SUMMARY
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) keeps evolving and mutating into newer variants over time, which gain higher transmissibility, disease severity, and spread in communities at a faster rate, resulting in multiple waves of surge in Coronavirus Disease 2019 (COVID-19) cases. A highly mutated and transmissible SARS-CoV-2 Omicron variant has recently emerged, driving the extremely high peak of infections in almost all continents at an unprecedented speed and scale. The Omicron variant evades the protection rendered by vaccine-induced antibodies and natural infection, as well as overpowers the antibody-based immunotherapies, raising the concerns of current effectiveness of available vaccines and monoclonal antibody-based therapies. This review outlines the most recent advancements in studying the virology and biology of the Omicron variant, highlighting its increased resistance to current antibody-based therapeutics and its immune escape against vaccines. However, the Omicron variant is highly sensitive to viral fusion inhibitors targeting the HR1 motif in the spike protein, enzyme inhibitors, involving the endosomal fusion pathway, and ACE2-based entry inhibitors. Omicron variant-associated infectivity and entry mechanisms of Omicron variant are essentially distinct from previous characterized variants. Innate sensing and immune evasion of SARS-CoV-2 and T cell immunity to the virus provide new perspectives of vaccine and drug development. These findings are important for understanding SARS-CoV-2 viral biology and advances in developing vaccines, antibody-based therapies, and more effective strategies to mitigate the transmission of the Omicron variant or the next SARS-CoV-2 variant of concern.
KEYWORDS: SARS-CoV-2 Omicron, booster, antibodies, resistance, entry inhibitors, fusion inhibitors, pan-CoV vaccines, T cell response, immune evasion
INTRODUCTION
The Coronavirus Disease 2019 (COVID-19) pandemic caused by novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been ongoing for over 2 years. Along with considerable interruptions of social and economic activities, the pandemic has posed global public health concerns and adverse financial impacts owing to its devastating effects on the survival of humanity (1–4). Presently, approximately 500 million confirmed cases and 6.2 million cases of death have been reported due to COVID-19 globally as of 18 April 2022 (https://covid19.who.int/) (1). Apart from original SARS-CoV-2, the emerging variants of this pandemic virus have altogether caused consequent waves of the pandemic over the time, and a massive and rapid surge in confirmed COVID-19 cases is currently being seen predominantly due to the recently heavily mutated and highly transmissible Omicron (B.1.1.529) variant. Several SARS-CoV-2 variants of concern (VOC) have been defined and designated by the World Health Organization (WHO), named Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1), Delta (B.1.617.2), and Omicron. The Omicron variant has three main distinct sublineages (BA.1/BA1.1, BA.2, and BA.3), with BA.1 and BA.2 being the most prevalent sublineages found globally (5, 6). The vast majority of experimental studies were conducted with the original Omicron BA.1 variant, while only limited studies on the Omicron BA.2/BA.3 lineages have been published recently (7–12). Therefore, the Omicron variant mentioned here refers to BA.1, unless otherwise stated. As the most recently identified VOC, the Omicron was initially detected in Botswana on 11 November 2021 and classified as a VOC by the WHO on 26 November 2021. The variant rapidly became the driving force behind the fourth wave of the epidemic in South Africa (13). Such newly emerging SARS-CoV-2 variants can show higher transmissibility and increase disease severity, morbidity, and mortality associated with COVID-19. These variants can potentially acquire features of immune escape from protection provided by vaccine-induced immunity and/or natural infection, and even modified monoclonal antibody (MAb)-based treatment regimens, which can result in breakthrough infections in vaccinated peoples, reinfection in COVID experienced recovered patients, and high and rapid increase in infected cases worldwide (14–17). The strong genomic surveillance system identified the Omicron variant within a week after a notification of a remarkable increasing confirmed COVID-19 cases in South Africa (13). Thereafter, the Omicron variant spread very rapidly to nearly 150 countries within a short time span of few weeks and drove the fourth peak of the confirmed COVID-19 cases across the globe (17, 18). Unsurprisingly, Omicron has now outcompeted the previous VOCs and has the largest prevalence in the world by replacing the previously dominating Delta variant. Omicron infections have increased dramatically, despite massive COVID-19 vaccination campaigns going on worldwide. The Omicron variant causes less severe disease and is believed to be associated with less pathogenic outcomes. For example, one clinical study showed that the proportion of patients that require oxygen supply dropped significantly in the fourth wave of infection caused by the Omicron variant spreading in South Africa compared to the previous waves (19). Furthermore, patients infected with the Omicron variant in southern California had a 53% reduced risk of being symptomatic hospitalization and a 74% reduction in risk of intensive care unit (ICU) admission, and there has been a 91% decreased risk of mortality compared to those with a Delta variant infection (20). The ability of the Omicron variant to breakthrough vaccine-elicited immunity results from elevated viral titers compared to the Delta variant, as well as an increased infectiousness (21). In addition, superspreading and regional outbreak of Omicron variant infections even occur in triple-vaccinated health care workers (22).
SARS-CoV-2, as a positive-sense RNA virus, harbors an ~30-kb large single-stranded genome and the whole genome encodes 4 structural proteins (membrane [M], envelope [E], nucleocapsid [N], and spike [S]), 16 nonstructural proteins, and 9 accessory proteins (23). The SARS-CoV-2 virion surface displays an average of 30 to 60 S trimers nearly 10 nm in length with an average of 15-nm distance from each other (24, 25). Entry of SARS-CoV-2 via cell surface fusion requires activation of the S protein by both host angiotensin converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2). The S protein contains two domains: S1 is responsible for receptor binding, and S2 is responsible for subsequent membrane fusion. S is primed by TMPRSS2 and drives the efficient fusion of the viral membrane with the cell membrane after the receptor-binding domain (RBD) in S binds to ACE2. Another route of entering cells by SARS-CoV-2 occurs via endosomal entry that also involves ACE2 binding, followed by endosomal acidification and cleavage of S2 by cathepsin L (26).
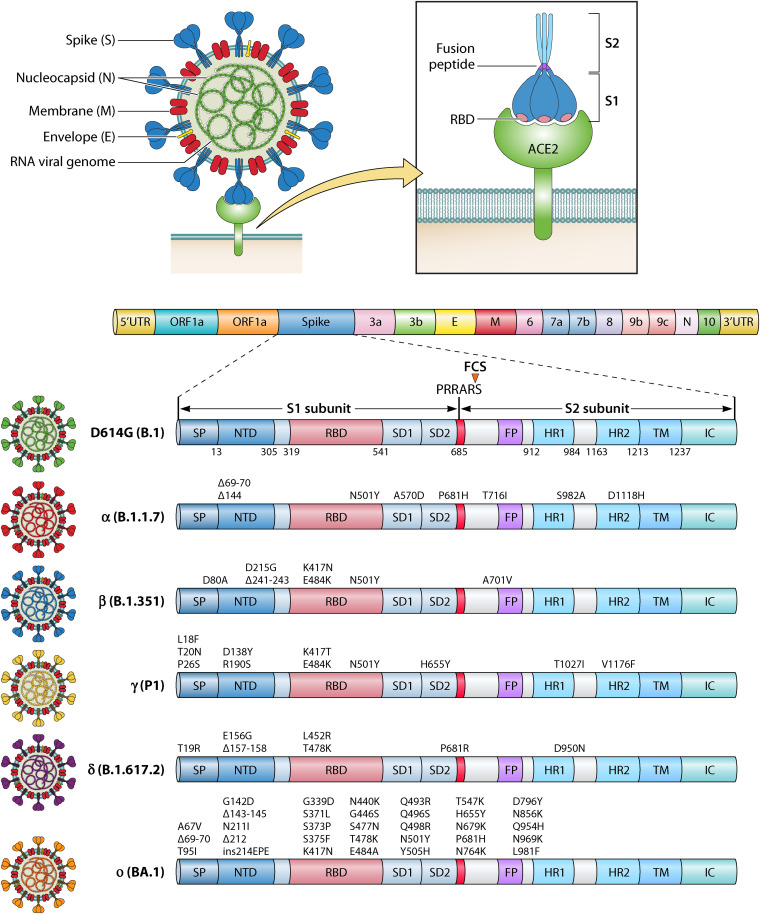
Surprisingly, the SARS-CoV-2 Omicron variant contains over 30 residual mutations in the S protein with 15 mutations located in the RBD (Fig. 1), which raises deep concerns regarding the efficacy of currently applied vaccines and therapeutic MAbs. VOC can be generated by mutations in the S gene, which improve “viral fitness” through several selected advantages such as increased affinity to ACE2 and accelerated viral replication rate, thereby increasing the risk of reinfection and severity of COVID-19 (27). Initial vaccines are designed and developed based on the ancestral SARS-CoV-2 S sequence, but mutations in the S RBD of VOCs are associated with humoral immune evasion. For example, one mutation, the N501Y mutation in the RBD, is universally common to all variants but is not present in the Delta variant (Fig. 1). This mutation increases the affinity of S for the ACE2 receptor, thus further enhancing viral attachment on the membrane and facilitating its subsequent entry steps into host cells (28). The ways additional mutations in the Omicron variant impact infection potential, the effectiveness of vaccines, and therapeutic MAbs are under investigation.
FIG 1.
Structural depiction of SARS-CoV-2 virion and genomic domains with mutations in the spike. Mutations are shown in spikes in VOCs from the Alpha to the Omicron (BA.1) variants. Viral genomic RNA, membrane (M), spike glycoprotein (S), nucleocapsid (N), envelope (E) are shown on the left. S binds to ACE2 and primes viral fusion. Schematic genomic structures are shown on top right. Domains in WT S and mutations in all current variants of concern are shown. The furin cleavage Site (FCS) PRRAS between S1 and S2 subunit is indicated in red. SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane region; IC, intracellular domain.
In this review, we summarize and demonstrate the most recent advancements and findings associated with the entry mechanisms, cellular infectivity, and transmissibility of the newly emerged SARS-CoV-2 Omicron variant. This variant confers immune escape by employing its evasion strategies against protective antibodies induced by vaccination or natural infection and overpowers antibody-based therapies. The significance of a vaccination booster to counter the outbreak is highlighted. Current advances in finding effective MAbs and powerful antiviral drugs that are suggested to be useful against Omicron and treating COVID-19 have been emphasized. Novel insights are presented on the entry mechanisms of Omicron, which is fundamentally distinct from that of the ancestral variant. Other highlights include cell tropism and entry preferences of the Omicron variant, infectivity and cell-to-cell fusion mechanisms, soluble receptor therapy, fusion, and entry inhibitor development, as well as the immune sensing and antagonism of SARS-CoV-2 against innate immunity.
INFECTIVITY, FUSION, AND CELLULAR TROPISM OF THE SARS-CoV-2 OMICRON VARIANT
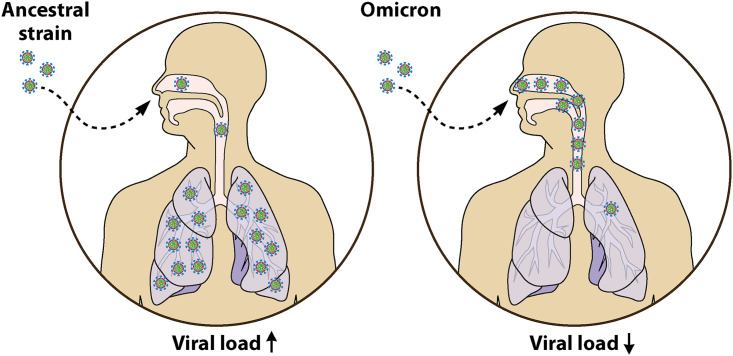
Although COVID-19 cases by the SARS-CoV-2 Omicron variant infection have skyrocketed, hospitalizations have increased only modestly. A recent study from Denmark demonstrated that increased fitness of Omicron appears to be attributed to the viral immune evasion rather increased intrinsic transmissibility (29). Strikingly, in several pilot studies in rodents, Omicron infections were often largely limited to the upper respiratory airway, including the nose, throat, and windpipe, but not the lungs, thereby causing less severe disease (Fig. 2). Distribution of the virus in this manner would allow viral particles to be exhaled easily from the nose or mouth as aerosols and allow for infection of new hosts. The Omicron variant was shown to be less fusogenic than the Delta variant and the D614G strain (30). In a recent study conducted by Abdelnabi et al. (31), a nearly 1,000-fold smaller amount of viral RNA was measured, with no detectable infectious virus in the lungs from hamsters after the Omicron infection compared to control animals infected with D614G. Furthermore, no signs of peribronchial inflammation and bronchopneumonia were found in the lungs of the Omicron virus-infected hamsters (31). Although the S protein of the Delta variant is effectively cleaved into two subunits, allowing for cell-cell fusion, the S protein of the Omicron variant is less efficiently processed (30). Formation of syncytium by the Omicron S was significantly reduced compared to the Delta S, owing to inadequate S1/S2 cleavage by furin and the inaccessibility of TMPRSS2 (32). In addition, the Omicron variant is not ravaging the lower respiratory systems like wild-type SARS-CoV-2. In fact, it was less successful at infecting lung cells and organoids than other variants (32). Similarly, in hamsters infected with the Omicron virus, fewer copies of the SARS-CoV-2 viral RNA were detected in the lungs compared to animals with the ancestral variant infection, and strikingly no infectious Omicron virus can be detected in the lungs (33). Consistently, attenuation of infection in different mouse strains such as 129, C57BL/6, and BALB/c infected with the Omicron variant also showed a reduced viral burden in both upper and lower respiratory tracts compared to previous SARS-CoV-2 variants (34, 35). In contrast to infection with previous SARS-CoV-2 Alpha, Beta, and Delta variants, infection with the Omicron variant results in the least amount of body weight loss and much lower mortality (36). Consistently, the Omicron variant replicated at a higher rate than prior SARS-CoV-2 strains in the bronchus but with less efficiency in the lung parenchyma (37). Although the determinants of severity are multifactorial, the decreased replication capabilities of the Omicron variant in human lungs may explain the decreased severity in Omicron variant-infected patients. The experiments from several Omicron isolates demonstrate attenuated lung pathogenesis in rodents, which are similar to preliminary clinical data in humans (38). Furthermore, the Omicron variant shows a lower replication rate in TMPRSS2-overexpressing VeroE6 cells and replicates poorly in Calu-3 cells (39). The SARS-CoV-2 Omicron variant has evolved into an upper-airway specialist, thriving in the throat and nose, but not in the low airway system, where the lung parenchyma is the major target of the prior SARS-CoV-2 variants (Fig. 2).
FIG 2.
Lower replication competence of the Omicron variant in human lungs. The SARS-CoV-2 Omicron variant has evolved into an upper-airway specialist, thriving in the throat and nose but not in the lower airway system, where the lung is the main target of the other prior SARS-CoV-2 variants.
RESISTANCE OF THE OMICRON VARIANT TO CONVALESCENT PLASMA
Antibodies can mediate protection against SARS-CoV-2 via viral neutralization. Possible sources of antibodies against SARS-CoV-2 are from convalescent-phase sera of humans who have previously experienced COVID-19 or certain genetically engineered animal hosts that can generate human antibodies (40). Early studies on the capacity of the antibody neutralization against the ancestral SARS-CoV-2 virus showed a positive correlation between serum neutralizing capacity and disease severity (41). Furthermore, the neutralizing potency of convalescent-phase sera against previous variants is significantly reduced compared to ancestral strains (42, 43).
Similarly, the neutralization of the Omicron variant from the serum samples of convalescents patients in Wuhan city 1 year after infection also revealed a marked (10-fold) reduction in titers compared to D614G (44) (Table 1). The average neutralization titer of convalescent-phase sera against the Omicron variant declined from 556 to 66 (8.4-fold) compared to that of D614G (45) (Table 1). Sera from convalescent individuals in Hong Kong showed a 10.6-fold decrease in neutralization titer against the Omicron variant compared to ancestral wild type (46). A sharp drop of >32-fold in neutralization titers was also observed against the Omicron variant, with only 2 of the 10 samples having barely detectable titers (47). Convalescent sera from hospitalized patients displayed a potent ability to neutralize the ancestral strain but a 37-fold reduction in neutralizing activity against the Omicron variant (48). Sera collected either 6 months or 1 year postsymptoms from COVID-19 convalescent patients showed low or even no neutralizing activity against the Omicron variant, indicating a high risk of Omicron variant reinfection in recovered patients (49). In addition, another study demonstrated that the neutralizing activity of the majority (73.3%) of convalescent-phase sera was undetectable when measured against the Omicron virus (50). Indeed, the Omicron variants do reinfect convalescent individuals during a SARS-CoV-2 Omicron infection wave.
