Abstract
The RNA genome of SARS-CoV-2 contains a 5′ cap that facilitates the translation of viral proteins, protection from exonucleases and evasion of the host immune response1–4. How this cap is made in SARS-CoV-2 is not completely understood. Here we reconstitute the N7- and 2′-O-methylated SARS-CoV-2 RNA cap (7MeGpppA2′-O-Me) using virally encoded non-structural proteins (nsps). We show that the kinase-like nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain5 of nsp12 transfers the RNA to the amino terminus of nsp9, forming a covalent RNA–protein intermediate (a process termed RNAylation). Subsequently, the NiRAN domain transfers the RNA to GDP, forming the core cap structure GpppA-RNA. The nsp146 and nsp167 methyltransferases then add methyl groups to form functional cap structures. Structural analyses of the replication–transcription complex bound to nsp9 identified key interactions that mediate the capping reaction. Furthermore, we demonstrate in a reverse genetics system8 that the N terminus of nsp9 and the kinase-like active-site residues in the NiRAN domain are required for successful SARS-CoV-2 replication. Collectively, our results reveal an unconventional mechanism by which SARS-CoV-2 caps its RNA genome, thus exposing a new target in the development of antivirals to treat COVID-19.
Subject terms: Kinases, SARS-CoV-2, Cryoelectron microscopy, Enzyme mechanisms, RNA
Reconstitution of the SARS-CoV-2 RNA 5′ cap reveals the unconventional mechanism by which SARS-CoV-2 caps its RNA genome, providing a new target in the development of antiviral agents to treat COVID-19.
Main
Coronaviruses (CoVs) are a family of positive-sense, single-stranded RNA viruses that cause disease in humans9, including SARS-CoV-2, the aetiological agent of the ongoing COVID-19 pandemic10,11. The 5′ proximal two-thirds of the SARS-CoV-2 RNA genome contains two open reading frames that are translated by host ribosomes to form two large polyproteins2. These polyproteins are cleaved by viral proteases to form 16 non-structural proteins (nsp1–16), some of which make up the replication–transcription complex (RTC)2. At the core of the RTC is the nsp12 RNA-dependent RNA polymerase (RdRp), which is the target of antiviral agents used to treat COVID-19, including remdesivir12 and molnupiravir13. nsp12 also contains an N-terminal NiRAN domain5. The NiRAN domain shares sequence and structural similarity with the pseudokinase selenoprotein O (SelO), which transfers AMP from ATP to protein substrates (a process termed AMPylation)14–16. Notably, the active-site kinase-like residues of the NiRAN domain are highly conserved in Nidovirales (Extended Data Fig. 1) and are required for the replication of equine arteritis virus (EAV) and SARS-CoV-1 in cell culture5. Several hypotheses for the function of the NiRAN domain have been proposed, including roles in protein-primed RNA synthesis, RNA ligation and mRNA capping5,17.
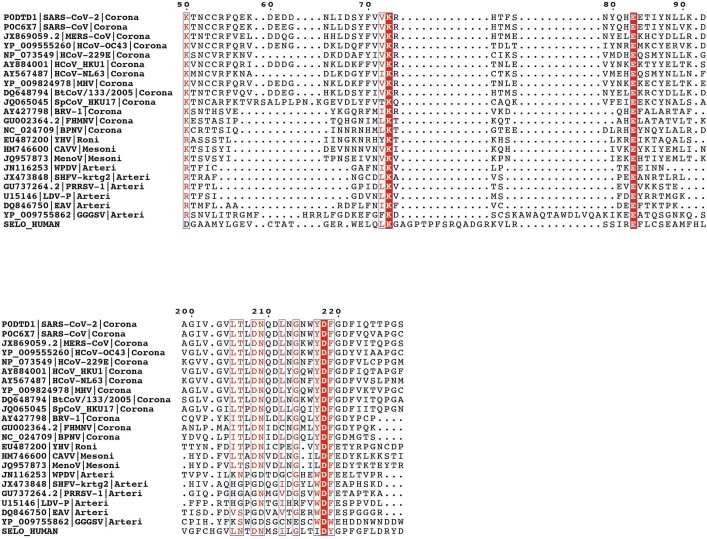
Extended Data Fig. 1. Sequence alignment of the NiRAN domain reveals similarity to the pseudokinase selenoprotein O (SELO).
Multiple sequence alignment highlighting conserved kinase-like active site residues in the NiRAN domain among several CoVs, other selected Nidovirales (Arteri-, Mesoni- and Roniviruses) and the human SELO pseudokinase. Top: amino acid sequence surrounding the Lys-Glu ion pair. Bottom: and the amino acid sequence surrounding the active site and the “DFG” motif.
The CoV RNA genome, like eukaryotic mRNAs, contains a methylated guanosine linked to the first nucleotide of the RNA through a reverse 5′ to 5′ triphosphate linkage1,4. Methylation of the ribose 2′-OH position of the first nucleotide completes the cap and protects the RNA from the host immune system18,19. This 5′ cap is important for RNA stability, initiation of mRNA translation and protection from exonucleases20. Thus, formation of the RNA cap is crucial for successful replication and transcription of the viral genome.
The 5′ cap structure was originally identified on RNAs of reovirus21 and vaccinia virus22, and the molecular mechanisms underlying the formation of the 5′ cap were subsequently determined in these systems23,24. This canonical capping pathway, which is conserved among most viruses as well as all eukaryotes, entails (1) an RNA triphosphatase (RTPase), which removes the γ-phosphate from the nascent 5′-triphosphorylated RNA (5′-pppRNA) to yield a 5′-diphosphorylated RNA (5′-ppRNA); (2) a guanylyltransferase (GTase), which transfers GMP from GTP to 5′-ppRNA to form the core cap structure GpppN-RNA; (3) a (guanine-N7)-methyltransferase (N7-MTase), which methylates the cap guanine at the N7 position; and (4) a (nucleoside-2′-O)-methyltransferase (2′-O-MTase), which methylates the ribose-2′-OH position on the first nucleotide of the RNA (Extended Data Fig. 2). In CoVs, the nsp13, nsp14 and nsp16 proteins have RTPase25, N7-MTase6 and 2′-O-MTase7 activities, respectively. Thus, it was presupposed that the CoV capping mechanism occurs in a similar manner to the eukaryotic capping pathway, with the NiRAN domain functioning as the GTase3,5,26. However, evidence to support this claim is lacking.
Extended Data Fig. 2. The canonical eukaryotic mRNA capping mechanism.
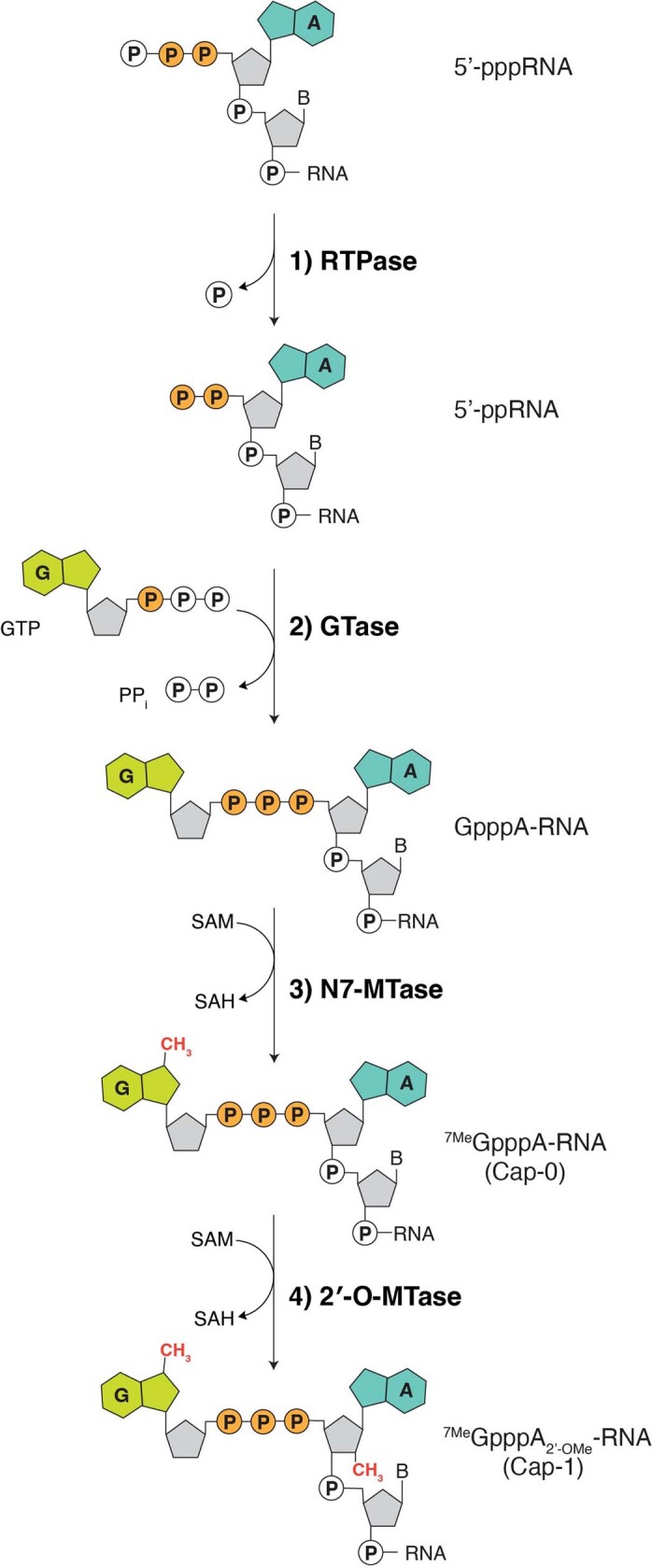
The 7MeGpppA2′-O-Me cap on eukaryotic RNA is formed co-transcriptionally by four enzymes: 1) an RNA triphosphatase (RTPase), which removes the γ-phosphate from the nascent 5′-triphosphorylated RNA (5′-pppRNA) to yield a 5′-diphosphorylated RNA (5′-ppRNA); 2) a guanylyltransferase (GTase), which transfers the GMP moiety from GTP to the 5′-ppRNA to form the core cap structure GpppN-RNA; 3) a (guanine-N7)-methyltransferase (N7-MTase), which methylates the cap guanine at the N7 position; and 4) a (nucleoside-2′-O)-methyltransferase (2′-O-MTase), which methylates the ribose-2′-OH position on the first nucleotide of the RNA. B denotes any base; GTP, Guanosine triphosphate; GDP, Guanosine diphosphate; PPi, pyrophosphate; SAM, S-Adenosyl methionine.
Here we show that the NiRAN domain transfers monophosphorylated RNA (5′-pRNA) from 5′-pppRNA to the N terminus of nsp9 as an intermediate step in cap synthesis. The NiRAN domain then transfers 5′-pRNA from RNAylated nsp9 to GDP to form the core cap structure GpppA-RNA. We demonstrate that the N terminus of nsp9 and the kinase-like active site residues in the NiRAN domain are required for SARS-CoV-2 replication.
The NiRAN domain NMPylates nsp9
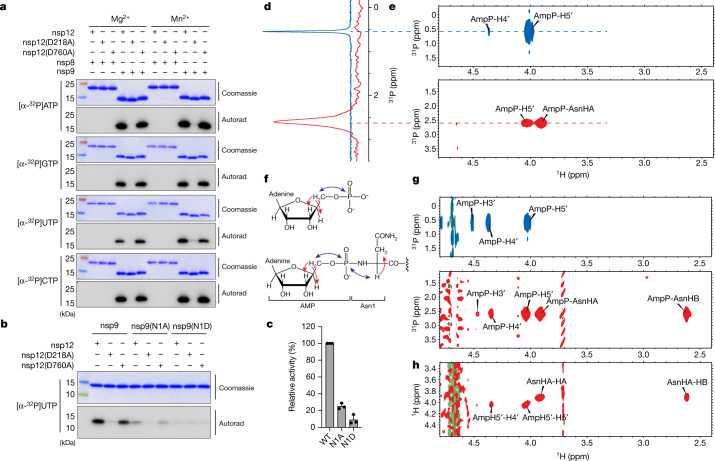
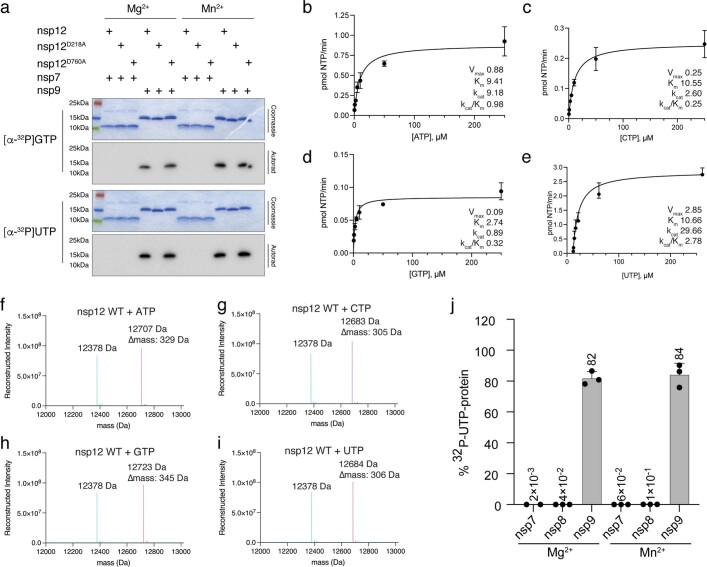
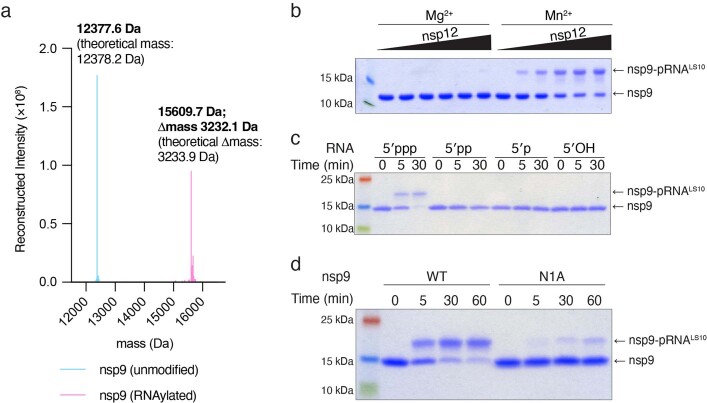
The NiRAN domain has been shown to transfer nucleotide monophosphates (NMPs) from nucleotide triphosphates (NTPs) (referred to as NMPylation) to protein substrates, including nsp917 and the nsp12 co-factors, nsp727 and nsp828. We observed NiRAN-dependent NMPylation of native nsp9, but not native nsp7 or nsp8 (Fig. 1a and Extended Data Fig. 3). Quantification of 32P incorporation and intact mass analyses suggests near stoichiometric incorporation of NMPs into nsp9 (Extended Data Fig. 3f–j). Mutation of nsp9 Asn1 to Ala or Asp reduced NMPylation of nsp9 (Fig. 1b,c), consistent with previous research suggesting that NMPylation occurs on the backbone nitrogen of nsp9 Asn117. To provide direct evidence that the amino terminus of nsp9 is NMPylated by the NiRAN domain, we performed nuclear magnetic resonance (NMR) spectroscopy of AMPylated nsp9. The two-dimensional (2D) 1H,31P heteronuclear single quantum coherence (HSQC) and 2D HSQC–total correlation spectroscopy (HSQC–TOCSY) spectra confirmed that the AMP is attached to the nitrogen backbone atom of Asn1 through a phosphoramidate linkage (Fig. 1d–h).
