Abstract
Organoids serve as a novel tool for disease modeling in three-dimensional multicellular contexts. Static organoids, however, lack the requisite biophysical microenvironment such as fluid flow, limiting their ability to faithfully recapitulate disease pathology. Here, we unite organoids with organ-on-a-chip technology to unravel disease pathology and develop therapies for autosomal recessive polycystic kidney disease. PKHD1-mutant organoids-on-a-chip are subjected to flow that induces clinically relevant phenotypes of distal nephron dilatation. Transcriptomics discover 229 signal pathways that are not identified by static models. Mechanosensing molecules, RAC1 and FOS, are identified as potential therapeutic targets and validated by patient kidney samples. On the basis of this insight, we tested two U.S. Food and Drug Administration–approved and one investigational new drugs that target RAC1 and FOS in our organoid-on-a-chip model, which suppressed cyst formation. Our observations highlight the vast potential of organoid-on-a-chip models to elucidate complex disease mechanisms for therapeutic testing and discovery.
Organoid-on-a-chip models unravel kidney disease pathology and bridge bench-to-bedside translation for drug discovery.
INTRODUCTION
Recent advances in stem cell biology have enabled the generation of mini-organs, known as organoids (1–3). Human organoids recapitulate cell types and three-dimensional (3D) tissue architecture found in human organs and have emerged as new platforms for disease modeling in translational medicine (4–6). However, there are still many challenges in replicating the complex physiology of native organs using static organoids, especially for modeling human diseases that require appropriate external stimuli from cellular microenvironments.
Autosomal recessive polycystic kidney disease (ARPKD) is a ciliopathy that develops a severe hepatorenal disorder characterized by enlarged kidneys with progressive loss of renal function. Polycystic kidney and hepatic disease 1 (PKHD1) is a causative gene for typical cases of ARPKD that encodes fibrocystin, which localizes to the primary cilia, basal bodies, and apical membrane in kidney and liver epithelial cells (7). The reported mortality rate in neonates is as high as 30% (8, 9), and 41% of ARPKD patients who survive infancy will require renal replacement therapy by 11 years of age (10). Despite substantial progress in understanding this disease, there is no U.S. Food and Drug Administration (FDA)–approved drug for these patients.
Limited treatment options may be attributed to the lack of physiologically relevant models of ARPKD. ARPKD cysts develop in the distal tubules and collecting ducts in humans (11). By contrast, a renal cystic phenotype is often absent in mouse models with Pkhd1−/− and, when present, is mild and progresses slowly in proximal tubule segments (12). The PCK (polycystic kidney) rats generally used to model ARPKD develop cysts in distal nephrons; however, the cyst progression is slow and focal, unlike human ARPKD, but rather similar to human autosomal dominant PKD (ADPKD) (11, 13). To overcome the limitations of animal models in clinical translation, organ-on-a-chip models are evolving for studying human cell biology under physiological conditions. To date, proximal tubule-on-a-chip models have been developed in planar (14–16) and 3D geometries (15, 16). However, since they are composed of proximal tubule cells seeded onto either a porous membrane or adherent extracellular matrix (ECM) housed within prefabricated microchannels, they cannot adequately recapitulate tubular enlargement in PKD. Hence, we hypothesized that the organoid-on-a-chip platform uniting an organoid model with organ-on-a-chip technology may provide appropriate dynamic cellular microenvironments to simulate patient kidney phenotypes and shed light on finding disease-specific therapeutic targets that remain unidentified in static models for clinical trials in patients with ARPKD.
RESULTS
Fluidic culture induces cysts in distal nephrons in PKHD1−/− organoids
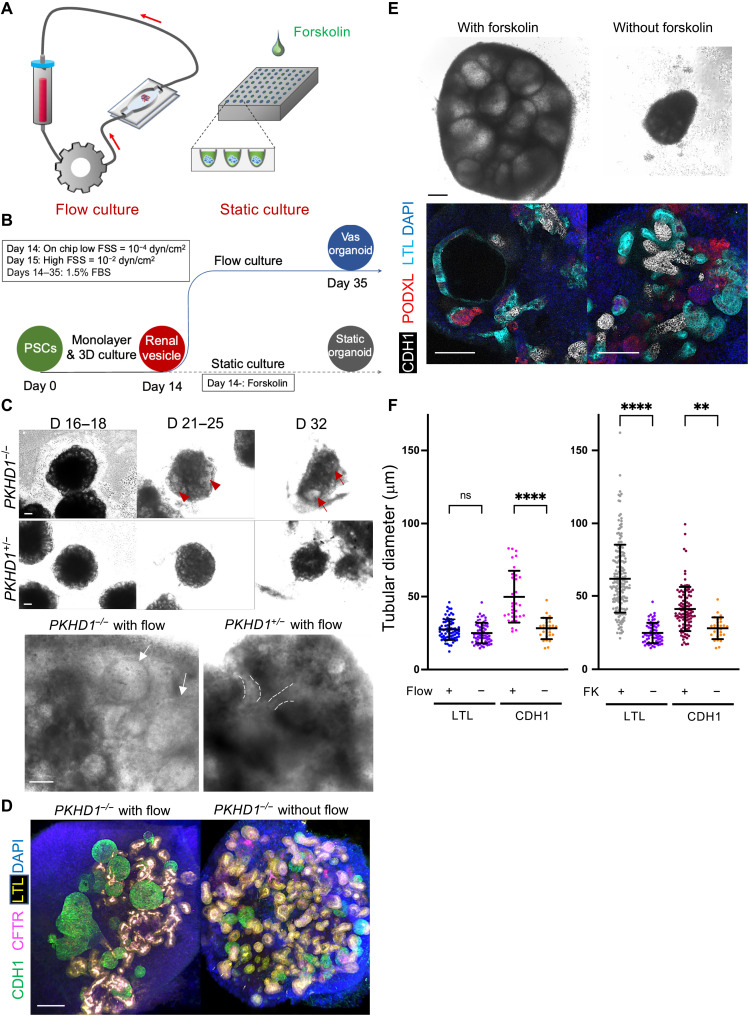
A prior ARPKD organoid model was developed using patient-derived induced pluripotent stem cells (iPSCs) (17); however, cysts were not formed in static culture without cyclic adenosine monophosphate (cAMP) stimulation. When treated with cAMP inducers, forskolin, or 8-bromoadenosine 3′,5′-cyclic monophosphate (8-Br-cAMP), LTL+ proximal tubules formed cystic structures, while CDH1+ distal nephrons rarely developed cysts. We also confirmed this nonphysiological result using forskolin, which induced cyst formation in LTL+ proximal tubules in static organoids derived from ARPKD patient’s iPSCs (fig. S1, A to G). However, ARPKD patients typically develop cysts in distal nephrons consisting of distal tubules and connecting tubules/collecting ducts (fig. S2) (11), which arise in part due to flow-mediated mechanosensitive signaling (18). Hence, we posit that creating an ARPKD organoid-on-a-chip model that enables fluid flow akin to the native tissue microenvironment may yield clinically relevant phenotypes that recapitulate cystogenesis occurring in ARPKD patients.
To test the effects of flow-induced signals on cystogenesis in PKHD1 mutants, we used 3D-printed millifluidic chips (19). Kidney organoids at the renal vesicle stage on day 14 of differentiation are transferred onto a gelatin-fibrin (gelbrin) matrix contained within each chip and then subjected to fluid flow from day 14 to day 35 (Fig. 1, A and B) (1). For perfusion experiments, we used a parental human embryonic stem (ES) line, H9, and its CRISPR-mutant lines with PKHD1 hetero/homozygous mutations to correlate genotypes with phenotypes (fig. S3, A to D). Cyst-like structures formed and gradually enlarged over time in PKHD1−/− organoids, whereas PKHD1+/− organoids did not exhibit such structures in the same fluidic culture (Fig. 1C, fig. S4A, and movie S1). For unbiased quantitative assessment of tubular enlargement, z-stack images are acquired from the top to bottom of each organoid after whole-mount immunostaining for LTL and CDH1 with sample clearing in flow- or forskolin-treated PKHD1−/− organoids (Fig. 1, D and E). Short diameters of each proximal tubule (LTL+CDH−) and distal nephron (LTL−CDH+) are measured (fig. S5). We find that the proximal tubular diameter in PKHD1−/− organoids subjected to fluid flow remains unchanged, while the distal nephron diameter increases from 33.9 ± 12.0 μm to 49.8 ± 17.9 μm (P = 0.0001), consistent with segment specificity in ARPKD patients (Fig. 1F, left, and fig. S4B). By contrast, forskolin increases the diameter in both LTL+CDH− and LTL−CDH1+ tubules (Fig. 1F, right, and fig. S3, E to H). To further validate whether our organoid-on-a-chip model recapitulates human ARPKD, we evaluated cAMP and mammalian target of rapamycin (mTOR) signaling, both of which have been implicated in ARPKD cystogenesis (20, 21). cAMP levels in PKHD1−/− organoids are significantly higher in organoids subjected to flow compared to those maintained under static conditions (fig. S4C). Flow also activates the mTOR pathway, as assessed by increased phospho-S6 ribosomal protein (pS6RP) in PKHD1−/− organoid compared to PKHD1+/− (fig. S4D). These findings suggest that the flow-induced ARPKD model recapitulates the requisite pathogenesis to enable identification of therapeutic targets.
Fig. 1. CDH1+ distal nephron dilates in PKHD1−/− kidney organoids cultured under flow.
(A) Schematic of kidney organoid-on-a-chip (flow) culture and static culture. (B) Diagram of differentiation days and culture conditions for flow and static culture. (C) Representative phase-contrast images of PKHD1−/− and PKHD1+/− organoids cultured under flow on days 16 to 18, days 21 to 25, and day 32 of differentiation. Red arrows highlight dilated tubules. Scale bars, 100 μm (top). Representative phase-contrast images of PKHD1−/− and PKHD1+/− organoids cultured under flow on day 35 of differentiation. White arrows highlight dilated tubules. Dashed white lines highlight normal tubules. Scale bar, 50 μm (bottom). (D) Immunocytochemistry with sample clearing for proximal tubule (LTL and CFTR) and distal tubule (CDH1) in PKHD1−/− organoids with and without flow on day 35. Scale bar, 100 μm. (E) Bright-field images of patient-derived ARPKD organoids with and without forskolin treatment in 3D culture on day 66. Scale bar, 250 μm (top). Immunocytochemistry for proximal tubule (LTL), distal tubule (CDH1), and podocyte (PODXL) in PKHD1−/− organoids with and without forskolin treatment on day 35. Scale bar, 100 μm (bottom). (F) Tubular diameters in nephron segments [proximal tubule (LTL) and distal tubule (CDH1)] in PKHD1−/− organoids with and without flow (left) and with and without forskolin treatment (right). Values represent means ± SD. **P < 0.01, ****P < 0.0001.
Mechanosensing signals are activated by fluid flow
Conflicting data exist on whether ciliary signals facilitate cystogenesis in ARPKD patients (22, 23). Loss of cilia suppressed renal cyst growth in rodent ADPKD models (24, 25). By contrast, disruption of cilia in Pkhd1-mutant mice did not suppress liver cyst formation or progression, calling into question whether primary cilia are implicated in ARPKD pathogenesis (26). To determine whether ciliary stress is involved in cyst formation in our model, we carried out microarray analysis using 3D Gene Human Oligo chips. Control organoids derived from PKHD1+/− mutants are subjected to fluid flow on chip from day 14 of differentiation and harvested on day 35. Direct comparison between flow and static culture conditions in control organoids revealed more mature nephron structures developed under flow, as demonstrated by a significant enrichment of hallmark gene sets for each segment, including proximal tubules, distal tubules, and cilia (fig. S6A). Differential expression gene (DEG) analysis identified 655 up-regulated and 225 down-regulated genes compared to the static control (Fig. 2A, top, and data S1). Genes up-regulated by flow are implicated in 111 gene ontology (GO) terms that are involved in cilia-related signals, including calcium and cAMP signals (Fig. 2A, bottom, and data S2 and S6) (27), suggesting that genes associated with the ciliary signals are activated in our kidney organoid-on-a-chip model.
