Abstract
The transcription of nhaA, encoding the main Na+/H+ antiporter of Escherichia coli, is induced by Na+, regulated by NhaR, and affected by H-NS. In this work the roles of the two nhaA promoters (P1 and P2) were studied by analysis of transcription both in vivo and in vitro and promoter mutations. We found that P1 is an NhaR-dependent, Na+-induced, and H-NS-affected promoter both in the exponential and stationary phases. An in vitro transcription assay demonstrated that P1 is activated by ς70-RNA polymerase and both NhaR and H-NS increase the specificity of P1. Remarkably, in marked contrast to P1, P2 exhibits very low activity during the exponential phase but is induced in the stationary phase to become the major promoter. Furthermore, P2 is activated by ςS and is neither induced by Na+ nor dependent on NhaR or affected by H-NS. Hence, this work establishes that nhaA has a dual mode of regulation, each involving a different promoter, and reveals that P2 and ςS together are responsible for the survival of stationary-phase cells in the presence of high Na+, alkaline pH, and the combination of high Na+ and alkaline pH, the most stressful condition.
Sodium proton antiporters are ubiquitous membrane proteins found in the cytoplasmic and organelle membranes of cells of many different origins, including plants, animals, and microorganisms. They are involved in cell energetics, playing primary roles in signal transduction and in regulation of intracellular pH, cell Na+ content, and cell volume (24, 26).
Escherichia coli has two antiporters, NhaA (11, 17) and NhaB (27), which specifically exchange Na+ or Li+ for H+ (25). nhaA is indispensable for adaptation to high salinity, for resistance to Li+ toxicity, and for growth at alkaline pH (in the presence of Na+ [23]). nhaB by itself confers a limited sodium tolerance to the cells but becomes essential when the lack of NhaA activity limits growth (28).
The pattern of regulation of nhaA during exponential growth, studied in a strain carrying an nhaA′-′lacZ fusion (16) and in the wild-type strain (10), reflects the importance of nhaA in salt adaptation. This system is specifically induced by intracellular Na+, and neither osmolarity nor ionic strength affect nhaA transcription (10). Only one additional regulatory system that is induced specifically by Na+ is known but has not been studied intensively yet (20).
The transcription of nhaA, which is positively regulated by NhaR (29), a member of the LysR-OxyR family (7, 14, 30, 34), is specifically induced by intracellular Na+. The regulation of nhaA is also affected by H-NS (10), a major DNA binding protein and a global regulator involved in salt stress in bacteria (8, 9, 15, 22, 35). An interplay between NhaR and H-NS in the regulation of nhaA has been suggested. Deleting hns causes derepression of nhaA, and both repression and Na+ inducibility are restored upon transformation with plasmidic nhaR (10).
Two promoters, P1 and P2, were previously identified for nhaA. They were located 30 and 172 bp upstream of the initiation codon, respectively (16). DNA gel retardation assay and DNase I footprinting analysis showed that NhaR binds a region of 92 bp located 18 to 119 bp upstream of the nhaA initiation codon (6). Dimethylsulfate methylation protection footprinting both in vivo and in vitro identified four bases in this region which form direct contact with NhaR. Furthermore, two of these bases were located around P1, and their binding to NhaR was found to be affected specifically by Na+.
These results suggest that P1 but not P2 is the Na+-specific promoter of nhaA and raises the question as to what the role of P2 is. We therefore studied the roles of P1 and P2 in nhaA transcription and found the following. (i) P1 but not P2 is the Na+- and NhaR-responsive promoter of nhaA. It is activated by ς70 and affected by H-NS. It is the major nhaA promoter in the exponential phase of growth but a minor promoter in the stationary phase. (ii) P2 has a very low constitutive activity in exponentially growing bacteria and is not affected by factors which affect P1 (Na+, NhaR, and H-NS). (iii) When the bacteria enter the stationary phase, P2 becomes the main promoter for nhaA and is activated by ςS in an Na+-independent fashion. (iv) In the stationary phase, P2 is responsible for survival of the cells under the stressful conditions of high Na+, alkaline pH, and alkaline pH in the presence of Na+, the most stressful combination.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are the E. coli K-12 derivatives described in Table 1. Cells were grown at 37°C in modified L broth (LBK) in which NaCl was replaced by KCl (87 mM [pH 7.5]). This medium was supplemented with 60 mM BTP {1,3-bis[tris(hydroxymethyl)-methylamino]propane}, and the pH was titrated to 7.5 with HCl. For plates, 1.6% agar was used. Antibiotic concentrations were 100 μg of ampicillin/ml, 50 μg of kanamycin/ml, and 12.5 μg of tetracycline/ml.
TABLE 1.
Characteristics of bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| TA15 | MelBLid nhaA+ nhaB+ lacZY | 11 |
| RK33Z | TA15 nhaA3:: kan (nhaA::lacZ)1 (hyb) thr-1 | 16 |
| W3313-2S | TA15 nhaR1 (NhaAup) | 5 |
| RK20 (ΔnhaA) | TA15 nhaA2:: kan thr-1 | 16 |
| OR100 (ΔnhaR) | TA15 nhaR1:: kan thr-1 | 29 |
| OR200 (ΔnhaA ΔnhaR) | TA15 nhaA4 nhaR2:: kan thr-1 | 29 |
| GM230 (hns) | MC4100 hns-205::Tn10 | 15 |
| PD32 (hns)a | MC4100 hns-206::amp | 8 |
| GSO15 (rpoS)b | rpoS::Tn10 | 1 |
| NDZ1 (ΔnhaR) | RK33Z nhaR1::kan thr-1 | 10 |
| TA15D2 | TA15 hns-206::amp | P1 transduction, PD32→TA15 |
| OR100D2 | OR100 hns-206::amp | P1 transduction, PD32→OR100 |
| OR200D1 | OR200 hns-205::Tn10 | P1 transduction, GM230→OR200 |
| TA15 rpoS | TA15 rpoS::Tn10 | P1 transduction, GSO15→TA15 |
| OR200 rpoS | OR200 rpoS::Tn10 | P1 transduction, GSO15→OR200 |
| TA15D2 rpoS | TA15D2 rpoS::Tn10 | P1 transduction, GSO15→TA15D2 |
Kindly provided by E. Bremer (Marburg, Germany).
Kindly provided by S. Altuvia (Jerusalem, Israel).
Plasmids.