TABLE 1.
Neutralization activities of convalescent-phase sera against the Omicron varianta
| No. of samples | Age range (yr) | Time after symptom onset | Neutralization titer |
Fold reduction (WT/Omicron) | Reference | |
|---|---|---|---|---|---|---|
| WT | Omicron | |||||
| 24 | NA | 1 yr | NA | NA | 10.1 | 44 |
| 28 | NA | 1–3 mo | ED50 = 556 | ED50 = 66 | 8.4 | 45 |
| 30 | 20–72 | 143–196 days | GMT = 85.7 | GMT = 8.1 | 10.6 | 46 |
| 10 | 45–79 | 9–120 days | IC50 = 4344 | IC50 < 135 | >32 | 47 |
| 17 | 25–78 | 14–28 days | NA | 23% samples above LOD | NA | 48 |
| 12 | 34–73 | 14–28 days (hospitalized) | NA | 75% samples above LOD | 37 | 48 |
| 16 | 32–77 | 6 mo | ED50 = 569 | 36% samples above LOD | NA | 49 |
| 22 | 23–82 | 12 mo | ED50 = 580 | 39% samples above LOD | NA | 49 |
| 15 | NA | 23–87 days | NA | 27% samples above LOD | >11.1 | 50 |
LOD, limit of detection; NA, not applicable.
RESISTANCE OF THE OMICRON VARIANT TO NEUTRALIZING ANTIBODIES ELICITED BY COVID-19 VACCINES
Pfizer-BioNTech (BNT162b2) and Moderna (mRNA-1273) mRNA Vaccines
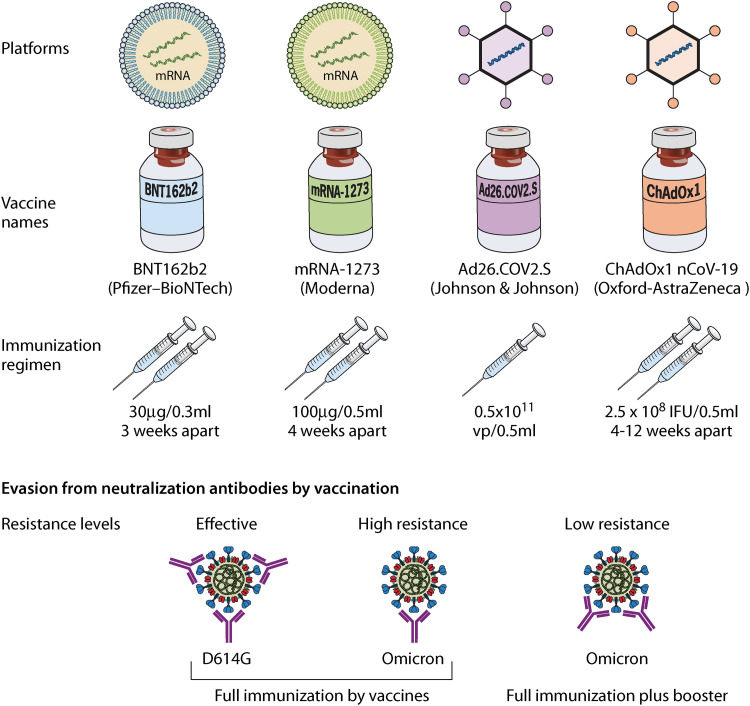
Correspondingly, the Omicron variant also bypasses the protection induced by vaccination. The Pfizer-BioNTech (BNT162b2) and the Moderna (mRNA-1273) vaccines are both potent mRNA vaccines. They have been shown to provide an extremely high level of protection against severe disease and death in relation to SARS-CoV-2. However, the Omicron variant shows obvious decreased sensitivity to neutralizing antibodies induced by these vaccines (46–51) (Fig. 3 and Table 2). The Pfizer-BioNTech vaccine’s effectiveness against hospitalization caused by the Omicron variant was estimated up to 70% (95% confidence interval [CI] = 62 to 76) in South Africa, whereas vaccine effectiveness was 93% (95% CI = 90 to 94) before the emergence of the Omicron variant (52). The titer of neutralizing antibodies induced by two doses of mRNA vaccines against the Omicron variant is significantly lower than that against the ancestral variant (Table 2).
FIG 3.
Resistance of the Omicron variant against to neutralizing antibodies elicited by COVID-19 vaccines. A representative vaccine based on mRNA and the viral vector platform and regimen of injection are shown at the top. The Omicron variant shows significant resistance against neutralization by vaccine-induced antibodies. A third booster increases the neutralization titer against the Omicron variant.
TABLE 2.
Resistance of Omicron variant to neutralizing antibodies induced by different platform-based vaccinesa
| Vaccine | Source | Platform | Resistance increase (WT/Omicron) | Reference |
|---|---|---|---|---|
| BNT162b2 | Pfizer-BioNTech | mRNA | 31.3-fold (2 doses) | 46 |
| 4.1-fold (3 doses) | 46 | |||
| >21-fold (2 doses) | 47 | |||
| >8.6-fold (2 doses) | 47 | |||
| 37-fold (2 doses) | 48 | |||
| Below the LOD (2 doses) | 49 | |||
| 17.6-fold (3 doses) | 49 | |||
| >23.3-fold (2 doses) | 50 | |||
| 7.5-fold (3 doses) | 50 | |||
| 22-fold (2 doses) | 51 | |||
| 33.8-fold (2 doses) | 53 | |||
| 8.1-fold (3 doses) | 53 | |||
| 33-fold (2 doses) | 54 | |||
| 36- to 40-fold (2 doses) | 55 | |||
| mRNA-1273 | Moderna | mRNA | 39-fold (2 doses) | 48 |
| 42.6-fold (2 doses) | 50 | |||
| 16.7-fold (3 doses) | 50 | |||
| 74-fold (2 doses) | 54 | |||
| Ad26.COV2.S | Johnson & Johnson | Adenovirus | Below the LOD (2 doses) | 47 |
| Below the LOD (92%) | 48 | |||
| 13.5-fold (1 dose+BNT162b2 booster) | 53 | |||
| ChAdOx1 nCoV-19 | Oxford-AstraZeneca | Adenovirus | Below the LOD (2 doses) | 47 |
| 21-fold (2 doses) | 48 | |||
| Below the LOD (2 doses) | 49 | |||
| 14-fold (2 doses) | 54 | |||
| 25.7-fold (2 doses) | 62 | |||
| Coronavac | Sinovac | Inactivated virus | 7.3-fold (3 doses) | 46 |
| 5.2-fold (2 doses+BNT162b2 booster) | 46 | |||
| Almost no titers against Omicron | 55 | |||
| 16.5-fold (3 doses) | 59 | |||
| Sputnik V | Gamaleya Research Institute | Adenovirus | Below the LOD (91%) | 48 |
| 11.76-fold (2 doses) | 60 | |||
| BBIBP-CorV | Sinoparm | Inactivated virus | Below the LOD (43%) | 48 |
| 20-fold (3 doses) | 61 | |||
| BBV152 | Bharat Biotech | Inactivated virus | 26.6-fold (2 doses) | 62 |
LOD, limit of detection.
Hoffmann et al. (53) have shown that the Omicron S protein evaded antibody neutralization in sera from individuals vaccinated with BNT162b2 with a value of 33.8-fold higher resistance than the ancestral S protein. Simultaneously, Willett and coauthors showed that vaccination with the two doses of BNT162b2 elicited neutralizing antibody with mean titer of 148 against the Omicron variant, about 33-fold lower than that against the ancestral strain (NT50: 4,978) (54). The Kelvin To’s group has shown that the geometric mean titer (GMT) against the Omicron variant in BNT162b2 recipients is 36- to 40-fold lower than that against the ancestral virus (55). Similarly, vaccinees with two doses of BNT162B2 had neutralizing antibody titer against the Omicron variant about 37-fold lower than that against the ancestral strain (48). BNT162b2 and the mRNA-1273 showed significantly decreased the half-maximal inhibitory concentration (IC50) by more than 21- and 8.6-fold, respectively (47). Vaccination with the two-dose mRNA1273 elicited neutralizing antibody with a mean titer of 285 against the Omicron variant, a 74-fold decrease, compared to the ancestral strain, with titer of 21,118 (54). Two doses of the mRNA vaccine resulted in limited neutralizing antibodies against Omicron but an increased antibody breadth over time, lasting up to 4.9 months after vaccination (56). More than a third of RBD-binding IgG+ resting memory B cells, on the other hand, bound the Omicron variant at first and gradually increased the BCR breadth over time (56).
AstraZeneca Vaccine (ChAdOx1) and Johnson & Johnson Vaccine (Ad26.COV2.S)
A similar pattern was also observed for the AstraZeneca-Oxford (ChAdOx1) vaccine. Samples collected from AstraZeneca vaccine recipients 5 months after full vaccination showed that neutralizing antibodies just barely inhibited the Omicron variant (49), with a 14- to 21-fold decrease in neutralization titer, compared to that of the ancestral variant (48, 54) (Table 2). On the other hand, an increase in neutralization titer against the Omicron variant was noticed upon three vaccinations, or heterologous vaccinations combining the AstraZeneca-Oxford and the Pfizer-BioNTech vaccines (53). Compared to the two doses vaccine regimens, the Johnson & Johnson vaccine (Ad26.COV2.S) is provided with one dose as the full vaccination program (Fig. 3). Plasma specimens from subjects with the single-dose of the Johnson & Johnson vaccine completely lacked detectable neutralization activity against the Omicron virus (48, 57), except for the samples from donors with a history of prior SARS-CoV-2 exposure (47). Although these findings demonstrate that the Omicron variant displays an unprecedented extent of immune escape from neutralizing antibody (Fig. 3), boosting and enhancing the breath and maturation of antibodies in convalescent or vaccinated people will provide another layer of protection against the Omicron variant.
Vaccines from Other Platforms
The CoronaVac recipients had almost no titers of neutralizing antibody against either the BA.1 or the BA.1.1 Omicron sublineage (55). CoronaVac immunized individuals maintain nonneutralizing antibodies that assist certain immune cells to engulf virus-infected cells (58). The average titer of the Omicron variant was 16.5-fold lower compared to the ancestral virus in patients (n = 254) who received three doses of CoronaVac vaccination (59). Two doses of Sputnik V showed a neutralization titer lower than detection limit (48). The neutralization antibodies from Sputnik V-vaccinated individuals decreased 11.76-fold against the Omicron variant after 6 to 12 months compared to the ancestral strain (60). The Omicron variant manifested a 20-fold lower neutralizing sensitivity than prior SARS-CoV-2 variants to the sera elicited by BBIBP-CorV (Sinopharm) after a booster (61). Vaccination with inactivated whole-virus vaccine (BBV152) induces a 27-fold lower geometric mean neutralization titer against the Omicron variant, whereas complete BBV152 vaccination plus prior SARSCoV-2 infection induced a 57-fold lower geometric mean neutralization titer (62).
BOOSTER VACCINE ENHANCES PROTECTIVE ANTIBODY TITERS AGAINST THE OMICRON VARIANT
Decreased neutralization against the Omicron variant was observed in most vaccinees receiving two-dose vaccines (see previous section). However, individuals boosted with an extra shot of the vaccine or in combination with the standard vaccination protocol and previously exposed to SARS-CoV-2 exhibited a remarkably enhanced level of neutralization (54, 57, 63). The emergence of the Omicron variant signifies the importance of booster vaccination to be incorporated into the ongoing vaccine campaigns worldwide since elevated levels of neutralizing antibodies provide better protection against VOCs and provide prevention from developing severe COVID-19 diseases (64, 65). The neutralization activity of sera from participants who have been exposed to the S protein more than three times is largely maintained, albeit at a reduced level (50). Indeed, a third dose of the Pfizer-BioNTech vaccine induced antibodies can efficiently neutralize the Omicron variant (GMT: 1.11 after the second shot versus 107.6 after the booster) (66). Neutralizing titers against the Omicron variant were substantial in all persons who previously had COVID-19 disease and then were fully vaccinated, as well as those who had received three regimens of an mRNA vaccine (57). In subjects previously vaccinated with two doses of either AstraZeneca-Oxford (ChAdOx1) or BNT162b2 vaccine, followed by a booster of either BNT162b2 or mRNA-1273, the absolute number of individuals displaying detectable neutralizing activity against the Omicron was higher in the ChAdOx1-primed group (13/21, 62%) compared to the BNT162b2-primed group (5/20, 25%) (54). Furthermore, a third dose of mRNA vaccine was reported to elicit potent variant cross-neutralization (67). A large reduction in Omicron neutralization titers is ameliorated by a third booster vaccine (63). In addition, a homologous booster by an inactivated vaccine or an enhanced heterologous boosting with a protein subunit vaccine can significantly reduce Omicron immune escape from neutralization (68). Serum samples from 101 CoronaVac-vaccinated donors showed no detectable neutralization against the Omicron variant before the booster vaccination, while 80% of analyzed samples showed some neutralization activity against Omicron after a booster with an mRNA vaccine (69). Receiving three doses of mRNA vaccine, as opposed to being unvaccinated or receiving two doses, was associated with the protection level against both Delta and Omicron viruses, although the protection is less for the Omicron variant than for the Delta variant (70). Omicron-neutralizing titers were 23-fold higher 1 month after the third vaccine dose compared to after the first two doses, with comparable neutralizing titers of the ancestral strain after two doses (71). After booster vaccination, all samples demonstrated detectable neutralization capacities against the Omicron virus, albeit with a marked drop (5.86- to 14.98-fold) in neutralization tiers against the Omicron variant compared to the prototype at 2 weeks after the heterologous or homologous boosters (72). The median antibody neutralization titer increased substantially from a level lower than 20 to 1,066 for the BA.1 variant and to 776 for the BA.2 variant 14 days after the BNT162b2 booster (10). In serum samples from individuals boosted with an mRNA vaccine, the neutralization profiles of the BA.2 variant were similar to those of the BA.1 variant (7, 12), indicating that the booster rescues neutralization ability against both Omicron sublineages, providing additional protection against COVID-19.
The Omicron variant remarkably evaded neutralization by antibodies from convalescent donors or those who received only two shots of Pfizer vaccine (BNT162b2), while neutralization by antibodies in individuals receiving three doses of BNT162b2 was more potent and efficient, supporting the concept that the neutralization capacity induced by vaccination plus boosting could achieve reasonable efficacy against the Omicron variant (51). However, this enhanced level of neutralization following a booster vaccination still shows a lower effect against the Omicron variant compared to the ancestral SARS-CoV-2 strain, both in convalescent patients and in individuals who had received three homologous mRNA vaccinations (47, 48). After mRNA-1273 full immunization or Ad26.COV.2 priming, BNT162b2 booster vaccination partially restored neutralization ability for the Omicron variant, but the responses were rising to 17-fold lower than the ancestral strain (73). Therefore, even with a third shot as a booster, it is conceivable that it may not provide adequate protection against Omicron infection (74). In summary, vaccines have weakened effect against the Omicron variant, but the booster shots may still enhance the neutralization level, providing protection against hospitalization and death, thereby making the booster option a desirable choice. More in-depth research and explorative investigations are necessary to assess the practical impacts of the Omicron variant on the efficacy of all available COVID-19 vaccines and to define both the kinetics and the durability of antibodies providing protection in recipients of booster vaccinations in light of the serious public health concerns posed by SARS-CoV-2 variants.
“Hybrid immunity” acquired from the combination of vaccinations and history of viral infection provides high level of protection against COVID-19-related disease and death. This enhanced hybrid immunity boosts the breadth of the humoral responses compared to only two doses of vaccine. The order of exposure to the viral antigen, meaning whether individuals are vaccinated after infection or experience breakthrough infection after vaccination, would reach a similar level of broadly neutralizing antibodies against multiple SARS-CoV-2 variants (75). A third dose of vaccine only provides a part of the viral antigen, whereas an authentic viral infection would induce broader cellular and humoral responses. However, it is not encouraged to infect individuals with viruses on purpose to achieve a robust immunity to infection since vaccination is a safer option than infection.
