Abstract
The E. coli RelA protein is a ribosome-dependent (p)ppGpp synthetase that is activated in response to amino acid starvation. RelA can be dissected both functionally and physically into two domains: The N-terminal domain (NTD) (amino acids [aa] 1 to 455) contains the catalytic domain of RelA, and the C-terminal domain (CTD) (aa 455 to 744) is involved in regulating RelA activity. We used mutational analysis to localize sites important for RelA activity and control in these two domains. We inserted two separate mutations into the NTD, which resulted in mutated RelA proteins that were impaired in their ability to synthesize (p)ppGpp. When we caused the CTD in relA+ cells to be overexpressed, (p)ppGpp accumulation during amino acid starvation was negatively affected. Mutational analysis showed that Cys-612, Asp-637, and Cys-638, found in a conserved amino acid sequence (aa 612 to 638), are essential for this negative effect of the CTD. When mutations corresponding to these residues were inserted into the full-length relA gene, the mutated RelA proteins were impaired in their regulation. In attempting to clarify the mechanism through which the CTD regulates RelA activity, we found no evidence for competition for ribosomal binding between the normal RelA and the overexpressed CTD. Results from CyaA complementation experiments of the bacterial two-hybrid system fusion plasmids (G. Karimova, J. Pidoux, A. Ullmann, and D. Ladant, Proc. Natl. Acad. Sci. USA 95:5752–5756, 1998) indicated that the CTD (aa 564 to 744) is involved in RelA-RelA interactions. Our findings support a model in which RelA activation is regulated by its oligomerization state.
The natural environments of bacteria are often characterized by changes in nutrient availability. When bacterial cells are deprived of an amino acid or a carbon source, changes in many cellular processes occur. This pleiotropic response, called the stringent response, was initially described for Escherichia coli in 1961 (30). The first observed feature of the stringent response was the accumulation of two unusual phosphorylated derivatives of GTP and GDP, called (p)ppGpp collectively, within a few seconds after amino acid starvation (4, 5, 8, 18). Other features of the stringent response include inhibition of rRNA and tRNA synthesis, inhibition of replication initiation and cell division, inhibition of the active transport of many metabolites, stimulation of the synthesis of enzymes involved in amino acid biosynthesis (6), and induction of the rpoS gene, which encodes the stationary phase sigma factor (10). The major effector of the stringent response is probably (p)ppGpp. In E. coli, the mutation causing the relaxed phenotype, in which cells failed to accumulate (p)ppGpp during amino acid starvation, was mapped to the relA gene at 59.2 min on the E. coli chromosome (1). This gene has been cloned, sequenced, and characterized: the relA gene encodes a protein of 744 amino acids of molecular mass 84 kDa (21). The RelA protein is a (p)ppGpp synthetase which is activated in response to amino acid starvation. It catalyzes the pyrophosphoryl group transfer of the beta and gamma phosphates from the ATP donor to the ribose 3′ hydroxyl of GTP (or GDP) (7, 15). For its reaction in vitro, purified RelA requires mRNA, functional ribosomes paused during elongation at a “hungry codon,” and uncharged cognate-tRNA bound at the acceptor site of that hungry codon (11, 12). In cell extracts, RelA is found bound to a small fraction of the ribosomes (about 1%) (26). A relA null mutant has been constructed that shows a relaxed phenotype upon amino acid starvation but can still respond to carbon source limitation (23).
The relA1 mutant allele which possesses weak residual (p)ppGpp synthetic activity was found to have an IS2 insertion between codons 85 and 86. The presence of the IS2 insertion creates two RelA fragments that complement each other in trans to yield residual (p)ppGpp synthetic activity. Neither fragment shows this activity when expressed alone (23). The fact that the two fragments could reconstitute RelA activity has led to the suggestion that the relA gene could be dissected into two domains both functionally and physically. To test this possibility, plasmids bearing the full-length relA or truncated relA fragments under the control of the Ptac promoter were constructed. Cells deleted for relA but which were overexpressing the various fragments were examined for their ability to accumulate (p)ppGpp. The induced full-length RelA protein was found to be a ribosome-dependent (p)ppGpp synthetase. In contrast, a truncated RelA protein, containing 455 N-terminal amino acids, acted as a ribosome-independent, constitutive, (p)ppGpp synthetase. This truncated protein was metabolically labile, with a half-life of about 7.5 min, compared to the stable full-length RelA (half-life, 2 to 3 h) (29). The remaining C-terminal fragment was found to be devoid of synthetic activity (29). These results indicated that the relA gene product can indeed be dissected, both functionally and physically, into two distinct domains: the N-terminal domain (NTD) (amino acids [aa] 1 to 455) is responsible for (p)ppGpp synthesis, and the C-terminal domain (CTD) (aa 456 to 744) is responsible for regulating RelA activity.
Here we report on our partial characterization of the sites involved in the synthetic activity of RelA. Our work has led to further insights about the manner in which the activity of RelA is regulated through the action of its CTD.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1 and Table 2.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| W3110 | RelA+ | Laboratory collection |
| W3110lacIq | A lacIq derivative of W3110; Knr | Laboratory collection |
| W3110ΔrelAlacIq | A ΔrelA251::kan lacIq derivative of W3110; Tetr | Laboratory collection |
| JF423lacIq | A lacIq derivative of relC; Rifr, Knr | Laboratory collection |
| W3110ΔrelApcnB | A pcnB derivative of W3110ΔrelA251::kan; Tetr | This study (17) |
| BTH101 | A cya strain | D. Ladant |
TABLE 2.
Plasmids used in this study
| Plasmid | Description [reference] |
|---|---|
| pALS10 | pBR322 derivative bearing the full-length relA; Apr [29, 32] |
| pALS13 | Like pALS10 but bearing the first 455 codons of relA [29, 32] |
| pALS14 | Like pALS10 but bearing the first 331 codons of relA [29, 32] |
| pALS10-G251E | Like pALS10 but containing the G251E mutation in relA |
| pALS13-G251E | Like pALS13 but containing the G251E mutation in relA |
| pALS10-H354Y | Like pALS10 but containing the H354Y mutation in relA |
| pNT8 | pKK-233-2 derivative encoding the truncated CTD of RelA (aa 497–744) |
| pYG4 | pKK-233-2 derivative encoding the truncated CTD of RelA (aa 564–675) |
| pYG4-C612G | Like pYG4 but containing the C612G mutation |
| pYG4-D637R | Like pYG4 but containing the D637R mutation |
| pYG4-C638F | Like pYG4 but containing the C638F mutation |
| pALS10-C612G | Like pALS10 but containing the C612G mutation |
| pALS10-D637R | Like pALS10 but containing the D637R mutation |
| pALS10-C638F | Like pALS10 but containing the C638F mutation |
| pT25 | Expression vector encoding the T25 fragment of CyaA in frame with a multi-cloning site; Cmr [14] |
| pT18 | Expression vector encoding the T18 fragment of CyaA in frame with a multicloning site; Apr [14] |
| PT25-RelA | PT25 derivative bearing the full-length relA (codons 1–744) |
| PT18-RelA | pT18 derivative bearing the full-length relA (codons 1–744) |
| pT25-CTD | pT25 derivative encoding the CTD of RelA (aa 564–744) |
| pT18-CTD | pT18 derivative encoding the CTD of RelA (aa 564–744) |
| pT18-NTD | pT18 derivative encoding the NTD of RelA (aa 1–456) |
| pT18-CTD-C612G | Like pT18-CTD but containing the C612G mutation |
| pT18-CTD-D637R | Like pT18-CTD but containing the D637R mutation |
| pT18-CTD-C638F | Like pT18-CTD but containing the C638F mutation |
| pT25-CTD long | pT25 derivative encoding the extended CTD of RelA (aa 564–744) |
| pT18-CTD long | pT18 derivative encoding the extended CTD of RelA (aa 564–744) |
Synthetic primers.
