Abstract
A chromosomal region encoding a two-component regulatory system, FlhRS, has been isolated from Paracoccus denitrificans. FlhRS-deficient mutants were unable to grow on methanol, methylamine, or choline as the carbon and energy source. Expression of the gene encoding glutathione-dependent formaldehyde dehydrogenase (fhlA) was undetectable in the mutant, and expression of the S-formylglutathione hydrolase gene (fghA) was reduced in the mutant background. In addition, methanol dehydrogenase was immunologically undetectable in cell extracts of FhlRS mutants. These results indicate that the FlhRS sensor-regulator pair is involved in the regulation of formaldehyde, methanol, and methylamine oxidation. The effect that the FlhRS proteins exert on the regulation of C1 metabolism might be essential to maintain the internal concentration of formaldehyde below toxic levels.
Paracoccus denitrificans is a nutritionally versatile bacterium found in soil, sewage, and sludge. The ability of the organism to adapt its metabolism to a variety of carbon and free energy sources may reflect the nature of its natural environment. P. denitrificans can grow heterotrophically on a variety of carbon sources and lithoautotrophically using hydrogen, thiosulfate, or reduced C1 compounds (methanol, methylamine, or formate) as free energy source. Expression of the genes encoding enzymes involved in C1 metabolism is tightly regulated. The synthesis of methanol dehydrogenase (MDH) and methylamine dehydrogenase (MADH), the enzymes that catalyze the oxidation of methanol and methylamine to formaldehyde, respectively, is induced when the cells grow on methanol or methylamine as the sole free energy source but is repressed in cells grown on energetically more favorable substrates (7). The synthesis of glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) and S-formylglutathione hydrolase (FGH), the enzymes that catalyze the oxidation of formaldehyde to formate, however, is not fully repressed under these conditions, since low but significant levels of both enzymes can be found in succinate-grown cells (14, 29). These low levels may help ensure a rapid response to small amounts of adventitiously formed formaldehyde or to the formaldehyde first generated during growth with methylotrophic substrates. Low levels of MDH were found in cultures grown on a variety of carbon sources during carbon limitation in a chemostat (7). Under these conditions, the genes involved in methanol oxidation are expressed at basal levels. Maximal expression was observed in cells grown on methanol, methylamine, and choline. Oxidation of all these compounds yields formaldehyde, so it has been postulated that formaldehyde is an important trigger in the regulation of expression of gene clusters involved in C1 metabolism (7). GD-FALDH, FGH, and cytochrome c553i are also synthesized to high levels in cultures grown on methanol, methylamine, and choline, so the expression of several gene clusters may respond to formaldehyde (14, 20, 21).
Genes involved in methanol oxidation are located in the mxa gene cluster of P. denitrificans (12, 30). The structural genes mxaF and mxaI encoding the subunits of MDH are located in the mxaFJGIR operon. Expression of these mxa genes is controlled by a specific two-component regulatory system, MxaYX, encoded in the mxaZYX operon (15). It has been hypothesized that the histidine kinase MxaY autophosphorylates in response to formaldehyde. The phosphoryl group is, presumably, then transferred to the cognate response regulator MxaX. Once phosphorylated, MxaX binds to the promoter region of mxaF and activates transcription. Surprisingly, however, a deletion of mxaY resulted in a wild-type phenotype with respect to methanol oxidation, suggesting the presence of a second regulatory system that is able to cross talk with MxaX (35). The MxaYX regulatory system appears to be specifically dedicated to the regulation of expression of the mxa genes since expression of the genes encoding MADH, GD-FALDH, FGH, and cytochrome c553i was unaffected by mxaYX mutations.
Genes involved in formaldehyde oxidation are located in the flh cluster of P. denitrificans (14, 21). GD-FALDH is encoded by flhA, while FGH is encoded by fghA. The cycB gene encoding cytochrome c553i is linked to the flhA and fghA genes. The expression of these three genes is up-regulated by formaldehyde, but this regulation is independent of the MxaYX proteins (35). Regulatory genes controlling the expression of the flh gene cluster have yet to be identified.
