Abstract
Bioengineering of viral vectors for therapeutic gene delivery is a pivotal strategy to reduce doses, facilitate manufacturing, and improve efficacy and patient safety. Here, we engineered myotropic adeno-associated viral (AAV) vectors via a semirational, combinatorial approach that merges AAV capsid and peptide library screens. We first identified shuffled AAVs with increased specificity in the murine skeletal muscle, diaphragm, and heart, concurrent with liver detargeting. Next, we boosted muscle specificity by displaying a myotropic peptide on the capsid surface. In a mouse model of X-linked myotubular myopathy, the best vectors—AAVMYO2 and AAVMYO3—prolonged survival, corrected growth, restored strength, and ameliorated muscle fiber size and centronucleation. In a mouse model of Duchenne muscular dystrophy, our lead capsid induced robust microdystrophin expression and improved muscle function. Our pipeline is compatible with complementary AAV genome bioengineering strategies, as demonstrated here with two promoters, and could benefit many clinical applications beyond muscle gene therapy.
Engineered AAV vectors with high specificity and potency in muscle are reported whose value is illustrated in two disease models.
INTRODUCTION
Adeno-associated virus (AAV) provides an exceptionally versatile basis for bioengineering of recombinant gene transfer vectors, owing to its low genetic complexity that facilitates the cloning, packaging, and delivery of therapeutic gene expression cassettes. Fostering AAV’s promise is the wealth of natural viral isolates with distinct properties that can be modulated or enhanced, including tissue specificity, efficiency of transgene expression, and escape from pre-existing or induced anti-AAV antibodies. Such repurposing is typically achieved in a high-throughput manner comprising the generation and iterative, top-down selection of libraries of synthetic AAV capsids, which were diversified using techniques such as DNA family shuffling, ancestral reconstruction, or peptide display (1, 2). Alternatively, specific vector properties can also be established in a rational bottom-up approach by introducing targeted changes in the capsid, as exemplified by the isolation of immunoevasive AAV variants in work from the Asokan and Agbandje-McKenna laboratories (3, 4).
While the power of these techniques is undisputed, it is also clear that further improvements are required to truly harness the full potential of AAV bioengineering. One reason is that, even when performed under stringent selection pressures, library screens often produce a collection of potentially interesting AAV capsid candidates rather than just a single one that would justify subsequent in-depth validation. To facilitate the identification of the most optimal synthetic AAV capsid variant, recent work by us and others proposes a solution in the form of DNA/RNA barcoding of lead candidates, which enables their concurrent head-to-head comparison and stratification in a single experiment in the same animal(s) (5–10). Another challenge for traditional directed AAV evolution approaches is posed by diseases that necessitate simultaneous transduction of multiple tissues in the body, ideally from a minimally invasive vector delivery route. Clinically relevant examples are monogenic hereditary muscle disorders that manifest during infancy and childhood, and that often cause substantial morbidity and mortality, such as X-linked myotubular myopathy (XLMTM) (11), glycogen storage disease type II (Pompe disease) (12), or Duchenne muscular dystrophy (DMD) (13).
Thus, the field urgently requires AAV capsids that, following systemic administration, effectively and selectively target the skeletal muscle, diaphragm, and heart that are affected in these diseases while sparing major off-targets such as the liver. A complication in the isolation of such capsids with multitarget transduction capability and specificity is that molecular evolution is mostly performed as an on-target selection. In this case, a capsid library is iteratively selected in a single cell or tissue type, in the absence of means to actively eliminate variants that also transduce unwanted off-targets. Until these challenges are solved, ongoing clinical trials primarily rely on and evaluate extant wild-type (WT) AAV serotypes, such as AAV8 for delivery of the MTM1 gene to treat XLMTM, or GAA-encoding AAV9 to cure Pompe disease. Others study AAV8, AAV9, or AAVrh74 for treatment of DMD, via expression of mini-/microdystrophin or CRISPR-mediated exon skipping and restoration of the dystrophin reading frame (14).
Alas, these and other WT AAVs are suboptimal for muscle gene therapy due to their broad tropism and their bias toward the liver, which enforces the use of high doses to achieve robust expression and therapeutic benefit in affected muscle types. This, in turn, imposes challenges for vector manufacturing and jeopardizes patient safety, as implied by dose-dependent toxicities observed in various cell types and organs, including the liver and dorsal root ganglia, in large mammals and humans (15–17). This comprises adverse events in several recent clinical trials such as IGNITE or C3391001 (DMD) and ASPIRO (XLMTM), in which administration of high doses of 2 × 1014 to 3 × 1014 AAV vector genomes (vg) per kilogram triggered kidney or liver injury and ultimately resulted in fatalities in several children. Preclinical use of the same AAV vector as in ASPIRO but at even higher doses of up to 8 × 1014 vg/kg was found to be safe in nonhuman primates, highlighting the species-specific differences in AAV toxicity and the need to dissect the underlying mechanisms in humans, possibly comprising a role of pre-existing hepatobiliary disease or anti-AAV immune responses (18).
In addition, a study investigating the long-term stability and safety of AAV-mediated human factor VIII expression in dogs yielded evidence for dose-dependent vector integration into the host genome, including near genes controlling cell growth (19). The propensity of AAV vectors to perturb host genome integrity is exacerbated in CRISPR applications, as implied by data from the long-term assessment of AAV8-CRISPR vectors in DMD mice showing unintended genome alterations in the liver, spleen, kidney, brain, and testes (20).
The current limitations of naturally occurring AAV serotypes and artificially selected capsid variants obtained through molecular evolution create a high demand for synthetic, and improved AAV vectors for muscle gene therapy (and others) that are more specific, safer, and more efficacious than the present AAV variants while remaining compatible with large-scale, clinical-grade manufacturing. Previously, we have achieved a seminal first step with the identification of the peptide-displaying AAV9 variant AAVMYO [also called AAV9P1 (21, 22), with P1 denoting the 7-mer peptide RGDLGLS displayed on AAV9], which robustly transduces the skeletal muscle, diaphragm, and heart in peripherally injected mice and outperforms numerous benchmarks, including gold standards in muscle gene therapy (6). Similarly, Tabebordbar et al. (23) have recently reported another set of AAV9 variants displaying RGD (arginine-glycine-aspartic acid)-containing peptides, which were called MyoAAV and likewise mediated robust transduction of the musculature from systemic injection in mice and also in nonhuman primates. Notably, several of the lead peptides in this latest study largely overlap with the P1 peptide used in our own preceding work (6, 21, 22) and particularly share an RGDL (arginine-glycine-aspartic acid-leucine) motif, supporting its critical role in muscle targeting.
Here, we aimed to build on our experience and to bioengineer a next generation of synthetic myotropic AAV vectors that maintains all the assets of AAVMYO, i.e., high efficiency in striated muscles and high vector yields, while exhibiting a further substantial reduction of off-targeting after peripheral intravenous delivery. To this end, we pursued a semirational, combinatorial bioengineering approach comprising de novo screens of two shuffled AAV capsid libraries in the murine musculature, a combination of the top hits with the myotropic peptide from AAVMYO, and massively parallel in vivo validation by vector barcoding and deep sequencing in two mouse strains. This led to the selection of two peptide-displaying chimeric AAV capsids called AAVMYO2 and AAVMYO3, which we then assessed in the diseased muscle in mouse models of myotubular myopathy or DMD. The two original AAV variants reported here represent exciting candidates for gene therapy of human muscle disorders and extend the rapidly growing collection of myotropic synthetic AAVs reported by us (6) and others (23–26). Concurrently, they exemplify the power of deliberately merging multiple, separately optimize features in a single AAV particle, and thus illustrate the great potential of semirational bioengineering of recombinant viruses for clinical use.
RESULTS
Creation of shuffled AAV libraries and in vivo cycling in murine musculature
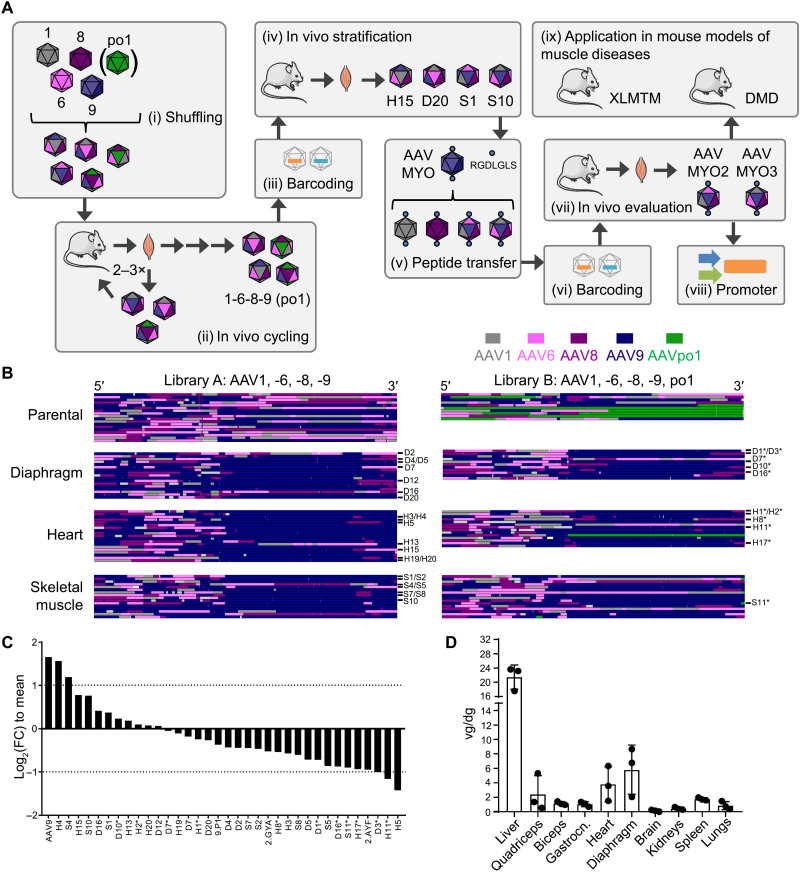
An overview of the workflow pursued in this study is depicted in Fig. 1A. Initially, we created two different AAV capsid libraries through DNA family shuffling of the capsid genes of AAV1, AAV6, AAV8, and AAV9 (library A) or the same set plus AAVpo1 [library B; Fig. 1A (i)]. These isolates were selected on the basis of data from the literature or our recent work (27) implying their high efficiency and/or specificity in different muscle types in vivo. Both libraries were injected independently via the tail vein into adult male C57BL/6J mice (n = 2 per library) at a dose of 1.25 × 1012 (library A) or 6.42 × 1011 (library B) particles per library and mouse. A week later, the animals were euthanized and whole AAV cap genes were polymerase chain reaction (PCR)–rescued from on-target tissues, i.e., quadriceps femoris, diaphragm, and heart. Enriched cap variants were then subcloned to generate a secondary library for the next in vivo screening round. In total, three such selection rounds were performed with library A and two with library B [Fig. 1A (ii)].
Fig. 1. Overview of AAV library generation, in vivo evolution, and validation.
(A) Workflow comprising nine consecutive steps: (i) creation and production of two AAV capsid libraries by DNA family shuffling of multiple cap genes; (ii) in vivo selection/cycling via systemic injection into mice, followed by vector genome amplification from on-target tissues (diaphragm, heart, and skeletal muscle), subcloning and production of secondary libraries for repeated injection; (iii and iv) barcoding and stratification in mice; (v) rational P1 peptide transfer into selected shuffled capsids; (vi) barcoding of the ensuing bioengineered capsids plus benchmarks; (vii) validation of the muscle specificity in WT mice; (viii) combination of the two best capsids (AAVMYO2 and AAVMYO3) with two myotropic promoters; and (ix) application in two mouse models of human muscle diseases. (B) Sequence analysis of shuffled AAV libraries. Top: Parental libraries composed of AAV1, -6, -8, and -9 (library A) or AAV1, -6, -8, -9, and po.1 (library B). Shown are representative examples. Each row depicts one clone. (C) Composition of the secondary barcoded library. Shown are log2(FC) values relative to the mean per barcoded AAV variant in the library. The two dotted lines mark a twofold change. Negative values indicate underrepresentation of a variant, and positive values imply overrepresentation. FC, fold change. (D) Viral DNA distribution of the library in multiple organs. Shown are vector genomes (individual mice and averages) per diploid genome (vg/dg; means + SD) from three C57BL/6J mice (each mouse is one dot, injected with 1 × 1012 vg, euthanized 1 week later). Vector genomes (EYFP probe) were normalized to RPP30 as a housekeeper. Gastrocn., gastrocnemius; Quadriceps, quadriceps femoris.
Sanger sequencing of individual clones after the final selection rounds revealed that, irrespective of starting library, the C-terminal portion of most of the enriched capsid protein sequences was nearly exclusively derived from one of the five input serotypes, namely, AAV9 (dark purple in Fig. 1B). In contrast, no enrichment or depletion was evident for any parental serotype in the N-terminal half or in the C-terminal end. AAVpo1 was largely underrepresented, probably because of its low sequence homology with the other four serotypes and the ensuing inability of chimeric capsid sequences containing AAVpo1 to assemble functional viral particles.
Stratification of selected capsid variants through DNA/RNA barcoding and NGS
We next selected 32 different cap sequences from all three tissues for further characterization, comprising 21 from library A and 11 from library B (marked in Fig. 1B). We added four benchmarks, i.e., one WT AAV9 and three AAV peptide display mutants, including AAVMYO (6) as a stringent control. The other two were based on AAV2 and previously reported to off-target the murine skeletal muscle in a library screen in the lung (AAV2.ESGHGYA and AAV2.ESGHAYF) (28). To enable their direct side-by-side comparison in the same animal, we produced them as barcoded vectors by harnessing our recently reported pipeline (6, 10). Briefly, each vector carried a CMV (cytomegalovirus) promoter–driven eyfp (enhanced yellow fluorescent protein) reporter containing a unique, capsid-specific, 15-nucleotide-long barcode in its 3′ untranslated region [Fig. 1A (iii)]. This design was chosen to permit the qualitative and quantitative tracking of each capsid variant on the DNA and mRNA levels in all organs of interest by next-generation sequencing (NGS). All 36 vectors were produced, purified, and titrated separately, and then mixed in equal amounts and concentrated together. NGS analysis of the resulting library confirmed a homogeneous contribution of all 36 variants with less than fourfold variation relative to the mean (Fig. 1C), which we consider to be within the typical error margin of AAV titration and purification and which, based on our experience (6, 10), is fully compatible with all downstream bioinformatic analyses.
We next peripherally injected this capsid pool into male C57BL/6J mice, followed by quantitative PCR (qPCR)–based measurement of its biodistribution in 10 major organs and tissues 1 week later [Fig. 1A (iv)]. As hoped for, this revealed an accumulation of vector DNA in all muscle types, next to the liver as a major off-target (Fig. 1D).