Fig. 1. The NiRAN domain NMPylates the N terminus of nsp9.
a, Incorporation of α-32P from [α-32P]ATP, GTP, UTP or CTP into nsp8 or nsp9 by WT nsp12, the NiRAN mutant (D218A) or the polymerase mutant (D760A). Reactions were performed in the presence of Mg2+ or Mn2+, and the products were resolved by SDS–PAGE and visualized by Coomassie staining (top) and autoradiography (autorad; bottom). b, Incorporation of α-32P from [α-32P]UTP into nsp9 or the indicated mutants by the NiRAN domain. The reaction products were analysed as described in a. c, Quantification of reaction products shown in b depicting the relative NiRAN-dependent UMPylation activity towards nsp9 or the indicated mutants. Radioactive gel bands were excised and quantified by scintillation counting. d, One-dimensional 31P spectrum of AMP–nsp9 (red) and AMP (blue) recorded in the same buffer as the reference. e, 2D 1H, 31P-HSQC spectra of AMP (top, blue) and AMP–nsp9 (bottom, red). f, The structure of AMP (top) and AMP–nsp9 (bottom) with arrows to indicate the magnetization transfer steps that result in the peaks observed in the 2D NMR spectra. The blue arrows indicate the transfer steps that yield the peaks in the HSQC spectra, and the red arrows show the additional magnetization transfer during the TOCSY that result in the additional peaks found in the HSQC–TOCSY spectra. g, 2D 1H,31P-HSQC–TOCSY spectra of AMP (top, blue) and AMP–nsp9 (bottom, red). h, 2D 1H,1H-HSQC–TOCSY spectra of AMP–nsp9. For a–c, the results are representative of at least three independent experiments. For c, data are mean ± s.d.
Extended Data Fig. 3. The NiRAN domain NMPylates nsp9.
a. Incorporation of α-32P from [α-32P]GTP or [α-32P]UTP into nsp7 or nsp9 by WT nsp12, the NiRAN mutant (D218A), or the polymerase mutant (D760A). Reactions were performed in the presence of Mg2+ or Mn2+ and the products were resolved by SDS-PAGE and visualized by Coomassie staining (top) and autoradiography (bottom). b–e. Kinetic analysis depicting the concentration dependence of (b) ATP, (c) CTP, (d) GTP, or (e) UTP on the rate of nsp9 NMPylation by the NiRAN domain. Km (μM), Vmax (pmol NTP/min), kcat (min−1) and kcat/Km (min−1/μM) values are indicated. Plots shown are the mean and SD of triplicate reactions. f–i. Intact mass LC/MS spectra of unmodified nsp9 (cyan) overlayed with NMPylated nsp9 (pink) following incubation with WT nsp12 and (f) ATP, (g) CTP, (h) GTP, or (i) UTP. The observed masses are shown in the insets. The theoretical mass of unmodified nsp9 is 12378.2 Da and the theoretical increase in mass with the addition of each NMP is as follows: AMP, 329 Da; CMP, 305 Da; GMP, 345 Da; UMP, 306 Da. j. Quantification of the stoichiometry of NMPylation of nsp7, nsp8 and nsp9. Reactions were performed in the presence of Mg2+ or Mn2+. Reaction products were resolved by SDS-PAGE, the corresponding protein bands were excised from the gel, and radioactivity was quantified by scintillation counting. Bars: mean ± SD of three independent experiments.
The NiRAN domain RNAylates nsp9
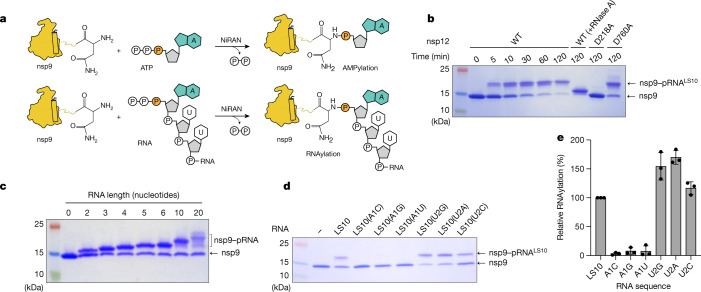
Given the ability of the NiRAN domain to transfer NMPs to nsp9 using NTPs as substrates, we wondered whether the NiRAN domain could also use 5′-pppRNA in a similar manner (Fig. 2a). We synthesized a 5′-pppRNA 10-mer corresponding to the first 10 bases in the leader sequence (LS10) of the SARS-CoV-2 genome (hereafter 5′-pppRNALS10) (Supplementary Table 1). We incubated 5′-pppRNALS10 with nsp9 and nsp12 and analysed the reaction products using SDS–PAGE. We observed an electrophoretic mobility shift in nsp9 that was time dependent, sensitive to RNase A treatment and required an active NiRAN domain, but not an active RdRp domain (Fig. 2b). Intact-mass analyses confirmed the incorporation of monophosphorylated RNALS10 (5′-pRNALS10) into nsp9 (Extended Data Fig. 4a). The reaction was dependent on Mn2+ and required a triphosphate at the 5′-end of the RNA (Extended Data Fig. 4b,c). Substituting Ala for Asn1 reduced the incorporation of RNALS10 into nsp9 (Extended Data Fig. 4d). We also observed RNAylation of nsp9 using LS RNAs ranging from 2 to 20 nucleotides (Fig. 2c). Mutation of the first but not the second nucleotide markedly reduced RNAylation (Fig. 2d,e). Thus, the NiRAN domain RNAylates the N terminus of nsp9 in a substrate-selective manner.
Fig. 2. The NiRAN domain RNAylates nsp9.
a, Schematic of the nsp9 AMPylation reaction (top) and the proposed nsp9 RNAylation reaction (bottom). b, Time-dependent incorporation of RNA into nsp9 by WT nsp12, the NiRAN mutant (D218A) or the polymerase mutant (D760A). The reaction products were analysed using SDS–PAGE and Coomassie staining. The samples were also treated with RNase A. c, Incorporation of different lengths of 5′-pppRNAs into nsp9 by the NiRAN domain. The reaction products were analysed as described in b. d,e, Incorporation of RNAs (with substitutions in the first and second base) into nsp9 by the NiRAN domain. Reaction products were analysed as described in b (d) and quantified by densitometry (e). For e, data are mean ± s.d. of three replicates. The results in this figure are representative of at least three independent experiments.
Extended Data Fig. 4. Characterization of NiRAN RNAylation activity.
a. Intact mass LC/MS spectra (overlayed) of unmodified nsp9 (cyan) or nsp9 after incubation with 5′-pppRNALS10 and WT nsp12 (pink). The theoretical and observed masses are shown in the insets. The Δmass of 3233.17 Da corresponds to monophosphorylated RNALS10 (5′-pRNALS10). b. Incorporation of RNA into nsp9 by nsp12 (0–4 µM) in the presence of Mg2+ or Mn2+. Reaction products were analysed as in Fig. 2b. c. Incorporation of RNA with the indicated 5′ ends into nsp9 by nsp12. Reaction products were analysed as in Fig. 2b. d. Time-dependent incorporation of 5′-pRNALS10 into nsp9 or the nsp9 N1A mutant. Reaction products were analysed as in Fig. 2b.
The NiRAN domain makes the core cap structure
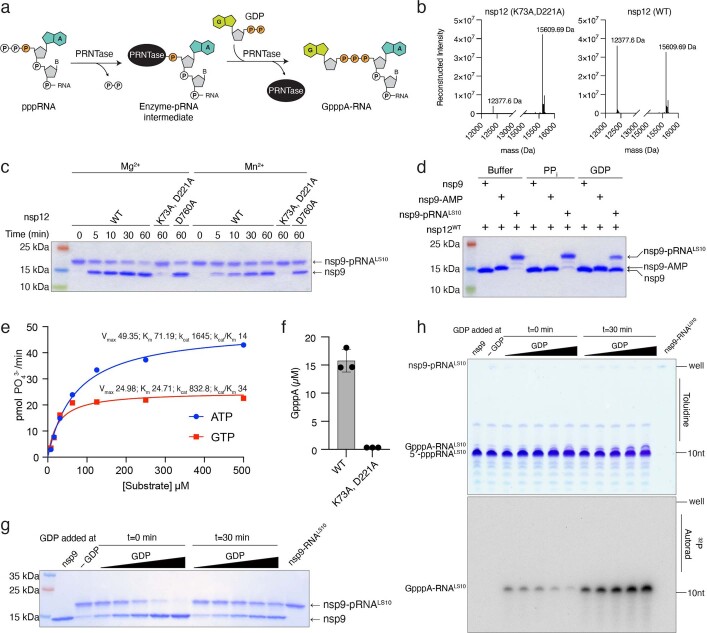
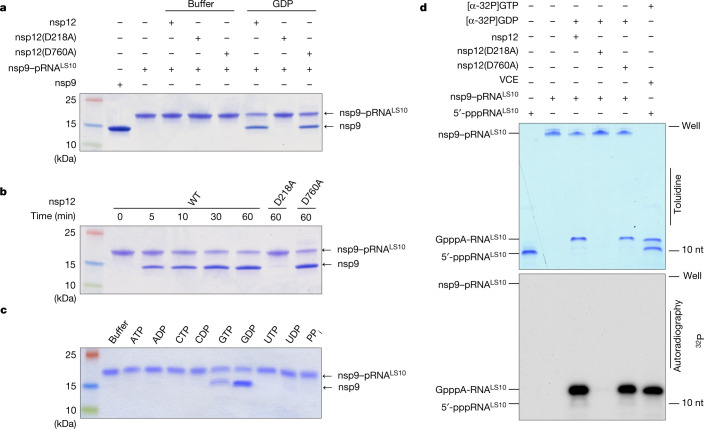
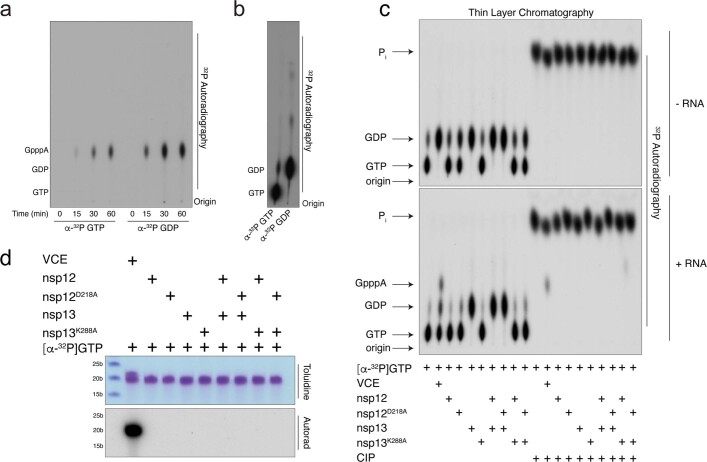
Negative-sense RNA viruses of the order Mononegavirales, including vesicular stomatitis virus (VSV), have an unconventional capping mechanism in which a polyribonucleotidyltransferase (PRNTase) transfers 5′-pRNA from 5′-pppRNA to GDP through a covalent enzyme–RNA intermediate29,30 (Extended Data Fig. 5a). As the NiRAN domain transfers 5′-pRNA to nsp9, we hypothesized that this protein–RNA species may be an intermediate in a similar reaction mechanism to that of the VSV system. To test this hypothesis, we purified nsp9–pRNALS10 and incubated it with GDP in the presence of nsp12. Treatment with GDP deRNAylated nsp9 in a NiRAN-dependent manner, as judged by the nsp9 electrophoretic mobility on SDS–PAGE (Fig. 3a) and its molecular mass based on intact-mass analysis (Extended Data Fig. 5b). The reaction was time dependent, (Fig. 3b), preferred Mg2+ over Mn2+ (Extended Data Fig. 5c) and was specific for GDP and—to some extent—GTP, but not the other nucleotides tested (Fig. 3c). Interestingly, although inorganic pyrophosphate (PPi) was able to deAMPylate nsp9–AMP, it was unable to deRNAylate nsp9–pRNALS10 (see Discussion and Extended Data Fig. 5d).
Extended Data Fig. 5. Characterization of nsp12 NiRAN GDP-PRNTase activity.
a. Schematic representation depicting the mechanism of core cap formation by vesicular stomatitis virus (VSV) polyribonucleotidyltransferase (PRNTase). b. Intact mass LC/MS spectra of nsp9–pRNALS10 after incubation with GDP and WT nsp12 (right) or the NiRAN mutant (left). The theoretical mass of nsp9 is 12378.2 Da and the theoretical mass of nsp9–pRNALS10 is 15611.5 Da. c. Time-dependent deRNAylation of nsp9–pRNALS10 by WT nsp12, the NiRAN mutant (K73A, D221A), or the polymerase mutant (D760A) in the presence of GDP and either Mg2+ or Mn2+. Reaction products were analysed as in Fig. 2b. d. NiRAN-dependent deAMPylation or deRNAylation of nsp9 in the presence of PPi or GDP. Reaction products were analysed as in Fig. 2b. e. Concentration dependence of ATP (blue) or GTP (red) on the rate of phosphate release catalysed by nsp13. Vmax (pmol PO43−/min), Km (μM), kcat (min−1) and kcat/Km (min−1/μM) values are indicated. f. HPLC/MS quantification of GpppA formed during the NiRAN-catalysed deRNAylation of nsp9–pRNALS10. Reaction products were digested with nuclease P1 prior to HPLC analysis. Reactions were performed in triplicate and error bars represent the standard deviation. g, h. NiRAN-catalysed capping reactions depicting the inhibitory effect of GDP on RNAylation. nsp9 was incubated with excess 5′-pppRNALS10 in presence of nsp12 with no GDP (-GDP), or increasing concentrations (6.25-100 µM) of [α-32P]GDP added either at time zero (t = 0 min), or after the RNAylation reaction was allowed to proceed for 30 min (t=30 min). Reaction products were analysed by SDS-PAGE (g), and urea–PAGE (h).
Fig. 3. The NiRAN domain catalyses the transfer of 5′-pRNA from nsp9 to GDP to form the core cap structure GpppA-RNA.
a, DeRNAylation of the covalent nsp9–RNALS10 species by WT nsp12, the NiRAN mutant (D218A) or the polymerase mutant (D760A) when incubated with buffer or GDP. The reaction products were analysed as described in Fig. 2b. b, Time-dependent deRNAylation of nsp9–pRNALS10 by WT nsp12, the NiRAN mutant (D218A) or the polymerase mutant (D760A). The reaction products were analysed as described in Fig. 2b. c, DeRNAylation of nsp9–pRNALS10 by nsp12 in the presence of different NTPs, NDPs or PPi. The reaction products were analysed as described in Fig. 2b. d, Incorporation of α-32P from [α-32P]GDP into nsp9–pRNALS10 by WT nsp12, the NiRAN mutant (D218A) or the polymerase mutant (D760A). VCE was used as a control but was incubated with [α-32P]GTP. The reaction products were resolved using urea–PAGE and visualized by toluidine blue O staining (top) and autoradiography (bottom). The results in this figure are representative of at least two independent experiments. nt, nucleotides.
