Abstract
The respiratory chain of the thermohalophilic bacterium Rhodothermus marinus contains an oxygen reductase, which uses HiPIP (high potential iron-sulfur protein) as an electron donor. The structural genes encoding the four subunits of this HiPIP:oxygen oxidoreductase were cloned and sequenced. The genes for subunits II, I, III, and IV (named rcoxA to rcoxD) are found in this order and seemed to be organized in an operon of at least five genes with a terminator structure a few nucleotides downstream of rcoxD. Examination of the amino acid sequence of the Rcox subunits shows that the subunits of the R. marinus enzyme have homology to the corresponding subunits of oxidases belonging to the superfamily of heme-copper oxidases. RcoxB has the conserved histidines involved in binding the binuclear center and the low-spin heme. All of the residues proposed to be involved in proton transfer channels are conserved, with the exception of the key glutamate residue of the D-channel (E278, Paracoccus denitrificans numbering). Analysis of the homology-derived structural model of subunit I shows that the phenol group of a tyrosine (Y) residue and the hydroxyl group of the following serine (S) may functionally substitute the glutamate carboxyl in proton transfer. RcoxA has an additional sequence for heme C binding, after the CuA domain, that is characteristic of caa3 oxidases belonging to the superfamily. Homology modeling of the structure of this cytochrome domain of subunit II shows no marked electrostatic character, especially around the heme edge region, suggesting that the interaction with a redox partner is not of an electrostatic nature. This observation is analyzed in relation to the electron donor for this caa3 oxidase, the HiPIP. In conclusion, it is shown that an oxidase, which uses an iron-sulfur protein as an electron donor, is structurally related to the caa3 class of heme-copper cytochrome c oxidases. The data are discussed in the framework of the evolution of oxidases within the superfamily of heme-copper oxidases.
In mitochondria, there is a single terminal enzyme, cytochrome c oxidase (aa3), in the respiratory electron transport system that couples the one-electron oxidation of cytochrome c to the four-electron reduction of dioxygen and to the translocation of protons across the membrane. In prokaryotes, the respiratory electron transport system is branched to different terminal oxidases which differ in oxygen affinity, electron donor specificity (e.g., quinol and cytochrome c), heme type, and metal compositions. This diversity can be partially correlated to the variability of growth conditions, namely, the availability of energy source and oxygen. Despite the differences, most of these terminal oxidases belong to the superfamily of heme-copper oxidases, which includes the mitochondrial aa3 cytochrome c oxidase (19, 82). However, bacterial and archaeal oxidases possess fewer protein subunits (generally three or four) than their mitochondrial counterpart.
The presence of a subunit homologous to the largest subunit of mammalian cytochrome c oxidase, subunit I, identifies the members of the superfamily. Subunit I contains a binuclear center, consisting of a heme and a copper ion (CuB) and a low-spin heme, which transfers electrons to the binuclear center (30, 79, 89). The presence of two hemes and CuB is common to all members of the superfamily of heme-copper oxidases (19, 82). Most of the prokaryote oxidases of the superfamily contain homologous proteins to subunits II and III of mitochondria, in addition to subunit I (69). As an example, the three-dimensional structures of the mitochondrial enzyme (79, 80) and Paracoccus denitrificans cytochrome aa3 (30, 52) show that their functional core structures are extremely similar with regard to subunits I, II, and III. Subunit II is, however, more diverse than subunit I, a fact that can be partially related to the type of electron donor. In the Cox subfamily, subunit II of cytochrome c oxidases contain a binuclear average-valence copper center, CuA, the initial electron acceptor from reduced cytochrome c (25), while quinol oxidases (82) do not have this center. Nevertheless, it was possible to engineer a CuA site in the subunit II of Escherichia coli bo3 quinol oxidase (83). In some cytochrome c oxidases, such as those from Thermus thermophilus (40) and several Bacillus species (62, 70), the C-terminal hydrophilic domain of subunit II has an extension containing a heme C binding site. These terminal enzymes constitute the class of caa3 and cao3 oxidases (19). As for subunit III of members of the Cox subfamily, its function is still controversial. Although the two-subunit enzyme is active in catalysis as well as in proton translocation, it has been reported that subunit III may be necessary for the correct assembling of the terminal oxidase (23) or for its stabilization (9). In the cbb3 cytochrome c oxidase subfamily, there are no mitochondrion-like subunits II and III. Instead, these oxidases have two subunits, containing one and two C-type heme centers, respectively (19).
Rhodothermus marinus is a strict aerobe and thermohalophile, one classified as a new genus of the group of Flexibacter-Bacteroides-Cytophaga (4). The R. marinus respiratory chain contains several unique features (55–59), namely, a novel quinone-oxidizing component: a multihemic bc complex, which is a functional analogue of the canonical bc1 complex, and an HiPIP (high potential iron-sulfur protein). The HiPIP is the electron carrier between the multihemic bc complex and the HiPIP:oxygen oxidoreductase, a terminal oxidase recently purified and characterized (55, 58, 59). This oxidase, isolated with four subunits with apparent molecular masses of 42, 35, 19, and 15 kDa is the first example of a terminal oxygen reductase having as its electron donor an iron-sulfur protein; it presents a higher turnover with HiPIP as an electron carrier than with mitochondrial or R. marinus cytochrome c (58). The oxidase contains hemes of the AS type, which have been reported only in T. thermophilus and in archaeal species (39).
HiPIPs are small soluble proteins containing a single tetranuclear cluster and are mainly found in purple photosynthetic bacteria (27, 45). It has been shown that HiPIPs can be oxidized upon photoexcitation of the photochemical reaction center through a specific redox interaction with the tetraheme cytochrome c (26, 29, 53). This finding indicates that HiPIPs have a role in the bacterial photosynthetic electron transfer chain. However, HiPIPs may also have a role in the respiratory oxidative processes, as first suggested by Pereira et al. (55) in R. marinus and by Hochkoeppler et al. (28) in the facultative phototroph Rhodoferax fermentans. Recently, with the purification of the caa3 oxidase and of the bc complex from R. marinus, it was possible to demonstrate that the HiPIP is indeed an electron carrier in the R. marinus respiratory chain, as stated above (58). Hence, in the last few years, evidence has been gathered indicating that HiPIP, a long-known iron-sulfur protein, functions as an analogue of mitochondrial cytochrome c, i.e., as an electron donor to a terminal oxygen reductase (8, 49, 58, 71).
In order to fully characterize the unique R. marinus HiPIP:oxygen oxidoreductase, it was essential to determine its complete primary structure. We report here the cloning and sequencing of the gene cluster encoding this enzyme. From the analysis of the deduced amino acid sequence of the subunits and both the previous and additional biochemical studies, we conclude that the HiPIP:oxygen oxidoreductase is clearly identified as a member of the superfamily of heme-copper terminal oxidases of the class caa3, in spite of the quite distinct electron donor. Some important new structural features, supported by modeling of the three-dimensional structure of this enzyme, are identified and are discussed in terms of the type of electron donor, proton transfer, and evolution of heme-copper oxidases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
R. marinus PRQ-62B was the source of the chromosomal DNA used to construct a lambda library in Lambda DASHRII replacement vector (Stratagene). E. coli strains XL1-Blue MRA (P2) and XL1-Blue MRA (Stratagene) were the host strains for the recombinant lambda phages. The first strain was used to amplify and to screen the genomic library. XL1-Blue MRA was used for the preparation of lysate from pure recombinant phage, followed by λ DNA extraction. Subcloning of lambda inserts into p-Zero-1 (Invitrogen) was performed in E. coli XL1-Blue (Stratagene). For further subcloning in pUC18 (see below), we used E. coli strain JM109 (Promega).
Media.
R. marinus PRQ-62B was grown in the medium described by Degryse et al. (15) supplemented with 0.25% yeast extract, 0.25% tryptone and 1% NaCl. Luria-Bertani (LB) (68) and LB low-salt (pH 7.5) (32) media were used for standard cultures of E. coli. Antibiotics were added at the following concentrations (in μg/ml) when necessary: ampicillin, 100; Zeocin, 50. XL1-Blue MRA (P2) and XL1-Blue MRA (Stratagene) strains were grown in LB medium containing 0.2% (wt/vol) maltose and 10 mM MgSO4.
Lambda stocks in SM buffer (68) were prepared from NZY plates (15 g of agar per liter) with NZY overlays containing 7 g of agarose per liter (68).
DNA techniques.
Genomic DNA was isolated with cetyltrimethylammonium bromide and chloroform extraction after cell lysis with sodium dodecyl sulfate (SDS) and proteinase K digestion (5). λ DNA was isolated essentially as described in Sambrook et al. (68), after 20% polyethylene glycol–2 M NaCl phage precipitation from 30- to 100-ml liquid lysates and treatment with DNase (10 μg/ml) and RNase (10 μg/ml) in order to degrade the bacterial DNA and RNA released during lysis. Plasmid DNA was prepared using a plasmid purification kit from Qiagen. For miniscale extractions, plasmid DNA was prepared from 2-ml overnight-grown cultures (7). In order to quick-check the size of E. coli plasmids, the procedure described by Akada (1) was used.