Thus, it is postulated that expression of genes involved in C1 metabolism in P. denitrificans involves at least two regulatory systems, both of which are activated by formaldehyde: (i) the MxaYX sensor regulator pair, which controls expression of the mxa locus, and (ii) an unidentified regulator of the flhA, fghA, and cycB genes. To investigate this hypothesis, we constructed an unmarked mxaY mutant, in which the activation of mxaX is dependent on another regulatory circuit. By introducing a cycB promoter-lacZ fusion into this new background, we were able to screen for mutants defective in activation of the cycB gene. Here we report the isolation of a gene cluster that encodes a two-component regulatory system, which controls the expression of that gene as well as the mxa, flhA, and fghA genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used are listed in Table 1. P. denitrificans and Escherichia coli were routinely grown aerobically either in brain heart infusion broth (GIBCO, Life Technologies Ltd., Paisley, United Kingdom) or mineral salt medium at 35°C (3). Carbon sources and their concentrations were as follows: 25 mM methanol (50 mM for solid media), 50 mM methylamine, 25 mM succinate, 50 mM formate, and 15 mM choline chloride. Autotrophic growth was on solid minimal medium without a carbon source, and plates were incubated in 4% carbon dioxide, 8% hydrogen, 3% oxygen, and 85% dinitrogen. Antibiotics were used at final concentrations of 40 μg ml−1 (rifampin), 25 μg ml−1 (kanamycin, streptomycin, and gentamicin), and 100 μg ml−1 (ampicillin).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| P. denitrificans | ||
| Pd1222 | Rifr, Specr, enhanced conjugation frequencies | 8 |
| Pd0921 | Pd1222, mxaX::Km | 15 |
| Pd0841 | Pd1222, ΔmxaY | 35 |
| Pd0841.hf | Pd0841, homology fragment | This study |
| Pd0841hf.611 | Pd0841hf, pPr611 integrated | This study |
| Pd6721 | Pd1222, flhA::Km | 21 |
| Pd6621 | Pd1222, fghA::Km | 14 |
| Pd6121 | Pd1222, cycB::Km | 20 |
| Pd5021 | Pd1222, flhR::Km | This study |
| Pd5121 | Pd1222, flhS::Km | This study |
| Pd9234 | Pd0841, flhR::Km | This study |
| Pd9235 | Pd0841, flhS::Km | This study |
| Pd13 | Pd0841hf.611, flhR A633G | This study |
| Pd1222::Pr071 | Pd1222, pPr071 integrated | This study |
| Pd1222::Pr501 | Pd1222, pPr501 integrated | This study |
| Pd5021::Pr071 | Pd5021, pPr071 integrated | This study |
| Pd5021::Pr501 | Pd5021, pPr501 integrated | This study |
| E. coli | ||
| S17-1 | Smr pro r− m+ RP4-2 integrated (Tc::Mu)(Km::Tn7) | 24 |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′ (traD36 proAB lacIq lacZ ΔM15) | 22 |
| Plasmids | ||
| pUC4K | Kmr (Tn903) | 32 |
| pGRPd1 | oriV (ColE1) AmproriT Smr | 31 |
| pRK2020 | Tcr, pRK2013 Km::Tn10 | 9 |
| pGEM7 | Ampr | Promega |
| pUT.hf | Ampr, mini-Tn5 homology fragment, Kmr | This study |
| pUT.Km | Ampr, mini-Tn5, Kmr | 16 |
| pLOF.hf | Ampr, mini-Tn10 homology fragment, Kmr | 16 |
| pEG400 | IncP, Smr, Spr, pUC12mcs | 10 |
| pEG400Gm | pEG400 derivative, Gmr | This study |
| pSK4 | pEG400Gm derivative, pcycA-flhA | This study |
| pBK11 | Smr, SprlacZ-promoter probe vector | 16 |
| pBK16 | pBK11, with amber codons in aadA and lacZ | |
| pJR62.1 | 3′ xoxF-cycB | |
| pNH15 | 5′ part of mxaF, mxaZ, 5′ part of mxaY 2.7-kbp genomic EcoRI fragment in pUC13 | |
| pPr611 | pBK16, pcycB-lacZ | This study |
| pPr071 | pBK11, pmxaZ-lacZ | This study |
| pPr501 | pBK11, pflhR-lacZ | This study |
| pR11.1 | pLAFR3 derivative, 23-kbp, chromosomal fragment of P. denitrificans | This study |
| pRTd5021 | pGRPd1, flhR::Km | This study |
| pRTd5121 | pGRPd1, flhS::Km | This study |
Plasmid construction.
Plasmid pUT.hf was constructed by ligation of the 6.4-kbp NotI fragment of pLOF.hf into the NotI vector fragment of pUT.Km. Plasmid pPr611 was constructed by ligation of an EcoRI-BspHI fragment, containing the promoter region of cycB, from pJR62.1 into the EcoRI-SmaI sites of pBK16. pPr071 was constructed by ligation of a 1.7-kbp EcoRI-SmaI fragment, harboring the mxaZ promoter of pNH15, into the EcoRI-SmaI site of pBK11. pPr501 was constructed by ligation of a PCR fragment harboring the flhR promoter into the EcoRI-BamHI sites of pBK11. The forward primer was CGGGGATCCCGGTCTTGCGACTGCATTTCG, and the reverse primer was CGGGAATTCCGATGCCGATCCTTTTGCCGC; cloning sites, indicated in bold, were introduced via the primers. To obtain pSK4, a SalI-EcoRI fragment containing the cycA promoter was cloned into pGEM7. The resulting plasmid was digested with SmaI, and a 1.6-kbp NruI-FspI fragment containing the flhA gene was inserted. The pcycA-flhA construct was subsequently transferred into the XbaI and HindIII sites of the broad-host-range vector pEG400Gm, a gentamicin-resistant derivative of pEG400, yielding pSK4. To obtain pRTd5021, a HincII-SphI fragment harboring flhR was inserted into the SmaI-SphI sites of pGRPd1. The resulting clone was subsequently digested with SmaI, and a HincII fragment of pUC4K containing the kanamycin resistance gene was inserted. To obtain pRTd5121, a BamHI-EcoRV fragment containing flhS was inserted into pGEM7. The resulting clone was digested with PstI, and a 1-kbp PstI fragment of flhS was replaced by the Pst fragment of pUC4K. The inactivated flhS gene was subsequently inserted into the BamHI-SphI sites of pGRPd1. The P. denitrificans gene library was obtained from Stephen Spiro (5).