We used urea–PAGE to analyse the fate of the RNALS10 during the deRNAylation reaction. Treatment of nsp9–pRNALS10 with nsp12 and [α-32P]GDP that was generated by treatment of [α-32P]GTP with nsp13 (Extended Data Fig. 5e) resulted in a [32P]-labelled RNA species that migrated similarly to GpppA-RNALS10 produced by the vaccinia capping enzyme (VCE; Fig. 3d). The reaction was dependent on a functional NiRAN domain but not an active RdRp domain. To confirm the presence of a GpppA-RNA cap, we digested the RNA produced from the nsp12 reaction with P1 nuclease and detected GpppA using high-performance liquid chromatography–mass spectrometry (HPLC–MS) analysis (Extended Data Fig. 5f). Thus, the NiRAN domain is a GDP-PRNTase that mediates the transfer of 5′-pRNA from nsp9 to GDP.
In our attempts to generate GpppA-RNALS10 in a ‘one pot’ reaction, we found that GDP inhibited the RNAylation reaction (Extended Data Fig. 5g). However, the formation of GpppA-RNALS10 could be achieved in one pot provided that the RNAylation occurs before the addition of GDP (Extended Data Fig. 5g,h).
nsp14 and nsp16 form cap-0 and cap-1 structures
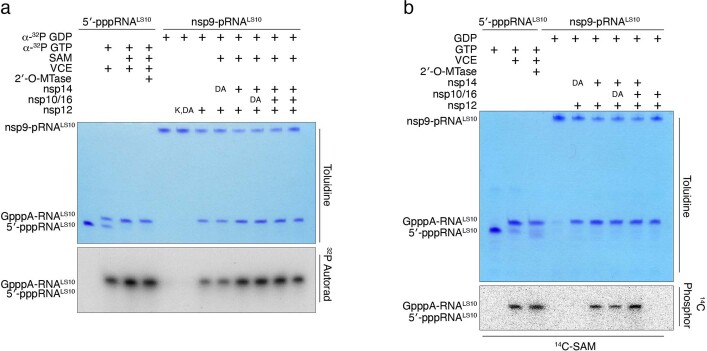
The SARS-CoV-2 genome encodes an N7-MTase domain within nsp146 and a 2′-O-MTase in nsp16, the latter of which requires nsp10 for activity7. nsp14 and the nsp10–nsp16 complex use S-adenosyl methionine (SAM) as the methyl donor. To test whether NiRAN-synthesized GpppA-RNALS10 can be methylated, we incubated 32P-labelled GpppA-RNALS10 with nsp14 and/or the nsp10–nsp16 complex in the presence of SAM and separated the reaction products using urea–PAGE (Extended Data Fig. 6a). We extracted RNA from the reaction, treated it with P1 nuclease and CIP, and then analysed the products using thin-layer chromatography (TLC) (Fig. 4a). As expected, the NiRAN-synthesized cap migrated similarly to the GpppA standard and the products from the VCE reaction (compare lanes 1 and 4). Similarly, reactions that included SAM and nsp14 migrated similarly to the 7MeGpppA standard and to the products from the VCE reaction after the addition of SAM (compare lanes 2 and 6). Furthermore, treatment of 7MeGpppA-RNALS10, but not unmethylated GpppA-RNALS10, with the nsp10–nsp16 complex produced the 7MeGpppA2′-O-Me-RNA cap-1 structure (compare lanes 3, 8 and 9). In parallel experiments, we incubated NiRAN-synthesized GpppA-RNALS10 with nsp14 and/or the nsp10–nsp16 complex in the presence of [14C]-labelled SAM (14C on the donor methyl group) and separated the reaction products using urea–PAGE. As expected, nsp14 and the nsp10–nsp16 complex incorporated 14C into GpppA-RNALS10 to form the cap-0 and cap-1 structures, respectively (Extended Data Fig. 6b). Thus, the SARS-CoV-2 7MeGpppA2′-O-Me-RNA capping mechanism can be reconstituted in vitro using virally encoded proteins.
Extended Data Fig. 6. SARS-CoV-2 nsp14 and nsp16 methyltransferases methylate the nsp12-synthesised core cap structure.
a. Incorporation of α-32P from [α-32P]GDP into nsp9–pRNALS10 by WT nsp12, or the NiRAN K73A, D221A mutant (K, DA). Reactions were subsequently incubated with SAM, nsp14 (or the D331A mutant; DA), and nsp10/16 (or the D130A nsp16 mutant; DA). The Vaccinia capping enzyme (VCE) was used as a positive control but incubated with [α-32P]GTP and the Vaccinia 2′-O-MTase. Reaction products were analysed as in Fig. 3d. b. Incorporation of 14C from [methyl-14C]SAM into GpppA-RNALS10 by nsp14 (WT or the D331A mutant; DA), and nsp10/16 (WT or the D130A nsp16 mutant; DA). The VCE and the Vaccinia 2′-O-MTase were used as positive controls. Reaction products were analysed as in Fig. 3d. The 14C signal was detected by phophorimaging.
Fig. 4. nsp14 and nsp16 catalyse the formation of the cap-0 and cap-1 structures.
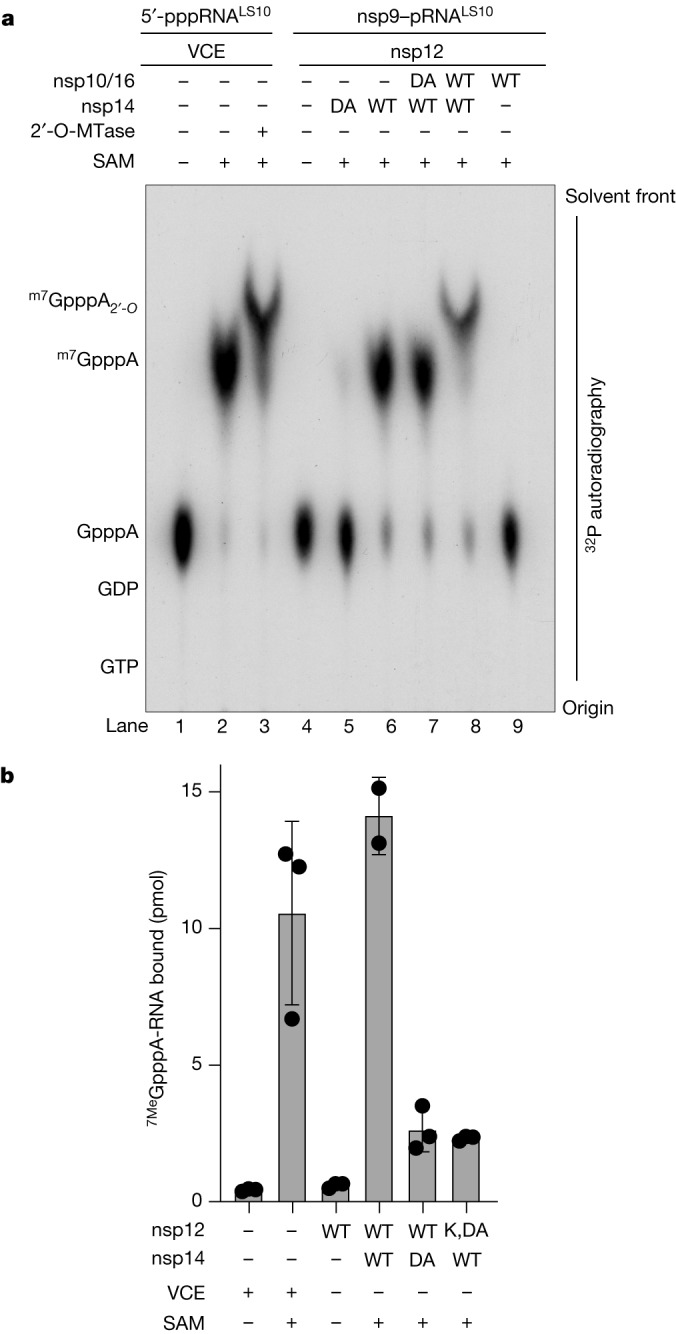
a, TLC analysis of the reaction products from Extended Data Fig. 6a, after extraction from the urea–PAGE gel and treatment with P1 nuclease and CIP. The location of the cold standards (left) was visualized by ultraviolet fluorescence and the reaction products were visualized by 32P autoradiography. b, Pull-down assays depicting the binding of [32P]7MeGpppA-RNALS10 to GST–eIF4E. [32P]7MeGpppA-RNALS10 was produced using SARS-CoV-2 virally encoded proteins or the VCE. Radioactivity in GST pull-down assays was quantified by scintillation counting. Data are mean ± s.d. The results shown in the figure are representative of three independent experiments. nsp14 DA, nsp14(D331A); nsp16 DA, nsp16(D130A).
Efficient translation of mRNAs is dependent on eIF4E binding to the 7MeGpppA-RNA cap31. To test whether the SARS-CoV-2 RNA cap is functional, we incubated [32P]-labelled 7MeGpppA-RNALS10 with GST-tagged eIF4E. We observed [32P]-labelled RNA in GST pull-downs of [32P]7MeGpppA-RNA but not the unmethylated derivative (Fig. 4b). Thus, the 7MeGpppA-RNA cap generated by SARS-CoV-2-encoded proteins is a substrate for eIF4E in vitro, suggesting that the cap is functional.
Structural insights into the capping reaction
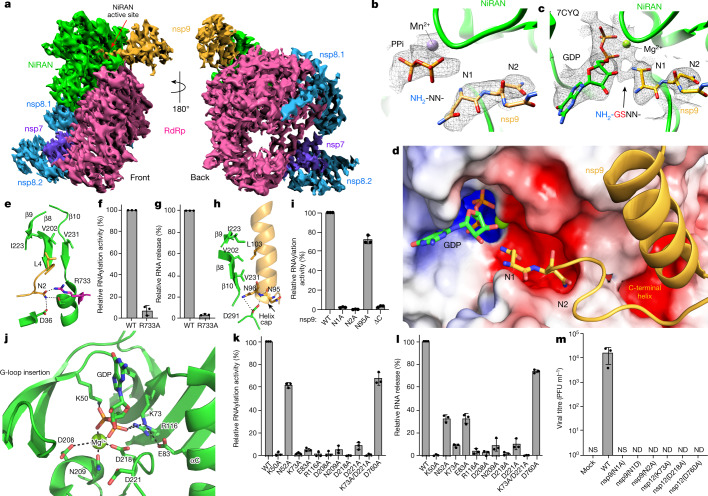
We determined a cryo-electron microscopy (cryo-EM) structure of the nsp7–nsp8–nsp9–nsp12 complex and observed a nsp9 monomer bound in the NiRAN active site (Fig. 5a, Extended Data Fig. 7 and Supplementary Table 2). The native N terminus of nsp9 occupies a similar position to previously reported structures using a non-native N terminus of nsp926 (Fig. 5b,c). Our cryo-EM analysis was hindered by the preferred orientation of the complex and sample heterogeneity, yielding final maps with high levels of anisotropy, with distal portions of nsp9 missing and weak density for the N-lobe of the NiRAN domain (Extended Data Fig. 7). Thus, we used the complex structure from ref. 26 (Protein Data Bank (PDB): 7CYQ) to study the structural basis of NiRAN-mediated RNA capping.
Fig. 5. Structural and genetic insights into RNA capping by the kinase domain.
a, Front and back views of nsp12, nsp7, nsp8 and nsp9 cryo-EM maps. Green, NiRAN; magenta, RdRp; violet, nsp7; light blue, nsp8; gold, nsp9. b,c, Density maps of the N terminus of nsp9 from this study (b) and from ref. 26 (PDB: 7CYQ) (c). The arrow in c indicates probable density of unmodelled Gly–Ser residues on non-native nsp9. d, Electrostatic surface of the NiRAN active site from PDB 7CYQ bound to nsp9, contoured at ±5 kT e−1. e, Cartoon representation of the nsp9 N terminus interacting with the β9–β8–β10 sheet in NiRAN, and Arg733 in RdRp (PDB: 7CYQ). f, Relative RNAylation activity of the nsp12(R733A) mutant. The reaction products were analysed as described in Fig. 2e. g, Relative GDP-PRNTase activity of the nsp12(R733A) mutant (measured as RNA release from nsp9–pRNALS10). The reaction products were separated by urea–PAGE, stained with SYBR Gold and quantified by densitometry. h, Cartoon representation of the nsp9 C-terminal helix interacting with the NiRAN domain (PDB: 7CYQ). i, Relative RNAylation activity of nsp12 towards indicated nsp9 mutants. ∆C, nsp91–92. The reaction products were analysed as described in Fig. 2e. j, Cartoon representation of the NiRAN active site (PDB: 7CYQ). Catalytic residues and GDP are shown as sticks, Mg2+ is shown as a green sphere and interactions are denoted by dashed lines. k, The relative RNAylation activity of nsp12 and the indicated mutants. The reaction products were analysed as described in Fig. 2e. l, The relative GDP-PRNTase activity of nsp12 and the indicated mutants. The reaction products were analysed as described in g. m, SARS-CoV-2 (and the indicated mutants) viral titres quantified as plaque-forming units in the culture medium (PFU per ml). Data are mean ± s.d. of n = 3 biological replicates. ND, not detected. For f, g, k and l, data are mean ± s.d. of one biological replicate.
