Abstract
Germinal centres are the engines of antibody evolution. Here, using human immunodeficiency virus (HIV) Env protein immunogen priming in rhesus monkeys followed by a long period without further immunization, we demonstrate germinal centre B (BGC) cells that last for at least 6 months. A 186-fold increase in BGC cells was present by week 10 compared with conventional immunization. Single-cell transcriptional profiling showed that both light- and dark-zone germinal centre states were sustained. Antibody somatic hypermutation of BGC cells continued to accumulate throughout the 29-week priming period, with evidence of selective pressure. Env-binding BGC cells were still 49-fold above baseline at 29 weeks, which suggests that they could remain active for even longer periods of time. High titres of HIV-neutralizing antibodies were generated after a single booster immunization. Fully glycosylated HIV trimer protein is a complex antigen, posing considerable immunodominance challenges for B cells1,2. Memory B cells generated under these long priming conditions had higher levels of antibody somatic hypermutation, and both memory B cells and antibodies were more likely to recognize non-immunodominant epitopes. Numerous BGC cell lineage phylogenies spanning more than the 6-month germinal centre period were identified, demonstrating continuous germinal centre activity and selection for at least 191 days with no further antigen exposure. A long-prime, slow-delivery (12 days) immunization approach holds promise for difficult vaccine targets and suggests that patience can have great value for tuning of germinal centres to maximize antibody responses.
Subject terms: Germinal centres, Protein vaccines, Antibodies
Using HIV Env protein immunogen priming in rhesus monkeys followed by a long period without further immunization, we demonstrate germinal centre B cells lasting at least 6 months, showing promise in regard to difficult vaccine targets.
Main
Antibodies serve as effective adaptive immunity frontline defences against most infectious diseases. As such, most efficacious vaccines aim to prophylactically elicit potent neutralizing antibodies and long-lasting immunological memory to the target pathogen. For rapidly mutating pathogens such as human immunodeficiency virus (HIV) there is an extra level of complication wherein an ideal vaccine should generate cross-reactive or broadly neutralizing antibodies that can protect against variants3,4; however, so far, no broadly neutralizing antibodies against HIV have been elicited in the serum of either humans or non-human primates (NHPs) by vaccination2.
High-affinity antibodies are typically the result of affinity maturation in germinal centres (GCs). GCs represent evolution in miniature, with proliferation (generations) accompanied by mutations and competition for limiting resources in the form of antigen and T cell help5–8. To accomplish this evolution, GC B cells (BGC cells) proliferate rapidly—every 4–6 h9,10. GCs are often observed for a few weeks following acute antigen exposure. Antigen-specific BGC cells have been widely observed for 14–28 days in most model systems, and such a time window can represent a substantial amount of antibody sequence space exploration by BGC due to their rapid cell cycle5,8. We previously showed that vaccine slow-delivery methods over a period of 7–14 days, such as the use of osmotic pumps or repeated small-dose injections, enhanced immune responses relative to traditional bolus immunizations11–13, with some evidence of increased durability of GCs for 2 months13. However, the full potential longevity of GCs, the biological programming of long-lasting GCs, antibody maturation under such conditions and the functionality and productivity of older GCs are minimally understood. Here, we used a 12 day, slow-delivery protein immunization strategy and antigen-specific molecular and cellular tools to explore the extent of GC durability after priming immunization, and the ensuing immunological outcomes.
Priming can fuel GCs for several months
Alum is a classic adjuvant used in many human vaccines14. A group of rhesus monkeys (RMs) were given bolus injections of recombinantly expressed stabilized HIV Env trimer MD39 (ref. 15) formulated with alum adjuvant (50 µg of protein and 500 µg of alhydrogel adjuvant per side), reflective of how most licensed human protein vaccines are formulated and administered as a bolus (group 1) (Fig. 1a). In an effort to generate more robust GCs, we immunized two groups of RMs with MD39 Env trimer formulated with the new immune-stimulating complex-type adjuvant saponin/MPLA nanoparticle (SMNP)16 (groups 2 and 3) (Fig. 1a). Priming immunization for these two groups was administered via a slow-delivery vaccination method termed escalating dose 11, in which the total dose of MD39 plus SMNP formulation (50 µg of protein and 375 µg of adjuvant per side) was split between seven gradually increasing doses, delivered every other day for a total of 12 days (Extended Data Fig. 1a). Group 3 was designed with an unusually long prime period to assess the durability of GCs after primary immunization. Each animal in the study was immunized bilaterally, thereby doubling the number of lymph nodes (LNs) and GCs that could be tracked over time. GCs were sampled every 2–3 weeks by fine-needle aspiration (FNA)17 of inguinal LNs (ILNs) (Fig. 1a).
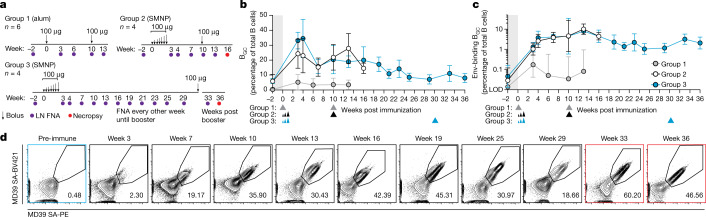
Fig. 1. GCs after priming immunization can last for more than 6 months.
a, Experimental schematic. b,c, Quantification of BGC cell kinetics. Triangles below indicate primary and booster immunizations. b, Quantification of total BGC cells as a percentage of total CD20+ B cells. c, Env-binding BGC cells as a percentage of total B cells. LOD, limit of detection. d, Representative flow cytometry plots of Env-binding BGC cells from left ILN of one long-prime group 3 animal over time. Red outline indicates post booster; gated on CD20+/CD71+CD38– BGC cells. Mean and s.d., or geometric mean and geometric s.d., are plotted depending on the scale in all figures unless otherwise stated. Left and right ILNs are graphed as independent data points (group 1, n = 12; groups 2 and 3, n = 8).
Extended Data Fig. 1. Escalating dose immunization strategy and representative flow cytometry analysis of FNA samples.
a, Priming escalating dose strategy and injection schedule. b, Gating strategy for the longitudinal analysis of GC-TFH and BGC cells from ILN FNA samples of Groups 2 and 3 animals. CD71+CD38−/ MD39+/+ cells were sorted for BCR sequencing and transcriptomic analyses. CD71+CD38−/MD39neg/neg cells were also sorted for weeks 3,4,7, and 10. c, Gating strategy for the longitudinal analysis BGC cells from ILN FNA samples of Group1 animals.
After priming immunization, conventional MD39 plus alum bolus-immunized monkeys (group 1) exhibited an increase in total BGC cell (CD71+CD38–) percentage at week 3 post immunization (Fig. 1b); the frequency of Env-binding BGC cells (CD71+CD38–Env+/+) peaked at week 3 and declined thereafter (Env-binding BGC cells as percentage of total B cells) (Fig. 1c). Both total and Env-binding BGC cells were substantially increased in MD39 plus SMNP ED-immunized RMs compared with those that received conventional protein plus alum bolus immunization (Fig. 1b,c and Extended Data Figs. 1b,c and 2a). Median peak BGC cell frequencies observed were 24–33% compared with 3.5% (P < 0.0001 by Mann–Whitney, week 3 groups 2 and 3 combined versus group 1; Fig. 1b). Median Env-binding BGC cell frequencies were around 7.8-fold greater at week 3 (P < 0.0025 by Mann–Whitney, groups 2 and 3 combined versus group 1) (Fig. 1c). Strikingly, in contrast to the conventionally primed group 1, frequencies of Env-binding BGC cells in groups 2 and 3 continued to increase, resulting in a 186-fold GC difference by week 10 (P < 0.0001 by Mann–Whitney, groups 2 and 3 combined versus group 1) (Fig. 1c).
Extended Data Fig. 2. BGC and GC-TFH kinetics.
a, Env-binding BGC cells as a percent of total CD20+ B cells. b, Env-binding BGC cells as a percent of total BGC cells. c, Ratio of PE geometric mean fluorescent intensity (gMFI) of MD39+/+ BGC cells to MD39neg/neg BGC cells calculated for Group 2 and 3 FNA samples at post prime time points. MD39+/+ (green gate) and MD39neg/neg BGC (magenta gate) gating is shown in Extended Data Fig. 1b. Data from only the LLN were analyzed for week 19-25 samples, since the RLN samples for these time points were analyzed on a different flow cytometer during the tracking period. d, Kinetics of GC-TFH cells for Group 2 & 3 animals. Gating strategy is shown in Extended Data Fig. 1b. e, Representative flow plot of an AIM assay to detect Env-specific GC-TFH cells after 18 hrs of ex vivo restimulation. Stimulation conditions: 15-mer overlapping Env peptide megapool (Env MP), staphylococcal enterotoxin (SEB), or unstimulated (unstim.). f, Env-specific GC-TFH cells (CXCR5hiPD-1hi, gated on FSC-A/SSC-A, FSC-H/FSC-W, SSC-H/SSC-W, Live/Dead Fixable e780−, CD4 AF700+B220 BV785−) detected at 6 wks post boost (Group 2: week 16, Group 3: week 36) quantified by AIM assay using the indicated pair of activation markers, or positive for any two of the three AIM markers (Or gates). Data shown is background (AIM+ signal from unstimulated samples) subtracted. Left and right ILNs are graphed as independent data points. Mean and SD or geometric mean and geometric SD are plotted depending on the scale. Limit of detection (LOD). Mann-Whitney test performed.