Restriction analysis of DNA was performed using restriction enzymes and buffers from Roche Molecular Biochemicals according to the conditions recommended by the manufacturer.
To prepare insert fragments for the library, genomic DNA was partially digested by Sau3A1 and submitted to sucrose gradient fractionation (68). The appropriate fractions (containing fragments in the range of 9 to 23 kb) were identified by agarose gel electrophoresis and combined. After dialysis and ethanol precipitation, the fractions were used in test ligations with T4 DNA ligase (Roche Molecular Biochemicals) to test the integrity of the λ DNA Sau3A extremities and in ligations to λ DASHRII–BamHI arms (Stratagene), which were followed by packaging and infection of XL1-Blue MRA (P2).
Cloning of the oxidase loci.
We used the degenerate primer 5′-TGGTKBTTYGGNCAYCCNGARGTNTAY-3′ and the degenerate reverse complementary primer 5′-NGTRWACATRTGRTGNRCCCANAC-3′ to amplify genomic DNA. These primers correspond to coding regions of the conserved sequences, established by the alignment of subunit I sequences of a representative group of terminal oxidases. After genomic DNA denaturation at 94°C for 5 min, conditions for the PCR were as follows: 94°C for 30 s, 50°C for 1 min, and 74°C for 1 min (35 cycles), followed by 1 cycle at 94°C for 30 s, 50°C for 1 min, and 74°C for 10 min.
The 174-bp reaction product of PCR was cloned in pGEMR-T vector (Promega) and sequenced. The NcoI-PstI fragment from this cloning was used as a homologous probe to screen the genomic library; labeling of this probe was performed with [α-32P]dCTP using the megaprime DNA labeling system (Amersham). Lambda plaques were transferred to nitrocellulose filters (68), where DNA was fixed by baking for 2 h at 80°C in an oven. The filters were then prehybridized and hybridized with the probe, at 50°C with 6× SSC (1× SSC is 0.15 M NaCl plus 15 mM trisodium citrate at pH 7.0), 5× Denhardt's reagent, 0.4% SDS (wt/vol), 20 mM NaH2PO4, and 500 μg of sonicated salmon sperm DNA per ml. Afterward, the filters were washed using 2× SSC and 2× SSC–0.1% SDS at the hybridization temperature.
After isolation and digestion of DNA from screened positive phages, Southern blot analysis (68) were performed, allowing us to select and subclone the lambda insert region containing the gene coding for oxidase subunit I. The probes were labeled nonradioactively with digoxigenin-11-labeled nucleotides (DIG-11-dUTP) using the DIG random-primer DNA labeling system (Roche Molecular Biochemicals). Hybridization with DIG-labeled probes and stringency washes were also performed as described by Roche Molecular Biochemicals.
The lambda insert region of interest, included in an approximately 6.4-kb HindIII-EcoRI fragment, was subcloned into p-Zero-1. Three independent fragments of this last construct, BamHI-SalI, BamHI-HindIII, and SalI-EcoRI, were subcloned into pUC18, yielding plasmids pMSAN1, pMSAN2, and pMSAN3, respectively. In the cloning procedure, dephosphorylation of plasmid vector was performed with Schrimp alkaline phosphatase (Amersham). The method described by Hanahan (24) was used to prepare and transform competent E. coli cells and resulted in transformation efficiencies of 107 CFU/μg of supercoiled plasmid DNA.
DNA sequencing and sequence analysis.
Inserts from plasmids pMSAN1, pMSAN2, and pMSAN3 were totally sequenced using LI-COR 4200 system (MWG-Biotech). The sequence strategy is represented in Fig. 1; the nucleotide sequence of both strands was obtained using standard pUC forward and reverse primers and synthetic oligonucleotide primers.
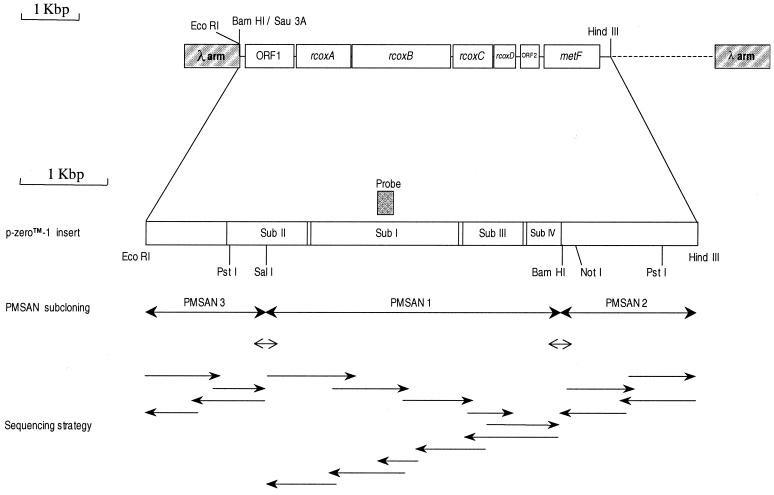
FIG. 1.
Map of R. marinus PRQ-62B genes sequenced in this work. The gene cluster includes ORF1, as well as rcoxA, rcoxB, rcoxC, and rcoxD, the structural genes encoding subunits II, I, III, and IV of HiPIP oxidoreductase, respectively. A terminator structure is found after rcoxD. ORF2 and metF (encoding methylenetetrahydrofolate reductase) are also represented. The restriction sites EcoRI, SalI, BamHI, and HindIII were used for subcloning. The nucleotide sequences of both strands were determined in each pMSAN insert, as represented by the arrows in the sequence strategy scheme. The PCR products used to sequence both strands across the SalI and BamHI junctions are also shown (see Materials and Methods).
In order to verify that no additional small fragments were generated when cutting the recombinant DNA with SalI and BamHI, we performed PCR across the junctions comprising these restriction sites, as indicated in Fig. 1. We used the primer 5′-GATGCCTACGAAATCCTGGTTCA-3′ and the reverse complementary primer 5′-AGTACTCCGTGCAGAAGACCGTG-3′ to obtain a 256-bp PCR product across the SalI junction. This product was sequenced on both strands using the same primers. Similarly, primer 5′-AACCTCAGCCCGGAGCCGATTCG-3′ and the reverse complementary primer 5′-GAATGCGTCCGGCCGTCACCACG-3′ were chosen in order to get a 255-bp product across the BamHI junction. The PCR templates were the genomic DNA and the p-Zero-1 insert.
Sequence data were analyzed using the Genetics Computer Group (GCG) program package provided by the Portuguese EMBnet Node. The search for homologous sequences was performed at the National Center for Biotechnology Information in the nonredundant protein library using the BLAST (3) network service. Sequence alignments were made with the CLUSTAL X program (77).
Protein techniques.
The HiPIP:oxygen oxidoreductase was purified according to the method of Pereira et al. (59). For peptide sequencing, the enzyme subunits were separated by SDS-polyacrylamide gel electrophoresis (PAGE) performed as described by Pereira et al. (59) and transferred to a polyvinylidene difluoride membrane. Each transblotted sample was submitted to N-terminal protein sequence analysis by automated Edman degradation (17). An Applied Biosystems model 477A protein sequencer was used.
Ionic strength dependence studies.
Cytochrome c oxidase activity was determined following the change in absorbance of cytochrome c at 550 nm (ɛ550 = 28,000 M−1 cm−1) at 25°C. Horse heart cytochrome c was prereduced by the addition of solid sodium dithionite followed by passage over a Sephadex G-10 size exclusion column. Different ionic strengths were obtained using 2 mM Tris-HCl (pH 8) and different concentrations of NaCl.
Homology-based modeling.
A preliminary three-dimensional model for subunit I of R. marinus oxidase was previously described (59). In the present work, the composite structure of subunit I and the truncated subunit II were modeled. The cytochrome oxidases from P. denitrificans (52) (PDB code 1AR1) and bovine (95) (PDB code 2OCC) were used for the derivation of subunit I. Note that the new bovine structure was used, since it was refined at a higher resolution. The first 20 residues and the last 41 residues of subunit I were excluded from the model due to the lack of homology with known structures. The derivation of subunit II is more complex, since this subunit, compared with known cytochrome oxidases, is not as conserved as subunit I. The transmembrane region shows a very low similarity with the cytochrome c oxidases and part of the sequence, corresponding to a loop in the CuA center region, is not present in the R. marinus enzyme. In addition, this oxidase has an extra carboxy-terminal domain, which is a C-type cytochrome domain. Thus, for the derivation of the subunit II model, different structures were used as templates for the different domains (Fig. 2A). For the transmembrane domain, the P. denitrificans and bovine cytochrome oxidases were used. For the CuA domain, the T. thermophilus ba3-type cytochrome c oxidase CuA domain (90) (PDB code 2CUA) and the E. coli membrane-exposed domain from the quinol oxidase with an engineered CuA center (91) (PDB code 1CYX) were also used. Subunits I and II (without the cytochrome domain) constitute a homology-derived model named model B.
