Abstract
Senescence is a cell fate that contributes to multiple aging-related pathologies. Despite profound age-associated changes in skeletal muscle (SkM), whether its constituent cells are prone to senesce has not been methodically examined. Herein, using single cell and bulk RNA-sequencing and complementary imaging methods on SkM of young and old mice, we demonstrate that a subpopulation of old fibroadipogenic progenitors highly expresses p16Ink4a together with multiple senescence-related genes and, concomitantly, exhibits DNA damage and chromatin reorganization. Through analysis of isolated myofibers, we also detail a senescence phenotype within a subset of old cells, governed instead by p2Cip1. Administration of a senotherapeutic intervention to old mice countered age-related molecular and morphological changes and improved SkM strength. Finally, we found that the senescence phenotype is conserved in SkM from older humans. Collectively, our data provide compelling evidence for cellular senescence as a hallmark and potentially tractable mediator of SkM aging.
Introduction
The progressive loss of skeletal muscle (SkM) mass, strength, and function with advancing age, termed sarcopenia, is evident in nearly all species. In humans, sarcopenia is a major threat to independence and quality of life 1. In the face of population aging, there is great interest in better understanding mechanisms of SkM aging to guide the development of therapeutic interventions.
Cellular senescence refers to the irreversible growth arrest that occurs when cells are exposed to various stressors 2. Senescent cells secrete pro-inflammatory cytokines, chemokines, and extra-cellular matrix degrading proteins, which are collectively known as the senescence-associated secretory phenotype (SASP) 3. The SASP contributes to tissue dysfunction by inducing paracrine senescence and inflammation, stem cell dysfunction, and alterations in the extracellular matrix 4. Senescent cells accumulate with age and at etiological sites of multiple chronic conditions 5. Importantly, genetic and pharmacological clearance of senescent cells alleviates several pathologies in mouse models of aging and age-related diseases 5.
Induction of senescence dramatically alters cell biology, including the quantity and functionality of organelles, architecture of chromatin and thereby transcriptional programming, and secretion of biologically active molecules 6. Despite profound molecular and morphological changes, there is no single, stand-alone, specific marker for senescent cell identification 7. Thus, experimental approaches that assess multiple markers, or core properties, simultaneously, are needed.
SkM is comprised of post-mitotic multinucleated cells, called myofibers, and a mixture of mitotically competent monocular cells, including satellite cells, fibroadipogenic progenitors (FAPs), endothelial cells, and macrophages. The susceptibility of SkM cell populations to senesce with advancing age is not clear. Earlier reports showed that mRNA levels of the senescence-associated genes p53 and p21Cip1 (p21 or Cdkn1a) were elevated in SkM of older mice, monkeys, and humans 8-10, while others failed to detect P16 (p16 or Cdkn2a gene) and senescence-associated beta-galactosidase (SA-β-Gal) in SkM from older adults 11. Notably, these studies relied on analyses of bulk tissues and lacked the resolution needed to define the state of specific cell types. To our knowledge, the constituent SkM cell-populations that may become senescent during aging and their core properties have not been methodically examined.
Thus, we conducted a comprehensive analysis of senescence in SkM of young and old mice and, importantly, identified the specific cell types that show a positive signature. We performed single-cell RNA sequencing (scRNA-seq) and single myofiber RNA-sequencing (RNA-seq) as well as spatially-resolved methods such as RNA in situ hybridization (RNA-ISH), immunofluorescence (IF), and immunohistochemistry (IHC). We then tested the impact of a senotherapeutic cocktail on SkM health in old mice. Finally, to address translational potential, we investigated senescence markers in SkM from younger and older humans.
Results
Aged SkM exhibits markers of cellular senescence
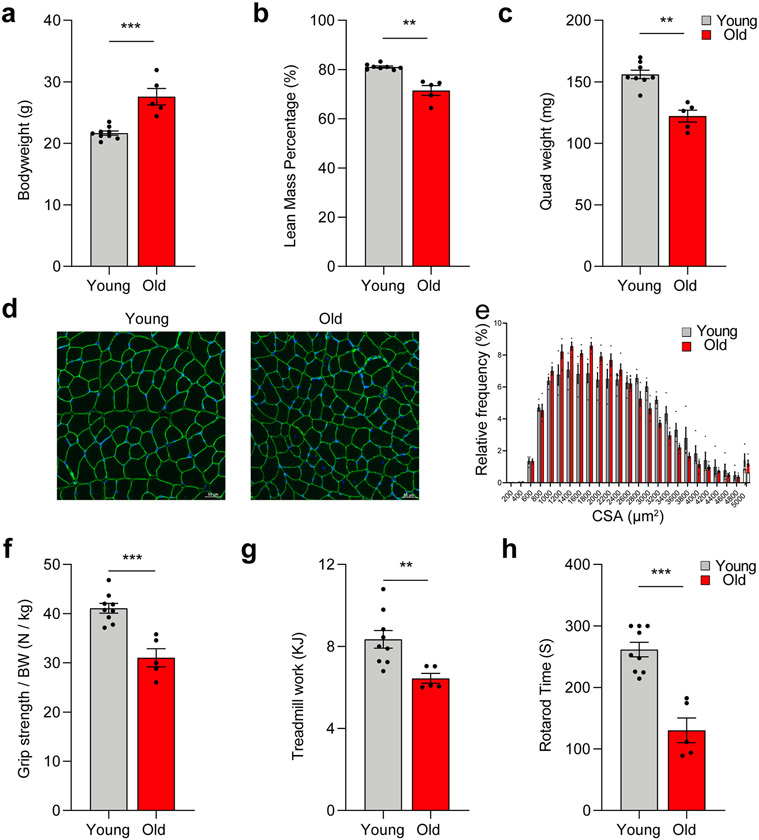
We first examined body composition and parameters of SkM health and function in young (6 months) and old (24 months) female and male mice. Old mice, regardless of sex, exhibited higher bodyweight but lower lean mass and SkM weight compared to young mice (Extended Data Fig. 1a-c). The age-associated loss of SkM mass was reflected in a reduction in myofiber cross-sectional area (CSA) (Extended Data Fig. 1d and 1e). Old mice also demonstrated significant deficits in measures of SkM performance and physical function, including grip strength, Rotarod time to failure, and treadmill work to exhaustion (Extended Data Fig. 1f-h). These data reflect the prominent phenotype of aged SkM 12.
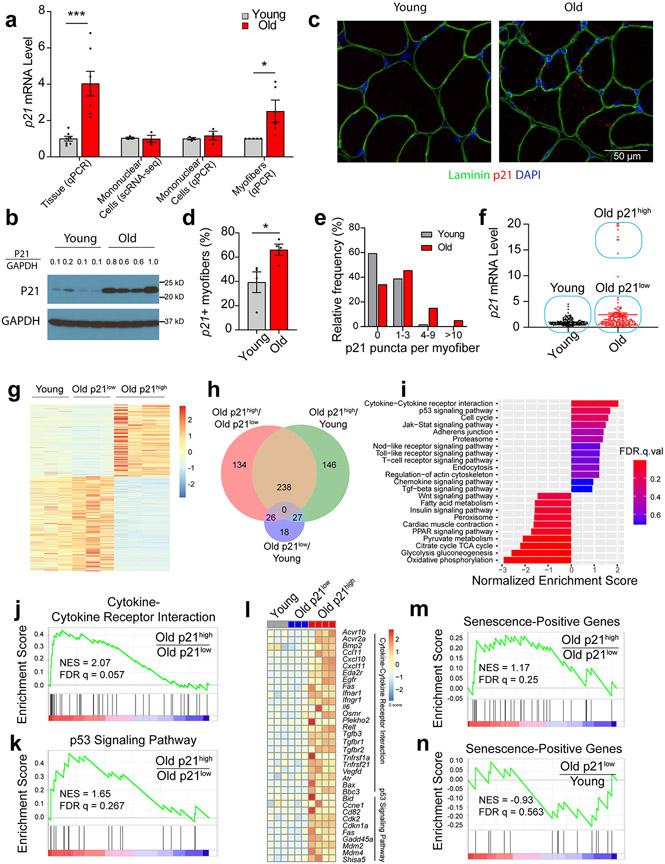
Given the absence of a stand-alone marker, we explored the presence of several core properties of senescence in SkM. First, analysis of the cyclin dependent kinase inhibitors (CDKIs) p16 and p21, which are key governors of the senescence program, by qPCR revealed significantly higher expression in SkMs isolated from old compared to young mice (Fig. 1a and 1b).
Fig. 1.
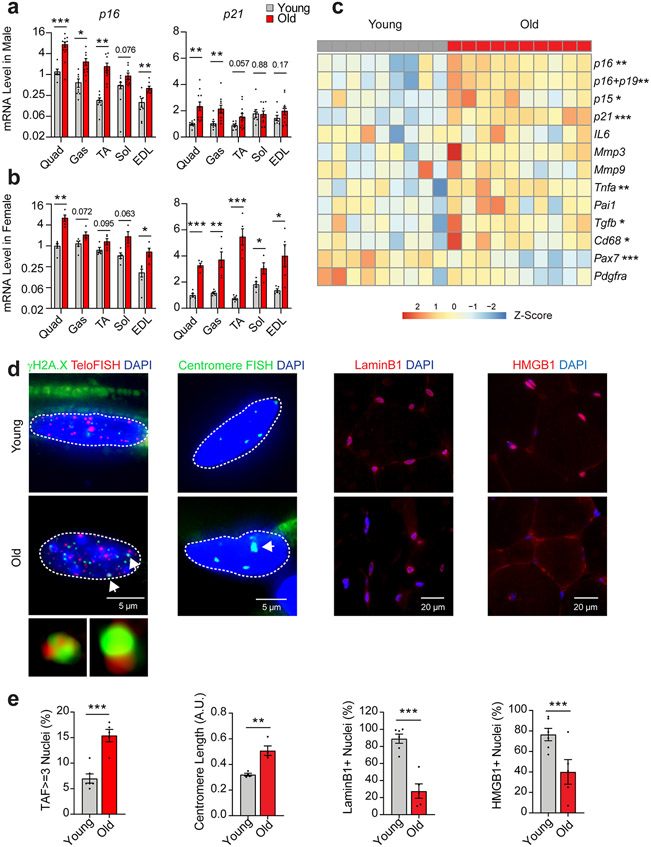
Hallmarks of cellular senescence in skeletal muscle. p16 and p21 gene expression in quadriceps (quad), gastrocnemius (gas), tibialis anterior (TA), soleus (sol), and extensor digitorum longus (EDL) muscles of (a) young (6-mo, n = 8) and old male mice (24-mo, n = 11) and (b) young (n = 5) and old female mice (n = 5; Two-tailed unpaired t tests). (c) Heatmap of senescence-related gene expression in quad muscle from young and old mice (female, young n=9, old n=10; Two-tailed unpaired t tests). Staining (d) and quantification (e) of telomere-associated DNA damage foci (TAF), senescence-associated distension of satellites (SADS), Lamin B1, and high-mobility group box protein 1 (HMGB1) in quadriceps muscle cross-sections of young (n = 6) and old (n = 5) male mice (Two-tailed unpaired t tests). Error bars represent the standard error of the mean. *, **, and *** denote p < 0.05, 0.01, and 0.001 respectively.
Higher p16 expression was most evident in SkMs of mixed contractile and metabolic properties (i.e., quadriceps (quad) and gastrocnemius as well as predominantly fast-twitch and more glycolytic SkMs (i.e., tibialis anterior and extensor digitorum longus) and was also more pronounced in old males compared to females. Age-related differences in p21 followed a similar pattern across SkM types, but were notably greater in old female mice, which also demonstrated increases in the slow-twitch oxidative soleus muscle (Fig. 1a and 1b).
Second, we compared the expression of prototypical components of the SASP between young and old quad muscle using qPCR. Old SkM had higher expression of the cytokines Il6, Tnfα, Pai-1, matrix remodeling proteins Mmp3 and Mmp9, and the growth factor Tgf-β compared to young SkM (Fig. 1c).
Third, we investigated telomere dysfunction by using immuno-FISH to quantify co-localization between DNA-damage response (DDR) protein γH2A.X and telomeres, hereafter referred to as telomere-associated foci (TAF). Telomere dysfunction has been proposed as a major inducer of senescence and results in a sustained DDR and activation of the senescence program 13. We found that a significantly higher proportion of nuclei in quad muscles of old male mice were positive for TAF (Fig. 1d and 1e).
