Abstract
A single-copy reporter system for Staphylococcus xylosus has been developed, that uses a promoterless version of the endogenous β-galactosidase gene lacH as a reporter gene and that allows integration of promoters cloned in front of lacH into the lactose utilization gene cluster by homologous recombination. The system was applied to analyze carbon catabolite repression of S. xylosus promoters by the catabolite control protein CcpA. To test if lacH is a suitable reporter gene, β-galactosidase activities directed by two promoters known to be subject to CcpA regulation were measured. In these experiments, repression of the malRA maltose utilization operon promoter and autoregulation of the ccpA promoters were confirmed, proving the applicability of the system. Subsequently, putative CcpA operators, termed catabolite-responsive elements (cres), from promoter regions of several S. xylosus genes were tested for their ability to confer CcpA regulation upon a constitutive promoter, PvegII. For that purpose, cre sequences were placed at position +3 or +4 within the transcribed region of PvegII. Measurements of β-galactosidase activities in the presence or absence of glucose yielded repression ratios between two- and eightfold. Inactivation of ccpA completely abolished glucose-dependent regulation. Therefore, the tested cres functioned as operator sites for CcpA. With promoters exclusively regulated by CcpA, signal transduction leading to CcpA activation in S. xylosus was examined. Glucose-dependent regulation was measured in a set of isogenic mutants showing defects in genes encoding glucose kinase GlkA, glucose uptake protein GlcU, and HPr kinase HPrK. GlkA and GlcU deficiency diminished glucose-dependent CcpA-mediated repression, but loss of HPr kinase activity abolished regulation. These results clearly show that HPr kinase provides the essential signal to activate CcpA in S. xylosus. Glucose uptake protein GlcU and glucose kinase GlkA participate in activation, but they are not able to trigger CcpA-mediated regulation independently from HPr kinase.
Carbon catabolite repression (CR) is a regulatory process in microorganisms, whereby the presence of a rapidly metabolizable carbon source inhibits expression of alternate catabolic functions. Although the final outcome of CR is uniform, reduced expression of certain genes and operons, the mechanisms leading to repression may be quite diverse (34). The presence of a repressing carbon source can result in lower concentrations of inducers specific for alternate routes of catabolism (35, 37), in altered activities of specific regulators (40), or in the activation of global control proteins, such as the cyclic AMP receptor protein in enteric bacteria (36) or the catabolite control protein CcpA in low-GC gram-positive bacteria (19). In many cases, genes are subject to multiple levels of control. It is therefore important to distinguish gene- or operon-specific regulatory processes from global control to be able to dissect the underlying mechanisms.
In low-GC, gram-positive bacteria, one consequence of the availability of a rapidly metabolizable carbon source is the regulation of gene expression by CcpA, the central transcriptional regulator of CR (19). CcpA, a member of the LacI/GalR family of transcription factors (29), shows a relatively weak affinity for its cognate operator sites, termed catabolite-responsive elements (cres) (20) and must therefore be activated in order to bind efficiently to cre. Several effectors, alone or in combination, have been found to enhance DNA binding of CcpA in vitro (12, 13, 16, 22, 23, 27), but only phosphorylated forms of HPr, the general phosphocarrier protein of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) (31) or Crh, an HPr-like protein detected in Bacillus subtilis (14), were shown to be of significance in vivo (7, 13, 26, 44). Accordingly, loss of HPr kinase function, the enzyme responsible for regulatory phosphorylation of serine at position 46 in HPr or Crh, resulted in relief from CR (15, 33). Evidence regarding the in vivo activation of CcpA so far had been available only from B. subtilis. More recently, HPr kinase mutants, which showed a pleiotropic loss of CR, have been constructed in Staphylococcus xylosus and Lactobacillus casei (8, 21). In addition, a glucose kinase in S. xylosus has been implicated in CR (42). Both kinases appeared to modulate CcpA activity, but it was not clear whether they could act upon CcpA independently from each other.
We are interested in CR in the nonpathogenic staphylococcal species S. xylosus, which is applied in food fermentation processes. In this communication, we describe the construction and evaluation of a single-copy reporter system based on the β-galactosidase gene of S. xylosus. The application of the system to study CcpA-dependent regulation is presented.
MATERIALS AND METHODS
Bacterial strains.
The S. xylosus lactose-negative derivatives of the wild-type strain C2a (17) that were used in this study are listed in Table 1. The glucose kinase mutant strain TX60 (ΔglkA) was constructed by introducing an internal glkA deletion into the genome of S. xylosus C2a. The resulting strain, TX60, showed the same regulatory phenotype as the original glkA::Tn917 insertion mutants (42).
TABLE 1.