DEVELOPMENT OF THE NEXT-GENERATION COVID-19 VACCINES
Most recently, Gagne et al. have developed an mRNA-1273-like vaccine by using mRNA encoding the Omicron S protein to replace that encoding the S protein of the wild-type SARS-CoV-2 WA1 strain in the mRNA-1273 vaccine (76). These researchers used either mRNA-1273 or the new mRNA-Omicron vaccine to boost mRNA-1273-vaccinated macaques. At 2 weeks after the boost, they found that titers of the neutralizing antibodies against D614G, as the wild-type SARS-CoV-2 control, and Omicron increased to 5,360 and 2,980, respectively, for the mRNA-1273 vaccine and to 2,670 and 1,930, respectively, for the mRNA-Omicron vaccine. However, after either boost, a comparable level of virus replication was observed in the lower respiratory tract, suggesting that boosting with the Omicron S protein-based vaccine may not provide better immunity or protection against Omicron infection than the current mRNA-1273 vaccine (76). Therefore, the best approach is the development of pan-sarbecovirus or pan-β-CoV vaccines (77). To address this need, Jiang and colleagues have developed a pan-sarbecovirus vaccine containing the STING agonist CF501 as an adjuvant and a recombinant subunit vaccine consisting of RBD of SARS-CoV-2 conjugated with a human IgG Fc fragment (CF501/RBD-Fc). These researchers found that this vaccine could elicit extremely potent neutralizing antibodies and T cell responses in mice, rabbits, and nonhuman primates (NHPs). Neutralizing antibodies can effectively neutralize infection by ancestral SARS-CoV-2, and 4 VOCs (Alpha, Beta, Gamma, and Delta), 5 previously identified variants of interests (VOI) (Epsilon, Zeta, Eta, Iota, and Kappa), and 41 mutants with individual point mutations in the S protein, as well as SARS-CoV- and SARS-related coronaviruses (SARSr-CoVs) from bats (78). More interestingly, these researchers found that the 50% neutralization titer (NT50) for neutralizing antibodies against pseudotyped Omicron variant infection in the sera of NHPs immunized with two doses of CF501/RBD-Fc vaccine reached to 6,469 by day 28 after the first immunization and maintained a high level from day 28 through day 113. Surprisingly, the neutralizing antibody titer increased dramatically after the third immunization, reaching up to 35,066 against pseudotyped Omicron variant. Consistently, the sera from NHPs immunized with CF501/RBD-Fc vaccine could neutralize authentic SARS-CoV-2 Omicron with an NT50 of 9,322 at day 122 after the first immunization (79). These findings suggest that CF501/RBD-Fc vaccine is a promising candidate as a pan-sarbecovirus vaccine to prevent infection by the current SARS-CoV-2 variants, including Omicron, and any possible future emerging VOCs (79).
RESISTANCE OF THE OMICRON VARIANT TO THERAPEUTIC MONOCLONAL ANTIBODIES
Monoclonal antibodies (MAbs) with high specificity and reliability have been utilized as powerful tools to treat COVID-19 patients. As an RNA virus with a very high mutation rate, SARS-CoV-2 evolves and mutates under strong evolutionary pressure of widely deployed prophylactic vaccines and therapeutic MAbs, and the use of antibody cocktails is an important alternative strategy to achieve effective COVID-19 management (80). However, several parallel studies show that the Omicron variant inevitably overpowers antibody treatments and antibody cocktails. For example, Regeneron MAbs Casirivimab (REGN10933) and Imdevimab (REGN10987) are absolutely knocked out by the Omicron variant (47), and their activity of neutralizing the Omicron variant is lost (49, 81), even at the highest concentration (82). At doses 2-fold greater than those demonstrated to be active against prior variants of the virus, Ronapreve reduced subgenomic RNA in lung and nasal turbinates of in K18-hACE2 mice only for the Delta variant but not the Omicron variant (83). These findings contribute to the growing evidence suggesting effectiveness of Ronapreve is impaired in the Omicron variant (83). Similarly, the Omicron variant is completely resistant to the Eli Lilly MAbs Bamlanivimab (LY-CoV555) and Etesevimab (LY-CoV016) (47, 49). In addition, AstraZeneca MAbs Cilgavimab (CoV2-2130) and Tixagevimab (CoV2-2196) lost their neutralizing potency against the Omicron variant (47, 82, 84), with a 43-fold reduction (84).
The GSK/Vir Biotechnology MAb Sotrovimab is ~3-fold less effective against the Omicron variant, compared to the ancestral SARS-CoV-2 virus (48, 82). Syrian Golden hamsters infected with SARS-CoV-2 and treated with Sotrovimab show a smaller extent of weight loss and decreased levels of infectious viral load in the lungs compared to those treated with a control MAb (85). S309 (a parent version of Sotrovimab) maintains similar IC50s against the D614G (202 ng/mL) and the Omicron (373 ng/mL) in TMPRSS2 overexpressed Vero cells (86). Sotrovimab retained some of its neutralizing activity against the Omicron variant (53, 84), possibly owing to its origin from a SARS-CoV-infected survivor. Sotrovimab appears to neutralize the SARS-CoV-2 virus by targeting a deeper and highly conserved epitope in RBD, but not an epitope in RBM (87). While the neutralizing potency of the other described MAbs was significantly impaired or completely lost, Sotrovimab still functions, albeit with decreased potency (88). LY-CoV1404 (Bebtelovimab) showed potent neutralizing activity, with an IC50 value of 5.1 ng/mL, against the Omicron variant (81). In general, the neutralization effects of the therapeutic antibodies are either abolished or impaired by the Omicron variant, although a few retain their effects.
MECHANISM BY WHICH THE OMICRON VARIANT RESISTS NEUTRALIZING ANTIBODIES
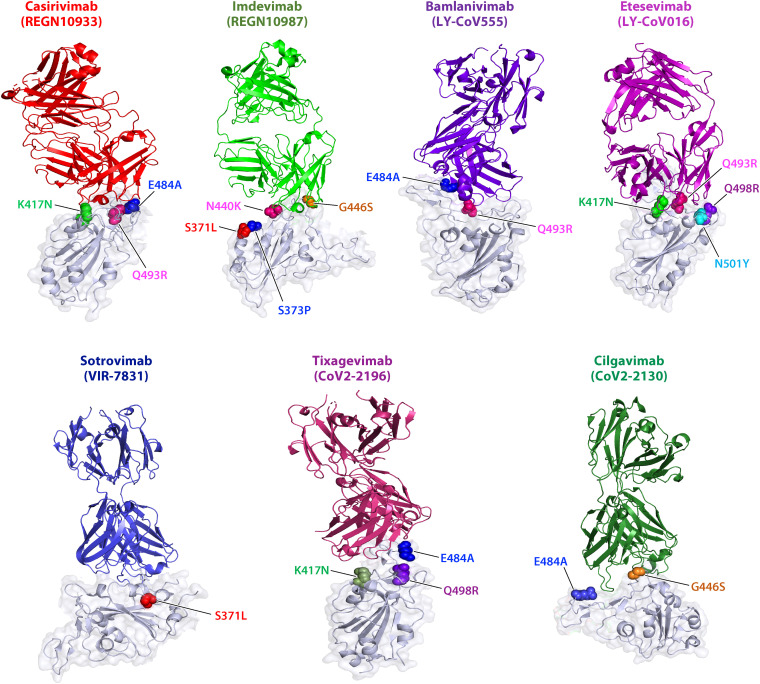
Specifically, several key mutations in the S protein that confer resistance to each antibody were mapped by structural analysis and experimental neutralization assays (Table 3 and Fig. 4). For instance, K417N, E484A, and Q493R are the key mutations that render resistance to REGN10933, whereas S371L, S373P, N440K, and G446S play essential roles in rendering resistance to REGN10987 (89). Therefore, both REGN antibodies lost their activities to neutralizing the Omicron variant (49). The Omicron variant is completely resistant to LY-CoV55, and this resistance is attributed to E484A and Q493R. RBD mutations in the Omicron variant were demonstrated to show reduced binding or decreased neutralization with antibodies based on the structural analysis (90). K417N, E484A, and Q493R were found for REGN10933, G446S for REGN10987, E484A, and Q493R for LY-CoV555, and K417N for LY-CoV016 (90). By causing an alteration in the spatial configuration of other residues, Q493R can lead to the elimination of hydrogen bonds or the collision of the antibody CDRH3 region, which could contribute to the inability of neutralization by LY-CoV016 and LY-CoV555. Consistently, the IC50 of each antibody against the Omicron was shown to be reduced, and key residues involved in neutralization evasion for representative antibodies were further mapped (8, 91). K417N, E484A, and Q493R were found for REGN10933, N440K and G446S for REGN10987, E484A and Q493R for LY-CoV555, and K417N, Q493R, and N501Y for LY-CoV016 (8, 91). Most mutations lie in the interface between antibodies and S protein to block the virus binding to the antibodies (Fig. 4). K417N, E484A, and Q493R would cause the loss of hydrogen binding with the heavy chain of antibodies. Furthermore, four residue mutations (S371L, S373P, S375F, and E484A) with completely opposite changes from hydrophilic to hydrophobic residues result in massive hydrophobic interactions (91). Notably, S477N, Q493R, G496S, Q498R, and Y505H that occurred in the Omicron variant, together with three already existing mutations—T478K, E484K, and N501Y—found in other VOCs, further modulate the antigenic properties and conformational features, leading to a noticeable immune evasion of the antibody (91). Substitutions such as S371L, S373P, and S375F also cause a distinct conformational change in another generally conserved antigenic site of sarbecoviruses (91–93), stabilizing two adjacent S RBDs with a one-up and one-down conformation (91). Substitution of G446S confers high resistance to antibodies that bind to the RBD right shoulder by changing the local conformation at the binding boundary (59). However, LY-CoV1404 maintains its neutralizing activity against the Omicron variant by minimizing interaction of side chain by changing mobility of the loop. The four alterations N440K, G446S, Q498R, and N501Y in the docking sites of LY-CoV1404 were kept at the end of its epitope, thus allowing conformational accessibility for the binding of LY-CoV1404 to RDB surfaces (81). LY-CoV1404 binds to a highly conserved epitope in the S proteins of different variants with high affinity in both “up” and “down” conformations, rendering its neutralization potency unaffected (94). For S309, two mutations, G339D and N440K, within or near the S309 recognition sites do not interfere with MAb binding and epitope (90). Even the abolished neutralization of S309 was acquired from the S371L point alteration; indeed, S309 retains its neutralization efficacy against the Omicron variant, indicating that other mutations would overcome the defects caused by S371L (81).
TABLE 3.
Omicron RBD mutations with reduced neutralization by therapeutic antibodiesa
| RBD mutation | Therapeutic Ab (alternate name) |
||||||
|---|---|---|---|---|---|---|---|
| Casirivimab (REGN10933) | Imadevimab (REGN10987) | Bamlanivimab (LY-CoV555) | Etesevimab (LY-CoV016) | Sotrovimab (VIR-7831) | Tixagevimab (CoV2-2196) | Cilgavimab (CoV2-2130) | |
| G339D | |||||||
| S371L | |||||||
| S373P | |||||||
| S375F | |||||||
| K417N | |||||||
| N440K | |||||||
| G446S | |||||||
| S477N | |||||||
| T478K | |||||||
| E484A | |||||||
| Q493R | |||||||
| G496S | |||||||
| Q498R | |||||||
| N501Y | |||||||
| Y505H | |||||||
The results observed for Regeneron MAbs (Casirivimab and Imdevimab), Eli Lilly MAbs (Bamlanivimab and Etesevimab), Sotrovimab, and AstraZeneca MAbs (Cilgavimab and Tixagevimab) are shown. Key residues that confer resistance to the antibody are highlighted by dark gray shading.
FIG 4.
Resistance of the Omicron variant to MAbs and responsible residues that confer resistance. Regeneron Abs (Casirivimab and Imdevimab), Eli Lilly Abs (Bamlanivimab and Etesevimab), and AstraZeneca Abs (Tixagevimab/CoV2-2196 and Cilgavimab/CoV2-2130), as well as Sotrovimab (VIR-7831), with each antibody binding to S protein, are shown via three-dimensional modeling. The binding motifs that confer resistance are highlighted in color.
OTHER MONOCLONAL ANTIBODIES AND NANOBODIES AGAINST THE OMICRON VARIANT
A number of antivirals and immunotherapeutic options have been employed for emergency use to lessen the severity of clinical disease in patients with COVID-19. Various other clinical therapeutics are being developed and undergoing clinical trials. However, any optimal choice of drug and treatment regimens are yet to become available (95, 96). Regdanvimab from Celltrion lost its neutralization activities against the Omicron variant (49). AX290 and AX677, two MAbs with nonoverlapping epitopes, bind to the RBD of the viral S protein with subnanomolar or nanomolar affinities (97). These antibodies neutralize Omicron S protein with the same efficiency as wild-type S protein. In a mouse model, the prophylactic administration of AX290 and AX677, either alone or in combination, successfully reduced viral load and inflammation in the lungs, as well as preventing illness after SARS-CoV-2 infection (97). The virus-neutralizing characteristics of chimeric mouse-human forms of the antibodies were fully replicated, indicating that they could be a promising approach for COVID-19 therapy (97). Despite having a nearly 46-fold lower efficacy than wild-type SARS-CoV-2, hu33, a non-ACE2-blocking neutralizing antibody, can still neutralize Omicron at a median neutralization dose of 154.3 ng/mL (98). Bovine-derived antibodies and camel-derived nanobodies have also been suggested to be highly effective against SARS-CoV-2 and its VOCs (99). Nanobodies could thus be an effective method of neutralizing SARS-CoV-2 VOCs, even if further variants with novel mutations emerge (100). The synergistic effect of nanobodies with antiviral treatments could be an effective tool in the COVID-19 armamentarium (101). Using either SAB-185 antibodies generated in trans-chromosomic bovines or Nb11-59 nanobodies from camels or combined as a cocktail containing two entities could be a very effective therapeutic options for mitigating newly emerging variants and attempting to prevent COVID-19 mortality. The ACE2-mimicking MAb S2K146 retains unaltered potency against Omicron (48). The broadly functional sarbecovirus MAbs S2X259 and S2H97 can also neutralize the Omicron variant by binding to the antigenic spot that is outside the receptor-binding motif (48).
ANTIVIRALS AGAINST THE OMICRON VARIANT
Some proposed antivirals have been investigated to figure out antiviral activities against Omicron, such as Camostat, Ensovibep, Molnupiravir, and Remdesivir. Omicron variant infections are very sensitive to Molnupiravir and Nirmatrelvir in human Calu-3 cells and airway organoids (102). The recent development of potent oral antiviral drugs, including Molnupiravir and Paxlovid (Nirmatrelvir/Ritonavir), and promising clinical results obtained indicating their practical utility in the clinics would facilitate the treatment of COVID-19 patients (103–105). Molnupiravir is an oral antiviral drug that contains the active ingredient β-d-N4-hydroxycytidine (NHC) and was developed collaboratively by Merck (Kenilworth, NJ) and Ridgeback (Miami, FL) (103). It deceives SARS-CoV-2 to integrate the compound into its genetic information, which is encoded by RNA, thereby mutating the virus to the point where it can no longer replicate (106). Molnupiravir and Nirmatrelvir, each administered for 5 days, which inhibit viral polymerase and protease, have been shown to lessen progression of the disease by 30 and 89%, respectively (107). A recent study has revealed Remdesivir, Molnupiravir, and Nirmatrelvir to be active against Omicron and other VOCs (108). Catlin et al. recently conducted nonclinical studies in rats and rabbits with Nirmatrelvir (1,000 mg/kg/day), and the existing safety data of Ritonavir show that there are no relevant risks related to Paxlovid administration during pregnancy and in people at a reproductive age (109). Therefore, the recently developed oral antiviral drugs provide additional options to ameliorate serious illness, providing effective protection from COVID-19 and thus reducing hospitalization. Hence, these could be useful in modulating the current course of the pandemic to a milder one and safeguard health from infection with SARS-CoV-2 Omicron and other variants.
