Abstract
Pseudomonas aeruginosa secretes a 29-kDa lipase which is dependent for folding on the presence of the lipase-specific foldase Lif. The lipase contains two cysteine residues which form an intramolecular disulfide bond. Variant lipases with either one or both cysteines replaced by serines showed severely reduced levels of extracellular lipase activity, indicating the importance of the disulfide bond for secretion of lipase through the outer membrane. Wild-type and variant lipase genes fused to the signal sequence of pectate lyase from Erwinia carotovora were expressed in Escherichia coli, denatured by treatment with urea, and subsequently refolded in vitro. Enzymatically active lipase was obtained irrespective of the presence or absence of the disulfide bond, suggesting that the disulfide bond is required neither for correct folding nor for the interaction with the lipase-specific foldase. However, cysteine-to-serine variants were more readily denatured by treatment at elevated temperatures and more susceptible to proteolytic degradation by cell lysates of P. aeruginosa. These results indicate a stabilizing function of the disulfide bond for the active conformation of lipase. This conclusion was supported by the finding that the disulfide bond function could partly be substituted by a salt bridge constructed by changing the two cysteine residues to arginine and aspartate, respectively.
The gram-negative bacterium Pseudomonas aeruginosa secretes an array of different enzymes via three distinct secretion pathways (40, 42, 43, 46). Most of them are exported in two consecutive steps through the so-called type II secretory pathway into the extracellular medium (11), among them a lipase LipA with a molecular mass of 29 kDa (39). This lipase shows remarkable enzymatic characteristics, e.g., a pronounced stereoselectivity (32) and a broad substrate specificity (23), making it an interesting candidate for biotechnological applications (24).
Enzymes being secreted via the type II secretion pathway contain a N-terminal signal sequence mediating the translocation across the cytoplasmic membrane by the Sec apparatus which has intensively been studied in Escherichia coli (8, 31). An additional machinery consisting of at least 12 so-called Xcp proteins is required for the secretion of the periplasmic intermediates through the outer membrane of P. aeruginosa (11). Although much effort has been expended to identify a potential recognition signal for the Xcp machinery within the extracellular proteins which distinguishes them from periplasmic proteins, no common linear recognition sequence has so far been identified. There is increasing evidence that extracellular proteins fold into a transport-competent conformation before translocation across the outer membrane (3, 5, 12–15, 30), suggesting that the formation necessary for secretion is a three-dimensional structural element composed of noncontiguous amino acid residues or parts of the primary structure. Many of the proteins secreted via the type II pathway contain a disulfide bond, and their secretion is reduced or abolished when the disulfide bond is broken. Examples include a cellulase from Erwinia chrysanthemi (3), the heat-stable enterotoxin II from E. coli (27), and the cholera toxin from Vibrio cholerae (28). On the other hand, there are also proteins and peptides which are secreted in the absence of intact disulfide bonds, examples being the proaerolysin from Aeromonas spp. (14), the lipase-acyltransferase from Aeromonas hydrophila (6), the heat-stable enterotoxin I from E. coli (47), and the pullulanase from Klebsiella oxytoca (35). Thus, it is impossible to ascribe to disulfide bonds in exported proteins a general function for secretion.
Lipases from different pseudomonads have been shown to depend on the presence of a specific additional protein to obtain a stable active conformation (for a review see reference 21). This protein seems to assist the folding process of the lipase in the periplasm, although the exact mechanism is still unknown. In P. aeruginosa this foldase is a 33-kDa periplasmic protein named LipH (or Lif) which is anchored to the cytoplasmic membrane (36).
The P. aeruginosa lipase is secreted via the type II secretion pathway (22, 25). The enzyme contains two cysteine residues which form a disulfide bond (20). Recently, it has been shown that this lipase is not secreted in a P. aeruginosa mutant strain devoid of the oxidoreductase DsbA, indicating the importance of the disulfide bond-forming system to the secretion of this enzyme (41). Here, we show by construction and characterization of cysteine-to-serine variants that the primary function of the disulfide bond in P. aeruginosa lipase is to stabilize the protein during and after translocation over the outer membrane.
MATERIALS AND METHODS
Bacterial strains.
E. coli JM 109, used for cloning experiments, was grown at 37°C in Luria-Bertani (LB) medium. E. coli BL21(DE3) pLysS was used for gene expression experiments which were performed as follows: 10 ml LB medium was inoculated to an optical density at 580 nm (OD580) of 0.05 and grown at 37°C to an OD580 of 0.6. Gene expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a concentration of 0.4 mM. Cells were grown for another 3 h and harvested by centrifugation for 15 min at 12,000 × g and 4°C. P. aeruginosa PABST7.1 (23), a lipase-deficient mutant of P. aeruginosa PAO1, was used as the homologous host for expression of lipase genes. Ten milliliters of 2× LB (20 g of trypton, 10 g of yeast extract, 10 g of NaCl, and 1 liter of distilled water) was inoculated with 50 μl of an overnight culture and grown for 16 h at 30°C. Gene expression was induced by addition of IPTG to a concentration of 0.5 mM. Cells were further grown for 24 h and were separated from the culture supernatant by centrifugation as described for E. coli. When appropriate, ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) were added to the medium for E. coli and carbenicillin (200 μg/ml) and tetracycline (50 μg/ml) were added to the medium for P. aeruginosa.
Plasmids.