S. xylosus β-galactosidase-deficient strains used in this study
| Strain | Genotypea | Derivation |
|---|---|---|
| TX300 | Wild type | C2ab (17) |
| TX400 | ΔglkA | TX60 (this work) |
| TX500 | ΔglcU | TX214 (11) |
| TX600 | ccpA::ermB | TX154 (9) |
| TX700 | hprK::ermB | TX300 (this work) |
Only the genotype relevant for regulation is listed. All strains are β-galactosidase and lactose permease deficient and harbor a truncated lacR gene (′lacR ΔlacP ′lacH).
The wild-type strain C2a is a derivative of S. xylosus DSM20267, cured of its endogenous plasmid pSX267 (17).
All plasmid constructions were performed in Escherichia coli DH5α [Φ80dlacZΔM15 Δ(lacZYA-argF) recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 deoR].
Plasmids.
The lactose utilization genes of S. xylosus were obtained from plasmids pBG303 and pBG304, containing the β-galactosidase gene lacH and the regulatory gene lacR, respectively (1). To construct the lac plasmids pLK1 and pLP1, the temperature-sensitive shuttle vectors pBT12 and pBT2 were used (3). Plasmid pBT12 is a derivative of pBT2, in which the multiple cloning site is replaced by the corresponding region of pBT1 (3). Plasmid pRB474, a chloramphenicol-resistant derivative of pRB374 (4), was used to isolate the B. subtilis vegII promoter and to clone the vegII(cre) promoter derivatives. Plasmid pKIN5E1 (21) was applied to inactivate the HPr kinase gene hprK in the S. xylosus chromosome.
DNA manipulations, sequencing, and transformation.
DNA manipulations, plasmid DNA isolation, transformation of E. coli, and preparation of media and agar plates for E. coli were done by standard procedures (38). Sequencing was performed with a Li-Cor automated sequencer. Plasmids were introduced in S. xylosus by electroporation with glycine-treated electrocompetent cells (3). PCR was carried out with Vent DNA polymerase (New England Biolabs). S. xylosus strains were cultivated in B medium, consisting of 1% peptone, 0.5% yeast extract, 0.5% NaCl, 0.1% KH2PO4, supplemented with 0.5% carbohydrate as indicated. To screen for the expression of β-galactosidase on agar plates, B medium agar was supplemented with 100 μg of 4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml.
Construction of the S. xylosus lac inactivation plasmid pLK1.
To use the endogenous β-galactosidase gene lacH as a reporter gene, the first step was to inactivate the chromosomal copy of the gene. For that purpose, the lac deletion plasmid pLK1 (Fig. 1) was constructed. On that plasmid, the lac operon of S. xylosus (1) as outlined in Fig. 1 was modified to yield ′lacR and ′lacH truncated at their 5′ ends and a complete deletion of lacP, the lactose permease gene including the lacPH promoter (Fig. 1). As the source for lac DNA, the plasmids pBG303 and -304 were used. The temperature-sensitive shuttle plasmid pBT12 served as the vector.
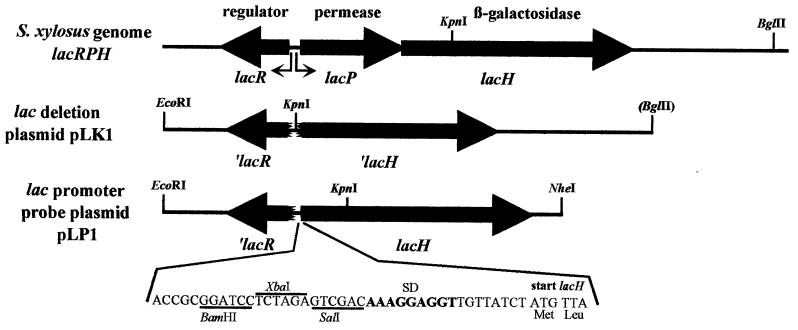
FIG. 1.
Genetic organization of the genomic reporter system for S. xylosus. The genetic organization of the wild-type S. xylosus lactose utilization genes lacRPH is shown in the first line. The locations of the two lac promoters are indicated by arrows. The lac deletion plasmid pLK1 harbors versions of ′lacR and ′lacH truncated at their 5′ ends and lacks lacP. The promoterless β-galactosidase genes lacH and ′lacR are contained on the promoter probe plasmid pLP1. The nucleotide sequence of the region preceding the reporter gene lacH is shown below pLP1. Three restriction sites, BamHI, XbaI, and SalI, are available for insertion of promoter fragments. The wild-type Shine-Dalgarno sequence (SD) of lacH (AAAGGGGT) has been changed to AAAGGAGGT on the promoter probe plasmid. The vectors used to construct pLK1 and pLP1 were the temperature-sensitive shuttle plasmids pBT12 and pBT2, respectively. After homologous recombination, the structure of the lac region in the genome of S. xylosus strains is as outlined for pLK1 and pLP1, except for the newly introduced EcoRI restriction site.
Construction of the lacH promoter probe plasmid pLP1.
