Abstract
Bacterial signal peptidase I is responsible for proteolytic processing of the precursors of secreted proteins. The enzymes from gram-negative and -positive bacteria are different in structure and specificity. In this study, we have cloned, expressed, and purified the signal peptidase I of gram-positive Streptococcus pneumoniae. The precursor of streptokinase, an extracellular protein produced in pathogenic streptococci, was identified as a substrate of S. pneumoniae signal peptidase I. Phospholipids were found to stimulate the enzymatic activity. Mutagenetic analysis demonstrated that residues serine 38 and lysine 76 of S. pneumoniae signal peptidase I are critical for enzyme activity and involved in the active site to form a serine-lysine catalytic dyad, which is similar to LexA-like proteases and Escherichia coli signal peptidase I. Similar to LexA-like proteases, S. pneumoniae signal peptidase I catalyzes an intermolecular self-cleavage in vitro, and an internal cleavage site has been identified between glycine 36 and histidine 37. Sequence analysis revealed that the signal peptidase I and LexA-like proteases show sequence homology around the active sites and some common properties around the self-cleavage sites. All these data suggest that signal peptidase I and LexA-like proteases are closely related and belong to a novel class of serine proteases.
Most proteins that are translocated across lipid bilayers are synthesized as precursors (preproteins) with an amino-terminal extension known as a signal (or leader) peptide. This signal sequence is involved in guiding the protein into the targeting and translocating pathway by interacting with the membrane and other components of the cellular secretory machinery (40). The final step in protein translocation and secretion is the release of the mature part of the protein from the membrane, which requires proteolytic removal of the signal peptide. This proteolytic processing occurs during or shortly after the translocation event and is catalyzed in both prokaryotes and eukaryotes by enzymes known as signal peptidases. Two major bacterial signal peptidases, signal peptidases I and II, with different cleavage specificities, have been identified. Signal peptidase I is responsible for processing the majority of secreted proteins (6, 31), whereas signal peptidase II exclusively processes glyceride-modified lipoproteins (12).
A number of genes encoding signal peptidase I have been cloned and sequenced from both gram-negative and gram-positive bacteria, including Escherichia coli (42), Salmonella enterica serovar Typhimurium (37), Haemophilus influenzae (9), Staphylococcus aureus (5), Bacillus subtilis (18, 30, 34), and Streptococcus pneumoniae (43). Sequence analysis demonstrated that some conserved motifs were present in both gram-negative and -positive signal peptidase-encoding genes. However, the genes from these two bacterial groups are significantly different. First, the primary sequences are quite different, and the deduced amino acid sequences have low sequence identities, 20 to 30%. Second, genes from gram-negative bacteria generally encode larger proteins, approximately 300 amino acids in size, as typified by lepB of E. coli (42). Genes from gram-positive bacteria, represented by sipS of B. subtilis (34) and spi of S. pneumoniae (43), generally encode smaller proteins, about 200 amino acids. Third, some of the interesting regions present in gram-negative signal peptidase I are missing in gram-positive enzymes, and one of these missing regions corresponds to an important membrane anchor of lepB of E. coli. Finally, from the substrate standpoint, the precursors of secreted proteins from gram-positive bacteria generally have longer and more hydrophobic signal peptides than those from gram-negative bacteria. It is thus speculated that these differences may reflect their differences in substrate specificities and other enzymatic properties.
To date, the biochemical characterization of signal peptidase I has concentrated on the enzyme from the gram-negative E. coli. This enzyme has been purified from native sources (31, 41, 45), and a truncated and catalytically active form has been overexpressed in E. coli and purified to homogeneity (14). E. coli signal peptidase I was able to cleave the precursors of many secreted proteins to generate mature products in vitro. In addition to naturally occurring precursor protein substrates, E. coli signal peptidase I was also capable of processing short and synthetic peptide substrates (7, 8, 14). The best substrate currently being used for E. coli signal peptidase I is a fusion protein consisting of the signal peptide of E. coli outer membrane protein A (OmpA) attached to Staphylococcus aureus nuclease A protein (4). In general, proteases are divided into four classes, serine, cysteine, metallo-, and aspartyl proteases, according to their mechanism of action. Signal peptidase I is not a member of any of these four classical groups due to its insensitivity to the characteristic protease inhibitors (6, 31). Evidence suggests that signal peptidase I is a special serine protease which utilizes a serine and lysine to form a catalytic dyad, unlike other serine proteases, such as trypsin, which hydrolyze peptide bonds by utilization of a catalytic triad consisting of serine, histidine, and aspartate (2, 3, 33). The active site of E. coli signal peptidase I resides in the periplasmic domain, which is anchored to the membrane by two transmembrane segments (1, 19, 42). Recently, the crystal structure of E. coli signal peptidase I in complex with a β-lactam inhibitor was solved. Structural analysis revealed that the catalytic serine acts as the nucleophile and lysine acts as a general base in the activation of the nucleophilic serine residue (20).
In this report, we describe the overexpression and purification of the S. pneumoniae signal peptidase I. The precursor of streptokinase has been identified as a substrate of the enzyme. We have demonstrated that S. pneumoniae signal peptidase I was able to catalyze an intermolecular self-cleavage reaction. The proteolytic activity of the enzyme was stimulated by phospholipids. Serine 38 and lysine 76 were identified as the active sites of the enzyme, indicating a serine-lysine catalytic dyad. The presence of this unique serine-lysine catalytic dyad and its specific self-cleavage as well as its primary sequence suggest that signal peptidases I and LexA-like proteases (17, 23, 27) belong to a novel class of serine proteases.
