated class C β-lactamase ACT-1. Acta Crystallogr Sect F Struct Biol Cryst Commun 64:334–337. doi: 10.1107/S1744309108008531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Plantan I, Selič L, Mesar T, Anderluh PS, Oblak M, Preželj A, Hesse L, Andrejašič M, Vilar M, Turk D, Kocijan A, Prevec T, Vilfan G, Kocjan D, Čopar A, Urleb U, Solmajer T. 2007. 4-substituted trinems as broad spectrum β-lactamase inhibitors: structure-based design, synthesis, and biological activity. J Med Chem 50:4113–4121. doi: 10.1021/jm0703237. [DOI] [PubMed] [Google Scholar]
- 92.Ruble JF, Lefurgy ST, Cornish VW, Powers RA. 2012. Structural analysis of the Asn152Gly mutant of P99 cephalosporinase. Acta Crystallogr D Biol Crystallogr 68:1189–1193. doi: 10.1107/S0907444912024080. [DOI] [PubMed] [Google Scholar]
- 93.Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, King P, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Clifton MC, Atkins K, Raymond A, Potts KT, Abendroth J, Boyer SH, Loutit JS, Morgan EE, Durso S, Dudley MN. 2015. Discovery of a cyclic boronic acid β-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J Med Chem 58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 94.Stojanoski V, Adamski CJ, Hu L, Mehta SC, Sankaran B, Zwart P, Prasad BV, Palzkill T. 2016. Removal of the side chain at the active-site serine by a glycine substitution increases the stability of a wide range of serine β-lactamases by relieving steric strain. Biochemistry 55:2479–2490. doi: 10.1021/acs.biochem.6b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pan X, He Y, Chen T, Chan KF, Zhao Y. 2017. Modified penicillin molecule with carbapenem-like stereochemistry specifically inhibits class C β-lactamases. Antimicrob Agents Chemother 61:e01288-17. doi: 10.1128/AAC.01288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crichlow GV, Nukaga M, Doppalapudi VR, Buynak JD, Knox JR. 2001. Inhibition of class C β-lactamases: structure of a reaction intermediate with a cephem sulfone. Biochemistry 40:6233–6239. doi: 10.1021/bi010131s. [DOI] [PubMed] [Google Scholar]
- 97.Crichlow GV, Kuzin AP, Nukaga M, Mayama K, Sawai T, Knox JR. 1999. Structure of the extended-spectrum class C beta-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry 38:10256–10261. doi: 10.1021/bi9908787. [DOI] [PubMed] [Google Scholar]
- 98.Nukaga M, Abe T, Venkatesan AM, Mansour TS, Bonomo RA, Knox JR. 2003. Inhibition of class A and class C β-lactamases by penems: crystallographic structures of a novel 1,4-thiazepine intermediate. Biochemistry 42:13152–13159. doi: 10.1021/bi034986b. [DOI] [PubMed] [Google Scholar]
- 99.Venkatesan AM, Gu Y, Dos Santos O, Abe T, Agarwal A, Yang Y, Petersen PJ, Weiss WJ, Mansour TS, Nukaga M, Hujer AM, Bonomo RA, Knox JR. 2004. Structure–activity relationship of 6-methylidene penems bearing tricyclic heterocycles as broad-spectrum β-lactamase inhibitors: crystallographic structures show unexpected binding of 1,4-thiazepine intermediates. J Med Chem 47:6556–6568. doi: 10.1021/jm049680x. [DOI] [PubMed] [Google Scholar]
- 100.Nukaga M, Kumar S, Nukaga K, Pratt RF, Knox JR. 2004. Hydrolysis of third-generation cephalosporins by class C β-lactamases: structures of a transition state analog of cefotaxime in wild-type and extended spectrum enzymes. J Biol Chem 279:9344–9352. doi: 10.1074/jbc.M312356200. [DOI] [PubMed] [Google Scholar]
- 101.Wouters J, Fonzé E, Vermeire M, Frère JM, Charlier P. 2003. Crystal structure of Enterobacter cloacae 908R class C β-lactamase bound to iodo-acetamido-phenyl boronic acid, a transition-state analogue. Cell Mol Life Sci 60:1764–1773. doi: 10.1007/s00018-003-3189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Michaux C, Charlier P, Frère JM, Wouters J. 2005. Crystal structure of BRL 42715, C6-(N1-methyl-1,2,3-triazolylmethylene)penem, in complex with Enterobacter cloacae 908R β-lactamase: evidence for a stereoselective mechanism from docking studies. J Am Chem Soc 127:3262–3263. doi: 10.1021/ja0426241. [DOI] [PubMed] [Google Scholar]
- 103.Powers RA, Swanson HC, Taracila MA, Florek NW, Romagnoli C, Caselli E, Prati F, Bonomo RA, Wallar BJ. 2014. Biochemical and structural analysis of inhibitors targeting the ADC-7 cephalosporinase of Acinetobacter baumannii. Biochemistry 53:7670–7679. doi: 10.1021/bi500887n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caselli E, Romagnoli C, Powers RA, Taracila MA, Bouza AA, Swanson HC, Smolen KA, Fini F, Wallar BJ, Bonomo RA, Prati F. 2018. Inhibition of Acinetobacter-derived cephalosporinase: exploring the carboxylate recognition site using novel β-lactamase inhibitors. ACS Infect Dis 4:337–348. doi: 10.1021/acsinfecdis.7b00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bouza AA, Swanson HC, Smolen KA, VanDine AL, Taracila MA, Romagnoli C, Caselli E, Prati F, Bonomo RA, Powers RA, Wallar BJ. 2018. Structure-based analysis of boronic acids as inhibitors of Acinetobacter-derived cephalosporinase-7, a unique class C β-lactamase. ACS Infect Dis 4:325–336. doi: 10.1021/acsinfecdis.7b00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Curtis BN, Smolen KA, Barlow SJ, Caselli E, Prati F, Taracila MA, Bonomo RA, Wallar BJ, Powers RA. 2020. Structural insights into inhibition of the Acinetobacter-derived cephalosporinase ADC-7 by ceftazidime and its boronic acid transition state analog. Antimicrob Agents Chemother 64:e01183-20. doi: 10.1128/AAC.01183-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Caselli E, Fini F, Introvigne ML, Stucchi M, Taracila MA, Fish ER, Smolen KA, Rather PN, Powers RA, Wallar BJ, Bonomo RA, Prati F. 2020. 1,2,3-Triazolylmethaneboronate: A structure activity relationship study of a class of β-lactamase inhibitors against Acinetobacter baumannii cephalosporinase. ACS Infect Dis 6:1965–1975. doi: 10.1021/acsinfecdis.0c00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeon JH, Hong MK, Lee JH, Lee JJ, Park KS, Karim AM, Jo JY, Kim JH, Ko KS, Kang LW, Lee SH. 2014. Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum β-lactamase isolated from Acinetobacter baumannii. Acta Crystallogr D Biol Crystallogr 70:2924–2936. doi: 10.1107/S1399004714019543. [DOI] [PubMed] [Google Scholar]
- 109.Kawai A, McElheny CL, Iovleva A, Kline EG, Sluis-Cremer N, Shields RK, Doi Y. 2020. Structural basis of reduced susceptibility to ceftazidime-avibactam and cefiderocol in Enterobacter cloacae due to AmpC R2 loop deletion. Antimicrob Agents Chemother 64:e00198-20. doi: 10.1128/AAC.00198-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim JY, Jung HI, An YJ, Lee JH, Kim SJ, Jeong SH, Lee KJ, Suh PG, Lee HS, Lee SH, Cha SS. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol Microbiol 60:907–916. doi: 10.1111/j.1365-2958.2006.05146.x. [DOI] [PubMed] [Google Scholar]
- 111.Kim MK, An YJ, Na JH, Seol JH, Ryu JY, Lee JW, Kang LW, Chung KM, Lee JH, Moon JH, Lee JS, Cha SS. 2017. Structural and mechanistic insights into the inhibition of class C β-lactamases through the adenylylation of the nucleophilic serine. J Antimicrob Chemother 72:735–743. [DOI] [PubMed] [Google Scholar]
- 112.Na JH, An YJ, Cha SS. 2017. GMP and IMP are competitive inhibitors of CMY-10, an extended-spectrum class C β-lactamase. Antimicrob Agents Chemother 61:e00098-17. doi: 10.1128/AAC.00098-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zavala A, Retailleau P, Elisée E, Iorga BI, Naas T. 2019. Genetic, biochemical, and structural characterization of CMY-136 β-lactamase, a peculiar CMY-2 variant. ACS Infect Dis 5:528–538. doi: 10.1021/acsinfecdis.8b00240. [DOI] [PubMed] [Google Scholar]
- 114.Oefner C, D'Arcy A, Daly JJ, Gubernator K, Charnas RL, Heinze I, Hubschwerlen C, Winkler FK. 1990. Refined crystal structure of β-lactamase from Citrobacter freundii indicates a mechanism for β-lactam hydrolysis. Nature 343:284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- 115.Powers RA, Blázquez J, Weston GS, Morosini MI, Baquero F, Shoichet BK. 1999. The complexed structure and antimicrobial activity of a non-β-lactam inhibitor of AmpC β-lactamase. Protein Sci 8:2330–2337. doi: 10.1110/ps.8.11.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Patera A, Blaszczak LC, Shoichet BK. 2000. Crystal structures of substrate and inhibitor complexes with AmpC β-lactamase: possible implications for substrate-assisted catalysis. J Am Chem Soc 122:10504–10512. doi: 10.1021/ja001676x. [DOI] [Google Scholar]
- 117.Caselli E, Powers RA, Blasczcak LC, Wu CY, Prati F, Shoichet BK. 2001. Energetic, structural, and antimicrobial analyses of β-lactam side chain recognition by β-lactamases. Chem Biol 8:17–31. doi: 10.1016/s1074-5521(00)00052-1. [DOI] [PubMed] [Google Scholar]
- 118.Tondi D, Powers RA, Caselli E, Negri MC, Blázquez J, Costi MP, Shoichet BK. 2001. Structure-based design and in-parallel synthesis of inhibitors of AmpC β-lactamase. Chem Biol 8:593–611. doi: 10.1016/s1074-5521(01)00034-5. [DOI] [PubMed] [Google Scholar]
- 119.Trehan I, Beadle BM, Shoichet BK. 2001. Inhibition of AmpC β-lactamase through a destabilizing interaction in the active site. Biochemistry 40:7992–7999. doi: 10.1021/bi010641m. [DOI] [PubMed] [Google Scholar]
- 120.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207–9214. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 121.Powers RA, Shoichet BK. 2002. Structure-based approach for binding site identification on AmpC β-lactamase. J Med Chem 45:3222–3234. doi: 10.1021/jm020002p. [DOI] [PubMed] [Google Scholar]
- 122.Beadle BM, Trehan I, Focia PJ, Shoichet BK. 2002. Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC β-lactamase. Structure 10:413–424. doi: 10.1016/s0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 123.Beadle BM, Shoichet BK. 2002. Structural bases of stability-function tradeoffs in enzymes. J Mol Biol 321:285–296. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 124.Powers RA, Morandi F, Shoichet BK. 2002. Structure-based discovery of a novel, noncovalent inhibitor of AmpC β-lactamase. Structure 10:1013–1023. doi: 10.1016/s0969-2126(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 125.Beadle BM, Shoichet BK. 2002. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob Agents Chemother 46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Trehan I, Morandi F, Blaszczak LC, Shoichet BK. 2002. Using steric hindrance to design new inhibitors of class C β-lactamases. Chem Biol 9:971–980. doi: 10.1016/S1074-5521(02)00211-9. [DOI] [PubMed] [Google Scholar]