For the construction of the promoter probe plasmid pLP1 (Fig. 1), the intact lacH gene without promoter was restored, concomitantly changing the wild-type lacH Shine-Dalgarno sequence AGGGGT to AGGAGGT, which should allow perfect pairing with 16S rRNA. The vector for the pLP1 construction was pBT2. Promoter fragments may be inserted into the SalI, XbaI, and BamHI sites located in front of the promoterless β-galactosidase gene lacH (Fig. 1).
Cloning of promoters into pLP1.
S. xylosus promoters that were analyzed with the promoter probe system are shown in Fig. 2. The promoters PmalRA and Pccpa were cloned on BamHI-SalI fragments in E. coli DH5α. Successful integration of promoter fragments into pLP1 could be detected on X-Gal-containing agar plates—however, only at 30°C. Although the β-galactosidase was active at 37°C in S. xylosus, it showed no activity above 30°C in E. coli. Promoter-containing plasmids were designated pLP3 (PmalRA) and pLP4 (Pccpa) (Fig. 2). To check the fidelity of the polymerase reactions, cloned promoters were sequenced prior to use in S. xylosus.
FIG. 2.
Nucleotide sequence of the promoters integrated in front of the β-galactosidase gene lacH. The promoter regions relevant for transcription initiation and regulation are shown. Putative RNA polymerase binding sites are underlined. Transcriptional start sites are shown in boldface. cres are underlined and shown in boldface. The elements were labeled according to the genes or operons where they originated from. Designation of the plasmids harboring the shown promoters and corresponding numbers to identify these promoters after their integration into the S. xylosus genome are shown in parentheses. Relevant restriction sites are indicated.
The B. subtilis vegII promoter (30) was excised from pRB474 by NheI and SalI and inserted into pLP1 cut with XbaI and SalI. The plasmid was designated pLP2 (Fig. 2).
Construction of cre-containing vegII promoter derivatives.
To convert the constitutive vegII promoter to a CcpA-regulated promoter, cre operators were placed between the HindIII and PstI restriction sites downstream of the vegII transcriptional start point (Fig. 2). To that end, overlapping pairs of oligonucleotides were designed that contained a cre in the double-stranded part and single-stranded extensions fitting into HindIII- and PstI-cut DNA. In addition, a SpeI restriction site was introduced downstream of cre to help to identify the small DNA fragment after cloning. The different cre sequences are shown in Fig. 2 in the context of the vegII(cre) promoters.
The annealed oligonucleotides were first cloned into pRB474. After confirmation of the promoter-cre structures by DNA sequencing, respective promoter (cre) fragments were moved as NheI-SalI fragments to pLP1 as described above for vegII. The vegII(cre) plasmids were designated pLP13 to pLP16 and pLP20 (Fig. 2).
Inactivation of the lac genes in the genome of S. xylosus.
The lac inactivation plasmid pLK1 (Fig. 1) was transferred by electroporation into S. xylosus wild-type C2a and the isogenic regulatory mutants listed in Table 1. Plasmid-containing colonies were selected with 20 μg of chloramphenicol per ml at 30°C. Subsequently, the transformants were patched onto B medium agar plates, supplemented with 100 μg of X-Gal per ml and 20 μg of chloramphenicol per ml and grown at 30°C for about 36 h. Although the strains appeared blue on these plates, the color was not equally distributed within the patches. Apparently, homologous recombination occurred with a high frequency introducing the lac gene deletions from plasmid pLK1 into the genome of many cells. After streaking the cells on X-Gal (100 μg/ml) agar plates without chloramphenicol and incubation at 37°C overnight, between 20 and 50% white colonies were detected. A second incubation of a white colony on agar without selective pressure efficiently cured the cells from the pLK1 plasmid, which does not replicate at 37°C. By this procedure, TX300, the β-galactosidase-deficient derivative of the wild-type strain and its isogenic regulatory mutants were produced (Table 1). The chromosomal organization of the lac region in these strains was confirmed by restriction analysis of PCR fragments obtained by primers annealing outside of the cloned lac region.
In the course of these constructions, it turned out that the hprK mutant strain TX66 (21) was not transformable with pLK1. Low transformation efficiency was already noticed during hprK complementation experiments (21), but the strain appeared transformable. The relatively large size of the pLK1 plasmid (15 kb) may have been the reason for the failure to introduce it into TX66. To circumvent this problem, the hprK inactivation plasmid pKIN5E1 was used in S. xylosus TX300 (′lacR ΔlacP ′lacH) to yield TX700 (hprK::ermB ′lacR ΔlacP ′lacH). Successful inactivation of hprK was confirmed by PCR and, phenotypically, by the appearance of the glucose-sensitive growth behavior (21).
Integration of promoters in front of the chromosomal β-galactosidase gene lacH.
