Abstract
In the initial process of coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects respiratory epithelial cells and then transfers to other organs the blood vessels. It is believed that SARS-CoV-2 can pass the vascular wall by altering the endothelial barrier using an unknown mechanism. In this study, we investigated the effect of SARS-CoV-2 on the endothelial barrier using an airway-on-a-chip that mimics respiratory organs and found that SARS-CoV-2 produced from infected epithelial cells disrupts the barrier by decreasing Claudin-5 (CLDN5), a tight junction protein, and disrupting vascular endothelial cadherin–mediated adherens junctions. Consistently, the gene and protein expression levels of CLDN5 in the lungs of a patient with COVID-19 were decreased. CLDN5 overexpression or Fluvastatin treatment rescued the SARS-CoV-2–induced respiratory endothelial barrier disruption. We concluded that the down-regulation of CLDN5 expression is a pivotal mechanism for SARS-CoV-2–induced endothelial barrier disruption in respiratory organs and that inducing CLDN5 expression is a therapeutic strategy against COVID-19.
Organ-on-a-chip technology elucidates the mechanism of SARS-CoV-2 infection–mediated disruption of respiratory vascular barrier.
INTRODUCTION
Airways and alveoli are the main organs infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 infects ciliated cells and type II alveolar epithelial cells using angiotensin-converting enzyme 2 (ACE2) (1). Because vascular endothelial cells (ECs) hardly express ACE2, SARS-CoV-2 rarely infects these cells (2). However, vasculitis and vascular barrier disruption are often observed in patients with severe coronavirus disease 2019 (COVID-19) (3). This barrier disruption promotes the transfer of SARS-CoV-2 from the respiratory organs to other organs through blood vessels. Vascular hyperpermeability in the lung induces the infiltration of immune cells and blood components there and respiratory failure, including pneumonia and acute respiratory distress syndrome (ARDS) (4). In this respect, elucidation of the mechanism for SARS-CoV-2–induced vascular permeability is an urgent issue, but the mechanism is poorly understood.
ECs cover the inner layer of blood vessels. In respiratory organs, ECs control the passage of substances and cells from epithelial cells to blood by regulating vascular permeability. The loss or impairment of this control can lead to severe and sometimes fatal organ dysfunction in pathological conditions, including inflammation and sepsis (5). Endothelial permeability is mainly regulated by cell-cell junctions, including adherens and tight junctions. Adherence junctions are regulated by cadherins and nectins, whereas tight junctions are composed of occludin, claudins, and junctional adhesion molecules (5, 6). Among these transmembrane proteins, vascular endothelial cadherin (VE-cadherin) and Claudin-5 (CLDN5), which are specifically expressed in ECs, are well studied as regulators of endothelial permeability. VE-cadherin binds to several proteins, including β-catenin, via its cytoplasmic domain. Pivotal roles of VE-cadherin for vascular integrity and permeability have been shown by antibody-mediated loss-of-function studies (7). On the other hand, CLDN5 has been shown to control the tight sealing of ECs in the blood-brain barrier (BBB). CLDN5 size-selectively inhibits the transfer of small molecules across the vasculature in the brain (8). However, little is known about the physiological function of CLDN5 in other organs.
A large number of patients with severe COVID-19 die from ARDS, pneumonia, and pulmonary edema, which are induced by vascular hyperpermeability in the lung (9). In patients with severe COVID-19, activated ECs produce inflammatory cytokines and adhesion molecules, including intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1), which induce cytokine storm, the infiltration of inflammatory cells, and vascular leakage (10, 11). This information suggests that SARS-CoV-2 also affects the inflammation and permeability of ECs in the lung. However, it has been reported that SARS-CoV-2 hardly infects ECs (2). In addition, there is limited information about how ECs are activated in patients with COVID-19. These studies would benefit from clarifying the effect of SARS-CoV-2 infection on EC functions in the lung, especially regarding the junction-mediated endothelial barrier function.
In patients with COVID-19, SARS-CoV-2 is found in the blood, and SARS-CoV-2 protein is expressed in many organs including respiratory organs (12). On the other hand, in the hamsters and ferrets widely used in COVID-19 studies, SARS-CoV-2 protein is hardly detected in organs other than respiratory organs (13). This difference is thought to be due to species differences of the SARS-CoV-2 distribution in the body. Therefore, a human model to investigate the respiratory-to-vascular transfer of SARS-CoV-2 and the SARS-CoV-2 infection–mediated disruption of the endothelial barrier is preferred. To examine the effects of SARS-CoV-2 replicated in airway epithelial cells on vascular ECs, in the present study, we used airway-on-a-chip technology (14, 15). An airway-on-a-chip is a coculture system of airway epithelial cells and ECs and mimics the in vivo dynamic microenvironment by flowing air and medium. In general, organ-on-a-chip technology makes it possible to generate a three-dimensional and dynamic in vitro model and to study interactions between multiple organs (e.g., respiratory organs and blood vessels).
In this study, a comprehensive gene expression analysis was performed of human lung microvascular ECs (HMVEC-L) in an airway-on-a-chip infected with SARS-CoV-2. We found that SARS-CoV-2 disrupts the respiratory endothelial barrier by decreasing CLDN5 expression and disorganizing the VE-cadherin–mediated junction. A loss of CLDN5 function by anti-CLDN5 antibody induced severe vascular leakage and edema specifically in the lungs of mice. Furthermore, we demonstrated that the respiratory endothelial barrier disruption caused by SARS-CoV-2 is suppressed by CLDN5 overexpression and Fluvastatin treatment. In conclusion, we successfully identified CLDN5 as a SARS-CoV-2–responsive gene in HMVEC-L and revealed the mechanism of SARS-CoV-2–mediated disruption of the respiratory vascular barrier in respiratory organs.
RESULTS
Investigating the causative gene of SARS-CoV-2 exposure–mediated vascular barrier disruption
To mimic the respiratory-to-vascular transfer of SARS-CoV-2 and SARS-CoV-2 infection–mediated disruption of the endothelial barrier, we used an airway-on-a-chip (Fig. 1A). An airway-on-a-chip consists of airway epithelial cells (ciliated, club, goblet, and basal cells) and HMVEC-L. By flowing air and medium through the airway and blood vessel channels, respectively, it is possible to create a mechanical stimulus that mimics the in vivo airway microenvironment. The viral genome copy number in the cell culture supernatant of the airway channel increased and reached a maximum at 4 days postinfection (dpi), while that in the blood vessel channel started increasing at 3 dpi and reached a maximum at 7 dpi (Fig. 1B). However, SARS-CoV-2 did not infect HMVEC-L in the absence of exogenous ACE2 expression (fig. S1). These results suggest that SARS-CoV-2 inoculated from the airway channel replicates behavior in airway epithelial cells before invading blood vessel channels. Vascular barrier disruption due to SARS-CoV-2 exposure was confirmed in eight viral variants but was highest in SARS-CoV-2 B.1.617.2 (fig. S2).
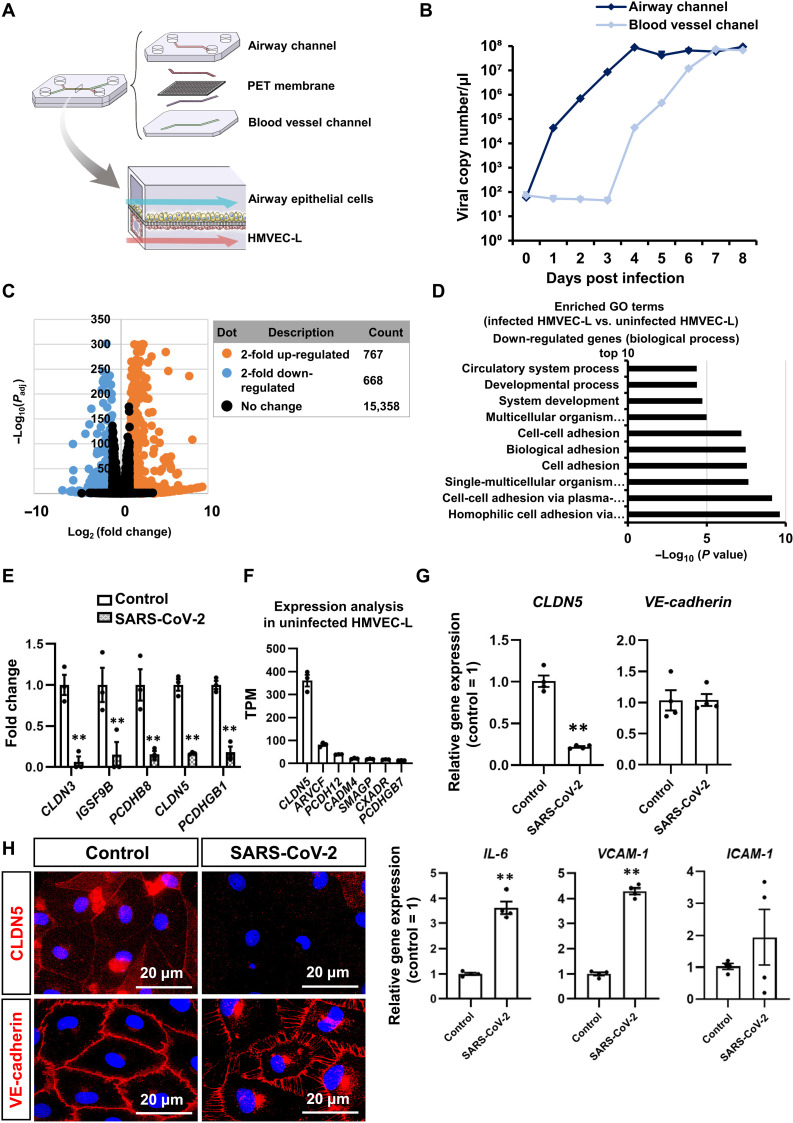
Fig. 1. SARS-CoV-2 down-regulates CLDN5 expression in HMVEC-L of the airway-on-a-chip.