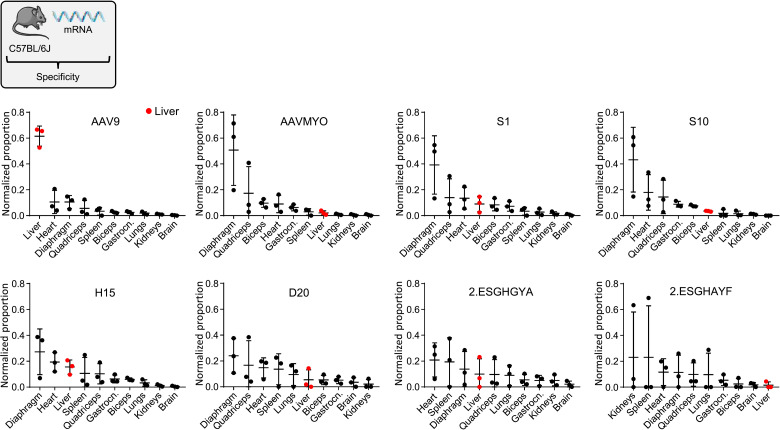
Subsequent deep sequencing and ranking of the individual 36 barcodes on the AAV mRNA level in each organ yielded a series of interesting findings. First, the stringent AAVMYO benchmark remained the top hit in terms of efficiency in all muscles, including three types of skeletal muscle as well as heart and diaphragm (dark purple in fig. S1; raw reads in fig. S2). Also concordant with our prior work (6) is the liver detargeting of AAVMYO as compared to its parental serotype AAV9; the latter was among the best performers in this organ. This is even more evident when the same mRNA data are sorted by the performance of each capsid in all 10 organs to better visualize in vivo capsid specificity (Fig. 2). Second, several chimeric capsids exhibited an mRNA biodistribution profile akin to AAVMYO, most notably, AAVS1, AAVS10, AAVH15, and AAVD20 (letters indicate the final library that each capsid was isolated from: D, diaphragm; H, heart; S, skeletal muscle). AAVS1 and AAVS10 were even less active than AAV9, AAVMYO, and most other chimeras in several off-targets including the liver, brain, spleen, kidney, and lungs (efficiency ranking in figs. S1 and S2). Similar patterns were also observed for most capsids on the AAV DNA level (figs. S2 to S4), with the exception of some that were found more abundantly in the spleen. The fact that the latter capsids gave low mRNA expression in the spleen (Fig. 2, also explaining the pronounced error bars) implies their nonspecific uptake by, e.g., phagocytosis in macrophages, resulting in their detection on the AAV DNA but not on the AAV mRNA level.
Fig. 2. Transcriptional specificity of selected AAVs in multiple tissues.
Shown is the transcriptional specificity (Tαβ) of 12 capsids as normalized proportion per cell (dg) in 10 tissues. Depicted are mean cDNA values with SD from three C57BL/6J mice. Liver samples are highlighted in red.
Combinatorial AAV engineering through rational peptide transfer
AAVS1 and AAVS10 showed a promising specificity profile (Fig. 2 and fig. S4); however, their efficiency was inferior to AAVMYO not only in the off-targets but also in all muscle types (figs. S1 and S3). Because their C-terminal portion stems from AAV9, it comprises the insertion site of the P1 peptide in our prior lead candidate in the musculature, AAVMYO (6). This tempted us to study whether the transfer of P1 into the same region in AAVS1 and AAVS10 would selectively improve their efficiency in the musculature without perturbing their specificity, particularly without concurrently increasing their unwanted expression in off-target tissues [Fig. 1A (v to vii)].
Therefore, we cloned these semirationally designed capsids—called AAVMYO2 (AAVS1P1) and AAVMYO3 (AAVS10P1)—and again harnessed our barcoding pipeline for their in vivo validation. Next to AAV9, we now also included AAV8 and AAVrh74 as controls as they are currently used in muscle gene therapy trials. Additional controls were taken from our previous work in which we had inserted the P1 peptide into 11 other AAV serotypes besides AAV9, namely, AAV1 through AAV8, AAVrh10, AAVpo1, and AAV12, and in which we had studied the resulting vectors in cultured cells (22). Last, because of the low but detectable activity of AAVMYO and other capsids in the brain (Fig. 2 and figs. S1 and S2), we added AAV-PHP.B as a further benchmark owing to its reported high efficiency and specificity in this tissue following peripheral delivery (29).
In total, this yielded 18 capsids [12 P1-modified WTs including AAVMYO (AAV9P1), as well as AAVMYO2, AAVMYO3, AAV8, AAV9, AAVrh74, and AAV-PHP.B] that we used to produce another library of barcoded CMV-eYFP vectors for side-by-side comparison in two mouse strains, C57BL/6J (as before) and NMRI (The Naval Medical Research Institute) (to detect potential strain differences). AAV6P1 was difficult to manufacture and thus excluded, leaving a final library of 17 capsid variants for in vivo analysis. Akin to the first library, all variants were homogeneously represented, as confirmed by NGS (Fig. 3, A and B). To focus our analysis of this library on the on-target tissues, we added tongue as another muscle type but omitted several off-targets that had yielded low barcode reads in the first screen, leaving seven tissues (quadriceps, triceps, heart, diaphragm, tongue, liver, and brain). After systemic library delivery, droplet digital PCR (ddPCR) quantification confirmed vector presence in all these tissues (Fig. 3C).
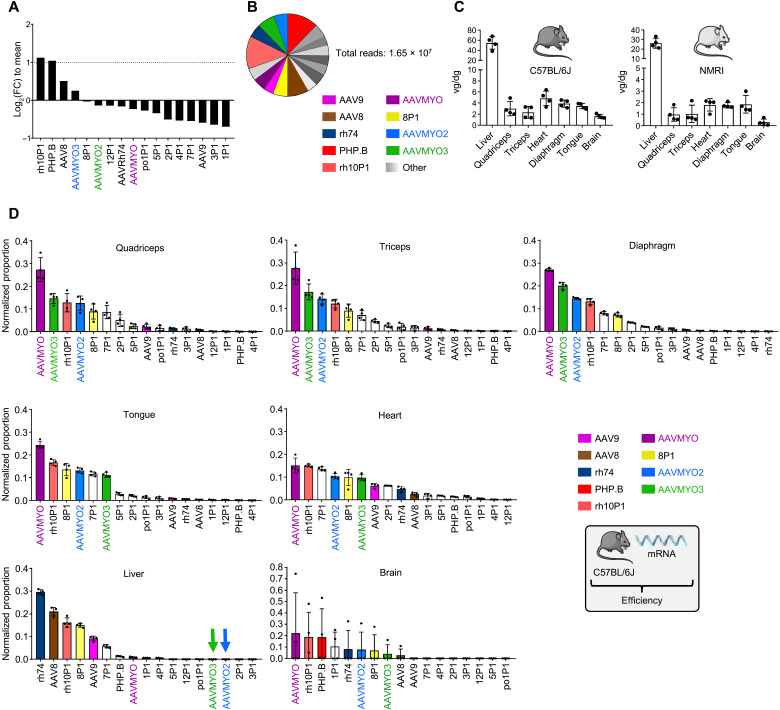
Fig. 3. Biodistribution of P1-displaying AAV variants.
(A) Composition of the barcoded library. Shown are log2(FC) values relative to the mean for each barcoded AAV variant in the library, with the dotted line marking a twofold change. Negative values indicate underrepresentation of the respective variant, and positive values imply overrepresentation. (B) Percentages of each barcoded AAV in the parental library based on unique recovered reads compared to the total reads (indicated next to the chart). (C) Viral DNA distribution of the library in seven organs. Shown are vg/dg (individual mice and averages) (means + SD) from four C57BL/6J or NMRI mice, respectively (each mouse is one dot). Detected vector genomes (eyfp probe) were normalized to RPP30 as a housekeeper. Mice were injected with 2.5 × 1012 vg and euthanized for tissue harvest 3 weeks later. (D) All 17 capsids were ranked by normalized transcriptional efficiency (Vαβ) in the shown seven tissues. Bars are mean values with SD from four C57BL/6J mice (each mouse is one dot). AAV variants of particular interest in this work are highlighted by colors.
Notably, the P1-modified chimeras AAVMYO2 and AAVMYO3 consistently ranked among the top performers on the AAV mRNA level in all muscle tissues and in both mouse strains [efficiency ranking in Fig. 3D (C57BL/6J) and fig. S5 (NMRI), specificity ranking in fig. S6 (C57BL/6J) and fig. S7 (NMRI)]. In these tissues, their barcode counts were always within a twofold range of those of AAVMYO, indicating an improved efficiency over the parental chimeras AAVS1 and AAVS10 (compare to Fig. 2 and figs. S1 to S3). Notably, AAVMYO2- or AAVMYO3-specific barcodes were nearly absent in the off-targets liver or brain. This suggested that the rational combination of two preselected myotropic components, i.e., shuffled capsid backbones enriched in the musculature and the P1 peptide, had acted synergistically and boosted on-target specificity without enhancing unwanted off-target activity.
We also noted that four other P1-displaying capsids (based on AAV2, AAV3, AAV5, or AAVpo1) showed a muscle specificity comparable to AAVMYO, AAVMYO2, and AAVMYO3 in both mouse strains (figs. S6 and S7). While their efficiencies were lower (Fig. 3D and fig. S5), this additionally confirms the inherent myotropic property of the P1 peptide in an AAV context. This peptide also shifted the tropism of AAV8 toward the musculature and away from the liver, albeit AAV8P1 remained relatively efficient in the liver (more than the other P1 variants). Another side observation was that, in our hands, AAVrh74 was inconspicuous in all muscle types but was the top hit in the liver on the AAV mRNA level (Fig. 3D and figs. S5 to S7). Last, we were intrigued by AAVrh10P1, which performed well in all seven tissues (Fig. 3D and figs. S5 to S7) and which even surpassed AAV9 that is known for its wide activity in various species including mice (30), suggesting the usefulness of AAVrh10P1 for future work requiring widespread in vivo transduction.
These trends were recapitulated on the AAV vector DNA level (figs. S8 to S10; raw data in fig. S11), with the notable exception of AAVrh74, which ranked higher in skeletal muscle as compared to the AAV mRNA data (Fig. 3D and figs. S5 to S7). This implies that only a limited portion of AAVrh74-encoded vector genomes is transcriptionally active in the murine muscle. We also note that AAVPHP.B was the top hit in the brain of C57BL/6 and, to a lesser extent, in NMRI mice, confirming its potency at crossing the blood-brain barrier especially in the C57BL/6 strain (29).
To additionally verify the increased efficiency and specificity of AAVMYO2 and AAVMYO3 with another reporter and in yet another mouse strain, we produced vectors encoding a firefly luciferase reporter under the control of either the human desmin (hDES) promoter or the synthetic SPc5-12 promoter [Fig. 1A (viii)]. Both cassettes were encapsidated in AAV9, AAVMYO2, or AAVMYO3 and intravenously injected into three mice per cohort at 1 × 1011 vg per animal. WT AAV9 was used as the benchmark in this experiment to detect the full range of complementary improvements, i.e., better on-targeting in the musculature combined with pronounced detargeting especially from the liver, that can be achieved by combinatorial engineering of a natural AAV serotype on the levels of transduction (capsid) and transcription (promoter).
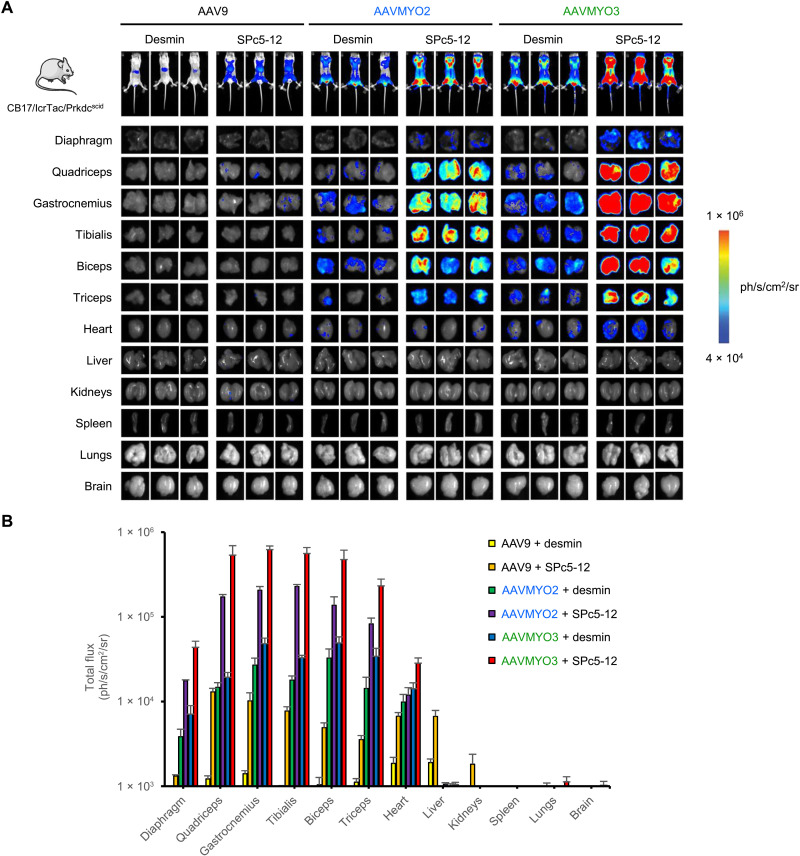
Whole-body imaging of luciferase expression 3 weeks later showed that, as compared to the respective AAV9 control and with both promoters, swapping to the AAVMYO2 or AAVMYO3 capsid substantially improved muscle-directed transgene expression (Fig. 4A). Imaging of luciferase expression in 12 different organs extracted from these mice confirmed that the increases observed on the whole-body level were due to a capsid- and/or promoter-mediated boost in all muscles, comprising the diaphragm, quadriceps femoris, gastrocnemius, tibialis anterior (TA), biceps brachii, triceps, and heart (Fig. 4B). The SPc5-12 promoter typically yielded significantly higher luciferase expression levels than the DES promoter across all skeletal muscle groups, heart, and diaphragm (Fig. 4, A and B). Combining SPc5-12 with AAVMYO3 resulted in a substantial, two- to three-log increase in luciferase expression in all skeletal muscle groups compared to the AAV9-hDES reference vector. Although low-level luciferase activity was apparent in the AAV9-transduced liver, due to leaky expression from the SPc5-12 or DES promoters, switching capsids to AAVMYO2 or AAVMYO3 resulted in effective liver detargeting consistent with the absence of any detectable luciferase expression in the liver.
Fig. 4. Biodistribution of AAVMYO2 and AAVMYO3.
CB17/IcrTac/Prkdcscid mice were intravenously injected with 1 × 1011 vg per mouse and analyzed 2 to 3 weeks later. (A) In vivo whole-body bioluminescence imaging of mice injected intravenously with the shown vectors, depicted on a color scale from 4 × 104 (blue) photons/s/cm2/sr to 1 × 106 (red) photons/s/cm2/sr at week 2 after injection. Photon emission was measured dynamically during 7 min in a supine position. Ex vivo bioluminescence imaging of individual organs harvested at week 3 after injection is represented on a color scale with luciferase intensities ranging from 2.5 × 104 (blue) photons/s/cm2/sr to 3.5 × 105 (red) photons/s/cm2/sr. Bioluminescence signals were quantified for 5 min. (B) A hand-drawn region of interest was used for every individual tissue. Luciferase expression from the individual muscles was measured as total flux, expressed in photons/s/cm2/sr (means ± SEM; n = 3).