MATERIALS AND METHODS
Cloning and expression of S. pneumoniae signal peptidase I gene.
The gene encoding S. pneumoniae signal peptidase I was cloned by PCR using genomic DNA as the template and two oligonucleotides as primers (5′-CCGGAATTCAGATCTCATATGAATTTATTTAAAAATTTCTTAAAAGAG-3′ and 5′-CCGGAATTCAGATCTTTAAAATGTTCCGATACGGGTGATTGGCCAG-3′). The primers were designed to contain NdeI and BglII restriction sites and initiator and stop codons at the 5′ ends, respectively, to enable cloning into the bacterial expression vector pET16b. The primers were synthesized in accord with the published sequence of a putative S. pneumoniae signal peptidase I (43). Expression vector pET16b-spi was constructed by replacing the NdeI-BamHI fragment of pET16b with the PCR-amplified fragment that had been purified and digested with NdeI and BglII. The identity of the cloned gene was confirmed by direct DNA sequencing. For expression of the signal peptidase I, E coli strain BL21(DE3)pLysS was transformed with pET16b-spi by standard techniques (26), grown, and induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C as described (28).
Cloning and expression of prestreptokinase.
The gene encoding Streptococcus pyogenes prestreptokinase was amplified by PCR utilizing genomic DNA as the template and two synthetic oligonucleotides as primers (5′-TTAGGAGGTTCATATGAAAAATTACTTATCT-3′ and 5′-TAGAAGATCGCTCGAGTTTGTCTTTAGGGTT-3′), which were synthesized according to the published sequence (11) and designed to contain NdeI and XhoI restriction sites at the 5′ ends, respectively. The PCR product was purified and cloned into the NdeI and XhoI sites of vector pET23b, resulting in pET23b-ska, a bacterial expression vector which directs the expression of a fusion protein containing a histidine tag at the carboxyl terminus of prestreptokinase. For the expression of prestreptokinase, E. coli strain BL21(DE3)pLysS was transformed with pET23b-ska, grown, and induced with IPTG as described (28).
Site-directed mutagenesis of signal peptidase I.
Two mutants of S. pneumoniae signal peptidase I, S38A and K76A, were generated utilizing the Quikchange site-directed mutagenesis kit from Stratagene. Briefly, two complementary oligonucleotides (5′-TTCGCGTAGAACATGCCATGGATCCGACCCTA-3′ and 5′-TAGGGTCGGATCCATGGCATGTCCTTCTACGCGAA-3′), designed for generating S38A, and another two complementary oligonucleotides (5′-GGCAATAAGGACATCGTCGCGCGCGTGATTGGAATGC-3′ and 5′-GCATTCCAATCACGCGCGCGACGATGTCCTTATTGCC-3′), designed for generating K76A, were synthesized according to the published sequence (43). The basic procedure utilized the purified pET16b-spi vector and a pair of synthetic oligonucleotide primers containing the desired mutation. The oligonucleotide primers, each complementary to the opposite strand of the vector, were extended by using PfuTurbo DNA polymerase. Incorporation of the oligonucleotide primers generated a mutated plasmid containing staggered nicks. Following temperature cycling, the product was treated with DpnI to digest the parental DNA template and to select for mutation-containing synthesized DNA. The nicked vector DNA incorporating the desired mutations was then transformed into E. coli strain XL1-Blue. The mutants were selected and confirmed by DNA sequencing.
Solubilization and purification of S. pneumoniae signal peptidase I.
One liter of IPTG-induced E. coli BL21(DE3)pLysS cells harboring pET16b-spi were harvested and resuspended in 20 ml of lysis buffer containing 50 mM Na2HPO4 and 300 mM NaCl (pH 8.0) and sonicated for 5 min on ice. The lysate was then centrifuged at 50,000 × g for 1 h at 4°C. The resulting supernatant was discarded, and the pellet was resuspended and sonicated for 5 min in 20 ml of lysis buffer with 1% Triton X-100. After centrifugation at 50,000 × g for 1 h at 4°C, the supernatant was diluted with 80 ml of lysis buffer and loaded onto a preequilibrated Ni-nitrilotriacetic acid (NTA) column, which was then washed with 50 ml of lysis buffer with 0.1% Triton X-100 and 15 mM imidazole. Finally, the protein was eluted with 10 ml of elution buffer containing 20 mM Tris-HCl (pH 8.0), 20% glycerol, and 100 mM imidazole.
Purification of prestreptokinase and streptokinase.
One liter of IPTG-induced E. coli BL21(DE3)pLysS cells containing expression vector pET23b-ska were harvested by centrifugation. The lysate was prepared by sonication as described above and centrifuged at 50,000 × g for 1 h at 4°C to result in supernatant (S1) and pellet (P1). S1 was saved for further purification, and P1 was solubilized with 20 ml of lysis buffer plus 1% Zwittergent 3-16. After sonication for 5 min on ice, the mixture was incubated at room temperature for 15 min and then centrifuged at 50,000 × g for 1 h. The resultant supernatant, S2, was collected and diluted with 80 ml of lysis buffer for further purification of prestreptokinase by Ni-NTA column chromotagraphy.
Two Ni-NTA columns were prepared and equilibrated with lysis buffer. Supernatants S1 and diluted S2 were each loaded onto one of the two Ni-NTA columns, which were then washed with 50 ml of lysis buffer with 15 mM imidazole (for S1) or 50 ml of lysis buffer with 15 mM imidazole and 0.1% Zwittergent (for S2). The streptokinase was eluted with 10 ml of elution buffer with 100 mM imidazole (for S1) or elution buffer with 100 mM imidazole and 0.1% Zwittergent (for S2). Streptokinase from S1 was the mature form that had been processed in vivo by endogenous E. coli signal peptidase I, and the protein purified from S2 was prestreptokinase, which was utilized as the substrate of the S. pneumoniae signal peptidase I.
