Abstract
IL-17 contributes to the pathogenesis of certain autoimmune diseases, but conversely is essential for host defense against fungi. Antibody-based biologic drugs that neutralize IL-17 are effective in autoimmunity but can be accompanied by adverse side effects. Candida albicans is a commensal fungus that is the primary causative agent of oropharyngeal and disseminated candidiasis. Defects in IL-17 signaling cause susceptibility to candidiasis in mice and humans. A key facet of IL-17 receptor signaling involves RNA binding proteins (RBP), which orchestrate the fate of target mRNA transcripts. In tissue culture models we showed that the RBP AT-rich interacting protein 5a (Arid5a) promotes the stability and/or translation of multiple IL-17-dependent mRNAs. Moreover, during OPC, Arid5a is elevated within the oral mucosa in an IL-17-dependent manner. However, the contribution of Arid5a to IL-17-driven events in vivo is poorly defined. Here, we used CRISPR/Cas9 to generate mice lacking Arid5a. Arid5a−/− mice were fully resistant to experimental autoimmune encephalomyelitis (EAE), an autoimmune setting in which IL-17 signaling drives pathology. Surprisingly, Arid5a−/− mice were resistant to OPC and systemic candidiasis, similar to immunocompetent WT mice and contrasting with mice defective in IL-17 signaling. Therefore, Arid5a-dependent signals mediate pathology in autoimmunity yet are not required for immunity to candidiasis, indicating that selective targeting of IL-17 signaling pathway components may be a viable strategy for development of therapeutics that spare IL-17-driven host defense.
Keywords: Arid5a, RNA Binding protein, IL-17, fungal infection, Candida albicans, autoimmunity
Introduction
IL-17 (IL-17A) is a member of a subclass of cytokines that are structurally and functionally distinct from other inflammatory cytokines. Though produced mainly by T lymphocytes (“Type 17” cells, including γδ- and αβ-T cells, ILC3, NKT cells), IL-17 predominantly activates downstream signals in non-hematopoietic cells to protect against mucosal pathogens, notably the commensal fungus Candida albicans (1, 2). Conversely, IL-17 and Type 17 cells have been implicated in the pathogenesis of many autoimmune and inflammatory diseases (3), highlighted in the success of anti-cytokine drugs targeting IL-17 or the IL-17 receptor. Though unquestionably these drugs have improved patients’ disease activity and quality of life, their use is associated with adverse side effects including worsening bowel inflammation in patients with inflammatory bowel disease (IBD) and the frequent development of opportunistic fungal infections, especially oral and esophageal candidiasis (4–6) (7).
IL-17 signals through a heterodimeric receptor composed of IL-17RA and IL-17RC, though some alternative configurations of the receptor have recently been described (8–10). Binding of IL-17 to the IL-17R complex promotes recruitment of the adaptor Act1 (11–13). In turn, Act1 binds to multiple TRAF factors that promote alternative downstream signaling cascades (14). TRAF6 activates several transcription factors, including NF-κB, IκBξ and C/EBP family members. Additionally, Act1 initiates recruitment of TRAF2 and TRAF5, which initiates a signaling cascade resulting in prolonged half-life of many IL-17-dependent mRNA transcripts. The effects of IL-17 signaling on mRNA fate is orchestrated by a complex array of RNA binding proteins (RBPs) (9). RBPs shape the output of inflammatory effector genes by stabilizing or degrading transcripts that encode inflammatory mediators. Some RBPs, including HuR (Elavl1) and IMP2 (Igf2bp2) act as positive regulators of mRNA expression, resulting in target mRNA stabilization or enhanced translation (15–17). Alternatively, RBPs such as Regnase-1, Splicing factor 2 (SF2) and Roquins negatively regulate mRNA expression, facilitating degradation of unstable transcripts (18–20). These pathways are interlinked because many transcription factors operative in the IL-17 pathway are encoded by RNAs that are subject to post-transcriptional control, for example, IκBξ (Nfkbiz) and C/EBPs (Cebpb, Cebpd).
AT-rich interaction domain 5A (Arid5a) is another RBP implicated in IL-17 signaling and Th17-driven diseases. Arid5a enhances target mRNA transcript stability and translation, in part by offsetting the destabilizing activity of the endoribonuclease Regnase-1 (15, 21–23). Arid5a controls expression of genes downstream of the Th17/IL-17R axis through control of Il6 and Nfkbiz, as well as Th17 cell responses via Il6, STAT3, TBX21 (Tbet), and OX40 transcripts (21, 23, 24). To date, the precise mechanisms by which IL-17 regulates Arid5a function in autoimmunity are not fully understood. Even less is known about Arid5a function in the context of host defense, though Arid5a mRNA is upregulated in an IL-17-dependent manner in oropharyngeal candidiasis (OPC) in the murine oral mucosa (15, 25). Here, we created a novel line of Arid5a−/− mice to assess the role of Arid5a in autoimmunity versus fungal infections. These data reveal a surprising distinction used between signaling pathways critical in host defense compared to those required in autoimmunity.
Material and Methods
Generation of Arid5a−/− mice
The Arid5a knockout allele was fortuitously generated during an attempt to create a conditional deletion with CRISPR/Cas9 (26, 27). Briefly, the targeting strategy was to introduce a LoxP site in intron 2 and a Myc-tag at the C-terminal with another loxP site just after the stop codon. In the process of genotyping potential founders, we identified a mouse carrying a deletion between the two SpyCas9 target sites, with a deletion of 3,359 bp deletion on chromosome 1 between position 36,316,840 and 36,320,199. Fertilized embryos (C57BL/6J, The Jackson Laboratory) produced by natural mating were microinjected in the cytoplasm with a mixture of 0.33 μM EnGen Cas9 protein (New England Biolabs, M0646T), Arid5a-guide2 and Arid5a-guide5 (21.23 ng/μl (~0.66 μM) and two single stranded oligonucleotides: Arid5a-Myc-HDR and Arid5a-guide5-HDR (0.5 μM, “Ultramers” from IDT). Injected zygotes were cultured overnight, and 2-cell embryos were transferred to pseudopregnant CD1 mice. The sgRNA templates were generated by PCR (26) and used for synthesis of sgRNAs using NEB HiScribe™ T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, E2050S). The sgRNAs purified using MEGAclear Kit (ThermoFisher Scientific). The target sequence of the sgRNAs are: ATAGGCTCTGGCCTACAGTTTGG for Arid5a-guide2 at chr1:36377037-36377060 and GTAAAAGCCAAATGCGCCCCAGG for Arid5a-guide5 at chr1:36373679-36373702 (coordinates from GRCm38/mm10 Assembly).
The sequence of the single-stranded oligonucleotides are: 5’-tcgaactcagagagttctgcctgcttctcctgagtgctaggattaaaggtgtgtgccaccactgcctgggAATTCATAACTTCGTATAATGTATGCTATACGAAGTTATgcgcatttggcttttaccatacggttgagggactctgaccctgccttccaggaactgagttggataa-3’ for Arid5a-guide5-HDR
5’-TGGCACATGCCACCCGTCACAACCTATGCGGCACCTCACTTCTTCCACCTCAACACCAAACTGGAGCAGAAACTCATCTCAGAAGAGGATCTGTAAGAATTCATAACTTCGTATAATGTATGCTATACGAAGTTATAGAGCCTATCCTGCTATGCTGTGGAGGATTTGATGGGCAGCTGCCGCCATTATCTCAGGCC-3’ for Arid5a-Myc-HDR.