Last, to verify that the properties of AAVMYO2 and AAVMYO3 are truly mediated by the P1 peptide and not by the modification in capsid loop VIII, we swapped P1 in AAVMYO3 with another peptide from our prior screen, P4 (NDVRSAN) (6, 31). In C57BL6/J mice, the ensuing variant AAVS10P4 showed a markedly different biodistribution, including robust targeting of the liver and much lower activity in the muscle as compared to AAVMYO3 or AAVMYO (fig. S12).
Efficient large-scale production and purification of AAVMYO2 and AAVMYO3
A pivotal prerequisite for clinical translation of bioengineered AAV vectors is compatibility with technology for large-scale vector manufacturing, including upstream production in suspension cells and downstream purification with scalable methodologies (32, 33). We thus compared the yields of AAVMYO2 or AAVMYO3 to those of the established, well-producing serotypes AAV8 and AAV9, by packaging five independent vector constructs (ID 1 to 5 in fig. S13) into all four capsids in human embryonic kidney (HEK) 293T cells grown in 250-ml suspension cultures in Erlenmeyer flasks. In all cases, yields for AAVMYO2 or AAVMYO3 were comparable to, or higher than, those obtained with the same vector construct in the AAV8 or AAV9 context (fig. S13A). Identical observations were made when the suspension cultures were scaled up to 10-liter bioreactors, which resulted in two- to fourfold higher titers for AAVMYO2 or AAVMYO3 as compared to AAV8 (fig. S13B).
Next, we assessed the compatibility of AAVMYO2 and AAVMYO3 with commercial affinity resins for AAV purification, i.e., AVB Sepharose and Poros AAV8, AAV9, or AAVX. These assays were performed in 96-well plates and AAV particle binding was determined via vector titration in the flow-through [% captured AAV = 100% – (feed titer/flow-through titer)]. We measured more than 80% binding of the AAV9 control to Poros AAV9 or AAVX but not to Poros AAV8, confirming the validity of the control and assay as well as its quantitative nature (fig. S13C). The two bioengineered capsids also bound very effectively (80 to almost 100%) to Poros AAV9 or AAVX, verifying their compatibility with latest-generation affinity resins. Last, we purified AAVMYO2 and AAVMYO3 vector stocks using Poros AAVX in a 10-liter scale and found that 95 to 100% of viral particles were recovered in the eluate (fig. S13D), proving that also downstream processing is scalable. With 31% (AAVMYO2) or 48% (AAVMYO3) full capsids measured by analytical centrifugation after affinity chromatography and without any capsid enrichment step, the two variants were again comparable to, or better than, AAV8 (32%).
Modeling of bioengineered capsids
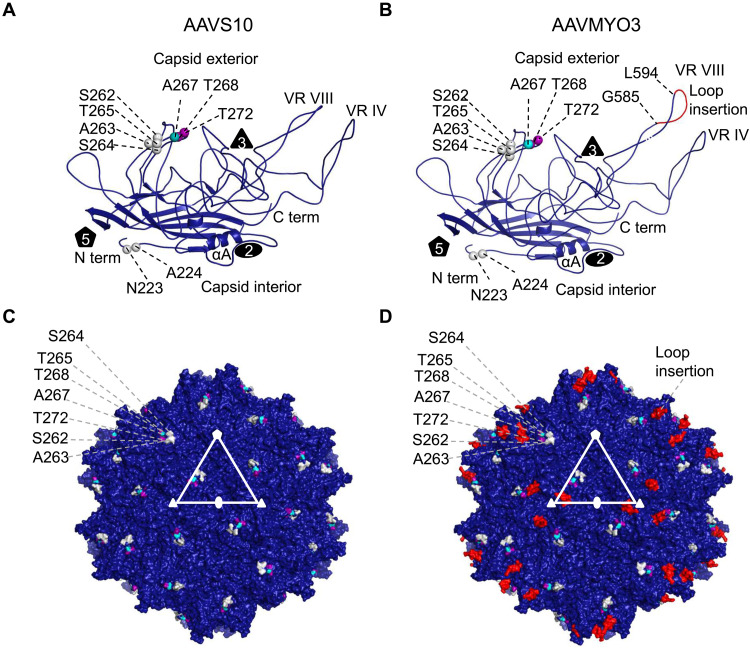
Modeling of the bioengineered capsids revealed that, consistent with the structure of other AAVs, AAVS1, AAVS10, AAVMYO, AAVMYO2, and AAVMYO3 monomers contain the conserved eight-stranded, antiparallel β barrel motif that forms the core of the virus capsid (Fig. 5, A and B, and fig. S14, A, B, E, and F). The interconnecting loops between the β strands form motifs whose sequence and structure differ between the AAV serotypes and that are designated variable regions (VRs), specifically VRs I to IX (34). These VRs are located predominantly on the capsid surface and they cluster around the icosahedral twofold, threefold, and fivefold axes (Fig. 5, C and D, and fig. S14, C, D, G, and H). Moreover, they play critical roles in receptor binding, transduction, and antigenicity (35). The VRs form the predominant features of the capsid exterior, for example, the threefold protrusion is assembled from the interaction of VRs VIII, IV, and V of a symmetry-related monomer, and five VR IIs form the inner wall of the fivefold pore. Furthermore, these VRs have been shown to tolerate structural modifications including loop swapping and peptide insertions (36, 37), which allow for virus targeting or detargeting of specific tissues. The AAVS10 capsid contains residues N223 and A224, which are the same residue types as AAV1 and AAV6 at these amino acid positions (Fig. 5A). This is similar to the sequence in AAVS1. In addition, the AAVS1 variant has M211 and T234 at the same residue positions and type as AAV1, AAV6, and AAV8. These residues are located under the fivefold pore of the ordered N-terminal region of VP3. As such, they are not seen on the surface representation of the AAVS10 60-mer (Fig. 5C). Previous reports showed the fivefold pore to be seminal for VP1 externalization and genome packaging (38, 39). The amino acid stretch 262 to 272 of AAVS10 is similar to residues found in VP3 of AAV1 and AAV6, and this region has been indicated to be important for AAV9 receptor (galactose) binding (40). Similarly, several amino acids between residues 264 and 274 of AAVS1 are the same as those found in AAV8. It is difficult to assign an AAV8 receptor binding profile to this region as there are currently no identified receptors for AAV8. The P1 peptide insertion G585-L594 is located within VR VIII, which is the most radially protruding motif of the capsid (Fig. 5D and fig. S14, B and F) (40). Insertion in this site has been shown to disrupt the natural laminin receptor binding domain of AAV9 (41) while exposing the foreign peptide for binding to alternative receptors. This is also true for the P1 peptide insertion in AAVMYO and AAVMYO2 (fig. S14).
Fig. 5. AAVS10 and AAVMYO3 variant models.
Ribbon diagram of (A) AAVS10 and (B) AAVMYO3 VP3 monomer. The eight-stranded β barrel motif βBIDG-βCHEF, N and C termini, αA helix, variable region VIII (VRVIII), and the capsid interior and exterior are as labeled. The position of the icosahedral two-, three-, and fivefold axes are shown as a black filled oval, triangle, and pentagon, respectively. Specific amino acids with the sequence and location equivalent to AAV9 are colored violet, and those similar to AAV1, AAV6, and AAV8 are colored gray, teal, and purple spheres, respectively. In addition, the peptide insertion P1 at VR VIII from G585-L594 is colored red in (B). (C and D) Surface representation of the (C) AAVS10 and (D) AAVMYO3 T = 1 icosahedral capsid, using the color scheme of (A) and (B). The surface features show the characteristic twofold depression at the twofold axes, threefold protrusions surrounding the threefold axes, the two-/fivefold wall, and the fivefold pore forming the fivefold axes. The twofold, threefold, and fivefold axes are labeled as white filled oval, triangle, and pentagon, respectively. The figures were generated using the program PyMOL.
Analysis of AAVMYO2 and AAVMYO3 seroprevalence
The presence of pre-existing anti-AAV antibodies represents a major hurdle for in vivo gene therapy as it precludes vector administration in a large portion of the population (42). Therefore, we evaluated the prevalence of antibodies against AAVMYO2 and AAVMYO3 in a cohort of human sera. AAV9, a serotype commonly used for muscle targeting and contributing extensively to the C-terminal portion of our engineered capsids, was used as a control. The two synthetic capsids showed a seroprevalence comparable to AAV9, as measured by enzyme-linked immunosorbent assay (ELISA) (fig. S15A) (43) and in vitro neutralization test (fig. S15B) (44). These results indicate that the serologic profile of the peptide-modified AAV9 capsid variants is equivalent to AAV9 and compatible with clinical translation.
Use of bioengineered myotropic AAVMYO2 and AAVMYO3 vectors for gene therapy of myotubular myopathy
XLMTM is a severe inherited disease caused by mutations in the MTM1 gene coding for the phosphoinositide phosphatase myotubularin and is characterized by generalized muscle weakness, respiratory insufficiency, and high mortality during the first years of life (45, 46). Animal models of the disease exist in various species, but Mtm1 knockout (KO) mice (47, 48) were pivotal in developing a gene replacement therapy by intravenous delivery of a recombinant AAV8 vector expressing myotubularin. This vector also showed efficacy in dogs with XLMTM, paving the way for an ongoing clinical trial in patients with XLMTM (49–51). We therefore used these mutant mice, which manifest reduced life expectancy, skeletal muscle weakness, and hypotrophy, to assess the therapeutic potential of the synthetic myotropic AAV capsids reported here [Fig. 1A (ix)].
Administration of AAVMYO2- or AAVMYO3-MTM1 vectors prolongs life span and increases strength of Mtm1-KO mice
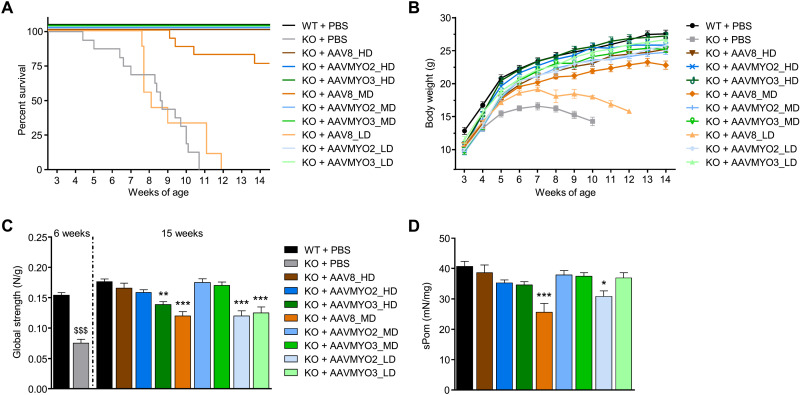
We performed a comparative dose-finding study over a 3-month period to define the therapeutic window of AAVMYO2, AAVMYO3, and AAV8 vectors expressing MTM1 under the control of a muscle-specific DES promoter by intravenous injection in 3-week-old Mtm1-KO mice. For each vector, three doses were tested: 1.0 × 1014 (high dose: HD), 2.0 × 1013 (mid dose: MD), and 4.0 × 1012 vg/kg (low dose: LD). WT and KO mice injected with phosphate-buffered saline (PBS) were used as controls. As shown in Fig. 6A, a single injection of AAV8-MTM1 at HD prolonged the life span of all mutant mice, whereas 75% of KO mice treated at MD and none at LD survived by the end of the study. In contrast, the administration of AAVMYO2 and AAVMYO3 vectors rescued the survival of all treated Mtm1-KO animals at the three different doses. In addition, the body weight of the mutant mice treated with AAVMYO2 and AAVMYO3 at LD and MD was similar to that of animals in the AAV8_HD group, and the mean body weight of AAVMYO2- and AAVMYO3-treated Mtm1-KO mice at HD reached the level of age-matched WT littermates (Fig. 6B). In contrast, the weight of mutant mice that received the MD and LD of AAV8-MTM1 was not rescued. Consistent with these results, the mass of a panel of skeletal muscles throughout the body (quadriceps, gastrocnemius, TA, extensor digitorum longus, soleus, biceps brachii, and triceps) in treated KO mice was similar to that of WT littermates. Exceptions comprised the AAV8_MD, AAVMYO2_LD, and AAVMYO3_LD groups, in which most analyzed muscles had lower weights (fig. S16A). As mutant mice injected with AAV8_LD did not survive until 3 months after injection, they were not included in these and the following analyses.
Fig. 6. Systemic gene therapy with AAVMYO2 or AAVMYO3 expressing myotubularin improves life span, body growth, and strength in Mtm1-KO mice at lower doses than AAV8.
(A) Survival rate and (B) body weight of WT (n = 19) and KO mice (n = 16) injected 3 weeks after birth with PBS or AAV8 and AAVMYO2 or AAVMYO3 vectors at 1.0 × 1014 vg/kg (HD, n = 10, 8, or 8, respectively), 2.0 × 1013 vg/kg (MD, n = 17, 9, or 11, respectively), or 4.0 × 1012 vg/kg (LD, n = 9). (C) Escape test measurements in WT (n = 5 at 6 weeks, n = 8 at 15 weeks) and KO mice (n = 6 at 6 weeks) injected with PBS or AAV8, AAVMYO2, or AAVMYO3 at HD (n = 7, 8, or 8, respectively), MD (n = 8), or LD (n = 8) at 15 weeks of age. (D) Specific tetanic force of TA muscles from WT or treated KO mice at 15 weeks of age (n = 8, except for KO + AAV8_HD and KO + AAVMYO3_HD n = 7). Data are means ± SEM. Statistical analyses were performed by using unpaired t test at 6 weeks ($$$P < 0.001) or one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison posttest, at 15 weeks; *P < 0.05; **P < 0.01; ***P < 0.001 versus WT + PBS.
We next assessed the effect of vector delivery on muscle strength of Mtm1-KO mice. The global force of animals was quantified using a noninvasive escape test, which was reduced by 50% in untreated mutant mice at 6 weeks of age compared to WT littermates (Fig. 6C). Three months after AAV vector injection, the global strength of MD AAVMYO2- and AAVMYO3-treated mutant animals was comparable to the AAV8_HD group and WT controls. Furthermore, the specific force of TA muscle from treated KO mice in these MD groups reached normal values (Fig. 6D).