Wild-type and variant lipase genes were expressed from derivatives of plasmid pUCPKS (44) constructed as follows: a BamHI/HindIII restriction fragment from pMALip4A, a pMa6 derivative (38) containing a recombinant lipase operon with a unique ApaI site between the genes lipA and lipH was ligated to pUCPKS opened with the same restriction enzymes. To eliminate a second ApaI site, this plasmid was digested with AflIII, deleting a 515-bp fragment, and the ends were blunted with T4 DNA polymerase and self-ligated, yielding plasmid pUCPL6A. For expression in E. coli the lipase gene encoding the mature protein was fused to the pelB signal sequence from Erwinia carotovora by ligating an EcoRI/Eco72I fragment from pUCPL6A and its cysteine variant derivatives to pELpelB-IR-O (S. Rösmann, unpublished), a pET22b derivative containing the lipase operon (Novagen, Bad Soden, Germany), which was cut with the same restriction enzymes.
Site-directed mutagenesis.
The cysteines in P. aeruginosa lipase, C183 and C235, were replaced by serines or by arginine and aspartate, respectively, by PCR mutagenesis using the method of Barettino et al. (2). An EcoRI/ApaI fragment of the gene lipA was ligated to pBluescript II KS (Stratagene, Heidelberg, Germany), yielding plasmid pMut3. This plasmid was digested with AflIII/KpnI and the ends were blunted with T4 DNA polymerase and self-ligated, yielding plasmid pMut4. These plasmids were used as the template DNA in the first and second PCR reactions respectively. For screening purposes, an endonuclease restriction site was either introduced or eliminated in each mutagenic primer in addition to the corresponding mutation. The following oligonucleotides were purchased from Eurogentec (Seraing, Belgium) (mutagenic base substitutions in bold and additional restriction site base substitutions in italics): C183S (TCGGCCAGCGGCGAAGGCGCGTACAAGGTC; ΔKasl), C235C (CTGGTCGGCACCAGCAGTTCGCACCTG; (ΔPstl) C183R (CCCACCTCGGCGCGCGGCGAAGGCGCC; BssHI), C235D (CTGGTCGGCACCGACAGTTCGCACCTG; ΔPstl), universal primer A (GCGCAATTAACCCTCACTAAAGGGAACAAA), and universal primer B (GCGTAATACGACTCACTATAGGGCAA). Amplification was performed using 1 ng of template DNA, 10 pmol each of mutagenic primer and universal primer A in the first PCR reaction and universal primers A and B in the second PCR reaction, and 2 U of Goldstar-Taq polymerase (Eurogentec) in a 50-μl reaction mixture volume containing 75 mM Tris-HCI (pH 9.0 at 25°C), 20 mM (NH4)2SO4, 0.01% (wt/vol) Tween 20, 1.5 mM MgCl2, 10% (vol/vol) dimethyl sulfoxide, and a 200 μM concentration of each deoxynucleoside triphosphate. To the second PCR reaction mixture 1 to 10 ng of the amplicon from the first PCR reaction was added as a megaprimer which was purified by agarose gel electrophoresis and subsequent gel elution using the Nucleospin extraction kit (Macherey & Nagel, Düren, Germany). Reaction conditions began with 2 min at 98°C, followed by the addition of the Taq polymerase and 30 cycles of 1 min at 94°C, 2 min at 64°C, and 1 min at 72°C, as well as a final incubation for 7 min at 72°C on a Robocycler 40 (Stratagene). After purification as described above, PCR products were digested with ApaI/EcoRI and ligated to pBluescript II KS (Stratagene), yielding pKSC183S, pKSC235S, pKSC183R, and pKSC235D. The identity of all PCR-amplified fragments was confirmed by DNA sequencing (34). The double variants C183S-C235S and C183R-C235D were constructed by replacement of the HindIII/ApaI fragments from pKSC183S and pKSC183R with the corresponding fragments obtained from pKSC235S and pKSC235D, respectively. Expression vectors containing variant lipase genes were constructed by ligating the corresponding EcoRI/ApaI fragment from the corresponding pKS derivatives to pUCPL6A.
In vitro refolding.
The in vitro refolding procedure will be described elsewhere. Briefly, cell lysates were obtained by suspension of cells in lysis buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA) to an OD580 of 20. After freezing and thawing, 600 μl of the suspension was treated with ultrasonication (10 min, 60% duty cycle, 50% power output) with a Branson Sonifier 250 equipped with a microtip. Subsequently, solid urea was added to a final concentration of 8 M and samples were incubated for 1 h at 37°C with agitation. Finally, samples were diluted in refolding buffer (50 mM Tris-HCl [pH 8.0], 3.5 mM CaCl2, 45% [vol/vol] glycerol, 0.7 mM laurylmaltoside, 0.5 mM oxidized glutathione, 10 ng of purified LipH per ml) and incubated for 3 h at 30°C with agitation.
Enzyme activity assay.
The photometric assay with p-nitrophenylpalmitate as the substrate has been described elsewhere (45). An ΔOD410 of 1.0 is equivalent to 0.161 nkat.
Immunoblotting.
