Abstract
Idiopathic pulmonary fibrosis (IPF) is a progressive interstitial lung disease, and the molecular mechanisms remain poorly understood. Our findings demonstrated that pyruvate kinase M2 (PKM2) promoted fibrosis progression by directly interacting with Smad7 and reinforcing transforming growth factor–β1 (TGF-β1) signaling. Total PKM2 expression and the portion of the tetrameric form elevated in lungs and fibroblasts were derived from mice with bleomycin (BLM)–induced pulmonary fibrosis. Pkm2 deletion markedly alleviated BLM-induced fibrosis progression, myofibroblast differentiation, and TGF-β1 signaling activation. Further study showed that PKM2 tetramer enhanced TGF-β1 signaling by directly binding with Smad7 on its MH2 domain, and thus interfered with the interaction between Smad7 and TGF-β type I receptor (TβR1), decreased TβR1 ubiquitination, and stabilized TβR1. Pharmacologically enhanced PKM2 tetramer by TEPP-46 promoted BLM-induced pulmonary fibrosis, while tetramer disruption by compound 3k alleviated fibrosis progression. Our results demonstrate how PKM2 regulates TGF-β1 signaling and is a key factor in fibrosis progression.
PKM2 tetramer interacts with Smad7 and reinforces TGF-β1 signaling and pulmonary fibrosis.
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and fatal interstitial lung disease, with an average life expectancy of 3 to 5 years after diagnosis (1). The etiology of IPF remains unclear. The current understanding of IPF pathogenesis involves the failure of alveolar epithelial repair, followed by excessive secretion of multiple cytokines and growth factors that promote myofibroblast differentiation (2, 3). These myofibroblasts, in turn, secrete exaggerated amounts of extracellular matrix (ECM) that subsequently remodel the lung architecture (4) and cause an irreversible loss of function (5).
Although the molecular mechanisms underlying IPF are not fully understood, considerable evidence indicates that transforming growth factor–β (TGF-β1) signaling plays a central role in fibrosis progression (6). Highly active TGF-β1 binds to a TGF-β type II receptor (TβR2) and induces its assembly with TβR1. TβR1, in turn, initiates an intracellular signaling by phosphorylating the receptor regulated Smads (R-Smads), Smad2 and Smad3, and then the phosphorylated R-Smad translocates into the nucleus and initiates gene expression (7–9). Thus, phosphorylation of R-Smad is a direct evidence for TGF-β1 signaling activation. Meanwhile, other members in the Smad family cooperate with Smad3 to regulate TGF-β1 signaling. Down-regulation of TGF-β1 signaling via Smad7-mediated ubiquitination has been extensively characterized. Smad7 binds to the activated TβR1 and serves as a scaffold to recruit the E3 ubiquitin ligase Smurf2 (SMAD specific E3 ubiquitin protein ligase 2) to target TβR1 for ubiquitination (10). This ubiquitination leads to a proteasome-mediated degradation of the TGF-β1 receptor complex and signaling attenuation (11). A Smad7-mediated ubiquitination is finely regulated and can be counteracted; for example, deubiquitylating enzymes (DUBs) such as ubiquitin specific peptidase 15 (USP15) and USP4 can facilitate TGF-β1 signaling by competing Smad7 and stabilizing TβR1 (12, 13). The key role of Smad7 in the negative feedback loop of TGF-β signaling makes it strongly involved in fibrogenesis. Mice that overexpressed Smad7 by intratracheal injection of adenovirus showed significantly less pulmonary fibrosis after bleomycin (BLM) treatment (14). Factors that enhance the function of Smad7 such as chitinase 1 (CHIT1) could promote pulmonary fibrosis progression (15). Thus, facilitating Smad7 function might be beneficial to fibrosis treatment.
Pyruvate kinase M2 (PKM2) catalyzes the final step in glycolysis. PKM2 is highly expressed by the embryonic cells and is subjected to complex allosteric regulation to execute different biological functions. The PKM2 tetramer is a highly active pyruvate kinase that regulates glycolysis (16) and oxidative phosphorylation (17), whereas the PKM2 dimer is a protein kinase that initiates gene transcription by translocating to the nucleus (18, 19). This characteristic enables PKM2 to both metabolically and nonmetabolically promote cellular proliferation; thus, PKM2 is preferentially expressed in most types of cancer (20–22) and some of the fibrotic diseases such as nonalcoholic steatohepatitis (23) and kidney fibrosis (24). Current studies focus on the metabolic and transcriptional functions of PKM2; however, the role of PKM2 in regulating signal transduction is poorly understood. Studies have reported that PKM2 can regulate a canonical and noncanonical TGF-β1 signaling (25, 26), but the details of the molecular mechanism are unclear.
Here, we identified PKM2 as an enhancer of TGF-β1 signaling through direct binding with Smad7 and promoting fibrogenesis. PKM2 was highly expressed during pulmonary fibrosis progression. Notably, the proportion of the PKM2 tetramer was elevated, along with its total expression. The loss of PKM2 markedly alleviated BLM-induced pulmonary fibrosis and myofibroblast activation. Mechanistically, PKM2 tetramer binding with Smad7 disrupted the interaction of Smad7 with TβR1, thereby decreasing the ubiquitination and degradation of TβR1. Pharmacologically regulating the conformational change in PKM2 into monomers protected mice against BLM-induced pulmonary fibrosis and might be a potential strategy for IPF treatment.
RESULTS
Elevation in PKM2 and its tetramer in pulmonary fibrosis
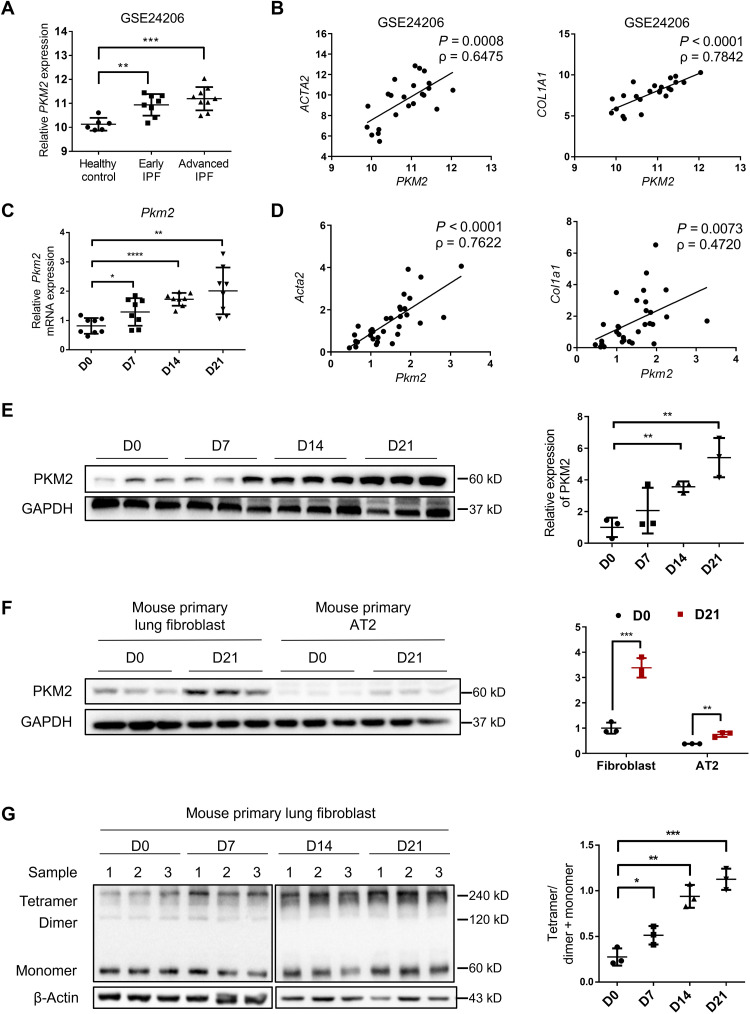
To determine the role of PKM2 in pulmonary fibrosis, we first analyzed PKM2 mRNA expression in a published Gene Expression Omnibus (GEO) dataset of IPF lungs (GSE24206) and found that PKM2 mRNA level was significantly increased with IPF progression (Fig. 1A). Independent IPF cohorts (GSE1999949 and GSE110147) from other sites were also analyzed to exclude heterogeneous patients with IPF, and the results showed that the PKM2 mRNA level was elevated in both datasets (fig. S1, A and B). Moreover, an analysis of this gene profile GSE24206 showed a statistically significant correlation between the mRNA expression of PKM2 and that of ACTA2 and COL1A1, two biomarkers of IPF (Fig. 1B). These findings indicate that PKM2 might play a role in fibrosis progression. To address this hypothesis, we further examined PKM2 expression in the lung tissues of mice with BLM-induced pulmonary fibrosis. BLM time dependently induced the Pkm2 mRNA expression (Fig. 1C), and the mRNA level was significantly correlated with that of Acta2 and Col1a1 (Fig. 1D). We also observed an overexpression of PKM2 at the protein level, and a sixfold up-regulation was observed in the fibrotic lungs following day 21 of BLM induction (Fig. 1E). To further confirm the cell type in which PKM2 was overexpressed, we isolated primary lung fibroblasts and alveolar type II (AT2) cells from mice treated with BLM, which are characterized by α-smooth muscle actin (α-SMA) and surfactant protein C (Prospc), respectively (fig. S2, A and B), and found that the increased expression of PKM2 was mainly in primary lung fibroblasts but not AT2 cells (Fig. 1F). It is known that a change in the ratio of PKM2 tetramers and dimers often triggers pathological changes (27). Thus, we assessed the status of PKM2 subunit association in primary lung fibroblasts by performing 3′,6-disinapoylsucrose (DSS) cross-linking. To our surprise, the level of the PKM2 tetramer was time dependently increased by a BLM challenge (Fig. 1G), which suggests that the PKM2 tetramer might be a key factor in fibrosis progression. We also stimulated human fetal lung fibroblast (HFL1 cells) with TGF-β1 and examined the levels of total and tetrameric PKM2 and found a similar result (fig. S3, A and B). These data collectively demonstrate that high expression of PKM2 in fibroblasts might have a significant role in fibrogenesis.
Fig. 1. Increase in PKM2 in patients with IPF and mouse model of BLM-induced pulmonary fibrosis.
(A) PKM2 expression in IPF lung tissues in a published GEO dataset of IPF lungs. Microarray analysis of PKM2 mRNA in lung samples of patients with early IPF (n = 8) and patients with advanced IPF (n = 9) compared with healthy controls (n = 6) (B) and the correlation between PKM2 mRNA expressions and those of ACTA2 and COL1A1 in lung tissues. The correlation coefficient (ρ) and the two-tailed significance are shown. (C) qRT-PCR analysis of Pkm2 mRNA expression in the lung tissues of mice following BLM induction (D) and the correlation between Pkm2 mRNA expressions and those of Acta2 and Col1a1 in the same lung tissues (n = 8 per group). Gapdh mRNA levels were used as an internal normalization control. (E) Representative results (n = 3 of Western blot with n = 10 mice per group) for Western blot analysis of PKM2 in the lung tissues of mice. (F) Representative results (n = 3 of Western blot with n = 8 mice per group) of PKM2 in primary lung fibroblasts and AT2 cells isolated from mice following BLM induction. (G) Representative results (n = 3 of Western blot with n = 8 mice per group) of PKM2 in the cross-linked primary lung fibroblasts. Data are represented as the means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Student’s t test.