FIG. 2.
(A) General topology of the caa3 terminal oxidase from R. marinus models derived in this work and the corresponding PDB structures used. (B) Alignment used to generate R. marinus model B: the two subunits are separated by a space in the alignment. (C) Alignment used to generate R. marinus model C.
For the derivation of a structural model of the subunit II carboxy-terminal domain (model C), molecules from two families (HOMSTRAD classification [48]) of cytochromes were used: the cytochrome c and c5 families. The first family included cytochrome c2 from Rhodopseudomonas viridis (73) (PDB code 1CRY), cytochrome c isozyme 1 from Saccharomyces cerevisiae (38) (PDB code 1YCC) and cytochrome c from Thunnus alalunga (76) (PDB code 5CYT). The members of the c5 family used were the cytochromes c-551 from Pseudomonas stutzeri (10) (PDB code 1COR) and Pseudomonas aeruginosa (42) (PDB code 351C). The nonstandard use of two families is a consequence of an overall homology with members of the cytochrome c family, whereas the region containing the heme-binding motif is highly similar to the members of the c5 family. In the sequence alignment (Fig. 2C), a gap was inserted in this heme region in order to exclude the c family information from this motif, which is therefore generated from the c5 family alone. This leads to an apparent low homology in the sequence alignment; however, when aligned with each family separately, the homology is much more evident.
In addition to models B and C, a model (model A) containing the full structure of subunits I and II was also tried. Although there is no homology in the junction region between the CuA and the heme domains, a small connection region was used. The template for this connection was the engineered quinol oxidase, which presents a longer carboxy terminus than the other structures.
The program Modeller version 4 (67) was used to generate the various models from the X-ray structures and the alignments presented in Fig. 2. For each model type, the program generated several models (from 10 to 20), and the one with the lowest value of the objective function was chosen.
Nucleotide sequence accession number.
The sequence has been deposited in the EMBL data library under accession no. AJ249578.
RESULTS AND DISCUSSION
Cloning and DNA sequence analysis.
A PCR-derived probe was used for the isolation of the oxidase genes from a genomic library constructed in a lambda vector (see Materials and Methods). We expected the probe, which was designed to hybridize with a sequence region of subunit I, to serve as a total-fit probe, since the other subunit genes could be located in the same operon, as in many other prokaryotes (11). Four positive recombinant phages were isolated (see Materials and Methods). Restriction DNA analysis showed that these recombinants have overlapping restriction patterns; the restriction site mapping of one of these recombinants is represented in Fig. 1. The absence of rearrangements within this recombinant lambda insert was checked by Southern blot, using R. marinus PRQ-62B chromosomal DNA as a control and the recombinant lambda phage DNA as a probe (results not shown). Southern blot analysis also allowed to select and subclone the lambda insert region containing the gene coding for oxidase subunit I; the HindIII-EcoRI fragment drawn in Fig. 1, which hybridizes to the PCR-derived probe, was then cloned into p-Zero-1. Further subcloning was performed, and the nucleotide sequence of the subcloned fragments was determined (see Fig. 1). The 6,346-bp segment of the DNA sequence extending from a Sau3A1 site to a HindIII site (EMBL accession no. AJ249578) is 61.1% G+C, i.e., slightly lower than the overall base contents of R. marinus R-10 (64.4%) and R-18 (64.7%) (2). After translation of the sequence in the six frames, the correct reading frames for translation were confirmed by codon usage analysis (22). Seven open reading frames (ORFs), located on the same DNA strand and showing a uniform codon usage and a third-position GC bias, were considered for further analysis. Among these ORFs, the coding sequences for cytochrome oxidase subunits I, II, and III could be easily identified since their products have clear homologies to the equivalent subunit proteins belonging to the superfamily of heme-copper oxidases. The relative positions of the structural genes for cytochrome oxidase subunits are indicated in Fig. 1; rcoxB (subunit I) and rcoxC (subunit III) are located between rcoxA (subunit II) and rcoxD (subunit IV).
ORF1, rcoxA, rcoxB, rcoxC, and rcoxD form, most probably, an operon, since few nucleotides separate each coding sequence. A putative terminator sequence for the operon is located 23 bp downstream of the TGA stop codon of rcoxD. A database search on ORF1, located upstream from 5′ rcoxA, suggests homology to the putative protein (GI-2984387) of Aquifex aeolicus. In this bacterium genome, this putative coding sequence is located between coxC (cytochrome c oxidase subunit III) and coxB (cytochrome c oxidase subunit II) (14). In some prokaryotes, additional genes required for the biogenesis of the oxidase, namely, genes involved in the biosynthesis of heme A, are found in the same cluster as the enzyme structural genes (see reference 78). It is therefore possible that ORF1 product is involved in Rcox biogenesis. Two more ORFs were identified downstream from rcoxD, after the putative terminator: ORF2 and ORF3. No significant homology to any sequence in databases was found for ORF2. Database searches suggest a homology of ORF3 to methylenetetrahydrofolate reductase, a protein that in prokaryotes catalyzes the penultimate step in the biosynthesis of methionine and in mammals is required for the regeneration of the methyl group of methionine (for a review, see reference 43).
Putative ribosomal binding sites (RBSs), complementary to the R. marinus 16S rRNA (4), precede the start codon of each ORF, except for ORF2, which could start at position 4832 or position 4838, due to the attribution of the putative RBS for this frame. Inspection for codon usage of deduced amino acid sequences reveals a similar codon usage for ORF1, ORF2, ORF3 (metF), and rcox genes (results not shown), which constitutes an argument for considering these ORFs to be proper genes. However, ORF2 is a short frame, and it is possible that the sequence between the rcoxD 3′ end and the ORF3 5′ end is noncoding. Also, sequences that might represent promoter elements are found at the end of ORF2 and in the following region upstream of the metF coding sequence. Thus, the ORF2 region could be involved in the control of expression of metF.
Characteristics of cytochrome oxidase subunits.
R. marinus HiPIP:oxygen oxidoreductase was isolated as a four-subunit complex. Tricine SDS-PAGE of the purified oxidase indicated the presence of four polypeptides with apparent molecular masses of 42, 35, 19, and 15 kDa (59). These values are close to the ones estimated from the deduced Rcox amino acid sequences except for subunits I and III. As observed for other oxidases, these subunits show anomalous migration, due to their high hydrophobicity (51, 62). Peptide amino acid sequences obtained from subunits III and IV argue for the functional expression of the rcox cluster, since these sequences are the same as the ones deduced from rcox DNA. Analysis of the amino acid sequence of each subunit and analysis of the modeled subunits (see below) reveals important structural and functional features of the R. marinus enzyme.
RcoxA.
RcoxA (316 residues, 36 kDa) has amino acid identities of 25 and 23% with subunit II from T. thermophilus and Bacillus subtilis caa3 oxidases, respectively (Table 1 and Fig. 3A). The amino acid identity with the other subunits II presented in the Fig. 3A is even lower (e.g., 19 and 14% identities with subunit II from Synechocystis sp. strain PCC 6803 and bovine aa3 oxidases, respectively). However, the hydropathy profile shows that subunit II is similar to other homologous bacterial subunits II, with two predicted transmembrane-spanning segments (data not shown).
TABLE 1.
Similarities between the deduced amino acid sequences of the four rcox gene products and homologous proteins from other microorganisms
| Gene | Subunit | Size (no. of residues) | % Identity (% similarity)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anabaena sp. strain PCC 7120 | Synechocystis sp. strain PCC 6803 | S. vulcanus | E. coli | B. subtilis | T. thermophilus | A. aeolicus | P. denitrificans | B. taurus | |||
| rcoxA | II | 316 | 20 (31) | 19 (35) | 20 (32) | 17 (30) | 23 (40) | 25 (46) | 19 (32) | 17 (28) | 14 (29) |
| rcoxB | I | 565 | 40 (59) | 39 (60) | 38 (61) | 28 (45) | 32 (53) | 38 (59) | 34 (52) | 35 (54) | 35 (54) |
| rcoxC | III | 226 | 30 (50) | 26 (42) | 25 (47) | 22 (43) | 19 (41) | 21 (36) | 17 (36) | 17 (33) | 21 (36) |
| rcoxD | IV | 122 | 13 (32) | 5 (21) | |||||||
Data are from the indicated accession numbers for Fig. 3.
FIG. 3.
Alignment of deduced amino acid sequences of subunits from R. marinus Rcox oxidase with homologous subunits from other oxidases was performed (accession nos. are in parentheses). Rmarinus, R. marinus; Asp, Anabaena sp. strain PCC 7120 (Z98264); Synsp, Synechocystis sp. strain PCC 6803 (P73261, P73262, and P73263); Svulcanus, S. vulcanus (D16254, P98054, and P50677); Ecoli, E. coli (J05492); Bsubtilis, B. subtilis (Z99111); Tthermophilus, T. thermophilus (P98005 and Q60020); Aaeolicus, A. aeolicus (067935, 067934, and 067932); Pdenitrificans, P. denitrificans (P98002, P08306, and P06030); Btaurus, B. taurus (bovine) (P00396, P00404, and P00415). (A) Subunit II. The ligands for CuA are indicated by a plus sign; the residues proposed to interact with cytochrome c (∗) and the consensus heme attachment site CXXCH (#) are also indicated. The residues A136 to P161 referred in the text are marked with an “x” (note that the mature P. denitrificans subunit II starts at Q29). (B) Subunit I. The conserved histidine residues indicated by an asterisk; the E278 residue of the D-channel and the YS motif are also marked (#). Also marked are the residues proposed to interact with cytochrome c (“+”; see the text). For both subunits, identical residues are shaded.