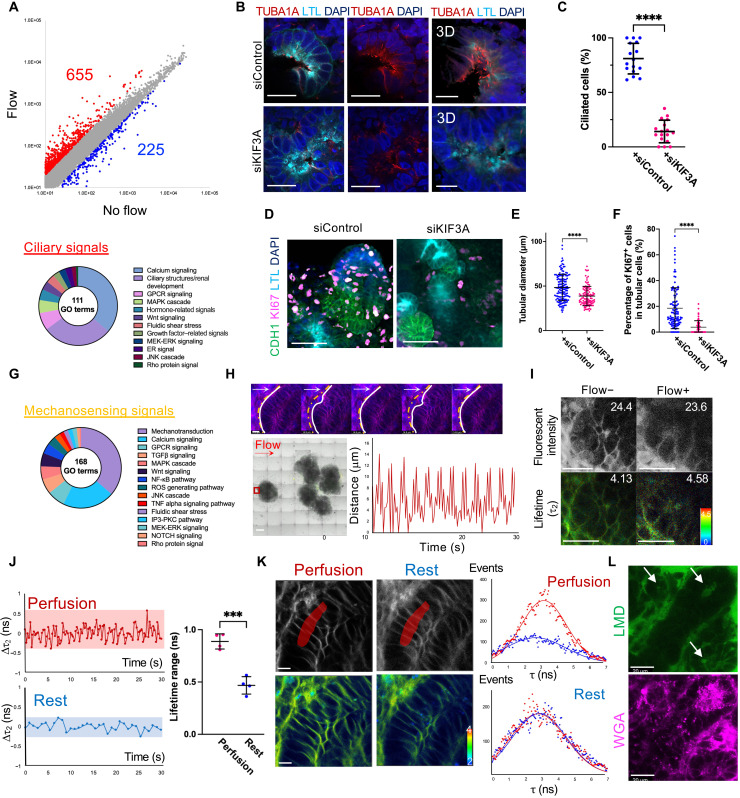
Fig. 2. Mechanosensing signals induced by flow.
(A) Scatterplots of gene expression in PKHD1+/− organoids under flow and no flow. Red spots: higher than twofold. Blue spots: lower than twofold. Disposition of GO terms related to ciliary signals (bottom). (B) Cross-sections (left and middle) and 3D rendering images (right) showing the ciliary marker TUBA1A in PKHD1−/− organoids transfected with siControl and siKIF3A under the flow condition on day 22. Scale bars, 20 μm (left and middle) and 10 μm (right). (C) Percentage of ciliated cells. (D) Immunocytochemistry for LTL, CDH1, and KI67 in PKHD1−/− organoids on day 22. Scale bars, 50 μm. (E) Tubular diameters in CDH1+ distal nephrons. (F) Percentage of KI67+ cells in CDH1+ distal nephrons. (G) Disposition of GO terms related to mechanosensing signals. (H) Time-lapse images of control organoids under flow. White lines: tubular basement membranes. Dashed yellow lines: the initial position of tubular basement membranes. White and red arrows: flow direction. Scale bar, 10 μm (top). Red square: imaged area. Scale bar, 500 μm (bottom left). Distance change of basement membranes from the initial position during flow (bottom right). (I) Fluorescent intensity and FLIM images in a tubule with and without flow. Values of mean fluorescent intensity and fitted τ2 lifetime are shown. Scale bars, 25 μm. (J) τ2 lifetime change (left) and lifetime ranges (right) during perfusion and rest. (K) Fluorescence (top left) and FLIM (bottom left) images of a tubule during perfusion and rest. Histograms (right) of overall lifetimes measured in the area highlighted by red. Scale bar, 10 μm. Red and blue curves indicate lifetimes of the highest and lowest, respectively, during the perfusion or rest. (L) Dextran perfusion imaging under flow. White arrow: tubular lumen filled with dextran. Scale bar, 20 μm.
Next, we used small interfering RNA (siRNA) to knock down primary cilia by targeting KIF3A that encodes one of the intraflagellar transports (24). siKIF3A is transfected twice on days 8 and 16 of differentiation, and PKHD1−/− organoids are subjected to flow from day 14 until the samples are harvested on day 22. A significant reduction of ciliated tubular cells from 81.0 ± 14.0% to 14.1 ± 10.4% is confirmed by z-stack imaging for acetylated α-tubulin (TUBA1A) (Fig. 2, B and C). While marked cyst formation is not observed in both siKIF3A and scrambled siRNA (siControl) samples at this early stage on day 22 (fig. S6B), ciliary knockdown significantly reduced KI67+ proliferating tubular cells from 18.5 ± 15.9% to 3.8 ± 5.0% in PKHD1−/− organoids with a significant reduction of tubular diameter compared to siControl under flow (Fig. 2, D to F). Together with GO term analysis, these results suggest the involvement of ciliary stress in cystogenesis in ARPKD organoids cultured under flow.
While the primary cilia serve as key mechanosensors, other cellular mechanosensing mechanisms such as the cytoskeleton-nucleus axis are also involved in the conversion of physical forces into biochemical and electrical signals for cellular responses (28). We find that fluid flow activated 168 GO terms related to mechanosensing signals in kidney organoids (Fig. 2G and data S2 and S6), including 61 GO terms related to cell adhesion, actin cytoskeleton, and membrane tension (29). Because flow is applied across whole kidney organoids in our model, we hypothesized that the induced mechanical stimuli might lead to a change in membrane tension within the organoids (30).
To determine whether cellular membrane tension is altered by mechanical forces in the kidney organoid-on-a-chip model, we used the fluorescent lipid tension reporter (FliptR) to visualize plasma membranes in live cells via fluorescent intensity. Simultaneously, membrane tension changes can be detected by fluorescence lifetime imaging microscopy (FLIM), where longer lifetimes (τ2) of the FliptR dye signify higher tension (31). Wild-type organoids cultured under flow are stained with FliptR and a live nuclear dye between days 16 and 21. Tubular nuclei and membranes are then imaged in live organoids using confocal microscopy, which revealed oscillatory motion of tubular basement membranes under fluidic shear stress (Fig. 2H; fig. S6, C to E; and movie S2) (19, 32). The FliptR lifetimes are captured first under superfusive flow and then after 1 hour of static conditions in the same tubular structures. The range of longer lifetime changes (Δτ2) in perfusion condition (0.89 ± 0.07 ns) is broader than in static condition (0.47 ± 0.08 ns), while the fluorescent intensity of FliptR is unchanged (Fig. 2, I and J, and fig. S6F). Notably, the overall lifetime distribution in cellular membranes is increased from 2.67 ± 1.81 ns to 3.17 ± 1.38 ns during flow, whereas it did not change during the rest period (2.77 ± 1.68 ns to 2.86 ± 1.65 ns) (Fig. 2K and fig. S6G). These data suggest that fluidic culture induces cellular stretching in organoid tubules, which may provide more physiological platforms for ARPKD modeling than those cultured under static conditions (33).
Next, we evaluate luminal flow by time-lapse live imaging of organoids-on-a-chip after live staining for fluorescently labeled wheat germ agglutinin (WGA) that binds to tubular epithelial membranes. Fluorescently labeled low–molecular weight dextran (LMD) is then slowly perfused by gravity to minimize the motion of tubules during live imaging. We initially observe LMD signals in the glomerular-like structures in 11 min of perfusion that are subsequently detected in the tubular lumens within 15 min after perfusion is initiated (Fig. 2L and fig. S7). These results suggest the presence of tubular luminal flow that activates mechanosensing in organoid tubules together with the interstitial flow. Although determining the influx route into the tubular lumens would require further study using reporter lines and high-resolution live imaging, we speculate that luminal flow may be initiated from glomerular vascular poles as LMD is first detected in those regions. We suspect that the glomerular vascular clefts may not fully closed at this early developmental stage. Given their flow-induced tubular enlargement, we posit that mechanosensing signals induced by flow might be essential for ARPKD pathogenesis.
Identification of novel therapeutic targets for ARPKD
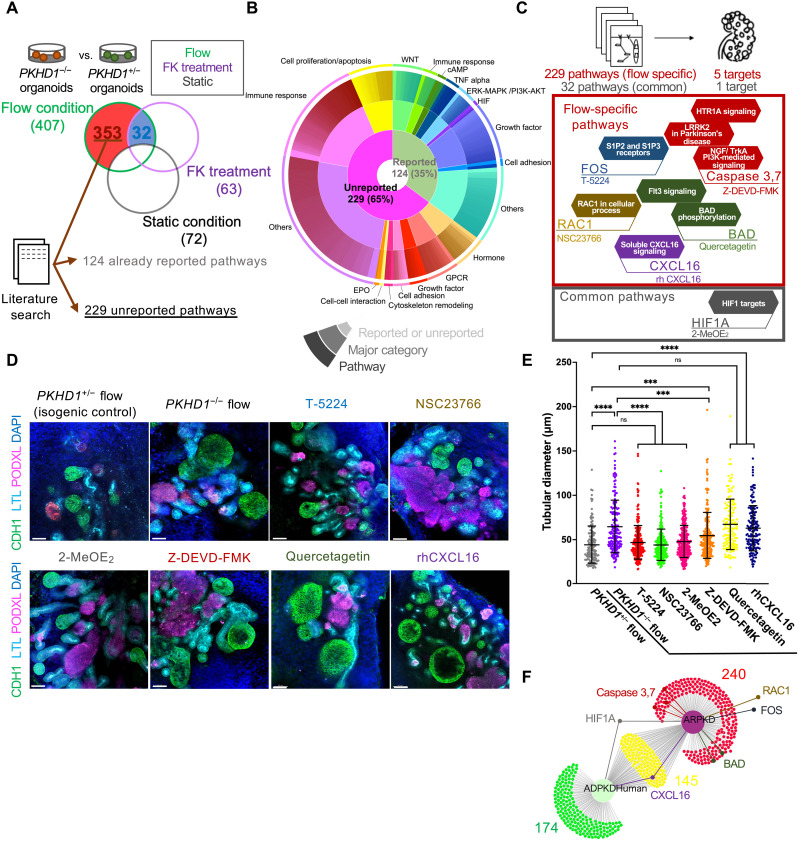
To investigate cystogenic signals, we compared the global gene expression of PKHD1−/− organoids to PKHD1+/− in each of flow, forskolin, or static conditions independently (Fig. 3A, top). DEG analysis between PKHD1−/− and PKHD1+/− identified 407 signals altered in the flow condition, 63 signals in forskolin, and 72 signals under static culture (Fig. 3A, bottom, and data S3 to S5). Since static organoids did not exhibit cysts in PKHD1−/−, we disregarded the 72 noncystogenic signal alterations seen in static culture. While 32 pathways are involved in both flow and forskolin conditions, 353 signal pathways are specific to organoids cultured under flow (Fig. 3A, bottom). Following a comprehensive literature search, we excluded 124 previously reported cytogenic signals identified in ADPKD/ARPKD studies (20, 27, 34, 35). Thus, we have identified 229 unreported pathways (data S6). We then categorized all signal pathways into biological processes for visualization (Fig. 3B), namely, cytoskeleton remodeling, immune response, G protein–coupled receptor (GPCR)–related, cell proliferation, growth factor, erythropoietin, hormone, cell adhesion, and others. Reported signals (124), including cAMP, WNT, tumor necrosis factor α (TNFα), and extracellular signal–regulated kinase (ERK)–mitogen-activated protein kinase (MAPK), validate the use of our kidney organoid-on-a-chip platform for ARPKD modeling.
Fig. 3. Identification of novel therapeutic targets for ARPKD and drug screening tests using commercially available compounds.
(A) Schematic illustrating the strategy to identify flow-specific pathways and common pathways by comparing PKHD1−/− versus PKHD1+/− organoids with flow, forskolin (FK) treatment, or static culture. The number of pathways is shown with parenthesis. (B) Proportions of pathways categorized in the three-level iterative analysis. (C) Schematic of the strategy to identify novel therapeutic targets for ARPKD. (D) Immunocytochemistry for proximal tubule (LTL), distal tubule (CDH1), and podocyte (PODXL) in PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without candidate compounds (T-5224, NSC23766, 2-MeOE2, Z-DEVD-FMK, quercetagetin, and rhCXCL16) under flow on day 35. Scale bars, 50 μm. (E) Distal tubular diameter in PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without each compound under flow on day 35. (F) DiVenn diagram that compares pathways in our ARPKD model and human ADPKD dataset. Values represent means ± SD. ***P < 0.001, ****P < 0.0001.
