Abstract
Acetate kinase, an enzyme widely distributed in the Bacteria and Archaea domains, catalyzes the phosphorylation of acetate. We have determined the three-dimensional structure of Methanosarcina thermophila acetate kinase bound to ADP through crystallography. As we previously predicted, acetate kinase contains a core fold that is topologically identical to that of the ADP-binding domains of glycerol kinase, hexokinase, the 70-kDa heat shock cognate (Hsc70), and actin. Numerous charged active-site residues are conserved within acetate kinases, but few are conserved within the phosphotransferase superfamily. The identity of the points of insertion of polypeptide segments into the core fold of the superfamily members indicates that the insertions existed in the common ancestor of the phosphotransferases. Another remarkable shared feature is the unusual, epsilon conformation of the residue that directly precedes a conserved glycine residue (Gly-331 in acetate kinase) that binds the α-phosphate of ADP. Structural, biochemical, and geochemical considerations indicate that an acetate kinase may be the ancestral enzyme of the ASKHA (acetate and sugar kinases/Hsc70/actin) superfamily of phosphotransferases.
Phosphoryl transfer is the most common enzymatic function encoded by the yeast genome (12), and the reaction is catalyzed by central regulatory enzymes, such as protein kinases, ATPases, and GTPases (7). A number of aspects of the mechanism of enzyme-catalyzed phosphoryl transfer are still incompletely understood and are a source of ongoing controversy (34). However, X-ray crystallographic studies of phosphotransferases are making a critical contribution to our understanding of this central reaction. They also ground evolutionary analyses of these enzymes (7, 23, 29).
Acetate kinase, discovered in 1944 by Lipmann (32) and isolated in 1954 by Ochoa et al. (41), is the prototypic carboxylate kinase and one of the earliest phosphoryltransfer enzymes to be recognized. Acetate kinase is widespread in both anaerobic and aerobic microbes of the Bacteria and Archaea domains and a central player in a major link in the global carbon cycle, the anaerobic decomposition of organic matter to methane, in which it performs a dual role (14). In the first step of methane production by microbial consortia, fermentative anaerobes from the Bacteria domain degrade complex organic matter to acetate. Acetate kinase catalyzes the final reaction in this process, conversion of acetyl phosphate and ADP into acetate and ATP. Anaerobes from the Archaea domain then convert the acetate into methane and carbon dioxide. In this second process, acetate kinase catalyzes the first reaction, activation of acetate to acetyl phosphate.
Acetyl phosphate is not only a precursor of important metabolic intermediates, such as acetyl coenzyme A (acetyl-CoA), but also a potential regulator of bacterial signal-transduction pathways. Bacterial responses to changes in environmental conditions are most commonly evoked through two-component regulatory systems consisting of a sensor kinase that autophosphorylates on a histidine residue and a response regulator (39). The response regulator is an enzyme that catalyzes the transfer of phosphate from the histidine residue of the sensor protein to its own active site asparate residue (42, 43). The active conformation of the response regulator for its regulatory function is the phosphoenzyme intermediate. It has been demonstrated that the response regulators can directly utilize acetyl phosphate but not ATP as a phosphoryl donor (13, 33). A number of studies have indicated that cellular levels of acetyl phosphate may regulate the in vivo function of response regulators through modulation of their phosphorylation state (6, 13, 21, 35, 36, 40, 44, 51).
One incompletely elucidated issue concerning the mechanism and evolution of acetate kinase is whether there are one or more covalent phosphoenzyme intermediates formed during catalysis by acetate kinase. In the presence of either ATP or acetyl-phosphate, Escherichia coli acetate kinase becomes phosphorylated on the side chain of one or more of its glutamate residues (49). The phosphoenzyme is relatively stable and can be isolated. The rate of phosphoenzyme formation is comparable to the rate of the overall reaction (19). The isolated phosphoenzyme is able to transfer its phosphoryl group to either of the normal substrates, ADP and acetate (2–4, 19), as well as to the active site of Enzyme I of the phosphotransferase system (18). This evidence argues that the acyl-phosphate form of the enzyme is a covalent intermediate in catalysis. However, it has been demonstrated that the phosphoryl group is transferred by E. coli acetate kinase with inversion of configuration (5). Such data are typically taken as evidence for a direct, in-line transfer of phosphate from substrate to product without an enzyme-linked covalent intermediate (28). Possible resolutions to the conflict in data have been discussed (10, 47, 48), but additional detailed structural and modern biochemical studies are required.
