Abstract
Potency analysis of mesenchymal stromal cells (MSCs) is required for their use in advanced clinical trials. Assay matrix strategy evaluating more than a single property of MSCs is an emerging strategy in potency analysis. Here we developed an assay matrix approach focusing on the secretory chemokine responses of MSCs using multiplex analytical method. MSCs’ innate fitness in secreting matrix of chemokines is correlated with their metabolic fitness in differential degrees. In addition, innately secreting chemokines are correlated among themselves in a unique pattern. MSC’s matrix chemokine responses to exogenous stimulation of IFNγ and/or TNFα are distinct. However, the combination of IFNγ and TNFα is superior than individual stimulations in eliciting robust and broad matrix chemokine responses of MSCs. Correlation matrix analysis has identified that chemokine responses to IFNγ and/or TNFα display unique correlative secretion patterns. MSC and peripheral blood mononuclear cells coculture analysis has identified the correlation matrix responses of chemokines that predicted immune suppression. In addition, MSC-mediated blocking of T-cell proliferation predominantly correlates with chemokines in an inverse manner. Knockdown of chemokines has demonstrated that MSC-sourced inherent chemokines do not actively play a role in T-cell suppression and thus are the bystander predictors of T-cell suppression. The present analysis of MSC’s matrix chemokine responses can be deployed in the advanced potency analysis of MSCs.
Keywords: mesenchymal stromal/stem cells, assay matrix, potency, interferon γ, tumor necrosis factor α, bone marrow, chemokines
Graphical Abstract
Graphical Abstract.
Significance Statement.
Potency analytical strategies are needed for the release criteria of MSCs in advanced clinical trials. In the present study, we have deployed an assay matrix strategy focusing on chemokine responses of MSCs to define their potency. Matrix chemokine responses and correlation of chemokines among each other predict MSC’s innate fitness, responsiveness to exogenous stimuli, and immunosuppressive potential. This chemokine matrix analytical method devoids universal ruler and thus can be used in the manufacturing of autologous and random donor MSC products.
Introduction
Mesenchymal stromal cells (MSCs) are widely being tested as cellular therapeutics in regenerative medicine due to their immunomodulatory and regenerative potential.1,2 MSCs are approved in Japan for the treatment of steroid-resistant graft versus host disease (GVHD) and in Europe for Crohn’s disease-associated perianal fistula.3,4 While clinical trials have demonstrated that MSC infusion is safe5 achieving consistent efficacy remains a concern in advanced clinical trials.6,7 Although variations in efficacy arise due to variabilities of donor and recipient, disease severity, dosing, and route of MSC delivery, cell manufacturing procedures also play a role.8 Cell manufacturing procedures evaluate sterility, viability, and identity of MSCs to determine release criteria as living cellular pharmaceuticals.4,9 Of these, sterility and viability are assured with standard monitoring and assessment technologies. Minimal criteria to define bone marrow-derived MSC identity includes the following: (1) plastic adherence and expansion, (2) trilineage (adipocyte, osteocyte, and chondrocyte) differentiation potential, (3) combination of positive (CD105, CD73, CD90) and negative (CD45, CD34, CD14, CD11b, CD79a, CD19, HL-DR) cell surface marker expression.10 Additionally, advanced phase clinical trials require potency analysis of MSCs as a component of release criteria beyond sterility, viability, and identity assessments.11,12 However, the development of potency assays has been challenging due to multiple factors including the fact that MSC’s mechanism of action in vivo in patients is yet to be understood.13 Potency analysis as a universal release criterion is complicated since cell manufacturing technologies differ from center to center due to the variations in tissue source, expansion, and handling protocols which alter MSCs’ critical quality attributes.14 International Society for Cell Therapy recommended the development of “in house” potency analytical strategies specific to the cell manufacturing facility which includes well-characterized MSC lots as references that are developed internally or from external sources.15 Although several studies have evaluated MSCs’ characteristics as a surrogate measure of potency that reflects their putative mechanism of action in vivo, further advanced potency testing strategies are required.16-23
Potency assays focusing on single-molecule/pathway of MSCs may not adequately represent their functionality since MSCs possess myriad of regenerative and immunomodulatory properties which may have synergistic and overlapping functions in vivo. This limitation is addressed with “assay matrix” strategy where the collection of MSC’s quality attributes is cumulatively evaluated to define its potency.15 These assay matrix analytical systems can include flow cytometric evaluation of functional molecules, quantification of an array of molecular transcripts relevant to functional proteins, and secretome evaluation of bioactive molecules.15 An example is the evaluation of CXCL5, CXCL8, and VEGF secretion by MSCs in conjunction with an angiogenic assay that defines the matrix potency of MSCs.24 Our own recent work has also demonstrated the potency of MSCs through an assay matrix analysis by evaluating the secretome of MSCs’ interaction with peripheral blood mononuclear cells (PBMCs) in combination with the evaluation of an array of MSCs’ molecular transcript responses to exogenous cues.25 In another assay matrix approach, we have evaluated an array of phosphorylation status of MSCs upon interaction with the secretome of activated PBMCs which defined the potency of MSCs.26 These studies collectively provide insights that assay matrix analysis of MSCs can be deployed to define the potency of MSCs; however, further characterization of MSCs’ attributes using this strategy is needed to inform advanced potency analysis.
MSC-sourced molecules, such as VEGF, IDO, TSG-6, TWIST-1, PD-L1, etc. can serve as the surrogate potency markers. Considering the versatile mechanism of action of infused MSCs in vivo in humans and the developing knowledge of the intricacy of MSC-derived functional molecules in executing cumulative pharmacological effects, it is significant to define additional relevant effector pathways such as chemokines of MSCs. Chemokines are classically defined as chemoattractants of leukocytes to the site of inflammation. However, recent evidences suggest that chemokines secreted from MSCs play a major role in immunomodulation and tissue regeneration.27-29 Preliminary biomarker studies have also demonstrated that GVHD patients responding to MSC therapy exhibits increasing levels of secretory chemokines in the plasma.30 In addition, it is emerging that MSCs’ regulatory functionality is dependent on the individual or combinatorial functions of chemokines.31-35 These indicate that MSC-derived chemokines can serve as the potency markers while the compendium matrix analysis of human MSC-derived chemokines is yet to be defined to accomplish this goal. Here we have rigorously evaluated the chemokinome of human bone marrow MSCs for more than 30 chemokines using multiplexed secretome technology which serve as chemokine assay matrix strategy in defining MSCs’ potency.
Materials and Methods
Human Bone Marrow MSC Isolation and Culture
Human bone marrow aspirates were obtained in accordance with the Institutional Review Board of Memorial Health Medical Center, Savannah and University of Wisconsin-Madison. Bone marrow aspirates were subjected to Ficoll density gradient to enrich mononuclear cells (MNCs). MNCs were plated at a density of 200,000-300,000 cells/cm2 in 1× α-minimum essential medium (MEM) containing 10% human platelet lysate and 100 IU/mL penicillin/streptomycin/amphotericin B. Three days post-culture, non-adherent cells were removed and replenished with fresh medium. Subsequently, cultures were maintained with media change at every 48-72 hours until colonies are observed. Once colonies are established, cells were trypsinized and reseeded at the density of 3000-5000 cells/cm2 in 1× alpha-MEM containing 5% human platelet lysate and 100 IU/mL penicillin/streptomycin/amphotericin B. MSCs were passaged regularly with the maximum confluence of 70%-80% and cells from the passages between 2 and 4 were used for all the assays. MSCs used in this study were culture expanded in human plate lysate until the experiments are performed. In the assays with ± exogenous stimulants, human platelet lysate media was used. However, FCS-containing media was used in the assays of MSCs with and without PBMC cocultures. MSC identity was confirmed based on the marker expression as defined in our earlier studies (CD45−CD105+ CD44+ CD90+ CD73+)36 CD45 FITC (Clone HI30), CD105 APC (Clone 266), CD44 APC (Clone G44-26), CD90 APC (Clone 5E10), CD73 PE (Clone AD2) (BD Biosciences, USA) (Supplementary Fig. S1).
MTT Assay
MSCs were seeded onto 96-well plates at desired densities. In some situations, cells were stimulated with IFNγ and/or TNFα in appropriate concentrations (ng/mL). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays were performed on day 3 or day 4 post-cultures. Supernatants were collected and cells were incubated with thiazolyl blue tetrazolium bromide for 5 hours. After 5 hours, formazan crystals were dissolved in DMSO, and optical densities were measured at the wavelength of 560 nm and subtracted with the values from reference wavelength of 670 nm.
MSC and PBMC Coculture
MSCs were seeded onto 96-well plates with appropriate densities. PBMCs from random donor were added into each well with the final concentration of 0.1 × 106 cells per well. 500 ng/mL staphylococcal enterotoxin B (SEB) (Toxin Technology, Sarasota, FL) was used to stimulate the T cells in the PBMCs. Ki67 proliferation assay was performed to measure T-cell proliferation after 4 days. For Ki67 proliferation assay, intracellular flow cytometry staining was performed with BD Cytofix and Cytoperm procedure with the antibodies CD3APCCy7 (Clone SK7) (Biolegend, USA) and Ki67 PE (Clone B56) (BD Biosciences, USA) according to manufacturer instructions (BD Biosciences, San Jose, CA). Samples were acquired in BD FACS Aria flow cytometer, and the results were analyzed in FlowJo software. For non-contact cocultures, MSCs are cultured on the bottom of the well and SEB-activated PBMCs were cultured on the transwell membrane (0.4 µM). Three days later MSCs were lysed and RNA was isolated. Total RNA is converted into cDNA (Qiagen, USA). Quantitative real-time PCR was performed to identify the expression of chemokines.
siRNA Knockdown on Human Mesenchymal Stromal Cells
MSCs were seeded in 96-well plates at a concentration of 20,000 cells per well on 1 day prior to transfection with non-targeting control siRNA or IDO, CCL7, CXCL16, CCL2, CXCL6, CXCL1, CXCL2 SMART Pool siRNA (Horizon Discovery, CO, USA). On the day of transfection, MSCs were conditioned with serum-free 10 mM HEPES (Corning, NY, USA) containing α-MEM for 30 minutes. Subsequently, siRNA was added to MSCs using the Dharmafect reagent 1 system. Transfected MSCs were then incubated for 5 h and the transfection medium was replaced with regular MSC culture medium. The next day, transfected MSCs were ready for coculture with PBMCs.