Tracking of priming immune response continued for group 3 monkeys beyond week 10 (group 1 and 2 monkeys were boosted at week 10) (Fig. 1a). GC responses were still active at weeks 13, 16, 21, 25 and 29 (Fig. 1b–d and Extended Data Fig. 2a,b). In group 2, the geometric mean fluorescent intensity of MD39 binding continued to increase beyond week 10, suggesting that MD39-specific BGC cells continued to gain affinity in the absence of a booster (Extended Data Fig. 2c). Env-binding BGC cell frequencies at week 29 were still 27-fold higher than the peak observed after conventional alum immunization, and they were also greater than the booster response to conventional alum immunization (Fig. 1c). At 191 days (27 weeks) after the end of the priming dose (29 weeks from day 0), median Env-binding BGC cell frequencies in ILNs were still around 49-fold higher than baseline (Fig. 1b–d and Extended Data Fig. 2a,b). Thus, GCs were present for more than 191 days with no further antigen introduced.
GC-T follicular helper (TFH) cells have a critical role in the recruitment and selection of BGC cells6. Total frequencies of GC-TFH cells in ILNs changed over the course of the priming period (Extended Data Fig. 2d). Env-specific GC-TFH cell frequencies were higher in group 3 after booster (Extended Data Fig. 2e,f)18; longitudinal quantitation was not possible due to the limiting availability of FNA samples. Long-lasting prime GCs may contribute to an improved antigen-specific GC-TFH response following booster immunization.
Enhanced antibody response quality
The Env-binding serum IgG titres of group 3 RMs remained stable from weeks 3 to 29 of the priming phase in the absence of a booster immunization (Fig. 2a). After boosting, group 2 and 3 monkeys generated similar peak binding antibody titres (2–3 weeks post booster) but group 3 monkeys maintained significantly higher Env-binding IgG titres at week 6 after the booster (Fig. 2b). The quality of the antibody response was evaluated for the ability to neutralize autologous tier-2 BG505 pseudovirus. Notably, autologous tier-2 neutralizing antibodies were detectable in all long-prime group 3 monkeys after only the priming immunization (geometric mean titre (GMT) about 170 at week 29) (Fig. 2c). All monkeys receiving escalting-dose immunization generated robust neutralizing antibody responses post booster (Fig. 2c,d); by contrast, only a single animal with conventional bolus immunization had detectable autologous neutralizing antibodies, which were of low titre (around 37) (Extended Data Fig. 3a). Peak observed autologous neutralizing GMTs in group 3 were all more than 2,000 (week 3 post booster) (Fig. 2c). Group 3 autologous neutralization titres were fourfold greater than those of group 2 at 6 weeks post booster (Fig. 2c,d). The titres described represent the most robust and consistent autologous tier-2 neutralizing antibody responses in RMs after two immunizations in any of our studies12,13,16.
Fig. 2. Long priming enhances antibody quality.
a, Env-binding serum IgG endpoint titres (ETs). b, Env-binding IgG titres following booster; triangles indicate the booster time point. c,d, BG505 HIV pseudovirus neutralization titres at 50% inhibition (ID50). Comparison of post-booster neutralization GMTs (d). GMT with geometric s.d. from four independent repeats, with duplicate wells per assay. b,d, Two-sided Mann–Whitney test was performed. e, Heterologous tier-2 virus neutralization titres. GMT with geometric s.d. from four independent repeats, with duplicate wells per assay. f, Heterologous tier-2 viruses neutralized (ID50 > 50) in the 12-virus global panel, after booster. Virus names are abbreviated (Methods). g, EMPEM of polyclonal plasma Fabs post booster. Group 1 at week 2 post booster; groups 2 and 3 at week 3 post booster; Env trimer is shown in grey. Fab colours coded as per epitope (h). h, Epitope sites recognized by EMPEM (g) for each animal (six possible sites per animal), tabulated for each animal group. Fisher’s exact test comparing epitopes recognized versus those not recognized between groups. Group 1, n = 6; groups 2 and 3, n = 4. **P < 0.01.
Extended Data Fig. 3. ED immunization using SMNP improves the quality of Env-specific antibody responses.
a, Group 1 autologous neutralization titers determined by assays performed by two independent labs (Scripps Research Institute [Scripps] or Duke) for week 12. ID50 < 20 is considered non-neutralizing (NN). b, Week 3 post-boost Group 2 and 3 serum neutralization ID50 titers determined independently from a second laboratory (Duke). NN: ID50 < 20. †Viruses for which serum neutralization was not determined for Group 3 animal DHHT due to limited serum availability. c, Number of heterologous tier-2 viruses neutralized out of 11 tested in (b). †Neutralization for only 6 viruses was tested for DHHT. d–e, Serum neutralization assays (Scripps) tested head-to-head with data shown in Fig. 2e using week 3 post-boost 2 sera from four RMs escalating-dose immunized with an ISCOM adjuvant (SMNP without MPLA) and BG505 Env (d)12, and week 2 post-boost 2 sera from six RMs bolus immunized with SMNP and MD39 Env (e)15. NN: ID50 < 30. f, Number of tier-2 heterologous viruses neutralized by post-boost 2 serum tested in d-e12,15. g, Number of tier-2 heterologous viruses neutralized with the indicated serum GMT titers (Scripps). h, Median tier-2 heterologous neutralization GMT across the 12-virus panel (Scripps).
Group 2 and 3 post-booster sera exhibited some of the broadest-observed tier-2 neutralizing antibody specificities elicited by Env trimer immunization, with most monkeys from group 3 exhibiting greater breadth than those in group 2 (Fig. 2e,f). The 12-virus global neutralization panel was repeated by an independent laboratory, with similar observation of neutralization breadth (Extended Data Fig. 3b,c). In head-to-head assays, limited tier-2 neutralization breadth was observed with serum from a previous RM study with escalating-dose immunization and two booster immunizations using a similar Env trimer (Olio6) and an earlier immune-stimulating complex adjuvant (SMNP without MPLA)13 (Extended Data Fig. 3d,f–h), and likewise for serum from an RM study with three bolus immunizations of MD39 Env trimer and SMNP (Extended Data Fig. 3e–h)16.
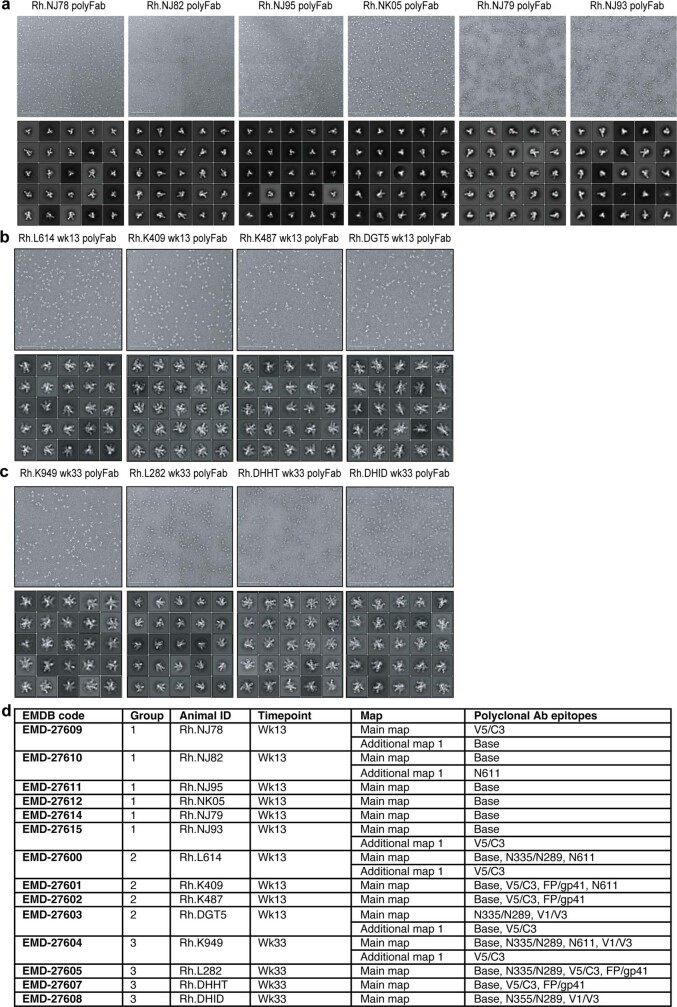
Most of the HIV Env-binding B cells and antibodies are usually directed to immunodominant non-neutralizing epitopes, such as the base of soluble recombinant Env trimers1,13,19–21. Electron microscopy polyclonal epitope mapping (EMPEM)21 of circulating antibodies showed that the number of targeted epitopes was significantly increased in groups 2 and 3 than in group 1 (P = 0.0066) (Fig. 2g,h and Extended Data Fig. 4), which correlated with autologous neutralizing titres (Fig. 2c (post booster) and Extended Data Fig. 3a). Group 2 and 3 monkeys generated antibody responses to V5/C3 and V1/V3 epitopes (Fig. 2g,h) associated with autologous BG505 SHIV protection22. Antibody responses in conventional bolus-plus-alum-immunized monkeys were largely restricted to the Env trimer base (Fig. 2g,h). In summary, the priming immunization strategy used was associated with substantially improved epitope breadth and quality of neutralizing antibodies.
Extended Data Fig. 4. EMPEM analysis of polyclonal antibodies.
a–c, negatively stained EM micrographs of MD39 Env trimer: polyclonal Fab complexes (top), and 2D-class averages (bottom). Data for RMs in Group 1 (a), Group 2 (b), and Group 3 (c). The scale bar shown in the lower left corner of each micrograph corresponds to 200 nm. d, Electron microscopy data bank (EMDB) codes corresponding to each map generated from EMPEM analysis.