Chimeric myotropic vectors AAVMYO2 and AAVMYO3 ameliorate the histological hallmarks of XLMTM muscle pathology
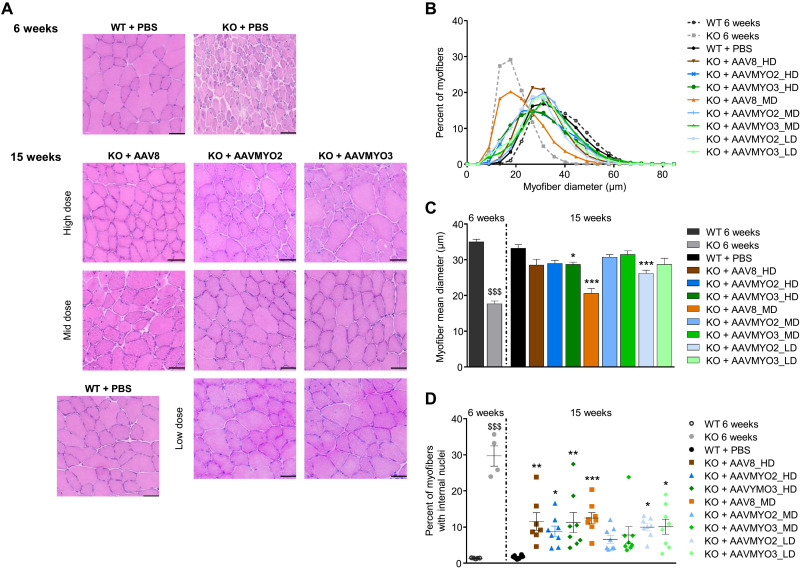
Skeletal muscles of mice deficient for myotubularin present hypotrophic fibers with mislocalization of internal organelles, such as nuclei and mitochondria (48). Therefore, we evaluated the potency of the AAVMYO2- and AAVMYO3-MTM1 vectors in correcting muscle morphology compared to the AAV8 vector in Mtm1-KO mice at 3 months after injection. Muscle cross sections from TA and biceps brachii were stained with hematoxylin and eosin (H&E) and nicotinamide adenine dinucleotide dehydrogenase–tetrazolium reductase (NADH-TR), which labels mitochondria, for analysis (Fig. 7A and fig. S16B). Whereas, at 6 weeks of age, muscles of untreated KO mice clearly present signs of the pathology, the mean myofiber size was normalized in TA of all AAV-treated groups, except in AAV8_MD, AAVMYO2_LD, and AAVMYO3_HD mice (Fig. 7, B and C). Abnormal small myofibers were observed in TA transduced with HDs of AAVMYO2- and AAVMYO3-MTM1 vectors, which may reflect an excessive level of myotubularin expression in these muscles. In the MD AAVMYO2 and AAVMYO3 groups, muscle fiber size and mitochondria positioning were normalized, and the percentage of fibers with internal nuclei was strongly reduced (Fig. 7, A, C, and D). In a less severely affected muscle of the forelimb, biceps brachii, remarkable histological improvements were achieved at 3 months in all AAV-treated Mtm1-KO mice (fig. S16, C to E). As previously described in XLMTM mice, but not found in dogs (50, 51), signs of cardiac lesions characterized by the presence of fibrotic areas in tissue cross sections were observed in KO mice overexpressing myotubularin in groups treated with high vector doses but were barely present in the AAVMYO2_MD and AAVMYO3_MD groups (fig. S16, F and G). In these regions of fibrosis, cellular infiltrates that could correspond to inflammatory cells were present (fig. S16H). Heart lesions following AAV-mediated gene therapy could be transgene-, species-, and/or disease-related and may differ between pathological conditions. Changing the balance of vector transduction and, in particular, vector-derived transgene expression toward skeletal muscle, while keeping expression in the heart, could be beneficial for disorders that require expression in both tissues, as opposed to other strategies such as microRNA-mediated transcript degradation (52). Collectively, these results show that intravenous administration of lower doses of AAVMYO2- and AAVMYO3-MTM1 vectors than AAV8-MTM1 can efficiently correct the signs of the disease in skeletal muscles with increased specificity in Mtm1-KO mice.
Fig. 7. AAVMYO2 or AAVMYO3 corrects skeletal muscle fiber hypotrophy and internal architecture of mutant mice at a lower dose than AAV8.
(A) Cross sections from TA stained with H&E from WT (at 6 and 15 weeks) and KO mice injected with PBS (at 6 weeks) or AAV8, AAVMYO2, or AAVMYO3 at 1.0 × 1014 (HD), 2.0 × 1013 (MD), or 4.0 × 1012 vg/kg (LD) at 15 weeks of age. Scale bars, 50 μm. (B) TA myofiber diameter frequency distribution (n = 5 per group). (C) Muscle fiber mean diameter from TA (n = 5 per group). (D) Percentage of myofibers with internal nuclei in TA (n = 5 per group at 6 weeks and n = 8 per group at 15 weeks, except for KO + AAV8_HD n = 7). Data are means ± SEM. Statistical analyses were performed by using unpaired t test at 6 weeks ($$$P < 0.001) or one-way ANOVA followed by Dunnett’s multiple comparison posttest: *P < 0.05; **P < 0.01; ***P < 0.001 versus WT + PBS.
Intravenous administration of AAVMYO2 and AAVMYO3 vectors results in increased transduction of skeletal muscles and liver detargeting in Mtm1 mutant mice
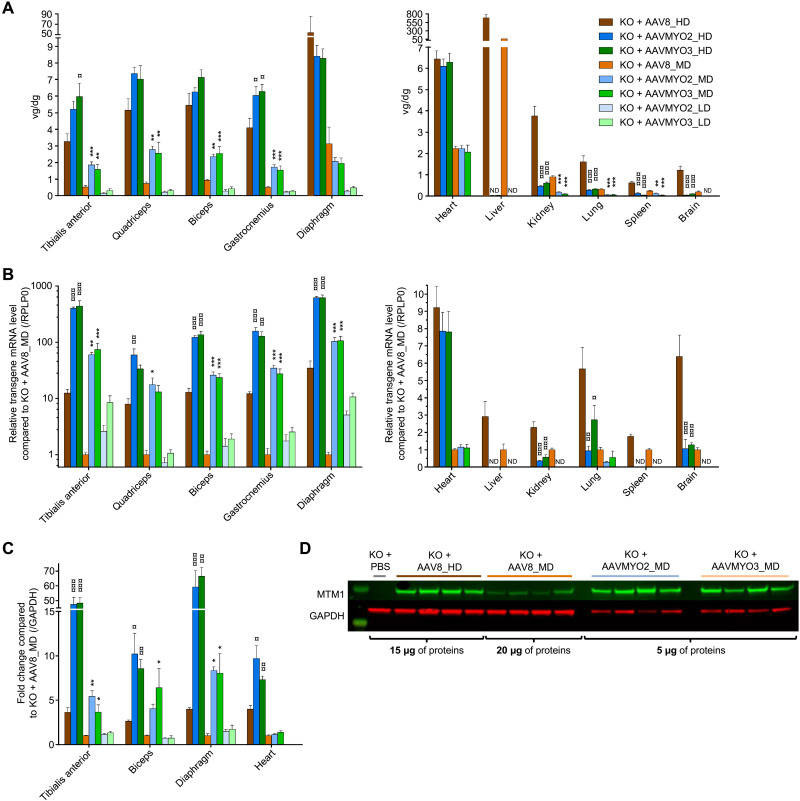
We additionally analyzed the biodistribution and transgene expression of AAVMYO2-, AAVMYO3-, and AAV8-MTM1 in KO mice 3 months after intravenous vector injection. Figure 8A shows that vector copy numbers were higher in skeletal muscles after AAVMYO2 and AAVMYO3 delivery, in particular at MDs, compared to AAV8, whereas transduction was similar in the heart. In addition and congruent with our data in WT mice (see above), the engineered capsids were also more specific as they were able to detarget main organs, such as liver or kidney. Transgene expression was largely superior to AAV8 in skeletal muscles, including the diaphragm, by AAVMYO2 and AAVMYO3 transduction (Fig. 8B), with MTM1 mRNA levels 10 to 100 times higher in the MD groups. This resulted in increased myotubularin levels in analyzed TA, biceps brachii, and diaphragm muscles, as assessed by Western blotting, reaching or exceeding the levels found with a fivefold higher AAV8 vector dose (Fig. 8, C and D).
Fig. 8. Biodistribution and transgene expression of AAVMYO2- or AAVMYO3-MTM1 vectors compared to AAV8 in XLMTM mice.
(A) Vector copy number (vg/diploid genome) of AAV8, AAVMYO2, or AAVMYO3 at 1.0 × 1014 (HD), 2.0 × 1013 (MD), or 4.0 × 1012 vg/kg (LD) in various muscles and organs of Mtm1-KO mice 3 months after injection (n = 8 per group, except for KO + AAV8_HD n = 7). (B) MTM1 mRNA levels normalized by KO + AAV8_MD values in muscles and organs of Mtm1-KO mice at 15 weeks of age (n = 8 per group, except for KO + AAV8_HD n = 7). (C) Myotubularin protein quantification by immunoblot in TA, biceps, diaphragm, and heart (n = 4 per group). (D) Immunoblot illustrating myotubularin (green) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as control in red in diaphragm of KO mice 3 months after vector injection (AAV8_HD, AAV8_MD, AAVMYO2_MD, and AAVMYO3_MD). Data are presented as means ± SEM. Statistical analyses were performed for each dose by using one-way ANOVA followed by Dunnett’s multiple comparison posttest: *P < 0.05; **P < 0.01; ***P < 0.001 versus KO + AAV8_HD or AAV8_MD depending on the group.
Last, we also evaluated the response of XLMTM mice to intravenous administration of AAV9-MTM1 at two doses (MD and LD) for a 3-month period. We found that at LD (4.0 × 1012 vg/kg), AAV9 was not as efficacious as AAVMYO2 and AAVMYO3 in prolonging the life span and increasing body weight, muscle mass, and strength in mutants (fig. S17).
Together, our results demonstrate that MTM1 vectors based on the AAVMYO2 and AAVMYO3 capsids are more potent and more specific than AAV8 and AAV9 for phenotype correction in mice with myotubular myopathy.
Application of bioengineered myotropic AAVMYO3 vector in a mouse model of DMD
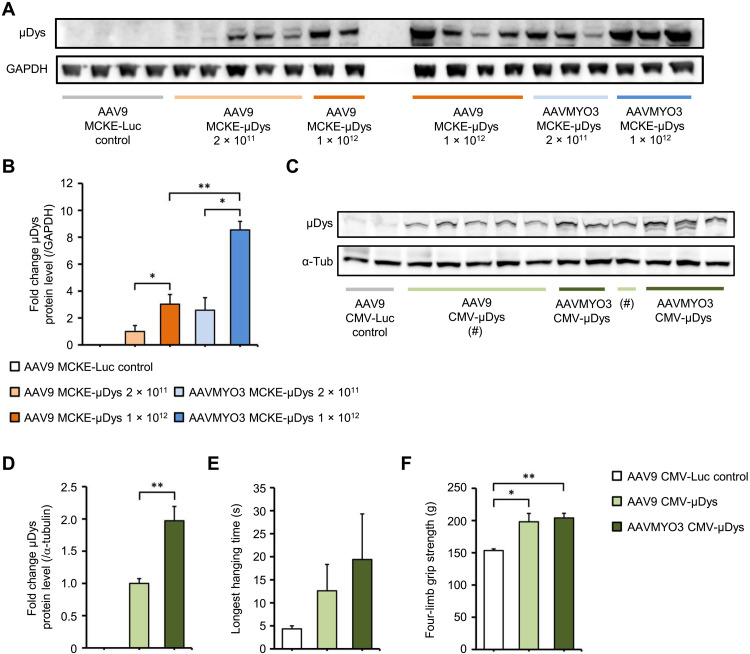
To additionally study the capacity of our bioengineered myotropic capsids for gene therapy of muscle disorders in an independent animal model, we analyzed the expression of a microdystrophin gene from the AAVMYO3 variant in comparison to AAV9 in dystrophin-deficient mdx mice. We found that expression from AAVMYO3 was significantly higher in skeletal muscle (quadriceps) at different doses (2 × 1011, 1 × 1012, or 2 × 1012 vg per mouse) when driven by the muscle creatine kinase (MCKE) promoter (Fig. 9, A and B) or the CMV promoter (Fig. 9, C and D). Similarly, we measured more robust expression from AAVMYO3 in the heart (fig. S18A) and diaphragm (fig. S18B). Using the CMV promoter–driven construct, we also noted functional benefits by quantifying the longest hanging time and by performing a four-limb grip strength assay 23 weeks after vector delivery. While the values did not reach statistical significance, we observed a trend toward better results with the AAVMYO3 vector over the AAV9 benchmark (Fig. 9, E and F).
Fig. 9. AAV9- and AAVMYO3-mediated overexpression of μDys in Mdx mice.
(A) Immunoblot with densitometry analysis (B) showing μDys expression in quadriceps femoris muscles of mdx mice 12 weeks after intravenous injection of 2 × 1011 or 1 × 1012 vg of a control vector (AAV9 Luc–expressing luciferase), or of AAV9 or AAVMYO3 encoding μDys under control of the muscle creatine kinase (MCKE) promoter (n = 3 to 5 per group; loading control GAPDH). (C) Immunoblot with densitometry analysis (D) showing μDys expression in quadriceps femoris muscles of mdx mice 23 weeks after intravenous injection of 2 × 1012 vg of a control vector (AAV9 Luc–expressing luciferase), or of AAV9 or AAVMYO3 encoding μDys under control of the CMV promoter (n = 5 to 6 per group; loading control α-tubulin). These mice were subjected to a four-limb hanging test (E) and four-limb grip strength testing (F) to evaluate muscle performance. Data are presented as means ± SEM. Statistical significance was calculated by unpaired two-tailed Student’s t test or one-way ANOVA. *P < 0.05; **P < 0.01.
DISCUSSION
In this work, we have pursued a complex approach at AAV vector bioengineering that merges two seminal technologies and constituents in a single particle, i.e., chimeric capsid scaffolds and a muscle-specific retargeting peptide recently identified by us by peptide library panning in the AAV9 context (6). Previously, we and many colleagues have already validated the power of each individual strategy, yet the current work advances the fields of AAV diversification, engineering, and screening by demonstrating the unique synergism that can be achieved through their combination. To our knowledge, this study represents the first combinatorial approach that merges iterative in vivo AAV capsid shuffling library screens with rational design via incorporation of a targeting peptide identified independently through peptide library screens.
Our key finding is that the top hits from the separate application of different AAV capsid evolution techniques (here, DNA family shuffling and peptide display) can be fused in a semirational fashion, yielding synthetic viral protein shells that can, in turn, be stratified in vivo using DNA/RNA barcoding. While we used a ubiquitously active CMV promoter for all in vivo screening experiments, the efficiency and specificity of these capsids in the musculature can be boosted even further via incorporation of potent muscle-specific promoters, as demonstrated here, or of synthetic genetic elements, such as muscle-specific cis-regulatory enhancers (53).
This conclusion has several important ramifications for the AAV vector and gene therapy fields, as it showcases the potential of merging multiple capsid evolution techniques and thus substantially expands our toolbox for vector bioengineering. Key for the success in this work was our original multistep AAV evolution and selection process, in which we first identified shuffled capsids with increased specificity but subpar efficiency in the on-target as compared to the AAV9 benchmark, before we further boosted both parameters in a second, rational step by engrafting a synthetic myotropic peptide onto selected candidates. To our best knowledge, this sequential and combinatorial process represents a conceptual advance that distinguishes our work from previous attempts at selecting myotropic capsids including our own (6), which had typically focused on a single AAV diversification technology, such as DNA family shuffling (25, 26, 54), peptide display in one WT serotype (23), or domain swapping between two serotypes (24).