Proteins from culture supernatants and cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12% T–3% C separation gel and blotted onto a polyvinylidene difluoride membrane (Bio-Rad, München, Germany) using carbonate buffer (10 mM NaHCO3, 3 mM Na2CO3 [pH 9.9]) containing 20% methanol (9). Lipase expressed in E. coli was detected using a polyclonal antiserum raised against the whole denatured P. aeruginosa lipase which was diluted 1:250,000 in Tris-buffered saline containing 0.2% (vol/vol) Tween 80 (TBST). Lipase expressed in P. aeruginosa was detected with a polyclonal serum diluted 1:80,000 in TBST raised against a peptide corresponding to amino acids 48 to 60 of the mature lipase. Horseradish peroxidase-labeled goat anti-rabbit antibody (Bio-Rad) was diluted 1:5,000 in TBST and used as second antibody. Detection was performed using the ECL system (Amersham, Braunschweig, Germany). Samples were prepared as follows: Aliquots of culture supernatant from P. aeruginosa corresponding to an OD580 of the culture of 0.05 were dried under vacuum and suspended in sample buffer containing 4% SDS. Cell lysates were prepared by boiling cells of P. aeruginosa cultures or concentrated and ultrasonified E. coli cultures in sample buffer. Aliquots of cultures corresponding to an OD580 of 0.15 were used. All samples were boiled for 10 min at 95°C. Densitometric analysis of immunoblots was performed with a video imager (MWG, Ebersberg, Germany) and the TINA 2.09 software (Raytest Isotopenmessgeräte GmbH, Straubenhardt, Germany).
Heat inactivation of lipase.
Aliquots of in vitro refolded lipase or samples from P. aeruginosa culture supernatant were incubated for the indicated time periods and temperatures in a temperature-controlled water bath and subsequently stored in an ice-water bath until determination of enzyme activity. Initial activities were chosen to yield a ΔOD410/15 min of 1 to 2.
Protease susceptibility.
To release intracellular proteases a culture of P. aeruginosa PABST7.1(pUCPKS) was adjusted to an OD580 of 20. Cells were sonified in 10 mM Tris-HCl (pH 8.0) as described above for E. coli. Samples of in vitro refolded lipase were mixed with P. aeruginosa cell lysate to give a protein mass ratio of 1:25, respectively, as determined by the Bradford assay (4), with bovine serum albumin as the standard. These samples were incubated at 37°C for the indicated time periods and kept in an ice-water bath until determination of enzyme activity.
Reduction of disulfide bond in wild-type lipase.
Samples of culture supernatant from P. aeruginosa PABST7.1(pUCPL6A) were first preincubated for 1 h at room temperature (RT) in presence of 100 mM dithiothreitol (DTT) (Sigma, Deisenhofen, Germany) or in the presence of 100 mM DTT and 100 mM iodoacetamide (IAA) (Sigma Aldrich) and then for 1 h at RT in the presence of 0.5% (wt/vol) SDS (Biomol, Hamburg, Germany) or for 1 h at 50°C. Subsequently, lipase activity was determined and compared to an untreated sample. When indicated, DTT and excess IAA were removed by gel permeation chromatography on PD 10 columns (Pharmacia, Freiburg, Germany) with 3.5 ml of 20 mM Tris-HCl buffer (pH 8.0) as the mobile phase. Fractions of 0.5 ml were collected. Lipase eluted in fractions 5 to 7 of which fraction 5 was used for further experiments.
RESULTS
Secretion by P. aeruginosa of wild-type and cysteine variant lipases.
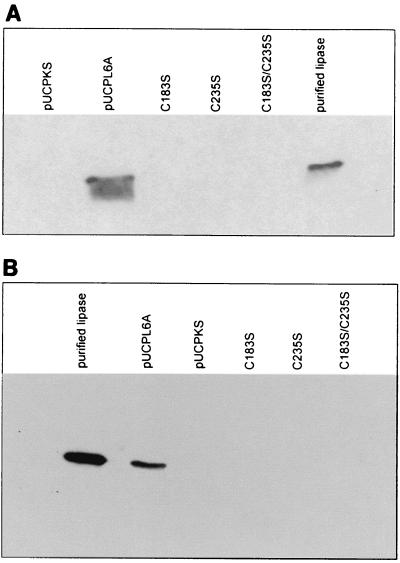
The genes of P. aeruginosa lipase (PAL) and of variant proteins C183S, C235S, and C183S-C235S were expressed in P. aeruginosa using a T7 RNA polymerase-based expression system (23). High lipolytic activity was obtained only in culture supernatants containing the wild-type protein (Table 1). Activities in supernatants from strains expressing the serine-for-cysteine variants were less than 0.05% of the wild-type activity. The variants with single cysteine-to-serine substitutions showed only one-third of the activity of the double variant. Immunoblotting of culture supernatants and cell lysates revealed that the reduced activities were due to a reduced amount of extracellular lipase protein, indicating that the disulfide bond is necessary for secretion (Fig. 1).
TABLE 1.
Extracellular lipolytic activity in culture supernatants of P. aeruginosa PABST7.1 expressing wild-type or cysteine variant lipase
| Lipase | Extracellular lipolytic activitya (nkat/ml of culture supernatant) | Relative activity (%) |
|---|---|---|
| Wild-type | 3,571 | 100 |
| C183S | 0.49 | 0.01 |
| C235S | 0.51 | 0.01 |
| C183SC235S | 1.61 | 0.05 |
| C183RC235D | 4.34 | 0.12 |
| Control | 0 | 0 |
Activity was assayed with p-nitrophenolpalmitate as the substrate and normalized to a cell density corresponding to an OD580 of 1.
FIG. 1.
