Summary
Extracellular vesicles (EVs) may mediate intercellular communication by carrying protein and RNA cargo. The composition, biology, and roles of EVs in physiology and pathology have been primarily studied in the context of biofluids and in cultured mammalian cells. The experimental tractability of C. elegans makes for a powerful in vivo animal system to identify and study EV cargo from its cellular source. We developed an innovative method to label, track, and profile EVs using genetically-encoded, fluorescent-tagged EV cargo and conducted a large-scale isolation and proteomic profiling. Nucleic acid binding proteins (~200) are over-represented in our dataset. By integrating our EV proteomic dataset with single-cell transcriptomic data, we identified and validated ciliary EV cargo: CD9-like tetraspanin TSP-6, ectonucleotide pyrophosphatase/phosphodiesterase ENPP-1, minichromosome maintenance protein MCM-3, and double stranded RNA transporter SID-2. C. elegans EVs also harbor RNA, suggesting that EVs may play a role in extracellular RNA-based communication.
In Brief / eTOC blurb:
Extracellular vesicles (EVs) are an ancient and conserved form of intercellular and inter-organismal communication. Nikonorova et al. used the conserved ciliary EV cargo PKD-2::GFP to enrich and profile the EV proteome of Caenorhabditis elegans. They identified and validated new EV cargo using a bioinformatic mining app MyEVome and live imaging of fluorescent reporters. Nucleic acid binding proteins SID-2 and MCM-3 are bona fide ciliary EV cargo. C. elegans EVs also carry RNA, suggesting that EVs may mediate extracellular RNA-based communication.
Introduction
Extracellular vesicles (EVs) may function as an ancient and conserved form of inter-cellular and inter-organismal communication in different physiological and pathological states, but a while ago were considered cellular debris [1,2]. EVs carry bioactive cargo to direct development and differentiation, initiate synapse formation in neurons, and modulate maturation and fertilization of gametes [3–5]. EVs may also spread toxic cargo, such as unfolded proteins in neurodegenerative diseases or signaling molecules to establish a metastatic niche for cancer cells [6]. While EVs are of profound medical importance, the field lacks a basic understanding of how EVs form, what cargo is packaged in different types of EVs originating from same or different cell types, and how different cargoes influence the range of EV targeting and bioactivities.
Our study focuses on EVs produced by cilia, the cellular antennae that receive and transmit signals for intercellular communication. Disrupted ciliary EV signaling is likely an important driver of the pathophysiology of many ciliopathies, such as polycystic kidney disease, retinal degeneration, and others [7]. Main discoveries in the field of ciliary EV signaling were pioneered in the green algae Chlamydomonas reinhardtii and the free-living nematode Caenorhabditis elegans [7]. Nematodes use EVs to communicate with mating partners and to modulate and evade host immune responses via specific EV cargo [7–13].
Here we used C. elegans to conduct a large-scale isolation and proteomic profiling of environmentally released EVs. The resultant dataset of 2,888 proteins captured ciliary and non-ciliary EV cargo candidates from multiple tissue sources. To predict and further validate ciliary EV cargo, we developed an online mining tool called MyEvome (https://myevome.shinyapps.io/evome-app/) that uses single-cell transcriptomic data available for C. elegans [14] to show enrichment of identified EV cargo candidates in cell types of interest. Using this strategy, we discovered four novel ciliary EV cargo - the dsRNA transporter SID-2, ectonucleotide pyrophosphatase/phosphodiesterase ENPP-1 (ortholog of human ENPP1), minichromosome maintenance protein MCM-3 (ortholog of human MCM3), and tetraspanin TSP-6 (ortholog of human CD9). We also found that ENPP-1 is transferred with seminal fluid from the male reproductive tract to hermaphrodite mating partners. We also demonstrate that EVs carry RNA. Combined, these data indicate that C. elegans produces a complex and heterogeneous mixture of EVs from multiple tissues in living animals and suggests that these environmental EVs play diverse roles in animal physiology.
Results
PKD-2, an evolutionarily conserved ciliary EV cargo, enables EV visualization and tracking during biochemical enrichment
Our strategy for biochemical EV enrichment relied on the use of a C. elegans strain with GFP-labeled polycystin-2, which is an evolutionarily conserved cargo of ciliary EVs [7,9]. EVs carrying PKD-2::GFP are shed from the cilia of male-specific sensory neurons (cephalic male CEM and tail ray RnB and hook HOB neurons) into the environment. PKD-2-carrying EVs function in inter-organismal communication by altering male locomotion. Males transfer PKD-2::GFP-carrying EVs to vulvae of their mating partners [9,10]. In addition to the male-specific PKD-2-expressing sensory neurons, C. elegans has a set of sex-shared ciliated neurons whose cilia are exposed and release EVs to the environment – six inner labial type 2 (IL2) neurons (Figure 1A). The male-specific PKD-2-expressing neurons and IL2s are collectively termed EV-releasing sensory neurons (EVNs) for their ability to release ciliary EVs with unique and/or common cargo, in contrast to the sex-shared major sensory organs in the head (amphids) and tail (phasmids), from which ciliary EV shedding had not been reported until very recently [15]. Since PKD-2 is a male-specific EV cargo, we used a strain with a high incidence of males (him-5) and cultured the mixed-sex and mixed-stage population on standard bacterial lawns of E. coli OP50; these conditions are favorable for mating which promotes the release of signaling ciliary PKD-2 EVs.
Figure 1. Buoyant density centrifugation enriches for ciliary PKD-2::GFP EVs more than 100-fold and resolves them into two populations.
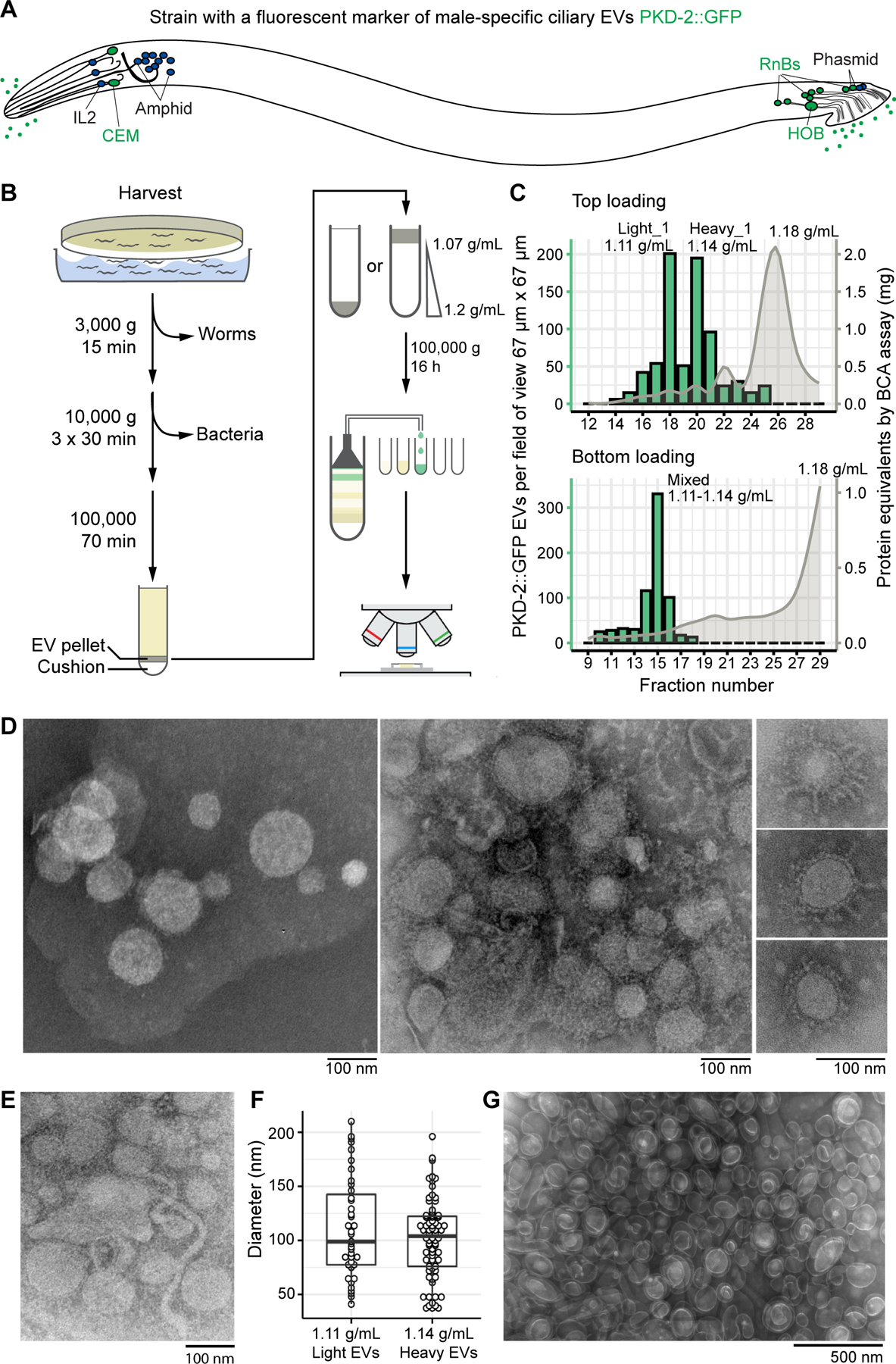
(A) Location of environmentally exposed cilia of sensory neurons. Male-specific cilia are labeled in green. Neurons: CEM – cephalic male, IL2 – inner labial type 2, HOB – hook type B, RnBs – ray neurons type B. (B) Schematic of the EV enrichment workflow. (C) Combination charts showing number of the PKD-2::GFP EVs and the amount of protein in the collected fractions. Centrifugation of the top-loaded sample resolved PKD-2 EVs in two types with densities 1.11 g/ml (light) and 1.14 g/mL (heavy), respectively. Bottom-loaded samples failed to resolve the PKD-2 EV subtypes but reached a similar level of enrichment (more than 100-fold). (D) Transmission electron microscopy (TEM) of the negatively stained PKD-2 EV enriched fractions revealed presence of vesicles with coatings. (E) Tubular structures of 17–20 nm in diameter reminiscent of the stem of a budding vesicle. (F) Diameters of EVs recovered from the light and heavy fractions fall in the range of 50 – 200 nm as measured by TEM. (G) EVs from the fraction with most protein, presumably bacterial OMVs. Refer to Figure S1 for images of PKD-2::GFP EVs as seen with the Airyscan detection system, protein and EV profiling of fractions for every replicate in the study, and other information on the samples.
EVs were collected using ultracentrifugation after serial enrichment by several differential centrifugation steps. The resulting crude EV mixture was then resolved using buoyant density centrifugation and high-resolution fractionation (Figure 1B). Collected fractions were profiled to determine their density, protein concentration, and the quantity of PKD-2::GFP EVs using Airyscan confocal imaging (Figure 1C, S1A).
In accordance with best practices for EV isolation [2], we tested two protocols of loading EVs for buoyant density centrifugation, top- and bottom-loading for top-down (pelleting) and bottom-up (flotation) gradient equilibration, respectively (Figure 1B). The top-loading method resolved PKD-2::GFP EVs into two sub-populations of 1.11 g/mL (termed “light”) and of 1.14 g/mL (termed “heavy”) densities. The bottom loading resulted in all PKD-2 EVs migrating as a single band that contained both light and heavy PKD-2 EVs (termed “mixed”). Analysis of protein distribution across gradients for both loading approaches showed that 99% of total protein was equilibrated at density of 1.18 g/mL and was not associated with the labeled EVs (Figure 1C), indicating that we achieved at least 100-fold enrichment for ciliary PKD-2::GFP EVs.
We confirmed the presence of EVs in the fractions-of-interest using transmission electron microscopy (Figure 1D-E). EVs from the PKD-2::GFP-enriched fractions ranged in size from 50 nm to 220 nm in diameter (Figure 1F), suggesting heterogeneity of the isolated environmental EVome. In addition to intact EVs, we also observed tubular structures reminiscent of budding vesicle stems [16] (Figure 1E).