In vitro transcription and translation of prestreptokinase.
In vitro transcription and translation of prestreptokinase gene were performed with the TNT coupled reticulocyte lysate system from Promega. Briefly, in 50 μl of reaction mixture, 1 μg of purified vector pET23b-ska DNA was mixed with 25 μl of TNT rabbit reticulocyte lysate, 10 U of T7 RNA polymerase, RNasin RNase inhibitor, 1 mM amino acid mixture, 2 μl of 35S-labeled methionine (>1,000 Ci/mmol at 10 mCi/ml), and 2 μl of TNT reaction buffer. The mixture was incubated at 30°C for 90 min. The in vitro-translated products were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
N-terminal peptide sequencing.
In order to determine the cleavage site of prestreptokinase and the internal cut site of S. pneumoniae signal peptidase I, the proteolytic products of reactions were fractionated by SDS-PAGE and transferred to a polyvinylidenedifluoride membrane by electroblotting. The membrane was then briefly stained with Coomassie brilliant blue and destained with 50% methanol. The expected protein bands were cut out and sequenced by N-terminal peptide sequencing.
Assay for processing of prestreptokinase.
Reaction mixture (20 μl) containing 0.1 μg of signal peptidase I was incubated at 37°C for 1 h with 2 μl of in vitro-translated prestreptokinase or 2 μg of purified prestreptokinase in 20 mM Tris-HCl (pH 8.0)–0.02% Triton X-100–5% glycerol–100 μg of phospholipid. Typically, the reactions were terminated by the addition of SDS sample buffer. Products of the reaction were separated on an SDS–12% polyacrylamide gel, and the gel was then subjected to autoradiography for in vitro-translated substrate or stained with Coomassie brilliant blue for purified substrate.
Self-cleavage of signal peptidase I.
Reaction mixtures (20 μl) containing signal peptidase I were incubated at 37°C for 1 h in 20 mM Tris-HCl (pH 8.0)–0.05% Triton X-100–5% glycerol–100 μg of E. coli lipid extract. The reactions were then terminated by addition of SDS sample buffer. Reaction products were separated on a 4 to 20% gradient polyacrylamide gel with SDS and stained with Coomassie brilliant blue. Densitometer analysis was performed using a Personal Densitometer SI and Image Quant 5.0 software from Molecular Dynamics.
RESULTS
Expression and purification of signal peptidase I.
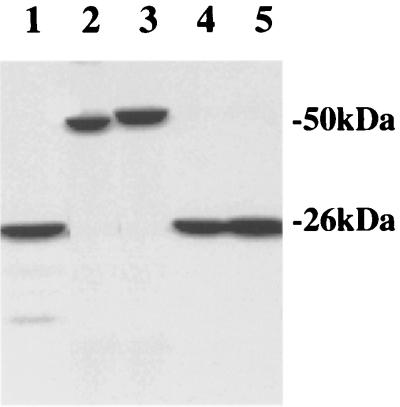
S. pneumoniae signal peptidase I is a membrane-bound protein, which contains one transmembrane segment near its N terminus (43). E. coli BL21(DE3)pLysS cells harboring the vector pET16b-spi were grown to an A600 of 0.7 to 0.9 at 30°C. After induction with 0.4 mM IPTG, a new protein band at the expected molecular mass of 26 kDa was visualized by SDS-PAGE followed by staining. The overexpressed signal peptidase I was not soluble by simple salt extraction (data not shown). However, it was solubilized by 1% Triton X-100 and purified by Ni-NTA column chromatography to homogeneity, as shown in Fig. 1 (lane 1). The purified enzyme was stable and active in buffer consisting of 20 mM Tris-HCl (pH 8.0), 0.1% Triton X-100, 20% glycerol, and 100 mM imidazole. About 10 to 15 mg of the purified protein was obtained from 1 liter of induced E. coli cells.
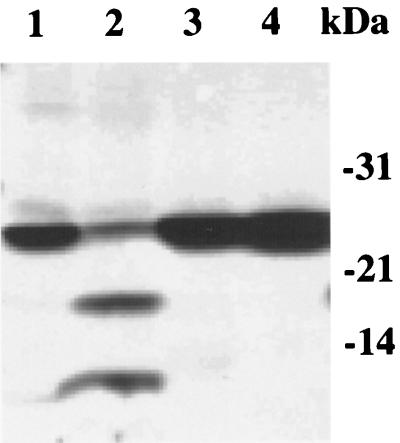
FIG. 1.
SDS-PAGE analysis of purified S. pneumoniae signal peptidase I, its mutants and substrate. The proteins were purified from E. coli as described in the text. Each protein (2 μg total) was separated on an SDS–12% polyacrylamide gel and stained with Coomassie brilliant blue. Lane 1, S. pneumoniae signal peptidase I; lane 2, mature streptokinase; lane 3, prestreptokinase; lane 4, mutant S38A; lane 5, mutant K76A.
Expression and purification of S. pyogenes streptokinase.
