Abstract
The Betta fish displays a remarkable variety of phenotypes selected during domestication. However, the genetic basis underlying these traits remains largely unexplored. Here, we report a high-quality genome assembly and resequencing of 727 individuals representing diverse morphotypes of the Betta fish. We show that current breeds have a complex domestication history with extensive introgression with wild species. Using a genome-wide association study, we identify the genetic basis of multiple traits, including coloration patterns, the “Dumbo” phenotype with pectoral fin outgrowth, extraordinary enlargement of body size that we map to a major locus on chromosome 8, the sex determination locus that we map to dmrt1, and the long-fin phenotype that maps to the locus containing kcnj15. We also identify a polygenic signal related to aggression, involving multiple neural system-related genes such as esyt2, apbb2, and pank2. Our study provides a resource for developing the Betta fish as a genetic model for morphological and behavioral research in vertebrates.
Betta genomics reveal its domestication history and genetic basis of enormous diversity in morphology and behavior.
INTRODUCTION
The Betta fish (Betta splendens) is indigenous to central Thailand and the lower Mekong (1) and is mostly known for its domesticated forms appreciated as an ornamental fish but was originally bred for its use in gambling matches similar to cock fights (2). Through captive breeding, a remarkable variety of behaviors and morphologies have emerged, including variation in aggressiveness, pigmentation, body size, and fin shape. B. splendens is easy to breed and maintain and provides a useful resource for exploration of the genetic basis of behavior and morphology in vertebrates, due to the high degree of intraspecific variability and the vast number of characterized phenotypes. It also provides a fascinating example of how direct and indirect selection imposed by humans has shaped a domesticated species. Although researchers have examined the inheritance of body color, fin length, and sex determination (SD) in classic crosses in the 1930s to 1940s (3–9), relatively little was known about the genetic basis of phenotypic variability in Betta fish until recent studies investigated the double tail, elephant ear, albino, fin spot, and SD phenotypes (10–12). Here, we report a high-quality chromosomal-level genome assembly of a female B. splendens, resequencing data of 727 domesticated individuals and 59 wild individuals from six other species in the B. splendens complex. We examine the evolutionary relationship and origins among breeds and use association mapping to identify the genetic basis of a number of different traits including SD, fin morphology, coloration, body size, and aggressiveness and other behaviors. More background on Betta fish and the samples used here can be found in text S2.
RESULTS
Genome assembly, annotation, and comparative analyses of the Betta fish
We generated a high-quality chromosomal-level assembly of the Betta fish using a multifaceted sequencing and assembling workflow, including PacBio reads, Illumina reads, Hi-C reads, 10x Genomics reads, and BioNano optical mapping (text S1 and table S1). The final assembled genome was 451.29 million base pairs (Mb) with contig and scaffold N50s reaching 4.07 and 19.63 Mb, respectively. A total of 93.6% of the scaffolds were placed onto 21 chromosomes, which is accordant with the chromosome karyotype reported previously (13). The assembly contains 119 Mb of repetitive sequences (tables S2 and S3 and fig. S1) and 25,104 annotated structural genes, 22,788 (90.77%) of which were functionally annotated (text S1, tables S4 to S6, and fig. S2). CEGMA (Core Eukaryotic Genes Mapping Approach) (14) analyses confirmed the presence of 239 of 248 (96.4%) complete core eukaryotic genes, and BUSCO (Benchmarking Universal Single-Copy Orthologs) (15) evaluation showed that 2522 of 2586 (97.5%) single-copy orthologous genes were annotated (table S7), indicating high completeness of the genome and gene annotation. We constructed a phylogeny using 465 single-copy orthologs shared by B. splendens and other 13 teleosts and showed that B. splendens diverged ~109.6 million years (Ma) ago from other Perciformes (Fig. 1A and fig. S3). The high-quality genome assembly coupled with comprehensive genome annotation represents an important addition and improvement to the existing reference genome resources for Betta fish research (table S7).
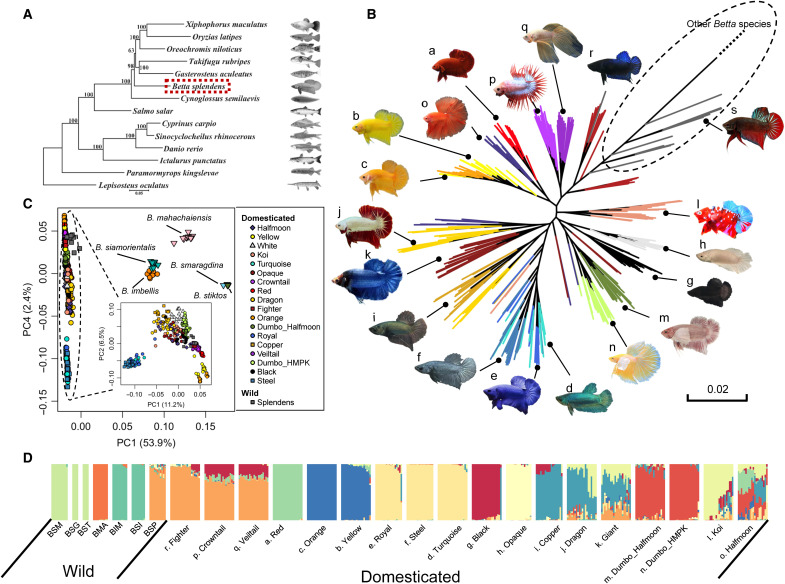
Fig. 1. Phylogeny and population structure of the Betta fish (B. splendens).
(A) The phylogeny of teleost including the Betta fish constructed with single-copy genes across the genome. (B) Maximum likelihood tree of concatenated genome-wide single-nucleotide polymorphisms (SNPs) for domesticated and wild forms of B. splendens. The tree is truncated at the branch connecting to individuals of other Betta species, and the untruncated tree can be found in fig. S5. The dashed-line circle indicates the wild Betta fish. The letters by the fish photos correspond to the letters in (D). (C) PCA of the Betta species complex. The inset is the PCA for all B. splendens individuals. (D) Admixture analysis of the B. splendens breeds and their closely related wild species. K = 12 is presented here, and the results with K varying from 2 to 15 are in fig. S7. Acronyms for wild species are as follows: BSP, B. splendens; BIM, B. imbellis; BSI, B. siamorientalis; BMA, B. mahachaiensis; BSM, B. smaragdina; BSG, B. smaragdina guitar; BST, B. stiktos.
Diversification of the Betta fish during domestication
We collected 14 breeds of Betta fish differing in tail type, coloration, sex, and body size (text S2, fig. S4, and table S8) and performed whole-genome resequencing of 727 individuals, resulting in ~2.7-Tb clean sequencing data with depths ranging from 3× to 34× (average 6.7×). To elucidate the population history of the B. splendens complex and the origins of the domesticated Betta fish, we further sequenced 59 individuals from six wild species of the B. splendens complex with an average depth of 24× (19× to 31×), to coanalyze with 20 randomly picked individuals from each breed. A maximum likelihood phylogeny (Fig. 1B) from concatenated sequences, representing the average genomic coalescent tree, showed that the domesticated breeds form a monophyletic group relative to other wild species (Fig. 1B and fig. S5). In addition, consistently, in principal components analysis (PCA), they form a close cluster, distinct from other wild individuals (Fig. 1C and figs. S7 and S8). These observations are compatible with the hypothesis that all current breeds of the Betta fishes were domesticated from the same group of wild B. splendens. The Betta siamorientalis individuals form a monophyletic clade within the cluster of Betta imbellis individuals in the tree (fig. S5). The lineages of Betta smaragdina guitar are interspersed with B. smaragdina, and Betta stiktos form a monophyletic clade within this cluster in the average genomic tree (fig. S5). These three species are also largely overlapped in PCA (Fig. 1C and fig. S7), suggesting that they all should be considered different varieties of the B. smaragdina species.
Several clades of the Fighter breed fall as outgroups to the rest of the domesticated breeds in the phylogenetic tree (Fig. 1B), which is compatible with the breeding record that the Fighter breed represents an early domesticated form and that the earliest domesticated Betta fish in fact were breeds selected for fighting (2). Other breeds falling toward the root of the tree include Veiltail and Crowntail. However, these observations can also be attributable to introgressions from the wild species into Fighter, Veiltail, and Crowntail breeds, as extensive gene flow signals are observed among different fish groups (text S3). Population structure, as revealed by the phylogeny (Fig. 1B), PCA (Fig. 1C and fig. S8), and admixture analyses (Fig. 1D and fig. S6), suggests that breeds defined by coloration and morphology are generally clustered together, although we note that this conclusion might be affected by the sampling strategy used here (text S2). The HMPK (Halfmoon Plakat) breeds, which are short-fin types distinct from the Fighter and wild B. splendens by a rounded tail shape and body shape, also cluster on the basis of appearance, mostly related to colors (Fig. 1B). The Red, Yellow, Orange, Turquoise-green, Royal-blue, Steel-blue groups of breeds carry substantial group-specific genetic drift that is notable in the PCA, structure analyses, and the genome-wide phylogeny (Fig. 1, B to D, and fig. S6), suggesting that these groups experienced strong bottleneck effects in their domestication history.