Integration of promoters in front of lacH was achieved by replacing the pLK1-generated copies of ′lacH with intact lacH from promoter-containing derivatives of pLP1. The β-galactosidase-deficient strains (Table 1) were transformed with the plasmids (Fig. 2), and transformants were selected with 20 μg of chloramphenicol per ml and grown for 36 h at 30°C. After patching onto chloramphenicol- and X-Gal-containing agar plates, the colonies appeared uniformly blue, due to β-galactosidase expression from plasmid. Plasmid curing on nonselective media at 37°C produced a mixture of white and blue colonies. Most of these blue colonies were chloramphenicol sensitive, indicating plasmid loss. Subsequent PCR analysis of the lac region confirmed the integration promoter-containing lacH genes into the chromosome. The resulting strains were designated according to the β-galactosidase-deficient parental strains (Table 1) and the numbers that were assigned to the promoters (Fig. 2). For example, PmalRA-harboring strains (pLP3) were designated S. xylosus TX303 (wild type) and TX603 (ccpA mutant).
Since the HPr kinase-deficient strain S. xylosus TX700 was not transformable with the pLP plasmids, hprK was inactivated in the TX300 series of S. xylosus strains to yield the TX700 collection, as described above for the TX700 construction. The strains were given the 700 designation numbers to aid their identification as being HPr kinase deficient, although they were not derived from TX700.
Integration of the promoterless β-galactosidase gene into the genome of S. xylosus.
To be able to determine the background level of β-galactosidase expression without promoter, the modified lacH gene of pLP1 was integrated into the wild-type strain C2a and the ccpA mutant TX154 to yield S. xylosus TX301 and TX601, respectively. Both strains appeared white on X-Gal-containing agar plates, and β-galactosidase activity was virtually undetectable. Therefore, the corresponding control strains with glkA, glcU, or hprK mutations were not constructed.
Determination of β-galactosidase activity in cell extracts.
The cells were grown at 37°C in B medium to an optical density at 578 nm (OD578) of 1. Sugars were added to a final concentration of 25 mM, when appropriate. After 1 h of further growth, the OD578 was determined, and the cells were harvested by centrifugation. The preparation of crude extracts and the assay of β-galactosidase activity have been described previously (1). Specific β-galactosidase activities are expressed in nanomoles of nitrophenol released per minute per milligram of protein. Protein concentrations were determined by the method of Bradford (2).
RNA preparation and primer extension analysis.
Preparation of RNA and primer extension reactions were done as described previously (1). The primer used to map the start point of PvegII was labeled at the 5′ end with an infrared dye, IRD700. Reverse transcripts were run on 8% polyacrylamide–urea gels and detected by a Li-Cor DNA sequencer.
RESULTS AND DISCUSSION
Analysis of CcpA-dependent regulation of the malRA and ccpA promoters to evaluate the single-copy promoter probe system.
To demonstrate that the promoter probe system is suitable for the investigation of gene regulation in S. xylosus, CcpA-mediated CR was chosen as an example. Two promoters, the promoter of the maltose utilization operon malRA (10) and one of the two ccpA promoters, had been previously shown to be subject to CcpA repression (9). In the malRA promoter region, a cre spanning positions −58 to −45 with respect to the transcriptional start point (Fig. 2) is essential for CcpA-mediated repression (9). In the ccpA promoter region, a cre overlaps the start point of the second promoter (Fig. 2) and is most likely responsible for the observed ccpA autoregulation (9). Both promoter regions were integrated in the S. xylosus genome in front of the promoterless β-galactosidase gene lacH, and β-galactosidase activities in the absence or presence of carbohydrates were determined in the resulting strains, TX303 (′lacR ΔlacP PmalRA-lacH) and TX304 (′lacR ΔlacP PccpA-lacH). In addition, the background level of β-galactosidase expression without a cloned promoter was determined in strain TX301 (′lacR ΔlacP lacH).
Both promoters were repressed between four- and twofold by the tested carbon sources, glucose and sucrose. Glucose exerted a slightly stronger negative effect on β-galactosidase expression than sucrose (data not shown). The fourfold repression by glucose of the malRA promoter in the lacH promoter probe system was less pronounced than the eightfold repression of malA-encoded α-glucosidase detected previously (9). Since malA, as the second gene of the malRA operon, is much further downstream than lacH in the promoter probe system, polar effects or minor cre sequences within malR may enhance malA repression. In addition, differences in the stability of α-glucosidase and β-galactosidase may also affect repression ratios. However, CR of the malRA promoter was clearly detectable in the promoter probe system.
The twofold glucose-mediated repression of β-galactosidase expression directed by the ccpA promoters is in accordance with the twofold reduction of CcpA production detected by Western blot analysis (9). Since only the second ccpA promoter, P2, is controlled by CcpA (9), overall repression of both ccpA promoters is moderate and less pronounced than PmalRA repression. The slight ccpA autorepression in the presence of carbon sources apparently reflects the need to balance CcpA production, when preformed CcpA is activated to carry out CR.