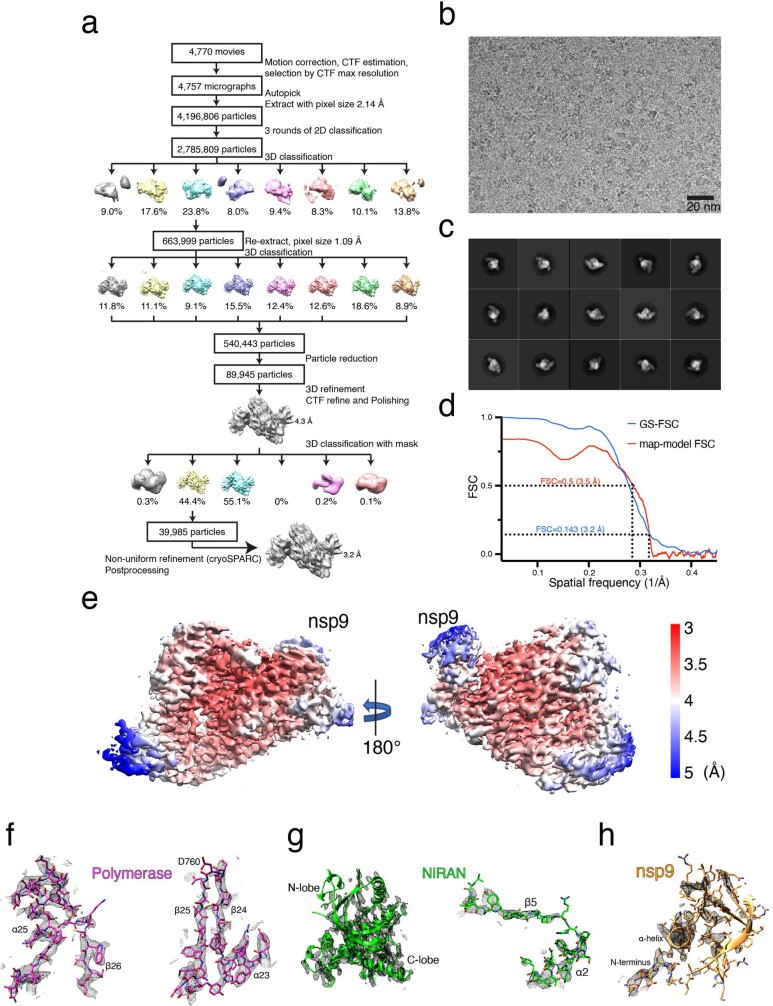
Extended Data Fig. 7. Cryo-EM analysis of the nsp7/8/9/12 complex.
a. Flow chart representing data processing for the nsp7/8/9/12 complex. b. A representative micrograph of the nsp7/8/9/12 complex grids. c. Representative 2D classes generated by RELION 2D-classification. d. Gold-standard FSC curve (blue), and map-model FSC curve (red). Curves were generated by cryoSPARC and Phenix suite, respectively. e. Local resolution of the nsp7/8/9/12 complex calculated by RELION from final cryoSPARC half-maps. Position of nsp9 is indicated. f-h. Exemplary cryo-EM density (black mesh) on (f) Polymerase (Magenta), (g) NiRAN (green) and (h) nsp9 (gold). Note weaker density in the N-lobe of the kinase-like NiRAN domain (left panel of g, compare top to bottom of the image), and poor density in nsp9 (h), in areas not in direct contact with the NiRAN domain.
The first four residues of nsp9 extend into the NiRAN active site, forming electrostatic and hydrophobic contacts in and around a groove near the kinase-like active site (Fig. 5d). Asn1 of nsp9 is positioned inside the active site, primed for transfer of 5′-pppRNA onto its N terminus. Although the terminal NH2 group of nsp9 is the substrate for RNAylation, the local quality of the structures is not high enough to distinguish its exact position. We modelled the nsp9 acceptor NH2 pointing towards what appears to be the phosphates of the nucleotide analogue UMP-NPP in the active site (Fig. 5b). In the structure in ref. 26, Asn1 was assigned an opposite conformation and there are unmodelled residues (non-native N terminus; NH2-Gly-Ser-) visible in the density maps, distorting local structural features32 (Fig. 5c (arrow)). Nevertheless, the presence of these additional residues does not affect the overall binding mode of nsp9 to the NiRAN domain.
Asn2 of nsp9 is in a negatively charged cleft around the NiRAN active site, and contacts Arg733, which extends from the polymerase domain and positions the N terminus of nsp9 (Fig. 5e). Alanine substitution of Arg733 and an nsp12 mutant lacking the RdRp domain (ΔRdRp; 1–326) markedly reduced both RNAylation and GDP-PRNTase activities (Fig. 5f,g and Extended Data Fig. 8a,b). Leu4 and the C-terminal helix of nsp9 form hydrophobic interactions with a β-sheet (β9–β8–β10) in the N-lobe of the NiRAN domain (Fig. 5e,h). The N-terminal cap of the nsp9 C-terminal helix also forms electrostatic interactions with a negatively charged pocket on the surface of the NiRAN domain (Fig. 5h). Removal of the C-terminal helix from nsp9 (ΔC; 1–92) and Ala substitutions of Asn1 and Asn2 abolished RNAylation by nsp12 (Fig. 5i).
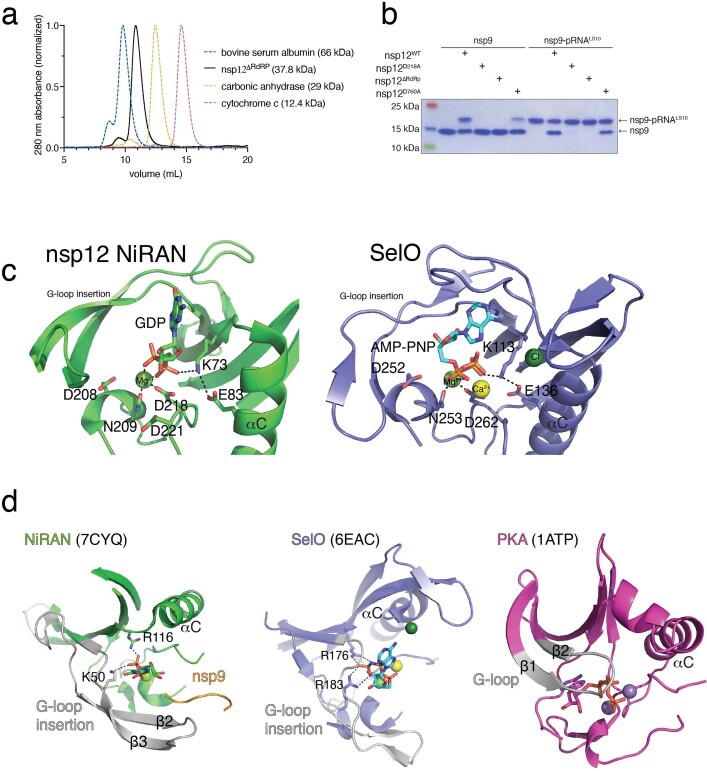
Extended Data Fig. 8. Biochemical analysis of the isolated NiRAN domain and comparison of the kinase-like domains of nsp12 and SELO.
a. Size exclusion chromatography analysis of the isolated NiRAN domain (residues 1–326; ∆RdRp). Standards are shown. b. Incorporation of 5′-pRNALS10 into nsp9 and deRNAylation of nsp9–pRNALS10 by WT nsp12, the NiRAN mutant (D218A), the polymerase mutant (D760A), or the ∆RdRP mutant. Reaction products were analysed as in Fig. 2b. c. Cartoon representation comparing the NiRAN active site catalytic residues (green) to the active site residues in SELO (purple). The divalent cations are shown as spheres. d. Comparison of the Gly-rich loop regions in NiRAN (PDB: 7CYQ; left), SELO (PDB: 6EAC), and the canonical kinase PKA (PDB: 1ATP; right). Green sphere – Mg2+, dark green sphere – Chloride, yellow sphere – calcium, violet sphere – Mn2+.
The NiRAN domain resembles SelO, with a root-mean-squared deviation of 5.7 Å over 224 Cα atoms (PDB: 6EAC)14 (Extended Data Fig. 8c). Lys73 (PKA nomenclature, Lys72) forms a salt bridge with Glu83 (PKA, Glu91) from the αC (α2) helix and contacts the phosphates of the nucleotide (Fig. 5j). As expected, the ‘DFG’ motif Asp218 (PKA, Asp184) binds to a divalent cation. Interestingly, the NiRAN domain lacks the catalytic Asp (Extended Data Fig. 1), (PKA, Asp166); however, like in SelO, Asp208 is next to the metal-binding Asn209 (PKA, Asn171) and may act as a catalytic base to activate the NH2 group on the N terminus of nsp9 (Fig. 5j). Alanine substitutions in these residues largely reduced both RNAylation (Fig. 5k) and GDP-PRNTase (Fig. 5l) activities.
In canonical kinases, the β1–β2 G-loop stabilizes the phosphates of ATP33. The NiRAN domain contains a β-hairpin insert (β2–β3) where the β1–β2 G-loop should be (Extended Data Fig. 8d). This insertion not only makes contacts with the N terminus of nsp9, but also contains a conserved lysine residue (Lys50) that stabilizes the phosphates of the bound nucleotide. Similarly, Arg116 also contacts the phosphates of the nucleotide. SelO contains a similar set of basic residues that accommodate the flipped orientation of the nucleotide to facilitate AMPylation (Extended Data Fig. 8d). Both Lys50 and Arg116 are required for RNAylation (Fig. 5k) and GDP-PRNTase (Fig. 5l) activities. Notably, Lys73, Arg116 and Asp218 in SARS-CoV-1 nsp12 are required for viral replication5.
NiRAN capping is required for viral replication
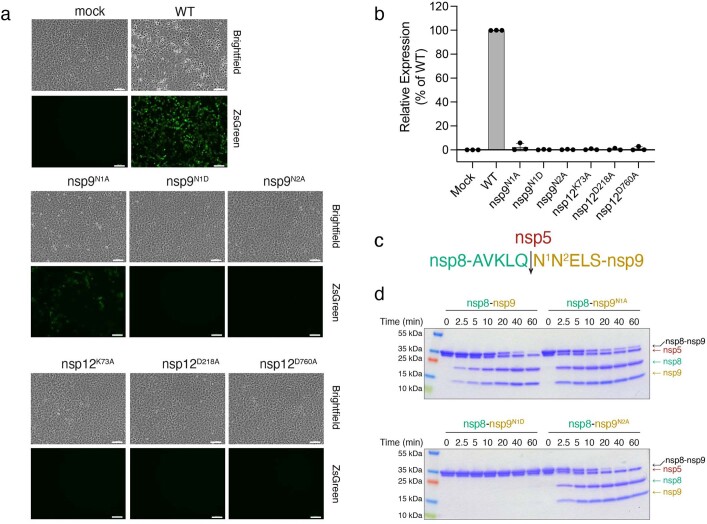
To determine the importance of the NiRAN domain and the N terminus of nsp9 in viral replication, we used a DNA-based reverse genetics system that can rescue infectious SARS-CoV-2 (Wuhan-Hu-1/2019 isolate) expressing a fluorescent reporter8 (Extended Data Fig. 9a). We introduced single point mutations in nsp9 (N1A, N1D and N2A) and nsp12 (K73A, D218A and D760A) and quantified viral titres by plaque assay. Notably, we could not detect plaque formation in cells incubated with undiluted supernatants for any of the mutants (Fig. 5m). We also quantified the virus in supernatants of producer cells using quantitative PCR with reverse transcription (RT–qPCR) to detect the viral N gene and observed more than a 99% reduction in viral load for all of the mutants compared with the wild type (WT) (Extended Data Fig. 9b). To account for the possibility of a proteolytic defect in the mutant viral polyprotein, we tested whether the main viral protease nsp5 (MPro) can cleave a nsp8–nsp9 fusion protein containing the Asn1/Asn2 mutations in nsp9. The N1D mutant was not cleaved by nsp5, suggesting that the replication defect observed for this mutant is a result of inefficient processing of the viral polyprotein. However, the N1A and N2A mutants were efficiently cleaved by nsp5 (Extended Data Fig. 9c,d). Collectively, these data provide genetic evidence that the residues involved in capping the SARS-CoV-2 genome are essential for viral replication.
Extended Data Fig. 9. Genetic insights into RNA capping by the NiRAN domain.
a. Microscopy images showing brightfield (upper) or fluorescence-based images (ZsGreen; lower) of SARS-CoV-2-ZsGreen production in VeroE6-C1008-TMPRSS2 cells. Mock-transfected panels were incubated with transfection reagents lacking DNA. The mutations engineered into either nsp9 or nsp12 are indicated above each set of images. Data are one set of images representative of three independent biological replicates. b. Relative viral yields from WT or mutant SARS-CoV-2 viruses bearing indicated mutations in nsp9 and nsp12. Viral RNA levels were quantified by RT-qPCR. Bars: mean ± SD of N = 3 biological replicates. c. The amino acid sequence between nsp8 (green) and nsp9 (gold) depicting the cleavage site for the nsp5 (dark red) protease. N1 and N2 of nsp9 are highlighted. The arrow denotes the location of cleavage. d. Time-dependent proteolysis of the nsp8-nsp9 fusion protein by nsp5. Reaction products were separated by SDS PAGE and visualized by Coomassie staining.
Discussion
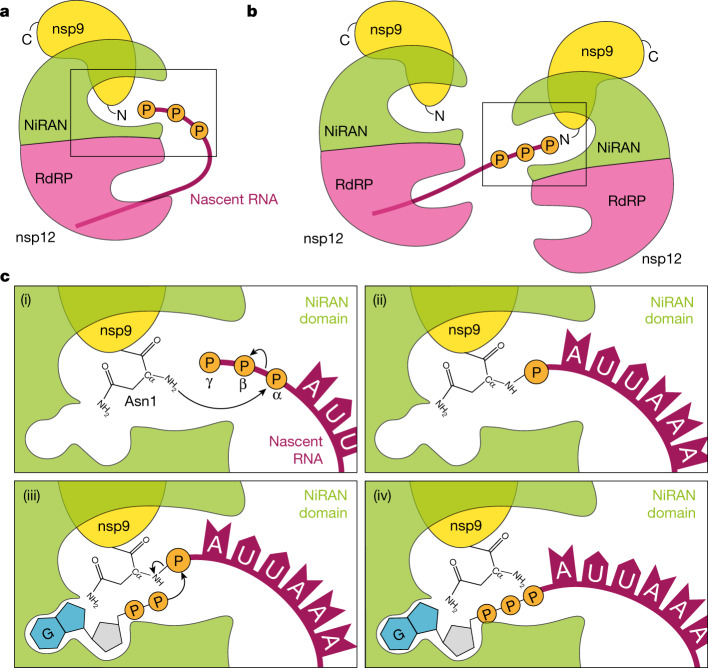
We propose the following mechanism of RNA capping by CoV: during transcription, the nascent 5′-pppRNA binds to the NiRAN active site, in either a cis (Fig. 6a) or trans (Fig. 6b) manner, and 5′-pRNA is subsequently transferred to the N terminus of nsp9, forming a phosphoramidate bond (Fig. 6c (i and ii)). The nsp13 protein produces GDP from GTP, which binds to the NiRAN active site and attacks RNAylated nsp9, releasing capped RNA and regenerating unmodified nsp9 (Fig. 6c (iii and iv)). Subsequently, nsp14 and nsp16 perform sequential N7 and 2′-O methylations, forming a fully functional 7MeGpppA2′-O-Me-RNA cap.
Fig. 6. Proposed model of the SARS-CoV-2 RNA-capping mechanism.
a,b, During transcription, the nascent 5′-pppRNA binds to the NiRAN active site in either a cis (a) or trans (b) manner. c, After binding, the N terminus of nsp9 attacks the α-phosphate of the nascent 5′-pppRNA (i), forming the covalent nsp9–pRNA species and releasing PPi (ii). After GDP binding to the NiRAN active site, the β-phosphate of the nsp13-generated GDP attacks the 5′-phosphate on the nsp9–pRNA (iii), releasing capped RNA and regenerating unmodified nsp9 (iv). Subsequent methylation events are carried out by nsp14 and nsp16 to generate the 7MeGpppA2′-O-Me-RNA cap.