DNA manipulations and analyses.
Routine methods for DNA manipulation were as previously described (2). Southern hybridizations employed positively charged nylon membranes as specified by the manufacturer (Boehringer GmbH, Mannheim, Germany). The nucleotide sequence was determined using the dideoxy chain termination method described by Sanger et al. (23) combined with the M13 cloning system, using an Automatic Sequenator (Applied Biosystems, Foster City, Calif.). For analysis of the sequences, we used the DNA-Strider and GeneWorks 2.3 programs. For homology studies on amino acid sequences, the international protein and DNA data banks were screened on-line by using GenBank (1, 11).
Gene transfer and mini-Tn5 transposition.
Wild-type chromosomal genes were replaced by homologous recombination as described previously (31). Fusions of lacZ to the mxaZ and flhR promoters were obtained by homologous recombination with a single crossover, leading to insertion of a complete plasmid in the chromosome (6). Transposon mutagenesis was carried out by triparental mating of a recipient P. denitrificans strain and two E. coli strains, one carrying the donor plasmid and the second carrying the helper plasmid pRK2020. The three strains were mixed on a brain heart infusion agar plate and incubated at 37°C for 24 h. The mating mixture was subsequently plated on minimal medium supplemented with choline and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
Insertion of a target for recombination.
This procedure was described by Kessler et al. (16) and allows the insertion into the chromosome of P. denitrificans of a recombinant transposon (the homology fragment) that carries DNA sequences homologous to the regions flanking the pcycB-lacZ fusion present on a multicopy promoter-probe-vector. Double recombination between the promoter-probe vector and the chromosomal homology region of the transposon is genetically selected by reconstitution and expression of wild-type sequences from truncated lacZ and aadA (streptomycin) resistance genes in the homology fragment. The double recombination event is confirmed by screening for loss of the transposon-encoded kanamycin resistance marker.
We cloned the recombination target cassette between the borders of a mini-Tn5 transposon and under the control of the Tn5 transposase. The resulting plasmid, pUT.hf, was transferred to strain Pd0841, and kanamycin-resistant colonies were obtained at a frequency of 10−6. Southern analysis showed that 25% of the colonies had received the transposon by a transposition event. One of these was designated Pd0841.hf. In the other 75% of cases, pUT.hf itself was integrated. Pd0841.hf was still able to grow on methanol, methylamine, and choline and was used for subsequent experiments.
Protein analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed in 13% (wt/vol) polyacrylamide, 1.5-mm-thick slab gels prepared by the method described by Laemmli (17). Samples were prepared for electrophoresis as described previously (20). Western blottings were performed essentially as described previously (13). For immunological detection of MDH, antibodies were used that had been raised against the holoenzyme. Proteins with covalently bound heme were stained with 3,3′,5,5′-tetramethylbenzidine by the method described by Thomas et al. (28).
FGH activity was measured as described previously (14). GD-FALDH activities were determined essentially as described by Van Ophem and Duine (29), but with a different buffer (250 mM Tris-HCl buffer, pH 8.8). β-Galactosidase activity was measured as described by Miller (19) with the minor modification that incubation with toluene was prolonged to 90 min. Protein determinations were performed by a modified Lowry method with bovine serum albumin as a standard (18).
Nucleotide sequence accession number.
The nucleotide sequences have been deposited with the EMBL Nucleotide Sequence Database under accession no. AJ223460.
RESULTS
Isolation of a regulatory mutant with a pleiotropic defect in C1 metabolism.
Our aim was to isolate a genetic locus involved in the regulation of flhA, fghA, and cycB expression and in activation of MxaX in an mxaY mutant. For this, we constructed a genomic reporter of cycB promoter activity by using a method that leaves the wild-type cycB gene intact (16). A DNA fragment containing the promoter region of cycB was cloned in the promoter-probe vector pBK16. The resulting plasmid, pPR611, was then integrated into the genome of the mxaY mutant Pd0841.hf at the site of the homology fragment (see Materials and Methods). Hybridization analysis confirmed the integration of the cycB promoter upstream of the lacZ gene in the homology fragment. The strain thus obtained, Pd0841hf611, expressed lacZ during growth on choline, but not on succinate, as judged by the appearance of blue and white colonies, respectively, on plates containing X-Gal.
We hypothesized that mutations in genes regulating C1 metabolism might also affect the expression of flhA, the gene that codes for GD-FALDH. Mutants with a defect in flhA are unable to grow on methanol, methylamine, and choline (21), and we reasoned that a regulatory mutant that is unable to express flhA would also not grow on these substrates. We therefore introduced a plasmid (pSK4) carrying a copy of the flhA gene downstream of the constitutive cycA promoter (31) into Pd0841hf611. Strains containing pSK4 expressed GD-FALDH constitutively as judged by (i) complementation of the flhA mutation in Pd6721 and (ii) expression of GD-FALDH to high levels, even under heterotrophic growth conditions (results not shown).
