Abstract
Viruses exploit host cell machinery to support their replication. Defining the cellular proteins and processes required for a virus during infection is crucial to understanding the mechanisms of virally induced disease and designing host-directed therapeutics. Here, we perform a genome-wide CRISPR-Cas9–based screening in lung epithelial cells infected with the PR/8/NS1-GFP virus and use GFPhi cell as a unique screening marker to identify host factors that inhibit influenza A virus (IAV) infection. We discovered that APOE affects influenza virus infection both in vitro and in vivo. Cell deficiency in APOE conferred substantially increased susceptibility to IAV; mice deficient in APOE manifested more severe lung pathology, increased virus load, and decreased survival rate. Mechanistically, lack of cell-produced APOE results in impaired cell cholesterol homeostasis, enhancing influenza virus attachment. Thus, we identified a previously unrecognized role of APOE in restraining IAV infection.
APOE inhibits IAV infection through regulating cellular cholesterol homeostasis.
INTRODUCTION
The influenza virus is responsible for almost half a million deaths every year and thus poses a serious global health threat. The latter is particularly true for the immunocompromised population, such as the elderly (>65 years) and the children, because these individuals are particularly prone to severe influenza virus–induced respiratory disease. In addition, influenza virus superinfection can produce antigenic shifting and lead to virus reassortment, providing evolutionary flexibility to sample different virus genotypes, some of which may be more pathogenic [for example, influenza strains such as the H1N1 strain or the related H7N9 and H5N1 strains that cause pandemic infection affecting a large proportion of the world population (1, 2)].
The host uses a variety of mechanisms to defend itself from virus infection. Much progress has been made in identifying host factors that are anti–influenza A virus (IAV) infection. Chief among these is the viral induction of interferons (IFNs), proteins that trigger the transcriptional up-regulation of many IFN-stimulated genes (ISGs). ISGs are critical for intrinsic cell defense against viruses: They encode typical restriction factors that target various stages of a viral life cycle [e.g., entry: IFN-inducible transmembrane 3 (IFITM3); nuclear import: myxovirus resistance 1 (MX1) and MX2; mRNA synthesis: apolipoprotein B mRNA editing enzyme and catalytic polypeptide-like families; protein synthesis: protein kinase R; replication: 2′,5′-oligoadenylate synthetase 1 (OAS1); egress: tetherin], which are capable of blocking viral infection in a variety of ways (3, 4). In addition, acute inflammation is also a critical host defense response during viral infection, but it will drive immunopathology and tissue damage when dysregulated. Moreover, obesity is an important factor that drives inflammation during IAV infection. Independent of other comorbidities or risk factors, individuals with obesity were at higher risk of death due to IAV infection during the 2009 pandemic (5). Even when influenza vaccination is accounted for, adults with obesity are still twice as likely to develop influenza or influenza-like illnesses than adults with healthy weight (6). Accordingly, obese mice infected with IAV have a sixfold increase in mortality compared to their nonobese counterparts (7). However, the exact mechanism of obesity versus enhanced IAV infection remains elusive. It is known that obesity is associated with dyslipidemia and metabolic syndrome (8, 9). Thus, one possibility is that obesity-induced dyslipidemia may lead to an enhanced IAV infection, but this has never been explored.
Apolipoprotein E (APOE) is a glycoprotein of 35 kDa that belongs to a class of cellular proteins involved in lipid metabolism. APOE also has an important role in cholesterol efflux, as documented by studies in patients and animal models with APOE deficiency or mutated APOE genes (10, 11). APOE plays an essential role in lipoprotein transport. For viral infection, it is known that apolipoproteins are exploited by the hepatitis C virus (HCV) to benefit virus entry, assembly, and transmission; in addition, APOE contributes to HCV pathogenesis and viral evasion from neutralizing antibodies (12–14).
On the other hand, viruses, as intracellular parasites, co-opt various host metabolic functions to facilitate their entry, replication, assembly, and egress. Identification of these functions has the potential to allow a better understanding of the mechanisms underlying the several phases of the virus life cycle. It could thus lead to the development of new antiviral compounds. Accordingly, various screening approaches have been used to identify host factors operative against influenza infection, such as small interfering RNA screening (15–17) and CRISPR-Cas9–based screening (18–22). Most of these screenings use cell survival as a screening marker. While this has resulted in the identification of hundreds of genes that have been shown to be important for influenza virus replication, it could not screen out antiviral genes. Thus, there is a large number of antiviral genes that remain to be found.
Here, using green fluorescent protein-high (GFPhi) cells as a selection marker after PR/8/NS1-GFP virus infection, we performed an extensive genome-wide CRISPR-Cas9 screening to uncover previously unidentified host factors participating in anti–influenza virus infection. Among the factors, one of the unknown factors for restricting IAV infection and that particularly drew our attention was APOE. We found that depletion of APOE resulted in markedly increased susceptibility to IAV infection in vitro and in vivo. The cholesterol level modulated by APOE restricted IAV attachment, suggesting that reduced cellular cholesterol is an essential host defense strategy against IAV infection.
RESULTS
Genome-wide CRISPR-Cas9 screening identified host factors that restrict IAV infection
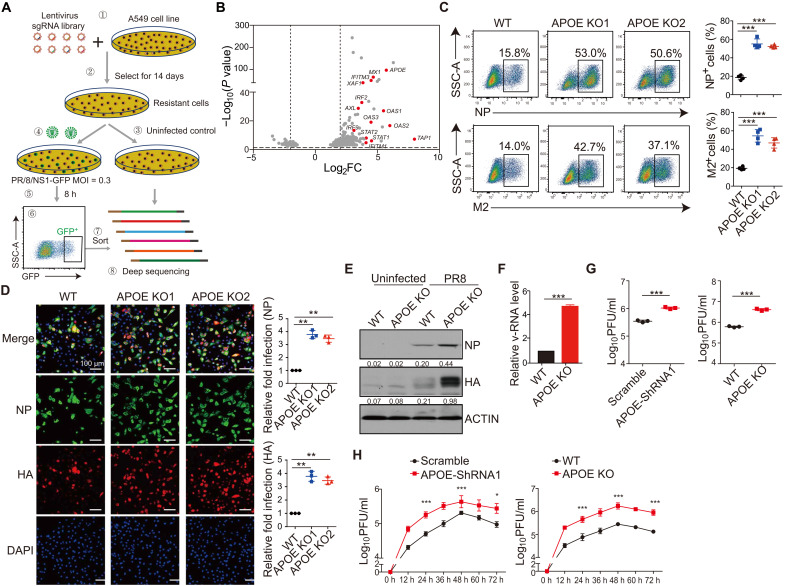
To search for host factors targeting IAV infection, we performed high-throughput genome-wide screening using a CRISPR-Cas9 lentiviral system. In this screening, human lung adenocarcinoma cells (A549) were transduced with a lentiviral single-guide RNA (sgRNA) library targeting 19,050 genes and subsequently treated with puromycin to obtain A549 cells that had undergone genome-wide knockout (A549-GeCKO) (Fig. 1A). The NS1 protein in influenza viruses is present in virions (albeit at low levels) (23) and is highly expressed when the virus replicates in cells; therefore, we used GFP expression as a screening marker after PR/8/NS1-GFP virus infection (24). Accordingly, A549-GeCKO cells were infected with PR/8/NS1-GFP, and live GFP-high cells (GFPhi) were sorted by flow cytometry. We assumed that GFPhi cells are cells with enhanced IAV infection after knocking out certain genes, especially host factors against virus infection. In contrast, total GFP+ cells will include cells where IAV infection is probably not affected by knocking out the gene. It should be noted that using live GFPhi cells as a screening marker is novel. It is different from the known strategy of using cell death (or survival) as an end point for CRISPR-Cas9 screening (18, 20). Using cell survival as a screening strategy can only enrich genes essential for IAV infection, but it could not enrich antiviral genes. sgRNAs in sorted GFPhi cells and control cells (A549-GeCKO cell without infection) were barcoded, amplified by polymerase chain reaction (PCR), and deep sequenced. The fold enrichment of the target gene in the GFPhi cell was calculated by comparing the gene reads of each guide RNA (gRNA) in the GFPhi with that in the control sample. We observed robust enrichment of >500 genes of sgRNA in the A549-GeCKO library (Fig. 1B and table S1). As expected, among the top hits, i.e., those that exhibited the most significant enrichment of their specific gRNAs, multiple genes have been reported to restrain IAV infection. These included MX1, IFNLR1, OAS1, and IFITM3 (Fig. 1B and fig. S1A) (3, 4), demonstrating that using GFPhi cells as a screening marker in the GeCKO system can reveal previously identified IAV host factors.
Fig. 1. Identification and validation of APOE as a factor in anti-IAV infection.
(A) Overview of GeCKO screen strategy in human lung epithelial (A549) cells. MOI, multiplicity of infection. SSC-A, side scatter parameter. (B) Volcanic mapping of genes enriched in GFP+ cells after PR/8/NS1-GFP infection of the A549-GeCKO library. Genes known for anti-IAV infection or genes with high enrichment are highlighted. Log2FC, log2 fold change. (C and D) A549 WT and APOE KO cells were infected with PR8 at a MOI of 0.5. NP+ and M2+ cells were assessed by flow cytometry (C) or immunofluorescence microscopy (D) 12 hours after infection (left). Relative fold infection from four independent experiments with SD was statistically analyzed (right). Scale bars, 100 μm. DAPI, 4′,6-diamidino-2-phenylindole. (E) A549 WT and APOE KO cells were infected as in (C), and NP and HA expression was assessed by Western blot. The data shown are representative of three independent experiments. (F and G) Supernatant from the A549 WT and APOE KD/KO cells infected as in (C) was collected, the v-RNA was quantified by qRT-PCR (F), and virus titer was detected by plaque assay (G). Data are shown as means ± SD from at least three independent experiments. (H) WT and APOE KD/KO A549 cells were infected with PR8 with a low MOI of 0.01, and supernatant was collected to detect the PR8 growth kinetics by plaque assay. The data shown are representative of three independent experiments. P values were determined using Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, we used the Gene Ontology (GO) analysis to identify enriched biological processes and determined that 130 (150 in GO analysis, MicroRNA excluded) genes from the screening (P < 0.05) mapped to known biological processes (fig. S1B). We observed enrichment for genes involved in the type I IFN pathway, important for anti–influenza virus infection. We also observed enrichment for genes involved in cholesterol and lipoprotein metabolism, Toll-like receptor and nuclear factor κB signaling pathways, etc. Therefore, the GeCKO screening confirmed previously described antiviral processes, such as the type I IFN pathway, and highlighted other metabolic processes, including cholesterol and lipoprotein, that may potentially be important for IAV replication.