Expression of β-galactosidase driven by the malRA and ccpA promoters was also tested in the CcpA-deficient S. xylosus strains TX603 and TX604. As predicted, repression by glucose or sucrose was virtually lost, substantiating the role of CcpA in CR of these promoters. In the absence of a functional CcpA, both promoter activities were higher than in the wild-type strain, even in complex B medium without an additional carbon source. These results strongly indicate that CcpA is not fully inactive under these growth conditions. Surprisingly, there was a marked difference in the consequences of ccpA inactivation on the activities of the tested promoters in the absence of repressing sugars. While PmalRA activity increased only 1.4-fold, PccpA-directed β-galactosidase expression was threefold higher. We are currently not able to offer a reasonable explanation for this observation.
Without promoters in front of lacH (TX301 and TX601), β-galactosidase activity was below the level of detection. Virtually no transcriptional readthrough occurs from neighboring chromosomal regions into lacH. Measured β-galactosidase activities do indeed reflect transcription directed by the cloned promoters. In conclusion, the new promoter probe system appears to be well suited to analyze CcpA-mediated gene regulation in S. xylosus. It will most likely also be valuable for the analysis of other regulatory events in this organism.
Evaluation of putative cre sequences.
To define whether a cre-like sequence indeed serves as a CcpA operator, cre function must be determined in the absence of sugar-specific regulators, because alternate mechanisms of CR may mimic CcpA regulation. If specific regulators are unknown or activators are involved, it is difficult and time-consuming to obtain constitutively expressed genes. An alternative approach to analyze cre function could be to move suspected cre sequences to a constitutive promoter. Subsequently, it can be determined whether the promoter is now regulated by CcpA.
To develop such a system, the constitutive vegII promoter from B. subtilis (30), which had been previously found to function efficiently in S. xylosus (10), was placed in front of lacH. The promoter was cloned on plasmid pLP2 and integrated into the genome of S. xylosus to yield strain TX302. Since the transcriptional start point of vegII in S. xylosus had not been determined, primer extension reactions were performed. Transcription starts at G within the HindIII restriction site (Fig. 2), at the same position as in B. subtilis (25). Unexpectedly, the signal with RNA prepared from glucose-grown cells was slightly stronger than that from cells grown in complex medium without glucose (data not shown). Likewise, β-galactosidase activity was about 1.1-fold higher under these conditions (Table 2). This marginal enhancement of transcription at the vegII promoter does not impair the intended cre evaluation. It will only lead to a slight underestimation of repression in glucose-grown cultures.
TABLE 2.
Regulation of β-galactosidase expression directed by the vegII promoter and cre-containing vegII promoter derivatives in S. xylosus wild-type and mutant strains altered in catabolite repression
| cre operatora | Strain | Genotypeb | β-Galactosidase activity (nmol of nitrophenol produced/ min/mg of protein)c
|
||
|---|---|---|---|---|---|
| B medium | B medium + glucose | Glucose repression ratiod | |||
| None | TX302 | Wild type | 139 | 150 | 0.9 |
| TX602 | ccpA::ermB | 125 | 131 | 0.9 | |
| TX402 | ΔglkA | 120 | 127 | 0.9 | |
| TX502 | ΔglcU | 110 | 122 | 0.9 | |
| TX702 | hprK::ermB | 124 | 97 | 1.3 | |
| malRA | TX313 | Wild type | 82 | 26 | 3.2 |
| TX613 | ccpA::ermB | 97 | 90 | 1.1 | |
| TX413 | ΔglkA | 89 | 53 | 1.7 | |
| TX513 | ΔglcU | 87 | 42 | 2.1 | |
| TX713 | hprK::ermB | 81 | 70 | 1.1 | |
| xylAB | TX314 | Wild type | 38 | 5 | 7.6 |
| TX614 | ccpA::ermB | 68 | 62 | 1.1 | |
| TX414 | ΔglkA | 33 | 9 | 3.7 | |
| TX514 | ΔglcU | 36 | 11 | 3.2 | |
| TX714 | hprK::ermB | 55 | 47 | 1.2 | |
| ccpA | TX315 | Wild type | 104 | 25 | 4.1 |
| TX615 | ccpA::ermB | 114 | 110 | 1.0 | |
| TX415 | ΔglkA | 110 | 57 | 1.9 | |
| TX515 | ΔglcU | 106 | 55 | 1.9 | |
| TX715 | hprK::ermB | 98 | 91 | 1.1 | |
| lacPH | TX316 | Wild type | 38 | 16 | 2.4 |
| TX616 | ccpA::ermB | 51 | 53 | 1.0 | |
| TX416 | ΔglkA | 36 | 24 | 1.5 | |
| TX516 | ΔglcU | 38 | 25 | 1.5 | |
| TX716 | hprK::ermB | 56 | 50 | 1.1 | |
| scrA | TX320 | Wild type | 128 | 71 | 1.8 |
| TX620 | ccpA::ermB | 118 | 115 | 1.0 | |
| TX420 | ΔglkA | 124 | 116 | 1.1 | |
| TX520 | ΔglcU | 108 | 94 | 1.2 | |
| TX720 | hprK::ermB | 103 | 91 | 1.2 | |
The sequences of the integrated cre operators are listed in Fig. 2; cre sequences were designated according to their corresponding genes.