- 127.Morandi F, Caselli E, Morandi S, Focia PJ, Blázquez J, Shoichet BK, Prati F. 2003. Nanomolar inhibitors of AmpC β-lactamase. J Am Chem Soc 125:685–695. doi: 10.1021/ja0288338. [DOI] [PubMed] [Google Scholar]
- 128.Meroueh SO, Minasov G, Lee W, Shoichet BK, Mobashery S. 2003. Structural aspects for evolution of β-lactamases from penicillin-binding proteins. J Am Chem Soc 125:9612–9618. doi: 10.1021/ja034861u. [DOI] [PubMed] [Google Scholar]
- 129.Roth TA, Minasov G, Morandi S, Prati F, Shoichet BK. 2003. Thermodynamic cycle analysis and inhibitor design against β-lactamase. Biochemistry 42:14483–14491. doi: 10.1021/bi035054a. [DOI] [PubMed] [Google Scholar]
- 130.Tondi D, Morandi F, Bonnet R, Costi MP, Shoichet BK. 2005. Structure-based optimization of a non-β-lactam lead results in inhibitors that do not up-regulate β-lactamase expression in cell culture. J Am Chem Soc 127:4632–4639. doi: 10.1021/ja042984o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Usher KC, Blaszczak LC, Weston GS, Shoichet BK, Remington SJ. 1998. Three-dimensional structure of AmpC β-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design. Biochemistry 37:16082–16092. doi: 10.1021/bi981210f. [DOI] [PubMed] [Google Scholar]
- 132.Chen Y, Minasov G, Roth TA, Prati F, Shoichet BK. 2006. The deacylation mechanism of AmpC β-lactamase at ultrahigh resolution. J Am Chem Soc 128:2970–2976. doi: 10.1021/ja056806m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Babaoglu K, Shoichet BK. 2006. Deconstructing fragment-based inhibitor discovery. Nat Chem Biol 2:720–723. doi: 10.1038/nchembio831. [DOI] [PubMed] [Google Scholar]
- 134.Venturelli A, Tondi D, Cancian L, Morandi F, Cannazza G, Segatore B, Prati F, Amicosante G, Shoichet BK, Costi MP. 2007. Optimizing cell permeation of an antibiotic resistance inhibitor for improved efficacy. J Med Chem 50:5644–5654. doi: 10.1021/jm070643q. [DOI] [PubMed] [Google Scholar]
- 135.Wyrembak PN, Babaoglu K, Pelto RB, Shoichet BK, Pratt RF. 2007. O-aryloxycarbonyl hydroxamates: new β-lactamase inhibitors that cross-link the active site. J Am Chem Soc 129:9548–9549. doi: 10.1021/ja072370u. [DOI] [PubMed] [Google Scholar]
- 136.Babaoglu K, Simeonov A, Irwin JJ, Nelson ME, Feng B, Thomas CJ, Cancian L, Costi MP, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. 2008. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against β-lactamase. J Med Chem 51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Morandi S, Morandi F, Caselli E, Shoichet BK, Prati F. 2008. Structure-based optimization of cephalothin-analogue boronic acids as β-lactamase inhibitors. Bioorg Med Chem 16:1195–1205. doi: 10.1016/j.bmc.2007.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tondi D, Calò S, Shoichet BK, Costi MP. 2010. Structural study of phenyl boronic acid derivatives as AmpC β-lactamase inhibitors. Bioorg Med Chem Lett 20:3416–3419. doi: 10.1016/j.bmcl.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen Y, McReynolds A, Shoichet BK. 2009. Re-examining the role of Lys67 in class C β-lactamase catalysis. Protein Sci 18:662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Teotico DG, Babaoglu K, Rocklin GJ, Ferreira RS, Giannetti AM, Shoichet BK. 2009. Docking for fragment inhibitors of AmpC β-lactamase. Proc Natl Acad Sci USA 106:7455–7460. doi: 10.1073/pnas.0813029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thomas VL, McReynolds AC, Shoichet BK. 2010. Structural bases for stability–function tradeoffs in antibiotic resistance. J Mol Biol 396:47–59. doi: 10.1016/j.jmb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eidam O, Romagnoli C, Caselli E, Babaoglu K, Pohlhaus DT, Karpiak J, Bonnet R, Shoichet BK, Prati F. 2010. Design, synthesis, crystal structures, and antimicrobial activity of sulfonamide boronic acids as β-lactamase inhibitors. J Med Chem 53:7852–7863. doi: 10.1021/jm101015z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eidam O, Romagnoli C, Dalmasso G, Barelier S, Caselli E, Bonnet R, Shoichet BK, Prati F. 2012. Fragment-guided design of subnanomolar β-lactamase inhibitors active in vivo. Proc Natl Acad Sci USA 109:17448–17453. doi: 10.1073/pnas.1208337109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hendershot JM, Mishra UJ, Smart RP, Schroeder W, Powers RA. 2014. Structure-based efforts to optimize a non-β-lactam inhibitor of AmpC β-lactamase. Bioorg Med Chem 22:3351–3359. doi: 10.1016/j.bmc.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 145.Barelier S, Eidam O, Fish I, Hollander J, Figaroa F, Nachane R, Irwin JJ, Shoichet BK, Siegal G. 2014. Increasing chemical space coverage by combining empirical and computational fragment screens. ACS Chem Biol 9:1528–1535. doi: 10.1021/cb5001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.London N, Miller RM, Krishnan S, Uchida K, Irwin JJ, Eidam O, Gibold L, Cimermančič P, Bonnet R, Shoichet BK, Taunton J. 2014. Covalent docking of large libraries for the discovery of chemical probes. Nat Chem Biol 10:1066–1072. doi: 10.1038/nchembio.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Barelier S, Cummings JA, Rauwerdink AM, Hitchcock DS, Farelli JD, Almo SC, Raushel FM, Allen KN, Shoichet BK. 2014. Substrate deconstruction and the nonadditivity of enzyme recognition. J Am Chem Soc 136:7374–7382. doi: 10.1021/ja501354q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lyu J, Wang S, Balius TE, Singh I, Levit A, Moroz YS, O'Meara MJ, Che T, Algaa E, Tolmachova K, Tolmachev AA, Shoichet BK, Roth BL, Irwin JJ. 2019. Ultra-large library docking for discovering new chemotypes. Nature 566:224–229. doi: 10.1038/s41586-019-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lang PA, Parkova A, Leissing TM, Calvopiña K, Cain R, Krajnc A, Panduwawala TD, Philippe J, Fishwick CWG, Trapencieris P, Page MGP, Schofield CJ, Brem J. 2020. Bicyclic boronates as potent inhibitors of AmpC, the class C β-lactamase from Escherichia coli. Biomolecules 10:899. doi: 10.3390/biom10060899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lang PA, Leissing TM, Page MGP, Schofield CJ, Brem J. 2021. Structural investigations of the inhibition of Escherichia coli AmpC β-lactamase by diazabicyclooctanes. Antimicrob Agents Chemother 65:e02073-20. doi: 10.1128/AAC.02073-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamaguchi Y, Sato G, Yamagata Y, Doi Y, Wachino J, Arakawa Y, Matsuda K, Kurosaki H. 2009. Structure of AmpC β-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix. Acta Crystallogr Sect F Struct Biol Cryst Commun 65:540–543. doi: 10.1107/S1744309109014249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Na JH, Cha SS. 2016. Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C β-lactamases AmpC BER and CMY-10. Acta Crystallogr D Struct Biol 72:976–985. doi: 10.1107/S2059798316011311. [DOI] [PubMed] [Google Scholar]
- 153.Lefurgy ST, Malashkevich VN, Aguilan JT, Nieves E, Mundorff EC, Biju B, Noel MA, Toro R, Baiwir D, Papp-Wallace KM, Almo SC, Frère JM, Bou G, Bonomo RA. 2016. Analysis of the structure and function of FOX-4 cephamycinase. Antimicrob Agents Chemother 60:717–728. doi: 10.1128/AAC.01887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lefurgy ST, Caselli E, Taracila MA, Malashkevich VN, Biju B, Papp-Wallace KM, Bonanno JB, Prati F, Almo SC, Bonomo RA. 2020. Structures of FOX-4 cephamycinase in complex with transition-state analog inhibitors. Biomolecules 10:671. doi: 10.3390/biom10050671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nukaga M, Papp-Wallace KM, Hoshino T, Lefurgy ST, Bethel CR, Barnes MD, Zeiser ET, Johnson JK, Bonomo RA. 2018. Probing the mechanism of inactivation of the FOX-4 cephamycinase by avibactam. Antimicrob Agents Chemother 62:e02371-17. doi: 10.1128/AAC.02371-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Oguri T, Furuyama T, Okuno T, Ishii Y, Tateda K, Bonomo RA, Shimizu-Ibuka A. 2014. Crystal structure of MOX-1, a unique plasmid-mediated class C β-lactamase with hydrolytic activity towards moxalactam. Antimicrob Agents Chemother 58:3914–3920. doi: 10.1128/AAC.02363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Oguri T, Ishii Y, Shimizu-Ibuka A. 2015. Conformational change observed in the active site of class C β-lactamase MOX-1 upon binding to aztreonam. Antimicrob Agents Chemother 59:5069–5072. doi: 10.1128/AAC.04428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Blizzard TA, Chen H, Kim S, Wu J, Young K, Park YW, Ogawa A, Raghoobar S, Painter RE, Hairston N, Lee SH, Misura A, Felcetto T, Fitzgerald P, Sharma N, Lu J, Ha S, Hickey E, Hermes J, Hammond ML. 2010. Side chain SAR of bicyclic β-lactamase inhibitors (BLIs). 1. Discovery of a class C BLI for combination with imipenem. Bioorg Med Chem Lett 20:918–921. doi: 10.1016/j.bmcl.2009.12.069. [DOI] [PubMed] [Google Scholar]
- 159.Chen H, Blizzard TA, Kim S, Wu J, Young K, Park YW, Ogawa AM, Raghoobar S, Painter RE, Wisniewski D, Hairston N, Fitzgerald P, Sharma N, Scapin G, Lu J, Hermes J, Hammond ML. 2011. Side chain SAR of bicyclic β-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712. Bioorg Med Chem Lett 21:4267–4270. doi: 10.1016/j.bmcl.2011.05.065. [DOI] [PubMed] [Google Scholar]
- 160.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Blizzard TA, Chen H, Kim S, Wu J, Bodner R, Gude C, Imbriglio J, Young K, Park YW, Ogawa A, Raghoobar S, Hairston N, Painter RE, Wisniewski D, Scapin G, Fitzgerald P, Sharma N, Lu J, Ha S, Hermes J, Hammond ML. 2014. Discovery of MK-7655, a β-lactamase inhibitor for combination with Primaxin. Bioorg Med Chem Lett 24:780–785. doi: 10.1016/j.bmcl.2013.12.101. [DOI] [PubMed] [Google Scholar]
- 162.Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. 2014. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 58:5704–5713. doi: 10.1128/AAC.03057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McKinney DC, Zhou F, Eyermann CJ, Ferguson AD, Prince DB, Breen J, Giacobbe RA, Lahiri S, Verheijen JC. 2015. 4,5-Disubstituted 6-aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles are broad-spectrum serine β-lactamase inhibitors. ACS Infect Dis 1:310–316. doi: 10.1021/acsinfecdis.5b00031. [DOI] [PubMed] [Google Scholar]
- 164.Morinaka A, Tsutsumi Y, Yamada M, Suzuki K, Watanabe T, Abe T, Furuuchi T, Inamura S, Sakamaki Y, Mitsuhashi N, Ida T, Livermore DM. 2015. OP0595, a new diazabicyclooctane: mode of action as a serine β-lactamase inhibitor, antibiotic and β-lactam “enhancer.” J Antimicrob Chemother 70:2779–2786. doi: 10.1093/jac/dkv166. [DOI] [PubMed] [Google Scholar]