Multiplex Analysis of Secretory Chemokines
MSC culture supernatants derived from ± stimulated conditions or cocultured with PBMCs were collected and stored at −80°C. Upon thawing, supernatants were centrifuged at 500g for 5 minutes to remove debris. Subsequently, supernatants were subjected to magnetic bead-based multiplex assay focused on indicated chemokines (R&D Biosystems) according to the manufacturer’s instructions using Luminex xMAP (multi-analyte profiling) technology. Results were plotted as picogram/milliliter (pg/mL).
Statistics
Data were analyzed with the GraphPad Prism 9.0 software. Correlation matrix and simple linear regression were performed with GraphPad Prism software to obtain correlation coefficient (r) and P values. Primary data were transformed to logarithmic values to best fit the regression line with the data points. Statistical significance was confirmed with the P value of <.05.
Results
Human Bone Marrow MSCs Innately Secrete Specific Set of Chemokines at Distinct Ranks
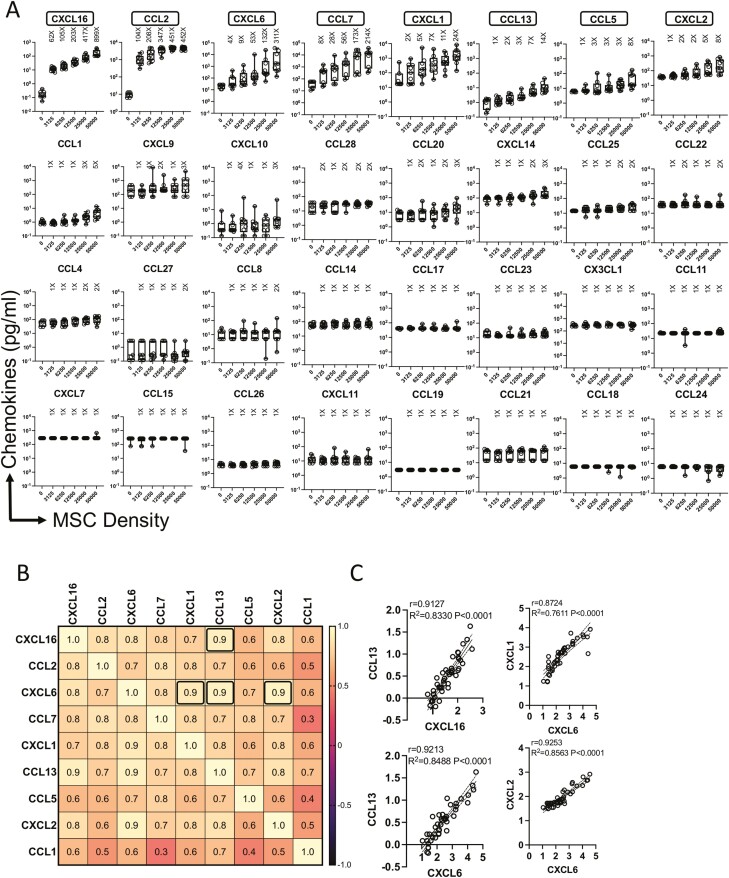
To identify the innate fitness and breadth of bone marrow MSCs in secreting chemokines, we performed a chemokine-focused multiplex secretome analysis. MSCs derived from the bone marrow of nine healthy donors were analyzed for secretory chemokines. Supernatants of MSC cultures from independent donors seeded at differential density were used for the multiplex secretory chemokine analysis. Our results show that 9 out of 32 chemokines are innately secreted by MSCs as we defined the criteria of their selection at least 5-fold above the background levels in the highest density cultures (Fig. 1A). Relative quantities of individual chemokines secreted by MSCs are different among each other as identified with the hierarchical differences in their fold change over the background. The ranking of chemokines is identified as follows, CXCL16, CCL2, CXCL6, CCL7, CXCL1, CCL13, CCL5, CXCL2, and CCL1. Of these, CCL13, CCL5, CXCL2, and CCL1 are considered as low secretors due to their relatively lower primary concentrations and fold change values over the background (Fig. 1A). None of the other 23 chemokines were detected with the threshold sensitivity of above 5-fold over the background (Fig. 1A). An important insight yet to be gained is the relationship among independent secretory molecules of MSCs for their correlative secretion which can validate the potency of MSCs with matrix functionalities. To identify the correlative secretory pattern among those 9 chemokines that are innately produced by MSCs, we used a correlation matrix strategy. In this approach, secretion levels of each chemokine are subjected to linear regression analysis, and the correlation coefficient values (r = 1 through r = 0 imply the best to no correlation, respectively) represent the degree of their coexpression. Correlation matrix was created among 9 chemokines with 81 pairwise combinations (Fig. 1B). We identified at least 4 best correlations with the r value of above 0.9 which includes CXCL16 and CCL13 (r = 0.91); CXCL6 and CXCL1 (r = 0.9); CXCL6 and CCL13 (r = 0.92); CXCL6 and CXCL2 (r = 0.92). All of these are statistically significant with a value of P < .0001 (Fig. 1B, 1C). Altogether these results demonstrated that human MSCs innately secrete a specific set of chemokines at distinct ranks with unique correlative pattern.
Figure 1.
Innate matrix chemokine signatures of human bone marrow MSCs. MSCs derived from nine independent donors were seeded at the indicated cell numbers in microwell plates. Ninety-six hours later, supernatants were centrifuged and stored. Chemokines were quantified in the supernatants using Luminex xMAP (multi-analyte profiling) technology. (A) Fold change in chemokine levels at each cell density over the background is calculated. Individual chemokines are hierarchically organized based on the cumulative fold change values over the background derived from nine independent donors. Mean and SD are shown with the cumulative data derived from nine independent MSC donors. Results are plotted as pg/mL. Cumulative fold change in chemokine secretion over the background at each cell density is shown. (B) Chemokines that are secreted at least 5-fold over the background at the highest density cultures were further evaluated for correlation matrix analysis. Chemokine concentrations were subjected to linear regression analysis among each other. Correlation coefficient (r) values are color-coded and bold black boxes indicate the best correlations with the r value of above 0.9. (C) Correlative plot with r and R2 values of above 0.9 are shown for the correlations (B) CXCL16 and CCL13, CXCL6 and CXCL1, CXCL6 and CCL13, CXCL6 and CXCL2. Correlation matrix and linear regression analysis were performed in GraphPad Prism to get r, R2, and P values. R2 = goodness of fit; P, Significance of the slope deviation from Zero.
Metabolic Activity of Human Bone Marrow MSCs Define the Secretion of Chemokines
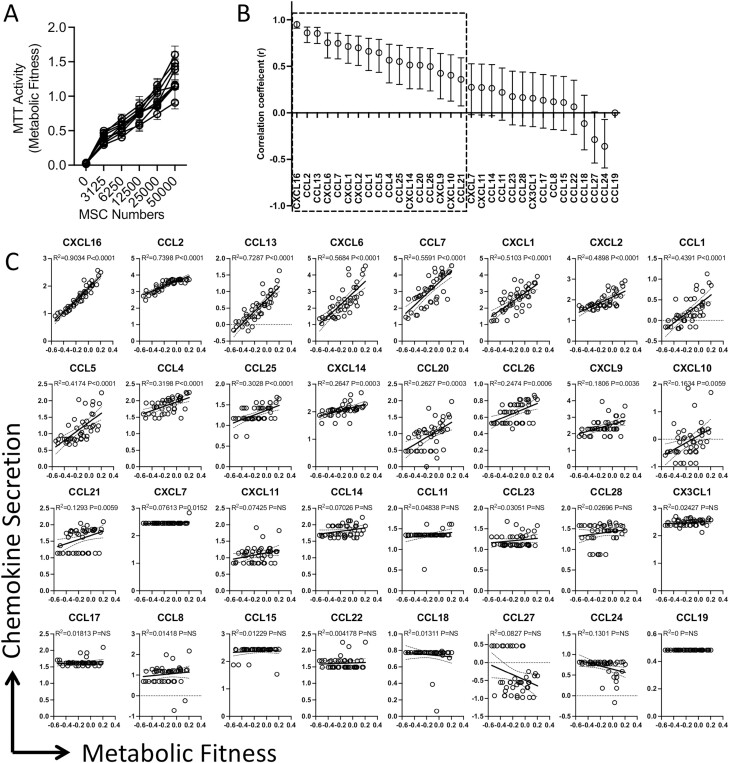
MSC’s secretome is largely quantified based on their seed densities normalized with viability assessments. However, viability is not equivalent to functionality. To overcome this limitation, we evaluated the metabolic fitness of MSCs as a base for chemokine secretion. We analyzed the fitness of chemokine secreting MSCs in reducing the tetrazolium dye MTT to its insoluble formazan. This reduction is dependent on cellular energetics and NAD(P)H-dependent oxidoreductase enzymes and thus represents the metabolic activity. MSCs derived from nine independent donors that were seeded at differential cell densities display dose-dependent MTT reduction (metabolic activity) (Fig. 2A). Next, we performed a linear regression analysis between metabolic activity and chemokine secretion levels. Our analysis identified a unique ranking based on the degree of their correlation as follows, CXCL16 (R2 = 0.90), CCL2 (R2 = 0.74), CCL13 (R2 = 0.73), CXCL6 (R2 = 0.57), CCL7 (R2 = 0.56), CXCL1 (R2 = 0.51), CXCL2 (R2 = 0.49), CCL1 (R2 = 0.44), CCL5 (R2 = 0.42) (Fig. 2B, 2C). All of these are statistically significant with values of P < .0001(Fig. 2C). Many other chemokines also showed statistically significant correlation albeit with reduced correlation coefficient values (Fig. 2B, 2C). However, based on the analysis of fold change over the background they were disqualified for consideration.