All plasmids were derived from pBR322. pKRZ2 is a plasmid bearing an nhaA′-′lacZ fusion containing the upstream regulatory sequence of nhaA (16). pKRZ1 is a derivative of pKRZ2 which contains only P1, the proximal nhaA promoter (16). pGM42 is a derivative of pBR322 bearing nhaA and nhaR (17). pGM42T is a derivative of pGM42 with nhaA inactivated (29). pGM36 is a derivative of pBR322 bearing nhaA (17). To construct pKRZ3, an NgoMI-Kpn2I fragment (2,138 bp) containing nhaR was obtained from pGM42 and ligated with Kpn2I-digested pKRZ2 (7,572-bp fragment). To construct pKRZ3tac, a BamHI-BamHI fragment (280 bp) containing the tac promoter was obtained from pKK223–3 (Pharmacia) and ligated with BglII-digested pKRZ3 (9,954 bp). To construct pKRZ4 and pKRZ1tac, the AatII-AatII fragment (2,076 bp) of pKRZ3 and pKRZ3tac, respectively, was replaced with the AatII-AatII fragment (1,270 bp) of PKRZ1, containing P1 only. To construct pKRZ2*, a plasmid carrying nhaA with a mutation in the −10 box of P1 (Fig. 1C, −10 P1) was mutated by PCR site-directed mutagenesis with the following mutagenic primers: Nir946–973, 5′-TGATTCGTGCGGGGCCCAAGAGTAAAAACGATCT-3′; Nir973–946, 5′-AGATCGTTTTTACTCTTGGGCCCCGCACGAATCA-3′. The mutated bases are underlined. The downstream end primer was P27 (5′-GGGAATAAGGGCGACACGGAAATGTTG-3′), and the upstream end primer was ARP1342 (5′-GCCAGTACACCAAGTGCAAAAGCAATGTCAGTAGCCG-3′). The PCR 1,558-bp fragment carrying the mutation was digested with NheI and BsmI, and the 1,052-bp fragment was ligated with pKRZ2 digested with the same enzymes (6,520-bp fragment).
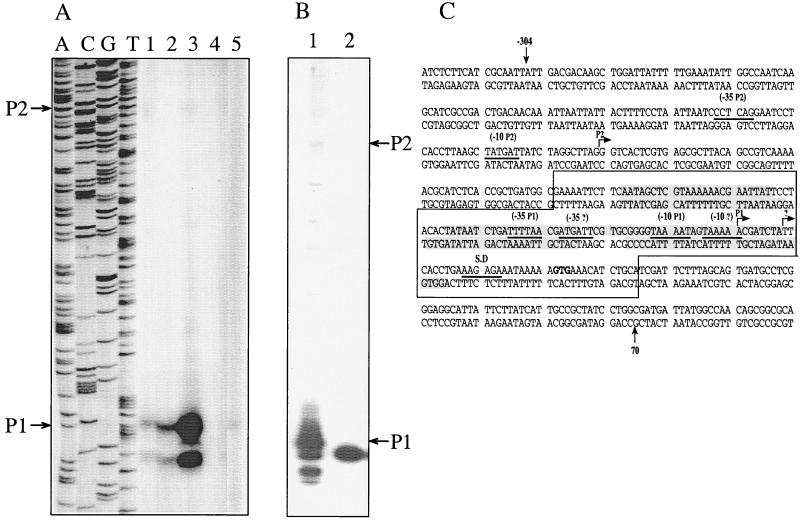
FIG. 1.
P1 is the Na+-sensitive promoter of nhaA. (A) Primer extension assay of nhaA transcription was conducted, as described in Materials and Methods, with total RNA (15 μg) isolated from cells grown exponentially for 2 h in LBK-BTP broth at pH 7.5, either in the presence or absence of 300 mM NaCl for induction. The same primer was used for the sequencing ladder, and the reaction mixtures were run on an 8% DNA sequencing gel. The cells used were TA15 (wild type) grown without addition of sodium (lane 1); TA15, sodium induced (lane 2); W3313–2S (nhaR1) cells grown without sodium (lane 3); RK20 (ΔnhaA), sodium added (lane 4); and OR100 (ΔnhaR), sodium added (lane 5). (B) Primer extension assay conducted as for panel A with RNA isolated from OR200 (ΔnhaA ΔnhaR) transformed with plasmid pGM42 (lane1) or pGM433 (lane 2). (C) Promoter elements of nhaA. Start sites (arrows), −10 and −35 sequences (underlined) are marked on the DNA sequence of nhaA. The putative third promoter elements of nhaA are indicated with question marks. Boxed area, binding site of NhaR (6). Shaded areas, nhaA sequences protected by NhaR from DNase I digestion (6).
To construct pKRZ3* and pKRZ3tac*, the AatII-AatII fragment (2,076 bp) from pKRZ3 and pKRZ3tac, respectively, was replaced with the same fragment from pKRZ2* carrying the mutation. To construct pKRZ1*, pKRZ4*, and pKRZ1tac*, the same procedures as those described above for pKRZ2*, pKRZ3*, and pKRZ3tac*, respectively, were used. To construct pKRZ33, the −10 box of the putative third promoter (Fig. 1C, −10?) was mutated by PCR site-directed mutagenesis with the following mutagenic primers: PETI1, 5′-GCGGGGTAAAATAGGACCAACGATCTATTCACC-3′; PETI2, 5′-GGTGAATAGATCGTTGGTCCTATTTTACCCCGC-3′. The mutated bases are underlined. The downstream end primer was P27 and the upstream end primer was ARP1342 (see sequences above). The PCR 1,558-bp fragment carrying the mutation was digested with NheI and BsmI, and the 1,052-bp fragment was ligated with pKRZ3 digested with the same enzymes (8,902-bp fragment).
To construct pGM433, the NheI-BsmI fragment (7,757 bp) obtained from pGM42 was ligated with the NheI-BsmI (1,052 bp) fragment obtained from pKRZ33. To construct pROB, fragment PflMI-PflMI (1,970 bp) obtained from pGM42, containing the upstream region of nhaA, was ligated with EcoRV-digested pBluescript KS+ (Stratagene) in the direction allowing transcription of a cRNA of nhaA from the T3 promoter. To construct pGM36*, the NheI-BsmI fragment (1,052 bp) obtained from pKRZ2*, containing the P1 mutation, was ligated with the NheI-BsmI fragment (4,696 bp) obtained from pGM36.
RNA isolation.
Total RNA was isolated by using the PUREscript RNA Isolation Kit of Gentra Systems, Inc., according to the manufacturer's instructions.
Primer extension.
An antisense primer, complementary to the region of DNA 138 to 161 bp downstream of the P1 start site (TCTCCAGAAAGTCGTGATACCATC) was 32P end labeled with T4 polynucleotide kinase (Amersham Pharmacia). Primer extension was conducted as described in reference 33. The RNA (15 μg of total cell RNA or 7 μl of in vitro-transcribed RNA) was mixed with the radioactively labeled primer (15,000 to 30,000 cpm) in water in a total volume of 9 μl. This mixture was heated at 75°C for 5 min and then cooled on ice for 10 min for annealing. Then, 3 μl of 5× buffer (0.25 M Tris-Cl [pH 8.3], 0.375 M KCl, 15 mM MgCl2), 1.5 μl of 0.1 M dithiothreitol, 0.5 μl of 25 mM deoxynucleoside triphosphates, and 0.5 μl of water were added and the mixture was warmed to 48°C for 5 min. Reverse transcriptase (0.5 μl of Superscript II [200 U/ml; Gibco]) was added, and the reaction mixture was incubated at 48°C for 60 min, dried, resuspended in 4 μl of formamide dye–1.5 μl of 0.1 M NaOH, heated at 90°C for 3 min, and loaded on an 8% DNA sequencing gel.
RPA.
For the RNase protection assay (RPA), the Ambion RPA II kit was used according to the instructions of the manufacturer. The cRNA probe was transcribed in vitro with the Riboprobe System kit (Promega). In order to create the template DNA, pROB was cut with Bsu36I and PvuII and the 596-bp fragment was purified. In vitro transcription from this fragment using T3 RNA polymerase (Promega) yielded, as expected, 394-base radiolabeled cRNA probe.
Na+.induction of nhaA transcription and nhaA′-′lacZ expression.