For rigor and reproducibility, we isolated EVs in two more biological replicates using the top-loading method. Each time, PKD-2::GFP-carrying EVs resolved into light and heavy sub-populations. Electrophoresis of proteins isolated from collected EVs revealed greater heterogeneity of the PKD-2 enriched fractions as compared to the fraction with the most protein (1.18 g/ml), where the predominant protein was 35–37 kD (Figure S1B). This size range and density of the band corresponded to bacterial outer membrane proteins (OMPs) [17,18], the major constituents of the outer membrane vesicles (OMVs) released by the E. coli culture lawn. OMPs were also present in the PKD-2::GFP enriched fractions, with more prominent presence in the heavy fraction. To increase the depth of protein identification in the subsequent mass spectrometry, we chose not to combine technical replicates (parts of the same biological sample that were run in different density gradient tubes) and treated each as separate samples (Figure S1C-S1E). In addition, we excluded the OMP band from heavy fractions prior to mass spectrometry to enrich for the presence of C. elegans proteins (Figure S1F). In total, we used 14 samples for protein identification analysis: one mixed fraction from the bottom-loading approach and three top-loaded biological replicates represented by one, two, and three technical replicates, respectively (Figure S1E-S1F).
Protein identification in the fractions enriched with PKD-2::GFP EVs
In total, we obtained 147,132 spectral counts that mapped to the C. elegans proteome. Among them 133,030 peptides unambiguously mapped to 2,888 C. elegans proteins and 14,102 peptides mapped to more than one C. elegans protein (Data S1). Pairwise comparison of the technical and biological replicates using Spearman correlation analysis showed the reproducibility of our approach. Correlation coefficients ranged within 0.72 – 0.88 between technical replicates and 0.43 – 0.76 between biological replicates (Figure S2A) and 60% of the EV cargo candidates were identified in at least 2 out of 3 biological replicates (Figure S2B-C). Qualitatively 58% of identified proteins were common between light and heavy fractions, whereas the mixed fraction shared 95% of its proteins with the light and heavy fractions (Figure S2D) consistent with presence of both light and heavy EVs. Both light and heavy EV proteomes had similar protein mass distribution profiles (Figure S2E), indicating that excision of the putative bacterial OMV protein band from the heavy fraction as a way of enrichment for the presence of C. elegans proteins did not result in a selective loss of C. elegans proteins of 30–40 kDa size (Figure S2E). Using this approach, we achieved the greatest depth of EV cargo identification available to date for C. elegans.
The light EV fraction was enriched in proteins involved in biosynthesis of amino acids, glycolysis, cysteine and methionine metabolism, amino sugar and nucleotide sugar metabolism, one-carbon metabolism, and other carbon metabolic processes (Benjamini-Hochberg adjusted p-value < 0.05). The heavy EV fraction was enriched in gene products involved in oxidative phosphorylation, proteasome, and glycerophospholipid metabolism (Benjamini-Hochberg adjusted p-value < 0.05). Overall, the identified EVome was enriched in proteins associated with endocytosis, lysosome, proteasome, phagosome, ribosome, SNARE machinery, TCA cycle, and glutathione metabolism (Data S2).
Of 2,888 identified EV cargo candidates, 2,489 were annotated with InterPro domains. Significant enrichment was established for proteins participating in nucleotide binding, vesicular dynamics, trafficking, glycoproteins, glycoprotein binding proteins, nucleic acid binding, post-translational modifiers, chaperones, redox proteins, proteases, and many proteins with domains known to play a role in protein-protein interactions in signaling cascades (Figure 2A, Data S2). Moreover, specific enrichment was observed in proteins carrying Snf7-like domains, major constituents of the endosomal sorting complex required for transport (ESCRT). Specifically, the Snf7-like domains sculpt membrane bilayers into filaments during vesicle budding [16]. Their enrichment in our EV preparations is consistent with observations of the tubular structures by TEM (Figure 1E). For the full list of the identified ESCRT-associated machinery and its human orthologs see Data S2.
Figure 2. Identified EV proteome is enriched in nucleic acid binding proteins and ciliary EV cargo.
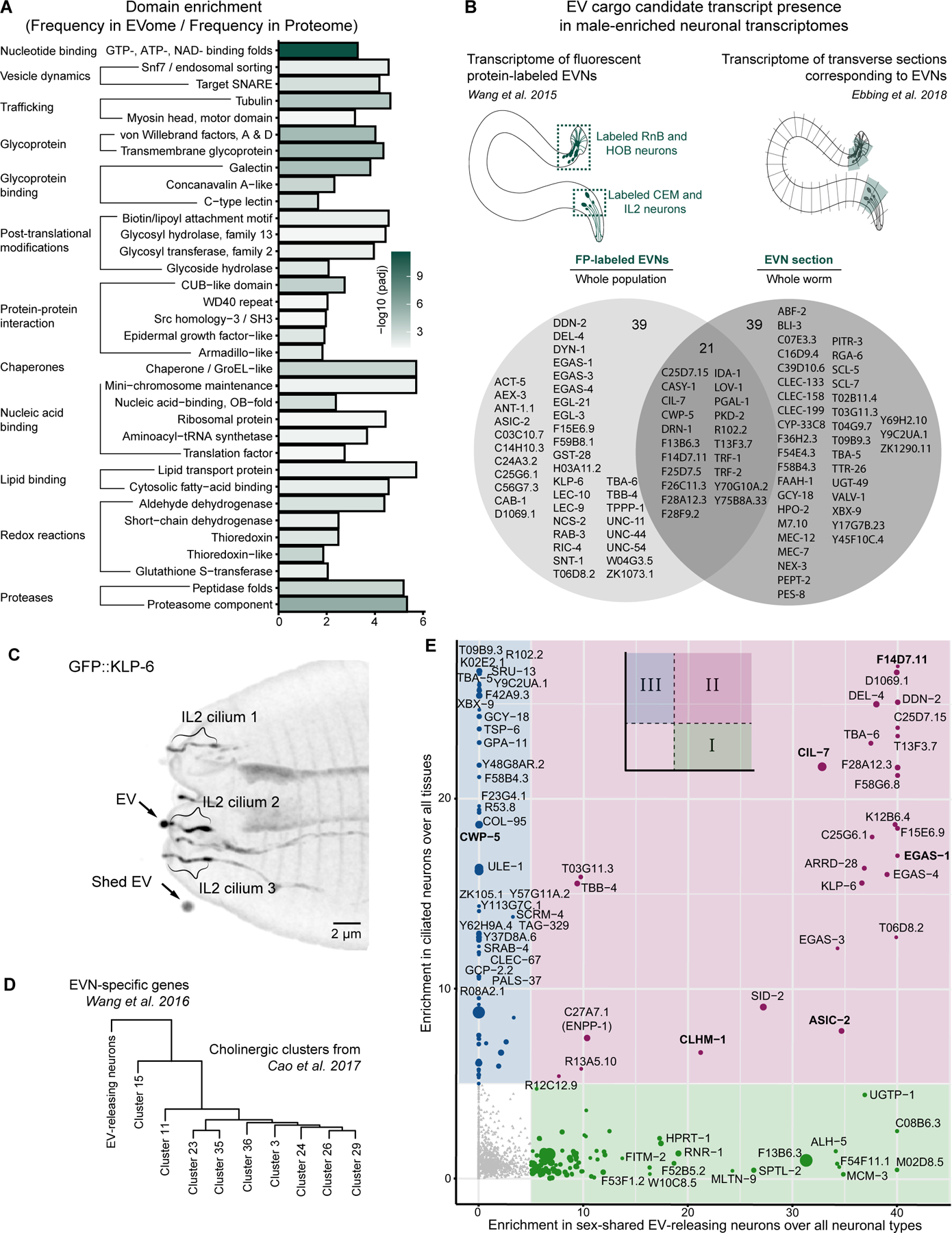
(A) Consolidated groups of InterPro domains enriched in the identified EV proteome (for the full list see Data S2). (B) Identified EVome contained proteins encoded by previously identified transcripts specific to EV-releasing neurons (EVNs). The Venn diagram shows EV cargo candidates that overlap with two transcriptomic datasets [8,26]. (C) GFP::KLP-6 EV shedding from the IL2 cilia of a wild-type hermaphrodite, orthogonal projection. GFP::KLP-6 was observed in EVs released outside the animal. (D) The transcriptome of EV-releasing neurons is most similar to the Cholinergic cluster 15 from [14] likely representing the IL2 neurons. (E) EV cargo candidates were categorized into three groups based on their relative enrichment in the IL2 sex-shared EV-releasing neurons (x-axis) and in the ciliated neurons (y-axis). Three categories are indicated by colors. Previously validated ciliary EV cargoes are bolded.
Refer to Figure S2 for comparative analysis between biological and technical replicates and protein size distribution histograms; and to Data S1-S3 for full list of identified proteins, enrichment analyses, and comparisons to other transcriptomic studies.
Integration with cell-specific transcriptomic datasets
Because the identified EV proteome contained 13% of the entire C. elegans proteome, we reasoned that the captured EV cargo candidates likely originated from multiple cellular sources and not solely from the ciliated EV-releasing neurons (EVNs). Nematodes produce EVs from the plasma membrane of embryonic cells [19], epithelial cells and cuticle [20], reproductive tract [21,22], nervous system [9,23,24], and intestine [25]. In these reports, compelling evidence for tissue-specific EV biogenesis was obtained using electron microscopy, whereas EV cargo composition remains unknown.
To identify EV cargo originating from the EVNs, we used cell-specific transcriptomic datasets reporting on EVN-enriched transcripts [8,26]. We found 99 gene products (Figure 2B, Data S3) in our EVome that are also enriched in EVNs, including previously identified and confirmed EV cargo such as the polycystins PKD-2 and LOV-1 (a homolog of human PKD1), a myristoylated coiled-coil protein CIL-7 required for environmental EV biogenesis [27], CWP-5 (a protein coexpressed with polycystins), acid-sensing sodium channels ASIC-2 and EGAS-1, an ion channel CLHM-1 [28], and a transmembrane cysteine-rich protein F14D7.11 that belongs to a family of proteins with CYSTM-like domains presumably involved in stress-resistance mechanisms [29]. The presence of these EVN-specific cargo in our EV preparations indicates a high degree of enrichment for ciliary EVs given the fact that EVNs occupy less than 0.01% of the whole-body volume [30] and are represented by 27 cells in an adult male and just 6 cells in the hermaphrodite out of almost 1000 somatic cells and 2000 germ cells per each animal.
Our dataset also contained confirmed EVN-specific ciliary proteins [8], including a TGFβ-like domain-containing protein F28A12.3, an ortholog of human ciliary protein CEP104 - T06D8.2, a tetraspanin-like protein T13F3.7, and mucin-domain containing proteins (F25D7.5, F26C11.3, F59A6.3, and Y70C10A.2). No spectral counts were assigned to pan-ciliary kinesin and dynein motors, IFT machinery, or BBSome components, suggesting that our preparations were not enriched in general ciliary proteins. We did observe EVN-specific ciliary kinesin-3 KLP-6, previously known as an EV cargo of mutants lacking EVN-specific tubulin TBA-6 [31]. Re-evaluation of the KLP-6 reporter in the wild-type animals revealed that KLP-6 was shed from the cilia of the IL2 neurons to the environment (Figure 2C), thereby providing experimental validation for KLP-6 as a member of the EVome. The KLP-6 mammalian homolog KIF13B undergoes EV-release from the ciliary tip in immortalized human retinal pigment epithelial (hTERT-RPE1) cells [32], indicating that cargo of our ciliary EVome are conserved.