Immunoblotting analysis of culture supernatants (A) and cell lysates (B) from P. aeruginosa PABST7.1 expressing wild-type or cysteine variant lipases. Aliquots of culture supernatants of P. aeruginosa PABST7.1 expressing wild-type (bearing plasmid pUCPL6A) or cysteine-to-serine variants of lipase were subjected to SDS-PAGE and immunoblotting. Cell lysates were prepared by boiling aliquots of the cultures corresponding to an OD580 of 0.15 in sample buffer. Samples were subjected to SDS-PAGE and immunoblotting. A polyclonal antiserum raised against a peptide consisting of amino acids 48 to 60 in mature P. aeruginosa lipase was used to detect lipase. P. aeruginosa PABST7.1(pUCPKS) and purified lipase served as negative and positive controls, respectively.
Enzymatic activities of cysteine variants.
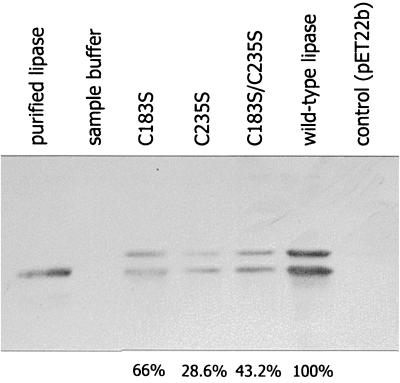
PAL can be refolded in vitro by its foldase after expression in E. coli and subsequent urea denaturation (37). We thus expressed the genes encoding PAL and the cysteine variants in E. coli and subjected the cell lysates to denaturation and in vitro refolding. Lipolytic activity could only be detected after fusing the genes encoding wild-type and variant lipases to the PelB signal sequence from E. carotovora pectate lyase. The expression of the fusion genes yielded a high amount of processed lipase in the periplasm (33). Both the signal sequence-containing 32-kDa precursor protein and the processed 29-kDa lipase were identified by immunoblotting in all strains tested (Fig. 2).
FIG. 2.
Expression of wild-type and cysteine variant lipases in E. coli. Samples of cell lysates from E. coli BL21(DE3)(pLysS) at an OD580 of 0.15 expressing wild-type or cysteine variant lipases were urea denatured and subjected to SDS-PAGE and immunoblotting. Lipase protein was identified with an antiserum raised against purified denatured lipase from P. aeruginosa. The relative intensities of immunoblot signals of mature lipase in these cell lysates were determined by densitometric analysis and are given below the corresponding lanes.
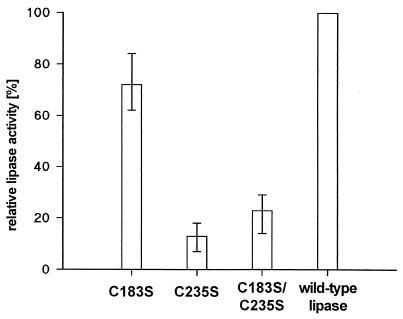
Lipolytic activity was detected with wild-type and all variant lipases. Therefore, an intact disulfide bond is required neither for the interaction of lipase with its foldase nor for reaching and maintaining an enzymatically active conformation. Although cell lysates from strains expressing wild-type and variant lipase genes differed in absolute lipolytic activity after refolding of the lipase (Fig. 3), the activities were correlated with the amount of lipase protein present in the cell lysates (Fig. 2) as determined by densitometric analysis of lipase signals on immunoblots.
FIG. 3.
Relative lipolytic activities of wild-type and variant lipases after heterologous expression in E. coli and in vitro refolding. Lipases from cell lysates of samples for which results are presented in Fig. 2 were urea denatured and in vitro refolded. Activities of the cysteine variants were compared to those of the wild-type enzyme. The results shown are means ± standard deviations (error bars) and were obtained from six different experiments.
Cysteine-to-serine replacements affect lipase stability.
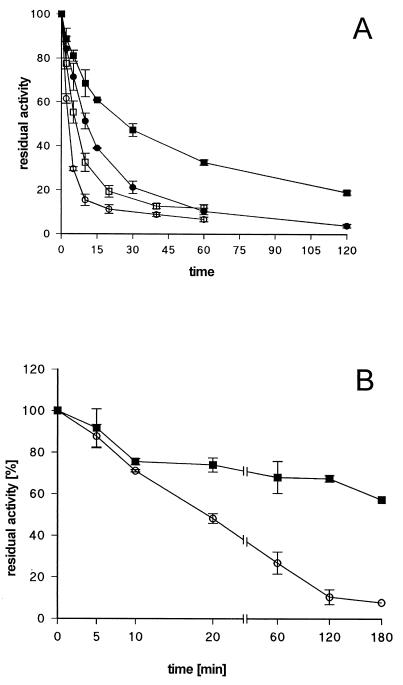
The relationship between activity and stability was investigated by recording heat denaturation profiles at different temperatures for the C183S-C235S variant lipase and comparing them to those of wild-type lipase. As shown in Fig. 4, variant lipase displayed a marked temperature sensitivity, with half-life values of 11.5 min and 2.6 min at 50°C and 60°C, respectively, whereas the corresponding values for the wild-type lipase were 29.1 min and 6.4 min. These results demonstrate that the disulfide bond stabilizes the active conformation of the enzyme.
FIG. 4.