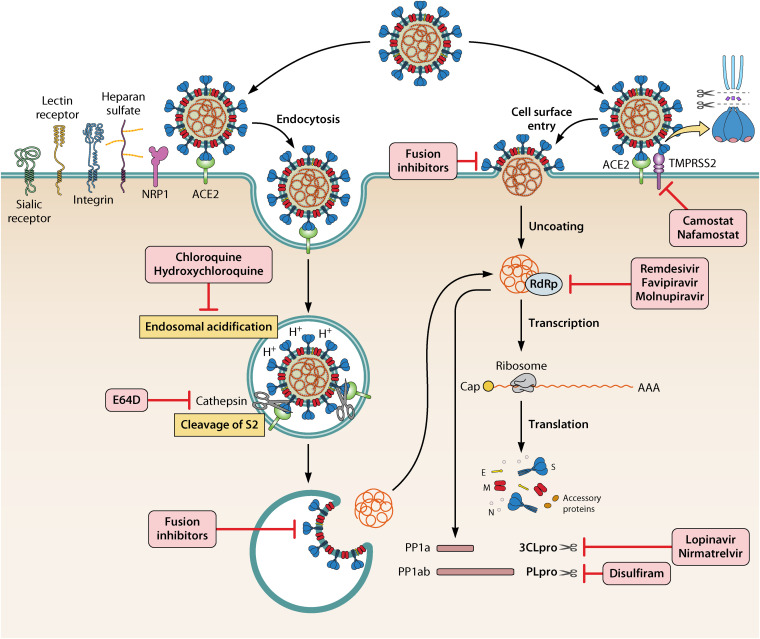
SENSITIVITIES TO ENTRY AND FUSION INHIBITORS
Sensitivity of the Omicron Variant to ACE2-Based SARS-CoV-2 Entry Inhibitors
Regardless of the number of mutated residues in the RBD of SARS-CoV-2 variants, including Omicron, in order to enter into and replicate inside the cells, the virus must still attach to the cell receptor through interaction between viral RBD in the viral S protein and ACE2 on the host cell in order to enter and replicate inside cells. Therefore, ACE2 can be utilized for the development of potential broad-spectrum SARS-CoV-2 entry inhibitors because the sequence of ACE2 generally does not change according to the mutations in viral RBD. Indeed, a series of SARS-CoV-2 entry inhibitors has been developed based on the soluble ACE2 protein and its mimics, such as ACE2.V2.4, IgG-ACE2, or nanobody. The S protein interacts with ACE2 on the cell membrane with high affinity, and membrane fusion occurs as a following step to enable viral entry. In contrast to the Delta variant, the Omicron variant displays a greater affinity for ACE2 (32). The binding affinity of the Omicron S protein (Omicron S-RBD) with human ACE2 was revealed to be 1.4-fold higher than the Delta variant and 2.7-fold higher than the Alpha variant by a molecular interaction energy assay (110). According to Kim et al. (111), the Omicron RBD displays enhanced binding affinity toward ACE2 via N501Y, Q493K/R, and T478K mutations, likely promoting greater viral binding and internalization. The results from single-cycle kinetic SPR analysis indicated that human ACE2 could bind the six mutated RBDs tested with enhanced affinity (48). Soluble recombinant human ACE2 APN01 effectively reduced the viral load in Vero E6 cells and blocked SARS-CoV-2 Omicron variant infection (112). Engineered ACE2 can neutralize a wide range of SARS-CoV-2 VOC, including previous Alpha, Beta, and Gamma variants. Even with the extensive additional mutations found in the Omicron S RBD, it still remains highly susceptible to blockade by three engineered high-affinity ACE2-Fc inhibitors: 3n39v4, 3j113v2, and 3j320v2 (113). In addition, the ACE2-mimicking MAb S2K146 retains unaltered potency against the Omicron variant (48). Integrin served as one of the coreceptors for SARS-CoV-2, and the pan-integrin inhibitor GLPG-0187 efficiently and significantly reduced both Omicron and Delta variant pseudovirus infections in HSAE cells (114). Inhibition of integrin signaling may help mitigate severity of COVID-19 through impaired TGF-β1 activation (114). Several other potential coreceptors of SARS-CoV-2 infection have been proposed as well, such as sialic receptors and Neuropilin 1 (NRP1). Whether these coreceptor-based inhibitors block infection requires further investigation.
Sensitivity of the Omicron Variant to Pan-CoV Fusion Inhibitors
Entry of a coronavirus into the host cell is initiated by binding of the RBD domain in S1 subunit of its S protein to its receptor, e.g., ACE2 for SARS-CoV and SARS-CoV-2. Both HR1 and HR2 domains in S2 subunit of the S protein then interact with each other to form a 6-helix bundle (6-HB), allowing the viral and cellular membranes to fuse, thereafter allowing viral RNA to enter the host cell for replication (Fig. 5) (115). Jiang and colleagues found that the peptide CP1 derived from the HR2 domain of SARS-CoV S could interact with the viral HR1 domain and block viral homologous 6-HB formation, thus inhibiting SARS-CoV S protein-mediated membrane fusion and infection (116). Using a similar approach, Jiang’s group also identified a potent short peptide derived from the MERS-CoV S protein with potent inhibitory ability against MERS-CoV infection (117). In 2019, Jiang’s team discovered the first pan-CoV fusion inhibitory peptide, termed EK1, which could interact with the HR1 domains of all five human coronaviruses (HCoVs) and effectively inhibit infection, including SARS-CoV, MERS-CoV, HCoV-229 E, HCoV-NL63, and HCoV-OC43, in addition to three SARSr-CoVs from bats (118, 119). By conjugating the EK1 peptide with polyethylene glycol (PEG) and cholesterol, Jiang and colleagues found that this PEGylated lipopeptide, termed EK1C4, exhibited improved stability and antiviral activity with >100-fold higher potency than that of its unmodified version (119). This can be attributed to the ability of EK1C4 to bind the viral membrane and enter the endosome together with viral virion, thus adding to its ability to inhibit endosomal membrane fusion (Fig. 5). Given that PEG linkers in a peptide drug may reduce its stability or induce anti-PEG antibodies in vivo, Jiang’s team also designed and synthesized a dePEGylated lipopeptide-based pan-CoV fusion inhibitor, termed EKL1C, and found that it could also potently inhibit infection of SARS-CoV-2 and its variants, other HCoVs, and some bat SARSr-CoVs. More importantly, EKL1C exhibited higher thermostability, higher resistance to proteolytic enzymes, and better druggability than EK1C4 (120).
FIG 5.
Fusion and entry mechanisms of SARS-CoV-2. SARS-CoV-2 enters the host cell via two different pathways: TMPRSS2-mediated cytoplasm membrane fusion and cathepsin-mediated endosomal membrane fusion. Peptide-based pan-CoV fusion inhibitors can inhibit both fusion pathways, while the inhibitors targeting TMPRSS2 and cathepsin L are expected to inhibit the cytoplasm and endosomal membrane fusion pathways, respectively. Chloroquine and hydroxychloroquine as agents targeting the endosomal acidification can inhibit SARS-CoV-2 S protein-mediated endosomal membrane fusion. Infection with the Omicron variant is not blocked by TMPRSS2 inhibitor (Camstat or Nafamostat) but is instead largely mediated via the endocytic pathway.
Considering that some novel mutations in Omicron S protein are located in its HR1 domain, including Q954H, N969K, and L981F, Jiang and colleagues evaluated the inhibitory activity of their pan-CoV fusion inhibitors, including EK1, EK1C4, and EKL1C peptides, against Omicron S-mediated membrane fusion. They found that all of these peptides could potently inhibit Omicron S-mediated cell to cell fusion between Caco2 cells and the Omicron S-EGFP overexpressed 293T cells and with IC50 values of 75, 0.9, and 6 nM, respectively. These researchers also effectively inhibited infection by pseudotyped and authentic Omicron infection (121), confirming that the Omicron variant is as sensitive to these potent and effective pan-CoV fusion inhibitors as both the original D614G and Delta strains and suggesting, in turn, that these peptides can be further developed as novel antiviral drugs for treating and preventing infection by SARS-CoV-2 and its variants, including Omicron, and other emerging and reemerging HCoVs in the future.
Sensitivity of the Omicron Variant to Enzyme Inhibitors Responsible for Viral Fusion
As mentioned above, SARS-CoV-2 enters the host cell to deliver its genome into the cytoplasm for replication through two different fusion pathways, cytoplasmic membrane fusion after cleavage of S protein by TMPRSS2 and endosomal membrane fusion mediated by the endosomal proteases cathepsin L or B (26) (Fig. 5). Both TMPRSS2 and cathepsin are host proteins, which should not mutate after encountering viral mutants, thus having broad-spectrum antiviral effects against SARS-CoV-2 and its variants. However, the host proteins generally have their own biological activity; therefore, an awareness of potential toxic effect of antiviral agents targeting host proteins is important. Low pH is the prerequirement for viral S protein-mediated endosomal membrane fusion, and agents targeting endosomal acidification, such as chloroquine and hydroxychloroquine, could inhibit SARS-CoV-2 S protein-mediated endosomal membrane fusion (122) (Fig. 5). RNA-dependent RNA polymerase can be targeted by Remdesivir, Favipiravir, and Molnupiravir, whereas the 3C-Like protease (3CL) and the Papain-Like Protease (PLPro) can be inhibited by Lopinavir, Nirmatrelvir, and Disulfiram (Fig. 5). Most recently, Zhao et al. have shown that the Omicron variant exhibits weaker cell-cell fusion activity and replicates more slowly than the Delta variant in TMPRSS2-overexpressing VeroE6 cells (39). Camostat, which inhibits the TMPRSS2 pathway, potently inhibited entry of the Delta variant, but not the Omicron variant, while both bafilomycin A1 and chloroquine, which inhibit the endocytic pathway, could inhibit both Omicron and Delta viruses, suggesting that infection with the Omicron variant is not largely enhanced by TMPRSS2, but instead is largely mediated via the endocytic pathway. Studies by Willett et al. also suggest that entry of the Omicron variant into host cells is mainly dependent on cathepsin L or B, but not TMPRSS2, because the Omicron variant shows high sensitivity to the drug E64d, an inhibitor of cathepsin L or B, but not to camostat, an inhibitor of TMPRSS2, as noted above. In contrast, the Delta variant is sensitive to camostat, but not E64d (54). Therefore, the inhibitors targeting cathepsin L or B, or endosomal acidification, are expected to be still effective against the Omicron variant.
T CELL RESPONSE TO THE SARS-CoV-2 OMICRON VARIANT
Neutralization antibodies from the humoral immune response serve as a defense against different viral variants; however, new emerging variants could often partially or completely escape the humoral responses when located inside host cells. Another important defense is the T cell response. Once stimulated and activated by the certain antigen, CD4+ T cells secrete cytokines as messengers, which stimulate the differentiation of B cells into mature plasma cells (antibody-producing cells) and CD8+ T cells (cytotoxic T cells), which are simultaneously activated by various cytokines, eventually bind to and kill infected cells. Naturally, booster vaccination or infection/reinfection induces a stronger T cell response since additional exposure to the antigen will induce a stronger and more specific T cell response. Moreover, natural infection can induce a broader T cell response against different viral proteins compared to vaccines, where only one viral protein is often presented. Caveats might differ, but overall, the T cell response is unlikely to let distinct SARS-CoV-2 variants escape based on the diversity of the T cell receptors. SARS-CoV-2 cross-reacts with a subgroup of T cells that have been primed against seasonal coronaviruses and may contribute to another layer of clinical protection, especially in the early stages of life. Memory T cells encompass a broad recognition of viral proteins, believed to be around 30 epitopes per person, and appear to be stable so far. Individual viral mutations may be limited by this breadth of identification, which is believed to underlie protection against serious disease caused by variants, including the Omicron variant. Current COVID-19 vaccinations elicit strong T cell responses, which are thought to provide extraordinary protection against hospitalization and death, and innovative or heterologous regimens have the potential to improve cellular responses (123).
Indeed, SARS-CoV-2 S protein-specific CD4+ and CD8+ T cells induced by prior natural infection or vaccination have been shown to recognize the Omicron variant, and the reactive T cells manifest functional and phenotypical similarities in response when we compare ancestral and Omicron variants (124). Furthermore, prior SARS-CoV-2 infection and vaccines induce robust human T cell responses and SARS-CoV-2 S protein-specific T cells are maintained in draining lymph nodes at least 6 months after vaccination (125), which is also observed for the Omicron variant (126). Indeed, one study even showed that only 28% of CD4+ and 14% of CD8+ epitopes are anticipated to be influenced by some mutations in the Omicron variant (127), which indeed suggests that the T cell response would largely remain intact and responsive also against the Omicron variant. Human CD8+ T cells can still recognize the epitopes of Omicron variant in both convalescent patients and vaccinated people even 6 months after exposure or vaccination (126). An additional booster induced a stronger T cell response against the Omicron variant (126). Several MHC-I and MHC-II epitopes were identified by ELISpot and cytolytic assays, and these epitopes are highly conserved across variants, including the Omicron variant, and compatible with presentation by most HLA alleles (128). Individuals after exposure to SARS-CoV-2 would induce anti-SARS-CoV-2 CD8+ T cell responses, which can recognize the Omicron variant (129). Keeton et al. investigated the ability of T cells to respond to the Omicron S protein in donors vaccinated with Ad26.CoV2.S, BNT162b2 vaccines, or unvaccinated convalescent patients (n = 70) and showed that 70 to 80% of the CD4+ and CD8+ T cell response to S protein was maintained (130). Naranbhai et al. also identified a subset of people with a >50% decrease in T cell reaction to the Omicron S protein (131). Measuring responsive CD4+ and CD8+ memory T cell responses revealed that diminished recognition to the Omicron S protein is mainly identified in the CD8+ T cell compartment (131). T cells specific for SARS-CoV-2 were found up to 6 months following all vaccination regimens, with specific CD4+ T cells being detected more consistently than CD8+ T cells (73). There were no significant variations in CD4+ or CD8+ T cell responses between original strain and variant-specific T cell responses, revealing low and minimal T cell immune escape (73). These findings reveal that those who have been vaccinated retain complete T cell immunity against the Omicron variant, perhaps compensating for the decreased effectiveness of neutralization antibodies in preventing infection or reducing severe COVID-19 (73). A total of >1,400 different SARS-CoV-2 CD4 and CD8 T cell epitopes were identified, highlighting a remarkable breadth of epitopes that are prevalently recognized in human populations (132). Therefore, owing to viral discrete immunodominant regions and those widely sensed T cell epitopes, it is unlikely that SARS-CoV-2 variants will completely escape T cell responses at large population levels (132).