(A) Schematic illustration of the airway-on-a-chip. Medium containing 0.1 multiplicity of infection (MOI) of SARS-CoV-2 was injected into the airway channel, which was then cultured for 8 days. (B) Viral copy numbers in the cell culture supernatant of the airway and blood vessel channels. (C) A volcano plot of differentially expressed genes between uninfected and infected HMVEC-L [log2 fold change > 2, adjusted P value (Padj) < 0.01]. Orange dots represent up-regulated genes, and blue dots represent down-regulated genes. (D) A Gene Ontology (GO) enrichment analysis of uninfected versus infected HMVEC-L in the airway-on-a-chip. (E) The top five down-regulated genes in homophilic cell adhesion via plasma membrane adhesion molecules and cell-cell adhesion via plasma membrane adhesion molecules in infected HMVEC-L compared with uninfected HMVEC-L. Control = 1. Two-way analysis of variance (ANOVA) with the Sidak post hoc test (**P < 0.01, control versus SARS-CoV-2). (F) The endogenous gene expression levels of the top seven genes in homophilic cell adhesion via plasma membrane adhesion molecules and cell-cell adhesion via plasma membrane adhesion molecules in infected HMVEC-L. (G) The gene expression levels of CLDN5, VE-cadherin, IL-6, VCAM-1, and ICAM-1 in uninfected and infected HMVEC-L. Control = 1. Unpaired two-tailed Student’s t test (**P < 0.01). (H) Immunofluorescent staining for VE-cadherin and CLDN5 in HMVEC-L in the airway-on-a-chip. Data are expressed as the means ± SEM (n = 3).
To search for the causative gene of the SARS-CoV-2 infection–mediated vascular barrier disruption, we performed an RNA sequencing (RNA-seq) analysis of HMVEC-L in the infected airway-on-a-chip. Although SARS-CoV-2 does not infect HMVEC-L, we expected that the gene expression profile of HMVEC-L would be significantly altered by contact with SARS-CoV-2 and cytokines secreted from infected airway epithelial cells (fig. S1). The infected airway-on-a-chip was cultured for 8 days, and HMVEC-L were collected for the RNA-seq analysis. SARS-CoV-2 infection up-regulated and down-regulated 767 and 668 genes more than twofold, respectively (Fig. 1C). An enrichment analysis showed that the up-regulated genes include innate immune response–, inflammation-, and viral response–related genes (fig. S3A), while down-regulated genes include cell adhesion–related genes (Fig. 1D). The top five down-regulated genes in the “homophilic cell adhesion via plasma membrane adhesion molecules” and “cell-cell adhesion via plasma membrane adhesion molecules” categories were CLDN3, CLDN5, PCDHB1, PCDHB8, and IGSF9B (Fig. 1E). Among the down-regulated genes in the homophilic cell adhesion via plasma membrane adhesion molecules and cell-cell adhesion via plasma membrane adhesion molecules categories, the endogenous gene expression of CLDN5 was the highest (Fig. 1F and fig. S3B). Because SARS-CoV-2 infection did not change the cell number of HMVEC-L, we confirmed that the decrease in CLDN5 expression level was not due to a decrease in cell viability (fig. S4). Thus, we concluded that CLDN5 is a potential causative gene of the SARS-CoV-2 infection–mediated respiratory vascular barrier disruption.
SARS-CoV-2 suppresses CLDN5 expression in lung microvascular ECs
To further investigate the SARS-CoV-2 infection–mediated respiratory vascular barrier disruption, we analyzed the expression of tight junction–related genes including CLDN5 in HMVEC-L isolated from the infected airway-on-a-chip. Consistent with the RNA-seq data, a quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis showed that the CLDN5 expression level was decreased by SARS-CoV-2 infection (Fig. 1G). In contrast, SARS-CoV-2 infection did not alter the VE-cadherin expression level but increased the expression levels of several inflammatory genes, including interleukin-6 (IL-6), VCAM-1, and ICAM-1 (Fig. 1G). In addition, immunofluorescent staining demonstrated a down-regulation of CLDN5 expression at the junctions between HMVEC-L (Fig. 1H). VE-cadherin staining revealed SARS-CoV-2 infection–mediated torn adherens junctions and free space between HMVEC-L without a significant change in the VE-cadherin expression. These results indicate that SARS-CoV-2 infection decreased the CLDN5 expression and altered the VE-cadherin localization in HMVEC-L, suggesting that SARS-CoV-2 regulates respiratory endothelial barrier function by disrupting both tight and adherens junctions.
SARS-CoV-2 exposure decreased CLDN5 expression independent of infected airway epithelial cells
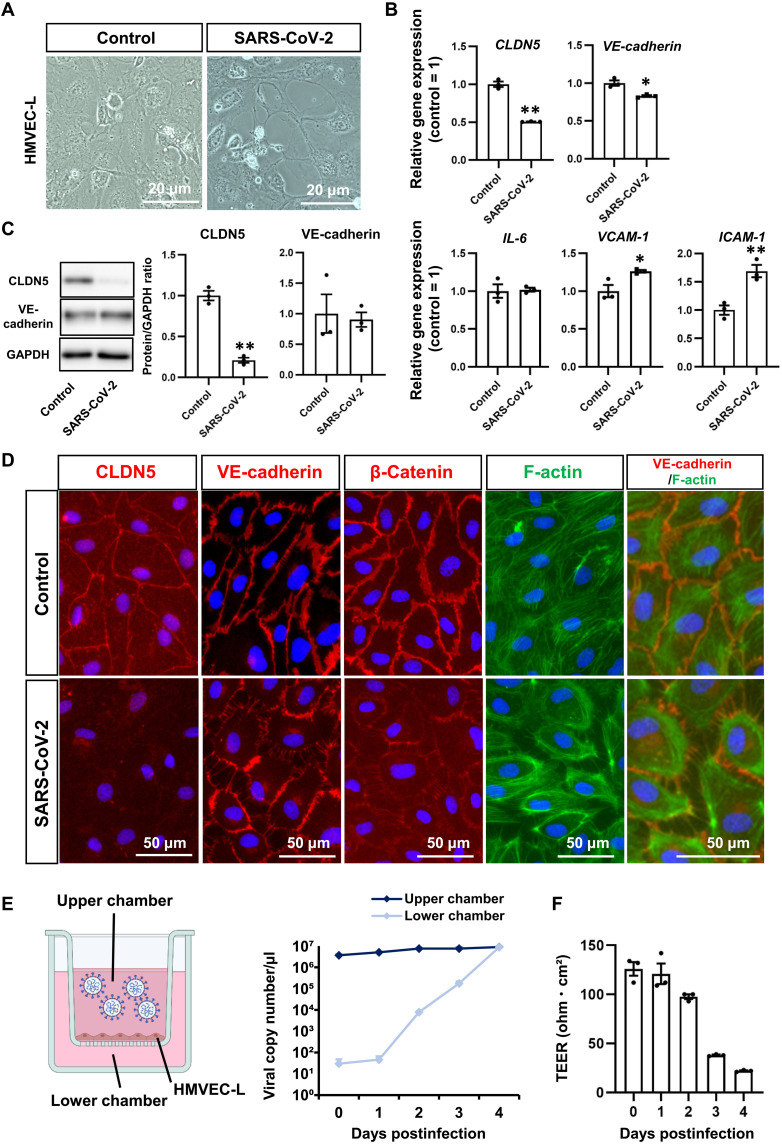
Infection experiments in the HMVEC-L monolayer were conducted to investigate whether the decreased CLDN5 expression in HMVEC-L of the infected airway-on-a-chip was caused by contact with SARS-CoV-2 or other secreted molecules derived from airway epithelial cells. When HMVEC-L were exposed to 0.1 multiplicity of infection (MOI) of SARS-CoV-2, CLDN5 down-regulation was not observed (fig. S5). We expected that HMVEC-L need to be continuously exposed to a high titer of SARS-CoV-2 to cause CLDN5 down-regulation, because HMVEC-L in the infected airway-on-a-chip are continuously exposed to a high titer of SARS-CoV-2 secreted from infected airway epithelial cells. To test this hypothesis, HMVEC-L were exposed to a high titer (1 MOI) of SARS-CoV-2 for four consecutive days. This treatment disrupted the HMVEC-L monolayer, suggesting weakened cell-cell adhesion (Fig. 2A). SARS-CoV-2 significantly decreased CLDN5 mRNA and protein expressions (Fig. 2, B and C, and fig. S5), while the down-regulation of VE-cadherin was hardly observed. In addition, SARS-CoV-2 exposure increased the expression levels of VCAM-1 and ICAM-1 but did not alter the expression level of IL-6. Immunofluorescent staining demonstrated decreased CLDN5 expression and disorganized VE-cadherin–mediated adherens junctions (Fig. 2D), consistent with observations in the infected airway-on-a-chip. Immunofluorescent staining also showed that SARS-CoV-2 exposure disrupted and decreased β-catenin localization at the cell junction. In addition, phalloidin staining demonstrated that SARS-CoV-2 exposure increased F-actin localization along the cell membrane and distances between cells. Consistently, SARS-CoV-2 can break through the endothelial monolayer (Fig. 2E), and transendothelial electrical resistance (TEER) was temporally decreased in the presence of SARS-CoV-2 (Fig. 2F). These results indicate that continuous exposure to abundant SARS-CoV-2 down-regulates CLDN5 and disrupts VE-cadherin–mediated cell junctions in HMVEC-L independent of infected airway epithelial cells.
Fig. 2. SARS-CoV-2 suppresses CLDN5 expression in lung microvascular ECs.
(A) Phase images of HMVEC-L cultured on a chamber slide in the presence or absence of 1 MOI of SARS-CoV-2 for 4 days. (B) The gene expression levels of CLDN5, VE-cadherin, IL-6, VCAM-1, and ICAM-1 in uninfected and infected HMVEC-L. Unpaired two-tailed Student’s t test (*P < 0.05 and **P < 0.01). (C) The protein expression levels of CLDN5 and VE-cadherin in uninfected and infected HMVEC-L. Unpaired two-tailed Student’s t test (**P < 0.01). (D) Immunofluorescent staining for CLDN5, VE-cadherin, β-catenin, and F-actin in uninfected and infected HMVEC-L. (E and F) The viral permeability assay using HMVEC-L monolayer. The medium containing 1 MOI of SARS-CoV-2 was injected into the upper chamber of the HMVEC-L monolayer. The viral transfer (E) and TEER (F) were evaluated. Data are expressed as the means ± SEM (n = 3).