SARS-CoV-2 nsp12 is thought to initiate transcription/replication starting with an NTP, or a short 5′-pppRNA primer34. Cryo-EM structures of the RTC suggest that the double-stranded RNA product makes its way out of the RdRp active site in a straight line, supported by the nsp8 helical stalks26,35,36. In a cis capping model, the helical duplex with nascent 5′-pppRNA would then need to unwind, flex 90° and extend into the NiRAN active site around 70 Å away (Fig. 6a). More likely, a separate RTC complex could perform capping in trans (Fig. 6b). Notably, ref. 37 proposed that the nascent RNA strand is separated from the template after passing through the proof-reading ExoN domain of nsp14 on a neighbouring RTC and threaded towards the NiRAN domain.
SARS-CoV-1 nsp13 has RNA helicase, nucleotide triphosphatase (NTPase) and RNA 5′-triphosphatase (RTPase) activities25. The RTPase activity implicated nsp13 in the first step of the capping mechanism; however, although nsp13 can act on 5′-pppRNA, this reaction is inhibited in the presence of cellular concentrations of ATP25. Thus, we favour the idea that the physiological functions for nsp13 are: (1) to use the energy from ATP hydrolysis to unwind double-stranded RNA (helicase), and (2) to hydrolyse GTP to GDP, which can then act as an acceptor for 5′-pRNA in the capping reaction.
The SARS-CoV-2 capping mechanism is reminiscent of the capping mechanism used by VSV, although there are some differences. The VSV large (L) protein is a multifunctional enzyme that carries out RdRp, PRNTase and methyltransferase activities to form the cap29,38,39. During the reaction, 5′-pRNA is transferred to a conserved His residue within the PRNTase domain, which is distinct from the protein kinases30. The presence of two different enzymatic mechanisms of capping, proceeding through covalent protein–RNA intermediates, in Mononegavirales and in Nidovirales is an example of convergent evolution.
Consistent with other reports17,32, we observed NiRAN-catalysed NMPylation of nsp9 (Fig. 1a and Extended Data Fig. 3) and did not detect NMPylation of nsp7 and nsp8. Yan et al.26 most likely did not observe NMPylation of nsp9 due to the presence of additional non-native residues at the N terminus of nsp9, which masked the modification site.
Although our results do not necessarily preclude a biologically relevant function for nsp9 NMPylation, it is worth noting that this modification is reversible in the presence of PPi32 (Extended Data Fig. 5d). PPi is produced during the RdRp reaction, making the stability of NMPylated nsp9 difficult to envision in vivo. RNAylated nsp9 was not reversible in the presence of PPi. Thus, RNAylation is probably the physiologically relevant modification of nsp9 during viral RNA capping.
Both GDP and GTP were able to deRNAylate nsp9–pRNALS10 (Fig. 3c) and both were able to form a GpppA cap structure in the presence of nsp9–pRNALS10 and nsp12 (Extended Data Fig. 10a). However, the GTP used in these assays contained GDP, probably due to GTP hydrolysis (Extended Data Fig. 10b). Thus, the NiRAN-mediated capping reaction is specific for GDP.
Extended Data Fig. 10. The NiRAN domain is specific for GDP in the capping reaction and does not transfer GMP from GTP to 5’-ppRNA.
a. Thin-layer chromatogram depicting the reaction products following incubation of nsp12 and nsp9–pRNALS10 with [α-32P]GTP or [α-32P]GDP for the indicated timepoints. Location of the cold standards (left) was visualized by UV fluorescence and the 32P by autoradiography. b. Thin-layer chromatogram depicting [α-32P]GTP or [α-32P]GDP used in the assays in a. Note the presence of GDP in the GTP sample. c. Thin-layer chromatograms depicting the reaction products resulting from the incubation of [α-32P]GTP with nsp12 or the inactive NiRAN mutant (D218A). Reactions were performed as described in26 with (lower) or without (upper) 5’-pppRNAA19C and included nsp13 or the inactive mutant (K288A) as indicated. Vaccinia capping enzyme (VCE) was used as a positive control. Reaction products were digested with nuclease P1, then treated with or without calf intestinal alkaline phosphatase (CIP) and analysed by PEI-cellulose thin-layer chromatography followed by autoradiography. The positions of the origin and standard marker compounds are indicated. d. RNA products from c were analysed by TBE urea–PAGE and visualized by toluidine blue O staining (upper) and autoradiography (lower). Markers indicate RNA size by base length.
Recent research suggested that the NiRAN domain is a GTase that transfers GMP from GTP to 5′-ppRNA, forming a GpppA-RNA cap intermediate3,26. In our efforts to reproduce these results, we did not detect nsp12-dependent GpppA cap formation by TLC or by urea–PAGE analysis of the RNA (Extended Data Fig. 10c,d), in contrast to our control, in which the VCE efficiently generated GpppA-RNA. As nsp13 and the VCE can hydrolyse GTP to GDP25, the cap reported previously26 appears to be GDP formed from nsp13- and VCE-dependent hydrolysis of GTP.
In summary, we have defined the mechanism by which SARS-CoV-2 caps its genome and have reconstituted this reaction in vitro using non-structural proteins encoded by SARS-CoV-2. Our results uncover new targets for the development of antiviral agents to treat COVID-19 and highlight the catalytic adaptability of the kinase domain.
Methods
Chemicals and reagents
Ampicillin sodium (A9518), ATP (A2383), ADP (A2754), chloramphenicol (C0378), CTP (C1506), CDP (C9755), dithiothreitol (DTT; D0632), EDTA (E5134), GTP (G8877), GDP (G7127), imidazole (I2399), IPTG (I5502), kanamycin sulfate (K1377), 2-mercaptoethanol (BME, M3148), Brilliant Blue R (B0149), magnesium chloride (MgCl2; M2670), manganese (II) chloride tetrahydrate (MnCl2; M3634), PEI-cellulose TLC plates (Z122882), potassium chloride (KCl; P9541), pyrophosphate (221368), urea (U6504), UTP (U6625) and UDP (94330) were obtained from Millipore-Sigma. Q5 DNA polymerase (M0492L), all restriction enzymes used for cloning, proteinase K (P8107S), yeast inorganic pyrophosphatase (M2403), Quick CIP (M0525S), nuclease P1 (M0660S), Vaccinia Capping System (M2080S), mRNA cap 2′-O-methyltransferase (M0366S), G(5′)ppp(5′)A RNA cap structure analogue (GpppA; S1406L) and m7G(5′)ppp(5′)A RNA cap structure analogue (m7GpppA; S1405S) were all obtained from New England Biolabs. Acetic acid (A38-212), RNase inhibitor (N8080119), 2× TBE-urea Sample Buffer (LC6876) and isopropanol (42383) were all obtained from Thermo Fisher Scientific. [α-32P]ATP (BLU003H250UC), [α-32P]CTP (BLU008H250UC), [α-32P]GTP (BLU006H250UC), [α-32P]UTP (BLU007H250UC) and S-[methyl-14C]-adenosyl-l-methionine (NEC363010UC) were all obtained from PerkinElmer. All 5′-triphosphorylated RNAs were custom synthesized by ChemGenes. Phenylmethylsulfonyl fluoride (PMSF; 97064-898) was obtained from VWR. 4–20% Mini-PROTEAN TGX Stain-Free Protein Gels (4568096) were obtained from Bio-Rad. Uridine-5′-[(α,β)-imido]triphosphate (UMP-NPP; NU-930L) was obtained from Sapphire North America.
Plasmids
SARS-CoV-2 nsp7, nsp8, nsp12, nsp13, nsp14 and nsp16 coding sequences (CDSs) were codon-optimized for bacterial expression and synthesized as gBlocks (Integrative DNA Technologies). The CDSs for nsp9 and nsp10 were amplified from mammalian expression vectors (a gift from N. Krogan)40. The CDSs were cloned into modified pET28a bacterial expression vectors containing N-terminal 6/8/10×His tags followed by the yeast Sumo (smt3) CDS amino acid mutations were introduced using QuikChange site-directed mutagenesis. In brief, primers were designed using the Agilent QuikChange primer design program to generate the desired mutation and were used in PCR reactions with PfuTurbo DNA polymerase. The reaction products were digested with Dpn1, transformed into DH5α cells and mutations were confirmed by Sanger sequencing.
For protein expression in Escherichia coli, ppSumo-SARS-CoV-2 nsps and mutants were cloned into a BamH1 site at the 5′ end, which introduced a Ser residue following the diGly motif in smt3. To make native N termini, the codon encoding the Ser was deleted using QuickChange mutagenesis. Thus, after cleavage with the ULP protease (after the diGly motif), the proteins contained native N termini. pGEX-2T-GST-eIF4E K119A41 was obtained from Addgene (112818).
Protein purification
nsp5, nsp7, nsp8, nsp10, nsp14 and nsp16
6×His-Sumo-nsp5/7/8/10/14/16 and the corresponding mutant plasmids (with native N termini following the diGly motif in Sumo) were transformed into Rosetta (DE3) E. coli or LOBSTR-BL21(DE3)-RIL cells under 50 μg ml−1 kanamycin exposure. LB Miller growth medium starter cultures (5 ml) containing 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol were grown for 2–4 h at 37 °C and then transferred to growth medium containing the same antibiotics. Typically, 2 l were grown for each protein. Protein expression was induced at an optical density (OD) of 0.7–1.0 by adding 0.4 mM IPTG and overnight incubation (16 h) at 18 °C. Cultures were centrifuged at 3,000g for 10 min and the bacterial pellet was resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 17.4 μg ml−1 PMSF, 15 mM imidazole pH 8.0 and 5 mM β-ME) and lysed by sonication. Lysates were centrifuged for 30 min at 30,000–35,000g and the supernatants incubated with Ni-NTA resin for 1–2 h at 4 °C. The Ni-NTA resin was washed with 50 mM Tris pH 8.0, 300 mM NaCl, 30 mM imidazole pH 8.0, 1 mM DTT and the protein was eluted in 50 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole pH 8.0 and 1 mM DTT. The eluted proteins were incubated with 5 μg ml−1 Ulp1 protease overnight at 4 °C. nsp7 was separated from 6×His–Sumo by anion exchange (Capto HiRes Q 5/50 column (Cytiva) equilibrated in 50 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT, eluted with 0–50% gradient of buffer containing 1 M NaCl). 6×His–SUMO–nsp8 was treated with Ulp1 on the Ni-NTA resin, to separate nsp8 and 6×His–SUMO in buffer with no imidazole. Proteins were further purified by size-exclusion chromatography using the Superdex 200 10/300 increase, Superdex 200 16/600, Superdex 75 10/300 increase or Superdex 75 16/600 column in 50 mM Tris pH 7.5–8.0, 150–300 mM NaCl and 1 mM DTT, depending on yield and size. The fractions containing proteins of interest were pooled, concentrated in an Amicon Utra-15 centrifugal filters (pore size, 3–50 kDa). The nsp16 protein was incubated with Ni-NTA resin post SEC to remove 6×His–SUMO. Purified proteins were aliquoted and stored at −80 °C.
nsp9
6×His-Sumo-nsp9 and respective mutant plasmids (with native N termini following the diGly motif in Sumo) were transformed into Rosetta (DE3) E. coli cells under 50 μg ml−1 kanamycin exposure. LB Miller growth medium starter cultures (5 ml) containing 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol were grown for 2–4 h at 37 °C and then transferred to Terrific Broth (TB) growth medium containing the same antibiotics and several drops of Antifoam B emulsion (Sigma-Aldrich, A5757). Protein expression was induced at an OD of 1.2 by adding 0.4 mM IPTG and incubating overnight (for 16 h) at 18 °C. The cultures were centrifuged at 3,000g for 10 min and the bacterial pellet was resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol 17.4 μg ml−1 PMSF, 15 mM imidazole pH 8.0 and 5 mM β-ME) and lysed by sonication. Lysates were centrifuged for 30 min at 30,000g and the supernatants incubated in Ni-NTA resin for 1–2 h at 4 °C. The Ni-NTA resin was washed with 50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 30 mM imidazole pH 8.0 and 1 mM DTT, and each protein was eluted in 50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 300 mM imidazole pH 8.0 and 1 mM DTT. The eluted protein was incubated with 5 μg ml−1 Ulp1 overnight at 4 °C. Proteins were diluted with 50 mM Tris pH 8.0, 10% glycerol, 1 mM DTT buffer to lower the NaCl concentration to 30 mM and subsequently ran through a Hi-Trap CaptoQ column where the flowthrough contained purified nsp9. NaCl was added to each protein to a final concentration of 150 mM, concentrated in an Amicon Ultra-15 with a 10 kDa molecular weight cut-off (MWCO), aliquoted and stored at −80 °C.
nsp12
8×His or 10×His-Sumo–nsp12 and respective mutant plasmids (with native N termini following the diGly motif in Sumo) were transformed into LOBSTR-BL21(DE3)-RIL E. coli cells under 50 μg ml−1 kanamycin exposure. LB Miller growth medium starter cultures (5 ml) containing 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol were grown for 2–4 h at 37 °C and then transferred to 1 l growth medium containing the same antibiotics. Protein expression was induced at an OD of 0.8–1.2 by adding 0.4 mM IPTG and incubating overnight (for 16 h) at 18 °C. The cultures were centrifuged at 3,000–3,500g for 10 min and the bacterial pellet was resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 17.4 μg ml−1 PMSF, 15 mM imidazole pH 8.0 and 5 mM β-ME) and lysed by sonication. The lysates were centrifuged for 30 min at 30,000–35,000g and the supernatants were incubated in Ni-NTA resin for 1–2 h at 4 °C. The Ni-NTA resin was washed with high-salt buffer (50 mM Tris pH 8.0, 1 M NaCl, 10% glycerol, 30 mM imidazole and 5 mM β-ME), followed by a high-imidazole wash (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 75 mM imidazole and 5 mM β-ME), and the protein was eluted in 50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 300 mM imidazole pH 8.0 and 1 mM DTT. The eluted proteins were incubated with 5 μg ml−1 Ulp1 overnight at 4 °C. Proteins were further purified by size-exclusion chromatography using the Superdex 200 10/300 increase column, or Superdex 200 16/600 in 50 mM Tris pH 8.0, 150–300 mM NaCl and 1 mM DTT. The fractions containing nsp12 were pooled, concentrated in an Amicon Ultra-15 with a 30–50 kDa MWCO centrifugal filter, aliquoted and stored at −80 °C.
nsp13
6×His-Sumo-nsp13 (used in Extended Data Fig. 10c) or 10×His-Sumo-nsp13 (used for [α32P]GTP conversion into GDP) and respective mutant plasmids (with native N termini following the diGly motif in Sumo) were transformed into Rosetta (DE3) E. coli cells under 50 μg ml−1 kanamycin exposure. LB Miller growth medium starter cultures (5 ml) containing 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol were grown for 2–4 h at 37 °C and then transferred to 1 l of growth medium containing the same antibiotics. Protein expression was induced at an OD of 1.0 by adding 0.4 mM IPTG and overnight incubation (16 h) at 18 °C. The cultures were centrifuged at 3,000–3,500g for 10 min and the bacterial pellet was resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 17.4 μg ml−1 PMSF, 15 mM imidazole pH 8.0 and 5 mM β-ME) and lysed by sonication. The lysates were centrifuged for 30 min at 30,000–35,000g and the supernatants were incubated in Ni-NTA resin for 1–2 h at 4 °C. Ni-NTA resin for 6×His–Sumo–nsp13 was washed with 50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 30 mM imidazole pH 8.0 and 1 mM DTT. 10×His–Sumo–nsp13 was washed with high-salt buffer (50 mM Tris pH 8.0, 1 M NaCl, 10% glycerol, 30 mM imidazole and 5 mM β-ME), followed by a high-imidazole wash (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 75 mM imidazole and 5 mM β-ME), and the protein was eluted in 50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 300 mM imidazole pH 8.0 and 1 mM DTT. The eluted protein was incubated with 5 μg ml−1 Ulp1 overnight at 4 °C. Proteins were buffer-exchanged or dialysed into a buffer containing 50 mM Bis-Tris pH 6.0, 30 mM NaCl, 10% glycerol and 1 mM DTT, and ion-exchange chromatography was then performed in a 5/50 MonoS column. The fractions containing nsp13 were pooled and further purified by size-exclusion chromatography using the Superdex 200 10/300 increase or Superdex 200 16/600 column in 50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol (only in 6×His), 1 mM DTT. The fractions containing nsp13 were pooled, concentrated in an Amicon Ultra-15 with a 30–50 kDa MWCO, aliquoted and stored at −80 °C.
eIF4E
For the production of GST–eIF4E(K119A), LOBSTR-BL21(DE3)-RIL cells were transformed with pGEX-2T-GST-eIF4E(K119A)41 and were grown in LB supplemented with 100 μg l−1 ampicillin. Protein expression was induced at an OD of 0.6 by adding 0.4 mM IPTG and incubating overnight (for 16 h) at 25 °C. The cultures were centrifuged at 3,000–3,500g for 10 min and the bacterial pellet was resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 17.4 μg ml−1 PMSF and 5 mM β-ME) and lysed by sonication. Lysates were centrifuged for 30 min at 35,000g and the supernatants incubated with Pierce Glutathione resin for 1–2 h at 4 °C. The resin was washed with lysis buffer, and the GST–eIF4E(K119A) eluted with 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM glutathione, 1 mM DTT. The protein was purified using size-exclusion chromatography on the Superdex 200 16/600 column in 50 mM Tris-HCl pH 8.0, 300 mM NaCl and 1 mM DTT, concentrated and stored as described above.