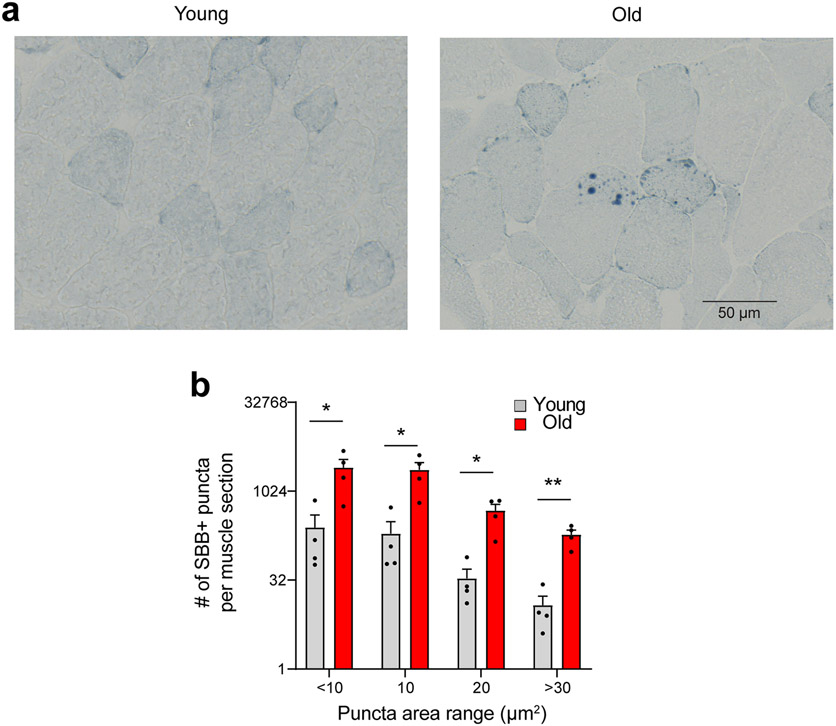
Finally, in SkM cross-sections, we assessed complementary markers strongly associated with the senescence program. We observed the loss of nuclear HMGB1 and Lamin B1 and increased centromere length in old compared to young SkM, indicative of senescence-associated satellite distension and decondensation (SADS) (Fig. 1d and 1e). Using the Sudan-Black-B histochemical stain, we also detected significantly more lipofuscin in SkM of old compared to young mice (Extended Data Fig. 2). No SA-β-Gal signal was detected, consistent with prior studies 11. Collectively, these data, in combination with upregulation of CDKIs, the SASP, and DNA damage markers, strongly suggest that SkM cell populations are prone to senesce with advancing age.
Aging, SkM mononuclear cell composition, and p16 and p21 expression
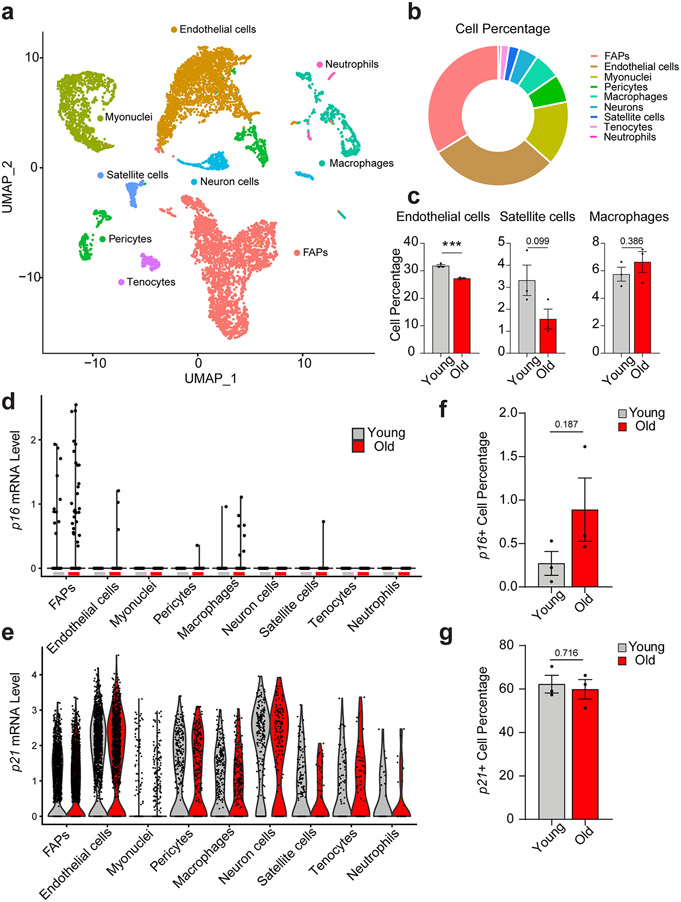
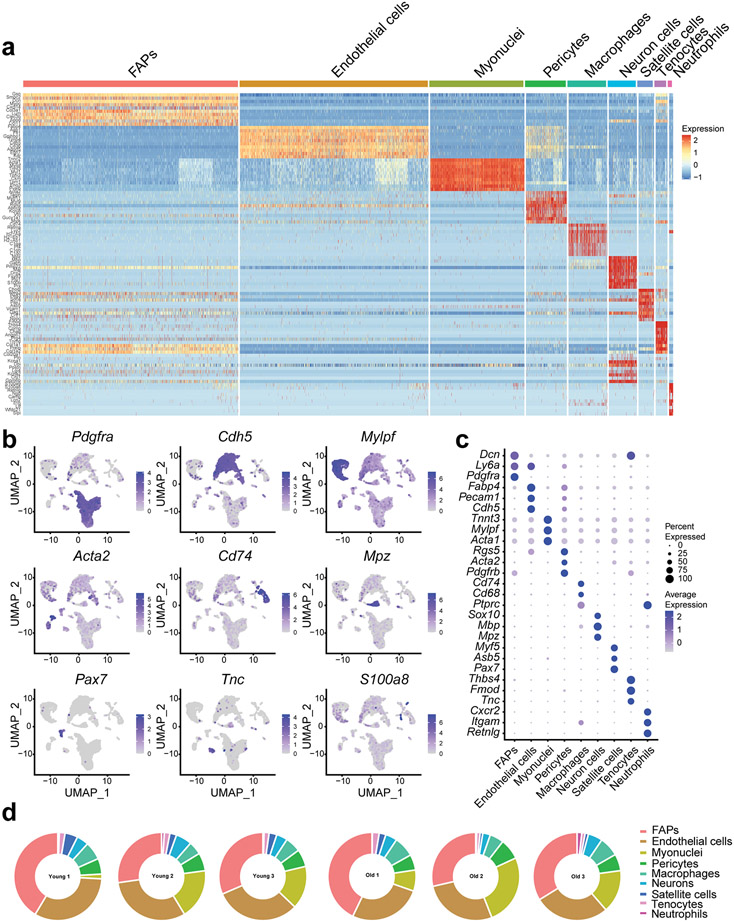
To investigate the cellular source of the age-dependent increase in senescence markers, we purified mononuclear cells from young and old female mouse SkM and performed single cell RNA-seq (scRNA-seq) using the 10X droplet-based method. We retrieved a total of 11,734 cells from 3 young and 3 old mice. Nine distinct clusters were identified based on their unique transcriptome (Fig. 2a and Extended Data Fig. 3a). Specific marker genes and canonical cell type markers identified endothelial cells (Cdh5), FAPs (Pdgfrα), macrophages (Cd74), pericytes (Acta2), neurons (Mpz), satellite cells (Pax7), tenocytes (Tnc), and neutrophils (S100a8) (Extended Data Fig. 3b and 3c) 14. Terminally differentiated, post-mitotic, myofibers, which comprise the largest component of SkM, were excluded from scRNA-seq due to mechanical/enzymatic digestion and size limitations for single cell isolation. Nevertheless, a small proportion of myonuclei (Mylpf) were ultimately still captured. Endothelial cells and FAPs were the most abundant mononuclear cell populations (Fig. 2b and Extended Data Fig. 3d). Endothelial and satellite cell abundance were lower in old compared to young SkM (Fig. 2c), as previously reported 15,16.
Fig. 2.
Single cell RNA-sequencing of skeletal muscle (SkM) reveals age-associated changes in mononuclear cell composition and p16 expression. (a) UMAP plot of the mononuclear cell populations identified by scRNA-seq in mouse SkM (data summary comprised of n = 3 young and n = 3 old female mice). (b) Relative abundance of constituent SkM cell populations. (c) Differences in the percentage of endothelial cells, satellite cells, and macrophages between young (n = 3) and old (n = 3) mice (Two-tailed unpaired Mann-Whitney tests were used). (d) Distribution of the p16 signal across cell populations revealed FAPs as the predominant source. Each dot represents the level of gene expression in a single cell, with young (gray) and old (red) samples shown in different colors. (e) The expression of p21 is evident in most SkM mononuclear cells. Each dot represents the level of gene expression in a single cell, with young (gray) and old (red) samples shown in different colors. Comparison of the percentage of (f) p16-positive cells and (g) p21-positive cells by scRNA-seq between young (n = 3) and (n = 3) old mice (Two-tailed unpaired t tests). Error bars represent the standard error of the mean. *** denote p < 0.001.
Examination of CDKI expression revealed that p16 was only detectable in specific cell populations, most prominently in FAPs and macrophages derived from SkM of old mice (Fig. 2d), while p21 was evident in most cell types of both young and old mice (Fig. 2e). Notably, we identified a trend for increased p16-positive cells in old SkM compared to young (Fig. 2f) while the percentage of p21-expressing mononuclear cells did not differ (Fig. 2g). These data suggest that p16, but not p21, increases in mitotically competent SkM cell populations, particularly FAPs, with age.
An interactive platform to explore single cell data has been developed and is available at https://mayoxz.shinyapps.io/Muscle/.
p16-expressing FAPs have a robust senescence phenotype
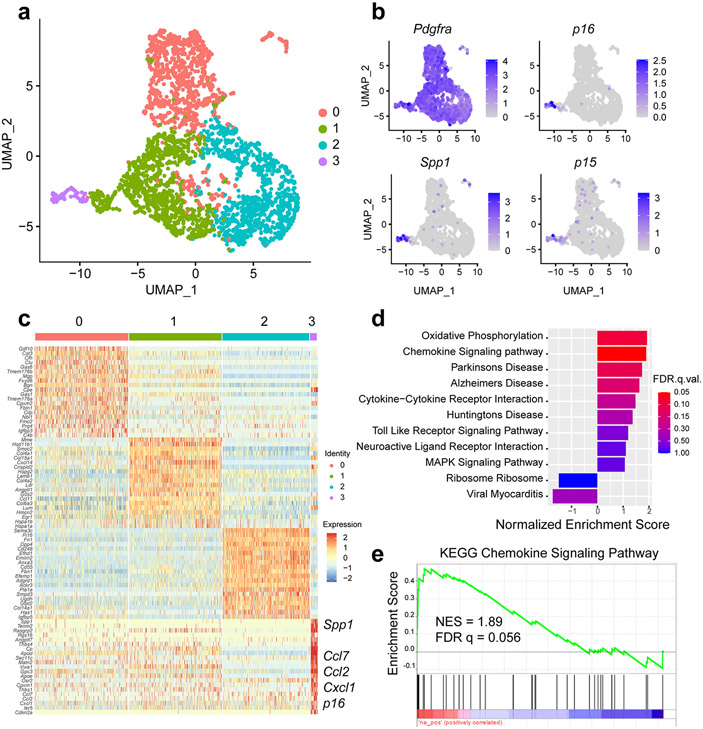
Further analysis of scRNA-seq data indicated that only a small subset of FAPs highly expressed p16 (39 (1.3%) of 3,063 cells). We hypothesized that these FAPs would have a unique transcriptional profile and, subsequently, clustered FAPs into four distinct subpopulations based on transcriptional differences (Fig. 3a).
Fig. 3.
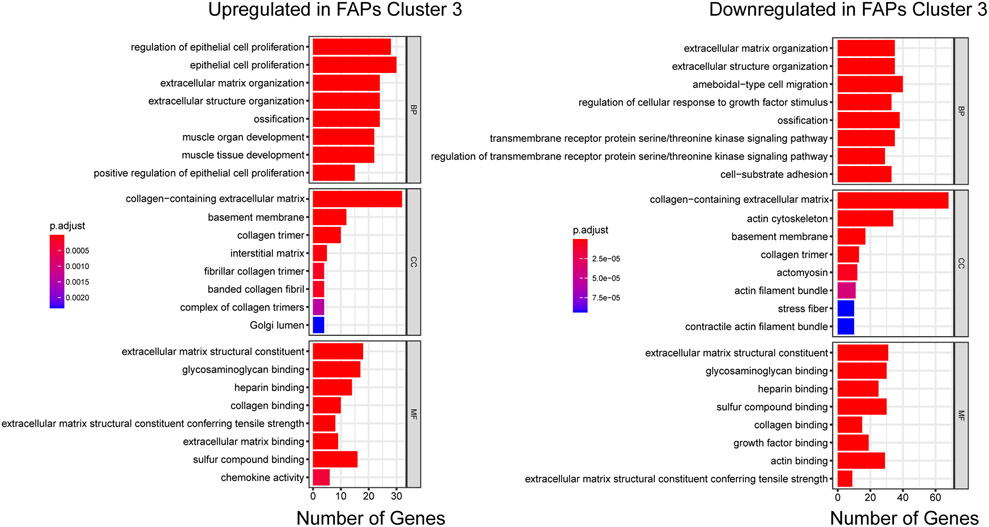
A distinct cluster of high p16-expressing FAPs in old skeletal muscle exhibits a senescence phenotype. (a) A UMAP plot of FAPs from 3 young and 3 old female mice reveals four distinct clusters. (b) The FAPs marker gene Pdgfra is distributed across all clusters, but senescence-related genes p16, p15, and Spp1 are enriched in cluster 3. (c) Heatmap of scRNA-seq-derived marker genes for the four FAP subclusters, with several labels for several senescence-related genes enriched in cluster 3. (d) KEGG pathway enrichment analysis of cluster 3 specific genes using GSEA. Top enriched pathways were selected for plotting. (e) GSEA enrichment plot of the chemokine signaling pathway in FAPs cluster 3.