Figure 2.
Human bone marrow MSCs’ metabolic fitness defines matrix chemokine secretions. MSCs derived from nine independent donors were seeded at the indicated cell numbers in microwell plates. Ninety-six hours later, MTT assay was performed and optical density was measured. (A) Dose-dependent increase in optical density values is shown with mean and SD. (B) Optical density values of MTT assays were subjected to linear regression analysis with the corresponding chemokine levels from the appropriate wells to obtain R2 values. Linear regression plots are organized based on the ranking of the degree of correlation. Data were transformed to logarithmic scale to fit the regression line. (C) Correlation coefficient values with appropriate range of 95% confidence intervals for each chemokine are shown. Linear regression analysis was performed in GraphPad Prism to get r, R2, and P values.
Combination of IFNγ and TNFα Is Superior than Individual Stimulations in Eliciting Robust and Broad Matrix Chemokine Responses of Human Bone Marrow MSCs
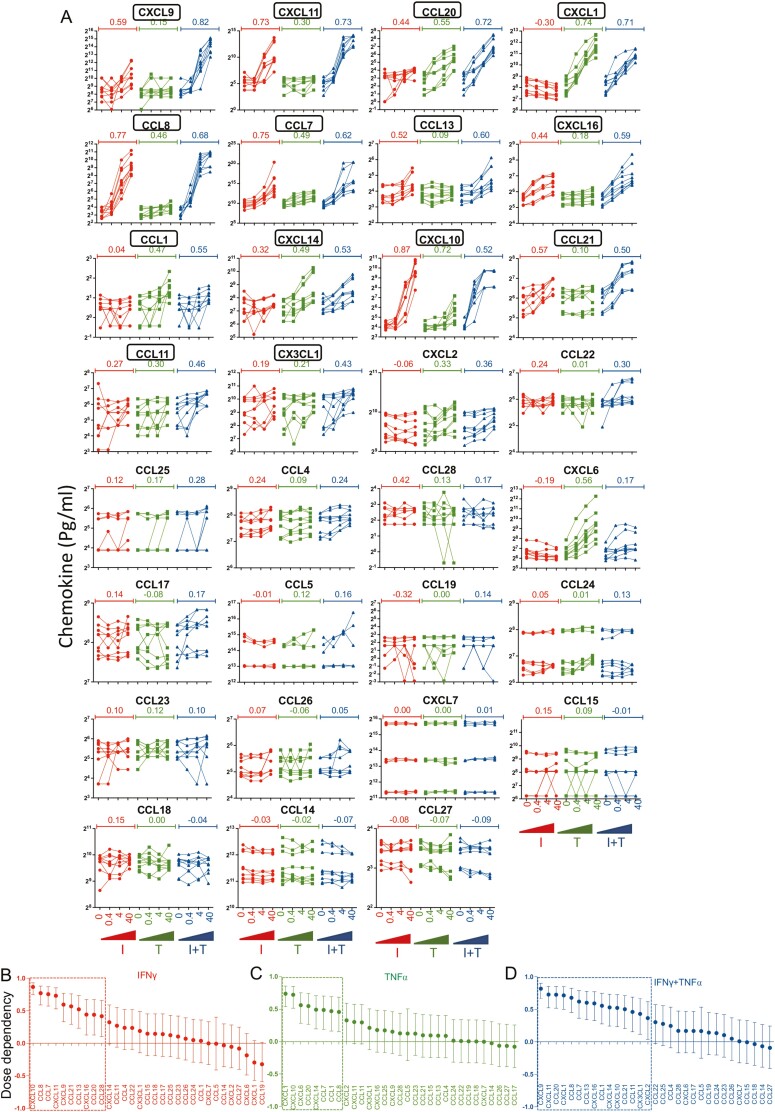
MSCs’ fitness to respond to exogenous stimulations could be defined as their surrogate measure of potency.37 In this strategy, effector molecules of MSCs, evoked by stimulants, such as IFNγ and/or TNFα, are enumerated and quantified as an assay matrix analytical system. We deployed this strategy to determine the fitness of MSCs in secreting chemokines upon exogenous stimulation. We stimulated MSCs (n = 9 donors) with dose-dependent concentrations (0, 0.4, 4, 40 ng/mL) of IFNγ and/or TNFα. Supernatants of MSCs stimulated with these conditions were subjected to secretory chemokine multiplex analysis. Our results identified that individual and combined stimulations with IFNγ and/or TNFα bestowed unique pattern of matrix chemokine responses and pronounced effects were observed with IFNγ and TNFα combined stimulations (Fig. 3A). At least 15 chemokines (CXCL9, CXCL11, CCL20, CXCL1, CCL8, CCL7, CCL13, CXCL16, CCL1, CXCL14, CXCL10, CCL21, CCL11, CX3CL1, and CXCL2) showed statistically significant dose-dependent upregulation with IFNγ and TNFα stimulations as determined by the r values (Fig. 3A, 3D). In addition, 10 chemokines (CXCL10, CCL8, CCL7, CXCL11, CXCL9, CCL21, CCL13, CXCL16, CCL20, CCL28) and 8 chemokines (CXCL1, CXCL10, CXCL6, CCL20, CXCL14, CCL7, CCL1, CCL8) showed statistically significant (P < .05) dose-dependent upregulation with either IFNγ or TNFα, respectively (Fig. 3A-3C). Our results also show that IFNγ and TNFα combined stimulation increases the magnitude of MSCs’ ability to secrete chemokines than individual stimulations (Supplementary Tables S1-S3). These results suggest that combined stimulation with IFNγ and TNFα is superior than individual stimulations in executing matrix chemokine responses of MSCs.
Figure 3.
Human Bone marrow MSCs’ distinct matrix chemokine responses to IFNγ and/or TNFα. MSCs derived from nine independent donors were seeded at the identical cell density (10 000 cells/well in 96-well plate) and stimulated with the indicated concentrations of IFNγ and/or TNFα. Forty-eight hours later, supernatants were centrifuged and stored. Chemokines were quantified in the supernatants using Luminex xMAP (multi-analyte profiling) technology. (A) Individual chemokine levels of independent MSC populations (n = 9 donors) that are stimulated with the indicated concentrations of IFNγ and/or TNFα are shown. Chemokines are hierarchically organized based on their dose-dependent response to the combined stimulation with IFNγ and TNFα while their relative responses to individual stimulations with IFNγ/TNFα are also shown in the same plots. Dose dependence values were obtained by linear regression analysis between chemokine levels and stimulating concentrations of IFNγ and/or TNFα. Hierarchical organization of chemokines based on their degree of dose-dependent responses to exogenous stimulations (B) IFNγ, (C) TNFα, and (D) IFNγ+ TNFα. Dose dependence values with statistical significance are shown with dotted line box.