The primers used for PCR are listed in Table 3.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′ → 3′) |
|---|---|
| G251E-5′ | GTTAAAGCGGAAGTGTATGAGCGTCCGAAACACATCa |
| G251E-3′ | GATGTGTTTCGGACGCTCATACACTTCCGCTTTAAC |
| EspI-5′ | GAGTGCTTAGCCGAAACCTGGb |
| NsiI-3′ | TTCATGCATCTGTTTGGTGCG |
| H354Y-5′ | GTTGGGTGTTGCTGCGTACTGGAAATATAAAG |
| H354Y-3′ | CTTTATATTTCCAGTACGCAGCAACACCCAAC |
| BamHI-3′ | CTCTAGAGGATCCCCTATGTAG |
| Yg4 (PstI) | TGCACTGCAGTTAATAAGCCGAGTGCCG |
| B3(HindIII) | GCGACGAAGCTTCTAACTCCCGTGCAACCG ACGCGC |
| C612G-5′ | GCACCACATCGCGCGCGGCTGCCAGCCGATTC |
| C612G-3′ | GAATCGGCTGGCAGCCGCGCGCGATGTGGTGC |
| D637R-5′ | GTATTTCAGTACACCGCGCCCGTTGCGAACAACTGGCGG |
| D637R-3′ | CCGCCAGTTGTTCGCAACGGGCGCGGTGTACTGAAATAC |
| C638F-5′ | CAGTACACCGCGCCGATTTCGAACAACTGGCGGAAC |
| C638F-3′ | CAGTTCCGCCAGTTGTTCGAAATCGGCGCGGTGTAC |
| NsiI-5′ | CAAATCCGCACCAAACAGATGCATGAAG |
| N-up | CTGCAGGTACCTATGGTTGCGGTAAGAAGT |
| N-down | TGAGGGTACCTGCAGCTGGTAGGTGAACGG |
| C-up | CTGCAGGTACCTTTTAATAAGCCGAGTGCCG |
| C-down | TGACGGTACCCGACTCCCGTGCAACCGACGC |
| C-long-up | CTGCAGGTACCTTACCAGCTGCAGATGGGCG |
| N-long-down | TGAGGGTACCTGATTAAATTGCGATTGCAGG |
Mutated bases are indicated in bold type.
Restriction sites are underlined.
Growth media.
The growth media used were Luria-Bertani (LB) and Super-LB (SLB) (1.6% tryptone, 1% yeast extract, 0.5% NaCl). Morpholinepropane sulfonic acid (MOPS) glucose minimal medium (25) was used in (p)ppGpp synthesis assays. Screening for the ability to ferment maltose was performed on MacConkey agar plates containing 1% maltose. Selection for resistance to 3-amino-1,2,4-triazole (AT) was performed on minimal M9 AT plates containing 15 mM AT, all amino acids except histidine, 1 mM adenine, and 1 mM thiamine (28). The antibiotics used for selections were used at the following concentrations: 100 μg of ampicillin/ml, 50 μg of kanamycin/ml, 20 μg of tetracycline/ml, 150 μg of rifampicin/ml, and 34 μg of chloramphenicol/ml.
Site-directed mutagenesis and plasmid construction.
Plasmid pALS10-G251E, bearing the full-length relA gene with the G251E mutation, was constructed as follows: the mutagenesis in codon 251 of the relA gene was performed by PCR on pALS10, bearing the full-length relA gene (29, 32), using four primers (Table 3): two complementary primers, G251E-5′ and G251E-3′, containing the G251E mutation, and two additional primers, EspI-5′ and NsiI-3′, containing unique restriction sites in the plasmid, upstream and downstream from the mutated nucleotides. Two reactions were carried out using the high-fidelity Pwo DNA polymerase (Boehringer, Mannheim, Germany) with pALS10 as a template. In the first reaction, primers G251E-5′ and NsiI-3′ were used, and in the second reaction, primers EspI-5′ and G251E-3′ were used. The amplified products of these reactions were two overlapping fragments, each containing the G251E mutation. In the next step, these fragments were used as templates for PCR with primers EspI-5′ and NsiI-3′ to obtain a continuous mutated fragment that was subsequently cleaved and cloned into the EspI and NsiI sites of pALS10. The mutagenesis was verified by sequencing. Plasmid pALS13-G251E, bearing the first 455 codons of relA with the G251E mutation, was constructed by deleting the 946-bp PstI fragment of pALS10-G251E. We used the same procedure that was carried out for plasmid pALS10-G251E to construct plasmid pALS10-H354Y, bearing the full-length relA gene with the H354Y mutation. Here we used primers H354Y-5′ and H354Y-3′ and primers EspI-5′ and BamHI-3′. To construct pYG4, we used PCR to amplify the region of relA corresponding to the truncated C-terminal fragment (aa 564 to 744) from pALS10 with primers YG4 and B3. The amplified fragment was cleaved and cloned into the PstI and HindIII sites of the expression vector pKK-233-2. Using pYG4, we mutagenized codons 612, 637, and 638 in the region corresponding to the C-terminal fragment of RelA by PCR with primers C612G-5′ and C612G-3′, D637R-5′ and D637R-3′, C638F-5′ and C638F-3′, and with primers YG4 and B3 containing the multilinker sites PstI and HindIII. This resulted in pYG4-C612G bearing the allele CTD-C612G, pYG4-D637R bearing CTD-D637R, and pYG4-C638F bearing CTD-C638F. These same three alleles corresponding to the RelA CTD were similarly incorporated into pALS10, which bears the full-length relA gene. In this case we used primers NsiI and BamHI-3′ containing unique restriction sites from pALS10. This resulted in plasmids pALS10-C612G, pALS10-D637R, and pALS10-C638F.