Validation of APOE as a factor in anti-IAV infection
As indicated in GO analysis, genes involved in cholesterol and lipoprotein metabolism were enriched. Among them, multiple sgRNAs targeting a well-known gene, APOE, appeared among the top hits (Fig. 1B) and were on the top hit in three independent experiments. APOE plays an essential role in lipoprotein transport. For viral infection, it is known that APOE benefits HCV infection. However, the possible antiviral function of APOE, especially in IAV infection, as implied by the findings above, is largely unreported.
Although APOE expression is relatively low in A549 cells, it is up-regulated upon IAV infection (fig. S2A). To validate APOE as a factor in anti-IAV infection, we knocked down APOE and two additional genes, IFITM3 and MX1, by short hairpin RNA (shRNA) to determine the effect of the genes on IAV infection; the latter two genes are both known to inhibit IAV infection and thus served as positive controls (15, 25). Accordingly, A549 cells were transduced with control, IFITM3, MX1, or APOE shRNAs and selected by puromycin to obtain stable knockdown (KD) cell lines. The KD efficiencies were estimated by quantitative reverse transcription (qRT)–PCR and immunoblot (fig. S2, B and C). The cells were then infected with PR/8/NS1-GFP and assayed for the percentage of infected cells. In line with previous reports, there were more GFP+ cells among cells subjected to IFITM3 and MX1 KD than control cells transduced with scrambled shRNA (fig. S2D). In addition, the fraction of GFP+ cells among cells with APOE KD was also increased compared with control cells. The number of GFP+ cells in APOE KD cells was similar to that in IFITM3 or MX1 KD cells and roughly correlated with its KD efficiency (fig. S2D). Furthermore, we infected control and APOE, IFITM3, and MX1 shRNA KD A549 cells with A/Puerto Rico/8/1934, H1N1 (PR8). We then stained the cells with anti-hemagglutinin (anti-HA), anti-nucleoprotein (anti-NP), or anti-M2 antibodies and detected them by either flow cytometry or confocal microscopy 12 hours after infection (15). We found that the percentage of NP+, M2+, or HA+ cells was greatly increased in APOE KD, IFITM3 KD, and MX1 KD cells compared with cells transduced with scrambled shRNA (fig. S2, E and F).
In complementing the above shRNA KD studies, we constructed A549 APOE knockout (KO) cells by APOE-specific CRISPR-Cas9 targeting. APOE KO in A549 cells was verified by sequencing and immunoblot (fig. S2, G and H). Two A549 APOE KO cell clones, as well as one wild-type (WT) cell clone, were infected with PR8 and then evaluated by either flow cytometry (Fig. 1C) or confocal microscopy (Fig. 1D) as above. We found that the proportion of NP+, M2+, or HA+ cells was significantly increased in A549 APOE KO cells compared with in WT cells. Moreover, cell lysate from the infected cell was detected by immunoblot to confirm viral protein expression. We found that HA and NP expression increased in APOE KO A549 cells compared with WT cells (Fig. 1E).
Although intracellular viral proteins are strongly correlated with the viral titer, detecting viral protein is not a direct measurement of IAV. Thus, we investigated whether APOE KD or APOE KO cells could generate more virus RNA (v-RNA) or infectious virus particles in the supernatant by qRT-PCR or classical plaque assay. APOE KD/KO and control cells were infected with PR8. The supernatant of the infected cells was collected at 12 hours after infection, RNA was extracted from the supernatant, complementary DNA (cDNA) was reverse-transcribed by IAV M1 vRNA primer, and the IAV v-RNA copy number was assessed by qRT-PCR. In line with the experiments above, the IAV v-RNA copy number in supernatants from APOE KO cells increased more significantly than in supernatants from control cells (Fig. 1F). Because the v-RNA copy numbers may not truly reflect the number of infectious virus particles, we next determined the infectious viral particles in the supernatants. Viral titers [plaque-forming units (PFU)/ml] were measured by plaque assays on Madin-Darby canine kidney (MDCK) cells. We found that the virus titer was increased in cells treated with supernatant from APOE KD/KO cells compared with that of control cells (Fig. 1G). Moreover, to detect IAV growth kinetics in WT and APOE KD/KO cells, we infected WT and APOE KD/KO cells with the H1N1 PR8 virus at a relatively low multiplicity of infection (MOI) of 0.01 and monitored virus production at 12, 24, 36, 48, 60, and 72 hours after infection by plaque assay. Virus production peaked after 48 hours after infection. We observed about a seven- to eightfold increase in virus titer in supernatant from APOE KD/KO cell compared to WT cells at almost all time points (Fig. 1H). Thus, these studies establish that cells lacking APOE increased actual IAV infection more than WT cells. Last, the increased IAV infection in APOE KO A549 cells noted above was irrelevant to immunological cytokine production in the cells because IFN-β and interleukin-6 expression increased in APOE KO after IAV infection, probably because of increased IAV virus load in the cells (fig. S3A). These findings thus prove that APOE plays a cell-intrinsic effect on the restriction of IAV infection.
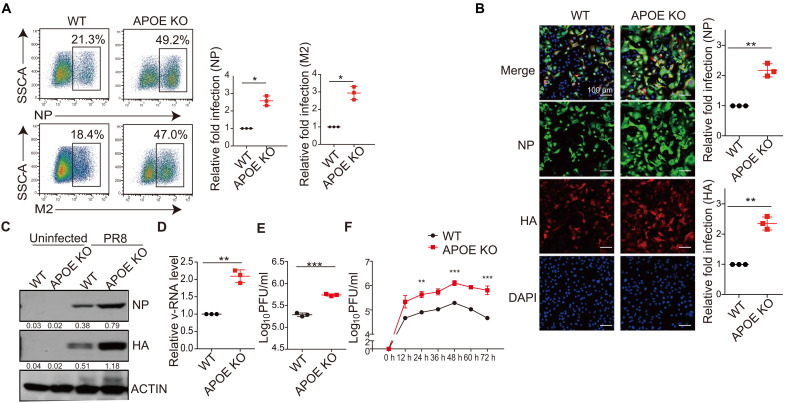
The APOE expression is relatively low in A549 cells. We next determined the effect of APOE on IAV infection in Huh7 cells that express somewhat higher levels of APOE (fig. S4A). We again used the lentivirus transduction of shRNA to construct Huh7 cell lines with stable KD of APOE, IFITM3, and MX1. The KD efficiency was estimated by qRT-PCR (fig. S4B) or immunoblot (fig. S4C). We found that the frequency of NP+ or HA+ cells was also significantly increased in APOE KD Huh7 cells (as well as in IFITM3 and MX1 KD cells) when compared to cells transduced with scrambled shRNA as detected by flow cytometry (fig. S4D) or by confocal microscopy (fig. S4E). Besides, APOE KO Huh7 cell lines were constructed by CRISPR-Cas9–mediated targeting, and APOE KO in the Huh7 cells was verified by either sequencing (fig. S4F) or immunoblot (fig. S4G). We created one clone of APOE KO in the Huh7 cell line, and a cell clone without any mutation was used as WT control. Huh7 WT and APOE KO cells were infected with PR8. We found that PR8 infection increased in Huh7 APOE KO cells detected by either flow cytometry (Fig. 2A) or confocal microscopy (Fig. 2B). Cell lysate from infected Huh7 cells was also detected by immunoblot, and HA or NP expression was increased in APOE KO Huh7 cells compared with that in WT cells (Fig. 2C). IAV v-RNA or viral particles were detected in the supernatant. The IAV v-RNA or infectious viral particles were increased in the supernatant of Huh7 APOE KO cells (Fig. 2, D and E) compared with that of WT Huh7 cells. A similar observation was obtained with the IAV growth kinetics study in WT and APOE KO Huh7 cells (Fig. 2F). We observed a seven- to eightfold increase in virus titer in Huh7 APOE KO cells after PR8 virus compared to WT Huh7 cells.
Fig. 2. Validation of APOE as a factor in anti-IAV infection.
(A and B) Huh7 WT and APOE KO cells were infected with PR8 at an MOI of 0.3. NP+ and M2+ cells were assessed by flow cytometry (A) or immunofluorescence microscopy (B) 12 hours after infection (left), and relative fold infection from three independent experiments with SD was statistically analyzed (right). Scale bars, 100 μm. (C) Huh7 WT and APOE KO cells were infected as in (A), and NP and HA expression was assessed by Western blot. The data shown are representative of three independent experiments. (D and E) Supernatant from the Huh7 WT and APOE KO cells infected as in (A) was collected, the v-RNA was quantified by qRT-PCR (D), and virus titer was detected by plaque assay (E). Data are shown as means ± SD from at least three independent experiments. (F) WT and APOE KD/KO Huh7 cells were infected with PR8 with a low MOI of 0.01, and supernatant was collected to detect the PR8 growth kinetics by plaque assay. The data shown are representative of at least three independent experiments. P values were determined using Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Last, to further detect whether APOE could protect the cell from IAV infection, we complement the loss-of-function APOE results by reinstating WT APOE in APOE KO cells and detecting whether WT APOE could reverse the increased virus infection in APOE KO cells. WT and APOE KO A549 or Huh7 cells were transduced with either an empty vector or lentiviral vector expressing APOE and selected by puromycin to obtain a cell line that stably expresses APOE. The complement of WT APOE in APOE KO cells was confirmed by immunoblot (fig. S5, A and B). WT, APOE KO, and APOE KO cells complemented with WT APOE were infected with PR8. We found that APOE KO cells complemented with APOE manifested reduced IAV infection, as indicated by decreased frequency of NP+ cells compared with APOE KO cells but comparable to that of WT cells (fig. S5, C and D). Moreover, a viral titer was also detected. We found that APOE KO cells complemented with APOE manifested comparable viral titer with that of WT cells (fig. S5E), suggesting that the reinstatement of WT APOE expression protected APOE KO cells from IAV infection.
Together, our screening identified a host factor APOE, whose loss rendered cells more susceptible to IAV infection.
