Summary
The cap‐binding protein eIF4E, through its interaction with eIF4G, constitutes the core of the eIF4F complex, which plays a key role in the circularization of mRNAs and their subsequent cap‐dependent translation. In addition to its fundamental role in mRNA translation initiation, other functions have been described or suggested for eIF4E, including acting as a proviral factor and participating in sexual development. We used CRISPR/Cas9 genome editing to generate melon eif4e knockout mutant lines. Editing worked efficiently in melon, as we obtained transformed plants with a single‐nucleotide deletion in homozygosis in the first eIF4E exon already in a T0 generation. Edited and non‐transgenic plants of a segregating F2 generation were inoculated with Moroccan watermelon mosaic virus (MWMV); homozygous mutant plants showed virus resistance, while heterozygous and non‐mutant plants were infected, in agreement with our previous results with plants silenced in eIF4E. Interestingly, all homozygous edited plants of the T0 and F2 generations showed a male sterility phenotype, while crossing with wild‐type plants restored fertility, displaying a perfect correlation between the segregation of the male sterility phenotype and the segregation of the eif4e mutation. Morphological comparative analysis of melon male flowers along consecutive developmental stages showed postmeiotic abnormal development for both microsporocytes and tapetum, with clear differences in the timing of tapetum degradation in the mutant versus wild‐type. An RNA‐Seq analysis identified critical genes in pollen development that were down‐regulated in flowers of eif4e/eif4e plants, and suggested that eIF4E‐specific mRNA translation initiation is a limiting factor for male gametes formation in melon.
Keywords: cucurbit, pollen, potyvirus resistance, resistance breaking, sexual development, translation initiation
Resistance to MWMV is associated with eIF4E editing and eif4e knock‐out plants show a post‐meiotic, sporophytic male sterility phenotype.

Introduction
Diseases caused by viruses affect food quality and reduce crop yields, and are very difficult to combat due to the scarcity of effective control measures. The use of genetic resistance, either natural or engineered, has often proved to be the only viable control strategy against a number of virus‐induced diseases. Therefore, the search for sources of genetic resistance to viruses, and their use in breeding programs, is considered one of the most desirable strategies to reduce losses caused by these pathogens (Fraser, 1990; Gómez et al., 2009; Revers and Nicaise, 2014). Plant viruses are obligate parasites that exploit the host cellular machinery to complete their life cycles. Susceptibility factor‐mediated resistance, more correctly defined as loss of susceptibility or passive resistance, can be achieved through the absence or modification of host factors that are essential for the virus. These factors are usually recessive, as a single copy of the allele is sufficient for the virus to complete its life cycle (Diaz‐Pendon et al., 2004; Fraser, 1990; Garcia‐Ruiz, 2018; Truniger and Aranda, 2009). Up to date, the best characterized recessive resistances against plant viruses are those based on the eukaryotic translation initiation factors (eIF) 4E and 4G and their isoforms (Julio et al., 2015; Revers and Nicaise, 2014; Saha and Mäkinen, 2020; Sanfaçon, 2015; Schmitt‐Keichinger, 2019; Truniger and Aranda, 2009).
eIF4E plays a critical role in the initiation of the canonical translation of eukaryotic mRNAs by binding to their 5′ m7G cap structures. Translation initiation is enhanced by the interaction of the poly(A)‐binding protein (PABP) with the mRNAs 3′ poly(A) tails and the binding of the eIF4G scaffold protein to eIF4E and PABP promoting circularization of the mRNA (Castellano and Merchante, 2021; Sanfaçon, 2015). In plants, two isoforms of eIF4F have been described as follows: eIF4F and eIFiso4F, which include eIF4E and eIF4G, and eIFiso4E and eIFiso4G, respectively (Browning, 2004; Roy and Arnim, 2013), and these are often encoded by small multi‐gene families (Patrick and Browning, 2012). Multiple studies have previously suggested that the eIF4E and eIF4G isoforms are highly selective in translating mRNAs, and therefore, a gradation of cap‐dependent to cap‐independent requirements to the selection may be observed (Martínez‐Silva et al., 2012; Mayberry et al., 2011). In melon, only one gene encoding eIF4E and one coding for eIFiso4E have been identified (Garcia‐Mas et al., 2012).
Natural mutations of eIF4E or eIFiso4E have been frequently associated with recessive resistance to potyviruses (Robaglia and Caranta, 2006; Yeam et al., 2007), with mutations shown to disrupt the eIF interactions with the potyviral protein that is covalently linked to the 5′‐end of the virus genome (VPg) (Charron et al., 2008; Gallois et al., 2010; Moury et al., 2014). Thus, it has been proposed that VPg may act as a cap surrogate, such that a specific eIF4E/VPg interaction may be required for translation initiation of potyvirus genomic mRNAs (Charron et al., 2008; Coutinho de Oliveira et al., 2019). Several virulent potyvirus isolates have been described to overcome recessive resistances conferred by eIF4E/iso4E alleles, and in most cases, the overcoming of resistance was due to mutations in the VPg protein (Ayme et al., 2007; Charron et al., 2008). However, other studies have suggested that the role of eIF4E in the potyvirus cycle may be distinct from its function in cellular mRNA translation initiation (Gallois et al., 2018).
Beyond their involvement in the canonical translation of cellular mRNAs, other biological functions have been demonstrated for eIFs. Animal and plant studies have shown that germ cell and embryonic fates are greatly affected by the eIF4 factor complexes unique to those cell types (Baker and Fuller, 2007; Callot and Gallois, 2014; Dinkova et al., 2005; Friday and Keiper, 2015; Shao et al., 2021). The use of genetics and biochemistry has also identified unique roles for eIF4E and eIF4G isoforms in reproduction. Furthermore, eIF4Es in plants, flies and frogs have shown unique roles in sexual development, judging by the reproductive phenotypes resulting from their deficiencies (Callot and Gallois, 2014; Ghosh and Lasko, 2015; Mazier et al., 2011; Patrick et al., 2014; Shao et al., 2021). Viruses could take advantage of these additional biological functions to control protein stability, regulate their replication and facilitate their intra‐ and intercellular movements.
Silencing eIF4E by RNA interference in transgenic plants has been successfully used to engineer resistance to one or several viruses (Rodríguez‐Hernández et al., 2012; Wang et al., 2013). Recently, eukaryotic genome editing has been incorporated into crop breeding, allowing precise genomic modifications (Biswas et al., 2021). CRISPR/Cas9 has been used to create genome editing mutants with resistance to various viral diseases in model and crop species, including cucurbits. In 2016, Pyott and colleagues employed CRISPR/Cas9 to knockout eIFiso4E in Arabidopsis. The resulting eifiso4e mutants were resistant to turnip mosaic virus (TuMV) infection without other traits affected (Pyott et al., 2016). Chandrasekaran et al. (2016) demonstrated that knocking out cucumber eIF4E resulted in resistance to a wide range of viruses (Chandrasekaran et al., 2016). Also, Gomez et al. (2019) showed that simultaneous CRISPR/Cas9‐mediated editing of cassava eIF4E isoforms nCBP‐1 and nCBP‐2 reduced cassava brown streak disease symptom severity and incidence (Gomez et al., 2019). The CRISPR/Cas technology has also contributed to the identification of genes and the study of the mechanisms underlying male sterility in plants, and once identified, the generation of male‐sterile lines through gene editing. In this regard, hxk5 rice mutants originated by CRISPR/Cas9 resulted in male sterility (Lee et al., 2020), and gene knockout of COPII components sarib and saric also altered pollen development and caused male sterility in Arabidopsis (Liang et al., 2020).
Here, we report CRISPR/Cas9 targeted eIF4E knockout in melon to attain virus resistance, and we describe, for the first time, the association between eIF4E editing and the development of male sterility in melon. Non‐transgenic, F2 generation homozygous plants harbouring a one base deletion in the eIF4E gene were inoculated with Moroccan watermelon mosaic virus (MWMV), and exhibited virus resistance compared with the susceptible heterozygous and non‐mutant F2 plants. Moreover, T0 and F2 homozygous mutant plants showed a male sterility phenotype as compared with the fertile heterozygous and non‐mutant F2 plants, displaying a perfect correlation between the segregation of the male sterility phenotype and the segregation of the eIF4E mutation. Microscopy and RNA‐Seq analyses were carried out to describe and better understand the male sterility phenotype observed in association with eIF4E knockout.
Results
Regeneration and transformation efficiencies of four melon genotypes
Melon organogenesis is highly dependent on the genotype (Nuñez‐Palenius et al., 2008). The regeneration ability of four melon accessions was evaluated. Significant differences (P < 0.05) were observed between accessions on the number of regenerated buds, with accessions M2 and M5 producing 1.7 and 2 buds per explant on average, respectively. All four accessions showed a high capacity for regeneration, especially M2 and M5, which showed rates of explants, with at least one regenerating bud higher than the rest of the accessions (Figure S1). We next used the fluorescent marker DsRed to evaluate the transformation efficiency of the four melon accessions (Table 1). Large foci of DsRed expression were detected in explants of all the accessions examined, with more abundant foci observed for M5, C46 and M9. However, these three accessions produced a lower number of rooted lines, which resulted in a lower transformation efficiency compared with M2 (Table 1) (Figure S1). We thus selected M2, which had the highest transformation efficiency (3%), for the transformation experiments with the CRISPR/Cas9 constructs.
Table 1.
Transformation efficiency and rooted plants produced from different melon accessions
| Genotype | Explants | Regeneration (%) 1 | dsRed + buds (%) 2 | Rooted lines | TE 3 |
|---|---|---|---|---|---|
| M2 | 300 | 288 (96) | 22 (7.3) | 10 | 3.33 |
| M9 | 300 | 250 (83.4) | 29 (9.7) | 6 | 2.00 |
| C‐46 | 300 | 215 (71.7) | 32 (10.7) | 2 | 0.67 |
| M5 | 300 | 262 (87.4) | 45 (15) | 4 | 1.33 |
Regeneration: number of explants with, at least, one regenerating bud. Between brackets the regeneration rate: regeneration/total number of infected explants × 100.
We used the vector pMOG800, which expresses dsRed after the 35S promoter. Consigned are buds with dsRed expression/total number explants × 100.
Transformation efficiency (TE): number of rooted lines/total number explants × 100.
CmeIF4E CRISPR/Cas9‐mediated editing
In the melon genome, eIF4E (MELO3C002698.2) and eIFiso4E (MELO3C023037.2) encode eIF4E (235 amino acids) and eIFiso4E (203 amino acids), respectively, sharing 61% of the coding nucleotides and 52% of the amino acids. gRNA1 was designed to target the first exon of eIF4E (Figure 1a) and had no sequence homology with eIFiso4E. Three independent transgenic T0 lines were generated by Agrobacterium‐mediated transformation. For two of the selected lines, sequencing of the PCR amplicons for the targeted region revealed a single‐nucleotide deletion, in homozygosis, three nucleotides upstream of the PAM sequence (Figure 1b); this mutation generates a premature stop codon downstream to the editing site (Figure 1c). The third line showed a WT sequence, although all the three lines tested positive for the Cas9 transgene.
Figure 1.