Pd0841hf611(pSK4) was then subjected to transposon mutagenesis, and mutants that were unable to express the cycB promoter-lacZ fusion during growth on choline were picked. Approximately 16 × 104 colonies were tested on choline–X-Gal plates. Seventeen white or light blue colonies were picked. One light blue colony, mutant Pd13(pSK4), was characterized further.
Characterization of the regulatory mutant.
Pd13(pSK4) is unable to grow on methanol and methylamine, while growth on choline, succinate, and formate and autotrophic growth are normal (Table 2). Curing of plasmid pSK4 resulted in a strain (Pd13) that is unable to grow on methanol, methylamine, or choline. A similar phenotype was also found for Pd6721, a mutant with an insertion in the flhA gene (21). Pd13 is able to express GD-FALDH in succinate grown cells to low levels, similar to those found in the wild-type strain (28 nmol of NADH min−1 mg of protein−1). Activities under inducing conditions (i.e., after growth on methanol, methylamine, or choline) could not be determined because the mutant does not grow on these substrates. The data indicate that the chromosomal flhA gene in Pd13 was not induced under C1 growth conditions above the basal level.
TABLE 2.
Growth characteristics of wild type and regulatory mutants of P. denitrificansa
| Strain | Relevant genotype | Presence of growth on:
|
|||||
|---|---|---|---|---|---|---|---|
| CH3OH | CH3NH2 | Choline | Formate | H2CO2 | Succinate | ||
| Pd1222 | Wild type | + | + | + | + | + | + |
| Pd0841 | ΔmxaY | + | + | + | + | + | + |
| Pd13(pSK4) | ΔmxaY flhR− pcycA::flhA | − | − | + | + | + | + |
| Pd13 | ΔmxaY flhR | − | − | − | + | + | + |
| Pd5021 | flhR::Km | − | − | − | ND | ND | + |
| Pd5021(pSK4) | flhR::Km pcycA::flAh | − | − | + | ND | ND | + |
| Pd5121 | flhS::Km | − | − | − | ND | ND | + |
| Pd5121(pSK4) | flhS::KM pcycA::flhA | − | − | + | ND | ND | + |
| Pd9234 | ΔmxaY flhR::Km | − | − | − | ND | ND | + |
| Pd9234(pSK4) | ΔmxaY flhR::Km pcycA::flhA | − | − | + | ND | ND | + |
| Pd9235 | ΔmxaY flhS::Km | − | − | − | ND | ND | + |
| Pd9235(pSK4) | ΔmxaY flhS::Km pcycA::flhA | − | − | + | ND | ND | + |
Growth was determined on solid media after 48 h of incubation at 35°C (choline or succinate) or 96 h of incubation (methanol, methylamine, formate, H2-CO2). +, growth; −, no growth; ND, not determined.
The constitutively expressed flhA gene on pSK4 complemented Pd13 for growth on choline, but not for growth on methanol or methylamine. This suggested that the mutation in Pd13 causes a pleiotropic defect in the expression of flhA, cycB, and other genes involved in C1 metabolism. It has been shown that a strain with a mutation in fghA is unable to grow on methanol and methylamine, while growth on choline is still possible (14). It is therefore possible that the expression of fghA is blocked by the mutation in Pd13. To investigate this hypothesis, we determined the FGH activity in Pd13(pSK4) grown on either choline or succinate. Pd13(pSK4) was able to synthesize FGH at levels comparable to the amount found in succinate-grown wild-type cells. This indicates that fghA can still be expressed at basal levels in Pd13, but that the strain has a defect in the choline-induced up-regulation (14).
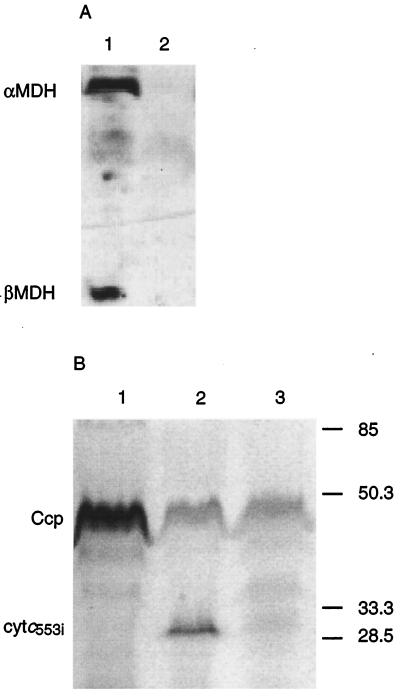
Although the phenotypes of Pd13 and Pd13(pSK4) could be explained by assuming that the expression of fghA was reduced by the mutation, it remained possible that expression of the mxa cluster was also affected. To investigate this, cell extracts were made from cells of Pd13(pSK4) grown on choline. Immunological analysis revealed that Pd13(pSK4) did not synthesize MDH under these conditions (Fig. 1a, lane 2), in contrast with the results obtained with the wild-type strain (Fig. 1a, lane 1), suggesting that regulation of the mxa gene cluster was indeed also affected by the mutation. In addition, the cycB promoter was not activated, as judged by the absence of cytochrome c553i (Fig. 1b, lane 3) and by the lack of expression of the pcycB-lacZ fusion (Table 3). The mutation in Pd13 appeared to be pleiotropic: it affected the metabolism of at least two substrates, methanol and formaldehyde. Since autotrophic growth with either hydrogen or formate as a free energy source was unaffected (Table 2), we concluded that the mutation does not control metabolism downstream of the formate oxidation pathway.