APOE-deficient mice exhibit enhanced IAV infection and severe disease pathology
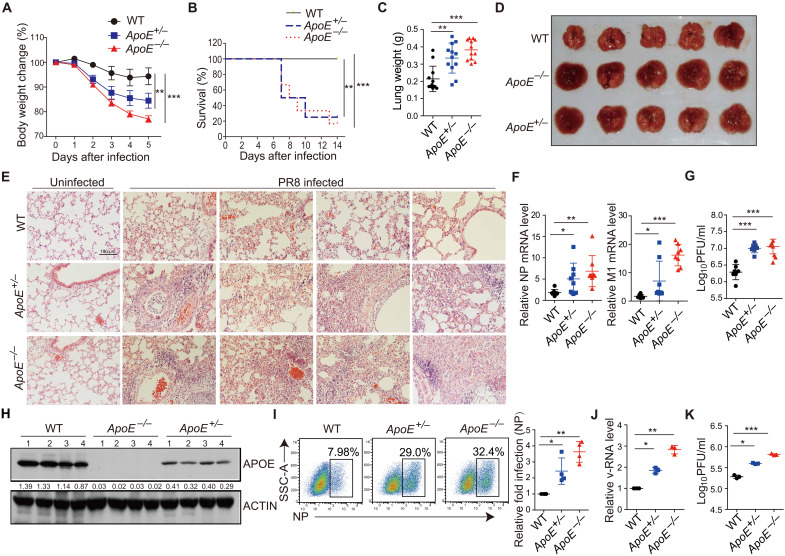
To further investigate the role of APOE during IAV infection in vivo, we first detected the immunological baseline of uninfected WT, ApoE+/−, and ApoE−/− mice. We found that the percentage and total number of neutrophils in the lung of uninfected ApoE−/− mice but not ApoE+/− mice were significantly decreased compared to WT mice (fig. S6A). In contrast, the expression of other cell subsets and proinflammatory cytokines is comparable among those mice (fig. S6, B to D). Dyslipidemia was only observed in ApoE−/− but not ApoE+/− mice (fig. S6E). We next infected ApoE+/−, ApoE−/−, and WT mice by intranasal administration of PR8 and then monitored the disease progression. PR8-infected ApoE+/− and ApoE−/− mice exhibited more significant weight loss (Fig. 3A) and a markedly decreased survival rate (Fig. 3B) than WT mice. In addition, the lung size or lung weight of ApoE+/− and ApoE−/− mice was significantly increased and exhibited enhanced necrosis and fibrosis compared with lungs from WT controls (Fig. 3, C and D). Bleeding into alveoli or neutrophil cell infiltration was enhanced in IAV-infected ApoE+/− and ApoE−/− lungs compared to WT lungs on day 4 or 7 after IAV infection (Fig. 3E and fig. S6F). These pathologic differences correlated with the presence of higher IAV mRNA levels in the ApoE+/− and ApoE−/− mice compared to that in WT mice at this time point (Fig. 3F), and the IAV titers in the lung at day 4 after infection were elevated as well (Fig. 3G). The percentage of neutrophils and proinflammatory cytokine expression was also increased in ApoE−/− and ApoE+/− mice on day 4 after infection (fig. S6, G and H), indicating enhanced inflammation. Thus, the finding in IAV-infected ApoE+/− mice and ApoE−/− mice provided further evidence supporting our conclusion that APOE plays an essential role in restriction IAV infection in vivo.
Fig. 3. APOE-deficient mice exhibit enhanced IAV infection and severe disease pathology.
(A) WT, ApoE+/−, and ApoE−/− mice were infected with 1000 PFU of PR8, and body weight was monitored. Each genotype of mice had 10 male and 5 female. (B) Survival rate of PR8-infected mice was detected. Each genotype of mice had nine male and three female. (C) The lung weight from mice infected as in (A) was measured on day 5 after infection. Data were pooled from three independent experiments with SD. (D) Representative images of infected lungs at day 5 after infection. (E) Pathology of lung from mice at day 7 after infection. Scale bars, 100 μm. (F) NP and M1 mRNA in lungs of infected mice were determined by qRT-PCR, each dot represents data from one mouse, and data were pooled from three independent experiments with SD. (G) Virus titers in lungs of infected mice at day 4 after infection were determined by plaque assay, each dot represents data from one mouse, and data were pooled from three independent experiments with SD. (H) APOE expression in the lung was determined by Western blot, and each lane represents data from one mouse. The data shown are representative of three independent experiments. (I) WT, ApoE+/−, and ApoE−/− MEFs were infected with PR8 at an MOI of 10. NP+ cells were assessed by flow cytometry 12 hours after infection. Relative fold infection from four independent experiments with an SD was statistically analyzed. (J and K) The supernatant from (I) was collected to determine v-RNA by qRT-PCR (J) or viral titer by plaque assay (K). Data are shown as means ± SD from at least three independent experiments. P values were determined using one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparisons test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Moreover, it should be noted that ApoE+/− mice exhibited an intermediate phenotype with respect to weight loss, histology, and virus titer as compared to ApoE−/− and WT mice, indicating a dose-dependent effect of APOE on IAV infection. The finding supporting this conclusion is that the protein level of APOE in the lung of ApoE+/− mice is intermediate between WT and ApoE−/− mice as detected by immunoblot (Fig. 3H).
To further confirm the in vivo phenotype observed in APOE-deficient mice, we evaluated infection in mouse embryonic fibroblasts (MEFs) derived from WT, ApoE+/−, and ApoE−/− embryos. We found that PR8 infection was enhanced in ApoE+/− and ApoE−/− MEF cells (Fig. 3I). The v-RNA and virus titer detected from IAV-infected ApoE+/− and ApoE−/− MEF cell supernatant were also increased compared to WT MEF cell supernatant (Fig. 3, J and K).
APOE broadly inhibits H5N1 and H7N9 IAV infection
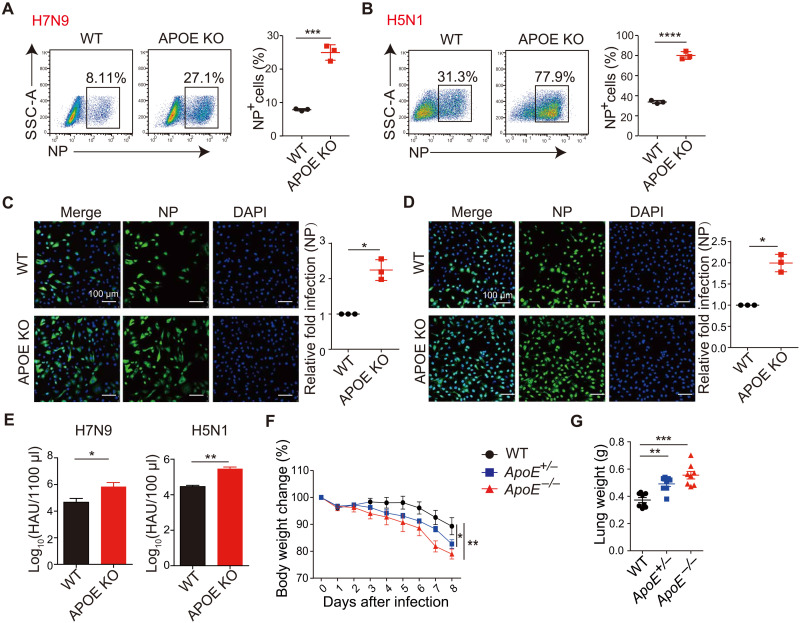
To establish that APOE-mediated restriction of IAV was not limited to the H1N1 PR8 strain studied above but was broadly applicable to IAV infection, we next tested APOE’s effect on two other highly pathogenic avian influenza virus strains: H7N9 and H5N1. We found that infection of A549 APOE KO cells with these viral strains led to a higher number of NP+ cells than that of WT cells, as determined by flow cytometry (Fig. 4, A and B) or confocal microscopy (Fig. 4, C and D). In addition, the culture supernatant of APOE KO cells that infected with H7N9 and H5N1 contained increased numbers of viral particles compared to WT supernatants, as detected by the chicken hemagglutination test (Fig. 4E). Last, to determine the effect of ApoE on H7N9 infection in vivo, WT, ApoE+/−, and ApoE−/− mice were intranasally infected with H7N9, and disease progression was monitored. As with H1N1 infection, H7N9-infected ApoE+/− and ApoE−/− mice exhibited more weight loss (Fig. 4F) than WT mice, and the lung weight of ApoE+/− and ApoE−/− mice was also increased (Fig. 4G) compared with that of WT mice. These findings further strengthen the physiological relevance of APOE acting as a regulator of IAV infection and a component of the host barrier that broadly restricts different strains of IAV infection in vitro and in vivo.
Fig. 4. APOE broadly inhibit H5N1 and H7N9 IAV infection.
(A and B) A549 WT and APOE KO cells infected with H7N9 (A) or H5N1 (B) at an MOI of 0.5 were collected 12 hours after infection and analyzed for NP+ cells by flow cytometry. Data are shown as means ± SD from at least three independent experiments. (C and D) A549 WT and APOE KO cells infected as in (A) or (B) were used for immunofluorescence microscopy by staining the cells with NP antibody (left), and relative fold infection from three independent experiments with SD was statistically analyzed (right). Scale bars, 100 μm. (E) Virus titer of cell supernatant from (A) and (B) was determined using chicken blood cell hemagglutination assay. Data are shown as means ± SD from four independent experiments. (F) H7N9 was intranasally administrated to WT, ApoE+/−, and ApoE−/− mice with 2000 PFU per mouse, and body weights were measured daily. Data were pooled from three independent experiments with SD (WT, n = 8; ApoE+/−, n = 10; ApoE−/−, n = 10). (G) Lungs were collected on day 8 after H7N9 infection, and the lung weight was measured. Each dot represents data from one mouse, and data were pooled from three independent experiments with an SD. P values were determined using Student’s t test or one-way ANOVA followed by Tukey’s multiple-comparisons test.*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
APOE deficiency affects viral attachment
To address where APOE block was occurring in the virus’s life cycle, we first used IAV pseudovirus particles to determine whether APOE affects the entry of IAV into the cells. The IAV pseudovirus particles contain an HIV genome encoding luciferase coated with envelope protein from IAV HA and NA [derived from the A/WSN/33(H1N1) viral strain]. APOE KO cells exhibited increased entry of the pseudovirus particles, as indicated by the increased luciferase signal observed in these cells (Fig. 5, A and B). Because the entrance of pseudovirus particles into a cell depends on its envelope’s composition, these data support the conclusion that APOE inhibits IAV entry into the cells (15). We next asked whether the APOE was required for IAV entry by allowing virus attachment to the cell surface membrane. We incubated WT and APOE KO cells with PR8 virus at 4°C for 60 min (to prevent viral fusion), washed and extracted RNA, and performed RT-PCR to detect the surface-bound virus. We found that v-RNA was higher in APOE KO cells than in WT cells, suggesting that APOE absence enhanced virus attachment (Fig. 5C). To detect whether APOE affects IAV fusion, a classic “acid bypass” assay was performed where the viral membrane is fused to the plasma membrane by a transient low-pH treatment, causing the viral ribonucleoprotein complex (vRNPs) to be deposited directly into the cytoplasm (26, 27). We found that fusion was not enhanced in APOE KO A549 cells in the acid bypass assay (fig. S7A), indicating that the lack of APOE does alter pH-dependent fusion. Last, it is important to functionally disentangle influenza’s normal receptor from APOE by demonstrating that treating cells with sialidase similarly abrogates virus attachment in WT or APOE KO cells. To this end, we treated WT and APOE KO cells with sialidase and performed an IAV attachment experiment as above. We found that sialidase treatment reduces viral attachment in both WT and APOE KO cells to a similar level as detected by RT-PCR (Fig. 5D). Collectively, our findings provided strong evidence that APOE inhibits IAV infection mainly through decreasing IAV attach to its receptors.