Six month BGC cells are highly functional
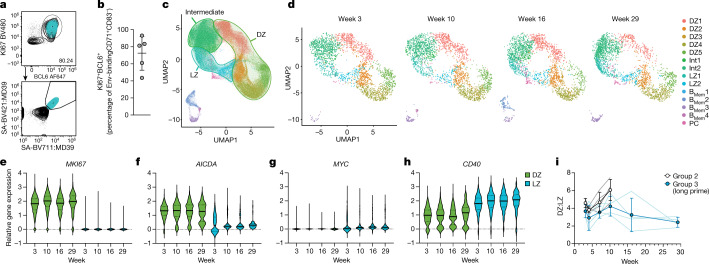
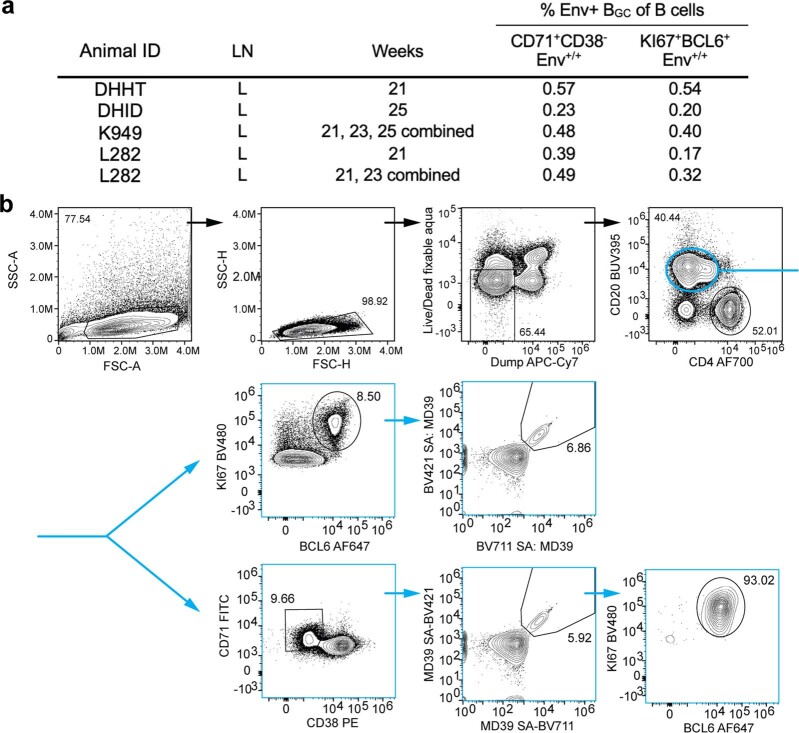
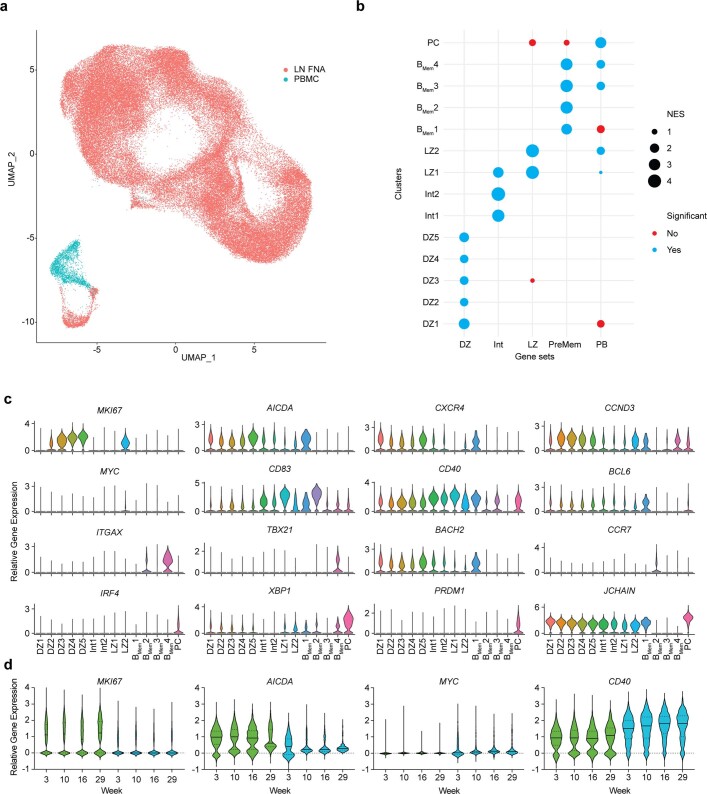
The characteristics of BGC cells after priming were interrogated in greater detail to assess their functionality over time. BCL6 is the lineage-defining transcription factor of BGC cells and is essential for their functionality7. KI67 (encoded by MKI67) marks rapidly dividing cells. LN B cells from months 5 to 6 (weeks 21–25) were stained for BCL6 and KI67 protein. On average, about 72% of Env-binding CD71+CD38– BGC cells were KI67+BCL6+ (Fig. 3a,b and Extended Data Fig. 5), indicating retained BGC programming and proliferation. To further ascertain the phenotypic and functional characteristics of BGC cells at different time points, single-cell transcriptional profiling was done for about 70,000 cells from LN FNAs of weeks 3, 4, 7, 10, 13, 16, 29 and 33, predominantly consisting of Env-binding BGC cells as well as peripheral blood-sorted Env-binding memory B (Bmem) cells from weeks 16 (group 2) and 36 (group 3). Dark-zone and light-zone cell clusters were clearly observed among LN B cells when analysing all time points together (Fig. 3c and Extended Data Fig. 6). We then examined BGC cell transcriptional profiles over the course of group 3 RM long priming period (weeks 3, 7, 16 and 29; Fig. 3d–h and Extended Data Fig. 6)23,24. Light- and dark-zone states were sustained across the 6 month period (Fig. 3d). Expression of key functional BGC genes MKI67, AICDA, MYC and CD40 was maintained over time, and was compartmentalized comparably between dark- and light-zone cell types at all time points (Fig. 3e–h and Extended Data Fig. 6d). The ratio of dark-zone to light-zone cells remained relatively consistent over the course of the priming period (Fig. 3i). Overall, antigen-specific BGC cells possessed stable phenotypic characteristics over a 6 month period, indicative of long-term maintenance of functional BGC cell properties in the absence of further immunization.
Fig. 3. BGC cell phenotypic and functional characteristics over the course of 6 months.
a, Representative flow cytometry gating showing CD71+CD38– and Ki67+BCL6+ BGC cells (see Extended Data Fig. 5a for sample information). Back-gating of CD71+CD38–Ki67+BCL6+Env+/+ BGC cells is shown in cyan. b, Frequency of KI67+BCL6+ cells among CD71+CD38–Env+/+ BGC cells. c, Uniform manifold approximation and projection (UMAP) visualization of single-cell gene expression profiles identifying clusters of B cell states from LN FNAs and peripheral blood mononuclear cells (PBMCs). d, Per time point UMAP plots extracted from c. e–h, Relative gene expression of MKI67 (e), AICDA (f), MYC (g) and CD40 (h) in dark-zone (DZ) and light-zone (LZ) subclusters representative of classical DZ and LZ phenotypes DZ5 and LZ1. Additional data given in Extended Data Fig. 6. i, The dark-zone to light-zone ratio, as determined by single-cell clustering in LNs after priming.
Extended Data Fig. 5. Intracellular staining of BGC markers.
a, Some left ILN samples were pooled for the intracellular staining panel, for higher cell numbers. b, Representative flow plot and complete gating strategy for BCL6 and KI67 staining.
Extended Data Fig. 6. Single cell transcriptional profiling of B cells.
a, Single cell transcripts of LN FNA BGC (Group 2 and 3 weeks 3, 4, 7,10 and 16, Group 2 week 13, Group 3 weeks 29 and 33) and PBMC Bmem cells (Group 2 week 16, Group 3 week 36) were assessed. b, A summary of the results from GSEA of upregulated gene profiles from single cell clusters using previously identified B cell subset gene signatures24. Size of the dots represent the normalized enrichment score (NES). Significant NES results are shown in blue, nonsignificant results in red. c, Expression levels of the displayed genes used to help identify clusters of B cell subsets. d, Relative gene expression of MKI67, AICDA, MYC, CD40 in combined DZ and LZ clusters.
BGC cell receptor evolution lasts for months
To directly assess the functionality of GCs over these extended time periods, multiple experimental approaches were used comparing group 2 and 3 RMs. B cell receptor (BCR) sequencing of Env-binding LN FNA-derived BGC cells was performed from nine time points to assess antibody somatic hypermutation (SHM) over time, as well as clonal diversity and mutational patterns in clonal lineages (Fig. 4 and Extended Data Figs. 7–10). Env-binding BGC cell heavy-chain (HC) nucleotide mutations increased significantly between weeks 3 and 10 (P < 0.0001 by Mann–Whitney for groups 2 and 3; Fig. 4a,b). Notably, BGC cells continued to accumulate mutations in the absence of further immunization through to week 29 in group 3 RMs, at which point the median number of HC mutations was 17, with the top 25% of BGC cells containing 22–45 HC mutations (Fig. 4a,b). The difference in SHM in the long prime (week 29) versus 10 week prime was highly significant (P < 0.0001 by Mann–Whitney, Fig. 4b; P < 0.0009, Fig. 4c), and the difference in median mutations between weeks 10 and 29 was as great as that between weeks 4 and 10, indicative of robust GC functionality continuing through to at least week 29 (Fig. 4a–c). The proportion of unmutated Env-binding BGC cells dropped over time, with 0.19–0.42% unmutated cells by week 7 (Fig. 4b). Substantial mutations were also observed in light chains (LC), with patterns comparable to HCs (Extended Data Fig. 7a–d). Env-binding BGC cells showed a gradual reduction in estimated clonal richness over time (Chao1) (Fig. 4d and Extended Data Fig. 7e), with sustained diversity (Simpson’s diversity index) (Extended Data Fig. 7f,g).
Fig. 4. Antigen-specific BGC and Bmem cell SHM and affinity maturation.