Comparison to cell-specific transcriptomic datasets captures only proteins that are enriched in the EVNs and depleted from other tissues. Thus, this comparison does not identify cargo produced by both EVNs and larger tissues, such as the intestine or reproductive tract, for vesicular trafficking and EV biogenesis. To overcome this limitation, we developed a relative scoring approach for EV cargo candidates where their expression in a cell-of-interest was compared to their mean expression value in the corresponding group of functionally related cells (e.g., EVNs compared to all neurons taken together). This relative scoring was made possible by single-cell transcriptomic data obtained from profiling hermaphrodites at the 2nd larval stage L2 [14]. Due to the lack of such data for C. elegans males, we used the sex-shared IL2 EVNs as a proxy for the male-specific EVNs, because of their similarities in transcriptional signatures including expression of components of the ciliary EV biogenesis machinery [8]. The IL2 cholinergic neurons were not explicitly identified in the study of Cao et al [14], however, all cholinergic neurons of C. elegans were segregated into nine subtypes. To determine which of the nine represented the IL2 neurons, we compared their transcriptional signatures to the EVN-specific transcriptome [8] and established that Cholinergic cluster 15 was most related to the EVNs (Figure 2D). We then scored our EV cargoes using two metrics: (x-axis) relative enrichment in the IL2 neurons over mean expression in all neuronal types and (y-axis) relative enrichment in ciliated sensory neurons over mean expression in all tissues (Figure 2E). Scoring EV cargo candidates using these two metrics segregated EV cargo into three categories-of-interest: (i) enriched in IL2 neurons, (ii) enriched in both IL2 and other ciliated neurons, and (iii) enriched in ciliated sensory neurons but excluded from IL2 neurons (Figure 2E). We proceeded with experimental validation of novel EV cargo candidates in all three categories defined by an arbitrary enrichment ratio of more than 5-fold (Figure S3A). We relied on the power of Airyscan super-resolution microscopy combined with fluorescent-protein tagged EV reporters to directly visualize EV shedding in vivo [33].
Cilia produce EVs that carry nucleic acid- and nucleotide- binding proteins SID-2, ENPP-1, and MCM-3
SID-2 (systemic RNA interference defective) is a double stranded RNA (dsRNA) transporter required for environmental RNAi in C. elegans. SID-2 internalizes ingested dsRNA from the intestinal lumen, the first step in the systemic silencing of gene expression [34–36]. The native function of SID-2 remains unknown. Intrigued by this and the abundance of SID-2 in our EV preparations, we examined SID-2 endogenous expression and protein localization patterns. We generated the C-terminal mScarlet fluorescent tag using CRISPR/Cas9-mediated insertion into the sid-2-coding region in the genome (Figure S3A). The resulting SID-2::mScarlet reporter was co-expressed with PKD-2::GFP in male-specific EVNs, as well as in the intestine in both males and hermaphrodites (Figure S3B-D). SID-2::mScarlet was abundant on the apical surface of the intestine (Figure S3B) and fully functional in environmental RNAi (Figure S3E), consistent with its known role in endocytosis of dsRNA [34–36].
In male-specific EVNs, SID-2::mScarlet was abundant at the ciliary base regions, suggesting that its primary function in these neurons is associated with the cilia. We also observed SID-2::mScarlet being environmentally released in the form of ciliary EVs (Figure 3B-C, S3F-G). Because SID-2 is a transmembrane RNA-binding protein, its presence in ciliary EVs suggests that ciliary EVs might be capable of transporting RNA. To our knowledge this is the first report of an RNA-binding protein as a cargo of ciliary EVs, suggesting that C. elegans cilia possess the molecular machinery for transport of extracellular RNA.
Figure 3. Nucleic acid binding transporter SID-2 is specifically enriched in the male-specific ciliated EV-releasing neurons (EVNs) and is shed in the form of EVs to the environment.
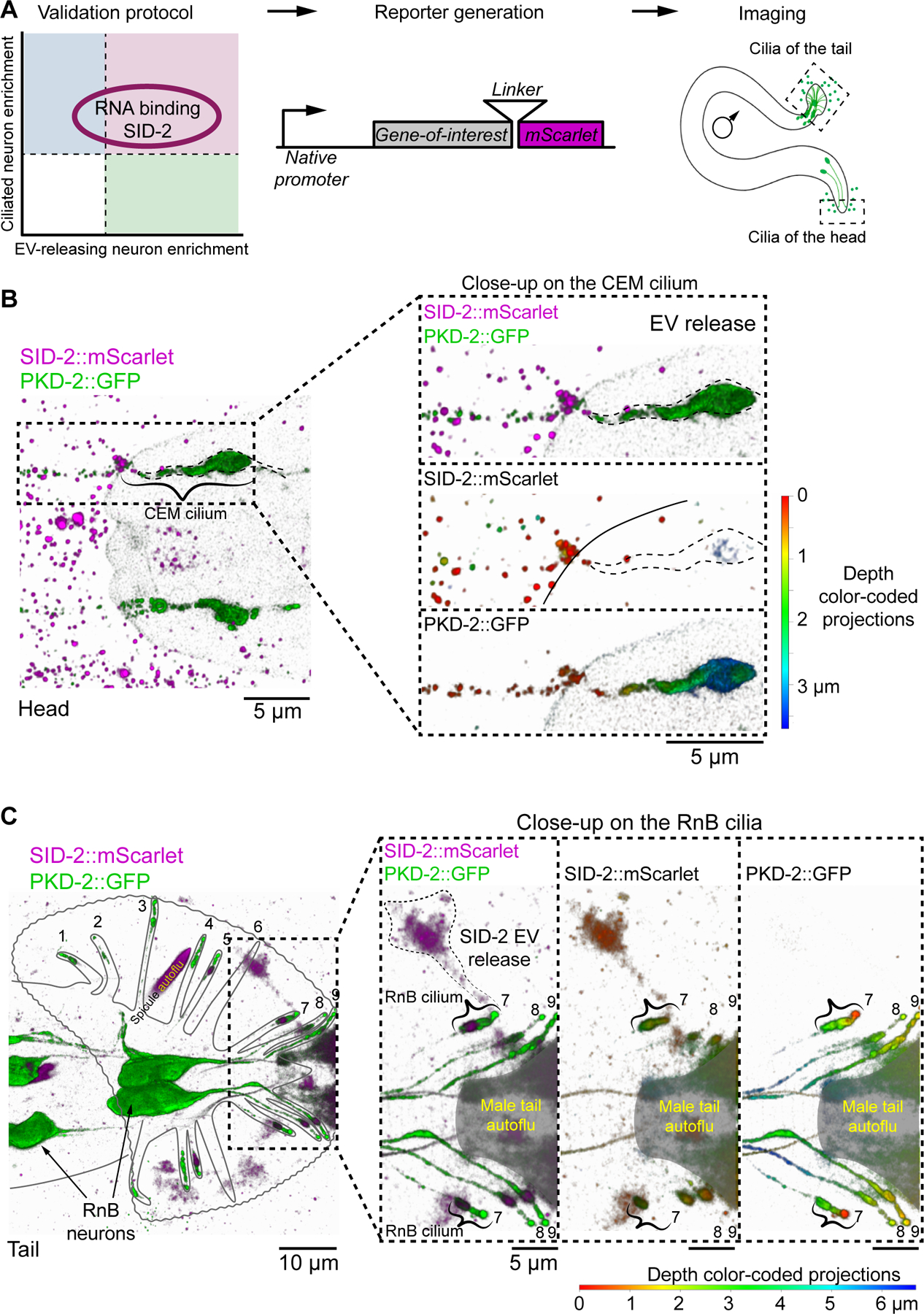
(A) Validation protocol for putative ciliary EV cargo.
(B-C) EV release of SID-2::mScarlet and PKD-2::GFP from cilia of CEM neuron of the head (B) and RnB neuron of the tail (C). Panel C shows ray #7 in the process of shedding SID-2::mScarlet as evidenced by continuous fluorescence signal spanning the ciliary base, ciliary shaft and the outside cloud of EVs. Other clouds on panel C are not labeled as EV release events, because it was not clear from which cilium they originated. Resolution of the imaging technique was 140 nm in lateral direction and 400 nm in axial direction, thus smaller EVs appear as clouds rather than individual EVs. Areas of intense inherent autofluorescence of the male tail are shaded with gray and labeled as autoflu. Images show 3D renderings: on merged images green and magenta colors represent different reporters, on images showing individual channels colors code for Z-depth - 3.5 μm for (B) and 6.5 μm for (C).
Ci- cilia, CEM – cephalic male neuron, HOB – hook neuron type B, RnB – Ray neuron type B. Black and white maximum intensity projections of (B-C) and other data related to SID-2::mScarlet expression are included in Figure S3.
ENPP-1 (C27A7.1), an ortholog of human ectonucleotide pyrophosphatase phosphodiesterases (ENPPs), is another candidate cargo that co-segregated with known ciliary EV cargo (Figure 2F). ENPPs are esterases that hydrolyze phosphodiester linkages in nucleotides and 2’−5’ connected nucleic acids (e.g. ENPP1) or lysophospholipids (e.g. ENPP2, aka Autotaxin) [37,38]. Comparative analysis of C27A7.1 to mammalian ENPP1 and ENPP2 showed that C27A7.1 possesses sequence determinants of substrate preference for nucleotides and nucleic acids, and not for lysophospholipids (Figure S4A). To reflect this homology we named C27A7.1 as ENPP-1 and generated an ENPP-1::mScarlet transgenic translational reporter.
ENPP-1::mScarlet showed a speckled presence in the EVNs, with accumulations in the cilia of the CEM and shared IL2 neurons in the head and ray RnB neurons in the male tail (Figure 4A-B, S4B-D). ENPP-1::mScarlet was released in environmental EVs from both CEMs and IL2s (Figure 4A-B, Video S1). Taken together our data establish the cilium as a cellular location for ENPP-1 EV shedding. Consistent with a role in cilia, mammalian ENPP1 loss-of-function manifests signs of ciliopathies, such as accelerated ossification, hearing impairment, and altered insulin metabolism and immune response [39,40].
Figure 4. Phosphodiesterase ENPP-1 (C27A7.1) is a ciliary EV cargo.

(A) Release of ENPP-1::mScarlet EVs from the IL2 (inner labial 2) cilia labeled as #1, 2 and 3. The IL2 cilia #1 and #2 are in a state of pre-EV release. The cilium #3 is captured in a post-EV release state with the released ENPP-1::mScarlet EVs forming a “stream” of smaller EVs (small arrowheads) that is preceded by 2 larger EVs located at the front end of the “EV stream” (large arrowheads). See Video S1 for time-lapse imaging of ENPP-1::mScarlet EV protruding from the IL2 cilia. (B) Simultaneous release of PKD-2::GFP EVs (green dashed circles) and ENPP-1::mScarlet EVs (magenta dashed circles) from the CEM (cephalic male) cilia. PKD-2::GFP serves as a marker of the CEM cilia and EVs; Ci – cilia. Images show 3D renderings: on merged images green and magenta colors represent different reporters, on images showing individual channels colors code for Z-depth - 2.5 μm for (A) and 4.0 μm for (B).
Black and white maximum intensity projections for (A-B) and other data related to ENPP-1:mScarlet expression are included in Figure S4. Refer to Video S1 for live imaging of ENPP-1::mScarlet EV release from the IL2 cilia.
To validate the IL2-enriched category of EV cargo candidates (Category I, Figure 2F), we focused on another nucleic acid binding protein, MCM-3 (minichromosome maintenance complex component 3 licensing factor) and generated transgenic animals expressing MCM-3::mScarlet driven by its native promoter. MCM-3::mScarlet showed nuclear localization in the developing embryo consistent with the known function of the MCM proteins in DNA replication licensing and DNA helicase activity of the MCM-2–7 complex [41] (Figure S5A). Strikingly, MCM-3::mScarlet was also specifically enriched in the cytoplasm of all EVNs in adult animals and was released from the cilia in the form of EVs (Figure 5A, S5B-D).
Figure 5. Different neuronal types carry distinct EV cargo; a single neuronal cilium may shed multiple cargoes.

