Introduction
Eustoma grandiflorum (lisianthus) is a Gentianaceae‐family ornamental plant. Because of its enormous rose‐like blossoms, long stems and extended vase life, its sales have increased dramatically in recent years, earning it the title of ‘next rose’. Selective breeding has produced commercial lisianthus with a wide range of flower colours and shapes (Figure S1a; Li et al., 2022). In polyploid crops including wheat, cotton, peanuts and others, polyploidy is critical for the development of high‐quality traits (Cheng et al., 2018). Polyploidy may also contribute to the development of desirable traits in cultivated lisianthus. Here, we report a high‐quality chromosome‐scale genome assembly for E. grandiflorum (2n = 6x = 72) using a combination of PacBio HiFi reads and Hi‐C scaffolding technology and reveal that polyploidy domestication of lisianthus contributes to ornamental traits in cultivated lisianthus.
A total of 32.05 Gb (~23.56X) of PacBio HiFi data and 140.14 Gb (~103.04X) Hi‐C data were generated for de novo whole‐genome sequencing. The total length of the assembly was 1.71 Gb, comprising 1056 contigs with a corresponding N50 of 7.33 Mb (Table S1), and 36 pseudo‐chromosomes were assembled (Figures S1 and S2, Table S2). BUSCO revealed a completeness rate of 94.7% and a duplication rate of 31.6% (Table S5). A total of 54 305 high‐quality protein‐coding genes were predicted (Tables S3–S8). In addition, 77.85% of the genome was annotated to be repeat sequences (Tables S9–S12).
Genome collinearity identified a large number of collinear blocks with a ratio of 6 : 1 between E. grandiflorum and Gelsemium sempervirens (Figure 1a), and a ratio of 2 : 1 between E. grandiflorum and E. grandiflorum (Figure S3), indicative of polyploidy events' existence. To this end, the 36 pseudo‐chromosomes could be divided into three subgenomes, then 12 homologous groups with three sets of monoploid chromosomes: A, B and C were obtained according to the transposable element profiles (Figure S4, Table S10–S12). The reasonable collinearity within subgenomes suggested that E. grandiflorum had experienced a whole‐genome duplication (WGD) event in the recent history of E. grandiflorum (Figure S5). The Ks distribution further confirmed that E. grandiflorum experienced a WGD event (Ks peak value = 0.93) and a whole‐genome triplication (WGT) event (Ks peak value = 0.21) after divergence from Calotropis gigantea, which is consistent with the ratio of 6 : 1 (E. grandiflorum: G. sempervirens) and the evolutionary relationships depicted by the phylogenetic tree (Figures 1a, S6–S7).
Figure 1.
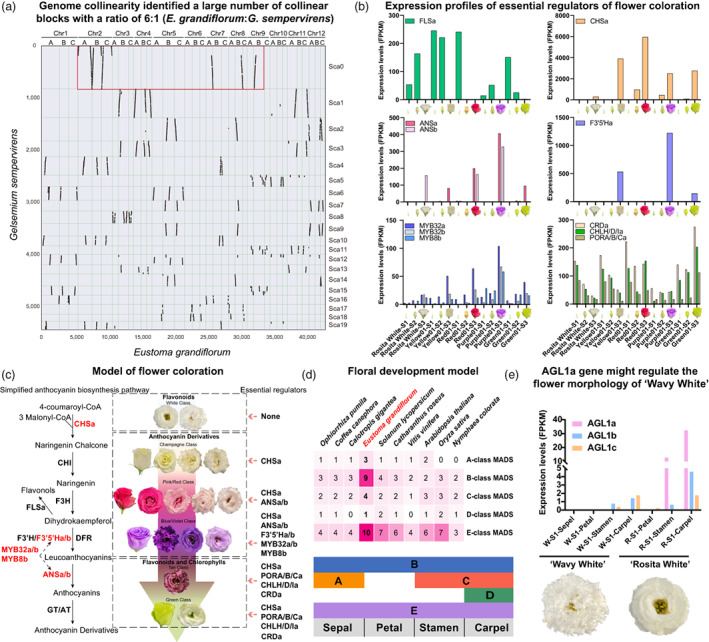
(a) Alignment of Eustoma grandiflorum chromosomes with Gelsemium sempervirens scaffolds. (b) Histograms showing expression profiles of essential regulators of flower coloration in lisianthus. S1, bud stage; S2, turning stage; S3, blooming stage. FLS, flavonol synthase; CHS, chalcone synthase; ANS, anthocyanidin synthase; F3′5′H, flavonoid 3′,5′‐hydroxylase; CRD, Mg‐protoporphyrin IX monomethyl ester cyclase; CHLH/D/I, Mg‐chelatase subunits H, D and I; PORA/B/C, protochlorophyllide reductase. (c) A proposed model of flower coloration in lisianthus. The potential simplified anthocyanin biosynthesis pathway in lisianthus is depicted. CHI, chalcone