Abstract
Polyketide synthases (PKSs) are predominantly microbial biosynthetic enzymes. They assemble highly potent bioactive natural products from simple carboxylic acid precursors. The most versatile families of PKSs are organized as assembly lines of functional modules. Each module performs one round of precursor extension and optional modification, followed by directed transfer of the intermediate to the next module. While enzymatic domains and even modules of PKSs are well understood, the higher-order modular architecture of PKS assembly lines remains elusive. Here, we visualize a PKS bimodule core using cryo–electron microscopy and resolve a two-dimensional meshwork of the bimodule core formed by homotypic interactions between modules. The sheet-like organization provides the framework for efficient substrate transfer and for sequestration of trans-acting enzymes required for polyketide production.
A microbial polyketide assembly line organizes into a sheet-like supramolecular structure of aligned and mobile domains.
INTRODUCTION
Polyketide synthases (PKSs) are biosynthetic factories, primarily occurring in microbes, for the production of bioactive secondary metabolites, which have applications in medicine as antibiotics, immunosuppressants, and anticancer drugs (1, 2). PKSs assemble polyketides by stepwise condensation and modification of acyl building blocks (3). The most versatile class of PKSs, type I PKSs, is organized into dimeric functional modules. Each module contains a condensing region comprising the condensing ketosynthase (KS) and a substrate-loading acyl transferase (AT) domain. Modules may further comprise an optional modifying region that often contains a double-hotdog-fold domain (DH). DH domains can have different enzymatic functions to act as dehydratases (3), isomerases (4), branching enzymes (5), or epimerases (6). Throughout stepwise polyketide synthesis, the intermediates are tethered to an acyl-carrier protein (ACP) domain. ACPs are integral parts of the multidomain PKS polypeptides and are flanked by flexible peptide linkers. Modular type I PKSs (modPKSs) consist of multiple modules acting as an assembly line. Each module performs one step of chain extension and optional modification of the added moiety and passes on the new intermediate to the KS of the downstream module for the next extension step. Individual modPKS proteins typically consist of two to four modules, and they carry docking domains at their termini, mediating interactions to upstream or downstream modules in the assembly line (Fig. 1A). A complete assembly line commonly consists of more than 10 modules (7, 8).
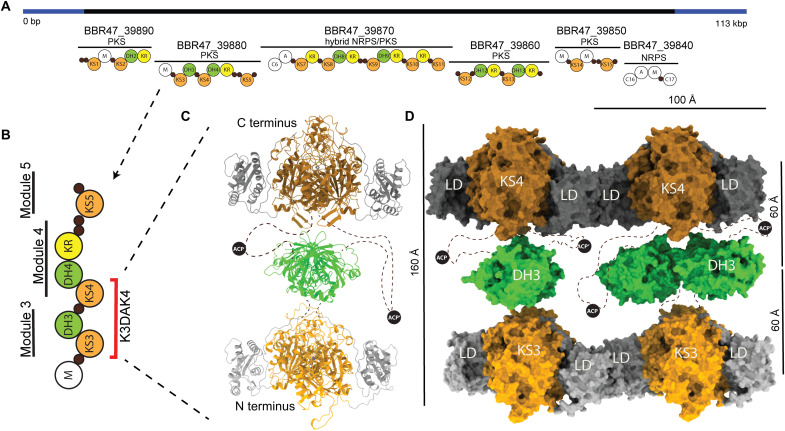
Fig. 1. The architecture of the K3DAK4 bimodule core of the B. brevis BGC11 PKS.
(A) Functional annotation of BGC11 as provided by Antismash (47). Regions coding for PKS and nonribosomal peptide synthetase (NRPS) core enzymes (black) or auxiliary enzymes (blue) are indicated. The domain organization of encoded PKS and NRPS proteins is shown schematically; PKS domains are labeled and shown in color; NRPS condensation (C), adenylation (A), and methylation (M) domains are shown as white circles; and small black circles represent carrier proteins. bp, base pair; kbp, kilo–base pair. (B) Domain and modular organization of the BBR47_39880 protein; the minimal K3DAK4 bimodular fragment containing two subsequent KS domains is indicated in red. (C) Pseudo-atomic model of an isolated dimeric K3DAK4 bimodule core assembled from individual high-resolution structures based on cryo-EM 2D classes (figs. S1B; S2, C to F; and S3B). The ACP is not resolved, and plausible positions are depicted schematically. (D) Surface representation of the K3DAK4 bimodule core (vertical) filament (horizontal) [Protein Data Bank (PDB) 7ZSK]. Individual KS and DH dimer models were rigid body–fitted into the overall intermediate resolution cryo-EM map (map 1 in fig. S1), with DH3 initial models positioned in two different orientations for visualization of rotational disorder, and the mobile ACP is indicated schematically. Distances between the center of mass of dimeric domains (right) and overall dimensions are indicated (left, top).
Two distinct types of modPKSs have evolved that either incorporate AT domains in each module (cis-AT PKSs) (9) or use trans-acting AT domains (trans-AT PKSs) (10). In many modPKSs, the organization of modules is colinear from gene order to protein interaction and biosynthetic reaction step (11), but trans-AT PKSs are known for exceptions to this rule of collinearity (12, 13). In cis-AT PKSs, the AT is connected to the KS by a ~130–amino acid linker domain (LD), the only common nonenzymatic scaffolding domain in modPKSs. Despite the absence of AT domains, trans-AT PKSs still contain LDs. On the basis of homotypic interactions of LDs in crystal structures of isolated KS domains, Keatinge-Clay and colleagues (14) proposed LDs of trans-AT PKSs to form lateral interactions between equivalent KS domains, termed laterally interacting KS sequences (LINKS). Structural data are available on individual PKS domains, two- to three-domain fragments (11) and individual modules of trans-AT (5) and iterative- or cis-AT (15–19) PKSs, but for the higher-order architecture of PKS multimodules, only conceptual models of serpentine (20), helical (21), sheet (14), and linear (22) arrangements have been proposed. Clusters of trans-AT PKSs of up to 0.2-μm size were identified by immunostaining electron microscopy (EM) in Bacillus subtilis (23). ModPKSs are important targets for natural product engineering, but efforts toward reengineering PKS assembly lines thus far are only partially successful (24). They are hampered by a lack of structural insights into the interplay of domains and modules and the architectural requirements for productive multimodular biosynthesis (11). Here, we resolve the organization of a PKS bimodule core by combining crystallographic and cryo-EM analysis of a fragment from a bacterial trans-AT PKS. Our analysis includes structures of isolated domains and of domains in the context of a bimodular assembly line and the higher-order organization of the bimodule core into a LINKS-bridged array.