(A) MCM-3::mScarlet is present in cilia of PKD-2-expressing and IL2 neurons. Circles indicate MCM-3::Scarlet (magenta) and PKD-2::GFP EVs (B-C) TSP-6::mScarlet is present in amphid channel and AWA neurons of the head (C) and is released in ciliary EVs into the environment (D). Amphid channel is outline with white dashed line. Images are shown as projected 3D objects with the use of actual pixel values in a transparent volume. Images show 3D renderings: on merged images green and magenta colors represent different reporters, on images showing individual channels colors code for Z-depth - 5.0 μm for (A) and 6.0 μm for (C). Black and white maximum intensity projections for (A-C) and other data related to MCM-3::mScarlet and TSP-6::mScarlet expression patterns are included in Figure S5.
Functions of MCM proteins outside of replication are known within Metazoa. For example, a paralog of MCM-3, the MCM2 protein, is required for normal cilia function during zebrafish development [42] and is enriched in sensory ciliated hair cells of the inner ear [43]. Mutations in human MCM2 are associated with autosomal dominant deafness. Our finding that mcm-3 is expressed in sensory ciliated EVNs of C. elegans suggests a conserved role for MCM components in the physiology of ciliated cells.
Finally, we explored EV cargo candidates enriched in the ciliated neurons but specifically excluded from IL2 neurons (Category III, Figure 2F). For this purpose, we examined the expression pattern of TSP-6, an ortholog of human tetraspanin CD9, a marker for human extracellular vesicles [2] (Figure 5B-C, S5E-F). TSP-6::mScarlet was present in the filament-like branches of the chemosensory AWA cilium, the rod-like amphid channel cilia of the head (Figure S5E), as well as the phasmid cilia of the tail, but was not visibly expressed in EVNs. We observed TSP-6-carrying EVs being released from a channel of the amphid sensillum (Figure 5C). While this work was being performed, TSP-6 was independently discovered by another group as an EV cargo that can also be taken up by surrounding amphid glial cells [15], suggesting a role of EVs in neuron-glia crosstalk. Collectively, these findings open a new field of studying ciliary EVs in diverse ciliary contexts in C. elegans and suggests that all cilia may have the potential to shed EVs.
Males transfer ENPP-1 to the hermaphrodite uterus during mating
We also observed that ENPP-1::mScarlet localized to the secretory cuboidal cells of the vas deferens, where it accumulated at the membranes of giant vesicles filled with seminal fluid (Figure 6A, Video S2). Because seminal fluid proteins originating from the cuboidal cells are transferred to hermaphrodites during mating [44], we tested whether ENPP-1 is transferred to hermaphrodites during copulation. By pairing an enpp-1::mScarlet-expressing male to a non-fluorescent hermaphrodite for copulation, we discovered that ENPP-1::mScarlet gets transferred from the male reproductive tract to the hermaphrodite uterus (Figure 6B-C, Video S3). Live imaging of the mating process revealed that in addition to ENPP-1::mScarlet transfer from the vas deferens to the uterus, males release ENPP-1::mScarlet into the environment when scanning for the vulva, presumably from the cilia of RnB neurons (Video S3). ENPP-1 localization to the male reproductive tract of C. elegans, along with its release into the environment, raises the possibility that ENPP-1 is an EV cargo, which would be consistent with reports of ENPP3 in EVs of epididymal fluid of mammals [45,46].
Figure 6. ENPP-1::mScarlet is transferred from the male reproductive tract to the hermaphrodite uterus during copulation.

(A) ENPP-1::mScarlet is present in the somatic part of the male reproductive tract in the vacuoles of cuboidal cells of the vas deferens. Zoomed inset shows a single plane through the giant vacuoles. Also see Video S2 for full Z-stack through the giant vacuoles showing appearance of speckled material with ENPP-1::mScarlet. (B) Male-to-hermaphrodite transfer experiment where ENPP-1::mScarlet males were mated to non-fluorescent hermaphrodites. (C) Imaging of the non-fluorescent hermaphrodite after copulation revealed presence of malederived ENPP-1::mScarlet in the uterus. Yellow asterisks indicate intestinal auto fluorescent droplets. See Video S3 for real-time imaging of male transferring ENPP-1::mScarlet into the hermaphrodite uterus. (D) Venn diagram showing overlap of the identified EV proteome with products of the male-enriched transcripts [47]. (E) Scatter plot showing EV cargo predicted to be of either neuronal or glial origin. See Figure S6 for enrichment of identified EV cargo candidates in other tissues (hypodermis and germline) and for RNA profiling of PKD-2::GFP EV fractions.
We proceeded to compare our dataset with the list of gene products overrepresented in males to identify potential EV cargo of seminal fluid [47] (Figure 6D, Data S3). We found 244 male-enriched or male-specific gene products, among which more than 50% were predicted to originate from the male reproductive tract (Data S3), whereas others were predicted to arise from male-specific neurons or glia (Figure 6E, Data S3). Overall, our study represents the first report of a male-specific EV proteome that largely expands the list of conserved and species-specific animal EV cargo.
Data mining tool to identify cell- and tissue- specific EVomes
Our dataset can be mined to predict tissue-specific EV cargo shed into the environment using the same scoring approach we used to identify new ciliary EV cargo. For example, among hypodermis-specific peptides (Figure S6A), we found proteins involved in maintenance of cuticle integrity. These include CPI-1, an inhibitor of cysteine proteinases, which are often used by plants as nematocidal agents [48], and NIT-1 nitrilase, a hydrolase of deaminated glutathione [49], that is likely a byproduct of glutathione-mediated cuticular shedding during molting [50]. We also found GRD-3, a hedgehog-related protein specific to seam cells. Seam cells are known to release exosomes to build alae [51], cuticular ridges running along the C. elegans body. Other hypodermis-enriched EV cargo candidates included proteins involved in cholesterol and one-carbon metabolism (SAMS-1, SAMT-1, PMT-1, PMT-2) [52,53], and melanin/tyrosine metabolism (PAH-1, HPD-1, TYR-4) [54–57]. Cuticle composition is regulated in response to different developmental and environmental cues [58,59]. We hypothesize that one way to achieve this would be to deliver matrix-modifying enzymes in the form of EVs to the cuticular extracellular space.
Another example of an EV source tissue that was highly represented in our EVome was the germline (Figure S6B). The germline-specific EV cargo candidates included proteins implicated in formation of extracellular matrix of the eggshell (EGG-1, EGG-2, EGG-6, PERM-4, CBD-1, CPG-1, CPG-2, GLD-1) [58] and RNA metabolism associated with germ granules: Y-box binding proteins, orthologs of YBX1 implicated in sorting RNAs into EVs (CEY-1, CEY-2, CEY-3, CEY-4) [60,61], Argonautes (CSR-1, PPW-1, PPW-2, WAGO-1, WAGO-4) [62], P-granule assembly proteins (CGH-1, PGL-1, PGL-3, DEPS-1, EGO-1, PID-2, PRG-1, ELLI-1, GLH-1, OMA-1, PUF-5) [63], and many other nucleic acid binding proteins, such as RUVB-1, M28.5, and all MCM family members (for the full list refer to Data S2).
The enrichment of nucleic acid binding proteins in our dataset and validation of the SID-2 dsRNA transporter as bona fide EV cargo prompted us to examine EV-enriched fractions for their RNA content. We conducted a pilot RNA isolation from fractions containing PKD-2::EVs (Figure S6C). We observed the presence of many RNA subtypes including ribosomal RNA (rRNA) in all the fractions. Lighter fractions had more C. elegans rRNA whereas heavier EVs were enriched in E. coli rRNA. This distribution is consistent with protein profiles (Figure S1B), indicating that lighter fractions are enriched in C. elegans EVs, and heavier EVs contain more E. coli cellular material, presumably OMVs. Overall, the presence of RNA in EV-enriched fractions and overrepresentation of nucleic acid binding proteins (Figure 2B, Data S2) in our dataset suggests that environmental EVs could serve as vectors for horizontal transfer of genetic information and regulation of gene expression in trans.
Discussion
This study presents a large-scale identification and a pipeline for validation of EV cargo candidates shed by C. elegans into the environment. We obtained the greatest depth of C. elegans EV protein identification to date [64,65]. Our isolation and identification strategy involved the use of a mixed-stage C. elegans population with males and hermaphrodites, enrichment via buoyant density centrifugations, and direct visualization of GFP-labeled EV fractions of interest. The proteogenomic mining approach that we applied to EV cargo candidates led to the discovery and validation of four novel ciliary EV cargoes. Our results show that EVs shed by different neuronal types carry distinct EV cargo and corroborate our previous report that a single neuronal cilium may shed multiple cargoes [11].
Ciliary EVs act as signaling devices in Chlamydomonas [66], C. elegans [9], and cultured mammalian cells [67]. Because EVs are membrane bound organelles, their sequestered content is inherently protected from environmental impact, making them ideal vehicles for targeted delivery of fragile cargo, such as RNAs. RNAs hold enormous capability to influence the physiology of the targeted tissue. The enrichment of nucleic acid binding proteins in our dataset is in agreement with findings of EVs carrying Argonaute proteins and abundant small RNAs in parasitic nematodes [12,68]. Our work demonstrates that ciliary EVs carry RNA- and DNA-binding proteins, SID-2 and MCM-3. This suggests that cilia and ciliary EVs may play a role in extracellular RNA communication and horizontal transfer of genetic information. MCM-3 is a soluble protein, whereas SID-2 is a single-pass transmembrane protein with its RNA-binding domain predicted to be extracellular. Whether and how nucleic acids are packaged in ciliary EVs are important questions that warrant further investigation.
The identified list of EV cargo candidates contains many potential regulators of EV biogenesis, including ESCRT-III components with Snf-7-like domains that sculpt membranes into tubular structures during ectocytosis. Consistent with a role for membrane-sculpting proteins in EV biogenesis, our EV preparations include similar tubular structures (Figure 1E, Data S2E). We hypothesize that such tubular structures might form when larger EVs bud from the ciliary tip as suggested by our ENPP-1::mScarlet and KLP-6::GFP reporter dynamics in IL2 cilia.
A role of the ESCRT machinery in ciliary EV biogenesis is suggested by studies in zebrafish [69,70] and Chlamydomonas [71,72], but remains largely unexplored in C. elegans and mammals. Our prior analysis of the involvement of the ESCRT machinery in ciliary PKD-2 EV biogenesis was limited to only three components - stam-1 (ESCRT-0), mvb-12 (ESCRT-I), and alx-1 (adaptor for ESCRT-III machinery). None of the mutants affected PKD-2 EV release, suggesting that these individual genes were not involved in PKD-2 EV biogenesis [9]. With access to robust single cell transcriptomic data [14], we learned that expression of various ESCRT components displays cell-specificity. Different neurons express different sets of ESCRT components. Thus, our dataset in conjunction with single cell transcriptomics will inform future endeavors to identify the general and cell-specific players of EV biogenesis in individual neurons and other cells.
Shedding ENPP-1::mScarlet from the cuboidal cells of the male reproductive tract suggests that C. elegans seminal fluid contains EVs. The ENPP-1::mScarlet reporter appears as large vacuoles filled with “dust specks”, in contrast to soluble TRY-5 and SWM-1 that fill vacuoles uniformly [44]. The presence of EVs in seminal fluid is documented for humans [73], and invertebrates [74]. In Drosophila, the secondary cells of the accessory glands of the male reproductive tract contain giant multivesicular bodies (MVBs) packed with exosomes that they release into females during ejaculation [74]. We hypothesize that the secondary cells of Drosophila are analogous to the cuboidal cells of C. elegans in that the “specks” of ENPP-1::mScarlet are exosomes and that the large vacuoles of the cuboidal cells are giant MVBs. Many of our identified EV cargo candidates are predicted to originate from the male reproductive tract and thus might play roles in regulation of gene expression [75], fitness of male spermatozoa, male fertility, and reproductive behavior [74,76].