Construction of the bacterial two-hybrid system fusion plasmids.
The regions of relA encoding the N-terminal (aa 1 to 455) and C-terminal (aa 564 to 744) fragments were amplified by PCR from pALS10 using the following primers (Table 3): N-up and N-down for the region corresponding to the N terminus and C-up and C-down for the region corresponding to the C terminus. The amplified fragments were cleaved and cloned into the Asp718 sites of pT25 and pT18 to yield plasmids pT25-NTD and pT18-NTD and plasmids pT25-CTD and pT18-CTD, encoding respectively the N-terminal and C-terminal domains of RelA fused in-frame into the T25 and T18 fragments of the adenylate cyclase protein (CyaA) (14). The region corresponding to the extended C-terminal fragment of RelA (aa 455 to 744) was amplified by PCR from pALS10 using primers C-long-up and C-down and then cleaved and cloned into the Asp718 sites of pT25 and pT18 to yield plasmids pT25-CTD long and pT18-CTD long, respectively. The region corresponding to the extended N-terminal fragment of RelA (aa 1 to 564) was amplified by PCR from pALS10 using primers N-up and N-long-down and then cleaved and cloned into the Asp718 site of pT18 to yield plasmid pT18-NTD long. pT18-RelA, encoding the full-length relA fragment, was constructed by ligation of the Asp718-PstI fragments of pT18-NTD long and pT18-CTD long into the Asp718 site of pT18. The full-length relA fragment was removed from pT18-RelA by cleavage with Asp718 and cloned into the Asp718 site of pT25 to yield plasmid pT25-RelA.
Measurements of (p)ppGpp accumulation.
Cells were grown in MOPS glucose minimal medium (25) supplemented with all amino acids except glutamine and glutamate. At an optical density at 600 nm (OD600) of 0.1, the cells were uniformly labeled with 33Pi, after which plasmid induction was performed by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). At an OD600 of 0.2, amino acid starvation was induced by adding 1 mg of serine-hydroxamate/ml. Samples were withdrawn at various time intervals and analyzed for their (p)ppGpp content by one-dimensional chromatography as described previously (3, 29). Nucleotides were quantified with a Fujix Bas 1000 imaging system. The amounts of (p)ppGpp were expressed as fractions of the total nucleotide pool. Measurements of (p)ppGpp accumulation after plasmid induction without starvation were performed by labeling the cells at an OD600 of 0.1 with 33Pi and adding 1 mM IPTG at an OD600 of 0.2. Samples were withdrawn after 30 min and analyzed for their (p)ppGpp content.
Ribosome preparation.
Ribosomes were prepared as described by Block and Haseltine (2) with the following modifications: cells were grown in SLB medium with shaking at 37°C. At an OD600 of 0.3, IPTG was added to a final concentration of 1 mM. After 1 h of incubation, 60 ml of the cell culture was pelleted at 4°C and resuspended in 15 ml of cold buffer containing 100 mM Tris-acetate (pH 8), 10 mM MgOAc, and 1 mM dithiothreitol. Lysozyme was added to a final concentration of 1 mg/ml, and cells were lysed in a Yeda-Press pressure cell for 2 min (twice). To remove the DNA, 15 U of RQ1 RNase-free Dnase (Promega, Madison, Wis.) and 3 ml of 10× ribosome buffer (0.5 M Tris-acetate [pH 8], 0.1 M MgOAc, 10 mM dithiothreitol, 0.27 M NH4OAc) were added to a final volume of 30 ml. Cell debris was removed by centrifugation at 27,000 × g at 4°C for 30 min. A sample of the supernatant representing the soluble proteins before fractionation was saved for further analysis. The rest of the supernatant was centrifuged in a Ti-60 rotor (100,000 × g) at 4°C for 3 h. After a sample of the resulting supernatant was saved for further analysis (nonribosomal fraction), the pellet was suspended in 0.5 ml of 1× ribosome buffer. The ribosomes were frozen and stored at −70°C. Protein concentrations in the soluble fraction, in the ribosomal fraction, and in the supernatant were determined using the Bio-Rad Protein Assay.
Western blot analysis.
Protein samples were loaded on sodium dodecyl sulfate polyacrylamide gels and separated by electrophoresis. The separated proteins were transferred to a nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany) using the Mini Trans-Blot Cell apparatus (Bio-Rad Laboratories, Richmond, Calif.). Proteins were detected using polyclonal antibodies against E. coli RelA and the ECL Western blotting detection system (Amersham Life Science, Inc., Buckinghamshire, United Kingdom).
β-galactosidase assay.
Cells were grown in LB medium supplemented with 0.5 mM IPTG at 30°C. β-galactosidase assays were performed as described by Miller (24).
RESULTS
Mutational analysis of the NTD of RelA. (i) Site-directed mutagenesis in the NTD.
To define sequences involved in RelA catalytic activity, we performed site-directed mutagenesis for two amino acids in the NTD. These two amino acids are found in regions that are conserved among E. coli and RelA homologues from other bacteria (20, 33). The first mutation that we inserted was G251E. A spontaneous Bacillus subtilis mutant that exhibited a relaxed phenotype has a mutation in codon 240 of its relA gene that is analogous to E. coli G251E (33). The second mutation that we inserted was H354Y. Since histidine is often found in the active sites of enzymes, we speculated that H354 might be located in the active site of RelA. The mutagenesis was performed on pALS10, an expression vector bearing the relA gene (29, 32), using PCR as described in Materials and Methods.
(ii) The mutated RelA proteins, RelA-G251E and RelA-H354Y, are impaired in their (p)ppGpp synthetic activity.
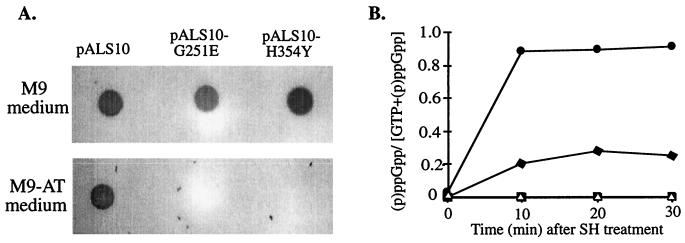
To examine the synthetic activity of RelA-G251E and RelA-H354Y, we used plasmids pALS10-G251E and pALS10-H354Y to transform the ΔrelAlacIq derivative of strain W3110. The transformed cells were tested for their ability to grow in the presence of 15 mM AT, a His analog that induces His starvation (13). Since in E. coli (p)ppGpp is a positive regulator of the his biosynthetic operon (31), cells can grow in the presence of AT only if their levels of (p)ppGpp are high enough (29). Strain W3110ΔrelAlacIq expressing the full-length normal RelA protein gained the ability to grow in the presence of AT. In contrast, the same strain expressing the mutated proteins RelA-G251E or RelA-H354Y remained AT sensitive (Fig. 1A).