Concurrently, our work illustrates the capacity of synthetic capsids to act as scaffolds for peptide display, which not only verifies our conclusions with the shuffled AAV-DJ capsid but also complements and extends the wealth of data obtained in the past with WT AAVs (1, 2). Together, this implies substantial benefit from performing future peptide library selections in a chimeric AAV capsid backbone, which is an as-of-yet underdeveloped approach with far-ranging potential also beyond the musculature that merits further investigation and application in the future. Particularly promising permutations of the strategy reported here that we envision include, for instance, the display of multiple different retargeting peptides in two separate loops of a preselected shuffled AAV capsid, akin to recent work by the Gradinaru laboratory that has improved upon an AAV9 peptide display mutant by insertion of a second, independent sequence in an alternative capsid loop (55). Another interesting approach to explore and expand the combinatorial strategy that was introduced here is to preselect shuffled or otherwise engineered capsids for evasion of neutralizing anti-AAV antibodies before semirational peptide modification, with the aim to enable in vivo vector readministration strategies in the same tissue that are typically impossible when using WT AAVs such as AAV9 as a single scaffold.
Until then, by selectively grafting a single, preselected peptide onto lead capsids from our separate screen and by obtaining superior particles, this work exemplifies the promise of semirational AAV bioengineering and its ability to accompany selection-driven, top-down screening approaches. This combinatorial strategy has great potential to fine-tune capsid specificity and efficiency and to thereby yield variants that concomitantly act in multiple tissues and to different degrees. This is illustrated by our notion that adding a myotropic peptide to two capsids that were inherently muscle specific resulted in nearly complete detargeting from the liver (the typical main off-target for AAVs), while maintaining the broad efficiency in the muscle that we had found for the P1 peptide in the AAV9 context (6). This further improvement in in vivo muscle specificity achieved here over our previously reported, first-generation AAVMYO capsid is seen in multiple datasets in this work (figs. S5 to S10), and it becomes particularly apparent in the direct comparison of AAVMYO versus AAVMYO3 shown in fig. S12. The AAVMYO2 and AAVMYO3 capsids resulted in an increased relative proportion of vector genomes and transgene expression in skeletal muscles versus the heart, as compared to analyzed natural serotypes or AAVMYO (Figs. 4 and 8 and fig. S5), which could be of particular interest for diseases that require expression of the transgene in both skeletal musculature and heart while reducing the risk of overexpression in cardiac tissue. Additionally advancing our prior work with the original AAVMYO capsid is our demonstration that in vivo muscle performance can be maximized by concurrent vector optimization on both the transduction (capsid) and transcription (promoter) levels (Fig. 4).
The possibility to detarget AAV vectors intended for muscle gene therapy from the liver, as verified here with the AAVMYO2 and AAVMYO3 capsids, is pivotal and encouraging in view of the aforementioned severe side effects including fatalities in recent HD AAV clinical trials, as it promises a better safety profile in future human vector recipients. Additional optimism is provided by our observation that such synthetic capsids can remain fully compatible with established up- and downstream AAV production processes. In combination with the elimination of liver as a typical sink for a major portion of the administered vector, this helps to alleviate the pressure on large-scale, clinical-grade AAV manufacturing and thus benefits future vector dissemination and application.
Curiously, in the present work, we found that the P1 peptide behaved differently in several other capsid scaffolds, which highlights the persisting gaps in our understanding of the complex AAV biology and strongly supports continued basic research, as a prerequisite for true rational vector engineering in the future. This also pertains to the two bioengineered capsids that we have introduced in this work, AAVMYO2 and AAVMYO3, as their biology remains enigmatic at this early point, including their unknown cellular receptor(s) and their activity in other species beyond the mouse. Notably, we have recently obtained several lines of experimental evidence supporting our prior notion (6) that the P1 peptide and other RGD-containing sequences interact with various types of integrins on the surface of target cells and harness these for transduction (56, 57). This is further supported by the latest work from the Sabeti laboratory, which also identified integrins as cellular receptors for their own AAV9-RGD capsid variants (23).
Some of the latter capsids showed efficiency and specificity in the musculature of healthy nonhuman primates, whereas others performed better in mice (23). While this raises optimism about the clinical translatability of these capsid variants including the chimeric AAVMYO2 and AAVMYO3 reported here, it also highlights pertaining general questions about the best preclinical model for selection of myotropic or other AAV capsids. Arguably, nonhuman primates are closer to humans than mice, yet this does not guarantee that the (integrin) receptors and other intracellular host factors targeted by the different RGD-displaying AAV variants are fully conserved and equally functional between all primate species including humans. Moreover, cellular gene expression signatures including surface receptors will vary between healthy and diseased musculature, implying the possibility that, regardless of species, selection in healthy animals may yield capsids that perform differently once applied in humans with a muscle disorder. Further exemplifying the challenges in identifying and harnessing an optimal animal model for in vivo selection or validation of AAV capsids for ultimate use in humans is the latest data reported at the 2022 Annual Conference of the American Society of Gene and Cell Therapy (ASGCT; https://annualmeeting.asgct.org/abstracts). For example, the Sabeti laboratory demonstrated that two AAV peptide display variants previously selected by the Gradinaru laboratory (55) for CNS (central nervous system) transduction in mice and shown to also function in marmosets (New World monkeys) failed to transduce the CNS in a different NHP model, i.e., macaques (Old World monkeys) (abstract 443). A second example was provided by the company AskBio, which reported an impressive performance of the synthetic AAV2i8 capsid (24) in the human heart in an ongoing clinical trial for NYHA class III nonischemic heart failure. Transduction of the human heart was 30-fold higher than that observed in preclinical studies in the porcine heart, despite the fact that pigs are widely considered and used as the physiologically most relevant large animal model for human heart disease and corresponding AAV vector development (abstract 1213). Last, as most recently illustrated by Chen et al. (58), it is possible to evolve synthetic AAV capsids in rodents that also perform well in multiple different NHP models, in this example, the peripheral nervous system of mice, rats, marmosets, and macaques. On the one hand, capsids that are widely active across small and large animal species offer benefits with respect to preclinical characterization including toxicity studies and are thus desirable. On the other hand, the sum of these latest data also raises important questions about the optimal animal models and the biological reasons underlying the notably different bioactivity of engineered AAV capsids that exclusively function in either small or large animals, respectively, whose performance differs between strains within the same mouse or NHP species or that do not translate well from animals to humans.
To begin to address these complex and comprehensive questions, an intriguing future task is to compare the lead candidates based on AAV9 or shuffled backbones from the identical dose and in healthy versus diseased, small or large animals, e.g., using barcoding technology, to gain further insights into the sequence and structure components governing muscle tropism across species up to humans and in normal versus affected tissue.
Notably, in this work, we have already verified the myotropic properties of these bioengineered capsids in healthy mice of four different strains, i.e., C57BL/6, NMRI, 129PAS, and CB17/lcrTac/Prkdcscid, and they were also maintained in two muscle disease models derived from C57BL/6 and 129PAS, supporting the broad applicability of these original AAVs. Accordingly, we propose that the two original AAV variants created and characterized here represent exciting tools for applications in the mouse that require highly specific, simple, and robust gene transfer to the entire musculature from systemic delivery for both basic research and preclinical evaluation of therapeutic concepts in healthy or diseased animals. For the reasons outlined above, these capsids or future derivatives with permutations in the amino acids surrounding the RGD motif likely hold equally great potential for muscle gene therapy in human patients, including those affected by DMD or XLMTM. While upcoming work will address and resolve this question of translatability into higher species including diseased humans, the major benefit of the present work is that it already illustrates the vast potential to derive unique gene therapy vectors by combining multiple AAV-directed evolution technologies, capsid and cargo engineering, and high-throughput in vivo screening via NGS and barcoding in a sequential and semirational fashion.
MATERIALS AND METHODS
Experimental design
The rationale and design of this work are depicted in Fig. 1A.
Generation of shuffled AAV capsid libraries
Details of the technology as well as all required plasmids and primers were previously described (59). In this work, two AAV capsid libraries were generated by shuffling the cap genes of AAV serotypes 1, 6, 8, and 9 (library A), or 1, 6, 8, 9, and po.1 (library B). All cap genes were PCR-amplified from corresponding plasmids as previously reported (59) and pooled in different ratios (1:1:2:2 or 1:1:2:2:3, respectively; AAV1 and AAV6 were purposely underrepresented because of their near sequence identity that favors their own recombination rather than with the other serotypes in the mix) to a final concentration of 4 μg. The two pools were fragmented using deoxyribonuclease (DNase I) (Invitrogen), and fragments ranging from 100 to 1000 base pairs (bp) were extracted following the separation of the reaction on 1% agarose gels. These fragments were then reassembled into full-length cap chimeras on the basis of partial homology by using a primerless PCR, followed by a large-scale PCR (18 reactions of 50 μl each, always using 2 μl of the first PCR as template). Per library, all 18 PCR reactions were concentrated on a PCR column (QIAquick PCR Purification Kit, Qiagen) and digested overnight at 37°C with Pac I and Asc I (cognate recognition sites flank all cap genes in our plasmids) using 20 U per enzyme in a total volume of 50 μl. The resulting 2.2-kb band was again gel-purified. A 40-μl ligation reaction was incubated overnight containing 1139 ng of chimeric cap genes and 861 ng of a replication-competent AAV plasmid containing AAV2 rep and ITRs (inverted terminal repeats) (59). One day later, 20 electroporations were performed using electrocompetent MegaXDH10B (Invitrogen) bacteria. To this end, the entire ligation reaction was mixed with 600 μl of bacteria and then aliquoted into 20 samples for electroporation. Immediately after transformation, the bacteria were resuspended in SOC (super optimal broth with catabolite repression) medium for recovery while shaking at 37°C. After pooling all reactions, 100 μl each of a 1:10 or a 1:100 dilution was plated on ampicillin-containing LB agar plates and the rest of the suspension was transferred into 300 ml of LB medium for overnight incubation at 37°C. On the following day, plasmid DNA was isolated using the NucleoBond Xtra Maxi kit (Macherey and Nagel). Library diversities were determined using the formula number of colonies × dilution × 10 (to correct from 100 μl to 1 ml) × number of electroporations.
Creation of secondary shuffled cap libraries
PCR rescue of cap genes from mouse tissue (25 to 100 ng) was done in the same manner as during library generation where large-scale PCR (16 reactions) was performed and cloned into plasmid pLib (59) to create secondary libraries. In the final library, rescued cap genes were cloned instead into pHelp after Pac I and Asc I digestion to produce individual vectors.
Sequencing of shuffled cap clones
Multiple clones (typically at least 10) were sent for Sanger sequencing (Eurofins) using three primers that cover the entire cap gene, i.e., SeqFor: 5′-GAT CTG GTC AAT GTG GAT TTG-3′ [compatible with plasmids pLib and pHelp (59)], SeqRev: 5′-GAC CGC AGC CTT TCG AAT GTC-3′ and LSeqRev2: 5′-GTC GCA AAA CAC TCA CGT GAC CTC-3′ (compatible with pLib or pHelp, respectively), and SeqMid: 5′-GAA ATT GGC ATT GCG ATT CC-3′ (compatible with all cap genes used here). The resulting partial sequences were manually reassembled into full-length cap sequences and then aligned with each other as well as with all parental WT DNA sequences using the AlignX tool in Vector NTI (Invitrogen) and ClustalX for multiple sequence alignments (www.clustal.org/). Alignments were subsequently saved as FASTA file and processed with SALANTO (59, 60), which assigns each amino acid of chimeric cap sequences to the parental AAVs. SALANTO output files were copied to Microsoft Excel, and distinct colors were assigned to the sequences using the conditional formatting function.
AAV capsid variants
The cap gene from AAV variant rh74 (GenBank: KH123010.1) was synthesized (IDT) with flanking Pac I and Asc I restriction sites to enable its insertion into the AAV helper plasmid pHelp (59). The shuffled AAV capsid variants bearing the P1 peptide (AAVMYO2 and AAVMYO3) were created by excising a Bsi WI/Spe I fragment of pHelp encoding P1-modified AAV9 cap and by inserting this fragment into pHelp carrying the respective shuffled capsid genes. Annealed oligonucleotides encoding peptide ESGHGYA (ESGHGYA_F: 5′-AGG CGA GAG CGG CCA CGG CTA CGC CGC CCA GG-3′ and ESGHGYA_R: 5′-GGG CGG CGT AGC CGT GGC CGC TCT CGC CTC TC-3′) were cloned into an AAV2 helper plasmid via two engineered Sfi I sites (22). Annealed oligonucleotides ESGHAYF_F (5′-AGG CGA GAG CGG CCA CGC CTA CTT CGC CCA GG-3′) and ESGHAYF_R (5′-GGG CGA AGT AGG CGT GGC CGC TCT CGC CTC TC-3′) encoding peptide ESGHAYF were cloned in the same manner. Capsid AAVS10P4 (P4: NDVRSAN) was created by first inserting the Sfi I site from AAV9 helper plasmid with the corresponding site (22) into the AAVMYO3 helper plasmid by digesting both plasmids with Bsi WI and Spe I. P4-encoding oligonucleotides P4F (5′-TGG CAA CGA TGT GCG CAG CGC GAA CGC CCA GG-3′) and P4R (5′-GGG CGT TCG CGC TGC GCA CAT CGT TGC CAC TC-3′) were annealed and ligated into the Sfi I site.
AAV barcoding and NGS
Protocols for AAV barcoding and NGS analysis as well as the associated in-house normalization strategy have all been described recently in detail (6). Briefly, 15-nucleotide-long DNA barcodes were placed after the stop codon of the eyfp coding region in the self-complementary AAV vector plasmid pscAAV-Pcmv-eyfp-BC-BGHpolyA (6, 10). Each barcode was then individually assigned to, and packaged into, a selected capsid variant including benchmarks. Following titration, all AAV variants were evenly pooled and concentrated on Amicon Ultra-15 100,000 NMWL (MERCK) before intravenous injection into the tail vein of mice. Barcoded vector genomes were amplified from genomic DNA (gDNA) and complementary DNA (cDNA) of muscle tissues and main organs. Amplicons with a length of 113 bp were purified with the QIAquick PCR purification kit (Qiagen), and the Ovation Library System for Low Complexity Samples Kit (NuGEN Technologies Inc.) was used to prepare NGS libraries. To this end, PCR repair, adaptor ligation, and amplification were all performed according to the manufacturer’s instructions. The quality and correct size of the amplicons were analyzed on an Agilent 2100 Bioanalyzer system using Agilent DNA 1000 Reagents (Agilent Technologies). Sample concentrations were measured by using the Qubit Fluorometer with the Qubit dsDNA HS Assay Kit (Invitrogen/Thermo Fisher Scientific). Samples were pooled and submitted to the EMBL sequencing facility (Heidelberg, Germany), where they were sequenced on an Illumina NextSeq 500 with the following parameters: Read 1 to 84 and Index 1 to 8. The resulting data were demultiplexed and processed using two scripts for Python 2.7. These exert multiple normalization steps to correct for deviations of total read counts from each flow cell, fluctuations in the composition of the initial viral library before injection, and variations in transduction efficiencies of the AAV variants in different tissues. Links to these scripts as well as details on the normalization strategy can be found in (6), particularly in the Supplementary Discussion and figure S18 of this former publication.