Our results indicate that environmental EVs are very heterogenous and that buoyant density centrifugation followed by fractionation worked well to resolve C. elegans EVs from bacterial OMVs, resulting in the greatest depth of identification of C. elegans EV cargo candidates to date. Nevertheless, our protein identification was limited by the two PKD-2::GFP-enriched fractions out of at least ten EV-containing fractions. All the EVs that did not co-migrate with PKD-2::GFP EVs in our enrichment pipeline, including larger EVs, escaped protein identification. Additionally, our data suggest that buoyant density equilibration alone is insufficient to resolve a complex EV mixture down to the level of individual EV subtypes. We hypothesize that similar results are likely to be achieved for many other complex biological fluids with heterogeneous EVs. For increased specificity toward EV subtypes-of-interest, our methodology can be combined with proximity-labeling approaches to define discrete EV-signaling units.
We acknowledge the possibility of false positives in our list of identified EV cargo candidates. Whether the analyzed material represents only EVs, or a mixture of EVs and other membrane/particles that are generally sloughed off cell surfaces cannot be resolved by this study alone. However, we provide effective validation strategies and compelling evidence for the presence of EV cargo in our dataset. A similar pipeline may be used to validate (or invalidate) other potential EV cargo from our dataset.
C. elegans offers many advantages for identifying EV cargo, mechanisms of EV biogenesis, and EV-based communication pathways, including genetic tractability, in vivo imaging and bioassays, and scalability necessary for identification of rare and/or signaling-induced EV cargoes. Our proteogenomic mining strategy can be applied to any cell- or tissue-of-interest using the MyEVome Web App (https://myevome.shinyapps.io/evome-app/). We envision that our dataset can be exploited for use as an experimental springboard in three different ways: (i) identification of conserved and species-specific drivers of EV-based communication in general, and ciliary EV biogenesis in particular, (ii) discovery of novel bioactivities that have not been associated with EVs, (iii) in combination with other identified EV proteomes, understanding of evolutionary specialization of EV-based communication within Nematoda clades, including parasites.
The proteogenomic approach that we used to score EV cargo based on their tissue-specific transcript enrichment is not absolute. The sole enrichment of a transcript encoding an EV cargo candidate in a certain cell type does not automatically mean that the cell sheds EVs with this cargo. Thorough experimental validation is needed to test predictions for each EV candidate and respective tissues of origin, especially in the cases where several tissues/cell types are identified to contain the transcript for the cargo. However, this methodology proved useful in our hands for identification of four novel EV cargo specific to different ciliated neurons, suggesting that the method might be employed by others with some adjustments for one’s unique research questions. Similar validation strategies are not possible in mouse or human yet. Additionally, our proteogenomic analysis was limited to the available transcriptomes [8,14,26,47]. The emergence of new single-cell transcriptomics datasets, especially for C. elegans males, will enable more accurate prediction for assigning EV cargo candidates to their potential source tissue.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maureen Barr (mmbarr@rutgers.edu)
Materials availability
Generated plasmids are available at Addgene, animal strains are available at the Caenorhabditis Genetics Center (University of Minnesota) and upon request from the lead contact. Accession numbers and identifiers for all the shared resources are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| E.coli, OP50 strain | Caenorhabditis Genetics Center (CGC), U of MN | RRID: WB-Strain: OP50 |
| Chemicals, peptides, and recombinant proteins | ||
| Levamisole | Acros | Cat # 187870100 |
| Agarose | Sigma-Aldrich | Cat # A9539 |
| Peptone | VWR | Cat # J636–500G |
| Agar | ThermoFisher | Cat # BP97442 |
| Cholesterol | Sigma-Aldrich | Cat # C8667–5G |
| OptiPrep (60% iodixanol) | Sigma-Aldrich | Cat # D1556 |
| Nano-W stain (Methylamine Tungstate) | Nanoprobes | Cat # 2018 |
| TRIzol LS Reagent | ThermoFisher | Cat # 10296010 |
| Trypsin | ThermoFisher | Cat # PI90058 |
| Critical Commercial Assays | ||
| Bicinchoninic acid (BCA) protein assay kit | ThermoFisher | Cat # 23225 |
| Experimental models: C. elegans strains | ||
| pha-1(e2123ts) III; him-5(e1490) myIs4[Ppkd-2::pkd-2::gfp + Punc-122::GFP] V; myEx888 [Pcil-7::cil-7::tagrfp + pBX] | [11] | PT3112 |
| pha-1(e2123) III; him-5(e1490) V; myEx686 [Pklp-6::gfp::klp-6::3’UTR + pBX] | [85] | PT2102 |
| pha-1 (e2123ts) III; myIs1 [pkd-2p::pkd-2::gfp + Punc-122::GFP] IV; him-5 (e1490) V; myEx944[pIN4 (Ptsp-6::tsp-6::mScarlet) 20 ng/ul + pBX 100 ng/ul] | This paper | PT3545 |
| sid-2 (my95[sid-2::mScarlet]) III; myIs4[Ppkd-2::pkd-2::gfp + Punc-122::GFP] him-5 (e1490) V | This paper | PT3562 |
| pha-1 (e2123ts) III; myIs1 [Ppkd-2::pkd-2::gfp + Punc-122::GFP] IV; him-5 (e1490) V; myEx951[pIN2 (Pmcm-3::mcm-3::mScarlet) 20 ng/ul + pBX 100 ng/ul] | This paper | PT3617 |
| pha-1 (e2123ts) III; myIs1 [Ppkd-2::pkd-2::gfp + Punc-122::GFP] IV; him-5 (e1490) V; myEx948[pIN3 (Penpp-1::enpp-1::mScarlet) 20 ng/ul + pBX 100 ng/ul] | This paper | PT3618 |
| sid-2 (my99 [Q163opal]) III; myIs4 [Ppkd-2::pkd-2::GFP + Punc-122::GFP] him-5 (e1490) V | This paper | PT3646 |
| unc-51 (e369) V | Caenorhabditis Genetics Center (CGC), U of MN | CB369 |
| Oligonucleotides | ||
| gRNA for mScarlet incorporation to the sid-2 locus: 5’-CCUUCGCUACAUUGGAAAGC-3’ | Integrated DNA Technologies | Custom synthesis |
| gRNA to introduce premature stop codon to the sid-2 locus: 5’-AAAUGUGCAACUGUUGAUUG-3’ | Integrated DNA Technologies | Custom synthesis |
| ssDNA donor template to introduce premature stop codon to the sid-2 locus: 5’-GCAGTGCCAACGCCGGCATCAACCACGCCGTTACCTTAATTAACAGTTGCACATTTGACGGTAAGCGCACAATTTTTT-3’ | Integrated DNA Technologies | Custom synthesis |
| Deposited data | ||
| Raw mass spectrometry data | This paper | MSV000087943 |
| Raw image files (.czi – Zeiss format) | This paper | 10.6084/m9.figshare.18130010 |
| Resource website with R code | This paper | https://zenodo.org/badge/latestdoi/302441198 |
| Code for MyEVome app | This paper | 10.5281/zenodo.6145714 |
| Recombinant DNA | ||
| pBX selection marker that rescues pha-1 | [78] | pBX |
| pIN1_GGGSx3_mScarlet | This work | Addgene #182936 |
| pIN2_mcm-3_mScarlet | This work | Addgene #182933 |
| pIN3_enpp-1_mScarlet | This work | Addgene #182934 |
| pIN4_tsp-6_mScarlet | This work | Addgene #182935 |
| Software and algorithms | ||
| MyEVome | Shiny App | https://myevome.shinyapps.io/evome-app |
| ImageJ (Fiji) | ImageJ, NIH | https://imagej.net/software/fiji/ |
| Zen Black 2.0 and ZenBlue 2.0 | Zeiss | https://www.zeiss.com/ |
| Other | ||
| Formvar/carbon coated copper grids 200 mesh | https://www.emsdiasum.com/ | Cat # FCF200-Cu |
| Whatman filter paper #1 | https://www.sigmaaldrich.com | Cat # WHA1001055 |
Data and code availability
Raw proteomics data generated in this study have been deposited at MassIVE and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original raw image Z-stacks have been deposited to Figshare and are publicly available as of the date of publication. The DOI is listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans animals were routinely maintained on 6-cm plates with normal growth medium (NGM) (3 g of NaCl, 2.5 g of peptone, and 20 g of agar per 1 L) seeded with 100 μL E. coli strain OP50 [77]. Wild-type strains were propagated at 20°C, whereas temperature sensitive strains carrying pha-1 (e2123 ts) alleles were grown at 15°C up until their transgenesis with a cocktail containing a functional pha-1 allele. After that, the transgenic animals were transferred to a restrictive temperature of 22°C to select for animals carrying the transgene-of-interest [78].
Strain identifiers and genotypes are cataloged in the Key Resources Table.
METHOD DETAILS
Propagation of C. elegans for EV enrichment
To initiate the culture, 180 hermaphrodites were transferred to 30 6-cm NGM plates (6 animals per plate) and grown for 2 generations. At day 6, each plate was chunked into quarters and the chunks were transferred into 15-cm plates with high growth medium (HGM) (3 g of NaCl, 2.5 g of peptone, and 20 g of agar per 1 L supplemented with 4 mL of cholesterol stock (5 mg/mL of ethanol), 1 mL 1M CaCl2, 1 mL 1M MgSO4, and 25 mL 1M potassium phosphate buffer pH 6.0) seeded with a full lawn of E. coli OP50 (1.8 mL/plate). Animals were allowed to propagate for 2 more generations until almost all bacteria were consumed, yet animals were not starved.
Animal harvest and EV enrichment procedure
Animals were collected into a shallow dish with the M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4 per 1 L of ultrapure water) by inverting the 15-cm plates and their gentle agitation against the surface of the buffer. A total of 200 mL of M9 buffer was used to release worms from 30 HGM plates. The collected worm suspension was centrifuged at 3,000 g for 15 minutes at 15°C in 15 mL conical tubes to pellet the worms and bacteria. The supernatant was transferred to fresh tubes and centrifuged at 10,000 g for 30 minutes 3 times at 4°C (in SS34 fixed angle rotor) to get rid of residual bacteria. Cleared supernatants were then transferred to fresh 12 mL Beckman Coulter round bottom tubes and layered with a 2 mL 36% iodixanol cushion. The cushion was prepared by mixing 3 parts of 60% iodixanol (OptiPrep, Sigma #D1556) and 2 parts of 8% sucrose to achieve final density of 1.2 g/L. EVs were collected onto the cushion via centrifugation in the SW41 Ti swinging bucket rotor at 30,000 rpm (110,000 g average) for 70 minutes at 4°C. EV pellets were collected with a thin glass Pasteur pipette, and further diluted either with M9 buffer to a final density of less than 1.07 g/mL for top loading or with 60% iodixanol to final density of 1.18 g/mL for bottom loading. The gradients were prepared ahead of time by a freeze-thaw method. Specifically, 12%, 16%, 20%, 24%, 28%, and 32% iodixanol solutions were made by mixing solution A (8% sucrose) with solution B (60% iodixanol) so that final osmolarities ranged from 270 to 320 mOsm and densities ranged from 1.08 to 1.20 g/mL. The solutions were then layered in a stepwise manner starting from the heaviest layer and freezing the content at −80°C after each added layer. The gradients were thawed at room temperature 1–2 hours prior to use and remained at 4°C up until loading. Loading was performed using a glass Pasteur pipette. Isopycnic centrifugation was carried out for 16 hours at 30,000 rpm using the SW41 Ti Beckman Coulter rotor at 4°C. Fractions were collected using the BioComp’s Piston Gradient Fractionator. Aliquots were taken from each fraction to measure density, protein concentration, and examine enrichment with PKD-2::GFP-carrying EVs. Density was measured by weighing out 150 μL of the examined solution on an analytical scale to calculate weight per volume ratio for each fraction. Protein concentration was measured using Bicinchoninic Acid (BCA) Protein Assay Kit (Pierce, Thermo Fisher #23225) according to manufacturer’s instructions in the presence of 0.2% SDS. PKD-2::GFP EV enrichment was assessed by scanning through an array of representative droplets from each fraction using a Zeiss LSM 880 confocal microscope and its Airyscan detector. Once the most enriched fractions were identified, total protein was isolated from them via methanol-chloroform extraction. Briefly, four volumes of methanol were added to one volume of the EV-enriched fraction, vortexed vigorously for 10 seconds, followed by addition of one volume of chloroform and repeated intense vortexing, followed by addition of three volumes of ultrapure water and vortexing. Phase separation was completed by centrifugation at 16,000 g in a tabletop centrifuge for 1 minute. The aqueous upper layer was discarded without disturbing the white flaky proteinaceous interphase. Protein was precipitated by adding four volumes of methanol and centrifugation at 16,000 g for 10 minutes. Protein pellets were dried and dissolved in 2x Laemmli buffer (0.125 M Tris HCl pH6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue) at 90°C for 5 minutes. Resulting proteins were analyzed by denaturing electrophoresis in polyacrylamide gel (SDS-PAGE) or sent for protein identification by mass spectrometry.