FIG. 1.
Accumulation of (p)ppGpp in a relA-null strain overexpressing the mutated N-terminal RelA proteins. (A) Strain W3110ΔrelAlacIq transformed with pALS10 (RelA), pALS10-G251E (RelA-G251E), or pALS10-H354Y (RelA-H354Y) was grown at 37°C with shaking overnight. Equal amounts of cells were washed twice with M9 medium and spotted either on minimal M9 AT plates containing 15 mM AT, all the amino acids except histidine, 1 mM adenine, and 1 mM thiamine or on the same medium but without AT. Growth was examined after incubation at 37°C for 16 h. (B) Strain W3110ΔrelAlacIq transformed with pALS10 (●), pALS10-G251E (▵), pALS10-H354Y (⧫), or pKK-223-2 (□) was grown in MOPS glucose minimal medium supplemented with all the amino acids except glutamine and glutamate. At an OD600 of 0.1, the cells were uniformly labeled with 33Pi, and plasmid induction was performed by adding 1 mM IPTG. At an OD600 of 0.2, amino-acid starvation was induced by the addition of 1mg of SH/ml. Samples were withdrawn at 10, 20, and 30 min and then analyzed for their (p)ppGpp content.
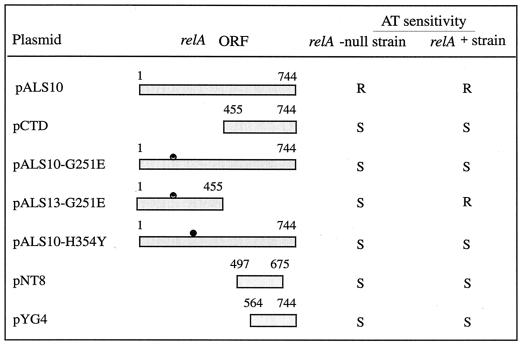
When we deleted the region encoding the C-terminal fragment from pALS10-G251E, we observed that strain W3110ΔrelAlacIq expressing the N-terminal truncated portion of RelA-G251E (pALS13-G251E) still failed to grow in the presence of AT (Fig. 2). Thus, the mutation in the NTD of RelA-G251E causes its loss of synthetic activity.
FIG. 2.
AT sensitivity of relA+ and relA-null cells expressing the various RelA truncated proteins. Strains W3110lacIq and W3110ΔrelAlacIq were transformed by pALS10 (RelA), pCTD (RelA [aa 455 to 744]), pALS10-G251E (RelA-G251E), pALS13-G251E (RelA [aa 1 to 455]-G251E), pALS10-H354Y (RelA-H354Y), pNT8 (RelA [aa 497 to 675]), or pYG4 (RelA [aa 564 to 744]). Growth and AT sensitivity tests were performed as described in the legend to Fig. 1A. S, sensitive; R, resistant.
Since sensitivity to AT can only discriminate qualitatively between high and low levels of (p)ppGpp, we carried out a quantitative assay of (p)ppGpp synthesis. The accumulation of (p)ppGpp during amino acid starvation was measured for strain W3110ΔrelAlacIq, which was induced to overexpress RelA (pALS10), RelA-G251E (pALS10-G251E), and RelA-H354Y (pALS10-H354Y). As a negative control, we used the vector plasmid pKK-223-2. Ten minutes after the addition of serine-hydroxymate (SH), we observed high levels of (p)ppGpp in strain W3110ΔrelAlacIq, in which the normal RelA protein was overexpressed. In contrast, cells in which RelA-G251E was overexpressed did not accumulate (p)ppGpp after SH treatment, and the levels of (p)ppGpp in cells in which RelA-H354Y was overexpressed were fourfold lower than in cells that expressed the wild-type RelA (Fig. 1B). Western blot analysis revealed that the normal and mutated RelA proteins were similarly expressed (data not shown). Therefore, the impairment in (p)ppGpp synthesis of the mutated proteins is not due to their instability.
In sum, the single-amino-acid replacement in codon 251 caused the RelA protein to lose its (p)ppGpp synthetic activity, while the single-amino-acid replacement in codon 354 only partially impaired the synthetic activity of RelA.
Deletion and mutational analysis of the CTD of RelA. (i) Expression of the CTD in relA+ cells negatively affects their ability to respond to amino acid starvation.
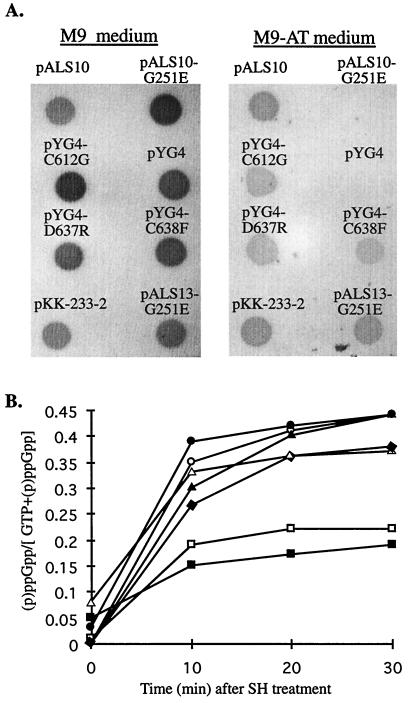
Previously in our laboratory, it was shown that the CTD of RelA (aa 456 to 744) is devoid of (p)ppGpp synthetic activity (29). Here we observed that the relA+ strain W3110lacIq became AT sensitive when it was transformed by the derivative of pKK-233-2 that bears the sequence corresponding to the C-terminal fragment (pCTD) (Fig. 2). We also observed a dominant-negative effect on (p)ppGpp accumulation in relA+ cells transformed by plasmids expressing the inactive mutated RelA proteins, RelA-G251E and RelA-H354Y (Fig. 2 and 3). In contrast, the same strain expressing the full-length RelA (pALS10) or the N-terminal truncated RelA with the G251E mutation (pALS13-G251E) remained AT resistant (Fig. 2 and 3). Thus, the negative effect on the response to amino acid starvation is due to the action of the CTD of RelA.
FIG. 3.
Accumulation of (p)ppGpp in a relA+ strain overexpressing the C-terminal mutated proteins. (A) Strain W3110lacIq was transformed by pALS10 (RelA), pALS10-G251E (RelA-G251E), pYG4 (CTD), pYG4-C612G (CTD-C612G), pYG4-D637R (CTD-D637R), pYG4-C638F (CTD-C638F), pALS13-G251E (RelA [aa 1 to 455]-G251E), or pKK-233-2. Growth and AT sensitivity tests were performed as described in the legend to Fig. 1A. (B) Strain W3110lacIq was transformed by pYG4 (□), pYG4-C612G (▴), pYG4-D637R (●), pYG4-C638F (⧫), pALS10-G251E (■), pALS13-G251E (▵), or pKK233-2 (○). Growth and the quantitative (p)ppGpp accumulation assay were performed as described in the legend to Fig. 1B.