Streptokinase is an extracellular protein produced by pathogenic streptococci, and it functions in the species-specific conversion of plasminogen to plasmin. Its precursor, prestreptokinase, contains a typical signal peptide (11). In order to keep the integrity of the N-terminal signal peptide of the pre-streptokinase, the gene was amplified by PCR and cloned into vector pET23b, resulting in pET23b-ska, which encodes prestreptokinase with a C-terminal histidine tag. Interestingly, after transformation into E. coli and induction with IPTG, part of the overexpressed prestreptokinase was processed into the mature streptokinase by endogenous E. coli signal peptidase I and secreted into the periplasm. This mature streptokinase, a hydrophilic protein, was easily solubilized by salt extraction and purified to homogeneity on an Ni-NTA column (Fig. 1, lane 2). The majority of the overexpressed prestreptokinase was not processed in E. coli and can be solubilized by 1% Zwittergent 3–16. The solubilized prestreptokinase was then purified to near homogeneity in the presence of Zwittergent on an Ni-NTA column (Fig. 1, lane 3). The purified prestreptokinase was used as the substrate for purified S. pneumoniae signal peptidase I.
Prestreptokinase is a substrate of S. pneumoniae signal peptidase I.
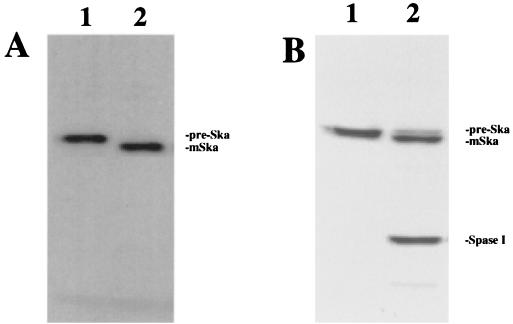
Signal peptidases from gram-positive and gram-negative bacteria are different in size and primary sequence. Our earlier effort to find a substrate for S. pneumoniae signal peptidase I was unsuccessful. We tested some known substrates of E. coli signal peptidase I, including the precursor of β-lactamase (36), a pre-OmpA fusion protein (10), and a modified peptide substrate (44). The results demonstrated that none of these substrates was effectively cleaved in vitro by the S. pneumoniae enzyme (data not shown) and indicated that different substrate specificities exist between signal peptidases from gram-positive and gram-negative bacteria. In order to establish an in vitro biochemical assay system to characterize the signal peptidase I of gram-positive bacteria, we examined if prestreptokinase could be hydrolyzed by the S. pneumoniae enzyme. As demonstrated in Fig. 2, the purified enzyme was able to cleave both the in vitro-translated prestreptokinase (Fig. 2A) and the purified protein (Fig. 2B). Incubation of the purified signal peptidase I with prestreptokinase resulted in a 3-kDa molecular mass shift, which confirmed the proteolytic activity of the putative signal peptidase I and suggested that prestreptokinase was a substrate of the enzyme. Peptide sequencing revealed that the N-terminal sequence of the proteolytic product is IAGYEWLLDRP, indicating that the cleavage site is between Ala-26 and Ile-27. This is the cleavage site predicted by the algorithms of von Heijne (39). However, this proteolytic activity was not inhibited significantly by classic protease inhibitors tested, including phenylmethylsulfonyl fluoride, antipain dihydrochloride, bestatin, chymostatin, E-64, leupeptin, pepstatin, phosphoramidon, aprotinin, and EDTA (data not shown). These data confirmed the substrate specificity and the enzymatic identity of S. pneumoniae signal peptidase I.
FIG. 2.
Prestreptokinase is a substrate of S. pneumoniae signal peptidase I. (A) Autoradiography of 35S-labeled prestreptokinase generated by in vitro translation and its cleavage by purified signal peptidase I. In vitro transcription and translation were performed as described in the text. (B) SDS-PAGE analysis of purified prestreptokinase and its cleavage by signal peptidase I. The proteolytic reactions were performed as described in the text. The reaction mixtures were separated on SDS–12% PAGE, and the gel was stained with Coomassie brilliant blue. Lane 1, prestreptokinase (pre-Ska); lane 2, prestreptokinase plus signal peptidase I (Spase I). Prestreptokinase was processed to mature streptokinase (mSka) upon incubation with signal peptidase I, as demonstrated in lane 2.
Signal peptidase I activity is stimulated by phospholipids.
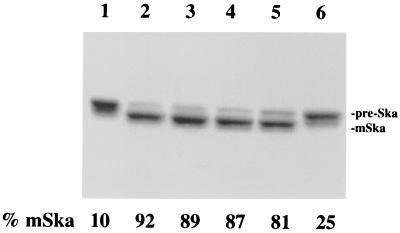
As a membrane-bound proteolytic enzyme, signal peptidase I is anchored to the cytoplasmic membrane by a transmembrane domain near its N terminus. Based upon this biochemical feature, E. coli lipid extract, composed mainly of phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin (24), was used to test its effect on enzymatic activity. Interestingly, the proteolytic activity of the signal peptidase I was stimulated by the E. coli lipid extract, as demonstrated in Fig. 3. In the absence of lipid extract, the enzyme activity was limited (lane 6); when lipid extract was included in the reaction mixture, the protease activity was enhanced by about fivefold (lane 2). To further define the lipid effects, three pure phospholipids, phosphatidylethenolamine, phosphatidylglycerol, and cardiolipin, were examined. As demonstrated in Fig. 3, all three pure phospholipids stimulated enzyme activity about four to fivefold (lanes 3 to 5).
FIG. 3.