PKM2 deficiency protects mice from BLM-induced pulmonary fibrosis
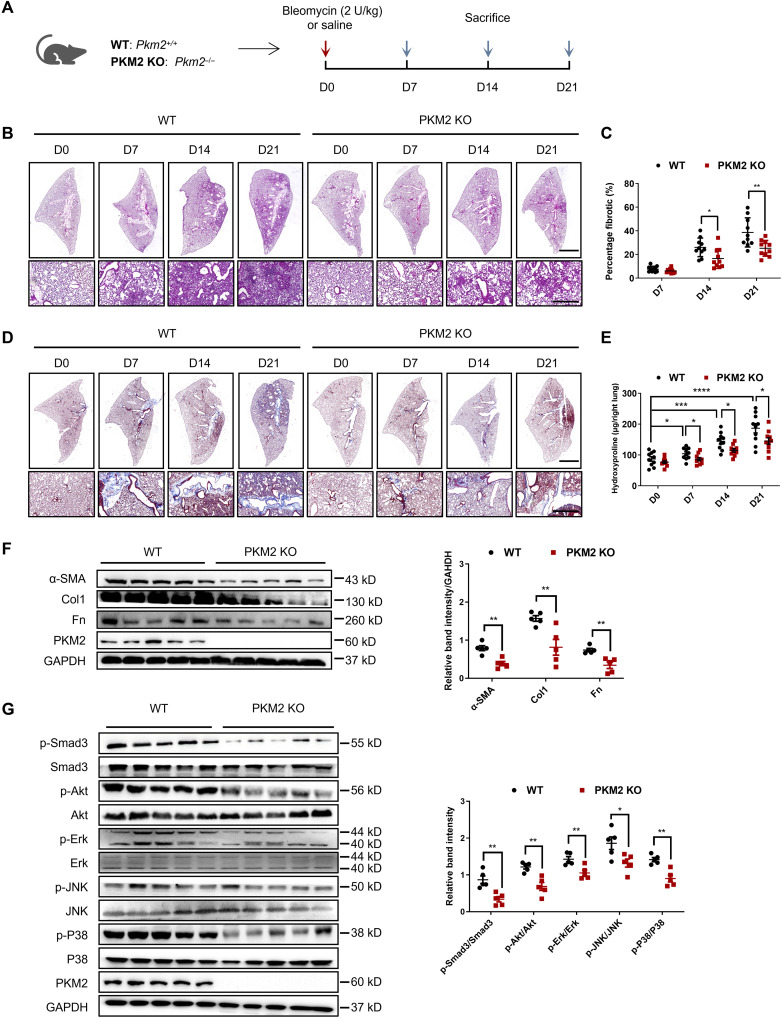
A previous study has generated a mouse model that allows conditional deletion of the PKM2 isoform-specific exon 10 while still allowing PKM1 splicing and protein expression (28). To investigate the role of PKM2 in the pathogenesis of pulmonary fibrosis, we bred mice with the previously described Pkm2 conditional allele (Pkm2flox/flox) with EIIa-Cre mice to generate a Pkm2–knockout (KO) mice (Pkm2−/−, PKM2-KO), and their littermates [Pkm2+/+, wild type (WT)] served as controls (Fig. 2A). The presence of the Pkm2fl and Cre alleles in adult mice was verified by Southern blot analysis of tail blood. PKM2-KO mice are healthy and have no overt phenotype, and pathological examination of main tissues also showed no obvious lesion (fig. S4). PKM2-KO and WT mice were challenged with BLM, and fibrosis severity was analyzed on the 7th, 14th, and 21st days. The degree of fibrosis in PKM2-KO mice was significantly reduced, as revealed by hematoxylin and eosin (H&E) staining (Fig. 2B), and this result was further confirmed by the reduced fibrotic scores in the PKM2-KO group (Fig. 2C). Collagen deposition was also markedly reduced in PKM2-KO mice, as seen by Masson’s staining (Fig. 2D) and the quantification of hydroxyproline levels (Fig. 2E). The protein expression of the profibrotic factors α-SMA, collagen 1 (Col1), and fibronectin (Fn) was also significantly decreased in the lungs of PKM2-KO mice (Fig. 2F). Given the critical role of TGF-β1 signaling in fibrosis progression, we further examined the activation of TGF-β1 signaling downstream of Smad3 and found that PKM2-KO mice had much lower levels of phosphorylated Smad3 (p-Smad3) than WT mice (Fig. 2G). The phosphorylation levels of the non-Smad factors Akt, extracellular signal–regulated kinase (Erk), c-Jun N-terminal kinase (JNK), and P38 were also decreased in the PKM2-KO group (Fig. 2H), which indicates that PKM2 KO might affect upstream TGF-β1 signaling. Together, these data suggest that PKM2 deletion protects mice against fibrogenesis, collagen deposition, and TGF-β1 signaling activation induced by BLM.
Fig. 2. PKM2 deletion attenuates BLM-induced pulmonary fibrosis.
(A) Schema illustrating generation of PKM2-KO mice. PKM2-KO and control mice were subjected to BLM treatment and euthanized at the indicated time points. (B) H&E staining of lung sections from control and PKM2-KO mice at various time points after BLM induction. Scale bars, 2 mm (top panel) and 100 μm (bottom panel). (C) Quantification of the severity of fibrosis. The fibrotic area is presented as percentage (n = 10 per group). (D) Masson’s staining of collagen on lung sections from control and PKM2-KO mice at various time points after BLM induction. Scale bars, 2 mm (top panel) and 100 μm (bottom panel). (E) Hydroxyproline content in lung tissues in control and PKM2-KO mice at various time points after BLM induction (n = 10 per group). (F to G) Representative results (n = 5 of Western blot with n = 10 mice per group) of α-SMA, Col1, and Fn (F); p-Smad3, Smad3, p-Akt, Akt, p-Erk, Erk, p-JNK, JNK, p-P38, and P38 (G) expression in lung tissues from control and PKM2-KO mice at various time points after BLM induction. Data are represented as the means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Student’s t test.
We also performed an in vivo RNA interference loss-of-function analysis to investigate whether Pkm2 knockdown could alleviate fibrosis progression during the fibrotic stage. A previous study has shown that selective organ targeting with lipid nanoparticles (LNPs) can deliver nucleic acids to a specific tissue (29). We developed a previously reported lung-specific LNP-based protocol and loaded these LNPs with a Pkm2–small interfering RNA (siRNA) that specifically targeted murine Pkm2 (30). To assess the distribution of LNPs, luciferase mRNA was loaded, and LNPs were intravenously injected into mice. An in vivo imaging system was used to measure the fluorescent signal. As expected, LNPs predominantly accumulated in the lung for at least 3 days (fig. S5A). Next, we confirmed the effect of Pkm2-siRNA–loaded LNPs on protein expression in lung tissue. We isolated lungs at 1, 2, 3, and 5 days after injection with Pkm2-siRNA–loaded LNPs and found that the expression of PKM2 was significantly decreased 3 days after LNP administration but recovered at day 5 (fig. S5B), which indicates that Pkm2-siRNA–loaded LNPs should be administered every 2 days to maintain their efficacy. Thus, we established a BLM-induced mouse pulmonary fibrosis model and injected Pkm2-siRNA–loaded LNPs (1.5 mg/kg) on days 14, 17, and 20 and euthanized the mice on day 21 (fig. S5C). LNPs loaded with scramble siRNA were used as controls. Pathological examination results showed that siRNA-mediated PKM2 knockdown markedly improved the BLM-induced lung architect destruction and collagen deposition (fig. S5D). The degree of fibrosis and hydroxyproline levels were analyzed and revealed similar improvements (fig. S5, E and F). Moreover, the expression of the profibrotic factors α-SMA, Col1, and Fn was decreased in the LNP group (fig. S5G). These data suggest that knocking down PKM2 during the fibrotic stage can protect against fibrosis progression.
PKM2 deficiency attenuates myofibroblast activation
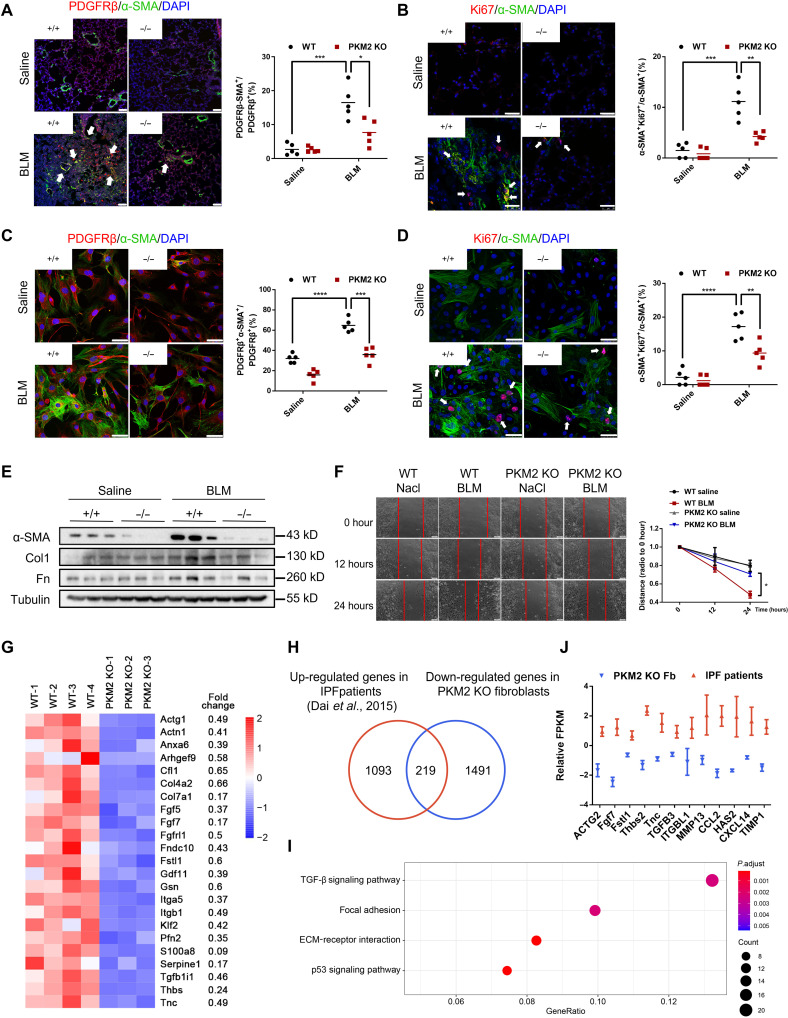
The accumulation of proliferating myofibroblasts contributes to fibrotic foci formation. Previous findings indicated that a β-type platelet-derived growth factor receptor (PDGFRβ) was highly expressed in myofibroblasts in fibrotic lungs (31). Thus, we costained lung sections with antibodies against PDGFRβ and α-SMA and found significantly fewer α-SMA–expressing myofibroblasts in PKM2-KO mice than in WT mice (Fig. 3A). We further costained lungs with antibodies against Ki67 and α-SMA to identify proliferating myofibroblasts and found few costained cells in PKM2-KO mice (Fig. 3B). We also harvested primary lung fibroblasts from PKM2-KO and WT mice and confirmed these results (Fig. 3, C and D), which indicated that fewer proliferating myofibroblasts were differentiated in PKM2-KO mice than in WT mice after BLM treatment. Consistently, Western blot analysis also verified reductions in the expression of the myofibroblast markers α-SMA, Col1, and Fn (Fig. 3E). A wound healing assay revealed a decline in the migration of fibroblasts from PKM2-KO mice (Fig. 3F). These results indicate that the loss of Pkm2 alleviates myofibroblast activation.