An overnight culture was grown in LBK-BTP broth (pH 7.5) for 16 to 20 h. For stationary-phase induction, the overnight culture was divided in two and incubation was continued for 2 h in the presence (induced) or absence (control) of 300 mM NaCl. For exponential-phase induction the overnight culture was diluted 1:300 and grown up to an optical density at 300 nm of 0.1 to 0.2. The culture was divided in two, and incubation was continued for 2 h in the presence (induced) or absence (control) of 300 mM NaCl. These conditions were found to cause maximal induction of nhaA transcription in the wild-type strain (10). At the end of each experiment total RNA was isolated. For nhaA′-′lacZ expression the procedure was done as above but the cells, containing the chromosomal nhaA′-′lacZ or transformed with various plasmids harboring nhaA′-′lacZ, were induced with 100 mM NaCl for 1 h. These conditions were found to cause maximal induction in the chromosomal fusion strain (RK33Z, NhaA− phenotype). β-Galactosidase activity was determined as previously described (16).
In vitro transcription.
For the protein-DNA binding reaction, 100 ng of supercoiled pGM42 plasmid and the indicated proteins were mixed in 50 ml of reaction buffer (40 mM Tris [pH 7.9], 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 5% [vol/vol] glycerol, 0.1% [vol/vol] Nonidet P-40) and incubated for 20 min at room temperature. Then, 1 U of E. coli ς70-RNA polymerase holoenzyme (Boehringer) was added and the reaction mixture was incubated for 10 min at 37°C. For the transcription reaction, 3.5 μl of a mixture of the four nucleoside triphosphates (2.5 mM each) and 40 U of RNase inhibitor (RNasin; Promega) were added and the reaction mixture was incubated for 30 min at 37°C. The reaction was terminated by incubation with 5 U of RQ1 DNase (Promega) for 10 min, the mixture was extracted twice with phenol-chloroform-isoamylalcohol (25:24:1) and once with chloroform and ethanol precipitated, and the precipitate was resuspended in 15 μl of double-distilled water (DDW). Purified H-NS was kindly provided by C. F. Higgins (Oxford, United Kingdom), and His-tagged NhaR was purified as previously described (6).
Nucleotide sequence accession numbers.
The nhaA and nhaR sequence data have been submitted to the GenBank database under accession numbers J03879 and L24072, respectively.
RESULTS
Analysis of nhaA transcripts by primer extension.
Primer extension was used to analyze transcription from each promoter (P1 or P2) of nhaA. The reaction was conducted with total RNA isolated from wild-type (TA15) cells grown exponentially for 2 h at pH 7.5 in the absence (Fig. 1A, lane 1) or presence (Fig. 1A, lane 2) of 300 mM NaCl. For a negative control both a ΔnhaA strain (RK20, Fig. 1A, lane 4) and a ΔnhaR strain (OR100, Fig. 1A, lane 5) were used, and for a positive control a strain overexpressing nhaA (W3313–2S [29], Fig. 1A, lane 3) was used. In samples derived from all reaction mixtures expressing nhaA, two major bands representing Na+-induced transcripts were observed (Fig. 1A, lanes 1 to 3 and Fig. 6A, lanes a and b). The most prominent band corresponds to a previously observed transcript (16; Fig. 1C) of a size expected for a start site of P1. A second band, less intense than that of P1 and not observed before, appeared 8 bases below P1. This short band still appeared when another primer was used for primer extension (data not shown).
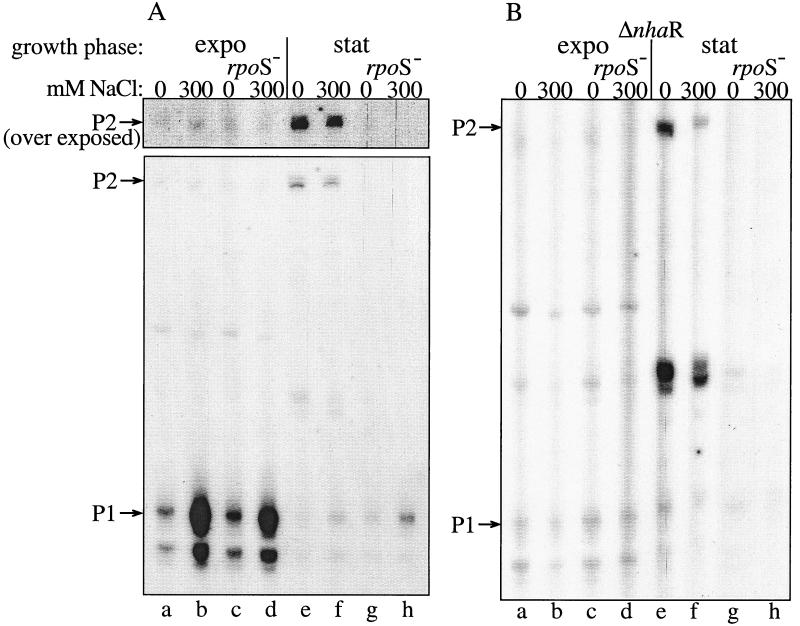
FIG. 6.
P2 is a stationary phase-induced RpoS-dependent promoter of nhaA, as evidenced by primer extension study. Cells were grown and Na+ induced in the exponential phase (expo) or stationary phase (stat) as described in Materials and Methods. RNA was isolated after 2 h of induction. Primer extension assay was conducted as described for Fig. 1. The relevant genotypes and NaCl concentrations are indicated. For the upper portion in panel A the gel was exposed 10-fold longer than the lower panel.
Figure 1A hardly shows a long transcript of a size expected for the start site of P2 (16; Fig. 1C). This transcript could easily be observed either after very long exposure of the primer extension products (Fig. 6A, lanes a and b) or by using a more sensitive method, the RPA (data not shown). In this assay, in which the hybridization probe is synthesized in the presence of radioactively labeled nucleotides, the larger the transcript, the stronger is its radioactivity.
P1 is the Na+-sensitive promoter.
While the activity of P1 was increased at least three- to fivefold after Na+ induction (compare lanes 1 and 2 of Fig. 1A), P2 activity was not increased by the ion (compare lanes a and b of Fig. 6). These results strongly suggest that P1 is the Na+-inducible promoter of nhaA.
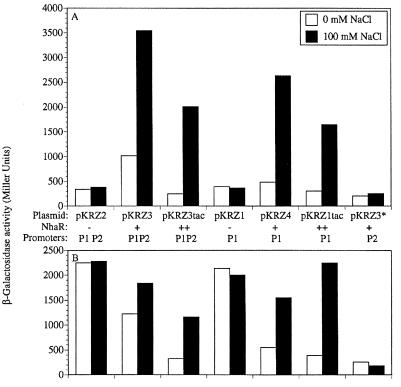
To further study the role of nhaA promoters in the Na+ induction of nhaA, it was important to mutagenize each of the promoters and study the effects of the mutations on Na+-dependent transcription. For this purpose, we have constructed two multicopy isogenic plasmids each carrying both nhaA′-′lacZ and nhaR, either with its native promoter (pKRZ3) or fused to the strong tac promoter (pKRZ3tac). Western blot analysis indicated that NhaR is expressed from pKRZ3 and overexpressed from pKRZ3tac (data not shown). The plasmids were transformed into OR200, a ΔnhaA ΔnhaR double mutant, and the β-galactosidase activity of the exponentially growing cultures was monitored under various growth conditions (Fig. 2A). As shown previously with a single-copy chromosomal nhaR (TA15/pKRZ2 [16]), Na+ did not induce the plasmidic nhaA′-′lacZ when no nhaR was present on the plasmid (Fig. 2A, pKRZ2). In marked contrast, plasmidic fusion in the presence of plasmidic nhaR (multicopy or both multicopy and strongly overexpressed) showed Na+ induction of the fusion (Fig. 2A, pKRZ3 and pKRZ3tac, respectively). The plasmidic induction ratio (the level of β-galactosidase activity observed in the presence of Na+ divided by that obtained in the absence of the ion) of 3 to 10 was very similar to the chromosomal induction ratio (16). These results confirmed the previous suggestion that the lack of Na+ induction of nhaA′-′lacZ on pKRZ2 was due to the very low concentration of NhaR in cells that contain a single chromosomal copy of nhaR (5); the low level of NhaR was readily titrated by binding to the regulatory sequences of nhaA when introduced into the cells on a multicopy plasmid like pKRZ2. Hence the new plasmidic system has become suitable to apply a genetic approach to study Na+-dependent expression of nhaA.