Interdependence of Chemokine Secretion Signature Evoked by Individual and Combined Cytokine Stimulations
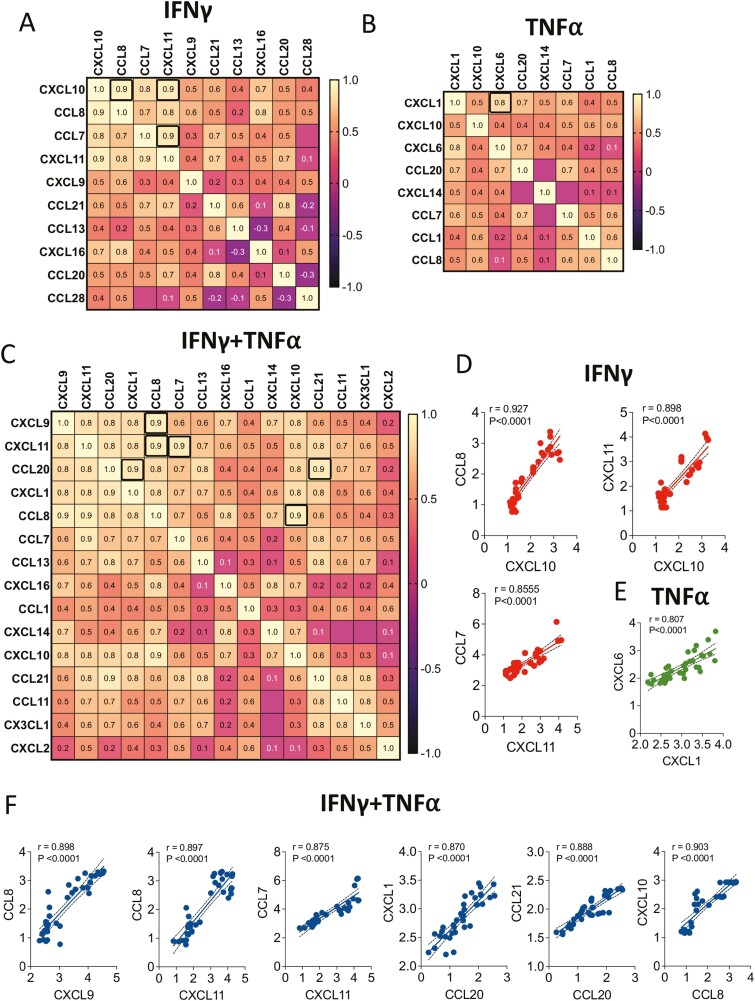
We then performed a correlation matrix analysis to determine the relationship among chemokine secretions, elicited by IFNγ and/or TNFα stimulations. In this strategy, chemokines that are upregulated by IFNγ and/or TNFα were subjected to linear regression analysis against each other to identify which chemokine secretions are correlated upon exogenous stimulations. Ten IFNγ-driven chemokines (CXCL10, CCL8, CCL7, CXCL11, CXCL9, CCL21, CCL13, CXCL16, CCL20, CCL28) were subjected to linear regression analysis among each other (100 combinations) to determine correlation coefficient values (Fig. 4A). Our results have identified that at least three top correlations, as defined with the r value of 0.9 and above which includes CXCL10 and CCL8 (r = 0.927), CXCL10 and CXCL11 (r = 0.898), CXCL11 and CCL7 (r = 0.855) (P < .0001 for all) (Fig. 4A, 4D). We analyzed 8 TNFα-driven chemokines (CXCL1, CXCL10, CXCL6, CCL20, CXCL14, CCL7, CCL1, CCL8) for correlative expressions with 64 combinations (Fig. 4B). We did not identify any single correlative expression with the coefficient value of 0.9 and above. However, CXCL1 and CXCL6 correlative expressions were identified as the top TNFα-induced correlated chemokines with the r value of 0.807 (P < .0001) (Fig. 4B, 4E). Next, we investigated 15 IFNγ- and TNFα-driven chemokines for their relationship among each other (Fig. 4C). Out of 225 combinations, 6 showed the highest correlations as defined with the r value of 0.9 and above which includes CXCL9 and CCL8 (r = 0.898), CXCL11 and CCL8 (r = 0.897), CXCL11 and CCL7 (r = 0.875), CCL20 and CXCL1 (r = 0.870), CCL20 and CCL21 (r = 0.888), CCL8 and CXCL10 (r = 0.903) (Fig. 4C, 4F). Altogether these results suggest that IFNγ and TNFα combined stimulation elicits pronounced matrix chemokine responses that are largely interdependent of each other in their secretion.
Figure 4.
Correlation matrix of responding chemokines to IFNγ and/or TNFα. Chemokines that displayed statistically significant dose-dependent responses to IFNγ and/or TNFα stimulations were subjected to correlation matrix analysis. Chemokine correlation matrix for (A) IFNγ, (B) TNFα, and (C) IFNγ and TNFα groups are shown. Correlation coefficient (r) values are color-coded and bold black boxes indicate the best correlations with the r value of above 0.9 for IFNγ and IFNγ and TNFα groups while 0.8 for TNFα group. Linear regression plots for the best-identified chemokine correlations from the groups of (D) IFNγ, (E) TNFα, and (F) IFNγ and TNFα are shown. Linear regression analysis and correlation matrix were plotted in GraphPad Prism to get r, R2, and P values.
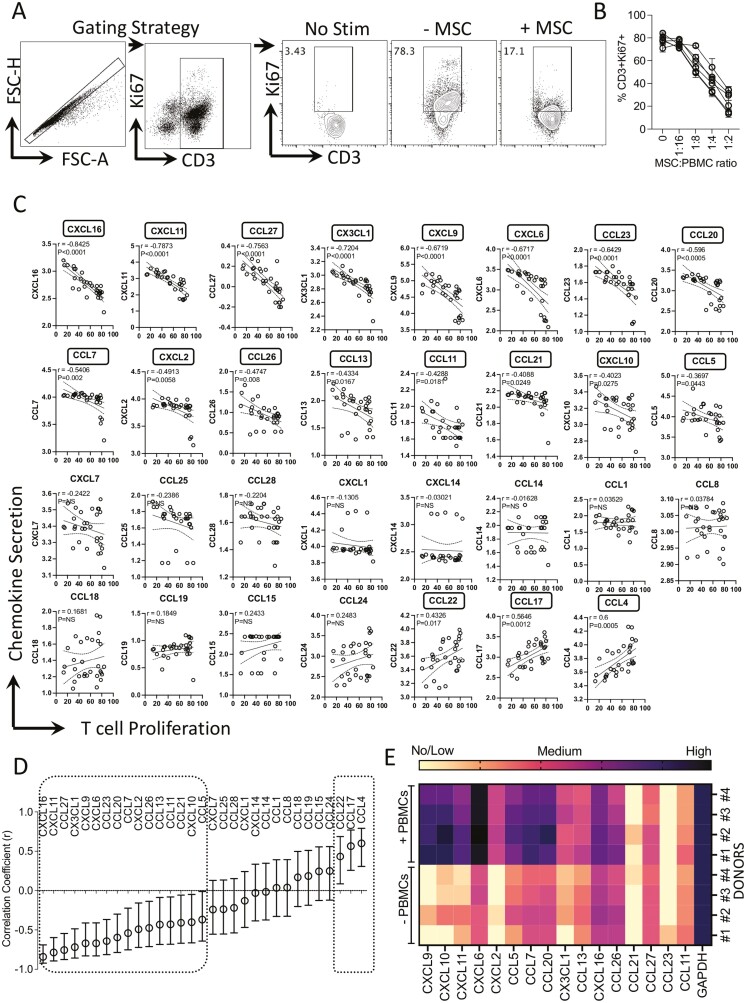
Human Bone Marrow MSC-mediated Blocking of T-cell Proliferation Predominantly Correlates with Chemokines in an Inverse Manner
To define the chemokine secretion signature that predicts MSC-mediated functional immune suppression, we analyzed the T-cell proliferation and secretory chemokine levels in the cocultures of MSCs and PBMCs. MSCs derived from 6 independent donors were cocultured with activated PBMCs from independent donors in different ratios. T-cell proliferation (%CD3+ Ki67+ T cells) was quantified using flow cytometry. Our results indicate that MSCs dose dependently inhibit T-cell proliferation as determined by the reduction in %CD3+ Ki67+ T cells (Fig. 5A, 5B). Next, we analyzed the supernatants of the MSC and PBMC culture conditions thereof for secretory chemokines using multiplex analysis. Chemokine levels of MSC and PBMC cocultures were subjected to linear regression analysis with appropriate %CD3+ Ki67+ values (Fig. 5C). Our results showed at least 16 chemokines that are inversely correlated with T-cell proliferation with a hierarchical degree as defined by the ascending negative correlation coefficient r values. The ranking and degree of this correlation are identified as CXCL16, CXCL11, CCL27, CX3CL1, CXCL9, CXCL6, CCL23, CCL20, CCL7, CXCL2, CCL26, CCL13, CCL11, CCL21, CXCL10, CCL5 (Fig. 5C, 5D). Our results have also identified that at least three chemokines are directly correlated with T-cell proliferation as defined with the positive correlation coefficient r values which includes CCL4, CCL17, and CCL22 (Fig. 5C, 5D). Altogether these results suggest that MSC-mediated inhibition of T-cell proliferation is profoundly and inversely correlated with a broad array of chemokine secretion. Next, to define MSC’s chemokine matrix responses to activated PBMCs in their cocultures, we used a two-chamber transwell non-contact coculture system. In this approach, MSCs (n = 4 independent donors) were cultured in the presence and absence of activated PBMCs which are separated by the metabolite permeable (0.4 µM) transmembrane. Three days later, MSCs were lysed and chemokine mRNA transcripts were assayed by real-time PCR. Analysis of 16 chemokine transcripts has identified that at least 11 chemokines (CXCL9, CXCL10, CXCL11, CXCL6, CXCL2, CCL5, CCL7, CCL20, CX3CL1, CCL13, CXCL16) were induced on MSCs upon coculture with activated PBMCs (Fig. 5E). These results validate that MSC sourced chemokines are the contributors of the profound inverse correlation between T-cell proliferation and chemokine secretion in MSC and PBMC cocultures.
Figure 5.
Matrix chemokine signatures of human bone marrow MSC and PBMC interactions. MSCs derived from six independent donors were cultured with SEB-activated PBMCs at the indicated ratios. PBMC numbers were kept constant with dose-dependent increase in MSC numbers. Four days post-culture, T-cell proliferation was measured by determining the percentage of CD3+ Ki67+ cells using flow cytometry. (A) Representative flow cytometry plot and gating strategy are shown for CD3 and Ki67 staining. NS indicates no stimulation of PBMCs. − and +MSCs indicate SEB-activated PBMCs cultured without and with MSCs. (B) Dose-dependent effect of MSCs from independent donors in inhibiting T-cell proliferation is cumulatively shown. Supernatants were collected from these cultures and quantitative levels (pg/mL) of chemokines were assayed through Luminex xMAP (multi-analyte profiling) technology. Individual chemokine levels of MSC and PBMC coculture were subjected to linear regression analysis with the corresponding T-cell proliferation values (%CD3+ Ki67+) to obtain correlation coefficient r values. r value of 1 and −1 indicates the best direct and inverse correlations, respectively, while 0 indicates no correlation. (C) Linear regression plots were organized hierarchically based on the correlation coefficient (r) ranking scale of −1 to +1. X-axis indicates %CD3+ Ki67+ cells (T-cell proliferation) and y-axis indicates chemokine levels. y-axis data are transformed to logarithmic scale to best fit linear regression lines. (D) Hierarchical organization of correlation coefficient values with appropriate range of 95% confidence intervals for each chemokine is shown in the scale of −1 to +1. Statistical significance (P < .05) or non-significance (NS) of the r values are shown. (E) MSCs with and without SEB-activated PBMCs were cocultured in a non-contact dependent two-chamber transwell assay system. Three days later, MSCs were harvested to isolate total RNA, and cDNA was prepared. Quantitative PCR was performed to determine the levels of chemokine expression. Heat map depicts the low, medium, and high expression of chemokines based on the relative cycle of threshold values. Cumulative results are shown from four independent experiments (n = 4 independent donors).
