Abstract
Introduction
Whole-exome sequencing (WES) has identified de novo variants in chromatin remodelling genes in patients with neurodevelopmental disorders (NDD). We report on a novel genetic discovery in chromatin remodelling in patients with NDD who also have corpus callosum (CC) anomalies.
Objective
To discover novel genes linked to both CC anomalies and NDD.
Methods
Clinical WES was performed for evaluation of NDD, identifying five patients with de novo variants in SUPT16H, a subunit of the FACT (facilitates chromatin transcription) complex. The clinical phenotypes, genetic results and brain MRIs were obtained and systematically reviewed. In silico protein function predictions were assessed and allele frequencies in control populations were compared.
Results
We identified four patients with de novo missense variants in SUPT16H and one patient with a de novo deletion including SUPT16H. These variants were not reported in the updated Genome Aggregation Database. When assayable, all protein products were predicted to be damaging. Symptoms included intellectual disability, autistic features, minor dysmorphic features and seizures. Anomalies of the CC were seen in all three patients with available brain imaging.
Conclusion
Our findings implicate the gene SUPT16H in a novel disorder characterised by neurodevelopmental deficits and CC anomalies.
INTRODUCTION
Corpus callosum (CC) anomalies are relatively common, seen in as many as 5% of children with neurodevelopmental disorders (NDD).1 Whole-exome sequencing (WES) studies have identified many genes with pathogenic de novo variants in children with NDD and suggest this approach will be informative for patients with CC anomalies. NDD-associated genes are found in transcription regulation and chromatin remodelling pathways.2 3
To understand the biology of disorders of CC development, WES and CNV data were analysed from clinical and research genetics of CC anomalies.4 Using similar approaches, we previously identified novel genes such as RERE, AKT3, ZBTB18 (ZNF238), DISC1, KIF1A and NF1A.5-9 We have now identified a cohort of individuals with de novo variants in the gene SUPT16H, encoding a subunit of the FACT (facilitates chromatin transcription) complex, a histone chaperone complex that regulates DNA replication, transcription and DNA repair. In our cohort, neurobehavioural challenges were prominent, including global developmental delay (GDD), autistic features and epilepsy. CC anomalies were noted in the three patients for whom brain MRIs were available for review. These consistent features in individuals with SUPT16H de novo variants point to a novel CC dysgenesis disorder.
METHODS
To investigate the cause of NDD, clinical WES and CNV analysis were performed. We recruited five patients who had de novo SUPT16H variants. Patients 1 and 5 were initially recruited through the University of California, San Francisco Brain Development Research Program (brain.ucsf.edu). Patients 2, 3 and 4 were identified through GeneMatcher.10 The clinical phenotypes of these individuals and their genetic information were determined through record review. Three patients had available brain MRIs, which were systematically evaluated by a paediatric neuroradiologist, as previously described.11 In silico pathogenicity predictions were assessed using CAAD score (Combined Annotation Dependent Depletion), MutationTaster, SIFT and PolyPhen-2. Allele frequencies in control populations were compared with the Genome Aggregation Database (gnomAD).
RESULTS
Patients
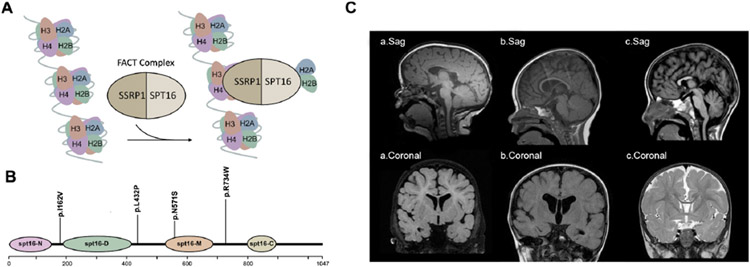
Detailed clinical features and identified genetic variants in our cohort are described and summarised in table 1. Representative brain imaging findings of the patients and a schematic of the protein encoded by SUPT16H are shown in figure 1.
Table 1.