Fig. 3. Loss of PKM2 suppresses myofibroblasts activation.
(A and B) On day 21 after BLM induction, lungs were stained with antibodies against α-SMA and PDGFRβ (A) or antibodies against α-SMA and Ki67 (B) (n = 5 per group). White arrowheads indicate double positive cells. Scale bars, 50 μm (A) and 25 μm (B). (C and D) On day 21 after BLM induction, primary lung fibroblasts were isolated and stained with antibodies against α-SMA and PDGFRβ (C) or Ki67 (D) (n = 5 per group). White arrowheads indicate double positive cells. Scale bars, 5 μm. (E) Western blot analysis of α-SMA, Col1, and Fn expressions in primary lung fibroblasts isolated from control and PKM2-KO mice 0 and 21 days after BLM treatment. (F) On day 21 after BLM induction, primary lung fibroblasts were isolated and seeded in a plate. When cells were fully confluent, a scratch was made in the center of the culture well. Images were obtained at 0, 12, and 24 hours. Three parallel lines were drawn, and images were analyzed using Image-Pro Plus software. (G) Heatmap of significantly down-regulated (<1.5-fold change) genes in PKM2-KO fibroblasts compared with WT fibroblasts. (H) Among 1710 down-regulated genes in PKM2-KO fibroblasts, 219 genes are up-regulated in patients with IPF. (I) Also shown is a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these 219 commonly genes. (J) Representative down-regulated genes in PKM2-KO fibroblasts that were up-regulated in patients with IPF. Data are represented as the means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Student’s t test.
We next investigated the signaling pathways by which PKM2 regulates myofibroblast activation. We used RNA sequencing (RNA-seq) to compare the gene expression of primary lung fibroblasts in BLM-treated PKM2-KO and control mice. A total of 5253 significantly differential genes (>1.5-fold change) were identified. Among the 1710 down-regulated genes in the PKM2-KO group, we characterized a series of genes that were highly involved in fibrosis progression (Fig. 3G), which confirmed that the loss of Pkm2 protected mice against fibrogenesis. To further confirm the key profibrotic genes among these down-regulated genes, we compared the down-regulated genes in PKM2-KO fibroblasts with the up-regulated genes in IPF lung tissues using the RNA-seq data from patients with IPF (32). A total of 219 common genes were identified (Fig. 3H), and the enrichment analysis of these 219 genes showed that many of them controlled TGF-β1 signaling, focal adhesion, and ECM-receptor interactions (Fig. 3, I and J). This RNA-seq analysis indicates that the effect of PKM2 on fibrosis progression might be highly associated with TGF-β1 signaling.
PKM2 regulates TGF-β1 signaling by directly interacting with Smad7
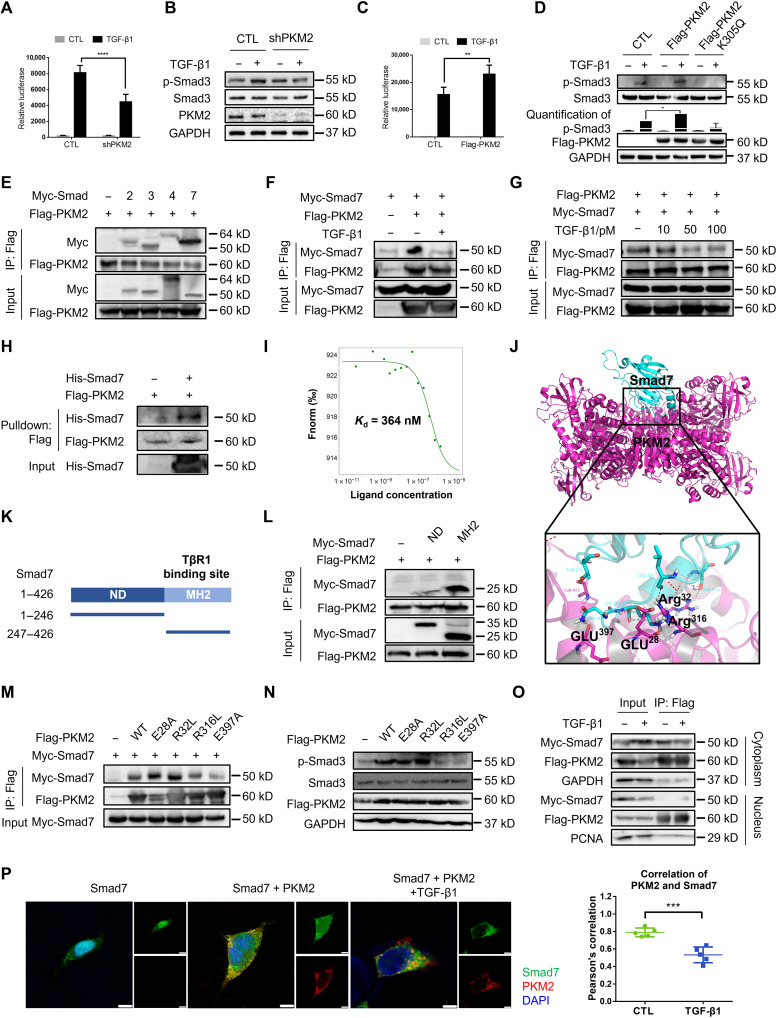
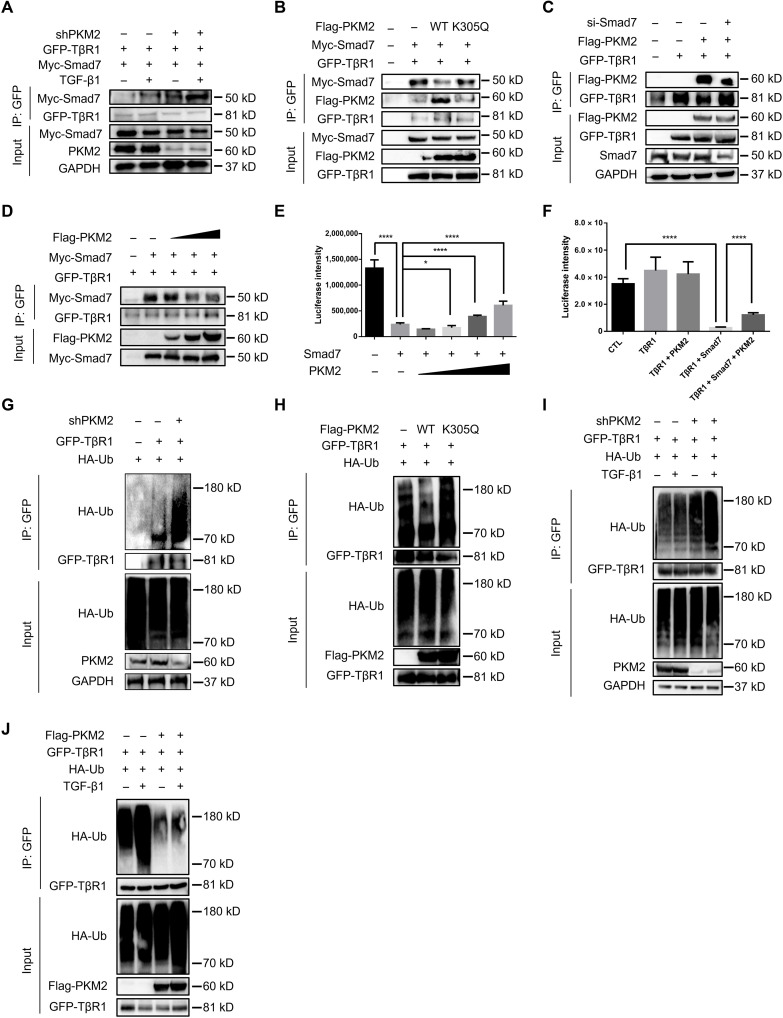
To investigate the role of PKM2 in TGF-β1 signal transduction, we constructed a short hairpin RNA (shRNA) vector specifically targeting human PKM2 (18) and efficiently decreased endogenous PKM2 expression in 293T cells (fig. S6). PKM2 knockdown repressed the activity of a TGF-β1–responsive luciferase reporter (CAGA-luc) to TGF-β1 (Fig. 4A) and decreased the amount of p-Smad3 (Fig. 4B). Moreover, overexpression of PKM2 enhanced CAGA-luc activity (Fig. 4C) and increased the level of p-Smad3 (Fig. 4D). These results suggest that PKM2 contributes to TGF-β1 signal transduction. Because we found that the proportion of the PKM2 tetramer was elevated during fibrosis progression and to further confirm the role of the PKM2 tetramer in TGF-β1 signaling, we established the PKM2 mutant K305Q that had been previously reported to maintain PKM2 mainly as a monomer (33). Unlike WT PKM2, K305Q failed to enhance TGF-β1–induced p-Smad3 (Fig. 4D), suggesting that PKM2 facilitates TGF-β1 signaling mainly via its tetramer.
Fig. 4. PKM2 enhances TGF-β1 signaling by directly interacting with Smad7.
(A) Luciferase assay in 293T cells transfected with the CAGA-luc reporter and shPKM2. (B) Immunoblot analysis in 293T cells transfected with the shPKM2 and treated with TGF-β1. (C) Luciferase assay in 293T cells transfected with the CAGA-luc reporter and Flag-PKM2. (D) Immunoblot analysis in 293T cells transfected with the Flag-PKM2 and Flag-PKM2-K305Q and treated with TGF-β1. (E) Immunoprecipitation with anti-Flag beads in 293T cells expressing Flag-PKM2 and Myc-tagged Smad2, Smad3, Smad4, and Smad7. (F and G) Immunoprecipitation with anti-Flag beads in 293T cells expressing Myc-Smad7 and Flag-PKM2. (H) Flag-tagged PKM2 and His-Smad7 were pulled down using anti-His beads. (I) The interaction of PKM2 with Smad7 was measured by MST. The Kd value was determined with MO.Affinity Analysis Software. (J) Molecular docking showed binding domain of human PKM2 and Smad7. (K) Domains of human Smad7. ND, N-terminal domain, amino acids 1 to 246; MH2, MH2 domain, amino acids 247 to 426. (L) Immunoprecipitation with anti-Flag beads in 293T cells transfected with Flag-PKM2 and different domains of Smad7. (M) Immunoprecipitation with anti-Flag beads in 293T cells transfected with Myc-Smad7 and Flag-PKM2 mutants. (N) Immunoblot analysis in 293T cells transfected with Flag-PKM2 mutants. (O) 293T cells were transfected with Flag-PKM2 and Myc-Smad7, and cytoplasmic and nuclear protein were separated and immunoprecipitated with anti-Flag beads. (P) 293T cells were transfected with Flag-PKM2 and Myc-Smad7 and stained with antibodies against Flag and Myc. Scale bars, 5 μm. Data are represented as the means ± SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001, Student’s t test.
Down-regulation of Smad3 activity strongly suggests that PKM2 might directly regulate TGF-β1 signaling activation. However, how PKM2 displays this effect remains unknown. Because PKM2 appears to mediate Smad3 phosphorylation, we reasoned that PKM2 might function through interacting with the main mediator of the TGF-β1 signaling, Smad family. Thus, to investigate the molecular mechanism underlying PKM2-mediated TGF-β1 signaling activation, we performed coimmunoprecipitation (co-IP) assays in 293T cells with Flag-tagged PKM2 (Flag-PKM2) and Myc-tagged Smads and found that PKM2 bound to Smad7 with higher affinity than to Smad2, Smad3, or Smad4 (Fig. 4E). In addition, the interaction between PKM2 and Smad7 was regulated by the intensity of TGF-β1 signaling, as the interaction of PKM2 with Smad7 can be decreased by TGF-β1 treatment (Fig. 4F), and PKM2 gradually disassociated from Smad7 in the presence of TGF-β1 in a dose-dependent manner (Fig. 4G).