The potential founders were identified by PCR and the sequence of the knockout allele was determined by sequencing. Arid5a−/− mice or littermates were genotyped with the following primers:
F31-5’-ACCTTCTGGCTACAACAGGC-3’
R31-5’-ACCTACACACTTGCTCCTGC-3’
F51-5’-ATATGGGTGCTAGGCAAGGC-3’
The WT allele is detected with primer F31 and R31 and produces a product of 564 bp. The knockout allele is detected with primer F51 and R31 and produces a product of 595 bp.
Candidiasis Models
OPC was induced in 7–9-week-old mice of both sexes by sublingual inoculation with C. albicans strain CAF2-1-saturated cotton balls for 75 minutes under anesthesia, as described (28, 29). Tongue homogenates were prepared on GentleMACS homogenizers (Miltenyi Biotec) with C-tubes, and CFU was determined by serial dilution plating on Yeast extract-peptone-dextrose (YDP) with ampicillin. Limit of detection is ~30 CFU/g. For systemic candidiasis, mice were injected i.v. with 2–7 × 105 C. albicans strain SC5412 in PBS. Three days post-infection, kidney tissues were homogenized in C-tubes, and CFU was determined as above.
Experimental autoimmune encephalomyelitis
Mice (males age 6–18 weeks) were immunized subcutaneously in 4 sites on the back with 100 μg MOG peptide 35–55 (Biosynthesis) emulsified with Complete Freund’s Adjuvant (CFA) with M. tuberculosis strain H37Ra (DIFCO laboratories) and administered 100 ng pertussis toxin (List Biological Laboratories) i.p. on day 0 and day 2, as described (30). Mice were scored daily as follows: 1. Flaccid tail; 2. Impaired righting reflex and hindlimb weakness; 3. Partial hindlimb paralysis; 4. Complete hindlimb paralysis; 5. Hindlimb paralysis with partial forelimb paralysis; 6. Moribund.
Flow Cytometry
Tongues were harvested and digested with Collagenase IV (0.7 mg/ml) in Hanks’ balanced salt solution for 30–45 mins at 37°C. Cells were separated by Percoll gradient centrifugation. Abs were from the following sources: anti-CD45 and anti-CD11b (BioLegend), anti-CD4 and anti-F4/80 (Invitrogen), anti-TCRβ, anti-CD8, and anti-CD19 (eBioscience), and anti-Ly6G and anti-Ly6C (BD Biosciences). Dead cells were excluded using Ghost Dye (eBioscience). Data were acquired with an LSRFortessa and analyzed using FlowJo software (TreeStar).
Collected single-cell suspensions from LN were filtered and dead cells were excluded using Ghost Dye (eBioscience). LN were cultured in complete medium (RPMI media containing 10% FCS, supplemented with L-glutamine and antibiotics) with 50 ng/ml PMA and 500 ng/ml ionomycin (Sigma-Aldrich) in the presence of Golgiplug (BD Biosciences) for 4 h, followed by staining with Ghost Dye, CD4 and IL-17 (Biolegend).
Immune Cell Isolation
Naïve splenocytes and thymocytes were isolated from 6-week old Arid5a−/− and Arid5a+/+ male mice. Flow cytometry was performed on single cell suspensions.
Western Blotting
Western blotting was performed as described (15). Abs against Arid5a were from Abcam (ab81149). Blots were imaged with a FluorChem E imager (ProteinSimple, Santa Clara CA).
Statistics
Data were analyzed on Graphpad Prism. Each symbol represents one mouse unless indicated. *P<0.05, **<0.01, ***<0.001, ****<0.0001. P values less than 0.05 were considered significant.
Study approval
All the experiments were conducted following NIH guidelines under protocols approved by the University of Pittsburgh IACUC.
Results
Characterization of Arid5a-deficient mice
To elucidate the role of Arid5a in vivo, CRISPR/Cas9 was used to create Arid5a-deficient mice with a deletion in exons 3–7 (Fig 1A, see Methods for details). Founders were validated by PCR (Fig 1B) and bred to homozygosity. Mice were born at expected Mendelian ratios and exhibited normal fertility and baseline body weights (data not shown). Arid5a is expressed at variable levels across tissues, with high expression in thymus, medium levels in spleen, and very low levels in kidney (Fig 1C). Confirming that mice lacked Arid5a protein expression, tissues from Arid5a−/− mice displayed undetectable levels of Arid5a compared to Arid5a+/+ mice.
Figure 1. Creation of Arid5a−/− mice by CRISPR/Cas9.
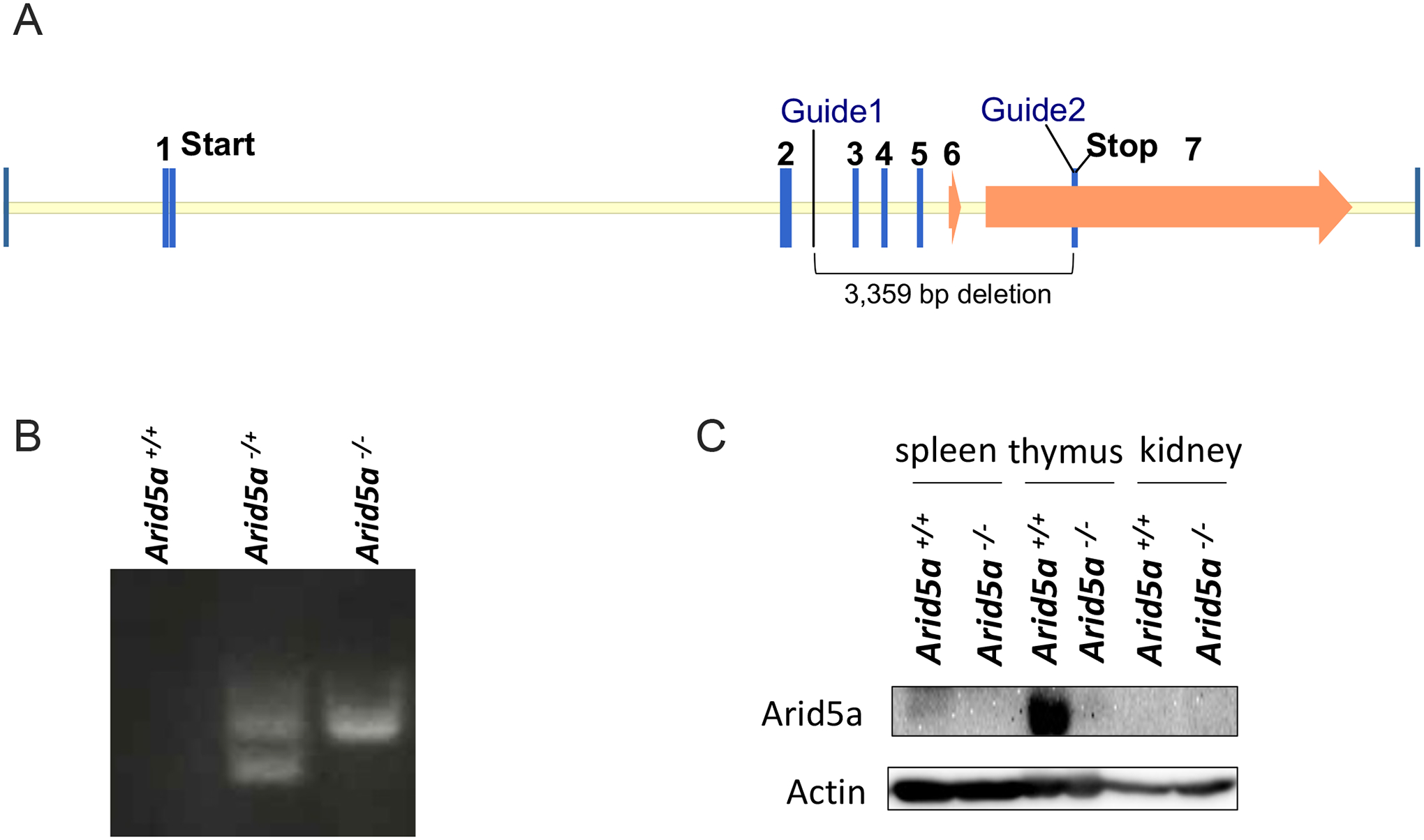
(A) Schematic diagram of Arid5a gene locus and area of deletion. Approximate locations of guide RNA target sites are indicated. (B) Representative genotyping validation of control littermates, heterozygous and homozygous Arid5a-knockout mice, performed on all mice used in experimentation. (C) Western blotting of spleen, thymus and kidney tissues from Arid5a−/− and Arid5a+/+ littermate controls. Data are representative of two independent experiments. Top: Arid5a. Bottom: β-actin loading control.