The amino acid residues, which are the ligands for CuA, are conserved in all of the cytochrome oxidases presented in Fig. 3A. In the case of E. coli bo3 quinol oxidase, the CuA center is absent (12). The CuA domain of R. marinus subunit II presents a substantial loop deletion corresponding to residues A136 to P161 of subunit II from P. denitrificans (see Fig. 2B, 3A, and also 4). At least one of the residues from this loop (D159) is important for the interaction with P. denitrificans cytochrome c (see below).
In the carboxy-terminal region of R. marinus HiPIP:oxygen oxidoreductase subunit II there is a cytochrome C domain containing a consensus heme attachment site, CXXCH. This motif is also present in the B. subtilis and in T. thermophilus caa3 oxidases (Fig. 3A), which are described as cytochrome c oxidases (40, 70). The 35-kDa subunit of the oxidase isolated from R. marinus contains a heme C, as shown by heme staining upon SDS-PAGE (59), and its molecular mass is in agreement with the expected value for the RcoxA product.
It is proposed that subunit II is threaded through the membrane by use of the general prokaryotic secretory pathway (sec) (for a review, see reference 16), in which the first hydrophobic domain is recognized as a signal sequence. The existence of subunit II N-terminal signal sequences has been already demonstrated for several subunits II of various oxidases, namely, those from P. denitrificans and B. subtilis (6, 75). R. marinus HiPIP:oxygen oxidoreductase subunit II might also have an N-terminal signal sequence, with a cleavage site located between positions 45 and 46 (VGA-MT). Signal sequences are approximately 20 amino acids in length and contain positively charged residues in the N terminus preceding a hydrophobic core. Some primary sequence constraints exist at position −3 (Ala, Gly, Ser, Val, and Ile) and −1 (Ala, Gly, and Ser) relative to the cleavage site, with alanine found especially frequently (84). Note that the rcoxA gene contains at least two putative start codons: AUG at position 951 and CUG at position 1029. A 19-residue signal peptide, in the range of sizes usually found for prokaryotic signal peptides, could be obtained using the second codon, suggesting that translation starts at the CUG codon (EMBL accession no. AJ249578).
RcoxB.
The rcoxB gene encodes a subunit of 565 residues with a molecular mass of 62 kDa which shows homology with subunit I of cytochrome oxidases. A hydropathy plot (not shown) suggests the presence of 12 transmembrane segments, as is the case for the subunit I of mitochondrial (79) and P. denitrificans (30) enzymes.
Among the Rcox subunits, subunits I shows the highest degree of identity with homologous oxidases subunits (39 and 40% amino acid identities with Synechocystis sp. strain PCC 6803 and Anabaena sp. strain PCC 7120 subunit I of cytochrome oxidases, respectively) (Table 1, Fig. 3B). Six totally conserved histidine residues (indicated by asterisks in Fig. 3B) are the ligands of the cytochrome a3-CuB center and cytochrome a of RcoxB.
Proton transfer pathways in subunit I were proposed based on sequence analysis and site-directed mutagenesis studies and were later corroborated by the crystal structures of the aa3 oxidases from P. denitrificans and from bovine heart (30, 79). Pathways D and K are named according to the conserved residues in each channel: aspartate and lysine, respectively (20, 31, 46). All residues proposed to be important for proton transfer in the channels are conserved in R. marinus oxidase, with the exception of the key glutamate residue of the D-channel (E278, P. denitrificans numbering). Other oxidases in Fig. 3B lack this glutamate residue, i.e., those from Anabaena sp. strain PCC 7120, Synechocystis sp. strain PCC 6803, Synechococcus vulcanus, T. thermophilus (caa3) and A. aeolicus (COXA1).
RcoxC.
The rcoxc gene is predicted to encode a 226-amino-acid protein (26 kDa). The hydropathy profile shows that RcoxC is a membrane protein with five predicted transmembrane segments (not shown). N-terminal sequencing was performed on this subunit. However, no information could be obtained on the sequence of the first residues. This fact could be due either to N-terminal blockage or degradation of subunit III. Nevertheless, a stretch of peptide sequence (HPPYLQH) matches the deduced amino acid sequence, confirming the expression of this subunit. The protein presents 30% amino acid identity with the Anabaena sp. strain PCC 7120 cytochrome oxidase subunit III and 26% amino acid identity with that of Synechocystis sp. strain PCC 6803 (Table 1). This percentage is lower for the caa3 oxidases from B. subtilis (19%) and T. thermophilus (21%) and even smaller for the aa3 enzyme from P. denitrificans and COXC from A. aeolicus (17%). However, in terms of amino acid similarity, the conservation is higher than for subunit II; subunit III has 50 and 47% amino acid similarities with the Anabaena sp. strain PCC 7120 and S. vulcanus corresponding subunits, respectively, whereas subunit II has 31 and 32% similarities.
RcoxD.
The presence of a fourth subunit in bacterial cytochrome oxidases was demonstrated for the P. denitrificans aa3 oxidase crystallized complex (30). Also, Bacillus PS3 caa3 oxidase (74), B. subtilis aa3 quinol oxidase (37), and E. coli bo3 quinol oxidase (33, 41, 47) are composed of four subunits. It was proposed that E. coli CyoD subunit IV is essential for CuB binding to subunit I, being a domain-specific molecular chaperone in the oxidase complex (65, 66). R. marinus caa3 oxidase has also a fourth subunit encoded by rcoxD, a 122-residue protein (13 kDa). The N-terminal sequence of this subunit A-H-A-T-H-H-I-I-P-R is identical to that predicted from the respective gene, except that the initial methionine is missing. It is then definitely demonstrated that subunits III and IV of the purified HiPIP:oxygen oxidoreductase and RcoxC and RcoxD are the same proteins. Subunit IV shows only 13 and 5% amino acid identities with subunit IV from the E. coli bo3 and the B. subtilis caa3 oxidases, respectively (Table 1). Although these values are low, the three subunits present a similar hydropathy profile with three hydrophobic segments (data not shown).
Analysis of the modeled structures: implications for electron and proton transfer mechanisms.
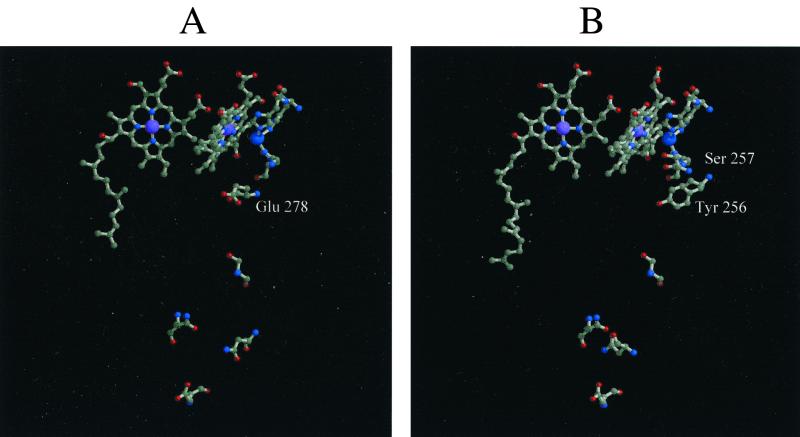
In this work, three models of R. marinus caa3 enzyme were derived. The results for model A (data not shown) clearly show that the available experimental data is not sufficient to determine the complete structure, since for each run a significantly different orientation of the cytochrome domain in relation with the rest of the molecule was obtained. In contrast to model A, model B is well defined (Fig. 4A). The structure is similar overall to that of the cytochrome c oxidases, but some important differences are found. A major difference is located in subunit I, in the D-proton channel. As stated above, with the exception of the E278 (P. denitrificans numbering) of the D-channel, all of the postulated important residues of the proton channels K and D are conserved in the R. marinus oxidase. Mutants obtained by substitution of E278 are unable to complete reduction of oxygen and consequently unable to translocate protons (20, 31, 46, 64, 88). However, the R. marinus caa3 oxidase performs the complete reduction of molecular oxygen to water (59) and is indeed a proton pump with the expected stoichiometry of 1 H+/e− (60). The carboxyl group of E278, which is located at the end of the channel, close to the catalytic center, is substituted in the same spatial position by the hydroxyl group of a tyrosine residue (Y256) in the R. marinus enzyme (Fig. 5). Another relevant difference in the D-channel is a glycine-to-serine substitution in the position close to the binuclear catalytic center. Based on theoretical calculations and Fourier transform infrared (FTIR) spectroscopy, it has been suggested that the glutamate residue acts as a proton shuttle, assuming two distinct conformations (“in” and “out”), related by a 180° rotation of the carboxylate group (61). Interestingly, in the R. marinus oxidase, the spatial positions equivalent to the in and out conformations of the glutamate side chain are occupied by the hydroxyl groups of the above-mentioned tyrosine and serine residues, Y256 and S257, respectively. Although the actual role of these residues has still to be established, both may be active elements in the proton pathway, either by maintaining an ordered chain of water molecules in the cavity or by being directly involved as part of the proton wire, where their OH groups could work in a hopping mechanism for proton transfer, assuring the proton connectivity between the end of the D-channel and the binuclear center. Additionally, a possible ionization of Y256 may allow a mechanism of coupling between proton and electron capture and release.
