Abstract
HtrA-type serine proteases participate in folding and degradation of aberrant proteins and in processing and maturation of native proteins. Mutation of the corresponding genes often confers a pleiotropic phenotype that can include temperature sensitivity, sensitivity to osmotic and oxidative stress, and attenuated virulence. There are three HtrA-type serine proteases, YkdA, YvtA, and YycK, encoded in the Bacillus subtilis genome. In this report we show that YkdA and YvtA display many similarities: their expression profiles during the growth cycle in wild-type and mutant backgrounds are very alike, with expression being directed by very similar promoters. Both are induced by temperature upshift and by heterologous amylases at the transition phase of the growth cycle. These characteristics are quite different for YycK, suggesting that it has a cellular function distinct from that of the other two proteases or that it performs the same function but under different conditions. We also show that inactivation of either ykdA or yvtA results in compensating overexpression of the other gene, especially during stress conditions, with a concomitant increase in resistance to heat and hydrogen peroxide stresses. Mutation of both ykdA and yvtA leads to growth defects and to thermosensitivity. The fact that their expression increases dramatically at the transition phase of the growth cycle under certain conditions suggests that the YkdA and YvtA proteases may function in the processing, maturation, or secretion of extracellular enzymes in B. subtilis.
Members of the HtrA family of serine proteases are widely distributed in nature, from bacteria to humans (10). The proteins are characterized by an amino-terminal domain that participates in protein localization, a catalytic domain containing an active serine residue, and a PDZ domain that functions in multimerization of the protein into the active dodecamer structure and perhaps also in identification of target proteins. Information derived from completely sequenced genomes shows that most eubacteria have a single HtrA-like serine protease. A recognizable member of the HtrA-protease family has been identified only in some archaebacteria, while the very small genomes of Mycoplasma pneumoniae and Mycoplasma genitalium do not appear to encode such a protease. However, a significant number of bacterial genomes encode more than one HtrA-like serine protease. Mycobacterium tuberculosis has four such genes; Escherichia coli, Bacillus subtilis, Treponema pallidum, Deinococcus radiodurans, and Synechocystis each have three copies, while Haemophilus influenzae and Pseudomonas aeruginosa each have two copies. These observations prompt questions about the physiological roles of each paralogue and of the extent to which their functions overlap.
A comparison of the amino-terminal and PDZ domain regions of paralogues from each bacterium suggests a significant functional divergence. E. coli and T. pallidum each have two HtrA-like proteases with two PDZ domains and one HtrA-like protease with a single PDZ domain. Similarly, H. influenzae and P. aeruginosa each have one HtrA-like protease with two PDZ domains and one HtrA-like protease with a single PDZ domain. Divergence within the amino-terminal regions suggests that paralogues may be located within different cellular compartments. Analysis of this protein region for the three paralogues in T. pallidum and D. radiodurans suggests that one protease is extracellular, having a cleavable signal sequence, a second has a transmembrane domain, suggesting that it is anchored within the cytoplasmic membrane, and the third has neither a signal sequence nor a transmembrane domain, and its location is predicted to be cytoplasmic. A clear functional distinction has been experimentally established for E. coli between DegS and the other two HtrA-like proteases, DegP and DegQ. Sigma factor SigE directs expression of an extracytoplasmic stress regulon that includes DegP. It has been shown that mutation of degS (encoding the HtrA-like protease with only one PDZ domain) stabilizes the anti-sigma factor RseA, thereby reducing the basal and induced levels of SigE activity, whereas mutation of degP and degQ has no effect on the levels of SigE activity (1, 8). Therefore, DegS has a distinct regulatory role in controlling induction of the SigE regulon that its paralogues DegP and DegQ do not. For P. aeruginosa, Boucher et al. have shown that mutation of mucD (encoding an HtrA-like protease) results in conversion to mucoidy with a concomitant increase in sensitivity to heat and hydrogen peroxide (3). Mutation of algW, encoding the second P. aeruginosa HtrA-like protease, also leads to increased sensitivity to heat and hydrogen peroxide, but with the additional phenotypes of paraquat sensitivity and increased alginate production in the presence of sublethal paraquat levels. These analyses show that individual HtrA-like proteases can have both distinct and overlapping regulatory and housekeeping functions in the cell.
There are three members of the HtrA-like serine protease family encoded in the B. subtilis genome, YkdA, YvtA, and YycK (7). Each of the three proteins has a single transmembrane domain positioned in the amino terminus and a single PDZ domain in the carboxy terminus. All three proteins are predicted to span the membrane, with the amino terminus located within the cytoplasm and the carboxy-terminal catalytic and PDZ domains located extracytoplasmically (16). The three proteases and their genes also differ in important respects. YkdA alone has a polyserine tract, which is characteristic of some cell wall-associated proteins, and each protein has an amino-terminal peptide of different length (for YkdA, 50 amino acids [aa]; for YvtA, 75 aa; for YycK, 24 aa) protruding into the cytoplasm. Promoter analysis suggests a partially overlapping expression profile for ykdA and yvtA, distinctive from that observed for yycK. The yycK encoding operon is expressed during exponential growth from the main operon promoter and is also expressed from a SigG-type promoter positioned immediately upstream of yycK that is active during sporulation (4). These analyses suggest that the three HtrA-like proteases encoded in B. subtilis may have distinctive but partially overlapping expression profiles and functions within the cell.