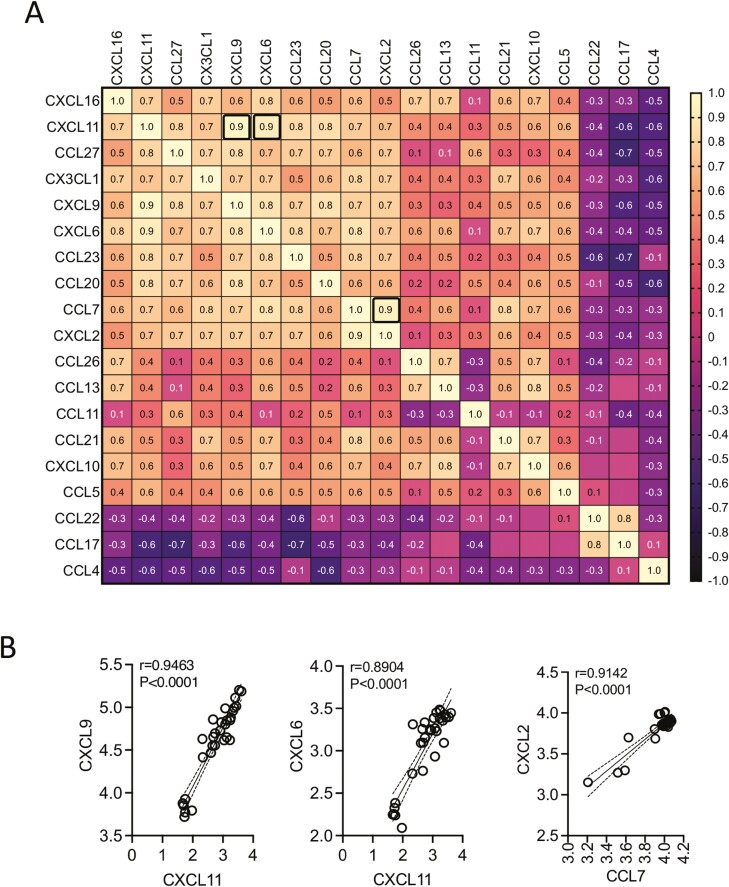
Correlation Matrix of Chemokines That Predicted Immune Suppression
To determine the correlative expressions among the 16 chemokines that predicted immune suppression, we then performed a correlation matrix analysis. Chemokines that are inversely (16 chemokines) and directly (3 chemokines) correlated with MSC-inhibited T-cell proliferation were subjected to linear regression analysis among each other in 361 combinations (Fig. 6A). This analysis identified at least three top correlations, as defined with the positive correlation r value of above 0.9, including CXCL11 and CXCL9 (r = 0.946), CXCL11 and CXCL6 (r = 0.890), CCL7 and CXCL2 (r = 0.914) (P < .0001 for all) (Fig. 6A, 6B). In addition, chemokines that are inversely and directly correlated with MSC-inhibited T-cell proliferation showed an inverse correlation among each other. However, none of them reached the r value of below −0.9 (Fig. 6A). Altogether, the present analysis has identified that chemokines of MSC and PBMC cocultures do exhibit direct correlation among themselves with a unique degree and pattern.
Figure 6.
Correlation matrix of chemokines that predict human bone marrow MSC’s immune suppression. Chemokines that displayed statistically significant correlation with MSC-inhibited T-cell proliferation were subjected to correlation matrix analysis. (A) Correlation matrix of chemokines that showed direct (n = 16 chemokines) and inverse (n = 3 chemokines) correlation with MSC-inhibited T-cell proliferation is shown. Correlation coefficient (r) values are color-coded and bold black boxes indicate the best correlations with the r value of above 0.9. (B) Linear regression plots for the best-identified chemokine correlations CXCL11 and CXCL9, CXCL11 and CXCL6, CCL7 and CXCL2 are shown. Linear regression analysis and correlation matrix were plotted in GraphPad Prism to get r, R2, and P values.
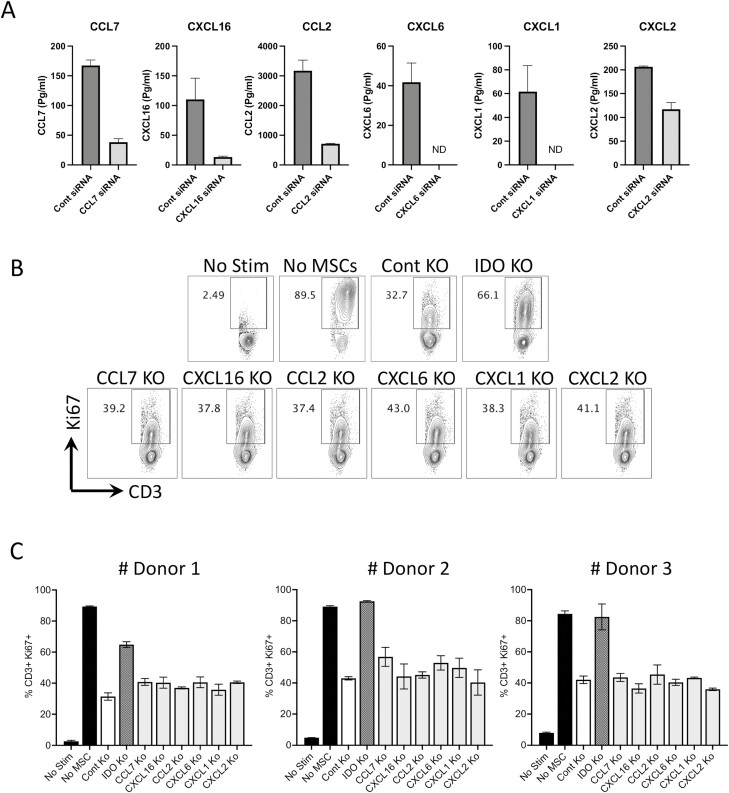
Human Bone Marrow MSC Sourced Inherent Chemokines Are the Bystander Predictors of T-cell Suppression
To define the immunosuppressive functional role of chemokines that are innately secreted by MSCs, we used a siRNA knockdown strategy. CCL7, CXCL16, CCL2, CXCL6, CXCL1, and CXCL2 that are innately secreted by MSCs are silenced by siRNA (Fig. 7A). Previous studies have identified that indoleamine 2,3-dioxygenase (IDO) play a major role in immune suppression.38 Hence, IDO silenced MSCs were also tested in parallel (Supplementary Fig. S2). We cocultured CCL7, CXCL16, CCL2, CXCL6, CXCL1, CXCL2, IDO, and control siRNA-transfected MSCs with activated PBMCs. Our results demonstrated that IDO siRNA-transfected MSCs fail to inhibit T-cell proliferation while CCL7, CXCL16, CCL2, CXCL6, CXCL1, CXCL2, and control siRNA-transfected MSCs still inhibit T-cell proliferation (Fig. 7B, 7C). These results suggest that MSC-secreted inherent chemokines are the bystander predictors of T-cell suppression and play no direct role in this process.
Figure 7.
Functionality of chemokines in modulating human bone marrow MSC’s inhibitory potential on T-cell proliferation. Control, CCL7, CXCL16, CCL2, CXCL6, CXCL1, and CXCL2 siRNA-transfected MSCs were cultured and supernatants were collected 72-hour post-transfection and assayed for appropriate chemokines. (A) Quantitative levels of CCL7, CXCL16, CCL2, CXCL6, CXCL1, and CXCL2 in the supernatants of control and appropriate siRNA-transfected MSCs are shown. (B) Control, IDO CCL7, CXCL16, CCL2, CXCL6, CXCL1, and CXCL2 siRNA-transfected MSCs were cultured with SEB-activated PBMCs. Four days post-culture, T-cell proliferation was measured in flow cytometry by Ki67 intracellular staining. Proliferation of T cells (%CD3+ Ki67+) in the presence and absence of siRNA-transfected MSCs is shown with (B) representative flow cytometry plots. (C) Cumulative data derived from three independent MSC donors are also shown with mean and SD. siRNA-transfected MSCs and PBMCs were cocultured in a ratio of 1:4 or 1:8.
Discussion
Assay matrix analytical system evaluating immunomodulatory and tissue regenerative pathways of MSCs has been an emerging platform for potency determination.25 In this strategy, potency is determined by evaluating more than a single effector molecule that is predicted to play role in MSCs’ therapeutic function. Assay matrix-based determination of MSCs’ potency can be deployed either through discovery-based broad omics approach or rigorous investigation of focused and selective pathways of significance.15 In the present study, we have deployed selective secretome-based approach focusing on chemokine matrix responses of MSCs. Our analysis has identified chemokine secretome signature of MSCs in their resting state, upon stimulation with inflammatory cues and coculture with immune responding PBMCs. In each of the categories, matrix sets of chemokines are identified with hierarchical ranking and degree of magnitude which collectively informed the potency of MSCs.