FIG. 2.
Na+ induction of plasmidic nhaA′-′lacZ fusion. (A) ΔnhaA ΔnhaR cells (OR200) transformed with the indicated plasmids were used. All plasmids carried an nhaA′-′lacZ fusion but differ in either the level of the encoded NhaR (−, none; +, moderate expression; ++, strong expression) and/or the nhaA promoters as indicated. Cells were grown exponentially for 1 h in LBK-BTP broth at pH 7.5 in the presence or absence of 100 mM NaCl. Expression was monitored as β-galactosidase activity in Miller units as described previously (16). (B) The plasmids and experimental procedure were as for panel A, but the host was OR200D1, a ΔnhaA ΔnhaR hns strain. The data from one representative experiment of five very similar independent repetitions are shown.
We first used these plasmids to inactivate P2 and obtained pKRZ1, pKRZ4, and pKRZ1tac plasmids, isogenic to pKRZ2, pKRZ3, and pKRZ3tac, respectively, but each containing only P1 of the two nhaA promoters (Fig. 2). Induction obtained with P1 alone was similar to that obtained in the presence of both promoters (Fig. 2A, compare pKRZ4 to pKRZ3 and pKRZ1tac to pKRZ3tac). In addition, this P1-mediated Na+ induction of nhaA was completely abolished in the absence of nhaR (pKRZ1).
A mutation replacing bases T−13, A−12, A−11, and T−8 with C−13, C−12, C−11, and G−8 at the −10 sequence of P1 (Fig. 1C) was then introduced into pKRZ3 to obtain pKRZ3*. Na+ inducibility and most of the β-galactosidase activity were lost by this mutation in P1 (Fig. 2A). Similar results were obtained when the mutation was introduced into pKRZ2 and pKRZ3tac (data not shown). Taken together these results indicate that in exponentially growing cells, P1 is the only NhaR-dependent and Na+-specific promoter of nhaA and P2 is responsible for a very low constitutive level of nhaA expression (Fig. 2A, pKRZ3*). Indeed, when both P1 and P2 promoters were inactivated, the level of β-galactosidase activity was further decreased (twofold) to the background level (data not shown).
In line with this conclusion, only the P1 activity was increased in W3313–2S, a strain that was shown to overexpress nhaA because of a point mutation in nhaR causing a change in the Na+ affinity of NhaR (5) (Fig. 1A, lane 3).
It is possible that the short transcript accompanying Na+ induction of P1 represents a second Na+-dependent promoter of nhaA. In line with this suggestion, at the −10 and −35 positions of the putative start site of the short transcript, a sequence resembling a promoter can be found (Fig. 1C, −35? and −10?). We therefore introduced a mutation at this −10 sequence of pGM42 or pKRZ3 and obtained plasmid pGM433 or pKRZ33, respectively. The mutation had no effect on the expression of nhaA whether tested by primer extension analysis with RNA isolated from OR200/pGM433 (compare lanes 1 and 2 of Fig. 1B) or by the Na+-induced expression of the reporter gene in OR200/pKRZ33, which showed Na+ induction very similar to that of pKRZ3 (data not shown).
hns derepression is mediated by P1.
We have previously reported that the expression of an nhaA′-′lacZ chromosomal fusion is derepressed in strains bearing hns mutations and that the degree of hns derepression is a function of the level of NhaR (10). NhaR in an hns+ strain is an Na+-dependent positive regulator, but in an hns mutant strain it (in multicopy) acts as a repressor which restores Na+ induction.
Since we now have a series of plasmids carrying nhaA′-′lacZ with different levels of expression of nhaR (pKRZ2, no nhaR; pKRZ3, multicopy nhaR; pKRZ3tac, overexpressed multicopy nhaR), we tested the expression of the plasmidic nhaA′-′lacZ fusion in an hns mutant strain with different amounts of NhaR. Figure 2B shows that, as shown before for the chromosomal fusion (10), the expression of the plasmidic nhaA′-′lacZ fusion is derepressed in an Na+-independent fashion in the hns mutant strain when no NhaR is present in the cells (pKRZ2). While the addition of nhaR to the plasmid (pKRZ3) increases the expression and the Na+ induction in the hns+ strain, it results in a decrease in the overall expression from the hns mutant strain but also restores part of the induction. This effect was more pronounced when nhaR was placed under the strong tac promoter (pKRZ3tac). The repressing capacity of NhaR can be seen, even in the hns+ strain, by manipulating its expression level (Fig. 2A); while increasing nhaR expression in a moderate way increases nhaA expression and Na+ induction (pKRZ3), a strong overexpression of nhaR results in a decrease in the extent of nhaA expression accompanied by a further increase in the induction ratio (pKRZ3tac).
Figure 2B also clearly shows that both the hns derepression (pKRZ1) and the nhaR repression and Na+ induction (pKRZ4 and pKRZ1tac) of nhaA in an hns mutant strain are mediated by P1. When P1 is destroyed (pKRZ3*) there is no effect of H-NS on nhaA expression.
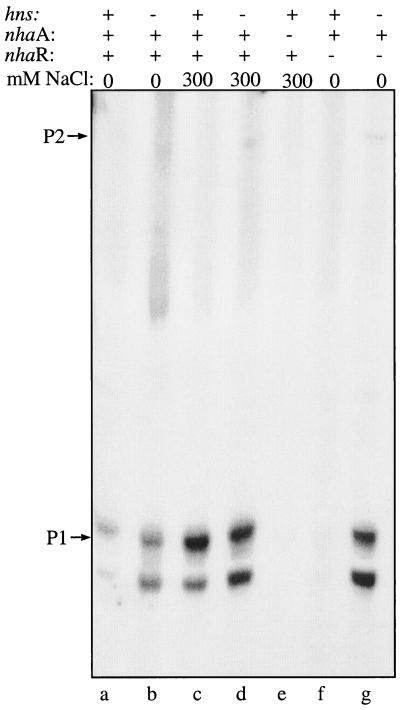
We also used primer extension to confirm that P1 is the promoter involved in hns derepression of nhaA during exponential growth (Fig. 3). Thus, whereas nhaA transcription from P2 remains very low, transcription from P1 is derepressed in an hns mutant strain (compare lanes a and b of Fig. 3). While in hns+ strains P1 transcription is dependent on nhaR (compare lanes a and f of Fig. 3) and Na+ (compare lanes a and c of Fig. 3), in an hns mutant strain it is nhaR independent (Fig. 3, lane g) and Na+ is not required (compare lanes a and b of Fig. 3). The hns derepression is increased in the ΔnhaR background (compare lanes b and g of Fig. 3). Taken together these results confirm that an interplay exists between NhaR and H-NS in the regulation of nhaA expression mediated by the P1 promoter.