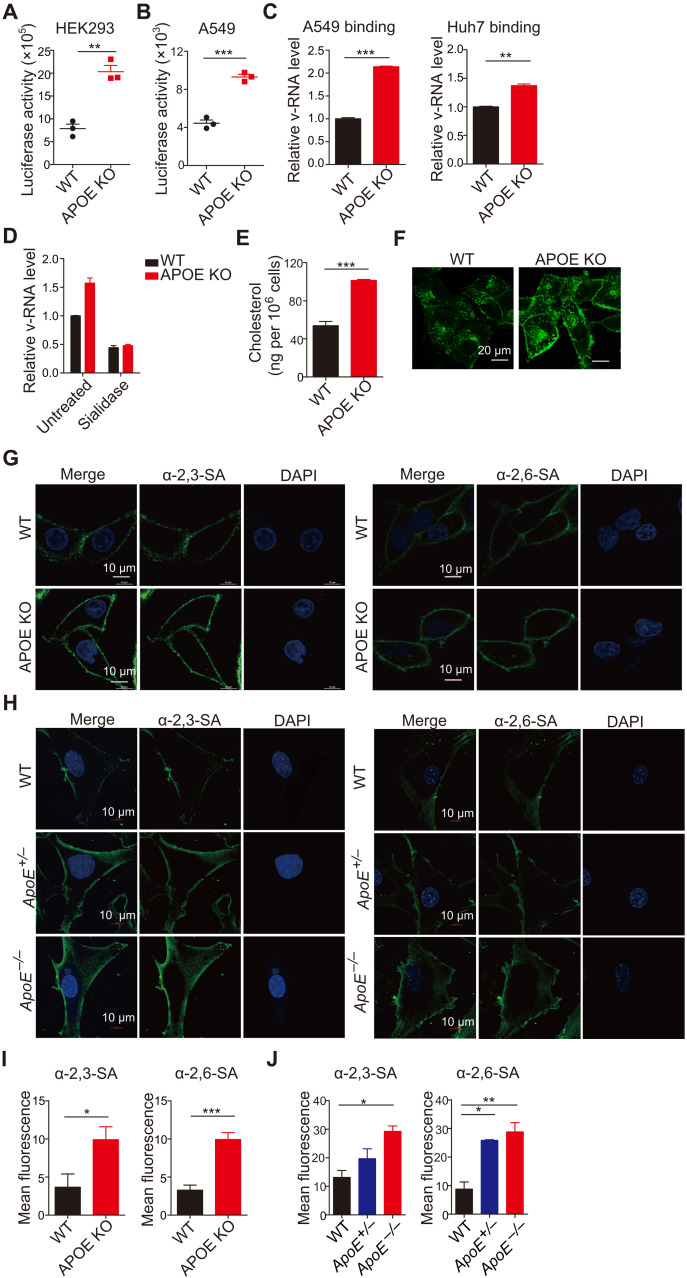
Fig. 5. APOE deficiency affects viral attachment and regulates the distribution of viral receptors on the cell surface.
(A and B) HEK293 (A) and A549 (B) WT and APOE KO cells were infected with pseudovirus. Virus entry was detected by luciferase activity. Data are shown as means ± SD from at least three independent experiments. (C) Viral attachment in WT and APOE KO A549 or Huh7 cells was determined by qRT-PCR. Relative v-RNA level from three independent experiments with SD was statistically analyzed. (D) WT and APOE KO A549 cells were pretreated with sialidase and infected as in (C). Viral attachment was determined by qRT-PCR. Relative v-RNA level from three independent experiments with SD was statistically analyzed. (E) Total cellular cholesterol in WT or APOE KO A549 cells was detected by an Amplex Red Cholesterol assay. Data are shown as means ± SD from at least three independent experiments. (F) Filipin staining was detected in A549 WT or APOE KO cells by confocal microscopy, and representative images are shown (scale bars, 20 μm) and are representative of at least three independent experiments. (G and H) A549 WT and APOE KO cells (G) and WT, ApoE+/−, and ApoE−/− MEFs (H) were stained with α-2,3–linked SA or α-2,6–linked SA and detected by confocal microscopy. Scale bars, 10 μm. Data are representative of at least three independent experiments. (I and J) Mean fluorescence of α-2,3–linked SA or α-2,6–linked SA in A549 (I) and MEF cells (J) from (G) and (H) was calculated by ImageJ. Data are shown as means ± SD from at least three independent experiments. P values were determined using Student’s t test or one-way ANOVA followed by Tukey’s multiple-comparisons test. *P < 0.05, **P < 0.01, and ***P < 0.001.
To determine whether APOE affects the IAV life cycle by regulating viral RNA synthesis, we constructed APOE KO human embryonic kidney (HEK) 293 cells by CRISPR-Cas9 technique and confirmed that these cells had undergone successful APOE KO by sequencing, immunoblot, and IAV infection (fig. S7, B to D). We then used a well-established minireplicon assay to examine whether the absence of APOE in these cells affected the polymerase activity of IAV (28, 29). In these studies, HEK293 WT and APOE KO cells were cotransfected with plasmids encoding various viral components, including plasmids encoding PB1, PB2, PA, and NP and a polymerase I (Pol I)-driven RNA expression plasmid encoding the HA v-RNA segment; HA expression was then estimated by immunoblot 24 hours after transfection. We found that WT and APOE KO cells exhibited comparable HA expression, indicating that H1N1 PR8 polymerase activity was similar in the two cells (fig. S7E). To further detect the effect of APOE on viral RNA replication, we used pPolINP-luc reporter, which is a polymerase I (PolI)–driven plasmid expressing influenza virus–like firefly luciferase RNA with a noncoding region of nucleoprotein gene of IAV on both ends (30–32) to quantify IAV polymerase activity in WT and APOE KO cells. We found that the luciferase activity in APOE KO cells was comparable with that of WT cells (fig. S7F). It was, therefore, apparent that APOE does not inhibit viral RNA replication. Collectively, the above studies provide evidence that APOE restricts IAV infection by inhibiting IAV attachment.
Cellular cholesterol homeostasis was regulated by APOE
We next explored the mechanism by which APOE restricts virus infection. Our starting point was that APOE played an important role in lipoprotein or cholesterol transportation and metabolism. APOE KO mice are dysregulated with systemic lipid metabolism (33). Thus, the absence of APOE is likely to lead to an increased cellular cholesterol/cholesterol derivative accumulation that is, in some way, responsible for enhanced IAV infection. We determined cellular cholesterol levels in APOE KO cells and found that A549 APOE KO (Fig. 5E) or Huh7 APOE KO cells (fig. S8A) exhibit increased levels of cholesterol when compared with WT A549 cells or WT Huh7 cells (Fig. 5E and fig. S8A). The accumulation of cholesterol in APOE KO cells was further confirmed by filipin staining; a reagent specifically binds to cholesterol ester (Fig. 5F). In addition, APOE KO cells complemented with WT APOE exhibited comparable cholesterol with that of WT cells (fig. S8B). As a consequence of cholesterol accumulation, APOE KO displayed a decreased expression of genes involved in cholesterol and fatty acid uptake [low-density lipoprotein (LDL) receptor (LDLR) and CD36] and cholesterol synthesis [hydroxyl methylglutaryl–coenzyme A (HMG-CoA) reductase (HMGCR) and 24-dehydrocholesterol reductase (DHCR24)] (fig. S8C). In contrast, the level of cholesterol 25-hydroxylase (CH25H), the enzyme that catalyzes cholesterol conversion to 25-OH, was higher in APOE KO cells (fig. S8D). We observed increased IAV membrane sialic acid (SA) receptor α-2,3–linked SA and α-2,6–linked SA distribution on the surface of A549 APOE KO cells compared with that of WT cells (Fig. 5, G and I, and fig. S8E). Besides, membrane SA receptor α-2,3–linked SA and α-2,6–linked SA expression on the surface of MEF cells from WT, ApoE+/−, and ApoE−/− mice was also detected. We found that there is more SA receptor distribution on the surface of both ApoE+/− and ApoE−/− MEF, and as expected, the SA expression on ApoE+/− MEF is intermediate compared with that of WT and ApoE−/− (Fig. 5, H and J). Collectively, these findings supported the concept that modulation of cellular cholesterol homeostasis by APOE plays a physiological role that affects virus receptor distribution and regulates virus entry.