Titration of WT or recombinant chimeric AAV
AAV yields were determined by quantitative real-time PCR (qRT-PCR), following alkaline lysis of the AAV particles to release packaged vector genomes (59). Libraries were titrated using primers rep2For: 5′-AAG TCC TCG GCC CAG ATA GAC-3′ and rep2Rev: 5’-CAA TCA CGG CGC ACA TGT-3′, and rep2Probe: 5′-FAM-TGA TCG TCA CCT CCA ACA-BHQ1-3′ against the AAV2 rep gene contained in plasmid pLib (59) and the SensimixII Probe kit (Bioline).
MTM1 and micro-DMD vectors
The recombinant AAV-MTM1 vector construct was generated by cloning the human MTM1 cDNA downstream of the DES promoter in the AAV2 expression plasmid pAAV2-pDes. The μDys-expressing vector genomes have been reported before (61). The AAVMYO2, AAVMYO3, AAV8, and AAV9 vectors for mouse studies were produced and purified as described below. Recombinant vectors were titrated by TaqMan real-time PCR assays and expressed as vector genomes per milliliter (vg/ml).
Cell culture
HEK293T cells were used for virus/vector productions. Cells were grown under standard sterile conditions in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% fetal calf serum (Biochrom AG), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine (all Life Technologies) at 37°C with 5% CO2. Cells were regularly transferred when reaching near 80% confluency.
Research-grade AAV production and purification
Shuffled AAV capsid libraries were produced via double transfection of HEK293T cells using the AAV library plasmid and an adenoviral helper plasmid. Single AAV capsid variants were produced as recombinant vectors via triple transfection using an AAV helper plasmid encoding the cap gene of interest (next to AAV2 rep), an AAV vector plasmid carrying a reporter or therapeutic gene, and an adenoviral helper plasmid. In both cases, adherent HEK293T cells were seeded in 15-cm dishes (Nunc/Thermo Fisher Scientific) at a density of 4 × 106 cells per plate 2 days before transfection. For production of the two parental shuffled libraries, either 55 dishes (library A, yield of 6.27 × 1012 vg/ml) or 50 dishes (library B, yield of 3.21 × 1012 vg/ml) were used. Secondary libraries for the second and third selection rounds were produced in 40 or 30 dishes, respectively, resulting in 1 × 1013 vg/ml on average. To produce the 36 recombinant vectors for the first barcoded AAV library that was eventually injected into three mice, three dishes were used per chimeric AAV capsid variant. Those that yielded less than 1.5 × 1011 vg/ml were excluded from the study. For the second barcoded AAV library comprising 17 capsid variants, more dishes (~15 dishes per variant) were used to enable infection of 10 mice. Last, for single validations of lead AAV candidates and benchmarks, 40 to 50 dishes were used per variant and experimental group (three mice). In all cases, a mixture of sodium chloride, polyethylenimine, and a total of 440 μg of DNA (evenly divided between the two and three constructs) was used for transfection, as described before in detail (59). Three days after transfection, cells were scraped off, pooled, and subjected to 5 cycles of freezing and thawing in liquid nitrogen and a 37°C water bath, respectively. Residual plasmid DNA and cellular DNA was digested by incubation with 75 U of benzonase (Merck Millipore) per milliliter. AAV particles were then purified using either iodixanol or cesium chloride density gradient centrifugation as previously described (31). When multiple capsids were compared within a single experiment, the identical purification method was used for all variants. All the aforementioned virus samples were concentrated on Amicon Ultra-15 100,000 NMWL (MERCK) filters and buffer-exchanged to PBS.
Large-scale production of AAV vectors in suspension cells
AAV vectors were produced in 250-ml single-use shake flasks and in 10-liter glass stirred tank bioreactors by transient triple transfection (AAV helper, AAV vector, and adenoviral helper plasmid, as above) of a subclone of HEK293T cells, following a modified version of the original method described by Xiao et al. (62). The suspension cells were grown in supplemented FreeStyle F17 Expression Medium (Thermo Fisher Scientific) and transfected using histidinylated polyethylenimine (PTG1plus, Polytheragene). Twenty-four hours after transfection, cell cultures were treated with benzonase and, 2 days later, cells were lysed with Triton X-100 (Sigma-Aldrich) and clarified by filtration. Vectors were purified by a single round of chromatography using the POROS CaptureSelect AAVX immuno-affinity resin (Thermo Fisher Scientific). The purified vectors were concentrated by tangential flow filtration, diafiltered against Ringer’s lactate solution containing 0.001% Pluronic (F68), and lastly stored at −80°C. Titers of AAV vectors were determined using qRT-PCR in several in-process samples (crude harvest, clarified harvest, after chromatography) and in the final concentrated and formulated product. Empty:full capsid ratios were measured by analytical ultracentrifugation (63).
Small-scale assessment of chromatography resins
Four commercial resins (AVB Sepharose from GE Healthcare and POROS AAV8, POROS AAV9, and POROS AAVX from Thermo Fisher Scientific) were distributed in AcroPrep 96-well filter plates (Pall). For each capsid to be tested, the crude clarified lysate from bioreactors was directly loaded on each resin and was allowed to pass through the resin by suction. Various particle amounts from 5 × 109 to 1 × 1011 vg were loaded per tested resin to be able to assess binding independently of the resin’s intrinsic capacity. Loaded material and flow-through were titrated for vector genomes by qPCR to determine the binding of each capsid to the resins.
Animals
Mouse strains used for the iterative selection of AAV libraries and for in vivo validation of specific AAV capsid variants include C57BL/6J (Janvier Labs) and NMRI (Charles River). All mice were intravenously injected via the tail vein with AAV doses that are indicated in the text. Mice were kept and handled in accordance with the animal protocol 35-9185.81/G-89/16 that was approved by the Regierungspräsidium Karlsruhe (Germany).
Mice deficient for myotubularin (Mtm1-KO, also named BS53d4) (47, 48) are bred in a 129PAS background. WT male littermates were used as controls. Mice were handled according to the French and European legislation on animal care and experimentation and approved by the institutional ethical committee (APAFIS #4250-2015121520109947). AAV vectors were injected into the tail vein of 3-week-old male Mtm1-KO mice at either 1.0 × 1014, 2.0 × 1013, or 4.0 × 1012 vg/kg. An equivalent volume of saline was administrated to either KO or WT animals as controls. At 8 to 10 weeks of age (KO + PBS) or 15 weeks for other groups, tissues were collected, weighed, and quickly frozen in nitrogen-cooled isopentane or liquid nitrogen for histological and biochemical assays, respectively.
Mice with a point mutation in exon 23 of the murine Dmd gene that impedes dystrophin expression (mdx-mice) were bred as described before (61). Animal experiments were carried out under the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes with the approval of the local authorities in Kiel (V 242-12956/2018). A single dose of 2 × 1011 or 1 × 1012 vg of AAV9 or AAVMYO3 expressing a miniaturized “microdystrophin” (μDys) (61) gene under the control of the MCKE promoter (6) was administered to 7-week-old male mdx mice by intravenous tail vein injection. After 12 weeks, quadriceps femoris muscles were collected and snap-frozen for expression profiling. In addition, a single dose of 2 × 1012 vg of AAV9 or AAVMYO3 expressing μDys under the control of the CMV promoter was injected into the tail vein of 5- to 10-week-old male mdx mice. Here, 23 weeks after AAV injection, muscle function was assessed before harvesting tissues for biochemical analysis. AAVs encoding firefly luciferase under control of the respective promoter served as controls.
In vivo bioluminescence imaging in CB17/IcrTac/Prkdcscid mice and luciferase measurement in extracted organs were conducted as previously described (27, 53) and were approved by the Animal Ethics Committee of Vrije Universiteit Brussel.
Muscle strength measurements
Mtm1-KO mice underwent an escape test to quantify global strength as previously described (64). The in situ force of TA muscle was measured as described by Hakim et al. (65) using an integrated muscle test (Dual-Mode Lever System with Bi-Phase Stimulator, Aurora Scientific) and analyzed with the Dynamic Muscle Control and Dynamic Muscle Analysis software (Aurora Scientific). sP0m corresponds to the maximal tetanic force relative to muscle mass.
Muscle strength in mdx mice was measured with the four-limb hanging test and four-limb grip strength test according to the standardized operating procedures from the Translational Research in Europe–Assessment and Treatment of Neuromuscular Diseases (www.treat-nmd.eu/research/preclinical/dmd-sops/; SOP numbers DMD_M.2.1.005 and DMD_M.2.1.005).
For the four-limb hanging test, mice were removed from the cage, weighed, and placed on a grid, which was then turned upside down above a cage filled with bedding. The holding time (or “hanging time” in seconds) was defined as the amount of time that it took the mouse to completely fall from the inverted screen. A fixed maximum time of 600 s was set but not achieved by any animal. The longest hanging time out of two sessions with three trials each was used for further analysis. All measurements were performed in a blinded fashion by a single examiner.
For the four-limb grip strength test, mice were removed from the cage, weighed, lifted by the tail, and lowered toward a metal grid connected to a grip strength meter (BIO-GS3; Bioseb) until all four limbs gained a hold. The mouse was then drawn along a straight line leading away from the sensor until its grip was broken by the investigator’s pull and the maximum amount of force displayed in grams could be recorded. This was repeated in two sessions with five trials each. The mean of the three best recorded values was calculated for analysis. All measurements were performed in a blinded fashion by a single examiner.
DNA/RNA isolation and cDNA synthesis
Tissues were homogenized using a Tissue Lyser LT (Qiagen). DNA and RNA were extracted using the DNeasy Blood and Tissue Kit (Qiagen) or the RNeasy Kit (Qiagen), respectively, following the manufacturer’s protocols. gDNA was removed from RNA samples before cDNA synthesis by DNase digestion using the RNase-Free DNase set (Qiagen). For cDNA synthesis, the High-Capacity cDNA Reverse Transcription Kit was used (Thermo Fisher Scientific).
Quantification of vector copy numbers and relative expression
AAV vector copy numbers per cell were measured by using a ddPCR QX200 system (Bio-Rad). Vector genomes were amplified and detected using primers egfpF/R and probe egfpProbe (FAM) (see above for sequences) and were normalized against the housekeeper rpp30, which was amplified using the 20× primers/probe mix (HEX) (Bio-Rad). The ddPCR supermix for probes (no deoxyuridine triphosphate, Bio-Rad) was used to prepare all samples. Alternatively, the StepOnePlus Real-Time PCR System (Applied Biosystems) was used for relative quantification of the egfp reporter gene transcript and housekeeper POLR2A, which was amplified using predesigned TaqMan primers and probes (Thermo Fisher Scientific). Likewise, SensiMix II Probe Kit with ROX solution (Bioline) was used for the preparation of all qPCR samples. Normalized transcriptional efficiencies (Vαβ) and transcriptional specificities (Tαβ) were calculated as previously reported (6).
For vector copy number analysis in Mtm1-KO mouse tissues, samples were prepared in Lysing Matrix A tubes (MP Biomedicals) and homogenized in cell lysis buffer (Qiagen) by the FastPrep-24 (MP Biomedicals) device for 40 s at the speed of 5 m/s. Total gDNA was then extracted using the Gentra Puregen Blood kit (QIAGEN) according to the manufacturer’s instructions. Total gDNA concentration was determined by spectrophotometry using the NanoDrop 8000 (Thermo Fisher Scientific). The number of vector genomes per diploid genome was quantified from 32 ng of total gDNA by TaqMan real-time PCR using a LightCycler 480 system (Roche). The murine Titin gene was used for standardization. Primers and probe used for vector genome (MTM1 cDNA) amplification were 5′-TTG GTT GTC CAG TTT GGA GTC TAC T-3′ (forward), 5′-CCG TCA CTG CAA TGC ACA AG-3′ (reverse), and 5′-ATA TCA AGC TCG TTT TGA C-3′ (probe). Primers and probe used for Titin amplification were 5′-TTC AGT CAT GCT GCT AGC GC-3′ (forward), 5′-AAA ACG AGC AGT GAC GTG AGC-3′ (reverse), and 5′-TGC ACG GAA GCG TCT CGT CTC AGT C-3′ (probe) (Applied Biosystems).
For quantitative real-time qRT-PCR, XLMTM mouse tissues were prepared in Lysing Matrix A tubes (MP Biomedicals) and homogenized in MagnaPure LC RNA Isolation Tissue Lysis Buffer (Roche) by the FastPrep-24 (MP Biomedicals). Total RNA was then extracted using the automated nucleic acid extraction instrument, MagNA Pure 96 (Roche), from MagNA Pure 96 Cellular RNA Large Volume kit, using the “RNA Tissue FF Standard LV3.1” protocol. Total RNA concentration was determined by spectrophotometry using the NanoDrop 8000 (Thermo Fisher Scientific). DNA was degraded using the DNA-free kit (Thermo Fisher Scientific) and cDNA was synthesized from 350 ng of total RNA with the RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific) in the presence of Random Hexamer Primers (Thermo Fisher Scientific) for all samples, except for the heart (100 ng of total RNA). Transgene expression was quantified by TaqMan real-time PCR using a LightCycler 480 system (Roche). The murine ribosomal protein large subunit P0 (Rplp0) gene was used for standardization. Primers and probe used for vector genome MTM1 cDNA amplification were as above. Primers and probe used for RplP0 amplification were 5′-CTC TGG AGA AAC TGC TGC CT-3′ (forward), 5′-CTG CAC ATC ACT CAG AAT TTC AA-3′ (reverse), and 5′-AGG ACC TCA CTG AGA TTC GGG ATA TGC-3′ (probe) (Applied Biosystems).
Western blot in mouse tissues
In the XLMTM model, muscle cryosections were sliced and proteins were extracted using a lysis buffer containing 150 mM NaCl, 10 mM tris-HCl (pH 7.4), 1 mM EGTA, 1 mM EDTA, 100 mM sodium fluoride, 4 mM sodium pyrophosphate, 2 mM sodium orthovanadate, 1% Triton X-100, 0.5% IGEPAL CA-630 (Sigma-Aldrich), and protease inhibitor cocktail (Roche) according to the manufacturer’s instruction. Proteins were separated in a NuPage Novex 4 to 12% Bis-Tris gel (Invitrogen) and transferred to an Amersham Protran Premium Nitrocellulose membrane (GE Healthcare). The accuracy of the transfer was checked by Ponceau staining of the membrane. After several washes, the membrane was blocked overnight at 4°C in a buffer containing 50% 1× PBS (Life Technologies) and 50% intercept-blocking buffer (LI-COR). Membranes were probed with a goat anti-hMTM1 polyclonal antibody (Abnova, PAB6061) or a mouse monoclonal antibody specific for glyceraldehyde-3-phosphate dehydrogenase (Millipore, MAB374) as internal control. Detection was performed using the IRDye 800CW donkey anti-goat immunoglobulin G (IgG) secondary antibody or the IRDye 680CW goat anti-mouse IgG secondary antibody (LI-COR) and images were acquired with the Odyssey CLx system. Quantification was done using the Image Studio software (Version 4.0).