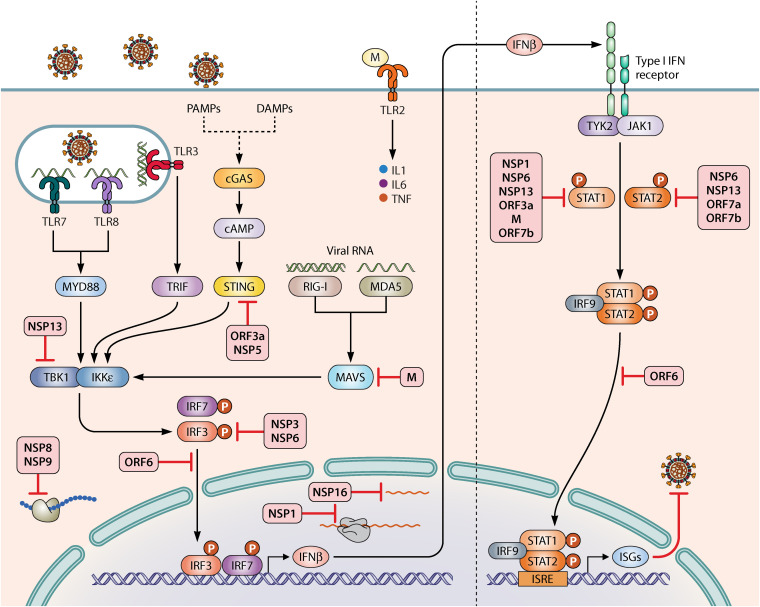
IMMUNE SENSING OF SARS-CoV-2 BY PATTERN RECOGNITION RECEPTORS
The innate immune system serves as the first line of defense that rapidly responds to pathogenic infection. Danger-associated molecular patterns (DAMPs) and/or pathogen-associated molecular patterns (PAMPs) can be recognized by a serious of host pattern recognition receptors (PRRs) (Fig. 6). These receptors are potent sensors to alarm host to respond to danger signals. For instance, retinoic acid-inducible gene (RIG-I)-like receptors, melanoma differentiation-associated gene 5 (MDA5), and cyclic GMP-AMP synthase (cGAS) localized in the cytosol and Toll-like receptors (TLR3/7/8) located in the endosomes can all sense DAMPs/PAMPs via distinct pathways. RIG-I sensors preferentially recognize short double-stranded RNAs with 5′ triphosphate moieties at their blunt ends (133–135). dsRNA recognition by MDA5 is independent of 5′ppp, and MDA5 preferentially recognizes longer dsRNA (136). In fact, MDA5 serves as a cytosolic RNA sensor to recognize SARS-CoV-2 infection in the lung cancer cell line Calu-3, which induces both type I and III IFN induction (137). Likewise, a deficiency in MDA5, RIG-I, or MAVS can enhance SARS-CoV-2 infection as SARS-CoV-2 activates a robust innate immune response and inflammatory mediators in lung epithelial cells by activation of both RIG-I and MDA5 (138). TLR3 recognizes both dsRNA and poly(I·C) and signals through TRIF to activate IFN responses. TLR7 and TLR8 are highly homologous and sense ssRNA to initiate downstream signal axis. The SARS-CoV-2 E protein sensed by TLR2 activates the production of proinflammatory cytokines (interleukin-1 [IL-1], IL-6, and tumor necrosis factor) and chemokines, and blocking TLR2 signals in vivo confers protection against viral pathogenesis (139). How TLRs play role in the process of SARS-CoV-2 sensing requires further investigations. cGAS is a double-stranded DNA (dsDNA) sensor and dsDNA activates catalytic activity of cGAS, which activates the production of 2′,3′-cyclic GMP-AMP (cGAMP), a second messenger molecule for activating the stimulator of IFN genes (STING). SARS-CoV-2 infection promotes cGAS-STING signal axis in macrophages and endothelial cells, which results in cell death and the production of type I IFNs (140). Pharmacological impairments of STING by highly potent STING agonists decreases inflammation in the lung caused by SARS-CoV2 and improves outcomes after infection in mice (140). A small-molecule STING agonist, diABZI, potently inhibits SARS-CoV-2 ancestral strain and Beta variant infection in mice by stimulation of IFN signaling (141). diABZI restricts SARS-CoV-2 replication in primary human bronchial epithelial cells (141). The administration of diamidobenzimidazole compound (diABZI-4) intranasally before or after SARS-CoV-2 challenge conferred complete protection against severe disease in ACE2 transgenic mice (142). These data provide important rationales for the development STING agonists as clinical therapeutic drugs against SARS-CoV-2 infection. The Omicron variant might initiate the innate immune response at an earlier time point in the upper respiratory system and enable the immune response to respond rapidly. Mannose-binding lectin (MBL) and long pentraxin 3 (PTX3) are two human-derived humoral fluid-phase pattern recognition molecules (PRMs) that can bind to the viral S and nucleocapsid proteins, respectively (143). MBL binds to trimeric S protein of VOC in a glycan-dependent manner and inhibits SARS-CoV-2 in vitro (143). MBL was believed to recognize the Omicron variant based on glycosylation site retention and modeling (143).
FIG 6.
Immune sensing and antagonism of SARS-CoV-2 to inhibit IFN responses. Immune sensing of SARS-CoV-2 by host pattern recognition receptors activates IFN expression and secretion (left). Secreted IFNs bind to IFN receptors to trigger IFN-stimulated gene production in an autocrine or paracrine manner (right). SARS-CoV-2 proteins that can target each layer of IFN pathway are shown as red inhibition signs.
ANTAGONISM OF SARS-CoV-2 AGAINST INNATE IMMUNITY
Viruses utilize diverse strategies to antagonize innate immunity by targeting or blocking innate immune proteins throughout whole signaling pathways (Fig. 6). IFN-I stands out as the first line of defense against viruses, and IFN treatment has been used in many therapeutic regimens. Therefore, understanding the interaction between the innate immunity system and SARS-CoV-2 is very important for therapeutics and vaccine development. SARS-CoV-2 encodes a number of nonstructural proteins that can exert these functions. SARS-CoV-2 ORF1a/b has overlapping parts, and these parts produce two polypeptides, pp1a and pp1ab, due to ribosomal frameshifting. PLpro and 3CLpro cleave pp1a and pp1ab polypeptides into 16 nonstructural proteins (NSPs). The viral subgenomic RNA also encodes eight accessory proteins. NSP1 is a very potent blocker of production of IFN-I by several mechanisms. SARS-CoV-2 NSP1 inhibits mRNA translation by interacting with the 40S subunit of ribosome complexes (144). A cryo-electron microscopy structure study also revealed that NSP1 binds to human 40S subunit and blocks the mRNA entry tunnel, thereby inhibiting innate immune response by shutting down host mRNA translation (145). Apart from turning off mRNA translation, SARS-CoV-2 NSP1 also inhibits phosphorylation of STAT1 and STAT2 to suppress the IFN signaling axis (146). SARS-CoV-2 NSP3, as PLpro, limits IFN expression by impairing IRF3 phosphorylation (147) and cleavage of IRF3 (148). SARS-CoV-2 NSP6 impairs IRF3 activation by interacting with TBK1 and impairs STAT1 and STAT2 phosphorylation (146). Similarly, as a helicase, NSP13 decreases TBK1 phosphorylation by binding to TBK1 (146, 149, 150). SARS-CoV-2 accessory proteins are also involved in the modulation of immune responses. For instance, SARS-CoV-2 ORF6 blocks the translocation of IRF3 and IRF9-STAT-STAT2 complex by hijacking nuclear importin karyopherin α2 (KPNA2) (146). It turns out that the inhibitory effects of ORF6 depend on its C terminus (151). Viral accessory protein ORF3a inhibits STING, and 3CL blocks K63-ubiquitin modification of STING to achieve effective innate immune suppression (152). SARS-CoV-2 M protein interacts with MAVS adaptor and impairs its aggregation, resulting in attenuation of the antiviral innate response (153). To suppress host defense, SARS-CoV-2 NSP8 and NSP9 bind to 7SL RNA to disrupt protein trafficking, whereas NSP16 interacts with the mRNA recognition regions of both U1 and U2 splicing RNAs to interfere with the process of mRNA splicing (154). The Omicron variant might also use its proteins to evade the immune response by similar mechanisms (Fig. 6).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
A strategy called “virus against virus” has been proposed as a potential solution since an asymptomatic Omicron variant could be developed as a promising live attenuated vaccine (155). Infection exposure to structural and nonstructural proteins, interhost sequence diversity, and a potentially longer presence of antigen in the host could be possible benefits from this approach. In contrast, a vaccine strategy using only one consensus version of the S protein would be narrower than natural induced infection immunity. It appears that the Omicron variant is less pathogenic but more transmissible. Two-dose immunization with vaccines, or even a booster vaccination, might not adequately protect from infection caused by the Omicron variant but will most likely lower the chances of severe disease and death. However, this variant still poses a high risk to unvaccinated people, the elderly, and those with comorbidities or immunosuppression. The administration of two vaccinations, along with an additional booster shoot, will help safeguard people from disease caused by SARS-CoV-2 and even its variants, where vaccines can still induce plentiful humoral and cellular immune responses. While neutralizing antibodies are shown to wane over time, the preservation of a T-cell-mediated humoral response is another potential mechanism of protection. The high resistance of the Omicron variant to neutralizing antibodies elicited by current COVID-19 vaccines and in convalescent-phase sera, as well as therapeutic monoclonal antibodies, narrows down the scope of efficacy provided by the current vaccines and therapeutics. While homologous or heterologous booster vaccines are expected to reduce the escape of SARS-CoV-2 Omicron variant, the durability, strength, and broadness of host immune immunity all require deeper understanding and extensive research (46, 68). The application of Omicron-specific vaccines for boost immunization does not seem to be very effective, while the development of pan-sarbecovirus vaccines looks very promising (77, 79, 92). The ongoing evolving scenario of the pandemic virus with newly emerging variants demands the design of highly efficacious vaccines, including variant-specific, mutation-proof, universal vaccines, to keep up the pace against emerging variants and to develop newer MAbs for treating patients with COVID-19 and to mitigate the ongoing pandemic. In addition to vaccine coverage, the development of efficient and simple therapeutic strategies to safeguard and cure predisposed persons is an extremely promising strategy that is urgently needed. The high sensitivity of the Omicron variant to the pan-CoV fusion inhibitors targeting the S2 subunit HR1 domain and the inhibitors of enzymes responsible for endosomal membrane fusion, as well as the ACE2-based entry inhibitors, offers hope in combating the newest wave of the COVID-19 pandemic caused by the Omicron variant and other emerging SARS-CoV-2 variants to come in the future.
ACKNOWLEDGMENTS
We thank our colleagues for proofreading of the manuscript. We thank Patrick Lane for graphics. We thank Vidya Chivukula for polishing the draft. We apologize to all colleagues whose contributions were not cited owing to space constraints.
H.Z., M.M., K.D., and S.J. conceived ideas and wrote the draft with contributions and efforts from all coauthors. H.Z. prepared final figures with suggestions from S.J. All coauthors revised the manuscript. All authors reviewed and approved the manuscript.
S.J. is one of the inventors in the applications of patents related to the pan-sarbecovirus vaccine and pan-CoV fusion inhibitors. Other authors declare there are no other competing interests.
Biographies

Hao Zhou, Ph.D., received his Ph.D. in molecular biology from Aarhus University in Denmark and then moved to the University of Massachusetts Medical School in the United States, where he studied innate immunity in infectious diseases. He currently works in the Department of Microbiology, Grossman School of Medicine, New York University, USA, investigating immune responses against highly emerging viral infections (SARS-CoV-2) and developing powerful preventive and therapeutic tools against SARS-CoV-2. He has published several research papers about resistance of vaccines and therapeutic monoclonal antibodies against emerging SARS-CoV-2 variants (mBio [2021], iScience [2021], Cell Reports [2021], Frontiers in Immunology [2022], and eBioMedicine [2022]).

Michelle Møhlenberg, Ph.D., received her Ph.D. from Aarhus University in Denmark, where she investigated the effect of genetic variants within the human IFN system upon viral liver and lung infections. She has published several articles, two of which focused on SARS-CoV-2 (EMBO Reports [2020] and Journal of Interferon and Cytokine Research [2021]). She is currently working as a postdoc employed at both Aarhus University, Denmark, and Ku Leuven, Belgium, within the field of angiogenesis and vascular heterogeneity with the aim of characterizing the immune modularly functions of the endothelium and the interplay between the immune system and the endothelia.

Jigarji C. Thakor, B.V.Sc./A.H. (Gold Medalist), M.V.Sc., is a Ph.D. student in the Division of Pathology, ICAR-Indian Veterinary Research Institute, Izatnagar, India. Upon his graduation, he received the Best Internee Award and five gold-plated silver medals for securing the highest overall grade point average in various subjects of the veterinary sciences. He has published 4 book chapters, 9 research articles, and 16 abstracts in a compendium published by the IAVP Congress. He is a young, dynamic, and enthusiastic researcher.

Hardeep Singh Tuli, Ph.D., is an assistant professor at the Department of Biotechnology, Maharishi Markandeshwar (Deemed to be University), Mullana, India. He previously served as a Lecturer at the Department of Applied Sciences, Institute of Science and Technology, Klawad, Yamunanagar, India (2009 to 2011). His research expertise includes natural bioactive molecules for their promising therapeutic potentials. He has actively published several articles on COVID-19 in leading journals. He has more than 10 years of teaching and research experience in pharmacology, mammalian physiology, and natural products. He has published 100 papers in peer-reviewed international journals and authored a number of book chapters. Dr. Tuli has been placed in the “Top 2% of Highly Cited Researchers in the World.” His Google scholar h-index is 29, and his Scopus h-index is 25.

Pengfei Wang, B.S., Ph.D., is a professor at the School of Life Sciences, Fudan University, Shanghai, China. He was a research scientist at Columbia University Medical Center and a postdoctoral fellow in Dr. David D. Ho’s lab in the Aaron Diamond AIDS Research Center, The Rockefeller University. He earned his Ph.D. in Genetics at Fudan University and a B.S. in Biology at Beijing Normal University in China. His research has focused on HIV and coronaviruses, and he has published more than 40 peer-reviewed journal articles. In recent years, he has published several papers to illustrate the resistance of SARS-CoV-2 variants to neutralization antibodies articles in prestigious journals (Nature, Cell Host & Microbe; Google scholar h-index 27, >5,000 citations).

Yehuda G. Assaraf, Ph.D., is Head of the Fred Wyszkowski Cancer Research Laboratory and Dean of the Faculty of Biology from 2012 to 2018, Technion-Israel Institute of Technology, Haifa, Israel. He is the incumbent Sylvia and Alexander Hassan Academic Chair. He conducted his postdoctoral research between 1986 and 1990 with Prof. Robert T. Schimke at Stanford University, Stanford, CA. His research focuses on deciphering the molecular mechanisms underlying multidrug resistance (MDR) in both hematological cancers and solid tumors. He is also developing novel targeted treatment modalities to overcome MDR in various human cancers. He authored more than 250 papers in leading peer-reviewed journals; he has an h-index of 60, with 13,000 citations. He holds many patents in biomedical research and cancer therapeutics. He has been the Editor-in-Chief of Drug Resistance Updates (IF 18.5) since 2015. He also serves on the editorial boards of many leading scientific journals and has received many prestigious research awards.

Kuldeep Dhama, M.V.Sc., Ph.D., is a Principal Scientist in the Division of Pathology, ICAR-Indian Veterinary Research Institute, Izatnagar, India. He has 27 years of research and teaching experience in areas of microbiology, immunology, virology, and biomedicine. He is recognized as an extremely productive researcher on COVID-19 research (2020 to 2022). Recently, he received the Highly Cited Researchers 2021 Award by Clarivate (Web of Science) and is listed among top 0.1% researchers across all disciplines in science. He is an NAAS (National Academy of Agricultural Science, India) Associate, Fellow-Royal Society of Medicine (FRSM), UK. He is currently serving as Editor in Chief (EIC), Co-EIC, Editor, Associate Editor, and/or Editorial Board Member of >15 scientific journals. His Google scholar h-index is 73 (24,000 citations); his Scopus h-index is 52 (12,500 citations).

Shibo Jiang, M.S., M.D., was educated in China for his M.S. and M.D. degrees, received postdoctoral training at The Rockefeller University in New York (1987 to 1990), worked in the LFK Research Institute of the New York Blood Center as Member and Laboratory Head (1990~2010), and joined Fudan University in Shanghai as a professor in late 2010. His major research interest is the research and development of antiviral therapeutics and vaccines. He pioneered the study of viral fusion and entry inhibitors, and he discovered the first pan-coronavirus fusion inhibitor and developed the first pan-sarbecovirus vaccine candidate. He has published 543 peer-reviewed papers in high-profile journals, such as Cell, Nature, Science, and The Lancet, with 35,814 citations and an h-index of 94 (Google Scholar). He has applied for 99 patents, 50 of which have been issued, and 11 of them have been licensed out. Three medical products have been developed and used in clinics.
Contributor Information
Hao Zhou, Email: hao.zhou@nyulangone.org.