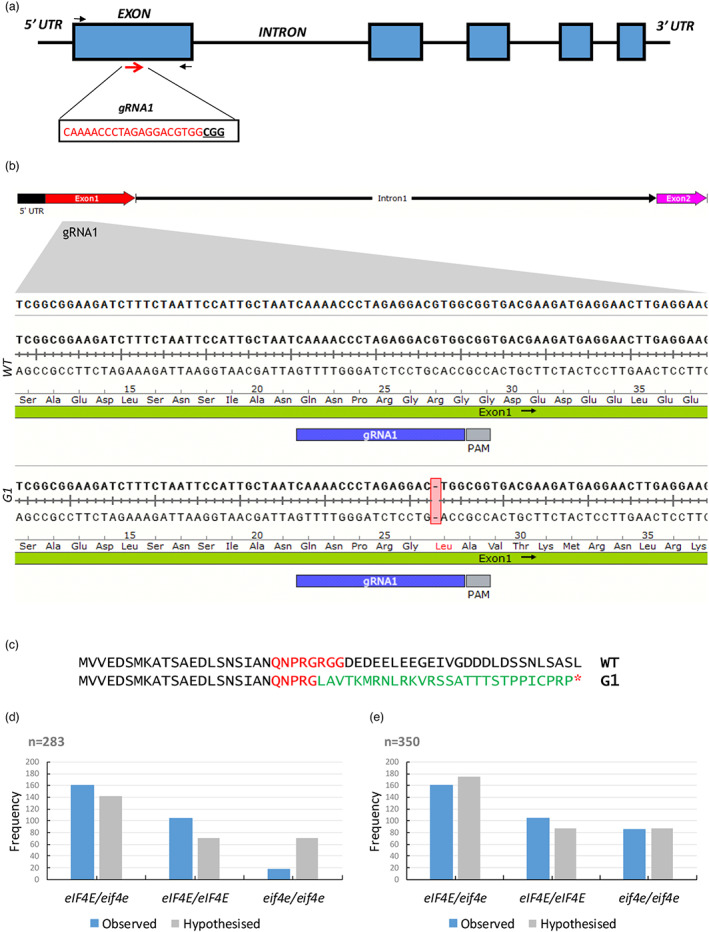
Gene editing of eIF4E mediated by CRISPR/Cas9 in melon plants. (a) Schematic representation of the melon eIF4E genomic map and the gRNA1 target site (red arrows). The target sequence is shown in red letters, and the protospacer adjacent motif (PAM) is marked in bold underlined letters. The black arrows indicate the primers flanking the target sites used to detect the mutations. (b) eIF4E genomic DNA alignment between WT and mutated T0 lines of fragments corresponding to a DNA fragment containing the gRNA1 sequence representative of the two independent lines obtained. A DNA deletion is highlighted in a red box. (c) eIF4E protein sequence alignments between WT and mutated T0 lines. The protein sequence downstream of the deletion site is shown in green; the target region in red and the red asterisk represents a premature stop codon. (d) Segregation and relative frequencies of the 283 F2 plants (excluding non‐germinated seeds) (N = 283, Ratio 1:2:1, degrees of freedom = 2, X2 = 57.6, P < 0.05) and (e) hypothesized segregation and relative frequencies of 350 F2 plants assigning non‐germinated seeds to the homozygous recessive group (N = 350, Ratio 1:2:1, degrees of freedom = 2, X2 = 4.3, P > 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
To test the heritability of the mutation, and to perpetuate the edited lines, we acclimatized edited T0 plants to ex vitro conditions, and grew them to adults in a greenhouse. During the flowering phase, we detected the lack of pollen in the eif4e knockout mutants, making selfing impossible. Nonetheless, female reproductive organs of hermaphrodite flowers appeared to be functional. We thus crossed eif4e/eif4e T0 plants and eIF4E/eIF4E WT plants to obtain the F1 generation (Figure S2a). The segregation of transgenic (with the Cas9 transgene) versus non‐transgenic (without the Cas9 transgene) plants in 14 plants of the F1 population was approximately 1:1, as expected for a single transgene insertion into the genome (Figure S2b). To evaluate the edition in F1 plants, PCR amplicons were sequenced for both transgenic and non‐transgenic plants. All non‐transgenic plants showed the same single‐nucleotide deletion as in T0, but in heterozygosis, as expected for a eif4e/eif4e x eIF4E/eIF4E cross. These results demonstrated that the induced mutations in melon can be stably transmitted through the germ line. In two of the transgenic plants, the chromatogram revealed a single‐nucleotide deletion in both alleles, likely indicating that gRNA1 and Cas9 were still active and able to edit the WT allele after fecundation. All remaining transgenic plants were heterozygous for the mutation in eIF4E (Figure S2c). Both transgenic and non‐transgenic plants were transferred to a greenhouse and grown to adults for seed multiplication. During the flowering phase, we checked the phenotype of each individual: all heterozygous mutants (transgenic and non‐transgenic) showed a fully fertile and normal growth phenotype. The two transgenic plants likely to be homozygous for the mutation showed male flowers with no or little pollen and reduced growth, phenotype that led, after attempting self‐pollination, to a failed fruit set or to reduced seed set and germination. On the contrary, we were able to normally self‐pollinate and obtain fruits from all heterozygous mutants.
To obtain the F2 generation, we exclusively used seeds obtained from self‐pollinations of non‐transgenic F1 individuals. Although we sowed 350 seeds, we were only able to obtain 283 F2 plants. The genotyping of each plant showed that the F2 progeny segregated into homozygous mutant (eif4e/eif4e), heterozygous (eIF4E/eif4e) and homozygous WT (eIF4E/eIF4E) (Figure S3). A X2 square test performed to analyse whether the frequency of each group was consistent with that expected for a 1:2:1 segregation rejected the null hypothesis, mainly due to a clear mismatch between the observed and the hypothesized frequencies for homozygous mutant plants (N = 283, ratio 1:2:1, degrees of freedom = 2, X2 = 57.6, P < 0.05) (Figure 1d), suggesting an adverse effect of the mutation. If non‐germinated seeds were arbitrarily assigned to the homozygous mutant category, a better fit with the frequencies of a 1:2:1 segregation could be deduced (N = 350, ratio 1:2:1, degrees of freedom = 2, X2 = 4.3, P > 0.05) (Figure 1e). The reduced growth phenotype observed in eif4e knockout mutants would also support the hypothesis of a negative effect of the eIF4E suppression resulting in poor seed germination.
Potential off‐targets were evaluated by using the CRISPR‐P program (Liu et al., 2017) to map the gRNA1 sequence against the melon genome. Five potential off‐target sites were identified (Table S1). Both amplicon sequencing and manual inspection of RNA‐seq data (see below) of alternative targets revealed WT sequences in non‐transgenic F2 edited plants (data not shown).
CmeIF4E edition associates with resistance to MWMV
We have demonstrated previously that transgenic RNAi eIF4E silenced melon plants show broad virus resistance, including resistance to the potyvirus MWMV (Rodríguez‐Hernández et al., 2012). To verify whether the knocking out of eIF4E conferred virus resistance in melon, all seedlings from the F2 progeny described above (n = 283) were mechanically inoculated with MWMV. Seedlings started to develop mosaic symptoms at 10 dpi, turning to severe leaf deformation at around 14 dpi in heterozygous and WT plants (Figure 2a). No symptoms were detected in any of the homozygous mutant plants at 20, 30 or 40 dpi. At 21 dpi, we determined the viral load in five plants taken randomly from each group (homozygous mutant, heterozygous and homozygous WT). No MWMV RNA was detected in homozygous mutant plants, whereas variable but significant viral loads were detected for heterozygous and WT plants. In this experiment, we introduced the RNAi eIF4E silenced melon line as control plants (Rodríguez‐Hernández et al., 2012), which showed no virus accumulation, as expected (Figure 2b).
Figure 2.
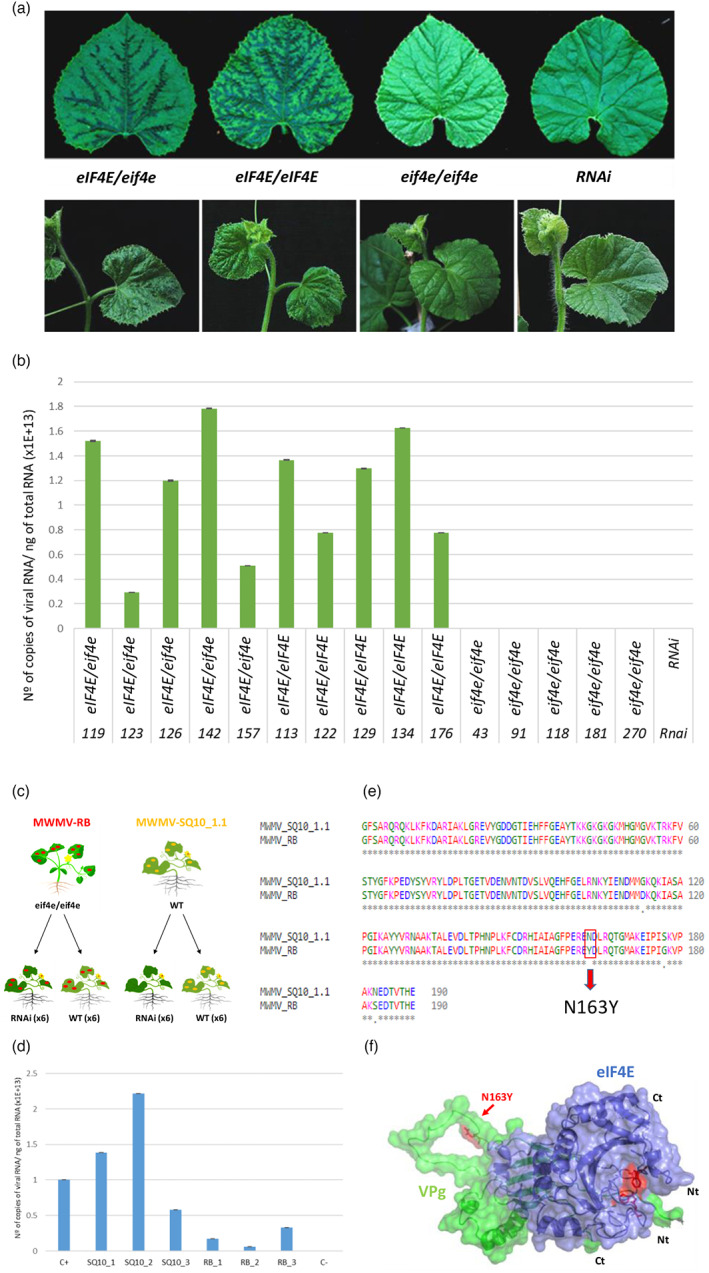
Homozygous eif4e mutant plants exhibited immunity to Moroccan watermelon mosaic virus (MWMV) infection. (a) Disease symptoms (leaves ‐upper panel‐ and plants ‐lower panel‐) of heterozygous (eIF4E/eif4e), homozygous wild type (WT) (eIF4E/eIF4E), and recessive homozygous (eif4e/eif4e) of the F2 edited generation and RNAi (control) plants (Rodríguez‐Hernández et al., 2012) at 14 days post inoculation (dpi). (b) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis of MWMV RNA accumulation at 21 dpi in five individuals heterozygous (eIF4E/eif4e), homozygous WT (eIF4E/eIF4E), recessive homozygous (eif4e/eif4e) and RNAi (control) (Rodríguez‐Hernández et al., 2012) plants. Error bars represent standard deviation. (c) Back‐inoculation assay: samples from an eif4e/eif4e resistant plant that presented late MWMV symptoms were used as a source of inoculum (red spots). MWMV‐SQ10_1.1 are WT plants infected with the original source of inoculum used to test the F2 mutants for resistance (yellow spots). (d) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis of MWMV RNA accumulation at 14 dpi in individual plants. SQ10_1, SQ10_2 and SQ10_3 correspond to WT plants inoculated with MWMV‐V. RB_1, RB_2 and RB_3 are RNAi lines infected with MWMV‐RB. C+ is a F2 susceptible plant infected with MWMV. C‐ is a mock inoculated plant. Error bars represent standard deviation. (e) Multiple alignment of amino acid sequences of VPg from MWMV‐RB and MWMV‐SQ10_1.1. The unique amino acid change between the two variants is underlined in red. (f) Simulated 3D surface model of the interaction between MWMV VPg (green) and eIF4E (blue) complex. Tryptophans W82 and W182 from eIF4E involved in the association with m7GTP Guanosine‐5′‐triphosphate cap analogue are shown in sticks and coloured red. The N163Y substitution responsible for the overcoming of the resistance to MWMV is indicated by a red arrow. [Colour figure can be viewed at wileyonlinelibrary.com]
Eighteen eif4e/eif4e plants from F2 were kept under observation for 6 months. Interestingly, 4 months after inoculation, symptoms compatible with MWMV infection were observed in two of the 18 plants, suggesting a resistance breaking event. To verify whether the symptoms were caused by MWMV, young symptomatic leaves from the two affected plants were sampled and RT‐qPCR was carried out to determine the presence of MWMV. Symptomatic plants tested positive for MWMV; therefore, a back‐inoculation assay (Figure 2c) was performed to confirm the presence of a resistance breaking (RB) MWMV isolate: We inoculated six WT and six RNAi plants with sap from the resistant plants that showed late symptoms, and another six WT and six RNAi plants with the original inoculum. After 14 dpi, WT plants inoculated with both sources of inocula showed symptoms, but only the isolate in the inoculum prepared from the late‐infected plants was able to infect the RNAi melon lines, overcoming resistance; from now on we will refer to this isolate as MWMV‐RB. An RT‐qPCR analysis to determine viral load in individual plants using RNA extracted from systemically infected leaves from plants inoculated with both sources of inocula confirmed that MWMV‐RB was able to overcome the resistance induced by eIF4E silencing. MWMV‐RB seemed to accumulate less in silenced plants (Figure 2d). It has been widely described that mutations in the viral VPg of potyviruses are frequently responsible for the overcoming of resistances associated with eIF4E (Gallois et al., 2018), we thus RT‐PCR‐amplified the entire VPg cistron and sequenced the DNA product. A sequence alignment showed a single‐nucleotide substitution leading to the single amino acid change N163Y (Figure 2e) in the MWMV‐RB isolate. In an attempt to further understand the interaction between eIF4E and viral VPg, we modelled the interaction between these two proteins, paying particular attention to residue 163; this modelling suggested that this residue was not located on the contact surface between eIF4E and VPg (Figure 2f). Contrastingly, a multiple sequence alignment of VPgs from different potyviruses showed that the amino acid 163 was embedded within two conserved motifs, and also, that a turnip mosaic virus (TuMV) RB isolate had an equivalent substitution responsible for overcoming resistance (Gallois et al., 2010) (Figures 2e; S4).