CLDN5 down-regulation induces severe vascular leakage specifically in the lung
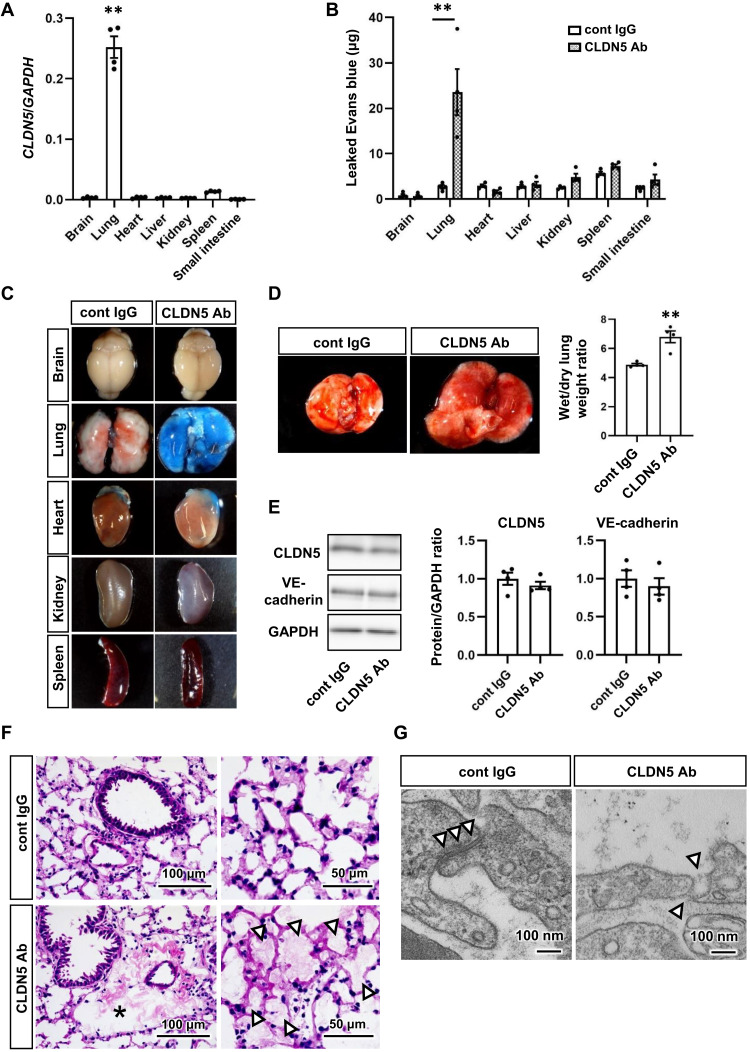
To investigate the effect of acute functional defects of CLDN5 on vascular leakage in vivo, an antibody-mediated loss-of-function study was conducted. We previously reported a well-characterized antibody against CLDN5 that specifically binds to the extracellular domain of primate CLDN5 and inhibits its function in the monkey brain (16, 17). To apply this efficient antibody to mice, we generated human CLDN5 knock-in mice (fig. S6). In these mice, CLDN5 expression was highest in the lung compared with other organs (Fig. 3A), which is similar to the expression pattern in human organs, as shown in the CLDN5 transcriptomics data of the Human Protein Atlas. Intravenous injection of CLDN5 antibody induced extravasation of Evans blue, which binds to albumin and behaves as a large molecule, in the lung but not in other organs (Fig. 3, B and C). In addition, CLDN5 antibody induced pulmonary edema and increased the lung wet/dry weight ratio (Fig. 3D) without changing the expression levels of CLDN5 or VE-cadherin (Fig. 3E). Further analysis using the lung sections indicated that CLDN5 antibody induces perivascular and alveolar edema (Fig. 3F) and disruption of the junction structure between pulmonary ECs (Fig. 3G and fig. S7). These findings indicate that the loss of CLDN5 function induces severe vascular leakage and edema specifically in the lung by disrupting endothelial junctions and that SARS-CoV-2 infection–mediated CLDN5 down-regulation is an important cause of the vascular barrier disruption in patients with COVID-19.
Fig. 3. CLDN5 inhibition induces severe lung-specific vascular leakage.
(A) The gene expression levels of CLDN5 in organs of human CLDN5 homozygous knock-in mice (hCLDN5-KI mice). One-way ANOVA followed by Dunnett’s post hoc test (**P < 0.01, lung versus other organs). (B and C) The Miles assay using hCLDN5-KI mice injected with anti-CLDN5 antibody (Ab) or control (cont) immunoglobulin G (IgG). Vascular leakage was assayed by measuring extravasated Evans blue (B). Two-way ANOVA with the Sidak post hoc test (**P < 0.01, cont IgG versus CLDN5 Ab). Images are representative organs used for the assay (C). (D) Quantification of the lung wet/dry weight ratio. Images are the lungs from hCLDN5-KI mice injected with anti-CLDN5 Ab or cont IgG. Unpaired two-tailed Student’s t test (**P < 0.01). (E) The protein expression levels of CLDN5 and VE-cadherin in the lungs of the antibody-injected mice. (F) Representative images of hematoxylin and eosin–stained lung sections from antibody-injected mice. The asterisk and white arrowheads indicate perivascular and alveolar edema, respectively. (G) Transmission electron microscopy (TEM) images of junctions between lung ECs (white arrowheads). Low-magnification TEM images are shown in fig. S7. Data are expressed as the means ± SEM (n = 4).
Mechanism of SARS-CoV-2 exposure–mediated CLDN5 down-regulation in lung microvascular ECs
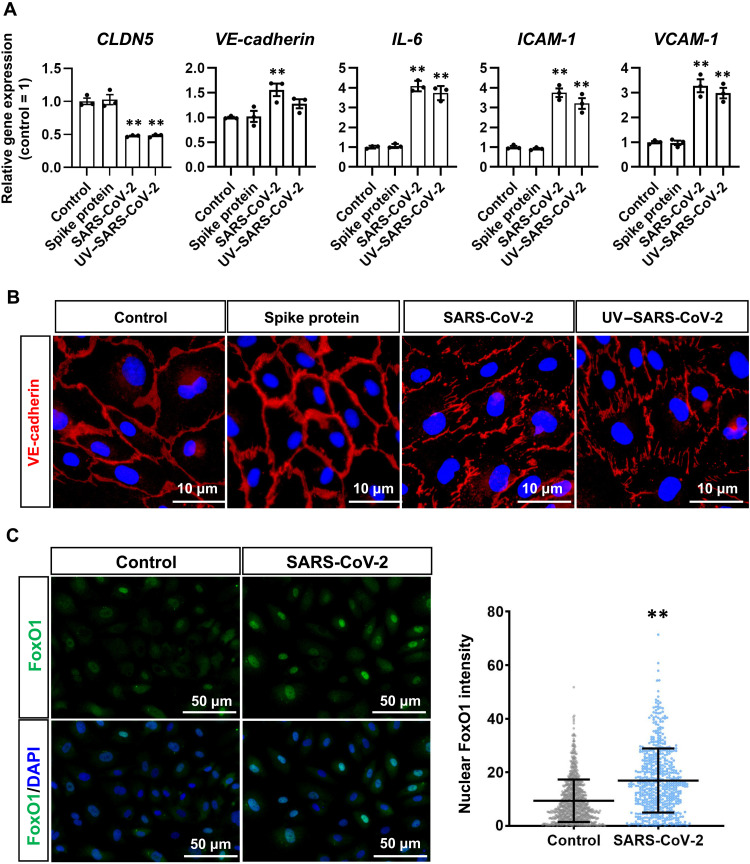
To investigate whether SARS-CoV-2 infection is necessary for the CLDN5 down-regulation in HMVEC-L, the HMVEC-L monolayer was exposed to ultraviolet-irradiated SARS-CoV-2 (UV–SARS-CoV-2), which lacks the ability to amplify in host cells. Both UV–SARS-CoV-2 and intact SARS-CoV-2 decreased the expression of CLDN5 but not of VE-cadherin (Fig. 4A). In addition, both UV–SARS-CoV-2 and intact SARS-CoV-2 increased the expression of inflammatory genes including IL-6, ICAM-1, and VCAM-1, while recombinant SARS-CoV-2 Spike protein did not (Fig. 4A). Consistently, UV–SARS-CoV-2 but not Spike protein induced the disorganized localization of VE-cadherin (Fig. 4B). These results indicate that SARS-CoV-2 replication is not essential for the CLDN5 down-regulation or endothelial barrier disruption in HMVEC-L and that these phenotypes cannot be induced by Spike protein alone.
Fig. 4. Mechanisms for SARS-CoV-2 exposure–mediated CLDN5 down-regulation and respiratory endothelial barrier disruption.
(A) The gene expression levels of CLDN5, VE-cadherin, IL-6, VCAM-1, and ICAM-1 in HMVEC-L in an airway-on-a-chip treated with Spike proteins, intact SARS-CoV-2 (0.1 MOI, exposed at 0 dpi), or UV-irradiated SARS-CoV-2 (UV–SARS-CoV-2; 1 MOI, exposed at 0, 1, 2, and 3 dpi) (control = 1). One-way ANOVA followed by Dunnett’s post hoc test (**P < 0.01, compared with “control”). Data are expressed as the means ± SEM (n = 3). (B) Immunofluorescent staining for VE-cadherin in HMVEC-L in airway-on-a-chip treated with recombinant Spike proteins, intact SARS-CoV-2, or UV–SARS-CoV-2. (C) Immunofluorescent staining for forkhead box protein O1 (FoxO1) in HMVEC-L treated with SARS-CoV-2. The nuclear FoxO1 intensity in the cells was quantified. Mann-Whitney U test (**P < 0.01).
To elucidate the mechanism that decreases CLDN5 mRNA and protein expression by SARS-CoV-2 exposure, we attempted to analyze transcription factors that can control CLDN5 expression. A previous report indicated that the transcription factor forkhead box protein O1 (FoxO1) suppresses CLDN5 expression by translocating from the cytoplasm to the nucleus (18). To investigate whether FoxO1 is involved in the SARS-CoV-2 exposure–mediated CLDN5 down-regulation, immunofluorescent staining was performed and showed that SARS-CoV-2 exposure increased the amount of FoxO1 translocated to the nucleus (Fig. 4C). Together, these results suggest that SARS-CoV-2 exposure–mediated CLDN5 down-regulation occurs through an increase in the amount of nuclear-localized FoxO1.