Dynamic regulation of cholesterol homeostasis by APOE mediates cell resistance to virus infection
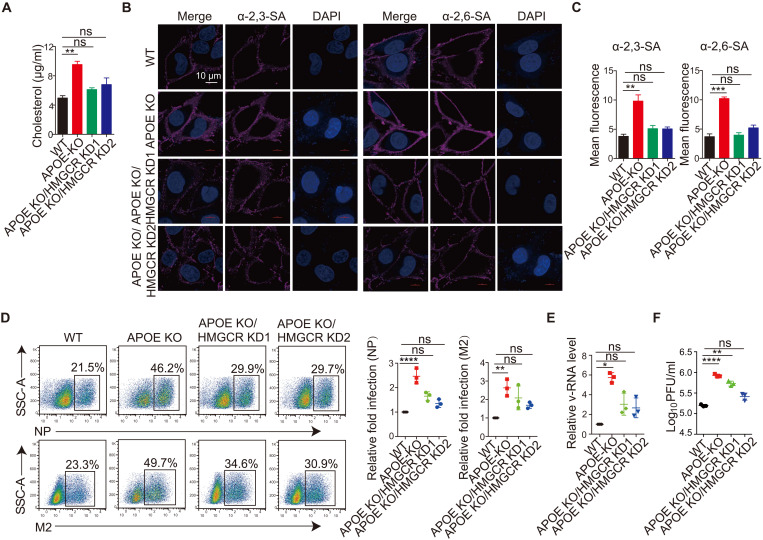
To determine whether the enhanced virus infection in APOE KO cells was indeed due to increased cholesterol accumulation in the cell, we next explored reducing cholesterol levels in APOE KO cells by knocking down the gene encoding HMG-CoA reductase (HMGCR), an enzyme involved in the cholesterol mevalonate biosynthetic pathway. The HMGCR shRNAs with about 25 or 50% KD efficiency were chosen (fig. S8F). HMGCR was knocked down in APOE KO cells to obtain APOE KO/HMGCR KD cells. We detected cholesterol levels or α-2,3–linked SA and α-2,6–linked SA distribution in the APOE KO/HMGCR KD cells. We found that both cholesterol levels (Fig. 6A) and α-2,3–linked SA or α-2,6–linked SA distribution (Fig. 6, B and C) in the APOE KO/HMGCR KD cell are comparable with that of WT cells. These findings demonstrated that intracellular cholesterol accumulation is responsible for the increased distribution of viral receptors. We next infected WT, APOE KO, and APOE KO/HMGCR KD cells with PR8 and then measured the frequency of virus-infected cells and the v-RNA or viral particles in the supernatant of infected cells as described above. We found that the frequency of virus-infected cells (NP+ cell or M2+ cell) (Fig. 6D), v-RNA (Fig. 6E), or infectious virus particles (Fig. 6F) in the supernatant of APOE KO/HMGCR KD cell was significantly reduced compared with that of APOE KO cells and was comparable to that of WT cells. Last, virus attachment was examined in APOE KO/HMGCR KD cells. We found the virus attachment was decreased significantly in KO/HMGCR KD cell than that of APOE KO cell but comparable with that of WT cells (fig. S8G). These results thus provided strong evidence demonstrating that cholesterol accumulation resulting from APOE deletion is a crucial contributor to enhanced virus infection in APOE KO cells.
Fig. 6. Dynamic regulation of cholesterol homeostasis by APOE mediates cell resistance to virus infection.
(A) Total cellular cholesterol in WT, APOE KO, and APOE KO/HMGCR KD A549 cells was detected by an Amplex Red Cholesterol assay. Data are shown as means ± SD from at least three independent experiments. (B) A549 WT, APOE KO, and APOE KO/HMGCR KD cells were stained with α-2,3–linked SA or α-2,6–linked SA and detected by confocal microscopy. Data are representative of at least three independent experiments. Scale bar, 10 μm. (C) Mean fluorescence of α-2,3–linked SA or α-2,6–linked SA in (B) was calculated by ImageJ. Data are shown as means ± SD from at least three independent experiments. (D) A549 WT, APOE KO, and APOE KO/HMGCR KD cells were infected with PR8 at an MOI of 0.5, and NP+ or M2+ cells (left) were analyzed by flow cytometry 12 hours after viral infection. The relative fold infection from three independent experiments with SD was statistically analyzed (right). (E and F) Supernatant from the A549 WT and APOE KO cells and APOE KO/HMGCR KD cells infected as in (D) was collected, and the v-RNA was quantified by qRT-PCR (E) or viral titer by plaque assay (F). Data are shown as means ± SD from at least three independent experiments. P values were determined using one-way ANOVA followed by Tukey’s multiple-comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
DISCUSSION
Although many host factors have been identified for modifying IAV infection, many factors remain to be found and less understood. Here, we executed a unique genetic screening using the PR/8/NS1-GFP reporter virus as a screening marker and identified APOE as an important host factor restricting IAV infection. APOE deficiency conferred increased human influenza virus H1N1 and high pathogenic avian influenza virus H5N1 and H7N9 infection in vitro and in vivo. Because one of the functions of APOE is to regulate cellular cholesterol homeostasis, our findings established that such homeostasis plays a critical role in inhibiting virus infection. A recent report identified cholesterol regulation as an important regulator of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry and/or replication by the CRISPR-Cas9 screening (34–36).
Genome-wide CRISPR-Cas9 screening has been widely used to identify vital host factors for IAV replication (18, 20–22). Most of these studies rely on cell survival or cell death as an end point to select essential genes for IAV replication. The limitation of this strategy is that it can only find factors essential for viral infection, but it could not find factors that inhibit IAV infection. Using GFP as a selection marker, we can enrich a live GFP relative high cell population in which IAV infection was potentially enhanced when certain genes were KO. We successfully enriched several well-known and important host factors that restrict IAV infection, such as MX1, interferon lambda receptor 1 (IFNLR1), OAS1, and IFITM3, suggesting that our strategy successfully screened host factors that limit IAV infection. We found APOE was on the top screening rank, and our series study demonstrated that APOE plays a critical role in anti-IAV infection both in vitro and in vivo.
APOE was initially described as a lipid transport protein and major ligand for LDL receptors with a role in cholesterol metabolism and cardiovascular disease. APOE is a component of certain plasma lipoproteins and mediates lipoprotein catabolism in the liver, mainly by binding the very-low-density lipoprotein (VLDL) receptor and the LDL receptor, on the hepatocyte surface (37, 38). APOE deficiency is associated with a decreased catabolism of atherogenic lipoproteins, thus favoring hypercholesterolemia and atherosclerosis development (39, 40). APOE is known to be involved in the pathogenesis of infectious diseases for virus infection. For example, APOE is required for infectivity and infectious viral particle production of HCV (41). APOE KO mice had significantly lower herpes simplex virus-1 concentrations in the nervous system than WT mice (42), suggesting that APOE is an essential component promoting these virus replications or replications infection. In contrast, and distinct from the above findings, APOE inhibits HIV infection by directly interacting with HIV gp160 and suppresses Env expression (43). Here, we found that APOE restricted IAV infection through modulating cell cholesterol homeostasis, which added a mechanism of APOE in limiting virus infection. Moreover, our data indicated that SARS-CoV-2 infection was greatly increased in APOE KO Huh7 cells and that the effect of APOE on SARS-CoV-2 infection is more marked than that of on IAV infection (fig. S9A). It is well established that APOE is involved in cholesterol homeostasis as part of the lipoprotein system that facilitates the transport of triglycerides and cholesterol between the liver and peripheral organs through the blood. In our study, APOE KO cells exhibited increased cholesterol accumulation compared with WT cells, suggesting that APOE might also play a role in maintaining cholesterol homeostasis at the cellular level. This observation is supported by the findings that APOE KO dendritic cells isolated from APOE KO mice also exhibited increased cholesterol in the cell (44).
Our studies of the mechanisms underlying the role of APOE in virus infection focused on identifying the stages of the viral life cycle. We showed that APOE restrains viral attachment to the cell. These studies set the second round of mechanistic studies focused on how APOE affects viral attachment. We showed that, as predicted by previous studies of apolipoprotein function, APOE regulates cholesterol homeostasis of the cells so that cells depleting these components manifested increased cholesterol concentrations in the cell. It is known that 70 to 80% of cholesterol is mainly located in the cell membrane (45), and cholesterol is a crucial constituent of the lipid rafts in the cell membrane that mediates membrane signaling (44). Thus, cholesterol has particular significance to the viral attachment. About the latter, we showed that APOE-deleted cells exhibit an increased surface distribution of IAV receptor SA. Although our high-resolution confocal microscopy data did not allow the identification of receptor clustering on APOE KO cells, this is predicted to occur in the presence of increased membrane cholesterol on the basis of the work of Goronzy et al. (46) and Liu and Boxer (47). However, it should be noted that the precise mechanisms connecting APOE, cholesterol, and changes in SA surface density remain unclear. Last, because cholesterol is important for many steps of the viral replication cycle, we cannot exclude the possibility that APOE may affect IAV infection other than the attachment step; that is, APOE might affect the intracellular transport of viral glycoproteins, assembly of virus particles, etc.
Influenza infection can cause increased inflammation in the vessel walls of APOE-deficient mice at plaque sites and thrombotic events. We are aware of previous studies on this phenomenon (48). It should be noted that these effects of infection have been conducted in old ApoE−/− mice to maximize visualization of effects on preexistent atherosclerosis. However, our studies were conducted on 8-week-old heterozygous and homozygous mice with minimal atherosclerosis. The pulmonary architecture of these mice before infection was equivalent to that of WT mice, except that the macrophage and neutrophil decreased significantly in ApoE−/− but not ApoE+/− mice, and bleeding into tissue was also normal at this time. Increased bleeding and inflammation in ApoE+/− and ApoE−/− mice were seen only after infection. However, it should be noted that basal cholesterol/lipid accumulation in ApoE−/− mice (fig. S6E) may also contribute to the disease severity and survival in ApoE−/− mice.
The antiviral function of APOE in our studies may have important clinical implications in that during the H1N1 pandemic. Patients with obesity were associated with an increased risk of influenza-associated intensive care and longer mechanical ventilation duration than nonobese (49). This observation is consistent with our findings that accumulated cholesterol increased IAV virus infection. It is known that influenza virus infection was associated with an increased risk of acute cardiovascular diseases, and during the flu season, hospital admissions due to acute exacerbations of various chronic diseases such as cardiovascular atherosclerotic disease are greatly increased (50–52), which may be caused (at least in part) by covert influenza infection in atherosclerotic plaque. The finding supporting the latter is that arterial segments associated with atherosclerotic plaques contained H1N1 and H3N3 influenza v-RNA, whereas segments free of plaques do not (53). Thus, the observations of APOE influencing IAV infection introduce the possibility that preexisting abnormalities of apolipoprotein/cholesterol metabolism may lead to local (endothelial) virus infection changes and thus regulate local inflammatory responses that enhance or inhibit the development of the disease. In summary, we identified APOE as an antiviral host factor and established that APOE functions as a defense strategy against IAV infection through regulating cellular cholesterol homeostasis.
MATERIALS AND METHODS
Cells and viruses
Human lung epithelial cells (A549), human hepatoma cells (Huh7), HEK (HEK293) cells, and MDCK cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS; HyClone), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Invitrogen). MEF cells were obtained by breeding ApoE+/− × ApoE+/− mice to generate WT, ApoE+/−, and ApoE−/− embryos, and MEF cells derived from the embryo at day 13.5 were prepared as described previously (17). All cells were cultured at 37°C in a 5% CO2 humidified atmosphere.