TreeMix analyses (figs. S9 and S10) suggest substantial gene flow between wild and domesticated species in Betta fish. An exhaustive D statistics analysis testing D (P1, domesticated; P2, domesticated; P3, wild; P4, Smaragdina) confirms these results and shows ubiquitous heterogeneity in the amount of gene flow between the domesticated and wild species, even noticeable among individuals from the same breed (text S3 and figs. S11 and S12). The high heterogeneity in admixture among individuals suggests not only that gene flow is common among wild and domesticated breeds but also that it is recent, likely because of anthropogenic influences. At least two wild species, Betta mahachaiensis and B. imbellis, have contributed to the genomic makeup of domesticated Siamese fighting fish (text S3 and figs. S9 and S12) and may, therefore, also have contributed to phenotypic variation in these breeds. A more detailed discussion of the phylogeny of the Betta species and the population structure of B. splendens can be found in text S3, as well as a detailed discussion of the pattern of admixture and gene flow.
Regulatory variants in dmrt1 mediate the male heterogamety of B. splendens
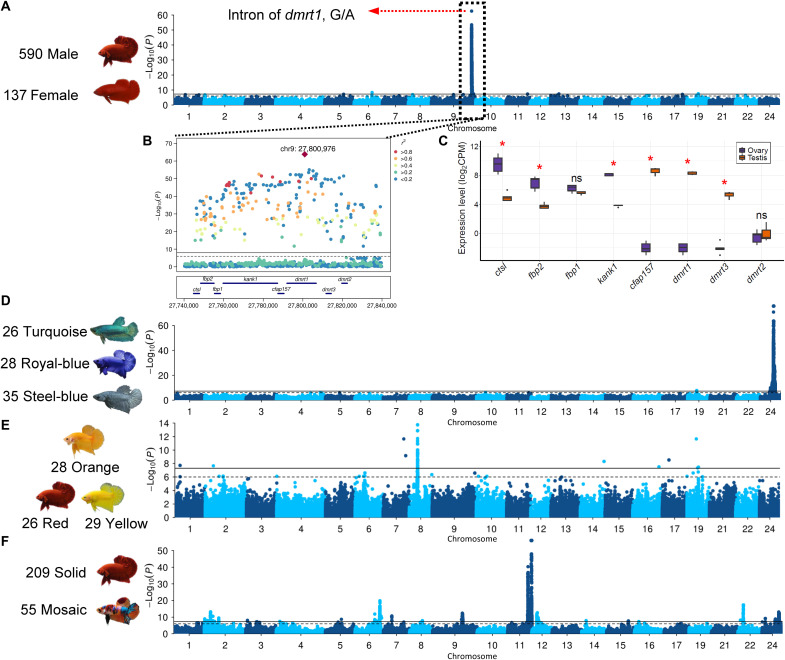
To map the SD locus, we performed association mapping on all 727 individuals consisting of 590 males and 137 females using a mixed linear model implemented in GEMMA (16). The kinship matrix and the first three principal components from PCA were used as covariates to minimize the effects of population stratification. We find a highly significant map location at position 27.75 to 27.81 Mb on chromosome 9 (P = 1.51 × 10−64; Fig. 2, A and B) and no evidence of statistical inflation (fig. S13). Mapping with different breeds consistently replicate the signal, supporting a common genetic basis of SD in all these breeds (fig. S14), which is also consistent with previous studies (11, 12). Five hundred thirty of the 537 individuals with heterozygous genotypes of the lead variant (chr9:27,800,976, G/A) are phenotypically males, and 130 of the 137 individuals homozygous for AA being female, which strongly supports the hypothesis of male heterogametic (XY/XX) in Betta fish (table S9).
Fig. 2. Genome-wide association studies and locus analysis of SD, body color, and pattern in B. splendens.
(A) Genome-wide association study (GWAS) of SD and expression profiles in testis and ovary for genes in the associated locus. A single genome-wide significant signal is identified, with a lead SNP located on the second intron of the dmrt1 gene. (B) LocusZoom plot on the associated peak region. (C) The expression levels of genes on the peak region in ovary and testis. CPM, counts per million. Manhattan plots for (D) the Royal-blue, Turquoise-green, and Steel-blue color phenotypes; (E) the Red, Orange, and Yellow color phenotypes; and (F) the Solid and Mosaic color pattern phenotypes. The horizontal lines on Manhattan plots represent genome-wide (solid line) and suggestive genome-wide (dashed line) significance level, respectively. ns, not significant.
The most strongly associated variants cluster in introns of dmrt1 and kank1 (Fig. 2B). kank1 has a role in cytoskeleton formation by regulating actin polymerization (17), which makes it a less likely candidate for SD. dmrt1, in contrast, is a well-known gene contributing to the SD in fish (18), bird (19), and reptiles (20). In addition, mRNA sequencing shows that dmrt1 is highly expressed in testis but barely detectable in ovary (Fig. 2C). dmrt1 in the Betta fish is evolutionarily close to the medaka fish dmy gene (fig. S15), which was the first sex-determining gene identified in teleosts (21). The presence of males with homogametic female genotype (52 of 590; table S9) may suggest that environmental factors, such as temperature, could also play a role in SD similarly to many other fish (22). Other wild species, including B. imbellis, B. siamorientalis, B. mahachaiensis, B. smaragdina, B. smaragdina guitar, and B. stiktos, probably have a different SD mechanism, as both female and male individuals are homozygous for G allele at the lead single-nucleotide polymorphism (SNP) (table S9), and genome-wide association study (GWAS) with these individuals did not recover the signal at the dmrt1 locus (fig. S16).
Colors and color patterning in the Betta fish
Crossing Royal-blue (fig. S4E) male and female individuals produces Turquoise-green (fig. S4D), Royal-blue (fig. S4E), and Steel-blue (fig. S4F) offspring, at a Mendelian ratio of 1:2:1 (5). A previous study comparing the composition of pigment cells in scales among these breeds showed that the Steel-blue differed from the others by lacking erythrophores on the lower layer of scales (23). A GWA analysis coding Steel-blue (n = 35), Royal-blue (n = 28), and Turquoise-green (n = 26) as 1, 2, and 3, identified a single locus (Fig. 2D) at 8.96 to 9.19 Mb on chromosome 24, and GWAS with other coding schemes consistently mapped to the same location (fig. S17). Genotype analysis at the peak SNP (chr24:9,191,247) show that the Turquoise-green and Steel-blue are homozygous for different alleles, and 27 of the 28 the Royal-blue individuals are heterozygous (table S10), which is highly consistent with the codominant inheritance pattern. This locus contains 13 protein-coding genes (fig. S18) including mthfd1l (methylenetetrahydrofolate dehydrogenase 1 like). mthfd1l is involved in the synthesis of tetrahydrofolate (24), which is engaged in the de novo assembly of purines, a key component of iridophores that generate iridescent colors including blue and green colors (25). Therefore, mthfd1l is a promising candidate that warrants in-depth functional investigation.
The Copper breed (fig. S4I) of Betta fish has a characteristic metallic appearance on scales, and breeding records indicate that it is derived by introducing the “metallic gene” into the genetic background of Steel-blue breed (fig. S19) (26). We performed a GWAS using Copper as cases (n = 43) and Steel-blue as control (n = 35) and identified two most significant peaks (fig. S20). The strongest one overlaps with the locus underlying the Royal-blue, Steel-blue, and Turquoise-green color variation, and the other is located at 10.37 Mb of chromosome 5 (fig. S21) with all peak variants residing in the srgap3 gene (fig. S21), a cytoskeleton regulator (27). Genotype analysis at the lead SNP of this locus (table S11) shows that Steel-blue is homozygous for the T allele, while Copper individuals carry one or two copies of the alternative allele (C), consistent with the known inheritance pattern (fig. S19). Therefore, we consider the associated region a strong candidate for the hypothesized Mendelian metallic gene.
In teleosts, erythrophores and xanthophores produce red and yellow pigments, respectively, and orange color can be conferred by either pteridine component or a mixture of red and yellow pigments (28). The case-control GWAS contrasting the Orange breed (n = 28; fig. S4C) with the Red (n = 26; fig. S4A) and Yellow (n = 29; fig. S4B) breeds identified a major locus at position 5.83 Mb of chromosome 8, harboring 93 variants, all residing in the rnf213 gene (Fig. 2E and figs. S22 and S23), which is involved in angiogenesis and the noncanonical Wnt signaling pathway in vascular development (29). The lead variant is a 1-bp (A) insertion/deletion variation in intron of rnf213. The Red and Yellow breeds are homozygous for the T (deletion) allele, while the Orange breed has a high frequency of the TA (A insertion) allele (table S12).
The mosaic color pattern (fig. S4L) is a spectacular phenotype in the Siamese fighting fish with multiple commercial names, including koi, candy, galaxy, lemon, and marble. Although the mosaic color pattern has also been observed in koi carp (30) and medaka (31), the underlying molecular basis has not been investigated. Here, we performed GWAS with a case-control design between solid (n = 209) and mosaic colors (n = 55; fig. S4L) and identified nine associated loci on eight chromosomes, suggesting a polygenic basis underlying this phenotype (Fig. 2F). The two strongest signals are found in two adjacent peaks on chromosome 11. On the most significant locus (16.20 to 16.67 Mb; fig. S24A), we highlight slc39a7, chs3, chs8, and col11a2, which are related to pigmentation, and tubb, which is involved in intracellular pigment mobilization (32). In addition, the adjacent locus (13.71 to 14.93 Mb; fig. S24B) also contains pigmentation-related genes including plec, eppk1, slc17a5, and slc52a2. Notably, there are eight copies of plec, a hub gene in a gene coexpression network analysis for “Pink-dark green” color alteration in cavefish (33), in the association peak.