Previous studies using an independently-generated Arid5a knockout mouse demonstrated a role for Arid5a in regulating several immune processes (21–24, 31–33). Therefore, we examined the baseline status of immune compartments in Arid5a+/+ and Arid5a−/− littermates in spleen (Fig 2A) and thymus (Fig 2B). As shown, no observable differences between Arid5a+/+ and Arid5a−/− mice were detected by flow cytometry in thymic or splenic hematopoietic cells, including TCRβ+ cells, B cells, neutrophils, monocytes or macrophages. Therefore, the absence of Arid5a does not appear to impact normal development of immune cells.
Figure 2. Immunophenotyping of Arid5a−/− mice.
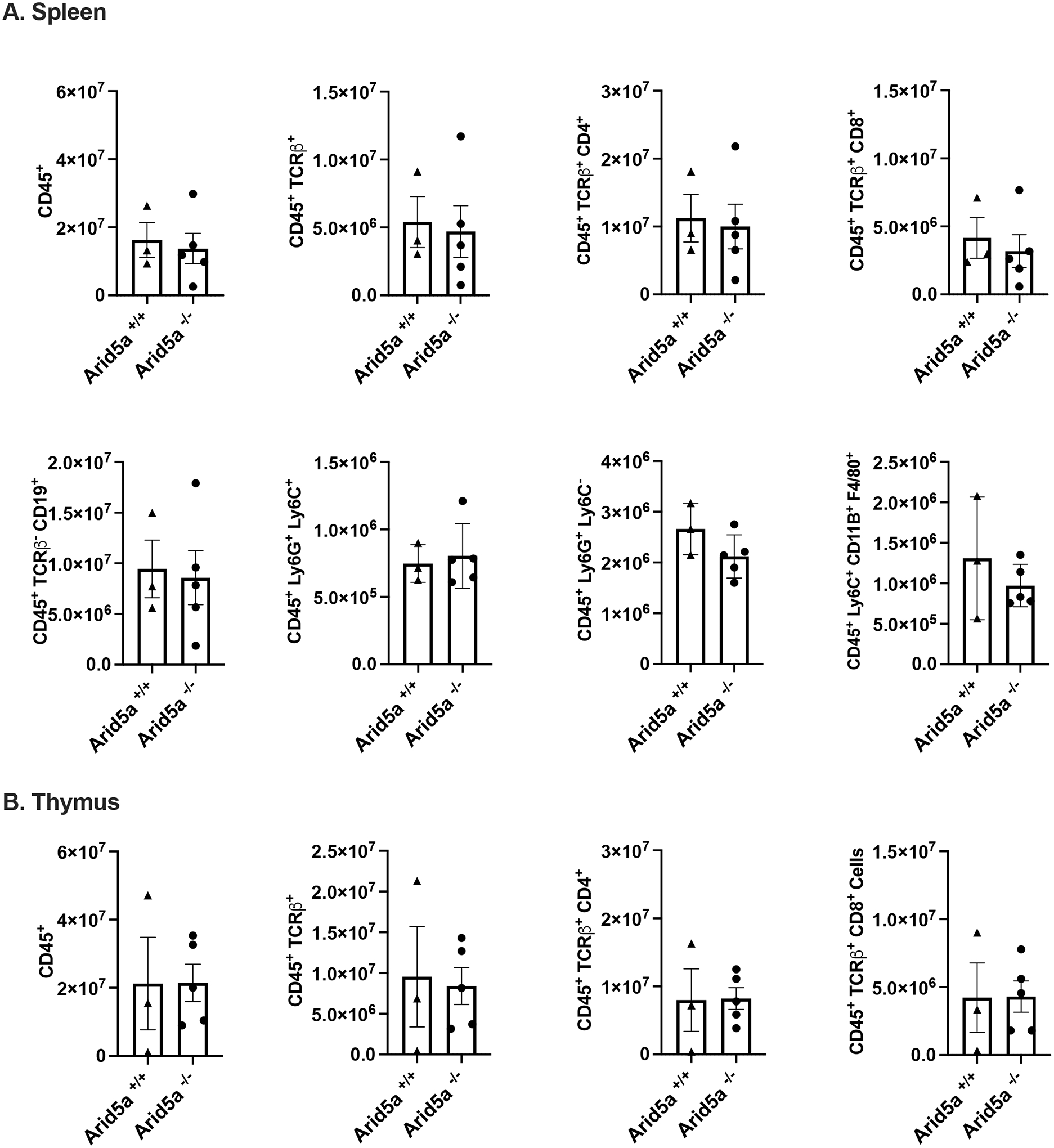
Spleen (A) or thymus (B) from Arid5a−/− or Arid5a+/+ littermate controls were stained with the indicated cellular markers and analyzed by flow cytometry. Y-axis indicates total cell numbers. Each symbol indicates one mouse (Arid5a+/+ n=3; Arid5a−/− n=5). Bars indicate mean + SEM. No significant differences between groups were identified by Student’s t-test.
Arid5a is required for pathogenesis of EAE
Several reports have described a role for Arid5a in driving pathogenesis in mouse models of autoimmunity, with emphasis on control of Th17 cell differentiation (21, 23). Accordingly, we subjected Arid5a−/− mice and Arid5a+/+ littermate controls to a standard model of experimental autoimmune encephalomyelitis (EAE), a strongly IL-17-dependent model a of multiple sclerosis (17, 34–37). Mice were injected with a myelin oligodendrocyte glycoprotein peptide (MOG 35–55) emulsified with complete Freund’s Adjuvant (CFA). Mice were assessed daily for signs of ascending paralysis on a standard scoring system. Arid5a+/+ control mice developed a typical onset and clinical course of EAE, peaking at 14–16 days post immunization. As expected, mice lacking the IL-17 receptor adaptor, Act1 and Arid5a−/− were resistant to disease (Fig 3A). Concomitant with reduced disease scores, there was marked reduction in EAE incidence in Arid5a−/− mice (Fig 3B). Th17 cells are pathogenic in EAE (3, 30, 38, 39). To determine if Arid5a affects Th17 cells during EAE, mice were immunized with MOG, and lymph nodes (LN) and spinal cords were harvested on day 12. Cells isolated from LN were stimulated with PMA and stained for CD4 and intracellular IL-17A. There were reduced numbers of CD4+ IL-17+ cells in Arid5a−/− compared with Arid5a+/+ mice (Fig 3C). Arid5a is known to regulate Il6 in many cell types (15, 21), and indeed there was a trend to decrease expression in CNS (Fig 3D). However, surprisingly, Stat3 mRNA expression was unchanged (Fig 3D) (22). These findings confirm prior work in an independent system that Arid5a is required for development of EAE (21), and also functionally verify that this line of Arid5a-knockout mice has the expected phenotype with respect to IL-17-driven autoimmunity.