In this paper we report on the induction stimuli and expression patterns of ykdA, yvtA, and yycK, the genes encoding the three HtrA-like serine proteases in B. subtilis. Expression of ykdA and yvtA is induced both by heat shock and by secretion stress using a common mechanism, whereas yycK is neither heat shock nor secretion stress inducible. We also show that inactivation of either ykdA or yvtA results in compensating overexpression of the other gene, with a concomitant increase in resistance to heat and hydrogen peroxide stresses, whereas inactivation of both genes leads to thermosensitivity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. E. coli and B. subtilis were maintained and propagated as previously described (9). Transformations of E. coli and B. subtilis were performed as previously described (9). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was included in solid media at a concentration of 40 μg/ml, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM. Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 3 μg/ml; erythromycin, 1 μg/ml; and kanamycin, 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source, reference, or derivationa |
|---|---|---|
| E. coli strain | ||
| TG-1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 12 |
| B. subtilis strains | ||
| 168 | trpC2 | Laboratory stock |
| DN2 | trpC2 amyE::PykdA-bgaB Cmr | 9 |
| DN3 | trpC2 Pspac-ykdA PykdA-lacZ Eryr | 9 |
| DN26 | trpC2 ykdAΔ439 | 9 |
| DN27 | trpC2 ykdAΔ439 amyE::PykdA-bgaB Cmr | 9 |
| DN40 | trpC2 yvtA::pDN40 (yvtA′-bgaB) Eryr | pDN40→168 |
| DN41 | trpC2 yycK::pDN41 (yycK′-bgaB) Eryr | pDN41→168 |
| DN110 | trpC 2 amyE::PyvtA-bgaB Cmr | pDN110→168 |
| DN111 | trpC2 yvtA::kan Kmr | ′yvtA′::kan→168 |
| DN112 | trpC2 yvtA::kan amyE::PyvtA-bgaB Kmr Cmr | pDN110→DN111 |
| DN113 | trpC2 ykdAΔ439 amyE::PyvtA-bgaB Kmr | pDN110→DN26 |
| DN114 | trpC2 yvtA::kan amyE::PykdA-bgaB Kmr Cmr | pDN2→DN111 |
| DN115supb | trpC2 ykdAΔ439 yvtA::kan sup Kmr | DN111→DN26 |
| DN116b | trpC2 ykdAΔ439 yvtA::kan amyE::PykdA-bgaB sup Kmr Cmr | pDN2→N115 sup |
| DN200 | trpC2 ykdA1[N289H S290M] yvtA::kan amyE::PykdA-bgaB Kmr Cmr Eryr | DN111→AH24 |
| DN201 | trpC2 amyE xylR-amyL yvtA::pDN40(yvtA′-bgaB) Cmr Eryr | DN40→KS408 |
| DN203 | trpC2 amyE xylR-amyL yycK::pDN41(yycK′-bgaB) Cmr Eryr | DN41→KS408 |
| AH22 | trpC2 ykdAΔ570 | pAH22→168 |
| AH23 | trpC2 ykdAΔ570 amyE::(PykdA-bgaB) Cmr | pDN2→AH22 |
| AH24 | trpC2 ykdA::pAH24(ykdA1 [N289H S290M]) amyE::PykdA-bgaB Eryr Cmr | pAH24→AH23 |
| KS408 | trpC2 amyE xylR-amyL Cmr | 14 |
| KS405b | trpC2 amyE xylR-amyLQS Cmr | 15 |
| RC010 | trpC2 amyE xylR-amyL Pspac-ykdA PykdA-lacZ Cmr Eryr | DN3→KS408 |
| RC011 | trpC2 amyE xylR-amyLQS Pspac-ykdA PykdA-lacZ Cmr Eryr | DN3→KS405b |
| Plasmids | ||
| pDL | Integration vector for the introduction of single-copy transcriptional fusions to bgaB by double crossover at the amyE locus (Apr Cmr) | 18 |
| pMOR60 | pBluescript SK with the erythromycin cassette from pE194 inserted into SacI/SmaI, allowing Campbell-type integration into the B. subtilis chromosome (Apr Eryr) | M. O'Reilly, unpublished data |
| pGhost4+ | Vector containing Ts replicon for conditional integration and excision, allowing construction of markerless deletions (Apr Eryr) | 2 |
| pUC19 | Cloning vector (Apr) | 17 |
| pDE | pMUTin-based integrating plasmid with the lacZ gene replaced with bgaB (Apr Eryr | E. Deuerling, unpublished data |
| pDN2 | pDL containing the full ykdA control region on a PCR-amplified fragment (Apr Cmr) | 9 |
| pDN110 | pDL containing the yvtA control region on a PCR-amplified fragment (Apr Cmr) | This work |
| pAH22 | pGhost4+ containing PCR-amplified juxtaposed fragments spanning the 3′ end of ykdA, inserted into EcoRI/HindIII sites (Apr Eryr) | This work |
| pAH24 | PCR-amplified fragment of the ykdA gene incorporating the site-directed mutations (N289H S290M) cloned into EcoRI/HindIII sites of pMOR60 (Apr Eryr) | This work |
| pDN40 | pDE containing a 326-bp PCR-amplified internal fragment of yvtA | This work |
| pDN41 | pDE containing a 191-bp PCR-amplified internal fragment of yycK | This work |
→indicates construction by transformation of the strain on the right of the arrow with the plasmid or chromosomal DNA on the left of the arrow.
The supressor mutation sup in these strains is uncharacterized.
Strain construction.