All four cell subsets expressed the FAP marker Pdgfrα (Fig. 3b). Interestingly, the high p16 signal was uniquely enriched in cluster 3 (p16high) (Fig. 3b). Examination of differentially expressed genes (DEGs) between clusters revealed enrichment of multiple senescence-related genes (e.g., p16, p15, Spp1, Ccl2, and Ccl7) in the p16high cluster (Fig. 3b and 3c). Gene Ontology (GO) analysis of 1,429 differentially expressed genes in this distinct cluster highlighted biological processes including cell proliferation regulation, collagen processing, and chemokine activity (Extended Data Fig. 4). Gene set enrichment analysis (GSEA) further emphasized upregulation of chemokine signaling, cytokine-cytokine receptor interactions, and the MAPK signaling pathway (Fig. 3d and 3e), which are all activated in senescent cells 17,18. Indeed, the transcriptional profile of the p16high FAPs is strikingly consistent with aspects of a senescence program.
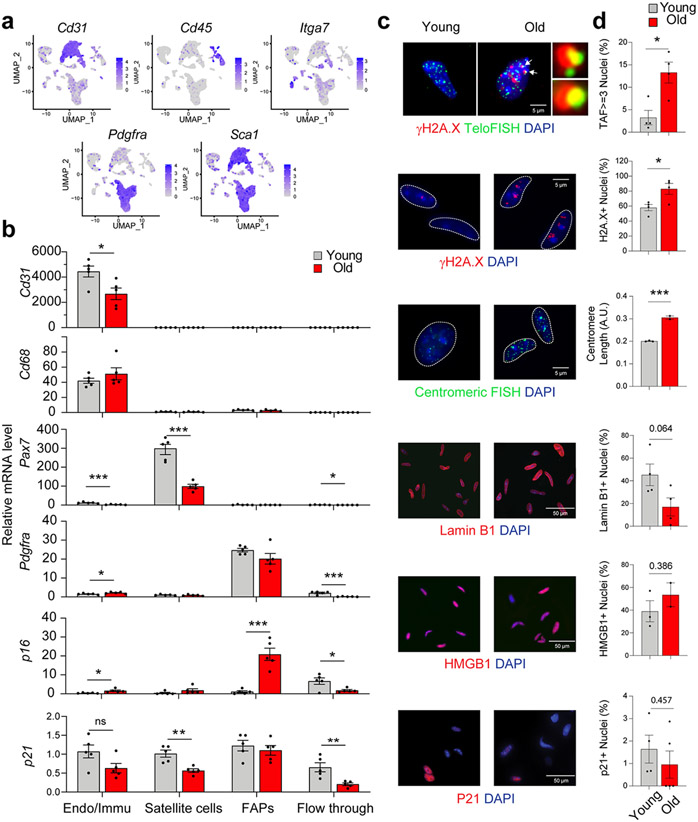
To corroborate scRNA-seq findings, four surface markers (CD31, CD45, Integrin α-7, and SCA1) were used to isolate and separate four cell populations of interest by magnetic activated cell sorting (MACS) (Fig. 4a). The specificity of endothelial cell (Cd31+), macrophage (Cd68+), satellite cell (Pax7+), and FAP (Pdgfrα+) isolations was confirmed by qPCR (Fig. 4b). Consistent with scRNA-seq data, FAPs were the predominant p16-expressing cell population, and p16 expression by qPCR was significantly higher in old compared to young FAPs. We also observed modest increases in p16 expression in old endothelial/macrophage populations and subtle, non-significant increases in old compared to young satellite cells (Fig. 4b), as reported prior 15. These data, in combination with scRNA-seq findings, point to FAPs as the predominant mononuclear cell population contributing to the p16 signal in SkM of old mice.
Fig. 4.
FAPs derived from old skeletal muscle (SkM) exhibit core properties of senescence. (a) UMAP plot of the markers used for MACS sorting. (b) Confirmation of effective endothelial/immune (Endo/Immu), satellite cell, and FAP separation by qPCR-based measures of cell type markers, and validation of FAPs as a major source of p16 signal in old SkM (n = 5 for both young and old samples, two-tailed unpaired t tests). Representative IF images (c) and quantification (d) of TAF, γH2A.X, SADS, Lamin B1-positive nuclei, HMGB1-positive nuclei, and P21 in FAPs isolated from young and old mice (2-5 male and female mice per group as detailed in Source Data, two-tailed unpaired t tests). Error bars represent the standard error of the mean. *, **, and *** denote p < 0.05, 0.01, and 0.001 respectively.
To further establish that p16-expressing FAPs are indeed senescent, complementary markers were measured in FAPs isolated from young and old mice. Consistently, FAPs derived from old mice exhibited significantly greater TAF, γH2A.X-positive nuclei, and centromere length by IF compared to FAPs derived from young mice (Fig. 4c and 4d). No significant age-related reductions in Lamin B1 and HMGB1 or increases in p21 expression were observed in isolated FAPs (Fig. 4c and 4d). Collectively, the coordinated increase in expression of p16, SASP factors, and SASP-effector pathways and marks of DNA damage and chromatin reorganization indicate that a distinct subset of FAPs become senescent with advancing age.
High p21-expressing myofibers exhibit hallmarks of senescence
Interestingly, the robust p21 expression observed in SkM of old compared to young mice (Fig. 1a) was not detectable in the mononuclear cell populations examined by either scRNA-seq or qPCR (Fig. 5a). The age-related increase in P21 was also evident by Western blot (Fig. 5b). We hypothesized that myofibers (myonuclei and sarcoplasm), were a plausible source of the high p21 mRNA detected in old SkM and, therefore, isolated single myofibers from EDL muscles of young and old mice and quantified p21 expression by qPCR. Indeed, higher levels of p21 were evident in myofibers from old compared to young mice (Fig. 5a). RNA-ISH staining of SkM cross-sections further demonstrated p21 was significantly increased within the myofiber boundaries of old compared to young SkM (Fig. 5d). Myofibers with 1-3 puncta were relatively common in both young and old SkM, but myofibers with 4-9 puncta were more frequent in old rather than young, and myofibers with more than 10 puncta were unique to old SkM (Fig. 5e). In combination, these data strongly suggest that myofibers are the primary source of the high p21 expression observed in SkM of old mice.
Fig. 5.
Skeletal muscles (SkM) of old mice have high p21-expressing myofibers with a senescence profile. (a) Comparison of p21 expression between young and old SkM tissue (n = 7 for both young and old samples) and isolated mononuclear cells (n = 7 for both young and old samples) by qPCR, mononuclear cells by scRNA-seq (n = 7 for both young and old samples), and isolated myofibers by qPCR (n = 5 for both young and old samples, two-tailed unpaired t tests). (b) Western blot of P21 and GAPDH in quadriceps muscle from young and old female mice (n = 4 per group). P21/GAPDH represents the ratio of the P21 signal intensity compared to GAPDH for each sample. (c) RNA-ISH staining of p21 and IF staining for Laminin in young and old quadriceps muscle cross-sections (n = 4 per group). (d) Quantification of the percentage of young and old myofibers staining positively for p21 puncta (two-tailed unpaired t test) and (e) the percentage of young and old myofibers with 0 to greater than 10 puncta. (f) Analysis of p21 expression by qPCR in 159 young and 159 old isolated, individual myofibers from female mice. (g) Differentially expressed genes (DEGs) between young, old p21low, and old p21high myofibers identified in the RNA-seq. (h) Venn diagram of DEGs between the three groups of myofibers. (i) Enriched pathways in old p21low compared to old p21high myofibers identified by GSEA, negative NES represent pathways enriched in old p21low myofibers. GSEA plot of (j) KEGG cytokine-cytokine receptor interactions and (k) the KEGG p53 signaling pathway based on DEGs between old p21high and old p21low myofibers. (l) Heatmap of the core DEGs in cytokine-cytokine receptor interactions and the p53 signaling pathway. GSEA plots of transcripts within the CellAge gene set that are positively correlated with senescence and are DEGs between (m) old p21high and old p21low myofibers, or (n) old p21low and young myofibers. Two-tailed unpaired t test was used; error bars represent the standard error of the mean. * and *** denote p < 0.05 and 0.001 respectively.
To further investigate age- and p21-associated changes in myofibers, we first isolated a total of 159 single myofibers from seven young and a total of 159 single myofibers from seven old mice and then quantified p21 expression by qPCR. Pax7 was undetectable, suggesting that satellite cells were a negligible source of RNA from the myofiber. Myofibers isolated from old mice separated into two different clusters: one large cluster with relatively low p21 expression (p21low) and one smaller cluster (10 (6.3%) of 159 cells) with notably higher p21 expression (p21high) (Fig. 5f). Of note, p21 levels were similar between young myofibers and old p21low myofibers.
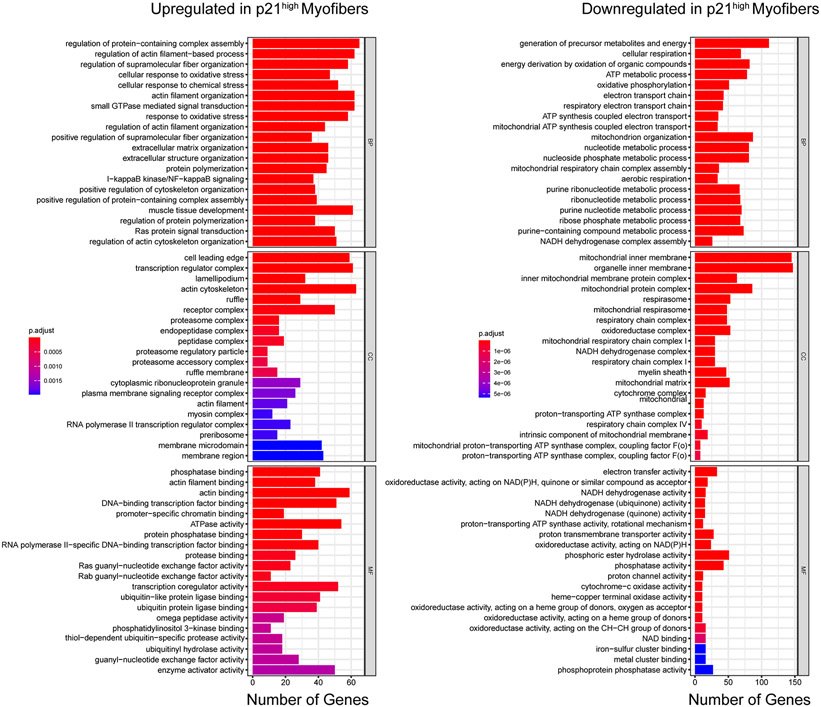
Next, we conducted RNA-seq on individual myofibers from the three different populations: young, old p21low, and old p21high. DEGs analysis showed that the transcriptional profile of old p21low myofibers was similar to young myofibers, while the old p21high myofibers were distinct from these two populations (Fig. 5g). We identified 398 DEGs (fold-change > 2.0 and p < 1 X 10−5) between old p21low and old p21high myofibers, 411 genes between young and old p21high myofibers, but only 71 genes between young and old p21low myofibers (Fig. 5h). GO analysis was performed to annotate the function of DEGs unique to old p21high myofibers and revealed upregulation of oxidative and chemical stress response pathways, the NFκB pathway, and Ras signaling (Extended Data Fig. 5). These cells also exhibited decreased metabolic respiration and oxidative phosphorylation pathways. Of note, p16 was undetectable in isolated myofibers by either RNA-seq or qPCR.
