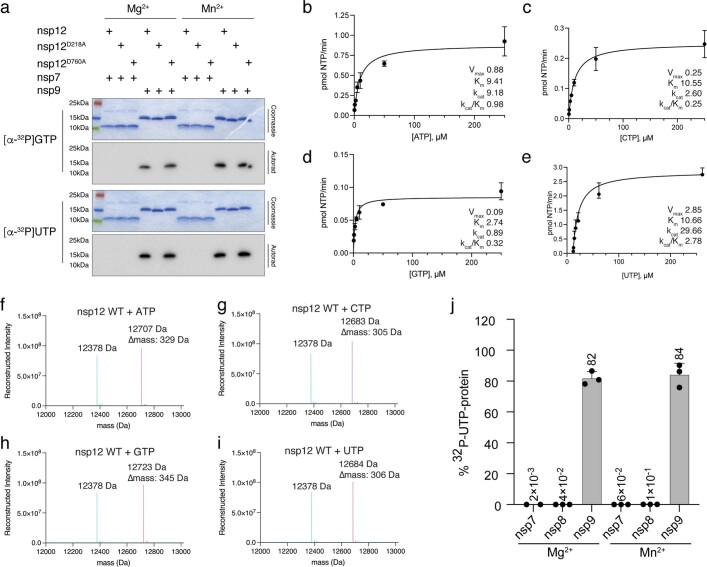
Extended Data Fig. 3. The NiRAN domain NMPylates nsp9.
a. Incorporation of α-32P from [α-32P]GTP or [α-32P]UTP into nsp7 or nsp9 by WT nsp12, the NiRAN mutant (D218A), or the polymerase mutant (D760A). Reactions were performed in the presence of Mg2+ or Mn2+ and the products were resolved by SDS-PAGE and visualized by Coomassie staining (top) and autoradiography (bottom). b–e. Kinetic analysis depicting the concentration dependence of (b) ATP, (c) CTP, (d) GTP, or (e) UTP on the rate of nsp9 NMPylation by the NiRAN domain. Km (μM), Vmax (pmol NTP/min), kcat (min−1) and kcat/Km (min−1/μM) values are indicated. Plots shown are the mean and SD of triplicate reactions. f–i. Intact mass LC/MS spectra of unmodified nsp9 (cyan) overlayed with NMPylated nsp9 (pink) following incubation with WT nsp12 and (f) ATP, (g) CTP, (h) GTP, or (i) UTP. The observed masses are shown in the insets. The theoretical mass of unmodified nsp9 is 12378.2 Da and the theoretical increase in mass with the addition of each NMP is as follows: AMP, 329 Da; CMP, 305 Da; GMP, 345 Da; UMP, 306 Da. j. Quantification of the stoichiometry of NMPylation of nsp7, nsp8 and nsp9. Reactions were performed in the presence of Mg2+ or Mn2+. Reaction products were resolved by SDS-PAGE, the corresponding protein bands were excised from the gel, and radioactivity was quantified by scintillation counting. Bars: mean ± SD of three independent experiments.