Effects of phospholipids on the activity of S. pneumoniae signal peptidase I. Prestreptokinase, the substrate, was purified from E. coli as described in the text and incubated with purified signal peptidase I in the presence of E. coli total lipid extract or different pure phospholipids. All samples were analyzed on an SDS–10% polyacrylamide gel and stained with Coomassie brilliant blue. Densitometer analysis was performed with a Personal Densitometer S1 and Image Quant 5.0 software from Molecular Dynamics. Lane 1, purified prestreptokinase only (10% of the mature streptokinase appearing in this lane was from purification), lanes 2 to 6, purified prestreptokinase incubated with purified signal peptidase I in the presence of E. coli lipid extract (lane 2), phosphatidylglycerol (lane 3), phosphatidylethenolamine (lane 4), cardiolipin (lane 5), and no phosphalipid (lane 6). Pre-Ska and mSka, prestreptokinase and mature streptokinase, respectively.
Residues serine 38 and lysine 76 are implicated in the catalytic mechanism of S. pneumoniae signal peptidase I.
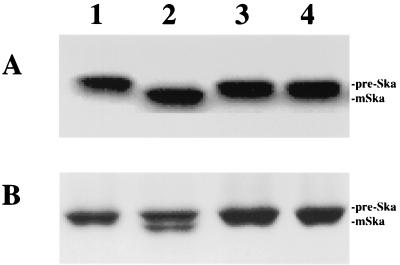
E. coli signal peptidase I contains a serine-lysine catalytic dyad, in which serine 90 and lysine 145 are involved in catalysis and are located within the active site (2, 29, 33). This catalytic dyad structure has been confirmed by recent structural analysis (20). In order to investigate the catalytic mechanism of S. pneumoniae signal peptidase I, the conserved serine 38 and lysine 76 residues have been identified by pairwise sequence alignment. Two mutants, S38A and K76A, were then generated by site-directed mutagenesis. The mutant proteins were expressed in E. coli and purified to homogeneity as shown in Fig. 1 (lanes 4 and 5). In vitro biochemical analysis revealed that these two mutants were unable to cleave purified prestreptokinse in vitro, as shown in Fig. 4. These data suggest that both serine 38 and lysine 76 are essential for enzyme function and involved in the active site of the enzyme. This indicates that S. pneumoniae signal peptidase I has a catalytic mechanism similar to that of E. coli signal peptidase I.
FIG. 4.
Implication of serine 38 and lysine 76 in the active site of S. pneumoniae signal peptidase I. (A) Autoradiography of 35 S-labeled prestreptokinase and its cleavage by wild-type and mutant signal peptidase I. (B). SDS-PAGE and Coomassie brilliant blue staining of purified prestreptokinase and its cleavage by wild-type and mutant signal peptidase I. The assays were performed as described in the text. Lane 1, prestreptokinase only; lane 2, prestreptokinase + wild-type signal peptidase; lane 3, prestreptokinase + mutant S38A; lane 4, prestreptokinase + mutant K76A. Pre-Ska and mSka, prestreptokinase and mature streptokinase, respectively.
Self-cleavage of S. pneumoniae signal peptidase I.
The highly purified S. pneumoniae signal peptidase I, when incubated at 37°C, resulted in cleavage of itself and the appearance of two products with molecular masses of 19 and 8 kDa (Fig. 5). Interestingly, this self-cleavage was also stimulated by E. coli lipid extract and was not inhibited significantly by any of the classical protease inhibitors tested (data not shown). To further confirm the specificity of this self-cleavage, the two active-site mutants S38A and K76A, described earlier, were treated under identical conditions. As shown in Fig. 5, purified S38A and K76A were unable to catalyze self-cleavage, confirming that the self-cleavage was specific and was not caused by possibly contaminating proteases. Strikingly, the self-cleavage and the signal peptide cleavage of the signal peptidase I have some common biochemical properties, i.e., insensitivity to classic protease inhibitors and stimulation by phospholipids. In addition, we mapped the self-cleavage site of the resulting products by amino acid sequencing analysis. The N-terminal sequence of the 19-kDa product was identified as HSMDPTLADG, corresponding to amino acids 37 to 46 of signal peptidase I, which suggested that the self-cleavage site of signal peptidase I was between G36 and H37. Signal peptidase I basically lost its activity after self-cleavage (data not shown).
FIG. 5.
SDS-PAGE analysis of in vitro self-cleavage catalyzed by S. pneumoniae signal peptidase I. Reactions (20 μl) containing 2 to 4 μg of wild-type signal peptidase I or its mutants were incubated at 37°C for 1 h in 20 mM Tris-HCl–0.05% Triton X-100–5% glycerol–100 μg of E. coli lipid extract. The samples were separated by 4 to 20% gradient SDS-PAGE, and the gel was stained with Coomassie brilliant blue. Lane 1, purified wild-type signal peptidase I before incubation; lane 2, wild-type signal peptidase I after incubation; lane 3, mutant S38A after incubation; lane 4, mutant K76A after incubation.
Self-cleavage of signal peptidase I is an intermolecular process.
To clearly define the self-cleavage mechanism, further biochemical analysis was performed. We found that the self-cleavage of signal peptidase I was a concentration-dependent event. The titration experiment suggested that specific activity of self-cleavage increased when the enzyme concentration was increased (Fig. 6A). In addition, the two active-site mutants S38A and K76A, which lost their ability to self-cleave, were cleaved by wild-type signal peptidase I (Fig. 6B). These two lines of evidence suggested that the self-cleavage of the enzyme was catalyzed through an intermolecular mechanism.
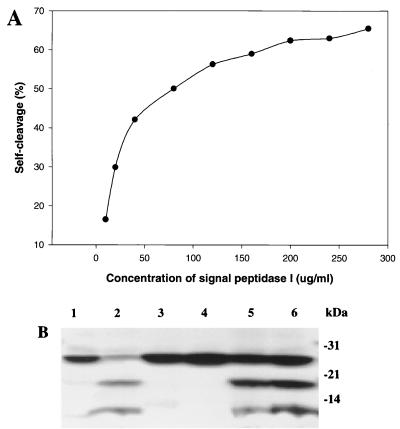
FIG. 6.