The Siamese fighting fish also show great diversity in their eye colors, and at least six color categories, including black, white, yellowish brown, yellow, light blue, and brown, can be visually identified. However, GWA mapping on each group did not reveal strongly associated loci (fig. S25), perhaps suggesting a complex genetic architecture or environmental cues underlying eye color variation.
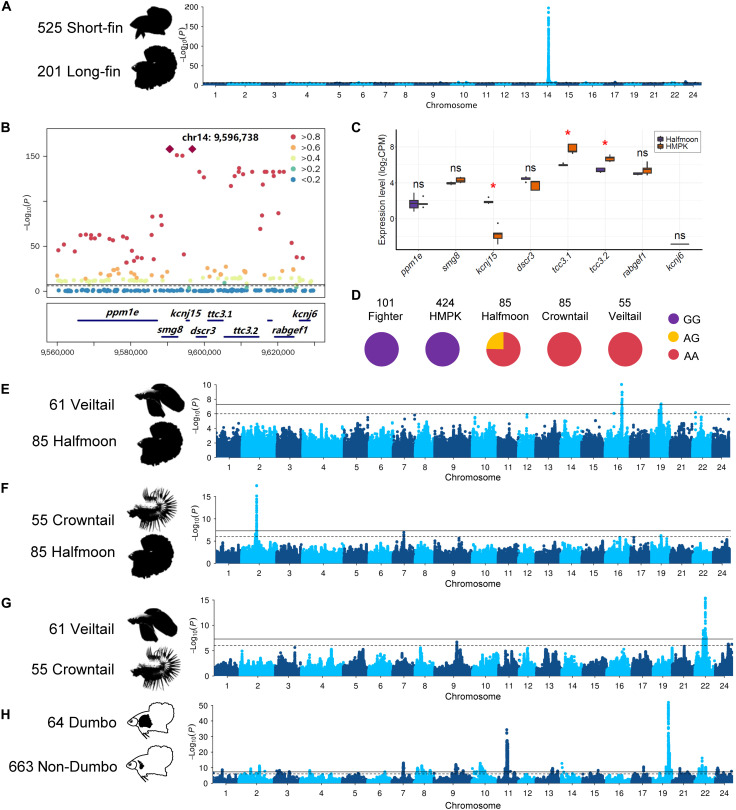
Gain of function of kcnj15 contributes to the overgrowth of fins
The most notable morphological difference among the Betta fish is in the fins, especially the caudal fin. Veiltail (n = 61; fig. S4Q), Crowntail (n = 55; fig. S4P), and Halfmoon (n = 85; fig. S4O) show a remarkable outgrowth in dorsal, anal, and caudal fins compared to the Fighter (n = 101; fig. S4R) and HMPK (n = 424; fig. S4) breeds (fig. S26). Interbreed crosses suggested a shared genetic basis for all long-fin varieties, and the long-fin phenotype was dominant over short-fin phenotype (34). To map the underlying genetic variation, we performed GWAS contrasting long-fin (case, n = 201) with short-fin individuals (controls, n = 525) (Fig. 3A and fig. S27). An extremely significant locus (P = 4.15 × 10−198) centered at position 9.60 Mb on chromosome 14 (Fig. 3A) was identified. There are three lead variants in complete linkage (Fig. 3B): one synonymous (9,590,677 bp, exon 3, C1551T), one intronic (9,590,546 bp, intron 2, G/A) located in smg8, and one in the 3′ untranslated region (3′UTR) of kcnj15 (9,596,738 bp, G/A). All three variants perfectly distinguish between long-fin and short-fin in domesticated individuals (Fig. 3D). The smg8 protein acts as a regulator of kinase activity involved in nonsense-mediated decay of mRNAs (35) and is an unlikely candidate of the long-fin phenotype. In contrast, kcnj15 encodes a potassium channel, which is a much better candidate, as several genes encoding potassium channels, including kcnk5b (36), kcnh2a (37), kcnj13 (38), and kcc4a (39), have been identified to cause various long-fin phenotypes in zebrafish. Moreover, when comparing the expression profiles of the two candidate genes in caudal fin between long-fin and short-fin individuals by RNA sequencing (RNA-seq), we found that the expression levels of smg8 were similar between different fish groups, while kcnj15 showed a significant difference as it is highly expressed in long-fin breeds (Fig. 3C and fig. S28A), and its transcripts were almost undetectable in short-fin breeds (Fig. 3C and fig. S28A).
Fig. 3. GWAS of fin morphology in B. splendens.
(A) Manhattan plot for the long-fin versus short-fin morphology. (B) LocusZoom plot for the most significant genome-wide association signal. (C) The expression profiles in the caudal fin for genes in the associated locus. The y axis is the normalized gene expression level measured using log2-transformed CPM reads. Expression differences between groups (n = 5 for each group) were tested using Mann-Whitney test, and red asterisks indicate adjusted P < 0.05 using the Benjamini-Hochberg method. (D) Genotype frequencies at the peak SNP in five different B. splendens breeds. (E to G) Manhattan plots of GWAS for different fin morphs among long-fin breeds. (H) Manhattan plot of GWAS for the Dumbo phenotype.
Most of the wild species, including B. splendens, B. imbellis, B. mahachaiensis, and B. stiktos, are considered short fin, and consistently, all these individuals are fixed for the G allele at the lead SNP in kcnj15, and few transcripts can be detected (table S13 and fig. S28B). In the B. smaragdina and B. smaragdina guitar populations, the variant is still segregating. In a heterozygous B. smaragdina individual, the predicted transcripts can be detected but with low expression levels (fig. S28B), which is probably caused by recent introgression from long-tail breeds during the in-captivity breeding effort in B. smaragdina. Given that the lead variant is located on a 3′UTR and the strong association between variant alleles and expression levels of kcnj15, we speculate that the variant might have affected the stability of the mRNA of kcnj15.
Veiltail, Crowntail, and Halfmoon phenotypes
Within the long-fin breeds, there is much additional phenotypic variation, with Veiltail, Crowntail, and Halfmoon breeds as some of the most distinctive breeds. Veiltail is the first long-fin breed recorded (8), while later selection for full 180° spread in caudal fin ray gave rise to the Halfmoon breed. The Crowntail breed differs from the Halfmoon breed by having reduced webbing tissue between the fin rays. GWAS contrasting Veiltail and Halfmoon identified a 300-kb region at 13.48 to 13.79 Mb on chromosome 16, possibly associated with the fin spread phenotype (Fig. 3E and fig. S29). In this locus, there are four genes (znf407, zadh2, tshz1, and znf516) that encode zinc finger proteins widely involved in transcriptional regulation and cellular functions (fig. S29) (40). GWAS comparing Halfmoon and Crowntail revealed a significant signal at 12.21 to 12.27 Mb on chromosome 2 (Fig. 3F and fig. S30), possibly associated with the webbing phenotype. Within the locus, a cluster of significantly associated variants is located in the intergenic region between a0zsk3 (neoverrucotoxin subunit α) and cep70 (centrosomal protein 70) (fig. S31), which do not have any function immediately connected to the phenotype, possibly suggesting a regulatory role of the underlying causal variant. Comparing the Crowntail and Veiltail breeds revealed a locus (8.25 to 8.40 Mb) on chromosome 22 (Fig. 3G and fig. S31) spanning eight genes including frmd6 (fig. S32), which is involved in actomyosin structure organization, and with loss of expression associated with epithelial-to-mesenchymal transition features (41), making this a strong candidate gene for explaining the phenotypic differences in fin morphology between the Crowntail and Veiltail breeds.
The “Dumbo” phenotype
The pectoral fin in teleosts is homologous to the anterior appendages in amphibians, reptiles, and mammals (42). One breed, the Dumbo, is characterized by the overgrowth of its paired pectoral fins, with more and elongated fin rays (Fig. 1B and fig. S4, M and N). GWAS using the Dumbo as cases (n = 64) and non-Dumbo as controls (n = 663) identified two strong signals (Fig. 3H), which are located on chromosomes 11 (8.87 to 9.72 Mb, P = 3.99 × 10−35; fig. S32) and 19 (4.25 to 4.47 Mb, P = 1.13 × 10−52; fig. S33), respectively. Wang et al. (10) investigated the same phenotype using an FST (genetic fixation index) scan with a smaller sample size (n = 47) and located a 1.3-Mb region on chromosome 11 (chromosome 9 in their assembly) that contains one of our GWAS peaks. They suggested, on the basis of gene expression analyses, that the causal gene could be kcnh8 (10). However, this gene resides outside of the associated locus on chromosome 11 identified in our study (fig. S32A). Moreover, no differential expression was detected in the pectoral fin tissue for kcnh8 when comparing Dumbo and non-Dumbo breeds (fig. S34). The lead SNP in our study is in a region containing a cluster of eight genes from the hoxa gene family that is essential for forming fin skeleton and digits in teleosts (43). For the signal on chromosome 19 (fig. S33B), one of the lead SNPs is located on the 3′UTR of fbxl15 (F-box and leucine-rich repeat protein 15), a gene involved in dorsal/ventral pattern formation and bone bass maintenance (44), rendering it a strong candidate gene.