Figure 3. Arid5a−/− mice are resistant to EAE.
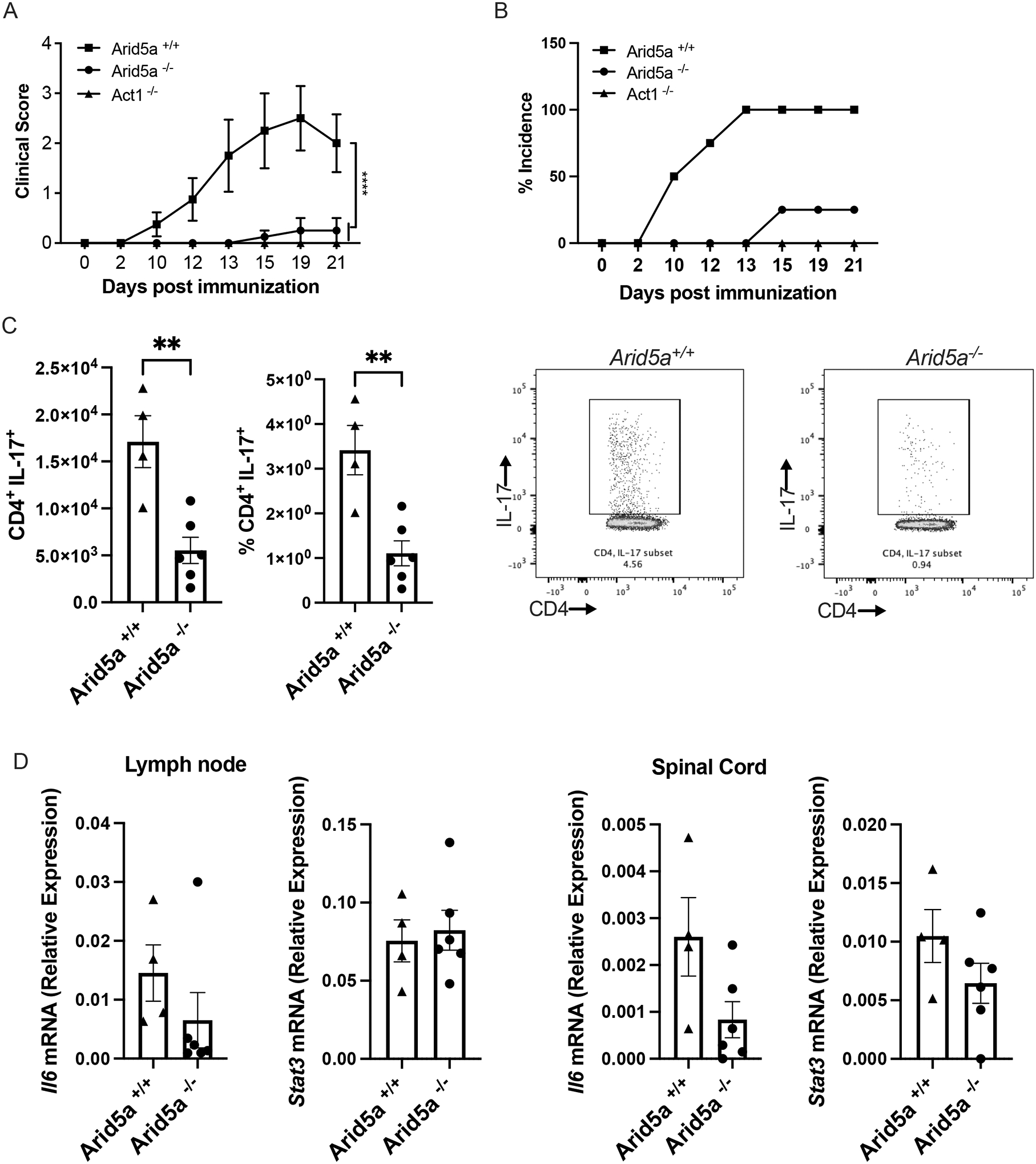
The indicated mice (Arid5a+/+ n=4; Arid5a−/− n=4; and Act1−/− n=3) were subjected to EAE by injection of MOG peptide. (A) Clinical score was assessed daily by investigators blinded to sample identity. *** P<0.001 by ANOVA with Mann-Whitney U analysis. (B) Percent incidence of EAE in each group is indicated. Experiment was performed once, reproducing published report (21). (C) Lymph node cells harvested on day 12 (Arid5a+/+ n=4 and Arid5a−/− n=6) were treated with PMA and ionomycin for 4 h. Cells were stained for CD4 and intracellular IL-17A and quantified by flow cytometry. Right: representative FACS plots. ** P<0.003 by Student t-test. (D) Il6 and Stat3 mRNAs were assessed by qPCR in inguinal lymph nodes and spinal cord at day 12, normalized to Gapdh. Data show relative expression ± SEM (Arid5a+/+ n=4; Arid5a−/− n=6). Experiment was performed twice.
Arid5a is dispensable for immunity to oral and systemic C. albicans infections
IL-17R signaling is well established to drive oral immunity to the commensal fungus C. albicans, (2, 28, 40). Moreover, we previously observed that Arid5a mRNA is rapidly upregulated in an IL-17-dependent manner in the oral mucosa of mice during OPC (15). Because Arid5a drives cellular responses to IL-17 through post-transcriptional control of mRNA in vitro (15, 41), we predicted that Arid5a−/− would likely be more susceptible to OPC than WT mice, which clear infection rapidly and do not exhibit overt signs of disease. To test this hypothesis, Arid5a−/− mice were subjected to OPC by sublingual exposure with a cotton ball soaked with 2 × 107 cells/ml C. albicans strain CAF2-1 (29). As a control, Act1−/− mice (which lack the ability to respond to IL-17 signaling (42, 43)) showed high fungal loads at 5 days after infection (Fig 4A) and progressively lost weight throughout the time course of infection (Fig 4B). In contrast, Arid5a+/+ and Arid5a−/− mice cleared the infection by day 5 and returned to normal body weights (Fig 4A, B).
Figure 4. Arid5a−/− mice show no alterations in susceptibility to mucosal or systemic candidiasis.