Structural analysis comparing male floral development in WT and eif4e knockout mutant plants
As described above, homozygous mutant T0 plants showed male sterility, while crossing with the WT restored fertility of the F1 individuals. The F2 showed a perfect correlation between the segregation of the male sterility phenotype and the segregation of the eIF4E mutation. Homozygous recessive mutant plants bolted 1–5 days later, and their flowers had smaller, paler petals than those of heterozygous and WT plants. The mutant anthers were small and white, without mature pollen grains, and did not dehisce, resulting in complete male sterility (Figure 3).
Figure 3.

Phenotype association of segregating F2 melon male flowers. Staminate and perfect flowers with viable pollen in the anthers of homozygous WT (eIF4E/eIF4E) and heterozygous (eIF4E/eif4e) F2 plants, small and not dehiscent anthers, without mature pollen grains in homozygous recessive (eif4e/eif4e) F2 plants at 40 days post germination. [Colour figure can be viewed at wileyonlinelibrary.com]
To identify structural differences during pollen development, longitudinal and transversal semithin sections of anthers at different stages of development from WT and eif4e mutant plants were examined. As for other cucurbits (Bai et al., 2004; Hao et al., 2003), male melon flower development can be divided into 12 stages, from the formation of the stamen primordium, to the release of mature pollen during anther dehiscence (Pechar, 2022). No significant differences were detected in anther structures in the mutant plants as compared with WT before stage 9 (data not shown). Therefore, the comparative analysis of anther development was carried out from stage 9 onwards (Figure 4). In stage 9, the WT anther primordia differentiated into a concentric structure, with microsporocytes (labelled as Ms in Figure 4) in the locule wrapped by a four‐layered anther wall, from surface to interior of the epidermis, endothecium, middle layer and tapetum (Figure 4a). The microsporocytes subsequently underwent meiosis, generating a tetrad by the end of stage 10. Meanwhile, tapetal cells went through their programmed cell death (PCD) (Figure 4c) until their complete degradation (Figure 4g). In contrast, during the differentiation of the eif4e mutant male flowers, both the anther walls and the microspores displayed an abnormal development: The anthers and locules were generally smaller in size and hosted a smaller number of microsporocytes (Figure 4b). The microsporocytes in the WT were characterized by a polyhedral morphology, with highly condensed chromatin nuclei and surrounded by highly organized tapetum cells (Figure 4a). On the contrary, in the mutant, microsporocytes with a more rounded shape were observed, surrounded by a tapetum with more disorganized and smaller cells (Figure 4b). At the tetrad stage, structural differences in pollen development became more evident: in the WT, tetrads were clearly formed and at least three of the four cells of the tetrad could be observed in the sections (Figure 4c). In the mutant, microsporocytes seemed to have undergone incomplete meiosis and microspore individualization, as anthers in that stage appeared to contain most of the cells in the dyad stage (Figure 4d). Although some microspore cells could be observed after meiosis in the mutant, their structure was somehow collapsed in comparison with the rounded shape of those from the wild type (compare Figure 4e,f). A remarkable distinction between WT and mutant anthers began to appear between stages 10 and 11: Normal vacuolated microspores with round shapes were uniformly distributed along the tapetum side, with a dark‐stained pollen wall corresponding to the exine in the WT (Figure 4e,g); in contrast, internal cavities of the mutant anthers became disorganized, instead of randomly distributed pollen grains inside the anther locules as in the WT, collapsed pollen grains in the mutant adhered to unstructured dark‐stained material, which could correspond to incompletely degenerated tapetum inside the locule walls (Figure 4h). The lumen of the anthers in the mutant at late stage 10 was filled with large amounts of dark‐stained material (Figure 5a–d), that could originate from droplets of sporopollenin secreted by the tapetum, as previously described in Arabidopsis and rice (Chang et al., 2016; Li et al., 2020; Sun et al., 2018). In stage 12, mature pollen was dark‐stained in the WT plants and the pollen grains appeared well structured, suggesting that the WT pollen grains were viable (Figures 4i; 5e and g; S5a and c). Contrastingly, mutant anthers showed a unstructured clumping of microspores and other stained/dark material of unknown origin in their anthers at stage 12 (Figures 4j; 5f,h; S5b and d).
Figure 4.
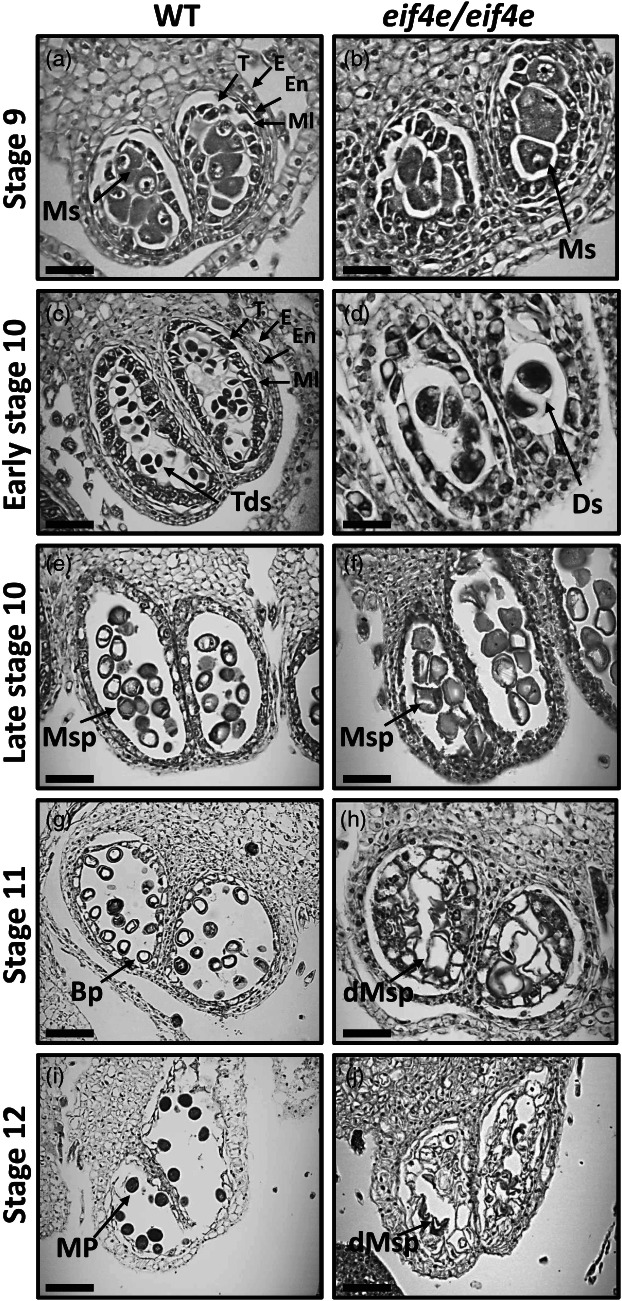
Transversal sections of anthers throughout development in wild‐type (WT) and eif4e mutant (eif4e/eif4e) observed by light microscopy. Locules from the WT (a, c, e, g, i) and eif4e mutant (b, d, f, h, j) anthers from stages 9 to 12 of development. BP, bicellular pollen; dMsp, degraded microspores; Ds, dyads; E, epidermis; En, endothecium; ML, middle layer; MP, mature pollen; Ms, microsporocyte; Msp, microspores; T, tapetum; Tds, tetrads. Scale bars = 50 μm (a, b, c, d, e, f), 100 μm (g, h, i, j).
Figure 5.