Self-cleavage of S. pneumoniae signal peptidase I is an intermolecular process. (A) Self-cleavage activity of signal peptidase I is dependent on protein concentration. Reaction mixtures (20 μl) containing different concentrations of signal peptidase I as indicated were incubated at 37°C for 30 min. The samples were separated on a 4 to 20% gradient SDS-polyacrylamide gel, and the gel was stained with Coomassie brilliant blue. Densitometer analysis was performed with a Personal Densitometer SI and Image Quant 5.0 software from Molecular Dynamics. (B) Signal peptidase I mutants S38A and K76A were cleaved by wild-type signal peptidase I. Reaction mixtures containing wild-type and/or mutant signal peptidase I were incubated at 37°C for 1 h. The samples were separated by 4 to 20% gradient SDS-PAGE, and the gel was stained by Coomassie brilliant blue. Lane 1, 2 μg of wild-type signal peptidase before incubation; lane 2, 2 μg of wild-type signal peptidase I after incubation; lane 3, 4 μg of mutant S38A after incubation; lane 4, 4 μg of mutant K76A after incubation; lane 5. 2 μg of wild-type signal peptidase I + 4 μg of S38A after incubation; lane 6, 2 μg of wild-type signal peptidase I + 4 μg of K76A after incubation.
DISCUSSION
Our biochemical knowledge of signal peptidase I was based almost exclusively upon investigation of the enzyme from the gram-negative E. coli, while biochemical characterization of gram-positive signal peptidase I was very limited and more speculative. In this study, we found that substrate specificities differ between gram-negative and gram-positive bacterial signal peptidases. The precursor of streptokinase, a well-characterized extracellular protein in pathogenic streptococci, has been shown to be a native substrate of the enzyme. We have established an in vitro reaction system using the purified S. pneumoniae signal peptidase I and a purified protein substrate to characterize the gram-positive signal peptidase I. This defined reaction system allows further comparative analysis of the biochemical properties and substrate specificities between gram-positive and gram-negative enzymes.
A number of genes encoding putative signal peptidases I have been cloned and sequenced from both gram-positive and gram-negative bacteria. Clearly, there are similarities between the two groups of enzymes. In fact, some of the conserved regions and critical residues involved in active sites are present in the enzymes of both bacterial groups. However, considerable differences also exist, as discussed earlier. These differences include the primary sequences, the size, and the topology of the enzymes. Why are signal peptidases from two bacterial groups so different although they catalyze a similar reaction in vivo? One of our hypotheses is that they may have different substrate specificities within the cells. Our data in this report support this hypothesis. We found that some known E. coli signal peptidase I substrates, including the precursor of β-lactamase (36), a pre-OmpA fusion protein (10), and a modified peptide substrate (44), were not effectively cleaved in vitro by purified S. pneumoniae signal peptidase I, although both enzymes were able to process prestreptokinase in vivo and in vitro (data not shown). Another piece of evidence to support this hypothesis is that the signal peptides from gram-positive bacteria are generally bigger and more hydrophobic than those from gram-negative bacteria. Recent structural analysis revealed that an unusually exposed hydrophobic surface extends across the E. coli signal peptidase I and includes the substrate-binding site and catalytic center (20). Therefore, we suspect that the signal peptidase I from S. pneumoniae and other gram-positive bacteria may have different hydrophobicity on the surface around the catalytic center and the substrate-binding site. Further investigation to address the different substrate specificities between the S. pneumoniae and E. coli signal peptidases is in process in our laboratory.
Interestingly, we have demonstrated that both E. coli lipid extract and pure phospholipids stimulated the proteolytic activity of the signal peptidase I. Similar stimulation has been reported for the truncated E. coli signal peptidase I (32). Considering that both signal peptidase I and the signal peptide of a secreted protein contain hydrophobic domains, the interactions of phospholipids with signal peptidase I or/and signal peptide may play an important physiological role in the catalytic mechanism of the enzyme. To date, it is not clear how phospholipids modulate the enzyme activity. Further investigation to address these issues is required. We have also investigated the influence of detergent on the activity of S. pneumoniae signal peptidase I. Because the enzyme is a membrane-bound protein, its solubilization and purification were expected to be detergent dependent. In this study, we found that 1% Triton X-100 could solubilize the protein, and the whole purification procedure was performed in the presence of the detergent. We also noticed that the detergent was required for storage of active enzyme (data not shown).
It has been suggested that E. coli signal peptidase I is a special serine protease that does not utilize a histidine as a catalytic base, but may instead employ a lysine side chain to fulfill this role (2, 3, 29, 33). In this regard, the hydroxyl group of the serine side chain acts as the nucleophile that attacks the scissile peptide bond of the preprotein cleavage site. The unprotonated form of the lysine ε-amino group serves to activate the hydroxyl group of the serine. In our study, a conserved serine 38 and lysine 76 have been identified. The lack of enzymztic activity of mutants S38A and K76A suggests that this serine and lysine are critical amino acids involved in the catalytic reaction of S. pneumoniae signal peptidase I. Thus, S. pneumoniae signal peptidase I is similar to the E. coli enzyme, which also contains a serine-lysine catalytic dyad.