Further evaluation of differences between old p21low and old p21high myofibers using GSEA analysis demonstrated significant enrichment of cytokine-related pathways, p53 signaling, the Jak-Stat pathway, chemokine signaling, and the TGF-β pathway (Fig. 5i), which are all linked to senescence 19,20. Enrichment score plots and a heatmap of cytokine-cytokine receptor interaction and p53 signaling genes illustrated the striking contrast between old p21low and old p21high myofibers as well as the remarkable similarity between young and old p21low myofibers (Fig. 5j-l). Finally, we queried the Database of Cell Senescence Genes (CellAge), a dataset that consists of 153 genes positively associated with senescence, to comprehensively analyze differential expression between the three cell populations 21. GSEA analysis showed clear enrichment of senescence-related genes in old p21high myofibers compared to old p21low myofibers (Fig. 5m), but no enrichment in old p21low myofibers compared to young p21low myofibers (Fig. 5n). Collectively, these data suggest that a subgroup of terminally differentiated myofibers in old mice acquires a senescence-like transcriptional profile, including high p21 expression and a significant increase in p53 signaling and cytokine-cytokine receptor interactions.
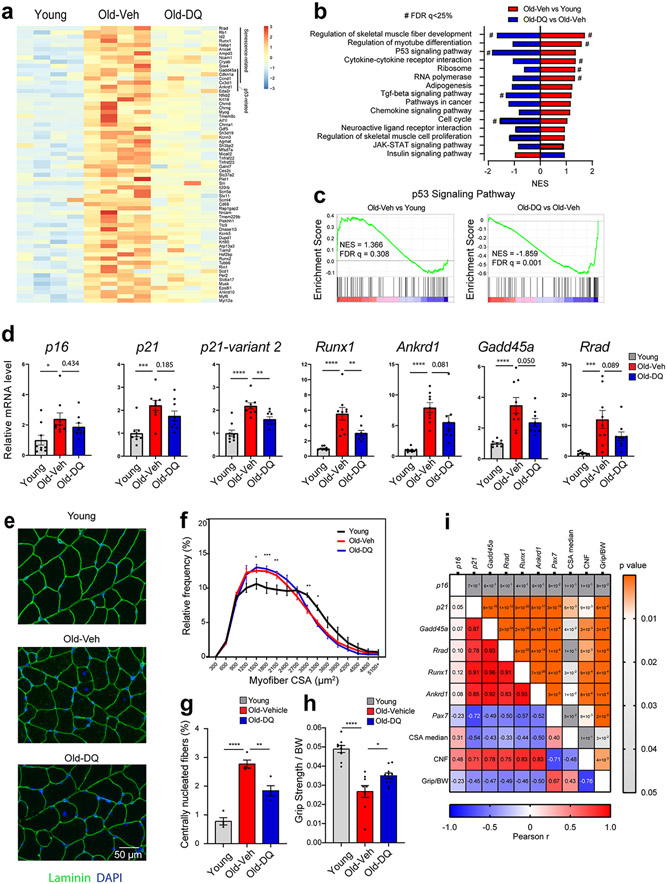
A senotherapeutic improves the molecular profile and function of SkM
To test whether senescent cells meaningfully contribute to hallmarks of SkM aging in vivo, we administered either vehicle (veh) or the senotherapeutic drug combination dasatinib plus quercetin (DQ) to 20-month-old mice for four months. Following the intervention, RNA-seq analysis revealed that the higher expression of a significant number of genes in the SkM of old-veh mice compared to young (6 month) mice was markedly attenuated in old-DQ mice (Fig 6a). Importantly, DQ countered the age-associated increase in multiple senescence and p53/p21-related genes, including Cdkn1a, Gadd45a 22 , Rrad 23, Runx1 24, Ampd325, Ncam1 26, Cryab, Ccnd1 27, Cx3cl1 28, and Ankrd1 29. Moreover, GSEA pathway analysis demonstrated that key pathways enriched in both p21high myofibers and SkM of old-veh mice (i.e., p53 signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway) were reversed in SkM of old-DQ mice. (Fig 6b and 6c). Additional analyses by qPCR (Fig 6d) did not show statistically significant differences in either p16 or p21 expression between the two groups of old mice; however, p21 variant 2 expression, which was recently reported to be more responsive to aging 30, was significantly lower in the SkM of old-DQ compared to old-veh mice (Fig 6d). Further analysis also confirmed DQ-driven reductions in the expression of additional elements of the p53 signaling pathway observed in RNAseq data, including Gadd45a, Rrad, Runx1, and Ankrd1.
Fig 6.
Senolytics improve the molecular phenotype and function of skeletal muscle (SkM) in female mice. (a) heatmap of the differentially expressed genes between young and old-vehicle mice which exhibited improvements in the old-DQ treated group. (b) GSEA enriched pathways between old-vehicle vs young groups (red) and between old-DQ vs old-vehicle groups (blue). X-axis shows the normalized enrichment score. Negative NES represent downregulation in the group. (c) GSEA plots of P53 signaling pathway between the two group comparisons. (d) qPCR confirmation of the RNAseq data in a larger cohort of mice (n = 9 per group; One way ANOVA test). (e) IF laminin staining on the SkM samples (Representative image of 4 mice per group). (f) CSA distribution in young, old-veh, and old-DQ samples. Asterisks denote the differences between young and old-veh groups (n = 4 per group, two way ANOVA). (g) Centrally nucleated fibers (CNF) in muscles from different groups (n = 4 per group). (h) Grip strength normalized to body weight of mice from different groups (n = 9-10 per group). (i) Pearson correlation analysis between gene expression and SkM morphological measures, and grip strength, with Pearson r value on the left and p value on the right. Error bars represent the standard error of the mean. *, **, and **** denote adjusted p value < 0.05, 0.01, and 0.0001 respectively. # denotes FDR q-value < 0.25 in GSEA analysis.
Age-related reductions in SkM mass and myofiber CSA were not attenuated by DQ (Fig 6e and f). However, the number of centrally nucleated fibers, which were highly prevalent in SkM of old-veh mice, was markedly reduced in old-DQ mice (Fig 6e and g). Importantly, we also detected significantly greater grip strength in old-DQ compared to old-veh mice (Fig 6h). Correlation analyses highlighted moderate to strong positive associations between p21 and other senescence markers and centrally nucleated fibers, and significant inverse associations with myofiber CSA and grip strength. In contrast, no significant correlations were observed with p16 (Fig. 6i). These data suggest that the senotherapeutic cocktail DQ partially counters senescent cell burden in SkM and improves some parameters of SkM quality in aged mice.
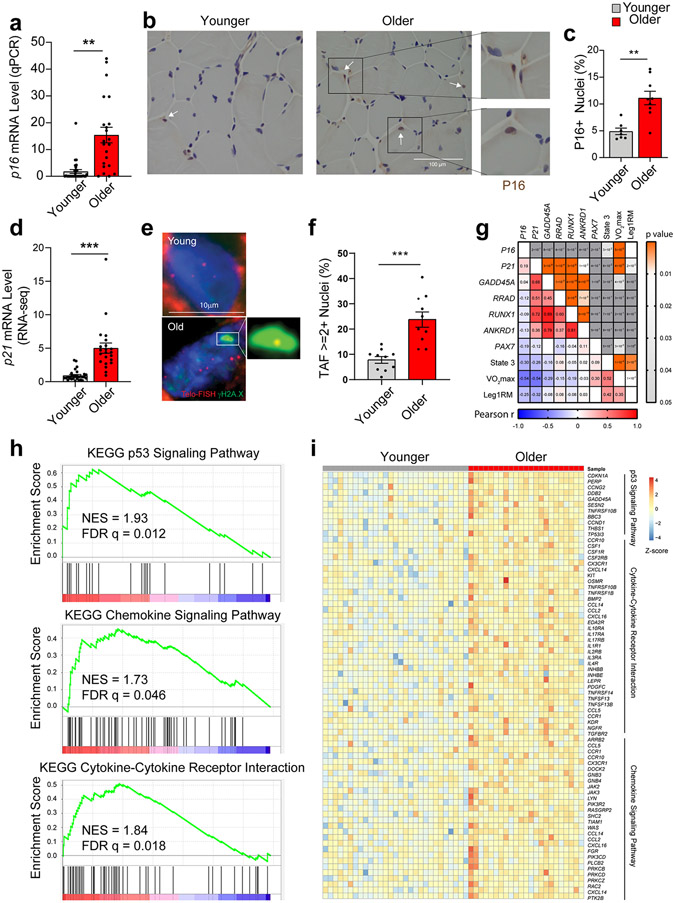
Age-associated increases in senescence markers in human SkM
To assess the translational potential of our preclinical findings, we first analyzed SkM biopsy RNA-seq data from younger (n = 30) and older (n = 22) women and men 31. We failed to detect a P16 transcript. Considering that bulk RNA-seq may not be sufficiently sensitive to detect low abundance transcripts, we performed qPCR. Consistent with mouse SkM, we observed higher P16 expression in tissue derived from older participants compared to younger participants (Fig. 7a). Immunohistochemical staining of SkM biopsies also revealed a higher abundance of P16-positive nuclei in older male and female individuals, which, on two-dimensional cross-sections, appeared to be both within and external to the myofibers (Fig. 7b and 7c). A significant age-related increase in P21-expression was evident by RNA-seq (Fig. 7d). We also observed significantly greater TAF-positive nuclei in SkM cross-sections of older compared to younger humans, in accordance with a senescence phenotype (Fig. 7f).
Fig. 7.
Aging and cellular senescence in human skeletal muscle (SkM). (a) Quantification of p16 expression by qPCR in SkM biopsy specimens from younger (n = 30) and older (n = 22) women and men (Two-tailed unpaired t test). (b) Representative images and (c) quantification of P16-positive nuclei by IHC staining (n = 5 per age group). (d) Comparison of p21 expression by qPCR between young (n = 29) and older (n = 22) SkM biopsies specimens. Representative images of TAF staining (e) and quantification of TAF-positive nuclei (f) (n=10 per age group, two-tailed unpaired t test). (g) Pearson correlation analysis between SkM gene expression and functional measurements including maximal oxygen consumption (VO2max, ml/kg/min), leg one-repetition maximum (AU) normalized to fat-free mass (kg) of the leg (Leg1RM), and maximal mitochondrial oxygen consumption (State 3) after the addition of saturating concentrations of adenosine diphosphate (pmol/s/mg tissue), with Pearson r value on the left bottom corner and p value on the right top corner. (h) GSEA plot of the p53 signaling pathway, the chemokine signaling pathway, and cytokine-cytokine receptor interactions that were significantly enriched based on DEGs by RNA-seq between younger and older human SkM. (i) Heatmap of expression of core genes in the enriched pathways. Two-tailed unpaired t test was used; error bars represent the standard error of the mean. ** and *** denote p < 0.01 and 0.001 respectively.
As in mice, correlation analysis highlighted significant associations between P21, but not P16, and the expression of several senescence markers affected by DQ, including GADD45A, RRAD, RUNX1, and ANKRD1, in human SkM (Fig. 7g). Further analysis RNA-seq data showed enrichment of the p53 signaling pathway, cytokine-cytokine receptor interactions, and chemokine pathways in older compared to younger human SkM (Fig. 7h and 7i), which is also strikingly consistent with our findings in mouse SkM and p21high myofibers (Fig. 5i-k). Regarding parameters of SkM quality and function, the expression of both P16 and P21 had modest inverse associations with SkM oxidative capacity (state 3 respiration), maximal oxygen consumption (VO2max), and leg strength (one-repetition maximum) (Fig. 7g). In total these data suggest that cellular senescence is a conserved mechanism of SkM aging between mice and humans, which may contribute to sarcopenia.
Discussion
In this study, we comprehensively analyzed and defined the SkM cell populations that are susceptible to cellular senescence with chronological aging. In mice with age-associated deficits in SkM mass, strength, and function, we identified a subpopulation of FAPs that exhibits a significant increase in p16 and other core properties of cellular senescence, including a SASP and markers of DNA damage and chromatin reorganization. Moreover, we discovered that a subset of terminally differentiated myofibers acquires an age-dependent increase in p21 and marked enrichment of senescence-associated pathways. Administration of a senotherapeutic intervention to old mice modestly countered these molecular features of SkM aging and improved grip strength. We confirmed core properties of the senescence program are evident in SkM of older humans and associate with parameters of SkM quality and function, which highlights the translational potential of these findings. Collectively, our data strongly suggest cellular senescence is a core feature of SkM aging and a potential contributor to sarcopenia.
Prior studies exploring cellular senescence in SkM have relied on analyses of bulk tissue, leaving questions regarding the origin and validation of possible signals (e.g., age-related increases in CDKI expression) unanswered 8-10. To address this gap, we first leveraged scRNA-seq to study the molecular phenotype of mitotically competent mononuclear cell populations from the SkM of young and old mice. Our data demonstrate that a distinct subset of the FAP population is the predominant source of p16 observed in old SkM. Old FAPs also exhibit a proinflammatory SASP, TAF, and increased centromere length, which signify the cell fate of senescence.