Ipp1
For the production of yeast inorganic pyrophosphatase (ipp1), The S. cerevisiae ipp1 CDS was cloned into pProEx2 containing an N-terminal 6×His-TEV linker and was transformed into Rosetta E. coli cells under 100 μg ml−1 ampicillin exposure. LB Miller growth medium starter cultures (5 ml) containing 100 μg ml−1 ampicillin were grown for 2–4 h at 37 °C and then transferred to 1 l growth medium containing the same antibiotics. Protein expression was induced at an OD of 0.8–1.2 by adding 0.4 mM IPTG and incubating (for 16 h) at 18 °C. The cultures were centrifuged at 3,000–3,500g for 10 min and the bacterial pellet was resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 17.4 μg ml−1 PMSF, 15 mM imidazole pH 8.0 and 5 mM β-ME) and lysed by sonication. The lysates were centrifuged for 30 min at 35,000g and the supernatants incubated in Ni-NTA resin for 1 h at 4 °C. Ni-NTA resin was washed with high-salt buffer (50 mM Tris pH 8.0, 1 M NaCl, 30 mM imidazole and 5 mM β-ME), and was eluted in 50 mM Tris pH 8.0, 50 mM NaCl, 300 mM imidazole pH 8.0 and 1 mM DTT. The eluted protein was loaded onto the Capto HiRes Q 5/50 column (Cytiva) equilibrated in 50 mM Tris pH 8.0, 50 mM NaCl and 1 mM DTT, eluted with a 0–50% gradient of buffer containing 1 M NaCl. Protein was further purified by size-exclusion chromatography using the Superdex 200 16/600 column in 25 mM Tris pH 7.5, 50 mM NaCl and 2 mM DTT. The fractions containing YIPP were pooled, concentrated and stored as described above.
nsp8–nsp9 fusion
The 6×His-Sumo-nsp8-nsp9 plasmid and mutants (N1A, N1D and N2A) were transformed into Rosetta (DE3) E. coli. Cells were grown in Terrific Broth medium in the presence of 50 μg ml−1 kanamycin and 25 μg ml−1 chloramphenicol to an OD of 1.0 and induced with 0.4 mM IPTG for 16 h at 18 °C. The cultures were centrifuged at 3,500g for 15 min, and the pellets were resuspended in lysis buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 25 mM imidazole and 10% glycerol) in the presence of 1 mM PMSF. Cells were lysed by sonication and the lysates were cleared by centrifugation at 25,000g for 1 h. The lysate was passed over Ni-NTA beads, which were washed with lysis buffer. The protein samples were eluted with elution buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 300 mM imidazole, 5% glycerol) and cleaved overnight at 4 °C with Ulp Sumo protease. The protein samples were further purified into cleavage assay buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5% glycerol) by size-exclusion chromatography using the Superdex 75 Increase 10/300 GL column.
GTase-activity assays
GTase-activity assays were performed as described previously26. Reactions were assembled in 20 µl containing 50 mM Tris pH 8.0, 5 mM KCl, 1 mM DTT, 0.005 U ml−1 inorganic pyrophosphatase, 10 μM 5′-pppACCCCCCCCCCCCCCCCCCC-3’ (5′-pppRNAA19C), 1.25 mM RNase inhibitor and, where indicated, 0.5 μM nsp12, nsp12(D218A), nsp13, nsp13(K288A) or 1 U ml−1 of VCE. The reactions were started with 1 mM MgCl2, 100 µM [α-32P]GTP (specific radioactivity = 1,000 cpm pmol−1) and incubated for 1 h at 37 °C. Half of the reaction was stopped by adding 0.8 U ml−1 proteinase K and was incubated for 30 min at 37 °C before the addition of 2× RNA loading dye (Novex) and incubated for 3 min at 95 °C. The reaction products were resolved in a 15% TBE-urea–PAGE gel. The gel was then stained with toluidine blue O and the 32P signal was detected by autoradiography.
The other half of the GTase reactions were treated with 10 U ml−1 P1 nuclease for 1 h at 37 °C. The reactions were then split in half again with one half treated with 1 U ml−1 Quick CIP for 30 min at 37 °C. The reactions were spotted on a PEI cellulose TLC plate and developed in a 0.4 M ammonium sulfate (NH4)2SO4 solvent system. The plate was dried and the 32P signal was detected by autoradiography.
NMPylation assays
NMPylation reactions were carried out in 20 µl containing 50 mM Tris (pH 7.5), 5 mM KCl, 1 mM DTT, 16 µM nsp7, nsp8 or nsp9 (and mutants), and 4.8 nM nsp12 (and mutants). The reactions were started with 1 mM MgCl2 or MnCl2, 200 µM [α-32P]ATP, [α-32P]UTP, [α-32P]GTP or [α-32P]CTP (specific radioactivity = 1,000 cpm pmol−1). The reactions were incubated at 37 °C for 5 min and stopped by adding 2 µl of 500 mM EDTA, followed by addition of 5× SDS–PAGE sample buffer with 10% β-ME and incubated for 3 min at 95 °C. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized by staining with Coomassie Brilliant Blue. The 32P signal was detected by autoradiography and scintillation counting.
For quantification of stoichiometry for nsp7, nsp8 and nsp9, [α-32P]UTP was used and the reactions were allowed to proceed for 16 h at 37 °C and processed as above.
nsp9 NMPylation kinetics
NMPylation reactions were carried out in a 20 µl reaction containing 50 mM Tris (pH 7.5), 5 mM KCl, 1 mM DTT, 16 µM nsp9, and 4.8 nM nsp12. The reactions were started by adding MnCl2 and [α-32P]ATP, CTP, GTP or UTP as indicated. The final concentration in the reaction was 0.5–200 µM (specific radioactivity of around 5,000 cpm pmol−1) of the indicated nucleotide triphosphate and 1 mM MnCl2. The reactions were incubated at 37 °C for 5 min and stopped by adding 2 µl of 500 mM EDTA, followed by the addition of 5× SDS–PAGE sample buffer + β-ME and boiling for 2–5 min. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized by staining with Coomassie Brilliant Blue. Incorporation of 32P was quantified by excising the nsp9 bands from the gel and scintillation counting. Background radioactivity was subtracted from each measurement. Rate measurements were fit to Michaelis–Menten kinetic models and Km and Vmax values for substrates were calculated by nonlinear regression using Prism v.9.3.0 for macOS (GraphPad Software; www.graphpad.com).
NMR
For NMR studies, non-isotopically enriched AMPylated nsp9 was dissolved in 50mM Tris buffer at pH 7.5, 150 mM NaCl, 1 mM DTT and 10% D2O for spectrometer locking. The final protein concentration of this solution was 0.5 mM. A total volume of 500 μl was then used with a 5 mm NMR tube to record all of the spectra.
All of the NMR experiments were run on the Bruker Avance III spectrometer operating at 600 MHz (1H) and equipped with a 5 mm proton-optimized quadruple resonance cryogenic probe. The temperature of the sample was regulated at 308 K throughout data collection.
A one-dimensional (1D) 31P spectrum was recorded with 8,192 scans and a repetition delay of 1.5 s for a total collection time of 3.5 h. The 31P spectral window and offset were set to 17 ppm and 2.6 ppm, respectively. Waltz16 decoupling was used on 1H during 31P acquisition.
To observe contacts between 31P and the nearest 1H nuclei, a 2D 1H, 31P-HSQC spectrum was recorded with 1,024 and 22 complex points in the direct 1H and indirect 31P dimensions, respectively. The spectral window and offset were set to 16.7 ppm and 4.7 ppm for the 1H dimension and 3.4 ppm and 2.6 ppm for the 31P dimension, respectively. Each FID was accumulated with 1,536 scans with a repetition delay of 1 s for a total recording time of approximately 21 h.
A 2D 1H, 31P-HSQC–TOCSY spectrum was recorded using spectral window and offset parameters for the 1H and 31P dimensions similar to the HSQC spectrum described above. A 60 ms long 1H-1H TOCSY pulse train using a DIPSI-2 sequence and a field strength of 10 kHz was tagged at the end of the HSQC sequence to observe signals from 1H nuclei that were further away from 31P. Given the lower sensitivity of this experiment, each FID was accumulated with 4,096 scans and a repetition delay of 1 s was used for a total recording time of 2 days and 14 h.
Using a similar pulse sequence, a 2D 1H,1H-HSQC–TOCSY spectrum was also recorded by evolving the indirect 1H dimension instead of 31P. The spectral window for the 1H indirect dimension was set to 4.2 ppm, whereas the offset was maintained at 4.7 ppm as for the direct 1H dimension. Forty complex points were recorded for then indirect 1H dimension, using 2,048 accumulations for each FID and a repetition delay of 1 s for a total recording time of 2 days and 14 h.
All 2D spectra were processed using NMRPipe42 and analysed using NMRFAM-SPARKY43.
Intact-mass analysis
Protein samples were analysed by LC–MS, using a Sciex X500B Q-TOF mass spectrometer coupled to an Agilent 1290 Infinity II HPLC. Samples were injected onto a POROS R1 reverse-phase column (2.1 mm × 30 mm, 20 µm particle size, 4,000 Å pore size) and desalted. The mobile phase flow rate was 300 μl min−1 and the gradient was as follows: 0–3 min, 0% B; 3–4 min, 0–15% B; 4–16 min, 15–55% B; 16–16.1 min, 55–80% B; 16.1–18 min, 80% B. The column was then re-equilibrated at the initial conditions before the subsequent injection. Buffer A contained 0.1% formic acid in water and buffer B contained 0.1% formic acid in acetonitrile.
The mass spectrometer was controlled by Sciex OS v.1.6.1 using the following settings: ion source gas 1, 30 psi; ion source gas 2, 30 psi; curtain gas, 35; CAD gas, 7; temperature, 300 °C; spray voltage, 5,500 V; declustering potential, 80 V; collision energy, 10 V. Data were acquired from 400–2,000 Da with a 0.5 s accumulation time and 4 time bins summed. The acquired mass spectra for the proteins of interest were deconvoluted using BioPharmaView v.3.0.1 (Sciex) to obtain the molecular mass values. The peak threshold was set to ≥5%, reconstruction processing was set to 20 iterations with a signal-to-noise threshold of ≥ 20 and a resolution of 2,500.
RNAylation assays
RNAylation reactions were typically carried out in a 10 µl volume containing 50 mM Tris (pH 7.5), 5 mM KCl, 1 mM DTT, 0.4 µg yeast inorganic pyrophosphatase, 20 µM nsp9 and 1 µM nsp12. The reactions were started by adding MnCl2 and 5′-pppRNALS10 to a final concentration of 1 mM and 100 µM, respectively. The reactions were incubated at 37 °C for the indicated time points and stopped by addition of 5× SDS-PAGE sample buffer + β-ME and boiling the samples for 5 min. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized by Coomassie staining and quantified with Fiji44 where indicated.
For the reactions testing RNA length specificity (Fig. 2c), 7.5 µl of a reaction master mix containing nsp9, nsp12, and yeast inorganic pyrophosphatase was added to 2.5 µl of start mix consisting of MnCl2 and the indicated 5′-pppRNA. The final reaction conditions were as follows: 50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT, 0.4 µg yeast inorganic pyrophosphatase, 20 µM nsp9, 2 µM nsp12, 1 mM MnCl2 and 100 µM of the indicated RNA. The reactions were incubated for 30 min at 37 °C, then stopped by addition of 5× SDS–PAGE sample buffer + β-ME and boiling the samples for 5 min. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized by Coomassie staining.
For RNAylation reactions comparing different RNA sequences (Fig. 2d,e) or nsp9 mutants (Fig. 5i), reactions were performed as described above, except using 1 µM nsp12. The reactions were incubated for 5 min and stopped by addition of 5× SDS–PAGE sample buffer + β-ME and boiling the samples for 5 min. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized by Coomassie staining.
For nsp12-mutant RNAylation comparisons, reactions contained 0.5 μM nsp12 WT or the indicated mutants and were conducted at room temperature for 60 min. Triplicate reactions were stopped with 5× SDS–PAGE loading dye, resolved on 15% SDS–PAGE gels, stained with Coomassie Brilliant Blue R and quantified with Fiji44.
Purification of nsp9–pRNALS10 species
Purified native nsp9 (0.8 mg ml−1, 65 μM) was incubated at room temperature overnight with 130 μM of 5′-pppRNALS10 and ~0.8 μM of nsp12 in presence of 0.05 mg ml−1 yeast inorganic pyrophosphatase and 1 mM MnCl2, in the reaction buffer (50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT). The samples were clarified by centrifugation to remove any precipitate and applied directly onto a Capto HiRes Q 5/50 column (Cytiva) equilibrated in 50 mM Tris pH 8.0, 50 mM NaCl and 1 mM DTT. An elution gradient of 0–50% with 1 M NaCl was applied over 30 column volumes. Under these conditions, RNA and nsp9–pRNALS10 bound to the column and unmodified nsp9 did not. nsp9–pRNALS10 and unreacted RNALS10 eluted as a peak doublet around 70 mS cm−1. The fractions were pooled, and further purified over Superdex 75 increase 10/300 GL (50 mM Tris pH 8.0, 300 mM NaCl, 1 mM DTT), separating nsp9–pRNALS10 from unreacted RNALS10 and nsp12. nsp9–pRNALS10 was quantified by spectrophotometry with an estimated extinction coefficient of ϵ260 = 130,650 M−1 cm−1. We also generated nsp9–pRNALS10 in the absence of inorganic pyrophosphatase. This nsp9–pRNALS10 was used in control reactions to test for PPi hydrolysis, necessary to ensure that the PPi-mediated deRNAylation reactions shown in Extended Data Fig. 5d were not affected by pyrophosphate hydrolysis. The results were similar to those presented in the figure, therefore confirming that there was no contaminating inorganic pyrophosphatase in the assays.