- 165.Cahill ST, Tyrrell JM, Navratilova IH, Calvopiña K, Robinson SW, Lohans CT, McDonough MA, Cain R, Fishwick CWG, Avison MB, Walsh TR, Schofield CJ, Brem J. 2019. Studies on the inhibition of AmpC and other β-lactamases by cyclic boronates. Biochim Biophys Acta Gen Subj 1863:742–748. doi: 10.1016/j.bbagen.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 166.Linciano P, Vicario M, Kekez I, Bellio P, Celenza G, Martín-Blecua I, Blázquez J, Cendron L, Tondi D. 2019. Phenylboronic acids probing molecular recognition against class A and class C β-lactamases. Antibiotics 8:171. doi: 10.3390/antibiotics8040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang SW, Pan J, Root Y, Scapin G, Xiao L, Su J. 2020. Serendipitous discovery of aryl boronic acids as β-lactamase inhibitors. Bioorg Med Chem Lett 30:126795. doi: 10.1016/j.bmcl.2019.126795. [DOI] [PubMed] [Google Scholar]
- 168.Pozzi C, Di Pisa F, De Luca F, Benvenuti M, Docquier JD, Mangani S. 2018. Atomic-resolution structure of a class C β-lactamase and its complex with avibactam. ChemMedChem 13:1437–1446. doi: 10.1002/cmdc.201800213. [DOI] [PubMed] [Google Scholar]
- 169.Arai S, Yonezawa Y, Okazaki N, Matsumoto F, Shibazaki C, Shimizu R, Yamada M, Adachi M, Tamada T, Kawamoto M, Tokunaga H, Ishibashi M, Blaber M, Tokunaga M, Kuroki R. 2015. Structure of a highly acidic β-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site. Acta Crystallogr D Biol Crystallogr 71:541–554. doi: 10.1107/S1399004714027734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Michaux C, Massant J, Kerff F, Frère JM, Docquier JD, Vandenberghe I, Samyn B, Pierrard A, Feller G, Charlier P, Van Beeumen J, Wouters J. 2008. Crystal structure of a cold-adapted class C β-lactamase. FEBS J 275:1687–1697. doi: 10.1111/j.1742-4658.2008.06324.x. [DOI] [PubMed] [Google Scholar]
- 171.Fisher JF, Mobashery S. 2014. The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg Chem 56:41–48. doi: 10.1016/j.bioorg.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Guérin F, Isnard C, Cattoir V, Giard JC. 2015. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother 59:7753–7761. doi: 10.1128/AAC.01729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Irazoki O, Hernandez SB, Cava F. 2019. Peptidoglycan muropeptides: release, perception, and functions as signaling molecules. Front Microbiol 10:500. doi: 10.3389/fmicb.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Juan C, Torrens G, González-Nicolau M, Oliver A. 2017. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol Rev 41:781–815. doi: 10.1093/femsre/fux043. [DOI] [PubMed] [Google Scholar]
- 175.Aguilera Rossi CG, Gómez-Puertas P, Ayala Serrano JA. 2016. In vivo functional and molecular characterization of the Penicillin-Binding Protein 4 (DacB) of Pseudomonas aeruginosa. BMC Microbiol 16:234. doi: 10.1186/s12866-016-0853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moya B, Zamorano L, Juan C, Pérez JL, Ge Y, Oliver A. 2010. Activity of a new cephalosporin, CXA-101 (FR264205), against β-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob Agents Chemother 54:1213–1217. doi: 10.1128/AAC.01104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Torrens G, Hernández SB, Ayala JA, Moya B, Juan C, Cava F, Oliver A. 2019. Regulation of AmpC-driven β-lactam resistance in Pseudomonas aeruginosa: different pathways, different signaling. mSystems 4:e00524-19. doi: 10.1128/mSystems.00524-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Babouee Flury B, Ellington MJ, Hopkins KL, Turton JF, Doumith M, Loy R, Staves P, Hinic V, Frei R, Woodford N. 2016. Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob Agents Chemother 60:2383–2390. doi: 10.1128/AAC.02835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kaneko K, Okamoto R, Nakano R, Kawakami S, Inoue M. 2005. Gene mutations responsible for overexpression of ampC β-lactamase in some clinical isolates of Enterobacter cloacae. J Clin Microbiol 43:2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kopp U, Wiedemann B, Lindquist S, Normark S. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother 37:224–228. doi: 10.1128/AAC.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Petrosino JF, Pendleton AR, Weiner JH, Rosenberg SM. 2002. Chromosomal system for studying AmpC-mediated β-lactam resistance mutation in Escherichia coli. Antimicrob Agents Chemother 46:1535–1539. doi: 10.1128/AAC.46.5.1535-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schmidtke AJ, Hanson ND. 2006. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob Agents Chemother 50:2030–2037. doi: 10.1128/AAC.01458-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hilty M, Sendi P, Seiffert SN, Droz S, Perreten V, Hujer AM, Bonomo RA, Mühlemann K, Endimiani A. 2013. Characterisation and clinical features of Enterobacter cloacae bloodstream infections occurring at a tertiary care university hospital in Switzerland: is cefepime adequate therapy? Int J Antimicrob Agents 41:236–249. doi: 10.1016/j.ijantimicag.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lindberg F, Lindquist S, Normark S. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J Bacteriol 169:1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pérez-Gallego M, Torrens G, Castillo-Vera J, Moya B, Zamorano L, Cabot G, Hultenby K, Albertí S, Mellroth P, Henriques-Normark B, Normark S, Oliver A, Juan C. 2016. Impact of ampC derepression on fitness and virulence: the mechanism or the pathway? mBio 7:e01783-16. doi: 10.1128/mBio.01783-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bagge N, Ciofu O, Hentzer M, Campbell JI, Givskov M, Høiby N. 2002. Constitutive high expression of chromosomal β-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob Agents Chemother 46:3406–3411. doi: 10.1128/AAC.46.11.3406-3411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Juan C, Maciá MD, Gutiérrez O, Vidal C, Pérez JL, Oliver A. 2005. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Langaee TY, Gagnon L, Huletsky A. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC β-lactamase expression. Antimicrob Agents Chemother 44:583–589. doi: 10.1128/AAC.44.3.583-589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schmidtke AJ, Hanson ND. 2008. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3922–3927. doi: 10.1128/AAC.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Balasubramanian D, Kumari H, Mathee K. 2015. Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathog Dis 73:1–14. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dik DA, Domínguez-Gil T, Lee M, Hesek D, Byun B, Fishovitz J, Boggess B, Hellman LM, Fisher JF, Hermoso JA, Mobashery S. 2017. Muropeptide binding and the X-ray structure of the effector domain of the transcriptional regulator AmpR of Pseudomonas aeruginosa. J Am Chem Soc 139:1448–1451. doi: 10.1021/jacs.6b12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Balcewich MD, Reeve TM, Orlikow EA, Donald LJ, Vocadlo DJ, Mark BL. 2010. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible AmpC β-lactamase. J Mol Biol 400:998–1010. doi: 10.1016/j.jmb.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 193.Nakano R, Nakano A, Yano H, Okamoto R. 2017. Role of AmpR in the high expression of the plasmid-encoded AmpC β-lactamase CFE-1. mSphere 2:e00192-17. doi: 10.1128/mSphere.00192-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kuga A, Okamoto R, Inoue M. 2000. ampR gene mutations that greatly increase class C β-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother 44:561–567. doi: 10.1128/AAC.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cabot G, López-Causapé C, Ocampo-Sosa AA, Sommer LM, Domínguez M, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Plesiat P, Oliver A. 2016. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Domitrovic TN, Hujer AM, Perez F, Marshall SH, Hujer KM, Woc-Colburn LE, Parta M, Bonomo RA. 2016. Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: a genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect Dis 3:ofw188. doi: 10.1093/ofid/ofw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tam VH, Schilling AN, LaRocco MT, Gentry LO, Lolans K, Quinn JP, Garey KW. 2007. Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 13:413–418. doi: 10.1111/j.1469-0691.2006.01674.x. [DOI] [PubMed] [Google Scholar]
- 198.Tsutsumi Y, Tomita H, Tanimoto K. 2013. Identification of novel genes responsible for overexpression of ampC in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 57:5987–5993. doi: 10.1128/AAC.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chang Q, Wu C, Lin C, Li P, Zhang K, Xu L, Liu Y, Lu J, Cheng C, Bao Q, Hu Y, Lu S, Li J. 2018. The structure of ampG gene in Pseudomonas aeruginosa and its effect on drug resistance. Can J Infect Dis Med Microbiol 2018:7170416. doi: 10.1155/2018/7170416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li P, Ying J, Yang G, Li A, Wang J, Lu J, Wang J, Xu T, Yi H, Li K, Jin S, Bao Q, Zhang K. 2016. Structure-function analysis of the transmembrane protein AmpG from Pseudomonas aeruginosa. PLoS One 11:e0168060. doi: 10.1371/journal.pone.0168060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mallik D, Pal S, Ghosh AS. 2018. Involvement of AmpG in mediating a dynamic relationship between serine β-lactamase induction and biofilm-forming ability of Escherichia coli. FEMS Microbiol Lett 365:fny065. [DOI] [PubMed] [Google Scholar]
- 202.Caroff N, Espaze E, Gautreau D, Richet H, Reynaud A. 2000. Analysis of the effects of -42 and -32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J Antimicrob Chemother 45:783–788. doi: 10.1093/jac/45.6.783. [DOI] [PubMed] [Google Scholar]
- 203.Tracz DM, Boyd DA, Hizon R, Bryce E, McGeer A, Ofner-Agostini M, Simor AE, Paton S, Mulvey MR, Canadian Nosocomial Infection Surveillance Program. 2007. ampC gene expression in promoter mutants of cefoxitin-resistant Escherichia coli clinical isolates. FEMS Microbiol Lett 270:265–271. doi: 10.1111/j.1574-6968.2007.00672.x. [DOI] [PubMed] [Google Scholar]