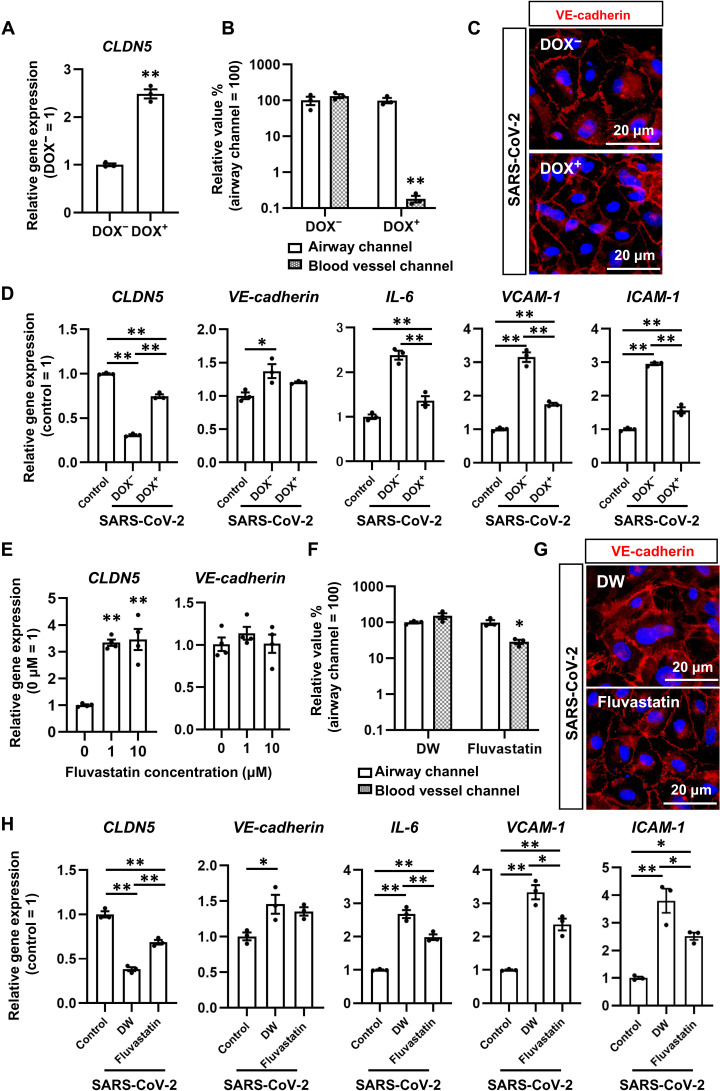
CLDN5 overexpression suppressed SARS-CoV-2 exposure–induced respiratory vascular barrier disruption
To investigate whether exogenous CLDN5 expression inhibits the SARS-CoV-2–induced respiratory endothelial barrier disruption, we established human CLDN5–expressing HMVEC-L (HMVEC-L–CLDN5) by transfecting doxycycline (DOX)–inducible CLDN5 vectors using the piggyBac system. We confirmed that DOX treatment increases the expression level of CLDN5 in HMVEC-L–CLDN5 (Fig. 5A). CLDN5 overexpression prevented SARS-CoV-2 entry into the blood vessel channel (Fig. 5B and fig. S8A) and the disruption of VE-cadherin–mediated adherens junctions (Fig. 5C). These results suggest that exogenous CLDN5 expression in HMVEC-L suppresses the SARS-CoV-2 exposure–mediated respiratory endothelial barrier disruption. In addition, exogenous CLDN5 expression did not alter the VE-cadherin expression, but it did suppress inflammatory responsive genes (IL-6, ICAM-1, and VCAM-1) and innate immunity–related genes (Fig. 5D and fig. S9A). Together, these results suggest that CLDN5 overexpression suppresses SARS-CoV-2 exposure–mediated endothelial inflammation by protecting respiratory endothelial barriers.
Fig. 5. CLDN5 overexpression and Fluvastatin treatment inhibit SARS-CoV-2–induced respiratory endothelial barrier disruption.
(A) The gene expression levels of CLDN5 in HMVEC-L treated with or without 1 μM DOX. HMVEC-L capable of overexpressing CLDN5 in response to DOX treatment were established using the piggyBac system. Unpaired two-tailed Student’s t test (**P < 0.01). (B) The viral copy numbers in the cell culture supernatant of the airway and blood vessel channels in the presence or absence of 1 μM DOX. Two-way ANOVA followed by the Sidak post hoc test (**P < 0.01). (C) Immunofluorescent staining for VE-cadherin in SARS-CoV-2–exposed HMVEC-L treated with or without 1 μM DOX. (D) The gene expression levels of CLDN5, VE-cadherin, IL-6, VCAM-1, and ICAM-1 in HMVEC-L treated with or without 1 μM DOX. One-way ANOVA followed by Tukey’s post hoc test (*P < 0.05 and **P < 0.01). (E) The gene expression levels of CLDN5 and VE-cadherin in HMVEC-L treated with or without 1 or 10 μM Fluvastatin. One-way ANOVA followed by Dunnett’s post hoc test (**P < 0.01, compared with “0 μM”). (F) The viral copy numbers in the cell culture supernatant of the airway and blood vessel channels in the presence or absence of 10 μM Fluvastatin. Two-way ANOVA with the Sidak post hoc test (*P < 0.05). DW, vehicle (distilled water)–treated cells. (G) Immunofluorescent staining for VE-cadherin in SARS-CoV-2–exposed HMVEC-L treated with or without 10 μM Fluvastatin. (H) The gene expression levels of CLDN5, VE-cadherin, IL-6, VCAM-1, and ICAM-1 in HMVEC-L treated with or without 10 μM Fluvastatin. One-way ANOVA followed by Tukey’s post hoc test (*P < 0.05 and **P < 0.01). Data are expressed as the means ± SEM (n = 3).
Fluvastatin treatment up-regulates CLDN5 expression and suppresses SARS-CoV-2 exposure–induced respiratory endothelial barrier disruption
A recent report found that patients with COVID-19 who were treated with statins showed decreased mortality (19). It was also reported that one of these statins, simvastatin, attenuates acute lung injury by increasing CLDN5 expression in pulmonary artery ECs (20). To investigate whether Fluvastatin treatment increased CLDN5 expression, HMVEC-L were treated with Fluvastatin, and CLDN5 expression was analyzed (Fig. 5E). Fluvastatin treatment increased the CLDN5 expression level. It also partially prevented the invasion of SARS-CoV-2 into the blood vessel channel (Fig. 5F and fig. S8B) and the disruption of VE-cadherin–mediated adherens junctions (Fig. 5G). In addition, Fluvastatin treatment increased CLDN5 expression and suppressed inflammatory genes (IL-6, ICAM-1, and VCAM-1) and innate immunity–related genes (Fig. 5H and fig. S9B). Together, Fluvastatin treatment increases CLDN5 expression and suppresses SARS-CoV-2 exposure–mediated respiratory endothelial barrier disruption.
CLDN5 expression in patients with COVID-19
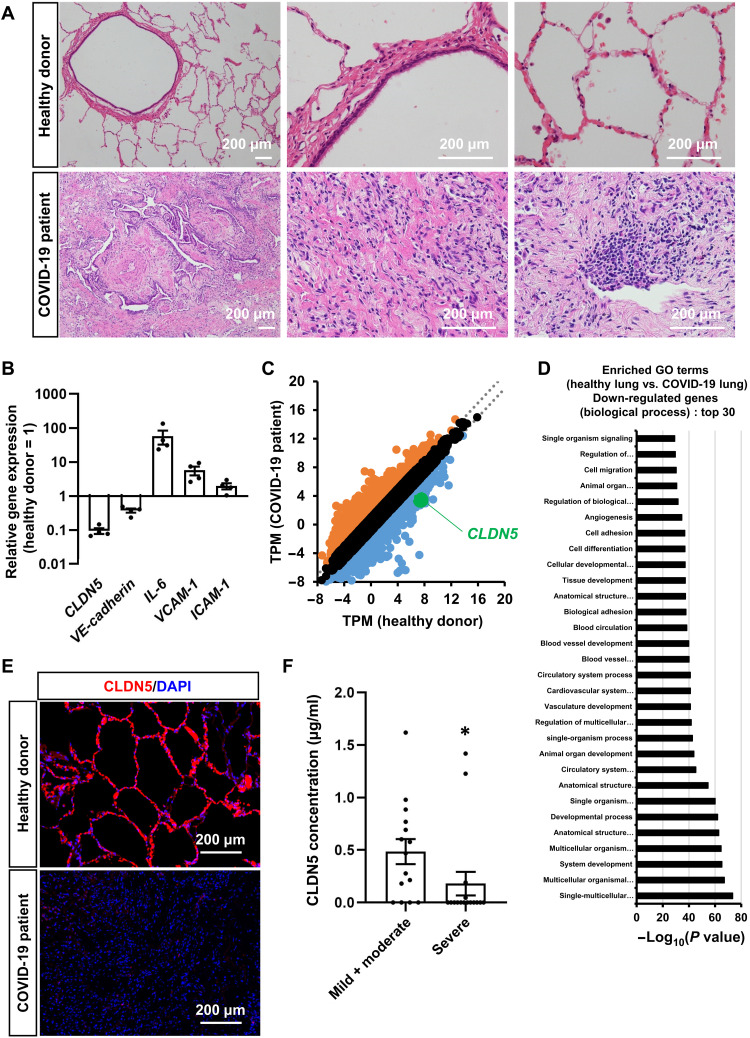
To examine the relationship between COVID-19 and CLDN5, we analyzed the CLDN5 expression in human lungs infected with or without COVID-19. In Fig. 6 (A to E), we used samples from the lungs of a COVID-19 patient with severe pneumonia (21) and surplus samples from the lungs of three living lung transplant donors. Fibrosis and immune cell infiltration were confirmed in the lungs of a patient with COVID-19 (Fig. 6A). In addition, the gene expression level of CLDN5 was decreased (Fig. 6B). This decrease held even when normalizing to the expression of conventional EC markers or aerocyte-specific markers (fig. S10), suggesting that it occurred in ECs. An RNA-seq analysis demonstrated that the expression levels of cell adhesion–, blood vessel morphogenesis–, and blood vessel development–related genes were decreased (Fig. 6, C and D). Immunofluorescent staining showed that CLDN5 expression was almost abolished in the lungs of a patient with COVID-19 (Fig. 6E). In addition, CLDN5 concentrations in the sera of patients with COVID-19 of different severity were measured (Fig. 6F). Because the main source of CLDN5 is vascular ECs, the amount of CLDN5 in serum is considered to correlate with the expression level of CLDN5 in those cells. Serum CLDN5 concentrations in patients with severe COVID-19 (180 pg/ml) were lower than those patients with mild and moderate COVID-19 (484 pg/ml). These observations confirmed that a decrease in the expression level of CLDN5 occurred not only in the infected airway-on-a-chip and HMVEC-L monolayer but also in the lungs of a patient with COVID-19.
Fig. 6. CLDN5 expression analysis of patients with COVID-19.