For detection of μDys expression in mdx mice, skeletal muscle tissue (quadriceps femoris) or cardiac tissue was transferred into lysis buffer containing 20 mM tris (pH 7.5), 500 mM NaCl, 12.5% (v/v) glycerol, 10 mM dithiothreitol, 1% (v/v) NP-40, protease inhibitor cocktail tablets (Roche), and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich) and homogenized using an Ultra-Turrax T25 tissue separator (Janke&Kunkel). Cell debris was removed by centrifugation. Subsequently, the concentration of total protein extracted from muscle tissues was measured with the DC Protein Assay (Bio-Rad Laboratories) according to the manufacturer’s guidelines. Proteins were resolved on 4 to 20% gradient gels (Bio-Rad) and transferred onto nitrocellulose membranes. Membranes were blocked in 3 or 5% dry milk prepared in 0.1% tris-buffered saline with Tween 20 (TBST) for 2 to 4 hours at room temperature and incubated with primary antibodies overnight at 4°C (rabbit polyclonal antibody RB-9024-P against the dystrophin C terminus, 1:800 dilution, Thermo Fisher Scientific; rabbit polyclonal antibody ab15277 against the dystrophin C terminus, 1:500 dilution; or mouse monoclonal antibody T5168 against α-tubulin, 1:5000 dilution, Sigma-Aldrich) in 3 to 5% dry milk in 0.1% TBST. After washing four times (10 min each) with 0.1% TBST, membranes were incubated with a horseradish peroxidase (HRP)–coupled secondary antibody (1:10,000; Santa Cruz Biotechnology). Following further four washes, bound antibodies were detected using the ECL Select chemiluminescence kit (GE Healthcare) and visualized on a FluorChem Q imaging system (Biozym). Quantification was performed by densitometry using AlphaView software version 3.0 (Alpha Inotec).
Immunofluorescence analysis
Diaphragm tissue was harvested and immediately embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Europe) and frozen on dry ice. Serial 6-μm diagonal-to-transverse cryosections were cut, washed with PBS, and fixed in 4% (w/v) paraformaldehyde (Sigma-Aldrich) in PBS for 10 min at room temperature. After blocking for 2 hours in 2.5% bovine serum albumin (BSA), sections were stained with rabbit polyclonal antibody 12715-1-AP (1:100 dilution in PBS with 2.5% BSA; Proteintech) against the dystrophin C terminus overnight at 4°C. After washing, the sections were incubated with Alexa Fluor 488–coupled chicken anti-rabbit secondary antibody (1:400 dilution; Invitrogen, Thermo Fisher Scientific, A21441) along with 4′,6-diamidino-2-phenylindole (DAPI; 1:1000 dilution; Vector Laboratories) and wheat germ agglutinin Alexa Fluor 647 conjugate (W32466; 1:400 dilution; Invitrogen, Thermo Fisher Scientific) for 1 hour at room temperature in the dark. Following washing with PBS containing 0.1% Tween 20 and embedding in FluorSave reagent (Merck Millipore), the sections were analyzed using the BZ-9000E HS all-in-one fluorescence microscope (Keyence) with the BZ-II Viewer software (version 2.1, Keyence).
Histological analysis of mouse muscles
Cross sections (8 μm) of isopentane-frozen muscles were stained with H&E or for NADH-TR reaction by incubation with nitroblue tetrazolium (1 mg/ml; Sigma-Aldrich) and β-NADH (0.4 mg/ml; Sigma-Aldrich) in 50 mM tris-HCl (pH 7.3) at 57°C. Heart cryosections were stained with Sirius red, and the percentage of collagen deposition per section area was quantified using ImageJ. The percentage of myofibers with internal nuclei was manually scored from HE-stained slides. For fiber size quantification, muscles were immunolabeled with rabbit anti-laminin antibody (DAKO Z0097) and EnVision+ System HRP-labeled polymer anti-Rabbit (DAKO K4002) and revealed with liquid DAB+ (DAKO K3468). The diameter of muscle fibers was quantified using the Ellix software (Microvision Instruments).
Assessment of seroprevalence in humans
Seroprevalence was measured by ELISA and using an in vitro neutralization assay. Anti-AAV IgG capture assay was performed as previously described (43). Briefly, recombinant AAV vectors were diluted in coating buffer [35 mM bicarbonate and 13 mM carbonate (pH >9.2)] to a final concentration of 2 × 1010 vector particles per milliliter. Fifty microliters was added to each well in a 96-well Nunc Maxisorp Immunoplate (Thermo Fisher Scientific). A standard curve made from purified human IgG (IVIg, Tégéline, LFB BIOMEDICAMENTS) was added directly to the plates, and the latter was coated overnight at 4°C. The next day, plates were washed three times with wash buffer (PBS, 0.05% Tween 20) and blocked with blocking buffer (PBS, 6% fat-free milk) for 2 hours at room temperature, then incubated with serum diluted in blocking buffer for 1 hour at 37°C. After three washes, HRP-conjugated antibody specific for human IgG (Southern Biotech) was incubated for 1 hour at 37°C and revealed with o-phenylenediamine dihydrochloride substrate (Sigma-Aldrich). Optical density was measured at 492 nm using a microplate reader (ENSPIRETM, Perkin Elmer). Anti-AAV IgG concentration was determined against a specific standard curve using four-parameter regression and results were expressed as micrograms per milliliter of IgG.
Serum samples were also analyzed for the presence of anti-AAV–neutralizing antibodies as previously described (44). Briefly, on day 1, 96-well plates were seeded with two V6.11 cells (American Type Culture Collection) at 2 × 104 cells per well for 24 hours in the presence of ponasterone A (Life Technologies). AAV9-Luc, AAVMYO2-Luc, or AAVMYO3-Luc vectors were then diluted in serum-free DMEM (Life Technologies) and incubated with semilog-fold serial dilutions of the serum samples for 1 hour at 37°C. Subsequently, the serum-vector mixtures were added to the cells at a multiplicity of infection of 200. After 24 hours, cells were lysed and the luciferase activity was measured in a luminometer (ENSPIRETM, Perkin Elmer). Luciferase expression was measured as relative light unit per second. The neutralizing titer was reported as the highest serum dilution that inhibited AAV transduction by at least 50% compared to the 100% transduction control.
Model building and structure visualization
The models of AAVS1, AAVS10, AAVMYO, AAVMYO2, and AAVMYO3 variant VP3 monomers were built using their respective sequences uploaded to the online program SWISS-MODEL with the coordinates of the x-ray structure of AAV9 [Protein Data Bank (PDB) ID: 3UX1] as the template (66). The program Coot2 was used to renumber the VP3 monomers’ coordinates generated starting at residue 209, which is the first ordered residue observed in the AAV9 structure. T = 1 icosahedral capsid models were generated for each of the VP3 monomers listed, using the subroutine Oligomer generator within the ViperDB online server (67). The VP3 monomers of AAVS1, AAVS10, AAVMYO, AAVMYO2, or AAVMYO3 were superposed with the available coordinates of the VP3 monomers of AAV1 (PDB ID: 3NG9), AAV6 (PDB ID: 3OAH), and AAV8 (PDB ID: 2QA0) in the program Coot (68). Amino acids of the VP3 variants that differed between AAV9, but are common to AAV1, AAV6, and AAV8 serotypes, were highlighted on the VP3 monomer and capsid variant images. Monomers and capsid surface representations were generated using the program PyMOL (www.pymol.org).
Statistical analysis
Statistical analyses in the context of the XLMTM and mdx mouse models were performed using GraphPad Prism 6 (XLMTM) or SigmaPlot 13.0 (μDys). Unpaired t test was used to compare two distinct groups, whereas a one-way analysis of variance (ANOVA) test followed by Dunnett’s multiple comparison posttest was selected for analysis of several groups (one symbol: P < 0.05; two symbols: P < 0.01; three symbols: P < 0.001). Results are expressed as means ± SEM.
Acknowledgments
We are very grateful to the platforms of Genethon for their excellent technical expertise and contribution to this work. We moreover thank J. Becker for helpful discussions during the revision of this manuscript.
Funding: We kindly acknowledge the support provided by the MYOCURE project to D.G., A.B.-B., M.K.C., T.V., F.M., and M.H. MYOCURE has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 667751. J.E.A., J.W., J.F., and D.G. moreover appreciate support by the German Research Foundation (DFG) through the Cluster of Excellence CellNetworks (EXC81). J.E.A. and D.G. were also funded by the ANR-DFG program KARTLE. D.G. is grateful for the support by the DFG Collaborative Research Centers SFB1129 (projektnummer 240245660) and TRR179 (projektnummer 272983813), as well as by the German Center for Infection Research (DZIF, BMBF; TTU-HIV 04.803, TTU-HIV 04.815, and TTU-HIV 04-819). J.F. and D.G. are thankful for funding from the Cystic Fibrosis Foundation (CFF; grant number GRIMM15XX0) and from the Heidelberg Biosciences International Graduate School HBIGS at Heidelberg University. T.V. and M.K.C. received grant support from the Vrije Universiteit Brussel Industrieel Onderzoeksfonds (IOF), Groups of Expertise in Advanced Research (GEAR), F.W.O. (Fonds Wetenschappelijk Onderzoek Vlaanderen), Koning Boudewijn Stichting (Fonds Walter Pyleman, Cremers-Opdebeeck, Richard Depasse), and Duchenne Parent Project. O.J.M. is grateful to the German Centre for Cardiovascular Research and the DFG (MU 1654/11-1). O.J.M. and D.G. are moreover thankful to the BMBF (Federal Ministry of Education and Research, Germany) for funding through the COMMUTE research consortium, as well as to the German Duchenne Foundation for support. This work was also supported by the “Association Française contre les Myopathies” (AFM-Telethon), the INSERM, and University of Evry Val d’Essonne.
Author contributions: This work was an integral part of the MYOCURE project that was scientifically coordinated by M.K.C. together with A.B.-B., F.M., T.V., D.G., and the other consortium partners. J.E.A., E.R.-G., W.T., J.W., L.M., Q.H.P., S.H., E.A., E.B., C.L., N.G., J.F., L.G., and M.T. designed, performed, and/or analyzed experiments. A.B., R.M., and M.A.-M. created and analyzed all capsid models. C.K. and E.W. provided essential technical help. A.B.-B., F.M., T.V., G.R., M.H., O.J.M., M.K.C., and D.G. designed the experiments and supervised the work in their laboratories. J.E.A., E.R.-G., A.B.-B., and D.G. were mainly responsible for writing the manuscript. All authors were involved in the preparation of figures and associated text, and all approved the final manuscript.
Competing interests: D.G. is a cofounder of AaviGen GmbH. J.E.A. now works for DiNAQOR, J.W. and L.G. are now employees of Novartis, M.T. now works for Merck, J.F. is now an employee of Roche Diagnostics, M.H. is now employed by LogicBio Therapeutics, and F.M. works at Spark Therapeutics. D.G., J.E.A., and J.W. are inventors on a patent application related to this work filed by the University of Heidelberg (PCT/EP2019/060790, filed 26 April 2019, published 31 October 2019 as WO2019/207132). All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The AAVMYO2 or AAVMYO3 helper plasmids reported in this work can be provided by D.G. pending scientific review and a completed material transfer agreement. Requests for these plasmids should be submitted to D.G. (dirk.grimm@bioquant.uni-heidelberg.de). The AAV vector plasmids expressing luciferase from the desmin or SPc5-12 promoter can be provided by T.V. pending scientific review and a completed material transfer agreement. Requests for these AAV vector plasmids should be submitted to T.V. (Thierry.Vandendriessche@vub.be).