Transcriptional fusions to the bgaB reporter gene were generated with plasmid pDL (18) or pDE. Plasmid pDN110 was constructed by cloning a 263-bp PCR-generated fragment (synthesized with primers YVTAPF [5′-GGAATTCGGCTCTTCACATCCTTTCAACG-3′] and YVTAPR [5′- CGGGATCCTAACGGTTATTCATTTATCG-3′]) into pDL. Sequences underlined in primers are restriction sites added for cloning purposes, and the sequences on the 5′ end of these sites are sequence clamps to aid in cutting the PCR-generated product. Strain DN110 was generated by transforming strain 168 with plasmid pDN110, selecting for chloramphenicol resistance. To generate strain DN111, a 326-bp fragment of the yvtA gene was amplified by PCR (synthesized with primers YVTAB1 [5′ ACGCGTCGACCATGTCGTTGAAAGGCGCG-3′] and YVTAB2 [5′-ACGCGTCGACCGGATTGATCGCTGCATC-3′]) and gel purified. The purified fragment was digested with HindIII, yielding two fragments that were subsequently ligated to a HindIII fragment from pDG780 that encoded the kanamycin resistance gene (5). An aliquot of the ligation mixture was transformed into strain 168 with selection for kanamycin resistance. That integration had occurred by a double crossover into the yvtA gene was confirmed by PCR. Strains DN112 and DN113 were generated by transforming strains DN111 and DN26, respectively, with plasmid pDN110. Strain DN114 was created by transforming strain DN111 with plasmid pDN2. The double-mutant strain DN115 was generated by transforming strain DN26 with chromosomal DNA of strain DN111 and selecting for kanamycin resistance. Confirmation that strain DN115 contained a ykdA null allele (ykdAΔ439) was confirmed by PCR amplification using primers YKDA6 and YKDADEL2F (9). Strain AH22 has a 570-bp deletion at the ykdA locus, extending from upstream of the active site serine residue to beyond the transcriptional terminator. It was generated by the method of Biswas et al. (2) using plasmid pAH22. Plasmid pAH22 was generated by cloning two fragments into pGhost4+: fragment 1 was synthesized using primers AL3 (5′-CGGGAATTCGAGGTTACTGCAAAGCTG-3′) and AL4 (5′-AAAACTGCAGGCTGCGTCTG TCTGAATG-3′), and fragment 2 was generated using primers AL5 (5′-AAAACTGCAGCTTTCTGACGAGATATCCG-3′) and AL6 (5′-CCCAAGCTTGCGTTATATTGGGGGCG-3′). Transformation of pAH22 into B. subtilis strain 168, followed by excision according to the method of Biswas et al., resulted in the generation of strain AH22 (2). Strain AH23 was constructed by transforming strain AH22 with plasmid pDN2 and selecting for chloramphenicol resistance. Strain AH24 contains a ykdA allele that has the active site serine residue of the protease mutated to a methionine. This was achieved by integration of plasmid pAH24 into the chromosome of AH23. Plasmid pAH24 was constructed as follows: a DNA fragment was amplified using primers AL3 (see above) and AL2 (5′-GGAATTCGCCcaTaTgACCTGGATTAATTGCTG-3′) and was cloned into EcoRI-digested pMOR60 to generate pAH24′. A second 819-bp fragment was synthesized using primers AL6 (see above) and AL1 (5′-GGAATTCGGTcAtAtgGGCGGTCCTTTGTTAA-3′) and was cloned into NdeI-HindIII-digested pAH24′ to generate pAH24 (the lowercase letters within primer sequences are the introduced site-directed mismatches that create an NdeI site and alter the active site). The complete insert in pAH24 was sequenced to verify that the construct contained only the desired site-directed mutations. The plasmid pAH24 was then transformed into strain AH23 by a Campbell-type event to generate strain AH24, thereby reconstituting an intact ykdA gene containing the desired point mutations. The correct integration was confirmed by PCR analysis. Strain DN200 was generated by transforming strain AH24 with chromosomal DNA from strain DN111, selecting for kanamycin resistance. Plasmid pDN40 was created by PCR amplifying of a 326-bp internal fragment of yvtA using primers YVTAB1 and YVTAB2 (see above) and ligating into the SalI site of pDE. Transformation of wild-type strain 168 with plasmid pDN40 with selection for erythromycin resistance generated strain DN40. Plasmid pDN41 was constructed by cloning a 191-bp PCR-amplified internal fragment of yycK (using primers YYCK1 [5′-AAGGGTCGACTGTTTGCAGGACTTCAGCG] and YYCK2 [5′-TGGGTCGACAAGCAGAGTCGCTGATTTCGG]) into the SalI site of pDE. Transformation of strain 168 with plasmid pDN41 with selection for erythromycin created strain DN41. Strains RC010 and RC011 were generated by transforming strains KS408 and KS405b with chromosomal DNA of strain DN3 with selection for erythromycin resistance. Strains DN201 and DN203 were generated by transformation of strain KS408 with chromosomal DNA of strains DN40 and DN41, respectively, and selecting for erythromycin resistance.
Stress induction and phenotypic analysis.
Heat stress induction of B. subtilis strains was carried out as described previously (9), with half the culture being transferred from 37 to 48 or 50°C. Induction by overexpression of amylases was undertaken by growth of appropriate strains in Luria-Bertani (LB) broth containing xylose at a final concentration of 1% (wt/vol). Amylase production and secretion into the medium was monitored by adding aliquots of culture supernatant (25 μl) to sterile discs, which were then placed on LB agar plates containing 0.2% (wt/vol) starch. Amylase activity was detected after overnight incubation at 37°C by staining plates with iodine solution (6).
Temperature-sensitive growth was measured by shifting exponentially growing cultures from 37 to 52°C and monitoring optical density at 550 nm (OD550). Quantification of survival after severe heat shock (54°C) or exposure to lethal concentrations (10 mM) of hydrogen peroxide was performed as previously described (9).
DNA manipulations.
Molecular biological procedures were performed according to the protocols described previously (12) except where stated. Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.), and T4 DNA ligase was purchased from Boehringer (Mannheim, Germany). The sequences of promoter fragments amplified by the PCR were verified by sequencing as described previously (9).
Transcriptional analysis.
Total RNA was prepared from B. subtilis cells as previously described (9) except that the cells were broken using a FastPrep shaker (Bio101). Primer extension analysis was performed using 25 μg of total RNA isolated from cells harvested at appropriate times. The RNA was annealed to radioactively end-labeled primer YVTART2 (5′-CGCATGTACAAGTTCAATTGTCC-3′) or primer YVTART1 (5′-GAATGTCCAATCAGCTTCTG G-3′).
Measurement of β-galactosidase activity.
β-galatosidase activity utilizing the LacZ or BgaB reporter enzymes was measured as previously described (9). The protein concentration was determined using the Bio-Rad microassay (Bio-Rad, Hercules, Calif.) according to the instructions of the manufacturer. One activity unit is defined as 1 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per μg of protein.
RESULTS
Temporal expression of the genes encoding the HtrA-like serine proteases.