An additional issue is the evolutionary relationship between acetate kinase and other phosphotransferases. The only other enzymes that are identified as similar to the acetate kinases by sequence comparison programs are the propionate and butyrate kinases (10, 20, 50). However, we have postulated, through secondary-structure prediction based upon comparative sequence analysis, that acetate kinase would possess a common topology with that of glycerol kinase, hexokinase, actin, and the 70-kDa heat shock cognate (Hsc70) (10).
In order to address these biological and biochemical questions, we have solved the structure of Methanosarcina thermophila acetate kinase by crystallography. The view of the active site of acetate kinase identifies residues for which roles in catalysis can be postulated. In addition, study of the structure has provided ideas about the early appearance of acetate kinase in evolution.
MATERIALS AND METHODS
Purification and crystallization.
Expression and purification of homogeneous acetate kinase from M. thermophila (1, 31, 37), as well as conditions for the crystallization of the native enzyme bound to ATP (10), have been described previously. Selenomethionyl crystals, also space group C2, required incubation at 20°C for a minimum of 3 weeks, followed by transfer to 37°C.
Data collection and model building.
Selenomethionyl data were collected at Advanced Photon Source beamline BM14D. The multiple isomorphous replacement (MIR) data were collected at 277K and the multiwavelength anomalous diffraction (MAD) data were collected at 100K. All data were processed with DENZO/SCALEPACK (38). Programs in the CCP4 suite (11) were used for phasing and phase refinement. Patterson maps were used to locate heavy atoms in MIR. To locate selenium atoms in MAD, a map was calculated with MIR phases and the anomalous difference from the peak MAD wavelength (0.9794 Å). Initial phases and positions of the MIR and MAD solutions were refined in the CCP4 program MLPHARE. Multicrystal density modification was performed using DMMULTI. The model was built using the program O (25). Refinement was carried out using XPLOR (8) followed by CNS (version 1.0) (9). Model quality was checked using Procheck (30). Noncrystallographic symmetry information was used in phase and model refinement.
Coordinates.
The coordinates and structure factors are deposited in the Protein Data Bank as 1G99.
RESULTS AND DISCUSSION
Architecture of acetate kinase.
The structure of M. thermophila acetate kinase was solved through the combination of two crystallographic methods (Tables 1 and 2). Electron density maps produced independently through either MIR or MAD using selenomethionine-substituted protein were incomplete. Multicrystal density modification was performed using the initial phases from both techniques and treating each domain as an independent group. This treatment significantly improved the quality of the electron density maps, allowing 96% of the structure to be built into the native-protein map during the first round of model building.
TABLE 1.
Crystallographic data collection statisticsa
| Parameter | Type of crystal and wavelength
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Native 1.5418 | EMP 1.5418 | IADP 1.5418 | EMP/IADP 1.5418 | TMA 1.5418 | Selenomethionyl acetate kinase
|
|||
| 0.9796 | 0.9794 | 0.9537 | ||||||
| a (Å) | 181.3 | 181.0 | 181.0 | 181.0 | 180.7 | 178.9 | ||
| b (Å) | 67.4 | 67.3 | 67.2 | 67.5 | 67.4 | 66.4 | ||
| c (Å) | 82.6 | 82.3 | 82.4 | 82.6 | 82.5 | 83.8 | ||
| β (°) | 102.9 | 103.0 | 102.9 | 102.7 | 103.2 | 102.9 | ||
| Resolution (Å) | 2.5 | 3.2 | 2.8 | 2.8 | 3.3 | 3.0 | ||
| No. of reflections | ||||||||
| Total | 239854 | 109479 | 71759 | 74469 | 78326 | 73070 | 73162 | 65482 |
| Unique | 65658 | 17719 | 23999 | 24337 | 14766 | 18745 | 18722 | 18625 |
| Completeness (%) | 91.2 | 99.4 | 93.7 | 86.2 | 100 | 99.7 | 99.7 | 99.6 |
| Rsym (%) | 6.6 | 10.3 | 9.1 | 8.4 | 11.0 | 8.9 | 9.3 | 11.4 |
| Riso (%) | 20.0 | 16.0 | 18.0 | 20.3 | ||||
| No. of sites | 3 | 2 | 5 | 5 | ||||
| Phasing power | 1.4 | 0.7 | 1.1 | 1.0 | ||||
EMP, ethylmercury phosphate; IADP, 2-iodo-adenosine-5′-diphosphate; TMA, trimethyl lead acetate. Rsym = Σ|I − <I>|/Σ<I>, where I is intensity. Riso = [Σ(|FPH| − |FP|)]/Σ|FP|), where FPH and FP are the structure factors for the heavy-atom derivative and the native protein, respectively. Phasing power = FH/|FPHi − |FP + FH||.
TABLE 2.