(A) Representative images of hematoxylin and eosin–stained lung sections from patients with or without COVID-19. (B) The gene expression levels of CLDN5, VE-cadherin, IL-6, VCAM-1, and ICAM-1 in the lungs of patients with or without COVID-19. (C) A scatterplot of differentially expressed genes between the lungs of a healthy donor and the lungs of a patient with COVID-19. Orange dots represent up-regulated genes (log2 fold change > 1), and blue dots represent down-regulated genes (log2 fold change > −1). (D) A GO enrichment analysis of the lungs of a healthy donor versus the lungs of a patient with COVID-19. (E) Immunofluorescent staining for CLDN5 in the lungs of patients with or without COVID-19. (F) CLDN5 concentration in sera of patients with mild/moderate and severe COVID-19 within 1 week after onset was measured by enzyme-linked immunosorbent assay (ELISA). Mann-Whitney U test (*P < 0.05). Data are expressed as the means ± SEM. The information of patients with COVID-19 is summarized in table S3.
DISCUSSION
Using an airway-on-a-chip, we demonstrated in this study that SARS-CoV-2 exposure induces vascular barrier disruption by decreasing the EC-specific tight junction protein CLDN5. This barrier disruption increased transport of SARS-CoV-2 across the endothelial monolayer. In addition, CLDN5 overexpression and Fluvastatin treatment suppressed SARS-CoV-2 exposure–mediated respiratory endothelial permeability. Therefore, we successfully revealed the mechanism by which SARS-CoV-2 exposure–mediated vascular hyperpermeability occurs in respiratory organs and provide a therapeutic approach by enhancing CLDN5 expression in lung microvascular ECs.
It is not well understood how SARS-CoV-2 regulates tight junctions. Some reports have shown a relationship between SARS-CoV-2 and CLDN5 in ECs. Because CLDN5 is known as a regulator of the BBB (8), the effects of SARS-CoV-2 on CLDN5 expression in brain ECs have been investigated (22, 23). These studies demonstrated that SARS-CoV-2 Spike protein or UV–SARS-CoV-2 treatment decreases CLDN5 mRNA and protein expressions (22, 23). In contrast, another study showed that SARS-CoV-2 infection does not alter CLDN5 expression in the brain or in isolated ECs from K18-hACE2 mice and hamsters (24). In addition, CLDN5 expression was shown to be decreased in the decidua and chorionic villi endothelium of female patients with severe COVID-19, implying a relationship between SARS-CoV-2 and CLDN5 (25). These findings suggest that SARS-CoV-2–mediated CLDN5 regulation may differ depending on the experimental conditions, including organs and species. In this study, we demonstrated that SARS-CoV-2 exposure decreases CLDN5 expression in human lung ECs and induces vascular barrier disruption. In addition, we showed the pivotal role of CLDN5 in regulating the vascular barrier in the respiratory organ.
CLDN5 was originally cloned as a transmembrane protein–deleted gene in velo-cardio-facial syndrome (TMVCF). TMVCF is one of the deleted genes in patients with 22q11 deletion syndromes, including velo-cardio-facial syndrome and DiGeorge syndrome (26). It was later identified as a member of CLDN family proteins and well-characterized as a regulator of the BBB. A recent study focusing on the haploinsufficiency of CLDN5 in 22q11 deletion syndrome indicated that CLDN5 expression level is important for BBB function (27). To the best of our knowledge, there are few papers showing a direct relationship between 22q11 deletion syndrome and pulmonary edema. It is suggested that the haploinsufficiency of CLDN5 gene does not induce a sufficient decrease in CLDN5 for pulmonary edema. An analysis of CLDN5 functions using CLDN5 knockout mice indicated that CLDN5 regulates tight sealing in brain blood vessels, and size selectively inhibits the transmigration of small molecules (8). However, in the brain blood vessels of the knockout mice, the structure of the tight junction was maintained, and no bleeding or edema was observed. These observations suggest that CLDN5 is not essential for the maintenance of the vascular structure in the brain. In addition, it is reported that the formation of the VE-cadherin–mediated adherens junction induces CLDN5 expression (18). These findings suggest that the VE-cadherin–mediated adherens junction is a major regulator of vascular integrity and that CLDN5 functions as a supportive protein that gives tight sealing in brain blood vessels. In contrast, CLDN5 functions in other organs, including the lung, have received little research attention despite the expression of CLDN5 in these tissues (28). In this study, we demonstrated that antibody-mediated CLDN5 inhibition induces severe vascular leakage in the mouse lung and alterations of the junction structure. These findings indicate the roles of CLDN5 in the vascular barriers and integrity of the lung.
We found that SARS-CoV-2 generated from infected airway epithelial cells disrupts endothelial junctions by decreasing CLDN5 expression and disrupting VE-cadherin–mediated junctions, allowing SARS-CoV-2 to leak into the blood vessel channel. In this process, CLDN5 down-regulation likely induces the disruption of VE-cadherin junctions, because CLDN5 overexpression neutralized the SARS-CoV-2 exposure–induced barrier disruption. Similarly, our results from the analysis using transmission electron microscopy (TEM) suggested the CLDN5 antibody–mediated disruption of endothelial junctions. However, because the disruption of VE-cadherin–mediated junctions can further decrease CLDN5 expression levels, further analysis of the molecular mechanism for CLDN5 down-regulation is needed. In addition, previous studies reported that EC-specific conditional knockout and antibody-mediated inhibition of VE-cadherin induces severe vascular leakage in the lung (29, 30). These results suggest that excessive down-regulation or inhibition of CLDN5 triggers the disruption of VE-cadherin junctions and induces severe vascular leakage in the lung. However, little is known about the effect of inhibiting CLDN5 on VE-cadherin functions. Further study of CLDN5 functions in the lung will contribute to understanding the ARDS and pneumonia induced by SARS-CoV-2.
In patients with severe COVID-19, inflammatory cytokines and immune cells invade epithelial tissue from the blood vessels after viral replication and vascular barrier disruption in respiratory organs (31). This invasion can lead to pneumonia and ARDS. However, the airway-on-a-chip we used does not contain immune cells. Thus, in this study, we only observed events immediately after the onset of severe COVID-19. An airway-on-a-chip that also contains immune cells should be used to study later stages.
We also showed that Fluvastatin can prevent SARS-CoV-2–induced vascular barrier disruption. Fluvastatin increases CLDN5 expression in lung ECs, but the mechanism is not fully understood. One report showed that Fluvastatin causes antiviral effects in epithelial cells (32). Fluvastatin has been used to treat many hypercholesterolemia patients; thus, its safety is well guaranteed. Because the organ distribution of SARS-CoV-2 is different between human and animal models (13, 33), the vascular barrier disruption induced by SARS-CoV-2 may also be different between human and animal models. To evaluate the efficacy of Fluvastatin as a therapeutic agent against COVID-19, infection experiments using a primate model are needed.
In conclusion, we demonstrated a mechanism for SARS-CoV-2–induced vascular barrier disruption in respiratory organs and proposed a therapeutic strategy against COVID-19. Our findings suggest that using airway-on-a-chip as SARS-CoV-2 infection models could be efficient for drug screening.
MATERIALS AND METHODS
Preparation of SARS-CoV-2
The SARS-CoV-2 strains B.1.1.214 [Global Initiative on Sharing Avian Influenza Data (GISAID) accession number: EPI_ISL_2897162] and B.1.617.2 (GISAID accession number: EPI_ISL_9636792) were isolated from a nasopharyngeal swab sample of a patient with COVID-19. This study was approved by the research ethics committee of Kyoto University (R2379-3). SARS-CoV-2 B was provided by the Kanagawa Prefectural Institute of Public Health; SARS-CoV-2 B.1 (NR-53514) was obtained from BEI Resources; SARS-CoV-2 B.1.1.7, B.1.351, and P.1 were provided by the National Institute of Infectious Diseases; and SARS-CoV-2 B.1.1529 (NR-56461) was obtained from BEI Resources. All figures except fig. S2 show data for SARS-CoV-2 B.1.1.214. The virus was proliferated in transmembrane serine protease 2 (TMPRSS2)/Vero cells (JCRB1818, JCRB Cell Bank) (34) and stored at −80°C.TMPRSS2/Vero cells were cultured with minimum essential medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin. All viral infection experiments were done in a biosafety level 3 (BSL-3) facility at Kyoto University, strictly following regulations.
Cell culture and airway-on-a-chip
HMVEC-L were obtained from Lonza and maintained in EGM-2 MV medium (Lonza). To prepare the airway-on-a-chip, first, the bottom channel of a polydimethylsiloxane (PDMS) device was precoated with fibronectin (3 μg/ml; Sigma-Aldrich). HMVEC-L were suspended at 5 × 106 cells/ml in EGM-2 MV medium. Suspension medium (10 μl) was injected into the fibronectin-coated bottom channel of the PDMS device. Then, the PDMS device was turned upside down and incubated for 1 hour. After 1 hour, the device was turned over, and EGM-2 MV medium was added into the bottom channel. After 4 days, human airway organoids (AOs) were dissociated and seeded into the top channel. AOs were dissociated into single cells and then suspended at 5 × 106 cells/ml in AO differentiation medium. Suspension medium (10 μl) was injected into the top channel. After 1 hour, AO differentiation medium was added to the top channel. The cells were cultured under a humidified atmosphere with 5% CO2 at 37°C.
Airway organoids
AOs were generated according to our previous report (35). Briefly, normal human bronchial epithelial (NHBE) cells (#CC-2540, Lonza) were used to generate AO. NHBE cells were suspended in cold Matrigel growth factor–reduced basement membrane matrix (10 mg/ml; Corning). Cell suspension (50 μl) was solidified on prewarmed cell culture–treated multidishes (24-well plates; Thermo Fisher Scientific) at 37°C for 10 min, and then 500 μl of expansion medium was added to each well. AOs were cultured with AO expansion medium for 10 days. To mature the AO, expanded AOs were cultured with AO differentiation medium for 5 days. Matured AOs were dissociated and plated into the PDMS device before the SARS-CoV-2 infection experiments.
Microfluidic device
The microfluidic device was generated according to our previous report (36). Briefly, the microfluidic device consisted of two layers of microchannels separated by a semipermeable membrane. The microchannel layers were fabricated from PDMS using a soft lithographic method. PDMS prepolymer (SYLGARD 184, Dow Corning) at a base to curing agent ratio of 10:1 was cast against a mold composed of SU-8 2150 (MicroChem) patterns formed on a silicon wafer. The cross-sectional size of the microchannels was 1 mm in width and 330 μm in height. To introduce solutions into the microchannels, access holes were punched through the PDMS using a 6-mm biopsy punch (Kai Corporation). Two PDMS layers were bonded to two semipermeable polyethylene terephthalate (PET) membrane containing 3.0-μm pores (#353091, Falcon) using a thin layer of liquid PDMS prepolymer as the mortar. PDMS prepolymer was spin-coated (4000 rpm for 60 s) onto a glass slide. Subsequently, both the top and bottom channel layers were placed on the glass slide to transfer the thin layer of PDMS prepolymer onto the embossed PDMS surfaces. The membrane was then placed onto the bottom layer and sandwiched with the top layer. The combined layers were left at room temperature for 1 day to remove air bubbles and then placed in an oven at 60°C overnight to cure the PDMS glue. The PDMS devices were sterilized by placing them under UV light for 1 hour before the cell culture.