RESULTS
Structural analysis of a trans-AT PKS bimodule core
We studied the KS3-DH3-ACP-KS4 (K3DAK4) fragment of a 438-kDa four-module protein (GenBank: BBR47_39880) from the orphan biosynthetic gene cluster 11 (BGC11) of Brevibacillus brevis strain NBRC [National Institute of Technology and Evaluation (NITE) Biological Resource Center] 100599 (Fig. 1, A and B). This fragment contains two subsequent KS domains; compared to a minimal PKS bimodule, it only lacks a flexibly tethered terminal ACP. We refer to this fragment as bimodule core. We combined x-ray crystallographic analysis of recombinantly expressed isolated enzymatic domains at 1.6 Å (KS3), 2.2 Å (DH3), and 3.2 Å (KS4) (Figs. 2 and 3 and table S1), with cryo-EM visualization of domains in the context of the bimodule core at 2.9-Å resolution (KS3 and KS4) (Fig. 1C, figs. S1 to S3, and table S2) and the entire bimodule core polymerized into filaments at 6.8-Å resolution with maps focused on subregions at 4.0- and 3.9-Å resolution, respectively (Fig. 1D and figs. S1 to S3). The mobile ACP domains with their linkers remain unresolved.
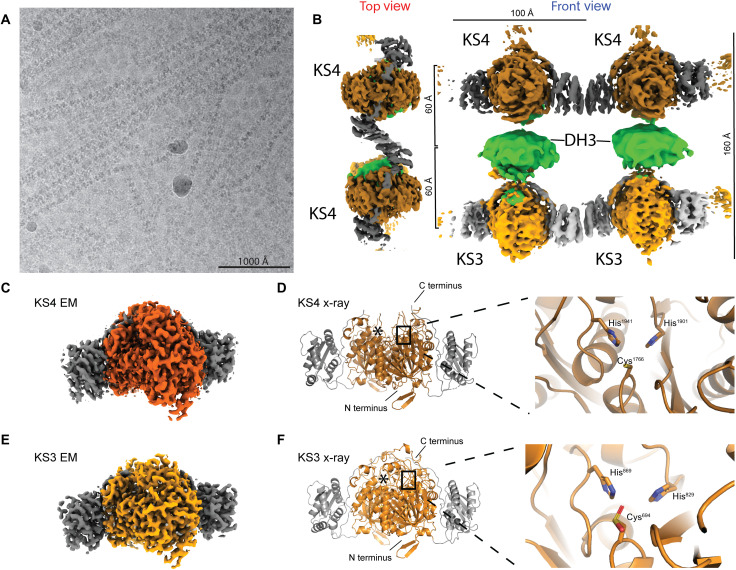
Fig. 2. Cryo-EM analysis of K3DAK4 filaments and individual bimodule cores.
(A) Cryo-EM micrograph from the filament dataset. (B) Hybrid representation of two adjacent bimodule cores within the filament assembled from two focused refinement maps for laterally linked pairs of KS3 (light gray/orange) and KS4 (dark gray/orange) dimers (maps 2 and 3 in fig. S1) and an excised region covering a pair of DH3 dimers (green) from an overall map of the bimodule core filament (map 1 in fig. S1). KS3 and KS4 form homotypic LINKS interactions. In the top view, the KS3 and DH domains are obscured by the KS4 domain (cf. also movie S1). (C to F) Structural analysis of dimeric KS3 and KS4 domains in isolation or in the context of K3DAK4 bimodule cores. (C) Cryo-EM map derived from focused refinement of KS4 in individual bimodule cores at 2.9-Å resolution. (D) X-ray crystallographic model of the isolated KS4 domain at 3.2-Å resolution. Notably, the area around the C terminus of KS4 is better ordered in the cryo-EM map (also see figs. S2 and S3A). (E) Cryo-EM map derived from focused refinement of KS3 in individual bimodule cores at 2.9-Å resolution. (F) X-ray crystallographic model of the KS3 domain at 1.6-Å resolution. The active site cysteine is oxidized in the crystal to a sulfinic acid. Asterisks in (D) and (F) depict position of the second protomers active site.
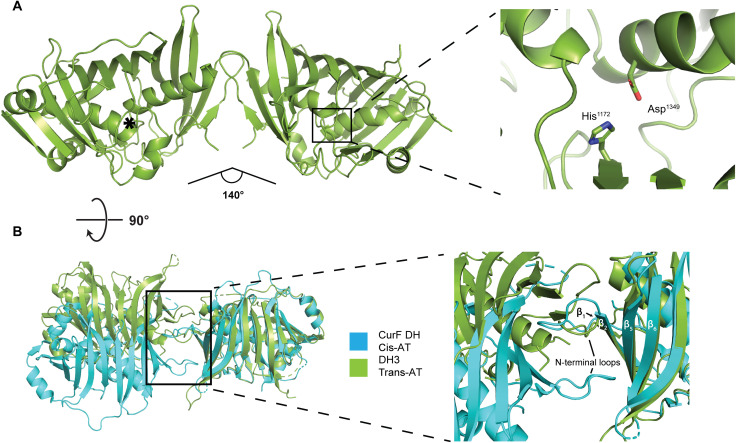
Fig. 3. The dimeric structure of the isolated DH3 domain of K3DAK4.
(A) Overview of the 2.2-Å resolution crystal structure of DH3 (left) and a close-up view of one active site (right). The active site of the second protomer is indicated by an asterisk. The angle between the two protomers is indicated. (B) Comparison of the dimer interface of DH3 (green) with the dehydratase domain of the cis-AT PKS CurF (turquoise). Models are aligned on the right protomer. The inset highlights the different topology of β strand and N-terminal loop interactions in the dimerization interfaces of DH3 and CurF DH.
Individual K3DAK4 bimodule cores adopt a single type of compact structure based on cryo-EM two-dimensional (2D) class analysis (figs. S1B and S3B), a linear arrangement of enzymatic domains (Fig. 1C) with substantial variability of domain positioning, in particular for DH3. We represent the individual bimodule core by a hybrid model assembled from high-resolution cryo-EM structures of KS3 and KS4 in the context of the bimodule core and a crystallographic model of DH3. The presence of alternative noncompact arrangements in solution cannot be ruled out. Folds and dimerization interfaces of individual domains (Fig. 3 and fig. S2, A to D) are preserved upon integration into the dimeric K3DAK4 bimodule core. The overall organization of domains in the third module with the DH3 domain positioned along the central dimer axis of KS3 (Fig. 1C) above the KS active sites agrees with previous models for iterative PKSs [LovB (19) and MAS-like PKS5 (18)] or trans-AT PKS didomains [RhiE (5)] but differs from a model proposed for the cis-AT PKS PikAIII (15).