As multi-potent progenitors with the ability to differentiate into fibroblasts and adipocytes, FAPs are critical for efficient SkM repair, myogenic commitment, and SkM homeostasis after damage caused by injury or exercise 32-34. It was recently shown that transplantation of young but not old FAPs restores the myogenic potential of old satellite cells 34, highlighting intrinsic changes in FAPs with age and the significance of cell-cell communication and FAP-derived autocrine and paracrine signals to SkM health 35. In this study, high p16 and p21 expression were not seen in the same senescent cells, emphasizing the heterogeneity of the senescence signature across different cell types. We did not observe an increase in p21 expression in FAPs with chronological aging, nor did prior studies of SkM injury and regeneration 36, suggesting p16 may be the major regulator of senescence in FAPs. While it has been reported that transient senescence in FAPs is beneficial to regeneration following exercise 36, chronic senescence evidenced by a deleterious SASP and resistance to apoptosis may lead to fibrosis or accumulation of fat and immune cells, which are characteristics of aged SkM. Indeed, FAPs that highly expressed p16 exhibited significant alterations in profibrotic factor Spp1 and chemokines including Ccl2 and Ccl7, consistent with playing a detrimental role in age-associated fibrosis and chronic inflammation.
The strengths of discovery by scRNA-seq are balanced by high cost and the potential for low abundance transcripts, like p16, not to be effectively converted into cDNA by 3’ end reverse transcription. Even so, we observed a trend for a greater percentage of p16-postive FAPs in SkM of three old compared to three young mice. Orthogonal analysis of SkM mononuclear cell populations from larger cohorts of young and old mice using MACS and qPCR corroborated scRNA-seq data and underscored the high p16 signal and senescent phenotype of old FAPs. We also noted subtle trends for age-associated increases in p16 expression in macrophages and satellite cells. Macrophages were not pursued further as p16 expression in this cell type may not be indicative of senescence 37. In satellite cells, a prior study reported an inverse relationship between p16 expression and regenerative capacity following injury in mice of advanced (28 months) age 15. Other properties of senescence in satellite cells are worthy of further exploration in this context, but in the case of sarcopenia, the significance of satellite cells remains unclear due to conflicting reports on the impact of their depletion on its onset and progression 38-40.
Bulk analysis of mouse and human SkM by RNA-seq revealed robust age-related increases in p21; however, to our surprise, no differences in expression were observed in the mononuclear cell populations of young and old mice. Instead, through RNA-ISH analysis of young and old SkM cross-sections and, in parallel, isolation and qPCR analysis of young and old myofibers, we discovered that a fraction of old myofibers highly express p21. We note that a recent preprint study that used single-nuclei RNA-seq also identified a cluster of high p21 expressing myonuclei in SkM of older adults 41. Here, we further demonstrate that old p21high myofibers are uniquely enriched with p53, NFκB, cytokine signaling pathways, and a repertoire of cataloged senescence-associated genes (CellAge). Notably, these enriched pathways were also evident in SkM of older compared to younger humans and countered by the administration of a senotherapeutic cocktail to old mice. We did not detect p16 activation by qPCR, RNA-seq, or IF in myofibers, further supporting a p21-governed senescence program. In knock-out cells, p16 or p21 alone is enough to drive senescence 42, suggesting the compensatory role of the two CDKIs during senescence development. However, in other contexts, p21 expression has been shown to be activated rapidly after stress followed by a gradual elevation in p16 expression in later stages of senescence 43, suggesting detection may be time-course dependent. As such, studying p16/p21 knock-out models under different biological conditions will be necessary to illustrate the distinct/collaborative roles of these two CDKIs in different cell types.
Traditionally, the field has maintained that only mitotically competent cells can undergo senescence; however, post-mitotic senescence is an emerging paradigm 44. Several studies have reported a senescent-like phenotype in terminally differentiated cells including neurons 45, cardiomyocytes 46 and osteocytes 47. Given that senescence can be induced by stressors that are not specific to replicating cells, it is plausible that post-mitotic cells could also be susceptible. For example, DNA damage in telomere regions, independent of replicative stress, can elicit a DDR and induce a senescence-like program in terminally differentiated cells 13. Through robust alterations in their transcriptional profile and corresponding changes in cellular activities (e.g., metabolic pathways and secretion of bioactive molecules), senescent post-mitotic cells are poised to exert deleterious effects on tissue health and function. In line with this, genetic and pharmacological clearance of p16-positive post-mitotic osteocytes resulted in an amelioration of age-related bone loss, potentially by reducing the SASP and, in turn, improving the microenvironment 47.
Prior studies have shown senescent cells can have a detrimental effect on SkM performance and physical function. For instance, transplantation of senescent preadipocytes into young mice was sufficient to compromise grip strength and exercise capacity 48. Moreover, pharmacological and genetic clearance of senescent cells have been reported to positively impact parameters of SkM performance and physical function in the context of senescence-inducing stressors, accelerated aging, and chronological aging 49,50. Although informative, in the absence of comprehensive analysis of core properties of senescence in constituent SkM cells, it is difficult to distinguish between SkM-specific and systemic effects of these study interventions on integrative measures of health.
To address this gap, we administered either vehicle or DQ to older mice to examine the effects on SkM molecular and functional phenotypes. Our data demonstrate that DQ significantly reversed the expression of genes and enrichment of pathways evident in the SkM of old versus young mice and associated with a senescence profile, especially as discovered in p21high myofibers. More modest effects were observed on the abundance of centrally nucleated fibers, which may be reflective of improved innervation or reduced degeneration 51, and grip strength, a primary measure of SkM performance.
Limitations of the study
One limitation of the current study is 10x scRNA-seq method fails to capture the whole length of mRNA, which is not ideal for low abundance transcripts like p16. The high cost of scRNA-seq also prohibits analysis of a larger number of samples. These issues emphasize the importance of orthogonal approaches (e.g., qPCR of MACS-sorted cells) and complementary methods, such as IF and IHC, for quantification of core senescent cell properties. We also acknowledge that the direct effects of DQ on either senescent FAPs or myofibers were not conclusively determined. D and Q were identified based on screens in human preadipocytes and endothelial cells and mouse embryonic fibroblasts, not the senescence-prone cell populations we have identified in SkM 49. Based on differences in the transcriptional profile between untreated and treated old mice (i.e., reductions in p53/p21-related genes and cytokine-cytokine receptor interaction and chemokine signaling pathways), we posit p21-expressing myofibers, more so than p16-expressing senescent FAPs, are affected by DQ; however, we did not directly assess cell death. Systemic effects of DQ, that may contribute to changes in measures of muscle function indirectly, cannot be ruled out. Additional studies that leverage genetic approaches to specifically target these cells independent of systemic effects will validate and guide the development of senotherapeutic candidate(s) that affect either the abundance (senolytics) or behavior (senomorphics) of p16high FAPs and/or p21high myofibers as a novel approach to improve late-life SkM health and function.
In summary, through methodical study of SkM cell populations from young and old mice, we identified core properties of cellular senescence, including CDKI upregulation, SASP, DNA damage, and chromatin reorganization, specifically enriched in subpopulations of SkM FAPs and myofibers. Administration of a senotherapeutic therapy attenuated the molecular features of senescence in SkM and improved features of SkM health. In total, these data suggest senescent cells may contribute to SkM aging. To this end, future studies are warranted to better understand the mechanistic role of senescent FAPs and myofibers in SkM aging and the therapeutic impact of their targeted elimination. Importantly, we observed conserved features of senescence in the SkM of older humans, underscoring the translational potential of our findings.
Methods
Mice
Male and female C57BL/6 mice aged 3-5 months and 22 months were either purchased from the National Institute on Aging and Harlan Laboratories (Blackthorn, UK) or bred in-house. Mice were group-housed in ventilated cages with a constant temperature of 25°C, 30-70% humidity, a 12-hour light/dark cycle, and provided standard chow. All animal experiments in this study were approved by the Mayo Clinic Institutional Animal Care and Use Committee or the Newcastle University Animal Welfare Ethical Review Board and were conducted in compliance with the UK Home Office (PPL60/3864). For senotherapeutic drug treatment, 20-month-old female mice were gavaged with vehicle (10% ethanol, 30% polyethylene glycol, and 60% Phosal 50) or dasatinib (5 mg/kg) (D) plus quercetin (50 mg/kg) (Q) for 3 continuous days, every 2 weeks, for 4 months. Thus, mice received a total of 24 doses of either vehicle or DQ. At the end of the study, mice were administered a lethal dose of pentobarbitol, exsanguinated, and perfused with PBS. Tissues were then harvested, processed, and/or stored for the analyses detailed below.
Body composition, SkM strength, and physical function measurements
Body composition (total body lean and fat mass) of young and old mice was assessed by quantitative MRI (EchoMRI-100; Houston, TX, USA) as described previously 52. Exercise capacity, defined by running time, distance, and work to exhaustion, was tested on a motorized treadmill (Columbus Instruments, Columbus, OH, USA) as reported prior 53. Forelimb grip strength was determined using a Grip Strength Meter (Columbus Instruments) 53. Neuromuscular coordination was measured using a Rotarod apparatus (TSE system, Chesterfield, MO, USA) as formerly described 48.
Preparation of mouse SkM sections
A subset of isolated mouse quad muscles from young and old mice were fixed in 4% formaldehyde aqueous solution (VWR, Radnor, PA, USA) and further processed into paraffin-embedded blocks (FFPE). Additional mouse quad muscles were embedded in Optimal Cutting Temperature compound (Sakura Finetek, Torrance, CA, USA), frozen in liquid nitrogen-cooled isopentane (Sigma, St. Louis, MO, USA), and stored at −80°C. Transverse 3-μm-thick sections were cut from FFPE blocks. Transverse 7 μm-thick frozen sections were cut with a Leica cryostat. Fresh and frozen sections were mounted onto SuperFrost Plus slides (Thermo Fisher Scientific, Waltham, MA, USA). Frozen SkM sections were dried for 2 hours at −20°C and then stored at −80°C until analysis.