MSCs inherently secrete at least 8 chemokines (CXCL16, CCL2, CXCL6, CCL7, CXCL1, CCL13, CCL5, CXCL2) without exogenous stimulation. Hence, the quantification of these chemokines describes MSCs’ innate fitness. Versatile fold changes of individual chemokine levels at differential MSC density over the background indicate MSC’s breadth and intensity in secreting chemokines. In addition, this analysis also indicates the potency of MSCs in secreting an array of chemokines with linearity and threshold sensitivity. However, the limitation of this strategy is variability in seeding cell densities which are defined based on dose (numbers) and viability. MSC dose and viability determination largely depends on classic trypan blue exclusion microscopic analysis and/or advanced methodologies to exclude pre-apoptotic and apoptotic cells.4 Although these methods identify cell dose and viability, it has been reported that MSC viability is not equal to their functionality. For example, senescent MSCs display high viability despite poor functionality, and similarly frozen-thawed MSCs display defective functionality despite high viability.39,40 These limitations are addressed in the present study where the metabolic activity of MSCs is set as common denominators in defining MSCs’ chemokine secretory fitness. This strategy identified the linear relationship between MSCs’ metabolic activity and chemokine secretion levels. Although this analysis identified the hierarchical organizations of the correlation values between MSC metabolic activity and chemokine secretions, fold change analysis needs to be considered contemporaneously for signal-to-noise validations. Thus, chemokine matrix analysis to define MSCs’ innate potency without exogenous stimulations can incorporate both metabolic fitness and fold change analysis.
It has been suggested that chemokine receptor and chemokine expression on MSCs varies depending on their origin tissue sources and may have impact on its potency.28 Although the present study did not evaluate chemokine receptor expression, we also confirmed that MSCs derived from visceral tissues such as adipose and umbilical cord also innately secrete six chemokines (CXCL6, CCL2, CCL7, CXCL16, CXCL1, CXCL2) that are in consensus with bone marrow MSCs (Supplementary Fig. S3). Nevertheless, further studies are warranted to define the regulation and functions of chemokines and chemokine receptors on MSCs from differential tissue resources.
Previous studies have shown that MSCs respond to the inflammatory cues produced by activated PBMCs.41 Utilization of this strategy mimic the in vivo situation post-MSC infusion. However, there is a limitation since activated PBMCs produce variable inflammatory cytokines, and assaying them as a stimulator is a confounder in potency analysis. Alternatively, recombinant cytokines such as IFNγ elicits immunosuppressive properties on MSCs, and assaying MSC’s responsiveness to IFNγ can be considered as a surrogate measure of potency.37 Previously, IFNγ-mediated upregulation of immunomodulatory effector molecules (IDO, PD-L1, ICAM-1, HLADR, etc.) has been incorporated in the assay matrix potency analysis.37,38 In the functionally relevant morphological profiling (FRMP) potency assays, stimulation with IFNγ results in the emergence of subpopulations of MSCs with unique morphological features that predict immune suppression.16 Similarly, TNFα-induced proteins such as TSG-6 play major a role in MSCs’ function, and thus MSCs’ responsiveness to TNFα also can be evaluated in the potency testing.42,43 In the present study, a rigorous comparison of MSCs stimulated with IFNγ and/or TNFα showed that combined stimulations elicit a broad and profound increase in chemokine matrix responses. Although stimulation with IFNγ and TNFα induces broad matrix chemokine responses, it also induces cytostatic responses on MSCs which results in attenuated metabolic activity (Supplementary Fig. S4). Hence, metabolic functionality of MSCs post-stimulation with IFNγ and TNFα does not correlate with chemokine secretion. Dendritic cells are known to undergo metabolic switch to glycolytic pathway upon activation which is associated with the secretion of immune active molecules.44 Although in the present study, MTT assay does not correlate with chemokine secretion upon activation with inflammatory cues, future investigations are warranted to define the glycolytic pathway of MSCs in predicting chemokine secretion.
The present results had shown that proinflammatory cytokine stimulations induce robust chemokine responses while anti-inflammatory cytokines such as TGF-β and IL-10 neither upregulate nor downregulate MSCs’ chemokine matrix responses (Supplementary Tables S4-S6). Altogether our results bring the significance of synergistic proinflammatory cytokines in evaluating matrix chemokine responses of MSCs. Animal model and human MSC studies have clearly indicated that cytokine primed/activated MSCs display enhanced efficacy/potency than their resting counterparts.41,45-52 In addition, cytokine priming strategy also rescues MSC functions that are impaired during cell manufacturing procedures such as cryopreservation and prolonged expansion associated with cellular senescence.53-55 Early phase clinical trials are on the way to test the safety of cytokine-primed MSCs in mitigating inflammatory disorders56 (NCT04328714). Our results also support the hypothesis that cytokine priming is an augmented strategy to enhance the potency of MSCs, since priming with IFNγ and TNFα enhances the secretion levels and repertoire of chemokines.
Inhibition of T-cell proliferation is the fundamental in vitro immunosuppressive property of MSCs. Previous studies have shown that MSCs’ in vitro immune suppression is directly correlated with the inhibition of proinflammatory cytokines secreted by activated PBMCs.17,25,57 Although these in vitro assessments identified that MSCs downregulate inflammatory cytokines, consistent reports are lacking to demonstrate the downregulation of serum inflammatory cytokines post-MSC infusion in patients that predict treatment outcome.8,58 Considering the complexity of in vivo mechanism of action of MSCs and the difficulty of recapitulating those as a whole in laboratory potency assays, in vitro inhibition of T-cell proliferation is still considered as a gold standard in predicting MSC functionality.15 Here in the present study, we have identified that MSC’s inhibition of T-cell proliferation is predominantly correlated with chemokine secretion in an inverse manner. Although some chemokines, such as CCL4, CCL17, and CCL22 are downregulated and directly correlated with MSC-induced suppression of T-cell proliferation, they are shadowed by a broad array of chemokines (16 chemokines) that are inversely correlated. Our results support the hypothesis that MSC-mediated increase in chemokine milieu evokes the regulatory properties of immune populations such as myeloid cells and provides the therapeutic outcome.27 Although robust biomarker studies that predict clinical outcome are yet to be available to support this hypothesis, serological increase in CXCL9 and CXCL0 has been reported in patients responding to MSC therapy.30 Our data further support the hypothesis that the inclination of chemokine milieu accumulated by MSCs largely bestows therapeutic function rather than propelling inflammation and injury.
CXCL8 (also known as IL-8) secretion is one of the significant characteristics of MSCs.51,52 Although we could not investigate the relative secretion of CXCL8 to other chemokines, single plex analysis confirmed that bone marrow-derived MSCs secrete CXCL8 which are further upregulated by IFNγ and TNFα (Supplementary Fig. S5). Our previous publication showed that CXCL8 is also secreted at high levels from activated PBMCs, and thus CXCL8 levels in the cocultures of MSC and PBMC did not predict T-cell suppression.25 Nevertheless, CXCL8 can be considered as one of the potency markers due to its proangiogenic properties.24
Our mechanistic knockdown experiments have demonstrated that MSC secreted chemokines CCL7, CXCL16, CCL2, CXCL6, CXCL1, and CXCL2 do not modulate the suppressive properties of MSCs on T cells unlike IDO. This suggests that innate secretion of chemokines is the bystander predictor of MSC-mediated T-cell inhibition. MSCs’ in vivo mechanism of action is multifactorial. Three major mechanisms have been proposed for the therapeutical benefit of MSCs based on the route of delivery which includes differentiation into mesoderm tissues, efferocytosis-based polarization of macrophages, and direct contact or soluble factor-mediated instruction of immune cells.13 Although in all these instances MSC-derived chemokines might play a role, our results provided insights on the last mechanism of action involving soluble factors predominantly chemokines. Despite MSC secreted chemokines do not mechanistically modulate T-cell suppression, their bystander existence and prediction of immune suppression warrant that matrix chemokine responses of MSCs are the surrogate measures of potency.
Correlation matrix analysis has identified interdependent synergistic correlations of chemokine secretions in three independent categories, namely in MSC’s resting stage, stimulation upon IFNγ and/or TNFα and upon coculture with activated PBMCs. The evaluation of matrix correlative chemokine secretions is amenable to potency analysis for use as comparative reference standards.25,26 In this strategy, the correlation of unique chemokine pairs displaying the highest correlation coefficient values can be compared with their correlation coefficient values with other chemokines. In an alternative strategy, correlation matrix of resting MSCs can be compared with their counterparts upon stimulation with IFNγ and/or TNFα and thus serve as the reference ruler. Although prior studies have suggested the usage of universal cellular standards or rulers for potency determination, it has been a challenge to identify apt cellular standards that entirely mimic the versatile properties of MSCs.59-62 The present strategy is independent of external cellular standards/rulers and can be adopted for the potency determination of autologous and random donor MSCs. Identification of the correlative matrix chemokine responses of MSCs suggests the synergistic involvement of chemokines in immunomodulation and tissue regeneration. However, mechanistic experiments had shown that individual knockdown of MSC’s innate chemokines does not modulate T-cell suppression. Thus, abruption of the chemokine synergy with individual knockdown does not dominate the inherent T-cell-suppressive properties of MSCs. Although previous studies had shown that IDO is the central dominant of MSCs’ in vitro T-cell-suppressive mechanism,25,63,64 future mechanistic animal model studies are needed to define the therapeutical significance of these correlative expressions.
Chemokines identified here have versatile functions in homeostatic immune responses, lymphoid development, tissue maintenance, and inflammation.65-67 Although chemokines are generally considered as the promoters of inflammation, it is emerging that MSC-derived chemokines execute pharmacological effects through unique mechanisms, such as chemoattraction, angiogenesis, immune modulation, and macrophage polarization.27,28 Thus, MSC-derived chemokines can attract leukocytes to the site of inflammation and tolerize them through immunosuppressive pathways such as IDO. In addition, MSC-derived chemokines can synergistically interact to promote macrophage polarization for immune suppression and tissue repair.34 Chemokines are known to play a role in angiogenesis and revascularization in tissue repair while they also perform anti-angiogenesis.68 However, it is clear that MSCs’ in vivo mechanism is not operated through a single pathway and thus narrowing to a single chemokine as the predictor of potency may not be informative considering the rich chemokinome of MSCs and their overlapping correlative expressions. The present assay matrix strategy identified that chemokine matrix responses of MSCs are broad, and the majority of them predict immunosuppression which warrants the consideration of matrix chemokine responses of MSCs as the surrogate measure of potency.