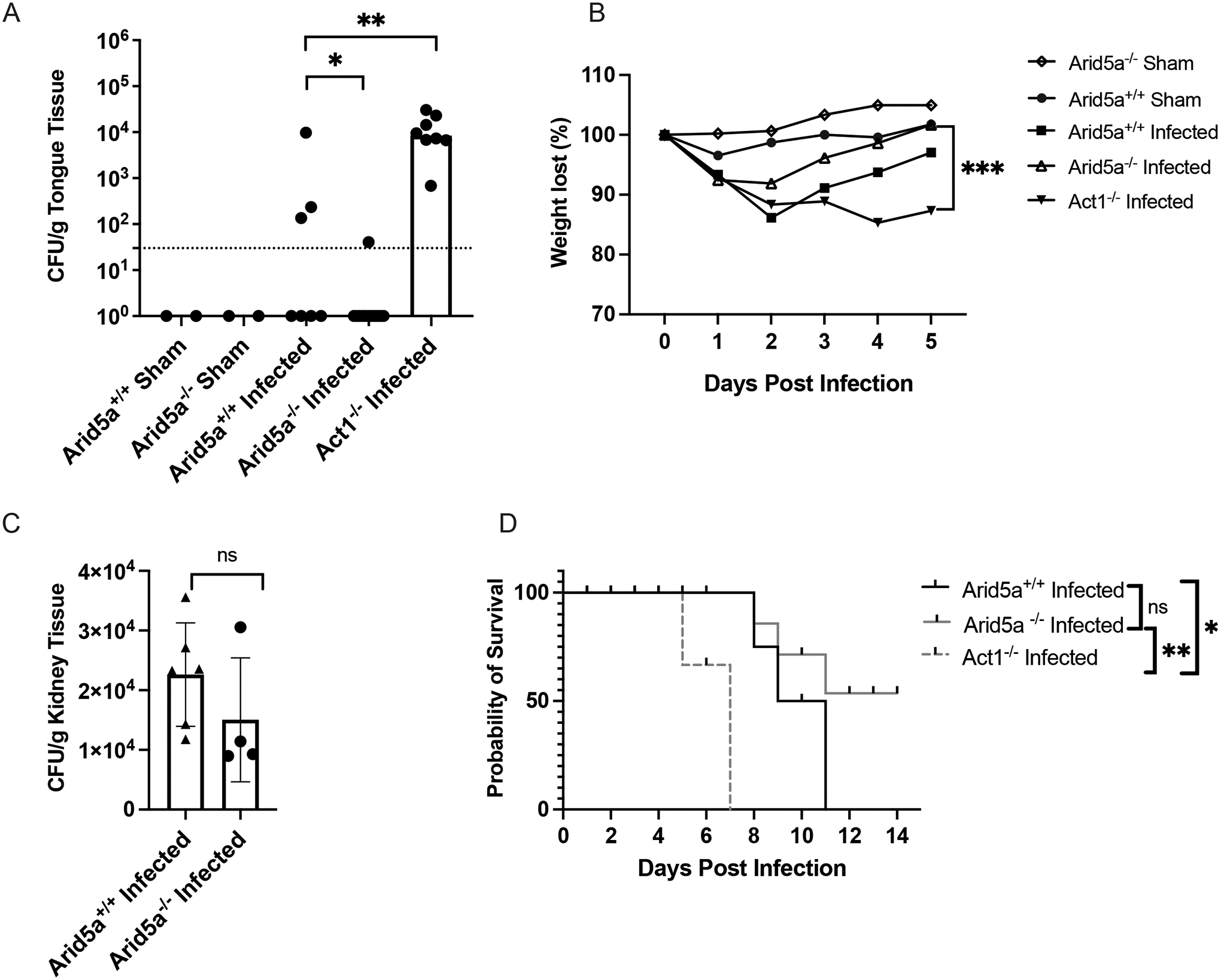
(A) The indicated mice were infected orally with PBS (Sham) or C. albicans strain CAF2-1 for 75 min. After 5 days, tongues were assessed for CFU by plating. Dashed line = limit of detection. Bars indicate geometric mean. *P<0.05, ****P<.0001 by ANOVA with Kruskal-Wallis test. Each symbol indicates one mouse. (Sham: n=2, reproducing extensive prior publications (28, 50). Infected mice: Arid5a+/+ n=7; Arid5a−/−n=12; Act1−/− n=8). Experiment was performed twice. (B) The mice in panel A were weighed daily and percent weight loss relative to day 0 is indicated. (C) The indicated mice (Arid5a+/+ n=6; Arid5a−/−n=4) were infected i.v. with C. albicans strain SC5314 and on day 3 fungal loads in kidney homogenates were assessed by serial dilution plating. Experiment was performed once. (D) The indicated mice (Arid5a+/+ n=12, Arid5a−/− n=15 and Act1−/− n=8) were infected i.v. with C. albicans strain SC5314 and time to lethal morbidity was monitored for 15 days. Data were analyzed by log-rank (Mantel-Cox) test. *P<0.05 **P<0.001. ns, not significant. Experiment was performed twice.
Disseminated candidiasis is a serious nosocomial infection associated with high rates of morbidity and mortality in humans (2). In mouse models, IL-17R signaling mediates immunity to candidiasis (44, 45). Because Arid5a controls genes that drive Th17 differentiation as well as genes downstream of the IL-17 receptor (14, 41), we postulated that Arid5a deficiency would render mice susceptible to disseminated candidiasis. Therefore, we subjected Arid5a−/− mice to disseminated candidiasis by intravenous inoculation of 2 × 105 C. albicans yeast cells (strain SC5314). Arid5a−/− mice had similar fungal loads as Arid5a+/+ mice (Fig 4C). Moreover, while Act1−/− mice succumbed to disease by day 7, most Arid5a+/+ and Arid5a−/− mice survived beyond day 10 and did not show statistically different survival patterns, indicating that Arid5a is not required for immunity to systemic C. albicans infections (Fig 4D). Collectively these data indicate that Arid5a is important for autoimmunity but does not play an important role in the response to C. albicans.
Discussion
Arid5a was originally described for its ability to interact with AT-rich DNA elements in DNA, modulating cellular proliferation, differentiation, and development (46, 47). Arid5a belongs to the ARID family that consists of 15 proteins, which can be divided into seven sub-groups based on the degree of similarity within the ARID domain. The ARID domain is a helix-turn-helix motif essential for binding to DNA elements (46, 47) and can also interact with cis-acting elements located in the 3’ UTR of certain inflammatory RNAs, such as Il6, Tbx21, Stat3, Ox40 (15, 21–24, 48). Binding of Arid5a to these transcripts promotes their longevity and/or facilitates their translation, though the underlying mechanisms are not well understood. Additionally, Arid5a offsets the action of a potent endoribonuclease, Regnase-1, which degrades target transcripts and thereby serves as a negative regulator of inflammation (21). Arid5a acts in hematopoietic cells (macrophages, T cells) and non-hematopoietic cells (fibroblasts, epithelial cells) (14, 41). Thus, Arid5a is a newly recognized RBP that is an important post-transcriptional modulator in multiple immune pathways.
The Arid5a−/− mice described here are not identical to the original published Arid5a−/− mouse strain (21). The latter lacks exons 1–3 and a major part of the DNA binding domain was replaced by a floxed Neo-cassette. In the Arid5a−/− mice described here, LoxP sites were introduced into intron 2 and immediately following the stop codon, creating a 3,359 bp deletion. Despite these differences, both Arid5a−/− lines are resistant to EAE (21), lending confidence that Arid5a plays an important role in driving Th17/IL-17-driven autoimmune inflammation.
IL-17 mediates effects through upregulation of target RNA accumulation, which can occur transcriptionally but also post-transcriptionally (9). Even so, the data presented here indicate that Arid5a−/− mice are, unexpectedly, resistant to oral and systemic candidiasis even though IL-17R signals are essential for immunity to this condition (44, 49, 50) and Arid5a functions downstream IL-17 to upregulate many IL-17 target genes (15, 25). Arid5a has been shown to act in T cells to stabilize genes such as Il6, STAT3, Nfkbiz, OX40, Tbet that drive Th17 differentiation and IL-17 production (22, 41). One explanation to reconcile these observations may be that the early responses to C. albicans in mice are innate in nature, as this fungus is not a commensal microbe in mice (51). Rather than conventional antigen-specific Th17 cells, IL-17 in these naïve settings comes primarily from a combination of γδ-T cells and innate-acting CD4+αβ-T cells; additionally, adaptive T cell memory responses can be established in mice that appear to reflect human T cells responses (52–58). In line with this, Il6−/− and Ccr6−/− mice are resistant to OPC, and Th1 cells (IL-12) are dispensable for protection against oral C. albicans (28, 52, 59). Thus, the genes regulated by Arid5a might not be important for protection against Candida albicans oral infections.