IF staining and quantification
For FFPE tissues, sections were first placed on a warming block to allow the wax to melt and the tissue to bond to the slide. Sections were then deparaffinized in 100% Histoclear and hydrated in 100, 90 and 70% ethanol and then incubated in distilled water. Antigen retrieval was performed by incubating sections in 0.01 M citrate buffer (pH 6.0) or EDTA (pH 7.0) and heated to boiling. Sections were blocked and then incubated with primary antibody overnight at 4°C and secondary antibody for 1 hour at room temperature. Sections were then incubated in Wheat Germ Agglutinin (WGA) (Invitrogen, Carlsbad, CA, USA) for 20 minutes at room temperature, washed, and mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen). When appropriate, a mouse-on-mouse detection kit (Vector Laboratories, Burlingame, CA, USA) was used to block endogenous mouse IgG staining to improve the specificity of secondary anti-mouse antibodies binding to primary mouse antibodies used on mouse sections. Frozen SkM sections were fixed, permeabilized, blocked, and then incubated in primary antibody overnight at 4°C, followed by secondary antibody for 2 hours in a humidified chamber at 4°C or 1 hour at room temperature. Sections were also mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen). Antibodies used for mouse tissue are listed in KEY RESOURCES TABLE.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-γH2AX (20E3) | Cell Signaling | Cat# 9718 (1:250 dilution for IF) |
| Rabbit anti-HMGB1 | Abcam | Cat# ab18256 (1:500 dilution for IF) |
| Rabbit anti-LaminB1 | Abcam | Cat# ab16048 (1:500 dilution for IF) |
| Rabbit anti-P21 | Abcam | Cat# ab109199 (1:200 dilution for IF) |
| Mouse anti-P21 | Santa Cruz | Cat# sc-53870 (1:500 for western blot) |
| Rabbit anti-GAPDH | Cell Signaling | Cat# 5174 (1:20,000 for western blot) |
| Rabbit anti-Laminin | Sigma | Cat# L9393 (1:100 dilution for IF) |
| Mouse anti-P16 | CINTec Histology, Roche | Cat# 9511 (IF, dilution as provided by manufacturer) |
| Wheat Germ Agglutinin, Alexa Fluo 488 Conjugate | Invitrogen | Cat# W11261 (1:750 dilution for IF) |
| Goat anti-rabbit Fluorescein-conjugated secondary antibody Alexa Fluor 488 | Invitrogen | Cat# A11008 (1:500 dilution for IF) |
| Goat anti mouse biotinylated secondary antibody | Vector Laboratories | Cat# AI-9200 (1:200 dilution for IHC) |
| Goat anti-rabbit biotinylated secondary anti-body | Vector Laboratories | Cat# PK-6101 (1:200) |
| m-IgGκ BP-HRP | Santa Cruz | Cat# sc-516102 (1:20,000 for western blot) |
| mouse anti-rabbit IgG-HRP | Santa Cruz | Cat# sc-2357 (1:20,000 for western blot) |
| Rabbit anti-rabbit Fluorescein-conjugated secondary antibody Alexa Fluor 594 | Invitrogen | Cat# A11012 (1:500 dilution for IF) |
| Rabbit anti-rabbit Fluorescein-conjugated secondary antibody Alexa Fluor 647 | Invitrogen | Cat# A21245 (1:500 dilution for IF) |
| Rabbit anti-mouse Fluorescein-conjugated secondary antibody Alexa Fluor 594 | Invitrogen | Cat# A44005 (1:500 dilution for IF) |
| Rabbit anti-guinea pig Fluorescein-conjugated secondary antibody Alexa Fluor 594 | Invitrogen | Cat# A11076 (1:500 dilution for IF) |
| CD31 MicroBeads, mouse | Miltenyi | Cat# 130-097-418 |
| CD45 MicroBeads, mouse | Miltenyi | Cat# 130-052-301 |
| Anti-Sca-1 (non-HSC) MicroBeads, mouse | Miltenyi | Cat# 130-106-641 |
| Anti-Integrin α-7 (Integrin Subunit Alpha 7) MicroBeads, mouse | Miltenyi | Cat# 130-104-261 |
| Primers for qPCR | ||
| Tbp | Integrated DNA Technologies | Cat# MM.PT.39a.22214839 |
| Gapdh | Integrated DNA Technologies | Cat# Mm.PT.39a.1 |
| p16Ink4a | Integrated DNA Technologies | Cat# MM.PT.58.42804808 |
| p21 | Integrated DNA Technologies | Cat# MM.PT.58.5884610 |
| p21 variant 2 | Integrated DNA Technologies | Cat# Mm.PT.58.31908645 |
| Ccl2 | Integrated DNA Technologies | Cat# MM.PT.58.42151692 |
| Tnfa | Integrated DNA Technologies | Cat# Mm.PT.58.12575861 |
| Tgfb | Integrated DNA Technologies | Cat# Mm.PT.58.11254750 |
| Il1a | Integrated DNA Technologies | Cat# Mm.PT.58.32778767 |
| Il6 | Integrated DNA Technologies | Cat# Mm.PT.58.10005566 |
| Pai1 | Integrated DNA Technologies | Cat# Mm.PT.58.6413525 |
| Mmp3 | Integrated DNA Technologies | Cat# Mm.PT.58.9719290 |
| Mmp9 | Integrated DNA Technologies | Cat# Mm.PT.58.10100097 |
| Cd31 | Integrated DNA Technologies | Cat# Mm.PT.58.12394061 |
| Cd68 | Integrated DNA Technologies | Cat# Mm.PT.58.32698807 |
| Pax7 | Integrated DNA Technologies | Cat# Mm.PT.58.12398641 |
| Runx1 | Integrated DNA Technologies | Cat# Mm.PT.58.5873245 |
| Ankrd1 | Integrated DNA Technologies | Cat# Mm.PT.58.11977470 |
| Gadd45a | Integrated DNA Technologies | Cat# Mm.PT.58.42316074 |
| Rrad | Integrated DNA Technologies | Cat# Mm.PT.58.10415826 |
| Pdgfra | Integrated DNA Technologies | Cat# Mm.PT.56a.5639577 |
| TBP (human) | Integrated DNA Technologies | Cat# Hs.PT.58.20792004 |
| P16Ink4a (human) | Integrated DNA Technologies | Sequence F: 5’ CCAACGCACCGAATAGTTACG 3’ R: 5’ GCGCTGCCCATCATCATG 3’ P: 5’ FAM - CCTGGATCGGCCTCCGAC - MGB 3’ |
| Chemicals, Peptides, and Recombinant Proteins, and Commercial Assays | ||
| Collagenase II | Worthington Biochemical Corporation | Cat# LS004176 |
| bFGF Recombinant Human Protein | Gibco | Cat# 13256029 |
| Chick Embryo Extract | Life Science Group | Cat# MD-004E-UK |
| TRIzol reagent | Invitrogen | Cat# 15596026 |
| miRNeasy Mini Kit | Qiagen | Cat# 217004 |
| Red Blood Cell Lysis Solution | Miltenyi | Cat# 130-094-183 |
| M-MLV Reverse Transcriptase | Invitrogen | Cat# 28025013 |
| PerfeCTa qPCR FastMix II, ROX, QuantaBio | Quantabio | Cat# 95119-05K |
| Skeletal Muscle Dissociation Kit | Miltenyi | Cat# 130-098-305 |
| RNAscope Multiplex Fluorescent v2 Assay | Advanced Cell Diagnostics | Cat# 323100-USM |
| RNAscope probe targeting p21 (Mm-Cdkn1a) | Advanced Cell Diagnostics | Cat# 408551 |
| Cy-3-labelled telomere-specific (CCCTAA) peptide nucleic acid probe | PANAGENE | Cat# F1002 |
| CENPB-FAM centromere specific probe | PANAGENE | Cat# F3001 |
| SMART-Seq v4 Ultra Low Input RNA Kit | Clontech | Cat# 634888 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat# FC-131-1096 |
| TruSeq Stranded mRNA Sample Prep Kit | Illumina | Cat# 20020594 |
| Chromium Single Cell 3’ Reagent Kits | 10X Genomics | Cat# PN-1000075 |
| M.O.M. (Mouse on Mouse) Immunodetection Kit, Basic | Vector Laboratories | Cat# BMK-2202 |
| Avidin/Biotin Blocking Kit | Vector Laboratories | Cat# SP-2001 |
| Vector NovaRED Peroxidase (HRP) Substrate Kit | Vector Laboratories | Cat# LS-J1084 |
| Fluorescein Avidin DCS | Vector Laboratories | Cat# SP-2011 |
| Sudan Black B | Frontier scientific | Cat# JK226545 |
| ProLong™ Gold Antifade Mountant with DAPI | Invitrogen | Cat# P36935 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific | Cat# 34577 |
| Protease inhibitor cocktail | Sigma | Cat# P8340 |
| PMSF | Thermo Scientific | Cat# 36978 |
| DC Protein Assay | Bio-Rad | Cat# 5000116 |
| PVDF membrane | Bio-Rad | Cat# 1620177 |
| Cell Lysis Buffer | Cell Signaling | Cat# 9803 |
| Deposited Data | ||
| RNA-seq data of isolated myofibers | GEO database | GEO: GSE172254 |
| RNA-seq data of skeletal muscle from young, old-Vehicle, and old-DQ mice | GEO database | GEO: GSE184348 |
| scRNA-seq data of skeletal muscle from young and old mice | GEO database | GEO: GSE172410 |
| Interactive website for scRNA-seq data | https://mayoxz.shinyapps.io/Muscle/ | |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT C57BL6/J | The Jackson Laboratory & NIA | Cat# JAX:000664 |
| Software and Algorithms | ||
| Prism | Graphpad | RRID:SCR_002798; V8.4.1 |
| ImageJ | https://imagej.net/ | RRID:SCR_003070; V2.0.0-rc-64/1.51s |
| Fiji | http://fiji.sc | RRID:SCR_002285; V2.0.0-rc-69/1.52p |
| Metamorph | http://www.moleculardevices.com/Products/Software/Meta-Imaging-Series/MetaMorph.html | RRID:SCR_002368; VV7.10.2 |
| Cell Ranger | 10X Genomics | RRID:SCR_017344; V3.0 |
| R | https://www.r-project.org/ | RRID: SCR_001905; V3.6.0 |
| Seurat | https://satijalab.org/seurat/ | RRID: SCR_016341; V3.1.3 |
| Tophat2 | http://ccb.jhu.edu/software/tophat/index.shtml | RRID:SCR_013035; V2.1.1 |
| RSeQC | http://code.google.com/p/rseqc/ | RRID:SCR_005275; V2.3.6 |
| edgeR | http://bioconductor.org/packages/edgeR/ | RRID:SCR_012802; V3.10 |
| Clusterprofiler | http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html | RRID:SCR_016884; V3.18.1 |
| GSEA | Broad Institute | RRID: SCR_003199; V4.0.3 |
| Cellchat | http://www.cellchat.org/ | V1.0.0 |
| Others | ||
| CellAge_Senescence_UP gene set | https://genomics.senescence.info/cells/ | |
| PanglaoDB | https://panglaodb.se/ |
For fluorescence microscopy, a Leica DM5500B widefield fluorescence microscope and Leica DMI8 were used. For light microscopy, a Nikon Eclipse E-800 Brightfield camera was used. Myofiber cross-sectional area, demarcated by Laminin staining, and centrally nucleated fibers were quantified using MuscleJ 54.
TAF staining and quantification
For FFPE tissues, immunohistochemistry was carried out as described above. After blocking, sections were rinsed with PBS and incubated in avidin for 15 min, followed by another PBS rinse and an incubation in Avidin/Biotin Blocking Kit (Vector Laboratories) for 15 minutes at room temperature. Sections were rinsed one more time and incubated with primary antibody overnight at 4°C. Following an overnight incubation with rabbit monoclonal anti-γH2A.X (Cell Signaling, Danvers, MA, USA), sections were then incubated with a goat anti-rabbit biotinylated secondary antibody (Vector Laboratories) for 1 hour at room temperature. Following three PBS washes, tissues were incubated with fluorescein avidin DCS (Vector Laboratories) for another 20 minutes at room temperature. Sections were then washed three times in PBS and cross-linked by incubation in 4% paraformaldehyde (Sigma) in PBS for 20 minutes. Sections were washed in PBS three times and then dehydrated in graded cold ethanol solutions (70, 90, 100%) for 3 minutes each. Tissues were then allowed to air-dry prior to being denatured in 10μl of PNA hybridization mix (70% deionized formamide (Sigma), 25 mM MgCl2, 1 M Tris pH7.2, 5% blocking reagent (Roche) containing 2.5 μg/ml Cy-3-labelled telomere-specific (CCCTAA) peptide nucleic acid probe (PANAGENE, Yuseong-gu, Daejeon, South Korea)) for 10 minutes at 80°C and then incubated overnight at 4°C in a dark humidified chamber to allow hybridization to occur. The following day, sections were washed in 70% formamide in 2×SCC for 10 min, followed by a wash in 2×SSC for 10 min, and a PBS wash for 10 min. Tissues were then incubated in WGA and washed as described previously and finally mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen). Sections were imaged using in-depth Z stacking (a minimum of 40 optical slices with 63×objective) followed by Huygens (SVI) deconvolution and ImageJ analysis. WGA was used to delineate myonuclei from interstitial and satellite cells. For telomere length analysis, Z projections were created for each individual image and the oval tool was used to measure the integrated density of each individual telomere signal in ImageJ. 100 myonuclei were analyzed per sample.
SADS staining and quantification
FFPE sections were deparaffinized in 100% Histoclear and hydrated in 100, 90, and 70% ethanol (twice for 5 minutes each) and incubated twice for 5 minutes in distilled water. Antigen retrieval was performed by incubating sections in 0.01 M citrate buffer (pH6.0) and boiling for 10 minutes. Sections were allowed to cool down to room temperature followed by two washes in distilled water and 1 wash in PBS for 5 minutes each. Sections were then cross-linked by incubation in 4% paraformaldehyde in PBS for 20 minutes. Sections were washed in PBS three times and then dehydrated in graded cold ethanol solutions (70, 90, 100%) for 3 minutes each. Tissues were then allowed to air-dry prior to being denatured in 10 μl of PNA hybridization mix (70% deionized formamide (Sigma), 25 mM MgCl2, 1 M Tris pH7.2, 5% blocking reagent (Roche) containing 2.5 μg/ml CENPB-FAM centromere specific probe (PANAGENE) for 10 minutes at 80°C and then incubated overnight at 4°C in a dark humidified chamber to allow hybridization to occur. The following day, sections were washed in 70% formamide in 2×SCC for 10 minutes, followed by a wash in 2×SSC for 10 minutes, and a PBS wash for 10 min. Tissues were then mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen). For centromere decondensation analysis, Z projections were created for each individual image and the oval tool was used to measure the integrated density of each individual centromere signal in ImageJ for 100 nuclei per sample.
Sudan Black B staining
Sudan Black B (SBB) staining was performed as previously described 55. Briefly, cryosections were thawed for 30 minutes at room temperature, fixed in 1% PFA in PBS for 5 minutes at room temperature, washed three times for 1 minute with deionized water, and then incubated in 100% ethylene glycol for 5 minutes. After this, three drops of SBB solution (7 mg/ml in ethylene glycol, Frontier scientific) were placed on each coverslip and the slides containing the cryosections were placed, face down, onto the coverslips with the cryosections in direct contact with the SBB solution and incubated for 3 hours at room temperature. The cryosections were washed with three brief rinses in deionized water. The cryosections were then incubated in 80% ethylene glycol for 3 minutes, followed by three brief rinses in deionized water. Slides were mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen). Images were captured at 40x magnification with ZEISS Axioscan 7 and analyzed using MetaMorph Microscopy Automation and Image Analysis Software.
