Abstract
The genetic diversity of group B Streptococcus in young, non-pregnant women is not well studied. Application of multilocus sequence analysis to 85 group B Streptococcus strains recovered from college students revealed similarities and differences in distribution of group B Streptococcus lineages, compared with that of previously studied pregnant populations, and revealed that strains of 1 clone were associated with antibiotic resistance.
Group B Streptococcus (GBS) colonization of pregnant women is a critical risk factor for invasive neonatal disease; ~20% of pregnant women [1] and 35% of young, nonpregnant women [2] are colonized, with rates varying by population and time. Among the 9 polysaccharide capsule serotypes of GBS, Ia, III, and V are most commonly associated with neonatal disease [3, 4]. The application of multilocus sequence typing (MLST) to GBS has shown that specific sequence types (STs) mark lineages that are usually associated with 1 serotype, although many lineages comprise a smaller percentage of other strains with different capsule types [5]. One GBS lineage, ST-17, was shown in previous studies [6, 7] to more frequently cause neonatal disease and has been suggested to have enhanced invasiveness [8]. Other lineages, such as ST-1, ST-19, and ST-23, appear to be more common among colonized women [7].
Little is known about the genetic variation and distribution of GBS lineages in nonpregnant women of childbearing age. Gaining a better understanding of the genotypes and resistance patterns of GBS strains circulating in this population is critical, because these women represent the future pregnant population and may be future targets for immunization with GBS vaccines. Here, we describe the distribution of GBS STs in nonpregnant female college students and report capsule type and antibiotic resistance frequencies.
Methods.
During September–December 2006, we enrolled 197 students who visited the Michigan State University Olin Health Center (East Lansing) for care. Approval to conduct the study was granted by the Biomedical and Health Institutional Review Board at Michigan State University, and informed consent was obtained from all participants. For a small monetary incentive, participants self-collected vaginal and rectal specimens using the Culture Swab Plus Collection System (Baltimore Biological Labs) and completed a questionnaire involving medical history and clinical symptoms.
Swabs were inoculated into Todd-Hewitt broth with gentamicin and nalidixic acid, were subcultured to trypticase soy agar with 5% sheep blood, and were incubated overnight at 37°C in carbon dioxide. Broths were screened by the PathoDx kit (Remel), which detects the group B antigen, and a Christie-Atkins-Munch-Peterson (CAMP) test was performed on suspect colonies. All GBS strains were characterized by PCR and sequencing of 7 housekeeping genes, as described elsewhere [5]. Sequence analysis and ST classification also were described elsewhere [5]. Strains were characterized by capsule (cps) genotyping [5] and for susceptibilities to penicillin, ampicillin, cefazolin, levofloxacin, erythromycin, clindamycin, and azithromycin by disk diffusion and Etest strips (AB Biodisk). Common interpretive guidelines [9] were used for all antibiotics except cefazolin, for which we used breakpoints described elsewhere [10]. Multivariate logistic regression analysis with adjustment for age, ethnicity, recent respiratory infection, recent urinary tract infection, vaginal symptoms, sexually transmitted infection history, and recent antibiotic use was used to identify predictors of resistance. Sexually transmitted infections were categorized as ⩾1 of the following: urinary tract infection, yeast infection, hepatitis, chlamydia, bacterial vaginosis, herpes, human papilloma virus infection, genital warts, or trichomonas.
Results.
Among the 197 women enrolled, 68% self-identified as Caucasian, 55% were 18–20 years of age, 34% reported respiratory symptoms, and 12% had taken antibiotics in the previous 2 weeks. Study participants who self-identified as Caucasian represented a slightly lower percentage (68%) when compared with the entire Michigan State University community at the time of the study (74%). Eighty (40.6%) of the 197 women were colonized with GBS; 12 (15%) and 14 (17.5%) of the 80 women had only vaginal or only rectal colonization, whereas 54 (67.5%) were colonized at both sites. Five of the 80 women had >1 GBS genotype, yielding 85 isolates for molecular characterization. MLST analysis identified 17 STs among the 85 strains (figure 1), with 4 clones accounting for 65% of the GBS strains. The predominant STs were ST-1 (20%), ST-22 (18%), ST-23 (14%), and ST-19 (13%). Six variants of previously described STs were identified: ST-23 variants differed in sdhA (2 variants) and tkt (1 variant), 1 ST-196 variant differed in sdhA, and 2 strains related to ST-88 and ST-22 had a unique combination of alleles. Overall, cps2 (27%), cps1a (24%), and cps5 (20%) predominated.
Figure 1.