Only the genotype relevant for regulation is shown. In all strains, the lac region is modified (′lacR ΔlacP P-lacH).
Mean values of three independent cultures are presented. The standard deviations did not exceed ±15%. Strains were grown in complex B medium to an OD578 of 1; 25 mM glucose was added as indicated. After 1 h of further growth, the OD578 was determined, and the cells were harvested and disrupted with glass beads. Extracts prepared from 30 ml of cells adjusted to an OD578 of 2 were used for the determination of β-galactosidase activities.
The glucose repression ratio was calculated by dividing the enzyme activity found in cultures in B medium by the enzyme activity detected in glucose-containing cultures.
To convert vegII into a regulatable promoter, cre sequences were inserted downstream of the transcriptional start point as described in Materials and Methods. Subsequently, the composite promoters were integrated into the S. xylosus genome. The sequences to be tested were derived from those S. xylosus genes (malRA, xylAB, ccpA, lacPH, and scrA) previously shown to be subject to CR (1, 9, 10, 39, 41). The respective cres were either known to be functional (malRA and xylAB) or suspected to be responsible for CcpA regulation (ccpA, lacPH, and scrA). The putative operators were placed at +3 (malRA, lacPH, and scrA) or +4 (xylAB and ccpA) (Fig. 2) within the transcribed region of vegII, a cre position that should lead to repression of transcription initiation (5, 18). The strains obtained in this way were designated TX313, -314, -315, -316, and -320 (Table 2).
As an example for a known functional cre, transcription of the promoter PvegII(malcre) containing the CcpA operator of the malRA operon (9) in S. xylosus TX313 was analyzed. Transcription initiation was repressed in the presence of glucose, and repression was lost in the isogenic ccpA mutant TX613 (data not shown). Therefore, this experimental approach is well suited to analyze cre and CcpA function in S. xylosus.
To determine the repression efficiency of the inserted cres, β-galactosidase activities were measured in cells grown in complex B medium without carbohydrate and in the presence of glucose or sucrose. As summarized in Table 2, all inserted operators conferred glucose repression upon vegII. The promoters were also repressed in the presence of sucrose, but to a lesser extent (data not shown). Since glucose repression is the focus of our interest, only these values are reported in Table 2. Inactivation of ccpA led to a complete relief of glucose repression (TX613, -614, -615, -616, and -620) (Table 2), proving that CcpA is responsible for the observed regulation.
Glucose repression mediated by different S. xylosus cres in vegII varied from hardly twofold (scrAcre) to almost eightfold (xylABcre) (Table 2). In a recent study of cres in B. subtilis, which were also tested in a chromosomal reporter system with a constitutive promoter, repression was found to be between 2- and 16-fold (28). However, the majority of the tested cres, 20 out of 22, mediated only two- to eightfold repression. Apparently, the range of CcpA repression is very similar in B. subtilis and S. xylosus. To explain varying repression ratios, one is tempted to attribute alterations exclusively to differences in the cre sequences and, consequently, to different affinities of CcpA for these sites. It has been shown in E. coli that the same operator inserted into various promoters may repress with different efficiency, indicating that the promoter strength influences repressor action (24). If one takes the unregulated β-galactosidase activities in the ccpA mutant background (TX600 series) (Table 2) as a measure for promoter strength, it is apparent that insertion of cres changed transcription efficiency up to threefold. Therefore, changing promoter strength by different cre sequences inserted into the early transcribed region of lacH may have also influenced the efficiency by which these operators direct CcpA for repression.
In the abovementioned B. subtilis cre evaluation, promoter strength varied up to fourfold (28), and even in the pioneering work of Weickert and Chambliss (43), where only point mutations have been introduced into an existing cre in front of the α-amylase gene, unrepressed promoter activities differed up to fivefold. It seems that changing sequences in promoter regions inevitably alters promoter strength, which in turn may influence the efficiency of operators. Detailed kinetic in vitro binding studies may be needed to exactly define affinities of CcpA for various cres.
While it may be difficult to draw definite conclusions about the affinity of CcpA for the tested S. xylosus cres, it appears reasonable to compare levels of repression of promoters of similar strengths. For example PvegII(xylcre) and PvegII(laccre) direct similar β-galactosidase activities in the absence of CcpA (68 and 51 U, respectively) (Table 2), but PvegII(xylcre) is repressed much stronger (7.6-fold) than PvegII(laccre) (2.4-fold). Therefore, cre of lacPH is less efficient directing CcpA repression than the xylAB cre, perhaps because the highly conserved C (20, 28, 43) at the right arm of cre is replaced by A (Fig. 2). Besides the clear identification of cre function, the results of this analysis illustrate how distinct promoter-cre combinations could establish a hierarchy of control by CcpA.