RNA in situ hybridization (RNA-ISH) and quantification
RNA in situ hybridization was performed on the quad muscle sections of young and old mice using the RNAscope Multiplex Fluorescent v2 Assay (Advanced Cell Diagnostics, Newark, CA, USA) following the manufacturer’s instructions. Briefly, sections were removed from −80°C and fixed in 4% paraformaldehyde for 15 minutes, washed in PBS and dehydrated with increasing concentrations of ethanol (50%, 70% and 100%). After drawing a hydrophobic barrier with the ImmEdge pen (Vector Laboratories), sections were incubated with hydrogen peroxide for 10 minutes at room temperature, washed with distilled water and incubated with Protease IV for 30 minutes at room temperature. Sections were then washed in PBS and incubated for 2 hours at 40°C with the RNAscope probe targeting p21 (Mm-Cdkn1a, 408551, Advanced Cell Diagnostics). After rinsing with the wash buffer, sections were stored overnight in 5x Saline Sodium Citrate (SSC) buffer at room temperature. The following day, SkM sections were incubated at 40°C with AMP1 (30 minutes), AMP2 (30 minutes) and AMP3 (15 minutes), with rinses in wash buffer at room temperature between each incubation step. Sections were next incubated at 40°C with HRP-C1 (15 minutes), Cyanine 3 TSA Plus Fluorophore (Perkin Elmer, Waltham, MA, USA) at a dilution of 1:750 (30 minutes), and HRP blocker (15 minutes), with rinses in wash buffer at room temperature between each incubation step. The RNA-ISH assay was followed by the IF staining for Laminin. Images were captured at 20x magnification with a Zeiss Axio Observer microscope. Image analysis was carried out using the open-source Fiji software 56.
Single-cell RNA-seq
Quadriceps (quad), gastrocnemius (gas), tibialis anterior (TA), and extensor digitorum longus (EDL) muscle from female mice were used for scRNA-seq. Single-cell suspensions were prepared from SkM using a Skeletal Muscle Dissociation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). After digestion, cells were filtered through 70 μm and then 40 μm strainers. Cells were treated with Red Blood Cell Lysis Solution (Miltenyi) for 2 minutes and then collected for further processing.
The dissociated cells were counted and measured for viability using the Vi-Cell XR Cell Viability Analyzer (Beckman-Coulter, Brea, CA, USA) and a basic hemocytometer and light microscope. About 5000 dissociated cells were loaded into the 10x Genomics Chromium system, where individual cells were captured into uniquely labeled GEMs (Gel Beads-In-Emulsion). GEMs were collected for reverse transcription using Chromium Single Cell 3’ Reagent Kits (10X Genomics, Pleasanton, CA, USA). Standard Illumina sequencing primers and a unique i7 Sample index were added to each cDNA for library construction and the libraries were then measured using Qubit High Sensitivity assays (Thermo Fisher Scientific), Agilent Bioanalyzer High Sensitivity chips (Agilent Technologies, Santa Clara, CA, USA), and Kapa DNA Quantification reagents (Kapa Biosystems, Wilmington, MA, USA). Libraries were sequenced at 60,000 fragment reads per cell following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit. The flow cells were sequenced as 100 x 2 paired end reads on an Illumina HiSeq 4000 using HiSeq 3000/4000 sequencing kit and HCS v3.3.52 collection software. Base-calling was performed using Illumina’s RTA version 2.7.3.
scRNA-seq data analysis
scRNA-seq raw data were processed with Cell Ranger (v3.0) (10X Genomics) and aligned to the mouse genome version mm10. Further analyses were carried out in R version 3.6.0 using Seurat version (v3.1.3) 57. Filtering: Cells with fewer than 500 detected genes or greater than 8000 genes were excluded. Cells with more than 40% of reads mapped to mitochondria were also excluded. After filtering, 11,734 cells from three young and three old SkMs were analyzed. Dimensionality reduction: Dimensionality reduction was performed using principal component analysis on the top 2000 variable genes between all cells. Top 20 principal components were selected based on an elbow plot analysis in Seurat. Clustering and sub-clustering: A Shared Nearest Neighbor (SNN) Graph was constructed using FindNeighbors function of Seurat with a resolution of 0.1. T-distributed stochastic neighbor embedding (t-SNE) was used to create a 2D map. For further sub-clustering of FAPs, the same procedure for finding variable genes, dimensionality reduction, and clustering were applied. Assessment of differentially expressed genes: The Wilcoxon Rank Sum test was used to identify differentially expressed genes between two groups of cells using the FindAllMarkers function of Seurat. Cell type identification: Marker genes of the different cell clusters were mapped to the Panglao database, which covers 178 cell types and 4679 gene symbols from 29 mouse and human tissues 58. Canonical cell type markers 14 were used to confirm the identification. The interactive website was generated using ShinyCell 59.
Bulk RNA-seq
For bulk RNA-seq, cDNA libraries were prepared according to the manufacturer’s instructions for the TruSeq Stranded mRNA Sample Prep Kit for the muscle tissue (Illumina, San Diego, CA) or SMART-Seq v4 Ultra Low Input RNA Kit for the single myofiber (Clontech). The concentration and size distribution of the completed libraries were determined using an Agilent Bioanalyzer DNA 1000 chip (Santa Clara, CA) and Qubit fluorometry (Invitrogen, Carlsbad, CA). Libraries were sequenced following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit on an Illumina HiSeq 4000.
Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) analysis
Differentially expressed genes were used for GO analysis using R package Clusterprofiler (v3.18.1) 60. For p16high FAPs scRNA-seq dataset, log fold change cutoff of 0.5 and p-value of 0.05 were used. For single myofiber and human SkM RNA-seq dataset, log fold change cutoff of 1 and p-value of 0.05 were used. Gene set enrichment analysis was done using software GSEA (v4.0.3) 61. LFC was used to rank the genes for analysis. For p16high FAPs scRNA-seq dataset, log fold change cutoff of 0.1 p-value of 0.05 were used. For single myofiber and human SkM RNA-seq dataset, p-value cutoff of 0.05 was used.
Magnetic-activated cell sorting (MACS)
For MACS, four antibodies were used: CD31 (Miltenyi), CD45 (Miltenyi), Integrin α-7 (Miltenyi), and SCA1 (Miltenyi) as previously described 62. Endothelial and immune cell populations were collected together as CD31+/CD45+. Satellite cells were selected as CD31−, CD45−, and Integrin α-7+. FAPs were selected as CD31−, CD45−, Integrin α-7−, and SCA1+. Pdgfra is more specific to FAPs than Sca1 in the scRNA-seq data, but the SCA1 antibody was more effective in MACS sorting.
Single myofiber isolation
Single myofibers were isolated as previously described 63, with minor modifications. Briefly, extensor digitorum longus (EDL) muscle was collected and digested in DMEM with 2.5% HEPES, 1% Penicillin/Streptomycin solution (Pen/Strep), and 400 U/ml of collagenase II for 2 hours in an incubator at 37°C and 5% of CO2. Single myofibers were flushed and transferred with sterile Pasture pipette into DMEM with 10% FBS, 1% Pen/Strep, 2% Chicken embryo extract, and 10 ng/ml bFGF. After washing for four times in medium, single myofibers were transferred to Trizol solution (Invitrogen) for RNA isolation. RNA was divided into two aliquots, one for qPCR to quantify p21 expression, and the other for RNAseq performed on a subset of young myofibers and subsets of old myofibers identified as p21low and p21high by qPCR.
RNA isolation, cDNA synthesis, and qPCR
RNA was isolated using Trizol according to manufacturer’s instructions (Invitrogen). RNA from single myofiber was isolated using miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA concentration was assessed by Nanodrop (Thermo Fisher Scientific). cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen), and real-time PCR was performed with PerfeCTa FastMix II (QuantaBio, Beverly, MA, USA) and the Applied Biosystems StepOne Plus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Gene expression was analyzed by delta-delta CT method and normalized to the reference gene, TATA-Box Binding Protein (TBP) or GAPDH. The primers and probes used are listed in KEY RESOURCES TABLE.
Western blotting
Total protein extracts were prepared from quadriceps muscles in cell lysis buffer (Cell Signaling Technology) with protease inhibitor cocktail (Sigma) and phenylmethylsulfonyl fluoride (PMSF). Protein concentration was determined by using the DC Protein Assay (Bio-Rad, Hercules, CA, USA). Equal amounts of protein (30 μg) were resolved by SDS-page and transferred to a PVDF membrane (Bio-Rad). The membrane was blocked with 5% non-fat dry milk and then incubated with primary and secondary antibodies. Antibodies are listed in KEY RESOURCES TABLE. Signal was developed using SuperSignal West Pico PLUS Chemiluminescent Substrate kit (Thermo Fisher Scientific) and quantified using ImageJ.
Human study
SkM biopsies (~350 mg) were collected from the vastus lateralis muscle of younger (n = 30, 24.7 ± 3.9 (mean ± SD) years of age, 45% female) and older (n = 22, average 69.8 ± 3.9 years of age, 48% female) women and men as we previously reported 31. The study was approved by the Mayo Clinic Institutional Review Board and registered under Clinical Trial #NCT01477164 (clinicaltrials.gov). All participants were informed of study procedures and provided written consent. Methods for SkM RNA isolation, RNA library preparation, data generation, and data analysis are detailed above and in our prior publication 31. Measures of mitochondrial function and clinical outcomes were also reported prior 31. FFPE sections were used for P16 IHC staining.
Statistics and Reproducibility
For mice experiments, between 3 and 10 mice were analysed per experiment. No power analyses were performed to determine sample sizes [optional] but our sample sizes are similar to those recommended by (47, 48, 50). . The sample size for the number of cells quantified was 100-250 cells per mouse, depending on the level of expression of the marker analysed. For human studies using RNAseq and qPCR, the sample size was determined detailed in previous study 31. For complementary analysis of senescence markers, no power analyses were performed to determine sample sizes but our sample sizes are similar to those reported in previous studies (47, 48, 50).
For differentially expressed gene analysis in scRNA-seq, a Wilcoxon Rank Sum test was used in the Seurat package. For differentially expressed gene analysis in bulk RNA-seq, a negative-binomial model was used in edgeR. For GSEA, Kolmogorov-Smirnov statistic was used in GSEA software. All other statistical analyses were carried out using GraphPad Prism 8, and a P <0.05 was considered statistically significant. All data were assessed for normality using D’Agostino & Pearson and Shapiro-Wilk normality tests (for n>7). The F test was used to compare variances. For comparisons between 2 groups, a two-tailed unpaired t-test (for parametric data) or a Mann-Whitney U test (for non-parametric data) was used. For comparisons between multiple groups, a One-way ANOVA with Tukey’s posthoc test was used. Correlations were analyzed using Pearson’s rank correlation test when the data was normally distributed or Spearman’s rank correlation test for datasets that were not normally distributed. All data are expressed as the mean ± S.E.M (standard error of the mean).
Extended Data
Extended Data Fig. 1.
The age-related loss of skeletal mass, strength, and function. Comparisons of (a) body weight, (b) lean mass (%), (c) and quadriceps (quad) muscle weight between young (n = 9) and old (n = 5) mice. (d) Representative images of quad muscle cross-sections stained for Laminin and (e) quantification of fiber size and distribution for young and old female mice (n = 4 per group). Comparison of (f) grip strength normalized to body weight, (g) treadmill exercise capacity, and (h) Rotarod endurance between young (n = 9) and old (n = 5) mice. Two-tailed unpaired t test was used; error bars represent the standard error of the mean. **, and *** denote p < 0.01 and 0.001 respectively.
Extended Data Fig. 2.
The age-related increase of lipofuscin in skeletal muscle (SkM). Representative images of Sudan Black B (SBB) staining (a) and quantification of the numbers of positive foci per quadriceps section (b) from young and old female mice (n = 4 per age group). Two-tailed unpaired t test was used; error bars represent the standard error of the mean. * and ** denote p < 0.05 and 0.01 respectively.
Extended Data Fig. 3.
Identification of skeletal muscle (SkM) cell populations by single cell RNA-sequencing. (a) Heatmap of the genes delineating 10 distinct cell populations in young and old female mice (n = 3 per group). (b) UMAP plot and (c) dot plot showing the main markers of each cell type. (d) Relative abundance of the distinct SkM cell populations in individual mice.
Extended Data Fig. 4.
Gene Ontology analysis of the upregulated and downregulated genes in high p16-expressing FAP cluster 3 compared to other FAPs. BP: biological process; MF: molecular function; CC: cellular component. Benjamini-Hochberg Procedure was used to calculate the FDR adjusted p value.
Extended Data Fig. 5.