On the basis of compound availability for these newly identified pathways and the reported literature, five target molecules from 229 flow-specific signals and one target from 32 common signals are selected for further evaluation (Fig. 3C, top). The selected compounds and target molecules are T-5224 for FOS/activator protein 1 (AP-1), Z-DEVD-FMK for caspase, quercetagetin for BCL2-associated agonist of cell death (BAD), NSC23766 for Rac family small guanosine triphosphatase (GTPase) 1 (RAC1), recombinant human C-X-C motif chemokine ligand 16 (rhCXCL16) for CXCL16 (Fig. 3C, bottom red), and 2-methoxyestradiol (2-MeOE2) for hypoxia-inducible factor-1 (HIF-1) (Fig. 3C, bottom gray). PKHD1−/− organoids are treated with those compounds from days 16 to 35 of differentiation in fluidic culture (fig. S8A). The optimal concentrations of all compounds are determined within the range used in previous studies by evaluating normal nephron formation in static culture (fig. S8B). Unbiased 3D imaging by tissue clearing of whole organoids revealed a significant reduction of CDH1+ tubular enlargement during treatment with T-5224, NSC23766, and 2-MeOE2 (Fig. 3, D and E). By contrast, Z-DEVD-FMK showed a marginal effect, while quercetagetin and rhCXCL16 did not affect cyst suppression. Closer examination of phenotypes, and associated molecular genetics, reveals convergent and divergent pathogenic mechanisms between ADPKD and ARPKD (27). To further investigate the utility of these candidates in ADPKD, we carried out global gene expression profiling comparing our ARPKD datasets with a meta-analysis–based dataset of human, rat, and mouse ADPKD tissues (36, 37). RAC1- and FOS-related mechanosensory signals are unchanged in large cysts when compared to noncystic tissue in human ADPKD (Fig. 3F and fig. S9, A and B) (38, 39). This may have anatomical explanation as urinary flow is absent in large ADPKD cysts that are noncontinuous with the tubular lumen, whereas ARPKD cysts are tubular dilatations (11).
Potential clinical translation from existing drugs for ARPKD
Validation is a critical step during drug discovery, since most candidate drugs are deemed ineffective or toxic (40). To mitigate safety concerns, we explored existing drugs that inhibit RAC1, FOS, or HIF-1 as potential new therapeutic targets. Of these, HIF-1 inhibitor is excluded from candidates, since HIF-1 suppression exacerbates renal anemia via impaired erythropoietin production (41). We focused on R-naproxen and R-ketorolac, two widely used FDA-approved nonsteroidal anti-inflammatory drugs (NSAIDs) that also have an inhibitory effect against RAC1 (42, 43), as well as a selective FOS inhibitor, T-5224 (44), chosen for favorable safety profile during clinical trials for arthritis (Fig. 4A) (45).
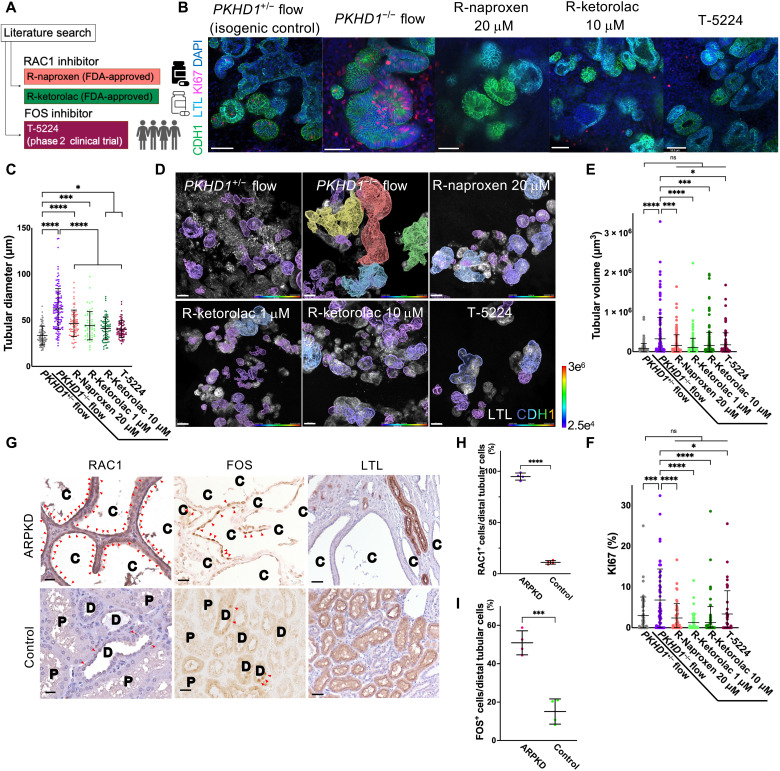
Fig. 4. Repurposing R-naproxen, R-ketorolac, and T-5224 as potential medications for clinical translation in ARPKD.
(A) Schematic of RAC1 and FOS inhibitors. (B) Immunocytochemistry for LTL, CDH1, and KI67 in PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without candidate medications (R-naproxen, R-ketorolac, and T-5224) under flow on day 35. Scale bars, 50 μm. (C) Distal tubular diameter in PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without each drug under flow on day 35. (D) Whole-organoid 3D confocal imaging stacks of representative samples [PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without candidate drugs (20 μM R-naproxen, 1 and 10 μM R-ketorolac, and 10 μM T-5224) under flow] used to measure distal tubular volumes in Imaris 3D surface rendering. Scale bars, 50 μm. (E) Distal tubular volume in PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without each drug under flow on day 35. (F) Percentage of KI67+ cells in distal tubules in PKHD1+/− organoids under flow and PKHD1−/− organoids treated with and without each drug under flow on day 35. (G) Immunostaining of RAC1 (left), FOS (middle), and LTL (right) in human kidney samples from patients with ARPKD and normal portions of tumor nephrectomy (control). Arrowheads indicate RAC1-positive (left) and FOS-positive (middle) cells. C, cyst; D, distal tubule; P, proximal tubule. Scale bars, 25 μm (left and middle) and 50 μm (right). (H) Percentage of RAC1+ cells in cystic epithelial cells (ARPKD, n = 4) and distal tubular cells (control, n = 4). (I) Percentage of FOS+ cells in cystic epithelial cells (ARPKD, n = 4) and distal tubular cells (control, n = 4).
PKHD1−/− organoids-on-a-chip are treated with these compounds from days 16 to 35 of differentiation in fluidic culture. The optimal concentration of each compound is determined on the basis of previous studies and their negligible impact on nephron formation (fig. S10, A and B) (46). Nephrotoxicity of each chemical compound is assessed by kidney injury molecule-1 (KIM-1) and double-stranded DNA breaks (DSBs) marked by γH2A histone family member X (γH2AX). Notably, none of these compounds resulted in any signs of toxicity to nephron epithelial and stromal cells at the concentration tested for cyst suppression (fig. S11). 3D imaging revealed significant reductions of CDH1+ tubular enlargement upon treatment by R-naproxen, R-ketorolac, and T-5224 (Fig. 4, B and C). To validate their therapeutic effects, distal tubular volume and KI67+ percentage in distal nephrons are measured by image analysis (Imaris) and shown to be significantly improved by these candidates (Fig. 4, D to F, and fig. S12A). To exclude the possibility of another therapeutic mechanism by an anti-inflammatory effect of R-naproxen, organoids are treated with S-naproxen, an optical isomer of R-naproxen, that lacks RAC1 inhibitory activity but has an anti-inflammatory effect (47). 3D imaging confirms that S-naproxen does not inhibit cyst formation in PKHD1−/− organoids cultured under flow, suggesting that the cyst suppression is not mediated by the anti-inflammatory effect of R-naproxen (fig. S12, B and C).
To determine whether RAC1 and FOS are activated in ARPKD patients, we immunostain for Rac1–guanosine triphosphate (Rac1-GTP; an activated form of RAC1) and FOS in kidney samples from four ARPKD patients and normal portions of tumor nephrectomy as a control. In human ARPKD samples, RAC1+ cells and FOS+ cells are 94.8 ± 3.5% and 50.9 ± 6.2%, respectively, in cystic epithelial cells, which are significantly higher than the controls with 11.1 ± 1.7% and 15.1 ± 6.6% positivity in distal tubules (Fig. 4, G to I). Of note, while RAC1+ cells are also observed in proximal tubules, FOS+ proximal tubules are rarely detected in ARPKD patients (fig. S13A). The increased expression of RAC1 in proximal and distal nephrons and FOS in enlarged distal nephrons is also confirmed in PKHD1−/− organoids cultured under flow (figs. S13B and S14, A to C).
On the basis of our observation of FOS activation in cystic epithelial cells in ARPKD patient kidneys, we postulate that FOS may be a crucial determinant of the cyst origins. To investigate the cell type–specific FOS expression in human kidneys, we analyze single-cell RNA sequencing (RNA-seq), regional transcriptomics, and the assay for transposase-accessible chromatin using sequencing (ATAC-seq) from the Kidney Precision Medicine Project (KPMP) and the Kidney Interactive Transcriptomic (KIT) (48). As expected from our immunostaining results in human ARPKD tissues (fig. S13A), FOS expression is predominantly detected in distal nephrons and only sparsely found in proximal tubules (fig. S15, A to C), which supports our observation (49). Conversely, mouse kidney datasets of single-cell RNA-seq from the KIT show FOS expression in both proximal tubules and collecting ducts in mice (fig. S15, D and E) (50), consistent with cyst formation in the S3 segment of proximal tubules in a mouse ARPKD model (12), and in proximal tubules and collecting ducts in TgN737Rpw mice (51). Given the FOS expression in specific tubular segments where cysts develop in human and mouse ARPKD (fig. S15F), FOS might be an important molecule that determines segment specificity of cystogenesis in ARPKD.
RAC1 and FOS involvement in ARPKD cystogenesis
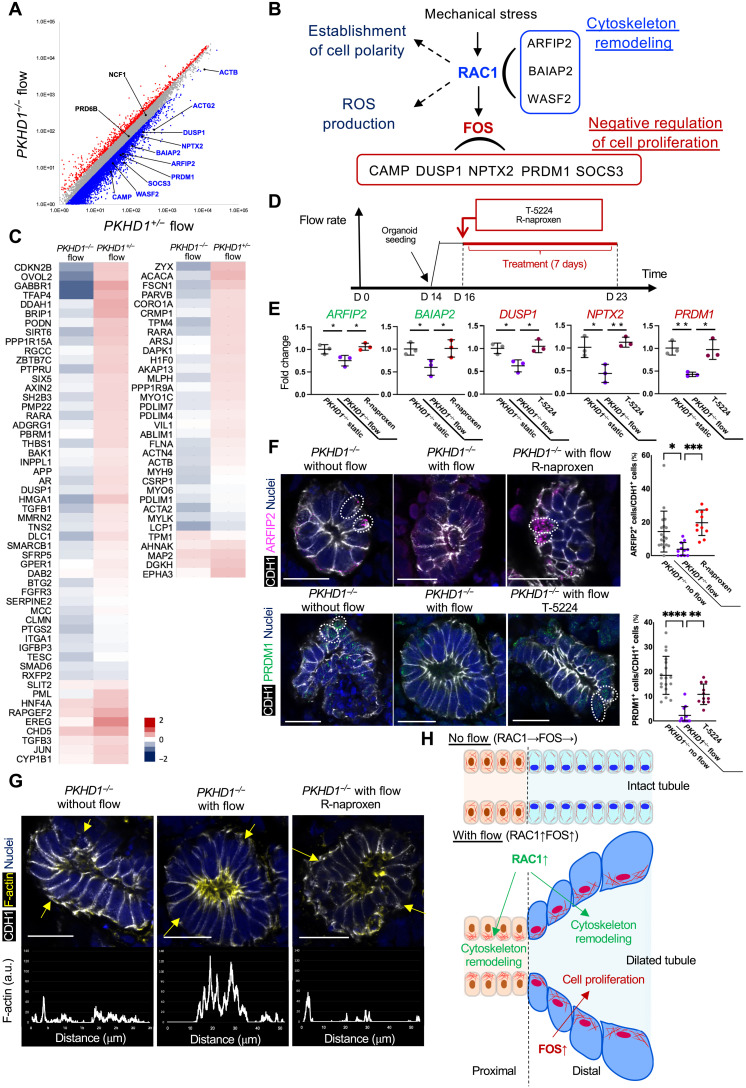
Prior studies demonstrated that mechanical stress activates RAC1 and FOS by stimulating basal adhesion proteins such as integrins involved in the activation of the Rho family of GTPase proteins including RAC1 (39, 52). FOS is one of downstream molecules of RAC1, regulating cell proliferation (53). To further investigate the cystogenic mechanisms initiated by mechanical stress in our ARPKD model, we analyzed gene expression involved in RAC1 and FOS pathways defined by MetaCore. Many downstream genes of RAC1 and FOS are down-regulated by flow in PKHD1−/− organoids, while flow activates those genes in the PKHD1+/− control (Fig. 5A and fig. S16, A and B). Among these genes, ARFIP2, BAIAP2, WASF2, ACTB, and ACTG2 are implicated in “cytoskeleton remodeling” under RAC1 signal according to MetaCore, and CAMP, DUSP1, NPTX2, PRDM1, and SOCS3 are involved in “negative regulation of cell proliferation,” which down-regulate cell cycle, in the FOS pathway (Fig. 5B) (54–58). In contrast, the expression of other RAC1 downstream molecules is not altered by PKHD1−/− mutations, which include NCF1 and PRD6B involved in “reactive oxygen species production” and “establishment of cell polarity,” respectively (Fig. 5B) (59, 60). Given that the inhibitors of RAC1 or FOS ameliorate cystogenesis in our ARPKD model, these results suggest that PKHD1−/− mutations conversely down-regulate cytoskeleton genes and negative cell cycle regulators upon activation of RAC1 and FOS and lead to cyst formation.