The IAV strains used were PR8 (A/Puerto Rico/8/1934, H1N1), PR/8/NS1-GFP (A/NS1-GFP/PR8 H1N1), low pathogenic version of H5N1 (A/Vietnam/1203/2004 VN04Low), and H7N9 (LPAI A/Anhui/2013). The virus was amplified using 10-day-old embryonic chicken eggs and then titrated by determining log10PFU/ml on MDCK cells or determining total virus particle with chicken hemagglutination test.
H1N1 experiments were performed in a biosafety level 2 (BSL-2) laboratory according to the protocol; all experiments with H5N1 and H7N9 virus were performed in a BSL-3 laboratory.
Mice
ApoE−/− mice were purchased from the Jackson Laboratory on a C57BL/6J background and were bred to generate WT, ApoE+/−, and ApoE−/− mice. All in vivo experiments were performed using age- and sex-matched littermate controls. Studies were carried out under animal care guidelines of the Institute of Microbiology, Chinese Academy of Science with license permit number of APIMCAS2017015.
A549-GeCKO library generation and H1N1 (PR/8/NS1-GFP) screening
A high-throughput CRISPR-Cas9–based screening was undertaken on an arrayed library (Addgene, cat. no. 1000000048) as previously reported (54) and was used to identify host factors against IAV infection. Briefly, to amplify the library, 2 μl of library DNA (50 ng/μl) was used to transform 25 μl of electro-competent Escherichia coli (TaKaRa). Transformed colonies (>6 × 107) were scraped off from the Luria-Bertani plates into the medium, and plasmids were extracted. To prepare the virus library, 293 T cells in the 15-cm dish were transfected with 25 μg of library DNA together with 15 μg of psPAX2 and 10 μg of pMD2.G. After 8 hours, 10 ml of complete medium (DMEM with 10% FBS; Invitrogen) was replaced, and the lentivirus supernatant was collected 48 hours after transfection. The supernatant was filtered with a 0.45-μm filter, and the viral titer was determined by plaque assay as described previously (55).
The sgRNA lentivirus library was then used to transduce A549 cells cultured in 15-cm dishes (10 dishes in total) at an MOI of 0.3 to obtain less than one sgRNA per cell (Fig. 1A, step 1), and puromycin (2 μg/ml) was used for 14 days to obtain whole genome-wide KO A549 cells (A549-GeCKO cells) (Fig. 1A, step 2) (56, 57). Half of the A549-GeCKO cells (15 dishes) were directly used for DNA extraction and sgRNA sequencing (Fig. 1A, step 3). The other half of the A549-GeCKO cells (15 dishes) were subjected to infection with the PR/8/NS1-GFP reporter virus at an MOI of 0.3 for 3 hours and incubated for another 8 hours, after which GFPhi cells among live cells were sorted on Biosciences FACSAria II (Fig. 1A, steps 4 to 7). The live infected GFPhi cells were sorted in the BSL-2 laboratory. The BD FACSAria II sorter was placed in a hood that the ceiling was mounted with a ducted hepa filter unit to protect users from aerosols generated during sorting. The rest of the five 15-cm culture dishes of the A549-GeCKO cells were left uninfected and used as control. DNA from the sorted GFPhi cells and control cells (uninfected A549-GeCKO cells) was extracted and dissolved in H2O (3 to 4 μg/μl) and used as the templates for amplification of the gRNA. sgRNAs in sorted GFPhi cells and control cells (A549-GeCKO cell without infection) were barcoded, amplified by PCR, and deep sequenced (Fig. 1A, step 8).
The sgRNAs were amplified using the rTaq DNA polymerase (TaKaRa Taq R001B) by a two-step PCR method. First, six 50-μl PCRs (each containing 50 μg of genomic DNA template) were performed with the forward primer v2Adaptor_F (sequence listed in table S2) and the reverse primer v2Adaptor_R (sequence listed in table S2); products of the first-step PCR were pooled together and used as the template for the second-step PCR. For the second step, six 50-μl PCRs (each containing 1 μl of the first-step PCR product) were performed with primer IlluminaF-01/02 and primer Illumina R-01/02 (primer 01 for the control group and primer02 for GFP+ group, sequence listed in table S2). Products of the second-step PCRs were subjected to electrophoresis, and the DNA [around 310–base pair (bp) band] was extracted and sequenced at the HiSeq 2500 instrument (Illumina) by using the 50-bp single-end sequencing protocol. The first 19 nucleotides from each sequencing read are the gRNA sequence recovered from the library. The fold of enrichment was calculated by comparing the read frequency of each sgRNA in the experiment sample with that in the control sample (table S1).
Enrichment of functions and signaling pathways analysis was performed on the basis of GO by Metascape online analyses. For functional enrichment analysis, all enriched genes were mapped to terms in the GO databases, and then, significantly enriched GO terms were searched for using P < 0.05 as the threshold.
Virus infection and viral titer detection
For virus infection, cells were washed with phosphate-buffered saline (PBS) twice and infected with H1N1 (PR8), H5N1, or H7N9 with different MOIs as indicated in the figure legends. The virus was diluted in DMEM without FBS, and viral infection was performed for 1 hour at 37°C. After viral removal, the cells were washed with PBS twice and cultured in 2% FBS DMEM unless otherwise mentioned. Twelve hours after infection, the cells were collected for flow cytometry analysis or immunofluorescence.
For viral titer plaque assay (PFU) or v-RNA determination, DMEM with 0.75% bovine serum albumin (BSA; VWR Life Science, no. 0332) were used as a culture medium after virus removal. Viral supernatant was harvested 12 hours after infection, and plaque assays were performed on MDCK cells to titrate virus titer as described previously (55). The v-RNA in cell supernatant was detected by qRT-PCR. For H5N1 and H7N9 viral titer detection, total titer of the virus was measured by HA assay (58). In particular, fresh chicken red blood cells were set at 2 × 107 cells/ml. HA titers were expressed as log10HA units per test volume (log10HAU/100 μl). Assuming one chicken red blood cell being sufficient for the agglutination of one virus particle, HA titers could be converted into concentration of total viruses.
For mouse infection, 8-week-old mice that were gender-matched were intranasally infected with H1N1 PR8 in 50 μl of PBS with 1000 PFU for body weight loss or survival or with 2000 PFU for virus titer detection in lungs at day 4 after infection.
Generation of shRNA KD cell line
APOE, IFITM3, MX1, or HMGCR KD A549 cells or Huh7 cells were constructed by using shRNA. Briefly, scramble shRNA sequence and shRNA sequence targeting the human APOE, IFITM3, MX1, or HMGCR gene (shRNA sequences are listed in table S3) were cloned into PLKO.1 lentiviral vector and applied for producing lentivirus on HEK293 cells. A549 or Huh7 cells were then infected with the scramble or target gene lentivirus. After 48 to 60 hours after infection, puromycin (2 μg/ml) was added to obtain stable KD cells. KD efficiency was confirmed by either qRT-PCR or Western blot.
Generation of APOE KO cell lines using the CRISPR-Cas9 system
APOE KO A549, Huh7, or HEK293 cells were generated by using the CRISPR-Cas9 system. The two sgRNAs are located in exons 1 and 2 of the APOE gene. The sgRNA sequence targeting the human APOE gene (sgRNA sequences are listed in table S4) was cloned into a lentiCRISPR v2 vector and applied for producing lentivirus. A549, Huh7, or HEK293 cells were infected with the lentiCRISPR v2 lentivirus. After 48 to 60 hours after infection, the single-cell clones acquired by using the limiting dilution method were expanded, and the KO of APOE was confirmed by either DNA sequencing or Western blot. Clones without any mutation were used as a control.
APOE plasmid construction
APOE-coding plasmid was obtained from Addgene and confirmed by sequencing. ApoE sequence was amplified by APOE gene–specific primers (listed in table S2) from the plasmid and cloned into FUIPW lentiviral vector. The cloned fragment was fully sequenced and confirmed (RefSeq NM_001302691).
Flow cytometry and immunofluorescence microscopy
For flow cytometry analysis, virus-infected cells were collected, fixed, and permed with 2% paraformaldehyde (PFA)/0.1% saponin for 10 min. Then, the cells were stained with the indicated primary antibody diluted in 0.1% saponin/PBS for 40 min at 4°C. The cells were washed with PBS and then stained with secondary antibody [Alexa Fluor 488/647 goat anti-rabbit/mouse immunoglobulin G (IgG) diluted in 1% BSA-PBS] at 4°C for 40 min. After washed with PBS, the cells were analyzed on FACSCanto II (BD Biosciences).
For immunofluorescence microscopy analysis, cells were grown on glass coverslips and infected with the virus. The cells were then fixed in 3.7% formaldehyde-PBS for 15 min and permeabilized with 0.2% Triton X-100–PBS containing 4′,6-diamidino-2-phenylindole (1 mg/ml) for 10 min. The cells were then blocked by 1% BSA-PBS for 30 min and incubated with primary antibody for 2 hours at room temperature. The cells were washed three times with PBS and stained with secondary antibody (Alexa Fluor 488/647 goat anti-rabbit/mouse IgG diluted in 1% BSA-PBS) at room temperature for 1 hour and again washed three times with PBS. The coverslips were mounted with mounting medium and then imaged by confocal microscopy (LSM700). Image-Pro was used to count NP- or HA-positive cells, and more than three fields of view (>200 cells) were counted. The percentage of positive cells in all counted fields was calculated.
RNA isolation and qRT-PCR
For qRT-PCR, cells were lysed with TRIzol reagent (Invitrogen), and total RNA was extracted according to the manufacturer’s instructions from cells, supernatant, or lung tissues of mice. The cDNA was generated using a high-capacity cDNA reverse transcription kit (Applied Biosystems) with random primers for host genes, IAV v-RNA (from M fragment) or IAV NP or M1 mRNA (primer sequences are listed in table S2). qRT-PCR was performed with technical duplicates using gene-specific primers and SYBR Green PCR Master Mix (Applied Biosystems). Delta cycle thresholds were calculated using hypoxanthine guanine phosphoribosyltransferase (HPRT) as the endogenous housekeeping gene control.