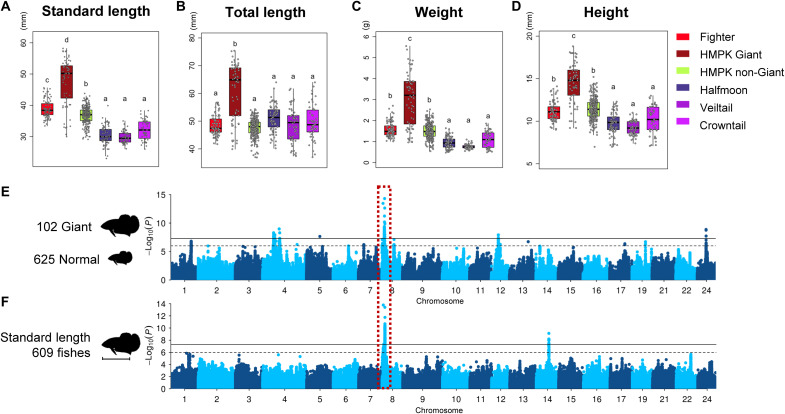
The Giant phenotype
Body size exhibits a polygenic inheritance in many organisms, including humans (45). In Betta fish, a Giant mutant shows significant body enlargement as indicated by the increase in total length, standard length (body length excluding tail), height, and weight compared to other breeds (Fig. 4, A to D). A case-control GWAS comparing Giant size with normal size breeds and GWAS using measurements of body length identified a shared significant locus at position 2.03 to 2.26 Mb of chromosome 8 (lead SNP chr8:2,130,270, P = 4.9 × 10−15 and 3.8 × 10−14), which explained 8.1 to 9.0% of phenotypic variance (Fig. 4, E and F). Scrutinizing the genes in the locus, we did not find a gene that encodes a global regulator of body development (figs. S35 and S43 and table S14), which is expected given the observation that the Giant mutation causes enlargement of many organs. Nevertheless, we highlight mrps34 and spsb3, which are associated with human height in the GWAS catalog (www.ebi.ac.uk/gwas/home).
Fig. 4. The phenotypic variation and GWAS of body size.
(A to D) Body measurements in five groups of the Betta fish. (E) The case-control GWAS comparing Giant and non-Giant individuals. (F) GWAS of standard body length in Betta fish. The shared genome-wide significant signal is highlighted with the red box.
Aggression
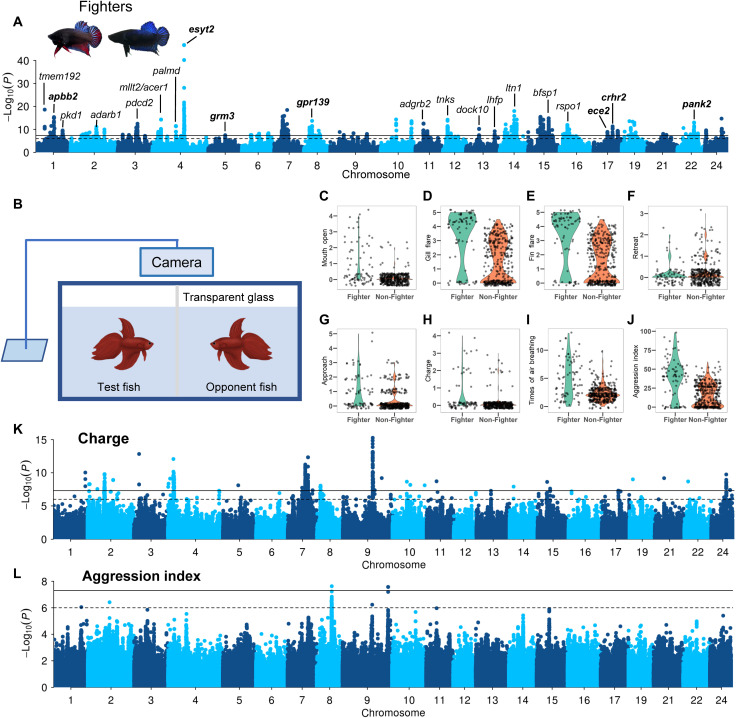
Aggressive behavior is a complex phenotype involving genetics, endocrinology, neurophysiology, and metabolism (46). Male winners of the Siamese fighting fish in one-versus-one fights have been selected by breeders for improved combating performance (47), thus producing the Fighter breed with stronger aggressiveness, longer fighting duration, and more active in motion. We performed a GWAS using the Fighters (n = 101) as cases and other breeds (n = 626) as controls and identified 36 association peaks distributed across 21 chromosomes (Fig. 5A, figs. S36 to S39, table S15), suggesting a polygenic basis for the behavioral differences between the Fighter and other breeds. The strongest association signal is found on chromosome 4, and the lead SNP (chr4:19,187,735, P = 1.83 × 10−47) is located in the vicinity of two copies of esyt2 (fig. S36), a promising candidate for the aggression phenotype as it is shown to promote neurotransmission and synaptic growth in Drosophila (48). We also highlight six other neural system–related genes that were tagged by the association peaks, including apbb2 (chr1:11,041,854, P = 4.08 × 10−16) that encodes β amyloid A4 precursor protein-binding family B member 2 protein and is associated with neurodegeneration and Alzheimer’s disease (49); pank2 (chr22:10,026,514, P = 9.69 × 10−14) that encodes pantothenate kinase and is associated with neurodegeneration and Parkinsonism (50); crhr2 (chr17:11,651,678, P = 4.79 × 10−12) that encodes corticotropin releasing hormone receptor 2 that mediates anxiety in mice (51); ece2 (chr17:8,494,762, P = 1.20 × 10−9) that encodes endothelin-converting enzyme-2 that regulates neurogenesis and neuronal migration in humans (52); gpr139 (chr8:4,905,640, P = 1.86 × 10−14) that encodes an orphan G protein–coupled receptor and is a central player in opioid modulation of brain circuits (53); and grm3 (chr5:10,257,908, P = 3.12 × 10−8) that is associated with bipolar disorder and schizophrenia (54, 55). Further investigation into these associations might provide mechanistic insights into the neural basis underlying aggressive behaviors.
Fig. 5. GWAS of aggression behaviors in B. splendens.
(A) GWAS of Fighter versus non-Fighter individuals. The genes tagged by lead SNPs in each peak are shown, and neural system–related genes are highlighted in bold. Two representative morphs of Fighter breed are shown as an inset. (B) Experimental setup to quantify the aggressiveness of B. splendens individuals. Aggressiveness indices of test fishes are recorded in the presence of an opponent fish. (C to J) Boxplots of eight aggressiveness phenotypes in Fighter versus non-Fighter individuals. (K and L) Manhattan plots of GWAS using the charge score and aggression index, respectively.
To study the behaviors associated with fighting, we phenotyped 10 different behaviors displayed during simulative fighting (Fig. 5B), including approach, charging, air breathing, gill flare, fin flare, jerk, mouth open, pacing, retreat, and shimmer (Fig. 5, C to J, and fig. S40), and performed association mapping to identify the genetic architecture for each behavior (Fig. 5, K and L, fig. S41, and table S16). A GWAS for the charging behavior during fighting (Fig. 5K) identified a strongly associated SNP (chr9:18,392,726, P = 5.36 × 10−16; fig. S42A) located close to two copies of gfra2, which plays a key role in the control of neuron survival and differentiation (53). Notably, the association signal is reproduced in GWAS for the mouth opening behavior (fig. S42B), suggesting a common genetic basis for these two correlated behaviors during fighting. We quantified the aggressiveness by synthesizing all the 10 behaviors in an aggression index (Fig. 5J) according to which the Fighters show 143% stronger aggression than the non-Fighters (P = 1.78 × 10−11, t test). GWAS on the aggression index identified a cluster of associated variants on chromosome 8, located on, or near, the atp5g2 gene, which is a subunit of mitochondrial adenosine triphosphate synthase (table S16) (56). Another signal on chromosome 9 tagged unc-13 homolog B gene (unc13b), which is associated with the risk of schizophrenia and partial epilepsy in humans (table S16) (57, 58).
DISCUSSION
The Betta fish has a complex domestication history involving introgression from other wild species and intensive selection on a variety of phenotypes. We here provide an extensive analysis of the history of domestication and the genetic basis of many phenotypes selected during domestication. A number of traits appear to have major effect loci, including the fin elongation, which we map to a locus containing kcnj15, and several coloration traits. The fact that this long-fin mutation is shared among Veiltail, Halfmoon, and Crowntail suggests perhaps that this mutation arose in their common ancestor before the mutations differentiating breeds arose. Presumably, these are traits that have been selected by breeders on the basis of individual de novo mutations that occurred in aquaculture and then were shared directly or indirectly among breeders. However, the behavioral traits associated with aggression show a more polygenic nature. We speculate that the heritability of these traits generally is more polygenic and selection affecting these traits may depend more on standing variations than de novo mutations of large effect, possibly due to constraints in the genetic architecture of these traits. We also found a major effect locus for SD, which we map to dmrt1. However, closely related wild species do not have the same sex-determining locus, suggesting that genetic SD has evolved de novo in B. splendens, making this species a great model system for understanding the evolution of genetic SD in vertebrates.