To further assess whether the interaction of PKM2 and Smad7 was direct, we expressed purified Flag-PKM2 in 293T cells and recombinant His-tagged Smad7 (His-Smad7) in BL2(DE3) cells. Pull-down assays showed that PKM2 directly bound to Smad7 in vitro (Fig. 4H). A microscale thermophoresis (MST) assay revealed a binding affinity for PKM2 and Smad7 at a dissociation constant (Kd) value of 346 nM (Fig. 4I). To identify the protein domain of Smad7 that was involved in the interaction with PKM2, we performed molecular docking analysis and found that the MH2 domain (amino acids 247 to 426) might have a key node (Fig. 4J). This hypothesis was confirmed as PKM2 bound to MH2 but not the N terminus (amino acids 1 to 246) of Smad7 (Fig. 4, K and L). Molecular docking also revealed multiple sites of PKM2 (Glu28, Arg32, Arg316, and Glu397) that might be involved in the interaction with Smad7. Thus, we constructed four PKM2 mutants (E28A, R32L, R316L, and E397A) to identify the residue that plays as a key node in this process. Because these four PKM2 mutants remained in tetrameric form and had similar pyruvate kinase activity compared with WT PKM2 (fig. S7), we reasoned that mutants of these four residues did not induce obvious allosteric change in the PKM2 tetramer. Thus, we performed co-IP analysis and found that R316L and E397A could partially abolish PKM2 binding with Smad7 (Fig. 4M). Besides, R316L and E397A could also reduce the PKM2-mediated enhancement in TGF-β1 signaling (Fig. 4N), suggesting that Arg316 and Glu397 might be essential for the binding of PKM2 and Smad7. These results indicate that PKM2 directly interacts with Smad7.
We next investigated the subcellular location at which PKM2 binds to Smad7. We harvested the nuclear and cytoplasmic proteins and performed co-IP analysis and found that PKM2 bound to Smad7 only in the cytoplasm and disassociated in the presence of TGF-β1 (Fig. 4O). We also confirmed this result using an immunostaining assay. PKM2 colocalized with Smad7 in the cytoplasm and disassociated upon TGF-β1 treatment (Pearson’s correlation ~0.8 versus ~0.5; Fig. 4P). Because we did not observe nuclear accumulation of the PKM2-Smad7 complex and only the PKM2 dimer was capable of nuclear translocation, we reason that in intact cells, the PKM2 tetramer interacts with Smad7 in the cytoplasm through a physical association.
PKM2 regulates TβR1 ubiquitination and stabilizes TβR1
PKM2 binds to Smad7 via its MH2 domain, through which Smad7 binds to TβR1 (34). We hypothesized that PKM2 could interfere with Smad7 binding to TβR1. PKM2 knockdown in 293T cells increased Smad7 binding to TβR1 (Fig. 5A). In contrast, PKM2 overexpression decreased the interaction between Smad7 and TβR1, whereas the PKM2 monomer with the K305Q mutation showed no obvious change (Fig. 5B). Notably, PKM2 binding did not completely abolish Smad7 interactions with TβR1. Some PKM2 was still found in the same complex as Smad7 and TβR1 (Fig. 5B), but Smad7 knockdown stopped PKM2 from forming a complex with TβR1 (Fig. 5C), suggesting that PKM2 does not directly bind with TβR1. In addition, PKM2 can compete for Smad7 binding to TβR1, as demonstrated by co-IP: At low concentrations of PKM2, most Smad7 was bound with TβR1, whereas as the concentration of PKM2 gradually increased, less Smad7 was able to form a complex with TβR1 (Fig. 5D). Consistently, PKM2 overexpression attenuated Smad7-mediated blockade of CAGA-luc activity (Fig. 5, E and F). These results suggest that PKM2 interferes with Smad7 binding with TβR1 and counteracts Smad7 activity.
Fig. 5. PKM2 counteracts Smad7 binding with TβR1.
(A) Immunoprecipitation with anti-GFP antibody in 293T cells transfected with GFP-TβR1, Myc-Smad7, and shPKM2. (B) Immunoprecipitation with anti-GFP antibody in 293T cells transfected with GFP-TβR1, Myc-Smad7, and Flag-PKM2 (WT or K305Q). (C) 293T cells were transfected with SMAD7 siRNA, then transfected with GFP-TβR1 and Flag-PKM2, and immunoprecipitated with anti-GFP antibody. (D) Immunoprecipitation with anti-GFP antibody in 293T cells transfected with GFP-TβR1, Myc-Smad7, and increasing concentrations of Flag-PKM2. (E) Luciferase assay in 293T cells transfected with the CAGA-luc reporter, Myc-Smad7, and increasing concentrations of Flag-PKM2. (F) Luciferase assay in 293T cells transfected with the CAGA-luc reporter, HA-TβR1, Myc-Smad7, and Flag-PKM2. (G) Immunoprecipitation with anti-GFP antibody in 293T cells transfected with GFP-TβR1, HA-ubiquitin, and shPKM2. Cells were treated with MG132 for 4 hours before harvest. (H) Immunoprecipitation with anti-GFP antibody in 293Tcells transfected with GFP-TβR1, HA-ubiquitin, and Flag-PKM2 (WT or K305Q). Cells were treated with MG132 for 4 hours before harvest. (I) Immunoprecipitation with anti-GFP antibody in 293T cells transfected with GFP-TβR1, HA-ubiquitin, and shPKM2. Cells were treated with MG132 for 4 hours before harvest. (J) Immunoprecipitation with anti-GFP antibody in 293T cells transfected with GFP-TβR1, HA-ubiquitin, and Flag-PKM2. Cells were treated with MG132 for 4 hours before harvest. Data are represented as the means ± SEM. *P < 0.05; ****P < 0.00001, Student’s t test.
Because Smad7 regulates TβR1 ubiquitination, we reasoned that PKM2 could affect the ubiquitination of TβR1. PKM2 knockdown in 293T cells increased TβR1 ubiquitination (Fig. 5G). In contrast, PKM2 overexpression reduced TβR1 ubiquitination, whereas K305Q showed no obvious changes (Fig. 5H). A previous study showed that TβR1 ubiquitination was promoted by TGF-β1 signaling (10). We found that PKM2 could regulate TβR1 ubiquitination even in the presence of TGF-β1 (Fig. 5, I and J), suggesting that PKM2 could protect TβR1 against TGF-β1–induced ubiquitination. Notably, PKM2 knockdown did not alter the ubiquitination of Smad7 (fig. S8).
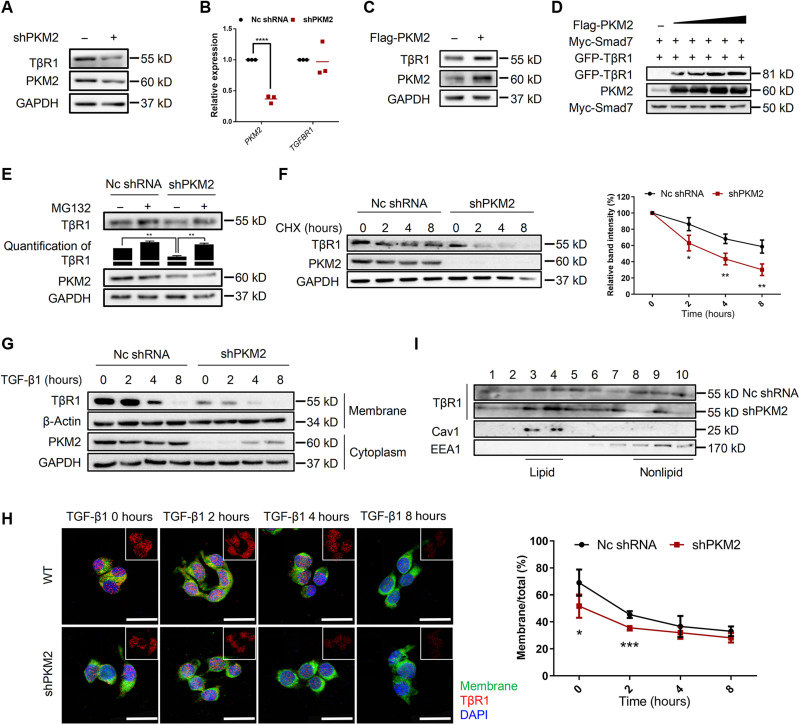
We next investigate the stability of TβR1. PKM2 knockdown in 293T cells reduced the endogenous protein level of TβR1 without affecting its mRNA expression (Fig. 6, A and B). Similar results were observed in mouse primary lung fibroblasts (fig. S9, A and B). Conversely, PKM2 overexpression increased both the endogenous protein levels of TβR1 without affecting its mRNA expression (Fig. 6C and fig. S9C). Meanwhile, PKM2 could counteract the effect of Smad7 on the TβR1, as demonstrated by increasing amounts of PKM2 that protected TβR1 from Smad7-mediated degradation in a dose-dependent manner (Fig. 6D). Moreover, the effect of PKM2 on TβR1 was mediated by proteasome, as proteasome inhibitor MG132 inhibited the turnover of TβR1 by PKM2 knockdown (Fig. 6E), and PKM2 knockdown decreased the half-life of TβR1 from 6 to <3 hours (Fig. 6F).
Fig. 6. PKM2 stabilizes TβR1 on the membrane.
(A) Western blot analysis of TβR1 expression in 293T cells transfected with shPKM2. (B) qRT-PCR analysis of TGFBR1 mRNA expression in 293T cells transfected with shPKM2. GAPDH mRNA levels were used as an internal normalization control (n = 3 per group). (C) Immunoblot analysis of TβR1 expression in 293T cells transfected with Flag-PKM2. (D) Immunoblot analysis of 293T cells transfected with Myc-Smad7, GFP-TβR1, and increasing amount of Flag-PKM2. (E) Representative immunoblot analysis of TβR1 in 293T cells transfected with shPKM2 in the presence or absence of MG132. (F) Immunoblot analysis of TβR1 in 293T cells transfected with shPKM2 in the presence or absence of cycloheximide. (G) 293T cells were transfected with shPKM2 and treated with TGF-β1, and membrane and cytoplasmic proteins were separated and subjected to immunoblot analysis. (H) 293T cells were transfected with shPKM2 and treated with TGF-β1. Membrane was stained with CellMask, and TβR1 was stained with anti-TβR1 antibody. Scale bars, 25 μm. The ratio of membrane TβR1 was shown as percentage. (I) TβR1 distribution in lipid and nonlipid fractions was analyzed in 293T cells transfected with shPKM2 and subjected to sucrose gradient subcellular fractionation to separate lipid rafts from other cellular components. An equal volume from each fraction was analyzed by immunoblot analysis. Data are represented as the means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Student’s t test.
The ubiquitination of TβR1 will promote its internalization and degradation (11). Thus, we lastly investigated whether PKM2 knockdown affected TβR1 levels at the membrane, where signaling is initiated. We separated the membrane from the plasma and found that PKM2 knockdown led to reduced cell surface TβR1 levels and accelerated degradation (Fig. 6G). Consistently, double staining of TβR1 and membrane also revealed a shorter half-life in PKM2 knockdown cells (Fig. 6H). The ubiquitination of TβR1 promotes its internalization into a raft-caveolar vesicular (11). We performed PKM2 knockdown before fractionated rafts by sucrose density centrifugation; PKM2 knockdown triggered more TβR1 distribution into rafts (Fig. 6I). Collectively, our results indicate that PKM2 regulates TβR1 ubiquitination and stabilization, thus enhancing TβR1 signaling.