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
REFERENCES AND NOTES
- 1.Grimm D., Zolotukhin S., E pluribus unum: 50 years of research, millions of viruses, and one goal–tailored acceleration of AAV evolution. Mol. Ther. 23, 1819–1831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D., Tai P. W. L., Gao G., Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havlik L. P., Simon K. E., Smith J. K., Klinc K. A., Tse L. V., Oh D. K., Fanous M. M., Meganck R. M., Mietzsch M., Kleinschmidt J., Agbandje-McKenna M., Asokan A., Coevolution of adeno-associated virus capsid antigenicity and tropism through a structure-guided approach. J. Virol. 94, e00976-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tse L. V., Klinc K. A., Madigan V. J., Castellanos Rivera R. M., Wells L. F., Havlik L. P., Smith J. K., Agbandje-McKenna M., Asokan A., Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. U.S.A. 114, E4812–E4821 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adachi K., Enoki T., Kawano Y., Veraz M., Nakai H., Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun. 5, 3075 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinmann J., Weis S., Sippel J., Tulalamba W., Remes A., El Andari J., Herrmann A. K., Pham Q. H., Borowski C., Hille S., Schonberger T., Frey N., Lenter M., VandenDriessche T., Muller O. J., Chuah M. K., Lamla T., Grimm D., Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 11, 5432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondratov O., Kondratova L., Mandel R. J., Coleman K., Savage M. A., Gray-Edwards H. L., Ness T. J., Rodriguez-Lebron E., Bell R. D., Rabinowitz J., Gamlin P. D., Zolotukhin S., A comprehensive study of a 29-capsid AAV library in a non-human primate central nervous system. Mol. Ther. 29, 2806–2820 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsic D., Mendez-Gomez H. R., Zolotukhin S., High-accuracy biodistribution analysis of adeno-associated virus variants by double barcode sequencing. Mol. Ther. Methods Clin. Dev. 2, 15041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westhaus A., Cabanes Creus M., Rybicki A., Baltazar G., Navarro R. G., Zhu E., Drouyer M., Knight M., Albu R. F., Ng B. H., Kalajdzic P., Kwiatek M., Hsu K., Santilli G., Gold W., Kramer B., Gonzalez-Cordero A., Thrasher A. J., Alexander I. E., Lisowski L., High-throughput in vitro,ex vivo,and in vivo screen of adeno-associated virus vectors based on physical and functional transduction. Hum. Gene Ther. 31, 575–589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kremer L. P. M., Cerrizuela S., Dehler S., Stiehl T., Weinmann J., Abendroth H., Kleber S., Laure A., El Andari J., Anders S., Marciniak-Czochra A., Grimm D., Martin-Villalba A., High throughput screening of novel AAV capsids identifies variants for transduction of adult NSCs within the subventricular zone. Mol. Ther. Methods Clin. Dev. 23, 33–50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungbluth H., Wallgren-Pettersson C., Laporte J., Centronuclear (myotubular) myopathy. Orphanet J. Rare Dis. 3, 26 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Ploeg A. T., Reuser A. J., Pompe’s disease. Lancet 372, 1342–1353 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Verhaart I. E. C., Aartsma-Rus A., Therapeutic developments for Duchenne muscular dystrophy. Nat. Rev. Neurol. 15, 373–386 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Mendell J. R., Al-Zaidy S. A., Rodino-Klapac L. R., Goodspeed K., Gray S. J., Kay C. N., Boye S. L., Boye S. E., George L. A., Salabarria S., Corti M., Byrne B. J., Tremblay J. P., Current clinical applications of in vivo gene therapy with AAVs. Mol. Ther. 29, 464–488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hordeaux J., Buza E. L., Dyer C., Goode T., Mitchell T. W., Richman L., Denton N., Hinderer C., Katz N., Schmid R., Miller R., Choudhury G. R., Horiuchi M., Nambiar K., Yan H., Li M., Wilson J. M., Adeno-associated virus-induced dorsal root ganglion pathology. Hum. Gene Ther. 31, 808–818 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Hordeaux J., Buza E. L., Jeffrey B., Song C., Jahan T., Yuan Y., Zhu Y., Bell P., Li M., Chichester J. A., Calcedo R., Wilson J. M., MicroRNA-mediated inhibition of transgene expression reduces dorsal root ganglion toxicity by AAV vectors in primates. Sci. Transl. Med. 12, eaba9188 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Hinderer C., Katz N., Buza E. L., Dyer C., Goode T., Bell P., Richman L. K., Wilson J. M., Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 29, 285–298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapti K., Grimm D., Adeno-associated viruses (AAV) and host immunity - a race between the hare and the hedgehog. Front. Immunol. 12, 753467 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen G. N., Everett J. K., Kafle S., Roche A. M., Raymond H. E., Leiby J., Wood C., Assenmacher C. A., Merricks E. P., Long C. T., Kazazian H. H., Nichols T. C., Bushman F. D., Sabatino D. E., A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat. Biotechnol. 39, 47–55 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson C. E., Wu Y., Gemberling M. P., Oliver M. L., Waller M. A., Bohning J. D., Robinson-Hamm J. N., Bulaklak K., Castellanos Rivera R. M., Collier J. H., Asokan A., Gersbach C. A., Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 25, 427–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunze C., Borner K., Kienle E., Orschmann T., Rusha E., Schneider M., Radivojkov-Blagojevic M., Drukker M., Desbordes S., Grimm D., Brack-Werner R., Synthetic AAV/CRISPR vectors for blocking HIV-1 expression in persistently infected astrocytes. Glia 66, 413–427 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Börner K., Kienle E., Huang L.-Y., Weinmann J., Sacher A., Bayer P., Stüllein C., Fakhiri J., Zimmermann K., Westhaus A., Beneke J., Beil N., Wiedtke E., Schmelas C., Miltner D., Rau A., Erfle H., Kräusslich H.-G., Müller M., Agbandje-McKenna M., Grimm D., Pre-arrayed pan-AAV peptide display libraries for rapid single-round screening. Mol. Ther. 28, 1016–1032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabebordbar M., Lagerborg K. A., Stanton A., King E. M., Ye S., Tellez L., Krunnfusz A., Tavakoli S., Widrick J. J., Messemer K. A., Troiano E. C., Moghadaszadeh B., Peacker B. L., Leacock K. A., Horwitz N., Beggs A. H., Wagers A. J., Sabeti P. C., Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell 184, 4919–4938.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asokan A., Conway J. C., Phillips J. L., Li C., Hegge J., Sinnott R., Yadav S., DiPrimio N., Nam H. J., Agbandje-McKenna M., McPhee S., Wolff J., Samulski R. J., Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat. Biotechnol. 28, 79–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L., Jiang J., Drouin L. M., Agbandje-McKenna M., Chen C., Qiao C., Pu D., Hu X., Wang D. Z., Li J., Xiao X., A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. U.S.A. 106, 3946–3951 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulk N. K., Pekrun K., Charville G. W., Maguire-Nguyen K., Wosczyna M. N., Xu J., Zhang Y., Lisowski L., Yoo B., Vilches-Moure J. G., Lee G. K., Shrager J. B., Rando T. A., Kay M. A., Bioengineered viral platform for intramuscular passive vaccine delivery to human skeletal muscle. Mol. Ther. Methods Clin. Dev. 10, 144–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tulalamba W., Weinmann J., Pham Q. H., El Andari J., VandenDriessche T., Chuah M. K., Grimm D., Distinct transduction of muscle tissue in mice after systemic delivery of AAVpo1 vectors. Gene Ther. 27, 170–179 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Korbelin J., Sieber T., Michelfelder S., Lunding L., Spies E., Hunger A., Alawi M., Rapti K., Indenbirken D., Muller O. J., Pasqualini R., Arap W., Kleinschmidt J. A., Trepel M., Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol. Ther. 24, 1050–1061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deverman B. E., Pravdo P. L., Simpson B. P., Kumar S. R., Chan K. Y., Banerjee A., Wu W. L., Yang B., Huber N., Pasca S. P., Gradinaru V., Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zincarelli C., Soltys S., Rengo G., Rabinowitz J. E., Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Borner K., Niopek D., Cotugno G., Kaldenbach M., Pankert T., Willemsen J., Zhang X., Schurmann N., Mockenhaupt S., Serva A., Hiet M. S., Wiedtke E., Castoldi M., Starkuviene V., Erfle H., Gilbert D. F., Bartenschlager R., Boutros M., Binder M., Streetz K., Krausslich H. G., Grimm D., Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 41, e199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Andari J., Grimm D., Production, processing, and characterization of synthetic AAV gene therapy vectors. Biotechnol. J. 16, e2000025 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Ayuso E., Mingozzi F., Bosch F., Production, purification and characterization of adeno-associated vectors. Curr. Gene Ther. 10, 423–436 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Govindasamy L., Padron E., McKenna R., Muzyczka N., Kaludov N., Chiorini J. A., Agbandje-McKenna M., Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 80, 11556–11570 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mietzsch M., Penzes J. J., Agbandje-McKenna M., Twenty-five years of structural parvovirology. Viruses 11, 362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burg M., Rosebrough C., Drouin L. M., Bennett A., Mietzsch M., Chipman P., McKenna R., Sousa D., Potter M., Byrne B., Jude Samulski R., Agbandje-McKenna M., Atomic structure of a rationally engineered gene delivery vector, AAV2.5. J. Struct. Biol. 203, 236–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guenther C. M., Brun M. J., Bennett A. D., Ho M. L., Chen W., Zhu B., Lam M., Yamagami M., Kwon S., Bhattacharya N., Sousa D., Evans A. C., Voss J., Sevick-Muraca E. M., Agbandje-McKenna M., Suh J., Protease-activatable adeno-associated virus vector for gene delivery to damaged heart tissue. Mol. Ther. 27, 611–622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonntag F., Bleker S., Leuchs B., Fischer R., Kleinschmidt J. A., Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80, 11040–11054 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bleker S., Sonntag F., Kleinschmidt J. A., Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79, 2528–2540 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell C. L., Gurda B. L., Van Vliet K., Agbandje-McKenna M., Wilson J. M., Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J. Virol. 86, 7326–7333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S. H., Yang J. Y., Madrakhimov S., Park H. Y., Park K., Park T. K., Adeno-associated viral vector 2 and 9 transduction is enhanced in streptozotocin-induced diabetic mouse retina. Mol. Ther. Methods Clin. Dev. 13, 55–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdera H. C., Kuranda K., Mingozzi F., AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 28, 723–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mingozzi F., Anguela X. M., Pavani G., Chen Y., Davidson R. J., Hui D. J., Yazicioglu M., Elkouby L., Hinderer C. J., Faella A., Howard C., Tai A., Podsakoff G. M., Zhou S., Basner-Tschakarjan E., Wright J. F., High K. A., Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci. Transl. Med. 5, 194ra192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meliani A., Leborgne C., Triffault S., Jeanson-Leh L., Veron P., Mingozzi F., Determination of anti-adeno-associated virus vector neutralizing antibody titer with an in vitro reporter system. Hum. Gene Ther. Methods 26, 45–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beggs A. H., Byrne B. J., De Chastonay S., Haselkorn T., Hughes I., James E. S., Kuntz N. L., Simon J., Swanson L. C., Yang M. L., Yu Z. F., Yum S. W., Prasad S., A multicenter, retrospective medical record review of X-linked myotubular myopathy: The recensus study. Muscle Nerve 57, 550–560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Annoussamy M., Lilien C., Gidaro T., Gargaun E., Che V., Schara U., Gangfuss A., D’Amico A., Dowling J. J., Darras B. T., Daron A., Hernandez A., de Lattre C., Arnal J. M., Mayer M., Cuisset J. M., Vuillerot C., Fontaine S., Bellance R., Biancalana V., Buj-Bello A., Hogrel J. Y., Landy H., Servais L., X-linked myotubular myopathy: A prospective international natural history study. Neurology 92, e1852–e1867 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A. H., Allard B., Mandel J. L., Laporte J., Jacquemond V., Buj-Bello A., T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl. Acad. Sci. U.S.A. 106, 18763–18768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buj-Bello A., Laugel V., Messaddeq N., Zahreddine H., Laporte J., Pellissier J. F., Mandel J. L., The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 99, 15060–15065 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buj-Bello A., Fougerousse F., Schwab Y., Messaddeq N., Spehner D., Pierson C. R., Durand M., Kretz C., Danos O., Douar A. M., Beggs A. H., Schultz P., Montus M., Denefle P., Mandel J. L., AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum. Mol. Genet. 17, 2132–2143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Childers M. K., Joubert R., Poulard K., Moal C., Grange R. W., Doering J. A., Lawlor M. W., Rider B. E., Jamet T., Daniele N., Martin S., Riviere C., Soker T., Hammer C., Van Wittenberghe L., Lockard M., Guan X., Goddard M., Mitchell E., Barber J., Williams J. K., Mack D. L., Furth M. E., Vignaud A., Masurier C., Mavilio F., Moullier P., Beggs A. H., Buj-Bello A., Gene therapy prolongs survival and restores function in murine and canine models of myotubular myopathy. Sci. Transl. Med. 6, 220ra210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mack D. L., Poulard K., Goddard M. A., Latournerie V., Snyder J. M., Grange R. W., Elverman M. R., Denard J., Veron P., Buscara L., Le Bec C., Hogrel J. Y., Brezovec A. G., Meng H., Yang L., Liu F., O’Callaghan M., Gopal N., Kelly V. E., Smith B. K., Strande J. L., Mavilio F., Beggs A. H., Mingozzi F., Lawlor M. W., Buj-Bello A., Childers M. K., Systemic AAV8-mediated gene therapy drives whole-body correction of myotubular myopathy in dogs. Mol. Ther. 25, 839–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roudaut C., Le Roy F., Suel L., Poupiot J., Charton K., Bartoli M., Richard I., Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation 128, 1094–1104 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Sarcar S., Tulalamba W., Rincon M. Y., Tipanee J., Pham H. Q., Evens H., Boon D., Samara-Kuko E., Keyaerts M., Loperfido M., Berardi E., Jarmin S., In’t Veld P., Dickson G., Lahoutte T., Sampaolesi M., De Bleser P., VandenDriessche T., Chuah M. K., Next-generation muscle-directed gene therapy by in silico vector design. Nat. Commun. 10, 492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C., Wu S., Albright B., Hirsch M., Li W., Tseng Y. S., Agbandje-McKenna M., McPhee S., Asokan A., Samulski R. J., Development of patient-specific AAV vectors after neutralizing antibody selection for enhanced muscle gene transfer. Mol. Ther. 24, 53–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goertsen D., Flytzanis N. C., Goeden N., Chuapoco M. R., Cummins A., Chen Y., Fan Y., Zhang Q., Sharma J., Duan Y., Wang L., Feng G., Chen Y., Ip N. Y., Pickel J., Gradinaru V., AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci. 25, 106–115 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Bauer A., Puglisi M., Nagl D., Schick J. A., Werner T., Klingl A., El Andari J., Hornung V., Kessler H., Gotz M., Grimm D., Brack-Werner R., Molecular signature of astrocytes for gene delivery by the synthetic adeno-associated viral vector rAAV9P1. Adv. Sci. 9, e2104979 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flitsch L. J., Borner K., Stullein C., Ziegler S., Sonntag-Buck V., Wiedtke E., Semkova V., Yeung S. W. C. A., Schlee J., Hajo M., Mathews M., Ludwig B. S., Kossatz S., Kessler H., Grimm D., Brustle O., Identification of adeno-associated virus variants for gene transfer into human neural cell types by parallel capsid screening. Sci. Rep. 12, 8356 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X., Ravindra Kumar S., Adams C. D., Yang D., Wang T., Wolfe D. A., Arokiaraj C. M., Ngo V., Campos L. J., Griffiths J. A., Ichiki T., Mazmanian S. K., Osborne P. B., Keast J. R., Miller C. T., Fox A. S., Chiu I. M., Gradinaru V., Engineered AAVs for non-invasive gene delivery to rodent and non-human primate nervous systems. Neuron 110, 2242–2257.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrmann A. K., Bender C., Kienle E., Grosse S., El Andari J., Botta J., Schurmann N., Wiedtke E., Niopek D., Grimm D., A robust and all-inclusive pipeline for shuffling of adeno-associated viruses. ACS Synth. Biol. 8, 194–206 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Schurmann N., Trabuco L. G., Bender C., Russell R. B., Grimm D., Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. 20, 818–826 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Schinkel S., Bauer R., Bekeredjian R., Stucka R., Rutschow D., Lochmuller H., Kleinschmidt J. A., Katus H. A., Muller O. J., Long-term preservation of cardiac structure and function after adeno-associated virus serotype 9-mediated microdystrophin gene transfer in mdx mice. Hum. Gene Ther. 23, 566–575 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Xiao X., Li J., Samulski R. J., Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72, 2224–2232 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collaud F., Bortolussi G., Guianvarc’h L., Aronson S. J., Bordet T., Veron P., Charles S., Vidal P., Sola M. S., Rundwasser S., Dufour D. G., Lacoste F., Luc C., Wittenberghe L. V., Martin S., Le Bec C., Bosma P. J., Muro A. F., Ronzitti G., Hebben M., Mingozzi F., Preclinical development of an AAV8-hUGT1A1 vector for the treatment of Crigler-Najjar Syndrome. Mol. Ther. Methods Clin. Dev. 12, 157–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson C. G., Makiejus R. V., A noninvasive procedure to detect muscle weakness in the mdx mouse. Muscle Nerve 13, 480–484 (1990). [DOI] [PubMed] [Google Scholar]
- 65.Hakim C. H., Li D., Duan D., Monitoring murine skeletal muscle function for muscle gene therapy. Methods Mol. Biol. 709, 75–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T., SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrillo-Tripp M., Shepherd C. M., Borelli I. A., Venkataraman S., Lander G., Natarajan P., Johnson J. E., Brooks C. L. III, Reddy V. S., VIPERdb2: An enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 37, D436–D442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18