Many of our association mappings are based on a case-control design comparing different breeds, which shares some similarities with classical FST scans for identifying selection. The limitation of the design is that it confounds the interpretation of the association signal, as the associated loci identified are probably related to the focal phenotype that differentiates the case and control groups, but it can also be attributable to other cryptic phenotypes that are selected in the focal breed during domestication. Further studies are needed to establish more specific genotype-phenotype connections for these association signals.
Overall, our results demonstrate that the Betta fish is a great model for understanding the genetic basis of a host of different traits in vertebrates, including coloration and patterning, skeletal development of fin/limbs, and behavioral traits such as aggression. As an easy breeder in captivity, the Betta fish has substantial potential as a model system to augment existing vertebrate models such as zebrafish and mice.
MATERIALS AND METHODS
Sampling and phenotyping
All the 727 domesticated individuals in the study were bought from the Yuexiu pet market in Guangzhou, China. The 59 “wild” individuals from the B. splendens complex were brought from fish farms in Bangkok, Thailand, through commercial exporters, and these phenotypically wild individuals had been kept and bred in fish farms. Two female HMPK Siamese fighting fishes used for de novo genome assembly and annotation came from the same brood of an F2 crossbred between two inbred solid red individuals. All the management of fishes are under the approval of the Animals Care and Use Committee of Nanchang University, China. More details on the provenance of the samples can be found in text S2.
Body weight was measured with a digital scale, and body height, standard length, and total length of each fish were measured with digital calipers. Body color, eye color, and fin shape were visually phenotyped. Sex was determined by manual gonadal dissection. Fin shape was phenotyped on the basis of photographs (deposited in https://doi.org/10.6084/m9.figshare.14398565.v1) and categorized as long fin if their morphotypes were Veiltail, Halfmoon, or Crowntail or short fin if their morphotypes were HMPK or Fighter.
To quantify aggressiveness, we set up a simulative fighting experiment and recorded the behaviors of the test fish with a sport camera for 1 min. The experiment is performed in a fish tank equally divided into halves using a transparent glass plate (Fig. 5B). An opponent fish is put on one side of the tank, and the test fish is put on the other side of the tank. Before simulative fighting, an opaque plate was inserted between the two fishes, and all fishes were acclimated to the tank for at least 10 and up to 20 min. We excluded those individuals that did not seem to be acclimated to the tank, on the basis of observation of their behaviors, i.e., if the fishes were standing still or behaved nervously. Then, the opaque plate was removed, and the behaviors of the test fish were recorded for 1 min. In total, 467 individuals were recorded by video. On the basis of these videos, the aggressiveness of each fish is scored on a range of 0 to 5 on nine indices: charge, mouth open, gill flare, fin flare, shimmer, jerk, approach, pacing, and retreat, which are explained in detail below. The number of times of air breathing within 1 min was also counted for each fish. On the basis of the nine behaviors recorded, we assign an overall aggressiveness rating, i.e., aggression index, with the equation: charge (×5) + mouth open (×5) + gill flare (×4) + fin flare (×3) + shimmer (×2) + jerk (×2) + approach (×2) + pacing (×1) − retreat (×1). The nine indices composing the aggression index are defined as follows: charge: swims toward the transparent barrier rapidly and repeatedly; mouth open: opens mouth and locks jaw not for eating purposes; gill flare: operculum flare under extension; fin flare: raises the dorsal fin; shimmer: displays one side of the body with shimmering color within ~3 cm of the opponent; jerk: rapid twitching movement resulting in direction change; approach: swims in the direction of the opponent; pacing: circular swimming around the edge of the tank; and retreat: swims in the direction opposite to the opponent.
DNA extraction, library construction, and sequencing
Genomic DNA was extracted from fish fin clips that were harvested from anaesthetized fish and preserved in ethanol in a −80°C freezer, except for genome assembly, where muscle tissue was used to extract high–molecular weight DNA using a MagAttract HMW DNA kit (QIAGEN, Germany) from the two samples mentioned above, obtaining DNA fragments with an average length of ~100 kb. Specifically, for the two fish individuals, one fish sample was used for generating PacBio, Illumina, and 10x Genomics data and the other fish for Hi-C and BioNano data. For PacBio Sequel sequencing, two 20-kb-insert-size SMRTbell libraries were prepared and sequenced on the PacBio Sequel platform. For 10x Genomics sequencing, a GEM (gel bead-in emulsion) reaction and library preparation were conducted using size-selected DNA with a length of approximately 50 kb. For Illumina long-range paired-end sequencing, libraries were barcoded and paired-end sequenced with the Rapid method on an Illumina HiSeq X Ten platform. For BioNano optimal map construction, the library was constructed using the BspQl enzyme (New England Biolabs) with an appropriate label density (14.5 labels per 100 kb) to digest long-range DNA fragments. For Hi-C, the library was constructed following standard protocols (59), and the Hi-C sequencing libraries were amplified by polymerase chain reaction for 12 to 14 cycles before being sequenced on the Illumina HiSeq X Ten platform (paired-end, 150 bp). The detailed information regarding insert sizes, data output, and genome coverage is listed in table S1. For population genomic resequencing, genomic DNA was extracted from fin tissue using the proteinase-K/phenol-chloroform method. Sequencing libraries with insert size 350 bp were constructed and then sequenced on an Illumina HiSeq X Ten platform to generate ~5× coverage data for each fish with 150-bp paired-end reads. All the library preparation and sequencing procedures described here were conducted by Novogene Bioinformatics Institute (China).
Genome assembling and annotation
To generate a high-quality genome assembly of the Betta fish (B. splendens), we applied the PacBio long reads and Illumina short reads, assisted with long-range scaffolding techniques including Hi-C, 10x Genomics sequencing, and BioNano optical mapping. We used a customized pipeline to assemble and annotate the final reference genome of the Siamese fighting fish. Detailed information about the methods of genome assembling and annotation is presented in text S1.
Identification of orthologs in teleost
Gene families were identified by OrthoMCL (v1.4) (60). First, nucleotide and protein data of 13 species representative of different teleost families (Cyprinus carpio, Danio rerio, Sinocyclocheilus rhinocerous, Cynoglossus semilaevis, Oryzias latipes, Salmo salar, Gasterosteus aculeatus, Oreochromis niloticus, Takifugu rubripes, Paramormyrops kingsleyae, Ictalurus punctatus, Xiphophorus maculatus, and Lepisosteus oculatus) were downloaded from Ensembl (Release 70) and National Center for Biotechnology Information to coanalyze with the B. splendens genome assembly. The longest transcript of a gene was retained among the different alternative splicing transcripts, genes with ≤30 amino acids were discarded, and then an “all against all” BLASTP comparison was performed followed by filtering using “E-value ≤ 1E-07” cutoff. The blastp alignments were clustered using OrthoMCL (v1.4) (60) with a 1.5 inflation index. After clustering, 24,159 gene clusters and 465 single-copy orthologs were detected across the 14 teleost species including B. splendens.
Phylogenetic tree construction and divergence time estimation
The aforementioned 465 shared single-copy orthologs were used to estimate a teleost phylogeny. Coding sequences (CDSs) of these orthologs were aligned by MUSCLE (v3.7, “-maxiters 2”) (61). With these CDS alignments, a maximum likelihood phylogenetic tree was constructed using RAxML (v7.2.3, “-m GTRGAMMA -p 12345 -x 12345 -f ad”) (62). Then, the program MCMCTree of PAML (v4.5) (63) (http://abacus.gene.ucl.ac.uk/software/paml.html) was applied to estimate divergence times among 14 species with parameters “burn-in=100,000, sample-number=100,000, and sample-frequency=2.” Seven calibration points were selected from the TimeTree website (www.timetree.org) as normal priors to restrain the age of the nodes, including 76 to 111 Ma between X. maculatus and O. latipes, 87 to 151 Ma between O. niloticus and O. latipes, 101 to 136 Ma between T. rubripes and G. aculeatus, 88 to 114 Ma between C. semilaevis and G. aculeatus, 186 to 227 Ma between B. splendens and S. salar, 17 to 51 Ma between C. carpio and S. rhinocerous, and 87.4 to 124.7 Ma between D. rerio and S. rhinocerous.
Variant calling
Clean reads were mapped onto the newly generated fighting fish genome assembly with the Burrows-Wheeler Aligner (v0.7.8) (64). Duplicated reads were marked using the MarkDuplicates tool from the Picard software package (65) with default options. Local realignment around indels was performed using the IndelRealigner tool from the GATK software package (v3.3.0) (66). To prepare a genotype dataset for the GWAS, we called genotypes from the BAM files of the 727 domesticated individuals with ANGSD (version 0.929) (67). We included parameters “-uniqueOnly 1 -remove_bads 1 -minMapQ 30 -minQ 20 -only_proper_pairs 1” to only consider reads with high mapping quality and high-quality bases, “-minMaf 0.02” to include SNPs with minor allele frequency greater than 0.02, “-postCutoff 0.95 -geno_minDepth 1” to call genotypes for individual with at least one read and has posterior genotype probability greater than 0.95, and “-minInd 400” to include SNPs with at least 400 individuals who have nonmissing genotypes. The indels were called using SAMtools (68) “mpileup.” After genotype calling, we imputed the missing data with Beagle (version 4.1) (69). Population genetic analyses were performed on 410 individuals including 59 phenotypically wild individuals from the B. splendens complex and ~20 randomly selected individuals from each breed of the domesticated B. splendens. Admixture and PCA were based on genotype likelihoods that were generated using “-doGlf 2” command from ANGSD (version 0.929) (67). Only SNPs with minor allele frequency greater than 0.02 and missing data lower than 40% were included. TreeMix and maximum likelihood tree analyses were based on pseudo-haploid calls that were generated with “-doHaploCall 1” command from ANGSD by randomly sampling a read at each site.