FIG. 1.
(A) Western blot analysis of α and β subunits of MDH from P. denitrificans wild type (lane 1) and mutant Pd13(pSK4) (lane 2). (B) Heme stain analysis of the cycB mutant Pd6121 (lane 1), wild type (lane 2), and mutant Pd13(pSK4) (lane 3). Molecular mass markers are indicated in kilodaltons. Ccp, cytochrome c peroxidase; cytc553i, cycB gene product.
TABLE 3.
Activities of several C1 promoters of P. denitrificans
| Strains plus pSK4 | Promoter | Activity on growth substratea
|
|
|---|---|---|---|
| Succinate | Choline + methylamine | ||
| Pd13 | cycB | 4 | 7 |
| Pd0841hf.611 | cycB | 3 | 101 |
| Pd1222::Pr071 | mxaZ | 18 | 15 |
| Pd5021::Pr071 | mxaZ | 21 | 18 |
| Pd1222::Pr501 | flhR | 21 | 20 |
| Pd5021::Pr501 | flhR | 24 | 14 |
Promoter activities are indicated in Miller units (19). Values represent the averages of two independent experiments, with each point assayed in duplicate.
Cloning and sequencing of the flhRS region from P. denitrificans.
Isolation and analysis of the region in which the mini-Tn5 transposon had integrated revealed that the plasmid carrying the transposon had formed a cointegrate with plasmid pSK4 as a consequence of a single recombination event that had taken place at the oriT locus of both plasmids. Apparently, the mutation in Pd13 causing the defects in C1 metabolism was not caused by a transposon insertion but rather by a spontaneous mutation. In order to isolate the corresponding gene, we transformed Pd13 with chromosomal fragments present in a genomic library of P. denitrificans. After conjugation of the gene library, colonies that were able to grow on choline were selected. One of these clones was found to contain a plasmid with a 23-kbp chromosomal DNA fragment. This plasmid, pR11.1, was subcloned, and a 5.9-kbp PstI fragment that was able to complement the mutation of Pd13 with respect to growth on choline, methanol, and methylamine was isolated. In addition, the plasmid restored activity of the cycB promoter to wild-type levels, as Pd13(pR11.1P8) formed dark blue colonies on choline–X-Gal plates. This result confirmed that a single locus was mutated in Pd13. The physical map of this region is shown in Fig. 2.
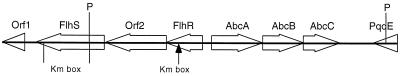
FIG. 2.
Physical map of the FlhRS region of P. denitrificans. Open arrows indicate ORFs. The 5.9-kbp fragment that is able to complement the mutation in Pd13 is indicated by a P. The positions of the kanamycin boxes in Pd5021 (flhR::Km) and Pd5121 (flhS::Km) are indicated.
Sequence analysis of the 5.9-kb complementing DNA fragment revealed eight open reading frames (ORFs). The protein sequence translated from orfl showed similarity (21% identity) with the N-terminal sequence of an uncharacterized protein (ORF7) found in Methylobacterium extorquens AM1 (4). The orf7 gene resides in a region separating two gene clusters involved in methylotrophic growth. ORF7 has no known role in C1 metabolism in M. extorquens AM1 (4). The deduced amino acid sequence encoded by orf8 has 48% identity with the C-terminal part of PqqE of M. extorquens AM1, one of the proteins that is involved in the biosynthesis of the PQQ cofactor of MDH. orf5, orf6, and orf7 translated into proteins that showed similarity with components of ATP-binding cassette (ABC)-type transporters. The highest degree of identity was found with the AbcABC proteins of M. extorquens AM1 (40, 46, and 33% identity, respectively) (4), and for this reason we used the same designation for the P. denitrificans counterparts.
The product of orf4 showed similarity over its entire length to proteins that belong to the family of aspartate kinase response regulators (27). The amino acid sequence has 26 to 28% identity with its closest relatives, which include E. coli NarL and NarP, Pseudomonas aeruginosa GlpR, and Burkholderia solanacearum VsrC (GenBank accession no. M24910, L11273, M60805, and U18134). We tentatively called this gene flhR, being the activator of, among others, flhA expression. FlhR showed, over its entire length, 23% identity with MxaX, the response regulator involved in methanol oxidation in P. denitrificans. The similarities with response regulators involved in methanol oxidation from other methylotrophs were lower (15 to 21% identity).