SARS-CoV-2 infection
SARS-CoV-2 (0.1 MOI) containing AO differentiation medium was injected into the top channel (airway channel) of an airway-on-a-chip. After 120 min, the culture medium containing SARS-CoV-2 was replaced with fresh medium. When exposing HMVEC-L to the SARS-CoV-2 once, the cells were treated with 0.1 or 1 MOI of SARS-CoV-2 for 120 min and then cultured with fresh medium for 4 days. When exposing HMVEC-L to SARS-CoV-2 four times, the cells were cultured for 4 days with daily medium change containing 0.1 or 1 MOI of SARS-CoV-2.
RNA sequencing
Total RNA was isolated from the airway-on-a-chip using the RNeasy Mini Kit (QIAGEN). RNA integrity was assessed with a 2100 Bioanalyzer (Agilent Technologies). The library preparation was performed using the TruSeq Stranded mRNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions. Sequencing was performed on an Illumina NextSeq 550. The fastq files were generated using bcl2fastq-2.20. Adapter sequences and low-quality bases were trimmed from the raw reads using Cutadapt version 3.4 (37). The trimmed reads were mapped to human reference genome sequences (hg38) using STAR version 2.7.9a (38) with the GENCODE (release 36, GRCh38.p13) (39) gtf file. The raw counts were calculated using htseq-count version 0.13.5 (40) with the GENCODE gtf file. Gene expression levels were determined as transcripts per kilobase million (TPM) values with DESeq2 v1.30.1 (41). Raw data concerning this study were submitted under gene expression omnibus (GEO) accession number GSE196113.
Total RNA was isolated from the lungs of patients with or without COVID-19 using the RNeasy Protect Mini Kit (QIAGEN). RNA integrity was assessed with a 2100 Bioanalyzer (Agilent Technologies). The library preparations for human lungs were performed using the TruSeq Stranded mRNA Sample Prep Kit and a Stranded Total RNA Prep, Ligation with Ribo-Zero Plus Kit according to the manufacturer’s instructions. Sequencing was performed on an Illumina NextSeq550. The fastq files were generated using bcl2fastq-2.20. Adapter sequences and low-quality bases were trimmed from the raw reads using Cutadapt version 3.4 (37). The trimmed reads were mapped to human reference genome sequences (hg38) using STAR version 2.7.9a (38) with the GENCODE (release 36, GRCh38.p13) (39) gtf file. The raw counts were calculated using htseq-count version 0.13.5 (40) with the GENCODE gtf file. Gene expression levels were determined as TPM values with DESeq2 v1.30.1 (41). Raw data concerning this study were submitted under GEO accession number GSE206635.
Quantification of viral RNA copy number
The cell culture supernatant was mixed with an equal volume of 2× RNA lysis buffer [distilled water containing SUPERase·In RNase inhibitor (0.4 U/μl; Thermo Fisher Scientific), 2% Triton X-100, 50 mM KCl, 100 mM tris-HCl (pH 7.4), and 40% glycerol] and incubated at room temperature for 10 min. The mixture was diluted 10 times with distilled water. Viral RNA was quantified using the One Step TB Green PrimeScript PLUS RT-PCR Kit (Perfect Real Time) (Takara Bio) on a QuantStudio 1 Real-Time PCR System (Thermo Fisher Scientific). The primers used in this experiment are shown in table S1. Standard curves were prepared using SARS-CoV-2 RNA (105 copies/μl) purchased from Nihon Gene Research Laboratories.
Quantitative reverse transcription polymerase chain reaction
Total RNA was isolated from cells and mouse tissues using ISOGEN (Nippon Gene) and the FastGene RNA Basic Kit (Nippon Genetics). cDNA was synthesized using total RNA with the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). Real-time RT-PCR was performed with SYBR Green PCR Master Mix (Thermo Fisher Scientific) or QuantiTect SYBR Green PCR Kit (QIAGEN) using a QuantStudio 1 (Thermo Fisher Scientific) or a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The relative quantitation of target mRNA levels was performed using the 2−ΔΔCt method or by calculating the copy number of the target transcripts from standard curves prepared using known amounts of plasmids containing the target sequences. The values were normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The PCR primer sequences are shown in table S1.
Western blot analysis for HMVEC-L and mouse lungs
Total cell extracts from HMVEC-L were prepared by dissolving the cells in sample buffer [100 mM tris-HCl (pH 6.8), 4% SDS, 8% 2-mercaptoethanol, 20% glycerol, and 0.001% bromophenol blue]. For the preparation of cell extracts from mouse lungs, the lungs were homogenized in radioimmunoprecipitation assay buffer [0.01 M sodium phosphate buffer (pH 7.2), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 2× cOmplete EDTA-free protease inhibitors (Roche)] and incubated for 3 hours at 4°C. The resulting lysates were centrifuged at 4°C for 30 min at 20,000g, and the supernatant was obtained as cell extracts. The cell extracts were separated by SDS–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were incubated with primary antibodies against CLDN5 (4C3C2, Thermo Fisher Scientific), VE-cadherin (F-8, Santa Cruz Biotechnology; AF1002, R&D Systems), or GAPDH (6C5, Merck Millipore) and with horseradish peroxidase (HRP)–conjugated secondary antibodies. Immunoreactive bands were detected using the ImageQuant LAS 4010 system (GE Healthcare). The primary antibodies used are summarized in table S2.
Immunofluorescent and actin staining
HMVEC-L cultured on a chamber slide (Thermo Fisher Scientific) with EGM-2 MV medium containing SARS-CoV-2 were used for immunofluorescent and actin staining. The PET membrane to which HMVEC-L were adhered was mechanically recovered from the airway-on-a-chip and then used for immunofluorescent staining. The resulting cells were fixed with 4% paraformaldehyde and permeabilized with phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and blocked with PBS containing 1% bovine serum albumin. The resulting cells were incubated with antibodies against CLDN5, VE-cadherin, β-catenin (E-5, Santa Cruz Biotechnology), and FoxO1 (C29H4, Cell Signaling Technology) followed by incubation with secondary antibodies conjugated with Alexa Fluor 555 or Alexa Fluor 488 (Thermo Fisher Scientific). F-actin was stained with Alexa Fluor 488 phalloidin (Thermo Fisher Scientific). The slides were mounted with VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and analyzed with a BZ-X700 (KEYENCE). The nuclear FoxO1 level was measured using Fiji/ImageJ. The primary antibodies used are summarized in table S2.
Measurement of TEER
HMVEC-L (3 × 104 cells) were seeded in cell culture inserts (for 24-well plate) with a pore size of 3.0 μm (BD Falcon) and incubated for 72 hours. The medium in the upper chamber was replaced with EGM-2 MV medium containing 1 MOI of SARS-CoV-2, and TEER was measured using a Millicell ERS-2 Voltohmmeter (Merck Millipore). The TEER value was calculated by the following formula: (resistance of experimental wells − resistance of blank wells) × 0.32 (the membrane area of the cell culture insert).
Generation of human CLDN5 knock-in mice and analysis of CLDN5 expression
A donor vector consisting of a homology arm sequence (2 kb) derived from the mouse genome linked to both ends of the human CLDN5 coding sequence (657 base pairs) was generated. Cas9 protein, single guide RNA, and the donor vector were introduced into mouse embryonic stem (ES) cells for homologous recombination. ES cells containing human CLDN5 in the appropriate locus were selected by genotyping by PCR and DNA sequencing. ES cells with normal karyotype were introduced into early embryos. The born chimeric mice were mated with C57BL/6N mice to establish human CLDN5 homozygous knock-in mice (hCLDN5-KI mice). For the genotyping, the DNA fragment was amplified using genomic DNA from the tail of hCLDN5-KI mouse and primers (table S1) and digested with Eag I (New England Biolabs) to check the genome DNA including the human CLDN5 sequence. To analyze the CLDN5 expression, organs were harvested from hCLDN5-KI mice (male, 7 to 9 weeks old) and used for the RNA preparation. For the western blotting, hCLDN5-KI mice (male and female, 7 to 8 weeks old) were injected with the anti-CLDN5 antibody (R9) or control rat immunoglobulin G (IgG). One and a half hours after the injection, the lungs were harvested and used for protein preparation. All animal experiments in this study were approved by the ethics committee of Osaka University (approval number 30-11-1) and performed in accordance with the relevant guidelines and regulations.
Vascular permeability assay (Miles assay)
hCLDN5-KI mice (male, 7 to 8 weeks old) were intravenously injected with anti-CLDN5 antibody (R9; 2.5 mg/kg) (16) or control rat IgG (Sigma-Aldrich). One hour later, the mice were intravenously injected with 100 μl of PBS containing 1% Evans blue (Tokyo Chemical Industry). Thirty minutes later, the mice were perfused with PBS containing 2 mM EDTA, and the organs were harvested, minced, and incubated in formamide at 55°C for 48 hours. The eluted Evans blue was quantified by measuring the optical density at 620 nm using the standard curve. Pictures for organs were taken under an SZX7 stereo microscope (Olympus) equipped with an MAS-500 USB camera (Marutsuelec).
Quantification of lung wet/dry weight ratio
hCLDN5-KI mice (male, 7 to 8 weeks old) were injected with anti-CLDN5 antibody (R9) or control rat IgG. One and a half hours after the injection, the lungs were harvested from the mice and immediately weighed (wet weight). The lungs were then dried in an incubator at 60°C for 48 hours and weighed (dry weight). Pulmonary edema was assessed by calculating the wet/dry weight ratio.
Hematoxylin and eosin staining for mouse and human lungs
hCLDN5-KI mice (male, 7 to 8 weeks old) were injected with anti-CLDN5 antibody (R9) or control rat IgG. Forty minutes after the injection, the mice were perfused and fixed with PBS containing 1% paraformaldehyde for 1 hour. The lungs were harvested and used for the preparation of frozen sections (5 μm). The resulting mouse sections and paraffin sections (5 μm) from the human lungs were stained with hematoxylin and eosin Y (Fujifilm Wako Pure Chemical Corporation) and analyzed with the BZ-X700.