Structural refinement statisticsa
| Parameter | Value |
|---|---|
| Final Rwork | 16.0% |
| Final Rfree | 19.2% |
| Highest resolution | 2.5 Å |
| Deviation from ideality | |
| rmsd in bond lengths | 0.006 Å |
| rmsd in bond angles | 1.2° |
| Ramachandran statistics | |
| Most favored phi-psi | 90.6% |
| Additionally allowed | 9.1% |
| Generously allowed | 0.3% |
| Average B values | |
| Protein (main chain) | 24.4 Å2 |
| Protein (side chains) | 28.9 Å2 |
| Ligands | 70.8 Å2 |
| Waters | 27.7 Å2 |
rmsd, root mean square deviation. Rwork = Σ|Fo − Fc|/Σ|Fo|, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is the cross-validation R factor calculated with 5% of the data omitted from the refinement.
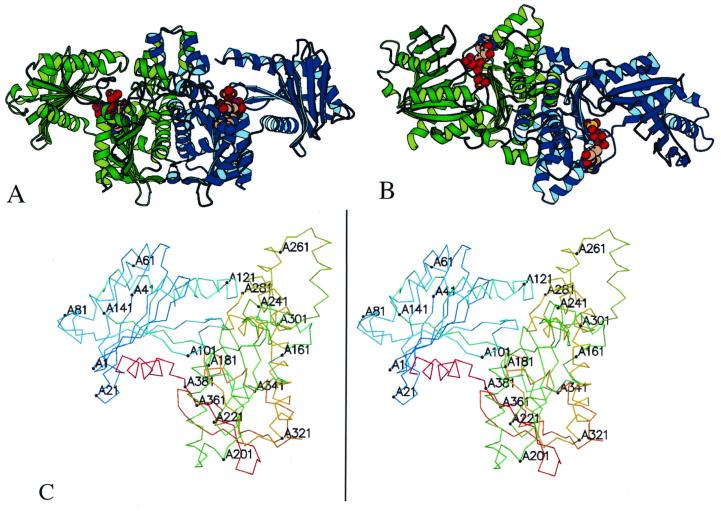
The overall structure of the acetate kinase dimer resembles a bird with its wings spread (Fig. 1A). The body of the bird is formed by the C-terminal domains of the monomers and contains the dimer interface, while the wings are composed of the N-terminal domains. The two monomers in the dimer are related by a noncrystallographic twofold rotation axis. Each monomer consists of two domains, each consisting of a central β-sheet surrounded by α-helices. The fold of the N-terminal domain consists of an eight-stranded β-sheet and eight helices. The C-terminal domain is composed of a seven-stranded β-sheet, eleven helices, and an additional small two-stranded β-sheet. The nucleotide-binding site is located in the cleft between the domains (Fig. 1B).
FIG. 1.
Structure of acetate kinase. The structure of the acetate kinase dimer (A) and a view with a 90° rotation around a horizontal axis (B) are shown. The two monomers of the dimer are shown in green and blue. The C-terminal domains, at the center, form the dimer interface. The ADP and sulfate molecules in the active site (between the N and C domains) are shown in space-filling models. The structure contains 801 of the 816 residues in the dimer, with the missing residues located at solvent-exposed regions following the C-terminal helix. (C) Stereoview of monomer A of acetate kinase, numbered every 20 residues.
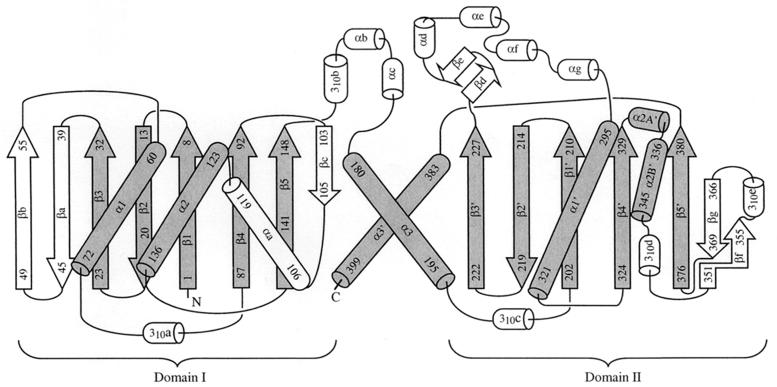
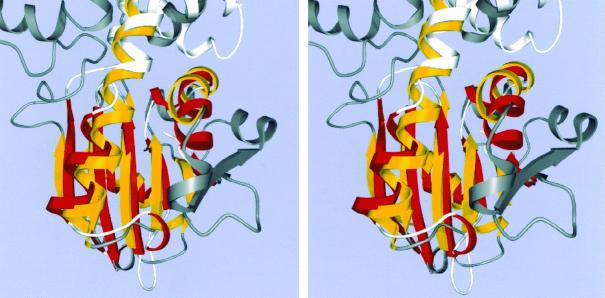
As we had predicted despite the absence of sequence identity (10), the fold of acetate kinase (Fig. 2) contains a core that is identical to that of the glycerol kinase/hexokinase/actin/Hsc70 superfamily (15–17, 23, 24, 26). Henceforth this family will be referred to as the ASKHA (acetate and sugar kinases/Hsc70/actin) superfamily of phosphotransferases. The ASKHA core consists of a duplicated βββαβαβα secondary structure with insertions of subdomains between particular elements of the β-sheet. As has been noted before, the C-terminal α-helix of each of these domains should properly be considered a part of the other domain (15). The alpha-carbon positions of 66 pairs of structurally equivalent amino acid residues within the C-terminal domains of acetate kinase and Hsc70 can be superimposed with a root mean square deviation of 2.08 Å (Fig. 3).