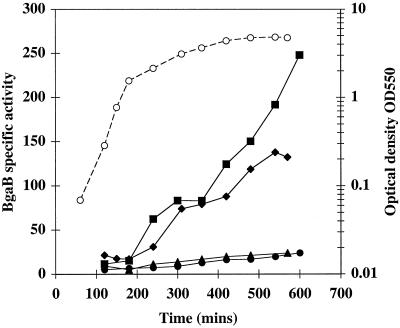
The expression profiles of ykdA, yvtA, and yycK were examined throughout the growth cycle in LB. In a wild-type background, both ykdA and yvtA were expressed at a low level during exponential growth, and their synthesis continued during stationary phase, with β-galactosidase accumulating to between 20 and 30 U at 8 h after the transition phase (Fig. 1). The temporal pattern of ykdA and yvtA expression was also established in their respective null mutant backgrounds. During exponential growth, expression of both genes was slightly increased over wild-type levels. However, the level of expression increased significantly when the culture entered the transition phase (at approximately 200 min) of the growth cycle and continued to increase for up to 6 h, the duration of the experiment (Fig. 1). This profile of expression was observed both for ykdA and yvtA, with the expression levels being slightly higher for ykdA than for yvtA. We also examined expression of yvtA in a ykdA null background during the growth cycle and found that the expression profile was similar to that observed in the yvtA null background (data not shown). In contrast, and in agreement with the results of Fabret and Hoch (4), yycK was expressed during exponential growth, and activity decreased after the transition stage of the growth cycle in LB (data not shown). These data show that the yycK gene can be distinguished from ykdA and yvtA on the basis of temporal expression and that inactivation of either ykdA or yvtA leads to a dramatic increase in their respective expression levels at the transition phase of the growth cycle.
FIG. 1.
Profiles of growth and β-galactosidase (BgaB) accumulation throughout the growth cycle in LB. Growth was monitored by measuring the OD550 and is indicated by open circles. The single growth curve shown is representative for all four strains. β-galactosidase accumulation is indicated by closed symbols for the following strains: circles, strain DN2 (amyE::PykdA-bgaB); triangles, strain DN110 (amyE::Pyvta-bgaB); squares, strain DN27 (ykdAΔ439 amyE::PykdA-bgaB); diamonds, strain DN112 (yvtA::kan amyE::Pyvta-bgaB).
Heat shock induction of the genes encoding the HtrA-like serine proteases.
The ykdA gene is induced by heat shock at 48°C, with the level of induction being greatly increased in a ykdA mutant background (9). To determine if yvtA and yycK were heat shock inducible, transcriptional fusions were generated using the bgaB reporter gene. For strain DN110, which has a yvtA-bgaB transcriptional fusion positioned at the amyE locus, the level of β-galactosidase accumulation was very low (1 U) in cells grown at 37°C. There was a significant increase in β-galactosidase activity accumulation (11 U) after growth at 48°C for 1 h (Table 2). These activity levels were approximately one-third those observed with the ykdA-bgaB fusion in the wild-type background at each temperature (Table 2). To investigate whether the YvtA protease affected expression of the yvtA gene, as was observed for the YkdA protease, expression of a yvtA-bgaB transcriptional fusion was examined in a yvtA mutant background (strain DN112). The results for DN112 (Table 2) show that there was a 12-fold increase in accumulated β-galactosidase activity when cells were grown at 37°C and a 16-fold increase in activity levels during growth at 48°C. When strain DN41, containing a yycK-bgaB fusion generated by a Campbell-type integration, was grown at 37 and 48°C, expression of the fusion was low, and no induction was observed for up to 1 h after temperature upshift (data not shown). Therefore, expression of yvtA is both heat shock inducible and negatively autoregulated, similar to that observed for its paralogue ykdA. In contrast, expression of yycK is not thermoinducible under these conditions.
TABLE 2.
Expression of transcriptional fusions between the ykdA and yvtA promoters and the bgaB reporter gene in various genetic backgrounds at 37 and 48°C
| Construct expressed | Background (strain) | BgaB sp act at a:
|
|
|---|---|---|---|
| 37°C | 48°C | ||
| PykdA-bgaB | 168 WTb (DN2) | 4 | 34 |
| ykdΔ439 (DN27) | 12 | 723 | |
| yvtA::kan (DN114) | 9 | 127 | |
| ykdAΔ439 yvtA::kan sup (DN116) | 80 | 1,200 | |
| ykdA(S290M) yvtA::kan (DN200) | 215 | 880 | |
| PyvTA-bgaB | 168 WT (DN110) | 1 | 11 |
| ykdAΔ439 (DN113) | 17 | 268 | |
| yvtA::kan (DN112) | 12 | 184 | |
All values presented are those found 60 min after the culture was split. One activity unit is defined as 1 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per μg of protein.
WT, wild type.
Since mutation of ykdA and yvtA leads to increased expression of the ykdA-bgaB and yvtA-bgaB transcriptional fusions, respectively, both at 37 and at 48°C, we examined whether mutation of ykdA would affect expression of the yvtA-bgaB fusion and similarly whether mutation of yvtA would influence expression of the ykdA-bgaB fusion. The results (Table 2) for strain DN114 (yvtA::kan ykdA-bgaB) show that mutation of yvtA leads to a twofold increase in β-galactosidase levels (4 to 9 U) at 37°C and an approximatly fourfold increase in β-galactosidase levels (34 to 127 U) at 48°C. These increased levels of ykdA-bgaB expression in the yvtA null background were lower than those observed in the ykdA null background. The results (Table 2) for strain DN113 (ykdAΔ439 yvtA-bgaB) show that mutation of ykdA leads to an increase in yvtA-bgaB expression: β-galactosidase levels increased 17-fold (from 1 to 17 U) at 37°C and 24-fold (from 11 to 268 U) at 48°C. These levels of yvtA-bgaB expression in a ykdA null background were higher than those observed in a yvtA null background. Therefore, the level of expression of ykdA affects the level of expression both of ykdA and of yvtA at 37 and 48°C. Similarly, the level of expression of yvtA influences the expression of yvtA and ykdA at both temperatures. It is also apparent, at least with regard to thermoinduction, that deletion of ykdA has a greater effect on the expression of both genes at both temperatures than has deletion of yvtA.