Ultrathin section TEM
hCLDN5-KI mice (male, 10 weeks old) were injected with anti-CLDN5 antibody (R9) or control rat IgG. One and a half hours after the injection, the lungs were harvested and fixed in cacodylate buffer with 2.5% glutaraldehyde and subsequently postfixed in 1% osmium tetraoxide at 4°C. After the fixation, the lungs were dehydrated in a graded series of ethanol and embedded in epoxy resin. Ultrathin sections were cut, stained with uranyl acetate and lead staining solution, and examined using an electron microscope (Hitachi H-7700) at 80 kV.
Generation of HMVEC-L–CLDN5
HMVEC-L were electroporated with pPB-TRE3G-CLDN5-V2 and pHL-EF1a-hcPBase-A (42) vectors using an NEPA21 electroporator (Nepa Gene) and then selected with blasticidin (Fujifilm Wako Pure Chemical Corporation). The piggyBac-based CLDN5-expressing plasmid, pPB-TRE3G-CLDN5-V2, was constructed by inserting human CLDN5 gene into multiple cloning sites of pPB-TRE3G-MCS(A)-P2A-MCS(B)V2, which was a gift from T. Maruyama (Waseda University). The piggyBac transposase-expressing plasmid, pHL-EF1a-hcPBase-A, was a gift from A. Hotta (Kyoto University).
Samples of patients with COVID-19
The current study was performed in accordance with the Declaration of Helsinki and conducted with the approval of the Kyoto University Graduate School and Faculty of Medicine, ethics committee (R2379-3). Informed consent was obtained in the form of an opt-out form on the institution’s website. The institutional ethics committee approved this informed consent plan. Archived, residual serum samples from patients with COVID-19 referred to Kyoto University Hospital, Kyoto, Japan, were used in this study. These clinical specimens were previously archived for future studies to identify biomarkers for disease progression but were repurposed for the present study. The severity of the patients was defined according to the criteria used by World Health Organization guidance for the clinical management of COVID-19 released 27 May 2020 (https://apps.who.int/iris/bitstream/handle/10665/332196/WHO-2019-nCoV-clinical-2020.5-eng.pdf?sequence=1&isAllowed=y). The information of patients with COVID-19 used for Fig. 6 (A to E) can be obtained from our previous report (21). The information of patients with COVID-19 used for Fig. 6F is summarized in table S3.
Immunofluorescent staining of human lungs
Human lungs were harvested and used for the preparation of paraffin sections (5 μm). The sections were permeabilized using tris-buffered saline with 0.1% Tween 20 detergent (Nacalai Tesque) for 10 min and were blocked using Blocking One Histo (Nacalai Tesque) for 45 min. Then, the sections were incubated in tris-buffered saline with 0.1% Tween 20 detergent with or without primary antibodies overnight at 4°C. The sections were then washed with PBS and incubated in tris-buffered saline with 0.1% Tween 20 detergent containing Alexa Fluor 595–conjugated secondary antibodies for 45 min. Last, the sections were washed and mounted with Fluoro-KEEPER antifade reagent, nonhardening type with DAPI (Nacalai Tesque) and analyzed with the BZ-X700. The primary antibodies are summarized in table S2.
Measurement of serum CLDN5 levels in patients with COVID-19
Clear flat-bottom immuno nonsterile 96-well plates (Thermo Fisher Scientific) were coated with a mixture of anti-CLDN5 antibodies (R9) and incubated at room temperature for 1 hour and then blocked with 1% bovine serum albumin containing PBS. The samples and standard CLDN5 proteins were diluted twofold with dilution buffer (0.05% Tween 20 in PBS), and 100 μl of samples was added to each well, followed by incubation at room temperature for 1 hour. After three washes with a washing buffer, 100 μl of anti-CLDN5 antibody (Abcam) was added to each well, followed by incubation at room temperature for 1 hour. After three washes with a washing buffer, 100 μl of Biotin-SP-AffiniPure Donkey Anti-Rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories) was added to each well, followed by incubation at room temperature for 1 hour. After three washes with a washing buffer, 100 μl of Pierce high-sensitivity streptavidin-HRP (Thermo Fisher Scientific, 21130) was added as a substrate to each well, followed by incubation at room temperature for 30 min. After three washes with a washing buffer, 100 μl of trimethylboron substrate solution (R&D Systems) was added as a substrate to each well, followed by incubation at room temperature for 30 min. The reaction was stopped by adding 100 μl of stop solution to each well. The optical density of each sample at 450 nm was then measured using TriStar LB 941 (Berthold).
Statistical analysis
Data are expressed as mean values ± SEM. P values were calculated using the unpaired t test, Mann-Whitney U test, or one-way or two-way analysis of variance (ANOVA) followed by Tukey’s test, Dunnett’s test, or Sidak correction. All statistical analyses were performed using Prism 9 software (GraphPad Software Inc.). The statistical significance of differences in the means was determined using the tests indicated in the figure legends.
Acknowledgments
We thank Y. Koyanagi, N. Misawa, and K. Shimura (Kyoto University) for the setup and operation of the BSL-3 laboratory at Kyoto University; A. Sakamoto, E. Sano, N. Mimura, N. Yasuhara, and Y. Matsubara (Kyoto University) for technical assistance with the cell culture, qRT-PCR analysis, immunofluorescent staining, plasmid construction, and enzyme-linked immunosorbent assay (ELISA); K. Okita and S. Sakurai (Kyoto University) for technical assistance with the RNA-seq experiments at the Single-Cell Genome Information Analysis Core (SignAC), ASHBi, Kyoto University; P. Karagiannis (Sofia Scientific Writing) for critical reading of the manuscript; S. Nishioka for technical assistance with the generation of hCLDN5-KI mice; and Y. Hashimoto for important technical advice.
Funding: This research was supported by the iPS Cell Research Fund, the Mitsubishi Foundation, the Senri Life Science Foundation, the Japan Agency for Medical Research and Development (AMED) (JP20fk0108533 and JP22gm1610005) (to K.Tak.), by JSPS KAKENHI [20H03382, 20K21481, and JP 16H06276 (AdAMS)] and AMED (JP21am0101084 and JP21am0101123), the Takeda Science Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Nippon Foundation–Osaka University Project for Infectious Disease Prevention (to Y.O.), by the AMED (JP22hm0102084) (to M.K.), by the JSPS Core-to-Core Program A, the Advanced Research Networks, the JST Core Research for Evolutional Science and Technology (JPMJCR20HA), and the Joint Usage/Research Center program of Institute for Frontier Life and Medical Sciences, Kyoto University (to T.N.).
Author contributions: R.H. prepared SARS-CoV-2, performed all the experiments using SARS-CoV-2–infected cells and the statistical analysis, and wrote the manuscript. J.T. analyzed SARS-CoV-2–infected cells and hCLDN5-KI mice and wrote the manuscript. K.S. generated the hCLDN5-KI mice, interpreted the data, and wrote the manuscript. R.F. generated the hCLDN5-KI mice and performed the Miles assay. K.K. fabricated the PDMS devices. S.D. performed the RNA-seq analysis. M.Y. collected the clinical samples and wrote the manuscript. Y.T. performed the TEM analysis. M.M. collected the mouse samples and performed the Miles assay. K.M. propagated the antibody and performed qRT-PCR using hCLDN5-KI mice. M.T. performed the immunofluorescent staining. T.K. performed the qRT-PCR. S.Tam. propagated the anti-CLDN5 antibody. N.T. collected the mouse samples. A.K. performed experiments using the Spike protein. S.Tan. analyzed and interpreted the data. M.I. generated the hCLDN5-KI mice. K.Tac. propagated the anti-CLDN5 antibody and generated the CLDN5 ELISA Kit. M.K. propagated the anti-CLDN5 antibody and generated the CLDN5 ELISA Kit. M.O. performed qRT-PCR and analyzed and interpreted the data. C.O. performed experiments using the Spike protein. Y.M. conceived the study and interpreted the data. A.O. collected the clinical samples and wrote the manuscript. T.N. performed the TEM analysis. T.Y. performed the RNA-seq analysis and wrote the manuscript. Y.Y. propagated the anti-CLDN5 antibody and interpreted the data. Y.-s.T. fabricated the PDMS devices. H.D. collected the clinical samples and wrote the manuscript. Y.F. conceived the study and interpreted the data. M.N. collected the clinical samples. K.Tak. conceived and designed the study, analyzed and interpreted the data, and wrote the manuscript. Y.O. conceived and designed the study, analyzed and interpreted the data, and wrote the manuscript.
Competing interests: R.H., J.T., Y.Y., M.O., Y.F., K.Tak., and Y.O. are inventors on a patent application related to this work filed by Kyoto University and Osaka University (no. 2022-064486, filed 8 April 2022). Y.Y. and N.T. are employed by the Research Foundation for Microbial Diseases of Osaka University. The other authors declare that they have no competing interest.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The hCLDN5-KI mice can be provided by Y.O. pending scientific review and a completed material transfer agreement. Requests for these mice should be submitted to Y.O. (okadabos@phs.osaka-u.ac.jp). The AOs, airway-on-a-chip, microfluidic devices, SARS-CoV-2 strains B.1.1.214 (GISAID accession number: EPI_ISL_2897162) and B.1.617.2 (GISAID accession number: EPI_ISL_9636792), and CLDN5-expressing vectors can be provided by K.Tak. pending scientific review and a completed material transfer agreement. Requests for above materials should be submitted to K.Tak. (kazuo.takayama@cira.kyoto-u.ac.jp).