FIG. 2.
Topology diagram of acetate kinase. Secondary structures conserved in the ASKHA family (the duplicated βββαβαβα core) are rendered gray, and the inserts are shown in white. In standard nomenclature, the βββαβαβα subdomains are denoted IA (left) and IIA (right). In acetate kinase, the core β strands in domain IA are numbered 1 to 5, whereas those in domain IIA are numbered 1′ to 5′.
FIG. 3.
Stereoview of the superposition of the C-terminal domains of acetate kinase and of Hsc 70. The conserved ASHKA core is colored yellow for acetate kinase (with the remainder dark gray) and red for Hsc 70 (with the remainder white). The graphics program O was used to calculate the superposition matrix. The terminal helix, which extends into the N-terminal domain, is not shown.
Similar architectural plans of related enzymes.
A detailed analysis of the secondary structure of acetate kinase is central to an understanding of this diverse family of proteins. Acetate kinase contains subdomains inserted between the third strand of each of the βββαβαβα cores and the first α-helix (the sites of subdomains IB and IIB in actin and Hsc70) (Fig. 2). In domain I, this insertion consists of a pair of β-strands that extend the sheet. Our analysis of the sequences of the E. coli acetate and propionate kinases and those of the butyrate kinases of various species predicts that these strands would be either absent or profoundly shortened in the structures of those proteins. An insertion in domain II (subdomain IIB) largely forms the dimer interface as is true for the analogous insertions in the other ASKHA polypeptides (15–17, 23, 24, 26). In acetate kinase it is associated with subdomain IC, which is inserted before α3 of domain I. An insertion at this site is unique to acetate kinase among the ASKHA family.
Acetate kinase, similarly to glycerol kinase and hexokinase, contains an insertion between β4 and α2. It consists of an additional β-strand that extends the subdomain IA β-sheet and an additional α-helix. Between α2′ and β5′ are inserted a β-strand, which forms the edge of the β-sheet, an α-helix, and a β-strand that borders β5′. This insertion is unique to the acetate kinases. Our structural prediction indicates that the elements inserted between α2′ and β5′ will not be present in the butyrate kinases.
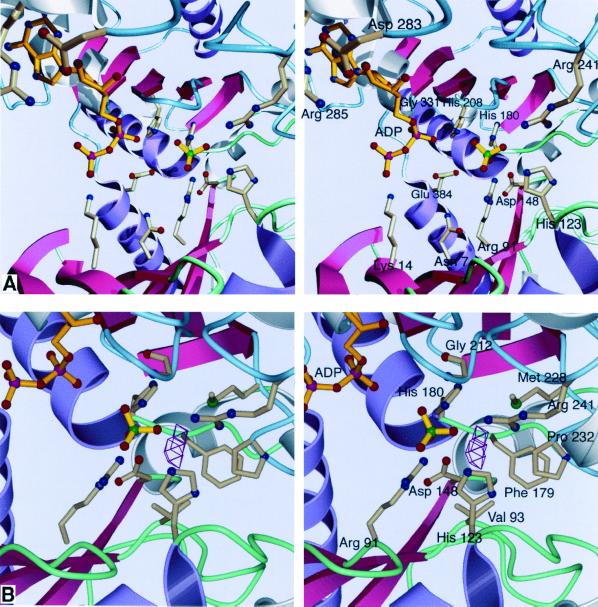
In the crystal, the active-site clefts in the two monomers of the acetate kinase dimer (monomers A and B) are closed to different extents. In addition, there is a sulfate ion in the active site resulting from the crystallization conditions. Nevertheless, we can examine some aspects of the active site that are revelatory of the mechanism and evolution of the ASKHA phosphotransferases. Although the crystals were grown in the presence of ATP, only two phosphates can be observed in the electron density. The β-phosphate of ADP appears to point away from the active site, perhaps resulting from the binding of a sulfate ion in the active site, which repels the β-phosphate from a site it would normally occupy. We propose that the sulfate ion occupies the site of the phosphate of acetyl phosphate. The sulfate ion binds to the side chains of arginine-91, histidine-123, histidine-180, and arginine-241 and the amide proton of glycine-212 (Fig. 4A); all are conserved within the acetate kinase-butyrate kinase family.