- 204.Coolen JPM, den Drijver EPM, Verweij JJ, Schildkraut JA, Neveling K, Melchers WJG, Kolwijck E, Wertheim HFL, Kluytmans J, Huynen MA. 2021. Genome-wide analysis in Escherichia coli unravels a high level of genetic homoplasy associated with cefotaxime resistance. Microb Genom 7:e000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ceccarelli D, Kant A, van Essen-Zandbergen A, Dierikx C, Hordijk J, Wit B, Mevius DJ, Veldman KT. 2019. Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli from Dutch livestock in 2007–2017. Front Microbiol 10:76. doi: 10.3389/fmicb.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pardon B, Smet A, Butaye P, Argudín MA, Valgaeren B, Catry B, Haesebrouck F, Deprez P. 2017. Nosocomial intravascular catheter infections with extended-spectrum β-lactamase-producing Escherichia coli in calves after strain introduction from a commercial herd. Transbound Emerg Dis 64:130–136. doi: 10.1111/tbed.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pérez A, Pérez-Llarena FJ, García P, Kerff F, Beceiro A, Galleni M, Bou G. 2014. New mutations in ADC-type β-lactamases from Acinetobacter spp. affect cefoxitin and ceftazidime hydrolysis. J Antimicrob Chemother 69:2407–2411. doi: 10.1093/jac/dku163. [DOI] [PubMed] [Google Scholar]
- 208.Nakano R, Okamoto R, Nakano Y, Kaneko K, Okitsu N, Hosaka Y, Inoue M. 2004. CFE-1, a novel plasmid-encoded AmpC β-lactamase with an ampR gene originating from Citrobacter freundii. Antimicrob Agents Chemother 48:1151–1158. doi: 10.1128/AAC.48.4.1151-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Reisbig MD, Hanson ND. 2002. The ACT-1 plasmid-encoded AmpC β-lactamase is inducible: detection in a complex β-lactamase background. J Antimicrob Chemother 49:557–560. doi: 10.1093/jac/49.3.557. [DOI] [PubMed] [Google Scholar]
- 210.Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother 50:607–617. doi: 10.1128/AAC.50.2.607-617.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Verdet C, Gautier V, Chachaty E, Ronco E, Hidri N, Decré D, Arlet G. 2009. Genetic context of plasmid-carried blaCMY-2-like genes in Enterobacteriaceae. Antimicrob Agents Chemother 53:4002–4006. doi: 10.1128/AAC.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Doloy A, Verdet C, Gautier V, Decré D, Ronco E, Hammami A, Philippon A, Arlet G. 2006. Genetic environment of acquired blaACC-1 β-lactamase gene in Enterobacteriaceae isolates. Antimicrob Agents Chemother 50:4177–4181. doi: 10.1128/AAC.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Reisbig MD, Hanson ND. 2004. Promoter sequences necessary for high-level expression of the plasmid-associated ampC β-lactamase gene blaMIR-1. Antimicrob Agents Chemother 48:4177–4182. doi: 10.1128/AAC.48.11.4177-4182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mazel D. 2006. Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 215.Collis CM, Hall RM. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 39:155–162. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lee YT, Chen TL, Siu LK, Chen CP, Fung CP. 2011. Impact of derepressed ampC β-lactamase ACT-9 on the clinical efficacy of ertapenem. Antimicrob Agents Chemother 55:4440–4442. doi: 10.1128/AAC.00271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hemarajata P, Amick T, Yang S, Gregson A, Holzmeyer C, Bush K, Humphries RM. 2018. Selection of hyperproduction of AmpC and SME-1 in a carbapenem-resistant Serratia marcescens isolate during antibiotic therapy. J Antimicrob Chemother 73:1256–1262. doi: 10.1093/jac/dky028. [DOI] [PubMed] [Google Scholar]
- 219.Nordmann P, Mammeri H. 2007. Extended-spectrum cephalosporinases: structure, detection and epidemiology. Future Microbiol 2:297–307. doi: 10.2217/17460913.2.3.297. [DOI] [PubMed] [Google Scholar]
- 220.Bogaerts P, Rodriguez-Villalobos H, Bauraing C, Deplano A, Laurent C, Berhin C, Struelens MJ, Glupczynski Y. 2010. Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered at two Belgian hospitals. Pathol Biol (Paris) 58:78–83. doi: 10.1016/j.patbio.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 221.Crémet L, Caroff N, Giraudeau C, Dauvergne S, Lepelletier D, Reynaud A, Corvec S. 2010. Occurrence of ST23 complex phylogroup A Escherichia coli isolates producing extended-spectrum AmpC β-lactamase in a French hospital. Antimicrob Agents Chemother 54:2216–2218. doi: 10.1128/AAC.01580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Haenni M, Châtre P, Madec JY. 2014. Emergence of Escherichia coli producing extended-spectrum AmpC β-lactamases (ESAC) in animals. Front Microbiol 5:53. doi: 10.3389/fmicb.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mammeri H, Eb F, Berkani A, Nordmann P. 2008. Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered in a French hospital. J Antimicrob Chemother 61:498–503. doi: 10.1093/jac/dkm538. [DOI] [PubMed] [Google Scholar]
- 224.Mammeri H, Nazic H, Naas T, Poirel L, Léotard S, Nordmann P. 2004. AmpC β-lactamase in an Escherichia coli clinical isolate confers resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother 48:4050–4053. doi: 10.1128/AAC.48.10.4050-4053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Doi Y, Wachino J-i, Ishiguro M, Kurokawa H, Yamane K, Shibata N, Shibayama K, Yokoyama K, Kato H, Yagi T, Arakawa Y. 2004. Inhibitor-sensitive AmpC β-lactamase variant produced by an Escherichia coli clinical isolate resistant to oxyiminocephalosporins and cephamycins. Antimicrob Agents Chemother 48:2652–2658. doi: 10.1128/AAC.48.7.2652-2658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Le Turnier S, Nordmann P, Eb F, Mammeri H. 2009. Potential evolution of hydrolysis spectrum for AmpC β-lactamase of Escherichia coli. J Antimicrob Chemother 63:216–218. doi: 10.1093/jac/dkn443. [DOI] [PubMed] [Google Scholar]
- 227.Kohlmann R, Bähr T, Gatermann SG. 2018. Species-specific mutation rates for ampC derepression in Enterobacterales with chromosomally encoded inducible AmpC β-lactamase. J Antimicrob Chemother 73:1530–1536. doi: 10.1093/jac/dky084. [DOI] [PubMed] [Google Scholar]
- 228.Kohlmann R, Bähr T, Gatermann SG. 2019. Effect of ampC derepression on cefepime MIC in Enterobacterales with chromosomally encoded inducible AmpC β-lactamase. Clin Microbiol Infect 25:1158.e1151–1158.e1154. doi: 10.1016/j.cmi.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 229.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moyà B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Martín-Pena ML, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. doi: 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 231.Horii T, Arakawa Y, Ohta M, Ichiyama S, Wacharotayankun R, Kato N. 1993. Plasmid-mediated AmpC-type β-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum β-lactams, including moxalactam. Antimicrob Agents Chemother 37:984–990. doi: 10.1128/AAC.37.5.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Alba J, Bauvois C, Ishii Y, Galleni M, Masuda K, Ishiguro M, Ito M, Frère JM, Yamaguchi K. 2003. A detailed kinetic study of MOX-1, a plasmid-encoded class C β-lactamase. FEMS Microbiol Lett 225:183–188. doi: 10.1016/S0378-1097(03)00448-8. [DOI] [PubMed] [Google Scholar]
- 233.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhelm R, Chong Y. 1996. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother 40:1926–1930. doi: 10.1128/AAC.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Raskine L, Borrel I, Barnaud G, Boyer S, Hanau-Berçot B, Gravisse J, Labia R, Arlet G, Sanson-Le-Pors MJ. 2002. Novel plasmid-encoded class C β-lactamase (MOX-2) in Klebsiella pneumoniae from Greece. Antimicrob Agents Chemother 46:2262–2265. doi: 10.1128/AAC.46.7.2262-2265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Doi Y, Paterson DL, Adams-Haduch JM, Sidjabat HE, O'Keefe A, Endimiani A, Bonomo RA. 2009. Reduced susceptibility to cefepime among Escherichia coli clinical isolates producing novel variants of CMY-2 β-lactamase. Antimicrob Agents Chemother 53:3159–3161. doi: 10.1128/AAC.00133-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wachino J, Kurokawa H, Suzuki S, Yamane K, Shibata N, Kimura K, Ike Y, Arakawa Y. 2006. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob Agents Chemother 50:534–541. doi: 10.1128/AAC.50.2.534-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lee SH, Jeong SH, Park YM. 2003. Characterization of blaCMY-10 a novel, plasmid-encoded AmpC-type β-lactamase gene in a clinical isolate of Enterobacter aerogenes. J Appl Microbiol 95:744–752. doi: 10.1046/j.1365-2672.2003.02040.x. [DOI] [PubMed] [Google Scholar]
- 238.Chen YT, Lauderdale TL, Liao TL, Shiau YR, Shu HY, Wu KM, Yan JJ, Su IJ, Tsai SF. 2007. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 β-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 51:3004–3007. doi: 10.1128/AAC.00167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ye Y, Xu XH, Li JB. 2010. Emergence of CTX-M-3, TEM-1 and a new plasmid-mediated MOX-4 AmpC in a multiresistant Aeromonas caviae isolate from a patient with pneumonia. J Med Microbiol 59:843–847. doi: 10.1099/jmm.0.016337-0. [DOI] [PubMed] [Google Scholar]
- 240.Mammeri H, Guillon H, Eb F, Nordmann P. 2010. Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC β-lactamases. Antimicrob Agents Chemother 54:4556–4560. doi: 10.1128/AAC.01762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Pérez-Llarena FJ, Kerff F, Zamorano L, Fernández MC, Nuñez ML, Miró E, Oliver A, Navarro F, Bou G. 2013. Characterization of the new AmpC β-lactamase FOX-8 reveals a single mutation, Phe313Leu, located in the R2 loop that affects ceftazidime hydrolysis. Antimicrob Agents Chemother 57:5158–5161. doi: 10.1128/AAC.00818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bauernfeind A, Wagner S, Jungwirth R, Schneider I, Meyer D. 1997. A novel class C β-lactamase (FOX-2) in Escherichia coli conferring resistance to cephamycins. Antimicrob Agents Chemother 41:2041–2046. doi: 10.1128/AAC.41.9.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Marchese A, Arlet G, Schito GC, Lagrange PH, Philippon A. 1998. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob Agents Chemother 42:464–467. doi: 10.1128/AAC.42.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Queenan AM, Jenkins S, Bush K. 2001. Cloning and biochemical characterization of FOX-5, an AmpC-type plasmid-encoded β-lactamase from a New York City Klebsiella pneumoniae clinical isolate. Antimicrob Agents Chemother 45:3189–3194. doi: 10.1128/AAC.45.11.3189-3194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mallo S, Pérez-Llarena FJ, Kerff F, Soares NC, Galleni M, Bou G. 2010. A tripeptide deletion in the R2 loop of the class C β-lactamase enzyme FOX-4 impairs cefoxitin hydrolysis and slightly increases susceptibility to β-lactamase inhibitors. J Antimicrob Chemother 65:1187–1194. doi: 10.1093/jac/dkq115. [DOI] [PubMed] [Google Scholar]