Transmission electron microscopy
Fractions most enriched with PKD-2::GFP carrying EVs were diluted 10–20 times with M9 buffer and applied to discharged formvar/carbon coated copper grids (200 mesh, Electron Microscopy Sciences # FCF200-Cu). Five microliters of the diluted fraction were dispensed on the grid and allowed on for 30–60 seconds followed by wicking the liquid away from the grid with Whatman filter paper #1. The grid was then transferred through 3 droplets of PBS (5 min in each droplet) to wash away residual sucrose and iodixanol. The excess of PBS was wicked away with the filter paper after each wash. For the unfixed preparation, following PBS washes the grid was exposed to the Nano-W stain (Methylamine Tungstate solution, Nanoprobes #2018) for 1 minute sharp. The stain was removed with filter paper and the grid was left to dry out completely for 30 minutes at room temperature. Imaging was performed on Philips CM12 electron microscope with AMT-XR11 digital camera. Best results were achieved with imaging the samples on the day of their staining.
Liquid chromatography tandem mass spectrometry
Mass spectrometry and protein identification were outsourced at the Rutgers Proteomics Core. Samples were allowed to run in an acrylamide gel for a distance of 3–5 cm. Regions-of-interest were excised for further protein processing. Each piece was subjected to reduction with 10 mM dithiothreitol for 30 min at 60°C, alkylation with 20 mM iodoacetamide for 45 min at room temperature in the dark, and digestion with sequencing grade trypsin (ThermoFisher #90058) overnight at 37°C. Peptides were extracted twice with 5% formic acid, 60% acetonitrile and dried under vacuum.
Samples Light_1 and Heavy_1 were analyzed using Dionex UltiMate 3000 RLSCnano System, ThermoFisher) interfaced with QExactive HF (ThermoFisher). Samples were loaded onto a fused silica trap column Acclaim PepMap 100, 0.075 mm x 200 mm (ThermoFisher). After washing for 5 min at 5 µl/min with 0.1% trifluoroacetic acid, the trap column was brought in-line with an analytical column (nanoEase M/Z Peptide BEH C18 column, 130 Å, 1.7 μm, 0.075 mm x 250 mm, Waters) for LC-MS-MS. Peptides were fractionated at 300 nL/min using a segmented linear gradient of 4–15% solution “B” in solution “A” for 30 min (where A is a polar solvent 0.2% formic acid, and B is an organic solvent of 0.16% formic acid and 80% acetonitrile), 15–25% for 40 min, 25–50% for 44 min, and 50–90% B for 11 min. The column was re-equilibrated with 4% solution “B” in solution “A” for 5 minutes prior to the next run. Cyclic series of full scan acquired in Orbitrap with resolution of 120,000 were followed by MS-MS (HCD relative collision energy 27%) of the 20 most intense ions at resolution 30,000 and dynamic exclusion duration of 20 sec.
For the samples Mixed, Light_2, Heavy_2, Light_3, Heavy_3, Light_4, Heavy_4, Light_5, Heavy_5, Light_6, Light_7, Heavy_6, LC-MSMS were done using Dionex UltiMate 3000 RLSCnano System interfaced with Eclipse Orbitrap tribrid (ThermoFisher). The LC method is the same as described above. For mass spectrometry, the scan sequences began with MS1 spectrum (Orbitrap analysis, resolution 120,000, scan range from M/Z 375–1500, automatic gain control (AGC) target 1E6, maximum injection time 100 ms). The top S (3 sec) and dynamic exclusion of 60 sec were used for selection of parent ions of charge 2–7 for MS-MS. Parent masses were isolated in the quadrupole with an isolation window of 1.2 m/z, AGC target 1E5, and fragmented with higher-energy collisional dissociation with a normalized collision energy of 30%. The fragments were scanned in Orbitrap with resolution of 15,000. The MS-MS scan range were determined by charge state of the parent ion but lower limit was set at 110 amu.
Protein identification
The peak list of the LC-MS-MS were generated by Thermo Proteome Discoverer (v. 2.1) into MASCOT Generic Format (MGF) and searched against databases of C. elegans (Ensembl), E. coli K12 substrain MG1655 (NCBI), and Contaminant Repository for Affinity Purification Mass Spectrometry Data (CRAP) using in house version of X!Tandem GPM (The Global Proteome Machine) Fury [79]. Search parameters were as follows: fragment mass error 20 ppm, parent mass 5 error +/− 7 ppm, fixed modification - carbamidomethylation on cysteine, variable modifications - oxidation on methionine, protease specificity - trypsin (C-terminal of R/K unless followed by P), with 1 miss-cut at preliminary search and 5 miss-cuts during refinement. To validate the protein and peptides, false positive rate (FPR) [80] value was used to decide the log(e) cutoff. Only spectra with log(e) < −2 were included in the final report.
Transgenesis for EV cargo validation
Pieces of genomic DNA were cloned into the vector backbone encoding the C-terminally linked flexible linker (amino acid sequence is GGGSGGGSGGGSGGG) fused to the mScarlet coding sequence and unc-54 3’UTR (pIN1_GGGSx3_mScarlet, Addgene plasmid #182936) from the pPD95.75 backbone (Addgene plasmid #1494). Promoter lengths for the generated transgenes were as follows: Pmcm-3 – 1005 bp, Penpp-1 – 1174 bp, and Ptsp-6 – 2017 bp upstream of the start codon of the longest isoform (if applicable).
All the transgenes were fully sequenced prior to transgenesis. Respective plasmids were injected with a co-injection selective marker pha-1(+) (pBX plasmid) [78] into pha-1 (e2123 temperature sensitive) III; him-5 V strain. Further maintenance of the transgenic worms at 22°C ensured selection for and retention of the extrachromosomal transgenic arrays.
The sid-2::mScarlet reporter was generated using CRISPR/Cas9-mediatead editing as described in [81] using a guide RNA (5’-CCUUCGCUACAUUGGAAAGC-3’) and a double stranded DNA donor template containing a sequence encoding the flexible linker of 15 amino acids (GGGGS)3 fused to the mScarlet coding sequence (Figure S3A). The mScarlet coding sequence was intron-less to not interfere with native splicing regulation within the dyf-2 gene. The sid-2 null mutant was generated using a guide RNA (5’-AAAUGUGCAACUGUUGAUUG-3’) and a single-stranded DNA (5’-GCAGTGCCAACGCCGGCATCAACCACGCCGTTACCTTAATTAACAGTTGCACATTTGACGGTAAGCGCACAATTTTTT-3’) to introduce a nonsense mutation encoding a stop codon instead of Gln163 - sid-2 (my99 [Q163opal]) III.
Mounting animals for live imaging
Animals were anesthetized with 10 mM levamisole solution in the M9 buffer. To completely immobilize the worm, we used thin agarose pads, prepared from 10% melted agarose (Sigma #A9539) and ultrapure water. Melting of the 10% agarose was achieved by pulse heating in a microwave in 3 second intervals for a total of 1 minute or until dry agarose pieces were no longer visible. Once melted, the 10% agarose was incubated at 95°C for about 10 minutes to allow air bubbles to escape. The melted agarose was dispensed onto glass slides in 100 μL droplets and the droplets were immediately pressed with a second glass slide to make a thin gel pad. The agarose pads were made in batches and stored in a sealed glass slide container at room temperature for up to a week. Animals were mounted by their placement in 1 µL droplet of 10 mM levamisole dispensed on an 18 × 18 cm coverslip, followed by flipping of the coverslip onto the center of the agarose pad. Animals were imaged within 30 minutes of their mounting.
Super-resolution microscopy and image processing
Super-resolution imaging was performed on a Zeiss LSM880 confocal system equipped with Airyscan Super-Resolution Detector, Single Photon Lasers (488 nm for GFP and 561 nm for mScarlet), motorized X-Y Stage with Z-Piezo and T-PMT. Planes of captured super-resolution Z-stacks were distanced 0.185 µm apart. Image analysis and processing was performed using ZenBlue 2.0 and ZenBlack 2.0. All the images underwent automated Airyscan processing and adjustment of brightness that was applied to the entire image. For Figure 3–5, images of cilia and EVs show 3D renderings of entire Z-stacks in a transparent volume (Transparency Render feature of ZenBlack 2.0). On these figures, merged images use two colors to indicate different reporters, whereas individual channels are color-coded for Z depth with red color assigned to structures closest to the objective and blue colors for structures most distant from the objective. This representation allows one to see EVs that are positioned outside of the animal in the Z-axis, but whose XY position overlaps with the body of the animal. Such EVs appear as signal coming from the inside of the animal on maximum intensity projections but are easily visible on 3D depth color-coded projections. The color of such EV on the depth color-coded projection would significantly differ from the color of internal anatomical structures of the animal because they positioned at different depth of the Z-stack. For all the main figures presented as 3D projections we include images of their individual channels as black and white maximum intensity projections in the Supplemental Data. We also deposited all the raw Z-stacks to https://dx.doi.org/10.6084/m9.figshare.18130010 from where they can be downloaded and analyzed using the Fiji open-source image processing package.
Imaging of EV release during mating
To visualize EV release during mating three unc-51 hermaphrodites were placed with ten males of the PT3618 strain in the center of an unseeded NGM plate, some bacteria were transferred on the pick to feed the hermaphrodites. The piece of agar with the placed worms (approximately 24 × 40 mm) was transferred onto a 24 × 60 coverslip with the worm side facing the coverslip. Then the agar chunk was covered with a 28 × 48 mm saran wrap, with a 10 mm- diameter hole in the center, so that the spot with hermaphrodites was exposed to air through the window. This covering method drove the males toward the center with the hermaphrodites. Mating started within 15 minutes. Imaging was performed using 40x objective.
RNAi experiment
The E. coli strain HT115 expressing either embryonic lethal mex-3 dsRNA or an empty vector [82] was cultured at 37 °C overnight and seeded the NGM plates containing carbenicillin (50 μg/mL) and IPTG (Isopropyl ß-D-1-thiogalactopyranoside, 1 mM). The plates were kept in the dark at room temperature. For the RNAi experiment, three L4 hermaphrodites were placed on the lawn and left for 24 hours to lay eggs, followed by daily transfer to a fresh RNAi plate with the corresponding lawn for another 24h of egg laying. Total progeny of three hermaphrodites was used as a measure of the ability to internalize environmental dsRNA.
Isolation of RNA from EV-enriched fractions
RNA isolation was performed using TRIzol LS Reagent (Thermo Fisher #10296010) using manufacturer instructions. Pellets were dissolved in 6 ul of nuclease-free water and analyzed using TapStation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Differential abundance analysis
For the differential abundance analysis of protein presence in light and heavy EVomes, the “Mixed” dataset was omitted as these EVs did not resolve in two populations. Proteins were quantified using the number of unique spectral counts mapped to the protein. Batch corrections were applied using the R package msmsEDA. For differential expression analysis, we considered only proteins that were identified by unique unambiguous peptides in more than one replicate for both heavy and light subpopulations. For these proteins, unique spectral counts were log2-transformed, quantile-normalized, and missing values were imputed using the QRILIC method as implemented in the R package DEP [83]. Differential expression analysis was performed using the R package limma [84], using the trend and robust hyperparameter estimation settings. P-values were adjusted using the Benjamini-Hochberg procedure. Proteins were considered differentially expressed between the heavy and light EVs if they had an absolute log2 fold changes greater or equal to 1.5 and an adjusted p-value < 0.05.