PKM2 regulates TGF-β1 signaling in human pulmonary fibroblasts
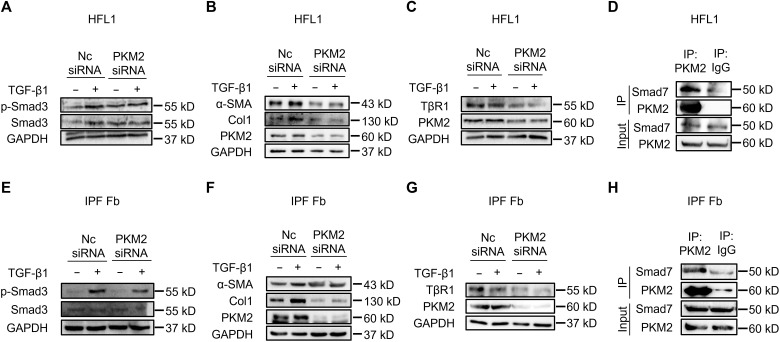
Given that PKM2 regulates TGF-β1 signaling in 293T cells and PKM2 is highly expressed in BLM-induced mouse primary lung fibroblasts, we next investigated whether PKM2 also regulated TGF-β1 signaling activation in human pulmonary fibroblasts. PKM2 knockdown in HFL1 decreased the expressions of p-Smad3 (Fig. 7A) and TGF-β1–induced profibrotic factors α-SMA, Col1, and Fn (Fig. 7B). In addition, PKM2 knockdown also resulted in a reduction in the endogenous level of TβR1 (Fig. 7C). Moreover, endogenous PKM2 bound to endogenous Smad7 (Fig. 7D), which is consistent with our previous findings. PKM2-induced alteration of TβR1 ubiquitination was also confirmed, as overexpression of PKM2 decreased the endogenous level of TβR1 ubiquitination (fig. S10A). We also confirmed these results using primary lung fibroblasts derived from patients with IPF and obtained similar outcomes (Fig. 7, E to H, and fig. S10B). These results suggest that PKM2 regulates TGF-β1 signaling in human pulmonary fibroblasts.
Fig. 7. PKM2 regulates TGF-β1 signaling in HFL1 and IPF fibroblasts.
(A to C) Western blot analysis of indicated proteins in HFL1 cells transfected with PKM2 siRNA for 48 hours and treated with TGF-β1 (5 ng/ml) for 1 hour (A) or 24 hours (B and C). (D) Immunoprecipitation of PKM2 with anti-PKM2 antibody in HFL1 cells. Western blot analysis of indicated proteins is shown. (E to G) Western blot analysis of indicated proteins in the IPF lung fibroblasts transfected with PKM2 siRNA for 48 hours and treated with TGF-β1 (5 ng/ml) for 1 hour (E) or 24 hours (F and G). (H) Immunoprecipitation of PKM2 with anti-PKM2 antibody in the IPF lung fibroblasts. Western blot analysis of indicated proteins is shown.
Pharmacologically regulating the conformation of PKM2 affects fibrosis progression
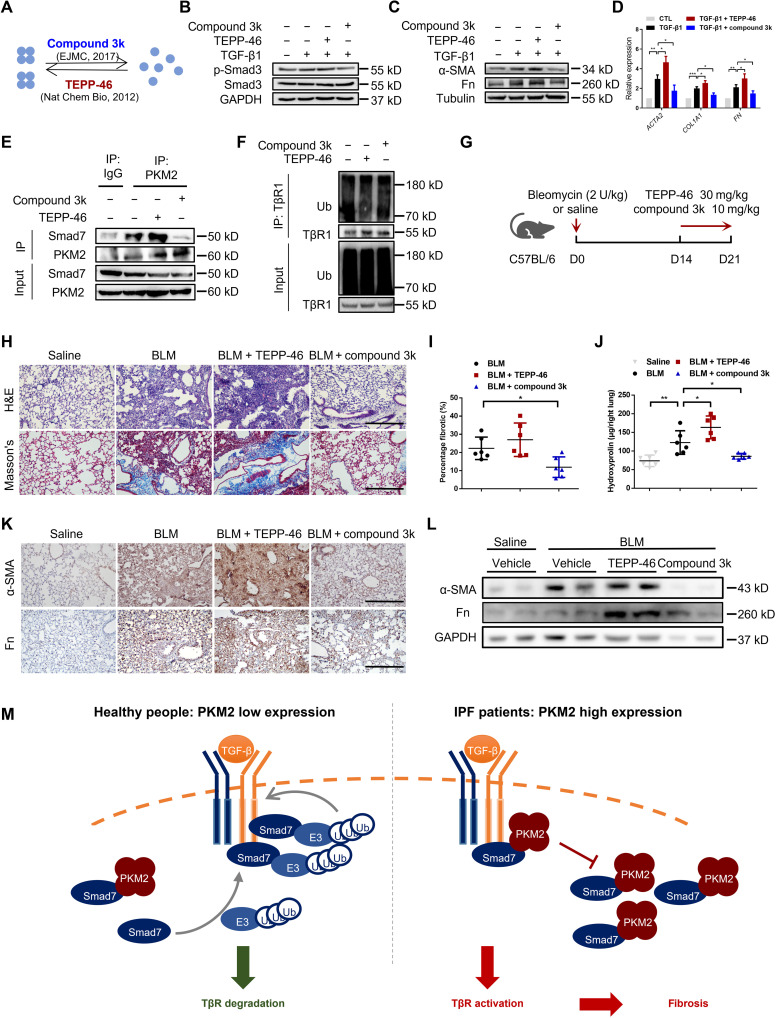
Several compounds have been reported to be capable of regulating PKM2 polymerization (Fig. 8A). TEPP-46 activates PKM2 by stabilizing its tetrameric form (35), while compound 3k antagonizes PKM2 by disrupting the PKM2 tetramer to form a monomer (36). Thus, we chose TEPP-46 and compound 3k to further test whether pharmacologically regulating PKM2 polymerization could affect fibrosis progression in vitro and in vivo. Through DSS cross-linking assay, we verified that TEPP-46 (10 μM) could promote PKM2 tetramer formation, while compound 3k (1 μM) could disrupt PKM2 tetramer into monomer (fig. S11). We first tested the effects of TEPP-46 and compound 3kin TGF-β1–induced HFL1 cells and found that TEPP-46 slightly increased the levels of p-Smad3 and the profibrotic factors α-SMA and Fn, whereas compound 3k markedly decreased the levels of p-Smad3, α-SMA, Col1, and Fn (Fig. 8, B to D). We further examined whether these two compounds could affect the interaction between PKM2 and Smad7 in vitro. We treated HFL1 cells with TEPP-46 or compound 3k for 24 hours and performed a co-IP analysis. Endogenous PKM2 interacted with endogenous Smad7, and TEPP-46 treatment enhanced the interaction between PKM2 and Smad7, whereas compound 3k decreased their interaction (Fig. 8E). In addition, TEPP-46 decreased the incorporation of ubiquitin into TβR1. In contrast, compound 3k facilitated an endogenous TβR1 ubiquitination (Fig. 8F). These findings suggest that pharmacologically regulating the PKM2 conformation can interfere PKM2 binding to Smad7 and TβR1 ubiquitination in vitro.
Fig. 8. Pharmacologically regulating the conformation of PKM2 affects fibrosis progression.
(A) Two compounds that were used for regulating PKM2 conformation. (B) HFL1 cells were treated with TEPP-46 or compound 3k for 24 hours then treated with TGF-β1 for 1 hour. (C and D) HFL1 cells were treated with TGF-β1 for 24 hours then treated with TEPP-46 or compound 3k for another 24 hours before harvest. Western blot (C) and qRT-PCR analysis (D) are shown. (E) HFL1 cells were treated with TEPP-46 or compound 3k and immunoprecipitated with anti-PKM2 antibody. (F) HFL1 cells were treated with TEPP-46 or compound 3k and then treated with MG132 for 4 hours before immunoprecipitation with anti-TβR1 antibody. (G) Schema of intervention study design using BLM-induced C57BL/6 mice. (H) Histological analysis of the severity of lung fibrosis in mice after BLM induction. Representative images for H&E (top) and Masson’s staining (bottom). Scale bars, 50 μm. (I) Quantification of the severity of fibrosis. The fibrotic area is presented as percentage (n = 6 per group). (J) Hydroxyproline content in lung tissues (n = 6 per group). (K) Representative images of the immunohistochemical analysis of α-SMA and Fn in lung sections. Scale bars, 50 μm. (L) Representative results (n = 2 of Western blot with n = 6 mice per group) of α-SMA and Fn in lung tissues. (M) Working model showing that the overexpression of PKM2 causes sustained stabilization of TβR1 and progression of pulmonary fibrosis. Data are represented as the means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
We lastly studied the effects of TEPP-46 and compound 3k on mice with BLM-induced pulmonary fibrosis. TEPP-46 (30 mg/kg) and compound 3k (10 mg/kg) were administrated by oral gavage from the 14th to the 21st day after BLM challenge (Fig. 8G). H&E and Masson’s staining revealed more severe pathological changes and collagen deposition in the TEPP-46–treated group than the control group, while compound 3k treatment markedly alleviated pulmonary construction collapse and collagen deposition (Fig. 8H). These results were also supported by quantification of the fibrotic score and hydroxyproline content (Fig. 8, I and J), coupled with a significant alteration in the expression of profibrotic factors α-SMA and Fn (Fig. 8, K and L). Together, our results demonstrated that pharmacologically regulating the conformation of PKM2 may be a potential strategy to ease lung fibrosis progression.
DISCUSSION
In this study, we found that PKM2 promoted pulmonary fibrosis progression by enhancing TGF-β1 signaling by directly interacting with Smad7. PKM2 was highly expressed in patients with IPF (GSE24206) and in the lungs and primary lung fibroblasts isolated from BLM-challenged mice. Notably, the proportion of PKM2 tetramers was elevated, along with total PKM2 expression. The loss of PKM2 reduced the severity of BLM-induced pulmonary fibrosis and myofibroblast activation. Through Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment and co-IP analysis, we identified that PKM2, especially its tetrameric form, could act as a regulator of TGF-β1 signaling. The PKM2 tetramer can form a complex with Smad7, interrupting Smad7 binding with TβR1 and leading to decreased TβR1 ubiquitination and sustained stabilization, thus facilitating TGF-β1 signaling and promoting fibrogenesis (Fig. 8M). In human fibroblasts, we were able to obtain the same result: PKM2 knockdown inhibited TGF-β1–induced myofibroblast activation. Last, we confirmed that pharmacologically controlling PKM2 to form monomers could alleviate BLM-induced pulmonary fibrosis. This finding suggests that controlling the biological form of PKM2 is a potential therapeutic strategy for IPF.
IPF is commonly known to be the result of repeated lung injury to the alveolar epithelium and recovery, leading to the proliferation and differentiation of lung fibroblasts, which ultimately become myofibroblasts (3). Therefore, we isolated primary lung fibroblasts and AT2 cells to identify the specific cell type involved in PKM2 alterations. The RNA and protein levels of PKM2 were significantly elevated in myofibroblasts but not AT2 cells, which is consistent with the high expression of PKM2 in persistently proliferating cells (27). Unlike the preference of the dimeric form in tumor cells (18, 37), we found tetrameric PKM2 as the preferred form during fibrosis progression. Previous studies have shown that the PKM2 dimer can act as a protein kinase and undergo phosphorylation and nuclear translocation to initiate oncogene transcription (18, 38, 39). However, in fibroblasts derived from mouse with BLM-induced pulmonary fibrosis (fig. S12, A and B) or HFL1 stimulated with TGF-β1 (fig. S12, C and D), we did not notice obvious changes in the phosphorylation or nuclear translocation of PKM2. These results collectively show that in pulmonary fibrosis progression, the PKM2 tetramer may have a more significant role than the PKM2 dimer.