Clinical and genetic findings in patients with SUPT16H variants
| Patient | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| SUPT16H-related variants | c.2200 C>T (p.R734W) | c.1712A>G (p.N571S) | c.1295T>C (p.L432P) | c.484A>G (p.I162V) | 2.05 Mb chr14q.11.2 del |
| Variant type | Missense | Missense | Missense | Missense | Deletion |
| Inheritance | De novo | De novo | De novo | De novo | De novo |
| Polyphen-2 | Probably damaging | Probably damaging | Probably damaging | Benign | NA |
| Polyphen-2 score | 0.998 | 1 | 0.997 | 0 | NA |
| SIFT | Damaging | Damaging | Damaging | Tolerated | NA |
| SIFT score | 0 | 0.04 | 0.01 | 1 | NA |
| MutationTaster | Disease-causing | Disease-causing | Disease-causing | Disease-causing | NA |
| CAAD raw score | 3.92263 | 3.773879 | 4.03 | 1.051617 | NA |
| CAAD PHRED score | 27.3 | 26.3 | 28.3 | 13.61 | NA |
| Alleles in updated gnomAD | None | None | None | None | NA |
| Other variants | ~336 kb 20q13.2 microdeletion—paternally inherited | − | 4q13.3 microduplication—maternally inherited | − | 30.17 Mb18p11.32q12.1dup |
| Sex | Female | Male | Male | Female | Female |
| Age (years) | 8 | 12 | 5 | 2 | 14 |
| Gross motor delay | + | + | + | + | + |
| Speech | Non-verbal | Non-verbal | Speech delay | Speech delay | Non-verbal |
| Cognitive delay/intellectual disability | Severe | Severe | Mild-moderate | Mild-moderate | Severe |
| Autistic features | + | + | − | Not assessed | − |
| Seizures | − | − | − | Simple partial | Complex partial |
| Spine | Spina bifida, tethered cord | − | − | − | Scoliosis |
| Sleep disturbance | + | + | − | − | + |
| Callosal anomalies | Thin CC | NA | NA | Thin CC | Partial ACC (absent posterior body and splenium) |
| Decreased white matter volume | + | NA | NA | + | + |
| Head | Frontal bossing, dolichocephaly | − | − | Plagiocephaly | Tall forehead, frontal bossing |
| Eye | Hypertelorism, epicanthal folds | Down-slanting palpebral fissures | Strabismus | − | Hypertelorism, down-slanting palpebral fissures |
| Ear | Thick helices, slightly posteriorly rotated and low-set | − | − | Cupped, dysplastic ears | Dysplastic ears |
| Nose | − | Broad nasal bridge | − | − | Wide nasal bridge and tip |
| Mouth | − | Bifid uvula, prominent cupid’s bow | − | − | Small mouth with full lips |
| Other findings | − | Tapered fingers | − | Right facial palsy | − |
| Precocious puberty | − | + | − | − | + |
| Congenital heart defects | − | − | − | + | + |
| Gastrointestinal | Constipation | History of feeding difficulties | − | G-tube placement | Constipation, GERD, congenital choledochal cyst/cholecystectomy, caecostomy tube placement, ileostomy |
| Others | Dermatographia | − | Asthma, pyramidal syndrome | Torticollis, hydronephrosis, mixed hearing loss | SOD, HGH deficiency, CVID |
ACC, agenesis of the corpus callosum; CC, corpus callosum; CIVD, common variable immunodeficiency; GERD, gastro-oesophageal reflux disease;gnomAD, Genome Aggregation Database; HGH, human growth hormone;NA, not applicable; PHRED, combined annotation dependent depletion, Phil's read editor ; SOD, septo-optic dysplasia.
Figure 1.
Molecular and clinical findings in patients with SUPT16H mutations. (A) A schematic model of the FACT complex interacting with the nucleosome. (B) Diagram of Spt16 protein indicating the location of principal domains and the variants identified. The mutations are grouped according to their locations on Spt16 domains. (C) Callosal abnormalities in selected patients with SUPT16H changes: (a) T1-weighted sagittal and coronal images demonstrating a thin corpus callosum and diminished white matter volume in patient 1; (b) T1-weighted sagittal and coronal images demonstrating a thin corpus callosum and enlarged lateral ventricles in patient 4; (c) T1 sagittal and T2 FLAIR coronal images demonstrating partial agenesis of the corpus callosum in patient 5. FACT, facilitates chromatin transcription; FLAIR, fluid-attenuated inversion recovery.