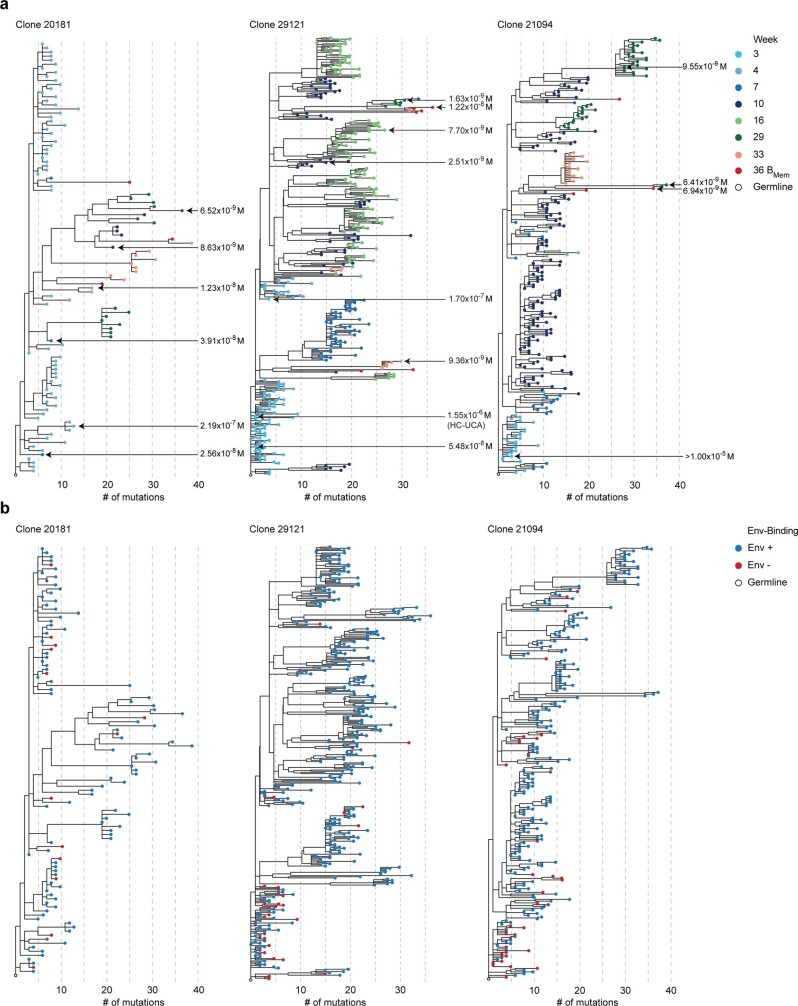
a, Nucleotide mutations in the V- and J-gene region of HC sequencesderived from Env-binding BGC cells after priming; spaghetti plots track mutations per animal (n = 8). b, Pre-booster (lighter colour) and post-booster (darker colour) nucleotide mutations in Env-binding BGC cells. The median and quartiles are denoted by thick and dotted lines, respectively. c, BGC mutations at the last pre-booster time point and 3 weeks post booster. Two-way analysis of variance (ANOVA) multiple comparisons test. d, BGC clonal richness at post-prime time points (Chao1). e, Frequency of Env-binding Bmem cells in blood. Two-way ANOVA multiple comparisons test. f, Clonal richness of Bmem cells after boosting (Chao1). g, HC mutations in week 6 post-booster Bmem cells. h, Env-binding Bmem cells that do not bind the trimer base. i, Serum ELISA titres of Env non-base-directed IgG. b,f,g,i, Two-sided Mann–Whitney test; c,e,h, two-way ANOVA multiple comparisons test. j, Representative clonal lineage trees from two group 3 monkeys (DHTT and L282), colour coded by time points (top) and Env binding (bottom). k, Clonal lineage 20181, colour coded by time point (top) and sample anatomical location (bottom). Clone 20181 has a 14-amino acid H-CDR3 with two or more N additions, and was represented by ten or more cells per LN. Each ring indicates five estimated HC nucleotide mutations from the most recent common ancestor.
Extended Data Fig. 7. BCR sequences of BGC and Bmem cells.
a–b, Number of nucleotide (NT) mutations in the V and J-gene region of LC sequences (Kappa: LC κ, Lambda: LC λ) derived from Env-specific BGC cells after priming. Spaghetti plots track the number of mutations in each animal. c, Number of HC mutations at week 6 post-boost. d, Number of LC κ and LC λ nucleotide mutations in BGC cells, respectively. There are two outliers among Group 3 week 3 LC κ sequences, each with 108 and 105 mutations. e, BGC cell pre-boost clonal richness. Pre-boost; week 10 and 29 for Group 2 & 3 respectively (Chao1). f, BGC population diversity at post-prime time points (Simpson’s diversity). g, BGC cell pre-boost population diversity. Pre-boost; week 10 and 29 for Group 2 & 3 respectively (Simpson’s diversity). h, Gating strategy for the detection of Env-specific Bmem cells. i, Diversity of of Bmem cells after boosting. Post-boost; week 16 and 36 for Group 2 & 3 respectively (Simpson’s diversity). j, Number of LC κ and LC λ nucleotide mutations in wk 6 post-boost Bmem cells. Mann-Whitney tests are performed.
Extended Data Fig. 10. Examples of clonal lineages with unique features.
a, A clonal lineage where a substantial fraction of early BGC cells did not bind Env trimer by flow cytometry. Clone 5491: H-CDR3 15 AAs in length, >2 N additions, found in one LN. b, Another clonal lineage with a substantial fraction of BGC cells that did not detectable bind Env trimer by flow cytometry. Clone 29183: H-CDR3 8 AA in length, >2 N additions, >10 cells per LN. This clone was excluded from LN migration quantitation analysis as it did not meet the required H-CDR3 length criteria. c, An example of a clonal lineage that was almost exclusively IgM. Clone 29183: H-CDR3 14 AA in length, >2 N additions,>10 cells per LN. In (a), (b) and (c), each B cell is labeled according to observed time point (left column), Env-binding by flow cytometry (center column), and anatomical sampling location (right column). d, Lineage tree 21094 shown in Fig. 4j by anatomical sampling location. Clone 21094: H-CDR3 14 AAs in length, >2 N additions, >10 cells per LN. e, Lineage tree 29121 shown in Fig. 4j by sampling location. Clone 29121: H-CDR3 18 AAs in length, >2 N additions, found in one LN. f, H-CDR3 length distribution of clones found in one or both ILNs using a broader definition of clonal lineages (H-CDR3 length > 10 AA, > 5 cells in lineage). Lineages found in a single LN and two distal LNs exhibited similar H-CDR3 length distributions. One hypothetical concern about the definition of clonality was the possibility of two independent naïve B cells having identical H-CDR3 and L-CDR3 recombination events. If that were the driving phenomenon behind the observation of matching BGC cells in distant LNs, based on recombination event likelihoods it would be expected that length distributions would be skewed towards shorter lengths and no N-additions would be observed. g, iNext42 plots showing the results of rarefaction analysis to determine the extent of sequence coverage from the Env+/+ LN FNA and PBMC VDJ sequence data.
After booster immunization, HC mutations increased in Env-binding BGC cells of both group 2 and 3 RMs, with the highest overall number of mutations in long-primed RMs (week 3 post-booster HC median mutations 13 versus 20) (Fig. 4b,c and Extended Data Fig. 7c). Comparable observations were made for LC mutations (Extended Data Fig. 7d).
Pre-booster Env-binding Bmem cell (CD20+IgD–Env+/+) frequencies in peripheral blood were equivalent in RMs from groups 2 and 3 (Fig. 4e and Extended Data Fig. 7h). Boosting increased Env-binding Bmem frequencies in both groups (Fig. 4e). RMs with the long prime had more highly mutated Bmem cells and greater clonal richness among Bmem cells (Fig. 4f,g), with equivalent Simpson’s diversity indices (Extended Data Fig. 7i). This was also reflected in a significant shift away from immunodominant, base-binding, Env-specific Bmem cells following boosting of the long-primed group, which was not seen in group 2 (Fig. 4h), and this phenomenon was also reflected in circulating antibody titres (Fig. 4i).
Clonal lineage analysis of paired BCR sequences was used to examine GC duration and functionality over several months after single immunization. Many clonal lineages were identified, with BGC clones observed at the first and last LN FNA time points post prime and extensive SHMs between (Fig. 4j,k and Extended Data Figs. 8–10). Some clonal lineages were identified with BGC clones observed at every, or nearly every, LN FNA time point, providing direct evidence of BGC cell persistence over 29 weeks (Fig. 4j,k and Extended Data Figs. 8, 9 and 10a,c). Diversification of clones in clonal lineages was apparent. Daughter clones most evolutionarily divergent from the germline were typically present at late time points (mutations to time correlation: lineage 21094, R2 = 0.72, P < 0.001; lineage 29121, R2 = 0.68, P < 0.001) (Fig. 4j and Extended Data Figs. 8 and 9c,d).
Extended Data Fig. 8. Examples of BCR clonal lineages with increasing affinities.
a, Clonal lineages shown in Fig. 4j, k, observing clones over different LN FNA and PBMC sampling periods, represented as linear phylogenic trees. MD39 Env binding KDs (M) were evaluated by surface plasmon resonance (SPR) for select monoclonal antibodies indicated by the black arrows. HC-UCA in clone 29121 refers to the HC having an identical AA sequence to the UCA. b, Linear phylogenic trees shown in (a) labeled by Env-binding by flow cytometry. These phylogenies are also shown as circle plots in Fig. 4j. The grey dotted lines indicate 5 estimated HC nucleotide mutations from the most recent common ancestor.
Extended Data Fig. 9. Additional examples of longitudinally assessed BCR clonal lineages.
a, Example of a clonal lineage observed over multiple different LN FNA and PBMC sampling time points. Clone 29431 has a 9 AA H-CDR3, contains >2 N additions and <10 cells per LN (This clone was excluded from LN migration quantitation as it did not meet either the stringent or relaxed H-CDR3 length criteria). b, Another clonal lineage observed over different LN FNA and PBMC sampling time points. Clone 6911 has an 11 AA H-CDR3, contains >2 N additions and >10 cells per LN. In (a) and (b), each B cell is labeled according to observed time point (left column), Env-binding by flow cytometry (center column), and sampling location (right column). Each ring indicates 5 estimated HC nucleotide mutations from the most recent common ancestor. c–d, Pearson correlation calculated between number of estimated HC nucleotide mutations gained from the most recent common ancestor, and time post-prime. Correlation coefficients are calculated for clones 21094 (c) and 29121 (d) shown in Fig. 4j.