Fig. 5. The mechanisms of RAC1- and FOS-mediated cystogenesis.
(A) Scatterplots comparing expression profiles of PKHD1−/− and PKHD1+/− organoids under flow. Red and blue dots represent >1.5-fold up-regulated and down-regulated genes, respectively, in PKHD1−/− organoids when compared to PKHD1+/− under flow conditions. (B) Schematic illustration of RAC1- and FOS-mediated signals. (C) Heatmaps of microarray datasets for “negative regulation of cell population proliferation” (left) and “actin cytoskeleton” (right) in PKHD1−/− and PKHD1+/− organoids, cultured under flow, on day 35 of differentiation. (D) Schema of the culture and treatment protocol. (E) qPCR for RAC1 genes (ARFIP2 and BAIAP2) in green color and FOS genes (DUSP1, NPTX2, and PRDM1) in red color in PKHD1−/− organoids treated with R-naproxen or T-5224 under flow conditions on day 23 of differentiation. Values are normalized against the control, PKHD1−/− organoids cultured in static conditions. (F) Immunocytochemistry for ARFIP2 and PRDM1 in PKHD1−/− organoids with and without flow and PKHD1−/− organoids treated with R-naproxen or T-5224 treatment under flow conditions on day 23 (left). Scale bars, 20 μm. Percentage of ARFIP2+CDH1+ tubules/CDH1+ tubules in PKHD1−/− organoids without flow, with flow, and with flow and R-naproxen (top right). Percentage of PRDM1+CDH1+ tubules/CDH1+ tubules in PKHD1−/− organoids without flow, with flow, and with flow and T-5224 on day 23 of differentiation (bottom right). (G) Immunohistochemistry for F-actin and CDH1 in organoid tubules under static (left), flow (middle), and R-naproxen–treated flow conditions (right) on day 23. Scale bars, 20 μm. The plot profiles at the bottom show the intensity of F-actin across the tubule denoted by yellow arrows. a.u., arbitrary units. (H) Schematic illustration of RAC1- and FOS-mediated disease mechanism. Values shown are means ± SD. P values were determined by Student’s t test. *P < 0.05, **P < 0.01.
To validate whether cytoskeleton remodeling and negative regulation of cell proliferation are crucial for cystogenesis and contrarily regulated under RAC1 and FOS in PKHD1−/− mutants, we refer to another genome informatics database (www.informatics.jax.org). Genes associated with these biological processes are selected by GO terms, namely, “actin cytoskeleton” and “negative regulation of cell population proliferation.” The heatmap of these genes reveals a reduction of actin cytoskeleton gene expression in PKHD1−/− organoids compared to PKHD1+/− organoids when cultured under flow (Fig. 5C, right), which is consistent with actin cytoskeleton reduction in PKD1-deficient mouse kidneys (61, 62). Furthermore, the expression of genes related to negative regulation of cell proliferation is substantially reduced in PKHD1−/− organoids (Fig. 5C, left). These results validate that PKHD1−/− mutations result in converse regulation of genes associated with cytoskeleton and negative cell cycle regulation when stimulated by flow.
To understand the therapeutic mechanisms of R-naproxen and T-5224 via modulating RAC1 and FOS signals, PKHD1−/− organoids and ARPKD patient–derived organoids cultured under flow are treated with these drugs from day 16 of differentiation and harvested on day 23 (Fig. 5D). Quantitative polymerase chain reaction (qPCR) confirms flow-induced down-regulation of RAC1 downstream genes, ARFIP2 and BAIAP2, in both CRISPR-mutant and patient-derived organoids. As expected, R-naproxen rescues the expression of these two genes (Fig. 5E and fig. S17). Similarly, qPCR of the FOS-downstream genes, DUSP1, NPTX2, and PRDM1, demonstrates their down-regulation by flow and reverse expression by T-5224. The quantitative immunohistochemistry for ARFIP2 and PRDM1, the downstream genes of RAC1 and FOS, respectively, validates their decreased expression by flow and the rescue by R-naproxen and T-5224 (Fig. 5F).
To discern how flow and PKHD1−/− mutations affect cytoskeleton remodeling in CDH1+ organoid tubules, we examine F-actin localization (63). PKHD1−/− organoids under flow conditions are treated with R-naproxen for 7 days from day 16 and assessed by phalloidin staining. In PKHD1−/− organoids without flow, the actin fibers are widely distributed throughout the cytoplasm in CDH1+ tubules (Fig. 5G, left, and fig. S18, top), whereas PKHD1−/− organoids under flow show basal diminution and apical enrichment of actin fibers (Fig. 5G, middle, and fig. S18, middle). In control organoids derived from the parental human pluripotent stem cell (hPSC) line, this redistribution of actin fibers is not observed under flow (fig. S19A). The apical redistribution of actin fiber is ameliorated by R-naproxen treatment in PKHD1−/− mutants (Fig. 5G, right, and fig. S18, bottom), collectively suggesting that cytoskeleton remodeling observed in PKHD1−/− mutants is a disease phenotype and can be reversed by R-naproxen. This cytoskeleton remodeling is also observed in proximal tubules in PKHD1−/− organoids under flow conditions, which is not detected in static PKHD1−/− organoids or in control organoids under flow (fig. S19B). Together, RAC1 and FOS activation initiated by mechanical stress (38, 64, 65) induces cytoskeleton remodeling and cell proliferation in distal nephrons, which can be potentially treated by R-naproxen and T-5224 in ARPKD patients (Fig. 5H).
DISCUSSION
ARPKD represents a leading cause of pediatric dialysis dependency and kidney transplantation (10). This disease progresses far more rapidly than ADPKD, conveying significant morbidity and mortality in children. While rodent models have improved the understanding of pathomechanisms in ARPKD (11, 12), it has been challenging to create physiological human models that accelerate translation to patients. In this study, we showed that our kidney organoid-on-a-chip platform provides a physiologically relevant model for ARPKD, allowing the identification of mechanosensing signals as key drivers of cystogenesis.
In validation of our findings, FDA-approved NSAIDs that inhibit RAC1 (R-naproxen and R-ketorolac) as well as a clinically tested inhibitor of FOS (T-5224) are shown to have therapeutic effects in our model. Although naproxen is technically approved for children over 2 years of age permitting efficacy trials in afflicted children (66), there are concerns to using NSAIDs in patients with kidney disease. Unfortunately, ARPKD is a developmental disease leading 30% of these patients dying neonatally (8, 9), and although naproxen is not an absolute contraindication during pregnancy, its uncertain safety profile for neonates is an additional consideration for clinical translation. Importantly, our study demonstrates cyst suppression with R-naproxen, an enantiomer with limited cyclooxygenase (COX) inhibition and enhanced RAC1 inhibition (47, 67) to assuage concerns regarding NSAID-related kidney side effects. If R-naproxen was found to have an unreasonable safety profile, then specific RAC1 inhibitors may prove clinically advantageous. For more rapid clinical translation, T-5224 is a specific FOS inhibitor proven to be safe in humans (45). Clinical studies may find optimal and safe dosages of R-naproxen and T-5224 that slow cyst progression, without significant risk of renal toxicity in ARPKD patients of all ages. Future studies may also allow us to develop new inhibitors of RAC1 and FOS based on R-naproxen and T-5224 structures, which can be used for ARPKD patients more safely and effectively.
Our observations shed light on two important questions regarding the disease mechanisms of ARPKD. The segment specificity of cyst formation has been an unresolved question for a long time. The segment specificity of cyst formation differs among species, especially between humans and mice, raising concern about the translatability of animal studies into patient care. Single-cell transcriptomics show that FOS is more localized to distal nephrons in humans compared to mice, where proximal tubules also express FOS. Since mouse ARPKD models develop proximal tubular cysts, FOS might be the crucial determinant of segment-specific cyst formation leading to species-dependent differences. Our model also provides new insights on why mutations in PKHD1 lead to cyst formation. Transcriptomic analysis demonstrates the converse regulation of genes associated with cytoskeleton remodeling and negative cell cycle regulation when PKHD1 homozygous mutations are introduced to human kidney organoids. However, the upstream molecules, RAC1 and FOS, are not down-regulated but up-regulated in PKHD1−/− organoids, suggesting that PKHD1 is involved in gene expression regulation downstream of RAC1 and FOS. On the basis of our findings, we hope that future studies will elucidate the precise mechanisms of the segment specificity and the gene regulation by PKHD1. In summary, kidney organoid-on-a-chip models offer a powerful platform for recapitulating physiologically relevant microenvironments in human organ-specific tissues, paving the way toward bench-to-bedside translation.
MATERIALS AND METHODS
Cell culture and maintenance
H9 (WiCell) human ES cells, H9-derived PKHD1 mutants, and ARPKD patient-derived iPSCs (68) were maintained on hESC-qualified Geltrex (Thermo Fisher Scientific)–coated plates using StemFit Basic02 (Ajinomoto Co. Inc.) supplemented with fibroblast growth factor 2 (FGF2; 10 ng/ml; PeproTech), as previously reported (69).
Nephron organoid differentiation
Nephron organoids were differentiated and maintained as established in previous protocols (1, 69). Quality check was performed by immunostaining for SIX2 on day 8 and for LTL, CDH1, and PODXL, segment-specific markers of nephron on day 21. For static culture, the basal medium consisting of Advanced RPMI (ARPMI; Thermo Fisher Scientific) and GlutaMAX (Thermo Fisher Scientific) was changed every 2 to 3 days.
Generation of heterozygous and homozygous PKHD1-mutant hPSCs
Heterozygotic (PKHD1+/−) and compound homozygotic (PKHD1−/−) mutant lines were generated using methodology previously outlined (70), which generated clones of isogenic progeny via clonal expansion from single cells. PX459 plasmid encoding Streptococcus pyogenes Cas9 and single-guide RNA (sgRNA) targeting the third exon of PKHD1 (5′-TCATTTTTGATGGTAGGTTG-3′) was transfected into H9 hESCs to generate the heterozygotic mutant line, which then served as the parental line to generate the compound homozygotic mutant. Transfected cells were selected by transient puromycin treatment. CRISPR-Cas9–induced mutations of individual clones were identified by deep sequencing.
Targeted deep sequencing
Each target area was PCR-amplified from the isolated single clones for next-generation sequencing (NGS) targeted deep sequencing at the Massachusetts General Hospital Center for Computational & Integrative Biology DNA Core (MGH CCIB DNA Core; Cambridge, MA). Briefly, target-specific primers were designed to amplify 200 to 280 base pairs (bp) from each iPSC clone, including the target site of CRISPR modification, and subjected to NGS deep sequencing. CRISPR-induced variants were determined by alignment to the public reference genome for comparison to unmodified iPSC clones.