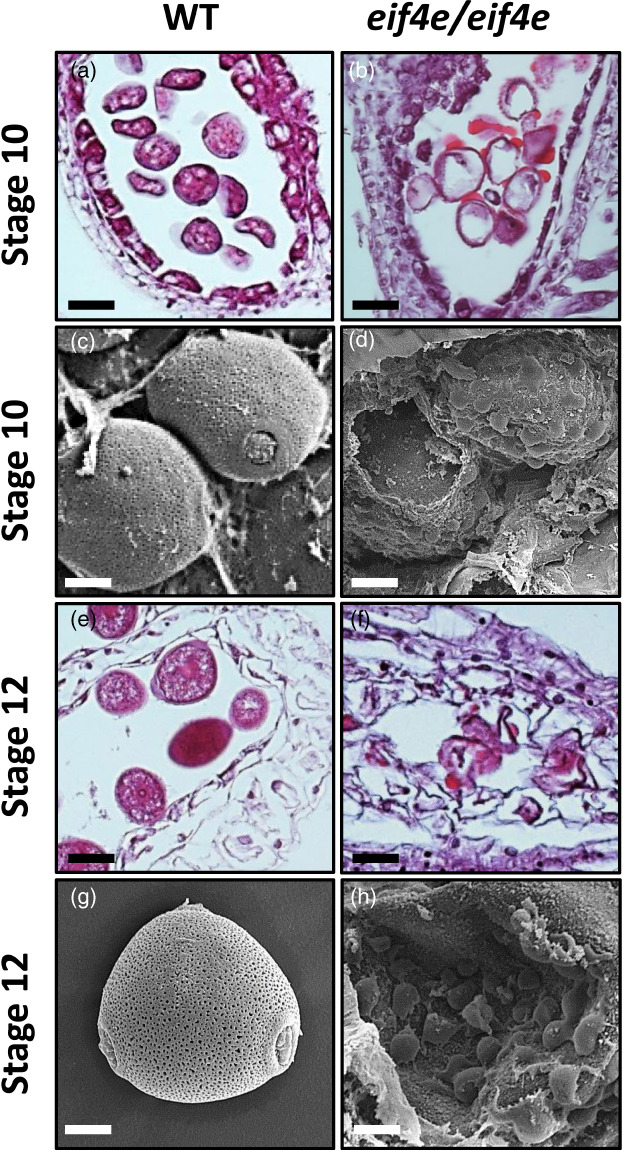
Light microscopy (a, b, e, f) and scanning electron microscopy (c, d, g, h) images of transversal sections throughout anther development in the wild‐type (WT) and eif4e mutant (eif4e/eif4e). (b, d) Locules are filled by large amounts of stained/dark material that could correspond to sporopollenin in late stages 10. (f, h) Locules showing clumping of microspores unstructured as well as other stained/dark material of unknown origin in early stage 12. Scale bars = 25 μm, (a, b, e, f), 10 μm (c, d), 5 μm (g, h). [Colour figure can be viewed at wileyonlinelibrary.com]
RNA‐seq analysis comparing male floral development in WT and eif4e knockout mutant plants
We next compared the transcriptomes of WT and mutant male flowers along four episodes during floral development: The floral structures formation (FS) episode, when male and hermaphrodite flowers are not yet differentiated and flower buds are less than 2 mm in length; it includes stage 9, in which we started to see morphological differences between WT and mutant plants. The gamete initiation (GI) episode, when floral buds are 2–5 mm in size, corresponding to flowers in stage 10, in which already important differences between WT and mutant were morphologically seen. The gamete maturation (GM) episode, when buds are 8–10 mm, corresponding to stage 11, when WT versus mutant differences were obvious. And the anthesis (AN) episode, when floral buds are larger than 2 cm, corresponding to stage 12, in which once again obvious morphological differences in WT versus mutant male flowers were observed (see above). Thus, we collected flowers in eight groups, named FS/FSmut, GI‐M/GI‐Mmut, GM‐M/GM‐Mmut and AN‐M/AN‐Mmut. Fifty floral buds were pooled per biological replicate, and three replicates were prepared per each of the groups. RNA was extracted, depleted in plant rRNAs and subjected to next‐generation sequencing. RNA‐seq reads were mapped to the melon reference genome. For all comparisons, read counts were normalized to fragments per kilo base of transcript per million mapped fragments (FPKM) to obtain the relative levels of gene expression. Genes were considered as expressed if the FPKM value was higher than 1 in the three biological replicates for each sample. Thus, ~14 500 genes were expressed in each episode (Figure 6a). A hierarchical clustering analysis showed that gene expression patterns of biological replicates were highly related (Figure S6). A PCA including all biological replicates showed a clear separation in gene expression patterns between WT and mutant, as well as a clear separation in the gene expression patterns across floral developmental episodes within both WT and mutant (Figure 6b).
Figure 6.
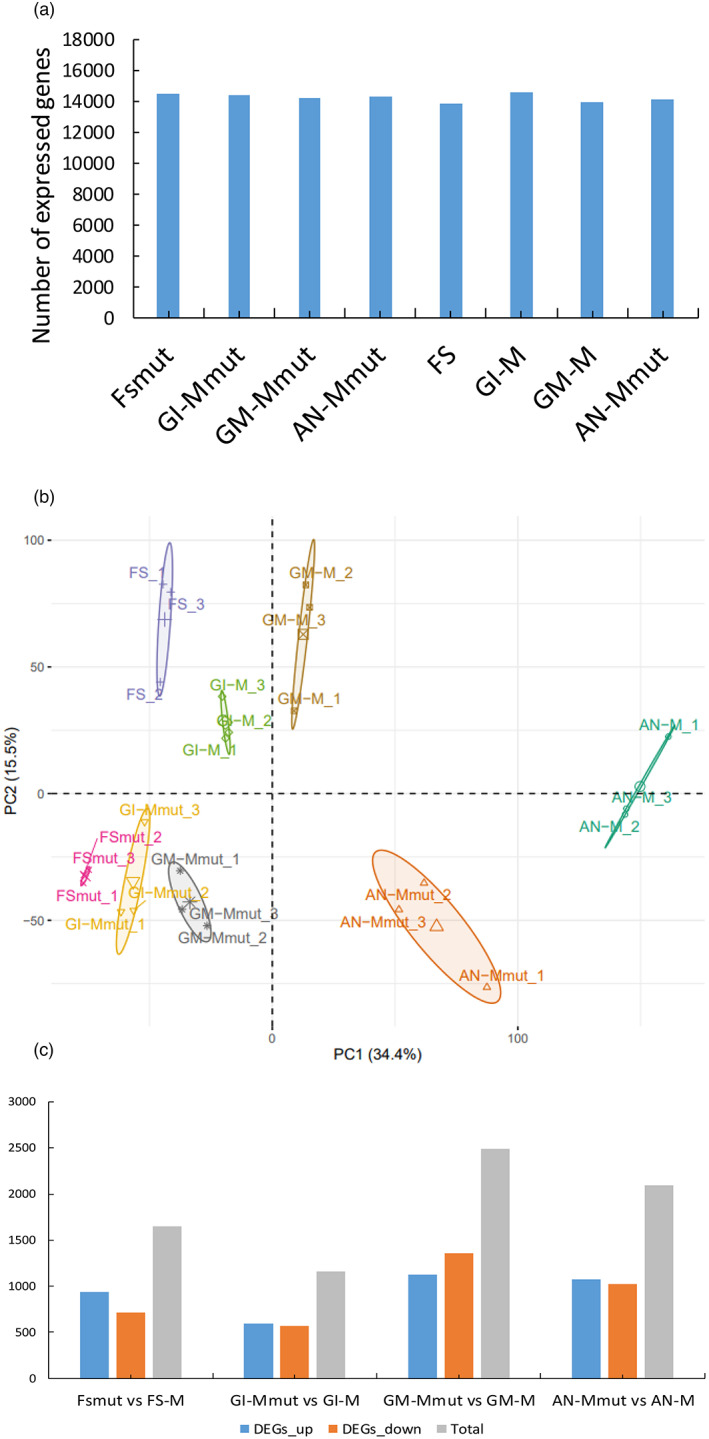
RNA‐seq along flower developmental episodes for wild‐type (WT) and eif4e mutant melon plants. Four episodes during floral development were considered: Floral structures formation (FS), gamete initiation (GI), gamete maturation (GM), and anthesis (AN) episodes. (a) Number of expressed genes per floral developmental episode. Expressed genes are considered those with an average value of FPKM (fragments per kilo base of transcript per million mapped fragments) > 1 in the three replicates in at least one episode. (b) Principal Component Analysis (PCA) scores plotted for eif4e mutant and WT floral development episodes. PCA was computed using expressed genes. PC1, principal component 1; PC2, principal component 2. The percentage of variance explained by PC1 and PC2 are 34.4% and 15.5%, respectively. Confidence ellipses were plotted around group mean points. (c) Number of DEG (up‐regulated (blue), down‐regulated (orange) and total (grey) in mutant versus WT in different episodes of flower development (adjusted P value < 0.01 and Log2 fold change >1 and <−1). [Colour figure can be viewed at wileyonlinelibrary.com]
A comparative analysis of differentially expressed genes (DEGs) between WT and mutant for each of the episodes was carried out. Only genes with an adjusted P value <0.01 and log2 fold change higher than 1 and lower than −1 were considered as DEGs. In total, 7397 DEGs were identified. The comparisons of FSmut versus FS, GI‐Mmut versus GI‐M, GM‐Mmut versus GM‐M, and AN‐Mmut versus AN‐M provided 1649, 1162, 2490 and 2096 DEGs, respectively, with GM‐Mmut versus GM‐M being the comparison in which the largest number of DEGs was found (Figure 6c). A GO enrichment functional category analysis was carried out with all the DEGs. The DEGs in the FS episode were enriched for ‘plastid’, ‘photosynthesis’, ‘ribosome’, ‘translation’ and ‘chloroplast’, among others (Figure 7a). The DEGs in the GI episode were enriched in the categories ‘MCM complex’, ‘enzyme inhibitor activity’, and ‘photosystem’ (Figure 7a). The most representative GO terms enriched categories for the GM episode were ‘membrane’, ‘metabolic process’, ‘rRNA binding’, ‘cell wall’, ‘catalytic activity’, ‘pectin catabolic process’, ‘transporter activity’, ‘extracellular region’, ‘transmembrane transporter activity’ and ‘cellular amino acid biosynthetic process’ (Figure 7a). Finally, the AN episode comparison included many GO terms, with ‘membrane’, ‘hydrolase activity’, ‘carbohydrate metabolic process’, ‘transporter activity’, ‘proteasome complex’, ‘calcium ion transport’ and ‘enzyme inhibitor activity’ being the most relevant (Figure 7a). In general, the GO enrichment analysis showed a shift between photosynthesis‐, ribosome‐ and translation‐related in the early FS episode towards membrane‐ and metabolic processes‐related GO terms shared by GM and AN, the late episodes.
Figure 7.