The K3DAK4 bimodule core forms filaments
Filaments of the K3DAK4 bimodule core form spontaneously and co-occur on cryo-EM grids with individual bimodule cores in varying populations. The average visualized filament (Fig. 2, A and B) contains around 20 dimeric bimodule cores with dimensions of 100 Å in length (in filament direction) and 160 Å in height (Fig. 1D), equivalent to a total filament length of 200 nm. Filaments can bend by up to approximately 12° per module. Our structural analysis does not identify any specific mechanism of filament length determination. Filament formation occurs via homotypic LINKS interactions (Fig. 1D and fig. S4), providing lateral connections of each KS domain with two identical domains of the two neighboring bimodule cores. The linear organization of stacked KS3, DH3, and KS4 domains resembles that observed for individual bimodule cores, but LINKS interactions reduce the overall conformational variability. In the filament, the two KS domains of all bimodule cores are locked in the same orientation, while the DH3 domains maintain a higher degree of rotational freedom (Fig. 2, A and B).
The distances between the centers of mass of KS3-DH3 and DH3-KS4 in the filaments are highly similar at around 60 Å. Given the discrepancy between the KS3-DH3 linker length of only nine amino acids and the linkers between DH3 and KS4 comprising 89 amino acids plus the ACP domain, the distance similarity between the DH and both KS domains is intriguing. No fully ordered rigid contact site between DH3 and KS4 or local binding of linker regions enforcing proximity is observed, suggesting that transient dynamic interactions are responsible for maintaining this organization. An obvious candidate is the transient binding of ACP to individual enzymatic domains distributed across the filaments. In addition, a loop region in KS4 (residues 1994 to 2006), termed here the clamp loop (figs. S5 and S6), protrudes toward DH3 and is posed to mediate further transient interactions. This loop is more prominent in trans-AT KSs (figs. S5 and S6) although also present in a small subset of less than 1% of cis-AT KSs (fig. S5D). Given its preferential occurrence in trans-AT PKS, it may also serve as a docking point for trans-acting enzymes. We hypothesize that the avidity of multiple transient ACP and clamp loop interactions across extended filaments results in the observed fixed intermodular distance, which determines the spatial constraints for substrate transfer.
The lateral interactions of KS domains
The two KS domains of the bimodule core adopt a canonical thiolase fold with complete active sites based on the structural conservation of the Cys-His-His motif (Cys694-His829-His869 for KS3 and Cys1766-His1901-His1941 for KS4) (Fig. 2, C to F). Both KS3 and KS4 clade with active KSs in transATor, a computational prediction tool for trans-AT PKS BGC products (25). The crystal structures of the separately expressed KS domains and the cryo-EM structures of the KS domains in the context of the isolated bimodule core closely resemble each other (fig. S2, A to G). The largest difference is observed for a region around the active site of KS4, which contains an extended ~20–amino acid loop absent from KS3. This region remains disordered in the crystal structure of isolated KS4 but is ordered in the cryo-EM analysis of the bimodule core (figs. S2, E to G, and S3A). KS3 and KS4 feature an LD domain with a lateral trigonal three–α helix LINKS motif (14). Homotypic LINKS interactions of these motifs combined with the dimeric structure of KS domains are the sole basis for filament formation; LINKS-based contacts are also involved in crystal lattice formation in our crystal structures of both isolated KS domains (fig. S4, A to C). The LINKS interaction in KS3 covers the sequence region of Asn1061 to Lys1096 with 16 amino acids directly contributing to the interface. In KS4, the interaction region is between residues Asn2132 to Gln2167 with 18 amino acids involved in contacts (fig. S4, A to C). At 38.9%, the sequence identity between KS3 and KS4 is lower in this region than across the entire KS domains. Distinct amino acid differences in the LINKS interfaces, such as Phe2139 of KS4 occupying the position equivalent to Leu1068 in KS3, explain the homospecificity of LINKS interactions (fig. S4, A to C). The integration of functional domains into the context of a multimodular PKS provides strong avidity of dimeric interactions across modules. The multimodular context thus reduces the evolutionary pressure on maintaining stable dimerization interfaces of individual domains (fig. S7). Consequently, KS domains derived from modular PKS are more likely to dissociate into monomers in solution than monofunctional dimeric KS enzymes, and this behavior is observed here for KS3 (fig. S7). In cryo-EM analysis, we also observed the isolated KS3 domain to dimerize via its LINKS interface without formation of the canonical thiolase fold KS dimerization interface (fig. S1A).
The dimeric organization of the K3DAK4 DH3 domain
The double-hotdog-fold DH3 is the central domain in the bimodule core. It defines the connecting region between KS3 and KS4 of the bimodule core, may engage in transient interactions across module borders, and provides the N-terminal anchoring point for the substrate-shuttling ACP domain. Still, DH3 retains a large degree of conformational variability within the bimodule core, which we interpret as rotational flexibility modeled by positioning the well-ordered crystal structure of the dimeric DH3 domain in two extreme orientations in the complete filament composite model (Figs. 1D and 2B). On the basis of crystallographic analysis, the DH3 domain forms a canonical double-hotdog fold observed in PKS dehydratases, isomerases, and product template domains and contains active site residues (Asp1349-His1172) characteristic of enzymatically active dehydratase domains (Fig. 3A) (6). The overall shape of the DH3 dimer is the result of a distinct mode of dimerization, different from those observed in cis-AT PKS or the iteratively acting PKS LovB (Fig. 3B and fig. S8). Trans-AT PKS and cis-AT PKS use 15 to 20 residues within the first 100 N-terminal amino acids for forming the DH dimer interface. DH domains of cis-AT PKS dimerize via N-terminal loops interacting with the solvent facing side of strands β1-β2 and β5-β6 of the double-hotdog fold. In DH3, however, the same loop between β1 and β2 is involved in homotypic dimerization with its counterpart of the second protomer. The interprotomer angle in trans-AT PKS and cis-AT PKS DHs is similar, in the range of ~140° to 190° (fig. S8E), but as a consequence of the different interaction topologies, the overall shape of the DH3 dimer is clearly different from those of cis-AT PKS DH domains (fig. S8, C and D). It also strongly differs from the DH dimer of the iterative LovB PKS, which is bent in opposite direction (fig. S8, A and B). To analyze the conservation of DH dimer organization within BGC11 and, more generally, the family of trans-AT PKS, we investigated crystal structures of DH4 [Protein Data Bank (PDB) 5HQW] from BGC11 and of the MlnD DH (PDB 5IL5) from the macrolactin trans-AT PKS of Bacillus amyloquefaciens, which we have determined previously. Both MlnD and DH4 use the same mode of dimer formation as DH3 (fig. S8F), supporting the existence of a third fundamental type of dimeric DH architecture specific to trans-AT PKS, reflecting the distinct evolutionary path of trans-AT PKSs, iterative fatty acid synthase–like PKSs, such as LovB, and cis-AT PKSs as previously inferred from phylogenetic and structural analysis (19, 26, 27).