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
REFERENCES
- 1.Wang H, Paulson KR, Pease SA, Watson S, Comfort H, Zheng P, Aravkin AY, Bisignano C, Barber RM, Alam T, Fuller JE, May EA, Jones DP, Frisch ME, Abbafati C, Adolph C, Allorant A, Amlag JO, Bang-Jensen B, Bertolacci GJ, Bloom SS, Carter A, Castro E, Chakrabarti S, et al. 2022. Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020-21. Lancet 399:1513–1536. 10.1016/S0140-6736(21)02796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Voorneveld M, de Koster R. 2022. Business transformation in an age of turbulence: lessons learned from COVID-19. Technol Forecast Soc Change 176:121452. 10.1016/j.techfore.2021.121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amankwah-Amoah J, Khan Z, Wood G. 2021. COVID-19 and business failures: the paradoxes of experience, scale, and scope for theory and practice. Eur Management J 39:179–184. 10.1016/j.emj.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaiser JC, Stathopoulos GT. 2020. Socioeconomic correlates of SARS-CoV-2 and influenza H1N1 outbreaks. Eur Respir J 56:2001400. 10.1183/13993003.01400-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahase E. 2022. Covid-19: what do we know about omicron sublineages? BMJ 376:o358. 10.1136/bmj.o358. [DOI] [PubMed] [Google Scholar]
- 6.Desingu PA, Nagarajan K, Dhama K. 2022. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol 94:1808–1810. 10.1002/jmv.27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iketani S, Liu L, Guo Y, Liu L, Chan JFW, Huang Y, Wang M, Luo Y, Yu J, Chu H, Chik KKH, Yuen TTT, Yin MT, Sobieszczyk ME, Huang Y, Yuen K-Y, Wang HH, Sheng Z, Ho DD. 2022. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604:553–556. 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai J, Wang X, He X, Zhao X, Zhang Y, Jiang Y, Li M, Cui Y, Chen Y, Qiao R, Li L, Yang L, Li Y, Hu Z, Zhang W, Wang P. 2022. Antibody resistance of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2 and BA.3 sub-lineages. bioRxiv. 10.1101/2022.04.07.487489. [DOI] [PMC free article] [PubMed]
- 9.Takashita E, Kinoshita N, Yamayoshi S, Sakai-Tagawa Y, Fujisaki S, Ito M, Iwatsuki-Horimoto K, Halfmann P, Watanabe S, Maeda K, Imai M, Mitsuya H, Ohmagari N, Takeda M, Hasegawa H, Kawaoka Y. 2022. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. N Engl J Med 386:1475–1477. 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J, Collier AY, Rowe M, Mardas F, Ventura JD, Wan H, Miller J, Powers O, Chung B, Siamatu M, Hachmann NP, Surve N, Nampanya F, Chandrashekar A, Barouch DH. 2022. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 variants. N Engl J Med 386:1579–1580. 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruel T, Hadjadj J, Maes P, Planas D, Seve A, Staropoli I, Guivel-Benhassine F, Porrot F, Bolland W-H, Nguyen Y, Casadevall M, Charre C, Péré H, Veyer D, Prot M, Baidaliuk A, Cuypers L, Planchais C, Mouquet H, Baele G, Mouthon L, Hocqueloux L, Simon-Loriere E, André E, Terrier B, Prazuck T, Schwartz O. 2022. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med. 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- 12.Bowen JE, Sprouse KR, Walls AC, Mazzitelli IG, Logue JK, Franko NM, Ahmed K, Shariq A, Cameroni E, Gori A, Bandera A, Posavad CM, Dan JM, Zhang Z, Weiskopf D, Sette A, Crotty S, Iqbal NT, Corti D, Geffner J, Grifantini R, Chu HY, Veesler D. 2022. Omicron BA.1 and BA.2 neutralizing activity elicited by a comprehensive panel of human vaccines. bioRxiv. 10.1101/2022.03.15.484542. [DOI] [PMC free article] [PubMed]
- 13.Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, Choga WT, Colquhoun R, Davids M, Deforche K, Doolabh D, Du Plessis L, Engelbrecht S, Everatt J, Giandhari J, Giovanetti M, Hardie D, Hill V, Hsiao NY, Iranzadeh A, et al. 2022. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603:679–686. 10.1038/d41586-021-03832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raman R, Patel KJ, Ranjan K. 2021. COVID-19: unmasking emerging SARS-CoV-2 variants, vaccines and therapeutic strategies. Biomolecules 11:993. 10.3390/biom11070993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rophina M, Pandhare K, Shamnath A, Imran M, Jolly B, Scaria V. 2022. ESC: a comprehensive resource for SARS-CoV-2 immune escape variants. Nucleic Acids Res 50:D771–D776. 10.1093/nar/gkab895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thye AY, Law JW, Pusparajah P, Letchumanan V, Chan KG, Lee LH. 2021. Emerging SARS-CoV-2 variants of concern (VOCs): an impending global crisis. Biomedicines 9:1303. 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandia R, Singhal S, Alqahtani T, Kamal MA, El-Shall NA, Nainu F, Desingu PA, Dhama K. 2022. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res 209:112816. 10.1016/j.envres.2022.112816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor L. 2022. Covid-19: omicron drives weekly record high in global infections. BMJ 376:o66. 10.1136/bmj.o66. [DOI] [PubMed] [Google Scholar]
- 19.Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. 2022. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA 327:583. 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. 2022. Clinical outcomes among patients infected with Omicron (B.1.1.529) SARS-CoV-2 variant in southern California. medRxiv. 10.1101/2022.01.11.22269045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puhach O, Adea K, Hulo N, Sattonnet P, Genecand C, Iten A, Jacquérioz BF, Kaiser L, Vetter P, Eckerle I, Meyer B. 2022. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, Delta and Omicron. medRxiv. 10.1101/2022.01.10.22269010. [DOI] [PubMed] [Google Scholar]
- 22.Helmsdal G, Hansen OK, Møller LF, Christiansen DH, Petersen MS, Kristiansen MF. 2021. Omicron outbreak at a private gathering in the Faroe Islands, infecting 21 of 33 triple-vaccinated healthcare workers. Clin Infect Dis ciac089. 10.1093/cid/ciac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone B, Urakova N, Snijder EJ, Campbell EA. 2022. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat Rev Mol Cell Biol 23:21–39. 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, Zhang J, Weng T, Zhang Z, Wu Z, Cheng L, Shi D, Lu X, Lei J, Crispin M, Shi Y, Li L, Li S. 2020. Molecular architecture of the SARS-CoV-2 virus. Cell 183:730–738.e13. 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, Lu JM, Peukes J, Xiong X, Krausslich HG, Scheres SHW, Bartenschlager R, Briggs JAG. 2020. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588:498–502. 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson CB, Farzan M, Chen B, Choe H. 2022. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirabara SM, Serdan TDA, Gorjao R, Masi LN, Pithon-Curi TC, Covas DT, Curi R, Durigon EL. 2021. SARS-CoV-2 variants: differences and potential of immune evasion. Front Cell Infect Microbiol 11:781429. 10.3389/fcimb.2021.781429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, Ku Z, An Z, Scharton D, Schindewolf C, Widen SG, Menachery VD, Shi P-Y, Weaver SC. 2022. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602:294–299. 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyngse FP, Mortensen LH, Denwood MJ, Christiansen LE, Møller CH, Skov RL, Spiess K, Fomsgaard A, Lassaunière MM, Rasmussen M, Stegger M, Nielsen C, Sieber RN, Cohen AS, Møller FT, Overvad M, Mølbak K, Krause TG, Kirkeby CT. 2021. SARS-CoV-2 Omicron VOC transmission in Danish households. medRxiv. 10.1101/2021.12.27.21268278. [DOI] [Google Scholar]
- 30.Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, Kosugi Y, Tsuda M, Orba Y, Sasaki M, Shimizu R, Kawabata R, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Suganami M, Oide A, Chiba M, Ito H, Tamura T, Tsushima K, Kubo H, Ferdous Z, Mouri H, Iida M, Kasahara K, Tabata K, Ishizuka M, Shigeno A, Tokunaga K, Ozono S, Yoshida I, Nakagawa S, Wu J, Takahashi M, Kaneda A, Seki M, Fujiki R, Nawai BR, Suzuki Y, Kashima Y, Abe K, Imamura K, Shirakawa K, The Genotype to Phenotype Japan (G2P-Japan) Consortium, et al. 2022. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603:700–705. 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdelnabi R, Foo CS, Zhang X, Lemmens V, Maes P, Slechten B, Raymenants J, André E, Weynand B, Dallmeier K, Neyts J. 2022. The omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. Antiviral Res 198:105253. 10.1016/j.antiviral.2022.105253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, Zepeda SK, Papa G, Kemp SA, Ikeda T, Toyoda M, Tan TS, Kuramochi J, Mitsunaga S, Ueno T, Shirakawa K, Takaori-Kondo A, Brevini T, Mallery DL, Charles OJ, Baker S, Dougan G, Hess C, Kingston N, Lehner PJ, Lyons PA, Matheson NJ, Ouwehand WH, Saunders C, Summers C, Thaventhiran JED, Toshner M, Weekes MP, Maxwell P, Shaw A, Bucke A, Calder J, Canna L, Domingo J, Elmer A, Fuller S, Harris J, Hewitt S, Kennet J, Jose S, Kourampa J, The CITIID-NIHR BioResource COVID-19 Collaboration, et al. 2022. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature 603:706–714. 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan KA, Watson RJ, Bewley KR, Burton C, Carnell O, Cavell BE, Challis A, Coombes NS, Emery K, Fell R, Fotheringham SA, Gooch KE, Gowan K, Handley A, Harris DJ, Humphreys R, Johnson R, Knott D, Lister S, Morley D, Ngabo D, Osman KL, Paterson J, Penn EJ, Pullan ST, Richards KS, Shaik I, Summers S, Thomas SR, Weldon T, Wiblin NR, Vipond R, Hallis B, Funnell SGP, Hall Y. 2021. Convalescence from prototype SARS-CoV-2 protects Syrian hamsters from disease caused by the Omicron variant. bioRxiv. 10.1101/2021.12.24.474081. [DOI] [PMC free article] [PubMed]
- 34.Diamond M, Halfmann P, Maemura T, Iwatsuki-Horimoto K, Iida S, Kiso M, Scheaffer S, Darling T, Joshi A, Loeber S, Foster S, Ying B, Whitener B, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Li R, Sakai Y, Liu Y, Larson D, Osorio J, Hernandez-Ortiz J, ÄŒiuoderis K, Florek K, Patel M, Bateman A, Odle A, Wong LY, Wang Z, Edara VV, Chong Z, Thackray L, Ueki H, Yamayoshi S, Imai M, Perlman S, Webby R, Seder R, Suthar M, Garcia-Sastre A, Schotsaert M, Suzuki T, Boon A, Kawaoka Y, Douek D, Moliva J, Sullivan N, et al. 2021. The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res Sq 10.21203/rs.3.rs-1211792/v1. [DOI] [Google Scholar]
- 35.Bentley EG, Kirby A, Sharma P, Kipar A, Mega DF, Bramwell C, Penrice-Randal R, Prince T, Brown JC, Zhou J, Screaton GR, Barclay WS, Owen A, Hiscox JA, Stewart JP. 2021. SARS-CoV-2 Omicron-B.1.1.529 variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. bioRxiv. 10.1101/2021.12.26.474085. [DOI]
- 36.Shuai H, Chan JF, Hu B, Chai Y, Yuen TT, Yin F, Huang X, Yoon C, Hu JC, Liu H, Shi J, Liu Y, Zhu T, Zhang J, Hou Y, Wang Y, Lu L, Cai JP, Zhang AJ, Zhou J, Yuan S, Brindley MA, Zhang BZ, Huang JD, To KK, Yuen KY, Chu H. 2022. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603:693–699. 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 37.Hui KPY, Ho JCW, Cheung MC, Ng KC, Ching RHH, Lai KL, Kam TT, Gu H, Sit KY, Hsin MKY, Au TWK, Poon LLM, Peiris M, Nicholls JM, Chan MCW. 2022. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603:715–720. 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 38.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, Darling TL, Joshi A, Loeber S, Singh G, Foster SL, Ying B, Case JB, Chong Z, Whitener B, Moliva J, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Warang P, Gagne M, Li R, Sakai-Tagawa Y, Liu Y, Larson D, Osorio JE, Hernandez-Ortiz JP, Henry AR, Ciuoderis K, Florek KR, Patel M, Odle A, Wong L-YR, Bateman AC, Wang Z, Edara V-V, Chong Z, Franks J, Jeevan T, Fabrizio T, DeBeauchamp J, Kercher L, Seiler P, Gonzalez-Reiche AS, Sordillo EM, Chang LA, van Bakel H, Consortium Mount Sinai Pathogen Surveillance (PSP) study group, et al. 2022. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603:687–692. 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H, Lu L, Peng Z, Chen LL, Meng X, Zhang C, Ip JD, Chan WM, Chu AW, Chan KH, Jin DY, Chen H, Yuen KY, To KK. 2022. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 11:277–283. 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beigel JH, Voell J, Kumar P, Raviprakash K, Wu H, Jiao JA, Sullivan E, Luke T, Davey RT, Jr.. 2018. Safety and tolerability of a novel, polyclonal human anti-MERS coronavirus antibody produced from transchromosomic cattle: a phase 1 randomised, double-blind, single-dose-escalation study. Lancet Infect Dis 18:410–418. 10.1016/S1473-3099(18)30002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Pan Z, Yue S, Yu F, Zhang J, Yang Y, Li R, Liu B, Yang X, Gao L, Li Z, Lin Y, Huang Q, Xu L, Tang J, Hu L, Zhao J, Liu P, Zhang G, Chen Y, Deng K, Ye L. 2020. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther 5:180. 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tada T, Zhou H, Dcosta BM, Samanovic MI, Cornelius A, Herati RS, Mulligan MJ, Landau NR. 2022. High-titer neutralization of Mu and C.1.2 SARS-CoV-2 variants by vaccine-elicited antibodies of previously infected individuals. Cell Rep 38:110237. 10.1016/j.celrep.2021.110237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR, Tada T. 2021. B.1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies. mBio 12:e01386-21. 10.1128/mBio.01386-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma C, Chen X, Mei F, Xiong Q, Liu Q, Dong L, Liu C, Zou W, Zhan F, Hu B, Liu Y, Liu F, Zhou L, Xu J, Jiang Y, Xu K, Cai K, Chen Y, Yan H, Lan K. 2022. Drastic decline in sera neutralization against SARS-CoV-2 Omicron variant in Wuhan COVID-19 convalescents. Emerg Microbes Infect 11:567–572. 10.1080/22221751.2022.2031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Li Q, Liang Z, Li T, Liu S, Cui Q, Nie J, Wu Q, Qu X, Huang W, Wang Y. 2022. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect 11:1–5. 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng SMS, Mok CKP, Leung YWY, Ng SS, Chan KCK, Ko FW, Chen C, Yiu K, Lam BHS, Lau EHY, Chan KKP, Luk LLH, Li JKC, Tsang LCH, Poon LLM, Hui DSC, Peiris M. 2022. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med 28:486–489. 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L, Iketani S, Guo Y, Chan JFW, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KKH, Yuen TTT, Yoon C, To KKW, Chen H, Yin MT, Sobieszczyk ME, Huang Y, Wang HH, Sheng Z, Yuen K-Y, Ho DD. 2022. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602:676–681. 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- 48.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Bassi J, Silacci-Fregni C, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini O, Ceschi A, Ferrari P, Cippà PE, Franzetti-Pellanda A, Garzoni C, Halfmann PJ, Kawaoka Y, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Diamond MS, et al. 2022. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602:664–670. 10.1038/d41586-021-03825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, Péré H, Veyer D, Puech J, Rodary J, Baele G, Dellicour S, Raymenants J, Gorissen S, Geenen C, Vanmechelen B, Wawina-Bokalanga T, Martí-Carreras J, Cuypers L, Sève A, Hocqueloux L, Prazuck T, Rey F, Simon-Loriere E, Bruel T, Mouquet H, André E, Schwartz O. 2022. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602:671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 50.Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin AJ, Kawabata H, Sominsky LA, Clark JJ, Adelsberg DC, Bielak DA, Gonzalez-Reiche AS, Dambrauskas N, Vigdorovich V, Alburquerque B, Amoako AA, Banu R, Beach KF, Bermúdez-González MC, Cai GY, Ceglia I, Cognigni C, Farrugia K, Gleason CR, van de Guchte A, Kleiner G, Khalil Z, Lyttle N, Mendez WA, Mulder LCF, Oostenink A, Rooker A, Salimbangon AT, Saksena M, Paniz-Mondolfi AE, Polanco J, Srivastava K, Sather DN, Sordillo EM, Bajic G, van Bakel H, Simon V, Krammer F, PSP-PARIS Study Group. 2022. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602:682–688. 10.1038/d41586-021-03846-z. [DOI] [PubMed] [Google Scholar]