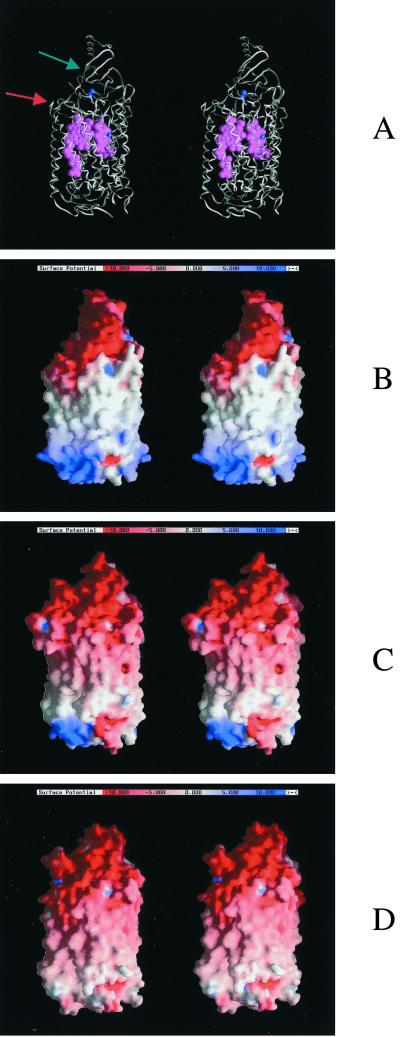
FIG. 4.
Comparison between the modeled structure (model B) and the structures of the known cytochrome c oxidases. All structures are within the same relative orientation. The stereo pictures were produced with GRASP (50). The potential values in the molecular surfaces range from −10 to +10 kT/e. (A) Fold of the R. marinus oxidase, with the hemes (in purple) and the copper atoms (in blue). The red arrow marks the region of subunit I where a loop deletion is observed; the green arrow marks the region of the loop deletion in subunit II (see text). (B, C, and D). Molecular surfaces of the oxidases colored by electrostatic potential: R. marinus (B), P. denitrificans (C), and bovine (D) cytochrome c oxidase.
FIG. 5.
D-channel in P. denitrificans (A) and R. marinus (B). The heme groups from subunit I, the CuB center, the histidine ligands, and the residues corresponding to the D-channel in both proteins are represented. Glu-278 in P. denitrificans and Tyr-256 and Ser-257 in R. marinus are labeled in panels A and B, respectively.
The electrostatic characteristics of the model B of R. marinus oxidase and the cytochrome c oxidases are compared in Fig. 4B, C, and D. These surfaces seem to be very similar, but R. marinus is closer to P. denitrificans than to the structure from the bovine enzyme due to the existence of a similar positive potential area in the surface of the molecule, which should be facing the cytoplasm. If the cytochrome domain is excluded, on the opposite side of the molecule, a simple reasoning would imply that the protein should behave as a cytochrome c oxidase, with negatively charged areas on the exposed copper center side, leading to a favorable interaction with positively charged areas, e.g., the heme edge of a mitochondrion-type cytochrome c. However, a more careful analysis of the residues considered to be important for the interaction of P. denitrificans oxidase and cytochrome c (36, 92–94) shows that not all are conserved in R. marinus caa3 oxidase. In subunit II, specifically in the CuA domain, a long loop (A136 to P161, P. denitrificans numbering) is missing (see Fig. 4A, segment deletion, marked with a green arrow). The absence of this loop results in a reduced distance between the CuA center and the surface of this domain and also in the absence of one important residue for interaction with cytochrome c (D159 in P. denitrificans). Also in this region, D135, which seems to be one of the most important residues for cytochrome c binding (92), is substituted by a threonine residue (see Fig. 3A). E126 (P. denitrificans numbering), D178, and W121, with this last considered essential for electron transfer (93), are nevertheless conserved. In subunit I, D156 is absent (segment deletion, marked with a red arrow in Fig. 4A), but D257 is present (see also Fig. 3B). Therefore, in the absence of the cytochrome domain, it could be considered that part of the cytochrome c binding site is present in this caa3 oxidase. However, the cytochrome domain present in this oxidase may change the properties for cytochrome c binding, in this case by reducing the efficiency of cytochrome c in transferring electrons to the oxidase (see reference 58).
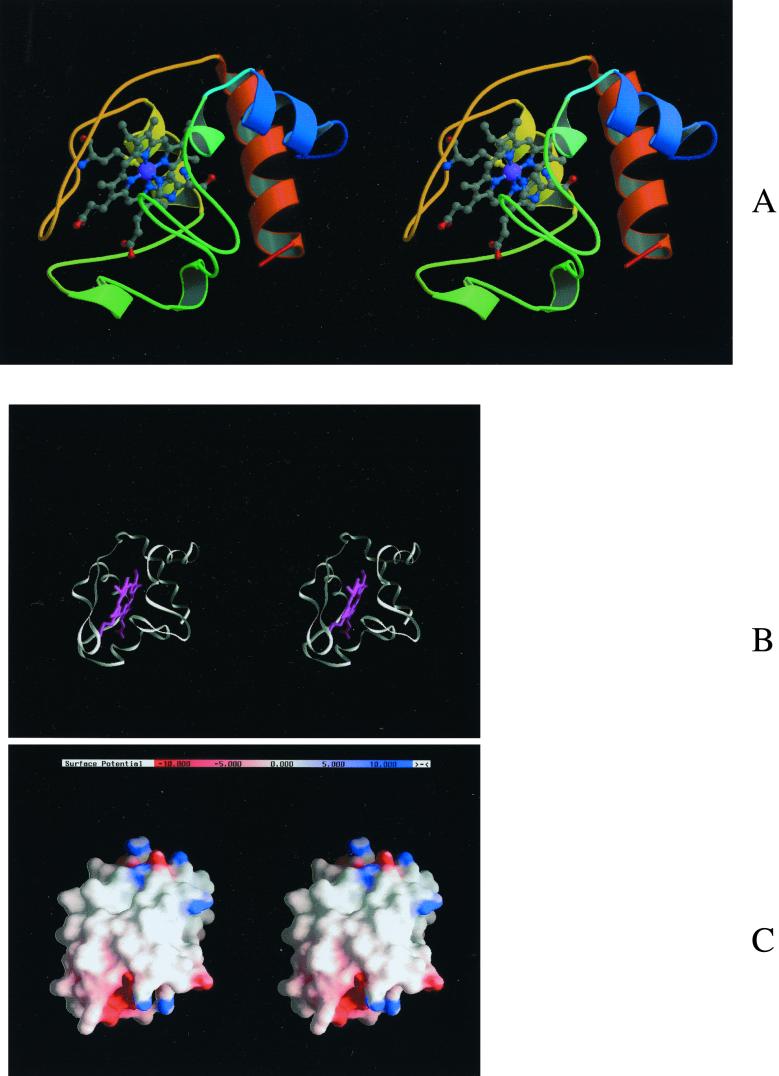
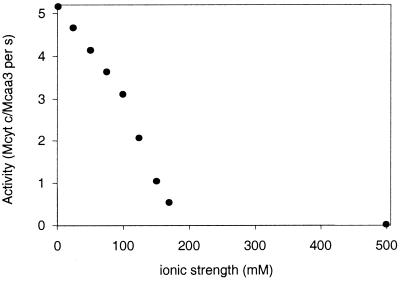
In order to study the cytochrome domain, model C (Fig. 6A) was created. The model is well defined, although less accurate in two loop structures (an upper loop and a lower loop) surrounding the propionate groups of the heme, since these are the places where the c and c5 families are most different (data not shown). In the model these two loops are long, while in the c and c5 families only the lower loop or the upper loop is long, respectively. The modeled cytochrome domain does not show a marked electrostatic character (see Fig. 6B and C), especially around the heme edge region, which is in general one of the most exposed regions involved in electron transfer. This is true for the cytochrome c family, for example, where the heme edge has been suggested to be involved in the electron transfer with flavodoxins (13, 72, 86), cytochrome b5 (87), cytochrome c peroxidase (54), and plastocyanin (81). In the actual case of the mitochondrion-type cytochrome c, the electron donor to cytochrome c oxidases, the heme edge has long been considered to be the entry and exit route for electrons. The lysine residues shown to be involved (18, 63) in complex formation with cytochrome c oxidases are located around this edge. These positively charged residues of cytochrome c interact with a number of important acidic residues located in subunits I and II at the surface of the oxidase, as mentioned above (36, 92, 94). In the case of R. marinus oxidase, if the cytochrome domain of the oxidase exposes the heme edge, its interaction with a redox partner will not be electrostatic in character, since this region is rather neutral (Fig. 6B and C). Therefore, molecules similar to mitochondrial cytochrome c would not form stable complexes. In fact, and in contrast to what is observed for the bovine and P. denitrificans aa3 oxidases, R. marinus cytochrome c oxidase activity decreases with the increase of ionic strength (Fig. 7), i.e., it does not follow a bell-shape curve (92). For those enzymes, the very low activity at low ionic strength was attributed to the formation of a stable electrostatic complex. A behavior similar to that of the R. marinus enzyme was observed for the ba3 oxidase from T. thermophilus (21). Unlike the mitochondrial cytochrome c family, which presents surfaces with positive electrostatic potential character, HiPIPs can have very different electrostatic characteristics. This suggests that in HiPIPs electron transfer is less dependent on electrostatic interactions and explains the higher electron transfer rate observed with HiPIP compared to the one observed for cytochrome c (58). A relevant example of such behavior is the photosynthetic reaction center of Rubrivivax gelatinosus. This center is able to receive electrons from both a cytochrome c and an HiPIP, showing a remarkable preference for the latter electron donor. Mutagenesis studies provide evidence of an electrostatic-interaction-driven binding of cytochrome c to the reaction center, while for HiPIP the binding is driven by hydrophobic interactions. Nevertheless, the binding regions overlap in the edge of the lower redox potential heme (53). Like this photosynthetic reaction center, the HiPIP interaction with the cytochrome domain of the caa3 oxidase of R. marinus may be mediated by hydrophobic interactions since, whatever the proposed orientation in the three-dimensional structure of the cytochrome domain, this does not show a marked electrostatic character.