DISCUSSION
The organization of a single complete PKS assembly as a linear but flexible stack of modules is obtained by propagating the module-to-module stacking observed here along the central dimer axis. The conformational variability of DH domains in these models increases the space that the ACP can access. Trans-AT PKS modules mediating different steps of modifications of intermediates differ in their domain composition and may contain a ketoreductase (KR) (3) or DH-KR segment instead of the single-DH domain. Because of the absence of defined rigid DH interactions, the linear assembly line architecture proposed here could accommodate all variations of the modifying domains and would even be compatible with the modular structure of cis-AT PKSs, which lack LINKS elements for lateral filament formation.
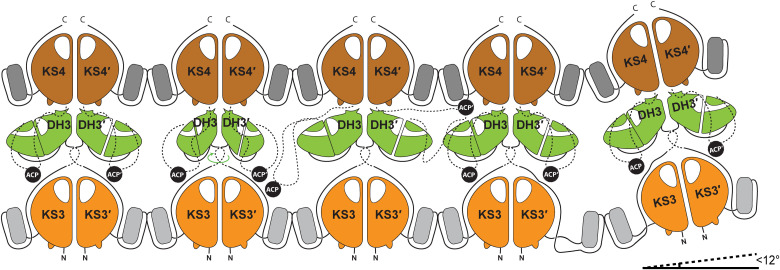
Horizontal filament formation between assembly lines via the LD (14) domains at each KS complements the vertical stacking of modules (Fig. 4). Association of extended assembly lines is favored by the avidity of multiple docking domains and LINKS-mediated interactions, even if individual domains in an assembly line would exhibit weak or no homotypic association, e.g., for individual trans-AT KS domains without LINKS motifs (14), or if the affinity of interpolypeptide docking elements are low (8). We hypothesize that multivalent transient interactions in two dimensions will result in module alignment and distance restraints between modules beyond the direct effect of peptide linker length to provide the framework for ACP-mediated substrate shuttling. Homospecific LINKS interactions ensure that assembly lines are always zipped up into filaments in register with product extension. Together with the moderate distance between neighboring assembly lines, this architecture could support ACP-mediated hopping of intermediates between neighboring assembly lines (see ACP positions in Fig. 4) as a strategy to overcome road blocks and to increase efficiency of precursor extension.
Fig. 4. Schematic depiction of a K3DAK4 filament.
Each bimodule core forms LINKS interactions with both its neighboring bimodule cores. The DH dimers in the middle portray a large degree of rotational freedom that together with flexible linkers (schematically indicated by dashed lines) may enable the ACP to reach also laterally to neighboring bimodule cores in the filament. The filament bends by up to 12° in direction of filament extension, and twisting in other directions occurs with smaller magnitude.
Extending the architecture of K3DAK4s filaments observed in cryo-EM with an average length of around 20 bimodules onto the entire BGC11 with 17 modules yields an extended 2D sheet structure, with dimensions that would be consistent with in vivo size estimates of 0.1 to 0.2 μm obtained for a complex of many copies of the bacillaene trans-AT PKS (14, 23). Local condensation of entire assembly lines into a 2D mesh would support sequestration of trans-acting enzymes, in particular, the essential trans-acting AT-AT-enoyl reductase (ER) domains by avidity of a multitude of weak interactions.
Notably, trans-AT PKS and, in particular, the BGC11 studied here form hybrid assembly lines with monomeric nonribosomal peptide synthetase (NRPS) modules to link amino acid and carboxylic acid building blocks. NRPSs require large-scale conformational flexibility for their multistep reactions (28–31). A 2D meshwork organization of trans-AT PKS not only provides different options for linking to or integrating NRPS modules (fig. S9) but also imposes steric restraints. Understanding the interplay between these two types of biosynthetic modules will be a crucial step toward engineering hybrid systems to massively expand the chemical product space.
Our analysis of a trans-AT PKS bimodule core covers all levels of organization from individual domains to filaments of multimodular PKS systems (Fig. 4). The visualization of a sheet-like organization of a trans-AT PKS assembly line provides a comprehensive basis for probing the role of trans-AT PKS assembly formation in vivo and for engineering bioinspired enzymatic systems.
MATERIALS AND METHODS
Cloning and protein expression
B. brevis (strain 47/JCM 6285/NBRC 100599 obtained from NBRC) was cultured at 30°C for 2 days in LB. Cells were pelleted and washed using lysis buffer [10 mM tris-HCl (pH 8.0), 25 mM EDTA, and 100 mM NaCl]. Cells were resuspended in lysis buffer and incubated for 1 day at 37°C after addition of ribonuclease (RNase) and lysozyme, followed by incubation for 1 day at 50°C supplemented with 0.028% of SDS and proteinase K (0.22 mg/ml). DNA was purified using phenol-chloroform extraction (32) and dissolved in tris-EDTA buffer [10 mM tris-HCl (pH 8.0) and 1 mM EDTA]. The BbC0ZGQ6 (B. brevis strain 47/JCM 6285/NBRC 100599, amino acids 532 to 2220, construct K3DAK4) bimodule core construct was cloned into the Gateway entry vector pDONR221 and transferred via Gateway-cloning into a modified pACEBac1 coding for an N-terminal His10-myc-Flag-tag and pETG10A-N-H6. The baculovirus expression system was used for protein production (33). Bacmids were transfected into, and virus generation was carried out in Sf21 insect cells (Expression Systems, IPLB-Sf-21-AE), grown in HyClone insect cell medium (GE Healthcare Life Sciences). Baculovirus-infected Sf21 cells (BICs) were harvested at generation 2 after transfection and stored at −80°C. The individual domains KS3 (amino acids 532 to 1135), DH3 (amino acids 1135 to 1434), and KS4 (amino acids 1587 to 2220) were each cloned out of the pETG10A-N-H6 vector into p7XNH3 vectors using FX cloning (34).