- 246.Fournier B, Arlet G, Lagrange PH, Philippon A. 1994. Klebsiella oxytoca: resistance to aztreonam by overproduction of the chromosomally encoded β-lactamase. FEMS Microbiol Lett 116:31–36. doi: 10.1111/j.1574-6968.1994.tb06671.x. [DOI] [PubMed] [Google Scholar]
- 247.Bou G, Martínez-Beltrán J. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother 44:428–432. doi: 10.1128/AAC.44.2.428-432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rodríguez-Martínez JM, Nordmann P, Ronco E, Poirel L. 2010. Extended-spectrum cephalosporinase in Acinetobacter baumannii. Antimicrob Agents Chemother 54:3484–3488. doi: 10.1128/AAC.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wu CJ, Wang HC, Chen PL, Chang MC, Sunny Sun H, Chou PH, Ko WC. 2013. AQU-1, a chromosomal class C β-lactamase, among clinical Aeromonas dhakensis isolates: distribution and clinical significance. Int J Antimicrob Agents 42:456–461. doi: 10.1016/j.ijantimicag.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 250.Girlich D, Naas T, Bellais S, Poirel L, Karim A, Nordmann P. 2000. Biochemical-genetic characterization and regulation of expression of an ACC-1-like chromosome-borne cephalosporinase from Hafnia alvei. Antimicrob Agents Chemother 44:1470–1478. doi: 10.1128/AAC.44.6.1470-1478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Binfiglio G, Stefani S, Nicoletti G. 1994. In vitro activity of cefpirome against β-lactamase-inducible and stably derepressed Enterobacteriaceae. Chemotherapy 40:311–316. doi: 10.1159/000239212. [DOI] [PubMed] [Google Scholar]
- 252.Vasoo S, Barreto JN, Tosh PK. 2015. Emerging issues in Gram-negative bacterial resistance: an update for the practicing clinician. Mayo Clin Proc 90:395–403. doi: 10.1016/j.mayocp.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 253.Nordmann P, Poirel L. 2019. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin Infect Dis 69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sawa T, Kooguchi K, Moriyama K. 2020. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care 8:13. doi: 10.1186/s40560-020-0429-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Then RL, Angehrn P. 1982. Trapping of nonhydrolyzable cephalosporins by cephalosporinases in Enterobacter cloacae and Pseudomonas aeruginosa as a possible resistance mechanism. Antimicrob Agents Chemother 21:711–717. doi: 10.1128/AAC.21.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Gutmann L, Williamson R. 1983. A model system to demonstrate that β-lactamase-associated antibiotic trapping could be a potential means of resistance. J Infect Dis 148:316–321. doi: 10.1093/infdis/148.2.316. [DOI] [PubMed] [Google Scholar]
- 257.Sanders CC. 1984. Inducible β-lactamases and non-hydrolytic resistance mechanisms. J Antimicrob Chemother 13:1–3. doi: 10.1093/jac/13.1.1. [DOI] [PubMed] [Google Scholar]
- 258.Goessens WH, van der Bij AK, van Boxtel R, Pitout JD, van Ulsen P, Melles DC, Tommassen J. 2013. Antibiotic trapping by plasmid-encoded CMY-2 β-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob Agents Chemother 57:3941–3949. doi: 10.1128/AAC.02459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Pfaendler HR, Schmidt HU, Freidank H. 2020. The novel CarbaLux test for carbapenemases and carbapenem deactivating AmpC β-lactamases. Front Microbiol 11:588887. doi: 10.3389/fmicb.2020.588887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.van Boxtel R, Wattel AA, Arenas J, Goessens WH, Tommassen J. 2017. Acquisition of carbapenem resistance by plasmid-encoded-AmpC-expressing Escherichia coli. Antimicrob Agents Chemother 61:e01413-16. doi: 10.1128/AAC.01413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bonnin RA, Jousset AB, Emeraud C, Oueslati S, Dortet L, Naas T. 2020. Genetic diversity, biochemical properties, and detection methods of minor carbapenemases in Enterobacterales. Front Med (Lausanne) 7:616490. doi: 10.3389/fmed.2020.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jacoby GA, Mills DM, Chow N. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3203–3206. doi: 10.1128/AAC.48.8.3203-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Robert J, Pantel A, Mérens A, Lavigne JP, Nicolas-Chanoine MH, ONERBA's Carbapenem Resistance Study Group. 2014. Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011–12. J Antimicrob Chemother 69:2706–2712. doi: 10.1093/jac/dku208. [DOI] [PubMed] [Google Scholar]
- 264.Jousset AB, Oueslati S, Bernabeu S, Takissian J, Creton E, Vogel A, Sauvadet A, Cotellon G, Gauthier L, Bonnin RA, Dortet L, Naas T. 2019. False-positive carbapenem-hydrolyzing confirmatory tests due to ACT-28, a chromosomally encoded AmpC with weak carbapenemase activity from Enterobacter kobei. Antimicrob Agents Chemother 63:e02388-18. doi: 10.1128/AAC.02388-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Clark RB. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J Antimicrob Chemother 38:245–251. doi: 10.1093/jac/38.2.245. [DOI] [PubMed] [Google Scholar]
- 266.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 267.Liu B, Liu L. 2021. Molecular epidemiology and mechanisms of carbapenem-resistant Acinetobacter baumannii isolates from ICU and respiratory department patients of a Chinese university hospital. Infect Drug Resist 14:743–755. doi: 10.2147/IDR.S299540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Limansky AS, Mussi MA, Viale AM. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J Clin Microbiol 40:4776–4778. doi: 10.1128/JCM.40.12.4776-4778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.del Mar Tomás M, Beceiro A, Pérez A, Velasco D, Moure R, Villanueva R, Martínez-Beltrán J, Bou G. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 49:5172–5175. doi: 10.1128/AAC.49.12.5172-5175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mussi MA, Relling VM, Limansky AS, Viale AM. 2007. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett 581:5573–5578. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 271.Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 272.Meletis G. 2016. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis 3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Moyá B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pai H, Kim J, Kim J, Lee JH, Choe KW, Gotoh N. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 45:480–484. doi: 10.1128/AAC.45.2.480-484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Rodríguez-Martínez JM, Poirel L, Nordmann P. 2009. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Papp-Wallace KM. 2019. The latest advances in β-lactam/β-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert Opin Pharmacother 20:2169–2184. doi: 10.1080/14656566.2019.1660772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Nicolas-Chanoine MH, Mayer N, Guyot K, Dumont E, Pagès JM. 2018. Interplay between membrane permeability and enzymatic barrier leads to antibiotic-dependent resistance in Klebsiella pneumoniae. Front Microbiol 9:1422. doi: 10.3389/fmicb.2018.01422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Barnes MD, Taracila MA, Rutter JD, Bethel CR, Galdadas I, Hujer AM, Caselli E, Prati F, Dekker JP, Papp-Wallace KM, Haider S, Bonomo RA. 2018. Deciphering the evolution of cephalosporin resistance to ceftolozane-tazobactam in Pseudomonas aeruginosa. mBio 9:e02085-18. doi: 10.1128/mBio.02085-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.MacVane SH, Pandey R, Steed LL, Kreiswirth BN, Chen L. 2017. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 61:e01183-17. doi: 10.1128/AAC.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Skoglund E, Abodakpi H, Rios R, Diaz L, De La Cadena E, Dinh AQ, Ardila J, Miller WR, Munita JM, Arias CA, Tam VH, Tran TT. 2018. In vivo resistance to ceftolozane/tazobactam in Pseudomonas aeruginosa arising by AmpC- and non-AmpC-mediated pathways. Case Rep Infect Dis 2018:9095203. doi: 10.1155/2018/9095203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wi YM, Greenwood-Quaintance KE, Schuetz AN, Ko KS, Peck KR, Song JH, Patel R. 2018. Activity of ceftolozane-tazobactam against carbapenem-resistant, non-carbapenemase-producing Pseudomonas aeruginosa and associated resistance mechanisms. Antimicrob Agents Chemother 62:e01970-17. doi: 10.1128/AAC.01970-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhanel GG, Chung P, Adam H, Zelenitsky S, Denisuik A, Schweizer F, Lagacé-Wiens PR, Rubinstein E, Gin AS, Walkty A, Hoban DJ, Lynch JP, 3rd, Karlowsky JA. 2014. Ceftolozane/tazobactam: a novel cephalosporin/β-lactamase inhibitor combination with activity against multidrug-resistant Gram-negative bacilli. Drugs 74:31–51. doi: 10.1007/s40265-013-0168-2. [DOI] [PubMed] [Google Scholar]
- 283.ECDC. 2018. Rapid risk assessment: emergence of resistance to ceftazidime-avibactam in carbapenem-resistant Enterobacteriaceae. European Centre for Disease Prevention and Control, Stockholm, Sweden. https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-emergence-resistance-ceftazidime-avibactam-carbapenem. [Google Scholar]
- 284.Biagi M, Wu T, Lee M, Patel S, Butler D, Wenzler E. 2019. Searching for the optimal treatment for metallo- and serine-β-lactamase producing Enterobacteriaceae: aztreonam in combination with ceftazidime-avibactam or meropenem-vaborbactam. Antimicrob Agents Chemother 63:e01426-19. doi: 10.1128/AAC.01426-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Humphries RM, Hemarajata P. 2017. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother 61:e00537-17. doi: 10.1128/AAC.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. doi: 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of β-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Porres-Osante N, Dupont H, Torres C, Ammenouche N, de Champs C, Mammeri H. 2014. Avibactam activity against extended-spectrum AmpC β-lactamases. J Antimicrob Chemother 69:1715–1716. doi: 10.1093/jac/dku002. [DOI] [PubMed] [Google Scholar]
- 289.Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Ω-loop. J Antimicrob Chemother 70:2279–2286. doi: 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Nelson K, Hemarajata P, Sun D, Rubio-Aparicio D, Tsivkovski R, Yang S, Sebra R, Kasarskis A, Nguyen H, Hanson BM, Leopold S, Weinstock G, Lomovskaya O, Humphries RM. 2017. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother 61:e00989-17. doi: 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Alm RA, Johnstone MR, Lahiri SD. 2015. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J Antimicrob Chemother 70:1420–1428. doi: 10.1093/jac/dku568. [DOI] [PubMed] [Google Scholar]