isomerase; F3H, flavanone 3‐hydroxylase; F3′H, flavonoid 3′‐hydroxylase; DFR, dihydroflavonol‐4‐reductase; GT, glucosyltransferase; AT, acyltransferase. Essential regulators of flower coloration in lisianthus are in red fonts. The main pigments refer to the reference (Gao and Li, 2020). (d) A proposed floral development model of lisianthus based on the expression patterns of floral identity genes. Heatmap shows the number of MADS genes across various species. (e) Histograms showing the expression levels of C‐class MADS genes in lisianthus. In all samples, AGL1d is zero in FPKM, hence it is not displayed. [Colour figure can be viewed at wileyonlinelibrary.com]
A total of 15 436 genes belonging to syntenic gene groups resulting from the WGT event were found based on collinearity across E. grandiflorum subgenomes, which were enriched in processes of external stimulus response, anatomical structure morphogenesis and biosynthetic process (Figure S8, Table S13). Substantial copy number variations of transcriptional factors (TFs) and structural genes participating in flavonoid/anthocyanin biosynthesis were discovered as a result of WGT including MYBs, bHLHs, CHIs, CHSs, DFRs, GTs and FLSs (Figures S9–S15). Thus, it was hypothesized that the polyploidy event would enable the breeding of colourful lisianthus varieties by providing enhanced genetic materials for anthocyanin production. We also found that numerous TFs regulating organ/flower development were generated by WGT, including floral identity genes in the MADS family, and members in the TCP family and HD‐Zip III family, which might influence the formation of E. grandiflorum's flower morphology (Figures 1d, S16–S17).
To examine the genetic pathways regulating flower pigmentation in lisianthus, we performed RNA‐seq of petals in the purple lisianthus (Purple01), the green lisianthus (Green01), the yellow lisianthus (Yellow01), the red lisianthus (Red01) and ‘Rosita White’ at different stages (bud stage, S1; turning stage, S2; blooming stage, S3) and conducted weighted correlation network analysis (WGCNA) (Figure S18). Genes involved in flavonoid biosynthesis were identified, and the putative anthocyanin biosynthesis pathway was depicted (Figures 1c, S11–S15, S19). We found that FLSa was highly expressed in S1 and S2, and was barely expressed in S3 (Figure 1b). In S3, we found that CHSa expression level was significantly higher in all coloured cultivars than in white cultivars, and ANSa/b displayed much higher expression levels in both pink/red and blue/violet cultivars (Figures 1b, S20). Moreover, compared to pink/red lisianthus, blue/violet lisianthus showed higher F3′5′Ha/b levels (Figures 1b, S20). Co‐expression network reconstruction of the module specific to S3 of lisianthus Purple01 (‘darkorange2’ module in Figure S18) identified several MYBs co‐expressed with F3′5′Ha and ANSa/b; among them, MYB32a, MYB32b and MYB8b showed high expression levels in S3 of lisianthus Purple01 similar to those of F3′5′Ha and ANSa/b during flower development (Figures 1b, S21). As a result, we suggested that in lisianthus CHSa, ANSa/b, F3′5′Ha/b, MYB32a/b and MYB8b codetermine blue/violet flower coloration, CHSa and ANSa/b codetermine pink/red flower coloration and CHSa determines yellow flower coloration (Figure 1c).