Patient 1 is an 8-year-old girl born at term via planned caesarean section to a non-consanguineous couple following an uncomplicated pregnancy. Her birth weight was 3629 g (67th percentile), length was 51 cm (69th percentile) and head circumference was 35 cm (54th percentile). Her 4-day neonatal hospital course was complicated by thick meconium, pneumothorax and respiratory distress. Over time, she was noted to have GDD: sitting at 10 months, walking at 29 months and no language acquisition by 3 years. Brain (figure 1C) and spine MRI at 7 months reported a thin CC with a dysmorphic splenium, diminished supratentorial white matter volume (although less reduction than the CC), fatty filum at L5–S1 with spina bifida and tethered cord. She had early accelerated head growth that returned to normal in ensuing months. She was also noted to have sleep disturbance not further specified. At her last evaluation at 5 years of age, growth parameters included weight of 21.3 kg (68th percentile), height of 113 cm (48th percentile) and head circumference of 52.5 cm (94th percentile). Physical exam revealed dolichocephaly, frontal bossing, hypertelorism, bilateral epicanthal folds, thickened helices and slightly posteriorly rotated and low-set ears, and dermatographia. In terms of development, she was only able to follow simple two-step directives and walk independently. In terms of her behaviour, she made no eye contact and had repetitive obsessions with very minimal language. A microarray revealed a 336kb deletion at 20q13.2 inherited from her asymptomatic father. WES revealed a de novo [NM_007192.3] c.2200C<T, p.(Arg734Trp) missense variant in SUPT16H.
Patient 2 is a 12-year-old boy born at 36 weeks’ gestation via caesarean section with a birth weight of 2460 g (27th percentile) to a non-consanguineous couple complicated by maternal preeclampsia. His neonatal course included difficulties feeding and sucking, initially requiring nasogastric tube feeds. He was then diagnosed with GDD, walking at 24 months and had minimal language development that subsequently regressed. Later he was noted to have intellectual disability, autistic-like behaviours and sleeping difficulties. His craniofacial features included bifid uvula, broad nasal bridge, slight down-slanting palpebral fissures, marked cupid’s bow with abrupt stop of the upper and lower vermillion borders laterally, and tapered fingers. Brain MRI performed at 2.5 years indicated increased extra-axial fluid and prominent sulci, consistent with cerebral atrophy. However, images were not available for our review. The last clinical evaluation at 8 years of age revealed precocious puberty and growth parameters including a head circumference of 56 cm (95th percentile) and length of 142 cm (99th percentile). His diagnostic work-up included a negative microarray and negative methylation-sensitive MLPA (Multiplex Ligation-dependent Probe Amplification) testing for Angelman syndrome. WES revealed a de novo [NM_007192.3] c.1712A>G, p.(Asn571Ser) missense variant in SUPT16H.
Patient 3 is a 5-year-old boy born at 38 weeks’ gestation via vaginal delivery with a birth weight of 4020 g (>90th percentile; 50th percentile for 1 month of age) to a non-consanguineous couple. Pregnancy was complicated by gestational diabetes. A mild pyramidal syndrome was confirmed. He presented with GDD with delays in walking (18 months), speech and cognition. He was also noted to have mild strabismus and asthma. Brain MRI was not available for our review. Chromosomal microarray showed a 4q13.3 microduplication which was inherited from his healthy mother. Metabolic screening was normal. WES revealed a de novo c.1295T>C, p.(Leu432Pro) missense variant in SUPT16H.
Patient 4 is a 2-year-old girl born to non-consanguineous parents at 36 weeks’ gestation via vaginal delivery in the setting of premature rupture of membranes. The pregnancy was notable for abnormal ultrasound findings of a two-vessel cord and congenital heart defect. Her birth weight was 2353 g (28th percentile), length was 46 cm (39th percentile) and head circumference was 30 cm (5th percentile).