Stability of in vitro refolded wild-type and variant lipases. (A) Heat denaturation profiles. Cell lysates of E. coli BL21(DE3)(pLysS) expressing wild-type or C183S-C235S variant lipases were urea denatured and in vitro refolded. Samples were incubated for the indicated time periods at 50 and 60°C. The activity is expressed as residual activity relative to that of an untreated sample. ■, wild-type lipase at 50°C; ●, C183S-C235S lipase at 50°C; □, wild-type lipase at 60°C; ○, C183S-C235S lipase at 60°C. (B) Protease treatment. Cell lysates of E. coli BL21(DE3)(pLysS) expressing wild-type (■) or C183S-C235S variant lipases (○) were urea denatured and in vitro refolded. Samples were mixed with P. aeruginosa PABST7.1(pUCPKS) cell lysate at a ratio of 1:25 based on mass of protein. These mixtures were incubated at 37°C for the indicated time periods. Lipolytic activity is expressed as residual activity relative to that of an untreated sample. Activities were corrected for the activity determined for the P. aeruginosa cell lysate.
Cysteine variant lipases unable to form a disulfide bond could not be detected in cell lysates of P. aeruginosa (Fig. 1B), suggesting a rapid degradation of these proteins. This hypothesis was tested by incubating in the presence of P. aeruginosa cell lysates wild-type and C183S-C235S variant lipases obtained by in vitro refolding. As shown in Fig. 4, inactivation of the variant protein proceeded much faster than that of the wild-type protein. Although both lipases lost their activity to nearly the same extent during the first 10 min of incubation with the P. aeruginosa cell lysate, the activity of wild-type lipase further decreased by only 16% (from 76 to 60%) of inital activity during the next 3 h, whereas the cysteine variant showed a residual activity of only 8% under the same experimental conditions (Fig. 4B).
Further evidence for a stabilizing function of the disulfide bond was obtained by construction of the double variant C183R-C235D, which allows for the formation of a salt bridge. Strains producing this variant exhibited a threefold increase in extracellular lipolytic activity as compared to the strain producing the variant C183S-C235S (Table 1).
Reduction of disulfide bond in wild-type lipase requires additional destabilizing factors.
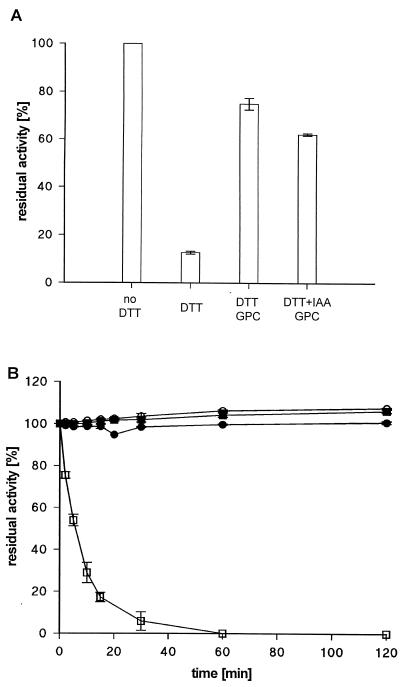
Wild-type lipase was treated with DTT, resulting in inactivation of the enzyme only in the presence of 0.5 % (wt/vol) SDS (Fig. 5A), which itself had no inactivating effect (data not shown). At this detergent concentration the activity of DTT-treated lipase was markedly reduced within 1 h of incubation at room temperature. Removal of DTT by gel permeation chromatography followed by SDS treatment had no inactivating effect, indicating that the disulfide bond is accessible to DTT only in the presence of the detergent. The addition of IAA during the DTT treatment did not alter this result.
FIG. 5.
Effect of DTT treatment on activity of extracellular P. aeruginosa lipase. Samples of lipase from P. aeruginosa PABST7.1(pUCPL6A) were preincubated with 100 mM DTT or 100 mM DTT and 100 mM IAA for 1 h at room temperature and subsequently incubated for 1 h at RT in the presence of 0.5% (wt/vol) SDS (A) or at 50°C (B) for the indicated time periods. Where indicated, DTT was removed by gel permeation chromatography (GPC) prior to SDS or heat treatment. Lipase activity is expressed as residual activity relative to that of an untreated sample.
We also investigated the effect of elevated temperatures on DTT-treated lipase. Again, the lipolytic activity was lost only when DTT exerted its inactivating effect in the presence of another destabilizing factor, e.g., elevated temperature (Fig. 5B). Removal of DTT by gel permeation chromatography after 1 h of incubation at RT and subsequent incubation of the lipase at 50°C had no inactivating effect, indicating that the disulfide bond is not accessible to DTT under these conditions. The addition of IAA did not alter this result.
DISCUSSION
The function of the disulfide bond in P. aeruginosa lipase was investigated by replacing each of the two cysteine residues with a serine. A cysteine-to-serine double variant was also constructed since we expected intermolecular disulfide bond formation during folding in the periplasm of single cysteine variants. Such incorrectly formed disulfide bonds might then hinder transport across the outer membrane. Indeed, the activity of the double variant was almost three times higher than that of the single variants although the absolute lipase activity in culture supernatants of all cysteine-to-serine variants was low.
Lipase protein was detectable by immunoblotting neither in cell lysates nor in culture supernatants from strains expressing any of the cysteine variants. This result indicates that an intact disulfide bond is important to ensure the transport of the enzyme across the outer membrane into the culture supernatant.