Admixture and PCA
The admixture and PCA analyses were performed with the genotype likelihood dataset described above. The admixture analysis was conducted with PCAngsd (v.1.02) (70) with parameters “-minMaf 0.05 -admix -admix_alpha 50,” and for each K, we ran 10 random seeds and took the one with the highest likelihood. PCA was performed with the “eigen” function in R (version 3.6.3) (71) on the covariance matrix generated by PCAngsd (v1.02) (70).
Maximum likelihood phylogenetic tree
To construct the maximum likelihood tree, we concatenated the haploid calls at the SNP sites and constructed the tree with IQTREE (version 1.6.12) (72) with the parameter “-alrt 1000 -m GTR+ASC.”
TreeMix
Each species or breed was grouped as a population. The three outlier individuals in the admixture analysis, one each from Yellow, Royal-blue, and Black breeds, were removed from TreeMix analysis (73). The allele frequency of each group at each SNP site was estimated with haploid calls for each individual. TreeMix was run with “-k 500” to account for linkage disequilibrium, and B. smaragdina, B. smaragdina guitar, and B. stiktos were set as outgroups.
D statistics
The D statistics (ABBA-BABA) analysis was conducted using ANGSD (v0.929) (67) with the “-doAbbababa” argument on bam files with a block size of 500 Kb. This procedure is based on sampling single reads and is not subject to genotype calling errors. We randomly sample three individuals from each breed or species for the analysis.
Genome-wide association study
Genome-wide association mapping was conducted using GEMMA (v.0.96) (16) with a mixed linear model. The Wald test is used to determine the association strength (P values). The genome-wide significance level is set as 5 × 10−8, and the suggestive significance level is set as 1 × 10−6. The effect of population stratification was corrected with a kinship matrix and the first three principal components from the PCA. The sex information was also included as a covariate for all association mapping except the SD GWAS. The proportion of phenotypic variance explained for associations was calculated using the equation provided in the GEMMA manual and corrected for winner’s curse using the false discovery rate inverse quantile transformation method implemented in the R package “winnerscurse” (https://amandaforde.github.io/winnerscurse/) (74).
RNA sequencing
For the RNA-seq analysis, 2-month-old fish individuals were used. Four independent RNA libraries were prepared and sequenced: (i) Library of pooled RNA samples of 11 tissues (brain, liver, muscle, eye, skin, scale, fin, intestine, testis, ovary, and embryo) of one HMPK individual was constructed and subjected to both full-length transcriptome sequencing using the PacBio Sequel platform and short-reads sequencing with the Illumina HiSeq 2500 platform. (ii) Libraries of RNA samples of the caudal fin tissues from HMPK (n = 5), Halfmoon (n = 5), Crowntail (n = 1), Veiltail (n = 1), wild B. splendens (n = 1), B. imbellis (n = 1), B. mahachaiensis (n = 1), and B. smaragdina (n = 1) individuals were prepared and sequenced with the Illumina HiSeq 2500 platform. (iii) Libraries of RNA samples of the pectoral fin from Dumbo (n = 5) and non-Dumbo (n = 5) individuals were prepared and sequenced with the Illumina HiSeq 2500 platform. (iv) Libraries of ovary RNA samples from female (n = 5) and testis samples from male (n = 5) individuals were prepared and sequenced with the Illumina HiSeq 2500 platform.
The raw sequencing data were first processed by removing adapters and low-quality reads. The resulting clean paired-end reads were mapped to our assembly using Hisat2 (v.2.0.5) (75). Then, the read number mapped to each gene was counted using featureCounts (v1.5.0-p3) (76) before the FPKM (expected number of fragments per kilobase of transcript sequence per millions of base pairs) was calculated. Transcriptomic reads used for genome annotation were performed with the edgeR package (3.18.1) (77). Differential gene expression analysis was conducted with DESeq2 (1.26.0) (78). The P values were adjusted for multiple testing using the Benjamini-Hochberg method. Genes with adjusted P value < 0.05 and fold change greater than 2 were considered as significantly differentially expressed.
Acknowledgments
We thank R. Tang, G. Deng, and D. Xu for providing high-performance computing for this project. We also thank all the Betta fish hobbyists around the world who provide useful information to us. We acknowledge H. Haryono for advice in sample selection and J. Wu, S. Rain, H. Sutrisno, H. Haryanto, and B. Sun for logistic support. We are grateful to J. Dai and Y. Ji for technical support in sample processing and maintenance.
Funding: This study was financially supported by the earmarked fund from the Jiangxi Agricultural Research System (JXARS-10) and the Natural Science Foundation of Jiangxi Province (20202BAB215012).
Author contributions: R.N., W.Z., Y.H., and J.R. conceived and initiated the project. W.Z., H.W., and R.N. designed the project. W.Z., W.S., and S.K. collected all the samples. W.Z., B.H., J.S., K.P., D.Z., S.J., D.W., G.Z., J.W., and J.H.M.v.E. conducted the morphological measurements and examined the sample attributions. W.Z., M.W., H.L., S.G., B.S., and Q.Z. took all the photographs and videos. W.Z. recorded and analyzed the behavioral phenotypes. G.Z., D.Z., and B.S. carried out the experiments. W.Z. and H.W. performed the bioinformatics analyses. Y.L. conducted the RNA-seq analyses. W.Z., H.W., D.Y.C.B., and R.N. interpreted the results. W.Z., H.W., and D.Y.C.B. wrote the manuscript. R.N. and Y.H. revised the manuscript. All authors have read and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The PacBio sequencing, Illumina sequencing, BioNano, Hi-C, and pool RNA-seq data associated with the genome assembly and annotation are available at BioProject PRJNA629633. The BioNano maps are available as Supplementary Files (SUPPF_0000003658) at BioProject PRJNA629633. The population genomic resequencing data for 727 domesticated and 59 wild individuals are available at BioProject PRJNA689926 and PRJNA809835. The RNA-seq data associated with the short-fin and long-fin morphology, Dumbo phenotype, and SD are available at BioProject PRJNA809309. The photos of the 727 domesticated fish are available at figshare (https://doi.org/10.6084/m9.figshare.14398565.v1). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Texts S1 to S3
Figs. S1 to S43
Tables S1 to S16
References
REFERENCES AND NOTES
- 1.W. J. Rainboth, Fishes of the Cambodian Mekong (Food and Agriculture Organization, 1996). [Google Scholar]
- 2.H. M. Smith, L. P. Schultz, The Fresh-water Fishes of Siam, or Thailand (Smithsonian Institution, U.S. National Museum Bulletin, Government Printing Office, 1945). [Google Scholar]
- 3.Lissmann H.-W., Die umwelt des kampffisches (Betta splendens Regan). Z. Vgl. Physiol. 18, 65–111 (1932). [Google Scholar]
- 4.Goodrich H., Mercer R. N., Genetics and colors of the Siamese fighting, Betta splendens. Science 79, 318–319 (1934). [DOI] [PubMed] [Google Scholar]
- 5.Umrath K., Über die Vererbung der Farben und des Geschlechts beim Schleierkampffisch, Betta splendens. Z.Ver-erbungslehre 77, 450–454 (1939). [Google Scholar]
- 6.Eberhardt K., Die Vererbung der Farben bei Betta Splendens Regan. Z.Ver-erbungslehre 79, 548–560 (1941). [Google Scholar]
- 7.Eberhardt K., Geschlechtsbestimmung und -Differenzierung bei Betta Splendens Regan I. Z.Ver-erbungslehre 81, 363–373 (1943). [Google Scholar]
- 8.Eberhardt K., Ein Fall von geschlechtskontrollierter Vererbung bei Betta splendens Regan. Z. Indukt. Abstammungs. Vererbungsl. 81, 72–83 (1943). [Google Scholar]
- 9.Svärdson G., Wickbom T., The chromosomes of two species of Anabantidae (Teleostei), with a new case of sex reversal. Hereditas 28, 212–216 (1942). [Google Scholar]
- 10.Wang L., Sun F., Wan Z. Y., Ye B., Wen Y., Liu H., Yang Z., Pang H., Meng Z., Fan B., Alfiko Y., Shen Y., Bai B., Lee M. S. Q., Piferrer F., Schartl M., Meyer A., Yue G. H., Genomic basis of striking fin shapes and colours in the fighting fish. Mol. Biol. Evol. 38, 3383–3396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon Y. M., Vranken N., Hoge C., Lichak M. R., Norovich A. L., Francis K. X., Camacho-Garcia J., Bista I., Wood J., McCarthy S., Chow W., Tan H. H., Howe K., Bandara S., von Lintig J., Ruber L., Durbin R., Svardal H., Bendesky A., Genomic consequences of domestication of the Siamese fighting fish. Sci. Adv. 8, eabm4950 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Sun F., Wan Z. Y., Yang Z., Tay Y. X., Lee M., Ye B., Wen Y., Meng Z., Fan B., Alfiko Y., Shen Y., Piferrer F., Meyer A., Schartl M., Yue G. H., Transposon-induced epigenetic silencing in the X chromosome as a novel form of dmrt1 expression regulation during sex determination in the fighting fish. BMC Biol. 20, 5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grazyna F.-S., Fopp-Bayat D., Jankun M., Krejszeff S., Mamcarz A., Note on the karyotype and NOR location of Siamese fighting fish Betta splendens (Perciformes, Osphronemidae). Caryologia 61, 349–353 (2008). [Google Scholar]
- 14.Parra G., Bradnam K., Korf I., CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Zhou X., Stephens M., Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanzo R. J., Twede H., Ho K. S., Prasad A., Martin M. M., South S. T., Wassman E. R., Clinical significance of copy number variants involving KANK1 in patients with neurodevelopmental disorders. Eur. J. Med. Genet. 62, 15–20 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Martínez P., Viñas A. M., Sánchez L., Díaz N., Ribas L., Piferrer F., Genetic architecture of sex determination in fish: Applications to sex ratio control in aquaculture. Front. Genet. 5, 340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., Doran T. J., Sinclair A. H., The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Ge C., Ye J., Weber C., Sun W., Zhang H., Zhou Y., Cai C., Qian G., Capel B., The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 360, 645–648 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C., Shimizu A., Shan Z., Haaf T., Shimizu N., Shima A., Schmid M., Schartl M., A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. U.S.A. 99, 11778–11783 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ospina-Alvarez N., Piferrer F., Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLOS ONE 3, e2837 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Yang N., Jiang F., Huang H., Inheritance of body colors of Siamese fighting fishes of different strains. Chinese J. Trop. Agric. 34, 109–113 (2014). [Google Scholar]