Engineered ECM preparation
The ECM was prepared as previously described (15, 19, 71). The matrix contained 300 bloom type A gelatin from porcine skin (Sigma-Aldrich) and fibrinogen (Millipore Sigma), which were enzymatically cross-linked using transglutaminase (Moo Glue, The Modernist Pantry) and thrombin (MPI Biologicals), respectively. Specifically, a solution of 2 weight % (wt %) gelatin, fibrinogen (10 mg/ml), 2.5 mM calcium chloride, and transglutaminase (2 mg/ml) in phosphate-buffered saline (PBS) was first prepared. The solution was mixed well and incubated at 37°C for 15 min. Following incubation, 1 ml of the solution was quickly mixed with thrombin by fast pipetting, resulting in a final thrombin concentration of 2 U/ml. The solution was then quickly cast onto the millifluidic chip (see below) and distributed evenly over the surface. This ECM solution gelled in ~5 min at room temperature (RT) and was further cured for 3 hours at 37°C in a cell culture incubator in a sterile container.
Millifluidic chip fabrication
Perfusion chip devices consisted of a 10-ml syringe medium reservoir (Nordson EFD), a leur lock connection nozzle (Nordson EFD), inlet perfusion tubing (2-Stop PharMed BPT tubing, Ismatec, Cole Parmer), perfusion pins (18 gauge, Nordson EFD), a custom machined stainless steel base, a 5-mm acrylic lid, outlet perfusion tubing (Cole-Parmer Peroxide-Cured Silicone Tubing, 1/32″ID × 3/32″OD), 4× 12-mm M4 screws (McMaster Carr), and a custom-designed millifluidic chip. The millifluidic chips were fabricated as previously described (19). Briefly, the millifluidic chip gasket was printed onto a 50 mm × 75 mm × 1 mm glass slide by a multimaterial 3D printer (AGB 10000, Aerotech Inc.) with a polydimethylsiloxane (PDMS) ink with a base–to–cross-linker ratio of 10:1 (SE1700, Dow Corning). A custom-written MatLab code was used to produce the G-code needed to print each gasket onto the top surface of a glass slide. After printing, each PDMS gasket was cured at 80°C for 2 hours. Next, the gasket was filled with a 1-mm-thick layer of ECM, as described above. To assemble the perfusion device, the medium reservoir was connected to the inlet and outlet tubing with a sterile syringe filter in line and the millifluidic chip via the perfusion pins. The chip was placed onto the stainless steel base, and the culture medium was allowed to advance via hose-pinch clamps in the gasket until about three-fourths of the hydrogel ECM was covered. Subsequently, the kidney organoids were placed into the indicated region (fig. S6C) and allowed to settle onto the ECM. Following the seeding of the kidney organoids, the lid was carefully placed on top of the gasket and secured in place by four screws. The chip was filled with culture medium and then placed inside of a cell culture incubator.
For live imaging of kidney organoids under flow conditions, the PDMS gasket was printed onto a 50 mm × 75 mm × 3 mm–thick acrylic substrate with a laser-cut rectangular window (32 mm × 45 mm) that was covered using a #1 cover glass (35 mm × 50 mm) and secured in place with cyanoacrylate. The PDMS was cured for 3 days at RT to avoid warping of the acrylic base. The millifluidic chip was assembled and filled with ECM. Kidney organoids were then placed on top of the ECM and cultured, as described above. For live imaging, the modified kidney organoid-on-a-chip assembly was placed onto the stage of a confocal microscope, where the objective was located under the imaging window.
Culturing kidney organoids-on-a-chip
The organoids were seeded onto the chip devices on day 14 of their differentiation and cultured on chip by flowing cell culture medium composed of ARMPI with GlutaMAX, 1% antibiotic/antimycotic solution (Gibco), and 1.5% heat-inactivated fetal bovine serum (Gibco). All cell culture medium was preequilibrated to the cell culture incubator environment for at least 2 hours before chip assembly and seeding. The kidney organoids-on-a-chip was then held in a cell culture incubator for 30 min under static conditions, before connecting the inlet perfusion tubing to an IPC-N peristaltic pump (Ismatec) and perfusing the cell culture medium at 42.7 μl/min overnight in the incubator. The following day (day 15 of organoid differentiation), the sterile filter was removed from the perfusion line to avoid bubble formation at high perfusion speeds, and the perfusion speed was ramped up to 4.27 ml/min. Cell culture medium was changed in the reservoir every 2 to 3 days.
Forskolin treatment of kidney organoids in static culture
Kidney organoids were cultured in static condition and treated with 20 μM forskolin (Sigma-Aldrich, # F6886), a cAMP inducer, from day 14 to day 35.
cAMP ELISA experiment
Kidney organoids were harvested on day 35 of differentiation, and cAMP measurement was determined using a Cyclic AMP Competitive ELISA (enzyme-linked immunosorbent assay) kit (Sigma-Aldrich, EMSCAMPL) according to the manufacturer’s instructions. The optical density was read at 405 nm with a Synergy H1 plate reader (BioSPX) and assessed with Gen5 software (BioTeK).
siRNA transfection
siKIF3A (Thermo Fisher Scientific, Silencer Select ID s21942) and siControl (Thermo Fisher Scientific, Silencer Select Negative Control No. 1) were obtained. According to the manufacturer’s instructions, the initial siRNA transfection was performed with Lipofectamine RNAiMax (Thermo Fisher Scientific) on day 8 of differentiation (corresponding to the nephron progenitor cell stage). Organoids on day 16 of differentiation were transfected again with siKIF3A and siControl in culture medium containing 1.5% heat-inactivated fetal bovine serum without antibiotics on chip (72). The medium was replaced 48 hours after each transfection.
FliptR and live nuclear dye labeling
Kidney organoids were seeded on modified chips (for living imaging) on day 14 of their differentiation and cultured under flow. FliptR (Spirochrome) and SPY700-DNA (Spirochrome) staining solution were applied to the organoids on days 16 to 21 of differentiation according to the manufacturer’s instructions and incubated at 37°C in a humidified atmosphere containing 5% CO2 for 15 min before imaging.
Lifetime measurements
FLIM imaging was performed using a Leica STELLARIS 8 confocal microscope. Excitation was performed using a pulsed 488-nm laser, and the emission signal was collected through a 575- to 650-nm bandpass filter. LAS X FLIM/FCS software (Leica) was then used to fit fluorescence decay data (from full images or regions of interest in organoid tubules). Fitted lifetime data were expressed as means ± SD. Δτ2 was calculated with the equation Δτ2 = τ2 − average τ2 during perfusion or rest.
Drug screening tests of kidney organoids
To determine optimal concentration of each compound, PKHD1-mutant organoids were treated with the following concentration from day 16 to day 35 of differentiation: T-5224 (Thermo Fisher Scientific, 50-115-1835) at 5, 10, and 20 μM; Z-DEVD-FMK at 1, 10, and 50 μM (R&D Systems, FMK004); quercetagetin at 10, 40, and 200 μM (Millipore Sigma, 551590); NSC23766 at 25, 50, and 100 μM (Abcam, ab142161); rhCXCL16 at 5, 10, and 25 ng/ml (R&D Systems, 976-CX-025); 2-MeOE2 at 1, 5, and 10 μM (Selleckchem, S1233); R-naproxen at 10, 20, 200, and 400 μM (Millipore Sigma, 82170); and R-ketorolac at 0.5, 1, 10, and 20 μM (Millipore Sigma, 1654) (45, 46, 73–77). Full medium exchanges were conducted every 2 to 3 days. Treatment of the kidney organoids-on-a-chip was started at day 16 of their differentiation until day 35. The compounds were added to the medium reservoir at every medium change in the following concentrations: T-5224 at 10 μM, Z-DEVD-FMK at 1 μM, quercetagetin at 10 μM, NSC23766 at 10 μM, rhCXCL16 at 1 ng/ml, 2-MeOE2 at 5 μM, R-naproxen at 20 μM, S-naproxen at 20 μM, and R-ketorolac at 1 and 10 μM.
Immunocytochemistry of 2D tissues
Nephron progenitor cells on day 8 of differentiation were rinsed with PBS and fixed with 4% paraformaldehyde (PFA) for 20 min at RT. Fixed cells were washed three times in PBS and blocked with blocking buffer (BB; 5 wt % donkey serum in PBS containing 0.3 wt % Triton X-100) for 1 hour at RT. The cells were then incubated with anti-SIX2 (Proteintech, 11562-1-AP) in antibody dilution buffer [ADB; 1% bovine serum albumin (BSA) containing 0.3% Triton X-100] overnight at 4°C. After washing three times in PBS, the cells were incubated with Alexa Fluor secondary antibodies (Life Technologies) in ADB for 1 hour at RT. 4′,6-Diamidino-2-phenylindole (DAPI; Sigma-Aldrich, #D8417) was added to stain the nucleus. Immunofluorescence was photographed using a Nikon C1 confocal microscope or a Leica STELLARIS 8 confocal microscope.
Immunocytochemistry of kidney organoids
Nephron organoids were fixed with 4% PFA for 1 hour at RT, incubated in 30% sucrose overnight at 4°C, and embedded in optimal cutting temperature (OCT) compound, and 10-μm frozen sections were cut using a cryostat. Samples were blocked with BB for 1 hour at RT and then incubated with primary antibodies in ADB overnight at 4°C. The following antibodies were used in these studies: anti-CDH1 (Abcam, ab11512), anti-LTL (Vector Laboratories, B-1325), anti-PODXL (R&D Systems, AF1658), anti-TUBA1A (Abcam, ab179484), anti-KI67 (Dako, m7248), anti-ATP1A1 (Abcam, ab76020), anti-PKHD1 [Abcam, ab204951(8G12A1)], anti-ARFIP2 (Bioss, BS-1442R), anti-PRDM1 (Novus Biologicals, NB600-235SS), anti-GATA3 (R&D Systems, AF2605), anti-BRN1 (Santa Cruz Biotechnology, sc-6028-R), anti–KIM-1 (R&D Systems, AF1750), and anti–phospho-histone H2AX (Cell Signaling Technology, #2577). For immunostaining with biotinylated LTL, the Streptavidin/Biotin Blocking Kit (Vector Laboratories, #SP-2002) was used according to the manufacturer’s protocol. After washing three times in PBS, samples were incubated with Alexa Fluor secondary antibodies in ADB for 1 hour at RT. Vectashield with DAPI (Vector Laboratories) was added to stain the nucleus. For phalloidin staining, Acti-stain 555 Phalloidin (Cytoskeleton Inc., #PHDH1-A) was used according to the manufacturer’s protocol. Images were taken using a Nikon C1 confocal microscope or a Leica STELLARIS 8 confocal microscope.
Whole-mount immunohistochemistry
Nephron organoids on day 21 of differentiation were fixed with 4% PFA for 1 hour at RT. Samples were blocked with BB for 1 hour at RT and then incubated with primary antibodies in ADB overnight at 4°C. The following antibodies were used in these studies: anti-CDH1 (Abcam, ab11512), anti-LTL (Vector Laboratories, B-1325), and anti-PODXL (R&D Systems, AF1658). For immunostaining with biotinylated LTL, the Streptavidin/Biotin Blocking Kit (Vector Laboratories, #SP-2002) was used according to the manufacturer’s protocol. After washing three times in PBS, samples were incubated with Alexa Fluor secondary antibodies in ADB for 1 hour at RT. Vectashield with DAPI (Vector Laboratories) was added to stain the nucleus. Images were taken using a Nikon C1 confocal microscope or a Leica STELLARIS 8 confocal microscope.
Whole-mount immunohistochemistry with sample clearing
The chip device was taken out of the incubator for fixation, and the lid was carefully removed in a sterile biosafety cabinet. The kidney organoids adherent on the surface of the ECM were rinsed with PBS, carefully overlaid with 4% PFA, and fixed for 45 min at RT. After fixation, the samples were washed three times with 0.1% Triton X-100 (Sigma-Aldrich) for 30 min. Organoids were stained and cleared with ethyl cinnamate (Sigma-Aldrich), as described before (78). The following antibodies were used in these studies: anti-CDH1 (Abcam, ab11512), anti-CFTR (Sigma-Aldrich, HPA021939), anti-LTL (Vector Laboratories, B-1325), anti-PODXL (R&D Systems, AF1658), anti-KI67 (Dako, m7248), anti-pS6RP (Cell Signaling Technology, 2211S), anti-RAC1 (Developmental Studies Hybridoma Bank, CPTC-RAC1-2), and anti-FOS (Abcam, ab190289). Samples were examined with a Zeiss TIRF/LSM 710 confocal microscope or a Leica STELLARIS 8 confocal microscope and by using Imaris 3D software (Bitplane).