MSCs have been tested in clinical trials over the past two decades despite conflicting results while consistent efficacy is yet to be accomplished.13 Nevertheless, marketing approval of MSC therapy has occurred for some clinical indications which encouraged further understanding of the attributes of MSCs to inform advanced potency testing strategies. In the absence of clinical studies that identify in vivo predictive biomarkers of MSC treatment responders vs non-responders,69 the potency analysis of MSCs largely depends on the comprehensive analysis of MSCs’ intrinsic and responding fitness in exhibiting an array of biologically functional molecules. Although discovery-based omics strategies identify new effector molecules of MSCs, focused putative pathways/molecules of MSC function can be rigorously investigated as a “selective omics” strategy. The present analysis focusing on the chemokine matrix responses of MSCs can be incorporated into the advanced potency testing of MSCs.
Supplementary Material
Contributor Information
Ariel Joy Lipat, Department of Biomedical Sciences, Mercer University School of Medicine, Savannah, GA, USA.
Chasen Cottle, Department of Biomedical Sciences, Mercer University School of Medicine, Savannah, GA, USA.
Bonnie M Pirlot, Department of Biomedical Sciences, Mercer University School of Medicine, Savannah, GA, USA.
James Mitchell, Diagnostic Radiology, Memorial Health University Medical Center, Savannah, GA, USA.
Brian Pando, Diagnostic Radiology, Memorial Health University Medical Center, Savannah, GA, USA.
Brian Helmly, Diagnostic Radiology, Memorial Health University Medical Center, Savannah, GA, USA.
Joanna Kosko, Department of Pathology, Memorial Health University Medical Center, Savannah, GA, USA.
Devi Rajan, Department of Biomedical Sciences, Mercer University School of Medicine, Savannah, GA, USA.
Peiman Hematti, Department of Medicine, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, USA.
Raghavan Chinnadurai, Department of Biomedical Sciences, Mercer University School of Medicine, Savannah, GA, USA.
Funding
This study was partly supported by the WES Leukemia Research Foundation (R.C.). This research was also supported by Mercer University School of Medicine’s research funds (R.C.) and generous support from the Landings Women’s Golf Association, Savannah, GA (R.C.). A.J.L. is supported by the graduate program of biomedical Sciences at Mercer University School of Medicine.
Conflict of Interest
The authors declared no potential conflicts of interest.
Author Contributions
A.J.L., C.C., B.M.P.: performed experiments related to MSCs. J.M., B.P., B.H., J.K., P.H.: provided bone marrow aspirates. D.R.: helped with Luminex Analysis. R.C.: conceived and designed the studies, performed experiments, analyzed and interpreted data, and wrote the manuscript. All authors approved the manuscript.
Data Availability
All the data are included in the figures and online supplementary materials of the manuscript.
References
- 1. Moll G, Hoogduijn MJ, Ankrum JA.. Editorial: safety, efficacy and mechanisms of action of mesenchymal stem cell therapies. Front Immunol. 2020;11:243. 10.3389/fimmu.2020.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cuende N, Rasko JEJ, Koh MBC, et al. . Cell, tissue and gene products with marketing authorization in 2018 worldwide. Cytotherapy. 2018;20(11):1401-1413. 10.1016/j.jcyt.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 3. Kabat M, Bobkov I, Kumar S, et al. . Trends in mesenchymal stem cell clinical trials 2004-2018: is efficacy optimal in a narrow dose range? Stem Cells Transl Med. 2020;9(1):17-27. 10.1002/sctm.19-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robb KP, Fitzgerald JC, Barry F, et al. . Mesenchymal stromal cell therapy: progress in manufacturing and assessments of potency. Cytotherapy. 2019;21(3):289-306. 10.1016/j.jcyt.2018.10.014 [DOI] [PubMed] [Google Scholar]
- 5. Lalu MM, McIntyre L, Pugliese C, et al. . Safety of cell therapy with mesenchymal stromal cells (SafeCell): a systematic review and meta-analysis of clinical trials. PLoS One. 2012;7(10):e47559. 10.1371/journal.pone.0047559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin I, Galipeau J, Kessler C, et al. . Challenges for mesenchymal stromal cell therapies. Sci Transl Med. 2019;11(480):eaat2189. 10.1126/scitranslmed.aat2189 [DOI] [PubMed] [Google Scholar]
- 7. Galipeau J, Sensebe L.. Mesenchymal stromal cells: clinical challenges and therapeutic opportunities. Cell Stem Cell. 2018;22(6):824-833. 10.1016/j.stem.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galipeau J, Krampera M, Leblanc K, et al. . Mesenchymal stromal cell variables influencing clinical potency: the impact of viability, fitness, route of administration and host predisposition. Cytotherapy. 2021;23(5):368-372. [DOI] [PubMed] [Google Scholar]
- 9. Radrizzani M, Soncin S, Lo Cicero V, et al. . Quality control assays for clinical-grade human mesenchymal stromal cells: methods for ATMP release. Methods Mol Biol. 2016;1416:313-337. [DOI] [PubMed] [Google Scholar]
- 10. Dominici M, Le Blanc K, Mueller I, et al. . Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315-317. [DOI] [PubMed] [Google Scholar]
- 11. Mendicino M, Bailey AM, Wonnacott K, et al. . MSC-based product characterization for clinical trials: an FDA perspective. Cell Stem Cell. 2014;14(2):141-145. [DOI] [PubMed] [Google Scholar]
- 12. Hematti P. Characterization of mesenchymal stromal cells: potency assay development. Transfusion. 2016;56(4):32S-35S. [DOI] [PubMed] [Google Scholar]
- 13. Krampera M, Le Blanc K.. Mesenchymal stromal cells: putative microenvironmental modulators become cell therapy. Cell Stem Cell. 2021;28(10):1708-1725. [DOI] [PubMed] [Google Scholar]
- 14. de Wolf C, van de Bovenkamp M, Hoefnagel M.. Regulatory perspective on in vitro potency assays for human mesenchymal stromal cells used in immunotherapy. Cytotherapy. 2017;19(7):784-797. [DOI] [PubMed] [Google Scholar]
- 15. Galipeau J, Krampera M, Barrett J, et al. . International Society for Cellular Therapy perspective on immune functional assays for mesenchymal stromal cells as potency release criterion for advanced phase clinical trials. Cytotherapy. 2016;18(2):151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klinker MW, Marklein RA, Lo Surdo JL, et al. . Morphological features of IFN-γ-stimulated mesenchymal stromal cells predict overall immunosuppressive capacity. Proc Natl Acad Sci USA. 2017;114(13):E2598-E2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maughon TS, Shen X, Huang D, et al. . Metabolomics and cytokine profiling of mesenchymal stromal cells identify markers predictive of T-cell suppression. Cytotherapy. 2022;24(2):137-148. [DOI] [PubMed] [Google Scholar]
- 18. Boregowda SV, Krishnappa V, Haga CL, et al. . A clinical indications prediction scale based on TWIST1 for human mesenchymal stem cells. EBioMedicine. 2016;4:62-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sherman SE, Kuljanin M, Cooper TT, et al. . High aldehyde dehydrogenase activity identifies a subset of human mesenchymal stromal cells with vascular regenerative potential. Stem Cells. 2017;35(6):1542-1553. [DOI] [PubMed] [Google Scholar]
- 20. Killer MC, Nold P, Henkenius K, et al. . Immunosuppressive capacity of mesenchymal stem cells correlates with metabolic activity and can be enhanced by valproic acid. Stem Cell Res Ther. 2017;8(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murgia A, Veronesi E, Candini O, et al. . Potency biomarker signature genes from multiparametric osteogenesis assays: will cGMP human bone marrow mesenchymal stromal cells make bone? PLoS One. 2016;11(10):e0163629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marklein RA, Lo Surdo JL, Bellayr IH, et al. . High content imaging of early morphological signatures predicts long term mineralization capacity of human mesenchymal stem cells upon osteogenic induction. Stem Cells. 2016;34(4):935-947. [DOI] [PubMed] [Google Scholar]
- 23. Bowles AC, Kouroupis D, Willman MA, et al. . Signature quality attributes of CD146+ mesenchymal stem/stromal cells correlate with high therapeutic and secretory potency. Stem Cells. 2020;38(8):1034-1049. [DOI] [PubMed] [Google Scholar]
- 24. Lehman N, Cutrone R, Raber A, et al. . Development of a surrogate angiogenic potency assay for clinical-grade stem cell production. Cytotherapy. 2012;14(8):994-1004. [DOI] [PubMed] [Google Scholar]
- 25. Chinnadurai R, Rajan D, Qayed M, et al. . Potency analysis of mesenchymal stromal cells using a combinatorial assay matrix approach. Cell Rep. 2018;22(9):2504-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chinnadurai R, Rajakumar A, Schneider AJ, et al. . Potency analysis of mesenchymal stromal cells using a phospho-STAT matrix loop analytical approach. Stem Cells. 2019;37(8):1119-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galipeau J. Macrophages at the nexus of mesenchymal stromal cell potency: the emerging role of chemokine cooperativity. Stem Cells. 2021;39(9):1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cuesta-Gomez N, Graham GJ, Campbell JDM.. Chemokines and their receptors: predictors of the therapeutic potential of mesenchymal stromal cells. J Transl Med. 2021;19(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hassanshahi G, Roohi MA, Esmaeili SA, et al. . Involvement of various chemokine/chemokine receptor axes in trafficking and oriented locomotion of mesenchymal stem cells in multiple sclerosis patients. Cytokine. 2021;148:155706. [DOI] [PubMed] [Google Scholar]