FIG. 4.
Stereoviews of the active site of acetate kinase. Conserved ASKHA secondary structures are pink and purple, and inserted secondary-structural elements are gray. Loops in the N-terminal domain are green, and those in the C-terminal domain are blue. (A) The binding site. It is likely that the sulfate occupies the position where the phosphate of acetyl phosphate would bind. No magnesium ion is apparent in our current structure despite the inclusion of 750 μM MgCl2 in the crystallization conditions. (B) The proposed site of acetate binding. VOIDOO (27) was used to locate solvent-accessible cavities. The cavity shown could easily accommodate the methyl group of acetate or acetyl phosphate, positioning the phosphate roughly where the sulfate is located. As shown, the center of the cavity is 4.2 Å away from the sulfate.
Design of the active site.
The adenine base of the nucleotide is bound in a hydrophobic pocket consisting of the aliphatic chains of arginine-285, isoleucine-332, and isoleucine-339. Arginine-285 is conserved throughout the acetate kinase family (the guanidinium group makes an electrostatic interaction with the carboxylate of the conserved aspartate-283), whereas the isoleucines can be substituted with other aliphatic residues in other family members. The isoleucine residues are from the same part of the polypeptide chain, a turn of a helix that follows β4′ and α2′, that forms the hydrophobic nucleotide base binding pocket in glycerol kinase, actin, and Hsc70. There are no single hydrogen bonds between the adenine and the protein. The adenine amino group is exposed to solvent. The lack of specific contacts with the base may explain the lack of specificity for a particular nucleotide triphosphate as the phosphoryl donor. The ribose ring is bound by phenylalanine-284 and the 2′ hydroxyl by the carboxylate of the absolutely conserved aspartate-283.
The α-phosphate is bound by the amide of glycine-331 (Fig. 4A); a similar interaction is seen with equivalent glycine residues in the other family members (G-411, G-302, and G-339 in glycerol kinase, actin, and Hsc70, respectively [15–17, 23, 24, 26]). Remarkably, this glycine is preceded by an alanine residue that is in the epsilon conformation (φ = 75.4°, ψ = 175.3°). This unusual conformation is also found in the residue that precedes the α-phosphate-binding glycine residues and initiates a turn of a helix in all of the ASKHA members. It is striking that the conformation of these residues, which has not been noted in previous publications, has been conserved over the course of the evolution of this superfamily.
The β-phosphate is bound by histidine-208 and the amides of asparagine-211 and glycine-212 (in monomer B), which are part of a loop between β1′ and β2′ that corresponds to a loop with a similar role in the other members of the ASKHA family. It is intriguing that the backbone carbonyl of asparagine-211 interacts through a water molecule with arginine-241, which binds the sulfate ion. Perhaps the binding of nucleotide is communicated through aspargine-211 to orient the conserved arginine-241 in the acetyl-phosphate binding site.
Two components of the interaction between the protein and the nucleotide that are observed in other ASKHA phosphotransferases but are absent in our structure are (i) the interaction between the loop between β1 and β2 and the β-phosphate of ATP and (ii) the conserved magnesium binding site. However, a movement of the loop of approximately 3 Å, similar to those observed in the other ASKHA phosphotransferases, would close the cleft between the two domains in monomer A and would position the amide protons of the residues in the loop, the ɛ-amino group of lysine-14, and the carbonyl group of asparagine-7 (an aspartate residue in the other ASKHA phosphotransferases that coordinates the nucleotide-bound magnesium ion) in the active site.