Instead of accumulating anthocyanins in the blooming stage, green lisianthus varieties synthesize and accumulate large amounts of chlorophylls, leading to green phenotypes. CHLMa, CRDa and PORA/B/Ca were assumed to be important regulators of chlorophyll biosynthesis in petals since they had the highest expression levels in S3 in lisianthus Green01 (Figure 1b). Multiple genes involved in photosynthesis, including those encoding Chlorophyll a/b‐binding proteins (CABs), were found to be co‐expressed with CHLMa, CRDa and PORA/B/Ca, suggesting that these CABs might also play important roles in green petal formation by inhibiting chlorophyll degradation via forming antenna complexes with free chlorophylls (Figures S22–S24). Based on the same expression patterns of genes involved in chlorophyll synthesis and photosynthesis (e.g. CABs) in the tan lisianthus with those in lisianthus Green01, it is possible that chlorophylls were also pigments in the tan lisianthus (Figures 1c, S24–S25).
In lisianthus, a total of 120 MADS‐box genes were identified, including 27 floral organ identity genes (three A‐class genes, nine B‐class genes, four C‐class genes, one D‐class genes and 10 E‐class genes; Figures 1d, S26–S28). The floral development model in E. grandiflorum was constructed according to floral identity genes' expression profiles (Figures 1d, S30). We found that multiple floral organ identity genes were generated by WGT, including B‐class AP3s, C‐class AGL1s and E‐class AGL9s, which might influence the evolution of E. grandiflorum's flower morphology (Li et al., 2022; Figure S29). ‘Rosita White’ and ‘Wavy White’ are two popular lisianthus cultivars, which have flat‐shaped petals and wave‐shaped petals respectively. We found that AGL1a, a C‐class MADS gene, was highly expressed in the stamen and carpel of ‘Rosita White’, while it was barely expressed in all the flower tissues of ‘Wavy White’, potentially due to differences in cis‐acting regulatory elements between their promoters (Figures 1e, S30–S31). It was reported that the absence of C‐class MADSs could result in the double‐flower phenotype. Thus, we speculated that AGL1a could be linked to the more apparent double‐flower phenotype with more petal numbers of ‘Wavy White’ (Figure S32).
In summary, we present the chromosome‐level genome of E. grandiflorum and identify the key candidate genes involved in flower coloration and morphology, which will speed up the molecular breeding of E. grandiflorum in the future.
Conflict of interest
The authors declare no conflicts of interest.
Authors’ contributions
Y. L. analysed the data and wrote the manuscript. F. L., J.W. and L. Z. conceived the study. Other authors carried out analysis and experiments.
Supporting information
Figure S1‐S32 Supplementary Figures.
Table S1‐S14 supportingsets.
Appendix S1 Plant materials.
Acknowledgements
We thank Yunnan Fundamental Research Projects (202201AU070168) for funding resources, and the High‐level Talent Introduction Program of Yunnan Province‐Industrial Talent Special Project (YNQR‐CYRC‐2020‐004).
Contributor Information
Xiaofan Zhou, Email: xiaofan_zhou@scau.edu.cn.
Liangsheng Zhang, Email: zls83@zju.edu.cn.
Jihua Wang, Email: wjh0505@gmail.com.
Data availability statement
All data are publicly available in the BIG Data Center (https://bigd.big.ac.cn/) under project number PRJCA010064. And the genome assembly sequences and gene annotations are available at https://figshare.com/s/5e570b76c63f159b8da1.
References
- Cheng, F. , Wu, J. , Cai, X. , Liang, J. , Freeling, M. and Wang, X. (2018) Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 4, 258–268. [DOI] [PubMed] [Google Scholar]
- Li, F. , Cheng, Y. , Yu, R. and Yang, C. (2022) Genome Size and Ploidy Level of Commercial Eustoma grandiflorum (Raf.) Shinners. J. Agric. Sci. Technol. 24, 739–748. [Google Scholar]
- Gao, S. and Li, T.L. (2020) Relationship between the composition of Anthocyanins and flower color variation in Lisianthus (Eustoma grandiflorum). J. Shenyang Agric. Univ. 4, 410–416 [in Chinese]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S32 Supplementary Figures.
Table S1‐S14 supportingsets.
Appendix S1 Plant materials.
Data Availability Statement
All data are publicly available in the BIG Data Center (https://bigd.big.ac.cn/) under project number PRJCA010064. And the genome assembly sequences and gene annotations are available at https://figshare.com/s/5e570b76c63f159b8da1.