Supplementary Materials
This PDF file includes:
Fig. S1 to S10
Tables S1 to S3
REFERENCES AND NOTES
- 1.Zhu N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., Deng Y., Mao L., Su C., Qiang G., Jiang T., Zhao J., Wu G., Song J., Tan W., Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 11, 3910 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nascimento Conde J., Schutt W. R., Gorbunova E. E., Mackow E. R., Recombinant ACE2 expression is required for SARS-CoV-2 to infect primary human endothelial cells and induce inflammatory and procoagulative responses. MBio 11, e03185-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varga Z., Flammer A. J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A. S., Mehra M. R., Schuepbach R. A., Ruschitzka F., Moch H., Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y., Ji W., Yang H., Chen S., Zhang W., Duan G., Endothelial activation and dysfunction in COVID-19: From basic mechanisms to potential therapeutic approaches. Signal Transduct. Target. Ther. 5, 293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dejana E., Orsenigo F., Lampugnani M. G., The role of adherens junctions and VE-cadherin in the control of vascular permeability. J. Cell Sci. 121, 2115–2122 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Cerutti C., Ridley A. J., Endothelial cell-cell adhesion and signaling. Exp. Cell Res. 358, 31–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corada M., Mariotti M., Thurston G., Smith K., Kunkel R., Brockhaus M., Lampugnani M. G., Martin-Padura I., Stoppacciaro A., Ruco L., McDonald D. M., Ward P. A., Dejana E., Vascular endothelial-cadherin is an important determinant of microvascular integrity in vivo. Proc. Natl. Acad. Sci. U.S.A. 96, 9815–9820 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitta T., Hata M., Gotoh S., Seo Y., Sasaki H., Hashimoto N., Furuse M., Tsukita S., Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J. Cell Biol. 161, 653–660 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G., Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teuwen L. A., Geldhof V., Pasut A., Carmeliet P., COVID-19: The vasculature unleashed. Nat. Rev. Immunol. 20, 389–391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pober J. S., Sessa W. C., Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., Yan B., Liu L., Hu Z., Wang M., Zhou Y., SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 7, 17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox R. M., Wolf J. D., Plemper R. K., Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 6, 11–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si L., Bai H., Rodas M., Cao W., Oh C. Y., Jiang A., Moller R., Hoagland D., Oishi K., Horiuchi S., Uhl S., Blanco-Melo D., Albrecht R. A., Liu W. C., Jordan T., Nilsson-Payant B. E., Golynker I., Frere J., Logue J., Haupt R., McGrath M., Weston S., Zhang T., Plebani R., Soong M., Nurani A., Kim S. M., Zhu D. Y., Benam K. H., Goyal G., Gilpin S. E., Prantil-Baun R., Gygi S. P., Powers R. K., Carlson K. E., Frieman M., tenOever B. R., Ingber D. E., A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat. Biomed. Eng. 5, 815–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benam K. H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H. H., Alves S. E., Salmon M., Ferrante T. C., Weaver J. C., Bahinski A., Hamilton G. A., Ingber D. E., Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 13, 151–157 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto Y., Shirakura K., Okada Y., Takeda H., Endo K., Tamura M., Watari A., Sadamura Y., Sawasaki T., Doi T., Yagi K., Kondoh M., Claudin-5-binders enhance permeation of solutes across the blood-brain barrier in a mammalian model. J. Pharmacol. Exp. Ther. 363, 275–283 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Tachibana K., Hashimoto Y., Shirakura K., Okada Y., Hirayama R., Iwashita Y., Nishino I., Ago Y., Takeda H., Kuniyasu H., Kondoh M., Safety and efficacy of an anti-claudin-5 monoclonal antibody to increase blood-brain barrier permeability for drug delivery to the brain in a non-human primate. J. Control. Release 336, 105–111 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Taddei A., Giampietro C., Conti A., Orsenigo F., Breviario F., Pirazzoli V., Potente M., Daly C., Dimmeler S., Dejana E., Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nat. Cell Biol. 10, 923–934 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Gupta A., Madhavan M. V., Poterucha T. J., DeFilippis E. M., Hennessey J. A., Redfors B., Eckhardt C., Bikdeli B., Platt J., Nalbandian A., Elias P., Cummings M. J., Nouri S. N., Lawlor M., Ranard L. S., Li J., Boyle C., Givens R., Brodie D., Krumholz H. M., Stone G. W., Sethi S. S., Burkhoff D., Uriel N., Schwartz A., Leon M. B., Kirtane A. J., Wan E. Y., Parikh S. A., Association between antecedent statin use and decreased mortality in hospitalized patients with COVID-19. Nat. Commun. 12, 1325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W., Sharma R., Rizzo A. N., Siegler J. H., Garcia J. G., Jacobson J. R., Role of claudin-5 in the attenuation of murine acute lung injury by simvastatin. Am. J. Respir. Cell Mol. Biol. 50, 328–336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohsumi A., Takamatsu J., Yuasa I., Tanaka S., Yutaka Y., Hamaji M., Nakajima D., Yamazaki K., Nagao M., Date H., Living-donor lung transplantation for dyskeratosis congenita. Ann. Thorac. Surg. 112, e397–e402 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Torices S., Cabrera R., Stangis M., Naranjo O., Fattakhov N., Teglas T., Adesse D., Toborek M., Expression of SARS-CoV-2-related receptors in cells of the neurovascular unit: Implications for HIV-1 infection. J. Neuroinflammation 18, 167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds J. L., Mahajan S. D., SARS-COV2 alters blood brain barrier integrity contributing to neuro-inflammation. J. Neuroimmune Pharmacol. 16, 4–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Zhou L., Bao L., Liu J., Zhu H., Lv Q., Liu R., Chen W., Tong W., Wei Q., Xu Y., Deng W., Gao H., Xue J., Song Z., Yu P., Han Y., Zhang Y., Sun X., Yu X., Qin C., SARS-CoV-2 crosses the blood-brain barrier accompanied with basement membrane disruption without tight junctions alteration. Signal Transduct. Target. Ther. 6, 337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores-Pliego A., Miranda J., Vega-Torreblanca S., Valdespino-Vazquez Y., Helguera-Repetto C., Espejel-Nunez A., Borboa-Olivares H., Espino Y. S. S., Mateu-Rogell P., Leon-Juarez M., Ramirez-Santes V., Cardona-Perez A., Villegas-Mota I., Torres-Torres J., Juarez-Reyes A., Rizo-Pica T., Gonzalez R. O., Gonzalez-Mariscal L., Estrada-Gutierrez G., Molecular insights into the thrombotic and microvascular injury in placental endothelium of women with mild or severe COVID-19. Cell 10, 364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirotkin H., Morrow B., Saint-Jore B., Puech A., Das Gupta R., Patanjali S. R., Skoultchi A., Weissman S. M., Kucherlapati R., Identification, characterization, and precise mapping of a human gene encoding a novel membrane-spanning protein from the 22q11 region deleted in velo-cardio-facial syndrome. Genomics 42, 245–251 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Greene C., Kealy J., Humphries M. M., Gong Y., Hou J., Hudson N., Cassidy L. M., Martiniano R., Shashi V., Hooper S. R., Grant G. A., Kenna P. F., Norris K., Callaghan C. K., Islam M. D., O’Mara S. M., Najda Z., Campbell S. G., Pachter J. S., Thomas J., Williams N. M., Humphries P., Murphy K. C., Campbell M., Dose-dependent expression of claudin-5 is a modifying factor in schizophrenia. Mol. Psychiatry 23, 2156–2166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morita K., Sasaki H., Furuse M., Tsukita S., Endothelial claudin: Claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J. Cell Biol. 147, 185–194 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frye M., Dierkes M., Kuppers V., Vockel M., Tomm J., Zeuschner D., Rossaint J., Zarbock A., Koh G. Y., Peters K., Nottebaum A. F., Vestweber D., Interfering with VE-PTP stabilizes endothelial junctions in vivo via Tie-2 in the absence of VE-cadherin. J. Exp. Med. 212, 2267–2287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duong C. N., Nottebaum A. F., Butz S., Volkery S., Zeuschner D., Stehling M., Vestweber D., Interference with ESAM (endothelial cell-selective adhesion molecule) plus vascular endothelial-cadherin causes immediate lethality and lung-specific blood coagulation. Arterioscler. Thromb. Vasc. Biol. 40, 378–393 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Tian W., Zhang N., Jin R., Feng Y., Wang S., Gao S., Gao R., Wu G., Tian D., Tan W., Chen Y., Gao G. F., Wong C. C. L., Immune suppression in the early stage of COVID-19 disease. Nat. Commun. 11, 5859 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zapatero-Belinchon F. J., Moeller R., Lasswitz L., van Ham M., Becker M., Brogden G., Rosendal E., Bi W., Carriqui-Madronal B., Islam K., Lenman A., Gunesch A. P., Kirui J., Pietschmann T., Overby A. K., Jansch L., Gerold G., Fluvastatin mitigates SARS-CoV-2 infection in human lung cells. iScience 24, 103469 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M. S., Oh B. K., Yang D., Oh E. Y., Kim Y., Kang K. W., Lim C. W., Koh G. Y., Lee S. M., Kim B., Ultra- and micro-structural changes of respiratory tracts in SARS-CoV-2 infected Syrian hamsters. Vet. Res. 52, 121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 117, 7001–7003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sano E., Suzuki T., Hashimoto R., Itoh Y., Sakamoto A., Sakai Y., Saito A., Okuzaki D., Motooka D., Muramoto Y., Noda T., Takasaki T., Sakuragi J. I., Minami S., Kobayashi T., Yamamoto T., Matsumura Y., Nagao M., Okamoto T., Takayama K., Cell response analysis in SARS-CoV-2 infected bronchial organoids. Commun. Biol. 5, 516 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deguchi S., Tsuda M., Kosugi K., Sakamoto A., Mimura N., Negoro R., Sano E., Nobe T., Maeda K., Kusuhara H., Mizuguchi H., Yamashita F., Torisawa Y. S., Takayama K., Usability of polydimethylsiloxane-based microfluidic devices in pharmaceutical research using human hepatocytes. ACS Biomater Sci. Eng. 7, 3648–3657 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Kechin A., Boyarskikh U., Kel A., Filipenko M., cutPrimers: A new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frankish A., Diekhans M., Ferreira A. M., Johnson R., Jungreis I., Loveland J., Mudge J. M., Sisu C., Wright J., Armstrong J., Barnes I., Berry A., Bignell A., Carbonell Sala S., Chrast J., Cunningham F., Di Domenico T., Donaldson S., Fiddes I. T., Garcia Giron C., Gonzalez J. M., Grego T., Hardy M., Hourlier T., Hunt T., Izuogu O. G., Lagarde J., Martin F. J., Martinez L., Mohanan S., Muir P., Navarro F. C. P., Parker A., Pei B., Pozo F., Ruffier M., Schmitt B. M., Stapleton E., Suner M. M., Sycheva I., Uszczynska-Ratajczak B., Xu J., Yates A., Zerbino D., Zhang Y., Aken B., Choudhary J. S., Gerstein M., Guigo R., Hubbard T. J. P., Kellis M., Paten B., Reymond A., Tress M. L., Flicek P., GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishida K., Xu H., Sasakawa N., Lung M. S. Y., Kudryashev J. A., Gee P., Hotta A., Site-specific randomization of the endogenous genome by a regulatable CRISPR-Cas9 piggyBac system in human cells. Sci. Rep. 8, 310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sano E., Deguchi S., Sakamoto A., Mimura N., Hirabayashi A., Muramoto Y., Noda T., Yamamoto T., Takayama K., Modeling SARS-CoV-2 infection and its individual differences with ACE2-expressing human iPS cells. iScience 24, 102428 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S10
Tables S1 to S3