When the P. aeruginosa lipase fused to the E. carotovora PelB signal sequence was expressed in E. coli, the lipase partly accumulated in the periplasm (33). This lipase can be refolded in vitro in the presence of its chaperone LipH after urea denaturation (36, 37). We have used this in vitro denaturation and refolding procedure to isolate and characterize the cysteine variants of lipase.
Several reports on lipases from pseudomonads belonging to groups I and II of family I (1) emphasize the dependence of these enzymes on the presence of intermolecular chaperones for correct folding and/or secretion (7, 12, 16, 18, 19, 26). A distinct specificity must exist for this interaction, since the chaperones of different species are not interchangeable, although the lipases have very similar three-dimensional structures (10). The mechanism by which these foldases recognize their cognate lipase and exert their function is still unknown. It is therefore conceivable that the disulfide bond may be part of such a recognition signal or that it stabilizes an essential folding intermediate, making it indispensable to the folding process. An important role during the folding process was suggested for the cysteine residues of Burkholderia cepacia lipase, which could not be refolded in the presence of DTT or β-mercaptoethanol (17). Such a folding function was recently confirmed by investigation of cysteine-to-serine variants of the B. cepacia lipase for which only low activities were obtained in an in vitro transcription-translation system (48). Also the wild-type lipase from P. aeruginosa was refolded with lower efficiency in the presence of β-mercaptoethanol (37). However, by refolding the disulfide bond variants of P. aeruginosa lipase expressed in E. coli we have demonstrated that this lipase can adopt an active conformation even in the absence of the disulfide bond. Thus, the disulfide bond is necessary neither for the recognition of lipase by its foldase nor for the interaction of both proteins. The effect of DTT on the refolding of the wild-type lipase must therefore be different from the reduction of cysteines. Furthermore, the formation of the disulfide bond is essential neither to the folding process nor to gaining enzymatic activity. Although we were unable to purify refolded wild-type and cysteine variant lipases and exactly determine specific activities, it seems likely that the absence of the disulfide bond did not change the activity markedly. The amount of lipase protein subjected to refolding was determined by densitometric analysis of immunoblots. Assuming that wild-type and variant lipases could be refolded with the same efficiency, we found that the amount of lipase protein correlated with the enzyme activity obtained after refolding.
Increasing experimental evidence suggests that secreted proteins must adopt a folded conformation to be released from the cell (3, 5, 13–15, 30). Since the cysteine variants were capable of folding into an active conformation, we conclude that it is not incorrect folding that blocked secretion. The fact that we could identify wild-type but no variant lipase in cell lysates of P. aeruginosa, even under overexpressing conditions, pointed to a rapid degradation of the cysteine variants. As extracellular lipase was found to be protease resistant, presumably due to a tight association of the protein with lipopolysaccharide (LPS) (39), we assume that proteolytic degradation of the lipase takes place in the periplasm. The enhanced susceptibility of the variant to proteolytic degradation by cell lysates of P. aeruginosa underscores this hypothesis. Therefore, the markedly reduced amount of secreted cysteine variant proteins is probably caused by lipase instability leading to rapid degradation by periplasmic proteases. Furthermore, the rapid inactivation of the cysteine double variant at elevated temperatures confirmed the stabilizing function of the disulfide bond.
Interestingly, attempts to chemically reduce the disulfide bond in wild-type lipase failed although the three-dimensional structure of the lipase revealed a surface-exposed (and thus accessible) location of the disulfide bond (26a). However, the simultaneous treatment of lipase with DTT and SDS or at elevated temperatures led to the inactivation of the enzyme, which was not observed when DTT was removed before incubation with SDS or exposure to elevated temperatures. This result indicates that additional destabilizing factors are necessary to render the disulfide bond accessible to the reductant. Brumlik et al. (6) found that the disulfide bond of the lipase-acyltransferase of A. hydrophila was reduced by DTT only in the presence of SDS. These authors suggested a buried location of the disulfide bond which became accessible to DTT only by the denaturing effect of SDS. We believe that the tight association of the P. aeruginosa lipase with LPS (39) might stabilize the enzyme and protect it from reduction by DTT. Such a stabilization could be weakened by the solubilizing effect of SDS. LPS association may also explain why lipase isolated from P. aeruginosa culture supernatant remained stable for at least 2 h at 50°C, which is unusual for an enzyme produced by a mesophilic organism. On the other hand, urea denatured and in vitro refolded wild-type lipase isolated from cell lysates of E. coli was inactivated at this temperature with a half-life of 29.1 min. In this case, an association with heterologous LPS might be less effective or even missing, leading to an increased susceptibility to heat denaturation.
The introduction of a salt bridge instead of a disulfide bond by substitution of the cysteines for arginine and aspartic acid did not result in a marked stabilization of the protein. Due to the very different pKa values of arginine (pKa = ∼12) and aspartic acid (pKa = 3.9 to 4.4) only a very small fraction of these amino acids may be present in a charged form under physiological conditions and thus capable of forming a salt bridge. Apparently, a change of the pKa values of the free amino acids caused by the local protein environment which has been reported for other enzymes (29) had not occured. Nevertheless, the lipase activity of this variant was about three times the activity of the cysteine-to-serine double variant, indicating that the salt bridge can at least partly compensate for the loss of the disulfide bond. This result again emphasizes that the only function of the disulfide bond is to stabilize the structure of the enzyme.