Immunostaining and lectin staining of human tissue
Immunohistochemistry of human ARPKD samples and normal portions of tumor nephrectomy were carried out as described previously (75, 79). Briefly, 4-μm-thick paraffin sections were blocked with 3% H2O2 and 1% BSA–PBS, after deparaffinization, hydration, and antigen retrieval in 0.01 M citrate buffer (pH 6.0) at 95°C for 30 min. The sections were incubated overnight at 4°C with primary antibodies: anti-human Rac1-GTP antibody (1:100; 26903, NewEast Biosciences) or anti-cFOS antibody (1:2000; Ab190289, Abcam). After washing with PBS three times, secondary reagents (Envision+ Rabbit IgG, DAKO, or HRP-Goat anti-Mouse IgM Antibody, Invitrogen) were applied, incubated at RT for 1 hour, and visualized with ImmPACT DAB (Vector Laboratories). The sections only for Rac1-GTP were counterstained with hematoxylin. The staining was evaluated in at least 500 cystic epithelial cells. The staining of biotin-labeled Lotus tetragonolobus lectin (Biotin-LTL; 1:200; B-1325, Vector Laboratories) was performed by a similar method, but without secondary reagent and with an additional blocking for endogenous streptavidin/biotin. This study was performed under the protocol (STUDY00001229) approved by Cedars-Sinai Medical Center Institutional Review Board.
Dextran perfusion assay
Organoids cultured on a chip were treated with WGA-conjugated 647 (5 μg/ml; Thermo Fisher Scientific, W32466), incubated at 37°C for 30 min under flow, and rinsed with live cell imaging solution (Thermo Fisher Scientific, A14291DJ) once. LMD (10 μg/ml; Thermo Fisher Scientific, D3306) was then perfused by gravity. Samples were examined with a Leica STELLARIS 8 confocal microscope.
DNA microarray and functional analysis
Total RNA was isolated from kidney organoids with TRIzol (Thermo Fisher) from three biological replicates in each condition (PKHD1−/− flow, forskolin treatment, or static and PKHD1+/− flow, forskolin treatment, or static). The quality of purified total RNA was analyzed with Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was amplified and labeled using the Amino Allyl MessageAmp II aRNA Amplification Kit (Thermo Fisher Scientific). Each sample of amplified RNA (aRNA) labeled with Cy5 was hybridized with a highly sensitive DNA chip, 3D-Gene Human Oligo Chip 25k (Toray Industries). Hybridization signals were scanned and analyzed using 3D-Gene Scanner (Toray Industries). All data were scaled by global normalization. The function of flow-associated DEGs in PKHD1+/− organoids was annotated and analyzed according to the three organizing principles of GO (Biological Process, Molecular Function, Cellular Component) by MetaCore analysis software (Clarivate). An integrated pathway enrichment analysis was performed using MetaCore analysis software. The heatmap was made by ExAtlas software (https://kolab.elixirgensci.com/exatlas/).
Quantitative reverse transcription PCR
Total RNA was isolated from kidney organoid samples using TRIzol (Invitrogen, #15596026). cDNA was synthesized from 500 ng of RNA using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, #4368814) according to the manufacturer’s instructions. Real-time PCR was performed using iTaq SYBR Green Supermix (Bio-Rad, #1725124) and QuantStudio3 Real-time PCR systems. The expression of mRNA was assessed by evaluating threshold cycle (CT) values. The CT values and relative expression levels were normalized by ribosomal protein S18 (RPS18), and the relative amount of mRNA specific to each of the target genes was calculated (n = 3 in each experiment). The primer sequences used for quantitative reverse transcription PCR (qRT-PCR) are listed in table S1.
Single-cell RNA-seq
Uniform manifold approximation and projection (UMAP) was made by using KPMP: DK114886, DK114861, DK114866, DK114870, DK114908, DK114915, DK114926, DK114907, DK114920, DK114923, DK114933, and DK114937. Abbreviations in UMAP are as follows: PC, principal cell; tPC-IC; principal-intercalated cell (transitional); CNT-IC-A, connecting tubule intercalated cell type A; IC-A, intercalated cell type A; VSMC/P, vascular smooth muscle cell/pericyte; MON, monocyte; MDC, monocyte-derived cell; CNT-PC, connecting tubule principal cell; aTAL2, thick ascending limb cell cluster 2 (adaptive/maladaptive/repairing); dCNT-PC, connecting tubule principal cell (degenerative); MAC-M2, M2-macrophage; cDC, classical dendritic cell; dPC, principal cell (degenerative); PEC, parietal epithelial cell; ncMON, nonclassical monocyte; dC-TAL, cortical thick ascending limb cell (degenerative); FIB, fibroblast; dVSMC, vascular smooth muscle cell (degenerative); DTL1, descending thin limb cell type 1; dEC-PTC, peritubular capillary endothelial cell (degenerative); pDC, plasmacytoid dendritic cell; EC-AEA, afferent/efferent arteriole endothelial cell; B, B cell; dCNT, connecting tubule cell (degenerative); C-TAL, cortical thick ascending limb cell; T-CYT, cytotoxic T cell; dDCT, distal convoluted tubule cell (degenerative); DCT1, distal convoluted tubule cell type 1; cycEC, endothelial cell (cycling); cycEPI, epithelial cell (cycling); aFIB, fibroblast (adaptive/maladaptive/repairing); EC-GC, glomerular capillary endothelial cell; dIC-A, intercalated cell type A (degenerative); IC-B, intercalated cell type B; EC-LYM, lymphatic endothelial cell; M-TAL, medullary thick ascending limb cell; MC, mesangial cell; cycMNP, mononuclear phagocyte (cycling); MyoF, myofibroblast; NK1, natural killer cell type 1; NK2, natural killer cell type 2; NKT, natural killer T cell; EC-PTC, peritubular capillary endothelial cell; PL, plasma cell; POD, podocyte; aPT, proximal tubule epithelial cell (adaptive/maladaptive/repairing); dPT, proximal tubule epithelial cell (degenerative); dPT/DTL, proximal tubule epithelial cell/descending thin limb cell (degenerative); PT-S1/S2, proximal tubule epithelial cell segment 1/segment 2; PT-S3, proximal tubule epithelial cell segment 3; Treg, regulatory T cell; REN, renin-positive juxtaglomerular granular cell; T, T cell; cycT, T cell (cycling); aTAL1, thick ascending limb cell cluster 1 (adaptive/maladaptive/repairing).
Statistical analysis and reproducibility
Data in all bar charts and dot plots are expressed as means ± SD. Statistical analysis was done in GraphPad Prism 9, and statistical significance was attributed to values of P < 0.05 as determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test unless otherwise noted. Different significance levels (P values) are indicated in each figure with asterisks: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
For transparency, we state here the number of times experiments were repeated independently, with similar results obtained, to produce the data shown. Figure 1D: Eight independent experiments were performed to confirm these phenotypes. Figure 1F: n = 74 LTL+ tubules with flow, n = 66 LTL+ tubules in static culture, n = 191 LTL+ tubules treated with forskolin, n = 32 CDH1+ tubules with flow, n = 27 CDH1+ tubules in static culture, and n = 104 CDH1+ tubules treated with forskolin. Figure 2C: n = 16 tubules transfected with siControl and n = 17 tubules transfected with siKIF3A. Eight organoids transfected with siControl and seven organoids transfected with siKIF3A from two independent chips were analyzed in Fig. 2C. Figure 2E: n = 145 tubules transfected with siControl and n = 102 tubules transfected with siKIF3A. Figure 2F: n = 107 tubules transfected with siControl and n = 58 tubules transfected with siKIF3A. Thirteen organoids transfected with siControl and 12 organoids transfected with siKIF3A from two independent chips were analyzed in Fig. 2 (E and F). Figure 2J: n = 4 (perfusion) and n = 4 (rest). Figure 2K: n = 3. Figure 2L: Three independent chips were analyzed for LMD perfusion experiment. Figure 3E: n = 141 tubules in PKHD1+/− organoids, n = 181 tubules in PKHD1−/− organoids, n = 289 tubules in PKHD1−/− organoids treated with T-5224, n = 235 tubules in PKHD1−/− organoids treated with NSC23766, n = 294 tubules in PKHD1−/− organoids treated with 2-MeOE2, n = 211 tubules in PKHD1−/− organoids treated with Z-DEVD-FMK, n = 140 tubules in PKHD1−/− organoids treated with quercetagetin, and n = 150 tubules in PKHD1−/− organoids treated with rhCXCL16. Three independent experiments were performed for Fig. 3E. Figure 4C: n = 90 tubules in PKHD1+/− organoids, n = 122 tubules in PKHD1−/− organoids, n = 76 tubules in PKHD1−/− organoids treated with 20 μM R-naproxen, n = 59 tubules in PKHD1−/− organoids treated with 1 μM R-ketorolac, n = 73 tubules in PKHD1−/− organoids treated with 10 μM R-ketorolac, and n = 76 tubules in PKHD1−/− organoids treated with T-5224. Figure 4E: n = 109 tubular clusters in PKHD1+/− organoids, n = 139 tubular clusters in PKHD1−/− organoids, n = 116 tubular clusters in PKHD1−/− organoids treated with 20 μM R-naproxen, n = 163 tubular clusters in PKHD1−/− organoids treated with 1 μM R-ketorolac, n = 171 tubular clusters in PKHD1−/− organoids treated with 10 μM R-ketorolac, and n = 83 tubular clusters in PKHD1−/− organoids treated with T-5224. Figure 4F: n = 48 tubular clusters in PKHD1+/− organoids, n = 55 tubular clusters in PKHD1−/− organoids, n = 48 tubular clusters in PKHD1−/− organoids treated with 20 μM R-naproxen, n = 48 tubular clusters in PKHD1−/− organoids treated with 1 μM R-ketorolac, n = 98 tubular clusters in PKHD1−/− organoids treated with 10 μM R-ketorolac, and n = 38 tubular clusters in PKHD1−/− organoids treated with T-5224. Figure 4H: n = 4 (control) and n = 4 (ARPKD). Figure 4I: n = 4 (control) and n = 4 (ARPKD). Figure 5E: n = 3 PKHD1−/− static organoids (no flow), n = 3 PKHD1−/− organoids with flow condition, n = 3 PKHD1−/− organoids treated with 20 μM R-naproxen under flow condition, and n = 3 PKHD1−/− organoids treated with 10 μM T-5224 under flow condition were analyzed. Figure 5F: n = 18 CDH1+ tubules in PKHD1−/− static organoids (no flow), n = 11 CDH1+ tubules in PKHD1−/− organoids with flow condition, and n = 11 CDH1+ tubules in PKHD1−/− organoids treated with 20 μM R-naproxen under flow condition were analyzed for ARFIP2 immunostaining. n = 18 CDH1+ tubules in PKHD1−/− static organoids (no flow), n = 11 CDH1+ tubules in PKHD1−/− organoids with flow condition, and n = 10 PKHD1−/− organoids treated with 10 μM T-5224 under flow condition were analyzed for PRDM1 immunostaining. Figure 5G: n = 3 CDH1+ tubules in PKHD1−/− static organoids (no flow), n = 3 CDH1+ tubules in PKHD1−/− organoids with flow condition, and n = 3 CDH1+ tubules in PKHD1−/− organoids treated with 20 μM R-naproxen under flow condition. Figure S1F: n = 5 untreated organoids (control), n = 6 organoids with 5 μM forskolin, n = 6 organoids treated with 10 μM forskolin, and n = 6 organoids treated with 20 μM forskolin. Figure S3F: n = 30 LTL+ tubules in control organoids without forskolin, n = 12 CDH1+ tubules in control organoids without forskolin, n = 29 LTL+ tubules in control organoids with forskolin, n = 13 CDH1+ tubules in control organoids with forskolin, n = 30 LTL+ tubules in PKHD1−/− organoids without forskolin, n = 10 CDH1+ tubules in PKHD1−/− organoids without forskolin, n = 26 LTL+ tubules in PKHD1−/− organoids with forskolin, and n = 12 CDH1+ tubules in PKHD1−/− organoids with forskolin. Figure S3H: n = 35 tubules in PKHD1+/− organoids without forskolin, n = 58 tubules in PKHD1+/− organoids with forskolin, n = 66 tubules in PKHD1−/− organoids without forskolin, and n = 69 in PKHD1−/− organoids with forskolin. Figure S4B: n = 74 LTL+ tubules in PKHD1−/− organoids with flow, n = 32 CDH1+ tubules in PKHD1−/− organoids with flow, n = 66 LTL+ tubules in PKHD1−/− organoids without flow, n = 27 CDH1+ tubules in PKHD1−/− organoids without flow, n = 33 LTL+ tubules in PKHD1+/− organoids with flow, n = 25 CDH1+ tubules in PKHD1+/− organoids with flow, n = 48 LTL+ tubules in PKHD1+/− organoids without flow, and n = 24 CDH1+ tubules in PKHD1+/− organoids without flow. Figure S4C: n = 5 (flow) and n = 8 (static). Figure S8B: n = 7 to 8 untreated organoids (control), n = 7 organoids with 1 μM 2-MeOE2, n = 4 organoids with 5 μM 2-MeOE2, n = 4 organoids with 10 μM 2-MeOE2, n = 5 organoids with 5 μM T-5224, n = 8 organoids with 10 μM T-5224, n = 9 organoids with 20 μM T-5224, n = 11 organoids with 1 μM Z-DEVD-FMK, n = 14 organoids with 10 μM Z-DEVD-FMK, n = 11 organoids with 50 μM Z-DEVD-FMK, n = 11 organoids with 10 μM quercetagetin, n = 12 organoids with 40 μM quercetagetin, n = 11 organoids with 200 μM quercetagetin, n = 11 organoids with 25 μM NSC23766, n = 9 organoids with NSC23766 50 μM, n = 7 organoids with NSC23766 100 μM, n = 8 organoids with rhCXCL16 (5 ng/ml), n = 8 organoids with rhCXCL16 (10 ng/ml), and n = 13 organoids with rhCXCL16 (25 ng/ml). Figure S10B: n = 5 untreated organoids (control), n = 5 organoids with 10 μM R-naproxen, n = 5 organoids with 20 μM R-naproxen, n = 5 organoids with 200 μM R-naproxen, n = 5 organoids with 400 μM R-naproxen, n = 6 organoids with 0.5 μM R-ketorolac, n = 6 organoids with 1 μM R-ketorolac, n = 6 organoids with 10 μM R-ketorolac, and n = 5 organoids with 20 μM R-ketorolac. Figure S12C: n = 120 tubules in PKHD1−/− organoids with no flow, n = 116 tubules in PKHD1−/− organoids with flow, and n = 139 tubules in PKHD1−/− organoids treated with 20 μM S-naproxen. Figure S14B: n = 7 organoids in PKHD1−/− organoids with flow condition, n = 6 organoids in PKHD1−/− organoids without flow condition, n = 6 organoids in PKHD1+/− organoids with flow condition, and n = 7 organoids in PKHD1+/− organoids without flow condition. Figure S14C: n = 37 tubules in PKHD1−/− organoids with flow condition, n = 81 tubules in PKHD1−/− organoids without flow condition, n = 51 tubules in PKHD1+/− organoids with flow condition, and n = 74 tubules in PKHD1+/− organoids without flow condition. Figure S17: n = 3 ARPKD patient-derived static organoids (no flow), n = 3 ARPKD patient-derived organoids with flow condition, n = 3 ARPKD patient-derived organoids treated with 20 μM R-naproxen under flow condition, and n = 3 ARPKD patient-derived organoids treated with 10 μM T-5224 under flow condition were analyzed. Figure S19A: n = 3 CDH1+ tubules in control organoids with flow condition were analyzed.