For expression of the full-length bimodule core K3DAK4, BICs were used to infect Sf21 cells, and cells were harvested after 3 days by centrifugation. Pellets were stored at −80°C. The isolated domains KS3, DH3, and KS4 were expressed in Escherichia coli BL21 (DE3) with induction using 0.1 mM isopropyl-β-d-thiogalactopyranoside at an optical density at 600 nm of 0.8, followed by overnight expression at 30°C. Cells were harvested using centrifugation and stored at −80°C.
Protein purification
Sf21 cells were resuspended in lysis buffer [50 mM NaPO4 (pH 7.5), 75 mM NH4SO4, 50 mM imidazole, 10% glycerol, 4 μM bestatin, 2 μM pepstatin A, 20 μM phenantroline, 2 μM phosphoramidon, 5 mM β-mercaptoethanol, 10 mM MgCl2, deoxyribonuclease, and RNase] and lysed by sonication. The lysate was cleared by centrifugation (158,420g for 1 hour at 4°C), filtered (0.45 μm), and loaded onto two serially connected 5-ml high-affinity Ni-charged resin column (GenScript) preequilibrated with lysis buffer. Unbound protein was eluted with 10–column volume (CV) lysis buffer 15-CV NiA [50 mM NaPO4 (pH 7.5), 75 mM NH4SO4, 100 mM imidazole, 10% (v/v) glycerol, 4 μM bestatin, 2 μM pepstatin A, 20 μM phenantroline, 2 μM phosphoramidon, and 5 mM β-mercaptoethanol]. The sample was eluted with 4 CV of NiB [20 mM tris-HCl (pH 8.0), 50 mM NH4Cl, 350 mM imidazole, 10% glycerol, 4 μM bestatin, 2 μM pepstatin A, 20 μM phenantroline, 2 μM phosphoramidon, and 5 mM β-mercaptoethanol] and diluted 1:4 with AIC-A [20 mM tris-HCl (pH 8.0), 20 mM NH4Cl, 10% glycerol, 4 μM bestatin, 2 μM pepstatin A, 20 μM phenantroline, 2 μM phosphoramidon, and 5 mM β-mercaptoethanol]. The sample was loaded onto a 6.5-ml anion exchange column (PL-SAX, 4000 Å, 10 μm, Agilent), followed by a wash step of 8 CV using AIC-A. Elution was performed with a gradient from 30% AIC-B [20 mM tris-HCl (pH 8.0), 500 mM NH4Cl, 10% glycerol, 4 μM bestatin, 2 μM pepstatin A, 20 μM phenantroline, 2 μM phosphoramidon, and 5 mM β-mercaptoethanol] to 100% AIC-B in 9 CV including a hold of 2.5 CV at 49%. Pure fractions were pooled and concentrated [50-kDa molecular weight cutoff (MWCO) Sartorius Vivaspin 20, 3220g, 5-min cycles], followed by size exclusion chromatography (Superdex 200 prep-grade 16/600, GE Healthcare) using GF buffer [50 mM NaPO4 (pH 7.5), 75 mM NaCl, 5% (v/v) glycerol, and 1 mM TCEP (Tris-(2-carboxyethyl)phosphin)]. Elution fractions were analyzed by SDS–polyacrylamide gel electrophoresis, pure fractions were combined, concentrated to 1.1 mg/ml (50-kDa MWCO Satorius Vivaspin 20, 3220g, 5-min intervals), and stored at −80°C.
E. coli cells were resuspended in lysis buffer 2 [50 mM Hepes-KOH (pH 8.0), 500 mM NaCl, 20 mM imidazole, 10% glycerol, 2 μM pepstatin A, 20 μM phenantroline, 4 μM E-64, 2 μM phosphoramidon, and 5 mM β-mercaptoethanol] and then lysed on ice using sonication. The lysate was cleared by ultracentrifugation (158,420g for 30 min at 4°C), filtered (0.8 μm), and loaded onto a with lysis buffer preequilibrated 5-ml high-affinity Ni-charged resin column (GenScript). Two wash steps were performed with 9 CV of lysis buffer, then 9 CV of NiA2 [50 mM Hepes-KOH (pH 8.0), 50 mM NaCl, 20 mM imidazole, 10% (v/v) glycerol, 1 μM pepstatin A, 10 μM phenantroline, 2 μM E-64, 1 μM phosphoramidon, and 5 mM β-mercaptoethanol], followed by elution using 8 CV of NiB2 [50 mM Hepes-KOH (pH 8.0), 50 mM NaCl, 400 mM imidazole, 10% (v/v) glycerol, 1 μM pepstatin A, 10 μM phenantroline, 2 μM E-64, 1 μM phosphoramidon, and 5 mM β-mercaptoethanol]. The sample was dialyzed overnight into 1.6 liters of dialysis buffer [12.5 mM Hepes-KOH (pH 8.0), 31.25 mM NaCl, 11.875% (v/v) glycerol, and 5 mM β-mercaptoethanol] under addition of 200 μg of C3 protease (homemade) to cleave the N-terminal His10 tag, followed by removal of cleaved tag and uncleaved protein using an orthogonal 5-ml column packed with high-affinity Ni-charged resin (GenScript). Subsequently, the sample was concentrated (10-kDa MWCO Satorius Vivaspin 20, 3220g at 4°C, 10-min intervals) and subjected to size exclusion chromatography (Superdex 200 prep grade 16/600, GE Healthcare) in GF buffer 2 [20 mM Hepes-KOH (pH 8.0), 250 mM NaCl, 5% (v/v) glycerol, and 5 mM dithiothreitol]. All isolated domains (KS3, DH3, and KS4) were purified following the same protocol but using a Superdex 75 prep grade 16/600 GE Healthcare column for the DH3 domain. All purification steps were carried out at 4°C.