FIG. 3.
P1, the Na+-sensitive promoter of nhaA is NhaR dependent and affected by H-NS. Cell growth and primer extension assays were conducted as described in the legend to Fig. 1. The relevant genotype and the NaCl concentrations are indicated.
Analysis of nhaA transcription in vitro
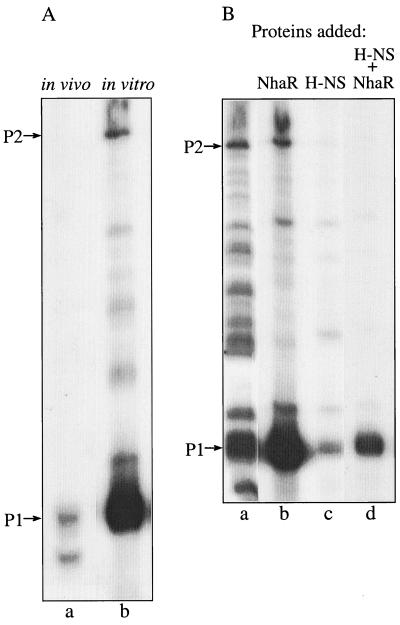
In vitro transcription was conducted with a supercoiled plasmid (pGM42) carrying nhaA and its regulatory sequences and the purified transcripts were analyzed by primer extension (Fig. 4). In a system containing both ς70-RNA polymerase holoenzyme and purified NhaR two prominent transcripts were formed. As in the in vivo primer extension study, a major transcript corresponding to the P1 promoter identified in vivo (Fig. 1A, lanes 1 to 3; Fig. 6A, lanes a and b) was observed (Fig. 4A, compare lanes a and b). However, note that the shorter transcript, which accompanies the P1 transcript in the in vivo primer extension studies (Fig. 1A, lanes 1 to 3; Fig. 6A, lanes a and b), did not form in the in vitro transcription system (Fig. 4A, lane b). The other transcript, which corresponds to P2, although less pronounced, was also identified in the in vitro system. When only RNA polymerase was added without NhaR the two transcripts were still detected but the transcription was much less specific and many additional bands appeared (Fig. 4B, lane a). As indicated above and in reference 10 we have previously shown that in vivo in an H-NS+ background NhaR acts as a positive regulator but in an H-NS− background it acts as a repressor. In the in vitro system only positive regulation by NhaR was detected. Thus, when only H-NS was added in addition to RNA polymerase the overall transcription was reduced (Fig. 4B, lane c). Remarkably, the combination of H-NS, NhaR, and RNA polymerase gave the most specific transcription, with only one major band corresponding to P1 (Fig. 4B, lane d). These results indicate that transcription from P1 occurs via ς70 and is positively regulated by NhaR in a fashion that is modulated by the level of H-NS. Addition of Na+ (up to 100 mM) did not have any consistent effect on the in vitro transcription (data not shown).
FIG. 4.
In vitro transcription of nhaA. The reactions were preformed using pGM42 (0.1 μg) as a template and E. coli ς70-RNA polymerase holoenzyme (1 U) alone or in the presence of either purified NhaR (6), H-NS, or both. Equal aliquots were removed and used as templates for a primer extension assay as for Fig. 1. (A) Comparison between in vivo (wild-type) and in vitro transcription. (B) In vitro transcription in the absence or presence of His-tagged NhaR (400 ng), H-NS (40 ng), or both as indicated.
P2 is induced in stationary phase.
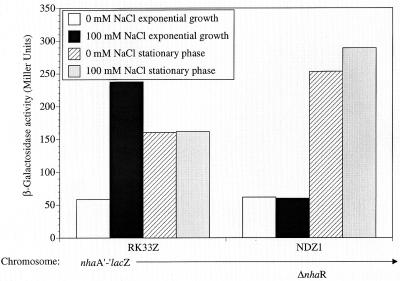
It is well established that while entering stationary phase, bacteria become more resistant to various stress conditions (18) including high levels of Na+ (13). We therefore studied the pattern of nhaA transcription in stationary phase. A comparison of the β-galactosidase activities of the nhaA′-′lacZ chromosomal fusion strain (RK33Z) and its ΔnhaR derivative (NDZ1) during exponential and stationary phases is shown in Fig. 5. As shown before (10), during exponential growth the β-galactosidase activity of RK33Z was low in the absence of added Na+ and was induced by the addition of the ion and no Na+ induction was observed in the ΔnhaR strain NDZ1. In the stationary phase, RK33Z bacteria showed an increase in the basal level (without addition of Na+) relative to the exponentially growing bacteria but this increased activity was not Na+ induced (Fig. 5). Surprisingly, in contrast to the very low basal level of β-galactosidase activity in the ΔnhaR strain observed during exponential growth, a dramatic increase was observed in the stationary phase. The maximal level of activity reached by the ΔnhaR strain in stationary phase was even higher than the maximal Na+-induced level reached by RK33Z in the exponential culture (Fig. 5). These results demonstrate that the pattern of regulation of nhaA expression in stationary phase differs from that in exponential phase; this pattern is dependent neither on Na+ nor on nhaR.
FIG. 5.
Expression of nhaA is dependent on growth phase. The cells, harboring the nhaA′-′lacZ fusion on the chromosome in a wild-type (RK33Z) or ΔnhaR (NDZ1) background, were grown and Na+ induced for 1 h as described in Materials and Methods. The nhaA′-′lacZ expression of the exponential and stationary-phase cultures was monitored as β-galactosidase activity in Miller units as described in the legend to Fig. 2. The data from one representative experiment of five very similar independent repetitions are shown.
The activities of the nhaA promoters in the exponential growth phase were then compared to that in stationary phase, using the primer extension assay. Total RNA was isolated from the wild-type strain (TA15) during the exponential and stationary phases and analyzed by primer extension (Fig. 6A, lanes a, b, e, and f). In both cases, Na+-induced and uninduced cultures were used. Figure 6A (lanes a and b) shows that, as reported above, during the exponential phase, P1 is the main and Na+-responsive nhaA promoter while P2 can hardly be detected. On the other hand, in the stationary phase (Fig. 6A, lanes e and f) transcription from P2 is dramatically increased. Note that also in the stationary phase P2 was not induced by Na+. On the other hand, P1 was still Na+ induced in the stationary phase albeit to a much lower level compared to the exponential-phase induction (Fig. 6, compare lanes e and f to a and b). Hence, in contrast to the exponential phase, after entering the stationary phase, P2 becomes the main nhaA promoter while much less active P1 is still the only Na+-responsive promoter.
P2 stationary-phase induction is dependent on RpoS (ςS).