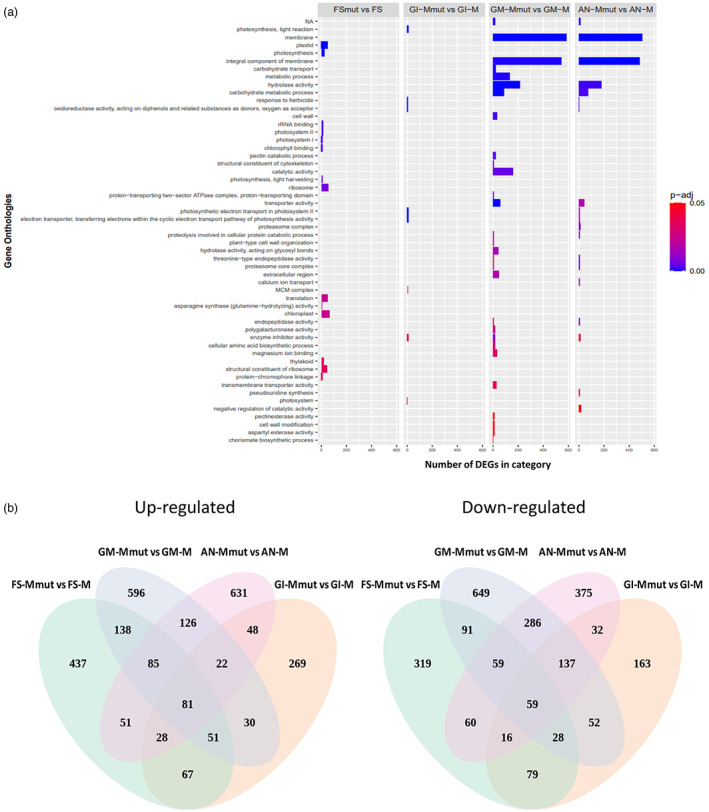
Differentially expressed genes along flower developmental episodes for wild‐type (WT) and eif4e mutant melon plants. (a) Mutant versus WT enriched GO terms among different episodes of flower development. The X‐axis indicates the enriched categories, and the Y‐axis indicates the differentially expressed gene number in each category. Enriched categories were considered those with a P‐adjust lower than 0.05, which is represented here in a coloured scale from 0 (blues) to 0.05 (reds). (b) Venn diagrams showing mutant versus WT differently expressed up (left) and down (right) regulated genes at different episodes of flower development. [Colour figure can be viewed at wileyonlinelibrary.com]
Inspection of DEGs along floral developmental episodes
We compiled a list of DEGs that were specifically up‐ or down‐regulated in each episode (Figure 7b) which may play a role in the genesis of the observed male sterility phenotype (Table S2). After comparing FSmut versus FS, we identified genes involved in ethylene synthesis (ethylene‐responsive transcription factor), stamen filament development (FHA domain‐containing protein FHA2), sporopollenin biosynthetic process (tetraketide alpha‐pyrone reductase 1), receptor‐like kinase related‐genes (pollen receptor‐like kinase 3), sugar transferase related‐genes and the genes encoding the eIF4G and eIF3 subunit G‐like, which were down‐regulated in the mutant. In contrast, genes related to gametophyte development and regulation of programmed cell death (apoptosis inhibitor 5‐like API5, male gametophyte defective 1, protein XRI1) were up‐regulated. The comparison of GI‐Mmut versus GI‐M identified genes encoding two proteins involved in floral development and flowering (MADS‐box protein SOC1 and Protein EARLY FLOWERING 3), and pollen wall formation‐related proteins (pollen‐specific leucine‐rich repeat extensin‐like protein 3) which were down‐regulated. eIF4G was still down‐regulated in this episode. Apoptosis inhibitor 5‐like API5 still showed the same up‐regulation pattern as in episode FS, whereas tetraketide alpha‐pyrone reductase 1, which is involved in the process of sporopollenin biosynthesis, appeared up‐regulated in GI, in contrast to what was observed in the previous episode. Another interesting up‐regulated gene was that encoding MACPF domain‐containing protein NSL1, involved in cell death and defence response by callose deposition. The comparison of GM‐Mmut versus GM‐M led to the identification of the down‐regulation of pollen sperm cell differentiation, generative cell mitosis/pollen development (F‐box/LRR‐repeat protein 17, pollen‐specific protein, pollen‐specific protein‐like) protein encoding genes, pollen tube development/polar nucleus fusion related‐encoding protein genes (O‐fucosyltransferase family protein), and pollen exine formation genes (polygalacturonase QRT3). Genes encoding proteins involved in pollen recognition (receptor‐like serine/threonine‐protein kinase SD1‐8, Serine/threonine‐protein kinase) and pollen exine formation (transmembrane protein) were up‐regulated in GM in the mutant. The comparison of AN‐Mmut versus AN‐M showed the down‐regulation of endopeptidase activity (Proteasome subunit alpha and beta type), pollen tube growth (Armadillo repeat only protein, DnaJ protein ERDJ3A), (1‐ > 3)‐Beta‐D‐glucan biosynthetic process (callose synthase 5‐like), structural constituent of cell wall (pollen‐specific leucine‐rich repeat extensin‐like protein 3), flower development (FRIGIDA‐like protein) and pollen development (Major pollen allergen Ole e 6, Pollen‐specific protein, pollen receptor‐like kinase 1, tetraketide alpha‐pyrone reductase 1) encoding genes, as well as eIF4G, eIF3 subunit G‐like, eIF1A, eIF2D, which were also down‐regulated in the mutant. In contrast, genes encoding proteins related to gametophyte development and regulation of programmed cell death (Apoptosis inhibitor 5‐like API5), recognition of pollen (Serine/threonine‐protein kinase) and eIF3 subunit A‐like were up‐regulated.
Discussion
In this study, we used CRISPR/Cas9 genome editing to generate melon eif4e knockout mutant melon lines. Major issues of concern while working with CRISPR/Cas are the editing efficiency and off‐target edition events. Here, we show that two the three mutated lines obtained had the same deletion of one nucleotide in homozygosis already from the T0 generation, while no editing of alternative targets was identified. Through the crossing of transgenic lines homozygous for the mutation with WT lines of the same genetic background, we were able to obtain an F1 generation, including about half of non‐transgenic individuals that presented the same single deletion as the T0, but in heterozygosis. These results indicated that the transgene insertion occurred in heterozygosis and was very likely single in T0 plants, and additionally demonstrated that induced mutations in melon can be stably transmitted through the germ‐line. Therefore, CRISPR/Cas editing appears to work well in melon, with regeneration and genetic transformation still being major limitations. In our study, differences in transformation efficiency were observed for the melon accessions tested, suggesting that assaying further melon genotypes could result in improved efficiencies.
Our group previously demonstrated that transgenic melon plants silenced in eIF4E by RNAi were resistant to cucumber vein yellowing virus, melon necrotic spot virus, zucchini yellow mosaic virus and MWMV (Rodríguez‐Hernández et al., 2012). Our results showed that CRISPR/Cas9‐mediated eIF4E knockout leads to resistance against MWMV; the finding that plants heterozygous for the mutation were susceptible to MWMV allows concluding that a single allelic copy of eIF4E is sufficient for virus multiplication. Significantly, 4 months after inoculation, we observed symptoms compatible with MWMV infection in two of the mutant plants, which were caused by a resistance breaking MWMV isolate that had a single‐nucleotide substitution in its VPg sequence, leading to the N163Y amino acid change. It has been described that mutations in the VPg of potyviruses are frequently responsible for overcoming resistances mediated by eIF4E (Ayme et al., 2007). It is worth noting that Gallois et al. (2010) described a homologous nucleotide change in TuMV VPg as being responsible for resistance breaking in A. thaliana mutants knocked out in eIFiso4E and eIFiso4G; these authors hypothesized that VPg‐N163Y may allow resistance breaking by interacting with an alternative, not yet identified partner in planta (Gallois et al., 2010). However, the same team showed later that the TuMV resistance breaking isolate had gained the ability to recruit either eIF4E1 or eIFiso4E (Bastet et al., 2018), which may also be the case for the MWMV‐RB isolate described here. In any case, our results indicate that further refinement of eIF4E editing, for example through the latest CRISPR base or prime editing applications (Bastet et al., 2019), needs to be tackled to obtain durable virus resistance, possibly by guiding editing with the information that can be derived from eIF4E and virus natural variation during evolution, as suggested by Bastet et al. (2018).
Our results showed that eif4e knockout plants had a reduced growth phenotype, in addition to being androsterile. Segregation of the mutation in the F2 generation showed a clear mismatch between observed and expected frequencies in the case of homozygous plants, which we attributed to a probable negative effect of eIF4E abolishment on the seed germination capacity. Differences in vegetative growth of the F2 plants were also observed from the flowering stage onwards, when the androsterility phenotype re‐emerged in homozygous mutants. Both the acclimatized T0 mutant plants and the homozygous F2 individuals presented a male sterility phenotype. Our RNAi plants, despite having partially impaired vegetative growth, were unaffected in pollen production (Rodríguez‐Hernández et al., 2012). A possible explanation for the phenotypic difference between the knockout and knockdown lines could be that the residual expression of eIF4E in the RNAi lines was sufficient to carry out its function in pollen development. Another possible explanation could be associated to a potential misfunction of the RNAi pathway in germ‐line cells which would determine the presence of pollen in the silenced plants. Despite the hopes placed in eIF4E editing, it does not seem to be a target free of limitations for virus resistance breeding. The study of plant–virus interactions and the identification of susceptibility factors are important activities for the establishment of new molecular targets and, therefore, for the breeding of new resistant varieties. The use of reverse genetics assays is a fundamental tool for the characterization of host genes involved in both the viral cycle and host developmental functions, such as reproduction. However, the most common approach used so far to identify sources of resistance to plant viruses is still based on direct genetic studies, in which aggressive isolates or resistance breaking isolates could be used to identify new sources of resistance. We believe that the combined use of these two strategies may be promising in the identification of new candidate proviral genes (Garcia‐Ruiz, 2019; Shopan et al., 2017).
With the structural analysis that we carried out, we did not detect significant differences in anther structures in mutant compared with WT plants before flowering in stage 9, and this was consistent with what has been previously described for male‐sterile rice lines generated by mutations in the Oryza sativa No Pollen 1 (OsNP1) nuclear male sterility gene (Chang et al., 2016). In melon, structural differences in pollen development became evident at the tetrad stage: In the WT, tetrads were clearly formed and at least three of the four cells of the tetrad could be observed in the flower sections (Figure 4c), while the mutant microsporocytes seemed to undergo incomplete meiosis and microspore individualization (Figure 4d). Therefore, the androsterility phenotype that we have described seemed to be a postmeiotic phenomenon. Furthermore, a remarkable distinction between WT and mutant anthers appeared between stages 10 and 11: Normal vacuolated, round shaped microspores were distributed along the tapetum side with a dark‐stained pollen wall corresponding to the exine in the WT (Figure 4g), whereas the internal cavities in the mutant anthers became disorganized, and collapsed pollen grains adhered to unstructured dark‐stained material that could correspond to incompletely degenerated tapetum inside the locule walls (Figure 4h).
Pollen development is synergistically controlled by sporophytic and gametophytic factors (McCormick, 2004). The sporophytic tapetum, adjacent to developing microspores, appears to be crucial for pollen development through its secretions in the early stages, and PCD in the late stages (Parish and Li, 2010). The functioning of the tapetum is controlled by an evolutionarily conserved transcriptional cascade (Wilson and Zhang, 2009; Zhu et al., 2015), as well as proteins involved in intercellular signalling. Many postmeiotic sporophytic mutants with affected pollen development have been isolated as male‐sterile in plants, and several male‐sterile mutations that disrupt tapetum function and affect exine and intine formation have been described (Ariizumi et al., 2004; Liang et al., 2020; Taylor et al., 1998; Zhang et al., 2021). In contrast, Callot and Gallois (2014) provided sound genetic evidence pointing towards eIF4E1 and eIFiso4E as essential for Arabidopsis male gametophyte development (Callot and Gallois, 2014). In our study, two lines of evidence strongly suggested that the male sterility phenotype that we have described was sporophyte‐associated. On the contrary, selfing heterozygous F1 individuals produced homozygous mutant progeny, albeit to a frequency lower than expected; therefore, gametes able to transmit the mutant allele are being produced by F1 individuals. On the contrary, the microscopy analysis of anther development showed the unusual secretion of protein material at early stages, as well as the differences in the timing of tapetum degradation at late stages between the mutant and the WT. Consistent with this finding, our transcriptomic analysis comparing WT versus eif4e mutant male flowers identified the down‐regulation of genes involved in cell growth, differentiation and cell cycle progression, coinciding with the FS episode. Furthermore, the down‐regulation of genes involved in ethylene synthesis, stamen filament development and sporopollenin biosynthetic processes was consistent with what was observed by Dai et al. (2019) in the comparison with DEGs between male fertile versus male‐sterile flowers in melon (Dai et al., 2019). In the GI episode, modifications in the expression patterns of genes related to ‘MCM complex’ and ‘DNA helicase’ may be associated with meiotic imbalances, while the down‐regulation of genes related to pectin metabolism could be due to impairments in tapetum development, as both phenomena have been associated with male sterility in Arabidopsis (Ma, 2005). Moreover, the misregulation of genes involved in programmed cell death, callose deposition and sporopollenin biosynthesis observed in this episode have often been linked to interferences in the timing of tapetal PCD (Khan et al., 2021). In the GM episode, the down‐regulation of genes related to membranes and cell wall may be associated with the gradual de‐structuring of the anther walls observed by microscopy between stages 10 and 11 of development. It has been described that the Arabidopsis nef1 mutant synthesizes sporopollenin, but it is not properly deposited on the pollen membrane (Ariizumi et al., 2004), just as we observed for the sporocytes of our melon mutant. The up‐regulation of numerous genes involved in sporopollenin production between the GI and GM episodes could be responsible for the invasion of droplets of apparently sporopollenin‐associated material observed by microscopy in the mutant. Regarding the AN episode, defects in the regulation of genes related to endopeptidase activity have often been associated with failures in protein ubiquitination and degradation by the proteasome. According to this, the presence of cell detritus in the anther lumen, the failure of anthesis and the irregular degradation of aborted grains observed in stages 11 and 12 could be explained by the deregulation of these genes observed in this episode. The role of the proteasome subunit alpha and beta type PSMB1 genes in determining this, has been extensively described in animal models and even in humans (Kapetanou et al., 2022; Sagi and Kim, 2012); however, their involvement in similar phenomena in plants has not been described yet.