Aspartate-148, which is in a loop following β5, is likely to participate in magnesium ion coordination as do the carbonyl-possessing side chains of residues at identical sites in the sequence of the other ASKHA phosphotransferases. Interestingly, it has been suggested that the aspartate residues at this position in glycerol kinase and hexokinase (aspartate-245 and aspartate-211, respectively) also function as the catalytic bases for sugar-substrate deprotonation (23, 24). In acetate kinase, following aspartate-148, there is a helical insert (domain IC) which is unique to acetate kinase and which projects away from the core polypeptide fold. This insert forms essentially a closed loop that positions the absolutely conserved histidine-180 adjacent to aspartate-148. As discussed above, histidine-180 is bound to the sulfate ion that we propose occupies the acetyl phosphate-binding site. It appears that during the course of the evolution of the kinases, two functions, one in catalysis and the other in magnesium ion binding, which are carried out by two different residues that are spatially adjacent in acetate kinase, were imposed on a single residue in glycerol kinase and hexokinase.
Glutamate-384 is absolutely conserved among the acetate and butyrate kinases, is essential for function (45), and is probably the site of phosphorylation that has been detected previously. In our structure it lies at the N-terminal end of α3′. The side chain carboxylate points towards the active site but is 5.9 Å from the sulfate ion and 6.5 Å from the β-phosphate (Fig. 4A). However, as we have noted above, α3′ is in fact a part of the N-terminal domain, and cleft closure would bring glutamate-384 into the active site, where it could participate directly in catalysis.
Acetyl phosphate binding site.
Our structure also permits us to model the acetate and acetyl phosphate binding site. We predict that the methyl group of acetate will be bound between the side chains of valine-93, phenylalanine-179, and methionine-228 and the cyclopentyl ring of proline-232 (Fig. 4B). These residues are virtually completely conserved in the acetate kinases but differ in the butyrate kinases. Interestingly, valine-93, which lies at the base of the proposed acetate-binding pocket is replaced with an alanine residue in the propionate kinases of E. coli and Salmonella enterica serovar Typhimurium. We suggest that this substitution creates the space for the additional methylene group in propionate as compared to acetate and is largely responsible for the altered substrate specificity of the propionate kinases. The orientation of the sulfate ion ligands suggests that arginine-241 will be directly involved in acetate binding and that arginine-91, histidine-123, and histidine-180 will be involved in binding to the phosphate moiety of acetyl phosphate. The results of site-directed mutagenesis and chemical rescue experiments support the essential roles of arginine-91 and arginine-241 in substrate binding, most probably to acetate and acetyl phosphate (46).
Acetate kinase, an ancient enzyme.
The ASKHA phosphotransferase family has undergone extensive divergent evolution. Nevertheless, a number of elements appear to have been conserved: (i) a two-domain structure containing duplicated core secondary structure elements, (ii) residues that bind the catalytic magnesium ion and the nucleotide α-phosphate (the unusual conformation of the residue preceding the glycine that binds the α-phosphate and the following turn of a helix are also conserved), and (iii) points of insertion of secondary-structure elements into the core fold.
The last item suggests a scenario for the evolution of the ASKHA enzymes. The common ancestor of these enzymes was a protein containing a duplication of the core βββαβαβa fold that had an insertion between β3 and α1, considering the universality of insertions at this position. Extending this analysis, we note that the kinases all possess inserted sequences between β4 and α2, whereas the ATPases in the ASKHA family do not. The superfamily probably originally evolved to transfer phosphoryl groups to substrates rather than to hydrolyze nucleotide triphosphates; therefore, this insertion was present in the common ancestor of the ASKHA enzymes and deleted during the evolution of the ATPases. The butyrate kinases provide an example of the deletion of sequences inserted into the core, for they have unquestionably evolved comparatively recently (as indicated by their limited phylogenetic distribution and high sequence identity) from the acetate kinases and have eliminated certain peripheral secondary-structure elements. Acetate kinase also has an insertion between β5 and α3, which brings into proximity two conserved residues (histidine-180 and aspartate-148) whose functions during further evolution of the enzymes were essentially incorporated into a single residue in glycerol kinase and hexokinase.
We hypothesize that the structure of acetate kinase that we have determined represents the best approximation of the common ancestor of the ASKHA superfamily. This hypothesis is consistent with the biochemistry of acetate kinase and the posited role of acetate in the early stages of the evolution of life. It possesses properties associated with “primitive” enzymes; the Km of M. thermophila acetate kinase for its substrates (2.8 mM for ATP and 22 mM for acetate) is quite high compared to that of the other ASKHA enzymes, and it lacks specificity for ATP relative to other nucleotide triphosphates (1).
Short-chain carboxylic acids were among the most common organic molecules in prebiotic and early biotic environments. Acetic acid has the interesting property of being capable of diffusing relatively freely across membranes; the retention of acetate as a biosynthetic precursor within a cell would require its phosphorylation. This also activates the acetate for biosynthetic reactions. Acetyl phosphate and CoA form acetyl-CoA in a reaction now catalyzed enzymatically by a phosphotransacetylase. Acetyl-CoA is the common precursor of fatty acid synthesis, so it is plausible that early cellular growth relied on retention and activation of acetate through phosphorylation.