Since it is now well established that the ςS subunit of RNA polymerase is a global regulator of many stationary-phase-inducible genes in E. coli (13), the possible involvement of ςS in the transcription of nhaA was tested. A TA15 rpoS mutant strain was constructed (Table 1), total RNA was isolated from exponential and stationary phases of Na+-induced and uninduced cultures, and primer extension analysis was conducted (Fig. 6A, lanes c, d, g, and h). rpoS had no effect on Na+-dependent transcription from P1 either in the exponential-phase (Fig. 6A, compare lanes c and d to a and b) or in the stationary-phase rpoS mutant cultures (Fig. 6A, compare lanes e and f to g and h). These results indicate that ςS does not seem to have any significant effect on the P1 promoter. rpoS also had no effect on the very low Na+-independent P2 transcription in the exponential phase (Fig. 6A, compare lanes c and d to a and b). On the other hand, in the stationary culture the strong P2-dependent transcription completely disappeared in the rpoS mutant strain (Fig. 6A, compare lanes e and f to g and h). These results show that ςS is involved in the transcription from the P2 promoter of nhaA only in the stationary phase.
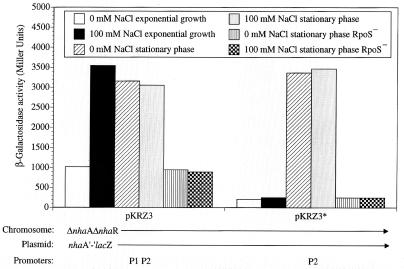
Using the plasmidic system expressing nhaA′-′lacZ from either P1 or P2 or both promoters, the stationary-phase induction of P2 was confirmed and further characterized (Fig. 7). When both promoters were present (pKRZ3) the stationary-phase induction was high, was Na+ independent, and was abolished in the rpoS mutant strain. When P2 was the only promoter (pKRZ3*) the entire stationary-phase induction was observed and this induction completely disappeared in the rpoS mutant strain (Fig. 7, pKRZ3*). When only the P1 promoter was present (pKRZ4) Na+ induction was observed in both growth phases, but the stationary-phase induction was absent (data not shown).
FIG. 7.
P2 is an RpoS-dependent promoter of nhaA, as evidenced by plasmidic nhaA′-′lacZ fusion. Cells were grown to the stationary phase and treated as in Fig. 2. The plasmids, bearing the nhaA′-′lacZ fusion, differ in nhaA promoters as indicated. The host was OR200 (ΔnhaA ΔnhaR) or OR200 rpoS as indicated. Expression was monitored as β-galactosidase activity in Miller units. The data from one representative experiment of five very similar independent repetitions are shown.
NhaR is not involved in ςS-dependent transcription of nhaA.
To study the involvement of NhaR in the stationary-phase transcription of nhaA, total RNA was isolated from exponential and stationary phases of Na+-induced and uninduced cultures of OR100, a ΔnhaR strain, and of the OR100 rpoS strain and primer extension was conducted. Figure 6B shows that while ΔnhaR totally eliminated P1 Na+ induction in both growth phases (compare lanes a, b, e, and f in Fig. 6A to those of Fig. 6B), it did not affect exponential-phase P2 transcription (compare lanes a and b in Fig. 6A to those in Fig. 6B) or the stationary-phase P2 transcription in the absence of Na+ addition (compare lane e in Fig. 5A to lane e Fig. 6B). In the presence of Na+, albeit somewhat reduced, there was still significant stationary-phase induction in the ΔnhaR strain (compare lane f in Fig. 6A to lane f in Fig. 6B). Taken together these results show that, in contrast to Na+ induction of P1, which is mediated by NhaR, stationary-phase induction of P2 is not dependent on NhaR.
Surprisingly, another band of a size intermediate between that of P1 and P2 and not reported before appeared only in the ΔnhaR bacteria grown to the stationary phase (Fig. 6B, lanes e and f). This extra band, reflecting P2 activity, is ςS dependent (compare lanes e and f to g and h in Fig. 6B). This transcript could have been produced by cleavage of the P2 transcript. However, we cannot exclude the possibility that it represents another start site of transcription which appears only in the ΔnhaR strain in stationary phase.
H-NS is not involved in ςS-dependent transcription of nhaA.
Since it was demonstrated that H-NS is an inhibitory component of the ςS stationary-phase induction network and also inhibits the expression of ςS itself (3) and since we now know that ςS also takes part in nhaA regulation, we examined the possible involvement of H-NS in rpoS-dependent stationary-phase induction of nhaA. Transcription from the nhaA promoters in various hns mutant strains (hns [TA15D2], hns rpoS [TA15D2 rpoS], and hns ΔnhaR [OR100D2]) in the presence or absence of Na+ induction and either during the exponential or stationary phase of growth was determined. hns had no effect on the pattern of expression from P2 either in the exponential or stationary phase of growth (data not shown).
The effect of H-NS was also tested by monitoring expression of nhaA′-′lacZ from P2 of plasmid pKRZ3* in either OR200 or OR200D1 (OR200 hns) at various optical density (OD) values during both the logarithmic and stationary phases of growth. Up to an OD of 3.5 there was no significant difference in expression between the two strains (data not shown).
Physiological role of P2.
Stationary-phase, ςS-dependent transcription via P2 revealed in this study raised the question as to the physiological significance of P2. To answer this question, we constructed a plasmid, pGM36*, which carries nhaA mutated in P1 but containing P2. As shown above, the same mutation in plasmid pKRZ3* totally inhibited induction of nhaA′-′lacZ in the logarithmic phase (Fig. 2) but did not affect the stationary-phase induction (Fig. 7). pGM36* was transformed into OR200, a strain devoid of nhaA and nhaR. OR200/pBR322 and OR200/pGM42 (wild-type nhaA) served as negative and positive controls, respectively. These strains were grown overnight to stationary phase in LBK, at which time they attained an OD at 600 nm of 3 to 4 and the medium pH was 8. Then the cells were exposed to various stress conditions with respect to salt and pH and the survival of the cells was determined (Table 2). It is apparent that an alkaline shift to pH 9.5 for 3 h reduced the survival of the cells lacking nhaA (OR200/pBR322) to 0.13%. The pH stress was drastically increased in the presence of increasing Na+ concentration. The survival was 0.014, 5 × 10−5, or 0% when the shift to pH 9.5 was conducted in the presence of 200, 400, or 600 mM NaCl, respectively. To assess whether the effect is specific to Na+, the pH shift was conducted in the presence of 400 mM KCl rather than NaCl. Table 2 shows that the stress effect of 400 mM Na+ was 1,300-fold stronger than that of 400 mM KCl.
TABLE 2.
Survival after alkaline and salt shock at stationary phase
| Row | Plasmida | Survival (%)b
|
||||
|---|---|---|---|---|---|---|
| NaCl (mM)
|
KCl (400 mM) | |||||
| 0 | 200 | 400 | 600 | |||
| 1 | pBR322 | 0.13 | 0.014 | 5 × 10−5 | 0 | 0.065 |
| 2 | pGM42 | 100 | 61 | 44 | 14 | 100 |
| 3 | pGM36* | 100 | 56 | 35 | 8 | 56 |
| 4 | pBR322 | 100 | 43.6 | 29.3 | 11.1 | 100 |
| 5 | pGM42 | 100 | 90 | 80 | 69.5 | 83 |
| 6 | pGM36* | 100 | 100 | 100 | 100 | 100 |
OR200 (ΔnhaA ΔnhaB) bacteria carrying the indicated plasmid were grown overnight (OD600 = 3 to 4) in LBK medium at 37°C and attained a pH value of 8. Rows 1 to 3, the cultures were shifted to pH 9.5 by titration with KOH with or without addition of the indicated salts and incubated for 3 h at 37°C. Rows 4 to 6, the same procedure as for rows 1 to 3 but without the pH shift.