To examine expression of ykdA in a double mutant background, strain DN115 was constructed with a deletion in the ykdA gene and a kanamycin resistance cassette inserted into the yvtA gene by a double crossover event. Strain DN115 grew extremely slowly, and colonies were small and round and had a mucoid appearance and consistency. This colony morphology was not stable, and rapidly growing cells with a normal colony appearance segregated from the small mucoid colonies. These cells have presumably accumulated a suppressor(s), compensating to some extent for the growth defect, mucoidy, or protease deficiency of ykdA yvtA double mutants. Further experiments were performed on the fast-growing stable double-mutant strains designated DN115sup. A ykdA-bgaB transcriptional fusion was inserted into the amyE locus of DN115sup to generate strain DN116, and expression levels are shown in Table 2. Expression of ykdA at 37°C rose in this strain, with an accumulated β-galactosidase activity level 20-fold higher than those observed in wild-type cells and six- to sevenfold higher than those seen in a ykdA background at this temperature. This trend was also observed at 48°C, where β-galactosidase activity levels were 40-fold higher than those observed in wild-type cells and almost 2-fold higher than those observed in cells carrying the single ykdA mutant. Expression of ykdA was also examined in strain DN200. In this strain, the yvtA gene has been inactivated as described for strain DN116, and the ykdA gene is intact but has the active site serine codon changed to a methionine codon, rendering the encoded protein protease negative. While colonies of strain DN200 had a mucoid appearance similar to that of DN116, this morphology was stable, and no faster-growing segregants were observed. The level of ykdA-bgaB expression in DN200 at 48°C was similar to that observed for the ykdA single mutant at this temperature. However, the level of ykdA expression at 37°C in strain DN200 was 50-fold higher than the wild-type level, 17-fold higher than the level for the single ykdA mutant, and 2 to 3 times higher than that of the double-mutant strain DN116. These data show that both ykdA and yvtA are heat shock inducible. Reciprocal cross-regulation is also evident in that expression of each gene was negatively regulated both by its own gene product and by the product of the other protease gene. Expression levels were further increased in strains in which both ykdA and yvtA are inactivated.
Induction of ykdA and yvtA expression in response to heterologous amylase production.
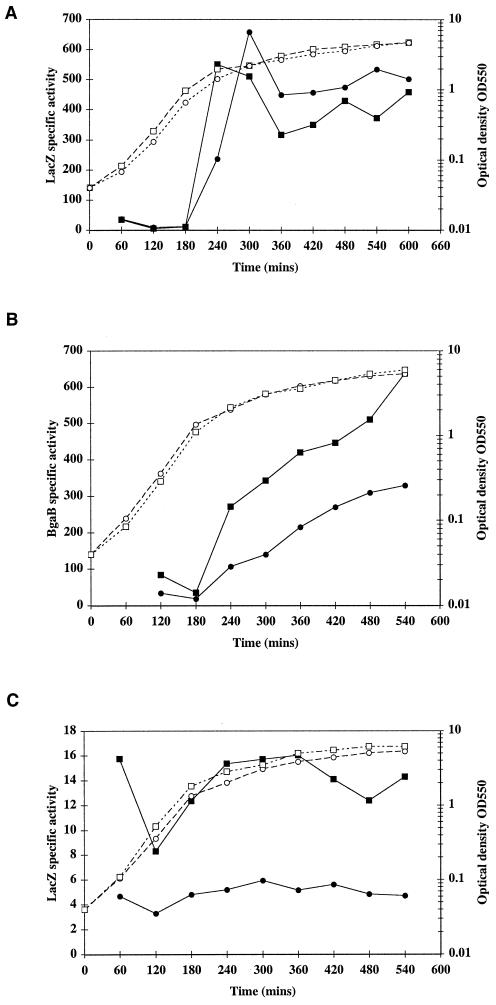
HtrA in E. coli degrades aberrant and misfolded proteins in the periplasm (10). We sought to establish if production of heterologous proteins in B. subtilis could play a role in induction of ykdA and/or yvtA expression. The heterologous proteins chosen were AmyL, the amylase from Bacillus licheniformis, and AmyLQS50.5, a modified amylase that has an increased net positive charge (15). Strains RC010 and RC011 were constructed, in which expression of amyL (RC010) and amyLQS (RC011) was under the control of a xylose-inducible promoter, and ykdA expression was monitored through the ykdA-lacZ transcriptional fusion resident on each chromosome. The results with the two amylases were essentially the same and are shown in Fig. 2A. In the absence of xylose, there was very little ykdA-lacZ expression, with approximately 4 U of β-galactosidase activity present during exponential growth, rising to approximately 10 to 14 U of activity during the stationary phase of the growth cycle (data not shown). In the presence of an inducer, there was still a very low level of expression during the exponential growth phase. However, a dramatic increase in ykdA-lacZ expression occurred at the transition phase of the growth cycle, with β-galactosidase levels rising to 500 to 700 U of activity (Fig. 2A). Activity levels dropped slightly thereafter to between 350 and 500 U for the remainder of the growth cycle. A similar experiment was performed to assess induction of yvtA by AmyL. Strain DN201 was constructed, in which amyL is under the control of a xylose-inducible promoter and yvtA expression is monitored by the resident yvtA-bgaB transcriptional fusion. It is important to note that strain DN201 is a null mutant for yvtA. The expression profile (Fig. 2B) shows that in the absence of the inducer (ie., no AmyL production), there was a low level of yvtA expression during exponential growth, but that expression rose sharply at the transition phase of the growth cycle, accumulating to a maximum of approximately 300 U over 6 h. In the presence of the inducer, expression of yvtA-lacZ increased even more sharply at the transition phase of the growth cycle, rising to 350 U within 2 h and continuing to increase to more than 600 U during the remainder of the experiment (Fig. 2B). We also assayed for the presence of amylase activity in the culture supernatant and showed that it is present extracellularly during both the exponential growth and stationary phases of the growth cycle (data not shown), confirming previously published results (15). To confirm that the observed induction of ykdA and yvtA is caused by amylase production and is not simply an effect of adding the xylose inducer, expression of a ykdA-lacZ fusion was examined throughout the growth cycle in strain DN3 in the presence and absence of xylose. The results (Fig. 2C) show that there was a low and constant level of β-galactosidase throughout the growth cycle in the absence of xylose, with an increase in activity levels to approximately 15 U at the late exponential and transition phases in the presence of xylose. Therefore, while xylose led to a small increase in ykdA expression, perhaps caused by induction of xylosidase, we conclude that the dramatic increase in both ykdA and yvtA expression at the transition stage of the growth cycle was caused by the presence of the heterologous amylases. Expression of a yycK-bgaB transcriptional fusion was also examined during the growth cycle in the presence and absence of xylose (strain DN203). There was no difference in the expression profiles of yycK in the presence and absence of AmyL, showing that in contrast to that of ykdA and yvtA, yycK expression was not induced by heterologous protein expression at the transition stage of the growth cycle (data not shown).