Crystallographic analysis of isolated domains
Crystallization experiments were performed using the sitting-drop vapor-diffusion technique. KS3 crystals were grown at 4°C with a mix of 0.2-μl drops of protein (12 mg/ml) in GF buffer 2 with 0.2 μl of reservoir solution [1% (w/v) polyethylene glycol (PEG) monomethyl ether (MME) 2000 and 1 M Na succinate]. DH3 crystals were grown at 20°C with a mix of 0.2-μl drops of protein (3.9 mg/ml) in GF buffer 2 with 0.2 μl of reservoir solution [20% (w/v) PEG MME 500; 10% PEG 2000, 0.1 M tris-bicine (pH 8.5), 0.06 M MgCl2 hexahydrate, and 0.06 M CaCl2 dihydrate]. KS4 crystals were grown at 20°C with a mix of 0.2-μl drops of protein (4 mg/ml) in GF buffer 2 supplemented by 10 mM MgSO4 with 0.15 μl of reservoir solution [0.2 M (NH4)2SO4, 0.1 M sodium citrate (pH 5.22), and 8% (w/v) PEG 3350]. All crystals were cryoprotected in mother liquor with ethylene glycol added to 25% (v/v) before flash freezing in liquid nitrogen. All data were collected at the Swiss Light Source (Paul-Scherrer Institute, Villigen, Switzerland) at 100 K. KS3 datasets were collected at PXIII, DH3, and KS4 datasets were collected at PXI, with wavelengths of 1.00, 1.00, and 0.98 Å, respectively. Data reduction was performed using XDS (35) and XSCALE (35), and phases were determined by molecular replacement in Phaser (36, 37). The final models were built using model building in Coot (38) and refinement in PHENIX (39) (table S1).
KS3 (7ZM9) crystallized in space group C2 with a unit cell of a = 126.78 Å, b = 92.69 Å, c = 99.27 Å, and β = 92.54°. Our earlier structure of the isolated KS2 domain from BGC11 was used for replacement (PDB 4Z37). The final model was refined to RWork/RFree of 0.15/0.17 at 1.62-Å resolution. Ramachandran values are 97.46%/2.54%/0.00% (favored/allowed/outliers).
DH3 (7ZMF) crystallized in space group of P21212 with a unit cell of a = 72.38 Å, b = 197.36 Å, and c = 39.17 Å. Our earlier structure of an isolated DH domain of another module (DH4) of the BGC11 served as a replacement model (PDB 5HQW). The final model was refined to a RWork/RFree of 0.22/0.25 at 2.21-Å resolution. Ramachandran values are 97.58%/2.42%/0.00% (favored/allowed/outliers).
KS4 (7ZMC) crystallized in space group C2221 with a unit cell of a = 73.56 Å, b = 191.79 Å, and c = 293.81 Å. Two datasets from a single crystal were merged using XSCALE. A search model was generated in Swiss model (40) using KS3 as a template, refined into the EM density of KS4 using PHENIX (39), and then used as a replacement model for molecular replacement in Phaser (37). The final model was refined to an RWork/RFree of 0.26/0.29 at 3.1-Å resolution. Ramachandran values are 93.14%/6.67%/0.19% (favored/allowed/outliers).
Cryo-EM sample preparation, data collection, and analysis
Samples were thawed, centrifuged (2100g for 30 min at 4°C), dialyzed overnight at 4°C into 37.5 mM NaPO4 with 5 mM TCEP, and centrifuged again (21,000g for 5 min at 20°C). Sample were diluted to 0.15 mg/ml (~1 μM) and incubated at 23°C for 2 to 6 hours before grid preparation. The 300-mesh Cu lacey grids (Agar Scientific) were glow-discharged (45 s) in air, a 4-μl sample was applied using a clipped pipette tip, adsorbed for 60 s, and plunge-frozen in liquid ethane using a Leica EM GP (3-s blot, 90% humidity, 20°C).
Three datasets were collected on a Titan Krios G1 (Thermo Fisher Scientific) electron microscope equipped with a Gatan LS Quantum energy filter and a K2 Summit detector (Gatan) in superresolution mode. Movies were collected with 50 frames over 10 s of exposure at a dose rate of ~6 e− s −1 Å−1 and a pixel size of 1.058 Å (0.529-Å superresolution pixel size) (table S2). Movies were motion-corrected using patch motion correction, followed by patch CTF correction in cryoSPARC (Structura Biotechnology Inc.). Particles were picked using the blob picker (filaments) and template picker (individual bimodule cores), followed by multiple rounds of 2D classification and sorting in cryoSPARC (fig. S1). For the filaments no symmetry was applied, whereas the individual bimodule core set had twofold symmetry applied in the final steps.
Ab initio classes for both the bimodule core dataset and the filament dataset were generated in cryoSPARC. After picking with an initial 2D class–derived template, a set of 99,566 particles of individual bimodule cores was selected from 2D classes. A large degree of flexibility between the individual dimeric domain prevented high-resolution reconstruction of the entire bimodule core. From the same dataset, using a template representing a single KS domain, a total of 353,464 particles after 2D classification were selected for further processing of individual KSs. An ab initio model was calculated in cryoSPARC, and nonuniform homogenous refinement, iterated with local and global CTF (contrast transfer function) refinement yielded a map at 2.9-Å resolution, which represents a combination of KS3 and KS4 domains as evident form the densities for nonidentical side chains and few differences of main chain regions. A 3D variability analysis (3DVA) on the whole set, followed by clustering into two clusters provided two starting models for heterogeneous refinement starting at a high initial resolution of 5 Å, leading to a split of 182,315 in the first and 171,149 particles in the second class, respectively. Map analysis confirmed successful sorting into KS3 and KS4 based on side-chain densities, few main-chain differences and relative position of residual density representing further domains of the bimodule core. Further nonuniform refinements for each subset applying C2 symmetry, followed by local and global CTF refinements and a final step of local refinement using a global mask and C2 symmetry, lead to final maps at 2.90-Å (KS4) and 2.93-Å (KS3) resolution. The maps are of excellent quality and clearly distinguish KS3 and KS4 at the sequence level (fig. S3). In both KSs, parts of the LINKS helices, which are not engaged in LINKS interactions, are less well resolved. Both KS3 and KS4 x-ray crystallographic models (PDB 7ZM9 and 7ZMC) were real space–refined into their respective maps using PHENIX, followed by manual model building in Coot and a final round of real-space refinement in PHENIX.
To determine an approximate average length of filaments, seven random micrographs were picked, all continuous filaments identified by having more than four repeat units were manually traced, and the repeat units were counted. Crossovers of filaments and the presence of filaments reaching beyond the field of view prevent an exact quantification of filament length.