Animal and plant studies have shown that germ cell and embryonic fates are greatly determined by the eIF4 factor complexes unique to those cells. Moreover, the coupled use of genetics and biochemistry identified unique roles for eIF4E and eIF4G isoforms in reproduction, so that eIF4Es in plants, flies, and frogs may play unique roles in sexual development (Keiper, 2019). Our transcriptomic results detected eIF‐encoding genes among those differentially expressed in all episodes; in particular, eIF4G was down‐regulated in the FS, GI, and AN episodes. Interestingly, eIFiso4E was not deregulated (Figure S7), pointing to a lack of functional redundancy between eIF4E isoforms. The interaction between eIF4E and eIF4G constitutes the core of the eIF4F complex (Merrick, 2015; Miras et al., 2017). Multiple studies have suggested that eIF4E and eIF4G isoforms are highly selective in translating mRNAs. Friday et al. (2015) proposed a model of selective and positive mRNA recruitment in germ cells based on the eIF4E and eIF4G isoforms in C. elegans (Friday et al., 2015). Furthermore, in plant studies, polysome and reporter analyses showed that each isoform preferentially recruits a unique subset of mRNAs for cap‐dependent translation (Dinkova et al., 2005). Based on these observations and on our results, we can hypothesize a specific role of the eIF4E‐eIF4G complex in mRNA translation in male germ cells. In this context, the absence of eIF4E in the mutant plants, and the consequent lack of interaction with eIF4G, would lead to a deficiency in mRNA translation in male germ cells, ultimately leading to the structural defects that characterize the male sterility phenotype. Of note, here is that melon eIF4E has been shown to be required for non‐canonical cap‐independent 3′‐CITE‐mediated translation initiation (Truniger et al., 2008, 2017), and also that down‐regulating eIFiso4E gene expression by RNAi in melon has proven impossible in our hands (unpublished results), suggesting that eIFiso4E may be playing a housekeeping role in cap‐dependent translation initiation in melon, while eIF4E might be fulfilling more specialized roles, including non‐canonical translation initiation during stress and, perhaps, male flower development.
Methods
eIF4E gRNA design and cloning
The eIF4E gene (MELO3C002698.2) of melon (Cucumis melo) was the editing target. The design of the RNA guide (gRNA1) was carried out using the online tool CRISPR‐P (http://crispr.hzau.edu.cn/CRISPR2/) (Lei et al., 2014). The Mfold tool (http://unafold.rna.albany.edu/?q=mfold) (Zuker, 2003) was used to check if the secondary structure prediction of the gRNA obeyed the parameters described by Liang et al. (2016). The pair of partially complementary primers AB338 (5′‐ATTGCAAAACCCTAGAGGACGTGG‐3′) and AB339 (5′‐ AAACCCACGTCCTCTAGGGTTTTG‐3′) was designed to form a duplex by adding the BbsI sequence at its ends. The duplex DNA was cloned into the plasmid pBS_KS_gRNA_BbsI (Abiopep S.L., Spain), previously digested with BbsI, and fused between the Arabidopsis ubiquitin promoter U6‐26 and the scaffold sequence. The resulting plasmid was digested with the restriction enzymes SpeI and KpnI. The products of the digestion were fractionated by agarose gel electrophoresis, the band of interest was cut out and purified on column (GeneClean Turbo Kit, MP Biomedicals). The purified DNA fragment was cloned into the binary vector pK7_CAS9‐TPC_MCS (Abiopep S.L.), which includes the coding sequence of the Cas9 protein and the neomycin phosphotransferase II (nptII) gene that confers resistance to kanamycin in plants, and sequenced to check the correct insertion of the fragment. Finally, the plasmid was used to transform electrocompetent A. tumefaciens cells strain GV3101 which was used for transformation of melon explants.
Plant materials and tissue collection
Four melon accessions (M2, M5, M9, C‐46) corresponding to the types ‘Cantaloupe’, ‘Piel de Sapo’ and ‘Amarillo’ were used for transformation in this study. The eif4e RNAi line described by Rodríguez‐Hernández et al. (2012) was used as control in the virus susceptibility assays, together with its parental line BGV‐130 (Rodríguez‐Hernández et al., 2012). Seeds from these accessions or from their transformed and/or edited progenies were pre‐germinated in Petri dishes at 25 °C in darkness and then sown in 1.5 L pots in a substrate composed of peat, coconut fibre and perlite in a 6:3:1 ratio. Pots were kept in a greenhouse or growth chamber with a temperature adjusted to 25 °C day and 18 °C night in a long cycle photoperiod (16 h light/8 h dark) for up to 3 months. For crossing and obtaining seeds, and/or when prolonged times of cultivation were necessary, plants were transplanted to coconut fibre sacs and grown in a greenhouse (Finca ‘La Matanza’, CEBAS‐CSIC, Murcia, Spain) under standard cultivation conditions until fruit maturation.
For the microscopy analysis of anther development, floral buds from wild‐type and eif4e mutant melon plants were collected by cutting the shoot tip containing young floral buds at various stages of development. For each episode, approximately 50 buds were pooled for each of three biological replicates, at a time interval of 15–20 days. One half of the samples was fixed in paraffin for light microscopy observations, whereas the other half was processed for scanning electron microscopy analysis. In both cases, flower buds were immersed in 70% alcohol and stored at 4 °C prior to fixation. For the collection of samples for RNA‐seq analysis, 50 flower buds at the different stages of development were sampled according to the procedure previously described for the microscopy analysis. Buds were hand‐dissected, immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
Agrobacterium tumefaciens‐mediated stable transformation of melon
The methodology used for the regeneration and stable transformation of melon was that described in García‐Almodóvar et al. (2017). Briefly, the A. tumefaciens GV3101 strain was transformed with p35SdsRed or pA60‐gRNA constructs. A single colony was inoculated into 50 mL of Luria‐Bertani (LB) medium containing the antibiotics required for the selection. The explants were immersed in the A. tumefaciens suspension for 20 min. After removing the bacterial solution, the explants were directly transferred to a regeneration medium (RM) containing 100 μM acetosyringone for a 72 h co‐cultivation period in the dark. The explants were then washed with liquid RM supplemented with 200 mg/L of vancomycin and 300 mg/L of cefotaxime and subsequently placed onto selective RM with 150 mg/L kanamycin and the same antibiotics. After 4 weeks, regenerated buds were excised and sub‐cultured to fresh selective RM for 2 weeks. The buds were then transferred to selective elongation RM (ERM) (150 mg/L kanamycin, 200 mg/L vancomycin, and 300 mg/L cefotaxime) for rooting. Additionally, rooted plants were further checked by PCR for the marker gene nptII with the specific primers CE1601 (5′‐ GAGGCTATTCGGCTATGACTG‐3′) and CE1602 (5′‐ ATCGGGAGCGGCGATACCGTA‐3′). Once the shoots produced roots, they were sub‐cultured in ERM and then acclimatized in a growth chamber.
PCR‐genotyping of transgenic edited lines
As a verification of the stable genetic transformation, a fragment of the coding sequence of Cas9 was PCR‐amplified. For this, a direct PCR on tissue from melon T0 plants was performed using the Phire Tissue direct PCR kit (ThermoFisher Scientific, USA) according to the manufacturer's specifications and the primer pair CE2176 (5′‐ CGCTAACAGAAACTTCATGCA‐3′) and CE2177 (5′‐ AGCGTTAAGGTAAGCATCGTGAG‐3′). Then, the targeted region of Cm‐eIF4E was amplified by direct tissue PCR using the primer pair AB385 (5′‐ GGGCGGTGCCATTCTTCTTC) and AB386 (5′‐ GAGTCGAGGTCGTCGTCGCC‐3′). DNA from WT plants of the same genotype was used as the negative control. For validation, the PCR products were purified and sequenced using an external service (STABVIDA, Portugal). The analysis of the sequences was performed using SnapGene software (GSL Biotech) and/or the bioinformatics tool CRISP‐ID V1.1 (Dehairs et al., 2016).
Melon pollinations
The crossing of the homozygous male‐sterile T0 lines, edited in eIF4E, by the WT lines of the same genotype (eif4e/eif4e x eIF4E/eIF4E) to obtain the F1 was performed manually. Male flowers were selected from WT plants, from which the pollen was collected. Subsequently, the hermaphrodite flowers of the T0 plants to be pollinated were selected. The flowers were emasculated and the stigmas were then pollinated, bagged with paper bags to avoid unwanted crosses and labelled. To obtain the F2 generation, the fertile heterozygous individuals were manually self‐fertilized. Once they had set about three fruits per plant, they were un‐bagged and the rest of the flowers were removed from the branch.
Virus inoculations
Moroccan watermelon mosaic virus mechanical inoculation was performed by rubbing recently expanded cotyledons and young fully expanded leaves with fresh extracts (in 0.03 M potassium phosphate buffer, pH 8.0) from MWMV‐infected squash plants (MWMV‐SQ10_1.1), following standard procedures (Miras et al., 2019). Plants were re‐inoculated 3 days later. After the inoculation, plants were monitored daily for the appearance of symptoms. A pool of systemically infected leaves was harvested from each individual plant at 21 dpi.
RNA isolation
For the analysis of viral accumulation, leaf samples were ground and cold‐homogenized using liquid N2 and adding TNA buffer (2% SDS, 100 mM Tris HCl pH8, 10 mM EDTA pH 8). Total RNA was extracted using Tri‐reagent (MRC, USA), purified by phenol‐chloroform, and treated with DNaseI (Sigma‐Aldrich, USA) following the manufacturer's recommendations. The integrity of the RNA was checked by 1% agarose gel separation, and RNA preparations were quantified using a NanoDrop One (ThermoFisher Scientific), normalized to 25 ng/μL, and used as a template to quantify the absolute accumulation of MWMV.
For RNA‐Sequencing analysis, flower tissue's total RNA was extracted using Tri‐reagent (MRC), purified by phenol‐chloroform extraction, and treated with DNaseI (Sigma‐Aldrich, USA). RNA preparations were quantified using a NanoDrop One (ThermoFisher Scientific), and normalized to equal amounts for each replicate. The RNA quality of the samples was analysed by agarose gel electrophoresis and with an Agilent 2100 Bioanalyser (Agilent Technologies, USA). The RNA integrity number (RIN) of every sample was above 7.
Viral load quantification by RT‐qPCR
The MWMV accumulation was estimated by measuring the viral RNA accumulation with absolute real‐time quantitative PCR (RT‐qPCR) with a StepOnePlus thermal cycler (Applied Biosystems, USA) using a One‐step NZYSpeedy RT‐qPCR Green kit, ROX plus (NZYTech, Portugal), and the primers CE2881 (5′‐ GGGACTTTCAGGGTTCCAA‐3′) and CE2882 (5’‐TGCCCTAGTGTTGGACAGG‐3′). For absolute quantification, a standard curve constructed from serial dilutions of a MWMV CP viral RNA in vitro transcript was used.
HADDOCK‐derived complex structure of VPg‐eIF4E
The HADDOCK2.4 webserver (Van Zundert et al., 2016) was used to generate restraint‐driven docking for the interaction between eIF4E and VPg using the standard protocols, with the VPg structure reported here and CmeIF4E (PDB ID code 5ME7). Default HADDOCK settings were used for the docking, generating a final number of 100 structures. The final models were clustered based on the fraction of common contacts using a 0.60 cut‐off.