Gene ontology and InterPro enrichment
KEGG enrichment was performed using clusterProfiler R package on the set of 2,888 proteins identified unambiguously. For enrichment of KEGG terms in light and heavy EVs, the heavy EVs included proteins that were upregulated relative to the light EVs and those proteins only detected in the heavy EVs. A similar procedure was performed for light EVs. The background for KEGG enrichments of differentially abundant proteins included only proteins detected unambiguously via LC-MS-MS. InterPro domain enrichment was performed using the DAVID Bioinformatics Resources (https://david.ncifcrf.gov/). Similar analyses were performed that included ambiguously identified proteins in the EVome dataset.
Mining with single cell transcriptomics data
EV cargo were assigned scores derived from single-cell transcriptomics datasets [14]. Enrichment in ciliated neurons was calculated as the ratio of transcript abundance (transcript per million, TPM) in ciliated neurons over mean transcript abundance across all the tissues. Enrichment in IL2 neurons was calculated as the ratio of transcript abundance in IL2 neurons over its mean abundance in all neurons. Identity of the IL2 neurons was established as Cholinergic clusters 15 from [14] based on similarities of their transcriptional profiles to the list of transcripts enriched in the EV-releasing neurons identified in [8]. The similarity was determined by hierarchical clustering analysis of the fold change enrichment values of EVN-specific transcripts from [8] to the fold change enrichment of respective cholinergic neurons transcripts over all identified cholinergic neuronal clusters of C. elegans. The values that were used for generation of the dendrogram on Figure 2E are indicated in Supplemental Data S3.
Supplementary Material
Data S1. Lists of identified peptides and proteins, related to Figure 2.
(A) Metadata on the overall numbers of peptides and identified proteins from C. elegans and E. coli. (B) Unique peptides with matched proteins. (C) Non-unique peptides matched with corresponding protein sets. (D) Summary of spectral counts of unique proteins for each gene product. Functions of the genes and human orthologs are indicated.
Data S2. Differential abundance and enrichment analyses, related to Figure 2.
(A-B) Differential abundance analyses of the light and heavy EV proteomes: KEGG pathway (A) and InterPro protein domain (B) enrichment analyses. (C) List of ESCRT proteins identified in the EV proteome. (D) List of nucleic acid binding proteins identified in the EV proteome.
Data S3. Mining of the EV proteome, related to Figure 2.
(A) Overlap of the EVome with the EVN-specific transcriptomes used to produce Figure 2C. (B) IL2 identification table contains relative enrichment values for cholinergic clusters from Cao et al. (2017). (C) EV cargo scores based on their absolute enrichment in ciliated sensory neurons and relative enrichment in the IL2 neurons of hermaphroditic L2 populations; these scores were used to generate the plot on Figure 2F. (D) Overlap of the identified EV proteome with the male-specific transcriptome from [47].
Video S1. ENPP-1::mScarlet EV release from the IL2 cilia into the environment, related to Figure 4 and S5.
During the time lapse focus changes from the cilium to EV plane over an axial distance of approximately 1 μm. EVs are connected to each other in a tandem manner and remain attached to the cilium during the course of the video.
Video S2. ENPP-1::mScarlet appearance in the vacuoles of the vas deferens, related to Figure 6.
Animated Z-stack.
Video S3. ENPP-1::mScarlet release from the male tail during mating to a hermaphrodite, related to Figure 6.
The footage shows two males mating with one hermaphrodite. Male #1 releases ENPP-1::mScarlet while backing along the body of the hermaphrodite scanning for vulva, whereas Male #2 releases ENPP-1::mScarlet during sperm transfer.
Highlights:
Large-scale identification and a pipeline for validation of EV cargo in C. elegans
Multiple tissues shed EVs and different neuronal cilia shed distinct EV cargo
Ciliary EVs carry nucleic acid-binding SID-2 and MCM-3 and phosphodiesterase ENPP-1
The male vas deferens transfers ENPP-1 to the hermaphrodite uterus during mating
Acknowledgments
We are grateful to Haiyan Zheng and Peter Loebel for assistance with the mass spectrometry analysis, Gloria Androwski and Helen Ushakov for outstanding technical assistance, Julie Claycomb, Rachel Kaletsky, Coleen Murphy, Tetsuya Nakamura, Joel Rosenbaum, Piali Sengupta, and Christopher Ward for discussions, members of the Boston and UCSF cilia supergroups and Rutgers C. elegans community for thought-provoking questions, Christine Maric-Bilkan for continued support and sound advice, and the three anonymous reviewers for constructive criticism. We also thank WormBase (release WS281) and WormBook that were used daily during this project.
Funding:
The work was funded by grants from the National Institutes of Health (NIH) DK116606, DK059418, and NS120745 to M.M.B, NSF DBI 1936046, NIH R35 GM124976, and start-up funds from HGINJ to P.S, NIH K12 GM093854 INSPIRE (IRACDA NJ/NY) Postdoctoral fellowships to J.D.W. and A.L.C, and Donald Stahlin Memorial Fund. Protein identification was performed in the Biological Mass Spectrometry Facility of Robert Wood Johnson Medical School at Rutgers University supported by the NIH instrumentation grants S10OD025140 and S10OD016400. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The Center for C. elegans Anatomy and WormAtlas provided valuable anatomical and ultrastructural resources (R24OD010943).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
ADDITIONAL RESOURCES
To aid in exploration of the proteomics dataset using our bioinformatical approach for prediction of source tissues for various EV cargo candidates, please access the web application called MyEVome (https://myevome.shinyapps.io/evome-app/).
References
- 1.Maas SLN, Breakefield XO, and Weaver AM (2017). Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol 27, 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung BH, Parent CA, and Weaver AM (2021). Extracellular vesicles: Critical players during cell migration. Dev. Cell 56, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foot NJ, and Kumar S (2021). The Role of Extracellular Vesicles in Sperm Function and Male Fertility. In Subcellular Biochemistry, pp. 483–500. [DOI] [PubMed]
- 5.Lizarraga-Valderrama LR, and Sheridan GK (2021). Extracellular vesicles and intercellular communication in the central nervous system. FEBS Lett 595, 1391–1410. [DOI] [PubMed] [Google Scholar]
- 6.Urabe F, Patil K, Ramm GA, Ochiya T, and Soekmadji C (2021). Extracellular vesicles in the development of organ‐specific metastasis. J. Extracell. Vesicles 10, 6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, and Barr MM (2018). Cell–cell communication via ciliary extracellular vesicles: clues from model systems. Essays Biochem 62, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Kaletsky R, Silva M, Williams A, Haas LA, Androwski RJ, Landis JN, Patrick C, Rashid A, Santiago-Martinez D, et al. (2015). Cell-Specific Transcriptional Profiling of Ciliated Sensory Neurons Reveals Regulators of Behavior and Extracellular Vesicle Biogenesis. Curr. Biol 25, 3232–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, and Barr MM (2014). C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr. Biol 24, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Nikonorova IA, Gu A, Sternberg PW, and Barr MM (2020). Release and targeting of polycystin-2-carrying ciliary extracellular vesicles. Curr. Biol 30, R755–R756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Nikonorova IA, Silva M, Walsh JD, Tilton PE, Gu A, Akella JS, and Barr MM (2021). Sensory cilia act as a specialized venue for regulated extracellular vesicle biogenesis and signaling. Curr. Biol 31, 3943–3951.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Q, He B, Weiberg A, Buck AH, and Jin H (2019). Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog 15, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drurey C, and Maizels RM (2021). Helminth extracellular vesicles: Interactions with the host immune system. Mol. Immunol 137, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science (80-. ) 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razzauti A, and Laurent P (2021). Ectocytosis prevents accumulation of ciliary cargo in C. elegans sensory neurons. Elife 10, e67670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, Mao Y, Fromme JC, and Emr SD (2015). Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. Elife 4, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee EY, Joo YB, Gun WP, Choi DS, Ji SK, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, et al. (2007). Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7, 3143–3153. [DOI] [PubMed] [Google Scholar]
- 18.Chutkan H, MacDonald I, Manning A, and Kuehn MJ (2013). Quantitative and Qualitative Preparations of Bacterial Outer Membrane Vesicles. In Methods in Molecular Biology, pp. 259–272. [DOI] [PMC free article] [PubMed]
- 19.Beer KB, Rivas-Castillo J, Kuhn K, Fazeli G, Karmann B, Nance JF, Stigloher C, and Wehman AM (2018). Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc. Natl. Acad. Sci. U. S. A 115, E1127–E1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liégeois S, Benedetto A, Michaux G, Belliard G, and Labouesse M (2007). Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics 175, 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosinski M, McDonald K, Schwartz J, Yamamoto I, and Greenstein D (2005). C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132, 3357–3369. [DOI] [PubMed] [Google Scholar]
- 22.Cohen JD, Sparacio AP, Belfi AC, Forman-Rubinsky R, Hall DH, Maul-Newby H, Frand AR, and Sundaram MV (2020). A multi-layered and dynamic apical extracellular matrix shapes the vulva lumen in Caenorhabditis elegans. Elife 9, 1–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, Taub D, Parker JA, Neri C, Gabel CV, et al. (2017). C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oren-Suissa M, Gattegno T, Kravtsov V, and Podbilewicz B (2017). Extrinsic repair of injured dendrites as a paradigm for regeneration by fusion in Caenorhabditis elegans. Genetics 206, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al. (2014). Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun 5, 5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebbing A, Vértesy Á, Betist MC, Spanjaard B, Junker JP, Berezikov E, van Oudenaarden A, and Korswagen HC (2018). Spatial Transcriptomics of C. elegans Males and Hermaphrodites Identifies Sex-Specific Differences in Gene Expression Patterns. Dev. Cell 47, 801–813.e6. [DOI] [PubMed] [Google Scholar]
- 27.Maguire JE, Silva M, Nguyen KCQ, Hellen E, Kern AD, Hall DH, and Barr MM (2015). Myristoylated CIL-7 regulates ciliary extracellular vesicle biogenesis. Mol. Biol. Cell 26, 2823–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clupper M, Gill R, Elsayyid M, Touroutine D, Jeffrey L, and Tanis JE (2021). Kinesin-II motors differentially impact biogenesis of distinct extracellular vesicle subpopulations shed from C. elegans sensory cilia. BioRxiv Available at: 10.1101/2021.12.19.473369. [DOI] [PMC free article] [PubMed]
- 29.Venancio TM, and Aravind L (2010). CYSTM, a novel cysteine-rich transmembrane module with a role in stress tolerance across eukaryotes. Bioinformatics 26, 149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Froehlich JJ, Rajewsky N, and Ewald CY (2021). Estimation of C. elegans cell- and tissue volumes. microPublication Biol 2021, 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva M, Morsci N, Nguyen KCQ, Rizvi A, Rongo C, Hall DH, and Barr MM (2017). Cell-Specific α-Tubulin Isotype Regulates Ciliary Microtubule Ultrastructure, Intraflagellar Transport, and Extracellular Vesicle Biology. Curr. Biol 27, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juhl AD, Anvarian Z, Berges J, Wüstner D, and Lotte B (2021). Transient accumulation and bidirectional movement of KIF13B in primary cilia. BioRxiv Available at: 10.1101/2021.07.09.451768v1.full. [DOI] [PubMed]