FIG. 6.
(A) Fold of the modeled structure of the cytochrome domain from R. marinus (model C). The stereo picture was produced with Molscript (34) and Raster 3D (44). (B and C) Fold and molecular surfaces (same orientation) of the cytochrome domain colored by electrostatic potential. The stereo pictures were produced with GRASP (50). The potential values in the molecular surface range from −10 to +10 kT/e.
FIG. 7.
Effect of ionic strength on the activity of the caa3-type oxidase from R. marinus, using horse heart cytochrome c as an electron donor, at 25°C.
Evolutionary aspects.
Subunit I from Rcox oxidoreductase contains the conserved residues of the proposed proton channels with the exception of the key glutamate (E278, P. denitrificans numbering) of the D-channel; homology modeling analysis suggests that tyrosine 256 and serine 257 could be the glutamate functional substitutes. Inspection of the performed sequence alignments (see Fig. 3B) reveals that these residues are also present in other oxidases. The correlation of these data with the 16S rRNA bacterial phylogenetic tree shows that enzymes containing the YS motif are present in organisms belonging to the deepest branches; on the other hand, the glutamate-containing oxidases are only present in purple bacteria, gram-positive bacteria, and mitochondria. These observations suggest that the R. marinus caa3 oxidase proton pathway, presumably involving the tyrosine and serine residues, is ancestral to the one involving the glutamate in the gram-positive and purple bacteria and reinforces the suggestion put forward by Castresana et al. (11) for lateral transfer of oxidase genes from gram-positive bacteria to purple bacteria, the older proton pathway being maintained in the early divergent branches.
R. marinus HiPIP:oxygen oxidoreductase is a true terminal oxidase (59, 60), being the first example of a member of the heme-copper superfamily using a HiPIP as an electron donor. The electron donor specificity of heme-copper oxidases is related to subunit II's structure. During evolution, mutations in the C-terminal domain would generate differences in the electron donor substrate specificity, this evolutionary change being parallel with the evolution of the type of electron donor. The first electron donor may have been an iron-sulfur protein, since these proteins are rather primordial molecules (85). If this hypothesis is correct, we will find caa3 oxidases to be ambivalent with respect to the electron donor (in parallel with what happens with the tetraheme cytochrome subunit bound to the photosynthetic reaction center of R. gelatinosus [53]), while others use only one type of electron donor, i.e., an iron-sulfur protein or a cytochrome c. A posterior step in the evolution of the C-terminal domain of caa3 oxidases would have been its own deletion, leading to the appearance of the aa3 cytochrome c oxidases.
We therefore propose that R. marinus oxidase arose from a common ancestor existing before the branching of the Bacteroides, Thermus, and Deinococcus genera, retaining characteristics of this primitive bacterial cytochrome c oxidase. Further mutagenesis studies on the C-terminal domain of subunit II will be important in order to evaluate the binding properties and to infer the proposed hypothesis for the subunit II evolution of cytochrome oxidases.
In conclusion, the study of this R. marinus oxidase expands the diversity of the members of the heme-copper superfamily, both in terms of the type of electron donor and in terms of the pathways for proton uptake and/or translocation. These characteristics bring new insight to the theory of terminal-oxidase evolution.
ACKNOWLEDGMENTS
M. Santana and M. Pereira are recipients of grants from Praxis XXI program (BPD/3382/96 and BPD/22054/99). This work was supported by Project BIO/37/96 to MT.
We are grateful to P. Fernandes and I. Marques, from the Bioinformatics Unit of the Instituto Gulbenkian de Ciência, for providing access and support in the use of GCG software for sequence analysis (Wisconsin Package). We thank Manuela Regalla for N-terminal sequencing.
REFERENCES
- 1.Akada R. Quick-check method to test the size of Escherichia coli plasmids. BioTechniques. 1994;17:58. [PubMed] [Google Scholar]
- 2.Alfredsson G A, Kristjansson J K, Hjorleifslottir S, Stetter K O. Rhodothermus marinus, gen. nov., sp. nov., a thermophilic halophilic bacterium from submarine hot springs in Iceland. J Gen Microbiol. 1988;134:299–306. [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipmun D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrésson O S, Fridjónsson O H. The sequence of the single 16S rRNA gene of the thermophilic eubacterium Rhodothermus marinus reveals a distant relationship to the group containing Flexibacter, Bacteroides, and Cytophaga species. J Bacteriol. 1994;176:6165–6169. doi: 10.1128/jb.176.20.6165-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 6.Bengtsson J, Tjalsma H, Rivolta C, Hederstedt L. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J Bacteriol. 1999;181:685–688. doi: 10.1128/jb.181.2.685-688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1522. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonora P, Principi I, Monti B, Ciurli S, Zannoni D, Hochkoeppler A. On the role of high-potential iron-sulfur proteins and cytochromes in the respiratory chain of two facultative phototrophs. Biochim Biophys Acta. 1999;1410:51–60. doi: 10.1016/s0005-2728(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 9.Bratton M R, Pressler M A, Hosler J P. Suicide inactivation of cytochrome c oxidase: catalytic turnover in the absence of subunit III alters the active site. Biochemistry. 1999;38:16236–16245. doi: 10.1021/bi9914107. [DOI] [PubMed] [Google Scholar]
- 10.Cai M, Bradford E, Timkovich R. Investigation of the solution conformation of cytochrome c-551 from Pseudomonas stutzeri. Biochemistry. 1992;31:8603–8612. doi: 10.1021/bi00151a030. [DOI] [PubMed] [Google Scholar]
- 11.Castresana J, Lübben M, Saraste M, Higgins D G. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chepuri V, Lemieux L, Au D C-T, Gennis R B. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem. 1990;265:11185–11192. [PubMed] [Google Scholar]
- 13.Cunha C A, Romão M J, Sadeghi S J, Valetti F, Gilardi G, Soares C M. Modelling of electron transfer complexes between flavodoxin and c-type cytochromes. J Biol Inorg Chem. 1999;4:360–374. doi: 10.1007/s007750050323. [DOI] [PubMed] [Google Scholar]
- 14.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swansson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 15.Degryse E, Glansdorff N, Piérard A. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch Microbiol. 1978;117:189–196. doi: 10.1007/BF00402307. [DOI] [PubMed] [Google Scholar]
- 16.Economou A. Following the leader: bacterial protein export through the Sec pathway. Trends Microbiol. 1999;7:315–320. doi: 10.1016/s0966-842x(99)01555-3. [DOI] [PubMed] [Google Scholar]
- 17.Edman P, Begg G. A protein sequenator. Eur J Biochem. 1967;1:80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson-Miller S, Brautigan D L, Margoliash E. Definition of cytochrome c binding domains by chemical modification. III Kinetics of reaction of carboxydinitrophenyl cytochromes c with cytochrome c oxidase. J Biol Chem. 1978;253:149–159. [PubMed] [Google Scholar]
- 19.García-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gennis R B. Cytochrome c oxidase: one enzyme, two mechanisms? Science. 1998;280:1712–1713. doi: 10.1126/science.280.5370.1712. [DOI] [PubMed] [Google Scholar]
- 21.Giuffrè A, Forte E, Antonini G, D'Itri E, Brunori M, Soulimane T, Buse G. Kinetic properties of ba3 oxidase from Thermus thermophilus: effect of temperature. Biochemistry. 1999;38:1057–1065. doi: 10.1021/bi9815389. [DOI] [PubMed] [Google Scholar]