- 24.Lee D., Xu I. M.-J., Chiu D. K.-C., Lai R. K.-H., Tse A. P.-W., Li L. L., Law C.-T., Tsang F. H.-C., Wei L. L., Chan C. Y.-K., Wong C.-M., Ng I. O.-L., Wong C. C.-L., Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J. Clin. Invest. 127, 1856–1872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagnara J. T., Matsumoto J., Ferris W., Frost S. K., Turner W. A. Jr., Tchen T. T., Taylor J. D., Common origin of pigment cells. Science 203, 410–415 (1979). [DOI] [PubMed] [Google Scholar]
- 26.J. H. M. v. Esch, Understanding metallic genetics (2008); http://www.bettaterritory.nl/BT-AABcoppergenetics.htm.
- 27.Bacon C., Endris V., Rappold G. A., The cellular function of srGAP3 and its role in neuronal morphogenesis. Mech. Dev. 130, 391–395 (2013). [DOI] [PubMed] [Google Scholar]
- 28.T. Hama, Chromatophores and iridocytes, in Medaka (Killifish): Biology and Strains, T. Yamamoto, Ed. (Keigaku Inc, 1975), pp. 138–153. [Google Scholar]
- 29.Liu W., Morito D., Takashima S., Mineharu Y., Kobayashi H., Hitomi T., Hashikata H., Matsuura N., Yamazaki S., Toyoda A., Kikuta K.-I., Takagi Y., Harada K. H., Fujiyama A., Herzig R., Krischek B., Zou L., Kim J. E., Kitakaze M., Miyamoto S., Nagata K., Hashimoto N., Koizumi A., Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLOS ONE 6, e22542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. Pietsch, P. Hirsch, Biology and Ecology of Carp (CRC Press, Taylor & Francis Group, 2015). [Google Scholar]
- 31.Tsutsumi M., Imai S., Kyono-Hamaguchi Y., Hamaguchi S., Koga A., Hori H., Color reversion of the albino medaka fish associated with spontaneous somatic excision of the Tol-1 transposable element from the tyrosinase gene. Pigment Cell Res. 19, 243–247 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Ahi E. P., Lecaudey L. A., Ziegelbecker A., Steiner O., Glabonjat R., Goessler W., Hois V., Wagner C., Lass A., Sefc K. M., Comparative transcriptomics reveals candidate carotenoid color genes in an East African cichlid fish. BMC Genomics 21, 54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C., Chen H., Zhao Y., Chen S., Xiao H., Comparative transcriptomics reveals the molecular genetic basis of pigmentation loss in Sinocyclocheilus cavefishes. Ecol. Evol. 10, 14256–14271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G. A. Lucas, “A study of variation in the Siamese fighting fish, Betta splendens, with emphasis on color mutants and the problem of sex determination,” thesis, Iowa State University (1968). [Google Scholar]
- 35.Zhu L., Li L., Qi Y., Yu Z., Xu Y., Cryo-EM structure of SMG1–SMG8–SMG9 complex. Cell Res. 29, 1027–1034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perathoner S., Daane J. M., Henrion U., Seebohm G., Higdon C. W., Johnson S. L., Nüsslein-Volhard C., Harris M. P., Bioelectric signaling regulates size in zebrafish fins. PLOS Genet. 10, e1004080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart S., Le Bleu H. K., Yette G. A., Henner A. L., Robbins A. E., Braunstein J. A., Stankunas K., longfin causes cis-ectopic expression of the kcnh2a ether-a-go-go K+ channel to autonomously prolong fin outgrowth. Development 148, dev199384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silic M. R., Wu Q., Kim B. H., Golling G., Chen K. H., Freitas R., Chubykin A. A., Mittal S. K., Zhang G., Potassium channel-associated bioelectricity of the dermomyotome determines fin patterning in zebrafish. Genetics 215, 1067–1084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanni J. S., Peal D., Ekstrom L., Chen H., Stanclift C., Bowen M. E., Mercado A., Gamba G., Kahle K. T., Harris M. P., Integrated K+ channel and K+Cl- cotransporter functions are required for the coordination of size and proportion during development. Dev. Biol. 456, 164–178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laity J. H., Lee B. M., Wright P. E., Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Angus L., Moleirinho S., Herron L., Sinha A., Zhang X., Niestrata M., Dholakia K., Prystowsky M. B., Harvey K. F., Reynolds P. A., Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene 31, 238–250 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Larouche O., Zelditch M. L., Cloutier R., Fin modules: An evolutionary perspective on appendage disparity in basal vertebrates. BMC Biol. 15, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura T., Gehrke A. R., Lemberg J., Szymaszek J., Shubin N. H., Digits and fin rays share common developmental histories. Nature 537, 225–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui Y., He S., Xing C., Lu K., Wang J., Xing G., Meng A., Jia S., He F., Zhang L., SCFFBXL15 regulates BMP signalling by directing the degradation of HECT-type ubiquitin ligase Smurf1. EMBO J. 30, 2675–2689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kemper K. E., Visscher P. M., Goddard M. E., Genetic architecture of body size in mammals. Genome Biol. 13, 244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niepoth N., Bendesky A., How natural genetic variation shapes behavior. Annu. Rev. Genomics Human Genet. 21, 437–463 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Ramos A., Gonçalves D., Artificial selection for male winners in the Siamese fighting fish Betta splendens correlates with high female aggression. Front. Zool. 16, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kikuma K., Li X., Kim D., Sutter D., Dickman D. K., Extended synaptotagmin localizes to presynaptic ER and promotes neurotransmission and synaptic growth in Drosophila. Genetics 207, 993–1006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Hollingworth P., Moore P., Foy C., Archer N., Powell J., Nowotny P., Holmans P., O’Donovan M., Tacey K., Doil L., van Luchene R., Garcia V., Rowland C., Lau K., Cantanese J., Sninsky J., Hardy J., Thal L., Morris J. C., Goate A., Lovestone S., Owen M., Williams J., Grupe A., Genetic association of the APP binding protein 2 gene (APBB2) with late onset Alzheimer disease. Human Mutat. 25, 270–277 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Seo J.-H., Song S.-K., Lee P. H., A novel PANK2 mutation in a patient with atypical pantothenate-kinase-associated neurodegeneration presenting with adult-onset parkinsonism. J. Clin. Neurol. 5, 192–194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kishimoto T., Radulovic J., Radulovic M., Lin C. R., Schrick C., Hooshmand F., Hermanson O., Rosenfeld M. G., Spiess J., Deletion of crhr2 reveals an anxiolytic role for corticotropin-releasing hormone receptor-2. Nat. Genet. 24, 415–419 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Buchsbaum I. Y., Kielkowski P., Giorgio G., O’Neill A. C., Di Giaimo R., Kyrousi C., Khattak S., Sieber S. A., Robertson S. P., Cappello S., ECE2 regulates neurogenesis and neuronal migration during human cortical development. EMBO Rep. 21, e48204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D., Stoveken H. M., Zucca S., Dao M., Orlandi C., Song C., Masuho I., Johnston C., Opperman K. J., Giles A. C., Gill M. S., Lundquist E. A., Grill B., Martemyanov K. A., Genetic behavioral screen identifies an orphan anti-opioid system. Science 365, 1267–1273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kandaswamy R., McQuillin A., Sharp S. I., Fiorentino A., Anjorin A., Blizard R. A., Curtis D., Gurling H. M. D., Genetic association, mutation screening, and functional analysis of a Kozak sequence variant in the metabotropic glutamate receptor 3 gene in bipolar disorder. JAMA Psychiatry. 70, 591–598 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Egan M. F., Straub R. E., Goldberg T. E., Yakub I., Callicott J. H., Hariri A. R., Mattay V. S., Bertolino A., Hyde T. M., Shannon-Weickert C., Akil M., Crook J., Vakkalanka R. K., Balkissoon R., Gibbs R. A., Kleinman J. E., Weinberger D. R., Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 101, 12604–12609 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyer M. R., Walker J. E., Sequences of members of the human gene family for the c subunit of mitochondrial ATP synthase. Biochem. J. 293, 51–64 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Egawa J., Hoya S., Watanabe Y., Nunokawa A., Shibuya M., Ikeda M., Inoue E., Okuda S., Kondo K., Saito T., Kaneko N., Muratake T., Igeta H., Iwata N., Someya T., Rare UNC13B variations and risk of schizophrenia: Whole-exome sequencing in a multiplex family and follow-up resequencing and a case-control study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 797–805 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Wang J., Qiao J.-D., Liu X.-R., Liu D.-T., Chen Y.-H., Wu Y., Sun Y., Yu J., Ren R.-N., Mei Z., Liu Y.-X., Shi Y.-W., Jiang M., Lin S.-M., He N., Li B., Bian W.-J., Li B.-M., Yi Y.-H., Su T., Liu H.-K., Gu W.-Y., Liao W.-P., UNC13B variants associated with partial epilepsy with favourable outcome. Brain 144, 3050–3060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belton J.-M., McCord R. P., Gibcus J. H., Naumova N., Zhan Y., Dekker J., Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L., Stoeckert C. J., Roos D. S., OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Li H., Durbin R., Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broad Institute, Picard toolkit (2019).