Pseudovirus entry assay
The pseudovirus assay was performed as below. Briefly, pseudovirus were produced by cotransfection of HEK293 cells with 6 μg each of pCAGGS-NA, pEWSN-HA, and HIV-pNL-Luc. At 8 hours after transfection, cell medium was replaced with DMEM supplemented with 10% FBS. After another 48 hours, cells and supernatants were all collected and frozen at −80°C for 1 cycle of freeze and thaw. The virus was aliquoted and stored at −80°C after filtered with a 0.45-μm filter. For pseudovirus infection, cells were infected with virus supernatant for 1 hour and then replaced with DMEM with 10% FBS and incubated for another 48 hours. The cells were then washed with PBS twice and lysed with passive lysis buffer (PLB) (Promega, E1941) for 20 min. The supernatant was used to detect luciferase activity.
Viral attachment assay
For virus attachment, A549 and Huh7 cells were infected with the PR8 for 1 hour at 4°C. Then, the cells were washed three times with cold PBS to remove the unbound virus. The virus attachment was determined by qRT-PCR (59).
Acid bypass assays
Entry assays were performed by incubating WT and APOE KO A549 cells with PR8 at an MOI of 0.1 in virus growth medium at 4°C for 1 hour followed by washing with cold PBS to remove unbound virus. Acid bypass assays were modified from previous approaches (27, 60). Following viral binding, cells were incubated in low-pH fusion buffer [50 mM citrate (pH 5.0) and 154 mM NaCl] or mock-treated with pH 7 buffer [20 mM Hepes (pH 7.4) and 154 mM NaCl] for 2 min at 37°C and washed with cold PBS. Last, prewarmed virus growth medium supplemented with 1 mM cycloheximide was added, and the cells were incubated at 37°C for 2.5 hours. Viral fusion was quantified using RT-PCR.
Minireplicon assay by immunoblot and luciferase reporter assay
Minireplicon assay was used to measure the polymerase activity. HEK293 WT and APOE KO cells were cotransfected with pCDNA3.1 plasmids encoding PB1, PB2, PA, and NP, as well as a POLI-driven RNA expression plasmid encoding the HA v-RNA segment. HA expression was detected by immunoblot 24 hours after transfection.
In addition, a dual-luciferase reporter assay system (Promega) was also used to compare the activities of viral RNP complexes. Briefly, the luciferase reporter plasmid pPolNP-Luc and the internal control plasmid Renilla were transfected into WT and APOE KO HEK293 cells together with the four protein expression plasmids pcDNA3.1-PB2, pcDNA3.1-PB1, pcDNA3.1-PA, and pcDNA3.1-NP. Cell lysates were analyzed 36 hours after transfection to measure firefly and Renilla luciferase activities.
Histological staining
One longitudinal section was obtained from each pair of lungs, fixed in 4% PFA, wax-embedded, sectioned, and stained with hematoxylin and eosin. Bleeding was scored semiquantitatively: 0, absent; 1, minimal; 2, mild; 3, moderate; and 4, marked.
Immunoblot
For immunoblot, cells were washed in PBS and subjected to lysis buffer [20 mM tris-HCl (pH 7.4), containing 150 mM NaCl, 0.5% (v/v) NP-40 10% (w/v) glycerol, 50 mM NaF, 1 mM Na3VO4, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and complete protease-inhibitor “cocktail” (Roche)] on ice for 45 min. The lysate was obtained after centrifugation, and the protein concentration was determined by the Bradford method. The cell lysate was subjected to SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes, blocked by PBS containing 5% nonfat dry milk and 0.1% Tween 20 for 1 hour at room temperature. The blocked membranes were then probed by incubation with antibodies against APOE (Cell Signaling Technology, 13366s), NP (Medix Biochemica, no. 100083), HA (GeneTex, GTX127357), and Actin (Santa Cruz Biotechnology, sc-47778) at 4°C overnight. After incubation with horseradish peroxidase (HRP)–conjugated secondary antibody for 1 hour at room temperature, the proteins were visualized by enhanced chemiluminesence (Thermo Fisher Scientific). Blot bands were quantified by ImageJ and represented the intensity normalized to actin or tubulin.
Cholesterol detection and filipin staining
The total cellular cholesterol was quantified using an Amplex Red cholesterol assay kit (Invitrogen A12216). Briefly, 5 million A549 cells or 2 million Huh7 cells were collected and washed three times with PBS, then the cell was diluted in 1× reaction buffer, and a volume of 50 μl was used for each reaction. Samples and controls were pipetted into separate wells of a microplate followed by adding 50 μl of Amplex Red reagent/HRP/cholesterol oxidase/cholesterol esterase working solution. After 30 min of incubation at 37°C protected from light, the samples were measured in a fluorescence microplate reader using an excitation of 540 nm.
Filipin staining was performed by using a cholesterol detection kit (Abcam, 133116). Briefly, the A549 cells were fixed with 3% PFA for 1 hour at room temperature and rinsed three times with PBS. A total of 1.5 mg of glycine/ml of PBS was then used to quench the PFA for 10 min. The cells were then stained with filipin at working solution (0.5 mg/ml) for 2 hours and then imaged by confocal microscopy (LSM700).
SA detection
A549 cells and MEF cells were used for SA staining and analyzed by immunofluorescence microscopy. The cells on the coverslips were first fixed with 4% formaldehyde for 10 min and blocked with blocking buffer. The cells were then stained with biotinylated Maackia amurensis lectin II (10 μg/ml) (which preferentially recognizes α-2,3–linked SA; Vector Laboratories) or with biotinylated Sambucus nigra lectin (5 μg/ml) (which preferentially recognizes α-2,6–linked SA; Vector Laboratories) for 2 hours, and then, the cells were washed three times with PBS and stained with 0.2 μl (in 20 μl of staining buffer) of allophycocyanin-conjugated streptavidin (BD Biosciences) for an additional 1 hour. The cells were washed three times with PBS. The coverslips were mounted with mounting medium and imaged by confocal microscopy (LSM700). The mean fluorescence of α-2,3–linked SA or α-2,6–linked SA was calculated by ImageJ.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5. Statistical significance was determined by two-tailed unpaired Student’s t test or one-way/two-way analysis of variance (ANOVA), and P values < 0.05 are considered significant and denoted as *P < 0.05, **P < 0.01, and ***P < 0.001. Nonsignificant values are denoted as ns.
Acknowledgments
We thank X. Zhang and Y. Xu for expert technical support.
Funding: This study was supported by the National Key Research and Development Project grant (2021YFC2300502 and 2016YFC1200302), the National Key Plan for Scientific Research and Development of China (2016YFD0500304), the National Science and Technology Major Project (2018ZX10101004), and the National Science and Technology Major Project (2017ZX10304402).
Author contributions: F.Z. conceived the original hypothesis and designed experiments. P.G., M.J., X.L., X.C., H.L. S.L., and C.L. performed experiments. P.G., M.J., X.L., X.C., and B.J. analyzed data. L.R., X.Z., Q.W., Y.B., and W.T. provided expertise for this research. F.Z. wrote, critically revised, and finalized the manuscript. X.T., B.H., X.Z., and T.D. helped critically revise the manuscript. J.W., G.F.G., and F.Z. supervised the study. All authors reviewed the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S4
REFERENCES AND NOTES
- 1.Mehrbod P., Omar A. R., Hair-Bejo M., Haghani A., Ideris A., Mechanisms of action and efficacy of statins against influenza. Biomed. Res. Int. 2014, 872370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maines T. R., Szretter K. J., Perrone L., Belser J. A., Bright R. A., Zeng H., Tumpey T. M., Katz J. M., Pathogenesis of emerging avian influenza viruses in mammals and the host innate immune response. Immunol. Rev. 225, 68–84 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Schneider W. M., Chevillotte M. D., Rice C. M., Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takaoka A., Yanai H., Interferon signalling network in innate defence. Cell. Microbiol. 8, 907–922 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Louie J. K., Acosta M., Samuel M. C., Schechter R., Vugia D. J., Harriman K., Matyas B. T.; California Pandemic (H1N1) Working Group , A novel risk factor for a novel virus: Obesity and 2009 pandemic influenza A (H1N1). Clin. Infect. Dis. 52, 301–312 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Neidich S. D., Green W. D., Rebeles J., Karlsson E. A., Schultz-Cherry S., Noah T. L., Chakladar S., Hudgens M. G., Weir S. S., Beck M. A., Increased risk of influenza among vaccinated adults who are obese. Int. J. Obes. (Lond) 41, 1324–1330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith A. G., Sheridan P. A., Harp J. B., Beck M. A., Diet-induced obese mice have increased mortality and altered immune responses when infected with influenza virus. J. Nutr. 137, 1236–1243 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Vekic J., Zeljkovic A., Stefanovic A., Jelic-Ivanovic Z., Spasojevic-Kalimanovska V., Obesity and dyslipidemia. Metabolism 92, 71–81 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Siegers J. Y., Novakovic B., Hulme K. D., Marshall R. J., Bloxham C. J., Thomas W. G., Reichelt M. E., Leijten L., van Run P., Knox K., Sokolowski K. A., Tse B. W. C., Chew K. Y., Christ A. N., Howe G., Bruxner T. J. C., Karolyi M., Pawelka E., Koch R. M., Bellmann-Weiler R., Burkert F., Weiss G., Samanta R. J., Openshaw P. J. M., Bielefeldt-Ohmann H., van Riel D., Short K. R., A high-fat diet increases influenza A virus-associated cardiovascular damage. J Infect Dis 222, 820–831 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Ghiselli G., Schaefer E. J., Gascon P., Breser H. B. Jr., Type III hyperlipoproteinemia associated with apolipoprotein E deficiency. Science 214, 1239–1241 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Plump A. S., Smith J. D., Hayek T., Aalto-Setälä K., Walsh A., Verstuyft J. G., Rubin E. M., Breslow J. L., Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell 71, 343–353 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Cun W., Jiang J., Luo G., The C-terminal alpha-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J. Virol. 84, 11532–11541 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rösch K., Kwiatkowski M., Hofmann S., Schöbel A., Grüttner C., Wurlitzer M., Schlüter H., Herker E., Quantitative lipid droplet proteome analysis identifies annexin A3 as a cofactor for HCV particle production. Cell Rep. 16, 3219–3231 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Wrensch F., Crouchet E., Ligat G., Zeisel M. B., Keck Z. Y., Foung S. K. H., Schuster C., Baumert T. F., Hepatitis C virus (HCV)-apolipoprotein interactions and immune evasion and their impact on HCV vaccine design. Front. Immunol. 9, 1436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brass A. L., Huang I.-C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J., The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlas A., Machuy N., Shin Y., Pleissner K.