- 22.Gribskov M, Devereux J, Burgess R R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984;12:539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haltia T, Finel M, Harms N, Nakari T, Raitio M, Wikström M, Saraste M. Deletion of the gene for subunit III leads to defective assembly of bacterial cytochrome oxidase. EMBO J. 1989;8:3571–3579. doi: 10.1002/j.1460-2075.1989.tb08529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Hill B C. The sequence of electron carriers in the reaction of cytochrome c oxidase with oxygen. J Bioenerg Biomembr. 1993;25:115–120. doi: 10.1007/BF00762853. [DOI] [PubMed] [Google Scholar]
- 26.Hochkoeppler A, Ciurli S, Venturoli G, Zannoni D. The high potential iron-sulfur protein (HiPIP) from Rhodoferax fermentans is competent in photosynthetic electron transfer. FEBS Lett. 1995;357:70–74. doi: 10.1016/0014-5793(94)01334-w. [DOI] [PubMed] [Google Scholar]
- 27.Hochkoeppler A, Kofod P, Ferro G, Ciurli S. Isolation, characterization, and functional role of the high-potential iron-sulfur protein (HiPIP) from Rhodoferax fermentans. Arch Biochem Biophys. 1995;322:313–318. doi: 10.1006/abbi.1995.1469. [DOI] [PubMed] [Google Scholar]
- 28.Hochkoeppler A, Kofod P, Zannoni D. HiPIP oxido-reductase activity in membranes from aerobically grown cells of the facultative phototroph Rhodoferax fermentans. FEBS Lett. 1995;375:197–200. doi: 10.1016/0014-5793(95)01188-k. [DOI] [PubMed] [Google Scholar]
- 29.Hochkoeppler A, Zannoni D, Ciurli S, Meyer T E, Cusanovich M A, Tollin G. Kinetics of photo-induced electron transfer from high-potential iron-sulfur protein to the photosynthetic reaction center of the purple phototroph Rhodoferax fermentans. Proc Natl Acad Sci USA. 1996;93:6998–7002. doi: 10.1073/pnas.93.14.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 31.Karpefors M, Ädelroth P, Aagaard A, Sigurdon H, Ek M S, Brzezinsky P. Electron-proton interactions in terminal oxidases. Biochim Biophys Acta. 1998;1365:159–169. doi: 10.1016/s0005-2728(98)00058-9. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy C K. Induction of colicin production by high temperature or inhibition of protein synthesis. J Bacteriol. 1971;108:10–19. doi: 10.1128/jb.108.1.10-19.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kranz R G, Gennis R B. Immunological characterization of the cytochrome o terminal oxidase from Escherichia coli. J Biol Chem. 1983;258:10614–10621. [PubMed] [Google Scholar]
- 34.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 35.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.Lappalainen P, Watmough N J, Greewood C, Saraste M. Electron transfer between cytochrome c and the isolate CuA domain: identification of substrate-binding residues in cytochrome c oxidase. Biochemistry. 1995;34:5824–5830. doi: 10.1021/bi00017a014. [DOI] [PubMed] [Google Scholar]
- 37.Lemma E, Simon J, Schägger H, Kröger A. Properties of the menaquinol oxidase (Qox) and of qox deletion mutants of Bacillus subtilis. Arch Microbiol. 1995;163:432–438. doi: 10.1007/BF00272132. [DOI] [PubMed] [Google Scholar]
- 38.Louie G V, Brayer G D. High-resolution refinement of yeast iso-1-cytochrome c and comparisons with other eukaryotic cytochromes c. J Mol Biol. 1990;214:527–555. doi: 10.1016/0022-2836(90)90197-T. [DOI] [PubMed] [Google Scholar]
- 39.Lübben M, Morand K. Novel prenylated hemes as cofactors of cytochrome oxidases: archaea have modified hemes A and O. J Biol Chem. 1994;269:21473–21479. [PubMed] [Google Scholar]
- 40.Mather M W, Springer P, Fee J A. Cytochrome oxidase genes from Thermus thermophilus. Nucleotide sequence and analysis of the deduced primary structure of subunit IIc of cytochrome caa3. J Biol Chem. 1991;266:5025–5035. [PubMed] [Google Scholar]
- 41.Matsushita K, Patel L, Kaback H R. Cytochrome o type oxidase from Escherichia coli. Characterization of the enzyme and mechanism of electrochemical proton gradient generation. Biochemistry. 1984;23:4703–4714. doi: 10.1021/bi00315a028. [DOI] [PubMed] [Google Scholar]
- 42.Matsuura Y, Takano T, Dickerson R E. Structure of cytochrome c-551 from Pseudomonas aeruginosa refined at 1.6 Å resolution and comparison of the two redox forms. J Mol Biol. 1982;156:389–409. doi: 10.1016/0022-2836(82)90335-7. [DOI] [PubMed] [Google Scholar]
- 43.Matthews R G, Sheppard C, Goulding C. Methylenetetrahydrofolate reductase and methionine synthase biochemistry and molecular biology. Eur J Pediatr. 1998;157:554–559. doi: 10.1007/pl00014305. [DOI] [PubMed] [Google Scholar]
- 44.Merritt E A, Bacon D J. Raster3D photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 45.Meyer T E, Cannac V, Fitsch J, Bartsch R G, Tollin G, Cusanovitch M A. Soluble cytochromes and ferredoxins from the marine purple phototrophic bacterium, Rhodopseudomonas marina. Biochim Biophys Acta. 1990;1017:125–138. doi: 10.1016/0005-2728(90)90143-r. [DOI] [PubMed] [Google Scholar]
- 46.Mills D A, Ferguson-Miller S. Proton uptake and release in cytochrome c oxidase: separate pathways in time and space? Biochim Biophys Acta. 1998;1365:46–52. doi: 10.1016/s0005-2728(98)00040-1. [DOI] [PubMed] [Google Scholar]
- 47.Minghetti K C, Goswitz V C, Gabriel N E, Hill J J, Barassi C, Georgiou C D, Chan S I, Gennis R B. Modified, large-scale purification of the cytochrome o complex of Escherichia coli yields a two heme/one copper terminal oxidase with high specific activity. Biochemistry. 1992;31:6917–6924. doi: 10.1021/bi00145a008. [DOI] [PubMed] [Google Scholar]
- 48.Mizuguchi K, Deane C M, Blundell T L, Overington J P. HOMSTRAD: a database of protein structure alignments for homologous families. Prot Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moschettini G, Hochkoeppler A, Monti B, Benelli B, Zannoni D. The electron transport system of the halophilic purple nonsulfur bacterium Rhodospirillum salinarum. 1. A functional and thermodynamic analysis of the respiratory chain in aerobically and photosynthetically grown cells. Arch Microbiol. 1997;168:302–309. doi: 10.1007/s002030050503. [DOI] [PubMed] [Google Scholar]
- 50.Nicholls A. GRASP: graphical representation and analysis of surface properties. New York, N.Y: Columbia University; 1992. [Google Scholar]
- 51.Nicoletti F, Witt H, Ludwig B, Brunori M, Malatesta F. Paracoccus denitrificans cytochrome c oxidase: a kinetic study on the two- and four-subunit complexes. Biochim Biophys Acta. 1998;1365:393–403. doi: 10.1016/s0005-2728(98)00092-9. [DOI] [PubMed] [Google Scholar]
- 52.Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 Å resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody Fv fragment. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osyczka A, Nagashima K V, Sogabe S, Miki K, Shimada K, Matsuura K. Comparison of the binding sites for high-potential iron-sulfur protein and cytochrome c on the tetraheme cytochrome subunit bound to the bacterial photosynthetic reaction center. Biochemistry. 1999;38:15779–15790. doi: 10.1021/bi990907d. [DOI] [PubMed] [Google Scholar]
- 54.Pelletier H, Kraut J. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 55.Pereira M M, Antunes A M, Nunes O C, Costa M S, Teixeira M. A membrane-bound HiPIP type center in the thermohalophile Rhodothermus marinus. FEBS Lett. 1994;352:337–330. doi: 10.1016/0014-5793(94)00985-6. [DOI] [PubMed] [Google Scholar]
- 56.Pereira M M, Carita J N, Anglin R, Saraste M, Teixeira M. Heme centers of Rhodothermus marinus respiratory chain. Characterization of its cbb3 oxidase. J Bioenerg Biomembr. 2000;32:143–152. doi: 10.1023/a:1005555829301. [DOI] [PubMed] [Google Scholar]
- 57.Pereira M M, Carita J N, Teixeira M. Membrane-bound electron transfer chain of the thermohalophilic bacterium Rhodothermus marinus: a novel multihemic cytochrome bc, a new complex III. Biochemistry. 1999;38:1268–1275. doi: 10.1021/bi9818063. [DOI] [PubMed] [Google Scholar]