An ORF designated orf2 was found downstream of flhR, the product of which does not show any similarity with sequences in the databases. The start of the coding region of orf2 overlaps with the end of flhR, suggesting that the two genes are translationally coupled. Computer analysis of ORF2 predicted three membrane-spanning helices, suggesting that the putative protein is located in the membrane. Only 3 bp downstream of orf2 is found another ORF, tentatively designated flhS. The deduced amino acid sequence of flhS showed similarity with proteins from the family of bacterial histidine kinases, and more specifically to the family of signal sensors that contain not only a signaling and a transmitter domain but also a receiver domain. The similarity was most significant over the central transmitter domain of 200 amino acids. The highest identity (36 to 38%) was found with the central domain of VsrB from B. solanacearum and of RcsC from E. coli (accession no. A36929 and P14376, respectively). FlhS has a rather short N-terminal signaling domain without any recognizable membrane-spanning regions. Since a signal sequence was not found, we assume that FlhS is a cytoplasmic protein.
Sequence analysis of the mutation in PD13.
Since the 5.9-kbp DNA fragment that complements the mutation in Pd13 contains the complete flhR gene and only the 5′ end of flhS, we assumed that flhR was mutated in Pd13. In order to test that hypothesis, we cloned the flhRS locus of Pd13 by using a PCR-based approach and determined the sequence over its entire length. Comparison of this sequence with that of the wild type revealed a single base pair change at position 633 of the flhR gene, which converted the wild-type ATG methionine codon into a GTG valine codon. The methionine residue is located in the predicted DNA binding helix-turn-helix motif of FlhR, suggesting that this residue is important for the DNA binding properties of the regulator.
Isolation and characterization of FlhR- and FlhS-deficient mutants.
To analyze the function of the gene products of flhR and flhS, the genes were mutated by insertion of a kanamycin resistance marker gene. Mutations were introduced into the wild-type Pd1222 (generating Pd5021 and Pd5121, respectively) and into the MxaY mutant Pd0841 (generating Pd9234 and Pd9235, respectively), after which the strains were tested for their ability to grow on various carbon sources. Strains Pd5021, Pd5121, Pd9234, and Pd9235 were all unable to grow on methanol, methylamine, or choline, like Pd13 (Table 2). This phenotype was apparently not the consequence of an inability to oxidize formaldehyde, since introduction of pSK4 (which directs the constitutive synthesis of GD-FALDH) into these strains restored their potential to grow on choline, but not on methanol or methylamine (Table 2). Hence, the pleiotropic effects of the mutations in these strains are best explained by assuming that the FlhRS proteins regulate metabolism of the latter two substrates. Indeed, we could demonstrate by Western analyses that the mutant strains were unable to synthesize MDH, just like Pd13 (results not shown).
Promoter-lacZ fusions were made to analyze whether the expression of the regulatory genes mxaZYX is under control of the FlhRS two-component regulatory system and whether the expression of the FlhRS couple is subject to autoregulation. The activities of both the mxaZ and the flhR promoter are low and constitutive, irrespective of whether the strains are grown on succinate or on choline (Table 3). The results of the flhR promoter activity studies indicate that the FlhRS two-component system does not control the promoter of its own genes. The results with the mxaZ promoter confirm the earlier data found with an mxaZ-lacZ fusion on a multicopy plasmid (15) and indicate that this promoter is regulated neither by a C1 substrate nor by the FlhRS two-component system. The mxaY gene is therefore normally expressed in the Flh mutants. Since the FlhS mutant (Pd5121) is unable to grow on C1 substrates, we can exclude the possibility that MxaY, the signal sensor of the MxaYX pair, is able to cross-activate FlhR. The data from our studies indicate that the FlhRS proteins are required for the expression of several loci involved in C1 metabolism of P. denitrificans.
DISCUSSION
Here we report on the isolation and sequencing of the flhRS genes of P. denitrificans that are involved in regulation of C1 metabolism. FlhS and FlhR show strong similarity with sensor-regulator proteins from the family of two-component regulatory systems. Since FlhS is apparently located in the cytoplasm, its cognate signal is likely to be sensed intracellularly. The FlhRS proteins are required for methanol and for formaldehyde oxidation. We base this conclusion on the fact that expression of the mxa, flhA-fghA, and cycB genes was abolished in the corresponding mutants, while they were also unable to grow on methanol, methylamine, and choline.
Expression of the mxa operon encoding methanol dehydrogenase in P. denitrificans is also regulated by a two-component regulatory system composed of the transcriptional activator MxaX and the sensor MxaY (15). It has been hypothesized that periplasmically formed formaldehyde is the signaling molecule for the latter protein. Surprisingly, however, an mxaY-deficient mutant had a wild-type phenotype with respect to methanol oxidation, suggesting the presence of another sensor able to activate MxaX (35). It is not known how the two sensor-regulator pairs, MxaZYX and FlhRS, which are both necessary for expression of the mxa genes, interact. In M. extorquens AM1 and in Methylobacterium organophilum XX, mxa gene expression is controlled by more than one sensor-regulator pair (26, 33, 34). In M. extorquens AM1, a regulatory hierarchy was found in which the sensor-regulator pair MxcQE controls expression of the sensor-regulator pair MxbDM (26). The latter in turn controls expression of a number of other genes involved in methanol oxidation (25), but not that of genes involved in formaldehyde oxidation. The situation in P. denitrificans is different in that the promoter upstream of mxaZ is constitutive and not regulated by FlhRS. Further, a mutation in mxaX did not affect growth on methylamine and choline (15), indicating that MxaX is not required for the expression of the flhR promoter. Thus, it may be that both MxaYX and FlhRS regulate mxa expression in concert. The most reasonable explanation is that FlhS is able to phosphorylate MxaX in response to intracellular formaldehyde. This would also explain why MDH is synthesized in cells grown on methanol and methylamine (which are oxidized to formaldehyde in the periplasm) as well as on choline (which yields cytoplasmic formaldehyde upon its oxidation). In this scenario, FlhS and MxaY might be cytoplasmic and periplasmic formaldehyde sensors, respectively.