PKM gene encodes both M1 and M2 isoforms and generates either the PKM1 or PKM2 by inclusion of exon 9 or exon 10, respectively, during mRNA splicing (40). Thus, PKM1 and PKM2 differ only by 22 amino acids (41), which makes it difficult to delete only PKM2 gene without affecting PKM1 when performing gene editing. A previous study has successfully generated a PKM2-specific deletion mouse model by introducing LoxP (locus of X-over P1) sites flanking exon 10 into the PKM locus of mouse embryonic stem cells using homologous recombination, then crossing it with Cre recombinase alleles, and generating a PKM2-specific deletion mouse without affecting the PKM1 expression (28). Thus, we used this mouse model to investigate the role of PKM2 in pulmonary fibrosis. PKM2 KO significantly alleviated BLM-induced pulmonary fibrosis, as shown by the fibrotic degree, myofibroblast activation, and profibrotic factor expression. The most studied function of PKM2 is the glycolysis-related function as pyruvate kinase. However, when we explored the key regulatory signaling by which PKM2 promotes fibrogenesis through comparing differential genes between PKM2-KO and WT primary fibroblasts, metabolic pathways did not appear to be the most significant process (fig. S13A). We also investigated glycolysis-related signals including lactate dehydrogenase (LDH) activity (fig. S13B) and lactate production (fig. S13C) in primary fibroblasts and found that PKM2 KO could mildly decrease LDH activity and lactate production induced by BLM treatment, but no significant difference was found. These results render us to focus more on nonmetabolic function of PKM2.
PKM2 knockdown significantly decreased TGF-β1–induced Smad3 activation. Because phosphorylation of Smad3 is affected by other members of the Smad family during TGF-β1 signaling transduction (7), we performed co-IP analysis between PKM2 and Smads to reveal the inner mechanism of PKM2 regulating TGF-β1 signaling and found that PKM2 interacted with Smad7 in vitro. A previous study reported that PKM2 knockdown alleviated TGF-β1–induced STAT3 activation and epithelial-mesenchymal transition (26), but no details have been revealed thus far. Our study demonstrated that PKM2 directly binds to Smad7 via its MH2 domain, interferes with the binding between Smad7 and TβR1, and decreases TβR1 ubiquitination. This result would explain why PKM2 knockdown decreases both canonical and noncanonical TGF-β1 signaling. PKM2 can disassociate from Smad7 when TGF-β1 signaling is getting stronger. PKM2 releases Smad7 and increases TβR1 ubiquitination and degradation, ultimately maintaining the balance in TGF-β1 signaling pathway. This newly identified PKM2 regulation of TGF-β1 signaling is similar to how DUBs tightly regulate TGF-β1 signaling (12, 13). Thus, when PKM2 is aberrantly overexpressed, more Smad7 will be blocked, leading to overactivation of TGF-β1 signaling. Meanwhile, we found that PKM2 binding with Smad7 cannot fully abolish the interaction between Smad7 and TβR1, thus facilitating the TβR1-Smad7-PKM2 complex assembly, which is similar to how Smad7 recruits E3 ligase to form the TβR1-Smad7-E3 complex. These facts indicate that PKM2 might display other roles in TGF-β1 signaling transduction except for counteracting Smad7. The recruitment of PKM2 on TβR1-Smad7 complex might further compete E3 ligase binding with Smad7 and decrease TβR1 ubiquitination, and the inner mechanism underlying this process deserves to be further elucidated.
Given that different conformations of PKM2 play distinct roles in TGF-β1 signaling transduction, we investigated whether pharmacologically regulating PKM2 allostery can affect fibrosis progression. PKM2 tetramer formation by TEPP-46 facilitated TGF-β1 signaling in lung fibroblasts and enhanced BLM-induced mouse pulmonary fibrosis, whereas PKM2 tetramer disruption by compound 3k markedly protected against pulmonary fibrosis. These findings are consistent with our previous results, suggesting that PKM2 is a potential therapeutic target in pulmonary fibrosis. Preliminary studies on PKM2 function in liver (42) and kidney (24) fibrosis indicate a more significant role of PKM2 dimer, and TEPP-46 showed protective effects on both the liver and kidney fibrosis model by decreasing glycolysis. Similar results are also found in cancer treatment, as PKM2 activation and inhibition can both suppress tumorigenesis (35, 43, 44). This fact impedes the clinical application of PKM2 allosteric regulator, because the tetramer-monomer equilibrium differs individually. Although the role of PKM2 in fibrotic diseases is not fully understood, both our study and the above studies showed that PKM2 knockdown efficiently alleviated fibrosis progression, which indicates the possibility of PKM2 inhibition in treating fibrotic diseases. However, considering PKM2 is a globally expressed enzyme, broad inhibition of PKM2 by small molecule might raise toxic effects on the human body. To this end, we performed a PKM2 conditional knockdown by delivering lung-specific LNPs carrying Pkm2 siRNA to BLM-treated mice and demonstrated a promising therapeutic effect on pulmonary fibrosis. Our results also raise the possibility that nucleic acid drug targeting PKM2 might be a potential strategy in treating pulmonary fibrosis. In conclusion, our results clearly suggest that PKM2 promotes pulmonary fibrosis progression by facilitating TGF-β1 signaling and is a previously unknown therapeutic target in treating pulmonary fibrosis.
MATERIALS AND METHODS
Generation of Pkm2−/− mice
Pkm2−/− mice were generated by crossing the Pkm2flox/flox mice [strain name: B6;129S-Pkm (tm1.1Mgvh)/J; no. 024048, the Jackson Laboratory] and EIIa-Cre transgenic mice [strain name: B6.FVB-Tg (EIIa-cre) C5379Lmgd/J; the Jackson Laboratory]. Pkm2flox/flox mice have LoxP sites on either side of exon 10 of the targeted gene. Pkm2flox/flox mice were mated to EIIa-Cre transgenic mice, in which the adenovirus EIIa promoter directs expression of the Cre enzyme in early mouse embryos (two- to eight-cell stage) to achieve homologous recombination between LoxP sites and triggers the deletion of exon 10 of PKM2 in all cells of the developing animal, including the germ cells that transmit the genetic alteration to progeny. Because of the mosaic activities of Cre recombinase, the first generation of EIIa-Cre; Pkm2flox/flox mice might be chimeric with different deletions. Therefore, the chimeric offspring were backcrossed with C57BL/6J to generate Pkm+/− mice, which then were intercrossed for production of PKM-deficient (Pkm2−/−) mice. Their WT littermates were used as control.
Mice and BLM induction of pulmonary fibrosis
All mice were housed and cared for in a pathogen-free facility at Nankai University. All animal experiments were approved by the Institutional Animal Care and Use Committee at Nankai University. All mice studies have followed Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines, and the checklist is provided with the manuscript. C57BL/6 male mice (6 to 8 weeks old) were intratracheally injected with BLM (2 U; Nippon Kayaku Co Ltd.) or the same volume of saline as control. Ten mice were used for each group to meet statistical significance, and mice were allocated randomly for study. Mice were euthanized at the 7th, 14th, and 21st day after BLM administration, and tissue samples were separated for further study. For evaluation of TEPP-46 and compound 3k, C57BL/6 mice (6 to 8 weeks old) were intratracheally injected with BLM and were gavaged for TEPP-46 [30 mg/kg; 0.5% O-carboxymethylcellulose (CMC)–Na and 5% dimethyl sulfoxide (DMSO)] or compound 3k (10 mg/kg; 0.5% CMC-Na and 5% DMSO) 14 to 21 days after BLM challenge, and mice in the BLM group were gavaged for the same vehicle. Mice were euthanized on day 21 after BLM treatment. No mice were excluded when analyzed, and the experimenters were blinded of group allocation during the conduct of the experiments.
Reagents and antibodies
CAGA-luc reporter was gifted by W. Ning (Nankai University). Flag-PKM2 was generated through subcloning human PKM2 cDNA (accession: NM_002654.6) into pCDNA3.1-Flag. Green fluorescent protein (GFP)–TβR1 was generated through subcloning human TβR1 cDNA (accession: NM_004612.4) into the vector pEGFP. The Smad7 fragments (amino acids 1 to 246 and MH2 domain) were cloned into pCDNA3.1. The PKM2 mutants K305Q, E28A, R32L, R316L, and E397A were generated using a site-directed mutagenesis kit (YEASEN) and confirmed by DNA sequencing. Myc-Smad2, Myc-Smad3, Myc-Smad4, Myc-Smad7, and human hemagglutinin (HA)-ubiquitin were purchased from MIAOLING. PKM2 shRNA was generated through inserting 5′-TTATTTGAGGAACTCCGCCGC-3′ targeting exon 10 of the PKM2 transcript into pLKO.1-puro as previously described (18), and its scramble control shRNA was generated through inserting 5′-TTCTCCGAACGTGTCACGT-3′ into pLKO.1-puro. The human PKM2 siRNA sequence used was 5 ′-CCAUAAUCGUCCUCACCAA-3′, and the murine Pkm2 siRNA sequence used was 5′-CAUCUACCACUUGCAAUUATT-3′ as previously described (30). The human SMAD7 siRNA sequence used was 5′-GAGGCTGTGTTG CTGTGAA-3′ as previously described (12). Human recombinant TGF-β1 was purchased from PeproTech. Antibodies against PKM2, TβR1, and EEA1 were purchased from Abcam. Antibodies against α-SMA and HA, were purchased from Santa Cruz Biotechnology. Antibody against Flag was purchased from Sigma-Aldrich. Antibodies against p-Smad3, Smad3, p-Akt, Akt, p-Erk1/2, Erk1/2, p-JNK, JNK, p-P38, and P38 were purchased from Cell Signaling Technology. Antibodies against Col1, Fn, Myc, GFP, His, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), tubulin, β-actin, and proliferating cell nuclear antigen (PCNA) were purchased from ProteinTech.
Hydroxyproline assay
Collagen contents in the lung homogenate were determined using a hydroxyproline kit (BioVision). The right lung of the mouse was isolated and placed into 5-ml ampoules, dried, hydrolyzed with hydrochloric acid, adjusted from pH 6.5 to 8.0, and filtered, and the samples were processed according to the hydroxyproline assay kit.
Histological and immunohistochemical analysis
The left lung was inflated and placed in fresh 10% neutral-buffered formalin for 24 hours to fix, then embedded in paraffin, and sectioned for histological analysis. Each field was scored for the fibrosis degree in a blinded fashion by two individuals. Digitized images were analyzed by Image-Pro Plus 6.0 software (Media Cybernetics). For immunohistochemical staining, sections were deparaffinized and antigen retrieved. Sections were stained with antibodies against α-SMA (Santa Cruz Biotechnology), p-Smad3, and Fn (Cell Signaling Technology) overnight, and the following steps were performed according to the instructions of the immunohistochemical staining kit (MAIXIN).