-P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., Mäurer A. P., Müller E., Wolff T., Rudel T., Meyer T. F., Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Konig R., Stertz S., Zhou Y., Inoue A., -Heinrich Hoffmann H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Gao Q., Andrews S. E., Bandyopadhyay S., De Jesus P., Tu B. P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., García-Sastre A., Young J. A. T., Palese P., Shaw M. L., Chanda S. K., Human host factors required for influenza virus replication. Nature 463, 813–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han J., Perez J. T., Chen C., Li Y., Benitez A., Kandasamy M., Lee Y., Andrade J., tenOever B., Manicassamy B., Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 23, 596–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heaton B. E., Kennedy E. M., Dumm R. E., Harding A. T., Sacco M. T., Sachs D., Heaton N. S., A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep. 20, 1503–1512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Clohisey S. M., Chia B. S., Wang B., Cui A., Eisenhaure T., Schweitzer L. D., Hoover P., Parkinson N. J., Nachshon A., Smith N., Regan T., Farr D., Gutmann M. U., Bukhari S. I., Law A., Sangesland M., Gat-Viks I., Digard P., Vasudevan S., Lingwood D., Dockrell D. H., Doench J. G., Baillie J. K., Hacohen N., Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat. Commun. 11, 164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran V., Ledwith M. P., Thamamongood T., Higgins C. A., Tripathi S., Chang M. W., Benner C., García-Sastre A., Schwemmle M., Boon A. C. M., Diamond M. S., Mehle A., Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat. Microbiol. 5, 1490–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Y., Huang H., Hu Y., Zhang J., Li F., Yin X., Shi J., Li Y., Li C., Zhao D., Chen H., A genome-wide CRISPR/Cas9 gene knockout screen identifies immunoglobulin superfamily DCC subclass member 4 as a key host factor that promotes influenza virus endocytosis. PLOS Pathog. 17, e1010141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchinson E. C., Denham E. M., Thomas B., Trudgian D. C., Hester S. S., Ridlova G., York A., Turrell L., Fodor E., Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLOS Pathog. 8, e1002993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez J. T., García-Sastre A., Manicassamy B., Insertion of a GFP reporter gene in influenza virus. Curr. Protoc. Microbiol. Chapter 15, Unit 15G.14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhelst J., Parthoens E., Schepens B., Fiers W., Saelens X., Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 86, 13445–13455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondal A., Dawson A. R., Potts G. K., Freiberger E. C., Baker S. F., Moser L. A., Bernard K. A., Coon J. J., Mehle A., Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery. eLife 6, e26910 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matlin K. S., Reggio H., Helenius A., Simons K., Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 91, 601–613 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fodor E., Crow M., Mingay L. J., Deng T., Sharps J., Fechter P., Brownlee G. G., A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76, 8989–9001 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L., Peng Y., Zhou K., Cao M., Wang J., Wang X., Jiang T., Deng T., New insights into the nonconserved noncoding region of the subtype-determinant hemagglutinin and neuraminidase segments of influenza A viruses. J. Virol. 88, 11493–11503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fodor E., Devenish L., Engelhardt O. G., Palese P., Brownlee G. G., García-Sastre A., Rescue of influenza A virus from recombinant DNA. J. Virol. 73, 9679–9682 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghanem A., Mayer D., Chase G., Tegge W., Frank R., Kochs G., García-Sastre A., Schwemmle M., Peptide-mediated interference with influenza A virus polymerase. J. Virol. 81, 7801–7804 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C., Ba Q., Wu A., Zhang H., Deng T., Jiang T., A peptide derived from the C-terminus of PB1 inhibits influenza virus replication by interfering with viral polymerase assembly. FEBS J. 280, 1139–1149 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Vonk A. G., Bont N. D., Netea M. G., Demacker P. N. M., van der Meer J. W. M., Stalenhoef A. F. H., Kullberg B. J., Apolipoprotein-E-deficient mice exhibit an increased susceptibility to disseminated candidiasis. Med. Mycol. 42, 341–348 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Daniloski Z., Jordan T. X., Wessels H.-H., Hoagland D. A., Kasela S., Legut M., Maniatis S., Mimitou E. P., Lu L., Geller E., Danziger O., Rosenberg B. R., Phatnani H., Smibert P., Lappalainen T., tenOever B. R., Sanjana N. E., Identification of required host factors for SARS-CoV-2 infection in human cells. Cell 184, 92–105.e1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider W. M., Luna J. M., -Heinrich Hoffmann H., Sánchez-Rivera F. J., Leal A. A., Ashbrook A. W., Pen J. L., Ricardo-Lax I., Michailidis E., Peace A., Stenzel A. F., Lowe S. W., MacDonald M. R., Rice C. M., Poirier J. T., Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184, 120–132.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R., Simoneau C. R., Kulsuptrakul J., Bouhaddou M., Travisano K. A., Hayashi J. M., Carlson-Stevermer J., Zengel J. R., Richards C. M., Fozouni P., Oki J., Rodriguez L., Joehnk B., Walcott K., Holden K., Sil A., Carette J. E., Krogan N. J., Ott M., Puschnik A. S., Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell 184, 106–119.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrow J. A., Segall M. L., Lund-Katz S., Phillips M. C., Knapp M., Rupp B., Weisgraber K. H., Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 39, 11657–11666 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Huang Y., Liu X. Q., Rall S. C. Jr., Taylor J. M., von Eckardstein A., Assmann G., Mahley R. W., Overexpression and accumulation of apolipoprotein E as a cause of hypertriglyceridemia. J. Biol. Chem. 273, 26388–26393 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Zhang S. H., Reddick R. L., Piedrahita J. A., Maeda N., Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science 258, 468–471 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Reddick R. L., Zhang S. H., Maeda N., Atherosclerosis in mice lacking apo E. Evaluation of lesional development and progression. Arterioscler. Thromb. 14, 141–147 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Chang K. S., Jiang J., Cai Z., Luo G., Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81, 13783–13793 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgos J. S., Ramirez C., Sastre I., Valdivieso F., Effect of apolipoprotein E on the cerebral load of latent herpes simplex virus type 1 DNA. J. Virol. 80, 5383–5387 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddiqui R., Suzu S., Ueno M., Nasser H., Koba R., Bhuyan F., Noyori O., Hamidi S., Sheng G., Yasuda-Inoue M., Hishiki T., Sukegawa S., Miyagi E., Strebel K., Matsushita S., Shimotohno K., Ariumi Y., Apolipoprotein E is an HIV-1-inducible inhibitor of viral production and infectivity in macrophages. PLOS Pathog. 14, e1007372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonacina F., Coe D., Wang G., Longhi M. P., Baragetti A., Moregola A., Garlaschelli K., Uboldi P., Pellegatta F., Grigore L., da Dalt L., Annoni A., Gregori S., Xiao Q., Caruso D., Mitro N., Catapano A. L., Marelli-Berg F. M., Norata G. D., Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat. Commun. 9, 3083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S. T., Kreutzberger A. J. B., Lee J., Kiessling V., Tamm L.-K., The role of cholesterol in membrane fusion. Chem. Phys. Lipids 199, 136–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goronzy I. N., Rawle R. J., Boxer S. G., Kasson P. M., Cholesterol enhances influenza binding avidity by controlling nanoscale receptor clustering. Chem. Sci. 9, 2340–2347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu K. N., Boxer S. G., Target membrane cholesterol modulates single influenza virus membrane fusion efficiency but not rate. Biophys. J. 118, 2426–2433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naghavi M., Wyde P., Litovsky S., Madjid M., Akhtar A., Naguib S., Siadaty M. S., Sanati S., Casscells W., Influenza infection exerts prominent inflammatory and thrombotic effects on the atherosclerotic plaques of apolipoprotein E-deficient mice. Circulation 107, 762–768 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Albashir A. A. D., The potential impacts of obesity on COVID-19. Clin. Med. (Lond.) 20, e109–e113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesonen E., Siitonen O., Acute myocardial infarction precipitated by infectious disease. Am. Heart J. 101, 512–513 (1981). [DOI] [PubMed] [Google Scholar]
- 51.Spodick D. H., Flessas A. P., Johnson M. M., Association of acute respiratory symptoms with onset of acute myocardial infarction: Prospective investigation of 150 consecutive patients and matched control patients. Am. J. Cardiol. 53, 481–482 (1984). [DOI] [PubMed] [Google Scholar]
- 52.Van Lenten B. J., Wagner A. C., Anantharamaiah G. M., Garber D. W., Fishbein M. C., Adhikary L., Nayak D. P., Hama S., Navab M., Fogelman A. M., Influenza infection promotes macrophage traffic into arteries of mice that is prevented by D-4F, an apolipoprotein A-I mimetic peptide. Circulation 106, 1127–1132 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Pleskov V. M., Bannikov A. I., Gurevich V. S., Pleskova I. V., Influenza viruses and atherosclerosis: The role of atherosclerotic plaques in prolonging the persistent form of influenza infection. Vestn. Ross. Akad. Med. Nauk , 10–13 (2003). [PubMed] [Google Scholar]
- 54.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F., Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Monaghan P., Simpson J., Murphy C., Durand S., Quan M., Alexandersen S., Use of confocal immunofluorescence microscopy to localize viral nonstructural proteins and potential sites of replication in pigs experimentally infected with foot-and-mouth disease virus. J. Virol. 79, 6410–6418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G., Zhang F., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang D., Peng W.-J., Ye Q., Liu X.-P., Zhao L., Fan L., Xia-Hou K., Jia H.-J., Luo J., Zhou L.-T., Li B.-B., Wang S.-L., Xu W.-T., Chen Z., Tan W.-S., Serum-free suspension culture of MDCK cells for production of influenza H1N1 vaccines. PLOS ONE 10, e0141686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmerman M. G., Quicke K. M., O’Neal J. T., Arora N., Machiah D., Priyamvada L., Kauffman R. C., Register E., Adekunle O., Swieboda D., Johnson E. L., Cordes S., Haddad L., Chakraborty R., Coyne C. B., Wrammert J., Suthar M. S., Cross-reactive dengue virus antibodies augment zika virus infection of human placental macrophages. Cell Host Microbe 24, 731–742.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banerjee I., Miyake Y., Nobs S. P., Schneider C., Horvath P., Kopf M., Matthias P., Helenius A., Yamauchi Y., Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346, 473–477 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S4