Acknowledgments
We thank E. P. Rhee and J. V. Bonventre for institutional support, K. Ida (MGH) for immunostaining for quality check in nephron progenitor cell stage, N. Lin and K. van Breugel (Wyss Institute) for help with organoid culture on a chip and harvesting organoids for enzyme-linked immunosorbent assay (ELISA) experiment, M. Mata (Wyss Institute) for help with organoid culture on a chip and treatment with chemical compounds, D. Tom (Leica Microsystems) for help with FLIM imaging analysis, B. Yoder (Department of Cell Biology, University of Alabama, Birmingham) and T. J. Watnick (Department of Nephrology, University of Maryland, Baltimore) for insightful discussion and expert advice, and TORAY for providing highly sensitive microarray kits.
Funding: This work was funded by a fellowship grant from the Uehara Memorial Foundation (K.H.), Harvard Stem Cell Institute Seed Grant (R.M.), NIH award DP2EB029388/DK133821 (R.M.), NIH grant U01EB028899/DK127587 (J.A.L. and R.M.), NIH grant UC2DK126023 (J.A.L. and R.M.), and NIH grant UH3TR002155 (M.T.V., J.A.L., and R.M.).
Author contributions: Methodology: K.H., T.M., K.T.K., N.R.G., M.T.V., T.F., M.Y., and R.M. Investigation: K.H., T.M., K.T.K., M.T.V., M.Y., and R.M. Visualization: K.H., T.M., K.T.K., M.T.V., M.Y., and R.M. Formal analysis: K.H. and T.M. Data curation: T.M. Resources: J.A.L. and R.M. Funding acquisition: J.A.L. and R.M. Project administration: M.Y., J.A.L., and R.M. Supervision: J.A.L. and R.M. Conceptualization: R.M. Writing—original draft: K.H. and R.M.
Competing interests: K.T.K., N.R.G., M.T.V., J.A.L., and R.M. are inventors on a patent related to this work filed by President and Fellows of Harvard College and Mass General Brigham (PCT/US2018/036677, filed on 8 June 2018, published on 13 December 2018). J.A.L. and R.M. are scientific advisory members in Trestle Biotherapeutics. The authors declare no other competing interests.
Data and materials availability: The microarray data have been deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE190272. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S19
Table S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
Data S1 to S7
REFERENCES AND NOTES
- 1.Morizane R., Lam A. Q., Freedman B. S., Kishi S., Valerius M. T., Bonventre J. V., Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wimmer R. A., Leopoldi A., Aichinger M., Wick N., Hantusch B., Novatchkova M., Taubenschmid J., Hämmerle M., Esk C., Bagley J. A., Lindenhofer D., Chen G., Boehm M., Agu C., Yang F., Fu B., Zuber J., Knoblich J., Kerjaschki D., Penninger J., Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koike H., Iwasawa K., Ouchi R., Maezawa M., Giesbrecht K., Saiki N., Ferguson A., Kimura M., Thompson W. L., Wells J. M., Zorn A. M., Takebe T., Modelling human hepato-biliary-pancreatic organogenesis from the foregut–midgut boundary. Nature 574, 112–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimori K., Ishikawa M., Otomo A., Atsuta N., Nakamura R., Akiyama T., Hadano S., Aoki M., Saya H., Sobue G., Okano H., Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 24, 1579–1589 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Dvela-Levitt M., Kost-Alimova M., Emani M., Kohnert E., Thompson R., Sidhom E.-H., Rivadeneira A., Sahakian N., Roignot J., Papagregoriou G., Montesinos M. S., Clark A. R., McKinney D., Gutierrez J., Roth M., Ronco L., Elonga E., Carter T. A., Gnirke A., Melanson M., Hartland K., Wieder N., Hsu J. C. H., Deltas C., Hughey R., Bleyer A. J., Kmoch S., Zivna M., Baresova V., Kota S., Schlondorff J., Heiman M., Alper S. L., Wagner F., Weins A., Golub T. R., Lander E. S., Greka A., Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell 178, 521–535.e23 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Gupta N., Matsumoto T., Hiratsuka K., Saiz E. G., Galichon P., Miyoshi T., Susa K., Tatsumoto N., Yamashita M., Morizane R., Modeling injury and repair in kidney organoids reveals that homologous recombination governs tubular intrinsic repair. Sci. Transl. Med. 14, eabj4772 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menezes L. F., Cai Y., Nagasawa Y., Silva A. M., Watkins M. L., Da Silva A. M., Somlo S., Guay-Woodford L. M., Germino G. G., Onuchic L. F., Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int. 66, 1345–1355 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Guay-Woodford L. M., Desmond R. A., Autosomal recessive polycystic kidney disease: The clinical experience in North America. Pediatrics 111, 1072–1080 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Hartung E. A., Guay-Woodford L. M., Autosomal recessive polycystic kidney disease: A hepatorenal fibrocystic disorder with pleiotropic effects. Pediatrics 134, e833–e845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgmaier K., Kilian S., Bammens B., Benzing T., Billing H., Büscher A., Galiano M., Grundmann F., Klaus G., Mekahli D., Michel-Calemard L., Milosevski-Lomic G., Ranchin B., Sauerstein K., Schaefer S., Shroff R., Sterenborg R., Verbeeck S., Weber L. T., Wicher D., Wühl E., Dötsch J., Schaefer F., Liebau M. C., Clinical courses and complications of young adults with autosomal recessive polycystic kidney disease (ARPKD). Sci. Rep. 9, 7919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann C., Guay-Woodford L. M., Harris P. C., Horie S., Peters D. J., Torres V. E., Polycystic kidney disease. Nat. Rev. Dis. Primers 4, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woollard J., Punyashtiti R., Richardson S., Masyuk T., Whelan S., Huang B., Lager D., Torres V., Gattone V., LaRusso N., Harris P. C., Ward C. J., A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int. 72, 328–336 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Mason S. B., Liang Y., Sinders R. M., Miller C. A., Eggleston-Gulyas T., Crisler-Roberts R., Harris P. C., Gattone V. H., Disease stage characterization of hepatorenal fibrocystic pathology in the PCK rat model of ARPKD. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 293, 1279–1288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang K.-J., Mehr A. P., Hamilton G. A., McPartlin L. A., Chung S., Suh K.-Y., Ingber D. E., Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 5, 1119–1129 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Homan K. A., Kolesky D. B., Skylar-Scott M. A., Herrmann J., Obuobi H., Moisan A., Lewis J. A., Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 6, 34845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musah S., Mammoto A., Ferrante T. C., Jeanty S. S., Hirano-Kobayashi M., Mammoto T., Roberts K., Chung S., Novak R., Ingram M., Tohid F.-D., Sandeep K., Weaver J. C., Church G. M., Ingber D. E., Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng. 1, 0069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low J. H., Li P., Chew E. G. Y., Zhou B., Suzuki K., Zhang T., Lian M. M., Liu M., Aizawa E., Esteban C. R., Yong K. S. M., Chen Q., Campistol J. M., Fang M., Khor C. C., Foo J. N., Izpisua Belmonte J. C., Xia Y., Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell 25, 373–387.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaika O., Mamenko M., Berrout J., Boukelmoune N., O’Neil R. G., Pochynyuk O., TRPV4 dysfunction promotes renal cystogenesis in autosomal recessive polycystic kidney disease. J. Am. Soc. Nephrol. 24, 604–616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homan K. A., Gupta N., Kroll K. T., Kolesky D. B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M. T., Ferrante T., Bonventre J. V., Lewis J. A., Morizane R., Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres V. E., Harris P. C., Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J. Am. Soc. Nephrol. 25, 18–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer D.-C., Jacoby U., Pape L., Ward C. J., Kuwertz-Broeking E., Renken C., Nizze H., Querfeld U., Rudolph B., Mueller-Wiefel D. E., Bergmann C., Haffner D., Activation of the AKT/mTOR pathway in autosomal recessive polycystic kidney disease (ARPKD). Nephrol. Dial. Transplant. 24, 1819–1827 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Dell K. M., The role of cilia in the pathogenesis of cystic kidney disease. Curr. Opin. Pediatr. 27, 212–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina F., Satlinb L. M., Polycystic kidney disease: The cilium as a common pathway in cystogenesis. Curr. Opin. Pediatr. 16, 171–176 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Ma M., Tian X., Igarashi P., Pazour G. J., Somlo S., Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat. Genet. 45, 1004–1012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao L., El-Jouni W., Kong F., Ramesh J., Kumar R. S., Shen X., Ren J., Devendra S., Dorschel A., Wu M., Barrera I., Tabari A., Hu K., Haque N., Yambayev I., Li S., Kumar A., Behera T. R., McDonough G., Furuichi M., Xifaras M., Lu T., Alhayaza R. M., Miyabayashi K., Fan Q., Ajay A. K., Zhou J., Genetic reduction of cilium length by targeting intraflagellar transport 88 protein impedes kidney and liver cyst formation in mouse models of autosomal polycystic kidney disease. Kidney Int. 98, 1225–1241 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Gallagher A. R., Somlo S., Loss of cilia does not slow liver disease progression in mouse models of autosomal recessive polycystic kidney disease. Kidney360 1, 962–968 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres V. E., Harris P. C., Mechanisms of disease: Autosomal dominant and recessive polycystic kidney diseases. Nat. Clin. Pract. Nephrol. 2, 40–55 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Martino F., Perestrelo A. R., Vinarský V., Pagliari S., Forte G., Cellular mechanotransduction: From tension to function. Front. Physiol. 9, 824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Discher D. E., Janmey P., Wang Y.-l., Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Kaarj K., Yoon J.-Y., Methods of delivering mechanical stimuli to organ-on-a-chip. Micromachines 10, 700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colom A., Derivery E., Soleimanpour S., Tomba C., Dal Molin M., Sakai N., González-Gaitán M., Matile S., Roux A., A fluorescent membrane tension probe. Nat. Chem. 10, 1118–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghavan V., Rbaibi Y., Pastor-Soler N. M., Carattino M. D., Weisz O. A., Shear stress-dependent regulation of apical endocytosis in renal proximal tubule cells mediated by primary cilia. Proc. Natl. Acad. Sci. U.S.A. 111, 8506–8511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W., Xu S., Woda C., Kim P., Weinbaum S., Satlin L. M., Effect of flow and stretch on the [Ca2+] i response of principal and intercalated cells in cortical collecting duct. Am. J. Physiol. Renal Physiol. 285, F998–F1012 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Malekshahabi T., Khoshdel Rad N., Serra A. L., Moghadasali R., Autosomal dominant polycystic kidney disease: Disrupted pathways and potential therapeutic interventions. J. Cell. Physiol. 234, 12451–12470 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Belibi F. A., Edelstein C. L., Novel targets for the treatment of autosomal dominant polycystic kidney disease. Expert Opin. Investig. Drugs 19, 315–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malas T. B., Formica C., Leonhard W. N., Rao P., Granchi Z., Roos M., Peters D. J., ‘t Hoen P. A. C., Meta-analysis of polycystic kidney disease expression profiles defines strong involvement of injury repair processes. Am. J. Physiol. Renal Physiol. 312, F806–F817 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Woo Y. M., Bae J.-B., Oh Y.-H., Lee Y.-G., Lee M. J., Park E. Y., Choi J.-K., Lee S., Shin Y., Lyu J., Jung H.-Y., Lee Y.-S., Hwang Y.-H., Kim Y.-J., Park J. H., Genome-wide methylation profiling of ADPKD identified epigenetically regulated genes associated with renal cyst development. Hum. Genet. 133, 281–297 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Curtis K. J., Coughlin T. R., Mason D. E., Boerckel J. D., Niebur G. L., Bone marrow mechanotransduction in porcine explants alters kinase activation and enhances trabecular bone formation in the absence of osteocyte signaling. Bone 107, 78–87 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Kroon J., Heemskerk N., Kalsbeek M. J., de Waard V., van Rijssel J., van Buul J. D., Flow-induced endothelial cell alignment requires the RhoGEF Trio as a scaffold protein to polarize active Rac1 distribution. Mol. Biol. Cell 28, 1745–1753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith C., Drug target validation: Hitting the target. Nature 422, 342–345 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Maxwell P., HIF-1: An oxygen response system with special relevance to the kidney. J. Am. Soc. Nephrol. 14, 2712–2722 (2003). [DOI] [PubMed] [Google Scholar]
- 42.A. N. Mahmoodi, P. Y. Kim, Ketorolac. StatPearls [Internet] (2021).