Immunofluorescence analysis
For tissue samples, sections were deparaffinized and antigen was retrieved before analysis. For cell samples, cells were fixed with 10% neutral-buffered formalin and permeabilized with 0.5% Triton X-100 (Invitrogen). Samples were stained with antibodies against Ki-67, TβR1 (1:100; Abcam), α-SMA (1:200; Santa Cruz Biotechnology), PDGFRβ, and caveolin-1 (1:200; R&D Systems). Fluorescein isothiocyanate– and fluorescein tetramethyl rhodamine isothiocyanate–conjugated secondary antibodies (1:200) were purchased from Jackson ImmunoResearch. The nucleus was labeled with 4′,6-diamidino-2-phenylindole (Santa Cruz Biotechnology). Membrane was stained with CellMask Green (Invitrogen). Pearson’s correlation was analyzed by Image-Pro Plus software.
Wound healing assay
Primary lung fibroblasts were seeded in plates. Before inflicting the wound, the cells were fully confluent. A scratch was made in the center of the culture well using a sterile 200-μl micropipette tip. The wounds were observed using an inverted optical microscope, and multiple images were obtained at areas flanking the intersections of the wound and the marker lines after the scratch at 0, 12, and 24 hours. Images were obtained for analysis using Image-Pro Plus software.
RNA-seq analysis
Mouse primary fibroblasts were harvested by TRIzol and sent to Genesky Technologies for analysis. Samples were sequenced on an Illumina HiSeq 2500 instrument using 2 × 150 base pair reads. Protein-coding genes with at least 2 RPKM (reads per kilobase of transcript, per million mapped reads) on average in either condition were used to perform the differential gene expression analysis using edgeR. The significance level was set at a false discovery rate <0.01 and |log2 fold change| >0.667. To compare these data with the previously described human IPF tissues RNA-seq data (GSE72073), homologous genes expressed in both human and mouse were selected. The differentially up-regulated genes in patients with IPF were obtained according to the significance level and are compared with the significantly down-regulated genes in PKM2-KO fibroblasts. The common genes were further undergoing KEGG pathway enrichment analysis.
Cell culture, transfection, and treatment
Primary lung fibroblasts from a patient with IPF were gifted by W. Ning (Nankai University). All cell lines were obtained from the American Type Culture Collection. 293T and NIH3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), and HFL1 cells were cultured in F12K, all supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37°C with 5% CO2. Cells were transfected using Lipofectamine 3000 or RNAiMAX (Invitrogen) for plasmid or siRNA transfection, respectively. For scramble control, Flag-PKM2 was controlled by pCDNA3.1-Flag, Myc-Smads were controlled by pCDNA3.1-Myc, GFP-TβR1 was controlled by pEGFP, human siRNA was controlled by a scramble siRNA sequence 5′-UUCUCCGAACGUGUCACGU-3′, and mouse siRNA was controlled by scramble siRNA sequence 5′-CUUACGCUGAGU ACUUCGA-3′. The treatments used were as follows: TGF-β1 (5 ng/ml), MG132 (5 μM in DMSO; Sigma-Aldrich), cycloheximide (CHX in DMSO; 20 μg/ml; Sigma-Aldrich), TEPP-46 [10 μM in DMSO; MedChemExpress (MCE)], and compound 3k (1 μM in DMSO; MCE).
Fibroblast isolation and culture
Primary lung fibroblasts were obtained as previously described (45). In brief, lungs were lavaged, minced, and digested with collagenase IV (2 mg/ml; Worthington) and dispase II (2 mg/ml; Roche) at 37°C for 30 min. The digested lung pieces were centrifuged and washed with DMEM three times, and cell pellet was resuspended with DMEM containing 10% FBS and cultured at 37°C with 5% CO2.
AT2 cell isolation and culture
Primary AT2 cells were obtained as previously described (45). In brief, lungs were lavaged three times with phosphate-buffered saline (PBS) containing 1% antibiotics, then digested with dispase II (2 mg/ml) and deoxyribonuclease I (0.3 U/ml) at 37°C for 45 min, and minced. The digested lung pieces were filtered through 100- and 40-μm cell strainers and centrifuged. Cell pellet was resuspended with DMEM containing 10% FBS and cultured at 37°C for 45 min in CD16/32- and CD45-coated dishes to remove inflammatory cells. The supernatant was further cultured at 37°C for 45 min to remove fibroblasts. Then, cells were resuspended with DMEM/F12 containing 10% FBS, seeded in collagen I (BD)–precoated plates, and cultured at 37°C with 5% CO2. AT2 cells were characterized by immunofluorescence staining using antibody against Prospc (Santa Cruz Biotechnology).
Luciferase assays
Luciferase assays were performed as previously described (45). In brief, 293T cells were transfected with the indicated plasmids for 48 hours and then serum starved for 24 hours before treatment with TGF-β1 (5 ng/ml). Cells were harvested 16 hours after stimulation to perform luciferase assays using a luciferase assay system (Promega) according to the instructions.
Immunoprecipitation and the in vitro deubiquitination assay
Cells were lysed in lysis buffer [50 mM tris, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40 (pH 7.5)]. Cell lysates were incubated with the indicated antibodies conjugated to protein A/G agarose (Santa Cruz Biotechnology) at 4°C overnight, washed six times with lysis buffer, and separated on SDS–polyacrylamide gel electrophoresis (PAGE) gels. For the ubiquitination detection, cells were pretreated with MG132 (5 μM) 4 hours before harvest.
DSS cross-linking
Cells were harvested using lysis buffer. DSS (2 mM; Abcam) was added to cross-link at room temperature for 30 min, and 10 mM tris was used to end the cross-link for 15 min. Samples were detected by SDS-PAGE.
Purification of recombinant proteins
His-Smad7 expression construct was generated by subcloning the full-length Smad7 into pET28a vectors. His-Smad7-pET28a plasmids were transformed into BL21 (DE3) cells. Cultures were grown at 37°C in fresh LB medium and grown at 37°C to an attenuance of 0.6 at 600 nm and induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside overnight at 16°C. BL21 cells were harvested using lysis buffer (500 mM NaCl, 10 mM TCEP, 10% glycerol, and 1× PBS) and purified with Ni–nitrilotriacetic acid agarose resin (GE Healthcare). Flag-PKM2 was generated by transfecting Flag-PKM2-pCDNA3.1 into 293T cells for 48 hours. Cells were harvested using lysis buffer and incubated with Flag-tag–conjugated beads (Sigma-Aldrich) at 4°C for 4 hours. Beads were washed six times with lysis buffer, and protein was eluted using Flag peptide (Sigma-Aldrich).
Pull-down assay
For pull-down assay, 20 μg of His-Smad7 was incubated with His-tag–conjugated beads at 4°C for 4 hours, and then Flag-PKM2 was added into the mixture and incubated for another 2 hours. Beads were washed six times with lysis buffer and analyzed by SDS-PAGE.
Microscale thermophoresis
Flag-PKM2 protein (20 μM) was labeled with fluorescence for 30 min and diluted into 166.7 nM for further testing. Recombinant His-Smad7 was gradient diluted 16 times from 1.49 μM, equivalently mixed with labeled Flag-PKM2, and incubated for 5 min. The mixture samples were loaded into capillaries and tested using Monolith NT.115 according to the manufacturer’s instructions.
Sucrose-gradient ultracentrifugation
To separate lipid and nonlipid components, we performed sucrose-gradient ultracentrifugation as previously described (11). In brief, 293T cells were harvested using 0.8 ml of 0.5 M Na2CO3 (pH 11), disrupted cell membranes by performing 15-s bursts of an ultrasonic disintegrator, and mixed with 0.8 ml 2× Hanks’ Balanced Salt solution (HBS). Homogenates were laid on a 35 and 10% sucrose gradient (0.25 M Na2CO3, 1× HBS, pH 11, 3.5 ml) and spun at 160,000g for 16 hours using a Beckman rotor (Optima XE-100). Fractions were equally separated into 10 portions and analyzed by SDS-PAGE.
Membrane, nuclear, and cytoplasmic protein extraction
For membrane protein extraction, a membrane and cytosol protein extraction kit (YESEAN) was used. For nuclear protein extraction, a nuclear and cytoplasmic protein extraction kit (YESEAN) was used. All steps followed the manufacturer’s instructions. Each fraction was separated by SDS-PAGE.
qRT-PCR analysis
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using the One Step RT-qPCR SYBR Green Kit (YESEAN), and the relative expression of each target gene was normalized by GAPDH as previously described. The following primers were used for each target gene: GAPDH, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and 5′-GGCTGTTGTCATACTTCTCATGG-3′; Gapdh, 5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-GGGGTCGTTGATGGCAACA-3′; ACTA2, 5′-GGCATTCACGAGACCACCTAC-3′ and 5′-CGACATGACGTTGTTGGCATAC-3′; Acta2, 5′-GTCCCAGACATCAGGGAGTAA-3′ and 5′-GTCCCAGAC ATCAGGGAGTAA-3′; COL1A1, 5′-GAGGGCCAAGACGAAGACATC-3′ and 5′-CAGATCACGTCATCGCACAAC-3′; Col1α1, 5′-ATGTGGACCCCTCCTGATAGT-3′ and 5′-ATGTGGACCCCTCCTGATAGT-3′; FN, 5′-TCGGATACTTCAGCGTCAGGA-3′ and 5′-TCGGATACTTCAGCGTCAGGA-3′; TGFBR1, 5′-ACGGCGTTACAGTGTTTCTG-3′ and 5′-GCACATACAAACGGCCTATCTC-3′; Tgfbr1, 5′-TCTGCATTGCACTTATGCTGA-3′ and 5′-AAAGGGCGATCTAGTGATGGA-3′; PKM2, 5′-ATGTCGAAGCCCCATAGTGAA-3′ and 5′-TGGGTGGTGAATCAATGTCCA-3′; and Pkm2, 5′-GCCGCCTGGACATTGACTC-3′ and 5′-CCATGAGAGAAATTCAGCCGAG-3′.
Preparation of siRNA-loaded LNPs
Pkm2-siRNA–loaded LNPs were prepared as reported (29). In brief, DLin-MC3-DMA (Selleck Chemicals), 1,2-dioleoyl-snglycero-3-phosphoethanolamine (Avanti Polar Lipids), cholesterol (Sigma-Aldrich), 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (Avanti Polar Lipids), and 1,2-dioleoyl-3-trimethylammonium-propane (Avanti Polar Lipids) were dissolved in ethanol at a molar ratio of 11.9:11.9:23.8:2.4:50. Pkm2 siRNA 5′-CAUCUACCACUUGCAAUUATT-3′ or scramble siRNA 5′-CUUACG CUGAGUACUUCGA-3′ (30) was dissolved in citrated buffer (10 mM, pH 3). The LNPs and the dissolved siRNA were then mixed at a molar ratio of 40:1 and a volume ratio of 1:3 and then unentrapped siRNA were excluded by ultrafiltration centrifugation. For in vivo imaging system, luciferase mRNA (0.5 mg/kg; APExBIO) was dissolved in citrated buffer (10 mM, pH 3) and mixed with the LNPs, intravenously injected into mice for 6 hours, and d-luciferin potassium salt (150 mg/kg; Beyotime) was injected before detection.
Statistical analysis
Data are expressed as the means ± SEM. Differences in measured variables between experimental and control groups were assessed by using Student’s t tests or a Student’s t test with Welch’s correction for unpaired data. Results were considered statistically significant at P < 0.05. GraphPad Prism software was used for statistical analysis. The numbers of technical replicates or biological replicates (independent experiments for cell culture or individual mouse for in vivo) in each group were stated in the figure legends.
Acknowledgments
We thank the patients and families for their involvement in this study. We thank C. Shan (Nankai University) for important comments. We thank W. Zhong (Nankai University) for assisting with the molecular docking analysis.