- 58.Pereira M M, Carita J N, Teixeira M. Membrane-bound electron transfer chain of the thermohalophilic bacterium Rhodothermus marinus: characterization of the iron-sulfur centers from the dehydrogenases and investigation of the high-potential iron-sulfur protein function by in vitro reconstitution of the respiratory chain. Biochemistry. 1999;38:1276–1283. doi: 10.1021/bi981807v. [DOI] [PubMed] [Google Scholar]
- 59.Pereira M M, Santana M, Soares C M, Mendes J, Carita J N, Carrondo M A, Saraste M, Teixeira M. The caa3 terminal oxidase of the thermohalophilic bacterium Rhodothermus marinus: a HiPIP:oxygen oxidoreductase lacking the key glutamate of the D-channel. Biochim Biophys Acta. 1999;1413:1–13. doi: 10.1016/s0005-2728(99)00073-0. [DOI] [PubMed] [Google Scholar]
- 60.Pereira M M, Verkhovskaya M L, Teixeira M, Verkhovsky M I. The caa3 terminal oxidase of Rhodothermus marinus lacking the key glutamate of the D-channel is a proton pump. Biochemistry. 2000;39:6336–6340. doi: 10.1021/bi992848+. [DOI] [PubMed] [Google Scholar]
- 61.Pomès R, Hummer G, Wikström M. Structure and dynamics of a proton shuttle in cytochrome c oxidase. Biochim Biophys Acta. 1998;1365:255–260. [Google Scholar]
- 62.Quirk P G, Hicks D B, Krulwich T A. Cloning of the cta operon from alkaliphilic Bacillus firmus OF4 and characterization of the pH-regulated cytochrome caa3 oxidase it encodes. J Biol Chem. 1993;268:678–685. [PubMed] [Google Scholar]
- 63.Rieder R, Bosshard H R. Comparison of the binding sites on cytochrome c for cytochrome c oxidase, cytochrome bc1 and cytochrome c1. Differential acetylation of lysyl residues in free and complexed cytochrome c. J Biol Chem. 1980;255:4732–4739. [PubMed] [Google Scholar]
- 64.Riistama S, Hummer G, Puustinen A, Dyer R B, Woodruff W H, Wikström M. Bound water in the proton translocation mechanism of the haem-copper oxidases. FEBS Lett. 1997;414:275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- 65.Saiki K, Mogi T, Tsubaki M, Hori H, Anraku Y. Exploring subunit-subunit interactions in the Escherichia coli bo-type ubiquinol oxidase by extragenic suppressor mutation analysis. J Biol Chem. 1997;272:14721–14726. doi: 10.1074/jbc.272.23.14721. [DOI] [PubMed] [Google Scholar]
- 66.Saiki K, Nakamura H, Mogi T, Anraku Y. Probing a role of subunit IV of the Escherichia coli bo-type ubiquinol oxidase by deletion and cross-linking analyses. J Biol Chem. 1996;271:15336–15340. doi: 10.1074/jbc.271.26.15336. [DOI] [PubMed] [Google Scholar]
- 67.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 69.Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990;23:331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- 70.Saraste M, Metso T, Nakari T, Jalli T, Laureus M, van der Oost J. The Bacillus subtilis cytochrome-c oxidase. Variations on a conserved protein theme. Eur J Biochem. 1991;195:517–525. doi: 10.1111/j.1432-1033.1991.tb15732.x. [DOI] [PubMed] [Google Scholar]
- 71.Schoepp B, Parot P, Menin L, Gaillard J, Richaud P, Vermeglio A. In vivo participation of a high potential iron-sulfur protein as electron donor to the photochemical reaction center of Rubrivivax gelatinosus. Biochemistry. 1995;34:11736–11742. doi: 10.1021/bi00037a010. [DOI] [PubMed] [Google Scholar]
- 72.Simondsen R P, Weber P C, Salemme F R, Tollin G. Transient kinetics of electron transfer reactions of flavodoxin: ionic strength dependence of semiquinone oxidation by cytochrome c, ferricyanide, and ferric ethylenediaminetetraacetic acid and computer modeling of reaction complexes. Biochemistry. 1982;21:6366–6375. doi: 10.1021/bi00268a008. [DOI] [PubMed] [Google Scholar]
- 73.Sogabe S, Miki K. Refined crystal structure of ferrocytochrome c2 from Rhodopseudomonas viridis at 1.6 Å resolution. J Mol Biol. 1995;252:235–247. doi: 10.1006/jmbi.1995.0491. [DOI] [PubMed] [Google Scholar]
- 74.Sone N, Shimada S, Ohmori T, Souma Y, Gonda M, Ishizuka M. A fourth subunit is present in cytochrome c oxidase from the thermophilic bacterium PS3. FEBS Lett. 1990;262:249–252. doi: 10.1016/0014-5793(90)80202-t. [DOI] [PubMed] [Google Scholar]
- 75.Steinrücke P, Steffens G C, Panskus G, Buse G, Ludwig B. Subunit II of cytochrome c oxidase from Paracoccus denitrificans. DNA sequence, gene expression and the protein. Eur J Biochem. 1987;167:431–439. doi: 10.1111/j.1432-1033.1987.tb13356.x. [DOI] [PubMed] [Google Scholar]
- 76.Takano T, Dickerson R. Redox conformation changes in refined tuna cytochrome c. Proc Natl Acad Sci USA. 1980;77:6371–6375. doi: 10.1073/pnas.77.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thony-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microb Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsukihara T, Aoyama H, Yamashita E, Tomizahi T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 80.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 81.Ubbink M, Bendall D S. Complex of plastocyanin and cytochrome c characterized by NMR chemical shift analysis. Biochemistry. 1997;36:6326–6335. doi: 10.1021/bi963199u. [DOI] [PubMed] [Google Scholar]
- 82.van der Oost J, de Boer A P N, de Gier J-W L, Zumft W G, Stouthamer A H, van Spanning R J M. The haem-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol Lett. 1994;121:1–10. doi: 10.1111/j.1574-6968.1994.tb07067.x. [DOI] [PubMed] [Google Scholar]
- 83.van der Oost J, Lappalainen P, Musacchio A, Warne A, Lemieux L, Rumbley J, Gennis R B, Aasa R, Pascher T, Malmstrom B G, et al. Restoration of a lost metal-binding site: construction of two different copper sites into a subunit of the E. coli cytochrome o quinol oxidase complex. EMBO J. 1992;11:3209–3217. doi: 10.1002/j.1460-2075.1992.tb05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weber P C, Tollin G. Electrostatic interactions during electron transfer reactions between c-type cytochromes and flavodoxin. J Biol Chem. 1985;260:5568–5573. [PubMed] [Google Scholar]
- 87.Wendoloski J J, Matthew J B, Weber P C, Salemme F R. Molecular dynamics of a cytochrome c-cytochrome b5 electron transfer complex. Science. 1987;238:794–797. doi: 10.1126/science.2823387. [DOI] [PubMed] [Google Scholar]
- 88.Wikström M. Proton translocation by the respiratory haem-copper oxidases. Biochim Biophys Acta. 1998;1365:185–192. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 89.Wikström M, Krab K, Saraste M. Cytochrome oxidase: a synthesis. London, England: Academic Press; 1981. [Google Scholar]
- 90.Williams P A, Blackburn N J, Sanders D, Bellamy H, Stura E A, Fee J A, McRee D E. The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 Å resolution. Nat Struct Biol. 1999;6:509–516. doi: 10.1038/9274. [DOI] [PubMed] [Google Scholar]
- 91.Wilmanns M, Lappalainen P, Kelly M, Sauer-Eriksson E, Saraste M. Crystal structure of the membrane-exposed domain from a respiratory quinol oxidase complex with an engineered dinuclear copper center. Proc Natl Acad Sci USA. 1995;92:11955–11959. doi: 10.1073/pnas.92.26.11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Witt H, Malatesta F, Nicoletti F, Brunori M, Ludwig B. Cytochrome-c-binding site on cytochrome oxidase in Paracoccus denitrificans. Eur J Biochem. 1998;251:367–373. doi: 10.1046/j.1432-1327.1998.2510367.x. [DOI] [PubMed] [Google Scholar]
- 93.Witt H, Malatesta F, Nicoletti F, Brunori M, Ludwig B. Tryptophan 121 of subunit II is the electron entry site to cytochrome-c oxidase in Paracoccus denitrificans. J Biol Chem. 1998;273:5132–5136. doi: 10.1074/jbc.273.9.5132. [DOI] [PubMed] [Google Scholar]
- 94.Witt H, Zickermann V, Ludwig B. Site-directed mutagenesis of cytochrome c oxidase reveals two acidic residues involved in the binding of cytochrome c. Biochim Biophys Acta. 1995;1230:74–76. doi: 10.1016/0005-2728(95)00050-s. [DOI] [PubMed] [Google Scholar]
- 95.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei M J, Libeu C P, Mizushima T, Yamaguchi H, Tomizaki T, Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]