FIG. 2.
Profiles of growth and β-galactosidase accumulation in the presence and absence of heterologous amylase production during the growth cycle in LB. Growth is represented by open symbols, and β-galactosidase accumulation is represented by closed symbols. (A) Growth and LacZ accumulation. Circles, strain RC010 (xylR-amyL Pspac-ykdA PykdA-lacZ); squares, strain RC011 (xylR-amyLQS Pspac-ykdA PykdA-lacZ). Both strains were grown in the presence of 1% xylose and 1 mM IPTG. (B) Growth and BgaB accumulation for strain DN201 (xylR-amyL yvtA::pDN40 yvtA′-bgaB) in the absence (circles) and presence (squares) of a 1% xylose inducer. (C) Growth and LacZ accumulation for strain DN3 (Pspac-ykdA PykdA-lacZ) in the absence (circles) and presence (squares) of a 1% xylose inducer and 1 mM IPTG.
Transcriptional analysis of yvtA.
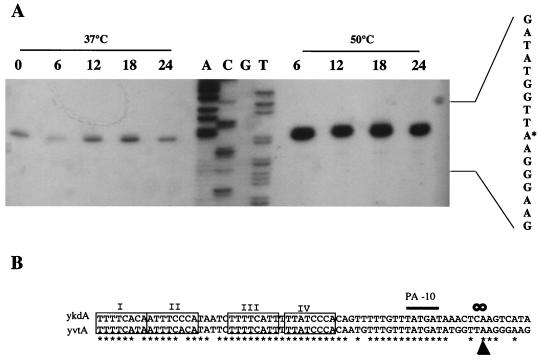
The control region of yvtA contains a sequence showing high homology to the region of the ykdA promoter that directs heat shock induction (9). Therefore, we wished to ascertain if this region also participated in yvtA heat shock induction. Primer extension analysis was performed on RNA samples isolated from cultures of strain DN111 grown at 37 and 50°C. The results show a low and constant level of transcript during the course of the experiment at 37°C (Fig. 3A). The level of transcript was greatly increased at 50°C, evident within 6 min of temperature upshift and remaining at this high level for up to 24 min. The initiation point of transcription is located at the same position relative to the conserved octamer repeats and putative −10 region as is the transcription initiation point of ykdA (Fig. 3B).
FIG. 3.
Primer extension analysis of yvtA. (A) RNA samples were prepared from an exponentially growing culture (OD550 = 0.3), strain DN111, at various times during growth at 37°C and after a temperature upshift to 50°C. Time is indicated in minutes, with 0 representing the sample taken before temperature upshift and 6, 12, 18, and 24 being the time in minutes after upshift. The signal is that obtained from 12.5 μg of total RNA. The sequencing ladder is shown (A C G T), with the sequence of the promoter indicated on the right. The base at which transcription initiation begins is indicated by an asterisk. (B) An alignment of the conserved regions of the ykdA and yvtA promoters. Nucleotide identity is indicated by an asterisk; the conserved octamer repeats (numbered I to IV) are boxed; the SigA-type −10 region is indicated by PA-10, and the initiation points of transcription are indicated by circles for ykdA and a closed triangle for yvtA.
In order to ascertain if heat shock and amylase induction of ykdA and yvtA are mediated by the same promoter, primer extension analysis was performed using ykdA- and yvtA-specific primers. RNA samples were prepared from cultures of strain KS408 growing in the presence and absence of xylose at specific times spanning the transition phase of the growth cycle. Results showed that the amylase induction of both genes occurred at the transcriptional level and that the initiation points of transcription of each gene were the same as those used during heat induction (data not shown). Therefore, heat shock and amylase induction of ykdA and yvtA expression are both mediated by the same promoter.
Phenotypic analysis of ykdA and yvtA mutants.
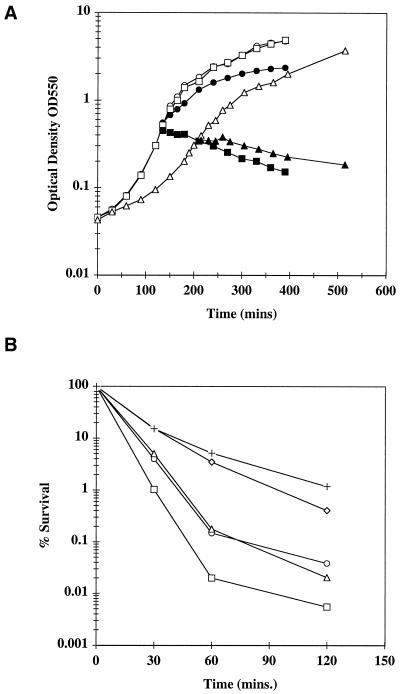
A strain carrying a null mutation in ykdA has a growth profile similar to that of wild-type cells at both 37 and 48°C. However, when these cells are shifted to 54°C (the killing temperature), the ykdA mutant cells show a 10-fold increase in survival levels compared to wild-type cells (9). To further investigate these phenomena, we decided to examine the growth and survival profiles of strains with single and double mutations in ykdA and yvtA after exposure to heat and oxidizing agents. The growth profiles at 37 and 48°C of strains carrying mutations in either ykdA or yvtA are similar to those of wild-type cells grown at these temperatures (data not shown). A comparison of the growth profiles of strains DN115sup and DN200 with that of the wild-type strain grown at 37°C and at the sublethal 52°C are shown (Fig. 4A). At 37°C, the growth profile of strain DN115sup (mutated in both ykdA and yvtA but carrying an unspecified suppressor mutation) was essentially identical to the wild-type profile. However, growth of strain DN200 was significantly slower than that of wild-type cells at this temperature. When exponentially growing cultures of these strains were shifted to 52°C, it was evident that the wild-type strain continued to grow, albeit at a slower rate. However when strains DN115sup and DN200 were shifted to 52°C, cell growth ceased immediately and cells slowly lysed during the remainder of the experiment. These experiments show that having a functional copy of either ykdA or yvtA allows cells to grow with essentially wild-type characteristics at an elevated temperature and that a temperature-sensitive growth phenotype is observed only when both these proteases are mutated.