For the filament dataset, all processing was performed in cryoSPARC in a single-particle workflow rather than by helical analysis due to the large size of the repeat unit of the flat filament and the variable connections between filament units. Four ab initio models were generated of which one representing a total of four bimodule core dimers was lastly selected as initial model. After two rounds of nonuniform refinement, a map region representing two adjacent bimodule core dimers was excised using chimera and expanded into a mask and used as model for all further reconstructions. Refinement of the whole filament using the mask around two bimodule core yielded a resolution of 6.8 Å in cryoSPARC. Overall resolution was limited by flexibility originating from the variety of filament conformations originating from the bend filament and an overrepresentation of top views. 3DVA in cryoSPARC revealed only minor shifts with regard to the positioning of KS domains, which are presumably linked to different bent angles of filaments. Local maps covering LINKS-bridged pairs of KS dimers were obtained from 260,601 particles of two bimodule core dimers in masked and local refinements of the corresponding regions for KS3 and KS4 (maps 2 and 3 in fig. S1, included as additional maps in EMDB-14945) from a set of local motion–corrected and reextracted particles. No improvements in resolution or map quality were observed using attempts of 3D classification in RELION 3.1 (41). Pronounced positional variability, predominantly rotation of individual DH dimers, precluded improvements of map quality in masked and local refinements of a lateral pair of DH3 dimers.
For obtaining an overall model of two laterally linked bimodule cores (PDB 7ZSK), we assembled start models for KS3, KS4, and DH based on the individual crystal and high-resolution cryo-EM structures (PDB 7ZMD, 7ZMA, 7ZM9, 7ZMC, and 7ZMF). Models for KS3 and KS4 were optimized by chain-, domain-, and segment-wise rigid-body fitting and overall coordinate and adenosine 5′-diphosphate (ADP) refinement into the respective maps of pairs of KS dimers from local refinement (maps 2 and 3 in fig. S1C, EMDB-14945 additional maps). A model of two laterally linked bimodule core dimers was then assembled from the refined KS3 and KS4 dimer pairs and DH3 dimers. Positional disorder prevents unambiguous identification of a single DH3 dimer orientation. To model this disorder, we manually defined two different starting orientations for rigid body refinement, representing divergent states of rotation of the DH3 dimer. Last, rigid-body and ADP refinement with one rigid group per KS3, KS4, or DH3 dimer against the overall intermediate resolution map (EMDB-14945, map 1 in fig. S1C) was carried out. The final results of rigid body refinement of DH3 are influenced by the choice of start model orientation. Refinement at intermediate resolution is not suited to optimize local side-chain interactions at the LINKS interface, which is best visualized in the respective higher resolution x-ray models of KS3 and KS4 dimers.
Sequence and structure analysis
All sequence comparisons were performed using BLAST (42) and visualized in Geneious Prime (version 2019-03-05). Clading analysis of the KSs was performed using TransATor (25). For module assignment, we used traditional module boundaries but acknowledged the relevance of alternate definitions based on evolutionary patterns (43, 44). Structural comparisons were performed using PDBeFold (45), interfaces were analyzed using PISA at the European Bioinformatics Institute (46) or QtPISA from CCP4 (36). For the comparison of DH dimer shape, one DH protomer was superposed on the basis of secondary structure elements. Angles between DH protomers were analyzed using phenix.angle based on ordered secondary structure elements only. BGC analysis was performed using antiSMASH (47). Sequence-based comparison of the clamp loop lengths was performed with a custom Python script (DOI: 10.5281/zenodo.6550895) to count the clamp loop lengths in amino acids for all entries in the antiSMASH database (48) that match both terms, “KS” and “type I PKS”. All sequences were aligned against KS3, and loop length was defined as distance between residues matching KS3 clamp loop anchor points 920 and 936. Analysis of clamp loop lengths in KSs with structural models in the PDB (Protein Data Bank) was performed on the basis of structural alignments using superpose (45) (secondary structure matching) from CCP4 instead of sequence alignments and manual counting of residues using the same anchor points.
Primers
The primers were used as follows: K3DAK4 gateway, GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAGAATCTGTATTTCCAGGGGTCGCCTTCATCGGTGGTGCG (forward 532) and GGGGACCACTTTGTACAAGAAAGCTGGGTACTACAAGGGATGCAGGTAAGCCGG (reverse 2220); KS3 FX cloning, TATATGCTCTTCGAGTTCGCCTTCATCGGTGGTGCG (forward) and TATATGCTCTTCATGCGCTGCTAACAGACCTGGCGTG (reverse); DH3 FX cloning, TATATGCTCTTCGAGTAGCACAGGTGTGGGTGTGATTC (forward) and TATATGCTCTTCATGCTGATCTAACAGATACATTTGCAATCTTGC (reverse); KS4 FX cloning, TATATGCTCTTCGAGTCCAAAAGGGGCGTTGGACATTG (forward) and TATATGCTCTTCATGCGGGATGCAGGTAAGCCGGC (reverse).
Acknowledgments
We thank J. Brunner and R. Dutzler for the donation of the FX-cloning plasmids used in this study (p7XNH3, Addgene, #47064); EMBL Heidelberg for providing the pETG10A vector; M. Chami, K. Goldie, and C. Alampi from the University of Basel BioEM facility and I. Mohammed for help in operating the electron microscopes; A. Mazur for help in coding the sequence comparisons; and the National Institute of Technology and Evaluation Biological Resource Center for providing the B. brevis NBRC 100599 strain. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing center at the University of Basel.
Funding: This study was supported by the Swiss National Science Foundation project funding 179323 and 159696 to T.M., the Swiss National Science Foundation R’EQUIP grant 177084 to T.M., and AL4BIOCH project of the H2020-MSCA-IF-2018 to H.M.-H.