DeRNAylation of nsp9–pRNALS10
DeRNAylation reactions were typically performed in a 10 µl reaction volume consisting of 50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT, 20 µM nsp9–pRNALS10, 500 nM nsp12, 1 mM MgCl2 and 500 µM GDP. The reactions were started by adding MgCl2/GDP and incubated at 37 °C for 5–60 min as indicated. The reactions were stopped by addition of 5× SDS–PAGE sample buffer + β-ME and boiling the samples for 5 min. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized with Coomassie staining.
For nsp12 mutant deRNAylation comparison, reactions contained 0.5 μM nsp12 WT or mutants and were conducted at room temperature for 45 min. Triplicate reactions were stopped with 2× formamide loading dye, resolved on 15% (19:1) TBE urea–PAGE gels, stained with SYBR Gold (Thermo Fisher Scientific) and quantified with Fiji44.
For deRNAylation reactions comparing various NTPs and NDPs, reactions were performed in 10 µl volume consisting of 50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT, 20 µM nsp9–pRNALS10, 500 nM nsp12, 1 mM MgCl2 and 500 µM of the indicated NTP or NDP. The reactions were incubated for 5 min at 37 °C and stopped by addition of 5× SDS–PAGE sample buffer with β-ME and boiling for 5 min. The reaction products were resolved by SDS–PAGE on a 4–20% gradient gel and visualized with Coomassie staining.
Generation of [α-32P]GDP using nsp13
To generate [α-32P]GDP from [α-32P]GTP, 0.3–1 mM of [α-32P]GTP (specific activity ~2,000 cpm pmol−1) was incubated with 0.5–1 mg ml−1 nsp13 (and in some cases yeast cet1 NTPase) in 20 µl reaction buffer (depending on the amount needed) consisting of 50 mM Tris (pH 7.5), 5 mM KCl, 1 mM DTT and 2 mM MgCl2. The reactions were started by addition of enzyme and allowed to proceed for 30 min at 37 °C. After incubation for 30 min, the reactions were boiled at 95 °C for 5 min to inactivate the nsp13 or cet1.
To compare [α-32P]GDP and [α-32P]GTP in RNA capping reactions (Extended Data Fig. 10a), [α-32P]GTP was treated as described above, except without the addition of nsp13.
Generation of radiolabelled GpppA-RNALS10 from nsp9–pRNALS10 and [α-32P]GDP
Reactions were performed in a 10 µl volume containing 50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT, 15 µM nsp9–pRNALS10, 377 nM nsp12, 1 mM MgCl2 and 500 µM [α-32P]GDP (specific radioactivity of around 2,000 cpm pmol−1). The reactions were started by addition of [α-32P]GDP/MgCl2 mixture (generated as described above) and incubated for 30 min at 37 °C. As a control, VCE was used but with [α-32P]GTP. VCE assays were generally performed as described in the NEB Capping Protocol (M2080) with the following modifications: the reaction contained 20 µM of 5′-pppRNALS10, 500 µM [α-32P]GTP (specific activity of around 2,000 cpm pmol−1), and did not contain SAM. The reactions were stopped by the addition of 2× TBE-urea sample buffer, boiled for 5 min and resolved by urea–PAGE (20%).
nsp13 ATPase and GTPase malachite green kinetics
nsp13 triphosphatase reactions were carried out in a 30 µl reaction volume containing 50 mM Tris (pH 7.5), 5 mM KCl, 1 mM DTT and 30 nM nsp13. The reactions were started by adding ATP/MgCl2 or GTP/MgCl2 as indicated. The final concentrations in the reactions were 8–500 µM of the indicated nucleotide triphosphate and 1 mM MgCl2. The reactions were incubated at room temperature for 10 min and stopped by adding 30 µl of malachite green reagent. Malachite green absorbance was measured with a Spectramax ID3 spectrometer (Molecular Devices). Background activity was subtracted from each measurement and a phosphate standard curve was generated with KH2PO4 to determine the amount of inorganic phosphate generated. Rate measurements were fit to Michaelis–Menten kinetic models and Km and Vmax values for substrates were calculated by nonlinear regression using Prism v.9.3.0 for macOS (GraphPad Software).
GDP inhibition of RNAylation (one-pot capping assays)
Reactions were performed in 50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT, 1 mM MgCl2 and 1 mM MnCl2, and contained 20 μM nsp9, 2 μM nsp12 and 100 μM 5′-pppRNALS10 in a volume of 20 μl. The [α-32P]GDP was prepared as described above (using 400 μM GTP, [α-32P]GTP at a specific activity of around 1,500 cpm pmol−1) and diluted to final reaction concentrations in the range of 6.25–100 μM. The reactions were started by the addition of nsp12. GDP was added either before the addition of nsp12 (t = 0), or after 30 min of preincubation. After an additional 30 min, the reactions were split in half and stopped by the addition of 5× SDS-PAGE or 2× formamide loading dyes and the products were analysed by 4–20% gradient SDS–PAGE gel, or 15% 19:1 TBE urea–PAGE gel, respectively.
LC–MS/MS analysis of GpppA
DeRNAylation reactions (in triplicate) were performed in 20 µl of buffer solution containing 50 mM Tris pH 7.5, 5 mM KCl, 20 µM nsp9–pRNALS10, 2 µM WT nsp12 or the D218A mutant, 1 mM MgCl2 and 100 µM GDP. The reactions were started by adding MgCl2 and GDP and were allowed to proceed for 1 h at 37 °C. After incubation for 1 h, the reactions were supplemented with 2 µl of 10× P1 buffer and 1 µl of nuclease P1 enzyme and allowed to proceed for an additional 30 min at 37 °C. The reactions were stopped by boiling for 5 min and submitted for LC–MS/MS analysis.
For standards, 20 µl of blank reaction buffer (50 mM Tris pH 7.5, 5 mM KCl, 1 mM DTT) plus 40 µl blank reaction buffer containing 0.33 µM (final) m7GpppA (m7G(5′)ppp(5′)A RNA Cap Structure Analogue, New England Biolabs, S1405S) as an internal standard (IS) was spiked with varying concentrations of GpppA (New England Biolabs, S1406L). The reaction samples (20 µl) were diluted with blank reaction buffer, containing 0.33 µM (final) m7GpppA IS, at 1:2 to a total volume of 60 µl. Standards and samples were mixed with 60 µl of 100% methanol, vortexed and then centrifuged for 5 min at 16,100g. The supernatant was removed and analysed by LC–MS/MS using a Sciex QTRAP 6500+ mass spectrometer coupled to a Shimadzu Nexera X2 LC. GpppA was detected with the mass spectrometer in positive multiple reaction monitoring mode by following the precursor to fragment ion transition 772.9 to 604.0. A Thermo Fisher Scientific BioBasic AX column (2.1 × 50 mm, 5 μm packing) was used for chromatography with the following conditions: buffer A: 8:2 dH2O:acetonitrile + 10 mM ammonium acetate, pH 6; buffer B: 7:3 dH2O:acetonitrile + 1 mM ammonium acetate, pH 10.5, 0.5 ml min−1 flow rate; 0–1 min, 0% B; 1–2.5 min, gradient to 35% B; 2.5–5 min, 35% B; 5–7 min, gradient to 65% B; 7–10 min, 65% B; 10–10.5 min, gradient to 100% B; 10.5–15 min, 100% B; 15–15.5 min, gradient to 0% B; 15.5–20.5 min, 0% B. m7GpppA (transition 787.1 to 508.0) was used as an internal standard. Peak areas were determined, and data were further analysed using the Sciex Analyst v.1.7.2 software package. Back-calculations of standard curve samples were accurate to within 15% for 100% of these samples at concentrations ranging from 0.001 µM to 10 µM. A limit of detection was defined as a level three times that observed in blank reaction buffer and the limit of quantification as the lowest point on the standard curve that gave an analyte signal above the limit of detection and within 20% of nominal after back-calculation. The limit of quantification for GpppA was 0.005 µM.
Methyltransferase assays
In 10 μl reactions, 40 μM nsp9–pRNALS10 was incubated with 2 μM nsp12 in the presence of 1 mM MgCl2 and 100 μM [32P]GDP (generated as described above) for 60 min at 37 °C. The reactions were filled to 15 μl with nsp14 and SAM (final 0.05 mg ml−1 and 100 μM, respectively), and incubated for another 30 min. Treatment with nsp10 and nsp16 can be done concurrently with nsp14; however, the nsp10–nsp14 complex partially processes RNALS10, resulting in a mobility shift45. Thus, before addition of nsp10 and nsp16, nsp14 exonuclease activity was removed by heat inactivation (5 min at 95 °C). For 2′-O methylation, the reactions were supplemented with nsp10 and nsp16 and fresh SAM to final concentrations of 0.05 mg ml−1 nsp10, 0.05 mg ml−1 nsp16, 100 μM SAM in a final volume of 20 μl. Vaccinia reactions were conducted according to the manufacturer’s instructions. Reactions were stopped by adding 2× formamide loading dye and were separated on 20% TBE-urea polyacrylamide gels (19:1). Radioactivity was visualized by autoradiography and RNA by toluidine staining. For TLC analysis, bands with detectible 32P signal were excised, fragmented and incubated overnight at 55 °C in elution buffer (1 M ammonium acetate, 0.2% SDS and 20 mM EDTA), rotating top over bottom. The solutions were filtered using 0.22 μm centrifugal filters, supplemented with 23 μg Glyco Blue co-precipitant (Invitrogen) and precipitated for 1 h at −20 °C by addition of isopropyl alcohol to a final concentration of 60%. The pellets were washed once with 70% ethanol and reconstituted in 10 μl of P1 buffer with P1 nuclease (NEB). After 30 min at 37 °C, the reactions were supplemented with Quick CIP (NEB) and rCut Smart buffer to a final volume of 12 μl. After further incubation for 30 min, the reactions were spotted onto PEI-Cellulose F TLC plates and resolved in 0.4 M ammonium sulfate mobile phase. Beforehand, TLC plates were prepared by development in water, removing yellow discoloration. The 32P signal was detected by autoradiography, and compared with cold standards of GTP, GDP, GpppA and m7GpppA detected by absorption of plate fluorescence, excitable with a ultraviolet lamp λ = 265 nm. The position of m7GpppA2′-O-Me was determined from the VCE and vaccinia 2′-O-methyltransferase control reaction.
Reactions with 14C-labelled SAM were conducted as described above, with two differences: cold GDP was used at 100 μM (Millipore Sigma, G7127) and 55 μM [14C]SAM (Perkin Elmer); SAM was used at the supplied radioactivity of 52.6 mCi mmol−1 (~117 cpm pmol−1), with no further dilution using cold SAM.
GST–eIF4E pulldown of 7MeGpppA-RNA
Capping reactions were set up in 20 μl and contained 2 μM nsp12, 0.04 mg ml−1 nsp14 WT or D331A, 30 μM nsp9–pRNALS10, 100 μM [α32P]GDP (specific radioactivity = 1,000 cpm pmol−1), 100 μM SAM, 2 mM MgCl2. The reaction buffer was 5 mM Tris pH 8.0, 5 mM KCl, 1 mM DTT. VCE controls were performed according to manufacturer’s instructions, with the same nucleotide and SAM concentrations as nsp12 reactions. After incubation for 90 min at 37 °C, 15.6 μg of GST–eIF4E(K119A) was added, along with 15 μl of Glutathione resin (Pierce, Thermo Fisher Scientific). The reactions were filled to 700 μl with 50 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT and nutated for 1 h. The resin was washed three times with 500 μl of 50 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT and radioactive signal was quantified by scintillation counting.
Cryo-EM grid preparation
To form nsp12–nsp7–nsp8 core complex (RTC), native nsp12, nsp7 and nsp8 were incubated at a 1:2:4 molar ratio and run over the Superdex 200 increase 10/300 GL column to separate unassociated monomers. The purified complex was concentrated using spin concentrators (Amicon, 10 kDa MWCO, Sigma-Millipore), and quantified by spectrophotometry. A 3× molar excess of nsp9 over RTC was added, followed by 0.05 mM final DDM detergent immediately before freezing. The final concentration of nsp12–nsp7–nsp8 was 2 mg ml−1. The buffer contained 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT, 2 mM MnCl2 and 1 mM UMP-NPP. Copper Quantifoil 1.2/1.3 mesh 300 grids were used to freeze 3.5 μl of sample at 100% relative humidity using Vitrobot Mark IV (Thermo Fisher Scientific).
Cryo-EM data collection
Before data collection, sample grids were screened on the Talos Artica microscope at the Cryo Electron Microscopy Facility (CEMF) at UT Southwestern. Cryo-EM data of nsp12–nsp7–nsp8–nsp9 complex were collected on the Titan Krios microscope at the Cryo-Electron Microscopy Facility (CEMF) at UT Southwestern Medical Center, with the post-column energy filter (Gatan) and a K3 direct-detection camera (Gatan), using SerialEM46. A total of 4,770 videos were acquired at a pixel size of 0.55 Å in super-resolution counting mode, with an accumulated total dose of 54 e− Å−2 over 50 frames. The defocus range of the images was set as −1.0 to −2.5 μm.
Image processing and 3D reconstruction
Unless described otherwise, all datasets were processed with Relion47. Videos were aligned and summed using MotionCor248, with a downsampled pixel size of 1.09 Å. The CTF parameters were calculated using Gctf49, and images with estimated CTF max resolution better than 5 Å were selected for further processing. A total of 4,196,086 particles were picked using crYOLO50 from 4,757 images, and extracted with a rescaled pixel size of 2.19 Å. A total of 663,999 particles were selected and re-extracted after multiple rounds of 2D and 3D classifications in Relion with the original pixel size of 1.09 Å. An additional round of 3D classification was carried out, followed by particle reduction with a custom script to remove particles from dominant orientations. The remaining 89,945 particles were processed for 3D refinement, CTF refinement and particle polishing sequentially. A final round of 3D classification with a reference mask led to 39,985 particles, which were then imported into cryoSPARC51 for one round of non-uniform refinement. The map resolution was reported at 3.18 Å from cryoSPARC with the gold standard FSC method.
nsp5 cleavage reactions
Concentrated protein samples were diluted in cleavage buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5% glycerol) and each reaction was performed in a total volume of 10 μl. Initial experiments measured the nsp5 concentration dependence of the nsp8–nsp9 cleavage reaction. To measure the time dependence, nsp5 (2.5 μΜ final concentration) was added to the nsp8–nsp9 fusion protein (12.5 μΜ final concentration). The reactions were incubated at 37 °C for varying amounts of time (0 to 80 min) and terminated by boiling the samples for 5 min in the presence of SDS–PAGE loading buffer. The reaction products were resolved on a 4–20% gradient tris-glycine gel and the products were visualized by Coomassie staining.