A precedent for a mechanism involving a serine-lysine dyad for a peptidase has been reported. The LexA protein, which is involved in the SOS response in E. coli, undergoes a self-cleavage reaction that inactivates the protein. Similar to signal peptidase I, the LexA protease employs a serine as the nucleophile that attacks the peptide bond (25) and a lysine that is deprotonated (15, 16). Moreover, X-ray crystallographic analysis has shown that the serine-lysine dyad is at the active site of UmuD protein, a member of the LexA peptidase family (21).
Van Dijl and his colleagues proposed that signal peptidase I and LexA-like proteases are structurally and functionally related enzymes, based upon structural analysis and site-directed mutagenesis of the sipS gene of Bacillus subtilis (35). Sequence analysis of S. pneumoniae signal peptidase I and LexA-like proteases revealed a conserved region around the active sites of these enzymes, and some critical residues involved in the active sites are identical among these proteases (Fig. 7A). Another striking common feature of S. pneumoniae signal peptidase I and LexA-like proteases is that they all catalyze self-cleavage. We have found that S. pneumoniae signal peptidase I cleaves itself to generate two products with molecular masses of 19 and 8 kDa. Self-cleavage has been found to be a unique property among LexA-like proteases. Sequence analysis demonstrated that the regions around self-cleavage sites of signal peptidase I and LexA-like proteases have some common properties, although there is no strong sequence homology. These self-cleavage sites, as compared in Fig. 7B, are similar to signal peptidase cleavage sites, with small neutral amino acid residues at the −1 (usually Ala, Gly, and Ser) and −3 (usually Ala, Gly, Ser, Val, and Cys) positions preceding a nonpolar amino acid stretch (13, 22, 38).
FIG. 7.
Similarities between. S. pneumoniae signal peptidase I and LexA-like proteases. (A) Sequence alignment around the active sites of S. pneumoniae signal peptidase I and LexA-like proteases. The alignment includes, with the EMBL/GenBank accession numbers in parentheses, LexA of E. carotovara (ER) (X63189), LexA of E. coli (EC) (P03033), ImpA of S. enterica serovar Typhimurium (P18641), SamA of S. enterica serovar Typhimurium (P23831), MucA of S. enterica serovar Typhimurium (P07376), UmuD of E. coli (P04153), DinR of B. subtilis (P31080), and signal peptidase I of S. pneumoniae (SpaseI). Identical residues are shown as black against a white background, and similar residues are shaded gray. The serine and lysine critical for the activity of LexA-like proteases and signal peptidase I are shown (∗). (B) Sequence comparison around self-cleavage sites among S. pneumoniae signal peptidase I and LexA-like proteases. Self-cleavage sites of signal peptidase I and LexA-like proteases are marked (▾). The −1 and −3 positions relative to the self-cleavage sites are highlighted. Nonpolar amino acid stretches are boxed.
Taken together, the data from this study and others (35) strongly suggest that bacterial signal peptidases I and LexA-like proteases are closely related enzymes and belong to a novel class of serine proteases which contain a conserved serine-lysine catalytic dyad. Self-cleavage seems to be common among signal peptidase I and LexA-like proteases. Whether all bacterial signal peptidases catalyze self-cleavage like LexA-like proteases remains to be determined. The physiological role of self-cleavage of signal peptidases also remains to be addressed.
ACKNOWLEDGMENTS
We thank JoAnn Hoskins for performing the DNA database search and Melvin G. Johnson for peptide sequencing.
REFERENCES
- 1.Bilgin N, Lee J I, Zhu H Y, Dalbey R E, von Heijne G. Mapping of catalytically important domains in Escherichia coli leader peptidase. EMBO J. 1990;9:2717–2722. doi: 10.1002/j.1460-2075.1990.tb07458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black M T. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J Bacteriol. 1993;175:4957–4961. doi: 10.1128/jb.175.16.4957-4961.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black M T, Munn J G R, Allsop A. On the catalytic mechanism of prokaryotic leader peptidase I. Biochem J. 1992;282:539–543. doi: 10.1042/bj2820539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee S, Suciu D, Dalbey R E, Kahn P C, Inouye M. Determination of Km and kcat for signal peptidase I using a full length secretory precursor, pre-OmpA-nuclease A. J Mol Biol. 1995;245:311–314. doi: 10.1006/jmbi.1994.0025. [DOI] [PubMed] [Google Scholar]
- 5.Cregg K M, Wilding E I, Black M T. Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. J Bacteriol. 1996;178:5712–5718. doi: 10.1128/jb.178.19.5712-5718.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of type I signal peptidase. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dev I K, Ray P H, Novak P. Minimum substrate sequence for signal peptidase I of Escherichia coli. J Biol Chem. 1990;265:20069–20072. [PubMed] [Google Scholar]
- 8.Dierstein R, Wickner W. Requirement for substrate recognition by bacterial leader peptidase. EMBO J. 1986;5:427–431. doi: 10.1002/j.1460-2075.1986.tb04228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, Mckenny K, Sutton G, Fitzhugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelly J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Verter J C. Whole genome random sequencing and assembly of Haemophilus influenzae. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Hsiung H M, Mayne N G, Becker G W. High-level expression, efficient secretion and folding of human growth hormone in Escherichia coli. Bio/Technology. 1986;4:991–995. doi: 10.1016/0014-5793(86)81403-x. [DOI] [PubMed] [Google Scholar]
- 11.Huang T T, Malke H, Ferretti J J. The streptokinase gene of group A streptococci: cloning, expression in Escherichia coli, and sequence analysis. Mol Microbiol. 1989;3:197–205. doi: 10.1111/j.1365-2958.1989.tb01808.x. [DOI] [PubMed] [Google Scholar]