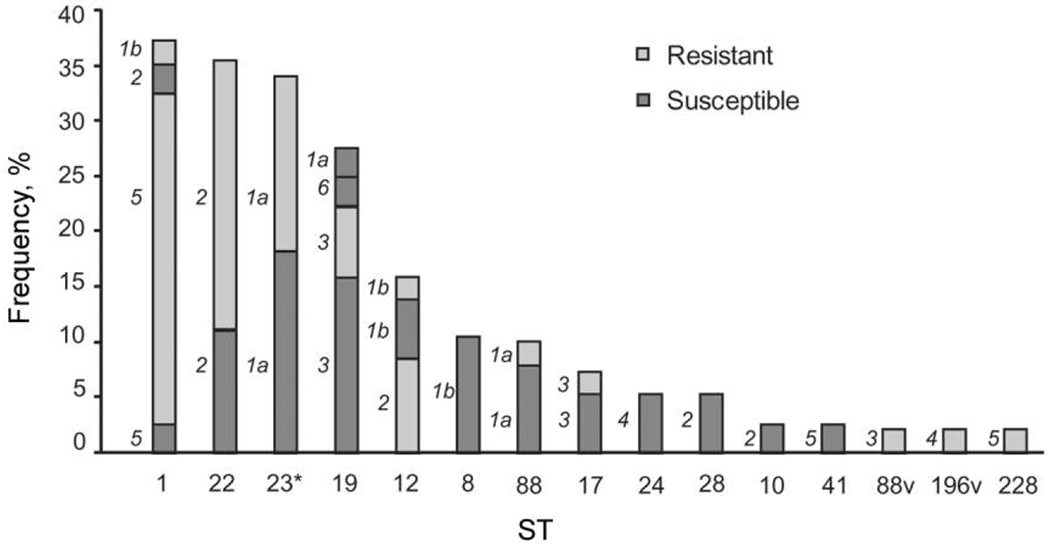
Distribution of antibiotic resistance in 85 group B Streptococcus strains isolated from nonpregnant women and stratified by multilocus sequence type (ST) and capsule (cps) genotype. The cps type is denoted as 1a, 1b, or 2–6 to the left of the representative portion of each bar. ST-1 cps5 (serotype V) strains were more frequently resistant, whereas ST-8 cps1b (serotype Ib) strains were more frequently susceptible, relative to all other ST and cps combinations. *ST-23 (n = 12) and ST-23 variants (n = 3) were included as 1 group; 1 ST-22 variant was identified and is included with the ST-22 group as well. v, ST variant.
Any resistance or intermediate resistance was detected in strains from 46 (57.5%) of the 80 women; 1 of these women had 2 GBS types that were resistant, resulting in 47 resistant strains. Resistance profiles included azithromycin, clindamycin, and erythromycin resistance (35 [74.5%] of the 47 strains); azithromycin and erythromycin resistance (9 [19.1%]); and clindamycin resistance only (3 [6.4%]). MLST data were used to assess whether certain STs were associated with resistance (figure 1). All but 1 of the 15 cps5 ST-1 strains were resistant, a statistically significant difference, compared with all other cps and ST combinations (χ2 = 10.6; 1 df; P = .001). Logistic regression analysis revealed that Caucasian women (OR, 4.4; 95% CI, 1.40–13.87; P = .01) and women reporting a history of sexually transmitted infections (OR, 3.4; 95% CI, 1.14–10.02; P = .03) were more frequently colonized with resistant strains.
Discussion.
The predominant STs (e.g., ST-1, ST-19, and ST-23) detected in earlier studies of strains from pregnant women [5, 7, 8] also occurred frequently in this sample of nonpregnant women. These clones likely represent strains that are well adapted to the vaginal mucosa. The second most prevalent clone identified in our nonpregnant population was ST-22 (n = 15; 18.8%), which occurred frequently in colonizing strains of pregnant women from Israel (11 [10.6%] of participants of that study) [8] but not Canada (5 [1%]) [5] or England (5 [2.6%]) [7]. The difference in ST-22 prevalence between this population and the Canadian and English populations separately and combined (χ2 = 33.6; 1 df; P < .001) was statistically significant, whereas no difference in ST-22 prevalence was observed between the nonpregnant women in our study and the Israeli population (χ2 = 3.2; 1 df; P = .07). In addition, the Canadian and English women together had a significantly lower prevalence of ST-22 colonization than did women from Israel (χ2 = 10.1; 1 df; P = .001). No statistically significant difference was observed in the prevalence of ST-17 colonization between the maternal and nonpregnant populations (χ2 = 1.7; 1 df; P = .2), but the small number of nonpregnant women with ST-17 colonization means that this finding must be interpreted with caution.
Antibiotic susceptibility testing revealed an unusually high frequency of antibiotic resistance in this group of nonpregnant women. Although the study was not designed to assess changes in resistance rates over time, the observed frequencies are higher than those from a similar population [10] and may reflect trends of increasing resistance. Genotyping by MLST revealed a significant preponderance of the cps5 ST-1 genotype among resistant strains, which is consistent with a previous report [11]; most ST-1 strains have been found to possess the type V capsule [5–8, 11]. These findings support the hypothesis that clonal dissemination and horizontal transfer of resistance genes, such as ermB, frequently contribute to macrolide resistance among serotype V strains [11, 12]. Large-scale studies exploring the distribution of resistant clones are warranted, as is an assessment of the differences in gene content between the ST-1 resistant lineage and the ST-8 susceptible lineage, to identify additional resistance mechanisms that may contribute to decreased susceptibility.