For some lineages, such as 5491 and 29183, most early BGC cell clones did not bind Env by flow cytometry (Env–/–) but BGC clones from later weeks and Bmem clones bound Env by flow cytometry (Env+/+), indicative of a clone that started with low affinity to Env and that subsequently matured (Extended Data Fig. 10a,b). To directly assess affinity maturation within these lineages, monoclonal antibodies from several time points in three large clonal lineages were generated. An early monoclonal antibody from lineage 20181 had relatively low affinity to MD39 (week 4, dissociation constant (Kd) = 2.19 × 10−7 M). An increase in affinity of roughly 17-fold (Kd = 1.23 × 10−8 M) was observed by week 16 and a further (roughly) twofold increase by week 29 (Kd = 6.52 × 10−9 M), representing around a 30-fold overall increase in affinity (Extended Data Fig. 8a). In lineage 29121, the antibody with the lowest affinity at week 3 (Kd = 1.55 × 10−6 M) used an HC identical to the amino acid sequence of the unmutated common ancestor (UCA) (Extended Data Fig. 8a). In this lineage, almost one-third of the cells found at week 3 did not bind MD39 via flow cytometry, including the UCA HC clone (Extended Data Fig. 8b), which indicates that this lineage started with a very weak affinity and was still able to participate substantially in the GC reaction. In the same lineage, antibodies were isolated at weeks 16 and 29 with affinities about 200-fold (Kd = 7.70 × 10−9 M) and 950-fold (Kd = 1.63 × 10−9 M) higher than the week 3 HC UCA clone (Extended Data Fig. 8a). Finally, a low-affinity lineage 21094 antibody (week 3, Kd > 1.00 × 10−5 M; Env–/– by flow cytometry) had gained over a 1,000-fold improvement in affinity by week 29 (Kd = 9.55 × 10−9 M, 6.49 × 10−9 M) (Extended Data Fig. 8a). Altogether, these data strongly support the conclusion that substantial affinity maturation occurs over the course of the 6 month priming period, leading in some cases to BGC cells with around 1,000-fold improved affinity.
Bmem cells were commonly represented in clonal lineage trees among multiple sublineages, including among the most mutated branches observed at late LN FNA BGC time points (Fig. 4j,k and Extended Data Figs. 8, 9a,b and 10a), demonstrating that GCs productively output highly mutated Bmem cells. Clonal lineages with other interesting features were observed among long-prime GCs, including lineage 21275, which was almost exclusively IgM from weeks 3 to 29 (Extended Data Fig. 10c).
There were clonal lineages with clones detected in both left and right ILN at different time points during the long-primed GCs (Fig. 4k and Extended Data Figs. 9a,b and 10b,c). Out of 169 clonal lineages passing stringent criteria (HC complementarity-determining region 3 (H-CDR3) length greater than 14, multiple N additions in H-CDR3, more than 20 cells in total and a matching LC), 11 lineages were observed in both LNs (ten or more cells per LN) at different time points, providing evidence that Bmem cells generated from GCs in one LN can exit, recirculate and enter ongoing GCs in another LN. Using less restrictive criteria of clonal lineages containing at least five cells with H-CDR3 length greater than 10 and a matching LC, we observed that 54 out of 889 clonal lineages could be found in bilateral LNs (Extended Data Fig. 10f).
In summary, key features observed in numerous antigen-specific clonal lineages provide direct evidence of BGC cell persistence over 29 weeks, continuous accumulation of somatic mutations at substantial rates, affinity maturation, clonal migration and seeding of the peripheral Bmem cell compartment which, together show that, under select conditions, GCs are able to undergo clonal evolution for extremely long durations without new antigen exposure.
Discussion
Long-lasting GCs have classically been observed in the context of chronic infections and gut microbiota exposure25,26, conditions known to have continuous live sources of renewed antigen. Recent reports of longer-lasting GCs in influenza infection27, SARS-CoV-2 infection28 and human RNA vaccines29–31 have raised renewed interest in the possibility of long-lasting GCs potentially under conditions of low or absent renewed antigen exposure. Here we demonstrate clearly that GCs can last for at least 191 days in the absence of new antigen, using an experimental system taking advantage of protein immunization, a 12-day slow-delivery immunization strategy, a robust adjuvant and the use of Env probes to identify antigen-specific BGC cells. Furthermore, we demonstrate that GCs are remarkably robust and functional for 6 months. BGC cells maintain proliferation, SHM and affinity maturation, and long-lasting GCs can produce high autologous tier-2 neutralizing antibody titres, heterologous neutralizing antibody titres and highly somatically mutated circulating Env-specific Bmem cells to non-Env base epitopes.
A 12- to 14-day slow-delivery immunization regimen can result in substantially greater capture of vaccine antigens by stromal follicular dendritic cells13. Observation of GCs for over 6 months indicates that endocytic recycling of immune complexes by follicular dendritic cells32 can be surprisingly efficient at maintaining proteins in GCs and protecting them from damage. One probable mechanism of slow-delivery enhancement of GCs is improved immune complex formation, due to antigen supply after the start of the antibody response. Given the immunodominance of antibody responses to the non-neutralizing base of the Env trimer after conventional immunizations, and epitope diversification to non-base epitopes in slow-delivery immunizations (Fig. 2g), we speculate that immune complexes with Env under slow-delivery conditions are primarily composed of base-binding antibodies, which shield the base of the Env trimer in GCs and orient the trimers to better demonstrate neutralizing epitopes antipodal to the base, thereby enriching for neutralizing antibody B cells. This is further illustrated by the shift away from base-directed immunodominance in Bmem cells during the long prime (Fig. 4h). Thus, the improved autologous and heterologous neutralization noted in group 3 monkeys is probably partly owing to the diversity of B cells recruited and partly due to increased affinity maturation from extensive GC responses. These outcomes match our prediction that immunodominance is a major factor that effects neutralizing antibody responses to candidate HIV vaccines, and the immunodominance of non-neutralizing antibody responses can be overcome by recruiting more diverse B cells to GCs and providing those cells with sufficient time for affinity maturation to become competitive as Bmem cells with non-neutralizing Bmem cells at the time of booster immunization1. It would be of interest to investigate the potential effects of extending the timing between bolus booster immunizations.
The appearance of neutralizing titres in the long-prime group 3 between weeks 20 and 29 after prime is indicative of continued affinity maturation, leading to GC output of new plasma cells that can produce neutralizing antibodies months after the initial immunization. Regarding the kinetics of B cell memory development, our data are inconsistent with a model of predominantly early Bmem cell production from continuing GCs. If that were the case, median Bmem cell HC SHMs would be low (for example, two to five mutations), corresponding to early BGC cells (3–4 weeks). By contrast, median Bmem cell HC SHM was 17 at week 36 (Fig. 4g) and less than 15% of Bmem cells had fewer than 10 mutations. The simplest interpretation of these observations is that early Bmem cells may re-enter GCs to undergo further SHM and affinity maturation. Our finding of shared BGC clones between left and right LNs and circulating Bmem cells is most consistent with this Bmem GC re-entry model. In addition, these data are inconsistent with a model of BGC cell extinction after around 1 month and dominant replacement by incoming naive B cells, because unmutated B cells were rare in GCs after 1 month (Fig. 4b) and median BGC cell SHM levels continued to increase over 6 months. BGC cell clonal lineages were also observed to span the full 6 months after priming. Overall, our data are consistent with ongoing production or recycling of Bmem cells and the production of plasma cells from ongoing GCs over the course of the priming period.
We have shown that GCs can persist for more than 6 months in response to a priming immunization, with several notable outcomes. These findings indicate that patience can have value in allowing antibody diversification and evolution in GCs over extended periods of time, and this long-prime, slow-delivery immunization approach holds promise for difficult vaccine targets.
Methods
Protein expression and purification
BG505 MD39 SOSIP Env trimers (MD39) were co-expressed with furin in HEK293F cells and expressed as previously described15. Trimers used for immunizations were expressed tag-free and quality checked for low endotoxin levels. BG505 MD39 SOSIP and BG505 MD39-base knockout trimers used as baits in flow cytometry were expressed with a C-terminal Avi-tag and biotinylated using a BirA biotinylation kit according to the manufacturer’s instructions (Avidity). The BG505 MD39-base knockout trimer had the following mutations relative to the BG505 MD39 SOSIP: A73C, R500A, P561C, C605T, S613T, Q658T, L660N, A662T and L663C.
Monoclonal antibodies were generated by cloning synthesized Fab variable-region genes into human antibody expression vectors. HCs were expressed as human IgG1. Antibody expression plasmids and recombinantly expressed monoclonal antibodies were produced by Genscript.
Animals and immunizations
For MD39 plus SMNP escalating-dose immunization groups, Indian rhesus macaques (Macaca mulatta) were housed at Alpha Genesis and treated in accordance with protocols approved by the Alpha Genesis Animal Care and Use Committee (IACUC). Two females and two males, matched for age and weight, were assigned to each experimental group. Monkeys were aged 2–3 years at the time of the priming immunization. All immunizations were given subcutaneously in the left and right mid-thigh with a total dose of 50 µg of MD39 and 375 µg of SMNP each side. For priming, a 12-day escalating dose strategy was used (Extended Data Fig. 1a)13.
For the MD39 plus alum bolus group, RMs were housed at the Tulane National Primate Research Center as part of a larger NHP study (I.P. et al., manuscript in preparation). This study was approved by the Tulane University IACUC. Animals were grouped together to match age, weight and gender. Animals were aged 3.5–5.0 years at the time of first immunization, with three females and three males in the study group. All immunizations were given subcutaneously in the left and right mid-thigh with 50 µg of MD39 and 500 µg of alum (alhydrogel adjuvant 2%; InvivoGen) per side. All animals were maintained in accordance with NIH guidelines.