Paraffin sections and light microscopy
Shoot tips and floral buds at various stages were fixed in FAA (formaldehyde: acetic acid: ethanol: H2O, 10:5:50:35, vol:vol) and then dehydrated through an ethanol series. Dehydrated plant material was embedded in paraffin and longitudinal and transverse semithin sections (1 μm) were cut using an ultra‐microtome (Leica RM2155). Samples were incubated for 2 h at 40 °C in an oven for dewaxing, and three steps were carried out in “Neo‐Clear®” (xylene substitute). Thin sections were then hydrated in three consecutive steps in ethanol at decreasing concentrations (100%, 96% and 70%), and finally left in distilled water for 5 min. At this point, staining with Mayer's haematoxylin was carried out. Finally, a second staining was carried out with alcoholic eosin, followed by three steps of dehydration in ethanol at increasing concentrations (70%, 96% and 100%). The procedure ended with a two‐step dealcoholisation in Neo‐Clear. The sections were observed, measured and photographed under a light microscope (Leica DMRB).
Scanning electron microscopy
Floral buds at various stages of development were fixed in FAA and subsequently subjected to several washes. Then, buds were fixed in 3% (v/v) glutaraldehyde diluted in 0.1 M cacodylate buffer, and washed in cacodylate buffer plus sucrose overnight. Subsequently, a postfixation procedure in 1% tetroxide was followed by a second wash in cacodylate buffer plus sucrose overnight. A serial dehydration was performed in acetone solution at increasing concentrations (30%, 50%, 70%, 90% and 100%,). Materials were critical‐point‐dried using 100% acetone and liquid CO2, mounted on aluminium stubs with double‐sided tape, 5 nm layer, platinum‐coated with a Leica EM ACE600 sputter coater and then examined through a field emission scanning electron microscope (ApreoS LoVac, ThermoFisher), set at 5 kV and 3.2 nA with a work distance of 15 mm and secondary electrons.
RNA‐Seq and data analysis
RNA‐Seq libraries were constructed according to the TrueSeq Stranded mRNA LT kit protocol (Illumina, USA) with ribosomal depletion using the Ribo‐Zero plant kit (Illumina, USA), and sequenced using the Illumina NovaSeq6000 platform (150 PE) (Macrogen Inc., South Korea). Approximately 66 to 85 million paired‐end reads were generated for each sample. Quality of raw reads was analysed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Poor‐quality reads (QC < 30 and length < 70 bp) and adapter sequences were filtered out, and the low‐quality nucleotides at the 5′ end of the reads were trimmed using Trimmomatic (Bolger et al., 2014). A second quality control of the filtered reads was performed again with FastQC, and reconstruction of paired‐reads was performed with BBMap (www.sourceforege.net/projects/bbmap). Reads were then mapped against the melon genome (DHL92/v3.6.1) using the MEM algorithm of BWA software (Li and Durbin, 2009). Mapping quality was analysed with Qualimap bamqc (García‐Alcalde et al., 2012).
The FeatureCounts function of the R package Rsubread was used to count the number of reads mapping to each mRNA (v4.0 of the gene models). Read counts were normalized to FPKM (fragments per kilobase per million mapped reads) using the DESeq2 R package. The PCA was drawn using the R package Factoextra. Venn diagrams were made using the VennDiagram R package. Differential expression analyses between WT and eif4e mutant episodes were performed using the DESeq2 R package. The Gene Ontology (GO) analysis was performed using the Goseq R Bioconductor package, using the list of DEG created previously. GO terms with a corrected false discovery rate below 0.05 were considered to be significantly enriched.
Statistical analyses
Transformation experiments were repeated at least three times (100 explants per experiment) for the genotypes tested. For the characterization of the male sterility phenotype in the F2 generation, a minimum of 10 flowers per plant along a time period of 60 days were examined; only homozygous mutant plants, all of them, showed the male sterility phenotype, whereas all other plants, either homozygous WT or heterozygous produced staminate and perfect flowers with viable pollen. Specific maximum likelihood contrasts were run to detect differences between genotypes. Differences were considered significant when P < 0.05. Segregation ratios of F2 progenies were contrasted with a Chi square to expected value.
Conflict of interest
Authors declare no conflict of interest.
Author contributions
MAA, LD, VT and GSP conceived the study. VT and GSP prepared the DNA constructs for transformation. CGA, BG and GSP transformed and edited the melon plants. GSP, LD and MAA performed the virus susceptibility analysis. GSP and MASP performed the microscopy study. LD and GSP performed the RNA‐Seq study, including analysing the data. GSP and MAA wrote the paper with input from all co‐authors.
Funding
This work was supported by grants AGL2015‐65838 and PLEC2021‐007715 from Ministerio de Ciencia e Innovación (Spain) and 2I16SA000057 of the RIS3Mur program from Consejería de empleo, universidades y empresa (Región de Murcia, Spain) and 20800/PI/18 from Fundación Seneca, Murcia. GSP was supported by grant BES‐2016‐077826 (Ministerio de Ciencia e Innovación; Spain).
Supporting information
Figure S1 Regeneration and transformation of four melon genotypes.
Figure S2 Genetic analysis of a F2 segregating generation.
Figure S3 Genotyping of eIF4E mutants in F2 progeny plants of the gRNA1 line.
Figure S4 Multiple amino acid sequence alignment of VPg's from different potyviruses.
Figure S5 Light microscopy images from longitudinal sections of wild‐type.
Figure S6 Cluster dendrogram of gene expression profiles between biological replicates and between different developmental episodes.
Figure S7 Expression of eIFiso4E and eIF4E across floral developmental episodes estimated as fragments per kilo base of transcript per million mapped fragments (FPKM).
Table S1 Putative eIF4E CRISPR/Cas9 gRNA1 off‐target sites.
Table S2 Selected differentially expressed genes during male flower development in male‐sterile eif4e mutants.
Acknowledgements
We thank José Antonio Esteban (Abiopep S.L., Spain) and Mari Carmen Montesinos (CEBAS‐CSIC, Spain) for their help with growing plants; Pau Bretó (Abiopep S.L., Spain) for teaching how to cross melon plants, and Mario Fon (mariogfon@gmail.com) for editing the manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Ariizumi, T. , Hatakeyama, K. , Hinata, K. , Inatsugi, R. , Nishida, I. , Sato, S. , Kato, T. et al. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39, 170–181. [DOI] [PubMed] [Google Scholar]
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. [DOI] [PubMed] [Google Scholar]
- Bai, S.L. , Peng, Y.B. , Cui, J.X. , Gu, H.T. , Xu, L.Y. , Li, Y.Q. , Xu, Z.H. et al. (2004) Developmental analyses reveal early arrests of the spore‐bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. [DOI] [PubMed] [Google Scholar]
- Baker, C.C. and Fuller, M.T. (2007) Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel elF4G homolog. Development 134, 2863–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet, A. , Lederer, B. , Giovinazzo, N. , Arnoux, X. , German‐Retana, S. , Reinbold, C. , Brault, V. et al. (2018) Trans‐species synthetic gene design allows resistance pyramiding and broad‐spectrum engineering of virus resistance in plants. Plant Biotechnol. J. 16, 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet, A. , Zafirov, D. , Giovinazzo, N. , Guyon‐Debast, A. , Nogué, F. , Robaglia, C. and Gallois, J.L. (2019) Mimicking natural polymorphism in eIF4E by CRISPR‐Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 17, 1736–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S. , Zhang, D. and Shi, J. (2021) CRISPR / Cas systems: opportunities and challenges for crop breeding. Plant Cell Rep. 40, 979–998. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, K.S. (2004) Plant translation initiation factors: It is not easy to be green. Biochem. Soc. Trans. 32, 589–591. [DOI] [PubMed] [Google Scholar]
- Callot, C. and Gallois, J.L. (2014) Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant signaling & behavior 9(2), e27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano, M.M. and Merchante, C. (2021) Plant Biology Peculiarities of the regulation of translation initiation in plants. Curr. Opin. Plant Biol. 63, 102073. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran, J. , Brumin, M. , Wolf, D. , Leibman, D. , Klap, C. , Pearlsman, M. , Sherman, A. et al. (2016) Development of broad virus resistance in non‐transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Z. , Chen, Z. , Wang, N. , Xie, G. , Lu, J. , Yan, W. , Zhou, J. et al. (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. 113, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, C. , Nicolaï, M. , Gallois, J.L. , Robaglia, C. , Moury, B. , Palloix, A. and Caranta, C. (2008) Natural variation and functional analyses provide evidence for co‐evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. [DOI] [PubMed] [Google Scholar]
- Coutinho de Oliveira, L. , Volpon, L. , Rahardjo, A.K. , Osborne, M.J. , Culjkovic‐Kraljacic, B. , Trahan, C. , Oeffinger, M. et al. (2019) Structural studies of the eIF4E–VPg complex reveal a direct competition for capped RNA: Implications for translation. Proceedings of the National Academy of Sciences 116(48), 24056–24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, D. , Xiong, A. , Yuan, L. , Sheng, Y. , Ji, P. , Jin, Y. , Li, D. et al. (2019) Transcriptome analysis of differentially expressed genes during anther development stages on male sterility and fertility in Cucumis melo L. line. Gene 707, 65–77. [DOI] [PubMed] [Google Scholar]
- Dehairs, J. , Talebi, A. , Cherifi, Y. and Swinnen, J.V. (2016) CRISP‐ID: Decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep. 6(June), 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Pendon, J.A. , Truniger, V. , Nieto, C. , Garcia‐Mas, J. , Bendahmane, A. and Aranda, M.A. (2004) Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 5, 223–233. [DOI] [PubMed] [Google Scholar]
- Dinkova, T.D. , Zepeda, H. , Martínez‐Salas, E. , Martínez, L.M. , Nieto‐Sotelo, J. and Sánchez De Jiménez, E. (2005) Cap‐independent translation of maize Hsp101. Plant J. 41, 722–731. [DOI] [PubMed] [Google Scholar]
- Fraser, R.S.S. (1990) The genetics of resistance to plant viruses. Annu. Rev. Phytopathol. 28, 179–200. [Google Scholar]
- Friday, A.J. , Henderson, M.A. , Morrison, J.K. , Hoffman, J.L. and Keiper, B.D. (2015) Spatial and temporal translational control of germ cell mRNAs mediated by the eIF4E isoform IFE‐1. J. Cell Sci. 128, 4487–4498. [DOI] [PubMed] [Google Scholar]
- Friday, A.J. and Keiper, B.D. (2015) Positive mRNA translational control in germ cells by initiation factor selectivity. Biomed. Res. Int. 2015, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois, J.L. , Charron, C. , Sánchez, F. , Pagny, G. , Houvenaghel, M.C. , Moretti, A. , German‐Retana, S. et al. (2010) Single amino acid changes in the turnip mosaic virus viral genome‐linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J. Gen. Virol. 91, 288–293. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L. , Moury, B. and German‐Retana, S. (2018) Role of the genetic background in resistance to plant viruses. Int. J. Mol. Sci. 19, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Alcalde, F. , Okonechnikov, K. , Carbonell, J. , Cruz, L.M. , Götz, S. , Tarazona, S. and Conesa, A. (2012) Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679. [DOI] [PubMed] [Google Scholar]
- García‐Almodóvar, R.C. , Gosalvez, B. , Aranda, M.A. and Burgos, L. (2017) Production of transgenic diploid Cucumis melo plants. Plant Cell Tissue Organ Cult. 130, 323–333. [Google Scholar]