Gene Ontology analysis of the upregulated and downregulated genes in old p21high myofibers compared to old p21low myofibers. BP: biological process; MF: molecular function; CC: cellular component. Benjamini-Hochberg Procedure was used to calculate the FDR adjusted p value.
Acknowledgments
We are grateful for the support of the National Institutes of Health, National Institute on Aging for grants P01 AG062413, R01 AG055529, and R56 AG060907 to N.K.L., and R01 AG068048 and UG3CA 268103 to J.F.P., and T32 AG049672 to D.A.E. This work was also supported by the Glenn Foundation for Medical Research and the Pritzker Foundation (N.K.L.). X.Z. was supported by a Robert and Arlene Kogod Center on Aging Career Development Award. L.H. was supported by NIHR Newcastle Biomedical Research Centre grant awarded to the Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University. R.A.F. and D.A.R. are partially supported by the US Department of Agriculture (USDA), under agreement No. 58-8050-9-004 and by and by NIH Boston Claude D Pepper Center (OAIC; 1P30AG031679). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the USDA. We would also like to thank members of P01 AG062413 (PI: SK) for helpful discussions, Dr. Julie M. Cunningham, and Dr. Eric Wieben within the Mayo Clinic Genome Analysis Core for RNA-seq, Dr. Ying Li within Division of Computational Biology and Department of Quantitative Health Sciences for assistance with bioinformatic analyses and the Optical Microscopy Core within the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30 DK084567) for guidance and use of imaging equipment.
Footnotes
Competing interests
The authors declare no competing interests.
Data Availability
The scRNA-seq (GSE172410), myofiber RNA-seq data (GSE172254), and SkM RNA-seq data (GSE184348) used in this study are deposited in Gene Expression Omnibus (GEO). Human SkM RNA-Seq data are publicly available at GEO (GSE97084).
An interaction website for the SkM scRNA-seq dataset can be found at https://mayoxz.shinyapps.io/Muscle/.
Code Availability
Codes and all other data are available from the corresponding author upon reasonable request.
References
- 1.Fielding RA et al. Sarcopenia: an undiagnosed condition in older adults. Current consensus definition: prevalence, etiology, and consequences. International working group on sarcopenia. J Am Med Dir Assoc 12, 249–256, doi: 10.1016/j.jamda.2011.01.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campisi J & di Fagagna FD Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Bio 8, 729–740, doi: 10.1038/nrm2233 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Coppe JP et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6, 2853–2868, doi: 10.1371/journal.pbio.0060301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acosta JC et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 15, 978–U221, doi: 10.1038/ncb2784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khosla S, Farr JN, Tchkonia T & Kirkland JL The role of cellular senescence in ageing and endocrine disease. Nat Rev Endocrinol 16, 263–275, doi: 10.1038/s41574-020-0335-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez-Segura A, Nehme J & Demaria M Hallmarks of Cellular Senescence. Trends Cell Biol, doi: 10.1016/j.tcb.2018.02.001 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Gorgoulis V et al. Cellular Senescence: Defining a Path Forward. Cell 179, 813–827, doi: 10.1016/j.cell.2019.10.005 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Welle S et al. Skeletal muscle gene expression profiles in 20-29 year old and 65-71 year old women. Exp Gerontol 39, 369–377, doi: 10.1016/j.exger.2003.11.011 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Kayo T, Allison DB, Weindruch R & Prolla TA Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. P Natl Acad Sci USA 98, 5093–5098, doi:DOI 10.1073/pnas.081061898 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards MG et al. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. Bmc Genomics 8, doi:Artn 80 10.1186/1471-2164-8-80 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dungan CM et al. In vivo analysis of gamma H2AX+cells in skeletal muscle from aged and obese humans. Faseb J 34, 7018–7035, doi: 10.1096/fj.202000111RR (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoji H & Miyakawa T Age-related behavioral changes from young to old age in male mice of a C57BL/6J strain maintained under a genetic stability program. Neuropsychopharmacol Rep 39, 100–118, doi: 10.1002/npr2.12052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewitt G et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 3, doi:ARTN 708 10.1038/ncomms1708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giordani L et al. High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol Cell 74, 609-+, doi: 10.1016/j.molcel.2019.02.026 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Sousa-Victor P et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316-+, doi: 10.1038/nature13013 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Groen BBL et al. Skeletal muscle capillary density and microvascular function are compromised with aging and type 2 diabetes. J Appl Physiol 116, 998–1005, doi: 10.1152/japplphysiol.00919.2013 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Coppe JP, Desprez PY, Krtolica A & Campisi J The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 5, 99–118, doi: 10.1146/annurev-pathol-121808-102144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anerillas C, Abdelmohsen K & Gorospe M Regulation of senescence traits by MAPKs. Geroscience 42, 397–408, doi: 10.1007/s11357-020-00183-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tominaga K & Suzuki HI TGF-beta Signaling in Cellular Senescence and Aging-Related Pathology. Int J Mol Sci 20, doi: 10.3390/ijms20205002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiley CD & Campisi J From Ancient Pathways to Aging Cells-Connecting Metabolism and Cellular Senescence. Cell Metab 23, 1013–1021, doi: 10.1016/j.cmet.2016.05.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avelar RA et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol 21, doi:ARTN 91 10.1186/s13059-020-01990-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tront JS, Hoffman B & Liebermann DA Gadd45a suppresses Ras-driven mammary tumorigenesis by activation of c-Jun NH2-terminal kinase and p38 stress signaling resulting in apoptosis and senescence. Cancer Res 66, 8448–8454, doi: 10.1158/0008-5472.CAN-06-2013 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Wei Z et al. Pan-senescence transcriptome analysis identified RRAD as a marker and negative regulator of cellular senescence. Free Radical Bio Med 130, 267–277, doi: 10.1016/j.freeradbiomed.2018.10.457 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Anderson G et al. RUNX-mediated growth arrest and senescence are attenuated by diverse mechanisms in cells expressing RUNX1 fusion oncoproteins. J Cell Biochem 119, 2750–2762, doi: 10.1002/jcb.26443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casella G et al. Transcriptome signature of cellular senescence. Nucleic Acids Res 47, 7294–7305, doi: 10.1093/nar/gkz555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemster BH et al. Induction of CD56 and TCR-independent activation of T cells with aging. J Immunol 180, 1979–1990, doi:DOI 10.4049/jimmunol.180.3.1979 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Hernandez-Segura A et al. Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr Biol 27, 2652–2660 e2654, doi: 10.1016/j.cub.2017.07.033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki M, Miyakoshi M, Sato Y & Nakanuma Y Modulation of the microenvironment by senescent biliary epithelial cells may be involved in the pathogenesis of primary biliary cirrhosis. J Hepatol 53, 318–325, doi:DOI 10.1016/j.jhep.2010.03.008 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Marques JCMT et al. Identification of new genes associated to senescent and tumorigenic phenotypes in mesenchymal stem cells. Sci Rep-Uk 7, doi:ARTN 17837 10.1038/s41598-017-16224-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Dominguez JA et al. Cdkn1a transcript variant 2 is a marker of aging and cellular senescence. Aging-Us 13, 13380–13392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MM et al. Enhanced Protein Translation Underlies Improved Metabolic and Physical Adaptations to Different Exercise Training Modes in Young and Old Humans. Cell Metab 25, 581–592, doi: 10.1016/j.cmet.2017.02.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heredia JE et al. Type 2 Innate Signals Stimulate Fibro/Adipogenic Progenitors to Facilitate Muscle Regeneration. Cell 153, 376–388, doi: 10.1016/j.cell.2013.02.053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuswanto W et al. Poor Repair of Skeletal Muscle in Aging Mice Reflects a Defect in Local, Interleukin-33-Dependent Accumulation of Regulatory T Cells. Immunity 44, 355–367, doi: 10.1016/j.immuni.2016.01.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukjanenko L et al. Aging Disrupts Muscle Stem Cell Function by Impairing Matricellular WISP1 Secretion from Fibro-Adipogenic Progenitors. Cell Stem Cell 24, 433-+, doi: 10.1016/j.stem.2018.12.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biferali B, Proietti D, Mozzetta C & Madaro L Fibro-Adipogenic Progenitors Cross-Talk in Skeletal Muscle: The Social Network. Front Physiol 10, 1074, doi: 10.3389/fphys.2019.01074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito Y, Chikenji TS, Matsumura T, Nakano M & Fujimiya M Exercise enhances skeletal muscle regeneration by promoting senescence in fibro-adipogenic progenitors. Nat Commun 11, doi:ARTN 889 10.1038/s41467-020-14734-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall BM et al. p16(Ink4a) and senescence-associated beta-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging-Us 9, 1867–1884, doi: 10.18632/aging.101268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fry CS et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat Med 21, 76–80, doi: 10.1038/nm.3710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keefe AC et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat Commun 6, doi:ARTN 7087 10.1038/ncomms8087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Englund DA et al. Depletion of resident muscle stem cells negatively impacts running volume, physical function, and muscle fiber hypertrophy in response to lifelong physical activity. Am J Physiol-Cell Ph 318, C1178–C1188, doi: 10.1152/ajpcell.00090.2020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez K et al. Single nuclei profiling identifies cell specific markers of skeletal muscle aging, sarcopenia and senescence. medRxiv, doi: 10.1101/2021.01.22.21250336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi S et al. Intrinsic cooperation between p16INK4a and p21Waf1/Cip1 in the onset of cellular senescence and tumor suppression in vivo. Cancer Res 70, 9381–9390, doi: 10.1158/0008-5472.CAN-10-0801 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Alcorta DA et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A 93, 13742–13747, doi: 10.1073/pnas.93.24.13742 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sapieha P & Mallette FA Cellular Senescence in Postmitotic Cells: Beyond Growth Arrest. Trends Cell Biol 28, 595–607, doi: 10.1016/j.tcb.2018.03.003 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Jurk D et al. Postmitotic neurons develop a p21-dependent senescence-like phenotype driven by a DNA damage response. Aging Cell 11, 996–1004, doi: 10.1111/j.1474-9726.2012.00870.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson R et al. Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. Embo J 38, doi:ARTN e100492 10.15252/embj.2018100492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farr JN et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat Med 23, 1072-+, doi: 10.1038/nm.4385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu M et al. Senolytics improve physical function and increase lifespan in old age. Nat Med 24, 1246–1256, doi: 10.1038/s41591-018-0092-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y et al. The Achilles' heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658, doi: 10.1111/acel.12344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker DJ et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236, doi: 10.1038/nature10600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Lee YI & Thompson WJ Changes in Aging Mouse Neuromuscular Junctions Are Explained by Degeneration and Regeneration of Muscle Fiber Segments at the Synapse. Journal of Neuroscience 31, 14910–14919, doi: 10.1523/Jneurosci.3590-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schafer MJ et al. Exercise Prevents Diet-Induced Cellular Senescence in Adipose Tissue. Diabetes 65, 1606–1615, doi: 10.2337/db15-0291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LeBrasseur NK et al. Myostatin inhibition enhances the effects of exercise on performance and metabolic outcomes in aged mice. J Gerontol A Biol Sci Med Sci 64, 940–948, doi: 10.1093/gerona/glp068 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Mayeuf-Louchart A et al. MuscleJ: a high-content analysis method to study skeletal muscle with a new Fiji tool. Skelet Muscle 8, 25, doi: 10.1186/s13395-018-0171-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.da Silva PFL et al. The bystander effect contributes to the accumulation of senescent cells in vivo. Aging Cell 18, doi:ARTN e12848 10.1111/acel.12848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682, doi: 10.1038/Nmeth.2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–U206, doi: 10.1038/nbt.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franzen O, Gan LM & Bjorkegren JLM PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database-Oxford, doi:ARTN baz046 10.1093/database/baz046 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouyang JF, Kamaraj US, Cao EY & Rackham OJL ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics 37, 3374–3376, doi: 10.1093/bioinformatics/btab209 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Yu GC, Wang LG, Han YY & He QY clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omics 16, 284–287, doi: 10.1089/omi.2011.0118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci USA 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marinkovic M et al. Fibro-adipogenic progenitors of dystrophic mice are insensitive to NOTCH regulation of adipogenesis. Life Sci Alliance 2, doi: 10.26508/lsa.201900437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallot YS, Hindi SM, Mann AK & Kumar A Isolation, Culture, and Staining of Single Myofibers. Bio Protoc 6, e1942, doi: 10.21769/BioProtoc.1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scRNA-seq (GSE172410), myofiber RNA-seq data (GSE172254), and SkM RNA-seq data (GSE184348) used in this study are deposited in Gene Expression Omnibus (GEO). Human SkM RNA-Seq data are publicly available at GEO (GSE97084).
An interaction website for the SkM scRNA-seq dataset can be found at https://mayoxz.shinyapps.io/Muscle/.
Codes and all other data are available from the corresponding author upon reasonable request.