Model building and refinement
The model was built using PDB 7CYQ as a template26. The model was manually rebuilt into the map using Coot52, and refined using Phenix real space refinement53. Model validation was performed using MolProbity software54.
Bioinformatics
A representative subset of NiRAN domain sequences, provided as a multiple alignment in the original NiRAN publication5, was supplemented by additional sequences used in this study (SARS-CoV-2, OC43, 229E strains). The human SELO sequence was added according to a FATCAT structural alignment55 between SARS-CoV-2 nsp12 and bacterial SELO (PDB: 7CYQ and 6EAC). The alignment was visualized using the ESPript server56.
SARS-CoV-2 infection experiments
Plasmid construction
To generate recombinant SARS-CoV-2 expressing ZsGreen mutants, the infectious clone pCC1-4K-SARS-CoV-2-Wuhan-Hu-1-ZsGreen was used as the parental backbone8. To generate nsp9(N1A, N1D or N2A) and nsp12(K73A) mutants, the mutations were introduced by overlap extension PCR. In brief, two fragments for each mutant were created by PCR using Herculase II Fusion DNA Polymerase (Agilent Technologies). These fragments shared homology at the 3′ end of fragment 1 and the 5′ end of fragment 2. The resulting 2 fragments were then used as a template for a third fragment to PCR a full-length amplicon containing the mutation flanked by Pac1 and MluI sites on the ends. Mutations were confirmed by DNA sequencing. To generate nsp12 mutants (D218A or D760A), a SARS-CoV-2 shuttle vector (ps1180.SARS-CoV-2-shuttle) was created using the ps1180.delXhoISacII plasmid as the backbone. Using Gibson cloning, a restriction enzyme linker that contained unique restriction sites specific to the SARS-CoV-2 genome was inserted. Smaller fragments of the SARS-CoV-2 genome were digested from the pCC1-4K-SARS-CoV-2-Wuhan-Hu-1-ZsGreen plasmid and ligated into ps1180.SARS-Cov-2-shuttle plasmid to create three new plasmids (MluI/SacI fragment for D218A region; SacI/Bsu36I fragment for D760A region). gBlocks containing mutations were synthesized by IDT and introduced into the SARS-CoV-2 shuttle vector by Gibson Assembly following standard protocols. To reassemble the full-length parental pCC1-4K-SARS-CoV-2-Wuhan-Hu-1-ZsGreen containing the new mutants, pCC1-4K-SARS-CoV-2-Wuhan-Hu-1-ZsGreen was digested with the PacI/MluI, MluI/SacI and SacI/Bsu36I restriction enzymes. An approximately 28–35 kb fragment was purified for each digest. PCR amplicons (nsp9N1A, nsp9N1D, nsp9N2A, nsp12K73A) were digested with PacI/MluI and the 5.3 kb fragment was purified. The SARS-CoV-2 shuttle plasmids were digested as follows: nsp12D218A (MluI/SacI releasing a 1.4 kb fragment); nsp12D760A (SacI/Bsu36I/PvuI releasing 3 kb fragments). All of the fragments were purified using the QIAexII Gel Purification Kit according to the standard protocol (Qiagen). The fragments were ligated together at a 3:1 ratio overnight. Ligated DNA was precipitated using 7.5 M ammonium acetate, glycogen and isopropanol, followed by a wash with ethanol. The DNA was electroporated into TransforMAX EPI300 electrocompetent E. coli (Lucigen). An overlapping eight-fragment PCR strategy was used to verify individual colonies by colony PCR. Confirmed colonies were grown in 10 ml tryptic soy broth (TSB; Sigma-Aldrich) containing 12.5 µg ml−1 chloramphenicol for 6–8 h, with shaking at 37 °C. A 10 ml culture was inoculated into 100 ml TSB/chloramphenicol culture and incubated overnight, with shaking at 37 °C. The overnight culture was diluted 1:5 into fresh TSB/chloramphenicol containing 0.1% arabinose and incubated for an additional 5 h. The bacteria were pelleted and the DNA was isolated using a homemade midi prep protocol followed by the Machery-Nagel NucleoBond Xtra Midi Kit (Thermo Fisher Scientific). Full-length infectious clone plasmid was confirmed by restriction digestion and eight-fragment PCR. A list of the oligonucleotides used is provided in Supplementary Table 3.
Virus production
To generate virus from DNA-based infectious clones, 3 µg of plasmid was transfected into 2 individual wells of a 6-well plate of 400,000 BHK-21J cells using X-treme Gene9 Transfection Reagent (Sigma-Aldrich). Three days after transfection, the supernatant from two individual wells was combined and 4 ml was transferred to a T25 flask containing 1 × 106 VeroE6-C1008-TMPRSS2 cells and incubated for 30 min. Inoculum was removed and 5 ml MEM supplemented with 10% FBS, 1× NEAA and 1× sodium pyruvate was added. After 4 days, 250 µl supernatant was added to 750 µl TriReagent for RNA extraction and RT–qPCR. The remaining supernatant was frozen for plaque assays. T25 flasks were fixed with 4% paraformaldehyde, imaged on the Nikon Eclipse Ti system and processed with ImageJ.
RNA extraction and iPCR
RNA was isolated using the Direct-zol RNA mini prep kit according to the manufacturer’s instructions (ZymoResearch). A 20 µl reaction contained 5 µl RNA, 5 µl TaqMan Fast Virus 1-Step Master Mix and 1.8 µl SARS-CoV-2 primer/probe set containing 6.7 µM of each primer and 1.7 µM probe (the final concentrations of primer and probe were 600 nM primer and 150 nM probe). SARS-CoV-2 primers and probes were designed as recommended by the Center for Disease Control (https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html). All oligonucleotides were synthesized by LGC Biosearch Technologies. RT was performed at 50 °C for 5 min, followed by inactivation at 95 °C for 2 min, and 40 cycles of PCR (95 °C for 3 s, 60 °C for 30 s) on the QuantStudio 3 (Applied Biosystems) system.
Cells
BHK-21J cells (a gift from C. Rice) were grown in MEM (Gibco) supplemented with 10% FBS and 1× NEAA. VeroE6-C1008 cells (ATCC) were transduced with lentiviral vector SCRBBL-TMPRSS2, selected with blasticidin (15 μg ml−1, Gibco) and maintained in MEM supplemented with 10% FBS, 1× NEAA, 1× sodium pyruvate and 8 µg ml−1 blasticidin.
Plaque assay
Viral titre was determined by plaque assay on Vero-C1008 cells (ATCC). Cells were plated at 650,000 cells per well and infected with 3 or 6 dilutions from a tenfold dilution series in 1% FBS/1× non-essential amino acids/MEM for 60 min at 37 °C, with rocking every 15 min. Overlay medium (2 ml) consisting of a 1:1 mix of 2× modified Eagle medium (Temin’s Modification) supplemented with 2 penicillin–streptomycin, 2× GlutaMAX, 10% FBS:2× Avicel RC-591 (Dupont) was added to cells. Cells were incubated for 3 days at 37 °C before the overlay medium was removed and cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 30 min at room temperature. The fixative was removed, and cells were stained for at least 5 min with crystal violet (0.2% crystal violet (Sigma-Aldrich), 20% ethanol), before removal and plaque enumeration.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-022-05185-z.
Supplementary information
Supplementary Fig. 1: uncropped gels. Supplementary Tables 1–3: lists of RNAs used in the study; data collection and refinement statistics; and the oligonucleotides used in the SARS-CoV-2 infection experiments.
Source Data Figs, 1, 2, 4 and 5 and Extended Data Figs. 3–5 and 9.
Acknowledgements
We thank the members of the Tagliabracci laboratory for discussions; D. Karlin for notifying us of the similarity between SELO and the NiRAN domain; B. Park for help with kinetics; A. Lemoff for help with intact-mass analysis; J. Kilgore, N. Williams and the staff at the UTSW Preclinical Pharmacology Core for detection and quantification of GpppA; S. Wilson and S. Rihn for the SARS-CoV-2 infectious clone and for technical guidance; and the staff at the Structural Biology Laboratory and the Cryo-Electron Microscopy Facility at UT Southwestern Medical Center (partially supported by grant RP170644 from the Cancer Prevention & Research Institute of Texas (CPRIT)) for cryo-EM studies. A portion of this research was supported by the W. M. Keck Foundation Medical Research Grant (to V.S.T., K.P. and J.W.S.), the National Institutes of Health grant R01GM135189 (to V.S.T.), 1DP1AI158124 (to J.W.S.), a Welch Foundation Grant I-1911 (to V.S.T.), a Life Sciences Research Foundation Fellowship (to G.H.) and a Polish National Agency for Scientific Exchange scholarship PPN/BEK/2018/1/00431 (to K.P.). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant R24GM141526. J.W.S. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease. V.S.T. is a Howard Hughes Medical Institute Investigator, a Michael L. Rosenberg Scholar in Medical Research, a CPRIT Scholar (RR150033) and a Searle Scholar.
Extended data figures and tables
Author contributions
G.J.P., A.O., G.H., J.W.S. and V.S.T. designed the experiments. G.J.P., A.O., G.H., J.L.E., A.M. and V.S.T. conducted experiments. G.J.P. discovered the RNAylation and GDP-PRNTase activity of the NiRAN domain. A.O., Z.C. and Y.L. performed the cryo-EM. A.O. performed methylation experiments. G.H. performed GTase and nsp9 NMPylation experiments. A.M. performed intact-mass data analysis. M.T. and K.H.-W. performed NMR experiments. K.P. performed the bioinformatics. G.J.P., A.O. and G.H. purified proteins. G.J.P., A.O. and V.S.T. performed cloning and site-directed mutagenesis. J.L.E. performed infectious SARS-CoV-2 experiments. G.J.P., A.O., K.P., J.W.S. and V.S.T. wrote the manuscript with input from all of the authors.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The data supporting the findings of this paper are available within the Article and its Supplementary Information, which contains raw images of gels. The atomic coordinates have been deposited at the PDB (7THM). The 3D cryo-EM map has been deposited in the Electron Microscopy Data Bank under the accession number EMD-25898. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gina J. Park, Adam Osinski
Extended data
is available for this paper at 10.1038/s41586-022-05185-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-022-05185-z.
References
- 1.Chen Y, Guo D. Molecular mechanisms of coronavirus RNA capping and methylation. Virol. Sin. 2016;31:3–11. doi: 10.1007/s12250-016-3726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2021;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker AP, et al. The SARS-CoV-2 RNA polymerase is a viral RNA capping enzyme. Nucleic Acids Res. 2021;49:13019–13030. doi: 10.1093/nar/gkab1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Vliet AL, Smits SL, Rottier PJ, de Groot RJ. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann KC, et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rihn SJ, et al. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021;19:e3001091. doi: 10.1371/journal.pbio.3001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.V’Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beigel JH, et al. Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, W. et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med.10.1126/scitranslmed.abl7430 (2021). [DOI] [PMC free article] [PubMed]
- 14.Sreelatha A, et al. Protein AMPylation by an evolutionarily conserved pseudokinase. Cell. 2018;175:809–821. doi: 10.1016/j.cell.2018.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudkiewicz M, Szczepinska T, Grynberg M, Pawlowski K. A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PLoS ONE. 2012;7:e32138. doi: 10.1371/journal.pone.0032138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slanina H, et al. Coronavirus replication-transcription complex: vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl Acad. Sci. USA. 2021;118:e2022310118. doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuichi Y, Morgan M, Muthukrishnan S, Shatkin AJ. Reovirus messenger RNA contains a methylated, blocked 5′-terminal structure: m-7G(5′)ppp(5′)G-MpCp. Proc. Natl Acad. Sci. USA. 1975;72:362–366. doi: 10.1073/pnas.72.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei CM, Moss B. Methylated nucleotides block 5′-terminus of vaccinia virus messenger RNA. Proc. Natl Acad. Sci. USA. 1975;72:318–322. doi: 10.1073/pnas.72.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin AJ. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J. Biol. Chem. 1976;251:5043–5053. doi: 10.1016/S0021-9258(17)33218-0. [DOI] [PubMed] [Google Scholar]
- 24.Ensinger MJ, Martin SA, Paoletti E, Moss B. Modification of the 5′-terminus of mRNA by soluble guanylyl and methyl transferases from vaccinia virus. Proc. Natl Acad. Sci. USA. 1975;72:2525–2529. doi: 10.1073/pnas.72.7.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov KA, et al. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L, et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conti BJ, Leicht AS, Kirchdoerfer RN, Sussman MR. Mass spectrometric based detection of protein nucleotidylation in the RNA polymerase of SARS-CoV-2. Commun. Chem. 2021;4:41. doi: 10.1038/s42004-021-00476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon, A. et al. Protein-primed RNA synthesis in SARS-CoVs and structural basis for inhibition by AT-527. Preprint at bioRxiv10.1101/2021.03.23.436564 (2021).
- 29.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Ogino T, Green TJ. RNA synthesis and capping by non-segmented negative strand RNA viral polymerases: lessons from a prototypic virus. Front. Microbiol. 2019;10:1490. doi: 10.3389/fmicb.2019.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonenberg N, Gingras AC. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 1998;10:268–275. doi: 10.1016/S0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Svetlov D, Artsimovitch I. NMPylation and de-NMPylation of SARS-CoV-2 nsp9 by the NiRAN domain. Nucleic Acids Res. 2021;49:8822–8835. doi: 10.1093/nar/gkab677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malone B, Campbell EA, Darst SA. CoV-er all the bases: structural perspectives of SARS-CoV-2 RNA synthesis. Enzymes. 2021;49:1–37. doi: 10.1016/bs.enz.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication–transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan L, et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184:3474–3485. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry JK, et al. An atomistic model of the coronavirus replication-transcription complex as a hexamer assembled around nsp15. J. Biol. Chem. 2021;297:101218. doi: 10.1016/j.jbc.2021.101218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino T, Yadav SP, Banerjee AK. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl Acad. Sci. USA. 2010;107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang B, et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotman JB, Giltmier AJ, Mukherjee C, Schoenberg DR. RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res. 2017;45:10726–10739. doi: 10.1093/nar/gkx801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogando NS, et al. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94:e01246-20. doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner T, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Jaroszewski L, Iyer M, Sedova M, Godzik A. FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 2020;48:W60–W64. doi: 10.1093/nar/gkaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: uncropped gels. Supplementary Tables 1–3: lists of RNAs used in the study; data collection and refinement statistics; and the oligonucleotides used in the SARS-CoV-2 infection experiments.
Source Data Figs, 1, 2, 4 and 5 and Extended Data Figs. 3–5 and 9.
Data Availability Statement
The data supporting the findings of this paper are available within the Article and its Supplementary Information, which contains raw images of gels. The atomic coordinates have been deposited at the PDB (7THM). The 3D cryo-EM map has been deposited in the Electron Microscopy Data Bank under the accession number EMD-25898. Source data are provided with this paper.