CcpA activity in the absence of glucose kinase GlkA and glucose uptake protein GlcU.
In previous studies aimed at isolating S. xylosus mutants altered in CR, a PTS-independent glucose utilization system consisting of a glucose uptake protein, GlcU, and a glucose kinase, GlkA, have been isolated (11, 42). In glkA or glcU mutant strains, glucose-mediated CR of several enzyme activities was reduced, but not abolished. The phenotypes of these mutant strains were very similar, suggesting that both mutations affect the same process in CR. It was not clear, however, to what extent the enzymatic activities measured in these studies reflected CcpA regulation. With the CcpA-dependent promoters at hand, it was therefore of interest to determine the consequences of glcU and glkA mutation for CcpA-dependent regulation.
Two isogenic sets of strains with mutations in glkA (TX400 series) or glcU (TX500 series) harboring the vegII(cre) promoters in front of lacH were constructed and analyzed for CR. As summarized in Table 2, glucose repression of four of the tested promoters is reduced to about 50% of the wild-type repression. In the least-repressed promoter, PvegII(scrcre), repression is practically lost upon glcU or glkA mutation. These results clearly show that GlcU and GlkA indeed affect CcpA-dependent regulation. Thus, both proteins participate in glucose-mediated CcpA activation. As found previously (11, 42), the effect of glcU or glkA inactivation was strictly glucose specific (data not shown). Hence, S. xylosus constitutes the first example that glucose, which was imported independently from the PTS, triggered CcpA-mediated CR. It remains to be determined whether this regulation also exists in other gram-positive bacteria. Considering that the GlcU-GlkA system operates independently from the PTS, the question arose of whether it could activate CcpA directly, perhaps bypassing the need for a functional HPr kinase.
CcpA activity in the absence of HPr kinase.
To clarify the interdependence of GlcU-GlkA and HPr kinase in CR, the HPr kinase gene, hprK (21), was inactivated in the TX300-derived wild-type strains, yielding the TX700 series of S. xylosus strains. As shown in Table 2, glucose repression of all promoters was abolished upon hprK inactivation. The slight reduction of β-galactosidase activity in glucose-containing cultures is also apparent for the vegII promoter (TX702) missing a cre. Therefore, it is not due to CcpA-mediated regulation. Complete loss of CcpA regulation without a functional HPr kinase clearly shows that GlcU and GlkA do not activate CcpA independently, strongly suggesting that HPr phosphorylated at serine 46 (P-Ser-HPr) is absolutely required to trigger CcpA-dependent CR in S. xylosus. To explain the role of GlcU and GlkA in this process, it is reasonable to assume that the proteins produce a surplus of glycolytic intermediates such as glucose-6-phosphate or fructose-1,6-diphosphate (FDP). Higher levels especially of FDP could have two conceivable consequences. First, HPr kinase activity would be stimulated, resulting in elevated levels of P-Ser-HPr (15, 21, 33). Second, FDP could act, together with P-Ser-HPr, as a second signaling molecule to enhance CcpA binding to cres. The latter would be consistent with a number of in vitro experiments, in which the concomitant presence of FDP and P-Ser-HPr greatly stimulated binding of CcpA to cre (13, 23, 32). It was also shown that FDP is able to enhance specific interaction of CcpA with P-Ser-HPr (6), which could explain its stimulatory effect on CcpA binding.
In conclusion, dependency of CcpA repression on a functional HPr kinase was clearly established by the genomic S. xylosus promoter probe system. In addition, it was demonstrated that the PTS-independent glucose utilization system GlcU-GlkA contributes significantly to regulation by providing glycolytic intermediates to modulate HPr kinase-dependent signal transduction. Therefore, S. xylosus constitutes an interesting system with which to study the contributions of PTS-dependent and -independent glucose uptake to CcpA-mediated catabolite repression.
ACKNOWLEDGMENTS
We thank F. Götz, in whose laboratory the work was carried out, for continuous support and J. Bassias for providing lac-containing plasmids.
The work was supported by the Deutsche Forschungsgemeinschaft within the priority program Molecular Analysis of Regulatory Networks in Bacteria (BR947/4-1).