- 43.J. C. Brutzkus, M. Shahrokhi, M. Varacallo, Naproxen. StatPearls [Internet] (2021). [PubMed]
- 44.Aikawa Y., Morimoto K., Yamamoto T., Chaki H., Hashiramoto A., Narita H., Hirono S., Shiozawa S., Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat. Biotechnol. 26, 817–823 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Makino H., Seki S., Yahara Y., Shiozawa S., Aikawa Y., Motomura H., Nogami M., Watanabe K., Sainoh T., Ito H., Tsumaki N., Kawaguchi Y., Yamazaki M., Kimura T., A selective inhibition of c-Fos/activator protein-1 as a potential therapeutic target for intervertebral disc degeneration and associated pain. Sci. Rep. 7, 16983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oprea T. I., Sklar L. A., Agola J. O., Guo Y., Silberberg M., Roxby J., Vestling A., Romero E., Surviladze Z., Murray-Krezan C., Waller A., Ursu O., Hudson L. G., Wandinger-Ness A., Novel activities of select NSAID R-enantiomers against Rac1 and Cdc42 GTPases. PLOS ONE 10, e0142182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duggan K. C., Walters M. J., Musee J., Harp J. M., Kiefer J. R., Oates J. A., Marnett L. J., Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen. J. Biol. Chem. 285, 34950–34959 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.21–21.29.29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muto Y., Wilson P. C., Ledru N., Wu H., Dimke H., Waikar S. S., Humphreys B. D., Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 12, 2190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H., Kirita Y., Donnelly E. L., Humphreys B. D., Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moyer J. H., Lee-Tischler M. J., Kwon H.-Y., Schrick J. J., Avner E. D., Sweeney W. E., Godfrey V. L., Cacheiro N. L., Wilkinson J. E., Woychik R. P., Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science 264, 1329–1333 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Fitzgerald J., Hughes-Fulford M., Mechanically induced c-fos expression is mediated by cAMP in MC3T3-E1 osteoblasts. FASEB J. 13, 553–557 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Sundberg L. J., Galante L. M., Bill H. M., Mack C. P., Taylor J. M., An endogenous inhibitor of focal adhesion kinase blocks Rac1/JNK but not Ras/ERK-dependent signaling in vascular smooth muscle cells. J. Biol. Chem. 278, 29783–29791 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Okumura K., Itoh A., Isogai E., Hirose K., Hosokawa Y., Abiko Y., Shibata T., Hirata M., Isogai H., C-terminal domain of human CAP18 antimicrobial peptide induces apoptosis in oral squamous cell carcinoma SAS-H1 cells. Cancer Lett. 212, 185–194 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Nettersheim D., Jostes S., Fabry M., Honecker F., Schumacher V., Kirfel J., Kristiansen G., Schorle H., A signaling cascade including ARID1A, GADD45B and DUSP1 induces apoptosis and affects the cell cycle of germ cell cancers after romidepsin treatment. Oncotarget 7, 74931–74946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Gao J., Li L., Li Z., Du Y., Gong Y., The neuronal pentraxin II gene (NPTX2) inhibit proliferation and invasion of pancreatic cancer cells in vitro. Mol. Biol. Rep. 38, 4903–4911 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Ma J., Nie K., Redmond D., Liu Y., Elemento O., Knowles D., Tam W., EBV-miR-BHRF1-2 targets PRDM1/Blimp1: Potential role in EBV lymphomagenesis. Leukemia 30, 594–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo F., Xu Z., Zhang Y., Jiang P., Huang G., Chen S., Lyu X., Zheng P., Zhao X., Zeng Y., Wang S., He F., FXR induces SOCS3 and suppresses hepatocellular carcinoma. Oncotarget 6, 34606–34616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Den Hartog G., Chattopadhyay R., Ablack A., Hall E. H., Butcher L. D., Bhattacharyya A., Eckmann L., Harris P. R., Das S., Ernst P. B., Crowe S. E., Regulation of Rac1 and reactive oxygen species production in response to infection of gastrointestinal epithelia. PLOS Pathog. 12, e1005382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jou T.-S., James Nelson W., Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J. Cell Biol. 142, 85–100 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao G., Su X., Nguyen V., Roberts K., Li X., Takakura A., Plomann M., Zhou J., Polycystin-1 regulates actin cytoskeleton organization and directional cell migration through a novel PC1-Pacsin 2-N-Wasp complex. Hum. Mol. Genet. 23, 2769–2779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo Y. M., Kim D. Y., Koo N. J., Kim Y.-M., Lee S., Ko J. Y., Shin Y., Kim B. H., Mun H., Choi S., Lee E. J., Shin J.-O., Park E. Y., Bok J., Park J. H., Profiling of miRNAs and target genes related to cystogenesis in ADPKD mouse models. Sci. Rep. 7, 14151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Locatelli L., Maier J. A., Cytoskeletal remodeling mimics endothelial response to microgravity. Front. Cell Dev. Biol. 9, 733573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu Y., Brown A. C., Myers D. R., Sakurai Y., Mannino R. G., Tran R., Ahn B., Hardy E. T., Kee M. F., Kumar S., Bao G., Barker T. H., Lam W. A., Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc. Natl. Acad. Sci. U.S.A. 111, 14430–14435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang X., Shen Y., Zhang Y., Wei L., Lai Y., Wu J., Liu X., Liu X., Rac1 mediates laminar shear stress-induced vascular endothelial cell migration. Cell Adh. Migr. 7, 472–478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.H. Kahsay, Assessment and treatment of pain in pediatric patients. Curr. Pediatr. Res. (2017).
- 67.Zhang J., Hou Z., Yao C., Yu Y., Purification and properties of lipase from a Bacillus strain for catalytic resolution of (R)-Naproxen. J. Mol. Catal. B Enzym. 18, 205–210 (2002). [Google Scholar]
- 68.Freedman B. S., Lam A. Q., Sundsbak J. L., Iatrino R., Su X., Koon S. J., Wu M., Daheron L., Harris P. C., Zhou J., Bonventre J. V., Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J. Am. Soc. Nephrol. 24, 1571–1586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morizane R., Bonventre J. V., Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nat. Protoc. 12, 195–207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta N., Susa K., Yoda Y., Bonventre J. V., Valerius M. T., Morizane R., CRISPR/Cas9-based targeted genome editing for the development of monogenic diseases models with human pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 45, e50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolesky D. B., Homan K. A., Skylar-Scott M. A., Lewis J. A., Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. U.S.A. 113, 3179–3184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morgan R. G., Chambers A. C., Legge D. N., Coles S., Greenhough A., Williams A. C., Optimized delivery of siRNA into 3D tumor spheroid cultures in situ. Sci. Rep. 8, 7952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuželová K., Grebeňová D., Hrkal Z., Labeling of apoptotic JURL-MK1 cells by fluorescent caspase-3 inhibitor FAM-DEVD-fmk occurs mainly at site (s) different from caspase-3 active site. Cytometry A 71, 605–611 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Alvarado-Sansininea J. J., Sánchez-Sánchez L., López-Muñoz H., Escobar M. L., Flores-Guzmán F., Tavera-Hernández R., Jiménez-Estrada M., Quercetagetin and patuletin: Antiproliferative, necrotic and apoptotic activity in tumor cell lines. Molecules 23, 2579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou K., Rao J., Zhou Z.-h., Yao X.-h., Wu F., Yang J., Yang L., Zhang X., Cui Y.-h., Bian X.-W., Shi Y., Ping Y.-f., RAC1-GTP promotes epithelial-mesenchymal transition and invasion of colorectal cancer by activation of STAT3. Lab. Invest. 98, 989–998 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Xiao G., Wang X., Wang J., Zu L., Cheng G., Hao M., Sun X., Xue Y., Lu J., Wang J., CXCL16/CXCR6 chemokine signaling mediates breast cancer progression by pERK1/2-dependent mechanisms. Oncotarget 6, 14165–14178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ryou M.-G., Choudhury G. R., Li W., Winters A., Yuan F., Liu R., Yang S.-H., Methylene blue-induced neuronal protective mechanism against hypoxia-reoxygenation stress. Neuroscience 301, 193–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hiratsuka K., Monkawa T., Akiyama T., Nakatake Y., Oda M., Goparaju S. K., Kimura H., Chikazawa-Nohtomi N., Sato S., Ishiguro K., Yamaguchi S., Suzuki S., Morizane R., Ko S. B. H., Itoh H., Ko M. S. H., Induction of human pluripotent stem cells into kidney tissues by synthetic mRNAs encoding transcription factors. Sci. Rep. 9, 913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pearson H. B., Li J., Meniel V. S., Fennell C. M., Waring P., Montgomery K. G., Rebello R. J., Macpherson A. A., Koushyar S., Furic L., Cullinane C., Clarkson R. W., Smalley M. J., Simpson K. J., Phesse T. J., Shepherd P. R., Humbert P. O., Sansom O. J., Phillips W. A., Identification of Pik3ca mutation as a genetic driver of prostate cancer that cooperates with Pten loss to accelerate progression and castration-resistant growth. Cancer Discov. 8, 764–779 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S19
Table S1
Movies S1 and S2
Data S1 to S7