(ii) Deletion analysis of the CTD.
We wished to localize the specific region in the CTD that is involved in the loss of AT resistance. For this purpose, we generated various C-terminal deletions by PCR with primers from random sites in the CTD and cloned them into pKK-233-2. We examined strain W3110lacIq transformed by plasmids encoding the truncated CTD fragments for its ability to grow on M9 AT plates. The shortest C-terminal fragments of RelA that were sufficient for interfering with (p)ppGpp accumulation contained either the 178 aa 497 to 675 (pNT8) or the 180 terminal aa 564 to 744 (pYG4) (Fig. 2 and 3A). We chose to use pYG4 for further studies.
(iii) Site-directed mutagenesis in the CTD.
The C-terminal fragment that is encoded by pYG4 contains a 27-aa-long sequence (aa 612 to 638) that is highly conserved between E. coli RelA and other RelA homologues (20, 33). This conserved sequence contains three cysteine residues that are known to play a special role in shaping the secondary structure of proteins by forming disulfide links. To further characterize amino acid sequences in the CTD that are important for its function, we replaced two of these cysteine residues, C612G and C638F. In addition, we inserted a mutation that does not disrupt the secondary structure of the protein into a conserved aspartate residue encoded by codon 637 (D637R). In all three cases, the mutagenesis was performed on plasmid pYG4 using PCR as described in Materials and Methods.
(iv) In relA+ cells, the mutated C-terminal proteins, CTD-C612G, CTD-D637R, and CTD-C638F, lose their negative effect on (p)ppGpp accumulation.
To study the activity of the proteins with the mutated CTDs, we transformed the relA+ strain W3110lacIq by pYG4 derivatives bearing the mutated C-terminal fragments, pYG4-C612G, pYG4-D637R, or pYG4-C638F, or the controls pKK-233-2, pYG4, pALS10-G251E, pALS13-G251E, or pALS10. The transformed cells were tested for their ability to grow on M9 AT plates. The relA+ cells harboring pYG4 or pALS10-G251E became AT sensitive. In contrast, relA+ cells transformed by pKK-233-2, pALS10, pALS13-G251E, or by plasmids bearing the three mutated C-terminal fragments remained AT resistant (Fig. 3A). Western blot analysis revealed that all the C-terminal proteins were similarly expressed (data not shown). Thus, the loss of function of the C-terminal mutated proteins was not due to their instability.
To assay quantitatively the effects of the normal and mutated C-terminal proteins on (p)ppGpp accumulation in relA+ cells during amino acid starvation, the transformed cells were induced to overexpress the various proteins as described in Materials and Methods. Within 10 min after the addition of SH, the levels of (p)ppGpp accumulated in the relA+ cells overexpressing the three mutated C-terminal proteins or the N-terminal truncated mutated RelA protein, NTD-G251E, were as high as the levels in the control cells harboring pKK-233-2 (Fig. 3B). The levels of (p)ppGpp in the relA+ cells overexpressing the normal C-terminal protein or the full-length N-terminal mutated RelA protein, RelA-G251E, were two- to threefold lower (Fig. 3B). Thus, the C-terminal proteins in which one of three specific amino acids has been replaced have lost their negative effect on (p)ppGpp accumulation in relA+ cells following amino acid starvation.
(v) The effect on RelA activity of point mutations in the CTD of the full-length RelA protein.
To ascertain how each of the three amino acid replacements in the CTD would affect the regulation of the full-length RelA protein, we carried out site-directed mutagenesis in codons 612, 637, and 638 in the full-length relA gene. The mutagenesis was performed on pALS10, using PCR as described in Materials and Methods.
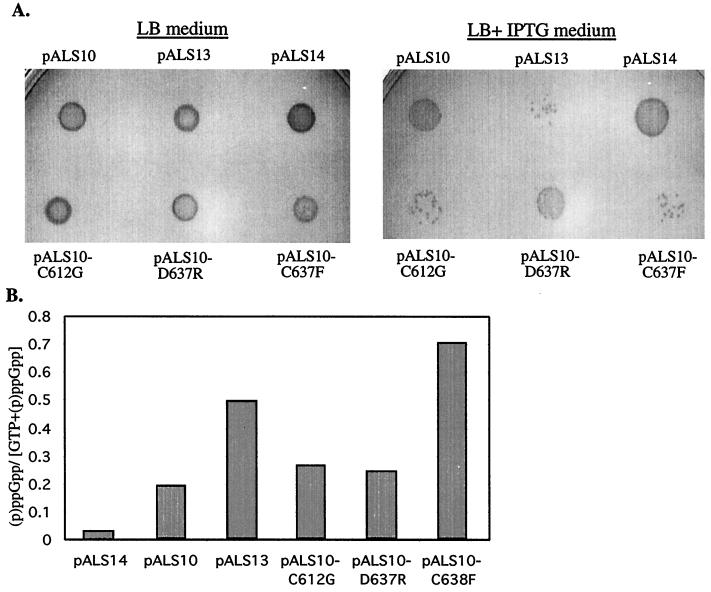
Here we used the relaxed mutant relC strain (JF423) that has a mutation in rplK, the structural gene for the ribosomal protein L11. In vitro, ribosomes from the relC strain have only 10% of the (p)ppGpp synthetic activity of ribosomes from the wild-type strain (9). Previously in our laboratory, it was shown that when they were overexpressed separately in the relC strain, the synthetic activity of the full-length RelA was severely impaired, while the truncated N-terminal RelA fragment was active (29). We therefore examined the phenotype of a lacIq derivative of strain JF423 transformed by plasmids bearing the various relA alleles. Since the truncated N-terminal RelA is an unstable protein (29), we could not use it in an AT sensitivity test. Instead, we examined the growth on LB medium supplemented with 1 mM IPTG of strain JF423lacIq in which the various relA alleles were overexpressed. The overaccumulation of (p)ppGpp is known to be toxic (6, 32); therefore, the deaths of the cells under these conditions would indicate that they synthesize (p)ppGpp. As we expected, in the presence of IPTG, strain JF423lacIq overexpressing either the full-length RelA protein (pALS10) or the short inactive RelA protein (pALS14) grew normally, while cells overexpressing the truncated N-terminal RelA fragment (pALS13) grew poorly because they synthesize (p)ppGpp constitutively (Fig. 4A) (29). The C-terminal mutated RelA proteins, RelA-C612G (pALS10-C612G) and RelA-C638F (pALS10-C638F), exhibited a phenotype resembling that of the truncated N-terminal RelA. Cells overexpressing these proteins grew poorly in the presence of IPTG, probably due to the accumulation of high levels of (p)ppGpp. The mutated protein RelA-D637R (pALS10-D637R) showed an intermediate phenotype: the growth of cells overexpressing this protein was affected by IPTG, but to a lesser degree than for the two other mutated proteins (Fig. 4A).