REFERENCES
- 1.Bassias J, Brückner R. Regulation of lactose utilization genes in Staphylococcus xylosus. J Bacteriol. 1998;180:2273–2279. doi: 10.1128/jb.180.9.2273-2279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:246–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 4.Brückner R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene. 1992;122:187–192. doi: 10.1016/0378-1119(92)90048-t. [DOI] [PubMed] [Google Scholar]
- 5.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dossonnet V, Monedero V, Zagorec M, Galinier A, Pérez-Martínez G, Deutscher J. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J Bacteriol. 2000;182:2582–2590. doi: 10.1128/jb.182.9.2582-2590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egeter O, Brückner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 10.Egeter O, Brückner R. Characterization of a genetic locus essential for maltose-maltotriose utilization in Staphylococcus xylosus. J Bacteriol. 1995;177:2408–2415. doi: 10.1128/jb.177.9.2408-2415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiegler H, Bassias J, Jankovic I, Brückner R. Identification of a gene in Staphylococcus xylosus encoding a novel glucose uptake protein. J Bacteriol. 1999;181:4929–4936. doi: 10.1128/jb.181.16.4929-4936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 13.Galinier A, Deutscher J, Martin-Verstraete I. Phosphorylation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J Mol Biol. 1999;286:307–314. doi: 10.1006/jmbi.1998.2492. [DOI] [PubMed] [Google Scholar]
- 14.Galinier A, Haiech J, Kilhoffer M C, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 17.Götz F, Zabielski J, Philipson L, Lindberg M. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid. 1983;9:126–137. doi: 10.1016/0147-619x(83)90015-x. [DOI] [PubMed] [Google Scholar]
- 18.Gralla J D. Activation and repression of E. coli promoters. Curr Opin Genet Dev. 1996;6:526–530. doi: 10.1016/s0959-437x(96)80079-7. [DOI] [PubMed] [Google Scholar]
- 19.Henkin T M. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 20.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 21.Huynh P L, Jankovic I, Schnell N F, Brückner R. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J Bacteriol. 2000;182:1895–1902. doi: 10.1128/jb.182.7.1895-1902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones B E, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 23.Kim J H, Voskuil M I, Chambliss G H. NADP, corepressor for the Bacillus catabolite control protein CcpA. Proc Natl Acad Sci USA. 1998;95:9590–9595. doi: 10.1073/pnas.95.16.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Grice S F, Shih C C, Whipple F, Sonenshein A L. Separation and analysis of the RNA polymerase binding sites of a complex Bacillus subtilis promoter. Mol Gen Genet. 1986;204:229–236. doi: 10.1007/BF00425503. [DOI] [PubMed] [Google Scholar]
- 26.Martin-Verstraete I, Deutscher J, Galinier A. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J Bacteriol. 1999;181:2966–2969. doi: 10.1128/jb.181.9.2966-2969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miwa Y, Nagura K, Eguchi S, Fukuda H, Deutscher J, Fujita Y. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 28.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 2000;28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen C C, Saier M H., Jr Phylogenetic, structural and functional analyses of the LacI-GalR family of bacterial transcription factors. FEBS Lett. 1995;377:98–102. doi: 10.1016/0014-5793(95)01344-x. [DOI] [PubMed] [Google Scholar]
- 30.Peschke U, Beuck V, Bujard H, Gentz R, Le Grice S. Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J Mol Biol. 1985;186:547–555. doi: 10.1016/0022-2836(85)90129-9. [DOI] [PubMed] [Google Scholar]
- 31.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Presecan-Siedel E, Galinier A, Longin R, Deutscher J, Danchin A, Glaser P, Martin-Verstraete I. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J Bacteriol. 1999;181:6889–6897. doi: 10.1128/jb.181.22.6889-6897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier M H J, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 34.Saier M H., Jr A multiplicity of potential carbon catabolite repression mechanisms in prokaryotic and eukaryotic microorganisms. New Biol. 1991;3:1137–1147. [PubMed] [Google Scholar]
- 35.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 36.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J J. Protein phosphorylation and regulation of carbon metabolism in gram-negative versus gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 37.Saier M H, Jr, Crasnier M. Inducer exclusion and the regulation of sugar transport. Res Microbiol. 1996;147:482–489. doi: 10.1016/s0923-2508(96)90150-3. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sizemore C, Wieland B, Götz F, Hillen W. Regulation of Staphylococcus xylosus xylose utilization genes at the molecular level. J Bacteriol. 1992;174:3042–3048. doi: 10.1128/jb.174.9.3042-3048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stülke J, Arnaud M, Rapoport G, Martin-Verstrate I. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 41.Wagner E, Götz F, Brückner R. Cloning and characterization of the scrA gene encoding the sucrose-specific enzyme II of the phosphotransferase system from Staphylococcus xylosus. Mol Gen Genet. 1993;241:33–41. doi: 10.1007/BF00280198. [DOI] [PubMed] [Google Scholar]
- 42.Wagner E, Marcandier S, Egeter O, Deutscher J, Götz F, Brückner R. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J Bacteriol. 1995;177:6144–6152. doi: 10.1128/jb.177.21.6144-6152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zalieckas J M, Wray L V, Jr, Fisher S H. trans-acting factors affecting carbon catabolite repression of the hut operon in Bacillus subtilis. J Bacteriol. 1999;181:2883–2888. doi: 10.1128/jb.181.9.2883-2888.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]