Since an FlhS-deficient mutant is unable to grow on C1 compounds and since the expression of the mxaZYX operon is not controlled by the FlhRS system, it can be concluded that MxaY is unable to complement the FlhS-deficient mutant and to activate FlhR. Whether cross-regulation between FlhS and MxaX occurs, however, could not be demonstrated by in vivo experiments carried out in this study. The reason for this is that an FlhS MxaY double mutant is unable to grow on methanol, since activation of both FlhR (by FlhS) and MxaX (by MxaY or FlhS) is necessary for growth on that substrate (Table 2).
Apart from the role of FlhRS in the regulation of formaldehyde formation, the FlhRS system is also necessary for expression of the formaldehyde oxidation system. A mutant lacking a functional FlhRS system, Pd13(pSK4) grown on choline, was able to express basal levels of FGH and GD-FALDH, but activation to wild-type levels was not found. The presence of basal levels of these enzymes in Pd13(pSK4) growing on succinate may be the result of the basal activity of the promoter that is only activated by FlhR∼P upon sensing of formaldehyde by FlhS.
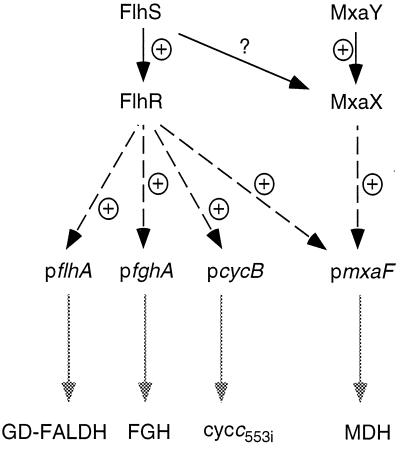
The FlhRS two-component regulatory system as described in this paper seems to have a key role in controlling the concentration of the toxic compound formaldehyde. For the type of pathways described here, and as shown in Fig. 3, one expects a regulatory system that can switch the pathways from “off” to “on” and in addition can control the activity of the formaldehyde producer (MDH) relative to that of the formaldehyde consumers (GD-FALDH, FGH). The latter regulation is necessary to avoid the accumulation of toxic amounts of formaldehyde. Indeed, the sensor-regulator pair FlhRS fulfills these roles and, consequently, mutations in this system have a pleiotropic effect. In contrast to this global regulation, MxaX has a specific role in regulation of the mxa gene cluster.
FIG. 3.
Model of the regulatory network controlling the expression of the structural genes encoding MDH, GD-FALDH, FGH, and cytochrome c553i. Possible protein-protein interactions are indicated by solid lines, protein-DNA interactions are indicated by dashed lines, and DNA expression is indicated by dotted lines. p, promoter; +, positive interactions; ?, possible cross talk between the two two-component regulatory systems.