- 292.Sato T, Ito A, Ishioka Y, Matsumoto S, Rokushima M, Kazmierczak KM, Hackel M, Sahm DF, Yamano Y. 2020. Escherichia coli strains possessing a four amino acid YRIN insertion in PBP3 identified as part of the SIDERO-WT-2014 surveillance study. JAC Antimicrob Resist 2:dlaa081. doi: 10.1093/jacamr/dlaa081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang Y, Kashikar A, Brown CA, Denys G, Bush K. 2017. Unusual Escherichia coli PBP 3 insertion sequence identified from a collection of carbapenem-resistant Enterobacteriaceae tested in vitro with a combination of ceftazidime-, ceftaroline-, or aztreonam-avibactam. Antimicrob Agents Chemother 61:e00389-17. doi: 10.1128/AAC.00389-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Shields RK, Iovleva A, Kline EG, Kawai A, McElheny CL, Doi Y. 2020. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin Infect Dis 71:2713–2716. doi: 10.1093/cid/ciaa355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Xu M, Zhao J, Xu L, Yang Q, Xu H, Kong H, Zhou J, Fu Y. 2022. Emergence of transferable ceftazidime-avibactam resistance in KPC-producing Klebsiella pneumoniae due to a novel CMY AmpC β-lactamase in China. Clin Microbiol Infect 28:136.e1-136–e6. doi: 10.1016/j.cmi.2021.05.026. [DOI] [PubMed] [Google Scholar]
- 296.Dahyot S, Broutin I, de Champs C, Guillon H, Mammeri H. 2013. Contribution of asparagine 346 residue to the carbapenemase activity of CMY-2 β-lactamase. FEMS Microbiol Lett 345:147–153. doi: 10.1111/1574-6968.12199. [DOI] [PubMed] [Google Scholar]
- 297.Mushtaq S, Vickers A, Ellaby N, Woodford N, Livermore DM. 2021. Selection and characterization of mutational resistance to aztreonam/avibactam in β-lactamase-producing Enterobacterales. J Antimicrob Chemother 77:98–111. doi: 10.1093/jac/dkab346. [DOI] [PubMed] [Google Scholar]
- 298.Lob SH, Karlowsky JA, Young K, Motyl MR, Hawser S, Kothari ND, Sahm DF. 2020. In vitro activity of imipenem-relebactam against resistant phenotypes of Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples—SMART Surveillance Europe 2015–2017. J Med Microbiol 69:207–217. doi: 10.1099/jmm.0.001142. [DOI] [PubMed] [Google Scholar]
- 299.Haidar G, Clancy CJ, Chen L, Samanta P, Shields RK, Kreiswirth BN, Nguyen MH. 2017. Identifying spectra of activity and therapeutic niches for ceftazidime-avibactam and imipenem-relebactam against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00642-17. doi: 10.1128/AAC.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Gomez-Simmonds A, Stump S, Giddins MJ, Annavajhala MK, Uhlemann AC. 2018. Clonal background, resistance gene profile, and porin gene mutations modulate in vitro susceptibility to imipenem-relebactam in diverse Enterobacteriaceae. Antimicrob Agents Chemother 62:e00573-18. doi: 10.1128/AAC.00573-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Gomis-Font MA, Cabot G, Sánchez-Diener I, Fraile-Ribot PA, Juan C, Moya B, Zamorano L, Oliver A. 2020. In vitro dynamics and mechanisms of resistance development to imipenem and imipenem/relebactam in Pseudomonas aeruginosa. J Antimicrob Chemother 75:2508–2515. doi: 10.1093/jac/dkaa206. [DOI] [PubMed] [Google Scholar]
- 302.Fraile-Ribot PA, Zamorano L, Orellana R, del Barrio-Tofiño E, Sánchez-Diener I, Cortes-Lara S, López-Causapé C, Cabot G, Bou G, Martínez-Martínez L, Oliver A. 2020. Activity of imipenem-relebactam against a large collection of Pseudomonas aeruginosa clinical isolates and isogenic β-lactam-resistant mutants. Antimicrob Agents Chemother 64:e02165-19. doi: 10.1128/AAC.02165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Petty LA, Henig O, Patel TS, Pogue JM, Kaye KS. 2018. Overview of meropenem-vaborbactam and newer antimicrobial agents for the treatment of carbapenem-resistant Enterobacteriaceae. Infect Drug Resist 11:1461–1472. doi: 10.2147/IDR.S150447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yamano Y. 2019. In vitro activity of cefiderocol against a broad range of clinically important Gram-negative bacteria. Clin Infect Dis 69:S544–S551. doi: 10.1093/cid/ciz827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zhanel GG, Golden AR, Zelenitsky S, Wiebe K, Lawrence CK, Adam HJ, Idowu T, Domalaon R, Schweizer F, Zhanel MA, Lagacé-Wiens PRS, Walkty AJ, Noreddin A, Lynch Iii JP, Karlowsky JA. 2019. Cefiderocol: a siderophore cephalosporin with activity against carbapenem-resistant and multidrug-resistant Gram-negative bacilli. Drugs 79:271–289. doi: 10.1007/s40265-019-1055-2. [DOI] [PubMed] [Google Scholar]
- 306.Iregui A, Khan Z, Landman D, Quale J. 2020. Activity of cefiderocol against Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii endemic to medical centers in New York City. Microb Drug Resist 26:722–726. doi: 10.1089/mdr.2019.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Burnard D, Robertson G, Henderson A, Falconer C, Bauer MJ, Cottrell K, Gassiep I, Norton R, Paterson DL, Harris PNA. 2021. Burkholderia pseudomallei clinical isolates are highly susceptible in vitro to cefiderocol, a siderophore cephalosporin. Antimicrob Agents Chemother 65:e00685-20. doi: 10.1128/AAC.00685-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. 2018. In vitro activity of the siderophore cephalosporin, cefiderocol, against carbapenem-nonsusceptible and multidrug-resistant isolates of Gram-negative bacilli collected worldwide in 2014 to 2016. Antimicrob Agents Chemother 62:e01968-17. doi: 10.1128/AAC.01968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Page MGP. 2019. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin Infect Dis 69:S529–S537. doi: 10.1093/cid/ciz825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Delgado-Valverde M, Conejo MDC, Serrano L, Fernández-Cuenca F, Pascual Á. 2020. Activity of cefiderocol against high-risk clones of multidrug-resistant Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Antimicrob Chemother 75:1840–1849. doi: 10.1093/jac/dkaa117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Grande Perez C, Maillart E, Miendje Deyi VY, Huang TD, Kamgang P, Dernier Y, Clevenbergh P. 2021. Compassionate use of cefiderocol in a pancreatic abscess and emergence of resistance. Infect Dis Now 51:399–401. doi: 10.1016/j.medmal.2020.10.022. [DOI] [PubMed] [Google Scholar]
- 312.Kohira N, Hackel MA, Ishioka Y, Kuroiwa M, Sahm DF, Sato T, Maki H, Yamano Y. 2020. Reduced susceptibility mechanism to cefiderocol, a siderophore cephalosporin, among clinical isolates from a global surveillance programme (SIDERO-WT-2014). J Glob Antimicrob Resist 22:738–741. doi: 10.1016/j.jgar.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 313.Streling AP, Al Obaidi MM, Lainhart WD, Zangeneh T, Khan A, Dinh AQ, Hanson B, Arias CA, Miller WR. 2021. Evolution of cefiderocol non-susceptibility in Pseudomonas aeruginosa in a patient without previous exposure to the antibiotic. Clin Infect Dis 73:e4472–e4474. doi: 10.1093/cid/ciaa1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tamma PD, Doi Y, Bonomo RA, Johnson JK, Simner PJ, Antibacterial Resistance Leadership Group. 2019. A primer on AmpC β-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin Infect Dis 69:1446–1455. doi: 10.1093/cid/ciz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Fernández L, Hancock RE. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Davin-Regli A, Lavigne JP, Pagès JM. 2019. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev 32:e00002-19. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kabra R, Chauhan N, Kumar A, Ingale P, Singh S. 2019. Efflux pumps and antimicrobial resistance: paradoxical components in systems genomics. Prog Biophys Mol Biol 141:15–24. doi: 10.1016/j.pbiomolbio.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Masi M, Winterhalter M, Pagès JM. 2019. Outer membrane porins. Subcell Biochem 92:79–123. doi: 10.1007/978-3-030-18768-2_4. [DOI] [PubMed] [Google Scholar]
- 319.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. 1999. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother 43:1924–1931. doi: 10.1128/AAC.43.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bradford PA, Urban C, Mariano N, Projan SJ, Rahal JJ, Bush K. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother 41:563–569. doi: 10.1128/AAC.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Mammeri H, Poirel L, Mangeney N, Nordmann P. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to β-lactams. Antimicrob Agents Chemother 47:1536–1542. doi: 10.1128/AAC.47.5.1536-1542.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Rasmussen BA, Keeney D, Yang Y, Bush K. 1994. Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob Agents Chemother 38:2078–2085. doi: 10.1128/AAC.38.9.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Payne DJ, Woodford N, Amyes SG. 1992. Characterization of the plasmid mediated β-lactamase BIL-1. J Antimicrob Chemother 30:119–127. doi: 10.1093/jac/30.2.119. [DOI] [PubMed] [Google Scholar]
- 326.Fosberry AP, Payne DJ, Lawlor EJ, Hodgson JE. 1994. Cloning and sequence analysis of blaBIL-1, a plasmid-mediated class C β-lactamase gene in Escherichia coli BS. Antimicrob Agents Chemother 38:1182–1185. doi: 10.1128/AAC.38.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Basu D, Narayankumar DV, Van Beeumen J, Basu J. 1997. Characterization of a β-lactamase from Mycobacterium smegmatis SN2. Biochem Mol Biol Int 43:557–562. doi: 10.1080/15216549700204361. [DOI] [PubMed] [Google Scholar]
- 328.Flores AR, Parsons LM, Pavelka MS. 2005. Genetic analysis of the β-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to β-lactam antibiotics. Microbiology (Reading) 151:521–532. doi: 10.1099/mic.0.27629-0. [DOI] [PubMed] [Google Scholar]
- 329.Fihman V, Rottman M, Benzerara Y, Delisle F, Labia R, Philippon A, Arlet G. 2002. BUT-1: a new member in the chromosomal inducible class C β-lactamases family from a clinical isolate of Buttiauxella sp. FEMS Microbiol Lett 213:103–111. doi: 10.1111/j.1574-6968.2002.tb11293.x. [DOI] [PubMed] [Google Scholar]
- 330.Ammenouche N, Dupont H, Mammeri H. 2014. Characterization of a novel AmpC β-lactamase produced by a carbapenem-resistant Cedecea davisae clinical isolate. Antimicrob Agents Chemother 58:6942–6945. doi: 10.1128/AAC.03237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Walsh TR, Hall L, MacGowan AP, Bennett PM. 1995. Sequence analysis of two chromosomally mediated inducible β-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J Antimicrob Chemother 36:41–52. doi: 10.1093/jac/36.1.41. [DOI] [PubMed] [Google Scholar]