FIG. 4.
Growth and survival profiles of strains grown at elevated temperatures. (A) Growth profiles of strains grown at 37 (open symbols) and 52°C (closed symbols). Growth was monitored by measuring the OD550. Circles, strain 168 (wild-type); squares, strain DN115sup (ykdAΔ439 yvtA::kan sup); triangles, strain DN200 (ykdA1[N289H S290M] yvtA::kan amyE::PykdA-bgaB). (B) Survival of strains after exposure to heat stress at 54°C for various time periods. Circles, strain 168 (wild-type); diamonds, strain DN26 (ykdAΔ439); crosses, strain DN111 (yvtA::kan); squares, strain DN115sup (ykdAΔ439 yvtA::kan sup); triangles, strain DN200 (ykdA1[N289H S290M] yvtA::kan amyE::PykdA-bgaB). Results shown are the averages of those from three separate experiments.
Quantitation of thermosensitivity was scored by exposing exponentially growing cultures of each strain to 54°C (lethal temperature) for increasing time periods. The results (Fig. 4B) confirm that strains mutated in either ykdA or yvtA are more thermotolerant than are wild-type cells. It is also evident that DN115sup cells are up to 10-fold more sensitive than wild-type cells after exposure to 54°C for more than 60 min. Surprisingly, we found that the thermosensitivity of DN200 cultures was similar to that of the wild type for up to 2 h of exposure at 54°C. Since the ykdA expression level is greatly increased in this background (Table 2), this result suggests that extreme overexpression of an intact YkdA protein devoid of protease activity may confer some thermoprotection on cells of this strain.
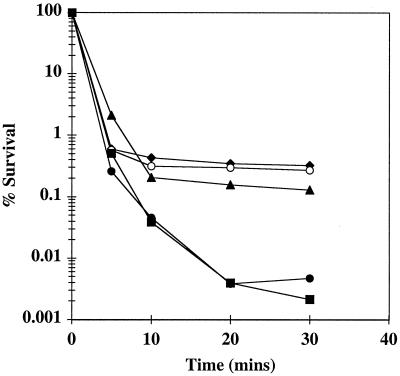
Previous results showed that mutation of ykdA also conferred resistance to lethal levels of hydrogen peroxide (9). To further investigate this phenomenon, we examined the resistance of strains both singly and doubly mutated in ykdA and yvtA to 10 mM hydrogen peroxide for increasing time periods. The results (Fig. 5) show that strains singly mutated in either ykdA or yvtA were up to 100-fold more resistant to 10 mM hydrogen peroxide than was the wild-type strain 168. Interestingly, strain DN115sup showed the same sensitivity as the wild-type strain. Strain DN200, while being slightly more sensitive to hydrogen peroxide than singly mutated strains, was still up to 100-fold more resistant than either the wild type or strain DN115sup. These data suggest that overproduction of either YkdA, YvtA, or a protease-negative form of YkdA confers some protection against lethal hydrogen peroxide levels during exposure periods of more than 10 min.
FIG. 5.
Survival of strains exposed to 10 mM hydrogen peroxide for various time periods. Strains were exposed to 10 mM hydrogen peroxide, and the percent survival for each strain was calculated as indicated in Materials and Methods. Closed circles, strain 168 (wild-type); diamonds, strain DN26 (ykdAΔ439); open circles, strain DN111 (yvtA::kan); squares, strain DN115sup (ykdAΔ439 yvtA::kan sup); triangles, strain DN200 (ykdA1[N289H S290M] yvtA::kan amyE::PykdA-bgaB).
DISCUSSION
In this study, we have investigated the relationship between the three HtrA-like serine proteases, YkdA, YvtA, and YycK, encoded in the B. subtilis genome. Phylogenetic analysis shows that YkdA and YvtA are similar to each other, with YycK being more distantly related (9). The results presented here support this classification. Expression profiles of the ykdA and yvtA genes are very similar. Both are inducible by heat shock during exponential growth and by heterologous amylases at the transition phase of the growth cycle. In cells with either ykdA or yvtA mutated, expression of the intact paralogue increases slightly during exponential growth but rises dramatically at the transition phase and continues to increase throughout an extended stationary phase. The phenotypes observed in strains carrying null mutations in either ykdA or yvtA are also very similar, showing increased tolerance to thermal stress and to lethal levels of hydrogen peroxide. The expression profile of yycK is quite different from that of ykdA or yvtA (4; our results). Expression is low during exponential growth, decreases at the transition phase of the growth cycle, and is not inducible by heat shock. In addition, expression of yycK is directed by a SigG-type promoter positioned immediately upstream of the gene under conditions conducive to sporulation. The negative autoregulation and reciprocal cross-regulation of ykdA and yvtA expression, and the similar phenotypes observed when either gene is mutated, suggest an overlap in the cellular roles of YkdA and YvtA. YycK, in contrast, either has a distinct cellular role or alternatively functions similarly to YkdA and YvtA but under different environmental or developmental conditions.