FIG. 4.
Accumulation of (p)ppGpp in the relClacIq strain overexpressing the C-terminal mutated RelA proteins. Strain JF423lacIq was transformed with pALS10 (RelA), pALS10-C612G (RelA-C612G), pALS10-D637R (RelA-D637R), pALS10-C638F (RelA-C638F), pALS13 (RelA [aa 1 to 455]), or pALS14 (RelA [aa 1 to 331). (A) The transformed cells were grown in LB at 37°C with shaking overnight. Equal amounts of cells were spotted on LB medium or on LB medium supplemented with 1 mM IPTG for plasmid induction. Growth was examined after incubation at 37°C for 16 h. (B) The transformed cells were grown in MOPS glucose minimal medium supplemented with all the amino acids except glutamine and glutamate. At an OD600 of 0.1, the cells were uniformly labeled with 33Pi. Plasmid induction was performed by adding 1 mM IPTG at an OD600 of 0.2. Samples were withdrawn at 30 min and then analyzed for their (p)ppGpp content.
To assay quantitatively the effects of the various relA alleles on (p)ppGpp accumulation in JF423lacIq cells, the transformed cells were induced to overexpress the various proteins as described in Materials and Methods. The results of the quantitative assay were in accordance with the results of the toxicity assay. Thirty minutes after the addition of IPTG, cells overexpressing either the short inactive RelA protein (pALS14) or the full-length RelA protein (pALS10) accumulated low levels of (p)ppGpp (Fig. 4B). The levels of (p)ppGpp in the cells overexpressing the truncated N-terminal RelA protein (pALS13) or the C-terminal mutated RelA protein, RelA-C638F, were high. Cells overexpressing RelA-C612G or RelA-D637R accumulated higher (p)ppGpp levels than cells with the full-length RelA protein but lower levels than cells with the truncated RelA protein (Fig. 4B). Thus, the single-amino-acid replacement in codon 638 caused the RelA protein to lose its ability to be regulated, while the single-amino-acid replacements in codons 612 and 637 only partially impaired the regulation of RelA.
The role of the CTD in controlling RelA activity.
In the next step, we carried out experiments to clarify the role of the CTD in the regulation of RelA activity and the involvement of amino acids Cys-612, Asp-637, and Cys-638 in this regulation.
(i) The negative effect of the CTD on (p)ppGpp accumulation in relA+ cells following amino acid starvation cannot be explained by competition for ribosomal binding.
RelA is a ribosome-associated protein that requires the presence of ribosomes for its reaction in vitro (11, 12). The results of induction experiments with a relC mutant strain revealed that when the CTD is deleted from RelA, the truncated protein becomes ribosome independent (29). These results suggested that the CTD is responsible for RelA binding to the ribosome and that this binding regulates RelA activity (6). However, due to its instability, the binding of the truncated N-terminal RelA protein to the ribosomes could not be examined (29). According to this model, the dominant-negative effect on relA+ cells of the overexpressed CTD is the result of competition with the normal RelA for ribosomal binding. On the other hand, because they can no longer bind the ribosome, the mutated C-terminal protein fragments will have lost their ability to interfere with RelA activity. This model also predicts that the full-length RelA proteins with point mutations in the CTD are constitutively active because they cannot bind to the ribosomes. As we will show below, our results did not support this model.
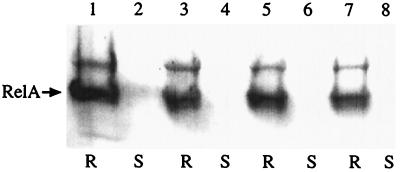
We carried out Western blot analysis with crude ribosome extracts from the relA+ strain W3110lacIq harboring plasmids bearing the various alleles for the CTD (pYG4, pYG4-C612G, pYG4-D637R, or pYG4-C638F). After the addition of IPTG, we found that the amounts of the ribosome-bound endogenous RelA protein were approximately the same in each case (Fig. 5). Moreover, Western blot analysis carried out with crude ribosome extracts from strain W3110ΔrelApcnB induced to express low levels of the mutated C-terminal RelA proteins (pALS10-C612G, pALS10-D637R, and pALS10-C638F) revealed that although the mutated RelA proteins were impaired in their regulation, they were still ribosome bound (data not shown). Thus, these results suggest that the negative effect of the overexpressed C-terminal fragment on RelA activity cannot be explained by competition for ribosomal binding.
FIG. 5.
Western blot analysis of ribosomal and nonribosomal fractions from a relA+ strain overexpressing the normal or mutated CTD proteins. Strain W3110lacIq harboring plasmids bearing the various alleles of the CTD was grown in SLB with shaking at 37°C until an OD600 of 0.3, at which time 1 mM IPTG was added. Protein lysates of these cultures were separated into the ribosomal (R) and nonribosomal (soluble) (S) fractions as described in Materials and Methods. Aliquots from these fractions, containing equal amounts of proteins, were loaded on sodium dodecyl sulfate polyacrylamide gels and electrophoresed, and Western blot analysis was performed using polyclonal antibodies against E. coli RelA. Lanes 1 and 2, pYG4; lanes 3 and 4, pYG4-C612G; lanes 5 and 6, pYG4-D637R; lanes 7 and 8, pYG4-C638F.
(ii) The CTD is involved in RelA-RelA interactions.
We considered the possibility that the CTD might be involved in RelA-RelA interactions. The CTD could regulate the activity of its own RelA molecule by interacting with the CTD of another RelA molecule. Alternatively, it could interact with its own NTD or with that of another RelA molecule. To study the interactions between the various RelA fragments, we used the bacterial two-hybrid system constructed by Karimova and colleagues (14). In this system, two proteins that are assumed to be interacting are genetically fused to two complementary fragments, T25 and T18, that constitute the catalytic domain of Bordetella pertussis adenylate cyclase (CyaA). The association of the two-hybrid proteins results in the functional complementation between the T25 and T18 fragments and leads to cyclic AMP (cAMP) synthesis. The transcriptional activation of catabolic operons, such as the maltose operon, is then triggered by cAMP. We cloned the gene fragments of the full-length RelA protein, NTD, CTD, and the mutated C-terminal fragments in-frame into pT25 and pT18 vector plasmids as described in Materials and Methods.