The number of CFU was determined, and survival is expressed as a percentage of that for the control.
Table 2 also shows that for each concentration of NaCl the stress caused without the alkaline pH shift was much less than that of the combination of pH 9.5 with NaCl. Remarkably, plasmidic nhaA with only the P2 promoter (pGM36*) was as good as wild-type plasmidic nhaA (pGM42) in maintaining the survival of OR200 in the presence of high Na+ levels (compare rows 5 and 6 of Table 2) or upon a pH shift to alkaline pH in the absence of Na+ (lane 1 of Table 2). Protection from pH shift in the presence of Na+ was slightly higher when both P1 and P2 were present (compare rows 2 and 3 of Table 2).
DISCUSSION
Using a chromosomal nhaA′-′lacZ fusion, we have previously shown that Na+ induction of nhaA is dependent on NhaR (29). Study of the NhaR-nhaA interaction with purified components identified the long binding site for NhaR on nhaA and showed that Na+ modifies the binding site (6). Hence, most puzzling were the findings that only P1 of the two promoters (P1 and P2) of nhaA (16) maps within the binding site for NhaR on nhaA (6). Therefore, in the present work two complementary approaches were undertaken to establish which is the Na+-dependent promoter of nhaA: (i) a study of the effect of Na+ on in vivo transcription of nhaA both by primer extension and RPA; (ii) mutagenic inactivation of each of the nhaA promoters to identify the role of each promoter in Na+ induction. The results show that in exponentially growing cells P1 is both the dominant promoter and the one which is NhaR dependent and Na+ induced.
The appearance of a third nhaA transcript 8 bp downstream of P1 (Fig. 1A) was a surprise, since for unknown reasons it was not detected before (16). We first considered it to reflect a secondary structure in the P1 transcript causing an interruption of the primer extension reaction. This was based on the observations that similar to the P1 transcript the short transcript is sodium induced (Fig. 1A), NhaR dependent (Fig. 1A), and derepressed in the hns mutant strain (Fig. 3). In addition, when the mutation deleting the −10 box of P1 was introduced, neither transcript could observed (data not shown). However, conducting the primer extension reaction at a high temperature (51°C) that is supposed to melt secondary structures of RNA had no effect on the results (data not shown). Furthermore, 8 bp downstream of the −10 box (TAAAAT) of the P1 promoter exists a DNA sequence which can be a −10 box (TAAAAA) of another promoter (Fig. 1C). These observations suggested that nhaA may have a second Na+-induced promoter. However, mutagenesis of this putative promoter within its putative −10 box eliminated the short transcript but caused only a slight shortening of the P1 transcript, an effect easily ascribed to the location of the mutation relative to P1 (Fig. 1B and C). Accordingly, this mutation only slightly reduced the Na+-induced level of the plasmidic nhaA′-′lacZ (data not shown).
The in vitro transcription study with the ς70-RNA polymerase holoenzyme (Fig. 4) clearly shows that the P1 promoter of nhaA operates with ς70. Most interestingly, the shortest transcript accompanying P1 transcription did not appear in the in vitro assay (Fig. 4A). These results suggest that a factor(s) and/or process occurring in vivo, such as RNA processing, may be responsible for the production of the short transcript. A factor(s) missing in the in vitro assay most probably also accounts for the lack of Na+ inducibility in the in vitro system.
As for many environment-responsive genes (2), nhaA transcription is derepressed in a hns mutant strain (10). In the case of nhaA this derepression is a function of the level of NhaR (10; this study). Most importantly, the present work shows that transcription from P1 but not P2 is affected by H-NS in an NhaR- and Na+-independent fashion (Fig. 3). The interplay between H-NS and NhaR in P1 transcription was further demonstrated in the in vitro transcription conducted with the purified components nhaA ς70-RNA polymerase, H-NS, and NhaR (Fig. 4B). In the assay containing only the RNA polymerase, the major transcript was from P1 but many other transcripts including one corresponding to that of P2 appeared. Addition of NhaR increased transcription from P1 and reduced that from P2 and nonspecific transcription. H-NS further increased the specificity to P1, but the most pronounced specific transcription from P1 was obtained by adding both H-NS and NhaR.
It is now well established that many genes that confer resistance to various stresses are induced at the stationary phase of E. coli (18). This stationary-phase induction involves a complex regulatory network controlled by the product of rpoS, the ςS subunit of RNA polymerase (13, 19). ςS also acts as a global regulator in various stress responses in the exponential phase. This includes the osmotic control of gene expression (13). Na+ stress cannot be separated from osmotic stress, yet our results show that in the exponential phase nhaA does not belong to the ςS osmotically regulated genes. In the exponential phase of growth, P2-mediated transcription was very low and constitutive (Fig. 1A and Fig. 2A, pKRZ3*) but was twofold above the background level observed when both P1 and P2 were inactivated by mutations (data not shown). However, 300 mM Na+ had no effect on the activity of P2. Furthermore, the activity of P2 remains very low in the exponential phase even in an hns mutant background, which is supposed to increase the level of RpoS (13). Therefore, the present work shows that the response of nhaA to Na+ stress in the exponential phase involves only the Na+-specific NhaR-dependent response via P1.
Interestingly, markedly reduced transcription of nhaA from the P1 promoter, which is dependent on NhaR and Na+ but not on rpoS, occurs in the stationary phase (Fig. 6A). The reduction in P1 transcription can be ascribed to a general reduction in transcription and mRNA content characteristic of the stationary phase (21).
We have previously shown that the level of expression of nhaA′-′lacZ is maintained in an Na+-independent fashion in stationary phase (10). However, for unknown reasons we could not show an effect of rpoS on this expression. The present work revealed that the P2 promoter of nhaA is a stationary phase-induced promoter which is activated by ςS and neither NhaR nor Na+ or H-NS is involved in this induction; in the stationary phase, transcription from P2 increased dramatically, and this increase was eliminated in an rpoS mutant strain (Fig. 6A) but not in a ΔnhaR strain (Fig. 6B). Mutation inactivation of P1 had no effect on the stationary-phase induction of the plasmidic nhaA′-′lacZ, and this induction was totally abolished in an rpoS mutant strain (Fig. 7). hns had an insignificant effect on the stationary-phase induction of P2.
Note that the P2 transcript in stationary phase is weaker than that of P1 in the presence of Na+ in the logarithmic phase (Fig. 6A, lanes b, e, and f); however, the β-galactosidase activity originating from P2 is equal to the activity of the P1 promoter when it is fully induced by Na+ (Fig. 7). This suggests that there might be posttranslational control; the two RNAs may differ in their secondary structure, lifetime, or processing mechanism and may have different translation abilities.
It should also be pointed out that in the stationary phase in the absence of NhaR, a new rpoS-dependent transcript (shorter than that observed in the presence of NhaR) appeared (Fig. 6, lanes e and f). A perfect Pribnow box located 46 bp upstream of P1 and within the binding site of NhaR might be responsible for this new start site (Fig. 1C).