Postnatal echocardiogram revealed an atrioventricular canal defect, ventricular septal defect, coarctation of the aorta, patent ductus arteriosus and atrial septal defect. A head ultrasound in infancy was notable for partial agenesis of the CC. A brain MRI obtained at 13 months of age showed a thin CC (figure 1C). She was noted to have outer-ear malformations bilaterally (cupped left ear with a flattened helix; absent superior two-thirds of the right helix), torticollis, unilateral grade 1 hydronephrosis, plagiocephaly and right lower facial palsy. Additionally, she had moderate to severe mixed hearing loss, speech delay, focal seizures and feeding difficulties requiring G-tube placement. At 15 months of age, her height was 73.5 cm (13th percentile), weight was 9.6 kg (24th percentile) and head circumference was 45 cm (26th percentile). She was taking steps with support and babbling at that time. Diagnostic work-up included WES which revealed a de novo [NM_007192.3] c.484A>G, p.(Ile162Val) missense variant in SUPT16H.
Patient 5 is a 14-year-old girl born at 35 weeks’ gestation with a birth weight of 1360 g (<1st percentile; 50th percentile for 30 weeks) to non-consanguineous parents via caesarean section due to fetal distress. She was hospitalised in the neonatal intensive care unit for 5 weeks due to growth and feeding difficulties, jaundice and cardiac anomalies. Over time, she was found to have septo-optic dysplasia, intractable complex partial epilepsy and mild cortical vision impairment. Additionally, she was noted to have congenital heart defects, neuromuscular scoliosis, common variable immunodeficiency, cyclical emesis syndrome, choledochal cyst and erosive oesophagitis associated with hiatal hernia.
She was noted to have GDD: sitting at 18 months, walking at 3 years and running at 5. Cognition and language were delayed with no appreciable words. Brain MRI obtained at 22 months revealed partial agenesis of the CC with absence of the splenium and inferior genu, and markedly diminished white matter volume (figure 1C).
Last evaluation at 13 years of age revealed weight of 36.4 kg (8th percentile), height of 153.8 cm (32nd percentile) and head circumference of 54 cm (62nd percentile). Craniofacial features included a tall forehead and frontal bossing, high anterior hair line, hypertelorism with slightly down-slanting palpebral fissures and intermittent left esotropia, slightly overfolded helices, wide nasal bridge and tip, and small mouth with full lips and mild underbite. She had difficulty with balance and ambulation and used an iPad device for augmentative communication. Diagnostic work-up included negative WES and a microarray revealing an unbalanced copy number loss of chromosome bands 14q11.1q11.2 of approximately 2.05 Mb in size (chr14: 20511672–22562282) including SUPT16H and a copy number gain of chromosome bands 18p11.32q12.1 of approximately 30.17 Mb in size.
DISCUSSION
We identified five individuals with NDD with de novo variants in the chromatin regulator gene, SUPT16H, three of whom had CC anomalies. All participants had developmental delay, including speech and cognitive delay, and later a number had intellectual disability and autistic-like behaviours. All brain MRIs available for our review showed CC anomalies. Most patients had minor dysmorphic features including tall forehead, down-slanting palpebral fissures, ear anomalies and broad nasal bridge. Other common clinical features included seizures, sleeping difficulties and precocious puberty (table 1).
Four individuals in our cohort had de novo missense variants in SUPT16H and one had a de novo 2.05 Mb deletion including SUPT16H. The variants identified in our study were not previously reported in gnomAD. In silico pathogenicity prediction tools such as CAAD score, MutationTaster, PolyPhen-2 and SIFT provided further causal evidence for these variants (table 1).