ACKNOWLEDGMENT
We thank S. Spiro for providing the genomic library of P. denitrificans and for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. 1 and 2. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1989. [Google Scholar]
- 3.Chang J P, Morris J G. Studies on the utilization of nitrate by Micrococcus denitrificans. J Gen Microbiol. 1962;29:301–310. doi: 10.1099/00221287-29-2-301. [DOI] [PubMed] [Google Scholar]
- 4.Chistoserdova L, Lidstrom M. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology. 1997;143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- 5.Crossman L C, Moir J W B, Enticknap J J, Richardson D J, Spiro S. Heterologous expression of heterotrophic nitrification genes. Microbiology. 1997;143:3775–3783. doi: 10.1099/00221287-143-12-3775. [DOI] [PubMed] [Google Scholar]
- 6.Delorme C, Huisman T T, Reijnders W N M, Chan Y-L, Harms N, Stouthamer A H, Van Spanning R J M. Expression of the mau gene cluster of Paracoccus denitrificans is controlled by MauR and a second transcription regulator. Microbiology. 1997;143:793–801. doi: 10.1099/00221287-143-3-793. [DOI] [PubMed] [Google Scholar]
- 7.De Vries G E, Harms N, Maurer K, Papendrecht A, Stouthamer A H. Physiological regulation of Paracoccus denitrificans methanol dehydrogenase synthesis and activity. J Bacteriol. 1988;170:3731–3737. doi: 10.1128/jb.170.8.3731-3737.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Vries G E, Harms N, Hoogendijk J, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a n(GATC)n DNA-modifying property. Arch Microbiol. 1989;152:52–57. [Google Scholar]
- 9.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X, Finlay D, Guiney D, Helinski D. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 10.Gerhus E, Steinrücke P, Ludwig B. Paracoccus denitrificans cytochrome c1 gene replacement mutants. J Bacteriol. 1990;172:2392–2400. doi: 10.1128/jb.172.5.2392-2400.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gish W, David J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 12.Harms N, De Vries G E, Maurer K, Hoogendijk J, Stouthamer A H. Isolation and nucleotide sequence of the methanol dehydrogenase structural gene from Paracoccus denitrificans. J Bacteriol. 1987;169:3969–3975. doi: 10.1128/jb.169.9.3969-3975.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms N, De Vries G E, Maurer K, Veltkamp E, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with defects in the metabolism of one-carbon compounds. J Bacteriol. 1985;164:1064–1070. doi: 10.1128/jb.164.3.1064-1070.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms N, Ras J, Reijnders W N M, Van Spanning R J M, Stouthamer A H. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J Bacteriol. 1996;178:6296–6299. doi: 10.1128/jb.178.21.6296-6299.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harms N, Reijnders W N M, Anazawa H, Van der Palen C J N M, Van Spanning R J M, Oltmann L F, Stouthamer A H. Identification of a two component regulatory system controlling methanol dehydrogenase synthesis in Paracoccus denitrificans. Mol Microbiol. 1993;8:457–470. doi: 10.1111/j.1365-2958.1993.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 16.Kessler B, de Lorenzo V, Timmis K N. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet. 1992;233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Markwell M K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Ras J, Reijnders W N M, Van Spanning R J M, Harms N, Oltmann L F, Stouthamer A H. Isolation, sequencing and mutagenesis of the gene encoding cytochrome c553i of Paracoccus denitrificans and characterization of the mutant strain. J Bacteriol. 1991;173:6971–6979. doi: 10.1128/jb.173.21.6971-6979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ras J, Van Ophem P W, Reijnders W N M, Van Spanning R J M, Duine J A, Stouthamer A H, Harms N. Isolation, sequencing and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J Bacteriol. 1995;177:247–251. doi: 10.1128/jb.177.1.247-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sanger F, Coulson R, Barrel B G, Smith J H, Roe B A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980;143:161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- 24.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulations of Gram-negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria-plant interactions. Berlin, Germany: Springer-Verlag KG; 1983. pp. 98–106. [Google Scholar]
- 25.Springer A L, Chou H-H, Fan W-H, Lee E, Lidstrom M E. Methanol oxidation mutants in Methylobacterium extorquens AM1: identification of new genetic complementation groups. Microbiology. 1995;141:2985–2993. doi: 10.1099/13500872-141-11-2985. [DOI] [PubMed] [Google Scholar]
- 26.Springer A L, Morris C J, Lidstrom M E. Molecular analysis of mxbD and mxbM, a putative sensor-regulator pair required for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology. 1997;143:1737–1744. doi: 10.1099/00221287-143-5-1737. [DOI] [PubMed] [Google Scholar]
- 27.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas P E, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P450 on SDS-polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 29.Van Ophem P W, Duine J A. NAD- and co-substrate (GSH or factor)-dependent formaldehyde dehydrogenases from methylotrophic microorganisms act as a class III alcohol dehydrogenase. FEMS Microbiol Lett. 1994;116:87–94. [Google Scholar]
- 30.Van Spanning R J M, Wansell C W, De Boer T, Hazelaar M J, Anazawa H, Harms N, Oltmann L F, Stouthamer A H. Isolation and characterization of the moxJ, moxG, moxI and moxR genes of Paracoccus denitrificans. Inactivation of the moxJ, moxG, and moxR genes and the resultant effect on methylotrophic growth. J Bacteriol. 1991;173:6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Spanning R J M, Wansell C W, Harms N, Oltmann L F, Stouthamer A H. Mutagenesis of the gene encoding cytochrome c550 of Paracoccus denitrificans and analysis of the resultant physiological effects. J Bacteriol. 1990;172:986–996. doi: 10.1128/jb.172.2.986-996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 33.Xu H H, Janka J J, Viebahn M, Hanson R S. Nucleotide sequence of the mxcQ and mxcE genes, required for methanol dehydrogenase synthesis in Methylobacterium organophilum XX: a two-component regulatory system. Microbiology. 1995;141:2543–2551. doi: 10.1099/13500872-141-10-2543. [DOI] [PubMed] [Google Scholar]
- 34.Xu H H, Viebahn M, Hanson R S. Identification of methanol-regulated promoter sequences from the facultative methylotrophic bacterium Methylobacterium organophilum XX. J Gen Microbiol. 1993;139:743–752. doi: 10.1099/00221287-139-4-743. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Reijnders W N M, Van Spanning R J M, Stouthamer A H, Harms N. Expression of the structural mox genes in Paracoccus denitrificans follows wild-type regulation in mutants with a deletion in mxaY, the gene encoding the signal sensor. Microbiology. 1995;141:825–830. [Google Scholar]