- 51.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, Hwa S-H, Giandhari J, Blackburn JM, Gosnell BI, Abdool Karim SS, Hanekom W, Davies M-A, Hsiao M, Martin D, Mlisana K, Wibmer CK, Williamson C, York D, Harrichandparsad R, Herbst K, Jeena P, Khoza T, Kløverpris H, Leslie A, Madansein R, Magula N, Manickchund N, Marakalala M, Mazibuko M, Moshabela M, Mthabela N, Naidoo K, Ndhlovu Z, Ndung’u T, Ngcobo N, Nyamande K, Patel V, NGS-SA, et al. 2022. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602:654–656. 10.1038/d41586-021-03824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collie S, Champion J, Moultrie H, Bekker LG, Gray G. 2022. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N Engl J Med 386:494–496. 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer AS, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck HM, Behrens GMN, Pöhlmann S. 2022. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185:447–456. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willett BJ, Grove J, MacLean OA, Wilkie C, Logan N, Lorenzo GD, Furnon W, Scott S, Manali M, Szemiel A, Ashraf S, Vink E, Harvey W, Davis C, Orton R, Hughes J, Holland P, Silva V, Pascall D, Puxty K, da Silva Filipe A, Yebra G, Shaaban S, Holden MTG, Pinto RM, Gunson R, Templeton K, Murcia P, Patel AH, Haughney J, Robertson DL, Palmarini M, Ray S, Thomson EC, investigators TC-DVCS, Consortium TC-GU, Consortium TGP-UNV, investigators TEoVADC-V. 2022. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. 10.1101/2022.01.03.21268111. [DOI] [Google Scholar]
- 55.Lu L, Mok BW, Chen LL, Chan JM, Tsang OT, Lam BH, Chuang VW, Chu AW, Chan WM, Ip JD, Chan BP, Zhang R, Yip CC, Cheng VC, Chan KH, Jin DY, Hung IF, Yuen KY, Chen H, To KK. 2021. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotaki R, Adachi Y, Moriyama S, Onodera T, Fukushi S, Nagakura T, Tonouchi K, Terahara K, Sun L, Takano T, Nishiyama A, Shinkai M, Oba K, Nakamura-Uchiyama F, Shimizu H, Suzuki T, Matsumura T, Isogawa M, Takahashi Y. 2022. SARS-CoV-2 Omicron-neutralizing memory B cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci Immunol 7:eabn8590. 10.1126/sciimmunol.abn8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2022. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med 386:599–601. 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartsch Y, Tong X, Kang J, Avendaño MJ, Serrano EF, García-Salum T, Pardo-Roa C, Riquelme A, Medina RA, Alter G. 2021. Preserved Omicron Spike specific antibody binding and Fc-recognition across COVID-19 vaccine platforms. medRxiv. 10.1101/2021.12.24.21268378. [DOI] [Google Scholar]
- 59.Wang K, Jia Z, Bao L, Wang L, Cao L, Chi H, Hu Y, Li Q, Zhou Y, Jiang Y, Zhu Q, Deng Y, Liu P, Wang N, Wang L, Liu M, Li Y, Zhu B, Fan K, Fu W, Yang P, Pei X, Cui Z, Qin L, Ge P, Wu J, Liu S, Chen Y, Huang W, Wang Q, Qin C-F, Wang Y, Qin C, Wang X. 2022. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 603:919–925. 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolzhikova I, Iliukhina A, Kovyrshina A, Kuzina A, Gushchin V, Siniavin A, Pochtovyi A, Shidlovskaya E, Kuznetsova N, Megeryan M, Dzharullaeva A, Erokhova A, Izhaeva F, Grousova D, Botikov A, Shcheblyakov D, Tukhvatulin A, Zubkova O, Logunov D, Gintsburg A. 2021. Sputnik Light booster after Sputnik V vaccination induces robust neutralizing antibody response to B.1.1.529 (Omicron) SARS-CoV-2 variant. medRxiv. 10.1101/2021.12.17.21267976. [DOI] [Google Scholar]
- 61.Yu X, Wei D, Xu W, Li Y, Li X, Zhang X, Qu J, Yang Z, Chen E. 2022. Reduced sensitivity of SARS-CoV-2 Omicron variant to antibody neutralization elicited by booster vaccination. Cell Discov 8:4. 10.1038/s41421-022-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medigeshi G, Batra G, Murugesan DR, Thiruvengadam R, Chattopadhyay S, Das B, Gosain M, Ayushi Singh J, Anbalagan A, Shaman H, Pargai K, Mehdi F, Das SJ, Kahlon N, Singh S, Kshetrapal P, Wadhwa N, Pandey AK, Bhatnagar S, Garg PK. 2022. Sub-optimal neutralization of Omicron (B.1.1.529) variant by antibodies induced by vaccine alone or SARS-CoV-2 infection plus vaccine (hybrid immunity) post 6-months. EBioMedicine 78:103938. 10.1016/j.ebiom.2022.103938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter T, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Ditse Z, Silva KD, Madhi S, Nunes MC, Malik T, Openshaw PJ, Baillie JK, Semple MG. 2021. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185:467–484.e15. 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burki TK. 2022. Omicron variant and booster COVID-19 vaccines. Lancet Respir Med 10:e17. 10.1016/S2213-2600(21)00559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pajon R, Doria-Rose NA, Shen X, Schmidt SD, O’Dell S, McDanal C, Feng W, Tong J, Eaton A, Maglinao M, Tang H, Manning KE, Edara V-V, Lai L, Ellis M, Moore KM, Floyd K, Foster SL, Posavad CM, Atmar RL, Lyke KE, Zhou T, Wang L, Zhang Y, Gaudinski MR, Black WP, Gordon I, Guech M, Ledgerwood JE, Misasi JN, Widge A, Sullivan NJ, Roberts PC, Beigel JH, Korber B, Baden LR, El Sahly H, Chalkias S, Zhou H, Feng J, Girard B, Das R, Aunins A, Edwards DK, Suthar MS, Mascola JR, Montefiori DC. 2022. SARS-CoV-2 Omicron variant neutralization after mRNA-1273 booster vaccination. N Engl J Med 386:1088–1091. 10.1056/NEJMc2119912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nemet I, Kliker L, Lustig Y, Zuckerman N, Erster O, Cohen C, Kreiss Y, Alroy-Preis S, Regev-Yochay G, Mendelson E, Mandelboim M. 2022. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N Engl J Med 386:492–494. 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Roederer AL, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2022. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 185:457–466.e4. 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Zhao X, Song J, Wu J, Zhu Y, Li M, Cui Y, Chen Y, Yang L, Liu J, Zhu H, Jiang S, Wang P. 2022. Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg Microbes Infect 11:477–481. 10.1080/22221751.2022.2030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, De la Cruz E, Jorge A, De los Santos M, Leon P, Breban MI, Billig K, Yildirim I, Pearson C, Downing R, Gagnon E, Muyombwe A, Razeq J, Campbell M, Ko A, Omer SB, Grubaugh ND, Vermund SH, Iwasaki A. 2021. Immunogenicity of heterologous BNT162b2 booster in fully vaccinated individuals with CoronaVac against SARS-CoV-2 variants Delta and Omicron: the Dominican Republic experience. medRxiv. 10.1101/2021.12.27.21268459. [DOI] [Google Scholar]
- 70.Accorsi EK, Britton A, Fleming-Dutra KE, Smith ZR, Shang N, Derado G, Miller J, Schrag SJ, Verani JR. 2022. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 Omicron and Delta variants. JAMA 327:639. 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muik A, Lui BG, Wallisch A-K, Bacher M, Mühl J, Reinholz J, Ozhelvaci O, Beckmann N, Güimil Garcia R. d l C, Poran A, Shpyro S, Finlayson A, Cai H, Yang Q, Swanson KA, Türeci Ö, Şahin U. 2022. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 375:678–680. 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ai J, Zhang H, Zhang Y, Lin K, Zhang Y, Wu J, Wan Y, Huang Y, Song J, Fu Z, Wang H, Guo J, Jiang N, Fan M, Zhou Y, Zhao Y, Zhang Q, Liu Q, Lv J, Li P, Qiu C, Zhang W. 2022. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect 11:337–343. 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.GeurtsvanKessel CH, Geers D, Schmitz KS, Mykytyn AZ, Lamers MM, Bogers S, Scherbeijn S, Gommers L, Sablerolles RSG, Nieuwkoop NN, Rijsbergen LC, van Dijk LLA, de Wilde J, Alblas K, Breugem TI, Rijnders BJA, de Jager H, Weiskopf D, van der Kuy PHM, Sette A, Koopmans MPG, Grifoni A, Haagmans BL, de Vries RD. 2022. Divergent SARS CoV-2 Omicron-reactive T- and B cell responses in COVID-19 vaccine recipients. Sci Immunol 7:eabo2202. 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuhlmann C, Mayer CK, Claassen M, Maponga T, Burgers WA, Keeton R, Riou C, Sutherland AD, Suliman T, Shaw ML, Preiser W. 2022. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 399:625–626. 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bates TA, McBride SK, Leier HC, Guzman G, Lyski ZL, Schoen D, Winders B, Lee J-Y, Lee DX, Messer WB, Curlin ME, Tafesse FG. 2022. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol 7:eabn8014. 10.1126/sciimmunol.abn8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gagne M, Moliva JI, Foulds KE, Andrew SF, Flynn BJ, Werner AP, Wagner DA, Teng I-T, Lin BC, Moore C, Jean-Baptiste N, Carroll R, Foster SL, Patel M, Ellis M, Edara V-V, Maldonado NV, Minai M, McCormick L, Honeycutt CC, Nagata BM, Bock KW, Dulan CNM, Cordon J, Todd J-PM, McCarthy E, Pessaint L, Van Ry A, Narvaez B, Valentin D, Cook A, Dodson A, Steingrebe K, Flebbe DR, Nurmukhambetova ST, Godbole S, Henry AR, Laboune F, Roberts-Torres J, Lorang CG, Amin S, Trost J, Naisan M, Basappa M, Willis J, Wang L, Shi W, Doria-Rose NA, Olia AS, Liu C. 2022. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits comparable B cell expansion, neutralizing antibodies and protection against Omicron. Cell 185:1556–1571.e18. 10.1016/j.cell.2022.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Su S, Li W, Jiang S. 2022. Developing pan-beta-coronavirus vaccines against emerging SARS-CoV-2 variants of concern. Trends Immunol 43:170–172. 10.1016/j.it.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Z, Zhou J, Xu W, Deng W, Wang Y, Wang M, Wang Q, Hsieh M, Dong J, Wang X, Huang W, Xing L, He M, Tao C, Xie Y, Zhang Y, Wang Y, Zhao J, Yuan Z, Qin C, Jiang S, Lu L. 2022. A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res 32:269–219. 10.1038/s41422-022-00612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Z, Chan JF-W, Zhou J, Wang M, Wang Q, Zhang G, Xu W, Chik KK-H, Zhang Y, Wang Y, Yuen K-Y, Lu L, Jiang S. 2022. A pan-sarbecovirus vaccine induces highly potent and durable neutralizing antibody responses in non-human primates against SARS-CoV-2 Omicron variant. Cell Res 32:495–497. 10.1038/s41422-022-00631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hwang YC, Lu RM, Su SC, Chiang PY, Ko SH, Ke FY, Liang KH, Hsieh TY, Wu HC. 2022. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J Biomed Sci 29:1. 10.1186/s12929-021-00784-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou T, Wang L, Misasi J, Pegu A, Zhang Y, Harris DR, Olia AS, Talana CA, Yang ES, Chen M, Choe M, Shi W, Teng I-T, Creanga A, Jenkins C, Leung K, Liu T, Stancofski E-SD, Stephens T, Zhang B, Tsybovsky Y, Graham BS, Mascola JR, Sullivan NJ, Kwong PD. 2021. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 376:eabn8897. 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aggarwal A, Stella AO, Walker G, Akerman A, Milogiannakis V, Brilot F, Amatayakul-Chantler S, Roth N, Coppola G, Schofield P, Jackson J, Henry JY, Mazigi O, Langley D, Lu Y, Forster C, McAllery S, Mathivanan V, Fichter C, Hoppe AC, Munier ML, Jack H-M, Cromer D, Darley D, Matthews G, Christ D, Khoury D, Davenport M, Rawlinson W, Kelleher AD, Turville S. 2021. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv. 10.1101/2021.12.14.21267772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tatham L, Sharp J, Kijak E, Herriott J, Neary M, Box H, Valentijn A, Cox H, Pertinez H, Curley P, Arshad U, Rajoli RK, Rannard S, Stewart J, Owen A. 2022. Lack of Ronapreve (REGN-CoV; casirivimab and imdevimab) virological efficacy against the SARS-CoV-2 Omicron variant (B.1.1.529) in K18-hACE2 mice. bioRxiv. 10.1101/2022.01.23.477397. [DOI] [PMC free article] [PubMed]
- 84.Touret F, Baronti C, Bouzidi HS, de Lamballerie X. 2022. In vitro evaluation of therapeutic antibodies against a SARS-CoV-2 Omicron B.1.1.529 isolate. Sci Rep 12:4683. 10.1038/s41598-022-08559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cathcart AL, Havenar-Daughton C, Lempp FA, Ma D, Schmid MA, Agostini ML, Guarino B, Di Iulio J, Rosen LE, Tucker H, Dillen J, Subramanian S, Sloan B, Bianchi S, Pinto D, Saliba C, Culap K, Wojcechowskyj JA, Noack J, Zhou J, Kaiser H, Chase A, Montiel-Ruiz M, Dellota E, Park A, Spreafico R, Sahakyan A, Lauron EJ, Czudnochowski N, Cameroni E, Ledoux S, Werts A, Colas C, Soriaga L, Telenti A, Purcell LA, Hwang S, Snell G, Virgin HW, Corti D, Hebner CM. 2021. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 10.1101/2021.03.09.434607. [DOI]
- 86.VanBlargan LA, Errico JM, Halfmann PJ, Zost SJ, Crowe JE, Purcell LA, Kawaoka Y, Corti D, Fremont DH, Diamond MS. 2022. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med 28:490–495. 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 88.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, Wang J, Wang Y, Niu X, Yang S, Liang H, Sun H, Li T, Yu Y, Cui Q, Liu S, Yang X, Du S, Zhang Z, Hao X, Shao F, Jin R, Wang X, Xiao J, Wang Y, Xie XS. 2022. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602:657–663. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tada T, Zhou H, Dcosta BM, Samanovic MI, Chivukula V, Herati RS, Hubbard SR, Mulligan MJ, Landau NR. 2021. Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. bioRxiv. 10.1101/2021.12.28.474369. [DOI] [PMC free article] [PubMed]
- 90.McCallum M, Czudnochowski N, Rosen LE, Zepeda SK, Bowen JE, Walls AC, Hauser K, Joshi A, Stewart C, Dillen JR, Powell AE, Croll TI, Nix J, Virgin HW, Corti D, Snell G, Veesler D. 2021. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 375:864–868. 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cui Z, Liu P, Wang N, Wang L, Fan K, Zhu Q, Wang K, Chen R, Feng R, Jia Z, Yang M, Xu G, Zhu B, Fu W, Chu T, Feng L, Wang Y, Pei X, Yang P, Xie XS, Cao L, Cao Y, Wang X. 2022. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 185:860–871. 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martinez DR, Schafer A, Gobeil S, Li D, De la Cruz G, Parks R, Lu X, Barr M, Stalls V, Janowska K, Beaudoin E, Manne K, Mansouri K, Edwards RJ, Cronin K, Yount B, Anasti K, Montgomery SA, Tang J, Golding H, Shen S, Zhou T, Kwong PD, Graham BS, Mascola JR, Montefiori DC, Alam SM, Sempowski G, Sempowski GD, Khurana S, Wiehe K, Saunders KO, Acharya P, Haynes BF, Baric RS. 2022. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci Transl Med 14:eabj7125. 10.1126/scitranslmed.abj7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lv Z, Deng YQ, Ye Q, Cao L, Sun CY, Fan C, Huang W, Sun S, Sun Y, Zhu L, Chen Q, Wang N, Nie J, Cui Z, Zhu D, Shaw N, Li XF, Li Q, Xie L, Wang Y, Rao Z, Qin CF, Wang X. 2020. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369:1505–1509. 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Westendorf K, Žentelis S, Wang L, Foster D, Vaillancourt P, Wiggin M, Lovett E, van der Lee R, Hendle J, Pustilnik A, Sauder JM, Kraft L, Hwang Y, Siegel RW, Chen J, Heinz BA, Higgs RE, Kallewaard NL, Jepson K, Goya R, Smith MA, Collins DW, Pellacani D, Xiang P, de Puyraimond V, Ricicova M, Devorkin L, Pritchard C, O’Neill A, Dalal K, Panwar P, Dhupar H, Garces FA, Cohen CA, Dye JM, Huie KE, Badger CV, Kobasa D, Audet J, Freitas JJ, Hassanali S, Hughes I, Munoz L, Palma HC, Ramamurthy B, Cross RW, Geisbert TW, Menacherry V, Lokugamage K, Borisevich V. 2022. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. bioRxiv. 10.1101/2021.04.30.442182. [DOI] [PMC free article] [PubMed]