LN FNA
FNAs were used to sample the left and right ILNs and were performed by a veterinarian. Draining LNs were identified by palpation. A 22-gauge needle attached to a 3 ml syringe was passed into the LN up to five times. Samples were placed in RPMI containing 10% (v/v) fetal bovine serum (FBS) and 1× penicillin/streptomycin (pen/strep). Samples were centrifuged, and ammonium chloride-potassium lysing buffer was used if the sample was contaminated with red blood cells. Samples were frozen down and maintained in liquid nitrogen until analysis.
Flow cytometry and sorting
Frozen FNA or PBMC samples were thawed and recovered in 50% (v/v) FBS in RPMI. Recovered live cells were enumerated and stained with the appropriate staining panel. MD39 and MD39-base knockout baits were prepared by mixing biotinylated MD39 with fluorophore-conjugated streptavidin (SA) in small increments at room temperature (RT) in an appropriate volume of 1× PBS over the course of 45 min. MD39 and streptavidin were added to the cells for 20 min, after which the antibody master mix was added for a further 30 min at 4 °C. Where knockout baits were used, these were first added to the cells for 20 min, then WT MD39 and streptavidin baits and were added for a further 20 min, followed by the addition of the remainder of the staining panel for a further 30 min at 4 °C, similar to a previously described protocol13. Fully supplemented RPMI (R10; 10% (v/v) FBS, 1× pen/strep, 1× GlutaMAX) was used as FACS buffer. For sorting, anti-human hashtag antibodies (BioLegend) were individually added to each sample at a concentration of 2.5 µg per 5 million cells at the time of addition of the master mix. Group 1 samples were sorted on a FACSFusion (BD Biosciences); those from groups 2 and 3 were either acquired or sorted on a FACSymphony S6 (BD Biosciences). Indexed V(D)J, Feature Barcode and GEX libraries of sorted LN FNA samples were prepared according to the protocol for Single Indexed 10X Genomics V(D)J 5' v.1.1, with Feature barcoding kit (10X Genomics). For sorted PBMC samples, Indexed V(D)J, Feature Barcode and GEX libraries were prepared using the Dual Indexed 10X Genomics V(D)J 5’ v.2 with Feature barcoding kit (10X Genomics). Custom primers were designed to target RM BCR constant regions. Primer set for PCR 1: forward, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC; reverse, AGGGCACAGCCACATCCT, TTGGTGTTGCTGGGCTT, TGACGTCCTTGGAAGCCA, TGTGGGACTTCCACTGGT, TGACTTCGCAGGCATAGA. Primer set for PCR 2: forward, AATGATACGGCGACCACCGAGATCT; reverse, TCACGTTGAGTGGCTCCT, AGCCCTGAGGACTGTAGGA, AACGGCCACTTCGTTTGT, ATCTGCCTTCCAGGCCA, ACCTTCCACTTTACGCT. Forward primers were used at a final concentration of 1 µM and reverse primers at 0.5 µM, each per 100 µl of PCR reaction. Libraries were pooled and sequenced on a NovaSeq Sequencer (Illumina) as previously described33.
During the long-prime tracking phase (weeks 16–25, right LNs), samples were stained as described above, fixed in BD Cytofix (BD Biosciences) then analysed on a FACSCelesta (BD Biosciences). For intracellular staining, cells were stained as described above then fixed with the Foxp3/Transcription factor staining kit (Invitrogen). Cells were washed with 1× diluted permeabilization buffer then stained for 1 h with antibodies targeting transcription factors of interest. Cells were washed and analysed on a Cytek Aurora (Cytek Biosciences). All flow cytometry data were analysed in Flowjo v.10 (BD Biosciences).
For LN FNA data inclusion in GC gating, a threshold of 250 B cells in the sample was used and, for Env-binding GC B cell gating, a threshold of 75 GC B cells was used. Any sample with fewer than 75 GC B cells but with a B cell count of more than 500 cells was set to a baseline of 0.001% Env+ GC B cells (percentage of B); otherwise, the limit of detection was calculated based on the median of (3/(number of B cells collected)) from pre-immunization LN FNA samples. Some group 2 and 3 LN FNA vials had barely detectable cells in the stained samples and were not analysed.
The following reagents were used for staining: Alexa Fluor 647 streptavidin (Invitrogen), BV421 streptavidin (BioLegend), BV711 streptavidin (BioLegend), PE streptavidin (Invitrogen), Live/Dead fixable aqua (Invitrogen), Propidium iodide (Invitrogen), eBioscience Fixable Viability Dye eFluor 780 (Invitrogen), mouse anti-human CD20 BV785, BUV395, Alexa Fluor 488, PerCP-Cy5.5 (2H7, BioLegend), mouse anti-human IgM PerCP-Cy5.5, BV605 (G20-127, BD Biosciences), mouse anti-human CD4 BV650, Alexa Fluor 700 (OKT4, BioLegend), mouse anti-human PD1 BV605 (EH12.2H7, BioLegend), mouse anti-human CD3 BV786, APC-Cy7 (Sp34-2, BD Biosciences), mouse anti-human CXCR5 PE-Cy7 (MU5UBEE, ThermoFisher), mouse anti-human CD71 PE-CF594 and FITC (L01.1), mouse anti-human CD38 PE, APC (OKT10, NHP Reagents), mouse anti-human CD8a APC-eFluor 780 (RPA-T8, ThermoFisher), mouse anti-human CD14 APC-Cy7 (M5E2, BioLegend), mouse anti-human CD16 APC-Cy7 (3G8, BioLegend), mouse anti-human CD16 APC-eFluor 780 (ebioCD16, Invitrogen), mouse anti-human IgG Alexa Fluor 700, BV510, and BV786 (G18-145, BD Biosciences), mouse anti-NHP CD45 BUV395 (D058-1283, BD Biosciences), mouse anti-human BCL6 Alexa Fluor 647 (K112-91, BD Biosciences), mouse anti-human KI67 BV480 (B56, BD Biosciences), mouse anti-human FoxP3 BB700 (236A/E7, BD Biosciences), mouse anti-human CD27 PE-Cy7 (O323, BioLegend), goat anti-human IgD FITC (polyclonal, Southern Biotech), Armenian hamster anti-mouse/human Helios PE/Dazzle 594 (22F6, BioLegend), TotalSeq-C anti-human Hashtag antibody 1-8 (LNH-94 and 2M2, BioLegend) and TotalSeq-C0953 PE Streptavidin (BioLegend).
Detection of antigen-specific GC-TFH cells
Antigen-induced marker-based identification of Env-specific GC-TFH cells was performed as previously described13,18. In summary, cells were thawed in 50% (v/v) FBS in RPMI and resuspended in 500 µl of DNase in R10 (100 µl of DNAse in 900 µl of R10) for 15 min at 37 °C in a CO2- and humidity-controlled incubator, 5 ml of R10 was added and cells were further rested for 3 h. Cells were enumerated and seeded at about 1 million per well in R10, and incubated with a final concentration of 2.5 µg ml–1 MD39 Env peptide pool, 10 pg ml–1 staphylococcal enterotoxin or media only (unstimulated) for 18 h at 37 °C in a CO2- and humidity-controlled incubator. Mouse anti-human CXCR5 PE-Cy7 (1:100, MU5UBEE; ThermoFisher) was added to each well at the start of stimulation. Cells were washed and stained for 45 min in the dark at 4 °C. After staining, cells were washed and fixed with BD Cytofix (BD Biosciences) and analysed on a BD FACSCelesta (BD Biosciences). The following antibodies were used in the flow panel: mouse anti-human CD4 Alexa Fluor 700 (OKT4, BioLegend), mouse anti-human CD20 BV785 (2H7, BioLegend), mouse anti-human PD1 BV605 (EH12.2H7, BioLegend), mouse anti-human CXCR5 PE-Cy7 (MU5UBEE, ThermoFisher), mouse anti-human CD134 PE (L106, BD BioLegend), mouse anti-human 4-1BB APC (4B4-1, BioLegend), mouse anti-human CD25 FITC (BC96, BioLegend), mouse anti-human CD16 APC-eFluor 780 (ebioCD16, Invitrogen), mouse anti-human CD8a APC-eFluor 780 (RPA-T8, ThermoFisher), mouse anti-human CD14 APC-Cy7 (M5E2, BioLegend) and eBioscience Fixable Viability Dye eFluor 780 (Invitrogen).
Neutralization assays
Pseudovirus neutralization assays at Scripps were performed as previously described12. BG505 pseudovirus neutralization was tested using the BG505.W6M.ENV.C2 isolate with the T332N mutation to restore the N332 glycosylation site. Assays were done with duplicate wells per assay and four independent repeats were performed. Heterologous neutralization breadth was tested on a panel of 12 cross-clade isolates, representative of larger virus panels isolated from diverse geography and clades34. In some figures virus names are abbreviated as follows: 398-F1_F6_20: 398F1; 246-F3_C10_2: 246F3; CNE55: not abbreviated; CNE8: not abbreviated; X2278-C2-B6: X2278; TRO.11: not abbreviated; BJOX002000.03.2: BJOX2000; CH119.10: not abbreviated; Ce703010217_B6: Ce0217; Ce1176_A3: Ce1176; 25710-2.43: 25710; X1632_S2_B10: X1632. Heterologous neutralization breadth assays at Scripps were performed with duplicate wells per assay, and four independent repeats of heterologous neutralization breadth assays. The cut-off for neutralizing serum dilution was set at 1:30 or 1:20 depending on the starting serum dilution. For Fig. 2c,d, an ID50 of less than or equal to 30 was considered non-neutralizing. Absolute ID50 values were calculated using normalized relative luminescence units and a customized nonlinear regression model:
with the bottom constraint set to 0 and top constraint set to the <100 model in Prism 8 (GraphPad). Pseudovirus neutralization assays at Duke were performed as previously described35. Positive controls (monoclonal antibodies) were included for every virus in every assay run and tracked as part of assay quality control, as well as a murine leukemia virus negative control. Duke samples were assayed in duplicate and assays were performed with good clinical laboratory practice compliance.