Anti-IL-17 therapy is not associated with risks to systemic candidiasis, likely because the major mechanisms of antifungal control at cutaneous/mucosal surfaces is through neutrophil recruitment and β-defensin production whereas circulating neutrophils are less IL-17-dependent (4, 50, 60, 61). In contrast, during systemic candidiasis IFN-γ, IL-12 as well as IL-17 signaling contribute to effective immunity against C. albicans (62–64). Previous studies showed that Arid5a regulates expression of IFN-γ in Th1 cells via control of the master Th1 transcription factor Tbet (24). Given this, we were surprised to find that Arid5a−/− mice showed the same susceptibility to systemic candidiasis as their wild type littermate counterparts. It is possible that other mRNA stabilizing factors either in T cells or in IL-17-responsive cells [likely, renal tubular epithelial cells (45), though NK cells have been reported (65)] can compensate for the loss of Arid5a during C. albicans infection, but more work will be needed to dissect this phenomenon.
The use of biologics targeting IL-17 or IL-17 receptor was a major advance in the treatment of psoriasis, psoriatic arthritis, and ankylosing spondylitis (10, 66, 67). Unexpectedly, IL-17 blockade failed in Crohn’s disease, which has been attributed to a vital tissue protective role of IL-17 in the intestine epithelium (5, 68–70). Intriguingly, IL-17 has been linked to controlling social behavior in mice (71), and blockade of IL-17RA was linked to a small risk of suicide in humans (72), though this is controversial (73). Thus, an ideal therapeutic approach to blocking IL-17 would inhibit its pathogenic effects while sparing host-defensive pathways. Based on these and other data, Arid5a may be an attractive candidate for selective blockade in IL-17-driven diseases, though achieving this is not trivial. Interestingly, recent advances in approaches to understand and target RNA binding proteins suggests potential ways to disrupt Arid5a (or other RBP) interactions with specific target transcripts (74). For example, an elegant study in the IL-17 system showed that aptamers that recognize the binding element of Act1 can interfere with its mRNA stabilizing capacity, and are effective at reducing inflammation in vivo (75). Indeed, the development and potential uses of RNA therapeutics is becoming increasingly appreciated (76, 77). Arid5a was reported to be a target of the antipsychotic drug chlorpromazine (22), though this remains to be confirmed. Targeting individual pathway components such as Arid5a potentially allows for a more targeted approach to treat IL-17-mediated inflammatory/autoimmune diseases while preserving the protective role of IL-17 in host defense.
Key Points:
We report creation of new Arid5a−/− mouse strain
Arid5a−/− mice are resistant to experimental autoimmune encephalomyelitis
Arid5a−/− mice retain immunity to oral and disseminated candidiasis
Acknowledgements
This work was supported by NIH grants to SLG (AI144436, AI147383, DE022550), PSB AI142354, AI159058) and MJM (AI148356). TCT was supported by T32-AI089443. We thank B.M. Coleman, N. Amatya and R. Bechara for valuable input. Graphical abstract was created with BioRender.com.
Footnotes
Conflicts of interest: SLG has consulted for Aclaris Therapeutics and Eli Lilly. There are no other conflicts of interest.
References
- 1.Conti HR, and Gaffen SL. 2015. IL-17-Mediated Immunity to the Opportunistic Fungal Pathogen Candida albicans. J Immunol 195: 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lionakis MS, and Levitz SM. 2018. Host Control of Fungal Infections: Lessons from Basic Studies and Human Cohorts. Annu Rev Immunol 36: 157–191. [DOI] [PubMed] [Google Scholar]
- 3.McGeachy MJ, Cua DJ, and Gaffen SL. 2019. The IL-17 Family of Cytokines in Health and Disease. Immunity 50: 892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson L, van den Reek J, Bruno M, van Hunsel F, Herings RMC, Matzaraki V, Boahen CK, Kumar V, Groenewoud HMM, van de Veerdonk FL, Netea MG, de Jong E, and Kullberg BJ. 2022. Risk of candidiasis associated with interleukin-17 inhibitors: A real-world observational study of multiple independent sources. Lancet Reg Health Eur 13: 100266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauny M, Moulin D, D’Amico F, Netter P, Petitpain N, Arnone D, Jouzeau JY, Loeuille D, and Peyrin-Biroulet L. 2020. Paradoxical gastrointestinal effects of interleukin-17 blockers. Ann Rheum Dis 79: 1132–1138. [DOI] [PubMed] [Google Scholar]
- 6.Saunte DM, Mrowietz U, Puig L, and Zachariae C. 2017. Candida infections in psoriasis and psoriatic arthritis patients treated with IL-17 inhibitors and their practical management. Br J Dermatol 177: 47–62. [DOI] [PubMed] [Google Scholar]
- 7.Smith MK, Pai J, Panaccione R, Beck P, Ferraz JG, and Jijon H. 2019. Crohn’s-like disease in a patient exposed to anti-Interleukin-17 blockade (Ixekizumab) for the treatment of chronic plaque psoriasis: a case report. BMC Gastroenterol 19: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Y, Huang J, Zhao X, Lu H, Wang W, Yang XO, Shi Y, Wang X, Lai Y, and Dong C. 2019. Interleukin-17 receptor D constitutes an alternative receptor for interleukin-17A important in psoriasis-like skin inflammation. Sci Immunol 4: eaau9657. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Bechara R, Zhao J, McGeachy MJ, and Gaffen SL. 2019. Interleukin 17 receptor-based signaling and implications for disease. Nature Immunology 20: 1594–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung SH, Ye XQ, and Iwakura Y. 2021. Interleukin-17 family members in health and disease. Int Immunol 33: 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonder SU, Saret S, Tang W, Sturdevant DE, Porcella SF, and Siebenlist U. 2011. IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem 286: 12881–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang SH, Park H, and Dong C. 2006. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem 281: 35603–35607. [DOI] [PubMed] [Google Scholar]
- 13.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang QW, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, and Li X. 2007. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol 8: 247–256. [DOI] [PubMed] [Google Scholar]
- 14.Amatya N, Garg AV, and Gaffen SL. 2017. IL-17 Signaling: The Yin and the Yang. Trends Immunol 38: 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amatya N, EE C, Cruz JA, Aggor F, Garg A, Berman A, Gudjonsson JE, Atasoy U, and Gaffen SL. 2018. IL-17 integrates multiple self-reinforcing, feed-forward mechanisms through the RNA-binding protein Arid5a. Science Signaling 11: eaat4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herjan T, Yao P, Qian W, Li X, Liu C, Bulek K, Sun D, Yang WP, Zhu J, He A, Carman JA, Erzurum SC, Lipshitz HD, Fox PL, and Hamilton TA. 2013. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J Immunol 191: 640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechara R, Amatya N, Majumder S, Zhou C, Li Y, Liu Q, McGeachy MJ, and Gaffen SL. 2022. The RNA-binding protein IMP2 drives a stromal-Th17 cell circuit in autoimmune neuroinflammation. JCI Insight 7: e152766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun D, Novotny M, Bulek K, Liu C, Li X, and Hamilton T. 2011. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing regulatory factor SF2 (ASF). Nat Immunol 12: 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg AV, Amatya N, Chen K, Cruz JA, Grover P, Whibley N, Conti HR, Hernandez Mir G, Sirakova T, Childs EC, Smithgall TE, Biswas PS, Kolls JK, McGeachy MJ, Kolattukudy PE, and Gaffen SL. 2015. MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 43: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL, Gloury R, Martin N, Lohs C, Lech M, Stehklein JE, Geerlof A, Kremmer E, Weber A, Anders HJ, Schmitz I, Schmidt-Supprian M, Fu M, Holtmann H, Krappmann D, Ruland J, Kallies A, Heikenwalder M, and Heissmeyer V. 2014. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol 15: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 21.Masuda K, Ripley B, Nishimura R, Mino T, Takeuchi O, Shioi G, Kiyonari H, and Kishimoto T. 2013. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci U S A 110: 9409–9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda K, Ripley B, Nyati KK, Dubey PK, Zaman MM, Hanieh H, Higa M, Yamashita K, Standley DM, Mashima T, Katahira M, Okamoto T, Matsuura Y, Takeuchi O, and Kishimoto T. 2016. Arid5a regulates naive CD4+ T cell fate through selective stabilization of Stat3 mRNA. J Exp Med 213: 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanieh H, Masuda K, Metwally H, Chalise JP, Mohamed M, Nyati KK, Standley DM, Li S, Higa M, Zaman MM, and Kishimoto T. 2018. Arid5a stabilizes OX40 mRNA in murine CD4(+) T cells by recognizing a stem-loop structure in its 3’UTR. Eur J Immunol 48: 593–604. [DOI] [PubMed] [Google Scholar]
- 24.Zaman MM, Masuda K, Nyati KK, Dubey PK, Ripley B, Wang K, Chalise JP, Higa M, Hanieh H, and Kishimoto T. 2016. Arid5a exacerbates IFN-gamma-mediated septic shock by stabilizing T-bet mRNA. Proc Natl Acad Sci U S A 113: 11543–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puel A, and Casanova J-L. 2018. Arid5a makes the IL-17A/F-responsive pathway less arid. Science Signaling 11: eaau8876. [DOI] [PubMed] [Google Scholar]
- 26.Pelletier S, Gingras S, and Green DR. 2015. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 42: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, and Cairns MJ. 2013. Gene set enrichment analysis of RNA-Seq data: integrating differential expression and splicing. BMC Bioinformatics 14 Suppl 5: S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti H, Shen F, Nayyar N, Stocum E, JN S, Lindemann M, Ho A, Hai J, Yu J, Jung J, Filler S, Masso-Welch P, Edgerton M, and Gaffen S. 2009. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 206: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solis NV, and Filler SG. 2012. Mouse model of oropharyngeal candidiasis. Nat Protoc 7: 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, and Cua DJ. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol 10: 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubey PK, Masuda K, Nyati KK, Uz Zaman MM, Chalise JP, Millrine D, Kai W, Ripley B, and Kishimoto T. 2017. Arid5a-deficient mice are highly resistant to bleomycin-induced lung injury. Int Immunol 29: 79–85. [DOI] [PubMed] [Google Scholar]
- 32.Nyati KK, Masuda K, Zaman MM, Dubey PK, Millrine D, Chalise JP, Higa M, Li S, Standley DM, Saito K, Hanieh H, and Kishimoto T. 2017. TLR4-induced NF-kappaB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res 45: 2687–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higa M, Oka M, Fujihara Y, Masuda K, Yoneda Y, and Kishimoto T. 2018. Regulation of inflammatory responses by dynamic subcellular localization of RNA-binding protein Arid5a. Proc Natl Acad Sci U S A 115: E1214–E1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komiyama Y, Nakae S, Matsuki T, Nambu A, Ishigame H, Kakuta S, Sudo K, and Iwakura Y. 2006. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J Immunol 177: 566–573. [DOI] [PubMed] [Google Scholar]
- 35.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, and Iwakura Y. 2009. Differential roles of interleukin-17A and −17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30: 108–119. [DOI] [PubMed] [Google Scholar]
- 36.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, and Weaver CT. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6: 1123–1132. [DOI] [PubMed] [Google Scholar]
- 37.Harrington LE, Mangan PR, and Weaver CT. 2006. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol 18: 349–356. [DOI] [PubMed] [Google Scholar]
- 38.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira SA, Gorman D, Kastelein RA, and Sedgwick JD. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421: 744–748. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, and Kuchroo VK. 2012. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13: 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Vinh DC, Casanova JL, and Puel A. 2017. Inborn errors of immunity underlying fungal diseases in otherwise healthy individuals. Curr Opin Microbiol 40: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyati KK, Zaman MM, Sharma P, and Kishimoto T. 2020. Arid5a, an RNA-Binding Protein in Immune Regulation: RNA Stability, Inflammation, and Autoimmunity. Trends Immunol 41: 255–268. [DOI] [PubMed] [Google Scholar]
- 42.Leonardi A, Chariot A, Claudio E, Cunningham K, and Siebenlist U. 2000. CIKS, a connection to Ikappa B kinase and stress-activated protein kinase. Proc Natl Acad Sci U S A 97: 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claudio E, Sonder SU, Saret S, Carvalho G, Ramalingam TR, Wynn TA, Chariot A, Garcia-Perganeda A, Leonardi A, Paun A, Chen A, Ren NY, Wang H, and Siebenlist U. 2009. The adaptor protein CIKS/Act1 is essential for IL-25-mediated allergic airway inflammation. J Immunol 182: 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W, Na L, Fidel PL, and Schwarzenberger P. 2004. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis 190: 624–631. [DOI] [PubMed] [Google Scholar]
- 45.Ramani K, Jawale CV, Verma AH, Coleman BM, Kolls JK, and Biswas PS. 2018. Unexpected kidney-restricted role for IL-17 receptor signaling in defense against systemic Candida albicans infection. JCI Insight 3: e98241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilsker D, Patsialou A, Dallas PB, and Moran E. 2002. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ 13: 95–106. [PubMed] [Google Scholar]
- 47.Patsialou A, Wilsker D, and Moran E. 2005. DNA-binding properties of ARID family proteins. Nucleic Acids Res 33: 66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masuda K, and Kishimoto T. 2018. A Potential Therapeutic Target RNA-binding Protein, Arid5a for the Treatment of Inflammatory Disease Associated with Aberrant Cytokine Expression. Curr Pharm Des 24: 1766–1771. [DOI] [PubMed] [Google Scholar]