Another scenario of the role of acetate kinase in the early stages of the evolution of life depends upon the chemoautotrophic theory of the origin of life. Early organisms fed through fixation of CO or CO2 at volcanic or hydrothermal sites with the formation of thioacetic acid in an analogous process to that carried out by the enzyme acetyl-CoA synthetase (22). If early organisms could synthesize acetyl phosphate from such thioesters, including acetyl-CoA itself, then ATP synthesis might have proceeded through the acetate kinase reaction. The secreted acetic acid could then be taken up by another early organism through acetate kinase-mediated phosphorylation in a primitive form of the metabolic cooperation observed between microbial species to the present day.
ACKNOWLEDGMENTS
Kathryn A. Buss and David R. Cooper contributed equally to this work.
We thank our colleagues in the Markey Structural Group at Purdue.
This work was supported by an NIH Biophysics Training Grant to K.A.B. and D.R.C. an American Cancer Society grant to D.C., a Department of Energy-Basic Energy Sciences grant to J.G.F., an NSF CAREER awards to D.A.S. and M.S.H., a March of Dimes and David and Lucille Packard Foundation Fellowship to M.S.H., and an NIH Cancer Center Support at Purdue University. The diffraction and computing facilities shared by the Structural Biology group at Purdue have been developed and supported by grants from NIH, NSF, the Lucille P. Markey Foundation, the Keck Foundation, and the office of the university executive vice president for academic affairs. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, and the Office of Energy Research. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources.
REFERENCES
- 1.Aceti D J, Ferry J G. Purification and characterization of acetate kinase from acetate-grown Methanosarcina thermophila. J Biol Chem. 1988;263:15444–15448. [PubMed] [Google Scholar]
- 2.Anthony R S, Spector L B. Exchange reactions catalyzed by acetate kinase. J Biol Chem. 1971;246:6129–6135. [PubMed] [Google Scholar]
- 3.Anthony R S, Spector L B. A phosphoenzyme intermediary in acetate kinase action. J Biol Chem. 1970;246:6730–6741. [PubMed] [Google Scholar]
- 4.Anthony R S, Spector L B. Phosphorylated acetate kinase. Its isolation and reactivity. J Biol Chem. 1972;247:2120–2125. [PubMed] [Google Scholar]
- 5.Blättler W A, Knowles J R. Stereochemical course of phosphokinases. The use of adenosine [γ-(S)-16O, 17O, 18O]triphosphate and the mechanistic consequences for the reactions catalyzed by glycerol kinase, hexokinase, pyruvate kinase, and acetate kinase. Biochemistry. 1979;18:3927–3933. doi: 10.1021/bi00585a013. [DOI] [PubMed] [Google Scholar]
- 6.Bouche S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol. 1998;27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 7.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 8.Brunger A T. XPLOR: A system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 9.Brunger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, Rice L M, Simonson T, Warren G L. Crystallography & NMR System. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 10.Buss K A, Ingram-Smith C, Ferry J G, Sanders D A, Hasson M S. Crystallization of acetate kinase from Methanosarcina thermophila and prediction of its fold. Protein Sci. 1997;6:2659–2662. doi: 10.1002/pro.5560061222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project Number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr Sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 12.Das S, Yu L H, Gaitatzes C, Rogers R, Freeman J, Bienkowska J, Adams R M, Smith T F, Lindellen J. Biology's new rosetta stone. Nature. 1997;385:29–30. doi: 10.1038/385029a0. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Atkinson M R, McCleary W, Stock J B, Wanner B L, Ninfa A J. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferry J G. Enzymology of the fermentation of acetate to methane by Methanosarcina thermophila. Biofactors. 1997;6:25–35. doi: 10.1002/biof.5520060104. [DOI] [PubMed] [Google Scholar]
- 15.Flaherty K M, DeLuca-Flaherty C, McKay D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–638. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 16.Flaherty K M, McKay D B, Kabsch W, Holmes K C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci USA. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletterick R J, Bates D J, Steitz T A. The structure of a yeast hexokinase monomer and its complexes with substrates at 2.7-A resolution. Proc Natl Acad Sci USA. 1975;72:38–42. doi: 10.1073/pnas.72.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox D K, Meadow N D, Roseman S. Phosphate transfer between acetate kinase and enzyme I of the bacterial phosphotransferase system. J Biol Chem. 1986;261:13498–13503. [PubMed] [Google Scholar]
- 19.Fox D K, Roseman S. Isolation and characterization of homogeneous acetate kinase from Salmonella typhimurium and Escherichia coli. J Biol Chem. 1986;261:13487–13497. [PubMed] [Google Scholar]