ELISA
Plasma samples were thawed, heat inactivated at 56 °C for at least 30 min and spun down. Corning 96-well half-area plates were coated overnight with streptavidin at 2.5 µg ml–1. Plates were washed three times with wash buffer (PBS, 0.05% (v/v) Tween-20) then coated with biotinylated MD39 or MD39-base knockout trimers at 1 µg ml–1. After washing three times with wash buffer, plates were blocked with blocking buffer (PBS, 3% (w/v) BSA) for 1 h at RT. Plasma serially diluted in blocking buffer was allowed to bind with trimers for 1 h at RT. Plates were washed three times and incubated with goat anti-rhesus IgG-HRP antibody (Southern Biotech, 1:10,000 in blocking buffer) for 1 h at RT. Plates were washed six times and developed with 1-Step Ultra TMB (ThermoFisher). The reaction was stopped with an equivalent volume of 2N H2SO4 (Ricca Chemical Co.), and the signal was read at optical density 450 nm on an EnVision plate reader (Perkin Elmer). Endpoint titres were interpolated from an Asymmetric Sigmoidal, 5PL X log(concentration) model in Prism 9 (GraphPad).
Surface plasmon resonance
The interactions of MD39 trimer analytes binding to IgGs captured on the surface plasmon resonance (SPR) sensor chip were assessed. The apparent affinities determined in these experiments do not reflect monovalent Fab–trimer interactions but rather include enhancement of binding due to avidity. Apparent kinetics and affinities of antibody–antigen interactions were measured on a Biacore 8K (Cytiva) using Series S CM5 Sensor Chip (Cytiva) and 1× HBS-EP+ pH 7.4 running buffer (20× stock from Teknova) supplemented with BSA at 1 mg ml–1. The Human Antibody Capture Kit was used according to the manufacturer’s instructions (GE) to immobilize about 7,000 response units of capture antibody on each flow cell. In a typical experiment, around 300–400 response units of monoclonal antibodies were captured on each flow cell, and trimer analytes were passed over them at 15 µl min–1 for 2 min followed by 5 min of dissociation time. Regeneration was accomplished using 3 M MgCl2 with 240 s contact time. The top concentration of the gp140 protomer was 10 µM, with three protomers per trimer. Five analyte concentrations were tested, with fourfold dilution. Raw sensorgrams were analysed using Biacore Insight Evaluation software v.3.0.12.15655 (Cytiva), including blank double referencing, and Kinetic fits with Langmuir model at 1:1 binding stoichiometry were used. Analyte concentrations were measured on a NanoDrop 2000c Spectrophotometer using an absorption signal at 280 nm.
EMPEM analysis
We performed polyclonal electron microscopy analysis as previously described21,36. Plasma antibodies were purified using Protein A Sepharose resin (GE Healthcare), eluted from the resin with 0.1 M glycine at pH 2.5 and buffer exchanged into 1× PBS. Fabs were generated using crystalline papain (Thermo Scientific), digested for 5 h at 37 °C and purified via size exclusion chromatography (SEC) using a Superdex 200 Increase 10/300 column (GE Healthcare). Complexes were assembled with 0.5 mg of polyclonal Fabs incubated overnight with 15 µg of MD39 Env trimers at RT, followed by purification to remove unbound Fab via SEC using a Superose 6 Increase 10/300 column (GE Healthcare). Complexes were diluted to 30–50 µg ml–1, immediately placed on 400-mesh Cu grids and stained with 2% (w/v) uranyl formate for 40 s. Images were collected via the Leginon automated imaging interface using either a Tecnai Spirit electron microscope, operated at 120 kV, or a Tecnai TF20 electron microscope, operated at 200 kV; for the former, nominal magnification was ×52,000 with a pixel size of 2.06 Å; the TF20 was operated at a nominal magnification of ×62,000 with a pixel size of 1.77 Å. Micrographs were recorded using a Tietz 4k × 4k TemCam-F416 CMOS camera. Particles were extracted via the Appion data processing package37 in which roughly 100,000 particles were auto-picked and extracted. Using Relion 3.0 (ref. 38), particles were two-dimensionally classified into 100 classes and those with antigen-Fab characteristics were selected for three-dimensional (3D) analysis. Initially 3D classification was done using 20–40 classes, with a low-resolution model of a non-liganded HIV Env ectodomain used as reference. Particles from similar-looking classes were combined and reclassified, with a subgroup of 3D classes processed using 3D auto-refinement. UCSF Chimera 1.13 was used to visualize and segment 3D-refined maps. Quantitation of epitope recognition in Fig. 2h was done by tabulating the number of epitopes recognized among each animal per group and then collation per group (for example, out of a total of six epitope sites × 4 animals = 24). Epitope sites not recognized per animal were also tabulated, to account for variation in animal group size. Fisher’s exact tests were used to test for significant differences in the number of epitopes recognized per group.
BCR sequencing and processing
A custom RM germline VDJ library was generated using references published previously13,39. CellRanger v.3.0 was used to assemble full-length V(D)J reads. The constants.py file in the CellRanger VDJ python library was modified to increase maximum acceptable CDR3 length to 110 nucleotides. CellRanger v.6 was used to obtain gene expression counts from sequenced GEX libraries. Libraries were aligned to the Ensemble Mmul10 reference genome, with the addition of mitochondrial genes from Mmul9. Sequences were de-multiplexed by hashtags using the MULTIseqDemux command in Seurat v.4 (ref. 40). For HC sequences in which both kappa and lambda light-chain (LC) contigs were detected, the B cell was assigned a lambda LC because lambda LC rearrangement occurs only if the kappa LC is not productive.
Longitudinal lineage and somatic mutation analysis of BCR sequences
The VDJ sequence output from CellRanger was further analysed using packages from the Immcantation portal41. An IgBLAST database was built from the custom RM germline VDJ Library and was then used to parse the 10× V(D)J output from CellRanger into an AIRR community standardized format using the Change-O pipeline, to allow for further downstream analysis with the Immcantation portal. Clonal lineages were determined for each animal with DefineClones.py using the appropriate clustering threshold as determined by the distToNearest command from the SHazaM package in R. Inferred germline V and J sequences from the reference library were added with CreateGermline.py. Because germline D gene sequences and N nucleotide additions cannot be accurately predicted, these were masked from further analysis. Rarefaction analysis for Env+ samples was conducted using the package iNext R42 to confirm acceptable sequence coverage (Extended Data Fig. 10g). The total number of mutations (V- and J-genes) for each HC and LC sequence was calculated by counting the number of nucleotide changes between the observed and predicted germline sequences with Shazam’s observed mutation command. For analysis of total HC mutations, all productive HC contigs were analysed; for LCs, only contigs paired with HCs were assessed. Sequences in which the VH or VL call aligned to alleles IGHV3-100*01, IGHV3-100*01_S4205, IGHV3-100*01_S4375, IGHV3-36*01_S5206, IGHV3-36*01_S6650, IGHV3-NL_11*01_S5714, IGHV4-79-a, IGHV4-NL_1*01_S0419, IGLV1-69, IGLV1-ACR*0 or IGLV2-ABX*01 were found to have an extremely high degree of substituted nucleotides at all time points compared with their inferred germline sequences, probably because of poor V-gene assignment due to an incomplete V(D)J reference library. These sequences were excluded from further analysis. Only clones that contained paired HC–LC BCR sequences were analysed when building clonal trees. Maximum-likelihood lineage trees were built for clonal families with Dowser43 using the pml method in the GetTrees function. For lineage trees, branch length represents the estimated number of total mutations that occurred in each HC sequence and its most recent common ancestor in lineage, rather than a simple count of nucleotide changes in the germline sequence. Strict criteria for identification of clonal lineages that could be found in both left and right LNs included the following requirements: H-CDR3 length greater than 14, multiple N additions in H-CDR3, greater than 20 cells in total and a matching LC within the lineage. A more lenient set of criteria was also used consisting of the following requirements: H-CDR3 length greater than 10, more than five cells in total and a matching LC within the lineage.
Diversity analysis
Chao1 estimation of clonal richness was calculated using the iNext R42 package according to the following formula:
where Sobs is the total number of observed species and F1 and F2 are the numbers of singletons and doubletons, respectively. Simpson diversity indices were computed using the alphaDiversity function from Alakazam, with uniform resampling down to 50 for LN FNA samples and 25 for PBMC samples to correct for sequencing depth, at diversity order (q) = 2. FNA samples containing fewer than 50 cells and PBMC samples containing fewer than 25 cells were excluded from diversity analysis. Simpson diversity (D) was calculated using the inverse Simpson index formula where R is richness (the total number of unqie lineages in sample) and Pi is the proportional abundance of the ith lineage:
Transcriptomics analysis
The package Seurat v.4 (ref. 40) was used for graph-based clustering and visualization of the gene expression data generated by CellRanger. Initial filtering was conducted on each sample to remove cells expressing fewer than 200 or more than 4,500 genes, as well as those with less than 10% of their transcriptome comprising mitochondrial DNA. Gene expression counts were log normalized via the NormalizeData command. A list of common variable genes across all samples was identified with the function SelectIntergrationFeatures. Expression of these common variable genes was scaled using principal component analysis (PCA) conducted with RunPCA. Next, all samples were integrated into a single dataset using a reciprocal PCA reduction to remove batch effects via the commands FindIntegrationAnchors and IntergrateData. Louvain clustering was conducted on the entire integrated dataset with the functions FindNeighbours and FindCluster. Clusters containing large numbers of cells with high levels of mtDNA, as well as those with low MS4A1 (CD20) and CD19 expression, were excluded from further analysis. Differentially expressed genes were identified in Seurat with the function FindMarkers by running a Wilcoxon rank-sum test for each cluster against all other clusters. Gene set enrichment analysis (GSEA) was conducted using the package fgsea in R44,45. Differentially expressed genes from previously identified human light-zone, dark-zone, intermediate, Bmem and plasma cell subsets were previously described24 and combined to create the gene sets used for GSEA. LZ1 and DZ5 were selected as representative light-zone and dark-zone clusters, respectively, based on GSEA scores.