- 332.Tokunaga H, Ishibashi M, Arakawa T, Tokunaga M. 2004. Highly efficient renaturation of β-lactamase isolated from moderately halophilic bacteria. FEBS Lett 558:7–12. doi: 10.1016/S0014-5793(03)01508-4. [DOI] [PubMed] [Google Scholar]
- 333.Müller A, Hächler H, Stephan R, Lehner A. 2014. Presence of AmpC β-lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb Drug Resist 20:275–280. doi: 10.1089/mdr.2013.0188. [DOI] [PubMed] [Google Scholar]
- 334.Ku YH, Lee MF, Chuang YC, Yu WL. 2018. Detection of plasmid-mediated β-lactamase genes and emergence of a novel AmpC (CMH-1) in Enterobacter cloacae at a medical center in Southern Taiwan. JCM 8:8. doi: 10.3390/jcm8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. 1996. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother 40:221–224. doi: 10.1128/AAC.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Gaillot O, Clément C, Simonet M, Philippon A. 1997. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J Antimicrob Chemother 39:85–87. doi: 10.1093/jac/39.1.85. [DOI] [PubMed] [Google Scholar]
- 337.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange PH, Philippon A. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob Agents Chemother 42:2352–2358. doi: 10.1128/AAC.42.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Barnaud G, Benzerara Y, Gravisse J, Raskine L, Sanson-Le Pors MJ, Labia R, Arlet G. 2004. Selection during cefepime treatment of a new cephalosporinase variant with extended-spectrum resistance to cefepime in an Enterobacter aerogenes clinical isolate. Antimicrob Agents Chemother 48:1040–1042. doi: 10.1128/AAC.48.3.1040-1042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Bonnet R, Chanal C, Ageron E, Sirot D, De Champs C, Grimont P, Sirot J. 2002. Inducible AmpC β-lactamase of a new member Enterobacteriaceae. Antimicrob Agents Chemother 46:3316–3319. doi: 10.1128/AAC.46.10.3316-3319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Naas T, Aubert D, Vimont S, Nordmann P. 2004. Identification of a chromosome-borne class C β-lactamase from Erwinia rhapontici. J Antimicrob Chemother 54:932–935. doi: 10.1093/jac/dkh446. [DOI] [PubMed] [Google Scholar]
- 341.Gonzalez Leiza M, Perez-Diaz JC, Ayala J, Casellas JM, Martinez-Beltran J, Bush K, Baquero F. 1994. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob Agents Chemother 38:2150–2157. doi: 10.1128/AAC.38.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cole M. 1982. Biochemistry and action of clavulanic acid. Scott Med J 27:S10–S16. doi: 10.1177/00369330820270S103. [DOI] [PubMed] [Google Scholar]
- 343.Knott-Hunziker V, Petursson S, Jayatilake GS, Waley SG, Jaurin B, Grundström T. 1982. Active sites of β-lactamases: the chromosomal β-lactamases of Pseudomonas aeruginosa and Escherichia coli. Biochem J 201:621–627. doi: 10.1042/bj2010621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Tzouvelekis LS, Tzelepi E, Mentis AF, Tsakris A. 1993. Identification of a novel plasmid-mediated β-lactamase with chromosomal cephalosporinase characteristics from Klebsiella pneumoniae. J Antimicrob Chemother 31:645–654. doi: 10.1093/jac/31.5.645. [DOI] [PubMed] [Google Scholar]
- 345.Tzouvelekis LS, Tzelepi E, Mentis AF. 1994. Nucleotide sequence of a plasmid-mediated cephalosporinase gene (blaLAT-1) found in Klebsiella pneumoniae. Antimicrob Agents Chemother 38:2207–2209. doi: 10.1128/AAC.38.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Lau SK, Ho PL, Li MW, Tsoi HW, Yung RW, Woo PC, Yuen KY. 2005. Cloning and characterization of a chromosomal class C β-lactamase and its regulatory gene in Laribacter hongkongensis. Antimicrob Agents Chemother 49:1957–1964. doi: 10.1128/AAC.49.5.1957-1964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Kimura H, Izawa M, Sumino Y. 1996. Molecular analysis of the gene cluster involved in cephalosporin biosynthesis from Lysobacter lactamgenus YK90. Appl Microbiol Biotechnol 44:589–596. doi: 10.1007/BF00172490. [DOI] [PubMed] [Google Scholar]
- 348.Papanicolaou GA, Medeiros AA, Jacoby GA. 1990. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 34:2200–2209. doi: 10.1128/AAC.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Jacoby GA, Tran J. 1999. Sequence of the MIR-1 β-lactamase gene. Antimicrob Agents Chemother 43:1759–1760. doi: 10.1128/AAC.43.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Horii T, Arakawa Y, Ohta M, Sugiyama T, Wacharotayankun R, Ito H, Kato N. 1994. Characterization of a plasmid-borne and constitutively expressed blaMOX-1 gene encoding AmpC-type β-lactamase. Gene 139:93–98. doi: 10.1016/0378-1119(94)90529-0. [DOI] [PubMed] [Google Scholar]
- 351.Nadjar D, Labia R, Cerceau C, Bizet C, Philippon A, Arlet G. 2001. Molecular characterization of chromosomal class C β-lactamase and its regulatory gene in Ochrobactrum anthropi. Antimicrob Agents Chemother 45:2324–2330. doi: 10.1128/AAC.45.8.2324-2330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Galleni M, Lindberg F, Normark S, Cole S, Honoré N, Joris B, Frère JM. 1988. Sequence and comparative analysis of three Enterobacter cloacae ampC β-lactamase genes and their products. Biochem J 250:753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Lodge JM, Minchin SD, Piddock LJ, Busby SJ. 1990. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem J 272:627–631. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 355.Feller G, Zekhnini Z, Lamotte-Brasseur J, Gerday C. 1997. Enzymes from cold-adapted microorganisms: the class C β-lactamase from the Antarctic psychrophile Psychrobacter immobilis A5. Eur J Biochem 244:186–191. doi: 10.1111/j.1432-1033.1997.00186.x. [DOI] [PubMed] [Google Scholar]
- 356.Pierrard A, Ledent P, Docquier JD, Feller G, Gerday C, Frère JM. 1998. Inducible class C β-lactamases produced by psychrophilic bacteria. FEMS Microbiology Lett 161:311–315. doi: 10.1111/j.1574-6968.1998.tb12962.x. [DOI] [Google Scholar]
- 357.Baumann M, Simon H, Schneider KH, Danneel HJ, Küster U, Giffhorn F. 1989. Susceptibility of Rhodobacter sphaeroides to β-lactam antibiotics: isolation and characterization of a periplasmic β-lactamase (cephalosporinase). J Bacteriol 171:308–313. doi: 10.1128/jb.171.1.308-313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Nomura K, Yoshida T. 1990. Nucleotide sequence of the Serratia marcescens SR50 chromosomal ampC β-lactamase gene. FEMS Microbiol Lett 58:295–299. doi: 10.1111/j.1574-6968.1990.tb13992.x. [DOI] [PubMed] [Google Scholar]
- 359.Matsumura N, Minami S, Mitsuhashi S. 1998. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob Agents Chemother 42:176–179. doi: 10.1128/AAC.42.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.De Luca F, Giraud-Morin C, Rossolini GM, Docquier JD, Fosse T. 2010. Genetic and biochemical characterization of TRU-1, the endogenous class C β-lactamase from Aeromonas enteropelogenes. Antimicrob Agents Chemother 54:1547–1554. doi: 10.1128/AAC.01252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Seoane A, Francia MV, García Lobo JM. 1992. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother 36:1049–1052. doi: 10.1128/AAC.36.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Mammeri H, Poirel L, Nazik H, Nordmann P. 2006. Cloning and functional characterization of the Ambler class C β-lactamase of Yersinia ruckeri. FEMS Microbiol Lett 257:57–62. doi: 10.1111/j.1574-6968.2006.00148.x. [DOI] [PubMed] [Google Scholar]
- 363.Tindall BJ, Sutton G, Garrity GM. 2017. Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13048) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980. Int J Syst Evol Microbiol 67:502–504. doi: 10.1099/ijsem.0.001572. [DOI] [PubMed] [Google Scholar]
- 364.Beaz-Hidalgo R, Martínez-Murcia A, Figueras MJ. 2013. Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martínez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst Appl Microbiol 36:171–176. doi: 10.1016/j.syapm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 365.Marathe NP, Salvà-Serra F, Karlsson R, Larsson DGJ, Moore ERB, Svensson-Stadler L, Jakobsson HE. 2019. Scandinavium goeteborgense gen. nov., sp. nov., a new member of the family Enterobacteriaceae isolated from a wound infection, carries a novel quinolone resistance gene variant. Front Microbiol 10:2511. doi: 10.3389/fmicb.2019.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Chen CM, Huang M, Wu HJ, Guo MK, Wu LT. 2018. Identification of CFE-2, a new plasmid-encoded AmpC β-lactamase from a clinical isolate of Citrobacter freundii. Int J Antimicrob Agents 52:421–424. doi: 10.1016/j.ijantimicag.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 367.Antonelli A, D'Andrea MM, Vaggelli G, Docquier JD, Rossolini GM. 2015. OXA-372, a novel carbapenem-hydrolysing class D β-lactamase from a Citrobacter freundii isolated from a hospital wastewater plant. J Antimicrob Chemother 70:2749–2756. doi: 10.1093/jac/dkv181. [DOI] [PubMed] [Google Scholar]
- 368.Guillon H, Tande D, Mammeri H. 2011. Emergence of ertapenem resistance in an Escherichia coli clinical isolate producing extended-spectrum β-lactamase AmpC. Antimicrob Agents Chemother 55:4443–4446. doi: 10.1128/AAC.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Crémet L, Caroff N, Giraudeau C, Reynaud A, Caillon J, Corvec S. 2013. Detection of clonally related Escherichia coli isolates producing different CMY β-lactamases from a cystic fibrosis patient. J Antimicrob Chemother 68:1032–1035. doi: 10.1093/jac/dks520. [DOI] [PubMed] [Google Scholar]
- 370.Mammeri H, Poirel L, Nordmann P. 2007. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J Antimicrob Chemother 60:490–494. doi: 10.1093/jac/dkm227. [DOI] [PubMed] [Google Scholar]
- 371.Dahyot S, Mammeri H. 2012. Hydrolysis spectrum extension of CMY-2-like β-lactamases resulting from structural alteration in the Y-X-N loop. Antimicrob Agents Chemother 56:1151–1156. doi: 10.1128/AAC.05630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Kotsakis SD, Miriagou V, Vetouli EE, Bozavoutoglou E, Lebessi E, Tzelepi E, Tzouvelekis LS. 2015. Increased hydrolysis of oximino-β-lactams by CMY-107, a Tyr199Cys mutant form of CMY-2 produced by Escherichia coli. Antimicrob Agents Chemother 59:7894–7898. doi: 10.1128/AAC.01793-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Mata C, Miró E, Rivera A, Mirelis B, Coll P, Navarro F. 2010. Prevalence of acquired AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes at a Spanish hospital from 1999 to 2007. Clin Microbiol Infect 16:472–476. doi: 10.1111/j.1469-0691.2009.02864.x. [DOI] [PubMed] [Google Scholar]