We used the various constructs to cotransform the cya strain BTH101. As a positive control, we used plasmids pT25-zip and pT18-zip bearing the leucine zipper region of the yeast protein GCN4 (14). The transformants were plated on MacConkey medium supplemented with 1% maltose; the color of the colonies was examined after incubation at 30°C for 36 to 72 h. Colonies of cells that could synthesize cAMP and thus were able to ferment maltose appeared red, while colonies of cells that were unable to ferment maltose appeared white. The efficiency of the complementation was quantified by measuring β-galactosidase activities in liquid cultures. The results of the complementation experiments revealed that CyaA activity was reconstituted with high efficiency when both parts of the vector pair were fused to the gene fragment encoding the full-length RelA protein. High efficiency of complementation was also obtained when the full-length RelA protein was coexpressed with the normal CTD (aa 564 to 744) but not when it was coexpressed with the normal NTD (aa 1 to 455) (Table 4). We observed high CyaA activity when both parts of the vector pair were fused to the normal CTD fragment. However, when the CTD was coexpressed with the mutated C-terminal fragments CTD-C612G, CTD-D637R, or CTD-C638F, CyaA activity was only partially reconstituted. The mutations in the CTD decreased the efficiency of complementation with the normal CTD by 52% (CTD-C638F), 39% (CTD-C612G), and 27% (CTD-D637R). We observed no CyaA activity when the normal NTD (aa 1 to 455) was coexpressed with the normal CTD (Table 4).
TABLE 4.
CyaA complementation of the bacterial two-hybrid system fusion plasmids
| Plasmids | Phenotype on MacConkey-maltosea at:
|
β-galactosidase activity (U/ml)b | ||
|---|---|---|---|---|
| 36 h | 48 h | 72 h | ||
| None | − | − | − | 101 |
| pT25-zip + pT18-zip | +++ | 3,961 | ||
| pT25-RelA + pT18-RelA | ++ | +++ | 3,310 | |
| pT25-CTD + pT18-RelA | ++ | +++ | 2,951 | |
| PT25-NTD + pT18-RelA | − | − | + | 1,371 |
| pT25-CTD + pT18-CTD | ++ | +++ | 3,606 | |
| pT25-CTD + pT18-CTD-C612G | − | − | + | 2,227 |
| pT25-CTD + pT18-CTD-D637R | − | ++ | +++ | 2,662 |
| pT25-CTD + pT18-CTD-C638F | − | − | + | 1,725 |
| pT25-CTD + pT18-NTD | − | − | − | 54 |
| pT25-CTD long + pT18-NTD | − | − | − | 111 |
| pT25-NTD + pT18-CTD long | − | − | − | 109 |
The intensity of the color of the colony: −, white; +, light red; ++, red; +++, dark red.
The results represent the average values obtained from at least four independent experiments.
Our results suggest that RelA forms homo-oligomers through its CTD. Furthermore, it appears that C638, and to a lesser extent C612 and D637, are involved in this interaction.
Since the C-terminal fragment that we used in our experiments contained only the last 180 amino acids of RelA (aa 564 to 744), and the N-terminal fragment contained the first 455 amino acids of the protein, we wished to examine the involvement in RelA-RelA interactions of the 109 remaining internal residues (aa 455 to 564). For this purpose, we constructed fusion plasmids containing an extended CTD (aa 455 to 744). The various constructs were cotransformed into strain BTH101. We found that lengthening the CTD had no effect on the efficiency of complementation. The extended C-terminal fragment was still unable to interact with the N-terminal fragment (Table 4). Thus, we found no evidence for interactions of the CTD with the NTD.
DISCUSSION
The E. coli RelA protein is a ribosome-dependent (p)ppGpp synthetase that is activated in response to amino acid starvation. RelA can be dissected both functionally and physically into two domains (23, 29). The NTD (aa 1 to 455) contains the catalytic domain of RelA. The CTD (aa 455 to 744), which is devoid of synthetic activity, is involved in regulating RelA activity. Here we used mutational analysis to localize sites important for RelA activity and regulation in these two domains.
We introduced two mutations into the NTD: G251E and H354Y. RelA-G251E lost its ability to synthesize (p)ppGpp, and RelA-H354Y partially lost its synthetic activity. Preliminary results from experiments recently carried out in our laboratory indicated that RelA-G251E is impaired in its ability to bind both ATP and GTP, while RelA-354 is impaired in its ability to bind ATP but can still bind GTP. The E. coli RelA and RelA homologues from other bacteria contain no conserved motifs for ATP and GTP binding (19, 22). The only exception is the relA homologue of Streptomyces coelicolor, which contains a putative P-loop motif (20). Thus, it seems that the RelA proteins must contain new unknown ATP and GTP binding domains. Note that Gly-251 and His-354 are highly conserved among the relA and spoT gene homologues that have been cloned and sequenced (19). Furthermore, a spontaneously relaxed mutant of B. subtilis has been found to have a mutation analogous to the G251E mutation (G240E) (33). Therefore, we hypothesize that these two amino acid residues are located in this new ATP and GTP binding domain(s) shared by the RelA proteins.
In our study we demonstrated that when the CTD of RelA was overexpressed, it negatively affected (p)ppGpp accumulation in relA+ cells starved for amino acids. We observed this dominant-negative effect in the presence of either the intact short C-terminal fragment (aa 564 to 744 or the full-length inactive RelA proteins (RelA-G251E and RelA-H354Y) (Fig. 2). By mutational analysis, we showed that the amino acids Cys-612, Asp-637, and Cys-638 found in the conserved sequence (aa 612 to 638) are essential for the negative effect of the CTD. Inserting the corresponding mutations into the full-length relA gene resulted in RelA proteins that were impaired to various degrees in their ability to be regulated (Fig. 4). Thus, amino acids Cys-612, Asp-637, and Cys-638 appear to be involved in regulating RelA activity. We speculate that the disulfide bonds formed by the cysteine residues in positions 612 and, particularly, 638 are essential for RelA control. When these disulfide bonds are disrupted, RelA may lose its ability to be regulated and thus would synthesize (p)ppGpp constitutively. Although the formation of disulfide bonds in the cytoplasm is rare, transient disulfide bonds have been detected in a few cytoplasmatic proteins, such as the transcription factor OxyR (34).
In attempting to clarify the mechanism through which the CTD regulates RelA activity, we found no evidence for competition for ribosomal binding between the normal RelA and the overexpressed CTD. Furthermore, the full-length RelA proteins with mutations in the CTD (RelA-C612G, RelA-D637R, and RelA-C638F) were still found to be ribosome associated. Our findings are supported by the fact that in relC mutants, while the endogenous RelA remained ribosome bound, it maintained only 10% of its normal activity (9). Thus, it seems that even though in these cells the ribosomes contain a mutant form of ribosomal protein L11, they can still bind RelA. Based on our results, we suggest that the regulation of RelA activity is not simply a question of whether or not RelA can bind the ribosome. It is possible that binding of RelA to the ribosome, and especially its association with the L11 protein, may be important for maintaining RelA in the “right” conformation for it to undergo activation in response to amino acid starvation.