SUPT16H encodes a subunit of FACT, a heterodimer protein complex implicated in DNA replication, transcription and repair12 (figure 1A). The two subunits of FACT, Spt16 and SSRP1, are both essential for histone regulation.13 Spt16, the subunit encoded by SUPT16H, interacts with the histone dimer H2A-H2B in the nucleosome during transcription, allowing RNA polymerase access to the previously coiled DNA. Both subunits of the FACT complex are highly conserved among all eukaryotes.14 15 As such, the Spt16 protein is predicted to be highly intolerant to loss-of-function (LoF) variants, with high probability of loss-of-function intolerance (pLI=1.0), and intolerant for missense mutations, with a calculated Z score of 5.53 for missense variants (exac.broadinstitute.org; Lek et al 201616).
Accordingly, deleterious effects are anticipated from disruptions in SUPT16H, providing further evidence for the role of SUPT16H in disorders of development. SSRP1, the other component of FACT, is also predicted to be highly intolerant to LoF and missense variants, respectively (pLI: 1:00; Z score=4.53; exac.broadinstitute.org; Lek et al 2016), although a role for SSRP1 in human disease has yet to be delineated.
No reports prior to this study linked SUPT16H to NDD. However, microdeletions and duplications of 14q11.2 are reported to be associated with ID (intellectual disability), ASD (autism spectrum disorder), macrocephaly and minor dysmorphic features.17-19 For CNV deletions previously reported in this region, CHD8 and SUPT16H have been proposed as the two candidate genes (as these two genes were the only ones found in a minimal critical region in this interval). CHD8 has also been proposed as a causative candidate gene because de novo point variations in CHD8 have been linked to both macrocephaly and ASD.20 21 In contrast, SUPT16H has been considered as a possible candidate gene because it is adjacent to CHD8 and for the established role of chromatin regulator in NDD. Interestingly, one of these prior patients with 14q deletions was reported to have CC hypoplasia. Our patient with a 2.05 Mb deletion of 14q11.2 also has a 30.17 Mb copy number gain of 18p11.32q12.1. Of note, partial trisomy 18p has not been reported in association with major malformations or complications. Rather, it is often associated with limited clinical consequence. While we cannot rule out its contribution to our patient’s phenotype, this patient notably had other anomalies including partial agenesis of CC. In our cohort, callosal malformations were a common finding but macrocephaly was not. Thus, we propose that disruptions in SUPT16H are sufficient to cause callosal abnormalities, while disruptions in CHD8 are associated with macrocephaly. As seen in other disorders of chromatin regulation, developmental delay and ASD are common associations.
In conclusion, our findings of five individuals with NDD and CC anomalies implicate de novo variants in SUPT16H as the cause of their findings and suggest a novel disorder of chromatin and transcription dysregulation.
Acknowledgements
We wish to thank the patients and their families for participating in this study.
Funding
This work was supported by a grant to ES from the National Institute of Neurological Disorders (NINDS) (5R01NS058721-10).
Footnotes
Ethics approval This study was approved by site-specific institutional review boards, and informed consent was obtained from all individuals.
The results were initially presented at the Society for Neuroscience Annual Meeting, 2018.