- 49.Puel A, Cypowji S, Bustamante J, Wright J, Liu L, Lim H, Migaud M, Israel L, Chrabieh M, Audry M, Gumbleton M, Toulon A, Bodemer C, El-Baghdadi J, Whitters M, Paradis T, Brooks J, Collins M, Wolfman N, Al-Muhsen S, Galicchio M, Abel L, Picard C, and Casanova J-L. 2011. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332: 65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conti H, Bruno V, Childs E, Daugherty S, Hunter J, Mengesha B, Saevig D, Hendricks M, Coleman BM, Brane L, Solis NV, Cruz JA, Verma A, Garg A, Hise AG, Naglik J, Naglik JR, Filler SG, Kolls JK, Sinha S, and Gaffen SL. 2016. IL-17RA signaling in oral epithelium is critical for protection against oropharyngeal candidiasis. Cell Host Microbe 20: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suegara N, Siegel JE, and Savage DC. 1979. Ecological determinants in microbial colonization of the murine gastrointestinal tract: adherence of Torulopsis pintolopesii to epithelial surfaces. Infect Immun 25: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conti H, Peterson A, Huppler A, Brane L, Hernández-Santos N, Whibley N, Garg A, Simpson-Abelson M, Gibson G, Mamo A, Osborne L, Bishu S, Ghilardi N, Siebenlist U, Watkins S, Artis D, McGeachy M, and Gaffen S. 2014. Oral-resident ‘natural’ Th17 cells and γδ-T cells control opportunistic Candida albicans infections. J Exp Med 211: 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma A, Richardson J, Zhou C, Coleman BM, Moyes D, Ho J, Huppler AR, Ramani K, McGeachy MJ, Mufazalov IA, Waisman A, Kane LP, Biswas P, Hube B, Naglik J, and Gaffen SL. 2017. Oral epithelial cells orchestrate innate Type 17 responses to Candida albicans through the virulence factor Candidalysin. Sci Immunol 2: eeam8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bär E, Gladiator A, Bastidas S, Roschitzki B, Acha-Orbea H, Oxenius A, and LeibundGut-Landmann S. 2012. A novel Th cell epitope of Candida albicans mediates protection from fungal infection. J Immunol 188: 5636–5643. [DOI] [PubMed] [Google Scholar]
- 55.Hernández-Santos N, Huppler AR, Peterson AC, Khader SA, KC M, and Gaffen SL. 2013. Th17 cells confer long term adaptive immunity to oral mucosal Candida albicans infections. Mucosal Immunol 6: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sparber F, Dolowschiak T, Mertens S, Lauener L, Clausen BE, Joller N, Stoitzner P, Tussiwand R, and LeibundGut-Landmann S. 2018. Langerin+ DCs regulate innate IL-17 production in the oral mucosa during Candida albicans-mediated infection. PLoS Pathog 14: e1007069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirchner FR, and LeibundGut-Landmann S. 2021. Tissue-resident memory Th17 cells maintain stable fungal commensalism in the oral mucosa. Mucosal Immunol 14: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Break TJ, Oikonomou V, Dutzan N, Desai JV, Swidergall M, Freiwald T, Chauss D, Harrison OJ, Alejo J, Williams DW, Pittaluga S, Lee CR, Bouladoux N, Swamydas M, Hoffman KW, Greenwell-Wild T, Bruno VM, Rosen LB, Lwin W, Renteria A, Pontejo SM, Shannon JP, Myles IA, Olbrich P, Ferre EMN, Schmitt M, Martin D, Genomics C Computational Biology, Barber DL, Solis NV, Notarangelo LD, Serreze DV, Matsumoto M, Hickman HD, Murphy PM, Anderson MS, Lim JK, Holland SM, Filler SG, Afzali B, Belkaid Y, Moutsopoulos NM, and Lionakis MS. 2021. Aberrant type 1 immunity drives susceptibility to mucosal fungal infections. Science 371: eaay5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farah C, Hu Y, Riminton S, and Ashman R. 2006. Distinct roles for interleukin-12p40 and tumour necrosis factor in resistance to oral candidiasis defined by gene targeting. Oral Microbiol Immunol 21: 252–255. [DOI] [PubMed] [Google Scholar]
- 60.Conti H, Baker O, Freeman A, Jang W, Li R, Holland S, Edgerton M, and Gaffen S. 2011. New mechanism of oral immunity to mucosal candidiasis in hyper-IgE syndrome. Mucosal Immunol 4: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lionakis MS, Lim JK, Lee CC, and Murphy PM. 2011. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun 3: 180–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cenci E, Mencacci A, Spaccapelo R, Tonnetti L, Mosci P, Enssle KH, Puccetti P, Romani L, and Bistoni F. 1995. T helper cell type 1 (Th1)- and Th2-like responses are present in mice with gastric candidiasis but protective immunity is associated with Th1 development. J Infect Dis 171: 1279–1288. [DOI] [PubMed] [Google Scholar]
- 63.Kaposzta R, Tree P, Marodi L, and Gordon S. 1998. Characteristics of invasive candidiasis in gamma interferon- and interleukin-4-deficient mice: role of macrophages in host defense against Candida albicans. Infect Immun 66: 1708–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balish E, Wagner RD, Vazquez-Torres A, Pierson C, and Warner T. 1998. Candidiasis in interferon-gamma knockout (IFN-gamma−/−) mice. J Infect Dis 178: 478–487. [DOI] [PubMed] [Google Scholar]
- 65.Bar E, Whitney PG, Moor K, Reis e Sousa C, and LeibundGut-Landmann S. 2014. IL-17 regulates systemic fungal immunity by controlling the functional competence of NK cells. Immunity 40: 117–127. [DOI] [PubMed] [Google Scholar]
- 66.Majumder S, and McGeachy MJ. 2021. IL-17 in the Pathogenesis of Disease: Good Intentions Gone Awry. Annu Rev Immunol 39: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McInnes IB, Mease PJ, Ritchlin CT, Rahman P, Gottlieb AB, Kirkham B, Kajekar R, Delicha EM, Pricop L, and Mpofu S. 2017. Secukinumab sustains improvement in signs and symptoms of psoriatic arthritis: 2 year results from the phase 3 FUTURE 2 study. Rheumatology (Oxford) 56: 1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin X, Gaudino SJ, Jang KK, Bahadur T, Singh A, Banerjee A, Beaupre M, Chu T, Wong HT, Kim CK, Kempen C, Axelrad J, Huang H, Khalid S, Shah V, Eskiocak O, Parks OB, Berisha A, McAleer JP, Good M, Hoshino M, Blumberg R, Bialkowska AB, Gaffen SL, Kolls JK, Yang VW, Beyaz S, Cadwell K, and Kumar P. 2022. IL-17RA-signaling in Lgr5(+) intestinal stem cells induces expression of transcription factor ATOH1 to promote secretory cell lineage commitment. Immunity 55: 237–253 e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J, Tato C, Joyce-Shaikh B, Gulan F, Cayatte C, Chen Y, Blumenschein W, Judo M, Ayanoglu G, McClanahan T, Li X, and Cua D. 2015. Interleukin-23-dependent IL-17 production regulates intestinal epithelial permeability. Immunity 43: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maxwell J, Zhang Y, Brown W, Smith C, Byrne F, Florino M, Stevens E, Bigler J, Davis J, Rottman J, Budelsky A, Symons A, and Towne J. 2015. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity 43: 739–750. [DOI] [PubMed] [Google Scholar]
- 71.Reed MD, Yim YS, Wimmer RD, Kim H, Ryu C, Welch GM, Andina M, King HO, Waisman A, Halassa MM, Huh JR, and Choi GB. 2020. IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature 577: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mullard A 2017. New plaque psoriasis approval carries suicide warning. Nat Rev Drug Discov 16: 155. [DOI] [PubMed] [Google Scholar]
- 73.Chiricozzi A, Romanelli M, Saraceno R, and Torres T. 2016. No meaningful association between suicidal behavior and the use of IL-17A-neutralizing or IL-17RA-blocking agents. Expert Opin Drug Saf 15: 1653–1659. [DOI] [PubMed] [Google Scholar]
- 74.Wu X, Gardashova G, Lan L, Han S, Zhong C, Marquez RT, Wei L, Wood S, Roy S, Gowthaman R, Karanicolas J, Gao FP, Dixon DA, Welch DR, Li L, Ji M, Aube J, and Xu L. 2020. Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis. Commun Biol 3: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herjan T, Hong L, Bubenik J, Bulek K, Qian W, Liu C, Li X, Chen X, Yang H, Ouyang S, Zhou H, Zhao J, Vasu K, Cockman E, Aronica M, Asosingh K, Licatalosi DD, Qin J, Fox PL, Hamilton TA, Driscoll D, and Li X. 2018. IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat Immunol 19: 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Damase TR, Sukhovershin R, Boada C, Taraballi F, Pettigrew RI, and Cooke JP. 2021. The Limitless Future of RNA Therapeutics. Front Bioeng Biotechnol 9: 628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crooke ST, Witztum JL, Bennett CF, and Baker BF. 2018. RNA-Targeted Therapeutics. Cell Metab 27: 714–739. [DOI] [PubMed] [Google Scholar]