Graphs, statistics and cell generation calculation
All statistics were calculated in Prism 9 or R unless stated otherwise. The statistical tests used are indicated in the respective figure legends and incorporated a two-tailed test. All graphs were generated in Prism 9 or R. Geometric mean and geometric s.d. are shown for data plotted on a log10 axis; mean and s.d. are plotted for data graphed on a linear axis. Median and quartiles 1 and 3 are shown for violin plots. UMAP plots were generated using Seurat v.4 (ref. 40). For comparison of total HC and LC mutations between groups 2 and 3, statistical significance was calculated only if mean mutations were significantly different according to per-animal comparisons. Extra statistical test information is provided in the individual Methods experimental approach sections. All Mann–Whitney tests are two-sided. P values are defined throughout as follows: not significant, P > 0.05; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P ≤ 0.0001.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-022-05216-9.
Supplementary information
Acknowledgements
We thank C. Kim and the Flow Cytometry Core at La Jolla Institute for Immunology (LJI) and the LJI Sequencing Core. We also thank G. Karlsson Hedestam and M. Corcoran from the Karolinska Institutet for provision of additional naive RM IGHV reference sequences before publication. This study was partly performed at AlphaGenesis, Inc. This work was supported in part by the National Institute of Allergy and Infectious Diseases of the NIH under award nos. CHAVI-ID UM1AI100663, CHAVD UM1AI144462, R01 AI125068, R01 AI136621 and P01AI048240; NIH NIAID SVEU contract no. HHSN272201300004I; and NIH NIAID NHP-CIL (contract nos. HHSN272201700022C and P51OD011104). This work was also supported by the Bill and Melinda Gates Foundation (no. BMGF OPP1203211) and the Collaboration of AIDS Vaccine Discovery (no. OPP1115782/INV-002916).
Extended data figures and tables
Source data
Source Data Fig. 1 and Extended Data Figs. 2 and 3.
Source Data Figs. 2 and 3 and Extended Data Fig. 3.
Source Data Extended Data Figs. 9 and 10.
Author contributions
J.H.L. and S.C. conceived and designed experiments. D.J.I. provided conceptual insights and SMNP. J.H.L. performed and analysed most experiments pertaining to group 2 and 3 LN FNA data, generated FNA BCR sequencing data and assisted with FNA BCR sequencing analysis. H.J.S. analysed BCR data, performed all experiments related to PBMC samples and generated and analysed single-cell transcriptomics data. J.H.L. and C.K. performed serum ELISA. R.N. and L.H. performed neutralization assays and analysis. X.S. and D.C.M. performed independent confirmation neutralization assays and analysis. S.T.R., L.M.S., J.L.T., W.-H.L. and G.O. generated and analysed EMPEM data. D.J.I. and M.S. supplied the SMNP adjuvant. A.L., C.A.C. and O.K. performed SPR experiments and analysis. C.A.C., E.G., M.K., S.H., T.-M.M., Y.A. and W.R.S. supplied immunogens and flow cytometry baits. C.A.C. and K.M.C. designed primers. I.P., A.K., C.A., M.F., B.F.G., J.P.D., F.S., P.P.A. and R.S.V. performed Env trimer plus alum rhesus studies. D.G.C. and G.S. provided technical guidance for studies performed at the Tulane National Primate Research Center. A.B.W. and D.R.B. provided supervision. J.H.L. and S.C. wrote the original draft. H.J.S. and L.H. contributed to figure generation. H.J.S., D.R.B., D.J.I., A.B.W. and W.R.S. provided comments and assisted with revision.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
Gene Expression data generated in the current study are available in the Gene Expression Omnibus database under accession code GSE209904. BCR sequencing data are available in the SRA under accession code PRJNA861839. 3D EM reconstructions are available from the Electron Microscopy Data Bank under the following EMDB codes: EMD-27600–27605, EMD-27607–27612 and EMD-27614–27615. Sequencing data and electron microscopy particle stacks are available on request. Source data are provided with this paper.
Competing interests
W.R.S. has a patent related to the MD39 immunogen (US patent no. 11,203,617). M.S., D.J.I. and S.C. have patent related to the SMNP adjuvant.
Footnotes
A list of affiliations appears at the end of the paper.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jeong Hyun Lee, Henry J. Sutton
Change history
1/30/2023
A Correction to this paper has been published: 10.1038/s41586-023-05741-1
Extended data
is available for this paper at 10.1038/s41586-022-05216-9.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-022-05216-9.
References
- 1.Havenar-Daughton C, Lee JH, Crotty S. Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol. Rev. 2017;275:49–61. doi: 10.1111/imr.12512. [DOI] [PubMed] [Google Scholar]
- 2.Lee JH, Crotty S. HIV vaccinology: 2021 update. Semin. Immunol. 2021;51:101470. doi: 10.1016/j.smim.2021.101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton DR, Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haynes BF, Burton DR, Mascola JR. Multiple roles for HIV broadly neutralizing antibodies. Sci. Transl. Med. 2019;11:eaaz2686. doi: 10.1126/scitranslmed.aaz2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mesin L, Ersching J, Victora GD. Germinal center B cell dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotty ST. Follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victora GD, Nussenzweig MC. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 8.Cyster JG, Allen CDC. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitlin, A. D. et al. T cell help controls the speed of the cell cycle in germinal center B cells. Science10.1126/science.aac4919 (2015). [DOI] [PMC free article] [PubMed]
- 10.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam HH, et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl Acad. Sci. USA. 2016;113:E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauthner M, et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46:1073–1088. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirelli KM, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177:1153–1171. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Giudice G, Rappuoli R, Didierlaurent AM. Correlates of adjuvanticity: a review on adjuvants in licensed vaccines. Semin. Immunol. 2018;39:14–21. doi: 10.1016/j.smim.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Steichen JM, et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva M, et al. A particulate saponin/TLR agonist vaccine adjuvant alters lymph flow and modulates adaptive immunity. Sci. Immunol. 2021;6:eabf1152. doi: 10.1126/sciimmunol.abf1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havenar-Daughton C, et al. Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 2016;17:2195–2209. doi: 10.1016/j.celrep.2016.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havenar-Daughton C, et al. Cytokine-independent detection of antigen-specific germinal center T follicular helper cells in immunized nonhuman primates using a live cell activation-induced marker technique. J. Immunol. 2016;197:994–1002. doi: 10.4049/jimmunol.1600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu JK, et al. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J. Virol. 2015;89:10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cottrell CA, et al. Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates. PLoS Pathog. 2020;16:e1008753. doi: 10.1371/journal.ppat.1008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianchi M, et al. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity. 2018;49:288–300.e8. doi: 10.1016/j.immuni.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao F, et al. Mapping neutralizing antibody epitope specificities to an HIV Env trimer in immunized and in infected rhesus macaques. Cell Rep. 2020;32:108122. doi: 10.1016/j.celrep.2020.108122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy DE, et al. Novel specialized cell state and spatial compartments within the germinal center. Nat. Immunol. 2020;21:660–670. doi: 10.1038/s41590-020-0660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes AB, et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J. Exp. Med. 2020;217:e20200483. doi: 10.1084/jem.20200483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowosad CR, et al. Tunable dynamics of B cell selection in gut germinal centres. Nature. 2020;588:321–326. doi: 10.1038/s41586-020-2865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovas C, et al. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yewdell WT, et al. Temporal dynamics of persistent germinal centers and memory B cell differentiation following respiratory virus infection. Cell Rep. 2021;37:109961. doi: 10.1016/j.celrep.2021.109961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon MML, et al. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci. Immunol. 2021;6:eabl9105. doi: 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner JS, et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lederer K, et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022;185:1008–1024. doi: 10.1016/j.cell.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim W, et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141–145. doi: 10.1038/s41586-022-04527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heesters BA, et al. Endocytosis and recycling of immune complexes by follicular dendritic cells enhances B cell antigen binding and activation. Immunity. 2013;38:1164–1175. doi: 10.1016/j.immuni.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH, et al. Vaccine genetics of IGHV1-2 VRC01-class broadly neutralizing antibody precursor naïve human B cells. NPJ Vaccines. 2021;6:113. doi: 10.1038/s41541-021-00376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.deCamp A, et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2014;88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarzotti-Kelsoe M, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogal B, et al. Mapping polyclonal antibody responses in non-human primates vaccinated with HIV Env trimer subunit vaccines. Cell Rep. 2020;30:3755–3765. doi: 10.1016/j.celrep.2020.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vázquez Bernat N, et al. Rhesus and cynomolgus macaque immunoglobulin heavy-chain genotyping yields comprehensive databases of germline VDJ alleles. Immunity. 2021;54:355–366. doi: 10.1016/j.immuni.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vander Heiden JA, et al. PRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods Ecol. Evol. 2016;7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 43.Hoehn KB, et al. Human B cell lineages associated with germinal centers following influenza vaccination are measurably evolving. eLife. 2021;10:e70873. doi: 10.7554/eLife.70873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv10.1101/060012 (2021).
- 45.Sergushichev, A. A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. Preprint at bioRxiv10.1101/060012 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene Expression data generated in the current study are available in the Gene Expression Omnibus database under accession code GSE209904. BCR sequencing data are available in the SRA under accession code PRJNA861839. 3D EM reconstructions are available from the Electron Microscopy Data Bank under the following EMDB codes: EMD-27600–27605, EMD-27607–27612 and EMD-27614–27615. Sequencing data and electron microscopy particle stacks are available on request. Source data are provided with this paper.