- 12.Innis M A, Tokunaga M, Williams M E, Loranger J M, Chang S Y, Wu H C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci USA. 1984;81:3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain R G, Rusch S L, Kendall D A. Signal peptide cleavage regions. Functional limits on length and topological implication. J Biol Chem. 1994;269:16305–16310. [PubMed] [Google Scholar]
- 14.Kuo D W, Chan H K, Wilson C J, Griffin P R, Williams H, Knight W B. Escherichia coli leader peptidase: production of an active form lacking a requirement for detergent and development of a peptide substrate. Arch Biochem Biophys. 1993;303:274–280. doi: 10.1006/abbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 15.Lin L L, Little J W. Autodigestion and RecA-dependent cleavage of Ind− mutant LexA proteins. J Mol Biol. 1989;210:439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- 16.Little J W. LexA cleavage and other self-processing reactions. J Bacteriol. 1993;175:4943–4950. doi: 10.1128/jb.175.16.4943-4950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markham B E, Little J W, Mount D W. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981;9:4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijer W J J, de Jong A, Bea G, Wiseman A, Tjalsma H, Venema G, Bron S, van Dijl J M. The endogenous B. subtilis plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol Microbiol. 1995;17:621–631. doi: 10.1111/j.1365-2958.1995.mmi_17040621.x. [DOI] [PubMed] [Google Scholar]
- 19.Moore K E, Miura S. A small hydrophobic domain anchors leader peptidase to the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1987;262:8806–8813. [PubMed] [Google Scholar]
- 20.Paetzel M, Dalbey R E, Strynadka N C J. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 21.Peat T S, Frank E G, McDonald J P, Levine A S, Woodgate R, Hendrickson W A. Structure of UmuD′ protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 22.Perlman D, Halvorson H O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- 23.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and umuAB operons whose products are required for UV-light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ractz C R H, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem. 1990;265:1235–1238. [PubMed] [Google Scholar]
- 25.Roland K L, Little J W. Reactions of LexA repressor with diisopropyl fluorophosphate: a test of the serine protease model. J Biol Chem. 1990;265:12828–12835. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Smith C M, Eisenstadt E. Identification of a umuDC locus in Salmonella typhimurium LT2. J Bacteriol. 1989;171:3860–3865. doi: 10.1128/jb.171.7.3860-3865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier F W, Rosenburg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 29.Sung M, Dalbey R E. Identification of potential active-site residues in the Escherichia coli leader peptidase. J Biol Chem. 1992;267:13154–13159. [PubMed] [Google Scholar]
- 30.Tjalsma H, Noback M A, Bron S, Venema G, Yamane K, van Dijl J M. Bacillus subtilis contains four closely related type I signal peptidase with overlapping substrate specificities. J Biol Chem. 1997;272:25983–25992. doi: 10.1074/jbc.272.41.25983. [DOI] [PubMed] [Google Scholar]
- 31.Tschantz W R, Dalbey R E. Bacterial leader peptidase I. Methods Enzymol. 1994;244:285–301. doi: 10.1016/0076-6879(94)44023-9. [DOI] [PubMed] [Google Scholar]
- 32.Tschantz W R, Paetzel M, Cao G, Suciu D, Inouye M, Dalbey R E. Characterization of a soluble, catalytically active form of Escherichia coli leader peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry. 1995;34:3935–3941. doi: 10.1021/bi00012a010. [DOI] [PubMed] [Google Scholar]
- 33.Tschantz W R, Sung M, Delgado-Partin V M, Dalbey R E. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase J. Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 34.van Dijl J M, de Jong A, Vehmaanpera J, Venema G, Bron S. Signal peptidase I of B. subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 1992;11:2819–2828. doi: 10.1002/j.1460-2075.1992.tb05349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Dijl J M, de Jong A, Venema G, Bron S. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis: structural and functional similarities with LexA-like proteases. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- 36.van Dijl J M, Smith H, Bron S, Venema G. Synthesis and processing of Escherichia coli TEM-beta-lactamase and Bacillus licheniformis alpha-amylase in E. coli: the role of signal peptidase I. Mol Gen Genet. 1988;214:55–61. doi: 10.1007/BF00340179. [DOI] [PubMed] [Google Scholar]
- 37.van Dijl J M, van den Bergh R, Reversma T, Smith H, Bron S, Venema G. Molecular cloning of the Salmonella typhimurium lep gene in Escherichia coli. Mol Gen Genet. 1990;223:233–240. doi: 10.1007/BF00265059. [DOI] [PubMed] [Google Scholar]
- 38.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;116:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 39.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickner W, Driessen A J M, Hartl F-U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]
- 41.Wolfe P B, Silver P, Wickner W. The isolation of homogeneous leader peptidase from a strain of Escherichia coli which overproduces the enzyme. J Biol Chem. 1982;257:7898–7902. [PubMed] [Google Scholar]
- 42.Wolfe P B, Wickner W, Goodman J M. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J Biol Chem. 1983;258:12073–12080. [PubMed] [Google Scholar]
- 43.Zhang Y, Greenberg B, Lacks S A. Analysis of a Streptococcus pneumoniae gene encoding signal peptidase I and overproduction of the enzyme. Gene. 1997;194:249–255. doi: 10.1016/s0378-1119(97)00198-4. [DOI] [PubMed] [Google Scholar]
- 44.Zhong W, Benkovic S J. Development of an internally quenched fluorescent substrate for Escherichia coli leader peptidase. Anal Biochem. 1998;255:66–73. doi: 10.1006/abio.1997.2471. [DOI] [PubMed] [Google Scholar]
- 45.Zwizinski C, Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980;255:7973–7977. [PubMed] [Google Scholar]