- 66.R. Poplin, V. Ruano-Rubio, M. A. DePristo, T. J. Fennell, M. O. Carneiro, G. A. Van der Auwera, D. E. Kling, L. D. Gauthier, A. Levy-Moonshine, D. Roazen, K. Shakir, J. Thibault, S. Chandran, C. Whelan, M. Lek, S. Gabriel, M. J. Daly, B. Neale, D. G. MacArthur, E. Banks, Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178 [Preprint]. 24 July 2018. 10.1101/201178. [DOI]
- 67.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browning B. L., Browning S. R., Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98, 116–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meisner J., Albrechtsen A., Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
- 72.Nguyen L.-T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pickrell J., Pritchard J., Inference of population splits and mixtures from genome-wide allele frequency data. PLOS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmer C., Pe’er I., Statistical correction of the winner’s curse explains replication variability in quantitative trait genome-wide association studies. PLOS Genet. 13, e1006916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marçais G., Kingsford C., A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chin C. S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., Clum A., Dunn C., O’Malley R., Figueroa-Balderas R., Morales-Cruz A., Cramer G. R., Delledonne M., Luo C., Ecker J. R., Cantu D., Rank D. R., Schatz M. C., Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E. E., Turner S. W., Korlach J., Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C. A., Zeng Q., Wortman J., Young S. K., Earl A. M., Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roach M. J., Schmidt S. A., Borneman A. R., Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19, 460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adey A., Kitzman J. O., Burton J. N., Daza R., Kumar A., Christiansen L., Ronaghi M., Amini S., Gunderson K. L., Steemers F. J., Shendure J., In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res. 24, 2041–2049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lam E. T., Hastie A., Lin C., Ehrlich D., Das S. K., Austin M. D., Deshpande P., Cao H., Nagarajan N., Xiao M., Kwok P. Y., Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30, 771–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burton J. N., Adey A., Patwardhan R. P., Qiu R., Kitzman J. O., Shendure J., Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benson G., Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.A. F. Smit, R. Hubley, RepeatModeler Open (2008).
- 89.Xu Z., Wang H., LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Price A. L., Jones N. C., Pevzner P. A., De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005). [DOI] [PubMed] [Google Scholar]
- 91.A. Smit, R. Hubley, P. Green, RepeatMasker Open v4.0 (2013).
- 92.Bao W., Kojima K. K., Kohany O., Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.A. Smit, R. Hubley, P. Green, RepeatMasker Open v3.0 (2004).
- 94.Chan P. P., Lowe T. M., tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nawrocki E. P., Kolbe D. L., Eddy S. R., Infernal 1.0: Inference of RNA alignments. Bioinformatics 25, 1335–1337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stanke M., Schöffmann O., Morgenstern B., Waack S., Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Salamov A. A., Solovyev V. V., Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parra G., Blanco E., Guigo R., GeneID in Drosophila. Genome Res. 10, 511–515 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Majoros W. H., Pertea M., Salzberg S. L., TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Korf I., Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Birney E., Clamp M., Durbin R., GeneWise and genomewise. Genome Res. 14, 988–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haas B. J., Delcher A. L., Mount S. M., Wortman J. R., Smith R. K. Jr., Hannick L. I., Maiti R., Ronning C. M., Rusch D. B., Town C. D., Salzberg S. L., White O., Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haas B. J., Salzberg S. L., Zhu W., Pertea M., Allen J. E., Orvis J., White O., Buell C. R., Wortman J. R., Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Apweiler R., Bairoch A., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O’Donovan C., Redaschi N., Yeh L.-S. L., UniProt: The universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S.-Y., Lopez R., Hunter S., InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pleeging C. C. F., Moons C. P. H., Potential Welfare issues of the Siamese fighting fish (Betta splendens) at the retailer and hobbyist aquarium. Vlaams Diergeneeskundig Tijdschrift 86, 213–223 (2017). [Google Scholar]
- 109.Lipson M., Loh P.-R., Levin A., Reich D., Patterson N., Berger B., Efficient moment-based inference of admixture parameters and sources of gene flow. Mol. Biol. Evol. 30, 1788–1802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hao Z., Lv D., Ge Y., Shi J., Weijers D., Yu G., Chen J., RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 6, e251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fan G., Chan J., Ma K., Yang B., Zhang H., Yang X., Shi C., Chun-Hin Law H., Ren Z., Xu Q., Liu Q., Wang J., Chen W., Shao L., Gonçalves D., Ramos A., Cardoso S. D., Guo M., Cai J., Xu X., Wang J., Yang H., Liu X., Wang Y., Chromosome-level reference genome of the Siamese fighting fish Betta splendens, a model species for the study of aggression. Gigascience 7, giy087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rhie A., McCarthy S. A., Fedrigo O., Damas J., Formenti G., Koren S., Uliano-Silva M., Chow W., Fungtammasan A., Kim J., Lee C., Ko B. J., Chaisson M., Gedman G. L., Cantin L. J., Thibaud-Nissen F., Haggerty L., Bista I., Smith M., Haase B., Mountcastle J., Winkler S., Paez S., Howard J., Vernes S. C., Lama T. M., Grutzner F., Warren W. C., Balakrishnan C. N., Burt D., George J. M., Biegler M. T., Iorns D., Digby A., Eason D., Robertson B., Edwards T., Wilkinson M., Turner G., Meyer A., Kautt A. F., Franchini P., Detrich H. W. III, Svardal H., Wagner M., Naylor G. J. P., Pippel M., Malinsky M., Mooney M., Simbirsky M., Hannigan B. T., Pesout T., Houck M., Misuraca A., Kingan S. B., Hall R., Kronenberg Z., Sović I., Dunn C., Ning Z., Hastie A., Lee J., Selvaraj S., Green R. E., Putnam N. H., Gut I., Ghurye J., Garrison E., Sims Y., Collins J., Pelan S., Torrance J., Tracey A., Wood J., Dagnew R. E., Guan D., London S. E., Clayton D. F., Mello C. V., Friedrich S. R., Lovell P. V., Osipova E., Al-Ajli F. O., Secomandi S., Kim H., Theofanopoulou C., Hiller M., Zhou Y., Harris R. S., Makova K. D., Medvedev P., Hoffman J., Masterson P., Clark K., Martin F., Howe K., Flicek P., Walenz B. P., Kwak W., Clawson H., Diekhans M., Nassar L., Paten B., Kraus R. H. S., Crawford A. J., Gilbert M. T. P., Zhang G., Venkatesh B., Murphy R. W., Koepfli K.-P., Shapiro B., Johnson W. E., Palma F. D., Marques-Bonet T., Teeling E. C., Warnow T., Graves J. M., Ryder O. A., Haussler D., O’Brien S. J., Korlach J., Lewin H. A., Howe K., Myers E. W., Durbin R., Phillippy A. M., Jarvis E. D., Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prost S., Petersen M., Grethlein M., Hahn S. J., Kuschik-Maczollek N., Olesiuk M. E., Reschke J.-O., Schmey T. E., Zimmer C., Gupta D. K., Schell T., Coimbra R., De Raad J., Lammers F., Winter S., Janke A., Improving the chromosome-level genome assembly of the Siamese fighting fish (Betta splendens) in a university master’s course. G3 (Bethesda) 10, 2179–2183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Texts S1 to S3
Figs. S1 to S43
Tables S1 to S16
References