- 20.Hesslinger C, Fairhurst S A, Sawers G. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades L-threonine to propionate. Mol Microbiol. 1998;27:477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- 21.Heyde M, Laloi P, Portalier R. Involvement of carbon source and acetyl phosphate in the external-pH-dependent expression of porin genes in Escherichia coli. J Bacteriol. 2000;182:198–202. doi: 10.1128/jb.182.1.198-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber C, Wachtershauser G. Activated acetic acid by carbon fixation on (Fe, Ni)S under primordial conditions. Science. 1997;276:245–247. doi: 10.1126/science.276.5310.245. [DOI] [PubMed] [Google Scholar]
- 23.Hurley J H. The sugar kinase/heat shock protein 70/actin superfamily-implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 24.Hurley J H, Faber H R, Worthylake D, Meadow N D, Roseman S, Pettigrew D W, Remington S J. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 25.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W, Mannherz H G, Suck D, Pai E F, Holmes K C. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt G J, Jones T A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 28.Knowles J R. Enzyme-catalyzed phosphoryl transfer reactions. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 29.Kull F J, Vale R D, Fletterick R J. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil. 1998;19:877–886. doi: 10.1023/a:1005489907021. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystyllogr. 1993;26:283–291. [Google Scholar]
- 31.Latimer M T, Ferry J G. Cloning, sequence analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J Bacteriol. 1993;175:6822–6829. doi: 10.1128/jb.175.21.6822-6829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipmann F. Enzymatic synthesis of acetyl phosphate. J Biol Chem. 1944;155:55–70. [Google Scholar]
- 33.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matte A, Tari L W, Delbaere L T. How do kinases transfer phosphoryl groups? Structure. 1998;6:413–419. doi: 10.1016/s0969-2126(98)00043-4. [DOI] [PubMed] [Google Scholar]
- 35.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 36.McCleary W R, Stock J B, Ninfa A J. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshima T. Early biochemical evolution: speculations on the biochemistry of primitive life. In: Osawa S, Honjo T, editors. Evolution of life: fossils, molecules, and culture. Tokyo, Japan: Springer-Verlag; 1991. pp. 353–363. [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Perraud A L, Weiss V, Gross R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 1999;7:115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- 40.Pruss B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 41.Rose I A, Grunberg-Manago M, Korey R S, Ochoa S. Enzymatic phosphorylation of acetate. J Biol Chem. 1954;211:737–756. [PubMed] [Google Scholar]
- 42.Sanders D A, Gillece-Castro B L, Burlingame A L, Koshland D E., Jr Phosphorylation site of NtrC, a protein phosphatase whose covalent intermediate activates transcription. J Bacteriol. 1992;174:5117–5122. doi: 10.1128/jb.174.15.5117-5122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders D A, Gillece-Castro B L, Stock A M, Burlingame A L, Koshland D E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, Che Y. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 44.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh-Wissmann K, Ingram-Smith C, Miles R D, Ferry J G. Identification of essential glutamates in the acetate kinase from Methanosarcina thermophila. J Bacteriol. 1998;180:1129–1134. doi: 10.1128/jb.180.5.1129-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh-Wissmann K, Miles R D, Ingram-Smith C, Ferry J G. Identification of essential arginines in the acetate kinase from Methanosarcina thermophila. Biochemistry. 2000;39:3671–3677. doi: 10.1021/bi991998h. [DOI] [PubMed] [Google Scholar]
- 47.Spector L B. Acetate kinase: a triple-displacement enzyme. Proc Natl Acad Sci USA. 1980;77:2626–2630. doi: 10.1073/pnas.77.5.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thatcher G R J, Kluger R. Mechanism and catalysis of nucleophilic substitution in phosphate esters. Adv Phys Org Chem. 1989;25:99–265. [Google Scholar]
- 49.Todhunter J A, Purich D L. Evidence for the formation of a γ-phosphorylated glutamyl residue in the Escherichia coli acetate kinase reaction. Biochem Biophys Res Commun. 1974;60:273–280. doi: 10.1016/0006-291x(74)90201-0. [DOI] [PubMed] [Google Scholar]
- 50.Walter K A, Nair R V, Cary J W, Bennett G N, Papoutsakis E T. Sequence and arrangement of two genes of the butyrate-synthesis pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;134:107–111. doi: 10.1016/0378-1119(93)90182-3. [DOI] [PubMed] [Google Scholar]
- 51.Wanner B L, Wilmes-Riesenberg M R. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J Bacteriol. 1992;174:2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]