- 30. Boberg E, von Bahr L, Afram G, et al. . Treatment of chronic GvHD with mesenchymal stromal cells induces durable responses: a phase II study. Stem Cells Transl Med. 2020;9(10):1190-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren G, Zhang L, Zhao X, et al. . Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell Stem Cell. 2008;2(2):141-150. [DOI] [PubMed] [Google Scholar]
- 32. Rafei M, Hsieh J, Fortier S, et al. . Mesenchymal stromal cell-derived CCL2 suppresses plasma cell immunoglobulin production via STAT3 inactivation and PAX5 induction. Blood. 2008;112(13):4991-4998. [DOI] [PubMed] [Google Scholar]
- 33. Lee HK, Kim HS, Kim JS, et al. . CCL2 deficient mesenchymal stem cells fail to establish long-lasting contact with T cells and no longer ameliorate lupus symptoms. Sci Rep. 2017;7:41258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giri J, Das R, Nylen E, et al. . CCL2 and CXCL12 derived from mesenchymal stromal cells cooperatively polarize IL-10+ tissue macrophages to mitigate gut injury. Cell Rep. 2020;30(6):1923-1934.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee SC, Lee YJ, Choi I, et al. . CXCL16/CXCR6 axis in adipocytes differentiated from human adipose derived mesenchymal stem cells regulates macrophage polarization. Cells. 2021;10(12). 10.3390/cells10123410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chinnadurai R, Copland IB, Ng S, et al. . Mesenchymal stromal cells derived from Crohn’s patients deploy indoleamine 2,3-dioxygenase-mediated immune suppression, independent of autophagy. Mol Ther. 2015;23(7):1248-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krampera M, Galipeau J, Shi Y, et al. . Immunological characterization of multipotent mesenchymal stromal cells—the International Society for Cellular Therapy (ISCT) working proposal. Cytotherapy. 2013;15(9):1054-1061. [DOI] [PubMed] [Google Scholar]
- 38. Guan Q, Li Y, Shpiruk T, et al. . Inducible indoleamine 2,3-dioxygenase 1 and programmed death ligand 1 expression as the potency marker for mesenchymal stromal cells. Cytotherapy. 2018;20(5):639-649. [DOI] [PubMed] [Google Scholar]
- 39. Chabot D, Tremblay T, Pare I, et al. . Transient warming events occurring after freezing impairs umbilical cord-derived mesenchymal stromal cells functionality. Cytotherapy. 2017;19(8):978-989. [DOI] [PubMed] [Google Scholar]
- 40. Loisel S, Dulong J, Menard C, et al. . Brief report: proteasomal indoleamine 2,3-dioxygenase degradation reduces the immunosuppressive potential of clinical grade-mesenchymal stromal cells undergoing replicative senescence. Stem Cells. 2017;35(5):1431-1436. [DOI] [PubMed] [Google Scholar]
- 41. Krampera M, Cosmi L, Angeli R, et al. . Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells. 2006;24(2):386-398. [DOI] [PubMed] [Google Scholar]
- 42. Prockop DJ. Inflammation, fibrosis, and modulation of the process by mesenchymal stem/stromal cells. Matrix Biol. 2016;51:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee RH, Yu JM, Foskett AM, et al. . TSG-6 as a biomarker to predict efficacy of human mesenchymal stem/progenitor cells (hMSCs) in modulating sterile inflammation in vivo. Proc Natl Acad Sci USA. 2014;111(47):16766-16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wculek SK, Khouili SC, Priego E, et al. . Metabolic control of dendritic cell functions: digesting information. Front Immunol. 2019;10:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chinnadurai R, Copland IB, Patel SR, et al. . IDO-independent suppression of T cell effector function by IFN-γ-licensed human mesenchymal stromal cells. J Immunol. 2014;192(4):1491-1501. [DOI] [PubMed] [Google Scholar]
- 46. Kim DS, Jang IK, Lee MW, et al. . Enhanced immunosuppressive properties of human mesenchymal stem cells primed by interferon-gamma. EBioMedicine. 2018;28:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boland L, Burand AJ, Brown AJ, et al. . IFN-γ and TNF-α pre-licensing protects mesenchymal stromal cells from the pro-inflammatory effects of palmitate. Mol Ther. 2018;26(3):860-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Noone C, Kihm A, English K, et al. . IFN-γ stimulated human umbilical-tissue-derived cells potently suppress NK activation and resist NK-mediated cytotoxicity in vitro. Stem Cells Dev. 2013;22(22):3003-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chinnadurai R, Bates PD, Kunugi KA, et al. . Dichotomic potency of IFNγ licensed allogeneic mesenchymal stromal cells in animal models of acute radiation syndrome and graft versus host disease. Front Immunol. 2021;12:708950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kanai R, Nakashima A, Doi S, et al. . Interferon-γ enhances the therapeutic effect of mesenchymal stem cells on experimental renal fibrosis. Sci Rep. 2021;11(1):850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thirlwell KL, Colligan D, Mountford JC, et al. . Pancreas-derived mesenchymal stromal cells share immune response-modulating and angiogenic potential with bone marrow mesenchymal stromal cells and can be grown to therapeutic scale under Good Manufacturing Practice conditions. Cytotherapy. 2020;22(12):762-771. [DOI] [PubMed] [Google Scholar]
- 52. Forbes S, Bond AR, Thirlwell KL, et al. . Human umbilical cord perivascular cells improve human pancreatic islet transplant function by increasing vascularization. Sci Transl Med. 2020;12(526). 10.1126/scitranslmed.aan5907 [DOI] [PubMed] [Google Scholar]
- 53. Chinnadurai R, Rajan D, Ng S, et al. . Immune dysfunctionality of replicative senescent mesenchymal stromal cells is corrected by IFNγ priming. Blood Adv. 2017;1(11):628-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chinnadurai R, Copland IB, Garcia MA, et al. . Cryopreserved mesenchymal stromal cells are susceptible to T-cell mediated apoptosis which is partly rescued by IFNγ licensing. Stem Cells. 2016;34(9):2429-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mendt M, Daher M, Basar R, et al. . Metabolic reprogramming of GMP grade cord tissue derived mesenchymal stem cells enhances their suppressive potential in GVHD. Front Immunol. 2021;12:631353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guess AJ, Daneault B, Wang R, et al. . Safety profile of good manufacturing practice manufactured interferon γ-primed mesenchymal stem/stromal cells for clinical trials. Stem Cells Transl Med. 2017;6(10):1868-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bloom DD, Centanni JM, Bhatia N, et al. . A reproducible immunopotency assay to measure mesenchymal stromal cell-mediated T-cell suppression. Cytotherapy. 2015;17(2):140-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheung TS, Bertolino GM, Giacomini C, et al. . Mesenchymal stromal cells for graft versus host disease: mechanism-based biomarkers. Front Immunol. 2020;11:1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Deans R. Towards the creation of a standard MSC line as a calibration tool. Cytotherapy. 2015;17(9):1167-1168. [DOI] [PubMed] [Google Scholar]
- 60. Viswanathan S, Keating A, Deans R, et al. . Soliciting strategies for developing cell-based reference materials to advance mesenchymal stromal cell research and clinical translation. Stem Cells Dev. 2014;23(11):1157-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salem B, Miner S, Hensel NF, et al. . Quantitative activation suppression assay to evaluate human bone marrow-derived mesenchymal stromal cell potency. Cytotherapy. 2015;17(12):1675-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tanavde V, Vaz C, Rao MS, et al. . Research using mesenchymal stem/stromal cells: quality metric towards developing a reference material. Cytotherapy. 2015;17(9):1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Meisel R, Zibert A, Laryea M, et al. . Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood. 2004;103(12):4619-4621. [DOI] [PubMed] [Google Scholar]
- 64. Boyt DT, Boland LK, Burand AJ Jr, et al. . Dose and duration of interferon gamma pre-licensing interact with donor characteristics to influence the expression and function of indoleamine-2,3-dioxygenase in mesenchymal stromal cells. J R Soc Interface. 2020;17(167):20190815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lopez-Cotarelo P, Gomez-Moreira C, Criado-Garcia O, et al. . Beyond chemoattraction: multifunctionality of chemokine receptors in leukocytes. Trends Immunol. 2017;38(12):927-941. [DOI] [PubMed] [Google Scholar]
- 66. Dyer DP. Understanding the mechanisms that facilitate specificity, not redundancy, of chemokine-mediated leukocyte recruitment. Immunology. 2020;160(4):336-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. David BA, Kubes P.. Exploring the complex role of chemokines and chemoattractants in vivo on leukocyte dynamics. Immunol Rev. 2019;289(1):9-30. [DOI] [PubMed] [Google Scholar]
- 68. Bosisio D, Salvi V, Gagliostro V, et al. . Angiogenic and antiangiogenic chemokines. Chem Immunol Allergy. 2014;99:89-104. [DOI] [PubMed] [Google Scholar]
- 69. Nolta JA, Galipeau J, Phinney DG.. Improving mesenchymal stem/stromal cell potency and survival: proceedings from the International Society of Cell Therapy (ISCT) MSC preconference held in May 2018, Palais des Congres de Montreal, Organized by the ISCT MSC Scientific Committee. Cytotherapy. 2020;22(3):123-126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are included in the figures and online supplementary materials of the manuscript.