- 374.Pope CE, Carter PE, Heffernan HM. 2009. CMY-29 and CMY-30, two novel plasmid-mediated AmpC β-lactamases. Antimicrob Agents Chemother 53:3178. doi: 10.1128/AAC.01586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Hentschke M, Kotsakis SD, Wolters M, Heisig P, Miriagou V, Aepfelbacher M. 2011. CMY-42, a novel plasmid-mediated CMY-2 variant AmpC β-lactamase. Microb Drug Resist 17:165–169. doi: 10.1089/mdr.2010.0137. [DOI] [PubMed] [Google Scholar]
- 376.Endimiani A, Doi Y, Bethel CR, Taracila M, Adams-Haduch JM, O'Keefe A, Hujer AM, Paterson DL, Skalweit MJ, Page MGP, Drawz SM, Bonomo RA. 2010. Enhancing resistance to cephalosporins in class C β-lactamases: impact of Gly214Glu in CMY-2. Biochemistry 49:1014–1023. doi: 10.1021/bi9015549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Pérez-Llarena FJ, Vázquez-Ucha JC, Kerff F, Zamorano L, Miró E, Cabral MP, Fleites A, Lantero M, Martínez-Martínez L, Oliver A, Galleni M, Navarro F, Beceiro A, Bou G. 2018. Increased antimicrobial resistance in a novel CMY-54 AmpC-type enzyme with a GluLeu217-218 insertion in the Ω-loop. Microb Drug Resist 24:527–533. doi: 10.1089/mdr.2017.0017. [DOI] [PubMed] [Google Scholar]
- 378.Tsukamoto K, Ohno R, Sawai T. 1990. Extension of the substrate spectrum by an amino acid substitution at residue 219 in the Citrobacter freundii cephalosporinase. J Bacteriol 172:4348–4351. doi: 10.1128/jb.172.8.4348-4351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Barlow M, Hall BG. 2003. Experimental prediction of the evolution of cefepime resistance from the CMY-2 AmpC β-lactamase. Genetics 164:23–29. doi: 10.1093/genetics/164.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Pires J, Taracila M, Bethel CR, Doi Y, Kasraian S, Tinguely R, Sendi P, Bonomo RA, Endimiani A. 2015. In vivo evolution of CMY-2 to CMY-33 β-lactamase in Escherichia coli sequence type 131: characterization of an acquired extended-spectrum AmpC conferring resistance to cefepime. Antimicrob Agents Chemother 59:7483–7488. doi: 10.1128/AAC.01804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. 2014. Reclaiming the efficacy of β-lactam-β-lactamase inhibitor combinations: avibactam restores the susceptibility of CMY-2-producing Escherichia coli to ceftazidime. Antimicrob Agents Chemother 58:4290–4297. doi: 10.1128/AAC.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Schneider I, Markovska R, Marteva-Proevska Y, Mitov I, Markova B, Bauernfeind A. 2014. Detection of CMY-99, a novel acquired AmpC-type β-lactamase, and VIM-1 in Proteus mirabilis isolates in Bulgaria. Antimicrob Agents Chemother 58:620–621. doi: 10.1128/AAC.01450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Ahmed AM, Shimamoto T. 2008. Emergence of a cefepime- and cefpirome-resistant Citrobacter freundii clinical isolate harbouring a novel chromosomally encoded AmpC β-lactamase, CMY-37. Int J Antimicrob Agents 32:256–261. doi: 10.1016/j.ijantimicag.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 384.Livermore DM, Mushtaq S, Barker K, Hope R, Warner M, Woodford N. 2012. Characterization of β-lactamase and porin mutants of Enterobacteriaceae selected with ceftaroline + avibactam (NXL104). J Antimicrob Chemother 67:1354–1358. doi: 10.1093/jac/dks079. [DOI] [PubMed] [Google Scholar]
- 385.Vakulenko SB, Golemi D, Geryk B, Suvorov M, Knox JR, Mobashery S, Lerner SA. 2002. Mutational replacement of Leu-293 in the class C Enterobacter cloacae P99 β-lactamase confers increased MIC of cefepime. Antimicrob Agents Chemother 46:1966–1970. doi: 10.1128/AAC.46.6.1966-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Morosini MI, Negri MC, Shoichet B, Baquero MR, Baquero F, Blázquez J. 1998. An extended-spectrum AmpC-type β-lactamase obtained by in vitro antibiotic selection. FEMS Microbiol Lett 165:85–90. doi: 10.1111/j.1574-6968.1998.tb13131.x. [DOI] [PubMed] [Google Scholar]
- 387.Tran QT, Dupont M, Lavigne JP, Chevalier J, Pagès JM, Sotto A, Davin-Regli A. 2009. Occurrence of efflux mechanism and cephalosporinase variant in a population of Enterobacter aerogenes and Klebsiella pneumoniae isolates producing extended-spectrum β-lactamases. Antimicrob Agents Chemother 53:1652–1656. doi: 10.1128/AAC.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Rodríguez-Martínez JM, Fernández-Echauri P, Fernández-Cuenca F, Diaz de Alba P, Briales A, Pascual A. 2012. Genetic characterization of an extended-spectrum AmpC cephalosporinase with hydrolysing activity against fourth-generation cephalosporins in a clinical isolate of Enterobacter aerogenes selected in vivo. J Antimicrob Chemother 67:64–68. doi: 10.1093/jac/dkr423. [DOI] [PubMed] [Google Scholar]
- 389.Raimondi A, Sisto F, Nikaido H. 2001. Mutation in Serratia marcescens AmpC β-lactamase producing high-level resistance to ceftazidime and cefpirome. Antimicrob Agents Chemother 45:2331–2339. doi: 10.1128/AAC.45.8.2331-2339.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Yatsuyanagi J, Saito S, Konno T, Harata S, Suzuki N, Kato J, Amano K. 2006. Nosocomial outbreak of ceftazidime-resistant Serratia marcescens strains that produce a chromosomal AmpC variant with N235K substitution. Jpn J Infect Dis 59:153–159. [PubMed] [Google Scholar]
- 391.Hidri N, Barnaud G, Decré D, Cerceau C, Lalande V, Petit JC, Labia R, Arlet G. 2005. Resistance to ceftazidime is associated with a S220Y substitution in the Ω loop of the AmpC β-lactamase of a Serratia marcescens clinical isolate. J Antimicrob Chemother 55:496–499. doi: 10.1093/jac/dki025. [DOI] [PubMed] [Google Scholar]
- 392.Mammeri H, Poirel L, Bemer P, Drugeon H, Nordmann P. 2004. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob Agents Chemother 48:716–720. doi: 10.1128/AAC.48.3.716-720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Papagiannitsis CC, Tzouvelekis LS, Tzelepi E, Miriagou V. 2007. Plasmid-encoded ACC-4, an extended-spectrum cephalosporinase variant from Escherichia coli. Antimicrob Agents Chemother 51:3763–3767. doi: 10.1128/AAC.00389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Doi Y, Shibata N, Shibayama K, Kamachi K, Kurokawa H, Yokoyama K, Yagi T, Arakawa Y. 2002. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 46:2427–2434. doi: 10.1128/AAC.46.8.2427-2434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Hee Lee S, Lee JH, Heo MJ, Bae IK, Jeong SH, Cha SS. 2007. Exact location of the region responsible for the extended substrate spectrum in class C β-lactamases. Antimicrob Agents Chemother 51:3778–3779. doi: 10.1128/AAC.00633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Sohn SG, Lee JJ, Sohn ES, Kang LW, Lee SH. 2008. Comment on: extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J Antimicrob Chemother 61:965–966. doi: 10.1093/jac/dkn023. [DOI] [PubMed] [Google Scholar]
- 397.Lee SJ, Kim JY, Jung HI, Suh PG, Lee HS, Lee SH, Cha SS. 2004. Crystallization and preliminary X-ray crystallographic analyses of CMY-1 and CMY-10, plasmidic class C β-lactamases with extended substrate spectrum. Acta Crystallogr D Biol Crystallogr 60:382–384. doi: 10.1107/S090744490302821X. [DOI] [PubMed] [Google Scholar]
- 398.Piotrowska M, Przygodzińska D, Matyjewicz K, Popowska M. 2017. Occurrence and variety of β-lactamase genes among Aeromonas spp. isolated from urban wastewater treatment plant. Front Microbiol 8:863. doi: 10.3389/fmicb.2017.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Kotsakis SD, Papagiannitsis CC, Tzelepi E, Tzouvelekis LS, Miriagou V. 2009. Extended-spectrum properties of CMY-30, a Val211Gly mutant of CMY-2 cephalosporinase. Antimicrob Agents Chemother 53:3520–3523. doi: 10.1128/AAC.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Bauvois C, Ibuka AS, Celso A, Alba J, Ishii Y, Frère JM, Galleni M. 2005. Kinetic properties of four plasmid-mediated AmpC β-lactamases. Antimicrob Agents Chemother 49:4240–4246. doi: 10.1128/AAC.49.10.4240-4246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Zhang Z, Yu Y, Musser JM, Palzkill T. 2001. Amino acid sequence determinants of extended spectrum cephalosporin hydrolysis by the class C P99 β-lactamase. J Biol Chem 276:46568–46574. doi: 10.1074/jbc.M102757200. [DOI] [PubMed] [Google Scholar]
- 402.Compain F, Debray A, Adjadj P, Dorchêne D, Arthur M. 2020. Ceftazidime-avibactam resistance mediated by the N346Y substitution in various AmpC β-lactamases. Antimicrob Agents Chemother 64:e02311-19. doi: 10.1128/AAC.02311-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Poirel L, Héritier C, Spicq C, Nordmann P. 2004. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J Clin Microbiol 42:3831–3833. doi: 10.1128/JCM.42.8.3831-3833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Mammeri H, Nordmann P, Berkani A, Eb F. 2008. Contribution of extended-spectrum AmpC (ESAC) β-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol Lett 282:238–240. doi: 10.1111/j.1574-6968.2008.01126.x. [DOI] [PubMed] [Google Scholar]
- 405.Armand-Lefèvre L, Leflon-Guibout V, Bredin J, Barguellil F, Amor A, Pagès JM, Nicolas-Chanoine MH. 2003. Imipenem resistance in Salmonella enterica serovar Wien related to porin loss and CMY-4 β-lactamase production. Antimicrob Agents Chemother 47:1165–1168. doi: 10.1128/AAC.47.3.1165-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother 50:3396–3406. doi: 10.1128/AAC.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Shi W, Li K, Ji Y, Jiang Q, Wang Y, Shi M, Mi Z. 2013. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 β-lactamase production. Braz J Microbiol 44:435–442. doi: 10.1590/S1517-83822013000200015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Chow JW, Shlaes DM. 1991. Imipenem resistance associated with the loss of a 40 kDa outer membrane protein in Enterobacter aerogenes. J Antimicrob Chemother 28:499–504. doi: 10.1093/jac/28.4.499. [DOI] [PubMed] [Google Scholar]
- 409.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 410.Raimondi A, Traverso A, Nikaido H. 1991. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and Proteus rettgeri lack porins. Antimicrob Agents Chemother 35:1174–1180. doi: 10.1128/AAC.35.6.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Quale J, Bratu S, Landman D, Heddurshetti R. 2003. Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin Infect Dis 37:214–220. doi: 10.1086/375821. [DOI] [PubMed] [Google Scholar]