We used the bacterial two-hybrid system constructed by Karimova and colleagues (14) to study the possible involvement of the CTD in RelA-RelA interactions. The results of our complementation experiments indicate that RelA forms homo-oligomers and that the CTD (aa 564 to 744) is involved in RelA-RelA interactions. The C-terminal proteins with amino-acid replacements in residues Cys-638 and, to a lesser extent, Cys-612 and Asp-637 are impaired in their ability to interact with the normal CTD, so it seems that these residues take part in these interactions. As mentioned above, we found no evidence for the interaction of the NTD (aa 1 to 455) with the CTD. However, the full-length RelA interacts weakly with the NTD (Table 4). We observed a weak CyaA activity also when both parts of the vectors were fused to the normal NTD (unpublished results). Thus, we suggest that the NTD is involved in weak RelA-RelA interactions.
Until now, generally it has been found that proteins that undergo homo-oligomerization are inactive when they are in a monomeric state and become activated upon dimerization. However, Livnah and colleagues (16) have shown that the erythropoietin receptor, once thought to be activated by ligand-induced homodimerization, is found as preformed dimers before being activated by a ligand. Furthermore, Remy and colleagues (27) have shown that in the presence of the ligand, the erythropoietin receptor homodimer undergoes a conformational change that activates it. We hypothesize that like the erythropoietin receptor, RelA is also in a homo-oligomer state even before it is activated.
Our results support a model in which RelA activation is controlled by its oligomerization state. We propose that RelA is inactive in its oligomeric state; following amino acid starvation, the oligomer dissociates, permitting RelA to become active. It is possible that instead of dissociation, there is a change in the conformation of the RelA oligomer (16, 27). According to our model, (i) when the C-terminal fragment is overexpressed in the cell, it forms an “incorrect” oligomer with the normal full-length RelA. This association causes the RelA protein to lose its ability to be activated; thus, the presence of the overexpressed CTD decreases the ability of relA+ cells to respond to amino acid starvation and (ii) the mutated C-terminal RelA proteins are impaired in their ability to form oligomers and therefore are impaired in their regulation.
ACKNOWLEDGMENTS
We thank M. Cashel and F. R. Warshaw-Dadon for critical readings of the manuscript. We thank D. Ladant and Hybrigenics for providing us with strain BTH101 and plasmids pT25 and pT18.
This work was supported by grant number 355/96 from the Israeli National Academy of Science foundation awarded to G. Glaser and by a fellowship from the Landowski foundation (number 034.4455) awarded to M. Gropp.
REFERENCES
- 1.Alfoldi L, Stent G S, Clowes R C. The chromosomal site for the RNA control (R.C.) locus in E. coli. J Mol Biol. 1962;5:348–355. doi: 10.1016/s0022-2836(62)80077-1. [DOI] [PubMed] [Google Scholar]
- 2.Block R, Haseltine W A. Purification and properties of stringent factor. J Biol Chem. 1975;250:1212–1217. [PubMed] [Google Scholar]
- 3.Bochner B R, Ames B N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 4.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 5.Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–884. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 6.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Resnikoff W, Riley M, Schaechter M, Umbarger A E, editors. E. coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 7.Cochran J W, Byrne R W. Isolation and properties of a ribosome-bound factor required for ppGpp and ppGpp synthesis in Escherichia coli. J Biol Chem. 1974;249:353–360. [PubMed] [Google Scholar]
- 8.Fiil N P, von Meyenburg K, Friesen J D. Accumulation and turnover of guanosine tetraphosphate in Escherichia coli. J Mol Biol. 1972;71:769–783. doi: 10.1016/s0022-2836(72)80037-8. [DOI] [PubMed] [Google Scholar]
- 9.Friesen J D, Fiil N P, Parker J M, Haseltine W A. A new relaxed mutant of Escherichia coli with an altered 50S ribosomal subunit. Proc Natl Acad Sci USA. 1974;71:3465–3469. doi: 10.1073/pnas.71.9.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase specific sigma factor ςs is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haseltine W A, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haseltine W A, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238:381–384. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 13.Hilton J L, Kearney P C, Ames B N. Mode of action of the herbicide, 3-amino-1,2,4-triazole(amitrole): inhibition of an enzyme of histidine biosynthesis. Arch Biochem Biophys. 1965;112:544–547. doi: 10.1016/0003-9861(65)90093-7. [DOI] [PubMed] [Google Scholar]
- 14.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipmann F, Sy J. The enzymic mechanism of guanosine 5′, 3′-polyphosphate synthesis. Prog Nucleic Acid Res Mol Biol. 1976;17:1–14. doi: 10.1016/s0079-6603(08)60063-x. [DOI] [PubMed] [Google Scholar]
- 16.Livnah O, Stura E A, Middleton S A, Johnson D L, Jolliffe L K, Wilson I A. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 17.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of E. coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 18.Lund E, Kjeldgaard N O. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur J Biochem. 1972;28:316–326. doi: 10.1111/j.1432-1033.1972.tb01916.x. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Costa O H, Arias P, Romero N M, Parro V, Mellado R P, Malpartida F. A relA/spoT homologous gene from Streptomyces coelicolor A3(2) controls antibiotic biosynthetic genes. J Biol Chem. 1996;271:10627–10634. doi: 10.1074/jbc.271.18.10627. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Costa O H, Fernandez-Moreno M A, Malpartida F. The relA/spoT-homologous gene in Streptomyces coelicolor encodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J Bacteriol. 1998;180:4123–4132. doi: 10.1128/jb.180.16.4123-4132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzger S, Dror I B, Aizenman E, Schreiber G, Toone M, Friesen J D, Cashel M, Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- 22.Metzger S, Sarubbi E, Glaser G, Cashel M. Protein sequences encoded by the relA and the spoT genes of Escherichia coli are interrelated. J Biol Chem. 1989;264:9122–9125. [PubMed] [Google Scholar]
- 23.Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 25.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen F S, Kjeldgaard N O. Analysis of the relA gene product of Escherichia coli. Eur J Biochem. 1977;76:91–97. doi: 10.1111/j.1432-1033.1977.tb11573.x. [DOI] [PubMed] [Google Scholar]
- 27.Remy I, Wilson I A, Michnick S W. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 28.Rudd K E, Bochner B R, Cashel M, Roth J R. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J Bacteriol. 1985;163:534–554. doi: 10.1128/jb.163.2.534-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 30.Stent G S, Brenner S. A genetic locus for the regulation of ribonucleic acid synthesis. Proc Natl Acad Sci USA. 1961;47:2005–2014. doi: 10.1073/pnas.47.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens J C, Artz S W, Ames B N. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp) positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc Natl Acad Sci USA. 1975;72:4389–4393. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svitil A L, Cashel M, Zyskind J W. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 33.Wendrich T M, Marahiel M A. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- 34.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]