There is a high level of identity between the promoters utilized for expression of ykdA and of yvtA under the conditions examined in this study. A comparison of these promoters (Fig. 3B) shows that they have a consensus SigA-type −10 region with a series of similarly spaced octamer repeats positioned at the −35 region. This suggests that both genes are under the control of the same regulator and that their expression is responsive to the same stimuli. Therefore, it is likely that under some conditions, YkdA and YvtA are both required to adequately deal with the cellular stress or condition that triggers their induction. The negative autoregulation and reciprocal cross-regulation observed in strains carrying either ykdA or yvtA null mutants is likely to be a manifestation of compensatory overexpression of YkdA and YvtA. Therefore, establishing the conditions under which such overexpression occurs gives an indication of the time and conditions when the inducing signal(s) are most prevalent. Two such conditions have been established in this study: (i) in single-mutant backgrounds, even under normal growth conditions, expression of both genes rises dramatically at the transition phase and continues throughout the stationary phase, and (ii) production of heterologous amylases leads to an additional increase in ykdA and yvtA expression at the transition phase over that observed during normal growth conditions. Interestingly, both events occur at the transition and stationary phases of the growth cycle. In B. subtilis, the transition phase coincides with the production and secretion of many extracellular enzymes. In view of these results and of those reported by Poquet et al. (11) showing that HtrA in L. lactis functions both to degrade abnormal proteins and to process natural propeptides, it is tempting to speculate that YkdA and YvtA may have similar functions in B. subtilis. Perhaps the gene duplication was triggered by the level and/or number of enzymes produced and secreted during stationary phase by this bacterium.
Further insight into the mechanism of ykdA and yvtA induction can be obtained from our expression data in response to heterologous amylase production. We and others (15) have demonstrated that heterologous amylases are produced and secreted during the exponential and stationary phases of the growth cycle in the presence of the xylose inducer. However, induction of ykdA and yvtA by such amylases was triggered only at the transition phase, continuing throughout the stationary phase of the growth cycle. Therefore, despite amylase production and secretion during exponential growth, expression of ykdA and yvtA was not significantly induced. These data suggest that neither the heterologous nature of the amylases nor their secretion per se is sufficient for ykdA and yvtA induction. Perhaps the induction stimulus is multifactorial, comprising (i) the secretion load (ie., the total number of proteins and the amount of each protein being processed and/or secreted), (ii) the level of protein maturation, and (iii) the level of aberrant protein degradation. Further experiments will be required to dissect the nature of the inducing signal.
Redundancy in the functions of YkdA and YvtA is also suggested by analysis of mutant phenotypes. In contrast to other bacteria, in which mutation of htrA leads to thermosensitivity, eg., htrA of E. coli, algW and mucD from P. aeruginosa, and htrA from Yersinia enterocolitica (10), mutation of either ykdA or yvtA results in strains that are more resistant to both heat and hydrogen peroxide. It is necessary to mutate both genes in order to observe thermosensitivity. Strains carrying mutations in both genes are slow growing and form mucoid colonies but segregate fast-growing colonies very rapidly. These cells grow normally at 37°C but still cannot grow at elevated temperatures, showing that the suppressor mutation affects growth but does not compensate for the loss of both YkdA and YvtA. Such suppressor mutations do not accumulate in a strain carrying a null mutation in yvtA and a ykdA allele encoding a protease-negative form of the YkdA protein (strain DN200). This strain has a severe growth defect at lower temperatures and is thermosensitive at elevated temperatures. Spiess et al. have shown that HtrA from E. coli has a chaperone activity that predominates at lower temperatures and a protease activity that predominates at elevated temperatures (13). The difference in the phenotypes of strains DN115 and DN200 may suggest that the protease-negative form of YkdA has such a chaperone activity that is sufficient to promote slow growth but that the protease activity is required for growth at higher temperatures. However, it is evident that the stimulus for ykdA induction is still extremely high in both these backgrounds, with expression levels at 37°C being 20- to 40-fold higher than those observed in the wild-type background.
In conclusion, our results show that two of the HtrA-like serine proteases in B. subtilis, YkdA and YvtA, show significant similarities in terms of expression profiles and of the phenotypes of mutant strains, suggesting that they perform similar functions within the cell.
ACKNOWLEDGMENTS
This work was supported by EU grants BIO4-CT96-0655 and QLG2-CT-1999-01455 (to K.M.D.) and by BioResearch Ireland (to D.N.) through the National Pharmaceutical Biotechnology Centre at Trinity College, Dublin.
We thank Colin Harwood for the gift of strains KS405b and KS408 and Elke Deuerling for plasmid pDE.
REFERENCES
- 1.Ades S E, Connolly L E, Alba B M, Gross C A. The Escherichia coli SigE-dependent extracytoplasmic response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion of mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologues of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerout-Fleury A M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 6.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, England: John Wiley and Sons; 1990. [Google Scholar]
- 7.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 8.Mecsas J, Rouviere P E, Erickson J W, Donohue T J, Gross C A. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 9.Noone D, Howell A, Devine K M. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J Bacteriol. 2000;182:1592–1599. doi: 10.1128/jb.182.6.1592-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 11.Poquet I, Saint V, Seznac E, Simoes N, Bolotin A, Gruss A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol. 2000;35:1042–1051. doi: 10.1046/j.1365-2958.2000.01757.x. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Spiess C, Biel A, Ehrmann M. A temperature dependent switch from chaperone to protease activity in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 14.Stephenson K, Harwood C R. Influence of a cell-wall-associated protease on production of alpha-amylase by Bacillus subtilis. Appl Environ Microbiol. 1998;64:2875–2881. doi: 10.1128/aem.64.8.2875-2881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson K, Carter N M, Harwood C R, Petit-Glatron M F, Chambert R. The influence of protein folding on late stages of the secretion of alpha-amylases from Bacillus subtilis. FEBS Lett. 1998;430:385–389. doi: 10.1016/s0014-5793(98)00698-x. [DOI] [PubMed] [Google Scholar]
- 16.Von Heijne G. Principles of membrane protein assembly and structure. Prog Biophys Mol Biol. 1996;66:113–139. doi: 10.1016/s0079-6107(97)85627-1. [DOI] [PubMed] [Google Scholar]
- 17.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 18.Yuan G, Wong S L. Regulation of groE expression in Bacillus subtilis: the involvement of the sigmaA-like promoter and the roles of the inverted repeat sequence (CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]