- Garcia‐Mas, J. , Benjak, A. , Sanseverino, W. , Bourgeois, M. , Mir, G. , González, V.M. , Hénaff, E. et al. (2012) The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. U. S. A. 109, 11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. (2018) Susceptibility genes to plant viruses. Viruses 10, 484. 10.3390/v10090484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. (2019) Host factors against plant viruses. Mol. Plant Pathol. 20, 1588–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S. and Lasko, P. (2015) Loss‐of‐function analysis reveals distinct requirements of the translation initiation factors eIF4E, eIF4E‐3, eIF4G and eIF4G2 in Drosophila spermatogenesis. PLoS ONE 10, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, M.A. , Lin, Z.D. , Moll, T. , Chauhan, R.D. , Hayden, L. , Renninger, K. , Beyene, G. et al. (2019) Simultaneous CRISPR/Cas9‐mediated editing of cassava eIF4E isoforms nCBP‐1 and nCBP‐2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, P. , Rodríguez‐Hernández, A.M. , Moury, B. and Aranda, M. (2009) Genetic resistance for the sustainable control of plant virus diseases: Breeding, mechanisms and durability. Eur. J. Plant Pathol. 125, 1–22. [Google Scholar]
- Hao, Y.J. , Wang, D.H. , Peng, Y.B. , Bai, S.L. , Xu, L.Y. , Li, Y.Q. , Xu, Z.H. et al. (2003) DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.). Planta 217, 888–895. [DOI] [PubMed] [Google Scholar]
- Julio, E. , Cotucheau, J. , Decorps, C. , Volpatti, R. , Sentenac, C. , Candresse, T. and Dorlhac de Borne, F. (2015) A Eukaryotic Translation Initiation Factor 4E (eIF4E) is Responsible for the “va” Tobacco Recessive Resistance to Potyviruses. Plant Mol. Biol. Rep. 33, 609–623. [Google Scholar]
- Kapetanou, M. , Athanasopoulou, S. and Gonos, E.S. (2022) Transcriptional regulatory networks of the proteasome in mammalian systems. IUBMB Life 74, 41–52. [DOI] [PubMed] [Google Scholar]
- Keiper, B.D. (2019) Cap‐Independent mRNA translation in germ cells. Int. J. Mol. Sci. 20, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R.M. , Yu, P. , Sun, L. , Abbas, A. , Shah, L. , Xiang, X. , Wang, D. et al. (2021) DCET1 controls male sterility through callose regulation, exine formation, and tapetal programmed cell death in rice. Front Genet 12, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.K. , Kim, H. , Cho, J.I. , Nguyen, C.D. , Moon, S. , Park, J.E. , Park, H.R. et al. (2020) Deficiency of rice hexokinase HXK5 impairs synthesis and utilization of starch in pollen grains and causes male sterility. J. Exp. Bot. 71, 116–125. [DOI] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: A web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.J. , Kim, Y.J. , Yang, L. , Liu, Z. , Zhang, J. , Shi, H. , Huang, G. et al. (2020) Grass‐specific EPAD1 is essential for pollen exine patterning in rice. Plant Cell 32, 3961–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G. , Zhang, H. , Lou, D. and Yu, D. (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9‐based plant genome editing. Sci. Rep. 6, 1–8. 10.1038/srep21451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Li, S.W. , Gong, L.M. , Li, S. and Zhang, Y. (2020) COPII components sar1b and sar1c play distinct yet interchangeable roles in pollen development. Plant Physiol. 183, 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Ding, Y. , Zhou, Y. , Jin, W. , Xie, K. and Chen, L.L. (2017) CRISPR‐P 2.0: an improved CRISPR‐Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. [DOI] [PubMed] [Google Scholar]
- Ma, H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. [DOI] [PubMed] [Google Scholar]
- Martínez‐Silva, A.V. , Aguirre‐Martínez, C. , Flores‐tinoco, C.E. , Alejandri‐Ramírez, N.D. and Dinkova, T.D. (2012) Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in Arabidopsis thaliana seedlings. PLoS One 7, e31606. 10.1371/journal.pone.0031606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry, L.K. , Allen, M.L. , Nitka, K.R. , Campbell, L. , Murphy, P.A. and Browning, K.S. (2011) Plant cap‐binding complexes eukaryotic initiation factors eIF4F and eIFISO4F. J. Biol. Chem. 286, 42566–42574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazier, M. , Flamain, F. , Nicolaï, M. , Sarnette, V. and Caranta, C. (2011) Knock‐down of both eIF4E1 and eIF4E2 genes confers broad‐spectrum resistance against potyviruses in tomato. PLoS One 6(12), e29595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. (2004) Control of male gametophyte development. Plant Cell 16(SUPPL), 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick, W.C. (2015) eIF4F: A retrospective *. J. Biol. Chem. 290, 24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras, M. , Juárez, M. and Aranda, M.A. (2019) Resistance to the emerging Moroccan watermelon mosaic virus in squash. Phytopathology 109, 895–903. [DOI] [PubMed] [Google Scholar]
- Miras, M. , Truniger, V. , Silva, C. , Verdaguer, N. , Aranda, M.A. and Querol‐Audí, J. (2017) Structure of eiF4E in complex with an eiF4G peptide supports a universal bipartite binding mode for protein translation. Plant Physiol. 174, 1476–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B. , Charron, C. , Janzac, B. , Simon, V. , Gallois, J.L. , Palloix, A. and Caranta, C. (2014) Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome‐linked protein (VPg): A game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 27, 472–480. [DOI] [PubMed] [Google Scholar]
- Nuñez‐Palenius, H.G. , Gomez‐Lim, M. , Ochoa‐Alejo, N. , Grumet, R. , Lester, G. and Cantliffe, D.J. (2008) Melon fruits: Genetic diversity, physiology, and biotechnology features. Crit. Rev. Biotechnol. 28, 13–55. [DOI] [PubMed] [Google Scholar]
- Parish, R.W. and Li, S.F. (2010) Death of a tapetum: A programme of developmental altruism. Plant Sci. 178, 73–89. [Google Scholar]
- Patrick, R.M. and Browning, K.S. (2012) The eIF4F and eIFiso4F complexes of plants: An evolutionary perspective. Comp Funct Genomics 2012, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, R.M. , Mayberry, L.K. , Choy, G. , Woodard, L.E. , Liu, J.S. , White, A. , Mullen, R.A. et al. (2014) Two arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E. Plant Physiol. 164, 1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechar, G. S. (2022). Impact of melon EIF4E editing on virus resistance and melon fertility. Impacto de la edición de EIF4E sobre la resistencia a virus y la fertilidad en plantas de melón. Retrieved from http://nadir.uc3m.es/alejandro/phd/thesisFinal.pdf%5Cnhttp://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:Universidad+de+murcia#0
- Pyott, D.E. , Sheehan, E. and Molnar, A. (2016) Engineering of CRISPR/Cas9‐mediated potyvirus resistance in transgene‐free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers, F. and Nicaise, V. (2014) Plant resistance to infection by viruses. eLS, 1–10. 10.1002/9780470015902.a0000757.pub3 [DOI] [Google Scholar]
- Robaglia, C. and Caranta, C. (2006) Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Hernández, A.M. , Gosalvez, B. , Sempere, R.N. , Burgos, L. , Aranda, M.A. and Truniger, V. (2012) Melon RNA interference (RNAi) lines silenced for Cm‐eIF4E show broad virus resistance. Mol. Plant Pathol. 13, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, B. and Arnim, A.G. (2013) Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 11, e0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, D. and Kim, S.K. (2012) An engineering approach to extending lifespan in C. elegans. PLoS Genet. 8, e1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S. and Mäkinen, K. (2020) Insights into the functions of eIF4E‐binding motif of VPg in potato virus A infection. Viruses 12, 197. 10.3390/v12020197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon, H. (2015) Plant translation factors and virus resistance. Viruses 7, 3392–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt‐Keichinger, C. (2019) Manipulating cellular factors to combat viruses: A case study from the plant eukaryotic translation initiation factors eIF4. Front. Microbiol. 10(FEB), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, L. , Fingerhut, J. M. , Falk, B. L. , Han, H. , Maldonado, G. , Qiao, Y. , Lee, V. , Hall, E. , Chen, L. , Polevoy, G. and Hernández, G. (2021) Eukaryotic translation initiation factor eIF4E‐5 is required for spermiogenesis in Drosophila melanogaster. bioRxiv. 10.1101/2021.12.19.473358 [DOI] [PubMed] [Google Scholar]
- Shopan, J. , Mou, H. , Zhang, L. , Zhang, C. , Ma, W. , Walsh, J.A. , Hu, Z. et al. (2017) Eukaryotic translation initiation factor 2B‐beta (eIF2Bβ), a new class of plant virus resistance gene. Plant J. 90, 929–940. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Xiang, X. , Yang, Z. , Yu, P. , Wen, X. , Wang, H. , Abbas, A. et al. (2018) OsGPAT3 plays a critical role in anther wall programmed cell death and pollen development in rice. Int. J. Mol. Sci. 19, 4017. 10.3390/ijms19124017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, P.E. , Glover, J.A. , Lavithis, M. , Craig, S. , Singh, M.B. , Knox, R.B. , Dennis, E.S. et al. (1998) Genetic control of male fertility in Arabidopsis thaliana: structural analyses of postmeiotic developmental mutants. Planta 61, 492–505. [DOI] [PubMed] [Google Scholar]
- Truniger, V. and Aranda, M.A. (2009) Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–231. 10.1016/s0065-3527(09)07504-6 [DOI] [PubMed] [Google Scholar]
- Truniger, V. , Miras, M. and Aranda, M.A. (2017) Structural and functional diversity of plant virus 3′‐cap‐independent translation enhancers (3′‐CITEs). Front. Plant Sci. 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger, V. , Nieto, C. , González‐Ibeas, D. and Aranda, M. (2008) Mechanism of plant eIF4E‐mediated resistance against a Carmovirus (Tombusviridae): Cap‐independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J. 56, 716–727. [DOI] [PubMed] [Google Scholar]
- van Zundert, G. , Rodrigues, J.P.G.L.M. , Trellet, M. , Schmitz, C. , Kastritis, P.L. , Karaca, E. , Melquiond, A.S.J. et al. (2016) The HADDOCK2.2 web server: user‐friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Kohalmi, S.E. , Svircev, A. , Wang, A. , Sanfaçon, H. and Tian, L. (2013) Silencing of the Host Factor eIF(iso)4E Gene Confers Plum Pox Virus Resistance in Plum. PLoS ONE 8, e50627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, Z.A. and Zhang, D.B. (2009) From arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 60, 1479–1492. [DOI] [PubMed] [Google Scholar]
- Yeam, I. , Cavatorta, J.R. , Ripoll, D.R. , Kang, B. and Jahn, M.M. (2007) Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19, 2913–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Chang, J. , Li, J. , Lan, G. , Xuan, C. , Li, H. , Ma, J. et al. (2021) Disruption of the bHLH transcription factor abnormal tapetum 1 causes male sterility in watermelon. Hort. Res. 8, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, E. , You, C. , Wang, S. , Cui, J. , Niu, B. , Wang, Y. , Qi, J. et al. (2015) The DYT1‐interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 83, 976–990. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Regeneration and transformation of four melon genotypes.
Figure S2 Genetic analysis of a F2 segregating generation.
Figure S3 Genotyping of eIF4E mutants in F2 progeny plants of the gRNA1 line.
Figure S4 Multiple amino acid sequence alignment of VPg's from different potyviruses.
Figure S5 Light microscopy images from longitudinal sections of wild‐type.
Figure S6 Cluster dendrogram of gene expression profiles between biological replicates and between different developmental episodes.
Figure S7 Expression of eIFiso4E and eIF4E across floral developmental episodes estimated as fragments per kilo base of transcript per million mapped fragments (FPKM).
Table S1 Putative eIF4E CRISPR/Cas9 gRNA1 off‐target sites.
Table S2 Selected differentially expressed genes during male flower development in male‐sterile eif4e mutants.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