Funding: This work is supported by the National Natural Science Foundation of China (82070060), the Tianjin Science and Technology Project (19YFZCSY00160), and the Foundation of Organ Fibrosis Druggability Joint Research Center of Nankai and Guokaixingcheng (735-F1040051).
Author contributions: S.G., X.L., and Q.J. were involved in the study design, the conduct of the experiments, and data analyses and wrote the manuscript. Q.L., S.L., F.Z., R.Z., J.L., and J.Z. were involved in Western blotting, histological analysis, immunostaining, and animal breeding. X.G. and T.X. assisted in data analysis and figure preparation. H.H. and S.C. contributed to the statistical analyses. W.N., G.Y., C.Y., and H.Z contributed to the study design and review of the manuscript. All authors were involved in drafting the article and revising it critically and gave approval for the final manuscript to be published.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data are available at https://datadryad.org/stash/share/X2Ary0F8whsoqVE50HRCQdu32GpcqLZB-ZwPQlLco_o.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
REFERENCES AND NOTES
- 1.Raghu G., Remy-Jardin M., Myers J. L., Richeldi L., Ryerson C. J., Lederer D. J., Behr J., Cottin V., Danoff S. K., Morell F., Flaherty K. R., Wells A., Martinez F. J., Azuma A., Bice T. J., Bouros D., Brown K. K., Collard H. R., Duggal A., Galvin L., Inoue Y., Jenkins R. G., Johkoh T., Kazerooni E. A., Kitaichi M., Knight S. L., Mansour G., Nicholson A. G., Pipavath S. N. J., Buendia-Roldan I., Selman M., Travis W. D., Walsh S., Wilson K. C.; E. R. S. J. R. S. American Thoracic Society; S. Latin American Thoracic , Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 198, e44–e68 (2018). [DOI] [PubMed] [Google Scholar]
- 2.King T. E. Jr., Pardo A., Selman M., Idiopathic pulmonary fibrosis. Lancet 378, 1949–1961 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Spagnolo P., Cottin V., Genetics of idiopathic pulmonary fibrosis: From mechanistic pathways to personalised medicine. J. Med. Genet. 54, 93–99 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Barkauskas C. E., Noble P. W., Cellular mechanisms of tissue fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am. J. Physiol. Cell Physiol. 306, C987–C996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellaye P.-S., Kolb M., Why do patients get idiopathic pulmonary fibrosis? Current concepts in the pathogenesis of pulmonary fibrosis. BMC Med. 13, 176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez F. J., Collard H. R., Pardo A., Raghu G., Richeldi L., Selman M., Swigris J. J., Taniguchi H., Wells A. U., Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers. 3, 17074 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Massagué J., Seoane J., Wotton D., Smad transcription factors. Genes Dev. 19, 2783–2810 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Kang J. S., Liu C., Derynck R., New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol. 19, 385–394 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Ikushima H., Miyazono K., TGFbeta signalling: A complex web in cancer progression. Nat. Rev. Cancer 10, 415–424 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L., Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6, 1365–1375 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L., Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat. Cell Biol. 5, 410–421 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Eichhorn P. J., Rodón L., Gonzàlez-Juncà A., Dirac A., Gili M., Martínez-Sáez E., Aura C., Barba I., Peg V., Prat A., Cuartas I., Jimenez J., García-Dorado D., Sahuquillo J., Bernards R., Baselga J., Seoane J., USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 18, 429–435 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Zhou F., Drabsch Y., Gao R., Snaar-Jagalska B. E., Mickanin C., Huang H., Sheppard K. A., Porter J. A., Lu C. X., ten Dijke P., USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat. Cell Biol. 14, 717–726 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Nakao A., Fujii M., Matsumura R., Kumano K., Saito Y., Miyazono K., Iwamoto I., Transient gene transfer and expression of Smad7 prevents bleomycin-induced lung fibrosis in mice. J. Clin. Invest. 104, 5–11 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C. M., He C. H., Park J. W., Lee J. H., Kamle S., Ma B., Akosman B., Cotez R., Chen E., Zhou Y., Herzog E. L., Ryu C., Peng X., Rosas I. O., Poli S., Bostwick C. F., Choi A. M., Elias J. A., Lee C. G., Chitinase 1 regulates pulmonary fibrosis by modulating TGF-β/SMAD7 pathway via TGFBRAP1 and FOXO3. Life Sci. Alliance 2, e201900350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Witney T. H., James M. L., Shen B., Chang E., Pohling C., Arksey N., Hoehne A., Shuhendler A., Park J. H., Bodapati D., Weber J., Gowrishankar G., Rao J., Chin F. T., Gambhir S. S., PET imaging of tumor glycolysis downstream of hexokinase through noninvasive measurement of pyruvate kinase M2. Sci. Transl. Med. 7, 310ra169 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., Lu Z., Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., Lyssiotis C. A., Aldape K., Cantley L. C., Lu Z., ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 14, 1295–1304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan Y. F., Huang Q. L., Ai Y. L., Chen Q. T., Zhao W. X., Wang X. M., Wu Q., Chen H. Z., Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene 39, 2408–2423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki A., Puri S., Leland P., Puri A., Moudgil T., Fox B. A., Puri R. K., Joshi B. H., Subcellular compartmentalization of PKM2 identifies anti-PKM2 therapy response in vitro and in vivo mouse model of human non-small-cell lung cancer. PLOS ONE 14, e0217131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Y., Zhai H., Ouyang Y., Lu Z., Chu C., He Q., Cao X., Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 19, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu F., Guo M., Huang W., Feng L., Zhu J., Luo K., Gao J., Zheng B., Kong L. D., Pang T., Wu X., Xu Q., Annexin A5 regulates hepatic macrophage polarization via directly targeting PKM2 and ameliorates NASH. Redox Biol. 36, 101634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Takagaki Y., Kumagai A., Kanasaki K., Koya D., The PKM2 activator TEPP-46 suppresses kidney fibrosis via inhibition of the EMT program and aberrant glycolysis associated with suppression of HIF-1α accumulation. J. Diabetes Investig. 12, 697–709 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Zheng C., Gao Z., Wang L., Chen C., Zheng Y., Meng Y., PKM2 promotes angiotensin-II-induced cardiac remodelling by activating TGF-β/Smad2/3 and Jak2/Stat3 pathways through oxidative stress. J. Cell. Mol. Med. 25, 10711–10723 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma R., Liu Q., Zheng S., Liu T., Tan D., Lu X., PKM2-regulated STAT3 promotes esophageal squamous cell carcinoma progression via TGF-β1-induced EMT. J. Cell. Biochem. 120, 11539–11550 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Zahra K., Dey T., Ashish, Mishra S. P., Pandey U., Pyruvate kinase M2 and cancer: The role of PKM2 in promoting tumorigenesis. Front. Oncol. 10, 159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Israelsen W. J., Dayton T. L., Davidson S. M., Fiske B. P., Hosios A. M., Bellinger G., Li J., Yu Y., Sasaki M., Horner J. W., Burga L. N., Xie J., Jurczak M. J., DePinho R. A., Clish C. B., Jacks T., Kibbey R. G., Wulf G. M., Di Vizio D., Mills G. B., Cantley L. C., Vander Heiden M. G., PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155, 397–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Q., Wei T., Farbiak L., Johnson L. T., Dilliard S. A., Siegwart D. J., Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg M. S., Sharp P. A., Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J. Exp. Med. 209, 217–224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun K. H., Chang Y., Reed N. I., Sheppard D., α-Smooth muscle actin is an inconsistent marker of fibroblasts responsible for force-dependent TGFβ activation or collagen production across multiple models of organ fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 310, L824–L836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng J., Huang X., Li Y., Xu X., Li S., Jiang D., Liang J., Jiang D., Wang C., Dai H., Down-regulation of USP13 mediates phenotype transformation of fibroblasts in idiopathic pulmonary fibrosis. Respir. Res. 16, 124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macpherson J. A., Theisen A., Masino L., Fets L., Driscoll P. C., Encheva V., Snijders A. P., Martin S. R., Kleinjung J., Barran P. E., Fraternali F., Anastasiou D., Functional cross-talk between allosteric effects of activating and inhibiting ligands underlies PKM2 regulation. eLife 8, e45068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanyu A., Ishidou Y., Ebisawa T., Shimanuki T., Imamura T., Miyazono K., The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J. Cell Biol. 155, 1017–1027 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anastasiou D., Yu Y., Israelsen W. J., Jiang J. K., Boxer M. B., Hong B. S., Tempel W., Dimov S., Shen M., Jha A., Yang H., Mattaini K. R., Metallo C. M., Fiske B. P., Courtney K. D., Malstrom S., Khan T. M., Kung C., Skoumbourdis A. P., Veith H., Southall N., Walsh M. J., Brimacombe K. R., Leister W., Lunt S. Y., Johnson Z. R., Yen K. E., Kunii K., Davidson S. M., Christofk H. R., Austin C. P., Inglese J., Harris M. H., Asara J. M., Stephanopoulos G., Salituro F. G., Jin S., Dang L., Auld D. S., Park H. W., Cantley L. C., Thomas C. J., Vander Heiden M. G., Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 8, 839–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ning X., Qi H., Li R., Li Y., Jin Y., McNutt M. A., Liu J., Yin Y., Discovery of novel naphthoquinone derivatives as inhibitors of the tumor cell specific M2 isoform of pyruvate kinase. Eur. J. Med. Chem. 138, 343–352 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Keller K. E., Tan I. S., Lee Y. S., SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science 338, 1069–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang P., Li Z., Fu R., Wu H., Li Z., Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell. Signal. 26, 1853–1862 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Hamabe A., Konno M., Tanuma N., Shima H., Tsunekuni K., Kawamoto K., Nishida N., Koseki J., Mimori K., Gotoh N., Yamamoto H., Doki Y., Mori M., Ishii H., Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 111, 15526–15531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi T., Inoue H., Tanaka T., The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J. Biol. Chem. 261, 13807–13812 (1986). [PubMed] [Google Scholar]
- 41.Mazurek S., Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int. J. Biochem. Cell Biol. 43, 969–980 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Zheng D., Jiang Y., Qu C., Yuan H., Hu K., He L., Chen P., Li J., Tu M., Lin L., Chen H., Lin Z., Lin W., Fan J., Cheng G., Hong J., Pyruvate kinase M2 tetramerization protects against hepatic stellate cell activation and liver fibrosis. Am. J. Pathol. 190, 2267–2281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J. H., Kundu A., Lee S. H., Jiang C., Lee S. H., Kim Y. S., Kyung S. Y., Park S. H., Kim H. S., Specific pyruvate kinase M2 inhibitor, compound 3K, induces autophagic cell death through disruption of the glycolysis pathway in ovarian cancer cells. Int. J. Biol. Sci. 17, 1895–1908 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng Z., Lan J., Lei S., Yang Y., He Z., Xue Y., Chen T., Simultaneous inhibition of ornithine decarboxylase 1 and pyruvate kinase M2 exerts synergistic effects against hepatocellular carcinoma cells. Onco. Targets. Ther. 13, 11697–11709 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong Y., Geng Y., Li L., Li X., Yan X., Fang Y., Li X., Dong S., Liu X., Li X., Yang X., Zheng X., Xie T., Liang J., Dai H., Liu X., Yin Z., Noble P. W., Jiang D., Ning W., Blocking follistatin-like 1 attenuates bleomycin-induced pulmonary fibrosis in mice. J. Exp. Med. 212, 235–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13