Author contributions: Conceptualization: D.A.H. and T.M.. Methodology: Y.U.T., D.A.H., R.P.J., and T.M. Software: Y.U.T. Investigation: Y.U.T., H.M.-H., S.F.X.M., and R.P.J. Visualization: Y.U.T. Project administration: T.M. Supervision: R.P.J. and T.M. Writing–original draft: Y.U.T., D.A.H., and T.M. Writing–review and editing: Y.U.T. and T.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All custom code is available under the Zenodo DOI provided in Materials and Methods. All data used in this work has been deposited in the PDB under 7ZM9 (KS3, x-ray crystallography), 7ZMA (KS4, cryo-EM), 7ZMC (KS4, x-ray crystallography), 7ZMD (KS3, cryo-EM), 7ZMF (DH3, x-ray crystallography), and 7ZSK (K3DAK4, cryo-EM). Cryo-EM maps have been deposited in the EMDB under EMD-14795 (KS3), EMD-14793 (KS4), and EMD-14945 (K3DAK4).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
Data S1
REFERENCES AND NOTES
- 1.Newman D. J., Cragg G. M., Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Piel J., Metabolites from symbiotic bacteria. Nat. Prod. Rep. 26, 338–362 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Hertweck C., The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Gay D. C., Spear P. J., Keatinge-Clay A. T., A double-hotdog with a new trick: Structure and mechanism of the trans-acyltransferase polyketide synthase enoyl-isomerase. ACS Chem. Biol. 9, 2374–2381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretschneider T., Heim J. B., Heine D., Winkler R., Busch B., Kusebauch B., Stehle T., Zocher G., Hertweck C., Vinylogous chain branching catalysed by a dedicated polyketide synthase module. Nature 502, 124–128 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Labonte J. W., Townsend C. A., Active site comparisons and catalytic mechanisms of the hotdog superfamily. Chem. Rev. 113, 2182–2204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broadhurst R. W., Nietlispach D., Wheatcroft M. P., Leadlay P. F., Weissman K. J., The structure of docking domains in modular polyketide synthases. Chem. Biol. 10, 723–731 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Zeng J., Wagner D. T., Zhang Z., Moretto L., Addison J. D., Keatinge-Clay A. T., Portability and structure of the four-helix bundle docking domains of trans-acyltransferase modular polyketide synthases. ACS Chem. Biol. 11, 2466–2474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodge G. J., Maloney F. P., Smith J. L., Protein-protein interactions in “cis-AT” polyketide synthases. Nat. Prod. Rep. 35, 1082–1096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helfrich E. J., Piel J., Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 33, 231–316 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Weissman K. J., The structural biology of biosynthetic megaenzymes. Nat. Chem. Biol. 11, 660–670 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Piel J., Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27, 996–1047 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T., Ishida K., Jenke-Kodama H., Dittmann E., Gurgui C., Hochmuth T., Taudien S., Platzer M., Hertweck C., Piel J., Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 26, 225–233 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Gay D. C., Wagner D. T., Meinke J. L., Zogzas C. E., Gay G. R., Keatinge-Clay A. T., The LINKS motif zippers trans-acyltransferase polyketide synthase assembly lines into a biosynthetic megacomplex. J. Struct. Biol. 193, 196–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta S., Whicher J. R., Hansen D. A., Hale W. A., Chemler J. A., Congdon G. R., Narayan A. R., Håkansson K., Sherman D. H., Smith J. L., Skiniotis G., Structure of a modular polyketide synthase. Nature 510, 512–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogan D. P., Zhang K., Li X., Li S., Pintilie G. D., Roh S. H., Craik C. S., Chiu W., Khosla C., Mapping the catalytic conformations of an assembly-line polyketide synthase module. Science 374, 729–734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagde S. R., Mathews I. I., Fromme J. C., Kim C. Y., Modular polyketide synthase contains two reaction chambers that operate asynchronously. Science 374, 723–729 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbst D. A., Jakob R. P., Zahringer F., Maier T., Mycocerosic acid synthase exemplifies the architecture of reducing polyketide synthases. Nature 531, 533–537 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Liang J., Chen L., Zhang W., Kong L., Peng C., Su C., Tang Y., Deng Z., Wang Z., Structural basis for the biosynthesis of lovastatin. Nat. Commun. 12, 867 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khosla C., Tang Y., Chen A. Y., Schnarr N. A., Cane D. E., Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 76, 195–221 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Staunton J., Caffrey P., Aparicio J. F., Roberts G. A., Bethell S. S., Leadlay P. F., Evidence for a double-helical structure for modular polyketide synthases. Nat. Struct. Biol. 3, 188–192 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Edwards A. L., Matsui T., Weiss T. M., Khosla C., Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J. Mol. Biol. 426, 2229–2245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straight P. D., Fischbach M. A., Walsh C. T., Rudner D. Z., Kolter R., A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 104, 305–310 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaus M., Grininger M., Engineering strategies for rational polyketide synthase design. Nat. Prod. Rep. 35, 1070–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Helfrich E. J. N., Ueoka R., Dolev A., Rust M., Meoded R. A., Bhushan A., Califano G., Costa R., Gugger M., Steinbeck C., Moreno P., Piel J., Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol. 15, 813–821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott T. A., Piel J., The hidden enzymology of bacterial natural product biosynthesis. Nat Rev Chem 3, 404–425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nivina A., Yuet K. P., Hsu J., Khosla C., Evolution and diversity of assembly-line polyketide synthases. Chem. Rev. 119, 12524–12547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimer J. M., Eivaskhani M., Harb I., Guarne A., Weigt M., Schmeing T. M., Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility. Science 366, (2019). [DOI] [PubMed] [Google Scholar]
- 29.Reimer J. M., Aloise M. N., Harrison P. M., Schmeing T. M., Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature 529, 239–242 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Drake E. J., Miller B. R., Shi C., Tarrasch J. T., Sundlov J. A., Allen C. L., Skiniotis G., Aldrich C. C., Gulick A. M., Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529, 235–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonhomme S., Dessen A., Macheboeuf P., The inherent flexibility of type I non-ribosomal peptide synthetase multienzymes drives their catalytic activities. Open Biol. 11, 200386 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D. W., Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006, pdb.prot4455 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald D. J., Berger P., Schaffitzel C., Yamada K., Richmond T. J., Berger I., Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021–1032 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Geertsma E. R., Dutzler R., A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry 50, 3272–3278 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boratyn G. M., Camacho C., Cooper P. S., Coulouris G., Fong A., Ma N., Madden T. L., Matten W. T., McGinnis S. D., Merezhuk Y., Raytselis Y., Sayers E. W., Tao T., Ye J., Zaretskaya I., BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vander Wood D. A., Keatinge-Clay A. T., The modules of trans-acyltransferase assembly lines redefined with a central acyl carrier protein. Proteins 86, 664–675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Hashimoto T., Qin B., Hashimoto J., Kozone I., Kawahara T., Okada M., Awakawa T., Ito T., Asakawa Y., Ueki M., Takahashi S., Osada H., Wakimoto T., Ikeda H., Shin-Ya K., Abe I., Characterization of giant modular PKSs provides insight into genetic mechanism for structural diversification of aminopolyol polyketides. Angew. Chem. Int. Ed. Engl. 56, 1740–1745 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., Medema M. H., Weber T., antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blin K., Shaw S., Kautsar S. A., Medema M. H., Weber T., The antiSMASH database version 3: Increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 49, D639–D643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madeira F., Pearce M., Tivey A. R. N., Basutkar P., Lee J., Edbali O., Madhusoodanan N., Kolesnikov A., Lopez R., Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 and S2
References
Movie S1
Data S1