- 95.Bae EY, Sanders JM, Johns ML, Lin K, Ortwine JK, Wei W, Mang NS, Cutrell JB. 2021. Therapeutic options for Coronavirus Disease 2019 (COVID-19): where are we now? Curr Infect Dis Rep 23:28. 10.1007/s11908-021-00769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zou H, Yang Y, Dai H, Xiong Y, Wang JQ, Lin L, Chen ZS. 2021. Recent updates in experimental research and clinical evaluation on drugs for COVID-19 treatment. Front Pharmacol 12:732403. 10.3389/fphar.2021.732403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kovacech B, Fialova L, Filipcik P, Skrabana R, Zilkova M, Paulenka-Ivanovova N, Kovac A, Palova D, Rolkova GP, Tomkova K, Csokova NT, Markova K, Skrabanova M, Sinska K, Basheer N, Majerova P, Hanes J, Parrak V, Prcina M, Cehlar O, Cente M, Piestansky J, Fresser M, Novak M, Slavikova M, Borsova K, Cabanova V, Brejova B, Vinař T, Nosek J, Klempa B, Eyer L, Hönig V, Palus M, Ruzek D, Vyhlidalova T, Strakova P, Mrazkova B, Zudova D, Koubkova G, Novosadova V, Prochazka J, Sedlacek R, Zilka N, Kontsekova E. 2022. Monoclonal antibodies targeting two immunodominant epitopes on the Spike protein neutralize emerging SARS-CoV-2 variants of concern. EBioMedicine 76:103818. 10.1016/j.ebiom.2022.103818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duan X, Shi R, Liu P, Huang Q, Wang F, Chen X, Feng H, Huang W, Xiao J, Yan J. 2022. A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants. Signal Transduct Target Ther 7:23. 10.1038/s41392-022-00879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saied AA, Metwally AA, Alobo M, Shah J, Sharun K, Dhama K. 2022. Bovine-derived antibodies and camelid-derived nanobodies as biotherapeutic weapons against SARS-CoV-2 and its variants: a review article. Int J Surg 98:106233. 10.1016/j.ijsu.2022.106233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun D, Sang Z, Kim YJ, Xiang Y, Cohen T, Belford AK, Huet A, Conway JF, Sun J, Taylor DJ, Schneidman-Duhovny D, Zhang C, Huang W, Shi Y. 2021. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat Commun 12:4676. 10.1038/s41467-021-24963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vallianou NG, Tsilingiris D, Christodoulatos GS, Karampela I, Dalamaga M. 2021. Anti-viral treatment for SARS-CoV-2 infection: a race against time amidst the ongoing pandemic. Metabol Open 10:100096. 10.1016/j.metop.2021.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li P, Wang Y, Lavrijsen M, Lamers MM, de Vries AC, Rottier RJ, Bruno MJ, Peppelenbosch MP, Haagmans BL, Pan Q. 2022. SARS-CoV-2 Omicron variant is highly sensitive to Molnupiravir, Nirmatrelvir, and the combination. Cell Res 32:322–324. 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fan H, Lou F, Fan J, Li M, Tong Y. 2022. The emergence of powerful oral anti-COVID-19 drugs in the post-vaccine era. Lancet Microbe 3:e91. 10.1016/S2666-5247(21)00278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jayk Bernal A, Gomes da Silva MM, Musungaie DB, Kovalchuk E, Gonzalez A, Delos Reyes V, Martín-Quirós A, Caraco Y, Williams-Diaz A, Brown ML, Du J, Pedley A, Assaid C, Strizki J, Grobler JA, Shamsuddin HH, Tipping R, Wan H, Paschke A, Butterton JR, Johnson MG, De Anda C. 2022. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med 386:509–520. 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W, Baniecki M, Hendrick VM, Damle B, Simón-Campos A, Pypstra R, Rusnak JM. 2022. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med 386:1397–1408. 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Couzin-Frankel J. 2021. Antiviral pills could change pandemic’s course. Science 374:799–800. 10.1126/science.acx9605. [DOI] [PubMed] [Google Scholar]
- 107.Soriano V, de-Mendoza C, Edagwa B, Trevino A, Barreiro P, Fernandez-Montero JV, Gendelman HE. 2022. Oral antivirals for the prevention and treatment of SARS-CoV-2 infection. AIDS Rev 24:41–49. 10.24875/AIDSRev.22000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vangeel L, Chiu W, De Jonghe S, Maes P, Slechten B, Raymenants J, Andre E, Leyssen P, Neyts J, Jochmans D. 2022. Remdesivir, Molnupiravir, and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res 198:105252. 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Catlin NR, Bowman CJ, Campion SN, Cheung JR, Nowland WS, Sathish JG, Stethem CM, Updyke L, Cappon GD. 2022. Reproductive and developmental safety of Nirmatrelvir (PF-07321332), an oral SARS-CoV-2 Mpro inhibitor in animal models. Reprod Toxicol 108:56–61. 10.1016/j.reprotox.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanai T. 2022. Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta 240:123206. 10.1016/j.talanta.2022.123206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim S, Liu Y, Ziarnik M, Cao Y, Zhang XF, Im W. 2022. Binding of human ACE2 and RBD of Omicron enhanced by unique interaction patterns among SARS-CoV-2 variants of concern. bioRxiv. 10.1101/2022.01.24.477633. [DOI] [PMC free article] [PubMed]
- 112.Monteil V, Stephanie D, Klingström J, Thålin C, Kellner MJ, Christ W, Havervall S, Mereiter S, Knapp S, Montserrat N, Braunsfeld B, Kozieradzki I, Ali OH, Hagelkruys A, Stadlmann J, Oostenbrink C, Wirnsberger G, Penninger JM, Mirazimi A. 2021. Clinical-grade ACE2 effectively inhibits SARS-CoV-2 Omicron infections. bioRxiv. 10.1101/2021.12.25.474113. [DOI]
- 113.Ikemura N, Taminishi S, Inaba T, Arimori T, Motooka D, Katoh K, Kirita Y, Higuchi Y, Li S, Itoh Y, Ozaki Y, Nakamura S, Matoba S, Standley DM, Okamoto T, Takagi J, Hoshino A. 2021. Engineered ACE2 counteracts vaccine-evading SARS-CoV-2 Omicron variant. bioRxiv. 10.1101/2021.12.22.473804. [DOI] [PMC free article] [PubMed]
- 114.Huntington KE, Carlsen L, So EY, Piesche M, Liang O, El-Deiry WS. 2022. Integrin/TGF-β1 inhibitor GLPG-0187 blocks SARS-CoV-2 Delta and Omicron pseudovirus infection of airway epithelial cells which could attenuate disease severity. medRxiv. 10.1101/2022.01.02.22268641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Xia S, Zhu Y, Lu L, Jiang S. 2021. Pan-coronavirus fusion inhibitors as the hope for today and tomorrow. Protein Cell 12:84–88. 10.1007/s13238-020-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante CR, Xiong H, Farmar J, Debnath AK, Tien P, Jiang S. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938–947. 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lu L, Liu Q, Zhu Y, Chan KH, Qin L, Li Y, Wang Q, Chan JF, Du L, Yu F, Ma C, Ye S, Yuen KY, Zhang R, Jiang S. 2014. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun 5:3067. 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, Wang Q, Du L, Tan W, Wilson IA, Jiang S, Yang B, Lu L. 2019. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv 5:eaav4580. 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. 2020. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30:343–355. 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou J, Xu W, Liu Z, Wang C, Xia S, Lan Q, Cai Y, Su S, Pu J, Xing L, Xie Y, Lu L, Jiang S, Wang Q. 2022. A highly potent and stable pan-coronavirus fusion inhibitor as a candidate prophylactic and therapeutic for COVID-19 and other coronavirus diseases. Acta Pharm Sin B 12:1652–1661. 10.1016/j.apsb.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xia S, Chan JF, Wang L, Jiao F, Chik KK, Chu H, Lan Q, Xu W, Wang Q, Wang C, Yuen KY, Lu L, Jiang S. 2022. Peptide-based pan-CoV fusion inhibitors maintain high potency against SARS-CoV-2 Omicron variant. Cell Res 32:404–406. 10.1038/s41422-022-00617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moss P. 2022. The T cell immune response against SARS-CoV-2. Nat Immunol 23:186–193. 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 124.Gao Y, Cai C, Grifoni A, Müller TR, Niessl J, Olofsson A, Humbert M, Hansson L, Österborg A, Bergman P, Chen P, Olsson A, Sandberg JK, Weiskopf D, Price DA, Ljunggren HG, Karlsson AC, Sette A, Aleman S, Buggert M. 2022. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med 28:472–476. 10.1038/d41591-022-00017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mudd PA, Minervina AA, Pogorelyy MV, Turner JS, Kim W, Kalaidina E, Petersen J, Schmitz AJ, Lei T, Haile A, Kirk AM, Mettelman RC, Crawford JC, Nguyen THO, Rowntree LC, Rosati E, Richards KA, Sant AJ, Klebert MK, Suessen T, Middleton WD, Wolf J, Teefey SA, O’Halloran JA, Presti RM, Kedzierska K, Rossjohn J, Thomas PG, Ellebedy AH, Estepp JH, Schultz-Cherry S, McGargill MA, Gaur A, Hoffman J, Mori M, Tang L, Tuomanen E, Webby R, Hayden RT, Hakim H, Hijano DR, Allison KJ, Allen EK, Bajracharya R, Awad W, Van de Velde L-A, Clark BL, Wilson TL, Souquette A, Castellaw A, et al. 2022. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell 185:603–613.e15. 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nguyen AT, Szeto C, Chatzileontiadou DSM, Tong ZWM, Dewar-Oldis MJ, Cooper L, Murdolo LD, Chew KY, Lineburg KE, Riboldi-Tunicliffe A, Williamson R, Gardiner BJ, Jayasinghe D, Lobos CA, Ahn YM, Grant EJ, Smith C, McMahon J, Good-Jacobson KL, Barnard PJ, Short KR, Gras S. 2022. COVID-19 vaccine booster induces a strong CD8+ T cell response against Omicron variant epitopes in HLA-A*02:01+ individuals. bioRxiv. 10.1101/2022.01.12.473243. [DOI]
- 127.Ahmed SF, Quadeer AA, McKay MR. 2021. SARS-CoV-2 T cell responses elicited by COVID-19 vaccines or infection are expected to remain robust against Omicron. Viruses 14:79. 10.3390/v14010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang S, Wu S, Zhao G, He Y, Guo X, Zhang Z, Hou J, Ding Y, Cheng A, Wang B. 2021. Identification of a promiscuous conserved CTL epitope within the SARS-CoV-2 spike protein. bioRxiv. 10.1101/2021.11.21.469172. [DOI] [PMC free article] [PubMed]
- 129.Redd AD, Nardin A, Kared H, Bloch EM, Abel B, Pekosz A, Laeyendecker O, Fehlings M, Quinn TC, Tobian AA. 2021. Minimal crossover between mutations associated with Omicron variant of SARS-CoV-2 and CD8+ T cell epitopes identified in COVID-19 convalescent individuals. mBio 13:e0361721. 10.1128/mbio.03617-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, Khan K, Cele S, Bernstein M, Karim F, Madzorera SV, Moyo-Gwete T, Mennen M, Skelem S, Adriaanse M, Mutithu D, Aremu O, Stek C, Du Bruyn E, Van Der Mescht MA, de Beer Z, de Villiers TR, Bodenstein A, van den Berg G, Mendes A, Strydom A, Venter M, Giandhari J, Naidoo Y, Pillay S, Tegally H, Grifoni A, Weiskopf D, Sette A, Wilkinson RJ, de Oliveira T, Bekker LG, Gray G, Ueckermann V, Rossouw T, Boswell MT, Bihman J, Moore PL, Sigal A, Ntusi NAB, Burgers WA, Riou C. 2022. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 603:488–492. 10.1038/s41586-022-04708-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Naranbhai V, Nathan A, Kaseke C, Berrios C, Khatri A, Choi S, Getz MA, Tano-Menka R, Ofoman O, Gayton A, Senjobe F, Denis KJS, Lam EC, Garcia-Beltran WF, Balazs AB, Walker BD, Iafrate AJ, Gaiha GD. 2022. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all prior infected and vaccinated individuals. medRxiv. 10.1101/2022.01.04.21268586. [DOI] [Google Scholar]
- 132.Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, Sette A. 2021. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe 29:1076–1092. 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rehwinkel J, Gack MU. 2020. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20:537–551. 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 135.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514:372–375. 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sampaio NG, Chauveau L, Hertzog J, Bridgeman A, Fowler G, Moonen JP, Dupont M, Russell RA, Noerenberg M, Rehwinkel J. 2021. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci Rep 11:13638. 10.1038/s41598-021-92940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thorne LG, Reuschl AK, Zuliani-Alvarez L, Whelan MVX, Turner J, Noursadeghi M, Jolly C, Towers GJ. 2021. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J 40:e107826. 10.15252/embj.2021107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng M, Karki R, Williams EP, Yang D, Fitzpatrick E, Vogel P, Jonsson CB, Kanneganti TD. 2021. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol 22:829–838. 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Di Domizio J, Gulen MF, Saidoune F, Thacker VV, Yatim A, Sharma K, Nass T, Guenova E, Schaller M, Conrad C, Goepfert C, De Leval L, von Garnier C, Berezowska S, Dubois A, Gilliet M, Ablasser A. 2022. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature 603:145–151. 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li M, Ferretti M, Ying B, Descamps H, Lee E, Dittmar M, Lee JS, Whig K, Kamalia B, Dohnalova L, Uhr G, Zarkoob H, Chen YC, Ramage H, Ferrer M, Lynch K, Schultz DC, Thaiss CA, Diamond MS, Cherry S. 2021. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci Immunol 6:eabi9007. 10.1126/sciimmunol.abi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Humphries F, Shmuel-Galia L, Jiang Z, Wilson R, Landis P, Ng SL, Parsi KM, Maehr R, Cruz J, Morales-Ramos A, Ramanjulu JM, Bertin J, Pesiridis GS, Fitzgerald KA. 2021. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci Immunol 6:eabi9002. 10.1126/sciimmunol.abi9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stravalaci M, Pagani I, Paraboschi EM, Pedotti M, Doni A, Scavello F, Mapelli SN, Sironi M, Perucchini C, Varani L, Matkovic M, Cavalli A, Cesana D, Gallina P, Pedemonte N, Capurro V, Clementi N, Mancini N, Invernizzi P, Bayarri-Olmos R, Garred P, Rappuoli R, Duga S, Bottazzi B, Uguccioni M, Asselta R, Vicenzi E, Mantovani A, Garlanda C. 2022. Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat Immunol 23:275–286. 10.1038/s41590-021-01114-w. [DOI] [PubMed] [Google Scholar]
- 144.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler L-A, Leibundgut M, Thiel V, Mühlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27:959–966. 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 145.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Stürzel CM, Fröhlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369:1249–1255. 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Müller S, Knobeloch K-P, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. 2020. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587:657–662. 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Moustaqil M, Ollivier E, Chiu H-P, Van Tol S, Rudolffi-Soto P, Stevens C, Bhumkar A, Hunter DJB, Freiberg AN, Jacques D, Lee B, Sierecki E, Gambin Y. 2021. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infect 10:178–195. 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583:459–468. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hoffmann HH, Sánchez-Rivera FJ, Schneider WM, Luna JM, Soto-Feliciano YM, Ashbrook AW, Le Pen J, Leal AA, Ricardo-Lax I, Michailidis E, Hao Y, Stenzel AF, Peace A, Zuber J, Allis CD, Lowe SW, MacDonald MR, Poirier JT, Rice CM. 2021. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe 29:267–280.e5. 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F, Zhao Z, Zhou Z, Xiang Z, Wang J. 2020. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11:3810. 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rui Y, Su J, Shen S, Hu Y, Huang D, Zheng W, Lou M, Shi Y, Wang M, Chen S, Zhao N, Dong Q, Cai Y, Xu R, Zheng S, Yu X-F. 2021. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct Target Ther 6:123. 10.1038/s41392-021-00515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fu Y-Z, Wang S-Y, Zheng Z-Q, Yi H, Li W-W, Xu Z-S, Wang Y-Y. 2021. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cellular and Molecular Immunology 18:613–620. 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees RM, Botten JW, Majumdar D, Guttman M. 2020. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183:1325–1339.e21. 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nguyen TM, Zhang Y, Pandolfi PP. 2020. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res 30:189–190. 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]