Bimodal control mechanisms, similar to the one described above for nhaA, were found in other environment-responsive genes. For example, the proU operon is transcribed from two promoters. As for nhaA, one promoter is stationary phase induced and ςS-dependent while the other is ς70 dependent. In contrast to nhaA, both proU promoters are induced by the operon's specific inducer (osmotic shock) and no specific trans-acting regulator was found (12). Like proU, the osmC gene is transcribed from two promoters (osmCp1 and osmCp2), but only osmCp2 is stationary phase induced and ςS dependent (4). A control mechanism, even more similar to that for nhaA, was described for the ftsQAZ gene cluster (31). In this system, one promoter is controlled by the specific activator SdiA, while the other promoter is stationary phase induced and ςS dependent but not SdiA dependent.
The dps gene, which is transcribed from a single promoter, is controlled by OxyR, which similar to NhaR, is a member of the LysR-OxyR family of positive activators. Interestingly, dps is controlled by OxyR and is ς70 dependent only during the exponential phase, while it becomes ςS dependent and OxyR independent in the stationary phase (1). These findings show that the same promoter can be recognized by more than one sigma factor. Indeed, according to our results the nhaA P2 promoter is transcribed by ς70-RNA polymerase during the exponential phase but it becomes totally dependent on ςS in the stationary phase (Fig. 6A).
Studying the effect of pH and Na+ on logarithmic-phase cells, we have previously shown that in addition to osmotic stress Na+ exerts a specific toxic effect on cells that is markedly increased with pH (24). This stressful combination of Na+ and alkaline pH was also documented with stationary-phase cells (32). We have also shown that the NhaA-Na+/H+ antiporter is essential for the adaptation of logarithmic-phase cells to the specific stress of Na+ and alkaline pH in the presence of Na+ (23). Most importantly, we show here that in the stationary phase nhaA via its P2 promoter becomes rpoS dependent and increases dramatically the survival of stationary cells in the presence of Na+, alkaline pH, and alkaline pH in the presence of Na+ (Table 2). It is suggested that this property of NhaA is of paramount importance for the survival of E. coli in its natural habitats such as the sea or the pylorus, where the combination of alkaline pH and sodium prevails.
ACKNOWLEDGMENTS
This work was supported by grants from the Israel Science Foundation, administered by the Israel Academy of Sciences and Humanities, and the BMBF and BMBF's International Bureau at the DLR (German-Israeli Project Cooperation on Future-Oriented Topics [DIP]). Thanks are also due to the Moshe Shilo Minerva Center for Marine Biogeochemistry and the Massimo and Adelina Della Pergolla Chair in Life Sciences.
REFERENCES
- 1.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of ςS and many ςS-dependent genes in Escherichia coli. J Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier J, Gordia S, Kampmann G, Lange R, Hengge-Aronis R, Gutierrez C. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol Microbiol. 1998;28:971–980. doi: 10.1046/j.1365-2958.1998.00855.x. [DOI] [PubMed] [Google Scholar]
- 5.Carmel O, Dover N, Rahav-Manor O, Dibrov P, Kirsch D, Schuldiner S, Padan E. A single amino acid substitution (G134-Ala) in NhaR1 increases the inducibility by Na+ of the product of nhaA, a Na+/H+ antiporter gene in Escherichia coli. EMBO J. 1994;13:1981–1989. doi: 10.1002/j.1460-2075.1994.tb06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmel O, Rahav-Manor O, Dover N, Shaanan B, Padan E. The Na+ specific interaction between the LysR-type regulator, NhaR and the nhaA gene, encoding the Na+/H+ antiporter of Escherichia coli. EMBO J. 1997;16:5922–5929. doi: 10.1093/emboj/16.19.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christman M, Storz G, Ames B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dersch P, Schmidt K, Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 9.Dorman C J. DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology. 1995;141:1271–1280. doi: 10.1099/13500872-141-6-1271. [DOI] [PubMed] [Google Scholar]
- 10.Dover N, Higgins C F, Carmel O, Rimon A, Pinner E, Padan E. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J Bacteriol. 1996;178:6508–6517. doi: 10.1128/jb.178.22.6508-6517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg B G, Arbel T, Chen J, Karpel R, Mackie G A, Schuldiner S, Padan E. Characterization of Na+/H+ antiporter gene of E. coli. Proc Natl Acad Sci USA. 1987;84:2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S, Haughn G, Calvo J, Wallace J. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E A. Physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 16.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 17.Karpel R, Olami Y, Taglicht D, Schuldiner S, Padan E. Sequencing of the gene ant which affects the Na+/H+ antiporter activity of Escherichia coli. J Biol Chem. 1988;263:10408–10414. [PubMed] [Google Scholar]
- 18.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 19.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 20.Murata T, Yamato I, Igarashi K, Kakinuma Y. Intracellular Na+ regulates transcription of the ntp operon encoding a vacuolar-type Na+-translocating ATPase in Enterococcus hirae. J Biol Chem. 1996;27:23661–23666. doi: 10.1074/jbc.271.39.23661. [DOI] [PubMed] [Google Scholar]
- 21.Neidhardt F C, Ingraham J L, Schaechter M. Growth rate and variable. In: Neidhardt F C, editor. Physiology of the bacterial cell, a molecular approach. Sunderland, Mass: Sinauer Associates, Inc.; 1990. pp. 418–441. [Google Scholar]
- 22.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S, Hinton J C, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 23.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. Deletion of ant in E. coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s) J Biol Chem. 1989;264:20297–20302. [PubMed] [Google Scholar]
- 24.Padan E, Krulwich T A. Sodium stress. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 117–130. [Google Scholar]
- 25.Padan E, Schuldiner S. Na+/H+ antiporters, molecular devices that couple the Na+ and H+ circulation in cells. J Bioenerg Biomembr. 1993;25:647–669. doi: 10.1007/BF00770252. [DOI] [PubMed] [Google Scholar]
- 26.Padan E, Schuldiner S. Bacterial Na+/H+ antiporters—molecular biology, biochemistry, and physiology. In: Konings W N, Kaback H R, Lolkema J, editors. The handbook of biological physics. II. Transport processes in membranes. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 501–531. [Google Scholar]
- 27.Pinner E, Padan E, Schuldiner S. Cloning, sequencing and expression of the nhaB gene, encoding a Na+/H+ antiport in E. coli. J Biol Chem. 1992;267:11064–11068. [PubMed] [Google Scholar]
- 28.Pinner E, Padan E, Schuldiner S. Kinetic properties of NhaB, a Na+/H+ antiporter from E. coli. J Biol Chem. 1994;269:26274–26279. [PubMed] [Google Scholar]
- 29.Rahav-Manor O, Carmel O, Karpel R, Taglicht D, Glaser G, Schuldiner S, Padan E. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in Escherichia coli. J Biol Chem. 1992;267:10433–10438. [PubMed] [Google Scholar]
- 30.Schell M. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 31.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storz G, Altuvia S. OxyR regulon. Methods Enzymol. 1994;234:217–223. doi: 10.1016/0076-6879(94)34088-9. [DOI] [PubMed] [Google Scholar]
- 34.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 35.Ussery D W, Hinton J C D, Jordi B J A M, Granum P E, Scirafi A, Stephen R J, Tupper A E, Berridge G, Sidebotham J M, Higgins C F. The chromatin-associated protein H-NS. Biochimie. 1994;76:968–980. doi: 10.1016/0300-9084(94)90022-1. [DOI] [PubMed] [Google Scholar]