In the present study, 13 of the 15 women colonized with ST-1 cps5 strains were Caucasian. The finding of Caucasian ethnicity in association with resistance was unexpected, because an earlier study of GBS resistance in pregnant women identified an association between resistance and African American ethnicity [13]. These results suggest that factors contributing to resistance vary by population, which may be related to antibiotic-use differences or variation in the prevalence of lineages associated with resistance. These findings require confirmation with studies of nonpregnant women of different ethnic backgrounds.
In summary, population genetic analysis by MLST of GBS strains from young, nonpregnant women has revealed that strains of the ST-1 lineage are more frequently resistant to antibiotics and that 3 common GBS lineages (ST-1, ST-19, and ST-23) identified in other populations also predominate in colonizing strains from this population [5, 7, 8]. The latter observation indicates that there is a widespread distribution of some GBS clones. However, we also observed a high prevalence of ST-22 strains, which occurred infrequently in 2 earlier maternal studies [5, 7] but in more (11%) of the colonizing strains isolated from mothers in Israel, an area with a low incidence of neonatal GBS disease [8]. This suggests that, similar to serotypes [4], the distribution of GBS clones may vary by population or time, which could have an impact on the efficacy of some GBS vaccines. Any capsule- or protein-based vaccine that does not cover the predominant GBS genotypes in circulation may have limited success, because it is possible that antibody responses vary because of differences in the clonal composition of colonizing strains. Future studies are required to assess whether these GBS-specific antibody responses differ among lineages.
Acknowledgments
We thank the staff of Olin Health Center at Michigan State University, particularly Barbara Forney; Richard Bortel of Sparty’s; Sarah Marzec; Drs.James Rudrik and Patricia Somsel of the Michigan Department of Community Health; and the participants.
Financial support.
National Institutes of Health Public Health Service (AI066081).
Footnotes
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Davies HD, Adair C, McGeer A, et al. Antibodies to capsular polysaccharides of group B Streptococcus in pregnant Canadian women: relationship to colonization status and infection in the neonate. J Infect Dis 2001; 184:285–91. [DOI] [PubMed] [Google Scholar]
- 2.Manning SD, Neighbors K, Tallman PA, et al. Prevalence of group B Streptococcus colonization and potential for transmission by casual contact in healthy young men and women. Clin Infect Dis 2004; 39:380–8. [DOI] [PubMed] [Google Scholar]
- 3.Zaleznik DF, Rench MA, Hillier S, et al. Invasive disease due to group B Streptococcus in pregnant women and neonates from diverse population groups. Clin Infect Dis 2000;30:276–81. [DOI] [PubMed] [Google Scholar]
- 4.Harrison LH, Elliott JA, Dwyer DM, et al. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J Infect Dis 1998; 177:998–1002. [DOI] [PubMed] [Google Scholar]
- 5.Manning SD, Lewis MA, Springman AC, Lehotzky E, Whittam TS, Davies HD. Genotypic diversity and serotype distribution of group B Streptococcus isolated from women before and after delivery. Clin Infect Dis 2008;46:1829–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luan SL, Granlund M, Sellin M, Lagergard T, Spratt BG, Norgren M. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J Clin Microbiol 2005; 43:3727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones N, Oliver KA, Barry J, et al. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin Infect Dis 2006;42:915–24. [DOI] [PubMed] [Google Scholar]
- 8.Bisharat N, Jones N, Marchaim D, et al. Population structure of group B Streptococcus from a low-incidence region for invasive neonatal disease. Microbiology 2005; 151:1875–81. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 17th informational supplement. January 2007.
- 10.Manning SD, Pearlman MD, Tallman P, Pierson CL, Foxman B. Frequency of antibiotic resistance among group B Streptococcus isolated from healthy college students. Clin Infect Dis 2001; 33:E137–9. [DOI] [PubMed] [Google Scholar]
- 11.Gherardi G, Imperi M, Baldassarri L, et al. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J Clin Microbiol 2007; 45:2909–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puopolo KM, Klinzing DC, Lin MP, Yesucevitz DL, Cieslewicz MJ. A composite transposon associated with erythromycin and clindamycin resistance in group B Streptococcus. J Med Microbiol 2007; 56:947–55. [DOI] [PubMed] [Google Scholar]
- 13.Manning SD, Foxman B, Pierson CL, Tallman P, Baker CJ, Pearlman MD. Correlates of antibiotic-resistant group B Streptococcus isolated from pregnant women. Obstet Gynecol 2003; 101:74–9. [DOI] [PubMed] [Google Scholar]