- 33.Verweij FJ, Balaj L, Boulanger CM, Carter DRF, Compeer EB, D’Angelo G, El Andaloussi S, Goetz JG, Gross JC, Hyenne V, et al. (2021). The power of imaging to understand extracellular vesicle biology in vivo. Nat. Methods 18, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEwan DL, Weisman AS, and Hunter CP (2012). Uptake of Extracellular Double-Stranded RNA by SID-2. Mol. Cell 47, 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braukmann F, Jordan D, and Miska E (2017). Artificial and natural RNA interactions between bacteria and C. elegans. RNA Biol 14, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuez I, and Félix M-A (2012). Evolution of Susceptibility to Ingested Double-Stranded RNAs in Caenorhabditis Nematodes. PLoS One 7, e29811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulsen JB, Andersen KR, Kjær KH, Vestergaard AL, Justesen J, and Martensen PM (2012). Characterization of human phosphodiesterase 12 and identification of a novel 2′−5′ oligoadenylate nuclease - The ectonucleotide pyrophosphatase/phosphodiesterase 1. Biochimie 94, 1098–1107. [DOI] [PubMed] [Google Scholar]
- 38.Stefan C, Jansen S, and Bollen M (2005). NPP-type ectophosphodiesterases: Unity in diversity. Trends Biochem. Sci 30, 542–550. [DOI] [PubMed] [Google Scholar]
- 39.Roberts F, Zhu D, Farquharson C, and Macrae VE (2019). ENPP1 in the Regulation of Mineralization and Beyond. Trends Biochem. Sci 44, 616–628. [DOI] [PubMed] [Google Scholar]
- 40.Coveney CR, Zhu L, Miotla‐Zarebska J, Stott B, Parisi I, Batchelor V, Duarte C, Chang E, McSorley E, Vincent TL, et al. (2022). Role of Ciliary Protein Intraflagellar Transport Protein 88 in the Regulation of Cartilage Thickness and Osteoarthritis Development in Mice. Arthritis Rheumatol 74, 49–59. Available at: 10.1002/art.41894. [DOI] [PubMed] [Google Scholar]
- 41.Sonneville R, Querenet M, Craig A, Gartner A, and Julian Blow J (2012). The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol 196, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casar Tena T, Maerz LD, Szafranski K, Groth M, Blätte TJ, Donow C, Matysik S, Walther P, Jeggo PA, Burkhalter MD, et al. (2019). Resting cells rely on the DNA helicase component MCM2 to build cilia. Nucleic Acids Res 47, 134–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Wang Q, Dong C, Chen S, Qi Y, and Liu Y (2015). Whole exome sequencing identified MCM2 as a novel causative gene for autosomal dominant nonsyndromic deafness in a Chinese family. PLoS One 10, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chavez DR, Snow AK, Smith JR, and Stanfield GM (2018). Soma-germ line interactions and a role for muscle in the regulation of C. elegans sperm motility. Development 145, dev167734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gatti JL, Métayer S, Belghazi M, Dacheux F, and Dacheux JL (2005). Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol. Reprod 72, 1452–1465. [DOI] [PubMed] [Google Scholar]
- 46.Mogielnicka-Brzozowska M, Prochowska S, Niżański W, Bromke MA, Wiśniewski J, Olejnik B, Kuzborska A, Fraser L, Młynarz P, and Kordan W (2020). Proteome of cat semen obtained after urethral catheterization. Theriogenology 141, 68–81. [DOI] [PubMed] [Google Scholar]
- 47.Kim B, Suo B, and Emmons SW (2016). Gene Function Prediction Based on Developmental Transcriptomes of the Two Sexes in C. elegans. Cell Rep 17, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phiri AM, De Pomerai D, Buttle DJ, and Behnke JMB (2014). Developing a rapid throughput screen for detection of nematicidal activity of plant cysteine proteinases: The role of Caenorhabditis elegans cystatins. Parasitology 141, 164–180. [DOI] [PubMed] [Google Scholar]
- 49.Peracchi A, Veiga-Da-Cunha M, Kuhara T, Ellens KW, Paczia N, Stroobant V, Seliga AK, Marlaire S, Jaisson S, Bommer GT, et al. (2017). Nit1 is a metabolite repair enzyme that hydrolyzes deaminated glutathione. Proc. Natl. Acad. Sci. U. S. A 114, E3233–E3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenvall J, Fierro-González JC, Swoboda P, Saamarthy K, Cheng Q, Cacho-Valadez B, Arnér ESJ, Persson OP, Miranda-Vizuete A, and Tuck S (2011). Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 108, 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liégeois S, Benedetto A, Garnier JM, Schwab Y, and Labouesse M (2006). The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol 173, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawasaki I, Jeong MH, Yun YJ, Shin YK, and Shim YH (2013). Cholesterol-responsive metabolic proteins are required for larval development in Caenorhabditis elegans. Mol. Cells 36, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Na K, Lee HJ, Lee EY, and Paik YK (2011). Contribution of sams-1 and pmt-1 to lipid homoeostasis in adult Caenorhabditis elegans. J. Biochem 149, 529–538. [DOI] [PubMed] [Google Scholar]
- 54.Loer CM, Davidson B, and McKerrow J (1999). A phenylalanine hydroxylase gene from the nematode C. elegans is expressed in the hypodermis. J. Neurogenet 13, 157–180. [DOI] [PubMed] [Google Scholar]
- 55.Loer CM, Calvo AC, Watschinger K, Werner-Felmayer G, O’Rourke D, Stroud D, Tong A, Gotenstein JR, Chisholm AD, Hodgkin J, et al. (2015). Cuticle integrity and biogenic amine synthesis in Caenorhabditis elegans require the cofactor tetrahydrobiopterin (BH4). Genetics 200, 237–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fisher AL, Page KE, Lithgow GJ, and Nash L (2008). The Caenorhabditis elegans K10C2.4 gene encodes a member of the fumarylacetoacetate hydrolase family: A Caenorhabditis elegans model of type I tyrosinemia. J. Biol. Chem 283, 9127–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esposito R, D’Aniello S, Squarzoni P, Pezzotti MR, Ristoratore F, and Spagnuolo A (2012). New insights into the evolution of metazoan tyrosinase gene family. PLoS One 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen JD, and Sundaram MV (2020). C. elegans apical extracellular matrices shape epithelia. J. Dev. Biol 8, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lažetić V, and Fay DS (2017). Molting in C. elegans. Worm 6, e1330246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnold A, Rahman MM, Lee MC, Muehlhaeusser S, Katic I, Gaidatzis D, Hess D, Scheckel C, Wright JE, Stetak A, et al. (2014). Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res 42, 13353–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youngman EM, and Claycomb JM (2014). From early lessons to new frontiers: The worm as a treasure trove of small RNA biology. Front. Genet 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sundby AE, Molnar RI, and Claycomb JM (2021). Connecting the Dots: Linking Caenorhabditis elegans Small RNA Pathways and Germ Granules. Trends Cell Biol 31, 387–401. [DOI] [PubMed] [Google Scholar]
- 64.Russell JC, Kim TK, Noori A, Merrihew GE, Robbins JE, Golubeva A, Wang K, MacCoss MJ, and Kaeberlein M (2020). Composition of Caenorhabditis elegans extracellular vesicles suggests roles in metabolism, immunity, and aging. GeroScience 42, 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duguet TB, Soichot J, Kuzyakiv R, Malmström L, and Tritten L (2020). Extracellular Vesicle-Contained microRNA of C. elegans as a Tool to Decipher the Molecular Basis of Nematode Parasitism. Front. Cell. Infect. Microbiol 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood CR, Huang K, Diener DR, and Rosenbaum JL (2013). The Cilium Secretes Bioactive Ectosomes. Curr. Biol 23, 906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volz A-K, Frei A, Kretschmer V, de Jesus Domingues AM, Ketting RF, Ueffing M, Boldt K, Krämer-Albers E-M, and May-Simera HL (2021). Bardet-Biedl syndrome proteins modulate the release of bioactive extracellular vesicles. Nat. Commun 12, 5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chow FWN, Koutsovoulos G, Ovando-Vázquez C, Neophytou K, Bermúdez-Barrientos JR, Laetsch DR, Robertson E, Kumar S, Claycomb JM, Blaxter M, et al. (2019). Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res 47, 3594–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jung E, Choi TI, Lee JE, Kim CH, and Kim J (2020). ESCRT subunit CHMP4B localizes to primary cilia and is required for the structural integrity of the ciliary membrane. FASEB J 34, 1331–1344. [DOI] [PubMed] [Google Scholar]
- 70.Ott C, Nachmias D, Adar S, Jarnik M, Sherman S, Birnbaum RY, Lippincott-Schwartz J, and Elia N (2018). VPS4 is a dynamic component of the centrosome that regulates centrosome localization of γ-tubulin, centriolar satellite stability and ciliogenesis. Sci. Rep 8, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diener DR, Lupetti P, and Rosenbaum JL (2015). Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol 25, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Long H, Zhang F, Xu N, Liu G, Diener DR, Rosenbaum JL, and Huang K (2016). Comparative Analysis of Ciliary Membranes and Ectosomes. Curr. Biol 26, 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Höög JL, and Lötvall J (2015). Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J. Extracell. Vesicles 4, 28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Corrigan L, Redhai S, Leiblich A, Fan SJ, Perera SMW, Patel R, Gandy C, Mark Wainwright S, Morris JF, Hamdy F, et al. (2014). BMP-regulated exosomes from Drosophila male reproductive glands reprogram female behavior. J. Cell Biol 206, 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, and Rando OJ (2018). Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev. Cell 46, 481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi C, and Murphy CT (2021). Sex and death. In Current Topics in Developmental Biology, pp. 353–375. [DOI] [PubMed]
- 77.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Granato M, Schnabel H, and Schnabel R (1994). pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res 22, 1762–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Craig R, and Beavis RC (2004). TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467. [DOI] [PubMed] [Google Scholar]
- 80.Gupta N, Bandeira N, Keich U, and Pevzner PA (2011). Target-decoy approach and false discovery rate: When things may go wrong. J. Am. Soc. Mass Spectrom 22, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust Genome Editing with Short Single-Stranded. Genetics 210, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, and Ahringer J (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 83.Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, and Vermeulen M (2018). Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc 13, 530–550. [DOI] [PubMed] [Google Scholar]
- 84.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morsci NS, and Barr MM (2011). Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of caenorhabditis elegans. Curr. Biol 21, 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Lists of identified peptides and proteins, related to Figure 2.
(A) Metadata on the overall numbers of peptides and identified proteins from C. elegans and E. coli. (B) Unique peptides with matched proteins. (C) Non-unique peptides matched with corresponding protein sets. (D) Summary of spectral counts of unique proteins for each gene product. Functions of the genes and human orthologs are indicated.
Data S2. Differential abundance and enrichment analyses, related to Figure 2.
(A-B) Differential abundance analyses of the light and heavy EV proteomes: KEGG pathway (A) and InterPro protein domain (B) enrichment analyses. (C) List of ESCRT proteins identified in the EV proteome. (D) List of nucleic acid binding proteins identified in the EV proteome.
Data S3. Mining of the EV proteome, related to Figure 2.
(A) Overlap of the EVome with the EVN-specific transcriptomes used to produce Figure 2C. (B) IL2 identification table contains relative enrichment values for cholinergic clusters from Cao et al. (2017). (C) EV cargo scores based on their absolute enrichment in ciliated sensory neurons and relative enrichment in the IL2 neurons of hermaphroditic L2 populations; these scores were used to generate the plot on Figure 2F. (D) Overlap of the identified EV proteome with the male-specific transcriptome from [47].
Video S1. ENPP-1::mScarlet EV release from the IL2 cilia into the environment, related to Figure 4 and S5.
During the time lapse focus changes from the cilium to EV plane over an axial distance of approximately 1 μm. EVs are connected to each other in a tandem manner and remain attached to the cilium during the course of the video.
Video S2. ENPP-1::mScarlet appearance in the vacuoles of the vas deferens, related to Figure 6.
Animated Z-stack.
Video S3. ENPP-1::mScarlet release from the male tail during mating to a hermaphrodite, related to Figure 6.
The footage shows two males mating with one hermaphrodite. Male #1 releases ENPP-1::mScarlet while backing along the body of the hermaphrodite scanning for vulva, whereas Male #2 releases ENPP-1::mScarlet during sperm transfer.
Data Availability Statement
Raw proteomics data generated in this study have been deposited at MassIVE and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original raw image Z-stacks have been deposited to Figshare and are publicly available as of the date of publication. The DOI is listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