REFERENCES
- 1.Paul LK, Brown WS, Adolphs R, Tyszka JM, Richards LJ, Mukherjee P, Sherr EH. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci 2007;8:287–99. [DOI] [PubMed] [Google Scholar]
- 2.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet 2013;14:347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimäki T, Lin C-F, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RKC, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD, TW Y, Study DDD, DDD Study, Homozygosity Mapping Collaborative for Autism, UK10K Consortium. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014;515:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sajan SA, Fernandez L, Nieh SE, Rider E, Bukshpun P, Wakahiro M, Christian SL, Rivière J-B, Sullivan CT, Sudi J, Herriges MJ, Paciorkowski AR, Barkovich AJ, Glessner JT, Millen KJ, Hakonarson H, Dobyns WB, Sherr EH. Both rare and de novo copy number variants are prevalent in agenesis of the corpus callosum but not in cerebellar hypoplasia or polymicrogyria. PLoS Genet 2013;9:e1003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fregeau B, Kim BJ, Hernández-García A, Jordan VK, Cho MT, Schnur RE, Monaghan KG, Juusola J, Rosenfeld JA, Bhoj E, Zackai EH, Sacharow S, Barañano K, Bosch DGM, de Vries BBA, Lindstrom K, Schroeder A, James P, Kulch P, Lalani SR, van Haelst MM, van Gassen KLI, van Binsbergen E, Barkovich AJ, Scott DA, Sherr EH. De novo mutations of RERE cause a genetic syndrome with features that overlap those associated with proximal 1p36 deletions. The American Journal of Human Genetics 2016;98:963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boland E, Clayton-Smith J, Woo VG, McKee S, Manson FDC, Medne L, Zackai E, Swanson EA, Fitzpatrick D, Millen KJ, Sherr EH, Dobyns WB, Black GCM. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase Akt3 in postnatal microcephaly and agenesis of the corpus callosum. The American Journal of Human Genetics 2007;81:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osbun N, Li J, O’Driscoll MC, Strominger Z, Wakahiro M, Rider E, Bukshpun P, Boland E, Spurrell CH, Schackwitz W, Pennacchio LA, Dobyns WB, Black GCM, Sherr EH. Genetic and functional analyses identify DISC1 as a novel callosal agenesis candidate gene. Am J Med Genet A 2011;155:1865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esmaeeli Nieh S, Madou MRZ, Sirajuddin M, Fregeau B, McKnight D, Lexa K, Strober J, Spaeth C, Hallinan BE, Smaoui N, Pappas JG, Burrow TA, McDonald MT, Latibashvili M, Leshinsky-Silver E, Lev D, Blumkin L, Vale RD, Barkovich AJ, Sherr EH. De novo mutations in KIF1A cause progressive encephalopathy and brain atrophy. Ann Clin Transl Neurol 2015;2:623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu W, Quintero-Rivera F, Fan Y, Alkuraya FS, Donovan DJ, Xi Q, Turbe-Doan A, Li Q-G, Campbell CG, Shanske AL, Sherr EH, Ahmad A, Peters R, Rilliet B, Parvex P, Bassuk AG, Harris DJ, Ferguson H, Kelly C, Walsh CA, Gronostajski RM, Devriendt K, Higgins A, Ligon AH, Quade BJ, Morton CC, Gusella JF, Maas RL. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet 2007;3:e80–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting Investigators with an interest in the same gene. Hum Mutat 2015;36:928–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hetts SW, Sherr EH, Chao S, Gobuty S, Barkovich AJ. Anomalies of the corpus callosum: an Mr analysis of the phenotypic spectrum of associated malformations. AJR Am J Roentgenol 2006;187:1343–8. [DOI] [PubMed] [Google Scholar]
- 12.Winkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 2011;286:18369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. Fact facilitates transcription-dependent nucleosome alteration. Science 2003;301:1090–3. [DOI] [PubMed] [Google Scholar]
- 14.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. Fact, a factor that facilitates transcript elongation through nucleosomes. Cell 1998;92:105–16. [DOI] [PubMed] [Google Scholar]
- 15.Orphanides G, Wu W-H, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor fact comprises human Spt16 and SSRP1 proteins. Nature 1999;400:284–8. [DOI] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, Langlois S, Martin H, Willatt L, Marra MA, Friedman JM. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet 2007;44:556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prontera P, Ottaviani V, Toccaceli D, Rogaia D, Ardisia C, Romani R, Stangoni G, Pierini A, Donti E. Recurrent ~100 Kb microdeletion in the chromosomal region 14q11.2, involving CHD8 gene, is associated with autism and macrocephaly. Am J Med Genet A 2014;164:3137–41. [DOI] [PubMed] [Google Scholar]
- 19.Smyk M, Poluha A, Jaszczuk I, Bartnik M, Bernaciak J, Nowakowska B. Novel 14q11.2 microduplication including the CHD8 and SUPT16H genes associated with developmental delay. Am J Med Genet A 2016;170:1325–9. [DOI] [PubMed] [Google Scholar]
- 20.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012;485:246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnard RA, Pomaville MB, O’Roak BJ, O’Roak BJ. Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front Neurosci 2015;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]