In conclusion, we have demonstrated here by replacing cysteine residues that the disulfide bond in P. aeruginosa lipase is not required for correct folding and interaction of lipase with its foldase. However, it confers to this enzyme a more stable conformation which is important not only in the bacterial culture supernatant but also during passage of the enzyme through the periplasm.
ACKNOWLEDGMENT
Purified foldase (LifH) from P. aeruginosa was kindly provided by Anja Seuter.
REFERENCES
- 1.Arpigny J-L, Jaeger K-E. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- 2.Barettino D, Feigenbutz M, Valcarcel R, Stunnenberg H G. Improved method for PCR-mediated site-directed mutagenesis. Nucleic Acids Res. 1994;22:541–542. doi: 10.1093/nar/22.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bortoli-German I, Brun E, Py B, Chippaux M, Barras F. Periplasmic disulfide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol Microbiol. 1994;11:545–553. doi: 10.1111/j.1365-2958.1994.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of micrograms quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;216:469–483. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Braun P, Tommassen J, Filloux A. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol Microbiol. 1996;19:297–306. doi: 10.1046/j.1365-2958.1996.381908.x. [DOI] [PubMed] [Google Scholar]
- 6.Brumlik M J, van der Goot F G, Wong K R, Buckley J T. The disulfide bond in the Aeromonas hydrophila lipase/acyltransferase stabilizes the structure but is not required for the secretion or activity. J Bacteriol. 1997;179:3116–3121. doi: 10.1128/jb.179.10.3116-3121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chihara-Siomi M, Yoshikawa K, Oshima-Hirayama N, Yamamoto K, Sogabe Y, Nakatani T, Nishioka T, Oda J. Purification, molecular cloning, and expression of lipase from Pseudomonas aeruginosa. Arch Biochem Biophys. 1992;296:505–513. doi: 10.1016/0003-9861(92)90604-u. [DOI] [PubMed] [Google Scholar]
- 8.den Blaauwen T, Driessen A J M. Sec-dependent preprotein translocation in bacteria. Arch Microbiol. 1996;165:1–8. doi: 10.1007/s002030050289. [DOI] [PubMed] [Google Scholar]
- 9.Dunn S D. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem. 1986;157:144–153. doi: 10.1016/0003-2697(86)90207-1. [DOI] [PubMed] [Google Scholar]
- 10.El Khattabi M, Ockhuijsen C, Bitter W, Jaeger K-E, Tommassen J. Specificity of the lipase-specific foldases of Gram-negative bacteria and the role of the membrane anchor. Mol Gen Genet. 1999;261:770–776. doi: 10.1007/s004380050020. [DOI] [PubMed] [Google Scholar]
- 11.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp-system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 12.Frenken LGL, Bos J W, Visser C, Müller W, Tommassen J, Verrips C T. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol Microbiol. 1993;9:579–589. doi: 10.1111/j.1365-2958.1993.tb01718.x. [DOI] [PubMed] [Google Scholar]
- 13.Frenken LGL, de Groot A, Tommassen J, Verrips C T. Role of the lipB gene product in the folding of the lipase of Pseudomonas glumae. Mol Microbiol. 1993;9:591–599. doi: 10.1111/j.1365-2958.1993.tb01719.x. [DOI] [PubMed] [Google Scholar]
- 14.Hardie K R, Schulze A, Parker M W, Buckley J T. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulfide bond formation. Mol Microbiol. 1995;17:1035–1044. doi: 10.1111/j.1365-2958.1995.mmi_17061035.x. [DOI] [PubMed] [Google Scholar]
- 15.Hirst T R, Holmgren J. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc Natl Acad Sci USA. 1987;84:7418–7422. doi: 10.1073/pnas.84.21.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobsen A H, Buckley C M, Aamand J L, Jørgensen S T, Diderichsen B, McConnell D J. Activation of a bacterial lipase by its chaperone. Proc Natl Acad Sci USA. 1993;90:5682–5686. doi: 10.1073/pnas.90.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobson A H, Buckley C M, Jørgensen S T, Diderichsen B, McConell D J. Interaction of the Pseudomonas cepacia DSM3959 lipase with its chaperone, LimA. J Biochem. 1995;118:575–581. doi: 10.1093/oxfordjournals.jbchem.a124948. [DOI] [PubMed] [Google Scholar]
- 18.Ihara F, Okamoto I, Nihira T, Oda J. Requirement in trans of the downstream limL gene for activation of lactonizing lipase from Pseudomonas sp. 109. J Ferment Bioeng. 1992;73:337–343. [Google Scholar]
- 19.Iizumi T, Nakamura K, Shimada Y, Sugihara A, Tominaga Y, Fugase T. Cloning, nucleotide sequencing, and expression in E. coli of a lipase and its activator genes from Pseudomonas sp. KWI-56. Agric Biol Chem. 1991;55:2349–2357. [PubMed] [Google Scholar]
- 20.Jaeger K-E, Ransac S, Koch H B, Ferrato F, Dijkstra B W. Topological characterization and modelling of the 3D strucure of the lipase from Pseudomonas aeruginosa. FEBS Lett. 1993;332:143–149. doi: 10.1016/0014-5793(93)80501-k. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger K-E, Ransac S, Dijkstra B W, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 22.Jaeger K-E, Schneidinger B, Liebeton K, Haas D, Reetz M T, Philippou S, Gerritse G, Ransac S, Dijkstra B W. Lipase of Pseudomonas aeruginosa: Molecular biology and biotechnical application. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: ASM Press; 1996. pp. 319–330. [Google Scholar]
- 23.Jaeger K-E, Schneidinger B, Rosenau F, Werner M, Lang D, Dijkstra B W, Schimossek K, Zonta A, Reetz M T. Bacterial lipases for biotechnological applications. J Mol Catal B. 1997;3:3–12. [Google Scholar]
- 24.Jaeger K-E, Reetz M T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- 25.Jaeger K-E, Dijkstra B W, Reetz M T. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 26.Johnson L A, Beacham I R, McRae I C, Free M L. Degradation of triglycerides by a Pseudomonad isolated from milk: molecular analysis of a lipase-encoding gene and its expression in E. coli. Appl Environ Microbiol. 1992;58:1776–1779. doi: 10.1128/aem.58.5.1776-1779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Nardini M, Lang D A, Liebeton K, Jaeger K E, Dijkstra B W. Crystal structure of Pseudomonas aeruginosa lipase in the open confirmation. The prototype for family I.1 of bacterial lipases. J Biol Chem. 2000;275:31219–31225. doi: 10.1074/jbc.M003903200. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto K T, Baba T, Yamanaka Y, Akashi N, Fujii Y. Disulfide bond formation and secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 1995;177:4579–4586. doi: 10.1128/jb.177.16.4579-4586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peek J A, Taylor R K. Characterization of a periplasmic thiol: disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc Natl Acad Sci USA. 1992;89:6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen MTN, Martel P, Petersen E, Drabløs F, Petersen S T. Surface and electrostatics of cutinases. Methods Enzymol. 1997;284:130–154. doi: 10.1016/s0076-6879(97)84009-8. [DOI] [PubMed] [Google Scholar]
- 30.Pugsley A P. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugsley A P. The complete general secretory pathway in Gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogalska E, Cudrey C, Ferrato F, Verger R. Stereoselective hydrolysis of triglycerides by animal and microbial lipases. Chirality. 1993;5:24–30. doi: 10.1002/chir.530050106. [DOI] [PubMed] [Google Scholar]
- 33.Rösmann S. Escherichia coli. Diplomathesis. Bochum, Germany: Ruhr-Universitaet Bochum; 1998. Bedeutung bakterieller Signalpeptide und der Lipase-spezifischen Foldase für die heterologe Expression der Lipase aus Pseudomonas aeruginosa PAO1 in. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauvonnet N, Pugsley A P. The requirement for DsbA in pullulanase is independent of disulfide bond formation in the enzyme. Mol Microbiol. 1998;27:661–667. doi: 10.1046/j.1365-2958.1998.00722.x. [DOI] [PubMed] [Google Scholar]
- 36.Schneidinger B. Überexpression und transkriptionelle Regulation des Lipaseoperons von Pseudomonas aeruginosa und funktionelle Charakterisierung der Lipase-spezifischen Foldase LipH. Ph.D. thesis. Bochum, Germany: Ruhr-Universitaet Bochum; 1997. [Google Scholar]
- 37.Seuter A. Molekulare Charakterisierung der Interaktion zwischen einer Lipase und einer Foldase aus Pseudomonas aeruginosa. Diplomathesis. Bochum, Germany: Ruhr-Universitaet Bochum; 1998. [Google Scholar]
- 38.Stansens P, McKeown Y, Friedrich K-H, Fritz H-J. Manual of the EMBO laboratory course: Directed mutagenesis and protein engineering. artinsried: Max-Planck-Institut für Biochemie; 1987. Oligonucleotide-directed construction of mutations by the gapped duplex DNA method using the pMa/c phasmid vectors; pp. 95–132. [Google Scholar]
- 39.Stuer W, Jaeger K-E, Winkler U K. Purification of extracellular lipase from Pseudomonas aeruginosa. J Bacteriol. 1986;168:1070–1074. doi: 10.1128/jb.168.3.1070-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tommassen J, Filloux A, Bally M, Murgier M, Lazdunzki A. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1992;103:73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- 41.Urban A, Leipelt M, Eggert T, Jaeger K-E. DsbA and DsbC affect extracellular enzyme formation in Pseudomonas aeruginosa. J Bacteriol. 2001;183:587–596. doi: 10.1128/JB.183.2.587-596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Gijsegem F, Genin S, Boucher C A. Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol. 1993;1:175–180. doi: 10.1016/0966-842x(93)90087-8. [DOI] [PubMed] [Google Scholar]
- 43.Wandersman C. Secretion across the bacterial outer membrane. Cell Mol Biol. 1996;1:955–966. [Google Scholar]
- 44.Watson A A, Alm R A, Mattick J S. Construction of improved vectors for protein production in Pseudomonas aeruginosa. Gene. 1996;172:163–164. doi: 10.1016/0378-1119(96)00026-1. [DOI] [PubMed] [Google Scholar]
- 45.Winkler U K, Struckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979;138:663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamanaka H, Nomura T, Fujii Y, Okamoto K. Extracellular secretion of Escherichia coli heat-stable enterotoxin I across the outer membrane. J Bacteriol. 1997;179:3383–3390. doi: 10.1128/jb.179.11.3383-3390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Kobayashi K, Iwasaki Y, Nakano H, Yamane T. In vitro analysis of roles of a disulfide bridge and a calcium binding site in activation of Pseudomonas sp. strain KWI-56 lipase. J Bacteriol. 2000;182:295–302. doi: 10.1128/jb.182.2.295-302.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]